Modified Nucleoside Triphosphates for In-vitro Selection Techniques
- 1Laboratorio de Química de Ácidos Nucleicos, INGEBI (CONICET), Ciudad Autónoma de Buenos Aires, Argentina
- 2Instituto de Ciencias, Universidad Nacional de General Sarmiento, Los Polvorines, Argentina
- 3Laboratorio de Biotransformaciones, Universidad Nacional de Quilmes, Bernal, Argentina
The development of SELEX (Selective Enhancement of Ligands by Exponential Enrichment) provides a powerful tool for the search of functional oligonucleotides with the ability to bind ligands with high affinity and selectivity (aptamers) and for the discovery of nucleic acid sequences with diverse enzymatic activities (ribozymes and DNAzymes). This technique has been extensively applied to the selection of natural DNA or RNA molecules but, in order to improve chemical and structural diversity as well as for particular applications where further chemical or biological stability is necessary, the extension of this strategy to modified oligonucleotides is desirable. Taking into account these needs, this review intends to collect the research carried out during the past years, focusing mainly on the use of modified nucleotides in SELEX and the development of mutant enzymes for broadening nucleoside triphosphates acceptance. In addition, comments regarding the synthesis of modified nucleoside triphosphate will be briefly discussed.
Introduction
Nowadays, nucleic acids are not only considered as genetic information messengers or repositories. New functions and applications of these molecules, like catalysis and molecular recognition have emerged in the last 25 years (Klussmann, 2006), impacting different fields like: therapeutics (Tei et al., 2015), target validation (Rodríguez et al., 2015), molecular biology (Lee et al., 2015), diagnostics (Wandtke et al., 2015), and analytical chemistry (Li and Lu, 2009; Mascini, 2009; Peinetti et al., 2015). The term “functional oligonucleotide” was coined in reference to these new functions.
Although some examples of these “non-traditional” oligonucleotide activities can be found in nature, as in the case of ribozymes, microRNAs and riboswitches (Li and Lu, 2009), a whole set of catalytic (RNA: ribozymes, DNA: DNAzymes) and molecular recognition oligonucleotides (aptamers) have been synthetically prepared, since they can be obtained using in vitro molecular evolution techniques. This methodology merges a combinatorial chemistry approach with a particular property of nucleic acids: amplification with the assistance of polymerases. This procedure was simultaneously developed in three independent laboratories in 1990 (Ellington and Szostak, 1990; Robertson and Joyce, 1990; Tuerk and Gold, 1990) and was denominated SELEX (Systematic Evolution of Ligands by Exponential Enrichment). Since then, many different variations of the technique were developed in order to achieve better selectivity and binding constants, and simpler experimental conditions (Sun and Zu, 2015; Yüce et al., 2015).
During the first years of aptamer development it was soon understood that this functional oligonucleotides could emulate monoclonal antibodies performance in diagnosis (Jayasena, 1999) and therapeutic (Schmid-Kubista et al., 2011) applications. Additionally, aptamers have some advantages over antibodies: they can be chemically synthesized without the assistance of animals; are thermally stable and can be easily fold and unfold. But also they have a major drawback (as DNAzymes and ribozymes); they have short lives in biological fluids due to the ubiquitous presence of endo and exonuclease activities. The strategy to overcome this limitation was the same that was employed in the development of antisense oligonucleotides (Iannitti et al., 2014): the structural chemical modification.
Modified functional oligonucleotides could be obtained according to two different strategies: post-selection modification or via modified-SELEX (mod-SELEX) techniques (Figure 1). The first approach has the advantage of dealing with natural oligonucleotide chemistry, but as consequence of the delicate relationship between structure and activity, post-selection modification without losing activity demonstrated to be a hard task in ribozymes (Pontiggia et al., 2010), DNAzymes (Robaldo et al., 2014), or aptamers (Bouchard et al., 2010; Förster et al., 2012).
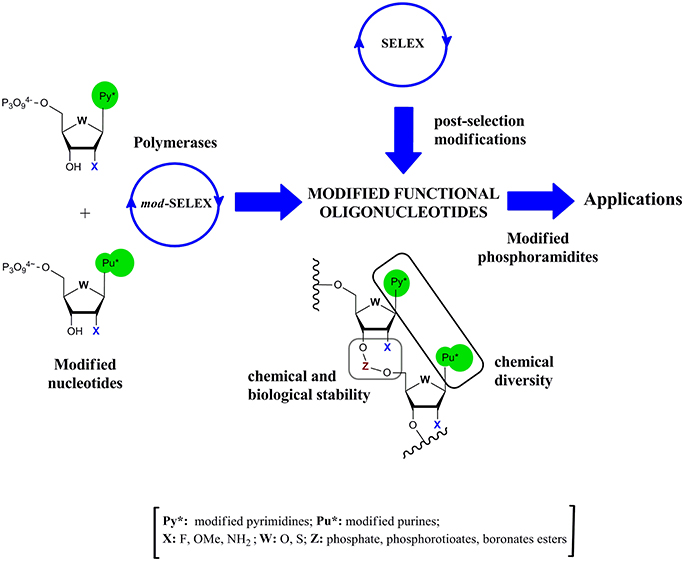
Figure 1. Schematic representation of the alternative routes to obtain modified functional oligonucleotides.
Taking into account this drawback, early efforts were done to modify the SELEX cycle in order to introduce modified nucleotides (Figure 1). The first report of mod-SELEX was done by Jayasena group (Lin et al., 1994) who succeeded in performing an in vitro selection of an RNA aptamer against human neutrophil elastase (HNE) using 2′-aminopyrimidine nucleotides. The resulting aptamer had a Kd of (6 ± 3) nM and a half life in serum of (9.3 ± 1.8) h, greatly improving the half life of the unmodified control sequence (4 min). Modifications at the 2′-position of the ribose moiety and phosphate internucleotide linkage were usually introduced in the oligonucleotide structure to enhance the chemical and biological stability (Figure 1).
In addition to stability, another important issue that drives the chemical modification of functional oligonucleotides is the augmentation of structural diversity. In this sense, one of the earliest examples was the obtainment of a DNA anti-thrombin aptamer using 5-pentinyl deoxyuridine by Toole group (Latham et al., 1994). Although the modified aptamer showed a weaker binding constant against thrombin compared to the natural aptamer obtained by the same group (Bock et al., 1992), they showed that the chemical structures of the modified and the natural aptamers were different.
It should be remarked that mod-SELEX and post-selection techniques are not excluding strategies as can be learned from the commercial anti-VEGF165 aptamer. Janjic group prepared a 2′-F-pyrimidine modified anti-VEGF165 aptamer using a mod-SELEX approach (Ruckman et al., 1998) and after the selection of the best sequence, 2′-OMe-ribopurine nucleotides were introduced in some positions without losing binding capacity [Kd = (49–130) pM]. This heavily modified aptamer became lately the first example of a therapeutic aptamer approved by the FDA against ocular vascular disease, Pegaptanib (Ng et al., 2006). In a variant of the mod-SELEX strategy Mayer and colleagues (Toole et al., 2015) obtained a modified aptamer against cycle 3 GFP (Kd = 18.4 nM) using 5-ethynyl-2′-deoxyuridine triphosphate which was further derivatized after polymerase amplification using click chemistry. A similar strategy, termed SELMA (Selection with Modified Aptamers), was used to generate DNA scaffolds containing ethynyl deoxyuridine moieties that were glycosylated employing glycan azides (Horiya et al., 2014). After selection of the most antigenic clusterings of the glycan, the bound sequences were amplified and reglycosylated to be used in the next selection step.
In the following sections the focus will be set on the different modified triphosphate nucleosides used in in vitro selection techniques but it should also be remarked that the modified phosphoramidites (Figure 1) are also essential in order to chemically prepare larger amounts of the modified functional oligonucleotides.
Although comprehensive and recent reviews about the different topics treated in this work are available (Diafa and Hollenstein, 2015; Lapa et al., 2016), the main purpose of this report is to connect all of them having in mind the application of modified nucleotides to in vitro selection techniques, focusing on recent related examples.
Modified Nucleotides for SELEX
The design of useful modified nucleotides appropriate for mod-SELEX has some restrictions that can be summarized in four conditions (Perrin et al., 1999):
1. should not disturb the base pair interactions (Watson-Crick and Hoogsteen);
2. must be substrates of the corresponding DNA or RNA polymerases;
3. the introduction of the modified nucleotide must be efficient at any position of the sequence;
4. the modified sequence must be a template for the corresponding polymerases.
In addition to the excellent available reviews in this field (Keffe and Cload, 2008; Hollenstein, 2012a; Kong and Bym, 2013) we intend in this section to summarize the main chemical nucleotide modifications that have been used for in vitro molecular selection, paying special attention to cases not reviewed earlier.
Regarding the modified nucleotides that have been used in SELEX, most examples are related to structural modifications of pyrimidine derivatives in three different positions: the α-phosphate, the 2′- and C5-positions (Figure 2). As consequence of the extraordinary sensitivity to degradation of RNA oligonucleotide libraries, the first mod-SELEX examples were motivated by the improvement of oligonucleotide stability against RNases, by substitution of the of 2′-hydroxyl of ribose by other functionalities. As mentioned above, the first reported example was developed by Jayasena group (Lin et al., 1994) who used 2′-aminouridine and 2′-aminocytidine triphosphates (3, Figure 2) to obtain a modified RNA aptamer against Human Neutrophil Elastase (HNE). When it was incubated in human serum and human urine, the HNE modified aptamer showed an enhanced stability compared to the unmodified sequence. Although the 2′-aminopyrimidine modification was also used to obtain a modified aptamer against the vascular permeability factor (VPF), the vascular endothelial growth factor (VEGF; Green et al., 1995) and the basic fibroblast growth factor (Jellinek et al., 1995), this modification was no longer used, probably due to the difficulties in the triphosphate preparation and the destabilizing effect of 2-amino groups in RNA duplexes. It was noticed that when the 2′-amino modification was compared to the 2′-fluoro analogs (4, Figure 2) for the same target, as in the case of the keratinocyte growth factor aptamer (Pagratis et al., 1997), both modifications have similar nuclease resistance but fluoro analogs have better affinities (two orders). As additional feature, compared to amine groups, 2′-fluoro modification does not require protection-deprotection steps along the oligonucleotide solid phase synthesis, reasons that could partially explain the popularity of this modification (Dupont et al., 2010; Svobodova et al., 2013).
