- Department of Endocrinology and Metabolism, First Affiliated Hospital of Jilin University, Changchun, China
The Caenorhabditis elegans genome possesses homologs of about two-thirds of all human disease genes. Based on its physiological aging characteristics and superiority, the use of C. elegans as a model system for studies on aging, age-related diseases, mechanisms of longevity, and drug screening has been widely acknowledged in recent decades. Lifespan increasing mutations in C. elegans were found to delay aging by impinging several signaling pathways and related epigenetic modifications, including the insulin/IGF-1 signaling (IIS), AMP-activated protein kinase (AMPK), and mechanistic target of rapamycin (mTOR) pathways. Interestingly, dietary restriction (DR) has been shown to increase the lifespan of numerous metazoans and protect them from multiple age-related pathologies. However, the underlying molecular mechanisms are unclear. In recent decades, C. elegans has been used as a unique model system for high-throughput drug screening. Here, we review C. elegans mutants exhibiting increased in lifespan and age-dependent changes under DR, as well as the utility of C. elegans for drug screening. Thus, we provide evidence for the use of this model organism in research on the prevention of aging.
Introduction
The eukaryotic multicellular organism Caenorhabditis elegans which has completely sequenced genetic profile, is an established genetic model organism (1), that can be used to study aging. The use of C. elegans as a model system to recapitulate most human diseases in recent decades is invaluable for experimental research at both the metabolic and genomic levels in vivo (2, 3). In addition, research studies on aging using C. elegans have provided desirable outcomes in identifying molecular signals, transcriptional regulators, and epigenetic modifications associated with longevity broadening our ability to understand how organisms age. In this review, we aim to provide an overview of the established and current novel concepts on transcriptional and epigenetic regulators in the field of research on aging using the model organism C. elegans and elucidate how dietary restrictions influence these specific regulators, as well as discuss the application of C. elegans in drug screening studies.
Background, Advantages, and Limitations of C. elegans for Studies on Aging
Basic Features and Age-Dependent Changes of C. elegans
C. elegans is a free-living, harmless nematode that feeds on microorganisms. It is particularly economical and easy to maintain in laboratory settings. Adult C. elegans are 1 mm long self-fertilizing hermaphrodites with a 2.5–4 days reproductive cycle at room temperature, and a mean lifespan of approximately 18–20 days when cultured at 20°C (4–7). After hatching, C. elegans can either develop directly to four larval stages (L1–L4) or proceed with the dauer larval stage after the L2 larval stage, instead of the L3 larval stage. The dauer larval stage is a developmentally arrested dispersal stage used to survive adverse conditions. Once the adverse conditions subside, C. elegans can recover and molt into the L4 larval stage, continuing normal development (8). In worms, features associated with aging could result in less active, uncoordinated movements, torpor, cessation of reproduction, and accumulation of auto-fluorescent deposits in cells (7, 9, 10)1. Clear age-dependent humanlike physiological changes at the tissue, cellular, and molecular levels make C. elegans a valuable model for research in the field of aging.
Aged C. elegans display a decline in their anatomical and functional features, including tissue integrity, motility, learning and memory, and immunity. The primary age-dependent changes at the tissue level include changes in the reproductive, nervous, and muscular systems. The rate of reproduction marked decreases and the structure of the reproductive system deteriorates with age. Oocyte size and quality also deteriorate with advancing age (11). C. elegans displays structural changes and functional deterioration of neurons during aging. Blebbing and branching structures can be seen in aged touch receptor neurons, indicating that the synaptic integrity degenerates with aging (12, 13). In addition, it has been widely reported that the loss of muscular integrity, sarcomeres density, and regular orientations in aged C. elegans result in impaired motility and an abnormal appearance (14).
At the cellular level, the primary age-dependent changes in C. elegans generally include diminishing integrity of nuclei and increased relative size of the nucleoli; however, these changes may vary depending on the tissue types. Moreover, mitochondria undergo age-dependent structural and functional changes, including mitochondrial fusion and increased mitochondrial fragmentation, which is consistent with changes in mitochondrial DNA copy numbers and oxygen consumption rates (15). The capacity of the unfolded protein response of endoplasmic reticulum (ERUPR) seems to be reduced in aged C. elegans. The ERUPR is activated and tasked with degrading the misfolded proteins under various stress conditions (16). Reduced ERUPR process result in misfolded proteins increasing and leading to age-related diseases.
The C. elegans genome possesses homologs of about two-thirds of all human disease genes. In a previous study, Zhao et al. performed functional analysis of 143 essential genes and found that 108 of them were human orthologs. Of these, 97 genes were associated with 1,218 different diseases (17). Age-associated molecular changes provide more information and serve as valuable biomarkers for aging. Many changes in aging-associated gene expression, which increase lifespan but decrease with age, have been identified during C. elegans aging (18). The quality of RNA control mechanisms, such as non-sense-mediated mRNA decay (NMD) activity in various organs and tissues, decline with advancing age in C. elegans. Also, the increased levels of introns and unannotated regions in the mRNAs denote a decrease of mRNA splicing fidelity in aged worms (19). Protein homeostasis associated with age-related diseases declines during aging (20). It has been reported that proteins involved in nucleosome assembly, ER nuclear signaling, and the response to unfolded proteins increase, whereas the abundance of proteins involved in metabolism (fatty acid, carbohydrate, and amino acid) decreases during aging (21). In addition, the levels of amino acid metabolites also change with age (22). The effects resulting from gene expression changes during aging are not yet fully understood. Further studies will be needed to comprehensively elucidate the roles of age-dependent changes in the levels of amino acid in aging and longevity.
Advantages of C. elegans on Aging Study
C. elegans is an excellent model organism used for aging research. The ease of its maintenance in the laboratory, transparent body for anatomical observation, high genetic homology (60–80%) with humans, availability of complete genome sequence, conserved biological molecular responses, high fertility rates (~250 eggs/worm within several days), and the availability of molecular biology tools (i.e., transgenic, gene knockouts, and RNAi knockdowns) make C. elegans a useful model for the study of aging mutations (23). In addition, the short lifespan of this organism (~3 weeks) and small size are favorable for the screening of anti-aging drugs due to the reduced experimental costs and their usability for a high throughput screening experiments (24). Moreover, experiments with C. elegans are free of ethical concerns. Many breakthrough discoveries in the field of aging research have been achieved using C. elegans because of these advantages (25).
