- Department of Cell and Molecular Biology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Serdang, Malaysia
Many algae are rich sources of sulfated polysaccharides with biological activities. The physicochemical/rheological properties and biological activities of sulfated polysaccharides are affected by the pattern and number of sulfate moieties. Sulfation of carbohydrates is catalyzed by carbohydrate sulfotransferases (CHSTs) while modification of sulfate moieties on sulfated polysaccharides was presumably catalyzed by sulfatases including formylglycine-dependent sulfatases (FGly-SULFs). Post-translationally modification of Cys to FGly in FGly-SULFs by sulfatase modifiying factors (SUMFs) is necessary for the activity of this enzyme. The aims of this study are to mine for sequences encoding algal CHSTs, FGly-SULFs and putative SUMFs from the fully sequenced algal genomes and to infer their phylogenetic relationships to their well characterized counterparts from other organisms. Algal sequences encoding CHSTs, FGly-SULFs, SUMFs, and SUMF-like proteins were successfully identified from green and brown algae. However, red algal FGly-SULFs and SUMFs were not identified. In addition, a group of SUMF-like sequences with different gene structure and possibly different functions were identified for green, brown and red algae. The phylogeny of these putative genes contributes to the corpus of knowledge of an unexplored area. The analyses of these putative genes contribute toward future production of existing and new sulfated carbohydrate polymers through enzymatic synthesis and metabolic engineering.
Introduction
Sulfates are found in algal proteins, carbohydrate, sulfolipids, and low molecular weight sulfated compounds (DeBoer, 1981). Many algae were reported to be rich sources of sulfated polysaccharides with biological activities (Hernandez-Sebastia et al., 2008). Sulfated fucans from brown algae and sulfated galactans from green and red algae have been reported to be potent anticoagulant agents (Pomin and Mourão, 2008). Some of these algal sulfated polysaccharides such as agar, agarose, and carrageenan, constitute the major component of algal extracellular matrix or cell wall, and have wide applications in food, cosmetics and pharmaceutical industries (McHugh, 2003).
Sulfur (normally in sulfate form) constitutes one of the nine essential macronutrients required by plants including algae (Yildiz et al., 1994). Sulfur assimilation in plants and algae begins with the activation of sulfate by ATP sulfurylase, which catalyzes the adenylation of sulfate to 5′-adenylylsulfate (APS). APS can either be phosphorylated by APS kinase or reduced by glutathione-dependent APS reductase. Both enzymes and pathways are important for cellular synthesis of sulfated and reduced sulfur compounds in algae, respectively (Gao et al., 2000). Sulfation is catalyzed by sulfotransferases (STs) which transfer a sulfuryl group (SO3) from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to a hydroxyl group of a substrate (Hernandez-Sebastia et al., 2008). In addition, activities of sulfatases which were assumed to be involved in the modification of sulfate moieties on sulfated polysaccharides have also been reported in various algae. The pattern and number of these substitutions not only affect the physicochemical/rheological properties of sulfated polysaccharides but also their biological activities (Opoku et al., 2006; Tuvikene et al., 2008).
Carbohydrate sulfotransferases (CHSTs) are of particular interest in algae because several genera of marine macroalgae synthesize sulfated polysaccharides that constitute the major component of their cell walls which chelate metallic ions and provide hydration to the cells. Mammals CHSTs are among the best characterized CHSTs. Most of them are Golgi-localized and membrane-bound, and are involved in the biosynthesis of sulfated oligosaccharides and glycosaminoglycans (Fukuda et al., 2001). In addition, CHSTs such as NodH and NoeE that are involved in the biosynthesis of nodulation factors have also been characterized from symbiotic rhizobacteria, Sinorhizobium melioti and Rhizobium sp. NGR234, respectively (Ehrhardt et al., 1995; Hanin et al., 1997). Characterization of algal candidate genes for CHSTs has not been reported.
Formylglycine-dependent sulfatase (FGly-SULF) (EC 3.1.5.6) belongs to a large protein family that catalyze the hydrolytic desulfation of sulfate ester and sulfamates from different sulfated substrates. These sulfated substrates include hydrophobic glucosinolates, steroids, tyrosine sulfates, amphiphilic sulfated carbohydrates found in glycosaminoglycans (GAGs), proteoglycans, glycolipids, and water-soluble mono- and disaccharide sulfates (Hanson et al., 2004). FGly-SULFs consist of a class of enzymes that share highly similar amino acid sequence (20–60% over the entire protein length), three-dimensional structure and catalytic site (Boltes et al., 2001; Hopwood and Ballabio, 2001).
The conserved catalytic site of FGly-SULFs consists a divalent metal ion located within a pocket in which substrates are bound, a highly conserved motif at the N-terminus (or “sulfatase signature”) which spans over a 12-mer linear sequence with a core motif C/S-X-P-X-R, and a unique active aldehyde residue, α-formylglycine (FGly) (Hanson et al., 2004). FGly is formed post-translationally by the oxidation of a cysteine (Cys) residue that is conserved in all eukaryotic and most prokaryotic sulfatases (Schmidt et al., 1995; Dierks et al., 1998b). Some bacterial species possess serine (Ser) residue instead of cysteine (Cys) residue at the same position of the catalytic site leading to the “Cys-type” or “Ser-type” prokaryotic sulfatases. The structural similarity amongst FGly-SULFs suggested that they shared a common ancestral gene (Meroni et al., 1996; Parenti et al., 1997).
Post-translational modification of Cys to FGly occurs at the endoplasmic reticulum at a stage the polypeptide is not yet folded into its native structure (Schirmer and Kolter, 1998). The enzyme that is involved in the post-translational modification of Cys to FGly in FGly-SULFs is known as sulfatase modifiying factor (SUMF) while the enzyme AtsB is responsible for the post-translational modification of bacterial Ser-type sulfatases (Dierks et al., 1998b; Schirmer and Kolter, 1998). SUMF1 was found to be responsible for the multiple sulfatase deficiency in human. SUMFs belong to a gene family that is highly conserved during evolution from bacteria to human (Dierks et al., 1997, 1998a; Landgrebe et al., 2003).
Despite the importance of these sulfated polysaccharides, the roles of CHSTs in their formation, and sulfatases in their modifications; little is known about the sequences and structures of algal CHSTs, FGly-SULFs and their SUMFs. In recent years, a few algal genomes have been fully sequenced (Armbrust et al., 2004; Merchant et al., 2007; Bowler et al., 2008; Cock et al., 2010; Bhattacharya et al., 2013; Collén et al., 2013) and can be used for the survey for algal candidate genes encoding algal FGly-SULFs and SUMFs. The aims of this study are to mine for sequences from the fully sequenced algal genomes and to infer their phylogenetic relationships to known CHSTs, FGly-SULFs, and SUMFs from other organisms.
Materials and Methods
Mining of Algal Sequences Encoding CHSTs, FGly-SULFs, and SUMFs
Search analyses for sequences encoding algal CHSTs, FGly-SULFs, and SUMFs across nine completed algal genomes, i.e., Chondrus crispus, Porphyridium cruetum (http://cyanophora.rutgers.edu/porphyridium/), Cyanidioschyzon merolae (http://merolae.biol.s.u-tokyo.ac.jp/blast/blast.html), Ectocarpus siliculosus (http://bioinformatics.psb.ugent.be/orcae/overview/Ectsi), Thalassiosira pseudonana, Phaeodactylum tricornutum, Ostreococcus tauri, Chlamydomonas reinhardtii, and Volvox carteri (JGI: http://genome.jgi-psf.org/); and algal ESTs/cDNAs from Porphyra umbilicalis, P. purpurea (http://dbdata.rutgers.edu/nori/blast.php), P. yezoensis, Laurentia dendroidea, Galderia sulphuraria, Gracilaria changii and G. salicornia; were performed using the BLASTX, BLASTP, or TBLASTX algorithms (Altschul et al., 1990). The search was performed using the known sequences encoding CHSTs, FGly-SULFs, and SUMFs from human and/or other eukaryotes, i.e., mouse, rat, yeasts (Neurospora crassa, Kluyveromyces lactis, Schizosaccharomyces pombe, Debaryomyces hansenii, Yarrowia lipolytica), Drosophila melanogaster and worm Caenorhabditis elegans (Sardiello et al., 2005). Homologous sequences from plants were also retrieved from Phytozome ver.3 (http://phytozome.jgi.doe.gov/). BLASTX and BLASTP analyses were performed on the retrieved sequences against the SwissProt database. Sequences that do not match with any sequences encoding CHSTs, FGly-SULFs, and SUMFs were removed upon the reciprocal search. Amino acid sequences that were incomplete without the translation start methionine and the sulfatase signature for FGly-SULFs were also discarded.
