One-Carbon Metabolism Regulates Embryonic Stem Cell Fate Through Epigenetic DNA and Histone Modifications: Implications for Transgenerational Metabolic Disorders in Adults
- 1Department of Biochemistry, Midwestern University, Downers Grove, IL, United States
- 2Department of Medical Humanities, Rocky Vista University, Parker, CO, United States
- 3Molecular Biology, Department of Biomedical Sciences, Rocky Vista University, Parker, CO, United States
Human (h) and mouse (m) embryonic stem (ES) cells need specific amino acids to proliferate. mES cells require threonine (Thr) metabolism for epigenetic histone modifications. Thr is converted to glycine and acetyl CoA, and the glycine is metabolized specifically to regulate trimethylation of lysine (Lys) residue 4 in histone H3 (H3K4me3). DNA methylation and methylation of other H3 Lys residues remain unimpaired by Thr deprivation in mES cell culture medium. Similarly, hES cells require methionine (Met) to maintain the Met-SAM (S-adenosyl methionine) cycle of 1-carbon metabolism also for H3K4me3 formation. H3K4me3 is needed specifically to regulate and maintain both mES and hES cell proliferation and their pluripotent states. Better understanding of this regulation is essential since treatment of human diseases and disorders will increasingly involve hES cells. Furthermore, since ES cells are derived from their progenitor cells in preimplantation blastocysts, they serve as models of 1-carbon metabolism in these precursors of all mammalian tissues and organs. One-carbon metabolism challenges, such as a maternal low protein diet (LPD) during preimplantation blastocyst development, contribute to development of metabolic syndrome and related abnormalities in adults. These 1-carbon metabolism challenges result in altered epigenetic DNA and histone modifications in ES progenitor cells and the tissues and organs to which they develop. Moreover, the modified histones could have extracellular as well as intracellular effects, since histones are secreted in uterine fluid and influence early embryo development. Hence, the mechanisms and transgenerational implications of these altered epigenetic DNA and histone modifications warrant concerted further study.
Introduction
Both sperm and egg cells contain DNA and histones that contribute to the genetic and epigenetic complements of mammalian conceptuses. The epigenetic complements of these gametes and early embryos are altered by environmental challenges to their one-carbon amino acid metabolism (Fleming et al., 2018; Clare et al., 2019; Goyal et al., 2019; Velazquez et al., 2019). Such challenges increase the prevalence of metabolic syndrome and related disorders in offspring that develop from the embryonic stem (ES) progenitor cells in these early conceptuses (Van Winkle and Ryznar, 2018).
Within days of conception, embryos give rise to ES progenitor cells and another type of cell in preimplantation blastocysts. A trophectoderm, one cell thick, forms the surface of blastocysts. This trophectoderm attaches to uterine epithelial cells to initiate implantation. Pluripotent cells, termed the inner cell mass (ICM), reside within the trophectoderm layer. After implantation, cells in the ICM differentiate into progenitors of all tissues in the offspring. Thus, environmentally induced epigenetic changes in ICM cells result in such changes in adult tissues and organs (Clare et al., 2019; Velazquez et al., 2019). Since the ICM is used to produce pluripotent ES cell lines, ES cells serve as an experimental model for environmental challenges to their progenitor cells in the ICM of blastocysts.
Mouse Embryonic Stem (mES) Cells Require Threonine (Thr) Metabolism to Remain Pluripotent
mES Cells Require Thr One-Carbon Metabolism for Specific, Epigenetic Histone Methylations
Mouse ES cells do not proliferate and begin to differentiate if Thr is not supplied in their culture medium (Wang et al., 2009). These cells catabolize Thr to glycine and acetyl CoA, which are used for and help to regulate epigenetic methylations and acetylations in mES cells (Jog et al., 2017). Di- and tri-methylation of lysine (Lys) residue 4 in histone H3 (H3K4me3) slows dramatically and selectively when Thr is not supplied in the medium, while methylation of DNA and other H3 Lys residues continues normally (Shyh-Chang et al., 2013). Mouse ES cells need H3K4me3 to proliferate and remain pluripotent (Ang et al., 2011; Kilberg et al., 2016; Chen and Wang, 2019). Hence, Thr, taken up from the culture medium by mES cells, seems somehow to be metabolized selectively to provide 1-carbon units for H3K4 methylation.
Might Membrane Transporters Feed Amino Acids to Specific Intracellular Metabolic Compartments?
In the preimplantation blastocyst, leucine taken up by the trophectoderm triggers mammalian target of rapamycin (mTOR) signaling, trophoblast motility, and penetration of the uterine epithelium (Van Winkle et al., 2006). This signaling requires leucine uptake via the B0,+ amino acid transporter. Leucine uptake by other transporters does not result in trophoblast motility. B0,+ either selectively directs leucine to sites of mTOR signaling, or it generates other signals needed to synergize with mTOR (González et al., 2012).
