- 1Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, Faculty of Life Sciences, Northwest University, Xi’an, China
- 2Department of Oral Biology and Department of Medical Microbiology, Rady Faculty of Health Sciences, University of Manitoba, Winnipeg, MB, Canada
Pseudomonas aeruginosa is an important human pathogen which uses the type III secretion system (T3SS) as a primary virulence factor to establish infections in humans. The results presented in this report revealed that the ATP-binding protein PA4595 (named ArtR, a Regulator that is an ATP-activated Repressor of T3SS) represses T3SS expression in P. aeruginosa. The expression of T3SS genes, including exoS, exoY, exoT, exsCEBA, and exsD-pscB-L, increased significantly when artR was knockout. The effect of ArtR on ExsA is at the transcriptional level, not at the translational level. The regulatory role and cytoplasm localization of ArtR suggest it belongs to the REG sub-family of ATP-binding cassette (ABC) family. Purified GST-tagged ArtR showed ATPase activity in vitro. The conserved aspartate residues in the dual Walker B motifs prove to be essential for the regulatory function of ArtR. The regulation of T3SS by ArtR is unique, which does not involve the known GacS/A-RsmY/Z-RsmA-ExsA pathway or Vfr. This is the first REG subfamily of ATP-binding cassette that is reported to regulate T3SS genes in bacteria. The results specify a novel player in the regulatory networks of T3SS in P. aeruginosa.
Introduction
Pseudomonas aeruginosa is a major Gram-negative opportunistic pathogen capable of causing a variety of infections in immuno-compromised individuals and cystic fibrosis patients (Richards et al., 2000; Kerr and Snelling, 2009). It uses an arsenal of virulence factors to establish wide range of human infections. Among these virulence factors, the type III secretion system (T3SS) plays a critical role in P. aeruginosa pathogenicity (Coburn et al., 2007; Engel and Balachandran, 2009). The T3SS system is composed of the type III secretion and translocation machinery, regulators, effectors and effector-specific chaperones. T3SS allows direct delivery of several bacterial effector proteins into eukaryotic host cells (Ghosh, 2004). Four effectors ExoS, ExoT, ExoU, and ExoY have been identified in different strains of P. aeruginosa (Frank, 1997; Yahr et al., 1998). The translocated effectors are important to the bacterium’s successful evasion of phagocytosis and dissemination (Kurahashi et al., 1999; Roy-Burman et al., 2001; Vance et al., 2005).
T3SS gene expression in P. aeruginosa is induced by the contact of host cells and low-calcium conditions (Frank, 1997; Vallis et al., 1999). All T3SS genes are under the direct transcriptional control of ExsA, a member of the AraC-type DNA binding protein (Hovey and Frank, 1995). ExsA auto-regulates its own expression by activating transcription of the exsCEBA operon. In addition, the T3SS is coordinately regulated through other regulatory pathways such as cAMP/Vfr and Gac/Rsm systems. Virulence factor regulator (Vfr) is a cAMP-dependent regulator of transcription. The Vfr regulon consists of hundreds of genes including quorum sensing, type IV pili, type II secretion system, and the T3SS (Wolfgang et al., 2003). Recent study shows that Vfr can regulate the expression of T3SS via directly activating exsA transcription (Marsden et al., 2016). The Gac/Rsm regulatory pathway is another central pathway in controlling T3SS expression. The global posttranscriptional regulatory protein RsmA can activate T3SS and repress biofilm formation, hence controlling the switch between acute and chronic infections (Brencic and Lory, 2009; Irie et al., 2010). Two small regulatory RNAs, RsmY and RsmZ, bind RsmA to antagonize its function. The GacS/GacA two-component regulatory system exclusively controls the expression of RsmY and RsmZ (Brencic et al., 2009). These studies indicate that the T3SS of P. aeruginosa is fine-tuned by complicated environmental cues and internal regulatory networks.
ATP binding cassette (ABC) transporters constitute one of the largest of paralogous protein families with a diversity of physiological functions and they are highly conserved in all species from microbes to human (Davidson et al., 2008). Although most known ABC ATPases such as the Classes 1 and 3 are involved in transmembrane transport of molecules, the superfamily also includes soluble ATPases (Class 2) performing diverse non-transport functions (Bouige et al., 2002; Davidson et al., 2008). The REG subfamily of Class 2 ABC ATPases has several members identified in bacteria (Bouige et al., 2002; Davidson et al., 2008). Proteins in this subfamily are devoid of transmembrane domains and consist of two ABC ATPase domains fused in tandem (Kerr, 2004). ChvD, in Agrobacterium tumefaciens has been found to be involved in the regulation of virulence gene expression (Liu et al., 2001). So far, the most investigated bacterial member of the REG subfamily is Uup in Escherichia coli. It is involved in transposon precise excision and bacterial competitiveness and ATP hydrolysis is essential for the function of the Uup (Hopkins et al., 1983; Reddy and Gowrishankar, 1997; Murat et al., 2006, 2008). Secondary structure and NMR resonance assignments of the C-terminal DNA-binding domain (CTD) of Uup protein suggests that Uup CTD is a DNA-binding coiled coil motif (Carlier et al., 2012a,b). There are three other REG subfamily proteins (YheS, YbiT, and YjjK) in E. coli (Murat et al., 2008). Recent studies demonstrate that the EttA (YjjK) protein in E. coli gates ribosome entry into the translation elongation cycle through a nucleotide-dependent interaction sensitive to ATP/ADP ratio (Boel et al., 2014; Chen et al., 2014).
In our search for regulatory genes of the T3SS in P. aeruginosa, a probable ABC ATPase, named ArtR, was found to repress the transcription and secretion of T3SS effectors in PAO1. ArtR is predicted to be a soluble ABC ATPase and has 74.77% similarity to the REG subfamily protein EttA in E. coli (Boel et al., 2014; Chen et al., 2014). So far, no member in this subfamily has been reported to be involved in regulation of T3SS. In this study, our results suggest that ArtR may not be involved in transport process, and that the ArtR protein could inhibit the transcription but not the translation of T3SS proteins, which seems to be functionally different from EttA in E. coli. These results add a novel regulatory factor to the T3SS expression in P. aeruginosa and provided a new thread for further study of the REG subfamily ATPases.
Results
Identification of a Putative ABC ATPase That Influences T3SS Transcription and Effector Secretion
T3SS is a primary virulence factor in P. aeruginosa, and alterations in T3SS expression often concur with disease progression. Previously, we have screened the P. aeruginosa genome for genes that affect T3SS expression as part of an attempt to investigate the regulatory mechanisms that govern P. aeruginosa infection. A transposon insertion library of wild-type PAO1 containing an exoS-lux transcriptional reporter on its chromosome was constructed and mutants with altered promoter activity of T3SS effector gene exoS were collected. A hybrid sensor kinase PA1611 was found to control biofilm formation and T3SS through direct interaction with RetS (Kong et al., 2013; Bhagirath et al., 2017). Among the rest of the transposon mutants that regulate T3SS, one mutant C1 with disrupted PA4595, which we designated as ArtR, a Regulator that is an ATP-activated Repressor of T3SS, was found to have significantly elevated exoS expression under both T3SS non-inducing and inducing conditions (Figure 1A). The artR gene encodes a putative soluble ABC ATPase. No ABC ATPase has previously been implicated in T3SS expression. Further characterization of this mutant was carried out to investigate the genetic mechanisms underlying the connection between ArtR and T3SS.
