- 1Department of Pomology, College of Horticulture Science and Engineering, Shandong Agricultural University, Taian, China
- 2Shandong Provincial Key Laboratory of Microbial Engineering, School of Biologic Engineering, Qi Lu University of Technology, Jinan, China
It has been well-demonstrated that the control of plasma membrane H+-ATPase (PM H+-ATPase) activity is important to plant salt tolerance. This study found a significant increase in PM H+-ATPase (PMA) activity in grape root exposed to NaCl. Furthermore, 7 Vitis vinifera PM H+-ATPase genes (VvPMAs) were identified within the grape genome and the expression response of these VvPMAs in grape root under salinity was analyzed. Two VvPMAs (VvPMA1 and VvPMA3) were expressed more strongly in roots than the other five VvPMAs. Moreover, roots exhibited diverse patterns of gene expression of VvPMA1 and VvPMA3 responses to salt stress. Interestingly, two transcripts of VvPMA1, which were created through alternative splicing (AS), were discovered and isolated from salt stressed root. Comparing the two VvPMA1 cDNA sequences (designated VvPMA1α and VvPMA1β) with the genomic sequence revealed that the second intron was retained in the VvPMA1β cDNA. This intron retention was predicted to generate a novel VvPMA1 through N-terminal truncation because of a 5′- terminal frame shift. Yeast complementation assays of the two splice variants showed that VvPMA1β could enhance the ability to complement Saccharomyces cerevisiae deficient in PM H+-ATPase activity. In addition, the expression profiles of VvPMA1α and VvPMA1β differed under salinity. Our data suggests that through AS, the N-terminal length of VvPMA1 may be regulated to accurately modulate PM H+-ATPase activity of grape root in salt stress.
Introduction
Salinity is one of the most important abiotic stresses limiting crop productivity, and the amount of land affected by salinity is increasing (Wakeel et al., 2010). Salt stress has been found to affect the activity of the plasma membrane H+-ATPase (PM H+-ATPase) in plant (Janicka-Russak et al., 2013). The PM H+-ATPase is part of a large family of ion transport proteins termed P-type ATPases, which are responsible for generating the electrochemical proton motive force across the plasma membrane of plant and fungal cells. The H+-ATPase is thus essential for energizing the plasma membrane (Palmgren, 2001; Arango et al., 2003) and is involved in many physiological roles, such as solute transport into the cell, cell pH homeostasis, and elongation (Duby and Boutry, 2009). Moreover, PM H+-ATPas provides the driving force for potassium retention and uptake through voltage-gated channels and for Na+ exclusion via Na+/H+ exchangers during salt stress (Kabała and Janicka-Russak, 2012; Hasegawa, 2013). Both of these traits are central to plant salinity tolerance. Therefore, the control of PM H+-ATPase activity is important to plant salt tolerance (Gévaudant et al., 2007; Janicka-Russak et al., 2013).
In plants, multiple mechanisms have evolved to regulate H+-ATPase expression and activity (Haruta et al., 2015; Falhof et al., 2016). At the posttranslational level, PM H+-ATPase activity is controlled by a C-terminal auto-inhibitory domain of about 100 residues (Piette et al., 2011). It is also well-established that protein kinase-mediated phosphorylation of the penultimate threonine can release the auto-inhibition of the C-terminal region, resulting in a hyperactive enzyme, with increased catalytic activity (Lecchi et al., 2008). In addition, other phosphorylated residues of the C-terminal region have been identified, and some have been shown to reduce or increase enzyme activity (Duby et al., 2009; Hsu et al., 2009). Unfortunately, the specific kinases that catalyze this reaction are still uncertain. Recently, the N-terminal region has also been proposed to be involved in enzyme auto-inhibition. Complete removal of the N terminus of the PM H+-ATPase in vitro results in activation of the PM H+-ATPase (Ekberg et al., 2010). However, as the enzyme hyperactivity caused by phosphorylation of the N-terminal region seems an ineffective regulatory mechanism, until now, the mechanisms though which regulation of the N-terminal region of PM H+-ATPase is achieved in vivo were unclear (Rudashevskaya et al., 2012).
Previous studies have shown that under saline conditions, fast posttranslational modification in the C-terminal region of PM H+-ATPase can occur (Kalampanayil and Wimmers, 2001; Janicka-Russak and Kłobus, 2007). In addition to posttranslational modification, increases in PM H+-ATPase transcription in response to NaCl exposure has been reported (Zhang et al., 1999; Chen et al., 2007; Sahu and Shaw, 2009).
In most plant species, the PM H+-ATPase is encoded by a multigene family. Extensive searching of cDNA and genomic libraries from Nicotiana plumbaginifolia results in the identification of nine PM H+-ATPase genes (Perez et al., 1992; Moriau et al., 1993; Oufattole et al., 2000). Complete sequencing of the Arabidopsis genome uncovered 12 genes (Palmgren, 2001). The existence of multiple isoforms of the enzyme might suggest considerable overlap of isoform expression in many cell types and tissues Gévaudant et al., 2001. Nonetheless, isoform diversity may also be related to cellular differentiation with individual isoforms exhibiting tissue, developmental, and environmental-specific expression and having slightly different biochemical and regulatory properties (Palmgren, 2001; Arango et al., 2003). The functional significance of the multiple isoforms of PM H+-ATPase, such as the possibility of their role in salt stress tolerance, is not well-understood, although PM H+-ATPase does respond to salt stress. For example, in rice cultivars, salt treatment induced expression of a new isoform of the PM H+ATPase gene (Sahu and Shaw, 2009).
Vitis vinifera (V. vinifera) is one of the world's oldest fruits and nowadays grows worldwide. Grapevine is adapted to semiarid environments, where drought and salinity are common problems. Grapevine is considered moderately tolerant to salt stress, and this moderate tolerance has been mainly attributed to salt exclusion or to restriction of toxic ions in the root system (Walker et al., 2002; Ma et al., 2015), which needs PM H+-ATPase of root to provide driving force. Up to now, research on PM H+-ATPases has mainly focused on annual herbs, while there is little information about the regulation of PM H+-ATPase activity in perennial woody plants, such as V. vinifera under saline conditions. Here the PM H+-ATPase isoforms within the grape genome (Jaillon et al., 2007) were identified and the effect of salinity on the expression of root specific isoforms was analyzed in grapevine.
