Revealing the Mechanisms of Synergistic Action of Two Magainin Antimicrobial Peptides
- 1University of Strasbourg/CNRS, UMR7177, Institut de Chimie de Strasbourg, Strasbourg, France
- 2Institut Universitaire de France (IUF), Paris, France
The study of peptide-lipid and peptide-peptide interactions as well as their topology and dynamics using biophysical and structural approaches have changed our view how antimicrobial peptides work and function. It has become obvious that both the peptides and the lipids arrange in soft supramolecular arrangements which are highly dynamic and able to change and mutually adapt their conformation, membrane penetration, and detailed morphology. This can occur on a local and a global level. This review focuses on cationic amphipathic peptides of the magainin family which were studied extensively by biophysical approaches. They are found intercalated at the membrane interface where they cause membrane thinning and ultimately lysis. Interestingly, mixtures of two of those peptides namely magainin 2 and PGLa which occur naturally as a cocktail in the frog skin exhibit synergistic enhancement of antimicrobial activities when investigated together in antimicrobial assays but also in biophysical experiments with model membranes. Detailed dose-response curves, presented here for the first time, show a cooperative behavior for the individual peptides which is much increased when PGLa and magainin are added as equimolar mixture. This has important consequences for their bacterial killing activities and resistance development. In membranes that carry unsaturations both peptides align parallel to the membrane surface where they have been shown to arrange into mesophases involving the peptides and the lipids. This supramolecular structuration comes along with much-increased membrane affinities for the peptide mixture. Because this synergism is most pronounced in membranes representing the bacterial lipid composition it can potentially be used to increase the therapeutic window of pharmaceutical formulations.
Introduction
Antimicrobial peptides (AMPs) are part of the innate immune system of higher organisms which provides a powerful and responsive first line of defense against a multitude of pathogenic microorganisms (1, 2). Several years after the discovery of penicillin in 1928 (3) gramicidin S was extracted from soil bacteria and used to treat gunshot wounds during world-war II (4–6). Furthermore, other peptidic compounds with antimicrobial activities have been detected in microorganisms (7, 8), but antimicrobial peptides also exist in many species of the plant and animal kingdom, including humans (9). The first of these peptides have been discovered decades ago and have been investigated ever since (4, 10–12). The list of amino acid sequences with antimicrobial activities is continuously increasing and they are accessible through various data bases (13–16). To understand their mechanisms of action several of them have been investigated by a variety of biological, biochemical, and biophysical approaches (17, 18).
During recent years the rapid increase in multiresistant pathogens (19) has brought back research on AMPs (20, 21), because their mechanisms of action has been shown to be less prone to microbial resistance when compared to conventional treatments (2, 22, 23). Although peptides are quickly digested by proteases (24–26) this limitation in their applicability can be overcome by unnatural building blocks, their protection inside nanostructures or when linked to surfaces (27–29). Furthermore, based on mechanistic studies of cationic amphipathic antimicrobial peptides small amphipathic molecules (30, 31), foldamer and pseudopeptides (32–40), and polymers (41) with potent antimicrobial properties have been designed.
Peptides have been found early on in the skin of toads and frogs (10, 42) and magainins from Xenopus laevis were among the first, for which the potential usefulness of their antimicrobial activity has been described (Table 1) (11). Soon after their discovery a multitude of investigations have been performed that reveal that magainins and other cationic amphipathic peptides interfere with the barrier function of bacterial membranes which by itself causes bacterial killing (43–45). Notably, related peptides have been shown to also enter the cell interior where further action can take place (46–48). Furthermore, many peptides are involved in modulating the immune response of the host organisms thereby adding an additional layer of efficiency and they are therefore also referred to as “host defense peptides” (49–52).
A membrane-active mechanism has been confirmed by the study of all-D analogs of AMPs which exhibit the same activity than their naturally occurring counter-part indicating that they do not interact with chiral proteinaceous receptors (47). Indeed, their amphipathic nature and in most cases an accumulation of cationic residues has been shown essential for membrane interaction and selectivity, rather than a specific amino acid composition (2). Peptides with helical (17, 18), cyclic (40, 53–55) and/or β-sheet arrangements have been investigated (56–60).
Magainins and related sequences have been developed up to phase IIb/III clinical trials (61, 62), and in parallel, have been explored in considerable detail by biophysical approaches [e.g., (63–66)]. The data from these studies were often unexpected and resulted in the need to introduce novel mechanisms to explain the activities of these peptides (67–69). Because the peptides and/or their biophysical investigations have been reviewed recently (20, 21, 70), this paper will only shortly summarize some of the key discoveries made with magainins and then focus on the synergistic interactions between PGLa and magainins. Combining antimicrobial peptides provides an interesting and little exploited alternative strategy to enhance their efficiency and to further reduce their susceptibility to bacterial resistance.
Magainins Form Membrane Openings
Magainins exhibit pore-forming and lytic activities when added to membranes which have also been studied by single-channel measurements (45, 71). On a macroscopic scale, magainin pore formation was investigated by fluorophore release experiments. For example, this allowed to measure the release kinetics from individual DOPC/DOPG giant unilamellar vesicles (GUV) at different peptide-to-lipid molar ratios (72). While after peptide addition it takes minutes before the release of fluorophores sets-in, once the pores have formed the micrometer vesicles empty within 30 s (73). It has been measured that fluorophore release is a two-stage process starting with the transient formation of very large pores corresponding to the equilibration of the peptide concentration between the outer and inner leaflets (74). Thereafter a slower but persistent release of fluorophore is observed through pores of 3 nm hydrodynamic radius, large enough to allow passage of globular proteins > 20 kDa (74).
The spatio-temporal events when antimicrobial peptides attach to live bacteria have been investigated by microscopic imaging techniques. Interestingly, the human peptide LL37 (Table 1) preferentially attacks septating E. coli cells at the septum and at the curved regions of the outer membrane (75). In non-septating cells, the peptide accumulates at one of the endcaps. As expected permeabilization starts with the outer membrane and after a short delay cytoplasmic membrane permeabilization occurs. The openings at both membranes occur in a localized and persistent manner (76). Related events are observed when cationic polymers, longer or shorter peptides such as LL37, cecropin A, or melittin are studied (Table 1) even though the exact details vary with the antimicrobial compound (77). Finally, the peptides enter the cells where they interact with anionic polymers including nucleic acids and proteins that are abundant in the cell interior (46, 48, 78).
Magainin Structural Investigations
Structural investigations show that magainins undergo a random coil to helix transition when they partition into the membrane (79). Interestingly the energies associated with this refolding have been identified to be one of the main driving forces for membrane association (80). During the same time period when the antimicrobial activity of magainins was investigated solid-state NMR approaches applied to uniaxially oriented lipid bilayers were under development (63). The latter technique measures angular constraints from polypeptides reconstituted into uniaxially oriented phospholipid bilayers to calculate their structure, topology, and dynamics (81–84). Because the 15N chemical shift of peptide bonds alone already provides an approximate tilt angle of helical domains (85) the very first experiments with magainin 2 and PGLa were indicative that these helices are oriented parallel to the membrane surface (63, 86–88). A membrane alignment parallel to the lipid bilayer surface has been confirmed for magainin 2 (89), for magainin analogs (90, 91) and for several other linear cationic antimicrobial peptides (22, 92–95). Later on oriented CD spectra and fluorescence quenching experiments confirmed such a topology of the magainin helix where the latter approach also shows an interfacial localization of the magainin 2 helix (64, 96). A parallel alignment of cecropin P1 (Table 1) using ATR FTIR was later on described by the “carpet model” (97). Notably, this peripheral membrane topology assures that the peptides can exchange between the membrane and the aqueous phase (89).
Whereas, the magainin 2 helix has been found to partition into the membrane parallel to its surface regardless of membrane lipid composition (64, 89), its relative PGLa (Table 1) exhibits a much wider range of topologies but only in membranes where all fatty acyl chains are saturated (89, 98, 99). In the presence of magainin 2 PGLa adopts transmembrane alignments (98–101) and early on this configuration has been suggested to be part of a synergistic complex between the two peptides (100, 102). However, in the presence of lipid unsaturations (such as palmitoyl-oleoyl-phosphatidylethanolamine), both PGLa and magainin 2 are aligned along the bilayer surface under all conditions so far investigated (98, 99, 101). Because unsaturations are an integral part of bacterial membranes it is highly likely that both magainin and PGLa exert their antimicrobial activity in a state with their amphipathic helices aligned within the membrane plane [e.g., data obtained with membranes made from E. coli lipid extracts (103)]. Correspondingly, alternative models explaining synergism have been suggested (69, 103) (Figure 1).
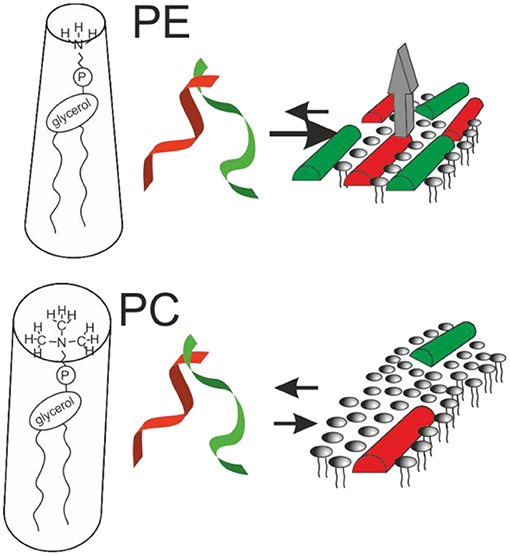
Figure 1. Sketches the structural findings made with magainin 2 (red) and PGLa (green). Both peptides adopt helical conformations that are oriented parallel to the membrane surface in membranes carrying lipid unsaturations. The peptides arrange in nematic mesophases which when added together result in increased membrane affinity and synergistic calcein release activity from POPE-containing liposomes (gray arrow; cf. text for details).
Notably, recent fluorescence self-quenching investigations have demonstrated the formation of nematic phases at the membrane surface by magainin, PGLa, and the LAH4 amphipathic designer peptide (Figure 1, Table 1). Formation of these supramolecular arrangements is dependent on the lipid composition and the salt concentration of the surrounding buffer suggesting that electrostatic but also lipophobic interactions contribute (69, 104).
An amphipathic peptide that partitions into the membrane interface takes up the place of a few lipids but penetrates only to the depth of the head group and glycerol region (64). Thereby the packing of the hydrophobic region becomes more disordered and the membrane thickness is reduced (105, 106). The resulting changes of the macroscopic phase properties, the bilayer packing and the dynamics of the lipids has been monitored by 2H and 31P solid-state NMR spectroscopy (107). Thereby magainin peptides have been shown to introduce curvature into membranes (106), to cause a considerable decreases in the fatty acyl chain order parameters in particular of lipid segments well into the bilayer interior (101, 108–110), and at higher peptide concentrations membrane disruption into micellar or bicellar structures (111). Such bilayer disordering has been estimated to reach over a diameter of 10 nm (112, 113).
Atomistic views how in-plane oriented peptides potentially form water-filled openings in lipid bilayers have been obtained from molecular dynamics simulations (114–116). A reoccurring limitation of the computational approaches is the limited time span covered by the simulations which does not allow to reach equilibrium of statistically relevant numbers or peptides. Therefore, at the present stage the results remain dependent on the starting conditions and careful comparison with experimental data an important control (115–117). Furthermore, for magainins the importance of the very details of the lipid composition has only become apparent about a decade ago (69, 98, 118, 119), therefore, only few such simulations have been performed with lipid compositions such as POPE/POPG 3/1 that closely mimic bacterial membranes. All-atom simulations covering 100 ns show a stable in-plane topology of magainin, some oligomerization, but no pore or supramolecular rearrangement within this time frame (120). Stable in-plane alignments and an interfacial localization in such membranes were also observed in more recent coarse-grain and all-atom simulations (116, 117). In summary, pores rather form through stochastic rearrangements of peptides and lipids rather than well-defined channel structures although some peptide-peptide interactions are sometimes apparent. Because of the relatively small size of the membrane patches and the short time frame covered by the simulations the membrane lytic nature of the peptides or the formation of large pores apparent in dye release experiments do not become visible (73, 74).
