- 1Laboratory of Cotton Disease, Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Department of Plant Pathology, University of California, Davis, Davis, CA, United States
- 3U.S. Agricultural Research Station, Salinas, CA, United States
- 4Crop Improvement and Protection Research Unit, United States Department of Agriculture, Agricultural Research Service, Salinas, CA, United States
Cysteine-rich receptor-like kinases (CRKs) are a large subfamily of plant receptor-like kinases that play a critical role in disease resistance in plants. However, knowledge about the CRK gene family in cotton and its function against Verticillium wilt (VW), a destructive disease caused by Verticillium dahliae that significantly reduces cotton yields is lacking. In this study, we identified a total of 30 typical CRKs in a Gossypium barbadense genome (GbCRKs). Eleven of these (>30%) are located on the A06 and D06 chromosomes, and 18 consisted of 9 paralogous pairs encoded in the A and D subgenomes. Phylogenetic analysis showed that the GbCRKs could be classified into four broad groups, the expansion of which has probably been driven by tandem duplication. Gene expression profiling of the GbCRKs in resistant and susceptible cotton cultivars revealed that a phylogenetic cluster of nine of the GbCRK genes were up-regulated in response to V. dahliae infection. Virus-induced gene silencing of each of these nine GbCRKs independently revealed that the silencing of GbCRK18 was sufficient to compromise VW resistance in G. barbadense. GbCRK18 expression could be induced by V. dahliae infection or jasmonic acid, and displayed plasma membrane localization. Therefore, our expression analyses indicated that the CRK gene family is differentially regulated in response to Verticillium infection, while gene silencing experiments revealed that GbCRK18 in particular confers VW resistance in G. barbadense.
Introduction
Plants are often infected by various pathogens including bacteria, fungi, oomycetes, and nematodes, but unlike animals, plants lack adaptive/acquired immune system, and rely entirely on innate immunity to resist numerous potential pathogens in the environment (Jones and Dangl, 2006; Boller and Felix, 2009; Zhou et al., 2017). To counteract pathogen invasion, plants have evolved different types of resistance proteins which activate immune responses and restrict pathogen proliferation (Chisholm et al., 2006; Jones and Dangl, 2006; Dodds and Rathjen, 2010), and these proteins can be classified into different types based primarily upon the presence of specific conserved structural motifs (Martin et al., 2003; Kruijt et al., 2005; Joshi and Nayak, 2011). Among these, a diverse array of plasma membrane-bound receptors are employed by plants to perceive endogenous and exogenous signals for regulation of immunity, and these cell surface receptors include receptor-like kinases (RLKs) and receptor-like proteins (RLPs) that harbor different extracellular domains for perception of distinct ligands (He et al., 2018).
The plant RLKs were first discovered in maize (Walker and Zhang, 1990) and represent the most abundant molecular recognition receptors found in plants. More than 600 genes encoding protein-like receptor kinases have been found in Arabidopsis, accounting for 2.5% of its gene pool (Shiu and Bleecker, 2001). Nearly 1300 genes in rice encode RLKs (Shiu et al., 2004). The typical RLK contains a signal peptide, a variable extracellular domain, a transmembrane domain, and an intracellular kinase domain (Van et al., 1994). RLKs are divided into different families according to their extracellular ligand binding domain, such as leucine-rich repeats (LRRs), lectin, lysine motif (LysM), and wall-associated kinases (WAK) (Tör et al., 2009). In general, the N-terminal extracellular region (the ectodomain) of the RLKs extends into the apoplast where it perceives the signals from the pathogen, whereas the C-terminal kinase domain resides within the cytoplasm and relays the perceived signals into the intracellular environment (Kimura et al., 2017).
Cysteine-rich receptor-like kinases (CRKs), also called domain of unknown function 26 (DUF26) RLKs, have a role in disease resistance and plant cell death (Chen et al., 2003). Extracellular domains of CRKs contain two copies of the DUF26 motif and have conserved Cys residues within the sequence C-X8-C-X2-C. The C-X8-C-X2-C domain is a novel motif structurally distinct from the Cys-rich region of S-locus glycoproteins or S-domain RLKs (Chen, 2001). These conserved Cys residues may participate in the formation of the three-dimensional structure of the protein through disulfide bonds or form zinc finger motifs, as in many DNA-binding transcription factors. Both disulfide bonds and zinc fingers are known to mediate protein–protein interactions, a critical step in the activation of many receptors, including those with protein kinase domains upon ligand binding (Hardie, 1999).
Cysteine-rich receptor-like kinases involved in plant defense responses have been well-studied in Arabidopsis and other model systems: overexpression of Arabidopsis CRK5, CRK6, CRK36, and CRK45 can significantly enhance resistance against Pseudomonas syringae pv. tomato DC3000, via rapid expression of the defense genes and ROS production (Chen et al., 2003; Zhang X. et al., 2013; Yeh et al., 2015); CRK4, CRK19, and CRK20 can be significantly induced by salicylic acid (SA) and pathogen infection (Chen et al., 2004; Ederli et al., 2011); CRK6 and CRK7 have an important role in extracellular ROS signaling (Idänheimo et al., 2014); up-regulation of CRK13 leads to the hypersensitive response associated with cell death, and induces defense against pathogens by causing increased accumulation of SA (Acharya et al., 2007); CRK28 and CRK29 can be induced in response to flagellin; and overexpression of CRK28 in Nicotiana benthamiana that results in cell death (Yadeta et al., 2017). Aside from these examples, the wheat CRK TaCRK1 is a resistance factor against Rhizoctonia cerealis (Yang et al., 2013). In barley, a CRK gene designated as HvCRK1, which encodes an endoplasmic reticulum-associated protein involved in the negative regulation of basal resistance in barley against Blumeria graminis f.sp. hordei (Rayapuram et al., 2012).
Verticillium wilt (VW) is caused by soil-borne Verticillium dahliae, a fungus with a broad host range of over 400 plant species, and this pathogen can survive in soil for many years (Tao et al., 2016). In cotton, VW is a devastating disease that results in major losses in yield and boll quality (Li et al., 2011). Improving host resistance is considered the most optimal method to manage VW, which entails identifying VW resistance genes in cotton germplasm and incorporating them into elite cotton varieties. To date, several genes have been characterized that contribute to defense responses against VW, including GbCAD1 and GbSSI2 (Gao et al., 2013), GbRLK (Zhao et al., 2015), GbSTK (Zhang Y. et al., 2013), GbTLP1 (Munis et al., 2010), GbSBT1 (Duan et al., 2016), GhPAO (Mo et al., 2015), GbNRX1 (Li et al., 2016), GbRVd (Yang et al., 2016), and GbVe/GbVe1/Gbvdr5 (Zhang et al., 2011; Zhang B. et al., 2012; Yang et al., 2015).
Currently, genomic analysis is an effective means of transferring knowledge from one taxon to another (Zhang Y. et al., 2012). At present, the genomes of diploid and allotetraploid cottons have been released and can facilitate our understanding of the expansion and functions of the CRK family genes in cotton. Except for one study (Zhang et al., 2018), no CRKs involved in disease resistance have been reported in cotton. The allotetraploid cotton G. barbadense is generally recognized as a VW-resistant species that contains abundant resistance gene resources (Zhang J. et al., 2012). The objectives of this current study were to: (1) identify members of the CRK gene family in G. barbadense genome; (2) elucidate the structure and characteristics of the G. barbadense CRK gene family; (3) investigate the candidate CRK genes involved in VW in cotton; (4) explore the relationships between plant hormones and defense responses mediated by the candidate CRK genes.
Materials and Methods
Identification of the Cotton CRK Gene Family
An available genomic database of G. barbadense was downloaded from http://database.chgc.sh.cn/cotton/index.html; and for the other three cotton species, Gossypium arboretum, Gossypium raimondii, and Gossypium hirsutum, data were all downloaded from https://www.cottongen.org/; data for Arabidopsis thaliana, Zea mays, and Oryza sativa were downloaded from http://www.plantgdb.org/. The Pfam protein family databases1 with the Stress-antifung (PF01657) and Pkinase (PF00069) (Finn et al., 2014) domains were used with the HMMER software version 3.0 (Eddy, 2011) to identify the CRK genes in these seven species. Putative gene sequences were further verified using the SMART database (Letunic and Bork, 2018) and InterproScan (Hunter et al., 2012). Stress-antifung domains of the CRK proteins were investigated statistically using MEME website2. Prediction of signal peptides and signal peptide cleavage sites of putative extracellular proteins were conducted using SignalP (version 4.1; D-Score cut-off set to 0.500) (Petersen et al., 2011). The CRK proteins with signal peptides were analyzed for the presence of transmembrane domains using TMHMM 2.0 (Krogh et al., 2001).
Mapping, Phylogenetic Tree Construction, and Structural Analysis
MapInspect3 was used to analyze the distribution of the CRK genes in the G. barbadense genome. The multiple sequence alignments of CRK proteins from seven species were carried out using ClustalX (version 1.83). A phylogenetic tree was built by MEGA 6 software4 using the neighbor-joining (NJ) method (bootstrap value was set 1000). The exon/intron structures were analyzed by aligning the genomic DNA sequences with their corresponding coding sequences using the online Gene Structure Display Server (GSDS)5. The similarity of 30 CRK genes was then compared and analyzed using local BLAST analysis.
Plant Materials and Treatments
Seedlings of G. barbadense cv. Hai 7124 and G. barbadense cv. GZ 57 were used for gene expression analyses under different stress treatments. Plants were grown in the same controlled environment chambers under 16 h light/8 h dark cycle at 28°C for 2 weeks. Seedlings of cv. Hai 7124 and cv. GZ 57, which exhibit, respectively, resistance and susceptibility to V. dahliae, were inoculated with the V. dahliae isolate Vd991 using the root dip-inoculation method (Wang et al., 2011). Vd991, a highly aggressive defoliating strain of V. dahliae, was cultured in complete medium (CM) at 25°C for 5 days. The concentration of conidia was adjusted to 5 × 106 conidia/ml using deionized water and subsequently used for inoculation of seedlings (Li et al., 2018). The seedling roots were harvested with three replicates at eight time intervals (0, 2, 6, 12, 24, 48, 72, and 120 h) after Vd991 inoculation. Each replicate consisted of five seedlings, which were quickly frozen in liquid nitrogen and stored at -80°C for RNA extraction.
