Bone Mesenchymal Stem Cells Promote Extracellular Matrix Remodeling of Degenerated Nucleus Pulposus Cells via the miR-101-3p/EIF4G2 Axis
- 1Department of Orthopedics, The Second Affiliated Hospital of Shandong First Medical University, Tai’an, China
- 2Department of Out-Patient, Tai’an Central Hospital Branch, Tai’an, China
The etiology of lumbocrural pain is tightly concerned with intervertebral disk degeneration (IDD). Bone mesenchymal stem cell (BMSC)-based therapy bears potentials for IDD treatment. The properties of microRNA (miRNA)-modified BMSCs may be altered. This study investigated the role and mechanism of BMSCs promoting extracellular matrix (ECM) remodeling of degenerated nucleus pulposus cells (NPCs) via the miR-101-3p/EIF4G2 axis. NPCs were collected from patients with IDD and lumbar vertebral fracture (LVF). The expressions of miR-101-3p and ECM-related proteins, Collagen-I (Col-I) and Collagen-II (Col-II), were detected using the reverse transcription-quantitative polymerase chain reaction. The expressions of Col-I and Col-II, major non-collagenous component Aggrecan, and major catabolic factor Matrix metalloproteinase-13 (MMP-13) were detected using Western blotting. BMSCs were cocultured with degenerated NPCs from patients with IDD. Viability and apoptosis of NPCs were measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay and flow cytometry. After the degenerated NPCs were transfected with the miR-101-3p inhibitor, the expressions of ECM-related proteins, cell viability, and apoptosis were detected. The targeting relationship between miR-101-3p and EIF4G2 was verified. Functional rescue experiments verified the effects of miR-101-3p and EIF4G2 on ECM remodeling of NPCs. Compared with the NPCs of patients with LVF, the degenerated NPCs of patients with IDD showed downregulated miR-101-3p, Col-II, and Aggrecan expressions and upregulated MMP-13 and Col-I expressions. BMSCs increased the expressions of miR-101-3p, Aggrecan, and Col-II, and decreased the expressions of MMP-13 and Col-I in degenerated NPCs. BMSCs enhanced NPC viability and repressed apoptosis. Downregulation of miR-101-3p suppressed the promoting effect of BMSCs on ECM remodeling. miR-101-3p targeted EIF4G2. Downregulation of EIF4G2 reversed the inhibiting effect of the miR-101-3p inhibitor on ECM remodeling. In conclusion, BMSCs increased the miR-101-3p expression in degenerated NPCs to target EIF4G2, thus promoting the ECM remodeling of NPCs.
Introduction
Approximately 84% of the population worldwide experiences low back pain in their lifetime, of which 10% develops into chronic disability, thus gravely affecting the quality of life and bringing huge financial burdens to families and society (Gore et al., 2012). Intervertebral disk degeneration (IDD) represents an extensively identified contributor to low back pain (Risbud and Shapiro, 2014). The intervertebral disk (IVD) is a complicated structure consisting of three anatomical substructures, namely, nucleus pulposus (NP), annulus fibrosus (AF), and cartilaginous endplates, which are essential to maintain the normal function of a disk (Qu et al., 2018). IDD is featured by the reduction of NP and degradation of proteoglycan, Aggrecan, and collagen in the extracellular matrix (ECM), which disrupts the homeostasis of NP and shifts IVD maintenance toward degeneration and catabolism (Wang et al., 2015). The current clinical interventions for IDD mainly include conservative medication and surgery (spinal fusion or total disk replacement); nevertheless, these therapeutic approaches only temporarily relieve the pain symptoms, instead of providing a permanent cure (Konovalov et al., 2016; Zhu et al., 2019). Hence, developing novel and potent therapies for patients with IDD remains an urgent issue to be solved.
Recently, biological therapeutic approaches such as stem cell transplantation for reversing IDD progression have aroused wide interest (Cai et al., 2019). Bone mesenchymal stem cells (BMSCs) have been extensively applied in transplantation research due to their considerable advantages such as high activity, low immunogenicity, and strong differentiation potential (Zhu et al., 2017; Gan et al., 2018). BMSC transplantation possesses the ability to facilitate IVD regeneration in the rabbit and pig models (Cai et al., 2015; Barczewska et al., 2018). BMSCs can enhance the synthesis of remaining ECM in NP and AF tissues and repress the expression of inflammatory mediators and the activity of matrix-degrading enzymes, thus maintaining the IVD structure (Hu et al., 2017). After BMSCs injection, the proteoglycan gene expression is upregulated, and ECM is gradually accumulated in NP (Yang et al., 2009). At present, the mechanism of BMSCs promoting ECM remodeling in IDD needs further exploration.
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules consisting of 18–23 nucleotides, which exert influences on posttranscriptional modulation of gene expression (Bartel, 2009; Shioya et al., 2010). Aberrant miRNA expression is implicated in the pathological processes of IDD, including NP cell (NPC) apoptosis, ECM degradation, and inflammatory response (Wang et al., 2015). The matured miR-101-3p is generated from miR-101-1 and miR-101-2 (the precursor miR) (Li et al., 2017b). miR-101-3p commonly acts as a tumor suppressor in human malignancies such as breast cancer (Li et al., 2017b) and hepatocellular carcinoma (Li et al., 2017a). Gao Z.X. et al. (2020) have revealed that miR-101-3p expression in degenerated NPCs is significantly lower than that in normal cells, and SNHG6 can suppress proliferation and promote apoptosis of NPCs by downregulating miR-101-3p expression and thus promote the occurrence of IDD. Whether miR-101-3p is implicated in the effect of BMSCs on ECM remodeling in IDD remains unknown. This study investigated the role and mechanism of BMSCs promoting ECM remodeling of degenerated NPCs via the miR-101-3p/EIF4G2 axis, which shall shed new insight into BMSC transplantation in the treatment of IDD.
Materials and Methods
Ethics Statement
This study was approved by the Ethical Committee of Affiliated Hospital of Shandong First Medical University (No. 2016-161). Informed consent was signed by each eligible participant.
Nucleus Pulposus Tissue Collection
The intervertebral disk tissue samples were collected from 14 patients with IDD-related low back pain (6 females and 8 males) who underwent surgery in Affiliated Hospital of Shandong First Medical University from March 2016 to June 2019 and 6 patients with lumbar vertebral fracture (LVF) without a history of low back pain (3 females and 3 males). Based on the Pfirrmann classification system, preoperative magnetic resonance imaging was performed for grading (Pfirrmann et al., 2001). The IDD samples were graded IV and V, and LVF samples were graded I and II.
Cell Culture
According to the previous study, NPCs were isolated and cultured (Gao Z.X. et al., 2020; Zhang et al., 2020). NP tissues were isolated from IVD tissues under the stereoscopic microscope. After washing with phosphate-buffered saline (PBS), NP tissues were sliced into small pieces and detached with 0.5% type II collagenase (Roche Diagnostics, Indianapolis, Indiana, United States) for 2 h. The tissue digestive solution was filtered through a cell filter, and the filtrate was collected and centrifuged at 100 g for 5 min. The supernatant was removed, and the precipitate was added into the F12 basic medium containing 10% of fetal bovine serum (FBS) and 100 U/ml of penicillin–streptomycin. The obtained cell suspension was transferred into the culture bottle with a density of about 1 × 105/ml in a humid incubator at 37°C with 5% CO2. On days 5–7, when the cells completely adhered to the wall, the culture medium was changed for the first time and then changed every 2–3 days. After more than 80% of the cells were covered in the bottom of the flask, the cells were detached with 0.25% of trypsin and passaged at a ratio of 1:2. The cells of the third generation were used in this study. Protein and mRNA levels of MMP-13, Collagen-I (Col-I), Collagen-II (Col-II), and Aggrecan, the specific indicators for degenerative and normal NPCs, were analyzed.
