- Department of Plant Sciences, University of Cambridge, Cambridge, United Kingdom
Homologous chromosomes must pair and recombine to ensure faithful chromosome segregation during meiosis, a specialized type of cell division that occurs in sexually reproducing eukaryotes. Meiotic recombination initiates by programmed induction of DNA double-strand breaks (DSBs) by the conserved type II topoisomerase-like enzyme SPO11. A subset of meiotic DSBs are resolved as crossovers, whereby reciprocal exchange of DNA occurs between homologous chromosomes. Importantly, DSBs are non-randomly distributed along eukaryotic chromosomes, forming preferentially in permissive regions known as hotspots. In many species, including plants, DSB hotspots are located within nucleosome-depleted regions. DSB localization is governed by interconnected factors, including cis-regulatory elements, transcription factor binding, and chromatin accessibility, as well as by higher-order chromosome architecture. The spatiotemporal control of DSB formation occurs within a specialized chromosomal structure characterized by sister chromatids organized into linear arrays of chromatin loops that are anchored to a proteinaceous axis. Although SPO11 and its partner proteins required for DSB formation are bound to the axis, DSBs occur preferentially within the chromatin loops, which supports the “tethered-loop/axis model” for meiotic recombination. In this mini review, we discuss insights gained from recent efforts to define and profile DSB hotspots at high resolution in eukaryotic genomes. These advances are deepening our understanding of how meiotic recombination shapes genetic diversity and genome evolution in diverse species.
1. Introduction
Meiosis is a specialized cell division program that is essential for sexual reproduction in eukaryotes. During this program, replication of chromosomal DNA to form sister chromatids is followed by two rounds of cell division. Maternal and paternal chromosomes (homologs) segregate at the first division and sister chromatids segregate at the second division. Chromosome number is thereby halved and, in diploid organisms, meiosis culminates in the production of haploid progeny cells (gametes). Chromosome segregation during meiosis is imperative for the continuation of the species, as it enables the formation of a zygote that inherits the full chromosome complement in the next generation by fusion of a male and a female gamete (Villeneuve and Hillers, 2001). DNA double-strand breaks (DSBs) occur at many genomic loci during early prophase I to initiate meiotic recombination, whereby pairing of and reciprocal DNA exchange (crossover) between homologous chromosomes promote their balanced segregation and genetic diversity (De Massy, 2013; Keeney et al., 2014). Meiotic DSBs form preferentially in permissive regions known as hotspots, giving rise to non-random DSB and crossover distributions that influence patterns of genetic linkage and genome evolution in eukaryotes (Baudat et al., 2013; Cooper et al., 2016). The genome-wide distribution and the resolution of a subset of DSBs as crossovers have immediate impacts on haplotype configurations in the recombinant gametes, as well as far-reaching, population-level consequences for locus-specific rates of genetic change over evolutionary time (Cooper et al., 2016).
Meiotic DSBs are catalyzed by SPO11 dimers in a type II topoisomerase-like reaction in which one SPO11 molecule becomes covalently bound to each 5′ end of the cleaved DNA (Bergerat et al., 1997; Keeney et al., 1997). To enable DSB repair as a crossover or a non-crossover, the two SPO11–oligonucleotide complexes are endonucleolytically released (Neale et al., 2005) and 5′–3′ resection exposes a 3′-overhanging, single-stranded DNA (ssDNA) tail at each end of the DSB (Cao et al., 1990; Sun et al., 1991; Zakharyevich et al., 2010). The meiotic recombinases DMC1 and RAD51 bind these ssDNA tails and promote the search for a homologous chromosome to provide a template for DNA repair (Bishop et al., 1992; Shinohara et al., 1992; Cloud et al., 2012). Following invasion of the homolog, strand-exchange intermediates can be processed via different DNA repair pathways to produce non-crossovers or crossovers (Hunter, 2015). Most non-crossovers are products of synthesis-dependent strand annealing (SDSA), whereby the homolog-invading DSB end initiates DNA synthesis and is subsequently displaced and annealed to the other end of the DSB (Pâques and Haber, 1999; McMahill et al., 2007). Non-crossovers can also result from dissolution of double Holliday junction joint molecules (dHJ-JMs) by combined helicase and topoisomerase activities (Cejka et al., 2010), or from unidirectional endonuclease cleavage of dHJ-JMs (Szostak et al., 1983; De Muyt et al., 2012). However, most if not all stable dHJ-JMs are resolved as crossovers during meiosis (Allers and Lichten, 2001; Hunter and Kleckner, 2001; Hunter, 2015).
Efforts to generate genome-wide, nucleotide-resolution maps of eukaryotic DSB landscapes have recently intensified with the advent of techniques to immunoprecipitate SPO11 and end-label, purify and sequence SPO11-bound oligonucleotides, which are a byproduct of DSB formation (Pan et al., 2011). SPO11-oligo mapping by these means has been applied in several budding yeast species, fission yeast, mouse and Arabidopsis thaliana (Pan et al., 2011; Fowler et al., 2014; Thacker et al., 2014; Lam and Keeney, 2015; Zhu and Keeney, 2015; Lange et al., 2016; Mohibullah and Keeney, 2017; Choi et al., 2018; Underwood et al., 2018). Parallel advances have been achieved by exploiting meiotic ssDNA formed at resected DSB ends for high-resolution mapping of recombination initiation sites in budding yeast, maize, mouse and human genomes (Blitzblau et al., 2007; Buhler et al., 2007; Borde et al., 2009; Smagulova et al., 2011; Brick et al., 2012; Khil et al., 2012; Pratto et al., 2014; Lange et al., 2016; He et al., 2017). Single-stranded DNA sequencing (SSDS) utilizes chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) of ssDNA bound by the strand-exchange proteins DMC1 and RAD51, thereby capturing chromosome fragments that are immediately adjacent to DSB sites (Smagulova et al., 2011; Khil et al., 2012). We discuss insights gained from these high-resolution physical maps of meiotic DSB landscapes, highlighting the genetic and epigenetic properties of hotspots for recombination initiation in different eukaryotic species.
2. Defining Meiotic DSB Hotspots
Maps describing recombination initiation profiles at nucleotide resolution have revealed that hotspots constitute one of several levels of DSB patterning (Cooper et al., 2016). The genome-wide DSB landscape is most accurately characterized as a continuous probability distribution, where DSB hotspots are defined as genomic loci with high local likelihoods of DNA cleavage by SPO11 (Pan et al., 2011). Most if not all genomic loci are sites of potential cleavege and many DSBs form in regions not defined as hotspots (Pan et al., 2011). The limits of DSB detection are determined by methodological constraints associated with quantifying the signal to noise ratio at each locus, which vary between organisms due to differences in both their underlying biology and the methodologies adopted. Thus, while the “hotspot” concept is useful for annotating preferred sites and identifying the possible determinants of recombination initiation activity, comparisons of quantitative measurements taken across species and methodologies should be considered with caution. In yeast species (Saccharomyces cerevisiae, S. kudriavzevii, S. mikatae, S. paradoxus, and Schizosaccharomyces pombe) and mouse, DSB hotspots have been defined as loci meeting or exceeding given thresholds for Spo11-oligo density and physical size (Pan et al., 2011; Fowler et al., 2014; Lam and Keeney, 2015; Zhu and Keeney, 2015; Lange et al., 2016; Mohibullah and Keeney, 2017). The accuracy of this method for DSB hotspot definition has been validated by comparing the spatial patterning of yeast Spo11-oligo maps with DSBs assayed directly by Southern blotting of genomic DNA from yeast meiocytes (Pan et al., 2011).
