Multifaceted Functions and Novel Insight Into the Regulatory Role of RNA N6-Methyladenosine Modification in Musculoskeletal Disorders
- 1Department of Orthopedics, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Hunan Key Laboratory of Tumor Models and Individualized Medicine, The Second Xiangya Hospital, Central South University, Changsha, China
- 3Department of Cardiovascular Surgery, The Second Xiangya Hospital, Central South University, Changsha, China
RNA modifications have emerged as key regulators of transcript expression in diverse physiological and pathological processes. As one of the most prevalent types of RNA modifications, N6-methyladenosine (m6A) has become the highlight in modulation of various diseases through interfering RNA splicing, translation, nuclear export, and decay. In many cases, the detailed functions of m6A in cellular processes and diseases remain unclear. Notably, recent studies have determined the relationship between m6A modification and musculoskeletal disorders containing osteosarcoma, osteoarthritis, rheumatoid arthritis, osteoporosis, etc. Herein, this review comprehensively summarizes the recent advances of m6A modification in pathogenesis and progression of musculoskeletal diseases. Specifically, the underlying molecular mechanisms, detection technologies, regulatory functions, clinical implications, and future perspectives of m6A in musculoskeletal disorders are discussed, with the aim to provide a novel insight into their association.
Introduction
Currently, a growing number of studies have shed light on a new hereditary manner, the epigenetics, which refers to changes in phenotype without DNA or RNA sequences alteration (Harvey et al., 2018). Several epigenetics manners have been identified, comprising the histone modification, DNA and RNA methylation, and noncoding RNA (ncRNA) modification. Of note, there have been more than 100 modifications demonstrated within RNAs over the past few years, including the N1-methyladenosine (m1A), m6A, 5-methylcytosine (m5C), 7-methylguanosine (m7G), m1G, m2G, m6G, etc. Among them, the m6A has received considerable attention because of its high abundance. It relates to a dynamic and reversible RNA modification that participate in a wide range of biological and pathological processes, such as the cancer progression (Lan et al., 2019) and inflammation (Zong et al., 2019). Specifically, it can manipulate the RNA splicing, export, translation, and degradation through methylation and demethylation mediated by multiple enzymes (Cao et al., 2016).
Musculoskeletal disorders are a set of prevalent diseases characterized by dysfunction of bone and skeletal muscle, including, but not limited to, osteosarcoma (OS), osteoarthritis (OA), rheumatoid arthritis (RA), osteoporosis (OP), etc. (Madan and Grime, 2015). Several epigenetics manners have been investigated in this field (Tu et al., 2019; van Wijnen and Westendorf, 2019). Among them, the alteration of m6A modification has been associated with the initiation and progression of musculoskeletal diseases.
In this review, we broadly summarize the functional repertoire of m6A in various musculoskeletal disorders, aiming to expand our understanding and discuss the putative perspective for adopting m6A as a novel biomarker and therapeutic target in musculoskeletal diseases.
RNA M6A Modification
As aforementioned, m6A modification is a dynamic and reversible epigenetic change (Zhang et al., 2020), which affects the stability and function of RNAs, thereby modulating the pathogenesis and progression of diseases (Qin et al., 2020; Wang et al., 2020; Zhu et al., 2020). M6A modification has been identified in more than 7,000 human genes, and it preferentially occurred at the site of stop codons and long internal exons within the RRACH sequence (R = G or A; H = A, C, or U) (Dominissini et al., 2012). Based on current evidences, m6A modification is able to interfere in RNA processing, splicing, export, degradation, and translation through the “writers,” “erasers,” and “readers” proteins (Chen et al., 2019). The detailed graphical description of the RNA m6A mechanism is presented in Figure 1.
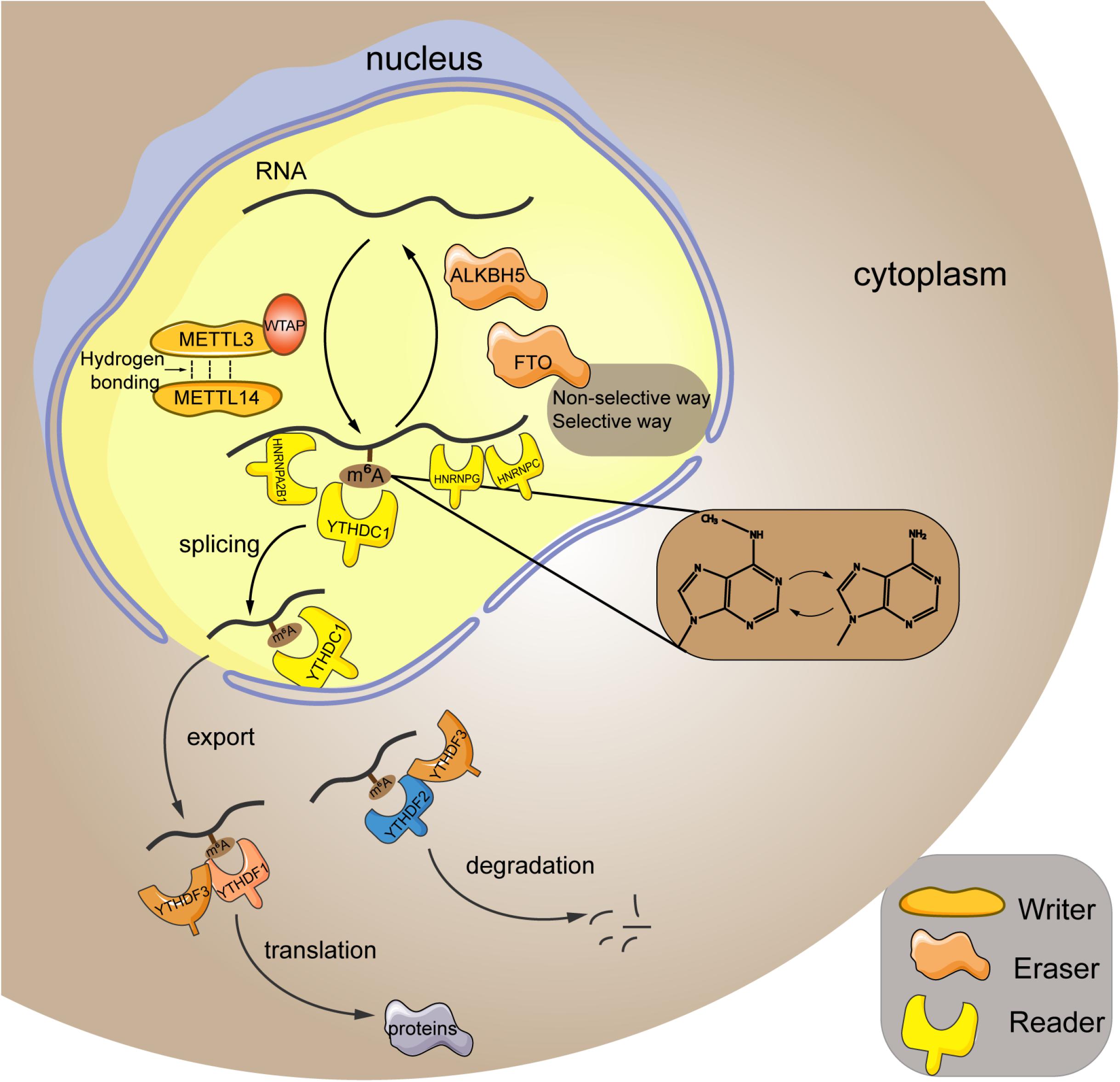
Figure 1. Mechanism of m6A RNA modification. M6A modification is a reversible process mediated by its regulatory proteins, including the writers (MELTT3, METTL14, WTAP, etc.), erasers (FTO, ALKBH5, etc.), and readers (YTHDF1, YTHDF2, YTHDF3, etc.). METTL3, methyltransferase-like 3; METTL14, methyltransferase-like 14; WTAP, Wilms tumor 1-associated protein; FTO, fat mass and obesity-associated protein; ALKBH5, alkB homolog 5; YTHDF1, YTH domain family; member 1; HNRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; HNRNPC, heterogeneous nuclear ribonucleoprotein C; HNRNPG, heterogeneous nuclear ribonucleoprotein G.
Writers refer to the RNA methyltransferases, including methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), methyltransferase-like 16 (METTL16), Wilms tumor 1-associated protein (WTAP), RNA-binding motif 15, etc. They are in charge of installing m6A to the RNA strand. In particular, METTL3 and METTL14 are the most studied key regulators in this process. They contain an S-adenosylmethionine–binding motif and are capable of adding methyl to the adenosine. Meanwhile, these two proteins can form a heterodimer core complex to modulate the cellular m6A deposition (Liu et al., 2014), in which they adopted a class I methyltransferase fold and interacted with each other through hydrogen bonding. Additionally, biochemical analysis revealed that METTL3 principally acted as the catalytic core, whereas METTL14 mainly functioned as an RNA-binding platform (Wang et al., 2016). Besides, the cofactor WTAP itself shows no methyltransferase activity, whereas it could interplay with the core complex to mediate its localization in nuclear spots (Ping et al., 2014). Furthermore, it has been reported that WTAP protein homeostasis in turn relied on the METTL3 protein levels (Sorci et al., 2018). Therefore, these writers cooperate to exert their function of m6A methylation within the specific RNA sites.
In contrast to the “writers,” the “erasers” are demethylases that function in methyl removal, canonically involving the fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5). FTO can oxidize m6A to generate the N(6)-hydroxymethyladenosine and N(6)-formyladenosine that have a half-life time less than 3 h (Fu et al., 2013). Generally, FTO erase methyl in either the selective or nonselective way. In the selective way, FTO recognizes and binds to the specific m6A-containing motif in cells (Li et al., 2019a). Because the association between FTO and RNA is weak, the additional cellular factors can corporately work with FTO to recognize and choose the target sites (Li et al., 2019a). In addition, the m6A itself can act as a “conformational marker” and interfere with the interaction between m6A and FTO via altering conformational outcomes in RNAs, which is the nonselective way of FTO-mediated demethylation (Zou et al., 2016). ALKBH5 is a 2-oxoglutarate and ferrous iron–dependent nucleic acid oxygenase that induces the demethylation of multiple RNAs. It was illuminated that ALKBH5 could occupy a similar region as L1 loop of the FTO protein that was associated with the single-strand RNA selectivity (Aik et al., 2014). Taken together, FTO and ALKBH5 are both the core regulators of m6A demethylation.
