- Department of Pharmaceutical & Biological Engineering, School of Chemical Engineering, Sichuan University, Chengdu, China
The aquatic bird, egret, could carry antibiotic resistance (AR) from a contaminated waterway (Jin River, Chengdu, China) into the surrounding environment (Wangjianglou Park). A systematic study was carried out on the unique environmental dissemination mode of AR mediated by birds. The minimum inhibitory concentrations of various antibiotics against the environmental Escherichia coli isolates were used to evaluate the bacterial AR at the environmental locations where these isolates were recovered, i.e., the Jin River water, the egret feces, the park soil, and the campus soil. The level of AR in the park soil was significantly higher than that in the campus soil that was seldom affected by the egrets, which suggested that the egrets mediated the transportation of AR from the polluted waterway to the park. Genotyping of the resistant E. coli isolates via repetitive-element PCR gave no strong correlation between the genotypes and the AR patterns of the bacteria. So, the transfer of resistant strains should not be the main mode of AR transportation in this process. The results of real-time PCR revealed that the abundance of antibiotic resistance genes (ARGs) and mobile genetic element (MGE) sequences (transposase and integrase genes) declined along the putative transportation route. The transportation of ARGs could be due to their linkage with MGE sequences, and horizontal gene transfer should have contributed to the process. The movable colistin-resistance gene mcr-1 was detected among the colistin-resistant E. coli strains isolated from the river water and the egret feces, which indicated the possibility of the environmental dissemination of this gene. Birds, especially the migratory birds, for the role they played on the dissemination of environmental AR, should be considered when studying the ecology of AR.
Introduction
The problem of environmental antibiotic resistance (AR) should be recognized from three levels. First, the discharge of antibiotics at sub-inhibition dosage raised from anthropogenic activities exerts long-standing selective pressure on environmental microbial community. Second, resistant bacteria may keep proliferating, disseminating, and persisting in environment. Third, antibiotic resistance genes (ARGs) transport among bacteria of different species via horizontal gene transfer (HGT) and persist in environmental microbial community. For the first level, we have already realized that it is necessary to restrict the environmental emission of antibiotics. Various methods have been developed for the detection of antibiotics in environment (Hirsch et al., 1999; Seifrtova et al., 2008; Yang et al., 2010), and the methods for efficiently eliminating antibiotics from polluted environments are also under development (Ji et al., 2009; Homem and Santos, 2011; Peterson et al., 2012; Fukahori et al., 2013; Bao et al., 2014; Shi et al., 2014; Zuo et al., 2016). But for the second and third levels, i.e., the dissemination of AR in environment and its intrinsic mechanism are not yet fully understood, and effective methods for controlling the dissemination need to be developed. The dominant mechanisms for AR transportation under certain environmental conditions are still unclear. Is it the transfer of resistant bacteria, the horizontal transfer of ARGs or even both of them that governs the transportation process? The question has not yet been answered. The scope and extent of the environmental AR pollution is also far from being fully understood.
Although the exact mechanism is not yet fully understood, most of the existing research results have revealed that the dissemination of environmental AR is closely related to anthropogenic factors. Bacteria seldom develop AR at regions free from antibiotic pressure and human activities (Thaller et al., 2010). The environmental discrepancies originated from anthropogenic factors structured the community differences in resistant bacteria of different regions (Skurnik et al., 2006). Nevertheless, Cristóbal-Azkarate et al. (2014) isolated Escherichia coli strains with resistance to clinic synthetic or semi-synthetic antibiotics from the feces of wildlife at northeast of Mexico. Bhullar et al. (2012) found in an isolated cave at New Mexico bacteria with high AR levels. Some strains were even resistant to 14 different commercially available antibiotics. All the information has indicated that the AR and its dissemination have already exceeded previous anticipations. More and more facts support the growing opinion that AR is a natural and ancient character of microbial community, and environmental microorganisms are reservoirs of ARGs (Allen et al., 2009; Forsberg et al., 2012). Meanwhile, scientists also have speculated that migratory animal, especially the migratory birds may carry resistant bacteria or genes and transport them to regions far from anthropogenic influences (Allen et al., 2010). The Enterobacteriaceae strains isolated from migratory birds feces on the island of Ustica, Sicily (Italy) showed high proportions of resistance against ampicillin, amoxicillin–clavulanic acid, and streptomycin (Foti et al., 2011). The migratory birds of prey at Germany and Mongolia carried similar proportion of extended-spectrum beta-lactamase (ESBL)-producing E. coli (Germany: 13.8%, Mongolia: 10.8%; Guenther et al., 2012). The dissemination of environmental AR mediated by the activities of birds, especially the migratory birds, has been gradually recognized (Dolejska et al., 2007, 2009; Literak et al., 2007; Poeta et al., 2008; Radimersky et al., 2010; Radhouani et al., 2012). Birds can be potential spreaders of environmental AR. Their activities can mediate remote transportation of environmental AR, interconnect environmental locations of different AR levels and patterns, and even bring the AR to unfrequented places. Billions of birds travel between their winter homes and summer breeding grounds each year, and the area of their migration covers all continents including Antarctica (Yogui and Sericano, 2009; Muller et al., 2016). As a special dissemination mode of AR in the environment, the bird-mediated transportation of AR and its intrinsic mechanism has not yet been systematically studied.
China is not only the biggest producer but also the largest consumer of antibiotics in the world (Zhang et al., 2015). Both antibiotics and bacterial AR have been routinely detected in different geographic regions of China (Xu et al., 2007; Luo et al., 2010; Tao et al., 2010; Yang et al., 2010; Su et al., 2011, 2012; Wei et al., 2011; Zou et al., 2011; Zhu et al., 2013; Xiong et al., 2015). The watershed (Jin River) at Chengdu suffered persistent pollution of AR from both suburban and urban sources according to our recent survey (data not shown). Dozens of egrets inhabit the river, drink and forage in the river; while at night, they reside in a park (Wangjianglou Park) nearby the river. It is reasonable to assume that the AR can be transported from the river to the park through the activities of these egrets. To prove this and try to find the intrinsic mechanism of the AR transportation therein, systematic study was carried out on this specific dissemination mode of environmental AR mediated by the activities of these egrets. The E. coli strains were isolated from the environmental samples nearby the habitation of egrets and the AR level was assessed by determining their antibiotic minimum inhibitory concentrations (MICs). By comparing the level of bacterial AR from different environmental locations, the direction and route of resistance transportation were determined. The genotypes and AR patterns of the E. coli isolates were analyzed in combination to disclose the intrinsic mechanism of the resistance transportation (the transfer of resistant bacteria or the horizontal transfer of resistance genes). The abundances of ARGs and mobile genetic element (MGE) sequences (transposase and integrase genes) in various environmental locations were determined via real-time PCR. The role of HGT on the environmental dissemination of AR was thus verified. The environmental dissemination of the newly discovered movable colistin resistance gene, mcr-1, was investigated via PCR survey of this gene among colistin-resistant E. coli isolates.
Materials and Methods
Sampling
Water samples (500 mL each) were collected from five sampling sites along the Jin River (Figure 1) on January 21, March 13, March 26, and April 1 of 2015, respectively. The GPS coordinates of the five sampling sites are as follows: 30° 38′ 23.8″ N and 104° 5′ 20.9″ E for site 1, 30° 38′ 13.1″ N and 104° 5′ 25.4″ E for site 2, 30° 38′ 3.4″ N and 104° 5′ 29″ E for site 3, 30° 37′ 52.6″ N and 104° 5′ 31″ E for site 4, and 30° 37′ 44″ N and 104° 5′ 21.8″ E for site 5. On the same sampling day, five soil samples were also collected from the nightly inhabit of the egrets (Wangjianglou Park) at the GPS coordinate around 30° 37′ 50.3″ N and 104° 5′ 24.4″ E. Additionally, five samples of fresh feces-droppings of egrets were collected in the same park on April 27, 2015. Five soil samples for control were collected on the campus of Sichuan University on September 7, 2015. The campus is separated by a road (the Wangjiang Road) from the park and is seldom affected by egrets. The GPS coordinates of five sampling sites on the campus are as follows: 30° 37′ 54″ N and 104° 4′ 34″ E for sample 1, 30° 37′ 54″ N and 104° 4′ 43″ E for sample 2, 30° 38′ 0″ N and 104° 4′ 48″ E for sample 3, 30° 37′ 57″ N and 104° 4′ 49″ E for sample 4, and 30° 38′ 1″ N and 104° 5′ 1″ E for sample 5. All samples were treated within 5 h after collection.
