- School of Life Sciences, Anhui Agricultural University, Hefei, China
Zinc finger-homeodomain (ZHD) genes encode a family of plant-specific transcription factors that not only participate in the regulation of plant growth and development but also play an important role in the response to abiotic stress. The ZHD gene family has been studied in several model plants, including Solanum lycopersicum, Zea mays, Oryza sativa, and Arabidopsis thaliana. However, a comprehensive study of the genes of the ZHD family and their roles in fiber development and pigmentation in upland cotton has not been completed. To address this gap, we selected a brown fiber cultivar for our study; brown color in cotton is one of the most desired colors in the textile industry. The natural colored fibers require less processing and little dying, thereby eliminating dye costs and chemical residues. Using bioinformatics approaches, we identified 37 GhZHD genes from Gossypium hirsutum and then divided these genes into seven groups based on their phylogeny. The GhZHD genes were mostly conserved in each subfamily with minor variations in motif distribution and gene structure. These genes were largely distributed on 19 of the 26 upland cotton chromosomes. Among the Gossypium genomes, the paralogs and orthologs of the GhZHD genes were identified and further characterized. Furthermore, among the paralogs, we observed that the ZHD family duplications in Gossypium genomes (G. hirsutum, G. arboreum, and G. raimondii) were probably derived from segmental duplication or genome-wide duplication (GWD) events. Through a combination of qRT-PCR and proanthocyanidins (PA) accumulation analyses in brown cotton fibers, we concluded that the candidate genes involved in early fiber development and fiber pigment synthesis include the following: GhZHD29, GhZHD35, GhZHD30, GhZHD31, GhZHD11, GhZHD27, GhZHD18, GhZHD15, GhZHD16, GhZHD22, GhZHD6, GhZHD33, GhZHD13, GhZHD5, and GhZHD23. This study delivers insights into the evolution of the GhZHD genes in brown cotton, serves as a valuable resource for further studies, and identifies the conditions necessary for improving the quality of brown cotton fiber.
Introduction
Zinc finger-homeodomain (ZHD) transcription factors (TFs) are major regulators of the body plan specification of higher plants and are especially involved in plant development (fiber development) and stress responses (Wang et al., 2015; Khatun et al., 2017). The first homeobox genes were identified in the fruit fly; however, these genes have since been isolated in many organisms, including fungi, plants, nematodes, and humans (Nam and Nei, 2005; Bhattacharjee et al., 2015). TFs can activate or repress target genes by directly binding to gene motifs or elements. Many TF families have evolved unique DNA-binding domains that direct their binding activities. The HD is a well-characterized DNA-binding domain that is encoded by a conserved 60 amino acid sequence (Mukherjee et al., 2009; Wang et al., 2014).
In plant and animal genomes, homeobox genes are part of a large gene family. Based on the number, nature, and spacing pattern, these genes can be categorized into different groups. Initially, zinc finger genes were categorized into the following groups: KNOX, ZM-HOX, BELL, AT-HB8, HAT, and GAL2 (Bharathan et al., 1997; Bhattacharjee et al., 2015). Over time, homeobox genes in rice were classified into ten subclasses: HD-Zip I, HD-Zip II, HD-Zip III, HD-Zip IV, KNOX I, KNOX II, BLH, WOX, PHD, and ZF-HD. Consequently, another systematic study on homeobox genes was carried out in which the genes were categorized into 14 subclasses, including the addition of some new classes, such as DDT, NDX, PHD, SAWADEE, LD, and PINTOX (Mukherjee et al., 2009). While some zinc fingers (C2H2, C2C2, and C3H) interact with one zinc ion, new approaches demonstrated that the animal Lin-11/Is1-1/Mec-3 (LIM) domain and plant RING finger domains interact with two zinc ions (Halbach et al., 2000; Englbrecht et al., 2004; Yanagisawa, 2004; Wang et al., 2014).
The first cluster of novel ZHD proteins was isolated from Flaveria as a potential regulator of the gene encoding C4 phosphoenolpyruvate carboxylase (PEPCase) (Windhovel et al., 2001). Windhovel et al. (2001) reported that the ZHD domain is capable of binding DNA, predominantly to the regulatory region of the C4 PEPCase genes. Subsequently, Windhovel et al. (2001) also described that ZF domains are not only intricately involved in DNA binding but also boost the protein–DNA interactions facilitated by the HD domain. A large number of studies on ZHD family genes have been performed in various plants, including Arabidopsis thaliana (Tan and Irish, 2006), Glycine max (Deng et al., 2002), Oryza sativa (Jørgensen et al., 1999), and Triticum aestivum (Bhattacharjee and Jain, 2013). Many members of the ZHD class are critical components in the regulation of blue light signaling, vascular development, biogenesis of the outer cell layer of plant organs, and the response to stress, in addition to controlling anthocyanin processes. While ZHD proteins were first reported to have a potential role in the regulation of floral development, it was later found that an Arabidopsis ZHD protein (AtZHD1) could bind to the promoter of EARLY RESPONSE TO DEHYDRATION STRESS 1 (ERD1). For example, the expression pattern of AtZHD1 is induced by abscisic acid, salt stress, and dehydration (Tran et al., 2007; Wang et al., 2014). In addition, ZHD proteins can interact with some NAC proteins and the simultaneous overexpression of ZHD and NAC genes increased drought tolerance in Arabidopsis (Tran et al., 2007; Hu et al., 2008). To date, 14 ZHD genes in Arabidopsis have been identified and characterized. Recently, the functions of ZHD genes in some other crops have been reported. For example, four rice ZHD genes have also been associated with gene regulation. Additionally, two soybean proteins, GmZHD1 and GmZHD2, have been found to bind to the promoter of the gene encoding calmodulin isoform 4 (GmCaM4) and increase its expression upon pathogen stimulation (Park et al., 2007; Hu et al., 2008; Wang et al., 2014). Hu et al. (2008) stated that MIF1 interacts with ZHD proteins and that the overexpression of MIF1 interfered with the normal functions of ZHD proteins. If this is true, ZHD proteins may play important roles in regulating plant physiology and development. However, while the function of ZHD genes has been elucidated in Arabidopsis and other model crops, the functions of these genes in fiber development in Gossypium hirsutum have not yet been identified. G. hirsutum is one of the most valuable agricultural crops in the world and has been extensively studied on the developmental and physiological levels. G. hirsutum is a heterologous tetraploid cotton containing AA and DD genomes, which were formed approximately 1–2 million years ago. It is widely believed that G. arboreum and G. raimondii were the donators of the A and D chromosomes, respectively (Paterson et al., 2012; Wang et al., 2012; Chen et al., 2017a,b). The availability of complete Gossypium genome sequences makes it possible to examine and identify transcriptomic differences, duplications, and family sizes on a genome-wide scale spanning a broad evolutionary distance in the plant kingdom. Here, we report the identification of ZHD genes using Gossypium genome sequences and describe their characteristics, including phylogenetic and syntenic analyses, gene duplications, chromosomal locations, evolutionary mechanism, PA content, and expression differences during various fiber development stages. Our results provide a valuable foundation for future studies on ZHD proteins in brown cotton to facilitate functional analysis.
Materials and Methods
Plant Material
The brown cotton plant line Zongcaixuan No. 1 was used in these experiments at the High Technology Agricultural Park of Anhui Agricultural University (Hefei, Anhui, China). In July 2017, 60 plants in the blooming stage with good growth characteristics were selected. The experimental material was frozen in liquid nitrogen and quickly transferred to the laboratory refrigerator. The RNA of cotton fibers at 6, 12, 18, 24, and 30 days post-anthesis (DPA) was isolated for this study.
Genomic Resources for the Screening of ZHD Genes in Gossypium Genomes
To identify the ZHD genes in Gossypium genomes, the genome sequences of the cotton species Gossypium arboreum (BJI, version 1.0), G. raimondii (JGI, version 2.0), and G. hirsutum (NAU, version 1.1) were downloaded from COTTONGEN1 (Yu et al., 2014), according to previously reported methods (Cao et al., 2017; Su et al., 2017; Abdullah et al., 2018b). Proteins with ZHD domains (PF04770) were retrieved from the Pfam database. HMMER software, which uses the hidden Markov model (HMM), was used on the Gossypium sequences with an e-value cut off of 0.001. Subsequently, we verified all sequences using various tools (Pfam, InterProScan database, NCBI, and SMART databases) (Zdobnov and Apweiler, 2001; Bateman, 2002; Letunic et al., 2012; Finn et al., 2014). The ExPASy program2 was used to determine the molecular weight and isoelectric points of the identified proteins (Gasteiger et al., 2003).
Phylogenetic and Gene Structure Analysis
CLUSTAL_X software was used to perform the alignments of all the ZHD amino acid sequences using default parameters (Thompson et al., 1997). The phylogenetic tree was generated with MEGA 5.1 software using full-length sequences by using the maximum likelihood (ML) method with 1,000 bootstrap replications. The map of exon–intron structures of the GhZHD genes was analyzed by the Gene Structure Display Server 2.0 (GSDS)3. The MEME online tool4 was used to search the conservative motifs of GhZHD proteins, with a maximum width of 200 amino acids, a limit of 20 motifs, and all other default parameters (Bailey et al., 2015). Additionally, Pfam, InterProScan, and SMART databases were used to annotate these motifs (Zdobnov and Apweiler, 2001; Bateman, 2002; Letunic et al., 2012).