A false discovery rate (FDR)-based peak-calling approach (Feng et al., 2011) was adopted to identify loci in the Arabidopsis genome with significantly higher-than-expected SPO11-1-oligo enrichment, using the binomial distribution to model enrichment relative to a control library derived from genomic DNA (Choi et al., 2018). Peaks identified in replicate SPO11-1-oligo libraries were ranked by their −log10-transformed FDR values and Arabidopsis DSB hotspots were defined as peaks with consistent rankings between replicates (i.e., peaks with irreproducible discovery rates [IDR] <0.05) (Li et al., 2011; Choi et al., 2018). Similar peak-calling approaches have been employed to define DSB hotspots derived from SSDS in maize, mouse and human genomes (Smagulova et al., 2011, 2016; Brick et al., 2012; Khil et al., 2012; Pratto et al., 2014; He et al., 2017), with confirmation of a sample of hotspots by qPCR or direct physical detection methods (Smagulova et al., 2011). Positive genome-wide associations between DSB maps and genetic or crossover maps provide further validation of these recombination initiation site mapping approaches (Smagulova et al., 2011; Pratto et al., 2014; Choi et al., 2018). Mouse DSB maps obtained by SPO11-oligo mapping and SSDS also show a high level of agreement (Lange et al., 2016).
Hotspot density in fission yeast is substantially lower than in budding yeast genomes (one hotspot per 20.9 kb compared with one hotspot per ~3 kb, respectively; Table 1), consistent with substantially longer chromosomes and lower recombination frequencies in fission yeast (Pan et al., 2011; Fowler et al., 2014; Thacker et al., 2014; Lam and Keeney, 2015; Zhu and Keeney, 2015; Mohibullah and Keeney, 2017). Another important difference to consider is the absence of crossover interference in fission yeast (Munz, 1994). It is possible that DSB formation in fission yeast is restricted by competition between potential DSB sites for a more limited pool of recombination-promoting factors, thereby obviating the requirement for a downstream mechanism such as crossover interference to regulate the spacing of recombination events (Cooper et al., 2016). A more conservative approach to DSB hotspot definition was applied in Arabidopsis, based on the identification of reproducible SPO11-1-oligo peaks across biological replicates (Choi et al., 2018). This method is useful for minimizing the occurrence of false positives in peak sets, but likely underestimates the number of hotspots in the Arabidopsis genome, suggesting that hotspot density lies somewhere between those of budding yeast and fission yeast (Table 1). In mouse genomes, comparable hotspot numbers and densities were obtained by peak calling using SSDS data (Brick et al., 2012; Khil et al., 2012) and by enrichment thresholding using SPO11-oligo data (Lange et al., 2016), although SSDS-derived hotspots are wider on average (2-3.4 kb vs. 281 bp, respectively; Table 1), consistent with the action of resection. While mouse and human genomes are of similar size, hotspot numbers and densities in humans are more than double those in mice (Table 1) (Pratto et al., 2014). This is consistent with a more than doubled genome-wide average crossover frequency in human (1.20 cM/Mb) compared with mouse (0.528 cM/Mb) (Jensen-Seaman et al., 2004).
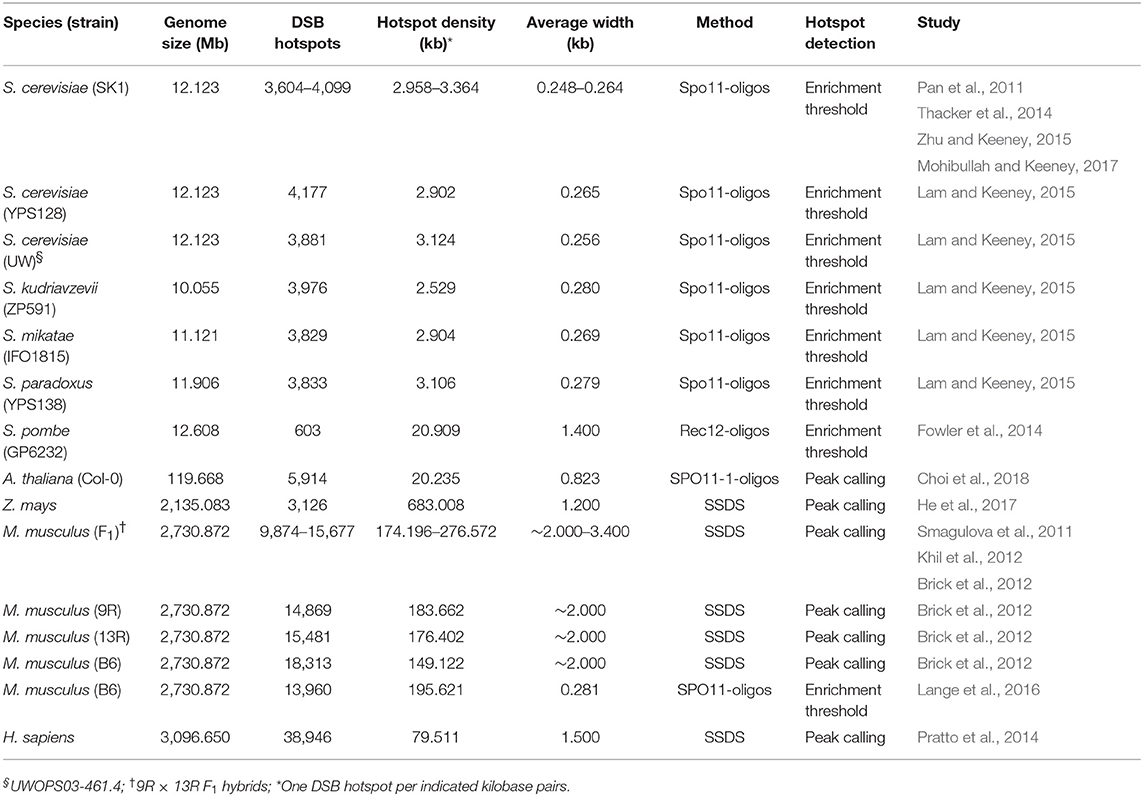
Table 1. Meiotic DNA double-strand break (DSB) hotspots identified in eukaryotes by SPO11-oligo mapping or single-stranded DNA sequencing (SSDS).
3. Chromatin Shapes the Meiotic DSB Landscape
3.1. Nucleosome Occupancy
DSB hotspot designation is controlled at multiple levels, with a hierarchy of “gatekeeper” factors acting in concert to determine the degree to which chromosome regions—at fine and broad scales—are conducive to DSB formation (Pan et al., 2011; De Massy, 2013; Cooper et al., 2016; Lange et al., 2016). Different strategies and mechanisms for the spatial regulation of DSB formation have evolved in different species, although commonalities exist (De Massy, 2013).
Genome-wide DSB maps for Saccharomyces species, Arabidopsis and maize have revealed that hotspots frequently occur within nucleosome-depleted regions (NDRs) in gene promoters (Figure 1), indicating that local chromatin accessibility contributes to DSB formation in these eukaryotes (Pan et al., 2011; Lam and Keeney, 2015; He et al., 2017; Choi et al., 2018). AT-sequence richness at budding yeast, Arabidopsis and tomato recombination hotspots is thought to exclude nucleosomes and thereby permit increased SPO11 recruitment to chromatin, promoting DNA cleavage and ultimately crossover formation (Pan et al., 2011; Choi et al., 2013, 2018; Wijnker et al., 2013; Shilo et al., 2015; Demirci et al., 2018). Indeed, elevated crossover recombination within gene promoters is conserved in several eukaryotes, including plants, canids and birds (Auton et al., 2013; Choi et al., 2013; Wijnker et al., 2013; Singhal et al., 2015; Demirci et al., 2017). The +1 nucleosomes of Arabidopsis genes whose promoters exhibit the highest crossover frequencies show greater deposition of the histone variant H2A.Z and enrichment of trimethylated lysine 4 on histone H3 (H3K4me3) (Choi et al., 2013), which are key determinants of transcriptional regulation (Deal and Henikoff, 2011; Coleman-Derr and Zilberman, 2012; Sura et al., 2017). Reduction of crossover frequency at crossover hotspots in the arp6 H2A.Z-deposition mutant confirmed a role for this histone variant in promoting recombination (Choi et al., 2013). Additionally, fewer RAD51 and DMC1 foci were observed in arp6 mutants, indicating that the recombination-promoting role of H2A.Z may include control of DSB numbers and localization (Choi et al., 2013). H2A.Z may also indirectly promote recombination by maintaining the boundaries of NDRs at which DSBs form.