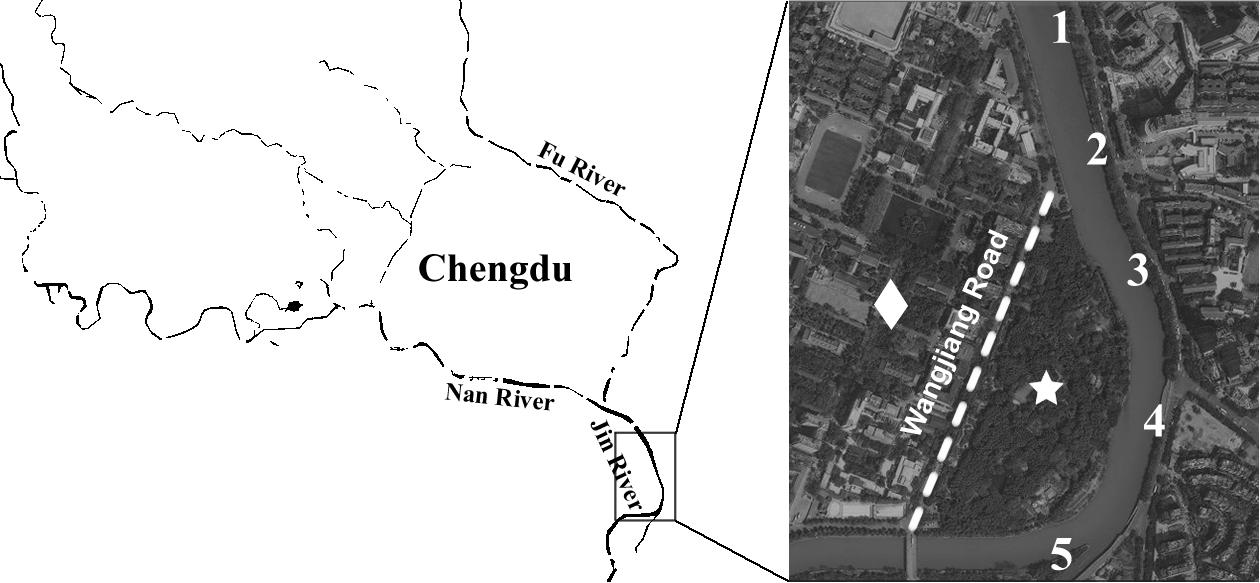
FIGURE 1. Illustration of sampling efforts. The numbers along the river marked the sites of water sampling. The star indicates the position of Wangjianglou Park, while the diamond indicates the Wangjiang campus of Sichuan University. The dashed line labels the Wangjiang Road that separates the park and the campus.
Escherichia coli Isolation
Solid samples (soil or feces) were first extracted using sterile distilled water to release E. coli cells in a procedure based on a previously described technique (Boehm et al., 2009; Cui et al., 2013). One milliliter of serial 10-fold dilutions of the solid sample extracts (10-1 and 10-2 for soil samples, and from 10-2 to 10-4 for feces samples) or the water samples (1, 10-1, and 10-2) were filtered through sterile 0.45 μm S-Pak® membranes (Millipore, Billerica, United States). For homogenous distribution of the bacteria, 20 mL of sterilized distilled water was applied to the membrane before exerting vacuum. The cell-bearing membranes were transferred onto membrane-Thermotolerant E. coli (mTEC) agar to selectively grow E. coli (USEPA, 2002).
Presumptive E. coli colonies were randomly picked from mTEC agar and streaked on Luria-Bertani (LB) agar for purification. After single colonies were picked from streak plates and inoculated in LB broth, the authenticity of the E. coli isolates was then verified by indole–methyl red–Voges–Proskauer–citrate (IMViC) tests, which resulted in verification ratios higher than 90%. The verified E. coli isolates were stored as glycerol stocks at -80°C for subsequent analysis.
Assessment of Antibiotic Resistance of Environmental E. coli Isolates
The MICs of 11 antibiotics against the E. coli isolates were determined in order to assess their AR. MIC data were collected via a modified broth micro-dilution method (Andrews, 2001). The E. coli isolates were first grown in 96-well plates with LB broth at 37°C overnight to reach stationary phase. The cell cultures were then used to inoculate test plates that contained LB broth with a range of antibiotic concentrations (0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, and 128 μg/mL). For the two antibiotics with higher antimicrobial potencies, ceftriaxone and ciprofloxacin, the range of concentrations in the broth was set at 0.0125, 0.025, 0.05, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, and 6.4 μg/mL. The inoculation was executed with a flame-sterilized 48-pin replicator to ensure the uniform inoculum density. The MIC endpoints were determined as the lowest concentration at which there was no visible growth after 20 h of incubation at 37°C. Duplicate tests for each antibiotic concentration were conducted, and the average number was calculated as the MIC value. Positive and negative controls were conducted in antibiotic-free LB to ensure growth of environmental E. coli isolates under lab conditions and sterility of the assay, respectively. Quality control of the procedure was conducted by using the susceptive E. coli standard strain ATCC 25922, which exhibited similar antibiotic MIC values as reported in the literature (Andrews, 2001).
Minimum inhibitory concentration breakpoint for each antibiotic was set at 16 times of the MIC of the standard strain. The isolates with MICs greater than the breakpoint were considered resistant. The AR level against an antibiotic was represented with corresponding percentage of resistant strains. The multiple antibiotic resistance index (MARI) was calculated to evaluate the multidrug resistance (MDR) for each type of environmental sample (Krumperman, 1983).
Where c is the number of individual strains isolated from specific type of environmental sample, ai is the number of antibiotics that strain i is resistant to, b is the number of antibiotics tested (11 in our case).
In order to compare the AR levels between different environmental locations, the antibiotic MIC50 and MIC90 values of the E. coli isolates from different environmental locations were normalized against the MIC values of the standard strain ATCC 25922, and thereafter were log2-transformed. The percentages of resistant strains against each antibiotic at different environmental locations were also log-transformed. The transformed MIC50/MIC90 values and resistance percentages against 11 antibiotics at two different environmental locations can be looked as pairs of observations. The paired-sample t-test was used to determine whether the difference in AR level at the two environmental locations was significant. A p-value of less than 0.05 was considered statistically significant.
In order to analyze the AR pattern of the E. coli isolated from different environmental samples, the MIC data were firstly normalized against the corresponding MICs of the standard strain. Then, the normalized MICs were multiplied with 100 and thereafter log2-transformed. The transformed MICs data were treated as a series of multidimensional vectors that represent the drug resistance patterns of the E. coli isolates. The transformed MICs were used to calculate the Euclidean distances between different strains. The generated Euclidean distance matrix was thereafter subjected to non-metric multidimensional scaling (NMDS) to compare the AR patterns of the E. coli isolated from different environmental samples. The NMDS was performed using 100 random starting configurations of sample points (E. coli isolates) with the built-in midscale function of Matlab_2016Ra (MathWorks, Natick, MA, United States); the accuracy of the NMDS representation was determined by calculating the Kruskal stress (Cui et al., 2013). A contour map of MDR was generated via interpolating the MDR data of these E. coli isolates using the scatteredInterpolant function of Matlab_2016Ra (MathWorks, Natick, MA, United States) and lined behind the NMDS plot.
Genetic Diversity of Environmental E. coli Isolates
Genomic DNA fingerprinting of E. coli isolates was performed using repetitive-element PCR (rep-PCR; Dombek et al., 2000). Briefly, fresh E. coli cells were collected via centrifugation from overnight 96-well plate pure culture in LB broth and then gently treated (60°C for 20 min) with 0.05 N NaOH to release total genomic DNA. The total genomic DNA in solution was separated from cell debris by centrifugation at 250 rpm for 10 min. The supernatants containing total genomic DNA were used as templates in PCR amplification for rep-PCR DNA fingerprinting. The BOX-A1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′) was used in this study. Following amplification, the PCR amplicons were electrophoresed, and the gel images were obtained using a Junyi gel imaging system JY04S-3E (Beijing Junyi Dongfang Electrophoresis Co., Ltd., Beijing). The DNA banding pattern (i.e., fingerprint) for each isolate was normalized for inter-gel comparisons using an external DNA size marker (100 bp DNA Marker; Beijing Dingguo Changsheng Biotechnology Co., Ltd., Beijing) that was loaded into both end and middle lanes of each gel. All fingerprint images were loaded into a database and processed using BioNumerics (Applied Maths, Kortrijk, Belgium). The dendrogram was created based on Pearson’s correlation using unweighted-pair group method with arithmetic means (UPGMA).
