- 1SciBerg e.Kfm, Mannheim, Germany
- 2Molecular Epidemiology C080, German Cancer Research Center, Heidelberg, Germany
- 3National Medical Research Center for Preventive Medicine, Moscow, Russia
- 4Department of Cell Biology, Higher School of Economics, Moscow, Russia
- 5Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry RAS, Moscow, Russia
- 6SRC BioClinicum, Moscow, Russia
Exosomes and microvesicles are two major categories of extracellular vesicles (EVs) released by almost all cell types and are highly abundant in biological fluids. Both the molecular composition of EVs and their release are thought to be strictly regulated by external stimuli. Multiple studies have consistently demonstrated that EVs transfer proteins, lipids and RNA between various cell types, thus mediating intercellular communication, and signaling. Importantly, small non-coding RNAs within EVs are thought to be major contributors to the molecular events occurring in the recipient cell. Furthermore, RNA cargo in exosomes and microvesicles could hold tremendous potential as non-invasive biomarkers for multiple disorders, including pathologies of the immune system. This mini-review is aimed to provide the state-of-the-art in the EVs-associated RNA transcriptome field, as well as the comprehensive analysis of previous studies characterizing RNA content within EVs released by various cells using next-generation sequencing. Finally, we highlight the technical challenges associated with obtaining pure EVs and deep sequencing of the EV-associated RNAs.
Introduction
The “Extracellular Vesicles (EVs)” is a general term used to describe various types of spheroid structures, encircled by a lipid membrane bilayer, which are secreted by mammalian cells either passively or upon certain stimuli (1). Since their initial discovery more than 30 years ago (2, 3) EVs have been purified from nearly all mammalian cell types including cells of the immune system (1). Furthermore, EVs have been detected in almost all human biological fluids, and shown to mediate cell-cell communication, thus playing a key role in the regulation of various physiological processes in the body (4) including the immune response (5–8). Finally, it becomes increasingly evident that EVs may contribute to carcinogenesis, as well as the spread of viruses, toxic proteins, and prions (1, 9).
There are three distinct types of EVs (as classified by their origin and biogenesis)—apoptotic bodies (ABs), microvesicles (MVs, also known as shedding vesicles), and exosomes (Figure 1A). The ABs are on average 1–5 μm in diameter and are by-products of cell disassembling during the apoptosis (10, 11). The MVs are formed by outward budding of the plasma membrane and are between 100 and 1,000 nm in diameter (8). The exosomes are the smallest type of EVs, having a diameter of 30–150 nm, and are primarily formed as intraluminal vesicles (ILVs) within multi-vesicular bodies (MVBs). Upon fusing of MVBs with the plasma membrane, the ILVs are released as exosomes into the extracellular space (8). Both MVs and exosomes contain various cytoplasmic and membrane-associated proteins as well as lipids, sugars, and nucleic acids (9), while ABs may in addition include nuclear fractions and cell organelles (1, 10, 11). Well-characterized protein markers for exosomes include various tetraspanins such as CD9, CD63, and CD81; while the MVs contain transmembrane proteins common for the plasmalemma such as integrins and selectins (8). Concurrently, ABs could be differentiated by the presence of histones (10, 11). Recently, large oncosomes (LOs) have been identified as the fourth type of EVs which are generated by shedding of membrane blebs from tumor cells and have a size similar to ABs (12).
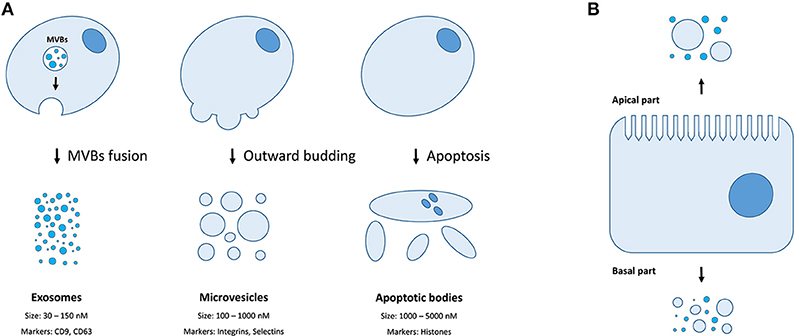
Figure 1. Extracellular membrane vesicles. (A) The mechanisms of generation, size distribution, and common protein markers of different EVs types; (B) Different populations of EVs could be released depending on the side of the plasma membrane of a polar cell.
