- 1Cologne Excellence Cluster on Cellular Stress Responses in Aging Associated Diseases, Köln, Germany
- 2Max-Planck-Institute for Neurological Research, Köln, Germany
- 3Department of Mouse Genetics and Metabolism and Center for Molecular Medicine Cologne, Institute for Genetics at the University Hospital of Cologne, University of Cologne, Cologne, Germany
- 4Center for Endocrinology, Diabetes and Preventive Medicine, University Hospital Cologne, University of Cologne, Cologne, Germany
Our understanding of genomic regulation was revolutionized by the discovery that the genome is pervasively transcribed, giving rise to thousands of mostly uncharacterized non-coding ribonucleic acids (ncRNAs). Long, ncRNAs (lncRNAs) have thus emerged as a novel class of functional RNAs that impinge on gene regulation by a broad spectrum of mechanisms such as the recruitment of epigenetic modifier proteins, control of mRNA decay and DNA sequestration of transcription factors. We review those lncRNAs that are implicated in differentiation and homeostasis of metabolic tissues and present novel concepts on how lncRNAs might act on energy and glucose homeostasis. Finally, the control of circadian rhythm by lncRNAs is an emerging principles of lncRNA-mediated gene regulation.
Introduction
The Non-Coding Genome
The canonical view of mammalian genomes revolves around the notion that the roughly 20,000 proteins within mammalian genomes are interspersed by somewhat conserved, yet functionally redundant non-coding regions with only limited regulatory potential. Regulatory properties of these “non-coding” regions were only attributed to cis-regulatory elements such as promoters or cis/trans-enhancer regions. This paradigm was fundamentally called into question by results obtained from whole-transcriptome sequencing efforts [e.g., by the ENCODE consortium (Birney et al., 2007; Thomas et al., 2007)] over the last decade that have revealed the pervasive transcription of mammalian genomes (Carninci et al., 2005; Birney et al., 2007; Derrien et al., 2012). Although the magnitude of pervasiveness remains under debate (van Bakel et al., 2010; Clark et al., 2011), recent meta-analyses of human ribonucleic acid-sequencing (RNA-Seq) datasets have confirmed that >80% of genomic sequences are rediscovered within RNA transcripts, often in a temporally and spatially specific manner (Hangauer et al., 2013). One logical consequence of pervasive transcription is the abundance of non-coding RNAs (ncRNAs) within mammalian genomes, a phenomenon which holds true for most eukaryotic species ranging from yeast (David et al., 2006), to Drosophila (Stolc et al., 2004), plants (Li et al., 2006) and humans (Hangauer et al., 2013). Given the predicted high number of ncRNAs within mammalian genomes, which probably surpasses that of coding genes, it is not surprising that a large conceptual void remains about the multifaceted role of ncRNAs in regulation of gene expression. Researchers have historically divided ncRNAs into small ncRNAs (sRNAs <200 nt length) such as microRNAs (miRNAs) and small nucleolar RNAs (snoRNAs) in contrast to so-termed long ncRNAs (lncRNAs; >200 nt length). Until today, the identification of biological processes, which are regulated by miRNAs as well as the elucidation of mRNA targets, which are posttranscriptionally regulated by disease-associated miRNAs remains an important focus of research (Bartel, 2009; Carthew and Sontheimer, 2009). It was demonstrated that the spectrum of biological processes, which are regulated by miRNAs, ranges from the development of organs, the homeostatic regulation of cellular metabolism to aging and neurodegenerative disorders. Although miRNAs are central to the understanding of the non-coding genome, the regulation of energy homeostasis and metabolism by miRNAs has been meticulously reviewed elsewhere (Lynn, 2009; Rottiers and Naar, 2012; Kim and Kyung Lee, 2013) and goes beyond the scope of this review. In contrast to miRNAs, the role of lncRNAs in control of metabolism and energy homeostasis remains rather elusive. Thus, we here review the known roles for lncRNAs, which probably constitute the numerical majority of ncRNAs encoded within mammalian genomes, during differentiation, homeostasis and metabolic regulation of tissues (Figure 1).
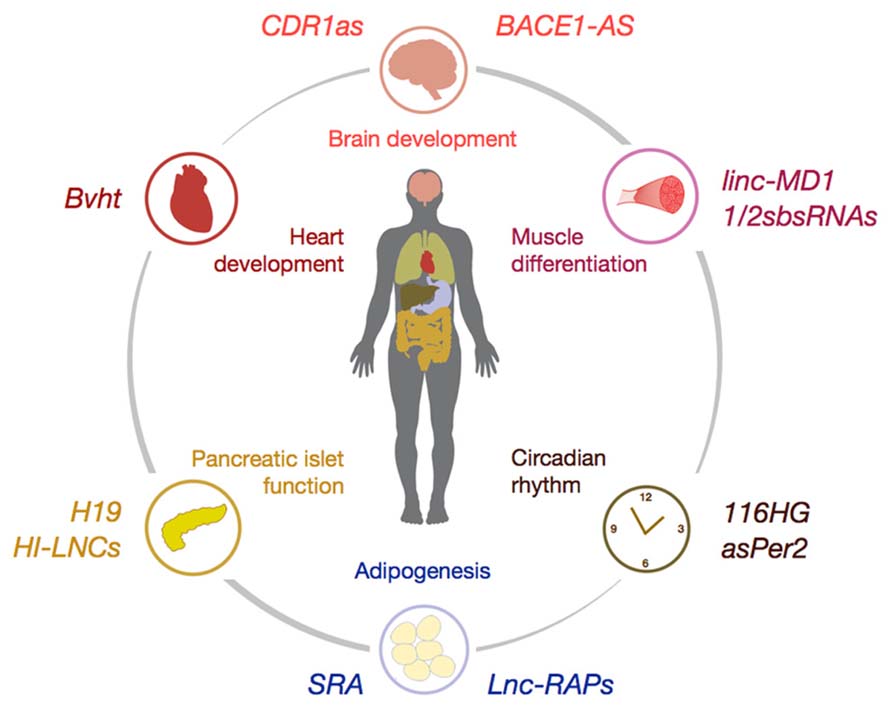
FIGURE 1. Schematic illustration of selected lncRNAs involved in the control of organ differentiation and development (e.g., CDR1as, Bvht, and linc-MD1), tissue homeostasis (e.g., Lnc-RAPs and HI-LNCs) and control of circadian rhythm (e.g., asPer2). For detailed description of lncRNA mode-of-action please refer to the main text.
