- 1State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Center for Crop Biotechnology, College of Agriculture, Anhui Science and Technology University, Fengyang, China
- 3College of Agriculture, Fujian Agriculture and Forestry University, Fuzhou, China
- 4State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangxi Key Lab of Sugarcane Biology, College of Agriculture, Guangxi University, Nanning, China
- 5College of Horticulture, Fujian Agriculture and Forestry University, Fuzhou, China
- 6Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 7Fujian Provincial Key Laboratory of Genetic Engineering for Agriculture, Biotechnology Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 8Pingtan Science and Technology Research Institute, Fujian Agriculture and Forestry University, Fuzhou, China
Sucrose non-fermenting 2 (Snf2) protein family, as chromatin remodeling factors, is an enormous and the most diverse protein family, which contributes to biological processes of replication, transcription, and DNA repair using the energy of adenosine triphosphate (ATP) hydrolysis. The members of Snf2 family proteins have been well characterized in Arabidopsis, rice, and tomato. Although this family received significant attention, few genes were identified uniquely for their roles in mediating reproductive development and stress tolerance in rice. In the present study, we comprehensively analyzed the expression profiling of Snf2 genes during reproductive development and biotic/abiotic stresses. Our results showed that five proteins (OsCHR712/715/720/726/739) were mainly localized in the nucleus, while OsCHR715/739 were also slightly expressed in the cell membrane. There were abundant cis-acting elements in the putative promoter of Snf2 genes, including dehydration, MeJA, MYB binding site for drought, ABA-responsive, and stress-responsive element. Most of the genes were induced immediately after Magnaporthe oryzae infection at 12 h post-infection (hpi). About 55% of the total genes were upregulated under salt and drought stresses during the entire time, and 22–35% of the total genes were upregulated at 3 h. It was noteworthy that the seven genes (OsCHR705, OsCHR706, OsCHR710, OsCHR714, OsCHR721, OsCHR726, and OsCHR737) were upregulated, and one gene (OsCHR712) was downregulated under salt and drought stresses, respectively. The deficiency of OsCHR726 mutations displayed a hypersensitive phenotype under salt stress. These results will be significantly useful features for the validation of the rice Snf2 genes and facilitate understanding of the genetic engineering of crops with improved biotic and abiotic stresses.
Introduction
Sucrose non-fermenting 2 (Snf2) family proteins consist of a sizeable and diverse class of multiprotein assemblies that remodel chromatin complexes to contribute to the biological processes of replication, transcription, and DNA repair using the energy of ATP hydrolysis. Chromatin remodeling factors (CHRs) are ATPases from the Snf2 family and modulate the position of nucleosomes on chromatin to enable dynamic access to packaged DNA (Song et al., 2021). During plant stress responses, altered transcriptional responses have been linked to chromatin-mediated inducible gene expression, which is performed by covalently modifying histone and/or the DNA, or non-covalently altering the nucleosome position, conformation, occupancy, and composition by chromatin remodeling ATPases (Li et al., 2007; Chinnusamy and Zhu, 2009; Kim et al., 2010). Snf2 family proteins, especially in a plant, are often classified by two conserved domains (SNF2_N/DExx and Helicase_C/HELICc) in the helicase-like region (Flaus et al., 2006; Knizewski et al., 2008). The Snf2 family members are identified and characterized as 41, 40, and 45 proteins in model plants Arabidopsis, rice, and tomato, respectively, and classified into 24 subfamilies of six groups (Song et al., 2021). The corresponding chromosome location, phylogenetic relationship, domain architectures, and expression pattern of partial genes have been clearly described (Knizewski et al., 2008; Hu et al., 2013; Zhang et al., 2019).
Many publications have insight into the critical roles of CHRs in regulating development, growth, and stress response in general plants (Han et al., 2015; Sarnowska et al., 2016; Song et al., 2021). As sessile organisms, the plants must depend on their capacity to cope with the adverse environment or situation, such as biotic stresses (pathogenic infections) and abiotic stresses (drought, salt stress, extreme temperatures, and heavy metals elements; Mohammadi et al., 2020; Han et al., 2021). When first exposed to stress, plants initiate rapid gene expression modulation by altering chromatin structures at promoters and other regulatory DNA regions using chromatin-remodeling enzymes (Mlynarova et al., 2007). MINU1/CHR12, an Snf2/Brahma-type chromatin-remodeling gene, participates in mediating the temporary growth arrest in Arabidopsis. The overexpression of MINU1/CHR12 leads to growth retardation under adverse stress conditions, and MINU1/CHR12 loss-function-of mutant displays tolerance to salt, drought, and heat stresses (Mlynarova et al., 2007). BRM (CHR2, an Snf2 subfamily member) directly represses the transcription of ABI5, and loss-of-function of BRM causes ABA hypersensitivity during post-germination development. In addition, brm was reported to increase drought tolerance (Han et al., 2012). PICKLE (PKL/CHR6, a Mi-2/CHD3 subfamily member) activates the expression of auxin and cell elongation-related genes IAA19 (INDOLE-3-ACETIC ACID INDUCIBLE 19) and IAA29 by repressing H3K27me3 deposition. The mutant of PKL displays hypocotyl reduction (Zha et al., 2017). In addition, a recent report shows that pkl mutant is hypersensitive to cold and freezing stresses (Yang et al., 2019).
Besides Arabidopsis, several Snf2 family proteins have been studied during biotic or abiotic stresses in other plant species. For example, the loss-function-of mutant of ZmCHB101 (the ortholog gene of SWI3D in Arabidopsis) is dramatically sensitive to osmotic and salt stress, and the transcriptional level of stress-responsive genes are affected by regulating RNA polymerase II association and nucleosome density near TSS (transcription start site) in the maize (Yu et al., 2018). Under salt stress, SlCHR1, the ortholog gene of MINU in Snf2 family, regulates the growth retardation in tomato, similar to CHR12 in Arabidopsis. OsCHR4/CHR729, a CHD3/Mi-2 subfamily member, affects the development of seedling and root via the signaling pathways of gibberellin and auxin, respectively, and decreases the contents of chloroplast in adaxial mesophyll cells in rice (Hu et al., 2012; Ma et al., 2015a; Wang et al., 2016). Functional deficiency of OsRFS (Rolled Fine Striped) affects leaf rolling, width, chloroplast development, reactive oxygen species (ROS) scavenging, and osrfs mutants exhibited accumulation of ROS due to the loss of H3K4me3 at the genomic loci of ROS-related genes (Cho et al., 2018; Mohammadi et al., 2021). OsALT1 (Alkaline Tolerance 1, OsCHR706), a Ris1 subfamily chromatin-remodeling ATPase, improves tolerance to alkaline stress by reducing ROS levels and alleviating oxidative stress damage (Guo et al., 2014).
Snf2 family proteins, as CHRs, is an enormous and most diverse protein family in rice. Previous researches have shown the function of some genes, such as OsCHR4, OsRFS, OsALT1, OsDDM1, OsCHR721, OsBRHIS1, OsENL1 (Higo et al., 2012; Hu et al., 2012; Guo et al., 2014; Hara et al., 2015; Li et al., 2015; Ma et al., 2015a; Wang et al., 2016; Cho et al., 2018; Zhang et al., 2020b). The systematical expression profiles of Snf2 family genes have not been analyzed during reproductive development and biotic/abiotic stresses. This study deeply investigated the expression profiling of the rice Snf2 genes in different organs/tissues, as well as the prediction of cis-acting elements in the promoter regions and responses under drought and salt treatments. Moreover, we report that the mutant lines of OsCHR726, which encodes a SMARCAL1 CHR, are sensitive to salinity stress. The results extend our understanding of Snf2 genes in plant development and are a valuable resource for further investigating stress tolerance in rice.
Materials and Methods
Plant Materials and Growth Conditions
The OsCHR726 T-DNA insertion mutant (PFG_2C-10296) in the Dongjin (Oryza sativa L.) background was obtained from Kyung Hee University, South Korea. The detailed information on the mutant can be searched at the SIGnAL database1. The OsCHR726 Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated proteins 9 (CRISPR/Cas9)-mediated mutant lines used in this study were in Zhonghua 11 (ZH11, Oryza sativa L.) background, which also was used as wild-type for analysis of Snf2 genes expression in specific tissues, as well as under biotic and abiotic stresses. All rice plants were grown in the greenhouse at Fujian Agriculture and Forestry University at 22–32°C and 80–90% humidity with a 14 h/10 h (light/dark) photoperiod.
To analyze the expression profile of representative Snf2 genes during vegetative and reproductive development, the tissues for expression pattern analysis were: 7-day-old seedling (YS), root (YR), and leaf (YL); 70-day-old leaf (ML); 0–3 cm panicles (P1); 3–5 cm panicles (P2); stigma (Sti) of mature ovary (OV) before pollination; and seed at 25 DAP (S5, day after pollination). The stamens and pistils at different developmental stages were carefully collected from young panicles of 60–75-day-old seedlings. Stamens and pistils from the different spikelets with 2–3, 3–4, 4–5, 5–6, 6–7 mm length and before flowering were defined as An1–6 and Ov1∼6, respectively.
For expression analysis of Snf2 genes in response to ABA, IAA, and other stress treatments, the seeds were germinated and sown on a plastic net floating on a nutrient solution in a growth chamber (Yoshida et al., 1971; Zhao et al., 2015). The seedling roots of 14-day-old ZH11 were submerged into a nutrient solution containing 200 mM NaCl, 100 mM ABA, 150 μM IAA, and nutrient solution treatment, respectively. For cold testing, the seedlings were treated at 4°C and the seedlings were carefully transferred onto paper as drought stress, in which they were air-dried (Zhao et al., 2012). The samples were collected at 0, 1, 3, 6, 12, and 24 h of H2O, salt, ABA, cold, and drought stresses. For biotic stress, 2-week-old seedlings were sprayed with a spore suspension (1 × 105 spores/mL) of the Magnaporthe oryzae (M. oryzae) isolate Guy11, and the leaf samples were taken 12, 24, 36, 48, 60, and 72 h post-infection (hpi), respectively. All materials contained three biological replicates and were immediately frozen in liquid nitrogen and stored at –80°C for RNA extraction.
Expression Data Analysis of Snf2 Family in Rice
The microarrays data were extracted from the Rice Functional Genomic Express Database (see footnote 1) and used to analyze the expression profiles of Snf2 genes in various tissues during different development stages (GSE6893, GSE6901, GSE7951; Ma et al., 2011). The microarray data of Sti and OV were from GSE7951 and YS was from GSE6901. The GSE6893 included the data of YR, ML, YL, SAM, P1-P6, and S1–S5. The data is provided in Supplementary Table 1. For microarray analysis, if more than one probe set was available for one gene, the average of the values was used for further study. The relative expression levels of Snf2 genes were determined by extracting their respective data from the total expression matrix using Genesis software.
The heatmaps were presented on expression data of Snf2 genes to drought and salt using the R package pheatmap. A lot of values are concentrated in a very small range, and suddenly there are a few very large values that are disproportionately large. The result is that most of the differences between the values are obscured to accommodate the few extreme values, leaving large areas of almost the same color. Standardization is to retain the law of data and scale the value in a relatively stable range after a certain proportion control, which is more convenient for showing.
RNA Isolation and qRT-PCR Analysis
Total RNA of all collected samples was extracted using Plant RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. A total of 1 μg of RNA was reverse transcribed using the PrimeScript RT-PCR kit (Takara, Kyoto, Japan; Cai et al., 2019). The relative expression level was detected by quantitative real time PCR (qRT-PCR) using the Bio-Rad QRT-PCR system (Foster City, CA, United States) and SYBR Premix Ex TaqII (TaKaRa Perfect Real Time; Zhang et al., 2020a). The qRT-PCR program was: 95°C for 30 s; 40 cycles of annealing at 95°C for 5 s, and extension at 60°C for 35 s; 95°C for 15 s (Zhang et al., 2020a; Zhao et al., 2021). The rice OsUBQ5 gene was used as an internal control (Jain et al., 2006). To evaluate the relative expression levels of the examined genes, we used the comparative ΔΔCT method (Su et al., 2017). The gene-specific primers are listed in Supplementary Table 2.
