Microbial Diversity Exploration of Marine Hosts at Serrana Bank, a Coral Atoll of the Seaflower Biosphere Reserve
- 1Centro de Bioinformática y Biología Computacional BIOS, Manizales, Colombia
- 2Vicerrectoría de Investigación, Universidad de los Andes, Bogotá, Colombia
Microorganisms represent nearly 90% of ocean biomass and are fundamental for the functioning and health of marine ecosystems due to their integral contribution to biogeochemical cycles and biological processes. In marine environments, microorganisms exist as microbial communities in the water column, benthonic substrates, and macroorganisms, where they establish symbiotic interactions and fulfill their ecological roles. Such interactions can have a harmful or beneficial impact on the hosts depending on the emergent properties of the communities, their taxonomic structure, and functionality. To evaluate these features, culture independent approaches like metabarcoding have been developed and have hugely contributed to the characterization of marine microbial diversity. The present study was aimed to explore the structure and metabolic functionality of microbial communities associated to marine hosts at the Serrana Bank, a coral atoll part of the Seaflower Biosphere Reserve (Archipelago of San Andrés, Old Providence and Saint Catalina, Colombia). We found a highly diverse microbial assemblage associated with the corals Siderastrea siderea, Colpophyllia natans, and Orbicella annularis, the sponge Haliclona sp. and sediment from Isla de los Pájaros lagoon. However, the coral Porites astreoides had significantly lower bacterial diversity and a different community composition. Proteobacteria was the most abundant phylum within bacterial communities in the evaluated hosts, except in P. astreoides, where Cyanobacteria was the predominant group. Firmicutes, Actinobacteria, Bacteroidetes, Acidobacteria, Chloroflexi, and Gemmatimonadetes were also identified within all microbiomes, but their dominance varied between hosts. Additionally, the most abundant group among the fungi communities associated with O. annularis, S. siderea, and C. natans was Ascomycota, but significant differences between clasess and order were observed among hosts. Finally, functional profiles revealed that the principal microbial functions were focused on membrane transport, carbohydrates, amino acids and energy metabolism, replication, and translation processes. A significant higher metabolic functionality was found in the sponge microbiome in comparison to the coral microbial communities.
Introduction
In marine environments, microorganisms represent nearly 90% of the biomass, are responsible for up to 98% of primary marine productivity, and are key players in mass and energy fluxes through biogeochemical processes (carbon, nitrogen, sulfur cycling, etc.) (Caron, 2005; Sogin et al., 2006). It is estimated that up to 3.5 × 1030 microorganisms live on the oceanic subsurface establishing complex communities. They can be found in the water column as plankton, in benthonic substrates, and adhered to macroorganisms (Glöckner et al., 2012; Grossart et al., 2013).
Among the macroorganisms that inhabit marine ecosystems, corals and sponges are a major component and they have existed for millions of years as structural and functional pieces. Coral reefs have an immense biodiversity, which can be compared to that of the tropical rain forest and contributes largely to the primary productivity in marine environments (Cárdenas et al., 2012). Sponges play a functional role in coral reefs in several ways: they enhance coral survival by binding corals to the reef frame, mediate regeneration of physical impacts on reefs by temporary stabilization of carbonate rubble, and take part in nutrient cycling through microbial symbionts (Bell, 2008). These organisms not only contribute to the global ocean productivity but also provide nutrients and shelter for other organisms (Wulff, 2006; Coker et al., 2014). Corals, sponges, and other marine organisms have been exposed to both natural and anthropogenic threats: in Colombia, 60% of coral reefs are at risk, 20% may disappear in the next decade, 19% have been destroyed, and 15% are in a critical conservation status (Galeano et al., 2016). These threats have led to a decline of their populations and changes in the associated microbiota living in their surface (Riegl et al., 2009).
The microbiome has an impact on its host, which could be harmful or beneficial depending on its microbial structure and functionality (Konopka, 2009; Kellogg et al., 2014). Microbial communities interact and establish symbiotic associations at the surface of macroorganisms. When there are shifts in the composition of these communities, hosts have several impacts including changes in the intake of nutrients, absorption of light (biofilm formation), and interaction with potential pathogens (Webster et al., 2008; Wahl et al., 2012; Zhang et al., 2015).
One of the major limitations to study structure and functionality of marine microbial communities has been the inability to cultivate around 99% of the microorganisms (Delmont et al., 2011). Advances in molecular biology and sequencing technologies have led to the development of new approaches including metabarcoding and metagenomics to overcome this limitation (Streit and Schmitz, 2004; Pavan-Kumar et al., 2015). These approaches have contributed to the knowledge that microbes represent a pivotal role on earth in terms of phylogenetic and functional diversity (Debnath et al., 2007; Dionisi et al., 2012; Glöckner et al., 2012). Metabarcoding studies on marine hosts like macro-algae, corals, and sponges have shown that microbial communities are not distributed randomly; their prevalence depends on a species-specific structure (Lachnit et al., 2011; Morrow et al., 2012) which can change in response to environmental factors or host features such as age, diseases, and physiological state (Lachnit et al., 2011; Lawler et al., 2016).
The Seaflower Biosphere Reserve, located in the Colombian Caribbean sea, covers an extension of 65.018 km2 of a marine protected area (MPA) created to preserve the biodiversity of the Archipelago of San Andrés, Old Providence, and Saint Catalina (Comisión Colombiana del Oceáno, 2015). The reserve encompasses three inhabited islands and several cays, banks, and atolls. One of these banks is Serrana Bank (14°23′00″ N, 80°16′00″ W), a reef complex located 150 km northwest of Old Providence Island (Figure 1). It has seven cays of which only the southwest cay “Serrana” (500 m × 200 m) has established vegetation (Díaz et al., 1996); another cay “Isla de los Pájaros” has a small extension, but it is an important station for hundreds of migrating birds species.
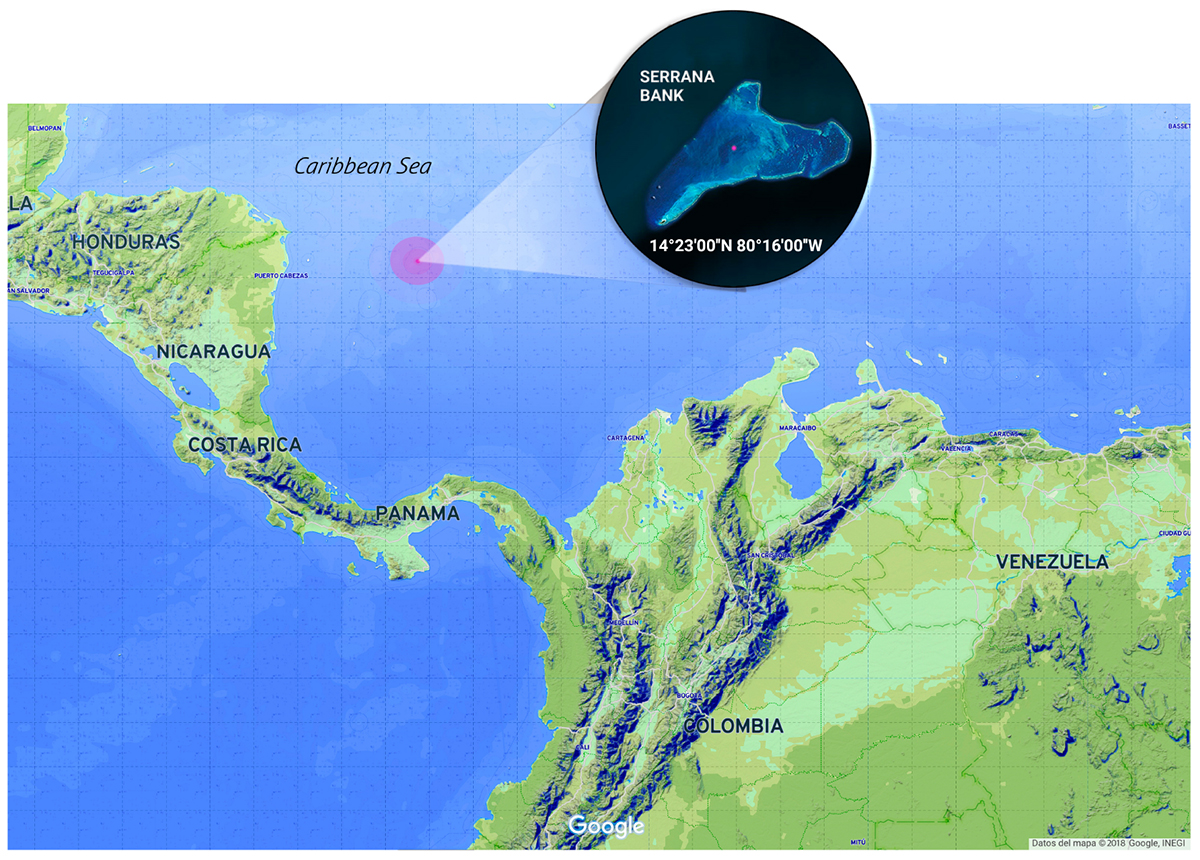
Figure 1. Serrana Bank location at the Seaflower Biosphere Reserve (Colombia). Samples collection area (red dot). Image edited from Google.
The reserve is a place largely unexplored within the third largest barrier reef of the world (Comisión Colombiana del Oceáno, 2015). Its great biodiversity includes more than 407 fish species, 48 hard corals, 54 soft corals, and 130 sponges. Starting in 2014, scientific explorations to generate knowledge about this diverse ecosystem have been launched. To date these explorations have contributed to the characterization of its flora and fauna, and the evaluation of the oceanographic and meteorological aspects of the reserve (Howard, 2006).
To our knowledge, no studies have been undertaken to characterize marine microbial diversity, or to evaluate microbiomes associated with specific macroorganisms on any ecosystem of the Seaflower Biosphere Reserve. The aim of this study was to assess the diversity, taxonomic structure, and potential metabolic functionality of microbial communities (bacteria and fungi) associated with two marine hosts by 16S rRNA and ITS gene metabarcoding on the Serrana Bank, one of the unexplored areas of the reserve.
Methodology and Tools
Experimental Design and Sampling
Serrana is an extensive reef complex, with a 37 km × 30 km dimension, including the insular platform. There is a lagoon with numerous seaweed patches and sea grass beds, which are highly productive. There is a secondary barrier, long and narrow, with the coral Acropora palmata as the dominant species, creating a quiet environment with reef patches (Montastraea) covering 60% of the deep bottom. Other coral species including Agaricia agaricites, Porites spp., Mycetophyllia ferox, Diploria spp., and Siderastrea siderea are also frequent in this coral reef habitat (Howard et al., 2004).
Our aim was to identify and analyze microbiomes associated with corals, algae, and sponges inhabiting the reserve. In addition, we collected a sample from sediment from a cay lagoon to compare its microbial composition and diversity with that of the macroorganisms sampled. All samples were collected by triplicate.
Eight microbial samples associated to corals were collected by scuba diving, at depths ranging from 7 to 12 m (Supplementary Table S1). Mucus on the surface of corals was collected using sterile syringes (10 ml). The mucus was gently shaken using the plastic tip of syringes to get rid of the viscous material and reduce pumping of seawater. Nitrile gloves were worn during collection to reduce human bacterial contamination and syringes were capped after collection to prevent further seawater contamination. The collected mucus was transferred to 2 ml tubes within the next hour and preserved in 70% alcohol. Samples were stored at 4°C for further analyses.