Leaves of 3-week-old cotton seedlings were sprayed with 10 mM SA, 10 mM ethephon (ETH), 10 mM methyljasmonate (MeJA), or 100 mM abscisic acid (ABA), respectively. Sterile distilled water served as a solvent control, and the leaves were collected at seven time points (0, 2, 6, 12, 24, 48, and 72 h) for RNA extraction. Three plants per replicate were harvested at each time point and stored at -80°C after flash-freezing in liquid nitrogen.
GbCRK18 was silenced in G. barbadense cv. Hai 7124 through virus-induced gene silencing (VIGS), and the expression level of GbCRK18 was assessed in the silenced plants to determine the efficiency of silencing. After determining the success of silencing protocol, wild-type and silenced plants were inoculated with V. dahliae Vd991 using a root dip-inoculation at 5 × 106 conidia/ml. Twelve hours after inoculation, cotton roots were harvested and washed with water for RNA extraction to detect the expression of marker resistance genes involved in the JA pathway. Empty vector pTRV2-silenced plants served as controls.
RNA Isolation and Analyses of Gene Expression
Total RNA was extracted from cotton seedling roots using a Plant RNA Purification Kit (Tiangen, Beijing, China). The cDNA was prepared using M-MLV reverse transcriptase and reverse transcription (RT)-qPCR analyses were conducted using the SYBR Premix Ex Taq kit (TaKaRa, Japan) with a QuantStudio 6 Flex qPCR System (Applied Biosystems, Foster City, CA, United States). Primers used for RT-qPCR amplification are listed in Supplementary Table S1 and were designed to avoid conserved regions within the members of the CRK family. The cotton 18S gene was used as an internal control to normalize the variance among samples. RT-qPCR conditions consisted of an initial denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 20 s. Relative expression levels were evaluated using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Cloning of the GbCRK18 Gene
Primers were designed according to the full open-reading frame (ORF) of the gene GOBAR_DD09713 in the G. barbadense reference genome (Liu et al., 2015; Supplementary Table S1). These primers were used to amplify the target fragment from G. barbadense cDNA. PCR conditions consisted of an initial 95°C denaturation step for 10 min, followed by 39 cycles at 95°C for 45 s, 55°C for 45 s, and 72°C for 2.5 min. All PCR products were cloned into the pGEM-T-Easy vector (Promega, Madison, WI, United States) and transformed into Escherichia coli DH5α. At least six clones were arbitrarily selected and sequenced.
Functional Characterization of Candidate CRK Genes via Virus-Induced Gene Silencing (VIGS) Analysis
For the VIGS assays, one fragment from each gene, GbCRK02, GbCRK03, GbCRK06, GbCRK07, GbCRK08, GbCRK18, GbCRK19, GbCRK22, and GbCRK23, was amplified from G. barbadense cDNA and integrated into the vector pTRV2 to construct pTRV2:CRK.n, which was introduced into Agrobacterium tumefaciens GV3101. Primers used for construction of the above VIGS vectors are listed in Supplementary Table S1. Agrobacterium strains harboring pTRV2:CRK.n plasmid were combined with Agrobacterium strains harboring the pTRV1 vector were mixed in a 1:1 ratio and co-infiltrated into the cotyledons of 2-week-old resistant cv. Hai 7124 plants, with cv. GZ 57 serving as a susceptible control. Control vector pTRV2:CLA1 was used to evaluate the effectiveness of VIGS since effective silencing of this gene leads to the development of white leaves. Two weeks later, white leaves were observed on plants in which the CLA1 gene had been targeted and silenced by VIGS, then plantlets were subjected to V. dahliae inoculation with 10 ml of conidial suspension (5 × 106 conidia/ml), and VW symptoms were investigated at 3 weeks post-inoculation. Seedling shoots were cut to investigate the vascular wilt symptom under a microscope (Liu et al., 2013; Chen et al., 2017). For fungal biomass quantification in planta, stems of three inoculated plants were harvested at 20 days post inoculation. qPCR was conducted with template genomic DNA isolated from the samples using the SYBR Premix Ex Taq kit (Takara, Japan). V. dahliae elongation factor 1-α (EF-1α) and cotton 18S genes were used to quantify fungal colonization by qPCR (Santhanam et al., 2013).
Subcellular Localization Analysis of the CRK18
To examine the subcellular localization of GbCRK18, we inserted the full-length GbCRK18 coding region into the pBin-GFP4 vector to generate a C-terminal fusion with the GFP gene under the control of a cauliflower mosaic virus (CaMV) 35S promoter (p35S:GbCRK18). Plasmids harboring GFP alone (empty vector, p35S:GFP) were used as controls. The p35S:GFP control and p35S:GbCRK18 vectors fused to GFP were transiently expressed in tobacco epidermal cells by infiltration with Agrobacterium. Subcellular localization of the p35S:GFP and p35S:GbCRK18 fusion proteins was observed by laser scanning confocal microscopy (LSMT-PMT) with excitation and emission wave lengths of 488 and 510 nm, respectively.
Results
Genome-Wide Identification of CRK Gene Family
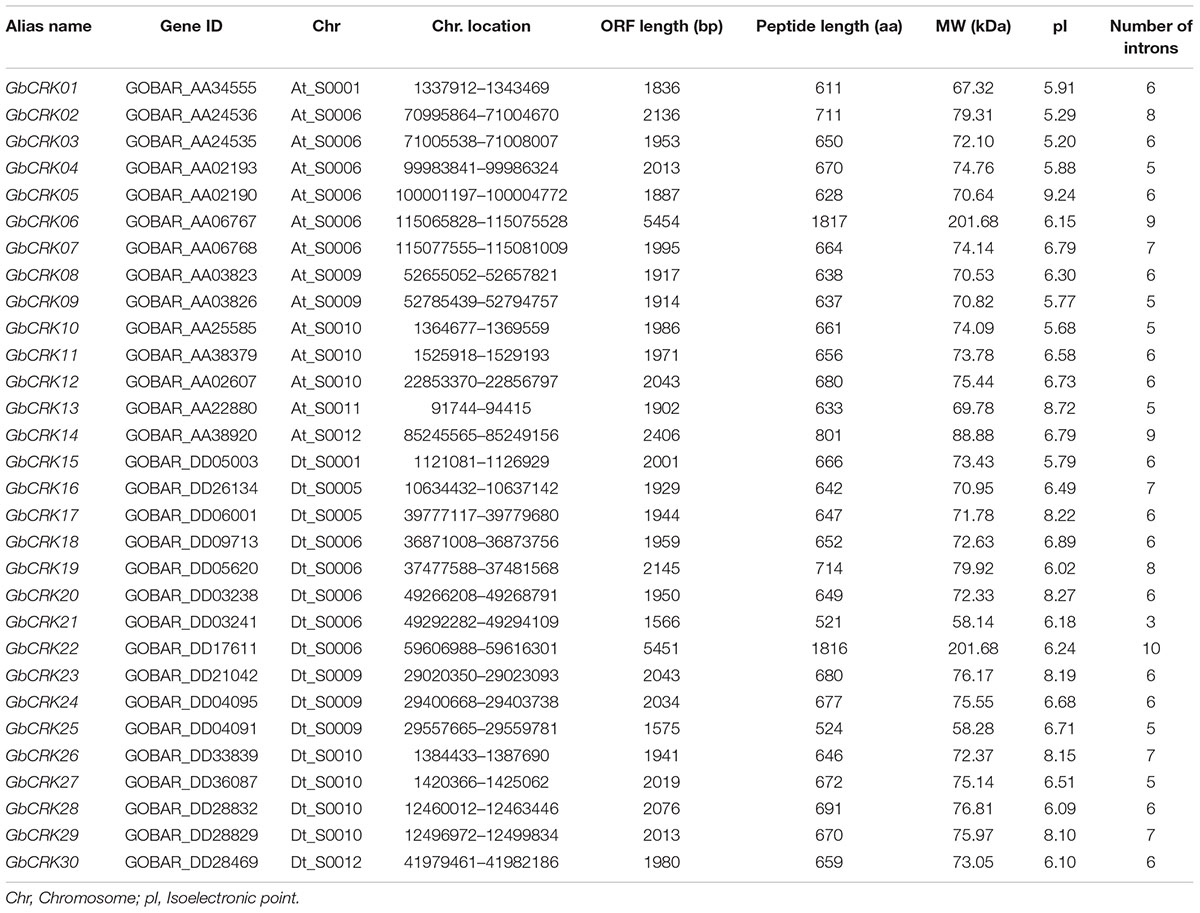
To evaluate the CRK gene family in a cotton genome, we performed an HMMER alignment search on the proteome of the G. barbadense genome with the hidden Markov model (HMM) profile of the Stress-antifung domain (PF01657) and Pkinase domain (PF00069). Manual selection resulted in the identification of 63 candidate CRK family members (Supplementary Table S2), and all of them were further subjected to domain analysis using SMART and InterproScan, which defined a set of typical CRK proteins that included a signal peptide, two stress-antifung domains, one transmembrane domain (TM), and one Pkinase domain (Figure 1A). A total of 30 CRK genes (GbCRK) were identified in the G. barbadense genome, which were numbered from GbCRK1 to GbCRK30 according to their localization on chromosomes (Table 1). The peptides encoded by GbCRKs consisted of 521–1817 amino acids, with putative molecular weights ranging from 58.14 to 201.68 kDa and isoelectric points ranging from 5.20 to 9.24 (Table 1). Multiple alignments revealed amino acid frequencies at each position were conserved in the homologous domain sequence (Figure 1B and Supplementary Figure S1). For instance, all cotton CRK family members displayed three conserved cysteine residues in two stress-antifung domains (Figure 1B), which is a typical characteristic of the stress-antifung domain (C-X8-C-X2-C; X represents random residues) (Chen, 2001).