Methods
Plant Materials and Growth Conditions
The table-grape cultivar Crimson seedless (C133–199 × Emperor) was used in this study. The sterilized seedlings were grown on 1/2 MS solid medium (Murashige and Skoog, 1962) in glass bottles (9 cm height × 6 cm diameter) at a 25/20°C growth room with white light illumination (120 μmol photons per m2 s1) under a 16/8 h light/dark photocycle. Shoot tips with a minimum of one bud were used to subculture every month. After 30 days, uniform plants were selected as the test material and were transferred into Hoagland solution (2 mM MgSO4, 5 mM Ca(NO3)2, 5 mM KNO3, 1 mM KH2PO4, and microelements contained 0.2 mM Fe-EDTA, 2 × 10−3 mM MnCl2, 2.5 × 10−4 mM H3BO3, 5 × 10−4 mM CuSO4, 2 × 10−3 mM ZnSO4 and 5 × 10−4 mM H2MoO4) for 7 days. The growth conditions were kept the same as above. The test seedlings were treated with 50 mM NaCl in Hoagland solution for 3, 6, 12, 24, 48, and 72 h, respectively. And the seedlings grown in Hoagland alone were used as the control (0 h). Samples were then harvested at the same time in the same day to minimize diurnal effects wherever possible, then frozen in liquid nitrogen and stored at −80°C.
Membrane Vesicle Isolation
Plasma membrane-enriched vesicles were isolated by discontinuous sucrose gradient centrifugation, according to the method of Wang et al. (2001) and Mandala and Taiz (1985) with modifications. Roots were washed with cold deionized water and homogenized in extraction medium containing 50 mM Tricine-Tris (pH 7.8), 3 mM EGTA, 3 mM MgSO4, 0.5% (V/V) PVP, 2 mM DTT, 0.2 mM PMSF, 5% (V/V) glycerol, and mannitol, which produced the same osmotic potential as within leaves. Two milliliters of the medium was used for each 2 g of fresh material. The homogenate was filtered through four layers of cheesecloth and centrifuged at 10,000 g for 20 min (Beckman JA-20). The supernatant was loaded on a 0%: 36%: 45% (W/W) sucrose gradient solution (5 mM HEPES adjusted to pH 7.5 with Tris, and 1 mM DTT) and centrifuged at 100,000 g for 2 h (Beckman SW40Ti). The plasma membrane-enriched vesicles were located at the 36%: 45% (W/W) sucrose interface. These vesicles were carefully collected and diluted 3–4-fold with dilution buffer (3 mM MgSO4, 10 mM HEPES–Tris pH 7.5, 1 mM DTT) and centrifuged at 100,000 g for 30 min (Beckman type 65). The pellets were suspended in a storage buffer (40% (V/V) glycerol, 2 mM DTT, 10 mM HEPES, adjusted to pH 7.0 with KOH).
Measurement of Protein Content
Protein content was determined using bovine serum album as a protein standard according to the Bradford method (Bradford, 1976).
ATPase Assays
PM H+-ATPase hydrolytic activity was calculated from the amount of inorganic phosphate released in the absence and presence of 0.1 mM Na3VO4. The enzyme reaction was run at 37°C for 30 min with 20 μg of membrane protein in an assay medium containing 30 mM Tris/Mes pH 6.5, 0.1 mM (NH4)2MoO4, 1 mM NaN3, 1 mM MgSO4, 0.03% (V/V) TritionX-100, 50 mM KCl, and 3 mM ATPNa2. Inorganic phosphate was assayed by using the method of Lin and Morales (1977).
Genome Wide Search and Identification of VvPMAs
The highly conserved domain “GDGVNDAPALK” was defined as the probe sequence according to the alignment results of published PM H+-ATPase amino acid sequences in plants (Palmgren, 2001). The non-redundant protein database of V. vinifera was retrieved through blastp on the NCBI website (National Center for Biotechnology Information, www.ncbi.nlm.nih.gov). Then the PM H+-ATPase functional domains of these candidate proteins were predicted using Pfam software (http://pfam.sanger.ac.uk; Finn et al., 2010). The corresponding gene sequences and cDNA sequences of these domain-verified PM H+-ATPases were acquired using accession numbers.
Phylogenetic Tree Construction and Multiple Sequence Alignment of VvPMAs
The phylogenetic tree was constructed using MEGA 5.0 software and making comparisons between candidate VvPMAs and previously identified AHA proteins from A. thaliana.
Total RNA Isolation from Grape
Total grape RNA was extracted using the CTAB method as previously described (Gambino et al., 2008), with a few modifications. Root samples (200 mg) were ground with mortar and pestle under liquid nitrogen. The powder was transferred to an RNase-free tube containing 2 mL CTAB extracting buffer (2% CTAB, 2.5% PVP-30, 2 M NaCl, 100 mM Tris-HCl (pH 8.0), 25 mM EDTA, and 2% 2-mercaptoethanol) and mixed. Then the samples were incubated at 65°C for 10 min. After centrifugation (4°C, 10, 000 g, 10 min), the supernatant was then transferred to an equal volume of extraction cocktail [chloroform/isoamyl alcohol (24/1)] in order to extract the RNA. The mixture was centrifuged at 10, 000 g for 10 min, after which, the supernatant was mixed with LiCl (3 M) and stored at −20°C for 12 h. After centrifugation (4°C, 10, 000 g, 20 min), the pellets were suspended in SSTE buffer [1 M NaCl, 1% SDS, 1 mM EDTA, and 10 mM Tris-HCl (pH 8.0)]. A chloroform/isoamyl alcohol (24/1) mixture was added, and then the samples were centrifuged at 10, 000 g for 10 min. The supernatant was transferred to a new RNase-free tube and mixed with 0.7 volume isopropanol. After centrifugation at 10,000 g for 20 min, the pellets were washed with 1 ml 75% alcohol, dried, and resuspended in RNase free water. The total RNA was quantified with a NanoDrop 2000C spectrophotometer (Thermo Scientific, USA) at wavelengths of 230, 260, and 280 nm and the 260/280 nm ratio was determined. RNA integrity was confirmed by running samples through 1% agarose gels. After that samples were stored at −80°C.
