Glycobiology of Eosinophilic Inflammation: Contributions of Siglecs, Glycans, and Other Glycan-Binding Proteins
- Division of Allergy and Immunology, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, United States
The historical focus on protein–protein interactions in biological systems, at the expense of attention given to interactions between other classes of molecules, has overlooked important and clinically relevant processes and points of potential clinical intervention. For example, the significance of protein–carbohydrate interactions, especially in the regulation of immune responses, has recently received greater recognition and appreciation. This review discusses several ways by which cell-surface lectin–glycan interactions can modulate eosinophil function, particularly at the levels of eosinophil recruitment and survival, and how such interactions can be exploited therapeutically. A primary focus is on discoveries concerning Siglec-8, a glycan-binding protein selectively expressed on human eosinophils, and its closest functional paralog in the mouse, Siglec-F. Recent advances in the synthesis of polymeric ligands, the identification of physiological ligands for Siglec-8 and Siglec-F in the airway, and the determination of the basis of glycan ligand discrimination of Siglec-8 are discussed. Important similarities and differences between these siglecs are outlined. Eosinophil expression of additional glycan-binding proteins or their glycan ligands, including interactions involving members of the selectin, galectin, and siglec families, is summarized. The roles of these molecules in eosinophil recruitment, survival, and inflammation are described. Finally, the modulation of these interactions and potential therapeutic exploitation of glycan-binding proteins and their ligands to ameliorate eosinophil-associated diseases are considered.
Eosinophils are innate immune cells that contribute to host defense responses against parasitic infections and appear to have been retained in evolution throughout vertebrate species (1–3). Yet, there is a sizable body of evidence that eosinophils, under other circumstances, can be pro-inflammatory and are thus thought to be major effector cells in allergic and other type 2 immune responses. These include common conditions such as asthma, often manifesting with comorbid upper airways diseases of chronic rhinosinusitis with or without nasal polyposis, disorders that similarly manifest prominent type 2 inflammatory signatures and features including elevated Th2 and ILC2 cells with IL-4, IL-5, IL-13, eotaxins, and other downstream mediators (4–7). Another common eosinophil-associated disease is atopic dermatitis, where eosinophils contribute to some but perhaps not all stages of the disease (8). Less common disorders where eosinophils are felt to play a major role include eosinophilic granulomatosis with polyangiitis (formerly known as Churg–Strauss syndrome); eosinophilic gastrointestinal disorders (EGID), namely eosinophilic esophagitis, gastritis, and colitis, existing alone or in combination (9–13); and other systemic and organ-specific hypereosinophilic syndromes and disorders (14).
Current treatments for these eosinophil-associated conditions include glucocorticosteroids, mediator receptor antagonists, and other anti-inflammatory drugs that reduce eosinophil numbers and activity, but they are neither fully effective nor curative or disease modifying, hence the need for additional therapies (15, 16). Advanced efforts to indirectly target eosinophils [e.g., with agents that antagonize TSLP (17) and IL-4 and IL-13 biology with FDA-approved dupilumab (18–21)] or more specifically target eosinophils (e.g., with the FDA-approved anti-IL-5 biologics mepolizumab and reslizumab, and perhaps someday with the anti-IL-5 receptor antibody benralizumab and the oral agent dexpramipexole) offer hope for improved management of these disorders (22–24). Despite these promising agents, and advancements in our understanding of the pathophysiology of each of these disorders, many patients remain refractory to treatment, or in the case of EGID, there are as yet no FDA-approved drugs. These and other unmet needs led to collaborative efforts to find additional eosinophil-selective targets, and in recent years, these have included the only known pure eosinophil-specific surface target EMR1 (25, 26), and Siglec-8, expressed not only on eosinophils but also on mast cells and weakly on basophils (27–29). The focus of this review is not only on the latter molecule but also includes discussions of other lectin–glycan interactions known to influence eosinophil responses.
Siglec-8
Receptor Discovery, Characteristics, and Expression Patterns
Siglec-8 [also originally named sialoadhesin family 2 (SAF-2)] is an I-type single pass transmembrane protein that was discovered from a human eosinophil cDNA library generated from a patient with hypereosinophilic syndrome and first described in the year 2000. Eosinophil mRNA was examined by random sequencing of expressed sequence tags, which led to the identification of a protein 431 amino acid residues (aa) in length that was highly homologous to others in the sialoadhesin/siglec family. The highest levels of homology were found with Siglec-7 (68%), Siglec-3 (49%), and Siglec-5 (42%). The extracellular region of Siglec-8 contains 358 aa with a hydrophobic signal peptide and three Ig-like domains, with the N-terminal Ig domain possessing an arginine at position 125 that is putatively necessary for sialic acid binding (27, 28). When originally described, the cytoplasmic domain was found to be unusually short, and no known signaling motifs were observed. Subsequent investigations by Foussias et al. led to the observation that Siglec-8 exists in two isoforms (the 431-aa originally identified Siglec-8 “short form” and a 499-aa Siglec-8 “long form”), both containing identical extracellular and transmembrane regions. However, like most other CD33-related siglecs, the long form of Siglec-8 contained two characteristic tyrosine-based motifs: a membrane-proximal immunoreceptor tyrosine-based inhibitory motif (ITIM) resembling a classical ITIM (ILVxYxxLV) and a membrane-distal immunoreceptor tyrosine-based switch motif (ITSM) resembling a motif (TxYxxIV) found in the intracellular region of signaling lymphocyte activation molecule (SLAM) (30). The Siglec-8 long form is now just called Siglec-8 because it was found to be the primary form of the receptor, with a molecular weight of ~54 kDa (30), although eosinophils usually but not always express the short form, the function of which remains unknown (31).
While quantitative PCR analysis for the Siglec-8 mRNA not surprisingly detected signals in hematopoietic organs, expression in lung was unexpected. Using monoclonal antibodies recognizing the extracellular region, it was soon discovered that Siglec-8 was not just an eosinophil marker. It was selectively expressed on the surface of eosinophils, mast cells, and at low levels on basophils, but not on any other cells, making it the first receptor to be exclusively expressed on these three allergic effector cell types (28). Using human CD34+ cell-derived culture systems, it was determined that Siglec-8 is a terminal differentiation marker in both eosinophils and mast cells, with maximum protein expression in each cell type occurring at 21 and 30 days of culture, respectively. In contrast, none of the eosinophilic cell lines express Siglec-8 and only modest expression was observed on the mast cell line HMC-1.2, furthering the concept that Siglec-8 is a terminal differentiation marker on these cell types (32, 33).
The SIGLEC8 gene, like other CD33-related siglecs, is located in the centromeric region of chromosome 19q13 (27, 30). However, little is known about regulation of SIGLEC8 expression at the transcriptional level. In a recent report, Hwang et al. identified Olig2, a basic helix-loop-helix transcription factor, as a potential regulator of SIGLEC8 gene expression. They showed that OLIG2 and SIGLEC8 are coexpressed late in eosinophil differentiation and that both proteins are expressed in terminally differentiated eosinophils. Furthermore, they showed that Olig2 siRNA reduced SIGLEC8 mRNA and Siglec-8 surface protein levels, suggesting that Olig2 is a transcriptional regulator of the SIGLEC8 gene (34). However, all of the currently available human eosinophilic cell lines express Olig2 protein but fail to express Siglec-8, as noted earlier. In addition, Olig2 is not expressed in cord blood-derived mast cells that express Siglec-8. Thus, it appears that SIGLEC8 gene expression is only partially regulated by Olig2 and further work is needed to determine the exact combination of transcription factors responsible for Siglec-8 expression (33, 34).
Ligands for Siglec-8
All siglecs contain an amino-terminal V-set Ig lectin domain that binds sialic acid, but each siglec has a characteristic specificity profile for binding only certain conformations of sialic acid. Most siglecs recognize α2-3- and α2-6-linked sialic acids, although some can also recognize α2-8-linked sialic acids (35, 36). Initial experiments to characterize Siglec-8 ligand-binding preferences demonstrated that Siglec-8 preferentially binds α2-3-sialic acids linked to Galβ1-4GlcNAc (27). Using a glycan array generated by the Consortium for Functional Glycomics, 172 glycan structures were screened, and it was discovered that Siglec-8 specifically bound 6′ sulfated sialyl LewisX (6′-sulfo-sLex or NeuAcα2-3Galβ1-4(Fucα1-3)(6-O-Sulfo)GlcNAcβ1). Siglec-8 did not bind sialyl LewisX, a common ligand for L-, P-, and E-selectins, demonstrating that the 6′-position sulfate on the galactose was absolutely necessary for Siglec-8 lectin-glycan binding (37). A subsequent re-screen of an expanded array containing over 600 structures revealed that the fucose was dispensible (38) (Figure 1). Experiments using heparinized whole blood showed that a polyacrylamide polymer decorated with 6′-sulfo-sLex bound only eosinophils in a Siglec-8-dependent manner, further demonstrating that this glycan is a specific ligand for Siglec-8 (39).
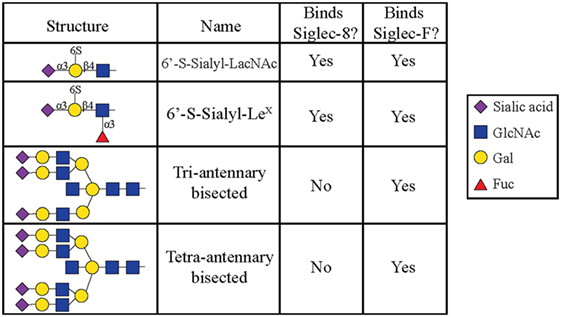
Figure 1. Glycans recognized by Siglec-8-Fc and Siglec-F-Fc, as determined by using glycan microarray analysis, are shown. Reproduced and modified from Ref. (38) with permission.
