- 1Department of Internal Medicine-Oncology, Fujian Cancer Hospital and Fujian Medical University Cancer Hospital, Fuzhou, China
- 2Department of Gastrointestinal Surgical Oncology, Fujian Cancer Hospital and Fujian Medical University Cancer Hospital, Fuzhou, China
- 3Department of Hepatopancreatobiliary Surgical Oncology, Fujian Cancer Hospital and Fujian Medical University Cancer Hospital, Fuzhou, China
- 4Department of Molecular Pathology, Fujian Cancer Hospital and Fujian Medical University Cancer Hospital, Fuzhou, China
Liquid biopsy, which generally refers to the analysis of biological components such as circulating nuclear acids and circulating tumor cells in body fluids, particularly in peripheral blood, has shown good capacity to overcome several limitations faced by conventional tissue biopsies. Emerging evidence in recent decades has confirmed the promising role of liquid biopsy in the clinical management of various cancers, including colorectal cancer, which is one of the most prevalent cancers and the second leading cause of cancer-related deaths worldwide. Despite the challenges and poor clinical outcomes, patients with metastatic colorectal cancer can expect potential clinical benefits with liquid biopsy. Therefore, in this review, we focus on the clinical prospects of liquid biopsy in metastatic colorectal cancer, specifically with regard to the recently discovered various biomarkers identified on liquid biopsy. These biomarkers have been shown to be potentially useful in multiple aspects of metastatic colorectal cancer, such as auxiliary diagnosis of metastasis, prognosis prediction, and monitoring of therapy response.
Introduction
One of the most prevalent cancers worldwide, colorectal cancer (CRC) is the second leading cause of cancer-related mortality (Bray et al., 2018). Metastatic CRC constitutes an inevitable challenge for clinical management because of the considerably worse survival of metastatic patients compared to patients with non-metastatic CRC (Dekker et al., 2019). Approximately one quarter of CRC patients have metastasis when they are diagnosed and a considerable number of non-metastatic patients postoperatively develop metastasis. Eventually, more than half of all CRC patients will progress to metastatic disease and require corresponding therapy to prolong survival (Labianca et al., 2013; Siegel et al., 2020). Thus, early detection of CRC patients at high risk of developing metastatic CRC is crucial for early intervention to improve patient outcomes and save treatment costs. Current therapies for metastatic CRC patients include surgical resection, chemotherapy, targeted therapy, and immunotherapy (Woo and Jung, 2017; Chakedis and Schmidt, 2018; Nappi et al., 2018; Wrobel and Ahmed, 2019). The individualized selection and sequence of therapies usually differ noticeably based on findings on multidisciplinary evaluation of CRC patients. Therefore, biomarkers, including diagnostic, prognostic, and predictive factors obtained from cancer biopsies are critically important for guiding the therapeutic strategy for patients with metastatic CRC.
Owing to the intratumoral heterogeneity and dynamics of cancer genome modifications of the treatment or the development of cancer (Yates and Campbell, 2012; McGranahan and Swanton, 2015), routine tissue biopsies have a limited capacity to comprehensively obtain real-time information. There is an urgent need for a biopsy method without the disadvantages of tissue biopsy such as patient discomfort, risk of tumor seeding, limited sample accessibility, and procedural complications. The development of precision medicine could potentially eliminate the abovementioned limitations by liquid biopsy, which generally refers to the testing of biological components obtained from body fluids, especially whole blood. Several recent excellent reviews have discussed the application and potential scenarios of liquid biopsy in CRC (Gargalionis and Papavassiliou, 2017; Klein-Scory et al., 2018; Normanno et al., 2018; Tarazona and Cervantes, 2018; Wills et al., 2018; Yamada et al., 2019; Ding et al., 2020). Herein, we focus on the current application and clinical perspectives of liquid biopsy in metastatic CRC, particularly on advances in the discovery of potential roles of liquid biopsy in diagnosing metastasis, prognosis assessment, and therapy response monitoring.
Methods of Liquid Biopsy
The analytes of liquid biopsy mainly refer to circulating tumor DNA (ctDNA), circulating tumor cells (CTC), and circulating non-coding RNAs, which include non-coding RNAs released by cancer cells into the circulation and those traveling inside exosomes (Poulet et al., 2019; Rapado-González et al., 2019). Following a brief introduction of each analyte, their performance consistency with tissue biopsy, especially in metastatic CRC, is reviewed and compared in this section.
The fragmented DNA released by tumor cells into the circulating system constitute CtDNA, which carry the genetic information of both primary-site and metastatic-site tumors, and are mainly detected by PCR and next-generation sequencing (NGS) in liquid biopsy. The most sensitive PCR-based methods (e.g., digital PCR) detect unique hotspots or well-identified mutations, whereas NGS-based methods have greater advantage in broad-range screening of mutations (Franczak et al., 2019). Several studies that evaluated the concordance between the detection of plasma-based ctDNA and tissue biopsy-based genomic DNA in metastatic CRC confirmed a high overall agreement (Grasselli et al., 2017; Bando et al., 2019; Galbiati et al., 2019; Kang et al., 2020; Yu et al., 2020). A prospective retrospective cohort study that enrolled 146 metastatic CRC patients compared the RAS mutational status by the plasma ctDNA-BEAMing (digital PCR) and tissue reference methods, and showed an 89.7% concordance rate (Grasselli et al., 2017). A similar agreement of 86.4% was recently demonstrated in another multicenter prospective study of 280 patients with metastatic CRC (Bando et al., 2019). A higher concordance rate of 92% between plasma ctDNA detected by digital PCR and tissue reference DNA was identified in common KRAS and BRAF mutations among 150 patients with metastatic CRC (Yu et al., 2020). However, another study reported a low concordance of 63.3% for the KRAS gene, which was explained by the different timings of liquid and tissue biopsies of the patients (Galbiati et al., 2019), and highlights the distinct mutational profiles among patients with different cancer stages. Conversely, ctDNA detected by NGS and tumor genomic DNA showed high agreement. Using ultra-deep target sequencing, Kang et al. (2020) investigated mutations in 10 genes (38-kb length) from ctDNA and genomic DNA derived from matched tumor tissues, and found an overall 93% concordance rate.
