- 1Aix-Marseille Univ, MEPHI, IRD, AP-HM, Marseille, France
- 2IHU-Méditerranée Infection, Marseille, France
- 3Aix-Marseille Univ, AP-HM, Hôpital Nord, Médecine Intensive-Réanimation, Marseille, France
- 4Aix-Marseille Univ, AP-HM, CHU Hôpital Nord, Service d'Anesthésie et de Réanimation, Marseille, France
- 5AP-HM, UF Immunologie, Marseille, France
The host defense against pathogens varies among individuals. Among the factors influencing host response, those associated with circadian disruptions are emerging. These latter depend on molecular clocks, which control the two partners of host defense: microbes and immune system. There is some evidence that infections are closely related to circadian rhythms in terms of susceptibility, clinical presentation and severity. In this review, we overview what is known about circadian rhythms in infectious diseases and update the knowledge about circadian rhythms in immune system, pathogens and vectors. This heuristic approach opens a new fascinating field of time-based personalized treatment of infected patients.
Introduction
The defense against pathogens varies among individuals. Several heritable and non-heritable influences may account for these inter-individual variations. It now appears that the most important variations of host response among individuals include those associated with circadian disruptions (1). The circadian rhythm (Latin origin: circa: almost; dies: day), also called biological clock, is found in all living organisms, including eukaryotes and prokaryotes, and is defined as a period of about 24 h, temperature-compensated and entrained by a Zeitgeber (German name for synchronizer) (2, 3). The idea that an intrinsic rhythmicity governs host adaptation to the environment was introduced three centuries ago with the movement of mimosa leaves and, one century later, by the demonstration of gene-encoded rhythm in Drosophila melanogaster (4–7). This fascinating field of research is expanding with the recent awarding of the Nobel Prize to Hall, Rosbash and Young (8).
The function of circadian rhythms is to anticipate the changes and cycles of the surrounding world, such as day/night cycle and earth rotation, in order to optimize the response of organisms to these changes (9, 10). In mammals, including humans and non-human primates, this biological rhythmicity is coordinated by a central molecular oscillator, the suprachiasmatic nucleus (SCN), which ensures synchronization with light/dark cycle through specialized neurons from retina that receive photonic signals and the SCN then sends projections to other regions of the brain with local clocks such as immune organs, leading to activation of peripheral clocks (11–14). Interestingly, the SCN triggers a circadian rhythm independently of any temporal reference and is an autonomous timekeeper (15). In peripheral tissues, most cells have an internal molecular clock (16), but the function of these specific oscillators is not fully understood.
The molecular mechanisms of circadian rhythms have been well-investigated so far. It involves the clock genes present in two main feedback loops (Figure 1). First, the positive loop consists of the genes encoding circadian locomotor output cycles kaput (CLOCK), brain and muscle arnt-like protein 1 (BMAL1), and retinoic acid-related orphan receptor α, β and γ (RORs) proteins. Second, the negative loop involves the genes encoding Period (PER) 1, 2, 3, cryptochrome (CRY) 1, 2 and REVERB-α, -β (also called NR1D1/2 or nuclear receptor subfamily 1 group D) proteins (17, 18). In most somatic cells, the transcription factor formed by the dimerization of BMAL1 and CLOCK proteins binds the E-box sequences of the promoters of the other clock genes (Per, Cry, Reverb, and ROR), inducing their expression and translation (19). The CLOCK/BMAL1 transcription factor controls a third circadian regulatory loop, DBP (D-box binding protein) by binding the D-box sequences of gene promoters (9). Following the action of the dimer CLOCK/BMAL1, the clock-controlled genes (Ccgs) are expressed. These latter control 30% of the mammalian genome and regulate numerous physiological functions (16), including body temperature, blood pressure, hormone concentrations, blood circulation, urine output, metabolism, hair growth and immune system (14). In this context, during pathological conditions the involvement of circadian rhythm is now admitted but its investigation remains to date complex (20).
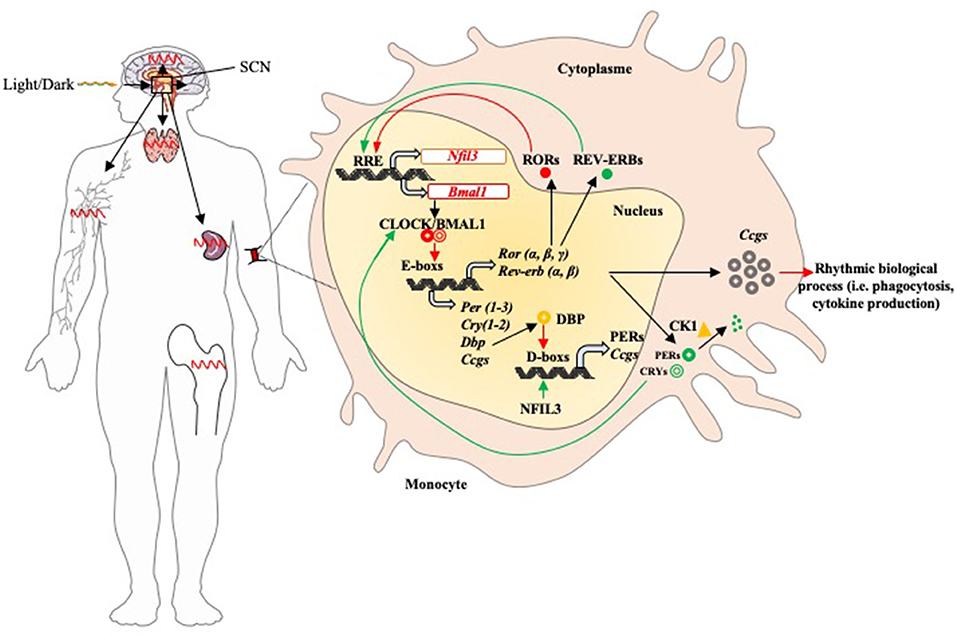
Figure 1. Circadian rhythm of the immune system components. The suprachiasmatic nucleus is the central oscillator that mediates all circadian variations in humans. It receives synchronization information with the day/night cycle via its retinal connection and then sends projections to other regions of the brain with local clocks such as immune organs (lymph nodes, spleen, thymus, and bone marrow) and activates peripheral clocks (activation of circadian feedback loops). In most immune cells (represented by functions such as phagocytosis and cytokine production), the feedback loops function as follows: once its gene is expressed, the BMAL1 protein dimerizes with CLOCK in the nucleus; the CLOCK/BMAL1 heterodimer binds to the E-box promoter sequences and induces expression of Pers, Crys, Reverbs, Rors, and Ccgs. The PERs and CRYs proteins dimerize in the presence of the casein kinase 1 (Ck1), then return to the nucleus to prevent the binding of the dimer CLOCK/BMAL1 on DNA. PERs proteins are phosphorylated by Ck1 and then degraded by the proteasome. REVERBS and RORs have antagonistic effects on Bmal1 expression: they bind the promoter of its gene and then induce (RORs) or inhibit (REVERBs) its expression. DBP protein induces the expression of Pers and Ccgs by binding D-box promoter sequences; it is inhibited by NFIL3. Positive and negative factors are represented by red and green arrows, respectively.
Here, we will summarize what is known about rhythmicity in infectious diseases. Then, we will update the knowledge about circadian rhythms in immune system, pathogens and vectors. Finally, we will translate this heuristic approach into a fascinating process for time-based personalized treatments of infected patients.
Circadian Rhythms of Immune Effectors, Pathogens and Vectors in Infections
The occurrence of infectious diseases results from the conjunction of different factors including the ability of the host to coordinate the immune response, the nature and the virulence of the microorganisms and, sometimes, the presence of vectors such as mosquitoes. If the rhythms of immune system have been a source of recent reviews, those of pathogens and vectors are less investigated. We will summarize what is known about circadian rhythms of the immune system, microbes and vectors with a special attention to microbiota.
Circadian Rhythms of Immune Effectors
It is well-established that the immune response varies according to circadian rhythms. These variations concern innate and adaptive immune responses at both quantitative and qualitative levels. At the quantitative level, the circulating number of hematopoietic stem and progenitor cells, and most mature leukocytes increases during the resting phase for rodents and the night for humans (21). The migration of immune cells to the tissues, a major phase of anti-microbial response, occurs preferentially during the active phase. This response also involves neuro-mediators released locally by sympathetic nerves, thus underlying the role of central pacemaker in addition to peripheral clocks (22). It has recently been shown that neutrophil traffic is regulated through a timer program. The deletion of aryl hydrocarbon receptor nuclear translocator-like protein 1 (Arntl) and Cxcr2 genes prevents diurnal rhythms, called neutrophil aging (23). The homing of circulating lymphocytes into lymph nodes and their egress into efferent lymphatic vessels obey to a rhythmic process. The egress of cells is based on circadian variations of CCR7 production and that of its ligand CCL21 in both T and B lymphocytes (24).
The immune functions are modulated by circadian control. Regarding the innate immune system, functions such as particle uptake and release of oxygen derivatives and cytokines exhibit circadian rhythms (25, 26). Regarding the adaptive immunity, there is evidence that the functions of T and B lymphocytes also exhibit circadian rhythms. Indeed, the production of antibodies in response to thymo-dependent and thymo-independent antigens oscillates with melatonin rhythm in mice; melatonin suppression is associated with increased levels of specific antibodies, which are corrected by the addition of melatonin and light (27). Similarly, immunoglobulin (Ig)-E levels and IgE-mediated allergic responses are regulated by molecular clock in mice (28) and human mast cells; similar results are obtained in human eosinophils activated with N-formyl-methionly-leucyl-phenylalanine, a canonical chemotactic peptide (29).
