- Laboratório de Genoma Funcional, Departamento de Genética, Evolução e Bioagentes, Instituto de Biologia, Universidade Estadual de Campinas, Campinas, São Paulo, Brazil
There is a growing demand for renewable energy, and sugarcane is a promising bioenergy crop. In Brazil, the largest sugarcane producer in the world, sugarcane plantations are expanding into areas where severe droughts are common. Recent evidence has highlighted the role of miRNAs in regulating drought responses in several species, including sugarcane. This review summarizes the data from miRNA expression profiles observed in a wide array of experimental conditions using different sugarcane cultivars that differ in their tolerance to drought. We uncovered a complex regulation of sugarcane miRNAs in response to drought and discussed these data with the miRNA profiles observed in other plant species. The predicted miRNA targets revealed different transcription factors, proteins involved in tolerance to oxidative stress, cell modification, as well as hormone signaling. Some of these proteins might regulate sugarcane responses to drought, such as reduction of internode growth and shoot branching and increased leaf senescence. A better understanding on the regulatory network from miRNAs and their targets under drought stress has a great potential to contribute to sugarcane improvement, either as molecular markers as well as by using biotechnological approaches.
Sugarcane: A Complex Grass
Sugarcane (Saccharum ssp.) belongs to the family Poaceae, subfamily Panicoideae and tribe Andropogoneae. The family contains 13 subfamilies (Sánchez-Ken et al., 2007), comprising a monophyletic clade that shows some particularities, such as the presence of a caryopsis fruit and a well-differentiated lateral embryo, a unique combination among monocotyledonous plants (GPWG, 2001). The tribe Andropogoneae contains species that are mostly polyploid and perennial and have a C4 photosynthetic mechanism (Clayton and Renvoize, 1982). It is one of the largest tribes of the Poaceae family and is widely distributed in tropical and subtropical regions of the world (Clayton and Renvoize, 1982; Sánchez-Ken and Clark, 2010).
Sugarcane, a crop of great worldwide economic importance, is responsible for approximately 75% of the global sugar production (Commodity Research Bureau, 2015), and is becoming increasingly relevant in the production of renewable energy. Sugarcane is a unique crop regarding the ability to accumulate sucrose, that can reach levels up to 50% of dry weight in its stalks (Botha and Black, 2000). Brazil stands out as the main producer of sugarcane, with a cultivated area estimated to be approximately 8.8 million hectares (as of the 2013/2014 harvest), producing more than 600 million tons, of which 46% was used for sugar production and 54% was used for ethanol production (Conab, 2013).
The cultivars that are grown worldwide are multi-species hybrids resulting from classical breeding, known as Saccharum spp. The species Saccharum officinarum was the basis for breeding programs, due to the high levels of sucrose in its stem, but this cultivar has low levels of resistance to diseases. S. spontaneum is another important species in the breeding program, mainly due to its characteristics of strength and resistance to pests (Miranda et al., 2008).
Sugarcane has one of the most complex genomes of all cultivated plant species, with a high chromosome number and high degree of aneuploidy (D'Hont et al., 1996). The modern hybrids have between 100 and 120 chromosomes; 70–80% of the chromosomes belong to S. officinarum, 10–23% of the chromosomes originated from S. spontaneum, and 8–13% of chromosomes are derived from recombination between the species (Piperidis et al., 2010). Despite this complexity, sugarcane has been subjected to genetic mapping using several types of molecular markers (Cordeiro et al., 2001; Ming et al., 2002a), which enabled the identification of QTLs, principally related to sugar yield, cane yield, fiber content and sucrose content (Ming et al., 2002b,c; Pastina et al., 2012). Comparative mapping between sugarcane and sorghum (Sorghum bicolor), another C4 grass, showed high synteny between the genomes, with 84% of the loci mapped using 242 probes presenting homology between these species, indicating their high conservation, that reaches 95.2% of sequence identity in the coding region (Ming et al., 1998; Asnaghi et al., 2000; Dillon et al., 2007; Wang et al., 2010b). Because the sugarcane genome has not been sequenced until now, sorghum has become a good reference genome for analyses involving sugarcane due to the high gene identity found between these two species (Jannoo et al., 2007; Wang et al., 2010b). Determining the genome sequence in complex organisms such as sugarcane is a complex task mainly due to the presence of a large fraction of repetitive DNA, that makes genome assembly very difficult (Setta et al., 2014). Therefore, the sequence of the sorghum genome can be used as reference, to guide the assembly of sugarcane sequences, providing a working draft of the entire sequence. Moreover, due to the high sequence identity between sugarcane and sorghum, it is possible to align to the sorghum genome partial sequences from sugarcane, allowing, for example, to identify the putative sequences of an entire mRNA or even miRNA precursors. Even though, the need of a reference genome from sugarcane is essential. This would allow, for example, to identify promoter regions, that usually are less conserved between species, and in the case of miRNAs, to clone the entire miRNA precursors. Also, as the sequencing costs decline, soon the sequencing by genotyping strategy will be feasible. This will allow the identification of single nucleotide polymorphisms (SNPs) that can be associated to several useful agronomical traits, generating new molecular markers for sugarcane breeding programs (Setta et al., 2014).
Drought Stress in Sugarcane
Sugarcane development can be divided into four stages: germination, tillering, grand growth and maturity (Gascho and Shih, 1983). Each of these stages is affected in different ways by water stress. Because a reduction in growth rates is one of the conserved evolutionary responses that plants activate in response to drought, the tillering and grand growth phases are known to be critically affected by water scarcity (Ramesh, 2000; Inman-Bamber and Smith, 2005). Unfortunately, up to 80% of sugarcane yield is produced during these two phases (Singh and Rao, 1987). The sugarcane crop cycle, from cane planting and harvest of mature cane, usually takes from 12 to 18 months. A plant crop refers to the plants grown for the first time after planting. Once the above ground plant part is harvested, it will regrowth, giving rise to a new plant that will be harvest again. This cycle usually has one plant crop and 3 to 4 ratoon (regrowth) crops. Typically, farmers have about 20% of their area with “new” plants (i.e., cane that will be harvest for the first time). Therefore, unlike other crops with shorter life cycle, such as maize, soybean and wheat, sugarcane farmers do not have any flexibility to avoid dry seasons, because sugarcane will face climate up and downs that take places along the entire year.
Drought causes several effects in sugarcane. There is evidence that stomata closure, intended to reduce water loss, is triggered by a combination of the water status of adjacent cells, intensity of photon flux (Assmann and Grantz, 1990) and the water deficit sensed by roots (Smith et al., 1999). As expected, drought reduces transpiration and photosynthesis and increases leaf temperature (Rodrigues et al., 2009, 2011; Graça et al., 2010). Sugarcane cultivars differ in their responses to drought stress. Usually, the assays to infer the tolerance to drought are done using different cultivars that are ranked according to their yield under drought stress (Kumar, 2005; Silva et al., 2007; Ribeiro et al., 2013). Stalk yield and the content of soluble solids in the stalk juice usually are the key parameters used by breeders to classify the degree of tolerance to drought in sugarcane genotypes. This is because sugarcane productivity is based in these two indexes. Therefore, although most cultivars show decreased yields under drought, some are more affected than others. Interestingly, a cultivar considered as sensitive to drought, i.e., reduced yield under drought stress, may be considered a useful cultivar. For example, Ribeiro et al., found that cultivar IACSP86-2042 had a 50% reduction in stalk yield under drought stress, much higher than IACSP94-2094 (29% reduction) and SP87–365 (no reduction). However, the absolute stalk yield of these cultivars were similar under drought conditions; i.e., IACSP86-2042 had a much higher productivity under non-stressful conditions (234% higher than SP87–365 and 50% higher than IACSP94-2094).
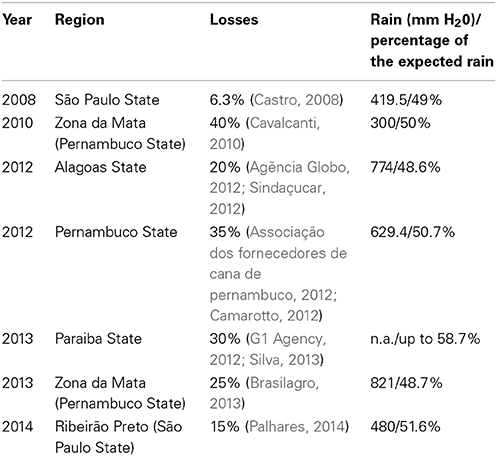
Losses due to drought are not unusual and almost every year, some sugarcane growing regions suffer mild to severe water shortages, as has been reported in Brazil (Table 1). Therefore, drought can cause major economic losses for sugarcane growers. Interestingly, a mild drought stress can have a positive impact on sugarcane yield. It is a common practice namely in countries that use irrigation, to apply a period of drying off (water withheld) at the end of the season. The drying off period has several benefits: save water and therefore costs associated with irrigation, reduces soil compaction during harvest and may even increase sucrose content (Robertson and Donaldson, 1998; Singels et al., 2000; Inman-Bamber, 2014). The increase in sucrose content may be due to the fact that growth is more affected than photosynthesis and therefore, assimilated CO2 can be diverted from leaf and culm growth to sucrose accumulation in the culm. Therefore, the regulation of sugarcane responses to drought certainly will have differences with those observed from other crops and model plants.
