Studying the Inflammatory Responses to Amyloid Beta Oligomers in Brain-Specific Pericyte and Endothelial Co-Culture From Human Stem Cells
- 1Department of Chemical and Biomedical Engineering, FAMU-FSU College of Engineering, Florida State University, Tallahassee, FL, United States
- 2FIT BEST Laboratory, Department of Chemical, Biological, and Bio Engineering, North Carolina A&T State University, Greensboro, NC, United States
- 3Department of Biological Science, Florida State University, Tallahassee, FL, United States
- 4Department of Materials Science and Engineering, University of Connecticut, Storrs, CT, United States
- 5Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL, United States
- 6Department of Industrial and Manufacturing Engineering, FAMU-FSU College of Engineering, Florida State University, Tallahassee, FL, United States
- 7The High-Performance Materials Institute, Florida State University, Tallahassee, FL, United States
Background: Recently, the in vitro blood–brain barrier (BBB) models derived from human pluripotent stem cells have been given extensive attention in therapeutics due to the implications they have with the health of the central nervous system. It is essential to create an accurate BBB model in vitro in order to better understand the properties of the BBB, and how it can respond to inflammatory stimulation and be passed by targeted or non-targeted cell therapeutics, more specifically extracellular vesicles.
Methods: Brain-specific pericytes (iPCs) were differentiated from iPSK3 cells using dual SMAD signaling inhibitors and Wnt activation plus fibroblast growth factor 2 (FGF-2). The derived cells were characterized by immunostaining, flow cytometry, and RT-PCR. In parallel, blood vessels organoids were derived using Wnt activation, BMP4, FGF2, VEGF, and SB431542. The organoids were replated and treated with retinoic acid to enhance the blood–brain barrier (BBB) features in the differentiated brain endothelial cells (iECs). Co-culture was performed for iPCs and iECs in the transwell system and 3D microfluidics channels.
Results: The derived iPCs expressed common markers PDGFRb and NG2, and brain-specific genes FOXF2, ABCC9, KCNJ8, and ZIC1. The derived iECs expressed common endothelial cell markers CD31, VE-cadherin, and BBB-associated genes BRCP, GLUT-1, PGP, ABCC1, OCLN, and SLC2A1. The co-culture of the two cell types responded to the stimulation of amyloid β42 oligomers by the upregulation of the expression of TNFa, IL6, NFKB, Casp3, SOD2, and TP53. The co-culture also showed the property of trans-endothelial electrical resistance. The proof of concept vascularization strategy was demonstrated in a 3D microfluidics-based device.
Conclusion: The derived iPCs and iECs have brain-specific properties, and the co-culture of iPCs and iECs provides an in vitro BBB model that show inflammatory response. This study has significance in establishing micro-physiological systems for neurological disease modeling and drug screening.
Introduction
Human brain organoids derived from human pluripotent stem cells provide attractive tools to study human brain tissue, which exhibits a distinct gene expression profile compared to that of mice (Qian et al., 2016; Song et al., 2020; Martinelli et al., 2022). Vascularization of human brain organoids not only provides the nutrients and oxygen to increase the organoid size but also recapitulates the complex cell–cell interactions that can promote neuronal differentiation and network formation (Cakir et al., 2019; Shi et al., 2020; Garreta et al., 2021; Matsui et al., 2021). Human brain endothelial cells (BECs), pericytes, and astrocytes form the blood–brain barrier (BBB) that expresses human-specific transporters, displays the heterogeneity among different brain regions as well as between human and other species and is altered during the pathological disease progression such as neurodegeneration, brain tumorigenesis, and Zika virus infection (Wilhelm et al., 2016; Blanchard et al., 2020; Lee et al., 2020; Chen W et al., 2021; Schaffenrath et al., 2021).
The functionality and the components of in vitro BBB models have been investigated in recent years. However, only a smattering of the mechanisms of the immune response and dysfunction has been known. It is necessary to establish an in vitro BBB model with complete structures and functionalities. In particular, brain pericytes are the cells that have multiple functions and reside along BEC in the BBB microvasculature (Brown et al., 2019; Heymans et al., 2020). Recently, it was discovered that pericytes have been implied in cerebral blood flow regulation and BBB maintenance (Brown et al., 2019). Mainly, the constant cross talk and interaction with pericytes and glial cells allow BECs to function properly and react accordingly to changes under physiological conditions (Daneman et al., 2010). The mice with no brain pericytes exhibited BEC hyperplasia, increased vascular permeability, and increased vessel diameter, thus leading to death at birth (Daneman et al., 2010). To date, the major challenge of establishing in vitro BBB models is to recapitulate the high transendothelial electrical resistance (TEER) value observed in vivo (Lauschke et al., 2017). The BBB properties are not intrinsic to endothelial cells, but rather to the complex interaction of BECs with brain pericytes, astrocytes, and neurons (Campisi et al., 2018; Gastfriend et al., 2018; Linville et al., 2019; Stebbins et al., 2019; Blanchard et al., 2020).
In vitro BBB models derived from human-induced pluripotent stem cells (hiPSCs) have shown promising results as micro-physiological systems to study BBB function and neurovascular unit (Campisi et al., 2018; Brown et al., 2019; Canfield et al., 2019; Nguyen et al., 2019; Browne et al., 2021). The most common BBB models would include a co-culture of two, three, or four cell types, which usually use primary brain pericytes due to the limitation in efficiently differentiating brain pericytes from hiPSCs (Lauschke et al., 2017). Recently, the differentiation of pericytes with brain tissue identity from hiPSCs became possible (Jeske et al., 2020). In addition, the traditional co-culture method is to use a transwell system where the two compartments represent blood (apical) and brain (basolateral), respectively. BEC can be seeded onto the apical side of the transwell membrane while pericytes and astrocytes are seeded onto the basolateral side. Other methods include aggregates in hydrogels or in microfluidic channels (Gastfriend et al., 2018). The three-dimensional (3D) tissue-engineered models of BBB in different biomaterials and scaffolds are summarized in our recent review (Chen X et al., 2021) and reported by others (Faley et al., 2019; Grifno et al., 2019). The advantage of the transwell model is that it allows for the high TEER values of up to 5,000 ohms × cm2 and the increased expression of tight junction proteins. The major downside is that it does not permit for an ideal amount of cell-to-cell contact and lacks the influence due to the physiological blood flow.
Our previous study assembled cortical spheroids with the isogenic vascular spheroids in the presence of human mesenchymal stem cells (hMSCs, i.e., pericyte-like) (Song et al., 2019a; Song et al., 2019b). However, the individual cell types (i.e., endothelial cells and hMSCs) used in the culture system are not optimal for BBB formation and function. Going one step further, this study focuses on the differentiation and characterizations of brain pericyte-like cells and brain endothelial cells that form the BBB from hiPSCs. It is hypothesized that the co-culture of hiPSC-derived brain pericyte-like cells and the isogenic endothelial cells promotes the inflammatory response compared to single cell type co-culture. Amyloid beta (Aβ) 42 oligomers were used in this study due to our continuous interests in studying Alzheimer’s disease pathology using hiPSC-based culture models (Yan et al., 2016; Bejoy et al., 2018a; Yan et al., 2018; Song et al., 2019c; Griffin et al., 2020). Both cells responded to the stimulation of Aβ42 oligomers. Co-culture of the two cell types differentiated from hiPSCs in a 2D transwell system were investigated. The co-culture in the transwell system showed the property of trans-endothelial electrical resistance. The endothelial cell culture was also demonstrated in 3D microfluidics channels, which can be used to study inflammatory response in the future more physiologically relevant co-culture models. This study has significance in establishing micro-physiological systems for neurological disease modeling and drug screening.
Materials and Methods
Culture of Human iPSK3 Cells and Human Adipose Tissue-Derived Stem Cells
Human iPSK3 cells were derived from human foreskin fibroblasts transfected with plasmid DNA encoding reprogramming factors OCT4, NANOG, SOX2, and LIN28 (kindly provided by Dr. Stephen Duncan, Medical College of Wisconsin) (Si-Tayeb et al., 2010a; Si-Tayeb et al., 2010b). Human iPSK3 cells were maintained in mTeSR serum-free medium (StemCell Technologies, Inc., Vancouver, BC, Canada) on growth factor reduced Geltrex- or Matrigel-coated surface (Life Technologies) (Yan et al., 2015a). The cells were passaged by Accutase every 7 days and seeded at 1 × 106 cells per well of a 6-well plate in the presence of 10 μM Y27632 (Sigma) for the first 24 h (Yan et al., 2015a; Song et al., 2016; Bejoy et al., 2018b).
Frozen human adipose tissue-derived stem cells (hASCs) at passage 1 were acquired from the Tulane Center for Stem Cell Research and Regenerative Medicine. The hASCs were isolated from the subcutaneous abdominal adipose tissue from three de-identified healthy donors that were younger than 45 years old with a body mass index lower than 25 (Bijonowski et al., 2020a; Bijonowski et al., 2020b). The isolated cells were characterized for their MSC properties through colony-forming unit (CFU) assays and tri-lineage differentiation potential (osteogenic, adipogenic, and chondrogenic differentiation) in vitro. The hASCs (1 × 106 cells/ml/vial) were frozen in media containing α-MEM, 2 mM L-glutamine, 30% fetal bovine serum (FBS), and 10% dimethyl sulfoxide and were thawed and cultured following the method described in our previous publications (Song et al., 2019a; Liu et al., 2019; Jeske et al., 2021). Briefly, hASCs were seeded at a density of 1,500 cells/cm2 in 150 mm diameter Petri dish (Corning Incorporated, Corning, NY, United States) in a standard 5% CO2 incubator. The cells were cultured in complete culture medium (CCM), containing αMEM (Life Technologies, Carlsbad, CA, United States) with 10% FBS (Atlanta Biologicals, Lawrenceville, GA, United States), sodium bicarbonate (1X, ThermoFisher Scientific), and 1% penicillin/streptomycin (ThermoFisher Scientific) undergoing media changes every 2–3 days. The cells were grown to 80% confluence and then harvested by incubation with 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA) (Invitrogen, Grand Island, NY, United States) for no greater than 7 min. The harvested cells were sub-cultured up to passage 10 for characterizations.