Readers are a group of proteins that discern the m6A modification and determine the functions of RNA transcripts. YTH domain (YTHD) family members (consisting of YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2) constitute a large class of m6A readers. They are located in nucleus or cytoplasm (specifically, YTHDF1 and YTHDF2 are located both in cytoplasm and nucleus, whereas YTHDF3 is only found in the cytoplasm) (Reichel et al., 2019), and characterized by containing the YT521B homology (YTH) domain that possesses an exquisite pocket with two conserved tryptophan residues (W377 and W428) for specific recognition of the methyl group (Xu et al., 2014). YTHDF1 is shown to promote translation of mRNA and enhance protein synthesis through impacting on the translation machinery, ensuring the sufficient protein generation is marked by m6A (Wang et al., 2015). Conversely, YTHDF2 mediates the degradation of its target m6A transcripts via reducing their stability (Li et al., 2018). Additionally, YTHDF3 participates in either promoting the protein synthesis in synergy with YTHDF1 or facilitating RNA degradation via interaction with YTHDF2. Thus, these three YTHDF proteins read the m6A modification in a cooperative way (Ni et al., 2019). Moreover, the nuclear YTHDC1 is associated with the RNA splicing (Kasowitz et al., 2018) and the export of m6A modified RNAs from nucleus to cytoplasm (Roundtree et al., 2017), whereas YTHDC2 engages in the elongation-promoting effect of m6A methylated RNA coding region (Mao et al., 2019). In addition to the YTHD family, the family of heterogeneous nuclear ribonucleoproteins (HNRNPs) is another set of m6A readers, which binds to pre-mRNA to interfere its stability and splicing (Geuens et al., 2016). HNRNPA2B1 has been well-accepted as a nuclear reader of m6A, binding to the RGm6AC containing sites on RNA to affect the alternative splicing and the processing of miRNAs (Alarcón et al., 2015). As to HNRNPC, because the m6A residues within RNA strand can destabilize the RNA duplexes (Kierzek and Kierzek, 2003), the structure of RNA may be altered when it is m6A modified. Given this, the m6A has been proposed to make the UUUUU tract within RNA to become more unfolded and accessible to HNRNPC (Liu et al., 2015), which is termed as the “m6A switch.” Moreover, HNRNPG also recognizes the m6A through “m6A switch.” HNRNPG has a low-complexity region that can discern a specific motif exposed by m6A-mediated RNA structural change (Liu et al., 2017), subsequently modulating the cotranscriptional pre-mRNA splicing (Zhou et al., 2019). Furthermore, the insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) can target the mRNA transcripts in an m6A-dependent way through recognizing their GG(m6A)C sequences, subsequently stabilizing the targeted RNAs under both the normal and stress conditions (Huang et al., 2018).
Advances in Technologies for M6A Detection
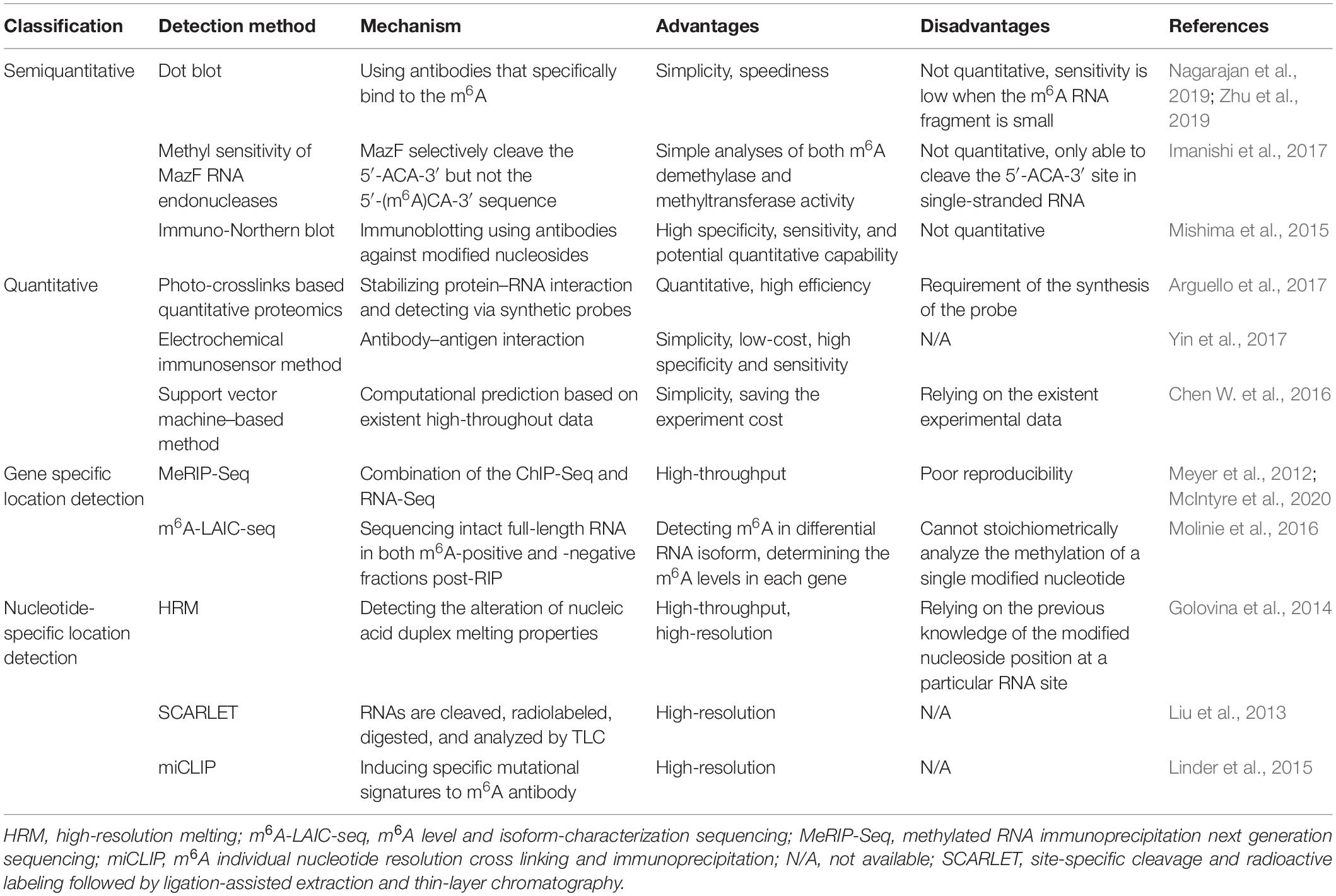
Currently, a massive number of technologies have been developed for m6A detection based on the immunohistochemistry or hybridization properties (Ovcharenko and Rentmeister, 2018). According to their detection performance, these technologies can be classified into the semiquantitative, quantitative, and precise location detection methods (Zhu et al., 2019; Table 1).
Semiquantitative Methods
Semiquantitative detection strategies, including the dot blot, methyl sensitivity of MazF RNA endonucleases, and immune-Northern blot, are used to determine the presence of m6A modification rather than the amount. Among them, dot blot is applied to detect the global change of m6A by using antibodies that specifically bind to the m6A site. It is relatively simple and fast but not quantitative (Zhu et al., 2019), with low sensitivity when the m6A RNA fragment is small in the samples (Nagarajan et al., 2019). A modified dot blot method has been adopted to increase the sensitivity through adding an immunoprecipitation step to enrich the m6A RNA before detection (Nagarajan et al., 2019). Further, Escherichia coli MazF is a sequence-specific endoribonuclease, which cleave the 5′-ACA-3′ sequence but not the 5′-(m6A)CA-3′ sequence within RNA strand, whereby it is an m6A-sensitive RNA cleavage enzyme. Based on this technology, a new high-throughput detection method for m6A has been established by researchers (Imanishi et al., 2017). Immuno-Northern blot is another way for semiquantitative detection of various types of RNA modifications. In this way, RNAs are separated and transferred onto a nylon membrane, followed by immunoblotting for measurement (Mishima et al., 2015).
Quantitative Methods
Unlike semiquantitative methods, the quantitative methods, including photo-crosslinks–based quantitative proteomics, electrochemical immunosensor method, and support vector machine–based method, can be utilized to determine the amount of m6A RNA. Arguello et al. (2017) have proposed that the photo-crosslinkers, a widely used method to stabilize the protein–RNA interaction, could combine with quantitative proteomics to detect m6A RNA. Meanwhile, a diazirine containing RNA probes has been recently synthesized to improve its efficiency (Arguello et al., 2017). Moreover, in the electrochemical immunosensor method (Yin et al., 2017), an anti-m6A antibody has been applied to recognize and capture the m6A-5′-triphosphate. Silver nanoparticles and amine-PEG3-biotin functionalized SiO2 nanospheres (Ag@SiO2) were used to amplify the signal, and phos-tag-biotin was employed as a bridge to connect the m6ATP and Ag@SiO2. This approach is convenient, low-cost, and of high specificity and sensitivity. Besides, the support vector machine–based method is a computational way to predict the m6A site within RNA strand based on the existent high-throughput experiment data (Chen W. et al., 2016).
M6A Location Detection Methods
To figure out the specific location of m6A within RNAs, several detection methods have been further developed by researchers (Ovcharenko and Rentmeister, 2018). Generally, these techniques can be divided into two sorts, namely, the gene-specific and the nucleoside-specific detection method. The former one comprises methylated RNA immunoprecipitation next-generation sequencing (MeRIP-Seq), m6A level and isoform-characterization sequencing (m6A-LAIC-seq), etc. While the latter contains high-resolution melting (HRM) analysis, site-specific cleavage, and radioactive labeling followed by ligation-assisted extraction and thin-layer chromatography (SCARLET), m6A individual nucleotide resolution cross-linking and immunoprecipitation (miCLIP), etc.
MeRIP-Seq combines the ChIP-Seq and RNA-Seq (Meyer et al., 2012), in which the anti-m6A antibody is incubated with the RNA fragments, and the precipitated fragments then are sequenced. Hence, it can determine the origin of m6A at a gene level. However, a more recent study reported that this method was of poor reproducibility because it was easily influenced by the noise (McIntyre et al., 2020). On the basis of MeRIP-seq, m6A-LAIC-seq (Molinie et al., 2016) is further developed to detect the dynamic range and isoform complexity of m6A content in a single gene, in which the intact full-length RNAs in both m6A-positive and m6A-negative fractions post-RIP are sequenced, and thus the differential isoform usages in each transcripts are detectable.
HRM (Golovina et al., 2014) is a high-throughput method for m6A detection at a specific site within the RNA strand. This method works by detecting the alteration of nucleic acid duplex melting properties caused by m6A modification. For instance, it has been observed that the melting temperature of RNA–DNA duplex was reduced by the presence of m6A modification (Golovina et al., 2014). SCARLET (Liu et al., 2013) is a method that enables detection of m6A status at any site in mRNA/long ncRNA (lncRNA), in which the m6A-containing candidate sites are cleaved, radiolabeled, and site-specific ligated, followed by complete nuclease digestion. The digested m6A residue is then measured by thin-layer chromatography (TLC). Moreover, MiCLIP can be used to determine the m6A site at a nucleotide-specific level (Linder et al., 2015). M6A antibodies bind to the m6A sites within RNA strands, and the m6A residues are then located by inducing specific mutational signatures after ultraviolet light–induced antibody-RNA cross-linking and reverse transcription.