Phylogenetic Analyses
Multiple sequence alignment of CHSTs, FGly-SULFs, and SUMF amino acid sequences were performed with Clustal W (Chenna et al., 2003), respectively. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007) using the Neighbor-Joining method (Saitou and Nei, 1987) with a bootstrap test performed on 1000 random combinations of the sequence alignment (Felsenstein, 1985).
Generation of Sulfatase Signature Logos
Logo analyses for FGly-SULF sequences were performed at the Berkeley Structural Genomics Center (http://weblogo.Berkeley.edu/) to visualize the information content associated with each position of a given motif shared by related sequences. In the graphical representation, the conservation at each position (expressed in bits) is represented by the overall height of each position whereas the relative frequencies of the symbols within a position are indicated by the relative sizes of the symbols. The reported values were computed as the rate between the information content of the given position and the information content of varying positions within the motif.
Results and Discussion
In total, 83, 41, and 14 algal sequences encoding CHSTs, FGly-SULFs, and SUMFs were retrieved, respectively (Tables 1–3). Human CHSTs, FGly-SULFs, and SUMFs were used for the mining and also phylogenetic analyses in this study mainly because sequences from human were the best characterized in terms of sequence and functions compared to those from other organisms.
Algal CHST Sequences
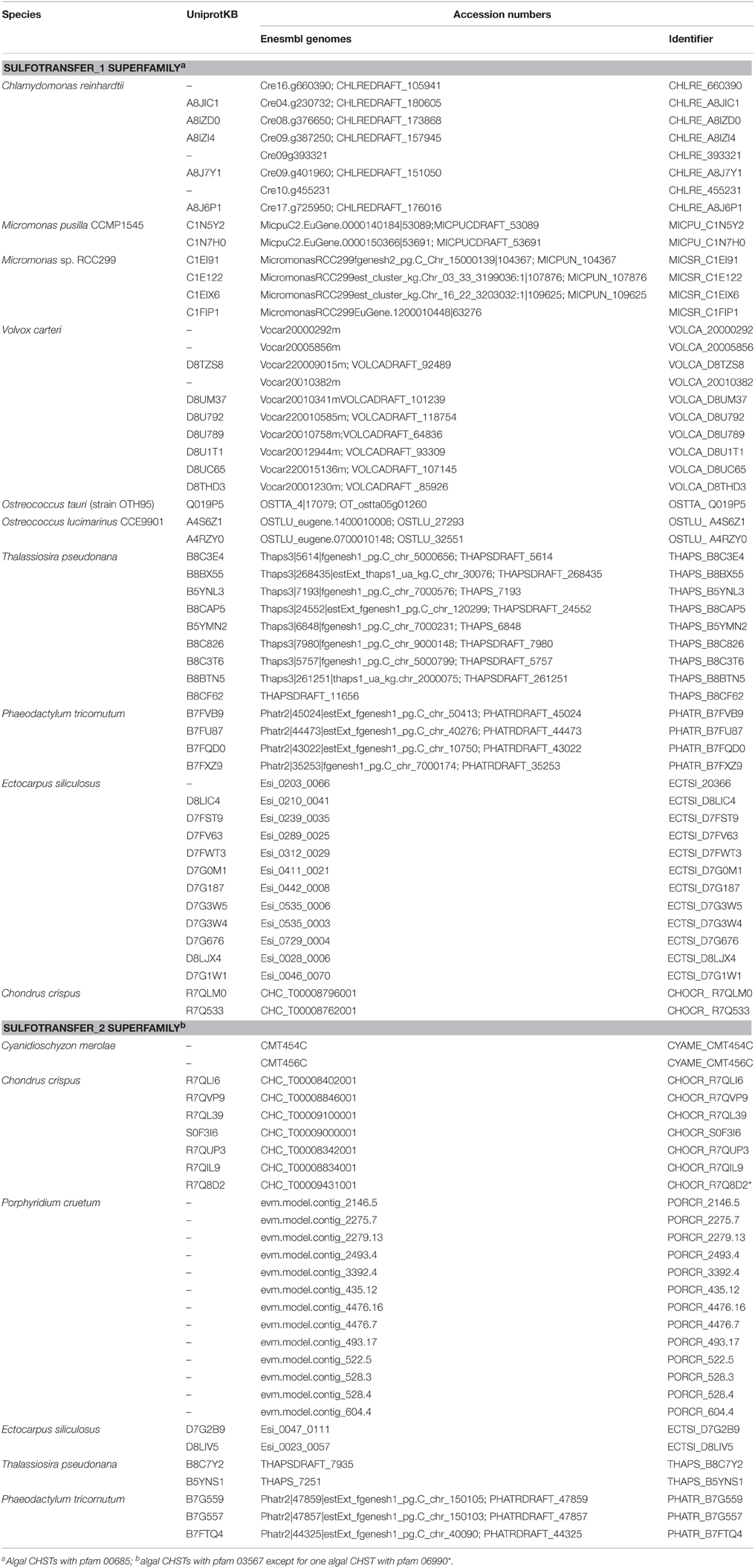
Human CHSTs can be divided into two groups based on the presence of two conserved domains for Superfamily Sulfotransferase 1 and 2, respectively (Figures 1, 2). All human CHSTs classified in the Superfamily Sulfotranferase 1 (CHSTs 1-7, mainly for Gal/N-acetylglucosamine/N-acetylglucosamine 6-O-STs; glucosamine N-deacetylase/N-ST or heparin sulfate STs, NDSTs; (heparan sulfate)- glucosamine 3-O STs, HS3S1, 2, 5, 6, A and B) were found to contain pfam 00685 for Sulfotransfer_1 domain, while most of those in the Superfamily Sulfotransferase 2 have pfam 03567 for Sulfotransfer_2 domain (CHSTs 8-15, for N-acetylgalactosamine 4-O STs and N-acetylgalactosamine 4-sulfate 6-O STs; heparan sulfate 6-O-STs, HS6ST 1-2; heparan sulfate 2-O ST, HS2ST and uronyl-2-O ST, UST) except for a few CHSTs such as galactose-3-O STs (G3ST1-4) that have pfam 06990 for Gal-3-O-Sulfotr domain. The findings on human CHSTs concur with the information published in the Interpro abstract for IPR005331 (www.ebi.ac.uk/interpro/) that Sulfotransfer_2 domain (pfam 03567) is present in a number of CHSTs that transfer sulfate to positions 3 (CHSTs 10), 4 (CHSTs 8, 9, 11 and 13; dermatan-4 ST, D4ST) and 6 (HS2ST, HS6ST, chondroitin-6 ST) of carbohydrate groups in glycoproteins and glycolipids. According to the Interpro abstract for IPR000863, Sulfotransfer_1 domain is found in flavonyl-3-STs, aryl STs, alcohol STs, and phenol STs. However, we found that many human CHSTs also contain this domain. The algal CHSTS (Table 1) were found to have either one of the pfams mentioned above or with no putative domain. All the green algal CHSTs were found to have pfam 00685 only while either pfam 00685 or pfam 03567 was found in the brown and red algal CHSTs. Only one red algal CHST from C. crispus (CHOCR_R7Q8D2) was found to have pfam 06990 (Figure 2, Table 1). The algal CHST sequences are generally very diverse. The use of phylogeney in assigning functions based on substrate specificity or pattern of sulfation requires further verification. Most of these algal CHSTs were clustered according to green, brown or red algae or even genera, except for a few clusters with green, red, and brown algal CHSTS (Figure 1). For examples, CHLRE 455231 was clustered among a group of brown algal CHSTs; THAPS B8C3T6 was in a group of green algal CHSTs; CHLRE A8IZD0 and CHOCR R7QLM0 were clustered with brown algal CHSTs; and CHOCR R7Q533 and PHATR B7FXZ9 were clustered with green algal CHSTs. These CHSTs could share similar functions in algae of different genera/species.