In mice, the peak of salivary IgA levels occurs during the night; this response seems to be under the control of a central pacemaker (30). The lessons from mice invalidated for clock genes underline the role of these genes in the ontogeny of B lymphocytes and plasma cells (31) The functions of T lymphocytes are also controlled in a circadian manner. The proliferative response of T lymphocytes to mitogens is strongly rhythmic and is impaired when clock genes are mutated (32). In addition, the clock genes are involved in the differentiation of Th17 cells. The Th17 cell development is suppressed by nuclear factor, interleulin-3 regulated (NFIL3), a circadian-regulated transcription factor; Nfil3 and related orphan receptor gamma (Rorγt) are expressed in CD4+ T cells rhythmically during dark phase and light phase, respectively (33, 34). The development of regulatory T cells producing interleukin (IL)-10 is controlled by melatonin, suggesting that all coordination of the immune response is under circadian control (34).
These results clearly show that the immune system is under the control of peripheral clocks. It is also regulated by hormones and neuro-mediators that reflect the activity of central pacemaker. The hypothalamic-pituitary-adrenal axis is activated in response to stress and appears synchronized to glucocorticoid circadian rhythms. Hence, oscillations of the lymphocyte number in humans are inversely correlated with diurnal rhythm of glucocorticoid production (35). It is likely that the rhythmicity of the immune response involves numerous mechanisms, including the contribution of light variation.
Circadian Rhythms in Microbes and Pathogens
For a long time, it was believed that circadian rhythms are not expressed by single-cell organisms such as prokaryotes. One reason for this belief was the idea that organisms duplicating more than once a day do not need a rhythm longer than their life cycle (36). In the 1980's, circadian rhythm was found present in cyanobacteria (37–39), photoautotrophic organisms that produce oxygen by photosynthesis as plants and eukaryotic algae (40, 41). In cyanobacteria, photosynthesis occurs in daylight, whereas nitrogen production peaks during the night (42). It has also been shown a circadian variation of the expression of most genes implicated in cell division, chromatin compaction and photosynthesis (43).
Mechanistic studies have enabled the identification of clock genes responsible for circadian rhythms have been recently discovered in most organisms including fungi, algae and photosynthetic bacteria; they are named Frq, Ibp, and Kai, respectively (44, 45). Regarding cyanobacteria, the central biochemical oscillator of their circadian rhythm is represented by three proteins encoded by the KaiABC cluster (10, 46, 47), SasA, CikA, and RepA, which control the timing training, and output of the cyanobacterial cell signaling process (48). Several studies have examined how functions the circadian loop of cyanobacteria (40, 42, 49–51). A theoretical model of the KaiABC oscillator suggests that ATP hydrolysis is a driving mechanism of phosphorylation oscillations and that the frequency of ATP hydrolysis in individual KaiC molecules is correlated with the circadian rhythm frequency (50) (Figure 2). The reconstitution of cyanobacterium circadian loop in vitro is associated with oscillations of Kai proteins (52, 53), validating the theoretical approach of the KaiABC oscillator.
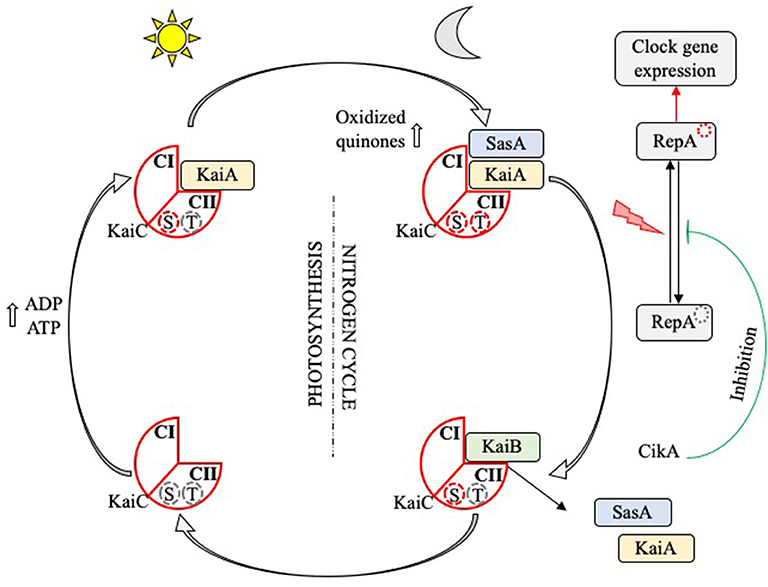
Figure 2. Circadian rhythm in cyanobacteria. The KaiC protein (red diamond) has two loop domains (CI and CII) and serine (S) and threonine (T) phosphorylation sites. At dawn, the increase in the ADP/ATP ratio, which reaches its maximum at noon, leads to the fixation of KaiA (yellow rectangle) on the CII domain, the phosphorylation of the T site and the induction of a training signal of photosynthesis elements. At nightfall, the increase in oxidized quinones following the initiation of the nitrogen cycle leads to the phosphorylation of the S site; KaiC is doubly phosphorylated. SasA (blue rectangle) is then fixed on the CI domain and activates the transcription factor RepA, which in turn induces expression of the target genes involved in the clock loop or others functions such as their multiplication. Finally, KaiB (green rectangle) releases KaiA and SasA, disable RepA via its interaction with the repressor CikA, and end up dephosphorylating KaiC which first finds itself serine phosphorylated.
The belief that circadian rhythms are specific of cyanobacteria and that eubacteria are “non-circadian” should be revisited. Using a bioinformatics approach, homologs of KaiB and KaiC genes have been found in non-circadian bacteria including Pseudomonas species and in archaea (46). The functions of non-cyanobacterial Kai proteins have not been described, and it remains unknown whether these proteins interact with input and output factors (54, 55). However, the KaiABC oscillator, when expressed into the “non-circadian” bacterium Escherichia coli, remains functional (56), suggesting that “non-circadian” bacteria possess the armaturium needed to circadian rhythms. We recently identified the RadA gene from E. coli as a KaiC structural homolog using a bioinformatic approach and showed the persistence of circadian rhythm in RadA−/− and RecA−/− mutants, suggesting that RadA is not the generator of circadian rhythm of E. coli. Hence, it does not play the same role than KaiC in cyanobacteria (unpublished data). To date, the identification of circadian rhythms in eubacteria remains challenging for the scientific community and opens new insights in terms of antibiotic resistance that could provide an interesting new therapeutic approach.
The circadian rhythms have been also described in parasites Plasmodium sp. (57). With a blood stage lasting 24 h or multiple of 24 h, its rhythm was found to be associated with recurrent fever. It has been reported that circadian variations of parasites are influenced by variations of glycemia from high levels during the night to low levels during the day (58, 59). Therefore, the links between rhythmicity of parasites and host homeostasis have been suggested. Similarly, the appearance of filarial parasites is also rhythmic (60). The parasites have their own circadian rhythm, as exemplified by Trypanosoma brucei that can generate circadian expression in the absence of host cells (61).
Circadian Rhythms in Vectors
The insect vectors have their own biological rhythm (62) that depends on the response to daily light/dark cycle and molecular clock activity. Anopheles mosquitoes that transmit malaria are active at dusk, so their bites occur at night during the host's resting phase. In contrast, Aedes mosquitoes that transmit dengue/yellow fever are day biters (63). The pathogens may affect vector circadian rhythms. The dengue virus is responsible for high amplitude of rhythmic locomotor activity of mosquitoes. Aedes aegypti exhibits diurnal rhythms that contribute to circadian transmission of Zika virus (64). The proteobacterium Wolbachia, an endosymbiont of arthropods, was reported to influence sleep time in flies (65). Recently, it has been shown that the daily activity of fly is affected by peripheral clocks and Wolbachia presence in flies (66).
The existence of biological rhythms in vectors may affect their resistance to toxic compounds. In Anopheles gambiae, the genes encoding the metabolic resistance to dichloro-diphenyl-trichloroethane (DDT) insecticide are rhythmically expressed (67, 68). Correlations between circadian enzymatic detoxification of insecticide and feeding time have been established (69). Thus, it appears that interactions between the circadian rhythms of the three partners involved in malaria transmission, namely mosquitoes, parasites and human host, lead to the development of the disease.
Rhythms of Intestinal Microbiota and Medical Consequences
At the intersection of microbial world, immune response and homeostasis, the intestinal microbiota exhibit variations of circadian rhythms (70, 71). The human gastrointestinal tract contains a large number of microorganisms, of which the most studied are the bacteria (72). They contribute to maintain various host physiological functions such as immune defense and/or tolerance (73). Recently, the circadian rhythm of the intestinal microbiome composition as well as the functions of this microbial community related to the circadian rhythm have been reviewed (74). Although intestinal bacteria are not exposed to the light/dark cycle, they are subjected to circadian changes associated to host activities, including food intake and exposure to antibiotics (74, 75). Nevertheless, light variations affect the abundance of some microbes such as Ruminococcus torques known to affect gut barrier integrity (76) and that of Clostridia sp. (77). Interestingly, it has been reported that Enterobacter aerogenes, a gram-negative bacterium, is sensitive to melatonin known to be a circadian rhythm synchronizer and expresses circadian swarming and motility (78). These findings strengthen the hypothesis of circadian rhythms in prokaryotes.
