- 1State Key Laboratory of Microbial Technology, Shandong University, Jinan, China
- 2Marine Biotechnology Research Center, Shandong University, Jinan, China
- 3Institute of Marine Science and Technology, Shandong University, Jinan, China
- 4Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Marine bacterial alginate lyases play a role in marine alginate degradation and carbon cycling. Although a large number of alginate lyases have been characterized, reports on alginate lyases with special characteristics are still rather less. Here, a gene alyPM encoding an alginate lyase of polysaccharide lyase family 7 (PL7) was cloned from marine Pseudoalteromonas sp. SM0524 and expressed in Escherichia coli. AlyPM shows 41% sequence identity to characterized alginate lyases, indicating that AlyPM is a new PL7 enzyme. The optimal pH for AlyPM activity was 8.5. AlyPM showed the highest activity at 30°C and remained 19% of the highest activity at 5°C. AlyPM was unstable at temperatures above 30°C and had a low Tm of 37°C. These data indicate that AlyPM is a cold-adapted enzyme. Moreover, AlyPM is a salt-activated enzyme. AlyPM activity in 0.5–1.2 M NaCl was sixfolds higher than that in 0 M NaCl, probably caused by a significant increase in substrate affinity, because the Km of AlyPM in 0.5 M NaCl decreased more than 20-folds than that in 0 M NaCl. AlyPM preferably degraded polymannuronate and mainly released dimers and trimers. These data indicate that AlyPM is a new PL7 endo-alginate lyase with special characteristics.
Introduction
Alginates are mainly produced by brown algae in the ocean, which are an important component of marine organic carbon. Alginates are liner unbranched copolymer consisting of β-D-mannuronate (M) and its C5 epimer, α-L-guluronate (G), arranged in block structures such as homopolymeric G block, M block, and heteropolymeric MG (GM) blocks (Gacesa, 1988). The specific conformation of the guluronic acid region allows preference to binding divalent cations and leading to better gelling capability and gel strength (Braccini et al., 1999). Therefore, the M/G ratio, sequence and G-block length are critical factors affecting the physical properties of alginate and its resultant hydrogels (Braccini and Perez, 2001). The alginate composition, sequence and molecular weight vary with the sources that alginates are extracted from Haug et al. (1959) and Tonnesen and Karlsen (2002). Determination of the fine structure of alginate helps us to produce and use alginate molecules with defined properties optimally suited for a given application (Aarstad et al., 2012; Lee and Mooney, 2012).
Alginate degradation, which is mainly performed by alginate lyases, is a part of marine carbon cycling. Alginate lyases have been reported from marine algae, mollusks, fungi, bacteria, bacteriophages, and viruses. Alginate lyases catalyze the degradation of alginate by a β-elimination of the 4-O-glycosyl bond to form a double bond between C-4 and C-5, producing 4-deoxy-L-erythro-hex-4-ene pyranosyluronate at the non-reducing end of the resulting oligosaccharides (Preiss and Ashwell, 1962). Alginate lyases are classified into three types based on their substrate specificities, the first type is specific toward G blocks (EC 4.2.2.11), the second type specific toward M blocks (EC 4.2.2.3), and the third type is bifunctional for G and M blocks (Wong et al., 2000). While a large number of alginate lyases have been characterized, reports on alginate lyases with special characteristics are still rather less. Some cold-adapted alginate lyases have been found to be secreted by bacteria from the Arctic (Dong et al., 2012) and other sea (Xiao et al., 2006; Li et al., 2015). A thermophilic alginate lyase (Inoue et al., 2016), a high-alkaline alginate lyase (Kobayashi et al., 2009) and several salt-activiated alginate lyases (Horikoshi, 1982; Lange et al., 1989; Brown and Preston, 1991; Kitamikado et al., 1992; Xiao et al., 2006; Uchimura et al., 2010) have also been reported.
Based on their amino acid sequences, alginate lyases are classified into seven polysaccharide lyase (PL) families (PL5, 6, 7, 14, 15, 17, 18) in the Carbohydrate-Active enZYmes (CAZy) database1 (Cantarel et al., 2009). Alginate lyases from family PL7 have been widely studied, and the crystal structures of six PL7 alginate lyases have been solved. Structural analysis shows that these lyases share a common β-sandwich fold consisting of two β-sheets, in which, conserved amino acid residues compose a deep active cleft that is covered by two flexible lid loops (Yamasaki et al., 2004, 2005; Osawa et al., 2005; Thomas et al., 2013). There are both endo-lytic and exo-lytic alginate lyases in this family. The substrate specificities of PL7 alginate lyases are also diverse, including G-specific, M-specific, and bifunctional enzymes.
Pseudoalteromonas sp. SM0524, isolated from rotten kelp collected from the seashore of Yantai, China, is a good producer of alginate lyases (Li et al., 2011). Aly-SJ02 produced by Pseudoalteromonas sp. SM0524 is a PL18 bifunctional alginate lyase that mainly releases dimers and trimers from alginate (Li et al., 2011). The N-terminal extension of aly-SJ02 plays a role in the correct folding of the catalytic domain (Dong et al., 2014). In this article, we characterized another alginate lyase, AlyPM, from Pseudoalteromonas sp. SM0524. The result showed that AlyPM was a new cold-adapted PL7 alginate lyase with a significant preference toward polyM. Moreover, AlyPM was shown to be a salt-activated endo-enzyme, which mainly released dimers and trimers from sodium alginate and polyM. These special characteristics of AlyPM indicate that AlyPM is well adapted to marine environment and may play a role in marine alginate degradation and carbon cycling.
Materials and Methods
Materials and Strains
Sodium alginate from brown algae (viscosity: 2% solution at 25°C, ~250) was purchased from Sigma. PolyM and PolyG (purity: about 95%) were kindly provided by Professor Wengong Yu in Ocean University of China. Glutathione was obtained from Sigma. Glutathione Sepharose 4B was purchased from GE Healthcare. Restriction enzymes were purchased from Thermo. Human Thrombin was purchased from Novagen.
Marine bacterium Pseudoalteromonas sp. SM0524 was previously isolated from rotten kelp which was collected from a kelp culture field at the seashore of Yantai, China (Li et al., 2011). This strain was grown on marine broth 2216 (Difco) at 15°C. Escherichia coli DH5α was used for plasmid construction, and E. coli BL21 (DE3) was used as the host for gene expression. These strains were grown in Luria-Bertani (LB) broth or on LB broth agar (LB broth supplemented with 1.5% agar) containing 100 μg/mL ampicillin.
