A Potential Combination Therapy of Berberine Hydrochloride With Antibiotics Against Multidrug-Resistant Acinetobacter baumannii
- 1School of Chemical Engineering and Technology, Key Laboratory of Systems Bioengineering (Ministry of Education), Frontiers Science Center for Synthetic Biology, Tianjin University, Tianjin, China
- 2Tianjin Modern Innovative TCM Technology Co. Ltd., Tianjin, China
- 3School of Engineering, University of Glasgow, Glasgow, United Kingdom
Multidrug-resistant (MDR) Acinetobacter baumannii strains can cause severe infections in intensive care units, and are rapidly developing resistance to the last-resort of existing antibiotics, posing a major global threat to health care system. Berberine hydrochloride (BBH), a kind of isoquinoline alkaloids extracted from Berberis and other plants, has been widely used as an antibacterial medicine for its reliable therapeutic efficiency. The in vitro synergistic effects of BBH with antibiotics against MDR A. baumannii were determined. BBH alone had weak antimicrobial activity (e.g., MIC≥256 mg/L) against MDR A. baumannii. However, it dramatically increased the susceptibility of MDR strains against antibiotics with FICI values <0.5, even reversed their resistance to antibiotics (e.g., tigecycline, sulbactam, meropenem and ciprofloxacin). In vivo study has suggested BBH with sulbactam had stronger antimicrobial efficiency than monotherapy in a neutropenic murine thigh infection model. The antibiotic-sensitizing mechanism of action of BBH was evaluated as well. BBH boosted adeB gene expression and bound to the AdeB transporter protein, resulting in low uptake of BBH, which may contribute to less extrusion of antibiotics by the AdeABC pump. Knockout of the adeB gene increased uptake of BBH and diminished the antibiotic sensitization and synergistic effects between antibiotics and BBH in MDR strains. Together, BBH effectively re-sensitizes this MDR pathogen to a range of antibiotics that have become barely effective due to antibiotic resistance, which indicates BBH may be a promising therapeutic adjuvant candidate to combat MDR A. baumannii.
Introduction
Acinetobacter baumannii is a Gram-negative coccobacillus and considered the most virulent among all Acinetobacter species (Wong et al., 2017). As an opportunistic nosocomial pathogen, it can cause various community-acquired and hospital-acquired infections including pneumonia, urinary tract, lower gastrointestinal tract and surgical site infections, especially in intensive care units (Lee et al., 2017; Watkins, 2018). It is estimated that there is a high mortality rate when A. baumannii is present in community-acquired pneumonia cases. (Antunes et al., 2014). Due to its intrinsic and acquired antibiotic resistance (e.g. by mutations and gene transfer) along with improper use of antibiotics (Yoon et al., 2013), more and more multidrug-resistant (MDR) A. baumannii strains have been discovered. MDR A. baumannii has been ranked first on the critical priority list of MDR pathogens by the World Health Organization to guide research and development of antibiotics due to its greatest threat to human health (WHO, 2017).
Among the resistance mechanisms, multidrug efflux pumps play an important role. Three efflux systems AdeABC, AdeFGH and AdeIJK, which belong to the resistance-nodulation-cell division (RND) family, are associated with drug-resistance of MDR A. baumannii (Yoon et al., 2013; Nowak et al., 2016; Yoon et al., 2016). It has been reported the AdeABC pump contributes to resistance by extruding aminoglycosides, some β-lactams, fluoroquinolones, tetracyclines, tigecycline, macrolides, chloramphenicol, and trimethoprim etc. (Coyne et al., 2011; Chen et al., 2014). These MDR strains display resistance to almost all of today’s commonly prescribed antibiotics, leaving limited treatment options, which is becoming a major global issue and threatening the whole healthcare systems worldwide. For a long time, tigecycline and colistin have been considered last-resort drugs to treat this pathogen. However, they are now challenged by strong side effects and increasingly emerging resistance (Cai et al., 2012; He et al., 2019). Therefore, there is an urgent need to discover new antibacterial components targeting MDR A. baumannii. Considering the fact that discovery of antibiotic may take many years, researchers have now set their sights on natural sources (Gao et al., 2018).
Berberine, a quaternary ammonium salt from the protoberberine group of isoquinoline alkaloids, can be found in many plants such as Coptis chinensis (a traditional Chinese herb, Huanglian), Hydrastis canadensis, Berberis aristata, Phellodendron amurense, etc. (Imenshahidi and Hosseinzadeh, 2016). Berberine hydrochloride (BBH) is the most common form of berberine. Their structures are shown in Figure 1. Both have been clinically used as an antibacterial medicine against gastroenteritis, abdominal pain and diarrhea in China for more than 2000 years (Tan et al., 2011; Gao et al., 2018). Relevant research has shown that berberine and its derivatives are efficient antibacterial agents against methicillin-resistant Staphylococcus aureus (MRSA) (Sun et al., 2019), Acinetobacter baumannii (Gao et al., 2018) and Pseudomonas aeruginosa (Morita et al., 2016). However, few studies have reported using BBH alone or in combination with antibiotics to treat MDR A. baumannii.
In this work, we evaluated the antimicrobial activity of BBH alone and in combination with several existing antibiotics against MDR A. baumannii in vitro and in vivo. Synergistic effects and antibiotic sensitization have been observed, and the mechanisms that BBH reserves resistance of MDR A. baumannii to the antibiotics have been investigated.
Materials and Methods
Bacterial Strains
Four strains of MDR A. baumannii (MDR-TJ, MDR-A, MDR-B and MDR-C) were isolated from the Second Hospital of Tianjin Medical University, China. Their drug-resistance profiles are listed in Table S1. In our previous study, we completed the whole genome sequence of MDR-TJ (GenBank accession number: NC_017847) and illustrated its mechanism of antibiotic resistance (Huang et al., 2012). The other two reference strains A. baumannii ATCC19606 and Escherichia coli ATCC 25922 were purchased from the American Type Culture Collection. All strains were stored in LB broth with 25% glycerol at -80°C.
Antimicrobial Agents
Antimicrobial agents include berberine hydrochloride (BBH), amikacin, ciprofloxacin (Solarbio, Beijing, China), sulbactam (Aladdin, Shanghai, China), tigecycline (BioVision, San Francisco, USA), meropenem, and tetracycline (National Institutes for Food and Drug Control, Beijing, China). They were dissolved in sterile distilled water and stored at -20°C before the test. Unless otherwise noted, all antimicrobial powders have a purity greater than 98%. Efflux pump inhibitors carbonyl cyanide m-chlorophenylhydrazone (CCCP) and reserpine were purchased from Solarbio (Beijing, China), and phenylalanine-arginine β-naphthylamide (PAβN) from MedChemExpress (Shanghai, China).
Determination of MICs
A two-fold broth microdilution method was used to determine the MICs of antimicrobials, following the guidance of the CLSI (M100) (Clinical and Laboratory Standards Institute, 2018). The MIC was defined as the lowest concentration of an antimicrobial agent that inhibits more than 90% bacterial growth from three independent tests, or as the lowest concentration with no visible bacterial growth (Table S2). A positive growth control, which contained bacterial suspensions in cation-adjusted Mueller-Hinton broth (CAMHB) without antimicrobials, and a negative control with broth only were included in a 96-well plate with a final volume of 200 μL. The plate was incubated for 20-24 h at 35°C in ambient air. E. coli ATCC 25922 was used as a quality control strain. The inhibition rate was calculated using the equation (1):
Up to now, there are no CLSI susceptibility breakpoints for tigecycline and sulbactam against A. baumannii. Therefore, we used the CLSI criterion of ampicillin/sulbactam combination for sulbactam breakpoint and the FDA criterion for tigecycline breakpoint (i.e. MIC value ≤ 2 mg/L, susceptible; 4-8 mg/L, intermediate; ≥ 8 mg/L, resistant) (Navon-Venezia et al., 2007).
Antimicrobial Combination Test by Checkerboard Assay
To determine the combinatory antimicrobial activities of antibiotics with BBH, the checkerboard assay was performed as previously described (Duarte et al., 2012). Briefly, a series of twofold dilutions of BBH and antibiotics were added in vertical and horizontal lines, respectively. The bacterial suspensions were added with a final concentration of about 5×105 CFU/mL. The plate was incubated for 20-24 h at 35°C in ambient air. Interactions between antibiotics and BBH were determined according to the fractional inhibitory concentration index (FICI), which is interpreted as follows: FICI ≤ 0.5, synergy; 0.5≤FICI ≤ 4, no interaction; FICI>4, antagonism (Odds, 2003).
