- Department of Microbiology and Immunology, Institute of Biomedicine, University of Gothenburg, Gothenburg, Sweden
It has been almost a decade since the 2009 influenza A virus pandemic hit the globe causing significant morbidity and mortality. Nonetheless, annual influenza vaccination, which elicits antibodies mainly against the head region of influenza hemagglutinin (HA), remains as the mainstay to combat and reduce symptoms of influenza infection. Influenza HA is highly antigenically variable, thus limiting vaccine efficacy. In addition, the variable HA head occupies the upper strata of the immunodominance hierarchy, thereby clouding the antibody response toward subdominant epitopes, which are usually conserved across different influenza strains. Isolation of monoclonal antibodies from individuals recognizing such epitopes has facilitated the development of recombinant vaccines that focus the adaptive immune response toward conserved, protective targets. Here, we review some significant leaps in recombinant vaccine development, which could possibly help to overcome B cell and antibody immunodominance and provide heterosubtypic immunity to influenza A virus.
Introduction
Influenza viruses belong to the family of Orthomyxoviridae and consists of A, B, C, and D types. Types A and B are currently circulating among humans (1–4). Influenza causes significant morbidity (30–50 million cases yearly) and mortality, with infection-associated respiratory deaths in the range of 4–8.8 per 100,000 individuals, posing heavy socioeconomic burden to society (5). Annual vaccination remains as the mainstay to prevent influenza infection, but, according to Centers for Disease Control and Prevention, it is effective only in 20–70% of the population, depending on season (6). Based on antigenic and phylogenetic properties of influenza surface glycoproteins, hemagglutinin (HA), and neuraminidase (NA), there are 18 HA (H1–H18), and 11 NA (N1–N11) Influenza A virus (IAV) serotypes and two influenza B of B/Victoria and B/Yamagata lineages (7, 8). HA is further divided into two phylogenetic groups. The current seasonal flu vaccines are either trivalent or quadrivalent containing HA from circulating H1N1, H3N2, and B/Victoria lineage or both influenza B lineages (9). IAV possess an error prone RNA polymerase, which results in mutations in surface antigens, leading to antigenic drift and antibodies being no longer effective. Therefore, it is necessary to update and administer vaccines every year by forecasting the drifted strains. In addition, the annual vaccination becomes ineffective during pandemic outbreaks, in which a new viral strain of zoonotic origin acquires the ability to replicate in humans (10, 11).
HA is the most abundant glycoprotein on the influenza virion surface and is crucial for host viral entry by binding to the terminal sialic acid residues on epithelial cells, resulting in fusion of viral and host cell membranes. HA is a trimer consisting of a globular head, harboring the receptor binding site, and an elongated stem region (12). Even though stem-specific B cells and antibodies are generated during infection and vaccination, the HA head is the main target of neutralizing antibodies. However, possibly due to its immunodominance (13), the head is subjected to higher rate of evolution (2.2–4.4 times) than the stem (14, 15). Intriguingly, while in animals, at least 12 mutations are necessary to drive full escape from immune sera (16), in humans, it appears that the polyclonal response can be extremely focused on one antigenic site (17–19). For example, in a circulating span of 35 years in humans, a single amino acid substitution at only seven sites in HA head beside the receptor binding site (RBS) was enough to drive major antigenic change in H3N2 (17, 20). HA stem, as a target for universal influenza vaccine, has gained enormous traction in recent years. One could argue that the stem region is inaccessible to B cells and antibodies (21). However, a study using a broad neutralizing antibody showed that nearly 75% of the HA on pandemic H1N1 is bound by a stem-specific mAb (22). There is an urgent need to introduce universal vaccines, targeting conserved regions and providing lifelong protection. This review focuses on possible strategies for developing universal influenza vaccines, mainly based on HA. Such strategies are summarized in Figure 1.
Figure 1. Summary of some promising strategies currently used to elicit broadly cross-reactive anti-HA B-cell responses.
Hemagglutinin Stem—A Promising Universal Vaccine Target
HA stem has been an important candidate for development of universal vaccines because the stalk region is relatively conserved and evolves much slower and accommodate less amino acid substitutions as compared to the head domain. This could be due to minimal antibody pressure from low amount of circulating anti-stem antibodies (23, 24) and low tolerance to mutations in the stalk domain, which can lead to loss of viral fitness (25, 26), even though partial escape mutations in the stem can be generated (27). However, amino acid substitutions in the stalk have been reported to minimally affect the neutralization capacity of human cross-reactive, anti-stalk monoclonal antibodies (14, 28).
HA stem antibodies protect by (i) preventing viral entry by blocking the fusion of host cell membrane and viral membrane (29), (ii) reducing viral egress by blocking neuraminidase activity through steric hindrance (30–32), and (iii) FcR-mediated induction of antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis and reactive oxygen species production (33–35). Several human-derived broadly neutralizing anti-stem antibodies have been identified against either influenza group 1 (36–39) or group 2 (38, 40–42) or both groups (40, 43–50) or even against both influenza A and B subtypes (51). The identification of these antibodies was an incentive to develop vaccines, which are discussed below.
Hemagglutinin Stem—Human Immune Responses
In humans, memory B cells (Bmem), and antibodies against HA stem are subdominant and present in low levels. Analysis of serum samples from 202 healthy individuals collected between 2004 and 2010 revealed that anti-stem antibodies of group 1 specificity is found in 84% of the population (52); however, their level, as measured in human Intravenous Immunoglobulin preparations, is very low (23).
Ellebedy and colleagues (53) found that immunization with H5N1, which is not currently circulating in humans, boosted cross-reactive antibody response toward HA stem, when compared to seasonal vaccines. Because of the existence of lower levels of H5 head-specific Bmem as compared to stem-specific Bmem, H5N1 vaccination led to recruitment of stem-specific Bmem, their expansion, and antibody production. On the contrary, boosting with the same HA favors anti-head responses (53). Another study found that nearly 6 out of 10 individuals have Bmem specific between group 1 and 2 HA (50). Indeed, it appears that baseline levels of H5–H1 cross-reactive Bmem and H1-specific Bmem are no more than 2-fold different (54). However, after priming with an H5 DNA plasmid vaccine and boosting with A/Indonesia/05/2005 monovalent-inactivated virus, both head and stem Bmem were expanded but only head-specific Bmem persisted, while stem-specific Bmem expanded and contracted rapidly (50, 54). Finally, Andrews et al. observed that immunization with group 1 virus (H5N1) elicited anti-stem memory responses exclusively against group 1, while group 2 (H7N9) induced high levels of cross-protective anti-stem memory B cell responses with diverse repertoire despite a lower overall response. This study in humans suggests the potential of group 2 based vaccines to provide a broader protection as compared to group 1 (55). While all these studies highlight the ability of individuals to generate stem-specific Bmem and plasmablasts, they all note a rapid contraction of stem-specific cells. This disconnect between cell numbers, longevity, and serum antibodies highlight the complexity of B-cell fate decision. Understanding how antigen specificity can influence cell differentiation is a crucial challenge for next generation vaccines.
