ADAR1-Mediated RNA Editing and Its Role in Cancer
- 1The United Innovation of Mengchao Hepatobiliary Technology Key Laboratory of Fujian Province, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fuzhou, China
- 2College of Life Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou, China
- 4Mengchao Med-X Center, Fuzhou University, Fuzhou, China
- 5Fujian Medical University Cancer Hospital, Fujian Cancer Hospital, Fuzhou, China
It is well known that the stability of RNA, the interaction between RNA and protein, and the correct translation of protein are significant forces that drive the transition from normal cell to malignant tumor. Adenosine deaminase acting on RNA 1 (ADAR1) is an RNA editing enzyme that catalyzes the deamination of adenosine to inosine (A-to-I), which is one dynamic modification that in a combinatorial manner can give rise to a very diverse transcriptome. ADAR1-mediated RNA editing is essential for survival in mammals and its dysregulation results in aberrant editing of its substrates that may affect the phenotypic changes in cancer. This overediting phenomenon occurs in many cancers, such as liver, lung, breast, and esophageal cancers, and promotes tumor progression in most cases. In addition to its editing role, ADAR1 can also play an editing-independent role, although current research on this mechanism is relatively shallowly explored in tumors. In this review, we summarize the nature of ADAR1, mechanisms of ADAR1 editing-dependent and editing-independent and implications for tumorigenesis and prognosis, and pay special attention to effects of ADAR1 on cancers by regulating non-coding RNA formation and function.
Introduction
A-to-I editing of RNA, a widespread co/post-transcriptional modification in mammals, is catalyzed by adenosine deaminases acting on RNA (ADARs) and has recently been recognized as an essential mechanism of cancer biology (Han et al., 2015; Baker and Slack, 2022). With the continuous increase of next-generation sequencing data, transcriptomics modifications, so-called RNA mutations, are becoming a significant force in promoting the transformation of normal cells into malignant tumors and providing tumor diversity to avoid immune attacks (Nishikura, 2010; Sagredo et al., 2018; Shevchenko and Morris, 2018). ADARs were first discovered in Xenopus laevis oocytes and embyros (Bass, 1987; Quin et al., 2021), confirming mediating editing of adenosine-to-inosine (A-to-I) in double-stranded (ds) RNA in mammals (Xu and Öhman, 2018). This edit acts as a functional A-to-G mutation by hydrolytic deamination at C6 of adenosine, changing it to inosine in the region of dsRNA (Figure 1A) (Nishikura, 2016).
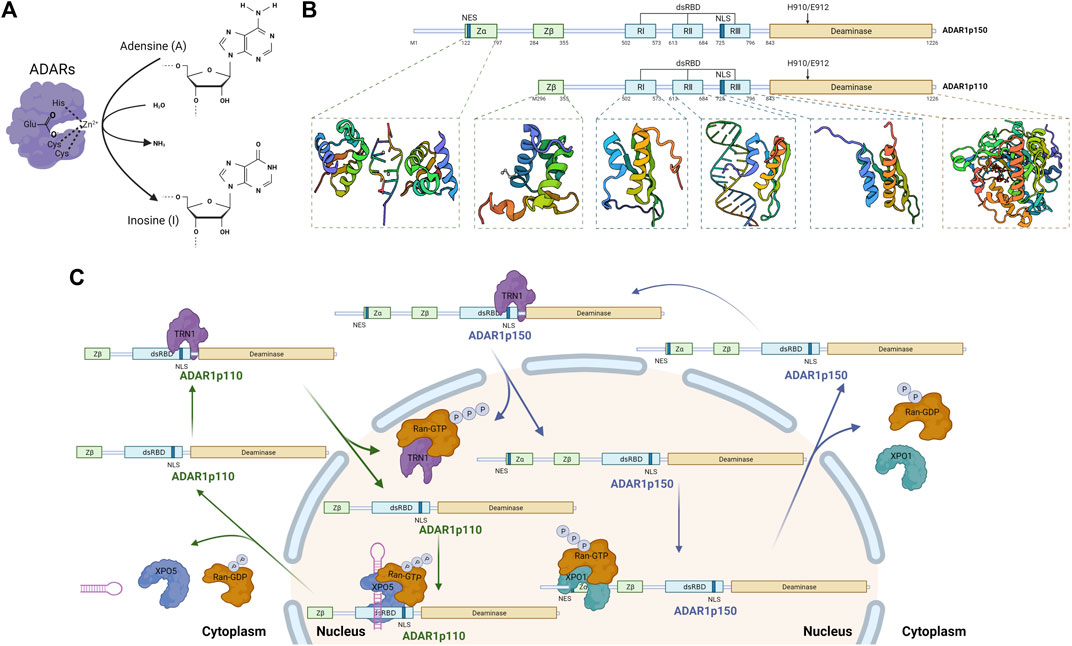
FIGURE 1. Catalytic function, domain organization, and shuttle mechanism of Adenosine Deaminase Acting on RNA 1 (ADAR1) (Created with BioRender.com). (A) RNA editing by ADARs. Alternative RNA editing by C6 deamination of adenosine (A) in the double-stranded RNA region to generate inosine (I), catalyzed by ADARs enzymes. (B) Schematic of the domain structure of two isoforms of ADAR1. ADAR1p150 and ADAR1p110 share identical sequence except for an additional N-terminal sequence for ADAR1p150, including an additional Zα domain (PDB: 2GDB) (Placido et al., 2007) and the NES which allows for both cytoplasmic and nuclear localization. Both contain a Zβ domain (PDB: 1XMK) (Athanasiadis et al., 2005) that mainly mediate Z-DNA/RNA binding, three dsRBDs domain (PDB: 2LJH, 2L2K, 2MDR) (Stefl et al., 2010; Barraud et al., 2012; Barraud et al., 2014) that mediate dsRNA binding and homodimerization, and a deaminase domain (PDB: 1ZY7) (Macbeth et al., 2005) that is the catalytic center of ADAR1. (C) Cellular localization of ADAR1. XPO1 binds to NES located within the Zα structural domain and regulates nuclear export of ADAR1p150 in the presence of RAN-GTP. Nuclear import of ADAR1p110 is mediated by binding of TRN1 to dsRBD3. NLS located in the third dsRBD, a domain common to both ADAR1 isoforms, binds with TRN1 to assist in the targeting of ADAR1 in the nucleus.
By hydrolytically deaminating adenosine’s 6-position, ADARs catalyze the deamination of adenosine to inosine (Polson et al., 1991). An important target for A-to-I RNA editing is a dsRNA derived from inverted Alu repetitive elements (Alu dsRNA) (Athanasiadis et al., 2004; Bazak et al., 2014). Since cells interpret inosine (I) as guanosine (G), A-to-I editing can lead to non-synonymous codon changes in the transcript and alternative splicing (Walkley and Li, 2017; Eisenberg and Levanon, 2018). In addition, ADARs can bind introns or 3′ untranslated regions (UTRs) to regulate the expression level of relative coding regions (Daniel et al., 2015; Sagredo et al., 2018). Still, it also affects targeting and disrupts the maturation of non-coding consequences, mainly microRNA, lncRNA, and circRNA (Velazquez-Torres et al., 2018; de Santiago et al., 2021; Shen et al., 2021). As a result of ADAR-mediated editing, which arises from mismatched A-to-I signatures after reverse transcription, more than 4 million edited positions have been identified (Bazak et al., 2014; Gallo et al., 2017).
Mammals have three ADAR genes, ADAR1 (also called ADAR, DRADA), ADAR2 (also called ADARB1), and ADAR3 (also called ADARB2), with specific structure, location, and function differences (Xu and Öhman, 2018; Shen et al., 2022). Currently, ADAR1 and ADAR2 have been shown to be expressed throughout most tissues and to be catalytically active, and the former mediates more editing events (Hogg et al., 2011). However, ADAR3 is specifically expressed in the brain and its function is only mentioned in a few articles (Melcher et al., 1996; Chen et al., 2000; Walkley and Li, 2017; Samuel, 2019). In glioblastoma, ADAR3 has been reported to be upregulated to competitively inhibit RNA editing at the Q/R site of GRIA2 by ADAR2 (Oakes et al., 2017; Wang et al., 2018). And these genes of the ADAR family have similar domain structures, mainly composed of a deaminase domain and two or three dsRNA binding domains (dsRBDs) (Erdmann et al., 2021; Quin et al., 2021). The deaminase domain of both ADAR1 and ADAR2 catalyzes the hydrolytic deamination of adenosine to inosine in dsRNA, while the catalytic activity of ADAR3 has not been reported so far (Chen et al., 2000; Nishikura, 2016; Oakes et al., 2017; Samuel, 2019). The dsRBD (∼65 amino acids) binds directly to dsRNA through its own structural domain. In spite of the fact that both ADAR1 and ADAR2 had been shown to be associated with tumor progression, editing events regulated by ADAR1 are more strongly correlated with cancer development (Maas et al., 2001; Paz-Yaacov et al., 2015). There could be two main reasons for this result. One is that the enrichment of ADAR1 far exceeds that of ADAR2. ADAR1 expression was seen in nearly all tissues, while ADAR2 was most expressed in the brain, but more minor in other tissues (Paz-Yaacov et al., 2015). Significantly, ADAR1 is absolutely required, and ADAR1-null mice die during embryonic life due to extensive apoptosis and defective hematopoiesis (Hartner et al., 2004; Wang et al., 2004). Furthermore, two is that human Adar1 maps to a single locus on chromosome 1q21, whose amplification is the most common cytogenetic abnormalities in multiple types of cancer and a poor prognostic factor (Knuutila et al., 1998; Jiang et al., 2012; Teoh et al., 2018).