Limitations of C. elegans on Aging Study
C. elegans shows many desirable features for aging studies, however, it still has some limitations as a model organism compared to other mammals. Firstly, C. elegans lack certain anatomical features of mammals, including a blood transport system, a blood-brain barrier, a first-pass metabolism process in the liver, and blood filtration in the kidney, which may be specific for certain signal pathways or epigenetic effects (26). As a model system to predict human research outcomes, the lack of DNA methylation, an epigenetic tag that possibly has a greater function in mammals than in nematodes, is another limitation of C. elegans (27). In addition, the lack of long-range transcriptional regulation makes it inadaptable for studying the relevant mechanism in other animal species; however, it is recommended as a simplified model for studies on signal mapping mechanisms (27).
Transcriptional Regulators and Epigenetic Regulation in C. elegans Aging
Previous studies have identified several loci that increase the lifespan of C. elegans when mutated. The molecular genetics of this organism is well-established and has been strongly supported by a fully sequenced genome, which provides insight regarding the entire 959 somatic cells that constitute it. Thus, far, over 50 genes that control aging in C. elegans have been identified (50). Of these, many have homologs in other organisms. Different upstream signals stimulate partially overlapping sets of downstream mediators and processes that ultimately extended the lifespan. Meanwhile, epigenetic regulation in cooperation with transcriptional regulators influence the functions of cells and the fate of organisms, and could act as markers of aging (51). Epigenetic regulation involves DNA methylation, chromatin remodeling, post-translational modifications (PTMs) of histones, and non-coding RNA transcription (28). Here we discuss longevity mechanisms related to transcriptional regulators of metabolic networks and epigenetic regulation in C. elegans.
The Insulin/IGF-1 Signaling Pathway
The insulin/IGF-1 signaling (IIS) pathway is one of the most studied longevity pathways (52, 53). This pathway has three key genes, namely daf-2, age-1, and daf-16. While daf-2 encodes a homolog of the mammalian insulin/IGF-I receptor (INSR) (54), age-1 encodes a homolog of the catalytic p110 subunit of mammalian phosphoinositide 3 kinase (PI3K) (55, 56). In C. elegans, daf-16 is widely expressed and encodes a homolog of human forkhead box O (FOXO) transcription factor (54). The greatest increases in lifespan due to mutations in single genes have been reported for daf-2 and age-1 (57–59). The mean survival of long-lived daf-2 and age-1 mutants was around 15% longer than that of the wild-type (60). Mutations in daf-2 and age-1 resulted in arrested larvae and forced larvae into the dauer stage, increasing the lifespan of the nematodes, as well as enhancing stress resistance (61). The prolonged lifespan of daf-2 and age-1 mutants were dependent on DAF-16(FOXO) (Figure 1). In worms with the daf-2 mutation, the activity of the IIS pathway decrease leading to the phosphorylation of DAF-16 by AKT-1/2 and the translocation of DAF-16 into the nucleus to bind and initiate expression of target genes, Consequently the lifespan of the worm is prolonged and stress-protective mechanisms, including the unfolded protein response and oxidative stress responses, are initiated (29). The transcription factor DAF-16(FOXO) induces juvenile C. elegans to develop into dauer larvae, which represents diapause that allows this organism to withstand harsh conditions. Reactive oxygen species (ROS) can modulate the import of DAF-16 into the nucleus via disulfide bond formation with transportin-1 (IMB-2) (62). The main role of DAF-2(AGE-1) signaling is to antagonize daf-16 (63, 64). It has been reported that the metabolic, longevity, and developmental defects caused by daf-2 and age-1 mutations are antagonized by daf-16 mutations (64–66). DAF-16 may directly regulate the transcription of the genes necessary for the increased longevity observed in age-1 and daf-2 mutants (29). As such, one research direction is to identify genes under the control of DAF-16. The strong association between FOXO expression and lifespan has been reported by several studies on humans and is considered a promising therapeutic target to promote longevity.
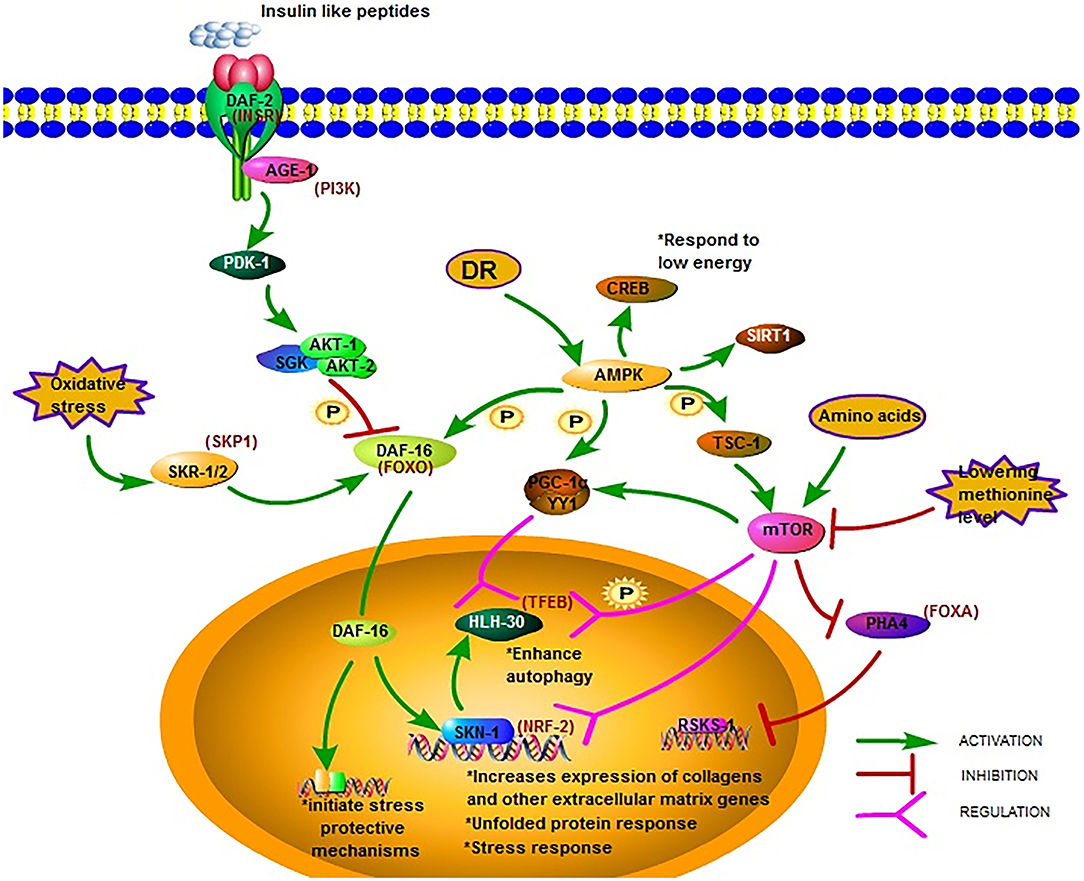
Figure 1. Major longevity pathways and longevity-associated transcription factors in C. elegans. The IIS pathway is one of the most studied longevity pathways. DAF-2, AGE-1, and DAF-16 are its three key genes. Mutations in DAF-2 and AGE-1 and low IIS activity prolong lifespan via DAF-16 and downstream gene expression. AMPK is another crucial metabolic energy sensor that links nutrient availability to lifespan by binding and phosphorylating a set of transcriptional (co)activators. The mTOR pathway is another critical pathway that links nutrient availability and metabolism to longevity; however, its mechanism remains to be fully elucidated (18, 28–49).