Quantitative RT-PCR (qRT-PCR) for Analysis of Gene Expression
First-strand cDNA was synthesized from approximately 3 μg of total RNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer's instructions. Quantitative PCR was carried out on a real-time PCR system (Bio-Rad iQ5) using the SYBR Green Kit (TaKaRa, Dalian, China). Thermal cycling conditions were 95°C for 10 min followed by 95°C for 10 s, 56°C for 10 s for 40 cycles, and a melting cycle was then performed from 65° to 95°C. A house-keeping gene actin was used as an internal control for qPCR (Reid et al., 2006), and it was previously used as a reliable internal reference gene in grape under salt stress (Ma et al., 2015). VvPMAs transcripts were quantified after normalization to actin. Results are reported as 2−ΔCT or 2−ΔΔCT, where ΔCT is the number of PCR cycles required for the log phase of amplification for the experimental gene minus the same measure for actin and ΔΔCT is the ΔCT of treatment minus the ΔCT of control (0 h) for the same gene (Livak and Schmittgen, 2001). Primer sequences used for the qPCR analyses were shown in Supplementary Table 1.
Cloning and Sequencing of the PM H+-ATPases Isoform VvPMA1
RNA was extracted from the root of a Crimson seedless grape plant and cDNA synthesis was carried out as described above. The full-length cDNA encoding VvPMA1 was isolated through PCR using Pfu DNA polymerase (TaKaRa, Dalian, China) and the primers listed in Supplementary Table 2. The generated PCR fragment was purified and directly inserted into the pEASY-Blunt vector following the manufacturer's instructions (Transgen Biotech. Co., China). The plasmids were transformed into E. coli DH5α cells. Finally, the 9 recombinant plasmids were prepared and then sequenced, respectively.
Expression of PM H+-ATPases in Saccharomyces cerevisiae
Plasmid pMP625 containing the promoter and terminator of PMA1 (yeast endogenous PM H+-ATPase) was kindly provided by Dr. Palmgren (University of Copenhagen, Denmark). The full-length coding regions of VvPMA1α and VvPMA1β were amplified from pEASY-Blunt-VvPMA1α and pEASY-Blunt-VvPMA1β using the primers VvPMA1α-P1, 5′CTCGAGATGGGAGGCGACAAATCC 3′, which includes an ud XhoI site, and VvPMA1-P2, 5′ GGGCCCTCATACTGTGTAATGCTGTTGGATT 3′, with an ud ApaI site; VvPMA1β-P1, 5′CTCGAGATGCGCATTTCATTGAATT 3′ and VvPMA1-P2, 5′ GGGCCCTCATACTGTGTAATGCTGTTGGATT 3′. The PCR products were subcloned into XhoI and ApaI sites of pMP625 to generate plasmid pMP625-VvPMA1α and pMP625-VvPMA1β. All PCR amplifications were carried out using Pfu DNA polymerase, which exhibits the lowest error rate of any thermostable DNA polymerase, and all constructs were sequenced to confirm their identity.
Yeast Strains and Culture Conditions
The yeast strain RS72 (also gifted from Dr. Palmgren) was cultured as described previously (Ekberg et al., 2010). In RS72 (MATa ade1–100 his4–519 leu2–3, 112), the natural constitutive promoter of the PMA1 has been replaced by the galactose-dependent GAL1 promoter. As PMA1 is essential for yeast growth, RS72 is only able to grow on galactose-containing medium. Using this strain, plasmid-borne plants' PM H+-ATPases under the control of the constitutive PMA1 promoter can be tested for their ability to rescue pma1 mutants on glucose-containing medium.
Yeast was made competent for plasmid uptake by treatment with lithium acetate and polyethyleneglycol according to Gietz et al. (1995). Positive transformants were selected on synthetic medium containing 2% (w/v) galactose, 0.7% (w/v) yeast nitrogen base without amino acids (Difco), 0.2 mM adenine, 0.4 mM histidine, without leucine after 3 days of growth at 30°C. And transformants were grown for 3 days at 30°C in liquid synthetic medium containing 2% galactose. Then, 5 ml transformed yeast cells were transferred to solid at two different concentrations (OD600 0.1 and OD600 0.01) or 20 ml liquid synthetic medium at OD600 0.01 containing either 2% galactose at pH 5.5 or 2% glucose at pH-values of 5.5, 4.5, and 3.5. The media were buffered with 50 mM succinic acid adjusted to various pH-values with Tris. Growth was recorded after incubation for 3 days at 30°C. Each experiment was replicated independently three times.
Statistical Analysis
Statistical analyses were performed using the software SigmaPlot 10.0 (Systat Software, Inc). Values were expressed as mean ± SD. All comparisons were done using Student's t-test for independent samples. In all cases, the confidence coefficient was set at 0.05.
Results
Effect of NaCl on PM H+-ATPase Activity
The plants were subjected to salt stress (50 mM NaCl) for 0, 3, 12, 24, or 72 h. ATP hydrolytic activity in PM vesicles obtained from plant roots increased under the NaCl treatments. Moreover, the enzyme activity was up regulated within 3 h of treatment (P < 0.01). The maximum increase in enzyme activity resulted in an activity about 2.7-fold greater than that of the control at 24 h. Although the enzyme activity decreased between 24 and 72 h, the activity level remained above the control activity level (P < 0.01) (Figure 1).
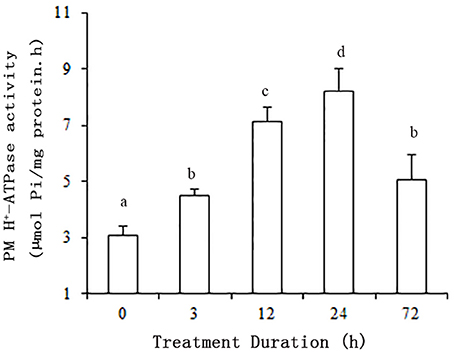
Figure 1. Effect of NaCl on the hydrolytic activity of PM H+-ATPase in grape root. The plasma membranes were isolated from roots treated with 50 mM NaCl for 0, 3, 12, 24, and 72 h. The values are means (n = 9) of three independent experiments ± SD. Values with the same letter are not significantly different (P > 0.05).