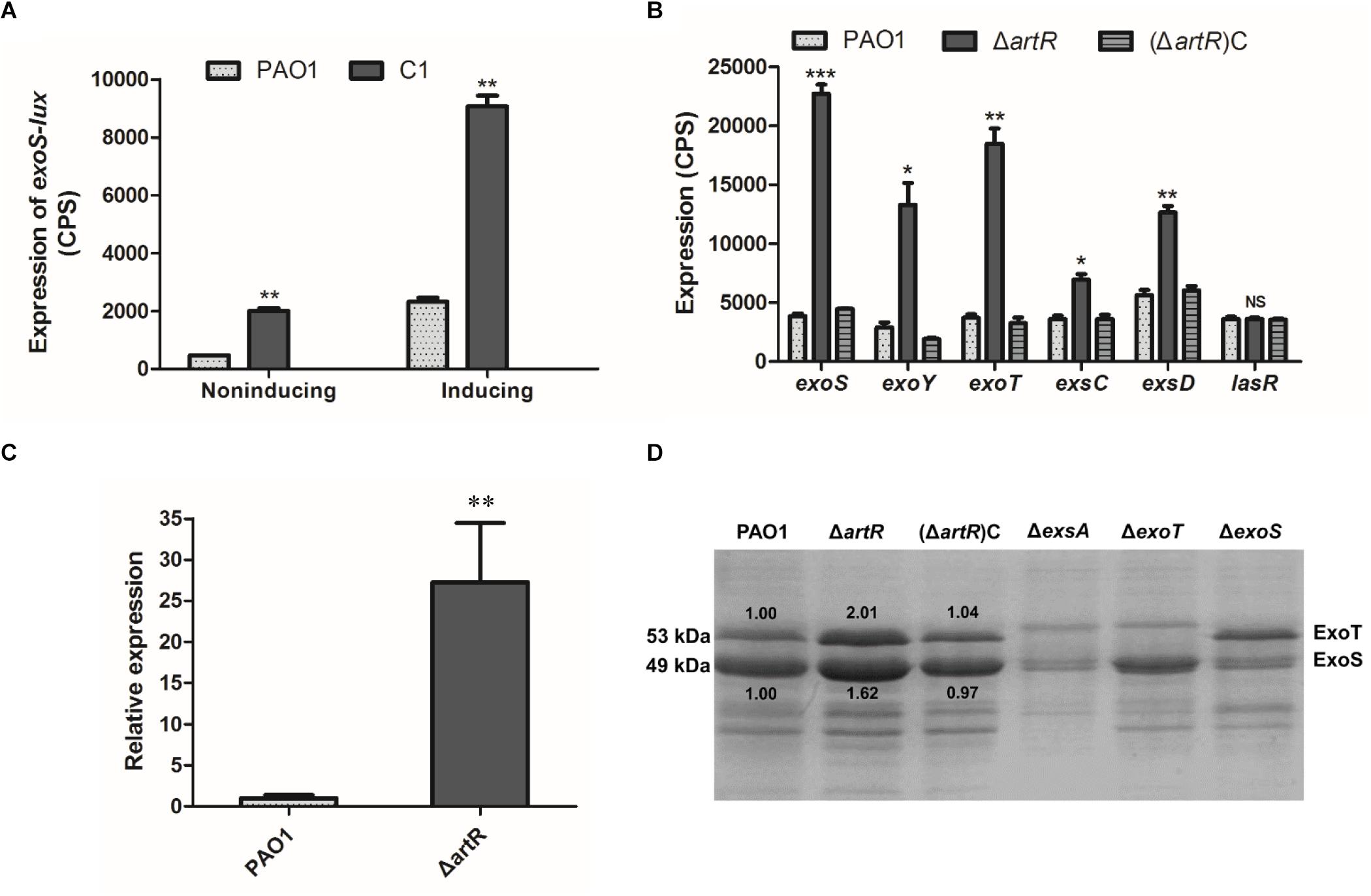
Figure 1. Effect of artR mutation on T3SS expression and secretion. (A) A CTX-exoS reporter fusion integrated on the chromosome was used to measure expression levels of exoS under T3SS non-inducing or inducing conditions in C1 mutant strain and wild-type PAO1 after 6 h growth. Error bars indicate one standard deviation. ∗∗p < 0.01 compared to the wild-type based on Student’s t-test. (B) Strains carried reporter plasmids pKD-exoS, pKD-exoY, pKD-exoT, pKD-exsC, or pKD-exsD were cultured under T3SS inducing conditions. Promoter activities were measured after 6 h growth. Error bars indicate one standard deviation. NS, p > 0.05, ∗p < 0.05, ∗∗p < 0.01, or ∗∗∗p < 0.001 compared to the wild-type. (C) The RT-qPCR analysis of the normalized mRNA expression level of the exoS gene in the ΔartR strain under T3SS inducing conditions. ∗∗p < 0.01. (D) Culture supernatants of various strains after 6 h of growth in T3SS inducing medium were concentrated and analyzed by SDS-PAGE, followed by staining with Coomassie blue. The bands of approximately 53 and 49 kDa were corresponding to ExoT and ExoS respectively. The relative grayscale value of ExoS and ExoT bands were quantified using ImageJ.
To verify the observed effect of artR disruption, a gene knockout mutant of artR was first constructed. A Gmr-lacZ cassette was inserted within artR, and the resultant mutant was named as ΔartR. The knockout strain ΔartR was also complemented by inserting an intact copy of artR with its promoter region at the attB site on the chromosome. The resultant strain was designated as (ΔartR)C. After introduction of reporter plasmid pKD-exoS into ΔartR, (ΔartR)C and the wild-type PAO1 respectively, the expression of exoS was compared. As shown in Figure 1B, the knockout mutant showed significant elevated exoS expression, and the level of exoS expression was restored to wild-type levels in the complementation strain, confirming that ArtR was responsible for the changed exoS expression.
To test if ArtR further influences the expression of other T3SS genes, the expression of exoY, exoT, exsCEBA and exsD-pscB-L was also compared in ΔartR, (ΔartR)C and PAO1. As shown in Figure 1B, the expression of these T3SS genes increased significantly in the artR knockout mutant and the levels of expression were restored to wild-type levels in the complementation strain. As a negative control, no difference in the expression level of quorum sensing regulatory gene lasR was observed between ΔartR mutant and PAO1. The result indicates that ArtR had a significant effect on T3SS.
To verify the transcriptional differences of these effector genes caused by artR deletion, RT-qPCR analysis of the mRNA level of the exoS gene in the ΔartR strain was carried out and the results indicate that the normalized exoS mRNA levels were greatly increased in the mutant comparing with that in the wild-type (Figure 1C). This confirms that artR had an effect on T3SS at the transcriptional level.
