- Department of Immunology, Center for Innate Immunity and Immune Diseases, University of Washington, Seattle, WA, United States
Type III interferons, also known as interferon lambdas (IFNλs), are the most recent addition to the IFN family following their discovery in 2003. Initially, IFNλ was demonstrated to induce expression of interferon-stimulated genes and exert antiviral properties in a similar manner to type I IFNs. However, while IFNλ has been described to have largely overlapping expression and function with type I IFNs, it has become increasingly clear that type III IFNs also have distinct functions from type I IFNs. In contrast to type I IFNs, whose receptor is ubiquitously expressed, type III IFNs signal and function largely at barrier epithelial surfaces, such as the respiratory and gastrointestinal tracts, as well as the blood–brain barrier. In further support of unique functions for type III IFNs, single nucleotide polymorphisms in IFNL genes in humans are strongly associated with outcomes to viral infection. These biological linkages have also been more directly supported by studies in mice highlighting roles of IFNλ in promoting antiviral immune responses. In this review, we discuss the current understanding of type III IFNs, and how their functions are similar to, and different from, type I IFN in various immune cell subtypes and viral infections.
Evolution of Type III IFN Genes
Type I IFN is produced and secreted rapidly following viral infection (1, 2). It subsequently signals to surrounding cells to initiate an antiviral state as a critical host defense mechanism. In humans, there are 13 subtypes of IFNα as well as IFNβ, IFNε, IFNκ, and IFNω [reviewed in Ref. (3)]. Type I IFNs are intronless genes clustered on chromosome 9 in humans and chromosome 4 in mice. In mammals, birds, reptiles, and amphibians, type I IFN genes lack introns, which suggests their origin may have been from retrotransposed genetic elements [reviewed in Ref. (4)]. However, type I IFNs in fish harbor introns and are thought to have arisen through a common ancestor of IL-10 family [reviewed in Ref. (5)]. Amphibians have been recently described to have both intron-containing and intron-less type I IFN genes (6). The current understanding of interferon evolution has not distinguished whether an independent or retrotransposition event led to the generation of intron-less type I IFN genes that may have been the ancestor of the intron-less type I IFN locus in reptiles, birds, and fish.
IFN lambda family members were initially named as interleukin-28 (IL-28) and IL-29 and classified into the IL-10 family genes as they signal through the common IL-10 receptor subunit 2 (IL-10R2) (7, 8). Humans have four IFNL genes, IFNL1 (IL29), IFNL2 (IL28A), IFNL3 (IL28B), and IFNL4. IFNL genes are present in tetrapods, but in contrast to the evolutionary diversity seen in type I IFNs, throughout vertebrates the type III IFN locus comprised of two to four family members, each containing introns (9). While IFN lambdas are most functionally similar to type I IFNs, they are structurally similar to members of the IL-10 family. Type III IFNs have a phase 0 intron–exon structure and utilize a component of IL-10R2 as a part of their receptor heterodimer complex for signaling (10). Sequence identities of type III IFNs when compared with type I IFNs (15–19% aa) or IL-10 (11–13% aa) are low (8). Among type III genes, IFNL1 and IFNL2 share 81% amino acid identity, whereas IFNL2 and IFNL3 share 96% amino acid identities. IFNL4 shares only ~28% amino acid identity with other IFNL genes, leading to speculation IFNL4 may have been introduced via a separate duplication event. While the evolutionary history of type III IFNs is still incomplete, a number of groups are working to understand the evolutionary constraints on type III IFNs [reviewed in Ref. (4, 11, 12)]. Utilizing an evolutionary genetics approach, Manry et al. demonstrated that type I and type III IFNs, and even individual genes within each of these types, have been subjected to distinct evolutionary pressures (11). This work suggests both redundant and specific, unique roles for these IFN families in pathogen defense.
In contrast to humans, in mice only Ifnl2 and Ifnl3 are functional; Ifnl1 and Ifnl4 are pseudogenes (13). Despite differences in human and murine Ifnl gene composition, murine studies have provided critical insights into the antiviral and immune modulatory functions that have relevant correlates to human infection. For example, in a murine asthma model, interferon lambda (IFNλ) treatment was demonstrated to lead to a Th1-biased immune response (14). In humans, IFNλ leads to enhanced Th1 responses during influenza virus vaccination (15). In addition, respiratory viral pathogens have evolved mechanisms to suppress IFNλ function or downstream signaling, highlighting the critical importance of IFNλ to respiratory immunity in particular, but also the contribution of IFNλ to infection at mucosal barriers in general (16, 17).
Expression IFN Lambda Genes During Viral Infection
IFNs are expressed following detection of pathogen-associated molecular patterns (PAMPs) by pattern-recognition receptors (PRRs) [reviewed in Ref. (18–20)]. Sensing of PAMPs by the RIG-I-like receptors results in the recruitment of mitochondrial antiviral signaling protein (MAVS) to mitochondrial associated membranes or peroxisomes, leading to activation of the transcription factors NF-κB and interferon regulatory factors (IRFs), which induce expression of both type I IFN and IFNλ (21). Multiple toll-like receptors induce expression of type I and III IFNs (22, 23). While the signals and pathways that induce type I and type III IFNs largely overlap, one notable exception does exist in the DNA sensing pathway. In HEK293 and THP-1 cells, binding of DNA to the cytosolic sensor Ku70 induces production of IFλ1 and IFNλ2/3 but not type I IFN (24). Following transfection of DNA or herpes simplex virus-2 infection, DNA binding to Ku70 leads to recruitment of STING and subsequent activation of IRF3 in addition to IRF1 and IRF7 (24, 25). Whether this novel, IFNλ-specific IFN induction exists in other cell types following DNA sensing is an interesting possibility that has not yet been investigated.
Although type I and III IFNs are all induced following infection, the transcription of these genes is temporally regulated. Type I IFNs are induced and resolved rapidly, followed by a delayed but sustained induction of IFNL genes (19, 26, 27). The mechanisms responsible for a distinct temporal induction pattern of type I and type III IFNs is currently unknown, but this could be due to utilization of different signaling molecules or transcription factors. The IFNL1 and IFNL3 promoters harbor binding sites for IRF1, IRF3, IRF7, and NF-κB (28). However, in contrast to type I IFNs, studies have suggested that transcription of IFNL is primarily dependent on NF-κB, and activation of both IRF and NF-κB signals is required for a robust induction of IFNL (29). The differential requirement for IRFs and NF-κB in the induction type I and type III IFNs following PAMP engagement by the PRRs could potentially contribute to the temporal difference in their transcriptional regulation of type III IFNs compared with type I IFNs.
Both type I and type III IFNs are produced following rotavirus infection in an adult murine model, but intestinal epithelial cells (IECs) respond preferentially to type III IFN (27, 30), suggesting a predominant role for IFNλ in antiviral defense in the intestine. In addition, type III IFNs are produced more abundantly at mucosal sites by epithelial and myeloid cells in response to viral infection (31). The mechanism for this preferential induction of type III IFN by IECs remains to be fully elucidated, but it might be due in part to the preferential induction of IFNλ upon MAVS localization to peroxisomes, which are highly abundant in epithelial cells, following PAMP sensing (21). Another possible mechanism is that undefined tissue-specific factors present at the epithelial barrier surfaces may promote IFNλ over type I IFN, similar to the IFNλ response in hepatocytes during hepatitis B and hepatitis C virus (HCV) infection (32). Further, IFNλ can be induced by type I IFN similar to an interferon-stimulated gene (ISG) in a feed-forward fashion (33). This type I IFN enhancement of IFNL is at least partially due to the ability of type I IFN to increase TLR expression; however, the functional consequences of this co-regulation remain to be tested.
Overall, a lack of IFNλ-specific mouse models and antibody detection reagents for ligands and receptors has slowed progress in determining the contribution of IFNλ to immunity. While whole body knockout mice lacking IFNλR exist, dissection of the role IFNλ signaling in various tissues and cell types in vivo will be advanced by studies in mice utilizing a recently reported floxed IFNλR model (34). In addition, an IFNλ2 cytokine reporter mouse has recently been developed (35). These new models will likely lead to a rapid advancement in understanding the unique functions of IFNλ in vivo.