Subcellular Localization Analysis of OsCHRs
The full-length coding sequences of OsCHR712/715/720/726/739 were amplified from WT cDNA using the primers listed in Supplementary Table 3. The PCR fragments were cloned into the pENTER/D-TOPO vector (Invitrogen, CA, United States), and pENTER/D-TOPO clones were recombined into the pGWB506 vector using LR ClonaseII enzyme (Invitrogen). The 35S:CDS-GFP recombinant construction and 35S:GFP (vector control) were transformed into Agrobacterium tumefaciens (GV3101) and infiltrated into tobacco leaves. The fluorescence signals were observed using a confocal microscope (SP8, Leica, Germany), and the excitation wavelength was 488 and 405 nm.
Analysis of the Putative Promoter Regions of Snf2 Genes
The cis-acting promoter elements are crucial for the coordinated expression of genes. We retrieved ≤ 2 kb upstream sequence of all 40 Snf2 genes and predicted the critical regulatory elements responsible for biotic and abiotic stresses by PlantCARE (Lescot et al., 2002).
Identification of the T-DNA Insertion and CRISPR/Cas9-Mediated Mutations
The T-DNA insertion in oschr726 was confirmed by PCR using primers 2C-10296-Lp and 2C-10296-Rp, and the T-DNA-specific primer LB (2717). In addition, we generated a gRNA construct with the gRNA (5′GCCGAGGGCTTCGCCTACCCCGG 3′), and plant-optimized Cas9 driven by rice OsU6a and maize pUBI-H promoters, respectively (Ma et al., 2015b). The plasmid was transformed into WT ZH11 callus, and the DNA isolated from transgenic plant leaves was amplified by PCR and sequencing analysis using the primer set P1 and P2. The primers sequences are described in Supplementary Table 8 for genotyping identification. The gene structure of OsCHR726 was searched from Gene Structure Display Server2.
Salt Stress of oschr726 Mutant Lines
To investigate the mutant salt response in a solid medium, seeds were disinfected with hypochlorous acid and germinated on half MS (Murashige and Skoog) medium for 2 days then transplanted to half MS with 0, 100, 150 mM NaCl for 5 days at 28°C in the growth chamber with a 14 h/10 h (light/dark) photoperiod.
Results
Expression Analysis of Snf2 Genes in Various Tissues Using Microarray Data
Snf2 family proteins, as CHRs, consisting of enormous protein members, play a crucial role in the process of plant development and growth, including transcription, replication, homologous recombination, and DNA repair. (Han et al., 2015; Sarnowska et al., 2016; Ojolo et al., 2018; Singh et al., 2020). The protein members of the Snf2 family have been well-characterized in rice (Hu et al., 2013). However, the spatial expression profiles of Snf2 family genes have not been analyzed.
To this end, the microarray data of various tissues were collected during vegetative and reproductive developmental stages from the Rice Functional Genomic Express Database (see footnote 1), including 7-day-old seedlings (YS), young root (YR), leaf (YL), mature leaf (ML), shoot apical meristem (SAM), panicles (P1-P6), stigma (Sti) of mature ovary (OV), and seed (S1–S5) development. The result showed that a total of 36 (90%) Snf2 genes were expressed in different tissues, while the rest of four genes (OsCHR720, OsCHR736, OsCHR737, OsCHR741) were not detected in the microarray data (Supplementary Table 1). A hierarchical cluster display average of log signal values of these genes was produced (Figure 1A). The microarray analysis revealed that 15 (37.5%) and 20 (50%) of Snf2 genes were predominately expressed in YL and SAM, respectively. OsCHR735, OsCHR746, OsCHR722, and OsCHR742 were distinctively expressed in SAM, and OsCHR704, OsCHR706 were uniquely expressed in YL. The expression levels of 13 (32.5%) Snf2 genes were highly expressed in OV. OsCHR731 and OsCHR733 are highly expressed in SAM, P1, and P2, while OsCHR711, OsCHR721, and OsCHR725 were highly expressed in SAM, P1, P2, and OV. Interestingly, OsCHR730, OsCHR715, OsCHR703, and OsCHR717 were uniquely expressed in OV, Sti, S2, and S3, respectively. It was noteworthy that the four genes (OsCHR715, OsCHR727, OsCHR731, and OsCHR739) marked with asterisks in Figure 1A were confirmed to be specifically expressed in Sti, YL and S5, P1 and P2, and YL using the qRT-PCR, respectively, which agreed well with the microarray data (Figure 1B). Taken together, we can conclude that most of the Snf2 genes were predominately expressed in YL, SAM, and OV, suggesting the participation of Snf2 genes in the development of young tissues and reproductive organs.
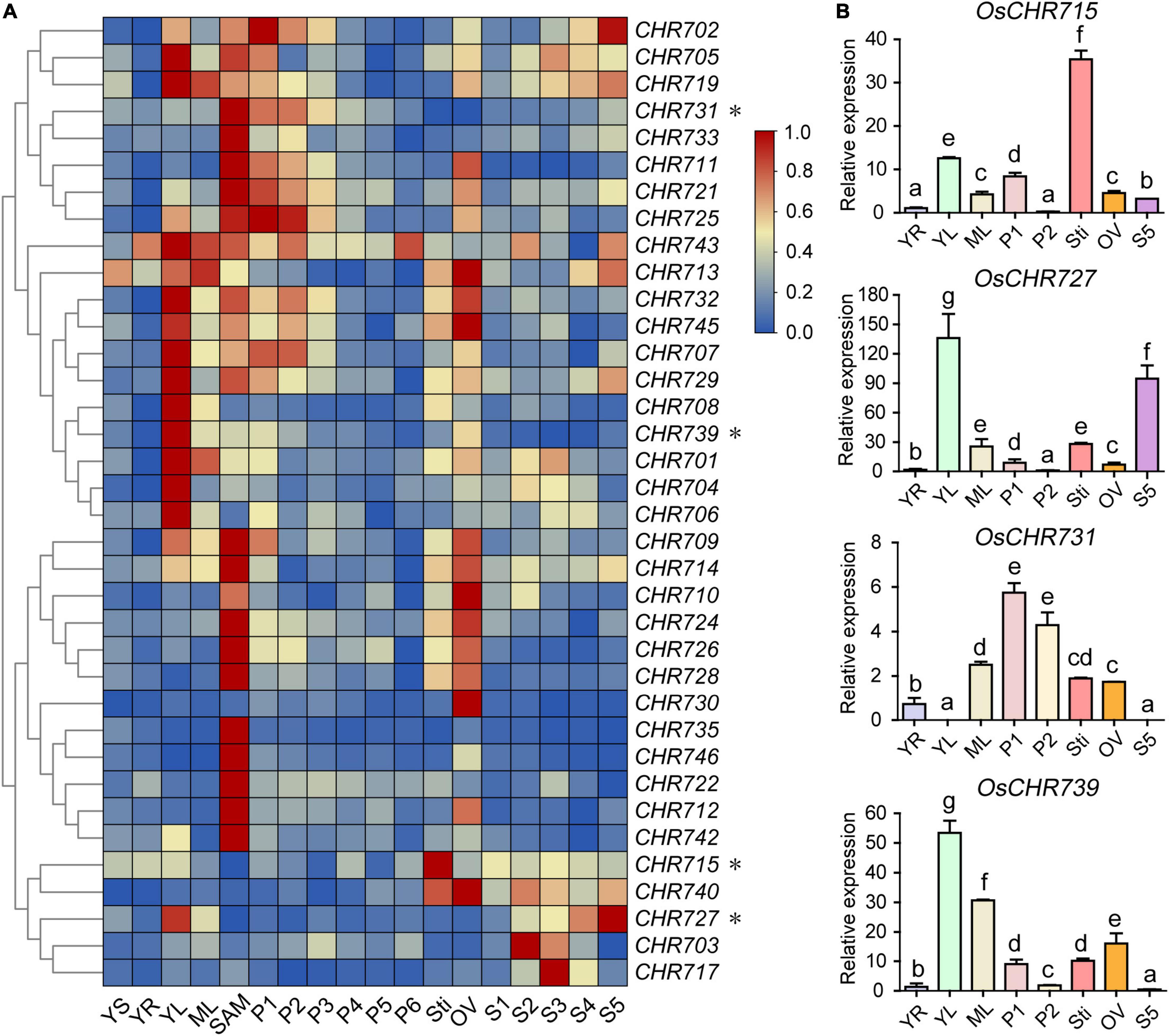
Figure 1. Expression profiles of Snf2 genes in various organs. (A) The microarray data (GSE6893, GSE6901, and GSE7951) of Snf2 genes’ expression in various organs at different developmental stages were reanalyzed. A heat map representing hierarchical clustering of average log2 expression values of Snf2 genes in various organs are generated (YS, 7-day-old seedlings; YR, roots from 7-day-old seedlings; YL, leaves from 7-day-old seedlings; ML, mature leaf; SAM, shoot apical meristem; different stages of panicle development: P1, 0–3 cm; P2, 3–5 cm; P3, 5–10 cm; P4, 10–15 cm; P5, 15–22 cm; P6, 22–30 cm; Sti, stigma of mature ovaries; OV, mature ovary; different stages of seed development: S1, 0–2 dap (day after pollination); S2, 3–4 dap; S3, 5–10 dap; S4, 11–20 dap; S5, 21–29 dap). The representative Snf2 genes are differentially expressed in various organs, for which real-time PCR analysis was performed, indicated by an asterisk (*) on the right. The color scale (representing average log signal values) is shown on the right side. (B) The representative Snf2 genes (asterisks indicate genes in A) are differentially expressed in various organs consistent with the samples of microarray using the qRT-PCR. Error bars indicate standard deviations of independent biological replicates (n = 3). Different letters denote significant differences at P < 0.05 according to ANOVA in combination with Duncan’s multiple range test.
Most Snf2 Genes Are Highly Expressed in the Late Development Stages of Stamen and Pistil
It has been reported that OsCHR721, a nuclear protein belonging to the SMARCAL1 subfamily in the Snf2 family, played a crucial role in both male and female reproductive development (Zhang et al., 2020b). To investigate the temporal expression profile of Snf2 family genes and find the candidate key genes that may have functions during the development of reproductive organs in rice, we performed a more comprehensive analysis of stamen and pistil at specific developmental stages using qRT-PCR. According to the length of spikelets, the stamens and pistils were divided into six developmental stages (An1–6 and Ov1–6). OsCHR703, OsCHR733, OsCHR735, OsCHR741, and OsCHR746 showed similar expression patterns, that is, they were preferentially expressed in both the early (An1) and late (An6) developmental stages of stamen (An1, spikelet length = 2∼3 mm, An6, mature spikelet, Figure 2A). OsCHR712, OsCHR714, OsCHR724, OsCHR727, OsCHR730, and OsCHR743 were preferentially expressed in the An1, An5, and An6 developmental stages of stamen (Figure 2B and Supplementary Figure 1A). OsCHR717 and OsCHR737 were preferentially expressed in the An4–6 (Supplementary Figure 1B). OsCHR704, OsCHR706, OsCHR709, OsCHR710, OsCHR719, OsCHR720, OsCHR729, OsCHR732, OsCHR736, and OsCHR739 were predominately expressed in the late developmental stages of stamen (An5 and An6, Figure 2C). There are several genes uniquely expressed in the An6, such as OsCHR701, OsCHR702, OsCHR705, OsCHR708, OsCHR713, OsCHR715, OsCHR721, OsCHR722, OsCHR725, and OsCHR728 (Figure 2D). These results exhibited that most Snf2 genes were predominately expressed in the late development stages of stamen, and suggested that Snf2 genes might play an important role in mature stamen development.
