- 1Institute of Biology, Leiden University, Leiden, Netherlands
- 2School of Life Sciences, University of Warwick, Coventry, United Kingdom
- 3Laboratory of Plant Physiology, Wageningen University, Wageningen, Netherlands
Arabidopsis PR1 is a salicylic acid (SA) inducible marker gene for systemic acquired resistance (SAR). However, the regulation of PR1 in plants is poorly understood. In this study, we showed that AtWRKY50 transcription factor binds to two promoter elements of PR1 via its DNA binding domain. Interestingly, the DNA-binding sites for AtWRKY50 deviate significantly from the consensus WRKY binding W-box. The binding sites are located in close proximity to the binding sites for TGA transcription factors. Transactivation experiments in Arabidopsis protoplasts derived from wild type, npr1-1 and tga256 mutant plants indicated that AtWRKY50 alone was able to induce expression of a PR1::β-glucuronidase (GUS) reporter gene, independent of TGAs or NPR1. However, co-expression of TGA2 or TGA5 with AtWRKY50 synergistically enhanced expression to high levels. Yeast-2-hybrid assays and bimolecular fluorescence complementation (BiFC) experiments revealed that AtWRKY50 could interact with TGA2 and TGA5. Using electrophoretic mobility shift assays (EMSA) it was established that AtWRKY50 and TGA2 or TGA5 simultaneously bind to the PR1 promoter. Taken together, these results support a role of AtWRKY50 in SA-induced expression of PR1.
Highlights: AtWRKY50 specifically binds to LS10 region of PR1 promoter and interacts with TGAs to synergistically activate PR1 expression.
Introduction
Upon pathogen attack plants mobilize inducible defense systems. A classic example is the systemic acquired resistance (SAR) which is effective against a broad range of pathogens. The signal transduction route leading to SAR involves the induced synthesis of the endogenous signal molecule salicylic acid (SA from here onward). SAR is accompanied by the de-novo synthesis of pathogenesis-related (PR) proteins of which many directly affect pathogen growth and disease proliferation. Although their exact function is still not fully characterized, the plant kingdom-wide conserved PR1 proteins are generally considered as marker proteins for SAR. Expression of the PR genes is tightly regulated to fine tune plant growth and defense (Linthorst, 1991; van Verk et al., 2008).
PR gene expression was shown to be enhanced by SA in Nicotiana tabacum by the group of Katagiri et al. (1989). This augmentation in expression was dependent on the presence of so called activation sequence-1 (as-1), a DNA element in the 90 bp core promoter consisting of two TGACG tandem repeats (Qin et al., 1994). The as-1 element is specifically bound by the tobacco ASF-1 protein complex, containing basic leucine zipper (bZIP) type TGACGTCA cis-element-binding protein (TGA) (Katagiri et al., 1989; Qin et al., 1994; Niggeweg et al., 2000a). TGA proteins are conserved among different plant species and bind to their cognate binding sites as dimers (Boyle et al., 2009).
Also, promoters of several PR genes, such as Arabidopsis thaliana PR1 and tobacco PR-1a contain as-1-(like) elements in promoter regions important for SA-inducible expression. In tobacco, the as-1-like element in the PR-1a promoter consists of a set of inverted TGACG motifs, which bind to TGA transcription factors (Strompen et al., 1998; Niggeweg et al., 2000b; Grüner et al., 2003). Likewise, a linker scanning analysis of the Arabidopsis PR1 promoter region responsible for induced expression by the SA analog 2,6-dichloroisonicotinic acid (INA) revealed the presence of an as-1 element with two TGACG direct repeats in inverted orientation, of which one is a positive regulatory element (-645 to -636 upstream of the transcription start site; for convenience this region will further be referred as LS7, the name of the linker that was used to mutate this element), while the other (LS5, -665 to -656) mediates negative regulation of PR1 expression (Lebel et al., 1998). Further studies revealed that additional elements other than LS5 and LS7 are also involved in INA-inducible PR1 expression (Pape et al., 2010). In these studies, mutation of LS5 resulted in a relatively small (less than 2-fold) enhancement of the level of inducible expression in comparison with the wild type promoter, while mutation of LS7 modestly reduced expression (approximately 3-fold). Moreover, when both LS5 and LS7 were mutated, PR1 promoter-driven expression was 2-fold higher than that of the wild type promoter, while in all these cases expression remained inducible by the SA-analog INA.
The ankyrin repeat protein NPR1 (NON-EXPRESSER OF PATHOGEN-RELATED GENE 1) plays a central role in PR gene induction and subsequent SAR establishment (Delaney et al., 1995; Cao et al., 1997; Wang et al., 2006). Pathogen-induced accumulation of SA induces a change of the redox state of the cell, resulting in the release of reduced NPR1 monomers from multimeric complexes residing in the cytoplasm, which subsequently translocate to the nucleus where they interact with TGA transcription factors to activate gene expression (Zhang et al., 1999; Després et al., 2000; Kinkema et al., 2000; Zhou et al., 2000; Mou et al., 2003). Through knockout analyses it was shown that Arabidopsis TGA2, TGA5, and TGA6 act as redundant but essential activators of PR1 expression and subsequently SAR activation (Zhang et al., 2003; Kesarwani et al., 2007). Interestingly, these TGAs are constitutive repressors required for basal repression of PR1 under non-stress conditions (Zhang et al., 2003; Rochon et al., 2006). It is believed that the interaction of the BTB/POZ domain of NPR1 neutralizes the repression domain of TGA2, while NPR1’s transactivation domain activates gene expression (Boyle et al., 2009). Recently, it was shown that coactivation by NPR1 occurs in a pulse-wise manner and is regulated by proteosomal degradation of NPR1 (Spoel et al., 2009; Fu et al., 2012).
In addition to TGAs, WRKY transcription factors are important for stress induced transcriptional reprogramming (Eulgem and Somssich, 2007; Pandey and Somssich, 2009). WRKY transcription factors are classified as a family of plant-specific DNA-binding proteins characterized by the presence of the peptide sequence Trp-Arg-Lys-Tyr (WRKY) followed by a Zn-finger domain (Rushton et al., 2010). An ever-increasing number of publications indicate the involvement of WRKY transcription factors in SAR. Unlike the TGA transcription factors that are present at steady state levels (Johnson et al., 2003), many of the WRKY genes are transcriptionally activated upon biotic and abiotic stress. Various WRKY proteins positively regulate resistance against necrotrophic pathogens, like AtWRKY33 (Zheng et al., 2006), others positively regulate defense against biotrophs, like AtWRKY53 and AtWRKY70 (Wang et al., 2006). In addition, WRKY proteins like AtWRKY18, -40 and -60 have dual effects n plant defense, either enhancing defense against biotrophic pathogens and diminishing defense against necrotrophs, or vice versa (Xu et al., 2006; Wang et al., 2006; Shen et al., 2007). Of the 74 WRKY genes in Arabidopsis, 49 are differentially expressed upon Pseudomonas syringae infection or treatment with SA (Dong et al., 2003). Most of the WRKY proteins bind to the W-box, a DNA motif with the core sequence TTGAC(T/C) and the overrepresentation of this motif in several defense related genes suggests that their expression is regulated by WRKY transcription factors (Eulgem and Somssich, 2007). Furthermore, for several WRKY genes, SA-induced expression is dependent on NPR1 and TGAs, suggesting a similar activation strategy as was originally proposed for PR1 (Dong et al., 2003; Wang et al., 2006). Despite the fact that extensive genetic information has been obtained on the physiological processes in which specific WRKYs are involved, surprisingly little is known about which specific genes they regulate.
In the same linker scanning study that identified the as-1-like regulatory element in the Arabidopsis PR1 promoter, a nearby consensus W-box motif (LS4, -675 to -666) with a strong negative effect was identified, suggesting that WRKY factors are important for SA-mediated PR1 gene expression (Lebel et al., 1998). The tobacco PR-1a promoter does not harbor a consensus W-box, however, NtWRKY12, was found to bind to a WK-box (TTTTCCAC) that was located 13 bp proximal to the as-1-like element in the tobacco PR-1a promoter (van Verk et al., 2008). Mutation of the WK-box sharply reduced SA-mediated PR-1a::GUS expression (van Verk et al., 2008). Furthermore, pull-down assays and Fluorescence Resonance Energy Transfer (FRET) analysis showed that NtWRKY12 specifically interacted with tobacco TGA2.2 (van Verk et al., 2011). These results indicate that NtWRKY12 and TGA2.2 interact in the regulation of tobacco PR-1a promoter activity.
In addition to the as-1 element and the W-box, the Arabidopsis PR1 promoter contains another nearby element that influences PR1 expression. Mutation of this LS10 element (-615 to -606) resulted in loss of INA-inducible expression, indicating the sequence as a positive regulatory element (Lebel et al., 1998; Pape et al., 2010). Based on the presence of the sequence TTTC, LS10 has been suggested to be a potential binding site for DOF transcription factors, although experimental data to support this are lacking (Yanagisawa, 2004). Taken together, these results suggest that TGA proteins are not the only transcription factors important for PR1 expression, but regulation of expression is mediated by additional transcription factors binding to the intricate mosaic of elements in the PR1 promoter, and especially underline the importance of transcription factors binding to sites in LS4 and LS10.
In the present study, we identified AtWRKY50 as an activator of PR1 gene expression and determined its binding sites in the promoter. In addition, we investigated its ability to interact with TGA2 and TGA5 and to synergistically enhance transcriptional activation.
Results
AtWRKY50 Is the Most Effective WRKY Activator of PR1 Gene Expression
Previously, we identified NtWRKY12 as a transcriptional activator of tobacco PR-1a gene expression (van Verk et al., 2008). To investigate if WRKY transcription factors are also involved in activation of Arabidopsis PR1 gene expression a protoplast transactivation assay (PTA) was established where 40 of the 74 Arabidopsis WRKYs proteins were screened (Wehner et al., 2011). Briefly, a fragment containing approximately 1000 bp upstream of the transcription start site of the PR1 gene was cloned in front of the coding sequence for firefly luciferase (LUC). Parallel co-transfections of Arabidopsis protoplasts with this reporter plasmid and expression vectors containing the panel of WRKY genes under control of the 35S promoter showed that AtWRKY50 was the most effective activator of the PR1::LUC reporter (Supplementary Table S1). AtWRKY50 is a small, 173 amino acid long protein that belongs to a small subgroup of WRKY proteins in which the domain that interacts with the DNA is characterized by the sequence WRKYGKK as opposed to WRKYGQK present in most other WRKY proteins (Yamasaki et al., 2005). Interestingly, NtWRKY12 also belongs to this GKK subgroup (van Verk et al., 2008). In addition to AtWRKY50, only two other Arabidopsis WRKY proteins, AtWRKY51 and AtWRKY59, possess the WRKYGKK sequence and of these three, AtWRKY50 has the highest homology to tobacco NtWRKY12 (68% sequence similarity). Since constructs corresponding to AtWRKY51 and AtWRKY59 were not present in the initial screen, separate transactivation assays in Arabidopsis protoplasts were done with 35S expression plasmids for these WRKYs co-expressed with a PR1::GUS reporter construct. While AtWRKY50 enhanced GUS expression approximately 5-fold, AtWRKY51 and AtWRKY59 did not increase expression over the background level (Figure 1A). The overexpression of AtWRKY50 also resulted in activation of endogenous PR1 gene expression as revealed by qRT-PCR (Figure 1B). In agreement with the co-expression experiment of Figure 1A, expression of AtWRKY51 and AtWRKY59 did not result in enhanced endogenous PR1 mRNA accumulation. Stable AtWRKY50 transgenic overexpressor plants also showed a significantly higher PR1 expression after 0.3 mM SA treatment than control (GUS) plants (Figure 1C). Taken together, we have shown that AtWRKY50 is the only Arabidopsis WRKY in the WRKYGKK group that specifically activates the PR1 gene expression.