Primary tumors or metastases shed CTC that circulate in peripheral blood and can be isolated by various methods based on epithelial markers expressed on the cell surface or on the physiological properties of cells (Pantel and Speicher, 2016; Cabel et al., 2017). The detection capacities of CTC and ctDNA for metastatic CRC were, respectively, evaluated in parallel in a cohort of 20 patients; ctDNA could be detected in all patients with metastatic CRC, whereas CTC were detectable in only one third of the patients (Germano et al., 2018). Buim et al. (2015) analyzed KRAS mutations in both CTC and matched primary tumor samples, and observed a concordance rate of 71% between CTC and tissues. Another study showed 50% KRAS mutation-status agreement between CTC and matched primary tumors of patients with metastatic colon cancer (Fabbri et al., 2013).
Studies have indicated that, besides ctDNA and CTC, circulating non-coding RNAs are emerging as important biomarkers for the clinical management of CRC patients (Rapado-González et al., 2019; Baassiri et al., 2020). In body fluids, circulating non-coding RNA mainly exist in either the cell-free state or the exosomal form. Exosomes are nano-sized extracellular vesicles secreted by various cell types and can be isolated using ultracentrifugation, density-based separation, and antibody-based immune-affinity capture (Greening et al., 2015). Furthermore, the intra-exosomal expression levels of non-coding RNAs could be evaluated. In a study of 326 CRC patients, the expression levels of exosomal miR-21 significantly correlated with those of the CRC tissue miR-21 (Tsukamoto et al., 2017). Similar correlations have been reported for the expression levels of miR-122 (Sun et al., 2020), miR-25-3p (Zeng et al., 2018), etc. The strengths and limitations of the abovementioned approaches are shown in Table 1 (Jia et al., 2017; Nordgård et al., 2018; Normanno et al., 2018; Burz and Rosca, 2019; Pantel and Alix-Panabières, 2019).
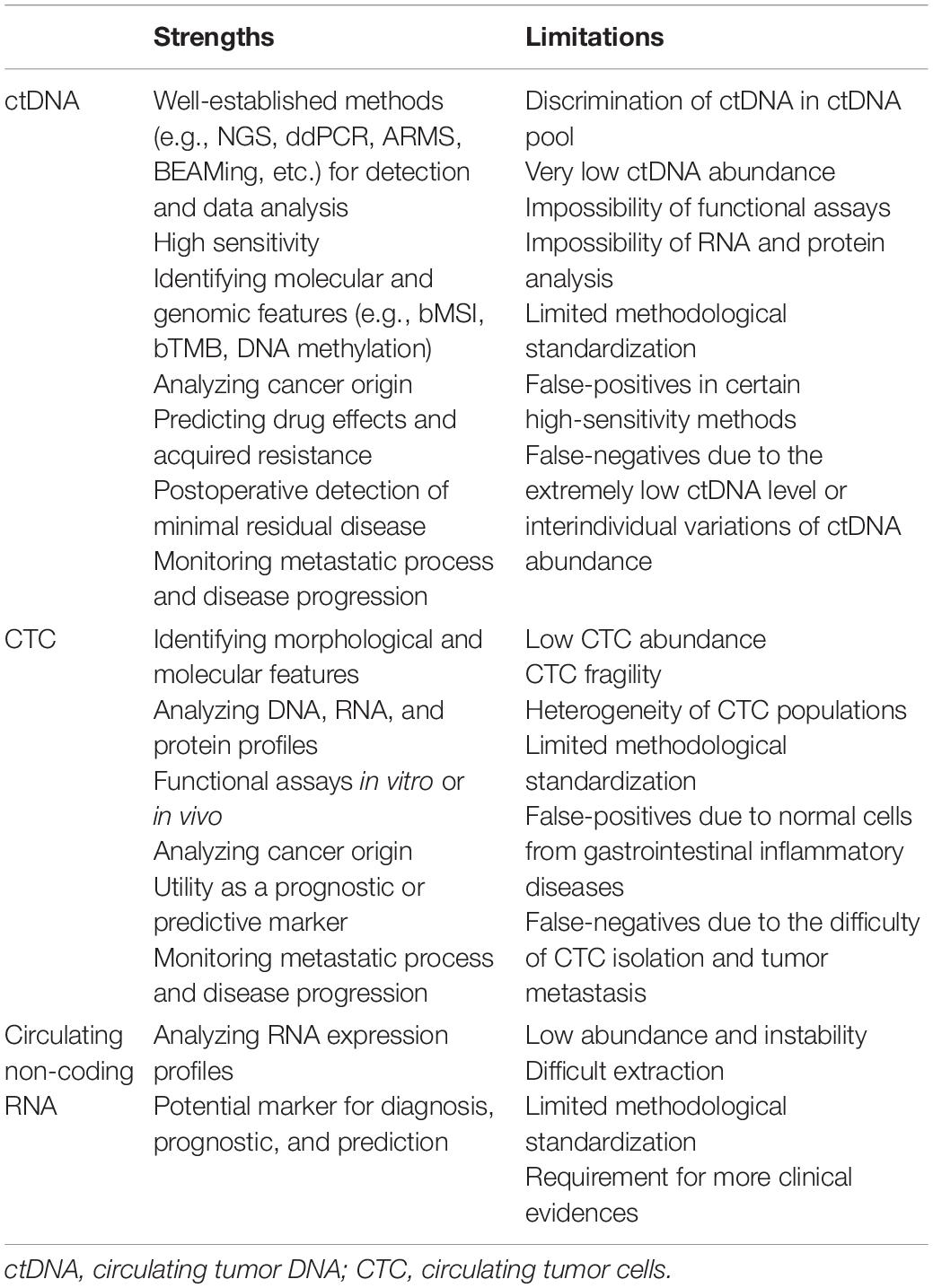
Table 1. Strengths and limitations of applying ctDNA, CTC, and non-coding RNA (Jia et al., 2017; Nordgård et al., 2018; Normanno et al., 2018; Burz and Rosca, 2019; Pantel and Alix-Panabières, 2019).