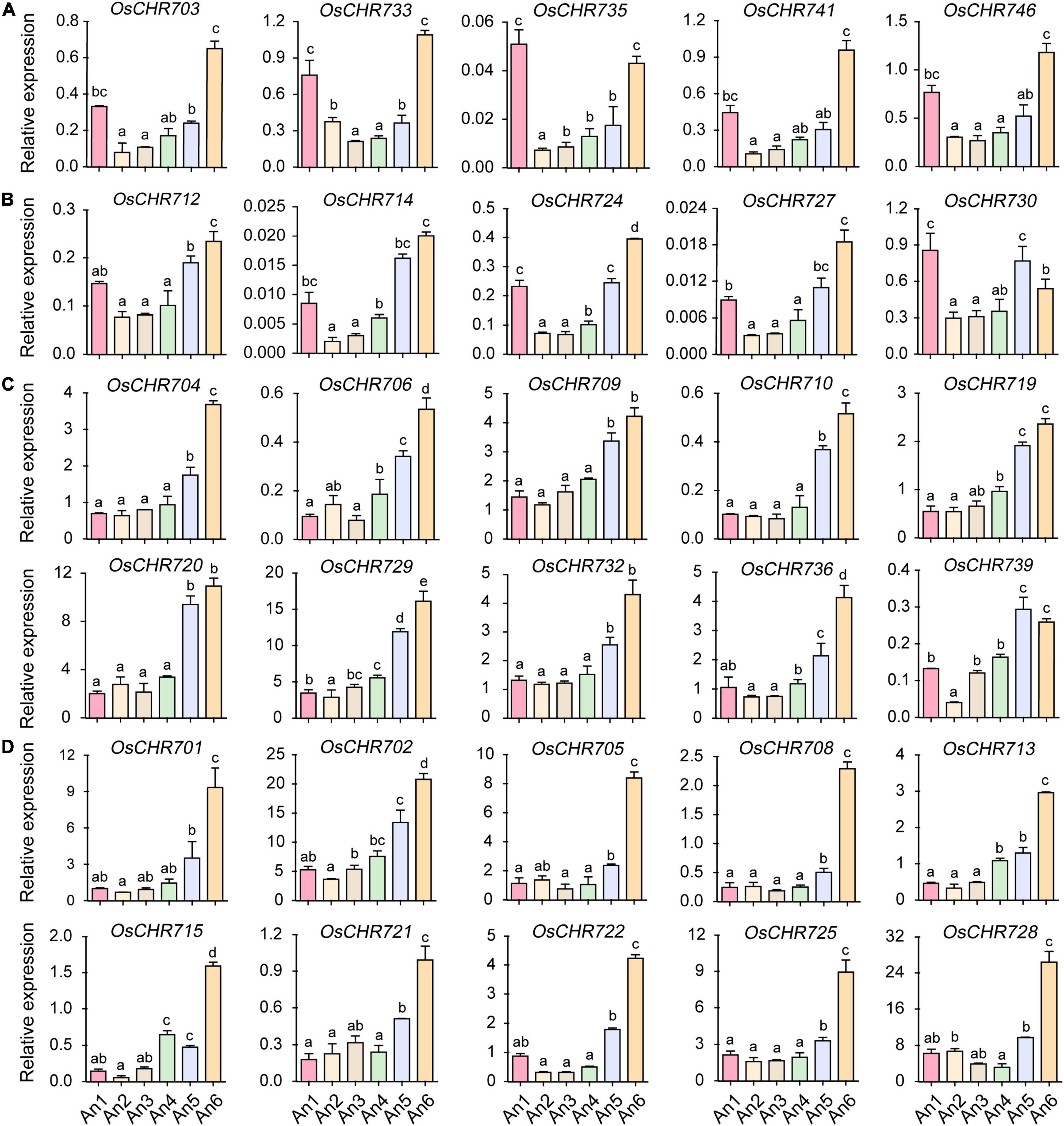
Figure 2. The relative expression level of Snf2 gene family in the stamen. (A) Predominately expressed Snf2 genes at the An1 and An6 stages. (B) Preferentially expressed Snf2 genes at the An1, An5, and An6 stages. (C) Preferentially expressed Snf2 genes at the An5 and An6 stages. (D) Predominately expressed Snf2 genes at the An6 stages. An1-5, the stamen of 2–3, 3–4, 4–5, 5–6, and 6–7 mm spikelet, respectively; An6, the stamen in the spikelet before flowering. The y-axis is the relative expression level of the gene compared to OsUBQ5 in different developmental stages of stamen using the qRT-PCR. The relative expression level of OsCHR701 to OsUBQ5 in An1 stage normalized as “1” was used as reference. Error bars indicate standard deviations of independent biological replicates (n = 3). Different letters denote significant difference at P < 0.05 according to ANOVA in combination with Duncan’s multiple range test.
Similarly, many genes displayed specific expression patterns during the pistil’s development. About 30 and 40% Snf2 genes were predominantly expressed in the early (Ov1, spikelet length = 2∼3 mm) and late (Ov6, mature spikelet) developmental stages of pistils, respectively. OsCHR709, OsCHR711, OsCHR720, OsCHR730, OsCHR735, OsCHR739, OsCHR742, OsCHR743, OsCHR745, and OsCHR746 were specifically expressed in the Ov1 (Figure 3A). OsCHR705, OsCHR706, OsCHR710, OsCHR721, and OsCHR725 were highly expressed in the late developmental stages of pistils (Ov5, spikelet length = 6–7 mm, Ov6, mature spikelet, Figure 3B), whereas, OsCHR704, OsCHR712, OsCHR713, OsCHR715, and OsCHR727 were preferentially expressed in both early and late developmental stages of the pistils (Supplementary Figure 2A). OsCHR703, OsCHR708, OsCHR719, OsCHR733, and OsCHR741 were predominately expressed in the Ov4 stages (Supplementary Figure 2B). OsCHR707, OsCHR714, OsCHR717, OsCHR722, OsCHR724, OsCHR726, OsCHR732, OsCHR736, OsCHR737, and OsCHR740 showed similar expression patterns, and their expression level gradually increased with the developmental period (Figure 3C). However, other gene transcripts were abundant in stamen and pistils (Supplementary Figures 1C, 2C). These data indicated that these genes might play various roles in specific reproductive tissues at different development stages and provided better reference to explore the key regulatory genes for reproductive organ development in crops.
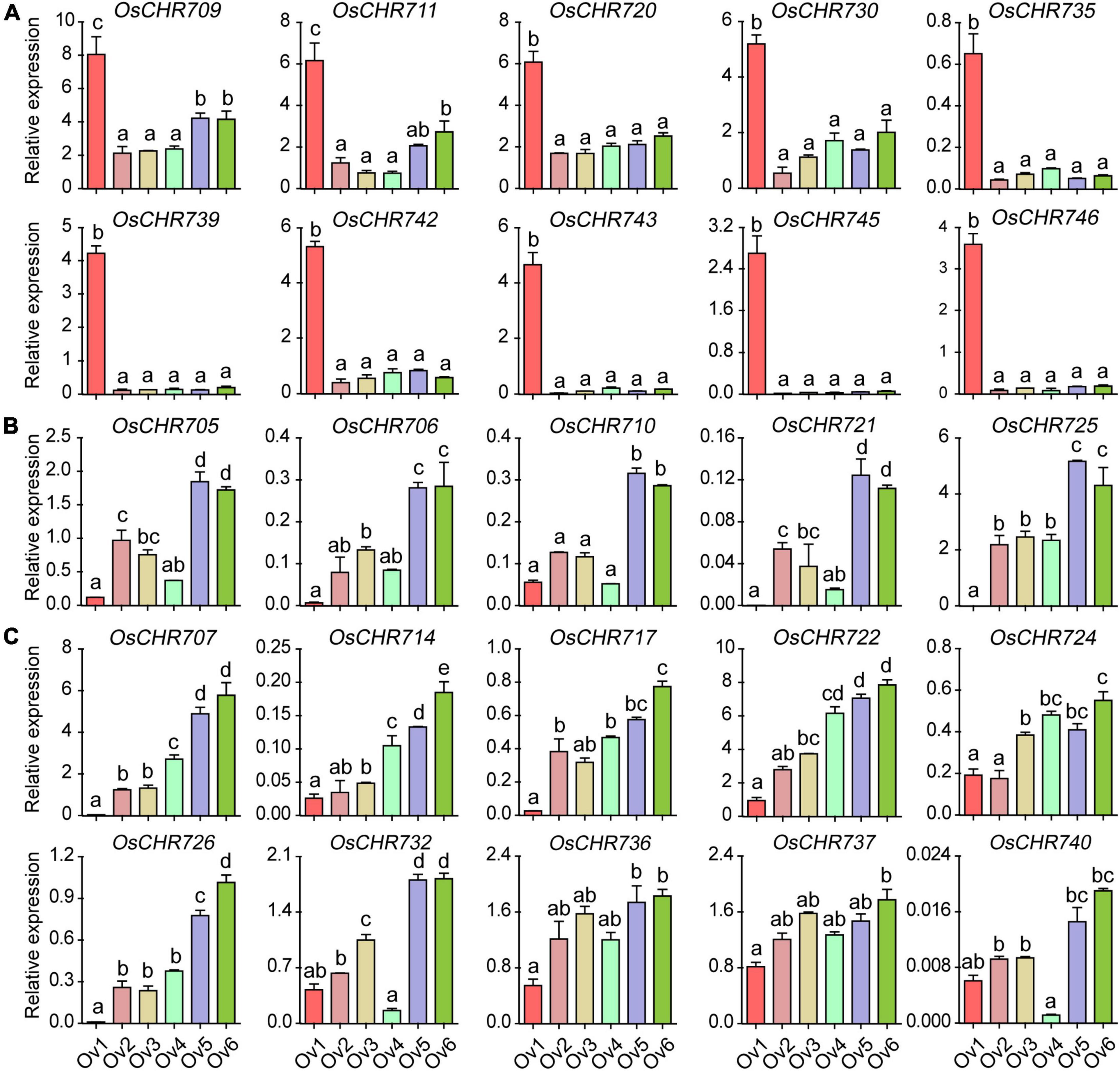
Figure 3. The relative expression level of Snf2 gene family in the ovary. (A) Specifically expressed Snf2 genes at the early developmental stages (Ov1) in the ovary. (B) Predominantly expressed Snf2 genes at the late developmental stages (Ov5 and Ov6) in the ovary. (C) The expression level of Snf2 gene gradually escalated from early to late developmental stages in the ovary. Ov1-5, the ovary of 2–3, 3–4, 4–5, 5–6, and 6–7 mm spikelet, respectively; Ov6, the ovary in the spikelet before pollination. The y-axis is the relative expression level of the gene compared to OsUBQ5 in different developmental stages of the ovary using the qRT-PCR. The relative expression level of OsCHR701 to OsUBQ5 in An1 stage normalized as “1” was used as reference. Error bars indicate standard deviations of independent biological replicates (n = 3). Different letters denote significant differences at P < 0.05 according to ANOVA in combination with Duncan’s multiple range test.
OsCHRs Are Mainly Located in the Nucleus
Snf2 proteins, as CHRs, control many aspects of DNA events (Hu et al., 2013) and serve as an integrative platform that translates various signals from the cellular environment into regulated responses from DNA (Knizewski et al., 2008). To evaluate the subcellular localization of OsCHRs proteins, the full-length coding sequences of OsCHR712/715/720/726/739, which were from different subfamilies, were severally fused with the sequence encoding green fluorescence protein (GFP), and the vectors were expressed transiently in tobacco epidermal cells. As the results are shown in Figure 4, the fluorescence signal from 35:GFP vector was detected throughout the whole cell and co-localized with DAPI signal in the nucleus. Then the 35S:OsCHR712/715/720/726/739-GFP were co-localized with the DAPI signal, suggesting that these five proteins were mainly localized in the nucleus. Meanwhile, OsCHR715/739 were also slightly expressed in the cell membrane (Figure 4). Taken together, our results suggested that Snf2 genes might play crucial roles in the nucleus and correspond to the predicted function of the family proteins.