Murine Thigh Infection Model
All animal procedures were performed at the Institute of Radiation Medicine Chinese and approved by the Animal Ethics and Welfare Committee of the Chinese Academy of Medical Science and Peking Union Medical College (Approval No. IRM-DWLL-2020106). The neutropenic mouse thigh infection model was utilized as previously described (Macvane et al., 2014; Barnes et al., 2019). Female ICR mice weighing 20 to 22 g were rendered transiently neutropenic via intraperitoneal injections of 150 and 100 mg/kg cyclophosphamide (National Institutes for Food and Drug Control, Beijing, China) at day 4 and day 1 prior to bacterial inoculation, respectively. Afterwards, 0.1 mL (~107 CFU/mL) of bacterial suspension (MDR-TJ strain) was injected to both thighs of the mice under isoflurane anesthesia. After 2 h following infection, mice were randomly divided into 5 groups (n=5 mice/10 thighs), and the baseline group was euthanized to obtain the initial bacterial burden. Then Berberine hydrochloride (BBH) was administrated at 20 mg/kg q12h (i.e., drugs were injected every 12 hours; 2 times total in a 24-h treatment period), sulbactam (SUL) at 400mg/kg q12h and combinations of BBH and SUL together as treatment regimens of monotherapy and combination therapy, respectively. All therapies were administered over a 24-h period by intraperitoneal injection of 0.2 mL doses each time, and total injection volume is 0.4 mL (0.2 mL BBH + 0.2 mL SUL) for combination therapy. Growth control animals were administered saline at the same volume and time interval. Subsequently, mice were euthanized, after which both thighs were excised aseptically, homogenized in 5 ml ice cold PBS (pH=7.2), and then plated on MHA plates by serial dilutions to determine bacterial burden expressed as Log10 CFU/thigh.
Detection of the adeABC System and Relative Expression of the adeB Gene
Genomic DNA from A. baumanii strains was extracted using the Bacteria Genomic DNA Extraction Kit (Solarbio, Beijing, China). Genes adeB, adeR, abeS, and 16S rRNA were detected by PCR. Expression of adeB was quantified by RT-PCR. In brief, A. baumannii isolates were grown in LB broth to reach mid-log phage with or without BBH, and then total RNA was extracted using the Trizol agent (Solarbio, Beijing, China). RT-PCR assays were performed using the SsoFast EvaGreen Supermix agent (Bio-Rad, USA) in the CFX96 Real-Time System (Bio-Rad, USA). The relative expression of the adeB gene was normalized against the housekeeping gene 16S rRNA and calculated using the 2−ΔΔCt method. A. baumannii ATCC 19606 was used as the reference strain, and RNase-free water was used as a negative control. All primers used in this study are listed in Table S3.
Molecular Docking Study
To simulate how the antimicrobials bind to the AdeB transporter protein (AdeB) in vitro, theoretical molecular docking studies were performed. The crystal structure of AdeB (PDB code: 6OWS) (Su et al., 2019) was obtained from the RCSB Protein Data Bank, and the 3D structures of the antimicrobials were taken from the ZINC or PubChem database. AutoDock 4.2 was used to predict their bindings (Bikadi and Hazai, 2009). The results of docking computations were ranked by the binding energy, where the one with the highest energy was recorded. Finally, the protein-ligand docking models were visualized by PyMOL (http://www.pymol.org).
adeB Gene Knockout
A marker-less gene deletion method was applied to delete the adeB gene (Amin et al., 2013). Briefly, a cross-over step was performed to select the MDR A. baumannii harboring the insertion plasmid Mo130-TelR-adeABC (with a 1.9kb Up/Down fragment of adeB). It was followed by a second cross-over step to replace the adeB gene. Knockout was confirmed by the absence (i.e. undetectable) of the adeB gene amplification products as well as the presence of a 1.9kb Up/Down fragment amplification products by PCR using specific primers (Table S3).
BBH Accumulation
BBH accumulation was measured as described previously with moderate changes (Wang et al., 2009). Briefly, bacteria were cultured into the exponential growth phase. They were centrifuged (8000×g, 2min), washed twice with 0.01M PBS (pH 7.2) and re-suspended in 1.5 mL PBS to OD600nm=1.0. BBH was added with a final concentration of 80 mg/L. 50 μM of an efflux pump inhibitor (i.e. CCCP, PAβN and reserpine) was added at an appropriate time (t = 60 min). Assays were performed in a 96-well black plate with a final volume of 100 μL per well. Fluorescence was measured every 5 min for 90 min in a microplate fluorometer (Thermo Scientific, USA) with an excitation wavelength of 355 nm and an emission wavelength of 538 nm.
Statistical Analysis
All results were shown as mean ± standard deviation from three independent experiments with at least triplicate in each experiment. Unless otherwise noted, statistical analysis was carried out using one-way analysis of variance (ANOVA) by GraphPad Prism 8 (San Diego, CA, USA). P values < 0.05 indicate statistically significant difference.
Results
Antibiotic Effects of BBH Alone and in Combination With Others
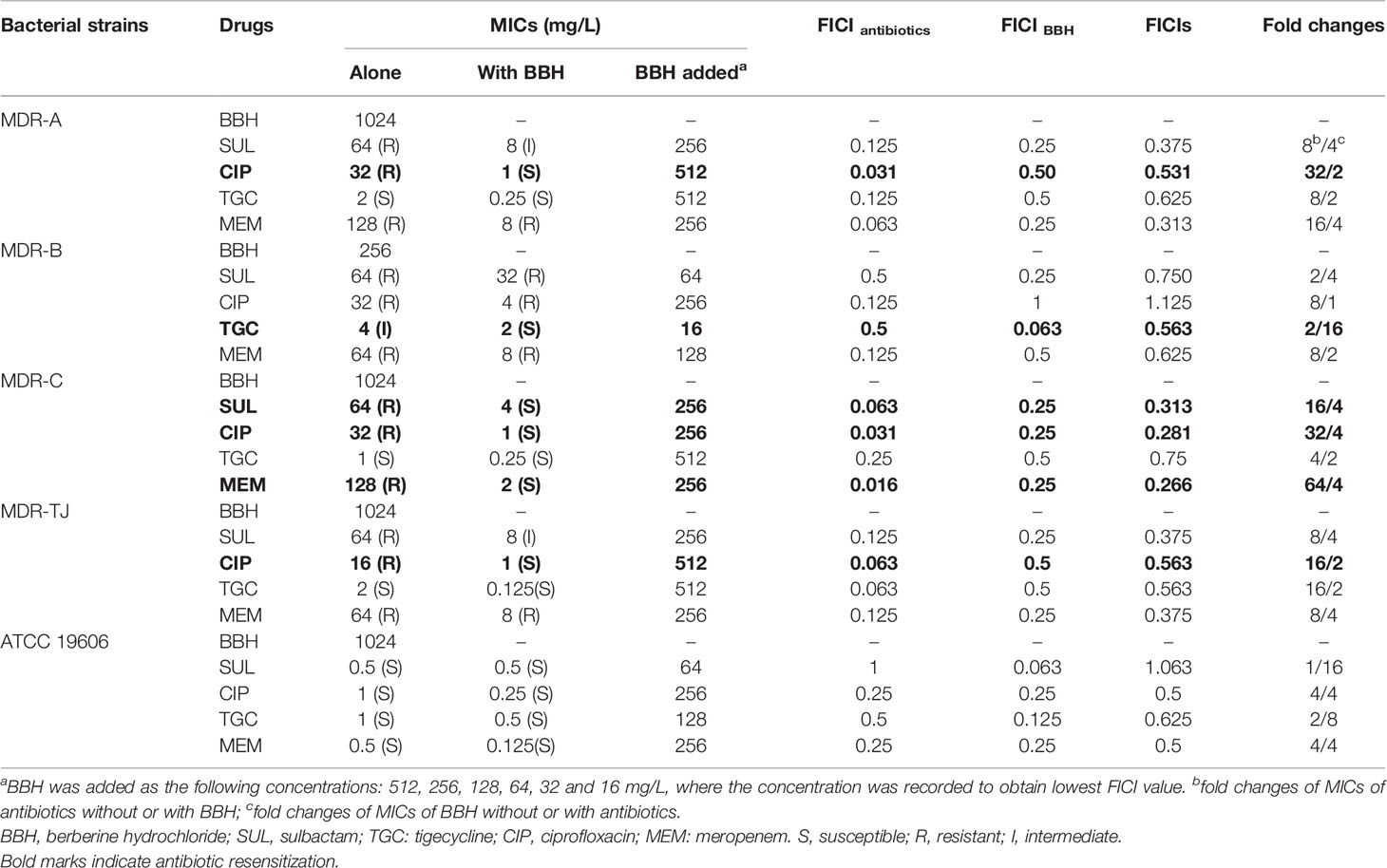
We firstly examined the antimicrobial potential of BBH alone and its combined effects with antibiotics against A. baumannii (Table 1). BBH, as a single antimicrobial agent, showed a slight antibacterial activity, with a higher inhibitory effect on MDR-B (i.e. MIC at 256mg/L) than on the other three MDR stains and type stains (i.e. both MICs at 1024mg/L). The MDR isolates showed higher resistance to sulbactam, ciprofloxacin and meropenem compared with the type strain ATCC 19606. The variation in the sensitivities against different antibiotics of the three MDR strains may be due to different resistant genes harbored as they have different antibiotic susceptibility profiles (See Table S1).
