Not so Dangerous After All? Venom Composition and Potency of the Pholcid (Daddy Long-Leg) Spider Physocyclus mexicanus
- 1Department of Biology, Lewis & Clark College, Portland, OR, United States
- 2Spider Pharm, Yarnell, AZ, United States
- 3Center for Genome Research and Biocomputing, Oregon State University, Corvallis, OR, United States
- 4Arizona Proteomics Consortium, University of Arizona, Tucson, AZ, United States
Pholcid spiders (Araneae: Pholcidae), officially “cellar spiders” but popularly known as “daddy long-legs,” are renown for the potential of deadly toxic venom, even though venom composition and potency has never formally been studied. Here we detail the venom composition of male Physocyclus mexicanus using proteomic analyses and venom-gland transcriptomes (“venomics”). We also analyze the venom's potency on insects, and assemble available evidence regarding mammalian toxicity. The majority of the venom (51% of tryptic polypeptides and 62% of unique tryptic peptides) consists of proteins homologous to known venom toxins including enzymes (astacin metalloproteases, serine proteases and metalloendopeptidases, particularly neprilysins) and venom peptide neurotoxins. We identify 17 new groups of peptides (U1−17-PHTX) most of which are homologs of known venom peptides and are predicted to have an inhibitor cysteine knot fold; of these, 13 are confirmed in the proteome. Neprilysins (M13 peptidases), and astacins (M12 peptidases) are the most abundant venom proteins, respectively representing 15 and 11% of the individual proteins and 32 and 20% of the tryptic peptides detected in crude venom. Comparative evidence suggests that the neprilysin gene family is expressed in venoms across a range of spider taxa, but has undergone an expansion in the venoms of pholcids and may play a central functional role in these spiders. Bioassays of crude venoms on crickets resulted in an effective paralytic dose of 3.9 μg/g, which is comparable to that of crude venoms of Plectreurys tristis and other Synspermiata taxa. However, crickets exhibit flaccid paralysis and regions of darkening that are not observed after P. tristis envenomation. Documented bites on humans make clear that while these spiders can bite, the typical result is a mild sting with no long-lasting effects. Together, the evidence we present indicates pholcid venoms are a source of interesting new peptides and proteins, and effects of bites on humans and other mammals are inconsequential.
Introduction
Pholcid spiders (Araneae: Pholcidae) are an example of disconnect between public perception, and scientific understanding. Commonly called “cellar spiders” some pholcids are also referred to as “daddy long-legs” and have been maligned in popular culture as having venoms with the highest mammalian potency of all spiders, that would be lethal if they only had fangs that could penetrate human skin. Pholcids, in reality, are a family of spiders that has been diversifying for over 200 million years (Dimitrov et al., 2012; Eberle et al., 2018) and are among the top six percent of the most diverse spider families (1,714 species in 94 genera, World Spider Catalog, February 2019). Reports of high mammalian potency emerge from rumor and non-scientific anecdote. We are not aware of any scientific work to date that has attempted to characterize venom composition of any pholcid species, and given their evolutionary depth and diversity, they provide remarkable opportunity to discover new toxins and lend insight into the broader patterns of venom diversity within spider venoms, while directly investigating a widespread urban myth.
Spiders of the family Pholcidae are interesting animals in their own right. Found on all continents except Antarctica, they range in size from minute to ~1 cm in adult body length. They tend to have long, thin (hair-like) legs that are readily dropped defensively and twitch to deter predators after being removed from the body (e.g., Johnson and Jakob, 1999). As generalist foragers of arthropods, they build tangly, non-adhesive webs in protected places, and diversification within the family has involved extensive diversification of web microhabitats (Eberle et al., 2018). In prey capture, they either wait for prey to encounter their web, or some species leave their web and enter webs of other spiders and capture them and/or consume their egg sacs or prey (Jackson and Brassington, 1987). When pholcids encounter prey, they immobilize them by first wrapping with silk, and then biting to envenomate and eventually feed (Jackson and Brassington, 1987). Many common pholcids readily feed on other spiders, including black widows (Latrodectus) (pers. obs.) that have highly potent venoms on both arthropods and vertebrates (Wang et al., 2007). Given that spiders are dangerous prey, this observation inspires the question of how potent pholcid venoms are on arthropods. While Kirchner and Opderbeck (1990) provide data suggesting venoms of Pholcus phalangiodes only weakly affect insect prey that are ultimately killed by digestive enzymes, we have much to learn about the details of pholcid venom composition and how the venoms affect prey. Although small, their fangs can certainly penetrate the cuticle of many insects.
Beyond insects, the fangs of adult male Physocyclus mexicanus are able to pierce uncalloused human skin (Kristensen, pers obs). In fact, the only recorded human bites we are aware of that are publically available are from Mythbusters (Episode 13, premiered 15 Feb 2004), in which male P. mexicanus bit from aggression resulting in a brief mild sting (https://www.discovery.com/tv-shows/mythbusters/videos/daddy-longlegs-minimyth). The same episode also reports the only direct tests of mammalian toxicity of daddy-long leg venom with injections of male P. mexicanus venom into mice. In comparative injections of the same volume (one μl, ~80 μg, Kristensen pers obs), venom from Western black widows Latrodectus hesperus, were more potent than P. mexicanus. Despite the fact that obtaining one μl required venom extraction from 50–100 adult males (Kristensen pers obs), the Mythbuster assays suggest that much larger amounts of venom would be required to obtain an LD50 for P. mexicanus venom on mice. Together, these first reports suggest bites from this species are not harmful to mammals. We expand on these observations in our discussion.
Shifting focus from potential risks to humans, venoms of pholcids and all spiders are exciting phenotypes because their rich chemical diversity contains a natural storehouse within which we can discover new molecular activities, and study patterns of diversity and mechanisms of evolution that have generated these phenotypes (e.g., Escoubas, 2006; King and Hardy, 2013). Venoms contain a complex mixture of bioactive components—proteins, peptides, and small molecules—that vary widely across the over 48,000 described species of spiders (Pineda et al., 2018; World Spider Catalog., 2019). A growing body of research characterizing venoms from diverse representatives of spiders is allowing inference of the phylogenetic distribution of venom components. Pholcids are in a higher lineage of spiders called Synspermiata (Michalik and Ramirez, 2014), and more specifically in the “lost trachea clade” (Wheeler et al., 2016). Within the last few years, select venoms in this clade have been characterized allowing inference of the presence and distribution of gene families that are expressed in venom. Venomic sampling to date includes venoms of Plectreurys tristis (Zobel-Thropp et al., 2014a), Scytodes thoracica (Zobel-Thropp et al., 2014b), and venoms within the family Sicariidae that includes brown recluse spiders (review in Chaves-Moreira et al., 2017). Adding a representative pholcid venom to this comparative set allows inference about the phylogenetic distribution and relative abundance of widespread gene families that contribute to spider venoms.
The goal of this study was to use transcriptomic and proteomic methods to characterize venom proteins of the North American pholcid, Physocyclus mexicanus. Our sampling includes venom from adult males, and transcriptomes with mRNAs from adult males and females sampled from the same population used in the Mythbusters episode. To our knowledge, this investigation is the first proteomic characterization of pholcid venom. We consider venom composition of this species in the context of what is known about venoms of other Synspermiata spiders, and discuss evidence for abundant and diverse astacins, comparable to their near relative Plectreurys tristis, and an expanded role of neprilysin homologs and diverse peptides with and without homology to known toxins. To better understand venom effects on natural prey, we use bioassays to evaluate potency on crickets. We also summarize information available on the propensity of these animals to bite humans and the effects of venom on mammals. Together, these results provide an important contribution to comparative venom analyses, and more formally address the urban myth of daddy long-leg spiders having exceptional venom potency on humans.
Materials and Methods
Spider and Venom Collection
All spiders used in this work were adult Physocyclus mexicanus Banks from a local population in a single building at Spider Pharm (Yarnell, AZ 34° 13′12.34” N; 112° 44′56.43” W; 4,796 ft elev). After spiders were unfed for at least 1 week, venom was extracted using electrostimulation as in Binford and Wells (2003). This was done under two circumstances. The first at Lewis & Clark College (L&C) was intended to induce upregulation of expression to optimize capture of venom mRNAs. In parallel, at Spider Pharm, spiders were milked for proteomics analyses and insect bioassays, taking care to avoid contamination with regurgitate. Electrostimulation did not yield sufficient, pure samples of venom from females in either lab. Rather, at L&C of 12 adult females, six yielded tiny drops of venom, three of these were contaminated by vomit. However between 500 and 600 adult males were milked at Spider Pharm to obtain sufficient uncontaminated venom for proteomics and bioassays. Therefore, all methods requiring crude venom were done with venom from males only collected at Spider Pharm, but venom-gland transcriptomes were obtained from a combination of glands from males and females as described below. Voucher specimens are retained in the L&C Natural History collections, and deposited in The National Museum of Natural History.
Proteomic Analyses
LC-MS/MS Detection of Tryptic Peptides From Crude Venom
Crude venom extracted at SpiderPharm (~80 μg from ~100 male spiders) was sent to the Arizona Proteomics Consortium (University of Arizona) for proteomic analysis. Proteins were analyzed from an in-gel tryptic digest (Figure 1A) with LC-MS/MS using an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) as in Zobel-Thropp et al. (2014a,b). The sample contained a mixture of two digests using trypsin (Princeton Separations) in the presence of ProteaseMAX (Promega, manufacturer's protocol). One digest was from the excision of an entire lane containing 10 μg of crude venom (12% Tris-glycine SDS-PAGE) and the other was an excised region of <10 kDa only from a separate gel (25 μg crude venom, 16.5% Tris-tricine SDS), thereby attempting to enrich the sample with small venom polypeptides and peptides which are prone to diffusing out of the protein gel. Samples were desalted using OMIX C18 tips (Agilent Technologies), injected onto a Proxeon Easy nano-HPLC (Proxeon, Odense) precolumn (100 μm, 2 cm, Fisher Scientific) and separated on a C18 column (75 μm, 10 cm, Fisher Scientific) at a flow rate of 300 nl/min over a 145 min gradient of 5–40% solvent B (A = H2O), B = acetonitrile, both with 0.1% (v/v) formic acid). Eluate was delivered by a nanospray source (Nanomate, Advion) with a voltage of 1.85 kV to the Orbitrap Velos. Survey scans were acquired at 60,000 resolution and the top 14m/z values were selected for CID (width = 2 amu) at 35% relative energy in the Velos linear trap and then placed on an exclusion list for 45s.