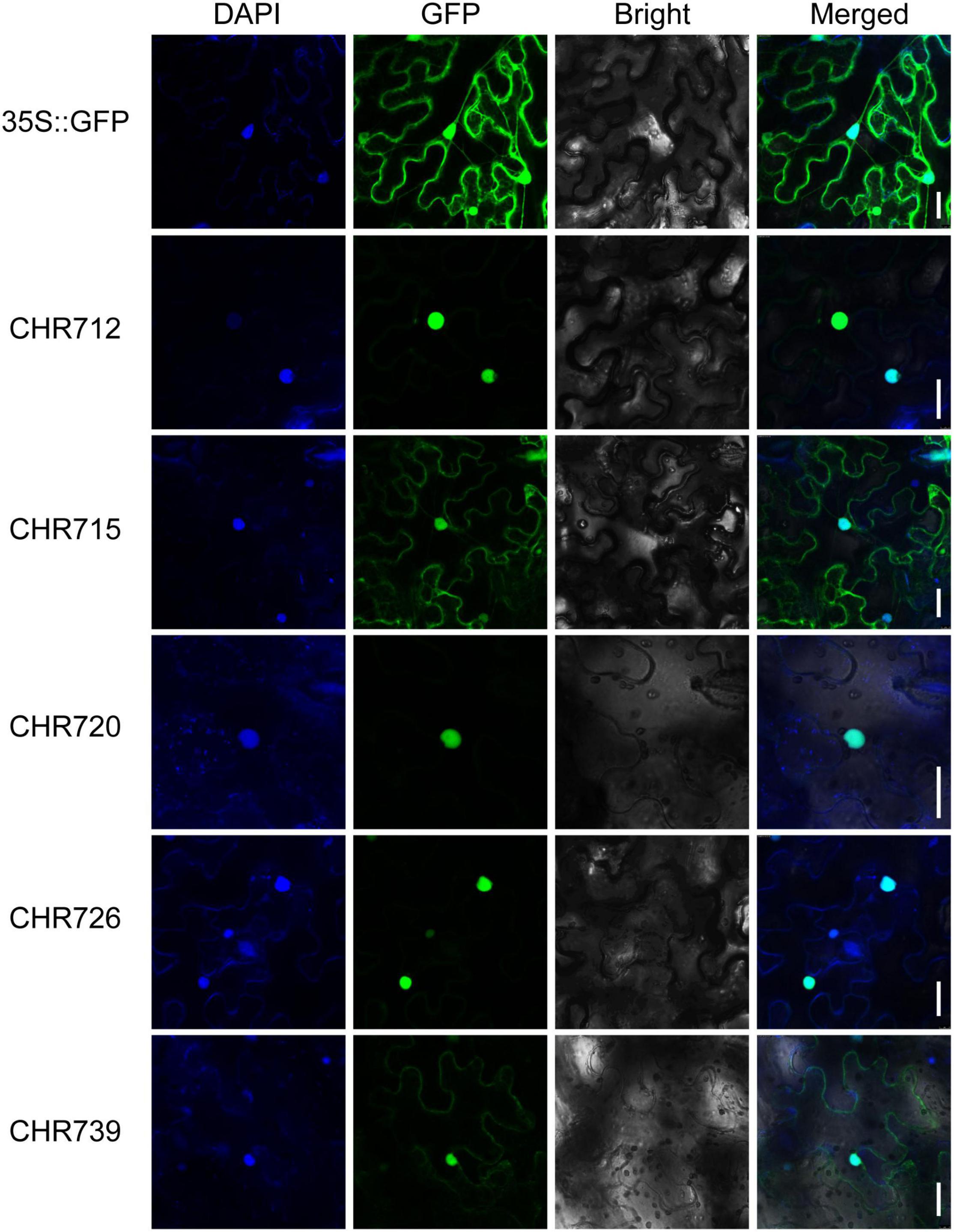
Figure 4. The subcellular localization of OsCHR712/715/720/726/739. The 35S:OsCHR712/715/720/726/739-GFP and control vector (35S:GFP) were transiently expressed in tobacco epidermal leaf cells. Confocal microscopy images were visualized after 24-h transformation. Bar = 30 μm.
Most Dehydration- and Drought-Responsive Elements Distributed in the Putative Promoter of Snf2 Genes
The cis-acting promoter elements are crucial for the coordinated expression of the genes. Then it is critical to characterize the regulatory elements to associate the expression profiles with the genetic components. We retrieved the promoter region (2 kb upstream sequence of the start codon) of all 40 Snf2 genes from the rice database. We predicted the cis-acting elements responsible for biotic and abiotic stress responses by PlantCARE (Lescot et al., 2002). A total of 11 stress-responsive elements were observed in the putative promoter regions of the Snf2 genes (Supplementary Table 4). These included DRE or binding site for MYC transcription factor for dehydration responsive elements (Narusaka et al., 2003; Tran et al., 2004). LTR for low temperature-responsive element, STRE or TC-rich motifs for stress responses, CCAAT box and MYB binding sites responsive to drought treatments, Box S and WUN motif for wounding and pathogen elicitation response elements (Xu et al., 2011; Yin et al., 2017). Several phytohormone responsive elements were also observed in the promoter regions, such as TCA element for salicylic acid responses, ERE as ethylene responses, CGTCA- or TGACG-motif for MeJA responsive element, ABRE as ABA-responsive element, TGA-element or AuxRR core or AuxRE for auxin responses, GARE-motif, and TATC-box for gibberellin responses.
The number and distribution of environmental stress-related and hormone-responsive elements in Snf2 genes are shown in Figures 5A,B, Supplementary Figures 3A,B, and Supplementary Table 5. The abundant elements were dehydration-responsive element, MeJA responsive element, stress-responsive element, MYB binding site for drought, and ABA-responsive elements. To explore the possible involvement of Snf2 genes in abiotic stress response, we analyzed the expression patterns of eight genes in the seedling after low temperature (4°C) and ABA treatments. The expressions of OsCHR711, OsCHR715, OsCHR727, OsCHR728, and OsCHR732, which have two or more LTR elements in each promoter region, were significantly increased within 3 h after cold stress treatment by qRT-PCR (Figure 5C). Similarly, the promoter regions of OsCHR703, OsCHR715, OsCHR728, OsCHR733, and OsCHR735 have four or more ABRE elements, and the expression level of these genes was remarkably increased within 6 h after ABA treatment (Figure 5D). However, the promoter regions of OsCHR707, OsCHR717, OsCHR724, OsCHR725, and OsCHR726 have one or no Auxin-responsive element, and these genes responded slowly or not to IAA treatment (Supplementary Figure 3C). These results suggested that Snf2 genes might be involved in plant response to abiotic and hormones stresses, and the cis-acting elements in the promoter regions were coordinated with the expression of genes.
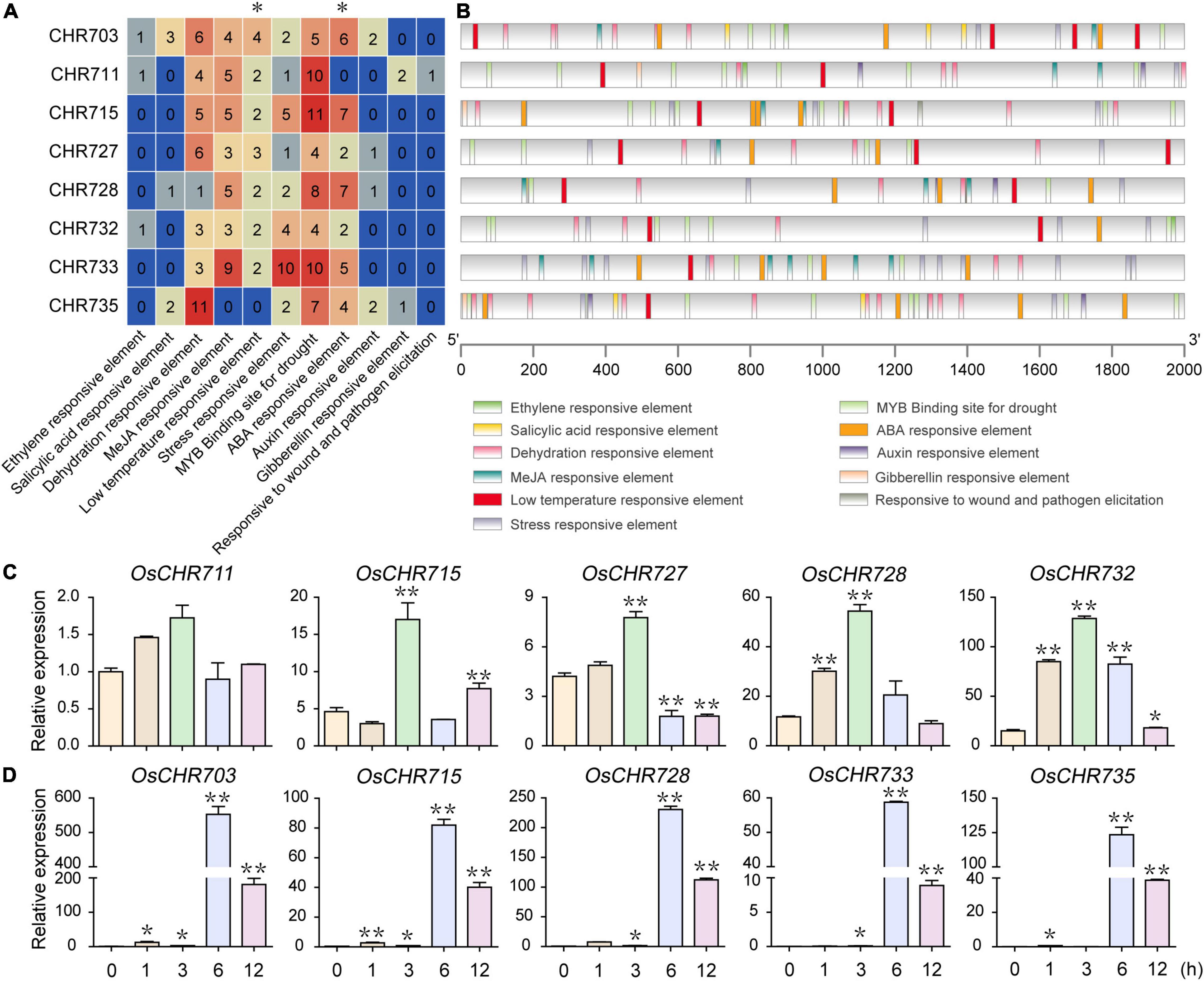
Figure 5. Prediction of cis-acting elements of environmental stress-related and hormone-responsive in Snf2 genes. (A) Number of cis-acting elements detected in the promoter regions (sequence retrieved from about ≤ 2 kb upstream region). The cis-acting elements were divided into 11 types. (B) Quantity, kind and position of environmental stress-related and hormone-responsive elements in the putative promoters of genes in Figure 5A. (C) The qRT-PCR analysis of Snf2 genes containing multiple LTR (low temperature-responsive) elements under 4°C treatment. (D) The qRT-PCR analysis of Snf2 genes containing multiple ABA-responsive elements under ABA treatment. The y-axis is the relative expression level of the gene compared to OsUBQ5 under different treatments using the qRT-PCR (C,D). The relative expression level of OsCHR711 to OsUBQ5 in an unstressed sample (0 h) normalized as “1” was used as a reference. Error bars indicate standard deviations of independent biological replicates (n = 3). Data are given as means ± SD. *p < 0.05, **p < 0.01 compared with unstressed samples using Student’s t-test.
Most Snf2 Genes Respond Quickly After the Magnaporthe oryzae Infection
The above result shows that Snf2 genes have the most abundant stress response elements in the putative promoter regions. Then what specific roles do Snf2 genes play in response to stress resistance? The previous researches revealed that OsCHR725 (OsBRHIS1) played a critical role in the SA-independent disease resistance by suppressing the innate immunity (Li et al., 2015), and OsCHR706 (OsALT1) negatively regulated alkaline tolerance (Guo et al., 2014). These observations had prompted us to analyze the differential expression pattern of Snf2 genes under biotic and abiotic stress conditions. We analyzed the expression levels of 40 Snf2 genes in two-week-old seedlings at seven different time points for biotic (M. oryzae) infection and at four different time points for abiotic (salt and drought) stresses.