Bone mesenchymal stem cells (BMSCs) and 293T cells were purchased from American Type Culture Collection (Manassas, VA, United States) and cultured in Dulbecco’s modified Eagle’s medium containing 10% of FBS and 100 U/ml of penicillin-streptomycin at 37°C with 5% of CO2. NPCs and BMSCs at passage 5 were used for subsequent experiments.
Cell Transfection
The miR-101-3p inhibitor, miR-101-3p mimic, and their corresponding negative control (NC) were purchased from RiboBio (Guangzhou, China); EIF4G2-siRNA and its corresponding NC were purchased from Genechem Co., Ltd. (Shanghai, China). NPCs in the logarithmic phase were seeded into the 6-well plates (2 × 105 cells/well) and cultured for 24 h. Then, cell transfection was performed using Lipofectamine 2000 (siRNA 10–100 nM and miRNA mimic 50 nM). The subsequent experiments were conducted after 48 h.
Coculture of NPCs and BMSCs
A 6-well Transwell system (0.4 μm pore size membrane; Corning, NY, United States) was used to evaluate the effect of BMSCs on NPCs. NPCs (1 × 105) were seeded into the basolateral chamber of Transwell plates, and BMSCs (1 × 105) were spread on the apical chamber. After 24 h, NPCs were harvested for further experiments. Specifically, NPC suspension in the basolateral chamber of a Transwell plate of the coculture system was collected into the centrifuge tube. The adherent cells were washed with PBS once, and then the cells were detached with appropriate amounts of trypsin solution. The adherent cells were incubated at room temperature for 5 min until they can be gently blown off. Then, the trypsin solution was sucked out, resuspended in PBS, and recovered to the cell suspension collected in the previous step. After centrifugation at 1,000 g for 5 min, the supernatant was discarded and the cells were collected. The cells were gently suspended with PBS and counted.
Western Blotting
After the coculture of NPCs and BMSCs, NPCs (1 × 107) were collected for the Western blotting analysis. The protein was extracted using the cell lysis buffer (Beyotime, Shanghai, China), and the bicinchoninic acid protein assay kit (Beyotime) was used for protein quantification. The protein was separated by 10% of SDS-PAGE (Beyotime) and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, United States). The membranes were blocked with PBS buffer containing 5% of skimmed milk for 2 h and then incubated with the primary antibodies at 4°C overnight. Later, the membranes were cultured with the secondary antibody for 2 h. The protein bands were detected using the enhanced chemiluminescence kit (Thermo Fisher Scientific Inc., Waltham, MA, United States) and quantitatively analyzed using Image J software. The antibodies MMP-13 (1:1,000, ab39012), Col-I (1:1,000, ab34710), Col-II (1:500, ab34712), Aggrecan (1:1,000, ab36861), and GAPDH (1:5,000, ab18160) were purchased from Abcam (Cambridge, MA, United States).
Reverse Transcription-Quantitative Polymerase Chain Reaction
After the coculture of NPCs and BMSCs, NPCs (2 × 106) were collected using the reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The total RNA was isolated from cells using TRIzol reagent (Invitrogen Inc., Carlsbad, CA, United States), purified using DNase, and reverse transcribed into cDNA using reverse transcription kit (Takara Bio Inc., Kyoto, Japan). RT-qPCR was performed on ABI 7500 (ABI, Foster City, CA, United States) using SYBR Premix Ex Taq (Takara, Dalian, China). The relative expression of genes was calculated by using the 2–ΔΔCT method, with GAPDH and U6 as internal references. Primer sequences are illustrated in Table 1.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide Assay
The cells were seeded into 96-well plates (5,000 cells/well) and cultured for 48 h. Then, the cells were cultured with 20 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) at 37°C with 5% of CO2 for 4 h. Later, the culture medium was removed, and 150 μl of dimethyl sulfoxide was added. The plates were shaken in the dark for 10 min. The absorbance value at 490 nm was evaluated by the microplate reader (Bio-Rad Laboratories, Hercules, CA, United States).
Flow Cytometry
After the coculture of NPCs and BMSCs, NPC suspensions (2 × 105) were centrifuged at 200 g for 5 min, and the supernatant was removed. The cell precipitates were resuspended in 500 μl of the binding buffer using the Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit I (556547, Becton, Dickinson and Company, Franklin Lakes, NJ, United States). The samples were mixed with 5 μl of Annexin V-FITC and propidium iodide and cultured for 15 min. The cell apoptosis was evaluated by the flow cytometer (FC500 MCL, Beckman Coulter, Chaska, MN, United States).
Dual-Luciferase Reporter Gene Assay
The binding site of miR-101-3p and EIF4G2 was predicted by Starbase. The binding sequence and mutant sequence were cloned into the luciferase vector pGL3 (Pro-mega, Madison, WI, United States) to construct wild-type and mutant-type (EIF4G2-WT and EIF4G2-MUT) luciferase plasmids. The constructed plasmids were co-transfected with miR-101-3p mimic or mimic NC into 293T cells using Lipofectamine 2000. After 48 h, the relative luciferase activity was detected using the dual-lucy assay kit (Solarbio, Beijing, China).
Statistical Analysis
Data analysis was analyzed and introduced using SPSS 21.0 (IBM Corp., Armonk, NY, United States) and GraphPad Prism 8.0 (GraphPad Software Inc., San Diego, CA, United States). First, the test of normality and homogeneity of variance was carried out, which showed that the data were consistent with normal distribution and homogeneity of variance. Data are expressed as mean ± SD. The independent sample t-test was adopted for comparison between two groups. One-way ANOVA was employed for the comparisons among multiple groups, following the Tukey’s multiple comparison test. The p-value was obtained from a two-tailed test, and p < 0.001 denoted the very statistical significance.
Results
Extracellular Matrix Degradation of NPCs in Patients With IDD
The miR-101-3p expression is downregulated in NPCs of rats with IDD and associated with the levels of Col-I and Col-II (Gao Z.X. et al., 2020). To determine the function of miR-101-3p in IDD, we analyzed the expressions of miR-101-3p, Col-I, and Col-II in NPCs of 14 patients with IDD and 6 patients with LVF using the RT-qPCR. Compared with NPCs of patients with LVF, NPCs of patients with IDD showed downregulated miR-101-3p and Col-II expressions and upregulated Col-I expression (all p < 0.001; Figure 1A). Western blotting exhibited that NPCs of patients with IDD had upregulated MMP-13 and Col-I and downregulated Aggrecan and Col-II (p < 0.001; Figure 1B). These results suggested the involvement of miR-101-3p in IDD progression and the degradation of ECM of NPCs in patients with IDD.
