- 1Institute of Plant Biology, National Taiwan University, Taipei, Taiwan
- 2Agriculture and Agri-Food Canada, London Research and Development Centre, London, ON, Canada
- 3Department of Biology, Western University, London, ON, Canada
- 4State Key Laboratory of Biocontrol and Guangdong Key Laboratory of Plant Resource, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
In Arabidopsis, the circadian rhythm is associated with multiple important biological processes and maintained by multiple interconnected loops that generate robust rhythms. The circadian clock central loop is a negative feedback loop composed of the core circadian clock components. TOC1 (TIMING OF CAB EXPRESSION 1) is highly expressed in the evening and negatively regulates the expression of CCA1 (CIRCADIAN CLOCK ASSOCIATED 1)/LHY (LATE ELONGATED HYPOCOTYL). CCA1/LHY also binds to the promoter of TOC1 and represses the TOC1 expression. Our recent research revealed that the histone modification complex comprising of LYSINE-SPECIFIC DEMETHYLASE 1 (LSD1)-LIKE 1/2 (LDL1/2) and HISTONE DEACETYLASE 6 (HDA6) can be recruited by CCA1/LHY to repress TOC1 expression. In this study, we found that HDA6, LDL1, and LDL2 can interact with TOC1, and the LDL1/2-HDA6 complex is associate with TOC1 to repress the CCA1/LHY expression. Furthermore, LDL1/2-HDA6 and TOC1 co-target a subset of genes involved in the circadian rhythm. Collectively, our results indicate that the LDL1/2-HDA6 histone modification complex is important for the regulation of the core circadian clock components.
Introduction
The circadian rhythm is an endogenous oscillation widely observed in plants, animals, fungi, and cyanobacteria (Edgar et al., 2012). The plant circadian rhythm is highly associated with multiple important biological processes, and maintained by multiple interconnected loops that generate robust rhythms. The circadian clock central loop is a negative feedback loop composed of the core circadian clock components such as TOC1 (TIMING OF CAB EXPRESSION 1) and CCA1 (CIRCADIAN CLOCK ASSOCIATED 1)/LHY (LATE ELONGATED HYPOCOTYL). TOC1 is highly expressed in the evening, but low expressed at dawn (Alabadi et al., 2001). Furthermore, TOC1 was identified as a repressor of CCA1 and LHY by binding to their promoters in the evening (Gendron et al., 2012; Huang et al., 2012). In contrast, CCA1 and LHY are highly expressed in the morning, but low expressed at nightfall (Schaffer et al., 1998; Wang and Tobin, 1998; Alabadi et al., 2001). CCA1 and LHY bind to the evening element (EE) on the promoter of TOC1 to inhibit its expression (Schaffer et al., 1998; Wang and Tobin, 1998; Alabadi et al., 2001; Nagel et al., 2015). CHE (CCA1 HIKING EXPEDITION) is an evening-expressed TCP-family transcription factor, which also targets the CCA1 promoter to repress its expression. Furthermore, CCA1 and LHY were shown to repress the CHE expression by targeting the CHE promoter (Pruneda-Paz et al., 2009).
Histone modifications play important roles in the regulation of gene expression. Histone methyltransferases and demethylases determine the methylation levels, whereas histone acetylation levels are regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs or HDAs). HDACs and the H3K4 demethylase LSD1 (Lysine-Specific Demethylase 1) are the core components of the Mi2/NuRD and CoREST protein complexes in yeast and animal cells (Khochbin et al., 2001; Lee et al., 2005; Wang et al., 2009). They act co-operatively to repress gene expression in mammals (Huang et al., 2011). The interactions among the core protein components of the HDAC complexes are relatively stable and the HDAC complexes can also interact with various transcription factors under different environmental conditions (Joshi et al., 2013; Liu et al., 2014). FLD (FLOWERING LOCUS D), LDL1 (Lysine-Specific Demethylase-LIKE 1), LDL2, and LDL3 are the LSD1 homologs in Arabidopsis (Jiang et al., 2007). LDL1 and LDL2 act redundantly to regulate FLC (FLOWERING LOCUS C) by H3K4 demethylation (Jiang et al., 2007). Furthermore, Arabidopsis HISTONE DEACETYLASE 6 (HDA6) directly interacts with FLD to repress FLC, MAF4, and MAF5 by reducing H3K4 methylation (H3K4me) and H3 acetylation (H3Ac) to regulate flowering time (Yu et al., 2011). In addition, HDA6 can also interact with LDL1 and LDL2 to regulate gene expression (Hung et al., 2018).
The HDAC inhibitor TSA treated plants show delayed phases and higher amplitudes of TOC1 expression (Perales and Más, 2007). In addition, the expression of Arabidopsis CCA1, LHY, and TOC1 is specifically associated with H3Ac and H3K4me changes (Hemmes et al., 2012; Malapeira et al., 2012), indicating that the expression of the core circadian clock components is associated with H3Ac and H3K4me level changes. Our recent study indicated that CCA1 and LHY can interact with the HDAC complex containing LDL1, LDL2, and HDA6. Furthermore, the LDL1/2-HDA6 complex can be recruit by the transcription repressors CCA1 and LHY to their target genes including TOC1. Since CCA1 and LHY are low expressed at nightfall, the expression of TOC1 is increased due to the release of LDL1/2-HDA6 from the TOC1 promoter (Hung et al., 2018). In this study, we demonstrated that LDL1/2-HDA6 can also interact with TOC1 to regulate the expression of CCA1 and LHY. Furthermore, LDL1/2-HDA6 and TOC1 co-target a subset of genes involved in the circadian rhythm.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana Columbia (Col-0) ecotype was used. Plants were grown at 22°C under 12/12 h light/dark conditions in growth chambers. The mutants used in this study were previously described, including ldl1/ldl2 (Jiang et al., 2007), hda6 (axe1-5) (Yu et al., 2011), hda6/ldl1/2 (Hung et al., 2018), toc1, and cca1/lhy (Wang et al., 2011). 35Spro::LDL1:GFP, 35Spro::GFP:HDA6, LDL1pro::LDL1:GFP and HDA6pro::HDA6:GFP transgenic plants were previously described (Yu et al., 2011; Hung et al., 2018).
The full-length coding sequence (CDS) fragment of TOC1 was PCR-amplified and cloned into the pCR8/GW/TOPO vector (Invitrogen), and then recombined into the PK7WGF2 binary vector or 3xFLAG Gateway vector (Invitrogen1). The 35S::TOC1:GFP vector was transformed into Col-0 WT or hda6/ldl1/2 by the floral dip method.
Bimolecular Fluorescence Complementation (BIFC) Assays
To generate the constructs for BiFC assays, the full-length coding sequence (CDS) fragment of TOC1 was amplified by PCR and cloned into the pCR8/GW/TOPO vector, and then recombined into the pEarleyGate201-YN (Lu et al., 2010). LDL1-YC and HDA6-YC were described in the previous studies (Yu et al., 2011; Hung et al., 2018). Constructed vectors were transformed into Arabidopsis protoplasts or tobacco (Nicotiana benthamiana) leaves for transient assays. Transformed protoplasts and tobacco leaves were then examined by confocal spectral microscope imaging system (NTU-TCS SP5, Leica2).
