The perspective of fish venom: An overview of the physiology, evolution, molecular and genetics
- 1Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals, Guangdong Research Center on Reproductive Control and Breeding Technology of Indigenous Valuable Fish Species, Fisheries College, Guangdong Ocean University, Zhanjiang, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), Zhanjiang, China
- 3Key Laboratory of Utilization and Conservation for Tropical Marine Bioresources of Ministry of Education, Yazhou Bay Innovation Institute, Hainan Tropical Ocean University, Sanya, China
- 4Hainan Key Laboratory for Conservation and Utilization of Tropical Marine Fishery Resources, Yazhou Bay Innovation Institute, Hainan Tropical Ocean University, Sanya, China
- 5Guangdong Havwii Agricultural Group Co., LTD, Zhanjiang, China
- 6Hainan Chenhai Aquatic Co., LTD, Sanya, China
Fish venom has several biological activities, including enzyme activity, cytotoxicity, neurotoxicity, muscular toxicity, haemolytic, and cardiotoxicity, when they enter other species or a human being, they disrupt the physiological systems. Transcriptomic analysis of the fish venom glands revealed a large number of proteins relevant to the pharmacological activity even though they are not well-studied. The limitations in studying fish venoms also have an impact on their molecular characterization. This is partly because of the nature of fish venoms, as they are extremely unstable at normal ambient temperatures making them difficult to study. Venomous fish inhabit both marine and freshwater environments, they have specialized venom-delivery apparatuses. Venom delivery systems have evolved in a various animal species, originally for different purposes including defense, competition, as well as predation. In coastal areas, fish stings are a major problem because they have a serious toxic effect on fishermen, local communities, and visitors. In this study, we have discussed the general perspective of fish venom from marine and freshwater species in different aspects basically in their molecular evolution, physiology, diversity, transcriptome, and proteomic studies. We expect that this paper will provide readers with a unique perspective on understanding the current status of fish venom research as well as working for future studies. Therefore, the gap of knowledge acquired from this study will play as a baseline for researchers discovering new studies and using fish venom in a broader range of biomedical applications, and their biological information that can be used to develop drugs for pharmaceutical uses.
1 Introduction to venom
Venom is a toxic substance produced by venomous animals including; snakes, fishes, scorpions, and bees resulting in injuries to their prey (Cameron and Endean, 1972; Liu et al., 2018). Usually, venom is produced by special glands being released to receptors (Isbister, 2001; Haddad and Lima, 2014). Venomous organisms have special structures for the release of venom, such as spines, barbs, and teeth or fangs, causing countless stings, bites, and barbs (Pandey and Upadhyay, 2020). Over 2.7 million injuries are caused by venom each year, prompting the study of venom as a potential source of novel physiological tools and pharmaceuticals. In the quest for new physiological tools and pharmaceuticals, venomous organisms have been the focus of numerous recent research (Pandey and Upadhyay, 2020).
Animal venoms have been considered an excellent source of novel, biologically active molecules (Bordon et al., 2020). There has been a lot of research into the bioactivities and components of terrestrial species venoms, including those from scorpions, snakes, and spiders, but there has been much less research into marine and aquatic venoms from fish (Terlau and Olivera, 2004; Koh et al., 2006; Sivan, 2009; Bordon et al., 2020). This is because terrestrial animals can be easily captured over marine specimens, and in part the threats posed by aquatic organisms are rarely considered. However, many venomous fish species found in marine habitats are capable of causing severe injuries in people, which can result in death. (da Silva et al., 2015; Gokulalakshmi et al., 2018). Many of these species are cnidarian and Conus invertebrates (Terlau and Olivera, 2004). Due to the extreme lability of some venom components, fish venom research is underrepresented in the literature. In addition, mucus in fish can contaminate with venom traces, posing a significant obstacle to their research (Pandey and Upadhyay, 2020). Nonetheless, fish venoms are a largely unexplored source of biologically significant compounds (Sivan et al., 2007).
1.1 A brief introduction to venomous fish
Studies have found that approximately 2500 fish species are venomous and have specialized structures related to teeth and fins such as fin spines, opercular spines, and cleithral spines (Wright, 2009; Wright, 2015; Harris and Jenner, 2019; Saggiomo et al., 2021). It has been reported that only 200 marine fish species, including stingrays, scorpionfish, zebrafish, stonefish, weeverfish, toadfish, stargazers, and some sharks, ratfish, catfish, surgeonfish, and blenny species, can sting humans, while the majority of venomous fish are non-migratory, slow-moving, and live-in shallow waters in protected habitats (Harris and Jenner, 2019). As a result, their inactivity is directly related to the evolution of venom and prey immobilization (Wright, 2015; Harris and Jenner, 2019). In a recent study examining phylogeny and venom evolution in ray-finned fishes, the number of venomous fish has raised that estimate to be more than 2000 (Saggiomo et al., 2021).
Studies conducted by Cameron and Endean (1970); Whitear (1975); Isbister (2001); and Wright (2015) reported, fish venom glands have been thought to be anti-predatory structures, but investigations into the potential benefits provided by these structures are relatively limited. For example, six specimens of Scatophagus argus, ranging in length from 62 mm to 296 mm, had a pair of venom glands located at anterolateral grooves in each fine spine, which were the first venom glands discovered in scatophagid fishes (Sivan et al., 2010; Wright, 2015).
Fish venom glands represent a common, putatively adaptive trait for which ecologically relevant comparative experiments have yet to demonstrate selective benefits. Regardless of its importance in the discovery of new drugs and biodiversity research, several studies have looked into the venom response of catfish (Isbister, 2001; Haddad and Martins, 2006; Wright, 2009; Smith et al., 2016). An effort to better understand fish venoms contributes not only to the development of drugs but also to more efficient biodiversity exploration (Sosa-Rosales et al., 2005; Atkinson et al., 2006; Pandey and Upadhyay, 2020).
Despite the severity of envenomation symptoms, the composition of stonefish venom has been relatively explored, stonustoxin (SNTX) is among the most studied toxins due to a number of the envenomation symptoms it causes and its applications in drug development (Ziegman et al., 2019). We used stonustoxin as a representative bioactive compound to construct the phylogenetic tree and sequence alignment (Figures 1, 2), despite having other known bioactive compounds. Since SNTX is a multifunctional lethal protein extracted from stonefish venom. It has two subunits, named alpha and beta which have molecular masses of 71 and 79 kDa respectively (Chun, 2004). It has been reported and known for inducing an array of biological responses both in vitro and in vivo (Chun, 2004; Ziegman and Alewood, 2015). In addition, SNTX is reported to be the first complete sequence of a protein derived from fish (Chun, 2004; Ziegman et al., 2019). Except for a short stretch of amino acids that makes up a B30.2 domain, which is present in many proteins of different functions and cellular locations, neither subunit-deduced amino acid sequence exhibits a significant homology with any known protein (Xie et al., 2017). Studies reported SNTX fusion proteins may be useful in the creation of antibodies against stonefish envenomation (Chun, 2004; Xie et al., 2017; Ziegman et al., 2019). Furthermore, SNTX-like MACPF/CDCs (Membrane Attack Complex Perforin/Cholesterol-Dependent Cytolysin) complexes are found throughout eukaryotic life and have a larger potentially immune-related function apart from venom (Xie et al., 2017).