When plants were exposed to environmental stresses, various physiological and biochemical responses were induced, leading to changes in gene expression (Ma et al., 2011). Briefly, after the plants were sprayed with M. oryzae GUY11 isolate, the spores of M. oryzae formed appressoria at 12 hpi and formed invasive hyphae at 24 hpi. Genes showing an up- and down-regulation were at least two-fold and significant difference compared with unstressed samples considered to further analysis compared to controls. Then, compared with the control plant, a total of 21 (52.5%) Snf2 genes were upregulated at 12 hpi, except for OsCHR720, which was upregulated at 24 hpi, and 12 (30%) genes were downregulated at all hpi (Figure 6 and Supplementary Figure 4). OsCHR725 (OsBRHIS1) was upregulated fourfold at 12 hpi, which could be consistent with its function in regulating innate immunity, and OsCHR707 was the most upregulated 12.6 folds than the control plants at 12 hpi, suggesting that most of Snf2 genes might play critical roles in regulating plant biotic stress and could respond quickly after the M. oryzae infection (Supplementary Table 6).
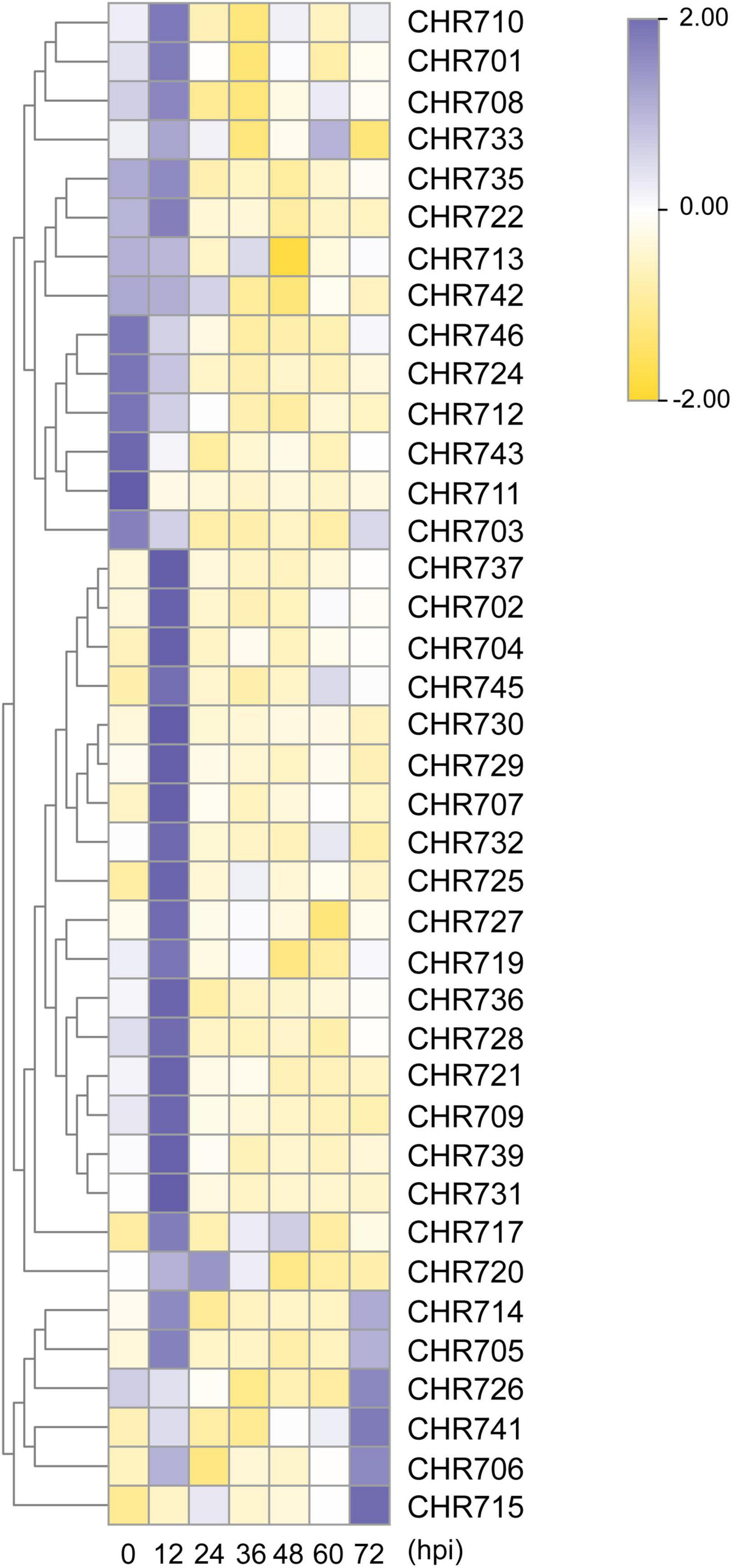
Figure 6. The relative expression level of Snf2 genes under M. oryzae stress. The expression analysis of Snf2 genes under the infection causal agent of rice blast. Heat map representing the temporal expression patterns was constructed by the heatmap packages in R. The data was normalized using OsUBQ5 as the internal reference gene. hpi, hours post-inoculation.
Most Snf2 Genes Were Upregulated Under Salt and Drought Treatments
Snf2 family proteins in Arabidopsis have been reported to play an important role in salt and drought stresses (Mlynarova et al., 2007). As described above, there were a lot of cis-acting elements related to stress response (dehydration-, drought-, and stress-responsive element) in the predicted promoters of Snf2 family genes (Figure 5 and Supplementary Figure 3). Then what is the expression patterns of Snf2 gene in response to salt and drought in rice ?
The expression profiles of Snf2 genes were displayed in Figure 7 under salt (200 mM) and drought stresses by qRT-PCR analysis. Genes showing an up- and downregulation were at least two-fold and significant difference compared with unstressed samples considered to further analysis compared to controls. Based on expression level, we divided the up-regulated genes into I (response at 3 h), II (response at 12 h), and III (response at 24 h) types (Figure 7). As indicated in the pie chart, about 55% of the total genes were upregulated under salt and drought treatments, and 22–35% genes were upregulated expression responses at 3 h (Type I). Salt, however, induced 10% type II and type III responsive genes’ expression, whereas, drought induced type II genes (30%) and type III genes (2.5%). The list of the expressed genes has been provided in Figures 7A,B. Most of the type I genes continued their expression till 24 h after the treatment, except for OsCHR702 and OsCHR732, which showed a split at subsequent time points (Supplementary Figures 5A,C). Both salt and drought stresses are associated with water management in plants, and then the results for these two stimuli were integrated. A total of 11 and 10 Snf2 genes were up-regulated during all experimental time points under salt and drought treatments, respectively. In addition, 9 and 4 genes were downregulated at all treatment time points, suggesting that they might play a negative role in salt and drought stresses (Supplementary Figures 5B,D). OsCHR721 and OsCHR714 were the most up-regulated, 22.5- and 39.5-folds than the control plants under salt and drought stresses, respectively (Supplementary Table 7). It was noteworthy that the seven genes (OsCHR705, OsCHR706, OsCHR710, OsCHR714, OsCHR721, OsCHR726, and OsCHR737) were upregulated and one gene (OsCHR712) was downregulated under both salt and drought stresses (Supplementary Figure 6). These results indicated that Snf2 genes might play a significant role in abiotic stress pathways and could be an excellent resource for evaluating stress tolerance in rice.
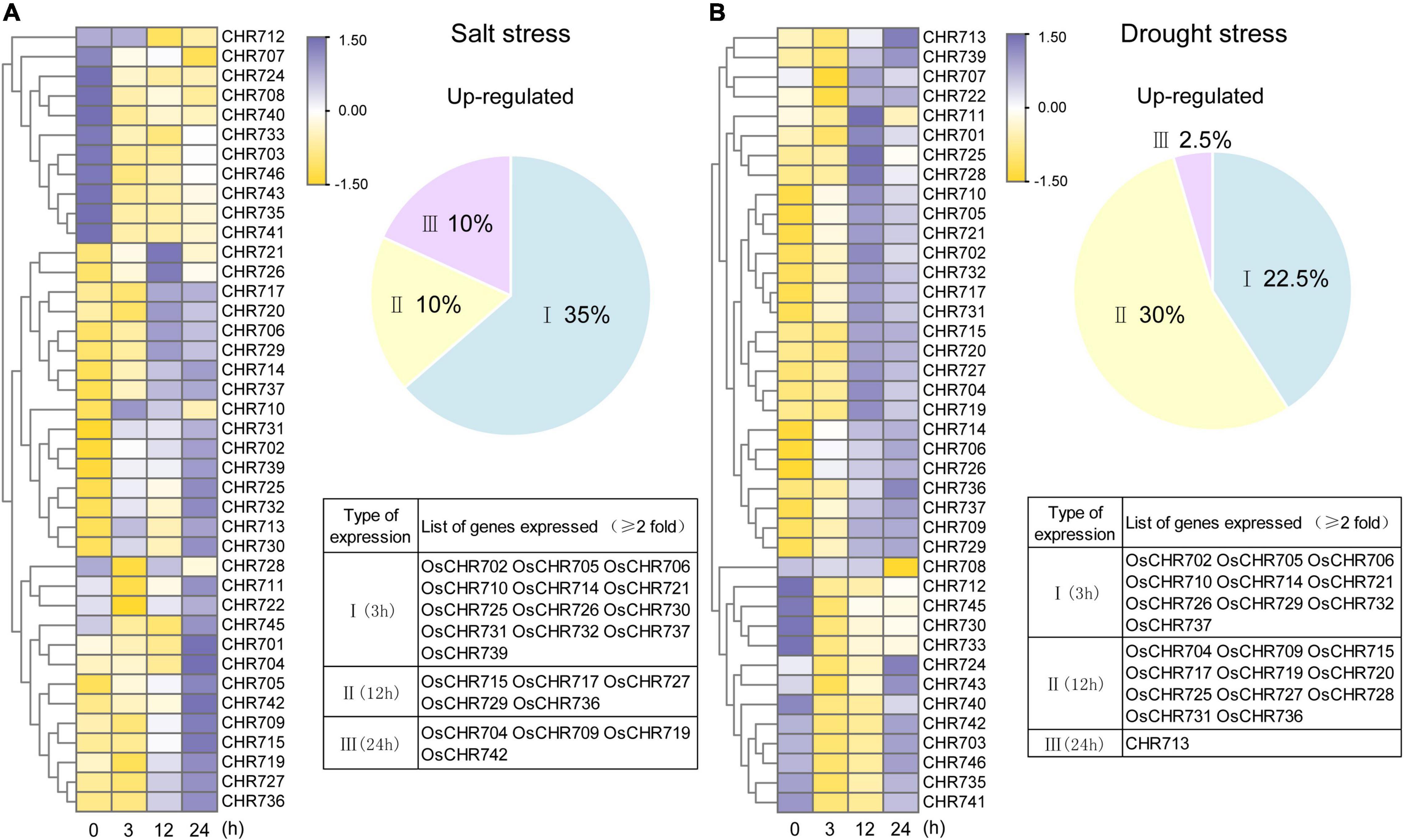
Figure 7. Expression analysis of Snf2 genes under salt and drought stresses. The temporal expression patterns of Snf2 genes under salt (A) and drought (B) stress treatments were presented by a heat map using the heatmap packages in R. The qRT-PCR values were normalized using OsUBQ5 as the internal reference gene and that of the unstressed samples using the ΔΔCT method. The percentage of genes upregulated (twofold and significant difference than the unstressed samples) under salt and drought treatments is represented in the pie chart beside the corresponding heat maps. The genes were annotated as I (3 h response), II (12 h response), and III (24 h response) types based on their time point of up-regulated expression. The names of the genes are provided in the forms.
Loss-Function of OsCHR726 Decreases Resistance of Transgenic Rice to Salinity Stress
According to the above results, OsCHR726 was upregulated under salt and drought stresses at all time points. Then what function does OsCHR726 play in response to stress resistance? Here, we characterized a T-DNA insertion mutant, oschr726-T (2C-10296.L), in Dongjin background (Figure 8), which was identified in SIGnAL database (see footnote 1). The homozygous plants for the T-DNA insertion were identified by PCR, and the OsCHR726 transcription level in oschr726-T was assessed by qRT-PCR (Supplementary Figure 7).
