- 1Department of Medical Microbiology, Radboud University Medical Center, Nijmegen, Netherlands
- 2Center of Expertise in Mycology Radboudumc/CWZ, Nijmegen, Netherlands
Aspergillus diseases are often caused by Aspergillus fumigatus. Azoles are the mainstay of therapy, but the management of aspergillosis is hampered by the emergence of azole resistance. Rapid detection of azole resistance might benefit treatment outcome by early treatment modifications. However, the yield of fungal culture in invasive aspergillosis is low and susceptibility testing requires several days to be completed. To overcome the low yield of fungal cultures and slow detection of resistance, it is possible to use molecular tools directly on clinical specimens in order to rapidly detect molecular markers of azole resistance. Molecular tools to detect resistant markers in the Cyp51A gene can be expected to be less sensitive compared to molecular tools to detect Aspergillus DNA as the Cyp51A gene is a single copy gene and the target for Aspergillus DNA is often a multi-copy gene. In this mini-review, we summarize the current molecular tools for detection of azole-resistant A. fumigatus directly in clinical material. Several in-house PCR assays have been applied directly on clinical material. Furthermore, two assays are commercial available; the AsperGenius and MycoGENIE. The amplification of resistance markers was successful in 70–100% of samples that were positive for Aspergillus DNA in BAL samples using the AsperGenius assay. Despite using several samples per patient, amplification of resistance markers was only successful in 33–57% of patients with Aspergillus DNA in blood. Furthermore, several sequence based methods have been applied with the benefit of the ability to detect other Cyp51A gene alterations.
Introduction
Aspergillus species cause a wide spectrum of disease, including invasive aspergillosis (IA), chronic pulmonary aspergillosis (CPA), Aspergillus bronchitis, and allergic bronchopulmonary aspergillosis (ABPA). The most common species involved in aspergillus diseases is Aspergillus fumigatus. If antifungal treatment is required, azoles are the first line of treatment, independently of the manifestation of disease (Kosmidis and Denning, 2015). However, the management of aspergillosis is hampered by the emergence of azole resistance (Verweij et al., 2016). Several case series indicate that detection of azole resistance is associated with treatment failure (van der Linden et al., 2011; Chong et al., 2016).
The detection of azole resistance in IA is challenging as the yield of fungal cultures is generally low and phenotypical evaluation of azole susceptibility is therefore often impossible. Furthermore, even if a culture is positive for A. fumigatus, susceptibility testing requires several days before results are available. A mature culture is recommended for MIC-testing, which requires up to 7 days of incubation and MICs can be read after an additional 48 h. As early treatment of patients with IA with an appropriate drug improves outcome (Hauggaard et al., 2002), inappropriate therapy due to azole resistance may increase treatment failure. Nowadays it is possible to use molecular tools directly on clinical specimens in order to rapidly detect resistance markers. In this review, we summarize the current molecular tools of azole resistance detection in A. fumigatus directly in clinical material.
Molecular Markers of Azole Resistance in A. fumigatus
The hot spot for resistance mutations in A. fumigatus is the Cyp51A gene, a gene important for the ergosterol biosynthesis pathway. Generally, two groups of resistance mutations can be acknowledged; the environmental resistance mutations and the resistance mutations due to long-term azole therapy. TR34/L98H and TR46/Y121F/T289A are the most common environmental resistance mutations which are now recovered globally. Furthermore, several less prevalent environmental resistance mutations have been reported including TR53, TR463, and TR464 (three and four repeats of TR46, respectively) (Zhang et al., 2017). The other group of resistance mutations consists of point mutations in the Cyp51A gene at various positions including G54, M220, P216, G138, and G448 with mutations at locus G54 and M220 leading to the most common amino acid substitutions (Meis et al., 2016; Verweij et al., 2016).
It is estimated that 10% of azole resistance is not Cyp51A mediated (Meis et al., 2016). Several other resistance mechanisms have been described including increased efflux pump activity and a substitution in the HapE gene (Camps et al., 2012; Fraczek et al., 2013). Moreover, it seems likely that several other, yet uncharacterized, mechanisms are in involved in azole resistance in A. fumigatus.