Brain Pericyte Differentiation From Human-Induced Pluripotent Stem Cells
The brain-specific pericyte differentiation from iPSK3 cells was performed using a modified protocol (Faal et al., 2019; Jeske et al., 2020). Human iPSK3 cells were seeded into Matrigel (1:30, Corning)-coated attachment 24-well plates (Corning Incorporated, Corning, NY, United States) at 3 × 105 cells/well in 1 ml of Dulbecco’s modified Eagle medium/nutrient mixture F-12 (DMEM/F-12) plus 2% B27 serum-free supplement (Life Technologies). Y27632 (10 μM) was added during the seeding and removed after 24 h. During day 1–15, the cells were treated with dual SMAD signaling inhibitors 10 μM SB431542 (SB, Sigma), 100 nM LDN193189 (LDN, Sigma), and 1 µM Wnt activator CHIR99021 (CHIR, Sigma) and 10 ng/ml fibroblast growth factor 2 (FGF2) (PeproTech) in DMEM/F12 plus 2% B27 (Stebbins et al., 2019). The cells were passaged when confluent (usually every 5–6 days) and replated onto Matrigel (1:30 diluted)-coated plates at 2.5 × 105 cells per well. After day 15, the cells were fed with DMEM/F12 plus 10% FBS and were used for characterization after day 22. The derived cells were referred as iPCs.
Brain Microvascular Endothelial Cell Differentiation From Human-Induced Pluripotent Stem Cells
Human iPSK3 cells were seeded into low attachment 24-well plates (Corning Incorporated) at 3 × 105 cells/well in 1 ml of DMEM/F-12 plus 2% B27 serum-free supplement. Y27632 (10 μM) was added during the seeding and removed after 24 h. At day 3, the cells were treated with 12 µM CHIR99021. The spent media were removed on day 5, 7, and 9 and replaced with media containing 30 ng/ml bone morphogenetic protein 4 (BMP4) (PeproTech), 30 ng/ml FGF2, and 30 ng/ml vascular endothelial growth factor A (VEGFA) (PeproTech) (Wimmer et al., 2019). At day 11, the media were supplemented with 10 µM SB431542 plus FGF2 and VEGFA to promote endothelial cell differentiation and suppress pericyte differentiation, and from then onward the media were changed to DMEM/F12 with 15% FBS containing VEGFA and FGF2. The media were replaced every 2–3 days. Vascular networks may be observed around day 18 and can be replated onto Matrigel (1:30)-coated plates. For comparison, retinoic acid (RA, 10 μM, Sigma) was added during day 11–18 to enhance the blood–brain barrier markers (Lippmann et al., 2014). The derived cells were characterized after day 25 and referred as iECs.
Treatment of Pericytes and Endothelial Cells With Amyloid Beta 42 Oligomers
To prepare oligomers of the Aβ42 peptide, biotinylated Aβ42 (Bachem) was fully dissolved at 0.5 mg/ml in hexafluor-2-propanole (HFIP, Sigma) (Yan et al., 2016; Bejoy et al., 2018a; Griffin et al., 2020). HFIP Aβ (1-42) solution (10 μl) was dispensed into a siliconized Snap-Cap microtube, placed in a desiccator to completely evaporate HFIP and thereafter stored at −80°C. Oligomer solutions were prepared freshly for each experiment. The stock was dissolved in 10 μl of DMSO (to 105 μM) and incubated for 3 h at room temperature. Oligomers of Aβ42 (at 1 μM) were added to the day 22–25 iPC or day 25–28 iEC cultures. After 3 days, the cells were harvested for mRNA isolation.
Immunocytochemistry
For biomarker detection, the cells were fixed using 4% paraformaldehyde (PFA) and permeabilized using 0.2% Trixton-X 100. The samples were blocked with 5% FBS in PBS and stained with the primary antibodies (Supplementary Table S1), followed by Alexa Fluor 488 goat antimouse IgG1 or Alexa Fluor 594 goat antirabbit IgG (Life technologies). Both primary and secondary antibody dilutions were made based on the manufacturer’s recommendations and were prepared in staining buffer (2% FBS in PBS). The samples stained with secondary antibody dilution only were used as the negative controls. The cell nuclei were stained with Hoechst 33342 (blue), and the images were taken under a fluorescence microscope (Olympus IX70).
Phagocytosis Assay
The day 25 derived iECs and iPCs in a 24-well plate were incubated with medium containing 0.5 × 108 fluorescent (0.86 μm, flash red, 660/690 nm) micron-sized particles of iron oxide (MPIO)/mL (Bangs Laboratories, Fishers, IN, United States, Part number ME03F/9772), corresponding to the concentration at 5 μg Fe/mL (Yan et al., 2015b; Sart et al., 2015). The attached cells that were not labeled with MPIO served as control. After 24-h incubation, the cultures labeled with MPIO were extensively washed (10 times) with phosphate buffer saline. The cultures were then harvested for flow cytometry to quantify the cells with MPIO.
Flow Cytometry
For marker quantification, trypsinized cells were fixed (4% PFA) and permeabilized with 100% cold methanol, blocked with 5% FBS in PBS, and then stained with the corresponding marker antibody overnight. The secondary Alexa Fluor 488 or 594 antibody was later used, incubated for 1 hour, then removed and rinsed with PBS twice, and then taken for flow cytometry measurement. The cells were acquired with BD FACSCanto II flow cytometer (Becton Dickinson, Franklin Lakes, NJ, United States) and analyzed against the negative isotype controls using FlowJo software.
Reverse Transcription Polymerase Chain Reaction Analysis
Total mRNA was isolated from different cell samples using the RNeasy Mini Kit (Qiagen, Valencia, CA, United States), according to the manufacturer’s protocol. The samples were further treated using DNA-free RNA kit (Zymo, Irvine, CA, United States) (Song et al., 2016). Reverse transcription was carried out according to the manufacturer’s instructions using 2 μg of total mRNA, anchored oligo-dT primers (Operon, Huntsville, AL, United States), and SuperScript III (Invitrogen, Carlsbad, CA, United States). The software Oligo Explorer 1.2 Primers (Genelink, Hawthorne, NY, United States) was used to design the primers specific for target genes (Supplementary Table S2). For normalization of expression levels, β-actin was used as an endogenous control. Using SYBR1 Green PCR Master Mix (Applied Biosystems), real-time reverse transcription polymerase chain reaction (RT-PCR) was performed on an ABI7500 instrument (Applied Biosystems, Foster City, CA, United States), The amplification reactions were performed as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 95°C for 15 s, 5°C for 30 s, and 68°C for 30 s. The Ct values of the target genes were first normalized to the Ct values of the endogenous control β-actin. The corrected Ct values were then compared for the treatment conditions to the experimental control. Fold changes in gene expression was calculated using the comparative Ct method:
Co-Culture of Endothelial Cells and Pericytes in a Transwell System
iECs were seeded onto the transwell inserts as described previously (Stone et al., 2019). Prior to seeding, Corning polyester transwell inserts (pore size 3 μm, for 12-well plates) were coated with Matrigel (1:30) for 1 hour at 37°C on the apical side of the membrane. iECs were incubated with Accutase for 5 min at 37°C. The dissociated cells were diluted with DMEM/F12 for counting and centrifuged for 5 min at 500 g. The supernatant/Matrigel was removed, and the cells were seeded on transwell inserts at a density of 2.0 × 106 cells/insert with 20 ng/ml FGF-2 and 20 ng/ml VEGFA in DMEM/F12 plus 15% FBS (iEC maturation media). Following seeding, media were changed every other day with 0.5 ml in the apical side and 1.5 ml in the basolateral side (no FGF2 or VEGFA). The cells became confluent after day 7 and can be used for further characterizations.
The co-culture of iECs and iPCs in a transwell insert was then performed. Initially, transwell inserts were inverted and placed in autoclaved cases to be set under UV light for 30 min. After 30 min, the basolateral membrane was coated with Matrigel (1:30) and carefully placed in the incubator for 1 h at 37°C. The harvested iPCs were gently placed onto inserts at 1.0 × 106 cells/insert so that they almost covered the membrane. The wells were filled with 1.5 ml of iPC media. The seeded inserts were placed back in the plates and incubated for 45 min. The cells were allowed to grow for 24 h before iEC seeding on the apical membrane. After both cells were seeded and attached to membrane, media were changed every other day with 1.5 ml of iPC media in the basolateral compartment and 0.5 ml of iEC media in the apical side. After 7 days, the cells can be used for characterizations.
Transendothelial Electrical Resistance Measurement
For barrier permeability quantification, TEER measurements of the cell cultures in a transwell plate set up were taken using STX2 electrodes attached to an EVOM volt/ohm meter (World Precision Instruments). The STX2 electrodes were placed in the transwell plate to make contact between the media on the upper compartment and the lower compartment. The resistance (Ω) values were measured five times in each transwell, including a cell-free control to account for the transwell membrane resistance. Later, the Ω values were adjusted with the surface growth area (cm2) of the transwells in the plate to obtain the TEER (Ω. cm2) values of the barrier formed by the iECs or the co-culture.
Device Preparation and Cell Culture in Microfluidic Channels
The microfluidic chip (OrganoPlate Graft, MIMETAS, Netherlands) was used to construct vascularized tissue. Briefly, the extracellular matrix (ECM) was prepared; 10 × phosphate-buffered saline (PBS) was added to concentrated rat tail collagen I solution (Corning, Collagen I Rat Tail, 10.21 mg/ml) diluted to 4 mg/ml with DMEM (without supplement), and the pH was adjusted to 7.0–7.4 with 20 mg/ml NaOH. Prepared collagen solution was injected into a gel inlet and polymerized in an incubator for 15 min. HUVEC were purchased from Lonza, thawed from liquid nitrogen, and cultured in T75 flasks (Nunc™ EasYFlask, Sigma F7552), with endothelial cell growth medium-2 (EGM™-2) and BulletKit™ (Lonza, CC-3162). We used at P3 until P9. Medium was replaced three times a week. All cell culture was performed in a humidified incubator at 37°C and 5% CO2. Suspended endothelial cells in EGM-2 medium were seeded into both perfusion inlet channels and placed them in an incubator to allow cells to attach. Additional medium (50 μl) was loaded into the inlet well and the plate was placed on an interval rocker (MIMETAS, Netherlands), allowing bi-directional flow for perfusion. Medium was refreshed every 2 days. Tissue chip was maintained in EGM-2 medium (changing every 2 days) in a rocker (OrganoFlow, MIMETAS) until it formed tubular structure. As a next step, angiogenic cocktail with VEGF (37.5 μg/ml), bFGF (10 μg/ml), MCP-1 (37.5 μg/ml), HGF (37.5 μg/ml), S1P (250 μM), and PMA (37.5 μg/ml) suspended in EGM-2 was added to the gel chamber only to generate concentration gradient. Medium was changed every 2 or 3 days.