In mES cells, selective use of Thr for H3K4 methylation may be more complex. Since Thr metabolism begins in perinuclear mitochondria in mES cells (Wanet et al., 2015; Lees et al., 2017; Woods, 2017), we propose that Thr is taken up selectively by a subpopulation of these organelles (Figure 1). Thr is then metabolized to formate, which leaves the mitochondria, and is converted to S-adenosyl methionine (SAM) methyl groups needed in the nucleus for H3K4 methylation (Scotti et al., 2013; Harvey, 2019). The formate from this subpopulation of mitochondria must be directed somehow to sites of H3K4 methylation in the nucleus, since only 1-carbon metabolism of Thr can be used to methylate this H3 residue (Shyh-Chang et al., 2013). Other 1-carbon donors cannot be used for this purpose unless supplied in excess to mES cells.
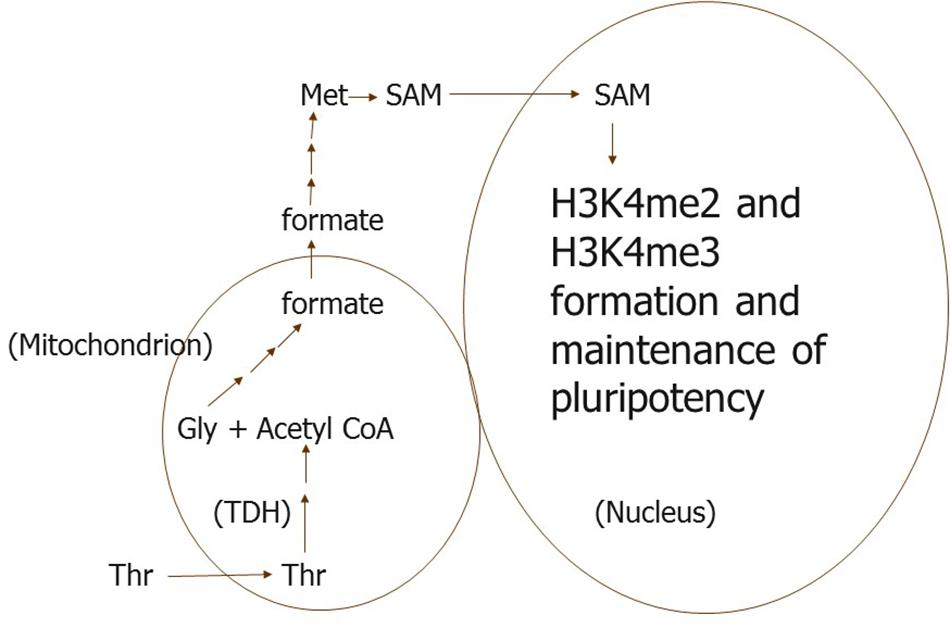
Figure 1. We propose that a subpopulation of perinuclear mitochondria is specialized to take up only Thr and metabolize it to formate in mouse ES and their progenitor cells. The formate is then converted to 1-carbon units in a pool of SAM used specifically for H3K4me di- and tri-methylation. TDH, Thr dehydrogenase.
Alternatively, Thr could be directed to the pertinent subpopulation of perinuclear mitochondria by one of at least three plasma membrane Thr transport proteins (Formisano and Van Winkle, 2016). One of these transporters could conceivably direct Thr to the subpopulation of perinuclear mitochondria much like the B0,+ transporter might selectively directs leucine to the site of mTOR signaling in the trophectoderm.
Does Thr Also Initiate Signaling by Interacting Directly With One of Its Transporters in Lipid Rafts?
In addition to providing metabolites for epigenetic DNA and histone methylation and acetylation (Jog et al., 2017), Thr itself is likely needed for proliferation and maintenance of an undifferentiated state in mES cells (Ryu and Han, 2011). Thr promotes expression of cMyc through processes mediated by mTOR in these cells. Intact lipid rafts containing a Thr transporter seems essential for this signaling, since the signaling cannot occur in mES cells when these rafts are disrupted. Either an intact raft supports Thr transport and resultant intracellular signaling by the amino acid itself, or raft caveolae initiate signaling during Thr interaction with its transporter in the plasma membrane. As an alternative to a present thesis (e.g., Figure 1), perhaps the latter signaling and Thr catabolism together foster H3K4 methylation by somehow selectively activating pertinent histone modifying enzyme(s).
