- 1Department of Plant Science and Crop Protection, University of Nairobi, Nairobi, Kenya
- 2International Crops Research Institute for the Semi-Arid Tropics-Kenya, Nairobi, Kenya
- 3Kenya Agricultural and Livestock Research Organization, Genetic Resources Research Institute, Kikuyu, Kenya
Striga hermonthica is the most important parasitic weed in sub-Saharan Africa and remains one of the most devastating biotic factors affecting sorghum production in the western regions of Kenya. Farmers have traditionally managed Striga using cultural methods, but the most effective and practical solution to poor smallholder farmers is to develop Striga-resistant varieties. This study was undertaken with the aim of identifying new sources of resistance to Striga in comparison with the conventional sources as standard checks. We evaluated 64 sorghum genotypes consisting of wild relatives, landraces, improved varieties, and fourth filial generation (F4) progenies in both a field trial and a pot trial. Data were collected for days to 50% flowering (DTF), dry panicle weight (DPW, g), plant height (PH, cm), yield (YLD, t ha−1), 100-grain weight (HGW, g), overall disease score (ODS), overall pest score (OPS), area under Striga number progress curve (ASNPC), maximum above-ground Striga (NSmax), and number of Striga-forming capsules (NSFC) at relevant stages. Genetic diversity and hybridity confirmation was determined using Diversity Arrays Technology sequencing (DArT-seq). Residual heterosis for HGW and NSmax was calculated as the percent increase or decrease in performance of F4 crossover midparent (MP). The top 10 best yielding genotypes were predominantly F4 crosses in both experiments, all of which yielded better than resistant checks, except FRAMIDA in the field trial and HAKIKA in the pot trial. Five F4 progenies (ICSVIII IN × E36-1, LANDIWHITE × B35, B35 × E36-1, F6YQ212 × B35, and ICSVIII IN × LODOKA) recorded some of the highest HGW in both trials revealing their stability in good performance. Three genotypes (F6YQ212, GBK045827, and F6YQ212xB35) and one check (SRN39) were among the most resistant to Striga in both trials. SNPs generated from DArT-seq grouped the genotypes into three major clusters, with all resistant checks grouping in the same cluster except N13. We identified more resistant and high-yielding genotypes than the conventional checks, especially among the F4 crosses, which should be promoted for adoption by farmers. Future studies will need to look for more diverse sources of Striga resistance and pyramid different mechanisms of resistance into farmer-preferred varieties to enhance the durability of Striga resistance in the fields of farmers.
Introduction
Sorghum [Sorghum bicolor (L.) Moench] is a diploid (2n = 2x = 20) cereal grass of the Gramineae family native to Africa (Doggett, 1988). It is the fifth most important cereal globally (Kiprotich et al., 2015) and a major staple food for more than 300 million inhabitants of Africa (Kidanemaryam et al., 2018). In Kenya, sorghum is ranked second after maize (Zea mays L.) in tonnage and production area, which is approximately 117,000 ha (FAOSTAT, 2016). Drought stress and poor soil fertility are the major abiotic factors affecting sorghum production in semi-arid areas (Ejeta and Knoll, 2007). Biotic stresses include diseases such as anthracnose (Colletotrichum graminicola) (Marley et al., 2005), leaf blight (Exserohilum turcicum) (Beshir et al., 2015), and the parasitic weed (Striga hermonthica).
The genus Striga comprises over 30 species, of which S. hermonthica, also known as the purple witchweed, is the most important in sub-Saharan Africa. S. hermonthica parasitizes several major cereal crops including maize, sorghum, rice (Oryza sativa L.), finger millet [Eleusine coracana (L.) Gaertn.], and pearl millet [Pennisetum glaucum (L.) R. Br.]. It remains one of the most devastating biotic factors affecting sorghum production in the western regions of Kenya (Khan et al., 2006) often characterized by low fertility and high moisture stress. The weed germinates on stimulation by a strigolactone (Bouwmeester et al., 2019; Aliche et al., 2020) induced by the host, or in some cases, non-host plants. The germinated Striga then attaches to the roots of the host plants, using a special invasive organ, the haustorium (Yoshida and Shirasu, 2009). The haustorium enables uptake of water and nutrients from the host plants for the growth and development of Striga, as well as the introduction of phytotoxins to the host (Van Hast et al., 2000). Consequently, the growth and development of the host plants become severely affected resulting in yield losses of up to 100% (Kim et al., 2002; Ejeta, 2007).
An adult Striga plant can produce up to 100,000 tiny seeds that can survive in the soil for 20 years or more (Pieterse and Pesch, 1983; Gurney et al., 2005), making it extremely difficult to control. Previous studies have reported Striga seed and plant densities in western Kenya at about 1,188 seeds per mature Striga seed capsule (Van Delft et al., 1997) and about 14 plants per m2 (MacOpiyo et al., 2010), respectively. In Kenya, the three crops most devastated by S. hermonthica are maize, finger millet, and sorghum. Traditionally, farmers have managed Striga in sorghum fields using cultural and mechanical methods including hand weeding (Frost, 1994), intercropping (Aasha et al., 2017), and crop rotations (Oswald and Ransom, 2001) with edible legumes such as common bean (Phaseolus vulgaris L.), pigeon pea [Cajanus cajan (L.) Millsp.], and mung bean [Vigna radiata (L.) R. Wilczek].
Pathogenic isolates of Fusarium oxysporum f. sp. Strigae have been reported to be effective as bioherbicides, especially when integrated with other control practices (Rebeka et al., 2013). The push–pull technology that involves the intercropping of cereals with a trap crop (pull), usually Napier grass (Pennisetum purpureum), and a forage legume, usually desmodium (Desmodium spp.), as a push crop (Khan et al., 2011) has resulted in low adoption due to the lack of use for desmodium by farmers. “Suicidal death” of Striga, which is achieved by inducing the germination of Striga by non-host legumes, has been employed in the reduction of Striga seed banks (Rubiales and Fernández-Aparicio, 2012). Chemical control has been tested in maize (Menkir et al., 2010) and sorghum (Bouréma et al., 2005; Dembélé et al., 2005; Tuinstra et al., 2009), but it is not environmentally friendly besides being unaffordable for the average sorghum farmer in Kenya. The most effective and practical solution for the smallholder sorghum farmers is to develop Striga-resistant sorghum varieties.
Sorghum germplasm screening against Striga is the first step toward the identification of Striga-resistant genotypes. Resistance has been reported among cultivated sorghum varieties including N13 (Haussmann et al., 2004), SRN39, FRAMIDA, and IS9830 (Ezeaku, 2005; Rodenburg et al., 2005). The resistance mechanism in N13 is a hypersensitive reaction characterized by thickening of the cell wall and silica deposition that limits xylem–xylem connection with the host plants (Maiti et al., 1984). SRN39, on the other hand, is known to harbor pre-attachment resistance that results in the production of a low germination stimulant, orobanchol (Satish et al., 2012; Mohemed et al., 2016). N13 and SRN39 have been used extensively as sources of resistance (Hess and Ejeta, 1992; Ngugi et al., 2015; Yohannes et al., 2015; Ali et al., 2016), and the quantitative trait loci (QTLs) responsible for resistance have been mapped (Haussmann et al., 2004; Satish et al., 2012). The outcrossing nature of Striga that results in different ecotypes with mixed responses to different genotypes (Fantaye, 2018) would require the pyramiding of multiple alleles from diverse sources into farmer-preferred varieties if the resistance were to be durable. Crop wild relatives (CWRs) and landraces of sorghum have been reported with significantly higher resistance to Striga than N13 and SRN39 (Mbuvi et al., 2017; Mallu et al., 2021). Such reports provide strong justification for more screening of CWRs and landraces toward the identification of additional sources of resistance to Striga.
Recommended methodologies for effective field screening include inoculation with Striga seeds, appropriate experimental designs with sufficient replications, quantitative data scoring, and inclusion of susceptible and resistant checks at regular intervals (Haussmann et al., 2000b; Rodenburg et al., 2005). A quantitative measure such as “area under Striga number progress curve” (ASNPC) alongside Striga count, Striga vigor, and grain yield has been used in past studies (Haussmann et al., 2012; Abate et al., 2017) with great success. The aim of this study was to screen for novel sources of resistance to Striga using both wild and landrace sorghum accessions, improved sorghum varieties, selected F4 progenies, and known Striga-resistant sources, such as N13, FRAMIDA, HAKIKA, IS9830, and SRN39, as checks.
Materials and Methods
Plant Material and Experimental Design
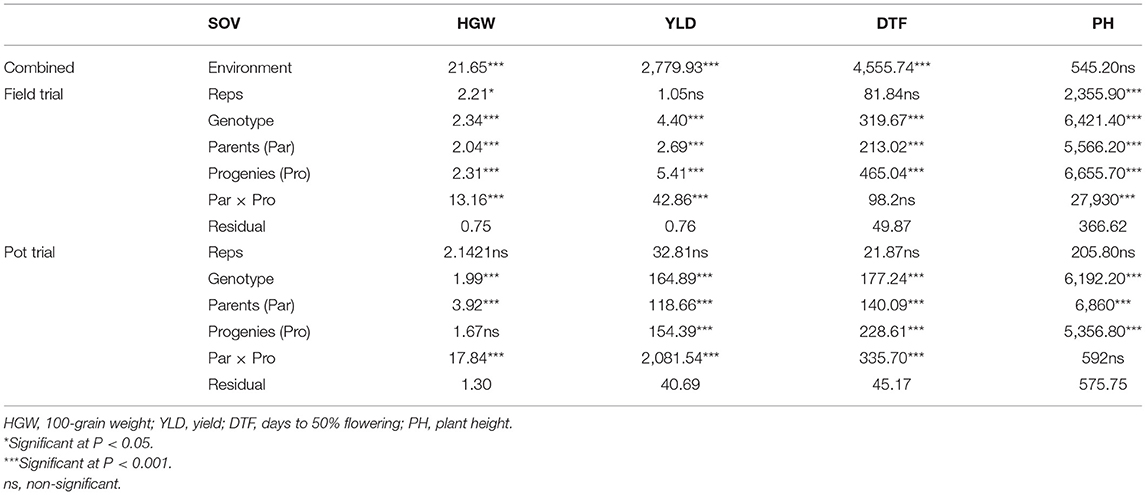
Sixty-four sorghum genotypes (Table 1) consisting of 17 wild relatives, 8 landraces, 13 improved varieties (high yielding), and 26 F4 progenies of selected parents were used in this study. The parental lines of the crosses included five improved varieties (B35, E36-1, MACIA, ICSVIII IN, and F6YQ212) and five landraces (LODOKA, OKABIR, IBUSAR, AKUOR-ACHOT, and LANDIWHITE). Successful crosses were selected morphologically at F1 and advanced to F4 using the bulk-population method.