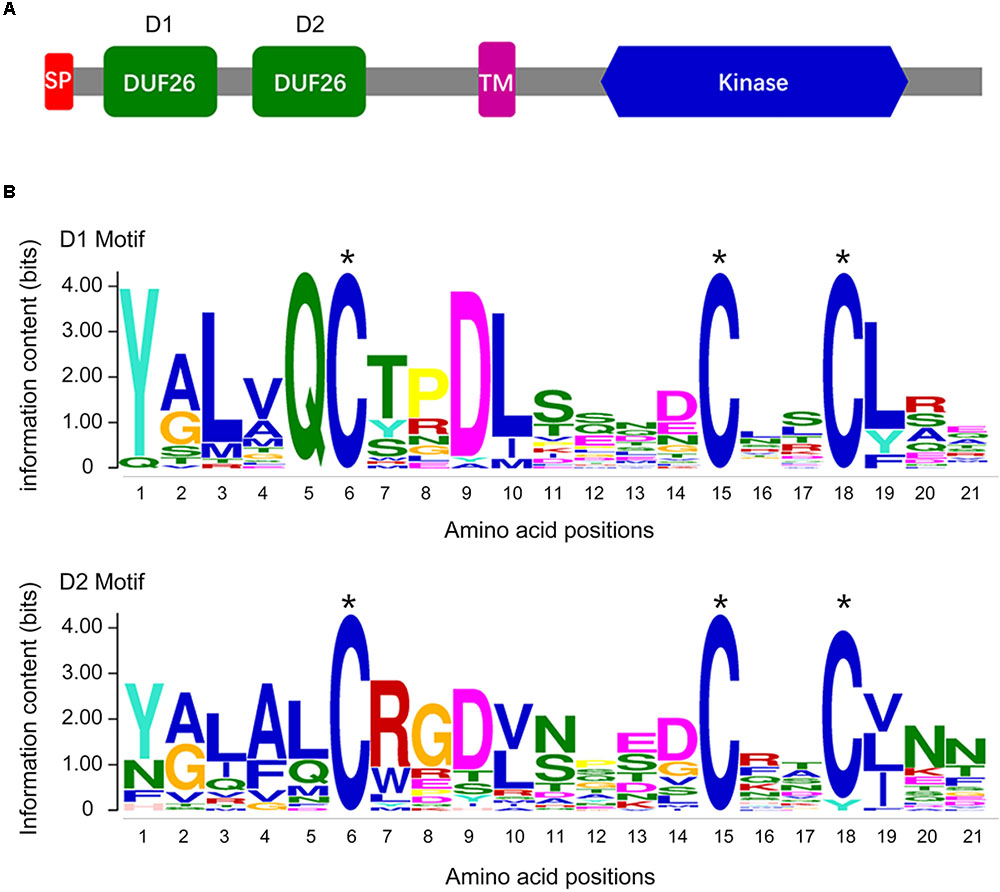
FIGURE 1. The DUF26 domain organization and the conserved structure of the CRK family. (A) Conserved structure of the CRK family: a signal peptide (SP), two stress-antifung domains (DUF26), a transmembrane domain (TM), and an intracellular kinase domain (Kinase); (B) conserved DUF26 domain.
Structural and Phylogenetic Analyses of the Cotton CRK Gene Family
Gene structure analysis of GbCRKs showed that the exon/intron organization varied in number and length (number of introns ranged from 3 to 10) (Supplementary Figure S1). Chromosome localization analysis revealed that 30 GbCRKs were distributed on 12 of the 26 chromosomes in cotton, and more than 30% of these genes (11 GbCRKs) were distributed on chromosomes A06 and D06 (Supplementary Figure S2). Similarity analysis revealed that 18 GbCRKs exhibited high identities (>80%), and that this similarity was observed pairwise, thus constituting nine different pairs (Figure 2), representative of nine paralogous pairs between the A and D sub-genome (Supplementary Figure S2). Chromosome physical location identification of the GbCRKs showed that most of them were present in tandem repeats (Supplementary Table S2); GbCRK23, GbCRK24, and GbCRK25 were located in the same position on chromosome A09, whereas GbCRK17 and GbCRK30 which shared a high sequence identity (>70%) were located on different chromosomes (A05 and A07) (Figure 2), suggesting that tandem duplications and segmental duplications have occurred in cotton.
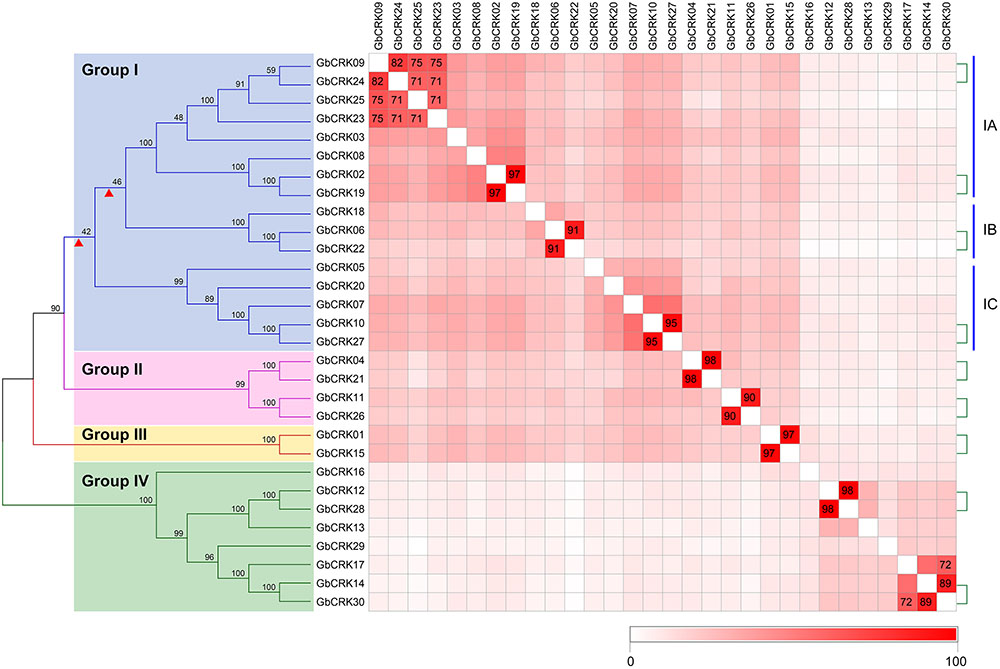
FIGURE 2. Phylogenetic analyses of CRK genes in Gossypium barbadense. Phylogenetic analyses divide the cotton CRK family into four subfamilies, here designated Group I–IV. Genes with similarities greater than 70% were plotted. The nine pairs of genes linked by the green-dotted lines are paralogous genes. Colored blocks from white to red represent the identity between genes from 0 to 100%; darker shades of red indicate higher levels of similarity.
To assess sequence polymorphisms of the CRK gene family in cotton, an unrooted NJ tree was constructed using full-length amino-acid sequences. The cotton CRK gene family was classified into four subgroups (Group I–IV) with 2–16 members in each subgroup (Figure 2). Bootstrap values of each subgroup were high, especially for Group II, III, and IV (Figure 2), suggesting that the GbCRKs may be derived from a small set of genes and that duplication events and subsequent sequence evolution occurred during cotton genome evolution. As expected, nine paralogous pairs of GbCRKs in tandem or segmental duplications were clustered in the same branch due to their high identities (Figure 2 and Supplementary Figure S2). The bootstrap values of 16 GbCRKs enriched in Group I were low among the different sub-clades (Group IA, IB, IC, ID) (Figure 2). Notably, all the GbCRKs in Group I are located on chromosomes 6 (A06 and D06) and 9 (A09 and D09) (11 and 5 CRKs, respectively) (Figure 2 and Supplementary Figure S2).
Investigations of the gene structures in the nine pairs of paralogous GbCRKs also showed that the numbers or lengths of introns were variable, except for those of the paralogous pair, GbCRK12 and GbCRK28 (Figure 2 and Supplementary Figure S1). Together, these results implied that the CRK gene family may have expanded by duplication from few common ancestral genes and followed by CRK sequence divergence in cotton.
Comparative Analysis of the CRK Genes in Plant Genomes
To gain insight into the evolution of the CRK gene family in cotton compared with other plant hosts, the phylogenetic relationships of CRK protein sequences from other six plant genomes were investigated by constructing an unrooted tree comprising representatives of three other cotton species, two monocotyledonous plants, O. sativa and Z. mays, and one dicotyledonous plant, A. thaliana. Using similar bioinformatics-driven methods, a total of 170 CRK genes were identified from these genome, including 21 in G. raimondii (GrCRK01–GrCRK21), 19 in G. arboreum (GaCRK01–GaCRK19), 37 in G. hirsutum (GhCRK01–GaCRK37), 33 in A. thaliana (AtCRK01–AtCRK33), 22 in O. sativa (OsCRK01–OsCRK33), and 38 in Z. mays (ZmCRK01–ZmCRK33) (Supplementary Table S3). Comparisons of the total number of CRK genes showed that the CRK gene family displayed expansion in the allotetraploid relative to the diploid cotton genome, but was not strictly associated with genome size, as the smallest genome, that of A. thaliana, encoded 33 CRK genes (Supplementary Tables S2, S3). The unrooted phylogenetic tree revealed that most of the CRKs were clustered according to dicot and monocot species. Three phylogenetic groups of CRK genes (M1–M3) clustered into monocot branches and two groups (D1 and D2) into dicot branches (Figure 3), respectively. The remaining CRK genes were clustered into two branches (DM-m1 and DM-m2), and contained genes from dicot and monocot species, while a small defined branch, DM-m2, contained CRK genes from all seven plant species (Figure 3). This finding suggested that dicot and monocot species shared the same set of ancient CRK genes, which later underwent diversification. Enrichment of CRKs in the phylogenetic branches was evident in branches shared by cotton and A. thaliana (Figure 3), suggesting that CRK duplication frequently occurs in dicots. Investigation of the CRKs from cotton showed that most of the CRKs can be clustered in small branches, concordant with the relationships between the four cotton species, and indicated that CRKs from G. raimondii and G. arboreum were donors to G. barbadense and G. hirsutum (Figure 3). Comparisons of unrooted trees showed that the GbCRKs were also clustered into four branches that correspond to the four groups of GbCRKs from G. barbadense (Figures 2, 3). The CRKs in the small clade of the D2 branch originated from the cotton species corresponding to Group II in the GbCRK phylogenetic tree (Figures 2, 3), forming a unique branch in cotton compared to A. thaliana, suggesting that these CRKs were specific to cotton. The CRK gene family displayed significant divergence between dicot and monocot plants, and tandem duplication of CRKs appears to be a major evolutionary mechanism in dicots.
