- 1Department of Microbiology and Immunology, Catholic University of Health and Allied Sciences, Mwanza, Tanzania
- 2Institute of Medical Microbiology, Justus-Liebig University, Giessen, Germany
- 3German Center for Infection Research, DZIF Partner Site Giessen-Marburg-Langen, Giessen, Germany
- 4Department of Microbiology/Immunology, Muhimbili University of Health and Allied Sciences, Dar es Salaam, Tanzania
- 5Southern African Centre for Infectious Disease Surveillance, Sokoine University of Agriculture, Morogoro, Tanzania
The increased presence of extended-spectrum beta-lactamase (ESBL)-producing bacteria in humans, animals, and their surrounding environments is of global concern. Currently there is limited information on ESBL presence in rural farming communities worldwide. We performed a cross-sectional study in Mwanza, Tanzania, involving 600 companion and domestic farm animals between August/September 2014. Rectal swab/cloaca specimens were processed to identify ESBL-producing Enterobacteriaceae. We detected 130 (21.7%) animals carrying ESBL-producing bacteria, the highest carriage being among dogs and pigs [39.2% (51/130) and 33.1% (43/130), respectively]. The majority of isolates were Escherichia coli [93.3% (125/134)] and exotic breed type [OR (95%CI) = 2.372 (1.460–3.854), p-value < 0.001] was found to be a predictor of ESBL carriage among animals. Whole-genome sequences of 25 ESBL-producing E. coli were analyzed for phylogenetic relationships using multi-locus sequence typing (MLST) and core genome comparisons. Fourteen different sequence types were detected of which ST617 (7/25), ST2852 (3/25), ST1303 (3/25) were the most abundant. All isolates harbored the blaCTX-M-15 allele, 22/25 carried strA and strB, 12/25 aac(6′)-lb-cr, and 11/25 qnrS1. Antibiotic resistance was associated with IncF, IncY, as well as non-typable plasmids. Eleven isolates carried pPGRT46-related plasmids, previously reported from isolates in Nigeria. Five isolates had plasmids exhibiting 85–99% homology to pCA28, previously detected in isolates from the US. Our findings indicate a pan-species distribution of ESBL-producing E. coli clonal groups in farming communities and provide evidence for plasmids harboring antibiotic resistances of regional and international impact.
Introduction
Extended-spectrum beta-lactamases (ESBL) are enzymes encoded on the chromosome or on plasmids, conferring resistance to penicillins, cephalosporins, and monobactams (Bradford, 2001). The burden of ESBL is currently of global concern not only to humans but also animals and the ecosystem at large (Hawkey and Jones, 2009; Dierikx et al., 2012; Geser et al., 2012; Lupindu et al., 2014). This is due to interfaces that promote and enable the transmission of strains and antibiotic resistance genes harbored therein (Lupindu et al., 2014; Rubin and Pitout, 2014). Different factors for ESBL fecal carriage have been reported in both humans and animals, such as previous exposure to antimicrobial agents, hygienic behavior, or microbial inherent genetic factors (Bester and Essack, 2008; Geser et al., 2012; Lupindu et al., 2014; Nelson et al., 2014). The prevailing evidence from hospital and community-based studies in Mwanza, Tanzania, suggests that CTX-M-15 is predominant in ESBL-producing clinical isolates and is associated with significant morbidity and mortality (Kayange et al., 2010; Mshana et al., 2011a). Moreover, despite evidence from other regions in Tanzania on the growing problem of antimicrobial resistance in animals associated with varying ESBL-alleles (Katakweba, 2014; Lupindu et al., 2014), there is still limited information regarding genetic diversity of ESBL isolates among companion and domestic farm animals in the Mwanza region. Therefore, this study aimed to address the magnitude and the risk factors associated with ESBL carriage, identify ESBL alleles and analyze the MLST types among companion and domestic farm animals in this region.
Materials and Methods
Study Design, Site, and Sampling Procedures
This cross-sectional study was conducted between August and September 2014, and involved 600 companion and domestic farm animals in three districts of the Mwanza region (Ilemela, Nyamagana, and Misungwi). Healthy dogs, sheep, goats, chickens, pigs, and cattle were sampled. In case of a herd of animals, 10% of animals were randomly selected and sampled.
Data Collection and Laboratory Procedures
Demographic data such as sex, residence, breed type, history of antibiotic use, and type of antibiotics used were collected. Rectal swabs from pigs, cattle, sheep, goats, and dogs and cloaca swabs from layers and local chickens were collected and placed in Amies transport medium (MAST GROUP Ltd., Bootle, UK). All samples were taken to the Catholic University of Health and Allied Sciences (CUHAS) laboratory for microbiological analysis.
Detection of ESBLs
Each fecal sample was plated on MacConkey agar (HI Media, Mumbai, India) supplemented with cefotaxime (2 μg/mL) and incubated at 35–37°C for 24–48 h. Predominant colonies were further identified using in-house biochemical tests as previously described (Koneman et al., 1997). Pure colonies were grown on ESBL ChromAgar (MAST GROUP Ltd.) for ESBL confirmation and further characterization based on different color of the colonies (Escherichia coli: dark pinkish to reddish colonies; Klebsiella spp./Enterobacter spp: metallic blue; Proteus spp: brown halo colonies).
Antimicrobial Susceptibility Testing
Drug susceptibility testing was done by Kirby–Bauer disk diffusion method on Mueller–Hinton agar (HI Media) based on recommendations of the Clinical Laboratory Standard Institute (Clinical Laboratory Standards Institute [CLSI], 2011). Resistance to the non-beta-lactam antimicrobials ciprofloxacin (5 μg), gentamicin (10 μg), tetracycline (30 μg) and trimethoprim/sulphamethoxazole (1.25/23.75 μg) was tested. For quality control E. coli ATCC 25922 and E. coli ATCC 35218 were used.