Fixation, Staining, and Imaging of Microfluidics Samples
The cell culture medium was aspirated and 50 µl of 4% paraformaldehyde (PFA) in phosphate-buffered saline (J61899, Alfa Aesar, MA, United States) was added to all the perfusion inlet and outlet wells. The device was placed on a rocker (OrganoFlow, MIMETAS) and incubated for 10 min at room temperature. After fixation, the PFA was aspirated from the wells, and the microfluidic chips were washed twice with 50 µl 1X PBS in all the perfusion inlets and outlets, followed by a permeabilization step of 0.3% Triton-X100 (VWR 28817295) and a second wash step with PBS. Nuclei were stained using 1:2000 Hoechst 33258 (H3569, Life Technologies, United States). Tissue construct was incubated with primary antibodies for overnight, αCD31 and αZO-1. Secondary antibodies with blocking solution are further incubated, AF568 (Invitrogen, A-11004) and AF488 (Invitrogen, A-11008). Image data were acquired and processed using a two-photon confocal microscope (ZEISS Multiphoton LSM 710) and ZEN software. Imaging depth was set at 16 bits, binning at one, and imaging resolution at 2048 × 2048. Autofocus was set at a 120 µm offset from channel bottom.
Statistical Analysis
The representative experiments were presented and the results were expressed as mean ± standard deviation. For the comparison of two groups (Figure 4 and partially Figure 5), the Student’s t-test was performed to determine if they have significant difference. For the comparison of more than two groups (Figure 2 and partially Figure 5), the statistical analysis methods for multiple group comparisons were requited. One-way ANOVA followed by the post hoc Tukey’s test (to control type I error) was performed to check if the difference between every two groups is statistically significant. A p-value < 0.05 was considered statistically significant.
Results
Characterizations of the Derived Brain-Specific Pericytes
Brain-specific differentiation was induced by dual SMAD signaling inhibitors SB and LDN and Wnt activator CHIR and FGF-2 (Figure 1A). This neuroectoderm induction should give rise to pericytes in the forebrain (Jeske et al., 2020). The derived pericytes (iPCs) were grown to confluency, showing mostly round-shaped cells with little cytoplasm (Figure 1B). The day 25 cells also secreted extracellular matrix proteins HA and CSPG. Verification of pericyte lineage was shown by positive signals of immunocytochemistry for the most commonly known pericyte markers NG2, PDGFRβ, and CD146 (Figure 1C), the surface proteins that have known to be essential for pericyte development of the vasculature (Jeske et al., 2020). The flow cytometry results indicate the highly positive signal for PDGFRβ (96.8%) and a fairly positive signal for NG2 (37.1%) (Figure 1D). The mesoderm progenitor marker KDR was minimal at this stage (day 25). For KDR expression, positive expression at day 6 of mesoderm differentiation of hiPSCs has been shown in our previous publication (Song et al., 2019c). KDR is only expressed in mesoderm progenitors and should be transiently expressed during mesoderm differentiation. Therefore, the negative KDR expression here should be the true observation. Comparison of the derived pericytes with primary hASCs was also performed (Figure 1E). PDGFRβ (72.8% vs. 96.8%) and NG2 (12.4% vs. 29.2%) show the less expression in hASCs than iPCs.
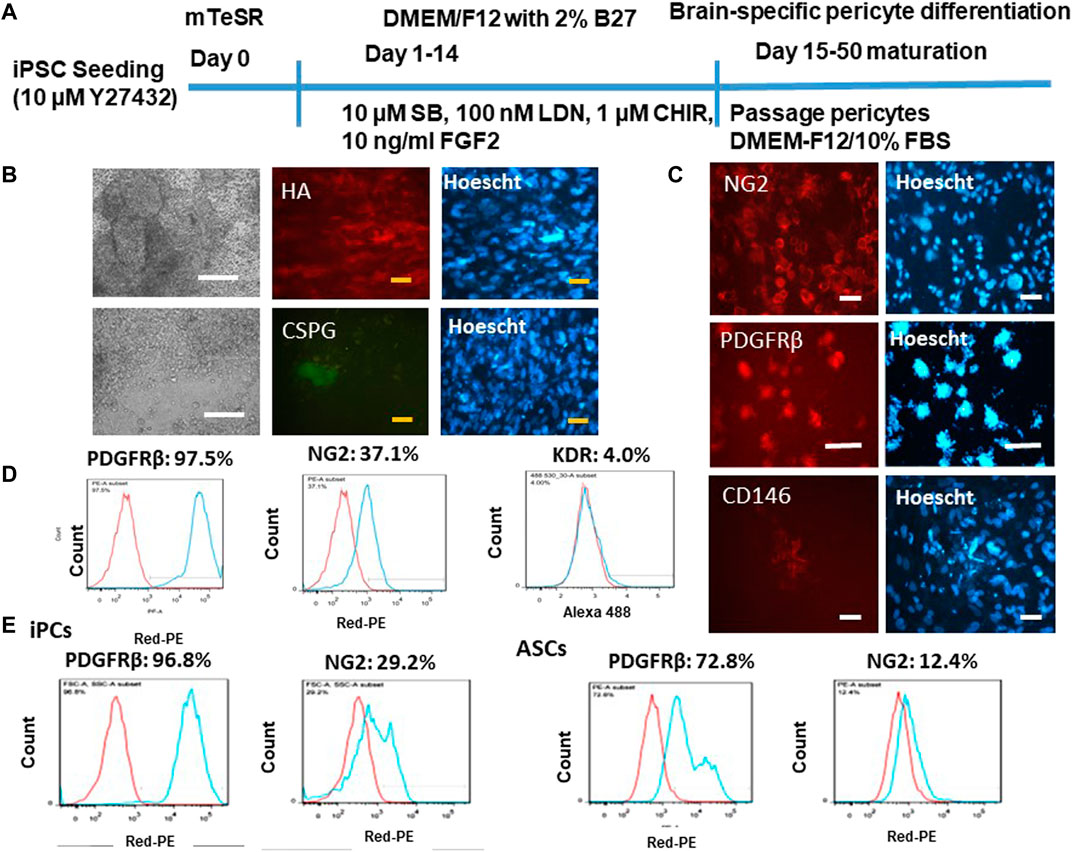
FIGURE 1. Characterization of brain-specific pericytes derived from iPSK3 cells (iPCs). (A) Schematic of protocol for differentiation of brain pericytes from iPSK3 cells; SB: SB431542; LDN: LDN193189; and CHIR: CHIR99021. (B) Cell morphology and fluorescent images for extracellular matrix proteins hyaluronic acid (HA) and chondroitin sulfate proteoglycan (CSPG) on day 25 pericytes. White scale bar: 200 μm, yellow scale bar: 50 µm. (C) Expression of pericytes receptor markers NG2, PDGFRβ, and CD146. Cell nuclei were counterstained with Hoescht. Scale bar: 50 µm. (D) Flow cytometry histograms for pericyte marker NG2, PDGFRβ, and mesoderm progenitor marker KDR. (E) Flow cytometry histograms for day 25 differentiated pericytes compared with human adipose-derived stem cells (ASCs). Red line: negative control; blue line: marker of interest.
In addition to the common pericyte markers, the brain identity of the iPCs was determined by RT-PCR for mRNA expression (Figure 2). The purpose is to observe if pericytes show the signs of BBB marker FOXF2 and genes that differentiate brain pericytes from other mural cells including ABCC9, KCNJ8, and ZIC1. The results show that the iPCs derived in the presence of CHIR had higher expression of ABCC9 (0.76 ± 0.22 vs. 0.33 ± 0.13) than the iPCs derived without CHIR, but the expression of FOXF2 did not have statistically significant difference. When compared to hASCs (P5), iPCs derived in the presence of CHIR had similar expression of FOXF2 and ABCC9. For KCNJ8, hASCs had the highest expression, and the presence of CHIR did not promote KCNJ8 expression. However, iPCs derived with or without CHIR both show much higher expression of ZIC1 than hASCs (0.52–0.91 vs. 0.12). The GAPDH expression shows large variations among different samples. Therefore, β-actin was used as the housekeeping gene. Taken together, differential expression of brain identity markers was observed in the iPCs, and the CHIR addition was used in the following study.
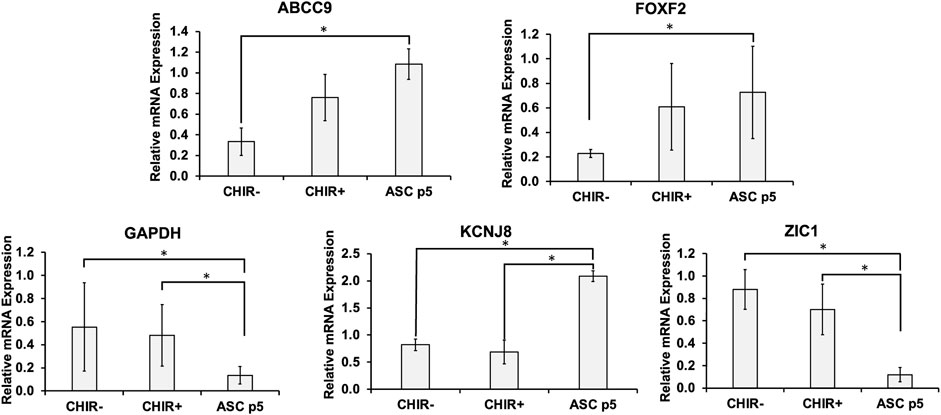
FIGURE 2. mRNA levels for brain-specific markers in the derived pericytes (iPCs). mRNA expression was determined by RT-PCR. Brain pericytes derived with Wnt activator CHIR99021 (CHIR) were compared to those derived in the absence of CHIR and human adipose-derived stem cells (ASCs). Gene markers tested include ABCC9, FOXF2, KCNJ8, ZIC1, and GAPDH. Expression was normalized to the non-CHIR pericytes. N = 6. The RT-PCR results were expressed as the mean ± standard deviation. Statistical analysis was performed by one-way ANOVA. * indicates p < 0.05 for the compared groups.