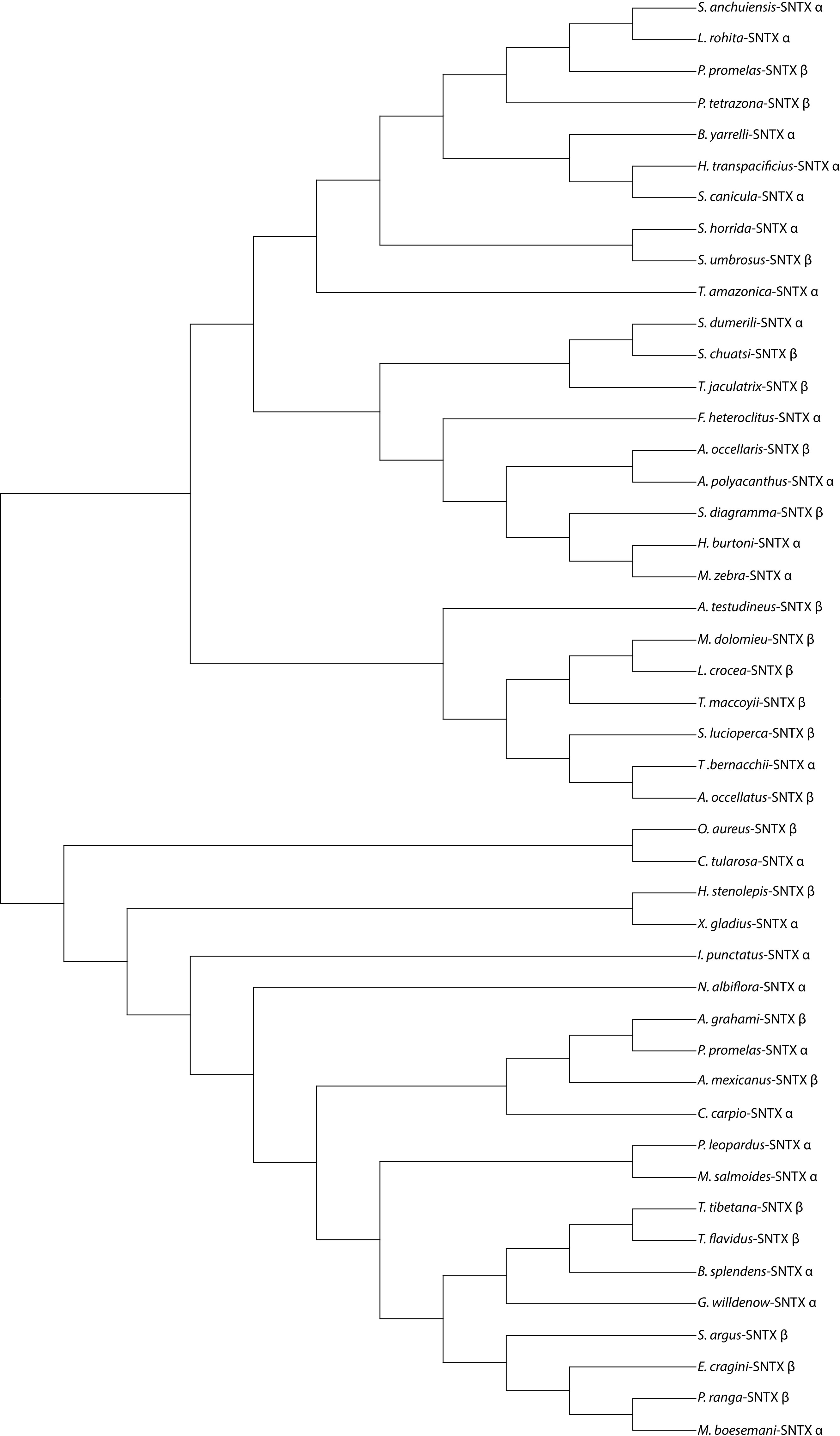
Figure 1 A phylogenetic tree showing representatives of some known venomous fishes that produces Stonustoxin (SNTX) as a venom bioactive compound. Sequences used to construct this phylogenetic tree were obtained from NCBI database. MEGA 11 software was used to construct this phylogenetic tree. Symbols α and β indicates the alpha and beta subunit of Stonustoxin. Full names of species and their accession numbers have shown in the table under Supplementary Material Table 1.

Figure 2 Sequence alignment of the α-subunit and β-subunit of Stonustoxin (SNTX) from some of venomous fish species. The FN3 domain is boxed in red borders. The PRY/SPRY domain are boxed in black borders. Amino acid numbers are shown on the right side and sequence discrepancies are highlighted in black and grey in a gradient based on similarity. Full names of species and their accession numbers from NCBI database have shown in the Table 1 under Supplementary Material.
This study is intended as a summary to provide the reader with an updated and unique perspective on the diversity of venomous fish, the evolution, physiology, molecular, and bioactivity of fish venom, and the application of fish venom in our daily life. We expect that gaps of knowledge will be exposed and stimulate future research on molecular characterization and transcriptomic analysis of fish venoms concerning their significance in our daily life, especially in the health sector.
2 Species diversity among venomous fish
To better comprehend the diversity of venomous fishes, it’s worth noting that the distribution of venomous fishes is evenly split between both freshwater and marine ecosystems (Haddad and Lima, 2014; Saggiomo et al., 2021). According to studies, freshwater ecosystems are home to about 58% of venomous fish (Haddad and Lima, 2014; Wright, 2015). Furthermore, while tropical oceans have the most diverse poisonous fish fauna, deadly species can also be found in temperate northern waters (Haddad and Lima, 2014). Freshwater stingrays live in larger rivers in South America, West Africa, and Southeast Asia. The diversity of separate venomous clades is not evenly distributed between marine and freshwater environments while having approximately equivalent species-level diversity (Haddad and Lima, 2014).
The enormous diversity of venomous catfishes explains a substantial part of the disparity (Harris and Jenner, 2019). Catfishes account for more than 95% of all venomous freshwater fishes and around 58% of all venomous fishes, according to current estimations based on several published studies of fish diversity (Table 1) (Cameron and Endean, 1970; Isbister, 2001; Haddad and Lima, 2014; Wright, 2015). Catfish are the most venomous freshwater fish and are found all over the world (Isbister, 2001; Wright, 2015). Their venom is contained in a sheath that surrounds three stingers in the dorsal and pectoral locations, and the envenomation is extremely painful compared to other species (Haddad and Martins, 2006). However, complications such as secondary infections and stinger breakage can occur. Stingrays from freshwater have one to four stingers on their tails, which can cause severe envenomation in humans, causing terrible agony and necrosis of the skin (Sosa-Rosales et al., 2005; Haddad and Martins, 2006; Fox and Serrano, 2007; Haddad and Lima, 2014).
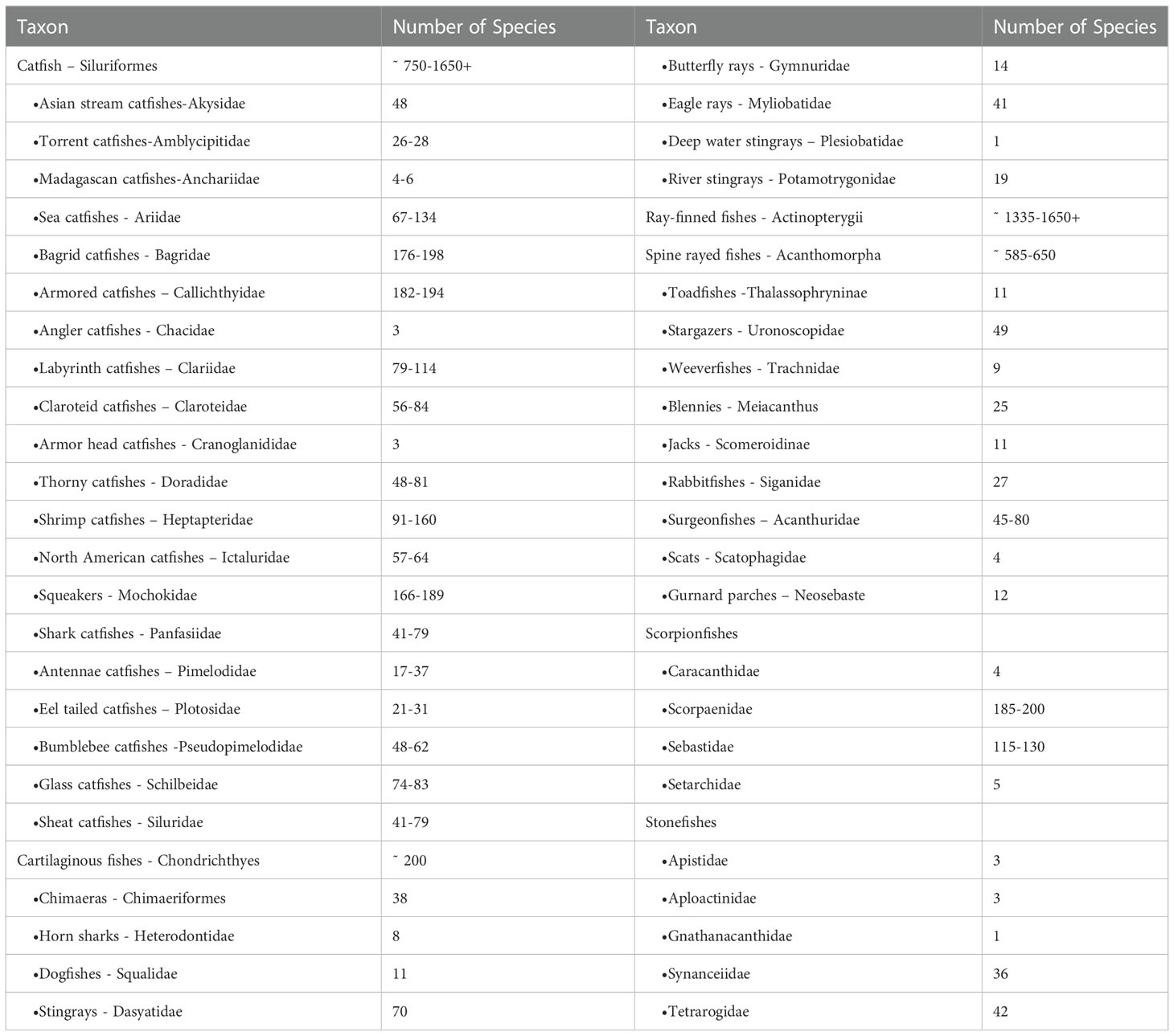
Table 1 Number and taxonomic distribution of venomous fish, estimates reproduced from (Wright, 2015; Smith et al., 2016).
3 Evolution of venom glands in fish
The production of toxic proteinaceous secretions, molecular and anatomical methods by the epidermis cells have been used to study and explore the evolution and variation within venomous fishes (Cameron and Endean, 1972; Novak et al., 1973; Whitear, 1975; Smith and Wheeler, 2006). Most ray-finned fishes have been defensive to themselves using pungent fin spines since the evolution of venom in glands involves the secretion of toxins in the vicinity of the puncturing apparatus (Harris and Jenner, 2019). A similar type of venom gland was described morphologically in catfish (Galeichthys felis), with two types of secretory cells, clavate cells, and mucous cells (Wright, 2015; Liu et al., 2018).