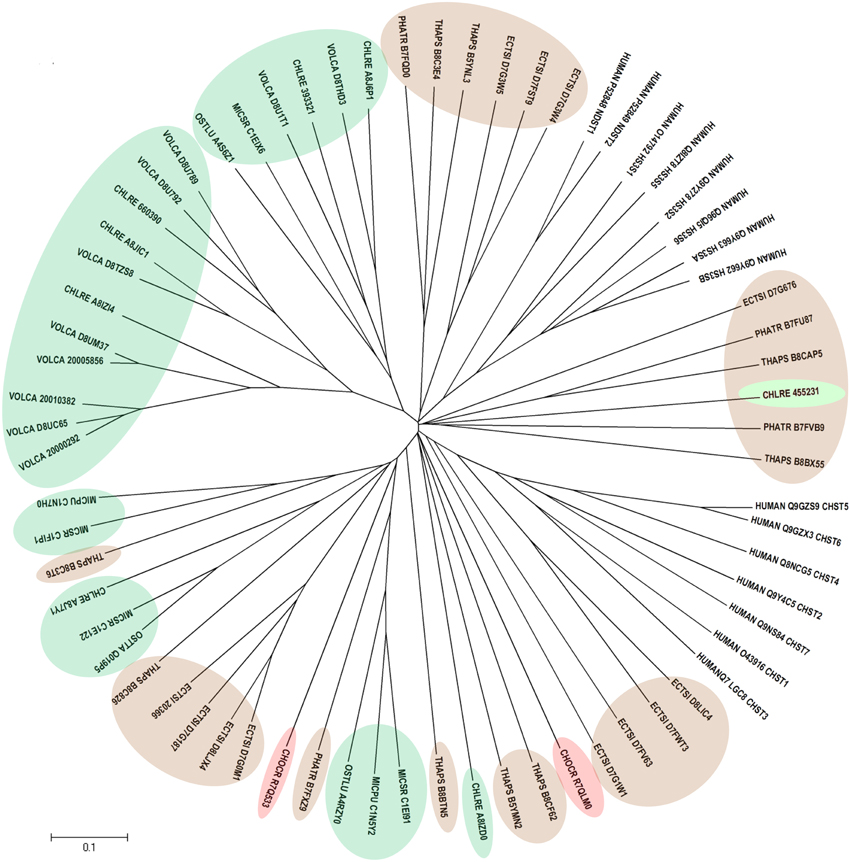
Figure 1. Phylogenetic relationship of algal CHSTs in Superfamily Sulfotransfer_1 (with pfam domain 00685). The evolutionary history was inferred using the Neighbor-Joining method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Sequences from green, brown, and red algae are shown by respective colors. The identifier of the sequence starts with the species abbreviation followed by the UNIPROT/Genbank accession number and annotation wherever possible (Table 1). HUMAN, Homo sapiens; CHSTs 1-7, for Gal/N-acetylglucosamine/N-acetylglucosamine 6-O-STs; NDST, glucosamine N-deacetylase/N-ST or heparin sulfate STs; and HS3S1, 2, 5, 6, A and B, (heparan sulfate)-glucosamine 3-O STs.
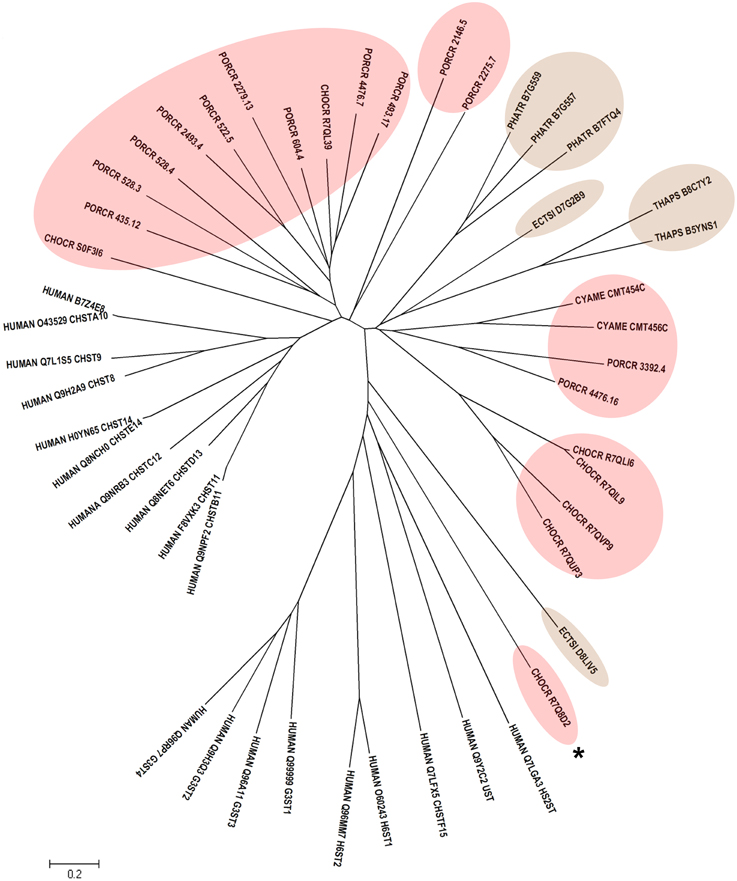
Figure 2. Phylogenetic relationship of algal CHSTs in Superfamily Sulfotransfer_2 (with pfam domains 03567 and 06990). The evolutionary history was inferred using the Neighbor-Joining method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Sequences from brown and red algae are shown by respective colors. The identifier of the sequence starts with the species abbreviation followed by the UNIPROT/Genbank accession number and annotation wherever possible (Table 1). HUMAN, Homo sapiens; CHSTs 8-15, for N-acetylgalactosamine 4-O STs, and N-acetylgalactosamine 4-sulfate 6-O STs; H6ST 1-2, heparan sulfate 6-O-STs; HS2ST, heparan sulfate 2-O ST; UST, uronyl-2-O ST; G3ST1-4, galactose-3-O STs. * represents algal CHST with pfam 06990.
