High Wolbachia Strain Diversity in a Clade of Dung Beetles Endemic to Madagascar
- 1Department of Bioinformatics and Genetics, Swedish Museum of Natural History, Stockholm, Sweden
- 2Department of Biology, Lund University, Lund, Sweden
- 3Organismal and Evolutionary Biology Research Program, University of Helsinki, Helsinki, Finland
Determining the drivers of diversity is a major topic in biology. Due to its high level of micro-endemism in many taxa, Madagascar has been described as one of Earth's biodiversity hotspot. The exceptional Malagasy biodiversity has been shown to be the result of various eco-evolutionary mechanisms that have taken place on this large island since its isolation from other landmasses. Extensive phylogenetic analyses have, for example, revealed that most of the dung beetle radiation events have arisen due to allopatric speciation, and adaptation to altitudinal and/or longitudinal gradients. But other biotic factors, that have yet to be identified, might also be at play. Wolbachia is a maternally transmitted endosymbiotic bacterium widespread in insects. The bacterium is well-known for its ability to modify its host reproductive system in ways that may lead to either discordance patterns between the host mitochondrial and nuclear phylogenies, and in some cases to speciation. Here, we used the MultiLocus Sequence Typing system, to identify and characterize five Wolbachia strains infecting several species within the Nanos clypeatus dung beetle clade. We discuss the implications of these Wolbachia strains for the evolution and diversification of their dung beetle hosts in Madagascar.
Introduction
With roughly 200,000 species of which at least 90% are endemic to the Island, Madagascar is one of the Earth's richest biodiversity hotspots (Paulian, 1987; Myers et al., 2000; Vences et al., 2009). For Malagasy dung beetles, the level of endemism reaches 96% (Miraldo et al., 2011). Like for many other organisms that have evolved on the island since its isolation from other land masses since the late Cretaceous era (Noonan and Chippindale, 2006), the high level of species radiation in dung beetles has been attributed to different eco-evolutionary mechanisms. Strong environmental gradients and ecosystem heterogeneity across the island have often been suggested as some of the main drivers of the Island's exceptional biodiversity (Vences et al., 2009; Miraldo and Hanski, 2014). In contrast, little is known about the potential influence of biotic factors, such as the presence of endosymbionts, in the diversification of Malagasy species.
The genus Nanos includes 42 species of dung beetles endemic to Madagascar (Montreuil et al., 2014). This clade is considered one of the most recent and ecologically successful dung beetle lineages in Madagascar (Wirta, 2009; Wirta et al., 2010). The phylogeny of this clade was recently revisited by combining morphological and molecular data from both mitochondrial and nuclear markers (Wirta, 2009; Montreuil et al., 2014). This work improved our understanding of the phylogenetic relationship between many morphologically similar Nanos species, but also highlighted potential introgression between species, especially within the N. clypeatus group (Wirta et al., 2008; Wirta, 2009; Montreuil et al., 2014). Furthermore, Wirta (2009) showed that the mitochondrial DNA of the six species from the N. clypeatus group was less variable than that of the rest of the Nanos species.
Maternally inherited endosymbiotic bacteria act as a selfish entities that can modify their host reproductive strategies to their own benefit (O'Neill et al., 1997; Dyson and Hurst, 2004; Engelstädter and Hurst, 2007). Text-book examples include the association between Wolbachia strains inducing cytoplasmic incompatibility (CI) in their hosts. Due to CI, the zygotes of uninfected female hosts mated to Wolbachia-infected males die, while zygotes from Wolbachia-infected females thrive in the environment (Carrington et al., 2011; Kriesner et al., 2013; Turelli et al., 2018). In these conditions, Wolbachia can obscure studies of mitochondrial variation in the host population. The mitochondrial selective sweep associated with the invasion of the inherited endosymbiotic bacteria can indeed reduce the mitochondrial diversity observed in infected lineages when compared to uninfected ones, and confuse conclusions from phylogenetic studies (Jiggins, 2003; Charlat et al., 2009; Jäckel et al., 2013; Kriesner et al., 2013; Mazur et al., 2016; Cariou et al., 2017). Furthermore, studies have suggested that the induction of CI can also select for premating isolation between matrilines carrying different infection status (Telschow et al., 2005a, 2007). The selection pressure for the evolution of mate-choice strategies toward individuals carrying the same infection status can lead to strong premating isolation, which in time will support reproductive isolation between the host lineages, and lead to speciation (Telschow et al., 2005a, 2007; Cariou et al., 2017).
Here we suggest that infections with Wolbachia strains might play a role in the loss of mitochondrial diversity observed in the N. clypeatus group, which includes six dung beetle species. We also hypothesize that incompatible Wolbachia infections could keep the different taxa as clear evolutionary units despite clear genetic introgression patterns between the N. dubitatus and N. vieittei species. We discuss the potential implications of these findings for the evolution and diversity of the dung beetles in Madagascar.
Materials and Methods
Material
The Nanos beetles used in this study were collected across Madagascar (Figure 1) between 2002 and 2008, for the purpose of several other studies investigating speciation in the Malagasy Nanos clade (Wirta, 2009; Miraldo and Hanski, 2014). Beetles were collected with standard baited pitfall-traps. The 1,5 dl plastic cups were filled to one third with water and dishwashing liquid to decrease water tension upon contact with beetles. A bait of raw fish (V = 3 cm3) was hanged over each cup to attract the beetles. Each trap was covered by a large leave or a plastic plate to prevent overflow with rain water. Duplouy et al. (2009) previously showed that Wolbachia cross-specimens contamination was not occurring during collection and storage in similar conditions.