Importantly, multiple research reports have demonstrated that various RNA species (including mRNA, miRNAs, and lncRNAs) entrapped within EVs can be transferred from donor to acceptor cells and interfere in gene expression in the latter (13, 14). This mini-review is aimed to provide a state-of-the-art in the EVs field, focusing primarily on the reported RNA cargo in different subtypes of EVs as well as the methodological challenges associated with purification of membrane vesicles and deep sequencing of their RNA content. Furthermore, we elaborate on a putative contribution of vesicular RNA to the functioning of the immune cells.
The Challenges in Purification of EVs and Characterization of Extracellular RNA
The techniques widely used so far for isolating EVs include ultracentrifugation, density gradient flotation, ultrafiltration, chromatography, polymer-based precipitation, and immunoprecipitation (15). Differential ultracentrifugation is the most commonly used approach for EVs purification and, in particular, for separating exosomes from ABs and MVs (16). A biological fluid is first depleted from living cells, cell debris, ABs, and MVs with a series of lower-speed centrifugation, and the exosomes fractions are ultimately pelleted by ultracentrifugation. However, the final exosomes pellets can be contaminated with low-sized MVs, large protein aggregates as well as viruses (17, 18). The method of density gradient flotation harnesses differences in size, shape, and density of different EVs types and allows much higher purity of isolated EVs especially when combined with ultracentrifugation. However, high-density lipoproteins (HDL) and low-sized MVs are ultimately co-isolated with exosomes when using density gradient ultracentrifugation (19). Size exclusion chromatography generally allows recovery of EVs populations free from ribonucleoproteins and other soluble contaminants, however, different EVs types with a similar size could co-elute (20). While, ultrafiltration can also efficiently remove soluble components from EVs preparations, the similarly sized particles (both membrane vesicles and protein aggregates) will co-purify (21). An alternative approach that is increasingly being applied is the use of co-precipitants such as polyethylene glycol combined with low-speed centrifugation to aggregate and pellet exosomes for subsequent processing (22). However, while precipitation techniques have generally very high exosomes recovery rates, they also co-precipitate various proteins (23). Finally, immunoprecipitation techniques utilize antibodies against certain proteins located on the surface of EVs and can specifically isolate CD63, CD9, and CD81 positive exosomes (24). However, large-scale separation of exosomes with immunoprecipitation is challenging due to their highly diluted state in the biological fluids.
While each of the above-mentioned approaches harnesses certain differences in biophysical or molecular properties of EVs (including the size, the density, and the content of the surface proteins), neither method can recover a pure material and allows only an enrichment for certain subpopulations of EVs in a sample (15, 25). As a result, the characterization of EV type-specific RNA cargo remains highly challenging and strongly depends on the purification method. In addition, the bovine serum that is used as a component of most cell culture media could be a source of contaminating extracellular RNAs in a sample that mask human-derived RNA species having a sequence similar to bovine RNAs (e.g., miR-122) (26). The accuracy of the subsequent analysis of EV-associated transcriptome is also highly dependent on an RNA qualification method, including a DNA library preparation for deep sequencing (15). For instance, widely used commercial kits for RNA sequencing, by default, capture only 5′-phosphorylated 3′-OH short RNA molecules representing only a fraction of total RNA in the sample (27). Likewise, most whole-transcriptome sequencing techniques can incorporate only relatively long RNAs and, thus, overlook small RNAs.