Characterizations of the Derived BBB-Related Endothelial Cells
To ensure that the derived BBB-related endothelial cells have the properties of endothelial cells, the mesoderm induction was used instead of neuro-ectodermal induction (Figure 3A) (Wimmer et al., 2019; Lu et al., 2021a). The differentiation was performed in both 3D and 2D cultures. The 3D blood vessel organoids have a morphology that is spherical in shape (Figure 3B). The blood vessel organoids were replated onto the Matrigel-coated surface. The cells can migrate out from the aggregate and grow into a monolayer. To confirm the endothelial cell identity, the flow cytometry quantification further shows that 70.7% CD31+ cells and 75% VE-cadherin+ cells were obtained (Figure 3C). Immunocytochemistry detected the expression of CD31, VE-cadherin, as well as the tight junction protein ZO-1 (Figure 3D). Tight junctions (TJ) including ZO-1, occludin, and claudin-5 allow the BECs to be held tightly together and limit paracellular flux (Daneman and Prat, 2015). The cells were also stained with CD45 (a hematopoietic cell marker) to assure that the iECs do not express a non-endothelial cell marker. ZO-1 flow cytometry shows high positive expression and CD45 flow cytometry shows little expression (Supplementary Figure S1). Phagocytosis ability of the iECs was shown by MPIO internalization as quantified by flow cytometry (∼90%) (Figure 3E).
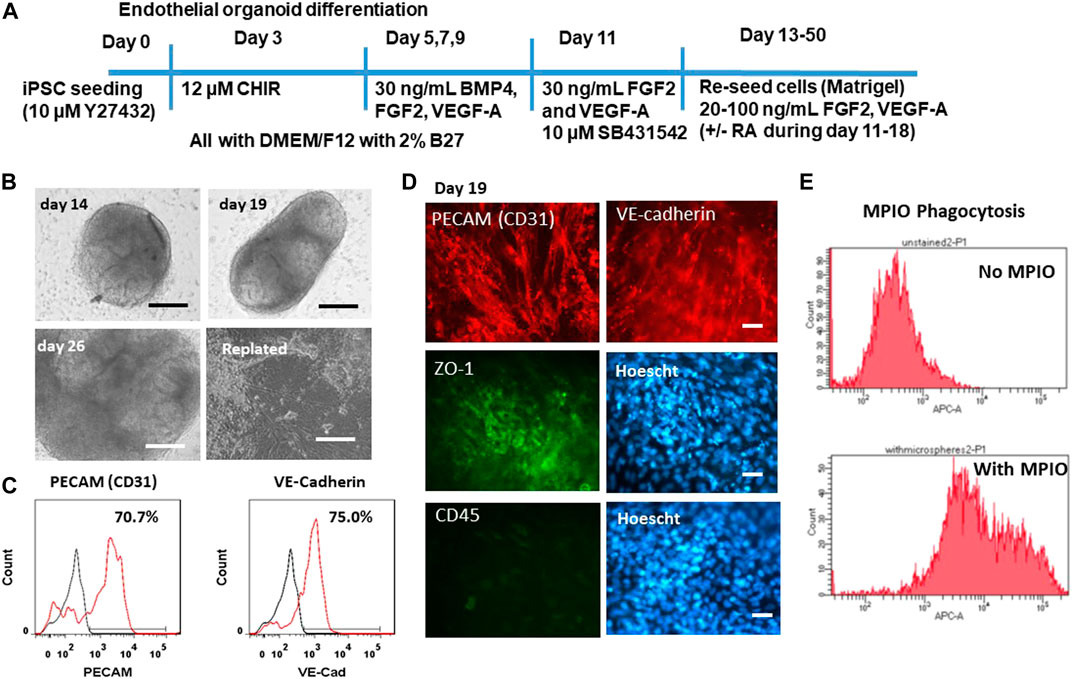
FIGURE 3. Characterization of endothelial cells derived from iPSK3 cells (iECs). (A) Schematic of protocol for differentiation of endothelial cells from iPSK3 cells. Brain-specific endothelial cells receive retinoic acid (RA) addition on day 11, 13, and 15. CHIR: CHIR99021. (B) Phase contrast images for the derived endothelial organoids at day 14, 19, and 26 as well as the replated cells. Scale bar: 200 µm. (C) Flow cytometry histograms for endothelial cell markers PECAM (CD31) and VE-cadherin. Black line: negative control; Red line: marker of interest. (D) Fluorescent images for endothelial cell markers PECAM (CD31), VE-cadherin, and ZO-1. CD45 is a hematopoietic cell marker. Cell nuclei were counterstained with Hoescht. Scale bar: 25 µm. (E) Phagocytosis ability of the replated iECs (day 25). The replated cells were exposed to MPIO at the concentration of 5 μg Fe/mL for 24 h. The cells were then harvested and fixed. The MPIO+ cells (Alexa 647) was quantified by flow cytometry.
To enhance the BBB property, RA was added during day 11–18 of differentiation to see if RA addition further pushes endothelial cells to a brain specific lineage. The BBB-related gene expression was determined by RT-PCR for mRNA expression of replicate derivations (Figure 4). The BRCP (an efflux transporter) expression was promoted by RA in the derivation 2 (0.73 ± 0.05 vs. 0.16 ± 0.06), but not for derivation 1. GLUT-1 (a glucose transporter) expression did not show statistical difference for both derivations. For PGP, that is, P-glycoprotein 1 that is also known as multidrug resistance protein 1, RA addition significantly enhanced the expression in both derivations (0.98 ± 0.07 vs. 0.29 ± 0.10, 0.28 ± 0.03 vs. 0.09 ± 0.01). Efflux transporters including P-glycoprotein work by allowing substrates up a concentration gradient to move from the luminal side to the blood side (Daneman and Prat, 2015). This limits diffusion of small lipophilic molecules into the luminal space. For ABCC1 that encodes multidrug resistance-associated protein 1, RA addition promoted the expression is derivation 1 (0.94 ± 0.05 vs. 0.49 ± 0.04), but no significant difference was observed for derivation 2. OCLN encodes occludin that is an enzyme oxidizing NADH. Together with claudins and zonula occludens-1 (ZO-1), occludin regulates the formation and function of tight junctions (Luissint et al., 2012). RA addition significantly enhanced the expression of OCLN in both derivations (0.80 ± 0.19 vs. 0.31 ± 0.18, 0.22 ± 0.07 vs. 0.05 ± 0.01). SLC2A1 encodes a major glucose transporter in the BBB. SLC2A1 or GLUT-1 is highly expressed in BEC compared to other endothelial cells and allows for a high amount of glucose to be taken up by the cells of the CNS (Daneman and Prat, 2015). RA addition also significantly enhanced the expression of SLC2A1 in both derivations (0.92 ± 0.12 vs. 0.42 ± 0.13, 0.42 ± 0.16 vs. 0.09 ± 0.03). ZO-1 expression for the iECs derived using RA or without RA was compared (Supplementary Figure S2). More than 65.7% of cells were ZO-1+ for both +/−RA groups, and the cells also expressed abundant VE-cadherin. Taken together, RA addition promoted the BBB characteristics of the iECs and was used in the following study.
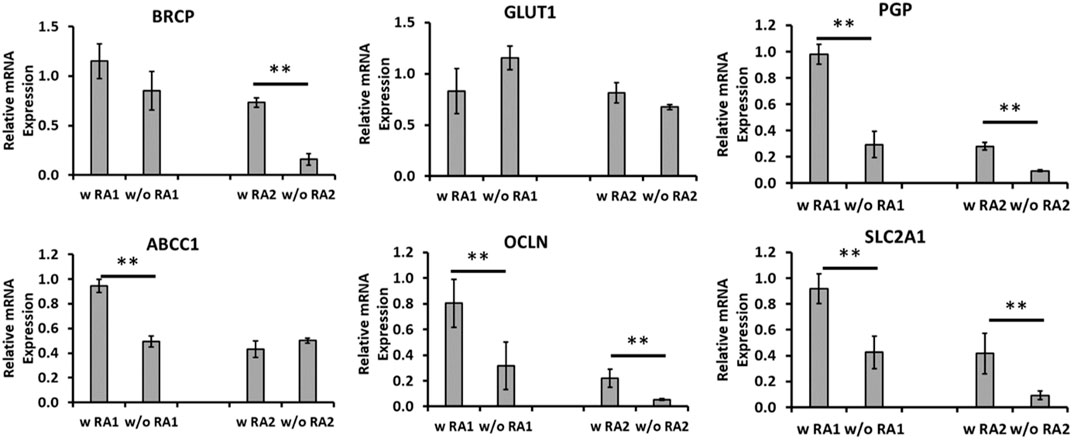
FIGURE 4. mRNA levels for blood–brain barrier (BBB) markers comparing endothelial cells (iECs) derived with and without retinoic acid (RA). mRNA expression was determined by RT-PCR for endothelial cells derived with RA compared to those without RA treatment in duplicate differentiations. mRNA expression was normalized to the non-RA–treated endothelial cells. Common BBB markers were chosen for comparison, including BRCP, GLUT-1, PGP, and ABCC1 encoding multidrug resistance-associated protein 1 (MRP1), OCLN, and SLC2A1 (different primer pair from GLUT-1). N = 3. The RT-PCR results were expressed as the mean ± standard deviation. ** indicates p < 0.05 for the compared groups.
Co-Culture in the Transwell System and Evaluation of Inflammatory Response Induced by Aβ42 Oligomers
The co-culture of iECs and iPCs in a transwell system was demonstrated (Figure 5A) and TEER values were determined along with iEC (with or without RA treatment) culture (4-week long) only (Supplementary Table S3). iECs culture showed the significantly higher TEER values (∼289–304, Ω × cm2) than blank control. However, the RA treatment cultures did show the enhancement in TEER values. The co-culture of iECs and iPCs in transwells for 1 week showed the TEER values of ∼293–311, Ω×cm2. High TEER values (740–4,100 Ω×cm2) can be achieved by differentiating hiPSCs into brain microvascular endothelial cells (iBMECs), using the co-differentiation of neuro-ectodermal and endothelial lineages, with or without co-culture of primary astrocytes (Supplementary Table S4) (Medina and Tang, 2022). Both iECs and iPCs exposed to Aβ42 oligomers showed the reduced phagocytosis ability of MPIO as determined by flow cytometry (Figure 5B).