Sites Of Vulnerability In Hemagglutinin Head
Although HA stem is an excellent candidate for the development of universal vaccines, anti-HA stem titers in human correlates only with reduced viral shedding but do not predict the severity of influenza infection (56, 57). A recent study in humans indicated that a 2-fold increase in hemagglutination inhibition titer gave a 23.4% reduction in H1N1 infection risk, while the same increase in HA-stem-specific antibodies conferred only 14.2% reduction (58).
The globular head of the HA is target for most of the neutralizing antibodies, which prevents the viral entry by blocking the binding of RBS to sialic acid residues on host cell membrane (59). RBS is a shallow depression on HA head and consists of 130 loop, 150 loop, 190 helix, which are relatively conserved, and 220 loop, which is diverse among IAV subtypes (60, 61). Amino acid substitutions in the RBS determine host tropism, and specific substitutions are connected to altered receptor binding within subtype (62). Some RBS binding, broadly neutralizing antibodies have been identified, such as C05, S139/1, and F045-092, which neutralize within groups; CH65, 5J8, 2G1, and H5.3, which neutralize within subtype (63–71); and C12G6 and CR8033, which neutralize both influenza B lineages (51, 72).
Apart from RBS, broadly neutralizing antibodies have been identified against other conserved sites on HA head (73). An antibody (F005-126) which neutralizes 12 H3N2 subtypes by occupying the cleft formed by two HA head monomers and cross-linking them is known to prevent viral entry by blocking pH-induced HA conformational change (74). Bajic et al. found that subdominant antibodies can target an occluded epitope located on the lateral surface on HA head between two monomers using an H3 immunogen, hyperglycosylated on dominant epitopes. These antibodies protected against H3N2 challenge in an Fc-dependent manner (75). Similarly, two independent studies identified broadly neutralizing antibody, which bind hidden epitopes at HA trimer interface. These antibodies do not neutralize the virus but are suspected to disrupt the HA trimer integrity. Passive transfer experiments revealed that they protect mice from groups 1 and 2 viruses by preventing cell-to-cell viral spread or by FcγR or complement mediated effector mechanism (76, 77). HA exhibits “breathing” phenomenon at neutral or low pH where reversible separation of HA monomers exposes hidden epitopes to these specific antibodies (78–80). Vestigeal esterase domain is another possible HA target; it is located at the base of the HA head and can be target of broadly protective antibodies, which protect within subtypes (81, 82) and both lineages of influenza B virus (83). Like stem-directed antibodies, they protect through various mechanisms such as blocking viral egress, fusion, or by ADCC.
For most of the epitopes described above, we are just at the first step of the reverse vaccinology pipeline. However, there is hope that using some advanced de novo protein design tools, we will be able to create effective immune-focusing antigens (84, 85).
Bioengineering Antigens To Refocus Immune Responses
Headless Hemagglutinins
One of the obvious ways to increase the anti-stem antibody response is to remove HA head to circumvent its immunodominance (13, 86). Even though this sounds simple, removing the head and the transmembrane domain may cause conformational changes in the stem leading to loss of recognition by anti-stem antibodies (87). One of many successful attempts was achieved by creating stable trimeric mini-HA from H1N1, which retained similar binding affinity to two broadly neutralizing antibodies, CR9114 and CR6261, as that of full-length HA. When used to immunize non-human primates, the elicited antibodies competed with CR9114, induced ADCC, and neutralized H5N1 virus (88). Another study used H1HA10-Foldon and H3HA10-Foldon mini-HA, which generated neutralizing antibodies cross-reactive to both groups 1 and 2 IAV in vitro but with limited cross-group protection in vivo (89, 90). In contrast to these mini-HAs, an H5-based mini-HA produced and purified from Escherichia coli elicited anti-stem antibody responses and protection against both IAV subtypes (91). Pre-existing immunity plays a role in subsequent immune response to viral infection and vaccination (92). When tested in non-human primates who have been exposed to flu infection, mini-HA were shown to induce higher cross-protective antibody response as compared to seasonal trivalent inactivated vaccine, indicating their potential as a universal vaccine (93).
Ferritin is an iron storage protein which self-assembles into a nanoparticle consisting of 24 repetitive polypeptides, which can induce strong immune response and antigen presentation (94). Based on this, Yassine et al. engineered a nanoparticle containing intact HA stem from H1 (H1-ss-np), which generated anti-stem response almost 2-fold higher than that of trivalent inactivated vaccine. When immunized with H1-ss-np, mice and ferrets elicited broad antibody response against group 1 IAV as well as group 2 IAV, demonstrating heterosubtypic protection (95). Based on this, influenza H1 stabilized stem ferritin vaccine (H1ssF_3928) has entered a phase I ongoing clinical trial to evaluate its tolerability and immunogenicity in healthy adults. However, a thermostable and immunogenic nanoparticle vaccine containing group 2 H3 and H7 stem conferred only protection against homosubtypic viral challenge. Given the fact that these stem immunogens activates IgM BCR of unmutated common ancestor of the cross-reactive human anti-stem antibodies, the authors speculate that the lack of cross-group protection in vivo might be due to difference in BCR repertoire in mice and human (96).
Chimeric Hemagglutinin
Chimeric HAs (cHA) are full-length HA with stem derived from human viruses and globular head from distinct, exotic HAs. Based on this concept, repeated immunization with cHA with head from different flu subtype and same H1 HA stem induced high titers of stem-reactive antibodies against homologous and heterologous viruses (97). Several such cHA constructs, which confer protection by eliciting stem Abs, have been developed for group 1, group 2, and Influenza B viruses, with some inducing long-lasting antibodies (98–105). An interim report on a clinical trial using a cHA prime boost strategy was recently released (106). In this study, healthy volunteers, with measurable baseline H1-stalk antibody levels, were boosted with cHAs. The sharpest stem-antibody level increase was obtained when challenging with cH8/1N1 in AS03 adjuvant intramuscularly. However, further immunization with other cHA did not additionally boost stem responses. To build upon these promising findings, more studies are needed to assess the longevity of these responses and their stability upon natural infection or seasonal immunization.