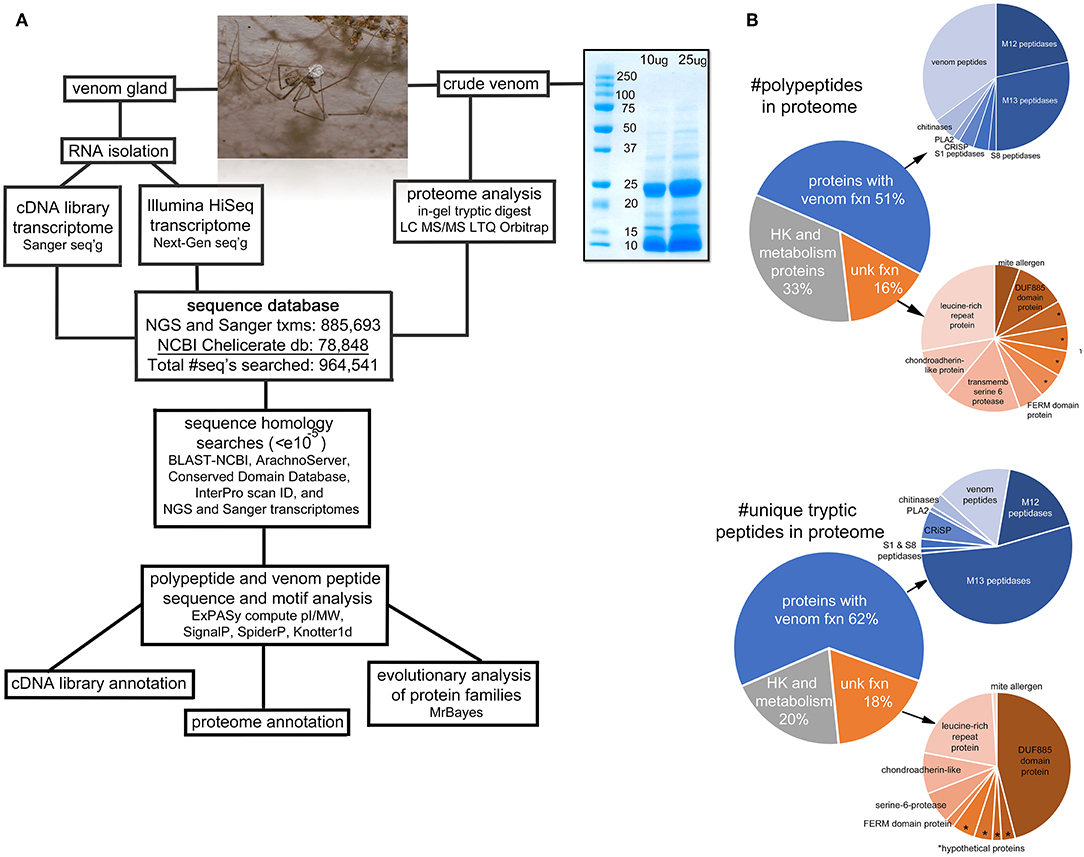
Figure 1. Overview of the project. (A) A flow-chart illustrating the sequence of steps of the procedures for transcriptomic and proteomic analyses. The photo is an adult male Physocyclus mexicanus carrying an egg sac. The gel image is an electrophoretic separation of male P. mexicanus crude venom used for the in-gel tryptic digest. (B) Pie charts of major categories of components identified in the proteome by overall transcript abundance, and proportion of unique tryptic peptides detected by Orbitrap. Asterisks (*) indicate hypothetical proteins with unknown function.
Resulting MS/MS data was searched using SEQUEST on Discoverer (v. 1.3.0.339, Thermo Fisher Scientific) against masses of theoretical fragments from two databases. The first included a combined set of sequences generated by the Binford lab including Illumina HiSeq NGS data from 12 Synspermiata species (described below), and Sanger cDNA library sequences from 10 species as in Zobel-Thropp et al. (2014b) (described below). The second included all chelicerate sequences in NCBI (downloaded 2/17/2016), totaling 964,541 sequences. Matches required <10 ppm precursor error. The reversed database was searched to provide quality scores for FDR filtering. The SEQUEST output was organized in Scaffold (version Scaffold_4.8.6, Proteome Software Inc., Portland, OR; Searle, 2010). Peptides were identified with 95% minimum threshold and 0% false discovery rate (FDR) and proteins were identified with 95% minimum threshold, two peptides minimum per protein, and 0% FDR. Details are in Table S2. While our goal for this analysis is thorough representation of peptides present in male crude venom, there are a number of standard reasons peptides or proteins may be missed. These include (but are not limited to) sample degradation or low concentration for mass spectrometry requirements, incomplete gel extraction, poor or no tryptic cleavage, lack of peptide binding to the C18 column, poor ionization or fragmentation, or posttranslational modification that not were not included in the data search. Proteome profiling data will be available through the PRIDE repository, and amino acid and cDNA sequences for all proteins detected in this study are deposited in GenBank (NGS accession numbers MN004877-MN004966) and included in Table S1.
Transcriptome Database
RNA Isolation
A single pool of RNA isolated from 12 adult P. mexicanus was used for generation of Sanger cDNA libraries, and Illumina 3000 transcriptomes. To up-regulate venom transcripts, venom was extracted from 12 spiders across two consecutive days. Six were milked on 1 day (three females and three males) and six spiders were milked the following day (three females and three males). Then, on the same day—three and 4 days after extraction—all 12 spiders were anesthetized with CO2 and venom glands were extracted and flash-frozen immediately in liquid nitrogen. Given differential success in obtaining venom, only glands from spiders from which venom was extruded were used for mRNA isolation. We pooled glands between sexes to optimize total mRNA, and generate a thorough within-species transcriptome with the goal of annotating the proteome. Transcriptional timing is not known for pholcids, however this window of time has been successful for isolating venom gland mRNAs from other Synspermiata spiders, and combining three and four-day transcripts likely increased the breadth of transcripts captured (Binford et al., 2005; Zobel-Thropp et al., 2014a,b). Total RNA was isolated using the ChargeSwitch Total RNA Cell kit (Invitrogen) following the manufacturer's protocol.
Sanger cDNA Library Construction and Screening
We constructed the venom gland cDNA library using the SMARTTM cDNA library construction kit (Clontech). Details of all procedures including library construction, cDNA packaging, screening and sequence analysis are the same as in Zobel-Thropp et al. (2014b). For this library, we used 94 ng of total RNA for first strand synthesis of cDNA and followed the manufacturer's protocol for library construction. The titer for this library was 1.4 × 106 pfu/ml. We screened 1,685 clones using PCR and sequenced 311 cDNAs ≥500 bp on an Applied Biosystems 3730 Analyzer at the Genomic Analysis and Technology Core (University of Arizona) using TriplEx2 vector-specified primers. We trimmed vector sequence and assembled sequences using Sequencher 5.1 (Gene Codes Corp.). Due to chromatogram ambiguity, we discarded 32 sequences, resulting in 279 high quality sequences for transcriptomic analysis.
Illumina 3000 Transcriptomes
From the same P. mexicanus RNA sample used for the Sanger cDNA library, we sequenced a transcriptome using Illumina 3000 at the Center for Genome Research and Biocomputing (CGRB) at Oregon State University. This was done as part of a larger project that included RNA samples from 23 transcriptomes from 11 species, All sequences resulting from these taxa were combined into a single dataset used to annotate the P. mexicanus proteome. The taxon representation in the dataset includes venom-gland and whole body transcriptomes from Plectreurys tristis and Scytodes thoracica (venom gland RNA is from the same samples used in Zobel-Thropp et al., 2014a,b, respectively), Austrarchaea mainae, Periegops suterii, Hexopthalma dolichocephalus, Drymusa serrana, Loxosceles spinulosa, L. rufescens, L. arizonica, and L. reclusa. For each of these samples, total RNA pools (ChargeSwitch, Invitrogen) were quantified and analyzed for quality using a Nano chip on the Agilent 2100 bioanalyzer at the CGRB. For NGS transcriptome construction, each total RNA pool was enriched for mRNA and prepared for multiplexed sequencing with PrepX reagents for RNA-Seq library construction (Wafergen). The final libraries were checked for quality and size distribution followed by qPCR analysis for quality control. Transcriptomes were multiplexed into two Illumina HiSeq 3000 150 bp paired end lanes.
Transcriptome Assembly
Each transcriptome sample was individually de novo assembled using Trinity (Grabherr et al., 2011). Coding regions were identified from the de-novo transcriptome assemblies with TransDecoder (Haas et al., 2013). The three-step TransDecoder process was used: first, TransDecoder.LongOrfs was used to identify the long open reading frames; second, BLASTP (Altschul et al., 1990) was used against a set of known proteins [spider proteins from GenBank (Benson et al., 2005), ArachnoServer (Pineda et al., 2018), previously isolated Sanger cDNAs from the Binford lab, Latrodectus hesperus (https://www.hgsc.bcm.edu/arthropods/western-black-widow-spider-genome-project), Loxosceles reclusa (https://www.hgsc.bcm.edu/arthropods/brown-recluse-spider-genome-project), Centruroides exilicauda (https://www.hgsc.bcm.edu/arthropods/bark-scorpion-genome-project)]; third, TransDecoder.Predict was used in conjunction with the TransDecoder.LongOrfs output and the protein output to predict the likely coding regions. Output contigs from TransDecoder were combined with the known proteins described above and the final two steps of TransDecoder were run a second time.
To make sure the TransDecoder steps did not remove any essential contigs a second comparison was done against the original Trinity assemblies. Contigs in the Trinity assembly not identified from the TransDecoder process were aligned to previously isolated Sanger venom-gland cDNAs (Binford lab, unpublished) to identify important but missing genes. Sequences identified here that had hits in the above alignment were then aligned back to the TransDecoder peptides, if no hit was found they were added to the final TransDecoder set.
Proteome Annotation
Our analysis pipeline for sequence annotation is illustrated in Figure 1. The final set of proteins was annotated into functional categories based on results from combined searches in the Conserved Domain Database (CDD v3.16, https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), BLAST searches in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and ArachnoServer (Pineda et al., 2018), http://www.arachnoserver.org) public databases, and our database containing sequences from several Synspermiata venom gland cDNA libraries constructed in the Binford lab (unpublished). Hits were recognized as significant if e-values were ≤ 10−5. All of the sequences that were detected by peptide spectra in mass spec analyses (116 unique polypeptides) were annotated manually using the following steps that largely follow those in Zobel-Thropp et al. (2014a).
We identified 38 unique polypeptides with close homology to proteins known to be involved in conserved metabolic, housekeeping, or maintenance functions. Hits had strong e-values (e10−20 to 0.0), and all matches agreed between hits in CDD, InterProScan, and NCBI (Table S1). Polypeptides that hit domains previously described as venom proteins were categorized as “proteins with venom function.” Sequences that had no hits in CDD, InterProScan, or NCBI (60) included peptides with characteristics consistent with venom peptides (secreted with characteristic disulfide patterns) and were categorized as “venom peptides.” Finally, 18 polypeptides were categorized as “proteins of unknown function,” because they either had strong matches to proteins that were not considered housekeeping/metabolic proteins (14, e10−29 to 0.0) or they did not have any hits in any databases (4), so potential function in venom could not be determined.
