- LIPM, Université de Toulouse, INRA, CNRS, Toulouse, France
The rhizobia-legume symbiosis is a mutualistic association in which bacteria provide plants with nitrogen compounds and the plant provides bacteria with carbon sources. A successful symbiotic interaction relies on a molecular dialog between the plant and the bacteria, and generally involves rhizobial lipo-chitooligosaccharide signals called Nod factors (NFs). In most cases, specific NF perception is required for rhizobia to enter root cells through newly formed intracellular structures called infection threads (ITs). Concomitantly to IT formation in root hairs, root cortical cells start to divide to create a new root organ called the nodule, which will provide the bacteria with a specific micro-environment required for symbiotic nitrogen fixation. During all these steps of plant–bacteria interaction, new plant cellular compartments and developmental programs are activated. This interaction is costly for the plant that tightly controls symbiosis establishment and functioning. Phytohormones are key regulators of cellular and developmental plasticity in plants, and they are influential endogenous signals that rapidly control plant responses. Although early symbiotic responses were known for decades to be linked to phytohormone-related responses, new data reveal the molecular mechanisms involved and links between phytohormones and the control of early symbiotic events. Reciprocally, NF signaling also targets phytohormone signaling pathways. In this review, we will focus on the emerging notion of NF and phytohormone signaling crosstalk, and how it could contribute to the tight control of symbiosis establishment in legume host plants.
Introduction
Legume plants can interact with soil bacteria, named rhizobia, to establish the rhizobium legume (RL) symbiosis. Legumes host Rhizobia in specific root organs called nodules, where they fix atmospheric nitrogen. This symbiosis provides the plant with nitrogen compounds (ammonium) and the plant provides the bacteria with carbon sources. The efficiency of this interaction relies for a great part on the massive intracellular infection of plant cells by rhizobia and the protective structure of the nodule. The plant host must invest a great deal of energy to both house and support nitrogen fixing bacteria, and as such, must tightly regulate this process.
Nod factors (NFs) are lipo-chitooligosaccharide (LCO) molecules produced by rhizobia in response to flavonoids present in root exudates. NFs are generally essential for the onset and the maintenance of the RL symbiosis (Dénarié, 1996). Intriguingly, NFs can also stimulate lateral root formation via the symbiotic signaling pathway implying that similar host mechanisms are involved in both nodule and lateral root development (Olah et al., 2005). Genetic pathways governing NF perception and signaling are now quite well understood (Gough and Cullimore, 2011). NFs are perceived by the plant at the root epidermis and are likely produced by rhizobia all along the infection process (Sharma and Signer, 1990).
Rhizobia first attach to root hairs (RHs) in a susceptible zone of host roots (Bhuvaneswari et al., 1981) and then enter RHs through a membrane invagination called an infection thread (IT). Even before the IT is formed, pericycle cells are activated and divide to start organogenesis of the nodule (Timmers et al., 1999). NFs are usually essential for the infection process and, in Medicago sativa, purified NFs can trigger pericycle activation and initiate nodule organogenesis (Truchet et al., 1991; Timmers et al., 1999). Defects in the infection process often result in defective nodule organogenesis. Infection and organogenesis events of the RL symbiosis are thus tightly coordinated. This tight coordination and signaling across root layers, and the events of cellular reactivation and cell division associated with symbiotic establishment strongly imply the involvement of hormonal pathways.
Recent transcriptomic data suggest that NF signaling regulates the host hormone biosynthesis and signal transduction pathways. Hormones function in the pico to nano-molar range (Gaspar et al., 2003). Their activity depends on a tight balance of activation (biosynthesis) and inactivation (conjugation, degradation) pathways, as well as transport and signaling. In this mini-review, we will focus on data showing positive and negative feedback regulatory mechanisms between NF signaling and hormonal pathways. We present data from legumes producing both indeterminate (Medicago truncatula, pea) and determinate [Lotus japonicus, Glycine max (soybean)] nodules. These two types of nodule differ in the site of initial cortical divisions, persistency of the nodule meristem and auxin sensitivity (Bensmihen, 2015; Ng and Mathesius, 2018). Effects of NF signaling on both hormone homeostasis and hormone signaling will be discussed.
NF Signaling and Regulation of Hormone Homeostasis
Phytohormones are key regulators of plant growth and responses to biotic and abiotic stresses. Several transcriptomic studies show that hormone biosynthesis, activation or degradation genes are differentially expressed upon NF treatment (see Table 1 and Figure 1).