Data Management and Analysis
Demographic data and laboratory results were entered into a log book, sorted, and transferred to Microsoft Excel. Analysis was done using STATA version 11.0 (STATA, College Station, TX, USA). Results were presented into percentages/proportions for categorical variables and median (IQR) for continuous variables. Univariate and multivariate logistic regression analyses were performed to identify independent predictors of ESBL carriage among the investigated animals. A p-value of < 0.05 was considered to be a statistical significant cut-off.
Whole-Genome Sequencing and Phylogenetic Analysis
Twenty-five ESBL-producing E. coli isolates were chosen for whole-genome sequencing (WGS). The isolates were confirmed to be ESBL-producers using the VITEK®2 compact system (bioMérieux, Nürtingen, Germany). DNA was isolated using the Purelink Genome DNA Mini kit (Invitrogen, Darmstadt, Germany). WGS was carried out on an Illumina MiSeq instrument (Illumina, San Diego, CA, USA) using an Illumina Nextera XT library with 2x300-bp paired-end reads. The data was assembled using SPAdes (version 3.0) (Bankevich et al., 2012). Contigs larger than 500-bp and a coverage higher than nine were ordered to E. coli MG1655 (accession number U00096.3) using MAUVE (Darling et al., 2004). The contigs were concatenated to generate pseudogenomes. Whole-genome phylogeny was determined by the software package Harvest Suite using E. coli MG1655 as reference (Treangen et al., 2014). The phylogenetic tree was drawn using MEGA5 (Hall, 2013).
The raw data of the sequenced E. coli are available at the European Nucleotide Archive (ENA), under the project number PRJEB12335.
In Silico Analyses