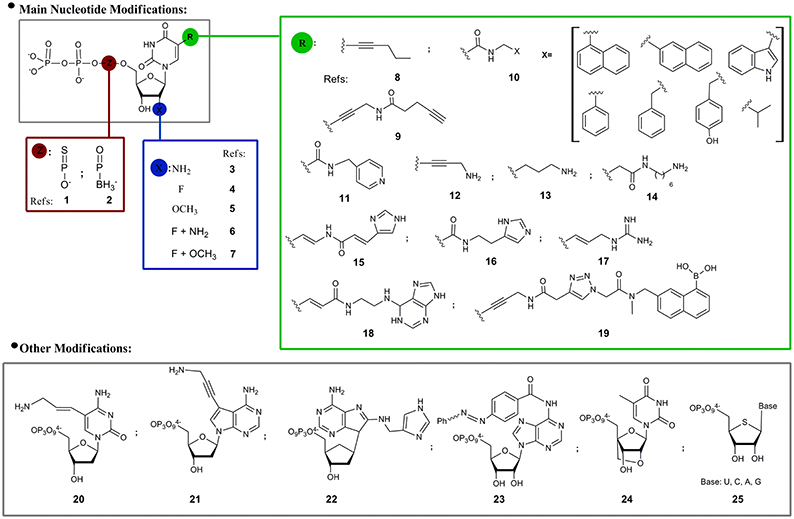
Figure 2. Modified nucleoside triphosphates used in SELEX experiments. References: 1: (Jhaveri et al., 1998; Somasunderam et al., 2010; Higashimoto et al., 2013); 2: (Lato et al., 2002); 3: (Lin et al., 1994; Green et al., 1995; Jellinek et al., 1995); 4: (Svobodova et al., 2013; Dupont et al., 2010); 5: (Burmeister et al., 2005); 6: (Pagratis et al., 1997); 7: (Friedman et al., 2015); 8: (Latham et al., 1994); 9: (Li et al., 2008); 10: (Vaught et al., 2010; Ochsner et al., 2013, 2014); 11: (Tarasow et al., 1997); 12: (Battersby et al., 1999); 13: (Vaish et al., 2003); 14: (Masud et al., 2004; Shoji et al., 2007); 15: (Santoro et al., 2000; Sidorov et al., 2004); 16: (Wiegand et al., 1997); 17: (Hollenstein et al., 2009); 18: (Imaizumi et al., 2013); 19: see 9; 20: (Hollenstein et al., 2009); 21: (Sidorov et al., 2004); 22: (Hollenstein et al., 2009); 23: (Liu et al., 2010); 24: (Kasahara et al., 2013); 25: (Minikawa et al., 2008).
Modified 2′-O-methylnucleotides (5, Figure 2) have also been used to get an aptamer against VEGF (Burmeister et al., 2005). Keefe and coworkers identified reaction conditions that allowed the incorporation of significant amounts of 2′-OMe-deoxyguanosine triphosphate into transcripts in the presence of ribosyl triphosphate. Guanosine positions must be finally checked to confirm the presence of natural or modified nucleotide. The obtained 23 nucleotide long aptamer (ARC245), showed a Kd of 2 nM and a fully modified structure which avoids degradation for at least 96 h in plasma. In the same sense, Li group (Friedman et al., 2015) recently developed a fully modified aptamer against Staphylococcus aureus Protein A (SpA) using 2′-F-deoxyguanosine, 2′-OMe-adenosine, -cytidine, and -uridine, an RNA library and a mutant (LAR) T7 RNA polymerase that circumvent the previously mentioned inconvenience of using small amounts of natural ribonucleotides.
A promising field is the aptamer obtainment using nucleotides with fully modified sugar moieties. Chaput and colleagues (Yu et al., 2012) succeeded in the preparation of a Threose Nucleic Acid (TNA) aptamer against human thrombin (Kd = 0.2–0.9 μM) with the assistance of an engineered variant of 9°N DNA polymerase.
Holliger and colleagues (Taylor and Holliger, 2015; Taylor et al., 2015) reported the preparation of four XNAzymes (catalyst from synthetic genetic polymers, XNA) based on four different sugar backbones: Arabino Nucleic Acids (ANA), 2′-Fluoroarabino Nucleic Acids (FANA), Hexitol Nucleic Acids (HNA) and Cyclohexene Nucleic Acids (CeNA). Mutant polymerases D4k for ANA and FANA, 6G12 for CeNA, and 6G12 I512L for HNA were employed for preparing the XNA libraries. Under these conditions RNA endonuclease (ANA, FANA, HNA, CeNA), RNA ligase and XNA ligase (FANA) activities were obtained.
Another position that has been modified with the objective of increasing nuclease resistance is the α-phosphate of nucleoside triphosphates. Ellington and coworkers (Jhaveri et al., 1998) obtained an anti basic fibroblast growth factor (bFGF) RNA aptamer in which all internucleotide linkages were modified with phosphorothioates (1, Figure 2). Although the yield of phosphorothiolated RNA following in vitro transcription and purification was low, it was enough for selection experiments and a 13 nucleotide consensus sequence with a Kd of 1.8 nM for bFGF was selected. Using the same modification Gorenstein and coworkers (Somasunderam et al., 2010) obtained a thiophosphate-modified DNA aptamer against the hyaluronic acid binding domain (HABD). In this case, all deoxyadenosine residues (with the exception of primers) were replaced by deoxyadenosine thiophosphates in the Sp configuration. More recently, Yamagishi and coworkers (Higashimoto et al., 2013) selected phosphorothioate-modified aptamers directed against advanced glycation end products (AGEs–thioaptamers) using a mixture of 2′-deoxyadenosine-5′-O-(1-thiotriphosphate) (dATP(αS)), 2′-deoxythymidine-5′-O-(1-thiotriphosphate) [dTTP(αS)] and a mixture of deoxycytidine and deoxyguanosine triphosphates. The obtained aptamers had Kd in the low nanomolar range. Another feasible modification in the α-phosphate moiety of the nucleotides is the replacement of oxygen by a borohydride group (boranophosphate, 2, Figure 2). Burke and coworkers (Lato et al., 2002) explored the use of uridine (bU) or guanosine (bG) 5′-(α-P-borane) in a SELEX process against ATP. Even if bU and bG are compatible with the selection process, when they were introduced in natural aptamer structures usually diminished or eliminated target recognition.
Although the 2′-position and the α-phosphate nucleotide modifications mainly pursue the increase of oligonucleotides stability, another important aspect of SELEX is the expansion of the chemical diversity of the nucleotide moiety. It was reasoned that a set of complementary functional groups, not originally available in the nucleotide structures, will enhance the chances of molecular recognition or catalytic phenomenon. In this regard, 5-position of pyrimidine, particularly uridine and deoxyuridine, was the most frequent modification option. Functional groups present in the 5-pyrimidine position could be classified in three main sets: hydrophobic, acid/base and others (boronic). In the first subset we can find examples such as compounds 8, 9, and 10 in Figure 2. Toole and coworkers (Latham et al., 1994) prepared DNA aptamers against thrombin using 5-(1-pentinyl)-2′-deoxyurindine (8, Figure 2) with Kd in the range of 400–1000 nM. Wang and coworkers (Li et al., 2008) introduced hydrophobic structures (9, Figure 2) in aptamers against fibrinogen. In a systematic way, Eaton and coworkers (Vaught et al., 2010) first and Janjic and coworkers later (Ochsner et al., 2014), explored the use of a set of hydrophobic moieties connected to the 5-position of deoxyuridine by an amide group (10, Figure 2). In Eaton's work an exploration of the best polymerases that accept the modified nucleotides as substrates was done, finding that the OD XL and D.Vent(exo-) were able to incorporate the modified deoxyuridine triphosphate derivatives with similar or better yields than with thymidine triphosphate. In this case the selected targets were the tumor associated calcium signal transducer 2 (TACSTD2) and the tumor necrosis factor receptor super family member 9 (TNFRSF9). In the first case benzyl and isopropyl moieties were used giving aptamers with Kds in the nM range. In the second case, where previous DNA selections failed, the benzyl moiety afforded an aptamer with a Kd of 100 nM.
As previously commented, Janjic and coworkers also explored the use of hydrophobic modifications. In this case the objective was to develop methods for the systematic isolation of aptamers that bind to different epitopes of proteins, allowing efficient pair-wise screening of multiple ligands (Ochsner et al., 2014; Rohloff et al., 2014). These aptamers, named slow off-rate modified aptamers (SOMAmer) were prepared using a benzyl moiety linked to the uracil by an amide group and were directed against human proteins like: ANGPT2, TSP2, CRDL1, MATN2, GPVI, C7, and PLG, some of them indicators of cardiovascular risk. The equilibrium dissociation constants ranged from 0.02 to 2.7 nM.
Considering now 5-pyrimidine nucleotide modifications with acid/base activity, a functional group usually present in this position is amine (or ammonium). Eaton and coworkers (Tarasow et al., 1997) synthesized a 5-pyridylmethylcarboxamide uridine (11, Figure 2) for the obtainment of a ribozyme with moderate dielsalderase activity (Diels Alder carbon-carbon bond formation). Benner and coworkers (Battersby et al., 1999) synthesized 5-(3″-aminopropynyl)-2′-deoxyuridine triphosphate (12, Figure 2) which was used to replace thymidine nucleotide in a SELEX process, with the assistance of the Vent DNA polymerase, against the ATP molecule as target. The consensus sequences were examined to determine if they matched or not the original Szostak sequence (Huizenga and Szostak, 1995), finding significant resemblance with the “Lin-Patel-Huizenga-Szostak motif” (Battersby et al., 1999).