The structural basis of how Siglec-8 interacts with its glycan ligand had been unexplored until a recent report by Propster et al., where they provide a detailed description of how Siglec-8 selectively recognizes its ligand, 6′-sulfo-sLex. First, using NMR spectroscopy, they determined the 3D structure of the lectin domain of Siglec-8. The structure is a V-set Ig-like β-sandwich of two antiparallel β-sheets formed by β-strands ABED and C′CFG, with the conserved arginine, responsible for providing a salt bridge interaction with sialic acid, located on β-strand F. Ligand specificity is mediated by two motifs, where the primary motif recognizes the terminal Neu5Ac, similar to other siglecs, and the secondary motif recognizes the subterminal Gal6S, which was found to be unique among siglecs. Although amino acid mutations failed to affect the overall structure of Siglec-8, a mutation in the conserved arginine eliminated Neu5Ac binding and completely abrogated Siglec-8–ligand interactions. In accordance with previous work done in our lab, this group also demonstrates that the sulfate modification was absolutely necessary for enhanced ligand affinity and revealed the key determinants for glycan specificity (40) (Figure 2).
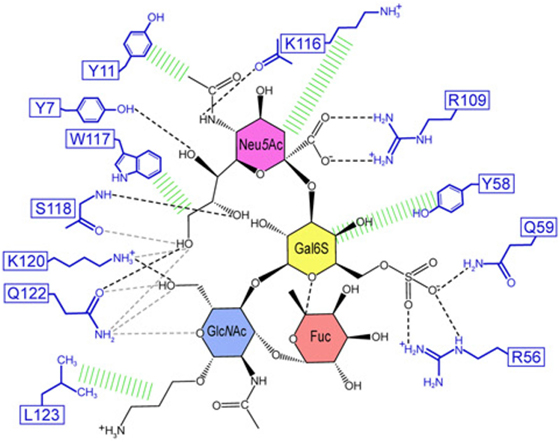
Figure 2. Structural basis for 6′ sulfated sLex recognition by human Siglec-8 illustrated by a representative structure (lowest energy) of the NMR ensemble. Schematic illustration of the Siglec-8–6′ sulfated sLex interaction network. Black dashed lines indicate hydrogen bonds in the depicted structure; gray dashed lines indicate hydrogen bonds abundantly observed in other structures of the ensemble. Hydrophobic contacts are shown in green. Reproduced from Ref. (40) with permission.
Siglec-8 Function on Human Eosinophils
Initial clues regarding how Siglec-8 might function came from an examination of its structural motifs. The cytoplasmic domain of Siglec-8 contains one ITIM and one ITSM, thought to be responsible for initiating downstream receptor function. Using peripheral blood human eosinophils, it was shown that multimeric engagement of Siglec-8 with a mouse monoclonal antibody (mAb) causes Siglec-8-dependent eosinophil apoptosis. However, this required the presence of a secondary anti-mouse cross-linking antibody; without secondary antibody, no cell death was seen (41). Further studies to delineate the mechanisms through which Siglec-8 induces eosinophil apoptosis revealed that Siglec-8 cross-linking promoted reactive oxygen species (ROS) production, loss of mitochondrial membrane potential and caspase cleavage (41, 42). Additionally, Siglec-8-dependent eosinophil apoptosis was paradoxically amplified under conditions of eosinophil priming with IL-5, GM-CSF, or IL-33, eliminating the need for secondary cross-linking antibody and changing the apoptotic mechanism to one dependent on ROS rather than caspase activity (41–44). Furthermore, incubation of IL-5-primed eosinophils with a synthetic polyacrylamide polymer decorated with 6′-sulfo-sLex induced eosinophil apoptosis (39), suggesting that Siglec-8 functions through different mechanisms in the presence or absence of cytokine priming.
In addition to studies using a mAb and a synthetic ligand, von Gunten et al. discovered that exposure of IL-5-primed eosinophils to intravenous immunoglobulin (IVIG), often used at high doses for the treatment of autoimmune and allergic diseases, resulted in eosinophil cytotoxicity. Further experiments revealed that IVIG contained autoantibodies against Siglec-8 that were responsible for this cytokine-dependent apoptotic effect of IVIG and that this effect was ROS-dependent (45), similar to what was observed when using mAbs to Siglec-8.
Although the intracellular signaling pathways for most siglecs are not well characterized, several studies have shown that engagement of CD33-related siglecs leads to downstream activation of Src family kinases (SFKs) that provide docking sites for Src-homology region 2 domain-containing phosphatases such as SHP-1 and SHP-2 that then propagate downstream functions (46–49). Indeed, ongoing work to further explore Siglec-8 signaling on eosinophils revealed that Siglec-8 engagement on IL-5-primed eosinophils leads to phosphorylation of SFKs, and use of SFK pharmacological antagonists inhibited Siglec-8-mediated eosinophil ROS production and apoptosis, although the SFKs involved in Siglec-8 function have yet to be determined (50). Furthermore, preliminary data show that Siglec-8 associates with SHP-2 and that pharmacological inhibition of protein tyrosine phosphatases inhibits Siglec-8-mediated eosinophil apoptosis (51). Together, these studies support the notion that Siglec-8 functions similar to other CD33-related siglecs.
The presence of an ITIM suggests that Siglec-8 should be involved in negative cell signaling; however, some of the latest observations suggest that Siglec-8 can, under certain circumstances, function as an activating receptor, such as after IL-5 priming. Initial evidence supporting this hypothesis showed that Siglec-8 cross-linking leads to enhanced phosphorylation of extracellular signal-regulated kinase (ERK) 1/2 and activation of ERK1/2 was necessary for Siglec-8-mediated eosinophil apoptosis (50).
Siglec-F
In view of the usefulness of mouse models for functional manipulations, it was imperative to identify a suitable mouse homolog of Siglec-8. In mice, there is no Siglec-8 ortholog, but Siglec-F has been found to be the closest functional paralog. Siglec-F is a 569-aa, CD33-related siglec that contains four Ig-like domains (Siglec-8 contains three) and, like Siglec-8, it contains both ITIM and ITIM-like motifs in its cytoplasmic tail. Using sequence homology comparisons, it was initially proposed that Siglec-F was the likely ortholog of human Siglec-5, but the homology was limited to the extracellular domains of both receptors (47). Furthermore, initial studies revealed that Siglec-F was predominantly expressed in bone marrow cells of the myelomonocytic lineage, and it was not expressed on mature neutrophils and monocytes (47), which have been shown to express Siglec-5 (52), further suggesting that Siglec-F may not be the true ortholog of Siglec-5.
Later efforts to fully characterize the expression pattern of Siglec-F and determine its functional counterpart in humans revealed that Siglec-F shared 38% similarity with human Siglec-8 (53). Using monoclonal antibodies to Siglec-F, it was found that Siglec-F, like Siglec-8, was predominantly expressed on the surface of mature eosinophils and on bone marrow eosinophils (54). However, Siglec-F is not expressed on mouse mast cells and surprisingly is instead expressed on mouse alveolar macrophages and subpopulations of intestinal epithelial cells (55–57) (Figure 3). Although they are structurally different and are expressed on different cell types, Siglec-F, like Siglec-8, has a binding preference for α2-3-linked sialic acids and recognizes 6′-sulfo-sLex (58). While subsequent studies have reproduced these findings, access to additional glycan structures for screening has allowed the identification of several multi-antennary structures that are recognized by Siglec-F but not Siglec-8 (38) (Figure 1). Indeed, this may explain why mouse lung ligands are recognized by Siglec-F but not by Siglec-8 (see below) (59). Based on these reports, it was concluded that Siglec-F and Siglec-8 are functionally convergent paralogs rather than orthologs.
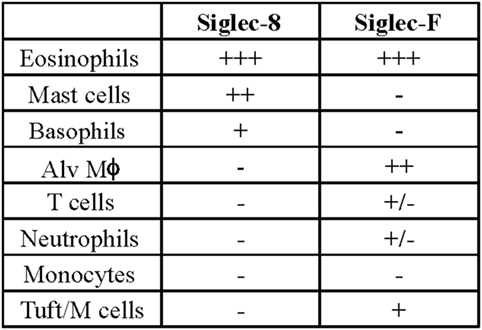
Figure 3. Comparison of cellular surface expression patterns for Siglec-8 and Siglec-F. Alv Mϕ, alveolar macrophage.
Subsequent studies to examine the biological roles of Siglec-F in vivo revealed that its expression is upregulated following allergen challenge in a mouse lung allergy model and the congenital deficiency via genetic deletion of the Siglec-F gene led to enhanced eosinophil numbers in the bone marrow, peripheral blood, and lungs during allergic inflammation but not at baseline. Furthermore, Siglec-F-null mice had diminished eosinophil death, suggesting a role for Siglec-F in mediating eosinophil apoptosis (60). Indeed, administration of anti-Siglec-F antibody reduced peripheral blood and tissue eosinophil numbers in wild-type mice, IL-5 transgenic mice, and in mouse models of hypereosinophilic syndrome and eosinophilic esophagitis, which was attributed to induction of eosinophil apoptosis. Additionally, the effect of the anti-Siglec-F antibody was specific to eosinophils and had no effect on other cells, not even Siglec-F-expressing alveolar macrophages (61–64). Despite our advances in understanding the role of Siglec-F in eosinophil survival in vivo and in vitro, little is known about the signaling mechanism of this receptor. A report by Mao et al. showed that Siglec-F engagement on mouse eosinophils led to caspase cleavage; however, unlike Siglec-8, there was no detectable ROS production, and Siglec-F function did not involve the activation of SFKs or SHP-1 (65). Therefore, further studies are needed to fully characterize the signaling pathways for Siglec-F.