ADARs editing is critical for survival in mammals, and its dysregulation may contribute to cancer development (Ramírez-Moya et al., 2021). The function of ADAR1-mediated RNA editing in immunity, especially innate immunity, has been summarized in many reviews (Lamers et al., 2019; Quin et al., 2021; Song et al., 2022), so this article will not address this aspect of the function. In addition to its editing role, ADAR1 can also play an editing-independent role. Regardless of whether the stability of ADARs regulatory RNAs is dependent on RNA editing, RNA binding is essential for their action (Wang I. X et al., 2013). Still, it will focus on the role of ADAR1 in cancer. For example, ADAR1 is more abundant in lung, liver, esophageal and chronic myelogenous leukemia, and with few exceptions, it promotes cancer progression (Jiang et al., 2012; Qin et al., 2014; Zhang et al., 2018; Sun et al., 2020). This review will focus on the structure and regulatory mechanisms of the ADAR1 enzymes and the relationship between aberrant editing of specific substrates and tumorigenesis, especially non-coding RNA.
Nature of Adenosine Deaminase Acting on RNA 1
The human ADAR1 gene spans approximately 40 kb and includes 17 exons, and the transcript level of ADAR1 is increased by IFN or pathogen stimulation (George et al., 2011). ADAR1 has two protein isoforms, full-length ADAR1p150 (150 kDa) and shorter ADAR1p110 (110 kDa) (Figure 1B) (Sun et al., 2021). Both contain a nuclear localization signal (NLS), allowing nuclear localization (Nishikura, 2016; Baker and Slack, 2022). In addition, ADAR1p150 contains a nuclear export signal (NES), accordingly shuttles between the nucleus and cytoplasm and is predominantly cytoplasmic (Xu and Öhman, 2018; Sun et al., 2021; Baker and Slack, 2022). Both isoforms contain a Zβ domain, three dsRNA-binding domains and a deaminase catalytic domain (George et al., 2011). 1) Zβ domain is a Z-DNA/RNA binding domain which NLS locates in (Athanasiadis et al., 2005); 2) dsRNA-binding domains (RI, RII, and RIII, ∼65 amino acids), which have an α-β-β-β-α configuration, make direct contact with dsRNA (Thomas and Beal, 2017); 3) deaminase domain is the catalytic center of ADAR1, in which the E912A point mutation in ADAR1p150 (E617A for ADAR1p110) disrupts catalytic deaminase activity (Wang I. X et al., 2013; Lamers et al., 2019; Baker and Slack, 2022). Moreover, besides Zβ domain, ADAR1p150 contains Zα domain, another Z-DNA/RNA binding domain, which may have affected its bonding preferences (Nishikura, 2016; Sun et al., 2021). Notably, shorter ADAR1p110 is constitutively expressed in ubiquitous types of cells, whereas full-length ADAR1p150 is expressed only when some activators stimulate cells, such as type I and type II IFN (Rice et al., 2012; Li et al., 2016). Among the inverted retrotransposon repeats of the long spacer nuclear element (LINE) and short spacer nuclear element (SINE) families are a number of endogenous long dsRNAs that are thought to be key-acting substrate RNAs for ADAR1 (Liddicoat et al., 2015). Moreover, the Alu motif mentioned above is a member of the SINE family and is also one of the sites found to be subject to the greatest extent and number of ADAR1 edits (Nishikura, 2010; Liddicoat et al., 2015; Nishikura, 2016).
Although ADAR1p150 is primarily localized in the cytoplasm and ADAR1p110 is predominantly localized in the nucleus, both ADAR1p150 and ADAR1p110 shuttle between the nucleus and cytoplasm (Figure 1C) (Strehblow et al., 2002; Desterro et al., 2003). The nuclear export factor exportin 1 (XPO1, also known as CRM1) binds to the nuclear export signal (NES) located within the Zα structural domain and regulates nuclear export of ADAR1p150 in the presence of RAN-GTP (Poulsen et al., 2001). The binding of transport protein 1 (TRN1) to dsRBD3 mediates nuclear import of ADAR1p110, a process that is inhibited by dsRNA binding (Barraud et al., 2014). The nuclear localization signal (NLS) located in the third dsRBD, a domain common to both ADAR1 isoforms, binds with TRN1 to assist in the targeting of ADAR1 in the nucleus (Fritz et al., 2009).
RNA Editing-Dependent of Adenosine Deaminase Acting on RNA 1 and Tumorigenesis and Progression Mechanisms
ADAR1 is frequently amplified in many diverse types of cancers with elevated activity (Fritzell et al., 2018), including hepatocellular carcinoma, non-small cell lung cancer, thyroid cancer, pancreatic cancer, esophageal cancer, cervical cancer, and multiple myeloma, and consistent with increased RNA editing levels of its substrates (Chen et al., 2013; Han et al., 2015; Paz-Yaacov et al., 2015; Chen Y et al., 2017; Hu et al., 2017; Sun et al., 2020). Conversely, decreased ADAR1 expression was observed in metastatic melanoma, invasive breast cancer, and kidney cancers (Shoshan et al., 2015; Gumireddy et al., 2016). Most RNA editing sites are located in noncoding regions or noncoding sequences and therefore do not result in changes in protein sequence or expression (Fritzell et al., 2018; Quin et al., 2021). However, a small number of coding events still occur in the coding region, thus affecting gene expression, or even if RNA editing occurs in non-coding sequences, it may indirectly regulates gene expression through mechanisms such as affecting the function of non-coding RNAs. The focus of this section is on this particular editing events by ADAR1 catalyzed, which contribute significantly to cancer development and metastasis. And the content of this section is divided into four main parts according to the differences in catalytic substrates, coding gene, intron, 3′ UTR, and three non-coding RNAs, including microRNA, lncRNA and circRNA. In most cases, increased ADAR1 expression and/or activity promotes cancer generation and progression; while in a few cancers, low expression and/or activity of ADAR1 mediating cancer phenotypes (Shoshan et al., 2015; Gumireddy et al., 2016). We have compiled and summarized the prior literature that experimentally confirms the substrate, editing site, and effects of ADAR1 editing on cancer phenotypes (Table 1).
Coding Genes
Early mechanistic studies on ADARs focused on causing protein recoding, which potentially modifies the amino acid sequence, thereby leading to decreased activity or acquisition of the encoded protein (Figure 2A). The coding sequence has a double-stranded structure on the exon, which is the region that may be subject to ADAR1-mediated A to I editing. Most A-to-I editing sites are generally found in introns and 3’ UTRs of coding genes, with 1% or fewer occurring in coding exons (Bahn et al., 2012; Wang I. X et al., 2013; Picardi et al., 2015).
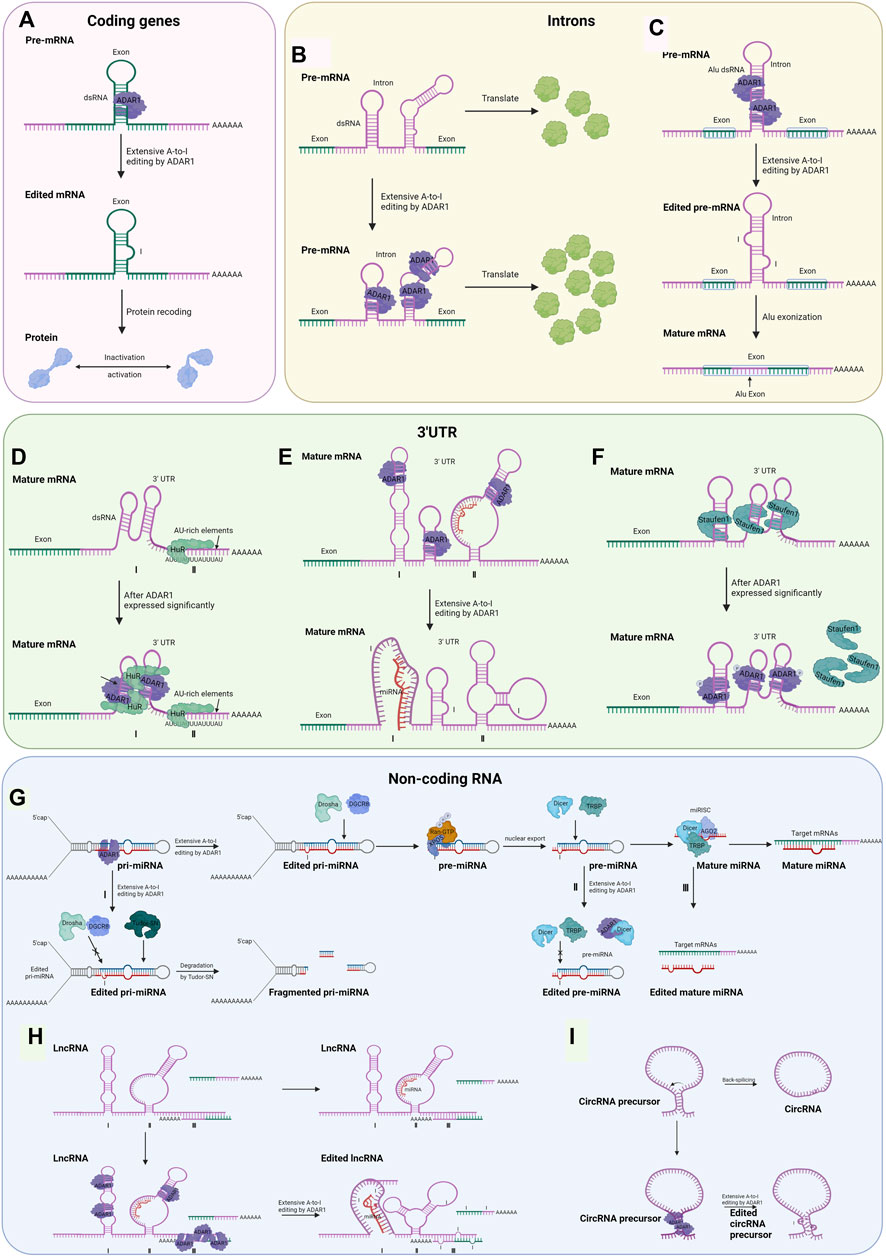
FIGURE 2. Possible regulatory mechanisms for RNA editing-dependent and editing-independent of ADAR1 involved in tumorigenesis and progression (Created with BioRender.com). (A) The dsRNA hairpin structures formed in the exonic region of the encoded gene are recognized by ADAR1. It undergoes A-to-I editing, and splicing machinery interprets inosine as guanosine, which leads to inactivation or activation of the final translation product. (B) ADAR1 binds to the intron of pre-mRNA, resulting in increased abundance of exon. (C) By editing intronic fold-back dsRNAs with A-to-I editing, splice sites could be created or deleted, leading to the inclusion or exclusion of Alu exons. (D) ADAR1 interacts with HuR proteins to coregulate common transcripts. (E) Editing of the 3′UTR can change the stability of the mRNA by creating or destroying the binding site of the miRNA. Ⅰ: acquired the ability to bind miRNA; Ⅱ: loss of the ability to bind miRNA. (F) ADAR1 competes with Staufen1 protein for the dsRNA binding site, thereby excluding Staufen1 binding and subsequent decaying of antiapoptotic genes in an editing-independent manner. (G) The Drosha/DGCR8 complex digests ADAR1-edited pri-miRNAs in the nucleus, generating approximately 70 nt of pre-miRNA intermediates that are translocated to the cytoplasm mediated by exportin-5 (Exp5) and Ran-GTP. The Dicer/TRBP complex undergoes a second cleavage to generate mature miRNAs which target and regulate downstream mRNAs. Because of the editing effect of ADAR1, the process may be aborted at three points. Ⅰ: in pri-mRNA, editing occurs at the RNA site that binds to Drosha and DGCR8, which causes the formation of fragmented pri-miRNA in the action of Tudor-SN; Ⅱ: in pre-miRNA, editing occurs at the RNA site that binds to Dicer and TRBP, or ADAR1 interacts with Dicer; Ⅲ: in mature miRNA, editing occurs at the RNA site that binds to mRNA. The mechanism of miRNA editing by ADAR1 is also applicable to the other two non-coding RNAs and will not be described below. (H) Regulatory mechanisms of ADAR1 Editing for dsRNA on lncRNA. Ⅰ: acquired the ability to bind miRNA; Ⅱ: loss of the ability to bind miRNA; Ⅲ: lncRNAs are recognized and edited by ADAR1 after complementary pairing with mRNA bases. (I) Editing of Alu repeats can antagonize circRNA formation.