Long Non-Coding RNAs
Those lncRNAs that were initially discovered in the late eighties had distinct, at that time considered exotic functions such as X chromosome inactivation in females by the lncRNA XIST (Penny et al., 1996). Another historical example was the imprinted lncRNA H19, which is involved in repression of Igf2 (Pachnis et al., 1988). After the discovery of pervasive genomic transcription it became clear that lncRNAs do not represent an exotic observation, but rather a prominent feature of the genome (Birney et al., 2007). Although the number of lncRNAs is still debated, recent meta-analyses posit the human genome to give rise to >60,000 lncRNA, albeit the majority is probably expressed at low levels (Derrien et al., 2012; Hangauer et al., 2013; for current lncRNA numbers consult NONCODE Version 4, www.noncode.org). Interestingly, lncRNAs on one hand exhibit many similarities with protein-coding transcripts: As true for mRNAs, lncRNAs are transcribed by RNA-polymerase (Pol) II (Guttman et al., 2009), spliced at canonical splicing sites (Chew et al., 2013), are partly polyadenylated (Cabili et al., 2011) and even associate with polysomes (Guttman et al., 2013). Further, lncRNAs harbor the same chromatin marks of H3K4 and H3K36 trimethylation as found in active promoters and transcribed regions of protein-coding transcripts, respectively, a phenomenon which aided in the identification of novel lncRNAs (Mikkelsen et al., 2007). It is noteworthy that the notion of distinct mRNA-like trimethylation marks on actively transcribed lncRNAs is incompatible with the criticism brought forward according to which lncRNAs are merely generated by unspecific Pol II activity which leads to low-level transcription of non-coding sequences (“transcriptional noise”; Derrien et al., 2012). On the other hand, certain features set lncRNAs apart from protein-coding genes: Generally, lncRNAs are expressed at lower levels, are less evolutionarily conserved and less frequently associate with ribosomes than protein-coding transcripts (Hangauer et al., 2013). Further, lncRNA are shorter than coding genes and are composed of a unique gene structure of usually 1–2 exons. Of note, some lncRNAs do give rise to small peptides and may act as both, coding and non-coding, transcript (reviewed here Dinger et al., 2008).
Principles of lncRNA-Mediated Gene Regulation
Currently, intensive research efforts are underway to better understand the molecular basis of gene regulation by lncRNAs. To date, four major paradigms have emerged on how lncRNA impinge on gene regulation:
(Epigenetic) Regulation of Gene Transcription
LncRNAs are able to bring gene-regulatory DNA-binding proteins and DNA sequences into close proximity and thus constitute an ideal docking platform for recruiting epigenetic modifiers to distinct genomic loci in cis- or trans. Indeed, early insights into lncRNA-based gene regulation have revealed the recruitment of the inhibitory polycomb repressive complex (PRC) 2 and the activating Trithrorax/MLL chromatin modifiers to specific genomic loci by the lncRNAs HOTAIR (Rinn et al., 2007) and HOTTIP (Wang et al., 2011), respectively. PRC2 and MLL then mark distinct lysine residues within histones via trimethylation, leading to inhibition or activation of gene transcription. In a similar fashion, the lncRNA ANRIL silences the INK4a tumor suppressor allele by H3K27 trimethylation via recruiting the Polycomb chromatin modifier CBX7 (Yap et al., 2010). The percentage of lncRNAs implicated in (epigenetic) gene regulation was systematically quantified by interrogating the PRC2 interactome using chromatin-state maps. This revealed the abundant interaction of Polycomb repressor proteins with up to 20% of expressed lncRNAs (Khalil et al., 2009). Thus, one prominent role of lncRNAs relates to writing and erasing chromatin marks, thereby controlling the epigenetic state of lncRNA-bound genomic loci (Spitale et al., 2011). In a systematic attempt to interrogate the function of 3019 human lncRNAs, Orom et al. (2010) revealed that a significant portion of the lncRNome possesses cis-regulatory enhancer properties (hitherto termed enhancer-like RNAs, eRNAs), which control the expression of neighboring protein-coding genes. Elegant follow-up studies using chromosome conformation capture (3C) technology revealed that the co-activator complex Mediator is involved in tethering eRNAs to their gene targets. Hence, lncRNAs regulate the three-dimensional (3D) structure of chromosomes via Mediator-dependent chromosome looping (Lai et al., 2013), thereby bridging large intra- and interchromosomal distances in order to activate distal promoters (reviewed in Orom and Shiekhattar, 2013). This study nicely complemented reports about the lncRNA-mediated regulation of HOXA genes, in which chromosomal looping brings the eRNA HOTTIP in proximity to its target genes, marks the chromatin by H3K4 trimethylation and thus activates gene transcription (Wang et al., 2011). Taken together, the translation of the information content that lies within higher-order (3D) structures of chromosomes into (epigenetic) modifications of chromatin and regulation of gene transcription seems to be an emerging principle of lncRNA function.