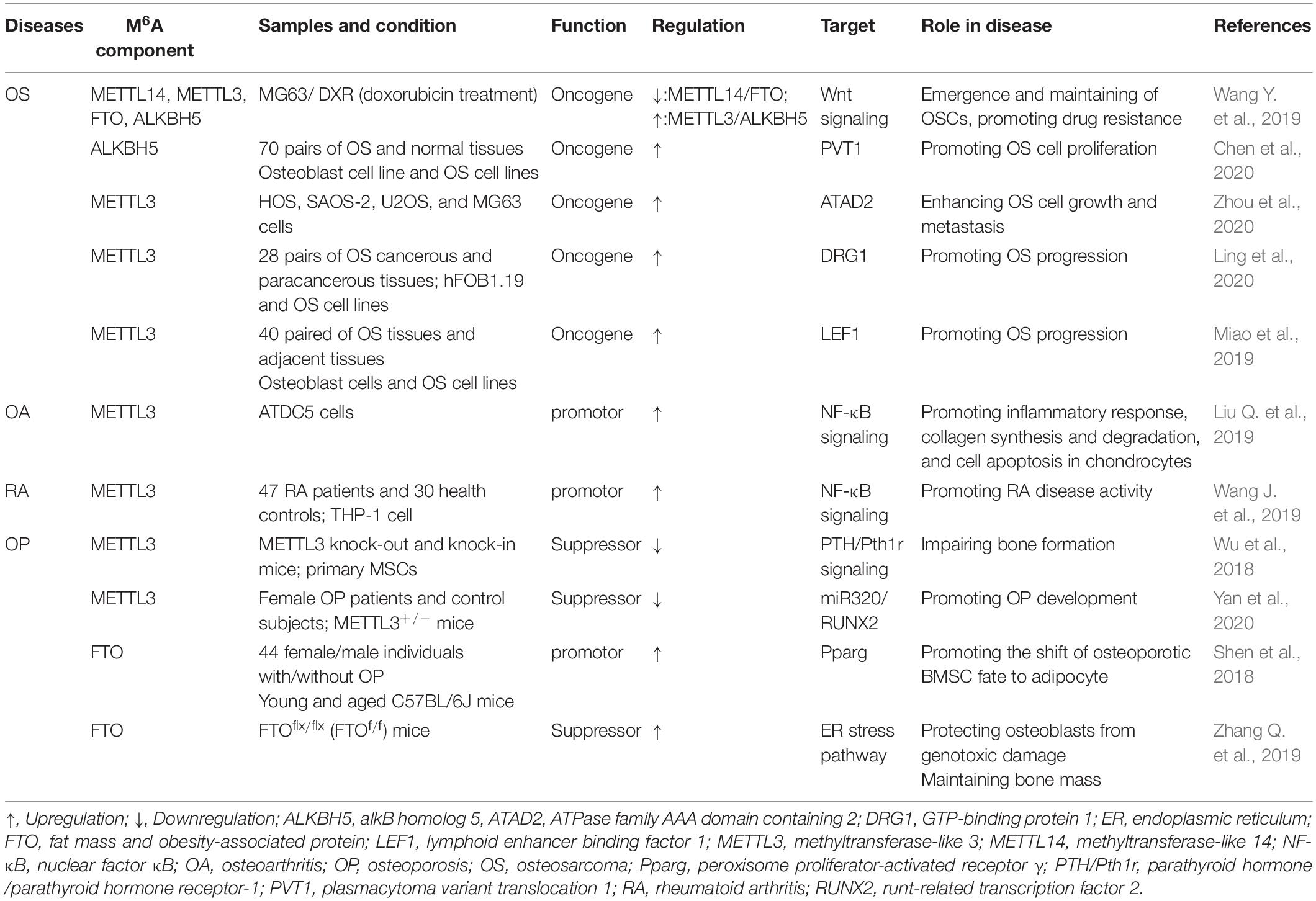
RNA M6A Methylation in Musculoskeletal Biology
Musculoskeletal system mainly consists of skeleton and skeletal muscle, which directly participates in the motor function of human body. Diverse transcriptional factors have been reported to involve in the genesis and maintenance of musculoskeletal system. M6A modification is a widely discovered and annotated epigenetic manner that takes part in the biology of musculoskeletal system. Herein, we discuss the mechanism and regulatory function of m6A in this process (Figure 2).
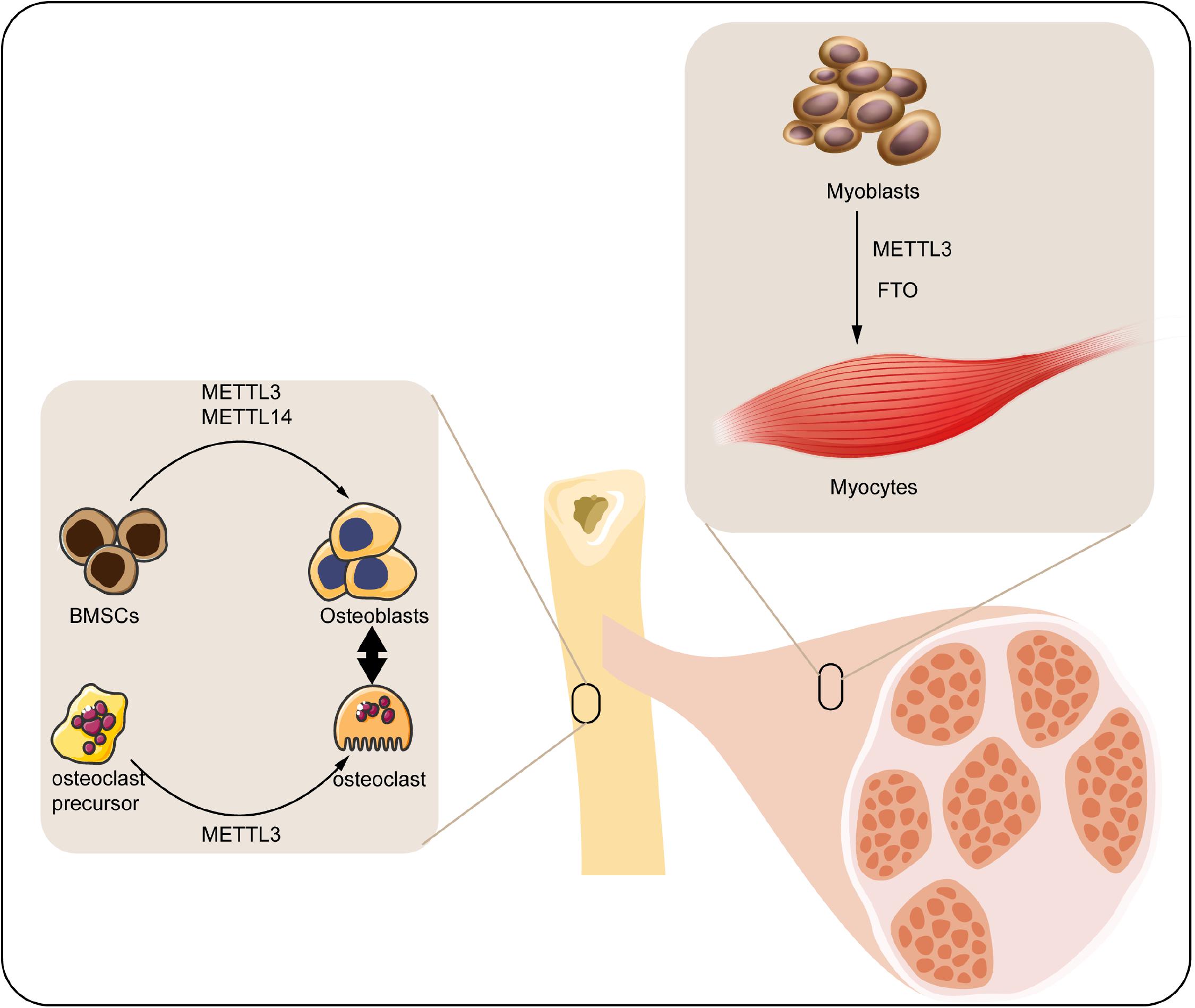
Figure 2. Roles of m6A modification in musculoskeletal biology. M6A regulatory proteins participate in differentiation of BMSCs, osteoclast precursors, and myoblasts. METTL3, methyltransferase-like 3; METTL14, methyltransferase-like 14; FTO, fat mass and obesity-associated protein; BMSCs, bone marrow mesenchymal stem cells.
M6A in Bone Remodeling
Bone is a connective tissue functioning in mechanical support, mineral homeostasis, hematopoiesis, etc., which maintains its metabolic balance principally through bone remodeling, a dynamic process involved in the formation of bone matrix through osteoblasts and removal of bone mass via osteoclasts (Hadjidakis and Androulakis, 2006). Osteoblast is derived from the bone marrow mesenchymal stem cells (BMSCs) under the impact of numerous regulators, which can produce matrix to form the bone tissue. With the development of bone matrix, some osteoblasts finally reorganized and embedded into the matrix as osteocytes (Blair et al., 2017). Accordingly, osteocytes play a vital role in monitoring the bone quality and sensing the mechanotransduction, as well as secreting regulatory factors associated with bone anabolism (Tresguerres et al., 2020). By contrast, osteoclasts are multinuclear cells derived from myeloid precursors, which function in degradation and resorption of bone through secreting proteolytic enzymes and acid (Charles and Aliprantis, 2014). The osteoblasts and osteoclasts couple with each other to maintain the dynamic homeostasis of bones (Weivoda et al., 2016). Any impairment of the homeostasis under pathological condition may contribute to the bone disorder.
As an indispensable part of epigenetic regulation, m6A modification is suggested as a crucial regulator participating in either the osteogenic or osteoclastogenic processes of bone. For instance, the expression of m6A methyltransferases (METTL3 and METTL14) was found significantly elevated in BMSCs undergoing osteogenic induction. Accordingly, knockdown of METTL3 reduced the mRNA level of genes related to BMSC proliferation and differentiation such as the Vegfa-164 and Vegfa-188 (Tian et al., 2019). Apart from this, MYD88 gene, a vital upstream regulator of nuclear factor κB (NF-κB) signaling, was methylated by METTL3, followed by activation of NF-κB and repression of osteogenic progression. Meanwhile, the METTL3-mediated osteogenic differentiation tendency could be reversed by demethylase ALKBH5 (Yu et al., 2020). Moreover, silencing METTL3 decreased the osteogenic markers, Smad signaling, and mineralized nodules in preosteoblast MC3T3-E1 cells, indicating the reduction of osteoblast differentiation (Zhang Y. et al., 2019). Hence, the METTL3-mediated m6A methylation significantly contributes to the maintenance of osteogenesis. Conversely, METTL3-mediated m6A methylation was also reported to facilitate the osteoclast differentiation. METTL3 level was elevated during osteoclastogenesis, and METTL3 depletion suppressed the differentiation and bone-resorbing ability of osteoclasts. Mechanistically, Atp6v0d2 mRNA, the principal regulator of osteoclast precursor cells fusion (Kim et al., 2009) was stabilized by the m6A-binding protein YTHDF2 when the METTL3 was abolished (Li et al., 2020).