Immune-Focusing Strategies
N-linked glycosylation on HA has been known to stabilize HA and shield virus from host immune response (107). Creating monoglycosylated HA or unmasking HA-stem of N-glycans could induce cross-strain neutralizing anti-stem antibodies (108, 109). Conversely, hyperglycosylating the HA head can also refocus response on stem (110). A recent study used “protect, modify, and deprotect” method to focus antibody response toward a stem epitope. In this strategy, after masking target epitope on stem with a monoclonal antibody, the surrounding undesired epitopes are chemically modified to reduce their antigenicity (111). On the other hand, other residues, outside stem, can also influence the neutralizing sensitivity to anti-stem antibodies by affecting HA-stalk stability and antibody access to stem epitopes (112).
Vaccine Engineering for Cross-Protection
Kanekiyo et al. used a ferritin nanoparticle displaying a repetitive array of HA hyper variable receptor binding domains (RBD from different H1N1 strains). Using this, they could subvert B cell response from strain-specific immunodominant epitopes to conserved, shared epitopes. Since cross-reactive B cells can recognize conserved epitope from an heterogenous array of RBD, these cells get preferentially activated due to avidity advantage and could selectively activate pre-existing cross-reactive Bmem. One of the antibodies generated after immunization, 441D6, binds to a conserved epitope, which lies opposite to RBS and protects against H1N1 strains spanning from 1977 to 2009 (113, 114). Anderson et al. generated cross-reactive antibody responses by injecting a mix of DNA vaccines containing HA genes from six members of group 1 IAV, which was further enhanced by inclusion of an antigen presenting cell targeting unit specific for MHCII, thus favoring BCR cross-linking (115). Another strategy to elicit broadly reactive antibodies within IAV subtypes is to use computationally optimized broadly reactive antigens (COBRA), which are displayed on virus like particles or expressed in live attenuated influenza vaccine. This approach uses multiple rounds of consensus generation to aggregate HA epitopes of IAV from different time periods. Such vaccines elicit polyclonal B cell responses and was shown to protect within subtypes (116–123). Combining several COBRA vaccines could confer heterosubtypic protection.
M2e-Based Vaccines
M2 is an ion channel with its ectodomain (M2e) exposed on virion and infected cell surfaces (124). M2e is quite conserved across IAV; therefore, it has historically been considered as an ideal universal vaccine candidate (125). The mechanism of M2e-mediated protection is debated with both antibodies and T cells being important players (126–128). Several approaches have been undertaken to increase M2e immunogenicity, using VLP or different adjuvants (129–131). Of note, it appears that M2e antibodies act via effector functions and thus are infection permissive, making M2e vaccines more suitable when used in combination with others.
Neuraminidase—The Emerging Player
IAV NA as vaccine target has been neglected for decades, despite early discovery of potent anti-viral activity of NA antibodies (132). Even more surprisingly, NA amount in licensed vaccines varies enormously and is not checked by manufactures or regulatory authorities (133). Exciting new studies strongly point to a major role for anti-NA antibodies in protecting from disease and as the best correlates of protection (56, 134, 135). Critically, Chen et al. identified a number of human NA antibodies that cross-protected mice in therapeutic and prophylactic setting (136). Even more promising, several broadly neutralizing anti-NA mAbs have been isolated from an infected patient. These mAbs, directed to NA active site, demonstrated an unusual breadth in binding several IAV and IBV NA and mediating cross-neutralization and cross-protection in vivo (137). Still, despite some early studies, we do not know enough about NA antigenicity and the immunodominance of its antigenic sites (138–141). By applying some of the methods that allowed us to study in detail anti-HA responses, we should be able to break down anti-NA responses and identify promising universal vaccine candidates.
Conclusions—Know What We Do Not Know
Bioengineering and design of epitope-focused immunogens is proceeding at an incredible speed in influenza and other fields. Several promising immunogens are now in clinical trials and, hopefully, will be available to the public soon, as long-lasting universal vaccines. It is, however, crucial to understand more about the basics of B cell responses to interpret results and inform on vaccination policies.
Introduction of pandemic H1N1 2009 virus showed that most individuals, with low serological anti-stem antibodies, were able to mount a stem-directed response, but repeated vaccinations skewed the immune response back to the immunodominant head (21). It will be critical to understand when, in which order and how often give universal vaccines to appropriately boost stem response. Andrews et al. demonstrated that novel B cells specific for variable epitopes have a different phenotype compared to reactivated Bmem specific for stem (142). To maximize success, efforts will need to be put in understanding how B-cell specificity can influence their programming and differentiation. Furthermore, it is still unclear how much of stem-specific antibodies are required for optimal protection from a drifted or heterologous virus. Table 1 summarizes what we know about antibody responses to the major targets on IAV and how seasonal vaccination is able to boost those responses.
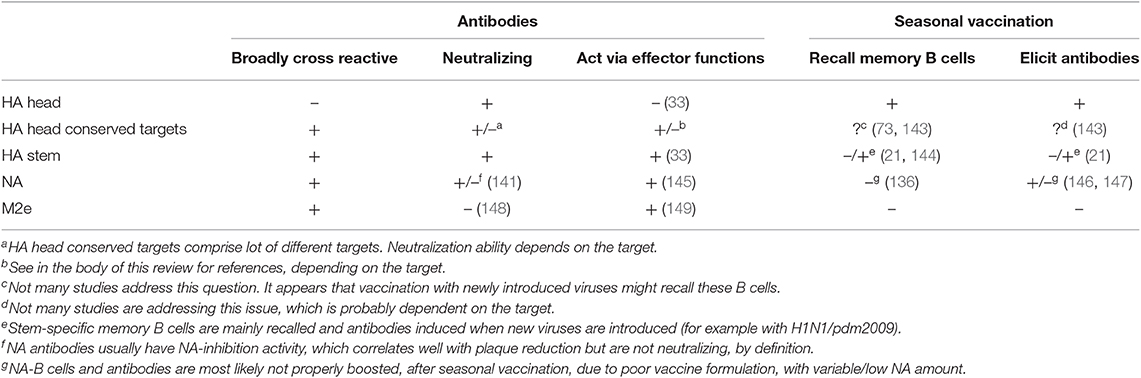
Table 1. Characteristics of antibody responses to current universal vaccine targets and ability of seasonal vaccination to recall memory B cells and specific antibodies.
Finally, but not less important, new vaccines platforms are constantly being tested. RNA-based vaccines have shown exciting results when expressing influenza proteins, at least in animals [reviewed in (150)]. Some of the engineered vaccines discussed in this review could be delivered as RNA vaccines, alone or in combination, possibly avoiding clearance from pre-existing antibodies. Other novel, slow-release, vaccine formulations could help refocusing immune responses to subdominant targets (151–153).
We are just entering an exciting season of clinical trials, and while expectations are really high, we should not be discouraged if some of the early attempts fail but rather persevere in the quest for a universal and long-lasting vaccine.