Interspecies Microsynteny and Cis-Acting Element Analysis
The Multiple Collinearity Scan toolkit (MCScanX package with default parameters) was used to determine microsynteny among the Gossypium genomes. The ZHD genes of G. hirsutum, G. arboreum, and G. raimondii were ordered according to their evolutionary tree classification. We examined the putative promoter sequence from each GhZHD coding sequence, which is defined as the 1,500 bp upstream of the start codon (TTS), and analyzed the cis-elements using the PLANT CARE program5.
Calculation of Non-synonymous (Ka) to Synonymous (Ks) Substitution Rates
DnaSP v5.0 software was used to determine the synonymous (Ks) and non-synonymous (Ka) nucleotide substitution rates. Each duplicated gene pair’s Ka/Ks ratio was calculated to determine the selection pressure. Sliding window analyses were performed for each duplicated gene pair to analyze the synonymous and non-synonymous substitution rates of encoding site paralogs.
Physical Localization and Expansion Patterns
Information on the specific location of all GhZHD genes was obtained from genome annotation data and the chromosomal location of each gene was mapped using MapInspect software6. The expansion patterns of GhZHD genes in G. hirsutum were examined using MCScanX software with default parameters (Wang et al., 2015).
Expression Analysis
The RNA-Seq data derived from the TM-1 transcriptome of the Cotton Functional Genomics Database7 (Zhu et al., 2017) were used to analyze the GhZHD gene expression profiles in G. hirsutum. In terms of expression level, we considered a gene expressed if its log2 (FPKM) value was higher than 1, and not expressed if its log2 (FPKM) value was equal to or less than 1. G. hirsutum GhZHD gene expression profiles were visualized using R software.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was extracted and reverse-transcribed from brown cotton fiber at 6, 12, 18, 24, and 30 DPA using the Tiangen plant RNA extraction kit (Beijing, China). We designed specific primers (Supplementary Table S3) using Primer Premier 6 software. All primers are listed in Supplementary Table S3. The qRT-PCR analysis was performed using SYBR Green Master Mix (Takara, Japan) and detected with a CFX96 TouchTM Real-time PCR Detection System (Singapore). Relative expression levels were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Proanthocyanidins (PA) Content
According to previously described methods (Ikegami et al., 2009), brown cotton fiber bolls at 6, 12, 18, 24, and 30 DPA were stripped, extracted with 80% methanol and placed into ultrasonic extraction for 30 min. Next, the samples were centrifuged for 15 min and the resulting supernatant was analyzed for soluble PAs. A methanol solution containing 1% HCl was added to the precipitate and the solution was placed in a 6°C water bath for 1 h. After centrifugation for 15 min, the supernatants contained the insoluble PAs and the PA content was determined by spectrophotometry according to a standard curve of catechins, which were used as controls (Li et al., 1996). For each experiment, three biological replicates were performed.
Results
Identification of ZHD Genes in Gossypium Genomes
To identify potential ZHD domain-encoding genes of G. hirsutum, G. arboreum, and G. raimondii, we obtained the ZHD domain (PF04770) from the Pfam database and generated an HMM profile with the HMMER 3.0 package. After removing repetitive sequences and any sequence lacking the ZHD domain, we obtained a total of 37 non-redundant GhZHD genes. The presence of 37 ZHD genes in G. hirsutum is greater than several in other species, including G. arboreum (22 ZHD genes), G. raimondii (19 ZHD genes), Chinese cabbage (31 ZHD genes) (Wang et al., 2015), grapes (13 ZHD genes) (Wang et al., 2014), and tomato (22 ZHD genes) (Khatun et al., 2017; Supplementary Table S1). We designated the genes GhZHD1–GhZHD37 according to their order on the chromosomes (Supplementary Figure S1). The lengths (aa) of the proteins encoded by all of the GhZHD family members varied from 396 to 1944 aa, with an average length of 818 aa. Similarly, the molecular weight (MW) and the isoelectronic point (IP) varied from 36646.47 to 15391.95 kDa and from 9.12 to 6.54, respectively. The details of all 37 GhZHD genes, including physicochemical characteristics such as chromosome location, gene identifier, protein length (aa), molecular weight (MW), and isoelectric point (pI) are reported in Supplementary Table S1.
Phylogenetic and Gene Structure Analysis of ZHD Genes
To gain insight into the evolutionary relationships of the upland cotton GhZHD gene family, a phylogenetic analysis was conducted from the 70 amino acid sequences of G. hirsutum, Arabidopsis (A. thaliana), maize (Zea mays), and rice (O. sativa) (Figure 1A). The phylogenetic tree was drawn based on sequence similarity and topology using Mega 5.1 software with the ML method and 1,000 bootstrap replications. After an examination of this phylogenetic tree, the GhZHD genes could be categorized into seven well-conserved clades (s1–s7) with bootstrap support. Clade s5 was the largest, containing 16 GhZHD members (approximately 22% of the total GhZHD genes), while clade s7 was the smallest, containing only two members. Numerous G. hirsutum GhZHD genes were not clustered with Arabidopsis ZHD genes. These GhZHD genes may have developed in upland cotton after diverging from the last common ancestor, or they may have been lost in Arabidopsis.
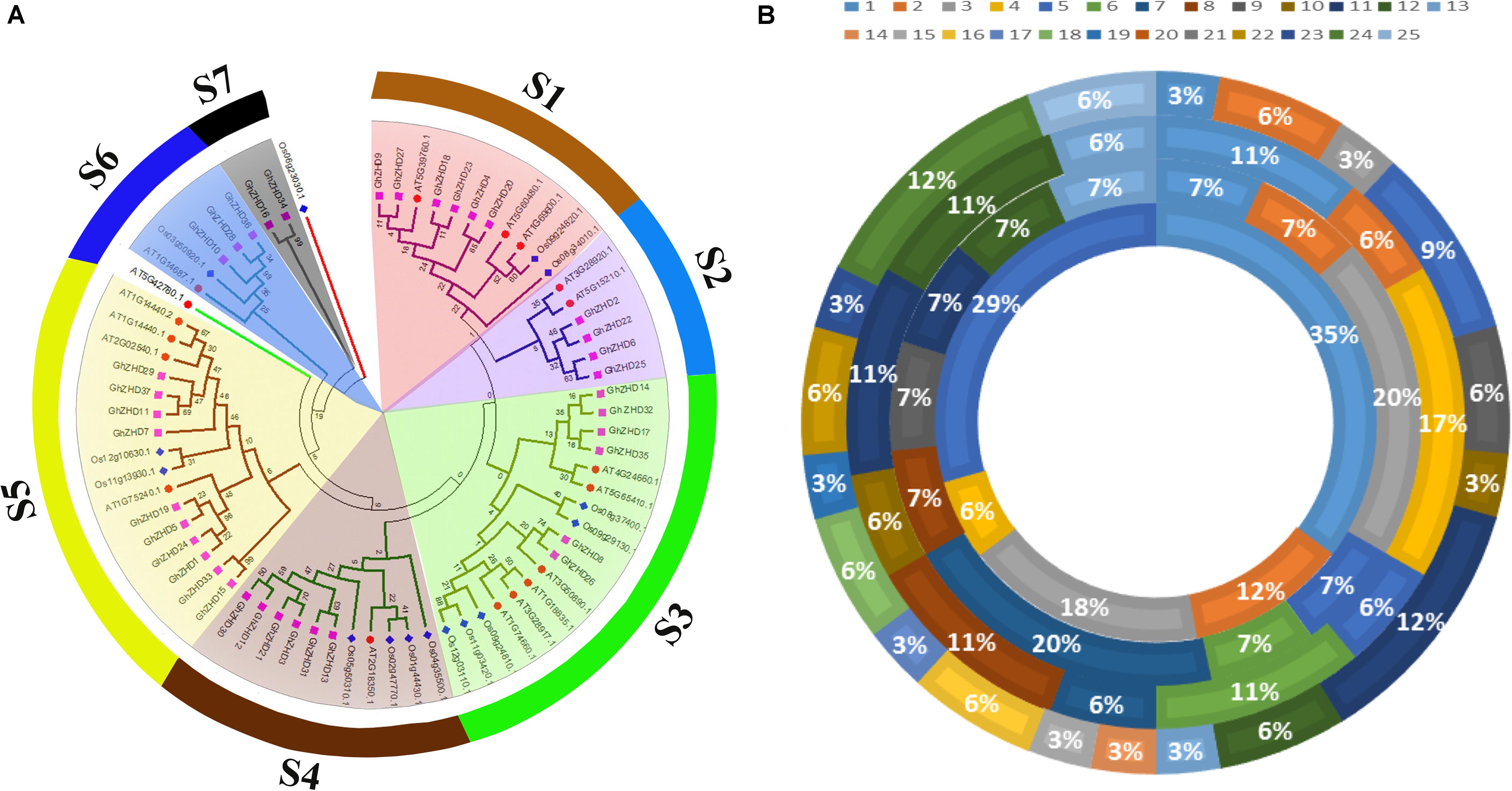
FIGURE 1. Phylogenetic tree of ZHD proteins from upland cotton, rice, maize, and Arabidopsis. The tree was generated with MEGA 5 software using the neighbor-joining method (A). The chromosomal distribution and percentage of ZHD family genes shared in G. hirsutum, G. arboreum, G. raimondii, and Arabidopsis. The outermost circle represents the chromosomes of G. hirsutum, followed by G. arboreum and G. raimondii, with the innermost circle representing Arabidopsis (B).