IFN Lambda Receptor Expression and Signaling
The general induction and signaling cascades of type I and type III IFNs are summarized in Figure 1. Type I and III IFNs each signal through distinct receptor heterodimer complexes [reviewed in Ref. (3, 17, 19, 36)]. Type I IFN binds to a receptor complex comprised of IFNAR1 and IFNAR2, which is broadly expressed on most cells [reviewed in Ref. (1, 2)]. IFNλ signals through a heterodimeric receptor comprised of IFNλR1 and IL-10R2 (7, 8); IL-10R2 is a receptor subunit that is broadly expressed and shared for signaling by members of the IL-10 cytokine family [reviewed in Ref. (37)]. By contrast, the expression of IFNλR1 is much more restricted to epithelial cells, subsets of myeloid cells, and neuronal cells. This limited expression likely explains the importance of IFNλ at mucosal sites and the blood/brain barrier [reviewed in Ref. (17, 38)]. Engagement of all IFNs with their receptors initiates downstream signaling events, namely, activation of the JAK–STAT signaling cascade. JAK1, TYK2, and potentially JAK2 are phosphorylated and activated, leading to subsequent phosphorylation and activation of STAT1 and STAT2, which then associate with IRF9. Together, the complex of STAT1, STAT2, and IRF9 is referred to as the interferon-stimulated gene factor 3 (ISGF3) transcriptional complex. Activated ISGF3 translocates to the nucleus and binds to the interferon-sensitive response element, initiating the transcription of a wide array of ISGs. SOCS1 can provide negative regulation of this JAK–STAT signaling pathway downstream of IFN in vitro and in vivo (39–41).
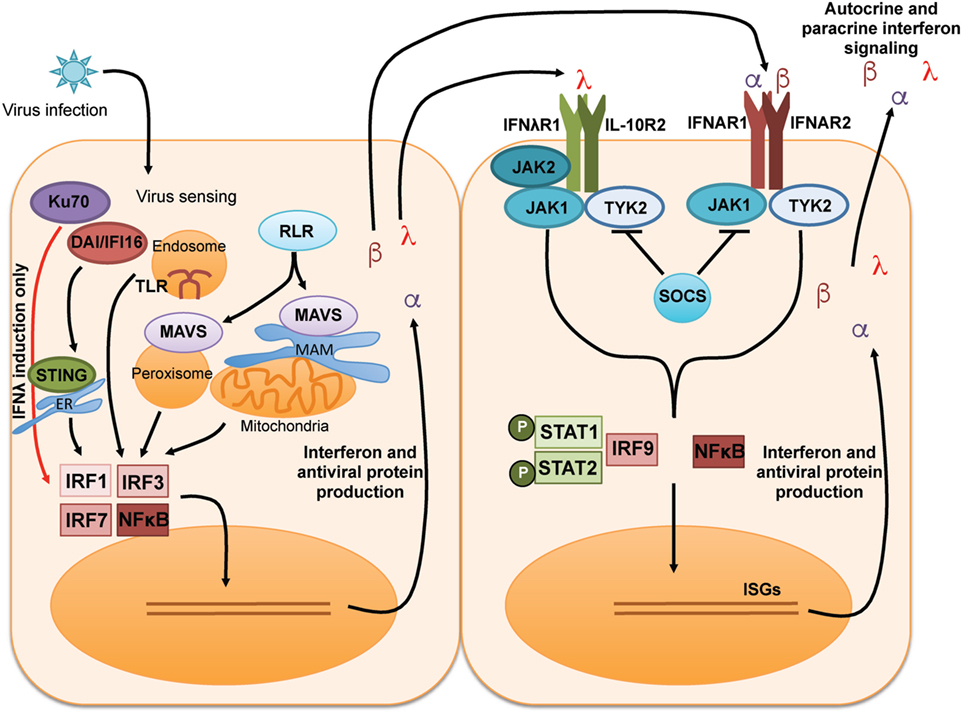
Figure 1. General induction and receptor signaling pathways of type I and type III IFNs. Recognition of virus by multiple pattern recognition receptor pathways leads to the activation of the transcription factors IRF1, IRF3, IRF7, and NF-κB to induce transcription, translation, and secretion of type I IFN (IFNα and IFNβ) and type III IFN [interferon lambda (IFNλ)]. Type I and type III IFNs signal to surrounding cells via distinct receptors to induce activation of the JAK–STAT pathway leading to the production of IFN-stimulated genes (ISGs) that can amplify the IFN signal and induce an antiviral state in infected cells/tissues.
In addition to activation of the JAK–STAT pathway, IFNs also activate PI3K and MAPK signaling cascades (1, 2). Perhaps the shared utilization of these signaling pathways between IFN and many other cytokines may help to explain the varied role of IFN in modulating antiviral and immune responses in various contexts and locations. Different affinities for their respective receptors exist among IFN subtypes, which may alter the signal strength upon receptor engagement, thus potentially adding another layer of regulation in control of immune responses by IFNs. Mendoza et al. developed a high-affinity IFNλ3 to discern the structure of the cytokine. When used in in vitro experiments this high-affinity IFNλ3 was found to have enhanced HCV and hepatitis B virus (HBV) antiviral activity (42). These results support the idea that enhancing the strength of the interaction of IFN with its receptor can modulate downstream functions. While this particular study investigated antiviral and anti-proliferative responses, it would be interesting to discern whether engineering of high-affinity IFNλ molecules can alter other facets of immunity. The recently solved IFNλ3/IFNλR1/IL-10R2 signaling complex structure could aid in answering these questions and in the development of IFNλ therapeutic agonists that have differential affinities for the receptor complex and downstream signaling strengths (42). Other mechanisms to regulate the response to IFNλ at the level of the IFNλR are conceivable. For example, in addition to the restricted nature of the IFNλR1 subunit, a soluble, secreted IFNλR1 has been described that could potentially sequester IFNλ as a regulatory mechanism (43). In summary, further studies are needed to dissect the intricate interplay of how IFN signaling pathways function in concert with stimulation by other cytokines that may activate similar or overlapping intracellular signaling pathways.
Antiviral Effects of Type III IFNs
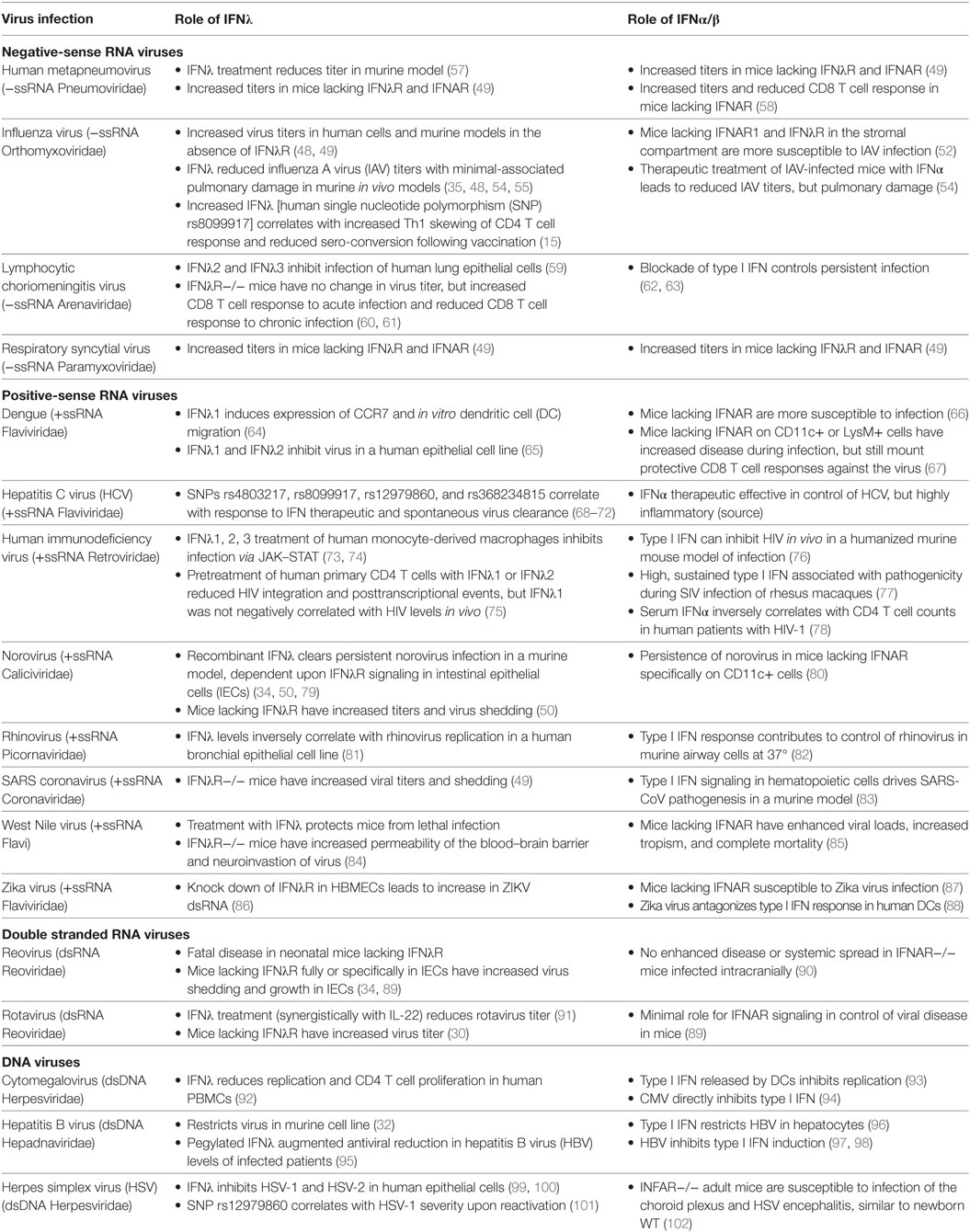
Interferon lambda is important in a wide variety of viral infections that including HCV, HBV, influenza virus, rhinovirus, respiratory syncytial virus (RSV), lymphocytic choriomeningitis virus (LCMV), rotavirus, reovirus, norovirus, and West Nile virus (WNV) [reviewed in Ref. (17, 19, 44–47)]. Many of these studies of IFNλ antiviral responses have been focused on viruses that infect the liver, the respiratory, and gastrointestinal mucosa, and, more recently, those that cross the blood–brain barrier (BBB) to cause a neuroinvasive viral infection. Experimental in vivo approaches using IFNλR knockout mice have highlighted the importance of IFNλ signaling in control of influenza A virus (IAV), SARS coronavirus, RSV, and human metapneumovirus levels in the lung as well as norovirus, reovirus, and rotavirus levels in the gastrointestinal tract (30, 48–50). It is also of note that type I and type III IFNs also have roles in cancer, parasitic infections, fungal infections, and several bacterial infections that include potential respiratory pathogens such as Staphylococcus aureus, Pseudomonas aeruginosa, and Mycobacterium tuberculosis, as well as Listeria monocytogenes and Salmonella typhimurium in addition to IFNs regulation of viral infections [reviewed in Ref. (47, 51)]. As the contribution of type I and type III IFNs in these other settings has been recently reviewed, we will not elaborate further herein.
