Tipping the Scales With Zebrafish to Understand Adaptive Tumor Immunity
- 1Department of Pharmacology & Experimental Therapeutics, Boston University School of Medicine, Boston, MA, United States
- 2Department of Medicine, Section of Hematology and Medical Oncology, Boston University School of Medicine, Boston, MA, United States
The future of improved immunotherapy against cancer depends on an in-depth understanding of the dynamic interactions between the immune system and tumors. Over the past two decades, the zebrafish has served as a valuable model system to provide fresh insights into both the development of the immune system and the etiologies of many different cancers. This well-established foundation of knowledge combined with the imaging and genetic capacities of the zebrafish provides a new frontier in cancer immunology research. In this review, we provide an overview of the development of the zebrafish immune system along with a side-by-side comparison of its human counterpart. We then introduce components of the adaptive immune system with a focus on their roles in the tumor microenvironment (TME) of teleosts. In addition, we summarize zebrafish models developed for the study of cancer and adaptive immunity along with other available tools and technology afforded by this experimental system. Finally, we discuss some recent research conducted using the zebrafish to investigate adaptive immune cell-tumor interactions. Without a doubt, the zebrafish will arise as one of the driving forces to help expand the knowledge of tumor immunity and facilitate the development of improved anti-cancer immunotherapy in the foreseeable future.
Introduction
Dr. Harold F. Dvorak wrote in 1986 that solid tumors are comprised of two discrete compartments, the malignant cells and the stroma in which they are dispersed, creating an environment that resembles a “wound that does not heal” (Dvorak, 1986). One challenge in cancer treatment–healing the “wound”–stems from the difficulty in fully understanding the mechanisms by which cancer cells escape immunosurveillance in the tumor microenvironment (TME). In recent years, immunotherapy has made revolutionary advances in our war against cancer. The successful development and application of a newer generation of cancer immunotherapy, such as immune checkpoint blockade and chimeric antigen receptor T cell (CAR-T), have led to improved outcomes in cancers including hematologic malignancies, melanoma, lymphomas, and lung cancers (Tang et al., 2017; Gong et al., 2018). Despite these progresses, immunologically “cold” tumors, encompassing a broad spectrum of solid tumors such as breast cancer, pancreatic cancer, neuroblastoma, and glioblastoma, present unique challenges for immunotherapy (Bonaventura et al., 2019). Some of the underlying causes are the lack of tumor antigens, a deficit in antigen-presenting B cells, and/or impaired trafficking of activated T cells into the TME (Bonaventura et al., 2019). For immunotherapies to become highly effective for cancers, the following challenges must be addressed: (1) enhancement of major histocompatibility complex (MHC) expression in tumor cells for sufficient presentation of tumor-associated antigens and (2) regulation of cytokines and manipulation of the TME to improve effector T-cell infiltration into “cold” tumors (Stambrook et al., 2017).
For decades, mice have represented the primary animal model and major contributor to cancer research including tumor immunity research (Callahan et al., 2014). However, this model has limitations. One major drawback is that drugs’ effectiveness and safety evaluated by pre-clinical murine models often cannot be reproduced in clinical trials (Rossa and D’Silva, 2019). This issue calls for the inclusion of additional animal models to obtain preclinical data that can be replicated in multiple systems in order to boost the success rates of clinical testing. Another drawback of the murine model is the difference in telomerase between human and mouse cells (Cheon and Orsulic, 2011). Specifically, most mouse cells have active telomerase throughout adulthood while human adult cells have largely inactive telomerase (Cheon and Orsulic, 2011). Interestingly, the zebrafish possesses human-like telomeres, which gradually decline with age, allowing its use to replicate similar genomic instabilities seen in humans (Carneiro et al., 2016). Another unique advantage of the zebrafish is that they are raised in non-sterile conditions, making them physiologically relevant to study immune responses (Iwanami et al., 2017). Coupled with their imaging capacities, the zebrafish enables real-time and non-invasive monitoring of tumor-immune cell interactions through differential fluorescent labeling of cells.
Through exhaustive comparison of the human and zebrafish genome assemblies, it was found that 82% of disease-causing human genes have at least one zebrafish ortholog (Howe et al., 2013). This level of conservation has led to the development of a wide array of cancer models in zebrafish (Casey and Stewart, 2018; Hason and Bartůněk, 2019; Casey and Stewart, 2020; Elliot, 2020). Through the years of ongoing research, the pathological similarities of cancer and preservation of the immune components have also been well established between zebrafish and humans. For instance, many oncogenic pathways are conserved in zebrafish along with similar development processes in hematopoiesis and the immune system (Traver et al., 2003; Davidson and Zon, 2004; White et al., 2013; Gore et al., 2018; Casey and Stewart, 2020). Hence, the zebrafish represents a physiologically relevant model system for tumor immunity research. Here, we summarize the suitability and unique advantages of the zebrafish in investigating adaptive tumor immunity. We also provide a few examples of early studies to demonstrate its feasibility in expanding the knowledge of this field and its great potentials in advancing cancer immunotherapy.
In the Same Sea: High Conservation of the Immune System
The suitability of using the zebrafish to study tumor immunity stems from the fact that hematopoiesis and most immune components are highly conserved between humans and zebrafish, beginning from the embryonic stage into adulthood (Figure 1). Similar to humans, the zebrafish immune system is divided into two main branches: innate and adaptive. Despite some differences in the locations and timing of immune cell development, many pathways in primitive and definitive hematopoiesis, lymphocyte differentiation, along with key markers that define specific groups of immune cells are shared between zebrafish and humans (Traver et al., 2003; Iwanami, 2014; Gore et al., 2018).
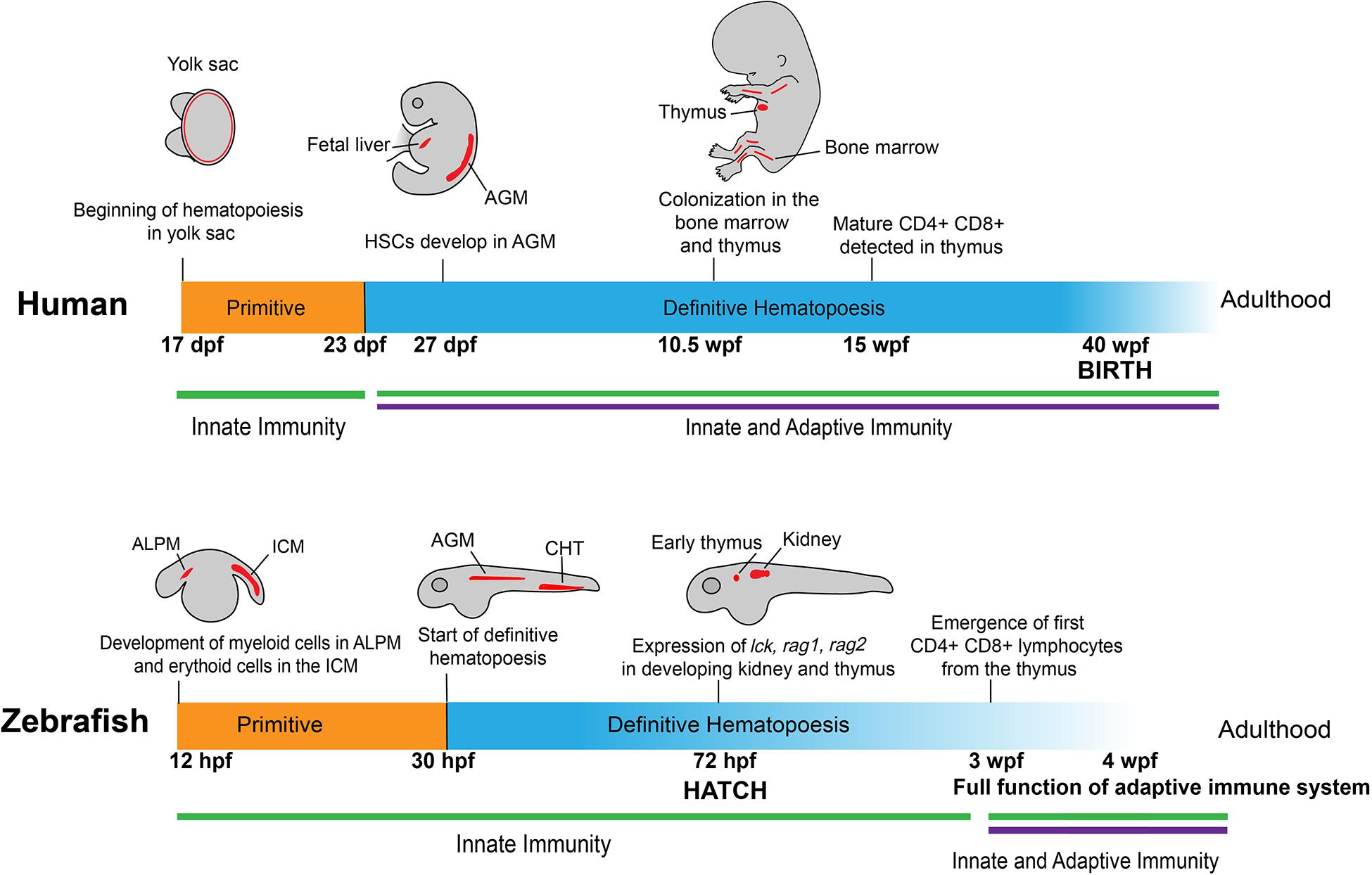
Figure 1. Development of human and zebrafish immune systems. The development of the immune system starts with hematopoiesis at ∼17 dpf in humans and 12 hpf in the zebrafish, with myeloid and erythroid cells arising in the ALPM and ICM, respectively (Jagannathan-Bogdan and Zon, 2013). In humans, myeloerythroid progenitor cells seed in the yolk sac before HSCs appear in the AGM at 27 dpf (Julien et al., 2016), a stage mirrored in zebrafish with the start of definitive hematopoiesis at 30 hpf in the AGM and transition into the caudal hematopoietic tissue (Jagannathan-Bogdan and Zon, 2013). At 72 hpf, vital markers for early lymphoid progenitors are present in developing immune organs, such as the early thymus and kidney in zebrafish (Willett et al., 1999; Langenau et al., 2004; Trede et al., 2004). This corresponds roughly to the colonization of immune cells in the bone marrow and thymus in the human fetus at 10.5 wpf (Kissa et al., 2008). Notably, at 72 hpf the zebrafish emerges from the chorion and into contact with the outside environment without fully developed CD4 + /CD8 + lymphocytes, which appear later at 3 wpf (Lam et al., 2004). This is contrasted to humans, in which lymphocytes are detectable at 12–13 wpf, well before birth at 40 wpf (Tavian et al., 2010). ALPM: anterior lateral plate mesoderm; ICM: intermediate cell mass; HSC: Hematopoietic stem cells; AGM: aorta-gonad-mesonephros.
Timing of Development
The development of the immune system begins with the emergence of erythroid and myeloid lineages (Figure 1). In humans, this occurs at the Carnegie Stage (CS) 7 and 9, which correspond to 16–18.5 days post-conception (Ivanovs et al., 2017). In zebrafish, primitive macrophages appear at around 12 hours post-fertilization (hpf) (Wattrus and Zon, 2018). Interestingly, these primitive macrophages from the yolk sac follow an expedited differentiation pathway, possessing the ability to engulf pathogens and patrol the entire organism (Herbomel et al., 2001; Novoa and Figueras, 2012). A portion of these primitive macrophages will become neutrophils by 33 hpf (Harvie and Huttenlocher, 2015). During this primitive wave of development, the zebrafish expresses multiple homologous genes as mammals, including lmo2, gata1a, scl, and cul4a in the erythroid lineage and pu.1 in the myeloid lineage (Yamada et al., 2001; Dooley et al., 2005; Galloway et al., 2005; Zhu et al., 2005; Yang et al., 2019).
Lymphoid cell development follows immediately after the appearance of erythroid and myeloid cells, similar to what has been observed at CS11 or 24 days post-conception in humans (Herbomel et al., 1999; Tavian et al., 2010; Carroll and North, 2014; Ivanovs et al., 2017). Markers of lymphoid progenitors, such as rag1, rag2, lck, and ikaros, present in the respective lymphoid progenitors and lymphocytes as those observed in humans, and are all detectable at the end of the primitive wave of hematopoiesis (Willett et al., 1997; Willett et al., 2001; Langenau et al., 2004; Jing and Zon, 2011). Although lymphoblast/lymphocyte markers arise as early as 3 days post fertilization (dpf), the zebrafish still rely on the innate immune system for defense against external threats. Starting at 4 dpf, four early hematopoietic markers, c-myb, ikaros, runx2, and scl, begin to express in the kidney of the zebrafish, marking the initiation of definitive hematopoiesis in this location (Murayama et al., 2006). Meanwhile, T cells develop in the thymus at a similar time and enter circulation at around 8 dpf (Page et al., 2013). Around 20 dpf, B progenitor cells develop in the dorsal aorta and posterior cardinal vein in zebrafish (Page et al., 2013). Adaptive immunity in the form of circulating lymphocytes does not fully mature until 3 weeks post-fertilization (wpf) (Willett et al., 1999; Trede et al., 2004; Novoa and Figueras, 2012). In humans, however, this lag does not exist. At 12–13 weeks, CD4 + and CD8 + T cells mature in the thymus of the developing human fetus and start circulating throughout the body before birth (Haynes et al., 1988; Jagannathan-Bogdan and Zon, 2013).
Locations of Development
The locations in which the hematopoietic lineages develop between zebrafish and humans also differ somewhat (Figure 1). In humans, hematopoiesis begins in the yolk sac before colonization in the fetal liver and the production of hematopoietic stem cells (HSCs) in the aorta-gonad-mesonephros (AGM) region at around 27 dpf (Mikkola and Orkin, 2006; Ivanovs et al., 2017). The definitive wave of hematopoiesis that produces the adult immune system in humans occurs in the bone marrow (Wattrus and Zon, 2018). In zebrafish embryos, early hematopoiesis originates in the intermediate cell mass (ICM) for erythroid cells and the anterior lateral plate mesoderm (ALPM) for myeloid cells (Willett et al., 1999; Berman et al., 2005; Hogan et al., 2006; Jing and Zon, 2011). Similar to humans, there is a transitional period in which zebrafish hematopoiesis occurs in the AGM. Following this stage, the HSCs move to the caudal hematopoietic tissue, which functions comparably to the mammalian fetal liver. Finally, the cells move into the definitive hematopoietic organs, the thymus and kidney, from where the lymphoid cells begin to develop and later emerge (Langenau et al., 2004; Burns et al., 2005; Murayama et al., 2006). The definitive wave of hematopoiesis that produces the adult immune system occurs in the zebrafish kidney as opposed to the bone marrow in humans (Jagannathan-Bogdan and Zon, 2013; Wattrus and Zon, 2018).
Conservation of Innate and Adaptive Immunity
Like humans, the zebrafish possess two main branches of immunity with fully fledged innate and adaptive components. In addition, the fish also possess two primary lymphoid organs, the kidney marrow (equivalent to the bone marrow in mammals) and thymus which shrinks in adult fish as in humans, as well as one secondary peripheral organ, the spleen in adult fish (Wattrus and Zon, 2018). However, one difference in terms of secondary immune organs is that the zebrafish, like other teleosts, lack lymph nodes. Instead, the vast majority of interactions between antigen-presenting cells (APCs) and lymphocytes occur in the spleen (Renshaw and Trede, 2012). Fish cells also express both MHC class I and II molecules, indicating the conserved interactions between the innate and adaptive immune systems (Fischer et al., 2013).
Zebrafish harbor the same fundamental system of innate immunity with leukocytes such as the entire macrophage lineage along with granulocytes like neutrophils and eosinophils (Traver et al., 2003; Novoa and Figueras, 2012). These cells mount a similar innate immune response as seen in mammals to local infections with the expression of characteristic cytokines such as TNF-α and IL-1β, which trigger the engulfment of pathogens by macrophages (Secombes et al., 2001; Traver et al., 2003). So far, 22 putative toll-like receptors (TLRs), including orthologs of all 10 human TLRs, have been discovered in zebrafish (Kanwal et al., 2014; Li et al., 2017).
In terms of adaptive immunity, as described above, zebrafish possess functional T and B cells after 3–6 weeks of development. T cell development occurs in the thymus of larval and adult fish, while mature T cells reside in the kidney marrow of adult fish. B cell development occurs in the pronephros of larval fish and kidney marrow of adult fish (Langenau et al., 2004; Trede et al., 2004). At 3 wpf, B cells become fully mature and active, in tandem with T cells as they emerge from the thymus (Lam et al., 2004). This indicates that the immune system in zebrafish reaches a mature stage at 3–6 wpf (Lam et al., 2004). However, one key difference exists between zebrafish and human B lymphocytes: three main classes of immunoglobulins have been discovered in zebrafish (IgD, IgM, and IgZ) versus five classes in humans (IgA, IgD, IgE, IgG, and IgM) (Zimmerman et al., 2011). Nevertheless, the overall similarities in the development of zebrafish and human immune systems allow for experimental modeling of human TME in zebrafish to visualize and understand the complex interactions between immune and tumor cells.