By interfering with Thr transport (Van Winkle et al., 2014; Formisano and Van Winkle, 2016) as well as catabolism (Wang et al., 2009), the Thr analog, 3-hydroxynorvaline (3-HNV), inhibits proliferation of mES cells. Similarly, this Thr transport and resultant signaling as well as catabolism in the ICM of blastocysts would help maintain pluripotency in these mES progenitor cells.
Human Embryonic Stem (HES) Cells Need Methionine (Met) Metabolism for Proliferation
Met Metabolism for Specific, Epigenetic Histone Methylations May Also Be Partially Compartmentalized in hES Cells
Naïve and primed mES cells are epigenetically different, and hES cells seem to better correspond to primed mES cells (Takahashi et al., 2018). Nevertheless, 1-carbon metabolism regulates hES cell proliferation. Met is required by hES cells, and their progenitors in the inner cell masses of blastocysts, for specific, epigenetic histone methylations. If Met is not in the culture medium, hES cells do not proliferate but undergo apoptosis (Shiraki et al., 2014). Exogenously supplied Met is metabolized to SAM in hES cells; these cells utilize SAM to carry out epigenetic methylations. A lack of Met (or supplemental SAM) in the medium nearly eliminates di- and tri-methylation of Lys residue 4 in histone H3 (H3K4me3). Global DNA methylation is also decreased by a modest amount through Met deprivation of hESC cells (Shiraki et al., 2014), but methylation of other H3 Lys residues remains unimpaired (Kilberg et al., 2016). As for mES cells, H3K4me3 maintains hES cell proliferation and pluripotency.
Met is needed in the culture medium apparently to maintain adequate concentrations of metabolites in the Met-SAM methylation cycle in hES cells (Figure 2). This cycle seems to be used preferentially by methyltransferases other than those catalyzing methylation of H3K4 when the concentration of metabolites in the cycle are low. Moreover, Met might be easily depleted from hES cells when not present in the culture medium owing to exchange of Met for other bulky zwitterionic amino acids in the medium. The only good transporter of Met in hES cell membranes appears to be one or more of those corresponding to the Na+-independent system L, which is used for such exchange (Van Winkle and Ryznar, 2019).
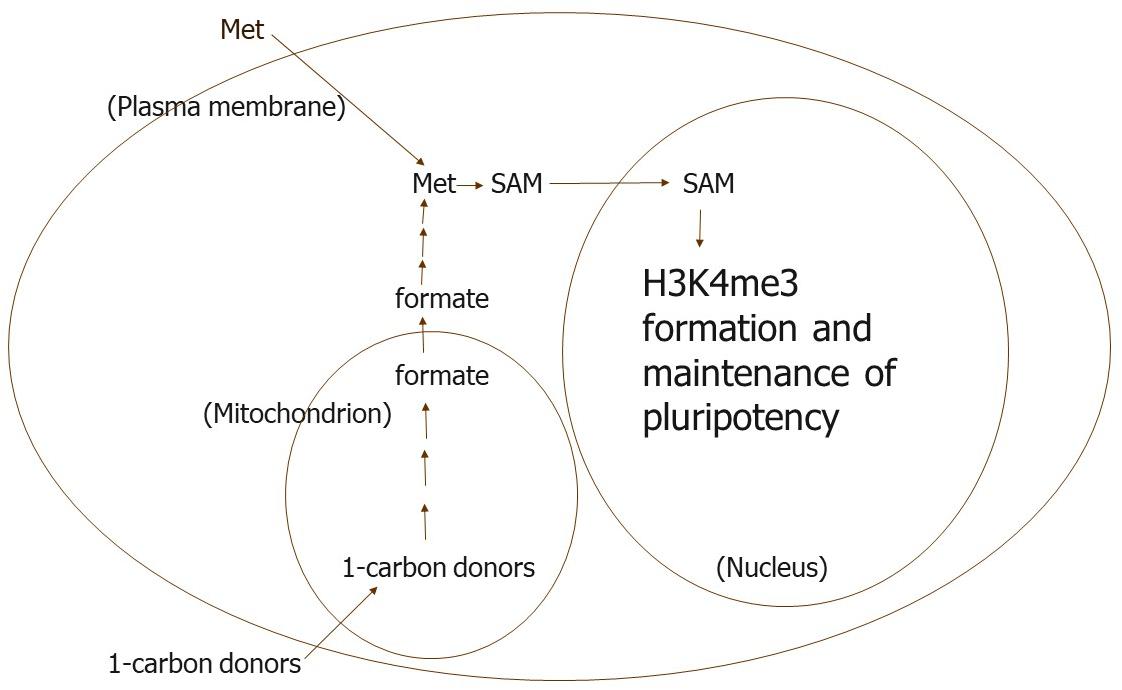
Figure 2. We propose that Met is needed in the culture medium to maintain the intracellular concentrations of metabolites in the Met-SAM methylation cycle that are needed for H3K4me3 formation in human ES and their progenitor cells. In our view, other methyl transferases are able to use SAM preferentially when its intracellular concentration is lower possibly because the Km values of these other transferases are lower than the transferases that methylate H3K4.