Processing/Degradation of mRNA
Every step of RNA metabolism is subjected to fine-tuned and complex regulation (reviewed in Moore, 2005). LncRNAs have recently been involved in the control of RNA stability, the processing of (pre)-mRNAs and the regulation of mRNA decay. Natural antisense transcripts (NATs) are lncRNAs which are characterized by their location antisense to other coding or non-coding transcripts (Faghihi and Wahlestedt, 2009). The upregulation of NATs often causes downregulation of protein-coding transcripts on the opposite strand by the formation of RNA duplexes and triggering of cellular RNAi, although recently the NAT-mediated upregulation of protein-coding transcripts on the opposite strand were reported (Carrieri et al., 2012). The repressive effect of NATs onto the opposite strand not only holds true for duplexes consisting of (i) a protein-coding mRNA and a non-coding lncRNA NAT, but also for (ii) duplexes between a lncRNA NAT and another lncRNAs as demonstrated for lncRNAs which base-pair with and target the PTEN pseudogene PTENpg1 (Johnsson et al., 2013). The lncRNA-mediated regulation of (pre)-mRNA processing was demonstrated for the nuclear retained lncRNA MALAT1, which modulates alternative splicing via assembly of serine/arginine splicing factors within subnuclear compartments called nuclear speckles (Tripathi et al., 2010). Finally, the timely degradation of mRNAs by a process called Staufen-mediated decay (SMD) involves lncRNA-recipient sequences in the 3′UTR of SMD target genes. Here, intermolecular base-pairing between lncRNAs and sequences within the 3′UTR of Staufen (Stau) target genes triggers a cellular SMD response and the ensuing degradation of the transcript (Gong and Maquat, 2011; Wang et al., 2013). The in vivo significance of lncRNA-mediated SMD decay was underscored by the observation that epidermal differentiation critically depends on lncRNA-elicited mRNA decay. Here, a lncRNA called TINCR is recruited to specific sequences called “TINCR box” within TINCR target genes and elicits the decay of TINCR-bound transcripts by SMD (Kretz et al., 2013; Kretz, 2013).
Posttranscriptional Gene Regulation
MiRNAs recognize their gene targets by binding to 6–8 nt sequences called “seeds” located in the 3′UTR region of protein-coding RNAs (Bartel, 2009). It was demonstrated that recognition, binding and degradation/translational inactivation of miRNA targets is not necessarily confined to protein-coding transcripts. A so-called “competing endogenous RNA (ceRNA) hypothesis” was brought forward according to which protein-coding RNAs, miRNAs, and lncRNAs transcripts form large-scale regulatory networks which impinge on the expression of other transcripts independently of protein translation via competing for a limited pool of miRNAs (Salmena et al., 2011). Here, transcripts, so-called ceRNAs regulate the expression of other transcripts based on the similarity of their 3′UTR miRNA response elements (MRE) profile. According to this notion, two transcripts with a strong degree of common MREs can crosstalk to each other by competing for a given pool of miRNAs. Upregulation of one ceRNA increasingly “sponges” a limited pool of miRNAs and relieves the miRNA-mediated repressive tone on ceRNA-linked transcripts. The experimental confirmation of a ceRNA-like interdependency of protein-coding transcripts was first demonstrated for the tumor suppressor gene PTEN (Karreth et al., 2011; Tay et al., 2011), which “crosstalks” to hitherto unknown tumor suppressors. PTEN loss-of-function during cancerogenesis is also controlled by the genomic loss of its (non-coding) pseudogene PTENP1 which acts as a ceRNA (Poliseno et al., 2010). An additional layer of posttranscriptional regulation by lncRNAs accordingly lies within the specific pattern of MREs within lncRNAs which allow it to influence the expression of coding and non-coding transcripts in a ceRNA-like fashion. This is exemplified by the upregulation of a non-coding antisense homologue of the beta-secretase BACE1 (BACE1-AS) that acts as BACE1 ceRNA and concomitantly increases Bace1 mRNA stability and leads to augmented deposition of Aβ-plaques in Alzheimer’s disease (AD; Faghihi et al., 2008, 2010). Finally, a novel, intriguing class of functional lncRNAs, which is encoded in eukaryotic genomes, is constituted by circular RNAs (circRNAs). CircRNAs are expressed at high levels, can act as ceRNAs and effectively sponge miRNAs as shown for the neuroendocrine miRNA miR-7 (Hansen et al., 2013; Memczak et al., 2013).
Regulation of Protein Activity
Ribonucleic acid possesses the unique biochemical property to recognize and bind most biomolecules including proteins with unprecedented affinity (Stoltenburg et al., 2007). Thus, lncRNAs can specifically bind proteins and elegant studies have attributed novel roles for lncRNAs in the control of tissue homeostasis via direct binding and modification of protein activity. For example, a lncRNA termed Evi2 was shown to form stable complexes with members of the Dlx/Dll family of transcription factors, which are crucial regulators of developmental timing in vertebrates, and thereby regulate their transcriptional output (Feng et al., 2006). Further, two lncRNAs termed PRNBR1 and PCGEM1 that are upregulated in aggressive prostate cancer, synergistically and coordinately bind the carboxyterminal part of the androgen receptor (AR) and are required for AR-dependent gene transcription. In androgen-refractory prostate cancer, PRNBR1 and PCGEM1 are robustly expressed and are implicated in the ligand-independent activation of AR signaling [AR “resistant” prostate cancer (Yang et al., 2013)]. Another emerging paradigm of lncRNA-mediated regulation of protein activity is the sequestration of transcription factors as exemplified by the lncRNA Gas5, which is induced under conditions of nutrient deprivation and cellular stress. Gas5 acts as glucocorticoid receptor (GR) decoy by competing with GR-responsive elements (GREs) in gene promoters for binding to the DNA-binding domain of the GR (Kino et al., 2010). Increased levels of Gas5 thus interfere with GR binding to the DNA and effectively inhibit transactivation of GR-dependent gene promoters. Another example is nuclear factor kappa b (NFkB) signaling, which translates extracellular, proinflammatory cues [e.g., by tumor necrosis factor alpha (TNF-α) receptor activation] into changes in gene expression. NFkB activation induces the transcription of a specific subset of lncRNAs, apart from the induction of classical inflammatory protein-coding genes. Among this subset of TNF-regulated lncRNAs, a lncRNA termed Lethe is recruited to the NFkB effector subunit RelA in an inducible fashion and inhibits RelA from DNA-binding and target gene activation (Rapicavoli et al., 2013). Finally, the hypoxia-regulated lncRNA linc-p21 was shown to physically interact with hypoxia-inducible factor (HIF) 1alpha transcription factors. This HIF1a-linc-p21 circuit controls the hypoxia-evoked increases of the glycolytic “Warburg effect” in tumor cells (Yang et al., 2014).