Six microbial samples associated to algae and one associated to sponges were collected by snorkeling and scuba diving, at depths ranging from 3 to 12 m (Supplementary Table S1). These specimens were collected by hand and stored in sterile plastic bags for 3 h. At the beach, samples were preserved in 70% alcohol and kept at 4°C for further analyses.
A sediment sample from the “Isla de los Pájaros” cay lagoon was collected during the expedition. This sample was collected at 0.5 m depth by hand, wearing nitrile gloves, stored immediately in 2 ml tubes with 70% alcohol, and cooled down for further analyses. The complete metadata associated with collected samples is described in Supplementary Table S1.
DNA Extraction and Sequencing
Total community DNA was extracted using the GeneJET Genomic DNA Purification Kit, GeneJET Plant Genomic DNA Purification Kit, and DNeasy Plant Mini Kit, according to manufacturer’s instructions with some required modifications (pressure changes, mechanical fragmentation, and cell lysis). DNA quality was evaluated by electrophoresis in agarose gels and spectrophotometry.
Amplicon library preparation and sequencing were performed by Novogene1, Chula Vista, CA, United States. The V4 hypervariable region of the 16S rRNA gene was amplified with the primers 515F/806R (5′-GTGCCAGCMGCCGCGGTAA-3′/5′-GGACTACHVGGGTWTCTAAT-3′), and for some samples, the ITS2 region was also amplified using the primers ITS3/ITS4 (5′-GCATCGATGAAGAACGCAGC-3′/5′-TCCTCCGCTTATTGATATGC-3′). All PCR reactions were carried out in 30 μl total volume with 15 μl of Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM of forward and reverse primers, and 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, elongation at 72°C for 30 s, and a final cycle at 72°C for 5 min. Libraries were sequenced using the Illumina HiSeq platform (Q30 > 80%) to obtain 250 bp paired reads (Table 1).
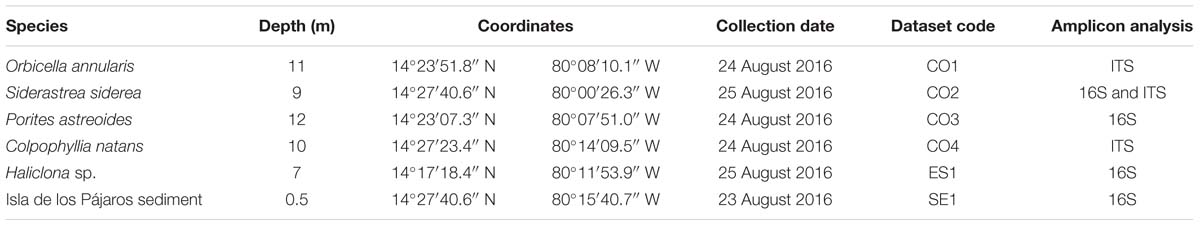
Table 1. Metadata associated to the samples collected and sequenced at the Seaflower Biosphere Reserve.
Amplicon Analyses
Taxonomic analyses were performed using QIIME 2 release 2017.92. The raw sequences quality control was ran with DEMUX plugin and reads were trimmed to a length of 250 bp using the DADA2 plugin. Filtered sequences were aligned and clustered into operational taxonomic units (OTUs) and assigned to clusters using a QIIME feature, which uses a Naive Bayes classifier to map each sequence to taxonomy. The classifier was trained on a qiime-compatible Silva database (release 119), Greengenes (Greengenes v13_5), and UNITES reference databases with 97% similarity. Statistical analyses were computed in R using the qiime2R v.0.99.13, and phyloseq (McMurdie and Holmes, 2013), a common tool for ecological analyses of microbiome data in R/Bioconductor; abundance plots were computed with ggplot2 (Wickham, 2016), retrieving the 20 most abundant classification groups (phylum, class, order, and family). Diversity indexes (Observed Shannon, Simpson) and rarefaction curves were calculated and constructed and, beta-diversity estimates were calculated within QIIME using Bray–Curtis distance between samples. Principal coordinate analyses (PCoA) and non-metric multidimensional scaling (NMDS) were computed from the resulting distance matrices to compress dimensionality into two-dimensions, enabling visualization of sample relationships.
Additionally, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to predict the functional profiles of microbial communities associated to hosts (Langille et al., 2013). Because PICRUSt uses “closed-reference” OTU for prediction, the filter_otus_from_otu_table.py script was used to get an output compatible with the OTUs table generated with QIIME 2. Function profiles were created based on the KEGG functional annotations and the output table was collapsed at KO level 2 to identify pathways significantly different among samples. One-way ANOVA with 999 permutations (P < (0.05), and pairwise Tukey’s tests were estimated. Relative abundances of KEGG level 2 gene ontologies were plotted in R using ggplot2.
Results
Sixteen samples were collected as part of the Seaflower Scientific Expedition 2016 (Supplementary Table S1), 8 from corals, 6 from algae, 1 from sponge, and 1 from the lagoon sediment. DNA extraction was unsuccessful for 10 of these samples; hence, the final 16S analysis was limited to four samples, and the ITS tests were restricted to three samples (Table 1). The sequenced microbiomes are in association with:
Orbicella annularis (CO1): O. annularis is a coral belonging to the family Merulinidae found between 1.5 and 82 m depth, and it is generally the most abundant coral up to 10 m of depth in semi-protected coral reefs, lagoons, and in the upper part of reef pendants (Reed, 1985). O. annularis colonies are massive, columnar, or flat; corallites can be flush with the surface or they can be conic. Its septa are uniformly arranged, alternating large and short septa, and its columellas are small and compact, and its color varies between brown and yellow (Ellis and Solander, 1786). This coral is considered an endangered species, its populations have decreased over 50% in the last 30 years through diseases and bleaching effects, and other factors of anthropogenic origin (Edmunds, 2015).
Siderastrea siderea (CO2): S. siderea, also known as the massive starlet coral, belongs to the family Siderastreidae found generally in depths between 0.5 and 2 m, forming reef patches toward the inner side of the reef. Its colonies are hemispheric or encrusting, some with >2 m in diameter, with more than 48 radial elements directed in a 45° pendant toward the columella. Its septa are highly compact shaping a soft surface; calices measure from 4 to 5 mm in diameter. It has a brown color with reddish tones (Ellis and Solander, 1786).
Porites astreoides (CO3): P. astreoides is a very common coral belonging to the family Poritidae that can be found in almost every reef environment, ranging from shallow to deep zones. This coral can be massive, laminar, or subnodular, covered by little protuberances or smooth; its colonies are hemispheric, flattened, or encrusting, generally not >60 cm in diameter; its calices measure from 1.25 to 1.5 mm in diameter, its 12 radial elements are porous, and its columella is also porous and very small with rough denticles in the septa. P. astreoides has a light green to yellowish brown color, and it adapts to different growth patterns; it is generally small but it can also cover extensions that reach several meters (Green et al., 2008).
Colpophyllia natans (CO4): C. natans belongs to the family Mussidae, it lives in large colonies which can measure >1 m in diameter. The largest colonies commonly have a regular hemispheric shape, while smaller colonies have a plane disc shape. In some cases, valleys of the coral extend completely across the colony or can be sub-divided into smaller series; valleys and walls are large measuring up to 2 cm width, walls usually present grooves along the cups, and there is a clear limit between the wall and the valley. Its septa have identical thickness and they have less than 12 septa per centimeter. The columella is discontinuous and thin, less than a quarter of the valleys thickness. This coral inhabits the upper part of reefs and slopes up to 50 m depth; ridges are generally brown and valleys are green, brown, or whitish (De-Kluijver et al., 2016).
Haliclona sp. (ES1): The genus Haliclona groups the encrusting-type sponges belonging to the family Chalinidae, which is normally found anchored to rocks and branches of coral reefs. Sponges have been widely studied and recognized as the largest sources of marine bioactive secondary metabolites, which represent up to 40% of all known natural marine products (Lee et al., 2001). Many of these metabolites have been linked with a microbial origin (Haygood et al., 1999; Lozada and Dionisi, 2015). In the case of Haliclona spp., at least 190 secondary metabolites have been identified exhibiting biological activities like antibiotic, antifungal, antimicrobial, antimalarial, cytotoxic activities, among others (Yu et al., 2006; Hoppers et al., 2015). Several of these compounds are produced by their associated microbiota, which make microbiomes the key players in the functional capabilities of sponges (Kennedy et al., 2009; Khan et al., 2011, 2014).
“Isla de los Pájaros” Sediment (SE1): Cayo Pájaro is a small island located at the Serrana reef complex and it is recognized as a stopover for thousands of migrating birds. It has a little lagoon with a rocky bottom, which is used as a hydration source for inhabitant birds of Serrana Bank and other Seaflower Reserve cays.
After sequencing, a total of 639,010 reads were obtained from the samples sequenced by 16S and 190,008 passed quality filters. Likewise, 517.769 reads were generated from the ITS sequencing of which 254,224 were quality accepted. These reads were used to perform the metabarcoding analysis.
Bacterial Profiling of Marine Hosts at Seaflower
The 16S reads were assigned to 35 different bacterial phyla, 95 classes, 134 orders, 196 families, and 282 genera (Supplementary Table S1). Proteobacteria was the most abundant phylum in S. siderea (39.35%), Haliclona sp. (38.88%), and the sediment microbiomes (65.43%), in contrast to P. astreoides, where bacteria from the phylum Cyanobacteria were predominant (32.17%). Firmicutes, Actinobacteria, Bacteroidetes, Acidobacteria, Chloroflexi, and Gemmatimonadetes were also identified, but they showed different dominance between hosts (Figure 2A). In addition, among the archaeal groups, three different phyla, five classes, seven orders, four families, and four genera were found. Crenarchaeota (0.16–1.21%) and Euryarchaeota (<1%) were identified as the predominant archaeal phyla.