Algal FGLy-SULF Sequences
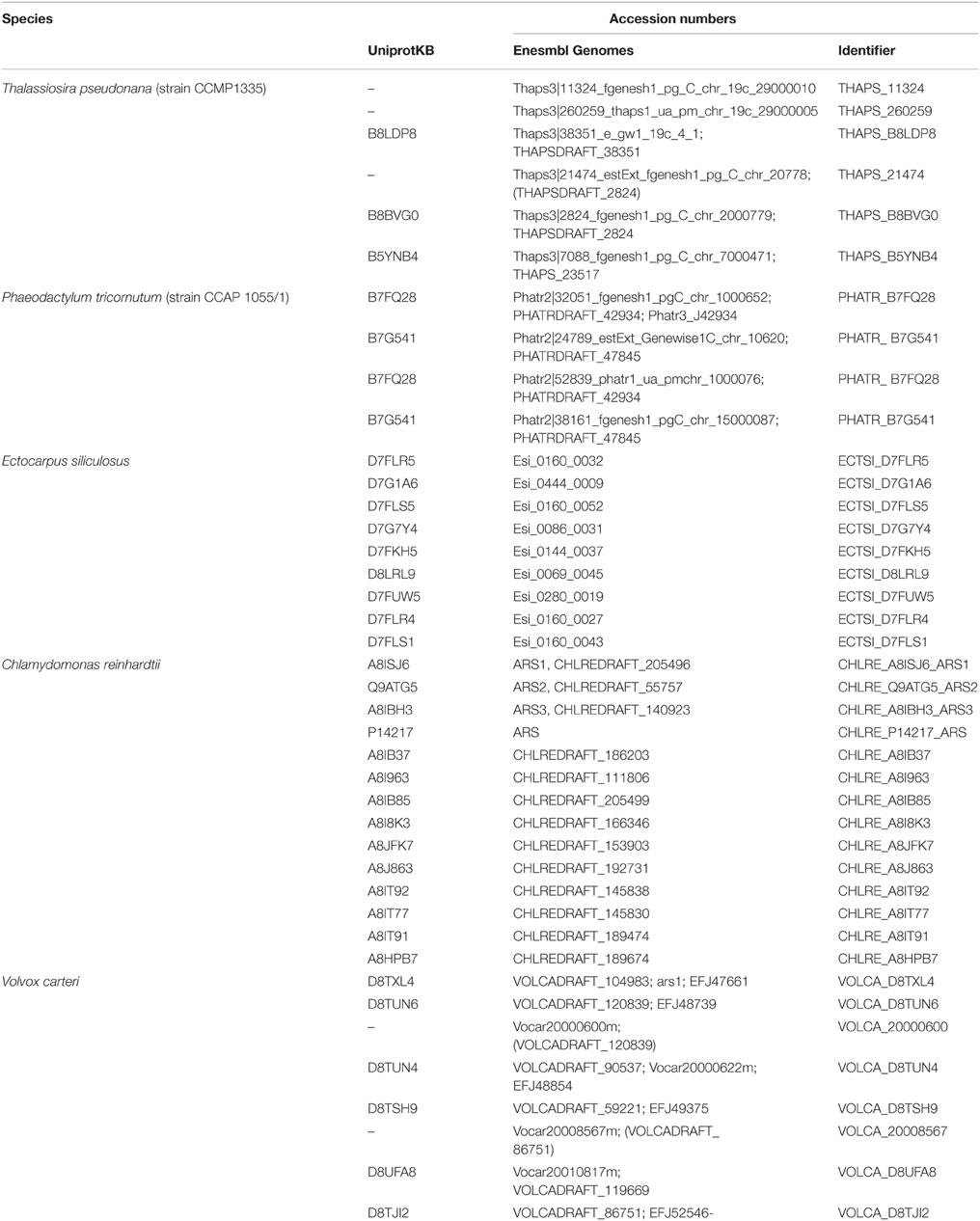
Sequences encoding putative algal FGly-SULFs were identified from complete green and brown algal genomes (Table 2). Although, sulfatase activities have been reported in a few red algae (Rees, 1961a,b; Wong and Craigie, 1978; Genicot-Joncour et al., 2009; Shukla et al., 2011; Qin et al., 2013; Wang et al., 2014), algal FGly-SULF was not retrieved from the red algal genomes i.e., C. crispus as reported by Collén et al. (2013), and red microalgal genomes from Por. purpureum (Bhattacharya et al., 2013) and Cy. merolae (Matsuzaki et al., 2004). Neither were these sequences detected among the available ESTs of red seaweeds from P. yezoensis (Nikaido et al., 2000; Kakinuma et al., 2006), Griffithsia okiensis (Lee et al., 2007) and G. changii (Teo et al., 2007). However, a few ESTs or incomplete cDNAs that are highly similar to sequences encoding FGly-SULFs were identified from P. purpurea. These sequences consist of partial coding sequences and share highly similar sequences to bacterial FGly-SULFs thus were not included in this analysis.
It is likely that the genes encoding FGly-SULF are absent from the genomes of red algae or at least in the red algal species examined. Since sulfate is not a limiting factor for marine algae that grow in seawater which has a high sulfate concentration (25–28 mM) compared to freshwater or land (10–50 μM) (Friedlander, 2001; Bochenek et al., 2013), it is possible that recycling of sulfate through FGly-SULFs may not be required. Furthermore, the biosynthesis of sulfated polysaccharides was proposed to be a possible result of physiological adaptation of macroalgae, marine angiosperms, and seagrasses (but not terrestrial plants) to marine environments (Aquino et al., 2005).
It is also possible that the red algal sulfatases belong to sulfatases other than the FGly-SULF type. Currently, three groups of sulfatases have been described: Group 1 which consists of the FGly-SULFs (Boltes et al., 2001); Group 2 with the Fe(II) α-ketoglutarate-dependent sulfatases (Müller et al., 2004); and Group 3 which consists of the zinc-dependent metallo β-lactamase superfamily or alkylsulfatases (Hagelueken et al., 2006). In addition, sulfatases (arylsulfatases) together with alkaline phosphatases and phosphoglycerate mutases were shown to belong to a superfamily of phospho-/sulfo-coordinating metalloenzymes that share the catalytic core of nucleotide pyrophosphatases/phosphodiesterases by homology searches and alignment-assisted mutagenesis (Gijsbers et al., 2001). The sulfatase genes may have also diverged to an extent that they cannot be readily identified using bioinformatic search tools. The red algal sulfatase could have novel sequences as reported for 12 sequences encoding putative D-galactose-2,6-sulfurylases I and II as revealed by the genome analyses of C. crispus (Collén et al., 2013). The galactose-2,6-sulfurylases I from C. crispus which share some identities to L-amino acid oxidase from C. reinhardtii (U78797) have no similarities to any reported sulfatases. Evidence on the enzyme activity of their recombinant proteins is crucial to show that they are indeed novel red algal sulfatases.
Sulfatase-like activities have also been reported previously in higher plants (Baum and Dodgson, 1957; Poux, 1966) although sequences encoding these enzymes have not been reported. Searching the complete plant genomes at the Phytozome revealed only one incomplete FGly-SULF-like sequence from Ricinus cucumis which contains a CSATR motif which resembles the sulfatase signature. However, this sequence was incomplete, short, and without introns. Further analyses revealed that similar sequences (orthologs) were absent in other plant species, thus was believed to be contaminated sequence from associated bacterial species.
Figure 3 shows the phylogeny of FGly-SULFs from human, yeasts, worm, fruitfly, and algae which has two main branches. The well characterized human FGly-SULFs were divided into two main branches with sulfatases SULF 1 and SULF 2, and glucosamine N-acetyl-6-sulfatase (GNS) in one branch while the remaining human FGly-SULFs (arylsulfatases, ARS A, B, C, D, E, F, G, H, I, J, K; N-galactosamine-6-sulfatase, GALNS; iduronate 2-sulfatase, IDS; and N-sulfoglucosamine sulfohydrolase, SGSH) are distributed in the other branch. The clustering of human FGly-SULFs may reflect their functions or substrate preference in general. The only two FGly-SULFs from worm were distributed one in each branch with SUL 1 in the same cluster as the human SULF 1 and SULF 2. The D. melanogaster SULF1, GNS, IDS, SGSH were grouped with their orthologs from human while another four uncharacterized FGly-SULFs formed a separate cluster which is unique for D. melanogaster. The FGly-SULF sequences from yeasts were clustered in the same branch except for that of ascomycetes Neurospora crassa which was found to be in a separate branch. It is likely that the FGly-SULFs from Saccharomycetes and Schizosaccharomycetes may have evolved after the divergence from Ascomycetes.