The thickened epidermis, which is separated from the epidermis connective tissue of the dermis by an increased component of proteinaceous secretory cells closer to the pungent spine, was discussed by Ghadessy et al. (1996); Ueda et al. (2006); Ziegman and Alewood (2015); Campos et al. (2016). The collagenous barrier attached laterally to the venom gland ensures that the venom is guided into the victim during envenomation by the spine. The spines tend to be grooved to accommodate the venom glands. In the stonefish, the spine grooves almost completely encasing the venom glands’ distal ends and transforming them into functional ducts. In venomous batrachoidids, the venom glands are located at the bases of their spines (Wright, 2015).
Dorsal spines are the most prevalent envenomed structures, according to anatomical investigations and phylogenetic reconstruction. Aside from envenomed spines, fishes have evolved venomous fangs, which accounts for around 2% of venomous fish species and two independent evolutions; cleithral spines, which accounts for around 2% of venomous fish species and one independent evolution; and opercular or sub-opercular spines, which account for only 1% of venomous fish species and three independent evolutions (Wright, 2012; Wright, 2015; Liu et al., 2018; Harris and Jenner, 2019). Understanding the variety and development of all venomous systems without the biased inherent nature of fish venom in focusing on charismatic venomous species requires studying many neglected venomous animals and their evolutionary background (Liu et al., 2018).
Given advances in our understanding of the anatomy of potentially envenomed structures, the discovery of venomous groups, and the interaction of fish, the time has come for the investigation of the evolution, phylogenetics, and distribution of venom across fish species (Sivan, 2009). We expect that the resulting phylogenetic framework will not only be important for studying the evolution across fishes, but will also provide an intensive and up-to-date baseline for studying venoms in future pharmaceutical, anatomical, and venomous studies (Sivan, 2009). In the section below the different aspects related to the evolution of venom glands in fish have been discussed.
3.1 Fish venom evolution for defense
Several fish species employ spines as a kind of defense against predatory assaults. Though not all spine defenses contain venom, non-venomous spines are ineffective against predators (Wright, 2012). The defensive strength of non-venomous spines varies depending on their shape, size, stoutness, and the addition of venom glands, which improves defense. The thickening and aggregation of epidermal cells that produce antiparasitic toxins near defensive spines are thought to have evolved into venom glands in fish.
The interactions between predator and prey as drivers of defense and the evolution of venom in fish are still largely unknown (Saggiomo et al., 2021). The only studies on predator responses to venomous fish prey that are available are on catfish, with other investigations on the pharmacological effects of fish venom toxins limited to mammal species that are not natural fish predators (Harris and Jenner, 2019). Sharks, rays, and sea snakes are the main predators of stone fish in their natural habitats, whereas lionfish are mainly predated on by eels, sharks, and groupers (Saggiomo et al., 2021). These predators may be susceptible to their prey’s defensive venom in different ways. Defensive venoms can kill a variety of organisms, including non-natural predators (Harris and Jenner, 2019). As a result, it is unlikely that a single predator species would drive defensive venom evolution. This diversity in the taxonomy of predators could explain why many defensive venoms, in terms of venom composition and delivery systems, are non-target specific (Wright, 2015). Although interactions between predators and venomous fish prey have not been deeply studied, there is evidence that such kind of interactions has resulted in the evolution of defensive spines, which are an essential structure for delivering venom (Smith and Wheeler, 2006; Wright, 2015).
Regarding these theories, it is very possible that in some fish species, host-parasite interactions paved the way for the evolution of skin toxins, while predator-prey interactions paved the way for the evolution of spines (Harris and Jenner, 2019). Skin toxins were then recruited into spine-associated venom due to selection pressures for increased anti-predator defenses. Even though antagonistic coevolution played a role in the evolution of spine defenses, the number of fish venom systems convergent theories suggests that venom evolution in fish is under strong selection pressure (Harris and Jenner, 2019).
3.2 Fish venom evolution for predation
Predation in fish is a major cause of venom evolution. The selection of venom specificity to prey has emerged from the interaction between predator and prey (Liu et al., 2018). These co-evolutionary cycles are the result of a constant conflict between increasing efficiency for subduing prey and prey toxin resistance, although there are few studies in this area. Venom delivery apparatuses close to an organism’s mouth, such as fangs, pincers, beaks, and proboscis, are most commonly associated with venom used for predation. Only two fish taxa jawed eels and lampreys, use fangs or teeth for venom-based feeding (Wright, 2015).
Most organisms are opportunistic hunters due to the scarcity of prey in deep-sea habitats. As a result, venom for predation in desolate environments would be very adaptable to prevent from escaping (Smith and Wheeler, 2006). Lampreys are jawless fish with a toothed sucking buccal cavity that dates back thousands of years (Liu et al., 2018). Most lamprey species engage in parasitic micro-predation, attaching their mouthparts to larger hosts and feeding on their blood for an extended period. Hematophagous organisms such as mosquitos, leeches, bats, and ticks have all evolved this type of predation (Ziegman and Alewood, 2015). Anticoagulants, as well as nociceptors and immune response inhibitors, have been identified as key components of their venom secretions in some studies (Sosa-Rosales et al., 2005).
Lampreys are renowned for their wide dispersal and habitat ranges, which they achieve by attaching themselves to migrating hosts and being carried to new areas. Research reported that the use of blood-feeding toxins and host attachment evolved simultaneously (Wright, 2015). The longer a lamprey can remain attached to a host to reach a specific destination while feeding, the better its chances of surviving the journey (Wright, 2015). This particular way of life probably influenced the evolution of this venom system, which has evolved numbing toxins that leads to a loss of sensation in part of the body to keep blood flowing and prevent detection by their hosts over long migration distances (Harris and Jenner, 2019). There has been very little research into how these micro-predatory venoms evolved, whether before or after the evolution of host attachment. Generally, lampreys are an excellent model for these studies because they are the only fish that use this technique (Harris and Jenner, 2019).
3.3 Fish venom evolution for competition
There is a minor difference between competitive and defensive competitive venom, as both serve the same purpose. The distinction is made based on the selective pressures that drive venom evolution and how it is used. Competition may have influenced the evolution of one venom system in fish (Smith and Wheeler, 2006; Harris and Jenner, 2019). Few animals, like the platypus and the slow loris, use venom for competition (Harris and Jenner, 2019). The striped fang blenny venom has a unique biological activity (Harris and Jenner, 2019).
Blennies are well known for their fierce territorial competition and aggressive combat with rivals (Liu et al., 2018). Their venom’s biological activities such as, cardiovascular activity and neuromuscular activity would be advantageous against competitors because disorientation and hindrance incoordination maximize the likelihood of the competitor becoming an easy target for predators, resulting in the competition being permanently removed from the habitat (Harris and Jenner, 2019). Due to the fierce competition for territory, blennies may have been under more pressure to develop fangs and venom (Liu et al., 2018).
4 Morphological identification and examination of venom glands in fish
Even within closely related species, the size and cellular morphology of fish venom glands can vary substantially (Liu et al., 2018). However, the majority of them appear to share some characteristics (Figure 3). The glands are typically made up of giant glandular cells surrounded by supporting cells that give strength and connect the gland to the tissue surrounding (Cameron and Endean, 1973). The venom appears to be secreted by a holocrine mechanism. Furthermore, histology frequently reveals the availability of cytoplasmic granules on the internal side of gland cell membranes, as well as smaller granules in the gland cell vacuoles (Smith and Wheeler, 2006). The serrated stings of stingrays are covered by epidermal tissue, which has both secretory cells specialized for the sting and secretory cells found throughout the animals’ epidermis, and the glandular epithelium found to cover the venom spine of the pacific ratfish (Cameron and Endean, 1973; Smith and Wheeler, 2006).

Figure 3 Representative venomous fish structure morphology: (A) Venomous dorsal spines from the lantern shark. (B) Venomous dorsal spine from the jack. (C) The barbed dorsal spine of the stingray. (D) The venomous dorsal spine with enlarged venom glands in the stonefish, (E) The barbed pectoral spine of the sea catfish. (F) The venomous opercular spine of the toadfish. (G) Cleithral spine of the stargazer. (H) The venomous fang of the one-jawed eel. Abbreviations: cs, cleithral spine; f, fang; pg, posterior groove; vg, venom gland. Source (Smith et al., 2016).
As previously reported, fish deliver venom by a variety of structures, including spines, barbs, teeth, and fangs, the position of which varies by species as indicated in (Figure 3). Venom spines are found on all fins, as well as around the operculum, while stingray barbs can be found on the tail (Smith and Wheeler, 2006; Harris and Jenner, 2019). Spines are frequently connected with the type of secretory cells or venom glands found close to the spines (Smith et al., 2016). Anterolateral grooves of the spine allow the venom to pass from the base to the tip of the spine in a hypodermic form, leading the toxins to enter the envenomed victim through a wound. Although this is the general form of species that uses venom spines, there are variations between species, according to their convergently evolved systems (Smith and Wheeler, 2006; Wright, 2012; Smith et al., 2016).