T3SS transcription is coupled with the effector secretion process in P. aeruginosa (Yahr and Wolfgang, 2006). To assess whether the increased T3SS transcription was reflected on the secretion of T3SS effectors, we grew these strains in T3SS inducing media and surveyed culture supernatants for secreted T3SS effectors. The ΔexsA mutant which does not produce any effectors was used as a control. SDS-PAGE analysis of supernatants of PAO1 showed bands of approximately 53 and 49 kDa, corresponding to ExoT and ExoS respectively, while the mutant strain ΔexoT and ΔexoS did not have the respective band corresponding to ExoT and ExoS (Kong et al., 2013). In contrast, ΔartR secreted more effectors than the wild-type strain (Figure 1D). We also quantified the change in ExoS and ExoT secretion in Figure 1D using ImageJ. The secretion of ExoS and ExoT was indeed increased in ΔartR. These results demonstrate that artR mutant indeed had increased T3SS secretion. Further verification of the effect of artR on T3SS was carried out by examining ExoS production in the cells of ΔartR, (ΔartR)C and PAO1 using Western blot. The results confirmed an increase of ExoS in the mutant (Supplementary Figure S1).
The Function of ArtR Is Neither Associated With the Adjacent Transmembrane Transport nor Involved in the Regulation via Vfr or RsmA
Most bacterial ABC ATPases in the transport systems are encoded within operons with membrane proteins, whereas artR is a single gene without association with any membrane protein. The operon (PA4594-PA4591) adjacent to artR in PAO1 genome is also predicted to encode components of ABC transporter.
To examine whether ArtR is associated with this operon and whether it functions as one of the components of the transporter to affect T3SS, a gene knockout mutant of PA4594 was constructed and the resultant strain was named ΔPA4594. Comparison of exoS and exsCEBA expression levels in ΔPA4594, ΔartR, and PAO1 indicates that PA4594 had no influence on the expression of exoS and exsCEBA (Figure 2A), indicating that the function of ArtR is distinct from the adjacent ABC transporter. Further comparison of the ExsA levels in the mutant and the wild type was performed by Western blot analysis of the ExsA-FLAG-C in these strains. As shown in Figure 2B, no difference was observed in the FLAG-tagged ExsA between them, confirming the adjacent transporter was not involved.
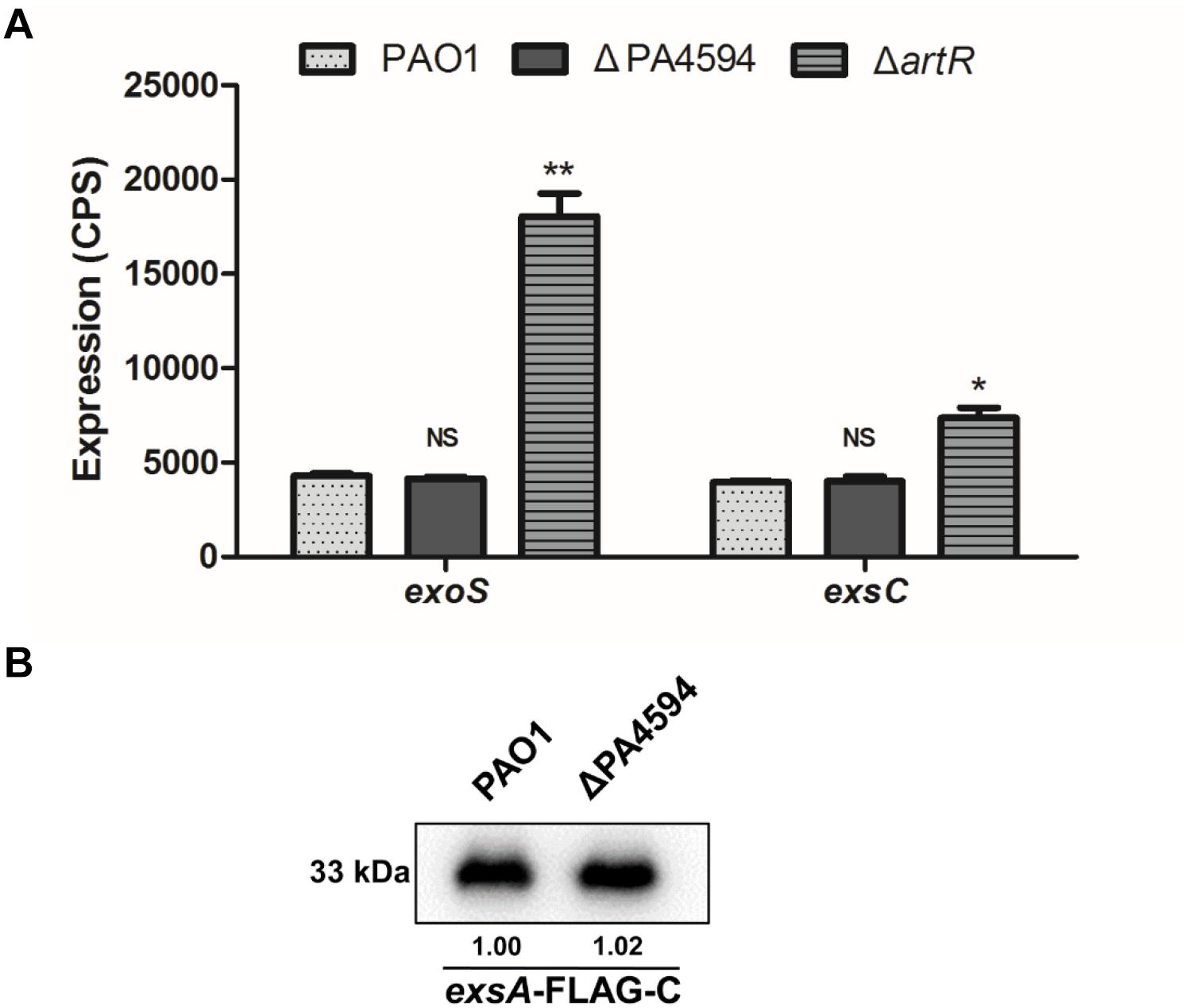
Figure 2. Expression of T3SS genes in ΔPA4594. (A) Comparison of the expression of exoS and exsC in PAO1, ΔartR and ΔPA4594. Strains carried reporter plasmids pKD-exoS or pKD-exsC were cultured under T3SS inducing conditions. Promoter activities were measured after 6 h growth. Unlike artR, deletion of the adjacent ABC transporter gene PA4594 had no effect on exoS expression. Error bars represent one standard deviation from the mean. NSp > 0.05, ∗p < 0.05, or ∗∗p < 0.01 compared to the wild-type. (B) The levels of ExsA-FLAG in PAO1 and ΔPA4594. Indicated strains carrying an exsA-FLAG-C fusion were grown to an OD600 of 1.0 in LB with 5 mM EGTA and 20 mM MgCl2. The whole-cell extracts from the designated strains were subjected to SDS-PAGE separation and subsequent immuno-blotting. The bands of approximately 33 kDa were corresponding to ExsA-FLAG. The relative grayscale value of the ExsA-FLAG bands was quantified using ImageJ and shown underneath.
Vfr is a cAMP-dependent DNA-binding protein that directly activates exsA transcription which is the central regulator of T3SS gene expression (Marsden et al., 2016). To investigate the regulation pathway of T3SS by ArtR, we measured vfr promoter activity in ΔartR mutant. No difference in the levels of vfr expression between ΔartR mutant and the wild-type strain was observed (Figure 3A).