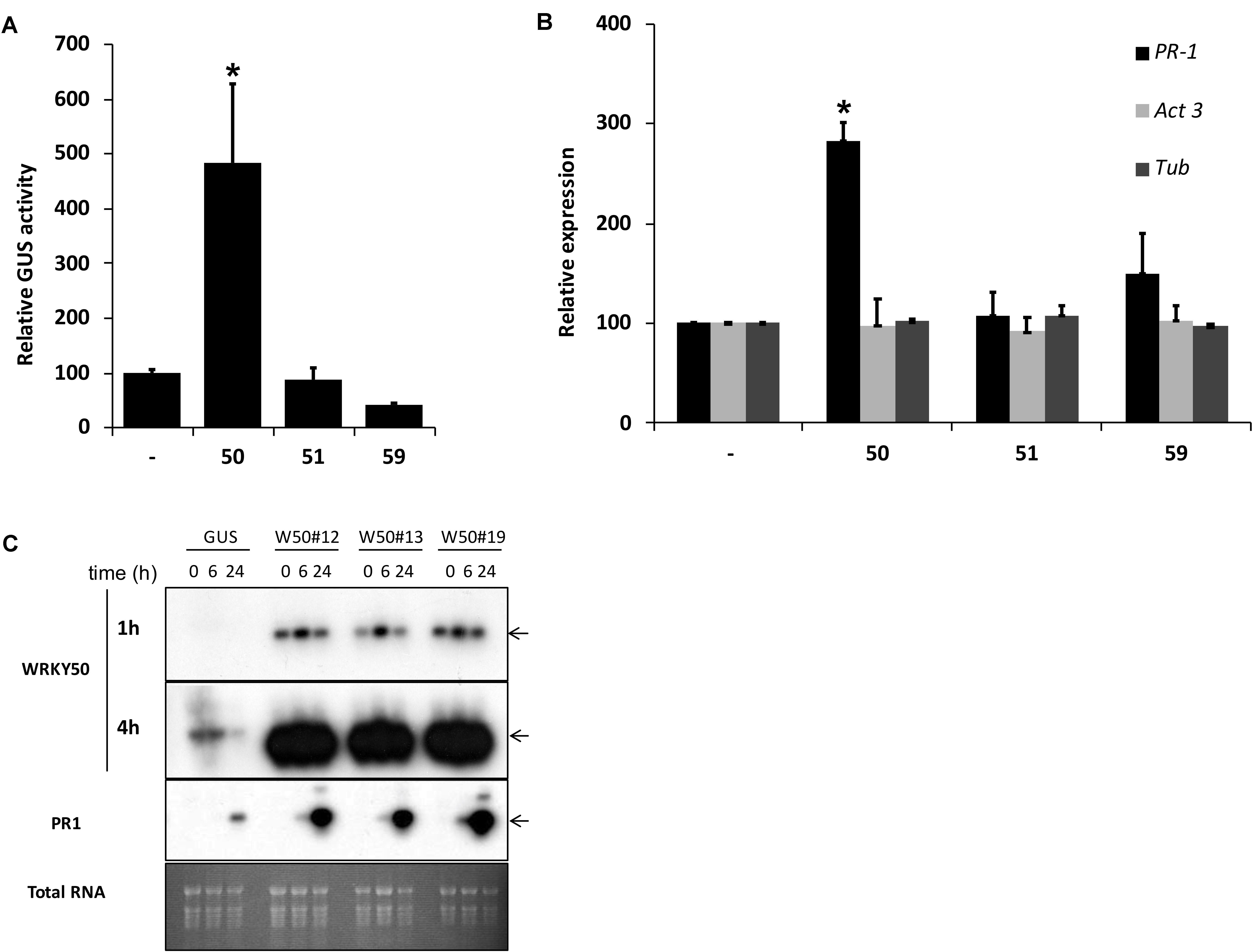
FIGURE 1. (A) AtWRKY50 activates the PR1 promoter. Arabidopsis protoplasts were co-transfected with a PR1::GUS construct together with empty pRT101 expression plasmid (minus sign) or with plasmids containing 35S::AtWRKY50 (50), 35S::AtWRKY51 (51) or 35S::AtWRKY59 (59). After incubation, GUS activity was measured spectrophotometrically. Expression levels (%) are given relative to the expression level without WRKY effector. (B) Effect of AtWRKY50, AtWRKY51, and AtWRKY59 on the expression of endogenous Arabidopsis genes. Expression of PR1, Act3 (actin) and Tub (tubulin) genes in Arabidopsis protoplasts was measured by qRT-PCR. Expression of each gene was measured in protoplasts transfected with the empty pRT101 vector (minus sign) or with the pRT101 vector containing the 35S::AtWRKY50 (50), 35S::AtWRKY51 (51), or 35S::AtWRKY59 (59) expression constructs. Bars represent the average level of mRNA accumulation observed in three experiments. mRNA levels in protoplasts transfected with the empty pRT101 vector were taken as 100%. Error bars represent the SEM. The experiment was repeated three times with similar results. Statistical differences among the samples is labeled with asterisk (p < 0.05). (C) Overexpression of AtWRKY50 enhances SA-induced expression of PR1 in plants. Three lines of transgenic seedlings overexpressing AtWRKY50 (W50#12, #13, #19) and a line expressing GUS were incubated for the indicated times (in hours) in liquid medium containing 0.3 mM SA, after which RNA was extracted and analyzed for accumulation of mRNA corresponding to genes AtWRKY50 and PR1. The blot hybridized with the AtWRKY50 probe was exposed for 1 and 4 h. A similar blot was hybridized with the PR1 probe. Arrows to the right of the blots indicate the expected positions of the respective mRNAs as deduced from the positions of the rRNAs. An Ethidium bromide stained gel showing total RNA is included as a loading control.
AtWRKY50-Induced PR1 Expression Is Independent of NPR1, TGA2, TGA5 or TGA6 Transcription Factors
SA-induced expression of PR1 genes in plants is dependent on NPR1 and it is generally assumed that NPR1 activates expression through its interaction with TGAs binding to the promoter. To investigate if the activation of the PR1 promoter by AtWRKY50 requires NPR1 or TGAs, transactivation assays were performed with protoplasts derived from npr1-1 mutant and tga256 triple mutant Arabidopsis plants. AtWRKY50 was able to activate the PR1::GUS reporter gene in these mutant backgrounds to similar levels as in wild type protoplasts (Figure 2A). Apparently, PR1 activation by AtWRKY50 did not require the TGAs or their co-activator NPR1.
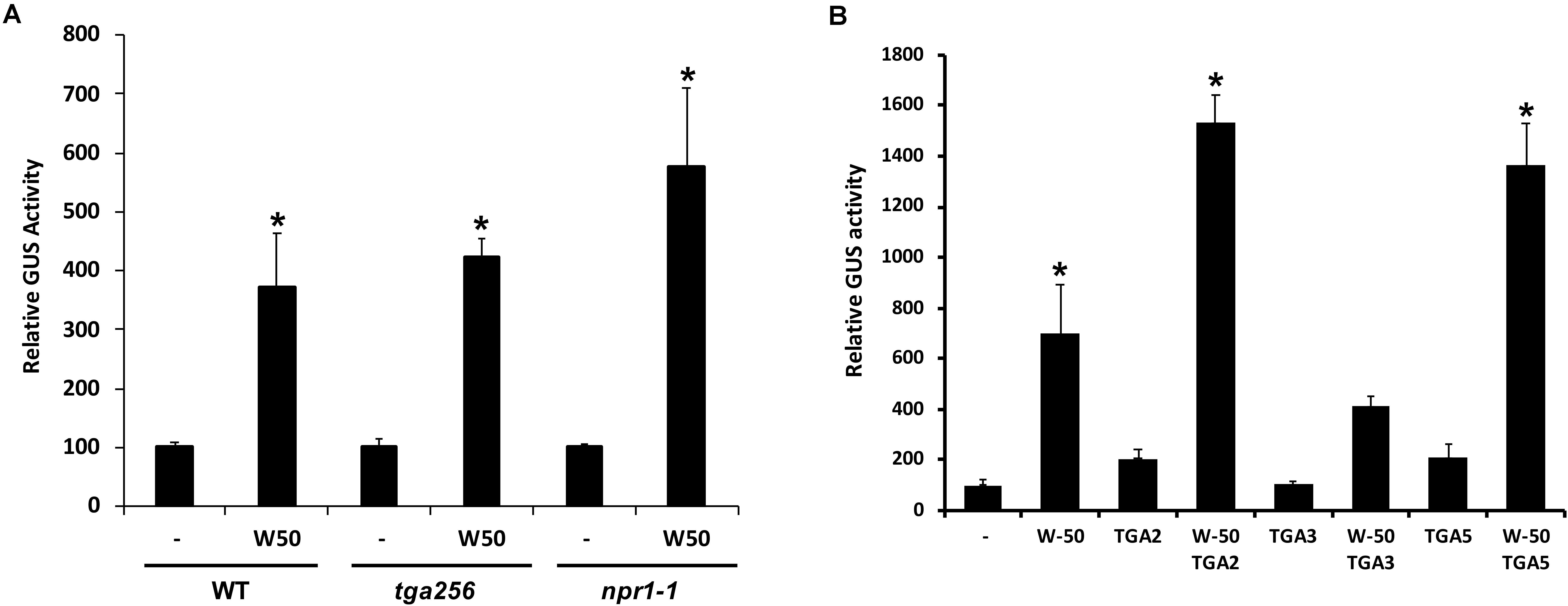
FIGURE 2. (A) AtWRKY50 induced expression is independent of NPR1 and TGAs. Protoplasts from wild type (WT), tga2-1 tga5-1 tga6-1 triple mutant (tga256) and npr1-1 mutant (npr1) plants were co-transfected with PR1::GUS reporter construct alone (minus sign) or together with expression plasmids containing 35S::AtWRKY50 (W50), as indicated. After incubation GUS activity was measured spectrophotometrically. The bars represent the percentage of GUS activity from triplicate experiments relative to that of the protoplasts co-transfected with the PR1::GUS construct and empty vector control. Error bars represent the SEM. Asterisk (∗) shows the statistical differences among samples (p < 0.05). (B) Synergistic effect of AtWRKY50 and TGA2 or TGA5 on PR1 expression. Arabidopsis protoplasts were co-transfected with PR1::GUS reporter construct alone (minus sign) or together with expression plasmids containing 35S promoter-controlled genes encoding AtWRKY50, TGA2, TGA3, TGA5, or combinations, as indicated. After incubation GUS activity was measured spectrophotometrically. Expression levels (%) are given relative to the expression the level without expression plasmid. The bars represent the percentage of GUS activity from triplicate experiments relative to that of the protoplasts co-transfected with the corresponding PR1::GUS construct and empty vector control. Error bars represent the SEM. Asterisk (∗) shows the statistical differences among samples (p < 0.05).
Although AtWRKY50 alone is able to activate the PR1 promoter, TGA factors may function in further modulation of gene expression. To determine the effect of TGA factors on AtWRKY50-activated PR1 expression, Arabidopsis protoplasts were co-transfected with the PR1::GUS reporter construct and plasmids containing 35S promoter-driven AtWRKY50, TGA2, TGA3 and TGA5 genes. While AtWRKY50 enhanced GUS expression approximately 7-fold, the TGA proteins alone could enhance expression only 2-fold (Figure 2B). However, combinations of AtWRKY50 and TGA2 or TGA5 boosted expression of the reporter gene up to 14-fold, while co-expression with TGA3 did not further enhance AtWRKY50-dependent GUS expression. The results indicate that TGA2 and TGA5 act synergistically with AtWRKY50 to maximize activation of the PR1 promoter.