The molecular shape concept provides a rationale for the bilayer disruptive properties of in-plane aligned amphipathic helices partitioning into the interface (121, 122). Geometrical considerations originally developed for lipids explain why PC lipids, that have the shape of cylinders, spontaneously arrange into extended bilayers. In contrast, PE exhibits a cone shape with a tendency to adopt hexagonal phases and detergents are inverted cones that arrange into micelles. In an analogous manner, surfactin, a cyclic peptide with a long fatty acyl chain (55), or the magainin 2 in-planar interfacial helix use up much more space in the head group than in the hydrophobic core region of the membrane, therefore, the molecular shape concept has recently been elaborated in some detail for amphipathic antimicrobial peptides (122).
Thus, magainins and other cationic amphipathic antimicrobial peptides appear to fulfill the criteria of the “carpet” model where peptides cover the membrane surface at alignments parallel to the surface (67) ultimately causing membrane lysis (111, 123). At lower peptide-to-lipid ratios the lipid membranes can adapt and compensate for the disruptive properties of the peptides, however from time to time openings form locally and transiently thereby resulting in channel-like recordings (124). The structural changes of the membrane have been monitored in the presence of magainin 2 in real-time revealing intermediate states, lysis and recovery (125). The peptides and lipids show a high degree of structural plasticity thus membranes can adopt a broad range of possible morphologies a behavior that is best described by phase diagrams which can take into account the peptide concentration, the membrane composition and other parameters (126). Such features are summarized in the SMART model where “Soft Membranes Adapt and Respond, also Transiently” (127). In line with this model, AMPs have been proposed to affect the membrane line tension (125, 128).
Magainin and PGLa carry several positive charges (nominal charge +4 to +5) and have been shown to interact better with negatively charged membranes. Such preferential association driven by electrostatic attraction is thought to be a reason for the selective killing of bacteria or tumor cells, that expose negative charges while they do not affect healthy eukaryotic cells which expose an overall charge neutral surface (129–133). Indeed, Joachim Seelig and co-workers have quantitatively dissected the interactions of membrane-association into an attractive electrostatic component and a hydrophobic insertion (with a partitioning constant around of the latter of 1,000 M−1) (130, 134). Notably, the apparent membrane association is more than an order of magnitude increased for bilayers mimicking the overall anionic composition of bacterial membranes (66). However, this relatively modest membrane association is boosted by electrostatic attraction to negatively charged surfaces which can dominate the association process (122, 135). Furthermore, the membrane association of multicationic antimicrobial peptides has been suggested to result in the interference of electrostatic attractions that keep peripheral membrane proteins in place and consequently antimicrobial action (68).
Magainin 2—PGLa Mixtures Show Synergistic Enhancement in Biological Assays
Interestingly combinations of antimicrobial compounds are sometimes much more potent than the individual components (49, 136). These observations have been made with mixtures of peptides (137), of peptides with conventional antibiotics (41, 138–142), with blood plasma components (143), or with ions (144). In particular magainin 2 has been shown to interact synergistically with PGLa (45) and with tachyplesin I a cyclic β-sheet peptide (145).
Because magainin and PGLa peptides (Table 1) are produced together and stored as a cocktail in the skin of Xenopus laevis frogs, very early on antimicrobial assays have been performed with mixtures of the two peptides. It was soon realized that the peptides exert cooperative behavior (26, 44, 45, 146, 147). These studies involved E. coli (24, 26, 43, 148), mitochondria (43), tumor cells (26), calcein loaded liposomes (26, 45), cytochrome oxidase liposomes (24), and hamster spermatozoa (44). Notably, in the latter case only the mixture exhibits detectable activity at all. More recent papers also report synergism of magainin and PGLa on a few selected bacterial species (E. coli, S. aureus, and S. epidermis) with somehow varying enhancement factors (102, 103, 119, 149).
In particular, the early investigations revealed the poration of membranes, loss of membrane potential, uncoupling of membranes concomitant with effects on respiratory control and thereby interference with energy production and cell survival [e.g., (26, 44, 45, 146)]. In a most recent investigation it was shown that both PGLa and magainin are capable to form fibers at physiological conditions (150). These fibers are somewhat less active in antimicrobial assays but maintain the synergy. Because the peptides are stored in the granules of the frog skin at high concentrations it was speculated that they form functional amyloids for protease protection and graded release (150).
In further studies these investigations have been extended to variants of the peptides and/or other bacteria and the combination index CI has been used as a quantitative measure of synergism (103, 151):
where MICi is the MIC value determined for peptide i alone (i = a or b), and MICa+b is the total peptide concentration at the MIC determined for the combination. For values <1 synergistic enhancement occurs whereas values > 1 correspond to antagonism (152). In other work the fractional inhibitory concentration index (FICI) has been used which follows a related definition (102).
In order to analyze bacterial growth curves in a quantitative manner, we have early on started to fit the dose-dependence curves to sigmoidal functions of the type:
where cp is the peptide concentration, G is the growth, m the slope, and Gmax is the maximal growth of the bacteria. In a subsequent step the additive curves (CI = 1) are simulated, which can then be compared to the observed dose-dependent bacterial killing when both peptides are present in the mixture (103). Notably during the transition, the normalized bacterial density (y-axis) is particularly sensitive to even small changes in peptide concentration. Because of the steep dependence of bacterial growth on peptide concentration a small error in peptide concentration (along the x-axis) translates into a pronounced standard deviation when the growth is measured in replicates [i.e., a big error bar in y-axis; cf. (103)].
Whereas, in case of a steep dose-response the slope m of the sigmoidal cannot be calculated from a 2-fold dilution series and was initially chosen arbitrarily (103) it turns out that this cooperativity index bears additional information of interest once more data points around the MIC50 reveal interesting differences between the peptides. Figure 2 and Table 2 present so far unpublished data and a more elaborate scheme for the investigation of antimicrobial activities and synergism. Instead of the often applied 2-fold dilution series more data points are included (1.5 dilution series) thus revealing the slope m of the transition.
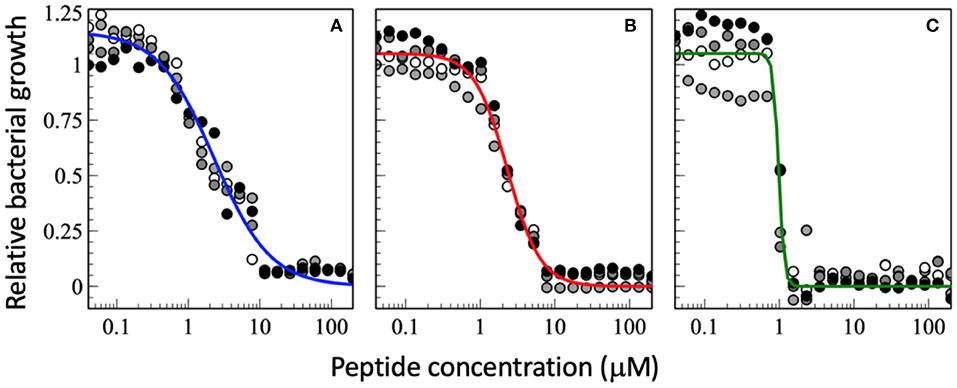
Figure 2. Dose-response curve of relative bacterial growth in the presence of (A) magainin 2a, (B) PGLa, and (C) the equimolar mixture of both. Bacterial supensions in MH medium (8.3 x 105 CFU/mL) are added to a serial dilution of peptides and the optical density at 600 nm is recorderd after an 18 h inclubation at 37°C. The experiments were performed on 96-well microplates (F-bottom sterile non-treated polystyrene, Thermo Scientific Nunc A/S, Roskilde, Denmark). Starting from a 200 μM peptide concentration a sequential dilution series was performed with a dilution factor of 1.5 in 22 steps yielding final peptide concentrations ranging from 200 to 0.040 μM (after addition of bacteria). Each condition was done in quadruplet and each experiment is represented by different symbols in the plots (i.e., white, gray, and black circles represent a different experiment). The sequential dilution series were normalized to the bacterial growth without treatment on the same plate. The data shown have not been published before.
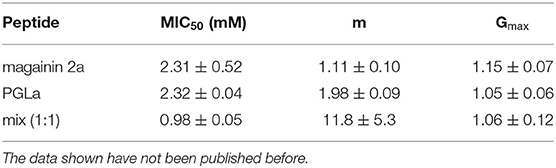
Table 2. This table shows the fit parameters of the blue, red, and green curves of Figure 2.
Four individual dilution series were fitted to Equation (2), from which averages for the MIC50, the slope m, and Gmax were calculated and these were used to calculate the fits represented by the continuous lines in blue, red, and green.
It has been shown that at sub-MIC only a fraction of bacteria is killed and their debris including anionic polymers from the cell interior neutralizes much of the peptide added (153, 154). As a consequence, with some delay the survivors catch up in such laboratory suspensions and may reach the full cell density (151). Given the complete coverage of bacteria with peptides at the cell densities used in standardized antimicrobial activity assays, it is unlikely that the fractional killing is due to statistical variations in peptide density. Therefore, it has been suggested that fractional killing is a consequence of phenotypic variations of single cells which makes them more or less susceptible to the antibacterial activity of AMPs (153). The same authors have estimated that under sub-MIC conditions shortly after peptide addition a relatively small number of bacteria persists in a well thus such fluctuations potentially become apparent.
Therefore, in the experiments shown in Figures 2, 3 the incubation of bacteria after addition of peptide was kept to 18 h which prevents that bacteria that are delayed in their growth by the peptides can catch up to full density. It is also interesting to note that the values Gmax > 1 indicate that at low peptide concentrations and low to standard inoculum (<106 CFU/mL) addition of peptide stimulates the bacterial growth.
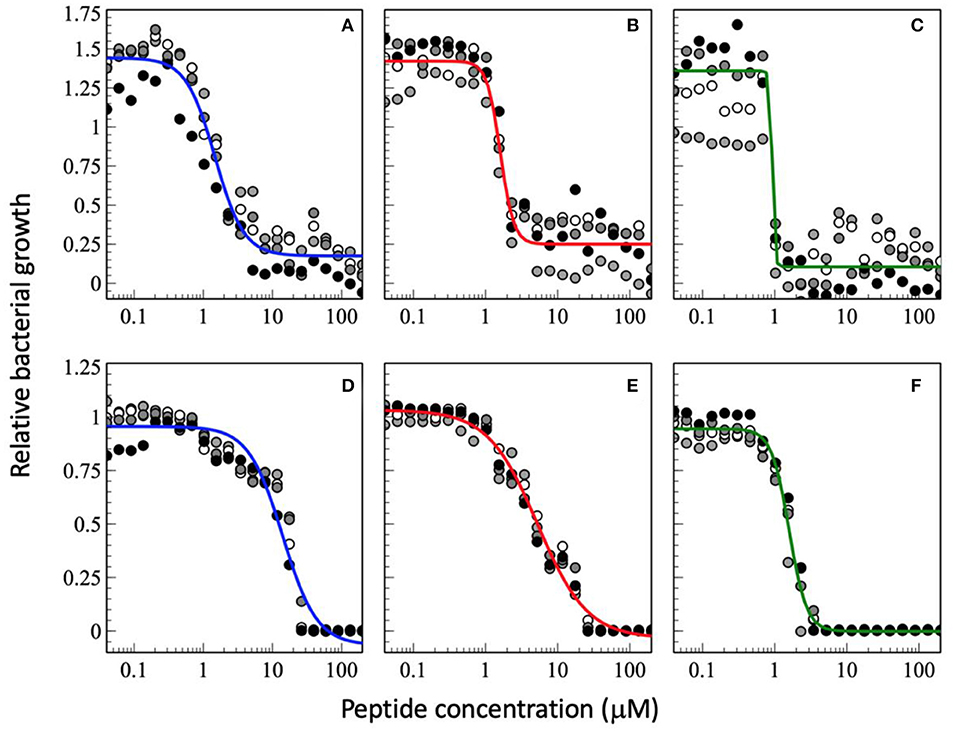
Figure 3. Dose-response curve of relative bacterial growth in the presence of antimicrobial peptides as a function of inocculum. The peptides were added to a bacterial supension in MH medium at low inocculum of 1.46 x 102 CFU/mL (A–C) or at high inocculum at 1.51 x 107 CFU/mL (D–F) and the optical density at 600 nm recorderd after an 18 h inclubation at 37°C. The peptides tested are magainin 2 (A,D), PGLa (B,E), and the equimolar mixture of both (C,F). The experimental conditions are those of Figure 2. The data shown have not been published before.