Regarding the existence of modified moieties found in RNA of biological sources, post-transcriptional modifications are implicated in catalytic or molecular recognition events (Limbach et al., 1994; Helm and Alfonzo, 2014). Although moieties with a positive charge are naturally rare, some examples are known, like in the case of archaeosine (Gregson et al., 1993), a modified purine that has a positive charge at physiological pH. This fact inspired McLaughlin and coworkers (Vaish et al., 2003) who used 5-(3-aminopropyl) uridine (13, Figure 2) to obtain an RNA aptamer against ATP. The resulting sequence had a Kd of 1.08 mM with several of the modified uridines critical for target recognition. Following the same concept, Sawai and coworkers (Masud et al., 2004) used the 5′-triphosphate of 5-N-(6-aminohexyl)carbamoylmethyl-2′-deoxyuridine (14, Figure 2) for the selection of a DNA aptamer (Kd = 4.9 μM) against sialyllactose, an oligosaccharide with a carboxy group that appears to be an essential receptor component of many animal viruses from different families, such as influenza A and C viruses. The same modified pyrimidine was lately used by this research group to develop an aptamer against the (R)-isomer of Thalidomide (Shoji et al., 2007) with a Kd of 1 μM.
Other nitrogen containing substituents were evaluated. Barbas and coworkers (Santoro et al., 2000) obtained by in vitro evolution a DNAzyme with RNase activity carrying an imidazolyl moiety at the 5-position of deoxyuridine (15, Figure 2). The DNAzyme has a minimum core of twelve nucleotides including three imidazole-functionalized nucleotides and requires μM concentration of Zn2+ and mM of Mg2+ and Na+ for being active. Lately, Williams and coworkers (Sidorov et al., 2004) used a combination of the 5-imidazolyl-modified deoxy uridine triphosphate (15, Figure 2) plus a 7-aminopropynyl modified 7-deaza-deoxyadenosine triphosphate analog (12, Figure 2), for the selection of a DNAzyme with RNA cleavage activity independent of divalent metal requirements. Following with the development of modified functional oligonucleotides with catalytic activity, Eaton and coworkers (Wiegand et al., 1997) used 5-imidazolyl uridine triphosphate analog (16, Figure 2) for the preparation of a ribozyme with amide-bond formation activity, pushing the hypothesis of the expansion of nucleic acid catalytic activity. With the same objective, Perrin and coworkers (Hollenstein et al., 2009) used a simultaneous combination of 8-histaminyl-deoxyadenosine (21, Figure 2), 5-guanidinoallyl-deoxyuridine (17, Figure 2) and 5-aminoallyl-deoxycytidine (20, Figure 2) triphosphates in an in vitro selection of a heavily modified metal2+ free DNAzyme with RNase activity. Pursuing a similar goal, Williams and coworkers (Sidorov et al., 2004) prepared and used a 3-(aminopropynyl)-7-deaza-deoxyadenosine triphosphate (21, Figure 2). In a recent example, Silverman and colleagues (Zhou et al., 2016) showed that the introduction of protein-like residues (5-thymidine primary alcohol, amine or carboxyl group) allowed the selection of modified DNAzymes with amide hydrolytic activity that was until then an elusive enterprise.
Sugimoto and coworkers (Imaizumi et al., 2013) prepared a (E)-5-(2-(N6-adeninyl)ethyl))carbamylvinyl)deoxyuridine triphosphate (23, Figure 2) that was used to obtain a modified DNA aptamer against camptothecin, a quinoline alkaloid that inhibits topoisomerase I. In this case, the modified aptamer showed a higher affinity constant than the unmodified captothecin aptamer, illustrating the hypothesis that additional aptamer functionalization can improve binding performance.
Returning to modified aptamer examples, Wang and coworkers (Li et al., 2008) prepared a boronic acid 5-modified thymidine triphosphate (19, Figure 2) with the aim of improving aptamer binding to glycoproteins (fibrinogen as model), based on the known ability of boron to coordinate glycol groups. The boronic acid modified aptamers selected against fibrinogen had a Kd in the low nM range, while aptamers prepared with natural nucleotides had Kds ca. 5 μM, confirming the proposed hypothesis.
Ito and coworkers (Liu et al., 2010) prepared a photoresponsive N6-azobencene-adenosine triphosphate (23, Figure 2) for the selection of a modified RNA aptamer against hemin with catalytic activity as peroxidase. The aim of this modification was the photocontrol of the recognition event and consequently of the catalytic activity. The visible and UV irradiation of the obtained modified aptamer controlled the cis-trans azobencene isomerization, which modified the hemin-aptamer catalytic ability as peroxidase.
Finally, some modifications in the ribose/deoxyribose moieties have also been explored. Kuwahara and coworkers (Kasahara et al., 2013) explored the use of 2′-O, 4′-C-methylene-bridged bicyclic ribonucleotides (24, Figure 2) for the selection of DNA aptamers against human thrombin. The aptamers were selected by capillary electrophoresis-SELEX (CE-SELEX) and had Kd in the low nM range. Matsuda and coworkers (Minikawa et al., 2008) used 4′-thioribonucleotides (25, Figure 2) for the selection of an RNA aptamer against human α-thrombin. The optimization of the SELEX conditions for the use of the four modified nucleotides (25, Figure 2) included the use of additional amounts of ATP and GTP and the assistance of mutant polymerases. The highly-modified 4′-thioRNA aptamer was also post-selection fully modified, having a Kd of 29.6 nM.
All the examples of modified nucleotides described so far have been successfully used in SELEX processes finally obtaining functional oligonucleotides. However, some examples of modified nucleotides, accepted by mutant polymerases and not applied yet to SELEX, have also been described. Just to mention two examples, Hollenstein (2012b) has described a set of 5-modified deoxyuridine, with moieties designed for organocatalysis (26, Figure 3). These modified nucleotides were substrates of Ven (exo-), Pwo DNA and E. coli Klenow fragment polymerases. More recently, Perrin and coworkers (Liu et al., 2015) reported the preparation of 5-aminomethyl deoxyuridine triphosphate derivatives (27, Figure 3) modified mainly with aromatic moieties. The authors successfully used all modified triphosphates as substrates of the Vent (exo-) polymerase.
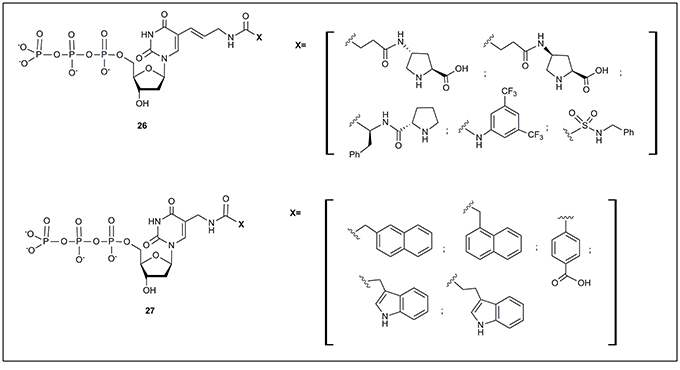
Figure 3. Modified nucleoside triphosphates accepted by modified polymerases but not yet used in SELEX protocols.
A mention to L- aptamers (Spiegelmers) will also be included. Although they were not strictly obtained using modified nucleoside triphosphates (Eulberg et al., 2006), the final result of this variant of the SELEX method is a fully modified L-aptamer. This technique has been applied when the enantiomer of the target was synthetically available, as in the case of peptides (Yatime et al., 2015) or RNA structures (Sczepanski and Joyce, 2013).
Another interesting approach is the use of the genetic alphabet expansion. Hirao and colleagues (Kimoto et al., 2013) have described the first example of modified aptamers against human VEGF-165 (Kd = 0.62 nM) and interferon-γ (Kd = 0.038 nM) using the four natural DNA bases plus an additional synthetic base pair in an expanded alphabet strategy, naming the technique as Expanded-SELEX (ExSELEX; Kimoto et al., 2016).
Modified Nucleoside Triphosphate Synthesis
When modified nucleoside triphosphates (NTPs) for in vitro selection techniques are needed they have to be synthesized, since only few of them are commercially available. Therefore, for the safe of completion, a brief summary of the main methods developed for this purpose are herein included.
The regioselective modification of nucleosides is a cumbersome goal due to their polyfunctional structure and in particular an effective and general methodology for the synthesis of NTPs is still a challenge without a proper solution. In addition, NTPs are polycharged and unstable molecules, characteristics that make difficult the work up processes associated with their purification.
The available protocols for NTPs synthesis were extensively reviewed (Burgess and Cook, 2000; Hollenstein, 2012a; Kore and Srinivasan, 2013), and here will be only briefly mentioned.
One of the pioneer methods was the one carried out by Ludwig (Ludwig, 1981) based on the regioselective 5′-phosphorylation designed by Yoshikawa (Yoshikawa et al., 1967). It consists in the one pot reaction of an unprotected nucleoside and phosphorus oxychloride using trimethyl phosphate as solvent. The reactive 5′-phosphorodichlorate intermediate is further treated with bis-tributylammonium pyrophosphate to generate a cyclic triphosphate which is finally hydrolysed to the corresponding NTP. This process is not suitable for modified nucleosides and in addition involves difficult purification processes due to the presence of secondary products. A recent contribution to this strategy (Korhonen et al., 2015) propose the use of tris{bis(triphenylphosphoranylidene) ammonium} (PPN) pyrophosphate as an alternative to the hygroscopic alkylammonium salts.
An alternative method developed by Ludwig and Eckstein (Ludwig and Eckstein, 1989) is still nowadays widely applied for the preparation of NTPs. It is also a one pot route but the nucleoside must be properly protected owing to the lack of regioselectivity of the process. This route involves the attack of salicyl phosphorochlorite to afford the corresponding 5′-phosphite derivative. The subsequent addition of bis-tributylammonium pyrophosphate produces the key cyclic phosphite intermediate, which is then oxidized in situ to yield the resultant NTP. Although the interfering nucleosidic functions need to be protected, the purification protocols are simpler since the presence of other polycharged species is minimized.
Another strategy, designed by Caton-Williams et al. (Caton-Williams et al., 2011a,b, 2013), also makes use of the salicyl phosphorochloridite reagent, but through and altered route. In this synthetic scheme the phosphitylating agent is first treated with pyrophosphate in DMF to generate the actual phosphitylating reagent that regioselectively reacts with the 5′-hydroxyl of an unprotected nucleoside to produce a similar cyclic intermediate to the one formed in the Ludwig-Eckstein strategy. Finally, the corresponding NTP is obtained by traditional iodine oxidation and hydrolysis. After performing a simple ethanol precipitation, the crude product can be directly used as polymerases substrate.