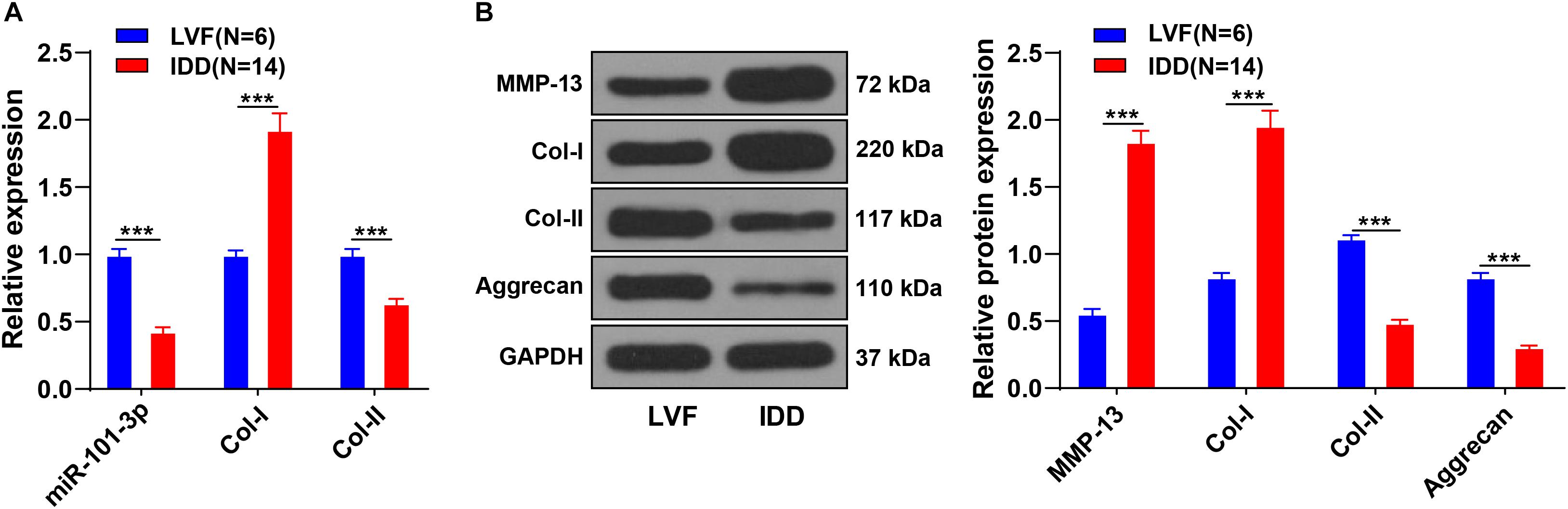
Figure 1. Extracellular matrix (ECM) degradation of nucleus pulposus cells (NPCs) in patients with intervertebral disk degeneration (IDD). NPCs were isolated from the intervertebral disk tissues of patients with IDD, with that of patients with LVF as control. (A) miR-101-3p, Collagen-I (Col-I), and Collagen-II (Col-II) expressions were detected using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and (B) MMP-13, Col-I/Col-II, and Aggrecan levels were detected using Western blotting. Data are presented as mean ± SD and analyzed using the independent sample t-test, ***p < 0.001.
Bone Mesenchymal Stem Cells Promoted ECM Remodeling of Degenerated NPCs
Bone mesenchymal stem cells (BMSCs) can ameliorate IDD by attenuating the degeneration of NPCs (Li X. et al., 2019). NPCs of one patient with IDD were extracted for the in vitro study. BMSCs were cocultured with NPCs of one patient with IDD, and then expressions of miR-101-3p, Col-I, and Col-II were measured using the RT-qPCR. Compared with NPCs, NPCs cocultured with BMSCs showed increased Col-II and miR-101-3p and decreased Col-I (all p < 0.001; Figure 2A). Western blotting exhibited that compared with the NPCs, NPCs cocultured with BMSCs had downregulated MMP-13 and Col-I and upregulated Aggrecan and Col-II levels (p < 0.001; Figure 2B). MTT assay and flow cytometry revealed that compared with the NPCs, the viability of NPCs cocultured with BMSCs was enhanced, and the apoptosis rate was repressed (p < 0.001; Figures 2C,D). Taken together, BMSCs enhanced the viability, suppressed apoptosis, and facilitated ECM remodeling of degenerated NPCs. miR-101-3p expression in degenerated NPCs was notably upregulated after coculture of BMSCs and NPCs, which indicated that miR-101-3p was implicated in the process of BMSCs, promoting ECM remodeling of degenerated NPCs.
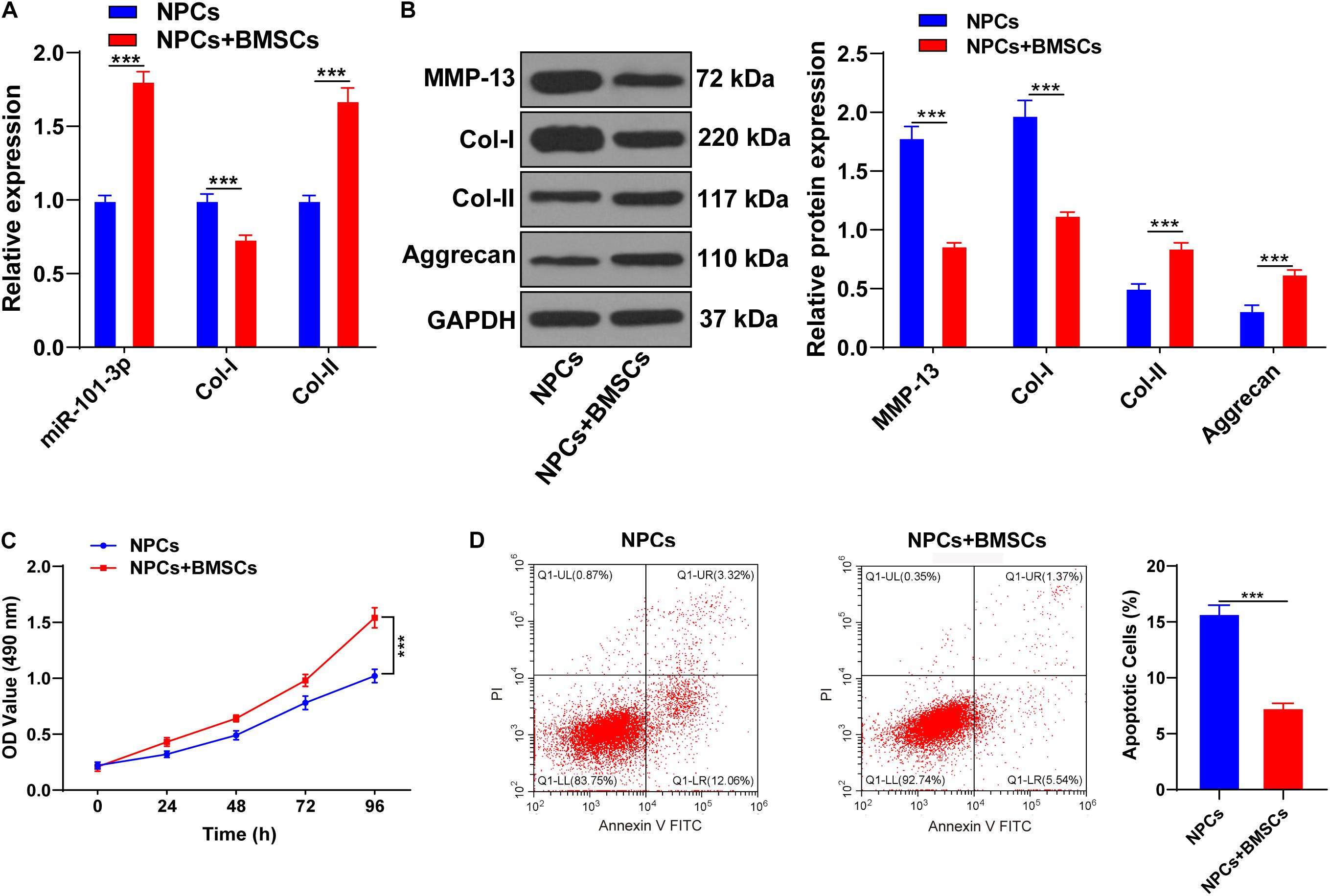
Figure 2. Bone mesenchymal stem cells (BMSCs) promoted ECM remodeling of NPCs. NPCs of patients with IDD were cocultured with BMSCs, with NPCs cultured alone as the control. (A) miR-101-3p, Col-I, and Col-II expressions were detected using the RT-qPCR; (B) MMP-13, Col-I, Col-II, and Aggrecan levels were detected using Western blotting; (C,D) the effects of BMSCs on the viability and apoptosis of NPCs were detected using MTT assay and flow cytometry. The cell experiments were repeated three times independently. Data are presented as mean ± SD. Data in panels (A–C) were analyzed using the independent sample t-test, ***p < 0.001.