As shown in Table 1, BBH in combination with antibiotics showed a more significant antimicrobial potential. Synergistic and antagonistic interactions between BBH and the tested antibiotics were indicated by FICIs. Several combinations showed 0.5<FICIs<1, suggesting a weak synergistic interaction between the BBH and antibiotics. Synergistic effects (FICI<0.5) were observed in the combinations of BBH/sulbactam and BBH/meropenem for the MDR strains. Furthermore, the addition of BBH reduced the MIC values of the majority of tested antibiotic-pathogen pairs and made the MDR strains re-sensitive to several antibiotics (Indicated by bold marks in Table 1). For example, the MIC of ciprofloxacin against MDR-A decreased from 32 to 1 mg/L, and the MIC of sulbactam and meropenem against MDR-C decreased from 64 to 4 mg/L and from 128 to 2 mg/L, respectively. This phenomenon demonstrates that BBH has the potential to reverse antibiotic resistance or enhance the susceptibility of A. baumannii to those no-longer-effective antibiotics. Therefore, BBH could be an effective adjuvant to bring those off-the-treatment-list antibiotics (e.g. sulbactam and ciprofloxacin) back to use for treating MDR A. baumannii infections.
Murine Thigh Infection Model of MDR A. baumannii
The MDR A. baumannii strain MDR-TJ was used to establish a murine thigh infection model for its board spectrum of drug-resistance profiles (See Table S1). This strain demonstrates resistant to sulbactam in vitro with an MIC of 64 mg/L, however, susceptibility is changed in the presence of BBH with a MIC of 8 mg/L (Table 1). The toxicity of BBH (pH=7.2) was evaluated prior to in vivo combination therapy tests, and the dose of 20 mg/kg administrated intraperitoneally was safe with in a 24-h period (See Figure S1). Monotherapy of SUL and BBH individually every 12 h with doses of 400 mg/kg and 20 mg/kg couldn’t inhibit bacteria growth, resulting increment of 1.56 and 2.11 Log10 CFU/thigh in 24h treatment compared with baseline value (Figure 2). Whereas, BBH can effectively boost the efficiency of SUL in vivo against MDR-TJ strain, making in a 2.53 Log10 CFU/thigh reduction, even shows a bactericidal action (-0.86 Log10 CFU/thigh reduction compared with baseline value).
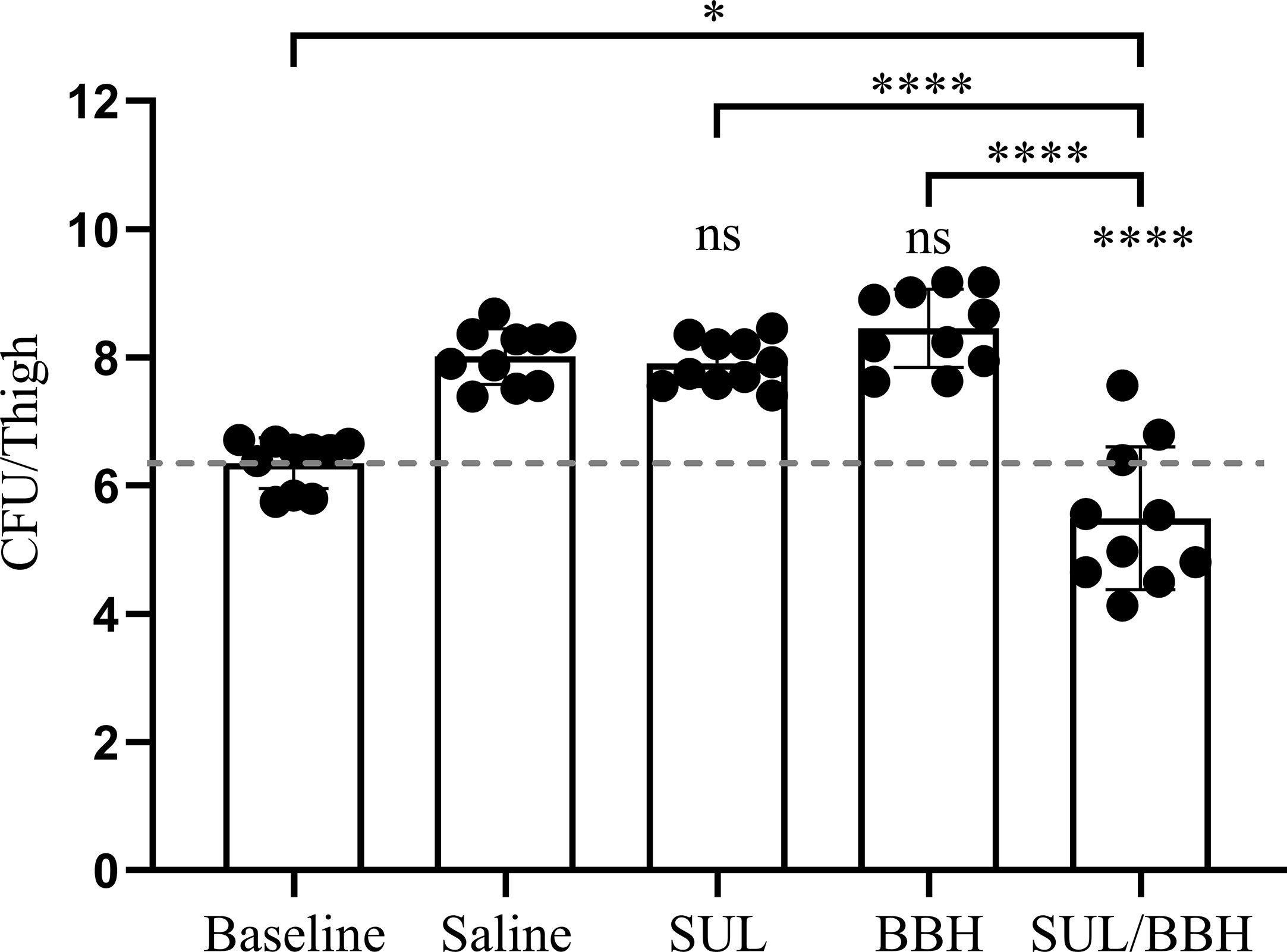
Figure 2 Efficiency of SUL/BBH combination therapy against MDR-TJ in murine thigh infection model (n = 5 mice, 10 thighs). The data were obtained from one single test and statistical analysis was carried out using one-way analysis of variance (ANOVA) with Tukey’s test by GraphPad Prism 8. ns, no significance; *P < 0.05; ****P < 0.0001.
Detection of the adeABC System and adeB Gene Expression
Given that BBH reduced more than 4-fold MICs of antibiotics in most combinations, it presents a similar effect of efflux pump inhibitors (Coyne et al., 2011). Therefore, we evaluated whether A. baumannii has the most prevalent efflux pump AdeABC system (multidrug transporter protein gene adeB and its two-component regulatory system genes adeR and adeS) (Coyne et al., 2011; Yoon et al., 2015). In all three strains, we detected the adeB and adeR-adeS genes by PCR (Figure S2) and the sequences of amplification products were highly identical with the target genes (≥95%). This proves that all strains in this study have the adeABC efflux pump system, including the sensitive strain ATCC19606.
The relative expressions of the adeB gene in the three strains with or without BBH are shown in Figure 3. Without BBH, there was no difference in adeB expression between the MDR strains and the ATCC 19606 strain. Following the addition of BBH, the expression of the adeB gene increased by 25.3, 4.3, 10.2 folds for ATCC 19606, MDR-B and MDR-TJ, respectively. The expression of multidrug pumps is typically subjected to induction by their substrates (Coyne et al., 2011). Thus, this phenomenon indicates that BBH is more likely to be recognized by the bacteria as a substrate for the AdeABC system rather than an inhibitor.