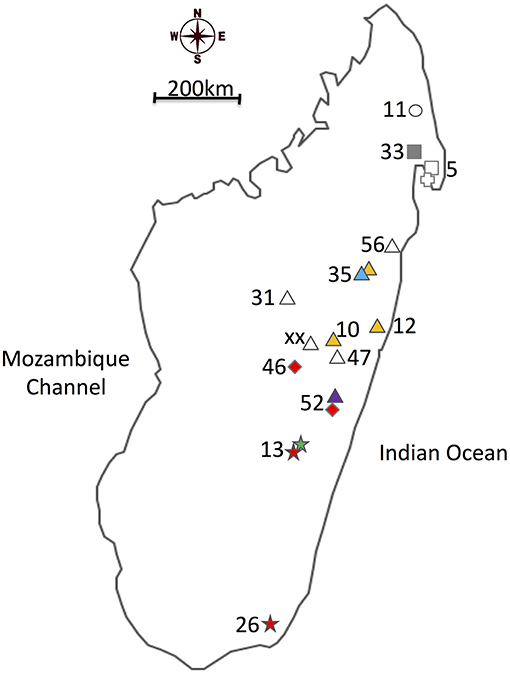
Figure 1. Map of the different sites where the Nanos beetles were collected colored according to the main Wolbachia strain infection. Collection sites are labeled as described in Table 1. Shapes are specific to host species, while colors inform on the Wolbachia infection status of the samples. (Cross): Nanos nitens, (Square): N. clypeatus, (Circle): N. mirjae, (Triangle): N. dubitatus, (Diamond): N. pseudoviettei, and (Star): N. viettei. (Yellow): strain A, (Red): strain B, (Green): strain C, (Blue): strain D, (Gray): strain E, and (White): Uninfected.
Our study focuses on the six species composing the “Nanos clypeatus clade,” namely N. clypeatus, N. dubitatus, N. veittei, N. pseudoviettei, N. mirjae, and N. nitens (Miraldo and Hanski, 2014; Montreuil et al., 2014). The species have differentiated at most 7 Mya, and show very little geographical overlap across Madagascar, due to strong niche competition between the species. The three most common species, N. veittei, N. dubitatus, and N. clypeatus, are, respectively, found in the South Eastern region, the Northern half region, and around the Masoala peninsula region of Madagascar (Wirta, 2009). One exception occurs in central Madagascar, where N. dubitatus, and N. pseudoviettei occur in sympatry in the locality of Vohitrombo-Marolambo. It is at this particular locality that specimens showing patterns of introgression between the two species were previously collected (Viljanen, 2009a; Wirta, 2009; Viljanen et al., 2010). Table 1 and Figure 1 provides sample size for each species and population sampled.
Molecular Work
We individually extracted the DNA from 119 dung beetles using a Qiagen DNAeasy blood and tissues extraction kit and following the manufacturer's protocol (Qiagen, Germany). The COI mitochondrial gene was amplified by PCR using the primer pairs LCO/HCO (Folmer et al., 1994). Although endosymbiotic bacteria are taxonomically diverse; including members of the bacterial genera Arsenophonus, Cardinium, Rickettsia, Spiroplasma, and Wolbachia; we here focused solely on infection by Wolbachia, which is considered the most common of them all, including in Coleopteran hosts (Kajtoch and Kotaskova, 2018; Kolasa et al., 2018). We screened for Wolbachia infection using five Wolbachia MLST markers (MultiLocus Sequence Typing genes: coxA, fbpA, ftsZ, gatB, and hcpA, using respective degenerate primers designed by Baldo et al. (2006). Amplicons from both reverse and forward strands were sequenced on an automated AB1-3730-DNA Analyser (Applied BiosystemsTM, USA). Sequences were checked for consistency between strands, and manually curated using Geneious R6 (http://www.geneious.com) (Kearse et al., 2012). All Wolbachia sequences were deposited to GenBank (GenBank # MK636654- MK636666). Unfortunately, as is often the case in many studies (Rodriguero et al., 2010; Roehrdanz and Wichmann, 2014), we failed to amplify each of the five MLST loci for several strains, and only assigned a full strain variant name when two or more of the MLST loci were sequenced. Variant names were assigned following the standard rules given on the pubMLST database (http://pubmlst.org/wolbachia) (Baldo et al., 2006). All sequences were compared to the pubMLST database using BLASTn (Baldo et al., 2006).