Taken together, the present studies have implicated the function of METTL3-mediated m6A in either the osteogenic or the osteoclastogenic differentiation of bone remodeling.
M6A in Skeletal Muscle Regulation
Skeletal muscle comprises almost 40% of the total body weight, which functions in both the mechanical and metabolic processes of the body such as force generation and heat production (Frontera and Ochala, 2015). Retaining skeletal muscle mass is crucial for its physiological function, which is principally determined by the size and amount of muscle fibers. Skeletal muscle fibers are generated from myoblasts via myoblasts fusion, a process named as myogenesis (Sampath et al., 2018). Epigenetic modifications involving the histone modification (Machado et al., 2017), ncRNA (Liu M. et al., 2019), DNA methylation (Miyata et al., 2015), etc. have been unraveled to play a part in this process. Among them, m6A was proposed as a concernful regulator.
In the previous analyses, m6A modification can modulate the activity of skeletal muscle via modifying the muscle mass and interfering with the myoblasts differentiation: (1) modifying the muscle mass. For instance, upregulation of FTO gene expression was closely associated with skeletal muscle mass increase in overweight individuals (Doaei et al., 2019). Moreover, maternal high-fat intake could even disturb the m6A modification and FTO gene expression in skeletal muscle of its offspring (Li et al., 2016). Wu et al. (2017) have confirmed that activation of AMPK decreases the lipid accumulation in skeletal muscle cells through inversely regulating FTO expression and FTO-mediated demethylation. (2) Interfering with the myoblasts differentiation. It has been elucidated that FTO downregulation inhibited the myoblasts differentiation of mice through affecting the activity of mTOR-PGC-1α-mitochondria axis, which suppressed the mitochondria biogenesis and energy production of skeletal muscle cells (Wang et al., 2017). In addition, myoblast differentiation was regulated by METTL3-promoted MyoD. Specifically, suppression of METTL3 downregulated the activation of MyoD by modifying the sites within 5’ untranslated region of MyoD mRNA (Kudou et al., 2017).
Roles of M6A in Musculoskeletal Disorders
Musculoskeletal disorders refer to abnormalities of skeleton or skeletal muscle, emerging as the tumor, inflammation, trauma, etc., which may lead to disability and paralysis. Although previous work has shed light on the roles of m6A in musculoskeletal disorders, the details of m6A function and its involvement in the pathogenesis and progression remain unclear. Here, we summarize the current evidences concerning the pleiotropic functions of m6A in musculoskeletal diseases, as presented in Figure 3 and Table 2.
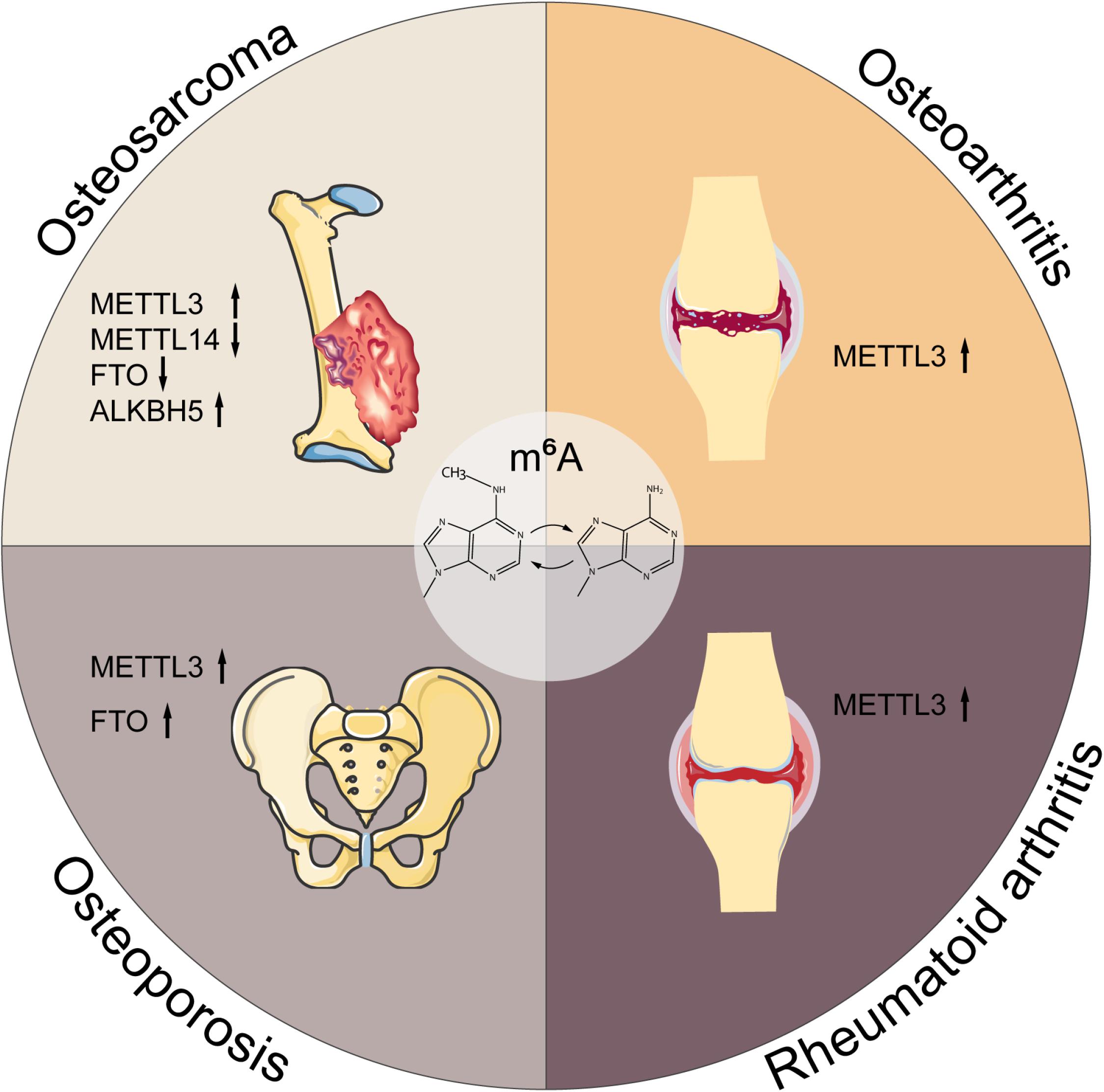
Figure 3. M6A in musculoskeletal disorders. M6A is associated with the progression of multiple musculoskeletal diseases, including osteosarcoma, osteoarthritis, rheumatoid arthritis, and osteoporosis. METTL3, methyltransferase-like 3; METTL14, methyltransferase-like 14; FTO, fat mass and obesity-associated protein; ALKBH5, alkB homolog 5; YTHDF1, YTH domain family, member 1.
M6A in OS
OS is the most common primary malignant bone tumor that mainly occurs in teenagers and adolescents with an annual incidence of 3.1 case per million (Gianferante et al., 2017). Although considerable advancement has been achieved in the past decades, the comprehensive mechanism network of OS has not yet been fully investigated. Currently, the standardized therapy for OS is limb salvage surgery or amputation combined with multiregimen-based chemotherapy. However, the survival rate of patients is unsatisfactory because of potential chemoresistance, lung metastasis, or tumor relapses. Recently, increasing studies have explored new therapeutic strategies for OS such as the molecular target therapy (Corre et al., 2020).
Epigenetic modification has been globally investigated in OS (Nebbioso et al., 2018). Almost all types of epigenetics, ranging from DNA methylation to histone modification, have been suggested to involve in the development and progression of OS (Cui et al., 2011). For instance, DNA methylation could downregulate miR-449c expression and eventually contributed to the tumorigenesis of OS (Li Q. et al., 2017). Meanwhile, histone methyltransferase has been discovered to regulate the chemosensitivity in OS (Jiang et al., 2018; He et al., 2019). Additionally, potential roles of multiple ncRNAs also have been validated in OS, such as the circRNA (Tu et al., 2020) and lncRNA (Ren et al., 2020; Xu et al., 2020). Li X. et al. (2017) reported that HOX transcript antisense intergenic RNA (HOTAIR) could enhance the development of OS via DNA methylation of CDKN2A gene. Notably, there have been some indications that m6A modification exerts pivotal functions in OS.
An integrative study performed by Wang J. et al. (2019) analyzing the transcriptome-wide m6A methylome enriched by chemotherapy in OS stem cells (OSCs) has revealed that several m6A-related enzymes (METTL3, METTL14, FTO, ALKBH5) were altered in OS cells compared with noncancerous counterparts. The aberrantly expressed genes were associated with pluripotency regulation of the OSCs (Wang J. et al., 2019). Further, it has been shown that lncRNA plasmacytoma variant translocation 1 (PVT1) transcript was upregulated because of m6A methylation decrease mediated by ALKBH5, which reduced the binding of reader protein YTHDF2 in PVT1, subsequently lessening the degradation of PVT1 and promoting tumorigenesis of OS (Chen et al., 2020). METTL3 was found localizing in both cytoplasm and nucleus of OS cells, which is also on the map of OS modulation. Downregulation of METTL3 was reported to suppress the expression of ATPase family AAA domain containing 2 (ATAD2), in conjunction with inhibition of OS cell growth and metastasis (Zhou et al., 2020). In addition, knockdown of METTL3 has been demonstrated to be associated with decreased m6A methylation of GTP-binding protein (DRG) 1. Concomitant with the decreased m6A methylation, the stability and expression level of DRG1 were reduced (Ling et al., 2020), resulting in suppression of the OS development, migration, and colony formation. Meanwhile, silence of METTL3 decreased the m6A methylation and expression of lymphoid enhancer binding factor 1 (LEF1), followed by the advent of Wnt/β-catenin signaling pathway deactivation. Consequently, aberration of Wnt/β-catenin pathway contributed to the development of OS (Miao et al., 2019).
Collectively, based on the current evidences, the m6A writers, mainly the METTL3, are found extensively involved in the tumorigenesis, progression, and migration of OS via methylating their target genes. However, the current recognition is still restricted because of the limited number and depth of studies.