Interestingly the slope m in the presence of PGLa is different from that of magainin (Figure 2). Most strikingly for the synergistic mixture the onset of activity is not that different but it has the deepest slope which brings the MIC100 from about 10 μM for the individual peptides to about 1 μM for the mixture. This 10-fold synergistic enhancement when the MIC100 is taken as an indicator decreases to a factor of 2.3 when the 50% killing is taken into consideration, i.e., the midpoint of the transition (Figure 2). Thus, a quantitative evaluation of synergy not only depends on the detailed experimental conditions but also the very details of how bacterial killing or growth are evaluated.
Because near the MIC50 only a fraction of the bacteria is killed the survivors are selected and take over, which may cause resistance development (153). It has been suggested that AMPs have a high Hill coefficient which explains why bacteria are less prone to develop resistance development (23). Here we show that the synergistic magainin 2/PGLa mixture exhibits even steeper slopes and hence adds an additional advantage (Figures 2, 3). It is possible that because the peptides affect bacterial subpopulations differently, there are less survivors that can fill the gap. The data also suggest that at least part of what is considered the synergistic interaction between the peptides is related to the steeper slope of the dose-response.
In a general sense m means some sort of cooperative or anti-cooperative behavior (i.e., non-linearity) (155) and in the case of a cooperative coefficient indicates that more than one molecule is required for a given activity. This can happen through oligomerization, but it could also mean that several peptides have to come into proximity on the bacterial surface (79), or that they form loosely packed lipid-peptide supramolecular structures (69). On the other hand, the slope of the curve can flatten should aggregation occur in solution at high peptide concentrations.
When the cell density is increased the MIC50 increases (Figures 3D–F, so far unpublished data). This is well-known phenomenon and has been associated with peptide adsorption to cell debris including anionic biopolymers and membranes (153, 156). Furthermore, cooperativity, especially of PGLa is reduced when the CFU per ml are increased. This probably reflects a larger phenotypic variability of cells with differences in AMP susceptibility (153). Furthermore, the synergy factor obtained by comparing the MIC50 values increases to 4.8 whereas it remains around 10 when the MIC100 are considered. Because at very high inoculum the peptides distribute on many more cells it may be more difficult to reach a high enough density to kill (154, 157, 158). Thereby, one may speculate that at high cell density the increased binding affinity and the mesophase structures when the two peptides are added conjointly (69) help to reestablish local hot spots and synergy. In this context it is noteworthy that the peptides have been shown to redistribute unevenly on the bacterial surface (159). Alternatively, it may also be possible that at high cell densities the ratio of surviving subpopulations increases thus it is easier to replenish the bacteria killed by a single antibiotic. This phenomenon would be less apparent when the combined action of two peptides keeps killing a large fraction of bacteria. Clearly, more experiments are needed to elaborate on these observations.
Interestingly, the best MICs are in the 1 μM range which agrees with observations made previously (103). As a consequence the synergistic factor is usually small for peptides that exhibit already high antimicrobial activity when investigated alone (103). This is also observed for the experimental series at a low CFU of 1.46 x 102 CFU/mL where the synergy factor by comparing the MIC50 is 1.7 but again higher for the MIC100 (Figures 3A–C, so far unpublished data). However, it was recently pointed out that peptides also stick to surfaces of the test equipment thus the available concentration is probably significantly lower especially at low peptide concentrations (158).
Magainin 2—PGLa Mixtures Show Synergistic Enhancement in Model Membranes
Such synergistic enhancements in antibacterial assays can occur through specific interactions (49, 160) but also when one compound helps the antimicrobial effector to reach its target (161). Thereby the PGLa/magainin 2 case is of particular interest because it works in pure lipid model membranes where the mode of action can be studied in biophysical detail (26, 119, 122, 162, 163). When the release of fluorophores from liposomal preparations was investigated these were made of well-defined lipid compositions thus revealing interesting lipid dependences.
First of all negative charges are important to assure a high local peptide concentration close to the membrane surface and thereby and increased membrane association (130, 134). Whereas, in the absence of negatively charged lipids the membrane partitioning coefficients of AMPs are in the 103 M−1 range they apparently increase by more than an order of magnitude due to negative membrane surface charges (66). In some cases the association of antimicrobial peptides seems solely driven by electrostatic attraction until charge compensation is achieved (122, 135). This observation explains the lack of structuration when the peptides were investigated by optical techniques in dilute suspension of zwitterionic liposomes (122, 164). Electrostatic effects also contribute to their selectivity in killing of bacteria which exhibit a negative surface over healthy eukaryotic cells which appear electrically neutral to the outside (129–133). Therefore, when association and pore formation to zwitterionic membranes is very low the peptides may be too dilute in the membrane to interact with each other (119).
When fluorophore release and peptide synergy are investigated in a P/L ratio-dependent manner it becomes clear that a quantitative evaluation of synergy also depends on the detailed conditions of the experiment (Figure 4). For example, inspection of Figure 4B shows the absence of synergy at low peptide concentrations, but this value continuously increases at ≥0.4 μM. Notably either the amount of leakage after a few minutes (102, 119, 162) or the initial leakage rate (45, 163) have been taken as indicators of synergy.
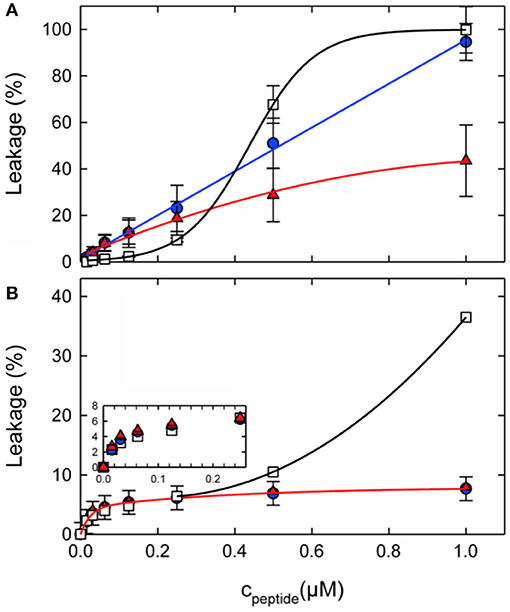
Figure 4. Calcein leakage from 100 nm unilamellar vesicles made from (A) POPC/POPG (3:1 mol/mol) or (B) POPE/POPG (3:1 mol/mol) as a function of peptide concentration. L18W-PGLa (blue circles), magainin 2a (red triangles), or their equimolar mixture (open squares) were added to 50 μM lipid. The lines were added to guide the eye. Taken from Leber et al. (119).
From a combination of fluorescence-based biophysical experiments Matsuzaki and co-workers suggested early on that the rate of pore formation is slower for magainin, but the openings are more stable than those of PGLa (162). Thereby, synergism arises from fast pore formation and moderate stability. A more recent paper by Heerklotz and co-workers suggests that the combination of making large enough pores that are well distributed among the liposomes causes synergism in vesicle dye release experiments (165). According to these models the size of the vesicles or of the bacterial cells and the heterogeneity of the peptide distribution have an effect on the observed synergism (165).
On Definitions to Quantitatively Define Synergistic Activity
A quantitative comparison of synergistic enhancement is further complicated not only due to differences in the experimental systems investigated but also by the different definitions used to quantitatively compare the activities of the mixture and individual peptide solutions. For example, Matsuzaki and co-workers compare the P/L ratio where 50% of the dye is released (162): .
In this definition the calculated values are the average of the P/L50 ratios obtained from the individual peptides. This approach is similar to the calculated MICs published by Glattard et al. (103).
In contrast, rather than comparing the peptide concentration needed to reach a defined functional activity (e.g., 50% leakage) Zerweck et al. use the (variable) leakage values (L) at a fixed 100 μM lipid concentration and a fixed P/L. Typically conditions are chosen where a good range of activities is observed (i.e., 3 μM peptide for PE/PG/CL 72/23/5 and 0.6 μM for PC/PG 3/1). From this data a synergy factor is calculated according to SF .
Furthermore, from the same team the P/L of the mixture was twice that of the individual peptides in one publication (102) but kept constant in a prior paper (163). Finally, Leber et al. use “Peptide Synergy” Σ = 1/SF at the “highest peptide concentration” (cf. Figure 4 showing data up to 1μM) and 50 μM lipid (119). The total peptide concentration when peptides are tested individually is half of that of the mixture. This definition of synergy is based on the assumption that a heterodimer complex forms (G. Pabst, personal communication). While all of these definitions have their justification comparing the data quantitatively becomes impossible. For example, for PE-rich membranes the values include Σ =0.4 (corresponding to SF=2.5) (119) and SF = 22 (102). Furthermore, the PGLa-driven calcein release from PC/PE/PS 2:5:3 vesicles requires 25 to 43-fold less PGLa when 3.7 μM magainin are present (50 μg/ml lipid in 0.5 M NaCl, 10 mM PIPES, pH 7) (164). At this concentration magainin exhibits no activity when added alone (164).
Correlating Synergism With Structural Investigations and Lipid Composition
Structural investigations were performed to develop a model how the peptides interact and thereby enhance their antimicrobial/pore-forming activities. When studied by solid-state NMR methods PGLa and magainin are both aligned along the surface of membranes when these carry at least one unsaturation per phospholipid (98, 99, 116, 166), including E. coli lipid extracts (103). Thus, in such membranes the helix topology resembles those of magainin or PGLa alone, i.e., in the absence of the other peptide (89, 98). In contrast, when magainin 2/PGLa mixtures are studied in fully saturated lipid bilayers magainin remains oriented along the membrane surface whereas PGLa adopts transmembrane alignments (98, 99). However, it should be noted that unsaturations are abundant in biological membranes thus a mechanism for synergistic antibacterial activities should consider an alignment of both peptides along the bilayer surface (98, 101, 103).
Not only the lipid fatty acyl chain but also the lipid head group composition seems of considerable importance for the synergistic enhancement to develop. Notably, in recent investigations it was demonstrated that for lipids with intrinsic negative curvature such as PE or PC/cholesterol the pore forming activity of the individual peptides is reduced when compared to PC/PG membranes. However, much of the activity was restored when adding the peptide mixture. This behavior results in a significant synergetic enhancement of activities in PE/PG but not in PC/PG membranes (Figure 4). In this context it should be noted that the antimicrobial activities of PGLa or magainin individually are much lower in PE/PG than in PC/PG, thereby synergetic enhancement in the bacterial membrane mimetic abolishes the differences observed for the individual peptides when investigated in membranes of different composition (cf. Figure 4 at 1 μM). This agrees with observations made when the antimicrobial activity of magainin and PGLa as well as derivatives thereof were investigated and the highest synergy was observed for peptides with intrinsically low activities (103).
Notably, a fluorescence quenching investigation has revealed the formation of mesophase structures along the membrane surface and correlated diffusion of both peptides (69). Fluorescence quenching occurs when the fluorophores, which were added to the amino-terminus of either magainin 2 or PGLa, approach each other within the nm range, i.e., closer than expected from a statistical distribution (104). These experimental observations are in-line with a recently published MD simulation of the peptide mixture where in stacked membranes a string of interacting peptides and lipids has been observed (117). Interestingly, the observed mesophases and diffusion correlation come along with a much increased membrane partitioning of the peptides when the mixture is investigated in PE/PG but not in the presence of PC/PG membranes (69). Because the membrane disruptive properties of magainin 2 extends over several layers of lipids (ca 0.8 nm in diameter) (112, 113) the formation of mesophases with interpeptide distances <1 nm (104) suggests a concerted action of several peptides in destabilizing the membrane (69). In this context is it interesting that microscopic imaging approaches have revealed an uneven distribution of antimicrobial peptides with preferential association with curved regions of the bacterial membrane such as pole caps or the septum of dividing cells (47, 77).