Yeast Two-Hybrid (Y2H) Assays and Co-immunoprecipitation (Co-IP) Assays
Yeast two-hybrid assays were performed based on the instruction for the Matchmaker GAL4-based two-hybrid system 3 (Clontech). The LDL1, LDL2, and TOC1 full length cDNA fragments were sub-cloned into pGADT7 and pGBKT7 vectors. All constructs were transformed into the yeast (Saccharomyces cerevisiae) strain AH109 by the lithium acetate method, and yeast cells were grown on a minimal medium/-Leu-Trp according to the manufacturer’s instructions (Clontech). Transformed colonies were grown on the medium containing X-α-gal for the α-galactosidase activity assay or minimal medium/-Leu-Trp-His (3DO) with 0.25 mM 3-amino- 1,2,4-triazole (3AT).
Co-immunoprecipitation assays were performed as previously described (Yu et al., 2011). The 35S::TOC1:3xFLAG plasmid was transformed into Arabidopsis protoplasts extracted from LDL1pro::LDL1:GFP or 35Spro::GFP transgenic plants. Total proteins were than extracted from the transformed protoplasts. Anti-GFP (Santa Cruz Biotechnologies, catalog no. SC-9996; 1:3000 dilution) and anti-FLAG (SIGMA catalog no. M2; 1:3000 dilution) antibodies were used as primary antibodies for Western blot. The resulting signals were detected by using a Pierce ECL Western blotting kit (Pierce3).
Quantitative Real-Time PCR (qRT-PCR) Analysis
The TRIZOL reagent (Invitrogen, 15596026) was used for total RNA isolation according to the manufacturer’s instructions. Total RNA treated with 2 μg of DNAse (Promega, RQ1 #M6101) were then used for cDNA synthesis (Promega, #1012891). The iQ SYBR Green Supermix solution (Bio-Rad, #170-8880) was used for real-Time quantitative PCR assays with the CFX96 real-time PCR Detection System (Bio-Rad Laboratories, Inc.). Cycling conditions were started with 95°C/10 min, followed by 45 cycles of 95°C/15 s, 60°C/30 s, and then fluorescent detection, and melting curve detection (65–95°C, incrementing 0.5°C for 5 s, and plate reading). Each sample was normalized by calculating delta quantification cycle (Cq) to the expression of the UBQ10 (Ubiquitin10) internal control and quantified at least in triplicate. The Cq and relative expression level are calculated by the Biorad CFX Manager 3.1 based on the MIQE guidelines (Bustin et al., 2009). Supplementary Table S1 listed the gene specific primers used for qRT-PCR. Standard deviations (SD) represent at least three technical and three biological replicates. The variance in average data is represented by standard error of the mean (SEM). The SD, SEM determination and P-value were calculated using Student’s paired t-test.
Protoplast Transient Assays
The CCA1pro::LUC plasmid construct was previously described (Wang et al., 2011). For transcriptional activity assays, the 35Spro::TOC1, 35Spro::LDL1, 35Spro::HDA6, or 35Spro::GFP effector constructs were co-transformed into protoplasts with CCA1pro::LUC, and the plant samples were collected at ZT0 after 12 h. The relative activities of LUC (luciferase) reporter were standardized by activities of co-expressed Renilla LUC. Experiments were repeated at least three times for each reporter-effector combination. The dual luciferase assay reagent (Promega) was used for Firefly LUC and Renilla LUC detection.
Chromatin Immunoprecipitation (ChIP) Assays and ChIP-seq Data Analyses
Chromatin immunoprecipitation assays were accomplished as previously described (Yu et al., 2011; Hung et al., 2018). Plant seedlings were treated with 1% formaldehyde for chromatin extraction. The extracted DNA was sheared to the mean length near 500 bp by sonication, proteins, and DNA fragments were then immunoprecipitated by the H3K9K14 (Millipore, catalog no. 06-599), H3K4me3 (Milipore, catalog no. 04-745), or GFP (Abcam, catalog no. ab290) antibodies. The cross-link between DNA with immunoprecipitated proteins were reversed, and then analyzed by real-time PCR using specific primers (Supplementary Table S1). The quantification cycle(Cq) was calculated by Biorad CFX Manager 3.1 based on the MIQE guideline (Bustin et al., 2009). Percent input was calculated as 2∧[Cq(IN)-Cq(IP)]X100. Each sample was quantified at least in triplicate, and normalized by calculating delta Cq to the expression of the internal control. Standard deviations (SD) represent at least three technical and three biological replicates. The variance in average data is represented by standard error of the mean (SEM). The SD, SEM determination and P-value were calculated using Student’s paired t-test.
ChIP-seq assays were performed based on previous research (Li et al., 2015, 2016; Hung et al., 2018). The LDL1 ChIP-seq data were deposited to NCBI-Gene Expression Omnibus (GEO) database (GSE118025) (Hung et al., 2018). The ChIP-Seq files from other research groups, GSE35952 (Huang et al., 2012) and (Kamioka et al., 2016), were downloaded from the NCBI-GEO database.
Results
LDL1 and HDA6 Interact With TOC1 and Directly Target on CCA1 and LHY
Our recent study indicated that CCA1/LHY can interact with the LDL1/2-HDA6 complex to repress TOC1 (Hung et al., 2018). In addition, the expression of TOC1, CCA1 and LHY is also associated with H3K4me and H3 acetylation changes (Hemmes et al., 2012; Malapeira et al., 2012). We further analyzed the functional correlation between TOC1 and the LDL1/2-HDA6 complex. TOC1 directly interacted with both LDL1 and LDL2 in BiFC assays by using Arabidopsis protoplasts and Agrobacterium-infiltrated tobacco leaves. The YFP fluorescence signal was detected in nucleus of the transformed cells (Figure 1A and Supplementary Figure S1). The interaction between LDL1, LDL2, and TOC1 was further confirmed by yeast two-hybrid assays (Figures 1B,C) and Co-IP assays using Arabidopsis protoplasts (Figure 1D and Supplementary Figure S1). Furthermore, TOC1 can also interact with HDA6 in BiFC assays (Supplementary Figures S2A,B). These results suggested that TOC1 may recruit the LDL1/2-HDA6 histone modification complex to its target genes such as CCA1 and LHY.