AtWRKY50’s C-terminal Half Binds to the PR1 Promoter
Previous work on the Arabidopsis PR1 promoter has shown that the region between approximately -700 and -600 bp upstream of the transcription start site is important for inducible gene expression upon treatment with the SA analog INA (Lebel et al., 1998). In addition to two inverted TGACG motifs (CGTCA in LS5 and LS7) comprising the as-1-like element, this region contains a consensus WRKY binding W-box (in LS4) and an additional sequence stretch (LS10). Mutational analyses revealed that all these elements are involved in INA-inducible expression. The schematic representation of the Arabidopsis PR1 promoter and a comparison to the tobacco PR-1a promoter is shown in Figure 3A.
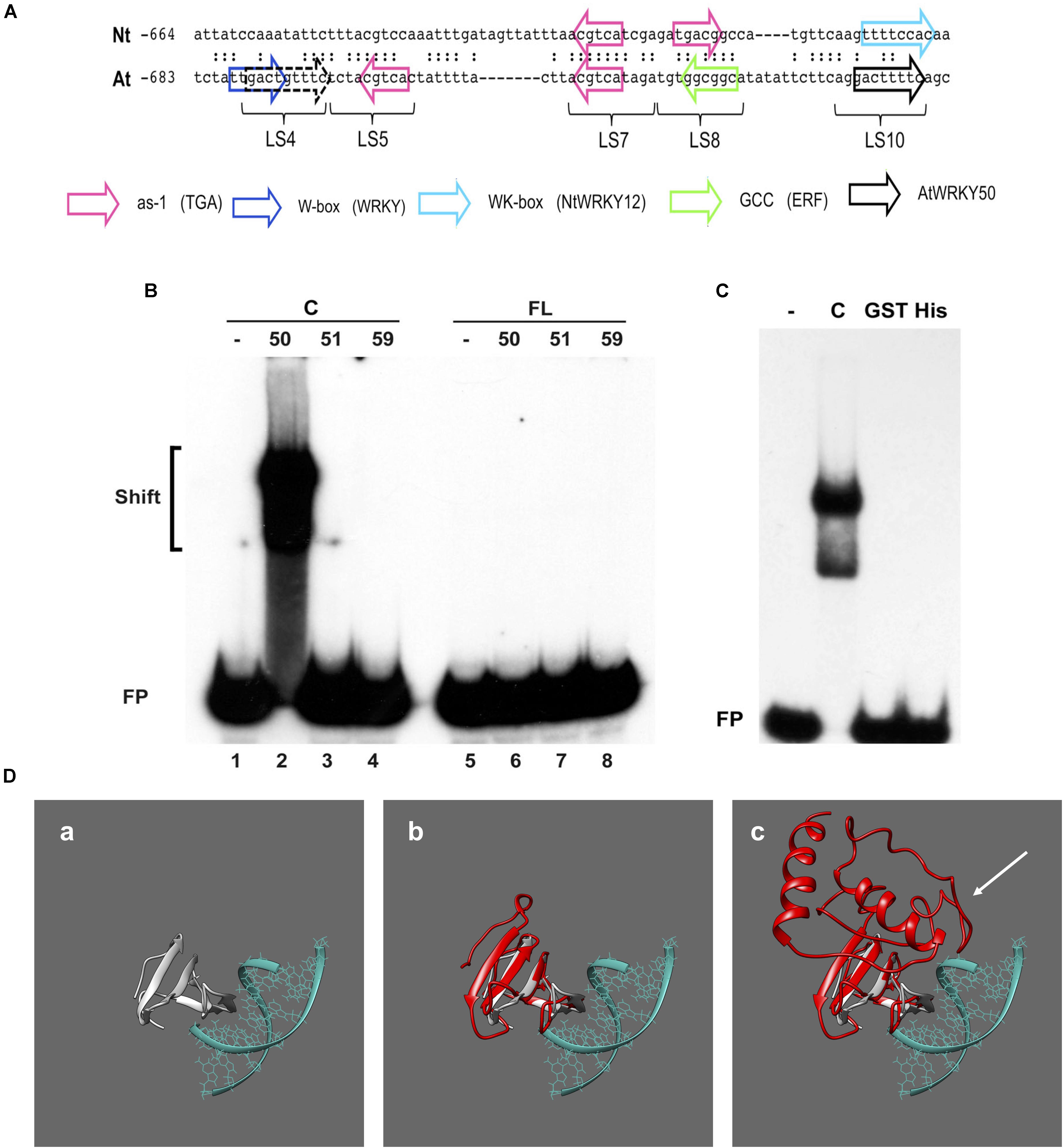
FIGURE 3. AtWRKY50 binds to the PR1 promoter. (A) Comparison of sequences in the promoters of tobacco PR1a (Nt) and Arabidopsis PR1 (At). Only the sequence of the top strands is given. The sequences of the promoter regions are shown with gaps to allow maximal alignment. The position of the leftmost nucleotide relative to the transcription start site is indicated. Corresponding nucleotides are indicated by colons. Colored block arrows mark consensus binding sites for various transcription factors, as indicated. The direction of the arrow indicates whether the consensus sequence is in the top (right-pointing arrow) or bottom strand. The dashed and solid black arrows mark the binding sites for AtWRKY50. The positions of sequence elements used in the linker scanning analysis of the PR1 promoter by Lebel et al. (1998) are indicated (LS). (B) EMSAs were performed with an 80-bp fragment of the PR1 promoter and GST-tagged C-terminal halves (Lanes C) or full-length (Lanes FL) versions of AtWRKY50, -51 and -59, as indicated above the lanes. (C) EMSAs were performed with the same probe together with the GST-tagged C-terminal half (Lane C) and GST-tagged (Lane GST) and His-tagged (Lane His) full-length versions of AtWRKY50. In (B,C), lanes labeled with the minus sign were loaded with the probe only. The positions of shifts and the unbound probe (FP) are indicated. (D) Modeling prediction analysis of C-terminal and full-length AtWRKY50 protein binding to DNA in silico. (a) shows the structure of C-terminal WRKY4-C domain bound to DNA from Yamasaki et al. (2012) onto which AtWRKY50 and its truncated form are superimposed. The overlap of the β-sheets and overall structure correlating with interactions is shown in (b). The full length AtWRKY50 sequence also demonstrates a superimposable C-terminal region (c) but the additional protein structure was predicted to possess a large disordered region (shown with white arrow).
To analyze if AtWRKY50, AtWRKY51, and AtWRKY59 are able to specifically bind to this region of the promoter, we set up electromobility shift assays (EMSA) with an 80-bp fragment of the PR1 promoter, corresponding to the region of -685 to -606, which covers all of the above elements. EMSAs were performed with affinity purified glutathione-S-transferase (GST)-coupled fusion products of the respective WRKY proteins expressed in Escherichia coli. Both full-length WRKYs and C-terminal halves containing both the WRKY- and Zn-finger domains were produced (Supplementary Figure S1). The GST-tagged, 88-amino acid long C-terminal half of AtWRKY50 (AtWRKY50-C) efficiently bound to the probe (Figure 3B, lane 2), but surprisingly, full-length GST-tagged AtWRKY50 did not produce a shift (Figure 3B, Lane 6). The GST-tagged full-length AtWRKY51 and AtWRKY59 did not produce band shifts as expected (Figure 3B, Lanes 7 and 8) and neither did their C-terminal halves (Figure 3B, Lanes 3 and 4), indicating that amino acids outside of the conserved WRKYGKK domain are also important determinants for specificity of binding to the PR1 promoter. This is similar to the observations where C-terminal of NtWRKY12 was shown to bind the tobacco PR-1a promoter much more efficiently than full-length NtWRKY12 (van Verk et al., 2008). It is unlikely that the relatively large GST-tag fused at the N-terminus of the full-length protein masks the WRKY’s DNA-binding domain for interaction with the DNA. However, when fused to the C-terminal half, it possibly leaves the binding domain exposed, since an EMSA with full-length AtWRKY50 fused to the much smaller His-tag also failed to produce a shift with the 80-bp promoter fragment (Figure 3C). In order to ascertain the absence of an interaction with the full-length protein we employed homology modeling for both full length and truncated proteins. We compared the models to a known structure for a related WRKY protein’s C-terminus with DNA bound to it (Yamasaki et al., 2012), where we observe a similar series of β-sheet features that interact with the major grove of the DNA (Figure 3Da). When the C-terminal AtWRKY50 model is overlaid the models are superimposable, sharing similar features and corroborating the DNA binding ability (Figure 3Db). This contrasts with the full-length model (Figure 3Dc), which although possessing similar C-terminal features, has an N-terminal region that is predicted to be bulky with some regions of disorder (white arrow, Figure 3Dc). From these comparisons we postulate that the N-terminal region possibly impedes the binding of DNA and could explain our observations.
Characterization of AtWRKY50’s Binding Site
To investigate if the W-box in LS4 is the binding site for AtWRKY50, a mutant version of the 80-bp fragment was constructed, in which the TTGACT sequence of the W-box was changed to TCAGCT (Figure 4A, probe Wm). While incubation of the wild type and mutant 80-bp probes with the C-terminal halves of AtWRKY51 and AtWRKY59 did not result in shifts (Figure 4B, Lanes 3, 4, 7, 8), AtWRKY50-C (C-terminal half) produced shifts with both probes (Figure 4B, Lanes 2, 6). Interestingly, a double shift is produced with the wild type probe, while with the mutant probe the higher shift is lost. This suggests that AtWRKY50-C has two binding sites in the 80-bp PR1 promoter fragment of which one overlaps with the W-box in LS4. The shift with the mutant probe indicates that AtWRKY50-C also binds to a second site in the 80-bp promoter fragment, which is different from the W-box consensus.