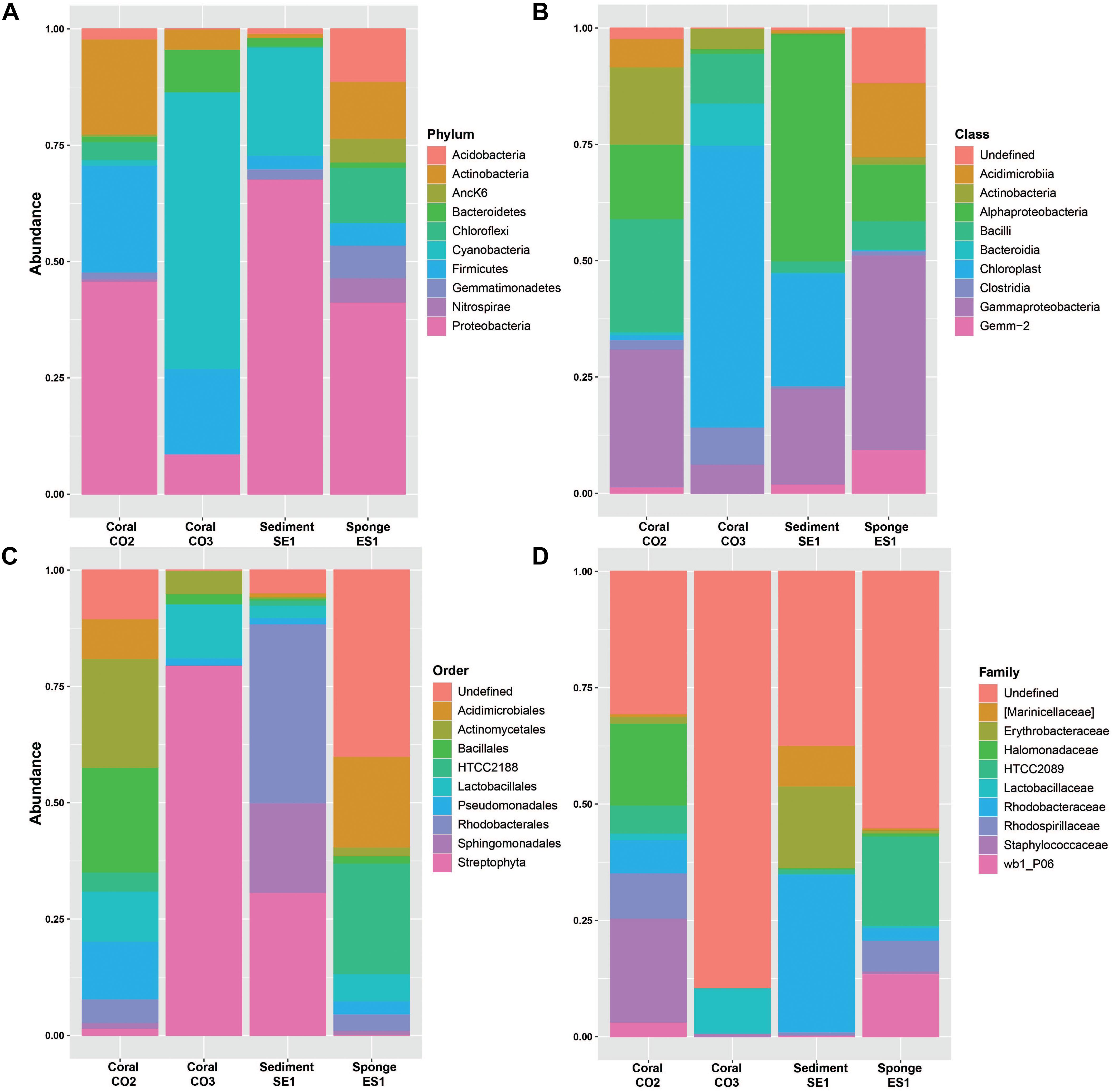
Figure 2. Relative abundances of bacterial microorganisms in marine hosts at the Seaflower Biosphere Reserve. (A) Phylum. (B) Class. (C) Order. (D) Family.
The taxonomic assignments, class and order, showed that dominant groups were very different between hosts (Figures 2B,C). The two more abundant classes within the Proteobacteria group were Gamma and Alphaproteobacteria. Bacilli and Clostridia were predominant within the Firmicutes group and Chloroplast was the most dominant class of the Cyanobacteria group. Actinobacteria and Acidimicrobiia were the predominant classes within Actinobacteria. The dominant class from Bacteroidetes was Bacteroidia, and Gemm-2 was identified as the most common class from the Gemmatimonadetes group. Additionally, the most represented classes within the Acidobacteria group were Sva0725 and Solibacteres, and Anaerolineae and SAR202 the most abundant classes within the Chloroflexi group.
Taxonomic assignments showed that the most predominant orders among the microbiomes associated to marine hosts were Streptophyta, Rhodobacterales, Unclassified Gemm-2, Sphingomonadales, Actinomycetales, Bacillales, HTCC2188, Acidimicrobiales, Pseudomonadales, and Lactobacillales. Rhodospirillales, Sva0725, Clostridiales, Rhizobiales, Caldilineales, Thiohalorhabdales, Marinicellales, and Oceanospirillales were also abundant among hosts. Family classification related to these groups is shown in Figure 2D.
Bacterial Community Composition Associated With Siderastrea siderea (CO2)
A total of 19 bacterial orders, 125 genera, and 34 species were identified to be specific for S. siderea (Supplementary Figure S1). Among the archaeal orders, vadinCA11 and Methanobrevibacter were associated to these groups. The metabarcoding analysis showed that Proteobacteria (39.35%), Firmicutes (21.13%), and Actinobacteria (20.18%) were the predominant phyla associated with this coral. A smaller proportion of Bacteroidetes (7.61) Chloroflexi (2.84%), Cyanobacteria (2.53%), Acidobacteria (1.71%), Gemmatimonadetes (1.01%), PAUC34f (0.95%), Planctomycetes (0.53%), Verrucomicrobia (0.48%), Nitrospirae (0.38%), and Spirochaetes (0.34%) were found within the microbiome. Gammaproteobacteria (21.58%), Bacilli (17.56%), Actinobacteria (16.66%), and Alphaproteobacteria (14.23%) were the dominant bacterial classes in association with this coral species, while Actinomycetales (19.86%) and Bacillales (14.08%) were the predominant orders in this host. Bacterial species included Shewanella algae, Janthinobacterium lividum, Candidatus Portiera, Acinetobacter johnsonii, Pseudomonas viridiflava, Bacteroides barnesiae, Pseudomonas fragi, Lactococcus garvieae, and Acinetobacter guillouiae.
Bacterial Community Composition Associated With Porites astreoides (CO3)
A total of 4 orders, 29 genera, and 6 species were found to be specific for P. porites (Supplementary Figure S1). Cyanobacteria (32.17%) and Firmicutes (21.99%) were the predominant phyla within the microbiome associated to this coral species, followed by Proteobacteria (21.24%) and Bacteroidetes (16.35%). Chloroplast (34.27%), Gammaproteobacteria (14.49%), Bacteroidia (12.65%), Bacilli (12.21%), and Clostridia (11.21%) were found as the main classes on this coral host. Streptophyta (41.96%), Bacteroidales (15.49%), Clostridiales (13.68%), and Lactobacillales (10.48%) were identified as the dominant orders. For this coral it was possible to identify 78 genera including, Streptophyta-related genera, Thermomonas, Flavisolibacter, Acinetobacter, Ramlibacter, Lactobacillus, and Vibrio and the species A. guillouiae, C. Portiera, Prevotella melaninogenica, Corynebacterium variabile, and Vibrio sp.
Bacterial Community Composition Associated With Haliclona sp. (ES1)
A total of two orders, four genera, and four species were found to be specific for Haliclona sp. (Supplementary Figure S1). For this group of sponges, Proteobacteria (38.88%) was the most abundant associated phylum, followed by Chloroflexi (12.36%), Actinobacteria (11.28%), Acidobacteria (10.33%), and Gemmatimonadetes (6.24%). A significant number of Nitrospirae (4.66%), Firmicutes (4.55%), and AncK6 (4.48%) was also identified. Gammaproteobacteria (28.54%), Acidimicrobia (10.72%), and Alphaproteobacteria (8.45%) were the dominant classes.
HTCC2188 (15.92%), Acidimicrobiales (13.09%), Sva0725 (6.45%), Caldilineales (6.44%%), Thiohalorhabdales (6.03%), Rhodospirillales (5.48%), and Chromatiales (4.98%) were identified as the predominant orders associated to this sponge species, HTCC2089 (14.14%), wb1_P06 (9.97%), Rhodospirillaceae (4.94%), and Rhodobacteraceae (1.99%) as the most abundant families, and A. johnsonii, S. algae, Sphingomonas yabuuchiae, Acinetobacter lwoffii, P. fragi, Bacteroides plebeius, P. viridiflava, and Prevotella copri as some of the species identified.
Bacterial Community Composition Associated With “Isla de los Pájaros” Sediment (SE1)
A total of 21 orders, 27 genera, and 2 species were found to be specific for the sediment (Supplementary Figure S1). Proteobacteria (65.43%) was the most abundant microbial group in association with the sediment, while less frequently Cyanobacteria (17.13%), Planctomyces (3.66%), Bacteroidetes (2.96%), Firmicutes (2.28%), and Gemmatimonadetes (1.91%) were found. Alphaproteobacteria (48.42%), Gammaproteobacteria (20.33%), and Chloroplast (18.19%) were identified as the predominant classes within the microbiome. Rhodobacterales (33.62%), Streptophyta (21.56%), Sphingomonadales (16.21%), Marinicellales (6.92%), Alteromonadales (4.13%), Rhizobiales (2.92%), and Thiotrichales (2.03%) were the predominant orders and Rhodopirellula baltica, Octadecabacte antarcticus, Tunicatimonas pelagia, A. johnsonii, Anaerospora hongkongensis, S. yabuuchiae, Brevibacterium aureum, and Rubidimonas crustatorum were some of the identified species.
Eukaryotic Profiling of Marine Hosts at Seaflower
ITS reads were assigned to 2 fungi phyla, 11 different classes, 28 orders, 46 families, 56 genera, and 42 species (Supplementary Table S3). The most abundant phylum among the microbial communities associated to O. annularis, S. siderea, and C. natans was Ascomycota (83.85%, 97.41%, and 99.40%, respectively) and Basidiomycota (3.64%, 2.26%, and 0.02%, respectively) had a very low representation among microbiomes (Figure 3A). Dothideomycetes, Sordariomycetes, Eurotiomycetes, Saccharomycetes, and Tremellomycetes were the predominant classes within the identified phyla (Figure 3B).
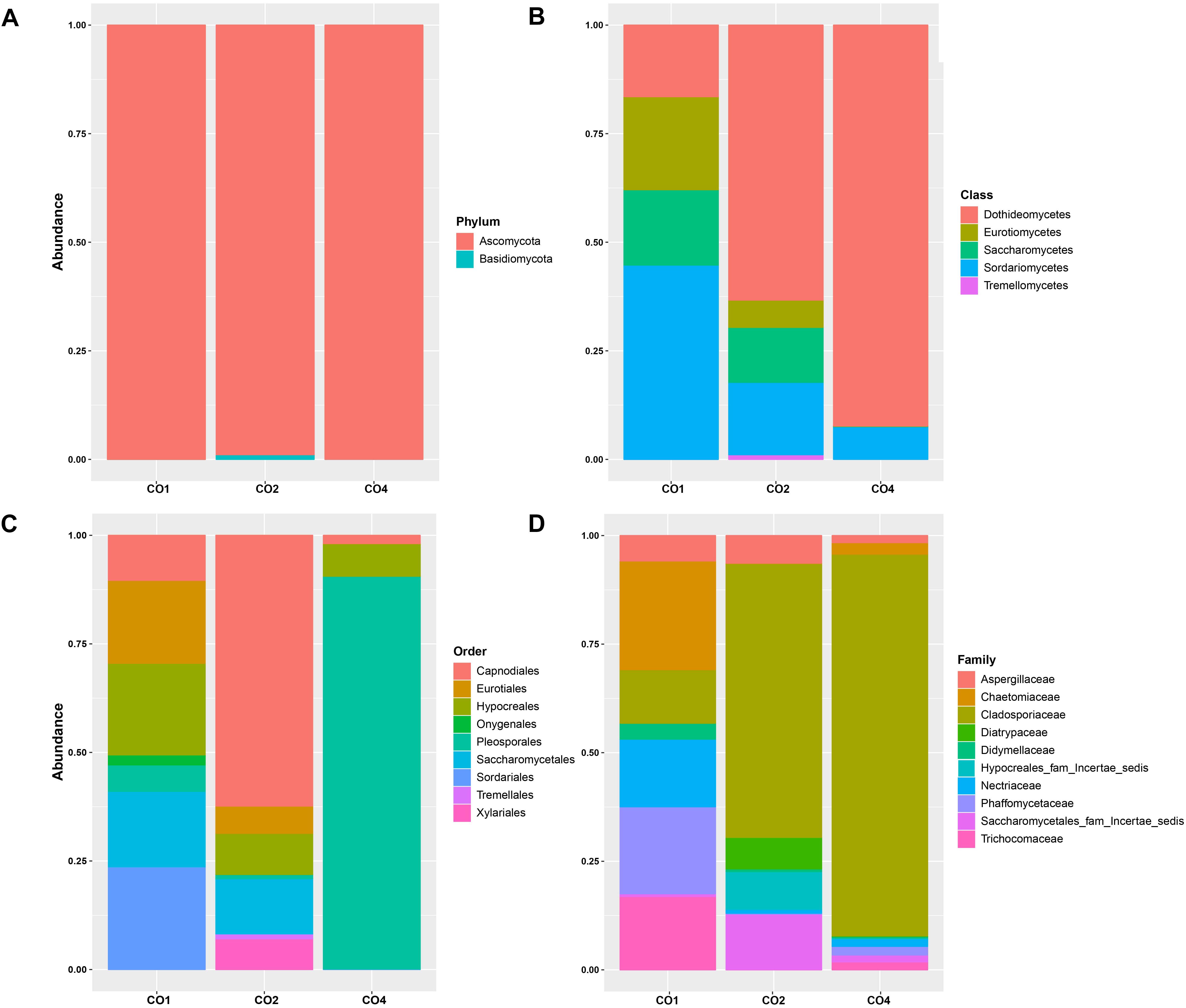
Figure 3. Relative abundances of eukaryotic microorganisms in marine hosts at the Seaflower Biosphere Reserve. (A) Phylum. (B) Class. (C) Order. (D) Family.