Reduced IIS promotes C. elegans longevity through the NF-E2-related factor (NRF2) ortholog SKN-1, which operates as a genetically distinct program from the dauer pathway and in parallel to DAF-16 (30). The transcription factor SKN-1(NRF2) is considered an important regulator of detoxification and oxidative stress responses in C. elegans; SKN-1 (NRF2) prominently increases the expression of collagens and other extracellular matrix genes when IIS level is decreased (18). The expression of the skn-1 target gene upon oxidative stress can also be promoted by SKR-1/2, which is the ortholog of the mammalian SCF-ubiquitin ligase complex member SKP1. Furthermore, it has been reported that DAF-16 can regulate SKN-1 transcription and that daf-16 is a target of SKR-1/2, indicating that SKN-1 mediated stress resistance may not be necessary associated with longevity (31) (Figure 1).
Epigenetic regulation on specific targets of metabolic signaling pathways can alter the lifespan. The demethylase UTX-1, which targets genes such as daf-2 on the IIS pathway, regulates the lifespan in C. elegans. It reduces DAF-16(FOXO) levels, as mentioned above, compromising cellular maintenance processes and weakening the ability to resist stress in C. elegans, thus inducing an aging-related decline in cellular functions (32). Besides, LncRNAs with a variable length spanning from 200 base pairs up to several kilobases are important for cell function, because it can target classic signal pathways such as IIS. For example, the LncRNA tts-1 extends lifespan by reducing ribosome levels in the daf-2 mutant C. elegans (28). IIS conserved in both insects and mammals; the genetic and biological characteristics of the IIS pathway in aging were successfully translated to mammals and humans (67).
AMP-Activated Protein Kinase Signaling Pathway
Different signal pathways could regulate each other by PTMs. For example, the post-translational modifications of DAF-16 include phosphorylation by AAK-2 (AMPK), which belongs to another important signal pathway related to metabolic energy. AMP-activated protein kinase (AMPK) is a crucial metabolic energy sensor linking nutrient availability with lifespan (68). The AMPK encoding gene is aak-2 in C. elegans. As a master regulator of cellular energy homeostasis, AMPK is required for the metabolic adjustment during the starved, developmentally quiescent diapause phase of C. elegans (69). The overexpression of AMPK extends lifespan, as shown in C. elegans (33). Upon activation, AMPK binds and phosphorylates a set of transcriptional coactivators, including PGC-1α, FOXO, and SIRT1, and the actions of AMPK activation at least partially overlap with sirtuin activation (34). Similar to the mechanism in the mammalian system, aak-2-mediated longevity requires the downregulation of the IIS pathway and the subsequent upregulation and translocation of DAF-16(FOXO). Post-translational modifications of DAF-16 include its phosphorylation by AMPK (70). AMPK modulation of lifespan has been shown to occur also via CREB-regulated transcriptional coactivators in response to low levels of energy (35) (Figure 1).
Starvation can induce long-term consequences through epigenetic change. AMPK is required for metabolic adjustment by blocking specific chromatin modifications and epigenetic changes in C. elegans larvae to resist starvation (69). The recovery of C. elegans after starvation in the early larval stage would be impaired with the absence of AMPK, and could become sterile. AMPK might affects heritable aspect including germline gene expression or genomic integrity, which need further research.
Recently, another of transgenerational lifespan regulation paradigm was shown in C. elegans. Lacking DNA methylation to activate histone modifications such as reduced methylation of Lys4 of histone H3 (H3K4me) are characters of actively transcribed genes (71). DNA methylation usually occurs at 5-methyl cytosine (5 mC) and result in transcriptional repression (72). In C. elegans, 5 mC methylation is rare, while methylation on N6 adenine (6 mA) is prevalent of silence DNA repeats (73). NMAD-1 (MT-A70 family) and DMAD (DNA 6 mA demethylase, TET ortholog) are 6 mA demethylases and can regulate 6 mA levels in C. elegans (74). DAMT-1 (AlKB family), a likely 6 mA methyltransferase, can also control the epigenetic inheritance of phenotypes which associated with the loss of the H3K4me2 demethylase SPR-5 (CoREST/LSD1 ortholog) (75). Deletion of the SPR-5, 6 mA increases across generations and can lead to a progressive transgenerational loss of fertility, the worms become sterile after several generations (76). Longevity can also be transmitted across generations by this kind non-genetic factors. Greer et al. demonstrated that deletion of the spr-5 in C. elegans causes a trans-generational increase in lifespan through mis-regulation and activation of lifespan-regulating signaling pathway (77). It is reported that SPR-5 has numerous consensus AMPK phosphorylation sites (69), but whether SPR-5 is yet another functional nuclear target of AMPK remains to be established.