Finally, certain cell types (e.g., cells of retinal pigmented epithelium and the intestine) exhibit a membrane polarity (Figure 1B). Therefore, EVs secreted by such cells might have distinct properties and molecular content depending on whether they derive from basal or apical parts of the membranes (28). While MVs and exosomes of polar cells have not yet been properly studied, the differences in structure, size, and lipid composition of apical and basal membranes could determine the features of the secreted EVs (28). The polarized trafficking machinery in certain cells suggests that additional care should be taken for isolating apical exosomes, including a careful control of the functional integrity of cell monolayers during preparation of conditioned media (29). On the contrary, an apical-only isolation approach risks missing important basolaterally released vesicles (28).
The Reported RNA Contents Within Different EVs Classes
The presence of mRNAs, miRNAs, and lncRNAs within exosomes and MVs have been consistently shown with microarrays and RT-qPCR techniques in multiple early (13, 30–36) as well as more recent reports (37–40). The application of more advanced high-throughput RNA sequencing methods revealed the presence of various other RNA species within subpopulations of EVs isolated from biological fluids and cell conditioned media (Table 1). Those RNA species include snRNA, snoRNA, piRNA, vault RNA, Y-RNA, scRNA, SRP-RNA, and 7SK-RNA; as well as short fragments originating from rRNA, tRNA, mRNA, lncRNAs, and various intergenic repeats (40–57).
In a pioneering work, Nolte-'t Hoen et al. characterized small RNA content in EVs released by the immune cells in culture using deep sequencing. Interestingly, the majority of total RNA isolated from EVs consisted of small RNAs (<200 nt), with minor amounts of 18S and 28S rRNA. Those short RNA fragments were primarily mapped to protein-coding regions and genomic repeats including SINE, LINE, and LTR sequences (Table 1). On the contrary, the majority of sequences present in the cellular small RNA population represented miRNAs, while the proportion of miRNAs in the daughter EVs was dramatically lower. Besides protein coding mRNA and repeats, the EV fractions contained all types of structural RNAs (such as vault RNA, Y-RNA, snRNA, snoRNA, SRP-RNA, and tRNA) as well as fragments deriving from lncRNAs and pseudogenes. Furthermore, many of the small non-coding transcripts were enriched in EVs relative to cellular RNA, indicated that cells might destine specific RNAs for extracellular release (41). A significant underrepresentation of miRNA over other RNA species in the exosomes released by various cultured cells have been also confirmed by multiple other studies (40, 42, 46, 50, 52, 55, 57). These data go in accordance with the previous observation that most individual exosomes does not carry any biologically significant number of miRNA copies (58). Nevertheless, other RNA sequencing experiments indicated that a significant proportion of small RNA-seq reads still correspond to miRNA in exosomes released by some cell lines (51, 53, 54, 57). Interestingly, several independent groups have observed a significant enrichment (15–50% of total reads) of RNA fragments mapped to genomic repeats comprising retroviral sequences, LTR, SINE, and LINE sequences (41, 42, 47, 50, 55). It has to be mentioned that the authors did not specify whether the small RNA library preparation protocols used in the above studies included the modifications to allow capturing 5′-OH and/or 3′-phosphorylated RNAs. Therefore, it remains unclear whether they actually characterized the full spectrum of small RNA in the corresponding EVs.