Phylogenetics
Phylogenetic trees were constructed using the online tree-building software Phylogeny.fr using the One Click mode with default settings (Dereeper et al., 2008). In brief, the One-Click method builds a maximum likelihood tree using PhyML (Guindon et al., 2010) with sequence alignment using MUSCLE (Edgar, 2004a,b). We rooted the COI gene tree using the COI sequences from N. manomboensis and N. binotatus (GenBank KF309798 and KF309750) (Miraldo et al., 2011). According to Montreuil et al. (2014), N. manobonensis and N. binotatus belong to the N. bimaculatus group, which falls outside the “N. clypeatus clade.” The respective MLST sequences from 14 additional Wolbachia strains (wPip, wRi, wMel, wBm, wBol2, wBol1, wBani_B, and wHho; GenBank #AM999887, CP001391, AE017196, AE017321, EF025179–183, EF078895, AB474245–249, AB094382, KY658553, KY658572, KY658591, KY658610, KY658630, KF722987-992; and wAu, wNo, wSni, wAsap_A, wCalt_B, wTcon_B; Wolbachia-PubMLST id#10, #27, #301, #3, #19 and #20, respectively), previously characterized as belonging to the A-, B-, or D-supergroup, were added to the Wolbachia phylogenetic trees. We first constructed a phylogenetic tree for each MLST locus separately (Figure S1), and then built a phylogeny based on the concatenated sequences (Figure 2). We rooted each tree using the D-supergroup strain, wBm, as the outgroup.
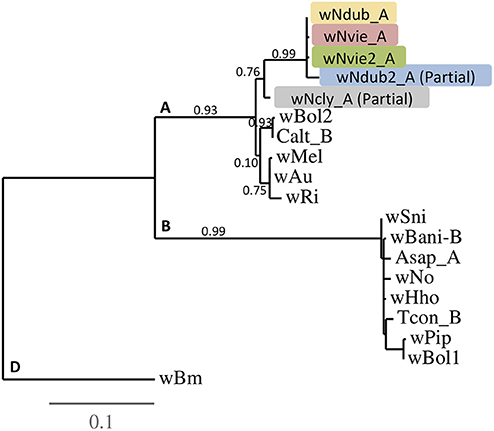
Figure 2. Phylogenetic tree of the five different Wolbachia strains characterized from the N. clypeatus clade, with bootstrap values. Input sequences were concatenated sequences from all five MLST (2,081 bp total), except for the partially characterized strains wNdub2_A (three MLSTs: 1,249 bp), and wNcly_A (five MLSTs, with partial hcpA sequence: 1,887 bp) (Table S1). Additional strains from the A-, B-, and D-supergroups were characterized from butterflies (wBol1, wBol2, wBani_B), fruitflies (wRi, wMel, wAu, wNo), mosquitoes (wPip), wasps (wHho), beetles (wSni, Asap_A, Calt_B, Tcon), and nematodes (wBm). The D-supergroup wBm strain was used as the outgroup for the tree.
Results
Wolbachia Infections
We characterized a total of five Wolbachia strains (Figure 2) from four of the six species screened (N. clypeatus, N. veittei, N. pseudoviettei, and N. dubitatus). All strain variants belong to the A-supergroup, and show little divergence between each other. The wNdub_A and wNvie_A strain variants differ by one substitution out of 2081bp sequenced (99.95% similarity), wNdub_A and wNvie2_A also differ by one substitution at a different position, while wNvie_A and wNvie2_A differ by two substitutions. Strains wNdub2_A and wNcly_A are only partial, but more divergent from the other three strain variants. There are 23 polymorphic sites between strain variants wNdub_A and wNdub2_A, and 42 polymorphic sites between strains wNdub_A and wNcly_A within the 1,887 bp sequenced.
The penetrance of the different strains varies greatly among Nanos species and populations (Table 1). The strain variant wNdub_A was common in three of the eight populations of N. dubitatus collected across the species range. The strain variant wNvie_A was found in all N. viettei and all N. pseudoviettei specimens included in this study. The strain variant wNvie2_A was found only in two N. veittei specimens collected in Ranomafana. Strikingly, the wNvie2_A-infected specimens are the same N. veittei specimens previously described as being the results of introgression events with N. dubitatus (Figure 3, Wirta, 2009). Strain variant wNdub2_A was found in one N. dubitatus sample collected in Zahamena. We also suggest multiple infections in four N. dubitatus samples collected in Zahamena. The sequences from those specimens consistently showed double peaks at several nucleotide positions, similar to double infection with strain variants wNdub_A, and wNdub2_A. Unfortunately, we could not confirm the identity of each variant independently for these multiply infected specimens, so we remain cautious with this particular result. Finally, the strain wNcly_A was described from one unique N. clypeatus specimen. The position of the five strain variants described in this study is similar between each phylogenies (Figure S1 and Figure 1), suggesting lack of recombination between the different Wolbachia of the N. clypeatus dung beetles clade.