Swimming Together: Fish Players in Adaptive Immunity
The status of adaptive immunity activation in the TME is a key predictor of prognosis for solid tumors (Bonaventura et al., 2019). Recent research seeks to understand the complex relationship among tumor cells, immune cells, fibroblasts, and endothelial cells, which together form the tumor mass (Hanahan and Coussens, 2012). In this section, we review the role of T and B lymphocytes in the TME and advocate for the utility of the zebrafish due to its high conservation in immune and oncogenic signaling cascades.
The Innate Immune System as a Helper to Adaptive Immune Responses
Dendritic cells (DCs), macrophages, and neutrophils are the three major players in human innate immunity, all capable of infiltrating into the TME. DCs play a key role in presenting exogenous tumor antigens to activate cytotoxic CD8 + T cells for anti-tumor responses, with recent evidence indicating that the suppressive TME can dampen the anti-tumor response of DCs (Fu and Jiang, 2018). Macrophages and neutrophils have been categorized into two broadly defined groups: M1/M2 and N1/N2 (Fridlender et al., 2009). In general, M1 macrophages and N1 neutrophils are associated with TLR-mediated responses/interferon signaling and exert strong pro-inflammatory response against pathogens (Italiani and Boraschi, 2014; Murray, 2017). On the other hand, M2 macrophages and N2 neutrophils are typically linked with T regulatory cell (Tregs) responses and thus participate in cell proliferation and tissue repair (Italiani and Boraschi, 2014; Murray, 2017; Orecchioni et al., 2019; Wu et al., 2019). M1 macrophages and N1 neutrophils secrete inflammatory cytokines that can activate the adaptive immune system for anti-tumor responses (Selders et al., 2017). In contrast, M2 macrophages and N2 neutrophils secrete immunosuppressive cytokines (e.g., IL-10 and TGF-β) and further suppress the immune system by producing CCL22 to recruit Tregs (Fridlender et al., 2009; Gajewski et al., 2013). Macrophages and neutrophils can shift between the subtypes and thus impact the TME in vastly different manners. In the initial stages of tumor development, macrophages are believed to have a prominent M1 profile, characterized by NF-κB expression and capable of attacking the malignant cells (Mantovani and Sica, 2010). However, in clinically detectable tumors, the presence of tumor-associated macrophages (TAMs) predict worse patient outcomes for several types of cancer including those with breast and pancreatic origin (Bingle et al., 2002; Campbell et al., 2011; Zhang et al., 2012; Di Caro et al., 2016). Similarly, the presence of tumor-associated neutrophils (TANs) has been linked to poor prognosis across a wide range of cancers (Gentles et al., 2015). TAMs and TANs promote both tumor initiation and progression by enhancing angiogenesis, suppressing anti-tumor immunity, and facilitating the migration and invasion of tumor cells (Qian and Pollard, 2010; Argyle and Kitamura, 2018; Wu et al., 2019). TAMs are generally believed to possess a M2 profile due to their low expression of MHC class II (MHC-II) which reduces the adaptive immune response and increases production of angiogenesis-promoting elements such as vascular endothelial growth factors (Mantovani et al., 2006; Mantovani and Sica, 2010).
Dendritic cells, macrophages, and neutrophils have all been identified and characterized in zebrafish. Histochemical and ultrastructural analyses confirmed that zebrafish DCs possess the same morphological features and key canonical activities such as antigen-presentation to T cells as its mammalian counterpart (Lugo-Villarino et al., 2010). Because the expression of macrophage expressed gene 1 (mpeg1) is tightly restricted to macrophages in humans, transgenic lines, which express the fluorescent reporter genes under the promoter of mpeg1, have been developed to study macrophage-like cells in zebrafish (Ellett et al., 2011). For instance, the Tg(mpeg1:mCherry) line was crossed to the transgenic line driving eGFP expression under the tumor necrosis factor-alpha promoter to identify and visually track M1 (mCherry+; eGFP+) and M2 (mCherry+; eGFP-) macrophages in zebrafish (Nguyen-Chi et al., 2015). Moreover, upon induced inflammation through fin-wounding, tumor transplantation, or Escherichia coli inoculation, zebrafish M1 and M2 macrophages recapitulate the activation and gene expression patterns as established in higher vertebrates (Nguyen-Chi et al., 2015; Sanderson et al., 2015; Hasegawa et al., 2017; Nguyen-Chi et al., 2017; Tsarouchas et al., 2018). Similar to what is observed in humans, zebrafish neutrophils possess polymorphic nuclei, granules, and myeloid-specific peroxidase coupled with NADPH oxidase (Lieschke et al., 2001; Henry et al., 2013). Taken together, the presence of DCs, macrophages, and neutrophils in zebrafish and their similarities to humans make the zebrafish suitable to study innate immunity and their interactions with adaptive immune cells in the TME.
T Lymphocytes
T lymphocytes are the major players in tumor immunity and include different subtypes characterized by their respective functions. Cytotoxic CD8 + T cells have become one of the most studied subtypes due to the recent success in checkpoint blockade therapies targeting CTLA-4 and PD-L1 signaling pathways (Gong et al., 2018). The anti-tumor response of CD8+T cells is canonically supported by CD4+Th1 helper cells (Ostroumov et al., 2018). Additionally, the TME also harbors other subtypes of CD4 + T lymphocytes, including Th2, Th17, and CD4 + /Foxp3 + Tregs that aid in immune evasion in most tumors (Speiser et al., 2016). T lymphocyte profiles within the TME of solid tumors vary greatly among patients. The ratio of one subtype versus another can predict treatment outcome and rates of disease relapse (Fridman et al., 2012). With similar subtypes and functions of T lymphocytes (Zhang and Wiest, 2016; Bajoghli et al., 2019), the zebrafish represents a useful model to facilitate understanding of the interplay among T lymphocyte subtypes within the TME. Translating this knowledge to the bedside can help improve immunotherapy and patient prognosis.
Cytotoxic CD8 + T Cells
Cytotoxic CD8 + T cells are derived from the αβ lineage of T-cell receptors (TCR) and recognize antigens presented by MHC-I molecules, serving as a major immune surveillance guard against tumors. Elevated numbers of activated CD8 + T cells within the TME are associated with positive outcomes among patients with breast cancer, colorectal cancer, renal cancer, and melanoma (Clemente et al., 1996; Tosolini et al., 2011; Gu-Trantien et al., 2013; Bohner et al., 2019; Ye et al., 2019). Cytotoxic CD8 + T cells recognize tumor antigens and physically engage tumor cells through spatial proximity to eliminate them (Pittet, 2009). Two main pathways are involved in this process: (1) granule exocytosis through the family of serine proteases: the perforin forms a pore on the membrane, allowing granule-associated enzymes (GZM) to access the target cytosol; or (2) apoptosis through cytotoxic effector ligands (e.g., TNFα or Fas) (Martínez-Lostao et al., 2015). However, as the TME becomes increasingly hostile over time, cytotoxic CD8 + T cells lose their ability to suppress tumor growth (Joyce and Fearon, 2015). For instance, lack of nutrients in the TME represents one disruptive factor, leading to exhaustion of these T cells (Chang and Pearce, 2016). Genetic alterations that deregulate oncogenic pathways, such as KRASG12D gain-of-function or TP53 loss-of-function mutations, can result in the recruitment of large numbers of suppressive myeloid cells into the TME to inhibit cytotoxic T cells (Anderson et al., 2017). Thus, more research efforts are needed to understand the TME beyond what activates and mobilizes CD8 + cytotoxic T cells.
Cytotoxic CD8 + T cells have been characterized in teleosts and demonstrated similarities to their human counterparts. Using rainbow trout, analysis of CD8α + and CD8α− cells outside the thymus indicate the existence of CD4-/CD8 + and CD4 + /CD8− lymphocyte populations, which correspond to CD8 + cytotoxic cells and CD4 + Th or Tregs (Takizawa et al., 2011a). In addition, these CD8α + cells also expressed high levels of perforin and granulysin, indicating their effective cytotoxic function. Research conducted in Ginbuna carp again indicates the presence of CD8α + cells with perforin-mediated cytotoxic activity (Toda et al., 2009; Toda et al., 2011). Upon allogeneic insult, a novel granzyme was found upregulated on CD8 + cells in Ginbuna carp (Matsuura et al., 2014). Moreover, the cell-extrinsic apoptosis pathway through the Fas ligand has been detected in the zebrafish, but whether it is expressed in T cells is yet to be determined (Eimon et al., 2006). In Japanese flounder, the Fas ligand has been identified in T-like lymphocytes (Yamaguchi et al., 2019). Overall, cell-cell contact is a key characteristic of cytotoxic CD8 + T cells in the TME and this feature is observed in teleosts (Toda et al., 2011). CD8α + cells have been observed infiltrating the TME of salmon with intestinal tumors, which were induced from chronic gut inflammation (Bjørgen et al., 2019).
CD4 + Lymphocytes
The functions of CD4 + T lymphocytes in the TME are diverse and have been characterized to varying degrees. This can be attributed to the fact that most non-hematological tumors lack the expression of MHC-II molecules, which CD4 + T cells utilize for antigen presentation. Moreover, CD4 + T cells include various subtypes: Th1, Th2, Th17, and Tregs, which exert different and even opposite roles. In teleosts, there are two cd4-like paralogs, cd4-1 and cd4-2. Their encoded proteins, which are widely coexpressed in zebrafish and rainbow trout (Yoon et al., 2015; Takizawa et al., 2016), differ in Ig domain structure. Cd4-1 exhibits a four Ig domain structure similar to mammalian CD4 (Castro et al., 2011). Cd4-2 has fewer Ig domains and its functional significance is currently unknown (Takizawa et al., 2016). Recent studies show that Cd4-1 + T cells in zebrafish infected with pathogens express Th1-, Th2-, and Th17-associated transcription factors and cytokines, indicating that T cell functions are well conserved in bony fish (Yoon et al., 2015; Maisey et al., 2016; Takizawa et al., 2016).
CD4 + Th1 Cells
The Th1 subtype produces large amounts of IFNγ, a cytokine that suppresses tumor growth by promoting proliferation and differentiation of CD8 + cytotoxic T cells and enabling the priming of APCs against tumor antigens (Shankaran et al., 2001; Huang et al., 2007; Ostroumov et al., 2018). Differentiation of Th1 cells is triggered by their exposure to the cytokines such as IFNγ and IL-12 and is characterized by the expression of the master transcription factor T-bet (Kanhere et al., 2012). IFNγ secreted by Th1 subset of cells can recruit natural killer cells and trigger cytotoxicity of tumor-infiltrating macrophages thus preventing tumor progression and angiogenesis (Haabeth et al., 2011; Kim and Cantor, 2014). This Th1 subset also aids in the clearance of pre-malignant, senescent hepatocytes working in conjunction with myeloid cells to prevent the development of liver cancer (Kang et al., 2011).
Both Th1 and Th2 subtypes are present in teleosts, similar to those observed in humans. T-bet, the transcription factor expressed in the Th1 subtype, has been identified and characterized in zebrafish (Mitra et al., 2010). Along with this, IFNγ, the characteristic cytokine of CD4 + Th1 cells, was identified for the first time outside of mammals in zebrafish (Igawa et al., 2006). Two experiments have been conducted in teleosts showing a generalized Th1-type response to antigen challenge. The zebrafish experiment showed that Cd4-1 + T cells, when exposed to a human antigen, induced T-bet and had higher expression of IFNγ (Yoon et al., 2015). A similar experiment conducted in rainbow trout revealed that Cd4 + lymphocytes expressed higher levels of IFNγ, IL-2, and IL-22 after being challenged with a bacterial pathogen (Takizawa et al., 2016). These results indicate a meaningful Cd4 + Th1 response in teleosts. Further research in zebrafish should seek to better understand Th1 cells beyond IFNγ production.
CD4 + Th2 Cells
The Th2 subtype secretes the cytokines IL-4, IL-5, and IL-13 and is characterized by the expression of the transcription factor GATA3 (Kanhere et al., 2012). Differentiation from naïve CD4 + into the Th2 subtype is regulated by IL-4 (Zheng and Flavell, 1997). Through studies in an airway hypersensitivity model, the transmembrane protein T cell immunoglobulin and mucin domain 1 (TIM-1) was found to be critical for Th2-type immune responses (Curtiss et al., 2012). Treatment with the TIM-1 antibody in the murine airway model resulted in the proliferation of Th2 cells, while the antibody against TIM-4, the ligand for TIM-1, induced T-cell proliferation in general (Meyers et al., 2005). Th2 cells can exert anti-tumor effects by recruiting both B cells and eosinophils into the TME (Nishimura et al., 1999; Mattes et al., 2003).
Compared to the Th1 subtype, Th2 cells in teleosts have been characterized with greater details. Dee et al. (2016) generated a zebrafish transgenic line, Tg(cd4-1:mCherry), to monitor Cd4-1 + cells in different organs. The authors identified Th2-like cells in the gills, which express gata3 and il-4/13b, consistent with what was found in salmonids (Takizawa et al., 2011b). They also found that Th2-like cells infiltrated into the TME of zebrafish melanoma. However, unlike the Th2 cells observed in the gills, these cells did not express il-4/13b, indicating heterogeneity of zebrafish Th2 subtype, a characteristic also observed in mammals. As previously seen in humans, blockade or knockdown of TIM-1 and TIM-4 in zebrafish significantly decreased the activation of Cd4 + T cells together with increased proliferation of Th2 subtype and B cells, indicating a key role of these two proteins in regulating CD4 + Th2 subtype (Xu et al., 2016).
CD4 + Th17 Cells
Differentiation from naïve CD4 + T cells into the Th17 subtype is positively regulated by the cytokines TGF-β, IL-6, and IL-23 (Zhu et al., 2010). Th17 cells express the transcription factor retinoid-related orphan receptor RORγt and produce the characteristic cytokines IL-17A, IL-17F, and IL-22. Th17 cells have been shown to induce auto-immune injury in tissues, a function opposite from Tregs (Bettelli et al., 2006). Although chronic inflammation is often carcinogenic, Th17 cells can exert powerful anti-tumor effects. A study using the murine model shows that Th17 polarized cells were highly effective in destroying tumors (Muranski et al., 2008). Additionally, adoptive T cell therapy utilizing Th17 polarized cells can induce cytotoxic CD8 + T cells (Martin-Orozco et al., 2009). In both studies, the anti-tumor effects of Th17 cells are even stronger than the control Th1 cells. Coupled with their long-lived and stem cell-like nature, Th17 cells are critical in anti-tumor responses (Muranski et al., 2011).
Evidence supports the presence of Th17 cells in teleosts, particularly in zebrafish. Firstly, the ROR family is present in zebrafish, including the RORγt transcription factor critical for Th17 cell differentiation (Flores et al., 2007; Monte et al., 2012; Zhang et al., 2013). Five different forms of IL-17 are also found in zebrafish, and they are upregulated in organs of the adaptive immune system such as the spleen and kidney marrow upon lipopolysaccharide (LPS) stimulation (Gunimaladevi et al., 2006). Transcriptome profiling analysis revealed that zebrafish vaccinated with an attenuated bacterial pathogen upregulated key Th17 cytokines, such as IL-17A and IL-22 together with RORγt (Zhang et al., 2013). This demonstrates a clear causal relationship between antigen challenges and the expression of these characteristic Th17 markers. Interestingly, large numbers of Th17-like cells expressing il17a/f1, il17a/f3, il22, and rorca are found recruited to the gut of a zebrafish model of autoimmune and inflammatory diseases, confirming their recognition of self-antigens (Coronado et al., 2019).
CD4 + Tregs
Tregs, another subtype of CD4 + T cells, produce immunosuppressive cytokines such as TGFβ and IL-10, and is characterized by the expression of the transcription factor Foxp3a (Kim and Cantor, 2014; Speiser et al., 2016). Tregs normally prevent excessive autoimmune responses and promote wound healing. In the TME, they suppress CD8 + cytotoxic T cells and support angiogenesis and metastasis of tumors (Zou, 2006). In fact, a significant proportion of tumor-infiltrating CD4 + cells are Tregs (Kim and Cantor, 2014). A high ratio of Tregs to cytotoxic CD8 + T cells is correlated with poor prognosis of patients with multiple cancer types, including pancreatic cancer, ovarian cancer, and colorectal cancer (Preston et al., 2013; Tang et al., 2014; Bencsikova et al., 2019). Hence, the key to the treatment of solid tumors is to suppress the recruitment of Tregs to the TME or to inhibit their immunosuppressive functions.
