- Department of Horticulture, Shenyang Agricultural University, Shenyang, China
Bolting is an important agronomic character of the Chinese cabbage, but premature bolting can greatly reduce its commercial value, yield, and quality. Here, early-bolting mutant 1 (ebm1) was obtained from a Chinese cabbage doubled haploid (DH) line “FT,” by using an isolated microspore culture and ethyl methanesulfonate (EMS) mutagenesis. The ebm1 was found to bolt extremely earlier than the wild type “FT.” Genetic analysis indicated that the phenotype of the ebm1 was controlled by a single recessive nuclear gene. Using a mapping population of 1,502 recessive homozygous F2 individuals with the ebm1 phenotype, the ebm1 gene was mapped to between the markers SSRhl-53 and SSRhl-61 on chromosome A04 by using SSR markers, and its physical distance was 73.4 kb. Seven genes were predicted in the target region and then cloned and sequenced; the only difference in the sequences of the ebm1 and “FT” genes was with Bra032169. Unlike that in “FT,” the Bra032169 in ebm1 had a novel 53 bp insertion that caused the termination of amino acid coding. The mutation was not consistent with EMS mutagenesis, and thus, may have been caused by spontaneous mutations during the microspore culture. Based on the gene annotation information, Bra032169 was found to encode the histone methyltransferase CURLY LEAF (CLF) in Arabidopsis thaliana. CLF regulates the expression of flowering-related genes. Further genotyping revealed that the early-bolting phenotype was fully co-segregated with the insertion mutation, suggesting that Bra032169 was the most likely candidate gene for ebm1. No significant differences were noted in the Bra032169 expression levels between the ebm1 and “FT.” However, the expression levels of the flowering-related genes FLC, FT, AG, and SEP3 were significantly higher in the ebm1 than in the “FT.” Thus, the mutation of Bra032169 is responsible for the early-bolting trait in Chinese cabbage. These results provide foundation information to help understand the molecular mechanisms of bolting in the Chinese cabbage.
Introduction
Bolting is a crucial stage in the transition from vegetative to reproductive growth in Cruciferous crops and is regulated by various internal regulatory and external environmental factors (Srikanth and Schmid, 2011; Andrés and Coupland, 2012; Cho et al., 2017). For crops to adapt to their climates, they must bolt at the appropriate time, and premature bolting as a result of environmental conditions and genetic factors, can seriously affect crop yields and quality.
For Brassica rapa crops, bolting is more important than flowering, because it can accurately represent the node of transition from vegetative growth to reproductive growth (Nishioka et al., 2005). Several models have been proposed that explain the molecular mechanisms underlying bolting, the main ones being the florigen theory, the nutrient diversion hypothesis, and the multifactorial control model (Yong et al., 2000). In Arabidopsis thaliana, studies have shown that a total of six regulatory pathways that are involved in flowering time affect bolting, including the vernalization, photoperiodism, gibberellin, autonomous, aging, and ambient temperature pathways (Fornara et al., 2010; Srikanth and Schmid, 2011; Whittaker and Dean, 2017). These pathways were shown to be mutually connected, forming an intricate regulatory network affecting flowering. Among these, the vernalization and photoperiodism pathways are the two most important regulatory pathways. New molecular regulatory mechanisms underlying flowering time are yet to be revealed (Chen and Penfield, 2018; Qin et al., 2019).
A total of 180 genes related to flowering time were found in A. thaliana (Fornara et al., 2010). The regulatory pathways involved in flowering are closely related and influence each other. They were all found to act on specific flowering-related genes to induce the transition of the plant from one growth stage to another. These flowering-related genes were named as floral integrator genes (Simpson and Dean, 2002) and included FT (Flowering Locus T), SOC1 (SUPPESSOR OF OVEREXPRESSION OF CONSTANS 1), and LFY (LEAFY) (Zhang Y. N. et al., 2014).
Previous studies on the genetic mechanisms of bolting in Chinese cabbage showed that naturally breeding materials were mostly used as research materials. Bolting is a complicated quantitative trait, controlled by multiple genes, and easily affected by environmental conditions (Elers and Wiebe, 1984; Nishioka et al., 2005; Wang and Zhu, 2011). Nowadays, quantitative trait locus (QTL) analysis or genome wide association study are carried out using segregated and natural populations, and numerous bolting-related QTLs have been identified (Wu et al., 2012; Li et al., 2015; Wang et al., 2018; Xi et al., 2018; Xiao et al., 2019). Three major genes affecting bolting, BrFLC1, BrFLC2, and BrVIN3.1, have previously been cloned and validated (Xiao et al., 2013; Dechaine et al., 2014; Su et al., 2018).
Chinese cabbage (Brassica rapa L. ssp. pekinensis) is part of the A genome class of the Brassica genus and is an important vegetable crop in China and other parts of east Asia. Bolting is an important agronomic trait in B. rapa crops. Premature bolting can result in non-heading, reduce the commercial value, and seriously affect the yield and quality of the Chinese cabbage. Therefore, the study of the genetic mechanisms controlling the bolting trait in Chinese cabbage is of great importance.
To explore new ways to elucidate the molecular mechanisms of the bolting trait, we created an early-bolting mutant 1 (ebm1) of Chinese cabbage via mutagenesis. The genetic linkage map of ebm1 was constructed using SSR molecular markers. Fine mapping of the mutated gene in ebm1 was conducted and a candidate gene was predicted. Thus, our results lay the foundation for elucidating the molecular mechanisms of the bolting trait in Chinese cabbage.
Materials and Methods
Plant Materials
The ebm1 plants was obtained from a doubled haploid (DH) line “FT” of Chinese cabbage by a combination of isolated microspore culture and ethyl methanesulfonate (EMS) mutagenesis; the mutant was diploid, as determined by flow cytometry, and exhibited stable inheritance after multiple generations (Huang et al., 2016a).
Mutant Trait Evaluation
In the spring of 2017, 30 wild-type “FT” plants (10 plants per replication, with three replications each) and 30 ebm1 plants (the same as that above) were selected for bolting trait identification under natural conditions. The BI (bolting index), DE (days to 5 cm elongated stalk), DE10 (days to 10 cm elongated stalk), and FT (flowering time) were recorded; the measurements were performed according to the method described by Yu et al. (2004) and Liu et al. (2016). The BI was an identification method for bolting, based on bud appearance, bolting, and flowering, and six grades for the bolting investigation and five grades for the bolting identification were established. The BI was evaluated when the first fully opened flower was visible in the wild-type “FT” and ebm1 plants, according to the method of Liu et al. (2016).
Genetic Analysis and Construction of Mapping Population
The ebm1 and its wild-type “FT” were used as the parents. The segregation of the F1, F2, and BC1 generations were observed and recorded at the bolting stage, based on the DE index in ebm1 and the wild-type “FT.” Finally, the genetic characteristics of the bolting were analyzed.
The ebm1 and a Pak-choi (Brassica rapa L. ssp. chinensis) DH line “13A516,” which exhibited late-bolting, were used as the parents to construct the mapping population. At the bolting stage, the recessive homozygous individuals with the early-bolting phenotype (based on the DE index in ebm1) in the F2 generation were selected as the mapping population under natural conditions.
DNA Extraction, PCR Amplification, and Development of Molecular Markers
The young leaves of the parents (ebm1 and Pak-choi DH line “13A516”) and the mapping population of the F2 generation were used for gene mapping, and the genomic DNA was extracted using the cetyltrimethyl ammonium bromide (CTAB) method, as described by Murray and Thompson (1980), with minor modifications.