The field and pot trials were set up during the long rainy season of 2019 at a S. hermonthica hotspot in Alupe, Busia County, Western Kenya. Alupe is located on 34° 07′ 28.6″ E and 00°30′ 10.1″ N with an annual rainfall range of 1,100–1,450 mm and daily mean temperatures of 24°C. The area has a bimodal rainfall pattern of long and short rains. Both experiments were laid out in a square lattice design with three replications, each block consisting of eight plots. The field experiment was planted in a Striga-infested field with a spacing of 75 cm between rows and 20 cm between plants in the row. Each row contained 21 plants. Striga inoculum was prepared by mixing 5 kg of sand with 15 g of Striga seeds that had been harvested from the same location in the previous season. A supplemental Striga inoculum of 15 g was spread along each row during planting to improve the consistency of Striga seed load across the plot. Phosphorus (P) was applied at the rate of 90 Kg ha−1 after thinning, while nitrogen (N) was applied at the rate of 92 kg ha−1 when the plants were 45–50 cm tall, which was around 30 days after germination. Insect pests, especially fall armyworm (Spodoptera frugiperda) and cutworms (Agrotis spp., Spodoptera spp., and Schizonycha spp.), were controlled using Voliam Targo® SC (Syngenta Crop Protection AG, Switzerland) containing active ingredients such as chlorantraniliprole and abamectin. The field experiment was purely rainfed.
For the pot experiment, pots of 30 cm diameter were filled with 20 kg of Striga-free soil obtained from Striga-free field. Each pot contained four plants and was used to represent a plot. The pot experiment was set up in the field alongside the field experiment and was not under any shelter. The pot experiment was rainfed as much as possible but due to the restricted pot size and high levels of evaporation from the pots, watering was done only when the plants were close to the permanent wilting point. Striga inoculation was done by adding 5 g of Striga inoculum to each pot. The application of fertilizer and insect pest control was carried out as already described.
Agronomic and Striga Data Collection
The data on agronomic traits were collected from six randomly selected plants from each plot, while the data on Striga response traits were collected per plot. The agronomic data were collected for days to 50% flowering (DTF), dry panicle weight (DPW, g), plant height (PH, cm), yield (YLD, t ha−1), and 100-grain weight (HGW, g), at relevant stages as per the recommendations of the International Board for Plant Genetic Resources (IBPGR) and the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) (1993). The date of first Striga emergence was recorded followed by Striga count at 2-week intervals until maturity. The number of Striga plants forming capsules (NSFC) per plot was recorded at 105 days after sowing. We used the overall disease score (ODS) and the overall pest score (OPS) on a scale of 1–9 to account for any diseases and pests observed, ranging from leaf blight (Helminthosporium turcicum), ladder leaf spot (Cercospora fuscimaculans), zonate leaf spot (Gloeocercospora sorghi), anthracnose (C. graminicola), spider mites (Oligonychus pratensis and Tetranychus urticae), sorghum midge (Contarinia sorghicola), and sorghum shoot fly (Atherigona soccata).
ANOVA and Striga Data Analysis
The maximum above-ground Striga (NSmax) was calculated as suggested by Rodenburg et al. (2006). The ASNPC was calculated according to the formula suggested by Haussman et al. (2000) as follows:
where n is the number of Striga assessment dates, Yi is the Striga count at the ith assessment date, and ti is the number of days after sowing at the ith assessment date.
ANOVA and means for quantitative traits were generated in the alpha lattice design using Genstat software version 19.1 (VSN International, 2017). Treatment means were compared using Fisher's protected least significant differences at P ≤ 0.05. The estimates of phenotypic and genotypic variance and the genotypic and phenotypic coefficients of variation were performed based on the formula proposed by Syukur et al. (2012).
Genotypic variance:
Phenotypic variance:
where = genotypic variance, = phenotypic variance, = environmental variance (i.e., error mean square from the ANOVA), MSg = mean square of genotypes, MSe = error mean square, and r = number of replications.
Genotypic coefficient of variation:
Phenotypic coefficient of variation:
where = genotypic variance, = phenotypic variance, and x is grand mean of a character.
Simple linear correlation coefficients were calculated to understand the relationship among the studied agronomic traits for each trial according to the formula given below:
where cov is the covariance, σx is the SD of x, and σy is the SD of Y.
Phenotypic correlations across the field and pot trials were estimated, and correlation plots were drawn by using R Version 4.0.4 according to the formula described by (Hallauer et al., 2010):
where rx,y is the correlation coefficient of each trait between the two sites, x (field trial), and y (pot trial); and ȳ are the means of the values of each of the traits in the field and pot trials, respectively. The significance of linear relationships in phenotypic correlation coefficients across the two trials was compared with r-coefficient values and the associated degrees of freedom (n = 2), at the probability levels of P = 0.05, 0.01, and 0.001.
Heritability Estimates
Estimations of broad-sense heritability (H2) for all traits were calculated based on parental and family means, respectively, according to the formula described by Allard (1960):
H2bs = heritability in broad sense, = genotypic variance, and = phenotypic variance. H2 scores were classified according to Robinson et al. (1949) as follows: 0–30% = low, 30–60% = moderate, and >60% = high.
Genotyping, Genetic Relatedness, and Confirmation of True Crosses
Molecular data of all the 37 parental genotypes in Table 1, which consisted of 17 wild accessions, 8 landraces, and 12 improved varieties, were generated for the genetic diversity analysis. DNA extraction, genotyping, and filtering of raw SNPs were performed as described by Ochieng et al. (2020). A neighbor-joining (NJ) dendrogram was generated using the Trait Analysis by aSSociation, Evolution, and Linkage (TASSEL) software version 5.2.67 (Bradbury et al., 2007). To undertake the hybridity confirmation, DNA was extracted from additional 115 F4 progenies that were representative of all the crosses, bringing the total number of individuals genotyped to 153. The filtered SNP variant call file for each of the genotyped F4 progenies was parsed through the GenosToABHPlugin in TASSEL 5.2.67 alongside the corresponding parents to obtain informative biallelic SNPs in the ABH format (i.e., female parent alleles as “A,” male parent allele as “B,” and heterozygotes as “H”).
Comparing the Agronomic Performance of Parents and Progenies Under Striga Field Trial Conditions
Crosses involving parents that failed to germinate were excluded from the analysis. The percentage increase or decrease in the performance of F4 crossover midparent (MP) was calculated to observe the residual heterotic effects for HGW and NSmax. The average F4-values per cross were used for the estimation of residual heterosis expressed in percentage over MP as described by Turner (1953).
where,
MP-value = (P1 + P2)/2
Residual heterosis = [(F4 – MP)/MP] × 100
Results
Field Trial and Pot-Screening Trial
All the trait means from the field and pot trials are provided in Supplementary Table 1. The two environments had an effect on all traits except PH (Table 2) and OPS (Table 3). Significant differences (P ≤ 0.001) were observed in the agronomic performance of genotypes for all traits across the field and pot trials (Table 2). There were also significant differences (P ≤ 0.001) in the performance of the parental lines when compared with the progenies, except for DTF in the field trial and PH in the pot trial (Table 2). We observed more consistency across replications in the pot trial for all agronomic traits than in the field trial, where differences across replications were observed for HGW and PH (Table 2).
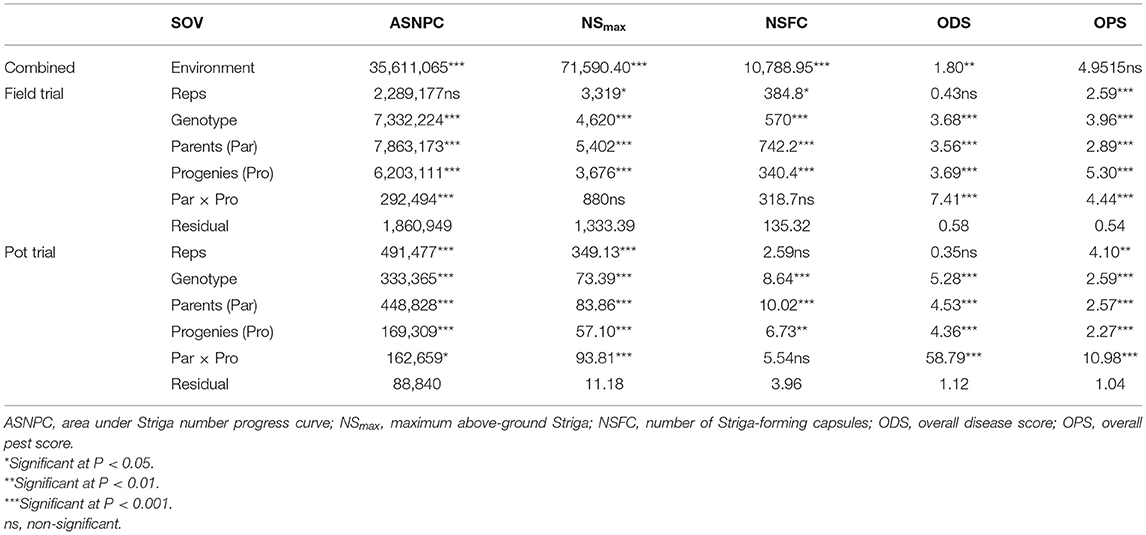
Table 3. Mean squares of Striga-, disease-, and pest-related traits measured under field trial and pot trial.