To further evaluate the diversity of upland cotton ZHD proteins, we used the MEME online program to predict conserved protein motifs. Five conserved motifs were identified in each comparison and labeled as Motif 1 through Motif 5 (Supplementary Figure S2). Interestingly, fewer motifs were observed in the GhZHD genes of upland cotton compared to other crops, including Arabidopsis, tomato, and Chinese cabbage (Wang et al., 2014; Khatun et al., 2017). Motif 1 and Motif 2 were the most common motifs and comprised the ZHD dimer domain. Most of the GhZHDs contained Motif 1 and Motif 2, suggesting that GhZHD genes have a conserved ZHD domain.
In addition to identifying conserved protein motifs, we also analyzed the structural diversity of upland cotton GhZHD genes. As shown in Supplementary Figure S2, most members of the same group have a similar exon–intron structure. We also highlighted domain positions (red color) within the exon–intron structure. Interestingly, after comparing the cDNA and genomic sequences of GhZHD genes, we observed that most of the GhZHD genes have only one exon and no introns. This unique pattern of GhZHD genes is different from other TFs. This intronless feature of this gene family indicates that GhZHD genes are less likely to undergo alternative splicing. This also indicates that the GhZHD gene family has a relatively fixed function compared to other TFs. Additionally, this feature significantly facilitates the annotation and identification of ZHD homologs in current and newly sequenced genomes.
Chromosomal Localization and Microsynteny Analysis
To determine the chromosomal localization of GhZHD genes in the genome of upland cotton, chromosome maps were constructed based on genome annotation, with the exception of three genes that were located on the scaffold (Supplementary Figure S1). We observed that the 37 GhZHD genes were spread across 20 chromosomes with a non-random distribution. Only one GhZHD gene each was located on chromosomes 1, 3, 10, 13, 14, 15, 17, 19, and 23, while the 11th and 24th chromosomes contained a maximum number of four GhZHD genes each (12%). We also illustrated the percentage of ZHD genes in G. hirsutum, G. arboreum, G. arboreum, and Arabidopsis (Figure 1B). Furthermore, relatively high densities of GhZHD genes were located at specific positions of some chromosomes, such as the bottom of Dt/chromosome 9 and the top of At/chromosome 5 (Supplementary Figure S1). We analyzed the ratio of non-synonymous substitutions (Ka) to synonymous substitutions (Ks) in the orthologous gene pairs (Supplementary Table S2) and found that most of the orthologous gene pairs had a Ka/Ks ratio of less than 1. According to the neutral theory, suggesting that their undergone purifying selection and the function was not clearly differentiated. In addition, a sliding window analysis was performed to determine the Ka/Ks ratios of the CDS sequences at different sites. This analysis demonstrated that the Ka/Ks ratios of some coding sites were greater than 1, suggesting that GhZHD genes have also undergone positive selection at some coding sites (Supplementary Figure S3).
Genomic comparisons are a comparatively quick and active way to transfer genomic knowledge acquired in one taxon to another (Lyons et al., 2008). In this study, synteny analysis was carried out using MCScanX software and whole genome sequences to visualize the locations of homologous or orthologous genes (Cao et al., 2016; Cheng et al., 2017). The identification of orthologous GhZHD genes will further define the evolutionary history of this gene family. Microsynteny analysis was performed across the G. hirsutum, G. arboreum, and G. raimondii genomes. Among G. hirsutum and G. raimondii, 32 collinear blocks were identified, while 45 orthologous gene pairs were found between G. hirsutum and G. arboreum (Figure 2A). These results suggest that a closer relationship exists between G. hirsutum and G. arboreum than between G. hirsutum and G. raimondii. Additionally, 39 collinear blocks were identified in G. hirsutum (Figures 2B,C). A total of 72 collinear gene pairs were identified between the cotton genomes due to an ancient tetraploid process. Four GhZHD genes had no collinear block, suggesting that, in addition to the whole genome duplication event, an independent duplication event also occurred during the evolution of these species.
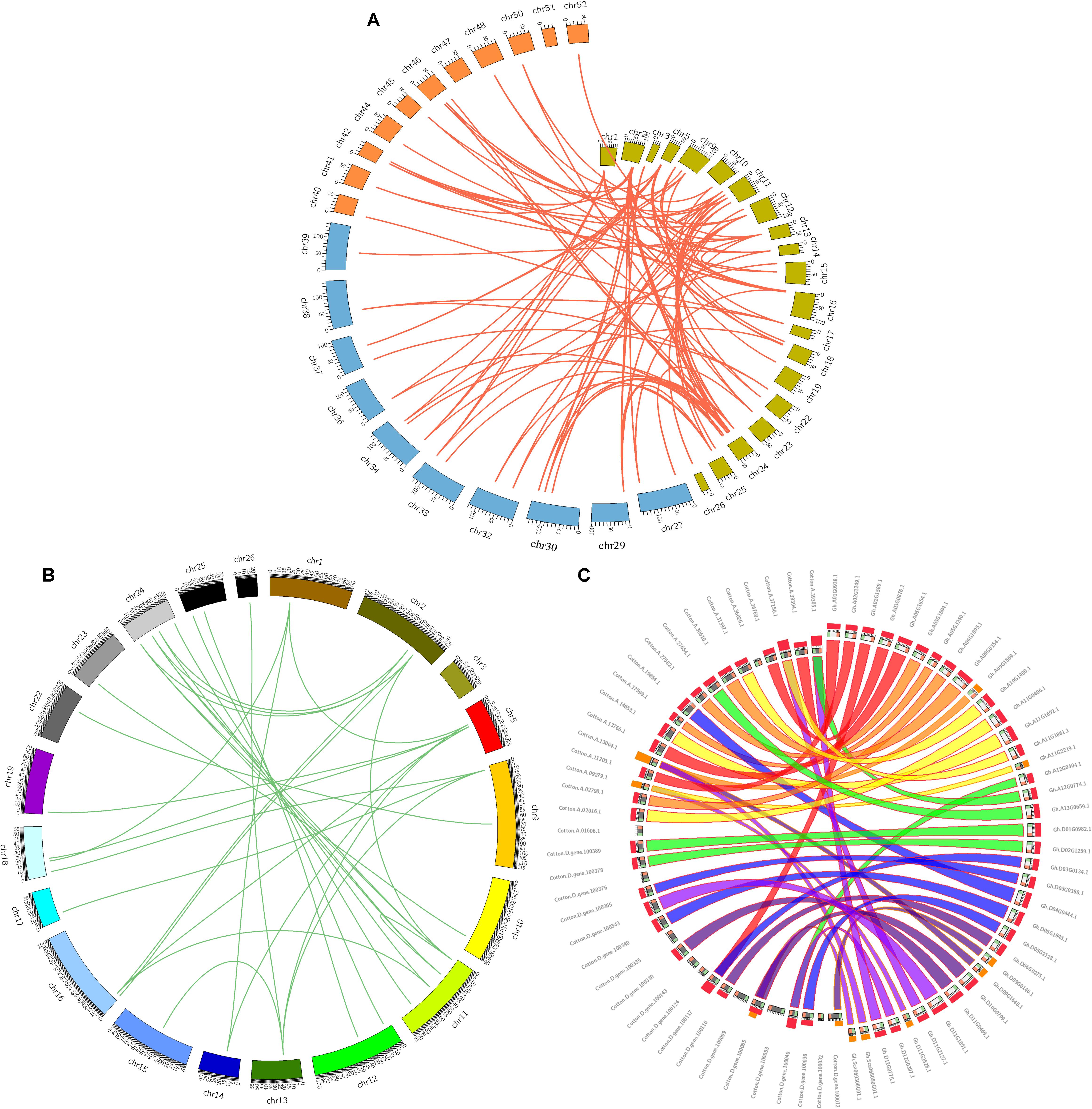
FIGURE 2. Microsynteny regions of ZHD genes among Gossypium genomes. (A) Synteny analyses of ZHD family genes between G. hirsutum, G. arboreum, and G. raimondii. The chromosome numbers of all three species are specified by different colors: olive green, blue, and orange represent the G. hirsutum, G. arboreum, and G. raimondii chromosomes, respectively. (B) Synteny of ZHD genes in the Gossypium hirsutum genome. The chromosome number is indicated on the outside by different colored boxes with the chromosome sequence lengths in megabases. Gene pairs with syntenic relationships are linked by a line. (C) Circles represent the ZHD gene sequence similarity among Gossypium genomes.