Downregulation of miR-101-3p Inhibited the Effect of BMSCs on ECM Remodeling of Degenerated NPCs
To explore the potential mechanism of miR-101-3p in BMSC-mediated IDD repair, the miR-101-3p inhibitor was transfected into the degenerated NPCs cocultured with BMSCs, with NPCs cocultured with BMSCs (blank group) and inhibitor NC as the control. miR-101-3p expression in miR-101-3p inhibitor-transfected NPCs was downregulated notably (p < 0.001; Figure 3A). Compared with the inhibitor NC group, the miR-101-3p inhibitor group showed promoted MMP-13 and Col-I expressions and decreased Aggrecan and Col-II expressions (all p < 0.001; Figure 3B). Additionally, compared with the inhibitor NC group, the miR-101-3p inhibitor group showed reduced cell viability (p < 0.001; Figure 3C) and enhanced apoptosis (p < 0.001; Figure 3D). In brief, the downregulation of miR-101-3p inhibited the effect of BMSCs on ECM remodeling of degenerated NPCs.
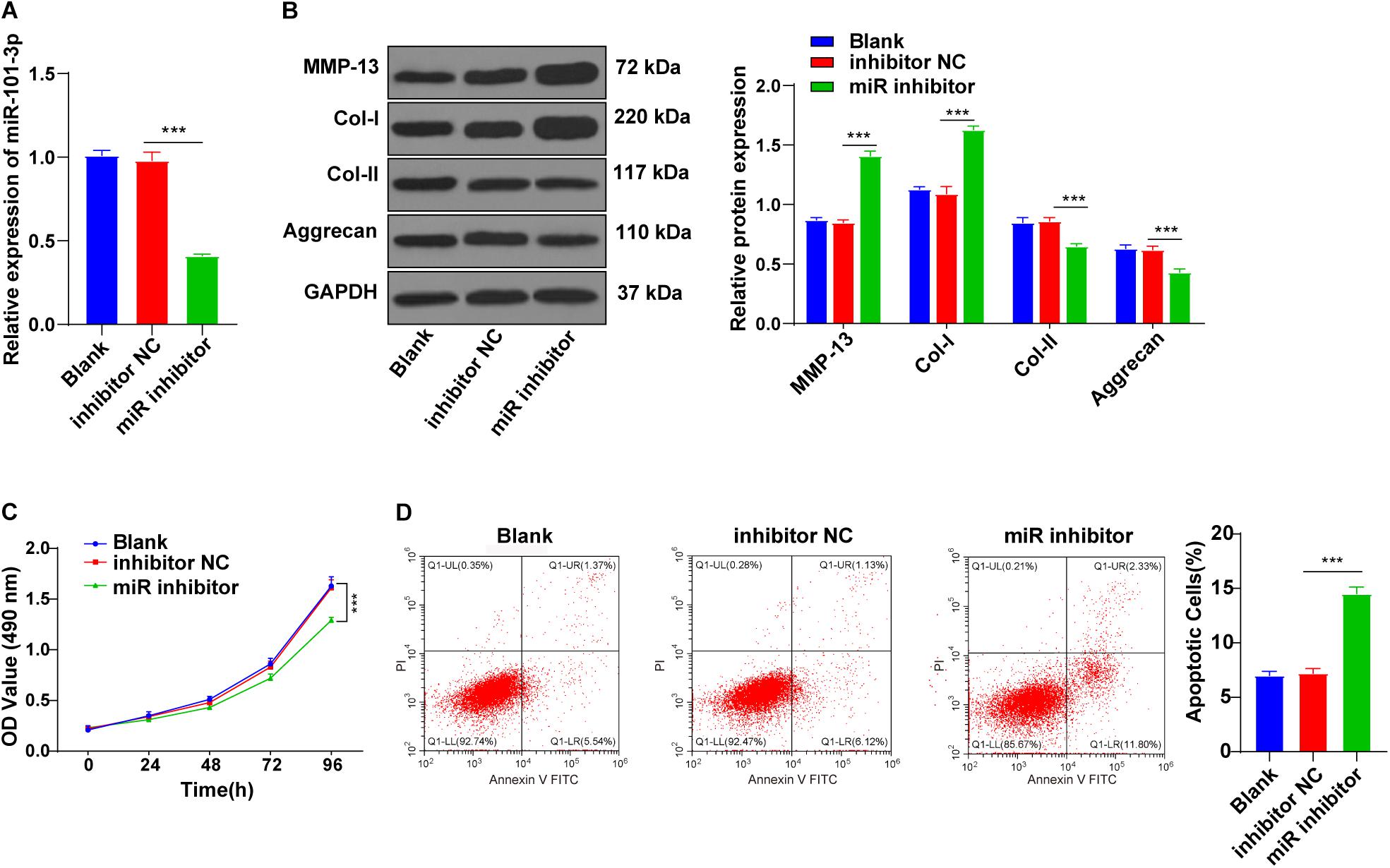
Figure 3. Downregulation of miR-101-3p inhibited the effect of BMSCs on ECM remodeling of NPCs. NPCs of patients with IDD were cocultured with BMSCs and then transfected with miR-101-3p inhibitor, with transfection of inhibitor negative control as control. (A) miR-101-3p expressions were detected using the RT-qPCR; (B) MMP-13, Col-I, Col-II, and Aggrecan levels were detected using Western blotting; and (C,D) viability and apoptosis of NPCs were detected using MTT assay and flow cytometry. The cell experiments were repeated three times independently. Data are presented as mean ± SD. Data were analyzed using one-way ANOVA, followed by the Tukey’s multiple comparison test, ***p < 0.001.
miR-101-3p Targeted EIF4G2
To further determine the downstream target of miR-101-3p-mediated ECM remodeling, we conducted follow-up experiments. EIF4G2 is abnormally expressed in osteoarthritis (Gao S. et al., 2020). The binding site between miR-101-3p and EIF4G2 was predicted by using the Starbase database (Figure 4A). The binding relationship between miR-101-3p and EIF4G2 was verified using dual-luciferase reporter gene assay (p < 0.001; Figure 4B). Compared with the inhibitor NC group, EIF4G2 expression in the miR-101-3p inhibitor group was notably upregulated (p < 0.001; Figure 4C). miR-101-3p negatively regulated EIF4G2 expression.