It has been demonstrated that Foxp3a-expressing Treg-like cells exist in zebrafish (Quintana et al., 2010; Kasheta et al., 2017). Along with a transgenic line that marks out these Treg-like cells by expressing eGFP-fluorescent reporter gene under the zebrafish foxp3a promoter, a mutant zebrafish line with loss-of-function of foxp3a has also been generated (Kasheta et al., 2017). Using these tools, Kasheta et al. (2017) found that foxp3a-/- mutant zebrafish exhibit overall inflammation in tissues, resembling severe human autoimmune disorders. Using the Tg(cd4-1:mCherry) transgenic zebrafish, Dee et al. (2016) demonstrated the existence of Treg-like cells in the gut mucosa. Research studying Tregs’ function in organ regeneration has revealed many potential roles of Tregs in tumorigenesis. When zebrafish were used to model spinal cord, heart, and retina regeneration, CD4 + Treg -like cells were found to quickly migrate to the damaged areas to aid in tissue regeneration (Hui et al., 2017). The ablation of these cells significantly impaired the regenerative capabilities of the tissue (Hui et al., 2017).
TCRγδ T Cells
The decision between the αβ and γδ fates is one of the first made by T cell progenitors in the thymus. The two groups diverge from the same progenitors based on the strength of TCR signaling: with the strong one generating γδ cells and the weak one generating αβ cells (Ciofani and Zuniga-Pflucker, 2010; Wong and Zuniga-Pflucker, 2010; Zarin et al., 2015). αβ cells are defined by successful rearrangement of the TCRβ loci and their progression through a CD4/CD8 double-positive stage, while the γδ subtype rearranges the TCRγ and TCRδ loci and avoids this double-positive stage (Kreslavsky et al., 2010; Zarin et al., 2015). The TCRαβ lineage comprises the vast majority of T cells in the body, while only 2–10% of the total T lymphocytes exhibit γδ characteristics (Sheridan et al., 2013). Unlike CD4+ or cytotoxic CD8 + T lymphocytes, these rare γδ T cells are not restricted to antigens presented by a particular class of MHC molecules and possess both cytotoxic and immune-stimulating properties. For instance, they express cytotoxic ligands (e.g., FasL) and have capacities of both phagocytosis and antigen presentation (Lawand et al., 2017; Zhao et al., 2018). Indeed, the high frequency of γδ cells is associated with positive prognostic outcomes across 25 different types of cancer, especially among solid tumors (Gentles et al., 2015). This association was even stronger than that observed for cytotoxic CD8 + T cells, highlighting the anti-tumor effect of γδ cells in the TME (Gentles et al., 2015).
Distinctive γδ T cells have been observed in teleosts and characterized in zebrafish. The TCRγ locus has been identified in the zebrafish genome assembly using conserved elements among species (Seelye et al., 2016). Wan et al. (2017) found that zebrafish γδ T cells exhibit the characteristic CD4-; CD8 + surface markers with similar flow cytometry scatter patterns/morphology as seen in human γδ cells. Moreover, both the non-specific phagocytic and antigen-presenting characteristics are also preserved in the zebrafish (Wan et al., 2017). The presence of TCRγ was also found in the gut mucosa of other teleosts, such as the sea bass (Picchietti et al., 2011). The above evidence indicates that the zebrafish is suitable for in-depth studies of γδ T cells in the TME.
B Lymphocytes
The role of B cells in the TME is less understood compared to T cells. Across all subtypes of breast cancer, nearly 60% of tumor-infiltrating lymphocytes within the TME were found to be B cells (Coronella-Wood and Hersh, 2003). A recent study, which examined the prognostic significance of tumor-infiltrating B and plasma cells, showed that both types were associated with either positive or neutral outcomes across a wide array of solid-tumor cancer types, including lung, colorectal, gastric, ovarian, and hepatocellular cancers (Wouters and Nelson, 2018). However, when the disease stage was taken into consideration for oro- and hypopharyngeal cancer, B cells were associated with a positive prognosis in patients with early disease but a negative prognosis for those with advanced disease (Wouters and Nelson, 2018).
Different subtypes of B cells can have opposite roles in the TME, either tumor-promoting or suppressing (Shen et al., 2016; Yuen et al., 2016; Largeot et al., 2019). Human mature B cells in the periphery include two main groups: follicular and marginal zone B cells. Follicular B cells can differentiate into IgG, IgE, and IgA antibody-secreting cells (ASCs) with the help of T cells, and can also form IgM ASCs in a T-cell independent manner (Allman and Pillai, 2008). Marginal zone B cells generally serve in an innate-like capacity, while pathogenic triggers such as LPS can induce them to become short-lived plasma cells (Allman and Pillai, 2008). In addition, specific subtypes of B cells expressing CD19, CD20, CD11c, and B220 can also function as APCs (Largeot et al., 2019). Beyond antigen presentation, CD19 + B cells can also express the death ligand FasL to directly induce cytotoxicity when stimulated by IL-17A (Lu et al., 2016). These same cells are negatively regulated by IL-10 (Tao et al., 2017). IL-21 can also stimulate B cells within the TME to produce granzyme B (Jahrsdörfer et al., 2006). Finally, B regulatory cells (Bregs) are a small population of B cells participating in immunomodulation. They exert immunosuppression by enhancing the activity of Tregs, secreting immunosuppressive cytokines such as IL-10, and suppressing the effector CD4+ and CD8 + T cells via the production of TNFα (Olkhanud et al., 2011; Sarvaria et al., 2017). Evidence from multiple studies indicates that Bregs are capable of shielding cancers from the immune system (Schioppa et al., 2011; Murakami et al., 2019). Bregs have also been shown to induce tissue heterogeneity in melanoma through crosstalk between tumor-produced fibroblast growth factor 2 and B-cell origin insulin growth factor 1 to reduce the effectiveness of kinase-inhibitor therapies (Somasundaram et al., 2017).
B cells have been characterized in zebrafish. While IgM and IgD are traditionally considered as the main surface markers for B cells in teleosts, another B cell marker, IgZ, was recently identified in zebrafish immune tissues such as the kidney, spleen, and gills after the LPS challenge (Hu et al., 2010). Page et al. (2013) characterized the development and behavior of IgM + B cells in zebrafish. Utilizing three fluorescent transgenic lines, they defined the existence of pro-B (Pax5+Rag2+IgM–) and immature/mature (Pax5+Rag2–/loIgM+) B cells in the kidney marrow of adult zebrafish (Page et al., 2013). Furthermore, they characterized plasma B cells and discovered a population of CD45-Blimp1 + cells that express plasma-based characteristic markers, such as xbp1, cd40, and irf4 (Page et al., 2013). Liu et al. applied a fluorescent cd79/cd79a transgenic reporter line to show that the pre-B cell stage does not exist in zebrafish, a key difference in B cell development between zebrafish and humans (Liu et al., 2017). Further work indicates that CD79a and CD22 can serve as meaningful markers to distinguish multiple stages of B cell development in teleosts (Liu et al., 2017; Peñaranda et al., 2019). Research examining gastrointestinal tumors in salmonids detected infiltration of IgM + B cells in the tumor stroma and also in metastatic outgrowths in the liver, mirroring what was observed in certain human cancers (Bjørgen et al., 2019). The above evidence supports the suitability of the zebrafish for studying B cell immunity in the TME.
Technological Advances for the Zebrafish
Among vertebrate models, the zebrafish possesses unique advantages for tumor immunity research. The high fecundity of female zebrafish provides hundreds of progeny ideal for statistical analysis, while their external reproduction provides unprecedented access to study the early development of the immune system and cancer (Zon and Peterson, 2005). The optical clarity of embryos and juvenile fish together with the development of highly transparent Casper adult fish enables live imaging studies of tumor-immune cell interactions. Moreover, significant advances have been made over the years to develop the technology needed for zebrafish research, including an assortment of transgenesis and mutagenesis techniques for gene modulations, as well as the ability to conduct large-scale genetic and chemical screens using zebrafish (Suster et al., 2009).
Transgenesis
Early transgenic zebrafish were created by microinjecting linearized plasmid DNA into one-cell-stage embryos (Stuart et al., 1988; Amsterdam et al., 1995). However, this technique was limited by low rates of transmission of the transgene to the progeny (Suster et al., 2009). In recent years, the development of meganuclease or transposon-mediated transgenesis has led to the generation of stable transgenic zebrafish (Michailidou et al., 2009; Chen et al., 2014). Furthermore, the application of conditional transgenesis allows for the inducible expression of target genes, such as oncogenes like xmrk and krasG12D (Li et al., 2012; Enya et al., 2018). One example is the Cre/lox system, which can be combined with the GAL4/Upstream activating system (GAL4/UAS) to regulate oncogene expression under tissue-specific promoters (Santoriello et al., 2010; Enya et al., 2018; Park and Leach, 2018). Another example is the LexPR system, which initiates the transcription of a specific reporter gene through binding to an operon (Kenyon et al., 2018). Finally, the Tet-On system allows the spatial induction of an oncogene under a tissue-specific promoter (Li et al., 2012). The combination of zebrafish transgenesis and its prolificacy has led to the generation of numerous transgenic lines rapidly in a cost-effective manner.
Targeted Gene Inactivation and Genome Editing
Gene inactivation and editing provide another means to modify gene expression in zebrafish. Endonucleases such as transcription activator-like effector nucleases have been utilized to induce targeted double-stranded DNA breaks in tumor suppressor genes to trigger tumorigenesis (Sander et al., 2011; Dahlem et al., 2012; Ignatius et al., 2018). Another approach to inactivate genes in zebrafish is the application of zinc-finger proteases to induce double-stranded breaks followed by non-homologous base-pairing (Payne and Look, 2009). Recently, the cluster regulatory interspaced short palindrome repeats CRISPR/Cas9 system has become the major method for zebrafish gene editing. This advanced technique can induce double-stranded DNA cleavages to delete DNA sequences or introduce genetic modifications in highly targeted locations within the genome. The high efficacy of the CRISPR/Cas9 system, combined with the ease of microinjection of the one-cell-stage embryos, allows for simultaneous targeting of multiple genes in zebrafish to provide insights into how each lineage develops. Furthermore, CRISPR/Cas9 manipulates the genome in a highly controlled manner with up to 50% accuracy (Liu et al., 2019). Collectively, these techniques have enabled the efficient procurement of genetic editing in zebrafish.
Xenografts
Xenografts are animals injected with human cells. While the murine xenografts take weeks or even months to establish, it only takes days to weeks to establish zebrafish xenograft (Xiao et al., 2020). Zebrafish are often transplanted as embryos with fluorescently labeled tumor cells, including patient-derived cancer cells (Au-Kemp et al., 2009; Veinotte et al., 2014; Hason and Bartůněk, 2019; Vargas-Patron et al., 2019). Due to the absence of the adaptive immune system at the early embryonic stage, tumor cell engraftment can occur in zebrafish easily, generating multiple models to study metastasis of human tumor cells including those from patients (Marques et al., 2009; Cabezas-Sainz et al., 2018; Xu et al., 2018). A recent example shows that zebrafish xenografts can serve as a fast, cost-effective preclinical tool with live imaging capabilities to investigate CAR T cell-mediated killing of human cancer cells in B cell malignancies (Pascoal et al., 2020). Although zebrafish are normally reared at an optimal temperature of 26–28°C, xenografted embryos can tolerate an upper limit of 37°C to simulate conditions within the human body (Cabezas-Sainz et al., 2018; Morgan et al., 2019; Yan et al., 2019). Despite the minimal effect on the proliferation of xenografted cells (Cabezas-Sainz et al., 2018; Xu et al., 2018), it is unknown whether these cells experience higher rates of mortality. A more detailed characterization is required to fully understand the properties of these transplanted tumor cells. Additionally, the field will benefit from fine-tuning husbandry techniques to improve zebrafish tolerance of higher temperatures.
For the adult zebrafish, human tumor cells can be introduced through intraperitoneal injection (Patton et al., 2005). However, successful transplantations can only occur in the immunocompromised zebrafish (Vargas-Patron et al., 2019). Similar to mammals, lymphocytes and APCs in adult zebrafish recognize the transplanted tumor as foreign cells and eliminate them (Langenau et al., 2004; Dee et al., 2016). APCs present tumor-antigens through MHC-II molecules to activate Cd4 + T cells and initiate anti-tumor immune responses (Wittamer et al., 2011; Dee et al., 2016). Hence, T cells must be ablated in recipient zebrafish through irradiation, chemical treatments, or mutagenesis to ensure engraftment (Langenau et al., 2004). To facilitate the use of zebrafish for cancer research, a panel of immunocompromised zebrafish mutants in the transparent Casper zebrafish background have been generated over the past few years (Table 3) (Moore et al., 2016; Tang et al., 2017). The zebrafish xenografts allow for therapeutic testing of compounds in a rapid and cost-effective manner. Moreover, coupled with the imaging technique, zebrafish xenografts represent valuable tools to study human tumor cell behavior, including proliferation, apoptosis, invasion, and interaction with host cells (Zhao et al., 2011; Jung et al., 2012; Drabsch et al., 2013; Pudelko et al., 2018; Vargas-Patron et al., 2019).
Small Molecule Screens
Due to their high fecundity and small size, zebrafish are suitable for small molecule screens to identify anti-cancer compounds, and zebrafish xenografts can even be used to screen for personalized therapeutics (Rennekamp and Peterson, 2013; Veinotte et al., 2014). The rapid development of zebrafish allows for the simultaneous assessment of drug efficacy and toxicity in a high-throughput manner (Williams and Hong, 2016; Hason and Bartůněk, 2019; Yan et al., 2019). One successful example of using zebrafish xenografts for small molecule screening is the identification of the drug, clotrimazole, for the treatment of melanoma (Precazzini et al., 2020). This drug, when co-administered with specific inhibitors targeting oncogenes such as Ras expression in melanoma, blocked transformed malignant cells from proliferating (Precazzini et al., 2020). Other researchers have also utilized zebrafish for small molecule screens to identify compounds that disrupt specific biological pathways important to cancer initiation, progression, or maintenance (Gore et al., 2018; Letrado et al., 2018; Lam and Peterson, 2019). Along with the discovery of novel oncogenic pathways that these compounds target, zebrafish chemical genetics have led to an improved understanding of tumor progression and treatment resistance (Gallardo et al., 2015; Dang et al., 2016).
Advanced Imaging Techniques
The optical clarity of the zebrafish during the embryonic to juvenile stage allows for high-resolution imaging. The generation of Casper fish, which lack pigmentation and have thin muscle tissues, extended this imaging capacity to adult zebrafish (White et al., 2008; Blackburn et al., 2011). A broad spectrum of fluorescent proteins can be exploited for differential labeling and visualization of stromal and tumor cells, including vasculatures and different types of immune cells (Oralova et al., 2019). Live imaging in zebrafish unfolds the process of neovascularization, morphology of tumor cells, and their movement, as well as the dynamic interactions with their surrounding tissues (Blackburn et al., 2011; Ignatius and Langenau, 2011). Low-resolution fluorescent macroscopes equipped with different LED lights and filters enable both still imaging and live-video recording of up to 30 zebrafish simultaneously (Blackburn et al., 2011). Laser confocal microscopy provides images with high resolution and contrast, but images are procured one pixel at a time, leading to photo-toxicity over time (Ignatius and Langenau, 2011). A spinning-disc confocal microscope that captures multiple pixels at once can ameliorate these drawbacks. Alternatively, the two-photon microscopy allows for time-lapse images of deep cell structures at high-resolution with long exposure times (Ignatius and Langenau, 2011).
Genetic Screens
Over the past two decades, a large body of work on zebrafish has been focused on genetic screens, uncovering numerous genes critical for tumorigenesis. The genetic screens were either based on chemical or insertional mutagenesis. For the former, the chemical ENU was often used as the mutagen, leading to the discovery of many zebrafish mutant lines for cancer research (Driever et al., 1996; Haffter et al., 1996; Shive et al., 2010). On the other hand, insertional mutagenesis screens utilize exogenous DNA or retroviruses as the mutagens (Sivasubbu et al., 2007). An additional screening method exploited by zebrafish is target-induced local lesions in genomes, a reverse genetic strategy that can identify point mutations of the gene of interest (Da Costa et al., 2014). More recently, screens through transgenic overexpression of genes, such as those using the MiniCoopR vectors, have been conducted to identify genetic modifiers of a known tumor phenotype (Iyengar et al., 2012; Ablain et al., 2018). All of these screening methods have led to the generation of a panel of zebrafish mutants that develop tumors and the identification of novel tumor suppressor genes, oncogenes, and genes that modify the existing tumor phenotype.