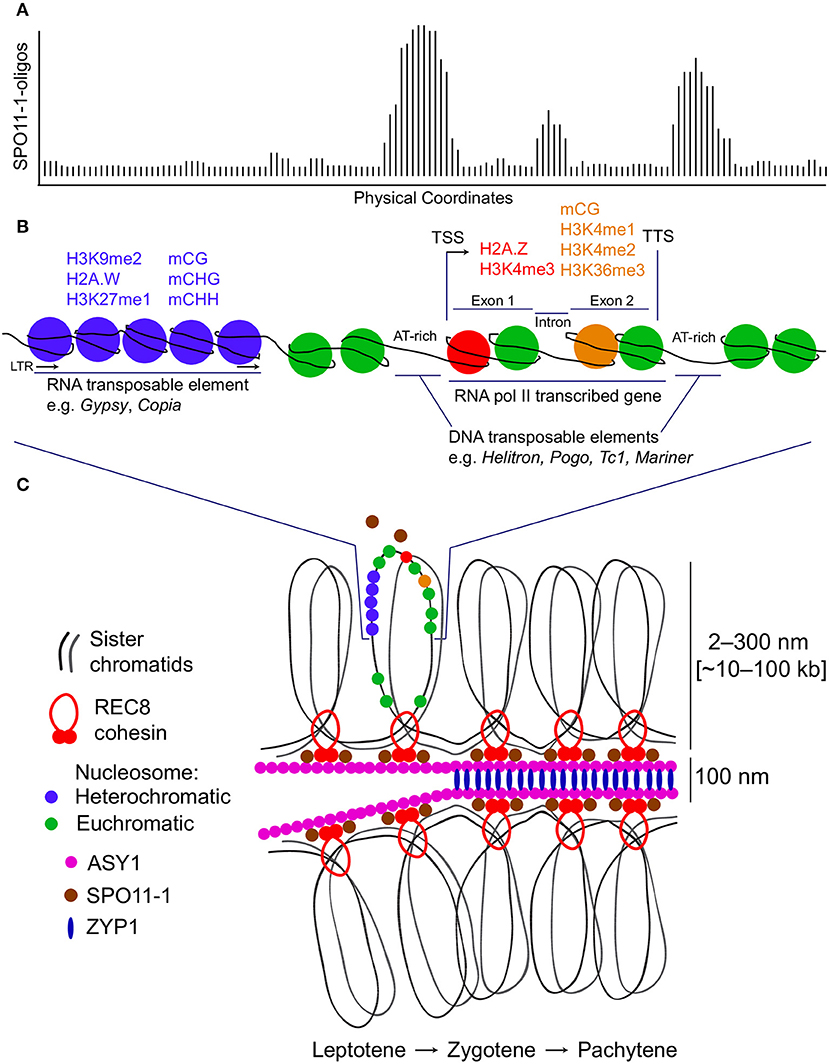
Figure 1. Meiotic DNA double-strand break hotspots, chromatin and chromosome architecture in plants. (A) A representative histogram showing relative levels of meiotic DNA double-strand breaks (DSBs) generated by SPO11-1. Physical coordinates along a hypothetical locus are represented on the x-axis and DSB signal intensity derived from SPO11-1-oligo mapping is indicated on the y-axis. The depicted hypothetical DSB topology maps on to the chromatin diagram in (B). (B) A representative chromosomal region is shown based on data from Arabidopsis thaliana. This region contains an LTR retrotransposon which has heterochromatic modifications (blue), including H3K9me2, H2A.W, H3K27me1, and DNA methylation in CG, CHG, and CHH sequence contexts. Adjacent is an RNA polymerase II transcribed gene with transcriptional start site (TSS) and termination site (TTS) indicated. The 5′ nucleosome within the gene contains H2A.Z and is H3K4me3 modified (red). Within the transcribed region, nucleosomes located toward the 3′ end are H3K4me1, H3K4me2, and H3K36me3 modified, and DNA methylated in the CG sequence context (orange). The regions of highest meiotic DSB formation correspond to gene promoter, terminator and intron regions, which tend to be AT-rich, nucleosome-depleted and contain insertions of DNA transposable elements. (C) The chromatin region shown in (B) is represented in the context of the tethered-loop/axis model for meiotic chromosomes. SPO11-1 is represented as both a freely diffusing pool and an axis/cohesin-associated pool. Paired sister chromatids organize as a linear loop array on an axial polymer, which includes ASY1. As meiosis progresses from leptotene to zygotene to pachytene, the homologs become synapsed at a distance of ~100 nm, with ZYP1 installed as transverse filaments of the synaptonemal complex. During this process, DSBs can undergo repair using a homologous chromosome, resulting in a crossover (not shown).
Arabidopsis SPO11-1-oligos also cluster in NDRs immediately downstream of transcription termination sites and in nucleosome-depleted introns (Figures 1A,B), suggesting a role for gene architecture in determining DSB positioning (Choi et al., 2018). Similarly, avian recombination rates toward the 5′ and 3′ ends of gene bodies (in both exons and introns) are elevated compared with more central regions (Singhal et al., 2015). In isolation, however, the presence of euchromatin does not adequately account for DSB hotspot locations, as NDRs immediately downstream of gene stop codons are not enriched for Spo11-oligos in budding yeast unless they overlap a promoter NDR (Pan et al., 2011). Furthermore, fission yeast DSBs do not form preferentially in NDRs, but rather at the boundaries between NDRs and well-positioned nucleosomes as inferred from population averages (Fowler et al., 2014). This might reflect preferential cleavage by the fission yeast Spo11 ortholog, Rec12, of DNA adjacent to or leaving a nucleosome (Fowler et al., 2014).
3.2. Meiotic Chromosome Architecture
Higher-order chromosome architecture plays an important role in governing DSB hotspot localization. Meiotic chromosomes are characterized by replicated sister chromatids organized into linear arrays of chromatin loops that emanate from a central chromosome axis (Figure 1C) (Blat et al., 2002; Borde and de Massy, 2013). This chromosome organization is dependent on cohesin rings that encircle the sister chromatids (Blat et al., 2002; Borde and de Massy, 2013). DSBs in budding yeast are known to occur primarily within the emanating chromatin loops, while most of the Spo11 accessory proteins that are essential for DSB formation are located on the cohesin-rich axis (Panizza et al., 2011). This is consistent with the “tethered-loop/axis complex” model, which proposes that meiotic recombination occurs at loci within chromatin loops that are tethered to the chromosome axis by recombination-promoting factors (Blat et al., 2002). Further supporting this model, the budding yeast PHD finger domain protein and Set1 complex member Spp1 binds to H3K4me2/3 near gene promoters in chromatin loops and interacts transiently with the axis-bound Spo11 accessory protein Mer2, forming a bridge between potential DSB sites and the recombination initiation machinery (Borde et al., 2009; Acquaviva et al., 2013; Borde and de Massy, 2013; Sommermeyer et al., 2013; Adam et al., 2018).
The meiotic cohesin subunit Rec8 shapes the distribution of Spo11 in budding yeast and is required for normal DSB distribution (Kugou et al., 2009). Spo11 has been observed to initially colocalize with Rec8 at axial cohesion sites and to subsequently associate with chromatin loops during DSB formation (Kugou et al., 2009; Ito et al., 2014). Translocation of Spo11 to chromatin loops is proposed to occur via Spp1-mediated tethering, giving rise to an anti-correlation between DSB formation and cohesin binding at hotspots and axis sites (Borde et al., 2009; Acquaviva et al., 2013; Sommermeyer et al., 2013). For example, lower-than-expected frequencies of budding yeast DSB hotspots are observed in proximity to Rec8 binding sites (Ito et al., 2014). Consistent with this, Spo11 enrichment is strongly diminished at DSB hotspots and increased at axis sites in spp1 mutants (Sommermeyer et al., 2013). Furthermore, inefficient induction of meiotic DSBs near axis sites in wild-type cells suggests that Spo11 may be inactivated or repressed by axial components (Ito et al., 2014). Preferential DSB formation within gene promoters, coupled with enrichment of cohesin toward and downstream of transcription termination sites in budding yeast (Ito et al., 2014; Sun et al., 2015), illustrates how gene organization may contribute to meiotic DSB hotspot localization (Cooper et al., 2016).