Author Contributions
NM and DA researched the literature and wrote the manuscript.
Funding
This work was supported by the Swedish Research Council, Vetenskapsrådet (2017-01439), the Jeanssons Foundations (JS2018-0011), and the Institute of Biomedicine at the University of Gothenburg.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
2. Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. (2013) 9:e1003176. doi: 10.1371/journal.ppat.1003176
3. Collin EA, Sheng Z, Lang Y, Ma W, Hause BM, Li F. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J Virol. (2015) 89:1036–42. doi: 10.1128/JVI.02718-14
4. Jackson D, Elderfield RA, Barclay WS. Molecular studies of influenza B virus in the reverse genetics era. J Gen Virol. (2011) 92(Pt 1):1–17. doi: 10.1099/vir.0.026187-0
5. Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, et al. Estimates of global seasonal influenza-associated respiratory mortality:a modelling study. Lancet. (2018) 391:1285–300. doi: 10.1016/S0140-6736(17)33293-2
6. Belongia EA, Skowronski DM, McLean HQ, Chambers C, Sundaram ME, De Serres G. Repeated annual influenza vaccination and vaccine effectiveness:review of evidence. Exp Rev Vaccines. (2017) 16:1–14. doi: 10.1080/14760584.2017.1334554
7. Hutchinson EC. Influenza Virus. Trends Microbiol. (2018) 26:809–810. doi: 10.1016/j.tim.2018.05.013
8. Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. (1990) 175:59–68. doi: 10.1016/0042-6822(90)90186-U
9. Grohskopf LA, Alyanak E, Broder KR, Walter EB, Fry AM, Jernigan DB. Prevention and control of seasonal influenza with vaccines:recommendations of the advisory committee on immunization practices - United States, 2019–20 influenza season. MMWR Recomm Rep. (2019) 68:1–21. doi: 10.15585/mmwr.rr6803a1
10. Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov. (2015) 14:167–82. doi: 10.1038/nrd4529
11. Past Pandemics. Centers for Disease Control and Prevention, National Center for Immunization and Respiratory Diseases (NCIRD). (2018). Available online at: https://www.cdc.gov/flu/pandemic-resources/basics/past-pandemics.html
12. Gaymard A, Le Briand N, Frobert E, Lina B, Escuret V. Functional balance between neuraminidase and haemagglutinin in influenza viruses. Clin Microbiol Infect. (2016) 22:975–83. doi: 10.1016/j.cmi.2016.07.007
13. Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, et al. Defining B cell immunodominance to viruses. Nat Immunol. (2017) 18:456–63. doi: 10.1038/ni.3680
14. Kirkpatrick E, Qiu X, Wilson PC, Bahl J, Krammer F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci Rep. (2018) 8:10432. doi: 10.1038/s41598-018-28706-1
15. Angeletti D, Yewdell JW. Understanding and manipulating viral immunity:antibody immunodominance enters center stage. Trends Immunol. (2018) 39:549–61. doi: 10.1016/j.it.2018.04.008
16. Das SR, Hensley SE, Ince WL, Brooke CB, Subba A, Delboy MG, et al. Defining influenza A virus hemagglutinin antigenic drift by sequential monoclonal antibody selection. Cell Host Microbe. (2013) 13:314–23. doi: 10.1016/j.chom.2013.02.008
17. Chambers BS, Parkhouse K, Ross TM, Alby K, Hensley SE. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014–2015 influenza season. Cell Rep. (2015) 12:1–6. doi: 10.1016/j.celrep.2015.06.005
18. Huang KY, Rijal P, Schimanski L, Powell TJ, Lin TY, McCauley JW, et al. Focused antibody response to influenza linked to antigenic drift. J Clin Invest. (2015) 125:2631–45. doi: 10.1172/JCI81104
19. Lee JM, Eguia R, Zost SJ, Choudhary S, Wilson PC, Bedford T, et al. Mapping person-to-person variation in viral mutations that escape polyclonal serum targeting influenza hemagglutinin. Elife. (2019) 8:e49324. doi: 10.7554/eLife.49324
20. Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. (2013) 342:976–9. doi: 10.1126/science.1244730
21. Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. (2015) 7:316ra192. doi: 10.1126/scitranslmed.aad0522
22. Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, et al. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci USA. (2013) 110:4592–7. doi: 10.1073/pnas.1214913110
23. Sui J, Sheehan J, Hwang WC, Bankston LA, Burchett SK, Huang CY, et al. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis. (2011) 52:1003–9. doi: 10.1093/cid/cir121
24. Hoa LNM, Mai LQ, Bryant JE, Thai PQ, Hang NLK, Yen NTT, et al. Association between Hemagglutinin Stem-Reactive Antibodies and Influenza A/H1N1 Virus Infection during the 2009 Pandemic. J Virol. (2016) 90:6549–56. doi: 10.1128/JVI.00093-16
25. Doud MB, Bloom JD. Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses. (2016) 8:155. doi: 10.3390/v8060155
26. Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci USA. (2013) 110:20248–53. doi: 10.1073/pnas.1320524110
27. Anderson CS, Ortega S, Chaves FA, Clark AM, Yang H, Topham DJ, et al. Publisher correction:natural and directed antigenic drift of the H1 influenza virus hemagglutinin stalk domain. Sci Rep. (2018) 8:276. doi: 10.1038/s41598-017-17926-6
28. Shafieyan Y, Hinz B, Doud MB, Lee JM, Bloom JD. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat Commun. (2018) 9:1386. doi: 10.1038/s41467-018-03665-3
29. Varecková E, Mucha V, Wharton SA, Kostolanský F. Inhibition of fusion activity of influenza A haemagglutinin mediated by HA2-specific monoclonal antibodies. Arch Virol. (2003) 148:469–86. doi: 10.1007/s00705-002-0932-1
30. Chen YQ, Lan LY, Huang M, Henry C, Wilson PC. Hemagglutinin stalk-reactive antibodies interfere with influenza virus neuraminidase activity by steric hindrance. J Virol. (2019) 93:e01526–18. doi: 10.1128/JVI.01526-18
31. Kosik I, Angeletti D, Gibbs JS, Angel M, Takeda K, Kosikova M, et al. Neuraminidase inhibition contributes to influenza A virus neutralization by anti-hemagglutinin stem antibodies. J Exp Med. (2019) 216:304–16. doi: 10.1084/jem.20181624
32. Wohlbold TJ, Chromikova V, Tan GS, Meade P, Amanat F, Comella P, et al. Hemagglutinin stalk- and neuraminidase-specific monoclonal antibodies protect against Lethal H10N8 influenza virus infection in mice. J Virol. (2016) 90:851–61. doi: 10.1128/JVI.02275-15
33. DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med. (2014) 20:143–51. doi: 10.1038/nm.3443
34. DiLillo DJ, Palese P, Wilson PC, Ravetch JV, DiLillo DJ. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest. (2016) 126:605–10. doi: 10.1172/JCI84428
35. Mullarkey CE, Bailey MJ, Golubeva DA, Tan GS, Nachbagauer R, He W, et al. Broadly neutralizing hemagglutinin stalk-specific antibodies induce potent phagocytosis of immune complexes by neutrophils in an Fc-dependent manner. MBio. (2016) 7:e01624–16. doi: 10.1128/mBio.01624-16
36. Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. (2009) 16:265–73. doi: 10.1038/nsmb.1566
37. Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. (2009) 324:246–51. doi: 10.1126/science.1171491