Gene Cloning, Expression, and Purification of AlyPM
Two degenerate primers were designed according to two conserved sequences of 30 kDa alginate lyases, SN1 (TGGCGNCAYGARTAYAARGT) according to the sequence WRHEYKV and SC1 (TANGCNCCNGCYTTRAARTA) according to the sequence YFKAGAY. A DNA fragment was amplified from the genomic DNA of Pseudoalteromonas sp. SM0524 by gradient PCR (annealing temperature from 44 to 56°C) with primers SN1 and SC1. Then, three specific primers were designed based on the 5′ terminal sequence, and three based on the 3′ terminal sequence of the amplified DNA fragment. Two general primers containing the protein initiation codon and the stop codon, respectively, were also designed. Thermal asymmetric interlaced (TAIL) PCRs (Liu and Whittier, 1995) were then carried out to amplify the neighboring sequences of the DNA fragment. As a result, a upstream sequence and a downstream sequence of the DNA fragment were amplified from the genomic DNA of Pseudoalteromonas sp. SM0524 and sequenced. Through assembly, a DNA sequence containing a 1,089 bp ORF that encodes gene alyPM was obtained. After verification by PCR-amplification and sequencing, the sequence of gene alyPM was submitted to the GenBank database under the accession number EU548076. The signal peptide of AlyPM was predicted by SignalP 3.0 server (Bendtsen et al., 2004) and the domains of AlyPM were predicted by the search in NCBI Conserved Domain Database (CDD2) (Marchler-Bauer et al., 2011). Multiple amino acid sequence alignment of the deduced catalytic domain was done by Clustal X 1.83 program with other characterized alginate lyases in family PL7 (Thompson et al., 1997).
The alyPM gene without the signal peptide sequence was cloned into the pGEX-4T-1 vector, which was then transferred into E. coli BL21 (DE3). The recombinant E. coli BL21 (DE3) cells harboring the constructed plasmid were cultured on a LB medium containing 100 μg/mL ampicilin to OD600 of 0.8–1.0 at 37°C, and then cultivated in a shaking incubator at 100 rpm for 24 h at 15°C under the induction of 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG). The cells were harvested and ultrasonically disrupted in 1 × GST binding buffer (5 mM phosphate buffer containing 137 mM NaCl, pH 7.3). After centrifugation at 12,000 g, 4°C for 15 min, the clear cell extract solution containing the GST-fused AlyPM was loaded on a Glutathione Sepharose 4B column equilibrated with 1 × GST binding buffer. The GST-tag of the fused AlyPM was cut off by Human thrombin at 15°C for 8 h. The released AlyPM was washed with 1 × GST binding buffer and then was stored at -20°C for further use. The purity of AlyPM was detected by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with the method of Laemmli (Laemmli, 1970). Protein concentration was determined by the bicinchoninic acid (BCA) method using a BCA protein assay kit (Thermo, USA) with bovine serum albumin as the standard.
Enzyme Assay
Alginate lyase activity was determined by monitoring the increase of absorbance at 235 nm (A235) caused by production of unsaturated uronic acids as the lyase cleaves glycosidic bonds in the polymer chain (Gacesa, 1992). Unless otherwise noted, the activity was measured at 30°C for 30 min in a mixture of 80 μl buffer (50 mM Tris-HCl, pH 8.0), 100 μl alginate substrate (5 mg/ml), and 20 μl enzyme extract. After incubation, the mixture was boiled for 5 min to terminate the reaction. One unit of enzyme activity was defined as the amount of enzyme that increased A235 by 0.1 per min.
Enzyme Characterization
Unless otherwise noted, sodium alginate was used as the substrate in enzyme assays for AlyPM characterization. To determine the optimum temperature for AlyPM catalysis, the alginate lyase activity of AlyPM was measured at 5–45°C in 5°C intervals. The optimum pH for AlyPM catalysis was measured in different buffers ranging from pH 5.0 to 11.0. The buffers used were 50 mM phosphate buffer (pH 5.0–7.0), 50 mM Tris-HCl (pH 7.0–9.5), and 50 mM NaHCO3-NaOH buffer (pH 9.5–11.0). The thermostability of AlyPM was determined by measuring the residual activity after incubating the enzyme at 5–50°C for 15 min. To analyze the effects of metal ions on the activity of AlyPM, different chemicals were added to the reaction mixture at a final concentration of 2 or 10 mM, respectively. NaCl of different concentrations in the range of 0–2 M was added to the reaction mixture to investigate the effect of NaCl on the activity of AlyPM. Kinetic parameters of AlyPM in 0 and 0.5 M NaCl were determined at 30°C by non-linear analysis based on the initial rates determined from 0.3 to 18 mg/ml of substrates and analyzed by the Michaelis–Menten equation using the Origin8 Pro SR4 software. Substrate specificity of AlyPM was tested by measuring its activities toward polyM, polyG, and sodium alginate. Oligomers released from polyM, polyG, and sodium alginate by AlyPM were analyzed by thin layer chromatography (TLC) and the solvent system was 1-butanol/acetic acid/water (4:6:1, v/v). The products were visualized by heating TLC plates at 90°C for 15 min after spraying with 10% (v/v) sulfuric acid in ethanol. Standard alginate di-, tri-, and tetrasaccharides, with molecular mass of 351, 527, and 703, respectively, were prepared as described previously (Li et al., 2011).
Circular Dichroism Spectra
AlyPM of 0.20 mg/ml in 20 mM Tris-HCl buffer (pH 8.0) with or without 0.5 M NaCl were subjected to circular dichroism (CD) spectroscopy assays at 25°C on a J-810 spectropolarimeter (Jasco, Japan). The CD spectra were recorded from 250 to 195 nm at a scan speed of 200 nm/min with a path length of 0.1 cm. Thermal unfolding (Tm) value of AlyPM was determined on the same CD spectrophotometer equipped with a Peltier type temperature controller with a heating rate of 1°C/min. The concentration of AlyPM was 0.20 mg/ml in 20 mM Tris-HCl buffer (pH 8.0). Spectra were recorded from 250 to 184 nm at 0.2 nm resolution. Data were collected from 20 to 70°C in 1°C intervals. CD signals at 222 nm were used for analysis of the unfolding curve.