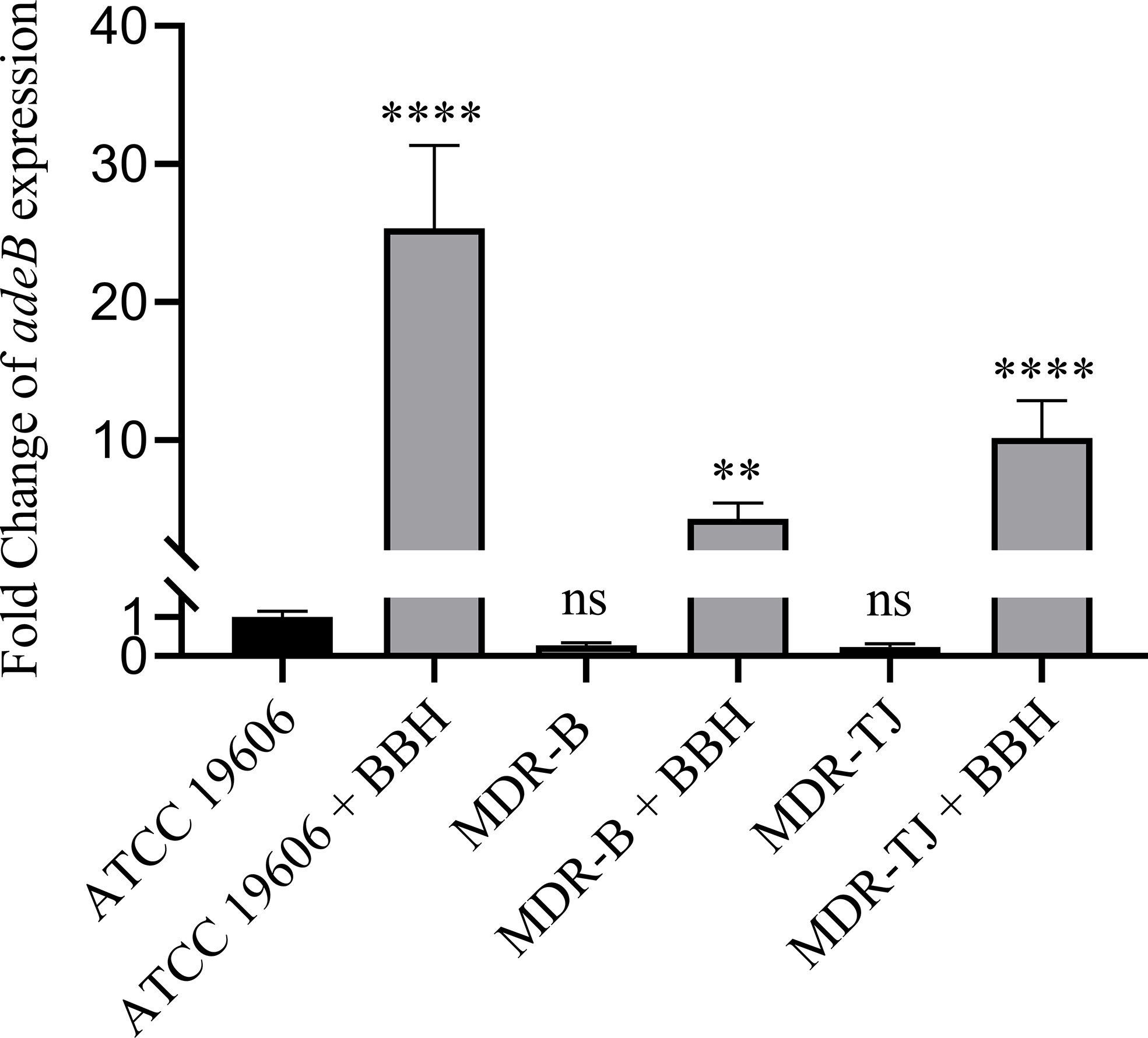
Figure 3 Quantification of adeB expression normalized with A. baumannii ATCC 19606. BBH was added at half MIC corresponding to each stain, which is 128 mg/L for MDR-B and 512 mg/L for MDR-TJ and ATCC 19606, respectively. Statistical analysis was carried out using one-way analysis of variance (ANOVA) with Tukey’s test by GraphPad Prism 8. ns, no significance; **P < 0.01; ****P < 0.0001.
Molecular Docking Study
To examine whether BBH could be a competitive substrate of AdeB protein, protein-ligand docking models were built (Figure 4), and their binding energies were computed (Table 2). Berberine was used as a substitute for BBH due to their identical 3D structure when dissolved. Six antibiotics (i.e. tigecycline, ciprofloxacin, sulbactam, tetracycline, meropenem, and amikacin) were also simulated, where tigecycline, ciprofloxacin, tetracycline and amikacin were proved as substrates of AdeB protein (Coyne et al., 2011).
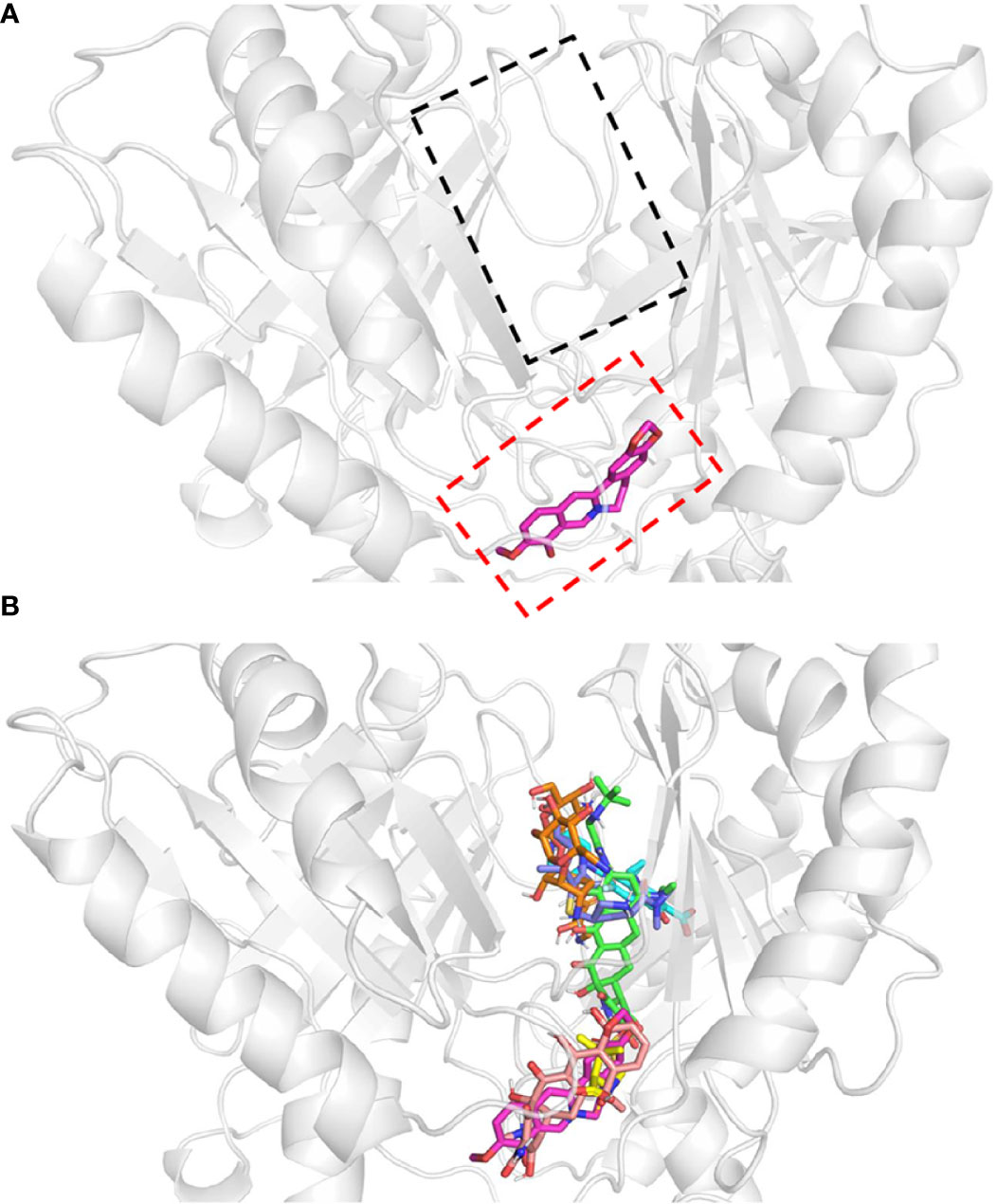
Figure 4 Protein-ligand docking models. Docking structure of (A) berberine, and (B) antibiotics. All antibiotics are close to the proximal multidrug binding site (F-loop, red frame; G-loop, black frame) of AdeB. Berberine, pink; tigecycline, green; ciprofloxacin, cyan; sulbactam, yellow; tetracycline, light salmon; meropenem, dark blue; amikacin, orange.