The use of mouse models in which the intestinal microbiota is modulated or mice with a clock gene deficiency underlines the association between the microbiota and circadian rhythm. Liang et al. (79) found that the absolute number of fecal bacteria in C57BL/6 mice follows a circadian rhythm and that mice deficient in Bmal1 gene show an altered daytime rhythm of microbiota in a sex-dependent manner. Mice deficient in Per1/2 and disrupted sleep cycle show an almost total loss of rhythmic fluctuations of microbiota and present intestinal dysbiosis (80). The circadian rhythms of microbiota over 24-h periods also depend on the host. Hence, this rhythmicity disappears in vitro when bacteria are cultivated, suggesting a regulation by the host (81). Leone et al. (82) showed that the relationship between microbiota and circadian rhythms is affected by the type of diet in germ-free mice. Indeed, bacteria from the family Lachnospiraceae exhibit circadian rhythms under regular low-fat chow, which are absent in mice with high fat diet. The diabetic mice exhibit a loss of diurnal rhythms of bacteria such as Akkermansia genus (83).
The association between circadian variations of microbiota and mechanisms of nutrition-associated disorders has been recently reported. Based on the observation that circadian misalignment promotes the occurrence of obesity, it has been shown that disruption of circadian rhythms impacts gut microbial composition and risk of obesity in rodents. In the absence of microbiota, the recruitment of histone deacetylase 3 is defective and cyclical histone acetylation is lost, suggesting a relationship between microbiota rhythms and epigenetic changes (84, 85). In addition, a new target of diurnal rhythms of microbiota, the group 3 innate lymphoid cells (ILC3), has been recently reported (86). Indeed, a dysregulation of brain rhythmicity affects circadian rhythms of ILC3 and microbiota, suggesting that environmental light directly regulates diurnal variations of enteric ILC3. In summary, a disruption of the circadian rhythm, by mutation of clock genes or disruption of the wake/sleep cycle, leads to changes in the composition of the intestinal microbiota and metabolic changes in host, opening a new fascinating field of investigations.
Circadian Rhythm in Infectious Diseases: From Experimental Studies to Clinics
The study of circadian rhythms in infectious diseases is more complex than the investigation of rhythmicity in immune effectors and microbes. It requires an integrated approach associating experimental investigations and clinical observations. The aim of this section is to discuss the impact of infection to host circadian rhythms, the role of circadian rhythms in the susceptibility and/or resistance of individuals to infection and finally the situation of septic syndromes.
Impact of Infection on Host Circadian Rhythm
The evidence of circadian variations in infectious diseases due to bacteria, parasites and virus is based on clinical observations and on the use of animal models including mice invalidated for clock genes (Table 1).
During infection, central and peripheral circadian rhythms of the host may be altered. In infected mice, the interaction of Streptococcus pneumonia with epithelial cells account for circadian variations of pulmonary inflammation including the release of inflammatory mediators and recruitment of inflammatory cells. In the absence of Bmal1 gene, the recruitment of neutrophils is increased in relation with a disruption of CXCL5-glucocorticoid receptor interaction (87). Moreover, the infection with Helicobacter pylori, that is responsible for gastritis and paves the way of gastric cancer, involves alterations in gastric acid secretion, which is regulated by circadian rhythm (89). H. pylori dysregulates the molecular clock in gastritis through the upregulation of Bmal1 gene expression in gastric epithelial cells. Importantly, the expression of the Bmal1 gene is also upregulated in gastric tissues from patients with atrophic gastritis and dramatically increased in patients with precancerous lesions, thus establishing a relationship between disrupted circadian rhythm and the severity of the infection. In addition, H. pylori disrupts the circadian rhythm of an important Bmal1 target, the gene encoding tumor necrosis factor (Tnf), exacerbating the TNF production (90).
Disruption of circadian rhythm was also observed during intracellular bacterial infection. Listeria monocytogenes is a foodborne gram-positive bacillus known to be pathogen when T-cell mediated responses are impaired. The number of inflammatory monocytes considered as the effectors of inflammation and anti-Listeria immunity vary according to a circadian rhythm. Their circadian traffic to the tissues is disrupted by the myeloid impairment of Bmal1 gene (91). Our team previously showed expression variations of clock genes in Q fever, a zoonosis due to Coxiella burnetii, an obligate intracellular bacterium. The analysis of microarrays performed to understand sexual dimorphism in C. burnetii infection shows that circadian genes (Bmal1, Clock, and Per2) are upregulated in the liver from infected female mice, as compared with healthy females and infected males (100). In patients, the Per2 gene is more expressed in males with acute Q fever than in healthy volunteers (101), suggesting that the unexpected relationship between circadian rhythm and gender dimorphism of Q fever may be extended to other human infectious diseases.
Interestingly, it was reported that parasites seem to be able to synchronize circadian rhythm and immune response. Indeed, in mice infected by Plasmodium chabaudi, the frequency of parasite replication is related to the circadian rhythm of TNF expression (59). Indeed, increased TNF is associated with a higher frequency of non-replicating cyclic forms of trophozoites. Infection alters circadian rhythms of blood glucose with a hypoglycemia at the end of the active phase and a direct impact on parasites. The oscillations of TNF and glucose levels seemed critical for parasite replication (58). It is established that malaria due to Plasmodium sp. exhibit a periodicity of fever that is secondary to the burst of Plasmodium-infected red blood cells every 24–72 h (58). Mice infected with T. brucei have a period shorter than 24 h and abnormal activity during the resting phase. This model reproduces sleeping sickness considered a circadian disorder in which infected individuals experience somnolence during the day and insomnia during the night (102).
Different viruses alter circadian-regulated biological processes such as CD4+ T cell numeration in human immunodeficiency virus infection (103). Other interferences have been described for simian immunodeficiency virus, coxsackievirus A16 (hand, foot and mouth disease) and human T-lymphotropic virus (96). Liver circadian-regulated genes that are likely targets for hepatitis C virus (HCV) may affect viral hepatitis. Patients with chronic HCV infection develop a disrupted circadian rhythm characterized by altered sleep patterns, although the association of sleep disorders and reduced survival in patients is debated (94). HCV infection also leads to decreased expression of Per2 and Cry2 genes. The Per2 gene is critical for the anti-viral response since its overexpression in a hepatocyte cell line reduces HCV replication and increases the expression of interferon-stimulated genes (95). On the other hand, liver-associated microRNA (miR)-122 is involved in HCV replication; miR122 is negatively regulated by Rev-erbα, strengthening the idea that HCV replication depends on the daily rhythm (104). The association between hepatitis and circadian rhythms has potential implications for the patient management. Hence, viral RNA levels rebound more frequently in patients infected with HCV and undergoing liver transplantation in the morning than in those transplanted in the afternoon (105). Hepatic circadian gene oscillation is associated with circadian rhythm and sleep in HCV infection (94).
Patients infected with respiratory viruses exhibit daily rhythms of clinical symptoms, such as increased nasal secretion and body temperature in the morning compared to the late evening in patients with cold or flu (106). Mice with deficient molecular clock are highly susceptible to virus infection, as shown by increased replication of influenza, respiratory syncytial and parainfluenza type 3 viruses. Hence, Bmal1 appears as a major clock gene controlling viral replication (107). The Bmal1 gene is also involved in the coordination of lung immune response to virus since the deletion of Bmal1 gene exacerbates acute bronchiolitis secondary to Sendai virus or influenza A virus (96). In the same extend, the severity of influenza A infection is directly controlled by clock genes expressed in lung epithelium and natural killer (NK)1.1+ cells, a subset of NK cells (108). The alteration of host circadian rhythm was also reported for Herpes simplex virus (HSV). The pathogenicity of HSV results from the interplay of viruses with circadian rhythms. The replication of HSV depends on histone deacetylation, and transcriptional machinery of HSV is associated with histone acetyltransferase, a clock-controlled gene (109).
Circadian Rhythms in Susceptibility and/or Resistance to Infection
Several studies reported that the susceptibility and the resistance to infections are associated with host circadian rhythm. More than 40 years ago, it was reported that susceptibility to S. pneumoniae is altered in blind and adrenalectomized mice with altered circadian rhythms (110). Recently, Kitchen et al. (88) reported that macrophages from S. pneumonia infected Bmal1-deficient-mice presented increase bacterial ingestion and cytoskeletal change both associated with an impaired function of RhoA pathway. The infection is more severe when mice are infected at the beginning of the resting phase than during the active phase (87). In contrast, an increased number of L. monocytogenes is found in mouse tissues at the onset of the resting phase as compared with the beginning of the active phase (91). Salmonella enterica serovar Typhimurium is a foodborne microorganism whose recovery depends on the cell-mediated immune response. In mice, bacterial colonization of the colon is more pronounced during the resting phase than during the active phase. The inflammation induced by Salmonella sp. varies similarly. This difference between resting and active time is abolished in clock mutant mice (93). The circadian rhythm pattern of S. typhimurium infection reflects circadian rhythm of the innate immune cells. Hence, the ingestion of S. typhimurium bacteria by peritoneal macrophages is maximum 16 h after serum-mediated synchronization (111).