PCR amplification was performed according to the method of Huang et al. (2016b), and the PCR products were analyzed on a 5% denaturing polyacrylamide gel and silver stained. A total of 176 SSR markers evenly distributed on 10 chromosomes across the Brassica A genome, and they were used to screen the polymorphisms between the ebm1 and “13A516.” After identification of the linkage markers, new SSR markers were further designed using Primer Premier 5.0 software; the sequences of the designed SSR markers are listed in Supplementary Table S1.
Linkage Analysis and Genetic Map Construction
Linkage analysis of the ebm1 with molecular markers was performed using JoinMap 4.0 and the Kosambi mapping function.
Cloning and Sequence Analysis of Candidate Genes
Within the target region, all the genes were identified using the Brassica Database1. The cloning and sequence analysis of the candidate genes were conducted as described by Huang et al. (2016b). Primers were used to amplify the coding sequence (CDS) and promoter (∼2000 bp) of the candidate gene; and the primers have been listed in Supplementary Tables S2–S4.
Phylogenetic Analysis
The amino acid sequence of the candidate gene was acquired from the Brassica Database, and those of its homologous genes in other species were further obtained from the National Center for Biotechnology Information database2. Based on the similarities of the amino acid sequences, the phylogenetic tree was constructed using the MEGA6 software.
Expression Analysis of the Candidate Genes
Quantitative reverse-transcription PCR (qRT-PCR) was used to identify the expression of the candidate gene in the wild-type “FT” and ebm1 plants. Total RNA was isolated from the leaves and flowers using the RNApure Total RNA kit (Aidlab, Beijing, China). The first-strand cDNA was synthesized using HiScript IIQ RT SuperMix (Vazyme, Nanjing, China). The Actin gene and 18S rRNA gene were used as the internal reference control (Huang et al., 2015; Chen et al., 2016). The qRT-PCR was performed using the QuantStudio 6 Flex Real-Time PCR System (ABI, United States).
The expression of flowering-related genes, such as FLC (Flowering Locus C), FT, AGAMOUS (AG), AGAMOUS LIKE 19 (AGL19), and SEPALLATA 3 (SEP3), was examined in the flowers and leaves of the wild-type “FT” and ebm1. The relative expression levels of the genes were calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001). All reactions were performed using three technical and biological replications. The differences in gene expression were analyzed using OriginPro8.0 software. The primers used for the qRT-PCR analysis are listed in the Supplementary Table S5.
Results
Morphological Characterization of ebm1
Compared with the wild-type “FT,” the ebm1 exhibited the early-bolting phenotype, with slightly curled leaves (Figure 1). The BI indicated that the ebm1 belonged to the extremely early-bolting type, and the three other indices of bolting, DE, DE10, and FT, differed significantly between the ebm1 and wild-type “FT” (Table 1).
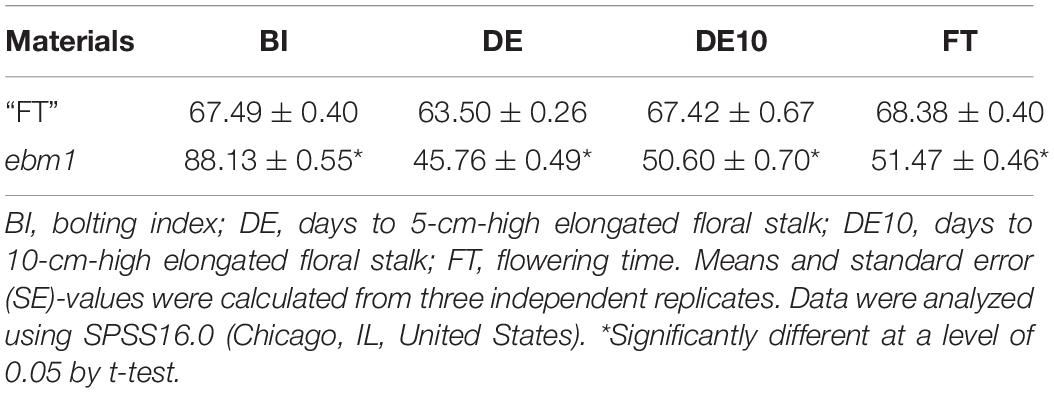
Table 1. Determination of the indicators for bolting traits between the wild type “FT” and ebm1 plants.
Genetic Analysis of ebm1
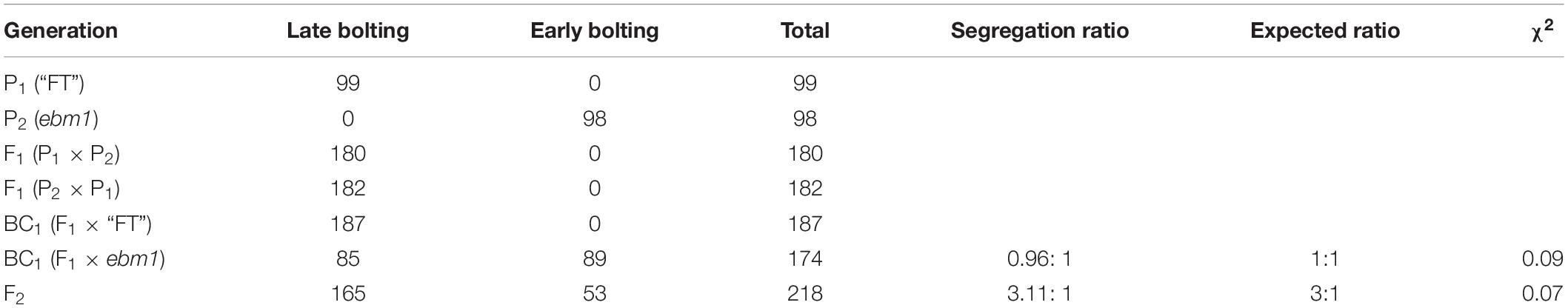
The reciprocal cross F1 plants exhibited the same phenotype as the wild-type “FT,” indicating that the early-bolting phenotype of the ebm1 was controlled by nuclear genes. The F2 population had late bolting, with an early-bolting ratio of 3.11:1 (χ2 = 0.07 < χ20.05 = 3.84), consistent with the Mendelian ratio of 3:1. In addition, the segregation ratio of the F1 × ebm1 in the BC1 progenies was ∼~1:1 (χ2 = 0.09 < χ20.05 = 3.84) (Table 2). These results demonstrated that the phenotype of ebm1 was controlled by a single recessive nuclear gene.
Identification of SSR Markers Linked to the ebm1 Gene
There were 176 SSR markers that were evenly distributed across 10 linkage groups of the Brassica A genome, which were used to detect the polymorphisms between the ebm1 and “13A516.” Of the 176 SSR markers, 81 exhibited polymorphisms between the two parents. Thirty recessive homozygous individuals from the mapping population of the F2 generation were selected to determine whether they were linked with the target gene. The marker cnu_m244a, located on chromosome A04, showed linkage with the ebm1 gene.