The most frequent orders were different among hosts, with Pleosporales, Capnodiales, Sordariales, Hypocreales, Eurotiales, Saccharomycetales, Onygenales, Xylariales, and Tremellames as the predominant orders within microbiomes (Figure 3C). Family classification related to these groups is shown in Figure 3D.
Eukaryotic Community Composition Associated With Orbicella annularis (CO1)
A total of two orders, seven genera, and four species were found to be specific for O. annularis (Supplementary Figure S2). Ascomycota (83.85%) was the predominant group within the microbiome of O. annularis. Sordariomycetes (42.35%), Saccharomycetes (15.32%) and Eurotiomycetes (12.73%) were identified as the dominant fungi classes, while Dothideomycetes (9.14%) was also identified as abundant. Among the orders Sordariales (28.41%), Hypocreales (21.06%), and Saccharomycetales (18.63%) were the dominant groups, and Cyberlindnera rhodanensis, Mycothermus thermophilus, Purpureocillium lilacinum, Trichoderma virens, and Fusarium solani were identified as common fungal species.
Eukaryotic Community Composition Associated With Siderastrea siderea (CO2)
A total of 10 fungi orders, 25 genera, and 23 species were found to be specific for S. siderea (Supplementary Figure S2). The eukaryotic communities identified in this study were limited to Ascomycota (97.41%) and Basidiomycota (2.26%) groups, in contrast to previous studies in which Microsporidia, Chytridiomycota, and Entomophthoromycota were detected as the most abundant phyla within the coral microbiome (Wang et al., 2018). Among the observed classes, Dothideomycetes (55.44%) and Sordariomycetes (18.13%) were the predominant classes but unlike other samples, Orbiliomycetes (0.47%) and Lecanoromycetes (0.08%) were also identified in association with this host. The more abundant eukaryotic orders were Capnodiales (49.90%), Saccharomycetales (12.29%), Hypocreales (9.36%), and Pleosporales (8.94%), with presence of the species like Acremonium alternatum, Leptographium bistatum, Cladosporium sphaerospermum, Sarocladium glaucum, and Meyerozyma guilliermondii.
Eukaryotic Community Composition Associated With Colpophyllia natans (CO4)
A total of one order, three genera, and two species were found to be specific for C. natans (Supplementary Figure S2). The analysis of the eukaryotic communities associated with this species revealed that Ascomycota was the most abundant phylum (99.40%), and Basidiomycota had a low representation within the microbiome (0.02%). Dothideomycetes was the dominant class with more that 90% of the microbial assignations. Pleosporales (89.82%) was the main order, and a minor representation of Hypocreales (7.23%) and Capnodiales (1.89%) was observed. ITS profiling revealed the presence of the species Cyberlindner rhodanensis, F. solani, Candida sake, Thermomyces lanuginosus, and Strelitziana eucalypti in this host.
Diversity of Microbial Communities at the Seaflower Biosphere Reserve
The observed OTUS and Simpson’s diversity index results showed a higher bacterial taxonomic diversity within the microbiome associated with S. siderea (0.99), followed by the sponge (0.98) and the sediment samples (0.96). P. astreoides showed microbial communities with lower diversity (0.92) (Figure 4A). Regarding eukaryotic characterization, O. annularis showed high diversity (0.97) while C. natans showed a very low diversity in comparison to the other coral species (0.26) (Figure 4B).
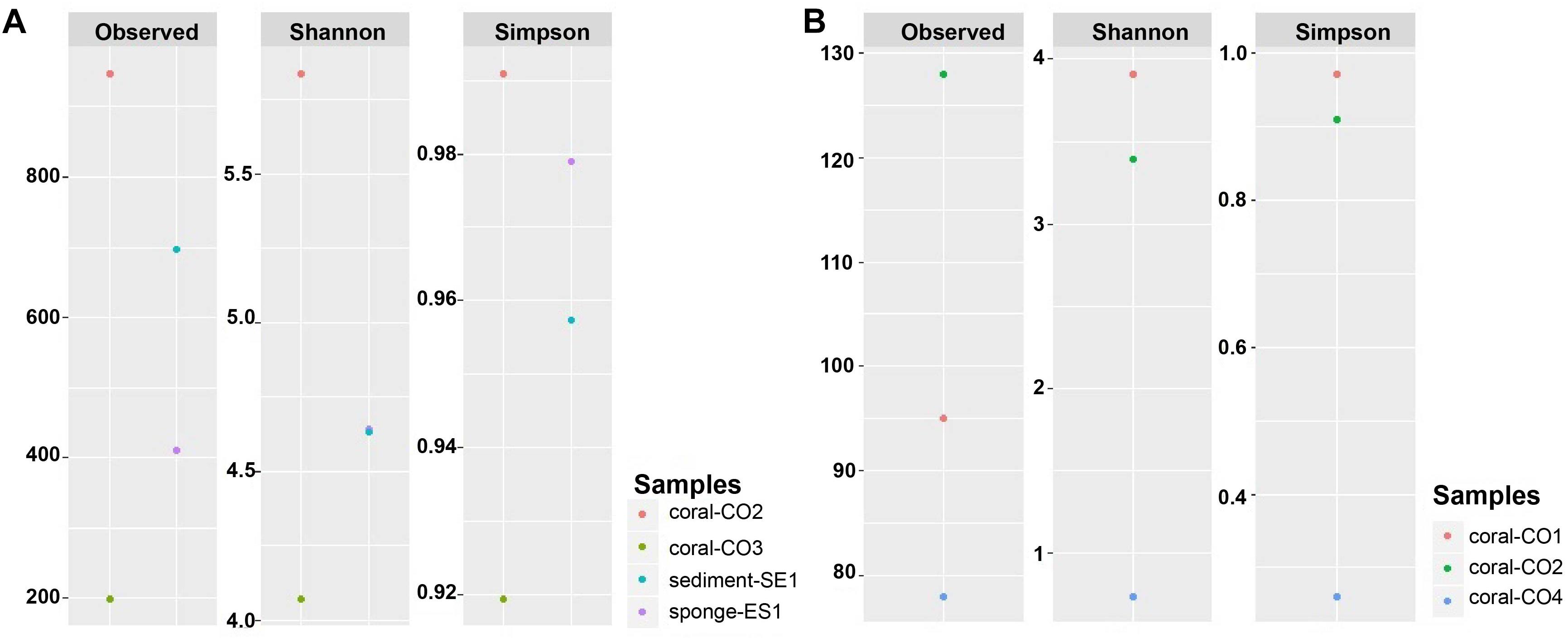
Figure 4. Alpha diversity indexes of microbial communities in marine hosts at the Seaflower Biosphere Reserve. (A) Bacterial and archaeal communities. (B) Eukaryotic communities.
Using Shannon’s index, higher evenness was observed in the microbiota associated with S. siderea (>5.83). P. astreoides showed the lowest evenness within its microbial diversity (4.07). Haliclona sp. and the sediment revealed a similar evenness associated to their taxonomic composition (4.64) (Figure 4A). Higher evenness of the eukaryotic communities was found in O. annularis (3.90), and C. natans showed the lowest evenness within its microbiome (0.74) (Figure 4B).
Alpha diversity rarefaction plots indicated that the abundance of bacterial or eukaryotic taxa associated to marine hosts has been widely surveyed at the maximum number of sequences read, except for P. astreoides, which did not reach an asymptote shape (Supplementary Figure S3). The diversity found in this coral was likely to be highly under-represented due to the low number of reads. Rarefaction plots also proved that the most diverse microbiota was associated with S. siderea.
The NMDS diagram showed that communities from Coral-CO3 were separated from the others, by the x-axis. However, the y-axis did not show a clear separation of the samples Coral-CO2 and Sponge-ES1. On the other hand, the PCoA showed a separation between four microbial community’s samples. The first axis divides the samples Coral-CO2 and Sponge-ES1 from other samples, containing 53.9% of the total variation, and the second axis explains the 26.4% of the variation, clustering the coral samples (Figure 5A). The PCoA and NMDS analyses for the fungi microbiomes showed a separation between three microbial community’s samples. For NMDS plot, communities from CO2 and CO4 were clearly separated from CO1, by the x-axis, on PCoA, the first axis divides the sample CO4 from CO2 and CO1 containing 56.5% of the total variation, while the second axis explains the 43.5% of the variation (Figure 5B). Because of the low number of samples that were compared, we consider that these analyses are not conclusive, but they do contribute to a possible understanding of the complexities of trophic interactions.