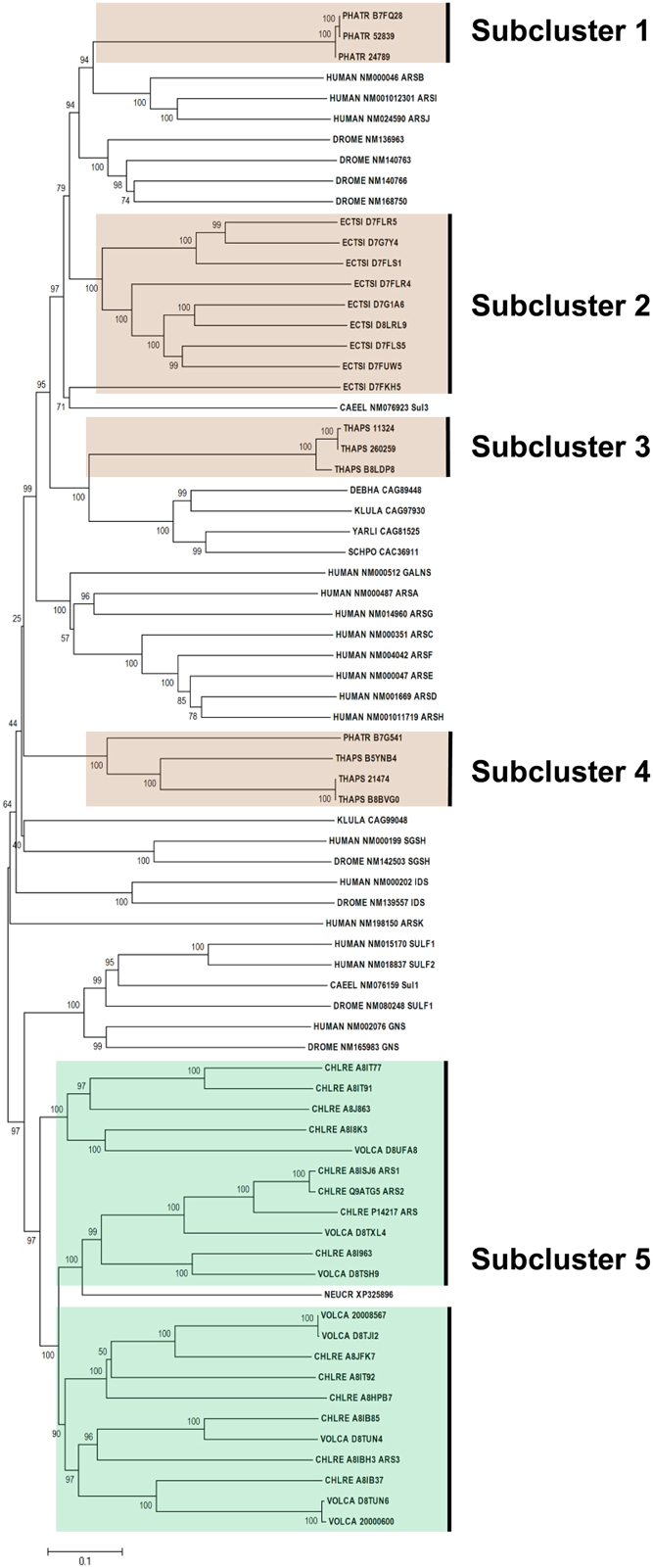
Figure 3. Phylogenetic relationship of algal FGly-SULFs. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Sequences from green and brown algae are shown by respective colors. The identifier of the sequence starts with the species abbreviation followed by the UNIPROT/Genbank accession number and annotation wherever possible (Table 2). HUMAN, Homo sapiens; CAEEL, Caenorhabditis elegans; DROME, Drosophila melanogaster; DEBHA, Debaryomyces hansenii; KLULA, Kluyveromyces lactis; YARLI, Yarrowia lipolytica; SCHPO, Schizosaccharomyces pombe; NEUCR, Neurospora crassa; ARS, arylsulfatase; GALNS, N-galactosamine-6-sulfatase; IDS, iduronate 2-sulfatase; and SGSH, N-sulfoglucosamine sulfohydrolase; Sulf, sulfatase; Sul, sulfatase.
All the green algal FGly-SULFs (Ch. reinhardtii and V. carteri) were distributed in the same branch as human SULF 1, SULT2, and GNS, while all the brown algal FGly-SULFs were divided into subclusters in the other branch (Figure 3), implying that FGly-SULFs from these two groups of algae could have evolved from different origins or from the same origin which has diversified before speciation of brown and green algae. The green algal subcluster (Subcluster 5) consists of sequences from both Ch. reinhardtii and V. carteri thus implying that these sequences may have originated from the same ancestral FGly-SULF which could have existed before speciation. The brown algal sequences were divided into four subclusters: Subcluster 1 which consists of three sequences from Ph. tricornutum whereby each contains an extra C-terminus; Subcluster 2 with nine sequences from E. siliculosus; Subcluster 3 which consists of three sequences from T. pseudonana with a gap in pfam 00884 and closely related to yeast FGly-SULFs (except for the sequence from N. crassa); Subcluster 4 which consists of three sequences from T. pseudonana, and a sequence from Ph. tricornutum; and Subcluster 5 which contains mainly green algal FGly-SULFs. The existence of highly identical sequences from each species suggests duplication of FGly-SULFs upon speciation (Figure 3).
All algal sequences analyzed contain the sulfatase domain (pfam 00884), with a few of them bearing a gap within this domain, mainly those from diatoms (three from T. pseudonana in Subcluster 2 and one from Ph. tricornutum in Subcluster 4). However, the presence of gap within these sequences has little consequence in affecting their phylogeny compared to their similarity within the same species. In addition, sequences from Ph. tricornutum in Subcluster 1 contain an extra C-terminus.
The comparison of amino acid at the active sites of algal sulfatases showed that the green and brown algae share only two conserved residues (C-X-X-X-R) at the same positions (Figure 4A), which is less conserved compared to the core motif for human FGly-SULFs (C-X-P-S-R). Both Ch. reinhardtii and V. carteri share the same core motif: C-C-P-(S/A)-R (Figures 4B,C), while the brown algae have more diverse core motif (C-X-X-X-R; Figures 4D–F) whereby only the first C residue and the last R residue are conserved. Within the FGly-SULFs from each brown algal species, the core motif C-T-P-(A/S)-R is conserved among those from E. siliculosus (Figure 4F), while C-(S/W)-(P/I)-(T/S)-R and C-(C/W)-(P/V/I)-S-R were shared by those in Ph. tricornutum and T. pseudonana, respectively (Figures 4D,E). The A residue immediately after the core motif is highly conserved in brown algae (Figures 4D–F).
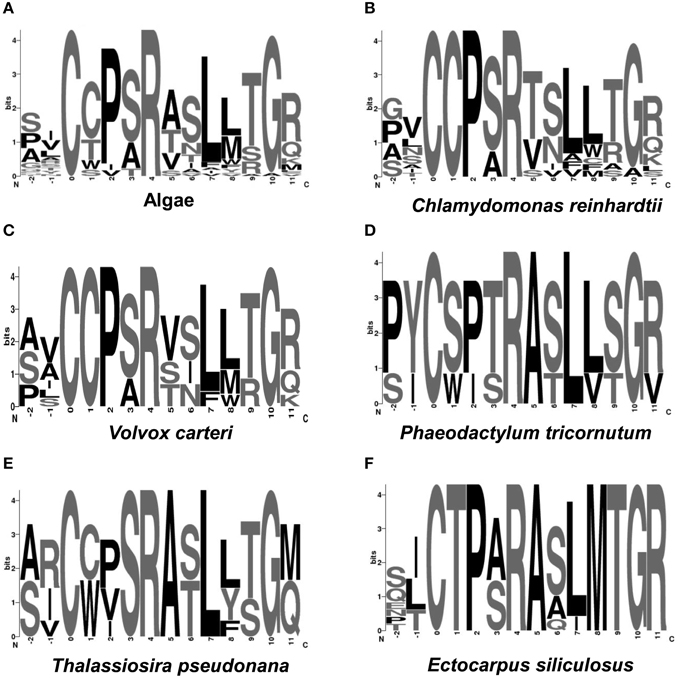
Figure 4. Logo representation of the catalytic cores of algal FGly-SULFs. The overall height of each column is proportional to the information content at that position, and within columns the conservation of each residue is visualized as the relative height of symbols representing amino acids. Position 1 indicates the residues directly involved in the enzymatic reaction. Position 1 of sulfatase cores indicates the amino acid (cysteine) to be modified into FGly. (A) Algae; (B) Chlamydomonas reinhardtii; (C) Volvox carteri; (D) Phaeodactylum tricornutum; (E) Thalassiosira pseudonana; (F) E. siliculosus.