Antizyme inhibitor 1 (AZIN1) is the most widely studied ADAR1 substrate in cancer, and edited AZIN1 promotes the development of hepatocellular carcinoma, non-small cell lung cancer, colorectal cancer, esophageal squamous cell carcinoma, and other cancers (Chen et al., 2013; Qin et al., 2014; Hu et al., 2017; Shigeyasu et al., 2018; Xu and Öhman, 2018). For instance, Under the action of deaminase of ADAR1, the serine (S) transforms glycine (G) substitution at residue 367, located in β-strand 15 of AZIN1 in HCC, resulting to its conformational change, inducing a cytoplasmic-to-nuclear translocation, and conferred “gain-of-function” phenotype, such as growth, colony formation, invasion, and migration (Chen et al., 2013). ADAR1 over-edited AZIN1 RNA is an independent risk factor for lymph node and distant metastasis and may serve as a prognostic basis for overall survival and disease-free survival (Shigeyasu et al., 2018). In several of these cancers, AZIN1 has the same editing site and causes similar phenotypic changes in cancer cells (Qin et al., 2014; Hu et al., 2017; Shigeyasu et al., 2018). However, in colorectal cancer, in addition to the above phenotypes, changes in cell stemness are also involved (Shigeyasu et al., 2018; Takeda et al., 2019).
Bladder cancer-associated protein (BLCAP) is a highly conserved gene that plays a tumor-suppressive role in different cancers and is a novel ADAR-mediated editing substrate in hepatocellular carcinoma and cervical cancer (Hu et al., 2015; Chen W et al., 2017). Over-editing of BLCAP was found in 40.1% HCCs compared to adjacent liver tissues. And RNA-edited BLCAP may stably promote cell proliferation by activating AKT/mTOR signal pathway (Hu et al., 2015). In cervical cancers, editing events by ADAR1 alter the genetically coded amino acid in BLCAP YXXQ motif, inducing BLCAP to lose the inhibition to signal transducer and activator of transcription 3 (STAT3) activation, which drives the carcinogenesis progression (Chen W et al., 2017). However, compared with the related normal tissues, BLCAP edited isoforms were reduced in astrocytoma, bladder cancer and colorectal cancer, indicating that the RNA editing level of BLCAP differs in different tumors (Meng et al., 2017).
However, in contrast to the above examples, in some cancers, such as breast cancer, melanoma, RNA editing of ADAR1 inhibits cancer development. In other words, low levels of ADAR1 editing in certain cancers lead to poor prognosis of patients (Gumireddy et al., 2016; Nishikura, 2016; Zhang et al., 2018; Baker and Slack, 2022). ADAR1-edited Gabra3 was only in non-invasive breast cancer and showed that edited Gabra3 reduced the abundance of wild-type Gabra3 on the cell surface and inhibited AKT activation, thereby suppressing breast cancer cell invasion and metastasis (Gumireddy et al., 2016). In melanoma, the ADAR1-edited cell cycle protein I (CCNI)R75G peptide activates tumor-infiltrating lymphocytes (TIL) and promotes TIL killing of cancer cells (Zhang et al., 2018).
To summarize, the above findings suggest that the expression of ADAR1 varies depending on the coding gene and cancer type. Whether ADAR1 overexpression or over-editing promotes or prevents cancer progression depends on the type of editing substrates, their level of expression, and how they participate respectively in regulating malignant changes in cancer cells.
Intron
The intron in certain coding regions has also been shown to recognize and bind to ADAR1 before being spliced and promote exon expression in an editing-dependent manner. A few articles have reported this approach, and the mechanisms have not been explored in detail (Figure 2B). ADAR1 post-transcriptionally increased the abundance of focal adhesion kinase (FAK) protein by binding and editing to a specific intronic site on chr8: 141,702,274 in FAK transcript, resulting in the increased stabilization of FAK mRNA, thereby promoting mesenchymal properties, migration, and invasion of lung adenocarcinoma (Amin et al., 2017).
RNA editing of Alu-rich intron regions may lead to Alu exonization, which controls mRNA levels and translation efficiency and potentially leads to disease (Figure 2C) (Lev-Maor et al., 2007; Sakurai et al., 2010). However, this modality is only found in G protein-coupled receptor 107 (Athanasiadis et al., 2004), nuclear prelamin A (Lev-Maor et al., 2007) and seryl-tRNA synthetase (Sakurai et al., 2010). However, there is no evidence that ADAR1 mediates cancer-associated gene exonization.
3′ Untranslated Region
There are two main ways ADAR1-catalyzed editing on the 3′ UTR influences gene expression regulation. A relatively common one is editing of Alu dsRNA on the 3′ UTR, which binds to and is regulated by microRNA when unedited. The other is dependent on editing to alter the stability of the mature mRNA, mainly achieved by the recruitment of HuR proteins with ADAR1 (Figure 2D) (Song et al., 2016). RNA editing events that occur in the 3′UTR of mRNAs may alter their interactions with miRNAs. Within the 3′ UTRs of mRNAs from nine different types of cancer, Pinto et al. identified over 63,000 editing sites that harbor Alu dsRNAs that serve as recruitment signals for ADAR (Pinto et al., 2018). Several studies have established that RNA editing that occurs in the 3′UTR can create or disrupt miRNA binding sites, thereby altering the mRNA stability of cancer-related genes (Figure 2E) (Liang and Landweber, 2007; Borchert et al., 2009).
For instance, 3′ UTR of Rho GTPase activating protein 26 (ARHGAP26) transcript undergoes ADAR1-catalyzed extensive A-to-I RNA editing (Wang Q et al., 2013). Known as a cancer suppressor, ARHGAP26 activates Rho GTPases (Zhong et al., 2019). Undergone A-to-I editing, the 3′ UTR of ARHGAP26 failed to pair with miR-30b-3p and miR-573, thus translating without interference, thereby promoting breast cancer growth and malignant transformation (Wang Q et al., 2013). As a counterexample, ADAR1, which is much less expressed in breast cancer, targets the 3′ UTR of DNA fragmentation factor subunit alpha (DFFA) transcript, therefore promoting cancer cell invasion (Roberts et al., 2018). DFFA is an inhibitor of caspase-activated DNase (ICAD) that triggers DNA fragmentation during apoptosis (Fawzy et al., 2017). In non-invasive, hormone-responsive breast cancer cell lines, editing of DFFA mRNA by ADFA1 rendered the mRNA unrecognized by miR-140-3p, thereby increasing DFFA levels and inducing apoptosis, whereas lack of editing in highly invasive, triple-negative breast cancer cell lines prevented DFFA from being regulated by miR-1403p (Roberts et al., 2018).
Altered RNA stability is an essential mechanism for ADAR1-mediated regulation of gene expression. ADAR1 recruits and interacts with human antigen R (HuR, gene name ELAVL1), a family of RNA-binding proteins selectively binds to single-stranded AU-rich RNA sequences (de Silanes et al., 2003; Meisner et al., 2004), to increase transcript stability (Figure 2D) (Wang I. X et al., 2013). It is estimated that 3′ UTR of up to 8% of all mRNAs contain AU-rich elements, including genes for cancer progression (Shaw and Kamen, 1986; Shaw et al., 2006). Among the 775 genes whose expression levels decreased after ADAR1 knockdown, the genes containing HuR binding sites showed significantly higher expression than the others (Wang I. X et al., 2013), such as MCM4 (Bagley et al., 2012), TMPO(Li et al., 2020), and GSR (McLoughlin et al., 2019).
Non-Coding RNA
Currently, dysregulated levels of non-coding RNAs (ncRNAs) appear to be reported for every type of cancer, and non-coding transcripts are expected to be the next class of diagnostic and therapeutic tools in oncology (Song et al., 2016; Shen et al., 2022). In addition to RNA editing mediated amino acid substitutions in specific genes contributing to cancer, elevated ADAR1 editing of non-coding RNAs, can also promote cancer progression (Qi et al., 2014; Ramírez-Moya et al., 2020). ADARs can also bind and edit some non-coding genes that contain inverted Alu repeats or LINE of non-coding genes, such as microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA) to suppress non-coding RNA maturation (Torsin et al., 2021).