Results
Sequence Analysis of AlyPM
The open reading frame of gene alyPM amplified from the genome of Pseudoalteromonas sp. SM0524 is 1,089 bp in length and encodes an alginate lyase contaning 362 amino acid residues. According to the results of signal peptide prediction by SignalP 3.0 and searching in NCBI CDD, AlyPM is a single-domain PL7 enzyme containing a 28-residue signal peptide (Met1-Ser28). Among the characterized alginate lyases, AlyPM had the highest identity (41%) to AlyVGI from Vibrio halioticoli IAM 14596T (Sugimura et al., 2000). Alginate lyases in family PL7 contain three highly conserved motifs, SA3 (RXEXR), SA4 (YXKAGXYXQ), and SA5 (QXH), which compose the active center and are crucial for substrate recognition and catalysis (Yamasaki et al., 2004; Kawamoto et al., 2006). Sequence alignment of AlyPM with AlyVGI and other PL7 alginate lyases that have crystal structures indicates that these conserved motifs in AlyPM are RSELR (SA3), YFKAGAYNQ (SA4), and QIH (SA5) (Figure 1).
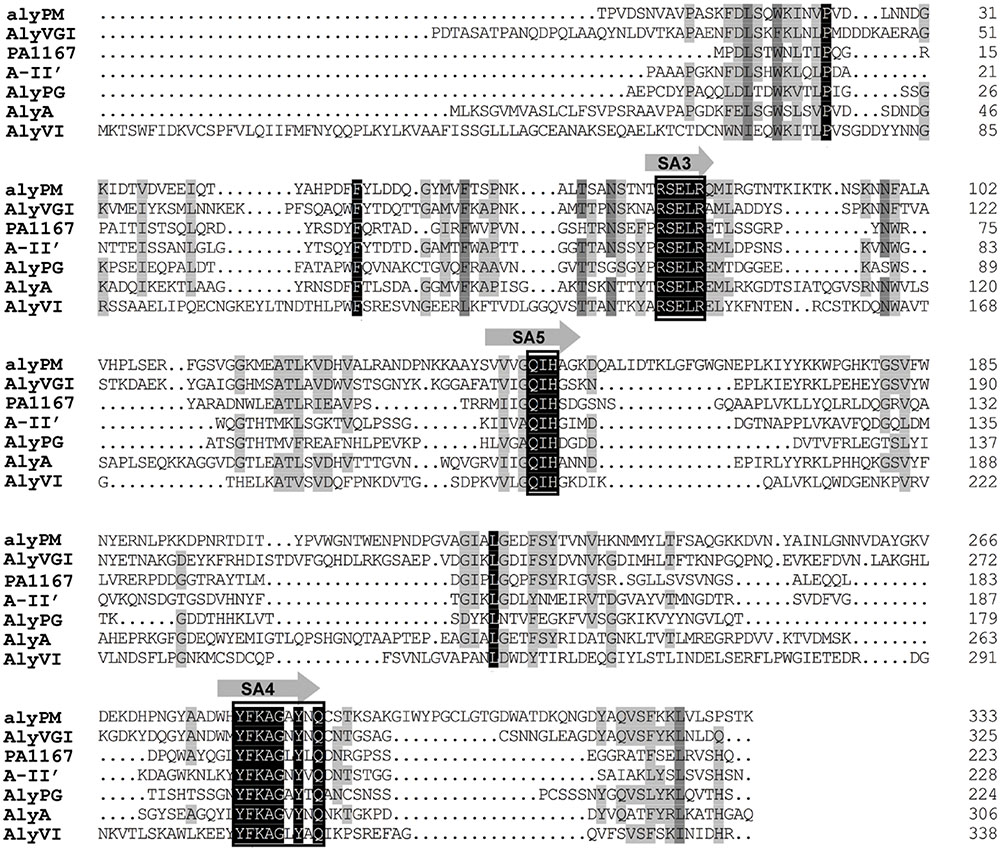
FIGURE 1. Amino acid alignment of AlyPM to other characterized PL7 alginate lyases. AlyPM (EU548076) from Pseudoalteromonas sp. SM0524 in this study; AlyVGI (AAF22512), an alginate lyase from Vibrio halioticoli IAM14596T; PA1167 (AAG04556), an alginate lyase from Pseudomonas aeruginosa PAO1; A1-II’ (BAD16656), an alginate lyase from Sphingomonas sp. A1; AlyPG (BAA83339), an alginate lyase from Corynebacterium sp. ALY-1; AlyA (AAA25049), an alginate lyase from Klebsiella pneumoniae SUBSP. AEROGENES; AlyVI (AAP45155), an alginate lyase from Vibrio sp. QY101. Identical and similar amino acid residues among the alginate lyases are shaded in black. Conserved regions are boxed.
Expression and Characterization of AlyPM
AlyPM without the signal peptide was expressed in E. coli BL21 (DE3) as a GST-fused protein, which was then bound to a Glutathione Sepharose 4B column. The recombinant AlyPM was cut off from the GST-fused protein bound to the column. SDS-PAGE analysis showed that the purified AlyPM displayed an apparent molecular weight of approximately 40 kDa, slight larger than the theoretical value (37.1 kDa) deduced from the predicted amino acid sequence (Figure 2A).
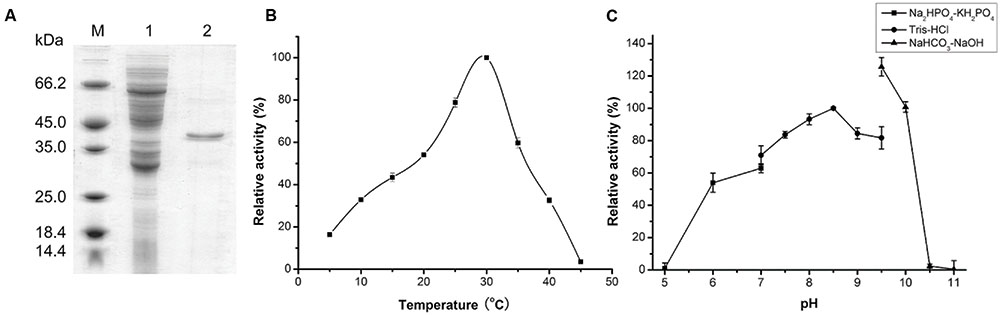
FIGURE 2. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of purified AlyPM alginate lyase and effect of temperature and pH on AlyPM activity. (A) SDS-PAGE analysis of purified AlyPM. Lane 1, Whole-cell protein extracts of transformed E. coli BL21 (DE3) cells; lane 2, purified AlyPM; lane M, protein mass markers. The proteins were subjected to electrophoresis on 12.5% acrylamide gel and stained with Coomassie Brilliant Blue R-250. (B) Effect of temperature on AlyPM activity. The assay was conducted at pH 8.0 in Tris-HCl buffer. Activity at 30°C was taken as 100%. (C) Effect of pH on AlyPM. Reactions were conducted at 30°C for 30 min in the following 50 mM buffers; PBS (closed squares), Tris-HCl (closed circles), and NaHCO3-NaOH (closed triangles). Activity at pH 8.5 in Tris-HCl was taken as 100%.