Identification of VvPMAs through Genome-Wide Analysis
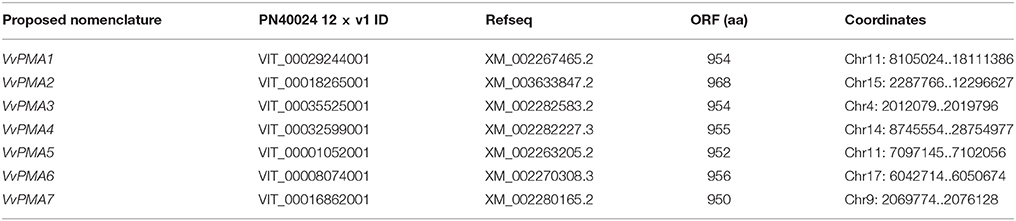
To explore the prevailing isoforms of PM H+-ATPase genes in grape root treated with salt stress, the VvPMA genes were screened first from genome of the near-homozygous PN40024 genotype of V. vinifera cv Pinot Noir. According to the retrieved candidate VvPMA proteins and Pfam software verification, we obtained putative 14 VvPMAs in silico. Subsequently, we eliminated overlapping protein sequences and short fragments based on the functional domain sequence. Finally, a total of 7 candidate genes were identified as possible grape PM H+-ATPase genes (Table 1).
In order to determine the evolutionary relatedness among the PM H+-ATPases from grape and Arabidopsis, a phylogenetic tree was constructed using MEGA 5.0 software and the deduced amino acid sequences. The phylogenetic analysis showed PM H+-ATPase protein from each of the two species to fall within the same clades with a high degree of similarity. Additionally, VvPMAs were classified into four subfamilies according to Arango et al. (2003; Figure 2).
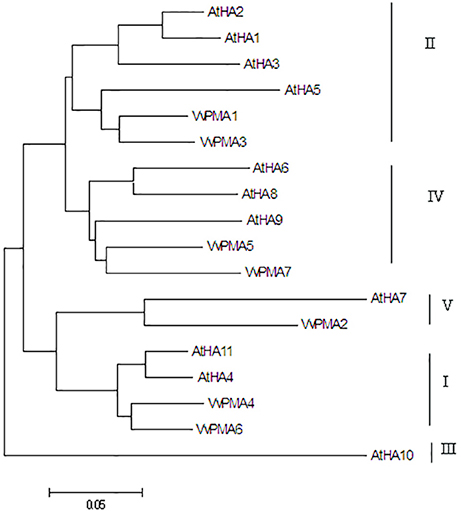
Figure 2. Phylogenetic tree of the PM H+-ATPase proteins from V. vinifera (VvPMA1-7) and A. thaliana (AtHA1-11). The phylogenetic tree was created using the MEGA 5.0 software. Roman numerals designated the subfamilies.
Expression of VvPMAs in Grape Root
In seedling roots grown under normal conditions, expression of VvPMA1 and VvPMA3 was markedly higher than VvPMA4 and VvPMA6 expression (P < 0.01), but expression levels did not significantly differ between VvPMA1 and VvPMA3 (P > 0.05). Additionally, although expression of VvPMA2, VvPMA5, and VvPMA7 was found in other tissues (Supplementary Figure 1), their expression was undetectable in roots using either of two different primer pairs (Figure 3).
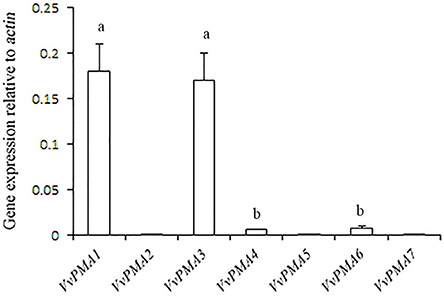
Figure 3. The relative expression levels of the different VvPMA isoforms in grape root. Vvactin was used as an internal standard. The data were analyzed according to 2−ΔCT method. The values are means ± SD (n = 5). Values with the same letter are not significantly different (P > 0.05).
As the predominant genes expressed in root, VvPMA1 and VvPMA3 were greatly up-regulated under 50 mM NaCl stress (Figure 4). Moreover, VvPMA1 expression was not up-regulated significantly until 12 h of treatment, and then reached its maximum level at 48 h, when it was as much as 3.6 times greater than that of the control (P < 0.01; Figure 4). Expression remained greater than control levels until 72 h of treatment. For VvPMA3, expression was highest at 6 h of treatment, when it was 3.3-fold greater than at 0 h. After that, expression reduced, but remained above 0 h levels until 72 h of treatment (P < 0.01; Figure 4).
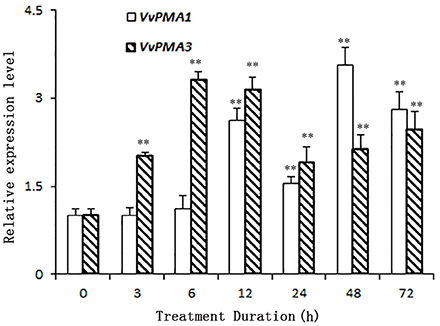
Figure 4. The relative expression levels of VvPMA1 and VvPMA3 in grape root under 50 mM NaCl stress for 0–72 h. Vvactin was used as an internal standard. The data were analyzed according to 2−ΔΔCT method. The values are means ± SD (n = 5). ** are significantly different from 0 h at P < 0.01.
Cloning and Alternative Splicing of VvPMA1
To study the function of VvPMA1, the full-length cDNA sequence of VvPMA1 was cloned and sequenced through reverse transcription PCR (RT-PCR). Interestingly, two different cDNA sequences of VvPMA1 were obtained. Comparison of the two VvPMA1 cDNA sequences (designated VvPMA1α and VvPMA1β) with the genomic sequence revealed that the second intron (93 bp) was retained in the VvPMA1β cDNA (Figure 5A, Supplementary Figure 2). Furthermore, RT-PCR using VvPMA1α and VvPMA1β specific primers (Figure 5A, Supplementary Table 1) detected both transcripts, respectively after 3 h of 50 mM NaCl stress (Figure 5B), proving that AS of VvPMA1 really occur.