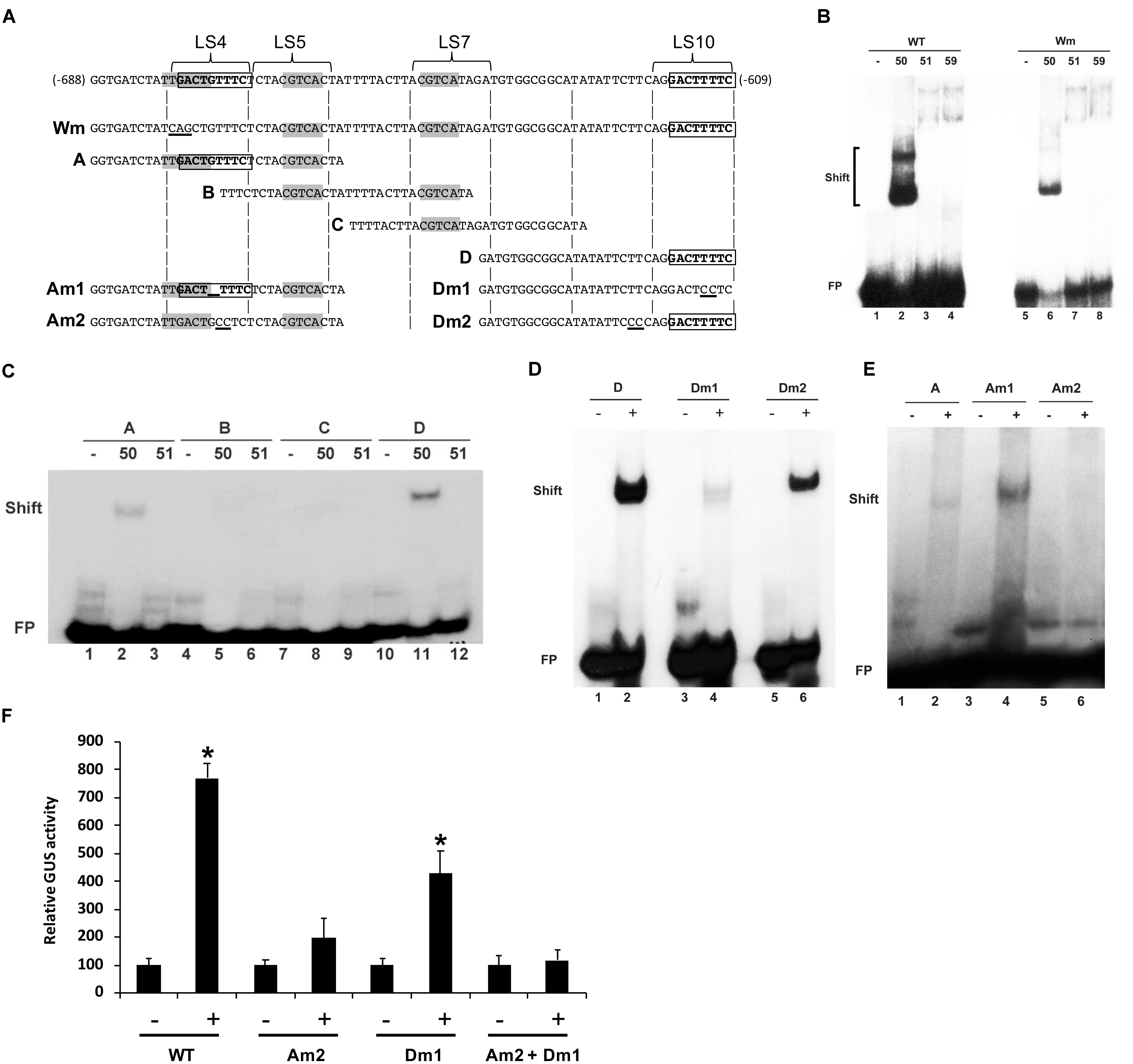
FIGURE 4. Characterization of AtWRKY50’s binding sites on the PR1 promoter (A) Sequences of PR1 promoter fragments used for EMSAs. Only the sequence of the upper strand is given. The top line displays the sequence of the 80-bp fragment corresponding to bp –688 to –609 upstream of the transcription start site. Regions LS4, LS5, LS7, and LS10, as used in the linker scanning analysis of Lebel et al. (1998), are indicated. Wm indicates an 80-bp fragment with a mutation (TTGACT to TCAGCT) in the W-box in LS4. Overlapping subfragments A, B, C, and D, and their mutant versions Am1, Am2, Dm1 and Dm2 are aligned with the sequence of the 80-bp fragment. The W-box (TTGACT) and the CGTCA boxes of the as-1 element are indicated in bold. Mutations in Wm, Am1, Am2, Dm1, and Dm2 are underlined. (B) EMSAs were performed with wild type 80-bp PR1 promoter fragment (WT) or with an 80-bp fragment with a mutation in the W-box (Wm) as probes together with the GST-tagged C-terminal halves of AtWRKY50, -51 and -59, as indicated above the lanes. The positions of band shifts and the unbound probe (FP) are indicated. Lanes labeled with the minus sign were loaded with probe only. (C) AtWRKY50 binds to the PR1 promoter at two positions. EMSAs were performed with overlapping PR1 promoter fragments A, B, C, and D as probes and GST-tagged AtWRKY50-C or the C-terminal half of AtWRKY51, as indicated above the lanes. The positions of band shifts and the unbound probe (FP) are indicated. Lanes labeled with the minus sign were loaded with probe only. (D,E) AtWRKY50 binds to the LS10 and LS4 element in the PR1 promoter. EMSAs were performed with wild type (D) and WT (A) and mutant versions (Dm1, Dm2) of fragment D and Am1, Am2 of fragment A of PR1 promoter as probes and GST-tagged AtWRKY50-C, as indicated above the lanes. The positions of band shifts and the unbound probe (FP) are indicated. Lanes labeled with the minus sign were loaded with probe only; lanes labeled with the plus sign were loaded with the probe and AtWRKY50-C. (F) PR1 activation by AtWRKY50 requires intact binding sites. Arabidopsis protoplasts were co-transfected with WT and mutant PR1::GUS construct alone (minus sign) or together with expression plasmids 35S::AtWRKY50. After incubation GUS activity was measured spectrophotometrically. Expression levels (%) are given relative to expression level without WRKY effector. Statistical differences among the samples is labeled with asterisk (p < 0.05).
To further delimit the AtWRKY50 binding sites in the 80-bp fragment, a series of overlapping subfragments (A to D) were generated as shown in Figure 4A. As expected, incubation with the AtWRKY51-C peptide did not result in shifts with any of the four subfragments (Figure 4C, Lanes 3, 6, 9, 12). However, AtWRKY50-C produced shifts with subfragments A and D (Figure 4C, Lanes 2 and 11, respectively). The shift with subfragment A supports the result from the EMSA shown in Figure 4B, suggesting that the sequence overlapping with the W-box in LS4 facilitates AtWRKY50-C binding. The shift with fragment D indicates the presence of an additional AtWRKY50 binding site, possibly in the LS10 element, which is different from the canonical W-box. To test this, double-stranded oligonucleotides corresponding to fragment D, containing mutations in the LS10 element (Dm1, Figure 4A) and upstream of the LS10 element (Dm2, Figure 4) were used as probes in EMSAs with AtWRKY50-C. Whereas the mutation of two nucleotides immediately upstream of the LS10 element (Dm2) did not change the ability of the probe to bind (compare Figure 4D, Lanes 2 and 6), mutation of two central T nucleotides in LS10 (Dm1) almost completely abolished binding of AtWRKY50-C (Figure 4D, Lane 4). This indicates that LS10 indeed contains a binding site for AtWRKY50, which is distinct from the consensus WRKY binding site (W-box).
Almost an exact copy of the sequence GACTTTTC of LS10 is present in LS4, partly overlapping with the W-box and with only a G inserted between the first and second T. An EMSA with the Am1 probe, in which this G was removed from subfragment A, shows that this results in enhanced binding (Figure 4E). We speculate that the binding of AtWRKY50-C to fragment A (Figure 4C, Lane 2) is actually caused by the presence of this LS10-like GACTGTTTC sequence, rather than by the W-box, as mutation of GACTGTTTC to GACTGCCTC (Am2, Figure 4A), which leaves the W-box intact, completely abolished binding to AtWRKY50-C (Figure 4E, Lane 6). The reduced binding of AtWRKY50-C observed upon mutation of the W-box (Figure 4B, Lane 6) could thus be caused by the fact that the W-box mutation changes the two left most nucleotides of the LS10-like element.
To test whether these promoter elements are necessary for activation of gene expression in planta, Am2 and Dm1 mutations (Figure 4A) were incorporated into the 1000 bp promoter of PR1::GUS reporter gene constructs, which were used in Arabidopsis protoplasts co-expression studies (Figure 4F). Mutation of the binding site in LS10 (Dm1) resulted in the reduction of GUS expression by approximately 50%, while mutation of the binding site in LS4 (Am2) reduced the expression to less than 20%. When both mutations were incorporated in the PR1 promoter, AtWRKY50 was no longer able to activate GUS expression (Figure 4F).
Taken together, the results of these experiments support the notion that GACT(G)TTTC is the binding site of AtWRKY50 and that both these sites in the PR1 promoter are required for maximal activation by AtWRKY50.
AtWRKY50 Interacts With TGA2 and TGA5
Previously, we found that the close proximity of the binding sites for NtWRKY12 and TGAs in the promoter of the tobacco PR1a gene may be functionally relevant for bringing both proteins together in order to direct full transcriptional activation. Further support for this came from studies that showed that NtWRKY12 interacted with TGA2.2 when expressed in yeast and in Arabidopsis protoplasts (van Verk et al., 2011). Similar to the PR1a promoter of tobacco, the AtWRKY50 binding sites in LS4 and LS10 and the TGA-binding as-1 element (in LS5 and LS7) of the Arabidopsis PR1 promoter are in close proximity (Figure 3A). To investigate if Arabidopsis TGAs and AtWRKY50 can interact, we performed yeast-two-hybrid assays. The full-length coding sequence of AtWRKY50 was fused to the binding domain (BD) of GAL4 and co-expressed with coding sequences for TGA2 and TGA5 fused to the GAL4 transcriptional activation domain (AD) in yeast containing a GAL1::HIS3 reporter gene. Growth of yeast was scored on media with and without histidine. TGA2 and TGA5 interacted weakly with AtWRKY50 as co-expression of TGA2-AD or TGA5-AD with AtWRKY50-BD could grow on a dropout medium (Figure 5A). The known interaction between TGA2 and NPR1 served as a positive control for this experiment. However, HIS3 expression was relatively low, since addition of 10 mM 3-amino-1,2,4-triazole (3AT) inhibited growth. Nevertheless, this indicates that AtWRKY50 and the TGAs interacted in the yeast system. Additional two-hybrid assays with N- and C-terminal halves of AtWRKY50 indicated that the TGA interaction domain is present in the N-terminal half of the protein and that this domain provides for a strong interaction resulting in high expression of HIS3 that was not inhibited by 10 mM 3AT. The C-terminal half of AtWRKY50 clearly did not interact with the TGAs (Figure 5B).