MicroRNAs
In addition to having conserved functions that extend beyond development, microRNAs (miRNAs) play crucial roles in the regulation of plant responses to several stimuli (Bartel, 2004), acting like a buffer for plant molecular dynamics. miRNAs are a class of small, non-coding RNAs of approximately 21 nucleotides in length that are endogenous to both plants and animals (Bartel, 2009; Carthew and Sontheimer, 2009) and function to regulate gene expression by sequence-specific interaction with target mRNAs (Bartel, 2004; Chapman and Carrington, 2007). Conserved miRNAs mainly regulate transcription factors involved in basic functions, such as cell division, hormonal control or meristem development (Garcia, 2008). miRNAs arose from genome duplications and rearrangements and, for this reason, frequently have many copies (Voinnet, 2009). However, some recent miRNAs are thought to be represented by single-copy genes that are not conserved in phylogenies (Zhang et al., 2008b).
In plants, most MIR genes possess their own transcriptional unit and are transcribed by RNA polymerase II (Pol II) into a primary miRNA (pri-miRNA) (Lee et al., 2004). The pri-miRNA forms an imperfect foldback structure, ranging from hundreds to thousands of bases (Zhang et al., 2009). These structures are stabilized by the addition of a 5′ 7-methylguanosine cap and a 3′ polyadenylated tail (Jones-Rhoades and Bartel, 2004; Xie et al., 2005; Zhang, 2005).
In nuclear processing centers called D-bodies (or SmD3/SmD3-bodies), the pri-mRNA is processed into a stem-loop precursor (pre-miRNA) and generates a double-stranded RNA duplex via a Dicer-like protein (DCL), a nuclear RNase III-like enzyme, and two RNA-binding proteins named HYPONASTIC LEAVES1 (HYL1) and the C2H2-zinc finger protein SERRATE (SE) (Kurihara et al., 2006; Lobbes et al., 2006; Fang and Spector, 2007).
The pre-miRNAs range from 60 to >400 nt in size (Xuan et al., 2011), and the double-strand RNA duplex, also called miRNA/miRNA*, ranges from 19 to 24 nt long (Reinhart et al., 2002; Bartel, 2004). The precise release of miRNA duplexes from the pre-miRNAs is both structure- and sequence-dependent (reviewed by Naqvi et al., 2012; Rogers and Chen, 2013), while the size is dependent on the action of the DCL family member (Margis et al., 2006); DCL1 produces small RNAs of 18–21 nt, while those of DCL2, DCL3, and DCL4 are 22 nt, 24 nt, and 21 nt, respectively (Voinnet, 2009). In plants, most miRNAs are processed by DCL1 (Reinhart et al., 2002) and are predominately 21 nt long (Chen et al., 2010).
The short double stranded RNAs (dsRNAs) that result from DCL processing have 2-nt 3′ overhangs and are methylated at their 3′ ends by the methyltransferase HEN-1 (Yu et al., 2005; Fang and Spector, 2007). This step protects dsRNAs from uridylation and subsequent degradation (Li et al., 2005).
The exact form in which miRNA/miRNA* duplexes are transported across the nuclear membrane is unclear. In the cytoplasm, one of the strands, called the mature miRNA, is incorporated into an Argonaute protein (AGO) to form the RNA-induced silencing complex (RISC), and the miRNA* strand is usually degraded (Reinhart et al., 2002; Bartel, 2004; Voinnet, 2009).
Similar to the DCL family, the Argonaute family also has several members, and AGO1 is generally associated with miRNA biogenesis (Vaucheret, 2008). The process of choosing the miRNA strand that incorporates the complex is dependent on the thermodynamic stability of the 5′ portion of the duplex; the strand with the lower stability is incorporated by AGO1 (Eamens et al., 2009). The incorporated mature miRNA guides the RISC to scan the cytoplasm to find a specific target mRNA by base pairing, leading to mRNA cleavage or translational repression (Bartel, 2004). Therefore, in most cases, miRNAs will reduce the expression of their target mRNAs.
miRNAs Modulated by Drought in Sugarcane
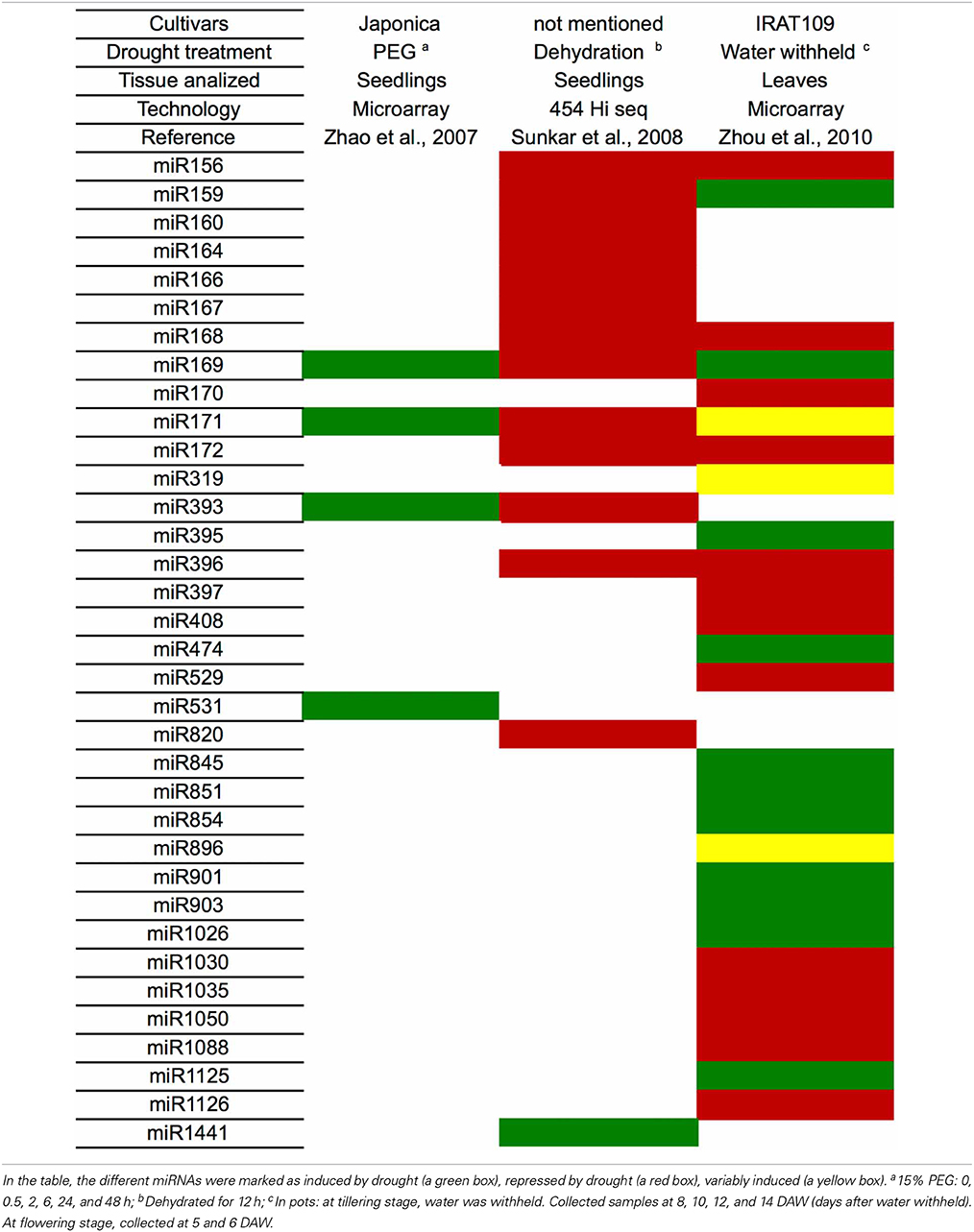
There are several miRNAs that have been identified in a wide array of species, but only a few studies have been performed to identify the mature miRNA sequences and analyze their expression in response to drought stress in sugarcane (Ferreira et al., 2012; Thiebaut et al., 2012; Gentile et al., 2013). Sugarcane is a complex polyploid and until now its genome sequence has not been obtained. Therefore, unlike model species with sequenced genomes such as Arabidopsis thaliana, miRNA characterization studies are much more complicated. For example, miRNA precursors are highly unstable, making their detection in the sugarcane EST collection (Vettore et al., 2003) very difficult. A sequenced genome would facilitate the discovery of these precursors. This is particularly relevant for the discovery of novel miRNA, since the finding of a precursor is a pre-requisite to consider a new sequence as a miRNA. Similarly, the discovery of miRNA targets is greatly facilitated when the complete genome is available. Therefore, the use of the genome of sorghum, a closely related species as mentioned above, is a key strategy to overcome this limitation.
Even though, in one study (Thiebaut et al., 2012), eight sugarcane cultivars were classified into two groups based on their tolerance to drought. Plants were grown in a greenhouse for three months and then submitted to drought stress by withholding irrigation for 24 h. Although the number of detected miRNAs was higher in the more tolerant cultivars, no miRNA was found to be induced by drought under these conditions (Thiebaut et al., 2012).
In the other two works, two sugarcane cultivars that differ in their tolerance to drought stress, RB867515 (higher tolerance, HT) and RB855536 (lower tolerance, LT), were either grown in a greenhouse for three months and then kept without water for 2 or 4 days (Ferreira et al., 2012) or field-grown for 7 months under irrigation or without irrigation (rainfed) (Gentile et al., 2013). Thirteen families of mature miRNAs were found in the two sugarcane cultivars studied (Table 2).