During early work on magainin/PGLa synergism little attention was given to the lipid compositions of the model systems investigated. With new data pointing to important effects of lipids exhibiting intrinsic negative membrane curvature it is of interest to review earlier publications. Indeed, the very early investigations were performed in PC/dicetylphosphate membranes of different (PC/DCA ratios) where dicetylphosphate is a small negatively charged “head group” made of phosphate carrying two C16 chains (26, 45). Thus, in a lipid membrane the molecule exhibits an inverted cone shape which is associated with negative curvature [cf. (122)]. Furthermore, experiments were performed with azolectin a soy bean lipid mixture which contains PC, PE, and PIs (45). Synergistic enhancements of fluorophore release were also observed with BBPS or egg-PC membranes (162), and thereby do not seem to fit the requirements of negative curvature observed by Leber et al. (119). However, it should be noted that the synergy factors published in this work are only 3.5 and thereby relatively low when compared to the antibacterial tests presented in the same paper where a factor of 8 was observed (162). For EYPC very high P/L ratios of 0.57, 0.14, and 0.05, respectively, were required to measure the S value. Thereby, although typically 100 nm vesicles are used for most of the dye release experiments (102, 119, 162) the detailed conditions are otherwise quite different and the values may not easily be compared, in particular as the dose response curves are not linear (Figure 4).
Biophysical Measurements OF PGLa—Magainin Interactions in Membranes
Spectral changes that occur upon membrane partitioning can be used to derive membrane association constants of polypeptides (69, 135, 167, 168). From such data the interaction between the membrane associated peptides can be derived including quantitative estimates of the energies involved. However, these values are only valid in the context of an assumed molecular model. In this manner favorable PGLa-magainin interaction energies were obtained in egg-PG membranes (162). Notably the quantitative evaluation of the data depends how many peptides are assumed to be involved in the interaction process. Energies for homo- and hetero-dimer formation have also been published for fully saturated membranes where PGLa exhibits a transition into the TM state (163). Therefore, this analysis probably includes many energy contributions (101, 169) which remain unimportant when synergy occurs between peptides that reside along the membrane surface of a bacterial membrane (103).
In a more recent investigation association of the two peptides with LUVs made of POPE/POPG 3/1 at pH 7 as a bacterial membrane mimetic were investigated by Isothermal Titration Calorimetry (ITC) (70) indicating strong membrane association with apparent membrane association constants in the 106 M−1 range (apparent stoichiometry P/L ≈ 1.7 mole%). Whereas, the membrane association of magainin and PGLa is characterized by an endothermic reaction enthalpy an additional exothermic contribution becomes apparent when the peptide mixture is titrated. Thus, these ITC data reveal an additional model-independent ΔH in the range of −2 kcal/mole in the magainin 2/PGLa mixture when compared to the peptides individually. This reaction enthalpy could be due to e.g., vesicle agglutination as observed by DLS experiments conducted on the same system (70). Peptide-driven intermembrane interactions were also apparent by a reduction in the bilayer repeat distance of mechanically oriented membranes (110, 117). Furthermore, the formation of loosely packed supramolecular assemblies also contributes to an energetic contribution in particular as these have been shown to correlate with an order of magnitude increased membrane affinity in the presence of POPE/POPG (69). Notably, when the proximity and thereby the interactions between magainin 2 and PGLa when associated to POPE/POPG 3/1 or POPC/POPS 3/1 membranes was tested by fluorescence spectroscopy a FRET effect was observed at high P/L ratios where close encounters of the peptides happen statistically (170). However, this effect diminishes and disappears when diluting the peptide with more lipid indicating that possible interactions between the peptides are rather weak.
Sequence Specificity of Magainin 2—PGLa Synergy
In order to define key structural elements of the synergistic interactions and residues that may be involved in the magainin 2—PGLa interactions the peptide sequences have been modified and tested. When the F16W or E19Q modifications of magainin 2 were studied for fluorophore release from egg PC/PG (1:1) liposomes a reduced synergistic activity was observed whereas the F5W alteration did not exhibit any effect (162). Introducing a positive charge at position 19 much abolished the synergistic interactions (102). In contrast, neutralizing the magainin 2 carboxyterminus by amidation has no effect on its synergism with PGLa (102, 103, 170, 171) although the antimicrobial activity of the peptide is increased (171, 172). Similar observations were made with the hydrophobic face of the magainin 2 helix (173).
Furthermore, when PGLa is modified the synergistic activity with magainin 2 is maintained even when a proline is added to the N-terminus and a negative charge to the C-terminus of PGL (171). When key residues of PGLa were searched exchanging the positive charges of lysines 15 and 19 by glutamines abolish synergistic enhancement whereas a more moderate reduction is measured for K5E and K12E (102) and L18W is tolerated (119, 162).
Such potential electrostatic interactions between the membrane-associated peptides have been suggested early on and are visualized in coarse grain and all-atom MD simulations even though the membranes used in the older simulations do not fulfill the criteria of a physiological composition (116, 117, 174, 175).
Furthermore, residues G7, G11, and L18 of PGLa have been shown important (102). Glycines 7 and 11 form a GxxxG motif which has been suggested to promote dimerization in highly apolar environments (176). However, the PGLa localization at the membrane interface (98, 103) may not be suitable for stable PGLa homodimer interactions. A detailed structural analysis of the peptide mixture in a lipid bilayer is required to resolve such ambiguities. In summary, the charges located at the carboxyterminal ends of the peptides have the strongest effects on synergy and an important role was also associated with G7, G11, and L18 of PGLa which needs further investigation.
Lessons Learned From Peptide Dimers
In previous publications the question how the oligomerization of peptides along the membrane surface influences activity was already discussed (122, 177). In the context of models proposed for the mechanisms of synergistic enhancement the comparison with covalently linked dimers is also of interest. Therefore, in a first step possible interactions between membrane-associated PGLa and magainin 2 were tested by preparing peptides carrying GGC extensions and cross linking experiments (178). In PC/PG (1/1) bilayers parallel dimers preferentially form (178), therefore, dimers linked through C-terminal GGC extensions were prepared and investigated. When compared to the same amount of unmodified peptides in a mixture all of, the (PGLa-GGC)2 and (magainin-GGC)2 homodimers as well as the magainin-GGC/PGLa-GGC heterodimer were all more active in calcein release experiments from POPE/POPG 3/1 liposomes (119). However, only the PGLa-homodimer and the PGLa-magainin heterodimer, but not the individual peptides or their mixture, showed significant dye release from POPC/Cholesterol 3/1 liposomes (119). When the antimicrobial activities of the wildtype magainin/PGLa mixture and the dimer of the GCC-derivatives are compared to each other a complication arises from the fact that the GGC extensions itself make the monomeric peptides more active (103, 149). Furthermore, a dimer linked through a carboxyterminal lysine extension was considerably more active than the monomer whereas the amino-terminal linkage through glutamic acid has no effect (179).
Furthermore, a cystine-linked magainin 2 dimer has been shown more active in membrane permeabilization and antimicrobial activities (180). From the ensemble of data it seems that the increased activity of the dimers is based on an increased membrane perturbation by the larger peptide aggregates rather than due to a particular structure of the PGLa-magainin 2 dimer (122, 180, 181). In line with such a model is the observation that the homotarsinin homo-dimer (2 x 24 residues) is more active in antibacterial assays than the corresponding amount of monomer (181).
Another dimer that has been studied is distinctin a heterodimer which forms a compact dimer of two dimers in solution which has been suggested to protect the protein from proteolytic digestion (182) thereby resulting in a slightly increased antimicrobial activity of the dimer when compared to the monomers (183). Its solution structure unfolds in the presence of membranes thus the 25-residue chain 2-helix partitions into the membrane parallel to the membrane surface. In contrast, the 22-residue chain 1 associates only loosely with the membrane (92, 184). Thereby the dimer acts similar to a monomeric linear cationic peptide.
Model for Synergistic PGLa—Magainin Interactions
In order to develop a model for antimicrobial synergism focus should be on structural data obtained in membranes carrying unsaturations and PE head groups such as they occur in bacterial membranes. Structural data indicate that the highly cationic peptides adopt amphipathic α-helical conformations and that these peptides are aligned parallel to the membrane surface (98, 101, 103). MD simulation and diffraction data show that the peptides partition into the interface of POPE/POPG 3/1 membranes with the large hydrophobic face of PGLa being inserted somewhat deeper than magainin 2 (103, 116, 117). Electrostatic interactions involving the dipolar charge distribution along the peptides (102, 103, 162) and interactions involving anionic lipids (104) and/or anions of the surrounding buffers (185) help in the formation of nematic phase arrangements along the membrane surface (Figure 1).
Furthermore, when the peptides partition into the membrane interface they have been shown to have a large disordering effect on the fatty acyl chains of the surrounding lipids (101). Interestingly, such and related changes in the membrane packing and structure have been postulated to result in lipid-mediated interactions over several molecular layers (186–188). The peptides disturb the finely tuned equilibrium of van-der Waals, hydrogen bonding and electrostatic interactions, entropic contributions of the lipid fatty acyl chains and of the membrane-associated water, that assure the lipid bilayer packing into well-defined supramolecular arrangements. Therefore, it is possible that the pronounced lipid disordering observed in the presence of magainin 2, PGLa, and the mixture constitutes an important driving force to bring peptides into closer proximity (Figure 5). Thereby new supramolecular arrangements of the lipids and peptides form which have been detected by fluorescence quenching techniques and MD simulations (69, 117). Notably, zones of high peptide density have also been observed when bacteria were imaged (159) but these measurements work on very different length scales and it is not clear if these observations correlate. Notably, the formation of supramolecular arrangements within POPE/POPG membranes and the correlation observed between PGLa and magainin 2 comes along with an order of magnitude increase in membrane partitioning of the peptides (Figure 5). The much-increased membrane affinity due to the presence of the other peptide by itself could explain the synergistic enhancement of activities, but the formation of peptide-lipid mesophases results in high local peptide concentrations which can also involve a modulation of activities.
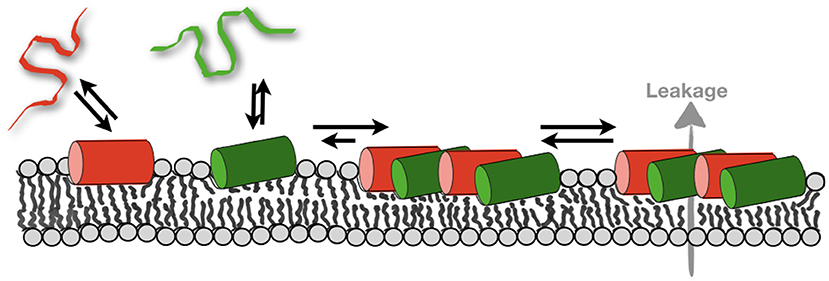
Figure 5. Schematically illustrates the membrane partitioning equilibria of magainin 2 (red) and PGLa (green). The formation of mesophases deletes the pool of monomeric peptides thereby more peptide can bind. As a consequence, the total amount of membrane-associated peptide increases and supramolecular structures that facilitate leakage form (69).
While extensive studies have been performed to define the range of pathogens and tumor cells susceptible to be killed by magainin antimicrobial peptides or the lack of toxicity against healthy human (or frog) cells (61, 62, 172, 189) much less data on toxicity or antimicrobial action are published about the synergistic mixture of PGLa and magainin 2 (26, 44, 190). For the individual peptides selectivity of bacteria over healthy eukaryotic cells has been explained by the negatively charged surface of bacteria and a high negative inside membrane potential of their plasma membrane assuring a high density of polycationic peptides at the bacterial membrane (129, 191). Furthermore, eukaryotic cells are protected from membrane lysis by these peptides due to the presence of cholesterol (62, 192). Because in this model the preferential killing of bacteria over healthy human cells is driven by the physico-chemical properties of the membrane similar considerations should also be applicable to the peptide mixtures. In this context it is notable that the synergistic enhancement of activities works for PE-rich (bacterial) membranes but not when this lipid is replaced by PC (eukaryotic membranes). Thereby, such biophysical findings suggest that the therapeutic window of these peptides potentially increase when added in combination.
Author Contributions
EG and DJ performed experiments. BB wrote the manuscript. All authors contributed to writing the manuscript and in the preparation of Figures.
Funding
The financial support by the Agence Nationale de la Recherche (projects MemPepSyn 14-CE34-0001-01, Biosupramol 17-CE18-0033-3 and the LabEx Chemistry of Complex Systems 10-LABX-0026_CSC), the University of Strasbourg, the CNRS, the Région Alsace and the RTRA International Center of Frontier Research in Chemistry is much appreciated.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Lorenzo Stella for valuable discussion.