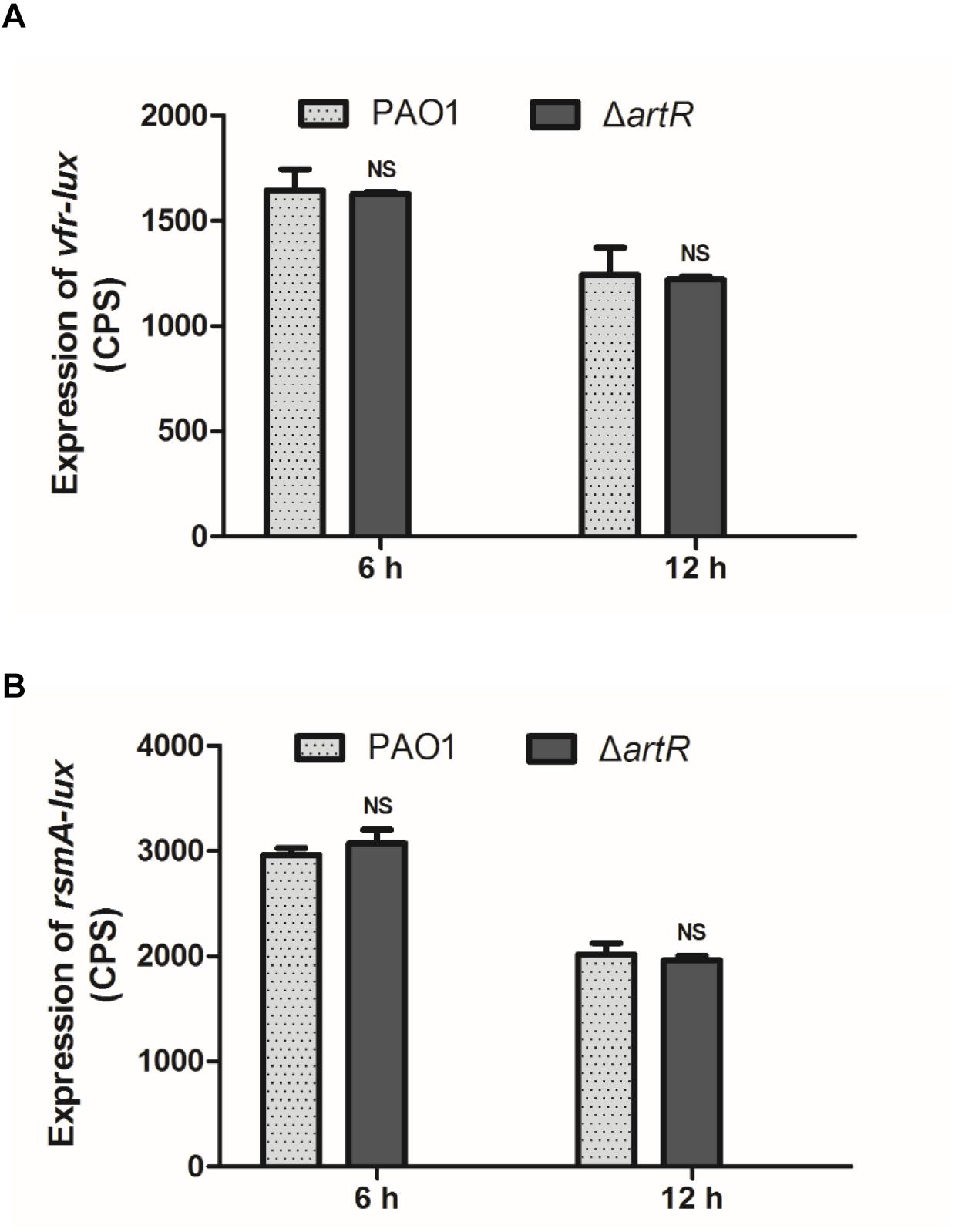
Figure 3. Expression of vfr and rsmA in PAO1 and ΔartR. (A) The expression of vfr in PAO1 and ΔartR. (B) The expression of rsmA in PAO1 and ΔartR. Strains carried reporter plasmids pKD-vfr or pKD-rsmA were cultured in LB medium. Promoter activities were measured after 6 and 12 h growth. Error bars represent one standard deviation from the mean. NSp > 0.05 compared to the wild-type.
The small protein RsmA in the Gac/Rsm regulatory pathway is a pleiotropic global posttranscriptional regulator in P. aeruginosa. The function of RsmA is regulated by two antagonists small RNA RsmY/RsmZ (Brencic and Lory, 2009; Brencic et al., 2009). RsmA regulates the T3SS via modulation of ExsA (Intile et al., 2014). We constructed an rsmA promoter-lux fusion (rsmA-lux) and measured its activity in the wild-type and ΔartR strains. The result, as shown in Figure 3B, indicates no difference in rsmA expression between these two strains. Consistently, neither the rsmY promoter-lux reporter nor rsmZ promoter-lux reporter showed changed activity in ΔartR comparing with the wild-type (data not shown).
The Regulatory Effect of ArtR on ExsA Is at the Transcriptional Level, Not at the Translational Level
ArtR is a putative soluble ABC ATPase which belongs to the REG subfamily. There are three other REG subfamily proteins in P. aeruginosa. To deduce ArtR action, a phylogenetic analysis of the REG subfamily ATPases of P. aeruginosa and E. coli K-12 was performed. ArtR showed stronger sequence similarity (74.77%) to EttA than to other soluble ABC proteins (Supplementary Figures S2, S3). Both EttA and ArtR harbor two tandem ABC ATPase domains but lack the C-terminal DNA-binding domain (CTD) of Uup (Supplementary Figure S4). The EttA protein in E. coli has been demonstrated to regulate protein synthesis probably in response to cellular energy status (Boel et al., 2014; Chen et al., 2014).
Given that the transcription of T3SS was affected by ArtR, we tested whether ArtR regulates T3SS via ExsA, the master regulator of T3SS. We reasoned that unless the effect is directly on individual T3SS genes such as exoS, exoT, exoY, and exsC, which is unlikely, any regulatory effect on T3SS would be reflected in the amount of ExsA protein in the cells. To differentiate the effect of ArtR on the transcription of exsCEBA or on the translation of ExsA, we cloned two exsA-FLAG fusions with different promoter regions. In the exsA-FLAG-C fusion, the promoter region of the exsCEBA operon and the exsA coding region were fused with FLAG tag. In the exsA-FLAG-A fusion, the tac promoter and the exsA coding region were fused with FLAG tag (Li et al., 2013). We transferred these two exsA-FLAG fusions into the artR mutant and PAO1. The expression levels of the ExsA-FLAG were then tested under T3SS inducing conditions.
As shown by results from the exsA-FLAG-C construct, the ExsA-FLAG protein level in the artR mutant was higher than that in PAO1 as expected. However, when the T3SS promoter was replaced by the tac promoter, the ExsA-FLAG protein levels were similar in both PAO1 and the artR mutant (Figure 4), demonstrating that the translation process of ExsA was not affected by the artR mutation.
