- 1Instituto de Microbiologia Paulo de Góes, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
- 2Laboratório Nacional de Computação Científica, Departamento de Matemática Aplicada e Computacional, Petrópolis, Brazil
- 3Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
- 4Instituto de Ciências Biomédicas, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
- 5School of Life Sciences, University of Nevada at Las Vegas, Las Vegas, NV, USA
Although magnetotactic bacteria (MTB) are ubiquitous in aquatic habitats, they are still considered fastidious microorganisms with regard to growth and cultivation with only a relatively low number of axenic cultures available to date. Here, we report the first axenic culture of an MTB isolated in the Southern Hemisphere (Itaipu Lagoon in Rio de Janeiro, Brazil). Cells of this new isolate are coccoid to ovoid in morphology and grow microaerophilically in semi-solid medium containing an oxygen concentration ([O2]) gradient either under chemoorganoheterotrophic or chemolithoautotrophic conditions. Each cell contains a single chain of approximately 10 elongated cuboctahedral magnetite (Fe3O4) magnetosomes. Phylogenetic analysis based on the 16S rRNA gene sequence shows that the coccoid MTB isolated in this study represents a new genus in the Alphaproteobacteria; the name Magnetofaba australis strain IT-1 is proposed. Preliminary genomic data obtained by pyrosequencing shows that M. australis strain IT-1 contains a genomic region with genes involved in biomineralization similar to those found in the most closely related magnetotactic cocci Magnetococcus marinus strain MC-1. However, organization of the magnetosome genes differs from M. marinus.
Introduction
Magnetotactic bacteria (MTB) are a morphologically, metabolically, and phylogenetically diverse group of prokaryotes that share the ability to synthesize intracellular, nano-sized magnetic particles called magnetosomes. Each magnetosome consists of a magnetite (Fe3O4) or greigite (Fe3S4) crystal enveloped by a lipid-bilayer membrane derived from the cytoplasmic membrane (Bazylinski and Frankel, 2004). Magnetosomes are generally organized in linear chains and orient the cell body along geomagnetic field lines while flagella actively propel the cells, resulting in so-called magnetotaxis (Bazylinski and Frankel, 2004; Schüler, 2008). MTB from the Southern Hemisphere swim antiparallel to the vertical component of the geomagnetic field toward the South and are termed South-seeking MTB (SS-MTB). In contrast, MTB from the Northern Hemisphere swim parallel to the vertical component of the geomagnetic field lines and are predominantly North-seeking (NS-MTB) (Blakemore et al., 1980). The inclination of the geomagnetic field lines is believed to direct cells downwards away from toxic concentrations of oxygen in surface waters, thereby helping them locate and maintain an optimal position in vertical gradients which is usually at or near the oxic-anoxic interface (OAI) (Blakemore, 1982; Frankel and Bazylinski, 1994; Bazylinski and Frankel, 2004). However, there are reports of SS-MTB and NS-MTB in both hemispheres (Simmons et al., 2006).
MTB are considered fastidious microorganisms (Schüler, 2008), although there has recently been a considerable increase in available cultures, including the first cultivation of a greigite producer (Lefèvre et al., 2011). The recent availability of MTB cultures has contributed to a better characterization of the physiology and biochemistry of these microorganisms. It has also contributed to an improved understanding of the evolution of MTB and of the biomineralization processes involved since differences in the sequences of magnetosome biomineralization genes in different MTB, particularly the mam genes, revealed a strong correlation between these magnetotaxis-related genes and phylogeny based on the 16S rRNA gene (Lefèvre et al., 2013a). Studies of magnetosome biomineralization genes in uncultivated MTB require unique approaches (Abreu et al., 2011; Jogler et al., 2011) that do not usually reveal the complete organization of biomineralization genes or genes involved in magnetotactic behavior unless the entire genome is sequenced. Moreover, because not all the magnetosome-related genes may be recognized, a direct correlation with phylogeny based on 16S rRNA gene sequences cannot be made with total accuracy.
The most characterized cultivated MTB strains are phylogenetically affiliated with the Alphaproteobacteria and include Magnetococcus marinus strain MC-1 (Bazylinski et al., 2013a), Magnetovibrio blakemorei strain MV-1 (Bazylinski et al., 2013b), the magneto-ovoid bacterium strain MO-1 (Lefèvre et al., 2009), Magnetospirillum magneticum strain AMB-1, Magnetospirillum gryphiswaldense strain MSR-1, Magnetospirillum magnetotacticum strain MS-1, Magnetospira thiophilla strain MMS-1 (Williams et al., 2012) and Magnetospira sp. QH-2 strain 1 (Ji et al., 2014). Cultivated strains belonging to Deltaproteobacteria include the sulfate-reducer Desulfovibrio magneticus strain RS-1, (Sakaguchi et al., 2002), Candidatus Desulfamplus magnetomortis strain BW-1 (Lefèvre et al., 2011) and enrichment cultures of the magnetotactic multicellular prokaryotes Candidatus Magnetoglobus multicellularis (Abreu et al., 2013). Two cultivated strains, BW-2 and SS-5, both belonging to Gammaproteobacteria, have also been reported (Lefèvre et al., 2012).
The biomineralization of magnetosomes is controlled by a set of highly conserved genes in magnetite-producing MTB (Richter et al., 2007; Jogler and Schüler, 2009; Jogler et al., 2009) and, as demonstrated more recently, in greigite-producing MTB as well (Abreu et al., 2011, 2013; Lefèvre et al., 2011, 2013b). In some species, the magnetosome biomineralization genes are clustered on a genomic magnetosome island (MAI), which partially supports the hypothesis of horizontal gene transfer (HGT) between various MTB presumably leading to the wide distribution of these genes among members of different phylogenetic groups (Jogler and Schüler, 2009; Jogler et al., 2009; Abreu et al., 2011). However, certain components of typical genomic islands (transposases, t-RNA sequences, integrases), such as those observed in M. magneticum strain AMB-1, M. gryphiswaldense strain MSR-1 and D. magneticus RS-1, are not universally shared within the MAI of all MTB (e.g., M. marinus; Schübbe et al., 2009). Moreover, phylogenetic analysis based on the amino acid sequences of magnetosome proteins from MTB are congruent with the phylogenetic tree based on the 16S rRNA gene sequences of the same microorganisms (Lefèvre et al., 2013a). Therefore, the evolution and divergence of magnetosome proteins and the 16S rRNA gene occurred similarly, suggesting that magnetotaxis originated monophyletically in the Proteobacteria phylum (Lefèvre et al., 2013a). Additional genome sequences and culture of MTB species are necessary to understand the evolution of biomineralization in Bacteria. Moreover, the availability of new cultures of MTB allows a better characterization of the physiology and biochemistry of these microorganisms, enabling the correlation of these features to magnetosome formation.
Despite being the most prevalent and diverse morphotype of MTB in the environment (Spring et al., 1998; Schübbe et al., 2009), there are currently only two cultivated strains of magnetotactic cocci: M. marinus strain MC-1 (Bazylinski et al., 2013a) and the magneto-ovoid bacterium strain MO-1 (Lefèvre et al., 2009). The complete genome sequence of the NS-MTB M. marinus has been reported (Schübbe et al., 2009), but further study is required to better understand the full diversity of the magnetotactic cocci as well as the ecological function and evolution of magnetosome biomineralization in the Alphaproteobacteria. Here, we describe both the isolation in axenic culture and the characterization of a new magnetotactic coccus, provisionally named Magnetofaba australis strain IT-1 that represents a new genus. We also conducted whole genome sequencing and functional annotation of genes related to magnetosome formation to gain insight into the phylogeny, physiology and biochemistry of this SS-MTB. This strain is the first cultivated SS-MTB, and the genomic data presented here are the first report of biomineralization genes in magnetotactic cocci capable of synthesizing elongated cuboctahedral magnetosomes.
Materials and Methods
Isolation and Cultivation of Magnetofaba australis Strain IT-1.