Many other different approaches have been explored in the search of a universal and efficient route for the preparation of NTPs, some of them employ nucleoside phosphites, phosphoramidites, diphosphates or displacement of 5′-O-leaving groups by triphosphate. A complete revision of these methods has already been reviewed (Burgess and Cook, 2000; Kore and Srinivasan, 2013).
The environmental friendly approaches offered by biocatalysis and biotransformations give an intestering alternative to the chemical preparation of NTPs since they provide regioselective reactions using mild conditions (Staffan, 2005). This process involves three steps being the first one catalyzed by specific nucleoside kinases. Therefore, alternative enzymatic procedures for the preparation of nucleoside monophosphates are relevant and have been recently reviewed (Iglesias et al., 2015).
As for chemical approaches, the biocatalyzed syntheses do not provide a general and efficient method for NTPs preparation and the appropriate route for each case has to be explored specifically.
Polymerases for Modified Nucleotides
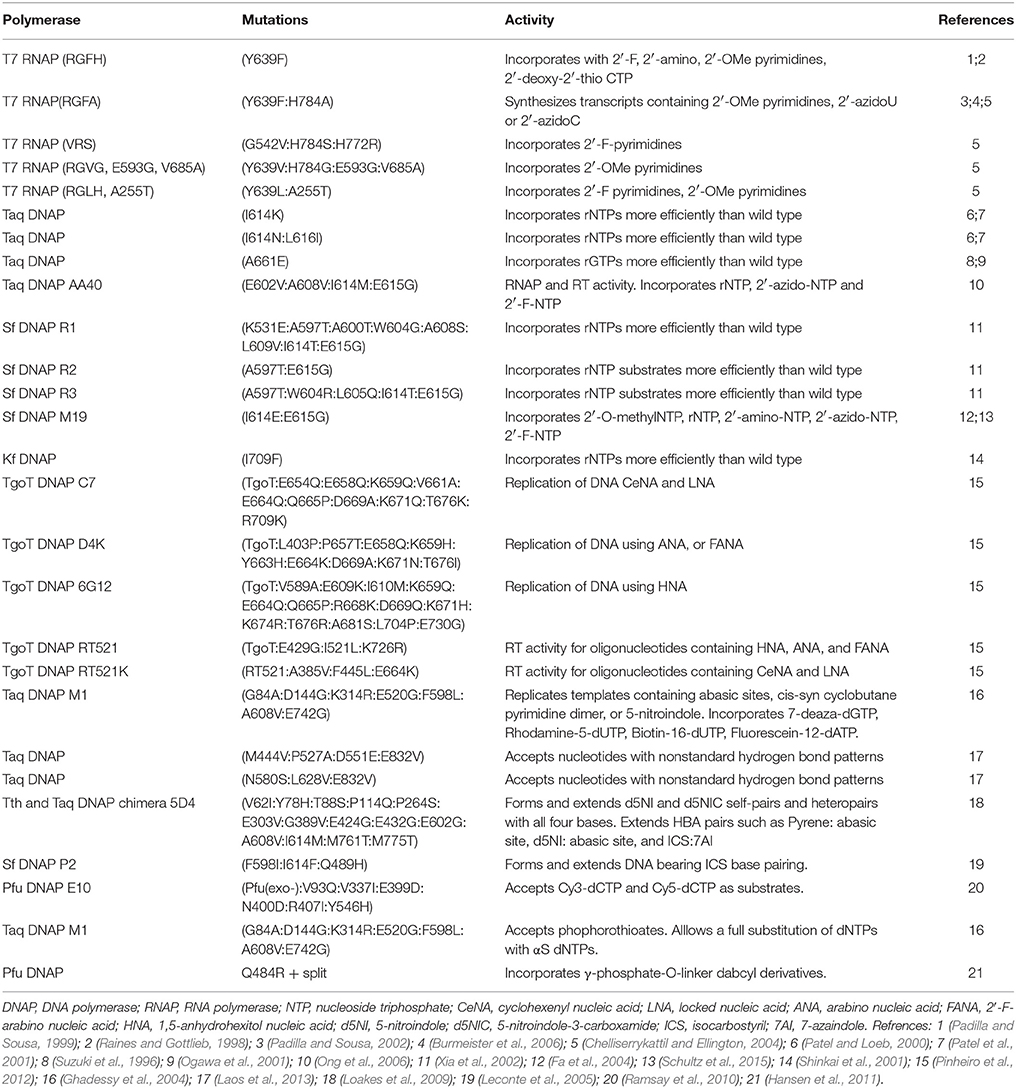
The increasing number of nucleotide analogs used for the development of functional oligonucleotides demands the access to polymerases with an expanded substrate repertoire. Over the years natural polymerases have been tested for their ability to accept unnatural nucleotides and many directed evolution experiments have been carried out to achieve this goal being these works properly reviewed in several papers (Henry and Romesberg, 2005; Lauridsen et al., 2012; Walsh and Beuning, 2012; Chen and Romesberg, 2014; Laos et al., 2014). A summary of polymerases evolved to accept modified nucleotides can be seen in Table 1.
Polymerases for Nucleotides Bearing Sugar Modifications
In the 1990s Aurup et al. demonstrated that wild type T7 RNA polymerase could accept 2′-fluoro-CTP, 2′-fluoro-ATP, 2′-fluoro-UTP, and 2′-amino-UTP as substrates in transcription reactions. Results showed that short templates could be transcribed to full-length RNA products in the presence of these modified nucleotides, although higher affinity was observed for the 2'-amino analogs (Aurup et al., 1992). This enzyme was also found to be capable of synthesizing quimeric nucleic acids composed of ribo and deoxyribonucleotides and of incorporating 2′-O-methylnucleotides in various positions (Conrad et al., 1995).
Since then, many directed evolution experiments have been performed to improve T7 RNA polymerase substrate repertoire and processivity. Rational design led to a T7 RNA polymerase bearing the Y639F mutation, which was able to synthesize transcripts with 2′-fluoro, 2′-amino, 2′-O-methylpyrimidines and 2′-deoxy-2′-thio-CTP more efficiently than the wild type (Raines and Gottlieb, 1998; Padilla and Sousa, 1999). A double mutant (Y639F:H784A) was later reported that showed improved use of 2′-O-methyl and 2′-azido pyrimidines as substrates (Padilla and Sousa, 2002). Using the autogene selection method many other variants were reported with better utilization of 2′-substitued NTPs (Chelliserrykattil and Ellington, 2004). Interestingly, mutations previously shown to increase the thermal tolerance of T7 RNA polymerase can also increase the activity of mutants with expanded substrate range (Meyer et al., 2015). T7 RNA polymerase was also tested for its ability to accept 2′-C-branched NTPs. Wild type enzyme was able to perform the reaction with 2′-α-hydroxymethyl-UTP while the Y639F mutant was unable to use any of the substrates (Pavey et al., 2004). In another work, the wild type enzyme was capable of synthesizing 4′-thioRNA using 4′-thioUTP and 4′-thioCTP (Kato et al., 2005).
Regarding accessible enzymes, it is worth mentioning that many commercially available DNA polymerases such as SequiTherm, Tht, AmpliTaq FS (exo-), Pfu (exo-), Vent (exo-), and deep Vent (exo-) as well as the human DNA polymerases α and γ, were qualified to accept 2′-fluoro-modified NTPs (Ono et al., 1997; Richardson et al., 2000).
Directed evolution efforts have also focused on the widely used Taq DNA polymerase to improve 2′-modified nucleotides recognition. Several variants of this enzyme have been identified to incorporate ribonucleotides more efficiently than the wild type (Suzuki et al., 1996; Patel and Loeb, 2000; Ogawa et al., 2001; Patel et al., 2001) whereas Taq DNAP AA40 was able to incorporate ribonucleotides as well as 2′-azido and 2′-fluoro NTPs (Ong et al., 2006). Stoffel fragment (Sf) DNA polymerase, a proteolytic fragment of Taq DNA polymerase, was also subjected to directed evolution obtaining the variants Sf R1, Sf R2 and Sf R3 with an increased efficiency to incorporate ribonucleotides (Xia et al., 2002), and Sf M19 that could also accept 2′-O-methyl NTP, 2′-amino-NTP, 2′-azido-NTP and 2′-F-NTP as substrates (Fa et al., 2004). A recent study improved the understanding of the mutational origins of 2′-modified substrate recognition, and also identified Sf M19 as the best candidate for further engineering (Schultz et al., 2015). The Klenow fragment (Kf), a proteolytic fragment of E. coli DNA polymerase I, bearing one mutation (I709F), presented better incorporation of ribonucleotides to the product than the wild type (Shinkai et al., 2001).
Another interesting finding regarding 2′-modified nucleotides was the ability to add 2′-C-methyl analogs into RNA by a hepatitis C virus RNA polymerase. Hepatitis C virus nonstructural protein 5B (HCV NS5B) accepted 2′-C-methyl-ATP and 2′-C-methyl-CTP as substrates while mutant NS5BΔ55 incorporated the modified nucleotides more efficiently (Carroll et al., 2003; Dutartre et al., 2006).
Enzymatic polymerization using LNA nucleotides was reported using many different polymerases. First, the group of Wengel (Veedu et al., 2007) reported that Phusion High-Fidelity DNA polymerase had the capacity of adding up to three LNA-TTPs and up to eight consecutive LNA-ATPs. Later, results showed that 9°NmTM DNA polymerase presented this activity as well, and was able to read across LNA residues in the template. In addition, T7 RNA polymerase was capable of transcription incorporating LNA-ATPs as substrates and also full length RNA transcripts were obtained when LNA and DNA templates were involved (Veedu et al., 2008). In another work, it was observed that using higher concentrations of KOD Dash, KOD (exo-) and Vent (exo-) DNA polymerases led to higher yields of full length products bearing LNA modifications (Kuwahara et al., 2008).