Algal SUMF and SUMF-like Sequences
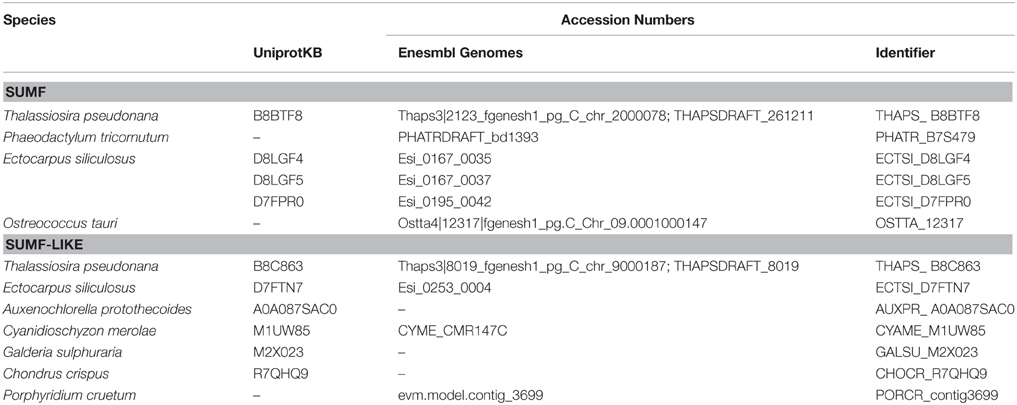
Since the sulfatase signature was identified in all algal FGly-SULF sequences (Figure 4), sequences that encode SUMFs which modify the C residue to FGly in the active site of FGly-SULFs were searched among the algal genomes. Table 3 shows that SUMF sequences that were highly similar to those of eukaryotic SUMFs were only retrieved from brown algae (Ph. tricornutum, T. pseudonana, and E. siliculosus) and a green microalga (plankton), Ostreococcus tauri. In addition to the SUMF sequences, SUMF-like sequences that are highly similar to the coding sequence of Meiotically Up-regulated Gene (MUG) 158 (also known as Egt1) from a yeast, Sch. pombe, were retrieved from green (Auxenochlorella protothecoides), brown (T. pseudonana and E. siliculosus) and red algae (C. crispus, Po. cruetum, Cy. merolae, and Ga. sulphuraria), as well as a moss (Bryophyte), Physcomitrella patens (Pp1s94_113V6 abbreviated as PHYPA_113V6) which represents the missing link between green algae and higher land plants (Figure 5).
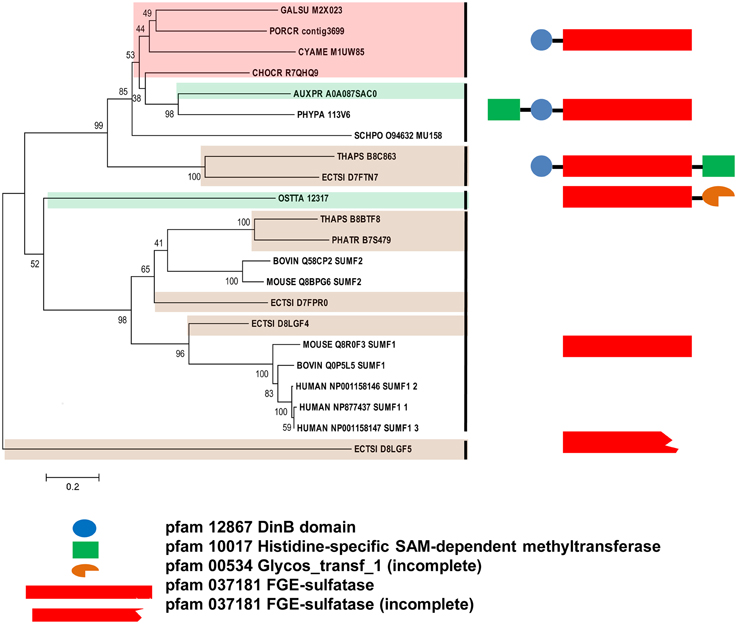
Figure 5. Phylogenetic relationship of algal SUMFs and SUMF-like sequences. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The identifier of the sequence starts with the species abbreviation followed by the UNIPROT/Genbank accession number and annotation wherever possible (Table 3). HUMAN, Homo sapiens; BOVINE, Bos taurus; MOUSE, Mus musculus; PHYPA, Physcomitrella patens; SCHPO, Schizosaccharomyces pombe. Sequences from green, brown, and red algae are shown by respective colors. The structure and pfam domains of each group are shown on the right panel.
The SUMF-like sequences were longer than the SUMF sequences. These two groups of sequences are only identical at the formyl-glycine generating enzyme (FGE)-sulfatase domain (pfam 03781). MUG 158 from Sch. pombe has an S-adenosyl-L-methionine (SAM)-dependent methyltransferase domain (pfam 10017; including DUF 2260, a domain with unknown function), an uncharacterized DinB_2 domain (pfam 12867; including an iron-binding motif, H-X(3)-H-X-E), in addition to the FGE-sulfatase domain (Pluskal et al., 2014). This protein was reported to be involved in cell division and its expression was up-regulated upon the entry of cell into meiosis (Mata et al., 2002). Highly identical to the sequences of NcEgt1 from N. crassa and MsEgtD from Mycobacterium smegmatis, MUG 158 was also reported to be involved in the first step of ergothioneine biosynthesis (Pluskal et al., 2014). Ergothioneine, an amino acid derived from thiourea that contains components associated with histidine, was reported to accumulate in oxidative-stress susceptible area in human body (Cheah and Halliwell, 2012) thus was believed to be able to scavenge oxidizing species that are not free radicals (Chaudière and Ferrari-Iliou, 1999). However, ergothioneine is only synthesized by a few filamentous fungi, actinobacteria, and cyanobacteria but not by higher plants and animals. The red alga, Po. purpureum SAG1380-1C, was reported to produce a small amount of ergothioneine (Saha et al., 2015). It is unknown whether the algal SUMF-like sequences share the same function as SUMF sequences, or have other functions in ergothionein biosynthesis or meiosis as in MUG 158. Alternatively, these sequences may possess both functions. At least three brown algae were found to have both SUMF and SUMF-like sequences, indicating that both types of sequences could have different functions.
The phylogeny of SUMF and SUMF-like sequences (Figure 5) shows two main clusters consisting of SUMF sequences and SUMF-like sequences, respectively; which may share the same ancestor. The SUMF cluster consists of human SUMF1-3, bovin SUMF 1-2, mouse SUMF 1-2 together with four SUMFs from three brown algae (one from Ph. tricornutum and T. pseudonana, respectively; two from E. siliculosus) and one from the green microalga O. tauri. Each of the SUMF sequences in this cluster contains a FGE-sulfatase domain except for one of the SUMF sequences from E. siliculosus (ECTSI_D8LGF4) which has an incomplete domain while the SUMF sequence from O. tauri has an additional but incomplete glycosyltransferase domain (pfam 00534). The SUMF-like cluster consists of MUG 158 from Sch. pombe, SUMF-like sequences from A. protothecoides, T. pseudonana, E. siliculosus, C. crispus, Po. cruetum, Cy. merolae, Ga. sulphuraria, and Phy. patens. The domains found in the SUMF-like sequences are more variable. The red algal SUMF-like sequences were found to contain DinB_2 domain (pfam 12867) at their N-termini, in addition to the FGE-sulfatase domain (Figures 5, 6). The SUMF-like sequences from the green lineage (moss and green alga), similar to MUG158, were found to have two additional domains, i.e., pfam 10017 (S-adenosyl-L-methionine (SAM)-dependent methyltransferase domain) and pfam 12867 at the N-terminus of the FGE-sulfatase domain; while the brown algal SUMF-like sequences have pfam 12867 and pfam 10017 at the N- and C-termini of FGE-sulfatase domain, respectively (Figure 6). One of the sequences from E. siliculosus (ECTSI_D8LGF5) which has an incomplete FGE-sulfatase domain could not be assigned to either group of sequences.

Figure 6. Multiple sequence alignment of algal SUMF-like sequences. The amino acid sequences were aligned by ClustalW. Identical and similar sequences were highlighted in black and gray, respectively. The pfam domains 10017 (Histidine-specific SAM-dependent methyltransferase), 12867 (DinB domain), and 037181 (FGE-sulfatase) are underlined with green (dotted line), blue (broken line) and red, respectively. The DinB_2 iron-binding motif is indicated by blue box while the red box shows the EgtB subfamily C-terminal sequences. The identifier of the sequence starts with the species abbreviation followed by the UNIPROT/Genbank accession number and annotation wherever possible (Table 2). PHYPA, Physcomitrella patens; SCHPO, Schizosaccharomyces pombe.