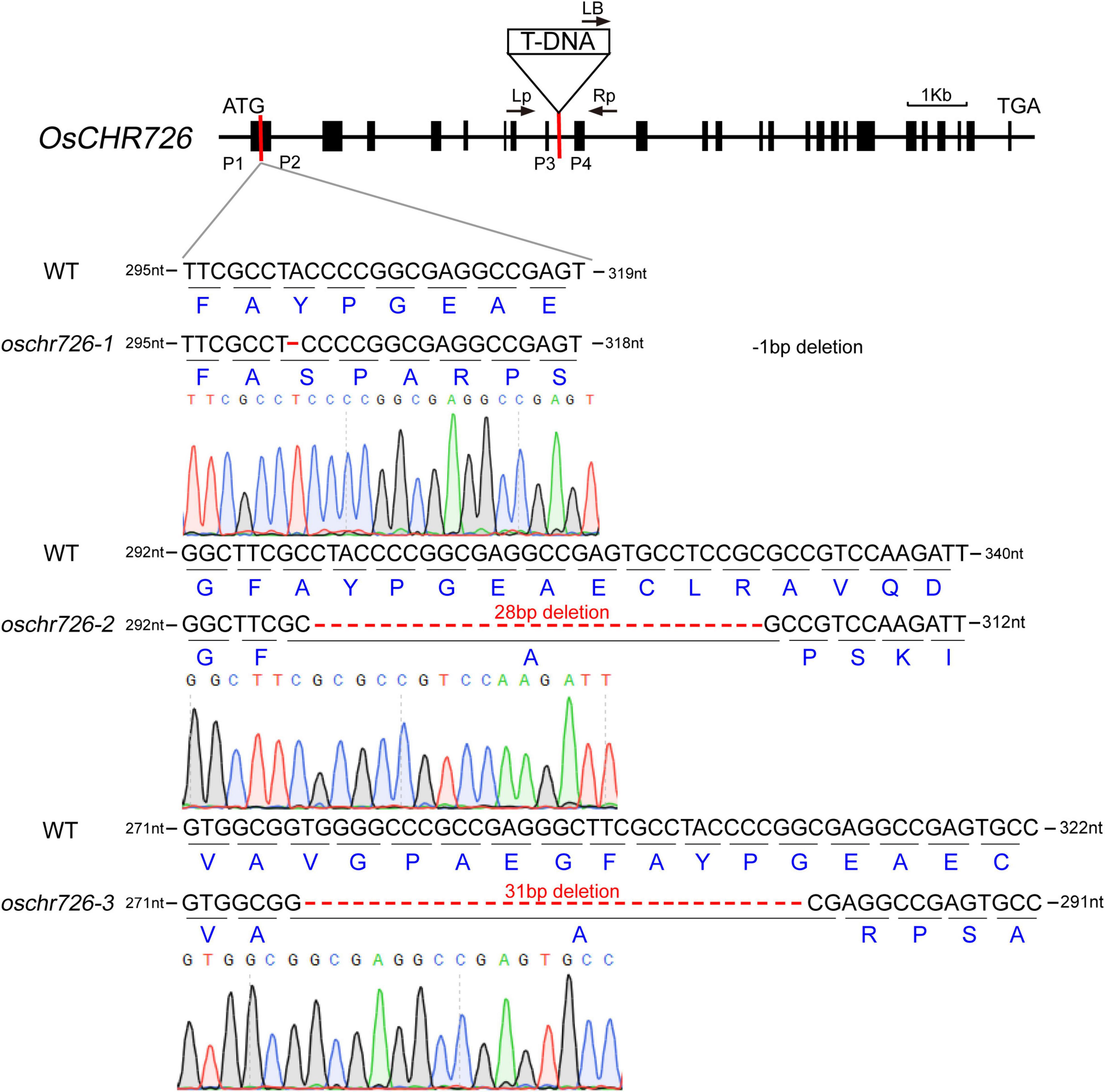
Figure 8. Genotyping analysis of oschr726. The triangle indicated the T-DNA insertion site in gene structure of OsCHR726. The arrows indicated the location of the primers for genotyping study of the T-DNA mutant. P3 and P4 were the positions of primers for identifying the transcription level of OsCHR726 in the T-DNA mutant by qRT-PCR. The primers of P1 and P2 were used for identifying the CRISPR/Cas9-mediated mutations in OsCHR726 in the T2 generation. The diagrams display the sequence results of the three CRISPR mutations. The black letters indicated the base pairs with WT in the target site. The dotted lines indicated nucleotide deletions, and dark blue letters indicated the amino acids encoded by the WT and oschr726 sequences.
Apart from this, we subsequently performed OsCHR726 CRISPR mutations using the CRISPR/Cas9 gene-editing system (Ma et al., 2015b). The primer P1 and P2 were used to identify whether mutations were introduced in the targeted sites by PCR and sequencing. Three homozygous mutant lines were obtained from T2 generation containing different mutations in the first exon of OsCHR726 and named oschr726-1, oschr726-2, oschr726-3, respectively (Figure 8). oschr726-1 generates a frameshift mutation resulting from a 1 bp (A) deletion at position C302 bp of the coding sequence (CDS). oschr726-2 contains a 28-bp deletion at positions C300-327 bp, and oschr726-3 contains a 31 bp deletion at positions C278-308 bp. The protein sequences of OsCHR726 were altered and its function was knocked out in the three mutant lines. The plant architecture of oschr726 mature plants were similar with the wild-type. However, the seed setting of oschr726 was reduced. However, it is unknown whether OsCHR726 has functions in the regulation of male or female gametophyte.
To confirm the effects of disrupted OsCHR726 function on salt tolerance, the salt responses of OsCHR726 mutation were examined on a solid medium. After germination for 2 days on half MS medium, seeds were transferred to a medium containing 0, 100, and 150 mM NaCl. The oschr726-T and oschr726-1/2/3 seedlings did not differ from DJ and ZH11 when grown on the control medium, respectively (Figure 9A). However, the OsCHR726 mutations showed greater growth inhibition with salt than wild-type seedlings depending on NaCl concentration (Figure 9A). The shoot and root length in the mutant lines were all significantly lower than the relevant wild-type (Figures 9B,C), indicating that OsCHR726 might play a positive role in salt tolerance.
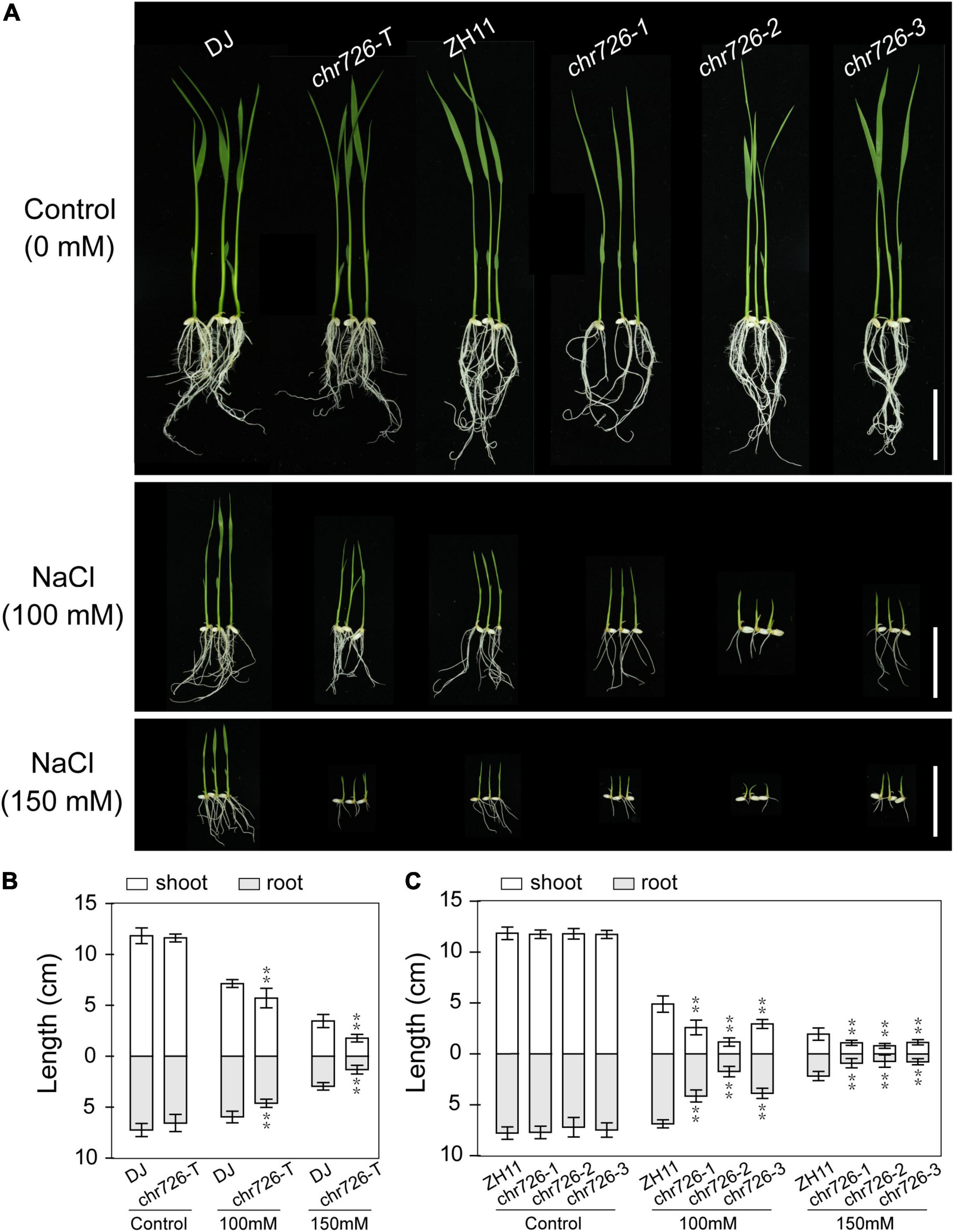
Figure 9. oschr726 is sensitive to salt stress treatment. (A) The performance of oschr726 seedlings compared with WT on media containing 0, 100, 150 mM NaCl. Bar = 5 cm. (B) The shoot and root lengths of 7-day-old seedlings were analyzed on WT (DJ) and T-DNA mutant. (C) The shoot and root lengths of 7-day-old seedlings were analyzed on WT (ZH11) and CRISPR/Cas9-mediated mutations. n = 20, Two asterisks (**p < 0.01, Student’s t-test) represent significant differences between the wild-type and mutants.
Discussion
Differential Expression Patterns of Snf2 Genes in Various Organs
In this study, we systematically analyzed the spatio-temporal expression patterns of Snf2 genes during reproductive development and stress treatments. Many genes displayed specific expression patterns at different developmental stages of various tissues. Most of the genes were highly expressed in YL, SAM, and OV (Figure 1). The loss-function of BRM reduced root length and plant height (Farrona et al., 2004), curly leaves (Hurtado et al., 2006), early flowering (Farrona et al., 2011) in Arabidopsis. OsCHR707, the homologous gene of BRM, was preferentially expressed in YL, P1, P2, and Ov5-6 (Figures 1A, 3C), suggesting that OsCHR707 might participate in leaf or reproductive development. The deficient function of AtCHR4 plants displayed faster leaf production in later rosette growth stages, more cauline leaves, and larger shoot apical meristems, and AtCHR4 played a positive regulator of floral transition (Sang et al., 2020). Similarly, OsCHR729, a homologous gene of AtCHR4, was predominantly expressed in YL, SAM, An5-6 (Figures 1A, 2B), and mutation of OsCHR729 exhibited rolled leaves, repressed stem elongation, and reduced chlorophyll contents (Hu et al., 2012). Apart from these, OsCHR729 regulated seedling and root development via gibberellin and auxin-related signaling pathways, respectively (Ma et al., 2015a; Wang et al., 2016). OsCHR721, a nuclear protein belonging to the SMARCAL1 subfamily in Snf2, was predominantly expressed in SAM, P1, P2, An5-6, and Ov5-6 (Figures 1A, 2B, 3B). OsCHR721 interacts with OsRPA1a (replication protein A), which is involved in DNA repair, and plays a crucial role in both male and female reproductive development (Zhang et al., 2020b). The above results suggest that the expression patterns of genes are coordinated and closely related with their function. OsCHR722, OsCHR742, OsCHR735, and OsCHR746 were specifically expressed in SAM, and OsCHR715, OsCHR703, OsCHR717 were specifically expressed in Sti, S2 and S3, respectively (Figures 1A,B). Interestingly, OsCHR709, OsCHR711, OsCHR720, OsCHR730, OsCHR735, OsCHR739, OsCHR742, OsCHR743, OsCHR745, and OsCHR746 were specifically expressed in the early development of pistil (Ov1). In general, the relative expression profiles of Snf2 genes extent our understanding in vegetative and reproductive development.