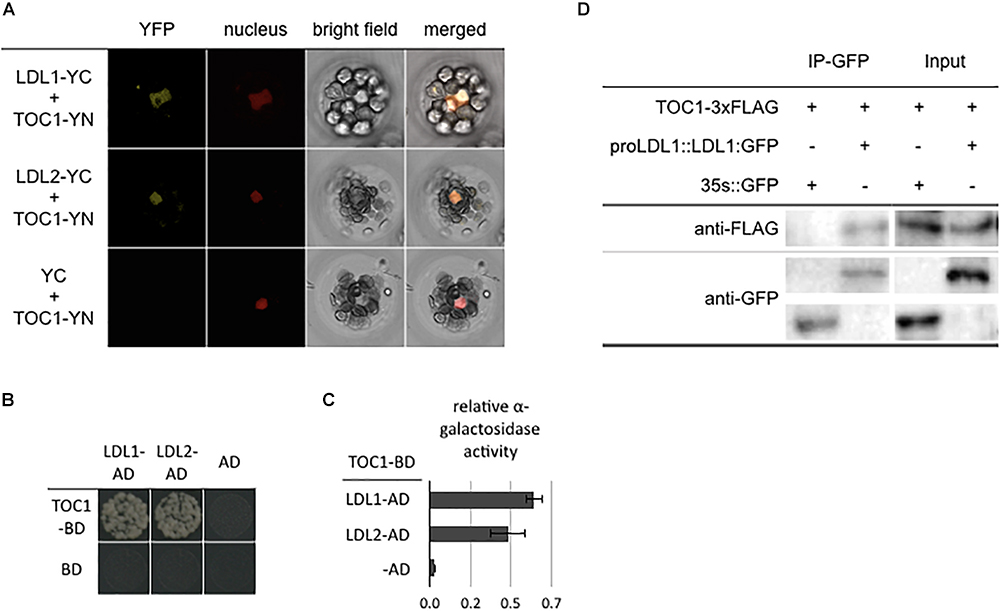
Figure 1. LDL1/LDL2 interact with TOC1. (A) BiFC assays in Arabidopsis protpplasts showing interaction between LDL1/LDL2 and TOC1 in living cells. LDL1, LDL2, and TOC1 fused with the N terminus (YN) or C terminus (YC) of YFP were co-delivered into Arabidopsis protpplasts. The nucleus was indicated by mCherry carrying a nuclear localization signal. (B,C) Yeast two hybrid analysis of the interaction of LDL1/LDL2 with TOC1. LDL1-BD/LDL2-BD with CCA1-AD or LHY-AD was co-transformed into the yeast strain AH109. The transformants were plated on the SD/-Leu-Trp-His medium. (C) Quantitative α-galactosidase assays for protein-protein interaction in yeast. Bars indicate SD from three biological replicates. (D) Co-IP of the native promoter driven LDL1:GFP with TOC1 in LDL1pro::LDL1:GFP transformed Arabidopsis protoplasts. Western blot (WB) was performed with the anti-FLAG and anti-GFP antibodies.
We further analyzed the binding of LDL1 and HDA6 to CCA1 and LHY by ChIP assays. The LDL1:GFP and HDA6:GFP transgenic plants were previously described (Yu et al., 2011; Hung et al., 2018). 14 days old plants grown under 12 h light/12 h dark condition were collected on Zeitgeber time 0 (ZT0) and ZT12. An anti-GFP antibody was used for ChIP assays, and the binding of LDL1 and HDA6 was analyzed by qPCR. We identified that both LDL1 and HDA6 can bind to the promoters of CCA1 and LHY. Furthermore, the binding of LDL1 and HDA6 to the promoters of CCA1 and LHY were significantly decreased on ZT0 compared to ZT12 (Figure 2 and Supplementary Figure S2C). The binding of LDL1 and HDA6 to the CCA1 and LHY promoters is correlated to TOC1 accumulation, since TOC1 is highly expressed at nightfall but low expressed in the morning (Alabadi et al., 2001).
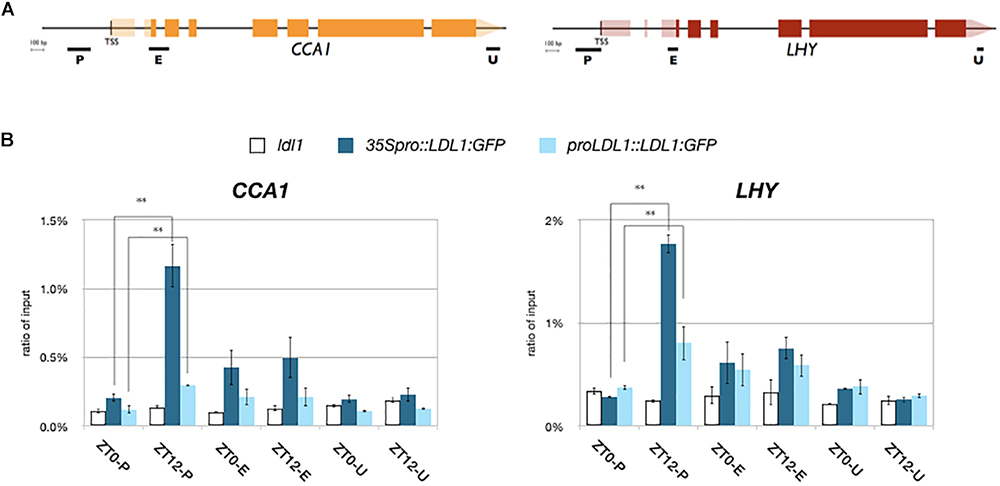
Figure 2. LDL1/LDL2 target on CCA1 and LHY. (A) Schematic diagram of CCA1 and LHY. P: promoter region, E: coding region, U: 3′ UTR. (B) LDL1 bindis to the CCA1 and LHY promoters. 35S pro::LDL1:GFP or LDL1pro::LDL1:GFP was transformed into ldl1. 14 days-old seedlings grown under 12/12: light/dark were harvested on ZT0 or ZT12. ChIP assays were performed with the anti-GFP antibody. The amount of immunoprecipitated DNA was quantified by qRT-PCR. Values represent the average immunoprecipitation efficiencies (%) against the total input DNA. Error bars correspond to standard deviations from three biological replicates. ∗P < 0.05, ∗∗P < 0.005 (Student’s t-test).
TOC1 and LDL1 Co-target Genes Involved in the Circadian Rhythm
Previously, we identified the global binding sites of LDL1 by ChIP-Seq assays (Hung et al., 2018). The GO-BP (Gene Ontology_Biological Process) analysis of LDL1-targeted genes revealed that LDL1 targets on a subset of circadian rhythm genes. Furthermore, LDL1 also binds to a cluster of circadian rhythm genes regulated by CCA1 (Hung et al., 2018). In this study, we further analyzed whether the LDL1 and TOC1 also co-target genes involved in the circadian rhythm.