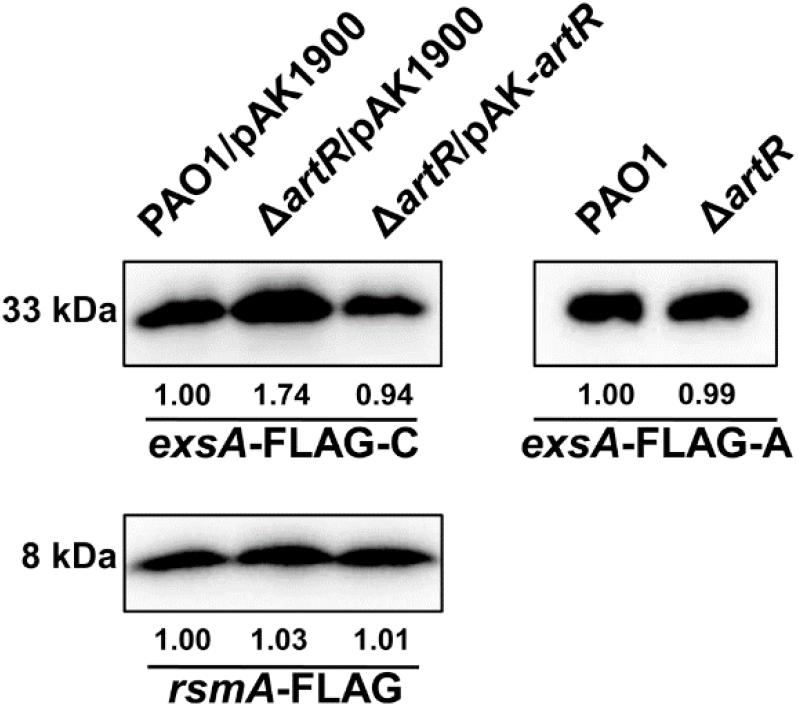
Figure 4. ArtR doesn’t influence the translation process of ExsA. Western-blotting showed the expression levels of ExsA-FLAG and RsmA-FLAG in PAO1 and the artR mutant. The exsA-FLAG-C fusion contained the promoter region of the exsCEBA operon and the exsA coding region. The exsA-FLAG-A fusion carried the tac promoter and the exsA coding region. The rsmA-FLAG fusion contained the promoter region and coding region of rsmA. The pAK1900 plasmid carrying the entire artR gene and the promoter region of artR was used for the complementation of ΔartR. The bands of approximately 33 and 8 kDa were corresponding to ExsA-FLAG and RsmA-FLAG respectively. The relative grayscale value of ExsA-FLAG and RsmA-FLAG bands were quantified using ImageJ.
We also constructed an rsmA-FLAG fusion with its own promoter region. As shown in Figure 4, RsmA-FLAG protein levels in PAO1 and the artR mutant were similar, suggesting that ArtR doesn’t affect either the transcription or translation of RsmA. These results indicate that despite the homology between ArtR and EttA, they seem to function through different mechanisms. The regulation of T3SS by ArtR does not appear to be through controlling the translation of T3SS protein.
To acquire a global view of the regulatory effect of ArtR in P. aeruginosa, we have performed RNA-seq analysis to compare the gene expression profiles between PAO1 and the artR mutant. The genes that were differentially expressed are listed in Supplementary Table S3. One of the most notable changes in the artR mutant was the significant higher expression level of the T3SS genes in the mutant. While total of 11 genes showed increased levels of expression in the mutant, 8 of them are T3SS genes, including exoS, exoT, exoY, pcrV, exsC and exsA, pscC and pscF. The RNA-seq result confirms the specific effect of ArtR on the transcription of T3SS genes. The effect of ArtR is apparently not a general effect on protein translation as how EttA functions in E. coli.
The Highly Conserved Aspartate Residues in Both Walker B Motifs Are Essential for the Function of ArtR
REG subfamily ATPases are consists of two ABC ATPase domains fused together. Each Walker B motif in the ATPase domain harbors a highly conserved aspartate residue which is important for ATP hydrolysis catalytic cycle of ABC ATPases (Kuhnau et al., 1991; Shyamala et al., 1991). To determine whether the conserved Asp residues are required for the function of ArtR, we introduced D187E and D469E single amino acid residue substitutions as well as double residue substitutions into ArtR. These derivatives of ArtR were then introduced into the artR knockout mutant, and the expression of exoS in these point mutants was compared. As shown in Figure 5, the constructs carrying either D187E or D469E or both abolished the complementation ability of ArtR to restore the exoS expression level. Only the complete ArtR was able to complement. Clearly, both Asp residues play critical roles in the function of ArtR.
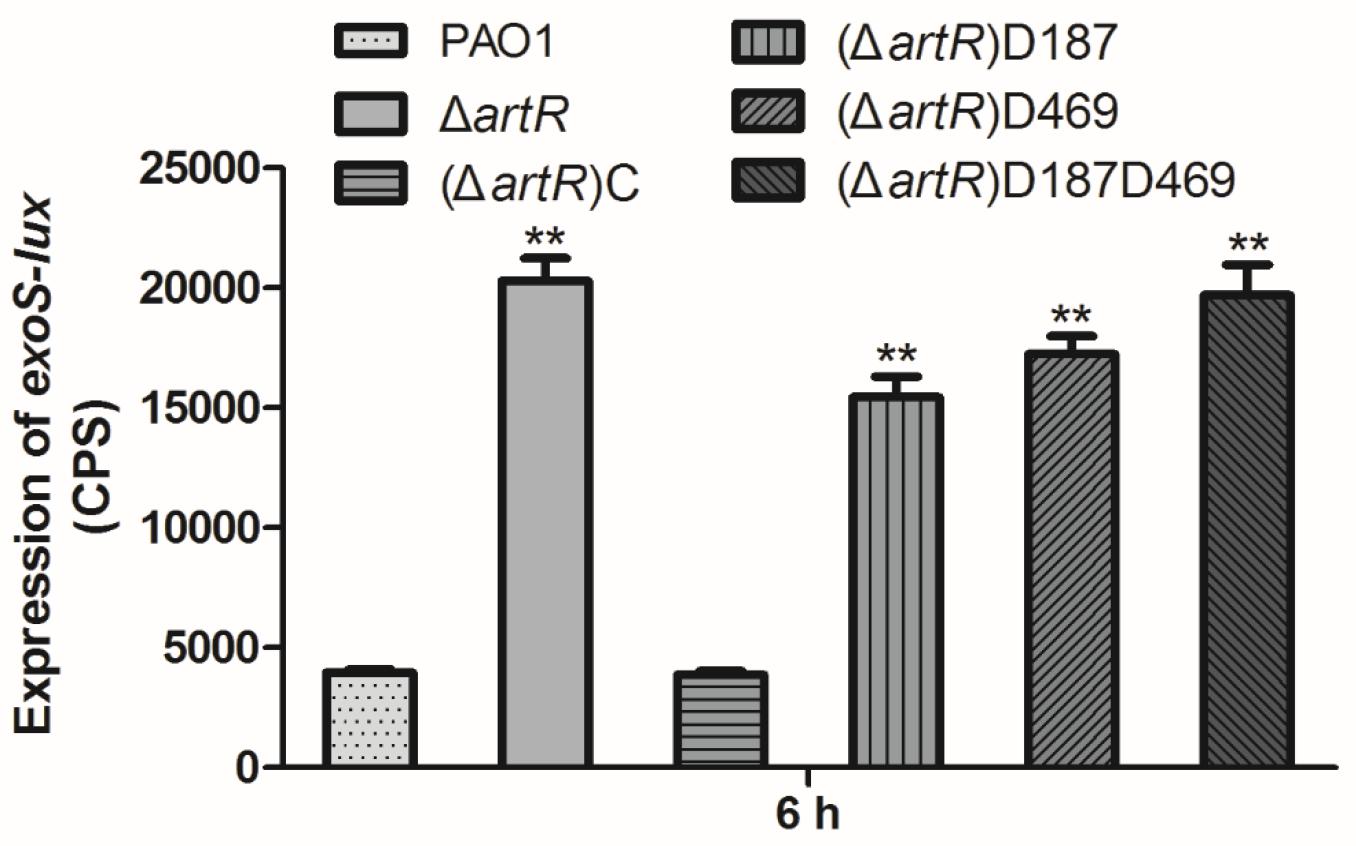
Figure 5. Expression of exoS in PAO1 and artR point mutants. Strains carried reporter plasmid pKD-exoS were cultured under T3SS inducing conditions. Promoter activities were measured after 6 h growth. Error bars represent one standard deviation from the mean. ∗∗p < 0.01 compared to the wild-type based on Student’s t-test.