Sequences were analyzed for MLST (according to Wirth et al., 2006), transferrable resistance genes, plasmid replicon types and pMLST using MLST 1.8, ResFinder, Plasmidfinder and pMLST software of the Center for Genomic epidemiology (Larsen et al., 2012; Zankari et al., 2012; Carattoli et al., 2014). Phylogenetic groups were determined as previously described (Clermont et al., 2000). The presence of chuA, yjaA and TSPE4C.2 sequences was examined using blastN (Altschul et al., 1990). The location of blaCTX-M-15 was determined by analyzing the contigs harboring blaCTX-M-15 using blastN. Plasmid comparisons were carried out using the software BRIG (Alikhan et al., 2011).
Study Approval and Ethical Considerations
Ethical clearance and approval to conduct this study was obtained from CUHAS/Bugando Medial Centre Ethics Review Board (CREC/043/2014). Permission to conduct the study was sought from Mwanza city authority. Voluntary and written informed consent was sought from every owner/keeper of the animals prior to inclusion in the study.
Study Limitation
The findings from this study are based on companion and domestic farm animals in the selected districts in the Mwanza region and may not be generalized to companion and domestic farm animals throughout the country.
Results
Baseline Characteristics
During the study period a total of 600 animals were sampled with each animal type constituting approximately 16.7% (100/600). Of the 600 animals, the majority was of local breed [66.6% (400/600)] (Supplementary Table S1).
ESBL Carriage Rates
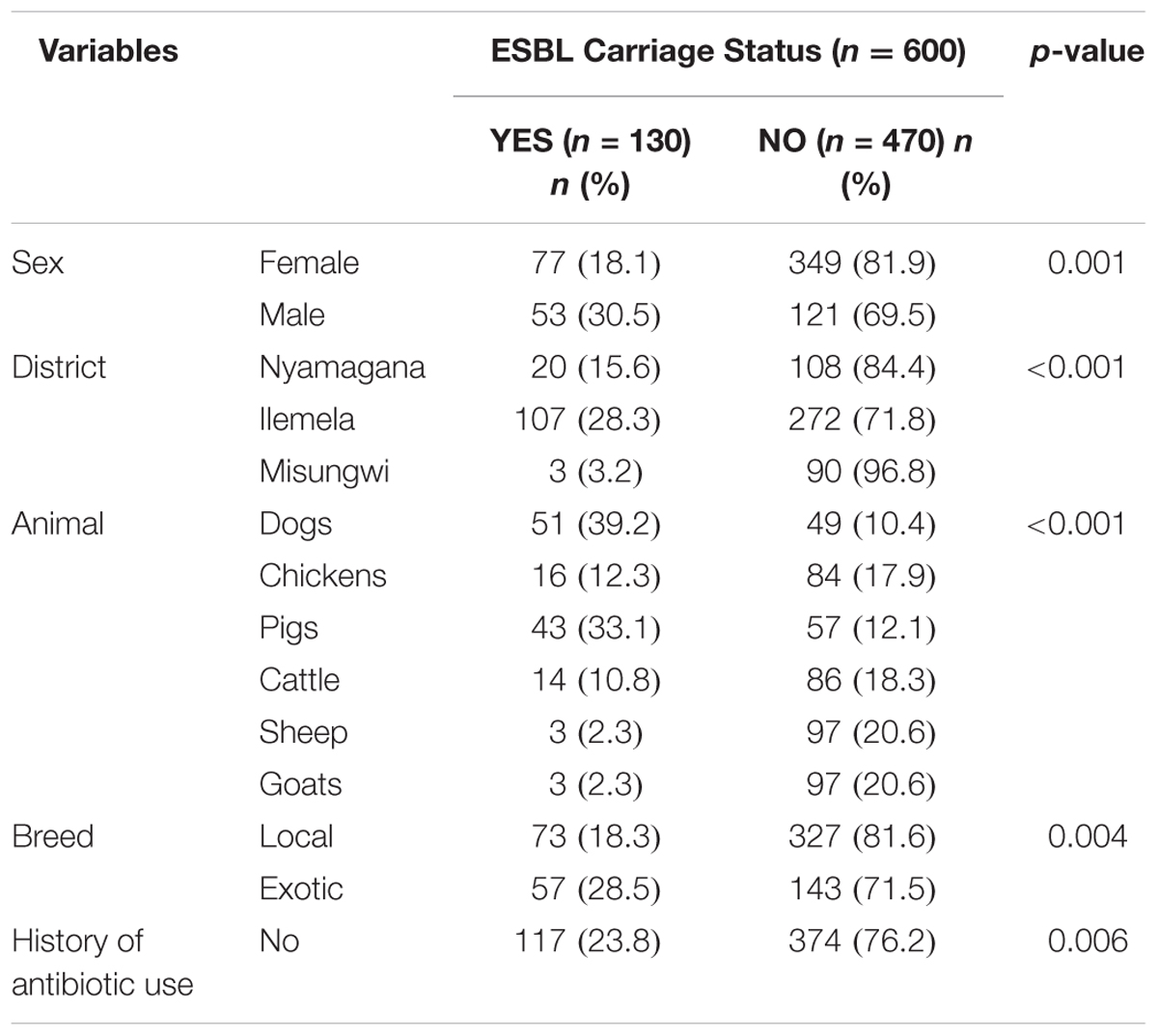
The ESBL carriage rate among companion and domestic farm animals was 21.7% (130/600) [95% CI; 18.4–24.9] with the highest carriage among dogs and pigs [39.2% (51/130) and 33.1% (43/130), respectively] (Supplementary Table S2). Four animals had double carriage. Thereby, the total number of ESBL-producing isolates was 134 (Table 1). The majority of the ESBL isolates were E. coli [93.3% (125/134)], whereas Klebsiella spp. and Proteus spp. accounted for the remaining 6.0% (8/134) and 0.7% (1/134), respectively.
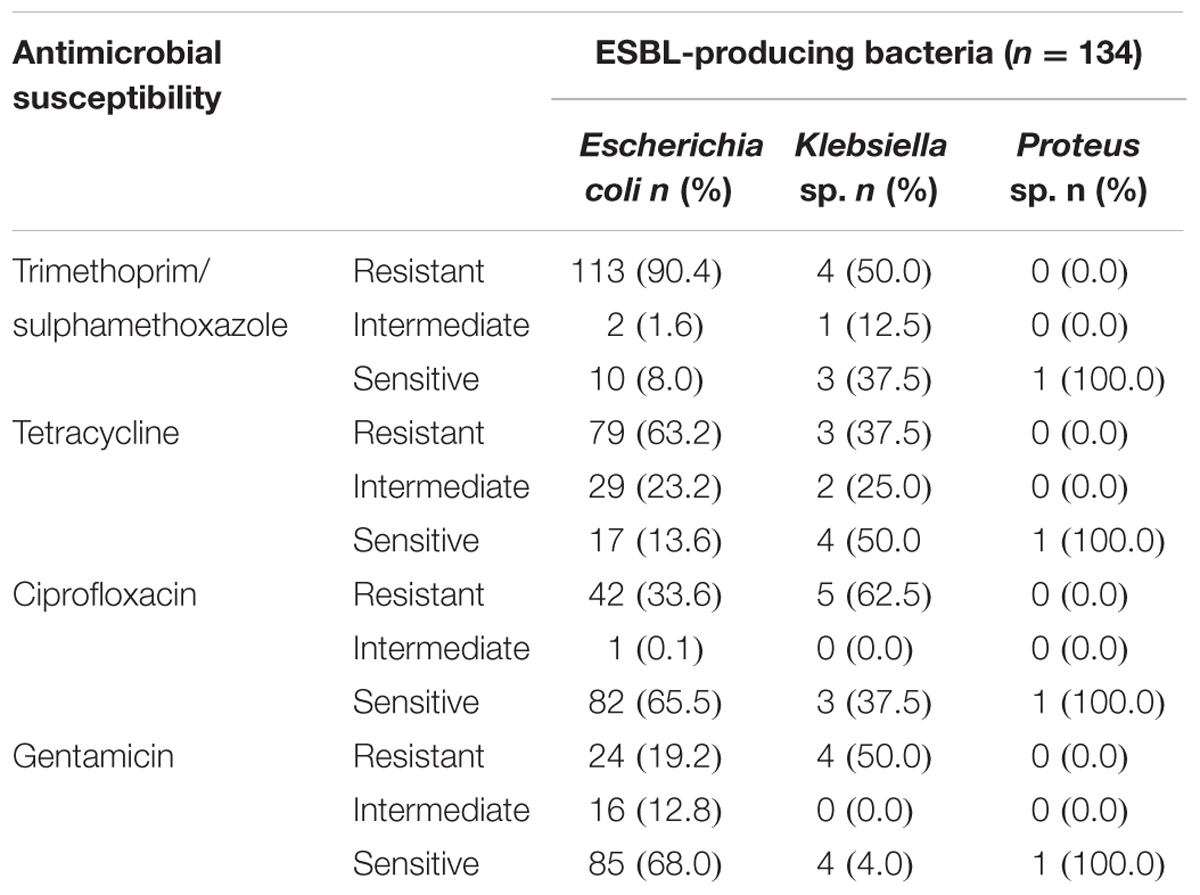
TABLE 1. Antimicrobial susceptibility profiles of ESBL isolates from companion and domestic farm animals.