Roles of Liquid Biopsy in Metastatic Crc
The clear advantages of liquid biopsy, compared with routine tissue-based methods, have generated interest in the application of non-invasive hematological methods to clinical oncology that has steadily increased in the past decades. From the clinical perspective of metastatic CRC, multiple aspects of liquid biopsy, primarily for the identification of diagnostic roles for metastasis, prognostic biomarkers, and monitoring the therapy response, have been explored.
Auxiliary Staging: Metastasis Diagnosis
Apart from the driver mutations shared with the primary tumor, metastatic cancers often carry new mutations, which may play essential roles in the metastatic process (Turajlic and Swanton, 2016; Robinson et al., 2017). The molecular landscape of metastatic tumors, rather than that of exclusive primary tumors, is preferably used to guide potential therapies or clinical trials for patients with metastatic cancer (Steeg, 2016). A review recently summarized the potential and methods of tumor metastasis prediction from genome sequencing data using various tools based on machine learning, protein–network, or biological pathways (Yuan et al., 2019). Liquid biopsy is capable of detecting emerging mutations in metastatic CRC. A study of 22 CRC patients with liver metastases compared NGS and digital PCR detection [sensitivity of 64% (23/36) and 89% (32/36)] of metastasis-related mutations in peripheral blood samples (Furuki et al., 2018). Another study conducted whole-exome sequencing of ctDNA in plasma samples, and used “differential presence of exon (DPE) analysis” to distinguish between metastatic and non-metastatic CRC, and the results imply that DPE characteristics might have a diagnostic value for CRC patients (Olmedillas-López et al., 2018). The association between the ctDNA level and CRC stage has been studied. Yang et al. (2018) analyzed ctDNA levels in 47 CRC patients in early or late cancer stages and found that Stage IV patients had significantly higher ctDNA concentrations than Stage I patients. These studies support the implication that ctDNA characteristics could facilitate the diagnosis of metastatic CRC.
The CTC are well known to play important roles in tumor metastasis (Massagué and Obenauf, 2016; Dasgupta et al., 2017; Micalizzi et al., 2017). Based 2 years of follow-up data and a microfluidic device utilizing antibody-conjugated non-fouling coating to capture and enumerate CTC, a multicohort study that included healthy control, non-metastatic, and metastatic CRC patients explored the correlation between neoplasm progression and CTC, and showed that a high CTC count was significantly associated with tumor progression, metastasis, and future occurrence of distant metastases in non-metastatic CRC patients (Tsai et al., 2016). An earlier study showed that liver metastasis in CRC patients was associated with apoptotic CTC, instead of intact CTC, in the peripheral blood (Allen et al., 2014).
Non-coding RNAs, especially the more studied microRNAs, are involved in tumor cell invasion, migration, and progression in CRC (Cekaite et al., 2016). The expression of circulating non-coding RNA generally reflects the profiles of both primary tumor and metastatic lesions, and non-coding RNA has potential application as a biomarker for the detection of metastatic cancer. Therefore, a study of CRC patients with or without liver metastases showed that the serum miR-29a expression level could, with 75% sensitivity and specificity, help to differentiate between patients with metastatic and non-metastatic CRC (Wang and Gu, 2012). A high serum miR-200c level was significantly associated with metastasis in CRC (Toiyama et al., 2014), and the serum miR-203 level significantly increased in relation to the tumor stage, especially in patients with liver or systemic metastasis (Hur et al., 2017). These results suggest the potential role of these serum microRNAs as metastasis-predictive biomarkers in CRC.
Besides the abovementioned cell-free microRNAs, potential roles of exosomal microRNAs in diagnosing metastasis have been extensively investigated. For example, the high expression of serum exosomal miR-203 correlated with distant metastasis, and xenograft mouse experiments further showed that miR-203-transfected CRC cells had more liver metastasis than controls (Takano et al., 2017). Exosomal miR-25-3p levels were significantly higher in metastatic CRC than in non-metastatic CRC (Zeng et al., 2018). Screening of differential exosomal microRNAs by sequencing and qPCR showed that exosomal miR-320d could distinguish, with an area under the ROC curve (AUC) of 0.633, a diagnosis of metastatic CRC (Tang et al., 2019). The expression of exosomal miR-139-3p was significantly decreased in metastatic CRC, and could facilitate a diagnosis of metastasis (AUC 0.766) (Liu et al., 2020). Furthermore, the level of exosomal miR-122 increased in CRC, especially in patients with liver metastasis, and its expression could help differentiate between patients with and without liver metastasis (AUC 0.81) (Sun et al., 2020).
Interestingly, other non-coding RNAs besides microRNA in liquid biopsy have emerged as potential biomarkers for metastatic CRC (Baassiri et al., 2020). Exosomal long non-coding RNA (lncRNA) CRNDE-h levels were significantly correlated with metastasis in CRC (Liu et al., 2016). Exosomal circular RNA (circRNA) hsa-circ0004771 was associated with distant metastasis (Pan et al., 2019). These findings suggest that circulating non-coding RNAs serve as novel potential diagnostic biomarkers for metastatic CRC.
Prognostic Biomarkers
With regard to the significance of providing prognostic and predictive information for cancer patients, liquid biopsy is promising as an indispensable clinical test for translational oncology. Increasingly, prognostic biomarkers for metastatic CRC patients have emerged with further research.
A study evaluating the prognostic value of ctDNA in 97 metastatic CRC patients showed that patients carrying more ctDNA had significantly decreased overall survival (OS), and the ctDNA fragmentation level was positively associated with shorter OS in the KRAS/BRAF-mutant cohort of patients, but not in the KRAS/BRAF-wild type cohort, indicating the roles of both qualitative and quantitative ctDNA analyses for prognostic assessment in metastatic CRC (El Messaoudi et al., 2016). This finding concurs with the conclusion of a meta-analysis that high pretreatment ctDNA levels correlated with shorter survival in patients with metastatic CRC (Spindler et al., 2017). A study of the prognostic value of pretreatment ctDNA in patients receiving first-line chemotherapy confirmed that increased ctDNA was associated with worse outcome in metastatic CRC patients (Hamfjord et al., 2019).