Multiple reports have suggested redundant roles for IFNα/β and IFNλ in response to infection (23, 28, 52). However, distinct contributions for IFNα/β and IFNλ to infection have begun to be appreciated. Table 1 summarizes viral infections where IFNλ has been demonstrated to contribute in comparison with the known role of IFNα/β in these infections in vitro and in vivo. While the differences between IFNλ and IFNα/β are still being investigated, studies have demonstrated the ISG response induced by IFNλ is reduced compared with IFNα/β, while in vivo IFNλ is much less inflammatory than IFNα/β (53–55). Interestingly, IFNλ retains many antiviral properties despite the less inflammatory response compared with type I IFNs. This has spurred development of IFNλ for clinical use as an alternative treatment to IFNα for HCV infection has been of recent interest (53). Enthusiasm within the HCV field for IFNλ as a therapeutic treatment has waned as a result of the availability of direct-acting antiviral drugs capable of clearing HCV infection (56). However, harnessing the potential antiviral and less inflammatory functions of IFNλ as a therapeutic may be useful in treatment of other hepatic viral infections.
More recent and broad hypotheses posit that IFNλ treatment could also be utilized to control respiratory viral infections. In several experimental studies, prophylactic and therapeutic treatment of mice with IFNλ2 or IFNλ3 was shown to control IAV pulmonary titers similarly to IFNα or IFNβ treatment (54, 55). Importantly, IFNλ treatment avoided excessive pulmonary inflammation associated with IFNα treatment (54). The authors of this study speculated treatment with either cytokine overcame the known IAV NS1 mediated block on the induction of both type I and type III IFNs. The IFNλ treatment used in this study also altered responses in pulmonary monocytes and antigen presenting cells; however, the potential direct effects of IFNλ on these specific cell populations have not been characterized in an antiviral therapeutic setting.
Genetic Association of IFN Lambda Locus to Viral Susceptibility
The function of IFNL genes and their ability to regulate immunity is further impacted by a number of single nucleotide polymorphisms (SNPs) that have been identified in genome-wide association studies and correlate strongly to infectious disease outcome. These have been described in great detail elsewhere [reviewed in Ref. (47)]. Here, we will briefly discuss more recent findings related to these SNPs where the mechanism of their function and direct outcome on immune responses has been described. There has been considerable progress in understanding the direct impact of these SNPs on immunity to infection and disease outside of correlative phenotypes.
Multiple SNPs in IFNL3 are associated with response to interferon-based therapeutics and natural clearance of the HCV (68–72), although until recently the mechanism of regulation provided by these SNPs had not been understood. Our group has recently described the mechanism of one IFNL3 SNP (rs4803217) where presence of the G allele correlates with HCV clearance, whereas the unfavorable T allele correlates with HCV persistence (103). Specifically, HCV was found to regulate expression of two microRNAs (miR-208b and miR-499a-5p) that target the 3′ untranslated region (UTR) of IFNL3 leading to its degradation, allowing for viral persistence. The T allele leads to changes in the 3′ UTR allowing for enhanced binding of these HCV-induced microRNAs and AU-rich element-mediated decay of IFNL3, impacting expression of the cytokine and the outcome of HCV infection. Intriguingly, these same microRNAs also dampen type I IFN signaling in HCV-infected hepatocytes by downregulating expression of IFNAR1, a mechanism distinct from miR-208b and miR-499a-5p regulation of type III IFN (104).
Mechanistic studies have also defined the immunological consequence of another SNP impacting the production of IFNλ4. Approximately, 40% of Caucasians have an intact open reading frame for IFNL4 gene (105). However, a frame-shift mutation (TT>dG at ss469415590) in IFNL4 renders it a pseudogene. Intriguingly, the G gene variant encoding full-length IFNL4 is strongly correlated with persistence of HCV. It was hypothesized, but not demonstrated, that IFNL4 may have an intracellular role for dampening the antiviral response. However, it is speculated that this effect could be at least in part an indirect one as the dG IFNL4 allele is linked with the less favorable IFNL3 genotype at rs12979860 and rs4803217 (106). This work confirmed IFNλ4 has similar antiviral function to IFNλ3. However, the functional full-length IFNL4 is induced at lower levels compared with IFNL3 and is poorly translated due to intron-retention splice isoforms and weak polyadenylation (polyA) signal. Interestingly, non-human primates do not contain the dG>TT frame-shift mutation, but still limit IFNλ4 translation by production of intron-retention splice isoforms and a weak polyA signal, suggesting the functional IFNλ4 isoform has been selected against before the arise of the pseudogene frame-shift mutation in humans (106). It is still currently unclear as to why IFNL4 is suppressed, and perhaps undergoing pseudogenization. Perhaps IFNL4 arose more recently through genetic duplication of IFNL3 but did not develop a specific function distinct from IFNλ3, similar to what has occurred for other IFNs. Future studies without the confounding factor of linkage of the unfavorable IFNL3 genotypes may reveal the function of bioactive IFNλ4 to antiviral immunity. In addition, more studies parsing out the mechanisms of IFNL SNPs regulation of disease could provide important insights for the development and functionality of IFNλ therapeutics.
Type I VS III IFNs in Autoimmunity
The contribution of type I IFNs to development and manifestation of autoimmunity is well established [reviewed in Ref. (1, 107, 108)]. Type I IFNs are commonly upregulated in systemic autoimmune diseases such as systemic lupus erythematosus (SLE), Aicardi–Goutieres syndrome, Sjogren’s syndrome, type I diabetes, and psoriasis. More than half of adult patients, and 90% of pediatric patients, with SLE have elevated peripheral IFNα (109). Mechanistic studies have identified plasmacytoid dendritic cells (pDC), which are a major source of type I IFN, to be enriched in SLE lesions in humans and mice (110–112). Interestingly, type III IFNs do not seem to be linked to exacerbation of autoimmune diseases. In fact, type III IFN has been demonstrated to remediate symptoms in a mouse model of arthritis (113). Further, in a murine model of colitis, a disease that can be autoimmune in humans, IFNλ signaling specifically in neutrophils leads to a reduction on the release of reactive oxygen species and prevention of intestinal pathology (114). In addition, mice lacking the IFNλR1 have exacerbated disease in a model of asthma (14). This potentially protective role of IFNλ in asthma has also begun to be explored in humans (81). One recent paper has identified a correlation between systemic sclerosis and elevated IFNλ1 levels (115), but mechanistic studies clearly identifying a role for IFNλ in autoimmune disease are lacking. Interestingly, pDC have been shown to express the IFNλR and respond directly to IFNλ (116, 117). Whether IFNλ signaling is altered in pDC in the context of immunity is an interesting question that could have implications for immune-mediated treatment.
IFNλ Immune Modulatory Effects
The optimal induction of interferon to control infection while simultaneously avoiding host immunopathology is critical for an effective immune response against pathogens. Although IFNλ is generally considered to be less inflammatory than type I IFN, a full understanding of IFNλ’s regulation of immune responses outside of direct antiviral action has remained largely unknown (Figure 2). Recent studies, predominantly in the context of viral infections, have begun to elucidate the contribution of IFNλ to the regulation of the broader innate and adaptive immune responses (see Table 1). While the role of IFNα/β and IFNλ is similar in many viral infections, some notable differences exist. For example, therapeutic treatment of influenza virus infected mice with IFNα leads to enhanced pulmonary inflammation and mortality, while IFNλ is protective (54, 55). In addition, IFNλ is critical for protection against intestinal viral pathogens such as reovirus and rotavirus (89, 90). This is likely due to the fact that IECs respond robustly to IFNλ, but not type I IFN, in vivo (30). However, there are also other important roles for IFNλ in other immune cell types in the intestine, such as murine neutrophils, that have only begun to be investigated (114). As part of the ongoing efforts to understand specific function of IFNs in host defense, more studies designed to examine specific effects on different tissues and cell types that contribute to innate and adaptive immunity will be informative.