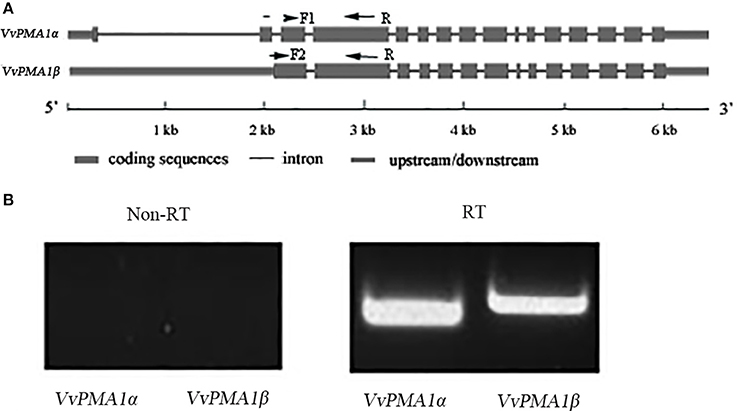
Figure 5. VvPMA1 alternative splicing events in grape root. (A) Genomic structure of VvPMA1 and the alternatively spliced variant. F1 and F2, forward primers; R, reverse primer. (B) Detection of alternatively spliced VvPMA1 by RT–PCR in grape root in 50 mM NaCl stress for 3 h.
The predicted VvPMA1α and VvPMA1β proteins had identical C domains, whereas a frame shift resulted in a loss of 35 amino acids at the N-terminus of VvPMA1β (Supplementary Figure 3).
Complementation of Yeast Mutants by Two VvPMA1 Splice Variants
To investigate whether both VvPMA1α and VvPMA1β encode functional proton pumps and test for their ability to complement pma1, we expressed them in mutant yeast RS72. The results were shown in Figure 6. The negative control (the strain MP625 with the yeast PMA1 under the GAL1 promoter) grew well in galactose medium only (Figure 6A). The strains with VvPMA1α and VvPMA1β both supported yeast growth on either glucose solid or liquid medium (Figures 6B,C), however, at pH 5.5 and more acidic pH levels, VvPMA1β increased yeast growth by a greater amount than VvPMA1α did (Figures 6B,C).
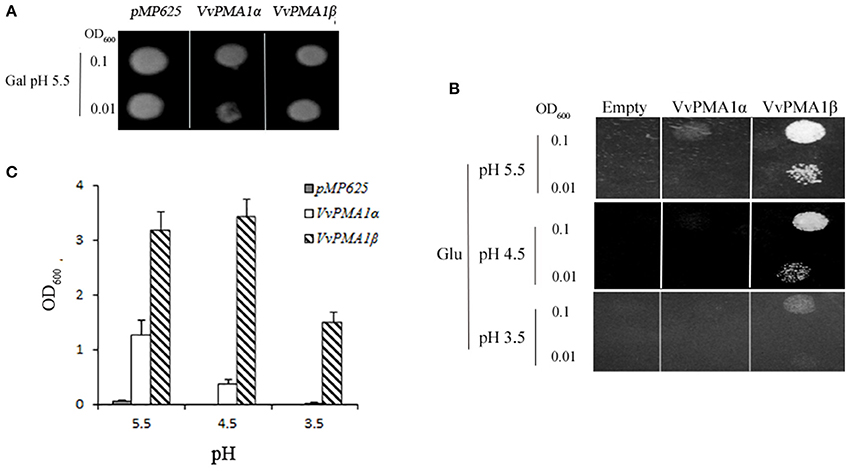
Figure 6. Activities of VvPMA1 spliced variants in a yeast complementation assay. Drop test for the growth of yeast strains on galactose (A) and glucose (B) solid medium or in glucose liquid medium (C). In the yeast strain RS72, the endogenous yeast PM H+-ATPase (PMA1p) has been placed under the control of a galactose promoter. The introduced, plasmid-borne VvPMA1 are under the control of the constitutive PMA1 promoter. Yeast growth on glucose is therefore dependent on a functional plasmid-borne VvPMA1. Transformed yeast cells, were spotted on either galactose-containing solid media (Gal) at pH 5.5 (A) or glucose-containing solid media (Glu) at different pH-values (B) at two different concentrations (OD600 = 0.1 and OD600 = 0.01). And same inoculated into glucose-containing liquid media at OD600 = 0.01 (C). Growth was recorded after 3–4 days at 30°C. The values are means ± SD (n = 3).
Gene Expression of the Two VvPMA1 Splice Variants in Grape Root under Salt Stress
To ascertain the regulatory mechanisms controlling the two VvPMA1 splice variants under salt stress, the expression levels of the two variants were examined in root under salt treatments. The results showed expression of VvPMA1α to be much higher than VvPMA1β expression in salt and fresh water treated roots (Figures 7A,B). However, VvPMA1α expression had not too much change until 48 h of treatment (Figure 7A), whereas, VvPMA1β expression increased rapidly and peaked at 12 h under salt treatment (Figure 7B).
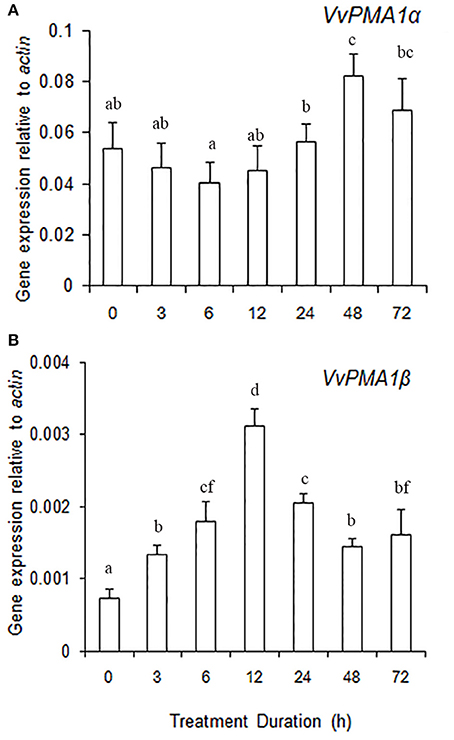
Figure 7. The relative expression levels of VvPMA1 spliced variants, VvPMA1α (A) and VvPMA1β (B), in grape root under 50 mM NaCl stress for 0–72 h. Vvactin was used as an internal standard. The data were analyzed according to 2−ΔCT method. The values are means ± SD (n = 5). Values with the same letter are not significantly different (P > 0.05).