Quantify the Abundance of ARGs and MGEs by Real-Time PCR (qPCR)
In order to quantify the abundance of ARGs and MGEs, we intensively collected the environmental samples once again in September 2015. The river water, egret feces, park soil, and campus soil were sampled in triplicate, respectively. Solid samples (soil and egret feces, 1 g each) and filtration retentates of water samples (from about 300 mL water sample each filtered through GN-6 0.45 μm membranes) were extracted the total genomic DNA with the FastDNA Spin Kit for Soil (MP Biomedicals, LLC., United States). DNA extracts of the same sample type were pooled together to make four composite DNA samples representing Jin River water, egret feces, park soil, and campus soil, respectively. The DNA samples were used as templates in qPCR to evaluate the abundances of ARGs and MGEs therein. The 16S rRNA gene was used as reference standard. All qPCRs were operated in triplicate. Quality control was obtained via checking the melting curves and electrophoresis image of the qPCR products after amplification. Confirmed specific amplifications were thereafter executed the data analysis. The abundances of ARGs and MGEs were expressed as the amount ratios of target genes to the reference gene. The acquisition of primer pairs, qPCR operation, and related data treatments were illustrated in Appendix 1 in Supplementary Material. The information of the target genes and related primer pairs were summarized in Supplementary Table S2. Averages, standard deviations, and relative abundance of ARGs and MGEs were determined using Excel 2013 (Microsoft Office 2013, Microsoft, United States). The clustered heatmap was performed in Matlab_2016Ra (MathWorks, Natick, MA, United States).
Survey of mcr-1 Gene Among Colistin-Resistant E. coli Isolates
Some colistin-resistant E. coli isolates (n = 6) were recovered from environmental samples in our case. Among these isolates, four were isolated from the river water, one from the egret feces and the other one from the park soil. Consequently, a PCR survey targeting mcr-1 gene among these colistin-resistant E. coli isolates was executed with the primer pair CLR5-F (5′-CGGTCAGTCCGTTTGTTC-3′) and CLR5-R (5′-CTTGGTCGGTCTGTAGGG-3′) (Liu et al., 2016). The PCR amplification products were sent to Sangon Biotech (Shanghai, China) for sequencing.
Results
Bacterial Antibiotic Resistance in Different Environmental Samples
Ninety-five E. coli isolates were recovered from river water, 94 from egret feces, 81 from park soil, and 91 from campus soil. The information of the E. coli collection was summarized in Supplementary Table S1. MICs data of 11 antibiotics against these E. coli isolates indicated the bacterial AR levels in different environmental samples. MIC50 and MIC90 of each group of E. coli strains isolated from different environmental samples (Table 1) showed that the AR level in Jin River water was the highest (paired-sample t-test, p < 0.05). The lowest level of AR occurred in the campus soil (p < 0.01). The AR in egret feces and park soil was at a similar level. The percentages of resistant E. coli among different environmental samples followed the same trend (Figure 2A). The E. coli strains isolated from Jin River water exhibited the highest resistance rates to 11 antibiotics (p < 0.01). The campus soil isolates gave the lowest resistance rates (p < 0.01). The resistance rates of the egret feces isolates to these antibiotics were similar to those of the park soil isolates. The resistance rates of the bird feces and park soil isolates to 11 antibiotics showed good exponential correlations with those of the Jin River water isolates, as demonstrated in Figure 2B (R2 > 0.77, p < 0.01). The detailed MIC distributions of the E. coli isolates from four environmental sample types are summarized in Supplementary Tables S3–S6, respectively. The antibiotic MICs of the susceptible standard E. coli strain, ATCC 25922, were also listed in the last column of Supplementary Table S3.
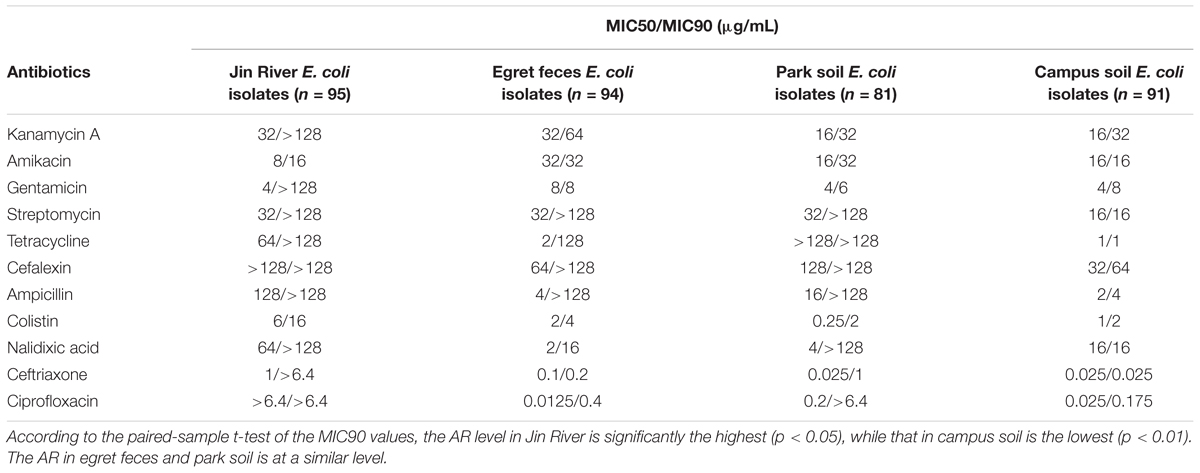
TABLE 1. MIC50 and MIC90 of different antibiotics against the E. coli strains isolated from different environmental samples.
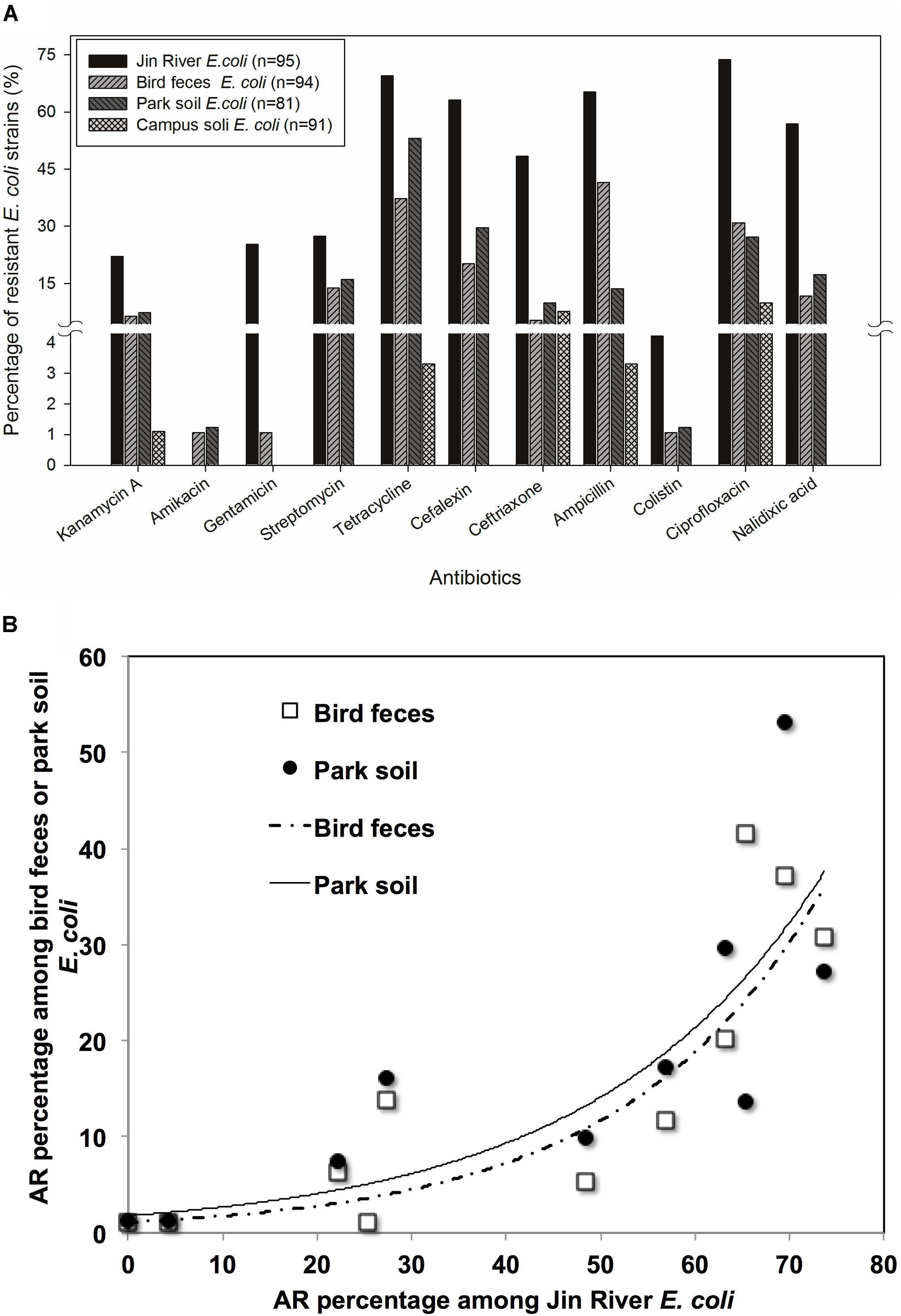
FIGURE 2. The role of bird in the AR transportation was indicated with the AR rates (percentage) of environmental E. coli isolates (A) and the correlation of AR rates among the E. coli strains isolated from the environmental samples in related to egret inhabitation (B). According to the paired-sample t-test of the AR rates (log-transformed) between the E. coli strains isolated from different environmental samples, the E. coli strains isolated from Jin River water exhibited the highest resistance rates to 11 antibiotics (p < 0.01). The campus soil isolates gave the lowest resistance rates (p < 0.01). The AR rates of the egret feces isolates were similar to those of the park soil isolates (A). The resistance rates of the bird feces and park soil isolates to 11 antibiotics showed good exponential correlations (R2 > 0.77, p < 0.01) with those of the Jin River water isolates (B).