M6A in OA
OA is the most prevalent chronic joint disease that mainly occurs in aging and obesity population. The prevalence of OA is still in continuous growth, and it is estimated that the proportion of OA in population 45 years or older will increase from 26.6 to 29.5% by 2032 (Hunter and Bierma-Zeinstra, 2019). Symptoms of OA typically include stiffness, pain, and movement restriction with a high risk of disability, which can bring substantial socioeconomic burden (Hunter et al., 2014). The pathogenesis of OA principally involves in degradation of cartilage matrix, which consists of collagen type II, minor collagen types IX and XI, and gel-like negatively charged proteoglycans (Kannu et al., 2009). Likewise, many inflammation-related molecules have been suggested to engage in this process such as the growth factors [transforming growth factor β, fibroblast growth factor 2 (FGF-2), and FGF-18], Wnt, β-catenin, HIF-2α, etc. (Xia et al., 2014). Targeting the inflammation pathways is regarded as a promising way for OA therapy. Particularly, epigenetic regulation has been reported to be in connection with the inflammatory factors and response (Shen et al., 2017). As an indispensable way of epigenetic regulation, m6A has also been partially studied in OA.
It was expounded that METTL3 regulated OA process via enhancing inflammatory response and extracellular matrix (ECM) synthesis (Liu Q. et al., 2019). Mechanistically, silencing METTL3 inhibited the inflammatory cytokines level and nuclear factor κB signaling in OA cells, thereby deactivating the progression of OA. Meanwhile, suppression of METTL3 boosted the degradation of chondrocytes ECM through downregulating the matrix metalloprotease-13 and collagenase type X, consequently promoting the development of OA (Liu M. et al., 2019). In addition, the association between OA and FTO has also been explored in some genome-wide association studies in which they clarified that FTO-mediated overweight increased the susceptibility of OA (Zeggini et al., 2012; Panoutsopoulou et al., 2014). However, another study performed by Dai et al. (2018) showed that the FTO polymorphism (rs8044769) was not linked to OA in the Chinese Han population, and their association may be mediated by other genes. Therefore, the correlation between FTO and OA remains elusive and requires further exploration.
M6A in RA
With a symptom of pain, swelling, and stiffness, RA is a common chronic inflammatory disease that primarily attacks the synovial joint (Littlejohn and Monrad, 2018). RA can bring substantial burden to both the individuals and socioeconomics because of its high morbidity and mortality (Hu et al., 2018). Besides, RA is in close linkage with the occurrence of cardiovascular diseases (Blum and Adawi, 2019). Autoimmune-mediated inflammation is a well-known cause of RA (Derksen et al., 2017), in which both the genetic regulation (Scott et al., 2010) and epigenetic regulation (Doody et al., 2017) are proposed to play indispensable roles.
The relationship between m6A modification and RA has been partially unraveled in several studies. A large-scale genome-wide association study identifying the m6A-associated SNPs (m6A-SNPs) that affected the progression of RA has been conducted. Thirty-seven RA-related m6A-SNPs were discovered, and 27 of them were verified to affect expression of 24 local genes in different RA cells or tissues, which indicated the potential roles of m6A-SNPs in RA (Mo et al., 2018b). Moreover, METTL3 was validated to significantly suppress the inflammatory response of macrophages in RA. Specifically, METTL3 inhibited the generation of IL-6 and TNF-α in macrophages via restraining the phosphorylation of NF-κB. Therefore, METTL3 may serve as a potential biomarker for diagnosis of RA (Wang J. et al., 2019). However, because of the limitation of current studies, we can only hypothesize that m6A can decelerate the progression of RA via regulating the inflammatory response of immune cells; studies are still needed to provide more evidences.
M6A in OP
Characterizing by depletion of bone mass and impairment of bone structure, OP is mostly a condition of postmenopausal women and aging, which may lead to disastrous fracture in some cases (Armas and Recker, 2012; Boyanov et al., 2014).
Several researches have enhanced our recognition of the roles of m6A modification in OP. A genome-wide identification study was indicative of the potential roles of m6A-SNPs in bone mineral density. The results revealed that 138, 125, and 993 m6A-SNPs were in linkage to femoral neck density disorders, lumbar spine density disorders, and heel density disorders, respectively (Mo et al., 2018a). Currently, it becomes obvious that factors associated with regulation of BMSCs differentiation are closely associated with OP. The imbalance between BMSC-derived osteoblasts and adipocytes was deemed underlying progression of OP (Chen Q. et al., 2016). It has been demonstrated that METTL3-mediated m6A methylation affected the function of BMSCs through several pathways. In the first place, knockdown of m6A methyltransferase METTL3 in mice induced pathological features related to the occurrence of OP via decreasing parathyroid hormone (PTH)/parathyroid hormone receptor-1 (Pth1r) signaling axis, interfering the PTH-induced osteogenic response of BMSCs (Wu et al., 2018). Moreover, downregulation of METTL3 in BMSCs inhibited the methylation of runt-related transcription factor 2 (RUNX2) and precursor (pre-) miR320 (Yan et al., 2020). RUNX2 is an essential regulator of osteoblast progenitor proliferation and osteogenic differentiation, which can enhance the bone mineralization, and multiple factors were reported to participate in the osteogenic process via targeting RUNX2 (Hou et al., 2019; Komori, 2019).
Conversely, the FTO, represented as the RNA demethylase, was reported to promote the shift of osteoporotic BMSC fate to adipocyte and impede the bone formation through a growth differentiation factor 11 (GDF11)-FTO-peroxisome proliferator-activated receptor γ (Pparg) axis, whereby high FTO expression indicated high risk of OP (Shen et al., 2018). Meanwhile, the expression of FTO could be repressed by overexpression of miR-149-3p, followed by a higher potential of BMSCs to differentiate into adipocytes (Li et al., 2019b). In addition, of SNPs in multitude utilizing the association analyses, it was shown that a FTO such as rs1421085, rs1558902 and rs1121980 were associated with bone mineral density and risk of fracture (Guo et al., 2011; Tran et al., 2014). Interestingly, although FTO inhibited the BMSCs from differentiating into osteoblasts, it could exert a protective role in differentiated osteoblasts. Osteoblasts with FTO suppression were prone to develop cell death, which was illustrated to be caused by the interruption of DNA repair pathway. Specifically, FTO was able to stabilize the endoplasmic reticulum stress pathway components, such as Hsp70, protecting osteoblasts from genotoxic damage (Zhang Q. et al., 2019).
Taken together, m6A modification is involved in the occurrence and development of OP via (1) METTL3-mediated differentiation of BMSCs to osteocyte, (2) FTO-mediated differentiation of BMSCs to adipocyte, and (3) FTO-mediated protection of osteoblasts from genotoxic damage. Herein, we assume that m6A may play a dual role in OP, by which it can either promote or decelerate the progression of OP via different modifications.
Clinical Utilizations of M6A in Musculoskeletal Disorders
RNA target therapy has become a hotspot and shown convincing prospects in treatment of many diseases with high specificity and efficacy (Crooke et al., 2018). The broad involvement of m6A in musculoskeletal disorders, as outlined previously, has driven extensive research efforts at m6A-based therapy. The functions of the m6A regulatory proteins including FTO, METTL3, ALKBH5, etc. have been determined in OS, OA, RA, and OP as mentioned previously. As a result, the possibility is then opened for developing inhibitors or promoters of them to control the diseases.
Some natural products have been discovered showing significant activity in FTO inhibition. The natural product rhein is the first identified small-molecule inhibitor of human FTO demethylase, which competitively binds to the FTO active site and inhibit the demethylation in vitro (Chen et al., 2012). Additionally, by using the structure-based hierarchical virtual screening, researchers have found that entacapone directly bound to FTO and subsided its activity (Peng et al., 2019). And the natural compound radicicol also has been recognized as a potent FTO inhibitor, which suppressed the FTO demethylation activity in a dose-dependent manner (Wang et al., 2018). Besides, the meclofenamic acid (MA), a nonsteroidal anti-inflammatory drug, was identified as a specific inhibitor of FTO. Mechanically, MA competed with FTO for the binding sites within the m6A modified RNA, reducing activity of FTO-mediated demethylation (Huang et al., 2015). Further, FTO may interfere the reaction of human body to other drugs. For instance, it has been elucidated that the rs7195994 variant at the FTO gene locus hereditarily impacted the TNF inhibitor response in RA patients, and the customized treatment based on the FTO genetic stratification of patients could improve the efficacy (Massey et al., 2018).
Similarly, because METTL3 up-regulation in OS, OA, and RA contributes to the progression of diseases, METTL3 may also be targeted for treatment. Although METTL3 inhibitor is not available so far, it provides us with a novel direction. The fact that several drug screening technologies for RNA-modifying enzymes such as the self-assembled monolayer desorption/ionization have been developed illustrates a promising future for METTL3-based drugs (Buker et al., 2020). Recently, the first series of small molecule inhibitors of METTL3 have been identified via high-throughput docking into the SAM binding site and protein X-ray crystallography, in which seven compounds belonging to N-substituted amide of ribofuranuronic acid analogs of adenosine or adenosine mimics with a six-member ring were uncovered to be the effective METTL3 inhibitors (Bedi et al., 2020).
In summary, m6A-modifying proteins can serve as potential targets for drugs with which the FTO and METTL3 inhibitor may have great prospect in the treatment of musculoskeletal disorders.
Discussion
Epigenetics has been widely illuminated in multiple diseases over the past decades, and researchers are consistently seeking for new remedy from this field (Prachayasittikul et al., 2017). Currently, a diverse set of RNA modifications has been identified and annotated. Of note, m6A is the most abundant among them (Linder et al., 2015). Even though the recent advances have highlighted the crucial role of m6A in a multitude of diseases, only a small percentage of them focus on the musculoskeletal disorders. In this review, we have discussed the molecular mechanisms, detection technologies, regulatory functions, and clinical implications of m6A in musculoskeletal diseases. As we summarized previously, m6A modification is of great potential in disease prediction and drug development, yet the current studies are insufficient.
The first issue is how to choose and optimize the laboratory used technology for m6A detection in clinical practice. Although a diverse set of methods have been continually developed in experiment to detect the m6A modification or analyze the m6A residue locations within RNA site over the past decades, the use in clinic has not been investigated yet. Here, we envision that we can associate the disease progression with the m6A presence patterns that may be presented as the presence of m6A, the proportion of m6A, or the specific site of m6A. With the appropriate detection method, doctors will be able to figure out the situation of musculoskeletal diseases through the m6A examination results.
Furthermore, it has remained a significant challenge to dissect the mechanism of m6A in musculoskeletal disorders because limited studies have been performed toward it, especially for OA and RA. Meanwhile, the current studies of m6A in musculoskeletal disorders focus only on the “writers” and “erasers,” whereas clues toward the “readers” are scarce. As a matter of fact, roles of the “readers” have been investigated in other diseases. For instance, high YTHDF1 expression was a significant predictor of malignant tumor behaviors and poor prognosis in colorectal cancer (Nishizawa et al., 2018). IGF2BPs was demonstrated to participate in suppression of glycolysis and stemness properties of breast cancer cells via a FGF13-AS1/IGF2BPs/Myc feedback loop (Ma et al., 2019). Thus, the “readers” may also engage in the pathophysiological processes of musculoskeletal disorders.