Antimicrobial Resistance of ESBL Isolates
The majority of E. coli ESBL-producing isolates were resistant to trimethoprim/sulphamethoxazole and tetracycline (90.4 and 63.2%, respectively). Resistance to ciprofloxacin and gentamicin accounted for 33.6 and 19.2%, respectively (Table 1). All E. coli isolates which underwent WGS exhibited a minimum inhibitory concentration for cefotaxime and ceftriaxone greater than 32 and 8 μg/mL, respectively.
Association of ESBL Carriage with Variables
On bivariate analysis, male animals were likely to be more colonized with ESBL isolates as compared to females (30.5% vs. 18.1%, p-value = 0.001). Exotic breeds were more colonized with ESBL isolates as compared to local breeds (28.5% vs. 18.3%, p-value = 0.004). Moreover, animals from Ilemela district (p-value < 0.001) were significantly more colonized than animals from other districts (p-value < 0.001; Table 2). Animals with previous history of non-beta-lactam antibiotics use were less likely to carry ESBL as compared to those with no history of non-beta-lactam antibiotics use [11.9% (13/109) vs. 23.8% (117/491), p-value = 0.006]. Eight of the 109 animals with a history of antibiotic use (7.3%) were treated with sulfadimidine, 23 (21.1%) with penstreptomycine, 63 (58%) with oxytetracycline, and 15 (14%) with thiopurine.
On multivariate logistic regression analysis, exotic breed type [OR (95% CI) = 2.372 (1.460–3.854), p-value < 0.001] was found to be a predictor of ESBL carriage among animals (Table 3).
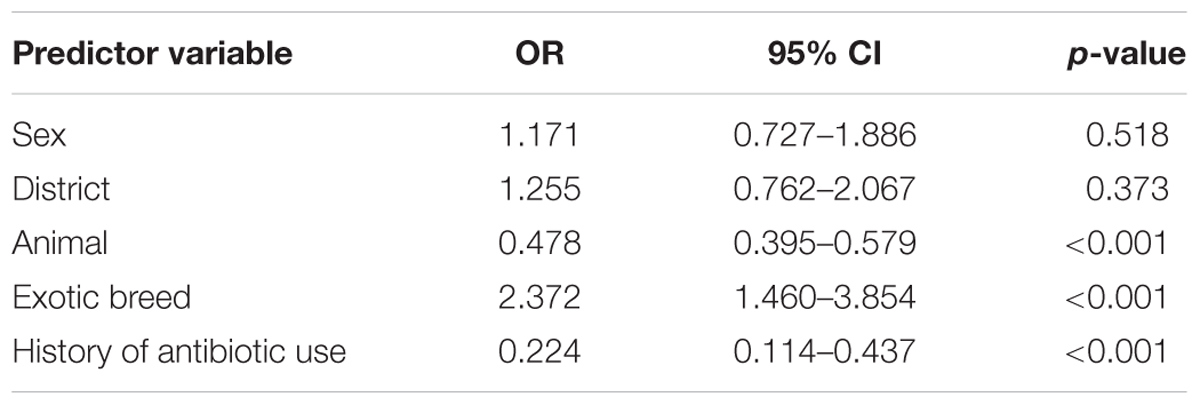
TABLE 3. Multivariate logistic regression analysis for predictor variables associated with ESBL carriage.
Phylogenetic Analysis of the Sequenced Isolates
Within the sequenced E. coli isolates four phylogenetic groups were detected. Phylogenetic group A was present in 17, B1 in five, B2 in two, and D in a single isolate. Fourteen different sequence types (ST) were detected (Supplementary Table S3). The most prevalent ST was ST617 (7/25), followed by ST1303 (3/25), ST2852 (3/25), and ST131 (2/25). E. coli ST617 was present in four different species (cattle, chicken, dog, pig). E. coli ST2852 and ST131 were present both in pigs and dogs, whereas E. coli ST1303 was present in cattle and a pig. E. coli ST617 and ST44 are members of the same clonal complex (CC) ST10, therefore this CC is predominant in this study.
Whole-genome phylogenetic analysis grouped the E. coli into two major clusters, one comprising only CC ST10 isolates and a second more diverse cluster with ST1303, ST2852, ST131, and other STs (Figure 1). STs that were present in more than one animal species were very closely related suggesting interspecies transfer of ESBL isolates.