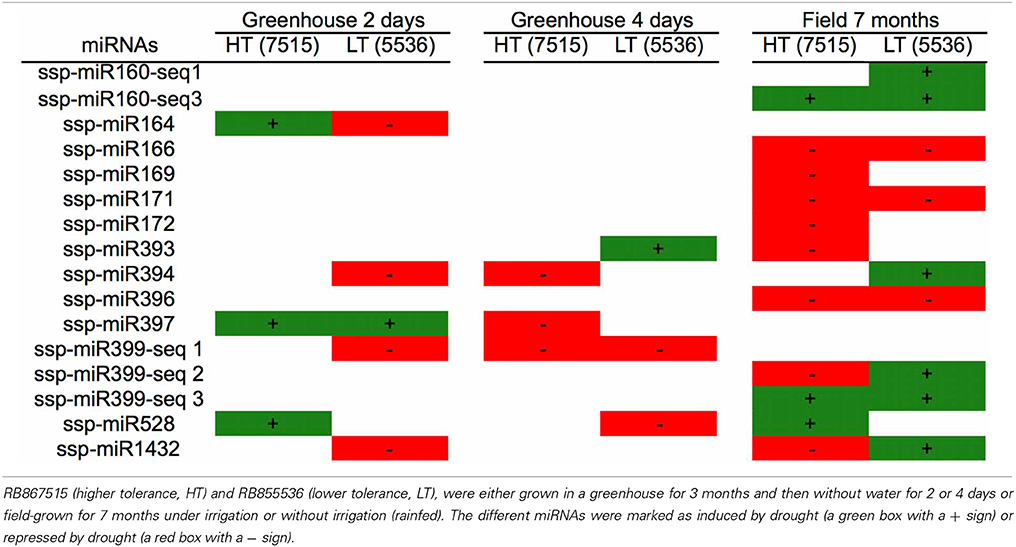
Table 2. miRNAs identified in two sugarcane cultivars differing in their tolerance to drought stress.
Different expression profiles of the miRNAs were observed, depending on the cultivar, the growth conditions and the type and duration of stress. Some miRNAs were found only in plants that grew in the greenhouse (ssp-miR164, ssp-miR397 and spp-miR399-seq1), while others were found only in field-grown plants (ssp-miR160-seq1, ssp-miR160-seq3, ssp-miR166, ssp-miR169, ssp-miR171 and ssp-miR172). This most likely reflects differences in the growth conditions, with field grown-plants giving a better picture of the real growth conditions that plants face in nature.
However, some miRNAs had opposing expression profiles depending on the cultivar, such as ssp-miR164, ssp-miR399-seq2 and ssp-miR1432 (up-regulated by drought in one cultivar and down-regulated in the other). In the experiment conducted in the field, a higher number of miRNAs were modulated by drought, and many of them showed a repressed profile (ssp-miR166, ssp-miR169, ssp-miR171 and ssp-miR172, among others), while others changed from down-regulated to up-regulated (spp-miR399-seq2 and ssp-miR1432) depending on the cultivar.
The timing of stress also affected the miRNA expression profile. For example, comparing greenhouse-grown plants that were stressed for 2 or 4 days, ssp-miR397 showed an altered expression profile, changing from induced to repressed, while ssp-miR399-seq1 remained invariantly down-regulated. The other miRNAs found in the greenhouse-grown plants had variable expression profiles, without a specific pattern. After 7 months, five miRNAs from the rainfed field-grown plants presented the same profile in both cultivars; two were induced (ssp-miR160-seq3 and ssp-miR399-seq3) and three were repressed (ssp-miR166, ssp-miR171 and ssp-miR396). Only two miRNAs (ssp-miR399-seq2 and ssp-miR1432) had opposite profiles among the different cultivars.
As a summary, by evaluating two sugarcane cultivars that differ in their level of drought tolerance according to their performance under field conditions (Gentile et al., 2013), a total of 16 mature miRNAs were found (Figure 1). Among the cultivars, we found that 15 mature miRNAs were differentially expressed and identified in the cultivar with higher tolerance to drought (HT, Figure 1A), while 14 were found in the cultivar with lower tolerance (LT, Figure 1B). We found that only two miRNAs (spp-miR394 and ssp-miR528) were shared among the different stress durations (2 days, 4 days, and 7 months) and two growing conditions (greenhouse and field) (Figure 1C).
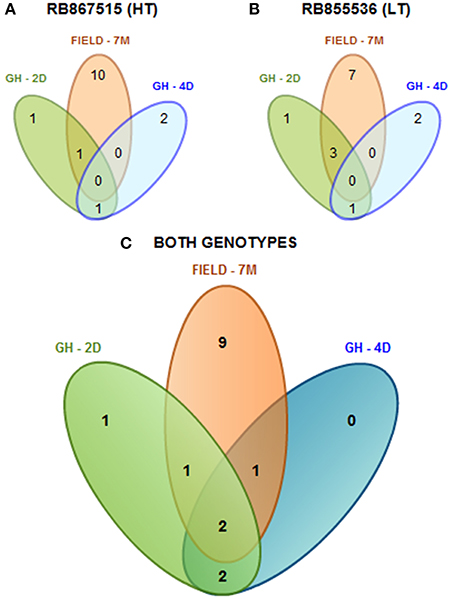
Figure 1. Diagram of all the differentially expressed mature miRNAs found in sugarcane. miRNAs found among the different stress times (2 days, 4 days, and 7 months) under greenhouse (GH) and field-grown (FIELD) conditions in the more tolerant cultivar (RB867515, HT) (A), in the less tolerant cultivar (RB855536, LT) (B) or in both genotypes together (C).
It is remarkable that the expression patterns of the majority of the miRNAs did not display clear correlations with the differences in drought tolerance observed in the two sugarcane cultivars. Only ssp-528 presented a consistent induction in the RB867515 cultivar, which has high tolerance to drought (Table 2). The miRNA expression profiles were influenced by the genetic background from the distinct sugarcane cultivars, and this was more evident under greenhouse conditions. These data suggest that miRNAs do not fully explain the different levels of drought tolerance observed in the sugarcane cultivars.
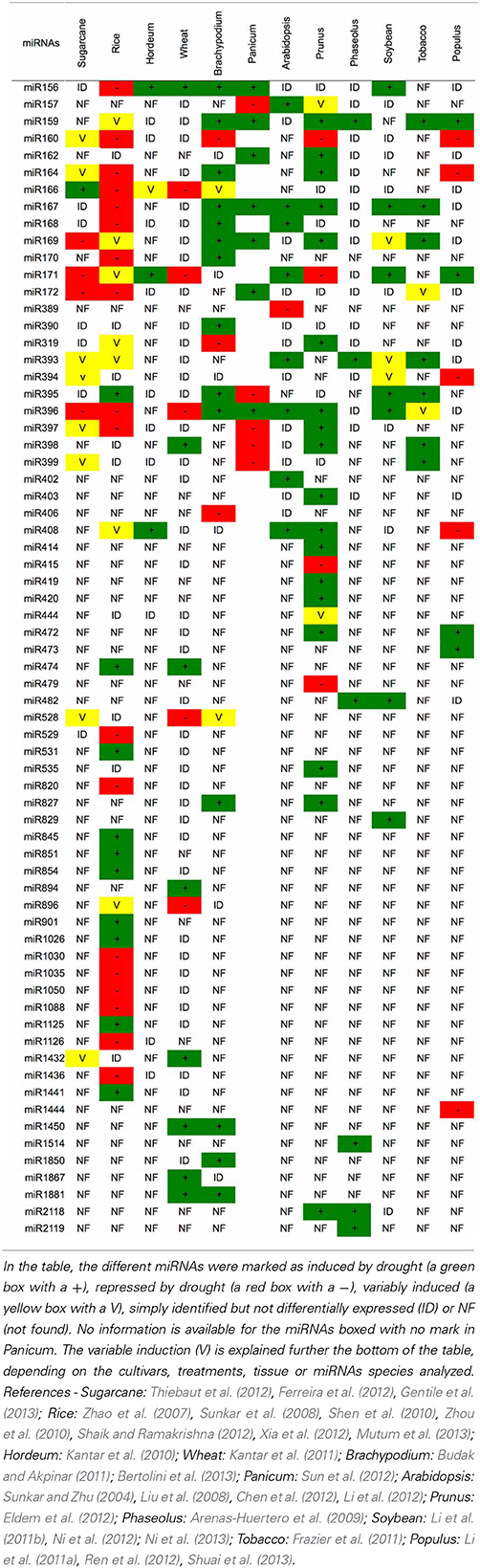
Several studies in other plants species also have identified miRNAs that are modulated by drought (Table 3). Until now, the majority of the miRNAs associated with this stress were induced by drought. Rice (Oryza sativa) is the plant with the largest number of identified miRNAs that are modulated by drought (35 miRNAs, Tables 3, 4). Barley (Hordeum vulgare) was the plant with the lowest number of identified miRNAs related to drought (4 miRNAs, Table 3). Bean (Phaseolus vulgaris) was the only plant species that had only induced miRNAs (6 miRNAs).
However, similar to the results in sugarcane, the expression pattern for a given miRNA was variable depending on the genetic background (species), type of drought treatment (PEG, dehydration, mannitol), tissue (leaves, seedlings, spikelets, roots), cultivar and growth condition (greenhouse, field, hydroponic). The miRNA that was differentially expressed in the most species was miR396 (9 species), followed by miR171, which is present in 8 plant species (Table 3). However, no miRNA was always induced or repressed in all the plant species analyzed. Moreover, no miRNA was differentially expressed in all plant species. Interestingly, sugarcane had the most variable miRNA expression profile, most likely reflecting the different cultivars, treatments and tissues that have been analyzed.
Sugarcane miRNA Targets
Plant miRNAs directly affect their target genes by nearly perfect base pairing complementarity, leading to cleavage or translation repression of these genes. Plant microRNAs can be classified into several different families, and the members of each family have very similar mature sequences. It has been reported that conserved miRNAs from the same family may have the same target genes in different species, as shown in the two model plants Arabidopsis thaliana and O. sativa (Cuperus et al., 2011). Combining data sets from high-throughput sequencing studies, Cuperus et al. (2011) identified eight miRNA families with a common ancestry in all embryophytes and a range of other families that share similar members between eudicots. The high similarity between the miRNA species in different plants allowed the development of tools for the prediction and validation of the target genes that are regulated by these miRNAs.