The sequencing of total (both long and small) RNAs in the EVs was reported by Jenjaroenpun et al. (46) and Miranda et al. (49) in the EVs present in conditioned media from MDA-MB cells and the urine, respectively, and showed a significant proportion of rRNA reads (87–97%) that was similar to the rRNA content in the cytoplasm. Out of the remaining 3–13% reads, approximately half was mapped to protein-coding transcripts while another half aligned to non-coding RNAs and genomic repeats. In another report by Beradrocco et al. the authors used both total RNA and small RNA sequencing protocols separately to characterize a spectrum of long RNAs encapsulated within the EVs released by four different liver cancer cell lines (55). The largest proportion (32–66%) of total RNA reads were mapped to rRNA, while 15–44% corresponded to the genomic repeats, and only 11–25% of reads were mapped to protein-coding and non-coding RNA genes. The small RNA sequencing performed on the same EVs preparations revealed only a slightly different distribution of RNA classes: rRNA (16–54%), genomic repeats (24–40%), and transcriptome (24–51%) (55). In another whole-transcriptome RNA-seq study paralleled with small RNA sequencing, Lasser et al. demonstrated that human mast and erythroleukemic cell lines release two exosomes populations (as separated by flotation on a density gradient into HD and LD fractions) (40). A clear lack of correlation between both long and short RNA cargo in HD and LD fractions suggests that extracellular RNA in these two fractions are associated with distinct pathways. Thus, reads mapped to mRNA transcripts were more abundant percentage-wise in the HD as compared to LD exosomes (75 vs. 20%), while the distribution of non-coding RNA reads was opposite (25 vs. 80%). In short RNA libraries, the HD fractions were enriched in mature miRNA (23%), while the LD fractions were dominated by tRNA (28%), and mature miRNA (10%) (40).
Another study investigated RNA content in three separate EVs types released by melanoma cells in culture and identified some non-coding RNAs to be enriched in every EV samples (53). Thus, RNA profiles indicated the presence of prominent 18S and 28S rRNA peaks in ABs and MVs with relatively moderate levels of small RNA. By contrast, exosomes contained predominantly small RNA and much less rRNA as compared to both ABs and MVs (53). Interestingly, a similar number of different miRNAs have been identified in every EV type. Overall, a close relationship between miRNA profiles was found in ABs and MVs (R = 0.91), MVs and exosomes (R = 0.86), as well as MVs and parental cells (R = 0.86). While a less strong correlation was found between ABs and exosomes (R = 0.79) and exosomes and cells (R = 0.75). Despite the fact that EVs subsets were different only to a minor degree from the aspect of their miRNA cargo, a significant number of miRNAs were detected only in exosomes and were absent in both ABs and MVs, supporting the concept of specific RNA loading into exosomes. It has to be mentioned that other ncRNA species were not only significantly more abundant as compared to miRNA but also selectively enriched in different EVs subtypes released by melanoma cells, which adds another level of complexity to investigating extracellular vesicle RNA cargo and its function (53).
Only a few reports have so far investigated small RNA cargo in EVs isolated from human biological fluids with next generation sequencing (44, 45, 48). These studies indicated that exosomes isolated from human plasma, saliva, and urine contained a significant proportion of miRNA reads (35–76% of total). The rest RNA species in the EVs from the above mentioned biofluids included fragments of rRNA, lncRNAs, tRNA, mRNA, repeated regions as well as small noncoding RNA such as piRNA, snRNA, and snoRNA. It is important to mention that exosome isolations from biofluids may contain much higher amounts of large protein aggregates, including miRNA-loaded AGO complexes that are normally released upon cell death (59), as compared to “few-days” cell conditioned media. Therefore, it remains to be validated whether the miRNAs detected in human biofluids were indeed associated with the EVs. Interestingly, deep sequencing of total RNA purified from urea exosomes (49) revealed drastically different transcripts distribution than that observed by Cheng et al. (48). Specifically, a substantial proportion (~87%) of total RNA reads was mapped to rRNA and only about 8% of reads aligned to non-coding RNA and DNA repeats, while the remaining ~5% of reads corresponded to protein-coding RNA (49). Conversely, the mapping statistics and reads distribution reported by Miranda et al. were similar to those obtained upon total RNA sequencing of exosomes from cell conditioned media (46, 55).
To conclude, the collective evidence evolving from the above mentioned studies (Table 1) argue that EVs released by most cells indeed carry significant amounts of non-coding and protein-coding transcripts, as well as their parts, that should be considered when studying the effects of extracellular RNA on recipient cells. The differences in EVs RNA cargo content among the reported studies might be explained in part by: (1) cell type-specific RNA expression differences; (2) different EVs and RNA isolation methods; and (3) the use of different NGS library preparation protocols and sequencing platforms.