Analysis of Cis-Elements in ZHD Genes
Cis-elements in the promoter regions of genes provide cues for determining the stress-responsive or tissue-specific expression patterns in different environmental conditions. Significant positive correlations have been reported between multistimulus response genes and the density of cis-elements in their upstream regions (Tran et al., 2007; Walther et al., 2007). The PlantCARE database was used to identify potential stress- and hormone-responsive cis-acting elements in the promoter regions of GhZHD genes. The promoter regions, consisting of the genomic DNA sequences 1,500 bp upstream of the transcriptional start site (TTS), were examined for 37 GhZHD family genes. We detected a large number of cis-elements in the promoter regions of GhZHD genes (Figure 3). Our results suggest that GhZHD family genes may have different functions due to different types of cis-acting elements in their promoter regions. The cis-acting elements identified in our study can be classified into three categories: plant growth and development, phytohormone response, and biotic/abiotic stress response (Abdullah et al., 2018a). In the growth and development category, cis-acting elements were placed widely throughout the promoter regions, including Box 4 and MRE (responsible for plant growth in response to light), CAT-box (involved in meristem expression), circadian (required for circadian control), O2-site (involved in the regulation of zein metabolism), and Skn-1-motif and GCN4-motif (critical for endosperm expression). Box-4 covered the largest portion (41%) of the first category of cis-acting elements, followed by O2-site (19%), circadian (13%), and Skn-1-motif (13%) (Figure 3). In the phytohormone response category, we identified cis-acting elements including the P-box and GARE-motif (gibberellin-responsive elements), ERE (ethylene response), and ABRE (related to ABA). Remarkably, of the hormone responsive motifs, the TGACG cis-acting element (involved in auxin response) was the most common (28%), followed by the TCA-element (related to salicylic acid responsiveness) (26%). In the biotic/abiotic stress response category, a series of stress-related cis-acting elements were identified, including ARE (involved in anaerobic induction), HSE (heat stress), Box-W1 (participates in fungal elicitors), TC-rich repeats (general stress responses), and the GC-motif (involved in anoxia). We observed higher expression patterns for GhZHD17, GhZHD22, and GhZHD26 in the RNA-Seq data and qRT-PCR analysis, and these particular genes also have a higher percentage of the cis-acting elements MBS (drought/salt stress responsiveness) and HSE (heat stress). Because the GhZHD genes contain MBS, HSE, and ABRE in their promoter regions, we propose that these genes are involved in salt, drought, and heat stress responses. These results indicate that GhZHDs have the potential to improve abiotic stress responses and may also respond to abiotic stresses (cold treatment, heat treatment, PEG-treatment, and salt treatment).
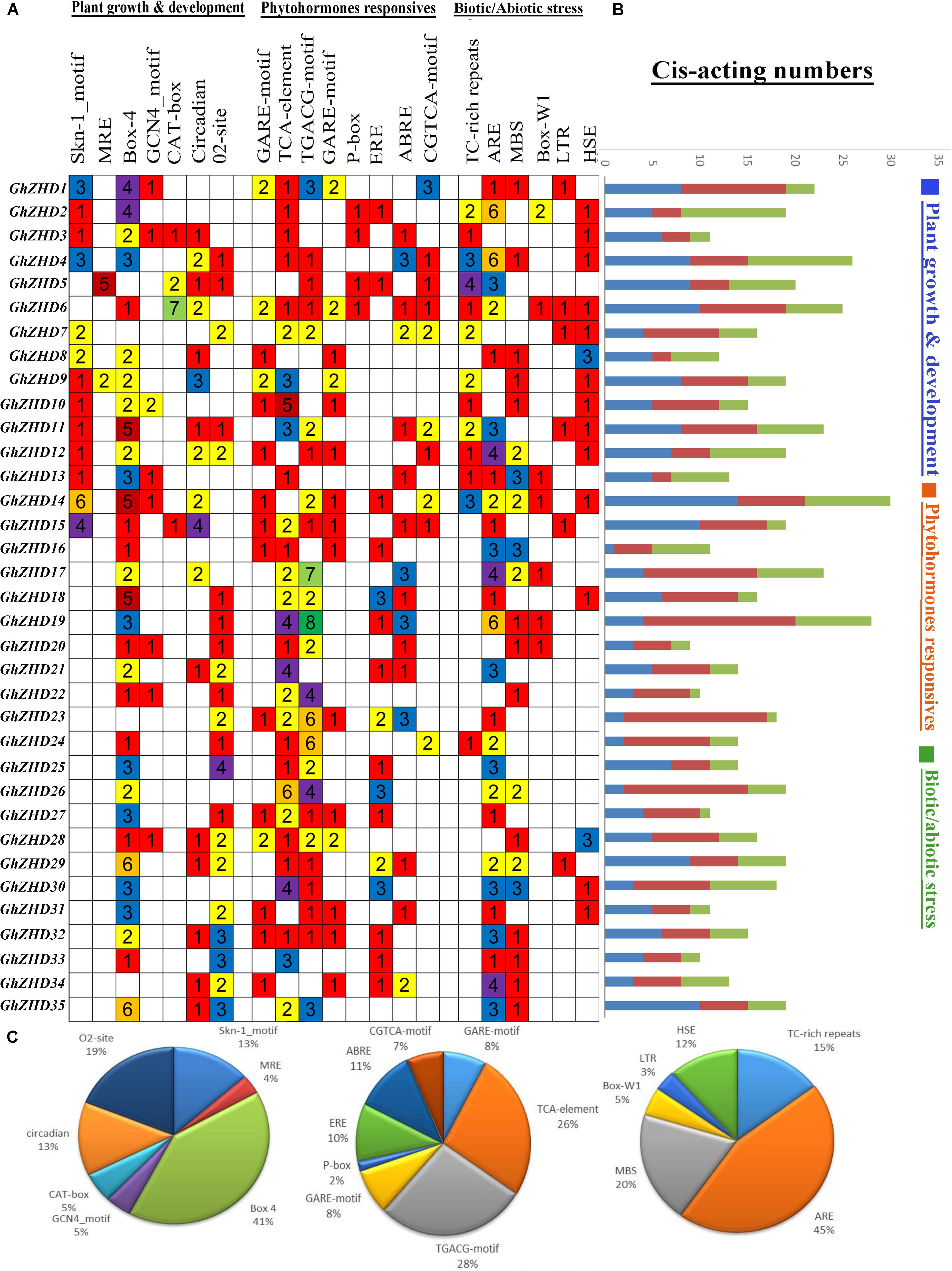
FIGURE 3. Identification of cis-acting elements in all GhZHD genes of G. hirsutum. (A) The different colors and numbers on the grid indicate the number of different promoter elements in each GhZHD gene. (B) The differently colored histograms represent the sum of cis-acting elements in each category. (C) Pie charts of different sizes indicate the ratio of each promoter element in each category.
To obtain further insights into the roles of GhZHD genes during abiotic stresses, we analyzed their correlation networks based on the PCCs of their relative gene expression (Figure 4; Huang et al., 2015; Wang et al., 2017). Several genes showed positive or negative correlation with these treatments (cold, hot, and salt treated) at the various time points evaluated. There was a close relationship between some GhZHD genes, such as GhZHD1 and GhZHD13. In addition, with the exception of GhZHD5, all of the other genes showed inverse correlations with GhZHD17. Surprisingly, we observed that duplicated GhZHD genes do not have closer relationships in terms of stress response compared to any other member of this gene family.