Tissue Ligands for Siglec-F and Siglec-8
Although both Siglec-F and Siglec-8 preferentially recognize the glycan 6′-sulfo-sLex, the identity of their natural ligands is still under investigation. Initial studies to identify endogenous tissue ligands in mice using Siglec-F-Fc chimeras and immunohistochemistry showed that Siglec-F ligands were constitutively expressed on airway epithelial cells and their expression was dependent on ST3Gal-III, a sialyltransferase that can add α2,3 terminal sialic acids to glycans (66). Expression of these ligands was increased upon induction of allergic airway inflammation (67, 68). Glycoproteomic analysis of material derived from mouse tracheal epithelial cells revealed that Siglec-F-Fc bound to glycans displayed on Muc5b and Muc4, but not Muc5ac. Mouse lungs deficient in Muc5b had reduced Siglec-F-Fc binding, and mice conditionally deficient in Muc5b showed enhanced eosinophilic inflammation in response to airway instillation of IL-13, further validating that Muc5b carries glycan ligands for Siglec-F and suggesting that only subsets of airway mucins display the glycan structures necessary for Siglec-F binding (38). Although less is known about Siglec-8 tissue ligands, a recent study showed that Siglec-8 ligands were expressed selectively on serous cells, a subpopulation of submucosal gland cells in the inferior turbinate, and inflammation that occurs in chronic sinusitis led to increased expression of Siglec-8 tissue ligands in the upper airways (69). Additional studies show that mouse airways do not express Siglec-8 ligands and Siglec-8-Fc binding in human tracheal sections is restricted to serous cells in submucosal glands and cartilage (59) (Figure 4). The exact identity of these ligands is still under investigation, but given the fact that the galactose 6-O-sulfotransferase CHST-1 is dispensable for generating Siglec-F ligands in the mouse, it appears that the 6′ sulfation needed for Siglec-8 binding is not required for Siglec-F binding (70).
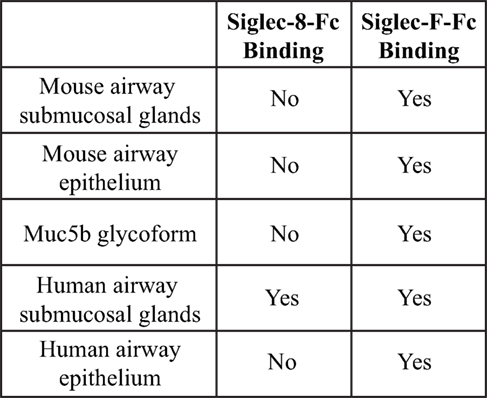
Figure 4. Comparison of binding of Siglec-F and Siglec-8 to mice and human tissue-based sialoside ligands. Based on data from Ref. (59).
Endocytosis of Siglec-F and Siglec-8
Beyond the physiological role of Siglec-8 in inducing cell death of eosinophils, Siglec-8 represents a promising target through which to deliver therapeutic payloads into eosinophils and other Siglec-8-expressing cells. Several studies have shown that siglecs are endocytic receptors and, once engaged, can carry their ligand—and presumably any associated cargo—into the cell (71–74). This strategy has been employed in the development of cancer therapeutics by targeting preferentially upregulated receptors such as the receptors for transferrin or folate or through antibodies targeting slightly more selectively expressed antigens, such as CD33 (Siglec-3) in acute myelogenous leukemia (75–77). An antibody-targeting CD22 (Siglec-2) is also under investigation to treat diseases involving B cells, such as B-cell acute lymphoblastic leukemia or systemic lupus erythematosus (78, 79). Due to the restricted expression pattern of Siglec-8, targeting of eosinophils through Siglec-8 offers promise. However, the capability to exploit Siglec-8 in this manner depends on whether Siglec-8 is endocytosed and is present and accessible on the surface of eosinophils in various circumstances. Siglec-F endocytosis has been studied in mouse eosinophils. Siglec-F is internalized in response to antibody ligation via a clathrin- and lipid raft-independent pathway that relies on ARF6 but not dynamin-1 (72). New data indicate that Siglec-8 is indeed internalized in response to antibody or synthetic ligand engagement on peripheral blood eosinophils and that this pathway can be exploited to deliver a toxin (the ribosome-inhibiting protein saporin) to eosinophils to induce cell death under conditions in which Siglec-8 engagement alone would be insufficient (i.e., in the absence of IL-5 priming) (80). Despite some similarities, including the lysosomal localization of the internalized siglec, the pathway utilized by Siglec-F internalization appears to be distinct from that of Siglec-8. The pathway of internalization can have profound effects on receptor function, leading to distinct signaling mechanisms and downstream functions or alterations in receptor turnover. For example, endocytosis of SR-A via a lipid raft/caveolae-dependent pathway is required for macrophage apoptosis in a ligand-dependent manner, whereas clathrin-mediated SR-A endocytosis is expendable for this effect (81). While there is abundant evidence linking endocytosis to the organization of signaling events (82), it remains to be determined whether the endocytosis of Siglec-8 affects its function.
Siglec-8 may also achieve part of its function by internalizing other surface proteins. Upon antigen stimulation, the B cell receptor (BCR) engages clathrin in lipid raft domains and thus is internalized via a mixed pathway (83, 84). While the siglec CD22 is initially excluded from lipid rafts, it colocalizes with the BCR and promotes its internalization when unmasked (85, 86). This downregulation of the BCR is thought to be one mechanism underlying the inhibitory function of CD22. Of note, the IL-5 receptor, which is critically important to the activation and survival of eosinophils, has been found to be internalized via distinct clathrin- and lipid raft-dependent pathways and is targeted for proteolytic degradation through the lipid raft-mediated endocytic pathway (87). It is an intriguing possibility that the endocytosis and trafficking of Siglec-8 and the IL-5 receptor may be linked in a way that influences the function of each receptor.
Other Siglecs Found on Mouse and Human Eosinophils
While Siglec-8 has garnered much attention as a cell-surface marker of eosinophils, there are a number of other glycans and glycan-binding proteins present on eosinophils that regulate their survival, trafficking, and adhesion and that may be useful markers of eosinophilic inflammation. In both mice and humans, eosinophils express siglec family members other than Siglec-F and Siglec-8, respectively.
CD22, Siglec-E, Siglec-G, and Mouse Eosinophils
Interestingly, mouse eosinophils in the gastrointestinal tract were found to express the siglec CD22 on their cell surface, a siglec previously thought to be restricted to B cells (88). CD22 expression was highest on eosinophils in the jejunum, although it was also found on eosinophils in the stomach, duodenum, or ileum. CD22 was not found on eosinophils in the blood or other tissues. The function of CD22 on these eosinophils is not yet clear, although CD22 ablation led to an increase in eosinophils in the jejunum, an effect that did not appear to be due to increased eosinophil differentiation from hematopoietic precursors or augmented IL-5 or eotaxin-2 signaling. In studies of mice overexpressing IL-5, which gives rise to eosinophilia, it was found that these eosinophils expressed mRNA for Siglec-E (orthologous to Siglec-9 in humans) and Siglec-G (orthologous to Siglec-10 in humans) (53). However, the surface expression and function of these siglecs on mouse eosinophils have not been studied.
CD33, Siglec-7, Siglec-10, and Human Eosinophils
Immature human eosinophils express low levels of CD33 (Siglec-3) and downregulate this receptor upon maturation (89). Human eosinophils also express modest levels of Siglec-7 both in the peripheral blood and in nasal polyps (90, 91). In addition, Siglec-10, which was identified by four different groups through genomic analysis and screens of cDNA libraries (including one from asthmatic eosinophils), is expressed by human eosinophils (92–95). Siglec-10 possesses three tyrosine-containing cytoplasmic motifs—a membrane-proximal Grb2 binding motif, a central ITIM, and a membrane-distal ITSM or ITIM-like motif—and has been found to interact with SHP-1 and SHP-2 but not with SLAM-associated protein (48, 94). While Siglec-10 was detected on B cells using polyclonal antibody (93), a mAb detected Siglec-10 expression only on eosinophils, neutrophils, and monocytes and failed to detect expression on B cells (94), suggesting either that the polyclonal antibody was not specific to Siglec-10 (perhaps binding to another siglec family member present on B cells) or that a unique variant of Siglec-10 is expressed on these cell populations and not on B cells. Interactions between Siglec-10 and CD24 (heat stable antigen) (96), vascular adhesion protein-1 (97), and CD52 (98) have been demonstrated in vitro or on other cell types; however, the function of Siglec-10 in eosinophils has not been described. Indeed, little is known about the functions of any of these three siglecs on human eosinophils. Antibody ligation of Siglec-7 on eosinophils failed to induce apoptosis or prevent chemotaxis under conditions in which Siglec-8 ligation produced these effects (91), and the role of CD33 on eosinophils has similarly not been determined. Given the lack of functional data and the broader cell expression patterns for these siglecs, there has been less interest in exploiting these receptors to address eosinophilic inflammation.
Selectins and Selectin Ligands on Eosinophils
Protein–glycan interactions are exceptionally important in the processes of cell adhesion and trafficking. In the initial steps of leukocyte extravasation, the cell must tether to and roll along the endothelium, which requires the interaction between selectins and their glycan ligands. Eosinophils depend to differing degrees on P-, E-, and L-selectin interactions in this process. Relative to neutrophils, eosinophils bind less well to E-selectin and bind to a greater extent to P-selectin through cell-surface glycan ligands (99), presumably due to increased levels of P-selectin glycoprotein ligand (PSGL)-1 (100). This same study found that L-selectin on the surface of eosinophils was important in tethering of eosinophils to human umbilical vein endothelial cells (HUVECs) but only due to establishing inter-eosinophil interactions rather than binding to the endothelial cells (99). However, others have found that diminished L-selectin expression or the use of blocking antibodies to L-selectin reduce eosinophil rolling and adhesion on HUVECs or on rabbit mesenteric venule endothelial cells under conditions of flow (101, 102). As demonstrated by the study by Sriramarao et al., human selectins are capable of interacting with ligands expressed in other species. Indeed, the selectin-binding sites in the best characterized P-selectin ligand, PSGL-1, are evolutionarily well conserved (103). However, distinct patterns of expression render cross-species comparisons more difficult. For example, P-selectin expression in mice but not in humans is induced by TNF-α or LPS, and cytokine regulation of human and primate P- and E-selectins is more selective than in mice (104, 105). However, mouse strains in which selectins have been knocked out have permitted elegant studies of their importance in mouse eosinophil migration. Using these mouse strains, several studies have shown that P-selectin is critical in eosinophil recruitment to the lung and peritoneum (106–108).