Samples of water and sediment were collected from the Itaipu Lagoon (22°57′51.90″ S 43°2′45.41″ W), a brackish to marine coastal lagoon near Rio de Janeiro, Brazil, and stored under dim light at room temperature. MTB were magnetically concentrated using a magnetic isolation apparatus described by Lins et al. (2003). After 20 min, cells were collected in a polypropylene tube. Concentrated South-seeking MTB were magnetically purified repeatedly using the racetrack technique (Wolfe et al., 1987) and inoculated at the OAI of culture tubes. Approximately 4/5 of the tubes were filled with an autotrophic semisolid oxygen concentration gradient ([O2]-gradient) medium. The medium was used to isolate M. marinus (Frankel et al., 1997) and contained bicarbonate as the major carbon source. The medium contained 5 mL of modified Wolfe's minerals elixir, 3.75 mM NH4Cl, 0.2 mL of 0.2% resazurin and 2 g of Bacto-Agar diluted in 1 L of artificial seawater (ASW). The medium was autoclaved, followed by the addition of 1.5 mL of 0.5 M KHPO4, pH 7.1, neutral fresh L-cysteine (final concentration of 0.2 g/L) and 2.68 mL of 0.8 M NaHCO3, 0.5 mL of vitamin solution and 2 mL of 0.01 M ferric quinate (final concentration of 20 μM). The pH was adjusted to 7.2. Cultures were incubated at 28°C until a microaerophilic band of cells was observed at the OAI and, subsequently, the bands were inoculated into a solid heterotrophic [O2]-gradient medium applying the dilution-to-extinction technique and shake-tubes (Seeley et al., 1991). Briefly, a band of cells were inoculated into the solid medium before it solidified (approximately at 45°C), followed by 7 serial 10-fold dilution steps. After inoculation and agitation by inversion, each tube was put on ice to solidify the medium quickly without killing a significant number of cells. Colonies grown on shake tubes were individually transferred to semi-solid heterotrophic medium. Each colony in culture was re-inoculated in fresh medium over 10 times to ensure that a pure culture was obtained. Purity of the culture was evaluated by light and electron microscopy and sequencing of the 16S rRNA gene.
The medium chosen for growth and maintenance of M. australis strain IT-1 was designed for heterotrophic growth, because cells grew faster and the number of magnetosome per cell was higher than in the autotrophic medium. The heterotrophic medium contained 5 mL of modified Wolfe's minerals (Frankel et al., 1997), 3.75 mM NH4Cl, 0.2 ml of 0.2% resazurin, 12 mM HEPES, 12 mM sodium acetate, 3.7 mM sodium succinate and 2 g of Bacto-Agar in 1 L of ASW. The medium was autoclaved, followed by the addition of 1.5 mL of 0.5 M KHPO4, pH 7.1, neutral fresh L-cysteine (final concentration of 0.2 g/L), 2.68 mL of 0.8 mM NaHCO3 and 4.8 mM Na2O3S2•5H2O. The pH was adjusted to 7.2, and 0.5 mL of a vitamin solution (Frankel et al., 1997) and 2.5 mL of 0.01 M ferric quinate were added. Cells were inoculated at the OAI, and the cultures were incubated at 28°C for at least 15 days.
Oxygen concentrations were measured using a Unisense OX 100 oxygen microsensor, with a detection limit of 0.3 μM, coupled to a micromanipulator MM33 (Unisense, Aarhus, Denmark). Measurements were carried in duplicate tubes at 24 h intervals for 8 days in semi-solid heterotrophic medium. Calibration was done by submerging the sensor in a 0.1 M of ascorbate and 0.1 M of NaOH solution (0% O2 saturation) and oxygenated water (100% O2 saturation). The oxygen concentration profile was determined to a depth of 11 mm from the culture medium surface in 200 μm steps taking 5 s for each measurement. The O2 microsensor was stabilized for 2–3 h before any measurement. Data were recorded in the software SensorTrace Pro v3.0.2 (Unisense)
Light and Electron Microscopy
For light microscopy imaging, drops of ASW containing magnetically-enriched MTB were placed onto coverslips and imaged with Zeiss Axioplan 2 or Zeiss Axioimager microscopes (Carl Zeiss, Göttingen, Germany), both equipped with differential interference optics. A bar magnet was used to direct MTB to the edge of the drop where they accumulated. Transmission electron microscope (TEM) imaging of cells and elemental analysis of both magnetosomes and cell inclusions were performed in unfixed and unstained samples with a Jeol 1200 EX transmission (Jeol, Peabody, MA, USA) electron microscope equipped with a Noran accessory for energy-dispersive X-ray analysis (EDS) (Thermo Scientific, Palm Beach, FL, USA). Cells were placed onto formvar-coated electron microscopy 300 mesh copper grids, rinsed with distilled water and air-dried. Observations were performed at 100 kV, and spectra were acquired using a spot size of approximately 80 nm in diameter. For magnetosome measurements, the grids were observed with a Morgagni TEM (FEI Company, Hillsboro, OR, USA) operating at 80 kV, and images were analyzed using ImageJ software (rsb.info.nih.gov/ij/). Crystal size and shape factor were calculated as (length + width)/2 and width/length, respectively. Analyses of variance were performed using Graphpad InStat version 3.0.
For energy-filtering transmission electron microscopy (EFTEM), unstained ultra-thin sections were imaged with a Zeiss EM902 (Carl Zeiss, Göttingen, Germany) TEM equipped with a mirror-prism. Iron and oxygen maps were calculated using the three-window method with iTEM-EFTEM software (Olympus Soft Imaging Solutions GmbH, Münster, Germany). For high-resolution TEM (HRTEM), ultra-thin sections obtained as described in Abreu et al. (2013) were placed on formvar-coated copper grids and imaged in a FEG-Titan 80-300 (FEI Company, Hillsboro, OR, USA) TEM operated at 300 kV. All images were digitized with a 2kx2k Gatan UltraScan 1000 CCD camera (Gatan, Pleasanton, CA, USA) using the Digital Micrograph software (Mitchell, 2008). FFT from magnification-calibrated images was obtained using the same software.
16S rRNA Phylogenetic Analysis, Genome Sequencing and Comparative Analysis of Genes Related to Magnetosome Formation
The 16S rRNA gene was amplified from M. australis strain IT-1 using bacterial specific primers 8bF (5′-GRGTTTGATCCTGGCTCAG-3′) and 1512uR (5′-ACGGHTACCTTGTTACGACTT-3′). PCR products were cloned into pGEM-T Easy vector (Promega Corporation, Madison, WI) and sequenced using Macrogen sequence service (Macrogen, Korea). The alignment of 16S rRNA gene was performed using CLUSTAL W with BioEdit sequence alignment editor (Hall, 1999). A phylogenetic tree was constructed using MEGA version 5.2 (Tamura et al., 2011). We used the maximum likelihood statistical method based on Kimura 2 parameters (Kimura, 1980) with Gamma distribution and invariant sites (K2 + G + I) for analyses. The bootstrap value was calculated with 1000 replicates.