We compared the previously published TOC1 ChIP-Seq data (Huang et al., 2012) with the LDL1 ChIP-Seq data (Hung et al., 2018). The genome browser views by Integrative Genomics Viewer (IGV) indicated that LDL1 bound to CCA1 and LHY, and the binding peaks of LDL1 are highly correlated with the TOC1 binding regions on CCA1 and LHY promoters (Figure 3A). Among 772 genes occupied by TOC1 (Huang et al., 2012), 195 of them are also co-occupied by LDL1 (P = 1.14e-16) (Figure 3B). Furthermore, the genomic binding regions of TOC1 are closed to the LDL1 binding regions (Figure 3C), indicating that TOC1 and LDL1 tend to bind to the similar genome sites. GO-BP analysis also indicated that LDL1 and TOC1 co-target on a subgroup of genes involved in circadian rhythm and response to cold (Figure 3D). In GO-BP analysis, the ratio of the circadian genes of LDL1/TOC1 co-targeted genes is increased when compared to the LDL1-targeted genes or the TOC1-targeted genes alone (Supplementary Figure S3). Interestingly, the ratio of the circadian rhythm genes is further increased in the LDL1/CCA1/TOC1 co-targeted genes (Supplementary Figure S3). Previous studies indicated that several cis-elements are enriched in the promoters of TOC1 regulated genes, including the (AG/CT)n repeat, G-box (CACGTG), Evening Element (EE)-like and TCP binding site (TBS, GGCCCA) (Gendron et al., 2012; Huang et al., 2012). Similar cis-elements are also enriched in the LDL1-targeted promoter regions (Hung et al., 2018).
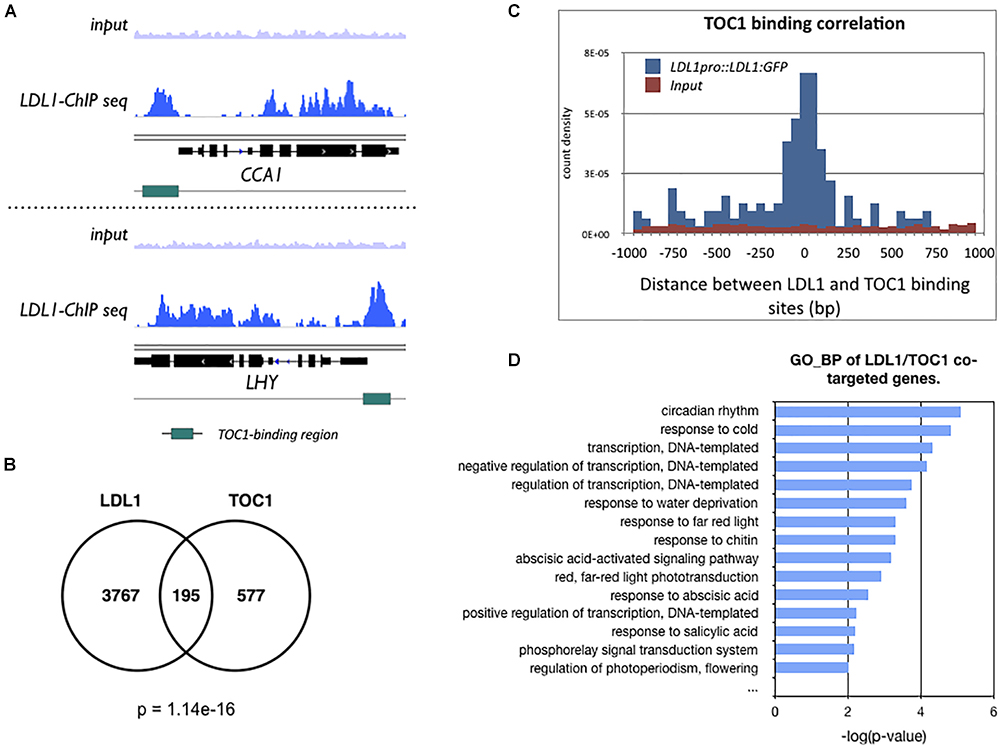
Figure 3. LDL1-occupied sites in the genome identified by ChIP-seq analysis. (A) Integrated genome view of LDL1 binding peaks on CCA1 and LHY. green BARS indicate the TOC1-binding regions form previous published data (Huang et al., 2012). (B) Overlap between TOC1 target genes (Huang et al., 2012) and LDL1 targeted genes (Hung et al., 2018) (hypergeometric distribution of TOC1 and LDL1 co-targeted genes: p = 1.14e–16). (C) Distribution of distances between the total binding sites of LDL1 and TOC1. (D) GO-BP annotation of LDL1/TOC1 co-occupied genes. Annotation terms with p-value < 0.01 were listed.
LDL1/2-HDA6 Is Involved in the Regulation of CCA1/LHY
TOC1 is a repressor and targets on the promoters of CCA1 and LHY. The expression of CCA1 and LHY is decreased in TOC1 over-expressing (TOC1-OE) plants (Gendron et al., 2012; Huang et al., 2012). Furthermore, additional TOC1 expression causes increased period length of CCA1 (Mas et al., 2003a). To investigate the functional relationship between TOC1 and LDL1/2-HDA6, we generated TOC1 over-expressing plants in WT (TOC1-OE) and the hda6/ldl1/2 background (TOC1-OE/hda6/ldl1/2). The binary vector containing CaMV 35S promoter driven GFP:TOC1 (35S::GFP:TOC1) was transformed into WT or hda6/ldl1/2. The expression patterns of CCA1 and LHY were compared by qRT-PCR in wild-type (WT), TOC1-OE and ldl1/2/hda6 plants grown under 12 h light/12 h dark for 14 days. As reported previously (Gendron et al., 2012; Huang et al., 2012), the expression of CCA1 and LHY was decreased in TOC1-OE plants. However, the expression of CCA1 and LHY was not significantly decreased in hda6/ldl1/2 compared to WT (Figure 4A and Supplementary Figures S4A,B). Furthermore, the decrease of CCA1 and LHY expression was recovered when TOC1 was over-expressed in hda6/ldl1/2 (Figure 4A and Supplementary Figures S4B,C). We also compared the daily expression patterns of CCA1, LHY, and TOC1 in ldl1/ldl2, hda6, hda6/ldl1/2, and WT grown under 12 h light/12 h dark conditions. The expression of CCA1 and LHY was not significantly decreased or shifted in ldl1/ldl2, hda6, and hda6/ldl1/2 compared to WT (Supplementary Figure S4A). The expression patterns of other TOC1 targets such as GI, PRR7 and PRR9 in ldl1/ldl2, hda6, and hda6/ldl1/2 were analyzed in our previous study (Hung et al., 2018). XTH27 and AT1G10020 were previously identified to be the target genes regulated by TOC1 (Gendron et al., 2012; Huang et al., 2012), which are also targeted by LDL1 (Hung et al., 2018). The expression of XTH27 and AT1G10020 was increased in ldl1/ldl2, hda6, and hda6/ldl1/2 compared to WT (Supplementary Figure S4C).
