- 1College of Horticulture, Northeast Agricultural University, Harbin, China
- 2Beijing Genomics Institute, Shenzhen, China
Gray leaf spot disease caused by Stemphylium lycopersici is a major disease in cultivated tomato plants and threatens tomato-growing areas worldwide. Sm is a single dominant gene that confers resistance to tomato gray leaf spot disease agent. However, the underlying molecular mechanism remains unclear. Here, resistant (cv. Motelle, containing the Sm gene) and susceptible (cv. Moneymaker) plants were inoculated with virulent Stemphylium lycopersici isolate at a time point at which both cultivars showed a strong response to S. lycopersici infection. Transcriptome analyses were performed in both cultivars using RNA-seq. The number of differentially expressed genes (DEGs) was higher in Motelle than Moneymaker. Functional classification revealed that most DEGs were involved in plant–pathogen interactions, plant hormone signal transduction, regulation of autophagy, glycerophospholipid metabolism, and α-linolenic acid metabolism. Moreover, the genes that were significantly up-regulated in Sm tomatoes were involved in plant–pathogen interaction pathways. A total of 26 genes were selected for confirmation of differentially expressed levels by quantitative real-time PCR. This knowledge will yield new insights into the molecular mechanism of Sm responses to S. lycopersici infection.
Introduction
Gray leaf spot disease is considered one of the most devastating diseases in plants such as pepper (Cho et al., 2001), cotton (Francovig et al., 1999), and spinach (Koike et al., 2001). Tomato gray leaf spot disease is caused by three species of Stemphylium: Stemphylium solani, Stemphylium floridanum, and Stemphylium lycopersici (Miranda et al., 2010). Gray leaf spot disease is considered a major disease in cultivated tomatoes and has threatened tomato-growing areas worldwide (Simmons, 2001). In the early stages, tomato gray leaf spot disease symptoms appear as brownish-black specks, which later expand to necrotic lesions with gray centers and dark brown borders. As the disease progresses, affected leaves became chlorotic, with perforated centers of lesions, ultimately leading to leaf drying and falling. S. lycopersici has been established as a cause tomato gray leaf spot disease based on morphology and molecular identification (Graham and Zeiders, 1960).
However, resistance is often governed by a ‘gene-for-gene’ interaction (Dangl and Jones, 2001), in which plants carrying a resistance (R) gene specifically recognize a pathogen carrying a corresponding avirulence (avr) gene. The avr gene is recognized by an effector protein after this protein is secreted into the apoplastic space during infection (Nekrasov et al., 2006), which induces either a compatible or incompatible interaction between the fungus and infected plant. An incompatible interaction (chlorosis) results in rapid cell death at the site of infection, which is called the hypersensitive response (HR), whereas a compatible interaction occurs when the pathogen can grow and ramify, causing necrosis in the infected cells (Hammond-Kosack and Jones, 1996; Soylu et al., 2003; Soylu et al., 2004; Pei et al., 2012). To date, only one tomato gray leaf spot disease resistance gene, the dominant gene Sm, has been identified and maps to chromosome 11 near two markers, TG110 and T10, in the tomato genome (Behare et al., 1991). Sm was derived from the wild tomato species S. pimpinellifolium, which was used to breed resistant tomato cultivars (Dennett, 1950). To the best of our knowledge, little is known about the mechanisms of tomato gray leaf spot disease resistance. Therefore, a comprehensive transcriptome analysis will provide a valuable resource for understanding tomato gray leaf spot disease resistance mechanisms.
With the development of second-generation sequencing technology, RNA-seq has become a useful tool for the comprehensive analysis of host–pathogen interactions in plants (Varshney et al., 2009; Haas and Zody, 2010; Du et al., 2014) such as wheat (Yang et al., 2015), rice (Bai et al., 2015), maize (Li et al., 2010), cabbage (Wang et al., 2016), cucumber (Zhang et al., 2014), and eggplant (Yang et al., 2017). Notably, the present study is the first to use Illumina RNA-seq to analyze the regulatory resistance mechanisms of the Sm tomato cultivar in response to S. lycopersici. This study may provide a basis for cloning Sm resistance genes, which will be useful for understanding the regulatory mechanisms involved in plant–pathogen interactions.
Materials and Methods
Plant Materials and S. lycopersici Inoculation
Two tomato cultivars, the resistant cv. Motelle containing the Sm gene (kindly provided by the Asian Vegetable Research and Development Center, AVRDC) and the susceptible cv. Moneymaker (kindly provided by the Chinese Academy of Agricultural Sciences), were used in this study. S. lycopersici was plated on potato dextrose agar (PDA) in Petri dishes. The isolated pathogen was incubated at 28°C for 5–10 days with a 12-h photoperiod. Tomato seedlings were sprayed with a conidial suspension (1 × 104 conidia/ml). Mock-treated plants were sprayed with sterilized water. All plants were maintained in a greenhouse at 28°C with relative humidity >85% (Sun et al., 2016).
Microscopy
To identify the interaction process of Sm-mediated HR and key time points involved in the mechanism, we used lactophenol trypan blue staining and scanning electron microscopy to provide a basis for the RNA-seq and RT-qPCR analyses (Franco et al., 2008; Zhao et al., 2015; Wang et al., 2016). Leaf samples of the resistant and susceptible cultivars were sampled at 0, 1, 2, 3, 4, 5, 6, 7, and 8 days after inoculation and observed under a light microscope. Moreover, the inoculated samples were cut into pieces of approximately 2 mm × 5 mm, soaked for 1.5 h in 2.5% glutaraldehyde (pH 6.8) at 4°C, and rinsed three times in 0.1 M phosphate buffer (pH 6.8) followed by a 50, 70, 90, and 100% ethanol dehydration series. The leaves were then soaked in 100% ethanol:tert-butyl alcohol at a 1:1 ratio followed by 100% tert-butyl alcohol. The leaves were placed in a refrigerator at -20°C for 30 min and then placed in a freeze-dryer (Hitachi ES-2030, Japan). Finally, scanning electron microscope (Hitachi S-3400, Japan) was used to observe the progress of Sm-mediated HR.
RNA Extraction and Illumina Sequencing
Total leaf RNA was collected at the 0 time point (mock-treatment, including resistant and susceptible cultivars), 5 days after incompatible interaction (resistant post-inoculation, RPI) and 5 days after compatible interaction (susceptible post-inoculation, SPI). Total RNA was extracted from three biological replicates for each treatment with three plants according to an RNeasy Plant Mini Kit extraction protocol and was then used in the RT-qPCR experiments (Schroeder et al., 2006; Fang et al., 2015). Total RNA was treated with DNase I, and oligo (dT) was used to isolate mRNA. After addition of the fragmentation buffer, mRNAs were fragmented. Then, cDNA was synthesized using the mRNA fragments as templates. The suitable fragments were selected for PCR amplification. RNA-seq library preparation and sequencing were performed by BGI Tech (Shenzhen, China). The libraries were generated using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States). Then, the library was sequenced using an Illumina HiSeq 4000, and 150-bp paired-end reads were generated.
Illumina Reads and Differentially Expressed Genes (DEGs)
SOAPnuke software1 was used to filter reads. Primary sequencing data (called raw reads) were cleaned by removing reads with adapters. A low-quality read was defined based on the percentage of bases in a read with a quality less than 15 or greater than 20%. Low-quality reads (sequencing quality less than 5) were also removed.
Clean reads were identified by filtering low-quality data and mapped to the S. lycopersicum reference genome sequence (Tomato Genome Consortium, 2012). Gene expression levels in terms of transcripts were quantified by RSEM (RNA-seq by expectation maximization) and FPKM (fragment per kilobase per million mapped; Trapnell et al., 2010; Li and Dewey, 2011). HISAT was used to align paired-end clean reads to the reference genome (Kim et al., 2015). Differentially expressed genes (DEGs) were detected using NOIseq methods with a Noisy Distribution Model (Anders and Huber, 2010; Tarazona et al., 2015) and are shown using a Venn diagram. Genes with a divergence probability (PNOI) ≥ 0.8 and log2 fold-change ≥ 2 were defined as significantly enriched (Benjamini and Hochberg, 1995). Novel transcripts were reconstructed using StringTie (Pertea et al., 2015).
Gene Ontology and KEGG Pathway Analysis of DEGs
The GO seq R package was used for Gene Ontology (GO) analysis of DEGs, and GO terms with an adjusted P-value < 0.05 were considered significantly enriched in DEGs (Chen et al., 2005). KOBAS was used for KEGG metabolic pathway analysis, and P-values ≤ 0.05 were defined as significantly enriched (Kanehisa et al., 2010).
Quantitative Real-Time PCR Analysis
A total of 26 DEGs were analyzed by RT-qPCR to verify the expression profiles obtained by RNA-seq. Reverse transcription was performed using the Reverse Transcriptase M-MLV (RNase H-) reverse transcription kit (TaKaRa) according to the manufacturer’s instructions. Data analysis was performed using the 2ΔΔCT method (Livak and Schmittgen, 2001) with EFa1 (R: 5′-CCACCAATCTTGTACACATCC-3′, S: 5′-AGACCACCAAGTACTACTGCAC-3′) as a reference gene for normalization (Supplementary Table S2).