The Role of the EVs RNA Content for the Immune System
While it was consistently shown that the exchange of exosomes and microvesicles among different immune cells contribute to both adaptive and innate immune response, the impact of the EVs RNA cargo onto immune cells function remains obscure (60). Mittelbrunn et al. demonstrated that exosomes originating from T cells are loaded with certain miRNAs (e.g., miR-335) can be internalized by the antigen-presenting cells APCs at the immune synapses and that subsequently led to the reduction of target mRNA SOX4 expression (61). Likewise, miRNA transfer from one dendritic cell (DC) to another via EVs led to alterations in recipient-cell gene expression (62), and regulatory T cells reduce Th1 (CD4+ IFNg+) inflammatory responses by EV transfer of miRNA (especially let-7d) to Th1 cells (63). The discovery of the variety of different RNA species in MVs and exosomes secreted by the immune cells as well (41), added another level of complexity to the theory of cell-cell communication via cell-free RNA. In particular, very marginal levels of EVs-encapsulated miRNAs as compared to other RNA species not only question the contribution of miRNA to cell-cell communication but also suggests that other RNAs might play much more determining biological role.
Thus, previous experiments revealed that extracellular miRNA can activate Toll-like receptor (TLR) 8 signaling, which induces cytokine secretion, presumably by mimicking viral RNA (64). The TLRs are a family of innate immune system receptors which recognize various molecular patterns of microbial pathogens and induce antimicrobial immune responses (65, 66). Specifically, both free-floating AGO protein bound miRNAs and miRNA encapsulated in EVs have been hypothesized to mediate communication between immune cells via binding to extracellular or intracellular Toll-like receptors (TLRs) (64, 67). Among the major effects of the nucleic acids-mediated activation of intracellular TLRs is the induction of certain cytokines essential for the innate immune response. While multiple other reports link activation of TLR pathways and exosomes, it remains unclear whether the observed effects were indeed mediated by the encapsulated RNAs. However, the finding that intracellular TLRs located within endolysosomal compartments can bind both double-stranded and single-stranded nucleic acids derived from viruses and bacteria (68) strongly suggest that various RNA classes incorporated within the EVs could also activate the corresponding TLRs. Due to the largely sequence-independent impact of nucleic acids on the TLRs, it is feasible that more abundant non-miRNA classes could significantly contribute to such activation. Overall, it remains feasible that combined interactions of vesicular RNAs and TLRs within and between diverse immune and non-immune cells could contribute to the regulation of the complex nexus of immune responses.
Conclusion and Future Perspectives
Massive parallel sequencing has enabled characterization of the whole spectrum of nucleic acids in a given sample, and was consistently applied to demonstrate the presence of the complex RNA cargo within EVs populations released by various cells. Interestingly, the intravesicular miRNAs (which were well-documented previously using microarray and qPCR-based methods) represented only a very marginal proportion as compared to other RNA species including various small non-coding RNAs, lncRNAs, and mRNA fragments. A putative biological impact of the EVs-associated transcriptomes remains to be validated; however, multiple studies indicated that, at least, exosomal miRNA could mediate communication among various cell types including the immune cells. In addition, EVs-encapsulated miRNAs have been shown to serve as highly specific biomarkers for various pathological conditions and correlate with the presence of malignant tumors. Indeed, exosomes carrying a tumor-specific miRNA repertoire have been consistently detected in the venous blood of cancer patients and mouse models. The collective finding that non-miRNA species are in fact much more abundant in the isolated EVs populations, suggests that they could serve as even more promising non-invasive biomarkers for cancer and/or other disorders.
Author Contributions
ATo and ATu conceived the study. ATu prepared the tables and figures and wrote the manuscript. ATo and OD participated in final manuscript design, and provided experts' opinion on the content and critical revision.
Funding
The study was partially funded by the Russian Science Foundation (project 17-14-01338).