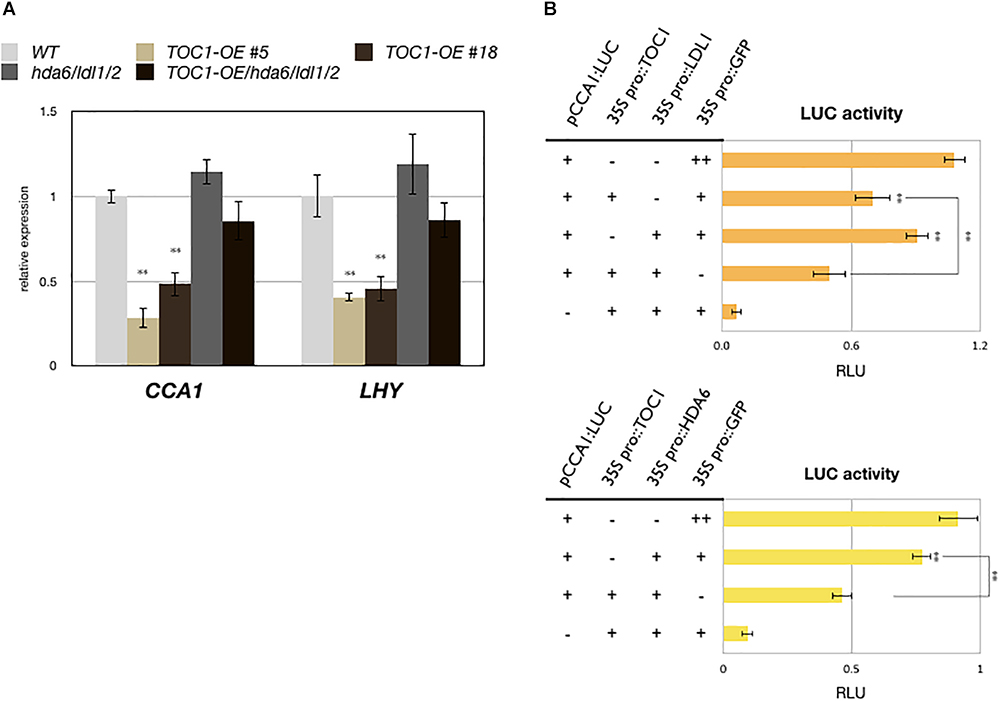
Figure 4. LDL1/2-HDA6 is involved in regulation of CCA1/LHY. (A) Expression of CCA1 and LHY in TOC1-OE plants, hda6/ldl1/2, and WT. Gene expression levels were determined by qRT-PCR and normalized to UBQ10. Plants were grown under 12/12 light/dark for 14 days and collected on ZT0. (B) Transient luciferase assays in CCA1pro::CCA1:LUC (pCCA1:LUC) transformed protoplasts. CaMV 35S promoter driven TOC1, HDA6, or LDL1 effector constructs were introduced into mesophyll protoplasts. Samples were collected on ZT0 after 12 h of transformation. Relative Light Units (RLU) represents firefly luciferase normalized by co-expressed 35S pro::Renilla luciferase. 35Spro::GFP transformed protoplasts were used as the negative control. Data points represent the average of three technical replicates. Error bars correspond to SD from three biological replicates. ∗P < 0.05, ∗∗P < 0.005 (Student’s t-test).
We further analyzed the functional correlation between LDL1, HDA6, and TOC1. CCA1pro::CCA1:LUC (pCCA1:LUC) was co-expressed with 35Spro::TOC1, 35Spro::LDL1, 35Spro::HDA6, or 35Spro::GFP in Arabidopsis protoplasts. Although the activity of CCA1:LUC was only slightly reduced when co-expressed with LDL1, and activity was further decreased when TOC1 was co-expressed with LDL1 (Figure 4B). Similar results were also observed when TOC1 was co-expressed with HDA6 (Figure 4C).
We also analyzed H3K4me and H3Ac levels of CCA1 and LHY in WT, TOC1-OE plants and hda6/ldl1/2. For ChIP-qPCR assays, 14-days old plants grown under 12 h light/12 h dark conditions were collected on ZT0. H3K4me and H3Ac of CCA1 and LHY were decreased in TOC1-OE plants (Figure 5), indicating that TOC1 affects the levels of H3K4me and H3Ac on CCA1 and LHY. We further analyzed H3Ac and H3K4me levels of CCA1 and LHY in 14 days old hda6, ldl1/ldl2, hda6/ldl1/2, and WT on ZT0 and ZT12. The H3Ac and H3K4me levels of CCA1 and LHY were not decreased in hda6, ldl1/ldl2, hda6/ldl1/2 (Supplementary Figure S4D). Interestingly, decreased H3K4me and H3Ac in TOC1-OE were recovered in TOC1-OE/hda6/ldl1/2, since the H3Ac and H3K4me levels of CCA1 and LHY were significant higher in TOC1-OE/hda6/ldl1/2 compared to the TOC1-OE plants (Figure 5). These results suggested that TOC1 is involved in regulation of H3K4me and H3Ac on CCA1 and LHY, and TOC1 repressed CCA1 and LHY expression is dependent on the function of LDL1/2-HDA6 complex.
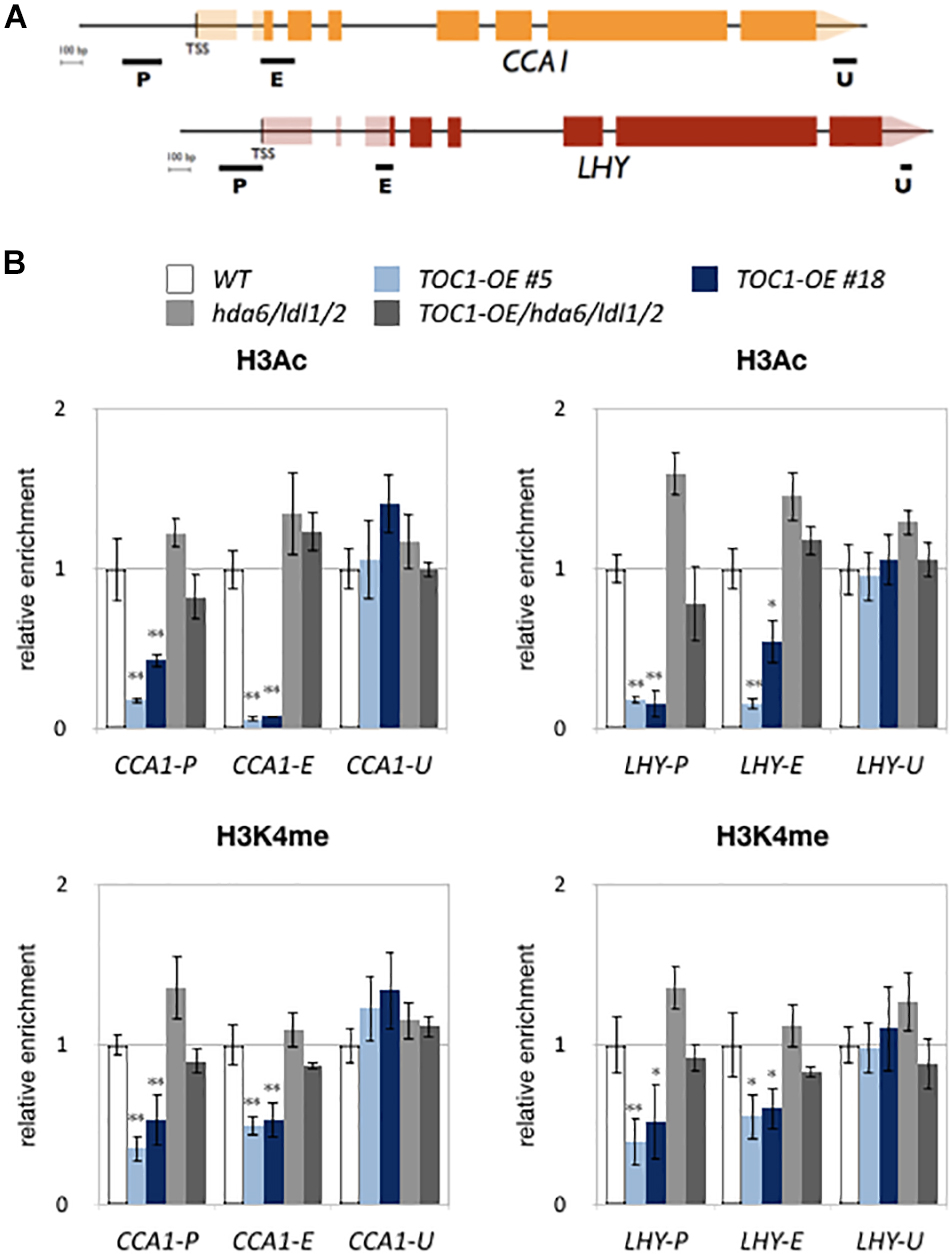
Figure 5. LDL1/2-HDA6 is involved in regulation of H3Ac and H3K4me of CCA1/LHY. (A) Schematic diagram of CCA1 and LHY. P: promoter region, E: coding region, U: 3′ UTR. (B) ChIP analysis of H3ac and H3K4me levels of CCA1 and LHY in TOC1-OE plants on ZT0. The amounts of DNA after ChIP were quantified by qRT-PCR and normalized to ACT2. Plants were grown under 12/12 : light/dark for 14 days. Data points represent the average of three technical replicates. Error bars correspond to SD from three biological replicates. ∗P < 0.05, ∗∗P < 0.005 (Student’s t-test).
Discussion
Arabidopsis HDA6 is a class I RPD3-like histone deacetylase associated with regulation of rRNA and transcription repression (Murfett et al., 2001; Probst et al., 2004; Earley et al., 2006; Liu et al., 2012; Yu et al., 2017). Different transcription factors can recruit HDA6 to regulate the gene expression involved in flowering, leaf development, abiotic stress response, and senescence (Wu et al., 2008; Chen et al., 2010; Yu et al., 2011; Luo et al., 2012; Liu et al., 2014). In animal and yeast cells, HDACs and LSD1 regulate gene expression cooperatively and they are both identified as the core components of Mi2/NuRD and CoREST complexes (Khochbin et al., 2001; Lee et al., 2005; Wang et al., 2009). Our recent study demonstrated that the Arabidopsis H3K4 demethylases LDL1 and LDL2 can interact with HDA6 to repress gene expression (Hung et al., 2018). The LDL1/2-HDA6 complex can also interact with CCA1/LHY and reduce H3Ac and H3K4me levels of the circadian core component TOC1 (Hung et al., 2018). Furthermore, a subset of genes involved in the circadian clock are co-targeted by LDL1 and CCA1 (Hung et al., 2018).