MicroRNA
According to their binding sites along with RNA transcripts, ADAR1 protects mRNA from degradation, regulates miRNA processing, and alters splicing patterns (Xu and Öhman, 2018).
It has been shown that ADAR1 interacts with Dicer or DCGR8 to mediate processing of pre-miRNAs, ultimately leading to microRNA destabilization (Bahn et al., 2015). In this way, the editing of certain microRNA precursors results in lower levels of expression or altered function of mature miRNAs, thereby leading to alterations in certain cancer phenotypes (Figure 2G). In thyroid cancer cells, ADAR1-dependent editing miR-200b exhibits a lower activity against its target ZEB1-3′ UTR, which facilitates the epithelial–mesenchymal transition, thus resulting in proliferation, invasion, and migration of thyroid tumor cells (Ramírez-Moya et al., 2020). In melanoma cells, ADAR1p150 directly interacted with Dicer, an enzyme cleave precursor miRNA (usually 70 nt) to form mature miRNA (around 22 nt), which increased the cleavage rate of pre-miRNA and promoted the loading and maturing of miRNA (Chiosea et al., 2006), thereby promoting the biosynthesis and function of miRNA-149-3p (Yujie Ding et al., 2020). The expression of GSK3α, a direct target of miR-149-3p, is decreased, eventually resulting in proliferation of melanoma cells and inhibited cell apoptosis (Yujie Ding et al., 2020).
Likewise, modification of certain microRNAs by ADAR1 can exert a suppressive effect on cancer, and such miRNAs tend to be less expressed in cancerous tissues with a high metastatic capacity (Shoshan et al., 2015; Velazquez-Torres et al., 2018; Xu and Öhman, 2018). For example, with high levels of A-to-I editing in low metastatic melanoma but not in high metastatic melanoma, edited miR-455-5p lost the binding site of its downstream cancer suppressor protein CPEB1, which resulted in the suppression of melanoma growth and metastasis (Shoshan et al., 2015). Only in non-metastatic melanoma cells, edited miR-378a-3p predominantly binds to and represses the expression of the 3′-UTR of the PARVA oncogene, thereby inhibiting the progression of melanoma to a malignant phenotype (Velazquez-Torres et al., 2018).
Long Non-Coding RNA
Recently, LncRNAs have been shown to be central regulators of gene expression in a variety of genes (Guo et al., 2020). The lncRNAs involved in oncogenes and tumor suppressors are modified by A-to-I RNA editing machinery, with modifications that are drastically altered in cancer cells (Silvestris et al., 2020). ADAR1 can change RNA expression levels by interacting with other RNA-binding proteins, such as Dicer (Deng et al., 2019) and HuR (Stellos et al., 2016), the same mechanism as the 3′ UTR and the miRNA described previously. In addition, editing of dsRNA on lncRNA by ADAR1 changes its structure, which affects the binding of downstream target miRNAs (Figure 2H). Exceptional cases are reported in the literature where lncRNAs are recognized and edited by ADAR1 after complementary pairing with mRNA bases (Figure 2H) (Bussemakers et al., 1999).
The expression of ADAR1-regulated lncRNA LINC00944 is immune-related in breast cancer cells. LINC00944 expression was positively correlated to tumor infiltrating T lymphocytes, the age at diagnosis, tumor size, and poor prognosis (de Santiago et al., 2021). In spite of the variations in LINC00944 expression corresponding to up and down regulation of ADAR1, it is unknown the regulatory mechanism of ADAR1 for this lncRNA (de Santiago et al., 2021).
Prostate cancer antigen 3 (PCA3), a long non-coding RNA, is upregulated in human prostate cancer (Bussemakers et al., 1999). PCA3 regulates the level of PRUNE2 through a unique regulatory mechanism that involves the formation of PRUNE2/ADAR1-edited PCA3 double-stranded RNA. Editing by ADAR1 at multiple sites in the PCA3/PRUNE2 duplex results in a reduction of PRUNE2, and an increase in PCA3 expression, therefore increasing in cancer cell the ability of cancer cell, such as proliferation, adhesion and migration (Bussemakers et al., 1999).
Circular RNA
As mentioned above, ADAR1 interacts with by binding to the inverse complementary dsRNA, such as Alu repeat region; circRNAs can also interact with it in this way (Chen and Yang, 2015). ADAR1 creates or disrupts splice sites by acting on dsRNA editing, thereby inhibiting circRNA cyclization (Figure 2I) (Zhang and Carmichael, 2001; Fritz et al., 2009; Shevchenko and Morris, 2018).
CircNEIL3 and ADAR1 are upregulated in pancreatic ductal adenocarcinoma (PDAC) cells and tissues, and circNEIL3 as a miRNA sponge leads to down-regulation of miR-432-5p, thereby suppressing the down-regulation effect caused by the 3′UTR interaction of this microRNA with ADAR1 (Xu et al., 2020; Shen et al., 2021). Subsequently, ADAR1 enhances GLI1 editing, which promotes the transcription of target genes, including cyclin D1, cyclin E and Snail, and the cyclin-dependent kinases CDK2, CDK4 and CDK6 afterward decreased (Olive et al., 2009; Wang et al., 2020; Shen et al., 2021). In summary, the circNEIL3/miR-432-5p/ADAR1/GLI1/cyclin D1/CDK axis regulates the proliferation and metastasis of PDAC via the downstream GLI1/cyclin D1 and EMT pathway (Olive et al., 2009; Shen et al., 2021).
In addition, ADAR1 has been described to repress circRNA production intensely, and its A-I editing process usually occurs near reverse complementary matches (RCMs) in circRNA flanking introns, A structure essential for circRNA cyclization (Shi et al., 2017; Ma et al., 2020). Through this editing, secondary structures between RCMs could be stronger or weaker respectively depending on whether correcting A:C was mismatched to I(G)-C pairs or I(G) U pairs (Shen et al., 2022).
Androgen receptor (AR), a transcriptional activator of ADAR1 promoter, could suppress circARSP91 expression by upregulating ADAR1 p110, eventually leading to HCC tumor growth both in vitro and in vivo (You et al., 2015; Shi et al., 2017). Circular RNA hsa_circ_0004872, dramatically downregulated in gastric cancer, molecular sponges miR-224 to upregulate the expression of the miR-224 downstream targets p21 and Smad4 by targeting the 3′-UTR (Ma et al., 2020). Smad4, as a transcription factor, could inhibit ADAR1 expression level by directly binding to the promoter region of ADAR1, thereby further upregulating hsa_circ_0004872 levels. In other words, Smad4/ADAR1/hsa_circ_0004872/miR-224/Smad4 axis regulated tumor size and local lymph node metastasis in gastric cancer (Hansen et al., 2013; Zhu et al., 2017; Ma et al., 2020).
ADAR1 can edit reverse complementary matches (RCM) of ADAR1-regulated circRNAs, altering the secondary structure formed between RCMs within the flanking introns and enhancing the binding of RNA-binding proteins (RBPs) to the site of action (Kristensen et al., 2019; Shen et al., 2022). And the different expression of ADAR1 leads to editing dysregulation of A-to-I RNA editome in multiple cancers, such as HCC, ESCC, CRC, and GC (Qin et al., 2014; Han et al., 2020; Song et al., 2021). These ADAR1-regulated circRNAs are not byproducts of reverse splicing but important molecules that influence cancer development and progression.
RNA Editing-Independent of Adenosine Deaminase Acting on RNA 1 and Tumorigenesis and Progression Mechanisms
The most direct role of ADAR1 relies on A-to-I editing, which can alter coding sequences, binding motifs, RNA structure, etc., to regulate substrate abundance. However, ADARs have also been shown to have an editing-independent role, as they can also function as RNA-binding proteins independent of catalytic activity (Nishikura, 2016).
In melanoma, ADAR1 controls the expression of integrin beta 3 (ITGB3), a cell surface protein associated with tumor invasion (Pinon and Wehrle-Haller, 2011; Orgaz and Sanz-Moreno, 2013), via miR-22 and PAX6 transcription factor at the post-transcriptional and transcriptional levels. The expression and functional output of both FOXD1, which controls miR-22 expression, and PAX6 are independent of RNA editing, therefore promoting growth and invasion of melanoma (Nemlich et al., 2018).
In cytoplasm, binding of the targeted 3′ UTR allows phosphorylated ADAR1p110 to preclude binding of Staufen1 and subsequent decay of the anti-apoptotic gene, thereby promoting survival of stressed cells in an editorially independent manner (Figure 2F) (Sakurai et al., 2017). It has been reported in the literature that Staufen1 has positive or negative effects on disease progression: in some malignancies, upregulated Staufen1 may act as an oncogene and promote cancer progression (Boulay et al., 2014; Xu et al., 2015; Crawford Parks et al., 2017); while in other cancers, Staufen1 may act as a tumor suppressor and inhibit disease progression (Gong and Maquat, 2011; Boulay et al., 2014; Su et al., 2020). Nevertheless, reports on the mechanism of ADAR1 for Staufen1 have not yet appeared in cancer. Interaction of lncRNA with Staufen1 has also been reported in a variety of cancers (Sakurai et al., 2017); however again no mechanism of action of ADAR1 for Staufen1 in tumors has been reported. In HCC, ADAR1 was detected to interact directly with Dicer without editing, resulting in the processing of pre-miR-27a (Qi et al., 2017). Mature miR-27a binds to the 3′-UTR of methyltransferase 7A (METTL7A), a known tumor suppressor, decreasing its expression level (Qi et al., 2017). In addition, ADAR1 can regulate miRNA processing in an RNA-binding but editing-independent manner. ADAR1 can indirectly affect miRNA biogenesis by regulating Dicer expression at the translational level through the Letal-7 gene (let-7) (Zipeto et al., 2016).