The MDR of these E. coli isolates are recorded and exhibited in Figure 3. The MARIs were calculated for four types of environmental samples and listed in Table 2. The situation of MDR was the most serious among the E. coli strains isolated from Jin River water, and followed by the strains isolated from egret feces and park soil. Most of the campus soil isolates (92.3%) exhibited no MDR.
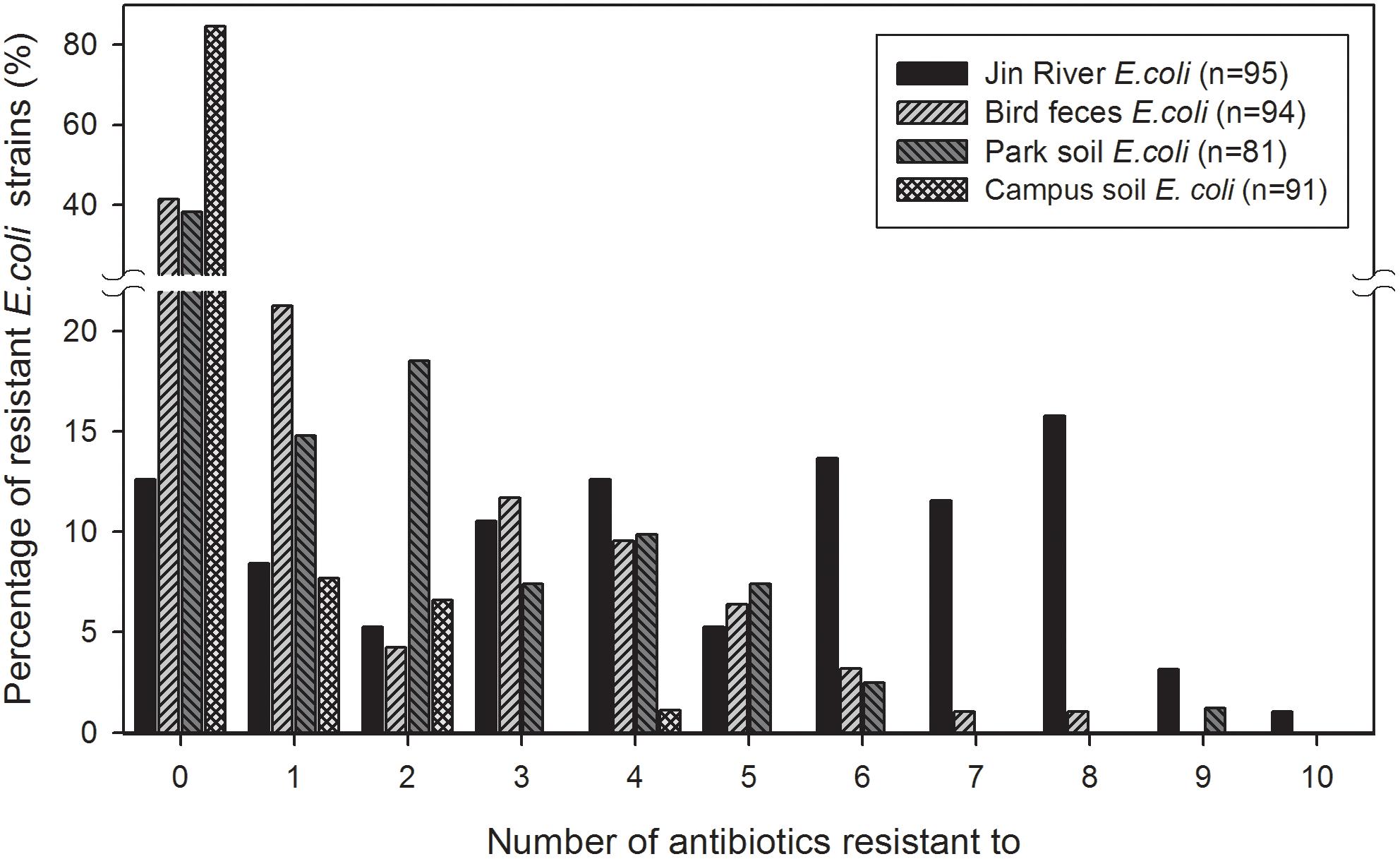
FIGURE 3. Multidrug resistance (MDR) among the E. coli isolates from different environmental samples. The E. coli isolates of Jin River water gave the highest level of MDR. Most of the campus soil isolates (92.3%) showed no MDR. The E. coli isolated from egret feces and park soil exhibited similar level of MDR.
Non-metric multidimensional scaling plot based on the MIC data of the E. coli isolates is exhibited in Figure 4. The E. coli strains isolated from different environmental samples exhibited various patterns of AR and MDR. The E. coli group recovered from Jin River water (red squares) contained strains of various AR patterns, which indicated their “multiple sources.” On the other hand, both of the E. coli groups isolated from egret feces (black diamonds) and park soil (pink dots) contained either strains of low-level AR or strains of various AR patterns with high degree of MDR. The E. coli group isolated from campus soil (white squares), however, exhibited mostly the low-level AR. Only few isolates gave weak MDR (resistant to two to four antibiotics).