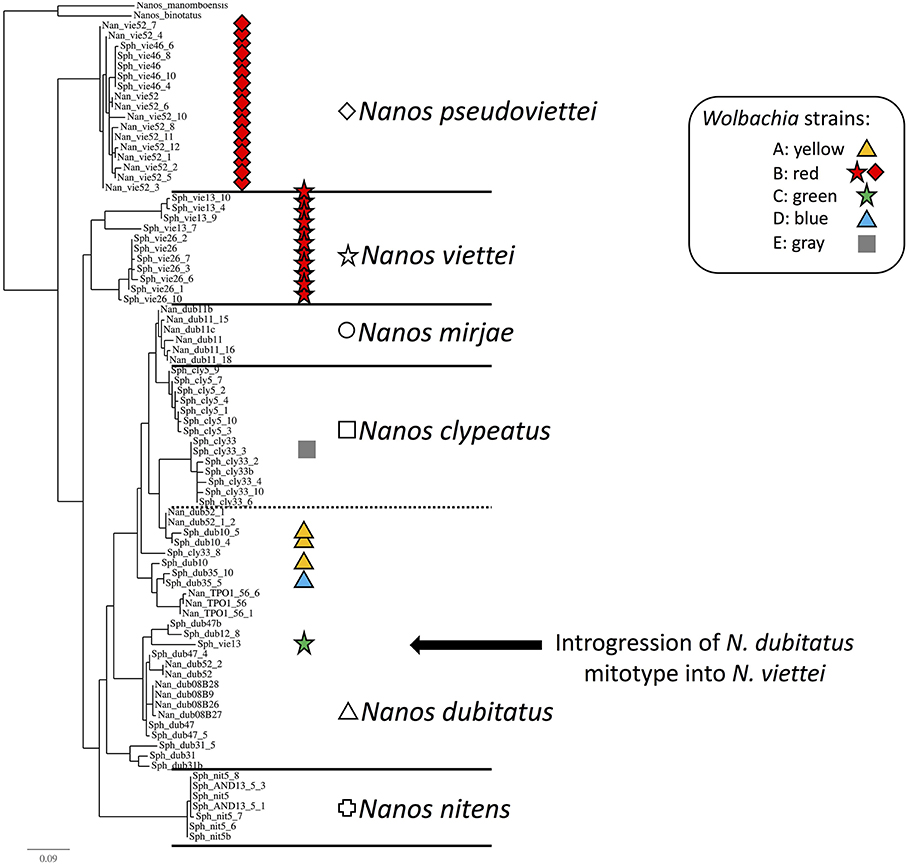
Figure 3. Phylogenetic tree of the Malagasy dung beetles based on the COI mitochondrial gene. Shapes are specific to host species, while colors inform on the Wolbachia infection status of the samples. (Cross): Nanos nitens, (Square): N. clypeatus, (Circle): N. mirjae, (Triangle): N. dubitatus, (Diamond): N. pseudoviettei, and (Star): N. viettei. (Yellow): strain A, (Red): strain B, (Green): strain C, (Blue): strain D, (Gray): strain E, and (White): Uninfected. The uncharacterized strain detected in three N. dubitatus is not included here. Nanos binotatus and N. manomboensis COI sequences from GenBank were used as outgroup for the tree. Introgression of N. dubitatus mtDNA in N. viettei is highlighted by the black arrow.
Mitochondrial Diversity
We characterized 67 mitochondrial DNA haplotypes (mitotype) from the 119 beetles included in this study (5 to 17 haplotypes per species). None of the mitotypes characterized from Wolbachia-free beetles were found in Wolbachia-infected beetles, with the exception of the mitotype associated with Wolbachia strain wNcly_A, which is also found in two uninfected individuals. The strain variant wNdub_A is found associated to three of the 17 mitotypes (18%) characterized from N. dubitatus. The strain variant wNvie_A is found in association to all 26 mitotypes described from both the N. veittei and N. pseudoveittei samples (N = 35 beetles). The strain variant wNvie2_A is found associated to the unique mitotype characterized from the two N. veittei specimens introgressed with N. dubitatus. Both strain wNdub2_A and strain wNcly_A were each characterized from one specimen, each carrying their own unique mitotype.
Discussion
Our study is the first to report on Wolbachia infections in Malagasy dung beetles. We detected Wolbachia infection in 56 specimens (47%) from four of the six dung beetle species screened (67%). This is consistent with current estimates proposing that over 40% of worldwide insect species (Stouthamer et al., 1999), and not so different from lower infection rate estimates detected in other families and species of Coleoptera (Kawasaki et al., 2016; Kajtoch and Kotaskova, 2018; Kolasa et al., 2018). Five of the Wolbachia variants we characterized belong to the Wolbachia A-supergroup, and are genetically very similar to each other (at most 42 polymorphic sites across 1,800 bp sequenced). The last infection remains to be characterized, or even confirmed. These results suggest the island is potentially not only a biodiversity hotspot for dung beetles, but also a diversity hotspot for the endosymbiotic bacteria they carry.
Each strain shows a very different prevalence within the respective host species. Studies have shown that variations in the prevalence of Wolbachia are not rare in insects (Sazama et al., 2019), and these patterns of infection can be the results of the host, the symbiont, or both (Charlat et al., 2005; Hornett et al., 2006). The successful spread of the Wolbachia strain wNvie_A in all N. pseudoviettei specimens, and almost all N. viettei specimens may suggest that the strain is beneficial to its host. As mentioned earlier, CI-inducing strains might rapidly spread in their host populations (Turelli et al., 2018), as the infected females produce viable offspring from mating with either infected or uninfected males, while survival of the offspring from uninfected females is restricted to the females mated to uninfected males (O'Neill et al., 1997; Telschow et al., 2005b). It is thus possible that wNvie_A is a CI-inducing strain. However, Wolbachia can also spread across their host populations through enhancing other aspect of its host's fitness. Recent studies have shown that Wolbachia can promote the survival of its host against pathogens such as viruses (Hedges et al., 2008; Teixeira et al., 2008; Osborne et al., 2012). Many dung beetles lay their eggs directly in patches of dung or carrions, or in underground chambers filled with these materials (Viljanen, 2009a,b). The Nanos beetles are all rollers, rolling away samples of dung or carrions on which their larvae later feed and develop (Hanski et al., 2008; Rahagalala et al., 2009; Viljanen, 2009b; Viljanen et al., 2010). These environments are often rich in pathogenetic micro-organisms, including bacteria, viruses, and fungi, that may threaten the survival of the offspring. Whether the common strain wNvie_A, or any strain described in this study, can protect their dung beetle hosts against pathogens from their environment remains to be investigated.