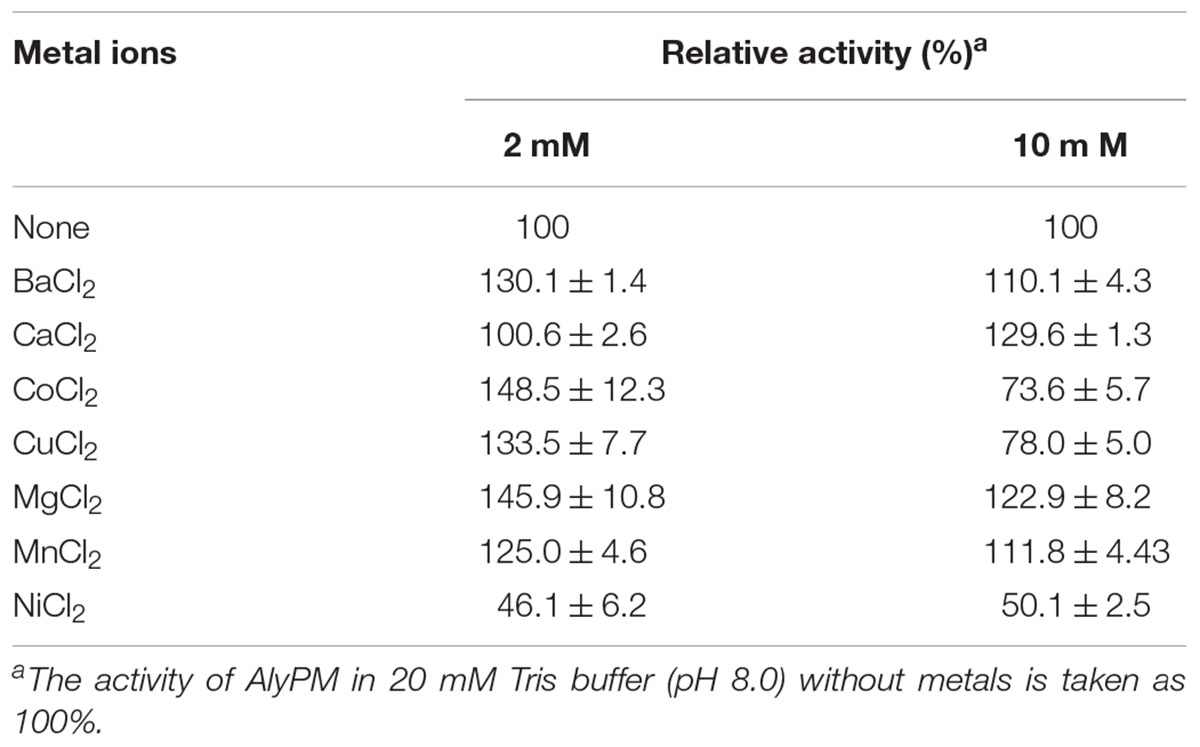
With sodium alginate as substrate, AlyPM showed the highest activity at 30°C (Figure 2B). In 50 mM Tris-HCl buffer, AlyPM had the highest activity at pH 8.5, and retained more than 70% of its highest activity between pH 7.0 and 9.5 (Figure 2C). AlyPM showed different activities in different buffers at the same pH, suggesting that buffer ions affect its activity. Thus, the effect of metal ions on the enzyme activity of AlyPM was measured (Table 1). Ni2+ at 2 and 10 mM both inhibited the enzyme activity by about 50%. Cu2+ and Co2+ increased the enzyme activity at 2 mM but inhibited it at 10 mM. The other metal ions, such as Ba2+, Ca2+, Mg2+, and Mn2+, had no or a little enhancing effect on the enzyme activity (Table 1).
Cold-Adapted Characters of AlyPM
Cold-adapted enzymes usually have lower optimum temperature, higher activity at low temperatures, and lower thermostability compared to their mesophilic homologs (Lonhienne et al., 2000; Feller and Gerday, 2003). As shown in Figure 2B, AlyPM had a low optimal temperature of 30°C, and remained 19% of the highest activity at 5°C, and no detectable activity at 45°C. This result suggests that AlyPM has cold-adapted characters. We further investigated the thermostability of AlyPM. As shown in Figure 3A, AlyPM was rapidly inactivated at temperatures above 30°C, and approximately 80% of its activity was lost after incubation at 40°C for 15 min. Moreover, the apparent melting temperature (Tm) of AlyPM determined by CD was only 37°C (Figure 3B). These results indicate that the thermostability of AlyPM is quite low, further supporting that AlyPM is a cold-adapted enzyme.