Expression Regulation of Snf2 Genes Under Biotic and Abiotic Stresses
Increasing evidence have demonstrated that some Snf2 genes are induced or inhibited expression by abiotic stress (Hu et al., 2013), and OsALT1 (Alkaline Tolerance 1, OsCHR706) improves tolerance to alkaline stress (Guo et al., 2014). We all know that high salinity and drought are two major stress factors that can seriously affect plant growth, development, and crop yield. When plants were exposed to environmental stresses, various physiological and biochemical responses were induced, and gene expression programs rapidly adapted to adverse situations. To investigate the expression pattern of Snf2 genes under biotic and abiotic stresses, we predicted the cis-acting elements of hormone-responsive and environmental stress-related in the promoter regions. We performed qRT-PCR analysis of M. oryzae infection, salt, and drought stress at different experimental timelines.
The most elements in Snf2 putative promoter were dehydration-responsive element, MeJA-responsive element, stress-responsive element, MYB-binding site for drought element, and ABA-responsive element. Interestingly, there were 15 stress-responsive elements in the promoter of OsCHR725 (Supplementary Figure 3), which was significantly upregulated at the 12 hpi of M. oryzae (Figure 6 and Supplementary Figure 4) and all treatment timelines of salt stress (Figure 7A and Supplementary Figure 5A). Previous studies have shown that OsCHR725 (OsBRHIS1) plays a critical role in the SA-independent disease resistance (Li et al., 2015), suggesting that the cis-acting elements in the promoter are coordinated with the gene expression and function. AtCHR19 acts as a transcriptional repressor and contributes to plant pathogen resistance (Kang et al., 2022). OsCHR714, the homologous gene of AtCHR19, was significantly upregulated at the 12 hpi of M. oryzae and all treatment timelines of salt and drought stress. This implied that this gene probably possesses similar characteristic functions on stress tolerance. A total of 20 genes were upregulated at 12 hpi but downregulated at subsequent time points under M. oryzae infection, indicating that these genes were possibly required for initial stress responses. OsCHR726 was upregulated under salt stress during the treatment timelines, and OsCHR726 loss-of-function mutants were hypersensitive to salt stress. Taken together, the expression profiles of Snf2 under biotic and abiotic stresses will provide a valuable resource for investigating stress tolerance in rice.
Chromatin Remodeling Factors With Epigenetic Regulation
In this study, the subcellular localizations of the representative Snf2 proteins were mainly in the nucleus (Figure 4). Then Snf2 family proteins, as CHRs, regulate diverse aspects of DNA events such as replication, transcription, homologous recombination, and DNA repair (Hu et al., 2013). Chromatin structure plays a significant platform in regulating gene expression, whose mechanisms are associated with epigenetic regulation: DNA methylation, histone modifications and variants, and chromatin remodeling (Knizewski et al., 2008; Clapier and Cairns, 2009). The epigenetic mechanisms in plant stress response have been discussed in multiple reviews (Chinnusamy and Zhu, 2009; Asensi-Fabado et al., 2017; Chang et al., 2020; Song et al., 2021). However, the function of chromatin remodeling in plant stress responses has not been in-depth analyzed recently.
DRD1, a Rad 54-like family member, and DDM1, a member of Snf2-like protein, are involved in DNA methylation (Kanno et al., 2004). Functional deficiency of DDM1 leads to promoting a decrease of methylation from some repeats and solid transcriptional activation of TEs (Lippman et al., 2004). OsDDM1 genes (OsDDM1a/OsCHR746 and OsDDM1b/OsCHR741) are essential for CHG and CG methylation in heterochromatic regions and CHH methylation in euchromatic regions (Tan et al., 2016). In addition, OsCHR729 recognizes and modulates the H3K4me3 and H3K27me3 levels to control target genes involved in plant development (Hu et al., 2012). OsCHR725 (OsBRHIS1), the ortholog to AtCHR28, can repress innate immunity for the reason of binding to specific histone H2A and H2B variants near the promoter regions of defense genes (Li et al., 2015). OsCHR726 is hypersensitive to salt stress, but it is still unknown which epigenetic mechanism is involved. Current research on CHRs is mainly performed in the model plant Arabidopsis and a few crop species. We believe that more studies will be conducted to analyze the stress adaptation of CHRs in crops. Here, we provide systematical expression profiles of Snf2 family genes during the reproductive development and stress response. Our data suggest that Snf2 family genes might play a significant role in plant development and stress responses and be valuable for developing new plant varieties with enhanced stress tolerance.
Accession Number
The cDNA and genomic DNA sequence data of OsCHR726 can be found in the Rice Genome Annotation Project data libraries under accession number LOC_Os07g40730.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YQ and HZ conceived the initial screening and research plans. HZ designed the experiments. MG, ZH, WZ, ZS, CS, and MY performed most of the experiments. WZ, CS, MY, and DT extracted the RNA. MM, MG, and WZ performed the qRT-PCR and analyzed the data. ZS performed the heatmap. ZH performed the CRISPR mutations. YQ, HZ, and MG wrote the manuscript with the contributions of all the authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Fujian Natural Science Foundation (2019J01425), the Science and Technology Program of Fujian Province (2019N5008), Scientific Research Foundation of Graduate School of Fujian Agriculture and Forestry University (324-1122yb069), College Students’ Innovative Entrepreneurial Training Plan Program (201510389081, 201610389062, and 201910389010), the National Natural Science Foundation of China (31970333), the Talent Introduction Start-up Fund Project of Anhui Science and Technology University (NXYJ202001), Science and technology innovation project of Pingtan Science and Technology Research Institute (PT2021007 and PT2021003), and a Guangxi Distinguished Experts Fellowship awarded to YQ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.910663/full#supplementary-material
Supplementary Figure 1 | The relative expression level of Snf2 gene family in the stamen. (A) Preferentially expressed Snf2 genes at the An1, An5, and An6 stages. (B) Preferentially expressed Snf2 genes at the An4∼An6 stages. (C) The irregularly expressed Snf2 genes in the different developmental stages of stamen. An1-5, the stamen of 2–3, 3–4, 4–5, 5–6, and 6–7 mm spikelet, respectively; An6, the stamen in the spikelet before flowering. The y-axis is the relative expression level of the gene compared to OsUBQ5 in different developmental stages of stamen using the qRT-PCR. Error bars indicate standard deviations of independent biological replicates (n = 3). Different letters denote significant difference at P < 0.05 according to ANOVA in combination with Duncan’s multiple range test.
Supplementary Figure 2 | The relative expression level of Snf2 gene family in the ovary. (A) The highly expressed Snf2 genes at the Ov1 and Ov4∼Ov6 stages. (B) The highly expressed Snf2 genes at the Ov4 stage. (C) The irregularly expressed Snf2 genes in all developmental stages of the ovary. Ov1-5, the ovary of 2–3, 3–4, 4–5, 5–6, and 6–7 mm spikelet, respectively; Ov6, the ovary of the spikelet before flowering. The y-axis is relative expression level of the gene compared to OsUBQ5 in different developmental stages of ovary using the qRT-PCR. Error bars indicate standard deviations of independent biological replicates (n = 3). Different letters denote significant difference at P < 0.05 according to ANOVA in combination with Duncan’s multiple range test.
Supplementary Figure 3 | Prediction of cis-acting elements of environmental stress-related and hormone-responsive in Snf2 genes. (A) Numbers of cis-acting elements detected in the promoter regions (sequence retrieved from about ≤ 2 kb upstream region). The cis-acting elements were divided into 11 types. (B) Quantity, kind and position of environmental stress-related and hormone-responsive elements in the putative promoters of genes in (A). (C) The qRT-PCR analysis of Snf2 genes under IAA treatment. Error bars indicate standard deviations of independent biological replicates (n = 3). Data are given as means ± SD. *p < 0.05, compared with unstressed samples using Student’s t-test.
Supplementary Figure 4 | Statistical analysis of Snf2 genes under biotic stress. The percentage of genes up- and down-regulated (at least twofold and significant difference) in 12 hpi than the unstressed samples, and others under the infection of Magnaporthe oryzae (M. oryzae) is represented in the pie chart. The corresponding number and the list of genes is mentioned in the figure. hpi, hours post-inoculation
Supplementary Figure 5 | Statistical analysis of Snf2 genes under abiotic stress. Venn diagrams using jvenn tools (http://jvenn.toulouse.inra.fr/app/usermanual.html) showing that the number of genes up-regulated (A,C) and down-regulated (B,D) were at least twofold and significant difference than the unstressed samples in different time points under salt and drought treatment. The corresponding number and the list of genes under each time point and in combination are mentioned in the figure.
Supplementary Figure 6 | Up- and down-regulated genes under both salt and drought stress. (A) Up-regulation genes under both salt and drought stress. (B) Down-regulation genes under both salt and drought stress.
Supplementary Figure 7 | Genotype identification of oschr726 T-DNA mutant. (A) Genotyping of T-DNA insertion plants. (B) Identifying the transcription level of OsCHR726 in the T-DNA insertion plants.
Footnotes
References
Asensi-Fabado, M. A., Amtmann, A., and Perrella, G. (2017). Plant responses to abiotic stress: the chromatin context of transcriptional regulation. Biochimica. Biophys. Acta-Gene Regul. Mech. 1860, 106–122. doi: 10.1016/j.bbagrm.2016.07.015
Cai, H. Y., Zhang, M., Chai, M. N., He, Q., Huang, X. Y., Zhao, L. H., et al. (2019). Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. New Phytol. 221, 295–308. doi: 10.1111/nph.15306
Chang, Y. N., Zhu, C., Jiang, J., Zhang, H. M., Zhu, J. K., and Duan, C. G. (2020). Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 62, 563–580. doi: 10.1111/jipb.12901
Chinnusamy, V., and Zhu, J. K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139.