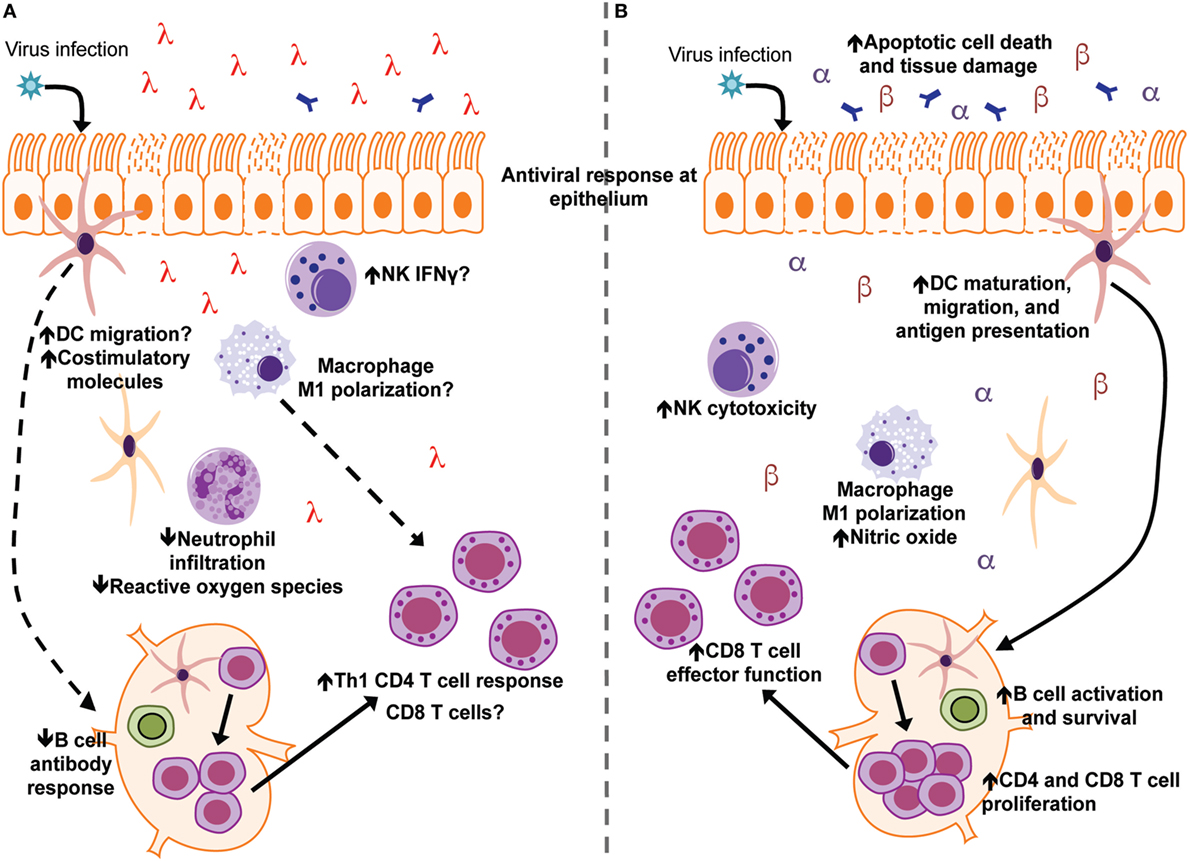
Figure 2. Interferon lambda (IFNλ) and IFNα/β differentially modulate immune responses during acute viral infection and tissue inflammation. (A) Following viral infection/tissue inflammation, IFNλ modulates functions of dendritic cells (DCs) neutrophils, CD4 T cells, and the B-cell antibody response. IFNλ signaling may also regulate macrophage, NK cell, and CD8 T cell function during infection/tissue inflammation (B) type I IFN (IFNα and IFNβ) have been the subject of a greater number of studies and have more defined roles during virus infection and tissue inflammation. Type I IFN enhances functions of DCs, macrophages, NK cells, B cells, CD4 T cells, and CD8 T cells toward an inflammatory/antiviral state.
Effects of IFN on Innate Immune Cells
IFN has been well described to have a direct antiviral effect in epithelial cells. Further, type I IFNs function on innate immune cells, such as DCs and macrophages. However, the functions of IFNλ on immune cells still remain largely unknown. For example, type I IFN signaling is well described to promote activation, survival, and cytotoxic function of NK cells during infection (118–120). By contrast, whether NK cells express IFNλR and respond directly to IFNλ to modulate NK cell function remains unclear (119, 121, 122). In this section, we will review the literature on the effect of IFNλs on innate immune cells.
IFN in Monocytic Cell Populations
Type I IFN promotes the polarization of macrophages to an inflammatory “M1” phenotype and increases the production of nitric oxide (73, 123–125). Type I IFN also enhances DC function by promoting their generation from monocytic precursors and leads to upregulation of MHC and costimulatory molecules in addition to increasing IL-12 production and enhancing DC migration (2, 126, 127). Conversely, the role of IFNλ in macrophages and DCs remains unresolved as studies have both supported and refuted the ability of these cells to directly respond to IFNλ (73, 128–130). Limited studies have indicated a role for IFNλ in the modulation of DC function. For example, it was demonstrated that human DC migration was enhanced as a direct response to IFNλ1 in the context of Dengue virus infection (64). In a separate report, IFNλ2 treatment increased IL-12 production and alteration of expression of the costimulatory molecule OX40L in CD11c+ cells (14). These changes, which may result in enhanced T cell immunity (131), indicate that IFNλ2 may also have a functional role at the interface of the innate and adaptive immune responses. While data implicating IFNλ regulation of DC functions are intriguing, further studies are needed to determine whether the defect in DCs in the absence of IFNλR signaling is intrinsic to these cells or influenced by the IFNλ response in epithelial cells at infection sites.
A possible explanation of the mixed reports regarding the contribution of IFN to DC function is that this could be due to differential responsiveness of various DC subsets to IFNλ and/or type I IFN. For example, pDC have been described to respond to both type I and type III IFNs to enhance their upregulation of ISGs, maturation, and antigen presentation function. We will not elaborate herein on the functions of IFN in pDC, as they have been well described in other recent reviews (132, 133). Given that there is a specific response of pDC to IFNλ that is not observed in the bulk heterogeneous DC population, it is possible other DC subsets may respond to IFNλ. During influenza and other viral infections in mice, CD103+ DCs are integral in delivery of antigen from the infected tissue to lung-draining lymph nodes where they can activate T cells (134–136). CD103+ DCs are less responsive to type I IFN, allowing for viral replication within these cells, and potentially leading to enhanced antigen presentation (137). Whether this difference could be due to preferential usage of IFNλR signaling to enhance antigen presentation has not been addressed. Interestingly, however, the ImmGen database indicates murine CD103+ DCs have higher levels of IFNλR compared with other DC subsets (138). However, as of this writing, the responsiveness of various DC and macrophage subsets to IFNλ signaling remains unclear. As T cells do not respond directly to IFNλ, it is likely that differential IFNα/β signaling compared with IFNλ in DCs could be modulating T cell responses (43, 61). Indeed, during Dengue virus infection, IFNλ leads to enhanced migration of DCs in vitro and increases CCR7 required for migratory function on DCs (64). Perhaps IFNλ signaling in DCs allows for optimal maturation and antigen presentation to T cells without excessive inflammation associated with IFNα/β signaling. It is also possible that at mucosal and barrier epithelial sites, epithelial cells themselves are regulating the alteration in DC response. Future studies in mice conditionally lacking IFNλR1 or IFNAR1 in DCs or epithelial cells specifically will delineate the role of IFNλ in these cell population.