Besides, m6A-based drugs remain poorly understood. Only a limited number of FTO and METTL3 inhibitors have been identified, yet their efficacy and safety are inconclusive. Notably, there are currently no m6A-based drugs developed for musculoskeletal disorders. Therefore, to address these limitations, we still have a long way to go.
Conclusion
As the most abundant RNA modification in eukaryotic cells, it is beyond doubt that m6A is a central node of the regulatory network of diseases. The illustration of m6A function has revealed its great importance in both the biological and pathological processes of bone and skeletal muscle. M6A modification has been partially studied in musculoskeletal disorders, including OS, OA, RA, OP, etc. Regarding its pivotal role in regulating the progression and development of diseases, m6A modification is of great potential to serve as the diagnostic biomarker or therapeutic target in musculoskeletal diseases, although more evidences are still warranted for validation in the future. Furthermore, a growing number of technologies have been developed for the m6A detection, and it has become evident that m6A is detectable and usable in disease prediction. Even with all of the effort over the recent years to figure out the detection methods for m6A, it should be noted that there is no clear indication for selecting the most suitable detection method for clinical application. Simplified m6A detection methods with high specificity/sensitivity and low costing, such as dot blot, immuno-Northern blot, and electrochemical immunosensor method, may be the promising methods to be utilized.
Author Contributions
WZ and CT conceived and designed the work. WZ, LH, and ZLiu contributed to material preparation and performed data collection and analysis. WZ wrote the first draft of the manuscript. XR, LQ, LW, and WW wrote sections of the manuscript. All authors commented on previous versions of the manuscript. CT revised the manuscript. ZLi contributed to manuscript drafting, critical revision, and final approval of the version to be published. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81902745), Natural Science Foundation of Hunan Province, China (2018JJ3716), and China Scholarship Council (201806375067).
Disclaimer
Figures of this review were created with the aid of Servier Medical Art (https://smart.servier.com/), reproduced under Creative Commons License attribution 3.0 Unported License.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aik, W., Scotti, J. S., Choi, H., Gong, L., Demetriades, M., Schofield, C. J., et al. (2014). Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 42, 4741–4754. doi: 10.1093/nar/gku085
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308. doi: 10.1016/j.cell.2015.08.011
Arguello, A. E., DeLiberto, A. N., and Kleiner, R. E. (2017). RNA Chemical Proteomics Reveals the N(6)-Methyladenosine (m(6)A)-Regulated Protein-RNA Interactome. J. Am. Chem. Soc. 139, 17249–17252. doi: 10.1021/jacs.7b09213
Armas, L. A., and Recker, R. R. (2012). Pathophysiology of osteoporosis: new mechanistic insights. Endocrinol. Metab. Clin. North Am. 41, 475–486. doi: 10.1016/j.ecl.2012.04.006
Bedi, R. K., Huang, D., Eberle, S. A., Wiedmer, L., Śledź, P., and Caflisch, A. (2020). Small-molecule inhibitors of METTL3, the major human epitranscriptomic writer. ChemMedChem 15, 744–748. doi: 10.1002/cmdc.202000011
Blair, H. C., Larrouture, Q. C., Li, Y., Lin, H., Beer-Stoltz, D., Liu, L., et al. (2017). Osteoblast differentiation and bone matrix formation in vivo and in vitro. Tissue Eng. Part B Rev. 23, 268–280. doi: 10.1089/ten.TEB.2016.0454
Blum, A., and Adawi, M. (2019). Rheumatoid arthritis (RA) and cardiovascular disease. Autoimmun. Rev. 18, 679–690. doi: 10.1016/j.autrev.2019.05.005
Boyanov, M., Shinkov, A., Psachoulia, E., Intorcia, M., and Petkova, R. (2014). Increased bone mineral density (Bmd) in postmenopausal women with osteoporosis (Op) receiving two denosumab injections in routine clinical practice in bulgaria. Value Health 17:A389. doi: 10.1016/j.jval.2014.08.2662
Buker, S. M., Gurard-Levin, Z. A., Wheeler, B. D., Scholle, M. D., Case, A. W., Hirsch, J. L., et al. (2020). A mass spectrometric assay of METTL3/METTL14 methyltransferase activity. SLAS Discov. 25, 361–371. doi: 10.1177/2472555219878408
Cao, G., Li, H. B., Yin, Z., and Flavell, R. A. (2016). Recent advances in dynamic m6A RNA modification. Open Biol. 6:160003. doi: 10.1098/rsob.160003
Charles, J. F., and Aliprantis, A. O. (2014). Osteoclasts: more than ‘bone eaters’. Trends Mol. Med. 20, 449–459. doi: 10.1016/j.molmed.2014.06.001
Chen, B., Ye, F., Yu, L., Jia, G., Huang, X., Zhang, X., et al. (2012). Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 134, 17963–17971. doi: 10.1021/ja3064149
Chen, Q., Shou, P., Zheng, C., Jiang, M., Cao, G., Yang, Q., et al. (2016). Fate decision of mesenchymal stem cells: Adipocytes or osteoblasts? Cell Death Differ. 23, 1128–1139. doi: 10.1038/cdd.2015.168
Chen, S., Zhou, L., and Wang, Y. (2020). ALKBH5-mediated m(6)A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 20:34. doi: 10.1186/s12935-020-1105-6
Chen, W., Feng, P., Ding, H., and Lin, H. (2016). Identifying N (6)-methyladenosine sites in the Arabidopsis thaliana transcriptome. Mol. Genet. Genomics 291, 2225–2229. doi: 10.1007/s00438-016-1243-7
Chen, X. Y., Zhang, J., and Zhu, J. S. (2019). The role of m(6)A RNA methylation in human cancer. Mol. Cancer 18:103. doi: 10.1186/s12943-019-1033-z
Corre, I., Verrecchia, F., Crenn, V., Redini, F., and Trichet, V. (2020). The osteosarcoma microenvironment: a complex but targetable ecosystem. Cells 9:976. doi: 10.3390/cells9040976
Crooke, S. T., Witztum, J. L., Bennett, C. F., and Baker, B. F. (2018). RNA-Targeted Therapeutics. Cell Metab. 27, 714–739. doi: 10.1016/j.cmet.2018.03.004
Cui, J., Wang, W., Li, Z., Zhang, Z., Wu, B., and Zeng, L. (2011). Epigenetic changes in osteosarcoma. Bull. Cancer 98, E62–E68. doi: 10.1684/bdc.2011.1400
Dai, J., Ying, P., Shi, D., Hou, H., Sun, Y., Xu, Z., et al. (2018). FTO variant is not associated with osteoarthritis in the Chinese Han population: replication study for a genome-wide association study identified risk loci. J. Orthop. Surg. Res. 13:65. doi: 10.1186/s13018-018-0769-2
Derksen, V., Huizinga, T. W. J., and van der Woude, D. (2017). The role of autoantibodies in the pathophysiology of rheumatoid arthritis. Semin. Immunopathol. 39, 437–446. doi: 10.1007/s00281-017-0627-z
Doaei, S., Kalantari, N., Mohammadi, N. K., Izadi, P., Gholamalizadeh, M., Eini-Zinab, H., et al. (2019). Up-regulation of FTO gene expression was associated with increase in skeletal muscle mass in overweight male adolescents. Arch. Med. Sci. 15, 1133–1137. doi: 10.5114/aoms.2019.87239
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Doody, K. M., Bottini, N., and Firestein, G. S. (2017). Epigenetic alterations in rheumatoid arthritis fibroblast-like synoviocytes. Epigenomics 9, 479–492. doi: 10.2217/epi-2016-0151
Frontera, W. R., and Ochala, J. (2015). Skeletal muscle: a brief review of structure and function. Calcif. Tissue Int. 96, 183–195. doi: 10.1007/s00223-014-9915-y
Fu, Y., Jia, G., Pang, X., Wang, R. N., Wang, X., Li, C. J., et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 4:1798. doi: 10.1038/ncomms2822
Geuens, T., Bouhy, D., and Timmerman, V. (2016). The hnRNP family: insights into their role in health and disease. Hum. Genet. 135, 851–867. doi: 10.1007/s00439-016-1683-5
Gianferante, D. M., Mirabello, L., and Savage, S. A. (2017). Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat. Rev. Endocrinol. 13, 480–491. doi: 10.1038/nrendo.2017.16
Golovina, A. Y., Dzama, M. M., Petriukov, K. S., Zatsepin, T. S., Sergiev, P. V., Bogdanov, A. A., et al. (2014). Method for site-specific detection of m6A nucleoside presence in RNA based on high-resolution melting (HRM) analysis. Nucleic Acids Res. 42:e27. doi: 10.1093/nar/gkt1160
Guo, Y., Liu, H., Yang, T. L., Li, S. M., Li, S. K., Tian, Q., et al. (2011). The fat mass and obesity associated gene, FTO, is also associated with osteoporosis phenotypes. PLoS One 6:e27312. doi: 10.1371/journal.pone.0027312
Hadjidakis, D. J., and Androulakis, I. I. (2006). Bone remodeling. Ann. N. Y. Acad. Sci. 1092, 385–396. doi: 10.1196/annals.1365.035
Harvey, Z. H., Chen, Y., and Jarosz, D. F. (2018). Protein-based inheritance: epigenetics beyond the chromosome. Mol. Cell 69, 195–202. doi: 10.1016/j.molcel.2017.10.030
He, C., Liu, C., Wang, L., Sun, Y., Jiang, Y., and Hao, Y. (2019). Histone methyltransferase NSD2 regulates apoptosis and chemosensitivity in osteosarcoma. Cell Death Dis. 10:65. doi: 10.1038/s41419-019-1347-1
Hou, Z., Wang, Z., Tao, Y., Bai, J., Yu, B., Shen, J., et al. (2019). KLF2 regulates osteoblast differentiation by targeting of Runx2. Lab. Invest. 99, 271–280. doi: 10.1038/s41374-018-0149-x
Hu, H., Luan, L., Yang, K., and Li, S. C. (2018). Burden of rheumatoid arthritis from a societal perspective: a prevalence-based study on cost of this illness for patients in China. Int. J. Rheum. Dis. 21, 1572–1580. doi: 10.1111/1756-185x.13028
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 43, 373–384. doi: 10.1093/nar/gku1276
Hunter, D. J., and Bierma-Zeinstra, S. (2019). Osteoarthritis. Lancet 393, 1745–1759. doi: 10.1016/s0140-6736(19)30417-9
Hunter, D. J., Schofield, D., and Callander, E. (2014). The individual and socioeconomic impact of osteoarthritis. Nat. Rev. Rheumatol. 10, 437–441. doi: 10.1038/nrrheum.2014.44
Imanishi, M., Tsuji, S., Suda, A., and Futaki, S. (2017). Detection of N(6)-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. 53, 12930–12933. doi: 10.1039/c7cc07699a
Jiang, C., He, C., Wu, Z., Li, F., and Xiao, J. (2018). Histone methyltransferase SETD2 regulates osteosarcoma cell growth and chemosensitivity by suppressing Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 502, 382–388. doi: 10.1016/j.bbrc.2018.05.176
Kannu, P., Bateman, J. F., Belluoccio, D., Fosang, A. J., and Savarirayan, R. (2009). Employing molecular genetics of chondrodysplasias to inform the study of osteoarthritis. Arthritis Rheum. 60, 325–334. doi: 10.1002/art.24251
Kasowitz, S. D., Ma, J., Anderson, S. J., Leu, N. A., Xu, Y., Gregory, B. D., et al. (2018). Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 14:e1007412. doi: 10.1371/journal.pgen.1007412
Kierzek, E., and Kierzek, R. (2003). The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 31, 4472–4480. doi: 10.1093/nar/gkg633
Kim, T., Ha, H. I., Kim, N., Yi, O., Lee, S. H., and Choi, Y. (2009). Adrm1 interacts with Atp6v0d2 and regulates osteoclast differentiation. Biochem. Biophys. Res. Commun. 390, 585–590. doi: 10.1016/j.bbrc.2009.10.010