Results
Microscopic Analysis of S. lycopersici Invasion in the Two Tomato Cultivars
After 3–5 days, symptoms of gray leaf spot disease appeared on the tomatoes. Figure 1 shows the tomatoes 5 days after inoculation. Plants carrying Sm resistance gene displayed strong HR post-inoculation as shown in Figure 1B, whereas susceptible plants had perforated centers of the lesions at 5 days after inoculation (Figure 1A).

FIGURE 1. Typical disease symptoms observed as necrotic leaf spot (arrows) on leaves of susceptible cv. Moneymaker (A) and lack of any visible symptoms on resistant cv Motelle (B) 5 days after inoculation.
To identify the interaction process involved in Sm-mediated HR and the key time points for this mechanism, lactophenol trypan blue staining and scanning electron microscopy were performed. Symptoms and different phenotypical responses triggered by S. lycopersici in resistant cv. Motelle (incompatible interaction) and susceptible cv. Moneymaker (compatible interaction) were observed. The microscopic analysis showed that germ tube extension occurred at 1–2 days (Figure 2). Conidiophore germination and hypha growth occurred at 2 or 3 days, and the hypha invaded the stomata at 3 days after inoculation in cv. Moneymaker. No difference was observed between Motelle and Moneymaker at 3 days after inoculation. In the compatible interaction, the hypha continued to invade and expand in cv. Moneymaker (Thomma et al., 2005) as the disease progressed, the affected areas of the leaves expanded to form necrotic lesions, and the centers of lesions became perforated. Furthermore, HR was observed at 4 days after inoculation in Motelle. At 5 days after inoculation, the cell wall of Motelle formed. Hyphal growth was restricted to the necrotic lesions on Motelle at 6 days after inoculation. Increasing necrotic lesions were apparent in the mesophyll cells and leaf veins at 7–8 days after inoculation.
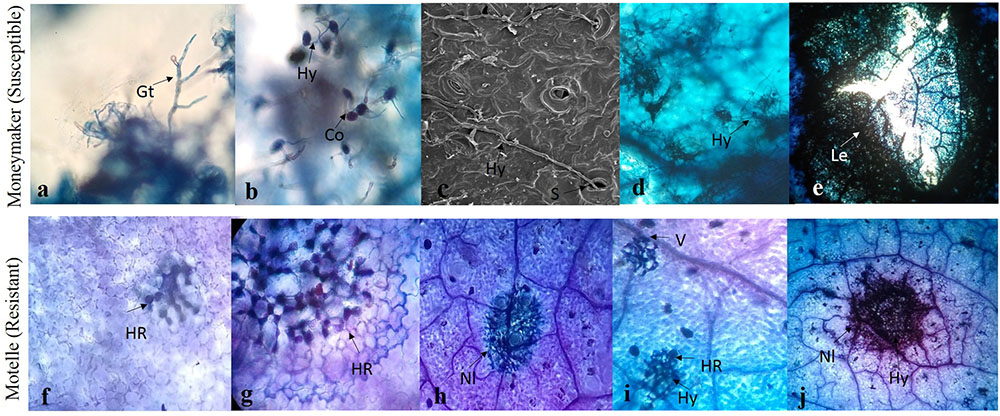
FIGURE 2. Trypan blue stained-tomato leaf samples inoculated with S. lycopersici. The S. lycopersici infection process (a–e) in Moneymaker (Susceptible). Germ tube extension were shown in (a). Conidiophores germinated and hypha growth at 2 or 3 dpi (b), the hypha invaded into stomata at 3 days after inoculation in Moneymaker and Sm tomato (c). The hypha continued to invade and expand in Moneymaker (d). As disease progressed, affected leaves expanded to necrotic lesions and the centers of lesions became perforated (e). The hypersensitive cell death response (f–j) in Motelle (Resistant). The hypersensitive-like symptom was found at 4 days after inoculation in resistant tomato (f). At 5 days after inoculation, the cell wall of resistant tomato were formed (g). Hyphal growth was restricted in the necrotic lesions on resistant tomato at 6 days after inoculation (h). The increasing necrotic lesions were showed in mesophyll cells and leaf veins at 7–8 days after inoculation (i,j). Hy, hyphae; S, stomata; Nl, Necrotic lesions; V, leaf veins; HR, hypersensitive response.
Summary of RNA-seq Data
In this study, an average of ∼8.78 Gb were generated from each sample using the Illumina HiSeq platform. The raw data were deposited in the NCBI Sequence Read Archive under the accession number SRP097450. Ultimately, 37,657 novel transcripts were generated with 16,388 unknown splicing events for known genes, 3,775 novel coding transcripts without any known features, and 17,494 transcripts for long non-coding RNA. Illumina quality scores of 20 (Q20) and 30 (Q30) represent the percentages of sequencing data with error rates less than 1 and 0.1%, respectively (Cock et al., 2010). In this study, more than 99% of reads were ≥Q20, and 97% of these clean reads were ≥Q30. Only data with a quality score ≥Q30 were used for next analyses. After filtering, 53.7–62.1 million clean reads were generated, and at least 86% of these reads were mapped to the tomato reference genome (Supplementary Table S1); of these, more than 85% of the clean reads were uniquely mapped reads, and 0.89% were multiply mapped to tomato chromosomes.
DEGs in Response to S. lycopersici
A gene was considered significantly differentially expressed when PNOI ≥ 0.8 and log2-fold ≥ 2. The two standards were used to identify DEGs in the R and S cultivars in response to S. lycopersici over 5 days after inoculation. All FPKM values for each gene and the fold-changes and PNOI for DEGs are shown in Supplementary Tables S3, S4, respectively. Overall, the number of DEGs was significantly higher in RPI compared with SPI at 5 days after inoculation. Additionally, the number of up-regulated genes was greater than the number of down-regulated genes in the two tomato cultivars. Overall, 1,603 and 977 genes were differentially expressed in the R and S cultivars, respectively, of which 569 and 219 genes were up- and down-regulated, respectively (Figure 3).
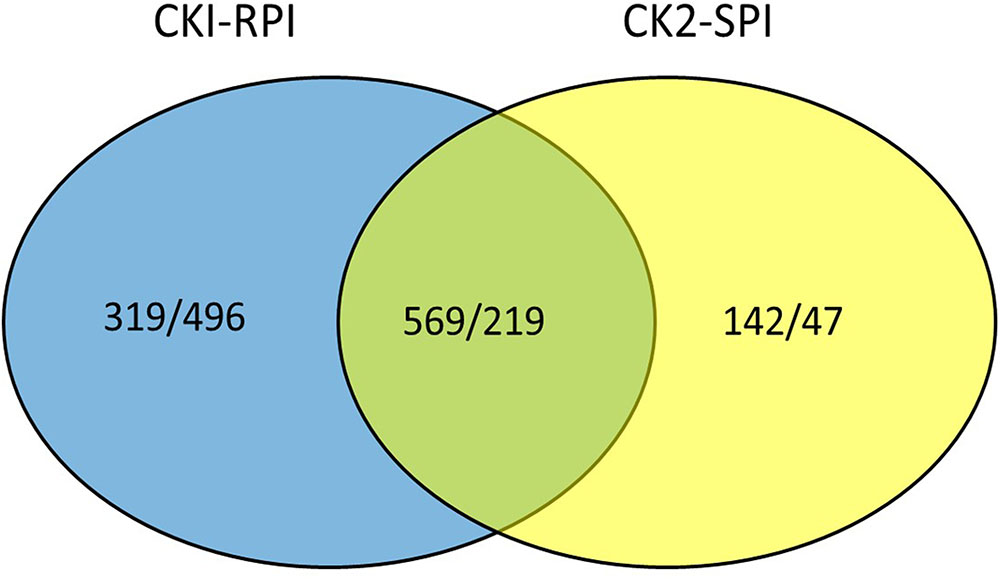
FIGURE 3. Venn diagram showing up/down-regulated in CK1-RPI and CK2-SPI post-inoculation Stemphylium lycopersici. CK1 and CK2: Resistant cultivar and susceptible cultivar were inoculated with water. RPI and SPI: Resistant cultivar and susceptible cultivar were inoculated with Stemphylium lycopersici.
GO and KEGG Enrichment Analysis of DEGs
To identify the functions of DEGs involved in the response to S. lycopersici, we determined GO assignments by using GO seq (Young et al., 2010). Most of the assigned functions of DEGs belonged to the biological process, cellular component and molecular function categories. In the biological process category, significantly enriched terms were metabolic process, cellular process, single organism process, response to stimulus, biological regulation, regulation of biological process and signaling, and these terms were related to disease resistance. In the cellular component category, significantly enriched terms included cell, cell part, organelle part, and membrane, which were found to be specific to the resistant cultivar. In the molecular function category, significantly enriched terms included catalytic activity, binding, nucleic acid binding transcription factor activity, transporter activity, and signal and transducer activity. Additionally, binding and catalytic activity terms were found to play a critical role in plant hormone signal transduction (Figure 4).