Several DNA polymerases were tested for their ability to synthesize 2′-deoxy-2′-fluoro-β-D-arabino nucleic acids (FANA). Experiments revealed that family B polymerases like Deep Vent (exo-), 9°Nm, Therminator and Phusion High-Fidelity were able to incorporate all four FANA analogs to yield full length products (Peng and Damha, 2007). A later investigation, proposed the development of polymerases that could synthesize Xeno nucleic acids (XNA, nucleic acids carrying different types of synthetic sugars) from a DNA template and polymerases that could reverse transcribe XNA back into DNA. Using compartmentalized self-tagging (CST) selection from a library of TgoT DNA polymerase, they obtained TgoT mutants that were able to replicate DNA using cyclohexenyl nucleic acids (CeNA), locked nucleic acids (LNA), arabino nucleic acids (ANA), 2′-fluoroarabinonucleic acids (FANA) and 1,5-anhydrohexitol nucleic acids (HNA) as substrates. By saturation mutagenesis based on statistical correlation analysis (SCA) and ELISA-like screening they evolved TgoT into variants capable of reverse transcription of templates containing HNA, ANA, FANA CeNA, and LNA (Pinheiro et al., 2012).
Recently, Holliger and co-workers (Cozens et al., 2015) engineered the polymerase TGLLK (TgoT: Y409G, I521L, F545L, E664K), which proved to be effective at fully replacing purine dNTPs and NTPs with their respective 3′-deoxy or 3′-O-methyl analogs with defined 2′-5′ linkages allowing template-directed synthesis and reverse transcription of oligonucleotides with mixed 2′-5′/3′-5′ backbone linkages, expanding the repertoire for future in vitro evolution experiments.
Polymerases for Nucleotides Bearing Nucleobase Modifications
Different thermostable DNA polymerases were tested using C5-substituted pyrimidines as substrates. Family A DNA polymerases, such as Taq, Tht and Thermosequenase, were able to accept propynyl dUTP, dUTP, and methyl dCTP whereas family B DNA polymerases, such as Pwo, Pfu, Vent (exo-), Deep Vent (exo-), were able to incorporate 7-amino-2,5-dioxaheptyl dUTP, 7-amino-2,5-dioxaheptyl dCTP, propynyl dUTP, and methyl dCTP (Kuwahara et al., 2003). Vent (exo-), Pwo and Kf DNA polymerases were also able to use NTPs carrying urea, proline, and sulfonamide groups in the C5 position (Hollenstein, 2012b). DNA molecules containing 5-vinyl-dUTP, 5-vinyl-dCTP, or 7-deaza-7-vinyl-dATP were prepared by polymerase incorporation using Pwo, Vent (exo-), and KOD XL DNA polymerases (Mačková et al., 2014).
KOD Dash DNA polymerase was used to perform SELEX of sialyllactose-binding DNA aptamers composed of several modified TTPs bearing a positively-charged amino group at the C5 position (Masud et al., 2004). Later, it was also showed the ability to successfully accept dUTP bearing different aminoacids at the C5 position (Kuwahara et al., 2006). Useful dCTP analogs for aptamer discovery via SELEX bearing a 5-(N-substituted-carboxamide) functional group were found to be suitable substrates of KOD DNA polymerase (Rohloff et al., 2015).
Directed evolution of Taq DNA polymerase led to more efficient enzymes that could bypassed blocking lesions such as an abasic site, a thymidine dimer or the base analog 5-nitroindol (Ghadessy et al., 2004; Loakes et al., 2009), and variants that could accept nucleotides with nonstandard hydrogen bond patterns therefore allowing the expansion of the genetic alphabet (Laos et al., 2013).
Using directed short-patch compartmentalized self-replication (spCSR) and the widely used fluorescent dye label Cy3-dCTP and Cy5- dCTP as substrates, a variant of Pfu DNA polymerase that was able to amplify double stranded DNA fragments incorporating these analogs was obtained (Ramsay et al., 2010).
Recent studies (Wyss et al., 2015) involving the ability to amplify DNA adducts showed that a mutant of KlenTaq DNA polymerase (KTqM747K), was able to incorporate an artificial nucleotide, BenziTP, opposite to a DNA alkylation adduct with high selectivity. In this way, the artificial nucleotide functions as a marker for the adduct in the original template being useful to investigate DNA damage levels.
Polymerases for Nucleotides Bearing Triphosphate Modifications
Nucleosides carrying 5′-O-(1-thiotriphosphate) moieties (NTP(αS)) are the most employed triphosphate modified substrates for natural and evolved enzymes used for the selection of aptamers. Taq DNA polymerase has been used to select a single-stranded DNA aptamer targeted to the transforming growth factor-b1 (TGF-b1) where all dATPs and dCTPs were substituted by phosphorothioates (Kang et al., 2008). In another study, the M1 variant of Taq DNA polymerase allowed the full substitution of dNTPs with phosphorothioates (Ghadessy et al., 2004). Regarding less frequent modifications, T7 RNA polymerase was used to select an ATP-binding aptamer containing 5′-(α-P-borano)-GTP and 5′-(α-P-borano)-UTP (Lato et al., 2002) and a mutated Pfu DNA polymerase was able to incorporate nucleotides bearing a bulky γ-phosphate-O-linker-dabcyl substituent (Hansen et al., 2011).
Conclusions and Perspectives
The vast number and types of applications that modified functional oligonucleotides offer to different fields such as diagnosis, therapy, analytical chemistry, target validation and molecular biology, makes of the use of modified nucleoside triphosphates for in vitro selection techniques a challenging research area. This strategy brings about a second generation of functional oligonucleotides with higher nuclease resistance and increased structural and chemical diversity allowing the selection of molecules with differential properties respect to the natural DNA or RNA oligomers such as improved binding or catalytic activities. In addition, other benefits such as better uptake for in vivo applications are expected.
Still many contributions and new developments are needed mainly from two disciplines: molecular biology and organic chemistry. The first one should generate a larger repertoire of evolved polymerases able to faithfully recognize a wider spectrum of modified nucleoside triphosphates. On the other side, although chemistry already made available a plethora of modified nucleosides, it still needs to provide a general and efficient synthetic route to modified nucleoside triphosphates and to propose improvements of the current complex purification protocols.
The research carried out in these areas, and summarized in this review, indicates that in the near future these issues will be successfully addressed expanding the scope of functional oligonucleotides as useful custom-made tools for in vivo and in vitro applications.
Author Contributions
JM mainly contributed to write the modified nucleotides for SELEX and introduction sections. MD mainly contributed to review the Polymerases for modified nucleotides section. AI mainly contributed to write the modified nucleoside triphosphate synthesis and Conclusions and perspectives sections and revised the whole paper.
Funding
JM and AI are research members of CONICET and MD has a doctoral fellowship from CONICET. This work has been partially supported by PICT 2011–2007.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aurup, H., Williams, D. M., and Eckstein, F. (1992). 2'-Fluoro- and 2'-amino-2'-deoxynucleoside 5'-triphosphates as substrates for T7 RNA polymerase. Biochemistry 31, 9636–9641. doi: 10.1021/bi00155a016
Battersby, T. R., Ang, D. N., Burgstaller, P., Jurczyk, S. C., Bowser, M. T., Buchanan, D. D., et al. (1999). Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J. Am. Chem. Soc. 121, 9781–9789. doi: 10.1021/ja9816436
Bock, L. C., Griffin, L. C., Latham, J. A., Vermaas, E. H., and Toole, J. J. (1992). Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355, 564–566. doi: 10.1038/355564a0
Bouchard, P. R., Hutabarat, R. M., and Thompson, K. M. (2010). Discovery and development of therapeutic aptamers. Annu. Rev. Pharmacol. Toxicol. 50, 237–257. doi: 10.1146/annurev.pharmtox.010909.105547
Burgess, K., and Cook, D. (2000). Syntheses of nucleoside triphosphates. Chem. Rev. 100, 2047–2059. doi: 10.1021/cr990045m
Burmeister, P., Lwis, S. D., Silva, R. F., Preiss, J. R., Horwitz, L. R., Pendergrast, P. S., et al. (2005). Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 12, 25–33. doi: 10.1016/j.chembiol.2004.10.017
Burmeister, P. E., Wang, C., Killough, J. R., Lewis, S. D., Horwitz, L. R., Ferguson, A., et al. (2006). 2′-Deoxy purine, 2′-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides 16, 337–351. doi: 10.1089/oli.2006.16.337
Carroll, S. S., Tomassini, J. E., Bosserman, M., Getty, K., Stahlhut, M. W., Eldrup, A. B., et al. (2003). Inhibition of hepatitis C virus RNA replication by 2'-modified nucleoside analogs. J. Biol. Chem. 278, 11979–11984. doi: 10.1074/jbc.M210914200
Caton-Williams, J., Hoxhaj, R., Fiaz, B., and Huang, Z. (2013). Use of a novel 5-regioselective phosphitylating reagent for one-pot synthesis of nucleoside 5-triphosphates from unprotected nucleosides. Curr. Protocol Nucleic Acid Chem. Chapter 1:Unit 1.30. doi: 10.1002/0471142700.nc0130s52
Caton-Williams, J., Lin, L., Smith, M., and Haung, Z. (2011a). Convenient synthesis of nucleoside 5-triphosphates for RNA transcription. Chem.Commun. 47, 8142–8144. doi: 10.1039/c1cc12201k
Caton-Williams, J., Smith, M., Carrasco, N., and Huang, Z. (2011b). Protection-free one-pot synthesis of 2-deoxynucleoside 5-triphosphatesand DNA polymerization. Org. Lett. 13, 4156–4159. doi: 10.1021/ol201073e
Chelliserrykattil, J., and Ellington, A. D. (2004). Evolution of a T7 RNA polymerase variant that transcribes 2'-O-methyl RNA. Nat. Biotechnol. 22, 1155–1160. doi: 10.1038/nbt1001
Chen, T., and Romesberg, F. E. (2014). Directed polymerase evolution. FEBS Lett. 588, 219–229. doi: 10.1016/j.febslet.2013.10.040
Conrad, F., Hanne, A., Gaur, R. K., and Krupp, G. (1995). Enzymatic synthesis of 2'-modified nucleic acids: identification of important phosphate and ribose moieties in RNase P substrates. Nucleic Acids Res. 23, 1845–1853. doi: 10.1093/nar/23.11.1845
Cozens, C., Mutschler, H., Nelson, G. H., Houlihan, G., Taylor, A. I., and Holliger, P. (2015). Enzymatic synthesis of nucleic acids with defined regioisomeric 2'-5'-linkages. Angew. Chem. Int. Ed Engl. 54, 15570–15573. doi: 10.1002/anie.201508678
Diafa, S., and Hollenstein, M. (2015). Generation of aptamers with an expanded chemical repertoire. Molecules 20, 16643–16671. doi: 10.3390/molecules200916643
Dupont, D. M., Madsen, J. B., Harmann, R. K., Tavitian, B., Ducougé, F., Kjens, J., et al. (2010). Serum-stable RNA aptamers to urodinase-ype plasminogen activator blocking receptor binding. RNA 16, 2360–2369. doi: 10.1261/rna.2338210
Dutartre, H., Bussetta, C., Boretto, J., and Canard, B. (2006). General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2'-modified nucleotide analogues. Antimicrob. Agents Chemother. 50, 4161–4169. doi: 10.1128/AAC.00433-06
Ellington, A. D., and Szostak, J. W. (1990). Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 346, 818–822. doi: 10.1038/346818a0
Eulberg, D., Jarosch, F., Vonhoff, S., and Klussmann, S. (2006). “Spiegelmers for therapeutic applications-use of chiral principles in evolutionary selection techniques,” in The Aptamer Handbook, ed S. Klussmann (Weinheim: Wiley-VCH), 417–442.