Multiple studies have confirmed the promising prognostic roles of baseline CTC for patients with metastatic CRC (Groot Koerkamp et al., 2013; Huang et al., 2015). In a prospective multicenter study comprising 430 metastatic CRC patients, Cohen et al. (2008) showed that patients with three or more baseline CTC had shorter median progression-free survival (PFS) (Cohen et al., 2008). Besides baseline CTC counts, the follow-up CTC levels persisted as strong predictors of PFS and OS during treatment for metastatic CRC patients. Importantly, the study further confirmed that the prognostic value of baseline CTC was unaffected by the characteristics of treatments or patients (Cohen et al., 2009). Besides CTC cell counts, the expression level of some genes in CTC may have prognostic effects. Ning et al. (2018) reported that the level of Akt-2 expression in CTC could predict PFS in metastatic CRC patients, Patients with Akt-2 expression in CTC had a significantly shorter PFS compared with those without Akt-2 expression in CTC.
Several circulating microRNAs are associated with survival in CRC patients (Rapado-González et al., 2019). For metastatic CRC, high plasma miR-141 levels are significantly associated with poor survival in metastatic CRC patients (Cheng et al., 2011). Increased plasma miR-200a and plasma miR-122 levels correlated with decreased OS in metastatic CRC (Maierthaler et al., 2017). Additionally, high levels of extracellular vesicle miR-222 were associated with shorter OS in patients with metastatic CRC (de Miguel Pérez et al., 2020).
Therapy Response Monitoring and Guiding
Efficient markers of therapeutic response in patients with metastatic CRC are very important for guiding individualized treatment optimization strategies. The real-time abilities of liquid biopsy to monitor the dynamics of the cancer disease promisingly meet this demand.
In a prospective study involving 53 metastatic CRC patients receiving standard first-line chemotherapy, Tie et al. (2015) assessed ctDNA levels of patients at several timepoints, corresponding to pretreatment, 3 days post-treatment, and before the next treatment cycle. They found that largely decreased ctDNA levels (≥ 10-fold) before the next cycle were significantly associated with the trend of increased PFS in these patients, and concluded that the response to later radiologic treatment could be predicted by the early dynamics of ctDNA level during first-line chemotherapy (Tie et al., 2015). Garlan et al. (2017) found a similar conclusion among consecutive patients receiving first-line or second-line chemotherapy for metastatic CRC. For metastatic CRC patients diagnosed with wild-type KRAS who received a standard FOLFIRI-cetuximab treatment, Toledo et al. (2017) observed that continued wild-type circulating DNA status was associated with prolonged response to anti-EGFR therapy, whereas clinical deterioration could be predicted by the explosion of mutation events. For patients with RAS/RAF mutation and metastatic CRC who received standard first-line treatment, changes in ctDNA levels during the treatment were significantly correlated with low or high risk of disease progression (Thomsen et al., 2018). In patients with metastatic CRC referred for potentially resectable liver metastasis, a recent study showed that poorer postoperative survival was associated with persistently detectable preoperative ctDNA levels (Bidard et al., 2019). In a multicenter study that enrolled 47 patients with metastatic CRC with indications for resection, 93% (26/28) of patients with R0 resection did not have postoperative ctDNA, and the remaining R0 cases with detectable ctDNA had recurrence after 6 months (Benešová et al., 2019). These findings indicate that the early dynamics of ctDNA concentration would be valuable and efficient for monitoring therapeutic efficacy in patients with metastatic CRC.
Besides the ctDNA concentration, the status of ctDNA modifications, such as methylation, are potential biomarkers for monitoring therapy response in cancer patients. A study to identify corresponding methylated biomarkers of metastatic CRC in liquid biopsy to monitor the therapy response (Barault et al., 2018) showed that the dynamics of five genes (EYA4, GRIA4, ITGA4, MAP3K14-AS1, and MSC) methylation panel, without being affected by chemotherapy or targeted therapy, were significantly associated with the treatment response and PFS in patients with metastatic CRC (Barault et al., 2018). Another study including 123 patients with metastatic CRC treated with combination chemotherapy showed that the levels of neuropeptide Y (NPY) promoter methylation in ctDNA after one cycle of chemotherapy significantly correlated with survival (Thomsen et al., 2020). Of note, an additional advantage of detecting ctDNA methylation is a possible benefit of the liquid biopsy-based follow-up of cancer patients without information on mutation status.
Sastre et al. (2012) demonstrated that metastatic CRC patients with low baseline CTC counts had significantly better survival (both median PFS and OS) than those carrying a high CTC count at baseline. A similar survival bias was associated with the CTC counts after three cycles of chemotherapy plus bevacizumab (Sastre et al., 2012). In patients with wild-type KRAS and metastatic CRC who received monoclonal antibody treatment, changes in CTC levels from baseline to the first follow-up timepoint were significantly correlated with PFS (Souza et al., 2016). Likewise, the dynamics of gene expression in CTC and changes in CTC counts are promising predictors of the outcome of patients with metastatic CRC. A study determined the level of CTC markers, including tissue-specific and epithelial-to-mesenchymal transition genes (GAPDH, VIL1, CLU, TIMP1, LOXL3, and ZEB2), from 50 patients with metastatic CRC at baseline and at follow-up after treatment onset. The authors observed that patients with decreased CTC markers during therapy had significantly longer PFS and OS than those with elevated markers (Barbazán et al., 2014). Notably, treatment-resistant patients were identified by detecting the abovementioned CTC markers corresponding to the expression levels of a six-gene panel, although they were not diagnosed by routine imaging techniques (Barbazán et al., 2014).