Arabidopsis circadian clock genes are regulated by a complicate feedback regulation network forming multiple interconnected loops. The central loop is comprised of the core clock components, such as TOC1 and CCA1/LHY (Schaffer et al., 1998; Wang and Tobin, 1998; Alabadi et al., 2001; Gendron et al., 2012; Huang et al., 2012; Nagel et al., 2015). The central loop is interlocked with the evening loop and morning loop. PRR5, PRR7, PRR9, and CCA1/LHY constitute the morning loop (Nakamichi et al., 2010; Salomé et al., 2010; Pokhilko et al., 2012), whereas PRR3, GI, ZTL (ZEITLUPE), and TOC1 comprise the evening loop (Kim et al., 2003; Mas et al., 2003b; Para et al., 2007; McClung and Gutiérrez, 2010). We found that LDL1 and CCA1 co-target to a subset of circadian genes, which are repressed by CCA1 in the morning. However, LDL1 also targets to the morning expressed circadian genes, which may not be repressed by CCA1 and LHY (Nagel et al., 2015; Kamioka et al., 2016; Hung et al., 2018). Although the binding of LDL1 on the LDL1/CCA1 co-targeted genes are reduced in the cca1/lhy mutant, their binding is not completely abolished (Hung et al., 2018). These results suggested that in addition to CCA1 and LHY, the LDL1/2-HDA6 complex may also functionally associate with other circadian clock genes. EC (Evening Complex) is also associated with regulation of the circadian genes, which is comprised of LUX (LUX ARRHYTHMO), ELF3 (EARLY FLOWERING3), and ELF4 (EARLY FLOWERING4) (Hazen et al., 2005; Nusinow et al., 2011). A previous study indicated that Arabidopsis HDACs are associated with PRR9 through direct interacting with TPL/TPR (TOPLESS/TOPLESS-RELATED) to regulate the expression of CCA1 (Wang et al., 2013). Further research is required to investigate the functional correlation among LDL1/2-HDA6, PRR9, and EC.
The central loop of Arabidopsis circadian clock is consisted of the core clock components including CCA1, LHY, and TOC1 (Schaffer et al., 1998; Wang and Tobin, 1998; Alabadi et al., 2001; Gendron et al., 2012; Huang et al., 2012; Nagel et al., 2015). Although CCA1 and LHY are low expressed at nightfall, they are highly induced at dawn (Schaffer et al., 1998; Wang and Tobin, 1998; Alabadi et al., 2001). Previously, we found that CCA1 interacts with LDL1 in the morning (Hung et al., 2018). The binding of LDL1 and HDA6 on promoter of TOC1 is higher in the morning but decreased in the evening (Hung et al., 2018). Furthermore, HDA6, LDL1, and LDL2 are constitutively expressed at different time periods. CCA1/LHY can therefore recruit the LDL1/2-HDA6 complex to suppress TOC1 expression at dawn (Hung et al., 2018). In this study, we found that LDL1/2 and HDA6 also interact with TOC1 to regulate the expression of CCA1 and LHY. In consistent with the fact that TOC1 is highly accumulated at nightfall (Alabadi et al., 2001), we found that the binding of LDL1 and HDA6 on the CCA1 and LHY promoters is higher in the evening but decreased in the morning. Since TOC1 is a repressor of CCA1 and LHY, the expression of CCA1 and LHY is decreased with increased TOC1 expression (Gendron et al., 2012; Huang et al., 2012). We found that histone acetylation and H3K4 methylation levels of CCA1 and LHY are decreased in TOC1-OE plants. However, the H3Ac, H3K4me and expression levels of CCA1 and LHY are significantly increased in TOC1-OE/hda6/ldl1/2 compared to the TOC1-OE plants, indicating that the LDL1/2-HDA6 complex is functionally associated with the regulation of CCA1 and LHY expression. Although the expression of TOC1 is highly increased in hda6/ldl1/2 compared to wild type, the expression of CCA1 and LHY is not decreased. It is possible that in addition to LDL1/2-HDA6, other unknown proteins may also be involved in the regulation of CCA1 and LHY expression.