Fa, M., Radeghieri, A., Henry, A. A., and Romesberg, F. (2004). Expanding the substrate repertoire of a DNA polymerase by directed evolution. J. Am. Chem. Soc. 126, 1748–1754. doi: 10.1021/ja038525p
Förster, C. H., Zydek, M., Rothkegel, M., Wu, Z., Gallin, C., Geßner, R., et al. (2012). Properties of an ‘LNA’-modified ricin RNA aptamer. Biochem. Biophys. Res. Commun. 419, 60–65. doi: 10.1016/j.bbrc.2012.01.127
Friedman, A. D., Kim, D., and Li, R. (2015). Highly stable aptamers selected from a 2'-fully modified fGmH RNA library for targeting biomaterials. Biomaterials 36, 110–123. doi: 10.1016/j.biomaterials.2014.08.046
Ghadessy, F. J., Ramsay, N., Boudsocq, F., Loakes, D., Brown, A., Iwai, S., et al. (2004). Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat. Biotechnol. 22, 755–759. doi: 10.1038/nbt974
Green, L. S., Jellinek, D., Bell, C., Beebe, L. A., Feistner, B. D., Gill, S. C., et al. (1995). Nuclease-resistant nucleic acid ligands to vascular permeability factor/vascular endothelial growth factor. Chem. Biol. 2, 683–695. doi: 10.1016/1074-5521(95)90032-2
Gregson, J. M., Crain, P. F., Edmonds, C. G., Gupta, R., Hashizume, T., Phillipson, D. W., et al. (1993). Structure of the archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro-4-oxo-7-beta-D-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (archaeosine)). J. Biol. Chem. 268, 10076–10086.
Hansen, C. J., Wu, L., Fox, J. D., Arezi, B., and Hogrefe, H. H. (2011). Engineered split in Pfu DNA polymerase fingers domain improves incorporation of nucleotide gamma-phosphate derivatives. Nucleic Acids Res. 39, 1801–1810. doi: 10.1093/nar/gkq1053
Helm, M., and Alfonzo, J. D. (2014). Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical legoland. Chem. Biol. 21, 174–185. doi: 10.1016/j.chembiol.2013.10.015
Henry, A. A., and Romesberg, F. E. (2005). The evolution of DNA polymerases with novel activities. Curr. Opin. Biotechnol. 16, 370–377. doi: 10.1016/j.copbio.2005.06.008
Higashimoto, Y., Matsui, T., Nishino, Y., Taira, J., Inoue, H., Takeuchi, M.i., et al. (2013). Blockade by phosphorothitate aptamers of advanced glycation en products-induced damage in cultured pericytes and endothelial cells. Microvasc. Res. 90, 64–70. doi: 10.1016/j.mvr.2013.08.010
Hollenstein, M. (2012a). Nucleoside triphosphates-building blocks for the modification of nucleic acids. Molecules 17, 13569–13591. doi: 10.3390/molecules171113569
Hollenstein, M. (2012b). Synthesis of deoxynucleoside triphosphates that include proline, urea, or sulfonamide groups and their polymerase incorporation into DNA. (2012). Chem. Eur. J. 18, 13320–13330. doi: 10.1002/chem.201201662
Hollenstein, M., Hipolito, C. H. J., Lam, C. H., and Perrin, D M. (2009). A DNAzyme with three protein-like functional groups: enhancing Catalytic Efficiency of M2+-Independent RNA Cleavage. Chembiochem 10, 1988–1601. doi: 10.1002/cbic.200900314
Horiya, S., MacPherson, I. S., Isaac, J., and Krauss, I. J. (2014). Recent strategies targeting HIV glycans in vaccine design. Nat. Chem. Biol. 10, 990–999. doi: 10.1038/nchembio.1685
Huizenga, D. E., and Szostak, J. W. (1995). Aptamer that binds adenosine and ATP. Biochemistry 34, 656–665. doi: 10.1021/bi00002a033
Iannitti, T., Morales-Medina, J. C., and Palmieri, B. (2014). Phosphorothioate oligonucleotides: effectiveness and toxicity. Curr. Drug Targets 15, 663–673. doi: 10.2174/1389450115666140321100304
Iglesias, L. E., Lewkowicz, E. S., Medici, R., Bianchi, P., and Iribarren, A. M. (2015). Biocatalytic approaches applied to the synthesis of nucleoside prodrugs. Biotechnol. Adv. 33, 412–434. doi: 10.1016/j.biotechadv.2015.03.009
Imaizumi, Y., Ksahara, Y., Fujita, H., Kitaduma, S., Ozaki, H., Endoh, T., et al. (2013). Efficacy of base-modification of target binding of small molecules DNA Aptamers. J. Am. Chem. Soc. 135, 9412–9419. doi: 10.1021/ja4012222
Jayasena, S. D. (1999). Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 45, 1628–1650.
Jellinek, D., Green, L. S., Bell, C., Lynott, C. K., Gill, N., Vargeese, C. H., et al. (1995). Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 34, 11363–11372. doi: 10.1021/bi00036a009
Jhaveri, S., Olwin, B., and Ellington, A. D. (1998). In vitro selection of phosphorothilated aptamers. Bioorg. Med. Chem. Lett. 8, 2285–2290. doi: 10.1016/S0960-894X(98)00414-4
Kang, J., Lee, M. S., Copland, J. A. III, Luxon, B. A., and Gorenstein, D. G. (2008). Combinatorial selection of a single stranded DNA thioaptamer targeting TGF-b1 protein. Bioorg. Med. Chem. Lett. 18, 1835–1839. doi: 10.1016/j.bmcl.2008.02.023
Kasahara, Y., Irisawa, Y., Ozaki, H., Obika, S., and Kuwahara, M. (2013). 2',4'-BNA/LNA aptamers: CE-SELEX using a DNA-based library of full-length 2'-O,4'-C-methylene bridged/linked bicyclic ribonucleotides. Bioorg. Med. Chem. Lett. 23, 1288–1292. doi: 10.1016/j.bmcl.2012.12.093
Kato, Y., Minakawa, N., Komatzu, Y., Harashima, H., and Matsuda, A. (2005). New NTP analogs: the synthesis of 4'-thioUTP and 4'-thioCTP and their utility for SELEX. Nucleic Acid Res. 33, 2942–2951. doi: 10.1093/nar/gki578
Keffe, A. D., and Cload, S. T. (2008). SELEX with modified nucleotides. Curr. Op. Chem. Biol. 12, 448–456. doi: 10.1016/j.cbpa.2008.06.028
Kimoto, M., Matsunaga, K., and Hirao, I. (2016). DNA Aptamer generation by genetic alphabet expansion SELEX (ExSELEX) using an unnatural base pair system. Methods Mol. Biol. 1380, 47–60. doi: 10.1007/978-1-4939-3197-2_4
Kimoto, M., Yamashige, R., Matsunaga, K., Yokoyama, S., and Hirao, I. (2013). Generation of high-affinity DNA aptamers using and expanded genetic alphabet. Nat. Biotechnol. 31, 453–457. doi: 10.1038/nbt.2556
Klussmann, S. (Ed.). (2006). The Aptamer Handbook. Functional Oligonucleotides and Their Applications. Weinheim: WILEY-VCH.