Several freely available tools are dedicated to the prediction of miRNA targets in plants and have been widely used (Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004; Fahlgren and Carrington, 2010). Ready-to-use online tools include RNAHybrid (Krüger and Rehmsmeier, 2006), UEA sRNA-tools (Moxon et al., 2008), Target-Align (Xie and Zhang, 2010), psRNAtarget (Dai and Zhao, 2011) and PMTED (Sun et al., 2013). All of these use algorithms that rely mostly on sequence complementarity analysis, implementing filtering criteria that aim to simulate the target recognition process that occurs within the RISC complex (Bartel, 2009). The first algorithms that was developed for target identification in plants required only that the miRNA:mRNA alignment did not exceed four mismatches, regardless of the position or nature of those mismatches. Recently, more sophisticated approaches have been successfully increasing target prediction results in several plant species, including sugarcane (Zanca et al., 2010). They were developed using a scoring system that differentiates gaps, simple mismatches, G:U pairing and consider the position at which these features occur in the alignment (Zhang et al., 2006). However, each of these tools uses different implementations of these criteria, which may be a source of divergent results.
The proliferation of target prediction tools over the last 10 years makes the choice and evaluation of their results a challenge. A recent study, by Srivastava et al. (2014), showed how 11 plant miRNA target prediction tools compare to each other. Most of the tested tools use modifications of the Smith-Waterman alignment algorithm (which is very precise, with low computational cost for short sequences). The best ranked tools make their predictions by coupling alignment results with programs that access secondary structure of RNA molecules and parameters based on the most recent findings on miRNA:Target recognition. The fact that all of the plant-specific tools have been developed and trained using Arabidopsis miRNAs explains the high overlap between their analysis, and also the lower success rate in predicting miRNA targets in non-model organism. Non plant-specific algorithms present large number of predictions with very low precision.
The results described by Srisvastava et al. also show that, although still highly skewed toward Arabidopsis, a number of tools perform well when predicting targets from other plant species, given that the user sets optimized parameters for the analysis. The most reliable and rapidly obtained results were observed with Targetfinder (Fahlgren et al., 2007), psRNAtarget (Dai and Zhao, 2011) and TapirHybrid (Bonnet et al., 2010).
It is noteworthy that algorithms that use prediction criteria beyond the concept of high sequence complementarity are amongst the best performers. But, as highlighted by the authors, the occurrence of false negatives suggests there are important target recognition details still to be uncovered. Therefore, when working with species other than Arabidopsis, users are advised to avoid default settings of those tools. The recommended approach is to use an experimentally validated dataset as control, from the same species or the closest relative, to adjust the parameters of the algorithm.
The degradation rate of a particular miRNA seems to be highly dependent on its target abundance and complementarity. After the cleavage of a target, the miRISC (miRNA and RISC complex) must survive; if this complex is not maintained, the released miRNA could form another miRISC, and another round of targeting would occur (Meng et al., 2011). Because the induction or repression of a particular miRNA may depend on stresses and cell type, it is expected that additional non-conserved miRNAs will be discovered as experiments with a wide array of conditions are performed.
Few studies have presented data on the expression of target genes in sugarcane (Zanca et al., 2010; Ferreira et al., 2012; Thiebaut et al., 2012; Carnavale-Bottino et al., 2013; Gentile et al., 2013; Ortiz-Morea et al., 2013), and only a fraction of these are related to drought stress (Ferreira et al., 2012; Thiebaut et al., 2012; Gentile et al., 2013). In fact, in most cases, researchers rely on RT-qPCR expression pattern analysis of possible targets that were previously predicted by in silico tools. Basically, these tools analyze the complementarity between a miRNA and a transcript and calculate the unpaired energy (UPE) that would be necessary to open the secondary structure around the small RNA target site on the mRNA (Zanca et al., 2010; Dai and Zhao, 2011). A recent approach for target validation is 5′ RLM-RACE (Llave et al., 2011), which provides the amplification of the 3′ cleavage product from the miRNA:target interaction events. However, the validation of the cleavage of miRNA targets under drought stress by this approach in sugarcane has not yet been reported. It is worth noting that miRNAs may interfere with gene expression by causing mRNA cleavage or by blocking mRNA translation.
In spite of the wide array of bioinformatics tools that can be used as first approach to identify putative targets, few reports in humans, animals and plants have used experimental tools to further prove the true targets of a particular miRNA. It is beyond the scope of this review to discuss the experimental strategies that can be used as well as their limitations, since this has been addressed by several reviews on this subject (Thomson et al., 2011; Ding et al., 2012; Moqadam et al., 2013). In general, a first approach is to check if the miRNA::target pair has opposed expression patterns, i.e., when the miRNA is up-regulated, the target is down regulated and vice-versa. In another approach, a construct containing the miRNA binding site in the coding region of the luciferase gene or in the 3′UTR is co-expressed with another construct overexpressing the miRNA. Decreased levels of luciferase can be observed in those cases where a true miRNA::target interaction takes place, as observed by Liu et al. (2014). The cleavage site in the mRNA can also provide an experimental evidence of miRNA action, by using RNA ligase mediated- 5′ rapid identification of cDNA ends (5′ RLM-RACE), as firstly observed for miR171 and a member of the Scarecrow-like (SCL) transcripton factor (Llave et al., 2002).
However, these and several other strategies aiming a single miRNA::target pair are work intensive. They are not suitable to address the challenge of evaluating the high number of miRNA::targets interactions predicted in miRNA expression profiling methods using DNA chips or RNAseq. A straightforward method to address this challenge is the analysis of the degradome. This strategy allows the sequencing of the entire set of cleavage products derived from all miRNAs in a sample, allowing the mapping of the exact cleavage site (Addo-Quaye et al., 2008; German et al., 2008). Clearly, with the decreasing costs of DNA sequencing, the research on sugarcane miRNAs will soon benefit from these high throughput technologies.
miRNAs and Sugarcane Responses to Drought
The miRNA expression patterns and the predicted targets described in the previous works with sugarcane under greenhouse and field conditions (Ferreira et al., 2012; Gentile et al., 2013) provide a working model of the defense strategies that might be regulated by miRNAs in sugarcane exposed to drought.
Plants grown under field conditions had increased levels of ssp-miR166 when stressed by drought. This miRNA targets transcription factors from the homeobox-leucine zipper. The overexpression of a transcription factor from this family caused reduced internodes in the model plant Arabidopsis thaliana. Interestingly, reduced stalk length is one of the most remarkable phenotype in sugarcane plants exposed to drought (Inman-Bamber and Smith, 2005; Silva et al., 2008, and references therein). Therefore, reduced ssp-miR166 would increase the levels of the transcription factor that is involved in shortening the internodes.
ssp-miR171, was also repressed by drought under field conditions, targets a sugarcane gene encoding a protein with high identity to members of the scarecrow-like transcription factor (SCL—GRAS domain protein) family. The Arabidopsis homologs of this protein induce shoot branching and are targets of miR171, and overexpression of miR171 caused reduced shoot branching in transgenic plants (Wang et al., 2010a). Shoot branching is reduced under drought stress in sugarcane, compromising plant survival and reducing crop productivity (Inman-Bamber and Smith, 2005; Silva et al., 2008; Kapur et al., 2011). Decreased levels of ssp-miR171 and conversely increased levels of the SCL transcription factor, could be a sugarcane response to counteract the deleterious effects of drought on tillering.
ssp-miR160 was up-regulated in response to drought in field-grown plants. This miRNA targets a sugarcane gene that has high identity to the VNI2 protein from Arabidopsis. This protein repress the activity of a transcription factor, VASCULAR-RELATED NAC-DOMAIN7 (VND7), that is a master inducer of xylem formation (Yamaguchi et al., 2010). Although, to our knowledge, there are no works showing xylem differentiation in response to drought in sugarcane, this response has been observed in poplar trees (Arend and Fromm, 2007). Therefore, ssp-miR160 induction could lead to decreased levels of VIN2, releasing the action of VND7, that would work in xylem differentiation. This could, in turn, improve the ability of sugarcane plants to transport water.
Delaying leaf senescence is an agronomical trait that has a positive impact on plant yield under drought stress, as observed in sorghum (Borrel et al., 2000). Moreover, increased levels of isopentenyltransferase, the rate-limiting step of cytokinin biosynthesis, delay senescence in transgenic tobacco plants and increase drought tolerance (Rivero et al., 2007). Leaf senescence is observed in sugarcane plants under drought stress and is correlated with decreased crop productivity (Inman-Bamber, 2004; Lopes et al., 2011). ssp-miR399 is induced by drought in field grown sugarcane and targets a protein associated with leaf senescence in maize. Increased levels of this miRNA could be a response to keep a green leaf phenotype, allowing sugarcane plans to sustain photosynthesis for a longer period under stress.