An Enigma: Thr Recuses hES Cells From Growth Inhibition by the Thr Analog, 3-HNV, but Not Because Thr Is Needed for One-Carbon Metabolism or Protein Synthesis
According to Wang and associates Wang et al. (2009), the Thr analog, 3-HNV, inhibits mES cell proliferation because it inhibits Thr dehydrogenase (TDH) and metabolism of Thr to glycine and acetyl CoA. We found, however, that 3-HNV also inhibits hES cell proliferation (Van Winkle et al., 2014). Unlike mouse cells, hES cells lack functional TDH and so do not catabolize Thr to the 1-carbon units needed for epigenetic H3K4 methylation and maintenance of pluripotency. Somewhat surprisingly, 3-HNV inhibits hES cell proliferation in a manner morphologically similar to the way it arrests mES cell growth (Van Winkle et al., 2014).
It has been proposed that 3-HNV inhibits proliferation of cells lacking TDH activity through 3-HNV incorporation into protein in place of Thr (Najafzadeh, 2018). But 3-HNV does not inhibit proliferation of other mouse cell lines or HeLa cells (a human cell line; Wang et al., 2009), so it is unlikely to inhibit hES cell growth owing to its incorporation into protein. An excess of Thr in the culture medium does, however, rescue hES proliferation from 3-HNV inhibition. These findings support the theory that Thr fosters hES and mES cell proliferation through signaling, such as that requiring intact lipid rafts, that is separate from (and in addition to) regulation of epigenetic DNA and histone modifications by 1-carbon metabolites of Thr in mES cells (Van Winkle et al., 2014).
Methyl Donor Perturbations, Including a Maternal Low Protein Diet, Likely Alter Transgenerational Epigenetic Dna and Histone Modifications in mEs Progenitor Cells
Does a More Invasive and Pinocytic Trophectoderm Supply More Thr to mES Progenitor Cells and Modify Placentation?
When female mice consume a LPD during preimplantation development of their conceptuses, trophoblast cells adapt in several ways in blastocysts. The trophectoderm exhibits more proliferation, motility during penetration of the uterine epithelium, and endocytic sequestration of uterine fluid proteins (reviewed by Fleming et al., 2018). These proteins are likely hydrolyzed to supply Thr to mES progenitor cells (Van Winkle and Ryznar, 2019), so increased accumulation of the proteins should result in delivery of more Thr to these pluripotent cells. More protein hydrolysis could also supply more leucine and arginine and, thus, augment trophoblast motility (González et al., 2012).
Other changes may also cause mES progenitor cells to accumulate more Thr. Mouse ES cells take up Thr via amino acid exchange (Formisano and Van Winkle, 2016). Thus, Thr can be accumulated against its chemical potential gradient only if other amino acids are first taken up against their gradients. For instance, mES cells should be able to accumulate alanine against its gradient via the Na+-dependent amino acid transport system A (Van Winkle and Ryznar, 2019). This alanine could then exit the cells via system ASC and drive uptake of Thr into mES cells. In this regard, alanine may accumulate in the environment of mES progenitor cells when their mothers are consuming a LPD. When blastocysts develop from the 2-cell stage in culture medium without added amino acids, their alanine concentration is six times higher than in morphologically similar blastocysts developing in vivo (Van Winkle and Dickinson, 1995). While this in vitro culture system is an extreme model, it might mimic the effects of LPD well enough to indicate how mES progenitor cells could accumulate more Thr in exchange for alanine. Greater Thr accumulation could alter the supply of 1-carbon units to SAM and its regulation of H3K4me di- and tri-methylation.
Maternal consumption of a LPD during the pre- and peri-implantation stages of development also results in development of more efficient placentas (Fleming et al., 2018). More vigorous trophoblast invasion of the uterine epithelium owing to maternal LPD consumption likely leads to this development. Likewise, greater delivery of nutrients by the yolk sac placenta occurs, owing to altered epigenetic histone modifications in the primitive endoderm that gives rise to this placenta (Fleming et al., 2018). In addition, switching from LPD to control diet consumption by pregnant mice at the time of blastocyst implantation causes the somatic cell lineages of mES progenitor cells to decrease then raise, respectively, the level of some of their epigenetic DNA modifications relative to the control rate. Such changes in the nutrient supply to conceptuses also likely change epigenetic histone modifications during early embryo development.