New SSR markers close to the cnu_m244a locus were designed and developed, and the results showed that the polymorphic markers SSRhl-1, SSRhl-7, SSRhl-20, SSRhl-30, and SSRhl-32 were tightly linked to the ebm1 gene. A total of 1,502 recessive homozygous individuals with the ebm1 phenotype in the F2 generation were investigated as the mapping population. The results indicated that the recombinant individuals selected using markers SSRhl-30 and SSRhl-32 were included in those for cnu_m244a, however, the recombinant individuals selected using markers SSRhl-1, SSRhl-7, and SSRhl-20 were completely different from those for cnu_m244a, suggesting that they were located on the other side of the ebm1 gene. Based on the recombination frequency statistics, the ebm1 gene was found to be located between the markers SSRhl-20 and SSRhl-30, at a genetic distance of 0.80 and 0.83 cM, respectively (Figure 2A and Supplementary Figure S1).
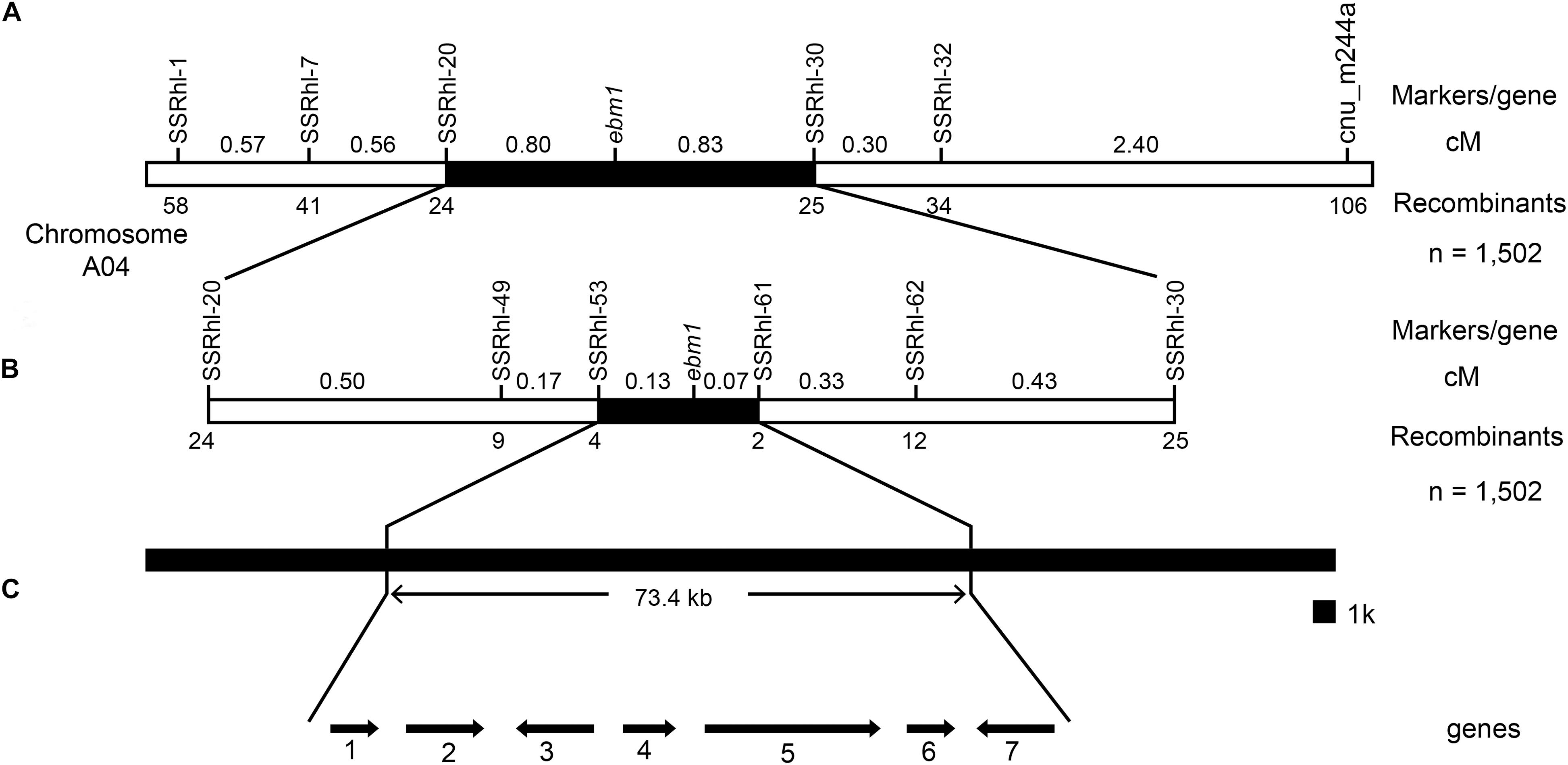
Figure 2. Genetic and physical maps of the ebm1 gene. (A) Preliminary mapping of the ebm1 gene locus. (B) Fine mapping of the ebm1 gene. (C) Candidate genes in the target region. 1–7: Bra032173–Bra032167. Arrows indicate the direction of gene expression. The number of recombinants between the markers and ebm1 is indicated under the linkage map; numbers above the linkage map present the genetic distance between the adjacent markers in centimorgans (cM).
Fine Mapping of the ebm1 Gene
To further narrow the target region, new SSR markers were designed and developed between the markers SSRhl-20 and SSRhl-30. Of these, markers SSRhl-49, SSRhl-53, SSRhl-61, and SSRhl-62 were tightly linked to the ebm1 gene, based on the distribution of the recombinant individuals, revealing that the ebm1 gene was located between the markers SSRhl-53 and SSRhl-61. The marker SSRhl-61 showed two recombinants with the ebm1 gene, and four recombinants were detected between SSRhl-53 and the ebm1 gene; the closest markers were SSRhl-61 and SSRhl-53 and they were 0.07 and 0.13 cM away from the ebm1 gene, respectively. The physical distance between these two markers was estimated to be ~73.4 kb (Figure 2B and Supplementary Figure S1).
Cloning and Sequencing Analysis of the Candidate Genes in the Target Region
Seven genes were predicted to be present in the target region using the Brassica Database (Figure 2C). These seven genes were cloned and sequenced, and the results showed that the only differences between the ebm1 and wild-type “FT” existed in Bra032169. Unlike that in the wild-type “FT,” a 53 bp insertion occurred in the ebm1 (Supplementary Figure S2), and the insertion site was located in the 12th exon (A04: 10,754,930) of Bra032169. Owing to this insertion, the codon at the insertion site was changed from TGC (Thr) to TTT (U), causing the termination of the amino acid coding in the ebm1. The CDS of Bra032169 is 2777 bp and encodes a protein of 686 amino acid residues in the ebm1 (Figure 3). The CDSs of the other genes evaluated in the mapping region showed no difference. In addition, the upstream 2.0 kb promoter sequence of Bra032169 was cloned, and the results showed that the promoter sequence did not differ between the ebm1 and wild-type “FT.”