Molecular Tools to Detect Azole Resistance Directly in Clinical Samples
Real-Time Resistance-PCR Tests
In-House Assays
The first report of the use of a qPCR for the detection of resistance mutations on clinical samples was in 2010 in a case of pulmonary and cerebral aspergillosis (van der Linden et al., 2010). A pulmonary isolate was phenotypically resistant to azoles. Brain biopsy showed septate hyphae but fungal cultures remained negative. Sequence-based analysis of the pulmonary isolates showed the TR34/L98H mutation and a real-time PCR was applied on the brain tissue. For detection of the L98H mutation, a 122-bp fragment was amplified, using a hybridization probe to detect the L98H mutation. Furthermore, a second amplification of a 110-bp fragment was used to detect the 34-bp insertion (TR34) using a TaqMan probe that was designed to bind to the last 13-bp and the first 9-bp of the 34-bp insertion. The real-time PCR revealed the presence of a L98H mutation as well as the 34-bp insertion (van der Linden et al., 2010). Further details of the published papers evaluating qPCR resistance detection can be found in Table 1.
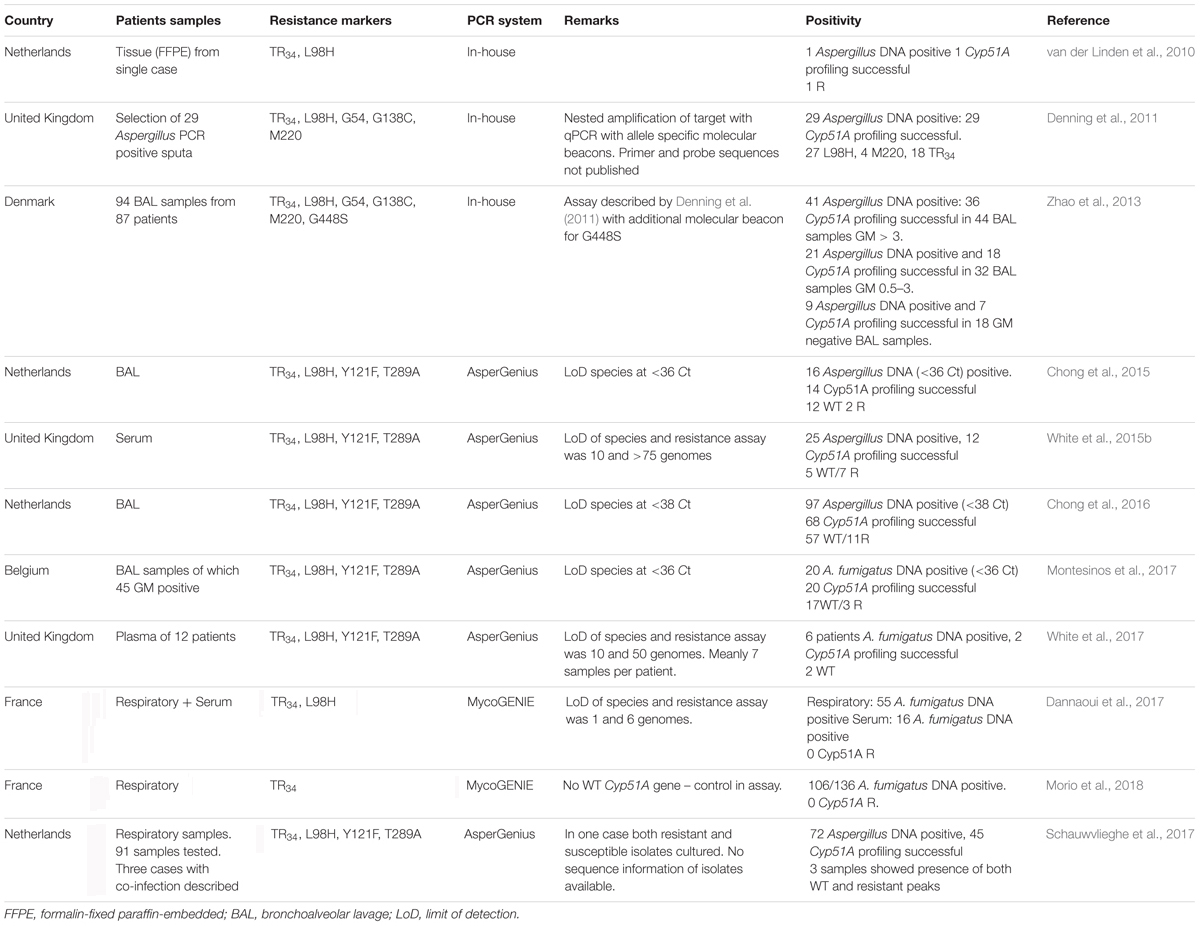
TABLE 1. Overview of studies that applied real-time PCR assays directly on clinical specimen to detect azole resistance markers in Aspergillus fumigatus.