IFN in Neutrophils
While few studies that have interrogated the direct effect of IFNα/β signaling on neutrophils, type I IFNs have been demonstrated to play a role in activation of neutrophil function (139). Murine neutrophils have recently been shown to express high levels of Ifnlr1 and respond directly to stimulation with IFNλ (114). Treatment of mice with arthritic symptoms with IFNλ2 was shown to prevent neutrophil infiltration into arthritic joints (113). While the potential therapeutic application of IFNλ to limit neutrophil-mediated pathology is interesting in this arthritis model, whether this paradigm is true should continue to be examined in the context of other inflammatory events. Neutrophils are known to significantly exacerbate disease severity during respiratory viral and bacterial infections and directly contribute to lung pathology [reviewed in Ref. (140)]. It is intriguing that IFNλ could potentially reduce or prevent neutrophil-mediated detrimental lung inflammation during respiratory infection via a similar mechanism. IFNλ has recently been demonstrated to act on neutrophils to control both influenza virus infection and DSS-induced colitis in murine models, indicating IFNλ directly alters neutrophil function in addition to recruitment as previously described (35, 114, 141). Interestingly, this IFNλ-specific dampening of neutrophil function is mediated in a non-transcriptional/translational fashion via Akt’s regulation of the release of reactive oxygen species (114). Importantly, this study represents the first reported such function of IFNλ and opens the intriguing possibility for IFNλ to yield changes in immune cells in a mechanism distinct from canonical JAK–STAT signaling. While these studies are intriguing, they have thus far only been validated in murine neutrophils. Future studies will be needed to determine whether human neutrophils respond to IFNλ in a similar fashion.
Effects of IFNs on Adaptive Immune Cells
Adaptive immunity is critical in controlling and providing long-term protection against infection. IFNs act at the interface of innate and adaptive immunity, by directly regulating innate as well as adaptive immune cells. For example, type I IFN promotes B-cell activation and class switching during acute viral infection [reviewed in Ref. (2)]. While there is currently no evidence demonstrating IFNλ has direct effects on the function of B cells, humans receiving influenza virus vaccination who had lower levels of circulating IFNλ correlated with increased sero-conversion (15). In addition, IFNλ has been reported to augment TLR-mediated activation and function of human B cells, but IFNλ could not directly and independently impact B-cell activation (142). Conversely, in a murine model of WNV infection, IFNLR1−/− mice had no effect on antibody responses compared with wild-type control mice (84). However, evidence supporting a role for IFNλ regulation of B cell functions is currently lacking but this still an area of active investigation.
IFN in T Cells
While T cells do not respond directly to IFNλ (43, 61), it is clear that IFNλ regulates function of T cells. IFNλ enhances T cell proliferation and Th1/Th17 cytokine production following treatment of peripheral blood mononuclear cells with IFNλ and in the context of asthma and influenza virus vaccination (14, 15). IFNλ has been shown to polarize the response toward a Th1 phenotype while suppressing Th2 and associated B cell responses. This is supported by studies in humans evaluating a SNP (rs8099917) in the IFNL locus, where individuals with the SNP that correlate to high IFNλ3 levels have lower sero-conversion rates following influenza virus vaccination, but a greater induction of Th1 CD4 T cells (15). Therefore, IFNλ-mediated effects on the T cell response might be indirect mediated by another cell subset known to express IFNλR. It is likely that IFNλ signaling in DCs is responsible for this alteration of the T cell response; however, the direct action of DCs in regulating Th1/Th2 responses is still unknown. In addition, whether IFNλ alters DCs to regulate CD8 T cell responses, which are critical for clearance of virus during many infections, is still unknown. Intriguingly, a report investigating acute and chronic LCMV responses in a murine model suggest IFNλ signaling negatively regulates virus-specific T cell responses during acute infection, but is required for the persistence of the T cell response during chronic infection (61). While the mechanism remains unclear, a study in macaques demonstrated that IFNλ3 drives cytotoxic ability of CD8 T cells, overall providing further evidence of the potential of IFNλ to function as an immune adjuvant or therapeutic agent to promote antiviral T cell responses (143).
In contrast to the absence of direct effects by IFNλ on CD4 and CD8 T cell activation, proliferation, and cytokine production, type I IFN directly regulates these T cell functions [reviewed in Ref. (144)]. Type I IFN signaling can regulate T cell responses via indirect effects on DCs or macrophages in addition to direct signaling effects on T cells themselves. This difference in mechanisms of T cell regulation is a major distinction between type I and type III IFNs that has not yet been fully evaluated. The potentially distinct, indirect mechanism of IFNλ regulation of T cell responses could yield interesting insights into the ability of IFNλ to be utilized as a therapeutic or vaccine adjuvant to augment the immune response against viral infections.
Role of Type III IFNs at the BBB
In addition to impacts on immune cells, IFNλ has also been described to regulate the BBB during WNV infection (84). Mice lacking the IFNλR1 show increased viral titers in central nervous system tissues and increased BBB permeability following WNV infection. Interestingly, IFNλ-mediated restriction of the endothelial tight junctions in an in vitro BBB model is independent of STAT1 or protein synthesis. These findings suggest there may be an undescribed, novel IFNλ signaling pathway that regulates endothelial cells. As endothelial cells are significant regulators of inflammatory responses, IFNλ could exert important effects on these cells types that would also be applicable to infection at the site of pulmonary and gastrointestinal barriers.
Outstanding Questions
While the functions of IFNλ and IFNα/β overlap in many infections and cell types, a growing number of notable differences are allowing for a better understanding of specialized roles of IFNs in regulation of immunity. In addition, the difference in IFNAR and IFNλR1 expression levels on various cell subsets and tissues could contribute to specific action of IFNλ vs type I IFN. This regulation along with SNPs in IFNL genes highlight that IFNλ has a unique role in antiviral immunity independent of type I IFN. While nearly 15 years of research has led to many insights in the function of IFNλ and its contribution to immunity, many questions remain to be answered. What are the distinct and redundant functions of each IFNL gene? Are there spatiotemporal effects that provide distinctions of IFNλ subtypes? Are there other signaling pathways that are active downstream of IFNλR differentially activate immune and epithelial cell subsets to modulate innate and adaptive immune response? Answering these questions with the help of newly available murine models will be critical to gain insights into the function of IFNλ, and to continue to develop IFNλ for use as a therapeutic against viral infections in the liver and at barrier surfaces.
Author Contributions
All authors contributed to the conceptualization, writing, and editing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Drs. Alison Kell and Matthew Long for their critical evaluation of this manuscript.
Funding
EH is supported by American Heart Association award 17POST33660907. The writing of this review was also supported by National Institutes of Health grants T32 AI007509 (EH), AI108765 (RS), AI127463, AI104002, and AI118916 (MG).
References
1. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol (2014) 14(1):36–49. doi:10.1038/nri3581
2. McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol (2015) 15(2):87–103. doi:10.1038/nri3787
3. Chow KT, Gale M Jr. SnapShot: interferon signaling. Cell (2015) 163(7):1808–1808.e1. doi:10.1016/j.cell.2015.12.008
4. Secombes CJ, Zou J. Evolution of interferons and interferon receptors. Front Immunol (2017) 8:209. doi:10.3389/fimmu.2017.00209
5. Boudinot P, Langevin C, Secombes CJ, Levraud JP. The peculiar characteristics of fish type I interferons. Viruses (2016) 8(11):1–17. doi:10.3390/v8110298
6. Sang Y, Liu Q, Lee J, Ma W, McVey DS, Blecha F. Expansion of amphibian intronless interferons revises the paradigm for interferon evolution and functional diversity. Sci Rep (2016) 6:29072. doi:10.1038/srep29072
7. Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol (2003) 4(1):69–77. doi:10.1038/ni875
8. Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol (2003) 4(1):63–8. doi:10.1038/ni873
9. Chen SN, Zhang XW, Li L, Ruan BY, Huang B, Huang WS, et al. Evolution of IFN-lambda in tetrapod vertebrates and its functional characterization in green anole lizard (Anolis carolinensis). Dev Comp Immunol (2016) 61:208–24. doi:10.1016/j.dci.2016.04.004
10. Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. J Biol Chem (2009) 284(31):20869–75. doi:10.1074/jbc.M109.002923
11. Manry J, Laval G, Patin E, Fornarino S, Itan Y, Fumagalli M, et al. Evolutionary genetic dissection of human interferons. J Exp Med (2011) 208(13):2747–59. doi:10.1084/jem.20111680
12. Hoffmann HH, Schneider WM, Rice CM. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol (2015) 36(3):124–38. doi:10.1016/j.it.2015.01.004
13. Lasfar A, Lewis-Antes A, Smirnov SV, Anantha S, Abushahba W, Tian B, et al. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res (2006) 66(8):4468–77. doi:10.1158/0008-5472.CAN-05-3653
14. Koltsida O, Hausding M, Stavropoulos A, Koch S, Tzelepis G, Ubel C, et al. IL-28A (IFN-lambda2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol Med (2011) 3(6):348–61. doi:10.1002/emmm.201100142
15. Egli A, Santer DM, O’Shea D, Barakat K, Syedbasha M, Vollmer M, et al. IL-28B is a key regulator of B- and T-cell vaccine responses against influenza. PLoS Pathog (2014) 10(12):e1004556. doi:10.1371/journal.ppat.1004556
16. Wei H, Wang S, Chen Q, Chen Y, Chi X, Zhang L, et al. Suppression of interferon lambda signaling by SOCS-1 results in their excessive production during influenza virus infection. PLoS Pathog (2014) 10(1):e1003845. doi:10.1371/journal.ppat.1003845
17. Lazear HM, Nice TJ, Diamond MS. Interferon-lambda: immune functions at barrier surfaces and beyond. Immunity (2015) 43(1):15–28. doi:10.1016/j.immuni.2015.07.001
18. Iversen MB, Paludan SR. Mechanisms of type III interferon expression. J Interferon Cytokine Res (2010) 30(8):573–8. doi:10.1089/jir.2010.0063
19. Levy DE, Marie IJ, Durbin JE. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol (2011) 1(6):476–86. doi:10.1016/j.coviro.2011.11.001
20. Durbin RK, Kotenko SV, Durbin JE. Interferon induction and function at the mucosal surface. Immunol Rev (2013) 255(1):25–39. doi:10.1111/imr.12101
21. Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol (2014) 15(8):717–26. doi:10.1038/ni.2915
22. Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, et al. Viral infection and toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol (2004) 34(3):796–805. doi:10.1002/eji.200324610
23. Siren J, Pirhonen J, Julkunen I, Matikainen S. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. J Immunol (2005) 174(4):1932–7. doi:10.4049/jimmunol.174.4.1932
24. Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol (2011) 186(8):4541–5. doi:10.4049/jimmunol.1003389
25. Sui H, Zhou M, Imamichi H, Jiao X, Sherman BT, Lane HC, et al. STING is an essential mediator of the Ku70-mediated production of IFN-lambda1 in response to exogenous DNA. Sci Signal (2017) 10(488): eaah5054. doi:10.1126/scisignal.aah5054
26. Bolen CR, Ding S, Robek MD, Kleinstein SH. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology (2014) 59(4):1262–72. doi:10.1002/hep.26657
27. Lin JD, Feng N, Sen A, Balan M, Tseng HC, McElrath C, et al. Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog (2016) 12(4):e1005600. doi:10.1371/journal.ppat.1005600
28. Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J Immunol (2007) 179(6):3434–42. doi:10.4049/jimmunol.179.6.3434
29. Thomson SJ, Goh FG, Banks H, Krausgruber T, Kotenko SV, Foxwell BM, et al. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc Natl Acad Sci U S A (2009) 106(28):11564–9. doi:10.1073/pnas.0904477106
30. Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A (2011) 108(19):7944–9. doi:10.1073/pnas.1100552108
31. Jewell NA, Cline T, Mertz SE, Smirnov SV, Flano E, Schindler C, et al. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J Virol (2010) 84(21):11515–22. doi:10.1128/JVI.01703-09
32. Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol (2005) 79(6):3851–4. doi:10.1128/JVI.79.6.3851-3854.2005
33. Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol (2006) 80(9):4501–9. doi:10.1128/JVI.80.9.4501-4509.2006
34. Baldridge MT, Lee S, Brown JJ, McAllister N, Urbanek K, Dermody TS, et al. Expression of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of interferon lambda against norovirus and reovirus. J Virol (2017) 91(7): e02079–16. doi:10.1128/JVI.02079-16
35. Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, et al. Interferon-lambda mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity (2017) 46(5):875–90.e6. doi:10.1016/j.immuni.2017.04.025
37. Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev (2008) 226:205–18. doi:10.1111/j.1600-065X.2008.00706.x
38. Lee S, Baldridge MT. Interferon-lambda: a potent regulator of intestinal viral infections. Front Immunol (2017) 8:749. doi:10.3389/fimmu.2017.00749
39. Piganis RA, De Weerd NA, Gould JA, Schindler CW, Mansell A, Nicholson SE, et al. Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon alpha receptor (IFNAR1)-associated tyrosine kinase Tyk2. J Biol Chem (2011) 286(39):33811–8. doi:10.1074/jbc.M111.270207
40. Liu B, Chen S, Guan Y, Chen L. Type III interferon induces distinct SOCS1 expression pattern that contributes to delayed but prolonged activation of Jak/STAT signaling pathway: implications for treatment non-response in HCV patients. PLoS One (2015) 10(7):e0133800. doi:10.1371/journal.pone.0133800
41. Blumer T, Coto-Llerena M, Duong FHT, Heim MH. SOCS1 is an inducible negative regulator of IFN lambda induced gene expression in vivo. J Biol Chem (2017) 292:17928–38. doi:10.1074/jbc.M117.788877
42. Mendoza JL, Schneider WM, Hoffmann HH, Vercauteren K, Jude KM, Xiong A, et al. The IFN-lambda-IFN-lambdaR1-IL-10Rbeta complex reveals structural features underlying type III IFN functional plasticity. Immunity (2017) 46(3):379–92. doi:10.1016/j.immuni.2017.02.017
43. Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W, et al. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun (2009) 10(8):702–14. doi:10.1038/gene.2009.72
44. Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res (2010) 30(8):555–64. doi:10.1089/jir.2010.0078
45. Odendall C, Kagan JC. The unique regulation and functions of type III interferons in antiviral immunity. Curr Opin Virol (2015) 12:47–52. doi:10.1016/j.coviro.2015.02.003
46. Wack A, Terczynska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol (2015) 16(8):802–9. doi:10.1038/ni.3212
47. Syedbasha M, Egli A. Interferon lambda: modulating immunity in infectious diseases. Front Immunol (2017) 8:119. doi:10.3389/fimmu.2017.00119
48. Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, et al. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog (2008) 4(9):e1000151. doi:10.1371/journal.ppat.1000151
49. Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol (2010) 84(11):5670–7. doi:10.1128/JVI.00272-10
50. Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science (2015) 347(6219):269–73. doi:10.1126/science.1258100
51. Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer (2016) 16(3):131–44. doi:10.1038/nrc.2016.14
52. Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog (2013) 9(11):e1003773. doi:10.1371/journal.ppat.1003773
53. Muir AJ, Arora S, Everson G, Flisiak R, George J, Ghalib R, et al. A randomized phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J Hepatol (2014) 61(6):1238–46. doi:10.1016/j.jhep.2014.07.022
54. Davidson S, McCabe TM, Crotta S, Gad HH, Hessel EM, Beinke S, et al. IFNlambda is a potent anti-influenza therapeutic without the inflammatory side effects of IFNalpha treatment. EMBO Mol Med (2016) 8(9):1099–112. doi:10.15252/emmm.201606413
55. Kim S, Kim MJ, Kim CH, Kang JW, Shin HK, Kim DY, et al. The superiority of IFN-lambda as a therapeutic candidate to control acute influenza viral lung infection. Am J Respir Cell Mol Biol (2017) 56(2):202–12. doi:10.1165/rcmb.2016-0174OC
56. D’Ambrosio R, Degasperi E, Colombo M, Aghemo A. Direct-acting antivirals: the endgame for hepatitis C? Curr Opin Virol (2017) 24:31–7. doi:10.1016/j.coviro.2017.03.017
57. Banos-Lara Mdel R, Harvey L, Mendoza A, Simms D, Chouljenko VN, Wakamatsu N, et al. Impact and regulation of lambda interferon response in human metapneumovirus infection. J Virol (2015) 89(1):730–42. doi:10.1128/JVI.02897-14
58. Hastings AK, Erickson JJ, Schuster JE, Boyd KL, Tollefson SJ, Johnson M, et al. Role of type I interferon signaling in human metapneumovirus pathogenesis and control of viral replication. J Virol (2015) 89(8):4405–20. doi:10.1128/JVI.03275-14