Evidence indicates that circulating microRNAs representing non-coding RNAs are promising potential biomarkers for clinical evaluation in the treatment of patients with CRC (Rapado-González et al., 2019; Baassiri et al., 2020). A study of metastatic CRC patients with or without tumor response to 5-fluorouracil- and oxaliplatin-based chemotherapy identified three plasma microRNAs (miR-106a, miR-484, and miR-130b) that are highly expressed in chemoresistant patients. The study found that high pretreatment plasma levels of these miRNAs were significantly associated with shorter PFS (Kjersem et al., 2014). Another study concluded that levels of exosomal miR-21-5p, miR-1246, miR-1229-5p, and miR-96-5p were elevated in patients with chemoresistant advanced CRC. More importantly, the panel of exosomal these four microRNAs could predict (AUC 0.804) the chemotherapy resistance of individuals with advanced CRC (Jin et al., 2019). In patients with metastatic CRC treated with bevacizumab, basal plasma levels of miR-20b-5p, miR-29b-3p, and miR-155-5p correlated with PFS and OS, respectively. This suggests the potential outcome predictive roles of circulating basal miRNA levels in bevacizumab-treated metastatic CRC (Ulivi et al., 2018).
Conclusion
Liquid biopsy, as a non-invasive and developing tool, could solve many limitations faced with conventional biopsies (Fernández-Lázaro et al., 2020). Studies have confirmed the promising roles of liquid biopsy in the clinical management of metastatic CRC. In the past decades, various biomarkers detected by liquid biopsy in metastatic CRC were found to be potentially useful in multiple areas, including auxiliary diagnosis of metastasis, prognosis prediction, and monitoring of therapy response. However, the practical clinical applications of these biomarkers in metastatic CRC, as well as other cancers, are very limited. One of the reasons for this is that most of the studies of the potential application of these biomarkers had small cohorts. Therefore, large, multicenter, and prospective clinical validation trials are needed for the implementation of these liquid biopsy-based biomarkers in clinical scenarios. Moreover, the standardization of the liquid biopsy process, for sample collection and subsequent analysis, is necessary in the clinical setting.
Author Contributions
WG designed the study. WG, YC, JY, and CZ collected the data and wrote the manuscript. SH, HZ, and YS reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Allen, J. E., Saroya, B. S., Kunkel, M., Dicker, D. T., Das, A., Peters, K. L., et al. (2014). Apoptotic circulating tumor cells (CTCs) in the peripheral blood of metastatic colorectal cancer patients are associated with liver metastasis but not CTCs. Oncotarget 5, 1753–1760. doi: 10.18632/oncotarget.1524
Baassiri, A., Nassar, F., Mukherji, D., Shamseddine, A., Nasr, R., and Temraz, S. (2020). Exosomal non coding RNA in liquid biopsies as a promising biomarker for colorectal cancer. Int. J. Mol. Cell. Med. 21:1398. doi: 10.3390/ijms21041398
Bando, H., Kagawa, Y., Kato, T., Akagi, K., Denda, T., Nishina, T., et al. (2019). A multicentre, prospective study of plasma circulating tumour DNA test for detecting RAS mutation in patients with metastatic colorectal cancer. Br. J. Cancer 120, 982–986. doi: 10.1038/s41416-019-0457-y
Barault, L., Amatu, A., Siravegna, G., Ponzetti, A., Moran, S., Cassingena, A., et al. (2018). Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut 67, 1995–2005. doi: 10.1136/gutjnl-2016-313372
Barbazán, J., Muinelo-Romay, L., Vieito, M., Candamio, S., Cano, A., Gómez-Tato, A., et al. (2014). A multimarker panel for circulating tumor cells detection predicts patient outcome and therapy response in metastatic colorectal cancer. Int. J. Cancer 135, 2633–2643. doi: 10.1002/ijc.28910
Benešová, L., Hálková, T., Ptáèková, R., Semyakina, A., Pudil, J., Ryska, M., et al. (2019). Significance of postoperative follow-up of patients with metastatic colorectal cancer using circulating tumor DNA. World J. Gastroenterol. 25, 6939–6948. doi: 10.3748/wjg.v25.i48.6939
Bidard, F. C., Kiavue, N., Ychou, M., Cabel, L., Stern, M. H., Madic, J., et al. (2019). Circulating tumor cells and circulating tumor dna detection in potentially resectable metastatic colorectal cancer: a prospective ancillary study to the unicancer Prodige-14 trial. Cells 8:516. doi: 10.3390/cells8060516
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Buim, M. E., Fanelli, M. F., Souza, V. S., Romero, J., Abdallah, E. A., Mello, C. A., et al. (2015). Detection of KRAS mutations in circulating tumor cells from patients with metastatic colorectal cancer. Cancer Biol. Ther. 16, 1289–1295. doi: 10.1080/15384047.2015.1070991
Burz, C., and Rosca, A. (2019). Liquid biopsy challenge and hope in colorectal cancer. Expert Rev. Mol. Diagn. 19, 341–348. doi: 10.1080/14737159.2019.1597708
Cabel, L., Proudhon, C., Gortais, H., Loirat, D., Coussy, F., Pierga, J. Y., et al. (2017). Circulating tumor cells: clinical validity and utility. Int. J. Clin. Oncol. 22, 421–430.
Cekaite, L., Eide, P. W., Lind, G. E., Skotheim, R. I., and Lothe, R. A. (2016). MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget 7, 6476–6505. doi: 10.18632/oncotarget.6390
Chakedis, J., and Schmidt, C. R. (2018). Surgical treatment of metastatic colorectal cancer. Surg. Oncol. Clin. N. Am. 27, 377–399.
Cheng, H., Zhang, L., Cogdell, D. E., Zheng, H., Schetter, A. J., Nykter, M., et al. (2011). Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One 6:e17745. doi: 10.1371/journal.pone.0017745
Cohen, S. J., Punt, C. J., Iannotti, N., Saidman, B. H., Sabbath, K. D., Gabrail, N. Y., et al. (2008). Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 26, 3213–3221. doi: 10.1200/jco.2007.15.8923
Cohen, S. J., Punt, C. J., Iannotti, N., Saidman, B. H., Sabbath, K. D., Gabrail, N. Y., et al. (2009). Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 20, 1223–1229. doi: 10.1093/annonc/mdn786
Dasgupta, A., Lim, A. R., and Ghajar, C. M. (2017). Circulating and disseminated tumor cells: harbingers or initiators of metastasis? Mol. Oncol. 11, 40–61. doi: 10.1002/1878-0261.12022
de Miguel Pérez, D., Rodriguez Martínez, A., Ortigosa Palomo, A., Delgado Ureña, M., Garcia Puche, J. L., Robles Remacho, A., et al. (2020). Extracellular vesicle-miRNAs as liquid biopsy biomarkers for disease identification and prognosis in metastatic colorectal cancer patients. Sci. Rep. 10:3974.
Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M., and Wallace, M. B. (2019). Colorectal cancer. Lancet 394, 1467–1480.
Ding, Y., Li, W., Wang, K., Xu, C., Hao, M., and Ding, L. (2020). Perspectives of the application of liquid biopsy in colorectal cancer. Biomed. Res. Int. 9:6843180.
El Messaoudi, S., Mouliere, F., Gillet, B., Nouaille, M., Fiess, C., Crapez, E., et al. (2016). Circulating DNA as a strong multimarker prognostic tool for metastatic colorectal cancer patient management care. Clin. Cancer Res. 22, 3067–3077. doi: 10.1158/1078-0432.ccr-15-0297
Fabbri, F., Carloni, S., Zoli, W., Ulivi, P., Gallerani, G., Fici, P., et al. (2013). Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Lett. 335, 225–231. doi: 10.1016/j.canlet.2013.02.015
Fernández-Lázaro, D., Hernández, J. L. G., García, A. C., Castillo, A. C. D., Hueso, M. V., and Cruz-Hernández, J. J. (2020). Clinical perspective and translational oncology of liquid biopsy. Diagnostics 10:443. doi: 10.3390/diagnostics10070443
Franczak, C., Filhine-Tresarrieu, P., Gilson, P., Merlin, J. L., Au, L., and Harlé, A. (2019). Technical considerations for circulating tumor DNA detection in oncology. Expert Rev. Mol. Diagn. 19, 121–135. doi: 10.1080/14737159.2019.1568873
Furuki, H., Yamada, T., Takahashi, G., Iwai, T., Koizumi, M., Shinji, S., et al. (2018). Evaluation of liquid biopsies for detection of emerging mutated genes in metastatic colorectal cancer. Eur. J. Surg. Oncol. 44, 975–982.
Galbiati, S., Damin, F., Burgio, V., Brisci, A., Soriani, N., Belcastro, B., et al. (2019). Evaluation of three advanced methodologies, COLD-PCR, microarray and ddPCR, for identifying the mutational status by liquid biopsies in metastatic colorectal cancer patients. Clin. Chim. Acta 489, 136–143. doi: 10.1016/j.cca.2018.12.004
Gargalionis, A. N., and Papavassiliou, A. G. (2017). Liquid biopsies in colorectal cancer: monitoring genetic heterogeneity. Trends Cancer 3, 166–168. doi: 10.1016/j.trecan.2017.01.003
Garlan, F., Sefrioui, D., Siauve, N., Didelot, A., Michel, P., Perkins, G., et al. (2017). Early evaluation of circulating tumor DNA as marker of therapeutic efficacy in metastatic colorectal cancer patients (PLACOL Study). Clin. Cancer Res. 23, 5416–5425. doi: 10.1158/1078-0432.ccr-16-3155
Germano, G., Mauri, G., Siravegna, G., Dive, C., Pierce, J., Bardelli, A., et al. (2018). Parallel evaluation of circulating tumor DNA and circulating tumor cells in metastatic colorectal cancer. Clin. Colorectal Cancer 17, 80–83. doi: 10.1016/j.clcc.2017.10.017
Grasselli, J., Elez, E., Matito, J., Santos, C., Macarulla, T., Vidal, J., et al. (2017). Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann. Oncol. 28, 1294–1301. doi: 10.1093/annonc/mdx112
Greening, D. W., Xu, R., Ji, H., Tauro, B. J., and Simpson, R. J. (2015). A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 1295, 179–209. doi: 10.1007/978-1-4939-2550-6_15
Groot Koerkamp, B., Rahbari, N. N., Büchler, M. W., Koch, M., and Weitz, J. (2013). Circulating tumor cells and prognosis of patients with resectable colorectal liver metastases or widespread metastatic colorectal cancer: a meta-analysis. Ann. Surg. Oncol. 20, 2156–2165. doi: 10.1245/s10434-013-2907-8
Hamfjord, J., Guren, T. K., Dajani, O., Johansen, J. S., Glimelius, B., Sorbye, H., et al. (2019). Total circulating cell-free DNA as a prognostic biomarker in metastatic colorectal cancer before first-line oxaliplatin-based chemotherapy. Ann. Oncol. 30, 1088–1095. doi: 10.1093/annonc/mdz139
Huang, X., Gao, P., Song, Y., Sun, J., Chen, X., Zhao, J., et al. (2015). Meta-analysis of the prognostic value of circulating tumor cells detected with the CellSearch System in colorectal cancer. BMC Cancer 15:202. doi: 10.1186/s12885-015-1218-9
Hur, K., Toiyama, Y., Okugawa, Y., Ide, S., Imaoka, H., Boland, C. R., et al. (2017). Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 66, 654–665. doi: 10.1136/gutjnl-2014-308737
Jia, S., Zhang, R., Li, Z., and Li, J. (2017). Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget 8, 55632–55645. doi: 10.18632/oncotarget.17184
Jin, G., Liu, Y., Zhang, J., Bian, Z., Yao, S., Fei, B., et al. (2019). A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother.Pharmacol. 84, 315–325. doi: 10.1007/s00280-019-03867-6
Kang, J. K., Heo, S., Kim, H. P., Song, H. S., Yun, H., Han, S. W., et al. (2020). Liquid biopsy-based tumor profiling for metastatic colorectal cancer patients with ultra-deep targeted sequencing. PLoS One 15:e0232754. doi: 10.1371/journal.pone.0232754 eCollection 2020
Kjersem, J. B., Ikdahl, T., Lingjaerde, O. C., Guren, T., Tveit, K. M., and Kure, E. H. (2014). Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol. Oncol. 8, 59–67. doi: 10.1016/j.molonc.2013.09.001
Klein-Scory, S., Maslova, M., Pohl, M., Eilert-Micus, C., Schroers, R., Schmiegel, W., et al. (2018). Significance of liquid biopsy for monitoring and therapy decision of colorectal cancer. Transl. Oncol. 11, 213–220. doi: 10.1016/j.tranon.2017.12.010