The multidrug transporter protein AdeB has three multidrug binding sites: the periplasmic cleft entrance, proximal multidrug binding site (F-loop and G-loop) and hydrophobic patch (Su et al., 2019). The F-loop and G-loop contain many multidrug binding sites (e.g. Trp-610, Phe-612, Ser-613, Ala-615, Pro-661, Ala-662, Ile-663, Asp-664, Glu-665, Leu-666, Thr-668), which can be found in other multidrug pumps, such as Cus A, AcrB, and MtrD pumps (Su et al., 2019). Therefore, these sites were selected as the docking sites. The docking model showed that all of these drugs were located in the proximal multidrug binding sites of AdeB (Figure 3). Binding energy calculations showed that berberine had higher binding energy (c.a. -7.42 kcal/mol) to AdeB than all the other antibiotics except tetracycline, which indicates that BBH is a positive substrate of the multidrug transporter protein AdeB (Table 2).
adeB Gene Deletion and MIC Reduction
The deletion of the adeB gene was confirmed by comparing PCR amplification products between the parental strains (MDR-B and MDR-TJ) and the mutant strains (△adeB MDR-B and △adeB MDR-TJ) (Figure S3). The parental strains generated the adeB gene amplification products and about 5.2kb Up/Down-fragment amplification products, whereas the mutants lacked the adeB gene products and generated about 1.9kb Up/Down-fragment products. These results showed that adeB knockout was successful in △adeB MDR-B and △adeB MDR-TJ mutants. Furthermore, sequencing results were identical to the target sequences.
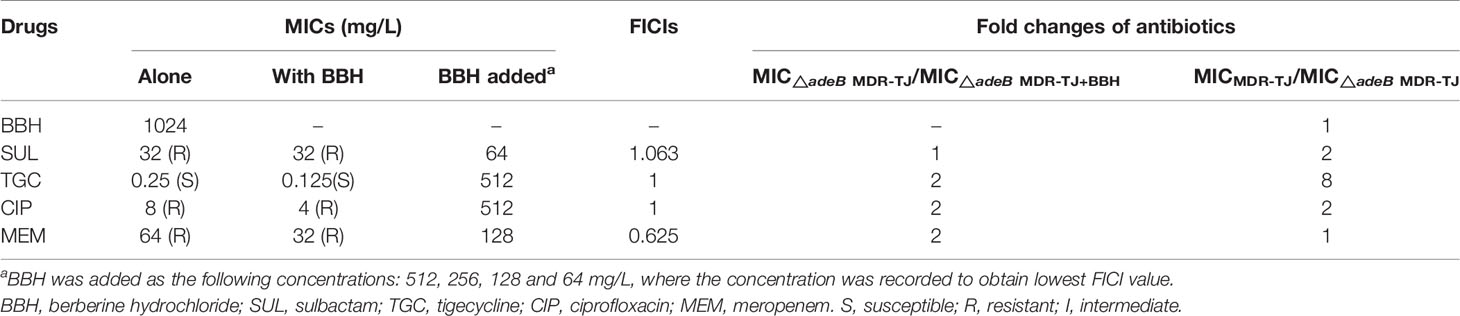
To verify the mechanism that BBH is a competitive substrate of the AdeB protein, the △adeB MDR-TJ was used to determine the MICs of the selected antibiotics with and without BBH (Table 3). The assumption is that synergistic effects between BBH and antibiotics would diminish or antibiotic sensitization would disappear in adeB deletion strains. As is shown in Table 3, compared to the parental MDR-TJ strain, △adeB MDR-TJ showed reduced MICs of sulbactam, tigecycline and ciprofloxacin, indicating that the lack of expelling these antibiotics by the AdeB protein can enhance the killing effect of the antibiotics. It should be noted that there was no difference in the MIC of BBH alone between the parental MDR-TJ strain and the △adeB MDR-TJ strain. This indicates that other efflux systems might also play an important role in the extrusion of BBH. For the △adeB MDR-TJ strain, the addition of BBH only induced a 2-fold decrease of the MICs for all antibiotics but sulbactam (no change with BBH) (Table 3). In contrast, the addition of BBH induced more than an 8-fold decrease of the MICs for the corresponding antibiotics in the parental MDR-TJ strain (Table 1). Furthermore, while the presence of BBH reversed the MDR-TJ strain susceptibilities to sulbactam and ciprofloxacin, the addition of BBH did not change the susceptibility of the △adeB MDR-TJ strain to any of the antibiotics tested. What’s more, the FICIs were more than 0.5, indicating no synergistic effects between antibiotics and BBH observed in △adeB MDR-TJ strain. If there is any other reason, in the deletion strain of MDR-TJ, the MIC of antibiotic with BBH will decrease. Whereas, as shown in Table 3, only one to two-fold MIC reduction was observed. Therefore, these data could be proved that the main target of BBH is AdeABC. Taken together, these data suggest that a key role of BBH is to compete against antibiotics for the binding sites of AdeB, which increased antibiotic susceptibility of MDR A. baumannii.
BBH Accumulation Study
The interior of bacteria is always negatively charged relative to the external environment. Therefore, BBH, as a cationic quaternary ammonium salt, can be easily retained within cells by membrane potential (Severina et al., 2001). It is reported that BBH, likely ethidium bromide, may bind to DNA or cytoplasmic membrane, resulting in increased fluorescence (Wang et al., 2009). Thereby, we utilized this phenomenon to estimate the uptake of BBH within bacteria, which were measured as relative fluorescence units (RFUs). As shown in Figure 5A, the uptake of BBH reached a stationary stage at about 60 min in the MDR-B, MDR-TJ and ATCC 19606 strains, suggesting that BBH may be extruded by bacteria through efflux pumps in general. For the △adeB MDR-B and △adeB MDR-TJ strains that did not produce any AdeB protein, significantly higher accumulation of BBH was observed in comparison with their parental strains, suggesting that part of BBH can be extruded by AdeB.
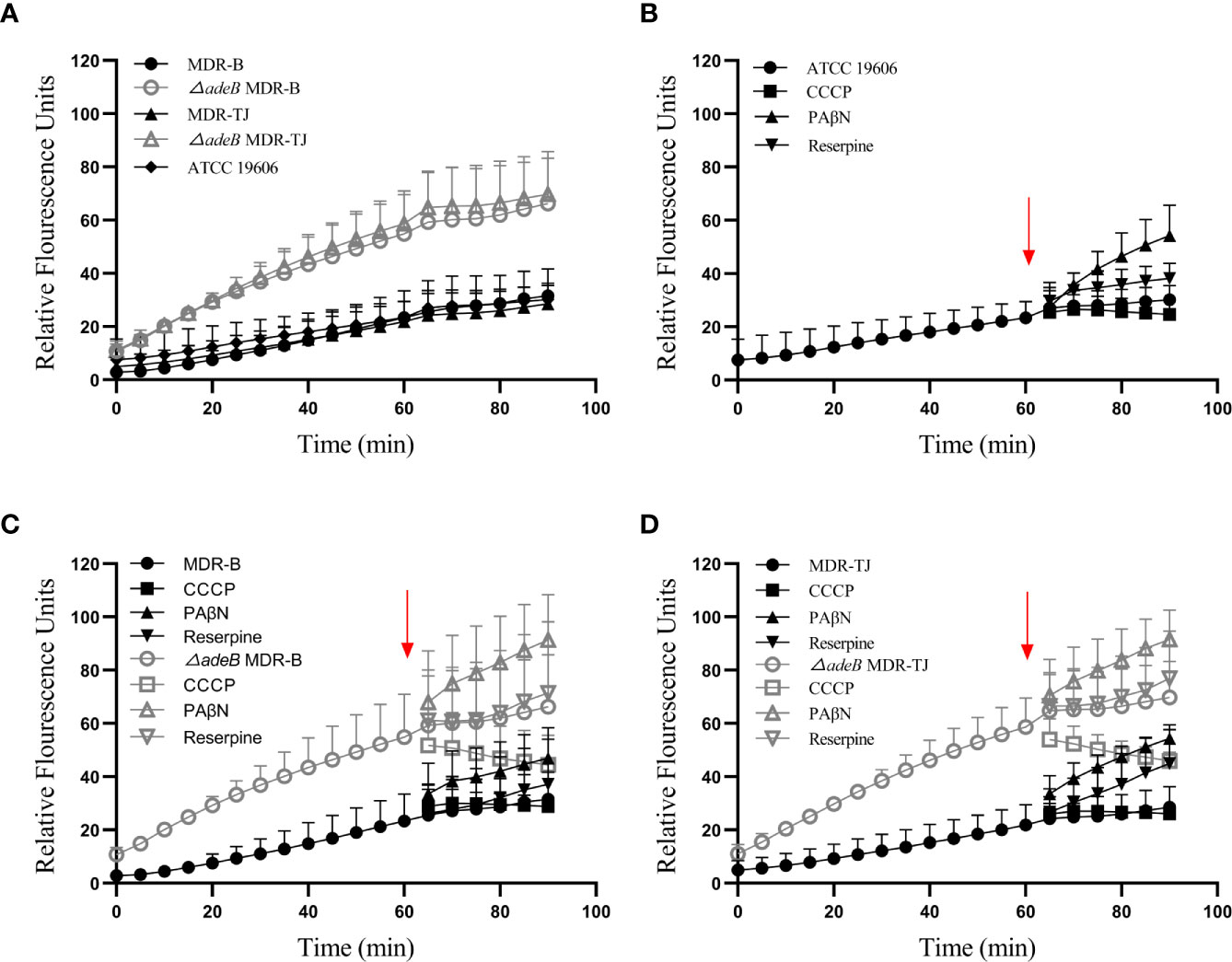
Figure 5 BBH accumulation in A. baumannii strains. The amount of BBH was determined as relative fluorescence units. (A) BBH uptake by A. baumannii strains in 90 min. BBH uptake with pump inhibitors CCCP, PAβN, and reserpine in (B) ATCC 19606 strain, (C) MDR-B and △adeB MDR-B strains, and (D) MDR-TJ and △adeB MDR-TJ strains. Red arrows indicate the pump inhibitors (50 µM) were added at 60 min. Grey letters indicate the inhibitors were added to the adeB knockout strains.