With the exception of sequences that hit housekeeping/metabolic proteins in our initial homology searches, we identified homologous sequences within venom proteomes across our Synspermiata dataset, and aligned those with homologs in NCBI using MAFFT followed with manual editing in Mesquite [Version 3.51 (build 898), http://mesquiteproject.org]. We identified conserved characteristics consistent with known venom toxins (e.g., cysteine-rich), including signal peptide cleavage sites with SpiderP (http://www.arachnoserver.org/spiderP.html) and knottin or ICK predicted folding patterns using Knotter1d (http://www.dsimb.inserm.fr/KNOTTIN/knoter1d.php) tools. All full-length sequences were analyzed for signal peptide cleavage sites using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/). Protein coding sequences were considered full-length if they contained a methionine and a stop codon.
Sanger cDNA Library Annotation
The cDNA library consists of 251 sequences that were aligned, analyzed, and separated into 24 clusters and 60 singletons (individual sequences) (Figure S1, Table S3). Of those in the original dataset (279), sequences from two clusters and 11 singletons were deemed “no open reading frame,” and 28 sequences were omitted from the dataset because they hit a sequence in NCBI, but when translated they were repetitive and did not contain an ORF. Our annotation of the Sanger cDNA sequences followed the same steps as those detailed above.
Given evidence of high levels of index swapping in our Illumina sequences, we only use our Illumina sequences as a database source for mass spec analyses. We limit our analysis of the transcriptome to sequences isolated from our Sanger cDNA library of P. mexicanus for which we are confident in the taxonomic and tissue source. The cDNA library includes combined transcripts of both male and female spiders and highly expressed homologs that we did not detect in the male only proteome may provide insight into female venom composition.
All non-redundant sequences >200 bp were submitted to GenBank and granted accession numbers (KP138796-KP138905).
Phylogenetic Analyses
We estimated phylogenetic relationships of the most abundant toxins detected in our proteomes to help interpret diversity within the group, and to infer how these toxins are related to homologs in NCBI and proteomes of other Synspermiata. To do this we identified sets of related sequences using BLAST with a threshold of e < 10−5, aligned them initially with MAFFT online server (http://MAFFT.cbrc.jp/alignment/server/) by manual alignment in Mesquite v3.2. Bayesian phylogenies were inferred using MrBayes XSEDE through the CIPRES portal. Analyses were run for 50 million generations, or until the average standard deviation of split frequencies decreased below 0.01.
Venom Potency Analysis
Insect Bioassays
To assess the potency of pholcid venom, we quantified the dose at which 50% of crickets (Acheta domesticus) injected were paralyzed after 60 min (PD50). We diluted venom in physiologically buffered lepidopteran saline solution (5 mM KH2PO4/100 mM KCl/4 mM NaCl/15 mM MgCl2/2 mM CaCl2, pH 6.5) and established a dosage range distributed around the PD50 by performing preliminary injections. Experimental doses were 0.25, 0.50, 1.0, 1.5, and 2.0 μg of crude venom protein with two replicate assays each on 15 crickets per dose by injection into the dorsal mesothorax using a PB-600 dispenser with a gastight syringe (Hamilton Co.). With each replicate, we injected an additional 15 control crickets with 0.4 μl of saline, a volume equal to the largest dose volume. Paralysis was scored every 10 min for 1 h based on the ability of a cricket to right itself. PD50 values were generated for both absolute amounts of venom injected (μg) and for weight standardized dose (μg/g cricket) using the EPA Probit Analysis Program for calculation of LC/EC values (version 1.5 SAS).
Results and Discussion
We detail below the chemical composition of P. mexicanus venoms with evidence that they include an expanded presence of neprilysin and astacin-like metalloproteases, and a diverse set of peptide toxins. The venom causes rapid, flaccid paralysis on crickets with potency that is within the norm of those estimated for other Synspermiata spiders. A collection of observations of bites on humans and subsequent effects indicate that P. mexicanus can bite through human skin, however the effect is minimal.
Venom Composition
A total of 2,021 tryptic peptides were detected via Orbitrap MS of our crude venom sample. These match 116 unique polypeptide sequences from our combined databases with >95% identity (Figure 1A). Of these, 33% have close homology with proteins that are likely involved in basic cellular function (Table S1), however these are represented by only 20% of the unique peptide spectra (Figure 1B). Within the remaining 78 polypeptides, the majority are homologs of known venom proteins (39) or venom peptides (21), and the remaining 18 are homologs of proteins of unknown function (Table S1; Figure 1B).
Toxic (and Potentially Toxic) Venom Proteins
Metallopeptidases are the most abundant components we detected in the venom, as quantified either by number of spectra, or unique polypeptides. These are primarily neprilysins (M13 peptidases) (17 polypeptides, 53% of the unique spectra) and astacin metalloproteases (M12 peptidases) (13 polypeptides, 18% of unique spectra) (Figure 1B). Other peptidases were present in lower abundance (1-2 polypeptides each) including neuroendocrine convertase (S8 peptidase), and chymotrypsin-like (S1 peptidase). Beyond peptidases, we also detected in low abundance (1-3 polypeptides each) chitinase, two cysteine rich secretory proteins (CRISP), and phospholipase A2.
Neprilysins (M13 peptidases)
The apparent dominance of neprilysins in P. mexicanus venom is unprecedented in spider venom proteomes, and likely indicates an important functional role. Neprilysins are thermolysin-like zinc metalloendopeptidases (Turner et al., 2001). They are typically membrane-bound proteins that can turn off peptide signaling events at the cell surface, and metabolize regulatory peptides (Turner et al., 2001). As venom components, they are predicted to degrade extracellular matrices around synapses, and possibly facilitate toxin access to targets (Undheim et al., 2013). Of the 17 unique sequences we detected in the venom proteome, only one was full length (MN004884) and was specifically hit uniquely by 110 spectra. This transcript has a signal peptide and codes for a mature protein of 697 residues with an estimated molecular weight of 79.7 kDa (Table S1). MN004884 aligns well with homologs from a broad set of taxonomic sources (Figure 2) and retains the conserved zinc binding motif HExxH…ExxA/GD (Turner et al., 2001) present in Gluzincins (Sterchi et al., 2008). The other sequence fragments range in length from 90 - 504 amino acids, and were all detected by anywhere from 4–24 unique spectra (Table S1).
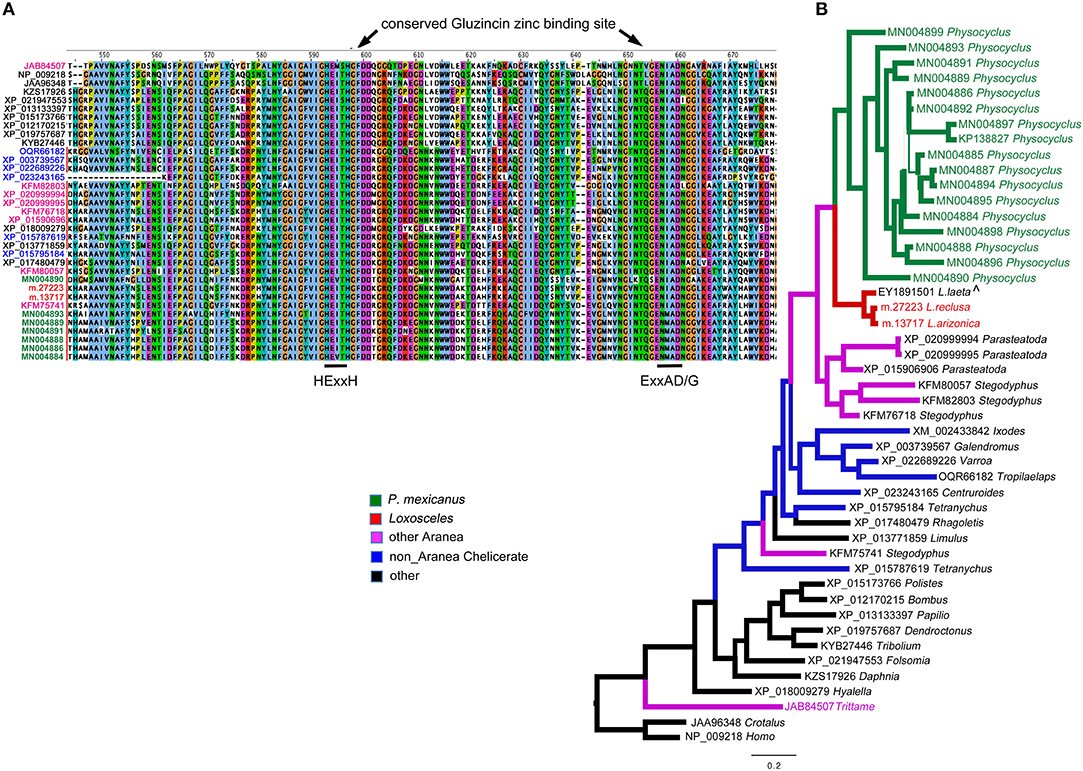
Figure 2. Neprilysins (M13 peptidases) identified in Physocyclus mexicanus venom are homologs of neprilysins from diverse organisms. (A) An alignment of the region of the conserved active sites of neprilysins with conserved binding sites labeled based on Sterchi et al. (2008). Sequence names are color coded by taxonomic source groups. (B) A Bayesian phylogeny of neprilysins with branches color coded by source group. Branch width correlates with posterior probability support for that branch. Names of taxa are in color when they have been confirmed to be in venom proteomes. The label ∧ indicates that sequence was identified in a venom gland EST.
Within spiders, neprilysins have been found in venoms of the barychelid mygalomorph Tritamme loki (Undheim et al., 2013), however in lower abundance than we see in P. mexicanus. We have detected single proteins in each of Loxosceles arizonica and L. reclusa venom proteomes, but in no other Synspermiata species. Interestingly, even with updated annotations (unpublished) based on our expanded NGS database, we have not detected neprilysin homologs in P. tristis venoms (Zobel-Thropp et al., 2014a), the closest relatives of P. mexicanus with characterized venoms.
Outside of spiders, homologs of neprilysins have been detected in snake venoms including timber rattlesnakes (annotated as SVMPs) Crotalus horridus (Rokyta et al., 2013, 2015), and cobras Naja kaouthia (Tan et al., 2017), however, they do not dominate any of the other venoms. Phylogenetic analysis with taxon inclusion of all venom expressed spider neprilysins, representative arthropod homologs detected in NCBI, the Crotalus venom toxin and a human homolog, indicate that the diverse set of homologs detected in P. mexicanus venom are monophyletic and sister to neprilysins in Loxosceles venoms (Figure 2A). The T. loki sequences are highly divergent from the Synspermiata clade. The latter shares a closer relationship with diverse chelicerate proteins detected in genomes than they do with the venom-expressed T. loki, making convergent recruitment into venoms from a diverse neprilysin lineage a reasonable explanation of this pattern. The detection of single proteins in Loxosceles suggests that neprilysins may be phylogenetically widespread in venoms at low levels with expansion of the diversity and perhaps functional role within a lineage including P. mexicanus.