As observed in many other species, drought induces oxidative stress in sugarcane, increasing H2O2 content and the levels of lipid peroxidation (Cia et al., 2012). A miRNA, ssp-miR169, repressed by drought in one sugarcane cultivar grown in the filed, targets a glutathione S-transferase (GST). These enzymes are involved in the detoxification of compounds generated during stress and transgenic plants overexpressing GSTs have increased tolerance to oxidative stress and water deficit (George et al., 2010; Ji et al., 2010). Reduced levels of ssp-miR169 could increase GST levels and therefore reduce the toxic effects of reactive oxygen species.
miRNA expression profiles also revealed a range of transcription factors that may be involved in plant responses and tolerance to drought stress (Table 5). We found many transcriptions factors, such as NAC domain, homeobox-leucine zipper, Nuclear Factor YA, GRAS/SCL, APETALA2 and bZIP transcription factors. All of these have been described as being related to drought stress and/or increasing tolerance to water stress when overexpressed in other plants (Dezar et al., 2005; Nelson et al., 2007; Stephenson et al., 2007; Li et al., 2008; Ma et al., 2010; Ditt et al., 2011; Golldack et al., 2011; Krishnaswamy et al., 2011). Other targets encode a wide array of proteins. A NSP-interacting kinase (NIK), which is a member of the serine/threonine kinase subfamily that is involved in plant development and responses to external stimuli. An auxin receptor that specifically binds to a repressor that is then degraded, allowing the expression of genes related to auxin, was also found (Dharmasiri et al., 2005). In addition to these targets, a GAPDH (glyceraldehyde-3-phosphate dehydrogenase), which is involved in generating more ATP, and a pyruvate dehydrogenase enzyme, involved in carbon balance during the stress (Chaves et al., 2009) were also identified. An inorganic pyrophosphatase 2-like was found among the targets and has already been reported to confer tolerance to drought stress when overexpressed in several plants (Gaxiola et al., 2001; Park et al., 2005; Zhang et al., 2011). Finally, some enzymes were identified as involved in cell modifications, such as laccases, that reduce cell elongation during drought stress (Cachorro et al., 1993). These results showed that the sugarcane miRNAs identified under drought stress could regulate different genes that function in several metabolic pathways, indicating the plasticity and the complexity of sugarcane responses to this stress.
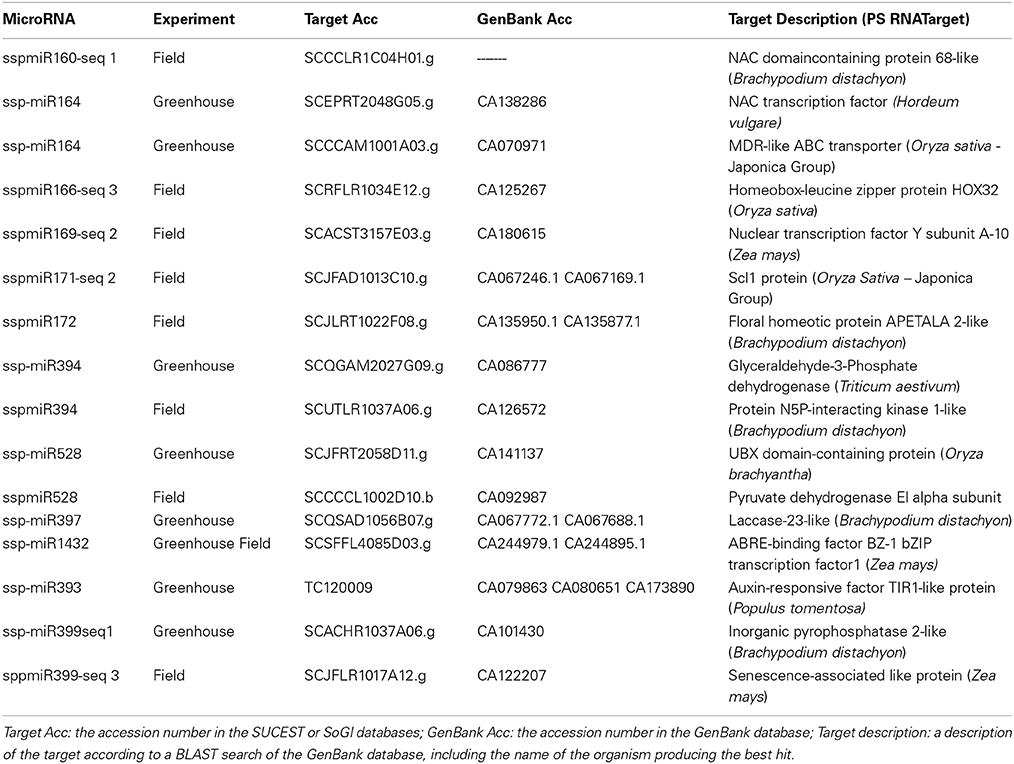
Table 5. Target prediction for the miRNAs that were differentially expressed in drought-stressed sugarcane plants.
Conclusions and Perspectives
In this review, we have evaluated studies describing the expression profiles of miRNAs from sugarcane under drought stress. Cultivars that differ in their level of drought tolerance were grown under different conditions and stressed in different ways. Our analysis provides insights into the complexity of the sugarcane miRNA regulatory network under drought stress. Few studies have evaluated plant responses under real field conditions, and we found that these responses differ considerably from those observed in the greenhouse. The different genetic background of the cultivars used in the sugarcane studies highlight a new layer of complexity in miRNA expression. Interestingly, this complexity observed in sugarcane was also detected in other plant species. Taken together, the data from miRNA expression under drought stress suggest that plants may adjust their microtransptome in a variety of ways to cope with different phases and intensity of drought stress and that these responses may be fine-tuned in particular genetic backgrounds.
This complexity of expression patterns urges us to move toward functional assays and the use of mutants with decreased or increased expression of selected miRNAs. These mutants could be produced, for example, by overexpressing or silencing miRNA precursors in transgenic plants, and will be extremely helpful in assessing the role of miRNAs in drought responses.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agẽncia Globo. (2012). Drought affects the development of sugarcane in Alagoas. (Original in portuguese: seca afeta desenvolvimento das lavouras de cana em AL.). Available online at: http://g1.globo.com/economia/agronegocios/noticia/2012/11/seca-afeta-desenvolvimento-das-lavouras-de-cana-em-al.html (Accessed on October 08, 2013).
Addo-Quaye, C., Eshoo, T. W., Bartel, D. P., and Axtell, M. J. (2008). Endogenous siRNA and microRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762. doi: 10.1016/j.cub.2008.04.042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arenas-Huertero, C., Perez, B., Rabanal, F., Blanco-Melo, D., De La Rosa, C., Estrada-Navarrete, G., et al. (2009). Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 70, 385–401. doi: 10.1007/s11103-009-9480-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arend, M., and Fromm, J. (2007). Seasonal change in the drought response of wood cell development in poplar. Tree Physiol. 27, 985–992. doi: 10.1093/treephys/27.7.985
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Asnaghi, C., Paulet, F., Kaye, C., Grivet, L., Deu, M., Glaszmann, J. C., et al. (2000). Application of synteny across Poaceae to determine the map location of a sugarcane rust resistance gene. Theor. Appl. Genet. 101, 962–969. doi: 10.1007/s001220051568
Assmann, S. M., and Grantz, D. A. (1990). Stomatal response to humidity in sugarcane and soybean: effect of vapour pressure difference on the kinetics of the blue light response. Plant Cell Environ. 13, 163–169.
Associação dos fornecedores de cana de pernambuco. (2012). Prolonged dry lets sugarcane growing cities in emergency (Original in Portuguese: Seca prolongada deixa cidades canavieiras em emerge ncia). Available online at: http://www.afcp.com.br/?p=3992 (Accessed on November 19, 2013).
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bertolini, E., Verelst, W., Horner, D. S., Gianfranceschi, L., Piccolo, V., Inzé, D., et al. (2013). Addressing the role of microRNAs in reprogramming leaf growth during drought stress in Brachypodium distachyon. Mol. Plant 6, 423–443. doi: 10.1093/mp/sss160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bonnet, E., He, Y., Billiau, K., and De Peer, Y. (2010). TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26, 1566–1568. doi: 10.1093/bioinformatics/btq233
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Borrel, A. K., Hammer, G. L., and Henzell, R. G. (2000). Does maintaining green leaf area in sorghum improve yield under drought? II. Dry matter production and yield. Crop Sci. 40, 1037–1048. doi: 10.2135/cropsci2000.4041037x
Botha, F. C., and Black, K. G. (2000). Sucrose phosphate synthase and sucrose synthase activity during maturation of internodal tissue in sugarcane. Funct. Plant Biol. 27, 81–85. doi: 10.1071/PP99098
Brasilagro. (2013). Pernambuco: drought devastates sugarcane production in the zona da Mata (Original in Portuguese: PE: seca devasta produção de cana-de-açúcar na Zona da Mata). Available online at: http://www.brasilagro.com.br/conteudo/pe-seca-devasta-producao-de-cana-de-acucar-na-zona-da-mata.html#.VGyPLpPF_Sk (Accessed on November 19, 2013).
Budak, H., and Akpinar, A. (2011). Dehydration stress-responsive miRNA in Brachypodium distachyon: evident by genome-wide screening of microRNAs expression. Omics 15, 791–799. doi: 10.1089/omi.2011.0073
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cachorro, P., Ortiz, A., Barcelo, A. R., and Cerda, A. (1993). Lignin deposition in vascular tissues of Phaseolus vulgaris roots in response to salt stress and Ca2+ ions. Phyton-Ann. Rei Bot. A 33, 33–40.
Camarotto, M. (2012). Drought will cause losses of 35% in the sugarcane harvest in Pernambuco, according to growers. (Original in Portuguese: seca causará perda de 35% da safra de cana em PE, estimam produtores). Available online at: http://www.valor.com.br/brasil/2904610/seca-causara-perda-de-35-da-safra-de-cana-em-pe-estimam-produtores (Accessed on October 08, 2013).
Carnavale-Bottino, M., Rosario, S., Grativol, C., Thiebaut, F., Rojas, C. A., and Farrineli, L., et al. (2013). High-throughput sequencing of small RNA transcriptome reveals salt stress regulated microRNAs in sugarcane. PLoS ONE 8:e59423. doi: 10.1371/journal.pone.0059423
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655. doi: 10.1016/j.cell.2009.01.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Castro, M. (2008). Cane: drought will reduce 6.3% of the harvest. (Original in Portuguese: cana: seca reduzirá 6,3% da safra). Available online at: http://www.estadao.com.br/noticias/suplementos,cana-seca-reduzira-63-da-safra,117410,0.htm (Accessed on October 08, 2013).