In infection due to Mycobacterium tuberculosis, it has been shown that cough, a major symptom of tuberculosis, is more frequent during daytime. This is related to highest sputum bacillary load (92). In a mice model, rhythmic melatonin release generated circadian rhythms in granulomatous lesions after inoculation with BCG, an attenuated strain of M. bovis used as a vaccine against M. tuberculosis infection (112). Moreover, it has been shown that the expression of metalloproteinases by peritoneal macrophages and spleen cells in response to M. tuberculosis infection is controlled by the circadian clock in a Bmal1-dependent manner (113). Surprisingly, although it is well-established that corticosteroids increase susceptibility to mycobacteria, there is no consensus on the relationship between cortisol circadian rhythms and tuberculosis (114, 115). Fever oscillations during daytime have been reported in Rocky Mountain spotted fever due to Rickettsia rickettsii. They consist of an elevated temperature in the evening with a decline early in the morning, while the number of R. rickettsii DNA copies is elevated in the blood of patients collected early in the morning, suggesting that rickettsemia has a profile release similar to that of cortisol (116).
Murine models of parasite infection confirm the involvement of circadian rhythm in the evolution of several parasitic infections (98). The moment of infection with the worm Trichuris muris affects the kinetics of worm expulsion. Hence, mice infected in the morning expel the parasite early, whereas the parasite persists in mice infected during the night. When the Bmal1 gene is deleted in antigen-presenting dendritic cells (DCs), the relationship of helminth expulsion and circadian rhythm disappears (99).
The relation between circadian rhythm and the regulation of the viral infection was also reported. Mouse models of infection exhibit daily variations in virus susceptibility: mice inoculated with Herpes virus at the beginning of the resting phase (in the daytime) exhibit increased virus load as compared with mice inoculated during the active phase (in the nighttime) (97). Recently it has been shown in a murine model that the severity of influenza An infection depends on the intrinsic pathogenicity of the virus and the uncontrolled inflammatory response. Mice infected before the onset of active phase exhibit higher mortality and morbidity than mice infected before the rest phase. When the Bmal1 gene is invalidated, the ability to fight the infection is lost through the hyper-inflammation induced by infection at the onset of the active phase (108).
Clinics: Septic Syndromes
Sepsis is an inflammatory response syndrome that appears after infection, or without documented infection (117). It is the leading cause of death of patients in intensive care units (118) and there is some evidence that circadian rhythm and sepsis are tightly associated (119).
Murine models of endotoxemia that mimic human sepsis provide important information on the mechanisms involved in sepsis and the association between sepsis and circadian rhythm. The administration of lipopolysaccharide (LPS) to mice induces a systemic inflammatory response in which cytokine and chemokine productions are critical. The production of inflammatory molecules including IL-6, IL-12p40, and chemokines, such as CCL2, CCL5, and CXCL1 in response to LPS is higher in mice inoculated early in the active phase than in mice inoculated during the resting phase (120). The endotoxemia in rats is associated with higher levels of inflammatory markers during nighttime than during daytime. In the presence of melatonin, daytime levels of inflammatory markers are increased, underlying the role of melatonin in circadian rhythm coordination in response to LPS (121).
The endotoxemia in young human volunteers is characterized by high levels of IL-10, an anti-inflammatory cytokine, during daytime and high levels of inflammatory cytokines (TNF, IL-1, and IL-6) and cytokine receptors during the nighttime (122). This is consistent with placebo-controlled design studies that show that males receiving LPS in the evening exhibit higher rectal temperature and inflammatory mediator production than those receiving LPS in the morning when cortisol production is the highest (123). There is evidence that LPS response depends on molecular oscillators. Hence, LPS injection induces a lesser production of cytokines in Clock−/− bone marrow-derived macrophages than in wild-type cells (93). The engagement of Toll-like receptor (TLR)-9 induces circadian rhythm of TNF and CCL2 with a peak during the active phase; cytokine rhythms are disrupted when the Per2 gene is invalidated (124). The genes of the molecular clock directly interact promoter regions of TLR genes, thus leading to their circadian rhythm (21).
Clinical studies suggest an association between circadian rhythm disruption and the development of sepsis. In a prospective study, our group assessed the circadian rhythm of cortisol and immune cells in 38 patients at day 2 after severe trauma. The trauma patients who develop an episode of sepsis later in their stay in the intensive care unit were compared to those who did not develop sepsis. The septic patients had higher levels of cortisol than the non-septic patients and delayed acrophases (ie., the peak of production during the period). The acrophases significantly differed between the two groups for lymphocytes, IL-10, and TNF (125). This suggests that the primary insult, represented here by trauma, is associated with disruption of circadian rhythm that affects the host response. In addition, admission to intensive care units is associated with sleep disturbance, sedative infusions and loss of the daily light/dark cycle (126). To counteract these adverse events, the use of melatonin was assessed in three randomized controlled trials (127–129). As the results of these three trials were disappointing, experts made no recommendation on melatonin use in intensive care units (130). Taken together, these findings suggest that circadian disruption is not only associated to sleep dependent of the melatonin, but is multifactorial, and that the initial inflammatory process seems much more critical than the hospitalization in intensive care units.
Circadian Rhythms and Management of Infected Patients
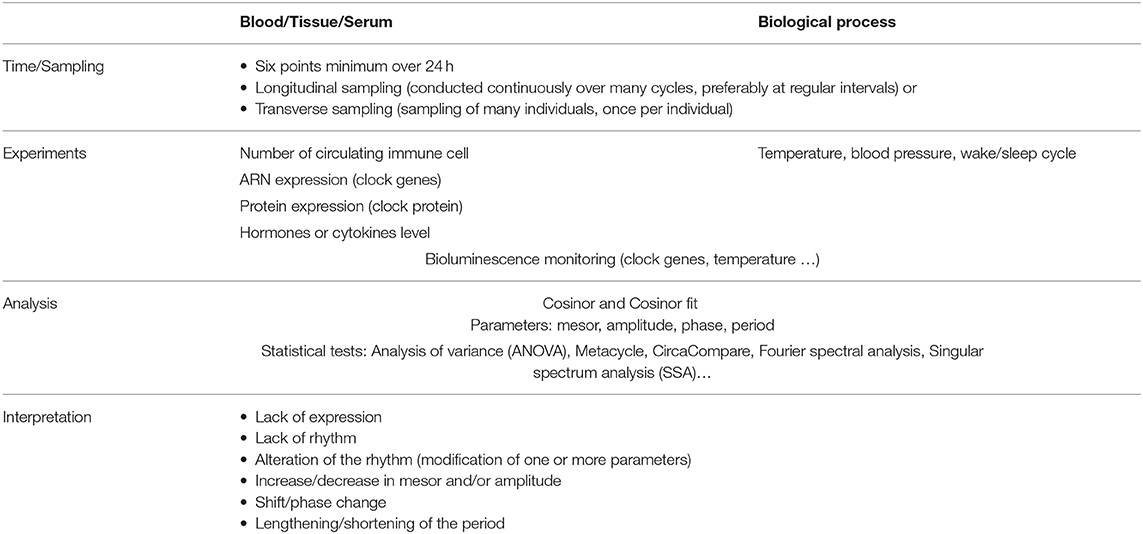
The investigation of circadian rhythms is well-established in basic research, but its relevance in the medical field remains limited. One of the challenges in the exploration of circadian rhythms in healthy individuals and patients is the high degree of interindividual variability. The circadian rhythm can be evaluated over a period of 24 h with a sampling of at least six points (Table 2). The sampling can be carried out in a longitudinal or transverse manner; longitudinal sampling is useful to obtain the time structure for one individual, whereas transverse or cross-sectional sampling is applied to a group of individuals. The circadian rhythm of individuals is evaluated through measurement of biological variables such as the number of circulating cells, ARN or protein expression of clock molecules or levels of hormones and cytokines. The biological processes associated with the circadian rhythm are also evaluated through physiological variables, such as body temperature (131), blood pressure or wake/sleep cycle. Finally, the circadian pacemaker is directly studied using a bioluminescent system associated with clock molecules. The analysis of biological or physiological variables during the day are based on the Cosinor method developed by Halberg et al. (132). This method enables the measurement of several variables including period, mesor, amplitude and phase of a cycle. These variables require an algorithm-dependent analysis such as Metacycle (133) or, more recently, CircaCompare (134) to estimate and statistically support differences in circadian rhythm (Table 2).
The use of chronobiology in the medical field has been well-documented in pharmacology. Indeed, circadian rhythm is used to determine the better-timed drug delivery, also named chronotherapy (135). This approach associates increased efficacy and reduced toxicity of the drug by the circadian evaluation of its absorption, its metabolism and its elimination (136, 137). The investigation of circadian oscillations in infected patients constitutes a challenge. In patients with sepsis, the measurement of body temperature or blood pressure at a given point instead of circadian manner has been debated (138, 139). We recently reported a circadian dysrhythmia of core body temperature in trauma patients at risk of sepsis, which is associated with increased mortality (131). In another cohort of trauma patients, we showed that the evaluation of the circadian rhythm through measurement of clock gene expression is critical for the identification of a circadian rhythm disruption associated with the occurrence of sepsis (125). The measurement of circadian variations of biological and clinical markers would permit a better stratification of patients, possibly enabling the definition of a chronotype for each infected patient (140).