Abbreviations
AMP, antimicrobial peptide; ATR FTIR, attenuated total reflection Fourier transform infrared; CFU, colony-forming unit; CD, circular dichroism; CL, cardiolipin; DCP, dicetylphosphate; DOPC, 1, 2-dioleoyl-sn-glycero-3-phosphocholine; DOPG, 1, 2-dioleoyl-sn-glycero-3- phospho-(1′-rac-glycerol); GUV, giant unilamellar vesicle; ITC, isothermal titration calorimetry; LUV, large unilamellar vesicle; MD, molecular dynamics; MIC, minimal inhibitory concentration; NMR, nuclear magnetic resonance; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PS, phosphatidylserine; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine; POPG, 1-palmitoyl-2-oleoyl -sn-glycero-3- phospho-(1′-rac-glycerol); POPS, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine; SMART, Soft Membranes Adapt and Respond, also Transiently; TM, transmembrane.
References
1. Boman HG. Peptide antibiotics and their role in innate immunity. Annu Rev Immunol. (1995) 13:61–92. doi: 10.1146/annurev.iy.13.040195.000425
2. Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. (2002) 415:389–95. doi: 10.1038/415389a
3. Aldridge S, Parascandola J, and Sturchio JL. The Discovery and Development of Penicillin 1928-1945. (1999). Royal Society of Chemistry, London and National Historic Chemical Landmarks Program of the American Chemical Society. Available online at: https://www.acs.org/content/acs/en/education/whatischemistry/landmarks/flemingpenicillin.html
4. Dubos RJ, and Hotchkiss RD. The production of bactericidal substances by aerobic sporulating bacilli. J Exp Med. (1941) 73:629–40. doi: 10.1084/jem.73.5.629
6. Gall YM, and Konashev MB. The discovery of Gramicidin S: the intellectual transformation of G.F. Gause from biologist to researcher of antibiotics and on its meaning for the fate of Russian genetics. Hist Philos Life Sci. (2001) 23:137–50.
7. Leitgeb B, Szekeres A, Manczinger L, Vagvolgyi C, and Kredics L. The history of alamethicin: a review of the most extensively studied peptaibol. Chem Biodivers. (2007) 4:1027–51. doi: 10.1002/cbdv.200790095
8. Rautenbach M, Troskie AM, and Vosloo JA. Antifungal peptides: to be or not to be membrane active. Biochimie. (2016) 130:132–45. doi: 10.1016/j.biochi.2016.05.013
9. Agerberth B, Gunne H, Odeberg J, Kogner P, Boman HG, and Gudmundsson GH. FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc Natl Acad Sci USA. (1995) 92:195–9. doi: 10.1073/pnas.92.1.195
10. Kiss G, and Michl H. Öber das Giftsekret der Gelbbauchunke Bombina variegata L. Toxicon. (1962) 1:33–9. doi: 10.1016/0041-0101(62)90006-5
11. Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA. (1987) 84:5449–53. doi: 10.1073/pnas.84.15.5449
12. Juhl DW, Van Rensburg W, Bossis X, Vosloo JA, Rautenbach M, and Bechinger B. Tyrocidine A interactions with saccharides investigated by CD and NMR spectroscopies. J Pept Sci. (2019) 25:e3163. doi: 10.1002/psc.3163
13. Pirtskhalava M, Gabrielian A, Cruz P, Griggs HL, Squires RB, Hurt DE, et al. DBAASP v.2: an enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res. (2016) 44:D1104–12. doi: 10.1093/nar/gkv1174
14. Wang G, Li X, and Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. (2016) 44:D1087–93. doi: 10.1093/nar/gkv1278
15. Liu S, Fan L, Sun J, Lao X, and Zheng H. Computational resources and tools for antimicrobial peptides. J Pept Sci. (2017) 23:4–12. doi: 10.1002/psc.2947
16. Aguilera-Mendoza L, Marrero-Ponce Y, Garcia-Jacas CR, Chavez E, Beltran JA, Guillen-Ramirez HA, et al. Automatic construction of molecular similarity networks for visual graph mining in chemical space of bioactive peptides: an unsupervised learning approach. Sci Rep. (2020) 10:18074. doi: 10.1038/s41598-020-75029-1
17. Sansom MSP. The biophysics of peptide models of ion channels. Prog Biophys Mol Biol. (1991) 55:139–235. doi: 10.1016/0079-6107(91)90004-C
18. Bechinger B. Structure and functions of channel-forming polypeptides: magainins, cecropins, melittin and alamethicin. J Membr Biol. (1997) 156:197–211. doi: 10.1007/s002329900201
19. Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. (2003) 348:1342–7. doi: 10.1056/NEJMoa025025
20. Koo HB, and Seo J. Antimicrobial peptides under clinical investigation. J Peptide Sci. (2019) 111:e24122. doi: 10.1002/pep2.24122
21. Mookherjee N, Anderson MA, Haagsman HP, and Davidson DJ. Antimicrobial host defence peptides: functions and clinical potential. Nat Rev Drug Discov. (2020) 19:311–32. doi: 10.1038/s41573-019-0058-8
22. Bechinger B, and Gorr SU. Antimicrobial peptides: mechanisms of action and resistance. J Dent Res. (2017) 96:254–60. doi: 10.1177/0022034516679973
23. Lazzaro BP, Zasloff M, and Rolff J. Antimicrobial peptides: Application informed by evolution. Science. (2020) 368:eaau5480. doi: 10.1126/science.aau5480
24. Juretic D, Chen HC, Brown JH, Morell JL, Hendler RW, and Westerhoff HV. Magainin 2 amide and analogues. Antimicrobial activity, membrane depolarization and susceptibility to proteolysis. FEBS Lett. (1989) 249:219–23. doi: 10.1016/0014-5793(89)80627-1
25. Resnick NM, Maloy WL, Guy HR, and Zasloff M. A novel endopeptidase from Xenopus that recognizes alpha-helical secondary structure. Cell. (1991) 66:541–54. doi: 10.1016/0092-8674(81)90017-9
26. Westerhoff HV, Zasloff M, Rosner JL, Hendler RW, De Waal A, Vaz G, et al. Functional synergism of the magainins PGLa and magainin-2 in Escherichia coli, tumor cells and liposomes. Eur J Biochem. (1995) 228:257–64. doi: 10.1111/j.1432-1033.1995.00257.x
27. Yang D, Zou R, Zhu Y, Liu B, Yao D, Jiang J, et al. Magainin II modified polydiacetylene micelles for cancer therapy. Nanoscale. (2014) 6:14772–83. doi: 10.1039/C4NR04405C
28. Yuksel E, and Karakecili A. Antibacterial activity on electrospun poly(lactide-co-glycolide) based membranes via Magainin II grafting. Mater Sci Eng C Mater Biol Appl. (2014) 45:510–8. doi: 10.1016/j.msec.2014.10.004
29. Reijmar K, Edwards K, Andersson K, and Agmo Hernandez V. Characterizing and controlling the loading and release of cationic amphiphilic peptides onto and from PEG-stabilized lipodisks. Langmuir. (2016) 32:12091–9. doi: 10.1021/acs.langmuir.6b03012
30. Arnusch CJ, Albada HB, Van Vaardegem M, Liskamp RMJ, Sahl HG, Shadkchan Y, et al. Trivalent ultrashort lipopeptides are potent pH dependent antifungal agents. J Med Chem. (2012) 55:1296–302. doi: 10.1021/jm2014474
31. Ghosh C, Harmouche N, Bechinger B, and Haldar J. Aryl-alkyl-lysines interact with anionic lipid components of bacterial cell envelope eliciting anti-inflammatory and anti-biofilm properties. ACS Omega. (2018) 3:9182–90. doi: 10.1021/acsomega.8b01052
32. Porter EA, Weisblum B, and Gellman SH. Mimicry of host-defense peptides by unnatural oligomers: antimicrobial beta-peptides. J Am Chem Soc. (2002) 124:7324–30. doi: 10.1021/ja0260871
33. Patch JA, and Barron AE. Helical peptoid mimics of magainin-2 amide. J Am Chem Soc. (2003) 125:12092–3. doi: 10.1021/ja037320d
34. Kuroda K, and Degrado WF. Amphiphilic polymethacrylate derivatives as antimicrobial agents. J Am Chem Soc. (2005) 127:4128–9. doi: 10.1021/ja044205+
35. Violette A, Fournel S, Lamour K, Chaloin O, Frisch B, Briand JP, et al. Mimicking helical antibacterial peptides with nonpeptidic folding oligomers. Chem Biol. (2006) 13:531–8. doi: 10.1016/j.chembiol.2006.03.009
36. Makovitzki A, Baram J, and Shai Y. Antimicrobial lipopolypeptides composed of palmitoyl Di- and tricationic peptides: in vitro and in vivo activities, self-assembly to nanostructures, and a plausible mode of action. Biochemistry. (2008) 47:10630–6. doi: 10.1021/bi8011675
37. Scott RW, Degrado WF, and Tew GN. De novo designed synthetic mimics of antimicrobial peptides. Curr Opin Biotechnol. (2008) 19:620–7. doi: 10.1016/j.copbio.2008.10.013
38. Rotem S, and Mor A. Antimicrobial peptide mimics for improved therapeutic properties. Biochim Biophys Acta. (2009) 1788:1582–92. doi: 10.1016/j.bbamem.2008.10.020
39. Palermo EF, and Kuroda K. Structural determinants of antimicrobial activity in polymers which mimic host defense peptides. Appl Microbiol Biotechnol. (2010) 87:1605–15. doi: 10.1007/s00253-010-2687-z
40. Laurencin M, Simon M, Fleury Y, Baudy-Floc'h M, Bondon A, and Legrand B. Selectivity modulation and structure of alpha/aza-beta(3) cyclic antimicrobial peptides. Chemistry. (2018) 24:6191–201. doi: 10.1002/chem.201800152
41. Rank LA, Walsh NM, Liu R, Lim FY, Bok JW, Huang M, et al. A cationic polymer that shows high antifungal activity against diverse human pathogens. Antimicrob Agents Chemother. (2017) 61:e00204-17. doi: 10.1128/AAC.00204-17
42. Giovannini MG, Poulter L, Gibson BW, and Williams DH. Biosynthesis and degradation of peptides derived from Xenopus laevis prohormones. Biochem J. (1987) 243:113–20. doi: 10.1042/bj2430113
43. Westerhoff HV, Juretic D, Hendler RW, and Zasloff M. Magainins and the disruption of membrane-linked free-energy transduction. Proc Natl Acad Sci USA. (1989) 86:6597–601. doi: 10.1073/pnas.86.17.6597
44. De Waal A, Gomes AV, Mensink A, Grootegoed JA, and Westerhoff HV. Magainins affect respiratory control, membrane potential and motility of hamster spermatozoa. FEBS Lett. (1991) 293:219–23. doi: 10.1016/0014-5793(91)81191-A
45. Vaz Gomes A, De Waal A, Berden JA, and Westerhoff HV. Electric potentiation, cooperativity, and synergism of magainin peptides in protein-free liposomes. Biochemistry. (1993) 32:5365–72. doi: 10.1021/bi00071a011
46. Moulay G, Leborgne C, Mason AJ, Aisenbrey C, Kichler A, and Bechinger B. Histidine-rich designer peptides of the LAH4 family promote cell delivery of a multitude of cargo. J Pept Sci. (2017) 23:320–8. doi: 10.1002/psc.2955
47. Savini F, Bobone S, Roversi D, Mangoni ML, and Stella L. From liposomes to cells: filling the gap between physicochemical and microbiological studies of the activity and selectivity of host-defense peptides. Pept Sci. (2018) 110:e24041. doi: 10.1002/pep2.24041
48. Zhu YY, Mohapatra S, and Weisshaar JC. Rigidification of the E. coli cytoplasm by the human antimicrobial peptide Ll-37 revealed by superresolution fluorescence microscopy. Biophys J. (2019) 116:138a. doi: 10.1016/j.bpj.2018.11.765
49. Mccafferty DG, Cudic P, Yu MK, Behenna DC, and Kruger R. Synergy and duality in peptide antibiotic mechanisms. Curr Opin Chem Biol. (1999) 3:672–80. doi: 10.1016/S1367-5931(99)00025-3
50. Holzl MA, Hofer J, Steinberger P, Pfistershammer K, and Zlabinger GJ. Host antimicrobial proteins as endogenous immunomodulators. Immunol Lett. (2008) 119:4–11. doi: 10.1016/j.imlet.2008.05.003
51. Diamond G, Beckloff N, Weinberg A, and Kisich KO. The roles of antimicrobial peptides in innate host defense. Curr Pharm Des. (2009) 15:2377–92. doi: 10.2174/138161209788682325
52. Steinstraesser L, Kraneburg U, Jacobsen F, and Al-Benna S. Host defense peptides and their antimicrobial-immunomodulatory duality. Immunobiology. (2010) 216:322–33. doi: 10.1016/j.imbio.2010.07.003
53. Cao P, Yang Y, Uche FI, Hart SR, Li WW, and Yuan C. Coupling plant-derived cyclotides to metal surfaces: an antibacterial and antibiofilm study. Int J Mol Sci. (2018) 19:793. doi: 10.3390/ijms19030793
54. Tsutsumi LS, Elmore JM, Dang UT, Wallace MJ, Marreddy R, Lee RB, et al. Solid-phase synthesis and antibacterial activity of cyclohexapeptide wollamide B analogs. ACS Comb Sci. (2018) 20:172–85. doi: 10.1021/acscombsci.7b00189
55. Zhao P, Xue Y, Gao W, Li J, Zu X, Fu D, et al. Bacillaceae-derived peptide antibiotics since 2000. Peptides. (2018) 101:10–6. doi: 10.1016/j.peptides.2017.12.018
56. Hong M, and Su Y. Structure and dynamics of cationic membrane peptides and proteins: insights from solid-state NMR. Protein Sci. (2011) 20:641–55. doi: 10.1002/pro.600
57. Salnikov E, Aisenbrey C, Balandin SV, Zhmak MN, Ovchinnikova AY, and Bechinger B. Structure and alignment of the membrane-associated antimicrobial peptide arenicin by oriented solid-state NMR spectroscopy. Biochemistry. (2011) 50:3784–95. doi: 10.1021/bi1018732
58. Rautenbach M, Troskie AM, Vosloo JA, and Dathe ME. Antifungal membranolytic activity of the tyrocidines against filamentous plant fungi. Biochimie. (2016) 130:122–31. doi: 10.1016/j.biochi.2016.06.008
59. Sychev SV, Sukhanov SV, Panteleev PV, Shenkarev ZO, and Ovchinnikova TV. Marine antimicrobial peptide arenicin adopts a monomeric twisted beta-hairpin structure and forms low conductivity pores in zwitterionic lipid bilayers. Biopolymers. (2017). doi: 10.1002/bip.23093. [Epub ahead of print].