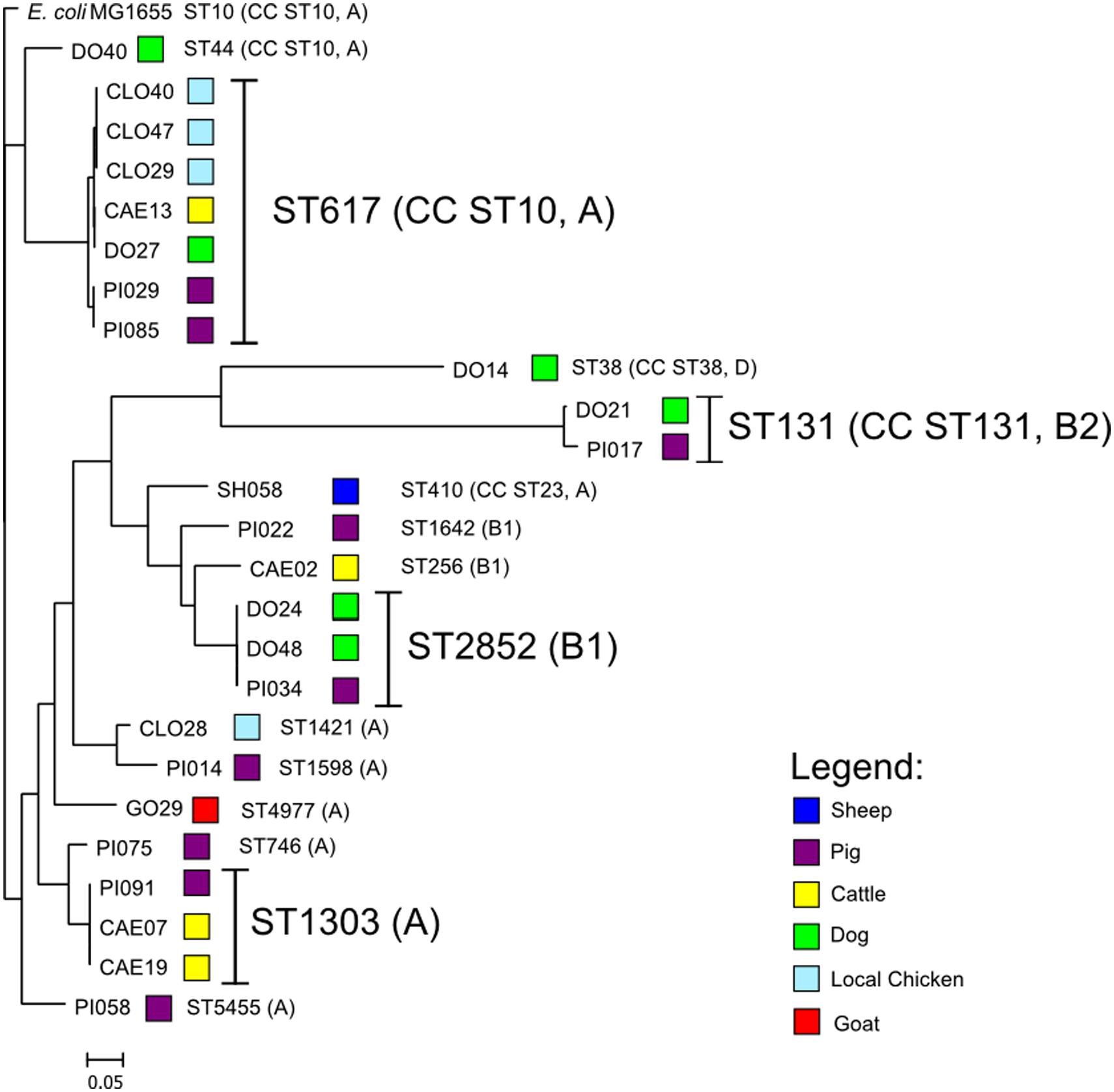
FIGURE 1. Depiction of the core-genome based phylogenetic tree of the ESBL-producing Escherichia coli rooted with E. coli MG1655 genome using the Harvest Suite and MEGA5 software.
ESBL and Antibiotic Resistance Gene Alleles
All sequenced E. coli isolates harbored the blaCTX-M-15 allele and carried up to two additional non-ESBL beta-lactamases. A total of 60% (15/25) isolates harbored only blaTEM-1B and two isolates (8%) carried only blaOXA-1. All other isolates displayed both blaOXA-1 and blaTEM-1B (32%, 8/25). A total of eight different aminoglycoside resistance genes were detected. All isolates harbored both strA and strB excepting CAE02, DO14 (no strA/strB), and PI017 (only strA). Other aminoglycoside resistance genes included aac(3)-IId (6/25), aac(3)-IIa (5/25), aadA5 (9/25), aadA1 (1/25), aadA2 (1/25), and aacA4 (1/25). Two different quinolone resistance genes were present: aac(6′)-Ib-cr (12/25 isolates), and qnrS1 (11/25). Most of the isolates had either sul1 or sul2, dfrA14 or dfrA17 and tet(A) or tet(B) genes conferring resistance to sulphonamides, trimethoprim or tetracycline (Supplementary Table S3).
Location of blaCTX-M-15, Plasmid Replicons, and Plasmid Similarity
The blaCTX-M-15 allele was present in a plasmid-like environment in 21/25 (84%) isolates (Supplementary Table S4). Four isolates harbored a chromosomally inserted blaCTX-M-15 into three different hypothetical proteins (DO21, PI017, PI075) or into a type III secretion system (CAE02). Plasmid replicon types detected were IncFIA, IncFIB, IncFII, and IncY (Supplementary Table S3). pMLST typing of the IncF plasmids revealed the presence of the common plasmid type F31:A4:B1 (n = 4) and F31:A6:B1 (n = 4).
Further analysis indicated that 11/25 (44%) isolates carry a multi-resistance region similar to that present in the plasmid pPGRT46 (accession no. KM023153), isolated from healthy pregnant women in Nigeria, harboring blaCTX-M-15 and the quinolone resistance protein QnrS1 (Fortini et al., 2015). Bioinformatic analysis revealed that these plasmids were highly related to pPGRT46 (accession no. CP009232) with an overall homology of 64 to 95% (Figure 2A). Eight plasmids where highly homologous to the plasmid pCA28, isolated in the USA (Li et al., 2015) and displayed an overlap of 86 to 99% (Figure 2B). Four isolates harbored a resistance cassette that was identical to the one previously described in pSTm-A54650 (accession no. LK056646).