Cavalcanti, J. (2010). Cane under drought. (Original in Portuguese: cana sob estiagem). Available online at: http://www.modclima.com.br/pdf/cana-sob-estiagem.pdf (Accessed on October 08, 2013).
Chapman, E. J., and Carrington, J. C. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Gen. 8, 884–896. doi: 10.1038/nrg2179
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chaves, M. M., Flexas, J., and Pinheiro, C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103, 551–560 doi: 10.1093/aob/mcn125
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, H., Li, Z., and Xiong, L. (2012). A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett. 586, 1742–1747. doi: 10.1016/j.febslet.2012.05.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, X., Zhang, Z., Liu, D., Zhang, K., Li, A., and Mao, L. (2010). SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J. Integr. Plant Biol. 52, 946–951. doi: 10.1111/j.1744-7909.2010.00987.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cia, M. C., Guimaraes, A. C. R., Medici, L. O., Chabregas, S. M., and Azevedo, R. A. (2012). Antioxidant responses to water deficit by drought-tolerant and -sensitive sugarcane varieties. Ann. Appl. Biol. 161, 313–324. doi: 10.1111/j.1744-7348.2012.00575.x
Clayton, W. D., and Renvoize, S. A. (1982). “Gramineae,” in Flora of Tropical East Africa, ed R. M. Polhill (Rotterdam: Balkema), Part 3, 700–767.
Commodity Research Bureau. (2015). The 2015 CRB commodity yearbook. Chicago, IL: Commodity Research Bureau.
Conab (Companhia nacional de abastecimento). (2013). Acompanhamento de safra brasileira: cana-de-açúcar, terceiro levantamento, abril/2013. Brasília: Companhia Nacional de Abastecimento.
Cordeiro, G. M., Casu, R., McIntyre, C. L., Manners, J. M., and Henry, R. J. (2001). Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci. 160, 1115–1123. doi: 10.1016/S0168-9452(01)00365-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cuperus, J. T., Fahlgren, N., and Carrington, J. C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23, 431–442. doi: 10.1105/tpc.110.082784
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
D'Hont, A., Grivet, L., Feldmann, P., Glaszmann, J. C., Rao, S., and Berding, N. (1996). Characterization of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol. Genet. Genomics 250, 405–413.
Dai, X., and Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dezar, C. A., Gago, G. M., Gonzalez, D. H., and Chan, R. L. (2005). Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 14, 429–440. doi: 10.1007/s11248-005-5076-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. doi: 10.1038/nature03543
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dillon, S. L., Shapter, F. M., Henry, R. J., Cordeiro, G., Izquierdo, L., et al. (2007). Domestication to crop improvement: genetic resources for Sorghum and Saccharum (Andropogoneae). Ann. Bot. 100, 975–989. doi: 10.1093/aob/mcm192
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ding, J., Zhou, S., and Guan, J. (2012). Finding MicroRNA targets in plants: current status and perspectives. Genomics Proteomics Bioinformatics 10, 264–275. doi: 10.1016/j.gpb.2012.09.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ditt, R. F., Gentile, A., Tavares, R. G., Camargo, S. R., Fernandez, J. H., Da, Silva, M. J., et al. (2011). Analysis of the stress-inducible transcription factor SsNAC23 in sugarcane plants. Sci. Agric. 68, 454–461. doi: 10.1590/S0103-90162011000400010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eamens, A. L., Smith, N. A., Curtin, S. J., Wang, M. B., and Waterhouse, P. M. (2009). The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15, 2219–2235. doi: 10.1261/rna.1646909
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eldem, V., Çelikkol Akçay, U., Ozhuner, E., Bakir, Y., Uranbey, S., and Unver, T. (2012). Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS ONE 7:e50298. doi: 10.1371/journal.pone.0050298
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fahlgren, N., and Carrington, J. C. (2010). miRNA target prediction in plants. Methods Mol. Biol. 592, 51–57. doi: 10.1007/978-1-60327-005-2_4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., et al. (2007). High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2:e219. doi: 10.1371/journal.pone.0000219
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fang, Y., and Spector, D. L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17, 818–823. doi: 10.1016/j.cub.2007.04.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ferreira, T. H., Gentile, A., Vilela, R. D., Costa, G. G. L., Dias, L. I., Endres, L., et al. (2012). microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS ONE 7:e46703. doi: 10.1371/journal.pone.0046703
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frazier, T. P., Sun, G., Burklew, C. E., and Zhang, B. (2011). Salt and drought stresses induce the aberrant expression of microrna genes in tobacco. Mol. Biotechnol. 49, 159–165. doi: 10.1007/s12033-011-9387-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
G1 Agency. (2012). Drought affects 195 municipalities and 2 cities in PB will have water rationing (Original in Portuguese: seca afeta 195 municípios da PB e 2 cidades vão ter racionamento de água). Available online at: http://g1.globo.com/pb/paraiba/noticia/2012/10/seca-afeta-195-municipios-da-pb-e-2-cidades-vao-ter-racionamento-de-agua.html (Accessed on November 19, 2014).
Garcia, D. (2008). A miRacle in plant development: role of microRNAs in cell differentiation and patterning. Garcia, D. Sem. Cell Dev. Biol. 19, 586–595. doi: 10.1016/j.semcdb.2008.07.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gascho, G. J., and Shih, S. F. (1983). “Sugarcane,” in Crop Water Relations eds I. D. Teare and M. M. Peet (New York, NY: John Wiley & Sons), 445–479.
Gaxiola, R. A., Li, J., Undurraga, S., Dang, L. M., Allen, G. J., et al. (2001). Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. U.S.A. 98, 11444–11449. doi: 10.1073/pnas.191389398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gentile, A., Ferreira, T. H., Mattos, R. S., Dias, L. I., Hoshino, A. A., Carneiro, M. S., et al. (2013). Effects of drought on the microtranscriptome of field-grown sugarcane plants. Planta 237, 783–798. doi: 10.1007/s00425-012-1795-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
George, S., Venkataraman, G., and Parida, A. (2010). A chloroplast-localized and auxin-induced glutathione S-transferase from phreatophyte Prosopis juliflora confer drought tolerance on tobacco. J. Plant Physiol. 167, 311–318. doi: 10.1016/j.jplph.2009.09.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
German, M. A., Pillay, M., Jeong, D. H., Hetawal, A., Luo, S., Janardhanan, P., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26, 941–946. doi: 10.1038/nbt1417
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Golldack, D., Luking, I., and Yang, O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30, 1383–1391. doi: 10.1007/s00299-011-1068-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
GPWG – Grass Phylogeny Working Group. (2001). Phylogeny and subfamilial classification of grasses (Poaceae). Ann. Missouri Bot. Gard. 88, 373–457.
Graça, J. P., Rodrigues, F. A., Farias, J. R. B., Oliveira, M. C. N., Hoffmann-Campo, C. B., and Zingaretti, S. M. (2010). Physiological parameters in sugarcane cultivars submitted to water deficit. Brazil. J. Plant Physiol. 22, 189–197. doi: 10.1590/S1677-04202010000300006
Inman-Bamber, N. G. (2004). Sugarcane water stress criteria for irrigation and drying off. Field Crops Res. 89, 107–122. doi: 10.1016/j.fcr.2004.01.018
Inman-Bamber, N. G., and Smith, D. M. (2005). Water relations in sugarcane and response to water deficits. Field Crops Res. 92, 185–202. doi: 10.1016/j.fcr.2005.01.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Inman-Bamber, G. (2014). “Sugarcane Yield and Yield-Limiting Processes” in Sugarcane: Physiology, Biochemistry & Functional Biology, eds P. H. Moore and F. C. Botha (Cambridge: Willey-Blackwell), 579–600.
Jannoo, N., Grivet, L., Chantret, N., Garsmeur, O., Glaszmann, J. C., Arruda, P., et al. (2007). Orhtologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J. 50, 574–585. doi: 10.1111/j.1365-313X.2007.03082.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ji, W., Zhu, Y., Li, Y., Yang, L., Zhao, X., Cai, H., et al. (2010). Over-expression of a glutathione S-transferase gene, GsGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnol. Lett. 32, 1173–1179. doi: 10.1007/s10529-010-0269-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jones-Rhoades, M. W., and Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799. doi: 10.1016/j.molcel.2004.05.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kantar, M., Lucas, S. J., and Budak, H. (2011). miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 233, 471–484. doi: 10.1007/s00425-010-1309-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kantar, M., Unver, T., and Budak, H. (2010). Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct. Integr. Genomics 10, 493–507. doi: 10.1007/s10142-010-0181-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kapur, R., Duttamajumder, S. K., and Rao, K. K. (2011). A breeder's perspective on the tiller dynamics in sugarcane. Curr. Sci. 100, 183–189.
Krishnaswamy, S., Verma, S., Rahman, M. H., and Kav, N. N. (2011). Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 75, 107–127. doi: 10.1007/s11103-010-9711-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Krüger, J., and Rehmsmeier, M. (2006). RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451–W454. doi: 10.1093/nar/gkl243
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, D. (2005). “Breeding for drought resistance,” in Abiotic stresses: Plant Resistance through Breeding and Molecular Approaches, ed M. Ashraf, P. J. C. Harris, (New York, NY: The Haworth Press), 145–175.