38. Nachbagauer R, Shore D, Yang H, Johnson SK, Gabbard JD, Tompkins SM, et al. Broadly reactive human monoclonal antibodies elicited following pandemic H1N1 influenza virus exposure protect mice against highly pathogenic H5N1 challenge. J Virol. (2018) 92:e00949–18. doi: 10.1128/JVI.00949-18
39. De Marco D, Clementi N, Mancini N, Solforosi L, Moreno GJ, Sun X, et al. A non-VH1–69 heterosubtypic neutralizing human monoclonal antibody protects mice against H1N1 and H5N1 viruses. PLoS ONE. (2012) 7:e34415. doi: 10.1371/journal.pone.0034415
40. Henry Dunand CJ, Leon PE, Kaur K, Tan GS, Zheng NY, Andrews S, et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Invest. (2015) 125:1255–68. doi: 10.1172/JCI74374
41. Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. (2011) 333:843–50. doi: 10.1126/science.1204839
42. Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G, et al. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci USA. (2014) 111:445–50. doi: 10.1073/pnas.1319058110
43. Sakabe S, Iwatsuki-Horimoto K, Horimoto T, Nidom CA, Le Mt, Takano R, et al. A cross-reactive neutralizing monoclonal antibody protects mice from H5N1 and pandemic (H1N1) 2009 virus infection. Antiviral Res. (2010) 88:249–55. doi: 10.1016/j.antiviral.2010.09.007
44. Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, et al. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell. (2016) 166:596–608. doi: 10.1016/j.cell.2016.05.073
45. Fu Y, Zhang Z, Sheehan J, Avnir Y, Ridenour C, Sachnik T, et al. A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve. Nat Commun. (2016) 7:12780. doi: 10.1038/ncomms12780
46. Tharakaraman K, Subramanian V, Viswanathan K, Sloan S, Yen HL, Barnard DL, et al. A broadly neutralizing human monoclonal antibody is effective against H7N9. Proc Natl Acad Sci USA. (2015) 112:10890–5. doi: 10.1073/pnas.1502374112
47. Marjuki H, Mishin VP, Chai N, Tan MW, Newton EM, Tegeris J, et al. Human monoclonal antibody 81.39a effectively neutralizes emerging influenza A viruses of group 1 and 2 hemagglutinins. J Virol. (2016) 90:10446–58. doi: 10.1128/JVI.01284-16
48. Wyrzucki A, Bianchi M, Kohler I, Steck M, Hangartner L, et al. Heterosubtypic antibodies to influenza A virus have limited activity against cell-bound virus but are not impaired by strain-specific serum antibodies. J Virol. (2015) 89:3136–44. doi: 10.1128/JVI.03069-14
49. Clementi N, De Marco D, Mancini N, Solforosi L, Moreno GJ, Gubareva LV, et al. A human monoclonal antibody with neutralizing activity against highly divergent influenza subtypes. PLoS ONE. (2011) 6:e28001. doi: 10.1371/journal.pone.0028001
50. Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell. (2016) 166:609–23. doi: 10.1016/j.cell.2016.06.043
51. Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, et al. Highly conserved protective epitopes on influenza B viruses. Science. (2012) 337:1343–8. doi: 10.1126/science.1222908
52. Yassine HM, McTamney PM, Boyington JC, Ruckwardt TJ, Crank MC, Smatti MK, et al. Use of hemagglutinin stem probes demonstrate prevalence of broadly reactive group 1 influenza antibodies in human sera. Sci Rep. (2018) 8:8628. doi: 10.1038/s41598-018-26538-7
53. Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci USA. (2014) 111:13133–8. doi: 10.1073/pnas.1414070111
54. Wheatley AK, Whittle JR, Lingwood D, Kanekiyo M, Yassine HM, Ma SS, et al. H5N1 Vaccine-elicited memory B cells are genetically constrained by the IGHV locus in the recognition of a neutralizing epitope in the hemagglutinin stem. J Immunol. (2015) 195:602–10. doi: 10.4049/jimmunol.1402835
55. Andrews SF, Joyce MG, Chambers MJ, Gillespie RA, Kanekiyo M, Leung K, et al. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol. (2017) 2:eaan2676. doi: 10.1126/sciimmunol.aan2676
56. Park JK, Han A, Czajkowski L, Reed S, Athota R, Bristol T, et al. Evaluation of preexisting anti-hemagglutinin stalk antibody as a correlate of protection in a healthy volunteer challenge with influenza A/H1N1pdm virus. MBio. (2018) 9:e02284–17. doi: 10.1128/mBio.02284-17
57. Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, et al. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. MBio. (2016) 7:e00417–16. doi: 10.1128/mBio.00417-16
58. Christensen SR, Toulmin SA, Griesman T, Lamerato LE, Petrie JG, Martin ET, et al. Assessing the protective potential of H1N1 influenza virus hemagglutinin head and stalk antibodies in humans. J Virol. (2019) 93:e02134–18. doi: 10.1128/JVI.02134-18
59. Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. (1988) 333:426–31. doi: 10.1038/333426a0
60. Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry:the influenza hemagglutinin. Annu Rev Biochem. (2000) 69:531–69. doi: 10.1146/annurev.biochem.69.1.531
61. Wu NC, Xie J, Zheng T, Nycholat CM, Grande G, Paulson JC, et al. Diversity of functionally permissive sequences in the receptor-binding site of influenza hemagglutinin. Cell Host Microbe. (2017) 21:742–53.e8. doi: 10.1016/j.chom.2017.05.011
62. Shi Y, Wu Y, Zhang W, Qi J, Gao GF. Enabling the 'host jump':structural determinants of receptor-binding specificity in influenza A viruses. Nat Rev Microbiol. (2014) 12:822–31. doi: 10.1038/nrmicro3362
63. Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. (2012) 489:526–32. doi: 10.1038/nature11414
64. Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, et al. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun. (2014) 5:3614. doi: 10.1038/ncomms4614
65. Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. (2011) 108:14216–21. doi: 10.1073/pnas.1111497108
66. Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. (2013) 87:12471–80. doi: 10.1128/JVI.01388-13
67. Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. (2011) 85:10905–8. doi: 10.1128/JVI.00700-11
68. Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci USA. (2012) 109:17040–5. doi: 10.1073/pnas.1212371109
69. Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, et al. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. (2009) 5:e1000350. doi: 10.1371/journal.ppat.1000350
70. Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Albrecht R, Blum DL, et al. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol. (2012) 86:6334–40. doi: 10.1128/JVI.07158-11
71. Winarski KL, Thornburg NJ, Yu Y, Sapparapu G, Crowe JE, Spiller BW. Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites. Proc Natl Acad Sci USA. (2015) 112:9346–51. doi: 10.1073/pnas.1502762112
72. Shen C, Chen J, Li R, Zhang M, Wang G, Stegalkina S, et al. A multimechanistic antibody targeting the receptor binding site potently cross-protects against influenza B viruses. Sci Transl Med. (2017) 9:eaam5752. doi: 10.1126/scitranslmed.aam5752
73. Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, et al. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci USA. (2018) 115:168–73. doi: 10.1073/pnas.1715471115
74. Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, Ikeda M, et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol. (2014) 88:7130–44. doi: 10.1128/JVI.00420-14
75. Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, et al. Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe. (2019) 25:827–35.e6. doi: 10.1016/j.chom.2019.04.003
76. Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, et al. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell. (2019) 177:1136–52.e18. doi: 10.1016/j.cell.2019.04.011
77. Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, et al. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell. (2019) 177:1124–35.e16. doi: 10.1016/j.cell.2019.03.048
78. Das DK, Govindan R, Nikić-Spiegel I, Krammer F, Lemke EA, Munro JB. Direct visualization of the conformational dynamics of single influenza hemagglutinin trimers. Cell. (2018) 174:926–37.e12. doi: 10.1016/j.cell.2018.05.050
79. Adachi Y, Tonouchi K, Nithichanon A, Kuraoka M, Watanabe A, Shinnakasu R, et al. Exposure of an occluded hemagglutinin epitope drives selection of a class of cross-protective influenza antibodies. Nat Commun. (2019) 10:3883. doi: 10.1038/s41467-019-11821-6
80. Yewdell JW, Taylor A, Yellen A, Caton A, Gerhard W, Bächi T, et al. Mutations in or near the fusion peptide of the influenza virus hemagglutinin affect an antigenic site in the globular region. J Virol. (1993) 67:933–42.
81. Bangaru S, Zhang H, Gilchuk IM, Voss TG, Irving RP, Gilchuk P, et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza. Nat Commun. (2018) 9:2669. doi: 10.3410/f.733623822.793548444
82. Paul SS, Mok CK, Mak TM, Ng OW, Aboagye JO, Wohlbold TJ, et al. A cross-clade H5N1 influenza A virus neutralizing monoclonal antibody binds to a novel epitope within the vestigial esterase domain of hemagglutinin. Antiviral Res. (2017) 144:299–310. doi: 10.1016/j.antiviral.2017.06.012
83. Chai N, Swem LR, Park S, Nakamura G, Chiang N, Estevez A, et al. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat Commun. (2017) 8:14234. doi: 10.1038/ncomms14234
84. Sesterhenn F, Bonet J, Correia BE. Structure-based immunogen design-leading the way to the new age of precision vaccines. Curr Opin Struct Biol. (2018) 51:163–9. doi: 10.1016/j.sbi.2018.06.002
85. Sesterhenn F, Yang C, Cramer JT, Bonet J, Wen X, Abriata LA, et al. Trivalent cocktail of de novo designed immunogens enables the robust induction and focusing of functional antibodies in vivo. bioRxiv. (2019) 685867. doi: 10.1101/685867
86. Angeletti D, Yewdell JW. Is it possible to develop a “Universal” Influenza virus vaccine? outflanking antibody immunodominance on the road to universal influenza vaccination. Cold Spring Harb Perspect Biol. (2018) 10:a028852. doi: 10.1101/cshperspect.a028852
87. Cohen J. Immunology. A once-in-a-lifetime flu shot? Science. (2013) 341:1171. doi: 10.1126/science.341.6151.1171
88. Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. (2015) 349:1301–6. doi: 10.1126/science.aac7263
89. Mallajosyula VV, Citron M, Ferrara F, Lu X, Callahan C, Heidecker GJ, et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci USA. (2014) 111:E2514–23. doi: 10.1073/pnas.1402766111
90. Mallajosyula VV, Citron M, Ferrara F, Temperton NJ, Liang X, Flynn JA, et al. Hemagglutinin sequence conservation guided stem immunogen design from influenza A H3 subtype. Front Immunol. (2015) 6:329. doi: 10.3389/fimmu.2015.00329
91. Valkenburg SA, Mallajosyula VV, Li OT, Chin AW, Carnell G, Temperton N, et al. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci Rep. (2016) 6:22666. doi: 10.1038/srep22666
92. Vatti A, Monsalve DM, Pacheco Y, Chang C, Anaya JM, Gershwin ME. Original antigenic sin: a comprehensive review. J Autoimmun. (2017) 83:12–21. doi: 10.1016/j.jaut.2017.04.008
93. van der Lubbe JEM, Huizingh J, Verspuij JWA, Tettero L, Schmit-Tillemans SPR, Mooij P, et al. Mini-hemagglutinin vaccination induces cross-reactive antibodies in pre-exposed NHP that protect mice against lethal influenza challenge. NPJ Vaccines. (2018) 3:25. doi: 10.1038/s41541-018-0063-7
94. Lee LA, Wang Q. Adaptations of nanoscale viruses and other protein cages for medical applications. Nanomedicine. (2006) 2:137–49. doi: 10.1016/j.nano.2006.07.009
95. Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. (2015) 21:1065–70. doi: 10.1038/nm.3927
96. Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, et al. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. MBio. (2019) 10:e02810–18. doi: 10.1128/mBio.02810-18
97. Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, et al. Influenza viruses expressing chimeric hemagglutinins:globular head and stalk domains derived from different subtypes. J Virol. (2012) 86:5774–81. doi: 10.1128/JVI.00137-12
98. Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. (2013) 87:6542–50. doi: 10.1128/JVI.00641-13
99. Nachbagauer R, Miller MS, Hai R, Ryder AB, Rose JK, Palese P, et al. Hemagglutinin stalk immunity reduces influenza virus replication and transmission in ferrets. J Virol. (2015) 90:3268–73. doi: 10.1128/JVI.02481-15
100. Nachbagauer R, Kinzler D, Choi A, Hirsh A, Beaulieu E, Lecrenier N, et al. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. NPJ Vaccines. (2016) 1:16015. doi: 10.1038/npjvaccines.2016.15
101. Krammer F, Hai R, Yondola M, Tan GS, Leyva-Grado VH, Ryder AB, et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J Virol. (2014) 88:3432–42. doi: 10.1128/JVI.03004-13
102. Ryder AB, Nachbagauer R, Buonocore L, Palese P, Krammer F, Rose JK. Vaccination with vesicular stomatitis virus-vectored chimeric hemagglutinins protects mice against divergent influenza virus challenge strains. J Virol. (2015) 90:2544–50. doi: 10.1128/JVI.02598-15
103. Margine I, Krammer F, Hai R, Heaton NS, Tan GS, Andrews SA, et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol. (2013) 87:10435–46. doi: 10.1128/JVI.01715-13
104. Ermler ME, Kirkpatrick E, Sun W, Hai R, Amanat F, Chromikova V, et al. Chimeric hemagglutinin constructs induce broad protection against influenza B virus challenge in the mouse model. J Virol. (2017) 91:e00286–17. doi: 10.1128/JVI.00286-17
105. Isakova-Sivak I, Korenkov D, Smolonogina T, Kotomina T, Donina S, Matyushenko V, et al. Broadly protective anti-hemagglutinin stalk antibodies induced by live attenuated influenza vaccine expressing chimeric hemagglutinin. Virology. (2018) 518:313–23. doi: 10.1016/j.virol.2018.03.013
106. Bernstein DI, Guptill J, Naficy A, Nachbagauer R, Berlanda-Scorza F, Feser J, et al. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates:interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect Dis. 20:80–91 (2020). doi: 10.1016/S1473-3099(19)30393-7.
107. Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. Playing hide and seek:how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses. (2014) 6:1294–316. doi: 10.3390/v6031294
108. Tseng YC, Wu CY, Liu ML, Chen TH, Chiang WL, Yu YH, et al. Egg-based influenza split virus vaccine with monoglycosylation induces cross-strain protection against influenza virus infections. Proc Natl Acad Sci USA. (2019) 116:3935–7. doi: 10.1073/pnas.1819197116
109. Liu WC, Jan JT, Huang YJ, Chen TH, Wu SC. Unmasking Stem-specific neutralizing epitopes by abolishing N-linked glycosylation sites of influenza virus hemagglutinin proteins for vaccine design. J Virol. (2016) 90:8496–508. doi: 10.1128/JVI.00880-16
110. Eggink D, Goff PH, Palese P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol. (2014) 88:699–704. doi: 10.1128/JVI.02608-13
111. Weidenbacher PA, Kim PS. Protect, modify, deprotect (PMD):A strategy for creating vaccines to elicit antibodies targeting a specific epitope. Proc Natl Acad Sci USA. (2019) 116:9947–52. doi: 10.1073/pnas.1822062116
112. Wang W, Song HS, Keller PW, Alvarado-Facundo E, Vassell R, Weiss CD. Conformational stability of the hemagglutinin of H5N1 influenza a viruses influences susceptibility to broadly neutralizing stem antibodies. J Virol. (2018) 92:e00247–18. doi: 10.1128/JVI.00247-18
113. Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat Immunol. (2019) 20:362–72. doi: 10.1038/s41590-018-0305-x
114. Krammer F. Fighting influenza through hemagglutinin diversity. Nat Immunol. (2019) 20:246–47. doi: 10.1038/s41590-019-0317-1
115. Anderson AM, Baranowska-Hustad M, Braathen R, Grodeland G, Bogen B. Simultaneous targeting of multiple hemagglutinins to APCs for induction of broad immunity against influenza. J Immunol. (2018) 200:2057–66. doi: 10.4049/jimmunol.1701088
116. Carter DM, Darby CA, Lefoley BC, Crevar CJ, Alefantis T, Oomen R, et al. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J Virol. (2016) 90:4720–34. doi: 10.1128/JVI.03152-15
117. Giles BM, Ross TM. Computationally optimized antigens to overcome influenza viral diversity. Exp Rev Vaccines. (2012) 11:267–9. doi: 10.1586/erv.12.3
118. Sautto GA, Kirchenbaum GA, Ecker JW, Bebin-Blackwell AG, Pierce SR, Ross TM. Elicitation of broadly protective antibodies following infection with influenza viruses expressing H1N1 computationally optimized broadly reactive hemagglutinin antigens. Immunohorizons. (2018) 2:226–37. doi: 10.4049/immunohorizons.1800044
119. Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, et al. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis. (2012) 205:1562–70. doi: 10.1093/infdis/jis232
120. Giles BM, Ross TM. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine. (2011) 29:3043–54. doi: 10.1016/j.vaccine.2011.01.100
121. Wong TM, Allen JD, Bebin-Blackwell AG, Carter DM, Alefantis T, DiNapoli J, et al. Computationally optimized broadly reactive hemagglutinin elicits hemagglutination inhibition antibodies against a panel of H3N2 influenza virus cocirculating variants. J Virol. (2017) 91:e01581–17. doi: 10.1128/JVI.01581-17
122. Allen JD, Owino SO, Carter DM, Crevar CJ, Reese VA, Fox CB, et al. Broadened immunity and protective responses with emulsion-adjuvanted H5 COBRA-VLP vaccines. Vaccine. (2017) 35:5209–16. doi: 10.1016/j.vaccine.2017.07.107
123. Crevar CJ, Carter DM, Lee KY, Ross TM. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum Vaccin Immunother. (2015) 11:572–83. doi: 10.1080/21645515.2015.1012013
124. Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. (2006) 281:8997–9000. doi: 10.1074/jbc.R500020200
125. Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Jou WM, Fiers W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med. (1999) 5:1157–63. doi: 10.1038/13484
126. De Filette M, Ramne A, Birkett A, Lycke N, Löwenadler B, Min Jou W, et al. The universal influenza vaccine M2e-HBc administered intranasally in combination with the adjuvant CTA1-DD provides complete protection. Vaccine. (2006) 24:544–51. doi: 10.1016/j.vaccine.2005.08.061
127. Eliasson DG, Omokanye A, Schön K, Wenzel UA, Bernasconi V, Bemark M, et al. M2e-tetramer-specific memory CD4 T cells are broadly protective against influenza infection. Mucosal Immunol. (2018) 11:273–89. doi: 10.1038/mi.2017.14
128. Fu TM, Freed DC, Horton MS, Fan J, Citron MP, Joyce JG, et al. Characterizations of four monoclonal antibodies against M2 protein ectodomain of influenza A virus. Virology. (2009) 385:218–26. doi: 10.1016/j.virol.2008.11.035
129. Bernasconi V, Bernocchi B, Ye L, Lê MQ, Omokanye A, Carpentier R, et al. Porous nanoparticles with self-adjuvanting M2e-fusion protein and recombinant hemagglutinin provide strong and broadly protective immunity against influenza virus infections. Front Immunol. (2018) 9:2060. doi: 10.3389/fimmu.2018.02060
130. Eliasson DG, El Bakkouri K, Schön K, Ramne A, Festjens E, Löwenadler B, et al. CTA1-M2e-DD:a novel mucosal adjuvant targeted influenza vaccine. Vaccine. (2008) 26:1243–52. doi: 10.1016/j.vaccine.2007.12.027
131. McMichael AJ, Picker LJ, Schepens B, De Vlieger D, Saelens X. Vaccine options for influenza:thinking small. Curr Opin Immunol. (2018) 53:22–9. doi: 10.1016/j.coi.2018.03.024
132. Kilbourne ED, Laver WG, Schulman JL, Webster RG. Antiviral activity of antiserum specific for an influenza virus neuraminidase. J Virol. (1968) 2:281–8.