Toadfishes of the subfamily Thalassophryninae have highly developed venom apparatus of the fishes which, the venomous dorsal and opercular spines appear as enclosed hollow tubes emerging from the venom glands, (Wright, 2012). The venom apparatus of the Fang blenny species is unique in that it uses venomous canine teeth, despite the buccal glandular tissue resembling that of other venomous species (Liu et al., 2018). Stonefish has a more advanced venom gland, with the distal end shortened to form a duct-like structure within the spine groove through which the venom can pass before being released (Smith et al., 2016). The venom mechanism when subjected to mechanical pressure, the venom is released through the canaliculated spines and into the victim, resulting in envenomation (Smith et al., 2016).
Catfish venom glands are located at the anterior margin of the dorsal and pectoral fins and are surrounded by sharp, bony spines as indicated in (Figure 3). When the fish is threatened, the bases of spines, as well as their associated musculature, are modified in such a way that the spines can be erected and locked into place. The pectoral spine stridulation is also responsible for producing sounds that appear to be important for intraspecific communication in many species (Wright, 2015).
Venomous fish are unable to control the discharge of their venom because they lack muscle connected with their venom apparatus (Haddad and Martins, 2006). Fish venom structures are assumed to have been acquired relatively recently in evolutionary history, and they serve primarily for defensive purposes, consistent with their involuntary expulsion mechanism (Harris and Jenner, 2019). Some venomous fish have been able to adopt a sedentary lifestyle by camouflaging themselves amid the rocks and debris on the seafloor and erecting their venomous spines when perceive threats are nearby, thanks to the evolution of such effective defense mechanism (Liu et al., 2018).
5 Adaptations and concerns related to evolving a venom system in fish
Adaptation research focuses on the most fundamental mechanisms driving evolution and, by extension, every other aspect of an organism’s biology. Individual traits and their use and variation in nature are generally examined in research focusing on a particular process. Several pieces of evidence have been used to infer the past action of adaptation on various traits, with the type of evidence varying depending on the adaptive trait definition to which the researcher adheres. Here we will discuss two adaptations and their challenges related to the evolution of the venom system in fish.
5.1 Aposematism
Aposematism is the use of a signal by an animal, especially a visual signal like bright colors or obvious markings, to alert predators that are poisonous or repulsive. Many chemically protected organisms have aposematic warning signals. They send out warning signals in the form of bright, contrasting patterns that alert predators to their chemical arsenal (Harris and Jenner, 2019). This type of defense goes together with defensive venom evolution, and venomous fishes are no exception. Predators in the early phases of predation have evolved to avoid these aposematic patterns. Predators can learn and avoid specific color patterns as warning coloration evolves, sparing them from getting envenomed defensively (Wright, 2012).
5.2 Mimicry
Mimicry is the evolution of similar appearances, behaviors, or scents that prey or predator connect with the species they are trying to catch or avoid (Whitear, 1975). The goal of a mimic is to deceive a predator or prey into thinking they are either a deadly or innocuous species. Mimicry can be parasitic on the model or mutualistic, with both parties benefiting at the same time. Many aposematic venomous species serve as attractive mimetic models for non-venomous species (Wright, 2015).
6 Intersexual variation in venom among fish species
Primarily for species like spiders, snakes, scorpions, and some fish, sexual variation in venom composition has been recorded (Lopes-Ferreira et al., 2016). Male venom from Cano toadfish has double the protein content than that of female venom with different bioactivities, with males seeming to have a higher target affinity for nociceptors where females have higher proteolytic activity (Sosa-Rosales et al., 2005; Ziegman and Alewood, 2015). From an ecological standpoint, these differences might have something to do with reproduction and brooding. The energy balancing of venom and reproduction may be connected to a decreased protein concentration in females (Lopes-Ferreira et al., 2016).
With a lesser yield, energy spent on venom maintenance can be diverted to other priorities, such as reproduction. On the other hand, males may have a higher protein production and potency when it comes to protecting eggs from predators and conspecifics. Other animals, such as spiders, have made similar observations. Females with eggs had lower venom production and lower proteins than females without eggs (Harris and Jenner, 2019). However, venom may be connected to other characteristics such as food, health, and size rather than being sex-specific. Size sexual dimorphism is frequent, and size may be fundamental in understanding why venom potency and yield differ across sexes. This, however, does not account for the differences in protein composition and bioactivities between males and females (Harris and Jenner, 2019).
7 Main components of fish venom
Because fish venoms are heat-labile, mucous-accessible, and difficult to extract, have received little attention (Church and Hodgson, 2002; Koh et al., 2006; Sivan et al., 2010; Ziegman and Alewood, 2015). Most of them are made up of a variety of chemicals and contain bioactive toxins that have a variety of neuromuscular, cardiovascular, cytotoxic, and nociceptive effects as discussed in chapter 8. Most fish venoms contain large pore-forming toxins like SNTX and verrucotoxin (VTX) as indicated in (Table 2) which are proteins in nature (Church and Hodgson, 2002; Ziegman and Alewood, 2015).
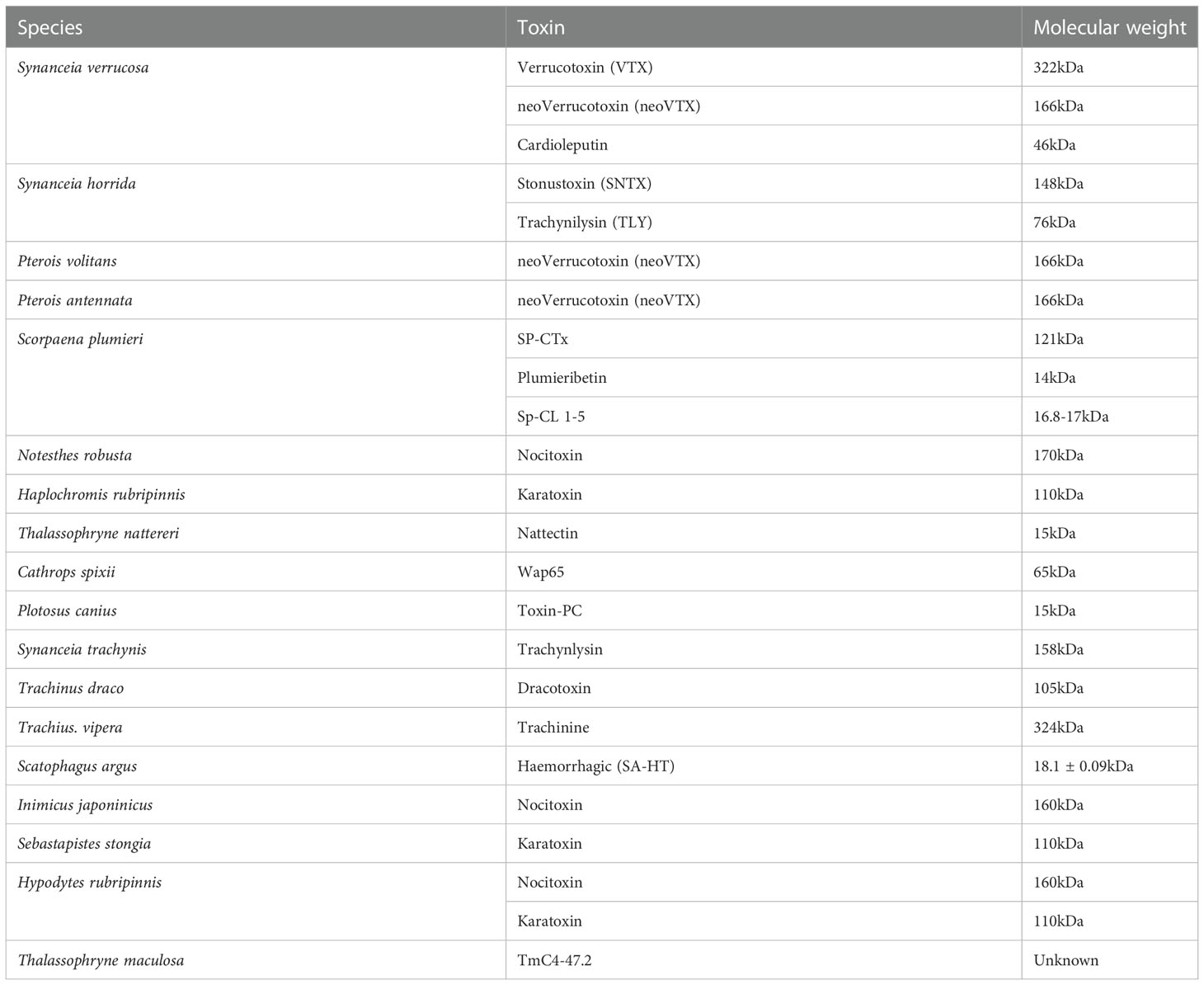
Table 2 Updated toxins found in some venomous fish with their molecular weight, and source (Sivan, 2009; Saggiomo et al., 2021).
Fish venom has yielded many types of toxins that have been identified (Ziegman and Alewood, 2015). In general, these toxins have all of the fatal properties as well as the majority of the other biological properties (Sivan, 2009). In contrast to the venoms of several terrestrial animals, piscine venoms appear to contain a small number of toxins (Koh et al., 2006; Ziegman and Alewood, 2015). At the same time, there are a variety of additional protein toxins, such as dracotoxin, trachinine, and nattectin, which have been identified from many species but are largely understudied (Ziegman and Alewood, 2015).