Conflict of Interest Statement
ATu is affiliated with the company SciBerg e.Kfm. ATo is affiliated with the company SRC BioClinicum.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. EL Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. (2013) 12:347–57. doi: 10.1038/nrd3978
2. Harding C, Heuser J, Stahl P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: demonstration of a pathway for receptor shedding. Eur J Cell Biol. (1984) 35:256–63.
3. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell (1983) 33:967–78. doi: 10.1016/0092-8674(83)90040-5
4. Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci. (2012) 37:460–5. doi: 10.1016/j.tibs.2012.08.003
5. Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. (1996) 183:1161–72. doi: 10.1084/jem.183.3.1161
6. Chaput N, Thery C. Exosomes: immune properties and potential clinical implementations. Semin Immunopathol. (2011) 33:419–40. doi: 10.1007/s00281-010-0233-9
7. Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic (2011) 12:1659–68. doi: 10.1111/j.1600-0854.2011.01225.x
8. Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. (2009) 9:581–93. doi: 10.1038/nri2567
9. Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. (2012) 21:R125–34. doi: 10.1093/hmg/dds317
10. Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol. (2014) 14:166–80. doi: 10.1038/nri3607
11. Atkin-Smith GK, Poon IKH. Disassembly of the dying: mechanisms and functions. Trends Cell Biol. (2017) 27:151–62. doi: 10.1016/j.tcb.2016.08.011
12. Di Vizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, et al. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. (2009) 69:5601–9. doi: 10.1158/0008-5472.CAN-08-3860
13. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. (2007) 9:654–9. doi: 10.1038/ncb1596
14. Wahlgren J, De LKT, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. (2012) 40:e130. doi: 10.1093/nar/gks463
15. Sadik N, Cruz L, Gurtner A, Rodosthenous RS, Dusoswa SA, Ziegler O, et al. Extracellular RNAs: a new awareness of old perspectives. Methods Mol Biol. (2018) 1740:1–15. doi: 10.1007/978-1-4939-7652-2_1
16. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. (2006) Chapter 3, Unit 3 22.
17. Jeppesen DK, Hvam ML, Primdahl-Bengtson B, Boysen AT, Whitehead B, Dyrskjot L, et al. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J Extracell Vesicles (2014) 3:25011. doi: 10.3402/jev.v3.25011
18. Coumans FAW, Brisson AR, Buzas EI, Dignat-George F, Drees EEE, El-Andaloussi S, et al. Methodological guidelines to study extracellular vesicles. Circ Res. (2017) 120:1632–48. doi: 10.1161/CIRCRESAHA.117.309417
19. Yuana Y, Levels J, Grootemaat A, Sturk A, Nieuwland R. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J Extracell Vesicles (2014) 3:23262. doi: 10.3402/jev.v3.23262
20. Boing AN, Van Der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles (2014) 3:23430. doi: 10.3402/jev.v3.23430
21. Vergauwen G, Dhondt B, Van Deun J, De Smedt E, Berx G, Timmerman E, et al. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci Rep. (2017) 7:2704. doi: 10.1038/s41598-017-02599-y
22. Rider MA, Hurwitz SN, Meckes DG Jr. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep. (2016) 6:23978. doi: 10.1038/srep23978
23. Gamez-Valero A, Monguio-Tortajada M, Carreras-Planella L, Franquesa M, Beyer K, Borras FE. Size-exclusion chromatography-based isolation minimally alters extracellular vesicles' characteristics compared to precipitating agents. Sci Rep. (2016) 6:33641. doi: 10.1038/srep33641
24. Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. (2016) 113:E968–77. doi: 10.1073/pnas.1521230113
25. Gardiner C, Di Vizio D, Sahoo S, Thery C, Witwer KW, Wauben M, et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles (2016) 5:32945. doi: 10.3402/jev.v5.32945
26. Shelke GV, Lässer C, Gho YS, Lötvall J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles (2014) 3:24783. doi: 10.3402/jev.v3.24783