For DNA preparation for pyrosequencing, M. australis strain IT-1 was grown in semisolid medium. After 15 days of growth, bands from different tubes were removed, concentrated by centrifugation and washed several times with sterile distilled water. DNA samples were prepared according to Chen and Kuo (1993). M. australis strain IT-1 DNA was sequenced on a 454 GS FLX System sequencer (Roche Diagnostics GmbH/454 Life Sciences Corporation, Branford, CT, USA). The DNA sequences were analyzed with the SABIA (System for Automated Bacterial Integrated Annotation) platform (Almeida et al., 2004). Amino acid sequences of the MAI proteins from M. magneticum strain AMB-1 (AP007255) (Matsunaga et al., 2005), M. gryphiswaldense strain MRS-1 (AM085146) (Lohβe et al., 2011), M. magnetotacticum strain MS-1 (NZ_AAAP01003731) (Bertani et al., 2001), M. marinus strain MC-1 (NC_008576) (Schübbe et al., 2009), M. blakemorei strain MV-1 (FP102531) (Jogler et al., 2009), Gammaproteobacteria strain SS-5 (AFX88983—AFX88992) and M. australis strain IT-1 were used for identity, positives and E-value analysis through Blastp. Other sequences used in this work include Ca. M. multicellularis (HQ336745 and HQ336746) (Abreu et al., 2011), D. magneticus strain RS-1 (AP010904) (Nakazawa et al., 2009), Ca. D. magnetomortis strain BW-1 (HF547348) and strains ML-1 (JX869936—JX869937) and FH-1 (KC196864—KC196902) (Lefèvre et al., 2013b). A phylogenetic tree of concatenated MamABEIKMPQ amino acid sequences was constructed using the maximum likelihood statistical method based on WAG (Whelan and Goldman, 2001) with frequencies and gamma distribution (WAG+G+F) for analyses. Bootstrap value was calculated with 1000 replicates. The sequence of the MAI region has been submitted to GenBank/NCBI under the accession number KF933436.
Results
Isolation, Growth and Phylogenetic Analyses of Strain IT-1
Magnetotactic cocci were the dominant MTB morphotype in the environmental samples. Occasionally, we detected magnetotactic multicellular prokaryotes, as previously described (Lins et al., 2007). After separation using the magnetic “racetrack” (Wolfe et al., 1987), magnetically-enriched cocci were inoculated at the OAI of the semisolid autotrophic medium. Four weeks later, microaerophilic bands of coccoid MTB were observed and were then inoculated in semisolid heterotrophic [O2] gradient medium in which the culture was maintained. Cells formed individual colonies in shake tubes of heterotrophic medium (see experimental procedures for details). Single colonies were re-inoculated in fresh semisolid medium and resulted in pure cultures of a magnetotactic coccus with an average size of 1.4 ± 0.3 × 1.1 ± 0.3 μm (n = 130) as observed by light microscopy (Figure 1A) and confirmed by TEM (Figure 1B). The morphology of cells observed by TEM resembles a “faba” bean, showing well-defined convex and concave surfaces (Figure 1B). Cells contain intracellular granules (Figure 1B) filled with phosphorus as detected by EDS (Figure 1E).
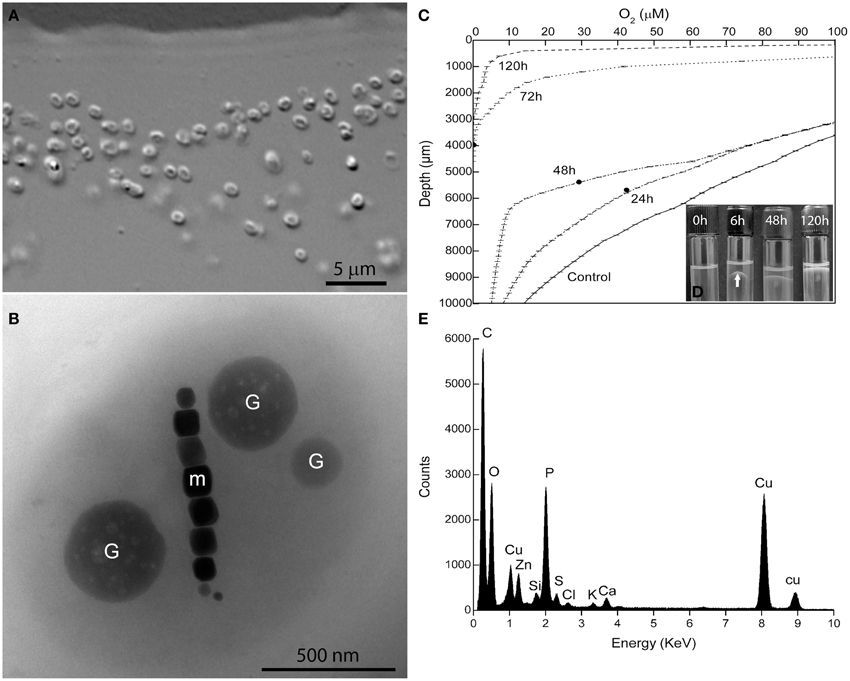
Figure 1. Characterization of Magnetofaba australis strain IT-1. (A) Differential interference contrast microscopy of a pure culture showing coccoid to ovoid cells. (B) Whole-mount transmission electron microscopy image of strain IT-1 showing a chain of elongated octahedral magnetosomes (m) and three conspicuous granules containing phosphorus (G). Oxygen concentration over time (C) and band formation (D) during strain IT-1 growth in semisolid heterotrophic medium. The points in the lines represent the position of the band in the culture medium at a given time. Control is represented by a non inoculated tube. Note the band with magnetotactic cells (arrow) after 6 h of inoculation. (E). Energy dispersive X-ray microanalysis spectrum of the phosphorus-rich granules. Ca, Zn, and K are cations associated with the granules. Cu peaks come from the supporting grid. The silicon peak is an artifact of the Si (Li) solid state detector used to collect X-rays.
The 16S rRNA gene of the culture was amplified, cloned, and sequenced for phylogenetic analyses. Approximately 50 clones were sequenced. These sequences were 99% similar, confirming the culture was pure. A consensus sequence was generated (accession number: JX534168) and phylogenetic analysis showed that strain IT-1 is phylogenetically affiliated with the Alphaproteobacteria (Figure 2). The 16S rRNA gene sequence of strain IT-1 is 93% similar to the sequence of an uncultured magnetotactic coccus collected from intertidal sediments of the Yellow Sea in China (Zhang et al., 2012; accession number JF421219) and 92% similar to sequences of the cultured species M. marinus strain MC-1 and MO-1 (accession numbers CP000471 and EF6435202, respectively). Thus, strain IT-1 represents a new genus of the magnetotactic cocci (and MTB in general). The name Magnetofaba australis gen. nov., sp. nov., is proposed for strain IT-1 (Ma. gne. to. faba Gr. n. magnês -êtos, a magnet; N.L. pref. magneto-, pertaining to a magnet; N.L. fem. N. faba, a faba bean; aus.tra'lis. L. masc. australis of Southern or of the south, which refers to the polar south-seeking magnetotaxis behavior and because the bacterium was isolated from South hemisphere).
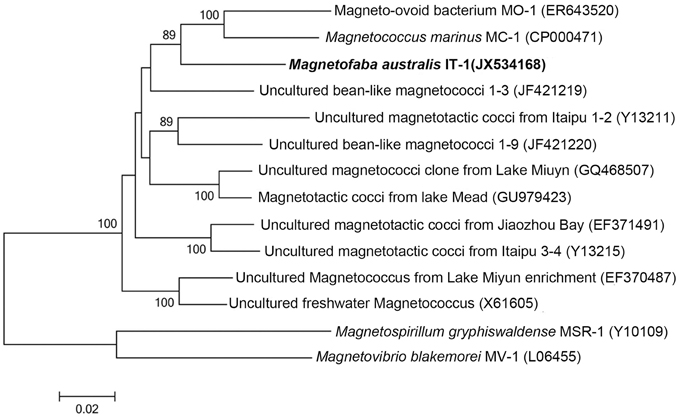
Figure 2. Phylogenetic analysis based on the 16S rRNA gene of strain IT-1 that forms a new genus in the Alphaproteobacteria. Bootstrap values at nodes are percentages of 1000 replicates; values higher than 70 are shown at the nodes. GenBank accession numbers are given in parentheses. The phylogenetic tree was constructed using the maximum likelihood method algorithm. The scale bar indicates 0.02 substitutions per nucleotide position.