Another in-house PCR assay was used on 29 Aspergillus DNA positive sputum samples from patients with CPA and ABPA. In this series a nested-PCR approach was used. The Cyp51A gene was amplified in two fractions and a qPCR with allele-specific molecular beacons was used to detect mutations at locus G54, L98, G138, and M220 and to detect the 34-bp insertion. Cyp51A was successfully amplified in all 29 Aspergillus PCR positive samples. Four samples had a mutation at M220, 27 of 29 had a L98H mutation of which 16 also had TR34. TR34 without L98H was found in two patients. In two patients with a M220R mutation, TR34 and L98H were also detected. Only four samples were culture positive, in three the molecular results did not correspond with the culture results (Denning et al., 2011). The detection of either TR34 or L98H alone and a TR34/L98H with an additional M220 mutation has not been reported in cultured isolates, to the best of our knowledge. It seems likely that due to fact that two genomic regions are amplificated independently for the detection of TR34 and L98H, the amplicons were from different strains, (e.g., a susceptible and resistant strain), resulting in the single detection of either TR34 or L98H. The frequency of resistance markers seems very high in this series and these results have not yet been confirmed by other studies.
The same nested PCR-assay, with the addition of a new molecular beacon for G448, was used to analyze 94 BAL samples. Sixty-one of 71 (86%) samples positive for a pan-Aspergillus PCR had a successful amplification of the Cyp51A gene. Four of the Cyp51A negative samples were culture positive for A. flavus. One sample contained M220V and another P216L. The authors did not describe the use of a molecular beacon for P216L and the mutation was thus presumably detected by subsequent sequence analysis (Zhao et al., 2013). The high rate of TR34/L98H mutations observed by Denning et al. (2011) was not observed in the latter study from Denmark.
Commercial Available Assays
Results from the first commercial available resistance PCR assay, the AsperGenius (PathoNostics, Maastricht, Netherlands) were published in 2015. The qPCR has the ability to detect Aspergillus DNA, and differentiates A. fumigatus and A. terreus. In addition, four mutations (TR34 and L98H to detect TR34/L98H, Y121F, and T289A to detect TR46/Y121F/T289A) can be detected using melting-curve analyses. The PCR was validated on stored BAL fluid samples from hematological and ICU patients. Of 19 BAL fluids, 16 were positive for Aspergillus DNA, whereas 14 of 16 were positive for A. fumigatus. All 14 samples were also positive for resistance targets and mutation analyses revealed 12 samples with WT Cyp51A DNA, one TR34/L98H and one TR46/Y121F/T289A (Chong et al., 2015).
The assay was validated prospectively in a multicenter study in 201 patients with hematological disease suspected for IA. The Aspergillus PCR was positive in 97 samples (74 samples with either galactomannan (GM)-index >1 or with a positive culture) and the resistance PCR was successful in 68 of 97 (70%) samples. In 7 samples TR34/L98H was detected, in one sample TR46/Y121F/T289A, while a combination of TR34/L98H and WT A. fumigatus was observed in three samples. Importantly, this study showed that the detection of resistance markers was associated with azole treatment failure; even in culture-negative cases (Chong et al., 2016).
Another study used the assay on 45 GM-positive BAL samples and 55 GM-negative BAL samples. Twenty-seven of 100 patients were positive for Aspergillus DNA and 20 for A. fumigatus DNA. The amplification of resistance markers appeared to be successful in all A. fumigatus positive samples and evidence for azole resistance was found in three (Montesinos et al., 2017).