Significant differences (P < 0.001) were observed between genotypes for all Striga-related traits, as well as for ODS and OPS (Table 3). The performance of parents against their progenies also revealed significant differences (P < 0.001) for ASNPC, ODS, and OPS. More consistency across replications was observed in the field trial for ASNPC and ODS and in the pot trial for NSFC and ODS (Table 3). Yield-related traits (i.e., YLD and HGW) were consistently higher in the pot trial than in the field trial, while Striga-related traits (i.e., ASNPC and NSmax) were lower in the pot trial in comparison with the field trial (Figure 1).
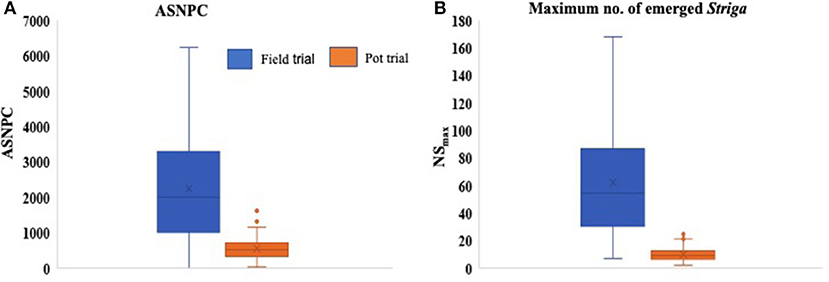
Figure 1. Box plots comparing the overall means of area under Striga number progress curve (ASNPC) (A) and maximum above-ground Striga (NSmax) (B) of genotypes in the field and pot trials.
Trait Correlations and Heritability
We observed positive and significant correlations between the yield-related data collected from the field and pot trials for all traits (i.e., PH, DPW, YLD, and HGW) except DTF (Figure 2). There was a weak non-significant correlation between the two trials for all the biotic stress-related traits (i.e., NSFC, ASNPC, NSmax, and ODS) except OPS (Figure 2).
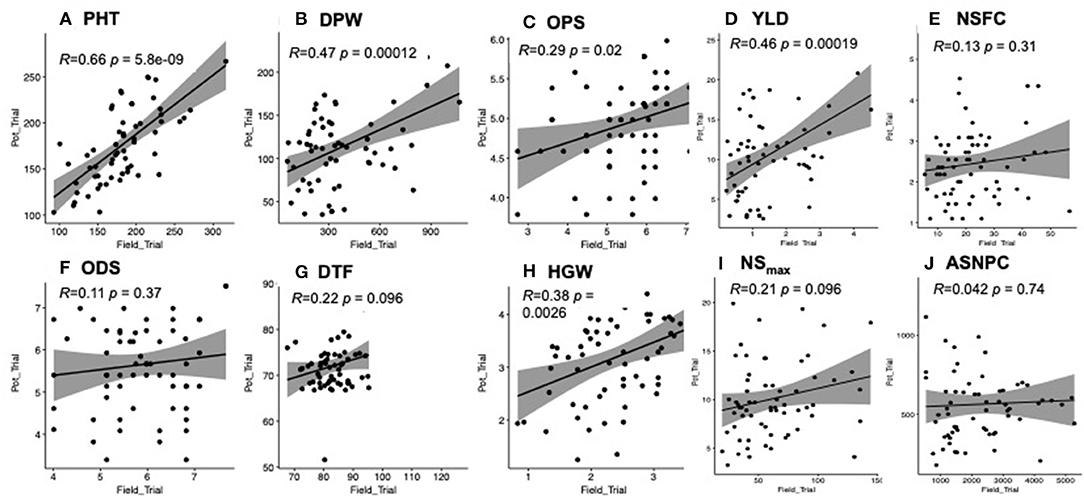
Figure 2. (A-J) Correlation of traits between field and pot trials. Significant correlations were observed for plant height (PH), dry panicle weight (DPW), overall pest score (OPS), yield (YLD), and 100-grain weight (HGW) at P < 0.05.
We also looked at trait correlations within each trial and recorded more significant trait correlations (P < 0.05) in the pot trial (21) than in the field trial (13) (Supplementary Table 1). Yield-related traits (i.e., HGW, YLD, DTF, and PH) were negatively correlated with ASNPC, NSmax, NSFC, ODS, and OPS, in both field and pot trials, although the correlation was largely non-significant (Supplementary Table 1). The highest positive significant correlations in the field trial were recorded between ASNPC and NSmax (r = 0.83; P < 0.001), ASNPC and NSFC (r = 0.77; P < 0.001), and NSmax and NSFC (r = 0.80; P < 0.001). The same traits were also highly positively correlated in the pot trial with comparable correlation values of r = 0.84 (i.e., ASNPC and NSmax) and r = 0.73 (i.e., NSFC and ASNPC), except for the correlation between NSFC and NSmax (r = 0.55).
Parents recorded high heritability values for all traits except DTF under the pot trial (Figure 3, Supplementary Table 1). Progenies displayed relatively lower heritability values for most traits in comparison with the parents, except for DTF in both environments (Figure 3).
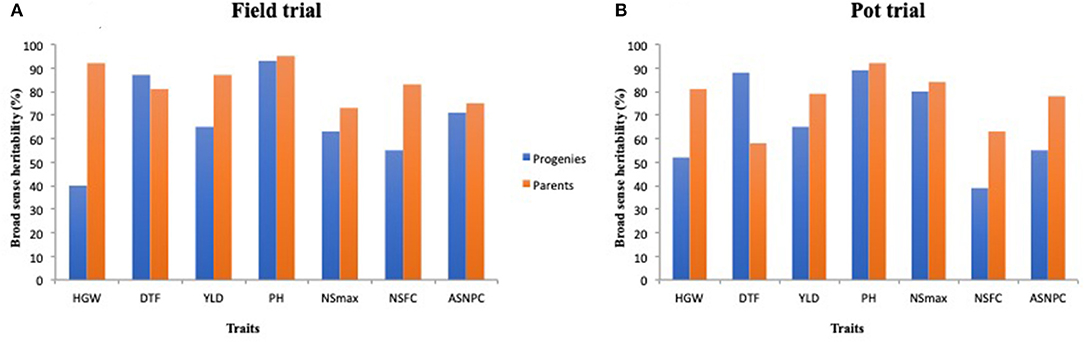
Figure 3. Broad-sense heritability estimates for select traits among parents and progenies evaluated under the field trial (A) and pot trial (B).
Agronomic Performance of the Germplasm Under Field and Pot Trials
We used HGW rather than YLD to compare the yield performance of genotypes between the two trials, given the differences in the trial conditions that would bias the total yield comparisons. Of note, 9 and 6 out of the 10 genotypes with the highest HGW in the field and pot trials, respectively, were F4 progenies (Figures 4A,B). Five (ICSVIII IN × E36-1, LANDIWHITE × B35, B35 × E36-1, F6YQ212 × B35, and ICSVIII IN × LODOKA) of the F4 progenies with the highest HGW were common in both trials (Figures 4A,B), revealing their potential stability for the trait. FRAMIDA and HAKIKA, which are both Striga-resistant and improved varieties, were the only resistant checks among the top 10 genotypes recording the highest HGW in the field and pot trials, respectively (Figures 4A,B). A landrace, AKUOR-ACHOT, and a wild accession, GBK044448, were also among the top 10 genotypes with high HGW of 4.5 and 4.7 g, respectively, in the pot trial (Figure 4B) but not in the field trial (Supplementary Table 1). Most of the genotypes with the lowest HGW were wild accessions or landraces in both trials, although some improved varieties (i.e., E36-1 in pot trial) and F4 progenies also fell into this category (Figures 4A,B).
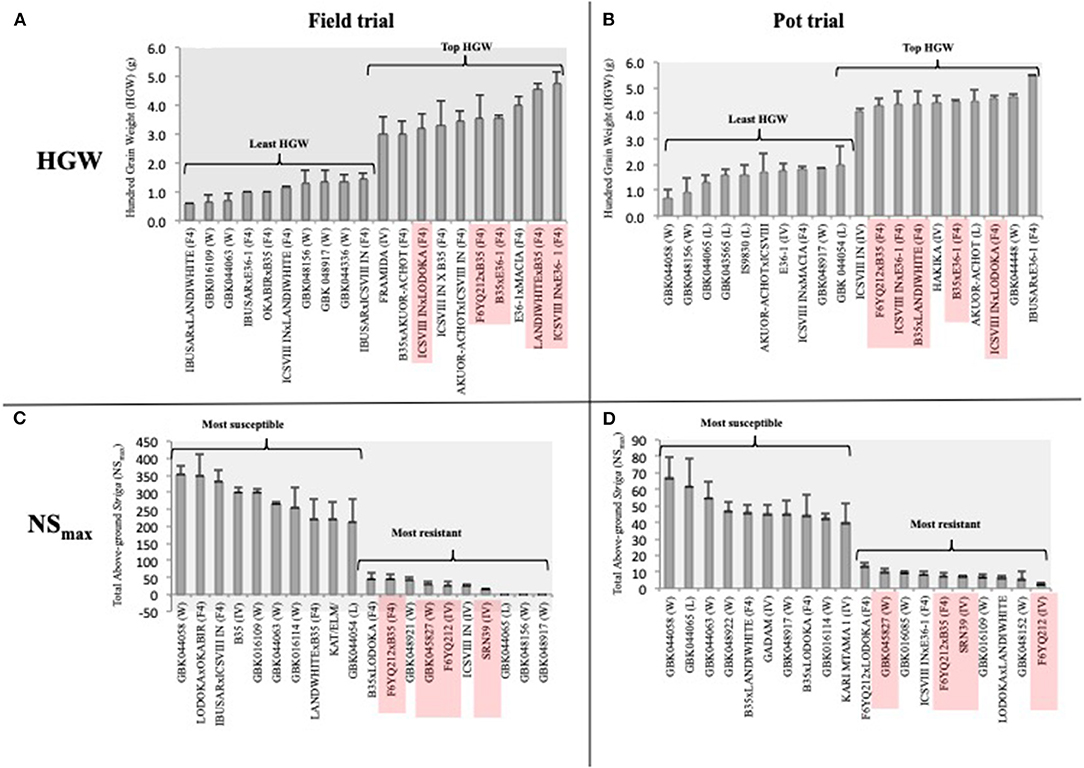
Figure 4. The top 10 best and worst performers for HGW and Striga (NSmax) in the field (A,C) and pot trial (B,D), respectively. Genotypes with consistent performance across the two environments are highlighted in red.