Labianca, R., Nordlinger, B., Beretta, G. D., Mosconi, S., Cervantes, A., Arnold, D., et al. (2013). Early colon cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 24(Suppl. 6), vi64–72. doi: 10.1093/annonc/mdt354
Liu, T., Zhang, X., Gao, S., Jing, F., Yang, Y., Du, L., et al. (2016). Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 7, 85551–85563. doi: 10.18632/oncotarget.13465
Liu, W., Yang, D., Chen, L., Liu, Q., Wang, W., Yang, Z., et al. (2020). Plasma Exosomal miRNA-139-3p is a novel biomarker of colorectal Cancer. J. Cancer 11, 4899–4906. doi: 10.7150/jca.45548
Maierthaler, M., Benner, A., Hoffmeister, M., Surowy, H., Jansen, L., Knebel, P., et al. (2017). Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int. J. Cancer 140, 176–187. doi: 10.1002/ijc.30433
Massagué, J., and Obenauf, A. C. (2016). Metastatic colonization by circulating tumour cells. Nature 529, 298–306. doi: 10.1038/nature17038
McGranahan, N., and Swanton, C. (2015). Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26. doi: 10.1016/j.ccell.2014.12.001
Micalizzi, D. S., Maheswaran, S., and Haber, D. A. (2017). A conduit to metastasis: circulating tumor cell biology. Genes Dev. 31, 1827–1840. doi: 10.1101/gad.305805.117
Nappi, A., Berretta, M., Romano, C., Tafuto, S., Cassata, A., Casaretti, R., et al. (2018). Metastatic colorectal cancer: role of target therapies and future perspectives. Curr. Cancer Drug Targets 18, 421–429. doi: 10.2174/1568009617666170209095143
Ning, Y., Zhang, W., Hanna, D. L., Yang, D., Okazaki, S., Berger, M. D., et al. (2018). Clinical relevance of EMT and stem-like gene expression in circulating tumor cells of metastatic colorectal cancer patients. Pharmacogenomics J. 18, 29–34.
Nordgård, O., Tjensvoll, K., Gilje, B., and Søreide, K. (2018). Circulating tumour cells and DNA as liquid biopsies in gastrointestinal cancer. Br. J. Surg. 105, e110–e120.
Normanno, N., Cervantes, A., Ciardiello, F., De Luca, A., and Pinto, C. (2018). The liquid biopsy in the management of colorectal cancer patients: current applications and future scenarios. Cancer Treat. Rev. 70, 1–8. doi: 10.1016/j.ctrv.2018.07.007
Olmedillas-López, S., García-Olmo, D. C., García-Arranz, M., Peiró-Pastor, R., Aguado, B., and García-Olmo, D. (2018). Liquid biopsy by NGS: differential presence of exons (DPE) in cell-free DNA reveals different patterns in metastatic and nonmetastatic colorectal cancer. Cancer Med. 7, 1706–1716. doi: 10.1002/cam4.1399
Pan, B., Qin, J., Liu, X., He, B., Wang, X., Pan, Y., et al. (2019). Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front. Genet. 10:1096. doi: 10.3389/fgene.2019.01096
Pantel, K., and Alix-Panabières, C. (2019). Liquid biopsy and minimal residual disease – latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–424. doi: 10.1038/s41571-019-0187-3
Pantel, K., and Speicher, M. R. (2016). The biology of circulating tumor cells. Oncogene 35, 1216–1224.
Poulet, G., Massias, J., and Taly, V. (2019). Liquid biopsy: general concepts. Acta cytologica 63, 449–455. doi: 10.1159/000499337
Rapado-González, Ó, Álvarez-Castro, A., López-López, R., Iglesias-Canle, J., Suárez-Cunqueiro, M., and Muinelo-Romay, L. (2019). Circulating microRNAs as promising biomarkers in colorectal cancer. Cancers 11:898. doi: 10.3390/cancers11070898
Robinson, D. R., Wu, Y. M., Lonigro, R. J., Vats, P., Cobain, E., Everett, J., et al. (2017). Integrative clinical genomics of metastatic cancer. Nature 548, 297–303.
Sastre, J., Maestro, M. L., Gómez-España, A., Rivera, F., Valladares, M., Massuti, B., et al. (2012). Circulating tumor cell count is a prognostic factor in metastatic colorectal cancer patients receiving first-line chemotherapy plus bevacizumab: a Spanish cooperative group for the treatment of digestive tumors study. Oncologist 17, 947–955. doi: 10.1634/theoncologist.2012-0048
Siegel, R. L., Miller, K. D., Fedewa, S. A., Butterly, L. F., Anderson, J. C., Cercek, A., et al. (2020). Colorectal cancer statistics, 2020. CA Cancer J. Clin. 70, 145–164. doi: 10.1016/j.clcc.2020.07.001
Souza, E. S. V., Chinen, L. T., Abdallah, E. A., Damascena, A., Paludo, J., Chojniak, R., et al. (2016). Early detection of poor outcome in patients with metastatic colorectal cancer: tumor kinetics evaluated by circulating tumor cells. Onco Targets Ther. 9, 7503–7513. doi: 10.2147/ott.s115268
Spindler, K. G., Boysen, A. K., Johansen, J. S., Tabernero, J., Jensen, B. V., Hansen, T. F., et al. (2017). Cell-free DNA in metastatic colorectal cancer: a systematic review and meta-analysis. Oncologist 22, 1049–1055. doi: 10.1634/theoncologist.2016-0178
Sun, L., Liu, X., Pan, B., Hu, X., Zhu, Y., Su, Y., et al. (2020). Serum exosomal miR-122 as a potential diagnostic and prognostic biomarker of colorectal cancer with liver metastasis. J. Cancer 11, 630–637. doi: 10.7150/jca.33022
Takano, Y., Masuda, T., Iinuma, H., Yamaguchi, R., Sato, K., Tobo, T., et al. (2017). Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget 8, 78598–78613. doi: 10.18632/oncotarget.20009
Tang, Y., Zhao, Y., Song, X., Song, X., Niu, L., and Xie, L. (2019). Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J. Clin. Lab. Anal. 33:e23004.