Kong, H. Y., and Bym, J. (2013). Nucleic acid aptamers: new methods for selection, stabilization and application in biomedical science. Biomol. Ther. 21, 423–434. doi: 10.4062/biomolther.2013.085
Kore, A. R., and Srinivasan, B. (2013). Recent advances in the syntheses of nucleoside triphosphates. Curr. Org. Synth. 10, 903–934. doi: 10.2174/15701794113109990001
Korhonen, H. J., Bolt, H. L., Vicente-Gines, L., Perks, D. C., and Hodgson, D. R. W. (2015). PPN pyrophosphate: a new reagent for the preparation of nucleoside triphosphates. Phosphorus Sulfur Silicon Relat. Elem. 190, 758–762. doi: 10.1080/10426507.2014.984032
Kuwahara, M., Hanawa, K., Ohsawa, K., Kitagata, R., Ozaki, H., and Sawai, H. (2006). Direct PCR amplification of various modified DNAs having amino acids: convenient preparation of DNA libraries. Bioorg. Med. Chem. 14, 2518–2526. doi: 10.1016/j.bmc.2005.11.030
Kuwahara, M., Obika, S., Nagashima, J., Ohta, Y., Suto, Y., Ozaki, H., et al. (2008). Systematic analysis of enzymatic DNA polymerization using oligo-DNA templates and triphosphate analogs involving 2',4'-bridged nucleosides. Nucleic Acids Res. 36, 4257–4265. doi: 10.1093/nar/gkn404
Kuwahara, M., Takahata, Y., Shoji, A., Ozaki, A. N., Ozaki, H., and Sawai, H. (2003). Substrate properties of C5-substituted pyrimidine 2'-deoxynucleoside 5'-triphosphates for thermostable DNA polymerases during PCR. Bioorg. Med. Chem. Lett. 13, 3735–3738. doi: 10.1016/j.bmcl.2003.08.001
Laos, R., Shaw, R., Leal, N. A., Gaucher, E., and Benner, S. (2013). Directed evolution of polymerases to accept nucleotides with nonstandard hydrogen bond patterns. Biochemistry 52, 5288–5294. doi: 10.1021/bi400558c
Laos, R., Thomson, J. M., and Benner, S. A. (2014). DNApolymerases engineered by directed evolution to incorporate non-standard nucleotides. Front. Microbiol. 5:565. doi: 10.3389/fmicb.2014.00565
Lapa, S. A., Chudinov, A. V., and Timofeev, E. N. (2016). The toolbox for modified aptamers. Mol. Biotechnol. 58, 79–92. doi: 10.1007/s12033-015-9907-9
Latham, J. A., Johnson, R., and Toole, J. J. (1994). The application of a modified nucleotide in aptamer selection: novel thrombin aptamers containing 5-(1-pentynyl)-2-0-deoxyuridine. Nucleic Acids Res. 22, 2817–2822. doi: 10.1093/nar/22.14.2817
Lato, S. M., Ozerova, N. D., He, K., Sergueevfe, Z., Shaw, B. R., and Burke, D. H. (2002). Boron-containing aptamers. Nucleic Acids Res. 30, 1401–1407. doi: 10.1093/nar/30.6.1401
Lauridsen, L. H., Rothnagel, J. A., and Veedu, R. N. (2012). Enzymatic Recognition of 2′-Modified Ribonucleoside5′-Triphosphates: towards the evolution of versatile aptamers. Chembiochem 13, 19–25. doi: 10.1002/cbic.201100648
Leconte, A. M., Chen, L., and Romesberg, F. E. (2005). Polymerase evolution: efforts toward expansion of the genetic code. J. Am. Chem. Soc. 127, 12470–12471. doi: 10.1021/ja053322h
Lee, K., Rafi, M., Wang, X., Aran, K., Feng, X., Lo Sterzo, C., et al. (2015). In vivo delivery of transcription factors with multifunctional oligonucleotides. Nat. Mater. 14, 701–706. doi: 10.1038/nmat4269
Li, M., Lin, N., Huang, Z., Du, L., Huang, Z., Du, L., et al. (2008). Selecting aptamers for a glycoprotein through the incorporation of the boronic acid moiety. J. Am. Chem. Soc. 130, 12636–12638. doi: 10.1021/ja801510d
Li, Y., and Lu, Y. (Eds.). (2009). Functional Nucleic Acids for Analytical Applications. New York, NY: Springer.
Limbach, P. A., Crain, P. F., and McCloskey, J. A. (1994). Summary: the modified nucleosides of RNA. Nucleic Acids Res. 22, 2183–2196. doi: 10.1093/nar/22.12.2183
Lin, Y., Qiu, Q., Gill, S. C., and Jayasena, S. D. (1994). Modified RNA sequence pools for in vitro selection. Nucleic Acids Res. 22, 5229–5234. doi: 10.1093/nar/22.24.5229
Liu, E., Lam, C. H., and Perrin, D. M. (2015). Synthesis and enzymatic incorporation of modified deoxyuridine triphosphates. Molecules 20, 13591–13602, doi: 10.3390/molecules200813591
Liu, M., Jinmei, H., Abe, H., and Ito, Y. (2010). In vitro selection of a photoresponsieve RNA aptamer to hemin. Bioorg. Med. Chem. Lett. 20, 2964–2967. doi: 10.1016/j.bmcl.2010.02.109
Loakes, D., Gallego, J., Pinheiro, V. B., Kool, E. T., and Holliger, P. (2009). Evolving a polymerase for hydrophobic base analogues. J. Am. Chem. Soc. 131, 14827–14837. doi: 10.1021/ja9039696
Ludwig, J. (1981). A new route to nucleoside 5'-triphosphates. Acta Biochim. Biophys. Acad. Sci. Hung. 16, 131–133.
Ludwig, J., and Eckstein, F. (1989). Rapid and efficient synthesis of nucleoside 5'-0-(1-thiotriphosphates), 5'-triphosphates and 2',3'-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 54, 631–635. doi: 10.1021/jo00264a024
Mačková, M., Pohl, R., and Hocek, M. (2014). Polymerase synthesis of DNAs bearing vinyl groups in themajor groove and their cleavage by restriction endonucleases. Chembiochem 15, 2306–2312. doi: 10.1002/cbic.201402319
Masud, M., Kuhahara, M., Ozaki, M., and Sawai, H. (2004). Sialyllactose-binding modified DNA aptamer bearing additional functionality by SELEX. Bioorg. Med. Chem. 12, 1111–1120. doi: 10.1016/j.bmc.2003.12.009
Meyer, A. J., Garry, D. J., Hall, B., Byrom, M. M., McDonald, H. G., Yang, X., et al. (2015). Transcription yield of fully 2'-modified RNA can be increased by the addition of thermostabilizing mutations to T7 RNA polymerase mutants. Nucleic Acids Res. 43, 7480–7488. doi: 10.1093/nar/gkv734
Minikawa, N., Sanji, M., Kato, Y., and Matsuda, A. (2008). Investigations toward the selection of a fully-modified 4'-thio RNA aptamers: optimization of in vitro transcription steps in the presence of 4'-thiNTPs. Bioorg. Med. Chem. 16, 9450–9456. doi: 10.1016/j.bmc.2008.09.048
Ng, E. W. M., Shima, D. T., Callias, P., Cunningham, E. T. Jr., Guyer, D. R., and Adamis, A.P. (2006). Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nature Revs. Drugs Discov. 5, 123–132. doi: 10.1038/nrd1955
Ochsner, U. A., Katilus, E., and Janjic, N. (2013). Detection of Clostridium difficile toxins A, B and binary toxin with slow off-rate modified aptamers. Diag. Micr. Infec. Dis. 76, 278–285. doi: 10.1016/j.diagmicrobio.2013.03.029
Ochsner, U. A., Green, L. S., Gold, L., and Janjic, N. (2014). Systematic selection of modified aptamer paris for diagnostic sandwich assays. Biotechniques 56, 125–133. doi: 10.2144/000114134
Ogawa, M., Tosaka, A., Ito, Y., Yoshida, S., and Suzuki, M. (2001). Enhanced ribonucleotide incorporation by an O-helix mutant of Thermus aquaticus DNA polymerase I. Mutat. Res. 485, 197–207. doi: 10.1016/S0921-8777(00)00081-1
Ong, J. L., Loakes, D., Jaroslawski, S., Too, K., and Holliger, P. (2006). Directed evolution of DNA polymerase, RNA polymerase and reverse transcriptase activity in a single polypeptide. J. Mol. Biol. 361, 537–550. doi: 10.1016/j.jmb.2006.06.050
Ono, T., Scalf, M., and Smith, L. M. (1997). 2'-Fluoro modified nucleic acids: polymerase-directed synthesis, properties and stability to analysis by matrix-assisted laser desorption/ionization mass spectrometry. Nucleic Acids Res. 25, 4581–4588. doi: 10.1093/nar/25.22.4581
Padilla, R., and Sousa, R. (1999). Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2'-groups using a mutant T7 RNA polymerase (RNAP). Nucleic Acids Res. 27, 1561–1563. doi: 10.1093/nar/27.6.1561
Padilla, R., and Sousa, R. (2002). A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 30:e138. doi: 10.1093/nar/gnf138
Pagratis, N. C., Bell, C., Chang, Y.-F., Jennings, S., Fitzwater, T., Jellinek, D., et al. (1997). Potent 2'-amino, and 2'-fluoro-2'-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 2215, 68–73. doi: 10.1038/nbt0197-68
Patel, P. H., Kawate, H., Adman, E., Ashbach, M., and Loeb, L. (2001). A single highly mutable catalytic site amino acid is critical for DNA polymerase fidelity. J. Biol. Chem. 276, 5044–5051. doi: 10.1074/jbc.M008701200
Patel, P. H., and Loeb, L. A. (2000). Multiple amino acid substitutions allow DNA polymerases to synthesize RNA. J. Biol. Chem. 275, 40266–40272. doi: 10.1074/jbc.M005757200
Pavey, J. B., Lawrence, A. J., O'Neil, I. A., Vortler, S., and Cosstick, R. (2004). Synthesis and transcription studies on 5'-triphosphates derived from 2'-C-branched-uridines: 2'-homouridine-5'-triphosphate is a substrate for T7 RNA polymerase. Org. Biomol. Chem. 2, 869–875. doi: 10.1039/B314348A
Peinetti, A. S., Ceretti, H., Mizrahi, M., González, G. A., Ramírez, S. A., Requejo, F., et al. (2015).Confined gold nanoparticles enhance the detection of small molecules in label free impedance aptasensors. Nanoscale 7, 7763–7769. doi: 10.1039/C5NR01429H
Peng, C. G., and Damha, M. J. (2007). Polymerase-directed synthesis of 2'-deoxy-2'-fluoro-beta-D-arabinonucleic acids. J. Am. Chem. Soc. 129, 5310–5311. doi: 10.1021/ja069100g
Perrin, D. M., Garestier, T., and Hélène, C. (1999). Expanding the catalytic repertoire of nucleic acids catalysts: simultaneous incorporation of two modified deoxyribonucleoside triphosphates bearing ammonium and imidazolyl functionalities. Nucleosides Nucleotides 18, 377–391.
Pinheiro, V. B., Taylor, A. I., Cozens, C., Abramov, M., Renders, M., Zhang, S., et al. (2012). Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344. doi: 10.1126/science.1217622
Pontiggia, R., Pontiggia, O., Simian, M., Montserrat, J. M., Engels, J. W., and Iribarren, A. M. (2010). 2'-Methyluridine modified hammerhead ribozyme against the Estrogen Receptor α. Bioorg. Med. Chem. Lett. 20, 2806–2808. doi: 10.1016/j.bmcl.2010.03.060
Raines, K., and Gottlieb, P. A. (1998). Enzymatic incorporation of 2'-thio-CTP into the HDV ribozyme. RNA 4, 340–345.