Response of Genotypes to Striga Under Field and Pot Trials
Despite the poor correlation of Striga-related traits between pot and field trials, we observed stability in the performance of four (F6YQ212, SRN39, F6YQ212xB35, and GBK045827) out of the 10 top Striga-resistant genotypes in both field and pot trials (Figures 4C,D) according to their NSmax ranking (Figures 4C,D). SRN39, a stable source of resistance to Striga, was the only resistant check among the topmost resistant genotypes in both trials (Figures 4C,D). Although the F4 progenies were the dominant best performers for HGW, they were the minority genotypes among the top 10 most Striga-resistant genotypes in both field and pot trials. F6YQ212, which showed resistance to Striga in both trials, was the common parent in two out of the three F4 progenies with resistance to Striga, suggesting it would be a good source of stable Striga resistance for future crosses. B35, a drought-tolerant variety, which was among the top 10 most Striga-susceptible genotypes in the field trial, was the common parent in two out of the three top Striga-resistant progenies in both trials (Figures 4C,D). The only genotype that was recorded among the top performers in both trials for HGW and Striga resistance (NSmax) was a cross between the Striga-resistant variety, F6YQ212, and the drought-tolerant variety, B35 (Figure 4).
Three genotypes that included two wild (GBK048156 and GBK048917) and a landrace (GBK044065) recorded no Striga germination in the field trial but supported the germination of significant amounts of Striga in the pot trial (Supplementary Table 1). A wild accession, GBK044058, was the most susceptible genotype to Striga in both trials. All genotypes tested for the Striga germination in the pot trial supported the germination of at least three Striga plants in at least one replicate (Supplementary Table 1). However, genotype GBK016109, which showed comparable resistance to Striga as SRN39 in the pot trial, was completely devastated in the field trial recording one of the worst performers (Figures 4C,D).
Genotype Relatedness and Confirmed Hybridity
A total of 26,291 raw SNPs were generated from DArT-seq of 64 genotypes (i.e., 17 wild, 8 landraces, 12 improved varieties, and 27 F4 progenies), of which 7,038 SNPs were retained after filtering. The NJ dendrogram resulted in three major clusters (Figure 5). The first cluster (A) (Figure 5) comprised of Striga-resistant genotypes including four resistant checks, namely, IS9830, SRN39, FRAMIDA, and HAKIKA. Other genotypes in cluster A were recorded as resistant to Striga in this study such as GBK048156 (field trial), GBK048152 (pot trial), F6YQ212 (field trial and pot trial), and GBK045827 (field trial and pot trial). The only susceptible genotype in this cluster was the improved variety KAT/ELM/2016PL1SD15. Cluster B comprised mostly of wild accessions and landraces. Two improved varieties, namely, B35 and MACIA, were also in cluster B, but different subclusters. The only Striga-resistant check in this cluster was N13, which was grouped in the same subcluster with the staygreen genotype, B35. Both B35 and N13 are known to have wild pedigrees.
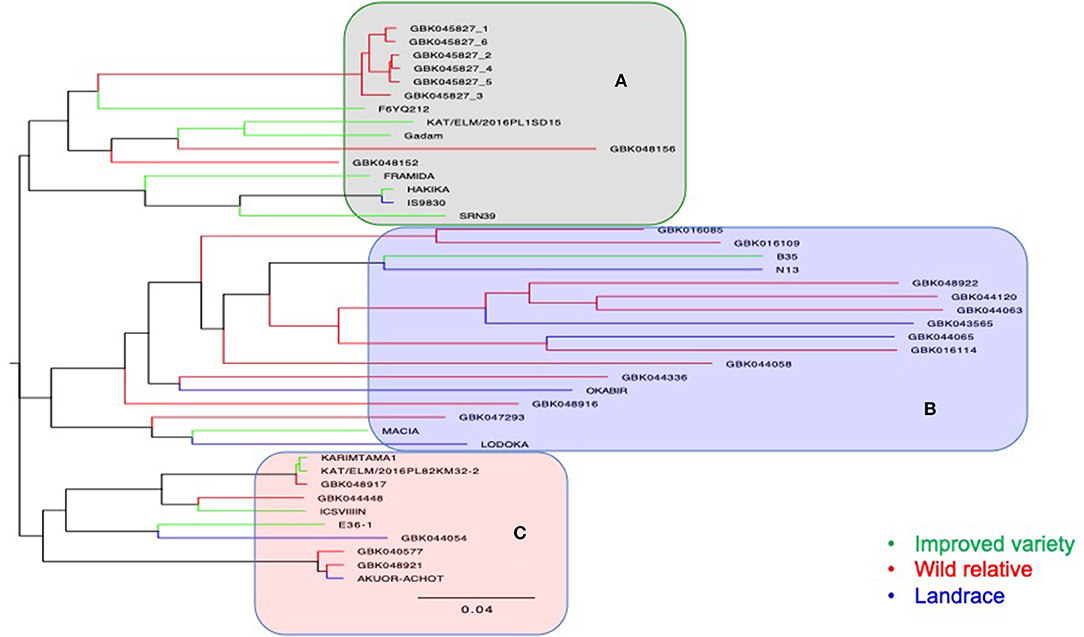
Figure 5. A neighbor-joining dendrogram showing genetic relatedness of 37 accessions that comprised of 17 wild accessions, 8 landraces, and 12 improved varieties. The main clusters generated are highlighted in gray (A), light blue (B), and pink (C).
Two wild genotypes (GBK016109 and GBK016085), which were among the top most Striga-resistant lines in the pot trial, and one landrace (GBK044065), which was among the most resistant lines in the field trial, were also grouped in cluster B (Figure 5). Cluster C was composed of four improved varieties, two landraces, and four wild accessions. Two genotypes (GBK048917 and ICSV III IN), which were also among the most resistant to Striga in the field trial, were also grouped under cluster C. Genotype E36-1, a well-known drought-tolerant (staygreen) material, was also grouped in cluster C alongside a landrace, GBK044054, which recorded one of the highest NSmax in the field trial (Figure 4C).
Crosses involving parental lines IBUSAR and LANDIWHITE were not included in the hybridity analysis as both parents failed to germinate in both trials. Hybridity of 16 F4 progenies was confirmed using biallelic SNP markers ranging from 1,204 to 2,868 that had been called from DArT-seq (Table 4). The highest proportion (22–46%) of heterozygous alleles were recorded in the progenies of the cross B35 × E36-1, while the lowest (<1%) were recorded in the crosses OKABIR × AKUOR-ACHOT and LODOKA × OKABIR. B35 and E36-1 are the improved varieties derived from wild backgrounds, while LODOKA and AKUOR-ACHOT are landraces.
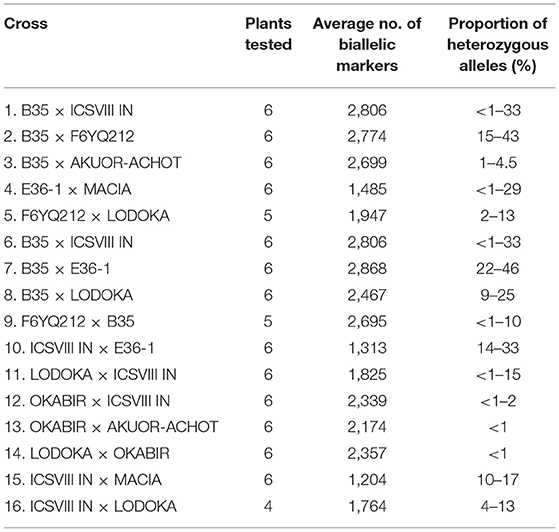
Table 4. Confirmation of hybridity among the fourth filial generation (F4) progenies using Striga number progress (SNP) markers.
Residual Heterosis for Yield and Resistance to Striga in the F4 Progenies
A complete record of the MP and residual heterosis values in the field and pot trials for HGW and NSmax is shown in Supplementary Table 2. Table 5 shows the residual heterosis values for HGW in each of the crosses in the field and pot trials ranked from the top to the lowest. Both the highest residual heterosis and the inbreeding depression for HGW were recorded in the field at 89.78% and −59.48%, respectively. LODOKA, a drought-tolerant landrace, was the common parent in the crosses with the highest residual heterosis in both trials (Table 5). Four (i.e., AKUOR-ACHOT × ICSVIII IN, B35 × AKUOR-ACHOT, ICSVIII IN × MACIA, LODOKA × ICSVIII IN) out of six crosses that recorded the inbreeding depression for HGW were consistent in both the field and pot trials (Table 5), suggesting they would be poor candidates for yield-related traits. Crosses involving ICSVIII IN, an improved variety, revealed some of the highest inbreeding depression for HGW in both field and pot trials (Table 5). In some cases, the highest residual heterosis recorded for HGW (86.19% for F6YQ212 × B35 and 52.94% for B35 × LODOKA) also corresponded to a low Striga count of −65.13 and −79.20% (Tables 5, 6), making these crosses good candidates for the future development of Striga-tolerant, high-yielding varieties.