Mechanistic Target of Rapamycin Signaling
Another critical pathway linking nutrient availability and metabolism to longevity is the mechanistic target of rapamycin (mTOR) pathway. This pathway is activated upon the increase of intracellular amino acids or during growth factor stimulation and modulates a set of downstream signaling pathways that manage cell proliferation, cell growth, motility, survival, and protein synthesis (78). Studies in C. elegans have shown that the inhibition of mTOR activity prolongs the worm lifespan (79). Longevity mediated by inhibiting the mTOR pathway is most likely distinct from the IIS pathway, and an overlapping mechanism may also occur between these two pathways.
mTOR regulates mitochondrial gene expression and control energy- and nutrient related mitochondrial respiration by activating the peroxisome proliferator-activated receptor coactivator-1 α (PGC-1 α) forming a complex with the transcription factor Yin-Yang 1 (YY1) to promote the expression of related genes (36) (Figure 1). One possible downstream pathway that serves as a shared longevity mechanism between the IIS and mTOR pathways is autophagy (31). Recent studies show that the effect of defective mitochondria on cells can be prevented by mTOR during aging via the mitophagy process, which is a kind of autophagy, which targets the mitochondria. Beside PGC-1 α signaling and mitophagy, mTOR may also influence mitochondria via SKN-1 signaling (37).
The transcription factor EB (TFEB), found in C. elegans as HLH-30, is an autophagy enhancer that regulate gene expression related to autophagy and lysosomal (38). In the nucleus, the localization of HLH-30(TFEB) is modulated via phosphorylation by mTOR and the function is regulated via competition with MXL-3/MAX and by its interaction with the Mondo-complex (Figure 1). Potential nuclear interactions between HLH-30 (TFEB) and DAF-16(FOXO) are perhaps required for longevity (39). The modulation of HLH-30 (TFEB) nuclear localization may be a promising strategy to improve somatic maintenance (32). Simultaneous mutations in the IIS and mTOR pathways that produce a synergistic effect was reported recently. Using genome-wide translational state analysis and genetic screening, Lan et al. identified ribosomal protein genes and cyc-2.1, which encodes one of the worm cytochrome orthologs, as negative regulators of longevity (80). Cyc-2.1 knockdown significantly extended lifespan by activating the intestinal mitochondrial unfolded protein response (UPRmt), mitochondrial fission, and AMPK (80). The influence of mitophagy is to extend the lifespan, thus, the role of mTOR mediated mitophagy in longevity needs further study.
Recent studies show that mTOR influence on lifespan also relies on epigenetic cues. Histone modification can regulate lifespan by acting on mTOR signaling pathways. In C. elegans, COMPASS H3K4me3 methyltransferase (methylation of Lys4 of histone H3) deficiency promotes fat accumulation and extends lifespan by targeting RSKS-1 (S6K) in the mTOR complex (28). H3K4me3 methyltransferases have homologs in humans. Mutations in the trithorax group (TrxG) can reduce the H3K4me3 level and, in turn, extend lifespan. Moreover, this influence is heritable for three generations even if the TrxG function is restored in the F1 progeny (81).
Dietary Restriction Extends Lifespan in C. elegans
Since the initial discovery in 1935 that animals feeding on less food lived substantially longer (82), dietary restriction (DR) has been shown to increase lifespan and delay the onset of multiple age-related pathologies in a wide variety of metazoans (83). DR extends lifespan not mediated by a single linear pathway but by multifactorial processes. There are two hypotheses postulated in C. elegans: (1) DR reduces insulin/IGF-I signaling, and (2) DR reduces the metabolic rate (84). In the model of C. elegans, Pandit elucidated the complexity of gene regulation following the initiation of DR in EAT-2 and defined the central role of PHA-4(FOXA) in this process, justifying its position as a robust genetic regulator of DR-induced longevity (40) (Figure 1). Siler also found that PHA-4 played a key role in regulating DR-mediated longevity in adult C. elegans. PHA-4(FOXA) is required for lifespan extension via DR, but not the extension resulting from reduced IIS via daf-2(insr) mutants (41), indicating that PHA-4(FOXA) may be a part of a pathway distinct from IIS (42). Increasing lifespan by reducing TOR signaling requires PHA-4(FOXA) and is mediated by the rsks-1 gene, encoding the homolog of the mammalian SK61. This indicates that FOXA is a necessary downstream component of a TOR-mediated increase in lifespan. However, the precise mechanism and intermediates that control this remain to be determined (43).
Since DR affects both IIS signaling and mTOR signaling, it is important to delineate the contribution of each to overall lifespan extension. In worms, a further reduction in TOR activity does not generate further lifespan extension under some DR regimens, nor protect from lifespan reduction by dietary enrichment, suggesting that mTOR may mediate an effect on lifespan under certain forms of DR (85). Lowering the methionine levels suppresses mTOR pathway activity and prolongs lifespan, suggesting that these types of diets can influence the aging process (86).
The nutrient-sensing pathway is regulated at the lysosomal membrane by several proteins, and the deficiency of which triggers widespread aging phenotypes. In response to environmental conditions, the lysosomal nutrient-sensing complex controls the autophagy process via several factors, including the transcription factors TFEB and FOXO, which have previously been shown to be associated with lifespan extension (Figure 1). A major regulator of autophagy and lysosomal gene expression is HLH-30 (TFEB). HLH-30 (TFEB) is required for innate immunity and lifespan extension in different long-lived nematode mutants via the autophagic response to starvation. The nuclear localization of HLH-30 (TFEB) is modulated via phosphorylation by mTOR (44). This key metabolic pathway strongly depends on nucleocytoplasmic compartmentalization, a cellular phenomenon that is gradually lost with aging.