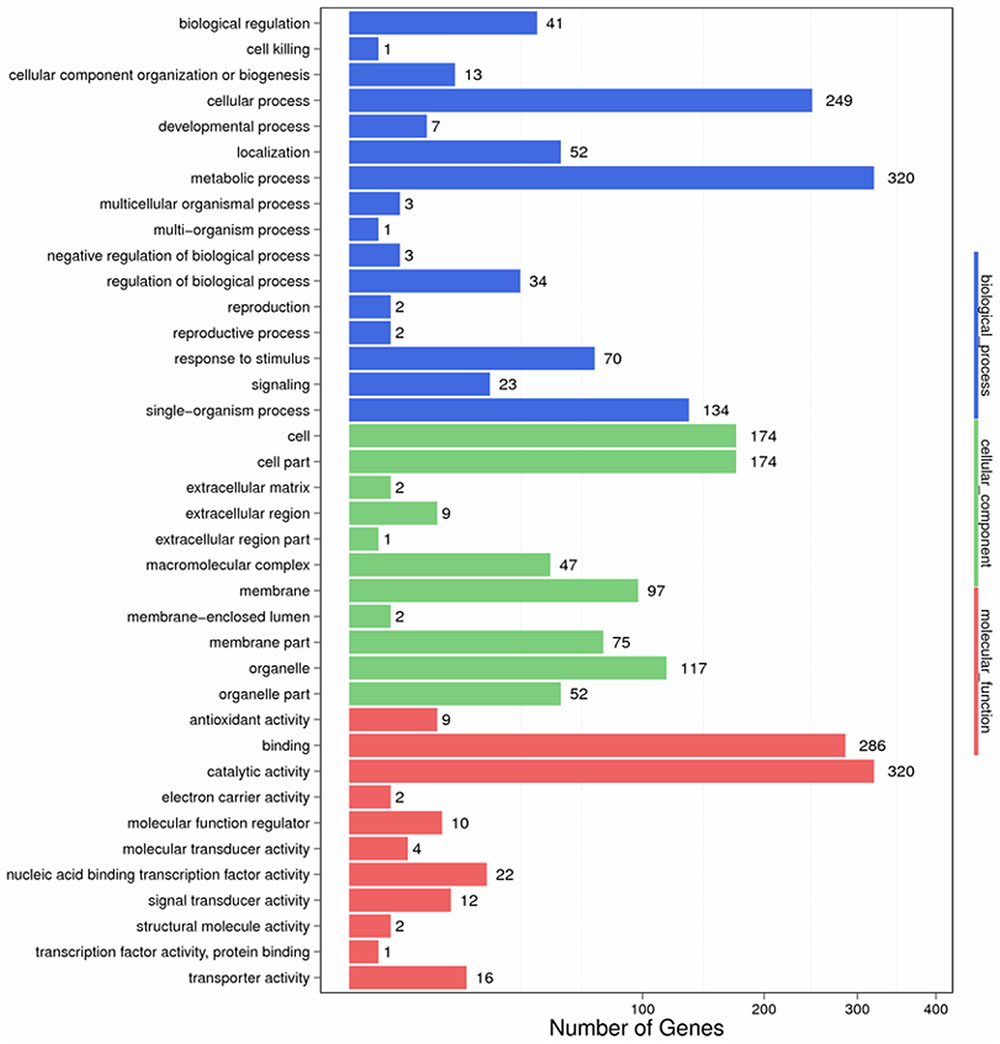
FIGURE 4. Gene ontology categories of differentially expressed genes (DEGs) in Sm tomato in response to S. lycopersici infection.
To investigate the biological pathways associated with DEGs, all DEGs were subjected to KEGG pathway analysis. In the CK1-RPI group, DEGs were significantly enriched in six metabolic pathways with Q- and P-values < 0.05: “Plant–pathogen interaction” (111 DEGs), “Regulation of autophagy” (23 DEGs), “Plant hormone signal transduction” (78 DEGs), “Glycerophospholipid metabolism” (24 DEGs), “alpha-Linolenic acid metabolism” (15 DEGs), and “Glycerolipid metabolism” (21 DEGs) (Table 1). These categories are shown in a scatter plot of the KEGG pathway enrichment of DEGs.
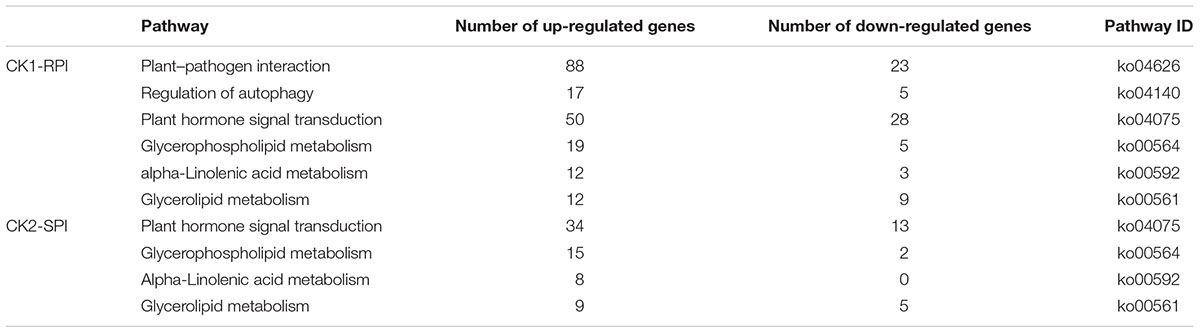
TABLE 1. Significantly enriched KEGG pathway of differentially expressed genes (DEGs) in response to S. lycopersici.
The enrichment factor is the ratio of the DEG number to the background number in a certain pathway (Figure 5). As shown in Figure 5, the number of genes and the enrichment factor in the pathways “Plant–pathogen interaction,” “Regulation of autophagy,” “Plant hormone signal transduction,” and “Biosynthesis of secondary metabolism” were significantly higher than in the other pathways. Many other disease-resistance pathways, including Photosynthesis-antenna proteins, Photosynthesis, Circadian rhythm-plant, Porphyrin and chlorophyll metabolism, Carotenoid biosynthesis, Ether lipid metabolism, and Aminoacyl-tRNA biosynthesis, were also enriched. In total, the most-enriched pathways, ‘Plant–pathogen interaction (111 DEGs)’ and ‘Plant hormone signal transduction (78 DEGs) (Tables 2, 3), may be the major metabolic pathways involved in the Sm tomato response to S. lycopersici infection.
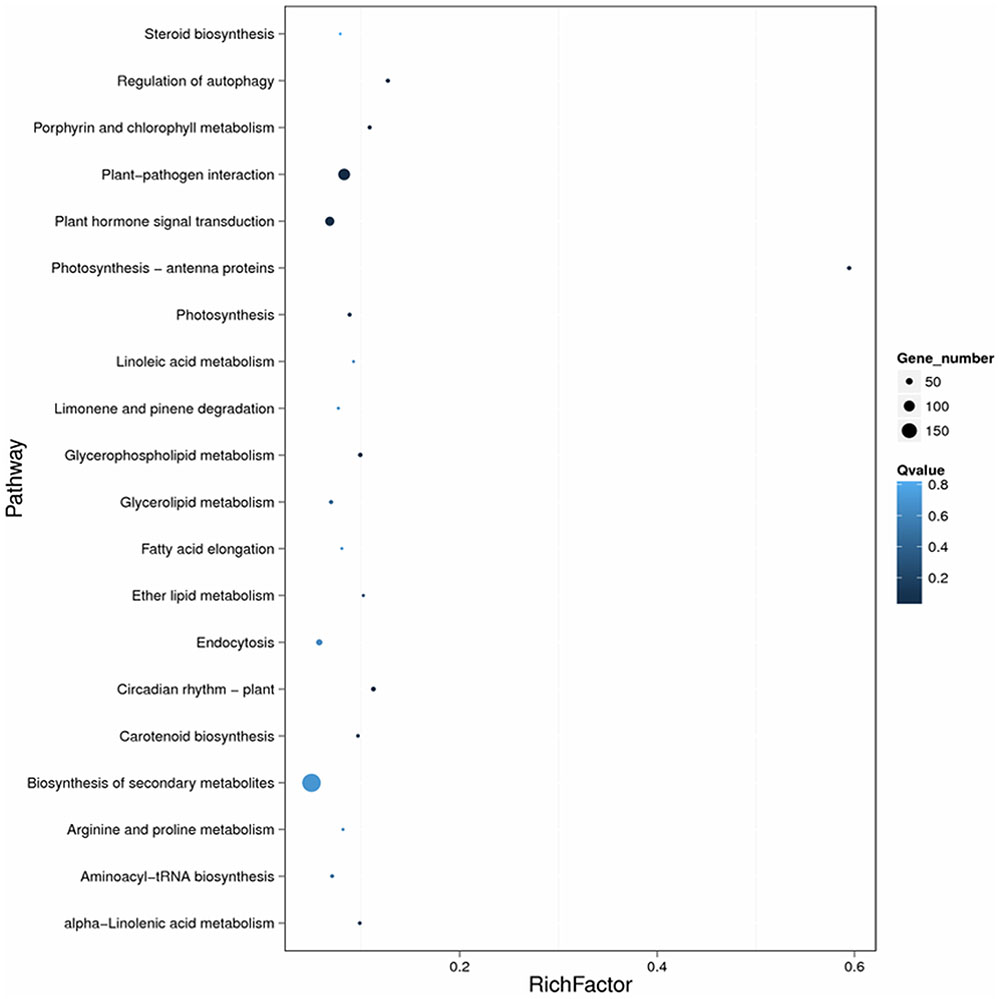
FIGURE 5. Scatter plot of the KEGG pathway enrichment of DEGs. Rich factor is the ratio of the DEG number to the background number in a certain pathway. The size of the dots represents the number of genes, and the color of the dots represents the range of the q-value.