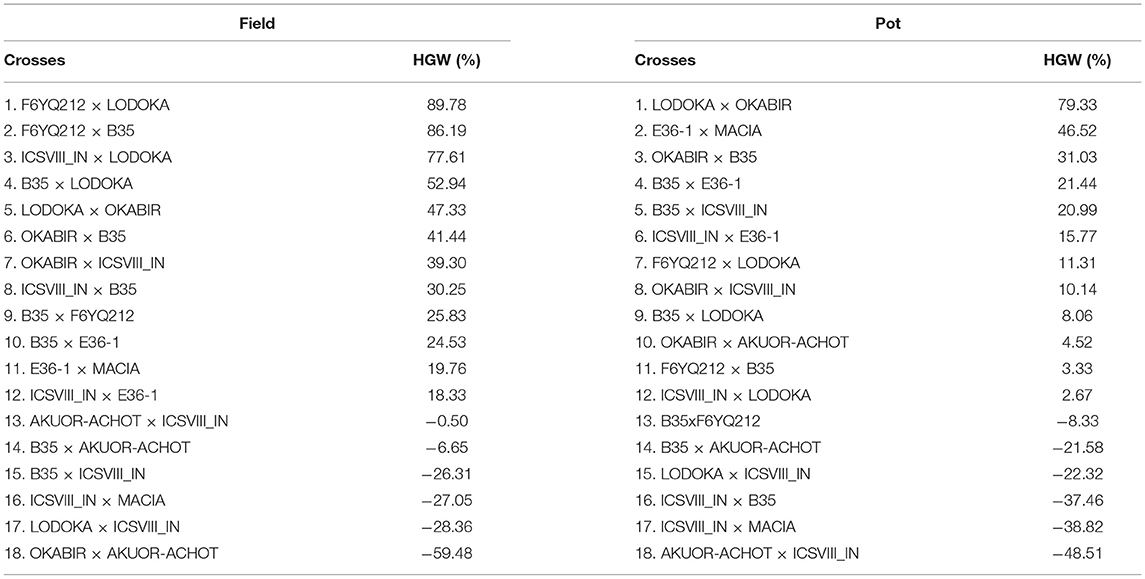
Table 5. Proportion of residual heterosis at F4 for 100-grain weight (HGW) for the pot and field trials.
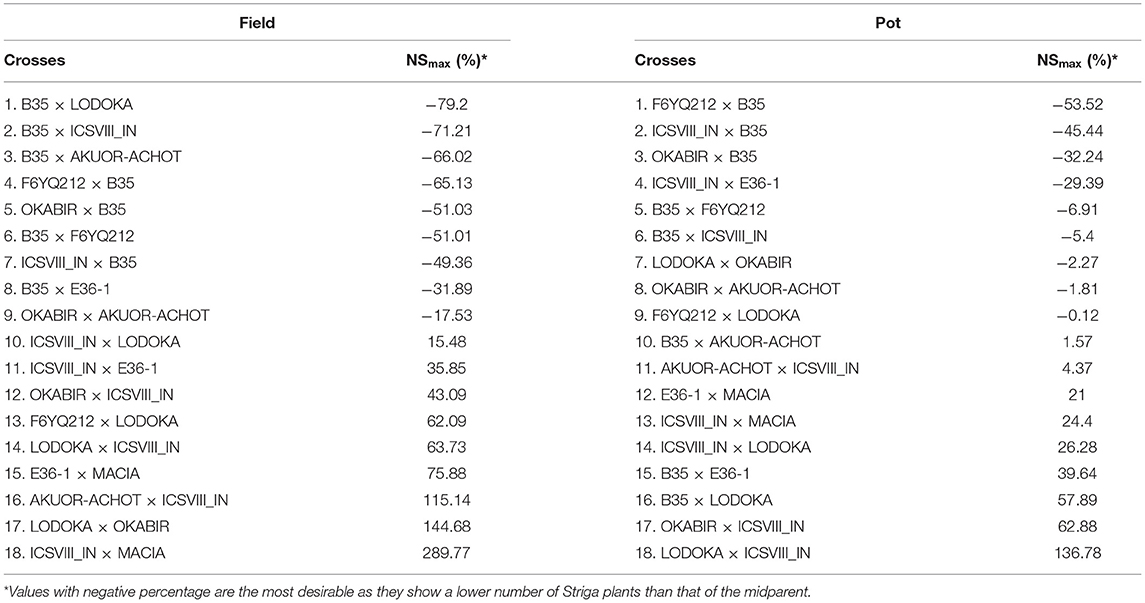
Table 6. Proportion of residual heterosis at F4 for maximum above-ground Striga (NSmax) for the pot and field trials.
Table 6 shows the residual heterosis values for NSmax in each of the crosses in the field and pot trials. Crosses involving B35, a drought-tolerant improved variety, had some of the highest inbreeding depression values for NSmax, which is desirable as it resulted in more resistance to Striga (Table 6). Like in HGW, crosses involving ICSVIII IN performed worse than the MP for their resistance to Striga, indicating that this parent may not be a good choice for improving either yield or Striga resistance (Table 6).
Discussion
Identification of Diverse High-Yielding Striga-Resistant Genotypes
The aim of this study was to identify new sources of Striga resistance in comparison with the conventional sources. The data collected found credible evidence that genotypes that were resistant to Striga had significantly lower ASNPC and NSmax-values in both trials. These parameters were considered alongside grain yield for the effective selection of superior Striga-resistant, as well as Striga-tolerant genotypes as recommended by Rodenburg et al. (2005). We identified three genotypes (F6YQ212, GBK045827, and F6YQ212 × B35) together with one check (SRN39) that were consistent in their response to Striga across both trials. SRN39 is known to harbor pre-attachment resistance that results in the production of a low germination stimulant, orobanchol (Satish et al., 2012; Mohemed et al., 2016). However, SRN39 is not high yielding and, therefore, not preferred by farmers and has been used mainly as a donor of Striga resistance to other improved varieties. Of specific interest is the cross between improved varieties, F6YQ212 × B35, which recorded consistency in resistance to Striga and was also high yielding. This particular cross is likely to perform well in the fields of farmers and would be a good genotype to advance for field trials.
The wild accession, GBK045827, which also showed consistency in Striga resistance across the two trials, did not only group together with F6YQ212 but also recorded comparable performance with F6YQ212 in both experiments. This observation strongly suggests that the resistance observed in F6YQ212 may have been introgressed from GBK045827. We know that Striga resistance is more abundant among wild relatives (Rich et al., 2004; Mbuvi et al., 2017), which tend to cross-pollinate with cultivated genotypes in open fields (Ohadi et al., 2017). Both F6YQ212 and GBK045827 clustered in group A with four resistant checks (FRAMIDA, HAKIKA, IS9830, and SRN39) but in a different subclade. This pattern of clustering suggests a narrow genetic base for Striga-resistant sources that are currently being used in breeding programs in Eastern Africa (Mohamed et al., 2010), except for N13. Genotype F6YQ212, therefore, provides a good alternative source of resistance as it was grouped in a different subclade. F6YQ212 has been previously screened for response to grain storage pests (Mwenda, 2019) but not for its resistance to Striga. Future studies will therefore need to determine the mode of resistance in F6YQ212, as well as in GBK045827. The mechanism of resistance in FRAMIDA, IS9830, and SRN39 is reported to be low germination stimulation (Haussmann et al., 2000b; Mohamed et al., 2010; Gobena et al., 2017), which is the most widely studied mechanism of resistance to Striga in sorghum. The only other resistant check that clustered differently was N13, a durra sorghum from India, which is known to stimulate Striga germination but forms a mechanical barrier to Striga penetration (Maiti et al., 1984; Mohemed et al., 2016; Mbuvi et al., 2017). Genotype N13 grouped together with B35, a drought-tolerant variety, which has its origins in Ethiopia (Ochieng et al., 2020).
Screening for Striga Resistance in Pot and Field Trial Conditions
The trials made use of an existing Striga-infested field that was supplemented with artificial Striga inoculation, as well as a pot trial with artificial Striga inoculation to represent a second environment. One of the major challenges of undertaking reliable field trials using Striga-infested fields is the lack of homogeneity of Striga infections across different points of the field. This is especially due to the outcrossing nature of Striga and the tendency of its seeds to remain dormant in the soil for up to 20 years (Teka, 2014). In our investigation, both field trials and inoculated pots were used to take care of natural infection conditions, as well as enhance uniformity of infection. There are a number of successful Striga studies that used both field and pot experiments together with supplemented artificial inoculation in the same way (Kountche et al., 2013; Rodenburg et al., 2015; Midega et al., 2016; Abate et al., 2017). Other studies also made use of pot experiments as Striga-free control (Samejima et al., 2016), or to enable the isolation of root exudates (Jamil et al., 2011; Hooper et al., 2015). The low correlation reported here between field and pot experiments for Striga-related traits has also been observed in other experiments (Haussmann et al., 2000a), suggesting that the use of pots for Striga screening should be completely discouraged.
Several factors could explain the observed differences in the pot and field experiments. First, the Striga infestation levels were significantly higher in the field trial than in the pot experiments, in which Striga-free soil was used before the addition of a standard amount of Striga in each pot. Second, it is likely that the field trial had a mixture of biotypes that had been accumulated over the years before the supplemented inoculation. Third, the Striga in the field trial was established exclusively under rainfed conditions, whereas the pot experiment was regularly watered to ensure sufficient moisture was maintained throughout the experiment. Furthermore, there could be rhizosphere differences that would affect the stimulation of Striga germination (Miché et al., 2000). Variable response to Striga under different test conditions has been observed in previous studies (Rao, 1984; Haussmann et al., 2000a).