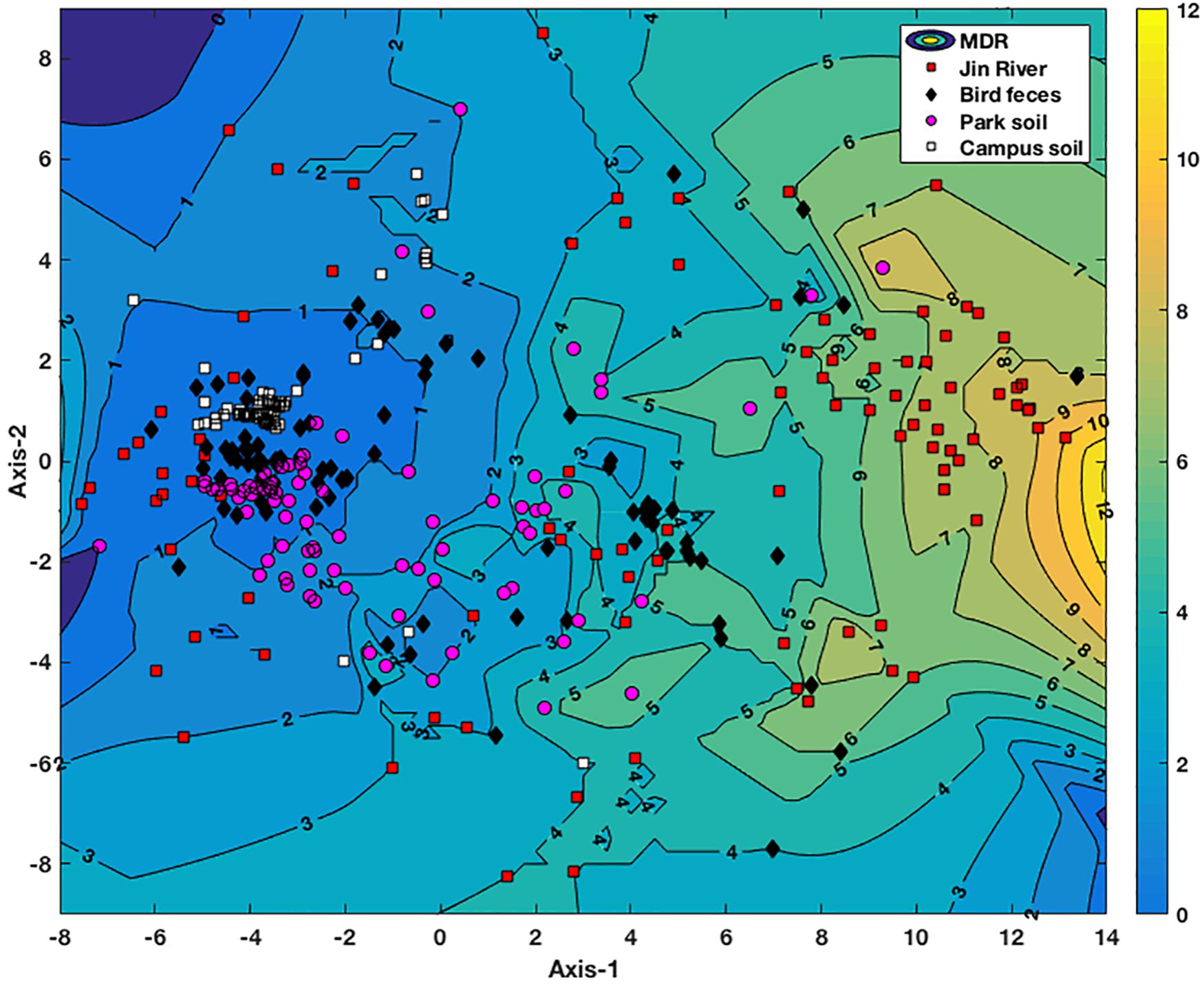
FIGURE 4. NMDS analysis of antibiotic resistance pattern of the E. coli isolates based on their antibiotic MIC data. The Kruskal stress is 0.1273. A contour map of multidrug resistance (MDR) of the E. coli isolates is generated via interpolating the MDR data of these strains and lined behind the NMDS plot. The numbers on the contour lines are the numbers of antibiotics that one strain is resistant to. The degree of the MDR is indicated by a color bar with warm color (yellow) representing high level of MDR and cool color (blue) for low MDR degree.
Genetic Diversity of the E. coli Isolates
Genetic diversity of environmental E. coli isolates was reflected via rep-PCR fingerprinting. According to the dendrogram of the DNA banding patterns (Supplementary Figure S1), most of the bacterial clusters of similar genotypes occurred among the E. coli strains isolated from the same environmental sample, while only minority included the isolates from different samples. At the threshold of 90% similarity in genomic structures (rep-PCR banding patterns), several typical clusters of E. coli isolates recovered from the same (F, G, and H) or different (A, B, C, D, and E) type of environmental samples were recorded the AR patterns of their members. The results are summarized in Supplementary Table S7.
Abundance of ARGs and MGEs in Environment
The result of AR assessment via qPCR coincided fairly well with that obtain via culture-based method (MIC data). A typical fact is that at the location where the phenotypes of resistance against certain antibiotics (tetracycline or cephalosporin) were detected (Figure 2A), the corresponding ARGs (tetW or blaTEM and blaCTX-M-14, respectively) were also mostly detected therein (Figure 5A). The transportation of different ARGs varied from one to another. Some ARGs were detected in all environmental samples in relation to the activities of egrets (Jin River water, bird feces, and the park soil) such as aac6ib and aadA. Some ARGs were only detected in river water and bird feces such as certain tetracycline resistance genes (tetL and tetO). The others were discovered only in Jin River water such as aac3ia, aac6iia, and blaCMY2. These ARGs were hardly transported among different environmental locations. The abundance of ARGs in the environment declined along the transportation route (i.e., from polluted waterway to egrets and then to the park soil affected by egrets, refer to Figure 5A). Most of the ARGs that are detected in the water samples from Jin River are absent (undetectable) in the campus soil. The MGE sequences (four transposase genes and one integrase gene) were also quantified. The abundance of these genes also declined along the AR transportation route. These genes were not detected in the campus soil (Figure 5A).
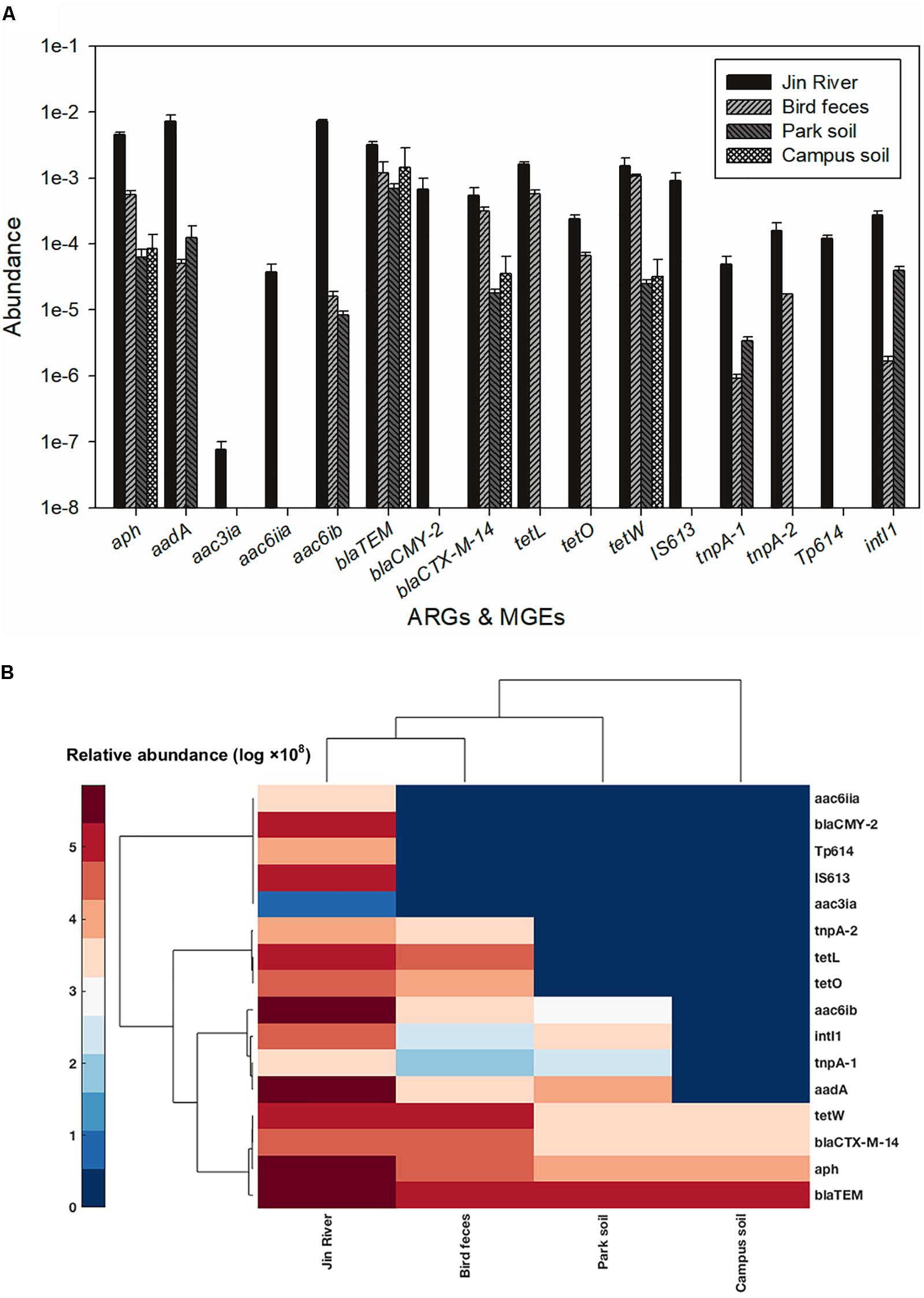
FIGURE 5. Abundance of ARGs and MGEs among different environmental samples (A) and the heatmap of the abundance (log-transformed) of ARGs and MGEs (B). The legend for the heatmap denotes corresponding log-transformed values of the abundances of these genes. Both columns (samples) and rows (genes) were clustered based on cosine similarity. None of the five MGEs was detected in the campus soil.