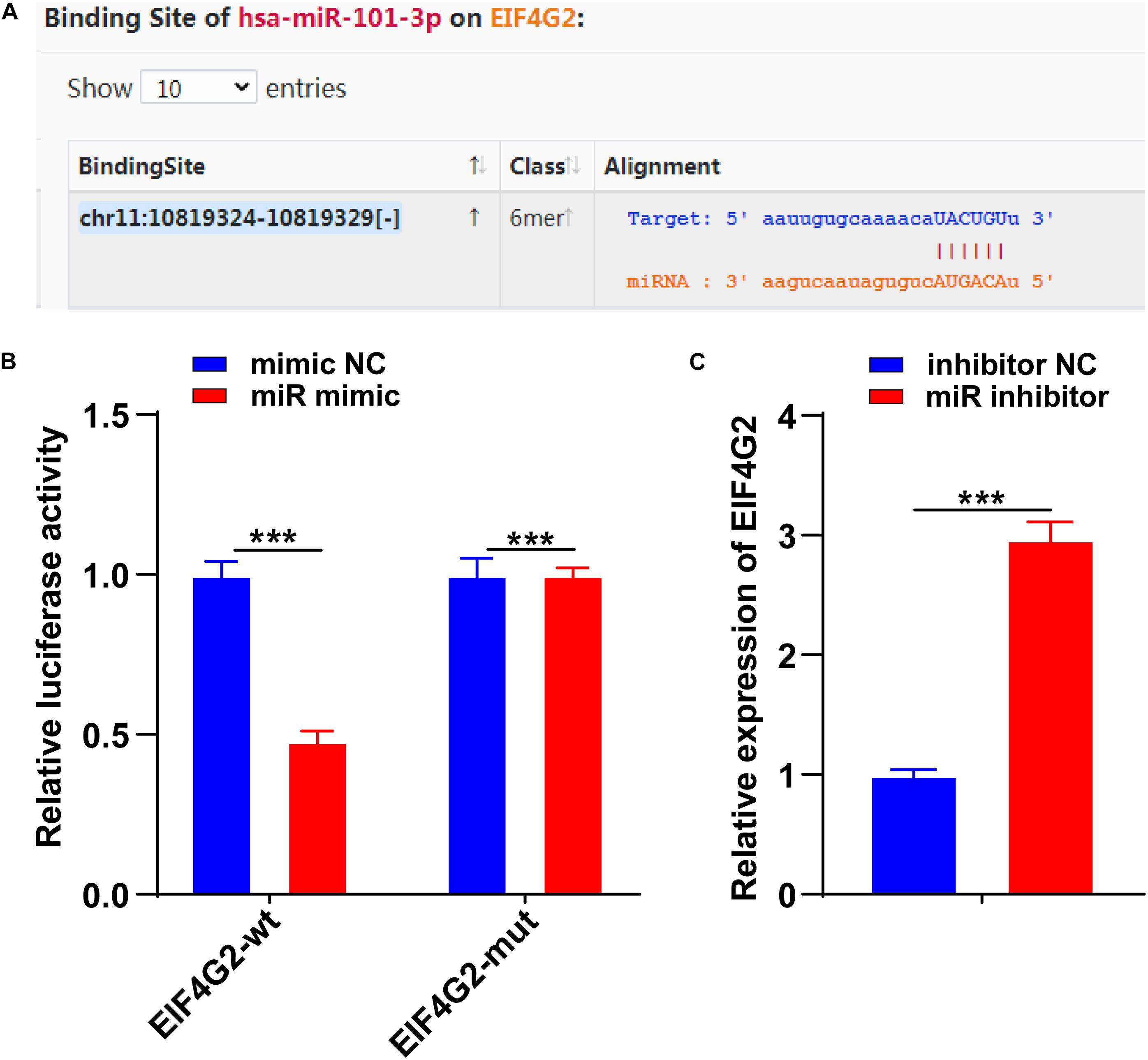
Figure 4. miR-101-3p targeted EIF4G2. (A) The binding site between miR-101-3p and EIF4G2 was predicted by using the Starbase database; (B) the binding relationship between miR-101-3p and EIF4G2 was verified using dual-luciferase reporter gene assay; and (C) expressions of EIF4G2 in miR-101-3p inhibitor-transfected NPCs were detected using the RT-qPCR. The cell experiments were repeated three times independently. Data are presented as mean ± SD. Data were analyzed using the independent sample t-test, ***p < 0.001.
miR-101-3p Enhanced the Effect of BMSCs on ECM Remodeling of Degenerated NPCs by Targeting EIF4G2
Functional rescue experiments were performed to verify whether miR-101-3p enhanced the promoting effect of BMSCs on ECM remodeling of degenerated NPCs by targeting EIF4G2. BMSC-cocultured NPCs were transfected with EIF4G2-siRNA, and the transfection efficiency was confirmed using the RT-qPCR (p < 0.001; Figure 5A). Downregulation of EIF4G2 reversed miR-101-3p inhibitor-mediated ECM protein levels in NPCs (p < 0.001; Figure 5B), as well as the viability and apoptosis of NPCs (p < 0.001; Figures 5C,D). miR-101-3p was confirmed to enhance the promoting effect of BMSCs on ECM remodeling of NPCs by targeting EIF4G2.
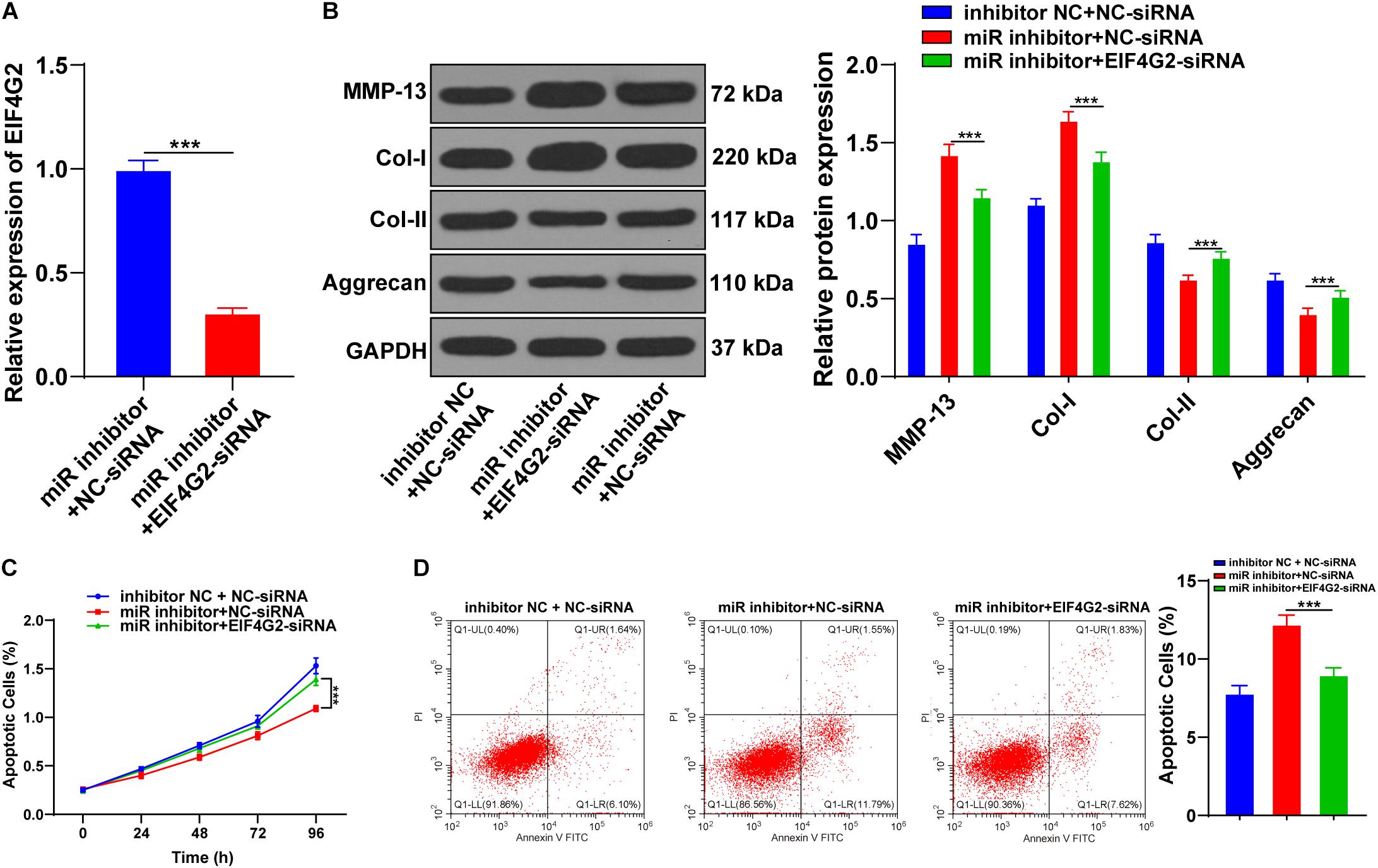
Figure 5. Downregulation of EIF4G2 reversed the inhibitory effect of miR-101-3p inhibitor on ECM remodeling of NPCs. (A) EIF4G2-siRNA transfection efficiency was confirmed using the RT-qPCR; (B) MMP-13, Col-I, Col-II, and Aggrecan levels were detected using Western blotting; (C,D) viability and apoptosis of NPCs were detected using MTT assay and flow cytometry. The cell experiments were repeated three times independently. Data are presented as mean ± SD. Data in panel (A) were analyzed using the independent sample t-test; and data in panels (B–D) were analyzed using one-way ANOVA, followed by the Tukey’s multiple comparison test, ***p < 0.001.