In budding yeast, proteins within the ZMM (Zip, Msh, and Mer) group participate in the assembly of the synaptonemal complex and promote crossing over (Lynn et al., 2007). ZMM proteins are thought to protect dHJ-JMs from disassembly by anti-crossover activity, including that of the RecQ helicase Sgs1 (Jessop et al., 2006; Oh et al., 2007), and are required for the formation of ~85% of crossovers (Lynn et al., 2007). However, more DSBs form globally and at hotspots in ZMM mutants (zip1, zip3, and msh5) than in wild-type budding yeast cells, indicative of a negative feedback loop in which homolog engagement following DSB formation suppresses Spo11 activity and prevents further breaks (Thacker et al., 2014). These mutants also exhibit increased noncrossover:crossover ratios at selected DSB hotspots (Thacker et al., 2014). Additionally, DSB hotspots with the greatest fold change in Spo11-oligo density in zip3 are more enriched for the axis proteins Hop1 and Red1, and for the axis-localized Spo11 partner proteins Rec114, Mei4 and Mer2 (Thacker et al., 2014). Thus, while Set1 is important for Spo11 targeting to sites for DSB formation (Borde and de Massy, 2013), the ZMM pathway includes feedback circuitry that controls DSB numbers while promoting crossover recombination (Thacker et al., 2014).
4. Meiotic DSB and Crossover Distributions
Budding yeast DSB and crossover densities are anti-correlated with chromosome size, with smaller chromosomes undergoing more DSBs and crossovers per kilobase than larger chromosomes (Pan et al., 2011). This chromosome-scale control of DSB density is thought to be dictated by a suppressive impact of homolog engagement upon the formation of further DSBs (Thacker et al., 2014), as it has been suggested that smaller chromosomes may engage their homologs more slowly on average, thereby extending the period during which breaks can accumulate (Thacker et al., 2014; Lam and Keeney, 2015). In view of the strong broad-scale correlation between DSB and crossover distributions in budding yeast, regulation of DSB density has been proposed to account for much of the variation in crossover density (Pan et al., 2011). Similarly, mouse chromosome size is negatively correlated with crossover density and, to a lesser extent, DSB density (Lange et al., 2016). The steeper slope observed for the relationship with crossover density suggests that chromosome size is a more important determinant of crossover density than regulation of DSB numbers in mouse (Lange et al., 2016).
Despite positive relationships between genome-wide DSB and crossover distributions, fine-scale correlations at Arabidopsis crossover hotspots are weaker and variable (Choi et al., 2018). Interhomolog sequence divergence near DSB hotspots may contribute to this discrepancy by inhibiting crossover formation in hybrids between diverged strains used to map crossovers (Choi et al., 2018). In mouse, for example, crossovers are repressed near indels within the A3 crossover hotspot (Cole et al., 2010). The absence of strong correlations between DSB levels and crossover frequencies at fine scale may also reflect the fact that a minority of DSBs mature into crossovers in plants and mice (3–10%; De Muyt et al., 2009; Cole et al., 2010). Most strand invasion recombination intermediates are resolved as non-crossovers via processes such as SDSA, dissolution of dHJ-JMs, unidirectional endonuclease cleavage of dHJ-JMs, or intersister repair (De Muyt et al., 2012; Hunter, 2015). For example, high-resolution mapping of meiotic recombination initiation sites in maize identified RAD51 ChIP-seq hotspots in all chromosome regions, whereas crossovers are largely confined to sub-telomeric gene-rich regions comprising open chromatin (Li et al., 2015; Rodgers-Melnick et al., 2015, 2016; He et al., 2017).
Fission yeast recombination landscapes are characterized by crossover invariance, which describes a near-uniform genome-wide distribution of crossovers between homologous chromosomes despite considerable variability in DSB levels (Hyppa and Smith, 2010). This mechanism of crossover control biases DSB repair toward the sister chromatid rather than the homologous chromosome, with intersister repair exceeding interhomolog repair ~3:1 at hotspots (Cromie et al., 2006; Hyppa and Smith, 2010). In DSB-cold regions, by contrast, interhomolog repair is favored (Hyppa and Smith, 2010). While the potential existence of crossover invariance in other eukaryotes remains to be investigated, this type of crossover control may help to explain varying DSB:crossover ratios in diverse species (Fowler et al., 2013).
5. PRDM9 and Histone H3 Lysine 4 Trimethylation
The histone-lysine trimethyltransferase PRDM9 dictates the position of the vast majority of DSB hotspots in mouse and primate genomes by conferring a dominant mechanism of DSB spatial regulation (Baudat et al., 2010; Berg et al., 2010; Brick et al., 2012). PRDM9 designates DSB hotspots by trimethylation of H3K4 at loci matching the DNA binding specificity of its zinc finger array (Buard et al., 2009; Grey et al., 2011; Smagulova et al., 2011; Diagouraga et al., 2018). A 12-bp motif matching part of a 36-bp PRDM9B6 binding sequence is enriched in DSB hotspots identified by SPO11-oligo mapping and SSDS, and adjacent to PRDM9-dependent H3K4me3 peaks in B6 mice (Brick et al., 2012; Baker et al., 2014, 2015; Lange et al., 2016). Interestingly, SSDS-derived DSB hotspots in Prdm9−/− mice occur at H3K4me3-marked gene promoters, sites at which DSBs form rarely in wild-type mice (Brick et al., 2012). This reveals a reversion to a masked, ancestral DSB hotspot designation mechanism analogous to that observed in eukaryotes lacking a PRDM9-like mechanism, including budding yeast, birds, dogs and plants (Cooper et al., 2016). PRDM9 thus diverts DSBs away from functionally conserved genomic elements and toward independent H3K4me3 and H3K36me3 markers deposited via its histone methyltransferase activity (Brick et al., 2012; Diagouraga et al., 2018). According to the “hotspot paradox” hypothesis, rapid loss of PRDM9 recognition sequences through biased gene conversion is predicted to result in the evolutionary erosion of hotspots in primate and mouse genomes (Myers et al., 2010; Cole et al., 2014; Baker et al., 2015). PRDM9 evolves rapidly, however, with the emergence of new allelic variants of its zinc finger motif causing DSB landscapes to be recast and the concomitant designation of new hotspots (Berg et al., 2010; Myers et al., 2010; Brick et al., 2012; Baker et al., 2015; Diagouraga et al., 2018).