To further understand the mechanism of action of BBH, efflux pump inhibitors CCCP, PAβN and reserpine were used to evaluate BBH accumulation (Figures 5B–D). PAβN mainly causes membrane permeabilization (Li et al., 2015), and it increased BBH accumulation in all the strains. Reserpine is a blocker of P-glycoprotein (Li et al., 2015), and caused a marginal decline of antibiotic resistance in MDR A. baumannii as a non-specific pump inhibitor (Pule et al., 2016). The addition of reserpine only slightly increased BBH accumulation, indicating other efflux pumps may contribute to BBH extrusion. CCCP as a proton uncoupler can inhibit the energy of efflux pumps by reducing transmembrane potential (Pule et al., 2016; Shi et al., 2018). It would increase BBH accumulation in this regard when BBH acts as a competitive substrate. However, our results showed a reduction of BBH in bacteria after the addition of CCCP, suggesting the occurrence of BBH leakage (Figures 5C, D). A plausible reason is that CCCP may inhibit the proton motive force on the outer membrane responsible for the penetration of BBH into the interior of bacteria (Severina et al., 2001).
Discussion
Nowadays, antibiotic resistance of A. baumannii has progressively increased, reducing the range of clinically available treatment options. The increasing cases of MDR A. baumannii accompanied by a scarcity of new antibiotics have led to monotherapy ineffective against this pathogen. It’s been reported transmission of MRD microorganisms accompanying with fecal microbiota transplantation (FMT) could lead to adverse infections, even causing fatal cases (DeFilipp et al., 2019). Combination therapy has been considered an effective approach to combat MDR A. baumannii (Worthington and Melander, 2013). It may offer a synergism even when MDR strains are not susceptible to the single antibiotics, but occasionally antagonism may be observed (Durante-Mangoni et al., 2014). In addition, in the process of combination therapy for infections with MDR or extensively drug-resistant (XDR) strains, colistin (polymyxin E) serves as one part of the mixtures most of the time, however, side effects including nephrotoxicity limit its clinical utility (Viehman et al., 2014; Wang et al., 2019). Tigecycline or tigecycline-based therapy has excellent activity against A. baumannii, but it has been associated with higher treatment failure rates than non-tigecycline therapy according to a study of 238 patients with carbapenem-resistant A. baumannii pneumonia (Liang et al., 2018). All in all, safe antimicrobial agents with excellent antibacterial activity should be discovered to address this dilemma. However, only five first-in-class antibiotics have been approved in the past two decades, none of which targets Gram-negative bacteria (Butler and Paterson, 2020). Although there are some β-lactamase/β-lactam inhibitor (BLI) combinations have been approved to against Gram-negative bacteria, the β-lactamases have been approved previously (Butler and Paterson, 2020).
Recently, researchers have begun to revitalize antibiotic development from natural herb compounds or clinically proven drugs against Gram-negative bacteria. For instance, halicin (Stokes et al., 2020) and SCH-79797 (Martin et al., 2020) were discovered to kill Gram-negative bacteria with a unique mechanism, which can provide novel promising antibiotic candidates. Similarly, we set our sights on Traditional Chinese Medicine for its time-honored clinical safety evaluation and effective treatment. Here, we discovered that BBH is a potential antimicrobial adjuvant against MDR A. baumannii when combined with antibiotics. We may conclude that BBH can attenuate β-lactamase or β-lactam inhibitor (meropenem and sulbactam) resistance, quinolone (ciprofloxacin) resistance and glycylcycline (tigecycline) resistance against MDR A. baumannii rather than aminoglycoside resistance of A. baumannii. For instance, the MIC values of amikacin (an aminoglycoside antibiotic) of MDR-B and MDR-TJ were >1024 mg/L with or without berberine hydrochloride (data did not show in this paper), which represented similar results with previous study (Morita et al., 2016). What’s more, combination therapy of BBH and SUL against MDR-TJ strain in the murine thigh infection model revealed stronger antimicrobial or bactericidal efficiency than monotherapy. Admittedly, the in vivo test was limited for we mainly focused on the combination of SUL/BBH. In future, a series of work may be carried out to investigate more combination therapies between different antibiotics and berberine hydrochloride against different A. baumannii isolates, accompanied with collecting data of pharmacokinetic/pharmacodynamic (PK/PD), which may provide more valuable data for clinical trials.
Genomic analysis based on whole-genome sequencing is a significant way to reveal the mechanism of antibiotic resistance in A. baumannii. Our previous research indicated A. baumannii strain MDR-TJ could harbor many resistance-associated genes inferring different resistance mechanisms, such as enzyme-mediated degradation, alteration of target sites, and efflux pumps (Huang et al., 2012). Among them, efflux pumps contribute to the extrusion of most antibiotics. In MDR-TJ, a specific efflux pump AdeABC belonging to the RND family has been characterized, which mediates the resistance to a broad spectrum of antibiotics, whereas AdeIJK, another efflux pump identified, is considered to be related to the inherent resistance of A. baumannii (Coyne et al., 2011). Therefore, we focused the target of BBH against MDR A. baumannii on pump AdeABC based on the identification of drug-resistance genes in MDR-TJ and the fact that BBH can resume antibiotic sensitivity. Previous studies indicated that cationic antibacterial agents (e.g. berberine) are ideal substrates of multidrug pumps and actively extruded by multidrug pumps in MDR strains (Hsieh et al., 1998; Morita et al., 1998; Stermitz et al., 2000; Severina et al., 2001; Tegos et al., 2002). Our results were consistent with those findings. BBH uptake reached a stationary stage at about 60 min, indicating a balance of accumulation and extrusion by membrane potential and multidrug pump, respectively. The significant increases in the amount of BBH accumulated in the △adeB strains or in the presence of pump inhibitor PAβN validated the competitive role of BBH in the efflux-mediated extrusion of antibiotics (Figure 5).
Some studies showed that the antibacterial activity of berberine and its derivatives is attributed to their role as a pump inhibitor (Morita et al., 2016; Pule et al., 2016). Our study demonstrated that BBH is more likely a pump competitor to resume antibiotic sensitivity than an inhibitor in the treatment of A. baumannii, because it significantly boosted the expression of pump gene adeB and had a higher affinity toward AdeB than antibiotics (Figure 3 and Table 2). In addition, the results that antibiotic-sensitizing activity and synergistic effects diminished in △adeB strain (Table 3) confirmed BBH could facilitate antibiotic efficacy by competing against antibiotics for the AdeB sites. However, the MICs of BBH did not decrease in the △adeB strains, and they changed slightly with pump inhibitors (Table S4), which implies that AdeB may not be the only efflux pump for BBH and other pumps may also contribute to BBH extrusion. Despite the poor absorption and bioavailability of BBH (Imenshahidi and Hosseinzadeh, 2016) as a single antimicrobial, it still has accumulated numerous reliable therapeutic data, and novel approach, like nanoparticulate delivery system (Tan et al., 2011), has been developed to improve its bioavailability. In contrast, for most efflux pump inhibitors (i.e. CCCP and PAβN), the strong side effects and toxicity limit their clinical applications (Li et al., 2015). Last but not least, berberine and berberine analogs which may have similar efficacy against MDR A. baumannii are worthy of further research and development in vitro and in vivo to alleviate imminent threats to human health posed by the continued emergence of multidrug-resistant bacteria.