Detection of mcr-1 Gene in Environmental E. coli Isolates
According to the results of PCR survey of mcr-1 gene, amplicons with the expected size were obtained from three out of the six colistin-resistant strains. The PCR products were submitted to sequencing and proved to be the mcr-1 gene. Obtained DNA sequences were deposited into GenBank under accession numbers KY218737 to KY218739. Two of the three mcr-1-positive E. coli strains were isolated from Jin River water samples, and the other one was isolated from egret feces.
Discussion
In this research, the common fecal indicator bacterium, E. coli, was isolated from environmental samples to assess the bacterial AR of the environment. E. coli as an indicator organism is currently used to monitor microbiological water quality and is well characterized in terms of acquired AR (Berendonk et al., 2015). This method may leave out some AR mechanism tied to special bacteria species. But it is still reasonable to a large extent if the ubiquitous HGT is taken into consideration (Rosen et al., 2015). Slight differences in the components of culture broth, the purities and potencies of antibiotics of different suppliers may affect the obtained MIC data. The introduction of the standard strain and the normalization of the MIC data against the MICs of the standard strain to some extend shielded these influences. Resistant breakpoints were set at 16 times of the MICs of the susceptible standard strain (ATCC 25922). The environmental isolates and the standard strain are of the same bacterial species, while the environmental isolates can survive at significantly higher antibiotic concentration. It is reasonable to believe that there must be considerable AR mechanisms in environmental isolates over the standard strain that endows them the capability to survive at the higher antibiotic concentration.
Jin River runs through wide-open irrigated agricultural area before flowing into the urban area of Chengdu. In China, there is no strict prohibition on antibiotic applications in animal husbandry and aquaculture. A dozen kinds of antibiotics are still on the permission list of feed additives. China shares the largest global antimicrobial consumption in food animal production (23% in 2010; Van Boeckel et al., 2015). The open discharge of wastewater from animal husbandry and aquaculture brings serious AR pollution into the water body. As combined sewer system is still in use at parts of the city, the river water carries both the agricultural and urban influences. It is predictable to observe high level of AR in the river. Although the park and the campus are geographically adjacent and are just separated by a road, the Wangjiang Road (Figure 1), the AR is significantly higher in the park soil (higher resistance percentages with p < 0.01). This probably relates to the influence of the egret on the park soil. According to our knowledge, there is no antibiotic selective pressure artificially introduced to the park soil via either manure fertilization or reclaimed water irrigation. As we did not analyze the antibiotic pollutants in the park soil, the possibility of selective pressure of antibiotics on the soil bacterial community could not be fully excluded. The resistance percentage of E. coli isolates against each antibiotic was the highest in river water, and the drug-resistance situation in the egret feces and the park soil was closely related to that in the river water (Figure 2A). More quantitatively, the AR rate among the E. coli isolates from bird feces and park soil exponentially correlated with the AR rate of the Jin River water isolates (Figure 2B). All these facts above suggested that egrets to some extent mediated the environmental transportation of AR.
For the environmental locations in relation to egret’s inhabitation, the percentages of AR in different locations were closely related to each other (Figure 2B). Some isolates recovered from different environmental samples did show similar AR patterns (the clusters formed by the points of different environmental samples in Figure 4), the number of the cases was, however, limited. More cases were that the bacteria isolated from different environmental samples exhibited distinct AR patterns, and MDR declined along the transportation route. There was no strong correlation between the genotypes and the AR patterns of the resistant E. coli isolates (Supplementary Table S7). Especially, the samples of river water and the park soil were associated in time, but the genotypic similarity of E. coli isolated from two types of environmental samples was fairly low (Supplementary Figure S1). It was to be expected that E. coli strains isolated from different environmental samples gave distinct genotypes. Rep-PCR genotyping had ever been used to track the source of environmental E. coli, and fairly good results were obtained (Dombek et al., 2000). The direct transfer of resistant bacteria among different environmental locations seemed rare herein. We cannot fully rule out the possibility of resistant bacteria transferring among environmental locations for the limited sampling scale and the temporal difference among the environmental E. coli isolates. Multilocus sequence typing and phylogenetic group designating ever revealed that wild bird and human share ESBL-producing E. coli population (Bonnedahl et al., 2009). The results herein, however, suggest that the transfer of resistant bacteria was not the dominant mechanism for the dissemination of AR in environment, at least under the specific transportation mode of AR in this study.
The abundance of ARGs in the environment declined along the transportation route (Figure 5A). This could be the reason why the level of MDR declined along the AR transportation route as shown in Figure 3. As the source of AR pollution, the Jin River water showed the highest level of ARGs, and as the deliverer and receptor, respectively, egret feces and park soil exhibited comparable abundance of ARGs. The campus soil, however, gave the lowest abundance. Some β-lactamase (i.e., blaTEM and blaCTX-M-14) and aminoglycoside O-phosphotransferase (aph) genes should be the indigenous genes of soil bacterial community (Forsberg et al., 2014; Johnson et al., 2016) and were even detected in campus soil. According to a previous study, mobility elements syntenic with ARGs were rare in soil by comparison with sequenced pathogens, suggesting that ARGs may not transfer between soil bacteria as readily as is observed between human pathogens. Therefore, it has been suggested that bacterial community composition primarily determines the soil ARG content and HGT cannot fully decouple the resistome from phylogeny therein (Forsberg et al., 2014). According to our qPCR results, the MGE sequences responsible for HGT such as tnpA-1 (transposase) and intI1 (integrase) were enriched in park soil under the effect of egrets in comparison with the campus soil, which implied that MGEs could mediate the transportation of AR therein. ARGs have been observed to co-localize with MGE sequences in the bacterial communities intensively affected by antibiotics (Partridge et al., 2009; Johnson et al., 2016), which drives the horizontal transfer of these ARGs. In our case, the transported ARGs (aac6ib and aadA) clustered with the MRGs (intI1 and tnpA-1) at the clustering analysis of the abundance data of these genes (Figure 5B). We searched the nucleotide database at the National Center for Biotechnology Information (NCBI) to ascertain if there was any precedent for the gene cluster that we observed. An E. coli plasmid (AF550679.1) and a Klebsiella pneumoniae plasmid (KC958437.1) were found carrying the gene cassettes containing genes aac6ib, intI1, tnpA, and aadA. Although various ARGs showed variant transferability, and the internal reasons are yet to be uncovered. The qPCR results presented here provided evidences that HGT contributed to the special transportation mode of environmental AR. The correlation of different ARGs with gene cassettes in relation to genetic mobility need to be clarified via intensive sequencing. As HGT may dominate the environmental transportation of AR, to block HGT can be an efficient method to control the environmental dissemination of AR. How to inhibit the transfer of resistance genes can be a good point of future study.
Colistin is one of the last-resort antibiotics for infection of multidrug-resistant bacteria. Its resistance was ever considered involving chromosomal mutations and cannot be transported among bacteria via HGT. But recently, plasmid-mediated transferable colistin resistance encoded by the gene mcr-1 was first described in China (Liu et al., 2016) and the discoveries of the same AR gene were reported successively almost all over the rest regions of the world (Skov and Monnet, 2016). China thereafter officially banned colistin as a feed additive for animals on November 1, 2016 (Walsh and Wu, 2016). This is a manifestation of the responsibility of great powers. We had tried to determine the abundance of mcr-1 in environmental samples via qPCR according to a reported method (Bontron et al., 2016). The abundance of this gene in the environment seems too low to be detected. It was not even detected in the Jin River water samples (data not shown), while we actually encountered several colistin-resistant E. coli isolates (n = 6). The movable colistin-resistant gene (mcr-1) was also detected in three colistin-resistant E. coli isolates among which two were isolated from Jin River water and one from egret feces. The genotypes of the three isolates are different from each other according to their rep-PCR banding patterns (E. coli isolates W_C08, W_H05, and B_A12 in Supplementary Figure S1). This should be the first case of the simultaneous detection of mcr-1 positive E. coli in wild birds and their habitat, which gave the possibility of environmental transportation of this gene even through birds’ activities. When tracking the source of mcr-1 gene in previous studies, the stream of people (entry–exit people) and goods (import and export commodities) were closely concerned (Arcilla et al., 2016; Fernandes et al., 2016; Kluytmans-van den Bergh et al., 2016; McGann et al., 2016), while environmental dissemination of this gene was rarely considered and much less the role that migratory birds might play in the process. In central parts of Chile, the ESBL-positive E. coli strains were isolated from fecal flora of wild birds (Franklin’s gulls) and the detection rate is even higher than that from local human (Hernandez et al., 2013). In Taif province, Saudi Arabia, MDR was more serious among Enterobacter strains isolated from migratory birds than local resident birds (Abo-Amer and Shobrak, 2015). The study in Sweden indicates that a potential of AR transfer between the human population and wild birds exists even in countries with a low level of AR (Bonnedahl et al., 2010). According to the study carried out along the northeastern coast of the United States, AR was more widespread in bacteria isolated from seabirds than those isolated from marine mammals (Rose et al., 2009). Birds, especially the migratory birds, due to their numerous amount and wide range of activities, need to be paid much more attention to on their roles in mediating the transportation of environmental AR. The effective control of AR should be based on universal worldwide strategies. The restraint of antibiotic application in local area cannot create “Peach Blossom Shangri-La” that is free from AR pollution.