It is intriguing that SUMF sequences were not found in the genomes of both green algae Ch. reinhardtii and V. carteri which have FGly-SULF sequences; and equally intriguing that SUMF sequence was found in O. tauri wherein FGly-SULF sequence was not detected. Similarly, SUMF or SUMF-like sequences were not reported in Saccharomyces cerevisiae and a few other yeasts which were shown to have FGly-SULFs. Could there be other sequences that are able to modify FGly-SULFs in Ch. reinhardtii and V. carteri? Alternatively, modification of Cys to FGly may not be necessary for these green algal FGly-SULFs. It is obvious that a group of SUMF-like sequences are present in green, brown, and red algae as well as moss, yeasts (at least Saccharomycetes and Ascomycetes), bacterium Mycobacterium, however, their functions are uncharacterized.
In general, the phylogeny of algal CHSTs, FGly-SULFs, and SUMFs or SUMF-like sequences revealed that many protein sequences were clustered according to their groups i.e., green (for CHSTs with pfam Sulfotransfer_1 domain), brown (for CHSTs, FGly-SULFs, and SUMFs or SUMF-like sequences), and red (for CHSTs with pfam Sulfotransfer_2 domain, FGly-SULFs and SUMFs or SUMF-like sequences) algae. Duplication/multiplication and functional divergence of these sequences could have happened after the divergence of these three groups of algae during evolution. Since only two green algal SUMFs or SUMF-like sequences were retrieved, the same trend was not observed. The clustering of a few CHSTs with pfam Sulfotransfer_1 domain from different groups of algae implied the existence of an ancestral sequence before the separation of these algal groups. The phylogenetic analyses of these putative genes contribute to the corpus of knowledge of an unexplored area. Algal CHSTs, FGly-SULFs, and SUMFs constitute a highly attractive target for future research to produce existing and new sulfated carbohydrate polymers through enzymatic synthesis and metabolic engineering.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was funded by Fundamental Research Grant Scheme (01-04-10-769FR) from the Ministry of Higher Education (MOHE) of Malaysia.
References
Altschul, S. F., Gish, W., Miller, W., Myres, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Aquino, R. S., Landeira-Fernandez, A. M., Valente, A. P., Andrade, L. R., and Mourão, P. A. S. (2005). Occurrence of sulfated galactans in marine angiosperms: evolutionary implications. Glycobiology 15, 11–20. doi: 10.1093/glycob/cwh138
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Baum, H., and Dodgson, K. S. (1957). Differentiation between myrosulphatase and the arylsulphatases. Nature 179, 312–313. doi: 10.1038/179312a0
Bhattacharya, D., Price, D. C., Chan, C. X., Qiu, H., Rose, N., Ball, S., et al. (2013). Genome of the red alga Porphyridium purpureum. Nat. Commun. 4, 1941. doi: 10.1038/ncomms2931
Bochenek, M., Etherington, G. J., Koprivova, A., Mugford, S. T., Bell, T. G., Malin, G., et al. (2013). Transcriptomic analysis of the sulfate deficiency response in the marine microalga Emiliania huxleyi. New Phytol. 199, 650–662. doi: 10.1111/nph.12303
Boltes, I., Czapinska, H., Kahnert, A., von Bülow, R., Dierks, T., Schmidt, B., et al. (2001). 1.3 A structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family. Structure 9, 483–491. doi: 10.1016/S0969-2126(01)00609-8
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the dynamic nature and multi-lineage evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Chaudière, J., and Ferrari-Iliou, R. (1999). Intracellular antioxidants: from chemical to biochemical mechanisms. Food Chem. Toxicol. 37, 949–962. doi: 10.1016/S0278-6915(99)00090-3
Cheah, I. K., and Halliwell, B. (2012). Ergothioneine; antioxidant potential, physiological function and role in disease. Biochim. Biophys. Acta 1822, 784–793. doi: 10.1016/j.bbadis.2011.09.017
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G., et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. doi: 10.1093/nar/gkg500
Cock, J. M., Sterck, L., Rouzé, P., Scornet, D., Allen, A. E., Amoutzias, G., et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465, 617–621. doi: 10.1038/nature09016
Collén, J., Porcel, B., Carré, W., Ball, S. G., Chaparro, C., Tonon, T., et al. (2013). Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on the evolution of the Archaeplastida. Proc. Natl. Acad. Sci. U.S.A. 110, 5247–5252. doi: 10.1073/pnas.1221259110
DeBoer, J. A. (1981). “Nutrients,” in The Biology of Seaweeds, eds C. S. Lobban and M. J. Wynne (Oxford: Blackwell Scientific), 359–391.
Dierks, T., Lecca, M. R., Schmidt, B., and von Figura, K. (1998a). Conversion of cysteine to formylglycine in eukaryotic sulfatases occurs by a common mechanism in the endoplasmic reticulum. FEBS Lett. 423, 61–65. doi: 10.1016/S0014-5793(98)00065-9
Dierks, T., Miech, C., Hummerjohann, J., Schmidt, B., Kertesz, M. A., and von Figura, K. (1998b). Posttranslational formation of formylglycine in prokaryotic sulfatases by modification of either cysteine or serine. J. Biol. Chem. 273, 25560–25564. doi: 10.1074/jbc.273.40.25560
Dierks, T., Schmidt, B., and von Figura, K. (1997). Conversion of cysteine to formylglycine: a protein modification in the endoplasmic reticulum. Proc. Natl Acad. Sci. U.S.A. 94, 11963–11968. doi: 10.1073/pnas.94.22.11963
Ehrhardt, D. W., Atkinson, E. M., Faull, K. F., Freedberg, D. I., Sutherlin, D. P., Armstrong, R., et al. (1995). In vitro sulfotransferase activity of NodH, a nodulation protein of Rhizobium meliloti required for host-specific nodulation. J. Bacteriol. 177, 6237–6245.
Felsenstein, J. (1985). Phylogenies and the comparative method. Am. Nat. 125, 1–15. doi: 10.1086/284325
Friedlander, M. (2001). Inorganic nutrition in pond cultivated Gracilaria conferta (Rhodophyta): nitrogen, phosphate and sulfate. J. Appl. Phycol. 13, 278–296. doi: 10.1023/A:1011139329415
Fukuda, M., Hiraoka, N., Akama, T. O., and Fukuda, M. N. (2001). Carbohydrate-modifying sulfotransferases: structure, function, and pathophysiology. J. Biol. Chem. 276, 47747–47750. doi: 10.1074/jbc.R100049200
Gao, Y., Schofield, O. M. E., and Leustek, T. (2000). Characterization of sulfate assimilation in marine algae focusing on the enzyme 5′-adenylylsulfate reductase. Plant Physiol. 123, 1087–1096. doi: 10.1104/pp.123.3.1087
Genicot-Joncour, S., Poinas, A., Richard, O., Potin, P., Rudolph, B., Kloareg, B., and Helbert, W. (2009). The cyclization of the 3,6-anhydro-galactose ring of ?-carrageenan is catalyzed by two ?-galactose-2,6-sulfurylases in the red alga Chondrus crispus. Plant Physiol. 151, 1609–1616. doi: 10.1104/pp.109.144329
Gijsbers, R., Ceulemans, H., Stalmans, W., and Bollen, M. (2001). Structural and catalytic similarities between nucleotide pyrophosphatases/phosphodiesterases and alkaline phosphatases. J. Biol. Chem. 276, 1361–1368. doi: 10.1074/jbc.M007552200
Hagelueken, G., Adams, T. M., Wiehlmann, L., Widow, U., Kolmar, H., Tümmler, B., et al. (2006). The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc. Natl. Acad. Sci. U.S.A. 103, 7631–7636. doi: 10.1073/pnas.0510501103
Hanin, M., Jabbouri, S., Quesada-Vincens, D., Freiberg, C., Perret, X., Promé, J. C., et al. (1997). Sulphation of Rhizobium sp. NGR234 Nod factors is dependent on noeE, a new host-specificity gene. Mol. Microbiol. 24, 1119–1129. doi: 10.1046/j.1365-2958.1997.3981777.x
Hanson, S. R., Best, M. D., and Wong, C. H. (2004). Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew. Chem. Int. Ed Engl. 43, 5736–5763. doi: 10.1002/anie.200300632
Hernandez-Sebastia, C., Varin, L., and Marsolais, F. (2008). “Sulfotransferases from plants, algae and phototrophic bacteria,” in Sulfur Metabolism in Phototrophic Organisms, eds R. Hell, C. Dahl, and T. Leustek (Dordrecht: Springer), 111–130.