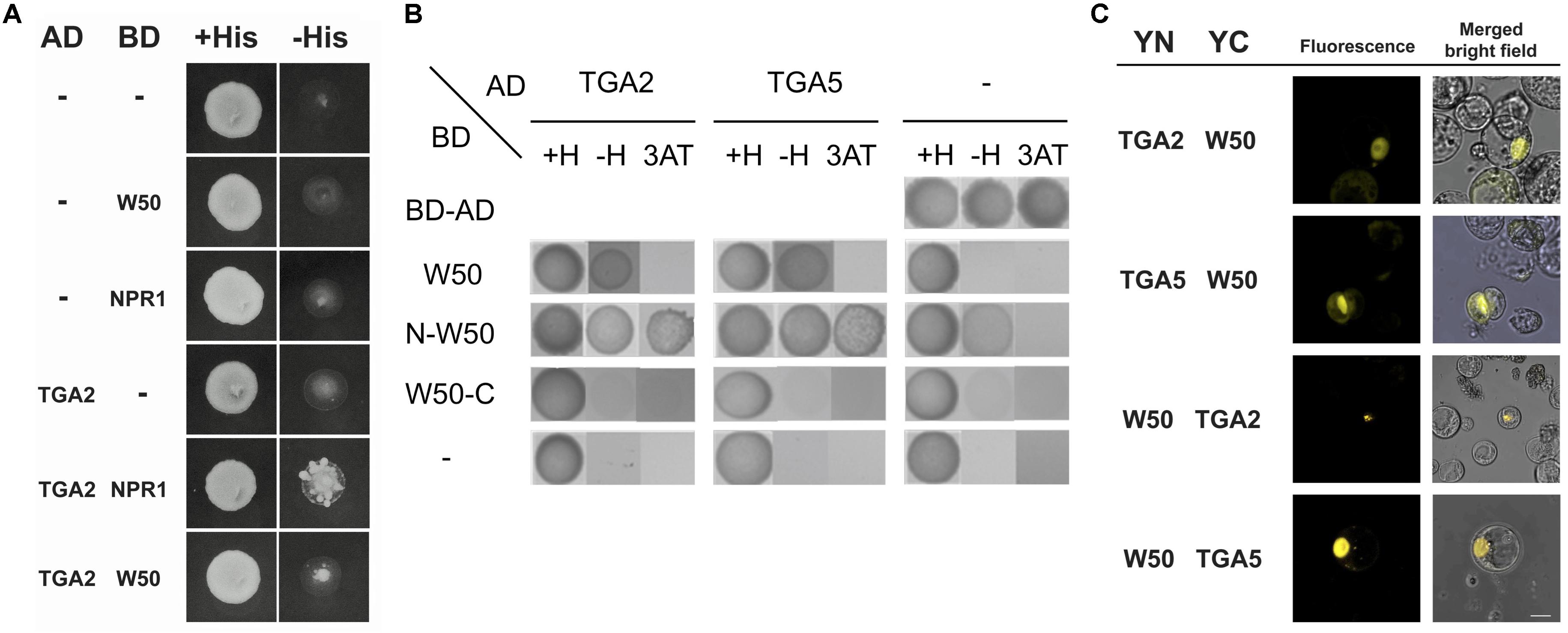
FIGURE 5. AtWRKY50 interacts with TGA2 and TGA5. (A) Yeast was transformed with expression plasmids pAS2.1 and pACT2, containing the coding regions of the binding domain (BD) and activation domain (AD) of GAL4, respectively. The BD domain was either fused to the coding regions of AtWRKY50 (W50) or NPR1, or was not fused (minus sign); the AD domain was not fused (minus sign) or was fused to the coding region of TGA2. Growth of transformed yeast was evaluated on medium containing histidine (+His) or minus histidine (–His). (B) The N-terminal of AtWRKY50 interacts with TGA2 and TGA5. Yeast was transformed with expression plasmids pAS2.1 and pACT2, containing the coding regions of the binding domain (BD) and activation domain (AD) of GAL4, respectively. The BD domain was either fused to the coding regions of full length AtWRKY50 or its N-terminal or C-terminal half. The AD domain was not fused (minus sign) or was fused to the coding region of TGA2 or TGA5. Growth of transformed yeast was evaluated on medium containing histidine (+H), minus histidine (–H) and minus histidine plus 10 mM 3AT (3AT). (C) AtWRKY50 interacts with TGA2 and TGA5 in Arabidopsis protoplasts. YFP fluorescence and merged bright field images of Arabidopsis protoplasts co-transformed with expression plasmids containing constructs encoding TGA2, TGA5, and AtWRKY50 (W50) fused to the N-terminus (YN) or the C-terminus (YC) of yellow fluorescent protein. Scale bar = 10 μm.
The interaction of AtWRKY50 with TGAs was confirmed using bimolecular fluorescence complementation (BiFC) assays. To this end, the full-length AtWRKY50 and TGA2 or TGA5 coding sequences were fused at the N- or the C-terminus to the N- (YN) or C- (YC) terminal halves of the yellow fluorescent protein (YFP), respectively. Protein-protein interaction was analyzed 16 h after co-transfection of Arabidopsis protoplasts with expression plasmids harboring these constructs by determining the fluorescence of reconstituted YFP using confocal laser scanning microscopy. For all combinations with YN- and YC-fused versions of AtWRKY50 and TGA2 or TGA5, fluorescence was visible most strongly in nuclei, indicating a predominant nuclear presence of the AtWRKY50 and TGA fusion proteins (Figure 5C). Controls with combinations of unfused YN and YC or with combinations of AtWRKY50 and TGA2 or TGA5 in which only one of the proteins was fused to a YFP half did not result in fluorescence (Supplementary Figure S2). The results indicate that AtWRKY50 interacts with both TGA2 and TGA5.
AtWRKY50-C and TGA2 or TGA5 Bind to the PR1 Promoter Simultaneously
Next, we investigated how combinations of AtWRKY50 and TGA2 or TGA5 influenced the binding to DNA. Therefore, EMSAs were performed with purified, E. coli-expressed GST-tagged AtWRKY50-C and His-tagged TGA2 and TGA5. A double shift resulting from the binding of one and two AtWRKY50-C peptides to the 80-bp PR1 promoter was observed (Figure 6A, Lanes 2 and 8). A number of band shifts were observed with the EMSAs with TGA2 and TGA5 of which the intensity increased with decreasing mobility (Figure 6A, Lanes 3 and 9). The presence of multiple shifts with TGA proteins (notably with TGA2 and TGA5) could be ascribed to possible different degrees of occupancy of the binding sites present in the probe (Miao et al., 1994; Miao and Lam, 1995; Pontier et al., 2001; Boyle et al., 2009). However, the possibility of aggregate formation during incubation, due to non-specific interactions of these TGAs could not be excluded. Nevertheless, we speculate that the weak bands in Lanes 3 and 9 (indicated by single black asterisks) represent complexes in which only one of the CGTCA binding sites in either LS5 or LS7 is occupied by a TGA dimer. The calculated molecular weights of complexes of the 80 bp probe with a TGA-His fusion protein dimer (125 kD) or with two GST-AtWRKY50-C fusion proteins (122 kD) are almost identical and this corresponds well with the similar mobilities of the respective bands in lanes 3/9 and 2/8. The more slowly migrating bands in Lanes 3 and 9 (indicated by white asterisks), represent higher order TGA shifts, possibly including shifts in which TGAs are bound at both the CGTCA sites in LS5 and LS7. Incubation of the 80-bp probe with combinations of AtWRKY50-C and TGA2 or TGA5 resulted in the formation of new bands indicated by the double asterisks in black and white (Figure 6A, Lanes 4 and 10). It is highly plausible that these new bands represent complexes of the probe with both AtWRKY50-C and the respective TGA dimers (black double asterisks, MW 161 kD) and TGA oligomers (white double asterisks).
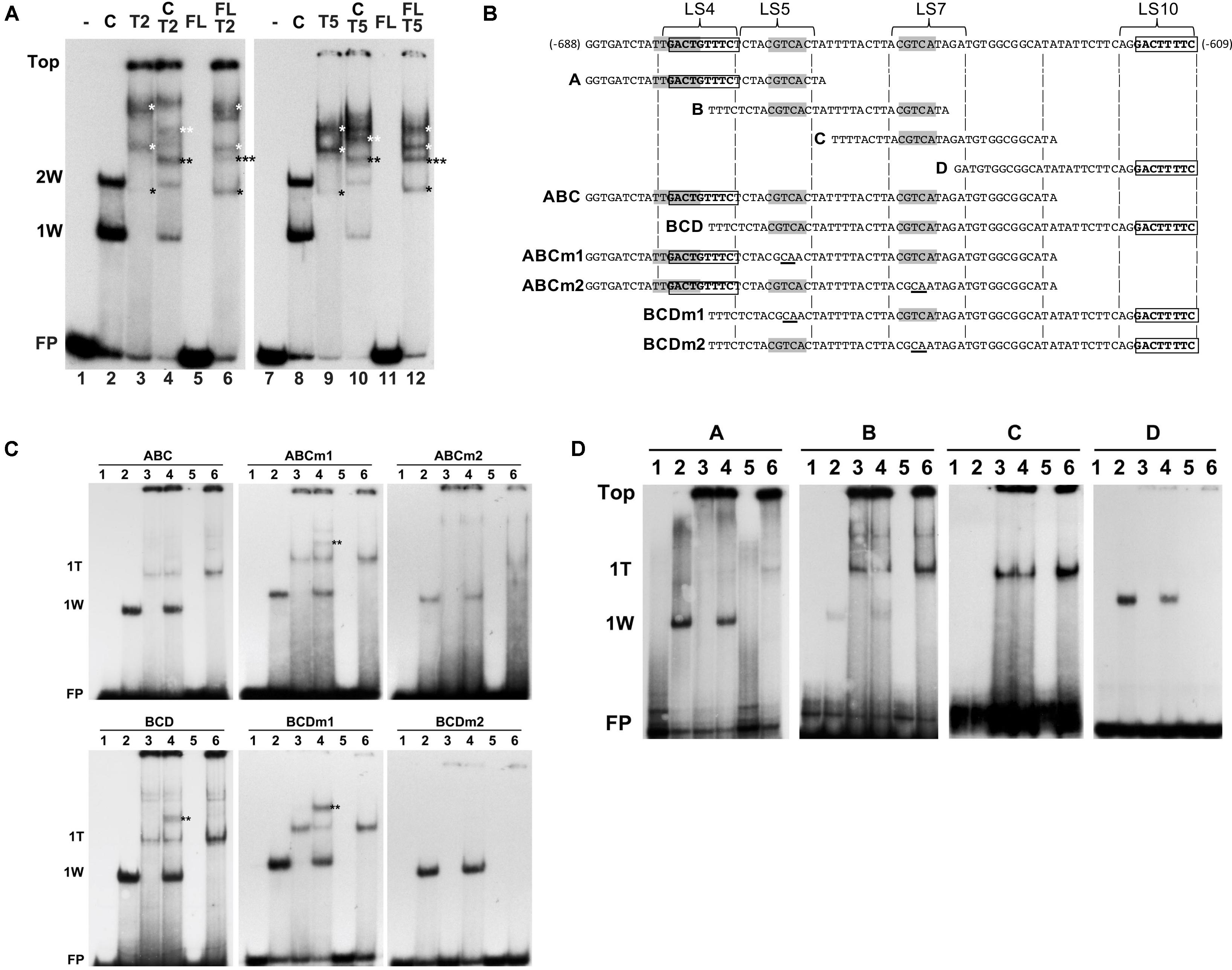
FIGURE 6. AtWRKY50 and TGA2 and TGA5 bind to the PR1 promoter. (A) EMSAs were performed with an 80-bp fragment of the PR1 promoter without protein (minus signs) or with GST-tagged C-terminal half (C) or full-length (FL) versions of AtWRKY50, and His-tagged TGA2 (T2) or TGA5 (T5), and combinations of these proteins, as indicated above the lanes. The positions of the unbound probe (FP), the top of the gel (Top), and of band shifts caused by one (1W) or two (2W) AtWRKY50-C proteins are indicated at the left. Shifts caused by binding of single (single black asterisks) or multiple (single white asterisks) TGA proteins and shifts caused by a combination of TGA and AtWRKY50 (double black asterisks) are indicated. (B) Sequences of PR1 promoter fragments used for electromobility shift assays. Overlapping subfragments A, B, C, and D, and subfragments ABC and BCD are aligned with the sequence of the 80-bp fragment. The overlapping W-box (TTGACT) and AtWRKY50 binding sequence (GACTGTTTC) in LS4, the CGTCA boxes of the as-1 element in LS5 and LS7, and the AtWRKY50 binding sequence (GACTTTTC) in LS10 are indicated in bold. Subfragments ABCm1, ABCm2, BCDm1 and BCDm2 represent variants of fragments ABC and BCD with mutations (underlined) in the CGTCA boxes in LS5 and LS7, respectively. (C) EMSAs were performed with probes corresponding to PR1 promoter fragments ABC and BCD and their mutated versions ABCm1, ABCm2, BCDm1, BCDm2, as indicated above the panels. EMSA incubation mixtures contained no protein (Lanes 1), AtWRKY50-C (Lanes 2), TGA2 (Lanes 3), AtWRKY50-C and TGA2 (Lanes 4), full-length AtWRKY50 (Lanes 5), and full-length AtWRKY50 and TGA2 (Lanes 6). The positions of the unbound probe (FP) and of band shifts caused by AtWRKY50-C (1W) or TGA2 (1T) are indicated at the left. Shifts caused by binding of a combination of TGA and AtWRKY50 (double black asterisks) are indicated. (D) EMSAs were performed with PR1 promoter fragments A, B, C, and D as probes, as indicated above the panels. EMSA incubation mixtures contained no protein (Lanes 1), AtWRKY50-C (Lanes 2), TGA2 (Lanes 3), AtWRKY50-C and TGA2 (Lanes 4), full-length AtWRKY50 (Lanes 5), and full-length AtWRKY50 and TGA2 (Lanes 6). The positions of the unbound probe (FP), the top of the gel (Top) and of band shifts caused by AtWRKY50-C (1W) or TGA2 (1T) are indicated at the left.