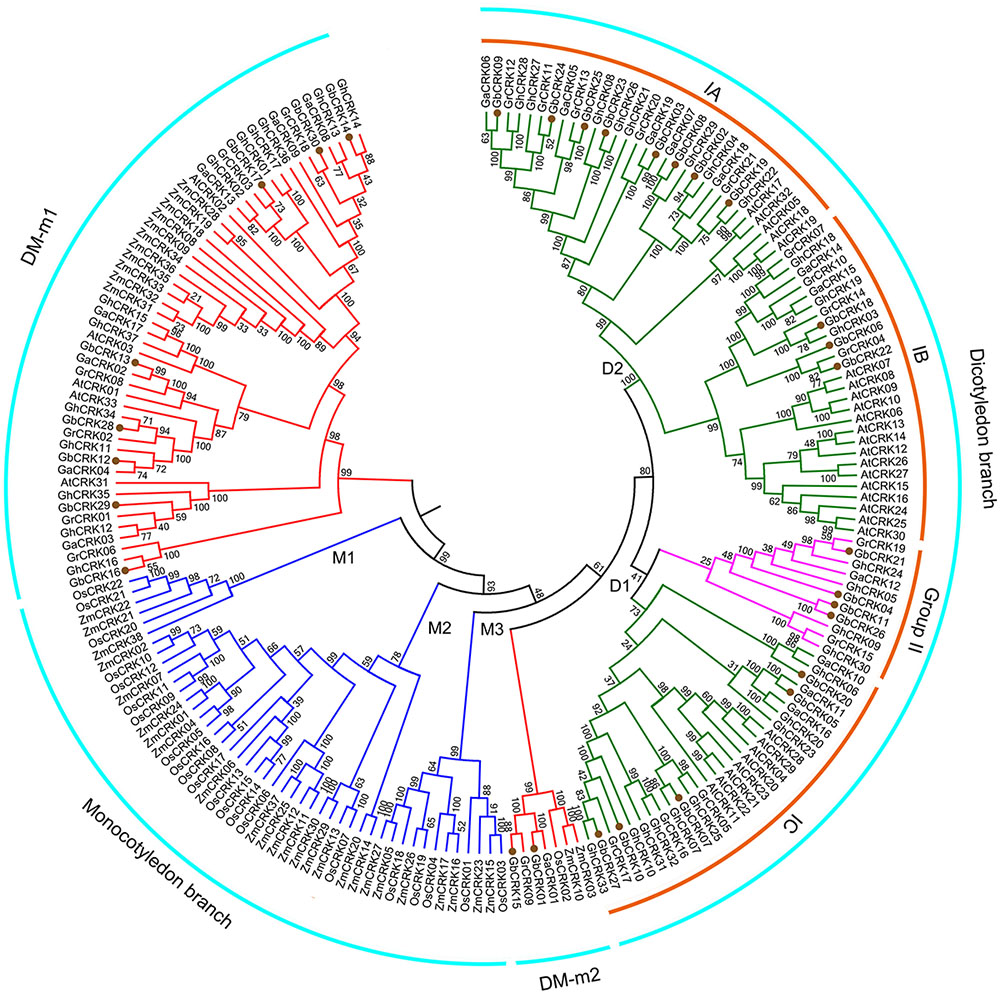
FIGURE 3. Phylogenetic relationships of CRK genes from seven plant species. An unrooted phylogenetic tree was constructed using the CRK nucleotide sequences from G. barbadense, G. hirsutum, G. arboretum, G. raimondii, Arabidopsis thaliana, Oryza sativa, and Zea mays using the NJ method with 1000 bootstrap replicates. Four groups are labeled as: dicotyledonous branch, DM-m1, DM-m2 and monocotyledonous branch. Subgroups are also labeled.
Expression Patterns of GbCRK Genes in Response to Verticillium dahliae
To gain insight into the role of the GbCRK genes in cotton disease resistance, the expression patterns of all 30 GbCRK genes were analyzed in response to V. dahliae. Two cultivars used in expression experiments were: G. barbadense cv. Hai 7124 and G. barbadense cv. GZ 57, which display resistance and susceptibility to VW, respectively. According to the reference RNA-seq data set of G. barbadense cv. Hai 7124 inoculated with V. dahliae (Chen et al., 2015), nine GbCRK genes were significantly up-regulated at the two evaluation points of 6 and 72 h after inoculation with V. dahliae, and all nine were clustered into Group I of the unrooted phylogenetic tree (Figure 4). RNA-seq analysis of the susceptible cotton cultivar G. barbadense cv. GZ 57 revealed that seven GbCRK genes were up-regulated at 6 and 72 h after inoculation with V. dahliae (Figure 4, unpublished RNA-seq data of the response of G. barbadense cv. GZ 57 to V. dahliae invasion). Interestingly, comparisons of the expression pattern of GbCRKs revealed that only four, GbCRK06, GbCRK08, GbCRK18, and GbCRK22 (three of them belong to Group I), were up-regulated at both evaluation points in G. barbadense cv. Hai 7124 but were either down-regulated or unchanged in G. barbadense cv. GZ 57 following inoculation with V. dahliae (Figure 4), suggesting that these GbCRK genes played a role in cotton defense response to VW. RT-qPCR analyses confirmed GbCRK gene expression patterns, as these were consistent with RNA-seq results with the exception of GbCRK08. The RT-qPCR analyses of GbCRK06, GbCRK18, and GbCRK22 that clustered in Group I revealed up-regulation of these genes at all evaluation points (2–120 h after inoculation with V. dahliae) in G. barbadense cv. Hai 7124 in response to V. dahliae (Figure 4). However, GbCRK06, GbCRK18, and GbCRK22 were down-regulated or only slightly up-regulated at each comparable evaluation point in G. barbadense cv. GZ 57 in response to V. dahliae (Figure 4). These results suggested that the cotton CRK genes can be activated by V. dahliae, and that a subset of genes contributes to VW resistance in G. barbadense.
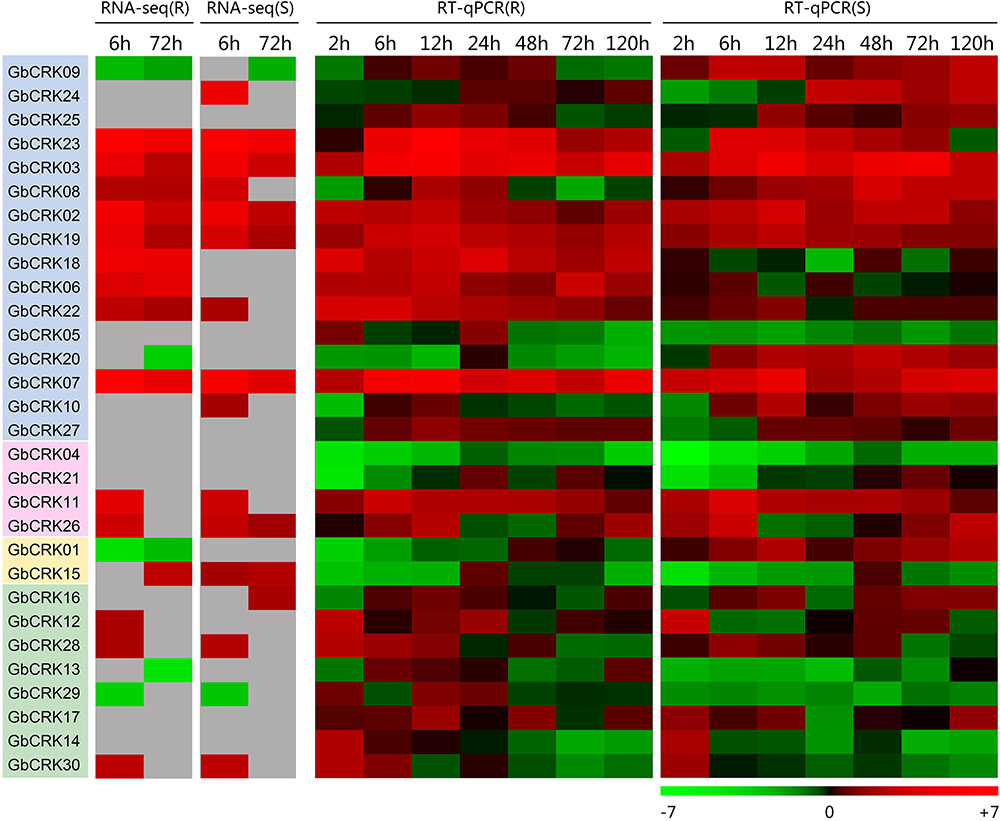
FIGURE 4. Expression patterns of CRK genes from Verticillium wilt resistant and susceptible G. barbadense cultivars. Both RNA-seq and RT-qPCR were used to examine the expression level of 30 CRK genes at different time points (6, 72 h) and (2, 6, 12, 24, 48, 72, and 120 h) after V. dahliae inoculation. A red box indicates up-regulation and green box indicates down-regulation. Color gradients indicate fold-change values (log2 values); with dark to light red representing increased transcript abundance and dark to light green indicating decreased transcript abundance. “R” indicates resistant G. barbadense cultivars and “S” indicates susceptible G. barbadense cultivars.
Functional Characterization of GbCRK Genes Involved in Cotton Verticillium Wilt Resistance
Expression patterns of GbCRK genes showed that nine GbCRKs in Group I were significantly up-regulated in cotton in response to V. dahliae infection (Figure 4). To identify the roles of these GbCRKs in VW resistance, tobacco rattle virus (TRV)-based VIGS was performed using the island cotton cv. Hai 7124 (VW-resistant cultivar). Fragments (∼500 bp) of all the nine GbCRKs amplified genes were separately integrated into the vector pTRV2 for generating gene-silenced cotton lines. Among the nine GbCRK genes in G. barbadense, the silencing of GbCRK18 significantly impaired VW resistance in cv. Hai 7124; i.e., GbCRK18-silenced plants displayed severe symptoms of wilting leaves (Figures 5A,C), and displayed serious vascular discoloration 21 days after inoculation with V. dahliae Vd991 (Figure 5B). However, silencing of the eight other genes independently in cv. Hai 7124 did not change the resistance phenotypes 21 days after inoculation with V. dahliae Vd991 (Figure 5). Interestingly, the expression patterns of GbCRK18 were sustainably up-regulated in resistant cotton, but down-regulated in the cotton susceptible to V. dahliae (Figure 4). Silencing GbCRK06 and GbCRK22 resulted in a similar expression pattern as that of GbCRK18 in resistant and susceptible cotton, but did not compromise resistance to V. dahliae (Figure 5A). The RT-qPCR analyses indicated that the gene silencing efficiency of all nine GbCRK genes was greater than 70% (Supplementary Figure S3). Additionally, qPCR quantification of fungal biomass in cotton plants demonstrated that the function of GbCRK18, but not GbCRK06 and GbCRK22, is associated with resistance, as there was increased fungal biomass associated with the GbCRK18-silenced plants that also displayed increased susceptibility to the pathogen (Figure 5D).