60. Usachev KS, Kolosova OA, Klochkova EA, Yulmetov AR, Aganov AV, and Klochkov VV. Oligomerization of the antimicrobial peptide Protegrin-5 in a membrane-mimicking environment. Structural studies by high-resolution NMR spectroscopy. Eur Biophys J. (2017) 46:293–300. doi: 10.1007/s00249-016-1167-5
61. Jacob L, and Zasloff M. Potential therapeutic applications of magainins and other antimicrobial agents of animal origin. [Review]. Ciba Found Sympos. (1994) 186:197–216; discussion 216. doi: 10.1002/9780470514658.ch12
62. Maloy WL, and Kari UP. Structure-activity studies on magainins and other host defense peptides. Biopolymers. (1995) 37:105–22. doi: 10.1002/bip.360370206
63. Bechinger B, Kim Y, Chirlian LE, Gesell J, Neumann JM, Montal M, et al. Orientations of amphipathic helical peptides in membrane bilayers determined by solid- state NMR spectroscopy. J Biomol NMR. (1991) 1:167–73. doi: 10.1007/BF01877228
64. Matsuzaki K, Murase O, Tokuda H, Funakoshi S, Fujii N, and Miyajima K. Orientational and aggregational states of magainin 2 in phospholipid bilayers. Biochemistry. (1994) 33:3342–9. doi: 10.1021/bi00177a027
65. Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, and Huang HW. Membrane pores induced by magainin. Biochemistry. (1996) 35:13723–8. doi: 10.1021/bi9620621
66. Wieprecht T, Apostolov O, Beyermann M, and Seelig J. Membrane binding and pore formation of the antibacterial peptide PGLa: thermodynamic and mechanistic aspects. Biochemistry. (2000) 39:442–52. doi: 10.1021/bi992146k
67. Shai Y. Mechanism of the binding, insertion, and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective lytic peptides. Biochim Biophys Acta. (1999) 1462:55–70. doi: 10.1016/S0005-2736(99)00200-X
68. Wenzel M, Chiriac AI, Otto A, Zweytick D, May C, Schumacher C, et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc Natl Acad Sci USA. (2014) 111:E1409–18. doi: 10.1073/pnas.1319900111
69. Aisenbrey C, Amaro M, Pospisil P, Hof M, and Bechinger B. Investigation of the supramolecular complexes formed by Magainin 2, PGLa and lipids. Sci Rep. (2020) 10:11652. doi: 10.1038/s41598-020-68416-1
70. Marquette A, and Bechinger B. Biophysical investigations elucidating the mechanisms of action of antimicrobial peptides and their synergism. Biomolecules. (2018) 8:18. doi: 10.3390/biom8020018
71. Duclohier H. Antimicrobial peptides and peptaibols, substitutes for conventional antibiotics. Curr Pharm Des. (2010) 16:3212–23. doi: 10.2174/138161210793292500
72. Islam MZ, Alam JM, Tamba Y, Karal MS, and Yamazaki M. The single GUV method for revealing the functions of antimicrobial, pore-forming toxin, and cell-penetrating peptides or proteins. Phys Chem Chem Phys. (2014) 16:15752–67. doi: 10.1039/C4CP00717D
73. Gregory SM, Cavenaugh A, Journigan V, Pokorny A, and Almeida PFF. A quantitative model for the all-or-none permeabilization of phospholipid vesicles by the antimicrobial peptide cecropin A. Biophys J. (2008) 94:1667–80. doi: 10.1529/biophysj.107.118760
74. Tamba Y, Ariyama H, Levadny V, and Yamazaki M. Kinetic Pathway of antimicrobial peptide Magainin 2-induced pore formation in lipid membranes. J Phys Chem B. (2010) 114:12018–26. doi: 10.1021/jp104527y
75. Barns KJ, and Weisshaar JC. Real-time attack of LL-37 on single Bacillus subtilis cells. Biochim Biophys Acta. (2013) 1828:1511–20. doi: 10.1016/j.bbamem.2013.02.011
76. Rangarajan N, Bakshi S, and Weisshaar JC. Localized permeabilization of E. coli membranes by the antimicrobial peptide Cecropin A. Biochemistry. (2013) 52:6584–94. doi: 10.1021/bi400785j
77. Yang Z, Choi H, and Weisshaar JC. Melittin-induced permeabilization, re-sealing, and re-permeabilization of E. coli membranes. Biophys J. (2018) 114:368–79. doi: 10.1016/j.bpj.2017.10.046
78. Scocchi M, Mardirossian M, Runti G, and Benincasa M. Non-membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr Top Med Chem. (2016) 16:76–88. doi: 10.2174/1568026615666150703121009
79. Bechinger B. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim Biophys Acta. (1999) 1462:157–83. doi: 10.1016/S0005-2736(99)00205-9
80. Wieprecht T, Apostolov O, Beyermann M, and Seelig J. Thermodynamics of the alpha-helix-coil transition of amphipathic peptides in a membrane environment: implications for the peptide-membrane binding equilibrium. J Mol Biol. (1999) 294:785–94. doi: 10.1006/jmbi.1999.3268
81. Das N, Dai J, Hung I, Rajagopalan MR, Zhou HX, and Cross TA. Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis, in lipid bilayers. Proc Natl Acad Sci USA. (2015) 112:E119–26. doi: 10.1073/pnas.1415908112
82. Gopinath T, Mote KR, and Veglia G. Simultaneous acquisition of 2D and 3D solid-state NMR experiments for sequential assignment of oriented membrane protein samples. J Biomol NMR. (2015) 62:53–61. doi: 10.1007/s10858-015-9916-9
83. Itkin A, Salnikov ES, Aisenbrey C, Raya J, Raussens V, Ruysschaert JM, et al. Evidence for heterogeneous conformations of the gamma cleavage site within the amyloid precursor proteins transmembrane domain ACS Omega. (2017) 2:6525–34. doi: 10.1021/acsomega.7b00619
84. Salnikov ES, Anantharamaiah GM, and Bechinger B. Supramolecular organization of apolipoprotein A-I - derived peptides within disc-like arrangements. Biophys J. (2018) 115:467–77. doi: 10.1016/j.bpj.2018.06.026
85. Bechinger B, and Sizun C. Alignment and structural analysis of membrane polypeptides by 15N and 31P solid-state NMR spectroscopy. Concepts Magn Reson. (2003) 18A:130–45. doi: 10.1002/cmr.a.10070
86. Bechinger B, Shon K, Eck H, Zasloff M, and Opella SJ. NMR studies of magainin peptide antibiotics in membranes. Biol Chem Hoppe Seyler. (1990) 371:758–758.
87. Bechinger B, Zasloff M, and Opella SJ. Structure and interactions of magainin antibiotic peptides in lipid bilayers: a solid-state NMR investigation. Biophys J. (1992) 62:12–4.
88. Bechinger B, Zasloff M, and Opella SJ. Structure and orientation of the antibiotic peptide magainin in membranes by solid-state NMR spectroscopy. Protein Sci. (1993) 2:2077–84.
89. Bechinger B. Insights into the mechanisms of action of host defence peptides from biophysical and structural investigations. J Pept Sci. (2011) 17:306–14. doi: 10.1002/psc.1343
90. Ramamoorthy A, Thennarasu S, Lee DK, Tan A, and Maloy L. Solid-state NMR investigation of the membrane-disrupting mechanism of antimicrobial peptides MSI-78 and MSI-594 derived from magainin 2 and melittin. Biophys J. (2006) 91:206–16. doi: 10.1529/biophysj.105.073890
91. Mason AJ, Moussaoui W, Abdelrhaman T, Boukhari A, Bertani P, Marquette A, et al. Structural determinants of antimicrobial and antiplasmodial activity and selectivity in histidine rich amphipathic cationic peptides. J Biol Chem. (2009) 284:119–33. doi: 10.1074/jbc.M806201200
92. Resende JM, Moraes CM, Munhoz VHDO, Aisenbrey C, Verly RM, Bertani P, et al. Membrane structure and conformational changes of the antibiotic heterodimeric peptide distinctin by solid-state NMR spectroscopy. Proc Natl Acad Sci USA. (2009) 106:16639–44. doi: 10.1073/pnas.0905069106
93. Resende JM, Verly RM, Aisenbrey C, Amary C, Bertani P, Pilo-Veloso D, et al. Membrane interactions of Phylloseptin-1,−2, and−3 peptides by oriented solid-state NMR spectroscopy. Biophys J. (2014) 107:901–11. doi: 10.1016/j.bpj.2014.07.014
94. Hayden RM, Goldberg GK, Ferguson BM, Schoeneck MW, Libardo MD, Mayeux SE, et al. Complementary effects of host defense peptides piscidin 1 and piscidin 3 on DNA and lipid membranes: biophysical insights into contrasting biological activities. J Phys Chem B. (2015) 119:15235–46. doi: 10.1021/acs.jpcb.5b09685
95. Sani MA, and Separovic F. Antimicrobial peptide structures: from model membranes to live cells. Chemistry. (2018) 24:286–91. doi: 10.1002/chem.201704362
96. Ludtke SJ, He K, Wu Y, and Huang HW. Cooperative membrane insertation of maginin correlated with its cytolytic activity. Biochim Biophys Acta. (1994) 1190:181–4.
97. Gazit E, Miller IR, Biggin PC, Sansom MSP, and Shai Y. Structure and orientation of the mammalian antibacterial peptide cecropin P1 within phospholipid membranes. J Mol Biol. (1996) 258:860–70.