Mouse SPO11-oligo-derived DSB hotspot midpoints are depleted of H3K4me3 and H3K36me3, while their flanking regions exhibit a continuum of left–right asymmetric enrichment of these marks, together with secondary SPO11-oligo peaks in adjacent valleys in histone H3 lysine trimethylation signal (Lange et al., 2016; Yamada et al., 2017). Arabidopsis DSB hotspots are similarly depleted of H3K4me3 and MNase-seq-derived nucleosome signal (Figures 1A,B) (Choi et al., 2018). This indicates that SPO11 preferentially forms DSBs between nucleosomes in mammalian and plant genomes, similar to its ortholog in budding yeast (Pan et al., 2011). Despite this, DSB formation is severely impaired in the absence of the H3K4 methyltransferase Set1 or the Set1 complex member Spp1, or following mutation of the H3K4 residue targeted by the Set1 complex in budding yeast (Borde et al., 2009; Acquaviva et al., 2013; Sommermeyer et al., 2013). Loci that exhibit the greatest reduction in DSB frequency in set1 mutants are also located within regions marked by high wild-type levels of H3K4me3 deposition (Borde et al., 2009). As discussed, H3K4me3 plays a role in tethering chromatin loops to the chromosome axis for DSB formation and recombination in nearby promoter NDRs (Borde et al., 2009; Acquaviva et al., 2013; Sommermeyer et al., 2013). Furthermore, SPO11-oligo frequency at mouse DSB hotspots is correlated with H3K4me3 signal in flanking regions (R2 = 0.40) (Lange et al., 2016). In Arabidopsis, by contrast, SPO11-1-oligo enrichment in gene promoters is uncorrelated with levels of H3K4me3 on the first nucleosome immediately downstream of gene transcriptional start sites (i.e., when genes are ordered by decreasing SPO11-1-oligo enrichment in gene promoters, there is no apparent relationship with the degree of H3K4me3 enrichment at the +1 nucleosome) (Choi et al., 2018). Similarly, a minority of maize DSB hotspots overlap H3K4me3 sites (He et al., 2017), and budding yeast DSB frequencies are not correlated with H3K4me3 signal (Tischfield and Keeney, 2012), MNase accessibility or transcriptional activity at hotspots (Zhu and Keeney, 2015). Taken together, these findings indicate that while H3K4me3 deposition is a key determinant of DSB frequency in some eukaryotes, additional factors are important for the local control of DSB numbers.
6. The Hotspot Paradox
The “hotspot paradox” predicts that recombination hotspots will be rapidly eliminated from populations in situations where there are strong cis-acting sequence determinants of hotspot activity (Boulton et al., 1997). Under this hypothesis, hotspot-activating alleles are rapidly replaced by hotspot-inactivating mutations via biased gene conversion, whereby DSB repair at an active hotspot allele uses an unbroken homolog bearing an inactive allele, conferring a transmission advantage to the recombination-suppressing allele (Úbeda and Wilkins, 2011). This is expected to give rise to dynamic genome-wide DSB landscapes, within which PRDM9-designated hotspots exist transiently in evolutionary time (Lam and Keeney, 2015). This prediction is reinforced by empirical studies and simulations of recombination hotspot activity and evolution in primates and mice (Pineda-Krch and Redfield, 2005; Coop and Myers, 2007; Friberg and Rice, 2008; Úbeda and Wilkins, 2011).
Conversely, the occurrence of DSB hotspots in the promoter NDRs of several other eukaryotes supports an alternative hypothesis, which proposes that hotspots can persist if DSBs form preferentially within genomic features that are conserved over extended evolutionary periods and whose functions and chromatin state are unrelated to their hotspot status (Lam and Keeney, 2015). This is supported by the strong conservation of DSB hotspot positions and intensities among Saccharomyces species (Lam and Keeney, 2015) and among Schizosaccharomyces species (Zanders et al., 2014), as well as by evolutionarily stable recombination hotspots in birds (Singhal et al., 2015). Many properties of chromatin structure, at both fine and broad scales, are likely constrained due to their functions in essential processes, including transcription, DNA replication, sister chromatid cohesion, chromatin compaction and chromosome segregation (Pan et al., 2011). As chromatin architecture shapes genome-wide DSB distributions, conservation of the DSB landscape is likely to be a common corollary of selective pressures on chromatin structures to maintain functions independent of meiotic recombination (Lam and Keeney, 2015).
7. Recombination Initiation in Repetitive Sequences
DSB formation within or adjacent to repetitive elements can lead to homologous recombination between non-allelic repeats, potentially resulting in harmful chromosomal rearrangements and copy-number instability in the germline (Yamada et al., 2017). DSBs are generally suppressed in budding yeast Ty elements, which may reflect a mechanism to preserve genome stability, although elevated DSB levels are associated with some Ty insertions (Pan et al., 2011; Sasaki et al., 2013). This overall trend is consistent with DSB and crossover repression and elevated transposon density within Arabidopsis pericentromeric heterochromatin (Choi et al., 2018). Loss of CG DNA methylation in Arabidopsis met1 mutants causes increased SPO11-1-oligo levels in EnSpm/CACTA and Gypsy elements and within pericentromeres generally, together with loss of pericentromeric nucleosome occupancy (Choi et al., 2018). Comparable impairment of DNA methyltransferase activity in mouse Dnmt3L−/− mutants also results in increased SPO11-dependent DSBs in retrotransposons (Zamudio et al., 2015). This is consistent with findings from epigenetic manipulations in Arabidopsis showing that RNA-directed DNA methylation (RdDM) targeted to meiotic hotspots suppresses crossover recombination (Yelina et al., 2015). Furthermore, the histone deacetylase Sir2 inhibits meiotic DSB formation and recombination in the repetitive ribosomal DNA (rDNA) array in budding yeast (Gottlieb and Esposito, 1989; Mieczkowski et al., 2007), indicating that DSB suppression by heterochromatin assembly on repetitive DNA is a conserved strategy to safeguard against genome destabilization.
Despite SPO11-1-oligo depletion in Arabidopsis pericentromeric regions, however, significant overlap was observed between DSB hotspots and transposable elements generally (Choi et al., 2018). Specifically, Arabidopsis DSB hotspots overlap DNA transposable elements within the Helitron, Pogo/Tc1/Mariner and MuDR families more than expected, whereas hotspots overlap DNA elements in the EnSpm/CACTA class and RNA elements in the Gypsy LTR (long terminal repeat), Copia LTR and LINE-1 classes less than expected (Choi et al., 2018). Helitron and Pogo/Tc1/Mariner transposition occurs preferentially in AT-rich gene regulatory sequences, at which nucleosome exclusion is thought to contribute to increased DSB frequencies (Figures 1A,B) (Kapitonov and Jurka, 2001; Guermonprez et al., 2008; Choi et al., 2018). Similarly, although SPO11-oligos are generally underrepresented in mouse repeats (including LINE-1 retrotransposons), elevated SPO11-oligo levels and functional PRDM9 binding sites were observed within DNA elements in the MULE-MuDR, TcMar-Mariner, hAT-Charlie and PiggyBac families (Yamada et al., 2017). By contrast, most maize DSB hotspots are located in repetitive sequences, although DSBs avoid heterochromatin, forming in transposon NDRs and exhibiting DNA hypomethylation (He et al., 2017). These DSB hotspots occur predominantly in Gypsy LTR retrotransposons, which are abundant in the maize genome (He et al., 2017). Similar to Arabidopsis and mouse, however, fewer-than-expected maize DSB hotspots occur in Copia LTR and LINE retrotransposons (He et al., 2017).
Citing the hotspot paradox hypothesis, Yamada et al. (2017) speculate that PRDM9 may target some repeat classes for biased gene conversion to inhibit the proliferation of selfish genetic elements. Rapid fixation of hotspot-inactivating mutations would reduce the copy number of PRDM9-targeted transposons in populations (Yamada et al., 2017). As a Krüppel-associated-box (KRAB)-zinc finger protein, PRDM9 may have derived functions to counteract transposon proliferation from an ancestral KRAB factor, many of which have roles in transposon silencing (Wolf et al., 2015; Yamada et al., 2017). Balancing this proposed transposon-antagonizing role of PRDM9 with mechanisms to prevent excessive DSB formation in repeats may be an important contributor to PRDM9 evolution and DSB hotspot designation in mammalian genomes (Yamada et al., 2017). In Arabidopsis, significant overlap occurs between comparable classes of DNA elements and DSB hotspots, many of which are located within functionally conserved sequences (Choi et al., 2018). This suggests that the hotspot paradox theory may not be applicable in this case and that Arabidopsis hotspots may be more evolutionarily stable. Nonetheless, comparisons between eukaryotes indicate that repeated sequences may influence meiotic recombination initiation landscapes in related ways.