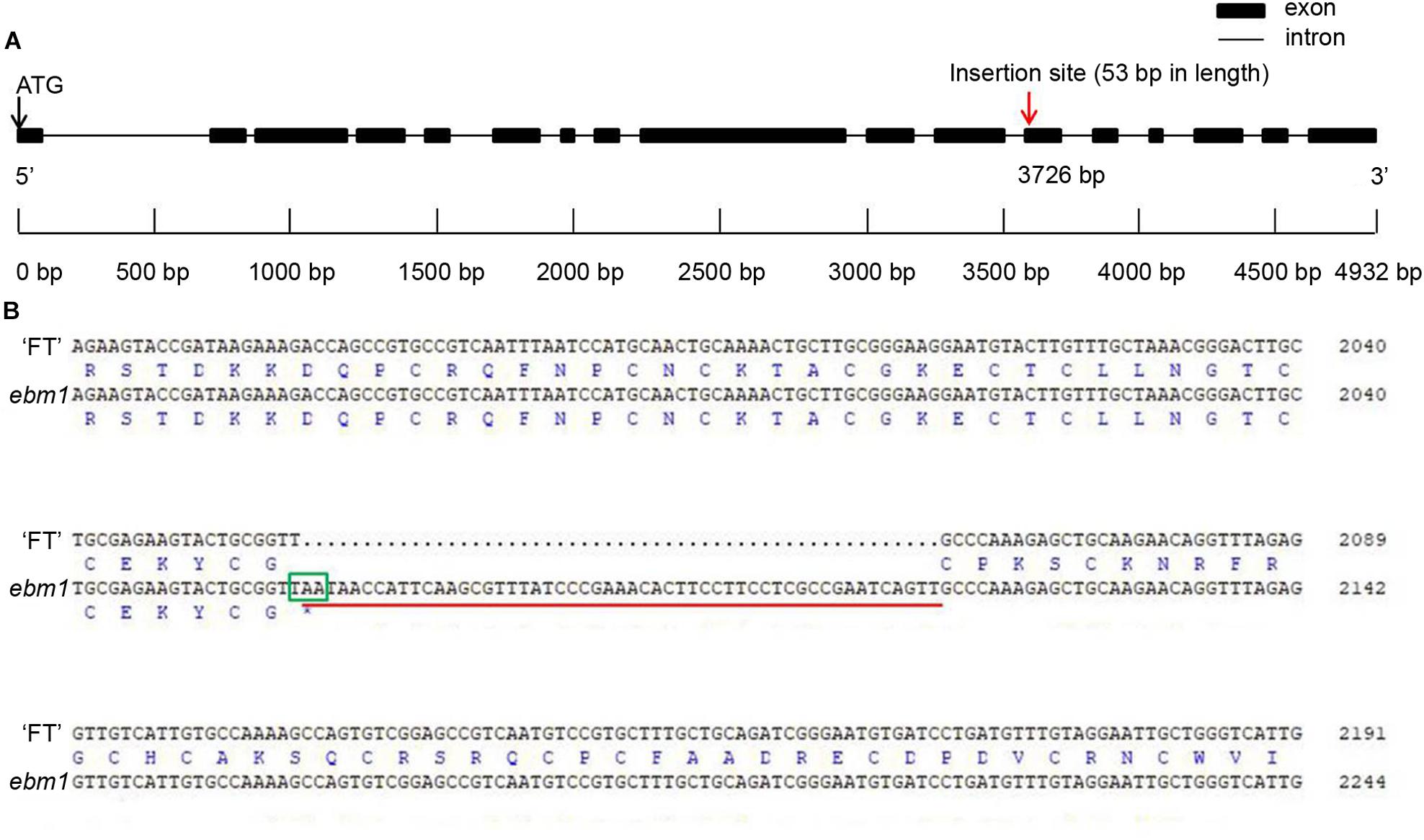
Figure 3. Gene structure and sequence alignment analysis. (A) Gene structure of Bra032169 and the insertion site. (B) Alignment of encoding sequences and amino acid sequences of Bra032169 between wild-type “FT” and ebm1 plants. Red underline, insertion points of the 53 bp sequences; Green frame, the position where amino acid coding was prematurely terminated.
Co-segregation and Candidate Gene Analysis
Annotation information for the seven genes in the target region was obtained from the Brassica Database (Supplementary Table S6). Bra032169 was determined to encode a histone methyltransferase CURLY LEAF (CLF), which is an important H3K27 methyltransferase (Schönrock et al., 2006; Zhang et al., 2007; Jiang et al., 2008; Hennig and Derkacheva, 2009). In A. thaliana, CLF (AT2G23380) is involved in the control of leaf and flower morphogenesis. Mutations in the CLF gene can result in an early-flowering phenotype with curled leaves (Berná et al., 1999; Serrano-Cartagena et al., 2000).
Genotype analysis of 49 recombinants between the two closely linked markers, SSRhl-20 and SSRhl-30, based on the results of primary mapping, indicated that the early-bolting phenotype was co-segregated with the insertion mutation (Supplementary Figure S3). Bra032169 was, thus, predicted to be the most likely candidate gene responsible for the ebm1 phenotype.
A phylogenetic tree was constructed to understand the phylogenetic relationship between these species (Supplementary Figure S4). Amino acid sequence identity between the Chinese cabbage CLF and other homologs varied from 98 to 100%. The amino acid sequence alignment showed a high similarity (98%) with the CLF in A. thaliana. Proteins that share significant homology with CLF are found in Brassica and other species. This sequence similarity among CLF and its homologs suggested that CLF is highly conserved and may serve a conserved function in various organisms.
Expression Analysis of the Candidate Gene
To examine the expression patterns of the Bra032169 gene in ebm1, qRT-PCR analysis was performed using the leaf and flower tissues, and no significant difference was noted between the ebm1 and wild-type “FT” (Figure 4).
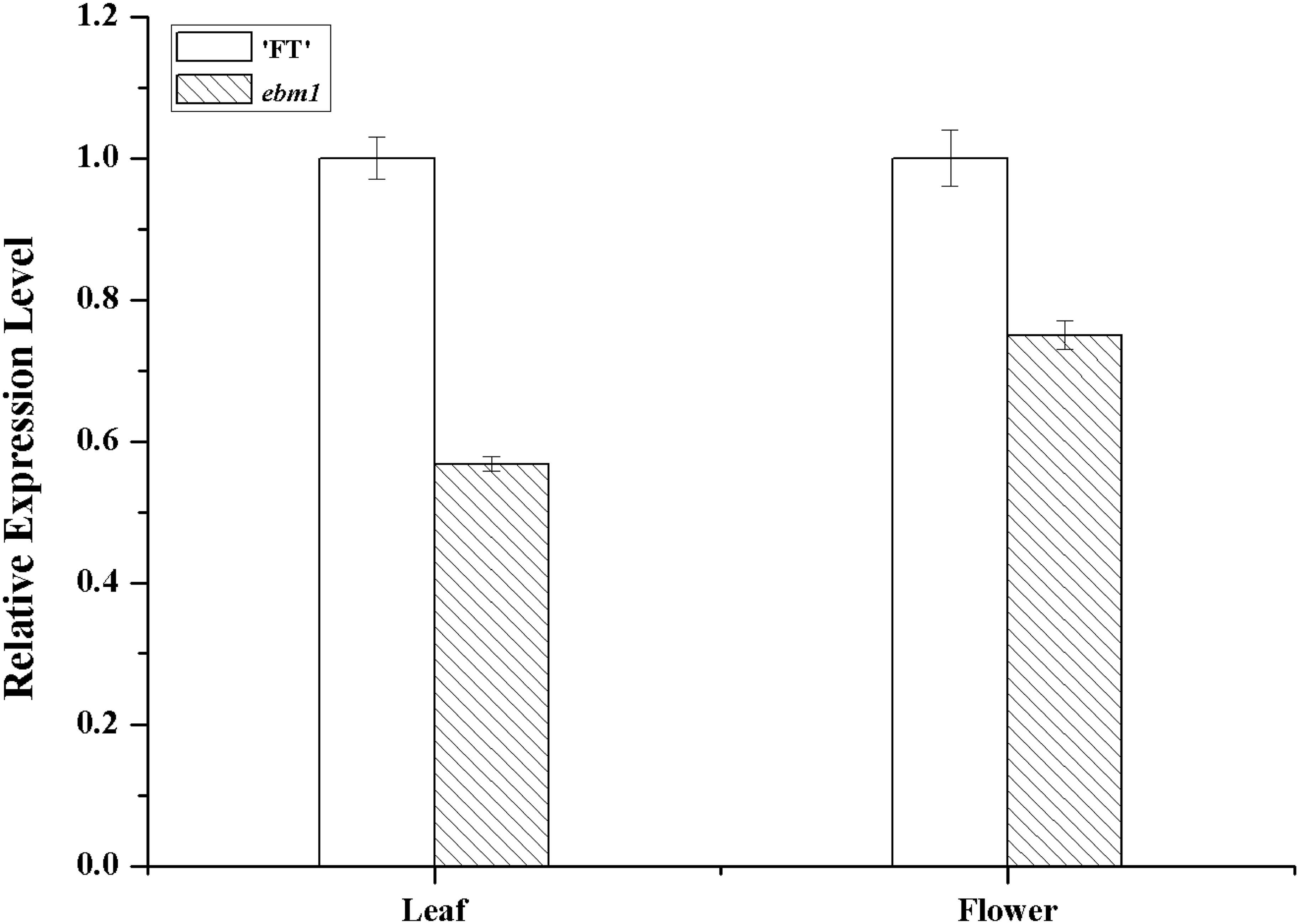
Figure 4. Expression level of Bra032169 in the leaf and flower of wild-type “FT” and ebm1 plants. Statistical analysis was performed using t-tests with significant differences at the 0.01 level.