Haussmann et al. (2004) hypothesized that abiotic stress, followed by ethylene production by microorganisms in the soil, could be responsible for the observed differences. In this study, drought stress could have been a factor in the field trial experiment, which was purely dependent on rain. Drought has been shown to induce strigolactone production in the roots (Haider et al., 2018), which in turn induces the germination of Striga (Cardoso et al., 1994). A significant decrease in rainfall in Kenya has been reported over the last four decades (Ayugi et al., 2016), suggesting that rainfed crops are highly likely to be exposed to drought during their growth periods. Rainfall data (not shown) from Alupe station during the growing season further confirmed the variability of rainfall that could have led to the exposure of the crops to drought. Nonetheless, we observed a number of genotypes showing consistency in their performance between the two trials for Striga-related traits.
Mechanism of Resistance to Striga in N13
Host plant resistance to Striga has been defined as the ability of the plant to reduce or prevent infection (Shew and Shew, 1994) through pre- and post-attachment mechanisms (Yoder and Scholes, 2010; Rodenburg et al., 2016). Tolerance refers to the extent to which effects of infection on the host plant are mitigated (Caldwell et al., 1958; Rodenburg et al., 2016). In this evaluation, N13 was not among the most resistant genotypes in both experiments. A study by Rodenburg et al. (2005) that incorporated N13, SRN39, IS9830, and FRAMIDA among other genotypes reported N13 as the most resistant genotype in the trial with ASNPC and NSmax-values significantly lower than all the other genotypes in the trial. Our results may suggest a possible breakdown of mechanical resistance that was initially recorded in N13, or potential contamination of seed source that may have led to the reduction in the resistance originally observed. However, these theories will need to be further investigated.
Although the genetic architecture of mechanical resistance to Striga in sorghum is yet to be understood, previous studies suggest that it is complex. QTL mapping studies in recombinant inbred populations using N13 as the resistant parent reported 11 and 9 QTLs in two different environments (Haussmann et al., 2004). The durability of mechanical resistance will therefore depend on the factors at play, which may range from cell wall thickening, lignification, and silica deposition (Maiti et al., 1984). A recent study of Striga resistance in rice reported that enhanced lignin deposition and maintenance of the structural integrity of lignin polymers deposited at the infection site are crucial for post-attachment resistance against S. hermonthica (Mutuku et al., 2019). Similar studies will be necessary in sorghum to enhance our understanding of both pre- and post-attachment resistance to Striga. Such an outcome will enable the pyramiding of genes responsible for both pre- and post-attachment resistance in order to enhance the durability of resistance in the fields of farmers.
Drought Tolerance and Yield Stability Under Striga Infestation
A majority of the best yielding (most tolerant) genotypes were derived from crosses. However, the most consistent performance among these top-performing crosses was observed when crosses were made with any of the drought-tolerant genotypes, such as LODOKA, B35, and E36-1. B35 and E36-1 are drought-tolerant improved varieties that have been used for decades in the region and globally, while LODOKA is a drought-tolerant landrace. The superior and consistent agronomic performance of crosses involving drought-tolerant genotypes under Striga conditions was not surprising. Both drought stress and S. hermonthica infestation result in the production of abscisic acid (ABA) (Frost et al., 1997; Sah et al., 2016), which triggers stomatal closure (Kim et al., 2010). However, drought-tolerant sorghum genotypes have been shown to adapt to drought stress by preventing excessive ABA responses (Varoquaux et al., 2019). Such an adaptation response of drought-tolerant sorghum would also benefit their response to S. hermonthica and enhance the production of photosynthates to sustain plant growth and development in the presence of both stresses. A recent study in maize reported up to 19% increase in yield under stress in hybrids simultaneously expressing drought tolerance and S. hermonthica resistance as compared with those expressing only one of the traits (Menkir et al., 2020). Similar studies will need to be undertaken in sorghum to fully establish the interaction of drought and S. hermonthica stresses.
The older sources of drought tolerance will, however, need to be replaced with new superior performing genotypes such as F6YQ212 × B35 and ICSVIII IN × LODOKA identified in the current study. OKABIR, AKUOR-ACHOT, and LODOKA are landraces, which have also been recently reported to perform better than B35 and E36-1 under drought conditions (Ochieng et al., 2020). While LODOKA clustered with MACIA, both OKABIR and AKUOR-ACHOT appeared to be distantly related to MACIA, B35, and E36-1 and will therefore be good alternative sources of both drought and Striga tolerance. Better still, more focused introgression of Striga resistance and drought tolerance into farmer-preferred varieties should be planned to ensure better replacement of some of these old varieties.
Molecular Breeding for Striga Resistance and Tolerance
The demonstrated residual heterosis in HGW and NSmax at F4 is great news for breeding programs as it shows the huge potential of enhancing the performance of varieties in response to Striga through improved genetics. Given the high variation in the ecotypes of Striga across different environments, the best breeding strategy would be genomic selection (GS) (Goddard, 2009). Our results provide a good basis for designing a GS strategy for developing Striga- and drought-tolerant sorghum varieties that will be suitable for the harsh environments typical of Striga-endemic ecologies. The available genomic resources in sorghum public databases will only enhance the ease with which GS is implemented in sorghum.
The DArT-seq technology (Sansaloni et al., 2011) that was used to characterize the germplasm proved to be a reliable and cost-effective technology for diversity analysis and confirming hybridity. While there are several studies reporting the use of DArT-seq for diversity analysis in sorghum (Kotla et al., 2019; Allan et al., 2020; Mengistu et al., 2020), this study is the first to use DArT-seq for hybridity testing. The unique SNP markers from this study will be useful for GS and for incorporation into marker panels that aim at the identification of successful hybrids from new crosses involving any of the parents. Future studies will also need to establish the specific molecular markers associated with new sources of resistance to Striga through genome-wide association studies (GWAS) or the characterization of biparental populations.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
KN and DO conceived the project, designed and supervised the experiment, and drafted the manuscript. NM did both the field and laboratory experiments and drafted the manuscript. EM provided some of the germplasm and supervised the fieldwork and data collection. WC supervised the fieldwork and data collection. MA supported the laboratory work and did part of the data analysis. DN and LW provided the genebank material and supervised the work, respectively. All authors approved the final version of the manuscript.
Funding
A postgraduate fellowship was provided to NM by the National Research Fund of Kenya (NRF) (https://researchfund.go.ke/). This study was partially funded by the CGIAR Research Program (CRP) on Grain Legumes and Dryland Cereals. The wild accessions were collected as part of the initiative Adapting Agriculture to Climate Change: Collecting, Protecting and Preparing Crop Wild Relatives, which is supported by the Government of Norway. The project is managed by the Crop Trust with the Millennium Seed Bank of the Royal Botanic Gardens, Kew and implemented in partnership with national and international gene banks and plant breeding institutes around the world. For further information, see the project website: http://www.cwrdiversity.org/.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the University of Nairobi for facilitation, technical support, and knowledge sharing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.671984/full#supplementary-material
References
Aasha, M. B., Amani, E. H., Ras, H., Abusin, M. A., and Khalil, N. A. (2017). Effects of intercropping pearl millet with some legumes on Striga hermonthica emergence. SSRG Int. J. Agric. Environ. Sci. 4, 64–72. doi: 10.14445/23942568/IJAES-V4I6P113
Abate, M., Hussien, T., Bayu, W., and Reda, F. (2017). Diversity in root traits of sorghum genotypes in response to Striga hermonthica infestation. Weed Res. 57, 303–313. doi: 10.1111/wre.12262
Ali, R., Hash, C. T., Damaris, O., Elhussein, A., and Mohamed, A. H. (2016). Introgression of striga resistance into popular Sudanese sorghum varieties using marker assisted selection. World J. Biotechnol. 1, 48–55. doi: 10.1234/wjb.v1i1.34
Aliche, E. B., Screpanti, C., Mesmaeker, A., Munnik, T., and Bouwmeester, H. J. (2020). Science and application of strigolactones. New Phytol. 227, 1001–1011. doi: 10.1111/nph.16489
Allan, V., Vetriventhan, M., Senthil, R., Geetha, S., Deshpande, S., Rathore, A., et al. (2020). Genome-wide DArTSeq genotyping and phenotypic based assessment of within and among accessions diversity and effective sample size in the diverse sorghum, pearl millet, and pigeonpea landraces. Front. Plant Sci. 11:587426. doi: 10.3389/fpls.2020.587426
Ayugi, B., Wen, W., and Chepkemoi, D. (2016). Analysis of spatial and temporal patterns of rainfall variations over Kenya. Environ. Earth Sci. 6, 69–83.
Beshir, M. M., Ahmed, N. E., Ali, A. M., Babiker, I. H., Rubaihayo, P., and Okori, P. (2015). Prevalence and severity of sorghum leaf blight in the sorghum growing areas of Central Sudan. Wudpecker J. Agric. Res. 4, 54–60. doi: 10.13140/RG.2.1.1785.1285
Bouréma, D., Daouda, D., and James, H. W. (2005). Herbicide seed treatments for control of purple witchweed (Striga hermonthica) in sorghum and millet. Weed Technol. 19, 629–635.