Kurihara, Y., Takashi, Y., and Watanabe, Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12, 206–212. doi: 10.1261/rna.2146906
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, Y., Kim, M., Han, J. J., Yeom, K. H., Lee, S., Baek, S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. Embo J. 23, 4051–4060. doi: 10.1038/sj.emboj.7600385
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, B., Qin, Y., Duan, H., Yin, W., and Xia, X. (2011a). Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J. Exp. Bot. 62, 3765–3779. doi: 10.1093/jxb/err051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, H., Dong, Y., Yin, H., Wang, N., Yang, J., Liu, X., et al. (2011b). Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol. 11:170. doi: 10.1186/1471-2229-11-170
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, J., Yang, Z., Yu, B., Liu, J., and Chen, X. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507. doi: 10.1016/j.cub.2005.07.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, W., Cui, X., Meng, Z., Huang, X., Xie, Q., Wu, H., et al. (2012). Transcriptional Regulation of Arabidopsis MIR168a and ARGONAUTE1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 158, 1279–1292. doi: 10.1104/pp.111.188789
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, W. X., Oono, Y., Zhu, J., He, X. J., Wu, J. M., Lida, K., et al. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, H. H., Tian, X., Li, Y., Wu, C., and Zheng, C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843. doi: 10.1261/rna.895308
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, Q., Wang, F., and Axtell, M. J. (2014). Analysis of complementarity requirements for plant MicroRNA targeting using a nicotiana benthamiana quantitative transient assay. Plant Cell 26, 741–753. doi: 10.1105/tpc.113.120972
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Llave, C., Franco-Zorrilla, J. M., Solano, R., and Barajas, D. (2011). “Target validation of plant microRNAs,” in MicroRNAs in Development: Methods and Protocols, Methods in Molecular Biology, Vol. 732, ed T. Dalmay (Cham: Springer International Publishing AG), 187–208.
Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002). Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. doi: 10.1126/science.1076311
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lobbes, D., Rallapalli, G., Schmidt, D. D., Martin, C., and Clarke, J. (2006). SERRATE: a new player on the plant microRNA scene. EMBO Rep. 7, 1052–1058. doi: 10.1038/sj.embor.7400806
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lopes, M. S., Araus, J. L., van Heerden, P. D. R., and Foyer, C. H. (2011). Enhancing drought tolerance in C4 crops. J. Exp. Bot. 62, 3135–3153. doi: 10.1093/jxb/err105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, H. S., Liang, D., Shuai, P., Xia, X. L., and Yin, W. L. (2010). The salt- and drought- inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 61, 4011–4019. doi: 10.1093/jxb/erq217
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Margis, R., Fusaro, A. F., Smith, N. A., Curtin, S. J., Watson, J. M., Finnegan, E. J., et al. (2006). The evolution and diversification of Dicers in plants. FEBS Lett. 580, 2442–2450. doi: 10.1016/j.febslet.2006.03.072
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meng, Y., Shao, C., Wang, H., and Chen, M. (2011). The regulatory activities of plant microRNAs: a more dynamic perspective. Plant Physiol. 157, 1583–1595. doi: 10.1104/pp.111.187088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ming, R., Liu, S. C., Lin, Y. R., Da SILVA, J., Wilson, W., Braga, D., et al. (1998). Detailed alignment of saccharum and sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics 150, 1663–1682.
Ming, R., Del Monte, T. A., Hernandez, E., Moore, P. H., Irvine, J. E., and Paterson, A. H. (2002a). Comparative analysis of QTLs affecting plant height and flowering among closely-related diploid and polyploid genomes. Genome 45, 794–803. doi: 10.1139/g02-042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ming, R., Liu, S. C., Bowers, J. E., Moore, P. H., Irvine, J. E., and Paterson, A. H. (2002b). Construction of a Saccharum consensus genetic map from two interspecific crosses. Crop Sci. 42, 570–583. doi: 10.2135/cropsci2002.5700
Ming, R., Wang, Y. W., Draye, X., Moore, P. H., Irvine, J. E., and Paterson, A. H. (2002c). Molecular dissection of complex traits in autopolyploids: mapping QTLs affecting sugar yield and related traits in sugarcane. Theor. Appl. Genet. 105, 332–345. doi: 10.1007/s00122-001-0861-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miranda, L. L. D., Vasconcelos, A. C. M., Landell, M. G., and Xavier, M. A. (2008). Viveiro de mudas. em: leila luci dinardo-miranda; antõnio carlos machado de vasconcelos; marcos guimarães de andrade landell. (org.). cana-de-açúcar. 1ed. Campinas 1, 535–546.
Moqadam, F. A., Pieters, R., and Den Boer, M. L. (2013). The hunting of targets: challenge in miRNA research. Leukemia 27, 16–23. doi: 10.1038/leu.2012.179
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moxon, S., Jing, R., Szittya, G., Schwach, F., Rusholme Pilcher, R. L., Moulton, V., et al. (2008). Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 18, 1602–1609. doi: 10.1101/gr.080127.108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mutum, R. D., Balyan, S. C., Kansal, S., Agarwal, P., Kumar, S., Kumar, M., et al. (2013). Evolution of variety-specific regulatory schema for expression of osa-miR408 in indica rice varieties under drought stress. FEBS J. 280, 1717–1730. doi: 10.1111/febs.12186
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naqvi, A. R., Sarwat, M., Hasan, S., and Roychodhury, N. (2012). Biogenesis, functions and fate of plant microRNAs. J. Cell Physiol. 227, 3163–3168. doi: 10.1002/jcp.24052
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nelson, D. E., Repetti, P. P., Adams, T. R., Creelman, R. A., Wu, J., Warner, D. C., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. U.S.A. 104, 16450–16455. doi: 10.1073/pnas.0707193104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ni, Z., Hu, Z., Jiang, Q., and Zhang, H. (2012). Overexpression of gma-MIR394a confers tolerance to drought in transgenic Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 427, 330–335. doi: 10.1016/j.bbrc.2012.09.055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ni, Z., Hu, Z., Jiang, Q., and Zhang, H. (2013). GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82, 113–129. doi: 10.1007/s11103-013-0040-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ortiz-Morea, F. A., Vicentini, R., Silva, G. F. F., Silva, E. M., Carrer, H., Rodrigues, A. P., et al. (2013). Global analysis of the sugarcane microtranscriptome reveals a unique composition of small RNAs associated with axillary bud outgrowth. J. Exp. Bot. 64, 2307–2320. doi: 10.1093/jxb/ert089
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palhares, I. (2014). Drought causes break up to 15% in crops in the region of Ribeirão Preto (Original in Portuguese: seca provoca quebra de até 15% em safras na região de Ribeirão Preto). Available online at: http://www1.folha.uol.com.br/cotidiano/ribeiraopreto/2014/10/1537883-seca-provoca-quebra-de-ate-15-em-safras-na-regiao-de-ribeirao-preto.shtml (Accessed on November 19, 2014).
Park, S., Li, J. S., Pittman, J. K., Berkowitz, G. A., Yang, H. B., Undurraga, S., et al. (2005). Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc. Natl. Acad. Sci. U.S.A. 102, 18830–18835. doi: 10.1073/pnas.0509512102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pastina, M. M., Malosetti, M., Gazaffi, R., Mollinari, M., Margarido, G. R. A., Oliveira, K. M., et al. (2012). A mixed model QTL analysis for sugarcane multiple-harvest-location trial data. Theor. Appl. Genet. 124, 835–849. doi: 10.1007/s00122-011-1748-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Piperidis, G., Piperidis, N., and D'hont, A. (2010). Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol. Genet. Genomics 284, 65–73. doi: 10.1007/s00438-010-0546-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ramesh, P. (2000). Effect of different levels of drought during the formative phase on growth parameters and its relationship with dry matter accumulation in sugarcane. J. Agron. Crop Sci. 185, 83–89. doi: 10.1046/j.1439-037x.2000.00404.x
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., and Bartel, D. P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. doi: 10.1101/gad.1004402
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ren, Y., Chen, L., Zhang, Y., Kang, X., Zhang, Z., and Wang, Y. (2012). Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Funct. Integr. Genomics 12, 327–339. doi: 10.1007/s10142-012-0271-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rhoades, M. W., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ribeiro, R. V., Machado, R. S., Machado, E. C., Machado, D. F. S. P., Magalhães Filho, J. R., and Landell, M. G. A. (2013). Revealing drought-resistance and productive patterns in sugarcane genotypes by evaluating both physiological responses and stalk yield. Exp. Agric. 49, 212–224. doi: 10.1017/S0014479712001263
Rivero, R. M., Kojima, M., Gepstein, A., Sakakibara, H., Mittler, R., Gepstein, S., et al. (2007). Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. U.S.A. 104, 19631–19636. doi: 10.1073/pnas.0709453104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Robertson, M. J., and Donaldson, R. A. (1998). Changes in the components of cane and sucrose yield in response to drying-off of sugarcane before harvest. Field Crops Res. 55, 201–208. doi: 10.1016/S0378-4290(97)00065-8
Rodrigues, F. A., Graça, J. P., Laia, M. L., Nhani, A. Jr., Galbiati, J. A., Ferro, M. I. T., et al. (2011). Sugarcane genes differentially expressed during water deficit. Biol. Plant. 55, 43–53. doi: 10.1007/s10535-011-0006-x
Rodrigues, F. A., Laia, M. L., and Zingaretti, S. M. (2009). Analysis of gene expression profiles under water stress in tolerant and sensitive sugarcane plant. Plant Sci. 176, 286–302. doi: 10.1016/j.plantsci.2008.11.007
Rogers, K., and Chen, X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25, 2383–2399. doi: 10.1105/tpc.113.113159
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sánchez-Ken, J. G., and Clark, L. G. (2010). Phylogeny and a new tribal classification of the Panicoideaes. l. (Poaceae) based on plastid and nuclear sequence data and structural data. Am. J. Bot. 97, 1732–1748. doi: 10.3732/ajb.1000024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sánchez-Ken, J. G., Clark, L. G., Kellogg, E. A., and Kay, E. E. (2007). Reinstatement and emendation of subfamily Micrairoideae (Poaceae). Syst. Bot. 32, 71–80. doi: 10.1600/036364407780360102
Setta, N., Monteiro-Vitorello, C. B., Metcalfe, C. J., Cruz, G. M. Q., Del Bem, L. E. V., Vicentini, R., et al. (2014). Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics 15:540. doi: 10.1186/1471-2164-15-540
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shaik, R., and Ramakrishna, W. (2012). Bioinformatic analysis of epigenetic and microRNA mediated regulation of drought responsive genes in rice. PLoS ONE 7:e49331. doi: 10.1371/journal.pone.0049331
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shen, J., Xie, K., and Xiong, L. (2010). Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol. Genet. Genomics 284, 477–488. doi: 10.1007/s00438-010-0581-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shuai, P., Liang, D., Zhang, Z., Yin, W., and Xia, X. (2013). Identification of drought-responsive and novel Populus trichocarpa microRNAs by high- throughput sequencing and their targets using degradome analysis. BMC Genomics 14:233. doi: 10.1186/1471-2164-14-233
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singels, A., Kennedy, A. J., and Bezuidenhout, C. N. (2000). The effect of water stress on sugarcane biomass accumulation and partitioning. SA Sugar Technol. Assoc. 74, 169–172.