Author Contributions
KY and YZ designed the research. DR isolated E. coli from environmental samples. YH collected the MIC data and operated the rep-PCR. JW and DR operated qPCR. JW did the PCR survey of mcr-1 gene and sent the PCR products for sequencing. YH, JW, and KY treated the data. KY and JW drafted the manuscript, which was edited by YZ.
Funding
This study was funded by the Initiating Research Fund for Talent Introduction of Sichuan University (No. YJ201355) and the National Natural Science Foundation of China (No. 21677104).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate Dr. Qian Zhang at Biotechnology Institute, University of Minnesota, for helping with the processing of rep-PCR fingerprinting data. We also thank Professor Tao Yan at the Department of Civil and Environmental Engineering, University of Hawai’i at Mānoa, for reviewing the manuscript and providing constructive advice. Finally, we acknowledge Dr. Zheng Yang at Chevron for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00745/full#supplementary-material
References
Abo-Amer, A. E., and Shobrak, M. Y. (2015). Antibiotic resistance and molecular characterization of Enterobacter cancerogenus isolated from wild birds in Taif province, Saudi Arabia. Thai J. Vet. Med. 45, 101–111.
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Allen, H. K., Moe, L. A., Rodbumrer, J., Gaarder, A., and Handelsman, J. (2009). Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 3, 243–251. doi: 10.1038/ismej.2008.86
Andrews, J. M. (2001). Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48(Suppl. 1), 5–16. doi: 10.1093/jac/48.suppl_1.5
Arcilla, M. S., van Hattem, J. M., Matamoros, S., Melles, D. C., Penders, J., de Jong, M. D., et al. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 147–149. doi: 10.1016/S1473-3099(15)00541-1
Bao, X., Qiang, Z., Chang, J. H., Ben, W., and Qu, J. (2014). Synthesis of carbon-coated magnetic nanocomposite (Fe3O4@C) and its application for sulfonamide antibiotics removal from water. J. Environ. Sci. 26, 962–969. doi: 10.1016/S1001-0742(13)60485-4
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317. doi: 10.1038/nrmicro3439
Bhullar, K., Waglechner, N., Pawlowski, A., Koteva, K., Banks, E. D., Johnston, M. D., et al. (2012). Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One 7:e34953. doi: 10.1371/journal.pone.0034953
Boehm, A. B., Griffith, J., McGee, C., Edge, T. A., Solo-Gabriele, H. M., Whitman, R., et al. (2009). Faecal indicator bacteria enumeration in beach sand: a comparison study of extraction methods in medium to coarse sands. J. Appl. Microbiol. 107, 1740–1750. doi: 10.1111/j.1365-2672.2009.04440.x
Bonnedahl, J., Drobni, M., Gauthier-Clerc, M., Hernandez, J., Granholm, S., Kayser, Y., et al. (2009). Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One 4:e5958. doi: 10.1371/journal.pone.0005958
Bonnedahl, J., Drobni, P., Johansson, A., Hernandez, J., Melhus, A., Stedt, J., et al. (2010). Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum beta-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J. Antimicrob. Chemother. 65, 1939–1944. doi: 10.1093/jac/dkq222
Bontron, S., Poirel, L., and Nordmann, P. (2016). Real-time PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. J. Antimicrob. Chemother. 71, 2318–2320. doi: 10.1093/jac/dkw139
Cristóbal-Azkarate, J., Dunn, J. C., Day, J. M., and Amabile-Cuevas, C. F. (2014). Resistance to antibiotics of clinical relevance in the fecal microbiota of Mexican wildlife. PLoS One 9:e107719. doi: 10.1371/journal.pone.0107719
Cui, H., Yang, K., Pagaling, E., and Yan, T. (2013). Spatial and temporal variation in enterococcal abundance and its relationship to the microbial community in Hawaii beach sand and water. Appl. Environ. Microbiol. 79, 3601–3609. doi: 10.1128/AEM.00135-13
Dolejska, M., Bierosova, B., Kohoutova, L., Literak, I., and Cizek, A. (2009). Antbiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106, 1941–1950. doi: 10.1111/j.1365-2672.2009.04155.x
Dolejska, M., Cizek, A., and Literak, I. (2007). High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from Black-headed Gulls in the Czech Republic. J. Appl. Microbiol. 103, 11–19. doi: 10.1111/j.1365-2672.2006.03241.x
Dombek, P. E., Johnson, L. K., Zimmerley, S. T., and Sadowsky, M. J. (2000). Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66, 2572–2577. doi: 10.1128/AEM.66.6.2572-2577.2000
Fernandes, M. R., Moura, Q., Sartori, L., Silva, K. C., Cunha, M. P., Esposito, F., et al. (2016). Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro Surveill. 21, 1–6. doi: 10.2807/1560-7917.ES.2016.21.17.30214
Forsberg, K. J., Patel, S., Gibson, M. K., Lauber, C. L., Knight, R., Fierer, N., et al. (2014). Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616. doi: 10.1038/nature13377
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Foti, M., Rinaldo, D., Guercio, A., Giacopello, C., Aleo, A., De Leo, F., et al. (2011). Pathogenic microorganisms carried by migratory birds passing through the territory of the island of Ustica, Sicily (Italy). Avian Pathol. 40, 405–409. doi: 10.1080/03079457.2011.588940
Fukahori, S., Fujiwara, T., Funamizu, N., Matsukawa, K., and Ito, R. (2013). Adsorptive removal of sulfonamide antibiotics in livestock urine using the high-silica zeolite HSZ-385. Water Sci. Technol. 67, 319–325. doi: 10.2166/wst.2012.513
Guenther, S., Aschenbrenner, K., Stamm, I., Bethe, A., Semmler, T., Stubbe, A., et al. (2012). Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS One 7:e53039. doi: 10.1371/journal.pone.0053039
Hernandez, J., Johansson, A., Stedt, J., Bengtsson, S., Porczak, A., Granholm, S., et al. (2013). Characterization and comparison of extended-spectrum beta-lactamase (ESBL) resistance genotypes and population structure of Escherichia coli isolated from Franklin’s gulls (Leucophaeus pipixcan) and humans in Chile. PLoS One 8:e76150. doi: 10.1371/journal.pone.0076150
Hirsch, R., Ternes, T., Haberer, K., and Kratz, K. L. (1999). Occurrence of antibiotics in the aquatic environment. Sci. Total Environ. 225, 109–118. doi: 10.1016/S0048-9697(98)00337-4
Homem, V., and Santos, L. (2011). Degradation and removal methods of antibiotics from aqueous matrices–a review. J. Environ. Manage. 92, 2304–2347. doi: 10.1016/j.jenvman.2011.05.023
Ji, L., Chen, W., Zheng, S., Xu, Z., and Zhu, D. (2009). Adsorption of sulfonamide antibiotics to multiwalled carbon nanotubes. Langmuir 25, 11608–11613. doi: 10.1021/la9015838
Johnson, T. A., Stedtfeld, R. D., Wang, Q., Cole, J. R., Hashsham, S. A., Looft, T., et al. (2016). Clusters of antibiotic resistance genes enriched together stay together in swine agriculture. mBio 7, e02214–e02215. doi: 10.1128/mBio.02214-15
Kluytmans-van den Bergh, M. F., Huizinga, P., Bonten, M. J., Bos, M., De Bruyne, K., Friedrich, A. W., et al. (2016). Presence of mcr-1-positive Enterobacteriaceae in retail chicken meat but not in humans in the Netherlands since 2009. Euro Surveill. 21:30149. doi: 10.2807/1560-7917.ES.2016.21.9.30149
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46, 165–170.