Other studies have evaluated the AsperGenius assay directly on blood samples, for early detection of Aspergillus disease in patients at risk for IA (White et al., 2015a). The assay was evaluated on serum samples from 14 patients with proven or probable IA (total 54 extracts). A. fumigatus DNA was detected in 25 of 54 samples. The amount of DNA in the serum, however, was low and only 12 samples of 7 cases had sufficient DNA-levels for the resistance PCR to be performed reflecting the lower sensitivity of the resistance PCR compared to the Aspergillus DNA assay. Samples from four patients (57%) had 3–4 loci (TR34, L98H, T289A, or Y121F) amplified and 50% of all patients had at least one loci amplified. Mutation analyses showed no mutations in these patients. Thus despite using multiple samples per patient; genotyping both TR34/L98H and TR46/Y121F/T289A was possible in only four of 14 patients with proven/possible IA. For successful amplification of resistance markers, a Ct value of <33 was needed in the A. fumigatus assay (White et al., 2015b).
As the performance of Aspergillus-PCR was shown to be superior on plasma compared to serum, it was hypothesized that plasma was also superior for the detection of resistance markers (White et al., 2015a). Eighty-six plasma samples from 10 patients with proven/probable IA and two patients with possible IA were tested. Eleven samples from six patients were positive for A. fumigatus. Amplification of resistance markers was successful in only two of six (33%) patients. Thus, using a mean of seven samples per patient, the resistance markers were successfully amplified in only two of 12 (17%) patients with IA. However, as the A. fumigatus PCR was only positive in six of 12 patients and all culture results were negative, it is not known whether the IA was due to A. fumigatus in the other patients (White et al., 2017). As the target for azole resistance detection is a single copy gene, the sensitivity can be expected to be lower than the Aspergillus DNA assay for which a multi-copy gene is targeted. The results of PCR used on blood samples clearly reflects this lower sensitivity and due to the low amount of DNA in blood samples, clinical utility in routine diagnostics is questionable.
An interesting advantage of the AsperGenius PCR is the simultaneous detection of both WT and mutant Cyp51A DNA. As the detection of resistance markers is based on melting curve analysis, mixed infections with both WT as well as DNA from resistant strains will result in two peaks in the assay. This was shown in three cases were evidence for both WT and L98H was found (Schauwvlieghe et al., 2017).
A report of a second commercial available assay, the MycoGENIE (Ademtech, Pessac, France), was published in 2017. The MycoGENIE is a multiplex assay detecting A. fumigatus DNA, the L98H mutation and TR34. The PCR was tested on a total of 55 serum samples from 16 patients and 88 respiratory samples from 62 patients. Fifty-Five respiratory and 16 serum samples were positive for A. fumigatus (Dannaoui et al., 2017). The same MycoGENIE was then used on stored clinical samples from 137 patients with fungal rhinosinusitis with a positive microscopy for fungal hyphae. The PCR was positive for A. fumigatus in 116 of 147 samples (78.9%), whereas 47 of 48 A. fumigatus culture positive samples were positive in the assay (Morio et al., 2018). In both studies no samples were positive for either TR34 or L98H.
An important limitation of the MycoGENIE is the absence of WT probes for the resistance marker. As a consequence, non-positivity in the resistance markers might be caused a WT genotype but might also be due to a missed mutation due to limited sensitivity compared to the probe for detection of A. fumigatus DNA. Furthermore, the less prevalent environmental TR46/Y121F/T289A mutation is not detected by the MycoGENIE.
Sequence Based Assays
In addition to direct qPCR assays, several other methods have been applied directly to clinical specimen (Table 2). Amplification with subsequent pyrosequencing was applied directly on blood samples to detect G54 mutations in the Cyp51A gene. A. fumigatus DNA was detected in two of 56 whole-blood samples targeting a 269-p Cyp51A gene region and Cyp51A sequence of locus G54 was determined. The study showed the utility of amplification and subsequent sequencing of clinical material for detection of Cyp51A-mutations (Trama et al., 2005). Most other loci were not covered by the targeted region but this can be overcome by sequencing of additional Cyp51A regions involved in azole resistance.