Discussion
To prevent accumulation of toxic Na+ amounts in the cytosol, active sodium efflux into the apoplast is one of the three strategies employed by plants to increase salt resistance (Greenway and Munns, 1980). Sodium ions are transported from the cytosol across the PM against the electrochemical gradient into the apoplast via Na+ /H+ antiporters. Secondary transporters in the PM of plant cells are activated by the PM proton pump, PM H+-ATPase, which is a major enzyme of the plant PM that plays a central role in physiological functions and adaptation of plants to changing conditions, especially stress. Thus, PM H+-ATPase may be important to environmental stress resistance in general (Palmgren, 2001; Palmgren and Nissen, 2011). Therefore, it can be assumed that PM H+-ATPases are controlled by multiple regulatory features that integrate signals from the environment. Our results were consistent with previous findings that saline conditions stimulate the hydrolytic activity of PM H+ -ATPase in root (Figure 1; Niu et al., 1996; Sibole et al., 2005). Moreover, in the present study, the enzyme responded rapidly, within 3 h, to salinity treatments. Therefore, rapid posttranslational modifications to PM H+-ATPase likely occurred in grape root under salt stress conditions. Posttranslational modification is the best-known mechanism for rapid regulation described to date, and involves the auto inhibitory action of the C-terminal domain of the enzyme (Hsu et al., 2009; Haruta et al., 2015). Further work will be needed to confirm whether phosphorylation of the C terminus of VvPMA occurs in grape root under salt stress.
In addition to posttranslational modifications, changes to PM H+-ATPase activity can partly result from changes in expression patterns of PM H+-ATPase genes. This enzyme is coded by a multigene family in grape, like in other plants (Figure 2; Arango et al., 2003). VvPMA1 and VvPMA3 (Figure 3) were the predominantly expressed isoforms found in grape root, and NaCl stress increased accumulation of their transcripts in the root (Figure 4). Moreover, the VvPMA3 expression increased significantly faster than VvPMA1 expression. These results suggested that transcription of VvPMA3 responded more quickly than VvPMA1 transcription in grape root under NaCl stress. Therefore, the root VvPMA3 appeared to play a critical role in early resistance to salt stress.
More interestingly, however, retention of the second intron in VvPMA1 mRNA was discovered under salt conditions (Figure 5A, Supplementary Figure 2). This intron retention produced a spliced isoform, VvPMA1β, shortened by 35 amino acids at the N-terminal domain (Supplementary Figure 3). Moreover, yeast complementation assays found that, compared with VvPMA1α, VvPMA1β enhanced the ability of VvPMA1 to support yeast growth, at pH 5.5 and more acidic pH levels (Figures 6B,C). This finding confirmed earlier findings that small deletions from the N terminus in vitro increased the ability of the plant PM H+-ATPase to support yeast growth. Ekberg et al. (2010) showed that destabilization or complete removal of the N terminus of PM H+-ATPase somehow results in an unmasking of the extreme C terminus, which makes the penultimate threonine residue accessible for protein kinase-mediated phosphorylation and subsequent activation of the PM H+-ATPase. We have shown, for the first time, that removal of the N terminus of PM H+-ATPase can be achieved in vivo through AS.
Recent genome-wide gene expression analyses have suggested that mRNA splicing is under the control of developmental and stress signals (Seo et al., 2011; Marquez et al., 2012). Here, the results showed that although expression of VvPMA1α was higher than VvPMA1β expression in root, regardless of treatment (Figures 7A,B), the expression level of VvPMA1β still greatly increased under salt stress, suggesting that alternative splicing of VvPMA1 was markedly induced under salt conditions. Previous results showed that splicing factors, such as serine/arginine-rich (SR), Ski-interacting protein (SKIP) etc., can control the AS of pre-mRNAs encoded by salt tolerance genes under salt stress in plants (Syed et al., 2012; Feng et al., 2015) and different growth conditions can modulate gene expression or protein modification of splicing factors causing dynamic changes in the splicing factor profile, further impacting expression of target genes. So we supposed that the expression of spliced variant VvPMA1β must be controlled by one or some specific splicing factors in grape root under salt conditions, moreover, VvPMA1β expression maintained low level in root under salt stress probably because the specific splicing factor has weak activity. Alternatively, PM H+-ATPase activity could be confined by an unknown feedback mechanism under different environment and overexpression of its gene might be disadvantageous to plant growth (Hashimoto-Sugimoto et al., 2013). Thus, it can be speculated that the amount of VvPMA1β protein with high activity is possible to be strictly controlled at posttranscriptional level in grape root under salt stress, to avoid wasting too much energy at feedback regulation of PM H+-ATPase activity. The specific splicing factor will be further screened from grape root and the function of VvPMA1β will be ascertained by transgenetic effectiveness in plant under salt stress.
Additionally, posttranscriptional regulation of VvPMA1 in response to salt stress occurred at 3 h of treatment as early as transcriptional regulation of VvPMA3 (Figures 4, 7). And both expression of VvPMA1β and VvPMA3 reached peak before 24 h. Meanwhile VvPMA1α expression was unchanged markedly. After that, VvPMA1α expression obviously increased, while the other two diminished. Combining these findings with the dynamic change of the activity of PM H+-ATPase and VvPMA1β activity (Figure 1, Figure 6B), we predicted that VvPMA1β would have a positive effect on PM H+-ATPase activity in salt stress, albeit with phenotypic lag. Therefore, subtle changes in the ratio of intact VvPMA1 and the alternatively spliced variant would likely have profound effects on PM H+-ATPase activity in grape root under saline conditions in spite of changes to the quantity of PM H+-ATPase present. Further work is required to determine changes in the quantity of each PM H+-ATPase isoform under salt stress.
Taken together, the results of this study revealed that NaCl has a stimulatory effect on PM H+-ATPase in grape root. Increases to enzyme activity were accompanied by transcriptional increases. Furthermore, cloning and sequencing of the PCR-amplified fragments of PM H+-ATPase transcripts revealed the expression of an N-terminal truncated splice variant in response to NaCl treatment. This variant had a higher activity rate than the intact variant. These findings suggest that alternative splicing, in coordination with gene transcriptional control, may help to maintain the appropriate level of the PM H+-ATPase activity in grape roots under saline condition. Furthermore, it is also possible that differential regulation of the PM H+-ATPase isoforms within the same cell could be an important mechanism for responding to a diverse range of environmental fluctuations.