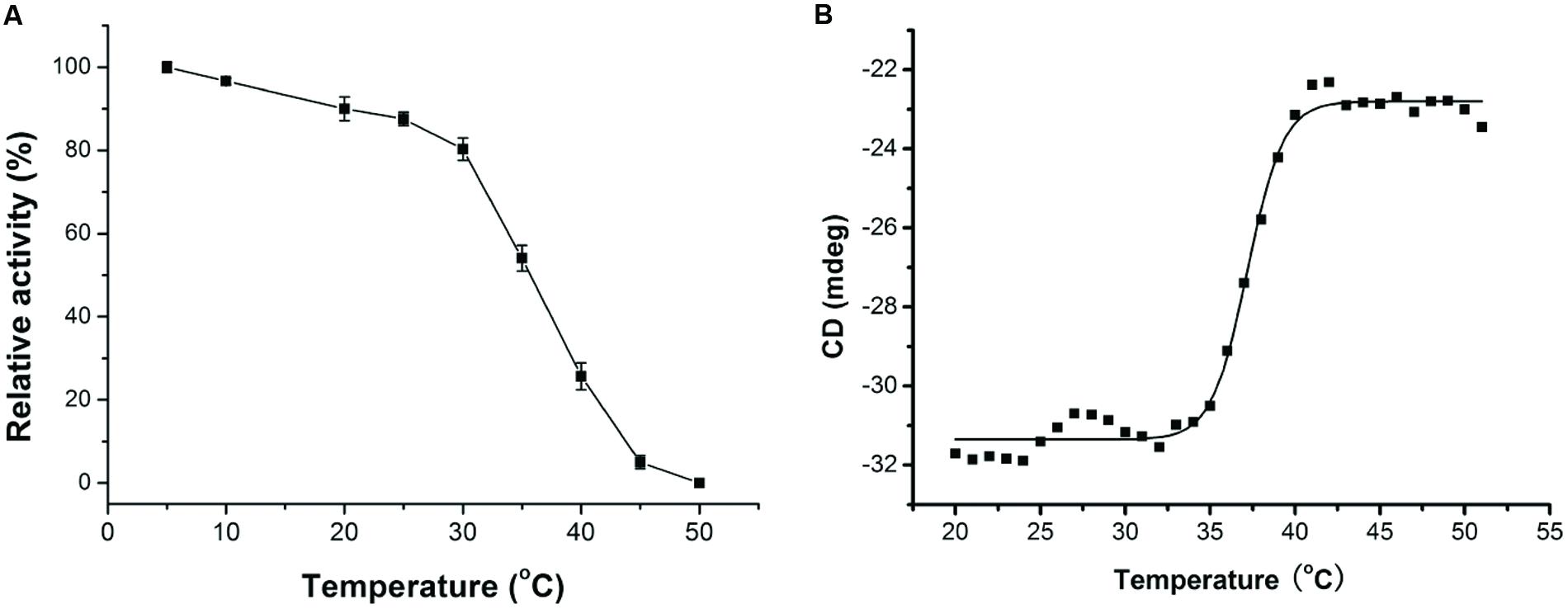
FIGURE 3. Thermostability and melting temperature (Tm) of AlyPM. (A) Thermostability of AlyPM. The enzyme was incubated at 5 to 50°C for 15 min. The remaining activity was measured under optimal conditions. (B) Tm valve of AlyPM. Data were collected from 20 to 70°C in 1°C intervals on a Jasco V810 Circular dichroism (CD) spectrophotometer. CD signals at 222 nm were used for analysis of the unfolding curve.
Salt-Activation of AlyPM
Because AlyPM is from a marine bacterium, we studied the effect of NaCl of different concentrations on the activity of AlyPM. Surprisingly, we found that the activity of AlyPM was significantly enhanced approximately sixfolds by 0.5–1.2 M NaCl (Figure 4A). This result indicates that AlyPM is a salt-activated enzyme (Kitamikado et al., 1992; Wong et al., 2000; Kobayashi et al., 2009).
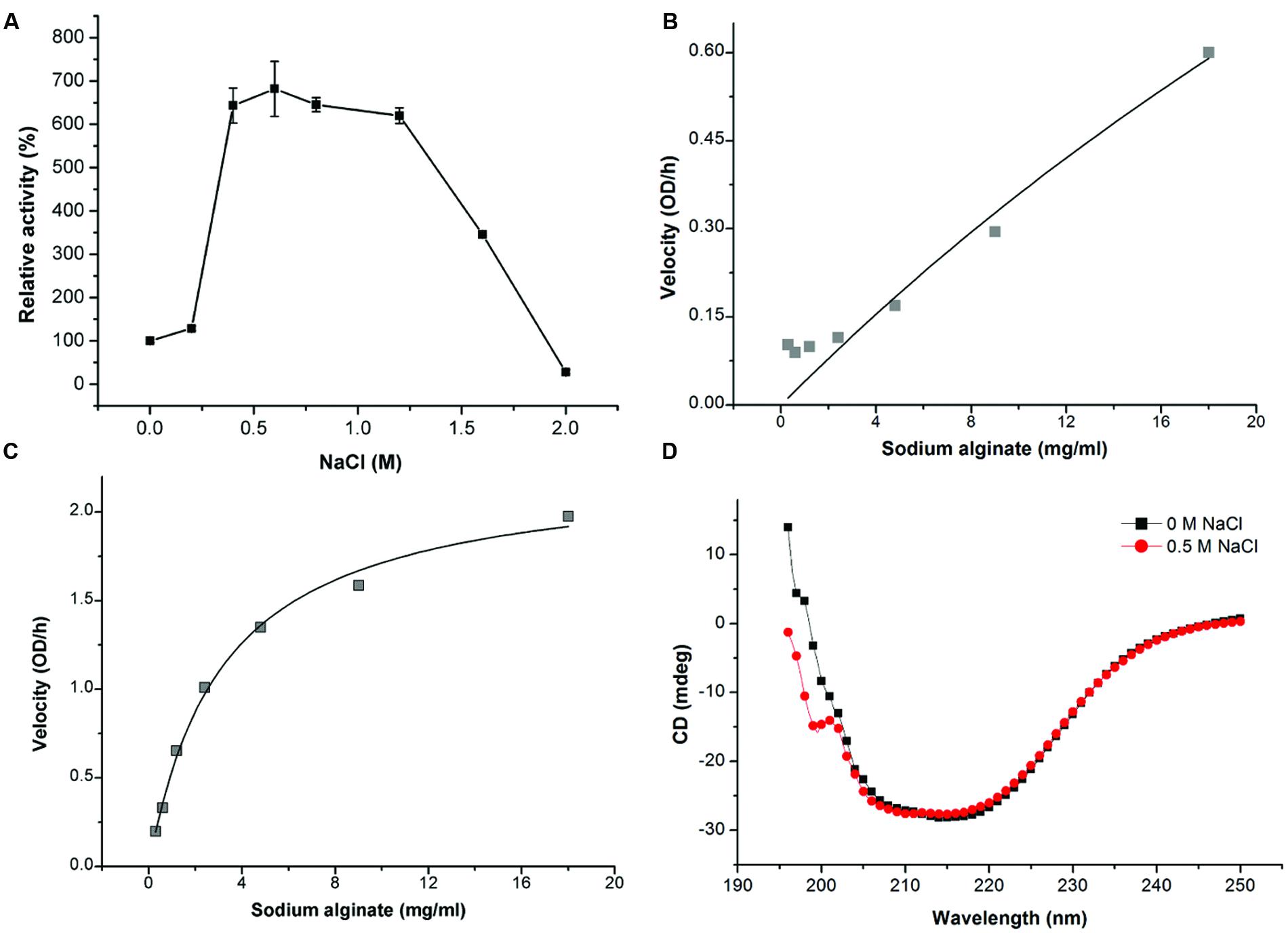
FIGURE 4. Effect of NaCl on AlyPM. (A) Effect of NaCl concentration on AlyPM activity. The activity was measured at 30°C for 30 min in 50 mM Tris-HCl buffer with the indicated NaCl concentrations. The activity in 0 M NaCl was taken as 100%. (B,C) Non-linear fit curves for the hydrolysis of sodium alginate by AlyPM in 0 M NaCl (B) and 0.5 M NaCl (C). (D) CD spectra of AlyPM in 0 and 0.5 M NaCl.