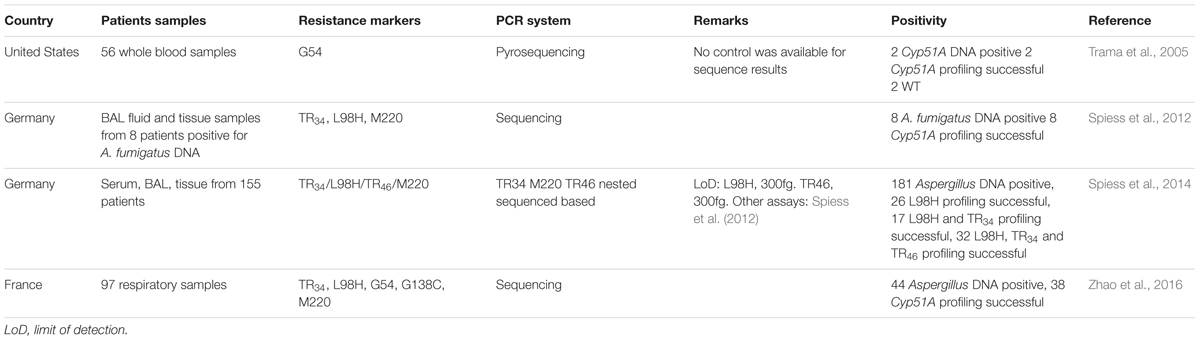
TABLE 2. Overview of studies that applied non-real-time molecular tools directly on clinical specimen to detect azole resistance markers in A. fumigatus.
Other authors generated additional primers for the specific amplification and subsequent sequencing of the A. fumigatus Cyp51A-gene. Three different primer sets were used for the amplification of the Cyp51A promoter region, L98 and M220 regions. Eight stored clinical samples were tested and TR34/L98H was successfully detected in a brain abscess. Interestingly, in one BAL fluid, L98H was found without the TR34 (Spiess et al., 2012). Using the same method with the addition of another target region for the detection of TR46, stored clinical samples of 155 immunocompromised patients previously tested positive for Aspergillus DNA were evaluated. In 75 of 181 (41%) investigated samples, the assay was positive for at least one region. The L98H region alone was successfully amplified and sequenced in 26 (14%), the regions for L98H and TR34 in 17 (9%); both revealed no mutations. In 22 of 181 (12%) samples the L98H, TR34 as well as the M220 PCR targets were successfully amplificated, showing two samples with both the TR34 and L98H mutations. One sample had L98H but not TR34 (Spiess et al., 2014). Higher rate of successful amplification was achieved by a nested approach. The sequence method described by Denning et al. (2011) was used but without the molecular beacons. The Cyp51A was successfully amplified in 38 of 44 (86%) pan-Aspergillus PCR positive samples. Using this technique, the authors found a sputum sample harboring TR34/L98H, which was confirmed by culture (Zhao et al., 2016).
Conclusion and Perspectives
The application of molecular tools has been shown to increase the sensitivity of detection of azole resistance in A. fumigatus compared to culture. Furthermore, the detection of resistance markers in BAL samples using the AsperGenius was associated with increased probability of azole treatment failure. The commercial available assays target TR34/L98H and TR46/Y121F/T289A mutations and are able to detect most of azole-resistant A. fumigatus in patients with acute IA. However, the clinical utility depends on regional resistance epidemiology. In patients with prolonged azole therapy, a greater diversity of resistance mutations should be taken into consideration. Detection of resistance markers in blood samples remains problematic; nonetheless if very low Ct value in the Aspergillus PCR is observed, resistance markers might be successfully detected.
Direct amplification and subsequent sequencing of the Cyp51A-gene showed the potential to cover most Cyp51A-mediated resistance mutations. However, compared to real-time assays, sequencing is relatively time-consuming, expensive and laborious, and might lack sensitivity due to low amounts of DNA in most clinical samples. Furthermore, compared to the commercial qPCR assay for detection of azole resistance, clinical validation studies are currently lacking.
New molecular assays should aim to increase sensitivity and extend the coverage of resistance markers to include both environmental as other azole resistance mutations. Sequencers have become portable, inexpensive and can be used in real time. When the capacity of whole genome sequencing increases in the near future, it seems likely that whole genome sequencing can be applied directly on clinical specimens to detect not only the pathogen, but also a wide range of resistance mechanisms.
Author Contributions
WM, JZ, and JB reviewed the available papers. JB wrote the first draft manuscript. WM and PV edited the manuscript. All authors gave their final approval.
Funding
This work was supported by internal funding.