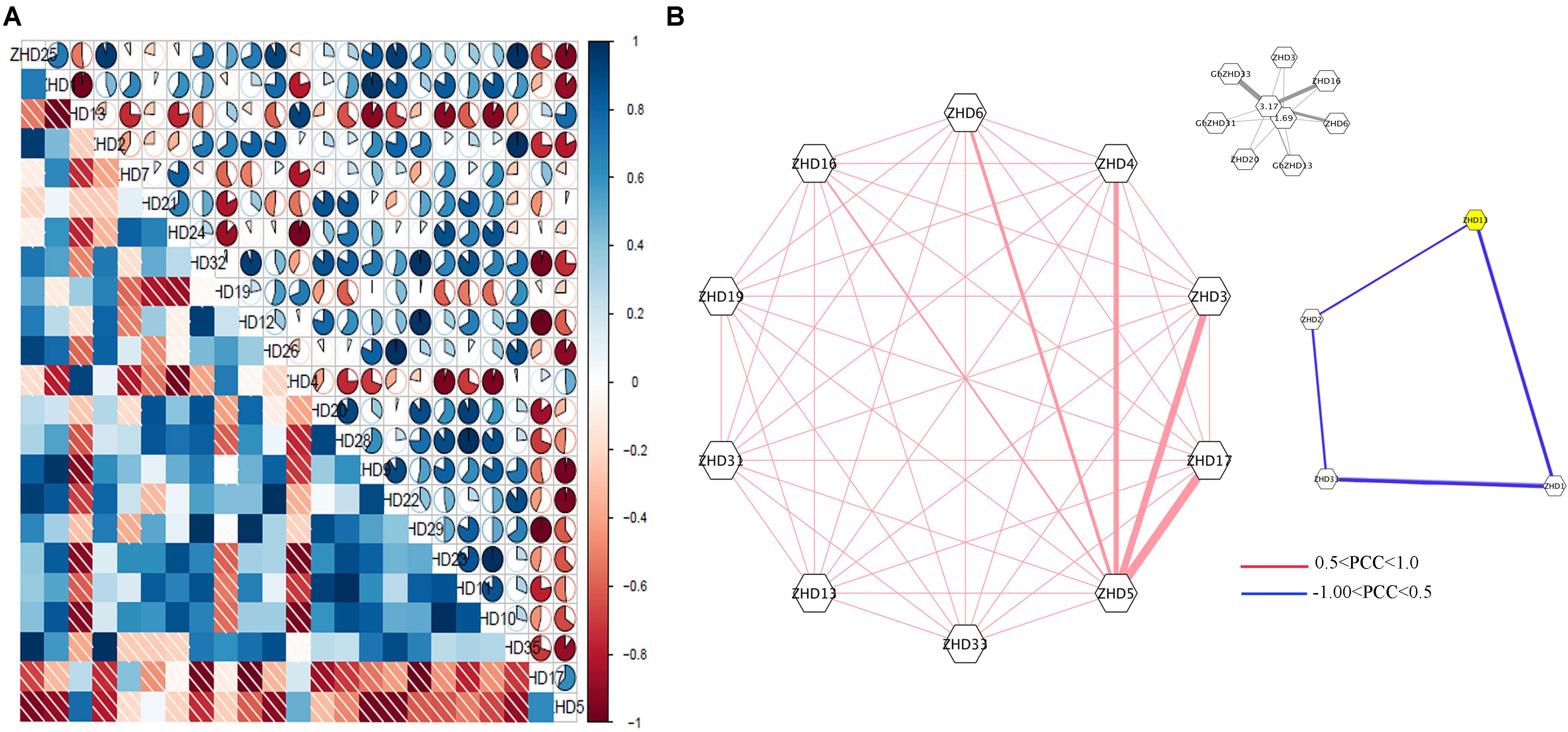
FIGURE 4. (A) Correlation analysis during abiotic stress. Correlation analysis was carried out using the R package program. Lower squares: correlations indicated by color and shading intensity. Upper circular symbols: each correlation is shown by the shades of blue and red and the size of the fan shape. Blue and red indicate a positive correlation and a negative correlation, respectively. (B) Co-regulatory networks. The co-regulatory networks of 37 GhZHD genes under stress treatments were established based on the Pearson correlation coefficients (PCCs) of these gene pairs using RNA-Seq data. The PCC of co-regulatory gene pairs was considered significant at the 0.05 significance level (p-value). Different color line styles indicate the different significance levels of the co-regulated gene pairs.
ZHD Gene Expression in Gossypium hirsutum
Upland cotton is widely cultivated around the world. Previous studies have shown that biotic and abiotic stresses adversely affect the normal growth and fiber quality of this crop (Dhandapani et al., 2015). To gain further insights into the expression patterns of GhZHD genes, we used the available RNA-Seq data for fiber development, abiotic/biotic stress, tissue and organ development, and developmental biology.
We analyzed the transcript levels of the 37 GhZHD genes from eight upland cotton tissues (calycle, leaf, petal, pistil, root, stamen, stem, and torus). The expression levels of these genes are presented in a circle heatmap (Figure 5). Of the 37 GhZHD genes in upland cotton, the transcript levels (FPKM values) of seven genes indicated that they were not expressed in any tissue, while the remaining 30 were expressed in at least one tissue. Among them, 10 GhZHD genes were differentially expressed in all the examined tissues. Some genes showed tissue-specific expression, for example, GhZHD6 was only expressed in the pistil. However, some genes were highly expressed in all of the examined tissues, including GhZHD9, GhZHD22, and GhZHD4. A large number of GhZHD genes showed preferential expression patterns in various tissues including the root (29), leaf (22), petal (16), pistil (32), stamen (20), stem (13), and torus (26). These results suggest that GhZHD genes may be involved in mediating plant growth and development. We also analyzed the RNA-Seq data collected during upland cotton fiber development. The transcript levels (FPKM values) of six genes indicated that they were not expressed in any fiber development stage, while the remaining genes were expressed in at least one fiber development stage. Based on this analysis, we selected candidate genes involved in fiber development. In brown cotton fibers, PAs are the key signs of pigment; hence, we examined whether such genes existed in brown cotton. We measured the PA content at different developmental stages of brown cotton fiber (Figure 6). Our results demonstrated that PA content gradually increased with the development of fiber and reached the highest level at 12 DPA, and then gradually decreased over time. Our results were consistent with previously published data on PA content during fiber development (Li et al., 2012; Feng et al., 2014). To identify the roles of GhZHD genes in PA accumulation during brown cotton fiber development, we designed primers based on the GhZHD gene sequences and performed qRT-PCR on 6, 12, 24, and 30 DPA brown cotton fiber (Figure 7). Interestingly, GhZHD29, GhZHD35, GhZHD30, GhZHD31, GhZHD11, GhZHD27, GhZHD18, GhZHD15, GhZHD16, GhZHD22, GhZHD6, GhZHD33, GhZHD13, GhZHD5, and GhZHD23 showed higher expression at the early stages of cotton fiber development and their transcript levels gradually decreased. This is similar to the accumulation of PAs in brown cotton fibers. Therefore, we hypothesize that some of these GhZHD genes may be involved in PA accumulation. The expression patterns we obtained for these candidate genes by qRT-PCR were similar to their expression patterns in the analyzed RNA-Seq data.
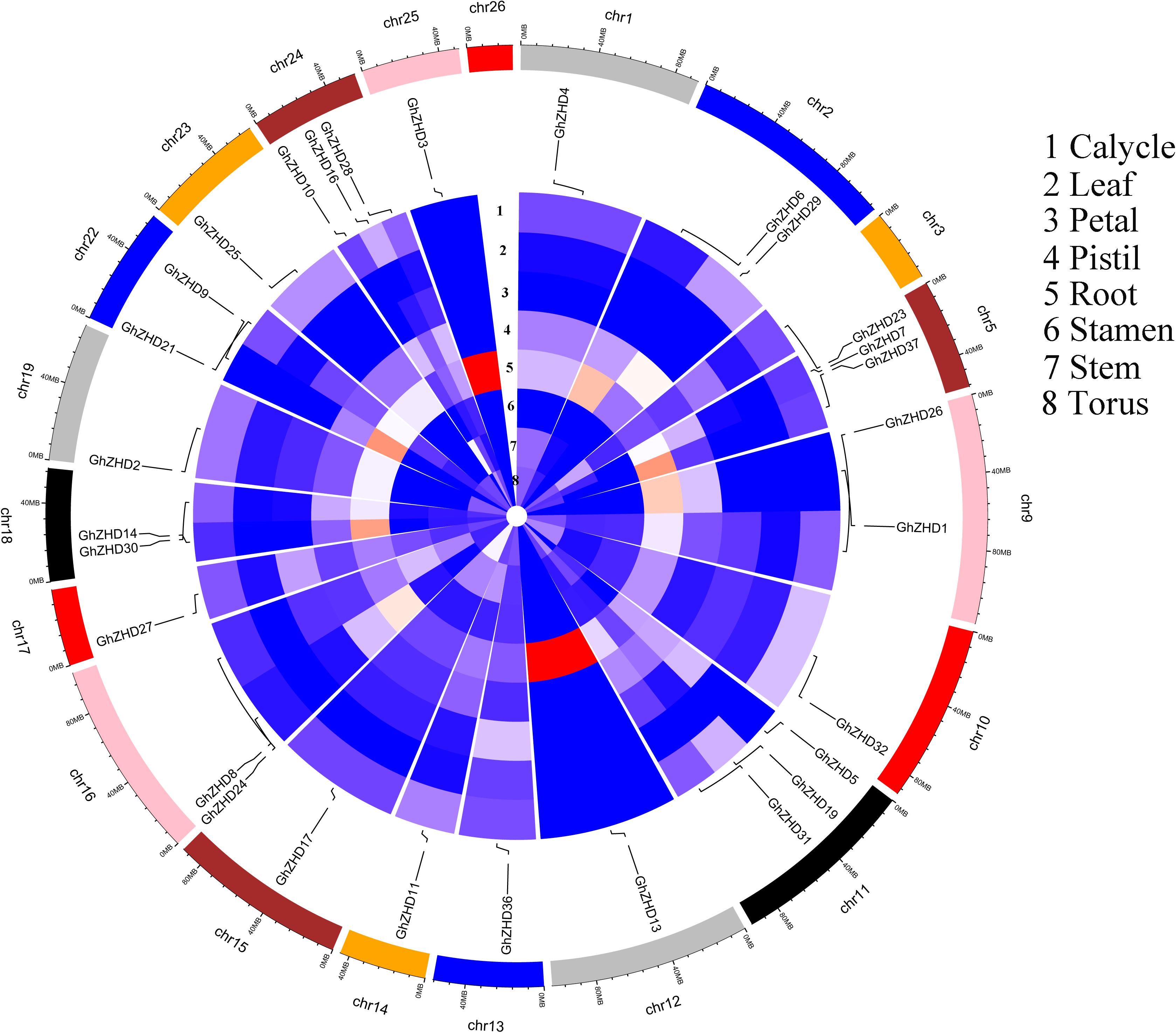
FIGURE 5. Circular heatmap (visualized using R software) depicting the stage-specific expression profiles of GhZHD genes. The FPKMs were calculated for expression values from RNA-Seq data.