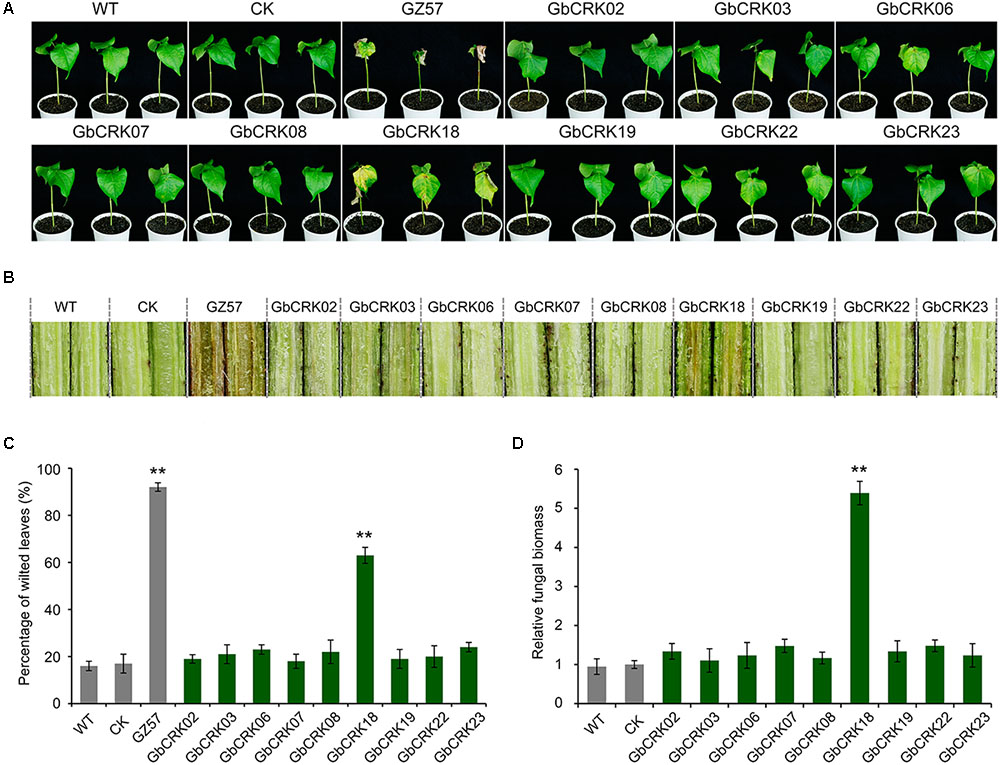
FIGURE 5. Virus-induced gene silencing of candidate CRKs implicated in VW resistance. (A) The VW phenotypes of wilting leaves in corresponding CRK gene-silenced plants post inoculation. Approximately 21 days after the VIGS procedure in the 2-week-old-resistant G. barbadense cv. Hai 7124, the pTRV2:00 (CK), the wild-type (WT), and the gene-silenced plants were inoculated with 20 mL of a 5 × 106 conidia/mL suspension of Verticillium dahliae strain Vd991. Experiments consisted of three replicates of 12 plants each, and were arranged in a complete random block design. Error bars were calculated based on three biological replicates using standard deviation while relative gene quantifications were determined using a comparative threshold 2-ΔΔCT method, ∗∗ indicates significant difference (P < 0.01). GbCRK02, GbCRK03, GbCRK06, GbCRK07, GbCRK08, GbCRK18, GbCRK19, GbCRK22, and GbCRK23 represent gene-silenced plants and G. barbadense cv. GZ 57 was used as a negative control. (B) Stem vascular discoloration of CK, WT, and gene-silenced plants. Photographs were taken 4 weeks after inoculation with V. dahliae strain Vd991. (C) Percentage of diseased leaves after V. dahliae strain Vd991 inoculation. (D) Quantitative PCR analyses of fungal biomass of CK, WT, and gene-silenced plants. At least 12 inoculated plants and the Verticillium EF-1α gene were used as reference genes.
Expression of GbCRK18 in Response to Verticillium dahliae Is Modulated by Jasmonic Acid
The expression of GbCRK18 was examined in the resistant G. barbadense cv. Hai 7124 after inoculation with the highly virulent V. dahliae strain Vd991, and quantified by RT-qPCR using the susceptible cotton lines G. barbadense cv. GZ 57 and G. hirsutum cv. JM 11 for comparison. Expression of GbCRK18 was quickly and strongly up-regulated (P < 0.001) in the resistant variety cv. Hai 7124 at seven time points until 120 h, relative to the lower levels observed at the comparable time points for cv. GZ 57 and cv. JM11 (Figure 6A). GbCRK18 was significantly up-regulated (P < 0.05) in cv. Hai 7124 after treatment with methyl jasmonic acid (MeJA) at several points within 120 h (Figure 6B), but was less affected by treatment with ethylene (ET) (Figure 6C) and not changed by treatment with SA or ABA (Figures 6D,E), which suggested that the GbCRK18 response of the resistant cultivar to V. dahliae is mediated by jasmonic acid (JA) signaling. Interestingly, RT-qPCR analysis of MeJA-related genes between the GbCRK18-silenced line cv. Hai 7124 and control showed that four of the genes tested, including GbAOS, GbOPR3, GbMYC2, and GbJAZ1, were significantly down-regulated in GbCRK18 silenced cotton that displayed susceptibility to V. dahliae (Figure 6F); moreover, the marker genes induced by MeJA including pathogenesis-related gene 3 (PR3), PR4, PR10, and PR12, were also significantly down-regulated in GbCRK18-silenced line cv. Hai 7124 after inoculation with V. dahliae (Figure 6G). These results suggested that GbCRK18 mediates a VW defense response through JA signaling.
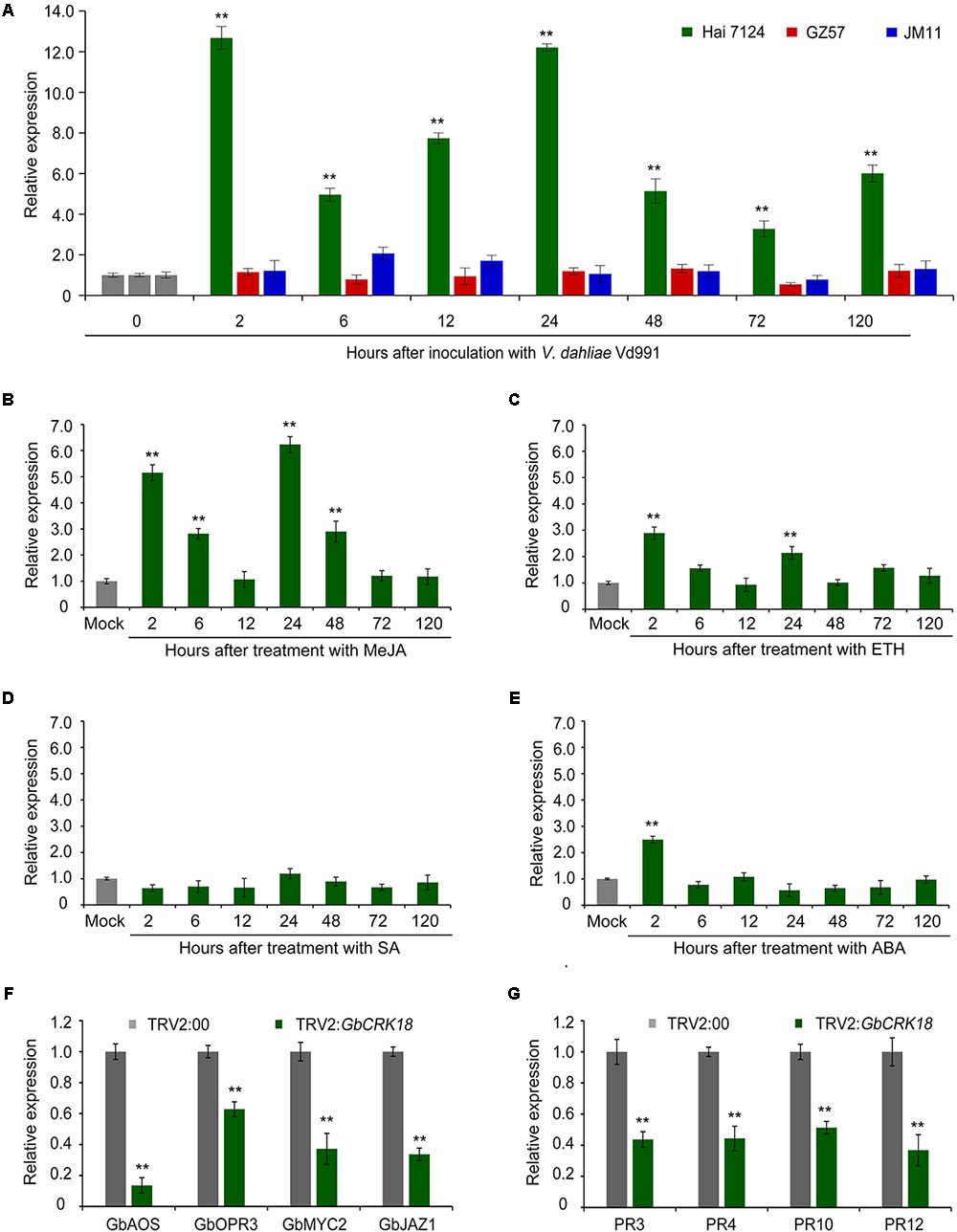
FIGURE 6. Expression pattern of CRK18 under different treatments and of the defense marker genes in the CRK18 transgenics. (A) Expression analysis of GbCRK18 in cotton varieties Hai 7124, GZ 57, and JM 11 after inoculation with V. dahliae Vd991 by RT-qPCR. (B–E) Expression analysis of GbCRK18 in G. barbadense cv. Hai 7124 in response to: (B) methyl jasmonic acid (MeJA), (C) ETH, (D) SA, and (E) ABA. Three-week-old G. barbadense plants were treated with 10 mM MeJA, 10 mM ETH, 10 mM SA, and 100 μM ABA, and harvested at 2, 6, 12, 24, 48, and 72 h, respectively. Relative expression analyses of GbCRK18 by RT-qPCRs were performed using the cotton 18S gene as a reference, and relative expression was compared with expression levels in cotton plants that were treated with sterile water (Mock). Transcript levels of genes related to (F) JA signaling pathways and (G) defense marker genes were analyzed in GbCRK18-silenced plants by RT-qPCR. Relative expression was calculated using the comparative threshold 2-ΔΔCT method, and values represent averages of three independent biological replicates of three plants each. Error bars were calculated based on three biological replicates using standard deviation; ∗∗ indicates significant difference (P < 0.01).