Subcellular Localization of ArtR
ATP-binding cassette ATPase are often embedded in the cell membrane. However, many REG subfamily ATPases are found in the cytoplasm in recent years (Murat et al., 2006, 2008; Boel et al., 2014; Chen et al., 2014). ArtR protein is also predicted by PSORT as a cytoplasm protein. To verify the cellular location of ArtR, solubilized His6-ArtR protein was purified to prepare polyclonal antibodies used to localize the ArtR protein in PAO1 cell fractions by Western blot. As shown in Figure 6A, ArtR was detected in the whole cell preparation and the cytoplasmic fraction of PAO1. No band was observed in the periplasmic or the membrane fractions of PAO1 or in the total cell extract of ΔartR, confirming that ArtR protein is a cytosolic protein.
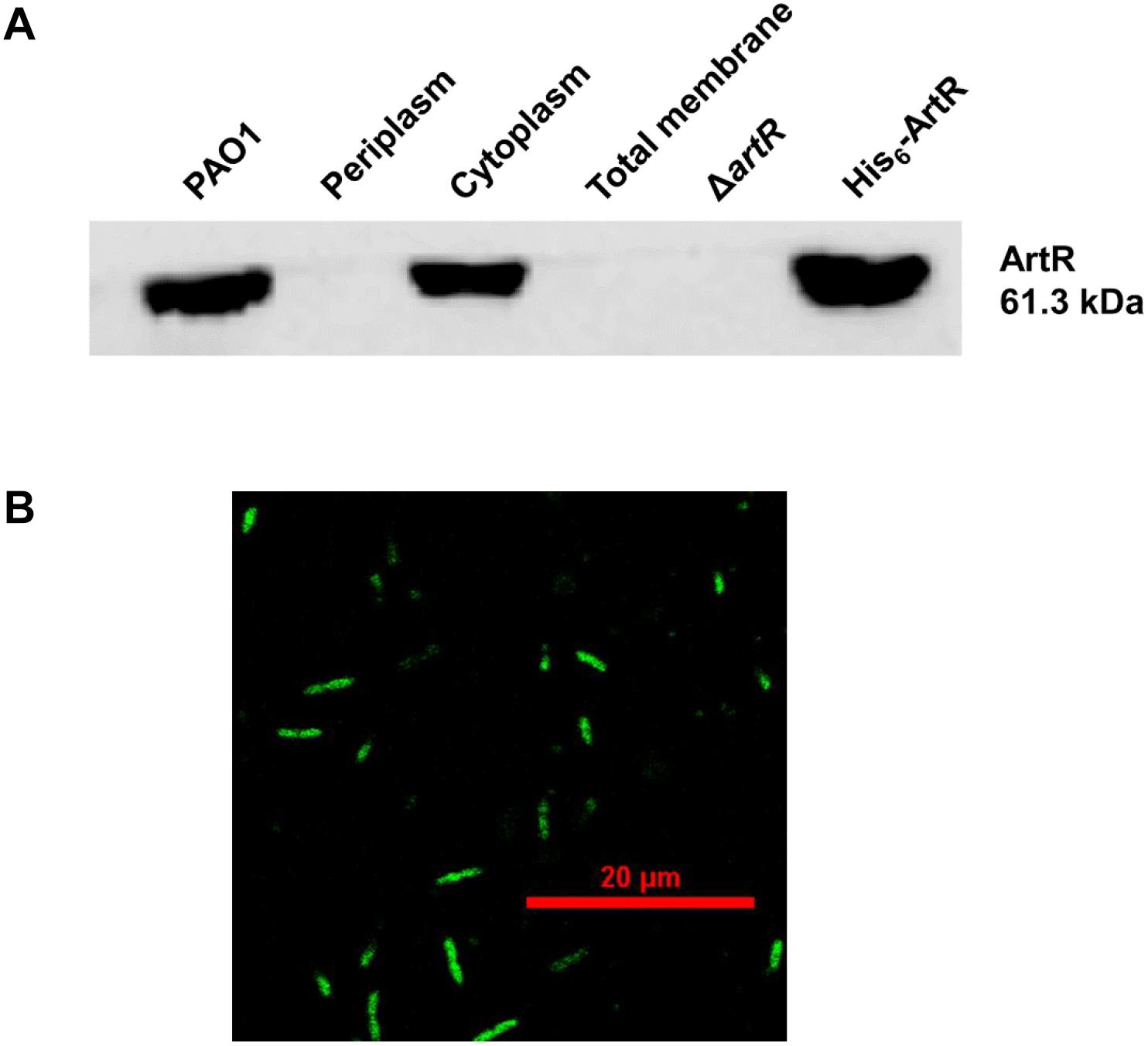
Figure 6. Subcellular localization of ArtR. (A) Localization of ArtR in the cell fractionations analyzed by Western blot. Immunoblot of crude extracts probed with the polyclonal antibody (1:3,000) directed against ArtR protein. Cells of a 50 ml culture in LB were fractionated. Specimens of the whole cell preparation of PAO1, periplasm, cytoplasm, total membrane, the total cell extract of ΔartR (negative control), and His6-ArtR (positive control) were separated by SDS-PAGE and probed by immunoblotting. (B) The localization of ArtR-GFP in PAO1. P. aeruginosa PAO1 carrying pME6032-artR-gfp was cultured with 0.5 mM IPTG at 37°C for 4 h. The cells were examined and imaged by Nikon A1 confocal laser scanning microscopy.
To verify the cytoplasmic location of ArtR, we also expressed GFP-tagged ArtR and examined the cellular distribution of ArtR-GFP in PAO1 by confocal laser scanning microscopy. As shown in Figure 6B, the GFP tagged ArtR is uniformly distributed in the cell, supporting its cytoplasmic location.
ArtR Has in vitro ATPase Activity
To verify the ATPase activity of ArtR, we carried out in vitro analysis. GST-tagged ArtR protein was purified by using a high-affinity GST resin column (Supplementary Figure S5). Spontaneous ATPase activity of ArtR was tested by analyzing the amount of inorganic phosphate liberated from the ATP substrate. The release of Pi occurred in a protein concentration-dependent manner (Figure 7) when purified GST-ArtR protein was incubated with ATP, demonstrating that purified GST-ArtR had ATPase activity and capable of hydrolyzing ATP.