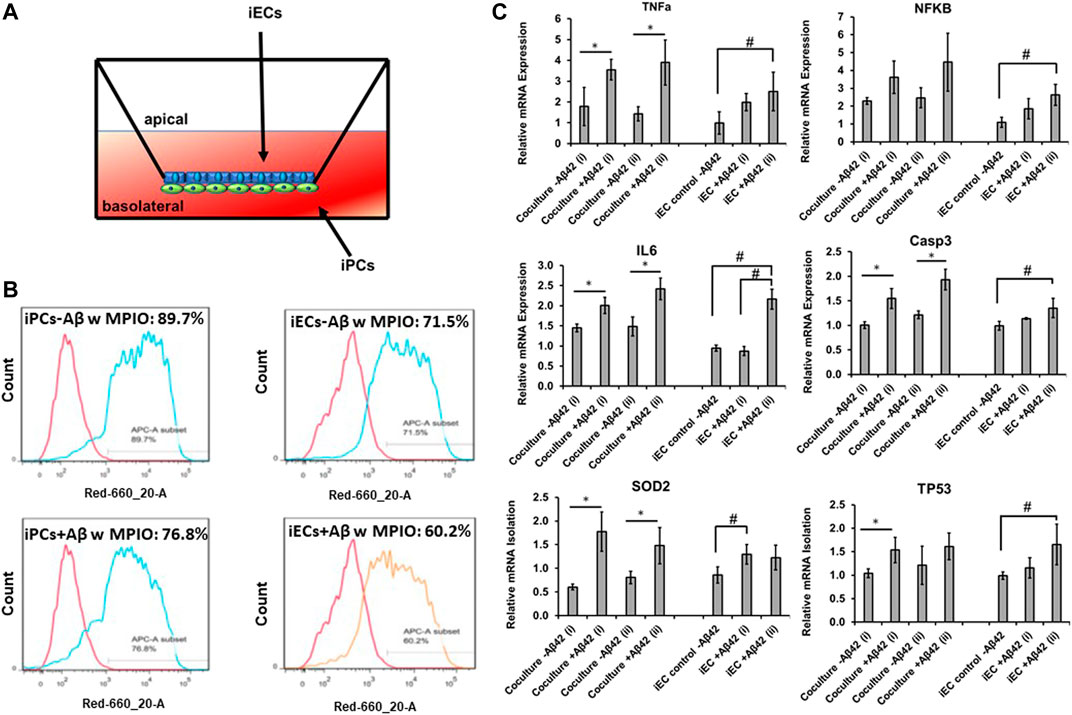
FIGURE 5. mRNA levels of inflammatory response genes for Aβ42 treated co-culture. (A) Illustration of the transwell of iEC and iPC co-culture. (B) Phagocytosis ability of the replated iPCs (day 25) and iECs determined by flow cytometry. The replated cells with or without Aβ42 oligomer treatment (48 h) were exposed to MPIO at 5 μg Fe/mL for 24 h. The MPIO+ cells (Alexa 647) was quantified by flow cytometry. To study the inflammatory response, the derived endothelial cells (iECs) were treated with Aβ42 oligomers and compared to those without Aβ42 treatment along with the transwell co-culture of iECs and iPCs with or without Aβ42 oligomers. (C) Relative mRNA expression of TNFa, NFKB, IL6, Casp3, SOD2, and TP53. The mRNA expression was normalized to the Aβ42 negative group of iECs. N = 3. The RT-PCR results were expressed as the mean ± standard deviation. The t-test was used to compare the co-culture−/+Aβ42 for run i and ii separately. * indicates p-value < 0.05 from the t-test. One-way ANOVA followed by the post hoc Tukey’s test was used to compare the three groups: iEC control-Aβ42, iEC + Aβ42 (i), and iEC + Aβ42 (ii). # indicates p-value < 0.05 from the Tukey’s test.
To evaluate the ability of inflammatory response of the co-culture in comparison to the derived iECs, the cells were subjected to Aβ42 oligomer treatment for 48 h. The expression of neural inflammation markers TNFα, NFKB, and IL-6, enzyme involved in the apoptosis process CASP3, enzyme involved in reactive oxygen species formation SOD2, and DNA TP53 was determined (Figure 5C). For the iECs exposed to Aβ42 oligomers, higher expression of the six genes was observed compared to no treatment control in general, that is, 2.0–2.5 fold for TNFa, 1.8–2.6 fold for NFKB, 2.2 fold for IL-6, 1.4 fold for CASP3, 1.2–1.3 fold for SOD2, and 1.2–1.7 fold for TP53. For transwell co-culture of iECs and iPCs, the no-treatment controls have a similar or higher baseline levels compared to the iECs (except SOD2). Exposure to Aβ42 oligomers upregulated all the six genes compared to no treatment control too. Specifically, TNFa expression was 3.6–3.9 fold vs. 1.4–1.8 fold. NFKB was 3.6–4.5 fold vs. 2.3–2.5 fold, IL-6 was 2.0–2.4 fold vs. 1.4–1.5 fold, CASP3 was 1.6–1.9 fold vs. 1.0–1.2 fold, SOD2 was 1.5–1.8 fold vs. 0.6–0.8 fold, and TP53 was 1.5–1.8 fold vs. 1.0–1.2 fold. Taken together, the iEC-iPC co-culture in a transwell shows inflammatory response to the Aβ42 oligomer stimulation.
Vascularization of Brain Tissue Models Using Microfluidics-Based Microchannels
For the potential development of a more in vivo-like BBB microenvironment, which cannot be provided by the transwell culture, the microfluidics-based microchannel model was explored. Figure 6A shows the schematic representation of different phases of vascularized tissue culture using the OrganoPlate Graft. The collagen hydrogel was selectively injected through the gel inlet. Then, endothelial cells were positioned in the two perfusion channels, forming vessels under gravity-driven flow. A vascular network was established by placing an angiogenic cocktail on top of the collagen hydrogel, which forms a gradient toward the perfusion channels, thereby attracting angiogenic sprouts. As shown in Figure 6B, the alignment of endothelial cells to perfusion direction of the vessel in the perfusion channel and endothelial cells in new vascularized tissue in the collagen hydrogel channel also formed tight junction (CD31: red and ZO-1: green). These results demonstrate our proof-of-concept strategy of vascularization of human brain-like tissues.
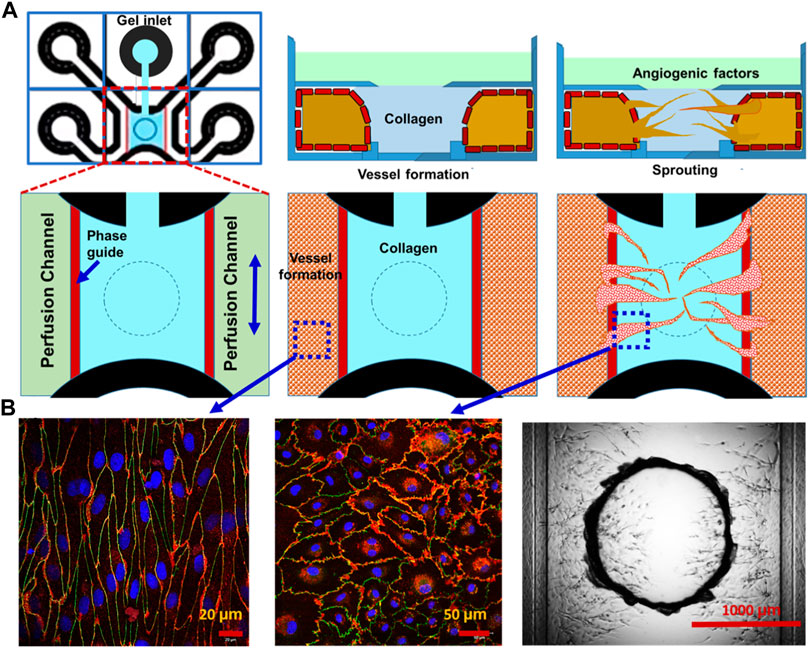
FIGURE 6. Vascularization of brain-like tissue model using microfluidics-based microchannels. Potential vascularization strategy of human brain-like tissue: (A) schematic representation of different phases of vascularized tissue culture using the OrganoPlate Graft. (B) Staining (CD31: red and ZO-1: green) and DIC images of vascularized tissue.
Discussion
Brain pericyte differentiation from hiPSCs have been a challenge due to the undefined identity of brain pericytes compared to other mural cells such as smooth muscle cells (Faal et al., 2019). Previous research suggests that pericytes have many similarities with human MSCs, although this has had some debate (da Silva Meirelles et al., 2016). For example, PDGFRβ and NG2 may also be expressed in human MSCs. At the protein level, there were no available cell lines in our laboratory that are known for negative expression of PDGFRβ and NG2. Therefore, negative controls in the staining were used for comparison. In addition, the immunocytochemistry results were verified by flow cytometry, which are usually more sensitive than the fluorescence microscope. In particular, additional pericyte markers for brain identity were evaluated at the molecular level by RT-PCR. Gene expression data from mice brain vasculature shows that PDGFRb, ABCC9, and CSPG4 are expressed in all mural cell types from brain vasculature (Vanlandewijck et al., 2018). Studies have suggested that FOXF2, ZIC1, and KCNJ8 are expressed in pericytes that have brain tissue identity (Reyahi et al., 2015). Our study compared the levels of these genes in hiPSC-derived brain pericytes with hASCs. The results indicate that the derived brain pericytes have similar expression of ABCC9 and FOXF2, but lower expression of KCNJ8 compared to hASCs. Higher expression of ZIC1 for the derived cells indicates a possible brain pericyte phenotype. Since the neuroectoderm gives rise to pericytes in the forebrain and the mesoderm progenitors gives rise to pericytes in the hindbrain/spinal cord (Etchevers et al., 2001; Korn et al., 2002; Kurz, 2009), our protocol should generate the perityces with forebrain identity. The transcriptome analysis of forebrain organoids reported in our previous study also showed the presence of various brain pericyte markers (Song et al., 2019c).