LncRNAs in Control of Metabolism
The regulation of metabolism and glucose homeostasis is orchestrated and fine-tuned by a complex interplay of tissues/organs. Currently, we are faced with an unprecedented rise of obesity in the civilized world and the concurrent increase in obesity-associated diseases such as insulin resistance and type 2 diabetes mellitus (T2D). Key to the understanding of whole-body metabolism are the pleiotropic effects of the anabolic master regulator insulin which simultaneously controls peripheral as well as central-nervous system-related aspects of metabolism (Kahn et al., 2006). Resistance toward the effects of insulin constitutes a key step in the development of metabolic disease. The exciting observation that insulin and insulin-like growth factor (IGF) 1 signaling also triggers distinct changes in lncRNA expression [e.g., of the lncRNA CRNDE (Ellis et al., 2013)] points to the fact that lncRNAs may also be implicated in the metabolic effects of insulin and the development of insulin resistance. Thus, a strong interest lies within the identification of lncRNA-mediated mechanisms governing energy and glucose homeostasis at the cell-intrinsic, organ and whole-body level.
Tissue-Specific Regulation of Metabolism by lncRNAs
Maintenance of Pancreatic Beta Cell Identity
The main function of pancreatic islets lies within the synthesis, storage and secretion of insulin and glucagon, two hormonal regulators of glucose homeostasis. The possible control of islet development and function by lncRNAs was first demonstrated in studies which reported that the lncRNA H19 is involved in the intergenerational transmission of diabetes mellitus [gestational diabetes mellitus (GDM)] and the GDM-associated impairments of islet infrastructure and function (Ding et al., 2012). Global lncRNA screening approaches conducted by Moran et al. (2012) systematically interrogated the lncRNA transcriptome in human pancreatic beta cells. Here, the dynamic, strand and tissue-specific regulation of >1,000 lncRNA was reported using integrated transcriptional and chromosomal maps. Utilizing RNA-Seq data of 16 non-pancreatic tissues, the aforementioned gene set of pancreatic lncRNAs was shown to be significantly more specific for islet cells (40–55% for intergenic and antisense lncRNAs, respectively) than protein-coding genes (9.4%). Furthermore, the upregulation of islet-specific lncRNAs during progenitor commitment, glucose-stimulated upregulation and the striking dysregulation of islet-specific lncRNAs in patients with T2D pointed to a pathophysiological role of lncRNAs in the homeostasis of pancreatic tissues. The fact that a significant percentage of mouse and human lncRNA orthologs display similar cell- and stage-specific expression patterns suggests that evolutionarily conserved properties of lncRNAs extend beyond their primary sequence. This study was corroborated by a publication from the McManus laboratory, which presented a new catalog of the human beta cell (non-coding) lncRNA transcriptome in which >1,000 lncRNA were expressed in an islet-specific fashion involving islet-specific splicing events and promoter utilization (Ku et al., 2012). However, the elucidation of the molecular mechanisms underlying lncRNA-mediated regulation of beta cell differentiation and function still await discovery.
Regulation of Adipogenesis and Adipose Tissue Plasticity
The body harbors two principal types of adipose tissues which possess key functions in regulating the equilibrium between nutrient deposition and energy expenditure: Whereas white adipose tissue (WAT) serves as storage organ for excess nutrients, brown adipose tissue (BAT) dissipates the proton gradient across mitochondrial membranes to generate heat via the BAT-intrinsic uncoupling protein 1 (UCP1; Bartelt and Heeren, 2012). The accumulation of excess lipids that leads to low-grade inflammation in WAT has been linked to the development of insulin resistance in obese patients (Saltiel and Kahn, 2001; Gregor and Hotamisligil, 2011; Glass and Olefsky, 2012). Also, impaired BAT thermogenesis can contribute to the development of insulin resistance and obesity (Connolly et al., 1982; Feldmann et al., 2009). The fact that lncRNAs are implicated in the differentiation of adipose tissues (adipogenesis) is exemplified by the lncRNA SRA, which is required for full transactivation of the proadipogenic transcription factor Peroxisome proliferator-associated receptor gamma (Pparg). Concomitantly, RNAi-mediated SRA loss-of-function interfered with in vitro differentiation of 3T3-L1 preadipocytes (Xu et al., 2010). In a seminal study by Sun et al. (2013), the systematic implication of lncRNAs during adipogenesis was addressed. Using global transcriptome profiling of undifferentiated and mature adipocytes from the WAT and BAT lineages, the significant and specific regulation of 175 lncRNAs during adipogenesis was reported (Sun et al., 2013) of which a significant portion were enriched within adipose tissues. Finally, subsets of newly identified lncRNAs termed lncRAPs (lncRNAs Regulated in AdiPogenesis) were depleted in vitro using siRNAs. Distinct lncRAPs, which were specifically upregulated during adipogenesis and were induced by the proadipogenic transcription factors Cebpa and Pparg, were required for timely and complete maturation of adipocyte progenitor cells. These studies provide first evidence for a crucial role of lncRNAs in the control of adipogenesis and fat cell metabolism.