Venoms from fish contain a variety of other active ingredients; for example, the venom of the greater weeverfish has significant levels of histamine and catecholamines, as well as cholinesterase activity. The 5-hydroxytryptamine, 5-nucleotidase, and phosphodiesterase are all found in the venom of the stingray (Koh et al., 2006; Ziegman and Alewood, 2015). Stonefish venom contains catecholamines as well as a variety of enzymic activities. Stonefish, lionfish, and freshwater stingray venoms are also having acetylcholine or a cholinomimetic (Ziegman and Alewood, 2015) which are useful for various biological activities.
8 Molecular genetics of fish venom
8.1 Sequencing and databases of fish venom toxins
It is well known that fish venom is still largely unexplored as reported by (Silva et al., 2018; Xie et al., 2019; Gorman et al., 2020; Campos et al., 2021; Lopes-Ferreira et al., 2022) and in previous sections of this paper. Because there are few fish toxin sequences available as references, most of them are from Scorpaenidae and Synanceiidae families, it is challenging to research fish toxins and to create quick and effective techniques for wide exploration of toxin proteins or genes (Xie et al., 2017). Some recent studies have isolated and provided several toxins sequences using different techniques (Smith and Wheeler, 2006; Xie et al., 2016; Costa et al., 2018; Xie et al., 2019; Campos et al., 2021; Lopes-Ferreira et al., 2022), but due to a given number of venomous fish (>2000 species) (Saggiomo et al., 2021), it would be just a small portion of it.
Tradition methods for venom analysis often use bioassay-guided fractionation, in which fractions that exhibits the required biological activity in a particular assay are purified and characterized for further investigation (Xie et al., 2017). However, this method takes a long time and uses a lot of crude venoms. Studies have shown, cDNA cloning and genomic sequencing were used to identify the full amino acids sequences of various toxins including SNTX (Chun, 2004), verrucotoxin (VTX) (Ziegman and Alewood, 2015), cytolysins (Campos et al., 2021), lectin (Nakagawa et al., 2015; Ziegman et al., 2019; Lopes-Ferreira et al., 2022), Sp-CTx (Costa et al., 2018), and hyaluronidases (Ziegman et al., 2019) and among the others in which most of them exhibit high sequence identity among and between species.
In contrast, most of the sequenced toxins that have been submitted along with publications are currently found in toxin-centered databases which are divided into two groups termed generalist and toxin-centered databases (Xie et al., 2017). Toxin-centered databases including, Tox-prot Program, the Animal Toxin Database (ATDB), ConoServer, and Arachnoserver in which sequences in these databases can be easily traced back to the original peer-reviewed papers or found in generalist databases (Xie et al., 2017). On the other hand, general databases including NCBI-RefSeq, NCBI-nucleotide, and UniProtKB/Swiss-Prot servers (Xie et al., 2017, Xie et al., 2019), these databases combine both toxin and non-toxin sequences, making it difficult to extract the required sequences.
8.2 Transcriptomic and proteomic analysis of fish venom
Our preliminary survey found that there is little transcriptomic and proteomic data from the venom of various fish species compared to scorpions, spiders, cone snails, and snakes. Over years now several putative venom proteins were identified in the venom gland transcriptomes and proteomes but no known fish toxins were found, even for the well-known pore-forming toxins related to stonustoxin that were recently identified in three scorpionfish transcripts (Xie et al., 2019). Currently, fewer venomous fish species have undergone transcriptome and proteome research. A ray species, catfish species, and fang blenny species are among those for whom there are comparable proteomic data (Xie et al., 2017; Xie et al., 2019; Lopes-Ferreira et al., 2022). Recently, expressed sequence tags were used to analyze the venom gland transcripts of toadfish (Ziegman and Alewood, 2015). According to Xie et al. (2017), a study on the fish venom gland reported that the fish gland is derived from the skin, and skin secretions are thought to be crucial for skin healing and immunity.
Fish venom research has become much more sensitive and effective in the 21st century as a result of the development of multi-omics approaches by employing next-generation sequencing (NGS) and liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Xie et al., 2017). These methods have been demonstrated to be effective in several fields including neuroendocrine research. The development of large-scale genomic and transcriptome sequencing projects to employ de novo assembling methods, which accurately assemble fragments into full-length transcripts, especially in the absence of reference genome sequence has been famous (Baumann et al., 2014; Júnior et al., 2016; Xie et al., 2017; Silva et al., 2018; Xie et al., 2019). For example, the Illumina MiSeq sequencing of the P. amandae and P. falkneri cDNA libraries generated ~14.5 and ~15.4 million paired-end reads, with average lengths of 231 and 220 bp, respectively. After adapter trimming and quality filtering, we obtained a total of ~13.1 and ~13.7 million high-quality reads, with 92.7 and 92.4% of all bases having Phred (Q) scores above 33, respectively, which indicates good sequencing quality (Júnior et al., 2016). Since there are so few venom genomes from fish, the assembly of transcriptome reads is difficult and should be approached with caution. Bioinformatics, based methods that combine transcriptome and proteome approaches can widely explore toxins present in fish venom.
Transcriptomic analysis of the venom glands of the toadfish and the stingray exposed a large number of proteins relevant to the pharmacological activity of these venoms, for example, Galectins, and C-type lectins, as well as some new to fish venom lectins (Sivan et al., 2010; Tamura et al., 2011; Baumann et al., 2014; da Silva et al., 2015). The presence of unknown genes of potential relevance in the venom gland was suggested by a preliminary analysis of expressed sequence tags obtained from spotted scorpionfish venom using a cDNA library (Gomes et al., 2010; Campos et al., 2016). Furthermore, antibodies against a lectin fraction from spotted scorpionfish venom were used to filter the library, revealing that lectin-like genes account for about 12% of all transcripts, a conclusion validated by comprehensive in silico analysis (Campos et al., 2016). These components are the initial steps toward understanding the molecular variety found in fish venoms (Kiriake and Shiomi, 2011).
According to a study on Chinese yellow catfish (Xie et al., 2016), a combination of transcriptome and proteome data revealed that (i) by using PTMs, alternative splicing, insertion, and premature transcription termination, different mature toxin sequences can develop from single toxin precursor, (ii) significance differences between the proteomic and transcriptomic data were observed in Chinese yellow catfish. It’s interesting to note that occasionally, toxins predicted based on transcriptome data cannot be validated by proteome data. On the other hand, some toxins in the proteome lack corresponding transcripts (Ziegman et al., 2019). The selective expression of venom peptides or proteins from the genes may cause these discrepancies. Also, sometimes lack enough venom in a single venom gland may lead to retrieving venom samples for next-generation sequencing in a single while proteomic material is only gathered from the other group (Xie et al., 2017; Lopes-Ferreira et al., 2022; Zancolli et al., 2022).
Generally, understanding the genetic basis of the diversification of venom-encoding genes in various species can offer fundamental biological insights into species evolution, ecological specialization, genetic novelties, and drug development as stated in previous sections.
9 Biological and pharmacological importance of venom from fish
Most piscine venoms have a similar mechanism of action, which involves depolarizing nerve and muscle cells (Church and Hodgson, 2002; Sivan, 2009). The majority of piscine venoms exhibit strong cytolytic activity, which appears to be the mechanism behind many of their neuromuscular and cardiovascular effects (Church and Hodgson, 2002; Sivan, 2009). Venom’s chemical features, which make them both harmful and fascinating, have the potential of being used as natural products for the development of useful medications and physiological functions if they can be harnessed and deeply investigated.
9.1 Cardiovascular activity
Hypotension is one of the clinical signs of significant fish envenomation in humans (Fox and Serrano, 2007). Experimental animals envenomed by fish venom showed increased cardiovascular activity. Cardiovascular action appears to be the most investigated and dominating activity of fish venom (Gomes et al., 2010). Both vascular and non-vascular smooth muscles are affected by piscine venoms (Saggiomo et al., 2021). Stonefish envenomation has been linked to possibly fatal cardiovascular consequences (Church and Hodgson, 2000; Isbister, 2001; Gomes et al., 2010). However, because it is unclear whether stonefish venom’s mortality is related to its cardiovascular effect or its neuromuscular activity, more research into the molecular genetics of this type of venom is needed. In isolated cardiac preparations, the soldierfish and stonefish venom pharmacological effects have been extensively investigated (Ziegman and Alewood, 2015; Saggiomo et al., 2021).
Several frequent cardiovascular symptoms associated with fish envenomation have been recorded in the literature (Church and Hodgson, 2002; Ziegman and Alewood, 2015). These include inotropic and chronotropic responses, as well as changes in endothelium-dependent smooth muscle relaxation and blood pressure. In experimental settings, fish venoms induced a wide range of blood pressure responses, many of which were multi-phasic. The animal models that are guinea pigs and rabbits died as a result of the continuously lowering blood pressure in the case of the catfish venoms (Sivan, 2009). Other sources of information include (Church and Hodgson, 2002; Baumann et al., 2014) attributed the responses to California scorpionfish venom to the release of endogenous acetylcholine from muscarinic receptors. Smith and Wheeler (2006); Sivan (2009); Ziegman and Alewood (2015); found the release of endogenous nitric oxide from endothelial cells was linked to the blood pressure response to the venom of the soldierfish.