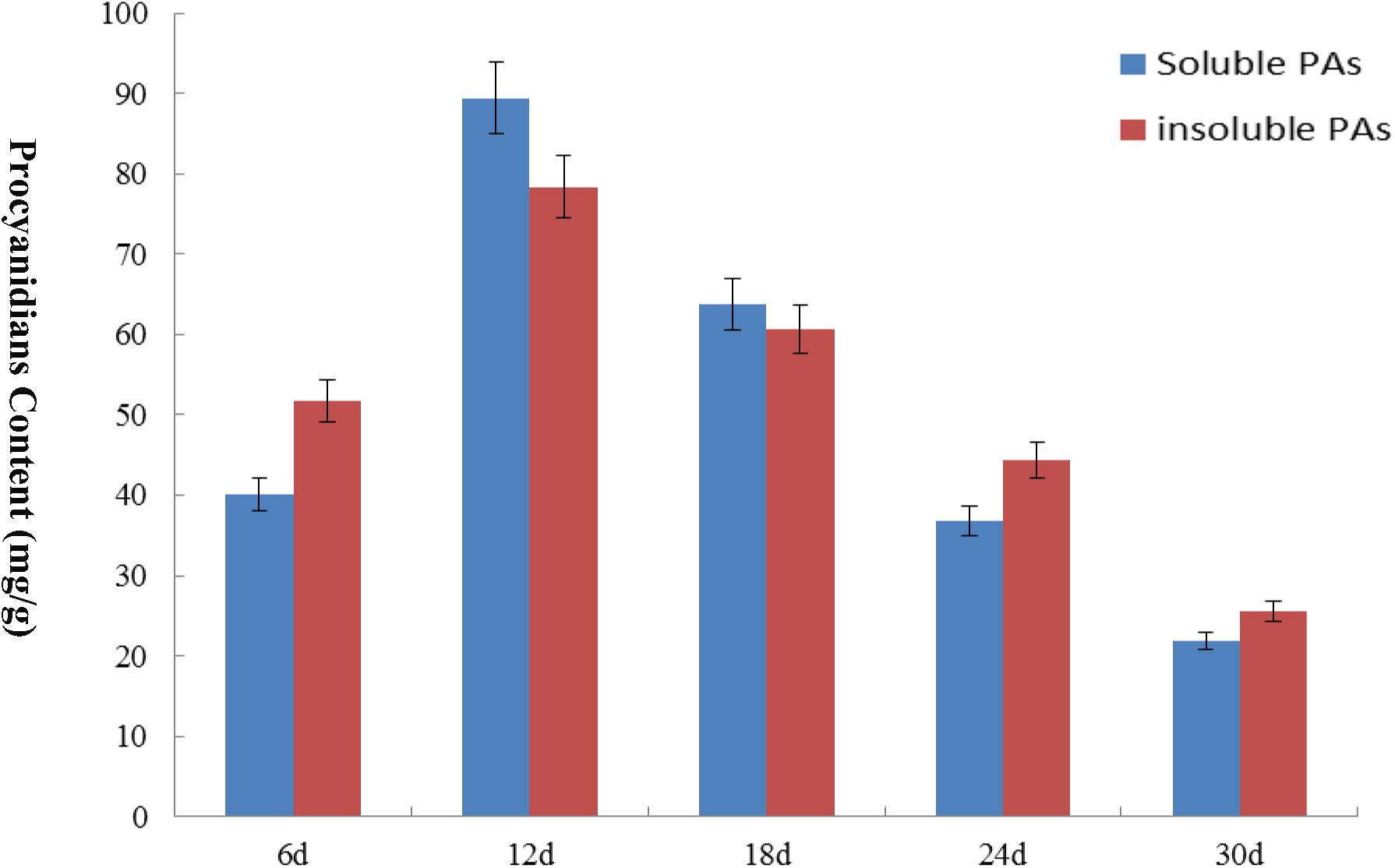
FIGURE 6. The PA content of brown cotton at different fiber development stages. The content levels of soluble PA and insoluble PA are expressed in different colors. The x-axis indicates different days of post-anthesis of cotton fibers, and the y-axis indicates the PA content. Error bars indicate SE.
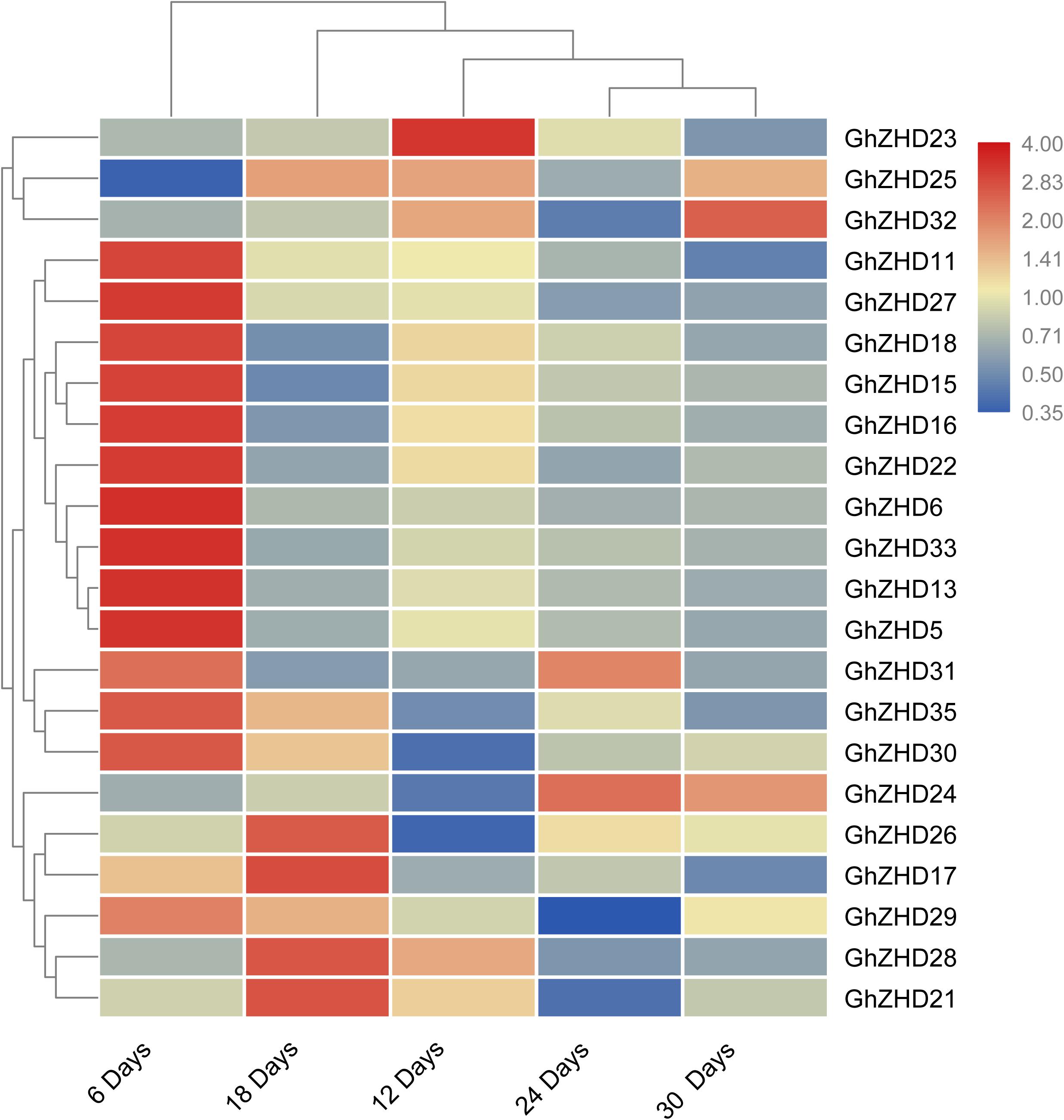
FIGURE 7. Expression patterns of GhZHD genes at different fiber developmental stages of brown cotton obtained by performing qRT-PCR. The relative expression levels were calculated using the 2-ΔΔCt method.
Discussion
Upland cotton is an important economic crop that is cultivated worldwide. The ZHD gene family is involved in a variety of processes, including plant development and physiological processes, as well as resistance to biotic/abiotic stresses. The plant-specific ZHD genes encode a family of TFs found in major groups of land plants, including vascular and non-vascular plants, but not found in prokaryotes, chlorophyte green algae, or fungi. The ZHD family genes may have evolved from a common ancestor or after the divergence of land plants from single-celled algae (Hu et al., 2008). The ZHD domain-containing gene family has been identified in many species, including Arabidopsis, maize, and rice. However, a systematic analysis of ZHD genes in upland cotton is still lacking. In this study, we aimed to complete a genome-wide survey of ZHD genes and their expression during fiber developmental processes and/or stress responses.