Bouwmeester, H., Schuurink, R. C., Bleeker, P. M., and Schiestl, F. (2019). The role of volatiles in plant communication. Plant J. 100, 892–907. doi: 10.1111/tpj.14496
Bradbury, P., Zhang, Z., Kroon, D., Casstevens, T., Ramdoss, Y., and Buckler, E. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Caldwell, R. M., Schafer, J. F., Compton, L. E., and Patterson, F. L. (1958). Tolerance to cereal leaf rust. Science 128, 714–715
Cardoso, C., Ruyter-Spira, C., Bouwmeester, H. J., Dawoud, D., and Sauerborn, J. (1994). Impact of drought stress and temperature on the parasitic weeds Striga hermonthica and Alectra vogelii in their early growth stages. Exp Agric. 30, 249–257. doi: 10.1017/S0014479700024182
Dembélé, B., Dembélé, D., and Westwood, J. (2005). Herbicide seed treatments for control of purple witchweed (Striga hermonthica) in sorghum and millet. Weed Technol. 19, 629–635. doi: 10.1614/WT-04-183R2.1
Ejeta, G. (2007). Breeding for Striga resistance in sorghum: exploitation of intricate host parasite biology. Crop Sci. 47, S-216–S-227. doi: 10.2135/cropsci2007.04.0011IPBS
Ejeta, G., and Knoll, J. E. (2007). “Marker-assisted selection in Sorghum,” in Genomics-Assisted Crop Improvement: Vol 2: Genomics Applications in Crops, eds R. K. Varshney and R. Tuberosa (Dordrecht: Springer), 187–205.
Ezeaku, I. (2005). Development of sorghum populations for resistance to Striga hermonthica in the Nigerian Sudan Savanna. African J. Biotechnol. 3, 324–329. doi: 10.5897/AJB2004.000-2059
FAOSTAT (2016). Database of Agricultural Production FAO Statistical Databases (FAOSTAT). Available online at: http://faostat.fao.org/default.aspx (accessed Aprile 26, 2021).
Frost, D. L., Gurney, A. L., Press, M. C., and Scholes, J. D. (1997). Striga hermonthica reduces photosynthesis in sorghum: the importance of stomatal limitations and a potential role for ABA? Plant Cell Environ. 20, 483–492. doi: 10.1046/j.1365-3040.1997.d01-87.x
Frost, H. M. (1994). Striga Research and Survey in Kenya. National Agricultural Research Project, KARI/ODA Crop Protection Project. Final Report. p. 61.
Gobena, D., Shimelis, M., Rich, P. J., Ruyter-spira, C., Bouwmeester, H., and Kanuganti, S. (2017). Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc. Natl. Acad. Sci. U.S.A. 114, 4471–4476. doi: 10.1073/pnas.1618965114
Goddard, M. (2009). Genomic selection: prediction of accuracy and maximisation of long-term response. Genetica 136, 245–257. doi: 10.1007/s10709-008-9308-0
Gurney, A. L., Slate, J., Press, M. C., and Scholes, J. D. (2005). A novel form of resistance in rice to the angiosperm parasite Striga hermonthica. New Phytol. 169, 199–208. doi: 10.1111/j.1469-8137.2005.01560.x
Haider, I., Andreo-Jimenez, B., Bruno, M., Bimbo, A., Floková, K., and Abuauf, H. (2018). The interaction of strigolactones with abscisic acid during the drought response in rice. J. Exp. Bot. 69, 2403–2414. doi: 10.1093/jxb/ery089
Hallauer, A. R., Carena, M. J., and Miranda Filho, J. B. (2010). Quantitative Genetics in Maize Breeding. New York, NY: Springer.
Haussmann, B. I., Hess, D. E., Omanya, G. O., Folkertsma, R. T., Reddy, B. V., Kayentao, M., et al. (2004). Genomic regions influencing resistance to the parasitic weed Striga hermonthica in two recombinant inbred populations of sorghum. Theor. Appl. Genet. 109, 1005–1016. doi: 10.1007/s00122-004-1706-9
Haussmann, B. I. G., Hess, D. E., Reddy, B. V. S., Mukuru, S. Z., Kayentao, M., Welz, H. G., et al. (2000a). Quantitative-genetic parameters of sorghum growth under Striga infestation in Mali and Kenya. Plant Breed. 120, 49–56. doi: 10.1046/j.1439-0523.2001.00546.x
Haussmann, B. I. G., Hess, D. E., Reddy, B. V. S., Welz, G., and Geiger, H. H. (2012). Analysis of resistance to Striga hermonthica in diallel crosses of sorghum. Euphytica 116, 33–40. doi: 10.1023/A:1004046001009
Haussmann, B. I. G., Hess, D. E., Welz, H. G., and Geiger, H. H. (2000b). Improved methodologies for breeding Striga-resistant sorghums. Field Crops Res. 66, 195–211. doi: 10.1016/S0378-4290(00)00076-9
Hess, D. E., and Ejeta, G. (1992). Inheritance of Resistance to Striga in Sorghum Genotype SRN39. Plant Breed. 109, 233–241. doi: 10.1111/j.1439-0523.1992.tb00178.x
Hooper, A. M., Caulfield, J. C., Hao, B., Pickett, J. A., Midega, C. A. O., and Khan, Z. R. (2015). Isolation and identification of Desmodium root exudates from drought tolerant species used as intercrops against Striga hermonthica. Phytochemistry 117, 380–387. doi: 10.1016/j.phytochem.2015.06.026
Jamil, M., Charnikhova, T., Cardoso, C., Jamil, T., Ueno, K., Verstappen, F., et al. (2011). Quantification of the relationship between strigolactones and Striga hermonthica infection in rice under varying levels of nitrogen and phosphorus. Weed Res. 51, 373–385. doi: 10.1111/j.1365-3180.2011.00847.x
Khan, Z., Midega, C., Pittchar, J., Pickett, J., and Bruce, T. (2011). Push—pull technology: a conservation agriculture approach for integrated management of insect pests, weeds, and soil health in Africa. Int. J. Agric Sustain. 9, 162–170. doi: 10.3763/ijas.2010.0558
Khan, Z. R., Pickett, J. A., Wadhams, L. J., Hassanali, A., and Midega, C. A. O. (2006). Combined control of Striga hermonthica and stemborers by maize–Desmodium spp. intercrops. Crop Prot. 25, 989–995. doi: 10.1016/j.cropro.2006.01.008
Kidanemaryam, W., Kassahun, B., and Taye, T. (2018). Assessment of Heterotic Performance and Combining Ability of Ethiopian Elite Sorghum (Sorghum bicolor (L.) Moench) Lines. Masters Thesis, Jimma University, Ethiopia.
Kim, S. K., Adetimirin, V. O., Thé, C., and Dossou, R. (2002). Yield losses in maize due to Striga hermonthica in West and Central Africa. Int. J. Pest Manag. 48, 211–217. doi: 10.1080/09670870110117408
Kim, T. H., Bohmer, M., Hu, H., Nishimura, N., and Schroeder, J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annual Rev. Plant Biol. 61, 561–591. doi: 10.1146/annurev-arplant-042809-112226
Kiprotich, F., Mwendia, M. C., Cheruiyot, K. E., and Wachira, N. F. (2015). Nutritional suitability of bred sorghum (Sorghum bicolor) accessions from East Africa. African J. Food Sci. 9, 326–334. doi: 10.5897/AJFS2015.1288
Kotla, K., Phuke, R., Hariprasanna, K., Mehtre, S. P., Rathore, A., Gorthy, S., et al. (2019). Identification of QTLs and candidate genes for high grain Fe and Zn concentration in sorghum [Sorghum bicolor (L.) Moench]. J. Cereal Sci. 90:102850. doi: 10.1016/j.jcs.2019.102850
Kountche, B. A., Hash, T. C., Dodo, H., Laoualy, O., Sanogo, M. D., Timbeli, A., et al. (2013). Development of a pearl millet Striga-resistant genepool: Response to five cycles of recurrent selection under Striga-infested field conditions in West Africa. Field Crops Res. 154, 82–90. doi: 10.1016/j.fcr.2013.07.008
MacOpiyo, L., Vitale, J., and Sanders, J. (2010). An Ex-Ante Assessment of a Striga Control Programme in East Africa. Kilimo Trust, p. 6–25. Available online at: http://www.parasite-project.org/wp-content/uploads/2014/02/Kilimo-trust-2009-Ex (accessed January 26, 2021).
Maiti, R. K., Ramaiah, K. V., Bisen, S. S., and Chidley, V. L. (1984). A comparative study of the haustorial development of Striga asiatica (L.) kuntze on Sorghum cultivars. Ann. Bot. 54, 447–457. doi: 10.1093/oxfordjournals.aob.a086816
Mallu, T. S., Mutinda, S., Githiri, S. M., Odeny, D., and Runo, S. (2021). New pre-attachment Striga resistant sorghum adapted to African agro-ecologies. Pest Manag. Sci. 77, 2894–2902. doi: 10.1002/ps.6325
Marley, P. S., Diourte, M., Neya, A., and Rattunde, F. W. (2005). Sorghum anthracnose and sustainable management strategies in West and Central Africa. J. Sustain. Agric. 25, 43–56. doi: 10.1300/J064v25n01_05
Mbuvi, D. A., Masiga, C. W., Kuria, E., Masanga, J., Wamalwa, M., Mohamed, A., et al. (2017). Novel sources of witchweed (Striga) resistance from wild sorghum accessions. Front. Plant Sci. 8:116. doi: 10.3389/fpls.2017.00116
Mengistu, D., Shimelis, H., Laing, M., Lule, D., Assefa, E., and Mathew, I. (2020). Genetic diversity assessment of sorghum (Sorghum bicolor (L.) Moench) landraces using SNP markers. South African J. Plant Soil 37, 220–226. doi: 10.1080/02571862.2020.1736346
Menkir, A., Adetimirin, V. O., Yallou, C. G., and Gedil, M. (2010). Relationship of genetic diversity of inbred lines with different reactions to Striga hermonthica (Del.) benth and the performance of their crosses. Crop Sci. 50, 602–611. doi: 10.2135/cropsci2009.05.0247
Menkir, A., Crossa, J., Meseka, S., Bossey, B., Muhyideen, O., Riberio, P. F., et al. (2020). Staking tolerance to drought and resistance to a parasitic weed in tropical hybrid maize for enhancing resilience to stress combinations. Front. Plant Sci. 11:166. doi: 10.3389/fpls.2020.00166
Miché, L., Bouillant, M. L., Rohr, R., Sallé, G., and Bally, B. (2000). Physiological and cytological studies on the inhibition of striga seed germination by the plant growth-promoting bacterium Azospirillum brasilense. Eur. J. Plant Pathol. 106, 347–351. doi: 10.1023/A:1008734609069
Midega, C. A., Pickett, J., Hooper, A., Pittchar, J., and Khan, Z. R. (2016). Maize landraces are less affected by Striga hermonthica relative to hybrids in western Kenya. Weed Technol. 30, 21–28. doi: 10.1614/WT-D-15-00055.1
Mohamed, A. H., Housley, T. L., and Ejeta, G. (2010). An in vitro technique for studying specific Striga resistance mechanisms in sorghum. Afr. J. Agric. Res. 5, 1868–1875. doi: 10.5897/AJAR.9000044
Mohemed, N., Charnikhova, T., Bakker, E. J., van Ast, A., Babiker, A. G., and Bouwmeester, H. J. (2016). Evaluation of field resistance to Striga hermonthica (Del.) Benth. in Sorghum bicolor (L.) Moench. The relationship with strigolactones. Pest Manag. Sci. 72, 2082–2090. doi: 10.1002/ps.4426
Mutuku, J. M., Cui, S., Hori, C., Takeda, Y., Tobimatsu, Y., Nakabayashi, R., et al. (2019). The structural integrity of lignin is crucial for resistance against Striga hermonthica parasitism in rice. Plant Physiol. 179, 1796–1809. doi: 10.1104/pp.18.01133
Mwenda, E. T. (2019). Phenotypic and Biochemical Screening of Sorghum Genotypes for Growth and Rice Weevil Resistance in Tanzania, Doctoral dissertation, Nelson Mandela African Institution of Science and Technology (NM-AIST), Tanzania.