9.2 Neuromuscular activity
The neuro-modulatory effects of venom are one of the most important topics of research in pharmacology and neurophysiology (Saggiomo et al., 2021). Fish venom’s neuromuscular characteristics have been extensively researched. When injected into a mouse, several piscine venoms cause neurotoxic symptoms such as paralysis, convulsions, muscle weakness, and at larger dosages, respiratory arrest, which leads to death (Sivan, 2009). To cause depolarization of cell membranes, all piscine venoms work both pre-junctionally and post-junctionally.
The separation and characterization of fish venom components with myotoxic and neuromodulatory activity, which is linked with the neuromuscular activity of venom, has proven to be a useful tool in the study of neuromuscular transmission (Sivan et al., 2010). Fish venom has been demonstrated to trigger a variety of different reactions in the laboratory, including cell depolarization and muscular contraction (Sivan, 2009).
9.3 Edematic activity
Edema is a common systemic sign of fish envenomation (Sivan, 2009). Edema is a symptom of cutaneous inflammatory processes, and it is caused by a synergistic interaction between mediators that increase vascular permeability and those that increase blood flow. In mice, fish venom causes a strong and long-lasting edematogenic reaction. Stonefish envenomation has been linked to severe edema at the sting site, according to reports (Smith and Wheeler, 2006). Even after antivenin medication, the edema usually lasts for 2-4 days. In keeping with this, stonustoxin causes significant inflammation in the mouse’s hind paw that lasts for more than 24 hours after injection (Ghadessy et al., 1996).
Neuropathic pain is evident immediately after venom injection, but the discomfort appears to diminish over the succeeding inflammatory period, indicating that the second phase of pain was caused by the inflammatory processes of leukocyte rolling and adhesion (Atkinson et al., 2006). In mouse models, venom from several different fish species induces considerable discomfort, according to similar studies. The predominant source of nociceptive action for crude venom has been identified as a single pain-causing protein fraction isolated from Notesthes robusta venom, although its mode of action is unknown (Sivan, 2009). Based on observed peaks from cation-exchange chromatography, the venom of the toadfish was divided into multiple fractions, each of which demonstrated nociceptive qualities (Sosa-Rosales et al., 2005).
The most common symptom of piscine envenomation is excruciating pain, Haddad and Martins (2006) reported that the venom of both Scatophagus argus and Pterois volitans contains antinociceptive chemicals. This was predicated on the fact that venom increased the activity of Na+, K+, and ATPase, which are known to modulate pain, in a dose-dependent manner in both cases. These findings add to our knowledge of fish venom’s nociceptive effects (Ziegman and Alewood, 2015). However, much more research is needed to fully comprehend this aspect of venom functioning.
9.4 Cytolytic activity
Venoms from animals are often complex combinations of bioactive chemicals, primarily proteins, and peptides (Koh et al., 2006; Ziegman and Alewood, 2015). These poisons interact with physiological targets, causing tissues to become immobile, die, or be digested. Because of their important function in homeostasis, the blood vasculature system is a relevant target, and many varieties of venom contain toxins that impact it (Sivan et al., 2010). Many species rely on cytolytic proteins and peptides to undertake offensive and defensive functions by lysing cells via enzymatic and non-enzymatic methods. Several marine peptides have shown anticancer activity of a high order (Ziegman and Alewood, 2015). The role of the venom’s active component, its isolation, molecular target, as well as the signaling mechanisms by which it promotes apoptosis in cancer cells are all still being researched (Ueda et al., 2006; Xie et al., 2019).
Necrosis and apoptosis are the two main mechanisms that induce cell death (Fox and Serrano, 2007; Baumann et al., 2014). Local tissue necrosis is the most common and major clinical outcome of fish envenomation. The venom of the fish has also been proven to induce apoptosis in cancer cell studies. The lionfish venom activates Caspase-3, causing apoptosis in Ehrlichs ascites carcinoma xenografted mice, as indicated by nuclear fragmentation (Ziegman and Alewood, 2015). On cultivated HEp2 and HeLa cells, a peptide termed 7.6 kDa from lionfish venom proved the role of apoptosis, and this peptide specifically targets cancer cells while sparing normal cells (Sivan, 2009). Furthermore, research on the mitogenic and cytotoxic effects of Synanceia verrucosa and Haplochromis rubripinnis on normal and tumor cell lines revealed mitogenic effects in normal cells, but considerable cytotoxicity in tumor cell lines was generated by crude venom and some separated fractions (Smith and Wheeler, 2006).
9.5 Enzymatic activity
Many enzymes other than proteases have been discovered in fish venom. The venom of S. argus contains alkaline and acid phosphatases, as well as phosphodiesterase activity (Sivan et al., 2010). All of these enzymes, as well as vesterase, were found in Gadopsis marmoratus and Synanceia horrida. Spotted scat venom had higher amounts of alkaline and acid phosphatase than G. marmoratus or S. horrida venom; however, S. horrida had significantly less phosphodiesterase activity than the other two species with comparable phosphodiesterase activity. Arius thalassinus venom also had esterase and alkaline phosphatase activity, although at much lower levels than G. marmoratus venom. S. verrucosa venom demonstrated activity for eight different esterases targeting various substrates, Trachydoras nattereri venom also contains angiotensin-converting enzyme activity, which adds to the venom’s inflammatory reaction (Pandey and Upadhyay, 2020).
10 Challenges of studying fish venom
Despite their medical significance and pharmacological appeal, fish venoms are challenging to research (Koh et al., 2006; Sivan, 2009). The limitations in studying fish venoms have an impact on their molecular characterization (Chhatwal and Dreyer, 1992; Sivan et al., 2010; Tamura et al., 2011). Currently, only a few publications have been published on the genetic investigation of fish venoms (Baumann et al., 2014; Campos et al., 2016; Xie et al., 2019). The venom collection itself is difficult because the venom gland in fish usually does not have a well-defined storage structure. Many species contain pectoral, caudal, cleithral, or even fang venom devices, which are often placed dorsally as expressed early in this paper. This helps to explain why fish venoms are so poorly understood at the molecular level (Sivan et al., 2010; Baumann et al., 2014).
Compared to other groups of venomous vertebrates, the use of next-generation sequencing techniques for investigating fish venoms is still in its infancy though is highly recommended (Ueda et al., 2006; Sivan et al., 2010; Xie et al., 2019). Some researchers were able to extract relatively pure venom from the blue-spotted stingray’s venom gland (Baumann et al., 2014) as a way forward for further research. Furthermore, they were able to annotate toxins with great accuracy and variety using bioinformatics tools. Both results suggested that transcriptomics is a useful method for identifying fish venoms (Baumann et al., 2014; Saggiomo et al., 2021).
11 Negative effects of fish venom on humans
Given that the majority of fish venoms appear to have developed as a protective tactic against other vertebrate species, envenomation occurrences in humans are likely to have serious consequences (Cameron and Endean, 1970; Atkinson et al., 2006; Haddad and Martins, 2006). Fish venom envenomation causes a wide range of symptoms, some of which have been reported to result in fatalities. The most noticeable sign is excruciating pain that is out of proportion to the severity of the damage (da Silva et al., 2015). Edema and erythema are also prevalent, and vesicles may form surrounding the lesion in rare cases (Sosa-Rosales et al., 2005; Sivan, 2009; Tamura et al., 2011). Ischemia, muscular spasms, tissue necrosis, persistent weakness, and nausea are among the systemic symptoms of fish stings, as a result, paralysis of the affected limb, hallucinations, hypotension, tachycardia, and respiratory distress may transpire (Magalhães et al., 2006; Ziegman and Alewood, 2015). Following envenomation, slow healing and necrosis have been noted as it is widely assumed that if death is going to happen, it will happen within the first few hours of contact (Ziegman and Alewood, 2015). The severity of envenomation damage varies depending on the species involved, the number and depth of envenomation sites, and individual sensitivities to the venom components.
When fishermen, divers, and bathers unintentionally step or mishandle venomous fish, their skin gets punctured by the spines, and they become envenomated (Haddad and Martins, 2006). Each year, an estimated 50,000 fish stings occur around the world. Hundreds of weeverfish stings have been reported along the United Kingdom and Adriatic coasts. Each year, an estimated 1500 ray stings and 300 scorpionfish stings occur in the United States. Freshwater ray stings are prevalent throughout the Amazon basin. Wading near the coast of coral reefs chances of stumbling upon, stonefish that have been partially hidden by sand, marine plants, and anemones are high (Novak et al., 1973; Whitear, 1975; Ghadessy et al., 1996; Adler, 2015; Saggiomo et al., 2021). Stingrays lash their tails, impaling the ankle in most cases.
12 Treatment from fish venom
The treatment is symptomatic and usually involves soaking the affected area in hot water that ranges from 45°C to 50°C) at least until the pain subsides, though the efficacy of this method is still debated (Atkinson et al., 2006). Though envenomation by poisonous fish is rarely fatal to humans, it has significant economic consequences because fishermen are the group most vulnerable to accidents (Isbister, 2001; Atkinson et al., 2006). According to official reports, accidents involving venomous fish vary by geographical area and habitat, and one should report the incident to the appropriate authorities for further investigation. The potential severity of the injuries caused by various venomous fish justifies the need for a deep investigation of these cases (Isbister, 2001).