Silva, M. A., Jifon, J. L., and Da Silva, J. A. G. (2007). Use of physiological parameters as fast tools to screen for drought tolerance in sugarcane. Brazil. J. Plant Physiol. 19, 193–201. doi: 10.1590/S1677-04202007000300003
Silva, M. A., Soares, R. A. B., Landell, M. G. A., and Campana, M. P. (2008). Agronomic performance of sugarcane families in response to water stress. Bragantia 67, 655–661. doi: 10.1590/S0006-87052008000300014
Silva, V. (2013). Drought will reduce by 30% sugarcane harvest in Paraíba. (Original in Portuguese: seca deve reduzir 30% da produção de cana na Paraíba). Available online at: http://ne10.uol.com.br/canal/cotidiano/economia/noticia/2013/04/12/seca-deve-reduzir-30_porcento-da-producao-de-cana-na-paraiba-412047.php (Accessed on October 08, 2013).
Sindaçucar. (2012). Drought punishes sugarcane region with only 5.7 mm of rain in November. (Original in Portuguese: seca castiga região canavieira com apenas 5,7 mm de chuva em novembro). Available online at: http://www.sindacucar-al.com.br/2012/12/seca-castiga-regiao-canavieira-com-apenas-57-mm-de-chuva-em-novembro/ (Accessed on November 19, 2014).
Singh, S., and Rao, P. N. G. (1987). Varietal differences in growth characteristics in sugarcane. J. Agri. Sci. 108, 245–247. doi: 10.1017/S0021859600064327
Smith, J. P., Lawn, R. J., and Nable, R. O. (1999). Investigations into the root:shoot relationship of sugarcane and some implications for rop productivity in the presence of sub-optimal conditions. Proc. Austr. Soc. Sugar Cane Technol. 21, 108–113.
Srivastava, P. K., Moturu, T. R., Pandey, P., Baldwin, I. T., and Pandey, S. P. (2014). A comparison of performance of planta miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genomics 15:348. doi: 10.1186/1471-2164-15-348
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stephenson, T. J., McIntyre, C. L., Collet, C., and Xue, G. P. (2007). Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 65, 77–92. doi: 10.1007/s11103-007-9200-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, G., Stewart, C. N., Xiao, P., and Zhang, B. (2012). Microrna expression analysis in the cellulosic biofuel crop switchgrass (Panicum virgatum) under abiotic stress. PLoS ONE 7:e32017. doi: 10.1371/journal.pone.0032017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, X., Dong, B., Yin, L., Zhang, R., Du, W., Liu, D., et al. (2013). PMTED: a plant microRNA target expression database. BMC Bioinformatics 14:174. doi: 10.1186/1471-2105-14-174
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sunkar, J., and Zhu, J. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. doi: 10.1105/tpc.104.022830
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sunkar, R., Zhou, X., Zheng, Y., Zhang, W., and Zhu, J. (2008). Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 8:25 doi: 10.1186/1471-2229-8-25
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thiebaut, F., Grativol, C., Carnavale-Bottino, M., Rojas, C. A., Tanurdzic, L. O. S., Farinelli, L., et al. (2012). Computational identification and analysis of novel sugarcane microRNAs. BMC Genomics 13:290. doi: 10.1186/1471-2164-13-290
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thomson, D. W., Bracken, C. P., and Goodall, G. J. (2011). Experimental strategies for microRNA target identification. Nucleic Acids Res. 39, 6845–6853 doi: 10.1093/nar/gkr330
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vaucheret, H. (2008). Plant argonautes. Trends Plant Sci. 13, 350–358. doi: 10.1016/j.tplants.2008.04.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vettore, A. L., Da Silva, F. R., Kemper, E. L., Souza, G. M., Da Silva, A. M., Ferro, M. I., et al. (2003). Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res. 13, 2725–2735. doi: 10.1101/gr.1532103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 699–687. doi: 10.1016/j.cell.2009.01.046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, L., Mai, Y. X., Zhang, Y. C., Luo, Q., and Yang, H. Q. (2010a). MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol. Plant 3, 794–806. doi: 10.1093/mp/ssq042
Wang, J., Roe, B., Macmil, S., Yu, Q., Murray, J. E., Tang, H., et al. (2010b). Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics. 11:261. doi: 10.1186/1471-2164-11-261
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xia, K., Wang, R., Ou, X., Fang, Z., Tian, C., Duan, J., et al. (2012). OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 7:e30039. doi: 10.1371/journal.pone.0030039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xie, F., and Zhang, B. (2010). Target-align: a tool for plant microRNA target identification. Bioinformatics 26, 3002–3003. doi: 10.1093/bioinformatics/btq568
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xie, Z., Allen, E., Wilken, A., and Carrington, J. C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. PNAS 102, 12984–12989. doi: 10.1073/pnas.0506426102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xuan, P., Guo, M., Huang, Y., Li, W., and Huang, Y. (2011). MaturePred: efficient identification of microRNAs within novel plant pre-miRNAs. PLoS ONE 6:e27422. doi: 10.1371/journal.pone.0027422
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamaguchi, M., Ohtani, M., Mitsuda, N., Kubo, M., Ohme-Takagi, M., Fukuda, H., et al. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in arabidopsis. Plant Cell 22, 1249–1263. doi: 10.1105/tpc.108.064048
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R. W., et al. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935. doi: 10.1126/science.1107130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zanca, A. S., Vicentini, R., Ortiz-Morea, F. A., Del Bem, L. E., da Silva, M. J., Vincentz, M., et al. (2010). Identification and expression analysis of microRNAs and targets in the biofuel crop sugarcane. BMC Plant Biol. 10:260. doi: 10.1186/1471-2229-10-260
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, B., Pan, X., Wang, Q., Cobb, G. P., and Anderson, T. A. (2006). Srisvastava Computational identification of microRNAs and their targets. Comput. Biol. Chem. 30, 395–407. doi: 10.1016/j.compbiolchem.2006.08.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, H., Shen, G., Kuppu, S., Gaxiola, R., and Payton, P. (2011). Creating drought- and salt-tolerant cotton by overexpressing a vacuolar pyrophosphatase gene. Plant Signal. Behav. 6, 861–863. doi: 10.4161/psb.6.6.15223
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, L., Chia, J. M., Kumari, S., Stein, J. C., Liu, Z., Narechania, A., et al. (2009). A genome-wide characterization of microRNA genes in maize. PLoS Genet. 5:e1000716. doi: 10.1371/journal.pgen.1000716
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y. (2005). miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 33, W701–W704. doi: 10.1093/nar/gki383
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Z., Wei, L., Zou, X., Tao, Y., Liu, Z., and Zheng, Y. (2008b). Submergence-responsive MicroRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Ann. Bot. (Lond.) 102, 509–519. doi: 10.1093/aob/mcn129
Zhao, B., Liang, R., Ge, L., Li, W., Xiao, H., Lin, H., et al. (2007). Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 354, 585–590. doi: 10.1016/j.bbrc.2007.01.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, L., Liu, Y., Liu, Z., Kong, D., Duan, M., and Luo, L. (2010). Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 61, 4157–4168. doi: 10.1093/jxb/erq237
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: sugarcane, drought stress, miRNAs, transcription factors, drought tolerance, cross-species comparisons
Citation: Gentile A, Dias LI, Mattos RS, Ferreira TH and Menossi M (2015) MicroRNAs and drought responses in sugarcane. Front. Plant Sci. 6:58. doi: 10.3389/fpls.2015.00058
Received: 13 February 2014; Accepted: 22 January 2015;
Published online: 23 February 2015.
Edited by:
Rodrigo A. Gutierrez, Pontificia Universidad Catolica de Chile, ChileCopyright © 2015 Gentile, Dias, Mattos, Ferreira and Menossi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcelo Menossi, Laboratório de Genoma Funcional, Departamento de Genética, Evolução e Bioagentes, Universidade Estadual de Campinas, CP6109, 13083-875 Campinas, São Paulo, Brazil e-mail: menossi@unicamp.br
 Agustina Gentile
Agustina Gentile Lara I. Dias
Lara I. Dias Raphael S. Mattos
Raphael S. Mattos Thaís H. Ferreira
Thaís H. Ferreira Marcelo Menossi
Marcelo Menossi