AAK-2 (AMPK) is another crucial metabolic energy sensor that links nutrient availability to lifespan (18). AMPK regulates mTORC1 activity and shares downstream effectors of lifespan modulation with mTOR (87). AMPK can also regulate mammalian FOXO3 (88). In worms, the activation of AMPK and its downstream metabolic targets often relies on the level of DR and the composition of the restricted diets (68). The response of AMPK to glucose and oxidants is glycogen-dependent (89). Moreover, AMPK signaling may provide a link between glucose toxicity, glycogen accumulation, oxidative stress, and aging. When activated by a drop in energy status, AMPK binds to AMP or ADP to promote ATP production. AMPK can bind and phosphorylate PGC-1α, FOXO, and SIRT1 (33). Especially, sirtuins as a specific group of histone NAD dependent deacetylases are associated with longevity (32). Deletion or inhibition of sirtuin SIR-2.1 (C. elegans ortholog of human SIRT1) reduces lifespan. DR could stimulate SIR-2.1 (SIRT1) and upregulates autophagy in C. elegans and human cells to extend lifespan (45). Moreover, human SIRT1, together with AMPK, could induce autophagy by upregulating autophagic genes and inhibiting mTOR signaling (46). These outcomes indicated that epigenetic regulation of lifespan was closely linked to cell metabolism and nutritional status in C. elegans.
Drug Screening For Compounds That Extend Lifespan in Nematodes
The C. elegans model provides several advantages when performing chemical screening for the identification of drug candidates. This is especially true for primary drug screening, which involves relatively smaller spaces, lower costs, and time-consuming assessments. Nematodes can be inexpensively cultured in large quantities, and the relatively short lifespan of C. elegans ensures this organism provides high-throughput screening for anti-aging drug. Also, the effects of drugs can be tested directly in the whole organism, such that compounds that are toxic for development can be eliminated immediately. C. elegans can also be used for genetic analysis and investigations of chemical interventions for longevity. Moreover, a variety of assays suitable for high-throughput screening for anti-aging compounds are currently being developed (6, 90). Based on mutations in the age-1(PI3K) or daf-2(INSR) genes, and reduction in the daf-16(foxo) mutant, several compounds have been identified that significantly increase the lifespan of this nematode. Kumar demonstrated that C. elegans treated with 25 and 50 μM silymarin increased the mean lifespan of this organism by 10.1 and 24.8%, respectively, as compared to untreated controls (91). Another study demonstrated that fullerenol attenuated the endogenous levels of ROS and provided protection to C. elegans by up-regulating stress-related genes under stress conditions, which was in a DAF-16-dependent manner, thus improving lifespan (92).
Many potential chemical candidates for extending lifespan are currently being investigated, including the following aging modulating compounds: (1) metformin (biguanide anti-glycemic agent for AMPK activation), (2) rapamycin (immunosuppressing agent and mTOR inhibitor), (3) resveratrol (polyphenol and sirtuin activator), (4) spermidine (polyamine and inductor of autophagy), (5) aspirin (COX inhibitor, antithrombosis, and antioxidant), and (6) masoprocol (catechol with antioxidant and anti-inflammatory properties) (93). Active AMPK downregulates mTORC1 activity indirectly by phosphorylating the serine sites on TSC2, and directly by phosphorylating Raptor. The AMPK activating drug metformin (commonly prescribed to diabetic patients) was shown to increase lifespan in C. elegans (47). Similarly, metformin has been shown to act on mTOR signaling via Redd1, also independently of AMPK (94).
The toxicity ranking screening of C. elegans has been repeatedly found to be as predictive of rat LD50 ranking as mouse LD50 ranking. Additionally, many instances of the conservation of the mode of toxic action have been reported between mammals. These consistent correlations make a case for the inclusion of C. elegans assays in early safety testing and as one component in tiered or integrated toxicity testing strategies (95). These findings indicate that C. elegans could be a bridge between in vitro assays and mammalian toxicity testing by combining established in vitro handling techniques and cost ratios with oral toxicity test data from an intact organism. Given that nematodes lack most mammalian organs, it is unrealistic to expect that any combination of C. elegans assays alone will replace in-depth descriptive toxicology analyses in mammals. However, although organismal toxicity endpoints often differ, many pathways of toxicity and modes of toxic action are conserved between worms and humans (95).
Besides acting as markers for the genetic regulation during aging, epigenetic mechanisms may also be targets for drug screening in aging or age-related diseases (28). Researchers have confirmed this promising application. For example, resveratrol, as an activator of SIR2.1 (SIRT1) and AMPK, extends the lifespan of C. elegans (48). The sir2 mutation could obliterate the effect of resveratrol. Natural compounds, such as curcumin or alkylresorcinols, enhance SIRT1 activity and have been confirmed to extend the lifespan (49). These findings indicated that SIR2.1 (SIRT1) could be a promising target for aging interventions. Overall, epigenetic research will be a powerful way for aging interventions of drugs. Lifespan extension for humans could be achieved by powerful genetic tools and further understanding of aging mechanisms in simple invertebrate models.
Conclusion and Future Challenges
As a model system, the nematode C. elegans could be used for studying genetic approaches to understand the aging process, age-related diseases, mechanisms of longevity, and drug screening for compounds that increase lifespan. Longevity studies on this lower organism have helped provide an outline of the signaling pathways involved in aging and predicting their behavior in complex organisms. However, which molecular pathways are causative and which accompany aging need further research. Also, the mechanisms of epigenetic regulation associated with aging are still on the way to be elucidated in depth. In the future, disease models including nematodes and C. elegans will definitely provide further insights into the aging process.
Author Contributions
ZL and GW: conceptualization and resources. SZ and TZ: methodology, literature arrangement, and writing - original draft preparation. SZ, ZL, and FL: writing - review and editing. ZL: funding acquisition. ZL, GW, and FL: supervision. All authors contributed to the article and approved the submitted version.
Funding
The present study was supported by grants from the Department of Science and Technology of Jilin Province (grant no. 20190701040GH was awarded to ZL), Special health project of Jilin Province (JLSWSRCZX2020-0044 was awarded to ZL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
1. ^What is C. elegans and Why Work on it? An Introduction for Those Unfamiliar With the Worm. Available online at: https://cbs.umn.edu/cgc/what-c-elegans.