Magnetofaba australis strain IT-1 grows as a microaerophilic band of cells in semisolid medium (Figure 1D). It grows slowly chemolithoautotrophically, using thiosulfate as electron donor and sodium bicarbonate as the major carbon source, forming a fine band of cells at the OAI at least 4 weeks after incubation. Under these conditions, cells biomineralize 6 ± 4 magnetosomes/cell (n = 100). Heterotrophic growth was also observed using sodium acetate and sodium succinate as the carbon source; cells grown under these conditions produced 9 ± 4 and 7 ± 3 magnetosomes per cell, respectively (n = 100 for both). When cells were grown in heterotrophic medium containing both sodium acetate and succinate, a band of magnetotactic cells, which contained 10 ± 3 magnetosomes/cell (n = 100), was observed at the OAI after 24 h. This band gradually moved toward the surface of the culture medium after 8 days of incubation (Figure 1D).
The oxygen concentration in the band was measured over 8 days (Figure 1C) in the heterotrophic medium; cells were initially inoculated at the OAI, in which [O2] was less than 3 μM. During the first 6 h after inoculation, cells moved up approximately 3 mm, forming a “bell-shaped” band (Figure 1D) in the medium ([O2] = 50 ± 5 μM). After 24 h, the band was positioned between 24.6 ± 0.7 and 43 ± 1 μM O2, with a less bent bell-shape. 48 h later, the band was located in 29.2 ± 2.7 μM of [O2]. After 72 h, the bell-shaped band became a flat band positioned at [O2] between 9.4 ± 1.5 μM. Until this time, the band did not reach the meniscus of the culture medium. As the cells grew (up to 168 h), O2 was consumed, and the dense population of cells reached the meniscus, presumably to use oxygen present in the headspace of the tube (Figure 1D). At 72 h of incubation the cells of M. australis have consumed near 90% of oxygen ([O2] < 9.4 ± 1.5 μM), and the band appears thicker than in 24 and 48 h. With 72 h, it is likely that the magnetite production also increased, given the higher number of cells and that the population remained responding to the magnetic field at the end of the experiment. Therefore, M. australis strain IT-1 can grow and synthetize magnetite with [O2] below 10 μM, similar to the Magnetospirillum species, which requires microaerobic conditions (2–7 μM O2) to grow and synthesize magnetite (Schüler and Baeuerlein, 1998).
In hanging drop assays under oxic conditions, M. australis strain IT-1 exhibited South-seeking polar magnetotaxis swimming under the magnetic field of a bar magnet with a fast back and forth swimming pattern near the edge of the drop. M. australis swims at average speeds of 186 μm.s−1 ± 63 (n = 50) and can reach 300 μm.s−1. Cells are propelled by two bundles of lophotrichous flagella, each at one extremity of the cell. A helical trajectory was observed when movement was recorded with a CCD camera using dark-field microscopy.
Magnetosomes
Cells of M. australis strain IT-1 each produce a single chain of magnetosomes (see Figure 1B). Each chain consists of 10 ± 3 magnetosomes (n = 100) in cells grown heterotrophically in semi-solid [O2] gradient medium. Energy-dispersive X-ray analysis (Figure 3A) and elemental mapping by EFTEM (Figure 3B) confirmed that the magnetosomes contain iron (Figure 3C) and oxygen (Figure 3D). Electron diffraction (Figure 3E) of isolated magnetosome crystals (Figure 3F) were indexed based on standard cubic system for magnetite. Distances and angles between spots were consistent with magnetite (Fe3O4). Approximately 4% defective twins and multiple twin magnetosomes are observed in M. australis strain IT-1. The crystals are octahedral particles elongated along the <111> axis. Figure 3G shows the size distribution of magnetosomes (n = 100), estimated by calculating the best fit of an ellipse (major axis = length; minor axis = width). The average size [(length + width)/2] was 78 ± 24 nm (average length = 83 ± 26 nm; average width 74 ± 23 nm). Figure 3H shows the shape factor distribution (average of width/length = 0.89 ± 0.05), and Figure 3I shows the scatter plot of length and width (adjustment r2 = 0.962). Magnetosomes in M. australis strain IT-1 are each enveloped by a membrane, as shown in TEM images of ultra-thin sections (Figure 4A).
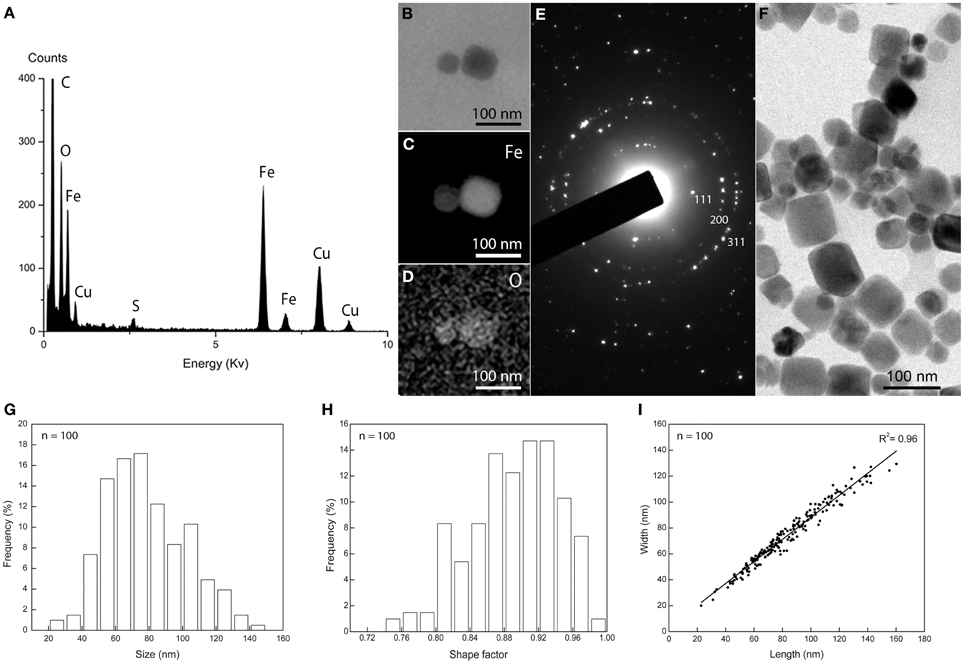
Figure 3. Magnetosome biomineralization in Magnetofaba australis strain IT-1. (A) Energy dispersive X-ray spectrum showing Fe and O as the main elements in the magnetosomes. Cu originates from the grid bar. (B) Elemental mapping by EFTEM of a magnetosome showing the distribution of iron (C) and oxygen (D). (E) Electron diffraction pattern of isolated magnetosomes shown in (F). (G) Size distribution, (H) shape factor distribution, and (I) scatter plot of length and width of magnetosomes in Magnetofaba australis strain IT-1 grown in heterotrophic medium.