Komori, T. (2019). Regulation of proliferation, differentiation and functions of osteoblasts by Runx2. Int. J. Mol. Sci. 20:1694. doi: 10.3390/ijms20071694
Kudou, K., Komatsu, T., Nogami, J., Maehara, K., Harada, A., Saeki, H., et al. (2017). The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation. Open Biol. 7:170119. doi: 10.1098/rsob.170119
Lan, Q., Liu, P. Y., Haase, J., Bell, J. L., Hüttelmaier, S., and Liu, T. (2019). The critical role of RNA m(6)A methylation in cancer. Cancer Res. 79, 1285–1292. doi: 10.1158/0008-5472.Can-18-2965
Li, D., Cai, L., Meng, R., Feng, Z., and Xu, Q. (2020). METTL3 modulates osteoclast differentiation and function by controlling RNA stability and nuclear Export. Int. J. Mol. Sci. 21:1660. doi: 10.3390/ijms21051660
Li, M., Zhao, X., Wang, W., Shi, H., Pan, Q., Lu, Z., et al. (2018). Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 19:69. doi: 10.1186/s13059-018-1436-y
Li, Q., Li, H., Zhao, X., Wang, B., Zhang, L., Zhang, C., et al. (2017). DNA methylation mediated downregulation of miR-449c controls osteosarcoma cell cycle progression by directly targeting oncogene c-Myc. Int. J. Biol. Sci. 13, 1038–1050. doi: 10.7150/ijbs.19476
Li, X., Lu, H., Fan, G., He, M., Sun, Y., Xu, K., et al. (2017). A novel interplay between HOTAIR and DNA methylation in osteosarcoma cells indicates a new therapeutic strategy. J. Cancer Res. Clin. Oncol. 143, 2189–2200. doi: 10.1007/s00432-017-2478-3
Li, X., Yang, J., Zhu, Y., Liu, Y., Shi, X., and Yang, G. (2016). Mouse maternal high-fat intake dynamically programmed mRNA m6a modifications in adipose and skeletal muscle tissues in offspring. Int. J. Mol. Sci. 17:1336. doi: 10.3390/ijms17081336
Li, Y., Wu, K., Quan, W., Yu, L., Chen, S., Cheng, C., et al. (2019a). The dynamics of FTO binding and demethylation from the m(6)A motifs. RNA Biol. 16, 1179–1189. doi: 10.1080/15476286.2019.1621120
Li, Y., Yang, F., Gao, M., Gong, R., Jin, M., Liu, T., et al. (2019b). miR-149-3p regulates the switch between adipogenic and osteogenic differentiation of BMSCs by targeting FTO. Mol. Ther. Nucleic Acids 17, 590–600. doi: 10.1016/j.omtn.2019.06.023
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772. doi: 10.1038/nmeth.3453
Ling, Z., Chen, L., and Zhao, J. (2020). m6A-dependent up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth, migration, and colony formation in osteosarcoma. Biosci. Rep. 40:BSR20200282. doi: 10.1042/bsr20200282
Littlejohn, E. A., and Monrad, S. U. (2018). Early diagnosis and treatment of rheumatoid arthritis. Prim. Care 45, 237–255. doi: 10.1016/j.pop.2018.02.010
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, M., Li, B., Peng, W., Ma, Y., Huang, Y., Lan, X., et al. (2019). LncRNA-MEG3 promotes bovine myoblast differentiation by sponging miR-135. J. Cell. Physiol. 234, 18361–18370. doi: 10.1002/jcp.28469
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. doi: 10.1038/nature14234
Liu, N., Parisien, M., Dai, Q., Zheng, G., He, C., and Pan, T. (2013). Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856. doi: 10.1261/rna.041178.113
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063. doi: 10.1093/nar/gkx141
Liu, Q., Li, M., Jiang, L., Jiang, R., and Fu, B. (2019). METTL3 promotes experimental osteoarthritis development by regulating inflammatory response and apoptosis in chondrocyte. Biochem. Biophys. Res. Commun. 516, 22–27. doi: 10.1016/j.bbrc.2019.05.168
Ma, F., Liu, X., Zhou, S., Li, W., Liu, C., Chadwick, M., et al. (2019). Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 450, 63–75. doi: 10.1016/j.canlet.2019.02.008
Machado, L., Esteves de Lima, J., Fabre, O., Proux, C., Legendre, R., Szegedi, A., et al. (2017). In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993. doi: 10.1016/j.celrep.2017.10.080
Madan, I., and Grime, P. R. (2015). The management of musculoskeletal disorders in the workplace. Best Pract. Res. Clin. Rheumatol. 29, 345–355. doi: 10.1016/j.berh.2015.03.002
Mao, Y., Dong, L., Liu, X. M., Guo, J., Ma, H., Shen, B., et al. (2019). m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 10:5332. doi: 10.1038/s41467-019-13317-9
Massey, J., Plant, D., Hyrich, K., Morgan, A. W., Wilson, A. G., Spiliopoulou, A., et al. (2018). Genome-wide association study of response to tumour necrosis factor inhibitor therapy in rheumatoid arthritis. Pharmacogenomics J. 18, 657–664. doi: 10.1038/s41397-018-0040-6
McIntyre, A. B. R., Gokhale, N. S., Cerchietti, L., Jaffrey, S. R., Horner, S. M., and Mason, C. E. (2020). Limits in the detection of m(6)A changes using MeRIP/m(6)A-seq. Sci. Rep. 10:6590. doi: 10.1038/s41598-020-63355-3
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’, UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Miao, W., Chen, J., Jia, L., Ma, J., and Song, D. (2019). The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem. Biophys. Res. Commun. 516, 719–725. doi: 10.1016/j.bbrc.2019.06.128
Mishima, E., Jinno, D., Akiyama, Y., Itoh, K., Nankumo, S., Shima, H., et al. (2015). Immuno-northern blotting: detection of RNA modifications by using antibodies against modified nucleosides. PLoS One 10:e0143756. doi: 10.1371/journal.pone.0143756
Miyata, K., Miyata, T., Nakabayashi, K., Okamura, K., Naito, M., Kawai, T., et al. (2015). DNA methylation analysis of human myoblasts during in vitro myogenic differentiation: de novo methylation of promoters of muscle-related genes and its involvement in transcriptional down-regulation. Hum. Mol. Genet. 24, 410–423. doi: 10.1093/hmg/ddu457
Mo, X. B., Zhang, Y. H., and Lei, S. F. (2018a). Genome-wide identification of m(6)A-associated SNPs as potential functional variants for bone mineral density. Osteoporos Int. 29, 2029–2039. doi: 10.1007/s00198-018-4573-y
Mo, X. B., Zhang, Y. H., and Lei, S. F. (2018b). Genome-Wide Identification of N(6)-Methyladenosine (m(6)A) SNPs associated with rheumatoid arthritis. Front. Genet. 9:299. doi: 10.3389/fgene.2018.00299
Molinie, B., Wang, J., Lim, K. S., Hillebrand, R., Lu, Z. X., Van Wittenberghe, N., et al. (2016). m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods 13, 692–698. doi: 10.1038/nmeth.3898
Nagarajan, A., Janostiak, R., and Wajapeyee, N. (2019). Dot Blot Analysis for Measuring Global N(6)-Methyladenosine Modification of RNA. Methods Mol. Biol. 1870, 263–271. doi: 10.1007/978-1-4939-8808-2_20
Nebbioso, A., Tambaro, F. P., Dell’Aversana, C., and Altucci, L. (2018). Cancer epigenetics: moving forward. PLoS Genet. 14:e1007362. doi: 10.1371/journal.pgen.1007362
Ni, W., Yao, S., Zhou, Y., Liu, Y., Huang, P., Zhou, A., et al. (2019). Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer 18:143. doi: 10.1186/s12943-019-1079-y
Nishizawa, Y., Konno, M., Asai, A., Koseki, J., Kawamoto, K., Miyoshi, N., et al. (2018). Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget 9, 7476–7486. doi: 10.18632/oncotarget.23554
Ovcharenko, A., and Rentmeister, A. (2018). Emerging approaches for detection of methylation sites in RNA. Open Biol. 8:180121. doi: 10.1098/rsob.180121
Panoutsopoulou, K., Metrustry, S., Doherty, S. A., Laslett, L. L., Maciewicz, R. A., Hart, D. J., et al. (2014). The effect of FTO variation on increased osteoarthritis risk is mediated through body mass index: a Mendelian randomisation study. Ann. Rheum. Dis. 73, 2082–2086. doi: 10.1136/annrheumdis-2013-203772
Peng, S., Xiao, W., Ju, D., Sun, B., Hou, N., Liu, Q., et al. (2019). Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 11:eaau7116. doi: 10.1126/scitranslmed.aau7116
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Prachayasittikul, V., Prathipati, P., Pratiwi, R., Phanus-Umporn, C., Malik, A. A., Schaduangrat, N., et al. (2017). Exploring the epigenetic drug discovery landscape. Expert Opin. Drug Discov. 12, 345–362. doi: 10.1080/17460441.2017.1295954
Qin, L., Min, S., Shu, L., Pan, H., Zhong, J., Guo, J., et al. (2020). Genetic analysis of N6-methyladenosine modification genes in Parkinson’s disease. Neurobiol. Aging. 93, 143.e9–143.e13. doi: 10.1016/j.neurobiolaging.2020.03.018
Reichel, M., Köster, T., and Staiger, D. (2019). Marking RNA: m6A writers, readers, and functions in Arabidopsis. J. Mol. Cell Biol. 11, 899–910. doi: 10.1093/jmcb/mjz085
Ren, X., He, J., Qi, L., Li, S., Zhang, C., Duan, Z., et al. (2020). Prognostic and clinicopathologic significance of long non-coding RNA opa-interacting protein 5-antisense RNA 1 in multiple human cancers. Artif. Cells Nanomed. Biotechnol. 48, 353–361. doi: 10.1080/21691401.2019.1709854
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 6:e31311. doi: 10.7554/eLife.31311
Sampath, S. C., Sampath, S. C., and Millay, D. P. (2018). Myoblast fusion confusion: the resolution begins. Skelet. Muscle 8:3. doi: 10.1186/s13395-017-0149-3
Scott, D. L., Wolfe, F., and Huizinga, T. W. (2010). Rheumatoid arthritis. Lancet 376, 1094–1108. doi: 10.1016/s0140-6736(10)60826-4
Shen, G. S., Zhou, H. B., Zhang, H., Chen, B., Liu, Z. P., Yuan, Y., et al. (2018). The GDF11-FTO-PPARγ axis controls the shift of osteoporotic MSC fate to adipocyte and inhibits bone formation during osteoporosis. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3644–3654. doi: 10.1016/j.bbadis.2018.09.015
Shen, J., Abu-Amer, Y., O’Keefe, R. J., and McAlinden, A. (2017). Inflammation and epigenetic regulation in osteoarthritis. Connect Tissue Res. 58, 49–63. doi: 10.1080/03008207.2016.1208655
Sorci, M., Ianniello, Z., Cruciani, S., Larivera, S., Ginistrelli, L. C., Capuano, E., et al. (2018). METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 9:796. doi: 10.1038/s41419-018-0843-z
Tian, C., Huang, Y., Li, Q., Feng, Z., and Xu, Q. (2019). Mettl3 regulates osteogenic differentiation and alternative splicing of vegfa in bone marrow mesenchymal stem cells. Int. J. Mol. Sci. 20:551. doi: 10.3390/ijms20030551
Tran, B., Nguyen, N. D., Center, J. R., Eisman, J. A., and Nguyen, T. V. (2014). Association between fat-mass-and-obesity-associated (FTO) gene and hip fracture susceptibility. Clin. Endocrinol. 81, 210–217. doi: 10.1111/cen.12335
Tresguerres, F. G. F., Torres, J., López-Quiles, J., Hernández, G., Vega, J. A., and Tresguerres, I. F. (2020). The osteocyte: a multifunctional cell within the bone. Ann. Anat. 227:151422. doi: 10.1016/j.aanat.2019.151422
Tu, C., He, J., Chen, R., and Li, Z. (2019). The emerging role of exosomal non-coding RNAs in musculoskeletal diseases. Curr. Pharm. Des. 25, 4523–4535. doi: 10.2174/1381612825666191113104946
Tu, C., He, J., Qi, L., Ren, X., Zhang, C., Duan, Z., et al. (2020). Emerging landscape of circular RNAs as biomarkers and pivotal regulators in osteosarcoma. J. Cell. Physiol. doi: 10.1002/jcp.29754 [Epub ahead of print].