Astacin metalloproteinases (M12 peptidases)
Homologs of astacin metalloproteinases (M12 peptidases) are the second most abundant venom proteins detected in P. mexicanus proteomes. Among the 13 distinct polypeptides, seven are full length and all have signal peptides (Table S1). The diversity of homologs includes seven polypeptide subgroups with <95% aa identity and separate into four distinct clades (Figure 3). Similar to neprilysins, M12 peptidases represent a gene family that is widespread in spider venoms, and homologs have been well described as contributors to venom function in Loxosceles (annotated as Loxosceles astacin-like proteases – LALPs; da Silveira et al., 2007; Trevisan-Silva et al., 2013), with up to three distinct proteins detected in proteomes of South American species (reviewed in Chaves-Moreira et al., 2017). The diversity of astacin-like metalloproteinases in the P. mexicanus venom proteome is comparable to that detected in P. tristis (Zobel-Thropp et al., 2014a). There are scattered M12 peptidase homologs in other proteomes of Synspermata (Figure 3), however, only P. tristis and P. mexicanus have more than three distinct homologs. Phylogenetic analyses including all venom proteome-confirmed M12 peptidase homologs across Synspermiata resolve into a monophyletic P. mexicana group that is derived from a paraphyletic P. tristis set of proteins (Figure 3). This pattern is consistent with an evolutionary expansion of the role of astacins in venom before the common ancestor of these two species, with further diversification in the pholcid lineage after the divergence from the shared ancestor with Plectreurys.
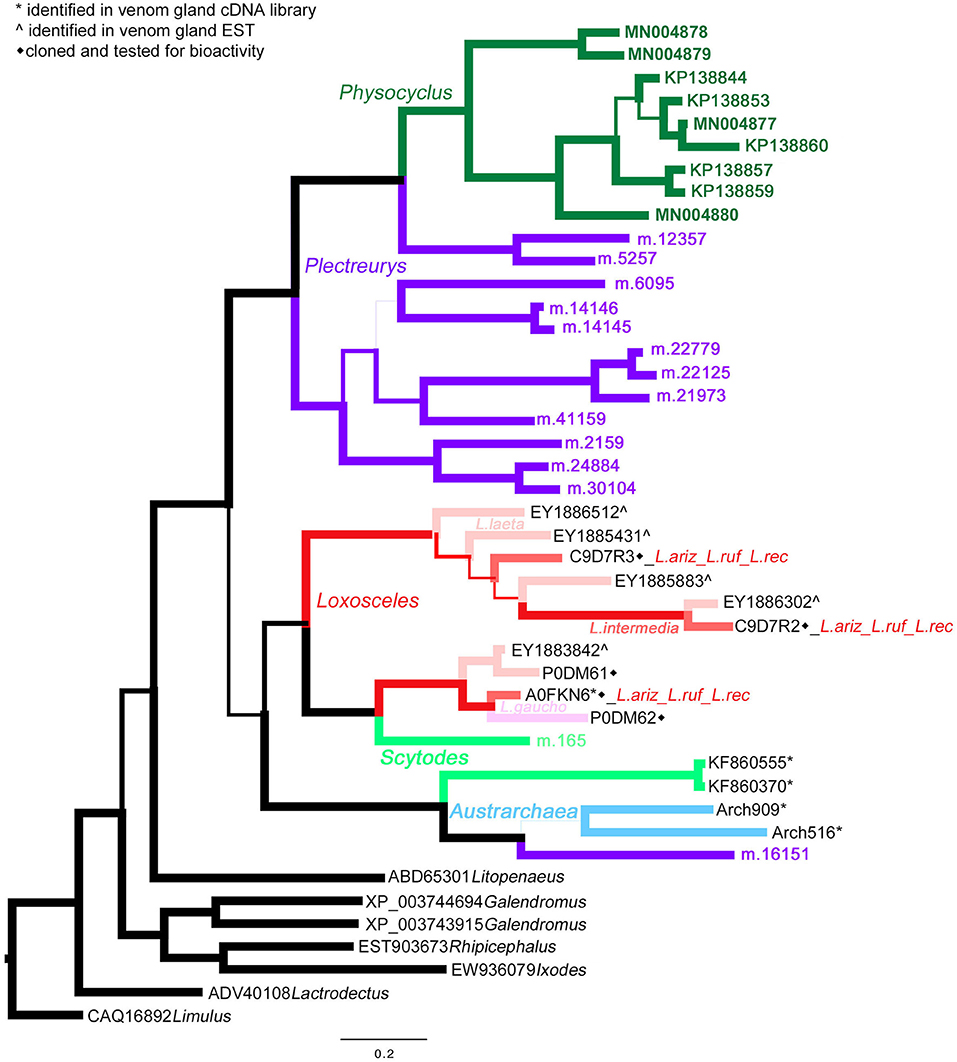
Figure 3. A Bayesian phylogeny of astacin metalloproteinases (M12 peptidases) supports evolutionary expansion in the venom of male Physocyclus mexicanus. Branch width correlates with posterior probability support for that branch. Branches are color coded by source taxon. Sicariids are different shades of pink for different species in the genus Loxosceles. Taxon names are in color when that polypeptide sequence has been confirmed to be present in a venom proteome and are otherwise labeled by symbols according to the legend.
All of the sequences we detect have the conserved catalytic domain HEXXHXXGXXHEXXRXDR- and a MXY region that is involved with a sequence turn and Zn-dependent activity (da Silveira et al., 2007). LALPs have proteolytic effects on gelatin, fibronectin, fibrinogen and extracellular matrices, and thus, as venom toxins, they are hypothesized to be spreading factors that act in synergy with other toxins (Trevisan-Silva et al., 2013). The hydrolytic effects of LALPs are suspected to contribute to hemostatic disturbance during mammalian envenomation, thereby influencing the dermonecrotic or systemic effects of Loxosceles bites (Trevisan-Silva et al., 2013).
Astacin-like metalloproteinases have also been detected as major components in digestive fluids secreted from the mouths of the mygalomorph Acanthoscurria geniculata (Walter et al., 2017) and araneomorphs Stegodyphus mimosarum (Walter et al., 2017), Nephilengys cruentata (Fuzita et al., 2016), and Argiope aurantia (Foradori et al., 2006) indicating a phylogenetically widespread presence in spider digestate. We are confident that the homologs we detect in venom proteomes represent presence in venom rather than contamination by regurgitate, because our venom collection techniques carefully exclude digestate. Expanded phylogenetic analyses including a broad taxonomic set of proteins obtained in oral digestate and venom resolve a monophyletic clade of venom-expressed homologs in P. mexicanus that is sister to all but one divergent venom-expressed homolog in P. tristis (data not shown), reflecting potential diversification in this gene family in the context of venom expression.
Other venom proteins
The remaining two peptidases in P. mexicanus venom are serine proteases and homologs of known venom toxins. Chymotrypsin-like homologs (S1 peptidases) were present with low diversity and abundance. The two homologs are fragments that each hit proteins with CUB domains. The other is a homolog of neuroendocrine convertase 1 (S8 peptidase) which are furin-like proteases, homologs of which cleave N- and C-termini in processing mature latrotoxins (Ushkaryov et al., 2004) and may be common in venoms across multiple lineages.
We detect three other non-peptidase toxic proteins that are homologs of well-established and widespread venom toxins. These include two CRiSP homologs, one that is full length (425 aa, 48.8 kDa); three chitinase homologs, one full length and corresponds to a 493 aa mature protein (est 54.7 kDa), and a single fragment of a bee venom-like phospholipase A2 (PLA2) with weak support (only 4 tryptic peptides). Of these, the chymotrypsins, chitinase, and PLA2 have not been detected in P. tristis, perhaps because of their low abundance, or, they may have a dynamic presence in venom extracts. However, the presence of CRiSP homologs adds to evidence that these are predictable widespread venom components in spider venoms (Fry et al., 2009; Undheim et al., 2013).
Proteins of Unknown Function
Beyond proteins with homology to established venom components, there are proteins in the proteome, some well-represented with respect to peptide spectra, that are homologs of uncharacterized and unnamed genes. Given high rates of discovery of new venom components, we carefully assessed these as potential candidates for venom activity. An intriguing protein is abundant (combined 239 peptide spectra, 62 of which are unique peptides) but not diverse; it is represented by two identical polypeptides, one full length (599 aa), with an estimated size 67.7 kDa. This protein has clear homology to a domain of unknown function (DUF885), however Fold Function and Alignment Searches (FFAS, http://ffas.godziklab.org/ffas-cgi/cgi/ffas.pl) detect sequence elements of lipoproteins, and folds similar to M32 carboxypeptidases, which are metalloproteases. High sequence similarity across arachnid genomes, suggests they perform a conserved, but undescribed function, which could be involved in venom polypeptide processing.
Of the remaining polypeptides with limited homology to known proteins, four are full length. One is a homolog of a protein we have previously detected in Scytodes venom (RP23-like) that is annotated as “mite allergen.” This basic protein (est pI 9.97) has a signal peptide, is estimated 20.7 kDa (188 aa) and has homologs across arachnid genomes. Curiously, proteins referred to as allergens and “mite allergens” in Loxosceles venoms are not homologs of this toxin (MN004921). The two remaining proteins are both secreted and do not have any cysteines. The first (MN004924) is acidic (pI 4.14) estimated to be 22.5 kDa, and is leucine and glutamic acid rich. The other two (MN004934 and MN004937) are from a group of five listed to have leucine-rich repeat domains, based on results from the CDD and InterProScan searches. Of the five, three are homologs (MN004924 is estimated to be 34.9 kDa with a pI of 4.95), and the other two are not homologous. One is estimated to be 34.4 kDa (pI 9.05). Four distinct polypeptides had no hits in any databases (KP138800, KP138796, MN004925, and MN004926); they are distinct from each other, and we have named them “hypothetical proteins.” None of these are found in other venom proteomes within Synspermiata, so we conclude they are not abundant or widespread contributors to venoms in this higher lineage.
Venom Peptide Toxins
While we limited our discussion of venom proteins to those confirmed in the proteome, we discuss the venom peptides in the proteome in the context of homologous sequences in our transcriptomes. We do this because patterns emerge in comparisons between these data sets that potentially reflect sexual dimorphism given that our proteome is male specific and our transcriptome combines male and female gland extracts.