Tarazona, N., and Cervantes, A. (2018). Liquid biopsy: another tool towards tailored therapy in colorectal cancer. Ann. Oncol. 29, 7–8. doi: 10.1093/annonc/mdx641
Thomsen, C. B., Hansen, T. F., Andersen, R. F., Lindebjerg, J., Jensen, L. H., and Jakobsen, A. (2018). Monitoring the effect of first line treatment in RAS/RAF mutated metastatic colorectal cancer by serial analysis of tumor specific DNA in plasma. J. Exp. Clin. Cancer Res. 37:55.
Thomsen, C. B., Hansen, T. F., Andersen, R. F., Lindebjerg, J., Jensen, L. H., and Jakobsen, A. (2020). Early identification of treatment benefit by methylated circulating tumor DNA in metastatic colorectal cancer. Ther. Adv. Med. Oncol. 12:1758835920918472.
Tie, J., Kinde, I., Wang, Y., Wong, H. L., Roebert, J., Christie, M., et al. (2015). Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 26, 1715–1722. doi: 10.1093/annonc/mdv177
Toiyama, Y., Hur, K., Tanaka, K., Inoue, Y., Kusunoki, M., Boland, C. R., et al. (2014). Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann. Surg. 259, 735–743. doi: 10.1097/sla.0b013e3182a6909d
Toledo, R. A., Cubillo, A., Vega, E., Garralda, E., Alvarez, R., Sarno, F., et al. (2017). Clinical validation of prospective liquid biopsy monitoring in patients with wild-type RAS metastatic colorectal cancer treated with FOLFIRI-cetuximab. Oncotarget 8, 35289–35300. doi: 10.18632/oncotarget.13311
Tsai, W. S., Chen, J. S., Shao, H. J., Wu, J. C., Lai, J. M., Lu, S. H., et al. (2016). Circulating tumor cell count correlates with colorectal neoplasm progression and is a prognostic marker for distant metastasis in non-metastatic patients. Sci. Rep. 6:24517.
Tsukamoto, M., Iinuma, H., Yagi, T., Matsuda, K., and Hashiguchi, Y. (2017). Circulating exosomal MicroRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology 92, 360–370. doi: 10.1159/000463387
Turajlic, S., and Swanton, C. (2016). Metastasis as an evolutionary process. Science 352, 169–175. doi: 10.1126/science.aaf2784
Ulivi, P., Canale, M., Passardi, A., Marisi, G., Valgiusti, M., Frassineti, G. L., et al. (2018). Circulating plasma levels of miR-20b, miR-29b and miR-155 as predictors of bevacizumab efficacy in patients with metastatic colorectal cancer. Int. J. Mol. Sci. 19:307. doi: 10.3390/ijms19010307
Wang, L. G., and Gu, J. (2012). Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 36, e61–e67.
Wills, B., Gorse, E., and Lee, V. (2018). Role of liquid biopsies in colorectal cancer. Curr. Probl. Cancer 42, 593–600. doi: 10.1016/j.currproblcancer.2018.08.004
Woo, I. S., and Jung, Y. H. (2017). Metronomic chemotherapy in metastatic colorectal cancer. Cancer Lett. 400, 319–324. doi: 10.1016/j.canlet.2017.02.034
Wrobel, P., and Ahmed, S. (2019). Current status of immunotherapy in metastatic colorectal cancer. Int. J. Colorectal Dis. 34, 13–25. doi: 10.1007/s00384-018-3202-8
Yamada, T., Matsuda, A., Koizumi, M., Shinji, S., Takahashi, G., Iwai, T., et al. (2019). Liquid biopsy for the management of patients with colorectal cancer. Digestion 99, 39–45.
Yang, Y. C., Wang, D., Jin, L., Yao, H. W., Zhang, J. H., Wang, J., et al. (2018). Circulating tumor DNA detectable in early- and late-stage colorectal cancer patients. Biosci. Rep. 38:BSR20180322.
Yates, L. R., and Campbell, P. J. (2012). Evolution of the cancer genome. Nat. Rev. Genet. 13, 795–806.
Yu, H., Han, L., Yuan, J., and Sun, Y. (2020). Circulating tumor cell free DNA from plasma and urine in the clinical management of colorectal cancer. Cancer Biomark. 27, 29–37. doi: 10.3233/cbm-182344
Yuan, L., Guo, F., Wang, L., and Zou, Q. (2019). Prediction of tumor metastasis from sequencing data in the era of genome sequencing. Brief. Funct. Genomics 18, 412–418. doi: 10.1093/bfgp/elz010
Keywords: metastatic colorectal cancer, liquid biopsy, biomarker, prediction, therapy response
Citation: Gao W, Chen Y, Yang J, Zhuo C, Huang S, Zhang H and Shi Y (2021) Clinical Perspectives on Liquid Biopsy in Metastatic Colorectal Cancer. Front. Genet. 12:634642. doi: 10.3389/fgene.2021.634642
Received: 28 November 2020; Accepted: 04 January 2021;
Published: 28 January 2021.
Edited by:
Tao Huang, Shanghai Institute of Nutrition and Health (CAS), ChinaReviewed by:
Lihong Peng, Hunan University of Technology, ChinaJujuan Zhuang, Dalian Maritime University, China
Copyright © 2021 Gao, Chen, Yang, Zhuo, Huang, Zhang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Gao, 13960986882@163.com
 Wei Gao
Wei Gao Yigui Chen1
Yigui Chen1