Concluding Remarks and Future Perspectives
In conclusion, we show the structure and regulatory mechanisms of the ADAR1, and the significant role of ADAR1 in regulating many aspects of RNA function in cancers, including regulating the biogenesis of coding gene, intron, 3′ UTR, and three prevalent non-coding RNA, such as miRNA, lncRNA, circRNA. However, many important questions remain in the field including: 1) What are the factors that influence altered ADAR1 expression in cancer, and how do they play a role? 2) There are many editing-independent mechanisms of action in cancer that exist only in theory and for which there is no clear experimental evidence. 3) Up-regulation of ADAR1 expression promotes cancer development and progression in most cases, and down-regulation of ADAR1 in some cases can also achieve cancer progression. How do cancer cells control the abundance of ADAR1 so precisely to enhance their progression? Future investigation of these issues may lead to further discoveries in A-to-I RNA editing.
Author Contributions
BZ and JzL conceived the content and wrote the manuscript. FW and YZ contributed to the figures and the literature collection. BZ and JfL supervised the overall project and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Key R&D Program of China (SQ2020YFF0426513), the Scientific Foundation of Fuzhou Municipal Health commission (2021-S-wp1), the Backbone Talents Training Project of Fujian provincial Health Commission (2020GGA072).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amin, E. M., Liu, Y., Deng, S., Tan, K. S., Chudgar, N., Mayo, M. W., et al. (2017). The RNA-Editing Enzyme ADAR Promotes Lung Adenocarcinoma Migration and Invasion by Stabilizing FAK. Sci. Signal. 10, eaah3941. doi:10.1126/scisignal.aah3941
Athanasiadis, A., Placido, D., Maas, S., Brown, B. A., Lowenhaupt, K., and Rich, A. (2005). The Crystal Structure of the Zβ Domain of the RNA-Editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-Domains. J. Mol. Biol. 351, 496–507. doi:10.1016/j.jmb.2005.06.028
Athanasiadis, A., Rich, A., and Maas, S. (2004). Widespread A-To-I RNA Editing of Alu-Containing mRNAs in the Human Transcriptome. PLoS Biol. 2, e391. doi:10.1371/journal.pbio.0020391
Bagley, B. N., Keane, T. M., Maklakova, V. I., Marshall, J. G., Lester, R. A., Cancel, M. M., et al. (2012). A Dominantly Acting Murine Allele of Mcm4 Causes Chromosomal Abnormalities and Promotes Tumorigenesis. PLoS Genet. 8, e1003034. doi:10.1371/journal.pgen.1003034
Bahn, J. H., Ahn, J., Lin, X., Zhang, Q., Lee, J.-H., Civelek, M., et al. (2015). Genomic Analysis of ADAR1 Binding and its Involvement in Multiple RNA Processing Pathways. Nat. Commun. 6, 6355. doi:10.1038/ncomms7355
Bahn, J. H., Lee, J.-H., Li, G., Greer, C., Peng, G., and Xiao, X. (2012). Accurate Identification of A-To-I RNA Editing in Human by Transcriptome Sequencing. Genome Res. 22, 142–150. doi:10.1101/gr.124107.111
Baker, A. R., and Slack, F. J. (2022). ADAR1 and its Implications in Cancer Development and Treatment. Trends Genet. doi:10.1016/j.tig.2022.03.013
Bakheet, T., Kamen, R., and Bakheet, T. (2006). ARED 3.0: the Large and Diverse AU-Rich Transcriptome. Nucleic Acids Res. 34, D111–D114. doi:10.1093/nar/gkj052
Barraud, P., Banerjee, S., Mohamed, W. I., Jantsch, M. F., and Allain, F. H.-T. (2014). A Bimodular Nuclear Localization Signal Assembled via an Extended Double-Stranded RNA-Binding Domain Acts as an RNA-Sensing Signal for Transportin 1. Proc. Natl. Acad. Sci. U.S.A. 111, E1852–E1861. doi:10.1073/pnas.1323698111
Barraud, P., Heale, B. S. E., O’Connell, M. A., and Allain, F. H.-T. (2012). Solution Structure of the N-Terminal dsRBD of Drosophila ADAR and Interaction Studies with RNA. Biochimie 94, 1499–1509. doi:10.1016/j.biochi.2011.12.017
Bass, B. (1987). A Developmentally Regulated Activity that Unwinds RNA Duplexes. Cell 48, 607–613. doi:10.1016/0092-8674(87)90239-X
Bazak, L., Haviv, A., Barak, M., Jacob-Hirsch, J., Deng, P., Zhang, R., et al. (2014). A-to-I RNA Editing Occurs at over a Hundred Million Genomic Sites, Located in a Majority of Human Genes. Genome Res. 24, 365–376. doi:10.1101/gr.164749.113
Borchert, G. M., Gilmore, B. L., Spengler, R. M., Xing, Y., Lanier, W., Bhattacharya, D., et al. (2009). Adenosine Deamination in Human Transcripts Generates Novel microRNA Binding Sites. Hum. Mol. Genet. 18, 4801–4807. doi:10.1093/hmg/ddp443
Boulay, K., Ghram, M., Viranaicken, W., Trépanier, V., Mollet, S., Fréchina, C., et al. (2014). Cell Cycle-dependent Regulation of the RNA-Binding Protein Staufen1. Nucleic Acids Res. 42, 7867–7883. doi:10.1093/nar/gku506
Bussemakers, M. J., van Bokhoven, A., Verhaegh, G. W., Smit, F. P., Karthaus, H. F., Schalken, J. A., et al. (1999). DD3: a New Prostate-specific Gene, Highly Overexpressed in Prostate Cancer. Cancer Res. 59, 5975–5979.
Chen, C.-X., Cho, D.-S. C., Wang, Q., Lai, F., Carter, K. C., and Nishikura, K. (2000). A Third Member of the RNA-specific Adenosine Deaminase Gene Family, ADAR3, Contains Both Single- and Double-Stranded RNA Binding Domains. RNA 6, 755–767. doi:10.1017/S1355838200000170
Chen, L.-L., and Yang, L. (2015). Regulation of circRNA Biogenesis. RNA Biol. 12, 381–388. doi:10.1080/15476286.2015.1020271
Chen, L., Li, Y., Lin, C. H., Chan, T. H. M., Chow, R. K. K., Song, Y., et al. (2013). Recoding RNA Editing of AZIN1 Predisposes to Hepatocellular Carcinoma. Nat. Med. 19, 209–216. doi:10.1038/nm.3043
Chen, W., He, W., Cai, H., Hu, B., Zheng, C., Ke, X., et al. (2017). A-to-I RNA Editing of BLCAP Lost the Inhibition to STAT3 Activation in Cervical Cancer. Oncotarget 8, 39417–39429. doi:10.18632/oncotarget.17034
Chen, Y., Wang, H., Lin, W., and Shuai, P. (2017). ADAR1 Overexpression Is Associated with Cervical Cancer Progression and Angiogenesis. Diagn Pathol. 12, 12. doi:10.1186/s13000-017-0600-0
Chiosea, S., Jelezcova, E., Chandran, U., Acquafondata, M., McHale, T., Sobol, R. W., et al. (2006). Up-Regulation of Dicer, a Component of the MicroRNA Machinery, in Prostate Adenocarcinoma. Am. J. Pathology 169, 1812–1820. doi:10.2353/ajpath.2006.060480
Crawford Parks, T. E., Marcellus, K. A., Langill, J., Ravel-Chapuis, A., Michaud, J., Cowan, K. N., et al. (2017). Novel Roles for Staufen1 in Embryonal and Alveolar Rhabdomyosarcoma via C-myc-dependent and -independent Events. Sci. Rep. 7, 42342. doi:10.1038/srep42342
Daniel, C., Lagergren, J., and Öhman, M. (2015). RNA Editing of Non-coding RNA and its Role in Gene Regulation. Biochimie 117, 22–27. doi:10.1016/j.biochi.2015.05.020
de Santiago, P. R., Blanco, A., Morales, F., Marcelain, K., Harismendy, O., Sjöberg Herrera, M., et al. (2021). Immune-related IncRNA LINC00944 Responds to Variations in ADAR1 Levels and it Is Associated with Breast Cancer Prognosis. Life Sci. 268, 118956. doi:10.1016/j.lfs.2020.118956
de Silanes, I. L., Fan, J., Yang, X., Zonderman, A. B., Potapova, O., Pizer, E. S., et al. (2003). Role of the RNA-Binding Protein HuR in Colon Carcinogenesis. Oncogene 22, 7146–7154. doi:10.1038/sj.onc.1206862
Deng, S.-J., Chen, H.-Y., Zeng, Z., Deng, S., Zhu, S., Ye, Z., et al. (2019). Nutrient Stress-Dysregulated Antisense lncRNA GLS-AS Impairs GLS-Mediated Metabolism and Represses Pancreatic Cancer Progression. Cancer Res. 79, 1398–1412. doi:10.1158/0008-5472.CAN-18-0419
Desterro, J. M. P., Keegan, L. P., Lafarga, M., Berciano, M. T., O'Connell, M., and Carmo-Fonseca, M. (2003). Dynamic Association of RNA-Editing Enzymes with the Nucleolus. J. Cell Sci. 116, 1805–1818. doi:10.1242/jcs.00371
Dong, X., Chen, G., Cai, Z., Li, Z., Qiu, L., Xu, H., et al. (2018). CDK13 RNA Over-editing Mediated by ADAR1 Associates with Poor Prognosis of Hepatocellular Carcinoma Patients. Cell Physiol. Biochem. 47, 2602–2612. doi:10.1159/000491656
Eisenberg, E., and Levanon, E. Y. (2018). A-to-I RNA Editing - Immune Protector and Transcriptome Diversifier. Nat. Rev. Genet. 19, 473–490. doi:10.1038/s41576-018-0006-1
Erdmann, E. A., Mahapatra, A., Mukherjee, P., Yang, B., and Hundley, H. A. (2021). To Protect and Modify Double-Stranded RNA - the Critical Roles of ADARs in Development, Immunity and Oncogenesis. Crit. Rev. Biochem. Mol. Biol. 56, 54–87. doi:10.1080/10409238.2020.1856768
Fawzy, M. S., Toraih, E. A., Ibrahiem, A., Abdeldayem, H., Mohamed, A. O., and Abdel-Daim, M. M. (2017). Evaluation of miRNA-196a2 and Apoptosis-Related Target Genes: ANXA1, DFFA and PDCD4 Expression in Gastrointestinal Cancer Patients: A Pilot Study. PLoS ONE 12, e0187310. doi:10.1371/journal.pone.0187310