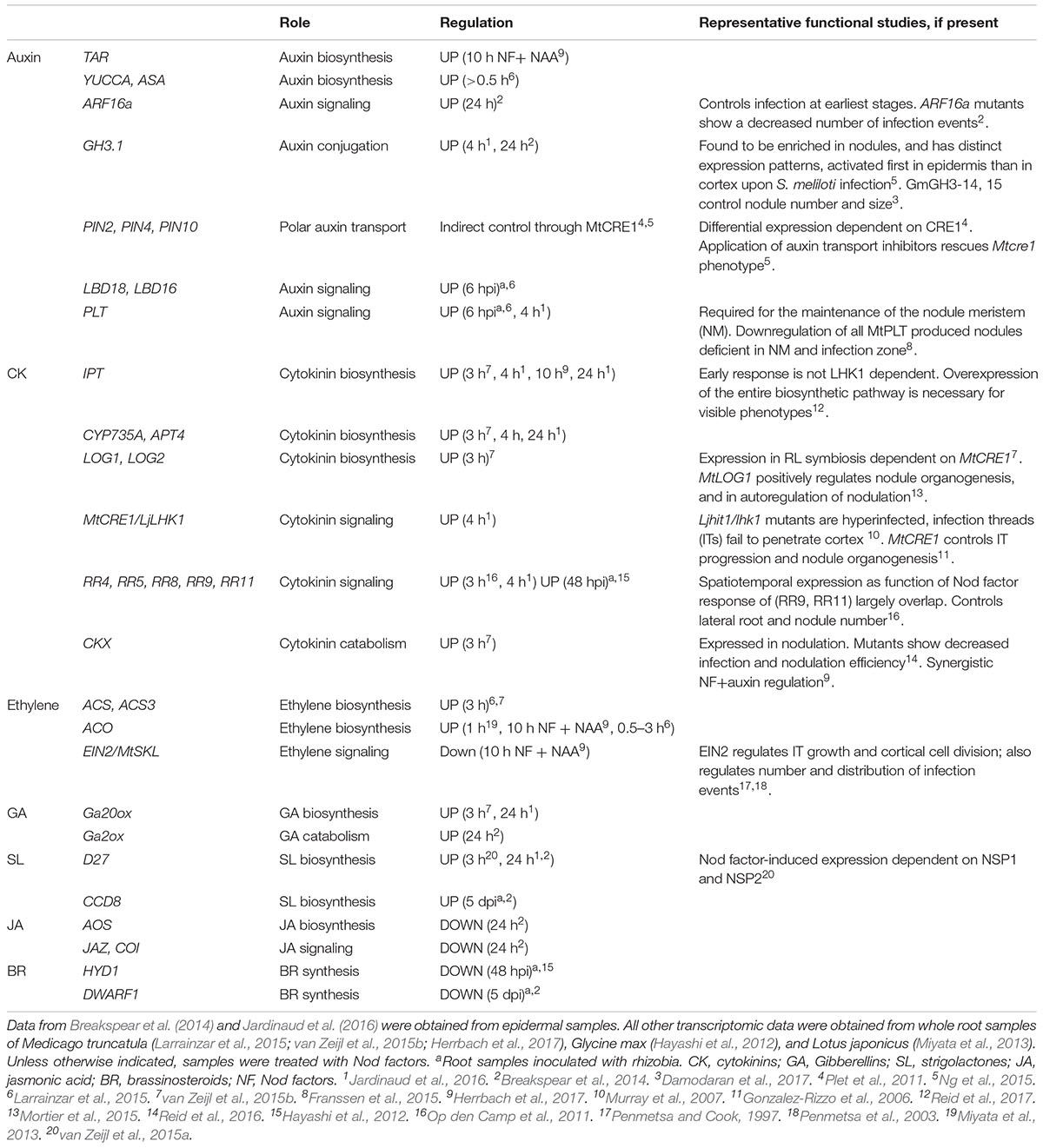
TABLE 1. Summary of major hormone homeostasis and signaling genes differentially regulated by Nod factors or rhizobia during symbiosis.
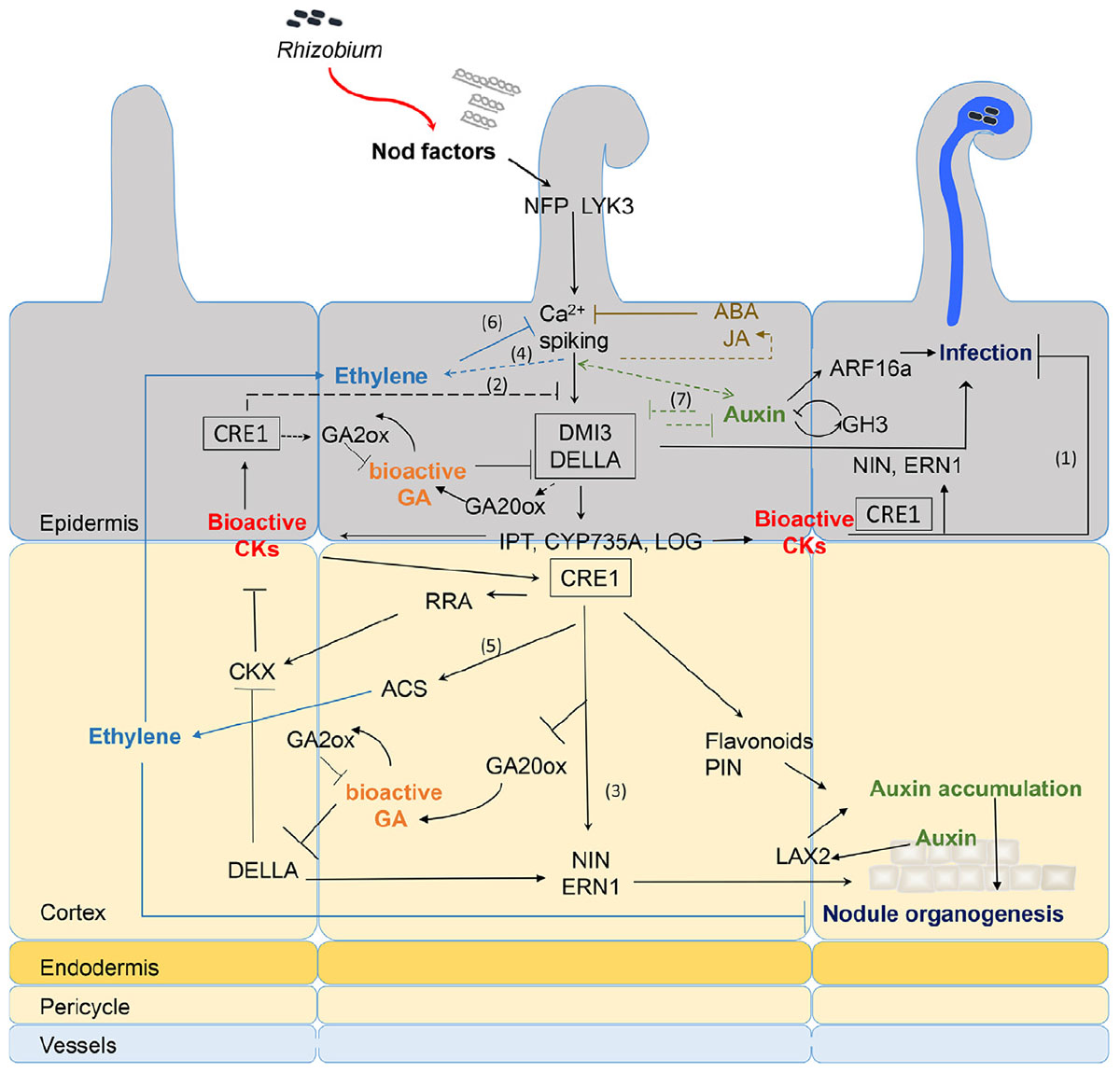
FIGURE 1. Recapitulative spatio-temporal scheme of the interactions between NF signaling and downstream hormonal pathways. Rhizobia produce NFs that are perceived in root hairs (RHs) through LysM-RLK receptors such as NFP and LYK3. NF perception leads to calcium spiking and activation of DMI3 that acts upstream of the CK receptor MtCRE1. The signaling cascade involving DMI3 and CRE1 is likely involved in both epidermal and cortical signaling (black boxes). NF treatment triggers early CK biosynthesis gene (IPT/CYP735A/LOG) expression and CK accumulation in M. truncatula roots downstream of DMI3 but independently of CRE1 signaling (van Zeijl et al., 2015b). Although tissue specificity of this CK production was not determined, evidence from L. japonicus and M. truncatula suggests that epidermal CK accumulation is a negative regulator of RH infection (1) and NF signaling (2) (Held et al., 2014; Jardinaud et al., 2016). In contrast, cortical CK is a positive regulator of nodule organogenesis (3) (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Reid et al., 2017). Bioactive CKs are perceived by CRE1 and induce expression of NIN and ERN1 (Ariel et al., 2012), which are positive regulators of both infection and nodule organogenesis (Andriankaja et al., 2007; Marsh et al., 2007). This induction might be partly through regulation of DELLA activities. GA is a negative regulator of DELLA protein stability. Bioactive GA pools are likely present in both epidermis and cortex early after NF treatment (Fonouni-Farde et al., 2016; Jardinaud et al., 2016; Herrbach et al., 2017). DELLAs play a positive regulatory role on symbiotic gene expression such as ERN1 and they negatively regulate CK degradation (Fonouni-Farde et al., 2016, 2017; Jin et al., 2016). In contrast, CK positively regulate GA inactivation enzymes and down-regulate expression of the GA20ox activation enzyme (Fonouni-Farde et al., 2018), suggesting a negative feedback of CK on GA active pools. NF signaling induces ethylene production, both independently of the LHK1/CRE1 CK pathway (4) (Reid et al., 2018) and downstream of CK perception (5) (van Zeijl et al., 2015b). Ethylene reduces CK production in M. truncatula roots, possibly through negative feedback on NF signaling (6) (van Zeijl et al., 2015b). Ethylene negatively regulates NF-induced calcium spiking, RH infection, and nodule organogenesis (Heidstra et al., 1997; Penmetsa et al., 2008). Regulation of auxin biosynthetic and conjugation enzyme (GH3) genes occurs in NF treated RHs and upon S. meliloti inoculation in an NF-dependent