Altered Epigenetic Histone Modifications in mES Progenitor Cells Likely Persist in Future Generations Without Further Methyl Donor Perturbations
Dysfunction of endothelial cells and elevated blood pressure occur in grandprogeny of rat dams consuming a LPD during pregnancy, in the absence of protein restriction of the F1 generation, probably owing to transgenerational epigenetic DNA and histone modifications (Torrens et al., 2008). For the same reasons, impaired placental function or older age of F0 mothers result in transmission of cardiorenal and metabolic alterations to their grandprogeny (Gallo et al., 2012; Master et al., 2015). Moreover, extracellular as well as intracellular actions of histones could have occurred, owing to uterine secretion of the modified histones by F1 females (Van Winkle and Ryznar, 2018).
Epigenetic methyl donor challenges can also be genetic. For example, impairment of folate metabolism through genetic mutation in F0 mice results in suppression of epigenetic methylation, with transgenerational effects on development of wild-type progeny mice, for at least five generations (Padmanabhan et al., 2013). Epigenetic modifications in the germline of F1 females cause congenital malformations, whereas alterations in the uterine environment of F1 mothers produce growth defects. The altered uterine environment likely includes epigenetically modified histones, since uterine cells of F1 wild-type mice exhibited hypomethylation of DNA and probably histones, and uterine fluid contains histones that likely influence early embryo development (Van Winkle, 2017; Van Winkle and Ryznar, 2018). Histones act both intracellularly and extracellularly, however, so the effects of their hypomethylation probably include both congenital malformations and growth defects.
The proposed actions of extracellular histones are also illuminated by methyl donor supplementation that prevents transgenerational obesity amplification in F3 agouti viable yellow mice (Waterland et al., 2008). In this study, germ cells of F0 mice were formed in the unsupplemented control environment since their parents received no methyl donor supplementation. As a result, mES progenitor cells that produced F1 mice had the epigenetic modifications associated with the control diet, so the organs that developed in F1 mice did not benefit from methyl donor supplementation. We propose that the resultant uterine secretion of altered histones by F1 mice (Van Winkle and Ryznar, 2018) contributed to development of obesity in the F2 generation. In contrast, methyl donor supplementation of F0 mice improved development of F1 germ cells, so organs, such as the uterus, also developed more normally in the F2 offspring of F1 mice. As a consequence, transgenerational obesity amplification was not observed in F3 mice, owing to methyl donor supplementation beginning in the F0 generation (Waterland et al., 2008).
Other Sources of Extracellular Histones in the Reproductive Tract
In addition to histones secreted in uterine fluid (Van Winkle, 2017; Van Winkle and Ryznar, 2018), other sources of extracellular histones include seminal fluid and dead sperm cells (Fung et al., 2004; Miller et al., 2010; Miller, 2018). Paternal as well as maternal environmental challenges adversely affect peri-implantation development of embryos through intergenerational epigenetic modifications (Clare et al., 2019; Su and Patti, 2019). These changes are likely present in extracellular and intracellular histones, thus providing one mechanism by which paternal diet could program offspring health through both seminal plasma- and sperm-specific pathways (Watkins et al., 2018).
Perturbations in Methyl Donor Supply Also Likely Alter Epigenetic DNA and Histone Modifications in Pre- and Peri-Implantation Human ES Progenitor Cells
Epigenetic DNA methylation marks change during formation of early embryos, and these changes foster normal tissue development. Human oocytes have high levels of the folate receptor FOLR1, suggesting that folate uptake for 1-carbom metabolism is important during the maternal to zygotic transition (Strandgaard et al., 2017). Babies born after IVF procedures have higher rates of disorders related to imprinting and epigenetic changes, maybe because commercial IVF media does not contain intermediates of 1-carbon metabolism, such as folates and methionine (Menezo et al., 2019). Moreover, epigenetic changes occurring during the activation of induced pluripotent stem cells require an optimal SAM pool for appropriate reprograming back to the pluripotent state (Fernández-Arroyo et al., 2015). This reprograming can be influenced by nutritional deficiencies of needed cofactors derived from vitamin B complexes. The altered methyl donor metabolism affects DNA methylation, histone modifications, and miRNAs. In addition, methyl donor challenges in humans, such as a LPD, alters epigenetically programed ES cell progenitor differentiation (Wu et al., 2019).
Furthermore, the differentially methylated CpG islands that regulate genomic imprinting may be highly susceptible to changes in diet during pre- and peri-implantation development. With over 150 imprinted genes identified in the human genome, and many human diseases associate with dysregulation of these areas, changes in diet affecting these regions could have profound effects on the offspring. For example, folic acid supplementation before versus during pregnancy showed changes in DNA methylation patterns specifically at the imprinted H19/IGF2 locus in infants. The authors proposed that since methylation marks regulating imprinted genes are acquired at differentially methylated DNA regions before gastrulation, their methylation patterns may reveal environmental exposures such as a change in diet or toxin exposure (Hoyo et al., 2011).