The number of GhZHD genes in Gossypium genomes was higher than in rice (15), Arabidopsis (17), Brassica rapa (31), or tomato (22). However, while the genome size of G. hirsutum (613 Mb) is smaller than that of tomato (950 Mb), it is larger than that of rice (441 Mb), Arabidopsis (164 Mb), and Brassica rapa (283.8 Mb). The number of ZHD family genes is relatively high in upland cotton, signifying that genome duplication events might have contributed to the expansion of GhZHD genes in this species. The identified protein characteristics and the conserved ZHD dimer domain of GhZHD family genes are consistent with those of other plant species, suggesting that the GhZHD proteins are structurally similar. The 37 GhZHD proteins were categorized into seven clades (s1–s7). Furthermore, we observed that most of the GhZHD genes were not closely related to the ZHD genes in Arabidopsis, consistent with the fact that upland cotton and Arabidopsis did not diverge from a recent common ancestor. The MEME server was used to identify the conserved motifs in the GhZHD proteins. Closely related members on the phylogenetic tree were found to have similar motifs, revealing the functional similarities between the proteins of the same subfamily. Intronless genes are very common in the genomes of higher eukaryotes (Louhichi et al., 2011). Gene structure analysis confirmed that all GhZHD genes of upland cotton are intronless (Supplementary Figure S2). Plant ZHD genes have previously been found to be intronless, and our data for upland cotton supports these findings (Wang et al., 2014, 2015; Khatun et al., 2017). Compared to other plants, the variable exon–intron structure of GhZHD family genes observed in upland cotton suggests that there is a structural divergence in the GhZHD gene family (Supplementary Figure S2). Furthermore, the similar exon–intron association among the different subfamilies suggests that these genes were highly conserved during evolution.
It is well known that gene duplication mechanisms (tandem/segmental duplication), transpositions, and whole genome duplications have a significant role in biological evolution (Xu et al., 2012). In our study, we observed that paralogous genes developed through segmental duplication, while no tandem duplications were observed for any gene pair, specifying that segmental duplication has played a significant role in the expansion of upland cotton ZHD family genes.
These plant-specific ZHD TFs are involved in various biological processes in plants including fiber development and responses to abiotic/biotic stress. The GhZHD genes contained specific DNA-binding motifs, such as the MYB motif, that are induced by several signals during stress conditions and various development processes (Yamaguchi-Shinozaki and Shinozaki, 2005). Recently, ZHD family proteins from Arabidopsis were shown to be induced by various stresses including salt and drought stresses (Nakashima and Yamaguchi-Shinozaki, 2006; Khatun et al., 2017). In our results, we observed that most GhZHD genes have preferential tissue expression patterns. Upland cotton fiber development is a complex biological process that ultimately leads to the production of crops for harvest. Fiber development is regulated by several transcriptional regulatory networks involving TFs. However, to date, the potential roles of GhZHD family genes in fiber development have not been characterized. Previous studies found that five key genes (CHI, F3H, DFR, ANS, and ANR) that participate in the PA synthesis pathway have higher expression levels in the early stages of fiber development (Xiao et al., 2007; Su et al., 2017; Chen et al., 2018), and after reaching their peak expression, they begin to gradually decrease. This result is consistent with the accumulation of PAs that we observed during the development of brown cotton fibers. Our results showed that the relative expression trends of GhZHD29, GhZHD35, GhZHD30, GhZHD31, GhZHD11, GhZHD27, GhZHD18, GhZHD15, GhZHD16, GhZHD22, GhZHD6, GhZHD33, GhZHD13, GhZHD5, and GhZHD23 in brown cotton fiber were similar to the level of PA accumulation (Figures 7). Consequently, we proposed that one or more of these genes may affect the accumulation of PAs in cotton fiber. In our study, many stress-responsive and growth-regulatory cis-elements were widely distributed in the promoter regions of upland cotton GhZHD genes. Further studies on these putative cis-elements in the GhZHD genes of upland cotton are needed to unravel their complex regulatory mechanism.
Conclusion
In summary, we conducted a genome-wide analysis of GhZHD genes in brown cotton, including gene structure analysis, chromosomal localization, conserved motif identification, phylogenetic relationship mapping, conserved microsynteny analysis, expression profiling during fiber development, PA contents, and functional divergence. The expression patterns of GhZHD genes during fiber development combined with PA synthesis analyses suggested that GhZHD genes may have functions in organ development and pigmentation. The identification and analysis of these GhZHD genes in brown cotton promotes a basic understanding of and provides a foundation for the extrapolation of the GhZHD gene function in future studies of brown cotton to improve fiber quality.
Author Contributions
MA designed and performed the experiments. XC and YuC analyzed the data. XS, JG, and MAM contributed to reagents, materials, and analysis tools. MA wrote the paper. YoC and YL provided guidance on the whole manuscript. All authors reviewed and approved the final submission.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 31640068 and 31672497).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers and editor for their careful reading and helpful comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00357/full#supplementary-material
FIGURE S1 | Chromosomal locations of the GhZHD genes in the genome of Gossypium hirsutum. The chromosome number is represented at the top of each chromosome and the left scale is in megabases (Mb).
FIGURE S2 | Gene structure and distribution of the conserved motifs in the ZHD genes in upland cotton. (A) Untranslated region (UTR), introns, and exons are indicated by the blue box, thin line, and green box, respectively. (B) Conserved motifs located in each gene with their relative combined p-values.
FIGURE S3 | Sliding window plots of candidate duplicated ZHD genes in upland cotton. The window size is 150 bp, and the step size is 9 bp. The x-axis indicates the synonymous distance within each gene.
TABLE S1 | Detailed information about GhZHD genes from upland cotton.
TABLE S2 | Synonymous and non-synonymous substitution rates for the duplication events in upland cotton.
TABLE S3 | The primers used in qRT-PCR.
Footnotes
- ^https://www.cottongen.org/
- ^http://www.expasy.org/tools/
- ^http://gsds.cbi.pku.edu.cn/
- ^http://meme-suite.org/
- ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^http://mapinspect.software.informer.com/
- ^https://cottonfgd.org/
References
Abdullah, M., Cao, Y., Cheng, X., Meng, D., Chen, Y., Shakoor, A., et al. (2018a). The sucrose synthase gene family in chinese pear (Pyrus bretschneideri Rehd.): structure, expression, and evolution. Molecules 23, 1–16. doi: 10.3390/molecules23051144
Abdullah, M., Cao, Y., Cheng, X., Shakoor, A., Su, X., Gao, J., et al. (2018b). Genome-wide analysis characterization and evolution of SBP genes in Fragaria vesca, Pyrus bretschneideri, Prunus persica and Prunus mume. Front. Genet. 9:64. doi: 10.3389/fgene.2018.00064
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bateman, A. (2002). The Pfam protein families database. Nucleic Acids Res. 30, 276–280. doi: 10.1093/nar/30.1.276
Bharathan, G., Janssen, B.-J., Kellogg, E. A., and Freeling, M. (1997). Did homeodomain proteins duplicate before the origin of angiosperms, fungi, and metazoa? Evolution 94, 13749–13753. doi: 10.1073/pnas.94.25.13749
Bhattacharjee, A., Ghangal, R., Garg, R., and Jain, M. (2015). Genome-wide analysis of homeobox gene family in legumes: Identification, gene duplication and expression profiling. PLoS One 10:e0119198. doi: 10.1371/journal.pone.0119198
Bhattacharjee, A., and Jain, M. (2013). “Homeobox genes as potential candidates for crop improvement under abiotic stress,” in Plant Acclimation to Environmental Stress, eds N. Tuteja and S. Singh Gill (New York, NY: Springer), 163–176. doi: 10.1007/978-1-4614-5001-6_7
Cao, Y., Han, Y., Jin, Q., Lin, Y., and Cai, Y. (2016). Comparative genomic analysis of the GRF genes in Chinese Pear (Pyrus bretschneideri Rehd), Poplar (Populous), grape (Vitis vinifera), Arabidopsis and rice (Oryza sativa). Front. Plant Sci. 7:1750. doi: 10.3389/fpls.2016.01750
Cao, Y., Han, Y., Meng, D., Li, G., Li, D., Abdullah, M., et al. (2017). Genome-wide analysis suggests the relaxed purifying selection affect the evolution of WOX genes in Pyrus bretschneideri, Prunus persica, Prunus mume, and Fragaria vesca. Front. Genet. 8:78. doi: 10.3389/fgene.2017.00078
Chen, W., Si, G. Y., Zhao, G., Abdullah, M., Guo, N., Li, D. H., et al. (2018). Genomic comparison of the P-ATPase gene family in four cotton species and their expression patterns in Gossypium hirsutum. Molecules 23:E1092. doi: 10.3390/molecules23051092
Chen, Z., Nie, H., Grover, C. E., Wang, Y., Li, P., Wang, M., et al. (2017a). Entire nucleotide sequences of Gossypium raimondii and G. arboreum mitochondrial genomes revealed A-genome species as cytoplasmic donor of the allotetraploid species. Plant Biol. 19, 484–493. doi: 10.1111/plb.12536
Chen, Z., Nie, H., Wang, Y., Pei, H., Li, S., Zhang, L., et al. (2017b). Rapid evolutionary divergence of diploid and allotetraploid Gossypium mitochondrial genomes. BMC Genomics 18:876. doi: 10.1186/s12864-017-4282-5
Cheng, X., Yan, C., Zhang, J., Ma, C., Li, S., Jin, Q., et al. (2017). The effect of different pollination on the expression of Dangshan Su Pear microRNA. Biomed Res. Int. 2017:2794040. doi: 10.1155/2017/2794040.