8. Beyond Hotspots: DSB-Dependent Spatial Regulation
Meiotic DSB hotspots are identified by mapping the DSB landscape in a population of cells. This landscape reveals a continuum of variation within which loci with high probabilities of DSB formation may be detected (Pan et al., 2011). However, spatial regulation that occurs as a consequence of DSB formation is largely obscured within the population average because low proportions of even the most active DSB hotspots are cleaved in individual meiocytes (~10–15%; Cooper et al., 2016). DSB interference, mediated by the DNA damage response (DDR) kinase Tel1ATM in budding yeast, suppresses the formation of clustered DSBs in cis over distances of ~70–100 kb (Garcia et al., 2015; Cooper et al., 2016). Loss of Tel1ATM activity allows DSBs to form independently of one another over ±20–100-kb distances, giving rise to DSB formation in neighboring regions at frequencies comparable to those expected by chance. Over distances of ± ~7.5 kb, by contrast, Tel1ATM inactivation permits the formation of adjacent DSBs significantly more frequently than expected, generating localized regions of “negative DSB interference” (Garcia et al., 2015). This short-range effect occurs only between DSB hotspots located within the same chromatin loop domain (Garcia et al., 2015; Cooper et al., 2016). Coincident DSB formation at adjacent intra-loop hotspots in the absence of cis-interference suggests that hotspots within the same loop domain are “primed” for cleavage. Cooper et al. (2016) speculate that tethering of a loop to the chromosome axis may pre-activate the loop and the hotspots within, an effect suggested to be concealed by Tel1ATM-dependent cis-interference, which restricts DSB formation to only one of the primed intra-loop hotspots. Spatial regulation of meiotic DSB formation also occurs in trans via a mechanism involving Tel1ATM and Mec1ATR, another DDR signal transduction kinase (Zhang et al., 2011). Following DSB formation on a chromatid, trans-interference inhibits DSB formation at the corresponding locus on its sister, its homolog or frequently both. This mechanism is thought to ensure that an intact template is available for DSB repair, and to prevent DSB formation at allelic loci on both homologs (Zhang et al., 2011). Meiotic DSB interference along and between chromatids is therefore likely important for ensuring even spacing of recombination events, thereby contributing to stable interhomolog interactions that facilitate proper chromosome pairing and successful completion of meiosis (Zhang et al., 2011; Garcia et al., 2015; Cooper et al., 2016).
9. Future Prospects
Genome-wide DSB mapping in different eukaryotes has revealed diversity with regard to the hierarchical combinations of factors that shape meiotic recombination landscapes and hotspots. These distinctions highlight the importance of studying DSB landscapes in diverse eukaryotes and beyond model organisms. Efforts to elucidate the mechanisms that determine DSB hotspot designation may inform genetic or epigenetic manipulations intended to reshape naturally constrained meiotic recombination landscapes. For example, the presence of hotspots in conserved genomic elements, such as nucleosome-depleted promoters, has relevance for targeting crossover recombination to specific loci in plants (Sarno et al., 2017). Manipulation of recombination has the potential to generate greater genetic diversity among gametes for accelerated crop improvement. Such approaches should be considered with caution, however, as forced recombination within repetitive heterochromatin also has the potential to compromise genome integrity in the germline.
Author Contributions
AT and IH wrote and edited the manuscript. AT created Table 1 and IH created Figure 1. Both authors approved the work for publication.
Funding
Research was supported by a European Research Council SynthHotSpot Consolidator Grant.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Henderson group and the reviewers for helpful comments and suggestions.
References
Acquaviva, L., Székvölgyi, L., Dichtl, B., Dichtl, B. S., de La Roche Saint André, C., Nicolas, A., et al. (2013). The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339, 215–218. doi: 10.1126/science.1225739
Adam, C., Guérois, R., Citarella, A., Verardi, L., Adolphe, F., Béneut, C., et al. (2018). The PHD finger protein Spp1 has distinct functions in the Set1 and the meiotic DSB formation complexes. PLoS Genet. 14:e1007223. doi: 10.1371/journal.pgen.1007223
Allers, T., and Lichten, M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57. doi: 10.1016/S0092-8674(01)00416-0
Auton, A., Rui Li, Y., Kidd, J., Oliveira, K., Nadel, J., Holloway, J. K., et al. (2013). Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 9:e1003984. doi: 10.1371/journal.pgen.1003984
Baker, C. L., Kajita, S., Walker, M., Saxl, R. L., Raghupathy, N., Choi, K., et al. (2015). PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 11:e1004916. doi: 10.1371/journal.pgen.1004916
Baker, C. L., Walker, M., Kajita, S., Petkov, P. M., and Paigen, K. (2014). PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res. 24, 724–732. doi: 10.1101/gr.170167.113
Baudat, F., Buard, J., Grey, C., Fledel-Alon, A., Ober, C., Przeworski, M., et al. (2010). PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840. doi: 10.1126/science.1183439
Baudat, F., Imai, Y., and Massy, B. D. (2013). Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 14, 794–806. doi: 10.1038/nrg3573
Berg, I. L., Neumann, R., Lam, K. W. G., Sarbajna, S., Odenthal-Hesse, L., May, C. A., et al. (2010). PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 42, 859–863. doi: 10.1038/ng.658
Bergerat, A., De Massy, B., Gadelle, D., Varoutas, P. C., Nicolas, A., and Forterre, P. (1997). An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386, 414–417. doi: 10.1038/386414a0
Bishop, D. K., Park, D., Xu, L., and Kleckner, N. (1992). DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69, 439–456. doi: 10.1016/0092-8674(92)90446-J
Blat, Y., Protacio, R. U., Hunter, N., and Kleckner, N. (2002). Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111, 791–802. doi: 10.1016/S0092-8674(02)01167-4
Blitzblau, H. G., Bell, G. W., Rodriguez, J., Bell, S. P., and Hochwagen, A. (2007). Mapping of meiotic single-stranded DNA reveals double-strand-break hotspots near centromeres and telomeres. Curr. Biol. 17, 2003–2012. doi: 10.1016/j.cub.2007.10.066
Borde, V., and de Massy, B. (2013). Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr. Opin. Genet. Dev. 23, 147–155. doi: 10.1016/j.gde.2012.12.002
Borde, V., Robine, N., Lin, W., Bonfils, S., Géli, V., and Nicolas, A. (2009). Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28, 99–111. doi: 10.1038/emboj.2008.257
Boulton, A., Myers, R. S., and Redfield, R. J. (1997). The hotspot conversion paradox and the evolution of meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 94, 8058–8063. doi: 10.1073/pnas.94.15.8058
Brick, K., Smagulova, F., Khil, P., Camerini-Otero, R. D., and Petukhova, G. V. (2012). Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645. doi: 10.1038/nature11089
Buard, J., Barthès, P., Grey, C., and De Massy, B. (2009). Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 28, 2616–2624. doi: 10.1038/emboj.2009.207
Buhler, C., Borde, V., and Lichten, M. (2007). Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 5, 2797–2808. doi: 10.1371/journal.pbio.0050324
Cao, L., Alani, E., and Kleckner, N. (1990). A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61, 1089–1101. doi: 10.1016/0092-8674(90)90072-M
Cejka, P., Plank, J. L., Bachrati, C. Z., Hickson, I. D., and Kowalczykowski, S. C. (2010). Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 17, 1377–1382. doi: 10.1038/nsmb.1919
Choi, K., Zhao, X., Kelly, K. A., Venn, O., Higgins, J. D., Yelina, N. E., et al. (2013). Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45, 1327–1336. doi: 10.1038/ng.2766
Choi, K., Zhao, X., Tock, A. J., Lambing, C., Underwood, C. J., Hardcastle, T. J., et al. (2018). Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 28, 1–15. doi: 10.1101/gr.225599.117
Cloud, V., Chan, Y. L., Grubb, J., Budke, B., and Bishop, D. K. (2012). Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225. doi: 10.1126/science.1219379
Cole, F., Baudat, F., Grey, C., Keeney, S., De Massy, B., and Jasin, M. (2014). Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat. Genet. 46, 1072–1080. doi: 10.1038/ng.3068
Cole, F., Keeney, S., and Jasin, M. (2010). Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol. Cell 39, 700–710. doi: 10.1016/j.molcel.2010.08.017
Coleman-Derr, D., and Zilberman, D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8:e1002988. doi: 10.1371/journal.pgen.1002988
Coop, G., and Myers, S. R. (2007). Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 3:e35. doi: 10.1371/journal.pgen.0030035
Cooper, T. J., Garcia, V., and Neale, M. J. (2016). Meiotic DSB patterning: a multifaceted process. Cell Cycle 15, 13–21. doi: 10.1080/15384101.2015.1093709
Cromie, G. A., Hyppa, R. W., Taylor, A. F., Zakharyevich, K., Hunter, N., and Smith, G. R. (2006). Single Holliday junctions are intermediates of meiotic recombination. Cell 127, 1167–1178. doi: 10.1016/j.cell.2006.09.050
De Massy, B. (2013). Spp1 links sites of meiotic DNA double-strand breaks to chromosome axes. Mol. Cell 49, 3–5. doi: 10.1016/j.molcel.2012.12.011
De Muyt, A., Jessop, L., Kolar, E., Sourirajan, A., Chen, J., Dayani, Y., et al. (2012). BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46, 43–53. doi: 10.1016/j.molcel.2012.02.020
De Muyt, A., Mercier, R., Mézard, C., and Grelon, M. (2009). “Meiotic recombination and crossovers in plants,” in Meiosis: Genome Dynamics, Vol 5, eds R. Benavente and J. N. Volff (Basel: Karger), 14–25.