Discussion
Intervertebral disk degeneration (IDD) is commonly caused by excessive degeneration of ECM, increased death of IVD cells, and impairment of biomechanical functions (Johnson et al., 2015). Transplantation of BMSCs can retard or reverse diseased IVD (Cao et al., 2015). Specific regulation of miRs also represents a promising approach for the management of IDD (Li et al., 2015). This study clarified that miR-101-3p enhanced the effect of BMSCs on ECM remodeling in IDD.
The pathogenesis of IDD concerns the degeneration of the water-binding ECM, mechanical overloading, and catabolic cell response (Vergroesen et al., 2015). With the degeneration of ECM, Col-II expression is gradually reduced, while Col-I expression is promoted (Vergroesen et al., 2015). Aggrecan represents the main non-collagenous component of ECM, which endows IVD with the ability to bear compressive load (Zhang et al., 2018). MMP-13 constitutes one of the primary catabolic factors in the ECM metabolism of NPCs (Wang et al., 2019). Patients with IDD showed elevated MMP-13 and Col-I expressions and reduced Col-II and Aggrecan expressions. Elevated MMP-13 expression is concerned with ECM degeneration (Liu et al., 2019), and the loss of Aggrecan causes the impairment of ECM function and initiation of IDD (Kepler et al., 2013). Deregulated miRs in NPCs functionally participate in IDD progression by acting on NPC proliferation, apoptosis, and ECM synthesis/degradation (Li Z. et al., 2019). miR-101 participates in ECM degradation in osteoarthritis (Dai et al., 2015; Lu et al., 2020). Still, the effect of miR-101-3p on ECM degradation of NPCs remains unclear. We showed that miR-101-3p expression was reduced in patients with IDD. Gao Z.X. et al. (2020) have unveiled that miR-101-3p is downregulated in degenerated NPCs, and miR-101-3p participates in the modulation of NPC proliferation and apoptosis. In brief, miR-101-3p is implicated in IDD progression.
Bone mesenchymal stem cells (BMSCs) are crucial tools for tissue repair, which can differentiate into NP-like cells in IVD and promote the proteoglycan content of ECM (Xu et al., 2016). Therefore, BMSCs were cocultured with NPCs of a patient with IDD. The BMSC-cocultured NPCs showed enhanced Aggrecan, Col-II, and miR-101-3p expressions and decreased Col-I and MMP-13 expressions. NPCs, the major cell type residing in NP, are responsible for the integrity of IVD via the role of producing ECM components (Wang et al., 2014; Li et al., 2015; Cheng et al., 2018). Deregulated phenotypes of NPCs, including aberrant apoptosis, autophagy, and proliferation, contribute to the initiation and progression of IDD (Li et al., 2018). The reduction of NPCs and the subsequent degeneration of ECM contribute to the progression of IDD (Makino et al., 2017; Yang et al., 2018). Coculture of NPCs with BMSCs can notably repress the degeneration and apoptosis of NPCs (Hu et al., 2015). Consistently, we revealed that NPCs cocultured with BMSCs had enhanced NPC viability and suppressed apoptosis. Yi et al. (2014) have clarified that BMSC transplantation enhances the ECM content by repressing ECM degradation and facilitating ECM synthesis. BMSCs possess an anti-apoptotic effect on IDD through the mitochondrial apoptotic pathway in AF cells (Hu et al., 2017). Taken together, BMSCs repressed NPC apoptosis and facilitated ECM remodeling of NPCs.
Additionally, miR-101-3p expression in NPCs was notably upregulated after coculture, which indicated that miR-101-3p was involved in the process of BMSCs, promoting ECM remodeling of NPCs. The promotion function of miR-101 on chondrogenic differentiation of BMSCs has been revealed previously (Gao et al., 2019). Then, we transfected miR-101-3p inhibitor into BMSC-cocultured NPCs. miR-101-3p inhibitor-transfected NPCs showed promoted MMP-13 and Col-I expressions and decreased Aggrecan and Col-II expressions. Additionally, miR-101-3p inhibitor-transfected NPCs showed reduced cell viability and enhanced apoptosis. In brief, the downregulation of miR-101-3p inhibited the effect of BMSCs on ECM remodeling of NPCs.
Thereafter, we further determined the downstream target of miR-101-3p. The binding site between miR-101-3p and EIF4G2 was predicted by using the Starbase database. Translation of most mRNAs is modulated at the initiation level, a process demanding the protein complex called EIF4F (including EIF4E, EIF4G, and EIF4A) (Caron et al., 2004; Xie et al., 2017). EIF4G has two functional homologs in mammals, namely, EIF4G1 and EIF4G2 (Caron et al., 2004). EIF4G2 expression is promoted in osteoarthritis cartilage tissues, and EIF4G2 may be implicated in the pathogenesis of osteoarthritis (Gao S. et al., 2020). Nevertheless, the role of EIF4G2 in IDD remained unknown, and this study filled this gap to a certain extent. EIF4G2 expression of miR-101-3p inhibitor-transfected NPCs was notably upregulated, indicating that miR-101-3p negatively regulated EIF4G2 expression. The functional rescue experiments were performed. BMSC-cocultured NPCs were transfected with EIF4G2-siRNA, and the results exhibited that the downregulation of EIF4G2 reversed miR-101-3p inhibitor-mediated ECM protein levels in NPCs, as well as viability and apoptosis of NPCs. It was confirmed that miR-101-3p enhanced the promoting effect of BMSCs on ECM remodeling of NPCs by targeting EIF4G2.
To sum up, miR-101-3p increased the promoting effect of BMSCs on ECM remodeling of degenerated NPCs by targeting EIF4G2. This study unveiled a new mechanism of BMSCs in the treatment of IDD. However, there are still some limitations and deficiencies in this study. In this study, we chose the commercial single mesenchymal stem cell (MSC) case to study and ignored the variation among individuals, which led to the oversimplification of the conclusion. Additionally, we only isolated and cultured NPCs from one patient with IDD for the in vitro study. Whether this regulatory mechanism is different in degenerated NPCs from patients with IDD with different clinical parameters, different clinical grading, and different genders remains to be further explored. This study did not explain what substances MSCs may secrete to drive the increase of miR-101-3p. The existing studies have demonstrated that MSC-derived exosomes can repair IDD by carrying miRNAs (Zhang et al., 2020; Zhu et al., 2020). Whether the increase of miR-101-3p was driven by MSC-derived exosomes remained unidentified. In this study, we selected degenerated NPCs from patients with IDD but did not study the effect of miR-101-3p on non-degenerated NPCs from patients with LVF. Whether the decrease of miR-101-3p can drive the catabolic phenotype of NPCs is also what we need to continue to explore. In addition, the mechanism of BMSCs promoting ECM remodeling via miR-101-3p/EIF4G2 axis also needs further functional experimental verification in vivo. In the future, we will continue to determine that whether MSCs can promote the ECM remodeling of degenerated NPCs by secreting exosomes to drive the increase of miR-101-3p and conduct functional experiments in vivo.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committee of The Second Affiliated Hospital of Shandong First Medical University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Ethical Committee of Shandong First Medical University.