Overview
In our combined transcriptome we detected a total of 103 sequences with homology to venom peptides, or cysteine motifs that are characteristic of venom peptides (91 in our Sanger cDNA library (taxon source confident, Table S3) and 12 NGS sequences detected as matches to our proteome). Among the 103 sequences, 49 are non-redundant and full length. These consist of 17 distinct groups, 13 of which are predicted knottins, and the other four with characteristics of spider venom peptides as described below. The peptides range in cysteine number from 7 to 10 and in predicted size from 3.6 kDa-11.9 kDa (mature, processed sequences) (Table S1). These sequences constitute over one-third (36%) of the venom gland transcriptome (Figure S1) and 18% of the proteome that is annotated as proteins with venom function (Figure 1B). We assigned the 17 groups of “pholcitoxins” with “unknown” functional notations (U1-U17) and a “PHTX-Pmx” name according to established nomenclatural convention (King et al., 2008). Suffixes of ascending letters distinguish distinct sequences within a group (e.g. 1a, 1b, 1c, etc.) and those with an underscore followed by a number differ only in the signal sequence region (accession numbers KP138863-KP1388905 and MN004909-MN004920).
Peptides detected in proteome
Of the 17 peptide groups identified in the transcriptome, 13 (with 36 distinct sequences) were detected in the venom proteome. These vary in abundance (as quantified by numbers of unique tryptic peptides) and diversity (as quantified by the numbers of non-redundant sequences) (Figure 4). Over 25% (61/220) of the tryptic peptides that hit peptide toxin matches were to U17 -PHTX-Pmx. The three 7.8 kDa peptides in this group are homologs of U1-HXTX-Iw1 that is abundant in the Australian hexathelid funnel-web spider Illawarra wisharti. While homologs appear to be widespread in spider venoms, the target of U1-HXTX-Iw1 remains unknown despite much experimental effort (reviewed in Wilson, 2015).
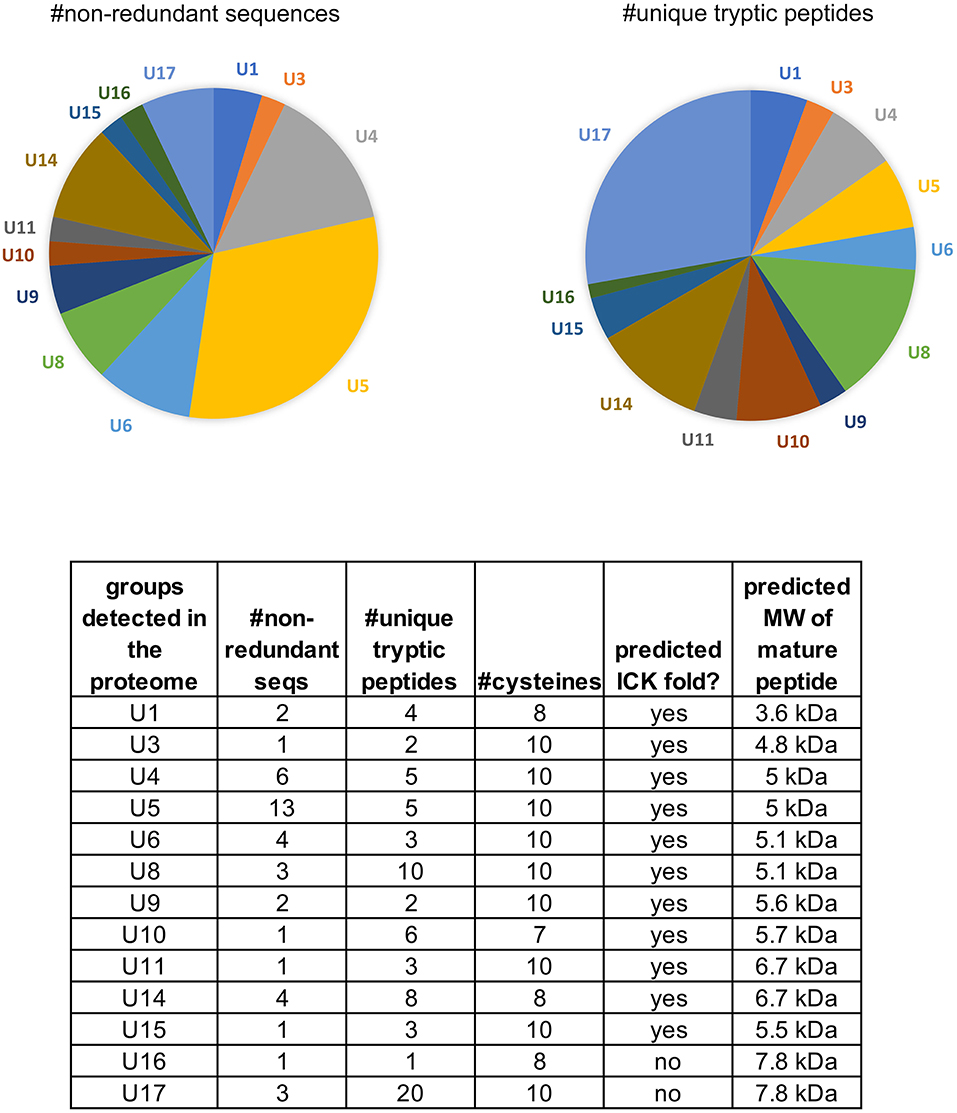
Figure 4. Peptides identified in the venom proteome of Physocyclus mexicanus summarized by homologous groups (families). The pie chart on the left indicates the proportions of non-redundant polypeptide sequences detected in the proteome for each group, and represents the sequence diversity within and among the peptide families. The pie chart on the right indicates the proportion of peptide families as measured by abundance of unique tryptic peptides in the proteome. The table details the number of non-redundant sequences (#nr seqs), the number of unique tryptic peptides, the number of cysteines in the mature peptide, whether or not the peptides are predicted to have an ICK fold, and the estimated molecular weight of the mature peptide.
The most diverse peptide group in P. mexicanus venom is the U5-PHTX set of 13 non-redundant sequences that correspond to ~5 kDa peptides and are homologs of ω-PLTX-Pt1a (Figure 4; Table S1). ω-PLTX-Pt1a blocks presynaptic voltage-gated Ca2+ channels in invertebrates [(King, 2007), PLTX-II in Branton et al. (1987)], specifically Dmca1A channels (Kuromi et al., 2004) and are the most abundant peptide toxins in venoms of P. tristis (Zobel-Thropp et al., 2014a). While ω-PLTX-Pt1a is the top hit in BLAST searches of U5-PHTX, ω-PLTX-Pt1a is homologous to two other well-characterized toxins from P. tristis, U1-PLTX-Pt1c and δ/ω-PLTX-Pt1a. U1-PLTX-Pt1c (Plt-XI) is a highly potent insecticidal toxin, however, the molecular mechanism of activity remains unknown (Quistad and Skinner, 1994). The activity of δ/ω-PLTX-Pt1a is better understood and is an excitatory toxin with activity on both Ca2+ and Na+ channels (Zhou et al., 2013). All of these toxins share a cysteine pattern of -C6C-CC-C2C1C-C1C-C- and some require O-palmitoylation for activity.
The P. mexicanus venom proteome is well represented in diverse homologs with the same cysteine pattern as ω-PLTX-Pt1a. These include U3-, U4-, U5-, U6-, U8-, U9- and U11-PHTX. While the top BLAST hits for these toxin families varied, they are alignable and share the same motif, and thus represent a superfamily (Figure 5). Collectively, the motif spacing is summarized with (1-4)C6C(4-10)CC(1-2)C2C1C(4-9)C1C(3-8)C(0-5). All of the sequences in this superfamily have residues that may be palmitoylized (threonine or serine, in addition to cysteine) near the C-terminus. Together, members of this superfamily make up a large proportion of the venom peptides in P. mexicanus venom (~45% of tryptic peptides, ~70% of the unique polypeptides). Moreover, given the similarities and homology with toxins of known activity, members of this superfamily in P. mexicanus are strong candidates for contributing to toxicity of these venoms and may include novel target specificities.
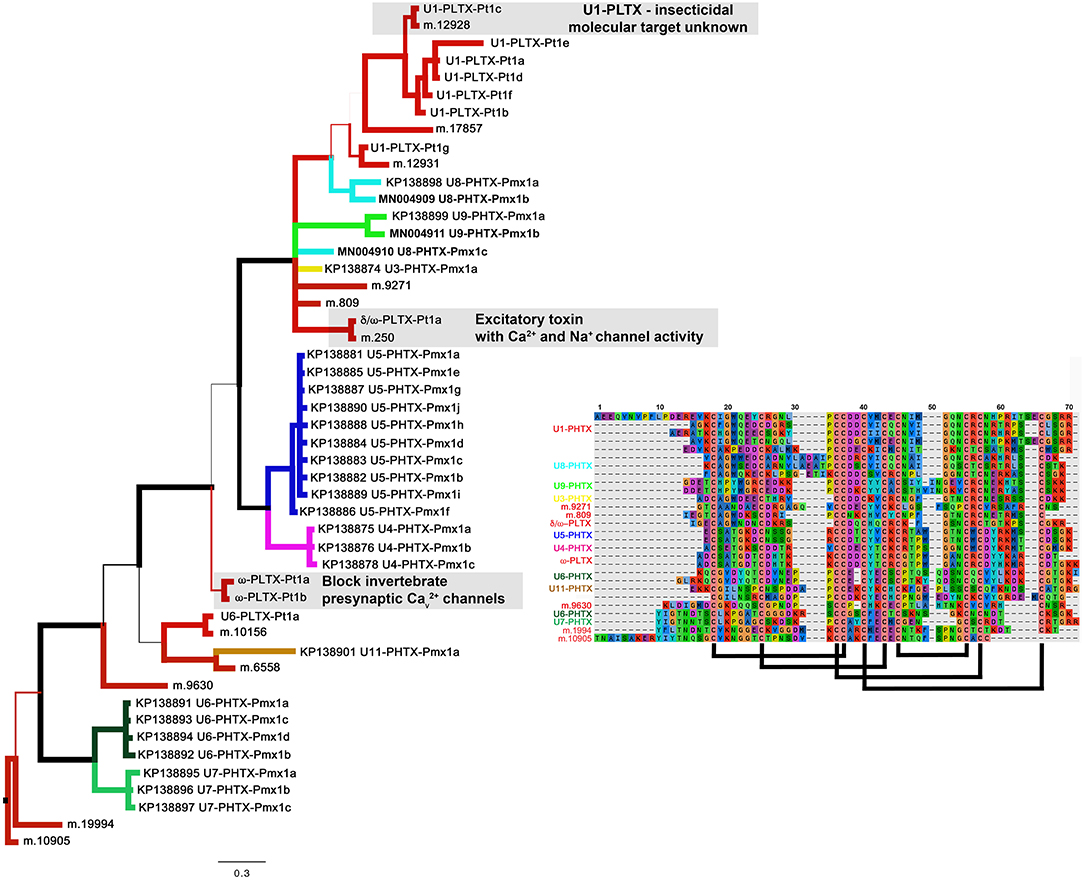
Figure 5. A Baysian phylogeny of a superfamily of homologous groups of ICK peptides from Plectreurys tristis and Physocyclus mexicanus. Branch width correlates with posterior probability support for that branch. All polypeptides included in this phylogeny have been confirmed to be present in the venom proteomes. Branches colored red are all from P. tristis, and all other colored branches represent groups of peptides identified in this work from P. mexicanus. Annotations of activity reflect the discussion in the text. The alignment includes a subset of the sequences included in this phylogeny, reduced to fit by eliminating highly similar sequences. Predicted disulfide bonding pattern is based on Quistad and Skinner (1994).