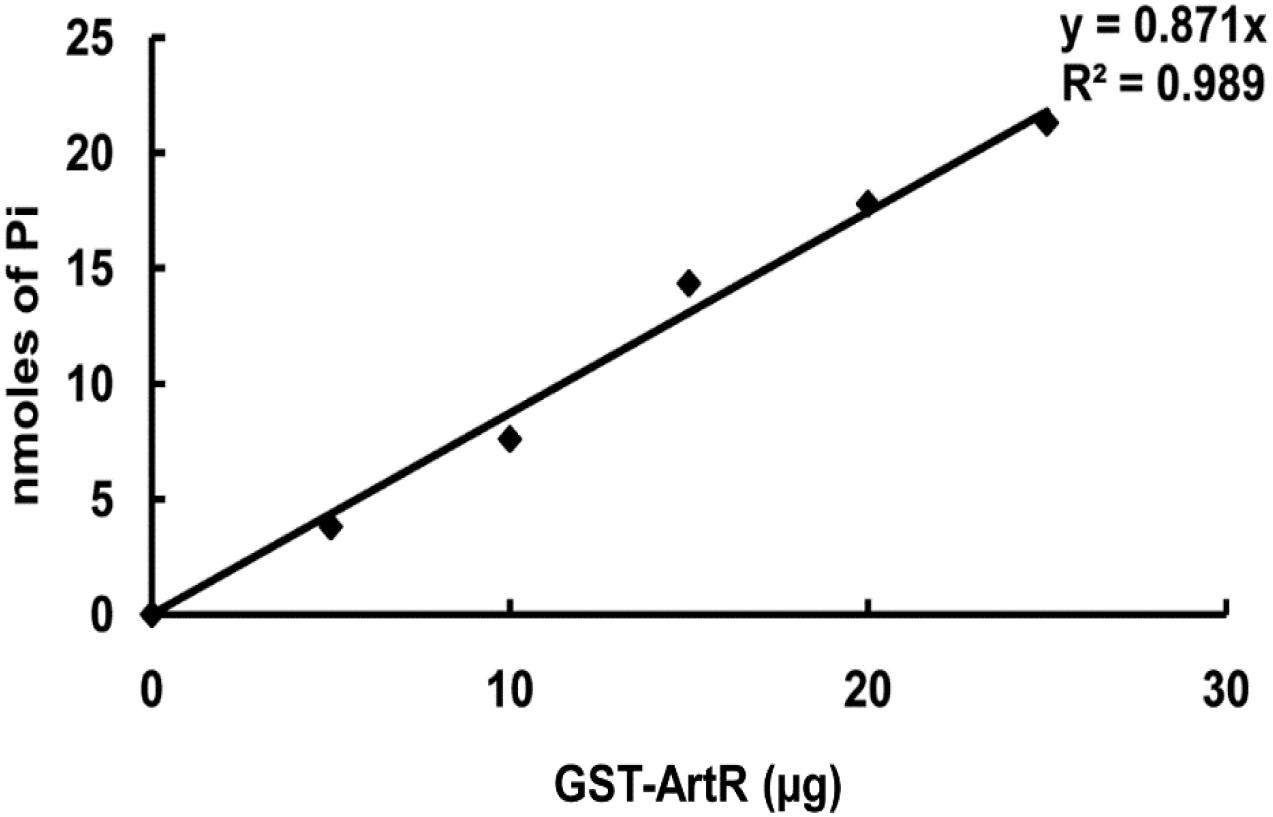
Figure 7. ATPase activity of purified GST-ArtR protein. The ATPase activity of GST-ArtR protein was detected by assaying the amount of inorganic phosphate liberated from ATP using NaH2PO4 as a standard. Purified GST-ArtR protein (0–25 μg) was added into the reaction mixtures. The reaction was started by adding 2 mM ATP and terminated after 1 h by the addition of the Malachite Green reagent. Purified GST tag was used as negative control and no ATPase activity was detected.
Discussion
Pseudomonas aeruginosa is a human pathogen that causes serious and often life-threatening infections. T3SS is a virulence factor in P. aeruginosa that is extremely important for its successful establishment of infection and evasion of phagocytosis. It is essential to understand the pathways and controlling mechanisms of the T3SS. T3SS also provides a novel target for the development of new therapeutic strategies against P. aeruginosa infections. Although dozens of genes implicated in the regulation of T3SS in P. aeruginosa have been identified in the past 10 years, important regulators of the T3SS are still to be discovered.
In this study, a probable ABC ATPase ArtR was found to repress T3SS expression at the level of transcription. The artR gene encodes a soluble ABC ATPase which belongs to the REG subfamily (Supplementary Figures S2, S3). There are only four similar soluble ATPases in P. aeruginosa which are respectively homologous to Uup, YheS, YbiT, and EttA in E. coli. So far, none of them has been investigated nor found to be connected with T3SS. Unlike most other ABC ATPases involving in transport process, artR is a single gene without membrane protein encoding genes in its operon. Our data demonstrate that ArtR has function different from the adjacent PA4594 operon which is predicted to encode components of ABC transporter. The influence of ArtR on the expression of T3SS is not due to the potential transport function. Neither the effect of ArtR on the T3SS was through the known pathways, GacS/A-RsmY/Z-RsmA-ExsA or Vfr pathways.
The regions of ArtR corresponding to Walker motif B were aligned with the Uup protein in E. coli. The invariant aspartate residues that are important for the catalytic cycle of ABC ATPases in Uup correspond to D187 and D469 in ArtR. The results presented here indicate that both residues D187 and D469 play critical roles in the function of ArtR. The negative regulation of ArtR on T3SS is dependent on its ATPase activity. Recombinant GST-ArtR protein acquired was found to display ATPase activity.
Members of REG subfamily display significant functional diversity in eukaryotes and prokaryotes. Although a few of them have been reported to participate in gene expression regulation, their exact mechanism of action and the role of ATP hydrolysis in their function have remained unknown. The yeast protein GCN20 and its human ortholog ABC50 have been implicated in the control of mRNA translation (de Aldana et al., 1995; Tyzack et al., 2000). The information collected on prokaryote proteins is quite descriptive and does not afford a clue for their actual physiological role and mechanism of action. In E. coli, one of the REG subfamily protein, Uup, was found to be involved in transposon precise excision and bacterial competitiveness (Murat et al., 2006, 2008). This protein is demonstrated to be a cytoplasmic ABC protein. Analysis of Walker motif B mutants suggests that ATP hydrolysis at the two ABC domains is strictly coordinated and is essential for the function of Uup in vivo (Murat et al., 2006). In addition, Uup carries a C-terminal DNA-binding coiled coil motif (Carlier et al., 2012a,b). Another REG subfamily protein EttA in E. coli has been demonstrated to regulate protein synthesis potentially in response to cellular energy status (Boel et al., 2014; Chen et al., 2014). In this study, we performed a phylogenetic analysis of the REG subfamily ATPases in P. aeruginosa and E. coli. The sequence of ArtR was similar to EttA and both lack the C-terminal DNA-binding domain of Uup (Supplementary Figure S4). We constructed two exsA-FLAG fusions to test whether ArtR regulates T3SS via controlling the translational process of ExsA. The results demonstrated that the translation process of ExsA is not affected by the artR mutation (Figure 4). ArtR and EttA seem to have different functional mechanisms. EttA has been shown to inhibit protein translation process in E. coli (Boel et al., 2014; Chen et al., 2014). It gates ribosome entry into the translation elongation cycle through a nucleotide-dependent interaction sensitive to ATP/ADP ratio (Boel et al., 2014). Despite its similarity to EttA, ArtR seems to specifically regulate the transcription of T3SS genes. This is in contrast to the effect of EttA on protein translation, as indicated by the unaffected translational levels of ExsA in ΔartR. However, it has been suggested that EttA may act preferentially on the translation of specific target mRNAs (Boel et al., 2014). It is possible that the translation of an unknown component in the T3SS regulatory network is specifically affected by ArtR, which resulted in the changed transcription of T3SS genes. Considering the activity of T3SS is energy demanding, it makes sense to couple the T3SS with the energy status of the cell. Further study is needed to confirm such possibilities.