Author Contributions
ZW: conceptualization and methodology. XD: data curation and writing—original draft preparation. FC: investigation. XZ: supervision and validation. JW: writing—reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Key Research and Development Program of Shandong Province, China (Grant No. 2017GSF218057), the Medical and Health Technology Development Program of Shandong Province, China (Grant No. 202004071169), and the Academic Promotion Programme of Shandong First Medical University (Grant No. 2019QL017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Barczewska, M., Jezierska-Wozniak, K., Habich, A., Lipinski, S., Holak, P., Maksymowicz, W., et al. (2018). Evaluation of regenerative processes in the pig model of intervertebral disc degeneration after transplantation of bone marrow-derived mesenchymal stem cells. Folia Neuropathol. 56, 124–132. doi: 10.5114/fn.2018.76616
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Cai, F., Hong, X., Tang, X., Liu, N. C., Wang, F., Zhu, L., et al. (2019). ASIC1a activation induces calcium-dependent apoptosis of BMSCs under conditions that mimic the acidic microenvironment of the degenerated intervertebral disc. Biosci. Rep. 39:BSR20192708. doi: 10.1042/BSR20192708
Cai, F., Wu, X. T., Xie, X. H., Wang, F., Hong, X., Zhuang, S. Y., et al. (2015). Evaluation of intervertebral disc regeneration with implantation of bone marrow mesenchymal stem cells (BMSCs) using quantitative T2 mapping: a study in rabbits. Int. Orthop. 39, 149–159. doi: 10.1007/s00264-014-2481-0
Cao, C., Zou, J., Liu, X., Shapiro, A., Moral, M., Luo, Z., et al. (2015). Bone marrow mesenchymal stem cells slow intervertebral disc degeneration through the NF-kappaB pathway. Spine J. 15, 530–538. doi: 10.1016/j.spinee.2014.11.021
Caron, S., Charon, M., Cramer, E., Sonenberg, N., and Dusanter-Fourt, I. (2004). Selective modification of eukaryotic initiation factor 4F (eIF4F) at the onset of cell differentiation: recruitment of eIF4GII and long-lasting phosphorylation of eIF4E. Mol. Cell Biol. 24, 4920–4928. doi: 10.1128/mcb.24.11.4920-4928.2004
Cheng, X., Zhang, G., Zhang, L., Hu, Y., Zhang, K., Sun, X., et al. (2018). Mesenchymal stem cells deliver exogenous miR-21 via exosomes to inhibit nucleus pulposus cell apoptosis and reduce intervertebral disc degeneration. J. Cell. Mol. Med. 22, 261–276. doi: 10.1111/jcmm.13316
Dai, L., Zhang, X., Hu, X., Liu, Q., Man, Z., Huang, H., et al. (2015). Silencing of miR-101 Prevents Cartilage Degradation by Regulating Extracellular Matrix-related Genes in a Rat Model of Osteoarthritis. Mol. Ther. 23, 1331–1340. doi: 10.1038/mt.2015.61
Gan, Y., Tu, B., Li, P., Ye, J., Zhao, C., Luo, L., et al. (2018). Low Magnitude of Compression Enhances Biosynthesis of Mesenchymal Stem Cells towards Nucleus Pulposus Cells via the TRPV4-Dependent Pathway. Stem Cells Int. 2018:7061898. doi: 10.1155/2018/7061898
Gao, F., Peng, C., Zheng, C., Zhang, S., and Wu, M. (2019). miRNA-101 promotes chondrogenic differentiation in rat bone marrow mesenchymal stem cells. Exp. Ther. Med. 17, 175–180. doi: 10.3892/etm.2018.6959
Gao, S., Liu, L., Zhu, S., Wang, D., Wu, Q., Ning, G., et al. (2020). MicroRNA-197 regulates chondrocyte proliferation, migration, and inflammation in pathogenesis of osteoarthritis by targeting EIF4G2. Biosci. Rep. 40:BSR20192095. doi: 10.1042/BSR20192095
Gao, Z. X., Lin, Y. C., Wu, Z. P., Zhang, P., Cheng, Q. H., Ye, L. H., et al. (2020). LncRNA SNHG6 can regulate the proliferation and apoptosis of rat degenerate nucleus pulposus cells via regulating the expression of miR-101-3p. Eur. Rev. Med. Pharmacol. Sci. 24, 8251–8262.
Gore, M., Sadosky, A., Stacey, B. R., Tai, K. S., and Leslie, D. (2012). The burden of chronic low back pain: clinical comorbidities, treatment patterns, and health care costs in usual care settings. Spine (Phila Pa 1976) 37, E668–E677. doi: 10.1097/BRS.0b013e318241e5de
Hu, J., Deng, G., Tian, Y., Pu, Y., Cao, P., and Yuan, W. (2015). An in vitro investigation into the role of bone marrowderived mesenchymal stem cells in the control of disc degeneration. Mol. Med. Rep. 12, 5701–5708. doi: 10.3892/mmr.2015.4139
Hu, J., Yan, Q., Shi, C., Tian, Y., Cao, P., and Yuan, W. (2017). BMSC paracrine activity attenuates interleukin-1beta-induced inflammation and apoptosis in rat AF cells via inhibiting relative NF-kappaB signaling and the mitochondrial pathway. Am. J. Transl. Res. 9, 79–89.
Johnson, Z. I., Schoepflin, Z. R., Choi, H., Shapiro, I. M., and Risbud, M. V. (2015). Disc in flames: roles of TNF-alpha and IL-1beta in intervertebral disc degeneration. Eur. Cell Mater. 30, 104–116. doi: 10.22203/ecm.v030a08
Kepler, C. K., Ponnappan, R. K., Tannoury, C. A., Risbud, M. V., and Anderson, D. G. (2013). The molecular basis of intervertebral disc degeneration. Spine J. 13, 318–330. doi: 10.1016/j.spinee.2012.12.003
Konovalov, N. A., Nazarenko, A. G., Asyutin, D. S., Zelenkov, P. V., Onoprienko, R. A., Korolishin, V. A., et al. (2016). [Modern treatments for degenerative disc diseases of the lumbosacral spine. A literature review]. Zh Vopr Neirokhir Im N N Burdenko 80, 102–108. doi: 10.17116/neiro2016804102-108
Li, C. Y., Pang, Y. Y., Yang, H., Li, J., Lu, H. X., Wang, H. L., et al. (2017a). Identification of miR-101-3p targets and functional features based on bioinformatics, meta-analysis and experimental verification in hepatocellular carcinoma. Am. J. Transl. Res. 9, 2088–2105.