manner (Breakspear et al., 2014; Larrainzar et al., 2015; Jardinaud et al., 2016; Herrbach et al., 2017). Reciprocal positive and negative feedback regulatory loops between some auxin and NF regulated genes (7) was shown by comparing the combined effect of auxin and NFs to either treatment alone (Herrbach et al., 2017). Downstream of CK perception, control of auxin transport in the cortex seems regulated by differential expression of PIN genes (Plet et al., 2011) or accumulation of flavonoid compounds. Auxin accumulation is also mediated by MtLAX2 that is induced upon S. meliloti infection in vasculature and early nodule primordia, and which is required for nodule organogenesis (Roy et al., 2017). In parallel, epidermal auxin signaling controls infection thread (IT) formation, at least partly through ARF16a (Breakspear et al., 2014). Exogenous application of high concentrations (μM range) of ABA or JA inhibits NF-induced calcium spiking (Sun et al., 2006; Ding et al., 2008) but this inhibition is so far not supported by any transcriptomic data. Plain bars represent negative and plain arrows positive regulations. Dashed lines are hypothetical relationships, and solid lines have evidence from the literature. Different hormones are highlighted by different colors. CK, cytokinins; GA, gibberellins; ABA, abscisic acid; JA, jasmonic acid; NF, Nod factors. Bacteria entrapped in curled root hair and IT are shown in blue.
Cytokinins
Biosynthesis and Activation
Numerous studies document the role of CKs as key regulators of nodule organogenesis (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007) and IT formation/progression (Held et al., 2014; Jardinaud et al., 2016). Recent transcriptomic data show that expression of CK biosynthesis genes such as isopentenyl transferases (IPT) and CYP735A that contribute to the biosynthesis of the bioactive CK form trans-zeatin, as well as CK activation genes such as LONELY GUYs (LOGs), can be induced by a 3 h NF treatment (van Zeijl et al., 2015b). Moreover, MtIPT1and MtCYP735A1 regulation is independent of the CK receptor MtCRE1, suggesting a direct (and possibly local) production of CK as an early event following NF signaling. Similarly, LjIPT2 and LjLOG4 are regulated by NF application independently of the CK receptor LjLHK1 in L. japonicus (Reid et al., 2017). Accumulation of bioactive CKs is also detected in the root susceptible zone following a 3 h NF treatment, and this is dependent on the NF signaling gene MtDMI3 (van Zeijl et al., 2015b). Several genes from the trans-zeatin metabolic network are regulated in RHs after a 24 h NF treatment, suggesting accumulation of the bioactive CK trans-zeatin upon NF treatment. However, bioactive CK levels have not been measured in NF treated RHs (Jardinaud et al., 2016).