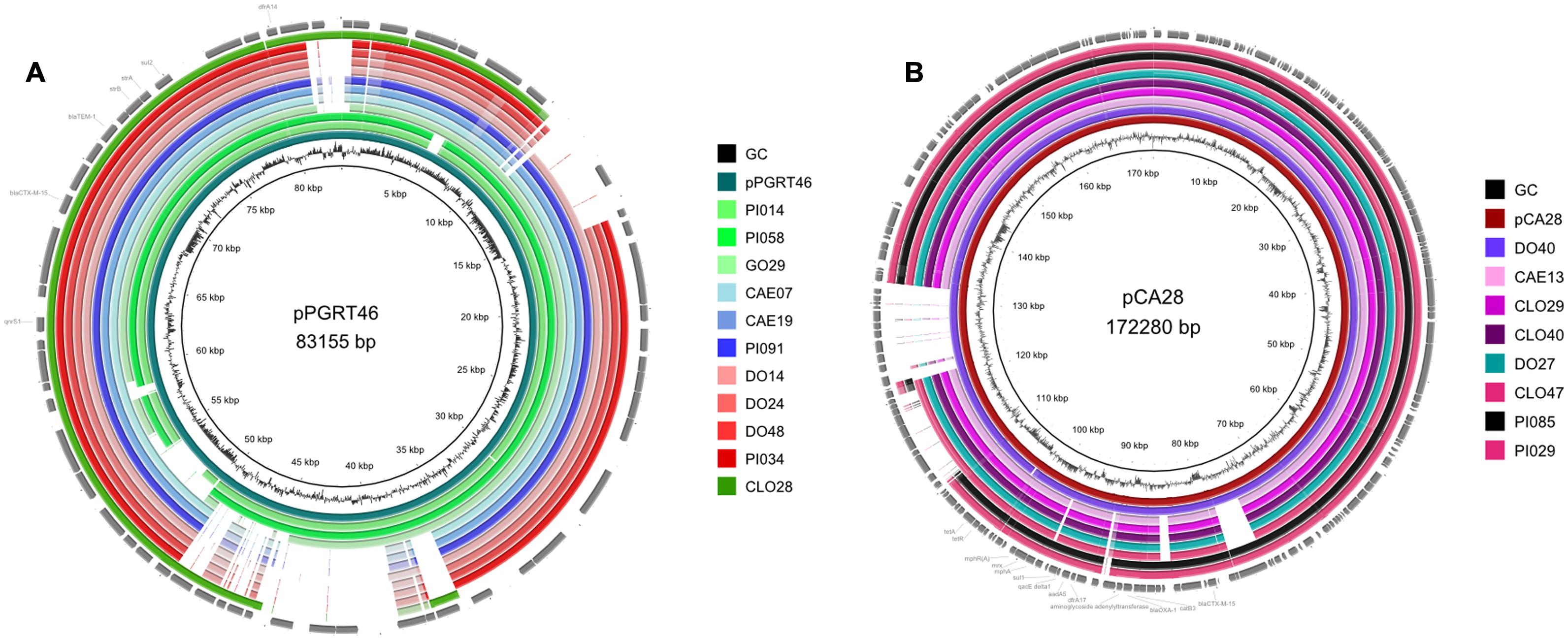
FIGURE 2. Comparative analysis of the whole genome sequences with plasmid pPGRT46 (A) and plasmid pCA28 (B) using BRIG software.
Discussion
The present study indicates high fecal colonization of companion and domestic farm animals with ESBL-producing Enterobacteriaceae, with dogs and pigs displaying the highest colonization rates. This study documents the situation present in rural farming communities in the Mwanza region and indicates circulation of related E. coli clones among different animal species.
In our study, similar to other studies, E. coli comprised the majority of ESBL-producing bacteria in companion and domestic farm animals (Madec et al., 2008; Ewers et al., 2012; Geser et al., 2012; Munang’andu et al., 2012). Over two thirds of E. coli ESBL-producing isolates in the present study were resistant to trimethoprim/sulphamethoxazole and tetracycline as compared to less than one third exhibiting resistance to ciprofloxacin and gentamicin. The wide-spread use of oxytetracycline (58%) among companion and domestic farm animals in this study and another similar study in Morogoro may account for this high resistance trend (Katakweba, 2014).
Despite the fact that animal type and history of non-beta-lactam antibiotics use were protective following multivariate analysis, exotic breed animals were 2.4 times more likely to carry ESBL-producing isolates as compared to local breed animals. This may be related to excessive use of antimicrobial agents in exotic breeds as compared to local breeds, thereby increasing the selective pressure, mutation rates and promoting evolution of resistance (Bester and Essack, 2008).
To identify, whether there are common clones or plasmids in humans and animals in Tanzania, we compared the results of our study with earlier studies on human’s isolates. Firstly, isolates from humans and animals share antibiotic resistance patterns, i.e., to gentamycin, tetracycline, and trimethoprim/sulfamethoxazole (Kayange et al., 2010). Secondly, all animal isolates examined in this study harbored the blaCTX-M-15 allele, which is also present in the majority of E. coli isolates circulating in the hospital in the same region (Mshana et al., 2011b; Katakweba, 2014). There is only a small overlap between clonal populations of E. coli present in either population. Thus it is likely that this distribution may be related to the presence of commonly occurring plasmids, rather than clonal isolates. This is in keeping with results obtained in a previous study (Mshana et al., 2011b), conducted in the same region, which observed that ST131 (CC ST131) of phylogroup B2 was the most common ST obtained from human clinical isolates. The majority of clones observed here, i.e., CC ST10 (ST617, ST44) of phylogroup A, is distinct from that seen in the human isolates. The presence of ST131 isolates in this study suggests close contact between humans and animals.