The vaccination is a relevant example of a situation in which circadian rhythms of host defense and microbes affect the management of patients. Indeed, the antibody response after vaccination depends on the time of administration. In a randomized controlled trial including 276 adults, a significant difference in adaptive response after influenza vaccine administration was observed, with increased antibody production when the vaccine is administered in the morning compared to the afternoon (141). The vaccination of mice with DCs loaded with antigen leads to the expansion of specific CD8+ T cells and a better efficiency for a bacterial challenge if the vaccination is done in the middle of the day as compared with other time points. This response is abrogated when the Bmal1 gene is invalidated in DCs (142) Even if these results are promising, confirmation in other types of vaccination remains required.
It is also likely that the circadian rhythms affect the response to antibiotics. Antibiotic resistance is becoming a major public health problem (143). Interestingly, it has been hypothesized that pathogenic bacteria have an intrinsic circadian rhythm that ensures antibiotic resistance. The resistance to ampicillin, oxacillin, ceftriaxone, meropenem, gentamycin, and ciprofloxacin in clinical strains of Enterobacteriaceae, non-fermenting Gram-negative bacilli, and Gram-positive Staphylococci has been investigating every 3 h for 24 h. The presence of periods of sensitivity is attested by significant changes in the minimum inhibitory concentrations of antibiotics (144). Similarly, in patients with surgical site infections, variations have been reported in Staphylococcus aureus coagulase activity and temporal expression of antibacterial resistance according to the moment of the sampling (145). Further studies will determine the role of circadian rhythm of either the host or the pathogen in this antibiotic resistance process, opening the way for a new strategy of antibiotics administration.
Conclusion
The analysis of literature has shown that circadian rhythms likely play a role in infectious diseases in terms of susceptibility, clinical expression and outcome. The use of animal models in which clock genes are invalidated has produced a large amount of data supporting the role of circadian pacemakers in the response to microbes. We have also analyzed the mechanisms of the rhythmicity in infectious diseases. It is clear that immune response to pathogens oscillate under the control of peripheral molecular clocks and SCN. The role of corticosteroids has to be considered according to the consequences of its use in patients. Other partners of infectious diseases present rhythms such as microbes and vectors. The identification of molecular rhythms in prokaryotes, particularly so-called “non-circadian” bacteria, will propose a new approach to understand antibiotic resistance. At the interface of immune response and host homeostasis, the rhythms of microbiota provide exciting prospects of understanding metabolic and inflammatory diseases. As circadian rhythms are poised to become biomarkers to assess the outcome of patients with infectious diseases including the risk of complications, new tools for investigating host circadian changes will be required. A clinical chronobiology is necessary to analyze circadian variations at the individual level in infectious diseases. This approach would pave the way for time-based treatment and for administration of molecules known to entrain rhythmicity.
Author Contributions
AD, BC, ML, SM, and J-LM conceived and wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
SM was supported by a Fondation pour la Recherche Médicale postdoctoral fellowship (Reference: SPF20151234951) and then the Fondation Méditerranée Infection. This work was supported by the French Government under the Investissements d'avenir (Investments for the future) programme managed by the Agence Nationale de la Recherche (Reference: 10-IAHU-03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Pr. Christian Capo for helping with the manuscript writing.
Abbreviations
ANOVA, analysis of variance; ARNTL, aryl hydrocarbon receptor nuclear translocator-like; BMAL1, brain and muscle arnt-like protein 1; Ccgs, clock-controlled genes; Ck1, casein kinase 1; CLOCK, circadian locomotor output cycles kaput; CRY, cryptochrome; DBP, d-box binding protein; DCs, dendritic cells; DDT, dichloro-diphenyl-trichloroethane; HC, hepatitis C virus; HSV, herpes simplex virus; Ig, immunoglobulin; IL, interleukin; ILC, innate lymphoid cells; LPS, lipopolysaccharide; miR, microRNA; NFIL3, nuclear factor, interleulin-3 regulated; NK, natural killer; NR1D, nuclear receptor subfamily 1 group D; PER, period; ROR, retinoic acid-related orphan receptor; RORγT, RAR-related orphan receptor gamma; SCN, suprachiasmatic nucleus; SSA, singular spectrum analysis; TLR, toll-like receptor; TNF, tumor necrosis factor.
References
1. Brodin P, Davis MM. Human immune system variation. Nat Rev Immunol. (2017) 17:21–9. doi: 10.1038/nri.2016.125
2. Yuan L, Li YR, Xu XD. Chronobiology-2017 Nobel Prize in physiology or medicine. Yi Chuan Hered. (2018) 40:1–11. doi: 10.16288/j.yczz.17-397
3. Peek CB, Ramsey KM, Levine DC, Marcheva B, Perelis M, Bass J. Circadian regulation of cellular physiology. Methods Enzymol. (2015) 552:165–84. doi: 10.1016/bs.mie.2014.10.006
4. Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. (1971) 68:2112–6. doi: 10.1073/pnas.68.9.2112
5. Paganelli R, Petrarca C, Di Gioacchino M. Biological clocks: their relevance to immune-allergic diseases. Clin Mol Allergy CMA. (2018) 16:1. doi: 10.1186/s12948-018-0080-0
6. De Candolle AP. Physiologie Végétale ou Exposition des Forces et Fonctions Vitales des Végétaux. Paris: Béchet (1832). doi: 10.5962/bhl.title.107506
7. De Mairan M. Observation Botanique. In: Histoire de l'Academie Royale de Sciences. Paris (1729). p. 35–6.
8. Huang RC. The discoveries of molecular mechanisms for the circadian rhythm: The 2017 Nobel prize in physiology or medicine. Biomed J. (2018) 41:5–8. doi: 10.1016/j.bj.2018.02.003
9. Curtis AM, Bellet MM, Sassone-Corsi P, O'Neill LAJ. Circadian clock proteins and immunity. Immunity. (2014) 40:178–86. doi: 10.1016/j.immuni.2014.02.002
10. Gan S, O'Shea EK. An unstable singularity underlies stochastic phasing of the circadian clock in individual cyanobacterial cells. Mol Cell. (2017) 67:659–72.e12. doi: 10.1016/j.molcel.2017.07.015
11. Colwell CS. Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci. (2011) 12:553–69. doi: 10.1038/nrn3086
12. Bedont JL, LeGates TA, Slat EA, Byerly MS, Wang H, Hu J, et al. Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep. (2014) 7:609–22. doi: 10.1016/j.celrep.2014.03.060
13. Lange T, Dimitrov S, Born J. Effects of sleep and circadian rhythm on the human immune system. Ann N Y Acad Sci. (2010) 1193:48–59. doi: 10.1111/j.1749-6632.2009.05300.x
14. Hastings MH, Maywood ES, Brancaccio M. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat Rev Neurosci. (2018) 19:453–69. doi: 10.1038/s41583-018-0026-z
15. Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. (2002) 111:919–22. doi: 10.1016/S0092-8674(02)01225-4
16. Tognini P, Murakami M, Liu Y, Eckel-Mahan KL, Newman JC, Verdin E, et al. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. (2017) 26:523–38.e5. doi: 10.1016/j.cmet.2017.08.015
17. Zhang SL, Yue Z, Arnold DM, Artiushin G, Sehgal A. A circadian clock in the blood-brain barrier regulates xenobiotic efflux. Cell. (2018) 173:130–9.e10. doi: 10.1016/j.cell.2018.02.017
18. Ella K, Mócsai A, Káldi K. Circadian regulation of neutrophils: control by a cell-autonomous clock or systemic factors? Eur J Clin Invest. (2018) 48(Suppl. 2):e12965. doi: 10.1111/eci.12965
19. Li Y, Li G, Wang H, Du J, Yan J. Analysis of a gene regulatory cascade mediating circadian rhythm in zebrafish. PLoS Comput Biol. (2013) 9:e1002940. doi: 10.1371/journal.pcbi.1002940
20. Rijo-Ferreira F, Takahashi JS. Genomics of circadian rhythms in health and disease. Genome Med. (2019) 11:82. doi: 10.1186/s13073-019-0704-0
21. Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. (2013) 13:190–8. doi: 10.1038/nri3386