Previous studies have shown that CLF can regulate the expression of flowering-related genes, such as FLC, FT, AG, AGL19, and SEP3, by methylation modification, thus, regulating flowering in A. thaliana (Bowman et al., 1989; Goodrich et al., 1997; Schubert et al., 2006; Schönrock et al., 2006; Saleh et al., 2007; Zhang et al., 2007; Jiang et al., 2008; Crevillén et al., 2014).
To investigate whether the ebm1 gene affected the expression of flowering-related genes, we evaluated the expression of FLC, FT, AG, AGL19, and SEP3 in the flowers and leaves of the ebm1 and wild-type “FT.” The expression of all these flowering-related genes was higher in the ebm1 than in the wild-type “FT.” The expression of FLC, FT, AG, and SEP3 was significantly higher, suggesting that these genes may influence the early-bolting phenotype in the ebm1 (Figure 5).
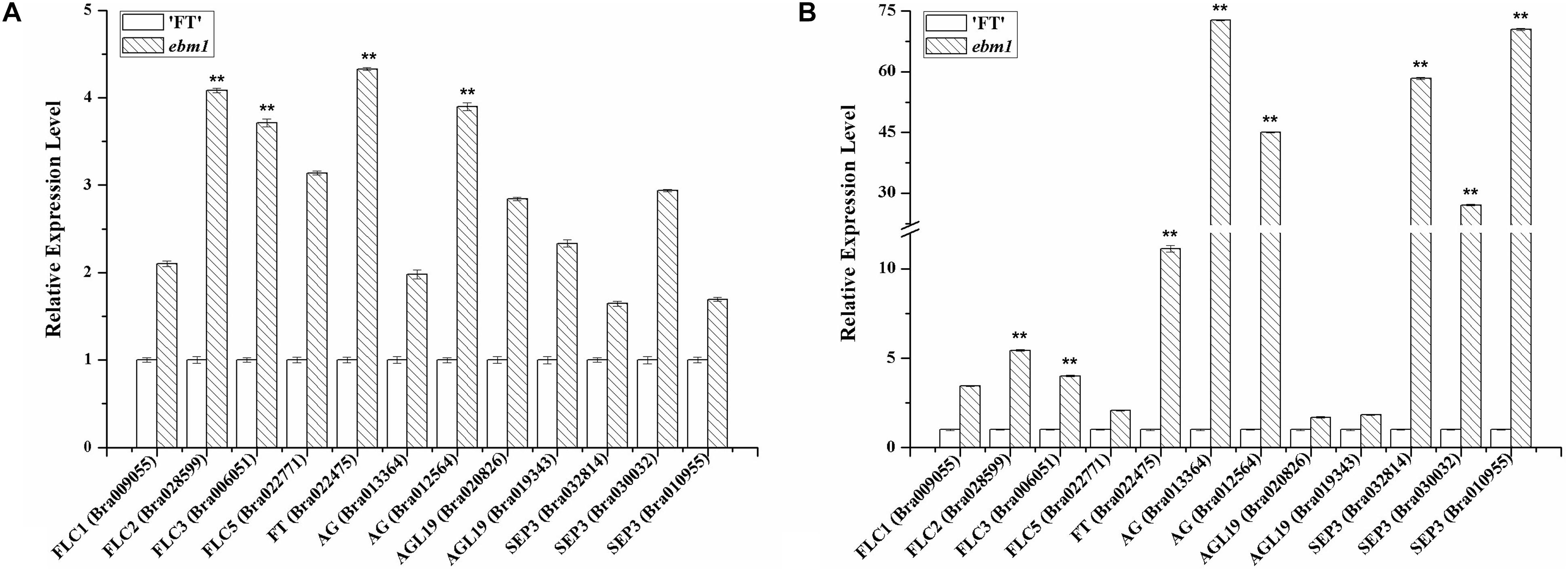
Figure 5. Expression levels of the flowering-related genes in the wild-type “FT” and ebm1 plants. (A) Flower. (B) Leaf. **Significantly different at the 0.01 level by t-test.
Discussion
Bolting is an important agronomic characteristic of Chinese cabbage, but premature bolting can seriously reduce its commercial value, yield, and quality. Therefore, understanding the molecular mechanisms that underlie bolting is important for Chinese cabbage. In the present study, we characterized the ebm1, which exhibited early bolting with slightly curled leaves, derived from a Chinese cabbage DH line “FT,” by using an isolated microspore culture combined with EMS mutagenesis. The genetic backgrounds of the ebm1 and wild-type “FT” were inferred to be highly consistent, and the differences mainly occurred at the mutation sites; thus, the ebm1 is an ideal material for the study of the molecular mechanisms underlying bolting in Chinese cabbage. Genetic analysis demonstrated that ebm1 was controlled by a single recessive nuclear gene. Using 1,502 recessive homozygous individuals with the ebm1 phenotype of the F2 generation as the mapping population, the ebm1 gene was found to be located between the markers SSRhl-61 and SSRhl-53 that were the closest, on chromosome A04. The physical distance was estimated to be 73.4 kb, containing seven predicted genes in the target region. The insertion mutation within the 12th exon of Bra032169 was co-segregated with the early-bolting trait; in conjunction with the results of the CDSs and the annotation information, Bra032169 was predicted to be the most likely candidate gene for the ebm1. Bra032169 encodes a histone methyltransferase CLF, the loss of which can result in the early-flowering phenotype with curled leaves in A. thaliana (Goodrich et al., 1997). To our knowledge, this is the first study to show that the histone methyltransferase CLF corresponds to the early-bolting trait in B. rapa.