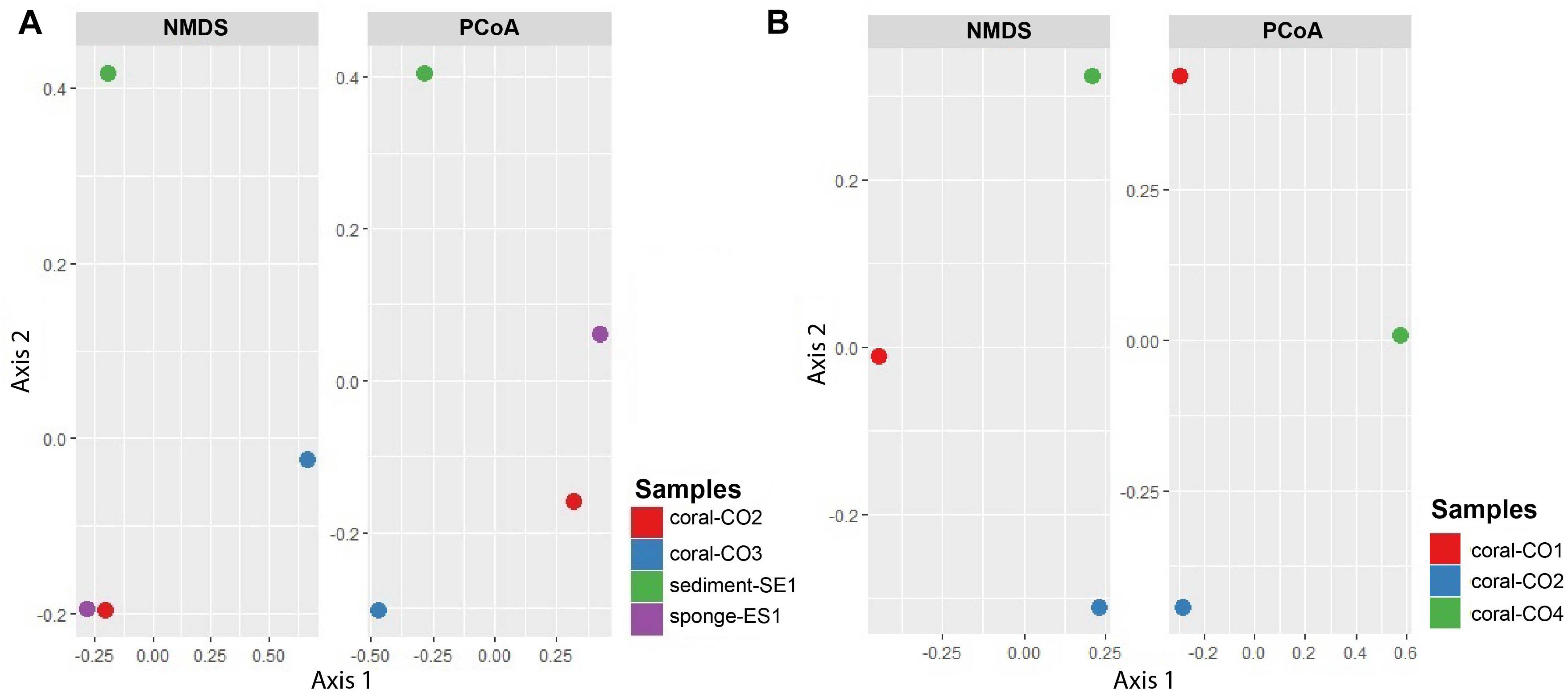
Figure 5. NMDS and principal coordinates analysis (PCoA) of metagenomes associated with hosts at the Seaflower Biosphere Reserve. (A) Bacterial and archaeal communities. (B) Eukaryotic communities.
Preliminary Metabolic Profiling of Microbial Communities Associated With Marine Hosts at Seaflower
Functional community profiles were obtained for metagenomes associated with S. siderea, P. astreoides, Haliclona sp., and the sediment. Predicted functional groups showed that there was not a significant change on metabolic functionality assignments between hosts except for amino acid (F = 2071, p < 0.05), and carbohydrate metabolism (F = 408, p < 0.05). For amino acid metabolism, the sponge had a larger number of metabolic processes, in comparison with the sediment and the corals. Among the carbohydrate metabolism, more pathways were assigned to the sponge and the sediment, while the corals had a lower assignment of pathways. Metabolic profiling revealed that the principal microbial functions are focused on membrane transport, carbohydrates, amino acids, and energy metabolism, as well as replication and translation processes (Figure 6).
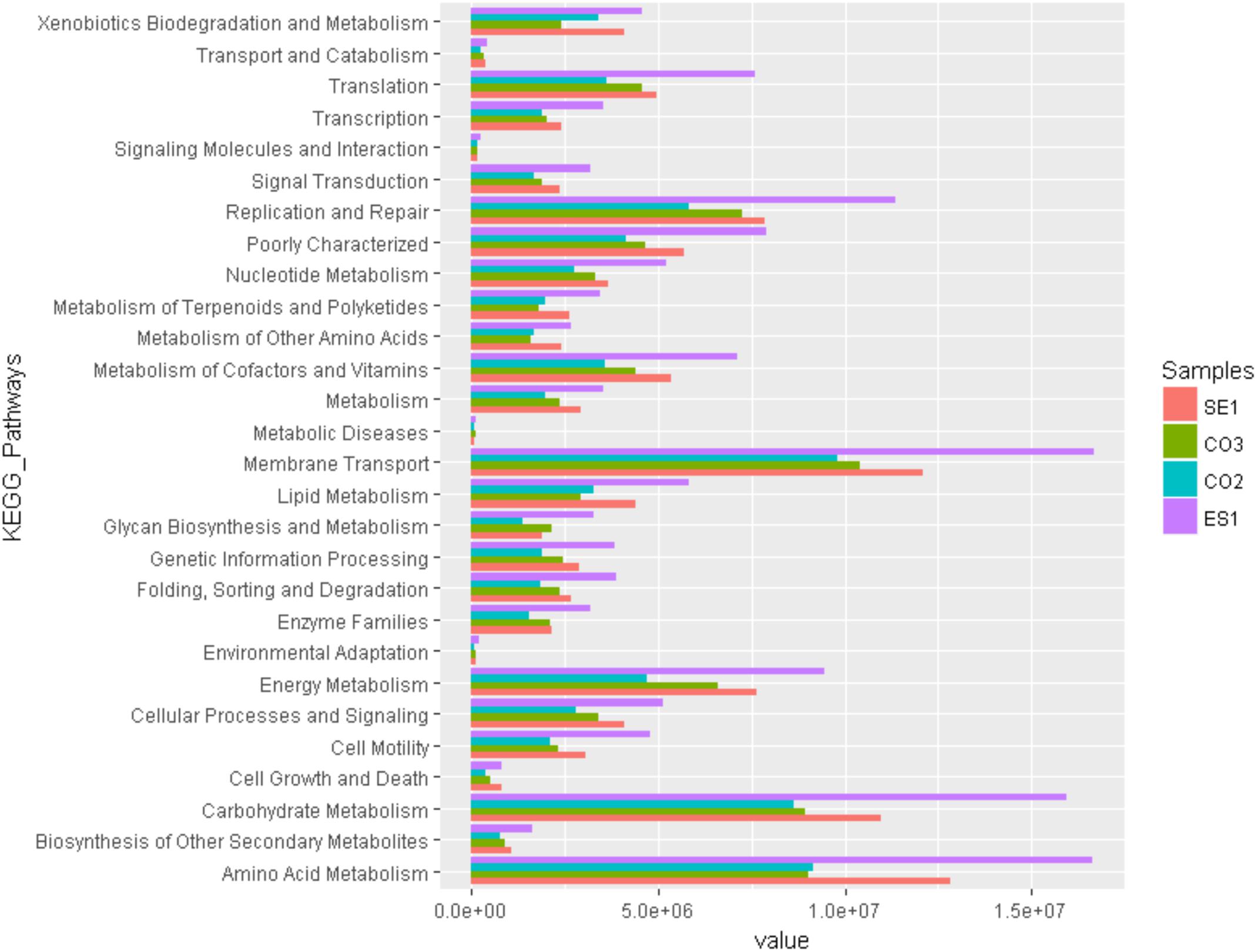
Figure 6. Predictive functional profiling of microbial communities associated with S. siderea, P. astreoides, Haliclona sp., and Isla de los Pájaros sediment at the Seaflower Biosphere Reserve.
Discussion
Metabarcoding using next-generation sequencing has been used for large-scale biodiversity analyses (Pavan-Kumar et al., 2015) and can be useful for biodiversity monitoring in environments that are undergoing rapid climate-related changes, such as ocean warming and eutrophication of coastal environments. We reported the first taxonomic survey of microbial communities associated to corals, a marine sponge, and sediment at Serrana Bank, an atoll of the Seaflower Biosphere Reserve. This survey provides an opportunity to investigate local patterns of diversity of sessile taxa. Although we collected 16 samples, it was unfortunate that just a small number produced DNA of high quality for sequencing. Ten samples did not fulfill DNA quality controls due to contamination with proteins (260/280 < 2), which can be a result of cross contamination with the host tissue residues. However, we decided to go ahead with the sequencing given that this was the first set of available samples with information on the microbiome composition of marine hosts at this reserve. We are certain that the study of the microbiomes associated to macroorganisms in Serrana Bank is an important complement to the collected information on the flora and the fauna that populates this ecosystem. We identified 1.776 OTUs assigned to bacteria and 227 OTUs assigned to fungi. Although this amount is not as big as in other studies performed in coral reefs (Rohwer et al., 2002; Hernandez-Agreda et al., 2017), it helped to gain insights about the microbial diversity associated to some marine hosts at the Serrana Bank.
We discovered differences in the presence/absence of microbial communities associated to corals, the sponge, and the sediment, with unique OTUS associated to every host (Supplementary Figures S1, S2). Most of the OTUs identified are common coral and sponge holobionts, for instance, alpha and gamma proteobacteria usually dominate the bacterial communities in corals, as well as Firmicutes, Bacteroidetes, Actinobacteria, and Acidobacteria. However, the species vary according to the host, age, health status, and the surrounding micro-environment (Hernandez-Agreda et al., 2017). Among the eukaryotic groups found within the microbiomes, Ascomycota was the major representative. This phyla is considered as an obligate fungi in corals (Raghukumar and Ravindran, 2012). Studies of fungi as marine symbionts have mainly focused in their role as pathogenic agents. However, they can also contribute to skeletal biomineralization, net bioerosion reduction, resistance to environmental stress, UV damage protection, among others (Ainsworth et al., 2017). The classes Dothideomycetes, Sordariomycetes, Eurotiomycetes, Saccharomycetes, and Tremellomycetes found in this study are commonly reported in marine environments (Xu et al., 2014). Important differences were identified in the microbiota of P. astreoides, where Cyanobacteria was the most abundant group. These groups have been reported in marine ecosystems fulfilling ecological roles, like nutrient cycling between macroorganisms and the water column (Dinsdale et al., 2008; Glöckner et al., 2012). Relative abundances of species within taxonomic phyla, classes, and orders differed among all samples.
The archaeal groups identified in the surveyed hosts included extremophile species (hyperthermophiles, mesophilic, psychrophilic, halophilic, methanogenic, etc.), acknowledged as the most abundant archaea in marine environments (Madigan and Martinko, 2005). Species of Candidatus, Nitrososphaera, and Nitrosopumilus, associated to the hosts, are widespread ammonia-oxidizing archaea (AOA), and key players in nitrification processes due to their metabolic versatility and adaptability (Spang et al., 2012).
Bacterial Community Composition Associated With Siderastrea siderea (CO2)
Our results match and complement the findings of previous diversity studies reported for S. siderea, where identical microbial phyla and classes were identified (Cárdenas et al., 2012; Kellogg et al., 2014). Within these groups, important bacterial species were found playing a common role in reducing nitrate to nitrite, degrading aromatic compounds, and oxidizing sulfur, ammonia, and carbon, among others (Dinsdale et al., 2008; Sun et al., 2016).