P-selectin glycoprotein ligand-1 is the best characterized P-selectin ligand on eosinophils, and its expression has been shown to be increased in patients with allergic asthma relative to healthy controls. This increase results in enhanced binding to P-selectin and IL-4-treated HUVECs (109). PSGL-1 contributes to eosinophil, but not neutrophil, adhesion to IL-13-activated HUVECs under conditions of physiological flow (110). Paradoxically, PSGL-1 expression is reduced on activated eosinophils and is shed from leukocytes (111). Although there have been reports that PSGL-1 can act as a ligand for E-selectin as well (112, 113), no changes in E-selectin binding were observed with eosinophils from allergic asthma patients in this study. There appear to be other significant ligands for P-selectin on eosinophils, however. In patients with atopic dermatitis, eosinophils are capable of binding substantially more soluble P-selectin than eosinophils from healthy donors but do not display more PSGL-1 on their surface (114).
Although the selectins bind to related sialylated glycans, these glycans can be present on a variety of different proteins or lipids that may be cell type- or tissue-specific. Eosinophils, for example, display far less sialyl LewisX antigen, a selectin ligand carbohydrate structure, than neutrophils, but a greater proportion is in the form of sialyl dimeric LewisX and is sensitive to endo-β-galactosidase treatment (115). Interestingly, neither the display of sialyl LewisX on eosinophils nor adhesion to immobilized E-selectin is protease-sensitive, indicating that these carbohydrate ligands may not be present on cell-surface proteins (115). Consistent with this, glycosphingolipids extracted from leukocytes were found to interact with E-selectin, and their biosynthesis was required for E-selectin-dependent, but not P-selectin-dependent, neutrophil adhesion (116). P-selectin ligands present on eosinophils clearly differ from those of E-selectin in that binding to immobilized P-selectin is protease-sensitive and endo-β-galactosidase-resistant (117). Indeed, although the expression of sialyl LewisX on eosinophils is not changed by cellular activation with platelet-activating factor (PAF), P-selectin binding is reduced following PAF activation and L-selectin is shed following transendothelial migration (117, 118).
Due to their critical role in eosinophil trafficking to peripheral tissues, selectins represent a potentially useful therapeutic target for diseases of eosinophilic inflammation. In fact, selectin antagonists interfere with eosinophil (and neutrophil) adhesion (119), and a pan-selectin antagonist glycomimetic agent is in clinical trials to modulate selectin-based adhesion in acute sickle cell crisis (120). Although lack of cell specificity is a concern, a more selective P-selectin antagonist or one that interacts with a eosinophil-selective P-selectin ligand would likely be effective in preventing further eosinophilic inflammation in the tissues with fewer potential complications.
Galectin Family Members and Their Glycan Ligands on Eosinophils
The galectin family of proteins, previously known as S-type lectins, has a binding preference, generally, for β-galactosides, although there appear to be exceptions to this. Most of the members of the family are secreted but can cross-link cell-surface receptors due to the presence of more than one carbohydrate recognition domain (CRD) or through multimerization of a monomer containing one CRD (galectin-3).
The galectin family member most commonly associated with eosinophils is galectin-10, also known as Charcot–Leyden crystal (CLC) protein. Galectin-10 makes up about 10% of the total protein content of human eosinophils (121), and CLC deposition in tissues has long been considered a marker of eosinophilic (or basophilic) inflammation (122). The protein localizes to both the cytosol and a subset of core-less granules (123). The CLC protein was initially believed to function as a lysophospholipase within the eosinophil (121); however, this enzymatic activity has since been ascribed not the CLC protein but to another enzyme that can associate to a degree with it (124). Due to sequence identity, structural homology, and genomic structure, CLC protein became known also as galectin-10 (125–127). Unlike other members of the galectin family, however, galectin-10 appears not to bind β-galactosides to any substantial degree but instead appears to bind to mannose-containing carbohydrate moieties (128). The natural ligand or ligands of galectin-10 and the functional significance of its ability to bind to carbohydrates remain undetermined. Despite the lack of functional data, galectin-10 mRNA and protein levels remain useful biomarkers for eosinophilic airway inflammation, active eosinophilic esophagitis, aspirin-exacerbated respiratory disease, CRTH2 activation, and celiac disease (129–133).
On the eosinophil cell surface, galectin–ligand interactions have been found to be important in eosinophil recruitment, activation, and survival. The granulocyte-specific and heavily glycosylated protein CD66b (also known as CEACAM8) is expressed on eosinophils and is upregulated in response to cellular activation (134, 135). Sialylated glycans on CD66b interact with E-selectin, and this interaction has been shown to be important for neutrophil adherence to activated endothelium (136). Glycans on CD66b also interact with galectin-3, and engagement of CD66b using either soluble galectin-3 or antibody induced ROS production and degranulation (137). Cross-linking of CD66b also caused the eosinophils to become more adherent, perhaps through the clustering of the integrin subunit CD11b.
Several other galectin interactions may be important in eosinophil adhesion and chemoattraction, including those with galectins-1, -3, and -9 (138–140). However, the role of galectins in eosinophil recruitment is covered in greater detail in a review in this volume by Sriramarao et al. and will not be discussed further here. It should be noted, however, that galectins play other important roles in eosinophil biology and are biomarkers of disease activity. High concentrations of galectin-1, for example, can induce eosinophil cell death (140), and levels of galectin-3 before treatment in patients with severe asthma predict treatment responses to omalizumab (141).
Eosinophil Glycomics
While many of the cell-surface and intracellular eosinophil proteins have been identified and extensively described, the glycans that coat the various cell-associated proteins and likely play important roles in numerous biological pathways remain largely shrouded in mystery. As part of an effort to characterize these glycans, the glycome of human eosinophils has been analyzed in cell lysates and compared to those of basophils and mast cells to elucidate the identities of these glycans, their relative abundances, and key differences between these cell types (142). Although mast cells possess substantial amounts of terminally sialylated epitopes on their various glycoproteins, eosinophils and basophils have far more part-processed terminal N-acetylglucosamine (GlcNAc)-containing structures. For example, the most abundant N-glycan by far in both eosinophils and basophils is a bi-antennary structure with two terminal non-extended GlcNAc sugars, which is far less abundant in mast cells. While the functional relevance of these patterns is unclear, it is unlikely that these modifications to the cell surface are random. In addition, it is uncertain how cytokine priming and other signals that may be present under inflammatory conditions may affect glycan synthesis and processing. However, such changes may well affect processes such as adhesion, activation, cell–cell interaction, and even survival.
Conclusion
There remains a clinical need to effectively and selectively treat diseases of eosinophilic inflammation. Due to their roles in recruitment, adhesion, activation, and survival, glycan–glycan-binding protein interactions are beginning to garner attention as therapeutic prospects. Siglec-8 and its ligands offer a cell-selective pathway to induce cell death in primed eosinophils and deliver therapeutic payloads into the cell. Antagonists of selectin interactions may help limit eosinophilic inflammation, but significant hurdles remain for achieving a safe, cell-selective effect. Monomeric β-galactosides or glycomimetics may also be clinically useful in antagonizing eosinophil lectin interactions that are involved in cell adhesion and activation. While targets and biomarkers have been identified, further studies are necessary to elucidate the functions of glycan-binding proteins on eosinophils, such as those of other members of the siglec family; to identify their natural glycan ligands and how they are modulated; and to determine the functional significance of the glycans displayed on the eosinophil cell surface.
Author Contributions
JO completed the sections on Siglec-8 endocytosis, selectins, galectins, and other members of the siglec family. DC completed all other sections regarding Siglec-8 as well as Siglec-F. BB organized the effort and wrote the introduction. All authors contributed to revisions of the manuscript.
Conflict of Interest Statement
BB has current or recent consulting or scientific advisory board arrangements with or has received honoraria from, Sanofi-Aventis, TEVA, AstraZeneca and Allakos, and owns stock in Allakos and Glycomimetics. He receives publication-related royalty payments from Elsevier and UpToDate™ and is a co-inventor on existing Siglec-8-related patents and thus may be entitled to a share of royalties received by Johns Hopkins University on the potential sales of such products. BB is also a co-founder of Allakos, which makes him subject to certain restrictions under University policy. The terms of this arrangement are being managed by the Johns Hopkins University and Northwestern University in accordance with their conflict of interest policies. The authors have no additional competing financial interests.
Funding
This work was supported by the National Heart, Lung, and Blood Institute (P01HL107151 to BB) and the National Institute of Allergy and Infectious Diseases (AI072265 to BB; T32AI083216 to JO).