Our study did not use the protocols that have been usually reported for the generation of brain microvascular endothelial cells for in vitro BBB models in most recent studies, which applied the co-differentiation of neuro-ectodermal and endothelial lineages or sequential activation of Wnt/beta-catenin and retinoic acid activation (Lippmann et al., 2012; Appelt-Menzel et al., 2017; Qian et al., 2017; Lu et al., 2021b). These cells were reported not to exhibit canonical endothelial response to inflammatory stimulus (Nishihara et al., 2020). It was recently revealed that these cells have a gene signature typical of neuro-ectodermal epithelial cells and require the ETS transcription factors to acquire vascular fate (Lu et al., 2021a). Our study modified the blood vessel organoid derivation and then used retinoic acid to enhance the BBB properties of the derived endothelial cells (Lippmann et al., 2014; Wimmer et al., 2019; Nguyen et al., 2021). This blood vessel organoid derivation is not specific for brain tissue, which may contribute to the lower TEER value observed in this study. This was verified in our iBMEC differentiation from hiPSCs with or without co-cultured primary astrocytes (with ∼740–4,100 Ω×cm2 TEER value). In addition to the common endothelial cell markers, the derived cells in our study exhibit BBB markers such as BRCP, GLUT-1, PGP, and OCLN, indicting the expression of tight junction proteins, small molecule transporters, and multidrug resistance proteins.
TEER values for the transwell-cultured monolayers of the hiPSC-derived BECs have been reported to vary, depending on the hiPSC source, the differentiation method, cell density, and the type of coating used for matrix (Lauschke et al., 2017). There are few options for coating transwell membranes including Matrigel, but more commonly collagen, laminin, and fibronectin are applied (Linville et al., 2019). TEER values in our study were low, which may be due to incomplete monolayer formation and the coating on the membrane. The use of collagen I and fibronectin needs to be examined. A co-culture that implements the BBB-inducing effects of the derived pericytes with the derived endothelial cells by direct and non-direct contact methods are shown to improve the TEER values. Cell–cell interaction signaling between BECs to pericytes should promote the BBB function of the BECs and increase TEER values (Stebbins et al., 2019). Evidence supports the idea that PDGF-B and PDGFRb signaling is highly important for pericyte health and the deletion of PDGFRb in mice leads to vascular leakage in the brain and death (Hill et al., 2014). Other important signaling pathways include TGF-β, Ang-1 and Ang-2, and Notch.
3-D microfluidics based BBB models have been reported in a few studies (Supplementary Figures S3–S5) (Linville et al., 2019; Vatine et al., 2019; Ahn et al., 2020; Chung et al., 2020; Chen X et al., 2021). These models can create a 3D tubular geometry (e.g., using collagen hydrogel) and introduce fluid flow in the tubular structure (e.g., using microfluidics), better mimicking the brain microvasculature. 3D mapping of nanoparticle distributions in the vascular and perivascular regions was demonstrated (Ahn et al., 2020), showing the distinct cellular uptakes and BBB penetrations through receptor-mediated transcytosis. Existing studies mainly investigated the permeability of the biomolecules, and few reports showed the inflammatory response of the 3D BBB models. Our 3D BBB model still needs a lot of improvements. For example, the third (isogenic astrocytes) and forth (isogenic neurons) cell types in the co-culture can be introduced. The seeding process and the effects of shear stress can be further illustrated.
To date, the hiPSC-derived in vitro BBB models were mostly used to study the permeability of the barrier, and very limited studies have been performed to examine the immune and inflammatory responses (Nishihara et al., 2020; Torok et al., 2021). Inflammatory responses of the BBB are highly important with evidence showing that many CNS diseases and injury are accompanied with BBB disfunction. More specifically in Alzheimer’s disease, Aβ42 oligomers have been shown to be a major cause of BEC damage and leakage that propagates the disease (Wan et al., 2015). Addition of Aβ42 oligomers in vitro would likely cause the same issues and BBB leakage. Inflammatory responses of endothelial cells and pericytes due to Aβ42 oligomers include the release of inflammatory cytokines IL-1β, IL-6, and TNF-α, which cause BEC leakage. It is necessary to evaluate the inflammatory response of the BBB models in vitro and an effective model should expand the contribution to disease modeling that can be applied to in vivo therapy. In our study, the iPC and iEC co-culture exposed to the Aβ42 oligomers upregulated the expression of TNF-α, IL-6, NK-KB, Casp3, SOD2, and TP53, showing the active inflammatory response, and the upregulation was more significant for the co-culture than the iEC alone, indicating that the co-culture was more responsive.
Brain regional heterogeneity of blood–brain barrier has been reported (Wilhelm et al., 2016). For example, BBB in the cerebral cortex and cerebellum showed the differences in inflammatory response caused by a pathogenic clearance of virus (Phares et al., 2006). The upregulation of TNF-α, IFN-γ, and macrophage inflammatory proteins (MIP)-1β and the loss of BBB integrity were more pronounced for BBB in the cerebellum than in the cerebral cortex (Phares et al., 2006). Our study mainly used the derived pericytes of forebrain identity. The derivation of pericytes of specific brain regions and establishing the brain region-specific BBB models still needs to be demonstrated. Our study used the healthy hiPSC-derived cells for establishing BBB models. Patient-derived hiPSCs from individuals with neurological diseases can predict disease-specific pathology due to lack of transporters and disruption of barrier integrity. By combining Organ-on-a-Chip technology and hiPSC-derived cells, the created neurovascular unit can recapitulate complex BBB functions, provide a platform for modeling inheritable neurological disorders, advance drug screening, and personalized medicines (Vatine et al., 2019).
Conclusion
This study characterized brain-specific pericytes and endothelial cells derived from hiPSCs. The co-culture of the derived pericytes and endothelial cells in a transwell system mimic the BBB structure at some aspects. Addition of Aβ42 oligomers has a significant effect on the inflammatory response of the co-culture and phagocytotic action of the derived pericytes and endothelial cells. The proof of concept strategy of vascularization of human brain-like tissues was also demonstrated in a microfluidics-based device. This study has significance in establishing micro-physiological systems for neurological disease modeling and drug screening.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
MM did most of the experiments and wrote the initial draft of the manuscript. XC and CZ helped in some co-culture experiments. AM and HT helped on the TEER analysis. TR and YY as well as ZW and XW contributed to the 3D microfluidic part of the study. TH and Q-XS helped performing flow cytometry analysis. YL facilitated data analysis, conceived the project, revised, and finalized the manuscript.
Funding
This work was partially supported by the National Science Foundation (CBET-1917618 and CMMI-2100987). Research reported in this publication was also partially supported by the National Institutes of Health (United States) under Award Number R01NS125016.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Dr. Brian K. Washburn and Kristina Poduch in Florida State University (FSU), Department of Biological Sciences, for their help with RT-PCR analysis and the support from the Flow Cytometry Core Facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fceng.2022.927188/full#supplementary-material
References
Ahn, S. I., Sei, Y. J., Park, H.-J., Kim, J., Ryu, Y., Choi, J. J., et al. (2020). Microengineered Human Blood-Brain Barrier Platform for Understanding Nanoparticle Transport Mechanisms. Nat. Commun. 11 (1), 175. doi:10.1038/s41467-019-13896-7
Appelt-Menzel, A., Cubukova, A., Günther, K., Edenhofer, F., Piontek, J., Krause, G., et al. (2017). Establishment of a Human Blood-Brain Barrier Co-culture Model Mimicking the Neurovascular Unit Using Induced Pluri- and Multipotent Stem Cells. Stem Cell Rep. 8 (4), 894–906. doi:10.1016/j.stemcr.2017.02.021
Bejoy, J., Song, L., Zhou, Y., and Li, Y. (2018). Wnt/Yes-Associated Protein Interactions during Neural Tissue Patterning of Human Induced Pluripotent Stem Cells. Tissue Eng. Part A 24 (7-8), 546–558. doi:10.1089/ten.TEA.2017.0153