Author Contributions
NH, XZ, and HZ participated in the study conception and design. XJ contributed to RNA extraction and qPCR. NH, YD, and XH contributed to data analysis. NH wrote the manuscript. YD and XH critically revised the manuscript. All authors approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (NSFC) No. 31501738, the National Modern Agricultural Research System Special Fund (CARS-30), and the Modern Agricultural Technology System of Shandong Province (SDAIT-06-14). We thank Dr. Michael G Palmgren at the University of Copenhagen for providing S. cerevisiae strain RS72, and we would also like to thank Dr. Christine Verhille at the University of British Columbia for her assistance with English language and grammatical editing of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00605/full#supplementary-material
Supplementary Figure 1. The relative expression levels of VvPMA2, VvPMA5, and VvPMA7 in leaf, stem and M-pericarp (mature pericarp) of grape. Vvactin was used as an internal standard. The data were analyzed according to 2−ΔCT method. The values are means ± SD (n = 5).
Supplementary Figure 2. ClustalW alignment of the 5′-terminal region of VvPMA1α and VvPMA1β. Gray line denoted start codon. Identical residues are shaded. Asterisks indicated highly conserved residues.
Supplementary Figure 3. ClustalW alignment of VvPMA1α and VvPMA1β. Identical residues are shaded. Amino acid residues not present within other sequences are denoted in dashes. Asterisks indicated highly conserved residues.
Supplementary Table 1. Primer pairs for real-time PCR.
Supplementary Table 2. Primer pairs for VvPMA1.
References
Arango, M., Gevaudant, F., Oufattole, M., and Boutry, M. (2003). The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216, 355–365. doi: 10.1007/s00425-002-0856-8
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of mocrogram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chen, Z., Pottosin, I. I., Cuin, T. A., Fuglsang, A. T., Tester, M., Jha, D., et al. (2007). Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol. 145, 1714–1725. doi: 10.1104/pp.107.110262
Duby, G., and Boutry, M. (2009). The plant plasma membrane proton pump ATPase: a highly regulated P-type ATPase with multiple physiological roles. Pflugers Arch. 457, 645–655. doi: 10.1007/s00424-008-0457-x
Duby, G., Poreba, W., Piotrowiak, D., Bobik, K., Derua, R., Waelkens, E., et al. (2009). Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. J. Biol. Chem. 284, 4213–4221. doi: 10.1074/jbc.M807311200
Ekberg, K., Palmgren, M. G., and Buch-Pedersen, M. J. (2010). A novel mechanism of P-type ATPase autoinhibition involving both termini of the protein. J. Biol. Chem. 285, 7918–17929. doi: 10.1074/jbc.M109.096123
Falhof, J., Pedersen, J. T., Fuglsang, A. T., and Palmgren, M. (2016). Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol. Plant 9, 323–337. doi: 10.1016/j.molp.2015.11.002
Feng, J., Li, J., Gao, Z., Lu, Y., Yu, J., Zheng, Q., et al. (2015). SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant 8, 1038–1052. doi: 10.1016/j.molp.2015.01.011
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., et al. (2010). The Pfam protein families' database. Nucleic. Acids. Res. 38, 211–222. doi: 10.1093/nar/gkp985
Gambino, G., Perrone, I., and Gribaudo, I. (2008). A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 19, 520–525. doi: 10.1002/pca.1078
Gévaudant, F., Duby, G., von Stedingk, E., Zhao, R., Morsomme, P., and Boutry, M. (2007). Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol. 144, 1763–1776. doi: 10.1104/pp.107.103762
Gévaudant, F., Petel, G., and Guilliot, A. (2001). Differential expression of four members of the H+-ATPase gene family during dormancy of vegetative buds of peach trees. Planta 212, 619–626. doi: 10.1007/s004250000438
Gietz, R. D., Schiestl, R. H., Willems, A. R., and Woods, R. A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. doi: 10.1002/yea.320110408
Greenway, H., and Munns, R. (1980). Mechanisms of salt tolerance in nonhalophytes. Annu. Rev. Plant Physiol. 31, 149–190. doi: 10.1146/annurev.pp.31.060180.001053
Haruta, M., Gray, W. M., and Sussman, M. R. (2015). Regulation of the plasma membrane proton pump (H+-ATPase) by phosphorylation. Curr. Opin. Plant Biol. 28, 68–75. doi: 10.1016/j.pbi.2015.09.005
Hasegawa, P. M. (2013). Sodium (Na+) homeostasis and salt tolerance of plants. Environ. Exp. Bot. 92, 19–31. doi: 10.1016/j.envexpbot.2013.03.001
Hashimoto-Sugimoto, M., Higaki, T., Yaeno, T., Nagami, A., Irie, M., Fujimi, M., et al. (2013). A Munc13-like protein in Arabidopsis mediates Hþ-ATPase translocation that is essential for stomatal responses. Nat. Commun. 4:2215. doi: 10.1038/ncomms3215
Hsu, J. L., Wang, L. Y., Wang, S. Y., Lin, C. H., Ho, K. C., Shi, F. K., et al. (2009). Functional phosphoproteomic profiling of phosphorylation sites in membrane fractions of salt-stressed Arabidopsis thaliana. Proteome Sci. 7:42. doi: 10.1186/1477-5956-7-42
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Janicka-Russak, M., Kabała, K., Wdowikowska, A., and Kłobus, G. (2013). Modification of plasma membrane proton pumps in cucumber roots as an adaptation mechanism to salt stress. J. Plant Physiol. 170, 915–922. doi: 10.1016/j.jplph.2013.02.002
Janicka-Russak, M., and Kłobus, G. (2007). Modification of plasma membrane and vacuolar H+-ATPases in response to NaCl and ABA. J. Plant Physiol. 164, 295–302. doi: 10.1016/j.jplph.2006.01.014
Kabała, K., and Janicka-Russak, M. (2012). Na+/H+ antiport activity in plasma membrane and tonoplast vesicles isolated from NaCl-treated cucumber roots. Biol. Plant. 56, 377–382. doi: 10.1007/s10535-012-0103-5