References
1. Lee JJ, Jacobsen EA, McGarry MP, Schleimer RP, Lee NA. Eosinophils in health and disease: the LIAR hypothesis. Clin Exp Allergy (2010) 40(4):563–75. doi:10.1111/j.1365-2222.2010.03484.x
2. Lee JJ, Jacobsen EA, Ochkur SI, McGarry MP, Condjella RM, Doyle AD, et al. Human versus mouse eosinophils: “that which we call an eosinophil, by any other name would stain as red”. J Allergy Clin Immunol (2012) 130(3):572–84. doi:10.1016/j.jaci.2012.07.025
4. Hirsch AG, Yan XS, Sundaresan AS, Tan BK, Schleimer RP, Kern RC, et al. Five-year risk of incident disease following a diagnosis of chronic rhinosinusitis. Allergy (2015) 70(12):1613–21. doi:10.1111/all.12759
5. Stevens WW, Lee RJ, Schleimer RP, Cohen NA. Chronic rhinosinusitis pathogenesis. J Allergy Clin Immunol (2015) 136(6):1442–53. doi:10.1016/j.jaci.2015.10.009
6. Stevens WW, Schleimer RP, Kern RC. Chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol Pract (2016) 4(4):565–72. doi:10.1016/j.jaip.2016.04.012
7. Schleimer RP. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu Rev Pathol (2017) 12:331–57. doi:10.1146/annurev-pathol-052016-100401
8. de Graauw E, Beltraminelli H, Simon HU, Simon D. Eosinophilia in dermatologic disorders. Immunol Allergy Clin North Am (2015) 35(3):545–60. doi:10.1016/j.iac.2015.05.005
9. Caldwell JM, Collins MH, Stucke EM, Putnam PE, Franciosi JP, Kushner JP, et al. Histologic eosinophilic gastritis is a systemic disorder associated with blood and extragastric eosinophilia, TH2 immunity, and a unique gastric transcriptome. J Allergy Clin Immunol (2014) 134(5):1114–24. doi:10.1016/j.jaci.2014.07.026
10. Wechsler JB, Bryce PJ. Allergic mechanisms in eosinophilic esophagitis. Gastroenterol Clin North Am (2014) 43(2):281–96. doi:10.1016/j.gtc.2014.02.006
11. Mehta P, Furuta GT. Eosinophils in gastrointestinal disorders: eosinophilic gastrointestinal diseases, celiac disease, inflammatory bowel diseases, and parasitic infections. Immunol Allergy Clin North Am (2015) 35(3):413–37. doi:10.1016/j.iac.2015.04.003
12. Travers J, Rothenberg ME. Eosinophils in mucosal immune responses. Mucosal Immunol (2015) 8(3):464–75. doi:10.1038/mi.2015.2
13. Davis BP, Rothenberg ME. Mechanisms of disease of eosinophilic esophagitis. Annu Rev Pathol (2016) 11:365–93. doi:10.1146/annurev-pathol-012615-044241
14. Simon D, Wardlaw A, Rothenberg ME. Organ-specific eosinophilic disorders of the skin, lung and gastrointestinal tract. J Allergy Clin Immunol (2010) 126:45–9. doi:10.1016/j.jaci.2010.03.042
15. Bochner BS, Book W, Busse WW, Butterfield J, Furuta GT, Gleich GJ, et al. Workshop report from the national institutes of health taskforce on the research needs of eosinophil-associated diseases (TREAD). J Allergy Clin Immunol (2012) 130(3):587–96. doi:10.1016/j.jaci.2012.07.024
16. Wechsler ME, Fulkerson PC, Bochner BS, Gauvreau GM, Gleich GJ, Henkel T, et al. Novel targeted therapies for eosinophilic disorders. J Allergy Clin Immunol (2012) 130(3):563–71. doi:10.1016/j.jaci.2012.07.027
17. Gauvreau GM, O’Byrne PM, Boulet LP, Wang Y, Cockcroft D, Bigler J, et al. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med (2014) 370(22):2102–10. doi:10.1056/NEJMoa1402895
18. Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med (2013) 368(26):2455–66. doi:10.1056/NEJMoa1304048
19. Beck LA, Thaci D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med (2014) 371(2):130–9. doi:10.1056/NEJMoa1314768
20. Bachert C, Mannent L, Naclerio RM, Mullol J, Ferguson BJ, Gevaert P, et al. Effect of subcutaneous dupilumab on nasal polyp burden in patients with chronic sinusitis and nasal polyposis: a randomized clinical trial. JAMA (2016) 315(5):469–79. doi:10.1001/jama.2015.19330
21. Thaci D, Simpson EL, Beck LA, Bieber T, Blauvelt A, Papp K, et al. Efficacy and safety of dupilumab in adults with moderate-to-severe atopic dermatitis inadequately controlled by topical treatments: a randomised, placebo-controlled, dose-ranging phase 2b trial. Lancet (2016) 387(10013):40–52. doi:10.1016/S0140-6736(15)00388-8
22. Bochner BS. Novel therapies for eosinophilic disorders. Immunol Allergy Clin North Am (2015) 35(3):577–98. doi:10.1016/j.iac.2015.05.007
23. Radonjic-Hoesli S, Valent P, Klion AD, Wechsler ME, Simon HU. Novel targeted therapies for eosinophil-associated diseases and allergy. Annu Rev Pharmacol Toxicol (2015) 55:633–56. doi:10.1146/annurev-pharmtox-010814-124407
24. Katial RK, Bensch GW, Busse WW, Chipps BE, Denson JL, Gerber AN, et al. Changing paradigms in the treatment of severe asthma: the role of biologic therapies. J Allergy Clin Immunol Pract (2017) 5(2S):S1–14. doi:10.1016/j.jaip.2016.11.029
25. Hamann J, Koning N, Pouwels W, Ulfman LH, van Eijk M, Stacey M, et al. EMR1, the human homolog of F4/80, is an eosinophil-specific receptor. Eur J Immunol (2007) 37(10):2797–802. doi:10.1002/eji.200737553
26. Legrand F, Tomasevic N, Simakova O, Lee CC, Wang Z, Raffeld M, et al. The eosinophil surface receptor epidermal growth factor-like module containing mucin-like hormone receptor 1 (EMR1): a novel therapeutic target for eosinophilic disorders. J Allergy Clin Immunol (2014) 133(5):1439–47. doi:10.1016/j.jaci.2013.11.041
27. Floyd H, Ni J, Cornish AL, Zeng Z, Liu D, Carter KC, et al. Siglec-8. A novel eosinophil-specific member of the immunoglobulin superfamily. J Biol Chem (2000) 275(2):861–6. doi:10.1074/jbc.275.2.861
28. Kikly KK, Bochner BS, Freeman SD, Tan KB, Gallagher KT, D’Alessio KJ, et al. Identification of SAF-2, a novel Siglec expressed on eosinophils, mast cells, and basophils. J Allergy Clin Immunol (2000) 105(6 Pt 1):1093–100. doi:10.1067/mai.2000.107127
29. Bochner BS. “Siglec”ting the allergic response for therapeutic targeting. Glycobiology (2016) 26(6):546–52. doi:10.1093/glycob/cww024
30. Foussias G, Yousef GM, Diamandis EP. Molecular characterization of a Siglec8 variant containing cytoplasmic tyrosine-based motifs, and mapping of the Siglec8 gene. Biochem Biophys Res Commun (2000) 278(3):775–81. doi:10.1006/bbrc.2000.3866
31. Aizawa H, Plitt J, Bochner BS. Human eosinophils express two Siglec-8 splice variants. J Allergy Clin Immunol (2002) 109(1):176. doi:10.1067/mai.2002.120550
32. Yokoi H, Myers A, Matsumoto K, Crocker PR, Saito H, Bochner BS. Alteration and acquisition of Siglecs during in vitro maturation of CD34+ progenitors into human mast cells. Allergy (2006) 61(6):769–76. doi:10.1111/j.1398-9995.2006.01133.x
33. Hudson SA, Herrmann H, Du J, Cox P, Haddad el B, Butler B, et al. Developmental, malignancy-related, and cross-species analysis of eosinophil, mast cell, and basophil Siglec-8 expression. J Clin Immunol (2011) 31(6):1045–53. doi:10.1007/s10875-011-9589-4
34. Hwang SM, Uhm TG, Lee SK, Kong SK, Jung KH, Binas B, et al. Olig2 is expressed late in human eosinophil development and controls Siglec-8 expression. J Leukoc Biol (2016) 100(4):711–23. doi:10.1189/jlb.1A0715-314RRR
35. Varki A, Angata T. Siglecs – the major subfamily of I-type lectins. Glycobiology (2006) 16(1):1R–27R. doi:10.1093/glycob/cwj008
36. Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol (2007) 7(4):255–66. doi:10.1038/nri2056
37. Bochner BS, Alvarez RA, Mehta P, Bovin NV, Blixt O, White JR, et al. Glycan array screening reveals a candidate ligand for Siglec-8. J Biol Chem (2005) 280(6):4307–12. doi:10.1074/jbc.M412378200
38. Kiwamoto T, Katoh T, Evans CM, Janssen WJ, Brummet ME, Hudson SA, et al. Endogenous airway mucins carry glycans that bind Siglec-F and induce eosinophil apoptosis. J Allergy Clin Immunol (2015) 135(5):1329–40. doi:10.1016/j.jaci.2014.10.027
39. Hudson SA, Bovin NV, Schnaar RL, Crocker PR, Bochner BS. Eosinophil-selective binding and proapoptotic effect in vitro of a synthetic Siglec-8 ligand, polymeric 6’-sulfated sialyl Lewis x. J Pharmacol Exp Ther (2009) 330(2):608–12. doi:10.1124/jpet.109.152439
40. Propster JM, Yang F, Rabbani S, Ernst B, Allain FH, Schubert M. Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8. Proc Natl Acad Sci U S A (2016) 113(29):E4170–9. doi:10.1073/pnas.1602214113
41. Nutku E, Aizawa H, Hudson SA, Bochner BS. Ligation of Siglec-8: a selective mechanism for induction of human eosinophil apoptosis. Blood (2003) 101(12):5014–20. doi:10.1182/blood-2002-10-3058
42. Nutku E, Hudson SA, Bochner BS. Mechanism of Siglec-8-induced human eosinophil apoptosis: role of caspases and mitochondrial injury. Biochem Biophys Res Commun (2005) 336(3):918–24. doi:10.1016/j.bbrc.2005.08.202
43. Nutku-Bilir E, Hudson SA, Bochner BS. Interleukin-5 priming of human eosinophils alters Siglec-8 mediated apoptosis pathways. Am J Respir Cell Mol Biol (2008) 38(1):121–4. doi:10.1165/rcmb.2007-0154OC
44. Na HJ, Hudson SA, Bochner BS. IL-33 enhances Siglec-8 mediated apoptosis of human eosinophils. Cytokine (2012) 57(1):169–74. doi:10.1016/j.cyto.2011.10.007
45. von Gunten S, Vogel M, Schaub A, Stadler BM, Miescher S, Crocker PR, et al. Intravenous immunoglobulin preparations contain anti-Siglec-8 autoantibodies. J Allergy Clin Immunol (2007) 119(4):1005–11. doi:10.1016/j.jaci.2007.01.023
46. Taylor VC, Buckley CD, Douglas M, Cody AJ, Simmons DL, Freeman SD. The myeloid-specific sialic acid-binding receptor, CD33, associates with the protein-tyrosine phosphatases, SHP-1 and SHP-2. J Biol Chem (1999) 274(17):11505–12. doi:10.1074/jbc.274.17.11505
47. Angata T, Hingorani R, Varki NM, Varki A. Cloning and characterization of a novel mouse Siglec, mSiglec-F: differential evolution of the mouse and human (CD33) Siglec-3-related gene clusters. J Biol Chem (2001) 276(48):45128–36. doi:10.1074/jbc.M108573200
48. Kitzig F, Martinez-Barriocanal A, Lopez-Botet M, Sayos J. Cloning of two new splice variants of Siglec-10 and mapping of the interaction between Siglec-10 and SHP-1. Biochem Biophys Res Commun (2002) 296(2):355–62. doi:10.1016/S0006-291X(02)00885-9
49. Avril T, Freeman SD, Attrill H, Clarke RG, Crocker PR. Siglec-5 (CD170) can mediate inhibitory signaling in the absence of immunoreceptor tyrosine-based inhibitory motif phosphorylation. J Biol Chem (2005) 280(20):19843–51. doi:10.1074/jbc.M502041200
50. Kano G, Bochner BS, Zimmermann N. Regulation of Siglec-8-induced intracellular reactive oxygen species production and eosinophil cell death by Src family kinases. Immunobiology (2017) 222(2):343–9. doi:10.1016/j.imbio.2016.09.006
51. Janevska D, O’Sullivan JA, Cao Y, Bochner BS. Specific subsets of kinases mediate Siglec-8 engagement -induced ROS production and apoptosis in human eosinophils. J Immunol (2016) 196(1 Suppl):191.5.