Bejoy, J., Song, L., Wang, Z., Sang, Q.-X., Zhou, Y., and Li, Y. (2018). Neuroprotective Activities of Heparin, Heparinase III, and Hyaluronic Acid on the Aβ42-Treated Forebrain Spheroids Derived from Human Stem Cells. ACS Biomater. Sci. Eng. 4, 2922–2933. doi:10.1021/acsbiomaterials.8b00021
Bijonowski, B. M., Fu, Q., Yuan, X., Irianto, J., Li, Y., Grant, S. C., et al. (2020). Aggregation‐induced Integrated Stress Response Rejuvenates Culture‐expanded Human Mesenchymal Stem Cells. Biotechnol. Bioeng. 117, 3136–3149. doi:10.1002/bit.27474
Bijonowski, B. M., Yuan, X., Jeske, R., Li, Y., and Grant, S. C. (2020). Cyclical Aggregation Extends In Vitro Expansion Potential of Human Mesenchymal Stem Cells. Sci. Rep. 10, 20448. doi:10.1038/s41598-020-77288-4
Blanchard, J. W., Bula, M., Davila-Velderrain, J., Akay, L. A., Zhu, L., Frank, A., et al. (2020). Reconstruction of the Human Blood-Brain Barrier In Vitro Reveals a Pathogenic Mechanism of APOE4 in Pericytes. Nat. Med. 26, 952–963. doi:10.1038/s41591-020-0886-4
Brown, L. S., Foster, C. G., Courtney, J.-M., King, N. E., Howells, D. W., and Sutherland, B. A. (2019). Pericytes and Neurovascular Function in the Healthy and Diseased Brain. Front. Cell. Neurosci. 13, 282. doi:10.3389/fncel.2019.00282
Browne, S., Gill, E. L., Schultheiss, P., Goswami, I., and Healy, K. E. (2021). Stem Cell-Based Vascularization of Microphysiological Systems. Stem Cell Rep. 16, 2058–2075. doi:10.1016/j.stemcr.2021.03.015
Cakir, B., Xiang, Y., Tanaka, Y., Kural, M. H., Parent, M., Kang, Y.-J., et al. (2019). Engineering of Human Brain Organoids with a Functional Vascular-like System. Nat. Methods 16, 1169–1175. doi:10.1038/s41592-019-0586-5
Campisi, M., Shin, Y., Osaki, T., Hajal, C., Chiono, V., and Kamm, R. D. (2018). 3D Self-Organized Microvascular Model of the Human Blood-Brain Barrier with Endothelial Cells, Pericytes and Astrocytes. Biomaterials 180, 117–129. doi:10.1016/j.biomaterials.2018.07.014
Canfield, S. G., Stebbins, M. J., Faubion, M. G., Gastfriend, B. D., Palecek, S. P., and Shusta, E. V. (2019). An Isogenic Neurovascular Unit Model Comprised of Human Induced Pluripotent Stem Cell-Derived Brain Microvascular Endothelial Cells, Pericytes, Astrocytes, and Neurons. Fluids Barriers CNS 16 (1), 25. doi:10.1186/s12987-019-0145-6
Chen, W., Foo, S.-S., Hong, E., Wu, C., Lee, W.-S., Lee, S.-A., et al. (2021). Zika Virus NS3 Protease Induces Bone Morphogenetic Protein-dependent Brain Calcification in Human Fetuses. Nat. Microbiol. 6 (4), 455–466. doi:10.1038/s41564-020-00850-3
Chen, X., Liu, C., Muok, L., Zeng, C., and Li, Y. (2021). Dynamic 3D On-Chip BBB Model Design, Development, and Applications in Neurological Diseases. Cells 10, 3183. doi:10.3390/cells10113183
Chung, B., Kim, J., Nam, J., Kim, H., Jeong, Y., Liu, H. W., et al. (2020). Evaluation of Cell-Penetrating Peptides Using Microfluidic In Vitro 3D Brain Endothelial Barrier. Macromol. Biosci. 20 (6), e1900425. doi:10.1002/mabi.201900425
da Silva Meirelles, L., Correa Bellagamba, B., Camassola, M., and Beyer Nardi, N. (2016). Mesenchymal Stem Cells and Their Relationship to Pericytes. Front. Biosci. 21, 130–156. doi:10.2741/4380
Daneman, R., and Prat, A. (2015). The Blood-Brain Barrier. Cold Spring Harb. Perspect. Biol. 7 (1), a020412. doi:10.1101/cshperspect.a020412
Daneman, R., Zhou, L., Kebede, A. A., and Barres, B. A. (2010). Pericytes Are Required for Blood-Brain Barrier Integrity during Embryogenesis. Nature 468 (7323), 562–566. doi:10.1038/nature09513
Etchevers, H. C., Vincent, C., Le Douarin, N. M., and Couly, G. F. (2001). The Cephalic Neural Crest Provides Pericytes and Smooth Muscle Cells to All Blood Vessels of the Face and Forebrain. Development 128 (7), 1059–1068. doi:10.1242/dev.128.7.1059
Faal, T., Phan, D. T. T., Davtyan, H., Scarfone, V. M., Varady, E., Blurton-Jones, M., et al. (2019). Induction of Mesoderm and Neural Crest-Derived Pericytes from Human Pluripotent Stem Cells to Study Blood-Brain Barrier Interactions. Stem Cell Rep. 12 (3), 451–460. doi:10.1016/j.stemcr.2019.01.005
Faley, S. L., Neal, E. H., Wang, J. X., Bosworth, A. M., Weber, C. M., Balotin, K. M., et al. (2019). iPSC-Derived Brain Endothelium Exhibits Stable, Long-Term Barrier Function in Perfused Hydrogel Scaffolds. Stem Cell Rep. 12 (3), 474–487. doi:10.1016/j.stemcr.2019.01.009
Garreta, E., Kamm, R. D., Chuva de Sousa Lopes, S. M., Lancaster, M. A., Weiss, R., Trepat, X., et al. (2021). Rethinking Organoid Technology through Bioengineering. Nat. Mat. 20 (2), 145–155. doi:10.1038/s41563-020-00804-4
Gastfriend, B. D., Palecek, S. P., and Shusta, E. V. (2018). Modeling the Blood-Brain Barrier: Beyond the Endothelial Cells. Curr. Opin. Biomed. Eng. 5, 6–12. doi:10.1016/j.cobme.2017.11.002
Griffin, K., Bejoy, J., Song, L., Hua, T., Marzano, M., Jeske, R., et al. (2020). Human Stem Cell-Derived Aggregates of Forebrain Astroglia Respond to Amyloid Beta Oligomers. Tissue Eng. Part A 26 (9-10), 527–542. doi:10.1089/ten.TEA.2019.0227
Grifno, G. N., Farrell, A. M., Linville, R. M., Arevalo, D., Kim, J. H., Gu, L., et al. (2019). Tissue-engineered Blood-Brain Barrier Models via Directed Differentiation of Human Induced Pluripotent Stem Cells. Sci. Rep. 9 (1), 13957. doi:10.1038/s41598-019-50193-1
Heymans, M., Figueiredo, R., Dehouck, L., Francisco, D., Sano, Y., Shimizu, F., et al. (2020). Contribution of Brain Pericytes in Blood-Brain Barrier Formation and Maintenance: a Transcriptomic Study of Cocultured Human Endothelial Cells Derived from Hematopoietic Stem Cells. Fluids Barriers CNS 17 (1), 48. doi:10.1186/s12987-020-00208-1
Hill, J., Rom, S., Ramirez, S. H., and Persidsky, Y. (2014). Emerging Roles of Pericytes in the Regulation of the Neurovascular Unit in Health and Disease. J. Neuroimmune Pharmacol. 9 (5), 591–605. doi:10.1007/s11481-014-9557-x
Jeske, R., Albo, J., Marzano, M., Bejoy, J., and Li, Y. (2020). Engineering Brain-specific Pericytes from Human Pluripotent Stem Cells. Tissue Eng. Part B Rev. 26, 367–382. doi:10.1089/ten.teb.2020.0091
Jeske, R., Yuan, X., Fu, Q., Bunnell, B. A., Logan, T. M., and Li, Y. (2021). In Vitro Culture Expansion Shifts the Immune Phenotype of Human Adipose-Derived Mesenchymal Stem Cells. Front. Immunol. 12, 621744. doi:10.3389/fimmu.2021.621744
Korn, J., Christ, B., and Kurz, H. (2002). Neuroectodermal Origin of Brain Pericytes and Vascular Smooth Muscle Cells. J. Comp. Neurol. 442 (1), 78–88. doi:10.1002/cne.1423
Kurz, H. (2009). Cell Lineages and Early Patterns of Embryonic CNS Vascularization. Cell Adhesion Migr. 3 (2), 205–210. doi:10.4161/cam.3.2.7855
Lauschke, K., Frederiksen, L., and Hall, V. J. (2017). Paving the Way toward Complex Blood-Brain Barrier Models Using Pluripotent Stem Cells. Stem Cells Dev. 26 (12), 857–874. doi:10.1089/scd.2017.0003
Lee, S. W. L., Campisi, M., Osaki, T., Possenti, L., Mattu, C., Adriani, G., et al. (2020). Modeling Nanocarrier Transport across a 3D In Vitro Human Blood-Brain-Barrier Microvasculature. Adv. Healthc. Mater 9, e1901486. doi:10.1002/adhm.201901486
Linville, R. M., DeStefano, J. G., Sklar, M. B., Xu, Z., Farrell, A. M., Bogorad, M. I., et al. (2019). Human iPSC-Derived Blood-Brain Barrier Microvessels: Validation of Barrier Function and Endothelial Cell Behavior. Biomaterials 190-191, 24–37. doi:10.1016/j.biomaterials.2018.10.023
Lippmann, E. S., Al-Ahmad, A., Azarin, S. M., Palecek, S. P., and Shusta, E. V. (2014). A Retinoic Acid-Enhanced, Multicellular Human Blood-Brain Barrier Model Derived from Stem Cell Sources. Sci. Rep. 4, 4160. doi:10.1038/srep04160
Lippmann, E. S., Azarin, S. M., Kay, J. E., Nessler, R. A., Wilson, H. K., Al-Ahmad, A., et al. (2012). Derivation of Blood-Brain Barrier Endothelial Cells from Human Pluripotent Stem Cells. Nat. Biotechnol. 30 (8), 783–791. doi:10.1038/nbt.2247
Liu, Y., Yuan, X., Muñoz, N., Logan, T. M., and Ma, T. (2019). Commitment to Aerobic Glycolysis Sustains Immunosuppression of Human Mesenchymal Stem Cells. Stem Cells Transl. Med. 8 (1), 93–106. doi:10.1002/sctm.18-0070
Lu, T. M., Barcia Durán, J. G., Houghton, S., Rafii, S., Redmond, D., and Lis, R. (2021). Human Induced Pluripotent Stem Cell-Derived Brain Endothelial Cells: Current Controversies. Front. Physiol. 12, 642812. doi:10.3389/fphys.2021.642812
Lu, T. M., Houghton, S., Magdeldin, T., Duran, J. G. B., Minotti, A. P., Snead, A., et al. (2021). Pluripotent Stem Cell-Derived Epithelium Misidentified as Brain Microvascular Endothelium Requires ETS Factors to Acquire Vascular Fate. Proc. Natl. Acad. Sci. U. S. A. 118 (8), e2016950118. doi:10.1073/pnas.2016950118
Luissint, A.-C., Artus, C., Glacial, F., Ganeshamoorthy, K., and Couraud, P.-O. (2012). Tight Junctions at the Blood Brain Barrier: Physiological Architecture and Disease-Associated Dysregulation. Fluids Barriers CNS 9 (1), 23. doi:10.1186/2045-8118-9-23