In the tropical butterfly Hypolimnas bolina, Duplouy et al. (2010) have previously shown that geographical variations in the prevalence of a Wolbachia strain across the South Pacific populations of the butterfly results from temporal variations in the history of the symbiosis on the different islands. Populations showing a low infection rate have thus more recently acquired the infection than populations showing a higher infection rate (Duplouy et al., 2010). The strain variant wNvie_A is found in almost all N. veittei and N. pseudoveittei specimens, and in association with a wide diversity of mitotypes in these species, we thus suggest that these host-symbiont associations might be rather old. The other five strains are much rarer in their respective host populations. This pattern might either suggest that those strains are new to their respective hosts and restricted to few mitotypes in the populations, or that the strains are being purge from their host populations, due to potential costs of the symbiont (Charlat et al., 2009; Duplouy et al., 2010).
Nanos viettei is a taxonomic unit that is, based on both the mitochondrial and nuclear DNA, very well-separated from all other species from the N. clypeatus group (Montreuil et al., 2014). Wirta (2009) calculated that the split between this species and the other five species in the group occurred at most 7.2 Mya. All strain variants characterized in these dung-beetle species are very similar to each other, but their phylogenetic tree is not congruent with that of their host. Such incongruence has been reported in many host-symbiont associations (Jäckel et al., 2013; Sontowski et al., 2015; Kolasa et al., 2019), including Wolbachia-infected insects of Madagascar (Linares et al., 2009; Zouache et al., 2011). Although it is possible that some of our Nanos dung-beetle species inherited their Wolbachia infections from a common ancestor, while others have lost or gained new infections during species radiation, our data also does not suggest radiation of Wolbachia in parallel to the radiation of N. clypeatus group.
Although mostly vertically transmitted from the mother to the offspring, Wolbachia is also known to transfer horizontally between host species (Vavre et al., 1999; Bailly-Bechet et al., 2017). Such transfer of infection may occur during hybridization events between species. For example, in Drosophila mauritania, different populations bear the mitotypes of another fruitfly species, D. simulans, and variants of the Wolbachia strain wMau, most likely due to transfer of both cytoplasmic entities during recent introgression events (Rousset and Solignac, 1995). However, other ecological links are likely to also support the horizontal transfer of symbiotic bacteria between host species. Species with similar resources, such as larval host plant, or species sharing parasites, such as parasitoid or mite infections, may thus also share similar Wolbachia infections (Stahlhut et al., 2010). Ecological studies on the Nanos species have shown that N. veittei mate with other Nanos species in the laboratory (Viljanen, 2009a,b). The survival of the offspring has however never been investigated. Furthermore, previous phylogenetic studies do not suggest introgression between the populations of N. pseudoveittei and N. viettei (Miraldo and Hanski, 2014). In contrast, the two species co-occur in the northern limit of N. viettei geographical range (Viljanen, 2009a; Miraldo et al., 2011; Montreuil et al., 2014). There, these two species, which are generalists in their choice of dung, may enter in contact and share dung patches or parasites. These ecological links may have facilitated the horizontal transfer of Wolbachia between the two species (Kolasa et al., 2019). Unfortunately, little is known about the parasitoid community associated to the Malagasy dung beetles, and no study has ever investigated the potential for dung as a suitable environment for symbiont transfer between carrion insects.
Strikingly, some of the rare strain variants characterized in this study are found in association with specimens showing clear genetic patterns of introgression. Strain variant wNvie2_A, a close variant of wNvie_A, is found in two N. viettei that carry the mitochondrial DNA from N. dubitatus. These particular specimens were collected from a region where extensive hybridization is known to happen between the two sympatric host species (Viljanen, 2009a; Wirta, 2009). Previous studies described these introgressed individuals as clear units within the N. viettei species that were however separated by unknown processes (Miraldo and Hanski, 2014). We suggest that sympatry of the two species may have facilitated the transfer of the Wolbachia between specimens. However, the divergence between strain variants wNvie_A and wNvie2_A may have led these specimens to become incompatible potentially due to CI. Such postzygotic incompatibility would have, with time, led to radiation of the introgressed lineage carrying wNvie2_A from the other N. viettei, wNvie_A-infected lineages. Simultaneously, because wNvie2_A-infected N. viettei individuals are morphologically similar to N. viettei, they might rarely mate with the locally occurring N. dubitatus specimens. Again, with time, prezygotic isolation may have also isolated this lineage from the other N. dubitatus lineages. The strain wNdub_A, a second close variant of wNvie_A, is found in N. dubitatus specimens. Both strains and their respective host species do not occur in sympatry in Madagascar. Although wNdub_A was identified in association to a rather wide diversity of mitotypes in N. dubitatus, our current sample size is unfortunately too small to further speculate on the potential phenotypic effects this strain might have on its hosts, or on the origin of the strain in N. dubitatus.