Next, we thought to investigate which of the two AtWRKY50 binding sites allows formation of a complex between AtWRKY50 and TGA. For this, promoter fragments consisting of the regions encompassing subfragments A, B and C (Figure 6B, ABC) and subfragments B, C and D (Figure 6B, BCD) were tested in EMSAs with AtWRKY50-C and TGA2. The ABC and BCD promoter fragments each contain only one of the AtWRKY50 binding sites. The single shifts (1W) observed in Lanes 2 correspond to AtWRKY50-C binding to the sites in LS4 of probe ABC and LS10 of probe BCD, respectively (Figure 6C). TGA2 predominantly binds to only one binding site in ABC and BCD (1T), while the presence of weak, high shifts in Lanes 3 suggests that binding of multiple TGA2 proteins occurred at low frequency. This is in contrast to the EMSAs with the longer 80-bp probe, which indicated that the TGAs preferentially bound as multimeric complexes (Figure 6A, Lanes 3 and 9). Yet, the ABC and BCD probes were only 18 bp shorter than the 80-bp probe, while all contained the same two CGTCA motifs. This suggests that the size of the probe determines the efficiency and number of TGA2 proteins it can bind. Obviously, the EMSAs with TGA2 alone resulted in a single prominent shift, suggesting that only one of the two CGTCA motifs in the fragments efficiently bound TGA2. To find out which of the CGTCA motifs is bound by TGA2, fragments ABCm1, BCDm1, ABCm2 and BCDm2 (Figure 6B), with mutations in LS5 (m1) and LS7 (m2), respectively, were used as probes in EMSAs (Figure 6C, middle and right panels). Apparently, mutation of the CGTCA box in LS7 interfered with TGA2 binding to the fragments (right panels), while mutation of the CGTCA motif in LS5 had no effect on binding of TGA2 (middle panels). These results were confirmed by the EMSAs of Figure 6D that show that fragment A, which contains the CGTCA motif in LS5, did not produce a shift upon incubation with TGA2, whereas predominantly single TGA shifts were present with probe B, containing both CGTCA motifs of LS5 and LS7, and with probe C that contains only the TGA binding site in LS7. As expected, probes A and D bind AtWRKY50-C. These results indicate that the CGTCA box in LS7 is the main binding site of TGA2. Furthermore, the shifts indicated by the double asterisks in Figure 6A, Lanes 4 in the panels with probes BCD and BCDm1 show that these fragments are able to bind AtWRKY50-C and TGA2 simultaneously and with higher efficiency than probe ABCm1.
AtWRKY50 Stimulates Binding of TGA2 and TGA5 to the PR1 Promoter
As was already shown, AtWRKY50-C binds highly efficiently and specifically to the DNA while full length AtWRKY50 was unable to bind the PR1 promoter. We speculated that a conformational change is required to release the N-terminal half of AtWRKY50 from blocking the C-terminal PR1 promoter binding domain. To investigate if such a release could be brought about by the interaction with TGA2 or TGA5, EMSAs were performed with the 80-bp probe and mixtures of full-length AtWRKY50 and TGA2 or TGA5. The results are shown in Figure 6A, Lanes 6 and 12. In addition to bands corresponding to TGA dimers and oligomers (black and white single asterisks, respectively; compare to Lanes 3 and 9), surprisingly, the EMSAs resulted in an extra band shift (indicated by the triple asterisks) that migrated to a similar position as the band shifts in Lanes 4 and 10, respectively, that correspond to the probe binding combinations of WRKY50-C and TGA (double asterisks). It is conceivable that these triple asterisks band shifts in Lanes 6 and 12 are the result of the binding of a combination of the TGA dimer and the WRKY protein as the molecular weight of such a complex (170 kD) would be close to the molecular weight of a complex of AtWRKY50-C with a TGA dimer (161 kD) as is visible in Lanes 4 and 10 (double black asterisks). The fusion product of full-length AtWRKY50 and GST (45 kD) used in these EMSAs is considerably smaller than the TGA-His dimer (76 kD), implicating that a band shift produced by the binding of a single AtWRKY50 protein to the probe would migrate to a position below the TGA dimer and intermediate of positions 1W and 2W.
Although binding of a complex of full-length AtWRKY50 with a TGA dimer is speculative, its addition to TGA2 or TGA5 resulted in an enhancement of the TGA dimer shifts (Figure 6A, compare the bands indicated by the single black asterisks in Lanes 6 and 12 with those in Lanes 3 and 9, respectively, and in Figure 6C, compare Lanes 6 and 3 in the leftmost panels). This effect was not observed with combinations of AtWRKY50-C and the TGAs (Figure 6A, Lanes 4 and 10; Figure 6C, left panels, Lanes 4). Apparently, full-length AtWRKY50 promotes binding of the TGA dimer to the 80-bp probe and to the ABC and BCD probes. This effect of AtWRKY50 on TGA binding does not require AtWRKY50’s binding site on the DNA. When combinations of full-length AtWRKY50 and TGA2 were incubated with promoter fragments lacking AtWRKY50’s binding site, the stimulating effect on TGA binding was still present. This can be seen in Figure 6D, where fragments B and C, containing two (LS5 and LS7) and one (LS7) CGTCA motifs, respectively, but lacking the AtWRKY50 binding site in LS4 or LS10, show an increased intensity of the TGA2 shifts in Lanes 6 of panels B and C.
Discussion
The PR1:LUC based screening with 40 members of the Arabidopsis WRKY protein family showed that WRKY50 was the best activator of the PR1 promoter. In earlier work, tobacco NtWRKY12 was shown to be involved in the expression of PR-1a gene (van Verk et al., 2008). Of all 74 Arabidopsis WRKY proteins, AtWRKY50 has the highest similarity to NtWRKY12, including the aberrant G-K-K sequence instead of G-Q-K immediately following the conserved W-R-K-Y sequence present in the majority of WRKY proteins. In the WRKY-DNA complex, the amino acids of the WRKY domain have been shown to be in direct contact with the DNA (Yamasaki et al., 2005). This could explain why the WK-box, NtWRKY12’s binding site in the DNA, is different from the consensus W-box. Also, AtWRKY59, one of the two other Arabidopsis WRKYs with a W-R-K-Y-G-K-K sequence was reported to lack binding specificity for the W-box (Dong et al., 2003). Here we found that AtWRKY50 binds to DNA sequences that are different from the W-box. We identified PR1 promoter fragments A and D to specifically bind the DNA-binding domain of AtWRKY50 (Figure 4C). Although we haven’t performed an extensive mutational analysis to determine the minimal binding sequence, changing the two central T-residues in the TTTTC stretch in LS10 or in the GTTTC stretch in LS4 to C’s significantly reduced the binding of AtWRKY50-C, indicating that these base pairs are important for AtWRKY50’s binding. It is worth to note that NtWRKY12 and AtWRKY50, although their binding sites are different (TTTTCCAC and GACT[G]TTTC, respectively), both contain a TTTC stretch.
Our results seem to contradict an earlier finding that a C-terminal region of AtWRKY50 binds to a W-box-containing probe (Brand et al., 2010). However, in that study a mutated version of the W-box probe was also bound with significant efficiency, while the probe also contained the sequence ACTTTT, which is identical to the central part of the binding sequence we characterized in LS10. Furthermore, the authors used a 77-amino acid long C-terminal peptide, while our AtWRKY50-C consists of the C-terminal 88 amino acids. Interestingly, the corresponding 11 amino acid overlap region of NtWRKY12 is important for binding to the promoter of tobacco PR-1a (van Verk et al., 2011). In this regard, it is highly plausible that the extra amino acids in AtWRKY50-C contribute to the binding specificity.
Intriguingly, the inability of full-length AtWRKY50 to bind the PR1 promoter was also observed previously with NtWRKY12 in tobacco (van Verk et al., 2008). The fact that the C-terminal His-tagged full-length AtWRKY50 protein didn’t produced a shift, it is unlikely that the binding capacity was masked by the relatively large GST-tag at the N-terminus of the protein. Also, it is plausible that the lack of correct structural folding in bacterial purified proteins show this effect. From our in silico modeling prediction analysis (Figure 3D), it is possible that the N-terminal halves of the full-length WRKYs themselves prevent binding to the DNA under EMSA conditions by stearic hindrance. The presence of bulky N-terminal region with predicted disorder (Figure 3Dc, white arrow) further supports our observation. We also note that work by Yamasaki et al. (2012) only studied the C-terminal region as opposed to the full-length protein. As both the tobacco and Arabidopsis homologs fail to bind full-length protein, it could indicate that this is a functionally relevant property, e.g., to prevent promiscuous binding of the WRKY protein to DNA regions with consensus binding sequences that are not in the correct structural context. It is possible that the interaction with other factors is required to change the configuration of the full-length WRKYs to release the binding domains to bind DNA. Likely candidates to bring about such a change could be TGA2 or TGA5, especially since they specifically interact with the N-terminal half of AtWRKY50 (Figure 5B).
Our studies in Arabidopsis protoplasts showed that AtWRKY50 enhanced expression of co-transfected PR1::LUC and PR1::GUS reporter genes and also of the endogenous PR1 gene, suggesting that the protein acts as a transcriptional activator. This was also the case for its tobacco homolog NtWRKY12 (van Verk et al., 2008). However, while the full-length NtWRKY12-GAL4BD fusion protein activated the His reporter gene in yeast, AtWRKY50 showed no transcriptional activity in this system. We speculate that either, the BD part of the fusion protein interferes with the correct folding of AtWRKY50, or that yeast lacks specific factors necessary for its activating function. The expression of PR1 in transgenic plants constitutively overexpressing AtWRKY50 was increased in comparison to control plants, but only after the plants were treated with SA (Figure 1C). This indicates that AtWRKY50 acts as a transcriptional enhancer, but that other activators are needed to trigger the SA response of the PR1 gene.
Previously, it was found that AtWRKY50 and AtWRKY51 are involved in repression of jasmonic acid (JA)-dependent defense responses, including PDF1.2 marker gene expression (Gao et al., 2010). Although it was not investigated if this was the effect of a direct interaction of the WRKYs with the PDF1.2 promoter, the authors contemplated that the WRKYs might act as transcriptional repressors, possibly by binding to specific binding sequences in the promoters of JA-responsive genes. In this context, it is significant to note that the PDF1.2 promoter lacks W-boxes, but contains the AtWRKY50 binding element GACTGTTTC.