To investigate whether the enhancement of AlyPM activity by NaCl was attributed to an increase in substrate affinity, we compared the Km values of AlyPM toward sodium alginate under 0 and 0.5 M NaCl. The Km values were determined by non-linear analysis based on the initial rates determined between 0.3 and 18 mg/ml of sodium alginate because 18 mg/ml was nearly the biggest solubility of sodium alginate in 50 mM Tris buffer. The result showed that the Km value of AlyPM in 0.5 M NaCl (3.15 mg/ml) decreased by more than 20-folds compared to that in 0 M NaCl (74.39 mg/ml) (Figures 4B,C). This indicated that the substrate affinity of AlyPM in 0.5 M NaCl was significantly increased, which probably resulted in the enhancement of AlyPM activity. We also compared the CD spectra of AlyPM in 0 M NaCl and 0.5 M NaCl, which showed no detectable changes between them (Figure 4D). This suggested that the increase in the substrate affinity of AlyPM in 0.5 M NaCl was not caused by structural changes.
Substrate Specificity and Action Mode of AlyPM
To determine the substrate specificity of AlyPM, the enzyme activities toward sodium alginate, polymannuronate (polyM), and polyguluronate (polyG) were measured. As shown in Figure 5A, AlyPM showed the highest activity toward polyM, and only a little activity toward polyG, indicating that AlyPM had a preference to polyM than to polyG. The oligosaccharides released from polyM, polyG, and sodium alginate through the action of AlyPM were analyzed by TLC (Figure 5B). The maximum oligosaccharide production generated by AlyPM was from polyM and no oligosaccharide was detected from polyG on the TLC plates, which agreed with the substrate specificity of AlyPM. Products from sodium alginate and polyM generated by AlyPM were mainly dimers and trimers, and also some tetramers and even larger oligosaccharides. Thus, AlyPM mainly acts on substrate endolytically. In addition, some products lower than dimers were also shown on the TLC plates. It remains to be further determined whether AlyPM has exotype activity.
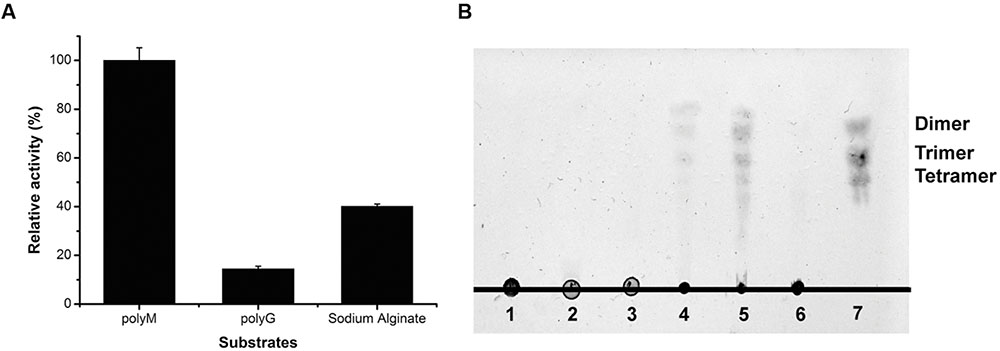
FIGURE 5. Substrate preference and action mode of AlyPM. (A) Substrate specificity of AlyPM evaluated with sodium alginate, polyM, and polyG. (B) TLC analysis of the oligomers released from polyM, polyG, and sodium alginate by AlyPM. A 500 μL reaction mixture containing 1 μg AlyPM and 2.5 mg polyM, polyG, or sodium alginate was incubated at 30°C for 2 h. Lane 1, sodium alginate; Lane 2, polyM; Lane 3, polyG; Lane 4, reaction products generated from sodium alginate. Lane 5, reaction products generated from polyM; Lane 6, reaction products generated from polyG. Lane 7, standard alginate oligomers.
Discussion
In this study, a gene encoding an alginate lyase AlyPM from marine bacterium Pseudoalteromonas sp. SM0524 was cloned. Sequence analysis showed that AlyPM belongs to the PL7 family. The PL7 family has the most alginate lyases among the seven PL families containing alginate lyases. There are now more than 535 PL7 alginate lyase sequences in the CAZy database3 (Cantarel et al., 2009). Moreover, various alginate lyases with different substrate specificity and characters have been characterized [for details see review of Zhu and Yin (2015)]. Among the characterized alginate lyases, AlyPM has the highest identity (41%) to AlyVGI from V. halioticoli IAM14596T (Sugimura et al., 2000), which indicates that AlyPM is a new member of the PL7 family.
A few cold-adapted alginate lyases have been characterized. Consistent with other cold-adapted enzymes investigated, the cold-adapted alginate lyases studied usually have lower temperature optimum, higher catalytic activities at low temperatures and lower thermostability than their mesophilic homologs (Xiao et al., 2006; Dong et al., 2012; Li et al., 2015). For an example, mesophilic alginate lyases usually have optimal temperatures around 50°C and generally stable at temperatures lower than 50°C (Kim et al., 2012; Park et al., 2012; Dou et al., 2013), whereas cold-adapted alginate lyases usually have optimal temperatures less than 35°C and generally unstable at temperatures higher than 30°C (Xiao et al., 2006; Li et al., 2015). We studied the effect of temperature on the activity and stability of AlyPM. The result showed that AlyPM had a low optimal temperature (30°C) for activity, and still remained 19% of the highest activity at 5°C. Moreover, AlyPM was quite thermolabile. It was unstable at temperatures beyond 30°C, and had a very low Tm (37°C). These results indicate that AlyPM is a cold-adapted alginate lyase. Pseudoalteromonas sp. SM0524 that secretes AlyPM was isolated from the seashore of the Yellow Sea where the temperature of seawater is usually less than 28°C (Fan and Huang, 2008; Payne et al., 2012). Therefore, the cold-adapted character of AlyPM reflects its adaptation to the marine environment where it functions.