59. Lukacikova L, Oveckova I, Betakova T, Laposova K, Polcicova K, Pastorekova S, et al. Antiviral effect of interferon lambda against lymphocytic choriomeningitis virus. J Interferon Cytokine Res (2015) 35(7):540–53. doi:10.1089/jir.2014.0083
60. Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol (2008) 180(4):2474–85. doi:10.4049/jimmunol.180.4.2474
61. Misumi I, Whitmire JK. IFN-lambda exerts opposing effects on T cell responses depending on the chronicity of the virus infection. J Immunol (2014) 192(8):3596–606. doi:10.4049/jimmunol.1301705
62. Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science (2013) 340(6129):207–11. doi:10.1126/science.1235214
63. Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science (2013) 340(6129):202–7. doi:10.1126/science.1235208
64. Hsu YL, Wang MY, Ho LJ, Lai JH. Dengue virus infection induces interferon-lambda1 to facilitate cell migration. Sci Rep (2016) 6:24530. doi:10.1038/srep24530
65. Palma-Ocampo HK, Flores-Alonso JC, Vallejo-Ruiz V, Reyes-Leyva J, Flores-Mendoza L, Herrera-Camacho I, et al. Interferon lambda inhibits dengue virus replication in epithelial cells. Virol J (2015) 12:150. doi:10.1186/s12985-015-0383-4
66. Orozco S, Schmid MA, Parameswaran P, Lachica R, Henn MR, Beatty R, et al. Characterization of a model of lethal dengue virus 2 infection in C57BL/6 mice deficient in the alpha/beta interferon receptor. J Gen Virol (2012) 93(Pt 10):2152–7. doi:10.1099/vir.0.045088-0
67. Zust R, Toh YX, Valdes I, Cerny D, Heinrich J, Hermida L, et al. Type I interferon signals in macrophages and dendritic cells control dengue virus infection: implications for a new mouse model to test dengue vaccines. J Virol (2014) 88(13):7276–85. doi:10.1128/JVI.03827-13
68. Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature (2009) 461(7262):399–401. doi:10.1038/nature08309
69. Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet (2009) 41(10):1100–4. doi:10.1038/ng.447
70. Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet (2009) 41(10):1105–9. doi:10.1038/ng.449
71. Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature (2009) 461(7265):798–801. doi:10.1038/nature08463
72. Sheahan T, Imanaka N, Marukian S, Dorner M, Liu P, Ploss A, et al. Interferon lambda alleles predict innate antiviral immune responses and hepatitis C virus permissiveness. Cell Host Microbe (2014) 15(2):190–202. doi:10.1016/j.chom.2014.01.007
73. Liu MQ, Zhou DJ, Wang X, Zhou W, Ye L, Li JL, et al. IFN-lambda3 inhibits HIV infection of macrophages through the JAK-STAT pathway. PLoS One (2012) 7(4):e35902. doi:10.1371/journal.pone.0035902
74. Wang Y, Li J, Wang X, Zhou Y, Zhang T, Ho W. Comparison of antiviral activity of lambda-interferons against HIV replication in macrophages. J Interferon Cytokine Res (2015) 35(3):213–21. doi:10.1089/jir.2014.0064
75. Tian RR, Guo HX, Wei JF, Yang CK, He SH, Wang JH. IFN-lambda inhibits HIV-1 integration and post-transcriptional events in vitro, but there is only limited in vivo repression of viral production. Antiviral Res (2012) 95(1):57–65. doi:10.1016/j.antiviral.2012.04.011
76. Lavender KJ, Gibbert K, Peterson KE, Van Dis E, Francois S, Woods T, et al. Interferon alpha subtype-specific suppression of HIV-1 infection in vivo. J Virol (2016) 90(13):6001–13. doi:10.1128/JVI.00451-16
77. Harris LD, Tabb B, Sodora DL, Paiardini M, Klatt NR, Douek DC, et al. Downregulation of robust acute type I interferon responses distinguishes nonpathogenic simian immunodeficiency virus (SIV) infection of natural hosts from pathogenic SIV infection of rhesus macaques. J Virol (2010) 84(15):7886–91. doi:10.1128/JVI.02612-09
78. Hardy GA, Sieg S, Rodriguez B, Anthony D, Asaad R, Jiang W, et al. Interferon-alpha is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS One (2013) 8(2):e56527. doi:10.1371/journal.pone.0056527
79. Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, et al. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science (2015) 347(6219):266–9. doi:10.1126/science.1258025
80. Nice TJ, Osborne LC, Tomov VT, Artis D, Wherry EJ, Virgin HW. Type I interferon receptor deficiency in dendritic cells facilitates systemic murine norovirus persistence despite enhanced adaptive immunity. PLoS Pathog (2016) 12(6):e1005684. doi:10.1371/journal.ppat.1005684
81. Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PA, Bartlett NW, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med (2006) 12(9):1023–6. doi:10.1038/nm1462
82. Foxman EF, Storer JA, Fitzgerald ME, Wasik BR, Hou L, Zhao H, et al. Temperature-dependent innate defense against the common cold virus limits viral replication at warm temperature in mouse airway cells. Proc Natl Acad Sci U S A (2015) 112(3):827–32. doi:10.1073/pnas.1411030112
83. Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe (2016) 19(2):181–93. doi:10.1016/j.chom.2016.01.007
84. Lazear HM, Daniels BP, Pinto AK, Huang AC, Vick SC, Doyle SE, et al. Interferon-lambda restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci Transl Med (2015) 7(284):284ra259. doi:10.1126/scitranslmed.aaa4304
85. Samuel MA, Diamond MS. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J Virol (2005) 79(21):13350–61. doi:10.1128/JVI.79.21.13350-13361.2005
86. Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ET Jr, et al. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe (2016) 19(5):705–12. doi:10.1016/j.chom.2016.03.008
87. Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, et al. A mouse model of Zika virus pathogenesis. Cell Host Microbe (2016) 19(5):720–30. doi:10.1016/j.chom.2016.03.010
88. Bowen JR, Quicke KM, Maddur MS, O’Neal JT, McDonald CE, Fedorova NB, et al. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog (2017) 13(2):e1006164. doi:10.1371/journal.ppat.1006164
89. Mahlakoiv T, Hernandez P, Gronke K, Diefenbach A, Staeheli P. Leukocyte-derived IFN-alpha/beta and epithelial IFN-lambda constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog (2015) 11(4):e1004782. doi:10.1371/journal.ppat.1004782
90. Dionne KR, Galvin JM, Schittone SA, Clarke P, Tyler KL. Type I interferon signaling limits reoviral tropism within the brain and prevents lethal systemic infection. J Neurovirol (2011) 17(4):314–26. doi:10.1007/s13365-011-0038-1
91. Hernandez PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, et al. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol (2015) 16(7):698–707. doi:10.1038/ni.3180
92. Egli A, Levin A, Santer DM, Joyce M, O’Shea D, Thomas BS, et al. Immunomodulatory function of interleukin 28B during primary infection with cytomegalovirus. J Infect Dis (2014) 210(5):717–27. doi:10.1093/infdis/jiu144
93. Holzki JK, Dag F, Dekhtiarenko I, Rand U, Casalegno-Garduno R, Trittel S, et al. Type I interferon released by myeloid dendritic cells reversibly impairs cytomegalovirus replication by inhibiting immediate early gene expression. J Virol (2015) 89(19):9886–95. doi:10.1128/JVI.01459-15
94. Paulus C, Krauss S, Nevels M. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc Natl Acad Sci U S A (2006) 103(10):3840–5. doi:10.1073/pnas.0600007103
95. Phillips S, Mistry S, Riva A, Cooksley H, Hadzhiolova-Lebeau T, Plavova S, et al. Peg-interferon lambda treatment induces robust innate and adaptive immunity in chronic hepatitis B patients. Front Immunol (2017) 8:621. doi:10.3389/fimmu.2017.00621
96. Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, et al. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest (2012) 122(2):529–37. doi:10.1172/JCI58847
97. Jiang J, Tang H. Mechanism of inhibiting type I interferon induction by hepatitis B virus X protein. Protein Cell (2010) 1(12):1106–17. doi:10.1007/s13238-010-0141-8
98. Chen J, Wu M, Zhang X, Zhang W, Zhang Z, Chen L, et al. Hepatitis B virus polymerase impairs interferon-alpha-induced STA T activation through inhibition of importin-alpha5 and protein kinase C-delta. Hepatology (2013) 57(2):470–82. doi:10.1002/hep.26064
99. Griffiths SJ, Koegl M, Boutell C, Zenner HL, Crump CM, Pica F, et al. A systematic analysis of host factors reveals a Med23-interferon-lambda regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog (2013) 9(8):e1003514. doi:10.1371/journal.ppat.1003514
100. Zhou L, Li JL, Zhou Y, Liu JB, Zhuang K, Gao JF, et al. Induction of interferon-lambda contributes to TLR3 and RIG-I activation-mediated inhibition of herpes simplex virus type 2 replication in human cervical epithelial cells. Mol Hum Reprod (2015) 21(12):917–29. doi:10.1093/molehr/gav058
101. Pica F, Volpi A, Gaziano R, Garaci E. Interferon-lambda in immunocompetent individuals with a history of recurrent herpes labialis. Antivir Ther (2010) 15(5):737–43. doi:10.3851/IMP1610