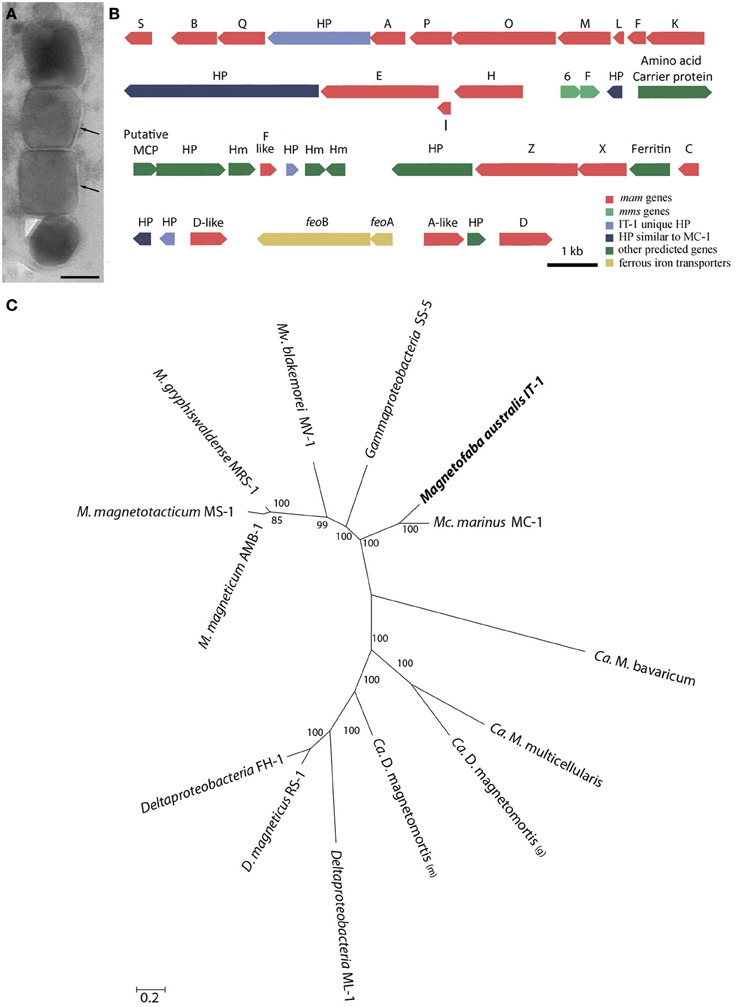
Figure 4. Mam genes in Magnetofaba australis strain IT-1. (A) Ultra-thin section image of a magnetosome chain showing the membrane (arrow) surrounding each particle. (B) Organization of open reading frames (ORFs) containing putative magnetosome-related genes. HP, hypothetical proteins. (C) Maximum likelihood phylogenetic analysis based on concatenated amino acid sequences encoded by conserved MamABEIKMPQ genes using the statistic method WAG+G+F. Note that M. australis strain IT-1 and M. marinus strain MC-1 share a common ancestor, indicating a similar biomineralization process. Bootstrap values at nodes are percentages of 1000 replicates. The scale bar indicates 0.2 substitutions per amino acid position.
Magnetosome Genes
Comparative genome analysis of M. australis strain IT-1 with other magnetotactic Alphaproteobacteria based on genes related to magnetotaxis and magnetosome synthesis revealed a genomic region of 40.399 Kb that contained both genes associated with magnetosome biomineralization as well as those that encode some hypothetical proteins present in the putative MAI of M. marinus. This region contains 39 genes, 22 of which show a high degree of similarity with the biomineralization-related genes of magnetotactic Alphaproteobacteria (mam genes and mms genes; Table 1). This region also contains eight hypothetical proteins, four with high similarity values to hypothetical proteins in the putative MAI of M. marinus and four that are not found in any known MTB. A feoAB gene cluster was also identified in this region, similar to that found in Magnetospira sp QH-2, Ca. Mg. multicellularis and Ca. D. magnetomortis. Figure 4B shows the organization of the genes in M. australis strain IT-1. The mamAB-like gene cluster has the same gene organization found in M. marinus (mamK, mamF, mamL, mamM, mamO, mamP, mamA, HP, mamQ, mamB and mamS) (Schübbe et al., 2009), except that mamT is absent. The mamC gene is located in the mamCXZ gene cluster, similar to M. blakemorei (Jogler et al., 2009), and not in the mamHIEC gene cluster as in M. marinus. The mms6 gene cluster and the 10 genes encoding MamF-like protein, chemotaxis protein, signal transduction proteins and three hypothetical proteins are located between the mamAB and mamCXZ gene clusters. At the end of the mamCXZ gene cluster, genes encoding MamD-like, FeoB, FeoA, MamA-like and MamD proteins are present. All predicted proteins related to biomineralization and magnetotaxis genes described here for M. australis strain IT-1 share the highest similarity to those of M. marinus, including MamA-like, MamD-like, MamF-like, FeoB and FeoA proteins. A notable exception is MamC, which is more related to that from M. magneticum strain AMB-1 (coverage 86%, identity 63%, E-value 5e-038).
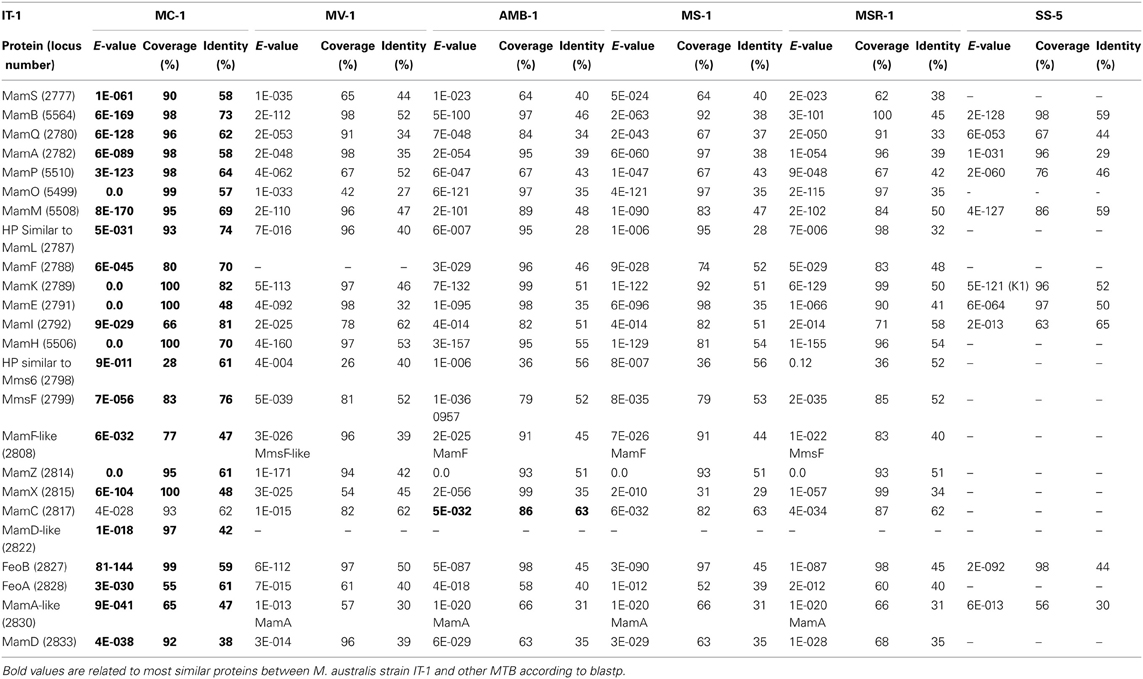
Table 1. Comparative analysis of biomineralization genes found in Magnetofava australis strain IT-1 and other cultivated magnetotactic bacteria with their genomes sequenced.
The coverage, identity and E-value of the Blastp analysis of predicted Mam proteins from magnetotactic Alphaproteobacteria and the Gammaproteobacteria strain SS-5 were analyzed (Table 1). M. australis MamA-like, MamD-like and MamF-like proteins were compared to MamA-like, MamD-like and MamF-like of M. marinus, and MamA, MamD and MamF proteins of other MTB. The M. australis MamA-like predicted protein is closely related to the M. marinus protein sequence (coverage 65%, identity 47%, E-value 9e-041), while MamD-like is only related to the MamD-like from M. marinus (coverage 97%, identity 42%, E-value 1e-018). For some MTB, M. australis MamF-like sequences were more similar to MmsF or MmsF-like proteins (e.g., from M. gryphiswaldense and M. blakemorei). MamF and MmsF of M. gryphiswaldense MSR-1 have already been reported to share 65% identity (Murat et al., 2012). M. australis strain IT-1 MamA-like predicted proteins with MamA proteins share 30% identity (coverage 57%, E-value 7E-018), while MamD-like predicted proteins share 73% identity with MamD (coverage 23%, E-value 7E-05), and MamF-like predicted proteins share 33% identity (coverage 89%, E-value 2E-016) and 39% identity (coverage 90%, E-value 1E-021) with MamF and MmsF, respectively.