Despite the fact that our restricted sample size is preventing a more comprehensive assessment of prevalence rates and any evidence of geographical variation between collection locations, our results still support the idea that Wolbachia might play a key role in the ecology and evolutionary biology of the endemic Malagasy dung beetles. Viljanen (2009b) showed that some Nanos species can mate in the laboratory, thus it is possible for future studies to confirm whether the different strains of Wolbachia induce CI in these insects. Only then will we be able to confirm whether Wolbachia play a key role in designing host species boundaries, or can in any ways enhance their host fitness and contribute to niche colonization and rapid geographical expansion of these ecological successful dung beetle species.
Data Availability
The datasets generated for this study can be found in GenBank, Accession codes are provided in the manuscript.
Ethics Statement
The study only include work on DNA extracts from samples collected under the Academy of Finland projects to late professor Ilkka Hanski. All samples were collected prior to 2014, no Nagoya protocol was then required.
Author Contributions
AD designed the research and wrote the manuscript. AM and AD collected the data, analyzed the data, and revised previous versions of the manuscript.
Funding
The project was funded by the Academy of Finland (#266021 to AD), and the Marie-Curie Sklodowska Individual fellowship (#790531, Host Sweet Home to AD). Samples were collected under the Academy of Finland funding (Finnish CoE Programme-Grant #133132, #256453, and #250444 to late professor Ilkka Hanski).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank T. Nyman for her help with the molecular work, and Prof. N. Wahlberg for comments on the manuscript. We would like to thank the two reviewers for their comments that help improve the quality of our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00157/full#supplementary-material
References
Bailly-Bechet, M., Martins-Simoes, P., Szöllosi, G. J., Mialdea, G., Sagot, M. F., and Charlat, S. (2017). How long does Wolbachia remain on board? Mol. Biol. Evol. 34, 1183–1193. doi: 10.1093/molbev/msx073
Baldo, L., Dunning Hotopp, J. C., Jolley, K. A., Bordenstein, S. R., Biber, S. A., and Choudhury, C., et al. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72, 7098–7110. doi: 10.1128/AEM.00731-06
Cariou, M., Duret, L., and Charlat, S. (2017). The global impact of Wolbachia on mitochondrial diversity and evolution. J. Evol. Biol. 30, 2204–2210. doi: 10.1111/jeb.13186
Carrington, L. B., Lipkowitz, J. R., Hoffmann, A. A., and Turelli, M. (2011). A re-examination of Wolbachia-induced cytoplasmic incompatibility in California Drosophila simulans. PLoS ONE 6:e22565. doi: 10.1371/journal.pone.0022565
Charlat, S., Duplouy, A., Hornett, E. A., Dyson, E. A, Davies, N., and Hurst, G. D., et al. (2009). The joint evolutionary histories of Wolbachia and mitochondria in Hypolimnas bolina. BMC Evol. Biol. 9:64. doi: 10.1186/1471-2148-9-64
Charlat, S., Hornett, E. A., Dyson, E. A., Ho, P. P., Loc, N. T., and Schilthuizen, N., et al. (2005). Prevalence and penetrance variation of male-killing Wolbachia across Indo-Pacific populations of the butterfly Hypolimnas bolina. Mol. Ecol. 14, 3525–3530. doi: 10.1111/j.1365-294X.2005.02678.x
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., and Dufayard, S., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue), W465–469. doi: 10.1093/nar/gkn180
Duplouy, A., Hurst, G. D., O'Neill, S. L., and Charlat, S. (2010). Rapid spread of male-killing Wolbachia in the butterfly Hypolimnas bolina. J. Evol. Biol. 23, 231–235. doi: 10.1111/j.1420-9101.2009.01891.x
Duplouy, A., Vermenot, C., Davies, N., Roderick, G., Hurst, G. D., and Charlat, S. (2009). Assessing risks of Wolbachia DNA cross-specimen contamination following mass collection and ethanol storage. Mol. Ecol. Resour. 9, 46–50. doi: 10.1111/j.1755-0998.2008.02421.x
Dyson, E. A., and Hurst, G. D. (2004). Persistence of an extreme sex-ratio bias in a natural population. Proc. Natl. Acad. Sci. U.S.A. 101, 6520–6523. doi: 10.1073/pnas.0304068101
Edgar, R. C. (2004a). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Edgar, R. C. (2004b). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Engelstädter, J., and Hurst, G. D. (2007). The impact of male-killing bacteria on host evolutionary processes. Genetics 175, 245–254. doi: 10.1534/genetics.106.060921
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Guindon, S., Dufayard, J.F, Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hanski, I., Wirta, H., Nyman, T., and Rahagalala, P. (2008). Resource shifts in Malagasy dung beetles: contrasting processes revealed by dissimilar spatial genetic patterns. Ecol. Lett. 11, 1208–1215. doi: 10.1111/j.1461-0248.2008.01239.x
Hedges, L. M., Brownlie, J.C, O'Neill, S.L, and Johnson, K. N. (2008). Wolbachia and virus protection in insects. Science 322:702. doi: 10.1126/science.1162418
Hornett, E. A., Charlat, S., Duplouy, A.M, Davies, N., Roderick, G. K., and Wedell, N., et al. (2006). Evolution of male-killer suppression in a natural population. PLoS Biol. 4:e283. doi: 10.1371/journal.pbio.0040283
Jäckel, R., Mora, D., and Dobler, S. (2013). Evidence for selective sweeps by Wolbachia infections: phylogeny of Altica leaf beetles and their reproductive parasites. Mol. Ecol. 22, 4241–4255. doi: 10.1111/mec.12389
Jiggins, F. M. (2003). Male-killing Wolbachia and mitochondrial DNA: selective sweeps, hybrid introgression and parasite population dynamics. Genetics 164, 5–12.