Inactivation and Degradation
Interestingly, CK inactivation enzymes adenyl phosphoribosides transferases (APT) and cytokinin oxidases (CKX) are also induced by a 3 h NF treatment but in an MtCRE1 dependent manner, suggesting downstream negative regulatory feedback loops on CK accumulation (see Figure 1). Indeed, expression of the AtCKX3 gene in M. truncatula plants under an epidermis specific promoter enhanced the number of ITs and nodules formed upon rhizobium inoculation. In contrast, expression of AtCKX3 under a cortical specific promoter reduced nodulation (Jardinaud et al., 2016). In L. japonicus, NFs also induce expression of the LjCKX3 gene but its promoter activity is specific to inner root tissues. Inactivation of LjCKX3 leads to enhanced production of bioactive CK forms and reduces nodule organogenesis and IT formation (Reid et al., 2016). Altogether, this suggests that reducing the levels of active CKs may increase the efficiency of the infection process. This fits with the model suggested by Held et al. (2014) that argues for an inhibitory feedback loop driven by CK accumulation in the epidermis at late stages of nodulation. This is also consistent with antagonistic effects of CK application on the induction of the NF responsive gene MtENOD11. Indeed, a 3 h CK pretreatment of M. truncatula roots significantly reduces the induction of MtENOD11 by NF. Conversely, epidermal expression of the AtCKX3 gene enhanced the NF induction of MtENOD11 (Jardinaud et al., 2016). In summary, growing evidence suggests that NFs induce CK production, which first controls nodule organogenesis, then rapidly activates negative feedbacks on NF signaling and infection processes, notably in the epidermis.
Gibberellins
Increasing evidence also suggests a role for gibberellins (GAs) in controlling early events of symbiosis. A 24 h NF treatment induces both GA metabolic and biosynthetic genes in M. truncatula RHs, suggesting accumulation of the bio-active GA precursor GA12 in the root epidermis upon NF treatment (Breakspear et al., 2014; Jardinaud et al., 2016). An important NF-dependent and transient activation of GA biosynthesis genes was observed in soybean roots 12 hpi with rhizobia (Hayashi et al., 2012). Interestingly, 10-7 M GA3 treatment was previously shown to suppress NF-induced expression of the transcription factors NSP2 and NIN in L. japonicus and 10-6 M GA3 suppressed NF induced RH deformation (Maekawa et al., 2009). Likewise, pretreatment of M. truncatula roots with 10-6 M GA3 suppressed NF dependent ENOD11 induction. NF treatment also induced expression of the GA catabolic and biosynthetic enzymes GA2 and GA20ox, respectively (Fonouni-Farde et al., 2016). Moreover, this GA2ox catabolic gene was positively regulated by a 10 h NF treatment (Herrbach et al., 2017) and down-regulated in RHs after a 24 h NF treatment (Zaat et al., 1989; Breakspear et al., 2014). These data suggest that NFs can rapidly induce the biosynthesis of bioactive GA in RHs, which consequently triggers a negative feedback leading both to the downregulation of NF signaling and the activation of GA catabolic enzymes (see Figure 1). Increasing evidence also shows that bioactive GAs negatively regulate nodulation and infection in both determinate and indeterminate nodules (Maekawa et al., 2009; Fonouni-Farde et al., 2016). Thus, NF induced GAs could help fine-tune NF signaling and rhizobium infection during symbiosis.
Ethylene
Previous genetic, physiologic, and pharmacologic studies highlight the key regulatory role ethylene plays in early symbiotic processes (Zaat et al., 1989; Heidstra et al., 1997; Oldroyd et al., 2001; Penmetsa et al., 2008). NFs can induce the expression of several ethylene biosynthetic genes, including two 1-aminocyclopropane-1-carboxylate (ACC) synthase genes (MtACS1 and MtACS2; van Zeijl et al., 2015b). Moreover, three ACS genes are upregulated by NF signaling at 6 hpi with rhizobium (Larrainzar et al., 2015). Similarly, NF dependent ethylene production is detected in L. japonicus roots as early as 6 hpi (Reid et al., 2018). MtACS3 is also synergistically regulated by a 10 h treatment with a combination of NFs and auxin (Herrbach et al., 2017). Promoter:GUS fusions showed RH expression of three ACS genes in M. truncatula (Larrainzar et al., 2015). This is consistent with previous data obtained in L. japonicus where a 1 h treatment with 10-8 M NF increased the expression of the ACC oxidase LjACO2, which was synergistically regulated by the combined application of NFs and ACC (Miyata et al., 2013). In contrast, NF-dependent downregulation of ethylene biosynthesis was observed at 48 hpi in the susceptible zone of soybean roots (Hayashi et al., 2012). Altogether, these data suggest that NF signaling rapidly and transiently induces the production of a negative regulator of infection, ethylene.
Auxin
Auxin Metabolism
Nod factor-dependent induction of genes encoding auxin biosynthetic enzymes such as YUCCA and ANTHRANILATE SYNTHASE (ASA) was observed early after Sinorhizobium meliloti inoculation in whole roots (Larrainzar et al., 2015). Several auxin signaling genes are activated in RHs after a 24 h NF treatment (Breakspear et al., 2014), which argues for an effect of NFs on auxin production and signaling (see section “Auxin Signaling”). Similarly, we recently described a synergistic effect of NFs and auxin on the transcription of a large number of hormone biosynthesis genes, including an auxin biosynthesis TRYPTOPHAN AMINOTRANSFERASE-related (TAR2) homolog (Herrbach et al., 2017). Interestingly, a GH3.1 gene, predicted to encode an auxin conjugation and inactivation enzyme (Yang et al., 2015) is expressed both in NF treated RHs as well as developing nodules (Breakspear et al., 2014; Ng et al., 2015; Jardinaud et al., 2016). We found this same GH3 gene to be synergistically upregulated by NFs and auxin (Herrbach et al., 2017). This suggests fine tuning of bioactive pools of auxin, in RHs and probably inner cortical tissues for indeterminate nodules, upon NF perception and rhizobium infection.
Auxin Transport
Several studies link NF signaling to auxin transport. Spot inoculation of compatible rhizobia (but not a non-host strain) or micro-targeting of specific LCOs can modify auxin gradients, as measured by the GH3:GUS reporter gene in white clover (Mathesius et al., 1998). Likewise, flavonoid application also inhibits auxin transport. NF application or S. meliloti infection inhibits auxin transport from shoot to root at 24 h, as well as regulating expression of some MtPIN auxin efflux transporter genes, in an MtCRE1 dependent manner (van Noorden et al., 2006; Plet et al., 2011; Ng et al., 2015). Moreover, MtCRE1 dependent pathways also control the accumulation of flavonoids in M. truncatula roots upon infection and flavonoid application can rescue the Mtcre1 nodulation phenotype. These data suggest that NF induced, CK signaling triggers flavonoid induction and the subsequent inhibition of polar auxin transport. The resulting accumulation of auxin initiates cortical cell division and nodule organogenesis (Figure 1).