The C-terminal half of AtWRKY50, containing the conserved amino acid sequence WRKYGKK and a proximal zinc-finger region, bound at two positions in the 80-bp region of the PR1 promoter essential for SA-inducible expression. This region also contains an as-1 element, consisting of two direct CGTCA motifs, that acts as a binding site for TGA transcription factors. Here we have shown that AtWRKY50 interacts with TGAs 2 and 5 in the nucleus. Using in vitro DNA binding assays, we showed that especially the rightmost CGTCA motif of the as-1 element was able to bind TGA2 with high efficiency and that simultaneous binding of AtWRKY50-C and TGA2 to the promoter occurred. Furthermore, although PTAs demonstrated that AtWRKY50 alone was able to activate the PR1 promoter, and that this did not require transcription factors TGA2, TGA5, or TGA6 or their co-activator NPR1, expression of the GUS reporter gene was greatly enhanced when TGA2 or TGA5 were present. Our finding that TGA2 did not efficiently bind to the CGTCA motif in LS5 is at variance with the results of Després et al. (2000) who found that TGA2 binds to both LS5 and LS7. However, the authors used DNA probes containing either the LS5 or the LS7 element, which precludes a comparison of the relative strengths with which the two elements are bound. LS5 has been identified as a DNA element conferring a negative effect on PR1 gene expression. The proximity of the TGA and AtWRKY50 binding sites in the PR1 promoter and the ability of the proteins to interact, suggest that such an interaction could also take place when the transcription factors are bound to the promoter and that this could be relevant for PR1 expression. Indeed, although AtWRKY50 expressed in protoplasts activates a co-transfected PR1::GUS gene, co-expression of TGA2 or TGA5 further enhanced GUS expression considerably. This synergistic effect was specific for TGA2 and TGA5, which both interacted with AtWRKY50, while TGA3, which did not strongly interact with AtWRKY50, was not able to enhance gene expression.
TGA2 on its own is not a transcriptional activator but requires binding of NPR1. In the absence of NPR1, TGA2 has been suggested to act as a repressor of PR1 gene expression (Zhang et al., 2003; Rochon et al., 2006; Boyle et al., 2009). TGA2, 5 and 6 belong to the same subclass of TGAs (clade II). There is accumulating evidence that in addition to NPR1, TGAs are able to interact with other proteins. Previously, glutaredoxin was shown to interact with Arabidopsis TGA2 and tobacco TGA2.2 (Ndamukong et al., 2007), while Arabidopsis TGA2 and TGA5 were found to interact with SCL14, a protein mediating regulation of genes involved in detoxification processes (Fode et al., 2008). Based on their findings, the authors suggested that clade II TGAs could act as sequence-specific anchor proteins to recruit other transcription regulatory proteins, like SCL14 and DELLA proteins, to the promoters of their target genes. In this perspective, we speculate that TGA2 could likewise assist in recruiting AtWRKY50 to the PR1 promoter. Also, WRKYs have been found to interact with other proteins. For example, Arabidopsis WRKY7 has been found to interact with calmodulin (CaM) through a CaM binding domain in the N-terminal half of the protein that is conserved in other members of the WRKY IId group (Park et al., 2005). Other examples are WRKY70 interacting with the EAR domain repressor ZAT7 (Ciftci-Yilmaz et al., 2007), WRKY53 interacting with mitogen activated protein kinase kinase kinase1 (MEKK1; Miao et al., 2007), WRKY33 interacting with mitogen activated protein kinase 4 (MAPK4; Andreasson et al., 2005), and WRKYs 38 and 62 that have been found to interact with histone deacetylase19 (HDA19; Kim et al., 2008). In our EMSAs, the new band shifts produced upon incubation of combinations of AtWRKY50-C and TGA2 or TGA5 with the 80-bp promoter fragment (Figure 6A, triple asterisks) or with probes ABC and BCD (Figure 6C, double asterisks) likely represent supershifts produced by the simultaneous binding of both proteins to the probe, possibly as a complex of interacting transcription factors.
In conclusion, we have shown that AtWRKY50 is an activator of Arabidopsis PR1 expression. Its C-terminal DNA-binding domain specifically binds to two GACT(G)TTTC elements that are located at -675 and -616 bp upstream of the transcription start site in the PR1 promoter. Complex formation between TGA2 or TGA5 and AtWRKY50 possibly functions in the regulation of PR1 expression in vivo. Our study sheds new light on the intricate regulation of the key SAR gene PR1. The working model of the current study is summerised in Supplementary Figure S3.
Materials and Methods
Reporter Vector Construction
The PR1:: LUC reporter was constructed by PCR amplification of 1000bp upstream region of PR1 (At2g14610) gene using Arabidopsis Col-0 genomic DNA as template (for primer sequences, see Supplementary Table S2). The DNA-fragments were inserted into the vector pBT10-LUC by using NcoI and EcoRI restriction enzymes. The screening was done according to Wehner et al. (2011). The PR1::GUS reporter was obtained by PCR on genomic DNA and cloned in front of the GUS coding region in pT7:GUS.
Bacterial Expression of AtWRKY50 Fusion Proteins
The full-length and C-terminal coding sequence of AtWRKY50, AtWRKY51 and AtWRKY59 were amplified by PCR (for primer sequences, see Supplementary Table S2) and cloned in-frame with the GST open reading frame of expression vector pGEX-KG (Guan and Dixon, 1991). These plasmids were transformed into E. coli BL21-DE3 cells. For induction of protein expression, cultures were grown to mid-log phase at 37°C, after which isopropyl-β-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM and incubation continued for 3 h at 22°C. The cells were harvested by centrifugation, resuspended in 1/20th volume sonication buffer (1x phosphate-buffered saline containing 2% [v/v] Tween 20, 0.1% [v/v] Triton X-100, 5 mM dithiothreitol [DTT], and 1 mg mL-1 lysozyme) and lysed by sonication (Vibracell). The fusion proteins were purified using glutathione-Sepharose 4B columns (Amersham), which were eluted overnight at 4°C with 10 mM reduced glutathione, after which 1/50th volume Complete (Roche) protease inhibitors were added. Expressed fusion proteins were analyzed using 12% SDS-PAGE.
Electrophoretic Mobility Shift Assay (EMSA)
Electrophoretic mobility shift assays were performed essentially as described by Green et al. (1989). DNA probes for the EMSA assays were obtained by slowly cooling down mixtures of equimolar amounts of complementary oligonucleotides from 95°C to room temperature. Annealed oligonucleotides were subsequently labeled using T4-nucleotide kinase and [γ-32P]ATP or using Klenow fragment and [α-32P]dCTP, after which unincorporated label was removed by Autoseq G-50 column chromatography (Amersham-Pharmacia Biotech). Different sets of oligonucleotides and their mutated versions are presented in Supplementary Table S2. EMSA reaction mixtures contained 0.5 μg purified protein, 3 μL 5x gel shift binding buffer [20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl, 50 mM Tris-HCl, pH 7.5, 0.25 mg ml-1 poly (dI-dC) x poly (dIdC) (Promega)] in a total volume of 14 μL. After 10-min incubation at room temperature, 1 μL containing 60,000 cpm of labeled probe was added and incubation was continued for 60 min on ice. The total mixture was loaded onto a 5% polyacrylamide gel in Tris-borate buffer and electrophoresed at 4°C. After electrophoresis, the gel was dried and auto radiographed.
qRT-PCR
Total RNA was isolated from pulverized frozen Arabidopsis tissue by phenol extraction and LiCl precipitation. Oligo (dT)-primed cDNA for PCR was obtained using M-MLV reverse transcriptase. Subsequently, qPCR was performed during 40 cycles with corresponding primers in Supplementary Table S2. The experiment was performed three times with three independent biological replicates and three technical replicates.
Plasmid Construction and Transactivation Experiments
The AtWRKY50 (At5g26170), AtWRKY51 (At5g64810), AtWRKY59 (At2g21900), TGA2 (At5g06950) and TGA5 (At5g06960) open reading frames were amplified by PCR using corresponding primer sets (Supplementary Table S2) from a cDNA library obtained from Arabidopsis plants 6 h after treatment with SA and cloned into pRT101. Protoplasts were prepared from Arabidopsis ecotype Columbia-0 cell suspension according to Axelos et al. (1992) with some modifications. A 5-days old cell suspension culture was diluted 5-fold in 50 mL medium (cell culture media-3.2 g/L Gamborg B5 basal medium with minimal organics [Sigma-Aldrich], 3% Sucrose, 1 μM naphthylacetic acid [NAA], pH 5.8) and incubated overnight at 25°C at 250 rpm. Cells were harvested, and cell walls digested with 20 mL of enzyme mix (0.4% macerozyme R-10, 1.5% cellulose R-10, 12% sorbitol, pH 5.8) for 3 h at 28°C with minimal shaking. The protoplasts were filtered with a 65-μm steel sieve and washed two times in 50 mL of protomedium (Gamborg B5 basal medium, 0.1 M Glc, 0.25 M mannitol, 1 μM NAA, pH 5.8). The volume of the protoplast suspension was adjusted to 4 × 106 cells/mL. Protoplasts were cotransfected with 2 μg of plasmid carrying one of the PR1 promoter::GUS constructs and 6 μg of effector plasmid pRT101 (Töpfer et al., 1987) carrying 35S::AtWRKY50, 35S::AtWRKY51 or 35S::AtWRKY59. As a control, co-transformation of PR1::GUS fusions with the empty expression vector pRT101 was carried out. Protoplasts were transformed using polyethylene glycol as described previously (Schirawski et al., 2000). The protoplasts were harvested 16 h after transformation and frozen in liquid nitrogen. For protoplast experiments, GUS activity was determined as described (van der Fits and Memelink, 1997), with minor modifications. GUS activities from triplicate experiments were normalized against total protein level.
Expression of TGA Fusion Proteins
The full-length coding sequence of Arabidopsis TGA2 and TGA5 were cloned in frame in front of the His-tag open reading frame of expression vector pASK-IBA45Plus (IBA Biotechnology, Göttingen, Germany). The PCR was amplified by sets of primer (Supplementary Table S2), digested with EcoRI and XhoI and cloned in pASK-IBA. These plasmids were transformed into E. coli BL21-(DE3) pLysS (Novagen). For induction of protein expression, cultures were grown to mid-log phase at 37°C, after which 2 mg/ml anhydrotetracyclin was added to a final concentration of 0.4 mM and incubation continued for 3 h at 29°C. The cells were harvested by centrifugation, resuspended in 20 ml binding buffer (5 mM imidazole, 0.5M NaCl, 20 mM Tris-HCl pH 8). The samples were sonicated until viscosity was low. The fusion proteins were purified using Ni-NTA agarose beads (Qiagen), which were eluted at 4°C with elution buffer (1M imidazole, 0.5M NaCl, 20 mM Tris-HCl, pH8), after which 1/50th volume Complete (Roche) protease inhibitors were added. Expressed fusion proteins were analyzed using 12% SDS-PAGE.
In Silico Protein Modeling Analysis
The AtWRKY50 full length and C-terminal truncated sequences were submitted to the Phyre2 protein folding prediction server (Kelley et al., 2015) and modeled independent of any reference structure. We retrieved the final models respectively for full-length and truncated proteins as structural “pdb” file. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311) (Pettersen et al., 2004).
Bimolecular Fluorescence Complementation Assays
In BiFC primer sets (Supplementary Table S2) used for WRKY50 cloning with SalI and BglII in pRTL2-YNEE and –YCHA; for WRKY50 cloning with SalI and NotI in pRTL2-EEYN and –HAYC; for TGA2 cloning with Sal1 and BglII in pRTL2-YNEE and –YCHA; for TGA2 cloning with SalI and NotI in pRTL2-EEYN and –HAYC; for TGA5 cloning with Sal1 and BglII in pRTL2-YNEE and –YCHA; for TGA5 cloning with SalI and NotI in pRTL2-EEYN and –HAYC. PCR-amplified inserts were digested with the restriction enzymes mention above and cloned in the respective pRTL2 derivates (Bracha-Drori et al., 2004) digested with the corresponding enzymes. Protoplasts were isolated and transformed with PEG as described above. Images of transfected protoplasts were acquired with a Leica DM IRBE confocal laser-scanning microscope equipped with an Argon laser line of 488 nm (excitation) and a band pass emission filter of 500–550 nm.