Fritz, J., Strehblow, A., Taschner, A., Schopoff, S., Pasierbek, P., and Jantsch, M. F. (2009). RNA-regulated Interaction of Transportin-1 and Exportin-5 with the Double-Stranded RNA-Binding Domain Regulates Nucleocytoplasmic Shuttling of ADAR1. Mol. Cell Biol. 29, 1487–1497. doi:10.1128/MCB.01519-08
Fritzell, K., Xu, L.-D., Lagergren, J., and Öhman, M. (2018). ADARs and Editing: The Role of A-To-I RNA Modification in Cancer Progression. Seminars Cell & Dev. Biol. 79, 123–130. doi:10.1016/j.semcdb.2017.11.018
Gallo, A., Vukic, D., Michalík, D., O’Connell, M. A., and Keegan, L. P. (2017). ADAR RNA Editing in Human Disease; More to it Than Meets the I. Hum. Genet. 136, 1265–1278. doi:10.1007/s00439-017-1837-0
Galore-Haskel, G., Nemlich, Y., Greenberg, E., Ashkenazi, S., Hakim, M., Itzhaki, O., et al. (2015). A Novel Immune Resistance Mechanism of Melanoma Cells Controlled by the ADAR1 Enzyme. Oncotarget 6, 28999–29015. doi:10.18632/oncotarget.4905
George, C. X., Gan, Z., Liu, Y., and Samuel, C. E. (2011). Adenosine Deaminases Acting on RNA, RNA Editing, and Interferon Action. J. Interferon & Cytokine Res. 31, 99–117. doi:10.1089/jir.2010.0097
Gong, C., and Maquat, L. E. (2011). lncRNAs Transactivate STAU1-Mediated mRNA Decay by Duplexing with 3′ UTRs via Alu Elements. Nature 470, 284–288. doi:10.1038/nature09701
Gumireddy, K., Li, A., Kossenkov, A. V., Sakurai, M., Yan, J., Li, Y., et al. (2016). The mRNA-Edited Form of GABRA3 Suppresses GABRA3-Mediated Akt Activation and Breast Cancer Metastasis. Nat. Commun. 7, 10715. doi:10.1038/ncomms10715
Guo, C.-J., Ma, X.-K., Xing, Y.-H., Zheng, C.-C., Xu, Y.-F., Shan, L., et al. (2020). Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 181, 621. doi:10.1016/j.cell.2020.03.006
Han, J., An, O., Hong, H., Chan, T. H. M., Song, Y., Shen, H., et al. (2020). Suppression of Adenosine-To-Inosine (A-To-I) RNA Editome by Death Associated Protein 3 (DAP3) Promotes Cancer Progression. Sci. Adv. 6, eaba5136. doi:10.1126/sciadv.aba5136
Han, L., Diao, L., Yu, S., Xu, X., Li, J., Zhang, R., et al. (2015). The Genomic Landscape and Clinical Relevance of A-To-I RNA Editing in Human Cancers. Cancer Cell 28, 515–528. doi:10.1016/j.ccell.2015.08.013
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA Circles Function as Efficient microRNA Sponges. Nature 495, 384–388. doi:10.1038/nature11993
Hartner, J. C., Schmittwolf, C., Kispert, A., Müller, A. M., Higuchi, M., and Seeburg, P. H. (2004). Liver Disintegration in the Mouse Embryo Caused by Deficiency in the RNA-Editing Enzyme ADAR1. J. Biol. Chem. 279, 4894–4902. doi:10.1074/jbc.M311347200
Hogg, M., Paro, S., Keegan, L. P., and O'Connell, M. A. (2011). RNA Editing by Mammalian ADARs. Adv. Genet. 73, 87–120. doi:10.1016/B978-0-12-380860-8.00003-3
Hu, X., Chen, J., Shi, X., Feng, F., Lau, K. W., Chen, Y., et al. (2017). RNA Editing of AZIN1 Induces the Malignant Progression of Non-small-cell Lung Cancers. Tumour Biol. 39, 101042831770000. doi:10.1177/1010428317700001
Hu, X., Wan, S., Ou, Y., Zhou, B., Zhu, J., Yi, X., et al. (2015). RNA Over-editing of BLCAP Contributes to Hepatocarcinogenesis Identified by Whole-Genome and Transcriptome Sequencing. Cancer Lett. 357, 510–519. doi:10.1016/j.canlet.2014.12.006
Jiang, A., Reece, D., and Chang, H. (2012). Genomic Stratification of Multiple Myeloma Treated with Novel Agents. Leukemia Lymphoma 53, 202–207. doi:10.3109/10428194.2011.608449
Knuutila, S., Björkqvist, A. M., Autio, K., Tarkkanen, M., Wolf, M., Monni, O., et al. (1998). DNA Copy Number Amplifications in Human Neoplasms: Review of Comparative Genomic Hybridization Studies. Am. J. Pathol. 152, 1107–1123.
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Lamers, M. M., van den Hoogen, B. G., and Haagmans, B. L. (2019). ADAR1: "Editor-In-Chief" of Cytoplasmic Innate Immunity. Front. Immunol. 10, 1763. doi:10.3389/fimmu.2019.01763
Lazzari, E., Mondala, P. K., Santos, N. D., Miller, A. C., Pineda, G., Jiang, Q., et al. (2017). Alu-dependent RNA Editing of GLI1 Promotes Malignant Regeneration in Multiple Myeloma. Nat. Commun. 8, 1922. doi:10.1038/s41467-017-01890-w
Lev-Maor, G., Sorek, R., Levanon, E. Y., Paz, N., Eisenberg, E., and Ast, G. (2007). RNA-editing-mediated Exon Evolution. Genome Biol. 8, R29. doi:10.1186/gb-2007-8-2-r29
Li, L., Qian, G., Zuo, Y., Yuan, Y., Cheng, Q., Guo, T., et al. (2016). Ubiquitin-dependent Turnover of Adenosine Deaminase Acting on RNA 1 (ADAR1) Is Required for Efficient Antiviral Activity of Type I Interferon. J. Biol. Chem. 291, 24974–24985. doi:10.1074/jbc.M116.737098
Li, Z., Feng, Y., Zhang, Z., Cao, X., and Lu, X. (2020). TMPO-AS1 Promotes Cell Proliferation of Thyroid Cancer via Sponging miR-498 to Modulate TMPO. Cancer Cell Int. 20, 294. doi:10.1186/s12935-020-01334-4
Liang, H., and Landweber, L. F. (2007). Hypothesis: RNA Editing of microRNA Target Sites in Humans? RNA 13, 463–467. doi:10.1261/rna.296407
Liddicoat, B. J., Piskol, R., Chalk, A. M., Ramaswami, G., Higuchi, M., Hartner, J. C., et al. (2015). RNA Editing by ADAR1 Prevents MDA5 Sensing of Endogenous dsRNA as Nonself. Science 349, 1115–1120. doi:10.1126/science.aac7049
Ma, C., Wang, X., Yang, F., Zang, Y., Liu, J., Wang, X., et al. (2020). Circular RNA Hsa_circ_0004872 Inhibits Gastric Cancer Progression via the miR-224/Smad4/ADAR1 Successive Regulatory Circuit. Mol. Cancer 19, 157. doi:10.1186/s12943-020-01268-5
Maas, S., Patt, S., Schrey, M., and Rich, A. (2001). Underediting of Glutamate Receptor GluR-B mRNA in Malignant Gliomas. Proc. Natl. Acad. Sci. U.S.A. 98, 14687–14692. doi:10.1073/pnas.251531398
Macbeth, M. R., Schubert, H. L., VanDemark, A. P., Lingam, A. T., Hill, C. P., and Bass, B. L. (2005). Inositol Hexakisphosphate Is Bound in the ADAR2 Core and Required for RNA Editing. Science 309, 1534–1539. doi:10.1126/science.1113150
McLoughlin, M. R., Orlicky, D. J., Prigge, J. R., Krishna, P., Talago, E. A., Cavigli, I. R., et al. (2019). TrxR1, Gsr, and Oxidative Stress Determine Hepatocellular Carcinoma Malignancy. Proc. Natl. Acad. Sci. U.S.A. 116, 11408–11417. doi:10.1073/pnas.1903244116
Meisner, N.-C., Hackermüller, J., Uhl, V., Aszódi, A., Jaritz, M., and Auer, M. (2004). mRNA Openers and Closers: Modulating AU-Rich Element-Controlled mRNA Stability by a Molecular Switch in mRNA Secondary Structure. ChemBioChem 5, 1432–1447. doi:10.1002/cbic.200400219
Melcher, T., Maas, S., Herb, A., Sprengel, R., Higuchi, M., and Seeburg, P. H. (1996). RED2, a Brain-specific Member of the RNA-specific Adenosine Deaminase Family. J. Biol. Chem. 271, 31795–31798. doi:10.1074/jbc.271.50.31795
Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y., Li, P., et al. (2017). CircRNA: Functions and Properties of a Novel Potential Biomarker for Cancer. Mol. Cancer 16, 94. doi:10.1186/s12943-017-0663-2
Nakano, M., Fukami, T., Gotoh, S., and Nakajima, M. (2017). A-to-I RNA Editing Up-Regulates Human Dihydrofolate Reductase in Breast Cancer. J. Biol. Chem. 292, 4873–4884. doi:10.1074/jbc.M117.775684
Nemlich, Y., Baruch, E. N., Besser, M. J., Shoshan, E., Bar-Eli, M., Anafi, L., et al. (2018). ADAR1-mediated Regulation of Melanoma Invasion. Nat. Commun. 9, 2154. doi:10.1038/s41467-018-04600-2
Nishikura, K. (2016). A-to-I Editing of Coding and Non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96. doi:10.1038/nrm.2015.4
Nishikura, K. (2010). Functions and Regulation of RNA Editing by ADAR Deaminases. Annu. Rev. Biochem. 79, 321–349. doi:10.1146/annurev-biochem-060208-105251
Oakes, E., Anderson, A., Cohen-Gadol, A., and Hundley, H. A. (2017). Adenosine Deaminase that Acts on RNA 3 (ADAR3) Binding to Glutamate Receptor Subunit B Pre-mRNA Inhibits RNA Editing in Glioblastoma. J. Biol. Chem. 292, 4326–4335. doi:10.1074/jbc.M117.779868
Olive, K. P., Jacobetz, M. A., Davidson, C. J., Gopinathan, A., McIntyre, D., Honess, D., et al. (2009). Inhibition of Hedgehog Signaling Enhances Delivery of Chemotherapy in a Mouse Model of Pancreatic Cancer. Science 324, 1457–1461. doi:10.1126/science.1171362
Orgaz, J. L., and Sanz-Moreno, V. (2013). Emerging Molecular Targets in Melanoma Invasion and Metastasis. Pigment. Cell Melanoma Res. 26, 39–57. doi:10.1111/pcmr.12041