Strigolactones
Strigolactones (SGLs) show a dose-dependent effect on nodulation in M. truncatula (Gomez-Roldan et al., 2008; De Cuyper et al., 2015). Interestingly, several studies (Larrainzar et al., 2015; van Zeijl et al., 2015a; Herrbach et al., 2017) showed a direct NF regulation of the expression of the SGL biosynthesis gene DWARF27 (D27). The promoter of the D27 gene is active in the root epidermis after a 3 h NF treatment and upon early steps of nodule organogenesis (van Zeijl et al., 2015a). Moreover, we observed that a combined auxin+NF treatment reduced the NF induction of D27 expression, suggesting that auxin can antagonize NF effects (Herrbach et al., 2017).
Jasmonic Acid (JA) and Brassinosteroids (BR)
Jasmonic acid (JA) and BR regulate plant stress responses and plant growth, but their role in RL symbiosis is not well understood. Conflicting reports indicate that host responses to these hormones vary depending on the legume species, hormone dose, or type of treatment studied (see Ferguson and Mathesius, 2014 for review and Conclusion and Perspectives section below). Some evidence suggests NF regulation of JA biosynthetic enzymes. Larrainzar et al. (2015) observed NF-dependent induction of two JA biosynthesis genes at 3–6 hpi and Breakspear et al. (2014) observed downregulation of a few JA biosynthetic genes in RHs after a 24 h NF treatment. In contrast, NF-dependent upregulation of several JA biosynthesis genes was observed in soybean at 48 hpi, while there seemed to be a reduction in expression of BR biosynthetic genes (Hayashi et al., 2012).
Hence, these data suggest that NF-signaling regulates several hormonal biosynthesis and activation pathways, with subsequent negative feedback loops that are rapidly activated after a brief NF treatment or upon rhizobium inoculation. Further evidence for a temporal regulation of bioactive hormone pools by NF signaling comes from the downstream regulation of many hormonal signaling genes (Table 1).
NF Regulation of Hormone Signaling Genes
CK Signaling
CK signaling utilizes a two-component signaling system comprised of a histidine-kinase (HK) receptor, phospho-transfer proteins (AHPs), and response regulators (RRs). Two types of RRs operate in CK signal transduction: A-type RRs are transcriptionally regulated upon CK perception but do not contain a DNA binding domain, and are generally considered as negative regulators of the CK response. In contrast, B-type RRs contain a Myb DNA binding domain, function as transcription factors and are generally considered as positive regulators (Kieber and Schaller, 2014). In M. truncatula and L. japonicus, the HK genes MtCRE1 and LjLHK1 have positive roles in cortical cell divisions and nodule organogenesis (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007). Recent transcriptomic and promoter fusion analyses demonstrate the induction of these CK receptor genes upon rhizobium infection or NF application; in Lotus, this epidermal expression inhibits infection (Held et al., 2014; Jardinaud et al., 2016).
Independent data suggest NF induced, RR gene expression occurs downstream of CK receptor activation. For instance, the A-type MtRR4, MtRR8, MtRR9, MtRR11, and the B-type MtRR1 are expressed in M. truncatula roots following inoculation with either NF or wild-type S. meliloti, but not with nod- S. meliloti mutants (Gonzalez-Rizzo et al., 2006; Op den Camp et al., 2011; van Zeijl et al., 2015b; Jardinaud et al., 2016). Likewise, an HK and an RR (GmRR5) gene are also induced in soybean in an NF-dependent manner (Hayashi et al., 2012). Finally, the CK signaling reporter construct, TCSn:GUS, can be induced in M. truncatula RHs upon a 4 h NF treatment (Jardinaud et al., 2016).
Altogether, an early activation of CK signaling probably occurs both in RHs and cortical cells, directly downstream of the NF signaling pathway (Figure 1).
Auxin Signaling
Accumulating evidence suggests that NFs regulate the expression of auxin signaling genes. NF application induces the auxin related transcription factors genes MtARF16a and MtPLETHORA3 in RHs, while several ARF genes are downregulated in RHs after a 24 h NF treatment (Breakspear et al., 2014; Jardinaud et al., 2016). These genes are also induced by auxin in M. truncatula roots (Herrbach et al., 2017). This suggests that NF perception could lead to auxin accumulation that would in turn activate some specific auxin signaling genes (Figure 1). Such genes could control cell divisions or IT formation (Breakspear et al., 2014).
GA Signaling
DELLAs are well-known negative regulators of GA signaling (Sun, 2011). Recent data showed a positive effect of DELLAs on nodulation, and more specifically on IT formation (Fonouni-Farde et al., 2016; Jin et al., 2016). DELLAs reside in the NSP1/NSP2/IPD3 transcription factor complex that mediates NF signaling (Fonouni-Farde et al., 2016; Jin et al., 2016). External application of GAs can repress NF induction of several genes, such as ERN1, ENOD11, NSP2, and NIN (Maekawa et al., 2009; Fonouni-Farde et al., 2016). However, NFs do not induce DELLA gene expression (Fonouni-Farde et al., 2016).
Ethylene and JA Signaling
Larrainzar et al. (2015) observed a biphasic regulation of expression of 47 AP2/ERF transcription factor genes in M. truncatula upon S. meliloti infection. The first wave of AP2/ERF expression takes place 1 hpi independent of NF perception, whereas the second wave of NF dependent, ethylene-regulated transcription occurs 6 hpi (Larrainzar et al., 2015). Although little is known about JA signaling during symbiosis, a 24 h NF treatment downregulates the expression of the putative JA receptor COI1 in M. truncatula RHs implying that NF perception may attenuate this signaling pathway (Breakspear et al., 2014).