Available Resources Enabling Zebrafish for Tumor Immunity Research
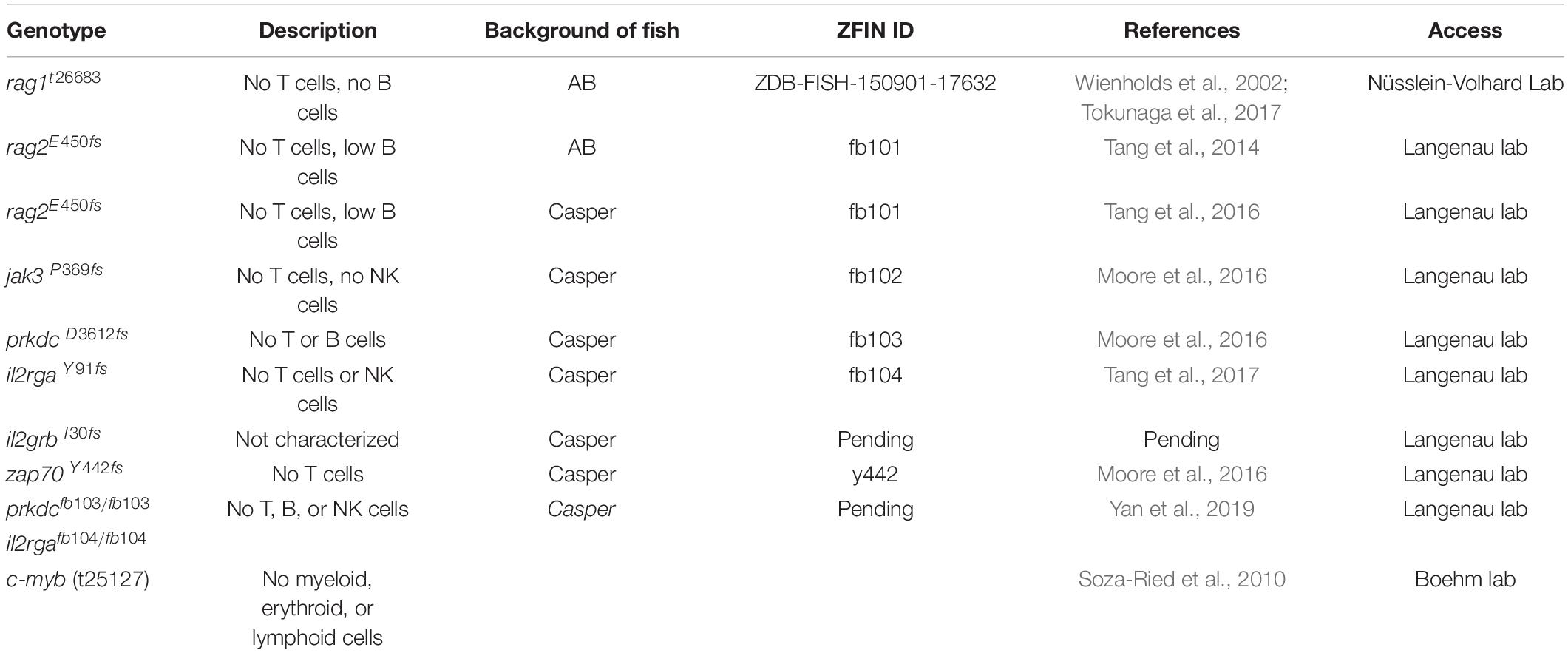
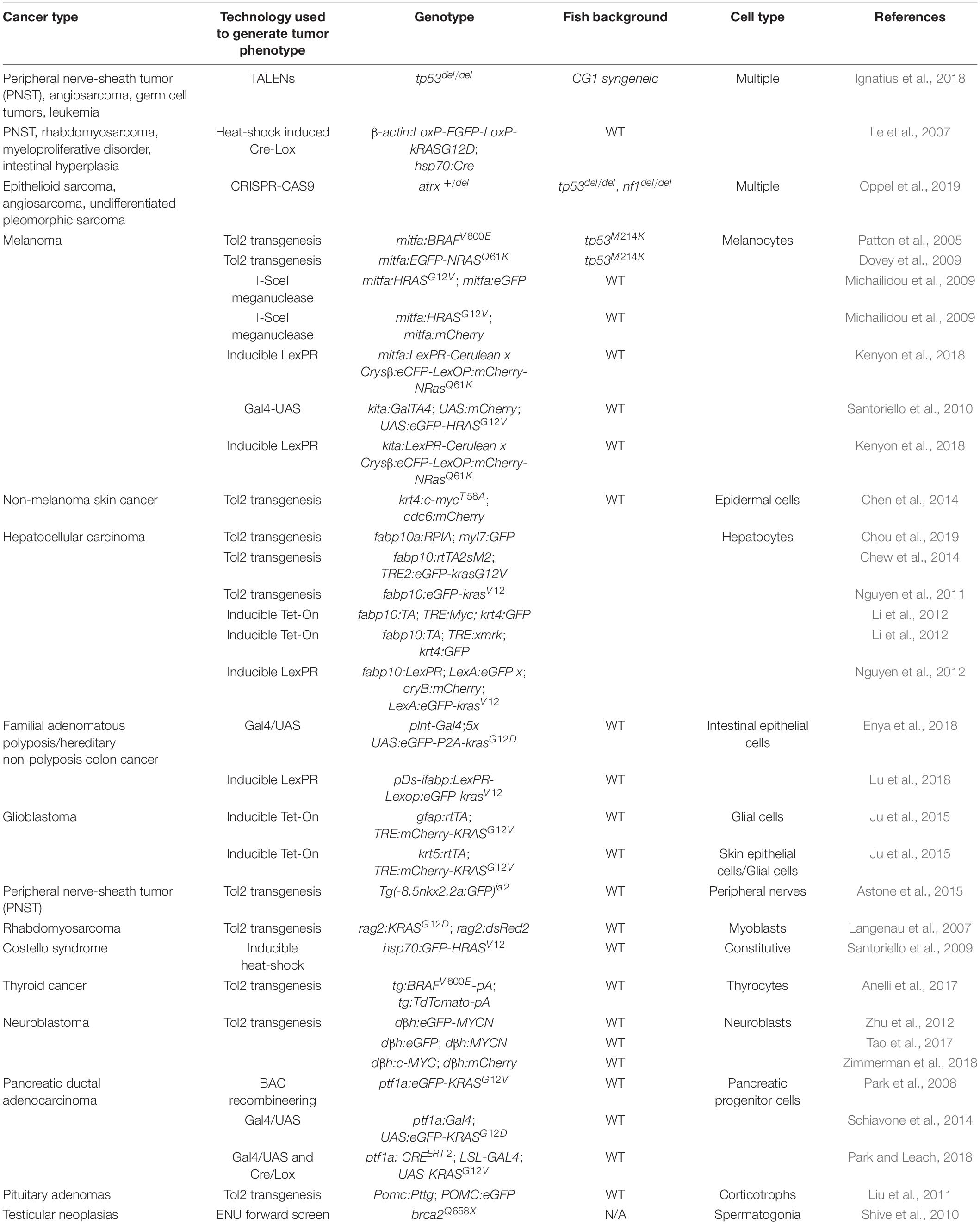
Along with the technological advances, there are a number of resources suitable for tumor immunity research using the zebrafish. Among them, transgenic fluorescent reporter lines help label and track different types of immune cells, especially those participating in adaptive immunity (Table 1). In addition, researchers have also generated and validated a panel of antibodies recognizing zebrafish T and B cells as well as mutant lines in which these adaptive immune cells are depleted (Tables 2, 3). Finally, two dozen zebrafish models of cancer have already showcased the power of this organism in understanding disease etiologies (Table 4). Together, these valuable resources opened the door for researchers to probe specific interactions between immune and tumor cells in various types of cancers, and to expand our understanding of the TME and adaptive tumor immunity.
Transgenic Lines for Tracking Adaptive Immune Cells
Transgenic fluorescent reporter lines represent powerful tools to track immune cells and their behaviors in the TME (Table 1). As the conserved genes governing the development of the adaptive immune system are identified, multiple fluorescent reporter lines have been rapidly generated to mark different immune components in zebrafish using their respective promoters. Similar to the reporter lines for the innate immune system, such as Tg(mpeg1:eGFP) for macrophage and Tg(mpx:GFP) for neutrophils (Ellett et al., 2011; Wang et al., 2014), reporter lines for the adaptive immune system have been utilized to understand the development of lymphocytes in vertebrates (Table 1) (Willett et al., 1999; Langenau et al., 2004). Since these reporter lines can label both lymphoid progenitors and mature lymphocytes, they represent valuable tools to investigate the role of T and B cells in the context of solid tumors and their differentiation in the TME. Interestingly, recent work has found that eGFP fluorescence levels in the Tg(lck:eGFP) line can distinguish B or T cells, indicating the usefulness of this transgenic line in simultaneously studying the behaviors of T and B cells (Borga et al., 2019). The Tg(cd4-1:mCherry) line can label both Cd4 + lymphocytes and macrophages, and has been used to document the conservation of zebrafish lymphocyte development and oncoimmunophenotypes (Dee et al., 2016).
Antibodies for the Study of the Adaptive Immune System in Zebrafish
The development of zebrafish antibodies is an ongoing process (Table 2). Prominent work from the Shao lab at the Zhejiang University has generated a wide variety of zebrafish antibodies against a number of key components in the adaptive immune system, such as IgM, CD40, TCRα/β/γ/δ, CD80/86, and CD83 (Gong et al., 2009; Zhu et al., 2014; Wan et al., 2017). In addition, cross-reactive antibodies developed for other species have helped expand this toolbox. For instance, CD4-1 and CD8α antibodies developed for Ginbuna carp have been validated for successful use in zebrafish (Miyazawa et al., 2018).
Immunocompromised Zebrafish Mutants
Immunocompromised zebrafish serve as the ideal host for tumor cell transplantation and are useful tools that help dissect the contribution of specific immune cells to tumorigenesis (Table 3). This line of research began with the development of the rag1-/- mutant (Wienholds et al., 2002), followed by the identification of c-myb(t25127) mutant fish (Soza-Ried et al., 2010). The Langenau laboratory later developed additional mutant fish with excellent survivability, leading to the establishment of multiple human cancer xenografts in which patterns of angiogenesis and metastasis are studied (Tang et al., 2014; Moore et al., 2016; Tang et al., 2016; Tang et al., 2017; Yan et al., 2019). Combined with their optical clarity, these immunocompromised zebrafish allows for the visualization of single-cell engraftments and the tracking of cancer cell proliferation and migration (Yan et al., 2019). In addition, when transplanted with human cancer cells at the adult stage, these mutant zebrafish are instrumental in evaluating and screening for compounds for personalized application (Hason and Bartůněk, 2019; Yan et al., 2019).
Zebrafish Models of Human Cancers
Beyond xenograft models mentioned above, zebrafish genetic models for human cancers represent one area with the most growth over the past 20 years. Pairing appropriate promoters with cancer-prone genetic alterations have led to the development of over two dozens of zebrafish cancer models (Table 4). Most of these models were generated through oncogene overexpression. Among them, the solid tumor models, such as melanoma and neuroblastoma, are well suited for investigating tumor-immune cell interactions in vivo. Examples of this type of research include studying the role of myeloid cells in early melanoma development utilizing the BRAFV600E transgenic fish as well as the LexPR-regulated conditional NRASQ61K zebrafish (Patton et al., 2005; Kenyon et al., 2018). The zebrafish expressing either N-MYC or C-MYC under the adrenal promoter dβh to model neuroblastoma recapitulates human phenotypes (Zhu et al., 2012; Tao et al., 2017; Zimmerman et al., 2018). C-MYC was also paired with a different promoter to generate a zebrafish model of hepatocellular carcinoma (Li et al., 2012).
Zebrafish models of human cancers have also been generated through inactivating tumor suppressors. An example of this is the zebrafish with tp53 mutations that primarily develop malignant peripheral nerve sheath tumors (MPNSTs) (Berghmans et al., 2005). Interestingly, a recent development of the zebrafish with deletion of tp53 revealed tumor development ranging from MPNSTs, angiosarcomas, and germ cell tumors to an aggressive natural killer cell-like leukemia (Ignatius et al., 2018). Since this model was generated in the CG1 syngeneic zebrafish strain, researchers can easily transplant tumors into this zebrafish to visualize metastatic and angiogenic capacities of tumor cells (Ignatius et al., 2018). CRISPR/Cas9 technology has also been used to delete tumor suppressor genes such as ATRX (Oppel et al., 2019). When ATRX was knocked out in the tp53 mutant fish background, the fish developed epithelioid sarcoma, angiosarcoma, and undifferentiated pleomorphic sarcoma (Oppel et al., 2019). ENU screens generated additional cancer models, such as the brca2Q658X mutant fish that develop testicular neoplasias (Shive et al., 2010). The availability and conservation of these zebrafish cancer models further expand the toolbox of the zebrafish for tumor immunity research.
Proof of Principle: A Small Fish DIP
Both genetic and xenograft cancer models of zebrafish are suitable for tumor immunity studies. The use of genetic models of cancer provides insights into how the immune system responds to malignant transformation and progression during animal development. The fact that zebrafish share conserved telomere regulation pathways and also live in a non-sterile environment as humans provides unique advantages for its utility in studying tumor immunity in comparison to mice (Carneiro et al., 2016). Xenograft assays, ranging from the transplantation of cultured human cell lines to primary patient cells, can complement the genetic models to quickly validate immunotherapies and other selected agents that can kill cancer cells but not the developing fish. In addition, some descriptive studies of the TME in teleosts have been conducted when cancer development is triggered by diet-mediated inflammation, such as metastatic intestinal adenocarcinoma (Bjørgen et al., 2019).
Zebrafish Genetic Models to Probe Tumor-Immune Interactions
Zebrafish genetic models of cancer are often paired with the fluorescent reporter lines to differentially label both the tumor and the immune system, especially the adaptive immune components (Tables 1, 4). Combined with the imaging capacities of the zebrafish, interactions between tumor and specific immune cells arising from the fish can be tracked and studied with high fidelity as seen in human cancers. Dee et al. studied the infiltration of melanoma by T cells and their findings recapitulate what is observed in mammalian systems (Dee et al., 2016). When melanoma develops from the radial growth phase into nodular tumors, T cell activities reduce in the TME of these fish as demonstrated by decreased transcript levels of lck, cd4-1, and cd8α (Dee et al., 2016). Further analysis of tumor-infiltrating Cd4-1 + cells showed that these cells expressed higher levels of gata3 and il-4/13a but not il-4/13b, indicating the presence of a specific subset of Th2 cells in the TME of melanoma, which are absent in normal mucosal environments such as the gills (Dee et al., 2016). Gómez-Abenza et al. sought to understand the TME using the zebrafish model of melanoma in the context of chronic inflammation (Gómez-Abenza et al., 2019). Through the use of a zebrafish line deficient of serine peptidase inhibitor kunitz type 1 (SPINT1), they showed that SPINT1 levels positively correlated with rates of macrophage infiltration into the TME, indicating its regulation of the crosstalk between tumor cells and the immune system. These findings demonstrate that zebrafish is suitable to investigate the interplays among cancer cells, innate, and adaptive immune system.
Zebrafish Xenografts to Screen for Immunomodulators and Test Cell-Based Immunotherapies
Similar to zebrafish genetic models of cancer, researchers also utilize differential labeling of immune and tumor cells in zebrafish xenograft models of cancer to visualize immune-tumor cell interactions in vivo. In addition, the small size and a large number of xenografted embryos enable rapid screens and evaluation of compounds that modulate tumor-immune cell interactions. In particular, the simultaneous introduction of both human cancer and immune cells from the same patient enables the evaluation and selection of optimal cell-based immunotherapy, such as CAR-T cells. For instance, Yan et al. have recently generated a Casper; prkdc-/-; il2rga-/- mutant fish that can be reared at 37°C to better replicate human conditions and support a wide variety of cancer cell engraftments, including patient-derived human cells (Yan et al., 2019). Their work demonstrated the feasibility of this model to assess the efficacy of CAR-T cells in eradicating cancer cells. Pascoal et al. showed that zebrafish xenografts can be used to screen optimal CAR-T cell therapies (Pascoal et al., 2020). Researchers have also utilized zebrafish xenografts to visualize the killing of dormant metastatic cancer cells by CAR-T cells in vivo at the single-cell resolution (He et al., 2020). Al-Samadi et al. (2017) demonstrated that extracellular vesicles isolated from human tongue carcinoma cells can trigger the diminishment of il-13 mRNA in zebrafish, which is an anti-inflammatory cytokine. These studies provide evidence to support the feasibility of zebrafish xenografts in immunomodulator screening and cell-based immunotherapy testing, demonstrating their potential to bridge in vitro studies and those conducted in mammals like mice (He et al., 2020; Pascoal et al., 2020).
Future Perspectives
In summary, the zebrafish has served as one of the key model organisms in recent decades to further our understanding of cancer etiologies and immune system development in vertebrates (Trede et al., 2004; Hason and Bartůněk, 2019; Rossa and D’Silva, 2019). Comparative studies of hematopoiesis demonstrated similarities in the genetic and cellular components of the immune system with only a few differences in the timing and location of the development. Furthermore, these studies revealed surprisingly high conservation in the adaptive immune system between zebrafish and humans. Distinct γδ T cells, cytotoxic CD8 + T cells, CD4 + T cells, and their subtypes are present in both species. In addition, zebrafish and humans share similar differentiation patterns and functions of B lymphocytes, with some differences in the classes and abundance of immunoglobulins as well as the absence of pre-B cells in zebrafish. The overarching similarities between the zebrafish and humans have encouraged the development of a myriad of technologies and tools available for tumor immunity research.