Conclusions
Collectively, our findings have demonstrated that BBH is a positive substrate of AdeB and possibly other efflux pumps, which may be responsible for the synergistic effects of BBH with antibiotics and antibiotic-sensitizing activity in MDR A. baumannii (Figure 6). In vivo study has suggested that BBH can dramatically boost the antimicrobial efficiency of sulbactam against MDR-TJ strain, which may provide a novel combination therapy to combat MDR A. baumannii infection. Meanwhile, despite of the substantial and clear results in vitro tests, more in vivo studies are needed to carry out in the future to reinforce the clinical effectiveness of the combination therapies.
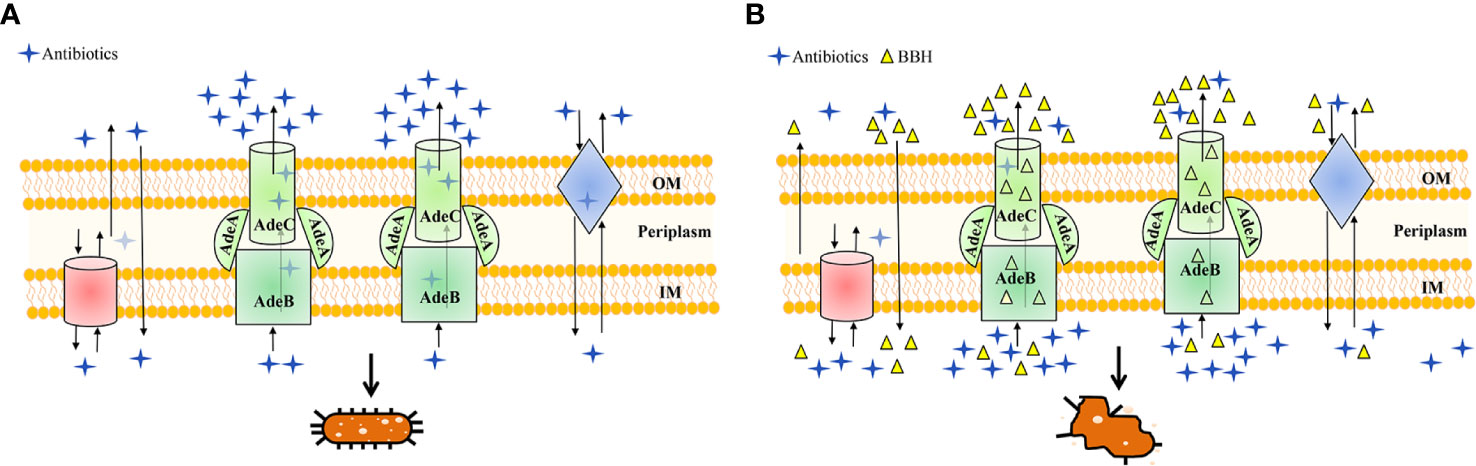
Figure 6 Schematic diagram highlighting the antibiotic efflux mechanisms with (A) BBH absent or (B) present. MDR A. baumannii may harbor abundant pump systems. For instance, AdeABC pump contributes to resistance by extruding a wide range of antibiotics into the extracellular environment, which causes a lower dose of antibiotics to accumulate within bacteria and leads to multidrug resistance and concomitant cell growth, as illustrated in (A). However, in the presence of BBH (B), multidrug resistance of antibiotics can be reversed for it could occupy the binding sites of AdeABC and other efflux proteins as a positive pump competitor. This way both the penetration of antibiotics from the outer membrane (OM) and accumulation inside the inner membrane (IM) are maintained, leading to the killing of bacteria.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by The Chinese Academy of Medical Science and Peking Union Medical College (Approval No. IRM-DWLL-2020106).
Author Contributions
HH, HY, PW and XL conceived and designed this experiment. XL, YS, LW and GK performed the experiments. XL, GK and YS analyzed the data. XL and YS wrote the draft manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from National Key Research and Development Project (No. 2019YFA0905600), Major State Basic Research Development Program of Natural Science Foundation of Shandong Province in China (No. ZR2020ZD11) and Science and Technology Program of Tianjin, China (No. 19YFSLQY00110). We thank partial support of NERC (NE/S008721/1).
Conflict of Interest
Authors XL and PW were employed by Tianjin Modern Innovative TCM Technology Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.660431/full#supplementary-material
References
Amin I. M., Richmond G. E., Sen P., Koh T. H., Piddock L. J. V., Chua K. L. (2013). A Method for generating marker–less gene deletions in multidrug–resistant Acinetobacter baumannii. BMC Microbiol. 13, 158. doi: 10.1186/1471–2180–13–158
Antunes L. C., Visca P., Towner K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301. doi: 10.1111/2049–632X.12125
Barnes M. D., Kumar V., Bethel C. R., Moussa S. H., O’donnell J., Rutter J. D., et al. (2019). Targeting Multidrug–Resistant Acinetobacter spp.: Sulbactam and the Diazabicyclooctenone beta–Lactamase Inhibitor ETX2514 as a Novel Therapeutic Agent. mBio 10 (2), e00159–19. doi: 10.1128/mBio.00159–19
Bikadi Z., Hazai E. (2009). Application of the PM6 semi–empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminform 1, 15. doi: 10.1186/1758–2946–1–15
Butler M. S., Paterson D. L. (2020). Antibiotics in the clinical pipeline in October 2019. J. Antibiot. (Tokyo) 73, 329–364. doi: 10.1038/s41429–020–0291–8
Cai Y., Chai D., Wang R., Liang B., Bai N. (2012). Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrobial. Chemother. 67, 1607–1615. doi: 10.1093/jac/dks084
Chen Q., Li X., Zhou H., Jiang Y., Chen Y., Hua X., et al. (2014). Decreased susceptibility to tigecycline in Acinetobacter baumannii mediated by a mutation in trm encoding SAM–dependent methyltransferase. J. Antimicrobial. Chemother. 69, 72–76. doi: 10.1093/jac/dkt319
Clinical and Laboratory Standards Institute (2018). Performance Standards for Antimicrobial Susceptibility Testing. Twenty–Eighth Informational Supplement. Document M100. CLSI (Wayne, PA: Clinical and Laboratory Standards Institute).