Phylogenetic analysis based on concatenated conserved Mam proteins (MamABEIKMPQ) of other MTB showed that M. australis strain IT-1 clusters with other Alphaproteobacteria that produce magnetite and is most related to M. marinus strain MC-1. The Gammaproteobacteria strain SS-5, which synthesizes cuboctahedral magnetite magnetosomes, groups with the Alphaproteobacteria. Interestingly, after M. marinus, MamB, MamQ and MamM of M. australis strain IT-1 have the most similarity with MamB of strain SS-5 (coverage 98%, E-value 2E-128, identity 59%), MamQ (coverage 67%, E-value 6E-053, identity 44%) and MamM (coverage 86%, E-value 4E-127, identity 59%). The phylogenetic tree (Figure 4C) with concatenated conserved mam genes does not show a clear evolutionary event that divides magnetotactic strains producing cuboctahedral and prismatic hexagonal crystals because bacteria such as M. blakemorei and M. marinus do not form a separate branch. The evolutionary relationship between M. australis and M. marinus suggests a recent divergence between the cellular magnetosome biomineralization machinery in these species.
Discussion
The number of MTB isolated in culture has recently increased (from 1978 to 2009, 11 MTB were available in axenic cultures; in 2012 this number was 25; Lefèvre and Long-Fei, 2013). However, all cultured MTB were isolated in the Northern Hemisphere and originally showed NS magnetotaxis. This work presents the first isolation of a SS-MTB from the Southern Hemisphere. The new isolate is phylogenetically affiliated with the Alphaproteobacteria class of the Proteobacteria phylum, a division that contains almost all known Fe3O4-producing MTB (DeLong et al., 1993; Spring et al., 1998), and clearly represents a new genus based on 16S rRNA gene sequence similarities. This new coccus represents a third phylogenetic group of MTB occurring in the Itaipu Lagoon (Spring et al., 1998). M. australis strain IT-1 is distinct from all the other cultivated magnetotactic cocci examined to date because of its South-seeking polar magnetotactic behavior, it has “faba bean” cell morphology and elongated cuboctahedral magnetite magnetosomes. Based on its 16S rRNA gene sequence, M. australis is more related to an uncultured magnetotactic coccus found in the intertidal sediments of the Yellow Sea in China (93% similarity; Zhang et al., 2012). This uncultured bacterium also shows a bean-like morphology and produces magnetite magnetosomes (Zhang et al., 2012). However, magnetosome crystal morphology, size, shape factor, magnetosome number and swimming speed in M. australis are different from the coccus described by Zhang et al. (2012). The close phylogenetic relationships may not be significantly associated to the biomineralization genes, which may result in variations in the regulation of crystal morphology between these MTB. Hopefully, physiological studies and genomic analysis of these MTB will result in information that advances the understanding of biomineralization in bean-like magnetotactic cocci.
Magnetofaba australis strain IT-1 has a swimming speed similar to that observed in strain MO-1 (Lefèvre et al., 2009), higher than speeds found in other magnetotactic cocci (Zhang et al., 2012). Possibly, a highly coordinated flagella rotation is necessary to allow this high swimming speed. The high swimming speed would be advantageous for the survival of M. australis strain IT-1 because it would enable the cell to escape quickly from unfavorable environment conditions. Most cells of M. australis strain IT-1 (over 80%) has a South-seeking behavior when observed in hanging drop assays under oxic conditions, but we have also found North-seeking cells in the culture flasks. Further studies are necessary to compare the swimming behavior and orientation of magnetotactic cocci, along with their flagellar apparatus at the genetic and structural levels. We believe that such studies can now be performed because of the available SS-MTB cultures.
The role of biomineralization and magnetotaxis genes in MTB is not only key in the determination of how magnetosomes are formed in MTB but also important in understanding the evolution of magnetotaxis (Lefèvre and Bazylinski, 2013). Although several recent reports have addressed this issue (Lefèvre et al., 2013a,b), only a relatively small number of MTB species have been considered thus far. However, advances in the culturing of new strains promises to improve the low number of species available for evolutionary studies. M. australis strain IT-1 is the first MTB isolated in axenic culture that produces cuboctahedral magnetite magnetosomes whose magnetosome biomineralization genes have been sequenced. New data on the magnetosome biomineralization genes of coccoid or ovoid MTB increases our understanding of the biomineralization processes in MTB in general. For example, M. marinus and M. australis share several hypothetical proteins, not found in other MTB that may have key functions in biomineralization or magnetotaxis like the hypothetical protein between MamE and MamK (locus 02790), the hypothetical protein between MmsF and the Amino acid carrier protein (locus 02801), the Amino acid carrier protein (locus 02803), a hemerythrin-like (locus 02811), and a ferritin-like (locus 02816).
The analysis of the putative functions of mam genes is also important in the interpretation of the evolution of magnetotaxis. Variations in both the order and sequence of mam genes between M. australis and the closely related M. marinus could explain differences between magnetosome crystal morphology in the two species. The MamC predicted protein sequence of M. australis is more similar to that of M. magneticum strain AMB-1, which is particularly interesting because cultivated Magnetospirillum species described thus far produce cuboctahedral magnetite crystals that are not elongated (Amann et al., 2007). Scheffel et al. (2008) showed that the protein MamC and other proteins in the same operon (mamGFDC) are not essential for magnetosome formation but are involved in controlling crystal size and morphology in M. gryphiswaldense. In M. australis, mamC is organized in a mamCXZ operon, similar to M. blakemorei. The other proteins involved in the size and shape of magnetosomes (MamD, MamF, Mms6, and MmsF) are more closely related to those found in M. marinus. Therefore, the fact that M. australis MamC is related to cuboctahedral magnetite-producing bacteria suggests that this protein might be responsible for crystal morphology in this case. Additionally, based on the similarity of mamXZC gene organization between M. australis and M. blakemorei, we speculate that gene organization and/or preferential expression of mamCXZ could be involved in crystal elongation. MmsF has been shown to be involved in the geometry of magnetosome maturation, as the deletion of mmsF resulted in elongated magnetosomes in M. magneticum strain AMB-1 (Murat et al., 2012). However, we did not identify a close similarity between MmsF from M. australis strain IT-1 and other MTB that synthesize elongated octahedral crystals. The expression level of MmsF may influence crystal morphology, which could explain how closely related mam genes from different species (i.e., M. australis and M. marinus) produce magnetosomes with different characteristics. Variation in the expression level of the mamGFDC operon in M. gryphiswaldense resulted in crystals exceeding the size of those of the wild-type (Scheffel et al., 2008). The absence of mamT in M. australis strain IT-1 reveals a new group of 19 genes common to cultivated magnetotactic Alphaproteobacteria: mamA, B, C, D, E, F, H, I, K, L, M, N, O, P, Q, R, S, X and Z, in addition to the mms6 and mmsF genes. Although mamT is present in the Alpha- and Deltaproteobacteria, it is not essential for biomineralization. Proteins with similar function (MamP or MamE) are likely sufficient to control the balance between Fe2+ and Fe3+ in the magnetosome. In M. magneticum (Murat et al., 2010) and M. gryphiswaldense (Lohβe et al., 2011) mamT is not essential for magnetosome synthesis.