Differentiation of Skeletal Muscle and Cardiomyocytes
The differentiation of skeletal muscle cells (myogenesis) is regulated by a complex, yet well understood, evolutionarily conserved circuitry of protein-coding genes which control the timely growth, morphogenesis, and terminal maturation of muscle progenitors (myoblasts; Buckingham and Vincent, 2009). Here, the implication of noncoding RNAs was first shown via the contribution of myogenic miRNAs (myomiRs like miR-1 and miR-133) during myoblast commitment (Chen et al., 2006; Liu et al., 2008). Gong and Maquat (2011) reported that in human cells the degradation of distinct, nascent coding transcripts by Staufen-mediated decay (SMD) was regulated by lncRNAs. Here, the intermolecular base-pairing between Alu elements located within the 3′UTR of an SMD target and an Alu site localized within a class of lncRNAs called 1/2sbsRNAs (1/2-STAU1-binding site RNAs) triggered SMD (Gong and Maquat, 2011). This process was interestingly conserved in rodents that lack canonical Alu repeats. Here, the mouse homologue of 1/2sbsRNA was shown to be implicated in terminal differentiation of myoblast cells (Wang et al., 2013), indicating a function of lncRNAs in myogenesis. Another lncRNA termed linc-MD1 is also critical for myogenesis. Here, increased levels of linc-MD1 trigger the muscle differentiation program by acting as a natural decoy for myomiRs miR-133 and miR-135 (Cesana et al., 2011). MiR-133 and -135 in turn repress the expression of two pro-myogenic transcription factors, MAML1 and MEF2C. Recent reports also revealed that linc-MD1 takes part in a molecular feedforward circuit involving the promyogenic protein HuR (Legnini et al., 2014). Collectively, linc-MD1 promotes terminal differentiation of myoblasts via acting as ceRNA for myogenic transcriptional regulators by sequestering anti-myogenic miRNAs. Interestingly, this complex ceRNA-based interplay of classical mRNAs, lncRNAs, and miRNAs was dysregulated in patients suffering from Duchenne muscular dystrophy (DMD), a condition of reduced terminal differentiation of myoblasts. Reinstating DMD-associated downregulation of linc-MD1 expression via lentiviral delivery led to improved maturation of DMD myoblasts. In a study published by Klattenhoff et al. (2013), the heart-intrinsic lncRNA Braveheart (Bvht) was demonstrated to be required for differentiation of mesodermal progenitors toward mature cardiomyocytes via interaction with PRC2 epigenetic modifiers. This report for the first time implicated a tissue-specific lncRNA in maintaining cell fate during mammalian organogenesis.
Regulation of Neurogenesis by lncRNAs
The discovery that peripherally secreted hormones such as insulin and leptin control energy homeostasis and glucose metabolism via CNS-acting neurocircuits expanded our understanding on how the body ingests, stores and dissipates energy (Belgardt and Bruning, 2010). In an approach to identify lncRNAs, which are implicated in brain development and neurogenesis, Aprea et al. (2013) utilized transgenic in vivo approaches to isolate neural stem cells, partially committed neuronal precursor cells as well as terminally differentiated neurons and quantified the expression of lncRNAs. Several lncRNAs were identified that were involved in neurogenesis, neuroblast commitment and neuron survival as shown for the neuroregulatory lncRNA Miat. Thus, maintenance of the neuron stem cell pool and terminal differentiation of neuron progenitors are also under lncRNA-mediated control. This will hopefully entail studies in the future specifically addressing the regulation of defined neuronal circuits, which regulate peripheral metabolic by lncRNAs.
Regulation of Circadian Rhythm by lncRNAs
The mammalian clock plays a fundamental role in the regulation of energy and glucose homeostasis. Dysregulation of the circadian rhythm underlies several metabolic pathologies like the development of insulin resistance and the metabolic syndrome (Marcheva et al., 2010; Hatori et al., 2013). In addition to the central clock located in the suprachiasmatic nucleus (SCN) of the pineal gland in the CNS, subordinate, tissue-specific clocks exist which are also key for the regulation of diurnal aspects of lipid metabolism, oscillations in core body temperature and timely insulin secretion from pancreatic beta cells (Cretenet et al., 2010; Marcheva et al., 2010; Gerhart-Hines et al., 2013). Interestingly, as found for plants (Hazen et al., 2009), lncRNAs are involved in the regulation of vertebrate circadian systems. A study published by Coon et al. showed that 112 lncRNAs are differentially expressed between day/night within the pineal gland of rats (Coon et al., 2012). An in-depth investigation of eight highly rhythmic lncRNA revealed the pivotal role of neuronal projections from the SCN as well as external zeitgebers like light exposure onto periodicity and amplitude of circadian lncRNAs. In addition, a report from Vollmers et al. (2012) observed that rhythmic expression of ncRNAs like NATs, lncRNAs and miRNAs leads to rhythmic chromatin modifications in the liver. Noteworthy, the circadian oscillator component Per2 itself is controlled by an antisense lncRNA termed asPer2. Reports about the brain-derived regulation of circadian metabolism remain scarce yet the pathogenesis of Prader–Willi syndrome (PWS), a CNS-controlled genetic disorder circadian rhythm with an associated dysregulation of metabolism and the development of obesity, was shown to be influenced by a PWS-associated lncRNA called 116G. After splicing, a lncRNA consisting of the remnants of 116G (termed 116HG) bound to the transcriptional activator RBBP5 and ensures a physiological circadian rhythm in the brain. Mice deficient for 116HG exhibit metabolic disorders due to the dysregulation of diurnally expressed circadian genes like Clock, Cry1, and Per2 in the CNS (Powell et al., 2013).
Emerging Concept: Intercellular Communication by Exosomal lncRNAs?
Exosomes are small vesicles generated by budding of the plasma membrane and constitute a specific vehicle for intercellular communication. Upon release from donor cells, exosomal surface motifs serve as “address codes” for binding and endocytosis on acceptor cells. Specific exosomal shuttling RNAs (esRNAs) such as miRNAs can be packaged into exosomes and released after binding to recipient cells, thus constituting a novel and intriguing way for ncRNAs to regulate systemic aspects of metabolism (Ramachandran and Palanisamy, 2012). Similarly, the intercellular transport of high-density lipoproteins (HDL)-bound miRNAs that are released by distinct donor cells influence the miRNA profile of acceptor cells and concomitantly alter the gene expression in HDL-recipient target tissues (Vickers et al., 2011). Of note, deep sequencing of human exosomes revealed that lncRNAs are localized within micro-vesicles and may emerge as novel means of cellular communication (Huang et al., 2013). Although experimental proof of concept is still lacking, the endocrine transfer of exosomal lncRNA might represent a novel facette relevant for lncRNA-mediated control of metabolism.