The predominance of E. coli ST617 suggests clonal distribution between different animal species. Eight (32%) of the sequenced isolates were members of the clonal complex CC ST10. The presence of CC ST10 ESBL-producing E. coli in humans has been commonly reported in Africa (Fam et al., 2011; Aibinu et al., 2012; Fortini et al., 2015; Rafaï et al., 2015; Sallem et al., 2015). In all countries these strains harbor blaCTX-M-15 genes in multiple IncF plasmids, but are also associated with a variety of other plasmid replicons, including rare plasmid types such as IncY and IncQ. In this study, eleven isolates were found to carry a plasmid with homology to the Inc-untypable plasmid pPGRT46 recently detected in Nigeria in E. coli isolates from humans. This indicates that homologous plasmids might have spread among different E. coli strains in the continent not only between humans, but also in animals. Moreover, these plasmids harbor adjacent blaCTX-M-15 and qnrS1 resistance genes and thereby confer both ESBL- and quinolone resistance resulting in a co-transfer of these resistances in the worst case, even if only one of these two antibiotic classes is used. Eight isolates carried a plasmid with homology to the pCA28 (F31:A4:B1) plasmid which has been recently found in the USA (Li et al., 2015). Interestingly, one canine isolate (DO40) harbored a plasmid that was not only 99% similar to the plasmid pCA28, but also had the same ST as the E. coli isolate, from which pCA28 was originally isolated (ST44). We note that CTX-M-15 was also present as chromosomal insertions of the blaCTX-M-15 allele in diverse strains of E. coli.
As in a previous study (Mshana et al., 2011b) conducted in the same region the majority of E. coli isolates had IncF plasmids with additional transferable resistances being gentamycin, tetracycline and trimethoprim/sulfamethoxazole. Unlike the previous study where no IncY plasmids were detected, isolates from animals 8/25 (32%) harbor IncY plasmids carrying blaCTX-M-15, blaTEM-1B, strA, strB, and qnrS1.
Conclusion
This study for the first time conducted in Tanzania examines the presence of CTX-M-15-producing E. coli in rural farming communities in Mwanza, Tanzania. Among these isolates, the CC ST10 (ST617, ST44) was predominant. The transmission of blaCTX-M-15 was probably plasmid driven and involved plasmid populations that are regionally present in East and West Africa. The high fecal carriage of ESBL-producing Enterobacteriaceae among companion and domestic farm animals harboring predominantly blaCTX-M-15 in combination with quinolone and aminoglycoside resistance genes underscores the need of an integrated antimicrobial surveillance system in veterinary and human medicine to investigate the potential sources and the dynamics of transmission.
Author Contributions
JS, MMM, MR, CI, TC, and SM conceived, designed and executed the study; JS, NS, and MMM collected the data and samples; JS, NS, and SM performed laboratory analysis; LF and TC performed WGS; JS, LF, NS, MMM, TC, and SM analyzed the data; JS, LF, TC, and SM wrote the manuscript which was critically reviewed by all authors. All authors have read and approved the final draft of the manuscript.
Funding
This study was supported by a grant from the Wellcome Trust (WT087546MA) to SACIDS and also by grants from the Bundesministerium fuer Bildung und Forschung (BMBF, Germany) within the framework of the RESET research network (contract no. 01KI1313G) and the German Center for Infection research (DZIF/grant number 8000 701–3 [HZI] to TC and CI and TI06.001 to TC).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to the technical support provided by Dr. Ganchwere and his colleague at the Tanzania Veterinary Laboratory Agency in Mwanza. We also appreciate the technical assistance by Mr. Vitus Silago, Hezron Bassu and other staff in the Department of Microbiology and Immunology, Dar es Salaam, and Christina Gerstmann at the Institute of Medical Microbiology in Giessen.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00142
References
Aibinu, I., Odugbemi, T., Koenig, W., and Ghebremedhin, B. (2012). Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin. Microbiol. Infect. 18, E49–E51. doi: 10.1111/j.1469-0691.2011.03730.x
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bester, L. A., and Essack, S. Y. (2008). Prevalence of antibiotic resistance in Campylobacter isolates from commercial poultry suppliers in KwaZulu-Natal, South Africa. J. Antimicrob. Chemother. 62, 1298–1300. doi: 10.1093/jac/dkn408
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/CMR.14.4.933-951.2001
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Clermont, O., Bonacorsi, S., Bingen, E., and Bonacorsi, P. (2000). Rapid and Simple Determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000.Updated
Clinical Laboratory Standards Institute [CLSI] (2011). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-first Informational Supplement. Document M100-S21s. Wayne, PA: Clinical and Laboratory Standards Institute.
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Dierikx, C. M., van Duijkeren, E., Schoormans, A. H. W., van Essen-Zandbergen, A., Veldman, K., Kant, A., et al. (2012). Occurrence and characteristics of extended-spectrum-β-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 67, 1368–1374. doi: 10.1093/jac/dks049
Ewers, C., Bethe, A., Semmler, T., Guenther, S., and Wieler, L. H. (2012). Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18, 646–655. doi: 10.1111/j.1469-0691.2012.03850.x
Fam, N., Leflon-Guibout, V., Fouad, S., Aboul-Fadl, L., Marcon, E., Desouky, D., et al. (2011). CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microbiol. Drug Resist. 17, 67–73. doi: 10.1089/mdr.2010.0063
Fortini, D., Fashae, K., Villa, L., Feudi, C., García-Fernández, A., and Carattoli, A. (2015). A novel plasmid carrying blaCTX-M-15 identified in commensal Escherichia coli from healthy pregnant women in Ibadan. Nigeria. J. Glob. Antimicrob. Resist. 3, 9–12. doi: 10.1016/j.jgar.2014.12.002
Geser, N., Stephan, R., and Hächler, H. (2012). Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 8:21. doi: 10.1186/1746-6148-8-21
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30, 1229–1235. doi: 10.1093/molbev/mst012
Hawkey, P. M., and Jones, A. M. (2009). The changing epidemiology of resistance. J. Antimicrob. Chemother. 64(Suppl. 1), i3–i10. doi: 10.1093/jac/dkp256
Katakweba, A. A. S. (2014). Prevalence of Antimicrobial Resistance and Characterization of Fecal Indicator Bacteria and Staphylococcus Aureus from Farm Animals, Wildlife, Pets and Humans in Tanzania, thesis Collection, Sokoine University of Agriculture, Morogoro.