GbCRK18 Is Localized to the Plasma Membrane
Conserved domain analysis of the GbCRK18 peptide sequence using SMART identified a typical CRK family protein with a signal peptide and TM domain, which was confirmed by SignalP4.1 (1–24 aa,) and TMHMM2.0 (290–312 aa) (Figure 7A), respectively. To test this hypothesis, we assessed the subcellular location of GbCRK18 by transient expression in tobacco and compared this localization of the control GFP fusion. As predicted, the fusion protein was clearly localized to plasma membrane (Figure 7B), in contrast to the fluorescence signal of GFP proteins (p35S:GFP), which was prevalent throughout the foliar cells in tobacco (Figure 7B). An extracellular domain of stress-antifung domain (27–239 aa) and an intracellular domain of Pkinase (347–596 aa) were also identified, suggesting that GbCRK18 is localized to the plasma membrane and may directly interact with pathogen-associated molecular patterns and mediate signal transduction via the intracellular domain.
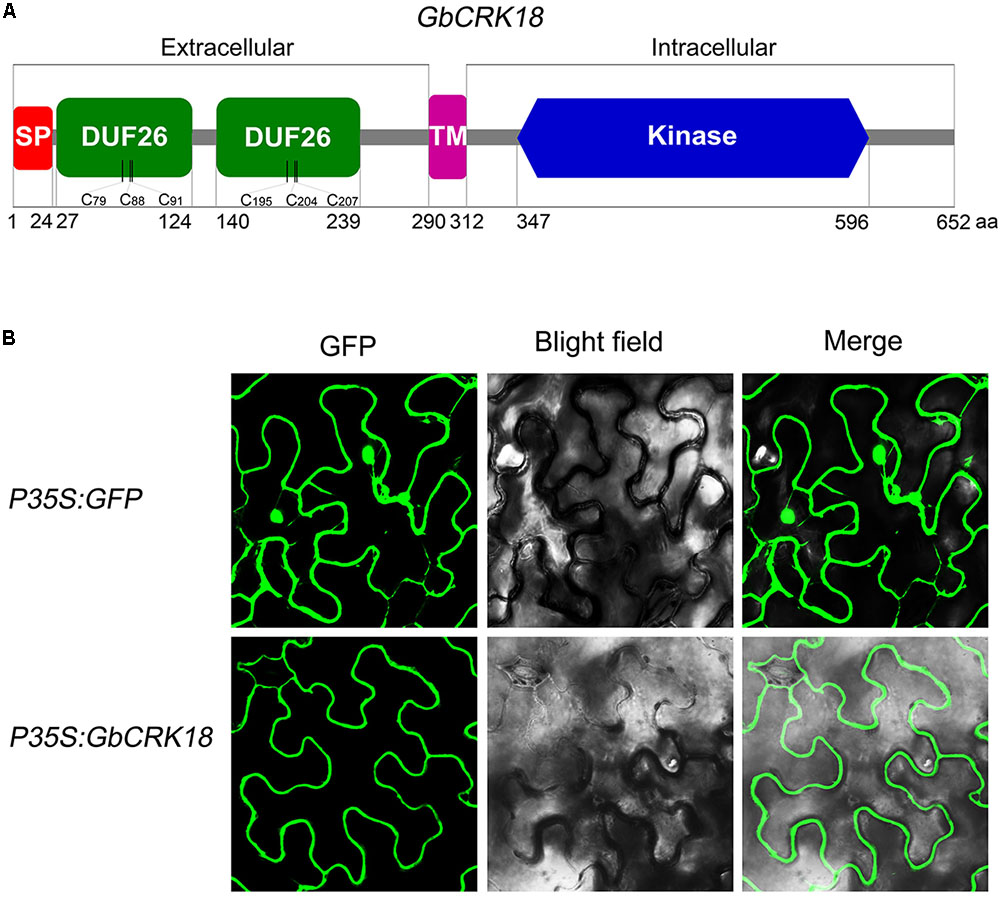
FIGURE 7. Protein structure model and subcellular localization of GbCRK18. (A) The conserved protein structure model of GbCRK18. Conserved domains of GbCRK18 including: a signal peptide (SP) from 1 to 24 amino acids; two stress-antifung domains (DUF26) from 27 to 124 and 140 to 239 amino acids; a transmembrane domain (TM) from 290 to 312 amino acids; and an intracellular kinase domain (Kinase) from 347 to 596 amino acids, respectively. (B) Subcellular localization of GbCRK18. For the subcellular localization of GbCRK18, a p35S:GbCRK18 containing a full-length coding sequence of GbCRK18 was inserted into the pBin-GFP4 vector and introduced into tobacco by Agrobacterium infiltration. The p35S:GFP empty vector was used as a control and transformants were viewed with laser scanning confocal microscopy ×200 magnification with excitation and emission wavelengths of 488 and 510 nm, respectively.
Discussion
Cotton (Gossipium spp.) is an important economic crop worldwide, and VW caused by V. dahliae, results in significant annual economic losses through reduced boll quality and quantity. Improving genetic resistance is the preferred method of managing VW in most crops (Schaible et al., 1951; Putt, 1964; Simko et al., 2004; Bolek et al., 2005; Zebrowska et al., 2006; Li et al., 2017), but identifying this resistance is difficult because of the unavailability of resistance genes against this pathogen. However, several genes that contribute to the defense response against VW have been reported in cotton, including GbCAD1 and GbSSI2 (Gao et al., 2013), GbaNA1 (Li et al., 2018), GbSTK (Zhang Y. et al., 2013), GbTLP1 (Munis et al., 2010), and GbNRX1 (Li et al., 2016), etc. In spite of this, the CRKs, with known roles in plant resistance (Chen et al., 2003; Acharya et al., 2007; Ederli et al., 2011; Zhang X. et al., 2013; Yeh et al., 2015; Yadeta et al., 2017), have not been cloned from cotton and examined for roles in VW resistance. In this study, we systematically investigated the CRK genes found in a representative genome of G. barbadense, which contains abundant resistance gene resources among the different cotton species. These CRKs and their respective translated products were characterized based on structures, conserved domains, phylogenetic relationships among plants, and expression profiles in response to V. dahliae. Notably, we identified a candidate resistance gene, GbCRK18 and its role in conferring VW resistance was confirmed by a gene silencing approach.
Plants that have been sequenced contain many copies of RLKs. For instance, over 600 and 1000 RLKs are contained in the genomes of Arabidopsis and rice, respectively (Shiu and Bleecker, 2003). RLKs have highly diverse extracellular regions, including one or more domains that are used to categorize RLKs into distinct subfamilies (Shiu and Bleecker, 2003). CRKs comprise a large subfamily of plant RLKs that possess an extracellular domain, a single-pass transmembrane domain, and an intracellular Ser/Thr protein kinase domain (Chen et al., 2003), important in disease resistance and apoptosis in plants (Chen et al., 2003; Acharya et al., 2007; Ederli et al., 2011). However, except for some well-studied examples in Arabidopsis (Chen et al., 2003; Acharya et al., 2007; Ederli et al., 2011; Zhang X. et al., 2013; Yeh et al., 2015; Yadeta et al., 2017), few CRK genes have been characterized for their role in plant disease resistance in cotton. Genomic analysis is an effective means of transferring knowledge from one taxon to another (Chen et al., 2003; Zhang Y. et al., 2012; Zhang et al., 2017; Xiao et al., 2017), allowing for a faster pace of gene discoveries associated with disease resistance.
In this study, we conducted genome-wide analyses of the CRK gene family in the four known genomes of cotton species, along with other three plant species; O. Sativa, Z. mays, and A. thaliana (Supplementary Tables S2, S3). Through data mining, CRKs were identified as transmembrane proteins characterized by the presence of two extracellular DUF26 domains (Figure 1A; Chen, 2001; Bourdais et al., 2015). Molecular phylogenetic analysis suggested a significantly divergent evolutionary history of CRK genes in dicot and monocot species, except for a common branch (DM-m1) enriched with the CRKs from sequenced plant genomes (Figure 3). CRKs from the same species tended to be clustered together in the same branch among dicot species but not monocot species, possibly implying that these CRKs originated from a few ancestral CRKs in dicot species and tandem or segmental duplication could have frequently occurred in dicot species relative to the monocot species.
The evolution of disease resistance genes is mediated by sequence exchange, tandem or segmental duplication events, or gene conversion (Bent et al., 1994; Hulbert et al., 2001; Leister, 2004). For instance, tandem and segmental duplications frequently occur in NBS-LRR gene clusters that have led to the formation of the phylogenetic lineage of NBS-LRR genes in the Arabidopsis genome (Baumgarten et al., 2003; Meyers et al., 2003), while sequence exchanges tend to homogenize the members of the Cf-9 gene cluster in tomato (Van der Hoorn et al., 2001). Among the 30 GbCRKs identified in G. barbadense in the current study, 14 and 16 genes were located in the A and D sub-genomes, respectively, while 18 other genes appeared to exist in orthologous pairs between the A and D sub-genomes (Figure 2 and Supplementary Figure S2). We interpret this observation to mean that few segmental duplications may have occurred between different non-allelic chromosomes of CRK gene family in cotton. Our investigation of the physical location of CRK genes revealed that some of them are located closely and also showed high sequence identities such as GbCRK23, GbCRK24, and GbCRK25 (Supplementary Figure S2), suggesting that tandem duplication is a main evolutionary mechanism of the CRK gene family in cotton. The total number of CRKs in the allotetraploid genomes were not more than twofold that of the diploid cotton genomes, and phylogenetic analysis also showed that most of the CRK genes had a corresponding ortholog in G. arboretum and G. raimondii, which are generally recognized as the donors of the allotetraploid cotton genome (Figure 3), suggesting that the CRK family expands through tandem duplication events.