It has been reported that in diseased S. siderea, there is an increase in Alphaproteobacteria and decrease in Gammaproteobacteria (Cárdenas et al., 2012). Furthermore, Corynebacterium and Acinetobacter are known as potential pathogens in corals due to their association with the dark spot syndrome in these organisms (Sweet et al., 2013). We found a high number of both groups of bacteria, as well as the presence of Corynebacterium and Acinetobacter, so at this point, it is not possible to make assumptions about the health of the coral, but it is likely that the coral could be in an early disease state. The number of bacteria genera identified in this study was lower than previously reported for this host (600 vs. 222) (Kellogg et al., 2014). However, this might be a consequence of the limitations of the software and databases used to do the taxonomic assignments.
Bacterial Community Composition Associated With Porites astreoides (CO3)
Our findings differ from some studies previously reported, in which Proteobacteria, Bacteroidetes, and Acidobacteria were the dominant phyla, while Oceanospirillales was the most abundant order (Wegley et al., 2007; Morrow et al., 2012). Our findings show in contrast that Cyanobacteria represented more than 30% of the microbial communities associated to P. astreoides. Cyanobacteria are oxy-photosynthetic bacteria that have dominated marine environments and play an essential role in modern coral reef ecosystems by forming a major component of epiphytic, epilithic, and endolithic communities, as well as of microbial mats. Theses bacteria also provide nitrogen to the coral reef ecosystems through nitrogen fixation. On the other hand, Cyanobacteria have been found forming pathogenic microbial consortia in association with other microbes on living coral tissues, causing coral tissue lysis and death, which have contributed to considerable declines in coral reefs (Charpy et al., 2010). It has been observed that a great abundance of Cyanobacteria, Bacteroidetes, Firmicutes, and Actinobacteria is related with black band disease (Barneah et al., 2007) and dark spot syndrome in corals (Sweet et al., 2013). However, when we searched for genera and species that have been associated specifically to dark spot syndrome (Oscillatoria) (Sweet et al., 2013) and black band disease (Cyanobacterium, Oscillatoria, Phormidium, and Pseudoscillatoria) (Frias-Lopez et al., 2003), none of these specific taxa were identified. It should be noted that a large number of OTUs do not present identification up to genus (Supplementary Tables S2, S3), so we need to be careful in the interpretation of the sequence comparison results, until the databases are better populated.
Among the identified genera, the usually dominant genus Endozoicomonas was not found in our study. Although there is not a clear consistency between community shifts in microbiomes associated with healthy and diseased corals (Kellogg et al., 2014), it has been suggested that Endozoicomonas is an obligate member of a healthy microbiome of P. astreoides and its loss and reduction has been observed in some specimens, along with a decrease in Oceanospirillales (Mouchka et al., 2010; Morrow et al., 2012; Meyer et al., 2014). This could suggest that this coral was in a diseased state. In addition, Vibrio was only found in P. astreoides, and not the other hosts. Acinetobacter and Vibrio have been commonly associated with marine organisms, and have also been recognized as pathogens in invertebrates and corals (Ben-Haim et al., 2003; Kellogg et al., 2014; Wang et al., 2015). A similar result was found in corals with unusual lesions, where a negative correlation was found between Endozoicomonas and members of the Vibrionaceae family which have been recognized as opportunistic pathogens (Meyer et al., 2014).
Overall, results revealed differences between the microbiome compositions previously described for P. astroides (Rodriguez-Lanetty et al., 2013). These differences are characterized by the higher abundance of Cyanobacteria within the microbial community, the lower representation of Oceanospirillales, the absence of Endozoicomonas, and the identification of Acinetobacter and Vibrio as possible opportunistic species in association with the coral. Our results could reflect the current health status of this marine host in Serrana bank, but also could be a consequence of the low number of reads that were obtained for this coral. Additional samples should be sequenced to fully characterize this microbiome and generate more accurate conclusions about their association with the coral host.
Bacterial Community Composition Associated With Haliclona sp. (ES1)
The bacterial phyla identified in this host have been previously found in association with different marine sponges and with Haliclona sp. (Khan et al., 2014; Jasmin et al., 2015) although their abundance is slightly different. These microorganisms are important because they fulfill critical ecological roles regarding nutrient acquisition, processing of metabolic waste, host defense, secondary metabolites production, and stabilization of the sponge skeleton (Hentschel, 2003). Interestingly, representatives of Acidobacteria, Chloroflexi, and Gemmatimonadetes are indicative of high microbial-abundant (HMA) sponges (Jiang et al., 2007; Khan et al., 2011), which suggest that the assessed sponge had a significant microbial abundance, where most of the identified taxa are related with healthy organisms. Also, marine Actinomycetes have been commonly associated to Haliclona species, where they represent an important microbial diversity, and an extensive metabolic potential for obtaining novel compounds such as polyketides (Jiang et al., 2007; Khan et al., 2011).
Because most of the taxonomic assignments for this host were not completed to the family level (database restrictions), it was difficult to complete assignations to specific genera and species. This limited our search for microorganisms that have been described as potential source of bioactive secondary metabolites (Streptomyces, Haliea, Nocardiopsis, Micromonospor, Cytophaga, Pseudoalteromonas, Verrucosispora, Pseudovibrio, and Bacillus) (Jiang et al., 2007; Kennedy et al., 2009).
Bacterial Community Composition Associated With “Isla de los Pájaros” Sediment (SE1)
The prevalence of Proteobacteria classes has been previously described on superficial lakes and marine sediment microbiomes as main drivers of biogeochemical cycling of carbon, nitrogen, and sulfur, and as the source of variations in nutrient content of water (Hongxiang et al., 2008; Franco et al., 2017). Cyanobacteria are found in lagoons and soft muddy floors forming associations with photosynthetic bacteria, sulfur bacteria, and other microorganisms. Biological N2 fixation has been described as one of the most important contribution of Cyanobacteria, where they also represent an organic source for planktonic and benthic heterotrophic organisms (Charpy et al., 2010).
Taxa identified in this sample show that the lagoon’s water contains microorganisms that are contributing to nutrient content and cycling in water, being a significant source of hydration for birds that use this place as a stopover in their migration. Nevertheless, the microbiome associated with the sediment sample showed the lowest bacterial taxonomic diversity. This was an unusual finding, since sediments of different origin seem to have a large microbial diversity in comparison with host-associated communities (Thompson et al., 2017). In this case, the high temperatures that are reached in the lake (>20°C), due to its shallowness, could expain this decreased richness (Milici et al., 2016). Since no general relationship between temperature and richness has been established (Hendershot et al., 2017), we would need to analyze additional samples to confirm this result and the factors that influence this lower microbial diversity on the sediment of this lagoon.
Eukaryotic Community Composition Associated With Orbicella annularis (CO1)
Several genera found in this study contain species described as saprophytes in corals including Aspergillus sydowii and C. sphaerospermum that have been reported in this species (Kendrick et al., 1982) but were not identified in this study. Additionally, the species F. solani has been reported as an important pathogen in marine organisms (Raghukumar and Ravindran, 2012) and it was found in this coral host. It would be premature to make inferences about the health status of this host, but it is important to highlight that some of the identified taxa have been associated to diseased organisms.
Eukaryotic Community Composition Associated With Colpophyllia natans (CO4)
Fungal biomass constituted up to 0.05% of the weight of the corals and are apparently an important component of this group of organisms (Raghukumar and Ravindran, 2012). Previous microscopic studies have shown that endolithic–Ascomycetes-like fungi are ubiquitously found across several genera of healthy corals. In contrast, other fungi such as Aspergillus spp. are known as coral pathogens (Kim et al., 2000, 2006; Alker et al., 2001). Fungi in coral holobionts are involved in carbon and nitrogen metabolism by conversion of nitrate and nitrite to ammonia, which enable nitrogen cycling within the host (Wegley et al., 2007). Coral-associated microbes have been involved in biogeochemical cycling, particularly nitrogen, sulfur, and phosphorus cycles, and they have been positively associated with coral health and the resilience of coral reef ecosystems (Sun et al., 2016). These features make fungi key players within the coral microbiomes and their functionality.
Diversity Analyses of Microbial Communities Associated With Marine Hosts
Results from observed OTUs and Simpson diversity indexes showed that the microbiota associated to the marine hosts is highly diverse, and it has a considerable species richness. The most diverse bacterial communities were found associated with S. siderea, and the least diverse were in association with P. porites. The most diverse eukaryotic community was found associated with S. siderea and the least diverse was in association with C. natans.
The Shannon index showed that bacterial communities associated with S. siderea had a random distribution in terms of species evenness. In contrast, communities associated with P. astreoides were dominated by a few abundant species, of Cyanobacteria. O. annularis and S. siderea microbiomes harbor eukaryotic communities with an even taxa representation. For the microbiome associated with C. natans, just a few eukaryotic groups were highly represented while others were underrepresented.
Rarefaction analysis was appropriate for the characterization of microbial diversity in the studied hosts. It would be convenient to identify rare species interacting in a smaller proportion with these macroorganisms (Gotelli and Colwell, 2001). For P. astreoides, additional sequencing efforts are required to fully characterize its microbiome.
Principal component analysis suggested that the composition of microbial communities was different between hosts but given the low sample number it is necessary to confirm this with a study that includes a larger number of specimens and replicates. This supports the idea that there are some species-specific relationships, as it has been suggested for some sponges and corals (Taylor et al., 2007; Morrow et al., 2012).
Potential Metabolic Functionality of Microbial Communities
Most functional subsystems were represented in all samples. The metabolic pathways identified within the marine microbiomes included membrane transport; transformation of carbon, nitrogen, phosphorus, and sulfur macromolecules (carbohydrate metabolism, amino acid metabolism, energy metabolism); and cell division processes (replication and repair, translation). Membrane transport systems are needed for the uptake of substrates that are present in the environment and can be used by microorganisms and marine hosts. Carbohydrate, amino acid, energy metabolism, replication, and translation processes are coupled because macromolecules are broken to produce energy and basic compounds for the synthesis of new essential biological structures (Karlsson, 2012). These processes also guarantee the correct mass fluxes through the biogeochemical cycles within the marine ecosystem (Caron, 2005; Glöckner et al., 2012). Functions of surveyed microbial communities in this study are involved in cofactors and vitamin metabolism, and in xenobiotics biodegradation. In vitamin metabolism, the production of porphyrin, chlorophyll, pantothenate, and CoA is fundamental for cellular development. Xenobiotic biodegradation includes the degradation of recalcitrant pools of organic matter such as benzoates, caprolactam, and polycyclic hydrocarbons, which are known to be toxic substances in different ecosystems (Dinsdale et al., 2008).
The described functional profiles can be related to the potential that has been attributed to marine microbes, in providing nutritional by-products, proteins, essential vitamins, and nitrogenous compounds to hosts (Xu and Gordon, 2003; Lesser, 2004; Croft et al., 2005), as well as their ability to transform polluting substances in association with macroorganisms (Wahl et al., 2012; Grossart et al., 2013).
Potential activities related to amino acid and carbohydrate metabolism were more abundant in sponges than in corals. This could be a consequence of the high potential of sponges to produce bioactive compounds, and to harbor a larger arsenal of microbial metabolic pathways. The range of compounds that can be isolated from Haliclona sp. and its microbiome includes steroids, terpenoids, polyacetylenes, alkaloids, cyclic peptides, unsaturated fatty acids, and polyketides (Yu et al., 2006). Previous studies had suggested that coral-associated microbes have a lower functional diversity because they rarely include processes associated with secondary metabolism, virulence pathways, cell signaling, and membrane transport pathways. These pathways did not show a significant difference in corals microbial communities identified in our study.