Deng, X., Phillips, J., Meijer, A. H., Salamini, F., and Bartels, D. (2002). Characterization of five novel dehydration-responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum. Plant Mol. Biol. 49, 601–610. doi: 10.1023/A:1015501205303
Dhandapani, G., Lakshmi Prabha, A., Kanakachari, M., Phanindra, M. L. V., Prabhakaran, N., Gothandapani, S., et al. (2015). GhDRIN1, a novel drought-induced gene of upland cotton (Gossypium hirsutum L.) confers abiotic and biotic stress tolerance in transgenic tobacco. Biotechnol. Lett. 37, 907–919. doi: 10.1007/s10529-014-1733-9
Englbrecht, C. C., Schoof, H., and Böhm, S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5:39. doi: 10.1186/1471-2164-5-39
Feng, H., Li, Y., Wang, S., Zhang, L., Liu, Y., Xue, F., et al. (2014). Molecular analysis of proanthocyanidins related to pigmentation in brown cotton fibre (Gossypium hirsutum L.). J. Exp. Bot. 65, 5759–5769. doi: 10.1093/jxb/eru286
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: The protein families database. Nucleic Acids Res. 42, 222–230. doi: 10.1093/nar/gkt1223
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Halbach, T., Scheer, N., and Werr, W. (2000). Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins. Nucleic Acids Res. 28, 3542–3550. doi: 10.1093/nar/28.18.3542
Hu, W., Depamphilis, C. W., and Ma, H. (2008). Phylogenetic analysis of the plant-specific zinc finger-homeobox and mini zinc finger gene families. J. Integr. Plant Biol. 50, 1031–1045. doi: 10.1111/j.1744-7909.2008.00681.x
Huang, Z., Tang, J., Duan, W., Wang, Z., Song, X., and Hou, X. (2015). Molecular evolution, characterization, and expression analysis of SnRK2 gene family in Pak-choi (Brassica rapa ssp. chinensis). Front. Plant Sci. 6:879. doi: 10.3389/fpls.2015.00879
Ikegami, A., Akagi, T., Potter, D., Yamada, M., Sato, A., Yonemori, K., et al. (2009). Molecular identification of 1-Cys peroxiredoxin and anthocyanidin/flavonol 3-O-galactosyltransferase from proanthocyanidin-rich young fruits of persimmon (Diospyros kaki Thunb.). Planta 230, 841–855. doi: 10.1007/s00425-009-0989-0
Jørgensen, J. E., Grønlund, M., Pallisgaard, N., Larsen, K., Marcker, K. A., and Jensen, E. O. (1999). A new class of plant homeobox genes is expressed in specific regions of determinate symbiotic root nodules. Plant Mol. Biol. 40, 65–77. doi: 10.1023/A:1026463506376
Khatun, K., Nath, U. K., Robin, A. H. K., Park, J.-I., Lee, D.-J., Kim, M.-B., et al. (2017). Genome-wide analysis and expression profiling of zinc finger homeodomain (ZHD) family genes reveal likely roles in organ development and stress responses in tomato. BMC Genomics 18:695. doi: 10.1186/s12864-017-4082-y
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, 302–305. doi: 10.1093/nar/gkr931
Li, T., Fan, H., Li, Z., Wei, J., Lin, Y., and Cai, Y. (2012). The accumulation of pigment in fiber related to proanthocyanidins synthesis for brown cotton. Acta Physiol. Plant. 34, 813–818. doi: 10.1007/s11738-011-0858-x
Li, Y. G., Tanner, G., and Larkin, P. (1996). The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J. Sci. Food Agric. 70, 89–101. doi: 10.1002/(SICI)1097-0010(199601)70:1<89::AID-JSFA470>3.0.CO;2-N
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Louhichi, A., Fourati, A., and Rebaï, A. (2011). IGD: A resource for intronless genes in the human genome. Gene 488, 35–40. doi: 10.1016/j.gene.2011.08.013
Lyons, E., Pedersen, B., Kane, J., Alam, M., Ming, R., Tang, H., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with Rosids. Plant Physiol. 148, 1772–1781. doi: 10.1104/pp.108.124867
Mukherjee, K., Brocchieri, L., and Bürglin, T. R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26, 2775–2794. doi: 10.1093/molbev/msp201
Nakashima, K., and Yamaguchi-Shinozaki, K. (2006). Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant. 126, 62–71. doi: 10.1111/j.1399-3054.2005.00592.x
Nam, J., and Nei, M. (2005). Evolutionary change of the numbers of homeobox genes in bilateral animals. Mol. Biol. Evol. 22, 2386–2394. doi: 10.1093/molbev/msi229
Park, H. C., Kim, M. L., Lee, S. M., Bahk, J. D., Yun, D. J., Lim, C. O., et al. (2007). Pathogen-induced binding of the soybean zinc finger homeodomain proteins GmZF-HD1 and GmZF-HD2 to two repeats of ATTA homeodomain binding site in the calmodulin isoform 4 (GmCaM4) promoter. Nucleic Acids Res. 35, 3612–3623. doi: 10.1093/nar/gkm273
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Su, X., Sun, X., Cheng, X., Wang, Y., Abdullah, M., Li, M., et al. (2017). Comparative genomic analysis of the PKS genes in five species and expression analysis in upland cotton. PeerJ 5:e3974. doi: 10.7717/peerj.3974
Tan, Q. K.-G., and Irish, V. F. (2006). The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 140, 1095–1108. doi: 10.1104/pp.105.070565
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tran, L. S. P., Nakashima, K., Sakuma, Y., Osakabe, Y., Qin, F., Simpson, S. D., et al. (2007). Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 49, 46–63. doi: 10.1111/j.1365-313X.2006.02932.x
Walther, D., Brunnemann, R., and Selbig, J. (2007). The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PLoS Genet. 3:e11. doi: 10.1371/journal.pgen.0030011
Wang, H., Yin, X., Li, X., Wang, L., Zheng, Y., Xu, X., et al. (2014). Genome-wide identification, evolution and expression analysis of the grape (Vitis vinifera L.) zinc finger-homeodomain gene family. Int. J. Mol. Sci. 15, 5730–5748. doi: 10.3390/ijms15045730
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Wang, W., Wu, P., Li, Y., and Hou, X. (2015). Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Mol. Genet. Genomics 291, 1451–1464. doi: 10.1007/s00438-015-1136-1
Wang, W., Wu, P., Liu, T., Ren, H., Li, Y., and Hou, X. (2017). Genome-wide Analysis and expression divergence of the Trihelix family in Brassica Rapa: insight into the evolutionary patterns in plants. Sci. Rep. 7:6463. doi: 10.1038/s41598-017-06935-0
Windhovel, A., Hein, I., Dabrowa, R., and Stockhaus, J. (2001). Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol. Biol. 45, 201–214. doi: 10.1023/A:1006450005648
Xiao, Y. H., Zhang, Z. S., Yin, M. H., Luo, M., Li, X. B., Hou, L., et al. (2007). Cotton flavonoid structural genes related to the pigmentation in brown fibers. Biochem. Biophys. Res. Commun. 358, 73–78. doi: 10.1016/j.bbrc.2007.04.084
Xu, G., Guo, C., Shan, H., and Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yanagisawa, S. (2004). Dof domain proteins?: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 45, 386–391. doi: 10.1093/pcp/pch055
Yu, J., Jung, S., Cheng, C. H., Ficklin, S. P., Lee, T., Zheng, P., et al. (2014). CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, 1229–1236. doi: 10.1093/nar/gkt1064
Zdobnov, E. M., and Apweiler, R. (2001). InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848. doi: 10.1093/bioinformatics/17.9.847
Keywords: genome-wide analysis, ZHD, fiber development, abiotic stress, qRT-PCR
Citation: Abdullah M, Cheng X, Cao Y, Su X, Manzoor MA, Gao J, Cai Y and Lin Y (2018) Zinc Finger-Homeodomain Transcriptional Factors (ZHDs) in Upland Cotton (Gossypium hirsutum): Genome-Wide Identification and Expression Analysis in Fiber Development. Front. Genet. 9:357. doi: 10.3389/fgene.2018.00357
Received: 13 November 2017; Accepted: 20 August 2018;
Published: 09 October 2018.
Edited by:
Susan Jones, James Hutton Institute, United KingdomReviewed by:
Mikhail P. Ponomarenko, Institute of Cytology and Genetics (RAS), RussiaXiaoya Chen, Shanghai Institutes for Biological Sciences (CAS), China
Copyright © 2018 Abdullah, Cheng, Cao, Su, Manzoor, Gao, Cai and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongping Cai, ypcaiah@163.com; swkx12@ahau.edu.cn Yi Lin, linyi1957@126.com
 Muhammad Abdullah
Muhammad Abdullah Xi Cheng
Xi Cheng Yunpeng Cao
Yunpeng Cao Xueqiang Su
Xueqiang Su Junshan Gao
Junshan Gao