Ngugi, K., Ngugi, A. J., Osama, S., and Mugoya, C. (2015). Combating Striga weed in sorghum by transferring resistance quantitative trait loci through molecular marker assisted introgression. J. Plant Breed. Genet. 3, 67–76.
Ochieng, G., Ngugi, K., Wamalwa, L. N., Manyasa, E., Muchira, N., Nyamongo, D. A., et al. (2020). Novel sources of drought tolerance from landraces and wild sorghum relatives. Crop Sci. 61, 104–118. doi: 10.1002/csc2.20300
Ohadi, S., Hodnett, G., Rooney, W., and Bagavathiannan, M. (2017). Gene flow and its consequences in Sorghum spp. Crit. Rev. Plant Sci. 36, 5–6. doi: 10.1080/07352689.2018.1446813
Oswald, A., and Ransom, J. (2001). Striga control and improved farm productivity using crop rotation. Crop Prot. 20, 113–120. doi: 10.1016/S0261-2194(00)00063-6
Pamer, E. G. (2016). Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352, 535–538. doi: 10.1126/science.aad9382
Pieterse, A. H., and Pesch, C. J. (1983). The witch weeds (Striga spp.) — a review. Abstr. Trop. Agric. 9:8.
Rao, M. J. V. (1984). “Patterns of resistance to Striga asiatica in sorghum and millets, with special reference to Asia,” in Workshop on the Biology and Control of Striga, 14-17 November 1983, (Dakar).
Rebeka, G., Hussein, S., Mark, L., Pangirayi, T., and Mandefro, N. (2013). Evaluation of sorghum genotypes compatibility with Fusarium oxysporum under Striga infestation. Crop Sci. 53, 385–393. doi: 10.2135/cropsci2012.02.0101
Rich, P. J., Grenier, C., and Ejeta, G. (2004). Striga resistance in the wild relatives of sorghum. Crop Sci. 44, 2221–2229. doi: 10.2135/cropsci2004.2221
Robinson, H. F., Comstock, R. E., and Harvey, P. H. (1949). Estimates of heritability and degree of dominance in corn. Agron. J. 41, 353–359. doi: 10.2134/agronj1949.00021962004100080005x
Rodenburg, J., Bastiaans, L., and Kropff, M. J. (2006). Characterization of host tolerance to Striga hermonthica. Euphytica 147, 353–365. doi: 10.1007/s10681-005-9030-2
Rodenburg, J., Bastiaans, L., Weltzien, E., and Hess, D. E. (2005). How can field selection for Striga resistance and tolerance in sorghum be improved? Field Crops Res. 93, 34–50. doi: 10.1016/j.fcr.2004.09.004
Rodenburg, J., Cissoko, M., Kayeke, J., Dieng, I., Khan, Z. R., Midega, C. A. O., et al. (2015). Do NERICA rice cultivars express resistance to Striga hermonthica (Del.) Benth. and Striga asiatica (L.) Kuntze under field conditions? Field Crops Res. 170, 83–94. doi: 10.1016/j.fcr.2014.10.010
Rodenburg, J., Demont, M., Zwart, S. J., and Bastiaans, L. (2016). Parasitic weed incidence and related economic losses in rice in Africa. Agric. Ecosyst. Environ. 235, 306–317. doi: 10.1016/j.agee.2016.10.020
Rubiales, D., and Fernández-Aparicio, M. (2012). Innovations in parasitic weeds management in legume crops. A review. Agron. Sustain. Dev. 32, 433–449. doi: 10.1007/s13593-011-0045-x
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. doi: 10.3389/fpls.2016.00571
Samejima, H., Babiker, A. G., Takikawa, H., Sasaki, M., and Sugimoto, Y. (2016). Practicality of the suicidal germination approach for controlling Striga hermonthica. Pest Manag. Sci. 72, 2035–2042. doi: 10.1002/ps.4215
Sansaloni, C., Petroli, C., Jaccoud, D., Carling, J., Detering, F., Grattapaglia, D., et al. (2011). Diversity Arrays Technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 5:P54. doi: 10.1186/1753-6561-5-S7-P54
Satish, K., Gutema, Z., Grenier, C., Rich, P. J., and Ejeta, G. (2012). Molecular tagging and validation of microsatellite markers linked to the low germination stimulant gene (lgs) for Striga resistance in sorghum [Sorghum bicolor (L.) Moench]. Theor. Appl. Genet. 124, 989–1003. doi: 10.1007/s00122-011-1763-9
Shew, H. D., and Shew, B. B. (1994). “Host resistance,” in Epidemiology and Management of Root Diseases, eds C. L. Campbell and D. M. Benson (Berlin: Springer Verlag), 244–275.
Syukur, M., Sujiprihati, S., and Yunianti, R. (2012). Teknik and Pemuliaan Tanaman. Jakarta: Penebar Swadaya.
Teka, H. B. (2014). Advance research on Striga control: a review. African J. Plant Sci. 8, 492–506. doi: 10.5897/AJPS2014.1186
Tuinstra, M., Soumana, S., Al-Khatib, K., Kapran, I., Toure, A., Ast, A., et al. (2009). Efficacy of Herbicide Seed Treatments for Controlling Striga Infestation of Sorghum. Crop Sci. 49, 923–921. doi: 10.2135/cropsci2008.06.0357
Turner, J. H. (1953). A study of heterosis in upland cotton II. Combining ability and inbreeding effects 1. Agron J. 45, 487–490. doi: 10.2134/agronj1953.00021962004500100008x
Van Delft, G. J., Graves, J. D., Fitter, A. H., and Pruiksma, M. (1997). Spatial distribution and population dynamics of Striga hermonthica seeds in naturally infested farm soils. Plant Soil 195, 1–15. doi: 10.1023/A:1004214015281
Van Hast, A., Bastiaans, L., and Kropff, M. J. (2000). A comparative study on Striga hermonthica interaction with a sensitive and a tolerant sorghum cultivar. Weed Res. 40, 479–493. doi: 10.1046/j.1365-3180.2000.00204.x
Varoquaux, N., Cole, B., Gao, C., Pierroz, G., Baker, C. R., Patel, D., et al. (2019). Transcriptomic analysis of field-droughted sorghum from seedling to maturity reveals biotic and metabolic responses. Proc. Natl. Acad. Sci. U.S.A. 116, 27124–27132. doi: 10.1073/pnas.1907500116
Yoder, J. I., and Scholes, J. D. (2010). Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr. Opin. Plant Biol. 13, 478–484. doi: 10.1016/j.pbi.2010.04.011
Yohannes, T., Abraha, T., Kiambi, D., Folkertsma, R., Tom Hash, C., Ngugi, K., et al. (2015). Marker-assisted introgression improves Striga resistance in an Eritrean farmer-preferred sorghum variety. Field Crops Res. 173, 22–29. doi: 10.1016/j.fcr.2014.12.008
Keywords: witchweed, residual heterosis, DArT-seq, CWR, pre-breeding, Striga hermonthica
Citation: Muchira N, Ngugi K, Wamalwa LN, Avosa M, Chepkorir W, Manyasa E, Nyamongo D and Odeny DA (2021) Genotypic Variation in Cultivated and Wild Sorghum Genotypes in Response to Striga hermonthica Infestation. Front. Plant Sci. 12:671984. doi: 10.3389/fpls.2021.671984
Received: 25 February 2021; Accepted: 03 June 2021;
Published: 08 July 2021.
Edited by:
Palak Chaturvedi, University of Vienna, AustriaReviewed by:
Therese Bengtsson, Swedish University of Agricultural Sciences, SwedenMuhammad Jamil, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2021 Muchira, Ngugi, Wamalwa, Avosa, Chepkorir, Manyasa, Nyamongo and Odeny. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kahiu Ngugi, kahiu@uonbi.ac.ke; Damaris A. Odeny, d.odeny@cgiar.org
 Nicoleta Muchira1,2
Nicoleta Muchira1,2 Kahiu Ngugi
Kahiu Ngugi Lydia N. Wamalwa
Lydia N. Wamalwa Eric Manyasa
Eric Manyasa Damaris A. Odeny
Damaris A. Odeny