Cho, S. H., Lee, C. H., Gi, E., Yim, Y., Koh, H. J., Kang, K., et al. (2018). The rice rolled fine striped (RFS) CHD3/Mi-2 chromatin remodeling factor epigenetically regulates genes involved in oxidative stress responses during leaf development. Front. Plant Sci. 9:364. doi: 10.3389/fpls.2018.00364
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Farrona, S., Hurtado, L., Bowman, J. L., and Reyes, J. C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131, 4965–4975. doi: 10.1242/dev.01363
Farrona, S., Hurtado, L., March-Diaz, R., Schmitz, R. J., Florencio, F. J., Turck, F., et al. (2011). Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLoS One 6:e17997. doi: 10.1371/journal.pone.0017997
Flaus, A., Martin, D. M. A., Barton, G. J., and Owen-Hughes, T. (2006). Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905. doi: 10.1093/nar/gkl295
Guo, M. X., Wang, R. C., Wang, J., Hua, K., Wang, Y. M., Liu, X. Q., et al. (2014). ALT1, a Snf2 Family Chromatin Remodeling ATPase, Negatively Regulates Alkaline Tolerance through Enhanced Defense against Oxidative Stress in Rice. PLoS One 9:e112515. doi: 10.1371/journal.pone.0112515
Han, S. K., Sang, Y., Rodrigues, A., Wu, M. F., Rodriguez, P. L., Wagner, D., et al. (2012). The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24, 4892–4906. doi: 10.1105/tpc.112.105114
Han, S. K., Wu, M. F., Cui, S. J., and Wagner, D. (2015). Roles and activities of chromatin remodeling ATPases in plants. Plant J. 83, 62–77. doi: 10.1111/tpj.12877
Han, X., Xi, Y., Zhang, Z., Mohammadi, M. A., Joshi, J., Borza, T., et al. (2021). Effects of phosphite as a plant biostimulant on metabolism and stress response for better plant performance in Solanum tuberosum. Ecotoxicol. Environ. Saf. 210:111873. doi: 10.1016/j.ecoenv.2020.111873
Hara, T., Katoh, H., Ogawa, D., Kagaya, Y., Sato, Y., Kitano, H., et al. (2015). Rice SNF2 family helicase ENL1 is essential for syncytial endosperm development. Plant J. 81, 1–12. doi: 10.1111/tpj.12705
Higo, H., Tahir, M., Takashima, K., Miura, A., Watanabe, K., Tagiri, A., et al. (2012). DDM1 (Decrease in DNA Methylation) genes in rice (Oryza sativa). Mol. Genet. Genom. 287, 785–792. doi: 10.1007/s00438-012-0717-5
Hu, Y. F., Liu, D. N., Zhong, X. C., Zhang, C. J., Zhang, Q. F., and Zhou, D. X. (2012). CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. U.S.A. 109, 5773–5778. doi: 10.1073/pnas.1203148109
Hu, Y. F., Zhu, N., Wang, X. M., Yi, Q. P., Zhu, D. Y., Lai, Y., et al. (2013). Analysis of rice Snf2 family proteins and their potential roles in epigenetic regulation. Plant Physiol. Biochem. 70, 33–42. doi: 10.1016/j.plaphy.2013.05.001
Hurtado, L., Farrona, S., and Reyes, J. C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62, 291–304. doi: 10.1007/s11103-006-9021-2
Jain, M., Nijhawan, A., Tyagi, A. K., and Khurana, J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. doi: 10.1016/j.bbrc.2006.04.140
Kang, H., Liu, Y., Fan, T., Ma, J., Wu, D., Heitz, T., et al. (2022). Arabidopsis CHROMATIN REMODELING 19 acts as a transcriptional repressor and contributes to plant pathogen resistance. Plant Cell 34, 1100–1116. doi: 10.1093/plcell/koab318
Kanno, T., Mette, M. F., Kreil, D. P., Aufsatz, W., Matzke, M., and Matzke, A. J. M. (2004). Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14, 801–805. doi: 10.1016/j.cub.2004.04.037
Kim, J. M., To, T. K., Nishioka, T., and Seki, M. (2010). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33, 604–611. doi: 10.1111/j.1365-3040.2009.02076.x
Knizewski, L., Ginalski, K., and Jerzmanowski, A. (2008). Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 13, 557–565. doi: 10.1016/j.tplants.2008.08.004
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, B., Carey, M., and Workman, J. L. (2007). The role of chromatin during transcription. Cell 128, 707–719.
Li, X. Y., Jiang, Y. X., Ji, Z. C., Liu, Y. G., and Zhang, Q. Y. (2015). BRHIS1 suppresses rice innate immunity through binding to monoubiquitinated H2A and H2B variants. EMBO Rep. 16, 1192–1202. doi: 10.15252/embr.201440000
Lippman, Z., Gendrel, A. V., Black, M., Vaughn, M. W., Dedhia, N., Mccombie, W. R., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476. doi: 10.1038/nature02651
Ma, H. L., Zhao, H. M., Liu, Z., and Zhao, J. (2011). The phytocyanin gene family in rice (Oryza sativa L.): genome-wide identification, classification and transcriptional analysis. PLoS One 6:e25184. doi: 10.1371/journal.pone.0025184
Ma, X. D., Ma, J., Zhai, H. H., Xin, P. Y., Chu, J. F., Qiao, Y. L., et al. (2015a). CHR729 Is a CHD3 Protein That Controls Seedling Development in Rice. PLoS One 10:e0138934. doi: 10.1371/journal.pone.0138934
Ma, X. L., Zhang, Q. Y., Zhu, Q. L., Liu, W., Chen, Y., Qiu, R., et al. (2015b). A Robust CRISPR/Cas9 System for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Mlynarova, L., Nap, J. P., and Bisseling, T. (2007). The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J. 51, 874–885. doi: 10.1111/j.1365-313X.2007.03185.x
Mohammadi, M. A., Cheng, Y., Aslam, M., Jakada, B. H., Wai, M. H., Ye, K., et al. (2021). ROS and oxidative response systems in plants under biotic and abiotic stresses: revisiting the crucial role of phosphite triggered plants defense response. Front. Microbiol. 12:631318. doi: 10.3389/fmicb.2021.631318
Mohammadi, M. A., Han, X., Zhang, Z., Xi, Y., Boorboori, M., and Wang-Pruski, G. (2020). Phosphite application alleviates Pythophthora infestans by modulation of photosynthetic and physio-biochemical metabolites in potato leaves. Pathogens 9:170. doi: 10.3390/pathogens9030170
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. doi: 10.1046/j.1365-313x.2003.01708.x
Ojolo, S. P., Cao, S. J., Priyadarshani, S. V. G. N., Li, W. M., Yan, M. K., Aslam, M., et al. (2018). Regulation of plant growth and development: a review from a chromatin remodeling perspective. Front. Plant Sci. 9:1232. doi: 10.3389/fpls.2018.01232
Sang, Q., Pajoro, A., Sun, H., Song, B., Yang, X., Stolze, S. C., et al. (2020). Mutagenesis of a quintuple mutant impaired in environmental responses reveals roles for CHROMATIN REMODELING4 in the Arabidopsis Floral Transition. Plant Cell 32, 1479–1500. doi: 10.1105/tpc.19.00992
Sarnowska, E., Gratkowska, D. M., Sacharowski, S. P., Cwiek, P., Tohge, T., Fernie, A. R., et al. (2016). The Role of SWI/SNF Chromatin Remodeling Complexes in Hormone Crosstalk. Trends Plant Sci. 21, 594–608. doi: 10.1016/j.tplants.2016.01.017
Singh, S., Singh, A., Singh, A., Yadav, S., Bajaj, I., Kumar, S., et al. (2020). Role of chromatin modification and remodeling in stem cell regulation and meristem maintenance in Arabidopsis. J. Exp. Bot. 71, 778–792. doi: 10.1093/jxb/erz459
Song, Z. T., Liu, J. X., and Han, J. J. (2021). Chromatin remodeling factors regulate environmental stress responses in plants. J. Integr. Plant Biol. 63, 438–450. doi: 10.1111/jipb.13064
Su, Z. X., Wang, L. L., Li, W. M., Zhao, L. H., Huang, X. Y., Azam, S. M., et al. (2017). Genome-Wide Identification of Auxin Response Factor (ARF) genes family and its tissue-specific prominent expression in pineapple (Ananas comosus). Trop. Plant Biol. 10, 86–96.
Tan, F., Zhou, C., Zhou, Q. W., Zhou, S. L., Yang, W. J., Zhao, Y., et al. (2016). Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiology 171, 2041–2054. doi: 10.1104/pp.16.00393
Tran, L. S. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Wang, Y. H., Wang, D., Gan, T., Liu, L. L., Long, W. H., Wang, Y. L., et al. (2016). CRL6, a member of the CHD protein family, is required for crown root development in rice. Plant Physiol. Biochem. 105, 185–194. doi: 10.1016/j.plaphy.2016.04.022
Xu, W. R., Yu, Y. H., Zhou, Q., Ding, J. H., Dai, L. M., Xie, X. Q., et al. (2011). Expression pattern, genomic structure, and promoter analysis of the gene encoding stilbene synthase from Chinese wild Vitis pseudoreticulata. J. Exp. Bot. 62, 2745–2761. doi: 10.1093/jxb/erq447
Yang, R., Hong, Y. C., Ren, Z. Z., Tang, K., Zhang, H., Zhu, J. K., et al. (2019). A Role for PICKLE in the regulation of cold and salt stress tolerance in Arabidopsis. Front. Plant Sci. 10:900. doi: 10.3389/fpls.2019.00900
Yin, X. J., Huang, L., Zhang, X. M., Guo, C. L., Wang, H., Li, Z., et al. (2017). Expression patterns and promoter characteristics of the Vitis quinquangularis VqSTS36 gene involved in abiotic and biotic stress response. Protoplasma 254, 2247–2261. doi: 10.1007/s00709-017-1116-x
Yoshida, S., Forno, D. A., Cock, J. H., and Gomez, K. A. (1971). Laboratory Manual for Physiological Studies of Rice. Los Baño: IRRI.
Yu, X. M., Meng, X. C., Liu, Y. T., Li, N., Zhang, A., Wang, T. J., et al. (2018). The chromatin remodeler ZmCHB101 impacts expression of osmotic stress-responsive genes in maize. Plant Mol. Biol. 97, 451–465. doi: 10.1007/s11103-018-0751-8
Zha, P., Jing, Y. J., Xu, G., and Lin, R. C. (2017). PICKLE chromatin-remodeling factor controls thermosensory hypocotyl growth of Arabidopsis. Plant Cell Environ. 40, 2426–2436. doi: 10.1111/pce.13049
Zhang, D. D., Gao, S. J., Yang, P., Yang, J., Yang, S. G., and Wu, K. Q. (2019). Identification and Expression Analysis of Snf2 Family Proteins in Tomato (Solanum lycopersicum). Int. J. Genom. 2019:5080935. doi: 10.1155/2019/5080935
Zhang, M., Liu, Y. H., Cai, H. Y., Guo, M. L., Chai, M. N., She, Z. Y., et al. (2020a). The bZIP Transcription Factor GmbZIP15 Negatively Regulates Salt- and Drought-Stress Responses in Soybean. Int. J. Mol. Sci. 21:7778. doi: 10.3390/ijms21207778
Zhang, Y. S., Chen, Q., Zhu, G. L., Zhang, F., Fang, X. H., Ren, H. B., et al. (2020b). CHR721, interacting with OsRPA1a, is essential for both male and female reproductive development in rice. Plant Mol. Biol. 103, 473–487. doi: 10.1007/s11103-020-01004-z
Zhao, H. M., Ma, H. L., Yu, L., Wang, X., and Zhao, J. (2012). Genome-Wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS One 7:e49210. doi: 10.1371/journal.pone.0049210
Zhao, H. M., Ma, T. F., Wang, X., Deng, Y. T., Ma, H. L., Zhang, R. S., et al. (2015). OsAUX1 controls lateral root initiation in rice (Oryza sativa L.). Plant Cell Environ. 38, 2208–2222. doi: 10.1111/pce.12467
Zhao, H. M., Maokai, Y., Cheng, H., Guo, M. L., Liu, Y. H., Wang, L. L., et al. (2021). Characterization of auxin transporter AUX, PIN and PILS gene families in pineapple and evaluation of expression profiles during reproductive development and under abiotic stresses. Peerj 9:e11410. doi: 10.7717/peerj.11410
Keywords: rice (Oryza sativa L.), Snf2 family, biotic and abiotic stress, OsCHR726, salinity stress
Citation: Guo M, Zhao H, He Z, Zhang W, She Z, Mohammadi MA, Shi C, Yan M, Tian D and Qin Y (2022) Comparative Expression Profiling of Snf2 Family Genes During Reproductive Development and Stress Responses in Rice. Front. Plant Sci. 13:910663. doi: 10.3389/fpls.2022.910663
Received: 01 April 2022; Accepted: 04 May 2022;
Published: 31 May 2022.
Edited by:
Qing-Yong Yang, Huazhong Agricultural University, ChinaReviewed by:
Xin He, Hunan Agricultural University, ChinaChuan Xia, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2022 Guo, Zhao, He, Zhang, She, Mohammadi, Shi, Yan, Tian and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heming Zhao, zhaohm@ahstu.edu.cn; Yuan Qin, yuanqin001@foxmail.com
 Mingliang Guo1
Mingliang Guo1 Zhimei He
Zhimei He Wenchao Zhang
Wenchao Zhang Zeyuan She
Zeyuan She Mohammad Aqa Mohammadi
Mohammad Aqa Mohammadi Maokai Yan
Maokai Yan Dagang Tian
Dagang Tian Yuan Qin
Yuan Qin