Several salt-activated alginate lyases have been reported, whose activities can be increased by cations such as sodium and calcium (Horikoshi, 1982; Lange et al., 1989; Brown and Preston, 1991; Kitamikado et al., 1992; Wong et al., 2000; Xiao et al., 2006; Uchimura et al., 2010). Among them, an alginate lyase from V. harveyi AL-128 (Kitamikado et al., 1992) and A9mT from Vibrio sp. JAM-A9m (Uchimura et al., 2010) are the most salt-activated, and their activities are both increased by 24 times at 1 and 0.4 M NaCl, respectively. However, the salt-activated mechanism of alginate lyases is still unclear. Cations are thought to decrease the surface density of the substrate charge, weakening the ionic interactions between alginate and the enzyme (Favorov et al., 1979). In this study, we found that AlyPM is a salt-activated alginate lyase, whose activity could be increased by six times at 0.5–1.2 M NaCl. Further analysis indicated that AlyPM has a much higher affinity to substrate in 0.5 M NaCl than in 0 M NaCl, which probably leads to the increase of its activity in 0.5 M NaCl. The salt-activated character of AlyPM reflects its adaptation to the salty seawater environment.
A large amount of alginates are annually produced by algae in the sea. Various alginate lyases secreted by marine bacteria are actively involved in marine alginate degradation, and therefore, play a role in marine carbon cycling. In this study, AlyPM secreted by marine bacterium Pseudoalteromonas sp. SM0524 was characterized as a new PL7 alginate lyase. AlyPM is cold-adapted and salt-activated, and can hydrolyze alginate to produce oligomers. These characteristics indicate that AlyPM is well adapted to marine environment and may play a role in marine alginate degradation and carbon cycling. In additon, the cold-adapted and salt-activated alyPM may have potentials in some biotechnology areas and industries, which merits further study.
Author Contributions
SD, FX, and FD performed all experiments. X-LC and B-BX directed the experiments. X-LC and SD wrote the manuscript. X-YZ and P-YL analyzed the data. B-CZ and Y-ZZ designed the research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Science Foundation of China (91228210, 91328208, 41276149, 31470001, 31270117), the Hi-Tech Research and Development Program of China (2014AA093509), and the Fundamental Research Funds of Shandong University (2014QY006, 2015JC001).
Footnotes
- ^ http://www.cazy.org/fam/acc\_PL.html
- ^ http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml
- ^ http://www.cazy.org/fam/acc_PL.html
References
Aarstad, O. A., Tondervik, A., Sletta, H., and Skjak-Braek, G. (2012). Alginate sequencing: an analysis of block distribution in alginates using specific alginate degrading enzymes. Biomacromolecules 13, 106–116. doi: 10.1021/bm2013026
Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Braccini, I., Grasso, R. P., and Perez, S. (1999). Conformational and configurational features of acidic polysaccharides and their interactions with calcium ions: a molecular modeling investigation. Carbohydr. Res. 317, 119–130. doi: 10.1016/S0008-6215(99)00062-2
Braccini, I., and Perez, S. (2001). Molecular basis of C(2+)-induced gelation in alginates and pectins: the egg-box model revisited. Biomacromolecules 2, 1089–1096. doi: 10.1021/bm010008g
Brown, B. J., and Preston, J. F. III. (1991). L-guluronan-specific alginate lyase from a marine bacterium associated with Sargassum. Carbohydr. Res. 211, 91–102. doi: 10.1016/0008-6215(91)84148-8
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Dong, S., Wei, T. D., Chen, X. L., Li, C. Y., Wang, P., Xie, B. B., et al. (2014). Molecular insight into the role of the n-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18. J. Biol. Chem. 289, 29558–29569. doi: 10.1074/jbc.M114.584573
Dong, S., Yang, J., Zhang, X. Y., Shi, M., Song, X. Y., Chen, X. L., et al. (2012). Cultivable alginate lyase-excreting bacteria associated with the Arctic brown alga Laminaria. Mar. Drugs 10, 2481–2491. doi: 10.3390/md10112481
Dou, W., Wei, D., Li, H., Li, H., Rahman, M. M., Shi, J., et al. (2013). Purification and characterisation of a bifunctional alginate lyase from novel Isoptericola halotolerans CGMCC 5336. Carbohydr. Polym. 98, 1476–1482. doi: 10.1016/j.carbpol.2013.07.050
Fan, H., and Huang, H. (2008). Response of coastal marine eco-environment to river fluxes into the sea: a case study of the Huanghe (Yellow) River mouth and adjacent waters. Mar. Environ. Res. 65, 378–387. doi: 10.1016/j.marenvres.2008.01.003
Favorov, V. V., Vozhova, E. I., Denisenko, V. A., and Elyakova, L. A. (1979). A study of the reaction catalysed by alginate lyase VI from the sea mollusc, Littorina sp. Biochim. Biophys. Acta 569, 259–266. doi: 10.1016/0005-2744(79)90061-5
Feller, G., and Gerday, C. (2003). Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1, 200–208. doi: 10.1038/nrmicro773
Gacesa, P. (1992). Enzymic degradation of alginates. Int. J. Biochem. 24, 545–552. doi: 10.1016/0020-711X(92)90325-U
Haug, A., Claeson, K., Hansen, S. E., Sömme, R., Stenhagen, E., and Palmstierna, H. (1959). Fractionation of alginic acid. Acta Chem. Scand. 13, 601–603. doi: 10.3891/acta.chem.scand.13-0601
Horikoshi, K. A. T. (1982). Alkalophilic microorganisms. Tokyo: Japan Scientific Societies Press, 137.
Inoue, A., Anraku, M., Nakagawa, S., and Ojima, T. (2016). Discovery of a novel alginate lyase from Nitratiruptor sp. SB155-2 thriving at deep-sea hydrothermal vents and identification of the residues responsible for its heat stability. J. Biol. Chem. doi: 10.1074/jbc.M115.713230 [Epub ahead of print].
Kawamoto, H., Horibe, A., Miki, Y., Kimura, T., Tanaka, K., Nakagawa, T., et al. (2006). Cloning and sequencing analysis of alginate lyase genes from the marine bacterium Vibrio sp. O2. Mar. Biotechnol. 8, 481–490. doi: 10.1007/s10126-005-6157-z
Kim, H. T., Ko, H. J., Kim, N., Kim, D., Lee, D., Choi, I. G., et al. (2012). Characterization of a recombinant endo-type alginate lyase (Alg7D) from Saccharophagus degradans. Biotechnol. Lett. 34, 1087–1092. doi: 10.1007/s10529-012-0876-9
Kitamikado, M., Tseng, C. H., Yamaguchi, K., and Nakamura, T. (1992). Two types of bacterial alginate lyases. Appl. Environ. Microbiol. 58, 2474–2478.