Other locations within the human genome may be affected by changes in epigenetic marks as the result of diet fluctuations. For example, active H3K4me3 and repressive H3K27me3 maintain a pluripotent epigenetic state in ES and their progenitor cells (Wu et al., 2019). In hES cells, there are also abundant non-CpG methylated cytosines that may be important to early development (Lister et al., 2009).
A maternal LPD also predisposition human as well as rodent offspring to metabolic syndromes by modifying genes including H19, IGF2, UBE3A, POMC, GR, and PPAR-α (Gong et al., 2010; Lillycrop and Burdge, 2011). These modified genes are related to imprinting, metabolism, or the stress response. Such changes in methylation patterns of the genome in the early embryo, after birth, or early in life provide evidence of how environment contributes to transgenerational human disease states.
Conclusion
Epigenetic DNA and histone modifications in ES and their progenitor cells in early embryos require membrane transport and compartmentalized metabolism of amino acids. One-carbon metabolites produced specifically to regulate H3 methylation in mES and hES cells require Thr and Met from the culture medium. ES cells require these 1-carbon units to produce H3K4me3 and remain undifferentiated. Further understanding of this regulation is essential. All adult tissues and organs arise from ES progenitor cells, and treatment of human diseases and disorders will increasingly involve hES cells.
Furthermore, metabolic syndrome and related disorders often result from altered 1-carbon metabolism in ES progenitor cells. This altered 1-carbon metabolism produces modified epigenetic DNA and histone methylations. Such epigenetically produced diseases and disorders can become transgenerational phenotypes in humans. Extracellular as well as intracellular histones may alter development beginning in early embryos when these histones are produced, through perturbation of 1-carbon methyl donor metabolism, and secreted in uterine fluid.
Author Contributions
Both authors created sections of the manuscript, reviewed and provided feedback to each other that facilitated its critical revision, and approved the final version of the manuscript for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ang, Y. S., Tsai, S. Y., Lee, D. F., Monk, J., Su, J., Ratnakumar, K., et al. (2011). Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–197. doi: 10.1016/j.cell.2011.03.003
Chen, G., and Wang, J. (2019). A regulatory circuitry locking pluripotent stemness to embryonic stem cell: interaction between threonine catabolism and histone methylation. Semin. Cancer Biol. 57, 72–78. doi: 10.1016/j.semcancer.2019.01.005
Clare, C. E., Brassington, A. H., Kwong, W. Y., and Sinclair, K. D. (2019). One-carbon metabolism: linking nutritional biochemistry to epigenetic programming of long-term development. Annu. Rev. Anim. Biosci. 7, 263–287. doi: 10.1146/annurev-animal-020518-115206
Fernández-Arroyo, S., Cuyàs, E., Bosch-Barrera, J., Alarcón, T., Joven, J., and Menendez, J. A. (2015). Activation of the methylation cycle in cells reprogrammed into a stem cell-like state. Oncoscience 2, 958–967.
Fleming, T. P., Watkins, A. J., Velazquez, M. A., Mathers, J. C., Prentice, A. M., Stephenson, J., et al. (2018). Origins of lifetime health around the time of conception: causes and consequences. Lancet 391, 1842–1852. doi: 10.1016/S0140-6736(18)30312-X
Formisano, T. M., and Van Winkle, L. J. (2016). At least three transporters likely mediate threonine uptake needed for mouse embryonic stem cell proliferation. Front. Cell Dev. Biol. 4:17. doi: 10.3389/fcell.2016.00017
Fung, K. Y., Glode, L. M., Green, S., and Duncan, M. W. (2004). A comprehensive characterization of the peptide and protein constituents of human seminal fluid. Prostate 61, 171–181. doi: 10.1002/pros.20089
Gallo, L. A., Tran, M., Moritz, K. M., Jefferies, A. J., and Wlodek, M. E. (2012). Pregnancy in aged rats that were born small: cardiorenal and metabolic adaptations and second-generation fetal growth. FASEB J. 26, 4337–4347. doi: 10.1096/fj.12-210401
Gong, L., Pan, Y. X., and Chen, H. (2010). Gestational low protein diet in the rat mediates Igf2 gene expression in male offspring via altered hepatic DNA methylation. Epigenetics 5, 619–626. doi: 10.4161/epi.5.7.12882
González, I. M., Martin, P. M., Burdsal, C., Sloan, J. L., Mager, S., Harris, T., et al. (2012). Leucine and arginine regulate trophoblast motility through mTOR-dependent and independent pathways in the preimplantation mouse embryo. Dev. Biol. 361, 286–300. doi: 10.1016/j.ydbio.2011.10.021
Goyal, D., Limesand, S. W., and Goyal, R. (2019). Epigenetic responses and the developmental origins of health and disease. J. Endocrinol. 242, T105–T119. doi: 10.1530/JOE-19-0009
Harvey, A. J. (2019). Mitochondria in early development: linking the microenvironment, metabolism and the epigenome. Reproduction 157, R159–R179. doi: 10.1530/REP-18-0431