Of the remaining four peptide families in the proteome, U14-PHTX is the most abundant and diverse with 28 tryptic peptides and four unique polypeptide sequences. These peptides have 8 cysteines with the pattern (2-4)C(6-7)C(5-7)CC(7-8)CRC(8-9)CHC(8-14). While they have no matches in databases, the cysteine pattern is similar to, but larger than, μ-agatoxins and they may share a similar fold with cysteine bonding of I-IV, II-V, III-VIII, VI-VII. The predicted molecular weight is ~6.65 kDa. U10, U15, and U16 are each represented by single polypeptides and U10 is well represented in the proteome with 17 tryptic peptide hits. Like U14-PHTX, U10 is unique to pholcids with no hits in databases, however it has seven cysteines and codes for a peptide with estimated molecular mass 5.7 kDa. U15- PHTX is a clear homolog of U3- PLTX-Pt1a, originally described as Plt-X (Quistad and Skinner, 1994) in P. tristis venom, which is insecticidal, however they are not instantly paralytic but lead to slow lethality in some lepidopteran larvae. U16- PHTX has significant homology with U1- PLTX, however, it has an extended N terminus that is lacking a cysteine that is present in the rest of the superfamily.
Not detected in proteome
Four peptide families in the transcriptome with characteristics of venom toxins were not detected in the proteome. One group U2-PHTX is among the most abundant and diverse peptides in the transcriptome (second only to U5-PHTX) with 16 total sequences, 7 non-redundant. These are homologs of a component in Lycosa singoriensis with unknown molecular target and function (U18-LCTX-Ls1a, Zhang et al., 2010). Nine transcripts (three non-redundant) in the U7-PHTX family are members of the superfamily described above, which are diverse and abundant in the male proteome (Figure 5). U12- and U13- PHTX are related to one another, are cysteine rich, and homologous to peptides isolated from a range of spider venom transcriptomes. They are distinctively large peptides (11.5–12 kDa), are not predicted knottins, and are remote homologs of Dickkopf-related proteins that are involved in development in a wide range of non-arthropod metazoans. Beyond standard analytical reasons why a protein may not be detected in a proteome (see Methods), lack of detection of these peptide families in the proteome could reflect them serving a non-venomous function in the cell, or potentially expression in female but not male P. mexicanus venoms.
Insecticidal Potency
The potency of male P. mexicanus venom, measured by PD50 on crickets, is comparable to that of venom from female P. tristis and other Synspermiata taxa (Figure 6, Zobel-Thropp et al., 2014a). Interestingly crude venom from all Synspermiata taxa tested to date on Acheta domestica are more potent than comparable estimates of effective doses on multiple taxa using a different cricket model, Grillus assimilis (Figure 6, Manzoli-Palma et al., 2003). However, there are distinct physiological symptoms of envenomation whereby crickets injected by P. mexicanus venom undergo rapid, irreversible flaccid paralysis in which the legs lay loosely extended and do not twitch. In contrast, crickets injected with venom of P. tristis (Zobel-Thropp et al., 2014a), and sicariid venoms (Zobel-Thropp et al., 2010, 2012) undergo rapid and irreversible excitatory paralysis with legs flexed and twitching. P. mexicanus envenomated crickets also develop darkened areas around the mouth and sternum, and on the ventral abdomen (Figure 6), symptoms we have not observed after injection of other Synspermiata venoms (Zobel-Thropp et al., 2010, 2012, 2014a).
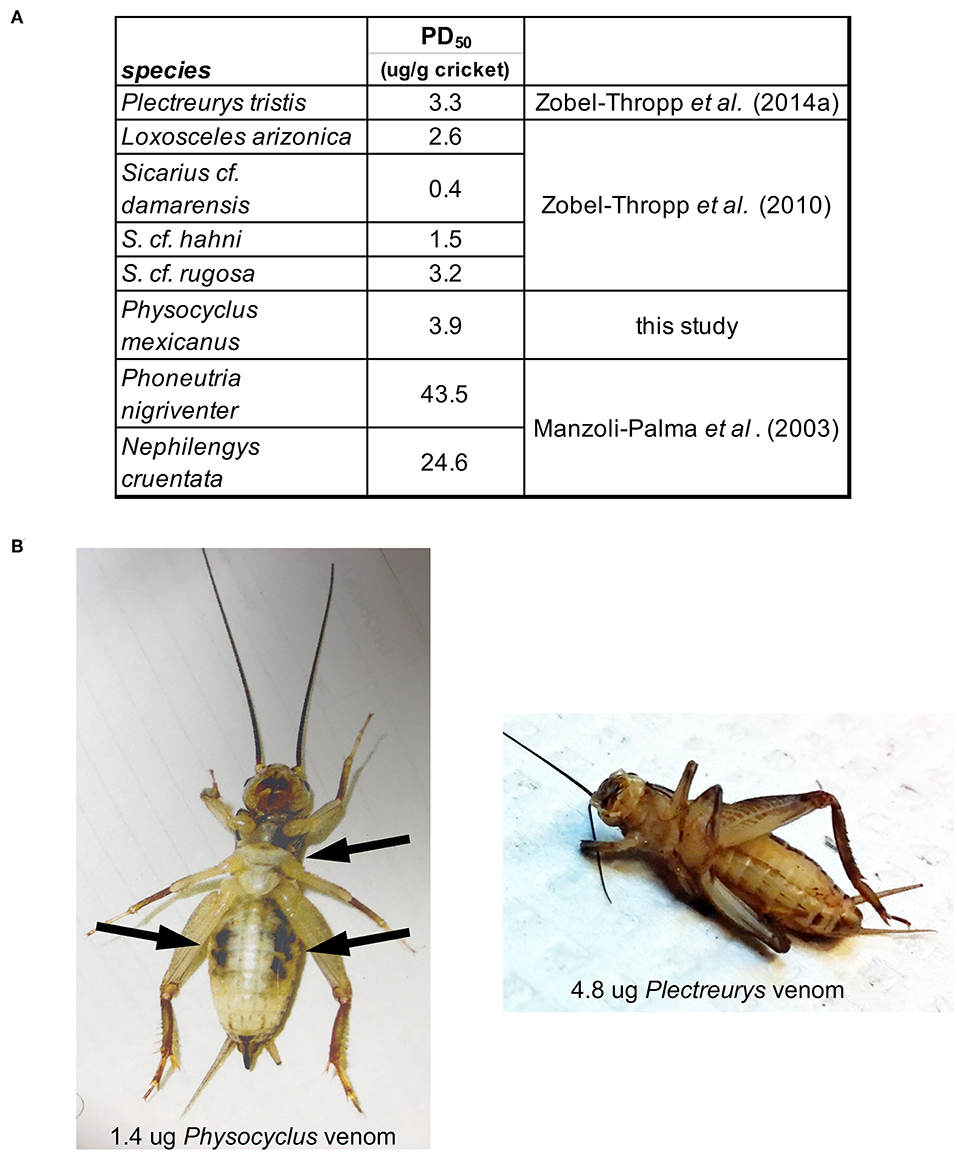
Figure 6. Physocyclus mexicanus bioassays on crickets (Acheta domestica) result in rapid, flaccid paralysis along with ventral darkened areas. (A) Table presenting the estimated dose at which 50% of crickets are paralyzed at 1 h post injection (PD50) for P. mexicanus and other Synspermiata spiders. (B) A ventral view of representative crickets at 1 h post injection by venom from P. mexicanus and P. tristis for comparison. Notable differences are the relaxed legs and darkened regions on the cricket injected by Physocyclus in contrast to the folded legs and lack of darkened regions on Plectreurys.
The components responsible for flaccid paralysis and areas of discoloration on crickets following injection with P. mexicanus venom are unknown. Neprilysins are candidates for contributing to these effects given their abundance in the venoms and uniqueness within Synspermiata. While melanism in crickets has been described as a symptom of intoxication by insecticides (Fisher and Brady, 1980) to our knowledge the mechanisms underlying this are unknown. Flaccid paralysis in insects can be induced by some polyamine toxins (Quistad et al., 1991), by Nav channel toxins (Bende et al., 2013, 2015) and a variety of peptide toxins that affect Ca2+ channels (e.g., Knaus et al., 1987; Troncone et al., 1995; Lipkin et al., 2002; King and Hardy, 2013). Characterization of the non-polypeptide and peptide components in P. mexicanus may identify homologous or convergent factors that are causing these symptoms.
Notes on P. mexicanus Behavior and Effects of Bites on Humans
While we have not performed controlled behavioral experiments a dense natural, local colony of P. mexicanus at Spider Pharm in Yarnell, Arizona, has allowed opportunity for anecdotal observations of their behaviors including their propensity to bite humans. We include an overview of observations, emphasizing those that inform the role of venom and may inspire future experimental work.
Propensity to Bite and Effects on Humans
Contrary to popular opinion about pholcid spiders, P. mexicanus readily bite humans when they are disturbed by moving or cleaning activities, but bites are rarely confirmed by direct observation (Kristensen, pers obs.). While working in the lab with an open colony of these animals, members of Spider Pharm staff are frequently bitten when their habitats are disturbed. Most bites are brief nips as the spiders are moving rapidly over exposed skin, possibly with a slight pinch or tug resulting in a mild and brief sting. All confirmed bites by this species have been by adult or subadult males.
In one case, an adult male P. mexicanus bite on the back of the neck produced a mild sting that became a small welt which blackened and eventually opened into a 1 mm diameter wound. Images of the wound were diagnosed by a dermatologist with expertise in spider envenomation (Van Stoeker, pers comm) as a typical reaction to foreign protein rather than necrosis resembling a triggered immune response comparable to loxoscelism. In a second case, three subadult males that were disturbed by moving trays of larvae walked down the fingers of the left hand and simultaneously bit the knuckle at the base of the little finger. This resulted in an immediate mild, temporary sting. The knuckle was sore and slightly inflamed for several days. Interestingly, the case of human bites recorded on Mythbusters (Episode 13, premiered 15 Feb 2004) involved adult males that were aggregated and engaged antagonistically with one another. In that case, when human arms were extended deeply into the enclosure spiders did not initially bite, though several spiders pressed their chelicerae against the skin without biting. A bite occurred on the back of the hand after the hand was pulled back closer to males fighting near the entrance (Kristensen pers obs).