Several CRK genes have been reported to play critical roles in disease resistance, including those from Arabidopsis (Acharya et al., 2007; Wrzaczek et al., 2010; Bourdais et al., 2015; Yeh et al., 2015; Yadeta et al., 2017), wheat (Yang et al., 2013), Medicago truncatula (Berrabah et al., 2014), barley (Rayapuram et al., 2012), and rice (Mawsheng et al., 2016). The functions of CRKs involved in disease resistance have been well studied in Arabidopsis, including CRK4, CRK5, CRK6, CRK13, CRK19, CRK20, CRK28, CRK36, and CRK45 (Lee et al., 2017). For example, over-expression of CRK5 and CRK13 and CRK28 in Arabidopsis led to enhanced resistance against Pseudomonas syringae pv. tomato (Pst) DC3000 (Chen et al., 2003; Acharya et al., 2007; Yadeta et al., 2017). However, involvement of CRK genes in VW resistance in cotton had not been reported except for the study by Zhang et al. (2018). In this study, we found that the members of the CRK gene family displayed up-regulation following infection by V. dahliae (Figure 4). One specific CRK gene (GbCRK18) participated in VW resistance and its silencing resulted in susceptibility to V. dahliae (Figure 5). Moreover, CRKs are also reported to participate in signal transduction of hormones such as ABA and abiotic stress (Zhu et al., 2007). In this study, we found that the expression of GbCRK18 in response to JA (Figure 6B) follows a temporal expression pattern similar to the expression pattern observed following V. dahliae inoculation (Figure 6A); the expression of genes associated with JA biosynthesis (Figure 6F), suggested that JA-mediated signaling is important for VW resistance via GbCRK18. Plants infected by pathogens initiate a variety of defense responses by hormone signaling pathways, which lead to activation of defined sets of target genes (Feys and Parker, 2000). As expected, the expression of defense response marker genes in JA signaling (Mazarei et al., 2007) was also significantly reduced after silencing the GbCRK18 in cotton (Figure 6G). The above results suggested that the CRK gene family, and particularly GbCRK18, plays an important role in VW resistance in cotton.
Conclusion
Genes in the CRK family from G. barbadense genome were systematically analyzed. Silencing of GbCRK18, encoding a protein that is localized in the plasma membrane, compromised resistance against VW resistance in G. barbadense. Our study also demonstrated that the CRK gene family plays an important role in defense responses, and that GbCRK18 is a functional gene that confers VW resistance in G. barbadense.
Author Contributions
X-FD, J-YC, and KS conceived the study and designed all experiments. J-YC performed the data analysis and interpretation. T-GL wrote the paper and performed the gene family analysis. KS, DS, SK, ND, and AH revised the manuscript. D-DZ, LZ, and DW performed the gene silencing and gene expression analysis. Z-QK, J-JL, B-LW, and C-MY performed the pathogenicity analysis.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFD0200601 and 2017YFD0201900), the National Natural Science Foundation of China (31671986, 31471759, 31772245, and 31501600), the Special Public Welfare Industry Research on Agriculture (201503109), the Young Elite Scientists Sponsorship Program by Cast (2016QNRC001), an Agricultural Science and Technology Innovation Program grant to X-FD, the Fundamental Research Funds for Central Non-profit Scientific Institution (Y2016CG11, S2016JC05, and S2016CG01).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01266/full#supplementary-material
Footnotes
- ^http://pfam.xfam.org/
- ^http://meme-suite.org/
- ^http://www.softsea.com/review/MapInspect.html
- ^http://www.megasoftware.net
- ^http://gsds1.cbi.pku.edu.cn/
References
Acharya, B. R., Raina, S., Maqbool, S. B., Jagadeeswaran, G., Mosher, S. L., Appel, H. M., et al. (2007). Overexpression of CRK13, an Arabidopsis cysteine rich receptor like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50, 488–499. doi: 10.1111/j.1365-313X.2007.03064.x
Baumgarten, A., Cannon, S., Spangler, R., and May, G. (2003). Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165, 309–319.
Bent, A. F., Kunkel, B. N., Dahlbeck, D., Brown, K. L., Schmidt, R., Giraudat, J., et al. (1994). RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. doi: 10.1016/0168-9525(94)90104-X
Berrabah, F., Bourcy, M., Eschstruth, A., Cayrel, A., Guefrachi, I., Mergaert, P., et al. (2014). A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol. 203, 1305–1314. doi: 10.1111/nph.12881
Bolek, Y., Elzik, K. M., Pepper, A. E., Bell, A. A., Magill, C. W., Thaxton, P. M., et al. (2005). Mapping of verticillium wilt resistance genes in cotton. Plant Sci. 168, 1581–1590. doi: 10.1016/j.plantsci.2005.02.008
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bourdais, G., Burdiak, P., Gauthier, A., Nitsch, L., Salojärvi, J., Rayapuram, C., et al. (2015). Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 11:e1005373. doi: 10.1371/journal.pgen.1005373
Chen, J., Li, N., Ma, X., Gupta, V. K., Zhang, D., Li, T., et al. (2017). The ectopic overexpression of the cotton Ve1 and Ve2-homolog sequences leads to resistance response to Verticillium wilt in Arabidopsis. Front. Plant Sci. 8:844. doi: 10.3389/fpls.2017.00844
Chen, J. Y., Huang, J. Q., Li, N. Y., Ma, X. F., Wang, J. L., Liu, C., et al. (2015). Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biol. 15:148. doi: 10.1186/s12870-015-0508-3
Chen, K., Du, L., and Chen, Z. (2003). Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 53, 61–74. doi: 10.1023/B:PLAN.0000009265.72567.58
Chen, K., Fan, B., Du, L., and Chen, Z. (2004). Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 56, 271–283. doi: 10.1007/s11103-004-3381-2
Chen, Z. (2001). A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol. 126, 473–476. doi: 10.1104/pp.126.2.473
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Duan, X., Zhang, Z., Jin, W., and Zuo, K. (2016). Characterization of a novel cotton subtilase gene GbSBT1 in response to extracellular stimulations and its role in Verticillium resistance. PLoS One 11:e0153988. doi: 10.1371/journal.pone.0153988
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Ederli, L., Madeo, L., Calderini, O., Gehring, C., Moretti, C., Buonaurio, R., et al. (2011). The Arabidopsis thaliana cysteine-rich receptor-like kinase CRK20 modulates host responses to Pseudomonas syringae pv. tomato DC3000 infection. J. Plant Physiol. 168, 1784–1794. doi: 10.1016/j.jplph.2011.05.018
Feys, B. J., and Parker, J. E. (2000). Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. doi: 10.1016/S0168-9525(00)02107-7
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, 222–230. doi: 10.1093/nar/gkt1223
Gao, W., Long, L., Zhu, L. F., Xu, L., Gao, W. H., Sun, L. Q., et al. (2013). Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteom. 12, 3690–3703. doi: 10.1074/mcp.M113.031013
Hardie, D. G. (1999). Plant protein serinr/threonine kinases: classification and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 97–131. doi: 10.1146/annurev.arplant.50.1.97
He, Y. X., Zhou, J. G., Shan, L. B., and Meng, X. Z. (2018). Plant cell surface receptor-mediated signaling – A common theme amid diversity. J. Cell Sci. 131:jcs209353. doi: 10.1242/jcs.209353
Hulbert, S. H., Webb, C. A., Smith, S. M., and Sun, Q. (2001). Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39, 285–312. doi: 10.1146/annurev.phyto.39.1.285
Hunter, S., Jones, P., Mitchell, A., Apweiler, R., Attwood, T. K., Bateman, A., et al. (2012). InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, 306–312. doi: 10.1093/nar/gkr948
Idänheimo, N., Gauthier, A., Salojärvi, J., Siligato, R., Brosché, M., Kollist, H., et al. (2014). The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 445, 457–462. doi: 10.1016/j.bbrc.2014.02.013
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/-nature05286
Joshi, R. K., and Nayak, S. (2011). Functional characterization and signal transduction ability of nucleotide-binding site-leucine-rich repeat resistance genes in plants. Genet. Mol. Res. 10, 2637–2652. doi: 10.4238/2011.October.25.10
Kimura, S., Waszczak, C., Hunter, K., and Wrzaczek, M. (2017). Bound by fate: the role of reactive oxygen species in receptor-like kinase signaling. Plant Cell 29, 638–654. doi: 10.1105/tpc.16.00947
Krogh, A., Larsson, B., Von, H. G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kruijt, M., De Kock, M. J., and de Wit, P. J. (2005). Receptor-like proteins involved in plant disease resistance. Mol. Plant Pathol. 6, 85–97. doi: 10.1111/j.1364-3703.2004.00264.x
Lee, D. S., Kim, Y. C., Kwon, S. J., Ryu, C. M., and Park, O. K. (2017). The Arabidopsis cysteine-rich Receptor-like kinase CRK36 regulates immunity through interaction with the cytoplasmic kinase BIK1. Front. Plant Sci. 8:1856. doi: 10.3389/fpls.2017.01856
Leister, D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20, 116–122. doi: 10.1016/j.tig.2004.01.007
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Li, X., Zhu, L., Tu, L., Liu, L., Yuan, D., Li, J., et al. (2011). Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. doi: 10.1093/jxb/err245
Li, Y., Han, L. B., Wang, H., Zhang, J., Sun, S., Feng, D., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Li, T., Ma, X., Li, N., Zhou, L., Liu, Z., Han, H., et al. (2017). Genome-wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 15, 1520–1532. doi: 10.1111/pbi.12734
Li, N., Ma, X., Short, D. P. G., Li, T., Zhou, L., Gui, Y., et al. (2018). The island cotton NBS-LRR gene GbaNA1 confers resistance to the non-race 1 Verticillium dahliae isolate Vd991. Mol. Plant Pathol. 19, 1466–1479. doi: 10.1111/mpp.12630
Liu, S. Y., Chen, J. Y., Wang, J. L., Li, L., Xiao, H. L., Adam, S. M., et al. (2013). Molecular characterization and functional analysis of a specific secreted protein from highly virulent defoliating Verticillium dahliae. Gene 529, 307–316. doi: 10.1016/j.gene.2013.06.089
Liu, X., Zhao, B., Zheng, H. J., Hu, Y., Lu, G., Yang, C. Q., et al. (2015). Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 5:14139. doi: 10.1038/srep14139
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/-meth.2001.1262
Martin, G. B., Bogdanove, A. J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. doi: 10.1146/annurev.arplant.54.031902.135035
Mawsheng, C., Xu, Q., Bart, R. S., Bai, W., Deling, R., Hoi, S. T. W., et al. (2016). A genetic screen identifies a requirement for cysteine-rich-receptor-like kinases in rice NH1 (OsNPR1)-mediated immunity. PLoS Genet. 12:e1006049. doi: 10.1371/journal.pgen.1006049
Mazarei, M., Elling, A. A., Maier, T. R., Puthoff, D. P., and Baum, T. J. (2007). GmEREBP1 is a transcription factor activating defense genes in soybean and Arabidopsis. Mol. Plant Microbe. Interact. 20, 107–119. doi: 10.1094/MPMI-20-2-0107
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Mo, H., Wang, X., Zhang, Y., Zhang, G., Zhang, J., and Ma, Z. (2015). Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 83, 962–975. doi: 10.1111/tpj.12941
Munis, M. F., Tu, L., Deng, F., Tan, J., Xu, L., Xu, S., et al. (2010). A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem. Biophys. Res. Commun. 393, 38–44. doi: 10.1016/j.bbrc.2010.01.069
Petersen, T. N., Brunak, S., Von, H. G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Putt, E. D. (1964). Breeding behaviour of resistance to leaf mottle or Verticillium in sunflowers. Crop Sci. 4, 177–179. doi: 10.2135/cropsci1964.0011183X000400020016x
Rayapuram, C., Jensen, M. K., Maiser, F., Shanir, J. V., Hornshøj, H., Rung, J. H., et al. (2012). Regulation of basal resistance by a powdery mildew-induced cysteine-rich receptor-like protein kinase in barley. Mol. Plant Pathol. 13, 135–147. doi: 10.1111/j.1364-3703.2011.00736.x
Santhanam, P., van Esse, H. P., Albert, I., Faino, L., Nürnberger, T., and Thomma, B. P. (2013). Evidence for functional diversification within a fungal NEP1-like protein family. Mol. Plant Microbe Interact. 26, 278–286. doi: 10.1094/MPMI-09-12-0222-R
Schaible, L., Cannon, O. S., and Waddoups, V. (1951). Inheritance of resistance to Verticillium wilt in a tomato cross. Phytopathology 41, 986–990.