References
2. Moreno-Arriola E, Cardenas-Rodriguez N, Coballase-Urrutia E, Pedraza-Chaverri J, Carmona-Aparicio L, Ortega-Cuellar D. Caenorhabditis elegans: a useful model for studying metabolic disorders in which oxidative stress is a contributing factor. Oxid Med Cell Longev. (2014) 2014:705253. doi: 10.1155/2014/705253
3. Alexander AG, Marfil V, Li C. Use of Caenorhabditis elegans as a model to study Alzheimer's disease and other neurodegenerative diseases. Front Genet. (2014) 5:279. doi: 10.3389/fgene.2014.00279
4. Schaffitzel E, Hertweck M. Recent aging research in Caenorhabditis elegans. Exp Gerontol. (2006) 41:557–63. doi: 10.1016/j.exger.2006.02.008
5. Golden TR, Melov S. Gene expression changes associated with aging in C. elegans. WormBook. (2007) 12:1–12. doi: 10.1895/wormbook.1.127.2
6. Hertweck M, Hoppe T, Baumeister R. C. elegans, a model for aging with high-throughput capacity. Exp Gerontol. (2003) 38:345–6. doi: 10.1016/S0531-5565(02)00208-5
7. Chew YL, Walker DS, Towlson EK, Vertes PE, Yan G, Barabasi AL, et al. Recordings of Caenorhabditis elegans locomotor behaviour following targeted ablation of single motorneurons. Sci Data. (2017) 4:170156. doi: 10.1038/sdata.2017.156
8. Chen Y, Scarcelli V, Legouis R. Approaches for studying autophagy in Caenorhabditis elegans. Cells. (2017) 6:27. doi: 10.3390/cells6030027
9. Olsen A, Vantipalli MC, Lithgow GJ. Using Caenorhabditis elegans as a model for aging and age-related diseases. Ann N Y Acad Sci. (2006) 1067:120–8. doi: 10.1196/annals.1354.015
10. Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. (2002) 161:1101–12.
11. Pickett CL, Dietrich N, Chen J, Xiong C, Kornfeld K. Mated progeny production is a biomarker of aging in Caenorhabditis elegans. G3 (Bethesda). (2013) 3:2219–32. doi: 10.1534/g3.113.008664
12. Chen CH, Chen YC, Jiang HC, Chen CK, Pan CL. Neuronal aging: learning from C. elegans. J Mol Signal. (2013) 8:14. doi: 10.1186/1750-2187-8-14
13. Liu J, Zhang B, Lei H, Feng Z, Liu J, Hsu AL, et al. Functional aging in the nervous system contributes to age-dependent motor activity decline in C. elegans. Cell Metab. (2013) 18:392–402. doi: 10.1016/j.cmet.2013.08.007
14. Podshivalova K, Kerr RA, Kenyon C. How a mutation that slows aging can also disproportionately extend end-of-life decrepitude. Cell Rep. (2017) 19:441–50. doi: 10.1016/j.celrep.2017.03.062
15. Dilberger B, Baumanns S, Schmitt F, Schmiedl T, Hardt M, Wenzel U, et al. Mitochondrial oxidative stress impairs energy metabolism and reduces stress resistance and longevity of C. elegans. Oxid Med Cell Longev. (2019) 2019:6840540. doi: 10.1155/2019/6840540
16. Martinez G, Duran-Aniotz C, Cabral-Miranda F, Vivar JP, Hetz C. Endoplasmic reticulum proteostasis impairment in aging. Aging Cell. (2017) 16:615–23. doi: 10.1111/acel.12599
17. Qin Z, Johnsen R, Yu S, Chu JS, Baillie DL, Chen N. Genomic identification and functional characterization of essential genes in Caenorhabditis elegans. G3 (Bethesda). (2018) 8:981–97. doi: 10.1534/g3.117.300338
18. Ewald CY, Landis JN, Porter Abate J, Murphy CT, Blackwell TK. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature. (2015) 519:97–101. doi: 10.1038/nature14021
19. Heintz C, Doktor TK, Lanjuin A, Escoubas CC, Zhang Y, Weir HJ, et al. Corrigendum: Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature. (2017) 547:476. doi: 10.1038/nature23313
20. Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P, et al. widespread proteome remodeling and aggregation in aging C. elegans. Cell. (2015) 161:919–32. doi: 10.1016/j.cell.2015.03.032
21. Son HG, Altintas O, Kim EJE, Kwon S, Lee SV. Age-dependent changes and biomarkers of aging in Caenorhabditis elegans. Aging Cell. (2019) 18:e12853. doi: 10.1111/acel.12853
22. Gao AW, Chatzispyrou IA, Kamble R, Liu YJ, Herzog K, Smith RL, et al. A sensitive mass spectrometry platform identifies metabolic changes of life history traits in C. elegans. (2017) 7:2408. doi: 10.1038/s41598-017-02539-w
23. Matsunami K. Frailty and Caenorhabditis elegans as a benchtop animal model for screening drugs including natural herbs. Front Nutr. (2018) 5:111. doi: 10.3389/fnut.2018.00111
24. Choi J, Ahn A, Kim S, Won CW. Global prevalence of physical frailty by fried's criteria in community-dwelling elderly with national population-based surveys. J Am Med Direct Assoc. (2015) 16:548–50. doi: 10.1016/j.jamda.2015.02.004
25. David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. (2010) 8:e1000450. doi: 10.1371/journal.pbio.1000450
26. Weinhouse C, Truong L, Meyer JN, Allard P. Caenorhabditis elegans as an emerging model system in environmental epigenetics. Environ Mol Mutagenesis. (2018) 59:560–75. doi: 10.1002/em.22203
27. Corsi AK, Wightman B, Chalfie M. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics. (2015) 200:387–407. doi: 10.1534/genetics.115.176099
28. Yu G, Wu Q, Gao Y, Chen M, Yang M. The epigenetics of aging in invertebrates. Int J Mol Sci. (2019) 20:4535. doi: 10.3390/ijms20184535
29. Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. (2016) 15:196–207. doi: 10.1111/acel.12427