102. Wilcox DR, Folmsbee SS, Muller WJ, Longnecker R. The type I interferon response determines differences in choroid plexus susceptibility between newborns and adults in herpes simplex virus encephalitis. MBio (2016) 7(2):e00437–16. doi:10.1128/mBio.00437-16
103. McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA, et al. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol (2014) 15(1):72–9. doi:10.1038/ni.2758
104. Jarret A, McFarland AP, Horner SM, Kell A, Schwerk J, Hong M, et al. Hepatitis-C-virus-induced microRNAs dampen interferon-mediated antiviral signaling. Nat Med (2016) 22(12):1475–81. doi:10.1038/nm.4211
105. Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet (2013) 45(2):164–71. doi:10.1038/ng.2521
106. Hong M, Schwerk J, Lim C, Kell A, Jarret A, Pangallo J, et al. Interferon lambda 4 expression is suppressed by the host during viral infection. J Exp Med (2016) 213(12):2539–52. doi:10.1084/jem.20160437
107. Di Domizio J, Cao W. Fueling autoimmunity: type I interferon in autoimmune diseases. Expert Rev Clin Immunol (2013) 9(3):201–10. doi:10.1586/eci.12.106
108. Niewold TB. Type I interferon in human autoimmunity. Front Immunol (2014) 5:306. doi:10.3389/fimmu.2014.00306
109. Postal M, Sinicato NA, Pelicari KO, Marini R, Lavras Costallat LT, Appenzeller S. Clinical and serological manifestations associated with interferon-alpha levels in childhood-onset systemic lupus erythematosus. Clinics (Sao Paulo) (2012) 67(2):157–62. doi:10.6061/clinics/2012(02)11
110. Farkas L, Beiske K, Lund-Johansen F, Brandtzaeg P, Jahnsen FL. Plasmacytoid dendritic cells (natural interferon- alpha/beta-producing cells) accumulate in cutaneous lupus erythematosus lesions. Am J Pathol (2001) 159(1):237–43. doi:10.1016/S0002-9440(10)61689-6
111. Rowland SL, Riggs JM, Gilfillan S, Bugatti M, Vermi W, Kolbeck R, et al. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J Exp Med (2014) 211(10):1977–91. doi:10.1084/jem.20132620
112. Sisirak V, Ganguly D, Lewis KL, Couillault C, Tanaka L, Bolland S, et al. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J Exp Med (2014) 211(10):1969–76. doi:10.1084/jem.20132522
113. Blazek K, Eames HL, Weiss M, Byrne AJ, Perocheau D, Pease JE, et al. IFN-lambda resolves inflammation via suppression of neutrophil infiltration and IL-1beta production. J Exp Med (2015) 212(6):845–53. doi:10.1084/jem.20140995
114. Broggi A, Tan Y, Granucci F, Zanoni I. IFN-lambda suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat Immunol (2017) 18:1084–93. doi:10.1038/ni.3821
115. Dantas AT, Goncalves SM, Pereira MC, de Almeida AR, Marques CD, Rego MJ, et al. Interferons and systemic sclerosis: correlation between interferon gamma and interferon-lambda 1 (IL-29). Autoimmunity (2015) 48(7):429–33. doi:10.3109/08916934.2015.1054028
116. Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, et al. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J Immunol (2012) 189(6):2735–45. doi:10.4049/jimmunol.1102038
117. Finotti G, Tamassia N, Calzetti F, Fattovich G, Cassatella MA. Endogenously produced TNF-alpha contributes to the expression of CXCL10/IP-10 in IFN-lambda3-activated plasmacytoid dendritic cells. J Leukoc Biol (2016) 99(1):107–19. doi:10.1189/jlb.3VMA0415-144R
118. Martinez J, Huang X, Yang Y. Direct action of type I IFN on NK cells is required for their activation in response to vaccinia viral infection in vivo. J Immunol (2008) 180(3):1592–7. doi:10.4049/jimmunol.180.3.1592
119. Paolini R, Bernardini G, Molfetta R, Santoni A. NK cells and interferons. Cytokine Growth Factor Rev (2015) 26(2):113–20. doi:10.1016/j.cytogfr.2014.11.003
120. Madera S, Rapp M, Firth MA, Beilke JN, Lanier LL, Sun JC. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J Exp Med (2016) 213(2):225–33. doi:10.1084/jem.20150712
121. Kramer B, Eisenhardt M, Glassner A, Korner C, Sauerbruch T, Spengler U, et al. Do lambda-IFNs IL28A and IL28B act on human natural killer cells? Proc Natl Acad Sci U S A (2011) 108(34):E519–20. doi:10.1073/pnas.1108850108
122. Souza-Fonseca-Guimaraes F, Young A, Mittal D, Martinet L, Bruedigam C, Takeda K, et al. NK cells require IL-28R for optimal in vivo activity. Proc Natl Acad Sci U S A (2015) 112(18):E2376–84. doi:10.1073/pnas.1424241112
123. Vadiveloo PK, Vairo G, Hertzog P, Kola I, Hamilton JA. Role of type I interferons during macrophage activation by lipopolysaccharide. Cytokine (2000) 12(11):1639–46. doi:10.1006/cyto.2000.0766
124. Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol (2014) 5:614. doi:10.3389/fimmu.2014.00614
125. Xie C, Liu C, Wu B, Lin Y, Ma T, Xiong H, et al. Effects of IRF1 and IFN-beta interaction on the M1 polarization of macrophages and its antitumor function. Int J Mol Med (2016) 38(1):148–60. doi:10.3892/ijmm.2016.2583
126. Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, et al. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood (2002) 99(9):3263–71. doi:10.1182/blood.V99.9.3263
127. Chen K, Liu J, Cao X. Regulation of type I interferon signaling in immunity and inflammation: a comprehensive review. J Autoimmun (2017) 83:1–11. doi:10.1016/j.jaut.2017.03.008
128. Liu BS, Janssen HL, Boonstra A. IL-29 and IFNalpha differ in their ability to modulate IL-12 production by TLR-activated human macrophages and exhibit differential regulation of the IFNgamma receptor expression. Blood (2011) 117(8):2385–95. doi:10.1182/blood-2010-07-298976
129. Dickensheets H, Sheikh F, Park O, Gao B, Donnelly RP. Interferon-lambda (IFN-lambda) induces signal transduction and gene expression in human hepatocytes, but not in lymphocytes or monocytes. J Leukoc Biol (2013) 93(3):377–85. doi:10.1189/jlb.0812395
130. de Groen RA, Boltjes A, Hou J, Liu BS, McPhee F, Friborg J, et al. IFN-lambda-mediated IL-12 production in macrophages induces IFN-gamma production in human NK cells. Eur J Immunol (2015) 45(1):250–9. doi:10.1002/eji.201444903
131. Mennechet FJ, Uze G. Interferon-lambda-treated dendritic cells specifically induce proliferation of FOXP3-expressing suppressor T cells. Blood (2006) 107(11):4417–23. doi:10.1182/blood-2005-10-4129
132. Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol (2015) 15(8):471–85. doi:10.1038/nri3865
133. Finotti G, Tamassia N, Cassatella MA. Interferon-lambdas and plasmacytoid dendritic cells: a close relationship. Front Immunol (2017) 8:1015. doi:10.3389/fimmu.2017.01015
134. del Rio ML, Bernhardt G, Rodriguez-Barbosa JI, Forster R. Development and functional specialization of CD103+ dendritic cells. Immunol Rev (2010) 234(1):268–81. doi:10.1111/j.0105-2896.2009.00874.x
135. Ho AW, Prabhu N, Betts RJ, Ge MQ, Dai X, Hutchinson PE, et al. Lung CD103+ dendritic cells efficiently transport influenza virus to the lymph node and load viral antigen onto MHC class I for presentation to CD8 T cells. J Immunol (2011) 187(11):6011–21. doi:10.4049/jimmunol.1100987
136. Neyt K, Lambrecht BN. The role of lung dendritic cell subsets in immunity to respiratory viruses. Immunol Rev (2013) 255(1):57–67. doi:10.1111/imr.12100
137. Moltedo B, Li W, Yount JS, Moran TM. Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS Pathog (2011) 7(11):e1002345. doi:10.1371/journal.ppat.1002345
138. Heng TS, Painter MW; Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol (2008) 9(10):1091–4. doi:10.1038/ni1008-1091
139. Kasimir S, Brom J, Konig W. Effect of interferon-alpha on neutrophil functions. Immunology (1991) 74(2):271–8.
140. Camp JV, Jonsson CB. A role for neutrophils in viral respiratory disease. Front Immunol (2017) 8:550. doi:10.3389/fimmu.2017.00550
141. Hemann EA, Schwerk J, Savan R. IFN-lambda ‘guts’ neutrophil-mediated inflammation. Nat Immunol (2017) 18(10):1061–2. doi:10.1038/ni.3834
142. de Groen RA, Groothuismink ZM, Liu BS, Boonstra A. IFN-lambda is able to augment TLR-mediated activation and subsequent function of primary human B cells. J Leukoc Biol (2015) 98(4):623–30. doi:10.1189/jlb.3A0215-041RR
143. Morrow MP, Yan J, Pankhong P, Shedlock DJ, Lewis MG, Talbott K, et al. IL-28B/IFN-lambda 3 drives granzyme B loading and significantly increases CTL killing activity in macaques. Mol Ther (2010) 18(9):1714–23. doi:10.1038/mt.2010.118
Keywords: interferon lambda, interferon, immunity, immune cells, infectious disease, virus
Citation: Hemann EA, Gale M Jr. and Savan R (2017) Interferon Lambda Genetics and Biology in Regulation of Viral Control. Front. Immunol. 8:1707. doi: 10.3389/fimmu.2017.01707
Received: 04 October 2017; Accepted: 20 November 2017;
Published: 06 December 2017
Edited by:
Ivan Zanoni, Harvard University, United StatesReviewed by:
Marco A. Cassatella, University of Verona, ItalyTImothy Nice, Oregon Health & Science University, United States
Copyright: © 2017 Hemann, Gale and Savan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ram Savan, savanram@uw.edu
 Emily A. Hemann
Emily A. Hemann Michael Gale Jr.
Michael Gale Jr. Ram Savan
Ram Savan