Naturally breeding materials have been used to investigate the genetic mechanisms of bolting in the Chinese cabbage. Bolting was considered to be a quantitative trait, and numerous QTLs related to bolting time have been located (Ajisaka et al., 1995; Zhang et al., 2006, 2015; Li et al., 2009; Kakizaki et al., 2011; Zhang Y. N. et al., 2014; Liu et al., 2016; Wang et al., 2018). In the present study, the early-bolting trait of ebm1 was found to be a quality character and controlled by a single gene, suggesting that the mutant gene of the ebm1 might be the major gene related to bolting. This is of vital significance for the study of node genes regulating flowering transition in Chinese cabbage.
Bra032169 was predicted to be the candidate gene for the ebm1, and the sequencing results showed that the insertion mutation occurred in the CDS of Bra032169, resulting in the termination of the amino acid coding. The common types of gene mutation resulting from EMS mutagenesis mainly include point mutation, base pair deletion, and chromosome fragment deletions (Greene et al., 2003). However, the mutation detected in the present study was clearly not consistent with what is expected after EMS mutagenesis. This may be explained by the fact that the ebm1 mutation was obtained via isolated microspore cultures, as some studies have shown that during this process, microspores develop to the callus by dedifferentiation, and the regenerated plants are then derived via redifferentiation. At the callus stage, the parenchyma cells are in the indeterminate status, and the callus cells are easily affected by external conditions and can be mutated during redifferentiation. Generally, the external causes of this type of spontaneous variation may be the types and proportions of hormones and carbon sources in the culture medium (Guo et al., 2014; Kadhimi et al., 2014) or the times of subculture (Nehra et al., 1992). These factors can cause gene mutations (Neelakandan and Wang, 2012), chromosomal abnormalities (Zhang D. et al., 2014), and transposon activation (Azman et al., 2014) when acting on the callus cells, thus, generating regenerated plants with mutant phenotypes. The mutation rate caused by gene mutations is relatively high in the spontaneous mutation type of tissue culture, while the injury of regenerated plants caused by gene mutation is relatively slight, facilitating stable inheritance in the offspring. Therefore, we speculated that the insertion mutation of Bra032169 in ebm1 may have been caused by a spontaneous mutation during the isolated microspore culture, rather than by the EMS mutagenesis.
Flowering is controlled by various epigenetic factors, which activate or repress the transcription of flowering genes (He, 2012). Previous studies have shown that not only was CLF involved in the leaf and flower morphology but that it also played an important role in the regulation of flowering time (Goodrich et al., 1997; Chen et al., 2010). In the present study, the qRT-PCR analysis showed that the expression of Bra032169 in the leaves and flowers had no significant difference between the ebm1 and wild-type “FT.” It has been reported that the variation in promoter sequences could affect the gene expression in plants (Wang et al., 2003; Xia et al., 2006; Li et al., 2019). The upstream 2.0 kb promoter sequence of Bra032169 showed no difference between the ebm1 and wild-type “FT,” which was consistent with the results of the qRT-PCR analysis. Since the insertion mutation in the Bra032169 caused the termination of the amino acid coding in the ebm1, we speculated that the mutation had no influence on the expression of Bra032169, however, loss-of-function proteins may be produced, thus, affecting the regulation of its downstream genes. In A. thaliana, loss of CLF causes early flowering, a reduction in global H3K27me3 levels, and increased expression of FLC and FT, as well as several floral homeotic genes, including AG, AGL19, and SEP3 (Schönrock et al., 2006; Schubert et al., 2006; Adrian et al., 2010; Lopez-Vernaza et al., 2012; Liu et al., 2018; Wu et al., 2018). Payá-Milans et al. (2019) isolated a tilling mutant line, braA.clf-1 in yellow sarson (Brassica rapa ssp. trilocularis), exhibiting the smaller and curled leaves and defective floral organs. ChIP (chromatin immunoprecipitation) and qRT-PCR analysis showed that the floral identity gene AG had high expression level due to reduced H3K27me3 in the leaves of braA.clf-1 mutant. Given this, we further analyzed the expression patterns of these flowering-related genes between the ebm1 and wild-type “FT.” FLC, FT, AG, and SEP3 were significantly higher in the ebm1 than in the wild-type “FT,” which was consistent with the results of previous studies (Schönrock et al., 2006; Schubert et al., 2006; Lopez-Vernaza et al., 2012). The AG, FT, SEP3, and FLC genes have been shown to be necessary for the early-flowering phenotype of clf mutants (Mizukami and Ma, 1992; Goodrich et al., 1997; Pelaz et al., 2001), and these target genes of CLF have antagonistic effects on the flowering time (Lopez-Vernaza et al., 2012). Of these, FT, AG, and SEP3 promote flowering, while FLC inhibits flowering in the clf mutants. Importantly, the effects of the elevated FLC expression are masked by increased FT activity in the clf mutants, causing the early-flowering phenotype. These results revealed that the phenotype of the clf mutants represented a balance between these antagonistic factors (Schönrock et al., 2006; Schubert et al., 2006; Lopez-Vernaza et al., 2012). Therefore, we speculated that the mutation of Bra032169 may affect the expression of FLC, FT, AG, and SEP3, and the antagonistic interactions of these genes influenced flowering time, eventually causing the early-bolting phenotype in the ebm1. Additional evidence is needed to elucidate the functions of CLF in controlling flowering time in the ebm1.
To conclude, the ebm1 gene was mapped to a 73.4 kb genomic region on the A04 chromosome, and Bra032169 was predicted to be the candidate gene that is mutated in the ebm1. Further exploration of the function of the candidate gene is required to characterize the molecular mechanisms underlying bolting in the Chinese cabbage.
Data Availability Statement
The datasets generated for this study can be found in the GenBank under accession numbers BankIt2283090 Seq1 MN708212 and BankIt2283090 Seq2 MN708213.
Author Contributions
HF and SH designed the experiments and supervised this study. SH and LH performed the research and wrote the manuscript. WF, CL, ZL, and XL participated in the research and data analysis. All authors approved the manuscript.
Funding
This research was supported by grants from the National Natural Science Foundation of China (31801854) and the Open Project of Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture of China (IVF201803).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Editage (www.editage.cn) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00547/full#supplementary-material
FIGURE S1 | Examples of markers tightly linked to ebm1 (TIF). (a) Polymorphisms screened by part of the SSR primers between the two parents. (b) Recombinant individuals with SSRhl-30 in the mapping population. (c) Recombinant individuals with SSRhl-20 in the mapping population. (d) Recombinant individuals with SSRhl-61 in the mapping population. (e) Recombinant individuals with SSRhl-53 in the mapping population. M, Marker; P1, “13A516”; P2, ebm1; *, recombinant individuals.
FIGURE S2 | Examination of the PCR products of the candidate gene Bra032169 by agarose gel electrophoresis (TIF). M, Marker D2000.
FIGURE S3 | Alignment of the cDNA sequences of Bra032169 in the 49 F2 recombinant individuals, “FT,” and mutant ebm1 (TIF). Forty-nine early-bolting individuals represent the F2 recombinants of the two closely linked markers, SSRhl-20 and SSRhl-30, based on the results of the primary mapping.
FIGURE S4 | Phylogenetic tree of the ebm1 protein in the Chinese cabbage and its homologs in other species (TIF).