98. Salnikov E, and Bechinger B. Lipid-controlled peptide topology and interactions in bilayers: structural insights into the synergistic enhancement of the antimicrobial activities of PGLa and magainin 2. Biophys J. (2011) 100:1473–80. doi: 10.1016/j.bpj.2011.01.070
99. Strandberg E, Zerweck J, Wadhwani P, and Ulrich AS. Synergistic insertion of antimicrobial magainin-family peptides in membranes depends on the lipid spontaneous curvature. Biophys J. (2013) 104:L9–11. doi: 10.1016/j.bpj.2013.01.047
100. Tremouilhac P, Strandberg E, Wadhwani P, and Ulrich AS. Synergistic transmembrane alignment of the antimicrobial heterodimer PGLa/magainin. J Biol Chem. (2006) 281:32089–94. doi: 10.1074/jbc.M604759200
101. Harmouche N, and Bechinger B. Lipid-mediated interactions between the amphipathic antimicrobial peptides magainin 2 and PGLa in phospholipid bilayers. Biophys J. (2018) 115:1033–44. doi: 10.1016/j.bpj.2018.08.009
102. Zerweck J, Strandberg E, Kukharenko O, Reichert J, Burck J, Wadhwani P, et al. Molecular mechanism of synergy between the antimicrobial peptides PGLa and magainin 2. Sci Rep. (2017) 7:13153. doi: 10.1038/s41598-017-12599-7
103. Glattard E, Salnikov ES, Aisenbrey C, and Bechinger B. Investigations of the synergistic enhancement of antimicrobial activity in mixtures of magainin 2 and PGLa. Biophys Chem. (2016) 210:35–44. doi: 10.1016/j.bpc.2015.06.002
104. Aisenbrey C, and Bechinger B. Molecular packing of amphipathic peptides on the surface of lipid membranes. Langmuir. (2014) 30:10374–83. doi: 10.1021/la500998g
105. Ludtke S, He K, and Huang H. Membrane thinning caused by magainin 2. Biochemistry. (1995) 34:16764–9. doi: 10.1021/bi00051a026
106. Kim C, Spano J, Park EK, and Wi S. Evidence of pores and thinned lipid bilayers induced in oriented lipid membranes interacting with the antimicrobial peptides, magainin-2 and aurein-3.3. Biochim Biophys Acta. (2009) 1788:1482–1496. doi: 10.1016/j.bbamem.2009.04.017
107. Bechinger B, and Salnikov ES. The membrane interactions of antimicrobial peptides revealed by solid-state NMR spectroscopy. Chem Phys Lipids. (2012) 165:282–301. doi: 10.1016/j.chemphyslip.2012.01.009
108. Hallock KJ, Lee DK, Omnaas J, Mosberg HI, and Ramamoorthy A. Membrane composition determines pardaxin's mechanism of lipid bilayer disruption. Biophys J. (2002) 83:1004–13. doi: 10.1016/S0006-3495(02)75226-0
109. Salnikov ES, Mason AJ, and Bechinger B. Membrane order perturbation in the presence of antimicrobial peptides by 2 H solid-state NMR spectroscopy. Biochimie. (2009) 91:743. doi: 10.1016/j.biochi.2009.01.002
110. Grage SL, Afonin S, Kara S, Buth G, and Ulrich AS. Membrane thinning and thickening induced by membrane-active amphipathic peptides. Front Cell Dev Biol. (2016) 4:65. doi: 10.3389/fcell.2016.00065
111. Bechinger B. Detergent-like properties of magainin antibiotic peptides: a 31P solid-state NMR study. Biochim Biophys Acta. (2005) 1712:101–8. doi: 10.1016/j.bbamem.2005.03.003
112. Chen FY, Lee MT, and Huang HW. Evidence for membrane thinning effect as the mechanism for Peptide-induced pore formation. Biophys J. (2003) 84:3751–8. doi: 10.1016/S0006-3495(03)75103-0
113. Mecke A, Lee DK, Ramamoorthy A, Orr BG, and Banaszak Holl MM. Membrane thinning due to antimicrobial peptide binding: an atomic force microscopy study of MSI-78 in lipid bilayers. Biophys J. (2005) 89:4043–50. doi: 10.1529/biophysj.105.062596
114. Vacha R, and Frenkel D. Simulations suggest possible novel membrane pore structure. Langmuir. (2014) 30:1304–10. doi: 10.1021/la402727a
115. Farrotti A, Bocchinfuso G, Palleschi A, Rosato N, Salnikov ES, Voievoda N, et al. Molecular dynamics methods to predict peptide location in membranes: LAH4 as a stringent test case. Biochim Biophys Acta. (2015) 1848:581–92. doi: 10.1016/j.bbamem.2014.11.002
116. Pachler M, Kabelka I, Appavou MS, Lohner K, Vacha R, and Pabst G. Magainin 2 and PGLa in bacterial membrane mimics I: peptide-peptide and lipid-peptide interactions. Biophys J. (2019) 117:1858–69. doi: 10.1016/j.bpj.2019.10.022
117. Kabelka I, Pachler M, Prevost S, Letofsky-Papst I, Lohner K, Pabst G, et al. Magainin 2 and PGLa in bacterial membrane mimics II: membrane fusion and sponge phase formation. Biophys J. (2020) 118:612–23. doi: 10.1016/j.bpj.2019.11.1985
118. Huang HW. Action of antimicrobial peptides: two-state model. Biochemistry. (2000) 39:8347–52. doi: 10.1021/bi000946l
119. Leber R, Pachler M, Kabelka I, Svoboda I, Enkoller D, Vácha R, et al. Synergism of antimicrobial frog peptides couples to membrane intrinsic curvature Strain. Biophys J. (2018) 114:1945–54. doi: 10.1016/j.bpj.2018.03.006
120. Amos ST, Vermeer LS, Ferguson PM, Kozlowska J, Davy M, Bui TT, et al. Antimicrobial peptide potency is facilitated by greater conformational flexibility when binding to gram-negative bacterial inner membranes. Sci Rep. (2016) 6:37639. doi: 10.1038/srep37639
121. Israelachvili JN, Marcelja S, and Horn RG. Physical principles of membrane organization. Q Rev Biophys. (1980) 13:121–200.
122. Aisenbrey C, Marquette A, and Bechinger B. The mechanisms of action of cationic antimicrobial peptides refined by novel concepts from biophysical investigations. Adv Exp Med Biol. (2019) 1117:33–64. doi: 10.1007/978-981-13-3588-4_4
123. Cruciani RA, Barker JL, Zasloff M, Chen HC, and Colamonici O. Antibiotic magainins exert cytolytic activity transformed cell lines through channel formation. Proc Natl Acad Sci USA. (1991) 88:3792–6.
124. Cruciani RA, Barker JL, Raghunathan G, Guy HR, Zasloff M, and Stanley EF. Magainin 2, a natural antibiotic from frog skin, forms ion channels in lipid bilayer membranes. Eur J Pharmacol. (1992) 226:287–96.
125. Hall K, Lee TH, Mechler AI, Swann MJ, and Aguilar MI. Real-time measurement of membrane conformational states induced by antimicrobial peptides: balance between recovery and lysis. Sci Rep. (2014) 4:5479. doi: 10.1038/srep05479
126. Bechinger B, and Lohner K. Detergent-like action of linear cationic membrane-active antibiotic peptides. Biochim Biophys Acta. (2006) 1758:1529–39. doi: 10.1016/j.bbamem.2006.07.001
127. Bechinger B. The SMART model: soft membranes adapt and respond, also transiently, to external stimuli. J Pept Sci. (2015) 21:346–55. doi: 10.1002/psc.2729
128. Henderson JM, Waring AJ, Separovic F, and Lee KYC. Antimicrobial peptides share a common interaction driven by membrane line tension reduction. Biophys J. (2016) 111:2176–89. doi: 10.1016/j.bpj.2016.10.003
129. Matsuzaki K, Harada M, Funakoshi S, Fujii N, and Miyajima K. Physicochemical determinants for the interactions of magainins 1 and 2 with acidic lipid bilayers. Biochim Biophys Acta. (1991) 1063:162–70.
130. Wenk M, and Seelig J. Magainin 2 amide interaction with lipid membranes: calorimetric detection of peptide binding and pore formation. Biochemistry. (1998) 37:3909–16.
131. Bechinger B. Membrane-lytic peptides. Crit Rev Plant Sci. (2004) 23:271–92. doi: 10.1080/07352680490452825
132. Klocek G, and Seelig J. Melittin interaction with sulfated cell surface sugars. Biochemistry. (2008) 47:2841–9. doi: 10.1021/bi702258z
133. Lohner K. New strategies for novel antibiotics: peptides targeting bacterial cell membranes. Gen Physiol Biophys. (2009) 28:105–16. doi: 10.4149/gpb_2009_02_105
134. Wieprecht T, Beyermann M, and Seelig J. Binding of antibacterial magainin peptides to electrically neutral membranes: thermodynamics and structure. Biochemistry. (1999) 38:10377–8.
135. Voievoda N, Schulthess T, Bechinger B, and Seelig J. Thermodynamic and biophysical analysis of the membrane-association of a histidine-rich peptide with efficient antimicrobial and transfection activities. J Phys Chem B. (2015) 119:9678–87. doi: 10.1021/acs.jpcb.5b04543
136. Acar JF. Antibiotic synergy and antagonism. Med Clin North Am. (2000) 84:1391–406. oi: 10.1016/S0025-7125(05)70294-7
137. Mangoni ML, and Shai Y. Temporins and their synergism against Gram-negative bacteria and in lipopolysaccharide detoxification. Biochim Biophys Acta. (2009) 1788:1610–9. doi: 10.1016/j.bbamem.2009.04.021
138. Chou S, Shao C, Wang J, Shan A, Xu L, Dong N, et al. Short, multiple-stranded beta-hairpin peptides have antimicrobial potency with high selectivity and salt resistance. Acta Biomater. (2016) 30:78–93. doi: 10.1016/j.actbio.2015.11.002
139. Bolosov IA, Kalashnikov AA, Panteleev PV, and Ovchinnikova TV. Analysis of synergistic effects of antimicrobial peptide arenicin-1 and conventional antibiotics. Bull Exp Biol Med. (2017) 162:765–8. doi: 10.1007/s10517-017-3708-z
140. Kim EY, Rajasekaran G, and Shin SY. LL-37-derived short antimicrobial peptide KR-12-a5 and its d-amino acid substituted analogs with cell selectivity, anti-biofilm activity, synergistic effect with conventional antibiotics, and anti-inflammatory activity. Eur J Med Chem. (2017) 136:428–41. doi: 10.1016/j.ejmech.2017.05.028
141. Payne JE, Dubois AV, Ingram RJ, Weldon S, Taggart CC, Elborn JS, et al. Activity of innate antimicrobial peptides and ivacaftor against clinical cystic fibrosis respiratory pathogens. Int J Antimicrob Agents. (2017) 50:427–35. doi: 10.1016/j.ijantimicag.2017.04.014
142. Sakoulas G, Kumaraswamy M, Kousha A, and Nizet V. Interaction of antibiotics with innate host defense factors against Salmonella enterica serotype newport. mSphere. (2017) 2:e00410–17. doi: 10.1128/mSphere.00410-17
143. Citterio L, Franzyk H, Palarasah Y, Andersen TE, Mateiu RV, and Gram L. Improved in vitro evaluation of novel antimicrobials: potential synergy between human plasma and antibacterial peptidomimetics, AMPs and antibiotics against human pathogenic bacteria. Res Microbiol. (2016) 167:72–82. doi: 10.1016/j.resmic.2015.10.002
144. Walkenhorst WF, Sundrud JN, and Laviolette JM. Additivity and synergy between an antimicrobial peptide and inhibitory ions. Biochim Biophys Acta. (2014) 1838:2234–42. doi: 10.1016/j.bbamem.2014.05.005
145. Kobayashi S, Hirakura Y, and Matsuzaki K. Bacteria-selective synergism between the antimicrobial peptides alpha-helical magainin 2 and cyclic beta-sheet tachyplesin I: toward cocktail therapy. Biochemistry. (2001) 40:14330–5. doi: 10.1021/bi015626w
146. Juretic D, Hendler RW, Zasloff M, and Westerhoff HV. Cooperative action of magainins in disrupting membrane-linked free-energy transduction. Biophys J. (1989) 55:W-Pos313.