Ramsay, N., Jemth, A. S., Brown, A., Crampton, N., Dear, P., and Holliger, P. (2010). CyDNA: synthesis and replication of highly Cy-dye substituted DNA by an evolved polymerase. J. Am. Chem. Soc. 132, 5096–5104. doi: 10.1021/ja909180c
Richardson, F. C., Kuchta, R. D., Mazurkiewicz, A., and Richardson, K. A. (2000). Polymerization of 2'-fluoro- and 2'-O-methyl-dNTPs by human DNA polymerase alpha, polymerase gamma, and primase. Biochem. Pharmacol. 59, 1045–1052. doi: 10.1016/S0006-2952(99)00414-1
Robaldo, L., Berzal-Herranz, A., Montserrat, J. M., and Iribarren, A. M. (2014). Activity of core modified 10-23 DNAzymes against HCV. Chem. Med. Chem. 9, 2271–2277. doi: 10.1002/cmdc.201402222
Robertson, D. L., and Joyce, G. F. (1990). Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344, 467–468. doi: 10.1038/344467a0
Rodríguez, L., Villalobos, X., Solé, A., Lliberós, C., Ciudad, C. J., and Noé, V. (2015). Improved design of PPRHs for gene silencing. Mol. Pharm. 12, 867–877. doi: 10.1021/mp5007008
Rohloff, J. C., Fowler, C., Ream, B., Carter, J. D., Wardle, G., and Fitzwater, T. (2015). Practical synthesis of cytidine-5-carboxamide-modified nucleotide reagents. Nucleosides Nucleotides Nucleic Acids 34, 180–198. doi: 10.1080/15257770.2014.978011
Rohloff, J. C., Gelinas, A. D., Jarvis, T. C., Ochsner, U. A., Schneider, D. J., Gold, L., et al. (2014). Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids 3, e201–e203. doi: 10.1038/mtna.2014.49
Ruckman, J., Green, L. S., Beeson, J., Waugh, S., Guillette, W. L., Henninger, D. D., et al. (1998). 2'-Fluoropyrimidine RNA-based Aptamers to the 165-amino acid from of vascular endothelial growth factor (VEFG165). J. Biol. Chem. 273, 20556–20567. doi: 10.1074/jbc.273.32.20556
Santoro, S. W., Joyce, G. F., Sakthivel, K., Gramatikova, S., and Barbas, C. F. III. (2000). RNA Cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 122, 2433–2439. doi: 10.1021/ja993688s
Schmid-Kubista, K. E., Krebs, I., Ansari-Shahrezaei, S., Haas, P., Hagen, S., and Binder, S. (2011). Comparing treatment of neovascular age-related macular degeneration with sequential intravitreal avastin and macugen versus intravitreal mono-therapy - A pilot study. Curr. Eye Res. 36, 958–963. doi: 10.3109/02713683.2011.597536
Schultz, H. J., Gochi, A. M., Chia, H. E., Ogonowsky, A. L., Chiang, S., Filipovic, N., et al. (2015). Taq DNA Polymerase mutants and 2′-modified sugar recognition. Biochemistry 54, 5999–6008. doi: 10.1021/acs.biochem.5b00689
Sczepanski, J. T., and Joyce, G. F. (2013). Binding of a Structured D-RNA Molecule by and L-RNA Aptamer. J. Am. Chem. Soc. 135, 13290–13293. doi: 10.1021/ja406634g
Shinkai, A., Patel, P. H., and Loeb, L. H. (2001). The conserved active site motif A of Escherichia coli DNA polymerase I is highly mutable. J. Biol. Chem. 276, 18836–18842. doi: 10.1074/jbc.M011472200
Shoji, A., Kuwahara, M., Ozaki, H., and Sawai, H. (2007). Modified DNA Aptamer that Binds the (R)-isomer of a thalidomide derivatie with high enantioselectivity. J. Am. Chem. Soc. 129, 1456–1464. doi: 10.1021/ja067098n
Sidorov, A. V., Grasby, J. A., and Williams, D. M. (2004). Sequence-specific cleavage of RNA in the absence of divalent metal ions by a DNAzyme incorporating imidazolyl and amino functionalities. Nucleic Acids Res. 32, 1591–1601. doi: 10.1093/nar/gkh326
Somasunderam, A., Thiviyanathan, V., Tanaka, T., Li, X., Neerathilingam, M., Lokesh, G. L. R., et al. (2010). Combinatorial Selection of DNA Thioaptamers Targeted to HA Binding Domain of CD44. Biochemistry 49, 9106–9112. doi: 10.1021/bi1009503
Staffan, E. (2005). “Enzymatic synthesis of nucleoside triphosphates,” in Nucleoside Triphosphates and Their Analogs, ed M. Vaghefi (Boca Raton, FL: CRC Press), 23–37.
Sun, H., and Zu, Y. (2015). A highlight of recent advances in aptamer technology and its application. Molecules 20, 11959–11980. doi: 10.3390/molecules200711959
Suzuki, M., Baskin, D., Hood, L., and Loeb, L. (1996). Random mutagenesis of Thermus aquaticus DNA polymerase I: concordance of immutable sites in vivo with the crystal structure. Proc. Natl. Acad. Sci. U.S.A. 93, 9670–9675. doi: 10.1073/pnas.93.18.9670
Svobodova, M., Bunka, D. H. H., Nadal, P., Stockley, P. G., and O'sullivan, C. K. (2013). Selection of 2'F-modified RNA aptamers against prostate-specific antigen and their evaluation for diagnostic and therapeutic applications. Anal. Bioanal. Chem. 405, 9149–9157. doi: 10.1007/s00216-013-7350-y
Tarasow, T. M., Tarasow, S. L., and Eaton, B. E. (1997). RNA-catalysed carbon-carbon bond formation. Nature 389, 54–57. doi: 10.1038/37950
Taylor, A. I., and Holliger, P. (2015). Direct evolution of artificial enzymes (XNAzymes) from diverse repertoires of synthetic genetic polymers. Nat. Protoc. 10, 1625–1642. doi: 10.1038/nprot.2015.104
Taylor, A. I., Pinheiro, V. B., Smola, M. J., Morgunov, A. S., Peak-Chew, S., Cozens, C., et al. (2015) Catalysts from synthetic genetic polymers. Nature 518, 427–430. doi: 10.1038/nature13982
Tei, S., Ishii, H. T., Mitsuhashi, H., and Ishiura, S. (2015). Antisense oligonucleotide-mediated exon skipping of CHRNA1 pre-mRNA as potential therapy for congenital myasthenic syndromes. Biochem. Biophys. Res. Commun. 461, 481–486. doi: 10.1016/j.bbrc.2015.04.035
Toole, F., Brändle, G. M., Matzner, D., and Mayer, G. A. (2015). Versatile approach towards nucleobase-modified aptamers. Angew. Chem. Int. Ed Engl. 54, 10971–10974. doi: 10.1002/anie.201503652
Tuerk, C., and Gold, L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510. doi: 10.1126/science.2200121
Vaish, N. K., Larralde, R., Fraley, A. W., Szostack, J. W., and McLaughlin, L. W. (2003). A novel, modification-dependent ATP binding aptamer selected from an RNA library incorporating a cationic functionality. Biochemistry 42, 8842–8851. doi: 10.1021/bi027354i
Vaught, J. D., Bock, C. H., Carter, J., Fitzwater, T., Otis, M., Schneider, D., et al. (2010). Expanding the Chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 132, 4141–4151. doi: 10.1021/ja908035g
Veedu, R. N., Vester, B., and Wengel, J. (2007). Enzymatic incorporation of LNA nucleotides into DNA strands. Chembiochem 8, 490–492. doi: 10.1002/cbic.200600501
Veedu, R. N., Vester, B., and Wengel, J. (2008). Polymerase chain reaction and transcription using locked nucleic acid nucleotide triphosphates. J. Am. Chem. Soc. 130, 8124–8125. doi: 10.1021/ja801389n
Walsh, J. M., and Beuning, P. J. (2012). Synthetic Nucleotides as Probes of DNA Polymerase Specificity. J. Nucl. Acids 2012:530963. doi: 10.1155/2012/530963
Wandtke, T., Wozniak, J., and Kopinski, P. (2015). Aptamers in diagnostics and treatments of viral infections. Viruses 7, 751–780. doi: 10.3390/v7020751
Wiegand, T. W., Janssen, R. C., and Eaton, B. (1997). Selection of RNA amide synthases. Chem. Biol. 4, 675–683. doi: 10.1016/S1074-5521(97)90223-4
Wyss, L. A., Nilforoushan, A., Eichenseher, F., Suter, U., Blatter, N., Marx, A., et al. (2015). Specific incorporation of an artificial nucleotide opposite a mutagenic DNA Adduct by a DNA Polymerase. J. Am. Chem. Soc. 137, 30–33. doi: 10.1021/ja5100542
Xia, G., Chen, L., Sera, T., Fa, M., Schultz, P. G., and Romesberg, F. E. (2002). Directed evolution of novel polymerase activities: mutation of a DNA polymerase into an efficient RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 99, 6597–6602. doi: 10.1073/pnas.102577799
Yatime, L., Maasch, C., Hoehling, K., Anderson, G. R., and Vater, A. (2015). Structural basis for the targeting of complement anaphylatoxin C5a using a mix L-RNA/L-DNA aptamer. Nat. Commun. 6:6481. doi: 10.1038/ncomms7481
Yoshikawa, M., Kato, T., and Takenishi, T. (1967). A Novel method for phosphorylation of nucleosides to 5'-nucleotides. Tet. Lett. 50, 5065–5068. doi: 10.1016/S0040-4039(01)89915-9
Yüce, M., Ullah, N., and Budak, H. (2015). Trends in aptamer selection methods and applications. Analyst 140, 5379–5399. doi: 10.1039/C5AN00954E
Yu, H., Zhang, S., and Chaput, J. (2012). Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 4, 183–187. doi: 10.1038/nchem.1241
Keywords: SELEX, modified nucleotides, functional oligonucleotides, aptamers, DNAzymes, ribozymes
Citation: Dellafiore MA, Montserrat JM and Iribarren AM (2016) Modified Nucleoside Triphosphates for In-vitro Selection Techniques. Front. Chem. 4:18. doi: 10.3389/fchem.2016.00018
Received: 16 February 2016; Accepted: 05 April 2016;
Published: 04 May 2016.
Edited by:
J. Carlos Penedo, University of St. Andrews, UKCopyright © 2016 Dellafiore, Montserrat and Iribarren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adolfo M. Iribarren, airibarren@unq.edu.ar
 María A. Dellafiore
María A. Dellafiore Javier M. Montserrat1,2
Javier M. Montserrat1,2  Adolfo M. Iribarren
Adolfo M. Iribarren