The size of the dots represents the number of genes, and the color of the dots represents the range of q-values. However, in the CK2-SPI group, DEGs were enriched for four major metabolic pathways with P-values < 0.05: “Plant hormone signal transduction” (47 DEGs), “Glycerophospholipid metabolism” (17 DEGs), “alpha-Linolenic acid metabolism” (8 DEGs), and “Glycerolipid metabolism” (14 DEGs). Therefore, DEGs related to disease-resistance pathways were significantly up-regulated in the Sm tomato cultivar at 5 days after inoculation with S. lycopersici (Table 2). Among the DEGs, 19 disease-resistance genes in the significantly enriched KEGG pathway “Plant–pathogen interaction” exhibited significant differences in expression after RPI compared with their expression after SPI in response to S. lycopersici infection in the Sm tomato cultivar and Moneymaker at 5 days post-inoculation. The network analysis of “Plant–pathogen interaction” predicted a response to S. lycopersici infection (Table 2). Finally, 20 DEGs in the significantly enriched KEGG pathway “Plant hormone signal transduction” were identified (Table 3).
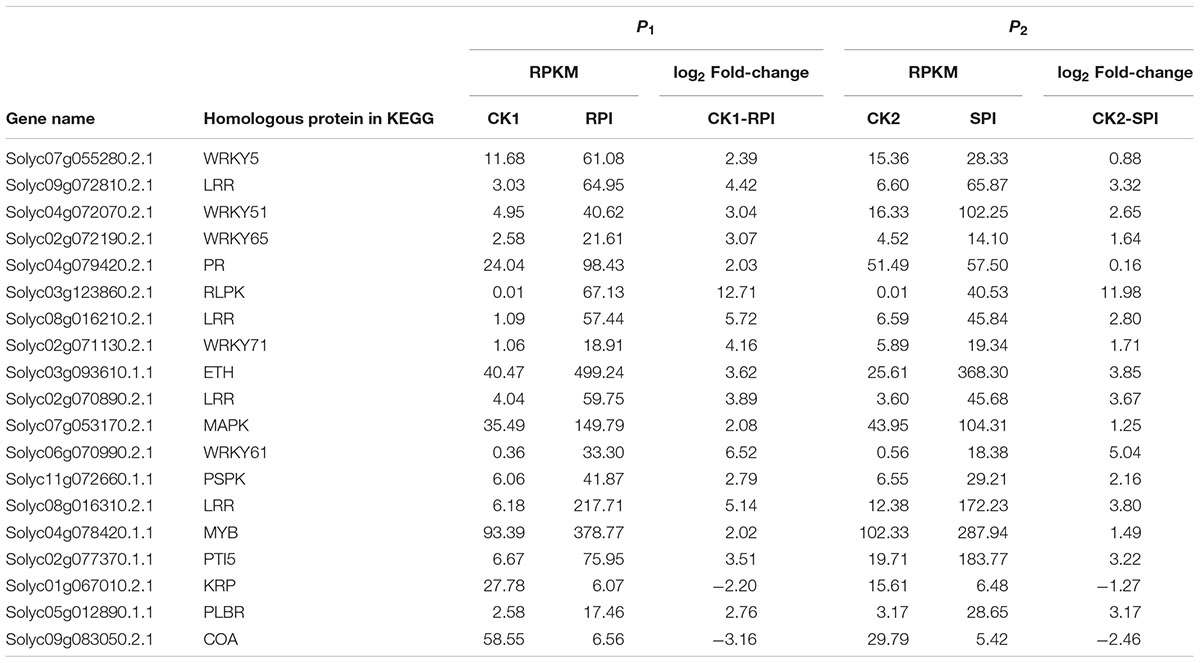
TABLE 2. Differentially expressed genes in the significantly enriched KEGG pathway “Plant–pathogen interaction” in tomato R cultivar and S cultivar at 5 days after inoculation (RPI, SPI).
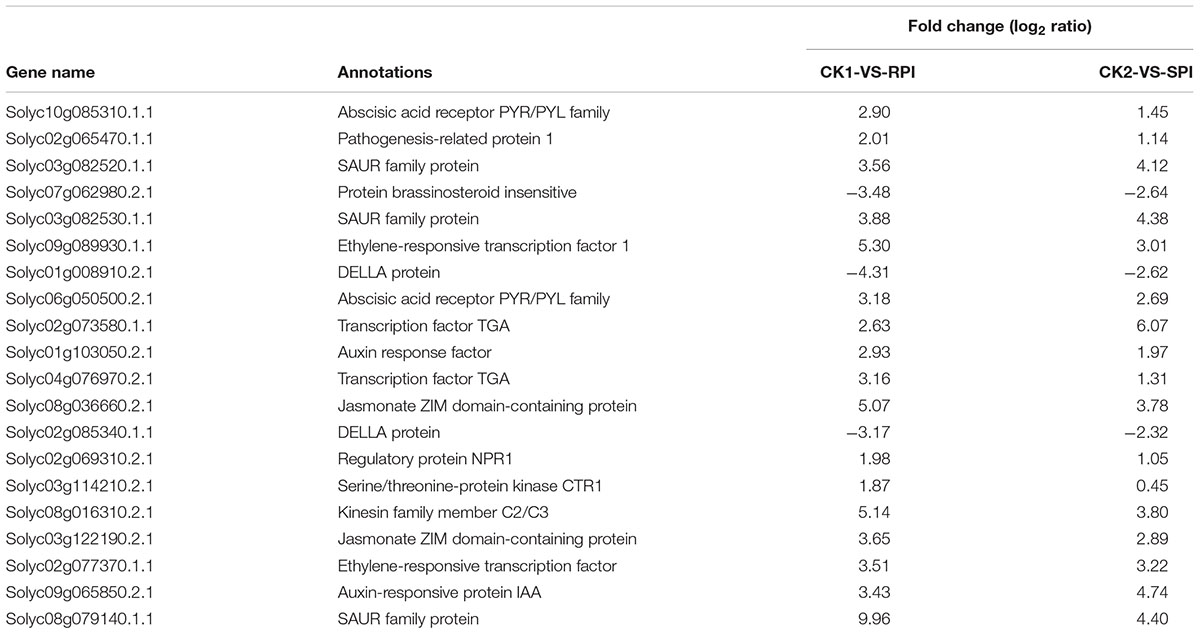
TABLE 3. Differentially expressed genes in the significantly enriched KEGG pathway “Plant hormone signal transduction” in tomato R cultivar and S cultivar at 5 days after inoculation (RPI, SPI).