22. Scheiermann C, Kunisaki Y, Lucas D, Chow A, Jang JE, Zhang D, et al. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. (2012) 37:290–301. doi: 10.1016/j.immuni.2012.05.021
23. Adrover JM, del Fresno C, Crainiciuc G, Cuartero MI, Casanova-Acebes M, Weiss LA, et al. A neutrophil timer coordinates immune defense and vascular protection. Immunity. (2019) 50:390–402.e10. doi: 10.1016/j.immuni.2019.01.002
24. Druzd D, Matveeva O, Ince L, Harrison U, He W, Schmal C, et al. Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity. (2017) 46:120–32. doi: 10.1016/j.immuni.2016.12.011
25. Geiger SS, Fagundes CT, Siegel RM. Chrono-immunology: progress and challenges in understanding links between the circadian and immune systems. Immunology. (2015) 146:349–58. doi: 10.1111/imm.12525
26. Stone EF, Fulton BO, Ayres JS, Pham LN, Ziauddin J, Shirasu-Hiza MM. The circadian clock protein timeless regulates phagocytosis of bacteria in drosophila. PLoS Pathog. (2012) 8:e1002445. doi: 10.1371/journal.ppat.1002445
27. Cernysiov V, Gerasimcik N, Mauricas M, Girkontaite I. Regulation of T-cell-independent and T-cell-dependent antibody production by circadian rhythm and melatonin. Int Immunol. (2010) 22:25–34. doi: 10.1093/intimm/dxp109
28. Nakamura Y, Nakano N, Ishimaru K, Hara M, Ikegami T, Tahara Y, et al. Circadian regulation of allergic reactions by the mast cell clock in mice. J Allergy Clin Immunol. (2014) 133:568–75. doi: 10.1016/j.jaci.2013.07.040
29. Baumann A, Feilhauer K, Bischoff SC, Froy O, Lorentz A. IgE-dependent activation of human mast cells and fMLP-mediated activation of human eosinophils is controlled by the circadian clock. Mol Immunol. (2015) 64:76–81. doi: 10.1016/j.molimm.2014.10.026
30. Wada M, Orihara K, Kamagata M, Hama K, Sasaki H, Haraguchi A, et al. Circadian clock-dependent increase in salivary IgA secretion modulated by sympathetic receptor activation in mice. Sci Rep. (2017) 7:8802. doi: 10.1038/s41598-017-09438-0
31. Silver AC, Arjona A, Hughes ME, Nitabach MN, Fikrig E. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immun. (2012) 26:407–13. doi: 10.1016/j.bbi.2011.10.001
32. Fortier EE, Rooney J, Dardente H, Hardy MP, Labrecque N, Cermakian N. Circadian variation of the response of T cells to antigen. J Immunol. (2011) 187:6291–300. doi: 10.4049/jimmunol.1004030
33. Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, et al. Th17 cell differentiation is regulated by the circadian clock. Science. (2013) 342:727–30. doi: 10.1126/science.1243884
34. Scheiermann C, Gibbs J, Ince L, Loudon A. Clocking in to immunity. Nat Rev Immunol. (2018) 18:423–37. doi: 10.1038/s41577-018-0008-4
35. Cain DW, Cidlowski JA. Immune regulation by glucocorticoids. Nat Rev Immunol. (2017) 17:233–47. doi: 10.1038/nri.2017.1
36. Golden SS, Canales SR. Cyanobacterial circadian clocks–timing is everything. Nat Rev Microbiol. (2003) 1:191–9. doi: 10.1038/nrmicro774
37. Huang TC, Grobbelaar N. The circadian clock in the prokaryote Synechococcus RF-1. Microbiology. (1995)535–40. doi: 10.1099/13500872-141-3-535
38. Huang TC, Lin RF. Circadian rhythm of cyanothece RF-1(Synechococcus RF-1). In: Ditty JL, Mackey SR, Johnson CH, editors. Bacterial Circadian Programs. Berlin; Heidelberg: Springer Berlin Heidelberg (2009). p. 39–61. doi: 10.1007/978-3-540-88431-6_3
39. Grobbelaar N, Huang TC, Lin HY, Chow TJ. Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol Lett. (1986) 37:173–7. doi: 10.1111/j.1574-6968.1986.tb01788.x
40. Arisaka S, Sukigara H, Osanai T. Genetic manipulation to overexpress rpaA altered photosynthetic electron transport in Synechocystis sp. PCC 6803. J Biosci Bioeng. (2018) 126:139–44. doi: 10.1016/j.jbiosc.2018.02.005
41. Herrero A, Flores E. The Cyanobacteria: Molecular Biology, Genomics, and Evolution. Sevilla: Caister Academic Press (2008).
42. Swan JA, Golden SS, LiWang A, Partch CL. Structure, function, and mechanism of the core circadian clock in cyanobacteria. J Biol Chem. (2018) 293:5026–34. doi: 10.1074/jbc.TM117.001433
43. Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA. (2009) 106:14168–73. doi: 10.1073/pnas.0902587106
44. Bhadra U, Thakkar N, Das P, Pal Bhadra M. Evolution of circadian rhythms: from bacteria to human. Sleep Med. (2017) 35:49–61. doi: 10.1016/j.sleep.2017.04.008
45. Husse J, Eichele G, Oster H. Synchronization of the mammalian circadian timing system: Light can control peripheral clocks independently of the SCN clock: alternate routes of entrainment optimize the alignment of the body's circadian clock network with external time. BioEssays News Rev Mol Cell Dev Biol. (2015) 37:1119–28. doi: 10.1002/bies.201500026
46. Schmelling NM, Lehmann R, Chaudhury P, Beck C, Albers SV, Axmann IM, et al. Minimal tool set for a prokaryotic circadian clock. BMC Evol Biol. (2017) 17:169. doi: 10.1186/s12862-017-0999-7
47. Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. (2002) 417:329–35. doi: 10.1038/417329a
48. Tseng R, Goularte NF, Chavan A, Luu J, Cohen SE, Chang YG, et al. Structural basis of the day-night transition in a bacterial circadian clock. Science. (2017) 355:1174–80. doi: 10.1126/science.aag2516
49. Brettschneider C, Rose RJ, Hertel S, Axmann IM, Heck AJR, Kollmann M. A sequestration feedback determines dynamics and temperature entrainment of the KaiABC circadian clock. Mol Syst Biol. (2010) 6:389. doi: 10.1038/msb.2010.44
50. Das S, Terada TP, Sasai M. Role of ATP hydrolysis in cyanobacterial circadian oscillator. Sci Rep. (2017) 7:17469. doi: 10.1038/s41598-017-17717-z
51. Diamond S, Rubin BE, Shultzaberger RK, Chen Y, Barber CD, Golden SS. Redox crisis underlies conditional light-dark lethality in cyanobacterial mutants that lack the circadian regulator, RpaA. Proc Natl Acad Sci USA. (2017) 114:E580–9. doi: 10.1073/pnas.1613078114
52. Kim YI, Boyd JS, Espinosa J, Golden SS. Detecting KaiC phosphorylation rhythms of the cyanobacterial circadian oscillator in vitro and in vivo. Methods Enzymol. (2015) 551:153–73. doi: 10.1016/bs.mie.2014.10.003
53. Murakami R, Mutoh R, Ishii K, Ishiura M. Circadian oscillations of KaiA-KaiC and KaiB-KaiC complex formations in an in vitro reconstituted KaiABC clock oscillator. Genes Cells. (2016) 21:890–900. doi: 10.1111/gtc.12392
54. Baca I, Sprockett D, Dvornyk V. Circadian input kinases and their homologs in cyanobacteria: evolutionary constraints versus architectural diversification. J Mol Evol. (2010) 70:453–65. doi: 10.1007/s00239-010-9344-0
55. Dvornyk V. Structure and molecular phylogeny of sasA genes in cyanobacteria: insights into evolution of the prokaryotic circadian system. Mol Biol Evol. (2004) 21:1468–76. doi: 10.1093/molbev/msh106
56. Chen AH, Lubkowicz D, Yeong V, Chang RL, Silver PA. Transplantability of a circadian clock to a noncircadian organism. Sci Adv. (2015) 1:e1500358. doi: 10.1126/sciadv.1500358
57. Carvalho Cabral P, Olivier M, Cermakian N. The complex interplay of parasites, their hosts, and circadian clocks. Front Cell Infect Microbiol. (2019) 9:425. doi: 10.3389/fcimb.2019.00425
58. Reece SE, Prior KF. Malaria makes the most of mealtimes. Cell Host Microbe. (2018) 23:695–7. doi: 10.1016/j.chom.2018.05.015
59. Hirako IC, Assis PA, Hojo-Souza NS, Reed G, Nakaya H, Golenbock DT, et al. Daily rhythms of TNFα expression and food intake regulate synchrony of plasmodium stages with the host circadian cycle. Cell Host Microbe. (2018) 23:796–808.e6. doi: 10.1016/j.chom.2018.04.016
60. Rijo-Ferreira F, Takahashi JS, Figueiredo LM. Circadian rhythms in parasites. PLOS Pathog. (2017) 13:e1006590. doi: 10.1371/journal.ppat.1006590
61. Rijo-Ferreira F, Pinto-Neves D, Barbosa-Morais NL, Takahashi JS, Figueiredo LM. Trypanosoma brucei metabolism is under circadian control. Nat Microbiol. (2017) 2:17032. doi: 10.1038/nmicrobiol.2017.32
62. Meireles-Filho ACA, Kyriacou CP. Circadian rhythms in insect disease vectors. Mem Inst Oswaldo Cruz. (2013) 108(Suppl. 1):48–58. doi: 10.1590/0074-0276130438
63. Rund SSC, O'Donnell AJ, Gentile JE, Reece SE. Daily rhythms in mosquitoes and their consequences for malaria transmission. Insects. (2016) 7:14. doi: 10.3390/insects7020014
64. Lima-Camara TN, Bruno RV, Luz PM, Castro MG, Lourenço-de-Oliveira R, Sorgine MHF, et al. Dengue infection increases the locomotor activity of Aedes aegypti females. PLoS ONE. (2011) 6:e17690. doi: 10.1371/journal.pone.0017690