Therapeutic Opportunities of lncRNA Inhibition
A high economic interest lies in the development of sequence-specific compounds for the inhibition of disease-associated ncRNAs. Short, chemically modified ribonucleic acid compounds like locked nucleic acids (LNAs) efficiently silence the expression of ncRNAs such as miRNAs and are generally well tolerated in vivo (Krutzfeldt et al., 2005; Esau et al., 2006; Elmen et al., 2008). These anti-RNA compounds were initially tested in mice (Krutzfeldt et al., 2005) and adopted to the non-human primate situation (Elmen et al., 2008) with unprecedented speed. This approach will hopefully be extended to disease-associated lncRNA in the near future. Most in vivo studies to date concentrated on disease-associated miRNAs, that were critically involved in the development of insulin resistance and the deterioration of metabolic health (Jordan et al., 2011; Trajkovski et al., 2011; Zhou et al., 2012; Kornfeld et al., 2013). In contrast, most insights concerning the metabolic functions of lncRNAs were inferred from in vitro studies. The rising numbers of lncRNA knockout models [exemplified by a recent report on 18 lncRNA loss-of-function mouse models (Sauvageau et al., 2013)] showcase that in order to convincingly assess, whether lncRNAs are implicated in the in vivo control of metabolism, further animal models for lncRNA loss- and gain-of-function are needed. This is of timely importance as systemic antisense oligonucleotide (ASO)-mediated inhibition of disease-associated lncRNAs (even in difficult to target organs like skeletal muscle) effectively improves degenerative diseases like myotonic dystrophy type 1 (DM1) in mice (Wheeler et al., 2012).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aprea, J., Prenninger, S., Dori, M., Ghosh, T., Monasor, L. S., Wessendorf, E., et al. (2013). Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 32, 3145–3160. doi: 10.1038/emboj.2013.245
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bartelt, A., and Heeren, J. (2012). The holy grail of metabolic disease: brown adipose tissue. Curr. Opin. Lipidol. 23, 190–195. doi: 10.1097/MOL.0b013e328352dcef
Belgardt, B. F., and Bruning, J. C. (2010). CNS leptin and insulin action in the control of energy homeostasis. Ann. N. Y. Acad. Sci. 1212, 97–113. doi: 10.1111/j.1749-6632.2010.05799.x
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigó, R., Gingeras, T. R., Margulies, E. H., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. doi: 10.1038/nature05874
Buckingham, M., and Vincent, S. D. (2009). Distinct and dynamic myogenic populations in the vertebrate embryo. Curr. Opin. Genet. Dev. 19, 444–453. doi: 10.1016/j.gde.2009.08.001
Cabili, M. N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. doi: 10.1101/gad.17446611
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. doi: 10.1126/science.1112014
Carrieri, C., Cimatti, L., Biagioli, M., Beugnet, A., Zucchelli, S., Fedele, S., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457. doi: 10.1038/nature11508
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and Mechanisms of miRNAs and siRNAs. Cell 136, 642–655. doi: 10.1016/j.cell.2009.01.035
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi: 10.1016/j.cell.2011.09.028
Chen, J. F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233. doi: 10.1038/ng1725
Chew, G. L., Pauli, A., Rinn, J. L., Regev, A., Schier, A. F., and Valen, E. (2013). Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 140, 2828–2834. doi: 10.1242/dev.098343
Clark, M. B., Amaral, P. P., Schlesinger, F. J., Dinger, M. E., Taft, R. J., Rinn, J. L., et al. (2011). The reality of pervasive transcription. PLoS Biol. 9:e1000625; discussion e1001102. doi: 10.1371/journal.pbio.1000625
Connolly, E., Morrisey, R. D., and Carnie, J. A. (1982). The effect of interscapular brown adipose tissue removal on body-weight and cold response in the mouse. Br. J. Nutr. 47, 653–658. doi: 10.1079/BJN19820077
Coon, S. L., Munson, P. J., Cherukuri, P. F., Sugden, D., Rath, M. F., Moller, M., et al. (2012). Circadian changes in long noncoding RNAs in the pineal gland. Proc. Natl. Acad. Sci. U. S. A. 109, 13319–13324. doi: 10.1073/pnas.1207748109
Cretenet, G., Le Clech, M., and Gachon, F. (2010). Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 11, 47–57. doi: 10.1016/j.cmet.2009.11.002
David, L., Huber, W., Granovskaia, M., Toedling, J., Palm, C. J., Bofkin, L., et al. (2006). A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U.S.A. 103, 5320–5325. doi: 10.1073/pnas.0601091103
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
Ding, G. L., Wang, F. F., Shu, J., Tian, S., Jiang, Y., Zhang, D., et al. (2012). Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes 61, 1133–1142. doi: 10.2337/db11-1314
Dinger, M. E., Pang, K. C., Mercer, T. R., and Mattick, J. S. (2008). Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 4:e1000176. doi: 10.1371/journal.pcbi.1000176
Ellis, B. C., Graham, L. D., and Molloy, P. L. (2013). CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim. Biophys. Acta 1843, 372–386. doi: 10.1016/j.bbamcr.2013.10.016
Elmen, J., Lindow, M., Schütz, S., Lawrence, M., Petri, A., Obad, S., et al. (2008). LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899. doi: 10.1038/nature06783
Esau, C., Davis, S., Murray, S. F., Yu, X. X., Pandey, S. K., Pear, M., et al. (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98. doi: 10.1016/j.cmet.2006.01.005
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730. doi: 10.1038/nm1784
Faghihi, M. A., and Wahlestedt, C. (2009). Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 10, 637–643. doi: 10.1038/nrm2738
Faghihi, M. A., Zhang, M., Huang, J., Modarresi, F., Van der Brug, M. P., Nalls, M. A., et al. (2010). Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11, R56. doi: 10.1186/gb-2010-11-5-r56
Feldmann, H. M., Golozoubova, V., Cannon, B., and Nedergaard, J. (2009). UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 9, 203–209. doi: 10.1016/j.cmet.2008.12.014