Validation of RNA-seq Data by RT-qPCR
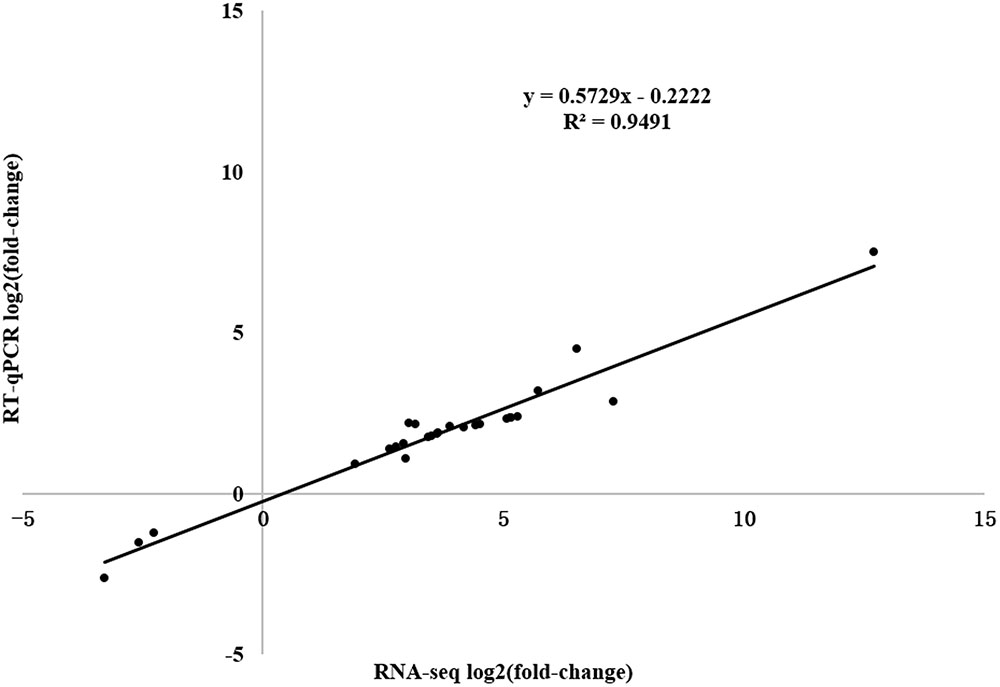
To verify the expression profiles obtained from RNA-seq and predict the defense-response process, we analyzed 26 DEGs [WRKY transcription factor, PR1 protein precursor, disease-resistance protein, receptor-like protein kinase, leucine-rich repeat (LRR) receptor-like serine/threonine-protein kinase, pathogenesis-related gene transcriptional activator, ethylene-responsive transcription factor, jasmonate ZIM domain-containing protein, and abscisic acid receptor PYL9] in the Plant–pathogen interaction, Regulation of autophagy, Plant hormone signal transduction, Glycerophospholipid metabolism, alpha-Linolenic acid metabolism, and Glycerolipid metabolism pathways using RT-qPCR (Supplementary Table S2). A significant positive correlation between the RT-qPCR results and RNA-seq data was detected, indicating that the RNA-seq data were reliable with a strong positive correlation coefficient (R2 = 0.9491).
Discussion
Sm is considered an effective gene for resistance to tomato gray leaf spot disease caused by S. lycopersici. In the present study, RNA-seq was used to verify the transcriptome profiles of Sm tomato in response to S. lycopersici infection. The reliability of the RNA-seq dataset was verified by the significant positive correlation between the RT-qPCR results and RNA-seq data. Ultimately, many significant DEGs were identified between the R and S cultivars in response to S. lycopersici infection. The overall number of DEGs was significantly higher in Sm tomatoes compared with cv. Moneymaker at 5 days after inoculation, and the number of up- and down-regulated genes in the Sm tomato cultivar was higher than that in cv. Moneymaker. Significantly enriched GO terms in the biological process category included metabolic process, cellular process, single response to stimulus, biological regulation, regulation of biological process, and signaling. Moreover, these terms were related to disease resistance. A total of 17 up-regulated genes in the plant–pathogen interaction pathway were analyzed in the R and S cultivars at 5 days after inoculation. Previous studies and functional annotations of genes showed that these up-regulated genes were related to defense responses against fungi (Van Loon et al., 2006).
Previous studies have indicated that the pathogenesis-related (PR) gene transcriptional activator PTI5 belongs to a specific family of defense-related proteins involved in defense against pathogens in plants (Jones and Dangl, 2006). These findings demonstrated a positive role of PTI5 in the regulation of defense genes and disease resistance, suggesting that a pathogen-activated post-transcriptional regulatory process is necessary for the pathogen-mediated induction of defense gene expression. Similar results were obtained in our study: Solyc02 g077370.1.1 (PR gene transcriptional activator PTI5) was shown to be involved in the enriched KEGG pathway “Plant–pathogen interaction,” and the up-regulated expression levels suggested that PTI5-type proteins in tomato may play specific roles in the response to S. lycopersici.
Plant hormones are known to regulate the expression of gene networks related to defense responses (Bari and Jones, 2009), among which jasmonic acid (JA), salicylic acid (SA), and ethylene (ET) play vital roles in resistance to biotrophic and necrotrophic pathogens, such as stress responses, oxide-reduction processes, and cell wall and wax biosynthesis processes (Grant and Lamb, 2006; Van Loon et al., 2006). Additionally, activation of signaling, such as by SA, JA, and ET, will induce defense responses that include most PR proteins. Previous studies have shown that ET responses are vital for B. cinerea resistance in tomato leaves. As Fu et al. (2014) demonstrated, ET and SA play important roles in the defense response of tomato against V. dahlia. In our study, based on KEGG analysis, 20 DEGs were identified in the significantly enriched KEGG pathway “Plant hormone signal transduction.” These results are consistent with previous studies (Shamrai, 2014) that showed that plant hormones and other defense-related proteins are involved in disease resistance. Interestingly, ERF1 (ethylene-responsive transcription factor1), JAZ1 (jasmonate ZIM domain-containing protein), and SAUR family proteins were identified in the KEGG pathway “Plant hormone signal transduction” in the present study, suggesting that JA, ET, and SAUR family proteins may play roles in the resistance of Sm tomato to S. lycopersici. Similarly, previous studies have shown that ERF transcription factors were the connecting factors of signal cross-linking pathway, which were involved in a variety of plant hormone signaling pathways and play an important role in resistance to biotic and abiotic stresses (Gutterson and Reuber, 2004).
Intracellular Ca2+ influx is considered a key and early event downstream of multiple pathogen-associated molecular pattern (PAMP) sensing, resulting in local and systemic acquired resistance (Lecourieux et al., 2005; Boudsocq et al., 2010). Accordingly, calcium-dependent protein kinase (CDPK) is immediately induced by the interaction of flg22 with Avr-Cf9 (Romeis et al., 2000). In addition, recent advances have identified CDPKs as central regulators of Ca2+-mediated immune and stress responses that are crucial signaling nodes mediating plant responses to both abiotic stress and pathogens (Boudsocq and Sheen, 2013). Interestingly, the results of our study showed that six CDPK genes (Solyc02g083850.2.1, Solyc10g050060.1.1, Solyc01g107740.2.1, Solyc10g079130.1.1, Solyc03g033540.2.1, and Solyc03g113390.2.1) were induced at 5 days after inoculation in the Sm tomato cultivar. Based on the results, we therefore propose that these CDPK genes are involved in JA- and SA-mediated defense responses of tomato against S. lycopersici (Figure 6). Similarly, Hu et al. (2016) demonstrated that CDPK genes play critical roles in plant responses to both abiotic stress and pathogens.
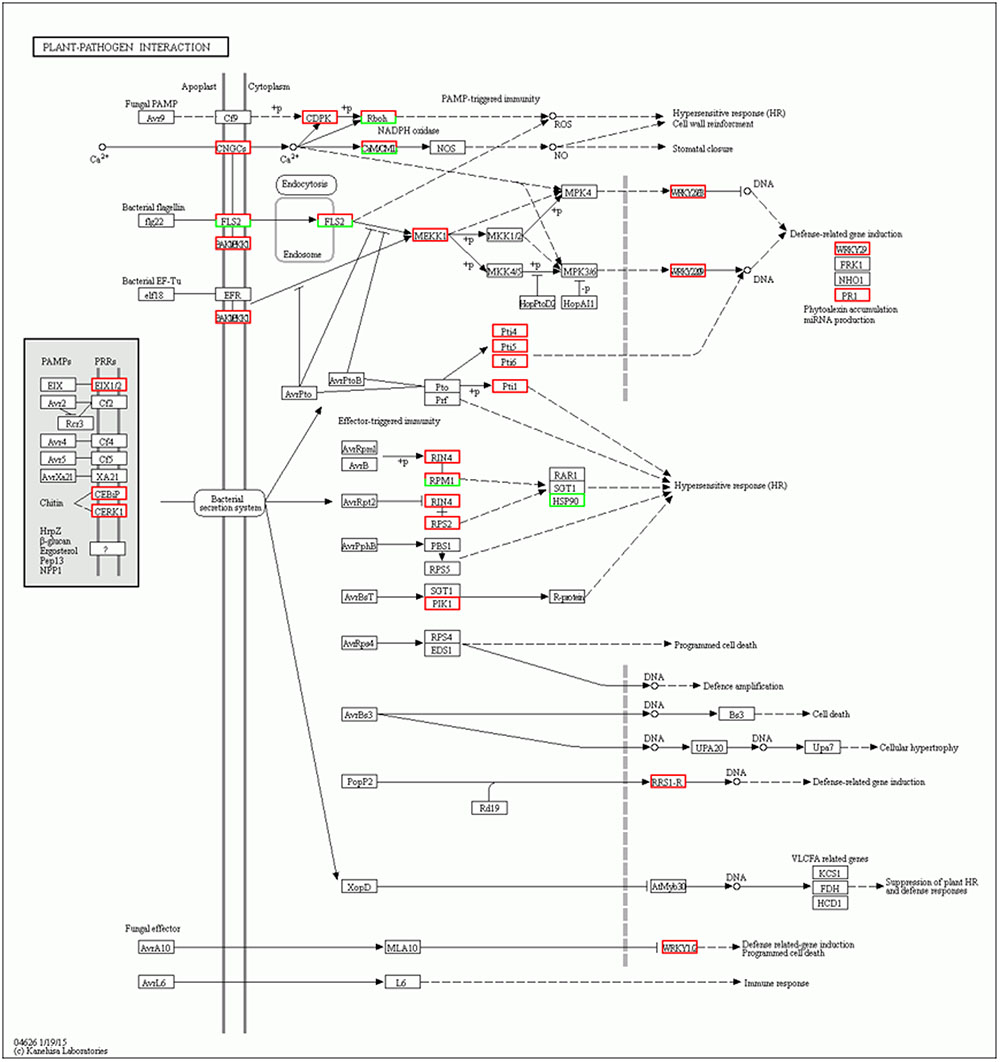
FIGURE 6. Network analysis of “Plant–pathogen interaction” predicts a response to Stemphylium lycopersici infection.