Deal, R. B., and Henikoff, S. (2011). Histone variants and modifications in plant gene regulation. Curr. Opin. Plant Biol. 14, 116–122. doi: 10.1016/j.pbi.2010.11.005
Demirci, S., Ridder, D. D., and Dijk, A. D. J. V. (2018). DNA sequence and shape are predictive for meiotic crossovers throughout the plant kingdom. Plant J. 95, 686–699. doi: 10.1111/tpj.13979
Demirci, S., van Dijk, A. D., Sanchez Perez, G., Aflitos, S. A., de Ridder, D., and Peters, S. A. (2017). Distribution, position and genomic characteristics of crossovers in tomato recombinant inbred lines derived from an interspecific cross between Solanum lycopersicum and Solanum pimpinellifolium. Plant J. 89, 554–564. doi: 10.1111/tpj.13406
Diagouraga, B., Clément, J. A. J., Duret, L., Kadlec, J., de Massy, B., and Baudat, F. (2018). PRDM9 methyltransferase activity is essential for meiotic DNA double-strand break formation at its binding sites. Mol. Cell 69, 853–865. doi: 10.1016/j.molcel.2018.01.033
Feng, X., Grossman, R., and Stein, L. (2011). PeakRanger: a cloud-enabled peak caller for ChIP-seq data. BMC Bioinform. 12:139. doi: 10.1186/1471-2105-12-139
Fowler, K. R., Gutiérrez-Velasco, S., Martín-Castellanos, C., and Smith, G. R. (2013). Protein determinants of meiotic DNA break hot spots. Mol. Cell 49, 983–996. doi: 10.1016/j.molcel.2013.01.008
Fowler, K. R., Sasaki, M., Milman, N., Keeney, S., and Smith, G. R. (2014). Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 24, 1650–1664. doi: 10.1101/gr.172122.114
Friberg, U., and Rice, W. R. (2008). Cut thy neighbor: Cyclic birth and death of recombination hotspots via genetic conflict. Genetics 179, 2229–2238. doi: 10.1534/genetics.107.085563
Garcia, V., Gray, S., Allison, R. M., Cooper, T. J., and Neale, M. J. (2015). Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature 520, 114–118. doi: 10.1038/nature13993
Gottlieb, S., and Esposito, R. E. (1989). A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56, 771–776. doi: 10.1016/0092-8674(89)90681-8
Grey, C., Barthès, P., Friec, G., Langa, F., Baudat, F., and de Massy, B. (2011). Mouse Prdm9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 9:e1001176. doi: 10.1371/journal.pbio.1001176
Guermonprez, H., Loot, C., and Casacuberta, J. M. (2008). Different strategies to persist: the pogo-like Lemi1 transposon produces miniature inverted-repeat transposable elements or typical defective elements in different plant genomes. Genetics 180, 83–92. doi: 10.1534/genetics.108.089615
He, Y., Wang, M., Dukowic-Schulze, S., Zhou, A., Tiang, C.-L., Shilo, S., et al. (2017). Genomic features shaping the landscape of meiotic double-strand-break hotspots in maize. Proc. Natl. Acad. Sci. 114, 12231-12236. doi: 10.1073/pnas.1713225114
Hunter, N. (2015). Meiotic recombination: the essence of heredity. Cold Spring Harbor Perspect. Biol. 7:a016618. doi: 10.1101/cshperspect.a016618
Hunter, N., and Kleckner, N. (2001). The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70. doi: 10.1016/S0092-8674(01)00430-5
Hyppa, R. W., and Smith, G. R. (2010). Crossover invariance determined by partner choice for meiotic DNA break repair. Cell 142, 243–255. doi: 10.1016/j.cell.2010.05.041
Ito, M., Kugou, K., Fawcett, J. A., Mura, S., Ikeda, S., Innan, H., et al. (2014). Meiotic recombination cold spots in chromosomal cohesion sites. Genes Cells 19, 359–373. doi: 10.1111/gtc.12138
Jensen-Seaman, M. I., Furey, T. S., Payseur, B. A., Lu, Y., Roskin, K. M., Chen, C.-F., et al. (2004). Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14, 528–538. doi: 10.1101/gr.1970304
Jessop, L., Rockmill, B., Roeder, G. S., and Lichten, M. (2006). Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2, 1402–1412. doi: 10.1371/journal.pgen.0020155
Kapitonov, V. V., and Jurka, J. (2001). Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 98, 8714–8719. doi: 10.1073/pnas.151269298
Keeney, S., Giroux, C. N., and Kleckner, N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. doi: 10.1016/S0092-8674(00)81876-0
Keeney, S., Lange, J., and Mohibullah, N. (2014). Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet. 48, 187–214. doi: 10.1146/annurev-genet-120213-092304
Khil, P. P., Smagulova, F., Brick, K. M., Camerini-Otero, R. D., and Petukhova, G. V. (2012). Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 22, 957–965. doi: 10.1101/gr.130583.111
Kugou, K., Fukuda, T., Yamada, S., Ito, M., Sasanuma, H., Mori, S., et al. (2009). Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol. Biol. Cell 20, 3064–3076. doi: 10.1091/mbc.e08-12-1223
Lam, I., and Keeney, S. (2015). Nonparadoxical evolutionary stability of the recombination initiation landscape in yeast. Science 350, 932–937. doi: 10.1126/science.aad0814
Lange, J., Yamada, S., Tischfield, S. E., Pan, J., Kim, S., Zhu, X., et al. (2016). The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell 167, 695–708. doi: 10.1016/j.cell.2016.09.035
Li, Q., Brown, J. B., Huang, H., and Bickel, P. J. (2011). Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779. doi: 10.1214/11-AOAS466
Li, X., Li, L., and Yan, J. (2015). Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat. Commun. 6, 1–9. doi: 10.1038/ncomms7648
Lynn, A., Soucek, R., and Börner, G. V. (2007). ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 15, 591–605. doi: 10.1007/s10577-007-1150-1
McMahill, M. S., Sham, C. W., and Bishop, D. K. (2007). Synthesis-dependent strand annealing in meiosis. PLoS Biol. 5:e299. doi: 10.1371/journal.pbio.0050299
Mieczkowski, P. A., Dominska, M., Buck, M. J., Lieb, J. D., and Petes, T. D. (2007). Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 104, 3955–3960. doi: 10.1073/pnas.0700412104
Mohibullah, N., and Keeney, S. (2017). Numerical and spatial patterning of yeast meiotic DNA breaks by Tel1. Genome Res. 27, 278–288. doi: 10.1101/gr.213587.116
Munz, P. (1994). An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 137, 701–707.