The zebrafish possesses unique advantages for tumor immunity research. The optical clarity of zebrafish together with their small size and low cost makes imaging studies of the TME and screening of immunomodulators feasible. Moreover, the ex-utero development of the zebrafish embryos allows easy and non-invasive access to image the TME, especially tumor-immune cell interactions, at the early stages of tumor development. An in-depth investigation of how the immune system evolves from the anti-tumor to pro-tumor stage holds the key to unlocking the mechanisms of immune evasion. For zebrafish embryos or juveniles under 3 weeks of age, they cannot mount an adaptive immune response (Rossa and D’Silva, 2019). This temporal lag in the development of innate and adaptive immunity in zebrafish is useful to study the differences in interactions between the innate and adaptive immune systems with tumor cells (Rossa and D’Silva, 2019). Moreover, the development of immunocompromised zebrafish mutants in which specific adaptive immune components are removed facilitates elegant probing of these individual component’s contribution to tumor development.
Like any other model system, the zebrafish also possesses some limitations. Gene duplication within the zebrafish genome, which can lead to redundancy in gene function, creates difficulties in gene knockout studies (Postlethwait et al., 2000; Taylor et al., 2003; Howe et al., 2013). As mentioned above, some differences also exist during immune cell development between zebrafish and humans. Importantly, preliminary work using zebrafish and other teleosts have provided fresh mechanistic insights into the regulation of the TME (Dee et al., 2016; Bjørgen et al., 2019; Gómez-Abenza et al., 2019), along with demonstrations of the feasibility of zebrafish xenografts for screening personalized treatments (Al-Samadi et al., 2017; Yan et al., 2019; Pascoal et al., 2020). With its technological advances, established research tools, and high levels of conservation, the zebrafish has joined murine models and human genetic studies for tumor immunology research to uncover novel immunotherapeutic strategies for improved cancer treatment.
Author Contributions
KM, GK, GM, XQ, and HF wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
HF acknowledges support from the National Institute of Health (1UL1TR001430 and CA215059), the American Cancer Society (RSG-17-204-01-TBG), and the National Science Foundation (1911253). KM acknowledges the predoctoral training support from the NIGMS T32GM008541 grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Rebecca Kang and Alexander Floru for proofreading of the manuscript. Due to space limitations, we regret that we could not include all of the work of many colleagues in the field. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
References
Ablain, J., Xu, M., Rothschild, H., Jordan, R. C., Mito, J. K., Daniels, B. H., et al. (2018). Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science 362, 1055–1060. doi: 10.1126/science.aau6509
Al-Samadi, A., Awad, S. A., Tuomainen, K., Zhao, Y., Salem, A., Parikka, M., et al. (2017). Crosstalk between tongue carcinoma cells, extracellular vesicles, and immune cells in in vitro and in vivo models. Oncotarget 8, 60123–60134. doi: 10.18632/oncotarget.17768
Amsterdam, A., Lin, S., and Hopkins, N. (1995). The Aequorea victoria green fluorescent protein can be used as a reporter in live Zebrafish embryos. Dev. Biol. 171, 123–129. doi: 10.1006/dbio.1995.1265
Anderson, K. G., Stromnes, I. M., and Greenberg, P. D. (2017). Obstacles Posed by the tumor microenvironment to T cell activity: a case for synergistic therapies. Cancer Cell 31, 311–325. doi: 10.1016/j.ccell.2017.02.008
Anelli, V., Villefranc, J. A., Chhangawala, S., Martinez-Mcfaline, R., Riva, E., Nguyen, A., et al. (2017). Oncogenic BRAF disrupts thyroid morphogenesis and function via twist expression. eLife 6:e20728.
Argyle, D., and Kitamura, T. (2018). Targeting macrophage-recruiting chemokines as a novel therapeutic strategy to prevent the progression of solid tumors. Front. Immunol. 9:2629. doi: 10.3389/fimmu.2018.02629
Astone, M., Pizzi, M., Peron, M., Domenichini, A., Guzzardo, V., Töchterle, S., et al. (2015). A GFP-tagged gross deletion on chromosome 1 causes malignant peripheral nerve sheath tumors and carcinomas in Zebrafish. PLoS One 10:e0145178. doi: 10.1371/journal.pone.0145178
Au-Kemp, H. A., Au-Carmany-Rampey, A., and Au-Moens, C. (2009). Generating chimeric zebrafish embryos by transplantation. J. Vis. Exp. 29:e1394.
Bajoghli, B., Aghaallaei, N., Hess, I., Rode, I., Netuschil, N., Tay, B.-H., et al. (2009). Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell 138, 186–197. doi: 10.1016/s9999-9994(09)20360-6
Bajoghli, B., Dick, A. M., Claasen, A., Doll, L., and Aghaallaei, N. (2019). Zebrafish and medaka: two teleost models of T-cell and thymic development. Int. J. Mol. Sci. 20:4179. doi: 10.3390/ijms20174179
Bencsikova, B., Budinska, E., Selingerova, I., Pilatova, K., Fedorova, L., Greplova, K., et al. (2019). Circulating T cell subsets are associated with clinical outcome of anti-VEGF-based 1st-line treatment of metastatic colorectal cancer patients: a prospective study with focus on primary tumor sidedness. BMC Cancer 19:687. doi: 10.1186/s12885-019-5909-5
Berghmans, S., Murphey, R. D., Wienholds, E., Neuberg, D., Kutok, J. L., Fletcher, C. D., et al. (2005). tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. U.S.A. 102, 407–412.
Berman, J. N., Kanki, J. P., and Look, A. T. (2005). Zebrafish as a model for myelopoiesis during embryogenesis. Exp. Hematol. 33, 997–1006. doi: 10.1016/j.exphem.2005.06.010
Bettelli, E., Carrier, Y., Gao, W., Korn, T., Strom, T. B., Oukka, M., et al. (2006). Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238. doi: 10.1038/nature04753
Bingle, L., Brown, N. J., and Lewis, C. E. (2002). The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J. Pathol. 196, 254–265. doi: 10.1002/path.1027
Bjørgen, H., Hellberg, H., Løken, O. M., Gunnes, G., Koppang, E. O., and Dale, O. B. (2019). Tumor microenvironment and stroma in intestinal adenocarcinomas and associated metastases in Atlantic salmon broodfish (Salmo salar). Vet. Immunol. Immunopathol. 214:109891. doi: 10.1016/j.vetimm.2019.109891
Blackburn, J. S., Liu, S., Raimondi, A. R., Ignatius, M. S., Salthouse, C. D., and Langenau, D. M. (2011). High-throughput imaging of adult fluorescent zebrafish with an LED fluorescence macroscope. Nat. Protoc. 6, 229–241. doi: 10.1038/nprot.2010.170
Bohner, P., Chevalier, M. F., Cesson, V., Rodrigues-Dias, S.-C., Dartiguenave, F., Burruni, R., et al. (2019). Double positive CD4+CD8+ T cells are enriched in urological cancers and favor T Helper-2 polarization. Front. Immunol. 10:622. doi: 10.3389/fimmu.2019.00622
Bonaventura, P., Shekarian, T., Alcazer, V., Valladeau-Guilemond, J., Valsesia-Wittmann, S., Amigorena, S., et al. (2019). Cold tumors: a therapeutic challenge for immunotherapy. Front. Immunol. 10:168. doi: 10.3389/fimmu.2019.00168
Borga, C., Foster, C. A., Iyer, S., Garcia, S. P., Langenau, D. M., and Frazer, J. K. (2019). Molecularly distinct models of zebrafish Myc-induced B cell leukemia. Leukemia 33, 559–562. doi: 10.1038/s41375-018-0328-1
Burns, C. E., Traver, D., Mayhall, E., Shepard, J. L., and Zon, L. I. (2005). Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19, 2331–2342. doi: 10.1101/gad.1337005
Cabezas-Sainz, P., Guerra-Varela, J., Carreira, M. J., Mariscal, J., Roel, M., Rubiolo, J. A., et al. (2018). Improving zebrafish embryo xenotransplantation conditions by increasing incubation temperature and establishing a proliferation index with ZFtool. BMC Cancer 18:3. doi: 10.1186/s12885-017-3919-8
Callahan, M. K., Masters, G., Pratilas, C. A., Ariyan, C., Katz, J., Kitano, S., et al. (2014). Paradoxical activation of T cells via augmented ERK signaling mediated by a RAF inhibitor. Cancer Immunol. Res. 2, 70–79. doi: 10.1158/2326-6066.cir-13-0160
Campbell, M. J., Tonlaar, N. Y., Garwood, E. R., Huo, D., Moore, D. H., Khramtsov, A. I., et al. (2011). Proliferating macrophages associated with high grade, hormone receptor negative breast cancer and poor clinical outcome. Breast Cancer Res. Treat. 128, 703–711. doi: 10.1007/s10549-010-1154-y
Carmona, S. J., Teichmann, S. A., Ferreira, L., Macaulay, I. C., Stubbington, M. J. T., Cvejic, A., et al. (2017). Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 27, 451–461. doi: 10.1101/gr.207704.116
Carneiro, M. C., De Castro, I. P., and Ferreira, M. G. (2016). Telomeres in aging and disease: lessons from zebrafish. Dis. Model. Mech. 9, 737–748. doi: 10.1242/dmm.025130
Carroll, K. J., and North, T. E. (2014). Oceans of opportunity: exploring vertebrate hematopoiesis in zebrafish. Genomics Model Organ. 42, 684–696. doi: 10.1016/j.exphem.2014.05.002
Casey, M., and Stewart, R. (2020). Pediatric cancer models in Zebrafish. Trends Cancer 6, 407–418. doi: 10.1016/j.trecan.2020.02.006
Casey, M. J., and Stewart, R. A. (2018). Zebrafish as a model to study neuroblastoma development. Cell Tissue Res. 372, 223–232. doi: 10.1007/s00441-017-2702-0
Castro, R., Bernard, D., Lefranc, M. P., Six, A., Benmansour, A., and Boudinot, P. (2011). T cell diversity and TcR repertoires in teleost fish. Fish Shellfish Immunol. 31, 644–654. doi: 10.1016/j.fsi.2010.08.016
Chang, C.-H., and Pearce, E. L. (2016). Emerging concepts of T cell metabolism as a target of immunotherapy. Nat. Immunol. 17, 364–368. doi: 10.1038/ni.3415
Chen, C.-H., Lin, D.-S., Cheng, C.-W., Lin, C.-J., Lo, Y.-K., Yen, C.-C., et al. (2014). Cdc6 cooperates with c-Myc to promote genome instability and epithelial to mesenchymal transition EMT in zebrafish. Oncotarget 5, 6300–6311. doi: 10.18632/oncotarget.2204
Cheon, D.-J., and Orsulic, S. (2011). Mouse models of cancer. Annu. Rev. Pathol. Mech. Dis. 6, 95–119.
Chew, T. W., Liu, X. J., Liu, L., Spitsbergen, J. M., Gong, Z., and Low, B. C. (2014). Crosstalk of Ras and Rho: activation of RhoA abates Kras-induced liver tumorigenesis in transgenic zebrafish models. Oncogene 33, 2717–2727. doi: 10.1038/onc.2013.240
Chou, Y.-T., Chen, L.-Y., Tsai, S.-L., Tu, H.-C., Lu, J.-W., Ciou, S.-C., et al. (2019). Ribose-5-phosphate isomerase A overexpression promotes liver cancer development in transgenic zebrafish via activation of ERK and β-catenin pathways. Carcinogenesis 40, 461–473. doi: 10.1093/carcin/bgy155
Ciofani, M., and Zuniga-Pflucker, J. C. (2010). Determining gammadelta versus alphass T cell development. Nat. Rev. Immunol. 10, 657–663. doi: 10.1038/nri2820
Clemente, C. G., Mihm, M. C. Jr., Bufalino, R., Zurrida, S., Collini, P., and Cascinelli, N. (1996). Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer 77, 1303–1310. doi: 10.1002/(sici)1097-0142(19960401)77:7<1303::aid-cncr12>3.0.co;2-5
Coronado, M., Solis, C. J., Hernandez, P. P., and Feijóo, C. G. (2019). Soybean meal-induced intestinal inflammation in Zebrafish is T cell-dependent and has a Th17 cytokine profile. Front. Immunol. 10:610. doi: 10.3389/fimmu.2019.00610
Coronella-Wood, J. A., and Hersh, E. M. (2003). Naturally occurring B-cell responses to breast cancer. Cancer Immunol. Immunother. 52, 715–738. doi: 10.1007/s00262-003-0409-4
Curtiss, M. L., Gorman, J. V., Businga, T. R., Traver, G., Singh, M., Meyerholz, D. K., et al. (2012). Tim-1 regulates Th2 responses in an airway hypersensitivity model. Eur. J. Immunol. 42, 651–661. doi: 10.1002/eji.201141581
Da Costa, M. M. J., Allen, C. E., Higginbottom, A., Ramesh, T., Shaw, P. J., and Mcdermott, C. J. (2014). A new zebrafish model produced by TILLING of SOD1-related amyotrophic lateral sclerosis replicates key features of the disease and represents a tool for in vivo therapeutic screening. Dis. Model. Mech. 7, 73–81. doi: 10.1242/dmm.012013
Dahlem, T. J., Hoshijima, K., Jurynec, M. J., Gunther, D., Starker, C. G., Locke, A. S., et al. (2012). Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8:e1002861. doi: 10.1371/journal.pgen.1002861
Dang, M., Fogley, R., and Zon, L. I. (2016). Identifying novel cancer therapies using chemical genetics and Zebrafish. Adv. Exp. Med. Biol. 916, 103–124. doi: 10.1007/978-3-319-30654-4_5
Davidson, A. J., and Zon, L. I. (2004). The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 23, 7233–7246. doi: 10.1038/sj.onc.1207943
Dee, C. T., Nagaraju, R. T., Athanasiadis, E. I., Gray, C., Fernandez Del Ama, L., Johnston, S. A., et al. (2016). CD4-Transgenic Zebrafish reveal tissue-resident Th2- and regulatory T cell-like populations and diverse mononuclear phagocytes. J. Immunol. 197, 3520–3530. doi: 10.4049/jimmunol.1600959
Di Caro, G., Cortese, N., Castino, G. F., Grizzi, F., Gavazzi, F., Ridolfi, C., et al. (2016). Dual prognostic significance of tumour-associated macrophages in human pancreatic adenocarcinoma treated or untreated with chemotherapy. Gut 65, 1710–1720. doi: 10.1136/gutjnl-2015-309193
Dooley, K. A., Davidson, A. J., and Zon, L. I. (2005). Zebrafish scl functions independently in hematopoietic and endothelial development. Dev. Biol. 277, 522–536. doi: 10.1016/j.ydbio.2004.09.004
Dovey, M., White, R. M., and Zon, L. I. (2009). Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish 6, 397–404. doi: 10.1089/zeb.2009.0606
Drabsch, Y., He, S., Zhang, L., Snaar-Jagalska, B. E., and Ten Dijke, P. (2013). Transforming growth factor-beta signalling controls human breast cancer metastasis in a zebrafish xenograft model. Breast Cancer Res. 15:R106.
Driever, W., Solnica-Krezel, L., Schier, A. F., Neuhauss, S. C., Malicki, J., Stemple, D. L., et al. (1996). A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123, 37–46. doi: 10.1242/dev.123.1.37
Dvorak, H. F. (1986). Tumors: wounds that do not heal. N. Engl. J. Med. 315, 1650–1659. doi: 10.1056/nejm198612253152606
Eimon, P. M., Kratz, E., Varfolomeev, E., Hymowitz, S. G., Stern, H., Zha, J., et al. (2006). Delineation of the cell-extrinsic apoptosis pathway in the zebrafish. Cell Death Differ. 13, 1619–1630. doi: 10.1038/sj.cdd.4402015
Ellett, F., Pase, L., Hayman, J. W., Andrianopoulos, A., and Lieschke, G. J. (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56.
Elliot, A. (2020). Inflammatory responses during tumour initiation: from Zebrafish transgenic models of cancer to evidence from mouse and man. Cells 9:1018. doi: 10.3390/cells9041018
Enya, S., Kawakami, K., Suzuki, Y., and Kawaoka, S. (2018). A novel zebrafish intestinal tumor model reveals a role for cyp7a1-dependent tumor–liver crosstalk in causing adverse effects on the host. Dis. Model. Mech. 11:dmm032383.
Fischer, U., Koppang, E. O., and Nakanishi, T. (2013). Teleost T and NK cell immunity. Fish Shellfish Immunol. 35, 197–206. doi: 10.1016/j.fsi.2013.04.018
Flores, M. V., Hall, C., Jury, A., Crosier, K., and Crosier, P. (2007). The zebrafish retinoid-related orphan receptor (ror) gene family. Gene Expr. Pat. 7, 535–543. doi: 10.1016/j.modgep.2007.02.001
Fridlender, Z. G., Sun, J., Kim, S., Kapoor, V., Cheng, G., Ling, L., et al. (2009). Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell 16, 183–194. doi: 10.1016/j.ccr.2009.06.017
Fridman, W. H., Pagès, F., Sautès-Fridman, C., and Galon, J. (2012). The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer 12, 298–306. doi: 10.1038/nrc3245
Fu, C., and Jiang, A. (2018). Dendritic cells and CD8 T cell immunity in tumor microenvironment. Front. Immunol. 9:3059. doi: 10.3389/fimmu.2018.03059
Gajewski, T. F., Schreiber, H., and Fu, Y. X. (2013). Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14, 1014–1022.