Footnotes
References
Adrian, J., Farrona, S., Reimer, J. J., Albani, M. C., Coupland, G., and Turck, F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22, 1425–1440. doi: 10.1105/tpc.110.074682
Ajisaka, H., Kuginuki, Y., Hida, K., Enomoto, S., and Hirai, M. (1995). A linkage map of DNA markers in Brassica campestris. Breed. Sci. 45(Suppl.):195.
Andrés, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Azman, A. S., Mhiri, C., Grandbastien, M. A., and Tam, S. (2014). Transposable elements and the detection of somaclonal variation in plant tissue culture. Malays Appl. Biol. 43, 1–12.
Berná, G., Robles, P., and Micol, J. L. (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152, 729–742.
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52.
Chen, D. H., Molitor, A., Liu, C. L., and Shen, W. H. (2010). The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20, 1332–1344. doi: 10.1038/cr.2010.151
Chen, J. J., Pang, W. X., Chen, B., Zhang, C. Y., and Piao, Z. Y. (2016). Transcriptome analysis of Brassica rapa near-isogenic lines carrying clubroot-resistant and -susceptible alleles in response to Plasmodiophora brassicae during early infection. Front. Plant Sci. 6:1183. doi: 10.3389/fpls.2015.01183
Chen, M., and Penfield, S. (2018). Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 360, 1014–1017. doi: 10.1126/science.aar7361
Cho, L. H., Yoon, J., and An, G. (2017). The control of flowering time by environmental factors. Plant J. 90, 708–719. doi: 10.1111/tpj.13461
Crevillén, P., Yang, H., Cui, X., Greeff, C., Trick, M., Qiu, Q., et al. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590. doi: 10.1038/nature13722
Dechaine, J. M. D., Brock, M. T., and Weinig, C. (2014). QTL architecture of reproductive fitness characters in Brassica rapa. BMC Plant Biol. 14:66. doi: 10.1186/1471-2229-14-66
Elers, B., and Wiebe, H. J. (1984). Flower formation of chinese cabbage I. Response to vernalization and photoperiods. Scientia Hortic. 22, 219–231. doi: 10.1016/0304-4238(84)90055-4
Fornara, F., de Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in Aradidopsis. Cell 141, 550.e1–550.e1. doi: 10.1016/j.cell.2010.04.024
Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E. M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. doi: 10.1038/386044a0
Greene, E. A., Codomo, A., Taylor, N. E., Henikoff, J. G., Till, B. J., Reynolds, S. H., et al. (2003). Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis thaliana. Genetics 164, 731–740.
Guo, L. F., Xue, F. D., Guo, J. F., and Na, R. (2014). Plant tissue culture: a recent progress and potential applications. Agric. Sci. Technol. 15, 2088–2095.
He, Y. (2012). Chromatin regulation of flowering. Trends Plant Sci. 17, 556–562. doi: 10.1016/j.tplants.2012.05.001
Hennig, L., and Derkacheva, M. (2009). Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 25, 414–423. doi: 10.1016/j.tig.2009.07.002
Huang, S. N., Liu, Z. Y., Li, D. Y., Yao, R. P., and Feng, H. (2016a). A new method for generation and screening of Chinese cabbage mutants using isolated microspore culturing and EMS mutagenesis. Euphytica 207, 23–33. doi: 10.1007/s10681-015-1473-5
Huang, S. N., Liu, Z. Y., Yao, R. P., Li, D. Y., Zhang, T., Li, X., et al. (2016b). Candidate gene prediction for a petal degeneration mutant, pdm, of the Chinese cabbage (Brassica campestris ssp. pekinensis) by using fine mapping and transcriptome analysis. Mol. Breed. 36:26. doi: 10.1007/s11032-016-0452-4
Huang, S. N., Liu, Z. Y., Yao, R. P., Li, D. Y., and Feng, H. (2015). Comparative transcriptome analysis of the petal degeneration mutant pdm in Chinese cabbage (Brassica campestris ssp. pekinensis) using RNA-seq. Mol. Genet. Genomics 290, 1833–1847. doi: 10.1007/s00438-015-1041-7
Jiang, D. H., Wang, Y. Q., Wang, Y. Z., and He, Y. H. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis polycomb repressive complex 2 components. PLoS One 3:e3404. doi: 10.1371/journal.pone.0003404
Kadhimi, A. A., Alhasnawi, A. N., Mohamad, A., Yusoff, W. M. W., and Zain, C. R. B. C. M. (2014). Tissue culture and some of the factors affecting them and the micropropagation of strawberry. Life Sci. J. 11, 484–493. doi: 10.7537/marslsj110814.63
Kakizaki, T., Kato, T., Fukino, N., Ishida, M., Hatakeyama, K., and Matsumoto, S. (2011). Identification of quantitative trait loci controlling late bolting in Chinese cabbage (Brassica rapa L.) parental line Nou 6 gou. Breeding Sci. 61, 151–159. doi: 10.1270/jsbbs.61.151
Li, F., Kitashiba, H., Inaba, K., and Nishio, T. (2009). A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 16, 311–323. doi: 10.1093/dnares/dsp020
Li, X., Huang, S. N., Liu, Z. Y., Hou, L., and Feng, H. (2019). Mutation in EMB1923 gene promoter is associated with chlorophyll deficiency in Chinese cabbage (Brassica campestris ssp. pekinensis). Physiol. Plant. 166, 909–920. doi: 10.1111/ppl.12979
Li, X. N., Wang, W. K., Wang, Z., Li, K. N., Lim, Y. P., and Piao, Z. Y. (2015). Construction of chromosome segment substitution lines enables QTL mapping for flowering and morphological traits in Brassica rapa. Front. Plant Sci. 6:432. doi: 10.3389/fpls.2015.00432
Liu, X., Yang, Y. H., Hu, Y. L., Zhou, L. M., Li, Y. G., and Hou, X. L. (2018). Temporal-specific interaction of NF-YC and CURLY LEAF during the floral transition regulates flowering. Plant Physiol. 177, 105–114. doi: 10.1104/pp.18.00296
Liu, Y. T., Li, C. Y., Shi, X. X., Feng, H., and Wang, Y. G. (2016). Identification of QTLs with additive, epistatic, and QTL × environment interaction effects for the bolting trait in Brassica rapa L. Euphytica 210, 427–439. doi: 10.1007/s10681-016-1710-6
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopez-Vernaza, M., Yang, S. X., Mller, R., Thorpe, F., de Leau, E., and Goodrich, J. (2012). Antagonistic roles of SEPALLATA3, FT and FLC genes as targets of the Polycomb group gene CURLY LEAF. PLoS One 7:e30715. doi: 10.1371/journal.pone.0030715
Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131. doi: 10.1016/0092-8674(92)90271-D
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Neelakandan, A. K., and Wang, K. (2012). Recent progress in the understanding of tissue culture-induced genome level changes in plants and potential applications. Plant Cell Rep. 31, 597–620. doi: 10.1007/s00299-011-1202-z
Nehra, N. S., Kartha, K. K., Stushnoff, C., and Giles, K. L. (1992). The influence of plant growth regulator concentrations and callus age on somaclonal variation in callus culture regenerants of strawberry. Plant Cell Tissue Organ Cult. 29, 257–268. doi: 10.1007/BF00034361
Nishioka, M., Tamura, K., Hayashi, M., Fujimori, Y., Ohkawa, Y., Kuginuki, Y., et al. (2005). Mapping of QTLs for bolting time in Brassica rapa (syn. campestris) under different environmental conditions. Breed. Sci. 55, 127–133. doi: 10.1270/jsbbs.55.127
Payá-Milans, M., Poza-Viejo, L., Martín-Uriz, P. S., Lara-Astiaso, D., Wilkinson, M. D., and Crevillén, P. (2019). Genome-wide analysis of the H3K27me3 epigenome and transcriptome in Brassica rapa. GigaScience 8, 1–13. doi: 10.1093/gigascience/giz147
Pelaz, S., Gustafson-Brown, C., Kohalmi, S. E., Crosby, W. L., and Yanofsky, M. F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26, 385–394. doi: 10.1046/j.1365-313x.2001.2641042.x
Qin, Z. R., Bai, Y. X., Muhammad, S., Wu, X., Deng, P. C., Wu, J. J., et al. (2019). Divergent roles of FT-like 9 in flowering transition under different day lengths in Brachypodium distachyon. Nat. Commun. 10:812. doi: 10.1038/s41467-019-08785-y
Saleh, A., Al-Abdallat, A., Ndamukong, I., Alvarez-Venegas, R., and Avramova, Z. (2007). The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 35, 6290–6296. doi: 10.1093/nar/gkm464
Schönrock, N., Bouveret, R., Leroy, O., Borghi, L., Köhler, C., Gruissem, W., et al. (2006). Polycombgroup proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 20, 1667–1678. doi: 10.1101/gad.377206
Schubert, D., Primavesi, L., Bishopp, A., Roberts, G., Doonan, J., Jenuwein, T., et al. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3lysine27. EMBO J. 25, 4638–4649. doi: 10.1038/sj.emboj.7601311
Serrano-Cartagena, J., Candela, H., Robles, P., Ponce, M. R., Pérez-Pérez, J. M., Piqueras, P., et al. (2000). Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156, 1363–1377.