Shiu, S. H., and Bleecker, A. B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768. doi: 10.1073/pnas.181141598
Shiu, S. H., and Bleecker, A. B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543. doi: 10.1104/pp.103.021964
Shiu, S. H., Karlowski, W. M., Pan, R., Tzeng, Y. H., Mayer, K. F., and Li, W. H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
Simko, I., Costanzo, S., Haynes, K. G., Christ, B. J., and Jones, R. W. (2004). Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor. Appl. Genet. 108, 217–224. doi: 10.1007/s00122-003-1431-9
Tao, Z., Yun, J., Zhao, J. H., Feng, G., Zhou, B. J., Fang, Y. Y., et al. (2016). Host-induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae. Mol. Plant 9, 939–942. doi: 10.1016/j.molp.2016.02.008
Tör, M., Lotze, M. T., and Holton, N. (2009). Receptor-mediated signalling in plants: molecular patterns and programmes. J. Exp. Bot. 60, 3645–3654. doi: 10.1093/jxb/erp233
Van, D. G. P., Hunter, T., and Lindberg, R. A. (1994). Receptor protein-tyrosine kinases and their signal transduction pathways. Ann. Rev. Cell Biol. 10, 251–331. doi: 10.1146/annurev.cb.10.-110194.001343
Van der Hoorn, R. A. L., Kruijt, M., Roth, R., Brandwagt, B. F., Joosten, M. H., and De Wit, P. J. (2001). Intragenic recombination generated two distinct Cf genes that mediate AVR9 recognition in the natural population of Lycopersicon pimpinellifolium. Proc. Natl. Acad. Sci. U.S.A. 98, 10493–10498. doi: 10.1073/pnas.181241798
Walker, J. C., and Zhang, R. (1990). Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature 345, 743–746. doi: 10.1038/345743a0
Wang, F. X., Ma, Y. P., Yang, C. L., Zhao, P. M., Yao, Y., Jian, G. L., et al. (2011). Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics 11, 4296–4309. doi: 10.1002/pmic.201100062
Wrzaczek, M., Brosché, M., Salojärvi, J., Kangasjärvi, S., Idänheimo, N., Mersmann, S., et al. (2010). Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 10:95. doi: 10.1186/1471-2229-10-95
Xiao, X. H., Yang, M., Sui, J. L., Qi, J. Y., Fang, Y. J., Hu, S. N., et al. (2017). The calcium-dependent protein kinase (CDPK) and CDPK-related kinase gene families in Hevea brasiliensis-comparison with five other plant species in structure, evolution, and expression. FEBS Open Biol. 7, 4–24. doi: 10.1002/2211-5463.12163
Yadeta, K. A., Elmore, J. M., Creer, A. Y., Feng, B., Franco, J. Y., Rufian, J. S., et al. (2017). A cysteine-rich protein kinase associates with a membrane immune complex and the cysteine residues are required for cell death. Plant Physiol. 173, 771–787. doi: 10.1104/pp.16.01404
Yang, J., Ma, Q., Zhang, Y., Wang, X., Zhang, G., and Ma, Z. (2016). Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene 575, 687–694. doi: 10.1016/j.gene.2015.09.046
Yang, K., Wei, R., Lin, Q., Li, J., Wei, X., and Zhang, Z. (2013). Isolation and characterization of a novel wheat cysteine-rich receptor-like kinase gene induced by Rhizoctonia cerealis. Sci. Rep. 3:3021. doi: 10.1038/srep03021
Yang, Y., Ling, X., Chen, T., Cai, L., Liu, T., Wang, J., et al. (2015). A cotton Gbvdr5 gene encoding a leucine-rich-repeat receptor-like protein confers resistance to Verticillium dahliae in transgenic Arabidopsis and Upland Cotton. Plant Mol. Biol. Rep. 33, 987–1001. doi: 10.1007/s11105-014-0810-5
Yeh, Y. H., Chang, Y. H., Huang, P. Y., Huang, J. B., and Zimmerli, L. (2015). Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 6:322. doi: 10.3389/fpls.2015.00322
Zebrowska, J., Hortyński, J., Cholewa, T., and Honcz, K. (2006). Resistance to Verticillium dahliae (Kleb.) in the strawberry breeding lines. Commun. Agric. Appl. Biol. Sci. 71, 1031–1036.
Zhang, B., Yang, Y., Chen, T., Yu, W., Liu, T., Li, H., et al. (2012). Island cotton Gbve1 gene encoding A receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS One 7:e51091. doi: 10.1371/journal.pone.0051091
Zhang, J., Sanogo, S., Flynn, R., Baral, J. B., Bajaj, S., Hughs, S. E., et al. (2012). Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to upland cotton (G. hirsutum). Euphytica 187, 147–160. doi: 10.1007/s10681-011-0549-0
Zhang, Y., Mao, L., Wang, H., Brocker, C., Yin, X., Vasiliou, V., et al. (2012). Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PLoS One 7:e32153. doi: 10.1371/journal.pone.0032153
Zhang, H., Wei, C., Yang, X., Chen, H., Yang, Y., Mo, Y., et al. (2017). Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon (Cucumis melo L.). PLoS One 12:e0176352. doi: 10.1371/journal.pone.0176352
Zhang, X., Han, X., Shi, R., Yang, G., Qi, L., Wang, R., et al. (2013). Arabidopsis cysteine-rich receptor-like kinase 45 positively regulates disease resistance to Pseudomonas syringae. Plant Physiol. Biochem. 73, 383–391. doi: 10.1016/j.plaphy.2013.10.024
Zhang, Y., Wang, X., Li, Y., Wu, L., Zhou, H., Zhang, G., et al. (2013). Ectopic expression of a novel Ser/Thr protein kinase from cotton (Gossypium barbadense), enhances resistance to Verticillium dahliae infection and oxidative stress in Arabidopsis. Plant Cell Rep. 32, 1703–1713. doi: 10.1007/-s00299-013-1481-7
Zhang, Y., Wang, X., Yang, S., Chi, J., Zhang, G., and Ma, Z. (2011). Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep. 30, 2085–2096. doi: 10.1007/s00299-011-1115-x
Zhang, Z. Q., Wang, J., Jin, W., Ge, D. D., Liu, K., Lv, F. N., et al. (2018). Identification and expression analysis of CRK gene family in upland cotton. Scientia Agricultura Sinica 51, 2442–2461. doi: 10.3864/j.issn.0578-1752.2018.13.002
Zhao, J., Zhang, Z., Gao, Y., Zhou, L., Fang, L., Chen, X., et al. (2015). Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 5:15048. doi: 10.1038/srep15048
Zhou, J. M., Tang, D., and Wang, G. (2017). Receptor kinases in plant pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Keywords: cotton, Verticillium wilt, cysteine-rich receptor-like kinases (CRKs), expression profiling, defense response
Citation: Li T-G, Zhang D-D, Zhou L, Kong Z-Q, Hussaini AS, Wang D, Li J-J, Short DPG, Dhar N, Klosterman SJ, Wang B-L, Yin C-M, Subbarao KV, Chen J-Y and Dai X-F (2018) Genome-Wide Identification and Functional Analyses of the CRK Gene Family in Cotton Reveals GbCRK18 Confers Verticillium Wilt Resistance in Gossypium barbadense. Front. Plant Sci. 9:1266. doi: 10.3389/fpls.2018.01266
Received: 20 March 2018; Accepted: 10 August 2018;
Published: 11 September 2018.
Edited by:
Jens Staal, Ghent University, BelgiumReviewed by:
Xiyin Wang, North China University of Science and Technology, ChinaBrigitte Mauch-Mani, University of Neuchâtel, Switzerland
Copyright © 2018 Li, Zhang, Zhou, Kong, Hussaini, Wang, Li, Short, Dhar, Klosterman, Wang, Yin, Subbarao, Chen and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krishna V. Subbarao, kvsubbarao@ucdavis.edu Jie-Yin Chen, chenjieyin@caas.cn Xiao-Feng Dai, daixiaofeng_caas@126.com
†These authors have contributed equally to this work
 Ting-Gang Li1†
Ting-Gang Li1† Lei Zhou
Lei Zhou Zhi-Qiang Kong
Zhi-Qiang Kong Adamu S. Hussaini
Adamu S. Hussaini Jie-Yin Chen
Jie-Yin Chen Xiao-Feng Dai
Xiao-Feng Dai