Conclusion
We report the first taxonomic survey of microbial communities associated with corals, a marine sponge, and sediment at Serrana Bank, an atoll of the Seaflower Biosphere Reserve. Although total OTUs assignment was lower compared to other studies performed in coral reefs, our results provide insights about the microbial diversity associated with several marine hosts at the Serrana Bank. We found that the bacterial and eukaryotic community structures are different between hosts. Despite the fact that the same phyla are dominant (Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, and Acidobacteria from bacteria; Ascomycota for Eukarya), the relative abundances of each group vary among corals, the sponge, and the sediment. This is recurrent at all taxonomic levels (class, order, family).
Our results showed that S. siderea harbors a higher bacterial diversity (richness and evenness) than the sponge and sediment. This finding was unsual, because host-associated communities have been found to be less diverse than sediments. Hence, additional analyses are necessary in order to confirm this result and the driving factors for this shift.
Additionally, important differences were identified in the microbiota of P. astreoides, where Cyanobacteria was the predominant phylum, and opportunistic species (Vibrio and Acinetobacter) were identified. These species could indicate a disease status for the coral, but concise conclusions cannot be drawn at this point. It is yet to be proved if human intervention, climate change, or other factors might be playing a role in the decline of this coral in the reserve. Since the diversity found in this coral was likely to be under-represented, due to the low number of reads, additional samples should be sequenced to fully characterize this microbiome and generate more accurate conclusions about their association with the coral host.
Regarding eukaryotic characterization, O. annularis showed a higher diversity than S. siderea and C. natans. Overall, the fungi found in association with each host are an important component of their holobionts, because they influence the health status, carbon and nitrogen cycling, and resilience of coral reef ecosystems.
Finally, potential metabolic functionality was evaluated for some hosts. Our results revealed that the sponge microbiome had a significantly higher number of metabolic activities (amino acid and carbohydrate metabolism) in comparison to corals. This result could be associated with the HMA found in the sponge. In general, we found functional evidence of energy production, cell reproduction, and nutrient cycling within the marine ecosystem.
The implementation of continuous monitoring of the wellbeing of this Caribbean ecosystem is crucial and it must include the study of microbial communities associated with its macroorganisms. In our study, metabarcoding data from these microbial communities generated a set of OTUs that could be potentially used for effective biodiversity monitoring through time. Extended surveys of biodiversity changes in the Seaflower Biosphere reserve will be a critical action for the application of proper management practices.
Author Contributions
MC and AA-Y participated in the conception and design of the study. AA-Y collected the data. AA-Y, JM-R, AN-P, and DL-A performed the analyses and interpretation of the results. AA-Y, AN-P, DL-A, and MC wrote the manuscript. All authors read, commented, and approved the manuscript.
Funding
This work was co-funded by the Department of Science, Technology, and Innovation (COLCIENCIAS) through the program Colombia BIO, the Comisión Colombiana del Oceáno under the program “Seaflower Scientific Expedition” and by the Centro de Bioinformática y Biología Computacional BIOS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors extend their appreciation to Valeria Pizarro from the Fundación Malpelo; Lizette Quan and Paula Quinceno from the Universidad de Antioquia; and to Juliana Sintura from the Comisión Colombiana del Océano for their collaboration during the species identification and sampling activities of this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00338/full#supplementary-material
Footnotes
References
Ainsworth, T. D., Fordyce, A. J., and Camp, E. F. (2017). The other microeukaryotes of the coral reef microbiome. Trends Microbiol. 25, 980–991. doi: 10.1016/j.tim.2017.06.007
Alker, A. P., Smith, G. W., and Kim, K. (2001). Characterization of Aspergillus sydowii (Thom et Church), a fungal pathogen of Caribbean sea fan corals. Hydrobiologia 460, 105–111. doi: 10.1023/A:1013145524136
Barneah, O., Ben-Dov, E., Kramarsky-Winter, E., and Kushmaro, A. (2007). Characterization of black band disease in Red Sea stony corals. Environ. Microbiol. 9, 1995–2006. doi: 10.1111/j.1462-2920.2007.01315.x
Bell, J. J. (2008). The functional roles of marine sponges. Estuar. Coast. Shelf Sci. 79, 341–353. doi: 10.1016/j.ecss.2008.05.002
Ben-Haim, Y., Thompson, F. L., Thompson, C. C., Cnockaert, M. C., Hoste, B., Swings, J., et al. (2003). Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 53, 309–315. doi: 10.1099/ijs.0.02402-2400
Cárdenas, A., Rodriguez-R, L. M., Pizarro, V., Cadavid, L. F., and Arévalo-Ferro, C. (2012). Shifts in bacterial communities of two caribbean reef-building coral species affected by white plague disease. ISME J. 6, 502–512. doi: 10.1038/ismej.2011.123
Caron, D. A. (2005). Marine microbial ecology in a molecular world?: what does the future hold? Scentia Mar. 69 (Suppl) 97–110. doi: 10.3989/scimar.2005.69s197
Charpy, L., Palinska, K. A., Casareto, B., Langlade, M. J., Suzuki, Y., Abed, R. M. M., et al. (2010). Dinitrogen-fixing cyanobacteria in microbial mats of two shallow coral reef ecosystems. Microb. Ecol. 59, 174–186. doi: 10.1007/s00248-009-9576-y
Coker, D. J., Wilson, S. K., and Pratchett, M. S. (2014). Importance of live coral habitat for reef fishes. Rev. Fish Biol. Fish. 24, 89–126. doi: 10.1007/s11160-013-9319-9315
Comisión Colombiana del Oceáno (2015). Aportes al conocimiento de la Reserva de Biósfera Seaflower. Bogota: Comisión Colombiana del Oceáno.
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J., and Smith, A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Debnath, M., Paul, A K., and Bisen, P. S. (2007). Natural bioactive compounds and biotechnological potential of marine bacteria. Curr. Pharm. Biotechnol. 8, 253–260. doi: 10.2174/138920107782109976
De-Kluijver, M., Gijswijt, G., De-Leon, R., and Da-Cunda, I. (2016). Marine Species Identification Portal. Boulder Brain Coral (Colpophyllia natans). Available at: http://species-identification.org/species.php?species_group=caribbean_diving_guide&id=305 (accessed June 20, 2018).
Delmont, T. O., Robe, P., Clark, I., Simonet, P., and Vogel, T. M. (2011). Metagenomic comparison of direct and indirect soil DNA extraction approaches. J. Microbiol. Methods 86, 397–400. doi: 10.1016/j.mimet.2011.06.013
Díaz, J. M., Díaz-Pulido, G., Garzón-Ferreira, J., Geister, J., Sánchez M J. A., and Zea, S. (1996). Atlas de los Arrecifes Coralinos del Caribe Colombiano. I. Complejos Arrecifales oceánicos. Santa Marta: Serie Publicaciones Especiales Invemar.
Dinsdale, E. A., Edwards, R. A., Hall, D., Angly, F., Breitbart, M., Brulc, J. M., et al. (2008). Functional metagenomic profiling of nine biomes. Nature 452, 629–632. doi: 10.1038/nature06810
Dionisi, H., Lozada, M., and Oliviera, N. L. (2012). Bioprospection of marine microorganisms: biotechnological applications and methods. Rev. Argent Microbiol. 44, 49–60. doi: 10.1590/S0325-75412012000100010
Edmunds, P. J. (2015). A quarter-century demographic analysis of the Caribbean coral, Orbicella annularis, and projections of population size over the next century. Limnol. Oceanogr. 60, 840–855. doi: 10.1002/lno.10075
Ellis, J., and Solander, D. C. (1786). The Natural History of Many Curious and Uncommon Zoophytes?: Collected from Various Parts of the Globe. London?: Printed for Benjamin White and Son... and Peter Elmsly.
Franco, D. C., Signori, C. N., Duarte, R. T. D., Nakayama, C. R., Campos, L. S., and Pellizari, V. H. (2017). High prevalence of gammaproteobacteria in the sediments of admiralty bay and north bransfield basin, Northwestern Antarctic Peninsula. Front. Microbiol. 8:153. doi: 10.3389/fmicb.2017.00153
Frias-Lopez, J., Bonheyo, G. T., Jin, Q., and Fouke, B. W. (2003). Cyanobacteria associated with coral black band disease in caribbean and indo-pacific reefs. Appl. Environ. Microbiol. 69, 2409–2413. doi: 10.1128/AEM.69.4.2409-2413.2003
Galeano, E., Gomez, D. I., Navas, R., Alonso, D., Zarza-González, E., Cano-Correa, M., et al. (2016). Reporte del Estado de los Arrecifes Coralinos y Pastos Marinos en Colombia (2014-2015) Proyecto COL75241, PIMS # 3997, Diseño e implementación de un Subsistema Nacional de Áreas Marinas Protegidas (SAMP) en Colombia. Santa Marta: Serie de publicaciones Generales del Invemar.
Glöckner, F. O., Stal, L., Sandaa, R. A., Gasol, J. M., O’Gara, F., Hernandez, F., et al. (2012). “Marine Microbial Diversity and its role in Ecosystem Functioning and Environmental Change. Marine Board Position Paper 17” in Marine Board Position Paper 17., eds N. McDonough and J.-B. Calewaert (Belgium: European Scientific Foundation), 84. doi: 10.13140/RG.2.1.5138.6400
Gotelli, N. J., and Colwell, R. K. (2001). Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4, 379–391. doi: 10.1046/j.1461-0248.2001.00230.x
Green, D. H., Edmunds, P. J., and Carpenter, R. C. (2008). Increasing relative abundance of Porites astreoides on Caribbean reefs mediated by an overall decline in coral cover. Mar. Ecol. Prog. Ser. 359, 1–10. doi: 10.3354/meps07454
Grossart, H. P., Riemann, L., and Tang, K. W. (2013). Molecular and functional ecology of aquatic microbial symbionts. Front. Microbiol. 4:59 doi: 10.3389/fmicb.2013.00059
Haygood, M. G., Schmidt, E. W., Davidson, S. K., and Faulkner, D. J. (1999). Microbial symbionts of marine invertebrates: opportunities for microbial biotechnology. J. Mol. Microbiol. Biotechnol. 1, 33–43.
Hendershot, J. N., Read, Q. D., Henning, J. A., Sanders, N. J., and Classen, A. T. (2017). Consistently inconsistent drivers of microbial diversity and abundance at macroecological scales. Ecology 98, 1757–1763. doi: 10.1002/ecy.1829
Hentschel, U. (2003). Microbial diversity of marine sponges. Prog. Mol. Subcell Biol. 68, 365–372. doi: 10.1007/978-3-642-55519-0_3
Hernandez-Agreda, A., Gates, R. D., and Ainsworth, T. D. (2017). Defining the core microbiome in Corals’ microbial soup. Trends Microbiol. 25, 125–140. doi: 10.1016/j.tim.2016.11.003
Hongxiang, X., Min, W., Xiaogu, W., Junyi, Y., and Chunsheng, W. (2008). Bacterial diversity in deep-sea sediment from northeastern Pacific Ocean. Acta Ecol. Sin. 28, 479–485. doi: 10.1016/S1872-2032(08)60026-60028
Hoppers, A., Stoudenmire, J., Wu, S., and Lopanik, N. B. (2015). Antibiotic activity and microbial community of the temperate sponge, Haliclona sp. J. Appl. Microbiol. 118, 419–430. doi: 10.1111/jam.12709
Howard, M. (2006). Evaluation Report Seaflower Biosphere Reserve Implementation: The First Five Years 2000–2005. Waltham, MA: Brandeis University.