In severe cases, commercially available antivenom of the stonefish can be used. Although tests have demonstrated that it does not cross-react with the venom of the scorpaeniform Bullrout, stonefish antivenom (SFAV) has shown cross-reactivity with numerous other scorpaeniform venoms (Church and Hodgson, 2002; Atkinson et al., 2006; Saggiomo et al., 2021). As a result, stonefish antivenom could be utilized to treat some cases (Atkinson et al., 2006). Although little is known about the mechanisms by which piscine venoms work, developing highly effective treatments against them is possible (Isbister, 2001; Smith and Wheeler, 2006; Saggiomo et al., 2021). A better understanding of the venom components and their modes of action is needed to aid in the creation of more effective envenomation treatment protocols, perhaps saving the lives of fishermen who are susceptible to fish venom.
13 Conclusion and future perspectives
We can begin to cross-check lines within toxicology by investigating the evolutionary ecology and deepen our understanding of transcriptomic and proteomic studies of fish venom and their related toxins. On a physiological and genetic level, understanding how venom has evolved on an ecological scale can help us better comprehend the purpose and function of venom. Investigation of individual toxic molecules to complex morphological delivery systems and behavioral adaptations to aposematic and mimetic colors and patterns, venom systems are integrated phenotypes with many components with interrelated functions across different levels of the organization is strongly recommended. The study of such systems in big groups, such as fish, could help us learn more about venomous taxa and their evolution, genetic diversity as well as applications in our daily life.
Despite all of the advances in science and technology, many concerns related to venomous fish remain unsolved, including physio-genomic studies, pharmacological effects, and the specific action mechanism of some components, as well as a large number of compounds yet to be discovered in the venom of various fish species. The study and exploration of the full potential contained in fish venoms can contribute to a better understanding of complex physiological processes like pain caused by envenomation, as well as the discovery of new drugs and the development of more effective ways to treat the injuries caused by these animals. Venomous catfishes are a diverse collection of species that may outnumber all other venomous vertebrates combined, with a great degree of variation in venom delivery mechanism morphology.
Due to their human impacts and potential untapped benefits, the evolutionary study of venom glands and their products presents an important understudied area. It also presents an important anti-predatory trait in a globally ubiquitous group of organisms that often makes up a significant portion of a given region’s aquatic vertebrate biodiversity. The anticipated venoms have evolved for the same goal, and the pharmacological properties of piscine venoms are strikingly comparable. In the majority of cases, a single venom toxin is responsible for the reported cardiovascular, neuromuscular, and cytolytic effects. Indeed, it appears likely that all of a toxin’s biological activities can be traced back to its cytolytic activity.
Author contributions
GL, HC, CBN, and UM designed the content and structure of the whole paper. CBN and DA designed the figures. CBN drafted the review, CBN UM, YW, DA GZ, CH, and RM wrote the final review. HH, GL, and HC proofread and revised the paper and Funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This article was supported by grants from this article was supported by the National Natural Science Foundation of China (Nos. 32273131), the Key Research and Development Program of Guangdong (2021B202020002), the South China Aquaculture Breeding Funding(2022RHDKFKT01), the talent team tender grant of Zhanjiang marine equipment and biology (2021E05035), the science and technology plan of Zhanjiang City (2022A01214), the science and technology plan of Yangjiang City (2022011) and the Major Science and Technology plan of Hainan Province (ZDKJ2021017).
Conflict of interest
Author YW was employed by Guangdong Havwii Agriculture Group Co., LTD. Authors GZ and CH were employed by Hainan Chenhai Aquatic Co., LTD.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1085669/full#supplementary-material
References
Adler E. M. (2015). Generally physiological of capturing fish, mobilizing enzymes, and a surprising source for serotonin. J. Gen. Physiol. 145 (3), 165–166. doi: 10.1085/jgp.201511372
Atkinson P. R. T., Boyle A., Hartin D., McAuley D. (2006). Is hot water immersion an effective treatment for marine envenomation? Emergency Med. J. 23 (7), 503–508. doi: 10.1136/emj.2005.028456
Baumann K., Casewell N. R., Ali S. A., Jackson T. N. W., Vetter I., Dobson J. S., et al. (2014). A ray of venom: Combined proteomic and transcriptomic investigation of fish venom composition using barb tissue from the blue-spotted stingray (Neotrygon kuhlii). J. Proteomics 109, 188–198. doi: 10.1016/j.jprot.2014.06.004
Bordon K. C. F., Cologna C. T., Fornari-Baldo E. C., Pinheiro-Júnior E. L., Cerni F. A., Amorim F. G., et al. (2020). From animal poisons and venoms to medicines: Achievements, challenges and perspectives in drug discovery. Front. Pharmacol. 11. doi: 10.3389/fphar.2020.01132
Cameron A. M., Endean R. (1970). Venom glands in scatophagid fish. Toxicon 8 (2), 171–174. doi: 10.1016/0041-0101(70)90156-X
Cameron A. M., Endean R. (1972). The venom glands of teleost fishes. Toxicon 10 (3), 301–303. doi: 10.1016/0041-0101(72)90018-9
Cameron A. M., Endean R. (1973). Epidermal secretions and the evolution of venom glands in fishes. Toxicon 11 (5), 401–410. doi: 10.1016/0041-0101(73)90115-3
Campos F. V., Menezes T. N., Malacarne P. F., Costa F. L. S., Naumann G. B., Gomes H. L., et al. (2016). A review on the Scorpaena plumieri fish venom and its bioactive compounds. J. Venomous Anim. Toxins Including Trop. Dis. 22 (1), 1–9. doi: 10.1186/s40409-016-0090-7
Campos F. V., Fiorotti H. B., Coitinho J. B., Figueiredo S. G. (2021). Fish cytolysins in all their complexity. Toxins 13 (12). doi: 10.3390/toxins13120877
Chhatwal I., Dreyer F. (1992). Isolation and characterization of dracotoxin from the venom of the greater weeverfish Trachinus draco. Toxicon 30 (1), 87–93. doi: 10.1016/0041-0101(92)90504-X
Chun L. H. (2004). Functional & structural studies of stonustoxin ( sntx ), a lethal factor from stonefish ( synanceja horrida ) venom (ScholarBank@NUS Repository). Available at: https://scholarbank.nus.edu.sg/handle/10635/14641.
Church J. E., Hodgson W. C. (2000). Dose-dependent cardiovascular and neuromuscular effects of stonefish (Synanceja trachynis) venom. Toxicon 38 (3), 391–407. doi: 10.1016/S0041-0101(99)00169-5
Church J. E., Hodgson W. C. (2002). The pharmacological activity of fish venoms. Toxicon 40 (8), 1083–1093. doi: 10.1016/S0041-0101(02)00126-5
Costa F. L. S., De Lima M. E., Figueiredo S. G., Ferreira R. S., Prates N. S., Sakamoto T., et al. (2018). Sequence analysis of the cDNA encoding for SpCTx: A lethal factor from scorpionfish venom (Scorpaena plumieri). J. Venomous Anim. Toxins Including Trop. Dis. 24 (1), 1–15. doi: 10.1186/s40409-018-0158-7
da Silva N. J., Ferreira K. R. C., Pinto R. N. L., Aird S. D. (2015). A severe accident caused by an ocellate river stingray (Potamotrygon motoro) in central Brazil: How well do we really understand stingray venom chemistry, envenomation, and therapeutics? Toxins 7 (6), 2272–2288. doi: 10.3390/toxins7062272
Fox J., Serrano S. (2007). Approaching the golden age of natural product pharmaceuticals from venom libraries: An overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications. Curr. Pharm. Design 13 (28), 2927–2934. doi: 10.2174/138161207782023739
Ghadessy F. J., Chen D., Kini R. M., Chung M. C. M., Jeyaseelan K., Khoo H. E., et al. (1996). Stonustoxin is a novel lethal factor from stonefish (Synancea horrida) venom. cDNA cloning and characterization. J. Biol. Chem. 271 (41), 25575–25581. doi: 10.1074/jbc.271.41.25575
Gokulalakshmi E., Vijayaraj R., Sri Kumaran N. (2018). A mini review on bioprospecting of fish venom. Int. J. Creative Res. Thoughts 6 (2), 1177–1181.