Coyne S., Courvalin P., Perichon B. (2011). Efflux–mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55, 947–953. doi: 10.1128/AAC.01388–10
DeFilipp Z., Bloom P. P., Torres Soto M., Mansour M. K., Sater M. R. A., Huntley M. H., et al. (2019). Drug–Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N. Engl. J. Med. 381, 2043–2050. doi: 10.1056/NEJMoa1910437
Duarte A., Ferreira S., Silva F., Domingues F. C. (2012). Synergistic activity of coriander oil and conventional antibiotics against Acinetobacter baumannii. Phytomedicine 19, 236–238. doi: 10.1016/j.phymed.2011.11.010
Durante-Mangoni E., Utili R., Zarrilli R. (2014). Combination therapy in severe Acinetobacter baumannii infections: an update on the evidence to date. Future Microbiol. 9, 773–789. doi: 10.2217/fmb.14.34
Gao W. W., Gopala L., Bheemanaboina R. R. Y., Zhang G. B., Li S., Zhou C. H. (2018). Discovery of 2–aminothiazolyl berberine derivatives as effectively antibacterial agents toward clinically drug–resistant Gram–negative Acinetobacter baumanii. Eur. J. Med. Chem. 146, 15–37. doi: 10.1016/j.ejmech.2018.01.038
He T., Wang R., Liu D., Walsh T. R., Zhang R., Lv Y., et al. (2019). Emergence of plasmid–mediated high–level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564–019–0445–2
Hsieh P. C., Siegel S. A., Rogers B., Davis D., Lewis K. (1998). Bacteria lacking a multidrug pump: A sensitive tool for drug discovery. Proc. Natl. Acad. Sci. U. S. A. 95, 6602–6606. doi: 10.1073/pnas.95.12.6602
Huang H., Yang Z. L., Wu X. M., Wang Y., Liu Y. J., Luo H., et al. (2012). Complete genome sequence of Acinetobacter baumannii MDR–TJ and insights into its mechanism of antibiotic resistance. J. Antimicrob. Chemother. 67, 2825–2832. doi: 10.1093/jac/dks327
Imenshahidi M., Hosseinzadeh H. (2016). Berberis Vulgaris and Berberine: An Update Review. Phytother. Res. 30, 1745–1764. doi: 10.1002/ptr.5693
Lee C.- R., Lee J. H., Park M., Park K. S., Bae I. K., Kim Y. B., et al. (2017). Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 7, 55. doi: 10.3389/fcimb.2017.00055
Li X. Z., Plesiat P., Nikaido H. (2015). The challenge of efflux–mediated antibiotic resistance in Gram–negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117–14
Liang C. A., Lin Y. C., Lu P. L., Chen H. C., Chang H. L., Sheu C. C. (2018). Antibiotic strategies and clinical outcomes in critically ill patients with pneumonia caused by carbapenem–resistant Acinetobacter baumannii. Clin. Microbiol. Infect. 24 (8), 908.e1–908.e7. doi: 10.1016/j.cmi.2017.10.033
Macvane S. H., Crandon J. L., Nicolau D. P. (2014). Characterizing In Vivo Pharmacodynamics of Carbapenems against Acinetobacter baumannii in a Murine Thigh Infection Model To Support Breakpoint Determinations. Antimicrobial Agents Chemother. 58, 599–601. doi: 10.1128/aac.02029–13
Martin J. K., 2., Sheehan J. P., Bratton B. P., Moore G. M., Mateus A., Li S. H., et al. (2020). A Dual–Mechanism Antibiotic Kills Gram–Negative Bacteria and Avoids Drug Resistance. Cell 181, 1518–1532.e1514. doi: 10.1016/j.cell.2020.05.005
Morita Y., Kodama K., Shiota S., Mine T., Kataoka A., Mizushima T., et al. (1998). NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782. doi: 10.1128/AAC.42.7.1778
Morita Y., Nakashima K., Nishino K., Kotani K., Tomida J., Inoue M., et al. (2016). Berberine Is a Novel Type Efflux Inhibitor Which Attenuates the MexXY–Mediated Aminoglycoside Resistance in Pseudomonas aeruginosa. Front. Microbiol. 7, 1223. doi: 10.3389/fmicb.2016.01223
Navon-Venezia S., Leavitt A., Carmeli Y. (2007). High tigecycline resistance in multidrug–resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 59, 772–774. doi: 10.1093/jac/dkm018
Nowak J., Schneiders T., Seifert H., Higgins P. G. (2016). The Asp20–to–Asn Substitution in the Response Regulator AdeR Leads to Enhanced Efflux Activity of AdeB in Acinetobacter baumannii. Antimicrob. Agents Chemother. 60, 1085–1090. doi: 10.1128/AAC.02413–15
Odds F. C. (2003). Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrobial Chemother. 52, 1–1. doi: 10.1093/jac/dkg301
Pule C. M., Sampson S. L., Warren R. M., Black P. A., Van Helden P. D., Victor T. C., et al. (2016). Efflux pump inhibitors: targeting mycobacterial efflux systems to enhance TB therapy. J. Antimicrob. Chemother. 71, 17–26. doi: 10.1093/jac/dkv316
Severina Ii, Muntyan M. S., Lewis K., Skulachev V. P. (2001). Transfer of cationic antibacterial agents berberine, palmatine, and benzalkonium through bimolecular planar phospholipid film and Staphylococcus aureus membrane. IUBMB Life 52, 321–324. doi: 10.1080/152165401317291183
Shi C., Li M., Muhammad I., Ma X., Chang Y., Li R., et al. (2018). Combination of berberine and ciprofloxacin reduces multi–resistant Salmonella strain biofilm formation by depressing mRNA expressions of luxS, rpoE, and ompR. J. Vet. Sci. 19, 808–816. doi: 10.4142/jvs.2018.19.6.808
Stermitz F. R., Lorenz P., Tawara J. N., Zenewicz L. A., Lewis K. (2000). Synergy in a medicinal plant: Antimicrobial action of berberine potentiated by 5 ‘–methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. U. S. A. 97, 1433–1437. doi: 10.1073/pnas.030540597
Stokes J. M., Yang K., Swanson K., Jin W., Cubillos–Ruiz A., Donghia N. M., et al. (2020). A Deep Learning Approach to Antibiotic Discovery. Cell 180, 688–68+. doi: 10.1016/j.cell.2020.01.021
Su C.- C., Morgan C. E., Kambakam S., Rajavel M., Scott H., Huang W., et al. (2019). Cryo–Electron Microscopy Structure of an Acinetobacter baumannii Multidrug Efflux Pump. Mbio 10 (4), e01295–19. doi: 10.1128/mBio.01295–19
Sun H., Ansari M. F., Battini N., Bheemanaboina R. R. Y., Zhou C. -H. (2019). Novel potential artificial MRSA DNA intercalators: synthesis and biological evaluation of berberine–derived thiazolidinediones. Organic Chem. Front. 6, 319–334. doi: 10.1039/c8qo01180j
Tan W., Li Y., Chen M., Wang Y. (2011). Berberine hydrochloride: anticancer activity and nanoparticulate delivery system. Int. J. Nanomed. 6, 1773–1777. doi: 10.2147/IJN.S22683
Tegos G., Stermitz F. R., Lomovskaya O., Lewis K. (2002). Multidrug pump inhibitors uncover remarkable activity of plant antimicrobials. Antimicrob. Agents Chemother. 46, 3133–3141. doi: 10.1128/aac.46.10.3133–3141.2002
Viehman J. A., Nguyen M. H., Doi Y. (2014). Treatment options for carbapenem–resistant and extensively drug–resistant Acinetobacter baumannii infections. Drugs 74, 1315–1333. doi: 10.1007/s40265–014–0267–8
Wang X., Yao X., Zhu Z., Tang T., Dai K., Sadovskaya I., et al. (2009). Effect of berberine on Staphylococcus epidermidis biofilm formation. Int. J. Antimicrob. Agents 34, 60–66. doi: 10.1016/j.ijantimicag.2008.10.033
Wang J., Niu H., Wang R., Cai Y. (2019). Safety and efficacy of colistin alone or in combination in adults with Acinetobacter baumannii infection: A systematic review and meta–analysis. Int. J. Antimicrob. Agents 53, 383–400. doi: 10.1016/j.ijantimicag.2018.10.020
Watkins R. R. (2018). A formidable foe: carbapenem–resistant Acinetobacter baumannii and emerging nonantibiotic therapies. Expert Rev. Anti Infect. Ther. 16, 591–593. doi: 10.1080/14787210.2018.1503054
World Health Organization (2017). List of Antibiotic Resistant Priority Pathogens. Available online at: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/.
Wong D., Nielsen T. B., Bonomo R. A., Pantapalangkoor P., Luna B., Spellberg B. (2017). Clinical and Pathophysiological Overview of Acinetobacter Infections: a Century of Challenges. Clin. Microbiol. Rev. 30, 409–447. doi: 10.1128/CMR.00058–16
Worthington R. J., Melander C. (2013). Combination approaches to combat multidrug–resistant bacteria. Trends Biotechnol. 31, 177–184. doi: 10.1016/j.tibtech.2012.12.006
Yoon E. J., Courvalin P., Grillot–Courvalin C. (2013). RND–type efflux pumps in multidrug–resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob. Agents Chemother. 57, 2989–2995. doi: 10.1128/AAC.02556–12
Yoon E. J., Chabane Y. N., Goussard S., Snesrud E., Courvalin P., De E., et al. (2015). Contribution of resistance–nodulation–cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 6 (2), e00359–15. doi: 10.1128/mBio.00309–15
Keywords: Acinetobacter baumannii, berberine hydrochloride, multidrug-resistance, antibiotic sensitization, synergistic effect, AdeABC
Citation: Li X, Song Y, Wang L, Kang G, Wang P, Yin H and Huang H (2021) A Potential Combination Therapy of Berberine Hydrochloride With Antibiotics Against Multidrug-Resistant Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 11:660431. doi: 10.3389/fcimb.2021.660431
Received: 29 January 2021; Accepted: 11 March 2021;
Published: 25 March 2021.
Edited by:
Milena Dropa, University of São Paulo, BrazilReviewed by:
Rafael Franco-Cendejas, National Institute of Rehabilitation Luis Guillermo Ibarra Ibarra, MexicoIsrael Castillo-Juárez, Colegio de Postgraduados (COLPOS), Mexico
Copyright © 2021 Li, Song, Wang, Kang, Wang, Yin and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huabing Yin, huabing.yin@glasgow.ac.uk; He Huang, huang@tju.edu.cn
 Xiaobo Li
Xiaobo Li Yanqing Song
Yanqing Song Lina Wang1
Lina Wang1  Guangbo Kang
Guangbo Kang Huabing Yin
Huabing Yin He Huang
He Huang