Kajtoch, L., and Kotaskova, N. (2018). Current state of knowledge on Wolbachia infection among Coleoptera: a systematic review. PeerJ 6:e4471. doi: 10.7717/peerj.4471
Kawasaki, Y., Schuler, H., Stauffer, C., Lakatos, F., and Kajimura, H. (2016). Wolbachia endosymbionts in haplodiploid and diploid scolytine beetles (Coleoptera: Curculionidae: Scolytinae). Environ. Microbiol. Rep. 8, 680–688. doi: 10.1111/1758-2229.12425
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., and Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kolasa, M., Kubisz, D., Gutowski, J.M, Scibior, R., Mazur, M.A, and Holecova, M., et al. (2018). Infection by endosymbiotic male-killing bacteria in Coleoptera. Folia Biol. Krakow 66, 165–177. doi: 10.3409/fb_66-4.18
Kolasa, M., Scibior, R., Mazur, M.A, Kubisz, D., Dudek, K., and Kajtoch, L. (2019). How hosts taxonomy, trophy, and endosymbionts shape microbiome diversity in beetles. Microb Ecol. doi: 10.1007/s00248-019-01358-y. [Epub ahead of print].
Kriesner, P., Hoffmann, A. A., Lee, S. F., Turelli, M., and Weeks, A. R. (2013). Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 9:e1003607. doi: 10.1371/journal.ppat.1003607
Linares, M. C., Soto-Calderón, I. D., Lees, D. C., and Anthony, N. M. (2009). High mitochondrial diversity in geographically widespread butterflies of Madagascar: a test of the DNA barcoding approach. Mol. Phylogenet. Evol. 50, 485–495. doi: 10.1016/j.ympev.2008.11.008
Mazur, M. A., Holecová, M., Lachowska-Cierlik, D., Lis, A., Kubisz, D., and Kajtoch, L. (2016). Selective sweep of Wolbachia and parthenogenetic host genomes - the example of the weevil Eusomus ovulum. Insect Mol. Biol. 25, 701–711. doi: 10.1111/imb.12255
Miraldo, A., and Hanski, I. A. (2014). Competitive release leads to range expansion and rampant speciation in malagasy dung beetles. Syst. Biol. 63, 480–492. doi: 10.1093/sysbio/syu011
Miraldo, A., Wirta, H., and Hanski, I. (2011). Origin and diversification of dung beetles in Madagascar. Insects 2, 112–127. doi: 10.3390/insects2020112
Montreuil, O., Viljanen, H., and Miraldo, A. (2014). Evolution of the Malagasy endemic genus Nanos Westwood, 1842 (Coleoptera, Scarabaeidae, Epilissini). Syst. Entomol. 39, 442–459. doi: 10.1111/syen.12063
Myers, N., Mittermeier, R. A., Mittermeier, C. G., da Fonseca, G. A. B., and Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853–858. doi: 10.1038/35002501
Noonan, B. P., and Chippindale, P. T. (2006). Vicariant origin of malagasy reptiles supports late cretaceous antarctic land bridge. Am. Nat. 168, 730–741. doi: 10.1086/509052
O'Neill, S., Hoffman, A., and Werren, J. (1997). Influential Passengers: Inherited Microorganisms and Arthropod Reproduction. Oxford: Oxford University Press.
Osborne, S. E., Iturbe-Ormaetxe, I., Brownlie, J. C., O'Neill, S. L., and Johnson, K. N. (2012). Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans. Appl. Environ. Microbiol. 78, 6922–6929. doi: 10.1128/AEM.01727-12
Paulian, R. (1987). The scarabaeidae beetles of some Tropical Islands. Bull. Soc. Zool. France-Evolu. Zool. 112, 255–275.