Paz-Yaacov, N., Bazak, L., Buchumenski, I., Porath, H. T., Danan-Gotthold, M., Knisbacher, B. A., et al. (2015). Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep. 13, 267–276. doi:10.1016/j.celrep.2015.08.080
Picardi, E., Manzari, C., Mastropasqua, F., Aiello, I., D’Erchia, A. M., and Pesole, G. (2015). Profiling RNA Editing in Human Tissues: towards the Inosinome Atlas. Sci. Rep. 5, 14941. doi:10.1038/srep14941
Pinon, P., and Wehrle-Haller, B. (2011). Integrins: Versatile Receptors Controlling Melanocyte Adhesion, Migration and Proliferation. Pigment Cell & Melanoma Res. 24, 282–294. doi:10.1111/j.1755-148X.2010.00806.x
Pinto, Y., Buchumenski, I., Levanon, E. Y., and Eisenberg, E. (2018). Human Cancer Tissues Exhibit Reduced A-To-I Editing of miRNAs Coupled with Elevated Editing of Their Targets. Nucleic Acids Res. 46, 71–82. doi:10.1093/nar/gkx1176
Placido, D., Brown, B. A., Lowenhaupt, K., Rich, A., and Athanasiadis, A. (2007). A Left-Handed RNA Double Helix Bound by the Zα Domain of the RNA-Editing Enzyme ADAR1. Structure 15, 395–404. doi:10.1016/j.str.2007.03.001
Polson, A. G., Crain, P. F., Pomerantz, S. C., McCloskey, J. A., and Bass, B. L. (1991). The Mechanism of Adenosine to Inosine Conversion by the Double-Stranded RNA Unwinding/modifying Activity: a High-Performance Liquid Chromatography-Mass Spectrometry Analysis. Biochemistry 30, 11507–11514. doi:10.1021/bi00113a004
Poulsen, H., Nilsson, J., Damgaard, C. K., Egebjerg, J., and Kjems, J. (2001). CRM1 Mediates the Export of ADAR1 through a Nuclear Export Signal within the Z-DNA Binding Domain. Mol. Cell Biol. 21, 7862–7871. doi:10.1128/MCB.21.22.7862-7871.2001
Qi, L., Chan, T. H. M., Tenen, D. G., and Chen, L. (2014). RNA Editome Imbalance in Hepatocellular Carcinoma. Cancer Res. 74, 1301–1306. doi:10.1158/0008-5472.CAN-13-3485
Qi, L., Song, Y., Chan, T. H. M., Yang, H., Lin, C. H., Tay, D. J. T., et al. (2017). An RNA editing/dsRNA Binding-independent Gene Regulatory Mechanism of ADARs and its Clinical Implication in Cancer. Nucleic Acids Res. 45, 10436–10451. doi:10.1093/nar/gkx667
Qin, Y.-R., Qiao, J.-J., Chan, T. H. M., Zhu, Y.-H., Li, F.-F., Liu, H., et al. (2014). Adenosine-to-Inosine RNA Editing Mediated by ADARs in Esophageal Squamous Cell Carcinoma. Cancer Res. 74, 840–851. doi:10.1158/0008-5472.CAN-13-2545
Quin, J., Sedmík, J., Vukić, D., Khan, A., Keegan, L. P., and O’Connell, M. A. (2021). ADAR RNA Modifications, the Epitranscriptome and Innate Immunity. Trends Biochem. Sci. 46, 758–771. doi:10.1016/j.tibs.2021.02.002
Ramírez-Moya, J., Baker, A. R., Slack, F. J., and Santisteban, P. (2020). ADAR1-mediated RNA Editing Is a Novel Oncogenic Process in Thyroid Cancer and Regulates miR-200 Activity. Oncogene 39, 3738–3753. doi:10.1038/s41388-020-1248-x
Ramírez-Moya, J., Miliotis, C., Baker, A. R., Gregory, R. I., Slack, F. J., and Santisteban, P. (2021). An ADAR1-dependent RNA Editing Event in the Cyclin-dependent Kinase CDK13 Promotes Thyroid Cancer Hallmarks. Mol. Cancer 20, 115. doi:10.1186/s12943-021-01401-y
Rice, G. I., Kasher, P. R., Forte, G. M. A., Mannion, N. M., Greenwood, S. M., Szynkiewicz, M., et al. (2012). Mutations in ADAR1 Cause Aicardi-Goutières Syndrome Associated with a Type I Interferon Signature. Nat. Genet. 44, 1243–1248. doi:10.1038/ng.2414
Roberts, J., Patterson, D., King, V., Amin, S., Polska, C., Houserova, D., et al. (2018). ADAR Mediated RNA Editing Modulates MicroRNA Targeting in Human Breast Cancer. Processes 6, 42. doi:10.3390/pr6050042
Sagredo, E. A., Blanco, A., Sagredo, A. I., Pérez, P., Sepúlveda-Hermosilla, G., Morales, F., et al. (2018). ADAR1-mediated RNA-Editing of 3′UTRs in Breast Cancer. Biol. Res. 51, 36. doi:10.1186/s40659-018-0185-4
Sakurai, M., Shiromoto, Y., Ota, H., Song, C., Kossenkov, A. V., Wickramasinghe, J., et al. (2017). ADAR1 Controls Apoptosis of Stressed Cells by Inhibiting Staufen1-Mediated mRNA Decay. Nat. Struct. Mol. Biol. 24, 534–543. doi:10.1038/nsmb.3403
Sakurai, M., Yano, T., Kawabata, H., Ueda, H., and Suzuki, T. (2010). Inosine Cyanoethylation Identifies A-To-I RNA Editing Sites in the Human Transcriptome. Nat. Chem. Biol. 6, 733–740. doi:10.1038/nchembio.434
Samuel, C. E. (2019). Adenosine Deaminase Acting on RNA (ADAR1), a Suppressor of Double-Stranded RNA-Triggered Innate Immune Responses. J. Biol. Chem. 294, 1710–1720. doi:10.1074/jbc.TM118.004166
Shaw, G., and Kamen, R. (1986). A Conserved AU Sequence from the 3′ Untranslated Region of GM-CSF mRNA Mediates Selective mRNA Degradation. Cell 46, 659–667. doi:10.1016/0092-8674(86)90341-7
Shen, H., An, O., Ren, X., Song, Y., Tang, S. J., Ke, X.-Y., et al. (2022). ADARs Act as Potent Regulators of Circular Transcriptome in Cancer. Nat. Commun. 13, 1508. doi:10.1038/s41467-022-29138-2
Shen, P., Yang, T., Chen, Q., Yuan, H., Wu, P., Cai, B., et al. (2021). CircNEIL3 Regulatory Loop Promotes Pancreatic Ductal Adenocarcinoma Progression via miRNA Sponging and A-To-I RNA-Editing. Mol. Cancer 20, 51. doi:10.1186/s12943-021-01333-7
Shevchenko, G., and Morris, K. V. (2018). All I's on the RADAR : Role of ADAR in Gene Regulation. FEBS Lett. 592, 2860–2873. doi:10.1002/1873-3468.13093
Shi, L., Yan, P., Liang, Y., Sun, Y., Shen, J., Zhou, S., et al. (2017). Circular RNA Expression Is Suppressed by Androgen Receptor (AR)-regulated Adenosine Deaminase that Acts on RNA (ADAR1) in Human Hepatocellular Carcinoma. Cell Death Dis. 8, e3171. doi:10.1038/cddis.2017.556
Shigeyasu, K., Okugawa, Y., Toden, S., Miyoshi, J., Toiyama, Y., Nagasaka, T., et al. (2018). AZIN1 RNA Editing Confers Cancer Stemness and Enhances Oncogenic Potential in Colorectal Cancer. JCI Insight 3, e99976. doi:10.1172/jci.insight.99976
Shimokawa, T., Rahman, M. F.-U., Tostar, U., Sonkoly, E., Ståhle, M., Pivarcsi, A., et al. (2013). RNA Editing of the GLI1 Transcription Factor Modulates the Output of Hedgehog Signaling. RNA Biol. 10, 321–333. doi:10.4161/rna.23343
Shoshan, E., Mobley, A. K., Braeuer, R. R., Kamiya, T., Huang, L., Vasquez, M. E., et al. (2015). Reduced Adenosine-To-Inosine miR-455-5p Editing Promotes Melanoma Growth and Metastasis. Nat. Cell Biol. 17, 311–321. doi:10.1038/ncb3110
Silvestris, D. A., Scopa, C., Hanchi, S., Locatelli, F., and Gallo, A. (2020). De Novo A-to-I RNA Editing Discovery in lncRNA. Cancers 12, 2959. doi:10.3390/cancers12102959
Song, B., Shiromoto, Y., Minakuchi, M., and Nishikura, K. (2022). The Role of RNA Editing Enzyme ADAR1 in Human Disease. WIREs RNA 13, e1665. doi:10.1002/wrna.1665
Song, C., Sakurai, M., Shiromoto, Y., and Nishikura, K. (2016). Functions of the RNA Editing Enzyme ADAR1 and Their Relevance to Human Diseases. Genes 7, 129. doi:10.3390/genes7120129
Song, Y., An, O., Ren, X., Chan, T. H. M., Tay, D. J. T., Tang, S. J., et al. (2021). RNA Editing Mediates the Functional Switch of COPA in a Novel Mechanism of Hepatocarcinogenesis. J. Hepatology 74, 135–147. doi:10.1016/j.jhep.2020.07.021
Stefl, R., Oberstrass, F. C., Hood, J. L., Jourdan, M., Zimmermann, M., Skrisovska, L., et al. (2010). The Solution Structure of the ADAR2 dsRBM-RNA Complex Reveals a Sequence-specific Readout of the Minor Groove. Cell 143, 225–237. doi:10.1016/j.cell.2010.09.026
Stellos, K., Gatsiou, A., Stamatelopoulos, K., Perisic Matic, L., John, D., Lunella, F. F., et al. (2016). Adenosine-to-inosine RNA Editing Controls Cathepsin S Expression in Atherosclerosis by Enabling HuR-Mediated Post-transcriptional Regulation. Nat. Med. 22, 1140–1150. doi:10.1038/nm.4172
Strehblow, A., Hallegger, M., and Jantsch, M. F. (2002). Nucleocytoplasmic Distribution of Human RNA-Editing Enzyme ADAR1 Is Modulated by Double-Stranded RNA-Binding Domains, a Leucine-Rich Export Signal, and a Putative Dimerization Domain. Mol. Biol. Cell 13, 3822–3835. doi:10.1091/mbc.e02-03-0161
Su, R., Ma, J., Zheng, J., Liu, X., Liu, Y., Ruan, X., et al. (2020). PABPC1-induced Stabilization of BDNF-AS Inhibits Malignant Progression of Glioblastoma Cells through STAU1-Mediated Decay. Cell Death Dis. 11, 81. doi:10.1038/s41419-020-2267-9
Sun, T., Yu, Y., Wu, X., Acevedo, A., Luo, J.-D., Wang, J., et al. (2021). Decoupling Expression and Editing Preferences of ADAR1 P150 and P110 Isoforms. Proc. Natl. Acad. Sci. U.S.A. 118, e2021757118. doi:10.1073/pnas.2021757118
Sun, Y., Fan, J., Wang, B., Meng, Z., Ren, D., Zhao, J., et al. (2020). The Aberrant Expression of ADAR1 Promotes Resistance to BET Inhibitors in Pancreatic Cancer by Stabilizing C-Myc. Am. J. Cancer Res. 10, 148–163.