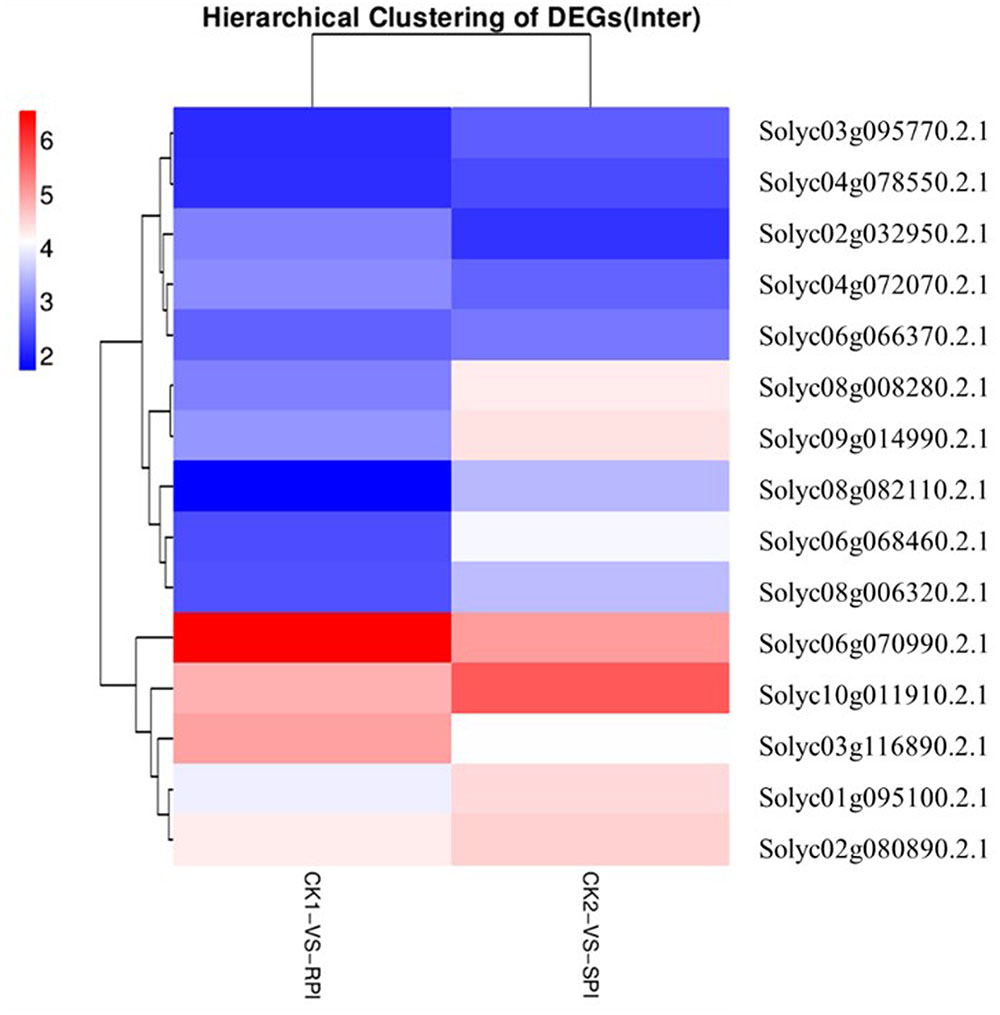
The recognition of PAMPs initiates downstream signaling pathways involving WRKY transcription factors to promote defense responses against bacterial and fungal pathogens and nematodes (Asai et al., 2002; Bhattarai et al., 2010). In this study, based on KEGG analysis, a total of 15 WRKY genes were differentially expressed as shown by the hierarchical clustering of DEGs in both tomato cultivars (Figure 7). Among them, 5 WRKY genes, Solyc07g055280.2.1, Solyc04g072070.2.1, Solyc02g072190.2.1, Solyc02g071130.2.1, and Solyc06g070990.2.1, which have been associated with defense responses against pathogens in previous studies (Spyropoulou et al., 2014; Du et al., 2015), were specifically up-regulated in the Sm tomato cultivar. In our study, these findings were also validated by RT-qPCR analysis, suggesting that WRKY transcription factors may play a role in the resistance of tomato Sm to S. lycopersici, similar to previous studies. These results suggest that the five WRKY genes may activate a series of downstream PR genes and play a key role in defense responses of tomato to S. lycopersici.
Furthermore, a series of PR proteins was activated at this stage. PR proteins are known to be induced upon pathogen invasion and can restrict pathogen growth (Van Loon et al., 2006). PR-1 proteins are a highly conserved family of plant proteins; however, the mechanism of PR-1 protein induction by pathogens has not been elucidated. PR-1 was recently shown to have antifungal activity (Stintzi et al., 1993). In our study, 19 DEGs in the significantly enriched KEGG pathway “Plant–pathogen interaction” were significantly up-regulated in the Sm tomato cultivar after inoculation. Solyc04 g079420.2.1, a resistance-related gene, was up-regulated. The results obtained from our study suggest the enrichment of KEGG pathway “Plant–pathogen interaction” and the higher expression levels indicate that the PR-1 type proteins of tomato may play significant roles in the response to S. lycopersici.
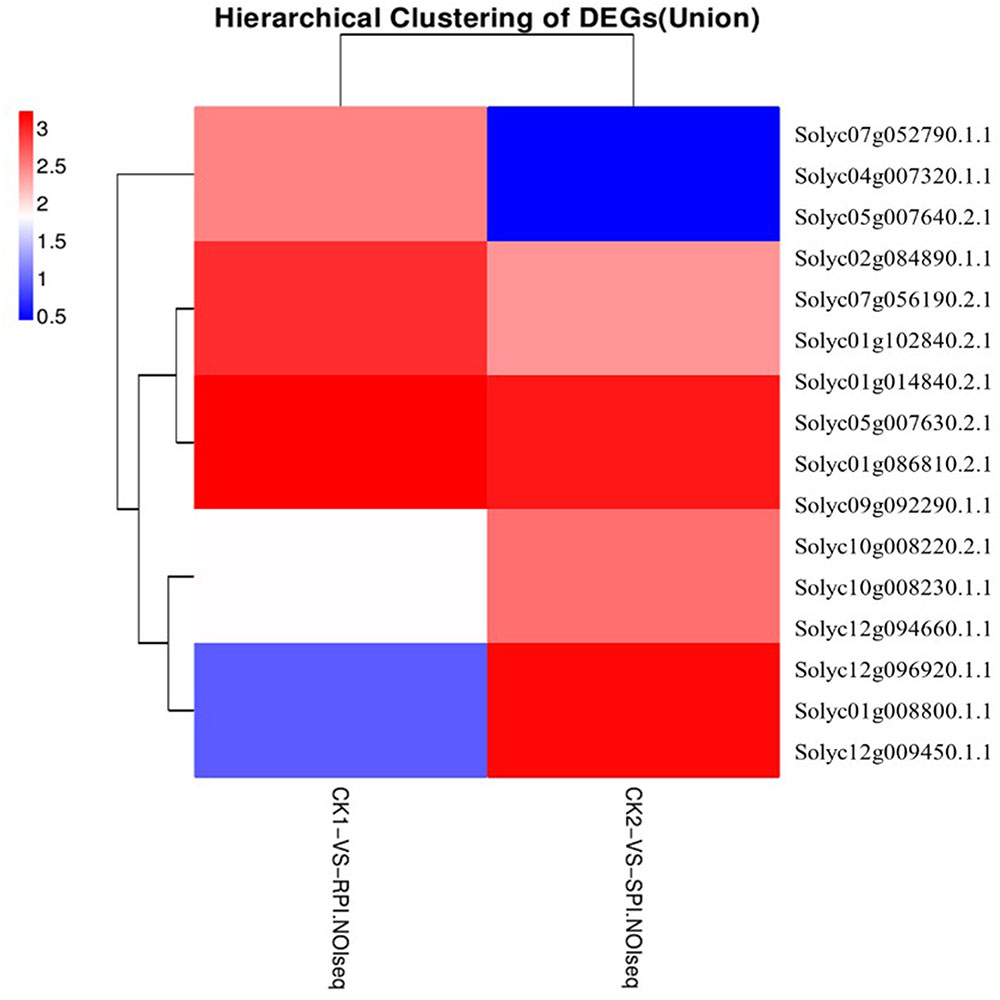
Plant disease resistance is generally dominated by the gene-for-gene hypothesis, which states that the AVR-encoding gene product of a pathogen is specifically recognized, either directly or indirectly, by plant disease R gene products. The R gene encodes a putative NBS-LRR class member that contains a nucleotide-binding site (NBS) and a carboxyl-terminal LRR (Vander Biezen and Jones, 1998). During plant–pathogen interactions, previous studies indicated that specific disease R proteins are generally related to downstream signaling transduction associated with pathogen resistance and are activated by specific effectors induced by R gene expression levels (De Young and Innes, 2006). Accordingly, additional efforts should be focused on putative R genes, including the NB-LRR domain, which may be induced by S. lycopersici infection. In our study, the number of up-regulated putative R genes was greater in the Sm tomato cultivar than in the tomato S cultivar. A total of 15 R genes were differentially expressed based on the hierarchical clustering of DEGs in both tomato cultivars (Figure 8). Moreover, two putative R genes encoding the TMV resistance N-like protein (Solyc07g052790.1.1 and Solyc04g007320.1.1) and RPM1 (Solyc05g007640.2.1 and Solyc05g007630.2.1) were specifically up-regulated (log2 fold change ≥ 2) in the tomato Sm cultivar at 5 days after inoculation. These results were validated by RT-qPCR analysis (Figure 9). Mackey et al. (2002) demonstrated that RPM1 is an NBS-LRR protein from Arabidopsis thaliana that confers resistance to Pseudomonas syringae expressing either avrRpm1 or avrB. RPM1 is also a peripheral membrane protein that likely resides on the cytoplasmic surface of the plasma membrane and is related to the onset of the HR (Mackey et al., 2002; Soylu et al., 2005). Consistent with these previous studies, the expression levels of these putative R genes were higher in the tomato Sm cultivar compared with cv. Moneymaker. These genes may play critical roles in the Sm tomato response to S. lycopersici infection and may be candidate genes induced by S. lycopersici infection.