Gomes H. L., Andrich F., Mauad H., Sampaio K. N., De Lima M. E., Figueiredo S. G., et al. (2010). Cardiovascular effects of scorpionfish (Scorpaena plumieri) venom. Toxicon 55 (2–3), 580–589. doi: 10.1016/j.toxicon.2009.10.012
Gorman L. M., Judge S. J., Fezai M., Jemaà M., Harris J. B., Caldwell G. S. (2020). The venoms of the lesser (Echiichthys vipera) and greater (Trachinus draco) weever fish– a review. Toxicon X, 6. doi: 10.1016/j.toxcx.2020.100025
Haddad V., Lima C. (2014). Marine and freshwater toxins. Mar. Freshw. Toxins 11 (1), 5–9. doi: 10.1007/978-94-007-6650-1
Haddad V., Martins I. A. (2006). Frequency and gravity of human envenomations caused by marine catfish (suborder siluroidei): a clinical and epidemiological study. Toxicon 47 (8), 838–843. doi: 10.1016/j.toxicon.2006.02.005
Harris R. J., Jenner R. A. (2019). Evolutionary ecology of fish venom: Adaptations and consequences of evolving a venom system. Toxins 11 (2), 1–21. doi: 10.3390/toxins11020060
Isbister G. K. (2001). Venomous fish stings in tropical northern Australia. Am. J. Emergency Med. 19 (7), 561–565. doi: 10.1053/ajem.2001.28325
Júnior N. G. D. O., Fernandes G. D. R., Cardoso M. H., Costa F. F., Cândido E. D. S., Neto D. G., et al. (2016). Venom gland transcriptome analyses of two freshwater stingrays (Myliobatiformes: Potamotrygonidae) from brazil. Sci. Rep. 6, 1–14. doi: 10.1038/srep21935
Kiriake A., Shiomi K. (2011). Some properties and cDNA cloning of proteinaceous toxins from two species of lionfish (Pterois antennata and pterois volitans). Toxicon 58 (6–7), 494–501. doi: 10.1016/j.toxicon.2011.08.010
Koh D. C. I., Armugam A., Jeyaseelan K. (2006). Snake venom components and their applications in biomedicine. Cell. Mol. Life Sci. 63 (24), 3030–3041. doi: 10.1007/s00018-006-6315-0
Liu S. Y. V., Frédérich B., Lavoué S., Chang J., Erdmann M. V., Mahardika G. N., et al. (2018). Buccal venom gland associates with increased of diversification rate in the fang blenny fish meiacanthus (Blenniidae; teleostei). Mol. Phylogenet. Evol. 125, 138–146. doi: 10.1016/j.ympev.2018.03.027
Lopes-Ferreira M., Sosa-Rosales I., Bruni F. M., Ramos A. D., Vieira Portaro F. C., Conceição K., et al. (2016). Analysis of the intersexual variation in Thalassophryne maculosa fish venoms. Toxicon 115, 70–80. doi: 10.1016/j.toxicon.2016.02.022
Lopes-Ferreira M., Sosa-Rosales I., Silva Junior P. I., Conceicao K., Maleski A. L. A., Balan-Lima L., et al. (2022). Molecular characterization and functional analysis of the nattectin-like toxin from the venomous fish thalassophryne maculosa. Toxins 14 (2). doi: 10.3390/toxins14010002
Magalhães G. S., Junqueira-de-Azevedo I. L. M., Lopes-Ferreira M., Lorenzini D. M., Ho P. L., Moura-da-Silva A. M. (2006). Transcriptome analysis of expressed sequence tags from the venom glands of the fish Thalassophryne nattereri. Biochimie 88 (6), 693–699. doi: 10.1016/j.biochi.2005.12.008
Nakagawa H., Nagasaka K., Sakai H., Edo K., Shinohara M., Ohura K. (2015). Isolation of a novel lectin from the dorsal spines of the devil stinger, inimicus japonicus. Int. Aquat. Res. 7 (2), 143–150. doi: 10.1007/s40071-015-0101-2
Novak V., Sket D., Cankar G., Lebez D. (1973). Partial purification of a toxin from tentacles of the sea anemone Anemonia sulcata. Toxicon 11 (5), 411–417. doi: 10.1016/0041-0101(73)90116-5
Pandey S., Upadhyay R. K. (2020). The fish venom toxins: natural source of pharmaceuticals and therapeutic agents “Pharmaceutical and therapeutic uses of fish venom toxins. Int. J. Pharm. Pharm. Sci. 12 (11), 1–14. doi: 10.22159/ijpps.2020v12i11.38215
Saggiomo S. L., Firth C., Wilson D. T., Seymour J., Miles J. J., Wong Y. (2021). The geographic distribution, venom components, pathology and treatments of stonefish (Synanceia spp.) venom. Mar. Drugs 19 (6), 1–23. doi: 10.3390/md19060302
Silva F., Huang Y., Yang V., Mu X., Shi Q., Antunes A. (2018). Transcriptomic characterization of the south american freshwater stingray potamotrygon motoro venom apparatus. Toxins 10 (12). doi: 10.3390/toxins10120544
Sivan G. (2009). Fish venom: Pharmacological features and biological significance. Fish Fisheries 10 (2), 159–172. doi: 10.1111/j.1467-2979.2008.00309.x
Sivan G., Venketasvaran K., Radhakrishnan C. K. (2010). Characterization of biological activity of Scatophagus argus venom. Toxicon 56 (6), 914–925. doi: 10.1016/j.toxicon.2010.06.014
Sivan G., Venketesvaran K., Radhakrishnan C. K. (2007). Biological and biochemical properties of Scatophagus argus venom. Toxicon 50 (4), 563–571. doi: 10.1016/j.toxicon.2007.05.002
Smith W. L., Stern J. H., Girard M. G., Davis M. P. (2016). Evolution of venomous cartilaginous and ray-finned fishes. Integr. Comp. Biol. 56 (5), 950–961. doi: 10.1093/icb/icw070
Smith W. L., Wheeler W. C. (2006). Venom evolution widespread in fishes: A phylogenetic road map for the bioprospecting of piscine venoms. J. Heredity 97 (3), 206–217. doi: 10.1093/jhered/esj034
Sosa-Rosales J. I., Piran-Soares A. A., Farsky S. H. P., Takehara H. A., Lima C., Lopes-Ferreira M. (2005). Important biological activities induced by Thalassophryne maculosa fish venom. Toxicon 45 (2), 155–161. doi: 10.1016/j.toxicon.2004.10.003
Tamura S., Yamakawa M., Shiomi K. (2011). Purification, characterization and cDNA cloning of two natterin-like toxins from the skin secretion of oriental catfish Plotosus lineatus. Toxicon 58 (5), 430–438. doi: 10.1016/j.toxicon.2011.08.001
Terlau H., Olivera B. M. (2004). Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 84 (1), 41–68. doi: 10.1152/physrev.00020.2003
Ueda A., Suzuki M., Honma T., Nagai H., Nagashima Y., Shiomi K. (2006). Purification, properties and cDNA cloning of neoverrucotoxin (neoVTX), a hemolytic lethal factor from the stonefish Synanceia verrucosa venom. Biochim. Biophys. Acta - Gen. Subj. 1760 (11), 1713–1722. doi: 10.1016/j.bbagen.2006.08.017
Wright J. J. (2009). Diversity, phylogenetic distribution, and origins of venomous catfishes. BMC Evolutionary Biol. 9 (1), 1–12. doi: 10.1186/1471-2148-9-282
Wright J. J. (2012). Adaptive significance of venom glands in the tadpole madtom Noturus gyrinus (Siluriformes: Ictaluridae). J. Exp. Biol. 215 (11), 1816–1823. doi: 10.1242/jeb.068361
Wright J. J. (2015). Evolutionary history of venom glands in the siluriformes. Evol. Venomous Anim. Their Toxins 14, 1–19. doi: 10.1007/978-94-007-6727-0_9-1
Xie B., Yu H., Kerkkamp H., Wang M., Richardson M., Shi Q. (2019). Comparative transcriptome analyses of venom glands from three scorpionfishes. Genomics 111 (3), 231–241. doi: 10.1016/j.ygeno.2018.11.012
Xie B., Huang Y., Baumann K., Fry B. G., Shi Q. (2017). From marine venoms to drugs: Efficiently supported by a combination of transcriptomics and proteomics. Mar. Drugs 15 (4), 1–10. doi: 10.3390/md15040103
Xie B., Li X., Lin Z., Ruan Z., Wang M., Liu J., et al. (2016). Prediction of toxin genes from chinese yellow catfish based on transcriptomic and proteomic sequencing. Int. J. Mol. Sci. 17 (4). doi: 10.3390/ijms17040556
Zancolli G., Reijnders M., Waterhouse R. M., Robinson-Rechavi M. (2022). Convergent evolution of venom gland transcriptomes across metazoa. Proc. Natl. Acad. Sci. United States America 119 (1). doi: 10.1073/pnas.2111392119
Ziegman R., Alewood P. (2015). Bioactive components in fish venoms. Toxins 7 (5), 1497–1531. doi: 10.3390/toxins7051497
Keywords: fish venom, venom glands, evolution, stonustoxin, envenomation, pharmaceutical activity
Citation: Brighton Ndandala C, Mustapha UF, Wang Y, Assan D, Zhao G, Huang C, Mkuye R, Huang H, Li G and Chen H (2023) The perspective of fish venom: An overview of the physiology, evolution, molecular and genetics. Front. Mar. Sci. 10:1085669. doi: 10.3389/fmars.2023.1085669
Received: 31 October 2022; Accepted: 03 February 2023;
Published: 17 February 2023.
Edited by:
Alaa El-Din Hamid Sayed, Assiut University, EgyptReviewed by:
Khaled Mohammed-Geba, University of Maryland, College Park, United StatesYong Zhang, Sun Yat-sen University, China
Copyright © 2023 Brighton Ndandala, Mustapha, Wang, Assan, Zhao, Huang, Mkuye, Huang, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huapu Chen, chenhp@gdou.edu.cn; Guangli Li, guangligdou@163.com
 Charles Brighton Ndandala1,2,3,4
Charles Brighton Ndandala1,2,3,4  Umar Farouk Mustapha
Umar Farouk Mustapha Daniel Assan
Daniel Assan Hai Huang
Hai Huang Guangli Li
Guangli Li Huapu Chen
Huapu Chen