Kalampanayil, B. D., and Wimmers, L. E. (2001). Identification and characterization of a salt stress- induced plasma membrane H+-ATPase in tomato. Plant Cell Environ. 24, 999–1005. doi: 10.1046/j.1365-3040.2001.00743.x
Lecchi, S., Nelson, C. J., Allen, K. E., Swaney, D. L., Thompson, K. L., Coon, J. J., et al. (2008). Tandem phosphorylation of Ser-911 and Thr-912 at the carboxy terminus of yeast plasma-membrane H+-ATPase leads to glucose-dependent activation. J. Biol. Chem. 282, 35471–35481. doi: 10.1074/jbc.M706094200
Lin, T., and Morales, M. F. (1977). Application of a one step procedure for measuring inorganic phosphate in the presence of proteins: the actomyosin ATPase system. Anal. Biochem. 77, 10–17. doi: 10.1016/0003-2697(77)90284-6
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-Time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Y., Wang, J., Zhong, Y., Geng, F., Cramer, G. R., and Cheng, Z. M. (2015). Subfunctionalization of cation/proton antiporter 1 genes in grapevine in response to salt stress in different organs. Hortic. Res. 2:15031. doi: 10.1038/hortres.2015.31
Mandala, S., and Taiz, L. (1985). Partial purification of a tonoplast ATPase from corn coleoptiles. Plant Physiol. 78, 327–333. doi: 10.1104/pp.78.2.327
Marquez, Y., Brown, J. W., Simpson, C., Barta, A., and Kalyna, M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22, 1184–1195. doi: 10.1101/gr.134106.111
Moriau, L., Bogaerts, P., Jonniaux, J. L., and Boutry, M. (1993). Identification and characterization of a second plasma membrane H+-ATPase gene subfamily in Nicotiana plumbaginifolia. Plant Mol. Biol. 21, 955–963. doi: 10.1007/BF00023594
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Niu, X., Damsz, B., Kononowicz, A. K., Bressan, R. A., and Hasegawa, P. M. (1996). NaCl-induced alterations in both cell structure and tissue-specific plasma membrane H+-ATPase gene expression. Plant Physiol. 111, 679–686. doi: 10.1104/pp.111.3.679
Oufattole, M., Arango, M., and Boutry, M. (2000). Identification and expression analysis of three new Nicotiana plumbaginifolia genes encoding isoforms of a plasma membrane H+-ATPase, one of which is induced by mechanical stress. Planta 210, 715–722. doi: 10.1007/s004250050672
Palmgren, M. G. (2001). Plant plasma membrane H+-ATPase: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–861. doi: 10.1146/annurev.arplant.52.1.817
Palmgren, M. G., and Nissen, P. (2011). P-type ATPases. Annu. Rev. Biophys. 40, 243–266. doi: 10.1146/annurev.biophys.093008.131331
Perez, C., Michelet, B., Ferrant, V., Bogaerts, P., and Boutry, M. (1992). Differential expression within a three-gene subfamily encoding a plasma membrane H+-ATPase in Nicotiana plumbaginifolia. J. Biol. Chem. 267, 1204–1211.
Piette, A. S., Derua, R., Waelkens, E., Boutry, M., and Duby, G. (2011). A phosphorylationin the C-terminal auto-inhibitory domain of the plant plasma membrane H+-ATPase activates the enzyme with no requirement for regulatory 14-3-3 proteins. J. Biol. Chem. 286, 18474–18482. doi: 10.1074/jbc.M110.211953
Reid, K. E., Olsson, N., Schlosser, J., Peng, F., and Lund, S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6:27. doi: 10.1186/1471-2229-6-27
Rudashevskaya, L. E., Ye, J., Jensen, O. N., Fuglsang, A. T., and Palmgren, M. G. (2012). Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. J. Biol. Chem. 287, 4904–4913. doi: 10.1074/jbc.M111.307264
Sahu, B. B., and Shaw, B. P. (2009). Salt-inducible isoform of plasma membrane H+ATPase gene in rice remains constitutively expressed in natural halophyte, Suaeda maritime. J. Plant Physiol. 166, 1077–1089. doi: 10.1016/j.jplph.2008.12.001
Seo, P. J., Kim, M. J., Ryu, J. Y., Jeong, E. Y., and Park, C. M. (2011). Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun. 2:303. doi: 10.1038/ncomms1303
Sibole, J. V., Cabot, C., Michalke, W., Poschenrieder, C., and Barcelo, J. (2005). Relationship between expression of the PM H+-ATPase, growth and ion partitioning in the leaves of salt-treated Medicago species. Planta 221, 557–566. doi: 10.1007/s00425-004-1456-6
Syed, N. H., Kalyna, M., Marquez, Y., Barta, A., and Brown, J. W. S. (2012). Alternative splicing in plants coming of age. Trends Plant Sci. 17, 616–623. doi: 10.1016/j.tplants.2012.06.001
Wakeel, A., Hanstein, S., Pitann, B., and Schubert, S. (2010). Hydrolytic and pumping activity of H+-ATPase from leaves of sugar beet (Beata vulgaris L.) as affected by salt stress. J. Plant Physiol. 167, 725–731. doi: 10.1016/j.jplph.2009.12.018
Walker, R. R., Blackmore, D. H., Clingeleffer, P. R., Boutry, M., and Duby, G. (2002). Rootstock effects on salt tolerance of irrigated field-grown grapevines(Vitis vinifera L. cv. Sultana). 1. Yield and vigour inter-relationships. Aust. J. Grape Wine Res. 8, 3–14. doi: 10.1111/j.1755-0238.2002.tb00206.x
Wang, B. S., Lüttge, U., and Ratajczak, R. (2001). Effect of salt treatment and osmotic stress on V-ATPase and V-PPase in leaves of the halophyte Suaeda salsa. J. Exp. Bot. 52, 2355–2365. doi: 10.1093/jexbot/52.365.2355
Keywords: grape root, alternative splicing, N-terminal regulation of PM H+-ATPase, expression profile, salt stress
Citation: Han N, Ji X-L, Du Y-P, He X, Zhao X-J and Zhai H (2017) Identification of a Novel Alternative Splicing Variant of VvPMA1 in Grape Root under Salinity. Front. Plant Sci. 8:605. doi: 10.3389/fpls.2017.00605
Received: 19 January 2017; Accepted: 03 April 2017;
Published: 21 April 2017.
Edited by:
Sergey Shabala, University of Tasmania, AustraliaReviewed by:
Stuart John Roy, University of Adelaide, AustraliaMagdalena Maria Julkowska, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2017 Han, Ji, Du, He, Zhao and Zhai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning Han, hn8265@163.com
Heng Zhai, zhaih@sdau.edu.cn
 Ning Han
Ning Han Xing-Long Ji1
Xing-Long Ji1 Heng Zhai
Heng Zhai