52. Cornish AL, Freeman S, Forbes G, Ni J, Zhang M, Cepeda M, et al. Characterization of Siglec-5, a novel glycoprotein expressed on myeloid cells related to CD33. Blood (1998) 92(6):2123–32.
53. Aizawa H, Zimmermann N, Carrigan PE, Lee JJ, Rothenberg ME, Bochner BS. Molecular analysis of human Siglec-8 orthologs relevant to mouse eosinophils: identification of mouse orthologs of Siglec-5 (mSiglec-F) and Siglec-10 (mSiglec-G). Genomics (2003) 82(5):521–30. doi:10.1016/S0888-7543(03)00171-X
54. Zhang JQ, Biedermann B, Nitschke L, Crocker PR. The murine inhibitory receptor mSiglec-E is expressed broadly on cells of the innate immune system whereas mSiglec-F is restricted to eosinophils. Eur J Immunol (2004) 34(4):1175–84. doi:10.1002/eji.200324723
55. Feng YH, Mao H. Expression and preliminary functional analysis of Siglec-F on mouse macrophages. J Zhejiang Univ Sci B (2012) 13(5):386–94. doi:10.1631/jzus.B1100218
56. Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature (2016) 529(7585):226–30. doi:10.1038/nature16527
57. Gicheva N, Macauley MS, Arlian BM, Paulson JC, Kawasaki N. Siglec-F is a novel intestinal M cell marker. Biochem Biophys Res Commun (2016) 479(1):1–4. doi:10.1016/j.bbrc.2016.08.055
58. Tateno H, Crocker PR, Paulson JC. Mouse Siglec-F and human Siglec-8 are functionally convergent paralogs that are selectively expressed on eosinophils and recognize 6’-sulfo-sialyl Lewis X as a preferred glycan ligand. Glycobiology (2005) 15(11):1125–35. doi:10.1093/glycob/cwi097
59. Yu H, Gonzalez-Gil A, Wei Y, Fernandes SM, Porell R, Vajn K, et al. Siglec-8 and Siglec-9 binding specificities and endogenous airway ligand distributions and properties. Glycobiology (2017) 27(7):657–68. doi:10.1093/glycob/cwx026
60. Zhang M, Angata T, Cho JY, Miller M, Broide DH, Varki A. Defining the in vivo function of Siglec-F, a CD33-related Siglec expressed on mouse eosinophils. Blood (2007) 109(10):4280–7. doi:10.1182/blood-2006-08-039255
61. Zimmermann N, McBride ML, Yamada Y, Hudson SA, Jones C, Cromie KD, et al. Siglec-F antibody administration to mice selectively reduces blood and tissue eosinophils. Allergy (2008) 63(9):1156–63. doi:10.1111/j.1398-9995.2008.01709.x
62. Song DJ, Cho JY, Lee SY, Miller M, Rosenthal P, Soroosh P, et al. Anti-Siglec-F antibody reduces allergen-induced eosinophilic inflammation and airway remodeling. J Immunol (2009) 183(8):5333–41. doi:10.4049/jimmunol.0801421
63. Rubinstein E, Cho JY, Rosenthal P, Chao J, Miller M, Pham A, et al. Siglec-F inhibition reduces esophageal eosinophilia and angiogenesis in a mouse model of eosinophilic esophagitis. J Pediatr Gastroenterol Nutr (2011) 53(4):409–16. doi:10.1097/MPG.0b013e3182182ff8
64. McMillan SJ, Richards HE, Crocker PR. Siglec-F-dependent negative regulation of allergen-induced eosinophilia depends critically on the experimental model. Immunol Lett (2014) 160(1):11–6. doi:10.1016/j.imlet.2014.03.008
65. Mao H, Kano G, Hudson SA, Brummet M, Zimmermann N, Zhu Z, et al. Mechanisms of Siglec-F-induced eosinophil apoptosis: a role for caspases but not for SHP-1, Src kinases, NADPH oxidase or reactive oxygen. PLoS One (2013) 8(6):e68143. doi:10.1371/journal.pone.0068143
66. Guo JP, Brummet ME, Myers AC, Na HJ, Rowland E, Schnaar RL, et al. Characterization of expression of glycan ligands for Siglec-F in normal mouse lungs. Am J Respir Cell Mol Biol (2011) 44(2):238–43. doi:10.1165/rcmb.2010-0007OC
67. Suzukawa M, Miller M, Rosenthal P, Cho JY, Doherty TA, Varki A, et al. Sialyltransferase ST3Gal-III regulates Siglec-F ligand formation and eosinophilic lung inflammation in mice. J Immunol (2013) 190(12):5939–48. doi:10.4049/jimmunol.1203455
68. Kiwamoto T, Brummet ME, Wu F, Motari MG, Smith DF, Schnaar RL, et al. Mice deficient in the St3gal3 gene product alpha2,3 sialyltransferase (ST3Gal-III) exhibit enhanced allergic eosinophilic airway inflammation. J Allergy Clin Immunol (2014) 133(1):240–7. doi:10.1016/j.jaci.2013.05.018
69. Jia Y, Yu H, Fernandes SM, Wei Y, Gonzalez-Gil A, Motari MG, et al. Expression of ligands for Siglec-8 and Siglec-9 in human airways and airway cells. J Allergy Clin Immunol (2015) 135(3):799–810. doi:10.1016/j.jaci.2015.01.004
70. Patnode ML, Cheng CW, Chou CC, Singer MS, Elin MS, Uchimura K, et al. Galactose 6-O-sulfotransferases are not required for the generation of Siglec-F ligands in leukocytes or lung tissue. J Biol Chem (2013) 288(37):26533–45. doi:10.1074/jbc.M113.485409
71. Biedermann B, Gil D, Bowen DT, Crocker PR. Analysis of the CD33-related Siglec family reveals that Siglec-9 is an endocytic receptor expressed on subsets of acute myeloid leukemia cells and absent from normal hematopoietic progenitors. Leuk Res (2007) 31(2):211–20. doi:10.1016/j.leukres.2006.05.026
72. Tateno H, Li H, Schur MJ, Bovin N, Crocker PR, Wakarchuk WW, et al. Distinct endocytic mechanisms of CD22 (Siglec-2) and Siglec-F reflect roles in cell signaling and innate immunity. Mol Cell Biol (2007) 27(16):5699–710. doi:10.1128/MCB.00383-07
73. Walter RB, Raden BW, Zeng R, Hausermann P, Bernstein ID, Cooper JA. ITIM-dependent endocytosis of CD33-related Siglecs: role of intracellular domain, tyrosine phosphorylation, and the tyrosine phosphatases, Shp1 and Shp2. J Leukoc Biol (2008) 83(1):200–11. doi:10.1189/jlb.0607388
74. Delputte PL, Van Gorp H, Favoreel HW, Hoebeke I, Delrue I, Dewerchin H, et al. Porcine sialoadhesin (CD169/Siglec-1) is an endocytic receptor that allows targeted delivery of toxins and antigens to macrophages. PLoS One (2011) 6(2):e16827. doi:10.1371/journal.pone.0016827
75. Daniels TR, Bernabeu E, Rodriguez JA, Patel S, Kozman M, Chiappetta DA, et al. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta (2012) 1820(3):291–317. doi:10.1016/j.bbagen.2011.07.016
76. Thol F, Schlenk RF. Gemtuzumab ozogamicin in acute myeloid leukemia revisited. Expert Opin Biol Ther (2014) 14(8):1185–95. doi:10.1517/14712598.2014.922534
77. Cheung A, Bax HJ, Josephs DH, Ilieva KM, Pellizzari G, Opzoomer J, et al. Targeting folate receptor alpha for cancer treatment. Oncotarget (2016) 7(32):52553–74. doi:10.18632/oncotarget.9651
78. Dorner T, Shock A, Goldenberg DM, Lipsky PE. The mechanistic impact of CD22 engagement with epratuzumab on B cell function: implications for the treatment of systemic lupus erythematosus. Autoimmun Rev (2015) 14(12):1079–86. doi:10.1016/j.autrev.2015.07.013
79. Kantarjian HM, DeAngelo DJ, Stelljes M, Martinelli G, Liedtke M, Stock W, et al. Inotuzumab ozogamicin versus standard therapy for acute lymphoblastic leukemia. N Engl J Med (2016) 375(8):740–53. doi:10.1056/NEJMoa1509277
80. O’Sullivan JA, Carroll DJ, Cao Y, Salicru AN, Bochner BS. Leveraging Siglec-8 endocytic mechanisms to kill human eosinophils and malignant mast cells. J Allergy Clin Immunol (2017). (in press).