Conclusion and Perspectives
Growing evidence shows that early events of NF signaling lead to the production and/or activation of hormones such as CK and GAs and the activation of their downstream signaling pathways. These hormones, in turn, interfere with other hormonal pathways, such as ethylene, ABA, and auxin, which can impinge upon NF signaling itself. For example, external application of CK or GA inhibits NF induction of MtENOD11 (Fonouni-Farde et al., 2016; Jardinaud et al., 2016). Similarly, JA, ABA, and ethylene are well-known negative regulators of NF signaling (Oldroyd et al., 2001; Sun et al., 2006; Ding et al., 2008). We observed that auxin both positively and negatively regulated different NF responsive genes in a combined auxin + NF treatment (Herrbach et al., 2017) providing further evidence for the regulation of symbiosis signaling pathways by hormones. These negative effects may underpin the mechanism by which the host controls nodulation following the initial NF/rhizobium recognition.
It should, however, be noted that the concentrations of hormones applied in these studies are often in the micro-molar range. In the absence of quantitative data reporting endogenous hormone levels during symbiotic responses, it is difficult to assess if these studies reflect biologically relevant responses to plant hormones and this field requires such analyses.
Furthermore, recent evidence suggests that hormones function in a tissue-specific manner during symbiotic responses (see Gamas et al., 2017) but the specificity of these hormonal pathways is only partly resolved. Future work should address the respective contribution of different root tissues in producing, sensing, and responding to NF induced hormones, using tissue specific tools or quantitative hormone biosensors (Jones et al., 2014; Liao et al., 2015).
Finally, the NF signaling pathway overlaps with other symbiotic and developmental pathways suggesting that hormones may also impinge upon these processes. Similar to Rhizobia, arbuscular mycorrhizal fungi produce LCOs and utilize the same core signaling pathway to initiate symbiosis (Maillet et al., 2011). Future work should address whether LCO/ hormone crosstalk regulates mycorrhization, where evidence already implicates a role for auxin, SGLs, GA, and LCO pathways (Lauressergues et al., 2012; Floss et al., 2013; Etemadi et al., 2014). Likewise, LCOs stimulate LR development and some NF induced genes are also involved in LR development (Herrbach et al., 2017). How LCO/hormone crosstalk could be involved in LCO stimulation of lateral root formation and how this could compare to its role in nodulation still requires further investigation.
Author Contributions
SB and WB drafted the review and produced Figure 1. SB wrote the review with help of WB. WB produced Table 1.
Funding
Research conducted by WB and SB at the LIPM was partly supported by the “Laboratoire d’Excellence” (LABEX) entitled TULIP (ANR-10-LABX-41). WB’s Ph.D. is funded by a joint French Embassy and Commission on Higher Education from the Philippines Government (CHED-Phil France) program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Clare Gough and Frédéric Debellé for critical reading of the manuscript. We apologize for colleagues whose work was not cited due to space limitation or our focus on crosstalk between NF and hormone signaling aspects only.
References
Andriankaja, A., Boisson-Dernier, A., Frances, L., Sauviac, L., Jauneau, A., Barker, D. G., et al. (2007). AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19, 2866–2885. doi: 10.1105/tpc.107.052944
Ariel, F., Brault-Hernandez, M., Laffont, C., Huault, E., Brault, M., Plet, J., et al. (2012). Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula. Plant Cell 24, 3838–3852. doi: 10.1105/tpc.112.103267
Bensmihen, S. (2015). Hormonal control of lateral root and nodule development in legumes. Plants (Basel) 4, 523–547. doi: 10.3390/plants4030523
Bhuvaneswari, T. V., Bhagwat, A. A., and Bauer, W. D. (1981). Transient susceptibility of root cells in four common legumes to nodulation by rhizobia. Plant Physiol. 68, 1144–1149. doi: 10.1104/pp.68.5.1144
Breakspear, A., Liu, C., Roy, S., Stacey, N., Rogers, C., Trick, M., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26, 4680–4701. doi: 10.1105/tpc.114.133496
Damodaran, S., Westfall, C. S., Kisely, B. A., Jez, J. M., and Subramanian, S. (2017). Nodule-enriched GRETCHEN HAGEN 3 enzymes have distinct substrate specificities and are important for proper soybean nodule development. Int. J. Mol. Sci. 18:2547. doi: 10.3390/ijms18122547
De Cuyper, C., Fromentin, J., Yocgo, R. E., De Keyser, A., Guillotin, B., Kunert, K., et al. (2015). From lateral root density to nodule number, the strigolactone analogue GR24 shapes the root architecture of Medicago truncatula. J. Exp. Bot. 66, 137–146. doi: 10.1093/jxb/eru404
Dénarié, J., Debellé, F., and Promé, J. C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65, 503–535. doi: 10.1146/annurev.bi.65.070196.002443
Ding, Y., Kalo, P., Yendrek, C., Sun, J., Liang, Y., Marsh, J. F., et al. (2008). Abscisic acid coordinates Nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20, 2681–2695. doi: 10.1105/tpc.108.061739
Etemadi, M., Gutjahr, C., Couzigou, J. M., Zouine, M., Lauressergues, D., Timmers, A., et al. (2014). Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 166, 281–292. doi: 10.1104/pp.114.246595
Ferguson, B. J., and Mathesius, U. (2014). Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 40, 770–790. doi: 10.1007/s10886-014-0472-7