Li, C. Y., Xiong, D. D., Huang, C. Q., He, R. Q., Liang, H. W., Pan, D. H., et al. (2017b). Clinical value of miR-101-3p and biological analysis of its prospective targets in breast cancer: a study based on The Cancer Genome Atlas (TCGA) and bioinformatics. Med. Sci. Monit. 23, 1857–1871. doi: 10.12659/msm.900030
Li, X., Wu, A., Han, C., Chen, C., Zhou, T., Zhang, K., et al. (2019). Bone marrow-derived mesenchymal stem cells in three-dimensional co-culture attenuate degeneration of nucleus pulposus cells. Aging (Albany NY) 11, 9167–9187. doi: 10.18632/aging.102390
Li, Z., Chen, X., Xu, D., Li, S., Chan, M. T. V., and Wu, W. K. K. (2019). Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 52:e12704. doi: 10.1111/cpr.12704
Li, Z., Li, X., Chen, C., Li, S., Shen, J., Tse, G., et al. (2018). Long non-coding RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 51:e12483. doi: 10.1111/cpr.12483
Li, Z., Yu, X., Shen, J., Chan, M. T., and Wu, W. K. (2015). MicroRNA in intervertebral disc degeneration. Cell Prolif. 48, 278–283. doi: 10.1111/cpr.12180
Liu, Y., Lin, J., Wu, X., Guo, X., Sun, H., Yu, B., et al. (2019). Aspirin-Mediated Attenuation of Intervertebral Disc Degeneration by Ameliorating Reactive Oxygen Species In Vivo and In Vitro. Oxid. Med. Cell. Longev. 2019:7189854. doi: 10.1111/cpr.12180
Lu, G., Li, L., Wang, B., and Kuang, L. (2020). LINC00623/miR-101/HRAS axis modulates IL-1beta-mediated ECM degradation, apoptosis and senescence of osteoarthritis chondrocytes. Aging (Albany NY) 12, 3218–3237. doi: 10.18632/aging.102801
Makino, H., Seki, S., Yahara, Y., Shiozawa, S., Aikawa, Y., Motomura, H., et al. (2017). A selective inhibition of c-Fos/activator protein-1 as a potential therapeutic target for intervertebral disc degeneration and associated pain. Sci. Rep. 7:16983. doi: 10.1038/s41598-017-17289-y
Pfirrmann, C. W., Metzdorf, A., Zanetti, M., Hodler, J., and Boos, N. (2001). Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 26, 1873–1878. doi: 10.1097/00007632-200109010-00011
Qu, Z., Quan, Z., Zhang, Q., Wang, Z., Song, Q., Zhuang, X., et al. (2018). Comprehensive evaluation of differential lncRNA and gene expression in patients with intervertebral disc degeneration. Mol. Med. Rep. 18, 1504–1512. doi: 10.3892/mmr.2018.9128
Risbud, M. V., and Shapiro, I. M. (2014). Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat. Rev. Rheumatol. 10, 44–56. doi: 10.1038/nrrheum.2013.160
Shioya, M., Obayashi, S., Tabunoki, H., Arima, K., Saito, Y., Ishida, T., et al. (2010). Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol. Appl. Neurobiol. 36, 320–330. doi: 10.1111/j.1365-2990.2010.01076.x
Vergroesen, P. P., Kingma, I., Emanuel, K. S., Hoogendoorn, R. J., Welting, T. J., van Royen, B. J., et al. (2015). Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthritis Cartilage 23, 1057–1070. doi: 10.1016/j.joca.2015.03.028
Wang, C., Wang, W. J., Yan, Y. G., Xiang, Y. X., Zhang, J., Tang, Z. H., et al. (2015). MicroRNAs: new players in intervertebral disc degeneration. Clin. Chim. Acta 450, 333–341. doi: 10.1016/j.cca.2015.09.011
Wang, J., Hu, J., Chen, X., Huang, C., Lin, J., Shao, Z., et al. (2019). BRD4 inhibition regulates MAPK, NF-kappaB signals, and autophagy to suppress MMP-13 expression in diabetic intervertebral disc degeneration. FASEB J. 33, 11555–11566. doi: 10.1096/fj.201900703r
Wang, S. Z., Rui, Y. F., Lu, J., and Wang, C. (2014). Cell and molecular biology of intervertebral disc degeneration: current understanding and implications for potential therapeutic strategies. Cell Prolif. 47, 381–390. doi: 10.1111/cpr.12121
Xie, X., Li, Y. S., Xiao, W. F., Deng, Z. H., He, H. B., Liu, Q., et al. (2017). MicroRNA-379 inhibits the proliferation, migration and invasion of human osteosarcoma cells by targetting EIF4G2. Biosci. Rep. 37:BSR20160542.
Xu, J., Xq, E., Wang, N. X., Wang, M. N., Xie, H. X., Cao, Y. H., et al. (2016). BMP7 enhances the effect of BMSCs on extracellular matrix remodeling in a rabbit model of intervertebral disc degeneration. FEBS J. 283, 1689–1700. doi: 10.1111/febs.13695
Yang, F., Leung, V. Y., Luk, K. D., Chan, D., and Cheung, K. M. (2009). Mesenchymal stem cells arrest intervertebral disc degeneration through chondrocytic differentiation and stimulation of endogenous cells. Mol. Ther. 17, 1959–1966. doi: 10.1038/mt.2009.146
Yang, Y., Wang, X., Liu, Z., Xiao, X., Hu, W., and Sun, Z. (2018). Osteogenic protein-1 attenuates nucleus pulposus cell apoptosis through activating the PI3K/Akt/mTOR pathway in a hyperosmotic culture. Biosci. Rep. 38:BSR20181708.
Yi, Z., Guanjun, T., Lin, C., and Zifeng, P. (2014). Effects of transplantation of hTIMP-1-expressing bone marrow mesenchymal stem cells on the extracellular matrix of degenerative intervertebral discs in an in vivo rabbit model. Spine (Phila Pa 1976) 39, E669–E675.
Zhang, B., Guo, W., Sun, C., Duan, H. Q., Yu, B. B., Mu, K., et al. (2018). Dysregulated MiR-3150a-3p promotes lumbar intervertebral disc degeneration by targeting aggrecan. Cell Physiol. Biochem. 45, 2506–2515. doi: 10.1159/000488269
Zhang, J., Zhang, J., Zhang, Y., Liu, W., Ni, W., Huang, X., et al. (2020). Mesenchymal stem cells-derived exosomes ameliorate intervertebral disc degeneration through inhibiting pyroptosis. J. Cell. Mol. Med. 24, 11742–11754. doi: 10.1111/jcmm.15784
Zhu, G., Yang, X., Peng, C., Yu, L., and Hao, Y. (2020). Exosomal miR-532-5p from bone marrow mesenchymal stem cells reduce intervertebral disc degeneration by targeting RASSF5. Exp. Cell Res. 393:112109. doi: 10.1016/j.yexcr.2020.112109
Zhu, J., Tang, H., Zhang, Z., Zhang, Y., Qiu, C., Zhang, L., et al. (2017). Kaempferol slows intervertebral disc degeneration by modifying LPS-induced osteogenesis/adipogenesis imbalance and inflammation response in BMSCs. Int. Immunopharmacol. 43, 236–242. doi: 10.1016/j.intimp.2016.12.020
Keywords: intervertebral disc degeneration, miR-101-3p, EIF4G2, bone mesenchymal stem cells, nucleus pulposus cells, extracellular matrix remodeling, Collagen-I, Collagen-II
Citation: Wang Z, Ding X, Cao F, Zhang X and Wu J (2021) Bone Mesenchymal Stem Cells Promote Extracellular Matrix Remodeling of Degenerated Nucleus Pulposus Cells via the miR-101-3p/EIF4G2 Axis. Front. Bioeng. Biotechnol. 9:642502. doi: 10.3389/fbioe.2021.642502
Received: 18 December 2020; Accepted: 30 June 2021;
Published: 27 August 2021.
Edited by:
Martin James Stoddart, AO Research Institute, SwitzerlandReviewed by:
Stephen Richardson, The University of Manchester, United KingdomFrancesco Grassi, Rizzoli Orthopedic Institute (IRCCS), Italy
Copyright © 2021 Wang, Ding, Cao, Zhang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingguo Wu, drwujingguo0723@163.com
 Zeng Wang1
Zeng Wang1  Jingguo Wu
Jingguo Wu