Conflict of Interest Statement
PV has received research grants from Gilead Sciences, Astellas, Merck Sharp & Dohme (MSD), F2G, and BioRad, is a speaker for Gilead Sciences and MSD, and is on the advisory boards for Pfizer, MSD, and F2G.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Camps, S. M., Dutilh, B. E., Arendrup, M. C., Rijs, A. J., Snelders, E., Huynen, M. A., et al. (2012). Discovery of a HapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One 7:e50034. doi: 10.1371/journal.pone.0050034
Chong, G. L., Van de Sande, W. W., Dingemans, G. J., Gaajetaan, G. R., Vonk, A. G., Hayette, M. P., et al. (2015). Validation of a new Aspergillus real-time PCR assay for direct detection of Aspergillus and azole resistance of Aspergillus fumigatus on bronchoalveolar lavage fluid. J. Clin. Microbiol. 53, 868–874. doi: 10.1128/JCM.03216-14
Chong, G. M., Van der Beek, M. T., Von dem Borne, P. A., Boelens, J., Steel, E., Kampinga, G. A., et al. (2016). PCR-based detection of Aspergillus fumigatus Cyp51A mutations on bronchoalveolar lavage: a multicentre validation of the AsperGenius assay® in 201 patients with haematological disease suspected for invasive aspergillosis. J. Antimicrob. Chemother. 71, 3528–3535.
Dannaoui, E., Gabriel, F., Gaboyard, M., Lagardere, G., Audebert, L., Quesne, G., et al. (2017). Molecular diagnosis of invasive aspergillosis and detection of azole resistance by a newly commercialized PCR kit. J. Clin. Microbiol. 55, 3210–3218. doi: 10.1128/JCM.01032-17
Denning, D. W., Park, S., Lass-Florl, C., Fraczek, M. G., Kirwan, M., Gore, R., et al. (2011). High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin. Infect. Dis. 52, 1123–1129. doi: 10.1093/cid/cir179
Fraczek, M. G., Bromley, M., Buied, A., Moore, C. B., Rajendran, R., Rautemaa, R., et al. (2013). The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 68, 1486–1496. doi: 10.1093/jac/dkt075
Hauggaard, A., Ellis, M., and Ekelund, L. (2002). Early chest radiography and CT in the diagnosis, management and outcome of invasive pulmonary aspergillosis. Acta Radiol. 43, 292–298.
Kosmidis, C., and Denning, D. W. (2015). The clinical spectrum of pulmonary aspergillosis. Thorax 70, 270–277. doi: 10.1136/thoraxjnl-2014-206291
Meis, J. F., Chowdhary, A., Rhodes, J. L., Fisher, M. C., and Verweij, P. E. (2016). Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150460. doi: 10.1098/rstb.2015.0460
Montesinos, I., Argudin, M. A., Hites, M., Ahajjam, F., Dodemont, M., Dagyaran, C., et al. (2017). Culture-based methods and molecular tools for azole-resistant Aspergillus fumigatus detection in a Belgian university hospital. J. Clin. Microbiol. 55, 2391–2399. doi: 10.1128/JCM.00520-17
Morio, F., Dannaoui, E., Chouaki, T., Cateau, E., Malard, O., Bonfils, P., et al. (2018). PCR-based detection of Aspergillus fumigatus and absence of azole resistance due to TR34 /L98H in a French multicenter cohort of 137 patients with fungal rhinosinusitis. Mycoses 61, 30–34. doi: 10.1111/myc.12702
Schauwvlieghe, A., Vonk, A. G., Buddingh, E. P., Hoek, R. A. S., Dalm, V. A., Klaassen, C. H. W., et al. (2017). Detection of azole-susceptible and azole-resistant Aspergillus coinfection by cyp51A PCR amplicon melting curve analysis. J. Antimicrob. Chemother. 72, 3047–3050. doi: 10.1093/jac/dkx262
Spiess, B., Postina, P., Reinwald, M., Cornely, O. A., Hamprecht, A., Hoenigl, M., et al. (2014). Incidence of Cyp51 A Key mutations in Aspergillus fumigatus-A study on primary clinical samples of immunocompromised patients in the period of 1995–2013. PLoS One 9:e103113. doi: 10.1371/journal.pone.0103113
Spiess, B., Seifarth, W., Merker, N., Howard, S. J., Reinwald, M., Dietz, A., et al. (2012). Development of novel PCR assays to detect azole resistance-mediating mutations of the Aspergillus fumigatus cyp51A gene in primary clinical samples from neutropenic patients. Antimicrob. Agents Chemother. 56, 3905–3910. doi: 10.1128/AAC.05902-11
Trama, J. P., Mordechai, E., and Adelson, M. E. (2005). Detection of Aspergillus fumigatus and a mutation that confers reduced susceptibility to itraconazole and posaconazole by real-time PCR and pyrosequencing. J. Clin. Microbiol. 43, 906–908.