Yeast Two Hybrid Assays
Full-length AtWRKY50 (At5g26170) and AtNPR1 (At1g64280) cloned in pAS2.1 with primer sets (Supplementary Table S2) digested with EcoRI and BamHI, cloned into pAS2.1 and were co-transformed to yeast strain PJ69-4A (James et al., 1996). The TGA2 (At5g06950) and the TGA5 (At5g06960) ORF was PCR-amplified from Arabidopsis cDNA library using the primer sets (Supplementary Table S2), digested with EcoRI and BamHI and cloned into pACT2. For auto activation assays, transformants were plated on minimal synthetic defined (SD)-glucose medium supplemented with Met/Ura/His and lacking Leu and Trp (-LT). Ability to activate transcription in yeast was evaluated by monitoring growth after 7 days on selective SD medium lacking Leu, Trp and His (-LTH). Interaction assays were performed by co-transformation of bait and prey plasmids into yeast strain PJ69-4A and plated on SD-LT medium. As control, empty pAS2.1 and pACT2 were used. Transformants were allowed to grow for 4–5 days. Subsequently, cells were incubated for 16 h in liquid SD-LT and 10 μL of 10-fold dilutions were spotted on SD-LTH medium. Yeast cells were allowed to grow for 7 days at 30°C.
Data Availability
The datasets and scripts generated during the current study are available from the corresponding author on request.
Author Contributions
RH designed and performed the experiments. RH, AS, IH, and HL wrote the manuscript. MQ helped with in silico modeling analysis. HL supervised this study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is part of the doctoral thesis of RH conducted at Leiden University, Netherlands (Hussain, 2012, Doctoral thesis, Leiden University). We thank Prof. Wolfgang Dröge-Laser for the protoplast transactivation assays. Dr. Mei-Liang Zhou and Ms. Nahye Kim are acknowledged for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00930/full#supplementary-material
References
Andreasson, E., Jenkins, T., Brodersen, P., Thorgrimsen, S., Petersen, N. H. T., Zhu, S., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO. J. 24, 2579–2589. doi: 10.1038/sj.emboj.7600737
Axelos, M., Curie, C., Mazzolini, L., Bardet, C., and Lescure, B. (1992). A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant. Physiol. Biochem. 30, 123–128.
Boyle, P., Le Su, E., Rochon, A., Shearer, H. L., Murmu, J., and Chu, J. Y. (2009). The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21, 3700–3713. doi: 10.1105/tpc.109.069971
Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., et al. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant. J. 40, 419–427. doi: 10.1111/j.1365-313X.2004.02206.x
Brand, L. H., Kirchler, T., Hummel, S., Chaban, C., and Wanke, D. (2010). DPI-ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 6:25. doi: 10.1186/1746-4811-6-25
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Ciftci-Yilmaz, S., Morsy, M. R., Song, L., Coutu, A., Krizek, B. A., Lewis, M. W., et al. (2007). The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 282, 9260–9268. doi: 10.1074/jbc.M611093200
Delaney, T. P., Friedrich, L., and Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606. doi: 10.1073/pnas.92.14.6602
Després, C., De Long, C., Glaze, S., Liu, E., and Fobert, P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12; 279–290. doi: 10.1105/tpc.12.2.279
Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. doi: 10.1023/A:1020780022549
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Fode, B., Siemsen, T., Thurow, C., Weigel, R., and Gatz, C. (2008). The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20, 3122–3135. doi: 10.1105/tpc.108.058974
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Gao, Q. M., Venugopal, S., Navarre, D., and Kachroo, A. (2010). Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155, 464–476. doi: 10.1104/pp.110.166876
Green, P. J., Kay, S. A., Lam, E., and Chua, N. H. (1989). “In vitro DNA footprinting,” in Plant Molecular Biology Manual B11, S. B. Gelvin and R. A. Schilperoort (Dordrecht: Kluwer Academic Publishers), 1–22.
Grüner, R., Strompen, G., Pfitzner, A. J. P., and Pfitzner, U. M. (2003). Salicylic acid and the hypersensitive response initiate distinct signal transduction pathways in tobacco that converge on the as-1-like element of the PR-1a promoter. Eur. J. Biochem. 270, 4876–4886. doi: 10.1046/j.1432-1033.2003.03888.x
Guan, K. L., and Dixon, J. E. (1991). Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192, 262–267. doi: 10.1016/0003-2697(91)90534-Z
Hussain, R. M. F. (2012). WRKY Transcription Factors Involved in PR-1 Gene Expression in Arabidopsis. Doctoral thesis, Leiden Universitym, Leiden.
James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two hybrid selection in yeast. Genetics 144, 1425–1436.
Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15, 1846–1858. doi: 10.1105/tpc.012211
Katagiri, F., Lam, E., and Chua, N. H. (1989). Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340, 727–730. doi: 10.1038/340727a0
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kesarwani, M., Yoo, J., and Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346. doi: 10.1104/pp.106.095299
Kim, K. C., Lai, Z., Fan, B., and Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371. doi: 10.1105/tpc.107.055566
Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. doi: 10.1105/tpc.12.12.2339
Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR1 gene expression in Arabidopsis. Plant J. 16, 223–233. doi: 10.1046/j.1365-313x.1998.00288.x
Linthorst, H. J. M. (1991). Pathogenesis-related proteins of plants. Crit. Rev. Plant Sci. 10, 123–150. doi: 10.1080/07352689109382309
Miao, Y., Laun, T. M., Smykowski, A., and Zentgraf, U. (2007). Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol. Biol. 65, 63–76. doi: 10.1007/s11103-007-9198-z
Miao, Z. H., and Lam, E. (1995). Construction of a trans-dominant inhibitor for members of the TGA family of transcription factors conserved in higher plants. Plant J. 7, 887–896. doi: 10.1046/j.1365-313X.1995.07060887.x
Miao, Z. H., Liu, X., and Lam, E. (1994). TGA3 is a distinct member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 25, 1–11. doi: 10.1007/BF00024193
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
Ndamukong, I. A., Abdallat, A., Thurow, C., Fode, B., Zander, M., Weigel, R., et al. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50, 128–139. doi: 10.1111/j.1365-313X.2007.03039.x
Niggeweg, R., Thurow, C., Kegler, C., and Gatz, C. (2000a). Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905. doi: 10.1074/jbc.M909267199
Niggeweg, R., Thurow, C., Weigel, R., Pfitzner, U., and Gatz, C. (2000b). Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol. 42, 775–788. doi: 10.1023/A:1006319113205
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Pape, S., Thurow, C., and Gatz, C. (2010). The Arabidopsis PR1 promoter contains multiple integration sites for the coactivator NPR1 and the repressor SNI1. Plant Physiol. 154, 1805–1818. doi: 10.1104/pp.110.165563
Park, C. Y., Lee, J. H., Yoo, J. H., Moon, B. C., Choi, M. S., Kang, Y. H., et al. (2005). WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 579, 1545–1550. doi: 10.1016/j.febslet.2005.01.057
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Pontier, D., Miao, Z. H., and Lam, E. (2001). Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. Plant J. 27, 529–538. doi: 10.1046/j.1365-313X.2001.01086.x
Qin, X. F., Holuigue, L., Horvath, D. M., and Chua, N. H. (1994). Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6, 863–874. doi: 10.1105/tpc.6.6.863
Rochon, A., Boyle, P., Wignes, T., Fobert, P. R., and Després, C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of its BTB/ POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18, 3670–3685. doi: 10.1105/tpc.106.046953
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schirawski, J., Planchais, S., and Haenni, A. L. (2000). An improved protocol for the preparation of protoplasts from an established Arabidopsis thaliana cell suspension culture and infection with RNA of a turnip yellow mosaic tymovirus: a simple and reliable method. J. Virol. 86, 85–94. doi: 10.1016/S0166-0934(99)00173-1
Shen, Q. H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103. doi: 10.1126/science.1136372
Spoel, S. H., Mou, Z., Tada, Y., Spivey, N. W., Genschik, P., and Dong, X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137, 860–872. doi: 10.1016/j.cell.2009.03.038
Strompen, G., Grüner, R., and Pfitzner, U. M. (1998). An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol. Biol. 37, 871–883. doi: 10.1023/A:1006003916284
Töpfer, R., Matzeit, V., Gronenborn, B., Schell, J., and Steinbiss, H. H. (1987). A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15:5890. doi: 10.1093/nar/15.14.5890
van der Fits, L., and Memelink, J. (1997). Comparison of the activities of CaMV 35S and FMV 34S promoter derivatives in Catharanthus roseus cells transiently and stably transformed by particle bombardment. Plant Mol. Biol. 33, 943–946. doi: 10.1023/A:1005763605355
van Verk, M. C., Neeleman, L., Bol, J. F., and Linthorst, H. J. M. (2011). Tobacco transcription factor NtWRKY12 interacts with TGA2.2 in vitro and in vivo. Front Plant Sci. 2:32. doi: 10.3389/fpls.2011.00032
van Verk, M. C., Pappaioannou, D., Neeleman, L., Bol, J. F., and Linthorst, H. J. M. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995. doi: 10.1104/pp.107.112789
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. doi: 10.1371/journal.ppat.0020123
Wehner, N., Hartmann, L., Ehlert, A., Böttner, S., Oñate-Sánchez, L., and Dröge-Laser, W. (2011). High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant J. 68, 560–569. doi: 10.1111/j.1365-313X.2011.04704.x
Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. doi: 10.1105/tpc.105.037523
Yamasaki, K., Kigawa, T., Inoue, M., Tateno, M., Yamasaki, T., Yabuki, T., et al. (2005). Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 17, 944–956. doi: 10.1105/tpc.104.026435
Yamasaki, K., Kigawa, T., Watanabe, S., Inoue, M., Yamasaki, T., Seki, M., et al. (2012). Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor. J. Biol. Chem. 287, 7683–7691. doi: 10.1074/jbc.M111.279844
Yanagisawa, S. (2004). Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 45, 386–391. doi: 10.1093/pcp/pch055
Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR1 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Zhang, Y., Tessaro, M. J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15, 2647–2653. doi: 10.1105/tpc.014894
Zheng, Z., Qamar, S. A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. doi: 10.1111/j.1365-313X.2006.02901.x
Zhou, J. M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., et al. (2000). NPR1differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13, 191–202. doi: 10.1094/MPMI.2000.13.2.191
Keywords: AtWRKY50, TGA, Arabidopsis, PR1, SAR, EMSA
Citation: Hussain RMF, Sheikh AH, Haider I, Quareshy M and Linthorst HJM (2018) Arabidopsis WRKY50 and TGA Transcription Factors Synergistically Activate Expression of PR1. Front. Plant Sci. 9:930. doi: 10.3389/fpls.2018.00930
Received: 11 March 2018; Accepted: 11 June 2018;
Published: 13 July 2018.
Edited by:
Jens Staal, Ghent University, BelgiumReviewed by:
Amit Bhardwaj, Langone Medical Center, New York University, United StatesBhaskar Gupta, Government General Degree College, Singur, India
Copyright © 2018 Hussain, Sheikh, Haider, Quareshy and Linthorst. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arsheed H. Sheikh, arsheedsheikh@gmail.com
 Rana M. F. Hussain1
Rana M. F. Hussain1 Arsheed H. Sheikh
Arsheed H. Sheikh Imran Haider
Imran Haider