Rahagalala, P., Viljanen, H., Hottola, J., and Hanski, I. (2009). Assemblages of dung beetles using cattle dung in Madagascar. Afr. Entomol. 17, 71–89. doi: 10.4001/003.017.0109
Rodriguero, M. S., Confalonieri, V. A., Guedes, J. V., and Lanteri, A. A. (2010). Wolbachia infection in the tribe Naupactini (Coleoptera, Curculionidae): association between thelytokous parthenogenesis and infection status. Insect Mol Biol. 19, 631–640. doi: 10.1111/j.1365-2583.2010.01018.x
Roehrdanz, R. L., and Wichmann, S. S. (2014). Wolbachia multilocus sequence typing of singly infected and multiply infected populations of Northern corn rootworm (Coleoptera: Chrysomelidae). Ann. Entomol. Soc. Am. 107, 832–841. doi: 10.1603/AN14006
Rousset, F., and Solignac, M. (1995). Evolution of single and double Wolbachia symbioses during speciation in the Drosophila simulans complex. Proc. Natl. Acad. Sci. U.S.A. 92, 6389–6393. doi: 10.1073/pnas.92.14.6389
Sazama, E. J., Ouellette, S. P., and Wesner, J. S. (2019). Bacterial endosymbionts are common among, but not necessarily within, insect species. Environ. Entomol. 48, 127–133. doi: 10.1093/ee/nvy188
Sontowski, R., Bernhard, D., Bleidorn, C., Schlegel, M., and Gerth, M. (2015). Wolbachia distribution in selected beetle taxa characterized by PCR screens and MLST data. Ecol. Evol. 5, 4345–4353. doi: 10.1002/ece3.1641
Stahlhut, J. K., Desjardins, C. A., Clark, M. E., Baldo, L., Russell, J. A., and Werren, J. H., et al. (2010). The mushroom habitat as an ecological arena for global exchange of Wolbachia. Mol. Ecol. 19, 1940–1952. doi: 10.1111/j.1365-294X.2010.04572.x
Stouthamer, R., Breeuwer, J. A., and Hurst, G. D. (1999). Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53, 71–102. doi: 10.1146/annurev.micro.53.1.71
Teixeira, L., Ferreira, A., and Ashburner, M. (2008). The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6:e2. doi: 10.1371/journal.pbio.1000002
Telschow, A., Flor, M., Kobayashi, Y., Hammerstein, P., and Werren, J. H. (2007). Wolbachia-induced unidirectional cytoplasmic incompatibility and speciation: mainland-island model. PLoS ONE 2:e701. doi: 10.1371/journal.pone.0000701
Telschow, A., Hammerstein, P., and Werren, J. H. (2005a). The effect of Wolbachia vs. genetic incompatibilities on reinforcement and speciation. Evolution 59, 1607–1619. doi: 10.1111/j.0014-3820.2005.tb01812.x
Telschow, A., Yamamura, N., and Werren, J. H. (2005b). Bidirectional cytoplasmic incompatibility and the stable coexistence of two Wolbachia strains in parapatric host populations. J. Theor. Biol. 235, 265–274. doi: 10.1016/j.jtbi.2005.01.008
Turelli, M., Cooper, B. S., Richardson, K. M., Ginsberg, P. S., Peckenpaugh, B., and Antelope, C. X., et al. (2018). Rapid global spread of wRi-like Wolbachia across multiple drosophila. Curr. Biol. 28, 963–971 e968. doi: 10.1016/j.cub.2018.02.015
Vavre, F., Fleury, F., Lepetit, D., Fouillet, P., and Boulétreau, M. (1999). Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 16, 1711–1723. doi: 10.1093/oxfordjournals.molbev.a026084
Vences, M., Wollenberg, K. C., Vieites, D. R., and Lees, D. C. (2009). Madagascar as a model region of species diversification. Trends Ecol. Evolu. 24, 456–465. doi: 10.1016/j.tree.2009.03.011
Viljanen, H. (2009b). Life history of Nanos viettei (Paulian, 1976) (Coleoptera: Scarabaeidae: Canthonini), a representative of an endemic clade of dung beetles in Madagascar. Coleopterists Bull. 63, 265–288. doi: 10.1649/1184.1
Viljanen, H., Wirta, H., Montreuil, O., Rahagalala, P., Johnson, S., and Hanski, I. (2010). Structure of local communities of endemic dung beetles in Madagascar. J. Trop. Ecol. 26, 481–496. doi: 10.1017/S0266467410000325
Wirta, H. (2009). Complex phylogeographical patterns, introgression and cryptic species in a lineage of Malagasy dung beetles (Coleoptera: Scarabaeidae). Biol. J. Linnean Soc. 96, 942–955. doi: 10.1111/j.1095-8312.2008.01156.x
Wirta, H., Orsini, L., and Hanski, I. (2008). An old adaptive radiation of forest dung beetles in Madagascar. Mol. Phylogen. Evolu. 47, 1076–1089. doi: 10.1016/j.ympev.2008.03.010
Wirta, H., Viljanen, H., Orsini, L., Montreuil, O., and Hanski, I. (2010). Three parallel radiations of Canthonini dung beetles in Madagascar. Mol. Phylogenet. Evolu. 57, 710–727. doi: 10.1016/j.ympev.2010.08.013
Zouache, K., Raharimalala, F. N., Raquin, V., Tran-Van, V., Raveloson, L. H., and Ravelonandro, P., et al. (2011). Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol. Ecol. 75, 377–389. doi: 10.1111/j.1574-6941.2010.01012.x
Keywords: endosymbiont, speciation, endemism, phylogeny, introgression
Citation: Miraldo A and Duplouy A (2019) High Wolbachia Strain Diversity in a Clade of Dung Beetles Endemic to Madagascar. Front. Ecol. Evol. 7:157. doi: 10.3389/fevo.2019.00157
Received: 14 March 2019; Accepted: 23 April 2019;
Published: 08 May 2019.
Edited by:
David Vieites, Spanish National Research Council (CSIC), SpainReviewed by:
Michał Robert Kolasa, Institute of Systematics and Evolution of Animals (PAN), PolandThomas Walker, London School of Hygiene and Tropical Medicine (LSHTM), United Kingdom
Copyright © 2019 Miraldo and Duplouy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:Anne Duplouy, anne.duplouy@biol.lu.se
 Andreia Miraldo
Andreia Miraldo Anne Duplouy
Anne Duplouy