Collectively, we propose a model to demonstrate how the core circadian clock components are regulated by H3K4 demethylation and histone deacetylation (Figure 6). The histone modification complex containing LDL1/2 and HDA6 can interact with both morning accumulated CCA1/LHY (Hung et al., 2018) and evening accumulated TOC1. The transcription repressors CCA1 and LHY can recruit the LDL1/2-HDA6 complex to their target loci including TOC1 in the morning (Hung et al., 2018). Furthermore, TOC1 can also recruit the histone modification complex to its targets such as CCA1 and LHY in the evening.
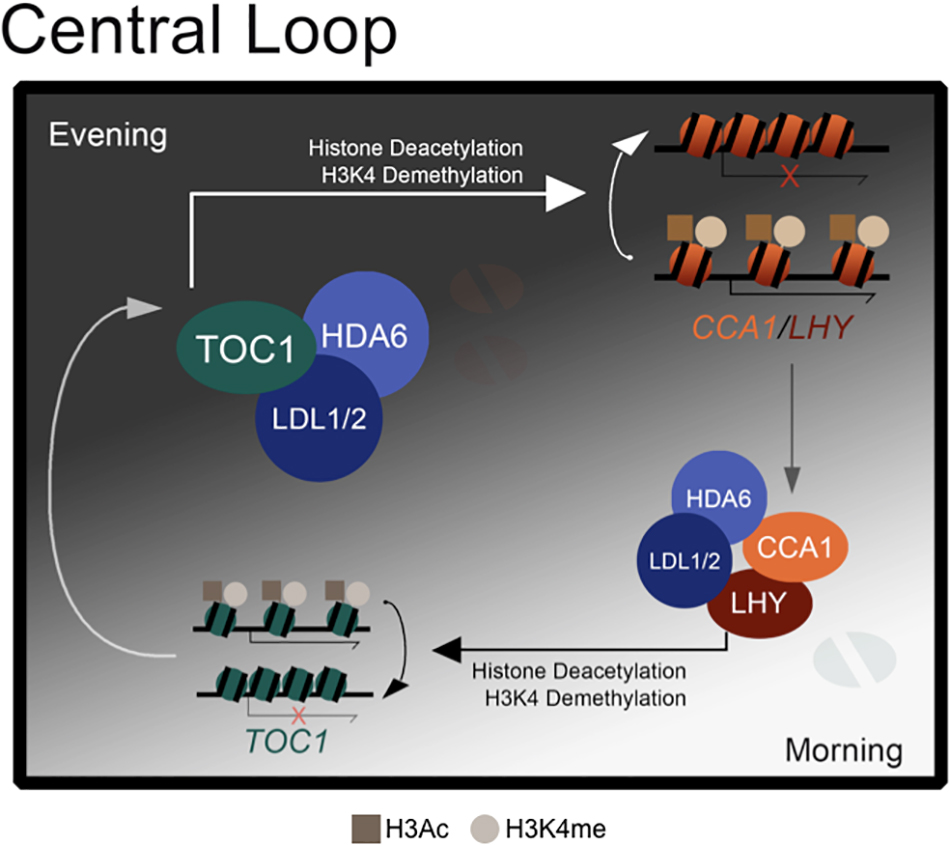
Figure 6. A model for LDL1/2 and HDA6 functions in the regulation of core circadian clock components. Both morning accumulated CCA1/LHY and evening accumulated TOC1 interact with the same histone modification complex containing LDL1/2 and HDA6. CCA1/LHY act as transcription repressors and recruit the histone modification complex to their target loci such as TOC1 in the morning. Meanwhile, TOC1 also recruits the histone modification complex to its targets such as CCA1 and LHY in the evening.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
F-YH, KW, and YC designed the research. F-YH, F-FC, and J-HC performed the research. F-YH, CL, CC, and KW analyzed the data. F-YH and KW wrote the article.
Funding
This work was supported by the Ministry of Science and Technology of the Republic of China (105-2311-B-002-012-MY3 and 106-2313-B-002-003- to KW) and National Taiwan University (106R891501 and 107L893101 to KW). This work was also supported by funding from the Agriculture and Agri-Food Canada A-base and the National Science and Engineering Research Council of Canada (RGPIN/04625-2017 to YC).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. S.-H. Wu. and J.-F. Wu (Academia Sinica) for sharing the toc1-101 mutant and CCA1pro::LUC plasmid construct. We are grateful to the Technology Commons, College of Life Science, National Taiwan University for the convenient use of the Bio-Rad real-time PCR system, the confocal spectral microscope imaging, and Delta-vision systems.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00233/full#supplementary-material
Footnotes
- ^https://www.psb.ugent.be/core-facilities/380-gateway-vectors
- ^https://www.leica-microsystems.com/products/confocal-microscopes/p/leica-tcs-sp5/
- ^https://www.lifetechnologies.com/
References
Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Chen, L. T., Luo, M., Wang, Y. Y., and Wu, K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Earley, K., Lawrence, R. J., Pontes, O., Reuther, R., Enciso, A. J., Silva, M., et al. (2006). Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20, 1283–1293. doi: 10.1101/gad.1417706
Edgar, R. S., Green, E. W., Zhao, Y., Van Ooijen, G., Olmedo, M., Qin, X., et al. (2012). Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464. doi: 10.1038/nature11088
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Hazen, S. P., Schultz, T. F., Pruneda-Paz, J. L., Borevitz, J. O., Ecker, J. R., and Kay, S. A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 102, 10387–10392. doi: 10.1073/pnas.0503029102
Hemmes, H., Henriques, R., Jang, I. C., Kim, S., and Chua, N. H. (2012). Circadian clock regulates dynamic chromatin modifications associated with Arabidopsis CCA1/LHY and TOC1 transcriptional rhythms. Plant Cell Physiol. 53, 2016–2029. doi: 10.1093/pcp/pcs148
Huang, P. H., Chen, C. H., Chou, C. C., Sargeant, A. M., Kulp, S. K., Teng, C. M., et al. (2011). Histone deacetylase inhibitors stimulate histone H3 lysine 4 methylation in part via transcriptional repression of histone H3 lysine 4 demethylases. Mol. Pharmacol. 79, 197–206. doi: 10.1124/mol.110.067702
Huang, W., Perez-Garcia, P., Pokhilko, A., Millar, A. J., Antoshechkin, I., Riechmann, J. L., et al. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79. doi: 10.1126/science.1219075
Hung, F. Y., Chen, F. F., Li, C., Chen, C., Lai, Y. C., Chen, J. H., et al. (2018). The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 46, 10669–10681. doi: 10.1093/nar/gky749
Jiang, D., Yang, W., He, Y., and Amasino, R. M. (2007). Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19, 2975–2987. doi: 10.1105/tpc.107.052373
Joshi, P., Greco, T. M., Guise, A. J., Luo, Y., Yu, F., Nesvizhskii, A. I., et al. (2013). The functional interactome landscape of the human histone deacetylase family. Mol. Syst. Biol. 9:672. doi: 10.1038/msb.2013.26
Kamioka, M., Takao, S., Suzuki, T., Taki, K., Higashiyama, T., Kinoshita, T., et al. (2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28, 696–711. doi: 10.1105/tpc.15.00737
Khochbin, S., Verdel, A., Lemercier, C., and Seigneurin-Berny, D. (2001). Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 11, 162–166. doi: 10.1016/S0959-437X(00)00174-X
Kim, W. Y., Geng, R., and Somers, D. E. (2003). Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. U.S.A. 100, 4933–4938. doi: 10.1073/pnas.0736949100
Lee, M. G., Wynder, C., Cooch, N., and Shiekhattar, R. (2005). An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435. doi: 10.1038/nature04021