Hoyo, C., Murtha, A. P., Schildkraut, J. M., Jirtle, R. L., Demark-Wahnefried, W., Forman, M. R., et al. (2011). Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics 6, 928–936. doi: 10.4161/epi.6.7.16263
Jog, R., Chen, G., Leff, T., and Wang, J. (2017). “Threonine catabolism: an unexpected epigenetic regulator of mouse embryonic stem cells,” in Handbook of Nutrition, Diet, and Epigenetics, eds V. B. Patel, and V. R. Preedy, (New York, NY: Springer International Publishing), 1–20. doi: 10.1007/978-3-319-31143-2_103-1
Kilberg, M. S., Terada, N., and Shan, J. (2016). Influence of amino acid metabolism on embryonic stem cell function and differentiation. Adv. Nutr. 7, 780S–789S. doi: 10.3945/an.115.011031
Lees, J. G., Gardner, D. K., and Harvey, A. J. (2017). Pluripotent stem cell metabolism and mitochondria: beyond ATP. Stem Cells Int. 2017:2874283. doi: 10.1155/2017/2874283
Lillycrop, K. A., and Burdge, G. C. (2011). Epigenetic changes in early life and future risk of obesity. Int. J. Obesity 35, 72–83. doi: 10.1038/ijo.2010.122
Lister, R., Pelizzola, M., Dowen, R. H., Hawkins, R. D., Hon, G., Tonti-Filippini, J., et al. (2009). Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322. doi: 10.1038/nature08514
Master, J. S., Thouas, G. A., Harvey, A. J., Sheedy, J. R., Hannan, N. J., Gardner, D. K., et al. (2015). Low female birth weight and advanced maternal age programme alterations in next-generation blastocyst development. Reproduction 149, 497–510. doi: 10.1530/REP-14-0619
Menezo, Y., Clément, P., and Dale, B. (2019). DNA methylation patterns in the early human embryo and the epigenetic/imprinting problems: a plea for a more careful approach to human assisted reproductive technology (ART). Int. J. Mol. Sci. 20:1342. doi: 10.3390/ijms20061342
Miller, D., Brinkworth, M., and Iles, D. (2010). Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 139, 287–301. doi: 10.1530/REP-09-0281
Miller, D. J. (2018). The epic journey of sperm through the female reproductive tract. Animal 12, s110–s120. doi: 10.1017/S1751731118000526
Najafzadeh, V. (2018). The Role of Amino Acids and the Threonine-SAM Pathway in the Development of Bovine Inner Cell Mass and Pluripotency, Doctoral dissertation, The University of Waikato, Hamilton.
Padmanabhan, N., Jia, D., Geary-Joo, C., Wu, X., Ferguson-Smith, A. C., Fung, E., et al. (2013). Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell 155, 81–93. doi: 10.1016/j.cell.2013.09.002
Ryu, J. M., and Han, H. J. (2011). L-threonine regulates G1/S phase transition of mouse embryonic stem cells via PI3K/Akt, MAPKs, and mTORC pathways. J. Biol. Chem. 286, 23667–23678. doi: 10.1074/jbc.M110.216283
Scotti, M., Stella, L., Shearer, E. J., and Stover, P. J. (2013). Modeling cellular compartmentation in one-carbon metabolism. Wiley Interdiscipl. Rev. 5, 343–365. doi: 10.1002/wsbm.1209
Shiraki, N., Shiraki, Y., Tsuyama, T., Obata, F., Miura, M., Nagae, G., et al. (2014). Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 19, 780–794. doi: 10.1016/j.cmet.2014.03.017
Shyh-Chang, N., Locasale, J. W., Lyssiotis, C. A., Zheng, Y., Teo, R. Y., Ratanasirintrawoot, S., et al. (2013). Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226. doi: 10.1126/science.1226603
Strandgaard, T., Foder, S., Heuck, A., Ernst, E., Nielsen, M. S., and Lykke-Hartmann, K. (2017). Maternally contributed folate receptor 1 is expressed in ovarian follicles and contributes to preimplantation development. Front. Cell Dev. Biol. 5:89. doi: 10.3389/fcell.2017.00089
Su, L., and Patti, M. E. (2019). Paternal nongenetic intergenerational transmission of metabolic disease risk. Curr. Diab. Rep. 19:38. doi: 10.1007/s11892-019-1163-0
Takahashi, S., Kobayashi, S., and Hiratani, I. (2018). Epigenetic differences between naïve and primed pluripotent stem cells. Cell. Mol. Life Sci. 75, 1191–1203. doi: 10.1007/s00018-017-2703-x
Torrens, C., Poston, L., and Hanson, M. A. (2008). Transmission of raised blood pressure and endothelial dysfunction to the F2 generation induced by maternal protein restriction in the F0, in the absence of dietary challenge in the F1 generation. Br. J. Nutr. 100, 760–766. doi: 10.1017/S0007114508921747
Van Winkle, L. J. (2017). Uterine histone secretion likely fosters early embryo development so efforts to mitigate histone cytotoxicity should be cautious. Front. Cell Dev. Biol. 5:100. doi: 10.3389/fcell.2017.00100
Van Winkle, L. J., and Dickinson, H. R. (1995). Differences in amino acid content of preimplantation mouse embryos that develop in vitro versus in vivo: in vitro effects of five amino acids that are abundant in oviductal secretions. Biol. Reprod. 52, 96–104. doi: 10.1095/biolreprod52.1.96
Van Winkle, L. J., Galat, V., and Iannaccone, P. M. (2014). Threonine appears to be essential for proliferation of human as well as mouse embryonic stem cells. Front. Cell Dev. Biol. 2:18.