30. Goh GY, Martelli KL, Parhar KS, Kwong AW, Wong MA, Mah A, et al. The conserved mediator subunit MDT-15 is required for oxidative stress responses in Caenorhabditis elegans. Aging Cell. (2014) 13:70–9. doi: 10.1111/acel.12154
31. Tullet JMA, Green JW, Au C, Benedetto A, Thompson MA, Clark E, et al. The SKN-1/Nrf2 transcription factor can protect against oxidative stress and increase lifespan in C. elegans by distinct mechanisms. Aging Cell. (2017) 16:1191–4. doi: 10.1111/acel.12627
32. Dall KB, Faergeman NJ. Metabolic regulation of lifespan from a C. elegans perspective. Genes Nutr. (2019) 14:25. doi: 10.1186/s12263-019-0650-x
33. Butler JA, Ventura N, Johnson TE, Rea SL. Long-lived mitochondrial (Mit) mutants of Caenorhabditis elegans utilize a novel metabolism. FASEB J. (2010) 24:4977–88. doi: 10.1096/fj.10-162941
34. Gao AW, Canto C, Houtkooper RH. Mitochondrial response to nutrient availability and its role in metabolic disease. EMBO Mol Med. (2014) 6:580–9. doi: 10.1002/emmm.201303782
35. Perez-Jimenez MM, Rodriguez-Palero MJ, Rodenas E, Askjaer P, Munoz MJ. Age-dependent changes of nuclear morphology are uncoupled from longevity in Caenorhabditis elegans IGF/insulin receptor daf-2 mutants. Biogerontology. (2014) 15:279–88. doi: 10.1007/s10522-014-9497-0
36. Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. (2011) 13:668–78. doi: 10.1016/j.cmet.2011.03.018
37. Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Felix AA, et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. (2016) 22:879–88. doi: 10.1038/nm.4132
38. Carranza ADV, Saragusti A, Chiabrando GA, Carrari F, Asis R. Effects of chlorogenic acid on thermal stress tolerance in C. elegans via HIF-1, HSF-1 and autophagy. Phytomedicine. (2020) 66:153132. doi: 10.1016/j.phymed.2019.153132
39. Seah NE, de Magalhaes Filho CD, Petrashen AP, Henderson HR, Laguer J, Gonzalez J, et al. Autophagy-mediated longevity is modulated by lipoprotein biogenesis. Autophagy. (2016) 12:261–72. doi: 10.1080/15548627.2015.1127464
40. Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Ann Rev Biochem. (2008) 77:727–54. doi: 10.1146/annurev.biochem.77.061206.171059
41. Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. (2007) 447:550–5. doi: 10.1038/nature05837
42. Pandit A, Jain V, Kumar N, Mukhopadhyay A. PHA-4/FOXA-regulated microRNA feed forward loops during Caenorhabditis elegans dietary restriction. Aging. (2014) 6:835–55. doi: 10.18632/aging.100697
43. Sheaffer KL, Updike DL, Mango SE. The target of rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. (2008) 18:1355–64. doi: 10.1016/j.cub.2008.07.097
44. Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. (2013) 4:2267. doi: 10.1038/ncomms3267
45. Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. (2010) 1:e10. doi: 10.1038/cddis.2009.8
46. Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, et al. AMPK and SIRT1: a long-standing partnership? Am J Physiol Endocrinol Metab. (2010) 298:E751–60. doi: 10.1152/ajpendo.00745.2009
47. Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. (2013) 153:228–39. doi: 10.1016/j.cell.2013.02.035
48. Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. (2004) 430:686–9. doi: 10.1038/nature02789
49. Kayashima Y, Katayanagi Y, Tanaka K, Fukutomi R, Hiramoto S, Imai S. Alkylresorcinols activate SIRT1 and delay ageing in drosophila melanogaster. Sci Rep. (2017) 7:43679. doi: 10.1038/srep43679
50. Tavernarakis N, Driscoll M. Caloric restriction and lifespan: a role for protein turnover? Mech Ageing Dev. (2002) 123:215–29. doi: 10.1016/S0047-6374(01)00341-4
51. Butler VJ, Gao F, Corrales CI, Cortopassi WA, Caballero B, Vohra M, et al. Age- and stress-associated C. elegans granulins impair lysosomal function and induce a compensatory HLH-30/TFEB transcriptional response. PLoS Genet. (2019) 15:e1008295. doi: 10.1371/journal.pgen.1008295
52. Clancy DJ, Gems D, Hafen E, Leevers SJ, Partridge L. Dietary restriction in long-lived dwarf flies. Science. (2002) 296:319. doi: 10.1126/science.1069366
53. Iser WB, Wolkow CA. DAF-2/insulin-like signaling in C. elegans modifies effects of dietary restriction and nutrient stress on aging, stress and growth. PLoS ONE. (2007) 2:e1240. doi: 10.1371/journal.pone.0001240
54. Murphy CT, Hu PJ. Insulin/insulin-like growth factor signaling in C. elegans. WormBook. (2013) 26:1–43. doi: 10.1895/wormbook.1.164.1
55. Wescott SA, Ronan EA, Xu XZ. Insulin signaling genes modulate nicotine-induced behavioral responses in Caenorhabditis elegans. Behav Pharmacol. (2016) 27:44–9. doi: 10.1097/FBP.0000000000000186
56. Karmacharya R, Sliwoski GR, Lundy MY, Suckow RF, Cohen BM, Buttner EA. Clozapine interaction with phosphatidyl inositol 3-kinase (PI3K)/insulin-signaling pathway in Caenorhabditis elegans. Neuropsychopharmacology. (2009) 34:1968–78. doi: 10.1038/npp.2009.35
57. Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. (1993) 366:461–4. doi: 10.1038/366461a0
58. Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. (1996) 382:536–9. doi: 10.1038/382536a0
59. Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA. (2002) 99:10417–22. doi: 10.1073/pnas.152161099