Simpson, G. G., and Dean, C. (2002). Arabidopsis, the rosetta stone of flowering time? Science 296, 285–289. doi: 10.1126/science.296.5566.285
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Su, T. B., Wang, W. H., Li, P. R., Zhang, B., Li, P., Xin, X. Y., et al. (2018). A genomic variation map provides insights into the genetic basis of spring Chinese cabbage (Brassica rapa ssp. pekinensis) selection. Mol. Plant 11, 1360–1376. doi: 10.1016/j.molp.2018.08.006
Wang, Y., Mai, W. J., Liang, C. Y., and Zhang, M. Y. (2003). Advances on studies of plant promoters. Acta Botanica Boreali Occident. Sin. 23, 2040–2048. doi: 10.3321/j.issn:1000-4025.2003.11.037
Wang, Y. G., Wang, X. S., Wang, X., Zhao, Q. N., Lv, X. X., and Feng, H. (2018). Construction of chromosome segment substitution lines of Chinese cabbage (Brassica rapa L. ssp. pekinensis) in the background of RcBr (B. rapa L. ssp. dichotoma) and characterization of segments representing the bolting trait. Mol. Breed. 38:35. doi: 10.1007/s11032-018-0794-1
Wang, Z. Z., and Zhu, Y. Y. (2011). Advance in studying bolting characteristics of Brassica vegetables. Acta Agric. Shanghai 27, 125–128.
Whittaker, C., and Dean, C. (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33, 555–575. doi: 10.1146/annurev-cellbio-100616-060546
Wu, H. W., Deng, S. L., Xu, H. Y., Mao, H. Z., Liu, J., Niu, Q. W., et al. (2018). A noncoding RNA transcribed from the AGAMOUS (AG) second intron binds to CURLY LEAF and represses AG expression in leaves. New Phytol. 219, 1480–1491. doi: 10.1111/nph.15231
Wu, J., Wei, K. Y., Cheng, F., Li, S. K., Wang, Q., Zhao, J. J., et al. (2012). A naturally occurring InDel variation in BraA.FLC.b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol. 12:151. doi: 10.1186/1471-2229-12-151
Xi, X., Wei, K. Y., Gao, B. Z., Liu, J. H., Liang, J. L., Cheng, F., et al. (2018). BrFLC5: a weak regulator of flowering time in Brassica rapa. Theor. Appl. Genet. 131, 2107–2116. doi: 10.1007/s00122-018-3139-x
Xia, J. D., Cheng, Z. Q., Wu, Y. S., and Ji, P. Z. (2006). Advances of the studies on function and composition of plant promoter. J. Yunnan Agric. Univ. 21, 7–14. doi: 10.1111/ppl.12822
Xiao, D., Shen, H. R., Zhao, J. J., Wei, Y. P., Liu, D. R., Hou, X. L., et al. (2019). Genetic dissection of flowering time in Brassica rapa responses to temperature and photoperiod. Plant Sci. 280, 110–119. doi: 10.1016/j.plantsci.2018.10.027
Xiao, D., Zhao, J. J., Hou, X. L., Basnet, R. K., Carpio, D. P. D., Zhang, N. W., et al. (2013). The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. J. Exp. Bot. 64, 4503–4516. doi: 10.1093/jxb/ert264
Yong, W. D., Zhong, K., Xu, Z. H., Tan, K. H., and Zhu, Z. Q. (2000). Studies on the gene regulation of flowering time in higher plants. Chin. Sci. Bull. 45, 455–466.
Yu, Y. J., Zhang, F. L., Zhao, X. Y., Zhang, D. S., and Zhang, Z. X. (2004). Rapid identification method of late bolting character in Chinese cabbage. China Vegetables 2004, 16–18.
Zhang, D., Wang, Z. H., Wang, N. N., Gao, Y., Liu, Y., Wu, Y., et al. (2014). Tissue culture-induced heritable genomic variation in rice, and their phenotypic implications. PLoS One 9:e96879. doi: 10.1371/journal.pone.0096879
Zhang, X. M., Meng, L., Liu, B., Hu, Y. Y., Cheng, F., Liang, J. L., et al. (2015). A transposon insertion in FLOWERING LOCUS T is associated with delayed flowering in Brassica rapa. Plant Sci. 241, 211–220. doi: 10.1016/j.plantsci.2015.10.007
Zhang, X. W., Wu, J., Zhao, J. J., Song, X. F., Li, Y., Zhang, Y. G., et al. (2006). Identification of QTLs related to bolting in Brassica rapa ssp. Pekinensis (syn. Brassica campestris ssp. pekinensis). Agr. Sci. China 5, 265–271. doi: 10.1016/S1671-2927(06)60048-5
Zhang, X. Y., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5:e129. doi: 10.1371/journal.pbio.0050129
Keywords: Chinese cabbage, bolting, insertion mutation, histone methyltransferase, isolated microspore culture
Citation: Huang S, Hou L, Fu W, Liu Z, Li C, Li X and Feng H (2020) An Insertion Mutation in Bra032169 Encoding a Histone Methyltransferase Is Responsible for Early Bolting in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 11:547. doi: 10.3389/fpls.2020.00547
Received: 14 November 2019; Accepted: 09 April 2020;
Published: 12 May 2020.
Edited by:
Ryo Fujimoto, Kobe University, JapanCopyright © 2020 Huang, Hou, Fu, Liu, Li, Li and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Feng, fenghuiaaa@syau.edu.cn
†These authors have contributed equally to this work and share first authorship
 Shengnan Huang
Shengnan Huang Li Hou†
Li Hou† Hui Feng
Hui Feng