65. Albertson R, Tan V, Leads RR, Reyes M, Sullivan W, Casper-Lindley C. Mapping Wolbachia distributions in the adult Drosophila brain. Cell Microbiol. (2013) 15:1527–44. doi: 10.1111/cmi.12136
66. Morioka E, Oida M, Tsuchida T, Ikeda M. Nighttime activities and peripheral clock oscillations depend on Wolbachia endosymbionts in flies. Sci Rep. (2018) 8:15432. doi: 10.1038/s41598-018-33522-8
67. Rund SSC, Hou TY, Ward SM, Collins FH, Duffield GE. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proc Natl Acad Sci USA. (2011) 108:E421–30. doi: 10.1073/pnas.1100584108
68. Rund SS, Gentile JE, Duffield GE. Extensive circadian and light regulation of the transcriptome in the malaria mosquito Anopheles gambiae. BMC Genomics. (2013) 14:218. doi: 10.1186/1471-2164-14-218
69. Shipp E, Otton J. Orcadian rhythms of sensitivity to insecticides in musca domestica (diptera, muscidae). Entomol Exp Appl. (1976) 19:163–71. doi: 10.1111/j.1570-7458.1976.tb02593.x
70. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. (2014) 20:1006–17. doi: 10.1016/j.cmet.2014.11.008
71. Dickson I. Intestinal microbiota oscillations regulate host circadian physiology. Nat Rev Gastroenterol Hepatol. (2017) 14:67. doi: 10.1038/nrgastro.2016.205
72. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. (2011) 473:174–80. doi: 10.1038/nature09944
73. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. (2013) 155:1451–63. doi: 10.1016/j.cell.2013.11.024
74. Liang X, FitzGerald GA. Timing the microbes: the circadian rhythm of the gut microbiome. J Biol Rhythms. (2017) 32:505–15. doi: 10.1177/0748730417729066
75. Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiol Read Engl. (2010) 156:3216–23. doi: 10.1099/mic.0.040618-0
76. Deaver JA, Eum SY, Toborek M. Circadian disruption changes gut microbiome taxa and functional gene composition. Front Microbiol. (2018) 9:737. doi: 10.3389/fmicb.2018.00737
77. Wu G, Tang W, He Y, Hu J, Gong S, He Z, et al. Light exposure influences the diurnal oscillation of gut microbiota in mice. Biochem Biophys Res Commun. (2018) 501:16–23. doi: 10.1016/j.bbrc.2018.04.095
78. Paulose JK, Wright JM, Patel AG, Cassone VM. Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS ONE. (2016) 11:e0146643. doi: 10.1371/journal.pone.0146643
79. Liang X, Bushman FD, FitzGerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA. (2015) 112:10479–84. doi: 10.1073/pnas.1501305112
80. Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. (2014) 159:514–29. doi: 10.1016/j.cell.2014.09.048
81. Takayasu L, Suda W, Takanashi K, Iioka E, Kurokawa R, Shindo C, et al. Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res Int J Rapid Publ Rep Genes Genomes. (2017) 24:261–70. doi: 10.1093/dnares/dsx001
82. Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. (2015) 17:681–9. doi: 10.1016/j.chom.2015.03.006
83. Beli E, Prabakaran S, Krishnan P, Evans-Molina C, Grant MB. Loss of diurnal oscillatory rhythms in gut microbiota correlates with changes in circulating metabolites in type 2 Diabetic db/db mice. Nutrients. (2019) 11:2310. doi: 10.3390/nu11102310
84. Bishehsari F, Keshavarzian A. Microbes help to track time. Science. (2019) 365:1379–80. doi: 10.1126/science.aaz0224
85. Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, et al. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science. (2019) 365:1428–34. doi: 10.1126/science.aaw3134
86. Godinho-Silva C, Domingues RG, Rendas M, Raposo B, Ribeiro H, da Silva JA, et al. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature. (2019) 576:1–5. doi: 10.1038/s41586-019-1579-3
87. Gibbs J, Ince L, Matthews L, Mei J, Bell T, Yang N, et al. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med. (2014) 20:919–26. doi: 10.1038/nm.3599
88. Kitchen GB, Cunningham PS, Poolman TM, Iqbal M, Maidstone R, Baxter M, et al. The clock gene Bmal1 inhibits macrophage motility, phagocytosis, and impairs defense against pneumonia. Proc Natl Acad Sci USA. (2020) 117:1543–51. doi: 10.1073/pnas.1915932117
89. Konturek PC, Brzozowski T, Konturek SJ. Gut clock: implication of circadian rhythms in the gastrointestinal tract. J Physiol Pharmacol. (2011) 62:139–50.
90. Li T, Shao W, Li S, Ma L, Zheng L, Shang W, et al. Helicobacter pylori infection induced BMAL1 expression and rhythm disorder aggravate gastric inflammation. EBioMedicine. (2019) 39:301–14. doi: 10.1016/j.ebiom.2018.11.043
91. Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. (2013) 341:1483–8. doi: 10.1126/science.1240636
92. Proaño A, Bravard MA, López JW, Lee GO, Bui D, Datta S, et al. Dynamics of cough frequency in adults undergoing treatment for pulmonary tuberculosis. Clin Infect Dis. (2017) 64:1174–81. doi: 10.1093/cid/cix039
93. Bellet MM, Deriu E, Liu JZ, Grimaldi B, Blaschitz C, Zeller M, et al. Circadian clock regulates the host response to Salmonella. Proc Natl Acad Sci USA. (2013) 110:9897–902. doi: 10.1073/pnas.1120636110
94. Shouval D. The impact of chronic hepatitis C infection on the circadian clock and sleep. J Hepatol. (2014) 60:685–6. doi: 10.1016/j.jhep.2014.01.004
95. Benegiamo G, Mazzoccoli G, Cappello F, Rappa F, Scibetta N, Oben J, et al. Mutual Antagonism between circadian protein period 2 and hepatitis C virus replication in hepatocytes. PLoS ONE. (2013) 8:e60527. doi: 10.1371/journal.pone.0060527
96. Zhuang X, Rambhatla SB, Lai AG, McKeating JA. Interplay between circadian clock and viral infection. J Mol Med. (2017) 95:1283–9. doi: 10.1007/s00109-017-1592-7
97. Edgar RS, Stangherlin A, Nagy AD, Nicoll MP, Efstathiou S, O'Neill JS, et al. Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc Natl Acad Sci USA. (2016) 113:10085–90. doi: 10.1073/pnas.1601895113
98. Rijo-Ferreira F, Carvalho T, Afonso C, Sanches-Vaz M, Costa RM, Figueiredo LM, et al. Sleeping sickness is a circadian disorder. Nat Commun. (2018) 9:62. doi: 10.1038/s41467-017-02484-2
99. Hopwood TW, Hall S, Begley N, Forman R, Brown S, Vonslow R, et al. The circadian regulator BMAL1 programmes responses to parasitic worm infection via a dendritic cell clock. Sci Rep. (2018) 8:3782. doi: 10.1038/s41598-018-22021-5
100. Textoris J, Ban LH, Capo C, Raoult D, Leone M, Mege JL. Sex-related differences in gene expression following Coxiella burnetii infection in mice: potential role of circadian rhythm. PLoS ONE. (2010) 5:e12190. doi: 10.1371/journal.pone.0012190
101. Mehraj V, Textoris J, Capo C, Raoult D, Leone M, Mege JL. Overexpression of the Per2 gene in male patients with acute Q fever. J Infect Dis. (2012) 206:1768–70. doi: 10.1093/infdis/jis600
102. Buguet A. Is sleeping sickness a circadian disorder? The serotonergic hypothesis. Chronobiol Int. (1999) 16:477–89. doi: 10.3109/07420529908998722
103. Duncan MJ, Bruce-Keller AJ, Conner C, Knapp PE, Xu R, Nath A, et al. Effects of chronic expression of the HIV-induced protein, transactivator of transcription, on circadian activity rhythms in mice, with or without morphine. Am J Physiol Regul Integr Comp Physiol. (2008) 295:R1680–7. doi: 10.1152/ajpregu.90496.2008
104. Zhuang X, Magri A, Hill M, Lai AG, Kumar A, Rambhatla SB, et al. The circadian clock components BMAL1 and REV-ERBα regulate flavivirus replication. Nat Commun. (2019) 10:377. doi: 10.1038/s41467-019-08299-7
105. Zhuang X, Lai AG, McKeating JA, Rowe I, Balfe P. Daytime variation in hepatitis C virus replication kinetics following liver transplant. Wellcome Open Res. (2018) 3:96. doi: 10.12688/wellcomeopenres.14696.1
106. Smith A, Tyrrell D, Coyle K, Higgins P, Willman J. Diurnal variation in the symptoms of colds and influenza. Chronobiol Int. (1988) 5:411–6. doi: 10.3109/07420528809067786
107. Majumdar T, Dhar J, Patel S, Kondratov R, Barik S. Circadian transcription factor BMAL1 regulates innate immunity against select RNA viruses. Innate Immun. (2017) 23:147–54. doi: 10.1177/1753425916681075
108. Sengupta S, Tang SY, Devine JC, Anderson ST, Nayak S, Zhang SL, et al. Circadian control of lung inflammation in influenza infection. Nat Commun. (2019) 10:4107. doi: 10.1038/s41467-019-11400-9
109. Kalamvoki M, Roizman B. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci USA. (2010) 107:17721–6. doi: 10.1073/pnas.1012991107
110. Shackelford PG, Feigin RD. Periodicity of susceptibility to pneumococcal infection: influence of light and adrenocortical secretions. Science. (1973) 182:285–7. doi: 10.1126/science.182.4109.285