Literak, I., Vanko, R., Dolejska, M., Cizek, A., and Karpiskova, R. (2007). Antibiotic resistant Escherichia coli and Salmonella in Russian rooks (Corvus frugilegus) wintering in the Czech Republic. Lett. Appl. Microbiol. 45, 616–621. doi: 10.1111/j.1472-765X.2007.02236.x
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Luo, Y., Mao, D., Rysz, M., Zhou, Q., Zhang, H., Xu, L., et al. (2010). Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ. Sci. Technol. 44, 7220–7225. doi: 10.1021/es100233w
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: First report of mcr-1 in the USA. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/AAC.01103-16
Muller, F., Taylor, P. D., Sjoberg, S., Muheim, R., Tsvey, A., Mackenzie, S. A., et al. (2016). Towards a conceptual framework for explaining variation in nocturnal departure time of songbird migrants. Mov. Ecol. 4:24. doi: 10.1186/s40462-016-0089-2
Partridge, S. R., Tsafnat, G., Coiera, E., and Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784. doi: 10.1111/j.1574-6976.2009.00175.x
Peterson, J. W., Petrasky, L. J., Seymour, M. D., Burkhart, R. S., and Schuiling, A. B. (2012). Adsorption and breakdown of penicillin antibiotic in the presence of titanium oxide nanoparticles in water. Chemosphere 87, 911–917. doi: 10.1016/j.chemosphere.2012.01.044
Poeta, P., Radhouani, H., Igrejas, G., Goncalves, A., Carvalho, C., Rodrigues, J., et al. (2008). Seagulls of the berlengas natural reserve of portugal as carriers of fecal Escherichia coli harboring CTX-M and TEM extended-spectrum beta-lactamases. Appl. Environ. Microbiol. 74, 7439–7441. doi: 10.1128/AEM.00949-08
Radhouani, H., Poeta, P., Goncalves, A., Pacheco, R., Sargo, R., and Igrejas, G. (2012). Wild birds as biological indicators of environmental pollution: antimicrobial resistance patterns of Escherichia coli and enterococci isolated from common buzzards (Buteo buteo). J. Med. Microbiol. 61, 837–843. doi: 10.1099/jmm.0.038364-0
Radimersky, T., Frolkova, P., Janoszowska, D., Dolejska, M., Svec, P., Roubalova, E., et al. (2010). Antibiotic resistance in faecal bacteria (Escherichia coli. Enterococcus spp.) in feral pigeons. J. Appl. Microbiol. 109, 1687–1695. doi: 10.1111/j.1365-2672.2010.04797.x
Rose, J. M., Gast, R. J., Bogomolni, A., Ellis, J. C., Lentell, B. J., Touhey, K., et al. (2009). Occurrence and patterns of antibiotic resistance in vertebrates off the Northeastern United States coast. FEMS Microbiol. Ecol. 67, 421–431. doi: 10.1111/j.1574-6941.2009.00648.x
Rosen, M. J., Davison, M., Bhaya, D., and Fisher, D. S. (2015). Microbial diversity. Fine-scale diversity and extensive recombination in a quasisexual bacterial population occupying a broad niche. Science 348, 1019–1023. doi: 10.1126/science.aaa4456
Seifrtova, M., Pena, A., Lino, C. M., and Solich, P. (2008). Determination of fluoroquinolone antibiotics in hospital and municipal wastewaters in Coimbra by liquid chromatography with a monolithic column and fluorescence detection. Anal. Bioanal. Chem. 391, 799–805. doi: 10.1007/s00216-008-2020-1
Shi, L., Ma, F., Han, Y., Zhang, X., and Yu, H. (2014). Removal of sulfonamide antibiotics by oriented immobilized laccase on Fe3O4 nanoparticles with natural mediators. J. Hazard. Mater. 279, 203–211. doi: 10.1016/j.jhazmat.2014.06.070
Skov, R. L., and Monnet, D. L. (2016). Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 21:30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155
Skurnik, D., Ruimy, R., Andremont, A., Amorin, C., Rouquet, P., Picard, B., et al. (2006). Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57, 1215–1219. doi: 10.1093/jac/dkl122
Su, H. C., Ying, G. G., Tao, R., Zhang, R. Q., Fogarty, L. R., and Kolpin, D. W. (2011). Occurrence of antibiotic resistance and characterization of resistance genes and integrons in Enterobacteriaceae isolated from integrated fish farms in South China. J. Environ. Monit. 13, 3229–3236. doi: 10.1039/c1em10634a
Su, H. C., Ying, G. G., Tao, R., Zhang, R. Q., Zhao, J. L., and Liu, Y. S. (2012). Class 1 and 2 integrons, sul resistance genes and antibiotic resistance in Escherichia coli isolated from Dongjiang River, South China. Environ. Pollut. 169, 42–49. doi: 10.1016/j.envpol.2012.05.007
Tao, R., Ying, G. G., Su, H. C., Zhou, H. W., and Sidhu, J. P. (2010). Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl rivers in South China. Environ. Pollut. 158, 2101–2109. doi: 10.1016/j.envpol.2010.03.004
Thaller, M. C., Migliore, L., Marquez, C., Tapia, W., Cedeno, V., Rossolini, G. M., et al. (2010). Tracking acquired antibiotic resistance in commensal bacteria of Galapagos land iguanas: no man, no resistance. PLoS One 5:e8989. doi: 10.1371/journal.pone.0008989
USEPA (2002). Method 1103.1: Escherichia coli (E. coli) in Water by Membrane Filtration Using Membrane-Thermotolerant Escherichia coli Agar (mTEC). Washington, DC: United States Environmental Protection Agency.
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U.S.A. 112, 5649–5654. doi: 10.1073/pnas.1503141112
Walsh, T. R., and Wu, Y. (2016). China bans colistin as a feed additive for animals. Lancet Infect. Dis. 16, 1102–1103. doi: 10.1016/S1473-3099(16)30329-2
Wei, R., Ge, F., Huang, S., Chen, M., and Wang, R. (2011). Occurrence of veterinary antibiotics in animal wastewater and surface water around farms in Jiangsu Province, China. Chemosphere 82, 1408–1414. doi: 10.1016/j.chemosphere.2010.11.067
Xiong, W., Sun, Y., Zhang, T., Ding, X., Li, Y., Wang, M., et al. (2015). Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in China. Microb. Ecol. 70, 425–432. doi: 10.1007/s00248-015-0583-x
Xu, W., Zhang, G., Li, X., Zou, S., Li, P., Hu, Z., et al. (2007). Occurrence and elimination of antibiotics at four sewage treatment plants in the Pearl River Delta (PRD), South China. Water Res. 41, 4526–4534. doi: 10.1016/j.watres.2007.06.023
Yang, J. F., Ying, G. G., Zhao, J. L., Tao, R., Su, H. C., and Chen, F. (2010). Simultaneous determination of four classes of antibiotics in sediments of the Pearl Rivers using RRLC-MS/MS. Sci. Total Environ. 408, 3424–3432. doi: 10.1016/j.scitotenv.2010.03.049
Yogui, G. T., and Sericano, J. L. (2009). Levels and pattern of polybrominated diphenyl ethers in eggs of Antarctic seabirds: endemic versus migratory species. Environ. Pollut. 157, 975–980. doi: 10.1016/j.envpol.2008.10.016
Zhang, Q. Q., Ying, G. G., Pan, C. G., Liu, Y. S., and Zhao, J. L. (2015). Comprehensive evaluation of antibiotics emission and fate in the river basins of china: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 49, 6772–6782. doi: 10.1021/acs.est.5b00729
Zhu, Y. G., Johnson, T. A., Su, J. Q., Qiao, M., Guo, G. X., Stedtfeld, R. D., et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U.S.A. 110, 3435–3440. doi: 10.1073/pnas.1222743110
Zou, S., Xu, W., Zhang, R., Tang, J., Chen, Y., and Zhang, G. (2011). Occurrence and distribution of antibiotics in coastal water of the Bohai Bay, China: impacts of river discharge and aquaculture activities. Environ. Pollut. 159, 2913–2920. doi: 10.1016/j.envpol.2011.04.037
Keywords: antibiotic resistance, environmental dissemination, wild birds, antibiotic resistance gene, horizontal gene transfer
Citation: Wu J, Huang Y, Rao D, Zhang Y and Yang K (2018) Evidence for Environmental Dissemination of Antibiotic Resistance Mediated by Wild Birds. Front. Microbiol. 9:745. doi: 10.3389/fmicb.2018.00745
Received: 17 November 2017; Accepted: 03 April 2018;
Published: 20 April 2018.
Edited by:
Patrícia Poeta, University of Trás-os-Montes and Alto Douro, PortugalCopyright © 2018 Wu, Huang, Rao, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun Yang, cookyoung@scu.edu.cn
†These authors have contributed equally to this work.
 Jiao Wu
Jiao Wu Ye Huang
Ye Huang Dawei Rao
Dawei Rao Yongkui Zhang
Yongkui Zhang Kun Yang
Kun Yang