When a specific part of the plant is infected by a pathogen, systemic acquired resistance is activated, and several downstream defense genes (such as PR1) and plant antitoxin may also be activated, inducing HR to prevent pathogen growth by cell death. HR is a common fungal defense response in tomato. In contrast to field resistance, HR is generally controlled by single dominant genes. Sm (which is found in the resistant cv. Motelle) is a dominant gene for resistance to tomato gray leaf spot disease caused by S. lycopersici (Behare et al., 1991). Morphological changes and microscopic observations were used to identify the interaction process in Motelle. The morphological changes that occurred in the interaction in Motelle, which showed a strong HR upon inoculation, at 5 days post-inoculation are shown in Figure 1B. By contrast, cv. Moneymaker exhibited perforated lesion centers (Figure 1A). These observations are consistent with our microscopic observation studies, which revealed no difference between Motelle and Moneymaker at 3 days after inoculation. The basal defense response of resistant genes was activated at this stage. Additionally, co-regulated genes in both resistant and susceptible tomato were shown to promote expression levels and increase the resistance response to S. lycopersici infection. Furthermore, HR-like symptoms were observed at 4 days after inoculation in Motelle. At 5 days after inoculation, the cell wall of Motelle formed, indicating a strong HR. To our knowledge, this report is the first to demonstrate the interaction process involved in Motelle.
In summary, microscopic and RNA-seq analyses were performed to observe interactions between S. lycopersici and Sm tomatoes. Microscopic analyses revealed hypersensitive-like symptoms at 4 days after inoculation in the resistant cv. Motelle plant at site of interactions. Network analysis was performed to identify S. lycopersici-responsive regulatory pathways. As the mycelium of S. lycopersici invades the stomata and mesophyll cells, some effector proteins secreted by S. lycopersici are rapidly recognized by the Sm tomato. This triggers downstream defense signaling transduction associated with the Ca2+ channel and several other pathways, including those involving JA, SA, and ET (ERF1). Subsequently, specific defense-related transcription factors, such as WRKY proteins (Moore et al., 2011; Van Verk et al., 2011; Puranik et al., 2012), are triggered, which activates the R genes and regulates a series of downstream resistance pathways. Finally, HR is induced, causing the death of cells surrounding the infection sites and limiting pathogen growth. These results suggest the potential mechanism of Sm tomato against S. lycopersici infection.
Database Link and Accessions
The clean data of all samples have been submitted to NCBI2 (Kodama et al., 2012), and each SRA accession corresponding to the treatment name was listed in Supplementary Table S5. The accession numbers is: SRP097450.
Author Contributions
HY, JL, XX, TZ, and JJ conceived and designed the experiments. XC, GL, DZ, CD, SW and HZ, performed the RNA isolation and qRT-PCR experiments. HY performed the data analysis and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by The National Key Research and Development Program of China (No. 2016YFD0101703), The Modern Agricultural Technology System of Special Funds (CARS-25-A-15), and Breeding of New Vegetable Varieties in Heilongjiang Province (GA15B103).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01257/full#supplementary-material
Footnotes
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Bai, B., Wu, J., Sheng, W. T., Zhou, B., Zhou, L. J., Zhuang, W., et al. (2015). Comparative analysis of anther transcriptome profiles of two different rice male sterile lines genotypes under cold stress. Int. J. Mol. Sci. 16, 11398–11416. doi: 10.3390/ijms160511398
Bari, R., and Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Behare, J., Laterrot, H., and Safatti, M. (1991). Restriction fragment length polymorphism mapping of Stemphylium resistance gene in tomato. Mol. Plant Microbe Interact. 4, 489–492. doi: 10.1094/MPMI-4-489
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate-a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300.
Bhattarai, K. K., Atamian, H. S., Kaloshian, I., and Eulgem, T. (2010). WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J. 63, 229–240. doi: 10.1111/j.1365-313X.2010.04232.x
Boudsocq, M., and Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Boudsocq, M., Willmann, M. R., Cormack, M., Lee, H., Shan, L. B., He, P., et al. (2010). Differential innate immune signaling via Ca2+ sensor protein kinases. Nature 464, 418–422. doi: 10.1038/nature08794
Chen, Z. Z., Xue, C. H., Zhu, S., Zhou, F. F., Ling, X. F. B., Liu, G. P., et al. (2005). Go pipe: streamlined gene ontology annotation for batch anonymous sequences with statistics. Prog. Biochem. Biophys. 32, 187–191.
Cho, H. J., Kim, B. S., and Hwang, H. S. (2001). Resistance to gray leaf spot in Capsicum peppers. Hortscience 36, 752–754.
Cock, P. J., Fields, C. J., Goto, N., Heuer, M. L., and Rice, P. M. (2010). The sanger fastq file format for sequences with quality scores, and the solexa/illumina fastq variants. Nucleic Acids Res. 38, 1767–1771. doi: 10.1093/nar/gkp1137
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
De Young, B. J., and Innes, R. W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249. doi: 10.1038/ni1410
Dennett, R. K. (1950). The association of resistance to Fusarium wilt and Stemphylium leaf spot in tomato, Lycopersicon esculentum. Proc. Am. Soc. Hortic. Sci. 56, 353–357.
Du, H., Wang, Y., Yang, J., and Yang, W. (2015). Comparative transcriptome analysis of resistant and susceptible tomato lines in response to infection by Xanthomonas perforans race T3. Front. Plant Sci. 6:1173. doi: 10.3389/fpls.2015.01173
Du, H. S., Li, H., Wang, Y. Q., and Yang, W. C. (2014). Identification of genes differentially expressed between resistant and susceptible tomato lines during time-course interactions with Xanthomonas perforans race T3. PLoS ONE 9:e93476. doi: 10.1371/journal.pone.0093476
Fang, S. M., Hu, B. L., Zhou, Q. Z., Yu, Q. Y., and Zhang, Z. (2015). Comparative analysis of the silk gland transcriptomes between the domestic and wild silkworms. BMC Genomics 16:60. doi: 10.1186/s12864-015-1287-9
Franco, F., Dario, M., Dario, C., and Marcello, I. (2008). Chemical-induced resistance against powdery mildew in barley: the effects of chitosan and benzothiadiazole. Biocontrol 53, 387–401. doi: 10.1007/s10526-007-9091-3
Francovig, P. C., Mehta, Y. R., Fonseca, N. S., and Reis, E. M. (1999). Sources of resistance to Stemphylium solani in cotton cultivars. Summa Phytopathol. 25, 217–222.
Fu, X., Wu, X., Zhou, X., Liu, S., Shen, Y., and Wu, F. (2014). Companion cropping with potato onion enhances the disease resistance of tomato against Verticillium dahliae. Front. Plant Sci. 6:726. doi: 10.3389/fpls.2015.00726
Graham, J. H., and Zeiders, K. E. (1960). Pathogenicity and morphology of some leguminicolous and related species of Stemphylium. Phytopathology 50, 757–760.