81. Zhu XD, Zhuang Y, Ben JJ, Qian LL, Huang HP, Bai H, et al. Caveolae-dependent endocytosis is required for class A macrophage scavenger receptor-mediated apoptosis in macrophages. J Biol Chem (2011) 286(10):8231–9. doi:10.1074/jbc.M110.145888
82. Polo S, Di Fiore PP. Endocytosis conducts the cell signaling orchestra. Cell (2006) 124(5):897–900. doi:10.1016/j.cell.2006.02.025
83. Stoddart A, Dykstra ML, Brown BK, Song W, Pierce SK, Brodsky FM. Lipid rafts unite signaling cascades with clathrin to regulate BCR internalization. Immunity (2002) 17(4):451–62. doi:10.1016/S1074-7613(02)00416-8
84. Stoddart A, Jackson AP, Brodsky FM. Plasticity of B cell receptor internalization upon conditional depletion of clathrin. Mol Biol Cell (2005) 16(5):2339–48. doi:10.1091/mbc.E05-01-0025
85. Collins BE, Smith BA, Bengtson P, Paulson JC. Ablation of CD22 in ligand-deficient mice restores B cell receptor signaling. Nat Immunol (2006) 7(2):199–206. doi:10.1038/ni1283
86. Grewal PK, Boton M, Ramirez K, Collins BE, Saito A, Green RS, et al. ST6Gal-I restrains CD22-dependent antigen receptor endocytosis and Shp-1 recruitment in normal and pathogenic immune signaling. Mol Cell Biol (2006) 26(13):4970–81. doi:10.1128/MCB.00308-06
87. Lei JT, Martinez-Moczygemba M. Separate endocytic pathways regulate IL-5 receptor internalization and signaling. J Leukoc Biol (2008) 84(2):499–509. doi:10.1189/jlb.1207828
88. Wen T, Mingler MK, Blanchard C, Wahl B, Pabst O, Rothenberg ME. The pan-B cell marker CD22 is expressed on gastrointestinal eosinophils and negatively regulates tissue eosinophilia. J Immunol (2012) 188(3):1075–82. doi:10.4049/jimmunol.1102222
89. Wood B. Multicolor immunophenotyping: human immune system hematopoiesis. Methods Cell Biol (2004) 75:559–76. doi:10.1016/S0091-679X(04)75023-2
90. Munitz A, Bachelet I, Eliashar R, Moretta A, Moretta L, Levi-Schaffer F. The inhibitory receptor IRp60 (CD300a) suppresses the effects of IL-5, GM-CSF, and eotaxin on human peripheral blood eosinophils. Blood (2006) 107(5):1996–2003. doi:10.1182/blood-2005-07-2926
91. Legrand F, Landolina NA, Levi-Schaffer F, Klion AD. Siglec-7 on peripheral blood eosinophils: surface expression and functional analysis. J Allergy Clin Immunol (2016) 137(2):Ab167. doi:10.1016/j.jaci.2015.12.680
92. Li N, Zhang W, Wan T, Zhang J, Chen T, Yu Y, et al. Cloning and characterization of Siglec-10, a novel sialic acid binding member of the Ig superfamily, from human dendritic cells. J Biol Chem (2001) 276(30):28106–12. doi:10.1074/jbc.M100467200
93. Munday J, Kerr S, Ni J, Cornish AL, Zhang JQ, Nicoll G, et al. Identification, characterization and leucocyte expression of Siglec-10, a novel human sialic acid-binding receptor. Biochem J (2001) 355(Pt 2):489–97. doi:10.1042/0264-6021:3550489
94. Whitney G, Wang S, Chang H, Cheng KY, Lu P, Zhou XD, et al. A new Siglec family member, Siglec-10, is expressed in cells of the immune system and has signaling properties similar to CD33. Eur J Biochem (2001) 268(23):6083–96. doi:10.1046/j.0014-2956.2001.02543.x
95. Yousef GM, Ordon MH, Foussias G, Diamandis EP. Molecular characterization, tissue expression, and mapping of a novel Siglec-like gene (SLG2) with three splice variants. Biochem Biophys Res Commun (2001) 284(4):900–10. doi:10.1006/bbrc.2001.5053
96. Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science (2009) 323(5922):1722–5. doi:10.1126/science.1168988
97. Kivi E, Elima K, Aalto K, Nymalm Y, Auvinen K, Koivunen E, et al. Human Siglec-10 can bind to vascular adhesion protein-1 and serves as its substrate. Blood (2009) 114(26):5385–92. doi:10.1182/blood-2009-04-219253
98. Bandala-Sanchez E, Zhang Y, Reinwald S, Dromey JA, Lee BH, Qian J, et al. T cell regulation mediated by interaction of soluble CD52 with the inhibitory receptor Siglec-10. Nat Immunol (2013) 14(7):741–8. doi:10.1038/ni.2610
99. Kitayama J, Fuhlbrigge RC, Puri KD, Springer TA. P-selectin, L-selectin, and alpha 4 integrin have distinct roles in eosinophil tethering and arrest on vascular endothelial cells under physiological flow conditions. J Immunol (1997) 159(8):3929–39.
100. Taylor ML, Brummet ME, Hudson SA, Miura K, Bochner BS. Expression and function of P-selectin glycoprotein ligand 1 (CD162) on human basophils. J Allergy Clin Immunol (2000) 106(5):918–24. doi:10.1067/mai.2000.110230
101. Knol EF, Tackey F, Tedder TF, Klunk DA, Bickel CA, Sterbinsky SA, et al. Comparison of human eosinophil and neutrophil adhesion to endothelial cells under nonstatic conditions. Role of L-selectin. J Immunol (1994) 153(5):2161–7.
102. Sriramarao P, von Andrian UH, Butcher EC, Bourdon MA, Broide DH. L-selectin and very late antigen-4 integrin promote eosinophil rolling at physiological shear rates in vivo. J Immunol (1994) 153(9):4238–46.
103. Baisse B, Galisson F, Giraud S, Schapira M, Spertini O. Evolutionary conservation of P-selectin glycoprotein ligand-1 primary structure and function. BMC Evol Biol (2007) 7:166. doi:10.1186/1471-2148-7-166
104. Pan J, Xia L, McEver RP. Comparison of promoters for the murine and human P-selectin genes suggests species-specific and conserved mechanisms for transcriptional regulation in endothelial cells. J Biol Chem (1998) 273(16):10058–67. doi:10.1074/jbc.273.16.10058
105. Yao L, Setiadi H, Xia L, Laszik Z, Taylor FB, McEver RP. Divergent inducible expression of P-selectin and E-selectin in mice and primates. Blood (1999) 94(11):3820–8.
106. Broide DH, Humber D, Sullivan S, Sriramarao P. Inhibition of eosinophil rolling and recruitment in P-selectin- and intracellular adhesion molecule-1-deficient mice. Blood (1998) 91(8):2847–56.
107. Broide DH, Sullivan S, Gifford T, Sriramarao P. Inhibition of pulmonary eosinophilia in P-selectin- and ICAM-1-deficient mice. Am J Respir Cell Mol Biol (1998) 18(2):218–25. doi:10.1165/ajrcmb.18.2.2829
108. Robinson SD, Frenette PS, Rayburn H, Cummiskey M, Ullman-Cullere M, Wagner DD, et al. Multiple, targeted deficiencies in selectins reveal a predominant role for P-selectin in leukocyte recruitment. Proc Natl Acad Sci U S A (1999) 96(20):11452–7. doi:10.1073/pnas.96.20.11452
109. Dang B, Wiehler S, Patel KD. Increased PSGL-1 expression on granulocytes from allergic-asthmatic subjects results in enhanced leukocyte recruitment under flow conditions. J Leukoc Biol (2002) 72(4):702–10.
110. Woltmann G, McNulty CA, Dewson G, Symon FA, Wardlaw AJ. Interleukin-13 induces PSGL-1/P-selectin-dependent adhesion of eosinophils, but not neutrophils, to human umbilical vein endothelial cells under flow. Blood (2000) 95(10):3146–52.