147. Juretić D. Antimicrobial peptides of the magainin family: membrane depolarization studies on E. coli and cytochrome oxidase liposomes. Stud Biophys. (1990) 138:79–86.
148. Juretic D, Hendler RW, Kamp F, Caughey WS, Zasloff M, and Westerhoff HV. Magainin oligomers reversibly dissipate delta microH+ in cytochrome oxidase liposomes. Biochemistry. (1994) 33:4562–70.
149. Nishida M, Imura Y, Yamamoto M, Kobayashi S, Yano Y, and Matsuzaki K. Interaction of a magainin-PGLa hybrid peptide with membranes: insight into the mechanism of synergism. Biochemistry. (2007) 46:14284–90. doi: 10.1021/bi701850m
150. Juhl DW, Glattard E, Lointier M, Bampilis P, and Bechinger B. The antimicrobial and synergistic activities of PGLa and magainin 2 fibrils. Front Cell Infect Microbiol. (2020) 10:526459. doi: 10.3389/fcimb.2020.526459
151. Yu G, Baeder DY, Regoes RR, and Rolff J. Combination effects of antimicrobial peptides. Antimicrob Agents Chemother. (2016) 60:1717–24. doi: 10.1128/AAC.02434-15
152. Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. (2006) 58:621–81. doi: 10.1124/pr.58.3.10
153. Jepson AK, Schwarz-Linek J, Ryan L, Ryadnov MG, and Poon WCK. What is the ‘minimum inhibitory concentration' (MIC) of pexiganan acting on Escherichia coli?—A cautionary case study. Adv Exp Med Biol. (2016) 915:33–48. doi: 10.1007/978-3-319-32189-9_4
154. Snoussi M, Talledo JP, Del Rosario NA, Mohammadi S, Ha BY, Kosmrlj A, et al. Heterogeneous absorption of antimicrobial peptide LL37 in Escherichia coli cells enhances population survivability. Elife. (2018) 7:e38174. doi: 10.7554/eLife.38174
155. Hill AV. The possible effects of the aggregation of the molecules of hemoglobin on its dissociation curves. J Physiol. (1910) 40:4–7.
156. Savini F, Loffredo MR, Troiano C, Bobone S, Malanovic N, Eichmann TO, et al. Binding of an antimicrobial peptide to bacterial cells: Interaction with different species, strains and cellular components. Biochim Biophys Acta. (2020) 1862:183291. doi: 10.1016/j.bbamem.2020.183291
157. Savini F, Luca V, Bocedi A, Massoud R, Park Y, Mangoni ML, et al. Cell-density dependence of host-defense peptide activity and selectivity in the presence of host cells. ACS Chem Biol. (2017) 12:52–6. doi: 10.1021/acschembio.6b00910
158. Loffredo MR, Savini F, Bobone S, Casciaro B, Franzyk H, Mangoni ML, et al. Inoculum effect of antimicrobial peptides. bioRxiv preprint. (2020). doi: 10.1101/2020.08.21.260620
159. Choi H, Rangarajan N, and Weisshaar JC. Lights, camera, action! antimicrobial peptide mechanisms imaged in space and time. Trends Microbiol. (2016) 24:111–22. doi: 10.1016/j.tim.2015.11.004
160. Mor A, Hani K, and Nicolas P. The vertebrate peptide antibiotics dermaseptins have overlapping structural features but target specific microorganisms. J Biol Chem. (1994) 269:31635–41.
161. Giacometti A, Cirioni O, Del Prete MS, Paggi AM, D'errico MM, and Scalise G. Combination studies between polycationic peptides and clinically used antibiotics against Gram-positive and Gram-negative bacteria. Peptides. (2000) 21:1155–60. doi: 10.1016/S0196-9781(00)00254-0
162. Matsuzaki K, Mitani Y, Akada K, Murase O, Yoneyama S, Zasloff M, et al. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry. (1998) 37:15144–53.
163. Zerweck J, Strandberg E, Burck J, Reichert J, Wadhwani P, Kukharenko O, et al. Homo- and heteromeric interaction strengths of the synergistic antimicrobial peptides PGLa and magainin 2 in membranes. Eur Biophys J. (2016) 45:535–47. doi: 10.1007/s00249-016-1120-7
164. Williams RW, Starmann R, Taylor KMP, Gable K, Beeler T, Zasloff M, et al. Raman spectroscopy of synthetic antimicrobial frog peptides magainin 2a and PGLa. Biochemistry. (1990) 29:4490–6.
165. Patel H, Huynh Q, Barlehner D, and Heerklotz H. Additive and synergistic membrane permeabilization by antimicrobial (Lipo)peptides and detergents. Biophys J. (2014) 106:2115–25. doi: 10.1016/j.bpj.2014.04.006
166. Salnikov ES, Aisenbrey C, Aussenac F, Ouari O, Sarrouj H, Reiter C, et al. Membrane topologies of the PGLa antimicrobial peptide and a transmembrane anchor sequence by Dynamic Nuclear Polarization/solid-state NMR spectroscopy. Sci Rep. (2016) 6:20895. doi: 10.1038/srep20895
167. Aisenbrey C, and Bechinger B. Equilibria governing the membrane insertion of polypeptides and their interactions with other biomacromolecules. In: Moreno Pirajan JC, editor. Thermodynamics/Book 2. Rijeka: IntechOpen (2011). p. 381–402.
168. Matsuzaki K, Nakamura A, Murase O, Sugishita K, Fujii N, and Miyajima K. Modulation of magainin 2-lipid bilayer interactions by peptide charge. Biochemistry. (1997) 36:2104–11.
169. Bechinger B. Towards membrane protein design: pH-sensitive topology of histidine-containing polypeptides. J Mol Biol. (1996) 263:768–75.
170. Marquette A, Salnikov E, Glattard E, Aisenbrey C, and Bechinger B. Magainin 2-PGLa interactions in membranes - two peptides that exhibit synergistic enhancement of antimicrobial activity. Curr Top Med Chem. (2015) 16:65–75. doi: 10.2174/1568026615666150703115701
171. Juhl DW, Glattard E, Aisenbrey C, and Bechinger B. Antimicrobial peptides: mechanism of action and lipid-mediated synergistic interactions within membranes. Faraday Discuss. (2020).
172. Cuervo JH, Rodriguez B, and Houghten RA. The Magainins: sequence factors relevant to increased antimicrobial activity and decreased hemolytic activity. Pept Res. (1988) 1:81–6.
173. Strandberg E, Zerweck J, Horn D, Pritz G, Berditsch M, Bürck J, et al. Influence of hydrophobic residues on the activity of the antimicrobial peptide magainin 2 and its synergy with PGLa. J Pept Sci. (2015) 21:436–45. doi: 10.1002/psc.2780
174. Han E, and Lee H. Synergistic effects of magainin 2 and PGLa on their heterodimer formation, aggregation, and insertion into the bilayer. RSC Adv. (2015) 5:2047–55. doi: 10.1039/C4RA08480B
175. Pino-Angeles A, Leveritt JM III, and Lazaridis T. Pore structure and synergy in antimicrobial peptides of the magainin family. PLoS Comput Biol. (2016) 12:e1004570. doi: 10.1371/journal.pcbi.1004570
176. Russ WP, and Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. (2000) 296:911–9. doi: 10.1006/jmbi.1999.3489
177. Bechinger B, Kinder R, Helmle M, Vogt TB, Harzer U, and Schinzel S. Peptide structural analysis by solid-state NMR spectroscopy. Biopolymers. (1999) 51:174–90.
178. Hara T, Kodama H, Kondo M, Wakamatsu K, Takeda A, Tachi T, et al. Effects of peptide dimerization on pore formation: antiparallel disulfide-dimerized magainin 2 analogue. Biopolymers. (2001) 58:437–46. doi: 10.1002/1097-0282(20010405)58:4<437::AID-BIP1019>3.0.CO;2-I
179. Lorenzon EN, Santos-Filho NA, Ramos MA, Bauab TM, Camargo IL, and Cilli EM. C-terminal lysine-linked magainin 2 with increased activity against multidrug-resistant bacteria. Protein Pept Lett. (2016) 23:738–47. doi: 10.2174/0929866523666160511150907
180. Dempsey CE, Ueno S, and Avison MB. Enhanced membrane permeabilization and antibacterial activity of a disulfide-dimerized magainin analogue. Biochemistry. (2003) 42:402–9. doi: 10.1021/bi026328h
181. Verly RM, Resende JM, Junior EFC, De Magalhães MTC, Guimarães CFCR, Munhoz VHO, et al. Structure and membrane interactions of the homodimeric antibiotic peptide homotarsinin. Sci Rep. (2017) 7:40854. doi: 10.1038/srep40854
182. Raimondo D, Andreotti G, Saint N, Amodeo P, Renzone G, Sanseverino M, et al. A folding-dependent mechanism of antimicrobial peptide resistance to degradation unveiled by solution structure of distinctin. Proc Natl Acad Sci USA. (2005) 102:6309–14. doi: 10.1073/pnas.0409004102
183. Dalla Serra M, Cirioni O, Vitale RM, Renzone G, Coraiola M, Giacometti A, et al. Structural features of distinctin affecting peptide biological and biochemical properties. Biochemistry. (2008) 47:7888–99. doi: 10.1021/bi800616k
184. Verardi R, Traaseth NJ, Shi L, Porcelli F, Monfregola L, De Luca S, et al. Probing membrane topology of the antimicrobial peptide distinctin by solid-state NMR spectroscopy in zwitterionic and charged lipid bilayers. Biochim Biophys Acta. (2011) 1808:34–40. doi: 10.1016/j.bbamem.2010.08.008
185. Vermeer LS, Hamon L, Schirer A, Schoup M, Cosette J, Majdoul S, et al. The transduction enhancing peptide vectofusin-1 forms pH-dependent α-helical coiled-coil nanofibrils, trapping viral particles. Acta Biomater. (2017) 64:259–68. doi: 10.1016/j.actbio.2017.10.009
186. Lague P, Zuckermann MJ, and Roux B. Lipid-mediated interactions between intrinsic membrane proteins: dependence on protein size and lipid composition. Biophys J. (2001) 81:276–84. doi: 10.1016/S0006-3495(01)75698-6
187. Duneau JP, and Sturgis JN. Lateral organization of biological membranes: role of long-range interactions. Eur Biophys J. (2013) 42:843–50. doi: 10.1007/s00249-013-0933-x
188. Bitbol A-F, Constantin D, and Fournier J-B. Membrane-mediated interactions. arXiv preprint arXiv:1903.05712 (2018).
189. Chen H-C, Brown JH, Morell JL, and Huang CM. Synthetic magainin analogues with improved antimicrbial activity. FEBS Lett. (1988) 236:462–6.
191. Wieprecht T, Apostolov O, and Seelig J. Binding of the antibacterial peptide magainin 2 amide to small and large unilamellar vesicles. Biophys Chem. (2000) 85:187–98. doi: 10.1016/S0301-4622(00)00120-4
Keywords: PGLa, membrane topology, membrane pore, membrane macroscopic phase, SMART model, carpet model, peptide-lipid interactions, molecular shape concept
Citation: Bechinger B, Juhl DW, Glattard E and Aisenbrey C (2020) Revealing the Mechanisms of Synergistic Action of Two Magainin Antimicrobial Peptides. Front. Med. Technol. 2:615494. doi: 10.3389/fmedt.2020.615494
Received: 09 October 2020; Accepted: 30 November 2020;
Published: 21 December 2020.
Edited by:
Miguel A. R. B. Castanho, University of Lisbon, PortugalReviewed by:
Octavio Luiz Franco, Catholic University of Brasilia (UCB), BrazilDavor Juretić, MEDILS - Mediterranean Institute for Life Sciences, Croatia
Copyright © 2020 Bechinger, Juhl, Glattard and Aisenbrey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Burkhard Bechinger, bechinge@unistra.fr
†Present address: Dennis Wilkens Juhl, Department of Chemistry, Aarhus University, Aarhus, Denmark
 Burkhard Bechinger
Burkhard Bechinger Dennis Wilkens Juhl1†
Dennis Wilkens Juhl1† 