Kobayashi, T., Uchimura, K., Miyazaki, M., Nogi, Y., and Horikoshi, K. (2009). A new high-alkaline alginate lyase from a deep-sea bacterium Agarivorans sp. Extremophiles 13, 121–129. doi: 10.1007/s00792-008-0201-7
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Lange, B., Wingender, J., and Winkler, U. K. (1989). Isolation and characterization of an alginate lyase from Klebsiella aerogenes. Arch. Microbiol. 152, 302–308. doi: 10.1007/BF00409667
Lee, K. Y., and Mooney, D. J. (2012). Alginate: properties and biomedical applications. Prog. Polym. Sci. 37, 106–126. doi: 10.1016/j.progpolymsci.2011.06.003
Li, J. W., Dong, S., Song, J., Li, C. B., Chen, X. L., Xie, B. B., et al. (2011). Purification and characterization of a bifunctional alginate lyase from Pseudoalteromonas sp. SM0524. Mar. Drugs 9, 109–123. doi: 10.3390/md9010109
Li, S., Yang, X., Zhang, L., Yu, W., and Han, F. (2015). Cloning, expression, and characterization of a cold-adapted and surfactant-stable alginate lyase from marine bacterium Agarivorans sp. L11. J. Microbiol. Biotechnol. 25, 681–686. doi: 10.4014/jmb.1409.09031
Liu, Y. G., and Whittier, R. F. (1995). Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. doi: 10.1016/0888-7543(95)80010-J
Lonhienne, T., Gerday, C., and Feller, G. (2000). Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim. Biophys. Acta 1543, 1–10. doi: 10.1016/S0167-4838(00)00210-7
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. doi: 10.1093/nar/gkq1189
Osawa, T., Matsubara, Y., Muramatsu, T., Kimura, M., and Kakuta, Y. (2005). Crystal structure of the alginate (poly alpha-l-guluronate) lyase from Corynebacterium sp. at 1.2 A resolution. J. Mol. Biol. 345, 1111–1118. doi: 10.1016/j.jmb.2004.10.081
Park, H. H., Kam, N., Lee, E. Y., and Kim, H. S. (2012). Cloning and characterization of a novel oligoalginate lyase from a newly isolated bacterium Sphingomonas sp. MJ-3. Mar. Biotechnol. 14, 189–202. doi: 10.1007/s10126-011-9402-7
Payne, M. C., Brown, C. A., Reusser, D. A., and Lee, H. II. (2012). Ecoregional analysis of nearshore sea-surface temperature in the North Pacific. PLoS ONE 7:e30105. doi: 10.1371/journal.pone.0030105
Preiss, J., and Ashwell, G. (1962). Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosac-charides and 4-deoxy-L-erythro-5-hexoseulose uronic acid. J. Biol. Chem. 237, 309–316.
Sugimura, I. I., Sawabe, T., and Ezura, Y. (2000). Cloning and sequence analysis of Vibrio halioticoli genes encoding three types of polyguluronate lyase. Mar. Biotechnol. 2, 65–73.
Thomas, F., Lundqvist, L. C., Jam, M., Jeudy, A., Barbeyron, T., Sandstrom, C., et al. (2013). Comparative characterization of two marine alginate lyases from Zobellia galactanovorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J. Biol. Chem. 288, 23021–23037. doi: 10.1074/jbc.M113.467217
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tonnesen, H. H., and Karlsen, J. (2002). Alginate in drug delivery systems. Drug Dev. Ind. Pharm. 28, 621–630. doi: 10.1081/DDC-120003853
Uchimura, K., Miyazaki, M., Nogi, Y., Kobayashi, T., and Horikoshi, K. (2010). Cloning and sequencing of alginate lyase genes from deep-sea strains of Vibrio and Agarivorans and characterization of a new Vibrio enzyme. Mar. Biotechnol. 12, 526–533. doi: 10.1007/s10126-009-9237-7
Wong, T. Y., Preston, L. A., and Schiller, N. L. (2000). ALGINATE LYASE: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54, 289–340. doi: 10.1146/annurev.micro.54.1.289
Xiao, L., Feng, H., Yang, Z., Lu, X. Z., and Yu, W. G. (2006). A novel alginate lyase with high activity on acetylated alginate of Pseudomonas aeruginosa FRD1 from Pseudomonas sp. QD03. World J. Microbiol. Biotechnol. 22, 81–88. doi: 10.1007/s11274-005-7713-4
Yamasaki, M., Moriwaki, S., Miyake, O., Hashimoto, W., Murata, K., and Mikami, B. (2004). Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: a novel alginate lyase with a beta-sandwich fold. J. Biol. Chem. 279, 31863–31872. doi: 10.1074/jbc.M402466200
Yamasaki, M., Ogura, K., Hashimoto, W., Mikami, B., and Murata, K. (2005). A structural basis for depolymerization of alginate by polysaccharide lyase family-7. J. Mol. Biol. 352, 11–21. doi: 10.1016/j.jmb.2005.06.075
Keywords: marine bacteria, alginate lyase, cold-adapted enzyme, salt-activated, polysaccharide lyase family 7
Citation: Chen X-L, Dong S, Xu F, Dong F, Li P-Y, Zhang X-Y, Zhou B-C, Zhang Y-Z and Xie B-B (2016) Characterization of a New Cold-Adapted and Salt-Activated Polysaccharide Lyase Family 7 Alginate Lyase from Pseudoalteromonas sp. SM0524. Front. Microbiol. 7:1120. doi: 10.3389/fmicb.2016.01120
Received: 29 April 2016; Accepted: 06 July 2016;
Published: 19 July 2016.
Edited by:
Senjie Lin, University of Connecticut, USAReviewed by:
Fu-Li Li, Chinese Academy of Sciences, ChinaFeng Han, Ocean University of China, China
Copyright © 2016 Chen, Dong, Xu, Dong, Li, Zhang, Zhou, Zhang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin-Bin Xie, xbb@sdu.edu.cn
†These authors have contributed equally to this work.
 Xiu-Lan Chen
Xiu-Lan Chen Sheng Dong1,2†
Sheng Dong1,2† Xi-Ying Zhang
Xi-Ying Zhang Yu-Zhong Zhang
Yu-Zhong Zhang Bin-Bin Xie
Bin-Bin Xie