Potential Venom Dimorphism, Male Aggression, and Feeding Biology
The difference between sexes in our ability to obtain venom by electrostimulation inspires consideration of potential sexual dimorphism in venom composition and/or functional role [repeated independently in the Binford Lab and the Kristensen Lab (Spider Pharm)]. The easier to obtain, higher yields of males could reflect larger volumes of venom in males, or different morphological/physiological circumstances of extrusion. There could also be seasonal differences in venom volume. Adult males and females are similar in size, but males have more ornate and armored or reinforced chelicerae. Cheliceral armor is a known dimorphic trait in pholcids that is a proposed adaptation for mating (Huber, 1999) but may also be an adaptation for fighting and defense. Males are aggressive to one another when at high densities. For example, after placement in enclosures for the Mythbuster staged encounters, males were concentrated on the cloth and interacted aggressively including biting that resulted in fatalities (Kristensen pers obs). There appears to be less aggression in natural stable colonies, and interactions are typically settled quickly and non-lethally. While most of the aggressive interactions observed were between males, females also exhibit aggression, especially when guarding clusters of hatchlings.
P. mexicanus readily capture and eat a diversity of arthropod prey, including spiders. The role of venom is not obvious and prey that are apparently envenomated often appear to be little affected. Like other pholcids, most prey are slowed or restrained by silk first and bitten several times during wrapping, frequently with no obvious immediate effect. In other instances, there is rapid paralysis or death after single bites. More rarely, spiders directly attack small, weak prey with an initial bite (frontal attack) without the use of silk. Single, short bites made during frontal attacks may result in paralysis or death. Given evidence that venom has a potent effect on some prey, the spiders may meter the amounts of venom injected. The ability to regulate the quantity of venom injected into prey as a function of size, activity levels, and differences in sensitivity to toxins, has been experimentally determined in Cupiennius salei (Wigger et al., 2002; Wullschleger and Nentwig, 2002). The observed differential effects of pholcid bites are consistent with a similar ability to regulate venom injection, and may indicate it is a general attribute of spider envenomation.
In many spider species males reduce or stop feeding when they reach adulthood. This is not the case for P. mexicanus (pers obs; Wilson et al., 2016). Not only do males hunt and capture prey but we have observed them presenting captured prey to females, with and without attempts to mate. We have also observed males with egg sacs in their chelicerae, which could reflect adoption of the egg sacs or infanticide by males. Together, these observations suggest there are interesting sexual dynamics in this species that could influence some degree of sexual dimorphism in venoms.
Conclusions
We conclude from the data and observations presented in this work that venoms of male P. mexicanus consist of a rich set of components, predominantly metalloproteinases and peptides. While activity of none of these components has been directly characterized, they include homologs of proteins and peptides that are demonstrated contributors to toxicity. Our evidence of potent toxicity in arthropods, in combination with homology to known toxins suggests that detailed activity characterizations have strong potential for discovering new or refined activities. The unprecedented extensive presence of neprilysins is intriguing with respect to contributions to immobilization or consumption of prey. While the information available on the effect of P. mexicanus venom on mammals is limited by lack of careful experimentation, we consider the documented accounts evidence that mouthparts of P. mexicanus are capable of piercing human skin, however envenomation results in a mild sting, and venom from a single P. mexicanus is not lethal to humans or other mammals. Given the tremendous diversity of pholcid spiders, we do not assume that venom composition in males of this species are representative of the entire family, and broader sampling within this group may uncover exciting differences that correlate with known ecological diversification within the group. Moreover, proteomic characterization of female venoms of P. mexicanus are needed to infer the full set of proteins identified in venom gland cDNA libraries that are present in adult members of this species, given the likelihood of at least some degree of sexual dimorphism. With expansive diversity to explore in pholcid venoms, we present these results as reason to be excited about potential discovery of new chemical activities and, by default, to change the public narrative and assume pholcid spiders are not harmful to humans.
Data Availability
The datasets generated for this study can be found in GenBank, UniProt Knowledgbase, KP138796-KP138905, and MN004877-MN004966.
Author Contributions
GB and PZ-T conceived of the project, wrote the manuscript, and oversaw all aspects of collecting and analyzing transcriptomic, proteomic, and insect bioassay data. JM generated and analyzed Sanger transcriptomes and conducted insect bioassays. BK generated the Illumina 3000 transcriptomes. CD and LB conducted proteomics analyses. CK provided spiders, collected venom, and documented behavior of the animals including human envenomation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by funding from National Institute of Health R15-GM-097696-01 to GB, and summer research support from Lewis & Clark College. Lewis & Clark students Sophia Horigan, Sasha Bishop, Katherine Delgado, Demi Glidden, and Kendra Autumn contributed to considerations of annotating the proteome. Matt Briggs and Owen Hart helped with the PD50 assays. We thank Matthew Cordes for help thinking about function of proteins with poor homology, Leslie Boyer for discussions about the manuscript, and Anita Kristensen for help with venom collection and observations of the natural colony of P. mexicanus at Spider Pharm. Careful and thoughtful comments from reviewers also improved the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00256/full#supplementary-material
Figure S1. Overview of cDNA library Sanger sequences from a combination of male and female P. mexicanus venom glands. (A) Proportionate representation of overall transcript abundance into five general categories based on database hits. (B) Table: the 99 and 85% identity threshold assemblies, respectively identify redundant and homologous sequences in the library. Flowchart: clusters and singleton sequences from the 85% threshold analysis were initially divided based on whether or not there were significant tBLASTx hits (e ≤ 10−5) in ArachnoServer or NCBI nt/nr databases. General function prediction, and analyses of sequences that hit nothing in databases was done as described in the text.
Table S1. Details of each of the unique polypeptides detected in the proteome of Physocyclus mexicanus. Sequences are categorized and color-coded as proteins with venom function (prey capture/feeding/defense)(blue), proteins of unknown function (orange), and proteins with housekeeping/metabolism function (gray). Polypeptide sequences detected in the proteome are in the far right column.
Table S2. Details of the proteomics analyses conducted at the University of Arizona Proteomics Consortium.
Table S3. Details of cDNAs identified in the cDNA library transcriptome of Physocyclus mexicanus. Sequences are categorized based on predicted proteins as in Table S1.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bende, N. S., Dziemborowicz, S., Herzig, V., Ramanujam, V., Brown, G. W., Bosmans, F., et al. (2015). The insecticidal spider toxin SFI1 is a knottin peptide that blocks the pore of insect voltage-gated sodium channels via a large-hairpin loop. FEBS J. 282, 904–920. doi: 10.1111/febs.13189
Bende, N. S., Kang, E., Herzig, V., Bosmans, F., Nicholson, G. M., Mobli, M., et al. (2013). The insecticidal neurotoxin Aps III ia an atypical knottin peptide that potentially blocks insect voltage-gated sodium channels. Biochem. Pharmacol. 85, 1542–1554. doi: 10.1016/j.bcp.2013.02.030
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Wheeler, D. L. (2005). GenBank. Nucleic Acids Res. 33, D34–D38. doi: 10.1093/nar/gki063
Binford, G. J., Cordes, M. H. J., and Wells, M. A. (2005). Sphingomyelinase D from venoms of Loxosceles spiders: evolutionary insights from cDNA sequences and gene structure. Toxicon 45, 547–560. doi: 10.1016/j.toxicon.2004.11.011
Binford, G. J., and Wells, M. A. (2003). The phylogenetic distribution of sphingomyelinase D activity in venoms of Haplogyne spiders. Comp Biochem Physiol Part B Biochem. Mol. Biol. 135, 25–33. doi: 10.1016/S1096-4959(03)00045-9
Branton, W. D., Kolton, L., Jan, Y. N., and Jan, L. Y. (1987). Neurotoxins from Plectreurys spider venom are potent presynaptic blockers in Drosophila. J. Neurosci. 7, 4195–4200. doi: 10.1523/JNEUROSCI.07-12-04195.1987
Chaves-Moreira, D., Senff-Ribeiro, A., Wille, A. C. M., Gremski, L. H., Chaim, O. M., and Veiga, S. S. (2017). Highlights in the knowledge of brown spider toxins. J. Venom Anim. Toxins Incl. Trop. Dis. 23:6. doi: 10.1186/s40409-017-0097-8
da Silveira, R. B., Wille, A. C., Chaim, O. M., Appel, M. H., Silva, D. T., Franco, C. R., et al. (2007). Identification, cloning, expression and functional characterization of an astacin-like metalloprotease toxin from Loxosceles intermedia (brown spider) venom. Biochem. J. 406, 355–363. doi: 10.1042/BJ20070363
Dimitrov, D., Lopardo, L., Giribet, G., Arnedo, M. A., Alvarez-Padilla, F., and Hormiga, G. (2012). Tangled in a sparse spider web: single origin of orb weavers and their spinning work unraveled by denser taxonomic sampling. Proc. R. Soc. B 279, 1341–1350. doi: 10.1098/rspb.2011.2011
Eberle, J., Dimitrov, D., Valdez-Mondragon, A., and Huber, B. A. (2018). Microhabitat change drives diversification in pholcid spiders. BMC Evol. Biol. 18:141. doi: 10.1186/s12862-018-1244-8
Escoubas, P. (2006). Molecular diversification in spider venoms: a web of combinatorial peptide libraries. Mol. Divers. 10:545–554. doi: 10.1007/s11030-006-9050-4
Fisher, C. W., and Brady, U. E. (1980). Increased rate of melanization in hemolymph of American cockroaches (Periplaneta Americana) and house crickets (Acheta domesticus) intoxicated by insecticides. Experientia 36, 93–94. doi: 10.1007/BF02003994
Foradori, M. J., Tillinghast, E. K., Smith, S., Townley, M. A., and Mooney, R. E. (2006). Astacin family metallopeptidases and serine peptidase inhibitors in spider digestive fluid. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 143, 257–268. doi: 10.1016/j.cbpb.2005.08.012
Fry, B. G., Roelants, K., Champagne, D. E., Scheib, H., Tyndall, J. D., King, G. F., et al. (2009). The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 10, 483–511. doi: 10.1146/annurev.genom.9.081307.164356
Fuzita, F. J., Pinske, M. W. H., Patane, J. S. L., Verhaert, P. D. E. M., and Lopes, A. R. (2016). High throughput techniques to reveal the molecular physiology and evolution of digestion in spiders. BMC Genomics 17:716. doi: 10.1186/s12864-016-3048-9
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protocols 8, 1494–1512. doi: 10.1038/nprot.2013.084
Huber, B. (1999). Sexual selection in pholcid spiders (Araneae, Pholcidae). J. Arachnol. 27, 135–141.