Gallardo, V. E., Varshney, G. K., Lee, M., Bupp, S., Xu, L., Shinn, P., et al. (2015). Phenotype-driven chemical screening in zebrafish for compounds that inhibit collective cell migration identifies multiple pathways potentially involved in metastatic invasion. Dis. Model. Mech. 8, 565–576. doi: 10.1242/dmm.018689
Galloway, J. L., Wingert, R. A., Thisse, C., Thisse, B., and Zon, L. I. (2005). Loss of Gata1 but not Gata2 converts erythropoiesis to myelopoiesis in Zebrafish embryos. Dev. Cell 8, 109–116. doi: 10.1016/j.devcel.2004.12.001
Gentles, A. J., Newman, A. M., Liu, C. L., Bratman, S. V., Feng, W., Kim, D., et al. (2015). The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945. doi: 10.1038/nm.3909
Gómez-Abenza, E., Ibáñez-Molero, S., García-Moreno, D., Fuentes, I., Zon, L. I., Mione, M. C., et al. (2019). Zebrafish modeling reveals that SPINT1 regulates the aggressiveness of skin cutaneous melanoma and its crosstalk with tumor immune microenvironment. J. Exp. Clin. Cancer Res. 38:405.
Gong, J., Chehrazi-Raffle, A., Reddi, S., and Salgia, R. (2018). Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J. Immunother. Cancer 6:8.
Gong, Y.-F., Xiang, L.-X., and Shao, J.-Z. (2009). CD154-CD40 interactions are essential for thymus-dependent antibody production in Zebrafish: insights into the origin of costimulatory pathway in helper T cell-regulated adaptive immunity in early vertebrates. J. Immunol. 182, 7749–7762. doi: 10.4049/jimmunol.0804370
Gore, A. V., Pillay, L. M., Venero Galanternik, M., and Weinstein, B. M. (2018). The zebrafish: a fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 7:e312. doi: 10.1002/wdev.312
Gunimaladevi, I., Savan, R., and Sakai, M. (2006). Identification, cloning and characterization of interleukin-17 and its family from zebrafish. Fish Shellfish Immunol. 21, 393–403. doi: 10.1016/j.fsi.2006.01.004
Gu-Trantien, C., Loi, S., Garaud, S., Equeter, C., Libin, M., De Wind, A., et al. (2013). CD4? follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Invest. 123, 2873–2892. doi: 10.1172/jci67428
Haabeth, O. A., Lorvik, K. B., Hammarstrom, C., Donaldson, I. M., Haraldsen, G., Bogen, B., et al. (2011). Inflammation driven by tumour-specific Th1 cells protects against B-cell cancer. Nat. Commun. 2:240.
Haffter, P., Granato, M., Brand, M., Mullins, M. C., Hammerschmidt, M., Kane, D. A., et al. (1996). The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123, 1–36. doi: 10.1016/j.molbrainres.2003.09.012
Hanahan, D., and Coussens, L. M. (2012). Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322. doi: 10.1016/j.ccr.2012.02.022
Harvie, E. A., and Huttenlocher, A. (2015). Neutrophils in host defense: new insights from zebrafish. J. Leukoc. Biol. 98, 523–537. doi: 10.1189/jlb.4mr1114-524r
Hasegawa, T., Hall, C. J., Crosier, P. S., Abe, G., Kawakami, K., Kudo, A., et al. (2017). Transient inflammatory response mediated by interleukin-1β is required for proper regeneration in zebrafish fin fold. eLife 6:e22716.
Hason, M., and Bartůněk, P. (2019). Zebrafish models of cancer-new insights on modeling human cancer in a non-mammalian vertebrate. Genes 10:935. doi: 10.3390/genes10110935
Haynes, B. F., Martin, M. E., Kay, H. H., and Kurtzberg, J. (1988). Early events in human T cell ontogeny. Phenotypic characterization and immunohistologic localization of T cell precursors in early human fetal tissues. J. Exp. Med. 168, 1061–1080. doi: 10.1084/jem.168.3.1061
He, X., Yin, X., Wu, J., Wickstrom, S. L., Duo, Y., Du, Q., et al. (2020). Visualization of human T lymphocyte-mediated eradication of cancer cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 117, 22910–22919. doi: 10.1073/pnas.2009092117
Henry, K. M., Loynes, C. A., Whyte, M. K. B., and Renshaw, S. A. (2013). Zebrafish as a model for the study of neutrophil biology. J. Leukoc. Biol. 94, 633–642. doi: 10.1189/jlb.1112594
Herbomel, P., Thisse, B., and Thisse, C. (1999). Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126, 3735–3745. doi: 10.1242/dev.126.17.3735
Herbomel, P., Thisse, B., and Thisse, C. (2001). Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 238, 274–288. doi: 10.1006/dbio.2001.0393
Hogan, B. M., Layton, J. E., Pyati, U. J., Nutt, S. L., Hayman, J. W., Varma, S., et al. (2006). Specification of the primitive myeloid precursor pool requires signaling through Alk8 in Zebrafish. Curr. Biol. 16, 506–511. doi: 10.1016/j.cub.2006.01.047
Howe, K., Clark, M., Torroja, C., Torrance, J., Berthelot, C., Muffato, M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. doi: 10.1038/nature12111
Hu, Y.-L., Xiang, L.-X., and Shao, J.-Z. (2010). Identification and characterization of a novel immunoglobulin Z isotype in zebrafish: implications for a distinct B cell receptor in lower vertebrates. Mol. Immunol. 47, 738–746. doi: 10.1016/j.molimm.2009.10.010
Huang, H., Hao, S., Li, F., Ye, Z., Yang, J., and Xiang, J. (2007). CD4+ Th1 cells promote CD8+ Tc1 cell survival, memory response, tumor localization and therapy by targeted delivery of interleukin 2 via acquired pMHC I complexes. Immunology 120, 148–159. doi: 10.1111/j.1365-2567.2006.02452.x
Hui, S. P., Sheng, D. Z., Sugimoto, K., Gonzalez-Rajal, A., Nakagawa, S., Hesselson, D., et al. (2017). Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 43, 659–672.e5.
Igawa, D., Sakai, M., and Savan, R. (2006). An unexpected discovery of two interferon gamma-like genes along with interleukin (IL)-22 and -26 from teleost: IL-22 and -26 genes have been described for the first time outside mammals. Mol. Immunol. 43, 999–1009. doi: 10.1016/j.molimm.2005.05.009
Ignatius, M. S., Hayes, M. N., Moore, F. E., Tang, Q., Garcia, S. P., Blackburn, P. R., et al. (2018). tp53 deficiency causes a wide tumor spectrum and increases embryonal rhabdomyosarcoma metastasis in zebrafish. eLife 7:e37202.
Ignatius, M. S., and Langenau, D. M. (2011). Fluorescent imaging of cancer in zebrafish. Methods Cell Biol. 105, 437–459. doi: 10.1016/b978-0-12-381320-6.00019-9
Italiani, P., and Boraschi, D. (2014). From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front. Immunol. 5:514. doi: 10.3389/fimmu.2014.00514
Ivanovs, A., Rybtsov, S., Ng, E. S., Stanley, E. G., Elefanty, A. G., and Medvinsky, A. (2017). Human haematopoietic stem cell development: from the embryo to the dish. Development 144, 2323–2337. doi: 10.1242/dev.134866
Iwanami, N. (2014). Zebrafish as a model for understanding the evolution of the vertebrate immune system and human primary immunodeficiency. Exp. Hematol. 42, 697–706. doi: 10.1016/j.exphem.2014.05.001
Iwanami, N., Hess, I., Schorpp, M., and Boehm, T. (2017). Studying the adaptive immune system in zebrafish by transplantation of hematopoietic precursor cells. Methods Cell Biol. 138, 151–161. doi: 10.1016/bs.mcb.2016.08.003
Iyengar, S., Houvras, Y., and Ceol, C. J. (2012). Screening for melanoma modifiers using a zebrafish autochthonous tumor model. J. Vis. Exp. 69:e50086.
Jahrsdörfer, B., Blackwell, S. E., Wooldridge, J. E., Huang, J., Andreski, M. W., Jacobus, L. S., et al. (2006). B-chronic lymphocytic leukemia cells and other B cells can produce granzyme B and gain cytotoxic potential after interleukin-21-based activation. Blood 108, 2712–2719. doi: 10.1182/blood-2006-03-014001
Jessen, J. R., Willett, C. E., and Lin, S. (1999). Artificial chromosome transgenesis reveals long-distance negative regulation of rag1 in zebrafish. Nat. Genet. 23, 15–16. doi: 10.1038/12609
Jing, L., and Zon, L. I. (2011). Zebrafish as a model for normal and malignant hematopoiesis. Dis. Model. Mech. 4, 433–438. doi: 10.1242/dmm.006791
Joyce, J. A., and Fearon, D. T. (2015). T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80. doi: 10.1126/science.aaa6204
Ju, B., Chen, W., Orr, B. A., Spitsbergen, J. M., Jia, S., Eden, C. J., et al. (2015). Oncogenic KRAS promotes malignant brain tumors in zebrafish. Mol. Cancer 14:18. doi: 10.1186/s12943-015-0288-2
Julien, E., El Omar, R., and Tavian, M. (2016). Origin of the hematopoietic system in the human embryo. FEBS Lett. 590, 3987–4001. doi: 10.1002/1873-3468.12389
Jung, D. W., Oh, E. S., Park, S. H., Chang, Y. T., Kim, C. H., Choi, S. Y., et al. (2012). A novel zebrafish human tumor xenograft model validated for anti-cancer drug screening. Mol. Biosyst. 8, 1930–1939. doi: 10.1039/c2mb05501e
Kang, T.-W., Yevsa, T., Woller, N., Hoenicke, L., Wuestefeld, T., Dauch, D., et al. (2011). Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551. doi: 10.1038/nature10599
Kanhere, A., Hertweck, A., Bhatia, U., Gökmen, M. R., Perucha, E., Jackson, I., et al. (2012). T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat. Commun. 3:1268.
Kanwal, Z., Wiegertjes, G. F., Veneman, W. J., Meijer, A. H., and Spaink, H. P. (2014). Comparative studies of Toll-like receptor signalling using zebrafish. Dev. Comp. Immunol. 46, 35–52. doi: 10.1016/j.dci.2014.02.003
Kasheta, M., Painter, C. A., Moore, F. E., Lobbardi, R., Bryll, A., Freiman, E., et al. (2017). Identification and characterization of T reg-like cells in zebrafish. J. Exp. Med. 214, 3519–3530. doi: 10.1084/jem.20162084
Kenyon, A., Gavriouchkina, D., Zorman, J., Chong-Morrison, V., Napolitani, G., Cerundolo, V., et al. (2018). Generation of a double binary transgenic zebrafish model to study myeloid gene regulation in response to oncogene activation in melanocytes. Dis. Model. Mech. 11:dmm030056.
Kim, H.-J., and Cantor, H. (2014). CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol. Res. 2, 91–98. doi: 10.1158/2326-6066.cir-13-0216
Kissa, K., Murayama, E., Zapata, A., Cortes, A., Perret, E., Machu, C., et al. (2008). Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood 111, 1147–1156. doi: 10.1182/blood-2007-07-099499
Kreslavsky, T., Gleimer, M., Garbe, A. I., and Von Boehmer, H. (2010). αβ versus γδ fate choice: counting the T-cell lineages at the branch point. Immunol. Rev. 238, 169–181. doi: 10.1111/j.1600-065x.2010.00947.x
Lam, P. Y., and Peterson, R. T. (2019). Developing zebrafish disease models for in vivo small molecule screens. Curr. Opin. Chem. Biol. 50, 37–44. doi: 10.1016/j.cbpa.2019.02.005
Lam, S. H., Chua, H. L., Gong, Z., Lam, T. J., and Sin, Y. M. (2004). Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 28, 9–28. doi: 10.1016/s0145-305x(03)00103-4
Langenau, D. M., Ferrando, A. A., Traver, D., Kutok, J. L., Hezel, J.-P. D., Kanki, J. P., et al. (2004). In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. U.S.A. 101, 7369–7374. doi: 10.1073/pnas.0402248101
Langenau, D. M., Keefe, M. D., Storer, N. Y., Guyon, J. R., Kutok, J. L., Le, X., et al. (2007). Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 21, 1382–1395. doi: 10.1101/gad.1545007
Langenau, D. M., Traver, D., Ferrando, A. A., Kutok, J. L., Aster, J. C., Kanki, J. P., et al. (2003). Myc-Induced T cell leukemia in transgenic Zebrafish. Science 299, 887–890. doi: 10.1126/science.1080280
Largeot, A., Pagano, G., Gonder, S., Moussay, E., and Paggetti, J. (2019). The B-side of cancer immunity: the underrated tune. Cells 8:449. doi: 10.3390/cells8050449
Lawand, M., Déchanet-Merville, J., and Dieu-Nosjean, M.-C. (2017). Key features of gamma-delta T-cell subsets in human diseases and their immunotherapeutic implications. Front. Immunol. 8:761. doi: 10.3389/fimmu.2017.00761
Le, X., Langenau, D. M., Keefe, M. D., Kutok, J. L., Neuberg, D. S., and Zon, L. I. (2007). Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc. Natl. Acad. Sci. U.S.A. 104, 9410–9415. doi: 10.1073/pnas.0611302104
Letrado, P., De Miguel, I., Lamberto, I., Diez-Martinez, R., and Oyarzabal, J. (2018). Zebrafish: speeding up the cancer drug discovery process. Cancer Res. 78, 6048–6058. doi: 10.1158/0008-5472.can-18-1029
Li, Y., Li, Y., Cao, X., Jin, X., and Jin, T. (2017). Pattern recognition receptors in zebrafish provide functional and evolutionary insight into innate immune signaling pathways. Cell. Mol. Immunol. 14, 80–89. doi: 10.1038/cmi.2016.50
Li, Z., Huang, X., Zhan, H., Zeng, Z., Li, C., Spitsbergen, J. M., et al. (2012). Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J. Hepatol. 56, 419–425. doi: 10.1016/j.jhep.2011.07.025
Lieschke, G. J., Oates, A. C., Crowhurst, M. O., Ward, A. C., and Layton, J. E. (2001). Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 98, 3087–3096. doi: 10.1182/blood.v98.10.3087.h8003087_3087_3096
Liu, K., Petree, C., Requena, T., Varshney, P., and Varshney, G. K. (2019). Expanding the CRISPR toolbox in zebrafish for studying development and disease. Front. Cell Dev. Biol. 7:13. doi: 10.3389/fcell.2019.00013
Liu, N.-A., Jiang, H., Ben-Shlomo, A., Wawrowsky, K., Fan, X.-M., Lin, S., et al. (2011). Targeting zebrafish and murine pituitary corticotroph tumors with a cyclin-dependent kinase (CDK) inhibitor. Proc. Natl. Acad. Sci. U.S.A. 108, 8414–8419. doi: 10.1073/pnas.1018091108
Liu, X., Li, Y.-S., Shinton, S. A., Rhodes, J., Tang, L., Feng, H., et al. (2017). Zebrafish B cell development without a Pre-B cell stage, Revealed by CD79 fluorescence reporter transgenes. J. Immunol. 199, 1706–1715. doi: 10.4049/jimmunol.1700552
Lu, J.-W., Raghuram, D., Fong, P.-S. A., and Gong, Z. (2018). Inducible intestine-specific expression of kras(V12) triggers intestinal tumorigenesis in transgenic Zebrafish. Neoplasia 20, 1187–1197. doi: 10.1016/j.neo.2018.10.002
Lu, L., Weng, C., Mao, H., Fang, X., Liu, X., Wu, Y., et al. (2016). IL-17A promotes migration and tumor killing capability of B cells in esophageal squamous cell carcinoma. Oncotarget 7, 21853–21864. doi: 10.18632/oncotarget.7869
Lugo-Villarino, G., Balla, K. M., Stachura, D. L., Bañuelos, K., Werneck, M. B. F., and Traver, D. (2010). Identification of dendritic antigen-presenting cells in the zebrafish. Proc. Natl. Acad. Sci. U.S.A. 107, 15850–15855.