Considering that both M. australis strain IT-1 and M. marinus strain MC-1 have a common magnetotactic ancestor and that biomineralization proteins apparently evolved together in both strains, it is reasonable to assume that a common ancestor exists among all freshwater and marine MTB from the Magnetococcales order. No non-MTB belonging to the Magnetococcales order has ever been reported, but this fact does not preclude HGT among Alphaproteobacteria because strains phylogenetically closer to Magnetospirillum do not have the magnetotactic phenotype (Jogler and Schüler, 2009). Thus, magnetosome biomineralization genes common to all MTB (mamABEIKMPQ) might have been acquired from an ancestor common to all MTB (Abreu et al., 2011; Lefèvre et al., 2013a). However, genes such as mamCDF, mamL, mamXZ, mms6, and mmsF could have been acquired by descent of magnetotactic Alphaproteobacteria and magnetotactic cocci, which appear to emerge as the most basal lineage of the Alpha- and Gammaproteobacteria (Singer et al., 2011; Lefèvre and Bazylinski, 2013). mamG, mamR, mamV, mamU, and mamY genes were likely acquired recently by Magnetospirillum species, given that the magnetotactic cocci studied so far, M. marinus strain MC-1 and M. australis strain IT-1, do not contain these genes. Differences observed in the biomineralization genes between M. australis strain IT-1, M. marinus strain MC-1 and the other Alphaproteobacteria are possibly a result of gene rearrangements, deletions or insertions of new genes through the evolution or a post-acquisition of the biomineralization genotype among MTB. Culture and sequencing of new species of magnetotactic cocci from freshwater or marine water are needed to improve the understanding the evolutionary events that occurred in the Alphaproteobacteria and magnetotactic cocci and will more precisely define the Magnetococcaceae family in the Magnetococcales order as either the earliest diverging order in the Alphaproteobacteria class or as a new class of Proteobacteria, as proposed by Singer et al. (2011). M. australis strain IT-1 is now the third cultivated magnetotactic coccus that represents a second new genus in the Magnetococcaceae family and is the first cultivated SS-MTB.
Author Contributions
All authors contributed to the analysis of data and composition of the paper; Viviana Morillo, Fernanda Abreu and Ana C. Araujo: experimental data acquisition and cultivation; Luiz G. P. de Almeida and Ana T. R. de Vasconcelos: pyrosequecing and bioinformatics; Alex Enrich-Prast: microeletrode measurements and interpretation; MF: high-resolution transmission electron microscopy, Viviana Morillo, Fernanda Abreu, Dennis A. Bazylinski and Ulysses Lins: analyzed data and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Dr. Andrea P. C. Campos and the DIMAT-INMETRO and LABNANO-CBPF for use of high-resolution transmission electron microscopy facilities. Financial support from the Brazilian agencies CAPES, CNPq and FAPERJ is acknowledged. Dennis A. Bazylinski is supported by US National Science Foundation (NSF) Grant EAR-0920718 and by subcontract SC-12-384 from US Department of Energy contract DE-AC02-07CH11358 issued to the Ames Laboratory at Iowa State University.
References
Abreu, F., Cantão, M. E., Nicolas, M. F., Barcello, F. G., Morillo, V., Almeida, L. G., et al. (2011). Common ancestry of iron oxide- iron-sulfide-based biomineralization in magnetotactic bacteria. ISME J. 5, 1634–1640. doi: 10.1038/ismej.2011.35
Abreu, F., Morillo, V., Nascimento, F. F., Werneck, C., Cantão, M. E., Ciapina, L. P., et al. (2013). Deciphering unusual uncultured magnetotactic multicellular prokaryotes through genomics. ISME J. doi: 10.1038/ismej.2013.203. [Epub ahead of print].
Almeida, L. G., Paixão, R., Souza, R. C., Costa, G. C., Barrientos, F. J., Santos, M. T., et al. (2004). A system for automated bacterial (genome) integrated annotation - SABIA. Bioinformatics 20, 2832–2833. doi: 10.1093/bioinformatics/bth273
Amann, R., Peplies, J., and Schüler, D. (2007). “Diversity and taxonomy of magnetotactic bacteria,” in Magnetoreception and Magnetosomes in Bacteria, ed D. Schüler (Berlin; Heidelberg: Springer-Verlag), 25–36. doi: 10.1007/7171_037
Bazylinski, D. A., and Frankel, R. B. (2004). Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2, 217–230. doi: 10.1038/nrmicro842
Bazylinski, D. A., Williams, T. J., Lefèvre, C. T., Berg, R. J., Zhang, C. L., Bowser, S. S., et al. (2013a). Magnetococcus marinus gen. nov., sp. nov., a marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. nov.; Magnetococcales ord. nov.) at the base of the Alphaproteobacteria. Int. J. Sys. Evol. Microbiol. 63, 801–808. doi: 10.1099/ijs.0.038927-0
Bazylinski, D. A., Williams, T. J., Lefèvre, C. T., Trubitsyn, D., Fang, J., Beveridge, T. J., et al. (2013b). Magnetovibrio blakemorei gen. nov. sp. nov., a magnetotactic bacterium (Alphaproteobacteria: Rhodospirillaceae) isolated from a salt marsh. Int. J. Syst. Evol. Microbiol. 63, 1824–1833. doi: 10.1099/ijs.0.044453-0
Bertani, L. E., Weko, J., Phillips, K. V., Gray, R. F., and Kirschvink, J. L. (2001). Physical and genetic characterization of the genome of Magnetospirillum magnetotacticum, strain MS-1. Gene 264, 257–263. doi: 10.1016/S0378-1119(01)00331-6
Blakemore, R. P. (1982). Magnetotactic bacteria. Ann. Rev. Microbiol. 36, 217–238. doi: 10.1146/annurev.mi.36.100182.001245
Blakemore, R. P., Frankel, R. B., and Kalmijn, A. J. (1980). South-seeking magnetotactic bacteria in the Southern Hemisphere. Nature 286, 384–385. doi: 10.1038/286384a0
Chen, W., and Kuo, T. (1993). A simple and rapid method for the preparation of Gram-negative genomic DNA. Nucleic Acids Res. 21, 2260. doi: 10.1093/nar/21.9.2260
DeLong, E. F., Frankel, R. B., and Bazylinski, D. A. (1993). Multiple evolutionary origins of magnetotaxis in bacteria. Science 259, 803–806. doi: 10.1126/science.259.5096.803
Frankel, R. B., and Bazylinski, D. A. (1994). Magnetotaxis and magnetic particles in bacteria. Hyperfine Interac. 90, 135–142. doi: 10.1007/BF02069123
Frankel, R. B., Bazylinski, D. A., Johnson, M. S., and Taylor, B. L. (1997). Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73, 994–1000. doi: 10.1016/S0006-3495(97)78132-3
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Ji, B., Zhang, S., Arnoux, P., Rouy, Z., Alberto, F., Philippe, F., et al. (2014). Comparative genomic analysis provides insights into the evolution and niche adaptation of marine Magnetospira sp. QH-2 strain 1. Environ. Microbiol. 16, 525–544. doi: 10.1111/1462-2920.12180
Jogler, C., Kube, M., Schübbe, S., Ullrich, S., Teellng, H., Bazylinski, D., et al. (2009). Comparative analysis of magnetosome gene clusters in magnetotactic bacteria provides further evidence for horizontal gene transfer. Environ. Microbiol. 11, 1267–1277. doi: 10.1111/j.1462-2920.2009.01854.x
Jogler, C., and Schüler, D. (2009). Genomics, genetics, and cell biology of magnetosome formation. Ann. Rev. Microbiol. 63, 501–521. doi: 10.1146/annurev.micro.62.081307.162908
Jogler, C., Wanner, G., Kolinko, S., Niebler, M., Amann, R., Petersen, N., et al. (2011). Conservation of proteobacterial magnetosome genes and structures in an uncultivated member of the deep-branching Nitrospira phylum. Proc. Natl. Acad. Sci. U.S.A. 108, 1134–1139. doi: 10.1073/pnas.1012694108