27. Turchinovich A, Surowy H, Serva A, Zapatka M, Lichter P, Burwinkel B. Capture and amplification by tailing and switching (CATS). An ultrasensitive ligation-independent method for generation of DNA libraries for deep sequencing from picogram amounts of DNA and RNA. RNA Biol. (2014) 11:817–28. doi: 10.4161/rna.29304
28. Klingeborn M, Stamer WD, Bowes Rickman C. Polarized exosome release from the retinal pigmented epithelium. Adv Exp Med Biol. (2018) 1074:539–44. doi: 10.1007/978-3-319-75402-4_65
29. Nikulin SV, Knyazev EN, Poloznikov AA, Shilin SA, Gazizov IN, Zakharova GS, et al. Expression of SLC30A10 and SLC23A3 transporter mRNAs in Caco-2 cells correlates with an increase in the area of the apical membrane. Mol Biol. (2018) 52:667–74. doi: 10.1134/S0026893318040131
30. Skog J, Wurdinger T, Van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. (2008) 10:1470–6. doi: 10.1038/ncb1800
31. Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE (2010) 5:e13515. doi: 10.1371/journal.pone.0013515
32. Xiao D, Ohlendorf J, Chen Y, Taylor DD, Rai SN, Waigel S, et al. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE (2012) 7:e46874. doi: 10.1371/journal.pone.0046874
33. Mitsuhashi M, Taub DD, Kapogiannis D, Eitan E, Zukley L, Mattson MP, et al. Aging enhances release of exosomal cytokine mRNAs by Abeta1-42-stimulated macrophages. FASEB J. (2013) 27:5141–50. doi: 10.1096/fj.13-238980
34. Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. (2013) 4:2980. doi: 10.1038/ncomms3980
35. Kogure T, Yan IK, Lin WL, Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer (2013) 4:261–72. doi: 10.1177/1947601913499020
36. Gezer U, Ozgur E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. (2014) 38:1076–9. doi: 10.1002/cbin.10301
37. Van Der Vos KE, Abels ER, Zhang X, Lai C, Carrizosa E, Oakley D, et al. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro Oncol. (2016) 18:58–69. doi: 10.1093/neuonc/nov244
38. Willms E, Johansson HJ, Mager I, Lee Y, Blomberg KE, Sadik M, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. (2016) 6:22519. doi: 10.1038/srep22519
39. Ahadi A, Brennan S, Kennedy PJ, Hutvagner G, Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep. (2016) 6:24922. doi: 10.1038/srep24922
40. Lasser C, Shelke GV, Yeri A, Kim DK, Crescitelli R, Raimondo S, et al. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. (2017) 14:58–72. doi: 10.1080/15476286.2016.1249092
41. Nolte-'T Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, Hoen TPA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. (2012) 40:9272–85. doi: 10.1093/nar/gks658
42. Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. (2012) 40:10937–49. doi: 10.1093/nar/gks832
43. Guduric-Fuchs J, O'connor A, Camp B, O'neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics (2012) 13:357. doi: 10.1186/1471-2164-13-357
44. Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics (2013) 14:319. doi: 10.1186/1471-2164-14-319
45. Ogawa Y, Taketomi Y, Murakami M, Tsujimoto M, Yanoshita R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol Pharm Bull. (2013) 36:66–75. doi: 10.1248/bpb.b12-00607
46. Jenjaroenpun P, Kremenska Y, Nair VM, Kremenskoy M, Joseph B, Kurochkin IV. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ (2013) 1:e201. doi: 10.7717/peerj.201
47. Li CC, Eaton SA, Young PE, Lee M, Shuttleworth R, Humphreys DT, et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol. (2013) 10:1333–44. doi: 10.4161/rna.25281
48. Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. (2014) 86:433–44. doi: 10.1038/ki.2013.502
49. Miranda KC, Bond DT, Levin JZ, Adiconis X, Sivachenko A, Russ C, et al. Massively parallel sequencing of human urinary exosome/microvesicle RNA reveals a predominance of non-coding RNA. PLoS ONE (2014) 9:e96094. doi: 10.1371/journal.pone.0096094
50. Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Perez Lanzon M, Zini N, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther. (2015) 6:127. doi: 10.1186/s13287-015-0116-z
51. Van Balkom BW, Eisele AS, Pegtel DM, Bervoets S, Verhaar MC. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J Extracell Vesicles (2015) 4:26760. doi: 10.3402/jev.v4.26760
52. Tosar JP, Gambaro F, Sanguinetti J, Bonilla B, Witwer KW, Cayota A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. (2015) 43:5601–16. doi: 10.1093/nar/gkv432
53. Lunavat TR, Cheng L, Kim DK, Bhadury J, Jang SC, Lasser C, et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells–Evidence of unique microRNA cargos. RNA Biol. (2015) 12:810–23. doi: 10.1080/15476286.2015.1056975
54. Ren J, Zhou Q, Li H, Li J, Pang L, Su L, et al. Characterization of exosomal RNAs derived from human gastric cancer cells by deep sequencing. Tumour Biol. (2017) 39:1010428317695012. doi: 10.1177/1010428317695012
55. Berardocco M, Radeghieri A, Busatto S, Gallorini M, Raggi C, Gissi C, et al. RNA-seq reveals distinctive RNA profiles of small extracellular vesicles from different human liver cancer cell lines. Oncotarget (2017) 8:82920–39. doi: 10.18632/oncotarget.20503
56. Conley A, Minciacchi VR, Lee DH, Knudsen BS, Karlan BY, Citrigno L, et al. High-throughput sequencing of two populations of extracellular vesicles provides an mRNA signature that can be detected in the circulation of breast cancer patients. RNA Biol. (2017) 14:305–16. doi: 10.1080/15476286.2016.1259061
57. Sork H, Corso G, Krjutskov K, Johansson HJ, Nordin JZ, Wiklander OPB, et al. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci Rep. (2018) 8:10813. doi: 10.1038/s41598-018-28485-9
58. Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA. (2014) 111:14888–93. doi: 10.1073/pnas.1408301111
59. Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. (2011) 39:7223–33. doi: 10.1093/nar/gkr254
60. Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. (2017) 27:172–88. doi: 10.1016/j.tcb.2016.11.003
61. Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. (2011) 2:282. doi: 10.1038/ncomms1285
62. Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood (2012) 119:756–66. doi: 10.1182/blood-2011-02-338004
63. Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T, et al. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity (2014) 41:89–103. doi: 10.1016/j.immuni.2014.05.019
64. Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. (2012) 109:E2110–6. doi: 10.1073/pnas.1209414109
65. Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. (2003) 21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126
66. Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity (2010) 32:305–15. doi: 10.1016/j.immuni.2010.03.012
67. Lehmann SM, Kruger C, Park B, Derkow K, Rosenberger K, Baumgart J, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. (2012) 15:827–35. doi: 10.1038/nn.3113
Keywords: apoptotic bodies, microvesicles, circulating RNA, next generation sequencing, exosomes, extracellular vesicle (EV)
Citation: Turchinovich A, Drapkina O and Tonevitsky A (2019) Transcriptome of Extracellular Vesicles: State-of-the-Art. Front. Immunol. 10:202. doi: 10.3389/fimmu.2019.00202
Received: 31 October 2018; Accepted: 23 January 2019;
Published: 28 February 2019.
Edited by:
Flavia Bazzoni, University of Verona, ItalyReviewed by:
Aldo Tagliabue, Institute for Genetic and Biomedical Research (IRGB), ItalyEdoardo Fiorillo, Institute for Genetic and Biomedical Research (IRGB), Italy
Copyright © 2019 Turchinovich, Drapkina and Tonevitsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrey Turchinovich, andrey.turchinovich@sciberg.com
 Andrey Turchinovich
Andrey Turchinovich Oxana Drapkina3
Oxana Drapkina3 Alexander Tonevitsky
Alexander Tonevitsky