Maisey, K., Montero, R., Corripio-Miyar, Y., Toro-Ascuy, D., Valenzuela, B., Reyes-Cerpa, S., et al. (2016). Isolation and characterization of Salmonid CD4+ T Cells. J. Immunol. 196, 4150–4163. doi: 10.4049/jimmunol.1500439
Mantovani, A., Schioppa, T., Porta, C., Allavena, P., and Sica, A. (2006). Role of tumor-associated macrophages in tumor progression and invasion. Cancer Metastasis Rev. 25, 315–322. doi: 10.1007/s10555-006-9001-7
Mantovani, A., and Sica, A. (2010). Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr. Opin. Immunol. 22, 231–237. doi: 10.1016/j.coi.2010.01.009
Marques, I. J., Weiss, F. U., Vlecken, D. H., Nitsche, C., Bakkers, J., Lagendijk, A. K., et al. (2009). Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer 9:128. doi: 10.1186/1471-2407-9-128
Martínez-Lostao, L., Anel, A., and Pardo, J. (2015). How do cytotoxic lymphocytes kill cancer cells? Clin. Cancer Res. 21, 5047–5056. doi: 10.1158/1078-0432.ccr-15-0685
Martin-Orozco, N., Muranski, P., Chung, Y., Yang, X. O., Yamazaki, T., Lu, S., et al. (2009). T helper 17 cells promote cytotoxic T cell activation in tumor immunity. Immunity 31, 787–798. doi: 10.1016/j.immuni.2009.09.014
Matsuura, Y., Yabu, T., Shiba, H., Moritomo, T., and Nakanishi, T. (2014). Identification of a novel fish granzyme involved in cell-mediated immunity. Dev. Comp. Immunol. 46, 499–507. doi: 10.1016/j.dci.2014.06.006
Mattes, J., Hulett, M., Xie, W., Hogan, S., Rothenberg, M. E., Foster, P., et al. (2003). Immunotherapy of Cytotoxic T cell–resistant tumors by T helper 2 cells: an eotaxin and STAT6-dependent Process. J. Exp. Med. 197, 387–393. doi: 10.1084/jem.20021683
Meyers, J. H., Chakravarti, S., Schlesinger, D., Illes, Z., Waldner, H., Umetsu, S. E., et al. (2005). TIM-4 is the ligand for TIM-1, and the TIM-1-TIM-4 interaction regulates T cell proliferation. Nat. Immunol. 6, 455–464. doi: 10.1038/ni1185
Michailidou, C., Jones, M., Walker, P., Kamarashev, J., Kelly, A., and Hurlstone, A. F. L. (2009). Dissecting the roles of Raf- and PI3K-signalling pathways in melanoma formation and progression in a zebrafish model. Dis. Model. Mech. 2, 399–411. doi: 10.1242/dmm.001149
Mikkola, H. K., and Orkin, S. H. (2006). The journey of developing hematopoietic stem cells. Development 133, 3733–3744. doi: 10.1242/dev.02568
Mitra, S., Alnabulsi, A., Secombes, C. J., and Bird, S. (2010). Identification and characterization of the transcription factors involved in T-cell development, t-bet, stat6 and foxp3, within the zebrafish, Danio rerio. FEBS J. 277, 128–147. doi: 10.1111/j.1742-4658.2009.07460.x
Miyazawa, R., Matsuura, Y., Shibasaki, Y., Imamura, S., and Nakanishi, T. (2018). Cross-reactivity of monoclonal antibodies against CD4-1 and CD8α of ginbuna crucian carp with lymphocytes of zebrafish and other cyprinid species. Dev. Comp. Immunol. 80, 15–23. doi: 10.1016/j.dci.2016.12.002
Monte, M. M., Wang, T., Costa, M. M., Harun, N. O., and Secombes, C. J. (2012). Cloning and expression analysis of two ROR-gamma homologues (ROR-gammaa1 and ROR-gammaa2) in rainbow trout Oncorhynchus mykiss. Fish Shellfish Immunol. 33, 365–374. doi: 10.1016/j.fsi.2012.05.023
Moore, F. E., Garcia, E. G., Lobbardi, R., Jain, E., Tang, Q., Moore, J. C., et al. (2016). Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish. J. Exp. Med. 213, 979–992. doi: 10.1084/jem.20152013
Morgan, R., Sundin, J., Finnøen, M. H., Dresler, G., Vendrell, M. M., Dey, A., et al. (2019). Are model organisms representative for climate change research? Testing thermal tolerance in wild and laboratory zebrafish populations. Conserv. Physiol. 7:coz036.
Murakami, Y., Saito, H., Shimizu, S., Kono, Y., Shishido, Y., Miyatani, K., et al. (2019). Increased regulatory B cells are involved in immune evasion in patients with gastric cancer. Sci. Rep. 9:13083.
Muranski, P., Boni, A., Antony, P. A., Cassard, L., Irvine, K. R., Kaiser, A., et al. (2008). Tumor-specific Th17-polarized cells eradicate large established melanoma. Blood 112, 362–373. doi: 10.1182/blood-2007-11-120998
Muranski, P., Borman, Z. A., Kerkar, S. P., Klebanoff, C. A., Ji, Y., Sanchez-Perez, L., et al. (2011). Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity 35, 972–985. doi: 10.1016/j.immuni.2011.09.019
Murayama, E., Kissa, K., Zapata, A., Mordelet, E., Briolat, V., Lin, H.-F., et al. (2006). Tracing hematopoietic precursor migration to successive hematopoietic organs during Zebrafish development. Immunity 25, 963–975. doi: 10.1016/j.immuni.2006.10.015
Nguyen, A. T., Emelyanov, A., Koh, C. H. V., Spitsbergen, J. M., Lam, S. H., Mathavan, S., et al. (2011). A high level of liver-specific expression of oncogenic KrasV12 drives robust liver tumorigenesis in transgenic zebrafish. Dis. Model. Mech. 4, 801–813. doi: 10.1242/dmm.007831
Nguyen, A. T., Emelyanov, A., Koh, C. H. V., Spitsbergen, J. M., Parinov, S., and Gong, Z. (2012). An inducible krasV12 transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis. Model. Mech. 5, 63–72. doi: 10.1242/dmm.008367
Nguyen-Chi, M., Laplace-Builhé, B., Travnickova, J., Luz-Crawford, P., Tejedor, G., Lutfalla, G., et al. (2017). TNF signaling and macrophages govern fin regeneration in zebrafish larvae. Cell Death Dis. 8:e2979. doi: 10.1038/cddis.2017.374
Nguyen-Chi, M., Laplace-Builhe, B., Travnickova, J., Luz-Crawford, P., Tejedor, G., Phan, Q. T., et al. (2015). Identification of polarized macrophage subsets in zebrafish. eLife 4:e07288.
Nishimura, T., Iwakabe, K., Sekimoto, M., Ohmi, Y., Yahata, T., Nakui, M., et al. (1999). Distinct role of antigen-specific T helper type 1 (Th1) and Th2 cells in tumor eradication in vivo. J. Exp. Med. 190, 617–628. doi: 10.1084/jem.190.5.617
Novoa, B., and Figueras, A. (2012). “Zebrafish: model for the study of inflammation and the innate immune response to infectious diseases,” in Current Topics in Innate Immunity II, eds J. D. Lambris and G. Hajishengallis (New York, NY: Springer), 253–275. doi: 10.1007/978-1-4614-0106-3_15
Olkhanud, P. B., Damdinsuren, B., Bodogai, M., Gress, R. E., Sen, R., Wejksza, K., et al. (2011). Tumor-evoked regulatory B cells promote breast cancer metastasis by converting resting CD4+ T Cells to T-regulatory cells. Cancer Res. 71, 3505–3515. doi: 10.1158/0008-5472.can-10-4316
Oppel, F., Tao, T., Shi, H., Ross, K. N., Zimmerman, M. W., He, S., et al. (2019). Loss of atrx cooperates with p53-deficiency to promote the development of sarcomas and other malignancies. PLoS Genet. 15:e1008039. doi: 10.1371/journal.pgen.1008039
Oralova, V., Rosa, J. T., Soenens, M., Bek, J. W., Willaert, A., Witten, P. E., et al. (2019). Beyond the whole-mount phenotype: high-resolution imaging in fluorescence-based applications on zebrafish. Biol. Open 8:bio042374.
Orecchioni, M., Ghosheh, Y., Pramod, A. B., and Ley, K. (2019). Macrophage polarization: different gene signatures in M1(LPS+) vs. Classically and M2(LPS–) vs. alternatively activated macrophages. Front. Immunol. 10:1084. doi: 10.3389/fimmu.2019.01084
Ostroumov, D., Fekete-Drimusz, N., Saborowski, M., Kühnel, F., and Woller, N. (2018). CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell. Mol. Life Sci. 75, 689–713. doi: 10.1007/s00018-017-2686-7
Page, D. M., Wittamer, V., Bertrand, J. Y., Lewis, K. L., Pratt, D. N., Delgado, N., et al. (2013). An evolutionarily conserved program of B-cell development and activation in zebrafish. Blood 122, e1–e11.
Park, J. T., and Leach, S. D. (2018). Zebrafish model of KRAS-initiated pancreatic cancer. Anim. Cells Syst. 22, 353–359. doi: 10.1080/19768354.2018.1530301
Park, S. W., Davison, J. M., Rhee, J., Hruban, R. H., Maitra, A., and Leach, S. D. (2008). Oncogenic KRAS induces progenitor cell expansion and malignant transformation in Zebrafish exocrine pancreas. Gastroenterology 134, 2080–2090. doi: 10.1053/j.gastro.2008.02.084
Pascoal, S., Salzer, B., Scheuringer, E., Wenninger-Weinzierl, A., Sturtzel, C., Holter, W., et al. (2020). A preclinical embryonic zebrafish xenograft model to investigate CAR T cells in vivo. Cancers 12:567. doi: 10.3390/cancers12030567
Patton, E. E., Widlund, H. R., Kutok, J. L., Kopani, K. R., Amatruda, J. F., Murphey, R. D., et al. (2005). BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol. 15, 249–254. doi: 10.1016/j.cub.2005.01.031
Payne, E., and Look, T. (2009). Zebrafish modelling of leukaemias. Br. J. Haematol. 146, 247–256. doi: 10.1111/j.1365-2141.2009.07705.x
Peñaranda, M. M. D., Jensen, I., Tollersrud, L. G., Bruun, J.-A., and Jørgensen, J. B. (2019). Profiling the Atlantic Salmon IgM+ B cell surface proteome: novel information on teleost fish B cell protein repertoire and identification of potential B cell markers. Front. Immunol. 10:37. doi: 10.3389/fimmu.2019.00037
Picchietti, S., Guerra, L., Bertoni, F., Randelli, E., Belardinelli, M. C., Buonocore, F., et al. (2011). Intestinal T cells of Dicentrarchus labrax (L.): gene expression and functional studies. Fish Shellfish Immunol. 30, 609–617. doi: 10.1016/j.fsi.2010.12.006
Pittet, M. J. (2009). Behavior of immune players in the tumor microenvironment. Curr. Opin. Oncol. 21, 53–59. doi: 10.1097/cco.0b013e32831bc38a
Postlethwait, J. H., Woods, I. G., Ngo-Hazelett, P., Yan, Y.-L., Kelly, P. D., Chu, F., et al. (2000). Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10, 1890–1902. doi: 10.1101/gr.164800
Precazzini, F., Pancher, M., Gatto, P., Tushe, A., Adami, V., Anelli, V., et al. (2020). Automated in vivo screen in zebrafish identifies Clotrimazole as targeting a metabolic vulnerability in a melanoma model. Dev. Biol. 457, 215–225. doi: 10.1016/j.ydbio.2019.04.005
Preston, C. C., Maurer, M. J., Oberg, A. L., Visscher, D. W., Kalli, K. R., Hartmann, L. C., et al. (2013). The ratios of CD8+ T cells to CD4+CD25+ FOXP3+ and FOXP3- T cells correlate with poor clinical outcome in human serous ovarian cancer. PLoS One 8:e80063. doi: 10.1371/journal.pone.0080063
Pudelko, L., Edwards, S., Balan, M., Nyqvist, D., Al-Saadi, J., Dittmer, J., et al. (2018). An orthotopic glioblastoma animal model suitable for high-throughput screenings. Neuro Oncol. 20, 1475–1484. doi: 10.1093/neuonc/noy071
Qian, B.-Z., and Pollard, J. W. (2010). Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51. doi: 10.1016/j.cell.2010.03.014
Quintana, F. J., Iglesias, A. H., Farez, M. F., Caccamo, M., Burns, E. J., Kassam, N., et al. (2010). Adaptive autoimmunity and Foxp3-based immunoregulation in Zebrafish. PLoS One 5:e9478. doi: 10.1371/journal.pone.0009478
Redd, M. J., Kelly, G., Dunn, G., Way, M., and Martin, P. (2006). Imaging macrophage chemotaxis in vivo: studies of microtubule function in zebrafish wound inflammation. Cell Motil. Cytoskeleton 63, 415–422. doi: 10.1002/cm.20133
Rennekamp, A. J., and Peterson, R. T. (2013). From phenotype to mechanism after zebrafish small molecule screens. Drug Discov. Today Dis. Model. 10, e51–e55.
Renshaw, S. A., and Trede, N. S. (2012). A model 450 million years in the making: zebrafish and vertebrate immunity. Dis. Model. Mech. 5, 38–47. doi: 10.1242/dmm.007138
Rossa, C. Jr., and D’Silva, N. J. (2019). Non-murine models to investigate tumor-immune interactions in head and neck cancer. Oncogene 38, 4902–4914. doi: 10.1038/s41388-019-0776-8
Sander, J. D., Cade, L., Khayter, C., Reyon, D., Peterson, R. T., Joung, J. K., et al. (2011). Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697–698. doi: 10.1038/nbt.1934
Sanderson, L. E., Chien, A.-T., Astin, J. W., Crosier, K. E., Crosier, P. S., and Hall, C. J. (2015). An inducible transgene reports activation of macrophages in live zebrafish larvae. Dev. Comp. Immunol. 53, 63–69. doi: 10.1016/j.dci.2015.06.013
Santoriello, C., Deflorian, G., Pezzimenti, F., Kawakami, K., Lanfrancone, L., D’adda Di Fagagna, F., et al. (2009). Expression of H-RASV12 in a zebrafish model of Costello syndrome causes cellular senescence in adult proliferating cells. Dis. Model. Mech. 2, 56–67. doi: 10.1242/dmm.001016
Santoriello, C., Gennaro, E., Anelli, V., Distel, M., Kelly, A., Köster, R. W., et al. (2010). Kita driven expression of oncogenic HRAS leads to early onset and highly penetrant melanoma in Zebrafish. PLoS One 5:e15170. doi: 10.1371/journal.pone.0015170
Sarvaria, A., Madrigal, J. A., and Saudemont, A. (2017). B cell regulation in cancer and anti-tumor immunity. Cell. Mol. Immunol. 14, 662–674. doi: 10.1038/cmi.2017.35
Schiavone, M., Rampazzo, E., Casari, A., Battilana, G., Persano, L., Moro, E., et al. (2014). Zebrafish reporter lines reveal in vivo signaling pathway activities involved in pancreatic cancer. Dis. Model. Mech. 7, 883–894. doi: 10.1242/dmm.014969
Schioppa, T., Moore, R., Thompson, R. G., Rosser, E. C., Kulbe, H., Nedospasov, S., et al. (2011). B regulatory cells and the tumor-promoting actions of TNF-α during squamous carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 10662–10667. doi: 10.1073/pnas.1100994108
Secombes, C. J., Wang, T., Hong, S., Peddie, S., Crampe, M., Laing, K. J., et al. (2001). Cytokines and innate immunity of fish. Dev. Comp. Immunol. 25, 713–723. doi: 10.1016/s0145-305x(01)00032-5
Seelye, S. L., Chen, P. L., Deiss, T. C., and Criscitiello, M. F. (2016). Genomic organization of the zebrafish (Danio rerio) T cell receptor alpha/delta locus and analysis of expressed products. Immunogenetics 68, 365–379. doi: 10.1007/s00251-016-0904-3
Selders, G. S., Fetz, A. E., Radic, M. Z., and Bowlin, G. L. (2017). An overview of the role of neutrophils in innate immunity, inflammation and host-biomaterial integration. Regen. Biomater. 4, 55–68. doi: 10.1093/rb/rbw041
Shankaran, V., Ikeda, H., Bruce, A. T., White, J. M., Swanson, P. E., Old, L. J., et al. (2001). IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410, 1107–1111. doi: 10.1038/35074122
Shen, M., Sun, Q., Wang, J., Pan, W., and Ren, X. (2016). Positive and negative functions of B lymphocytes in tumors. Oncotarget 7, 55828–55839. doi: 10.18632/oncotarget.10094
Sheridan, B. S., Romagnoli, P. A., Pham, Q. M., Fu, H. H., Alonzo, F. III, Schubert, W. D., et al. (2013). gammadelta T cells exhibit multifunctional and protective memory in intestinal tissues. Immunity 39, 184–195. doi: 10.1016/j.immuni.2013.06.015
Shive, H. R., West, R. R., Embree, L. J., Azuma, M., Sood, R., Liu, P., et al. (2010). brca2 in zebrafish ovarian development, spermatogenesis, and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 19350–19355. doi: 10.1073/pnas.1011630107
Sivasubbu, S., Balciunas, D., Amsterdam, A., and Ekker, S. C. (2007). Insertional mutagenesis strategies in zebrafish. Genome Biol. 8(Suppl 1):S9.