Myers, S., Bowden, R., Tumian, A., Bontrop, R. E., Freeman, C., MacFie, T. S., et al. (2010). Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327, 876–879. doi: 10.1126/science.1182363
Neale, M. J., Pan, J., and Keeney, S. (2005). Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057. doi: 10.1038/nature03872
Oh, S. D., Lao, J. P., Hwang, P. Y. H., Taylor, A. F., Smith, G. R., and Hunter, N. (2007). BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272. doi: 10.1016/j.cell.2007.05.035
Pâques, F., and Haber, J. E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404.
Pan, J., Sasaki, M., Kniewel, R., Murakami, H., Blitzblau, H. G., Tischfield, S. E., et al. (2011). A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731. doi: 10.1016/j.cell.2011.02.009
Panizza, S., Mendoza, M. A., Berlinger, M., Huang, L., Nicolas, A., Shirahige, K., et al. (2011). Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146, 372–383. doi: 10.1016/j.cell.2011.07.003
Pineda-Krch, M., and Redfield, R. J. (2005). Persistence and loss of meiotic recombination hotspots. Genetics 169, 2319–2333. doi: 10.1534/genetics.104.034363
Pratto, F., Brick, K., Khil, P., Smagulova, F., Petukhova, G. V., and Camerini-Otero, R. D. (2014). Recombination initiation maps of individual human genomes. Science 346, 1256442–1256442. doi: 10.1126/science.1256442
Rodgers-Melnick, E., Bradbury, P. J., Elshire, R. J., Glaubitz, J. C., Acharya, C. B., Mitchell, S. E., et al. (2015). Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc. Natl. Acad. Sci. U.S.A. 112, 3823–3828. doi: 10.1073/pnas.1413864112
Rodgers-Melnick, E., Vera, D. L., Bass, H. W., and Buckler, E. S. (2016). Open chromatin reveals the functional maize genome. Proc. Natl. Acad. Sci. U.S.A. 113, E3177–E3184. doi: 10.1073/pnas.1525244113
Sarno, R., Vicq, Y., Uematsu, N., Luka, M., Lapierre, C., Carroll, D., et al. (2017). Programming sites of meiotic crossovers using Spo11 fusion proteins. Nucleic Acids Res. 45:e164. doi: 10.1093/nar/gkx739
Sasaki, M., Tischfield, S. E., van Overbeek, M., and Keeney, S. (2013). Meiotic recombination initiation in and around retrotransposable elements in Saccharomyces cerevisiae. PLoS Genet. 9:e1003732. doi: 10.1371/journal.pgen.1003732
Shilo, S., Melamed-Bessudo, C., Dorone, Y., Barkai, N., and Levy, A. A. (2015). DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis. Plant Cell 27, 2427–2436. doi: 10.1105/tpc.15.00391
Shinohara, A., Ogawa, H., and Ogawa, T. (1992). Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69, 457–470. doi: 10.1016/0092-8674(92)90447-K
Singhal, S., Leffler, E. M., Sannareddy, K., Turner, I., Venn, O., Hooper, D. M., et al. (2015). Stable recombination hospots in birds. Science 350, 928–932. doi: 10.1126/science.aad0843
Smagulova, F., Brick, K., Pu, Y., Camerini-Otero, R. D., and Petukhova, G. V. (2016). The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 30, 266–280. doi: 10.1101/gad.270009.115
Smagulova, F., Gregoretti, I. V., Brick, K., Khil, P., Camerini-Otero, R. D., and Petukhova, G. V. (2011). Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472, 375–378. doi: 10.1038/nature09869
Sommermeyer, V., Béneut, C., Chaplais, E., Serrentino, M. E., and Borde, V. (2013). Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell 49, 43–54. doi: 10.1016/j.molcel.2012.11.008
Sun, H., Treco, D., and Szostak, J. W. (1991). Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64, 1155–1161. doi: 10.1016/0092-8674(91)90270-9
Sun, X., Huang, L., Markowitz, T. E., Blitzblau, H. G., Chen, D., Klein, F., et al. (2015). Transcription dynamically patterns the meiotic chromosome-axis interface. eLife 4, 1–23. doi: 10.7554/eLife.07424
Sura, W., Kabza, M., Karlowski, W. M., Bieluszewski, T., Kus-Slowinska, M., Paweloszek, L., et al. (2017). Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell 29, 791–807. doi: 10.1105/tpc.16.00573
Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J., and Stahl, F. W. (1983). The double-strand-break repair model for recombination. Cell 33, 25–35. doi: 10.1016/0092-8674(83)90331-8
Thacker, D., Mohibullah, N., Zhu, X., and Keeney, S. (2014). Homologue engagement controls meiotic DNA break number and distribution. Nature 510, 241–246. doi: 10.1038/nature13120
Tischfield, S. E., and Keeney, S. (2012). Scale matters: the spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle 11, 1496–1503. doi: 10.4161/cc.19733
Úbeda, F., and Wilkins, J. F. (2011). The Red Queen theory of recombination hotspots. J. Evol. Biol. 24, 541–553. doi: 10.1111/j.1420-9101.2010.02187.x
Underwood, C. J., Choi, K., Lambing, C., Zhao, X., Serra, H., Borges, F., et al. (2018). Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 28, 519–531. doi: 10.1101/gr.227116.117
Villeneuve, A. M., and Hillers, K. J. (2001). Whence meiosis? Cell 106, 647–650. doi: 10.1016/S0092-8674(01)00500-1
Wijnker, E., James, G. V., Ding, J., Becker, F., Klasen, J. R., Rawat, V., et al. (2013). The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2013, 1–22. doi: 10.7554/eLife.01426
Wolf, G., Greenberg, D., and Macfarlan, T. S. (2015). Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family. Mobile DNA 6, 1–20. doi: 10.1186/s13100-015-0050-8
Yamada, S., Kim, S., Tischfield, S. E., Jasin, M., Lange, J., and Keeney, S. (2017). Genomic and chromatin features shaping meiotic double-strand break formation and repair in mice. Cell Cycle 16, 1870–1884. doi: 10.1080/15384101.2017.1361065
Yelina, N. E., Lambing, C., Hardcastle, T. J., Zhao, X., Santos, B., and Henderson, I. R. (2015). DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 29, 2183–2202. doi: 10.1101/gad.270876.115
Zakharyevich, K., Ma, Y., Tang, S., Hwang, P. Y. H., Boiteux, S., and Hunter, N. (2010). Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell 40, 1001–1015. doi: 10.1016/j.molcel.2010.11.032
Zamudio, N., Barau, J., Teissandier, A., Walter, M., Borsos, M., Servant, N., et al. (2015). DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 29, 1256–1270. doi: 10.1101/gad.257840.114
Zanders, S. E., Eickbush, M. T., Yu, J. S., Kang, J.-W., Fowler, K. R., Smith, G. R., et al. (2014). Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. eLife 3:e02630. doi: 10.7554/eLife.02630
Zhang, L., Kim, K. P., Kleckner, N. E., and Storlazzi, A. (2011). Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc. Natl. Acad. Sci. U.S.A., 108, 20036–20041. doi: 10.1073/pnas.1117937108
Keywords: meiosis, recombination, DSB, crossover, hotspot, chromatin, nucleosomes, epigenetics
Citation: Tock AJ and Henderson IR (2018) Hotspots for Initiation of Meiotic Recombination. Front. Genet. 9:521. doi: 10.3389/fgene.2018.00521
Received: 05 July 2018; Accepted: 15 October 2018;
Published: 05 November 2018.
Edited by:
Carina Farah Mugal, Uppsala University, SwedenReviewed by:
Eugenio Mancera, Unidad Irapuato (CINVESTAV), MexicoValérie Borde, Centre National de la Recherche Scientifique (CNRS), France
Michael Lichten, National Institutes of Health (NIH), United States
Copyright © 2018 Tock and Henderson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ian R. Henderson, irh25@cam.ac.uk
 Andrew J. Tock
Andrew J. Tock Ian R. Henderson
Ian R. Henderson