111. Oliva-Ramírez J, Moreno-Altamirano MMB, Pineda-Olvera B, Cauich-Sánchez P, Sánchez-García FJ. Crosstalk between circadian rhythmicity, mitochondrial dynamics and macrophage bactericidal activity. Immunology. (2014) 143:490–7. doi: 10.1111/imm.12329
112. Lopes C, deLyra JL, Markus RP, Mariano M. Circadian rhythm in experimental granulomatous inflammation is modulated by melatonin. J Pineal Res. (1997) 23:72–8. doi: 10.1111/j.1600-079X.1997.tb00338.x
113. Lou J, Wang Y, Zhang Z, Qiu W. Activation of MMPs in macrophages by Mycobacterium tuberculosis via the miR-223-BMAL1 signaling pathway. J Cell Biochem. (2017) 118:4804–12. doi: 10.1002/jcb.26150
114. Raghupati Sarma G, Immanuel C, Ramachandran G, Krishnamurthy PV, Kumaraswami V, Prabhakar R. Adrenocortical function in patients with pulmonary tuberculosis. Tubercle. (1990) 71:277–82. doi: 10.1016/0041-3879(90)90041-6
115. York EL, Enarson DA, Robert EJ, Fanning FA, Sproule BJ. Adrenocortical function in patients investigated for active tuberculosis. Chest. (1992) 101:1338–41. doi: 10.1378/chest.101.5.1338
116. Kato C, Chung I, Paddock C. Estimation of Rickettsia rickettsii copy number in the blood of patients with Rocky Mountain spotted fever suggests cyclic diurnal trends in bacteraemia. Clin Microbiol Infect. (2016) 22:394–6. doi: 10.1016/j.cmi.2015.12.019
117. Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. (2013) 369:840–51. doi: 10.1056/NEJMra1208623
118. Deng W, Zhu S, Zeng L, Liu J, Kang R, Yang M, et al. The circadian clock controls immune checkpoint pathway in sepsis. Cell Rep. (2018) 24:366–78. doi: 10.1016/j.celrep.2018.06.026
119. Truong KK, Lam MT, Grandner MA, Sassoon CS, Malhotra A. Timing matters: circadian rhythm in sepsis, obstructive lung disease, obstructive sleep apnea, and cancer. Ann Am Thorac Soc. (2016) 13:1144–54. doi: 10.1513/AnnalsATS.201602-125FR
120. Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, et al. The nuclear receptor REV-ERB mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. (2012) 109:582–7. doi: 10.1073/pnas.1106750109
121. Alamili M, Klein M, Lykkesfeldt J, Rosenberg J, Gögenur I. Circadian variation in the response to experimental endotoxemia and modulatory effects of exogenous melatonin. Chronobiol Int. (2013) 30:1174–80. doi: 10.3109/07420528.2013.808653
122. Alamili M, Bendtzen K, Lykkesfeldt J, Rosenberg J, Gögenur I. Pronounced inflammatory response to endotoxaemia during nighttime: a randomised cross-over trial. PLoS ONE. (2014) 9:e87413. doi: 10.1371/journal.pone.0087413
123. Pollmächer T, Mullington J, Korth C, Schreiber W, Hermann D, Orth A, et al. Diurnal variations in the human host response to endotoxin. J Infect Dis. (1996) 174:1040–5. doi: 10.1093/infdis/174.5.1040
124. Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. (2012) 36:251–61. doi: 10.1016/j.immuni.2011.12.017
125. Coiffard B, Diallo AB, Culver A, Mezouar S, Hammad E, Vigne C, et al. Circadian rhythm disruption and sepsis in severe trauma patients. Shock. (2019) 52:29–36. doi: 10.1097/SHK.0000000000001241
126. Telias I, Wilcox ME. Sleep and circadian rhythm in critical illness. Crit Care. (2019) 23:82. doi: 10.1186/s13054-019-2366-0
127. Erland LAE, Saxena PK. Melatonin natural health products and supplements: presence of serotonin and significant variability of melatonin content. J Clin Sleep Med. (2017) 13:275–81. doi: 10.5664/jcsm.6462
128. Hatta K, Kishi Y, Wada K, Takeuchi T, Odawara T, Usui C, et al. Preventive effects of ramelteon on delirium: a randomized placebo-controlled trial. JAMA Psychiatry. (2014) 71:397–403. doi: 10.1001/jamapsychiatry.2013.3320
129. Nishikimi M, Numaguchi A, Takahashi K, Miyagawa Y, Matsui K, Higashi M, et al. Effect of administration of ramelteon, a melatonin receptor agonist, on the duration of stay in the ICU: a single-center randomized placebo-controlled trial. Crit Care Med. (2018) 46:1099–105. doi: 10.1097/CCM.0000000000003132
130. Devlin JW, Skrobik Y, Gélinas C, Needham DM, Slooter AJC, Pandharipande PP, et al. Clinical practice guidelines for the prevention and management of pain, agitation/sedation, delirium, immobility, and sleep disruption in adult patients in the ICU. Crit Care Med. (2018) 46:e825–73. doi: 10.1097/CCM.0000000000003259
131. Culver A, Coiffard B, Antonini F, Duclos G, Hammad E, Vigne C, et al. Circadian disruption of core body temperature in trauma patients: a single-center retrospective observational study. J Intensive Care. (2020) 8:4. doi: 10.1186/s40560-019-0425-x
132. Halberg F, Galicich JH, Ungar F, French LA. Circadian rhythmic pituary adrenocorticotropic activity, rectal temperature and pinnal mitosis of starving, dehydrated C mice. Proc Soc Exp Biol Med. (1965) 118:414–9. doi: 10.3181/00379727-118-29861
133. Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinforma Oxf Engl. (2016) 32:3351–3. doi: 10.1093/bioinformatics/btw405
134. Parsons R, Parsons R, Garner N, Oster H, Rawashdeh O. CircaCompare: a method to estimate and statistically support differences in mesor, amplitude and phase, between circadian rhythms. Bioinformatics. (2020) 36:1208–12. doi: 10.1093/bioinformatics/btz730
135. Mandal AS, Biswas N, Karim KM, Guha A, Chatterjee S, Behera M, et al. Drug delivery system based on chronobiology-A review. J Control Release. (2010) 147:314–25. doi: 10.1016/j.jconrel.2010.07.122
136. Dallman MF. Early life stress: nature and nurture. Endocrinology. (2014) 155:1569–72. doi: 10.1210/en.2014-1267
137. Ozturk N, Ozturk D, Kavakli IH, Okyar A. Molecular aspects of circadian pharmacology and relevance for cancer chronotherapy. Int J Mol Sci. (2017) 18:2168. doi: 10.3390/ijms18102168
138. Papaioannou V, Mebazaa A, Plaud B, Legrand M. ‘Chronomics’ in ICU: circadian aspects of immune response and therapeutic perspectives in the critically ill. Intensive Care Med Exp. (2014) 2:18. doi: 10.1186/2197-425X-2-18
139. Papaioannou VE, Sertaridou EN, Chouvarda IG, Kolios GC, Pneumatikos IN. Determining rhythmicity and determinism of temperature curves in septic and non-septic critically ill patients through chronobiological and recurrence quantification analysis: a pilot study. Intensive Care Med Exp. (2019) 7:53. doi: 10.1186/s40635-019-0267-9
140. McKenna H, van der Horst GTJ, Reiss I, Martin D. Clinical chronobiology: a timely consideration in critical care medicine. Crit Care. (2018) 22:124. doi: 10.1186/s13054-018-2041-x
141. Long JE, Drayson MT, Taylor AE, Toellner KM, Lord JM, Phillips AC. Morning vaccination enhances antibody response over afternoon vaccination: A cluster-randomised trial. Vaccine. (2016) 34:2679–85. doi: 10.1016/j.vaccine.2016.04.032
142. Carvalho MF, Gill D. Rotavirus vaccine efficacy: current status and areas for improvement. Hum Vaccines Immunother. (2019) 15:1237–50. doi: 10.1080/21645515.2018.1520583
143. Abat C, Rolain JM, Dubourg G, Fournier PE, Chaudet H, Raoult D. Evaluating the clinical burden and mortality attributable to antibiotic resistance: the disparity of empirical data and simple model estimations. Clin Infect Dis. (2017) 65:S58–63. doi: 10.1093/cid/cix346
144. Bukharin OV, Perunova NB, Fadeev SB, Timokhina TK, Iavnova SV. Biorhythms of antibiotic resistance of microorganisms. Zh Mikrobiol Epidemiol Immunobiol. (2008) 35–8.
Keywords: circadian rhythm, clock genes, microorganisms, microbiota, immune response, infectious diseases
Citation: Diallo AB, Coiffard B, Leone M, Mezouar S and Mege J-L (2020) For Whom the Clock Ticks: Clinical Chronobiology for Infectious Diseases. Front. Immunol. 11:1457. doi: 10.3389/fimmu.2020.01457
Received: 07 February 2020; Accepted: 04 June 2020;
Published: 09 July 2020.
Edited by:
Alexandre Corthay, Oslo University Hospital, NorwayReviewed by:
Axel Lorentz, University of Hohenheim, GermanyTina Sørensen Dalgaard, Aarhus University, Denmark
Copyright © 2020 Diallo, Coiffard, Leone, Mezouar and Mege. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soraya Mezouar, soraya.mezouar@univ-amu.fr; Jean-Louis Mege, jean-louis.mege@univ-amu.fr
†These authors have contributed equally to this work and share first authorship
 Aïssatou Bailo Diallo
Aïssatou Bailo Diallo Benjamin Coiffard
Benjamin Coiffard Marc Leone
Marc Leone Soraya Mezouar
Soraya Mezouar Jean-Louis Mege
Jean-Louis Mege