Floss, D. S., Levy, J. G., Lévesque-Tremblay, V., Pumplin, N., and Harrison, M. J. (2013). DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 110, E5025–E5034. doi: 10.1073/pnas.1308973110
Fonouni-Farde, C., Kisiala, A., Brault, M., Emery, R. J. N., Diet, A., and Frugier, F. (2017). DELLA1-Mediated gibberellin signaling regulates cytokinin-dependent symbiotic nodulation. Plant Physiol. 175, 1795–1806. doi: 10.1104/pp.17.00919
Fonouni-Farde, C., McAdam, E., Nichols, D., Diet, A., Foo, E., and Frugier, F. (2018). Cytokinins and the CRE1 receptor influence endogenous gibberellin levels in Medicago truncatula. Plant Signal. Behav. 13:e1428513. doi: 10.1080/15592324.2018.1428513
Fonouni-Farde, C., Tan, S., Baudin, M., Brault, M., Wen, J., Mysore, K. S., et al. (2016). DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat Commun 7:12636. doi: 10.1038/ncomms12636
Franssen, H. J., Xiao, T. T., Kulikova, O., Wan, X., Bisseling, T., Scheres, B., et al. (2015). Root developmental programs shape the Medicago truncatula nodule meristem. Development 142, 2941–2950. doi: 10.1242/dev.120774
Gamas, P., Brault, M., Jardinaud, M. F., and Frugier, F. (2017). Cytokinins in symbiotic nodulation: when, where, what for? Trends Plant Sci. 22, 792–802. doi: 10.1016/j.tplants.2017.06.012
Gaspar, T., Kevers, C., Faivre-rampant, O., Crèvecoeur, M., Penel, C., Greppin, H., et al. (2003). Changing concepts in plant hormone action. Soc. In Vitro Biol._ Plant 39, 85–106. doi: 10.1079/IVP2002393
Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pages, V., Dun, E. A., Pillot, J. P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189–194. doi: 10.1038/nature07271
Gonzalez-Rizzo, S., Crespi, M., and Frugier, F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18, 2680–2693. doi: 10.1105/tpc.106.043778
Gough, C., and Cullimore, J. (2011). Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol. Plant Microbe Interact. 24, 867–878. doi: 10.1094/MPMI-01-11-0019
Hayashi, S., Reid, D. E., Lorenc, M. T., Stiller, J., Edwards, D., Gresshoff, P. M., et al. (2012). Transient Nod factor-dependent gene expression in the nodulation-competent zone of soybean (Glycine max [L.] Merr.) roots. Plant Biotechnol. J. 10, 995–1010. doi: 10.1111/j.1467-7652.2012.00729.x
Heidstra, R., Yang, W. C., Yalcin, Y., Peck, S., Emons, A. M., van Kammen, A., et al. (1997). Ethylene provides positional information on cortical cell division but is not involved in Nod factor-induced root hair tip growth in Rhizobium-legume interaction. Development 124, 1781–1787.
Held, M., Hou, H., Miri, M., Huynh, C., Ross, L., Hossain, M. S., et al. (2014). Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell 26, 678–694. doi: 10.1105/tpc.113.119362
Herrbach, V., Chirinos, X., Rengel, D., Agbevenou, K., Vincent, R., Pateyron, S., et al. (2017). Nod factors potentiate auxin signaling for transcriptional regulation and lateral root formation in Medicago truncatula. J. Exp. Bot. 68, 568–582. doi: 10.1093/jxb/erw474
Jardinaud, M. F., Boivin, S., Rodde, N., Catrice, O., Kisiala, A., Lepage, A., et al. (2016). A laser dissection-RNAseq analysis highlights the activation of cytokinin pathways by Nod factors in the Medicago truncatula root epidermis. Plant Physiol. 171, 2256–2276. doi: 10.1104/pp.16.00711
Jin, Y., Liu, H., Luo, D., Yu, N., Dong, W., Wang, C., et al. (2016). DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 7:12433. doi: 10.1038/ncomms12433
Jones, A. M., Danielson, J. A., Manojkumar, S. N., Lanquar, V., Grossmann, G., and Frommer, W. B. (2014). Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3:e01741. doi: 10.7554/eLife.01741
Kieber, J. J., and Schaller, G. E. (2014). Cytokinins. Arabidopsis Book 12, e0168. doi: 10.1199/tab.0168
Larrainzar, E., Riely, B. K., Kim, S. C., Carrasquilla-Garcia, N., Yu, H. J., Hwang, H. J., et al. (2015). Deep Sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol. 169, 233–265. doi: 10.1104/pp.15.00350
Lauressergues, D., Delaux, P. M., Formey, D., Lelandais-Brière, C., Fort, S., Cottaz, S., et al. (2012). The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 72, 512–522. doi: 10.1111/j.1365-313X.2012.05099.x
Liao, C. Y., Smet, W., Brunoud, G., Yoshida, S., Vernoux, T., and Weijers, D. (2015). Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207–210, 2 p following 210. doi: 10.1038/nmeth.3279
Maekawa, T., Maekawa-Yoshikawa, M., Takeda, N., Imaizumi-Anraku, H., Murooka, Y., and Hayashi, M. (2009). Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 58, 183–194. doi: 10.1111/j.1365-313X.2008.03774.x
Maillet, F., Poinsot, V., André, O., Puech-Pages, V., Haouy, A., Gueunier, M., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63. doi: 10.1038/nature09622
Marsh, J. F., Rakocevic, A., Mitra, R. M., Brocard, L., Sun, J., Eschstruth, A., et al. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144, 324–335. doi: 10.1104/pp.106.093021
Mathesius, U., Schlaman, H. R., Spaink, H. P., Of Sautter, C., Rolfe, B. G., and Djordjevic, M. A. (1998). Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 14, 23–34. doi: 10.1046/j.1365-313X.1998.00090.x
Miyata, K., Kawaguchi, M., and Nakagawa, T. (2013). Two distinct EIN2 genes cooperatively regulate ethylene signaling in Lotus japonicus. Plant Cell Physiol. 54, 1469–1477. doi: 10.1093/pcp/pct095
Mortier, V., Wasson, A., Jaworek, P., De Keyser, A., Decroos, M., Holsters, M., et al. (2014). Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula. New Phytol. 202, 582–593. doi: 10.1111/nph.12681
Murray, J. D., Karas, B. J., Sato, S., Tabata, S., Amyot, L., and Szczyglowski, K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315, 101–104. doi: 10.1126/science.1132514