Takeda, S., Shigeyasu, K., Okugawa, Y., Yoshida, K., Mori, Y., Yano, S., et al. (2019). Activation of AZIN1 RNA Editing Is a Novel Mechanism that Promotes Invasive Potential of Cancer-Associated Fibroblasts in Colorectal Cancer. Cancer Lett. 444, 127–135. doi:10.1016/j.canlet.2018.12.009
Teoh, P. J., An, O., Chung, T.-H., Chooi, J. Y., Toh, S. H. M., Fan, S., et al. (2018). Aberrant Hyperediting of the Myeloma Transcriptome by ADAR1 Confers Oncogenicity and Is a Marker of Poor Prognosis. Blood 132, 1304–1317. doi:10.1182/blood-2018-02-832576
Thomas, J. M., and Beal, P. A. (2017). How Do ADARs Bind RNA? New Protein-RNA Structures Illuminate Substrate Recognition by the RNA Editing ADARs. BioEssays 39, 1600187. doi:10.1002/bies.201600187
Torsin, L. I., Petrescu, G. E. D., Sabo, A. A., Chen, B., Brehar, F. M., Dragomir, M. P., et al. (2021). Editing and Chemical Modifications on Non-coding RNAs in Cancer: A New Tale with Clinical Significance. Int. J. Mol. Sci. 22, 581. doi:10.3390/ijms22020581
Velazquez-Torres, G., Shoshan, E., Ivan, C., Huang, L., Fuentes-Mattei, E., Paret, H., et al. (2018). A-to-I miR-378a-3p Editing Can Prevent Melanoma Progression via Regulation of PARVA Expression. Nat. Commun. 9, 461. doi:10.1038/s41467-018-02851-7
Walkley, C. R., and Li, J. B. (2017). Rewriting the Transcriptome: Adenosine-To-Inosine RNA Editing by ADARs. Genome Biol. 18, 205. doi:10.1186/s13059-017-1347-3
Wang, F., Wu, P., Gong, S., Chen, Y., Gao, J., Wang, S., et al. (2020). Aberrant TRPM4 Expression in MLL-Rearranged Acute Myeloid Leukemia and its Blockade Induces Cell Cycle Arrest via AKT/GLI1/Cyclin D1 Pathway. Cell. Signal. 72, 109643. doi:10.1016/j.cellsig.2020.109643
Wang I. X, I. X., So, E., Devlin, J. L., Zhao, Y., Wu, M., and Cheung, V. G. (2013). ADAR Regulates RNA Editing, Transcript Stability, and Gene Expression. Cell Rep. 5, 849–860. doi:10.1016/j.celrep.2013.10.002
Wang Q, Q., Hui, H., Guo, Z., Zhang, W., Hu, Y., He, T., et al. (2013). ADAR1 Regulates ARHGAP26 Gene Expression through RNA Editing by Disrupting miR-30b-3p and miR-573 Binding. RNA 19, 1525–1536. doi:10.1261/rna.041533.113
Wang, Q., Miyakoda, M., Yang, W., Khillan, J., Stachura, D. L., Weiss, M. J., et al. (2004). Stress-induced Apoptosis Associated with Null Mutation of ADAR1 RNA Editing Deaminase Gene. J. Biol. Chem. 279, 4952–4961. doi:10.1074/jbc.M310162200
Wang, R., Zhang, S., Chen, X., Li, N., Li, J., Jia, R., et al. (2018). CircNT5E Acts as a Sponge of miR-422a to Promote Glioblastoma Tumorigenesis. Cancer Res. 78, 4812–4825. doi:10.1158/0008-5472.CAN-18-0532
Xu, L.-D., and Öhman, M. (2018). ADAR1 Editing and its Role in Cancer. Genes 10, 12. doi:10.3390/genes10010012
Xu, T.-p., Liu, X.-x., Xia, R., Yin, L., Kong, R., Chen, W.-m., et al. (2015). SP1-induced Upregulation of the Long Noncoding RNA TINCR Regulates Cell Proliferation and Apoptosis by Affecting KLF2 mRNA Stability in Gastric Cancer. Oncogene 34, 5648–5661. doi:10.1038/onc.2015.18
Xu, T., Lei, T., Li, S.-Q., Mai, E.-H., Ding, F.-H., and Niu, B. (2020). DNAH17-AS1 Promotes Pancreatic Carcinoma by Increasing PPME1 Expression via Inhibition of miR-432-5p. World J. Gastroenterol. 26, 1745–1757. doi:10.3748/wjg.v26.i15.1745
You, X., Vlatkovic, I., Babic, A., Will, T., Epstein, I., Tushev, G., et al. (2015). Neural Circular RNAs Are Derived from Synaptic Genes and Regulated by Development and Plasticity. Nat. Neurosci. 18, 603–610. doi:10.1038/nn.3975
Yu, J., Zhang, C., Yu, Q., Yu, H., and Zhang, B. (2019). ADAR1 P110 Enhances Adhesion of Tumor Cells to Extracellular Matrix in Hepatocellular Carcinoma via Up-Regulating ITGA2 Expression. Med. Sci. Monit. 25, 1469–1479. doi:10.12659/MSM.911944
Yujie Ding, M. M., Shi, X., Ji, J., and Su, Y. (2020). ADAR1p150 Regulates the Biosynthesis and Function of miRNA-149* in Human Melanoma. Biochem. Biophysical Res. Commun. 523, 900–907. doi:10.1016/j.bbrc.2019.12.110
Zhang, M., Fritsche, J., Roszik, J., Williams, L. J., Peng, X., Chiu, Y., et al. (2018). RNA Editing Derived Epitopes Function as Cancer Antigens to Elicit Immune Responses. Nat. Commun. 9, 3919. doi:10.1038/s41467-018-06405-9
Zhang, Z., and Carmichael, G. G. (2001). The Fate of dsRNA in the Nucleus. Cell 106, 465–476. doi:10.1016/s0092-8674(01)00466-4
Zhong, H., Wangxia, L., Fang, Y., Liu, Y., Zhao, Y., and Shi, Z. (2019). Circular RNA ARHGAP26 Is Over-expressed and its Downregulation Inhibits Cell Proliferation and Promotes Cell Apoptosis in Gastric Cancer Cells. Saudi J. Gastroenterol. 25, 119. doi:10.4103/sjg.SJG_283_18
Zhu, J., Ye, J., Zhang, L., Xia, L., Hu, H., Jiang, H., et al. (2017). Differential Expression of Circular RNAs in Glioblastoma Multiforme and its Correlation with Prognosis. Transl. Oncol. 10, 271–279. doi:10.1016/j.tranon.2016.12.006
Keywords: ADAR1, RNA editing, cancer, non-coading RNA, adenosine to inosine
Citation: Liu J, Wang F, Zhang Y, Liu J and Zhao B (2022) ADAR1-Mediated RNA Editing and Its Role in Cancer. Front. Cell Dev. Biol. 10:956649. doi: 10.3389/fcell.2022.956649
Received: 30 May 2022; Accepted: 23 June 2022;
Published: 11 July 2022.
Edited by:
Xiaoxing Li, The First Affiliated Hospital of Sun Yat-sen University, ChinaReviewed by:
Yao Lin, Fujian University of Traditional Chinese Medicine, ChinaHeather Hundley, Indiana University, United States
Copyright © 2022 Liu, Wang, Zhang, Liu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingfeng Liu, drjingfeng@126.com; Bixing Zhao, bixingzhao@gmail.com
 Jizhe Liu
Jizhe Liu Fei Wang1,4
Fei Wang1,4  Jingfeng Liu
Jingfeng Liu Bixing Zhao
Bixing Zhao