Kayange, N., Kamugisha, E., Mwizamholya, D. L., Jeremiah, S., and Mshana, S. E. (2010). Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatr. 10:39. doi: 10.1186/1471-2431-10-39
Koneman, E. W., Allen, S. D., Janda, W. M., and Schreckenberger, P. C. (1997). Color Atlas and Textbook of Diagnostic Microbiology. Washington, DC: Lippincott Williams & Wilkins.
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Li, J.-J., Spychala, C. N., Hu, F., Sheng, J.-F., and Doi, Y. (2015). Complete nucleotide sequences of bla(CTX-M)-harboring IncF plasmids from community-associated Escherichia coli strains in the United States. Antimicrob. Agents Chemother. 59, 3002–3007. doi: 10.1128/AAC.04772-14
Lupindu, A. M., Olsen, J. E., Ngowi, H. A., Msoffe, P. L. M., Mtambo, M. M., Scheutz, F., et al. (2014). Occurrence and characterization of Shiga toxin-producing Escherichia coli O157:H7 and other non-sorbitol-fermenting E. coli in cattle and humans in urban areas of Morogoro, Tanzania. Vector Borne Zoonotic Dis. 14, 503–510. doi: 10.1089/vbz.2013.1502
Madec, J.-Y., Lazizzera, C., Châtre, P., Meunier, D., Martin, S., Lepage, G., et al. (2008). Prevalence of fecal carriage of acquired expanded-spectrum cephalosporin resistance in Enterobacteriaceae strains from cattle in France. J. Clin. Microbiol. 46, 1566–1567. doi: 10.1128/JCM.02299-07
Mshana, S. E., Gerwing, L., Minde, M., Hain, T., Domann, E., Lyamuya, E., et al. (2011a). Outbreak of a novel Enterobacter sp. carrying blaCTX-M-15 in a neonatal unit of a tertiary care hospital in Tanzania. Int. J. Antimicrob. Agents 38, 265–269. doi: 10.1016/j.ijantimicag.2011.05.009
Mshana, S. E., Imirzalioglu, C., Hain, T., Domann, E., Lyamuya, E. F., and Chakraborty, T. (2011b). Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin. Microbiol. Infect. 17, 1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x
Munang’andu, H. M., Kabilika, S. H., Chibomba, O., Munyeme, M., and Muuka, G. M. (2012). Bacteria isolations from broiler and layer chicks in zambia. J. Pathog. 2012:520564. doi: 10.1155/2012/520564
Nelson, E., Kayega, J., Seni, J., Mushi, M. F., Kidenya, B. R., Hokororo, A., et al. (2014). Evaluation of existence and transmission of extended spectrum beta lactamase producing bacteria from post-delivery women to neonates at Bugando Medical Center, Mwanza-Tanzania. BMC Res. 7:279. doi: 10.1186/1756-0500-7-279
Rafaï, C., Frank, T., Manirakiza, A., Gaudeuille, A., Mbecko, J.-R., Nghario, L., et al. (2015). Dissemination of IncF-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from surgical-site infections in Bangui, Central African Republic. BMC Microbiol. 15:341.
Rubin, J. E., and Pitout, J. D. D. (2014). Extended-spectrum β-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet. Microbiol. 170, 10–18. doi: 10.1016/j.vetmic.2014.01.017
Sallem, R., Ben Slama, K., Ben Estepa, V., Cheikhna, E. O., Mohamed, A. M., Chairat, S., et al. (2015). Detection of CTX-M-15-producing Escherichia coli isolates of lineages ST410-A, ST617-A and ST354-D in faecal samples of hospitalized patients in a Mauritanian hospital. J. Chemother. 27, 114–116. doi: 10.1179/1973947814Y.0000000172
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Keywords: farming communities, companion animals, domestic farm animals, ESBL, Mwanza, Tanzania
Citation: Seni J, Falgenhauer L, Simeo N, Mirambo MM, Imirzalioglu C, Matee M, Rweyemamu M, Chakraborty T and Mshana SE (2016) Multiple ESBL-Producing Escherichia coli Sequence Types Carrying Quinolone and Aminoglycoside Resistance Genes Circulating in Companion and Domestic Farm Animals in Mwanza, Tanzania, Harbor Commonly Occurring Plasmids. Front. Microbiol. 7:142. doi: 10.3389/fmicb.2016.00142
Received: 02 December 2015; Accepted: 25 January 2016;
Published: 11 February 2016.
Edited by:
David W. Graham, Newcastle University, UKReviewed by:
Juan Wang, University College Dublin, IrelandSeamus Fanning, University College Dublin, Ireland
Copyright © 2016 Seni, Falgenhauer, Simeo, Mirambo, Imirzalioglu, Matee, Rweyemamu, Chakraborty and Mshana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen E. Mshana, mshana72@yahoo.com; Trinad Chakraborty, trinad.chakraborty@mikrobio.med.uni-giessen.de
†These authors have contributed equally to this work.
 Jeremiah Seni
Jeremiah Seni Linda Falgenhauer
Linda Falgenhauer Nabina Simeo1
Nabina Simeo1 Mariam M. Mirambo
Mariam M. Mirambo Trinad Chakraborty
Trinad Chakraborty Stephen E. Mshana
Stephen E. Mshana