Hopwood, J. J., and Ballabio, A. (2001). “Multiple sulfatase deficiency and the nature of the sulfatase family,” in The Metabolic and Molecular Bases of Inherited Disease, eds C. R. Scriver, A. L. Beaudet, W. S. Sly, D. Valle, B. Childs, K. W. Kinzler et al. (New York, NY: McGraw-Hill), 3725–3732.
Kakinuma, M., Kaneko, I., Coury, D. A., Suzuki, T., and Amano, H. (2006). Isolation and identification of gametogenesis-related genes in Porphyra yezoensis (Rhodophyta) using subtracted cDNA libraries. J. Appl. Phycol. 18, 489–496. doi: 10.1007/s10811-006-9052-8
Landgrebe, J., Dierks, T., Schmidt, B., and von Figura, K. (2003). The human SUMF1 gene, required for posttranslational sulfatase modification, defines a new gene family which is conserved from pro- to eukaryotes. Gene 316, 47–56. doi: 10.1016/S0378-1119(03)00746-7
Lee, H., Lee, H. K., An, G., and Lee, Y. K. (2007). Analysis of expressed sequence tags from the red alga Griffithsia okiensis. J. Microbiol. 45, 541–546.
Mata, J., Lyne, R., Burns, G., and Bähler, J. (2002). The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32, 143–147. doi: 10.1038/ng951
Matsuzaki, M., Misumi, O., Shin-I, T., Maruyama, S., Takahara, M., Miyagishima, S. Y., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. doi: 10.1038/nature02398
McHugh, D. J. (2003). “A guide to the seaweed industry,” in FAO Fisheries Technical Paper (Rome), 441.
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., Witman, G. B., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. doi: 10.1126/science.1143609
Meroni, G., Franco, B., Archidiacono, N., Messali, S., Andolfi, G., Rocchi, M., et al. (1996). Characterization of a cluster of sulfatase genes on Xp22.3 suggests gene duplications in an ancestral pseudoautosomal region. Hum. Mol. Genet. 5, 423–431. doi: 10.1093/hmg/5.4.423
Müller, I., Kahnert, A., Pape, T., Sheldrick, G. M., Meyer-Klaucke, W., Dierks, T., et al. (2004). Crystal structure of the alkylsulfatase AtsK: insights into the catalytic mechanism of the Fe(II) alpha-ketoglutarate-dependent dioxygenase superfamily. Biochemistry 43, 3075–3088. doi: 10.1021/bi035752v
Nikaido, I., Asamizu, E., Nakajima, M., Nakamura, Y., Saga, N., and Tabata, S. (2000). Generation of 10,154 expressed sequence tags from a leafy gametophyte of a marine red alga, Porphyra yezoensis. DNA Res. 7, 223–227. doi: 10.1093/dnares/7.3.223
Opoku, G., Qiu, X., and Doctor, V. (2006). Effect of oversulfation on the chemical and biological properties of kappa carrageenan. Carbohyd. Polym. 65, 134–138. doi: 10.1016/j.carbpol.2005.12.033
Parenti, G., Meroni, G., and Ballabio, A. (1997). The sulfatase gene family. Curr. Opin. Genet. Dev. 7, 386–391. doi: 10.1016/S0959-437X(97)80153-0
Pluskal, T., Ueno, M., and Yanagida, M. (2014). Genetic and metabolomic dissection of the ergothioneine and selenoneine biosynthetic pathway in the fission yeast, S. pombe, and construction of an overproduction system. PLoS ONE 9:e97774. doi: 10.1371/journal.pone.0097774
Pomin, V. H., and Mourão, P. A. (2008). Structure, biology, evolution and medical importance of sulfated fucans and galactans. Glycobiology 18, 1016–1027. doi: 10.1093/glycob/cwn085
Poux, N. (1966). Ultrastructural localization of aryl sulfatase activity in plant meristemic cells. J. Histochem. Cytochem. 14, 932–933. doi: 10.1177/14.12.932
Qin, X., Ma, C., Lou, Z., Wang, A., and Wang, H. (2013). Purification and characterization of ?-Gal-6-sulfurylasesfrom Eucheuma stratrium. Carbohyd. Polym. 96, 9–14. doi: 10.1016/j.carbpol.2013.03.061
Rees, D. A. (1961b). Enzymic synthesis of 3:6-andydro-L-galactose within porphyran from L-galactose 6-suphate units. Biochem. J. 81:347. doi: 10.1042/bj0810347
Saha, S. K., McHugh, E., Murray, P., and Walsh, D. J. (2015). “Chapter 12 Microalagae as a source of nutraceuticals,” in Phycotoxins: Chemistry and Biochemistry, 2nd Edn., eds L. M. Botana and A. Alfonso (Chichester: John Wiley & Sons, Ltd), 271.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sardiello, M., Annunziata, I., Roma, G., and Ballabio, A. (2005). Sulfatases and sulfatase modifying factors: an exclusive and promiscuous relationship. Hum. Mol. Genet. 14, 3203–3217 doi: 10.1093/hmg/ddi351
Schirmer, A., and Kolter, R. (1998). Computational analysis of bacterial sulfatases and their modifying enzymes. Chem. Biol. 5, R181–R186. doi: 10.1016/s1074-5521(98)90154-5
Schmidt, B., Selmer, T., Ingendoh, A., and von Figura, K. (1995). A novel amino acid modification in sulfatases that is defective in multiple sulfatase deficiency. Cell 82, 271–278. doi: 10.1016/0092-8674(95)90314-3
Shukla, M. K., Kumar, M., Prasad, K., Reddy, C. R. K., and Jha, B. (2011). Partial characterization of sulfohydrolase from Gracilaria dura and evaluation of its potential application in improvement of the agar quality. Carbohyd. Polym. 85, 157–163. doi: 10.1016/j.carbpol.2011.02.009
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: MOLECULAR Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Teo, S.-S., Ho, C.-L., Teoh, S., Lee, W.-W., Tee, J.-M., Raha, A. R., et al. (2007). Analyses of expressed sequence tags from an agarophyte, Gracilaria changii (Gracilariales, Rhodophyta). Eur. J. Phycol. 42, 41–46. doi: 10.1080/09670260601012461
Tuvikene, R., Truus, K., Kollist, A., Volobujeva, O., Mellikov, E., and Pehk, T. (2008). Gel-forming structures and stages of red algal galactans of different sulfation levels. J. Appl. Phycol. 20, 527–535. doi: 10.1007/s10811-007-9229-9
Wang, A., Islam, M. N., Qin, X., Wang, H., Peng, Y., and Ma, C. (2014). Purification, identification and characterization of D-galactose-6-sulfurylase from marine algae (Betaphycus gelatinus). Carbohydr. Res. 388, 94–99. doi: 10.1016/j.carres.2013.12.010
Wong, K. F., and Craigie, J. S. (1978). Sulfohydrolase activity and carrageenan biosynthesis in Chondrus crispus (Rhodophyceae). Plant Physiol. 61, 663–666. doi: 10.1104/pp.61.4.663
Keywords: algae, carbohydrate sulfotransferases, sulfatases, phylogeny, sulfatase modifying factors
Citation: Ho C-L (2015) Phylogeny of Algal Sequences Encoding Carbohydrate Sulfotransferases, Formylglycine-Dependent Sulfatases, and Putative Sulfatase Modifying Factors. Front. Plant Sci. 6:1057. doi: 10.3389/fpls.2015.01057
Received: 25 September 2015; Accepted: 13 November 2015;
Published: 26 November 2015.
Edited by:
Stanislav Kopriva, University of Cologne, GermanyReviewed by:
Fumiya Kurosaki, University of Toyama, JapanFrédéric Marsolais, Agriculture and Agri-Food Canada, Canada
Copyright © 2015 Ho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chai-Ling Ho, clho@upm.edu.my
 Chai-Ling Ho
Chai-Ling Ho