Grant, M., and Lamb, C. (2006). Systemic immunity. Curr. Opin. Plant Biol. 9, 414–420. doi: 10.1016/j.pbi.2006.05.013
Gutterson, N., and Reuber, T. L. (2004). Regulation of disease resistance pathways by ap2/erf transcription factors. Curr. Opin. Plant Biol. 7, 465–471. doi: 10.1016/j.pbi.2004.04.007
Haas, B. J., and Zody, M. C. (2010). Advancing RNA-Seq analysis. Nat. Biotechnol. 28, 421–423. doi: 10.1038/nbt0510-421
Hammond-Kosack, K. E., and Jones, J. D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8, 1773–1791. doi: 10.1105/tpc.8.10.1773
Hu, Z., Lv, X., Xia, X., Zhou, J., Shi, K., Yu, J., et al. (2016). Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 7:469. doi: 10.3389/fpls.2016.00469
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kanehisa, M., Goto, S., Furumichi, M., Tanabe, M., and Hirakawa, M. (2010). KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38, 355–360. doi: 10.1093/nar/gkp896
Kim, D., Langmead, B., and Salzberg, S. L. (2015). Hisat: a fast spliced aligner with low memory. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kodama, Y., Shumway, M., and Leinonen, R. (2012). The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 40, D54–D56. doi: 10.1093/nar/gkr854
Koike, S. T., Henderson, D. M., and Butlerre, E. (2001). Leaf spot disease of spinach in California caused by Stemphylium botryosum. Plant Dis. 85, 126–130. doi: 10.1094/PDIS.2001.85.2.126
Lecourieux, D., Lamotte, O., Bourque, S., Wendehenne, D., Mazars, C., Ranjeva, R., et al. (2005). Proteinaceous and oligosaccharidic elicitors induce different calcium signatures in the nucleus of tobacco cells. Cell Calcium 38, 527–538. doi: 10.1016/j.ceca.2005.06.036
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, P., Ponnala, L., Gandotra, N., Wang, L., Si, Y., Tausta, S. L., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42, 1060–1067. doi: 10.1038/ng.703
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T). Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mackey, D., Iii, B. F. H., Wiig, A., and Dangl, J. L. (2002). Rin4 interacts with Pseudomonas syringae type III effector molecules and is required for rpm1-mediated resistance in Arabidopsis. Cell 108, 743–754. doi: 10.1016/S0092-8674(02)00661-X
Miranda, B. E. C., Bolteux, L. S., and Reis, A. (2010). Identification of Solanum (section lycopersicon) accessions with resistance to Stemphylium solani and S. lycopersici. Hortic. Bras. 28, 178–184. doi: 10.1590/S0102-05362010000200007
Moore, J. W., Loake, G. J., and Spoel, S. H. (2011). Transcription dynamics in plant immunity. Plant Cell 23, 2809–2820. doi: 10.1105/tpc.111.087346
Nekrasov, V., Ludwig, A. A., and Jones, J. D. (2006). CITRX thioredoxin is a putative adaptor protein connecting Cf-9 and the ACIK1 protein kinase during the Cf-9/Avr9- induced defence response. FEBS Lett. 580, 4236–4241. doi: 10.1016/j.febslet.2006.06.077
Pei, C. C., Wang, H., Zhang, J. Y., Wang, Y. Y., Francis, D. M., and Yang, W. (2012). Fine mapping and analysis of a candidate gene in tomato accession PI128216 conferring hypersensitive resistance to bacterial spot race T3. Theor. Appl. Genet. 124, 533–542. doi: 10.1007/x00122-011-1726-1
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Romeis, T., Piedras, P., and Jones, J. D. G. (2000). Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12, 803–815. doi: 10.1105/tpc.12.5.803
Schroeder, A., Mueller, O., Stocker, S., Salowsky, R., Leiber, M., Gassmann, M., et al. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7:3. doi: 10.1186/1471-2199-7-3
Shamrai, S. N. (2014). Plant immune system: basal immunity. Cytol. Genet. 48, 258–271. doi: 10.3103/S0095452714040057
Soylu, E. M., Soylu, S., and Mansfield, J. W. (2004). Ultrastructural characterisation of pathogen development and host responses during compatible and incompatible interactions between Arabidopsis thaliana and Peronospora parasitica. Physiol. Mol. Plant Pathol. 65, 67–78. doi: 10.1016/j.pmpp.2004.12.002
Soylu, S., Brown, I., and Mansfield, J. W. (2005). Cellular reactions in Arabidopsis following challenge by strains of Pseudomonas syringae: from basal resistance to compatibility. Physiol. Mol. Plant Pathol. 66, 232–243. doi: 10.1016/j.pmpp.2005.08.005
Soylu, S., Keshavarzi, M., Brown, I., and Mansfield, J. W. (2003). Ultrastructural characterisation of interactions between Arabidopsis thaliana and Albugo candida. Physiol. Mol. Plant Pathol. 63, 201–211. doi: 10.1016/j.pmpp.2003.12.002
Spyropoulou, E. A., Haring, M. A., and Schuurink, R. C. (2014). RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genomics 15:402. doi: 10.1186/1471-2164-15-402
Stintzi, A., Heitz, T., Prasad, V., Wiedemann-Merdinoglu, S., Kauffmann, S., Geoffroy, P., et al. (1993). Plant ’pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie 75, 687–706. doi: 10.1016/0300-9084(93)90100-7
Sun, X. T., Zhang, L., and Zhang, J. Z. (2016). First report of tomato gray leaf spot caused by Stetnphyliutn lycopersici in Zhejiang province, China. Plant Dis. 100:227. doi: 10.1094/PDIS-05-15-0615-PDN
Tarazona, S., Furió-Tarí, P., Turrà, D., Pietro, A. D., Nueda, M. J., Ferrer, A., et al. (2015). Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 43:e140. doi: 10.1093/nar/gkv711
Thomma, B. P., Van Esse, H. P., Crous, P. W., and De Wit, P. J. (2005). Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6, 379–393. doi: 10.1111/j.1364-3703.2005.00292.x
Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Van Loon, L. C., Rep, M., and Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Van Verk, M. C., Bol, J. F., and Linthorst, H. J. (2011). Prospecting for genes involved in transcriptional regulation of plant defenses, a bioinformatics approach. BMC Plant Biol. 11:88. doi: 10.1186/1471-2229-11-88
Vander Biezen, E. A., and Jones, J. D. (1998). The NB-ARC domain: a novel signaling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, R226–R228.
Varshney, R. K., Nayak, S. N., May, G. D., and Jackson, S. A. (2009). Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 27, 522–530. doi: 10.1016/j.tibtech.2009.05.006
Wang, S., Wang, C., Zhang, X. X., Chen, X., Liu, J. J., Jia, X. F., et al. (2016). Transcriptome de novo assembly and analysis of differentially expressed genes related to cytoplasmic male sterility in cabbage. Plant Physiol. Biochem. 105, 224–232. doi: 10.1016/j.plaphy.2016.04.027
Yang, H. H., Xu, X. Y., Zhao, T. T., Jiang, J. B., Liu, G., and Li, J. F. (2017). First report of Stemphylium lycopersici causing gray leaf spot on eggplant in China. Plant Dis. 101:834. doi: 10.1094/PDIS-09-16-1343-PDN
Yang, Z., Peng, Z., Wei, S., Liao, M., Yan, Y., and Jang, Z. (2015). Pistillody mutant reveals key insights into stamen and pistil development in wheat (Triticum aestivum L.). BMC Genomics 16:211. doi: 10.1186/s12864-015-1453-0
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. doi: 10.1186/gb-2010-11-2-r14
Zhang, N., Zhang, H. J., Zhao, B., Sun, Q. Q., Cao, Y. Y., Li, R., et al. (2014). The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J. Pineal Res. 56, 39–50. doi: 10.1111/jpi.12095
Keywords: S. lycopersici, Sm tomato, RNA-seq, regulatory resistance mechanisms, differentially expressed genes
Citation: Yang H, Zhao T, Jiang J, Chen X, Zhang H, Liu G, Zhang D, Du C, Wang S, Xu X and Li J (2017) Transcriptome Analysis of the Sm-Mediated Hypersensitive Response to Stemphylium lycopersici in Tomato. Front. Plant Sci. 8:1257. doi: 10.3389/fpls.2017.01257
Received: 16 March 2017; Accepted: 03 July 2017;
Published: 19 July 2017.
Edited by:
Mahmut Tör, University of Worcester, United KingdomReviewed by:
David John Studholme, University of Exeter, United KingdomSoner Soylu, Mustafa Kemal University, Turkey
Copyright © 2017 Yang, Zhao, Jiang, Chen, Zhang, Liu, Zhang, Du, Wang, Xu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangyang Xu, xxy709@126.com Jingfu Li, huanyaya0126@sina.com
 Huanhuan Yang1
Huanhuan Yang1 Songbo Wang
Songbo Wang Jingfu Li
Jingfu Li