60. Munoz MJ, Riddle DL. Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics. (2003) 163:171–80.
61. Toker A, Cantley LC. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. (1997) 387:673–6. doi: 10.1038/42648
62. Putker M, Madl T, Vos HR, de Ruiter H, Visscher M, van den Berg MC, et al. Redox-dependent control of FOXO/DAF-16 by transportin-1. Mol Cell. (2013) 49:730–42. doi: 10.1016/j.molcel.2012.12.014
63. Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. (1997) 389:994–9. doi: 10.1038/40194
64. Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. (1997) 278:1319–22. doi: 10.1126/science.278.5341.1319
65. Mukhopadhyay A, Oh SW, Tissenbaum HA. Worming pathways to and from DAF-16/FOXO. Exp Gerontoly. (2006) 41:928–34. doi: 10.1016/j.exger.2006.05.020
66. Labuschagne CF, Stigter EC, Hendriks MM, Berger R, Rokach J, Korswagen HC, et al. Quantification of in vivo oxidative damage in Caenorhabditis elegans during aging by endogenous F3-isoprostane measurement. Aging Cell. (2013) 12:214–23. doi: 10.1111/acel.12043
67. López-Otín C, Galluzzi L, Freije JMP, Madeo F, Kroemer G. Metabolic control of longevity. Cell. (2016) 166:802–21. doi: 10.1016/j.cell.2016.07.031
68. Finkel T. The metabolic regulation of aging. Nature medicine. (2015) 21:1416–23. doi: 10.1038/nm.3998
69. Demoinet E, Roy R. Surviving starvation: AMPK protects germ cell integrity by targeting multiple epigenetic effectors. BioEssays. (2018) 40:1700095. doi: 10.1002/bies.201700095
70. Zhao L, Zhao Y, Liu R, Zheng X, Zhang M, Guo H, et al. The transcription factor DAF-16 is essential for increased longevity in C. elegans exposed to bifidobacterium longum BB68. Sci Rep. (2017) 7:7408. doi: 10.1038/s41598-017-07974-3
71. Kouzarides T. Chromatin modifications and their function. Cell. (2007) 128:693–705. doi: 10.1016/j.cell.2007.02.005
72. Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Ann Rev Biochem. (2005) 74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721
73. Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizábal-Corrales D, et al. DNA methylation on N6-Adenine in C. elegans. Cell. (2015) 161:868–78. doi: 10.1016/j.cell.2015.04.005
74. Luo GZ, Blanco MA, Greer EL, He C, Shi Y. DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes? Nat Rev Mol Cell Biol. (2015) 16:705–10. doi: 10.1038/nrm4076
75. Wasson JA, Simon AK, Myrick DA, Wolf G, Driscoll S, Pfaff SL, et al. Maternally provided LSD1/KDM1A enables the maternal-to-zygotic transition and prevents defects that manifest postnatally. Elife. (2016) 5:e08848. doi: 10.7554/eLife.08848
76. Greer EL, Beese-Sims SE, Brookes E, Spadafora R, Zhu Y, Rothbart SB, et al. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Rep. (2014) 7:113–26. doi: 10.1016/j.celrep.2014.02.044
77. Greer EL, Becker B, Latza C, Antebi A, Shi Y. Mutation of C. elegans demethylase spr-5 extends transgenerational longevity. Cell Res. (2016) 26:229–38. doi: 10.1038/cr.2015.148
78. Uno M, Nishida E. Lifespan-regulating genes in C. elegans. NPJ Aging Mech Dis. (2016) 2:16010. doi: 10.1038/npjamd.2016.10
79. Tullet JM. DAF-16 target identification in C. elegans: past, present and future. Biogerontology. (2015) 16:221–34. doi: 10.1007/s10522-014-9527-y
80. Lan J, Rollins JA, Zang X, Wu D, Zou L, Wang Z, et al. Translational regulation of non-autonomous mitochondrial stress response promotes longevity. Cell Rep. (2019) 28:1050–62.e6. doi: 10.1016/j.celrep.2019.06.078
81. Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. (2011) 479:365–71. doi: 10.1038/nature10572
82. McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size 1935. Nutrition. (1989) 5:155–71.
83. Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. (1996) 273:59–63. doi: 10.1126/science.273.5271.59
84. Zhou JJ, Chun L, Liu JF. A comprehensive understanding of dietary effects on C. elegans Physiology. Curr Med Sci. (2019) 39:679–84. doi: 10.1007/s11596-019-2091-6
85. Anderson RM, Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, et al. Caloric restriction improves health and survival of rhesus monkeys. Nat Commun. (2017) 8:14063. doi: 10.1038/ncomms14063
87. Zhang Y, Lanjuin A, Chowdhury SR, Mistry M, Silva-Garcia CG, Weir HJ, et al. Neuronal TORC1 modulates longevity via AMPK and cell nonautonomous regulation of mitochondrial dynamics in C. elegans. eLife. (2019) 8:e49158. doi: 10.7554/eLife.49158
88. Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. (2007) 282:30107–19. doi: 10.1074/jbc.M705325200
89. Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. (2007) 17:1646–56. doi: 10.1016/j.cub.2007.08.047
90. Gill MS, Olsen A, Sampayo JN, Lithgow GJ. An automated high-throughput assay for survival of the nematode Caenorhabditis elegans. Free Rad Biol Med. (2003) 35:558–65. doi: 10.1016/S0891-5849(03)00328-9
91. Kumar J, Park KC, Awasthi A, Prasad B. Silymarin extends lifespan and reduces proteotoxicity in C. elegans Alzheimer's model. CNS Neurol Disord Drug Targets. (2015) 14:295–302. doi: 10.2174/1871527314666150116110212
92. Cong W, Wang P, Qu Y, Tang J, Bai R, Zhao Y, et al. Evaluation of the influence of fullerenol on aging and stress resistance using Caenorhabditis elegans. Biomaterials. (2015) 42:78–86. doi: 10.1016/j.biomaterials.2014.11.048
93. Pan H, Finkel T. Key proteins and pathways that regulate lifespan. J Biol Chem. (2017) 292:6452–60. doi: 10.1074/jbc.R116.771915
94. Ben Sahra I, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P, et al. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res. (2011) 71:4366–72. doi: 10.1158/0008-5472.CAN-10-1769
95. Casey W, Jacobs A, Maull E, Matheson J, Clarke C, Lowit A. A new path forward: the interagency coordinating committee on the validation of alternative methods (ICCVAM) and national toxicology program's interagency center for the evaluation of alternative toxicological methods (NICEATM). J Am Assoc Lab Anim Sci. (2015) 54:170–3.
Keywords: IGF-1, AMPK, mTOR, dietary restriction, drug screening
Citation: Zhang S, Li F, Zhou T, Wang G and Li Z (2020) Caenorhabditis elegans as a Useful Model for Studying Aging Mutations. Front. Endocrinol. 11:554994. doi: 10.3389/fendo.2020.554994
Received: 23 April 2020; Accepted: 01 September 2020;
Published: 05 October 2020.
Edited by:
Alessandro Cellerino, Normal School of Pisa, ItalyCopyright © 2020 Zhang, Li, Zhou, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuo Li, zhuoli@jlu.edu.cn
 Siwen Zhang
Siwen Zhang Fei Li
Fei Li Tong Zhou
Tong Zhou Guixia Wang
Guixia Wang Zhuo Li
Zhuo Li