Feng, J., Bi, C., Clark, B. S., Mady, R., Shah, P., and Kohtz, J. D. (2006). The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484. doi: 10.1101/gad.1416106
Gerhart-Hines, Z., Feng, D., Emmett, M. J., Everett, L. J., Loro, E., Briggs, E. R., et al. (2013). The nuclear receptor Rev-erbalpha controls circadian thermogenic plasticity. Nature 503, 410–413. doi: 10.1038/nature12642
Glass, C. K., and Olefsky, J. M. (2012). Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 15, 635–645. doi: 10.1016/j.cmet.2012.04.001
Gong, C., and Maquat, L. E. (2011). lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470, 284–288. doi: 10.1038/nature09701
Gregor, M. F., and Hotamisligil, G. S. (2011). Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 29, 415–445. doi: 10.1146/annurev-immunol-031210-101322
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. doi: 10.1038/nature07672
Guttman, M., Russell, P., Ingolia, N. T., Weissman, J. S., and Lander, E. S. (2013). Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251. doi: 10.1016/j.cell.2013.06.009
Hangauer, M. J., Vaughn, I. W., and McManus, M. T. (2013). Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 9:e1003569. doi: 10.1371/journal.pgen.1003569
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Hatori, M., Vollmers, C., Zarrinpar, A., DiTacchio, L., Bushong, E. A., Gill, S., et al. (2013). Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860. doi: 10.1016/j.cmet.2012.04.019
Hazen, S. P., Naef, F., Quisel, T., Gendron, J. M., Chen, H., Ecker, J. R., et al. (2009). Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 10, R17. doi: 10.1186/gb-2009-10-2-r17
Huang, X., Yuan, T., Tschannen, M., Sun, Z., Jacob, H., Du, M., et al. (2013). Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14, 319. doi: 10.1186/1471-2164-14-319
Johnsson, P., Ackley, A., Vidarsdottir, L., Lui, W. O., Corcoran, M., Grandér, D., et al. (2013). A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 20, 440–446. doi: 10.1038/nsmb.2516
Jordan, S. D., Krüger, M., Willmes, D. M., Redemann, N., Wunderlich, F. T., Brönneke, H. S., et al. (2011). Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 13, 434–446. doi: 10.1038/ncb2211
Kahn, S. E., Hull, R. L., and Utzschneider, K. M. (2006). Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444, 840–846. doi: 10.1038/nature05482
Karreth, F. A., Tay, Y., Perna, D., Ala, U., Tan, S. M., Rust, A. G., et al. (2011). In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395. doi: 10.1016/j.cell.2011.09.032
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Nat. Acad. Sci. U.S.A. 106, 11667–11672. doi: 10.1073/pnas.0904715106
Kim, W., and Kyung Lee, E. (2013). Post-transcriptional regulation in metabolic diseases. RNA Biol. 9, 772–780. doi: 10.4161/rna.20091
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., and Chrousos, G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3, ra8. doi: 10.1126/scisignal.2000568
Klattenhoff, C. A., Scheuermann, J. C., Surface, L. E., Bradley, R. K., Fields, P. A., Steinhauser, M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. doi: 10.1016/j.cell.2013.01.003
Kornfeld, J. W., Baitzel, C., Könner, A. C., Nicholls, H. T., Vogt, M. C., Herrmanns, K., et al. (2013). Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 494, 111–115. doi: 10.1038/nature11793
Kretz, M. (2013). TINCR, staufen1, and cellular differentiation. RNA Biol. 10, 1597–1601. doi: 10.4161/rna.26249
Kretz, M., Siprashvili, Z., Chu, C., Webster, D. E., Zehnder, A., Qu, K., et al. (2013). Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 93, 231–235. doi: 10.1038/nature11661
Krutzfeldt, J., Rajewsky, N., Braich, R., Rajeev, K. G., Tuschl, T., Manoharan, M., et al. (2005). Silencing of microRNAs in vivo with ‘antagomirs.’ Nature 438, 685–689. doi: 10.1038/nature04303
Ku, G. M., Kim, H., Vaughn, I. W., Hangauer, M. J., Myung Oh, C., German, M. S., et al. (2012). Research resource: RNA-Seq reveals unique features of the pancreatic beta-cell transcriptome. Mol. Endocrinol. 26, 1783–1792. doi: 10.1210/me.2012-1176
Lai, F., Orom, U. A., Cesaroni, M., Beringer, M., Taatjes, D. J., Blobel, G. A., et al. (2013). Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501. doi: 10.1038/nature11884
Legnini, I., Morlando, M., Mangiavacchi, A., Fatica, A., and Bozzoni, I. A. (2014). Feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell 53, 506–514. doi: 10.1016/j.molcel.2013.12.012
Li, L., Wang, X., Stolc, V., Li, X., Zhang, D., Su, N., et al. (2006). Genome-wide transcription analyses in rice using tiling microarrays. Nat. Genet. 38, 124–129. doi: 10.1038/ng1704
Liu, N., Bezprozvannaya, S., Williams, A. H., Qi, X., Richardson, J. A., Bassel-Duby, R., et al. (2008). microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 22, 3242–3254. doi: 10.1101/gad.1738708
Lynn, F. C. (2009). Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol. Metab. 20, 452–459. doi: 10.1016/j.tem.2009.05.007
Marcheva, B., Ramsey, K. M., Buhr, E. D., Kobayashi, Y., Su, H., Ko, C. H., et al. (2010). Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631. doi: 10.1038/nature09253
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560. doi: 10.1038/nature06008
Moore, M. J. (2005). From birth to death: the complex lives of eukaryotic mRNAs. Science 309, 1514–1518. doi: 10.1126/science.1111443
Moran, I., Akerman, I., van de Bunt, M., Xie, R., Benazra, M., Nammo, T., et al. (2012). Human beta cell transcriptome analysis uncovers incRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 16, 435–448. doi: 10.1016/j.cmet.2012.08.010
Orom, U. A., Derrien, T., Beringer, M., Gumireddy, K., Gardini, A., Bussotti, G., et al. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58. doi: 10.1016/j.cell.2010.09.001
Orom, U. A., and Shiekhattar, R. (2013). Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154, 1190–1193. doi: 10.1016/j.cell.2013.08.028
Pachnis, V., Brannan, C. I., and Tilghman, S. M. (1988). The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 7, 673–681.