Van Winkle, L. J., and Ryznar, R. (2018). Can uterine secretion of modified histones alter blastocyst implantation, embryo nutrition, and transgenerational phenotype? Biomol. Concepts 9, 176–183. doi: 10.1515/bmc-2018-0017
Van Winkle, L. J., and Ryznar, R. (2019). Amino Acid Transporters: Roles for Nutrition, Signalling and Epigenetic Modifications in Embryonic Stem Cells and Their Progenitors. Chichester: John Wiley & Sons, Ltd.
Van Winkle, L. J., Tesch, J. K., Shah, A., and Campione, A. L. (2006). System B0,+ amino acid transport regulates the penetration stage of blastocyst implantation with possible long-term developmental consequences through adulthood. Hum. Reprod. Update 12, 145–157. doi: 10.1093/humupd/dmi044
Velazquez, M. A., Fleming, T., and Watkins, A. J. (2019). Periconceptional environment and the developmental origins of disease. J. Endocrinol. 242, T33–T49.
Wanet, A., Arnould, T., Najimi, M., and Renard, P. (2015). Connecting mitochondria, metabolism, and stem cell fate. Stem Cells Dev. 24, 1957–1971. doi: 10.1089/scd.2015.0117
Wang, J., Alexander, P., Wu, L., Hammer, R., Cleaver, O., and McKnight, S. L. (2009). Dependence of mouse embryonic stem cells on threonine catabolism. Science 325, 435–439. doi: 10.1126/science.1173288
Waterland, R. A., Travisano, M., Tahiliani, K. G., Rached, M. T., and Mirza, S. (2008). Methyl donor supplementation prevents transgenerational amplification of obesity. Int. J. Obesity 32, 1373–1379. doi: 10.1038/ijo.2008.100
Watkins, A. J., Dias, I., Tsuro, H., Allen, D., Emes, R. D., Moreton, J., et al. (2018). Paternal diet programs offspring health through sperm-and seminal plasma-specific pathways in mice. Proc. Natl. Acad. Sci. U.S.A. 115, 10064–10069. doi: 10.1073/pnas.1806333115
Woods, D. C. (2017). Mitochondrial heterogeneity: evaluating mitochondrial subpopulation dynamics in stem cells. Stem Cells Int. 2017:7068567. doi: 10.1155/2017/7068567
Keywords: one-carbon metabolism, folate, threonine metabolism, methionine metabolism, inner cell mass, embryonic stem cells, epigenetic histone modification, metabolic syndrome
Citation: Van Winkle LJ and Ryznar R (2019) One-Carbon Metabolism Regulates Embryonic Stem Cell Fate Through Epigenetic DNA and Histone Modifications: Implications for Transgenerational Metabolic Disorders in Adults. Front. Cell Dev. Biol. 7:300. doi: 10.3389/fcell.2019.00300
Received: 04 September 2019; Accepted: 08 November 2019;
Published: 21 November 2019.
Edited by:
Mireille Khacho, University of Ottawa, CanadaReviewed by:
Amar M. Singh, University of Georgia, United StatesJyotsna Dhawan, Centre for Cellular & Molecular Biology (CCMB), India
Copyright © 2019 Van Winkle and Ryznar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lon J. Van Winkle, lvanwinkle@rvu.edu; lvanwi@midwestern.edu
 Lon J. Van Winkle
Lon J. Van Winkle Rebecca Ryznar3
Rebecca Ryznar3