Somasundaram, R., Zhang, G., Fukunaga-Kalabis, M., Perego, M., Krepler, C., Xu, X., et al. (2017). Tumor-associated B-cells induce tumor heterogeneity and therapy resistance. Nat. Commun. 8:607.
Soza-Ried, C., Hess, I., Netuschil, N., Schorpp, M., and Boehm, T. (2010). Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc. Natl. Acad. Sci. U.S.A. 107, 17304–17308. doi: 10.1073/pnas.1004640107
Speiser, D. E., Ho, P.-C., and Verdeil, G. (2016). Regulatory circuits of T cell function in cancer. Nat. Rev. Immunol. 16, 599–611. doi: 10.1038/nri.2016.80
Stambrook, P. J., Maher, J., and Farzaneh, F. (2017). Cancer immunotherapy: whence and whither. Mol. Cancer Res. 15, 635–650. doi: 10.1158/1541-7786.mcr-16-0427
Stuart, G. W., Mcmurray, J. V., and Westerfield, M. (1988). Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development 103, 403–412. doi: 10.1242/dev.103.2.403
Suster, M. L., Kikuta, H., Urasaki, A., Asakawa, K., and Kawakami, K. (2009). “Transgenesis in Zebrafish with the Tol2 transposon system,” in Transgenesis Techniques: Principles and Protocols, ed. E. J. Cartwright (Totowa, NJ: Humana Press), 41–63. doi: 10.1007/978-1-60327-019-9_3
Takizawa, F., Dijkstra, J. M., Kotterba, P., Korytáø, T., Kock, H., Köllner, B., et al. (2011a). The expression of CD8α discriminates distinct T cell subsets in teleost fish. Dev. Comp. Immunol. 35, 752–763. doi: 10.1016/j.dci.2011.02.008
Takizawa, F., Koppang, E. O., Ohtani, M., Nakanishi, T., Hashimoto, K., Fischer, U., et al. (2011b). Constitutive high expression of interleukin-4/13A and GATA-3 in gill and skin of salmonid fishes suggests that these tissues form Th2-skewed immune environments. Mol. Immunol. 48, 1360–1368. doi: 10.1016/j.molimm.2011.02.014
Takizawa, F., Magadan, S., Parra, D., Xu, Z., Korytar, T., Boudinot, P., et al. (2016). Novel Teleost CD4-bearing cell populations provide insights into the evolutionary origins and primordial roles of CD4+ lymphocytes and CD4+ macrophages. J. Immunol. 196, 4522–4535. doi: 10.4049/jimmunol.1600222
Tang, Q., Iyer, S., Lobbardi, R., Moore, J. C., Chen, H., Lareau, C., et al. (2017). Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med. 214, 2875–2887. doi: 10.1084/jem.20170976
Tang, Q., Moore, J. C., Ignatius, M. S., Tenente, I. M., Hayes, M. N., Garcia, E. G., et al. (2016). Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nat. Commun. 7:10358.
Tang, Y., Xu, X., Guo, S., Zhang, C., Tang, Y., Tian, Y., et al. (2014). An increased abundance of tumor-infiltrating regulatory T cells is correlated with the progression and prognosis of pancreatic ductal adenocarcinoma. PLoS One 9:e91551. doi: 10.1371/journal.pone.0091551
Tao, T., Sondalle, S. B., Shi, H., Zhu, S., Perez-Atayde, A. R., Peng, J., et al. (2017). The pre-rRNA processing factor DEF is rate limiting for the pathogenesis of MYCN-driven neuroblastoma. Oncogene 36, 3852–3867. doi: 10.1038/onc.2016.527
Tavian, M., Biasch, K., Sinka, L., Vallet, J., and Péault, B. (2010). Embryonic origin of human hematopoiesis. Int. J. Dev. Biol. 54, 1061–1065. doi: 10.1387/ijdb.103097mt
Taylor, J. S., Braasch, I., Frickey, T., Meyer, A., and Van De Peer, Y. (2003). Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 13, 382–390. doi: 10.1101/gr.640303
Toda, H., Araki, K., Moritomo, T., and Nakanishi, T. (2011). Perforin-dependent cytotoxic mechanism in killing by CD8 positive T cells in ginbuna crucian carp, Carassius auratus langsdorfii. Dev. Comp. Immunol. 35, 88–93. doi: 10.1016/j.dci.2010.08.010
Toda, H., Shibasaki, Y., Koike, T., Ohtani, M., Takizawa, F., Ototake, M., et al. (2009). Alloantigen-specific killing is mediated by CD8-positive T cells in fish. Dev. Comp. Immunol. 33, 646–652. doi: 10.1016/j.dci.2008.11.008
Tokunaga, Y., Shirouzu, M., Sugahara, R., Yoshiura, Y., Kiryu, I., Ototake, M., et al. (2017). Comprehensive validation of T- and B-cell deficiency in rag1-null zebrafish: implication for the robust innate defense mechanisms of teleosts. Sci. Rep. 7:7536.
Tosolini, M., Kirilovsky, A., Mlecnik, B., Fredriksen, T., Mauger, S., Bindea, G., et al. (2011). Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 71, 1263–1271. doi: 10.1158/0008-5472.can-10-2907
Traver, D., Herbomel, P., Patton, E. E., Murphey, R. D., Yoder, J. A., Litman, G. W., et al. (2003). The zebrafish as a model organism to study development of the immune system. Adv. Immunol. 81, 253–330.
Trede, N. S., Langenau, D. M., Traver, D., Look, A. T., and Zon, L. I. (2004). The use of Zebrafish to understand immunity. Immunity 20, 367–379. doi: 10.1016/s1074-7613(04)00084-6
Tsarouchas, T. M., Wehner, D., Cavone, L., Munir, T., Keatinge, M., Lambertus, M., et al. (2018). Dynamic control of proinflammatory cytokines Il-1β and Tnf-α by macrophages in zebrafish spinal cord regeneration. Nat. Commun. 9:4670.
Vargas-Patron, L. A., Agudelo-Dueñas, N., Madrid-Wolff, J., Venegas, J. A., González, J. M., Forero-Shelton, M., et al. (2019). Xenotransplantation of Human glioblastoma in Zebrafish larvae: in vivo imaging and proliferation assessment. Biol. Open 8:bio043257.
Veinotte, C. J., Dellaire, G., and Berman, J. N. (2014). Hooking the big one: the potential of zebrafish xenotransplantation to reform cancer drug screening in the genomic era. Dis. Model. Mech. 7, 745–754. doi: 10.1242/dmm.015784
Wan, F., Hu, C.-B., Ma, J.-X., Gao, K., Xiang, L.-X., and Shao, J.-Z. (2017). Characterization of γδ T cells from zebrafish provides insights into their important role in adaptive humoral immunity. Front. Immunol. 7:675. doi: 10.3389/fimmu.2016.00675
Wang, X., Robertson, A. L., Li, J., Chai, R. J., Haishan, W., Sadiku, P., et al. (2014). Inhibitors of neutrophil recruitment identified using transgenic zebrafish to screen a natural product library. Dis. Model. Mech. 7, 163–169. doi: 10.1242/dmm.012047
Wattrus, S. J., and Zon, L. I. (2018). Stem cell safe harbor: the hematopoietic stem cell niche in zebrafish. Blood Adv. 2, 3063–3069. doi: 10.1182/bloodadvances.2018021725
White, R., Rose, K., and Zon, L. (2013). Zebrafish cancer: the state of the art and the path forward. Nat. Rev. Cancer 13, 624–636. doi: 10.1038/nrc3589
White, R. M., Sessa, A., Burke, C., Bowman, T., Leblanc, J., Ceol, C., et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189. doi: 10.1016/j.stem.2007.11.002
Wienholds, E., Schulte-Merker, S., Walderich, B., and Plasterk, R. H. (2002). Target-selected inactivation of the zebrafish rag1 gene. Science 297, 99–102. doi: 10.1126/science.1071762
Willett, C. E., Cortes, A., Zuasti, A., and Zapata, A. G. (1999). Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev. Dyn. 214, 323–336. doi: 10.1002/(sici)1097-0177(199904)214:4<323::aid-aja5>3.0.co;2-3
Willett, C. E., Kawasaki, H., Amemiya, C. T., Lin, S., and Steiner, L. A. (2001). Ikaros expression as a marker for lymphoid progenitors during zebrafish development. Dev. Dyn. 222, 694–698. doi: 10.1002/dvdy.1223
Willett, C. E., Zapata, A. G., Hopkins, N., and Steiner, L. A. (1997). Expression of ZebrafishragGenes during Early Development Identifies the Thymus. Dev. Biol. 182, 331–341. doi: 10.1006/dbio.1996.8446
Williams, C. H., and Hong, C. C. (2016). Zebrafish small molecule screens: Taking the phenotypic plunge. Comput. Struct. Biotechnol. J. 14, 350–356. doi: 10.1016/j.csbj.2016.09.001
Wittamer, V., Bertrand, J. Y., Gutschow, P. W., and Traver, D. (2011). Characterization of the mononuclear phagocyte system in zebrafish. Blood 117, 7126–7135. doi: 10.1182/blood-2010-11-321448
Wong, G. W., and Zuniga-Pflucker, J. C. (2010). gammadelta and alphabeta T cell lineage choice: resolution by a stronger sense of being. Semin. Immunol. 22, 228–236. doi: 10.1016/j.smim.2010.04.005
Wouters, M. C. A., and Nelson, B. H. (2018). Prognostic Significance of Tumor-Infiltrating B cells and plasma cells in human cancer. Clin. Cancer Res. 24, 6125–6135. doi: 10.1158/1078-0432.ccr-18-1481
Wu, L., Saxena, S., Awaji, M., and Singh, R. K. (2019). Tumor-associated neutrophils in cancer: going pro. Cancers 11:564. doi: 10.3390/cancers11040564
Xiao, J., Glasgow, E., and Agarwal, S. (2020). Zebrafish xenografts for drug discovery and personalized medicine. Trends Cancer 6, 569–579. doi: 10.1016/j.trecan.2020.03.012
Xu, W., Foster, B. A., Richards, M., Bondioli, K. R., Shah, G., and Green, C. C. (2018). Characterization of prostate cancer cell progression in zebrafish xenograft model. Int. J. Oncol. 52, 252–260.
Xu, X.-G., Hu, J.-F., Ma, J.-X., Nie, L., Shao, T., Xiang, L.-X., et al. (2016). Essential Roles of TIM-1 and TIM-4 homologs in adaptive humoral immunity in a Zebrafish model. J. Immunol. 196, 1686–1699. doi: 10.4049/jimmunol.1501736
Yamada, T., Abe, M., Higashi, T., Yamamoto, H., Kihara-Negishi, F., Sakurai, T., et al. (2001). Lineage switch induced by overexpression of Ets family transcription factor PU.1 in murine erythroleukemia cells. Blood 97, 2300–2307. doi: 10.1182/blood.v97.8.2300
Yamaguchi, T., Takizawa, F., Furihata, M., Soto-Lampe, V., Dijkstra, J. M., and Fischer, U. (2019). Teleost cytotoxic T cells. Fish Shellfish Immunol. 95, 422–439. doi: 10.1016/j.fsi.2019.10.041
Yan, C., Brunson, D. C., Tang, Q., Do, D., Iftimia, N. A., Moore, J. C., et al. (2019). Visualizing engrafted human cancer and therapy responses in immunodeficient zebrafish. Cell 177, 1903–1914.e14.
Yang, F., Hu, H., Liu, Y., Shao, M., Shao, C., and Gong, Y. (2019). Cul4a promotes zebrafish primitive erythropoiesis via upregulating scl and gata1 expression. Cell Death Dis. 10:388.
Ye, L., Zhang, T., Kang, Z., Guo, G., Sun, Y., Lin, K., et al. (2019). Tumor-infiltrating immune cells act as a marker for prognosis in colorectal cancer. Front. Immunol. 10:2368. doi: 10.3389/fimmu.2019.02368
Yoon, S., Mitra, S., Wyse, C., Alnabulsi, A., Zou, J., Weerdenburg, E. M., et al. (2015). First demonstration of antigen induced cytokine expression by CD4-1+ lymphocytes in a poikilotherm: studies in Zebrafish (Danio rerio). PLoS One 10:e0126378. doi: 10.1371/journal.pone.0126378
Yuen, G. J., Demissie, E., and Pillai, S. (2016). B lymphocytes and cancer: a love–hate relationship. Trends Cancer 2, 747–757. doi: 10.1016/j.trecan.2016.10.010
Zarin, P., Chen, E. L., In, T. S., Anderson, M. K., and Zuniga-Pflucker, J. C. (2015). Gamma delta T-cell differentiation and effector function programming, TCR signal strength, when and how much? Cell. Immunol. 296, 70–75. doi: 10.1016/j.cellimm.2015.03.007
Zhang, H., Fei, C., Wu, H., Yang, M., Liu, Q., Wang, Q., et al. (2013). Transcriptome profiling reveals Th17-like immune responses induced in ZEBRAFISH bath-vaccinated with a live attenuated Vibrio anguillarum. PLoS One 8:e73871. doi: 10.1371/journal.pone.0073871
Zhang, Q.-W., Liu, L., Gong, C.-Y., Shi, H.-S., Zeng, Y.-H., Wang, X.-Z., et al. (2012). Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One 7:e50946. doi: 10.1371/journal.pone.0050946
Zhang, Y., and Wiest, D. L. (2016). Using the Zebrafish model to study T cell development. Methods Mol. Biol. 1323, 273–292. doi: 10.1007/978-1-4939-2809-5_22
Zhao, C., Wang, X., Zhao, Y., Li, Z., Lin, S., Wei, Y., et al. (2011). A novel xenograft model in zebrafish for high-resolution investigating dynamics of neovascularization in tumors. PLoS One 6:e21768. doi: 10.1371/journal.pone.0021768
Zhao, Y., Niu, C., and Cui, J. (2018). Gamma-delta (γδ) T cells: friend or foe in cancer development? J. Transl. Med. 16:3.
Zheng, W.-P., and Flavell, R. A. (1997). The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596. doi: 10.1016/s0092-8674(00)80240-8
Zhu, H., Traver, D., Davidson, A. J., Dibiase, A., Thisse, C., Thisse, B., et al. (2005). Regulation of the lmo2 promoter during hematopoietic and vascular development in zebrafish. Dev. Biol. 281, 256–269. doi: 10.1016/j.ydbio.2005.01.034
Zhu, J., Yamane, H., and Paul, W. E. (2010). Differentiation of effector CD4 T cell populations (∗). Annu. Rev. Immunol. 28, 445–489. doi: 10.1146/annurev-immunol-030409-101212
Zhu, L.-Y., Lin, A.-F., Shao, T., Nie, L., Dong, W.-R., Xiang, L.-X., et al. (2014). B cells in teleost fish act as pivotal initiating APCs in priming adaptive immunity: an evolutionary perspective on the origin of the B-1 Cell Subset and B7 Molecules. J. Immunol. 192, 2699–2714. doi: 10.4049/jimmunol.1301312
Zhu, S., Lee, J.-S., Guo, F., Shin, J., Perez-Atayde, A. R., Kutok, J. L., et al. (2012). Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 21, 362–373. doi: 10.1016/j.ccr.2012.02.010
Zimmerman, A. M., Moustafa, F. M., Romanowski, K. E., and Steiner, L. A. (2011). Zebrafish immunoglobulin IgD: unusual exon usage and quantitative expression profiles with IgM and IgZ/T heavy chain isotypes. Mol. Immunol. 48, 2220–2223. doi: 10.1016/j.molimm.2011.06.441
Zimmerman, M. W., Liu, Y., He, S., Durbin, A. D., Abraham, B. J., Easton, J., et al. (2018). MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov. 8, 320–335. doi: 10.1158/2159-8290.cd-17-0993
Zon, L. I., and Peterson, R. T. (2005). In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 4, 35–44. doi: 10.1038/nrd1606
Keywords: cancer, immunotherapy, zebrafish, adaptive tumor immunity, TME, lymphocyte
Citation: Miao KZ, Kim GY, Meara GK, Qin X and Feng H (2021) Tipping the Scales With Zebrafish to Understand Adaptive Tumor Immunity. Front. Cell Dev. Biol. 9:660969. doi: 10.3389/fcell.2021.660969
Received: 30 January 2021; Accepted: 19 April 2021;
Published: 20 May 2021.
Edited by:
Liqing Zang, Mie University, JapanReviewed by:
Yu H. Sun, University of Rochester, United StatesCristina Maccalli, Sidra Medicine, Qatar
Copyright © 2021 Miao, Kim, Meara, Qin and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Feng, huifeng@bu.edu
†These authors have contributed equally to this work
 Kelly Z. Miao
Kelly Z. Miao Grace Y. Kim
Grace Y. Kim Grace K. Meara1†
Grace K. Meara1†  Hui Feng
Hui Feng