van Wijnen, A. J., and Westendorf, J. J. (2019). Epigenetics as a new frontier in orthopedic regenerative medicine and oncology. J. Orthop. Res. 37, 1465–1474. doi: 10.1002/jor.24305
Wang, J., Yan, S., Lu, H., Wang, S., and Xu, D. (2019). METTL3 attenuates LPS-induced inflammatory response in macrophages via NF-κB signaling pathway. Mediators Inflamm. 2019:3120391. doi: 10.1155/2019/3120391
Wang, R., Han, Z., Liu, B., Zhou, B., Wang, N., Jiang, Q., et al. (2018). Identification of natural compound radicicol as a Potent FTO inhibitor. Mol. Pharm. 15, 4092–4098. doi: 10.1021/acs.molpharmaceut.8b00522
Wang, T., Kong, S., Tao, M., and Ju, S. (2020). The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer 19:88. doi: 10.1186/s12943-020-01204-7
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578. doi: 10.1038/nature18298
Wang, X., Huang, N., Yang, M., Wei, D., Tai, H., Han, X., et al. (2017). FTO is required for myogenesis by positively regulating mTOR-PGC-1α pathway-mediated mitochondria biogenesis. Cell Death Dis. 8:e2702. doi: 10.1038/cddis.2017.122
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wang, Y., Zeng, L., Liang, C., Zan, R., Ji, W., Zhang, Z., et al. (2019). Integrated analysis of transcriptome-wide m(6)A methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics 11, 1693–1715. doi: 10.2217/epi-2019-0262
Weivoda, M. M., Ruan, M., Pederson, L., Hachfeld, C., Davey, R. A., Zajac, J. D., et al. (2016). Osteoclast TGF-β Receptor Signaling Induces Wnt1 Secretion and Couples Bone Resorption to Bone Formation. J. Bone Miner. Res. 31, 76–85. doi: 10.1002/jbmr.2586
Wu, W., Feng, J., Jiang, D., Zhou, X., Jiang, Q., Cai, M., et al. (2017). AMPK regulates lipid accumulation in skeletal muscle cells through FTO-dependent demethylation of N(6)-methyladenosine. Sci. Rep. 7:41606. doi: 10.1038/srep41606
Wu, Y., Xie, L., Wang, M., Xiong, Q., Guo, Y., Liang, Y., et al. (2018). Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 9:4772. doi: 10.1038/s41467-018-06898-4
Xia, B., Di, C., Zhang, J., Hu, S., Jin, H., and Tong, P. (2014). Osteoarthritis pathogenesis: a review of molecular mechanisms. Calcif. Tissue Int. 95, 495–505. doi: 10.1007/s00223-014-9917-9
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y., et al. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929. doi: 10.1038/nchembio.1654
Xu, S., Gong, Y., Yin, Y., Xing, H., and Zhang, N. (2020). The multiple function of long noncoding RNAs in osteosarcoma progression, drug resistance and prognosis. Biomed. Pharmacother. 127:110141. doi: 10.1016/j.biopha.2020.110141
Yan, G., Yuan, Y., He, M., Gong, R., Lei, H., Zhou, H., et al. (2020). m(6)A methylation of precursor-miR-320/RUNX2 controls osteogenic potential of bone marrow-derived mesenchymal stem cells. Mol. Ther. Nucleic Acids 19, 421–436. doi: 10.1016/j.omtn.2019.12.001
Yin, H., Wang, H., Jiang, W., Zhou, Y., and Ai, S. (2017). Electrochemical immunosensor for N6-methyladenosine detection in human cell lines based on biotin-streptavidin system and silver-SiO(2) signal amplification. Biosens. Bioelectron. 90, 494–500. doi: 10.1016/j.bios.2016.10.066
Yu, J., Shen, L., Liu, Y., Ming, H., Zhu, X., Chu, M., et al. (2020). The m6A methyltransferase METTL3 cooperates with demethylase ALKBH5 to regulate osteogenic differentiation through NF-κB signaling. Mol. Cell. Biochem. 463, 203–210. doi: 10.1007/s11010-019-03641-5
Zeggini, E., Panoutsopoulou, K., Southam, L., Rayner, N. W., Day-Williams, A. G., Lopes, M. C., et al. (2012). Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 380, 815–823. doi: 10.1016/s0140-6736(12)60681-3
Zhang, H., Shi, X., Huang, T., Zhao, X., Chen, W., Gu, N., et al. (2020). Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 19:88. doi: 10.1093/nar/gkaa347
Zhang, Q., Riddle, R. C., Yang, Q., Rosen, C. R., Guttridge, D. C., Dirckx, N., et al. (2019). The RNA demethylase FTO is required for maintenance of bone mass and functions to protect osteoblasts from genotoxic damage. Proc. Natl. Acad. Sci. U.S.A. 116, 17980–17989. doi: 10.1073/pnas.1905489116
Zhang, Y., Gu, X., Li, D., Cai, L., and Xu, Q. (2019). METTL3 regulates osteoblast differentiation and inflammatory response via Smad Signaling and MAPK Signaling. Int. J. Mol. Sci. 21:199. doi: 10.3390/ijms21010199
Zhou, K. I., Shi, H., Lyu, R., Wylder, A. C., Matuszek, Ż., Pan, J. N., et al. (2019). Regulation of Co-transcriptional Pre-mRNA Splicing by m(6)A through the Low-Complexity Protein hnRNPG. Mol. Cell 76, 70–81.e9. doi: 10.1016/j.molcel.2019.07.005
Zhou, L., Yang, C., Zhang, N., Zhang, X., Zhao, T., and Yu, J. (2020). Silencing METTL3 inhibits the proliferation and invasion of osteosarcoma by regulating ATAD2. Biomed. Pharmacother. 125:109964. doi: 10.1016/j.biopha.2020.109964
Zhu, W., Wang, J. Z., Xu, Z., Cao, M., Hu, Q., Pan, C., et al. (2019). Detection of N6-methyladenosine modification residues (Review). Int. J. Mol. Med. 43, 2267–2278. doi: 10.3892/ijmm.2019.4169
Zhu, Z. M., Huo, F. C., and Pei, D. S. (2020). Function and evolution of RNA N6-methyladenosine modification. Int. J. Biol. Sci. 16, 1929–1940. doi: 10.7150/ijbs.45231
Zong, X., Zhao, J., Wang, H., Lu, Z., Wang, F., Du, H., et al. (2019). Mettl3 deficiency sustains long-chain fatty acid absorption through suppressing Traf6-dependent inflammation response. J. Immunol. 202, 567–578. doi: 10.4049/jimmunol.1801151
Keywords: RNA N6-methyladenosine, METTL3, FTO, musculoskeletal disorders, epigenetics
Citation: Zhang W, He L, Liu Z, Ren X, Qi L, Wan L, Wang W, Tu C and Li Z (2020) Multifaceted Functions and Novel Insight Into the Regulatory Role of RNA N6-Methyladenosine Modification in Musculoskeletal Disorders. Front. Cell Dev. Biol. 8:870. doi: 10.3389/fcell.2020.00870
Received: 30 June 2020; Accepted: 11 August 2020;
Published: 02 September 2020.
Edited by:
Changjun Li, Central South University, ChinaReviewed by:
Zhengwei Yan, First Affiliated Hospital of Nanchang University, ChinaAmelia Eva Aranega, University of Jaén, Spain
Copyright © 2020 Zhang, He, Liu, Ren, Qi, Wan, Wang, Tu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Tu, tuchao@csu.edu.cn; Zhihong Li, lizhihong@csu.edu.cn
 Wenchao Zhang
Wenchao Zhang Lile He
Lile He Zhongyue Liu
Zhongyue Liu Xiaolei Ren
Xiaolei Ren Lin Qi
Lin Qi Lu Wan
Lu Wan Wanchun Wang
Wanchun Wang Chao Tu
Chao Tu Zhihong Li
Zhihong Li