Ng, J. L., Hassan, S., Truong, T. T., Hocart, C. H., Laffont, C., Frugier, F., et al. (2015). Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. Plant Cell 27, 2210–2226. doi: 10.1105/tpc.15.00231
Ng, J. L. P., and Mathesius, U. (2018). Acropetal auxin transport inhibition is involved in indeterminate but not determinate nodule formation. Front. Plant Sci. 9:169. doi: 10.3389/fpls.2018.00169
Olah, B., Briere, C., Bécard, G., Dénarié, J., and Gough, C. (2005). Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J. 44, 195–207. doi: 10.1111/j.1365-313X.2005.02522.x
Oldroyd, G. E., Engstrom, E. M., and Long, S. R. (2001). Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13, 1835–1849. doi: 10.1105/tpc.13.8.1835
Op den Camp, R. H., De Mita, S., Lillo, A., Cao, Q., Limpens, E., Bisseling, T., et al. (2011). A phylogenetic strategy based on a legume-specific whole genome duplication yields symbiotic cytokinin type-a response regulators. Plant Physiol. 157, 2013–2022. doi: 10.1104/pp.111.187526
Penmetsa, R. V., and Cook, D. R., (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275, 527–530. doi: 10.1126/science.275.5299.527
Penmetsa, R. V., Frugoli, J. A., Smith, L. S., Long, S. R., and Cook, D. R. (2003). Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 131, 998–1008. doi: 10.1104/pp.015677
Penmetsa, R. V., Uribe, P., Anderson, J., Lichtenzveig, J., Gish, J. C., Nam, Y. W., et al. (2008). The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 55, 580–595. doi: 10.1111/j.1365-313X.2008.03531.x
Plet, J., Wasson, A., Ariel, F., Le Signor, C., Baker, D., Mathesius, U., et al. (2011). MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65, 622–633. doi: 10.1111/j.1365-313X.2010.04447.x
Reid, D., Liu, H., Kelly, S., Kawaharada, Y., Mun, T., Andersen, S. U., et al. (2018). Dynamics of ethylene production in response to compatible nod factor. Plant Physiol. 176, 1764–1772. doi: 10.1104/pp.17.01371
Reid, D., Nadzieja, M., Novák, O., Heckmann, A. B., Sandal, N., and Stougaard, J. (2017). Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiol. 175, 361–375. doi: 10.1104/pp.17.00832
Reid, D. E., Heckmann, A. B., Novák, O., Kelly, S., and Stougaard, J. (2016). CYTOKININ OXIDASE/DEHYDROGENASE3 maintains cytokinin homeostasis during root and Nodule Development in Lotus japonicus. Plant Physiol. 170, 1060–1074. doi: 10.1104/pp.15.00650
Roy, S., Robson, F., Lilley, J., Liu, C. W., Cheng, X., Wen, J., et al. (2017). MtLAX 2, a functional homologue of the Arabidopsis auxin influx transporter AUX 1, is required for nodule organogenesis. Plant Physiol. 174, 326–338. doi: 10.1104/pp.16.01473
Sharma, S. B., and Signer, E. R. (1990). Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev. 4, 344–356. doi: 10.1101/gad.4.3.344
Sun, J., Cardoza, V., Mitchell, D. M., Bright, L., Oldroyd, G., and Harris, J. M. (2006). Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J. 46, 961–970. doi: 10.1111/j.1365-313X.2006.02751.x
Sun, T. P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21, R338–R345. doi: 10.1016/j.cub.2011.02.036
Timmers, A. C., Auriac, M. C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126, 3617–3628.
Tirichine, L., Sandal, N., Madsen, L. H., Radutoiu, S., Albrektsen, A. S., Sato, S., et al. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315, 104–107. doi: 10.1126/science.1132397
Truchet, G., Roche, P., Lerouge, P., Vasse, J., Camut, S., de Billy, F., et al. (1991). Sulphated lipo-oligosaccharide signals of Rhizobium meliloti elicit root nodule organogenesis in alfalfa. Nat. Rev. Genet. 351, 670–673. doi: 10.1038/351670a0
van Noorden, G. E., Ross, J. J., Reid, J. B., Rolfe, B. G., and Mathesius, U. (2006). Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 140, 1494–1506. doi: 10.1104/pp.105.075879
van Zeijl, A., Liu, W., Xiao, T. T., Kohlen, W., Yang, W. C., Bisseling, T., et al. (2015a). The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis. BMC Plant Biol. 15:260. doi: 10.1186/s12870-015-0651-x
van Zeijl, A., Op den Camp, R., Deinum, E., Charnikhova, T., Franssen, H., Op den Camp H., et al. (2015b). Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant 8, 1213–1226. doi: 10.1016/j.molp.2015.03.010
Yang, Y., Yue, R., Sun, T., Zhang, L., Chen, W., Zeng, H., et al. (2015). Genome-wide identification, expression analysis of GH3 family genes in Medicago truncatula under stress-related hormones and Sinorhizobium meliloti infection. Appl. Microbiol. Biotechnol. 99, 841–854. doi: 10.1007/s00253-014-6311-5
Zaat, S. A., Van Brussel, A. A., Tak, T., Lugtenberg, B. J., and Kijne, J. W. (1989). The ethylene-inhibitor aminoethoxyvinylglycine restores normal nodulation by Rhizobium leguminosarum biovar. viciae on Vicia sativa subsp. nigra by suppressing the ’Thick and short roots’ phenotype. Planta 177, 141–150. doi: 10.1007/BF00392802
Keywords: auxin, cytokinins, gibberellin, ethylene, strigolactone, infection, nodule organogenesis, LCOs
Citation: Buhian WP and Bensmihen S (2018) Mini-Review: Nod Factor Regulation of Phytohormone Signaling and Homeostasis During Rhizobia-Legume Symbiosis. Front. Plant Sci. 9:1247. doi: 10.3389/fpls.2018.01247
Received: 06 April 2018; Accepted: 06 August 2018;
Published: 03 September 2018.
Edited by:
Pascal Ratet, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Jeanne Marie Harris, University of Vermont, United StatesBrendan Keith Riely, University of California, Davis, United States
Copyright © 2018 Buhian and Bensmihen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandra Bensmihen, sandra.bensmihen@inra.fr
 William P. Buhian
William P. Buhian Sandra Bensmihen
Sandra Bensmihen