133. Kosik I, Yewdell JW. Influenza Hemagglutinin and Neuraminidase:Yin(-)Yang proteins coevolving to thwart immunity. Viruses. (2019) 11:346. doi: 10.3390/v11040346
134. Maier HE, Nachbagauer R, Kuan G, Ng S, Lopez R, Sanchez N, et al. Pre-existing anti-neuraminidase antibodies are associated with shortened duration of influenza A (H1N1)pdm virus shedding and illness in naturally infected adults. Clin Infect Dis. (2019) 2019:ciz639. doi: 10.1093/cid/ciz639
135. Ng S, Nachbagauer R, Balmaseda A, Stadlbauer D, Ojeda S, Patel M, et al. Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat Med. (2019) 25:962–7. doi: 10.1038/s41591-019-0463-x
136. Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, et al. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell. (2018) 173:417–29.e10. doi: 10.1016/j.cell.2018.03.030
137. Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science. (2019) 366:499–504. doi: 10.1126/science.aay0678
138. Laver WG, Air GM, Webster RG, Markoff LJ. Amino acid sequence changes in antigenic variants of type A influenza virus N2 neuraminidase. Virology. (1982) 122:450–60. doi: 10.1016/0042-6822(82)90244-6
139. Webster RG, Hinshaw VS, Laver WG. Selection and analysis of antigenic variants of the neuraminidase of N2 influenza viruses with monoclonal antibodies. Virology. (1982) 117:93–104. doi: 10.1016/0042-6822(82)90510-4
140. Air GM, Els MC, Brown LE, Laver WG, Webster RG. Location of antigenic sites on the three-dimensional structure of the influenza N2 virus neuraminidase. Virology. (1985) 145:237–48. doi: 10.1016/0042-6822(85)90157-6
141. Jiang L, Fantoni G, Couzens L, Gao J, Plant E, Ye Z, et al. Comparative efficacy of monoclonal antibodies that bind to different epitopes of the 2009 pandemic H1N1 influenza virus neuraminidase. J Virol. (2016) 90:117–28. doi: 10.1128/JVI.01756-15
142. Andrews SF, Chambers MJ, Schramm CA, Plyler J, Raab JE, Kanekiyo M, et al. Activation dynamics and immunoglobulin evolution of pre-existing and newly generated human memory B cell responses to influenza hemagglutinin. Immunity. (2019) 51:398–410.e5. doi: 10.1016/j.immuni.2019.06.024
143. Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, et al. Viral receptor-binding site antibodies with diverse germline origins. Cell. (2015) 161:1026–34. doi: 10.1016/j.cell.2015.04.028
144. Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci USA. (2012) 109:9047–52. doi: 10.1073/pnas.1118979109
145. Job ER, Ysenbaert T, Smet A, Van Hecke A, Meuris L, Kleanthous H, et al. Fcgamma receptors contribute to the antiviral properties of influenza virus neuraminidase-specific antibodies. MBio. (2019) 10:e01667–19. doi: 10.1128/mBio.01667-19
146. Couch RB, Atmar RL, Keitel WA, Quarles JM, Wells J, Arden N, et al. Randomized comparative study of the serum antihemagglutinin and antineuraminidase antibody responses to six licensed trivalent influenza vaccines. Vaccine. (2012) 31:190–5. doi: 10.1016/j.vaccine.2012.10.065
147. Laguio-Vila MR, Thompson MG, Reynolds S, Spencer SM, Gaglani M, Naleway A, et al. Comparison of serum hemagglutinin and neuraminidase inhibition antibodies after 2010–2011 trivalent inactivated influenza vaccination in healthcare personnel. Open Forum Infect Dis. (2015) 2:ofu115. doi: 10.1093/ofid/ofu115
148. Schotsaert M, Ysenbaert T, Neyt K, Ibañez LI, Bogaert P, Schepens B, et al. Natural and long-lasting cellular immune responses against influenza in the M2e-immune host. Mucosal Immunol. (2013) 6:276–87. doi: 10.1038/mi.2012.69
149. El Bakkouri K, Descamps F, De Filette M, Smet A, Festjens E, Birkett A, et al. Universal vaccine based on ectodomain of matrix protein 2 of influenza A:Fc receptors and alveolar macrophages mediate protection. J Immunol. (2011) 186:1022–31. doi: 10.4049/jimmunol.0902147
150. Scorza FB, Pardi N. New kids on the block:RNA-based influenza virus vaccines. Vaccines. (2018) 6:20. doi: 10.3390/vaccines6020020
151. Angeletti D, Kosik I, Santos JJS, Yewdell WT, Boudreau CM, Mallajosyula VVA, et al. Outflanking immunodominance to target subdominant broadly neutralizing epitopes. Proc Natl Acad Sci USA. (2019) 116:13474–9. doi: 10.1073/pnas.1816300116
152. Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell. (2019) 177:1153–71.e28. doi: 10.1016/j.cell.2019.04.012
Keywords: influenza A virus, immunodominance, vaccines, B cells, antibodies
Citation: Mathew NR and Angeletti D (2020) Recombinant Influenza Vaccines: Saviors to Overcome Immunodominance. Front. Immunol. 10:2997. doi: 10.3389/fimmu.2019.02997
Received: 18 October 2019; Accepted: 06 December 2019;
Published: 10 January 2020.
Edited by:
Giuseppe Andrea Sautto, University of Georgia, United StatesReviewed by:
Ali Ellebedy, Washington University in St. Louis, United StatesGene Tan, J. Craig Venter Institute, United States
Copyright © 2020 Mathew and Angeletti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Davide Angeletti, davide.angeletti@gu.se
 Nimitha R. Mathew
Nimitha R. Mathew Davide Angeletti
Davide Angeletti