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S., and Brockdorff, N. (1996). Requirement for Xist in X chromosome inactivation. Nature 379, 131–137. doi: 10.1038/379131a0
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J., Pandolfi, P. P., et al. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. doi: 10.1038/nature09144
Powell, W. T., Coulson, R. L., Crary, F. K., Wong, S. S., Ach, R. A., Tsang, P., et al. (2013). A Prader–Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum. Mol. Genet. 22, 4318–4328. doi: 10.1093/hmg/ddt281
Ramachandran, S., and Palanisamy, V. (2012). Horizontal transfer of RNAs: exosomes as mediators of intercellular communication. Wiley Interdiscip. Rev. RNA 3, 286–293. doi: 10.1002/wrna.115
Rapicavoli, N. A., Qu, K., Zhang, J., Mikhail, M., Laberge, R. M., Chang, H. Y., et al. (2013). A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2, e00762. doi: 10.7554/eLife.00762
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Rottiers, V., and Naar, A. M. (2012). MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 13, 239–250. doi: 10.1038/nrm3313
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Saltiel, A. R., and Kahn, C. R. (2001). Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806. doi: 10.1038/414799a
Sauvageau, M., Goff, L. A., Lodato, S., Bonev, B., Groff, A. F., Gerhardinger, C., et al. (2013). Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2, e01749. doi: 10.7554/eLife.01749
Spitale, R. C., Tsai, M. C., and Chang, H. Y. (2011). RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6, 539–543. doi: 10.4161/epi.6.5.15221
Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M. F., Rifkin, S. A., et al. (2004). A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306, 655–660. doi: 10.1126/science.1101312
Stoltenburg, R., Reinemann, C., and Strehlitz, B. (2007). SELEX–a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 24, 381–403. doi: 10.1016/j.bioeng.2007.06.001
Sun, L., Goff, L. A., Trapnell, C., Alexander, R., Lo, K. A., Hacisuleyman, E., et al. (2013). Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 3387–3392. doi: 10.1073/pnas.1222643110
Tay, Y., Kats, L., Salmena, L., Weiss, D., Tan, S. M., Ala, U., et al. (2011). Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147, 344–357. doi: 10.1016/j.cell.2011.09.029
Thomas, D. J., Rosenbloom, K. R., Clawson, H., Hinrichs, A. S., Trumbower, H., Raney, B. J., et al. (2007). The ENCODE Project at UC Santa Cruz. Nucleic Acids Res. 35, D663–D667. doi: 10.1093/nar/gkl1017
Trajkovski, M., Hausser, J., Soutschek, J., Bhat, B., Akin, A., Zavolan, M., et al. (2011). MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649–653. doi: 10.1038/nature10112
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
van Bakel, H., Nislow, C., Blencowe, B. J., and Hughes, T. R. (2010). Most “dark matter” transcripts are associated with known genes. PLoS Biol. 8:e1000371. doi: 10.1371/journal.pbio.1000371
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433. doi: 10.1038/Ncb2210
Vollmers, C., Schmitz, R. J., Nathanson, J., Yeo, G., Ecker, J. R., and Panda, S. (2012). Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 16, 833–845. doi: 10.1016/j.cmet.2012.11.004
Wang, J., Gong, C., and Maquat, L. E. (2013). Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 27, 793–804. doi: 10.1101/gad.212639.112
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi: 10.1038/nature09819
Wheeler, T. M., Leger, A. J., Pandey, S. K., MacLeod, A. R., Nakamori, M., Cheng, S. H., et al. (2012). Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 488, 111–115. doi: 10.1038/nature11362
Xu, B., Gerin, I., Miao, H., Vu-Phan, D., Johnson, C. N., Xu, R., et al. (2010). Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS ONE 5:e14199. doi: 10.1371/journal.pone.0014199
Yang, F., Zhang, H., Mei, Y., and Wu, M. (2014). Reciprocal regulation of HIF-1alpha and LincRNA-p21 modulates the Warburg effect. Mol. Cell 53, 88–100. doi: 10.1016/j.molcel.2013.11.004
Yang, L., Lin, C., Jin, C., Yang, J. C., Tanasa, B., Li, W., et al. (2013). lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500, 598–602. doi: 10.1038/nature12451
Yap, K. L., Li, S., Muñoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S., et al. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674. doi: 10.1016/j.molcel.2010.03.021
Keywords: lncRNAs, glucose homeostasis, metabolism and obesity, non-coding RNA (ncRNA), cell differentiation
Citation: Kornfeld J-W and Brüning JC (2014) Regulation of metabolism by long, non-coding RNAs. Front. Genet. 5:57. doi: 10.3389/fgene.2014.00057
Received: 28 January 2014; Accepted: 05 March 2014;
Published online: 25 March 2014.
Edited by:
Romano Regazzi, University of Lausanne, SwitzerlandReviewed by:
Bernard Mari, Centre National de la Recherche Scientifique, FranceZhengyu Jiang, Fox Chase Cancer Center, USA
Copyright © 2014 Kornfeld and Brüning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jens C. Brüning, Max-Planck-Institute for Neurological Research, Gleueler Strasse 50, 50931 Köln, Germany e-mail: ai023@uni-koeln.de
 Jan-Wilhelm Kornfeld
Jan-Wilhelm Kornfeld Jens C. Brüning1,2,3,4*
Jens C. Brüning1,2,3,4*