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Lefèvre, C. T., and Bazylinski, D. A. (2013). Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol. Mol. Biol. Rev. 77, 497–526. doi: 10.1128/MMBR.00021-13
Lefèvre, C. T., Bernadac, A., Yu-Zhang, K., Pradel, N., and Long-Fei, W. (2009). Isolation and characterization of a magnetotactic bacterial culture from the Mediterranean Sea. Environ. Microbiol. 11, 1646–1657. doi: 10.1111/j.1462-2920.2009.01887.x
Lefèvre, C. T., and Long-Fei, W. (2013). Evolution of the bacterial organelle responsible for magnetotaxis. Trends Microbiol. 21, 534–543. doi: 10.1016/j.tim.2013.07.005
Lefèvre, C. T., Menguy, N., Abreu, F., Lins, U., Pósfai, M., Prozorov, T., et al. (2011). A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate reducing bacteria. Science 334, 1720–1723. doi: 10.1126/science.1212596
Lefèvre, C. T., Trubitsyn, D., Abreu, F., Kolinko, S., Almeida, L. G. P., Vasconcelos, A. T., et al. (2013a). Monophyletic origin of magnetotaxis and the first magnetosomes. Environ. Microbiol. 15, 2267–2274. doi: 10.1111/1462-2920.12097
Lefèvre, C. T., Trubitsyn, D., Abreu, F., Kolinko, S., Jogler, C., Gonzaga, L., et al. (2013b). Comparative genome analysis of magnetotactic bacteria from the Deltaproteobacteria provides new insights into magnetite and greigite magnetosome genes required for magnetotaxis. Environ. Microbiol. 15, 2712–2735. doi: 10.1111/1462-2920.12128
Lefèvre, C. T., Viloria N., Schmidt, M. L., Pósfai, M., Frankel, R. B., and Bazylinski, D. A. (2012). Novel magnetite-producing magnetotactic bacteria belonging to the Gammaproteobacteria. ISME J. 6, 440–450. doi: 10.1038/ismej.2011.97
Lins, U., Freitas, F., Neumann, C. K., Barros, H. L., Esquivel, D. M., and Farina, M. (2003). Simple homemade apparatus for harvesting uncultured magnetotactic microorganisms. Braz. J. Microbiol. 34, 111–116. doi: 10.1590/S1517-83822003000200004
Lins, U., Keim, C., Evans, F., Farina, M., and Buseck, P. (2007). Magnetite (Fe3O4) and Greigite (Fe3S4) crystals in multicellular magnetotactic prokaryotes. Geomicrobiol. J. 24, 43–50. doi: 10.1080/01490450601134317
Lohβe, A., Ullrich, S., Katzmann, E., Borg, S., Wanner, G., Richter, M., et al. (2011). Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS ONE 6:e25561. doi: 10.1371/journal.pone.0025561
Matsunaga, T., Okamura, Y., Fukuda, Y., Wahyudi, A. T., Murase, Y., and Takemaya. H. (2005). Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 12, 157-166. doi: 10.1093/dnares/dsi002
Mitchell, D. R. G. (2008). DiffTools: software tools for electron diffraction in digital micrograph. Microsc. Res. Tech. 71, 588–593. doi: 10.1002/jemt.20591
Murat, D., Falahati, V., Bertineti, L., Csencsits, R., Kornig. A., Downing. K., et al. (2012). The magnetosome membrane protein, MmsF, is a major regulator of magnetite biomineralization in Magnetospirillum magneticum AMB-1. Mol. Microbiol. 85, 684–699. doi: 10.1111/j.1365-2958.2012.08132.x
Murat, D., Quilan, A., Vali, H., and Komeili, A. (2010). Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. U.S.A. 107, 5593–5598. doi: 10.1073/pnas.0914439107
Nakazawa, H., Arakaki, A., Narita-Yamada, S., Yashiro, I., Jinno, K., Aoki, N., et al. (2009). Whole genome sequence of Desulfovibrio magneticus strain RS-1 revealed common gene clusters in magnetotactic bacteria. Gen. Res. 19, 1801–1808. doi: 10.1101/gr.088906.108
Richter, M., Kube, M., Bazylinski, D., Lombardot, T., Reinhardt, R., and Schüler, D. (2007). Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J. Bacteriol. 189, 4899–4910. doi: 10.1128/JB.00119-07
Sakaguchi, T., Arakaki, A., and Matsunaga, T. (2002). Desulfovibrio magneticus sp. nov., a novel sulfate-reducing bacterium that produces intracellular single-domain-sized magnetite particles. Int. J. Syst. Evol. Microbiol. 52, 215–221.
Scheffel, A., Gärdes, A., Grünberg, K., Wanner, G., and Schüler, D. (2008). The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J. Bacteriol. 190, 377–386. doi: 10.1128/JB.01371-07
Schübbe, S., Williams, T. J., Xie, G., Kiss, H. E., Brettin, T. S., Martinez, D., et al. (2009). Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Appl. Environ. Microbiol. 75, 4835–4852. doi: 10.1128/AEM.02874-08
Schüler, D. (2008). Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol. Rev. 32, 654–672. doi: 10.1111/j.1574-6976.2008.00116.x
Schüler, D., and Baeuerlein, E. (1998). Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 180, 159–162.
Seeley, H. W., Vandemar, P. J., and Lee, J. J. (1991). Microbes in Action: a Laboratory Manual of Microbiology, 4th Edn. New York, NY: W. H. Freeman and Co.
Simmons, S. L., Bazylinski, D. A., and Edwards, K. J. (2006). South-seeking magnetotactic bacteria in the northern hemisphere. Science 311, 371–374. doi: 10.1126/science.1122843
Singer, E., Emerson, D., Webb, E. A., Barco, R. A., Kuenen, J. G., Nelson, W. C., et al. (2011). Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS ONE 6:e25386. doi: 10.1371/journal.pone.0025386
Spring, S., Lins, U., Amann, R., Schleifer, K. H., Ferreira, L. C., Esquivel, D. M., et al. (1998). Phylogenetic affiliation and ultrastructure of uncultured magnetic bacteria with unusually large magnetosomes. Arch. Microbiol. 169, 136–147. doi: 10.1007/s002030050553
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699. doi: 10.1093/oxfordjournals.molbev.a003851
Williams, T. J., Lefèvre, C. T., Zhao, W., Beveridge, T. J., and Bazylinski, D. A. (2012). Magnetospira thiophila, gen. nov. sp. nov., a new marine magnetotactic bacterium that represents a novel lineage within the Rhodospirillaceae (Alphaproteobacteria). Int. J. Sys. Evol. Microbiol. 62, 2443–2450. doi: 10.1099/ijs.0.037697-0
Wolfe, R. S., Thauer, R. K., and Pfennig, N. (1987). A capillary racetrack method for isolation of magnetotactic bacteria. FEMS Microbiol. Ecol. 45, 31–35. doi: 10.1111/j.1574-6968.1987.tb02335.x
Keywords: Magnetofaba australis strain IT-1, magnetite, magnetosome, South-seeking magnetotactic bacteria, biomineralization genes
Citation: Morillo V, Abreu F, Araujo AC, de Almeida LGP, Enrich-Prast A, Farina M, de Vasconcelos ATR, Bazylinski DA and Lins U (2014) Isolation, cultivation and genomic analysis of magnetosome biomineralization genes of a new genus of South-seeking magnetotactic cocci within the Alphaproteobacteria. Front. Microbiol. 5:72. doi: 10.3389/fmicb.2014.00072
Received: 30 November 2013; Accepted: 10 February 2014;
Published online: 25 February 2014.
Edited by:
Damien Faivre, Max Planck Society, GermanyReviewed by:
Concepcion Jimenez-Lopez, University of Granada, SpainDirk Schüler, Ludwig-Maximilians-Universität München, Germany
Copyright © 2014 Morillo, Abreu, Araujo, de Almeida, Enrich-Prast, Farina, de Vasconcelos, Bazylinski and Lins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ulysses Lins, Instituto de Microbiologia Paulo de Góes, Universidade Federal do Rio de Janeiro, Avenida Carlos Chagas Filho, 373, 21941-902 Rio de Janeiro, Brazil e-mail: ulins@micro.ufrj.br
 Viviana Morillo
Viviana Morillo Fernanda Abreu
Fernanda Abreu Ana C. Araujo
Ana C. Araujo Luiz G. P. de Almeida2
Luiz G. P. de Almeida2 Alex Enrich-Prast
Alex Enrich-Prast Dennis A. Bazylinski
Dennis A. Bazylinski Ulysses Lins
Ulysses Lins