111. Davenpeck KL, Brummet ME, Hudson SA, Mayer RJ, Bochner BS. Activation of human leukocytes reduces surface P-selectin glycoprotein ligand-1 (PSGL-1, CD162) and adhesion to P-selectin in vitro. J Immunol (2000) 165(5):2764–72. doi:10.4049/jimmunol.165.5.2764
112. Sako D, Chang XJ, Barone KM, Vachino G, White HM, Shaw G, et al. Expression cloning of a functional glycoprotein ligand for P-selectin. Cell (1993) 75(6):1179–86. doi:10.1016/0092-8674(93)90327-M
113. Asa D, Raycroft L, Ma L, Aeed PA, Kaytes PS, Elhammer AP, et al. The P-selectin glycoprotein ligand functions as a common human leukocyte ligand for P- and E-selectins. J Biol Chem (1995) 270(19):11662–70. doi:10.1074/jbc.270.19.11662
114. Satoh T, Kaneko M, Wu MH, Yokozeki H, Nishioka K. Contribution of selectin ligands to eosinophil recruitment into the skin of patients with atopic dermatitis. Eur J Immunol (2002) 32(5):1274–81. doi:10.1002/1521-4141(200205)32:5<1274::AID-IMMU1274>3.0.CO;2-Q
115. Bochner BS, Sterbinsky SA, Bickel CA, Werfel S, Wein M, Newman W. Differences between human eosinophils and neutrophils in the function and expression of sialic acid-containing counterligands for E-selectin. J Immunol (1994) 152(2):774–82.
116. Nimrichter L, Burdick MM, Aoki K, Laroy W, Fierro MA, Hudson SA, et al. E-selectin receptors on human leukocytes. Blood (2008) 112(9):3744–52. doi:10.1182/blood-2008-04-149641
117. Wein M, Sterbinsky SA, Bickel CA, Schleimer RP, Bochner BS. Comparison of human eosinophil and neutrophil ligands for P-selectin: ligands for P-selectin differ from those for E-selectin. Am J Respir Cell Mol Biol (1995) 12(3):315–9. doi:10.1165/ajrcmb.12.3.7532979
118. Ebisawa M, Bochner BS, Georas SN, Schleimer RP. Eosinophil transendothelial migration induced by cytokines. I. Role of endothelial and eosinophil adhesion molecules in IL-1 beta-induced transendothelial migration. J Immunol (1992) 149(12):4021–8.
119. Kim MK, Brandley BK, Anderson MB, Bochner BS. Antagonism of selectin-dependent adhesion of human eosinophils and neutrophils by glycomimetics and oligosaccharide compounds. Am J Respir Cell Mol Biol (1998) 19(5):836–41. doi:10.1165/ajrcmb.19.5.3032
120. Wun T, Styles L, DeCastro L, Telen MJ, Kuypers F, Cheung A, et al. Phase 1 study of the E-selectin inhibitor GMI 1070 in patients with sickle cell anemia. PLoS One (2014) 9(7):e101301. doi:10.1371/journal.pone.0101301
121. Weller PF, Bach DS, Austen KF. Biochemical characterization of human eosinophil Charcot-Leyden crystal protein (lysophospholipase). J Biol Chem (1984) 259(24):15100–5.
122. Dor PJ, Ackerman SJ, Gleich GJ. Charcot-Leyden crystal protein and eosinophil granule major basic protein in sputum of patients with respiratory diseases. Am Rev Respir Dis (1984) 130(6):1072–7. doi:10.1164/arrd.1984.130.6.1072
123. Dvorak AM, Letourneau L, Login GR, Weller PF, Ackerman SJ. Ultrastructural localization of the Charcot-Leyden crystal protein (lysophospholipase) to a distinct crystalloid-free granule population in mature human eosinophils. Blood (1988) 72(1):150–8.
124. Ackerman SJ, Liu L, Kwatia MA, Savage MP, Leonidas DD, Swaminathan GJ, et al. Charcot-Leyden crystal protein (galectin-10) is not a dual function galectin with lysophospholipase activity but binds a lysophospholipase inhibitor in a novel structural fashion. J Biol Chem (2002) 277(17):14859–68. doi:10.1074/jbc.M200221200
125. Ackerman SJ, Corrette SE, Rosenberg HF, Bennett JC, Mastrianni DM, Nicholson-Weller A, et al. Molecular cloning and characterization of human eosinophil Charcot-Leyden crystal protein (lysophospholipase). Similarities to IgE binding proteins and the S-type animal lectin superfamily. J Immunol (1993) 150(2):456–68.
126. Leonidas DD, Elbert BL, Zhou Z, Leffler H, Ackerman SJ, Acharya KR. Crystal structure of human Charcot-Leyden crystal protein, an eosinophil lysophospholipase, identifies it as a new member of the carbohydrate-binding family of galectins. Structure (1995) 3(12):1379–93. doi:10.1016/S0969-2126(01)00275-1
127. Dyer KD, Handen JS, Rosenberg HF. The genomic structure of the human Charcot-Leyden crystal protein gene is analogous to those of the galectin genes. Genomics (1997) 40(2):217–21. doi:10.1006/geno.1996.4590
128. Swaminathan GJ, Leonidas DD, Savage MP, Ackerman SJ, Acharya KR. Selective recognition of mannose by the human eosinophil Charcot-Leyden crystal protein (galectin-10): a crystallographic study at 1.8 a resolution. Biochemistry (1999) 38(42):13837–43. doi:10.1021/bi995093f
129. Devouassoux G, Pachot A, Laforest L, Diasparra J, Freymond N, Van Ganse E, et al. Galectin-10 mRNA is overexpressed in peripheral blood of aspirin-induced asthma. Allergy (2008) 63(1):125–31. doi:10.1111/j.1398-9995.2007.01558.x
130. De Re V, Simula MP, Cannizzaro R, Pavan A, De Zorzi MA, Toffoli G, et al. Galectin-10, eosinophils, and celiac disease. Ann N Y Acad Sci (2009) 1173:357–64. doi:10.1111/j.1749-6632.2009.04627.x
131. Lin TA, Kourteva G, Hilton H, Li H, Tare NS, Carvajal V, et al. The mRNA level of Charcot-Leyden crystal protein/galectin-10 is a marker for CRTH2 activation in human whole blood in vitro. Biomarkers (2010) 15(7):646–54. doi:10.3109/1354750X.2010.511266
132. Chua JC, Douglass JA, Gillman A, O’Hehir RE, Meeusen EN. Galectin-10, a potential biomarker of eosinophilic airway inflammation. PLoS One (2012) 7(8):e42549. doi:10.1371/journal.pone.0042549
133. Furuta GT, Kagalwalla AF, Lee JJ, Alumkal P, Maybruck BT, Fillon S, et al. The oesophageal string test: a novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut (2013) 62(10):1395–405. doi:10.1136/gutjnl-2012-303171
134. Torsteinsdottir I, Arvidson NG, Hallgren R, Hakansson L. Enhanced expression of integrins and CD66b on peripheral blood neutrophils and eosinophils in patients with rheumatoid arthritis, and the effect of glucocorticoids. Scand J Immunol (1999) 50(4):433–9. doi:10.1046/j.1365-3083.1999.00602.x
135. Zhao L, Xu S, Fjaertoft G, Pauksen K, Hakansson L, Venge P. An enzyme-linked immunosorbent assay for human carcinoembryonic antigen-related cell adhesion molecule 8, a biological marker of granulocyte activities in vivo. J Immunol Methods (2004) 293(1–2):207–14. doi:10.1016/j.jim.2004.08.009
136. Kuijpers TW, Hoogerwerf M, van der Laan LJ, Nagel G, van der Schoot CE, Grunert F, et al. CD66 nonspecific cross-reacting antigens are involved in neutrophil adherence to cytokine-activated endothelial cells. J Cell Biol (1992) 118(2):457–66. doi:10.1083/jcb.118.2.457
137. Yoon J, Terada A, Kita H. CD66b regulates adhesion and activation of human eosinophils. J Immunol (2007) 179(12):8454–62. doi:10.4049/jimmunol.179.12.8454
138. Matsumoto R, Matsumoto H, Seki M, Hata M, Asano Y, Kanegasaki S, et al. Human ecalectin, a variant of human galectin-9, is a novel eosinophil chemoattractant produced by T lymphocytes. J Biol Chem (1998) 273(27):16976–84. doi:10.1074/jbc.273.27.16976
139. Rao SP, Wang Z, Zuberi RI, Sikora L, Bahaie NS, Zuraw BL, et al. Galectin-3 functions as an adhesion molecule to support eosinophil rolling and adhesion under conditions of flow. J Immunol (2007) 179(11):7800–7. doi:10.4049/jimmunol.179.11.7800
140. Ge XN, Ha SG, Greenberg YG, Rao A, Bastan I, Blidner AG, et al. Regulation of eosinophilia and allergic airway inflammation by the glycan-binding protein galectin-1. Proc Natl Acad Sci U S A (2016) 113(33):E4837–46. doi:10.1073/pnas.1601958113
141. Mauri P, Riccio AM, Rossi R, Di Silvestre D, Benazzi L, De Ferrari L, et al. Proteomics of bronchial biopsies: galectin-3 as a predictive biomarker of airway remodelling modulation in omalizumab-treated severe asthma patients. Immunol Lett (2014) 162(1 Pt A):2–10. doi:10.1016/j.imlet.2014.08.010
Keywords: eosinophils, Siglec-8, Siglec-F, selectins, galectins, glycans
Citation: O’Sullivan JA, Carroll DJ and Bochner BS (2017) Glycobiology of Eosinophilic Inflammation: Contributions of Siglecs, Glycans, and Other Glycan-Binding Proteins. Front. Med. 4:116. doi: 10.3389/fmed.2017.00116
Received: 24 March 2017; Accepted: 10 July 2017;
Published: 02 August 2017
Edited by:
Florence Emmanuelle Roufosse, Free University of Brussels, BelgiumReviewed by:
Owen McCarty, Oregon Health & Science University, United StatesHirohito Kita, Mayo Clinic Minnesota, United States
Copyright: © 2017 O’Sullivan, Carroll and Bochner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bruce S. Bochner, bruce.bochner@northwestern.edu
†These authors have contributed equally to this work.
 Jeremy A. O’Sullivan
Jeremy A. O’Sullivan Daniela J. Carroll
Daniela J. Carroll Bruce S. Bochner
Bruce S. Bochner