Howard, M., Valeria, P., and Mow, J. M. (2004). Ethnic and biological diversity within the seaflower biosphere reserve. Int. J. Isl. Aff. 13, 109–114.
Jasmin, C., Anas, A., and Nair, S. (2015). Bacterial diversity associated with cinachyra cavernosa and haliclona pigmentifera, cohabiting sponges in the coral reef ecosystem of gulf of mannar, Southeast Coast of India. PLoS One 10:e0123222. doi: 10.1371/journal.pone.0123222
Jiang, S., Sun, W., Chen, M., Dai, S., Zhang, L., Liu, Y., et al. (2007). Diversity of culturable actinobacteria isolated from marine sponge Haliclona sp. Antonie van Leeuwenhoek 92, 405–416. doi: 10.1007/s10482-007-9169-z
Karlsson, T. (2012). Carbon and Nitrogen Dynamics in Agricultural Soils Model Applications at Different Scales in Time and Space. Uppsala: Acta Universitatis agriculturae Sueciae.
Kellogg, C. A., Piceno, Y. M., Tom, L. M., DeSantis, T. Z., Gray, M. A., and Andersen, G. L. (2014). Comparing bacterial community composition of healthy and dark spot-affected Siderastrea sidereain Florida and the Caribbean. PLoS One 9:e108767. doi: 10.1371/journal.pone.0108767
Kendrick, B., Risk, M. J., Michaelides, J., and Bergman, K. (1982). Amphibious microborers: bioeroding fungi isolated from corals. Bull. Mar. Sci. 32, 862–867.
Kennedy, J., Baker, P., Piper, C., Cotter, P. D., Walsh, M., Mooij, M. J., et al. (2009). Isolation and analysis of bacteria with antimicrobial activities from the marine sponge haliclona simulans collected from irish waters. Mar. Biotechnol. 11, 384–396. doi: 10.1007/s10126-008-9154-9151
Khan, S. T., Komaki, H., Motohashi, K., Kozone, I., Mukai, A., Takagi, M., et al. (2011). Streptomyces associated with a marine sponge Haliclona sp.; biosynthetic genes for secondary metabolites and products. Environ. Microbiol. 13, 391–403. doi: 10.1111/j.1462-2920.2010.02337.x
Khan, S. T., Musarrat, J., Alkhedhairy, A. A., and Kazuo, S. (2014). Diversity of bacteria and polyketide synthase associated with marine sponge Haliclona sp. Ann. Microbiol. 64, 199–207. doi: 10.1007/s13213-013-0652-7
Kim, K., Alker, A. P., Shuster, K., Quirolo, C., and Harvell, C. D. (2006). Longitudinal study of aspergillosis in sea fan corals. Dis. Aquat. Organ. 69, 95–99. doi: 10.3354/dao069095
Kim, K., Harvell, C. D., Kim, P. D., Smith, G. W., and Merkel, S. M. (2000). Fungal disease resistance of Caribbean sea fan corals (Gorgonia spp.). Mar. Biol. 136, 259–267. doi: 10.1007/s002270050684
Konopka, A. (2009). What is microbial community ecology? ISME J. 3, 1223–1230. doi: 10.1038/ismej.2009.88
Lachnit, T., Meske, D., Wahl, M., Harder, T., and Schmitz, R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665. doi: 10.1111/j.1462-2920.2010.02371.x
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotech. 31, 814–821. doi: 10.1038/nbt.2676
Lawler, S. N., Kellogg, C. A., France, S. C., Clostio, R. W., Brooke, D., and Ross, S. W. (2016). Coral-associated bacterial diversity is conserved across two deep-sea anthothela species. Front. Microbiol. 7:458. doi: 10.3389/fmicb.2016.00458
Lee, Y., Lee, J., and Lee, H. (2001). Microbial symbiosis in marine sponges. J. Microbiol. 39, 254–264
Lesser, M. P. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000. doi: 10.1126/science.1099128
Lozada, M., and Dionisi, H. M. (2015). “Microbial bioprospecting in marine environments,” in Springer Handbook of Marine Biotechnology. Springer Handbooks. ed. S. K Kim (Springer: Heidelberg).
Madigan, M. T., and Martinko, J. M. (2005). Brock Biology of Microorganisms. 11 ed. Upper Saddle River, NJ: Prentice Hall.
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Meyer, J. L., Paul, V. J., and Teplitski, M. (2014). Community shifts in the surface microbiomes of the coral porites astreoides with unusual lesions. PLoS One 9:e100316. doi: 10.1371/journal.pone.0100316
Milici, M., Tomasch, J., Wos-Oxley, M. L., Wang, H., Jáuregui, R., Camarinha-Silva, A., et al. (2016). Low diversity of planktonic bacteria in the tropical ocean. Sci. Rep. 6:19054. doi: 10.1038/srep19054
Morrow, K. M., Moss, A. G., Chadwick, N. E., and Liles, M. R. (2012). Bacterial associates of two caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 78, 6438–6449. doi: 10.1128/AEM.01162-1112
Mouchka, M. E., Hewson, I., and Harvell, C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674. doi: 10.1093/icb/icq061
Pavan-Kumar, A., Gireesh-Babu, P., and Lakra, W. S. (2015). DNA metabarcoding: a new approach for rapid biodiversity assessment. J. Cell Sci. Mol. Biol. 2:111
Raghukumar, C., and Ravindran, J. (2012). Fungi and their role in corals and coral reef ecosystems. Biol. Mar. Fungi 53 89–113. doi: 10.1007/978-3-642-23342-5_5
Reed, J. K. (1985). “Deepest distribution of Atlantic hermatypic corals discovered in the Bahamas,” in Proceedings of the Fifth International Coral Reef Congress, Tahiti.
Riegl, B., Bruckner, A., Coles, S. L., Renaud, P., and Dodge, R. E. (2009). Coral reefs. Ann. N. Y. Acad. Sci. 1162, 136–186. doi: 10.1111/j.1749-6632.2009.04493.x.
Rodriguez-Lanetty, M., Granados-Cifuentes, C., Barberan, A., Bellantuono, A. J., and Bastidas, C. (2013). Ecological inferences from a deep screening of the complex bacterial consortia associated with the coral, Porites astreoides. Mol. Ecol. 22, 4349–4362. doi: 10.1111/mec.12392
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Sogin, M. L., Morrison, H. G., Huber, J. A., Welch, D. M., Huse, S. M., Neal, P. R., et al. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120. doi: 10.1073/pnas.0605127103
Spang, A., Poehlein, A., Offre, P., Zumbrägel, S., Haider, S., Rychlik, N., et al. (2012). The genome of the ammonia-oxidizing candidatus nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ. Microbiol. 14, 3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x
Streit, W. R., and Schmitz, R. A. (2004). Metagenomics - the key to the uncultured microbes. Curr. Opin. Microbiol. 7, 492–498. doi: 10.1016/j.mib.2004.08.002
Sun, W., Anbuchezhian, R., and Li, Z. (2016). “Association of Coral-Microbes, and the Ecological Roles of Microbial Symbionts in Corals,” in The Cnidaria, Past, Present and Future.eds S. Goffredo and Z Dubinsky (Switzerland: Springer International Publishing)
Sweet, M., Burn, D., Croquer, A., and Leary, P. (2013). Characterisation of the bacterial and fungal communities associated with different lesion sizes of dark spot syndrome occurring in the coral stephanocoenia intersepta. PLoS One 8:e62580. doi: 10.1371/journal.pone.0062580
Taylor, M. W., Radax, R., Steger, D., and Wagner, M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347. doi: 10.1128/MMBR.00040-06
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
Wahl, M., Goecke, F., Labes, A., Dobretsov, S., and Weinberger, F. (2012). The second skin: ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 3:292. doi: 10.3389/fmicb.2012.00292
Wang, L., Chen, Y., Huang, H., Huang, Z., Chen, H., and Shao, Z. (2015). Isolation and identification of Vibrio campbellii as a bacterial pathogen for luminous vibriosis of Litopenaeus vannamei. Aquac. Res. 46, 395–404. doi: 10.1111/are.12191
Wang, L., Shantz, A. A., Payet, J. P., Sharpton, T. J., Foster, A., Burkepile, D. E., et al. (2018). Corals and their microbiomes are differentially affected by exposure to elevated nutrients and a natural thermal anomaly. Front. Mar. Sci. 5:101. doi: 10.3389/fmars.2018.00101
Webster, N. S., Xavier, J. R., Freckelton, M., Motti, C. A., and Cobb, R. (2008). Shifts in microbial and chemical patterns within the marine sponge Aplysina aerophoba during a disease outbreak. Environ. Microbiol. 10, 3366–3376. doi: 10.1111/j.1462-2920.2008.01734.x
Wegley, L., Edwards, R., Rodriguez-Brito, B., Liu, H., and Rohwer, F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x.
Wulff, J. L. (2006). Ecological interactions of marine sponges. Can. J. Zool. 84, 146–166. doi: 10.1139/z06-019
Xu, J., and Gordon, J. I. (2003). Honor thy symbionts. Proc. Natl. Acad. Sci. U.S.A. 100, 10452–10459. doi: 10.1073/pnas.1734063100
Xu, W., Pang, K. L., and Luo, Z. H. (2014). High fungal diversity and abundance recovered in the deep-sea sediments of the pacific ocean. Microb. Ecol. 68, 688–698. doi: 10.1007/s00248-014-0448-448
Yu, S., Deng, Z., Proksch, P., and Lin, W. (2006). Oculatol, oculatolide, and A-nor sterols from the sponge haliclona oculat a. J. Nat. Prod. 69, 1330–1334. doi: 10.1021/np0600494
Keywords: seaflower, microbial diversity, marine hosts, amplicons, symbiotic interactions
Citation: Alvarez-Yela AC, Mosquera-Rendón J, Noreña-P A, Cristancho M and López-Alvarez D (2019) Microbial Diversity Exploration of Marine Hosts at Serrana Bank, a Coral Atoll of the Seaflower Biosphere Reserve. Front. Mar. Sci. 6:338. doi: 10.3389/fmars.2019.00338
Received: 31 July 2018; Accepted: 03 June 2019;
Published: 26 June 2019.
Edited by:
Santiago Herrera, Lehigh University, United StatesReviewed by:
Rika Anderson, Carleton College, United StatesJames Davis Reimer, University of the Ryukyus, Japan
Copyright © 2019 Alvarez-Yela, Mosquera-Rendón, Noreña-P, Cristancho and López-Alvarez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Astrid Catalina Alvarez-Yela, catalina.alvarez@bios.co; catalinaalvarezy@gmail.com; Diana López-Alvarez, dianalopez430@gmail.com
†Present address: Diana López-Alvarez, Facultad de Ciencias Agropecuarias, Universidad Nacional de Colombia, Palmira, Colombia
 Astrid Catalina Alvarez-Yela
Astrid Catalina Alvarez-Yela Jeanneth Mosquera-Rendón
Jeanneth Mosquera-Rendón Alejandra Noreña-P
Alejandra Noreña-P Marco Cristancho
Marco Cristancho Diana López-Alvarez
Diana López-Alvarez