Li, C. L., Chen, C., Gao, L., Yang, S. G., Nguyen, V., Shi, X. J., et al. (2015). The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genet. 11:e1004944. doi: 10.1371/journal.pgen.1004944
Li, C. L., Gu, L. F., Gao, L., Chen, C., Wei, C. Q., Qiu, Q., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48, 687–693. doi: 10.1038/ng.3555
Liu, X., Yang, S., Zhao, M., Luo, M., Yu, C. W., Chen, C. Y., et al. (2014). Transcriptional repression by histone deacetylases in plants. Mol. Plant 7, 764–772. doi: 10.1093/mp/ssu033
Liu, X. C., Yu, C. W., Duan, J., Luo, M., Wang, K. C., Tian, G., et al. (2012). HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158, 119–129. doi: 10.1104/pp.111.184275
Lu, Q., Tang, X., Tian, G., Wang, F., Liu, K., Nguyen, V., et al. (2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61, 259–270. doi: 10.1111/j.1365-313X.2009.04048.x
Luo, M., Yu, C. W., Chen, F. F., Zhao, L., Tian, G., Liu, X., et al. (2012). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 8:e1003114. doi: 10.1371/journal.pgen.1003114
Malapeira, J., Khaitova, L. C., and Mas, P. (2012). Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 109, 21540–21545. doi: 10.1073/pnas.1217022110
Mas, P., Alabadi, D., Yanovsky, M. J., Oyama, T., and Kay, S. A. (2003a). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15, 223–236.
Mas, P., Kim, W. Y., Somers, D. E., and Kay, S. A. (2003b). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570.
McClung, C. R., and Gutiérrez, R. A. (2010). Network news: prime time for systems biology of the plant circadian clock. Curr. Opin. Genet. Dev. 20, 588–598. doi: 10.1016/j.gde.2010.08.010
Murfett, J., Wang, X. J., Hagen, G., and Guilfoyle, T. J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13, 1047–1061. doi: 10.1105/tpc.13.5.1047
Nagel, D. H., Doherty, C. J., Pruneda-Paz, J. L., Schmitz, R. J., Ecker, J. R., and Kay, S. A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, E4802–E4810. doi: 10.1073/pnas.1513609112
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua, N. H., and Sakakibara, H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605. doi: 10.1105/tpc.109.072892
Nusinow, D. A., Helfer, A., Hamilton, E. E., King, J. J., Imaizumi, T., Schultz, T. F., et al. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402. doi: 10.1038/nature10182
Para, A., Farre, E. M., Imaizumi, T., Pruneda-Paz, J. L., Harmon, F. G., and Kay, S. A. (2007). PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19, 3462–3473. doi: 10.1105/tpc.107.054775
Perales, M., and Más, P. (2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19, 2111–2123. doi: 10.1105/tpc.107.050807
Pokhilko, A., Fernandez, A. P., Edwards, K. D., Southern, M. M., Halliday, K. J., and Millar, A. J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8:574. doi: 10.1038/msb.2012.6
Probst, A. V., Fagard, M., Proux, F., Mourrain, P., Boutet, S., Earley, K., et al. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16, 1021–1034. doi: 10.1105/tpc.018754
Pruneda-Paz, J. L., Breton, G., Para, A., and Kay, S. A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485. doi: 10.1126/science.1167206
Salomé, P. A., Weigel, D., and Mcclung, C. R. (2010). The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22, 3650–3661. doi: 10.1105/tpc.110.079087
Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I. A., et al. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. doi: 10.1016/S0092-8674(00)81465-8
Wang, L., Kim, J., and Somers, D. E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 761–766. doi: 10.1073/pnas.1215010110
Wang, Y., Wu, J. F., Nakamichi, N., Sakakibara, H., Nam, H. G., and Wu, S. H. (2011). LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23, 486–498. doi: 10.1105/tpc.110.081661
Wang, Y., Zhang, H., Chen, Y., Sun, Y., Yang, F., Yu, W., et al. (2009). LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672. doi: 10.1016/j.cell.2009.05.050
Wang, Z. Y., and Tobin, E. M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. doi: 10.1016/S0092-8674(00)81464-6
Wu, K., Zhang, L., Zhou, C., Yu, C. W., and Chaikam, V. (2008). HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 59, 225–234. doi: 10.1093/jxb/erm300
Yu, C. W., Liu, X., Luo, M., Chen, C., Lin, X., Tian, G., et al. (2011). HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 156, 173–184. doi: 10.1104/pp.111.174417
Keywords: H3K4 demethylases, HDA6, circadian clock, CCA1/LHY, TOC1, Arabidopsis
Citation: Hung F-Y, Chen F-F, Li C, Chen C, Chen J-H, Cui Y and Wu K (2019) The LDL1/2-HDA6 Histone Modification Complex Interacts With TOC1 and Regulates the Core Circadian Clock Components in Arabidopsis. Front. Plant Sci. 10:233. doi: 10.3389/fpls.2019.00233
Received: 25 October 2018; Accepted: 12 February 2019;
Published: 26 February 2019.
Edited by:
Jean-Benoit Charron, McGill University, CanadaReviewed by:
R. Glen Uhrig, University of Alberta, CanadaJun Xiao, Institute of Genetics and Developmental Biology (CAS), China
Copyright © 2019 Hung, Chen, Li, Chen, Chen, Cui and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuhai Cui, Yuhai.cui@canada.ca Keqiang Wu, kewu@ntu.edu.tw
 Fu-Yu Hung
Fu-Yu Hung Fang-Fang Chen1
Fang-Fang Chen1 Chenlong Li
Chenlong Li Chen Chen
Chen Chen Yuhai Cui
Yuhai Cui Keqiang Wu
Keqiang Wu