Jackson, R. R., and Brassington, R. J. (1987). The biology of Pholcus phalangioides (Araneae, Pholcidae): predatory versatility, araneophagy and aggressive mimicry. J. Zool. 211, 227–238. doi: 10.1111/j.1469-7998.1987.tb01531.x
Johnson, S. A., and Jakob, E. M. (1999). Leg autonomy in a spider has minimal costs in competitive ability and development. Anim. Behav. 57, 957–965. doi: 10.1006/anbe.1998.1058
King, G. F. (2007). modulation of insect Cav channels by peptidic spider toxins. Toxicon 49, 513–530. doi: 10.1016/j.toxicon.2006.11.012
King, G. F., Gentz, M. C., Escoubas, P., and Nicholson, G. M. (2008). A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 52, 264–276. doi: 10.1016/j.toxicon.2008.05.020
King, G. F., and Hardy, M. C. (2013). Spider-Venom peptides: structure, pharmacology, and potential for control of insect pests. Annu. Rev. Entomol. 58, 475–496. doi: 10.1146/annurev-ento-120811-153650
Kirchner, W., and Opderbeck, M. (1990). Predation effect of venom and food intake of pholcus-phalangioides araneae pholicdae. Verhandlungen-des-Naturwissenschaftlichen-Vereins-in-Hamburg 31–32, 15–46.
Knaus, H. G., Striessing, J., Koza, A., and Glossman, H. (1987). Neurotoxic aminoglycoside antibiotics are potent inhibitors of [125I]-omega-conotoxin GVIA binding to guinea-pig cerebral cortex membranes. Naunyn. Schmiedebergs. Arch. Pharmacol. 336, 583–586. doi: 10.1007/BF00169318
Kuromi, H., Honda, A., and Kidokoro, Y. (2004). Ca2+ influx through distinct routes controls exocytosis and endocytosis at Drosophila presynaptic terminals. Neuron 41, 101–111. doi: 10.1016/S0896-6273(03)00815-8
Lipkin, A., Kozlov, S., Nosyreva, E., Blake, A., Windass, J. D., and Grishin, E. (2002). Novel insecticidal toxins from the venom of the spider Segestria florentina. Toxicon 40, 125–130. doi: 10.1016/S0041-0101(01)00181-7
Manzoli-Palma, M. F., Gobbi, N., and Palma, M. S. (2003). Insects as biological models to assay spider and scorpion toxicity. J. Venom Anim. Toxins incl. Trop. Dis. 9, 174–185. doi: 10.1590/S1678-91992003000200004
Michalik, P., and Ramirez, M. J. (2014). Evolutionary morphology of the male reproductive system, spermatozoa and seminal fluid of spiders (Araneae, Arachnida)—current knowledge and future directions. Arthropod Struct. Dev. 43, 291–322. doi: 10.1016/j.asd.2014.05.005
Pineda, S. S., Chaumeil, P. A., Kunert, A., Kaas, Q., Thang, M. W. C., Le, L., et al. (2018). ArachnoServer 3.0: an online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics 34, 1074–1076. doi: 10.1093/bioinformatics/btx661
Quistad, G. B., Reuter, C. C., Skinner, W. S., Dennis, P. A., Suwanrumpha, S., and Fu, E. W. (1991). Paralytic and insecticidal toxins from the funnel web spider, Hololena curta. Toxicon 29, 329–336. doi: 10.1016/0041-0101(91)90286-Z
Quistad, G. B., and Skinner, W. S. (1994). Isolation and sequencing of insecticidal peptides from the primitive hunting spider, Plectreurys tristis (Simon). J. Biol. Chem. 269, 11098–11101.
Rokyta, D. R., Wray, K. P., and Margres, M. J. (2013). The genesis of an exceptionally lethal venom in the timber rattlesnake (Crotalus horridus) revealed through comparative venom-gland transcriptomics. BMC Genomics 14:394. doi: 10.1186/1471-2164-14-394
Rokyta, D. R., Wray, K. P., McGivern, J. J., and Margres, M. J. (2015). The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus). Toxicon 98, 34–48. doi: 10.1016/j.toxicon.2015.02.015
Searle, B. C. (2010). Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10, 1265–1269. doi: 10.1002/pmic.200900437
Sterchi, E. E., Stöcker, W., and Bond, J. S. (2008). Meprins, membrane-bound and secreted astacin metalloproteinases. Mol. Aspects Med. 29, 309–328. doi: 10.1016/j.mam.2008.08.002
Tan, K. Y., Tan, C. H., Chanhome, L., and Tan, N. H. (2017). Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: elucidating geographical venom variation and insights into sequence novelty. PeerJ 5:e3142. doi: 10.7717/peerj.3142
Trevisan-Silva, D., Bednaski, A. V., Gremski, L. H., Chaim, O. M., Veiga, S. S., and Senff-Ribeiro, A. (2013). Differential metalloprotease content and activity of three Loxosceles spider venoms revealed using two-dimensional electrophoresis approaches. Toxicon 76, 11–22. doi: 10.1016/j.toxicon.2013.08.065
Troncone, L. R., Lebrun, I., Mangoli, F., and Yamane, T. (1995). Biochemical and pharmacological studies on a lethal neurotoxic polypeptide from Phoneutria nigriventer spider venom. Neurochem. Res. 20, 879–883. doi: 10.1007/BF00969702
Turner, A. J., Isaac, R. E., and Coates, D. (2001). The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays 23, 261–269. doi: 10.1002/1521-1878(200103)23:3<261::AID-BIES1036>3.0.CO;2-K
Undheim, E. A. B., Sunagar, K., Herzig, V., Kely, L., Low, D. H., Jackson, T. N., et al (2013). A proteomics and transcriptomics investigation of the venom from the Barychelid spider Trittame loki (brush-foot trapdoor). Toxins 5, 2488–2503. doi: 10.3390/toxins5122488
Ushkaryov, Y. A., Volynski, K. E., and Ashton, A. C. (2004). The multiple actions of black widow spider toxins and their selective use in neurosecretion studies. Toxicon 43, 527–542. doi: 10.1016/j.toxicon.2004.02.008
Walter, A., Bechsgaard, J., Scavenius, C., Dyrlund, T. S., Sanggaard, K. W., Enghild, J. J., et al. (2017). Characterisation of protein families in spider digestive fluids and their role in extra-oral digestion. BMC Genomics 18:600. doi: 10.1186/s12864-017-3987-9
Wang, X. C., Duan, Z. G., Yang, J., Yan, X. J., Zhou, H., He, X. Z., et al. (2007). Physiological and biochemical analysis of L. tredecimguttatus venom collected by electrical stimulation. J. Physiol. Biochem. 63:221–230. doi: 10.1007/BF03165785
Wheeler, W. C., Coddington, J. A., Crowley, L. M., Dimitrov, D., Goloboff, P. A., Griswold, C. E., et al. (2016). The spider tree of life: phylogeny of Araneae based on target-gene analyses from an extensive taxon sampling. Cladistics 33, 574–616. doi: 10.1111/cla.12182
Wigger, E., Kuhn-Nentwig, L., and Nentwig, W. (2002). The venom optomisation hypothesis: a spider injects large venom quantities only into difficult prey types. Toxicon 40, 749–752. doi: 10.1016/S0041-0101(01)00277-X
Wilson, D. E., Felt, K. D., Campbell, E. N., Cohen, O. R., Geisel, L. E., Graham, J., et al. (2016). Resource allocation in food-restricted male Physocyclus mexicanus Banks, 1898 spider does not favor proportionally larger testes (Araneae: Pholcidae). J. Arachnol. 44, 408–411. doi: 10.1636/M15-50.1
Wilson, D. T. R. (2015). “The venom of Australian spiders,” in Spider Venoms, eds P. Gopalakrishnakone, G. Corzo, M. E. de Lima, Diego-Garcia, E., Cham: Springer Dordrecht.1–20. doi: 10.1007/978-94-007-6646-4_21-1
World Spider Catalog. (2019). World Spider Catalog, Version 20.0. Bern: Natural History Museum. Available online at: http://wsc.nmbe.ch (accessed February 25, 2019).
Wullschleger, B., and Nentwig, W. (2002). Influence of venom availability on a spider's prey-choice behavior. Funct. Ecol. 16, 802–807. doi: 10.1046/j.1365-2435.2002.00686.x
Zhang, Y., Chen, C., Tang, X., Wang, F., Jiang, L., Xiong, X., et al. (2010). Transcriptome analysis of the venom glands of the Chinese wolf spider Lycosa singoriensis. Zoology 113, 10–18. doi: 10.1016/j.zool.2009.04.001
Zhou, Y., Zhao, M., Fields, G. B., Wu, C. F., and Branton, W. D. (2013). δ/ω-Plectoxin-Pt1a: an excitatory spider toxin with actions on both Ca2+ and Na+ channels. PLoS ONE 8:e64324. doi: 10.1371/journal.pone.0064324
Zobel-Thropp, P. A., Bodner, M. R., and Binford, G. J. (2010). Comparative analyses of venoms from American and African Sicarius spiders that differ in sphingomyelinase D activity. Toxicon 55, 1274–1282. doi: 10.1016/j.toxicon.2010.01.019
Zobel-Thropp, P. A., Correa, S. M., Garb, J. E., and Binford, G. J. (2014b). Spit and venom from Scytodes spiders: a diverse and distinct cocktail. J. Proteome Res. 13, 817–835. doi: 10.1021/pr400875s
Zobel-Thropp, P. A., Kerins, A. E., and Binford, G. J. (2012). Sphingomyelinase D in sicariid spider venom is a potent insecticidal toxin. Toxicon 60, 265–271. doi: 10.1016/j.toxicon.2012.04.350
Keywords: neprilysin, astacin metalloproteinase, insecticidal neurotoxin, proteome, PD50
Citation: Zobel-Thropp PA, Mullins J, Kristensen C, Kronmiller BA, David CL, Breci LA and Binford GJ (2019) Not so Dangerous After All? Venom Composition and Potency of the Pholcid (Daddy Long-Leg) Spider Physocyclus mexicanus. Front. Ecol. Evol. 7:256. doi: 10.3389/fevo.2019.00256
Received: 28 March 2019; Accepted: 18 June 2019;
Published: 12 July 2019.
Edited by:
Kartik Sunagar, Indian Institute of Science (IISc), IndiaReviewed by:
Volker Herzig, The University of Queensland, AustraliaDilza Trevisan Silva, Instituto Butantan, Brazil
Copyright © 2019 Zobel-Thropp, Mullins, Kristensen, Kronmiller, David, Breci and Binford. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Greta J. Binford, binford@lclark.edu
 Pamela A. Zobel-Thropp
Pamela A. Zobel-Thropp Jennifer Mullins1
Jennifer Mullins1  Charles Kristensen
Charles Kristensen Greta J. Binford
Greta J. Binford