van der Linden, J. W., Snelders, E., Arends, J. P., Daenen, S. M., Melchers, W. J., and Verweij, P. E. (2010). Rapid diagnosis of azole-resistant aspergillosis by direct PCR using tissue specimens. J. Clin. Microbiol. 48, 1478–1480. doi: 10.1128/JCM.02221-09
van der Linden, J. W., Snelders, E., Kampinga, G. A., Rijnders, B. J., Mattsson, E., Debets-Ossenkopp, Y. J., et al. (2011). Clinical implications of azole resistance in Aspergillus fumigatus, The Netherlands, 2007–2009. Emerg. Infect. Dis. 17, 1846–1854. doi: 10.3201/eid1710.110226
Verweij, P. E., Chowdhary, A., Melchers, W. J., and Meis, J. F. (2016). Azole resistance in Aspergillus fumigatus: Can we retain the clinical use of mold-active antifungal azoles? Clin. Infect. Dis. 62, 362–368. doi: 10.1093/cid/civ885
White, P. L., Barnes, R. A., Springer, J., Klingspor, L., Cuenca-Estrella, M., Morton, C. O., et al. (2015a). Clinical performance of Aspergillus PCR for testing serum and plasma: a study by the European Aspergillus PCR initiative. J. Clin. Microbiol. 53, 2832–2837. doi: 10.1128/JCM.00905-15
White, P. L., Posso, R. B., and Barnes, R. A. (2015b). Analytical and clinical evaluation of the PathoNostics AsperGenius assay for detection of invasive aspergillosis and resistance to azole antifungal drugs during testing of serum samples. J. Clin. Microbiol. 53, 2115–2121.
White, P. L., Posso, R. B., and Barnes, R. A. (2017). Analytical and clinical evaluation of the PathoNostics AsperGenius assay for detection of invasive aspergillosis and resistance to azole antifungal drugs directly from plasma samples. J. Clin. Microbiol. 55, 2356–2366. doi: 10.1128/JCM.00411-17
Zhang, J., Snelders, E., Zwaan, B. J., Schoustra, S. E., Meis, J. F., Van Dijk, K., et al. (2017). A novel environmental azole resistance mutation in Aspergillus fumigatus and a possible role of sexual reproduction in its emergence. mBio 8:e00791-17. doi: 10.1128/mBio.00791-17
Zhao, Y., Garnaud, C., Brenier-Pinchart, M. P., Thiebaut-Bertrand, A., Saint-Raymond, C., Camara, B., et al. (2016). Direct molecular diagnosis of aspergillosis and CYP51A profiling from respiratory samples of French patients. Front. Microbiol. 7:1164. doi: 10.3389/fmicb.2016.01164
Keywords: azole resistance, Aspergillus fumigatus, polymerase chain reaction, azole, antifungal, diagnostics
Citation: Buil JB, Zoll J, Verweij PE and Melchers WJG (2018) Molecular Detection of Azole-Resistant Aspergillus fumigatus in Clinical Samples. Front. Microbiol. 9:515. doi: 10.3389/fmicb.2018.00515
Received: 31 January 2018; Accepted: 06 March 2018;
Published: 21 March 2018.
Edited by:
Juergen Prattes, Medizinische Universität Graz, AustriaReviewed by:
Cornelia Lass-Flörl, Division of Hygiene and Medical Microbiology, AustriaTakahito Toyotome, Obihiro University of Agriculture and Veterinary Medicine, Japan
Copyright © 2018 Buil, Zoll, Verweij and Melchers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jochem B. Buil, jochem.buil@radboudumc.nl
 Jochem B. Buil
Jochem B. Buil Jan Zoll1,2
Jan Zoll1,2 Willem J. G. Melchers
Willem J. G. Melchers