Martinelli, I., Tayebati, S. K., Tomassoni, D., Nittari, G., Roy, P., and Amenta, F. (2022). Brain and Retinal Organoids for Disease Modeling: The Importance of In Vitro Blood-Brain and Retinal Barriers Studies. Cells 11 (7), 1120. doi:10.3390/cells11071120
Matsui, T. K., Tsuru, Y., Hasegawa, K., and Kuwako, K.-i. (2021). Vascularization of Human Brain Organoids. Stem Cells 39 (8), 1017–1024. doi:10.1002/stem.3368
Medina, A., and Tang, H. (2022). iPS Cell Differentiation into Brain Microvascular Endothelial Cells. Methods Mol. Biol. 2429, 201–213. doi:10.1007/978-1-0716-1979-7_13
Nguyen, E. H., Dombroe, M. J., Fisk, D. L., Daly, W. T., Sorenson, C. M., Murphy, W. L., et al. (2019). Neurovascular Organotypic Culture Models Using Induced Pluripotent Stem Cells to Assess Adverse Chemical Exposure Outcomes. Appl. Vitro Toxicol. 5 (2), 92–110. doi:10.1089/aivt.2018.0025
Nguyen, J., Lin, Y. Y., and Gerecht, S. (2021). The Next Generation of Endothelial Differentiation: Tissue-specific ECs. Cell Stem Cell 28, 1188–1204. doi:10.1016/j.stem.2021.05.002
Nishihara, H., Gastfriend, B. D., Soldati, S., Perriot, S., Mathias, A., Sano, Y., et al. (2020). Advancing Human Induced Pluripotent Stem Cell‐derived Blood‐brain Barrier Models for Studying Immune Cell Interactions. FASEB J. 34, 16693–16715. doi:10.1096/fj.202001507rr
Phares, T. W., Kean, R. B., Mikheeva, T., and Hooper, D. C. (2006). Regional Differences in Blood-Brain Barrier Permeability Changes and Inflammation in the Apathogenic Clearance of Virus from the Central Nervous System. J. Immunol. 176 (12), 7666–7675. doi:10.4049/jimmunol.176.12.7666
Qian, T., Maguire, S. E., Canfield, S. G., Bao, X., Olson, W. R., Shusta, E. V., et al. (2017). Directed Differentiation of Human Pluripotent Stem Cells to Blood-Brain Barrier Endothelial Cells. Sci. Adv. 3 (11), e1701679. doi:10.1126/sciadv.1701679
Qian, X., Nguyen, H. N., Song, M. M., Hadiono, C., Ogden, S. C., Hammack, C., et al. (2016). Brain-region-specific Organoids Using Mini-Bioreactors for Modeling ZIKV Exposure. Cell 165 (5), 1238–1254. doi:10.1016/j.cell.2016.04.032
Reyahi, A., Nik, A. M., Ghiami, M., Gritli-Linde, A., Pontén, F., Johansson, B. R., et al. (2015). Foxf2 Is Required for Brain Pericyte Differentiation and Development and Maintenance of the Blood-Brain Barrier. Dev. Cell 34 (1), 19–32. doi:10.1016/j.devcel.2015.05.008
Sart, S., Bejarano, F. C., Baird, M. A., Yan, Y., Rosenberg, J. T., Ma, T., et al. (2015). Intracellular Labeling of Mouse Embryonic Stem Cell-Derived Neural Progenitor Aggregates with Micron-Sized Particles of Iron Oxide. Cytotherapy 17 (1), 98–111. doi:10.1016/j.jcyt.2014.09.008
Schaffenrath, J., Wyss, T., He, L., Rushing, E. J., Delorenzi, M., Vasella, F., et al. (2021). Blood-brain Barrier Alterations in Human Brain Tumors Revealed by Genome-wide Transcriptomic Profiling. Neuro Oncol. 23 (12), 2095–2106. doi:10.1093/neuonc/noab022
Shi, Y., Sun, L., Wang, M., Liu, J., Zhong, S., Li, R., et al. (2020). Vascularized Human Cortical Organoids (vOrganoids) Model Cortical Development In Vivo. PLoS Biol. 18 (5), e3000705. doi:10.1371/journal.pbio.3000705
Si-Tayeb, K., Noto, F. K., Nagaoka, M., Li, J., Battle, M. A., Duris, C., et al. (2010). Highly Efficient Generation of Human Hepatocyte-like Cells from Induced Pluripotent Stem Cells. Hepatology 51 (1), 297–305. doi:10.1002/hep.23354
Si-Tayeb, K., Noto, F. K., Sepac, A., Sedlic, F., Bosnjak, Z. J., Lough, J. W., et al. (2010). Generation of Human Induced Pluripotent Stem Cells by Simple Transient Transfection of Plasmid DNA Encoding Reprogramming Factors. BMC Dev. Biol. 10, 81. doi:10.1186/1471-213x-10-81
Song, H. W., Foreman, K. L., Gastfriend, B. D., Kuo, J. S., Palecek, S. P., and Shusta, E. V. (2020). Transcriptomic Comparison of Human and Mouse Brain Microvessels. Sci. Rep. 10 (1), 12358. doi:10.1038/s41598-020-69096-7
Song, L., Wang, K., Li, Y., and Yang, Y. (2016). Nanotopography Promoted Neuronal Differentiation of Human Induced Pluripotent Stem Cells. Colloids Surfaces B Biointerfaces 148, 49–58. doi:10.1016/j.colsurfb.2016.08.041
Song, L., Yan, Y., Marzano, M., and Li, Y. (2019). Studying Heterotypic Cell-Cell Interactions in the Human Brain Using Pluripotent Stem Cell Models for Neurodegeneration. Cells 8, 299–323. doi:10.3390/cells8040299
Song, L., Yuan, X., Jones, Z., Griffin, K., Zhou, Y., Ma, T., et al. (2019). Assembly of Human Stem Cell-Derived Cortical Spheroids and Vascular Spheroids to Model 3-D Brain-like Tissues. Sci. Rep. 9, 5977. doi:10.1038/s41598-019-42439-9
Song, L., Yuan, X., Jones, Z., Vied, C., Miao, Y., Marzano, M., et al. (2019). Functionalization of Brain Region-specific Spheroids with Isogenic Microglia-like Cells. Sci. Rep. 9, 11055–11072. doi:10.1038/s41598-019-47444-6
Stebbins, M. J., Gastfriend, B. D., Canfield, S. G., Lee, M. S., Richards, D., Faubion, M. G., et al. (2019). Human Pluripotent Stem Cell-Derived Brain Pericyte-like Cells Induce Blood-Brain Barrier Properties. Sci. Adv. 5 (3), eaau7375. doi:10.1126/sciadv.aau7375
Stone, N. L., England, T. J., and O’Sullivan, S. E. (2019). A Novel Transwell Blood Brain Barrier Model Using Primary Human Cells. Front. Cell. Neurosci. 13, 230. doi:10.3389/fncel.2019.00230
Torok, O., Schreiner, B., Schaffenrath, J., Tsai, H. C., Maheshwari, U., Stifter, S. A., et al. (2021). Pericytes Regulate Vascular Immune Homeostasis in the CNS. Proc. Natl. Acad. Sci. U. S. A. 118 (10), e2016587118. doi:10.1073/pnas.2016587118
Vanlandewijck, M., He, L., Mäe, M. A., Andrae, J., Ando, K., Del Gaudio, F., et al. (2018). A Molecular Atlas of Cell Types and Zonation in the Brain Vasculature. Nature 554 (7693), 475–480. doi:10.1038/nature25739
Vatine, G. D., Barrile, R., Workman, M. J., Sances, S., Barriga, B. K., Rahnama, M., et al. (2019). Human iPSC-Derived Blood-Brain Barrier Chips Enable Disease Modeling and Personalized Medicine Applications. Cell Stem Cell 24 (6), 995–1005. doi:10.1016/j.stem.2019.05.011
Wan, W., Cao, L., Liu, L., Zhang, C., Kalionis, B., Tai, X., et al. (2015). Abeta(1-42) Oligomer-Induced Leakage in an In Vitro Blood-Brain Barrier Model Is Associated with Up-Regulation of RAGE and Metalloproteinases, and Down-Regulation of Tight Junction Scaffold Proteins. J. Neurochem. 134 (2), 382–393. doi:10.1111/jnc.13122
Wilhelm, I., Nyúl-Tóth, Á., Suciu, M., Hermenean, A., and Krizbai, I. A. (2016). Heterogeneity of the Blood-Brain Barrier. Tissue Barriers 4 (1), e1143544. doi:10.1080/21688370.2016.1143544
Wimmer, R. A., Leopoldi, A., Aichinger, M., Wick, N., Hantusch, B., Novatchkova, M., et al. (2019). Human Blood Vessel Organoids as a Model of Diabetic Vasculopathy. Nature 565, 505–510. doi:10.1038/s41586-018-0858-8
Yan, Y., Song, L., Bejoy, J., Zhao, J., Kanekiyo, T., Bu, G., et al. (2018). Modeling Neurodegenerative Microenvironment Using Cortical Organoids Derived from Human Stem Cells. Tissue Eng. Part A 24 (13-14), 1125–1137. doi:10.1089/ten.TEA.2017.0423
Yan, Y., Bejoy, J., Xia, J., Guan, J., Zhou, Y., and Li, Y. (2016). Neural Patterning of Human Induced Pluripotent Stem Cells in 3-D Cultures for Studying Biomolecule-Directed Differential Cellular Responses. Acta Biomater. 42, 114–126. doi:10.1016/j.actbio.2016.06.027
Yan, Y., Martin, L. M., Bosco, D. B., Bundy, J. L., Nowakowski, R. S., Sang, Q.-X. A., et al. (2015). Differential Effects of Acellular Embryonic Matrices on Pluripotent Stem Cell Expansion and Neural Differentiation. Biomaterials 73, 231–242. doi:10.1016/j.biomaterials.2015.09.020
Yan, Y., Sart, S., Calixto Bejarano, F., Muroski, M. E., Strouse, G. F., Grant, S. C., et al. (2015). Cryopreservation of Embryonic Stem Cell-Derived Multicellular Neural Aggregates Labeled with Micron-Sized Particles of Iron Oxide for Magnetic Resonance Imaging. Biotechnol. Prog. 31 (2), 510–521. doi:10.1002/btpr.2049
Keywords: stem cell derived brain-specific cells, co-culture, human pluripotent stem cells, brain pericytes, brain endothelial cells, microfluidics, inflammatory response
Citation: Marzano M, Chen X, Russell TA, Medina A, Wang Z, Hua T, Zeng C, Wang X, Sang Q-X, Tang H, Yun Y and Li Y (2022) Studying the Inflammatory Responses to Amyloid Beta Oligomers in Brain-Specific Pericyte and Endothelial Co-Culture From Human Stem Cells. Front. Chem. Eng. 4:927188. doi: 10.3389/fceng.2022.927188
Received: 23 April 2022; Accepted: 06 June 2022;
Published: 22 July 2022.
Edited by:
Xiang Zou, Southwest University, ChinaReviewed by:
Yoichi Morofuji, National Hospital Organization Nagasaki Medical Center, JapanNikolaos Dimitrakakis, Harvard University, United States
Copyright © 2022 Marzano, Chen, Russell, Medina, Wang, Hua, Zeng, Wang, Sang, Tang, Yun and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Li, yli4@fsu.edu
 Mark Marzano1
Mark Marzano1  Changchun Zeng
Changchun Zeng Xueju Wang
Xueju Wang Qing-Xiang Sang
Qing-Xiang Sang Hengli Tang
Hengli Tang Yeoheung Yun
Yeoheung Yun Yan Li
Yan Li