ArtR is found to be highly conserved in many bacteria through homology analysis, suggesting that the protein performs important functions in bacteria. This report reveals that ArtR is a unique component in the regulatory network of T3SS in P. aeruginosa and provides new path to further study the REG subfamily ATPases in bacteria.
Experimental Procedures
Strains, Plasmids and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. P. aeruginosa and E. coli were routinely grown on Luria-Bertani (LB) agar or in LB broth at 37°C unless otherwise specified. LB was used as T3SS non-inducing conditions and LB supplemented with 5 mM EGTA and 20 mM MgCl2 as T3SS inducing conditions. Antibiotics were used at the following final concentrations. For P. aeruginosa: gentamicin (Gm) at 50 μg/ml in LB or 150 μg/ml in Pseudomonas isolation agar (PIA), tetracycline (Tc) at 70 μg/ml in LB or 300 μg/ml in PIA, carbenicillin (Cb) at 250 μg/ml in LB and trimethoprim (Tmp) at 300 μg/ml in LB. For E. coli: kanamycin (Kn) at 50 μg/ml, ampicillin (Ap) at 100 μg/ml, Tc at 15 μg/ml, and Gm at 15 μg/ml in LB.
Construction of Gene Expression Detecting System
The plasmid pMS402 carrying a promoterless luxCDABE reporter gene cluster was used to construct promoter-luxCDABE reporter fusions as reported previously (Duan et al., 2003). Promoter regions (including the intergenic region and partial coding region) of genes were PCR amplified respectively using high-fidelity Pfu DNA polymerase (Fermentas) and primers designed according to the PAO1 genome data (Stover et al., 2000). The promoter regions were cloned into the BamHI-XhoI site upstream of the lux genes on pMS402. The resultant plasmids were transformed into PAO1 by electroporation. DNA manipulation, PCR, and transformation were performed following standard procedures. Cloned promoter sequences were confirmed by DNA sequencing.
Besides the plasmid-based reporter system, an integration plasmid CTX6.1 originated from plasmid mini-CTX-lux (Becher and Schweizer, 2000) was used to construct chromosomal fusion reporter system. This plasmid has all the elements required for integration, origin of replication, and tetracycline resistance. A DNA fragment of the pMS402-based reporters that contains the kanamycin resistance determinant, the multiple cloning sites, and the promoter-luxCDABE reporter was then ligated into CTX6.1. The plasmid generated was transferred into E. coli SM10-λpir (Simon et al., 1983) and the integration strain was constructed through biparental mating as reported previously (Hoang et al., 2000; Liang et al., 2008).
Using these lux-based reporters, gene expression in liquid cultures was examined as counts per second (cps) of light production in a Synergy 2 Multimode Microplate Reader (BioTek). Measurements were taken every 30 min for 24 h. Bacterial growth was monitored at the same time by measuring the OD600 using the Microplate Reader.
Screening for Regulatory Genes of the T3SS
The transposon mutagenesis library was constructed as previously described except some modifications (Kulasekara et al., 2005). Briefly, the donor strain E. coli SM10-λpir containing pBT20 and the P. aeruginosa PAO1 carrying the reporter exoS-lux on its chromosome were grown on solid media overnight and the cells were collected and spotted on LB agar plate at a ratio of 2:1. After incubation for 2 h, the mixed culture was diluted and spread on PIA containing gentamicin. A transposon mutant library was constructed by picking 10,000 colonies grown on the selective plates. After overnight incubation, colonies with changed activities of exoS-lux under a LAS-3000 imaging system (FUJIFILM) were collected. Verification of the mutant phenotype was carried out in liquid cultures. The expression of exoS in collected mutants was examined in Synergy 2 Multimode Microplate Reader. The sites of transposon insertion in the selected mutants were determined by arbitrary primed PCR and subsequent sequencing of the PCR products (Friedman and Kolter, 2004; Liang et al., 2008).
Construction of Gene Knockout Mutant
For gene knockout, the previously described sacB-based strategy was employed (Hoang et al., 1998). The regions of target genes were PCR amplified using primers listed in Supplementary Table S2. The PCR products were digested with restriction enzymes and cloned into pEX18Tc. The fragment containing Gmr-lacZ from the plasmid pZ1918-lacZGm (Schweizer, 1993) was then inserted into the target genes on pEX18Tc. P. aeruginosa knockout mutants were obtained following triparental mating between E. coli strain containing the helper plasmid pRK2013 (Ditta et al., 1980), the P. aeruginosa recipient and the E. coli donor harboring the pEX18Tc based knockout construct. The resultant mutants were verified by PCR the region containing the target gene and PCR products sequencing.
Complementation of the artR Knockout Mutant
For the complementation experiments, the entire gene together with upstream promoter region of artR was integrated into the attB site on the mutant’s chromosome using the mini-CTX-lacZ system (Becher and Schweizer, 2000). The DNA fragment was PCR amplified using primers pEX-artR-S and pEX-artR-A (Supplementary Table S2). The product was cloned into mini-CTX-lacZ. The generated plasmid was transferred into E. coli SM10-λpir and the complementary strain was obtained after biparental mating. The resultant strain was designated as (ΔartR)C.
Construction of ArtR Point Mutations
The construction of the D187E mutation in the first Walker B motif of ArtR was accomplished by PCR using oligonucleotide primers containing the point mutation and an XhoI restriction site. The XhoI restriction site was generated from the base alterations at D187 without affecting adjacent codons. The DNA fragment upstream of codon D187 was amplified using primers pEX-artR-S containing a KpnI site and artRD187-A containing an XhoI site; the downstream region of D187 was generated using the primers artRD187-S containing an XhoI site and pEX-artR-A containing a HindIII site (Supplementary Table S2). These two PCR fragments were cloned in turn into mini-CTX-lacZ. In the final construct, the aspartate residue D187 of ArtR was replaced by a glutamate residue. The same strategy was used to construct the D469E mutation in the second Walker B motif of ArtR. The primers used are listed in Supplementary Table S2. The D187E D469E double substitution mutations on ArtR were generated by swapping the wild-type fragment upstream of the D187E mutation with the fragment containing D469E mutation using the SphI and HindIII restriction sites. All these mutations were verified by sequencing. The plasmids generated were used to complement of the artR knockout mutant as above and the resultant strains were designated as (ΔartR)D187, (ΔartR)D469, and (ΔartR)D187D469 respectively.
RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
Total RNA was extracted from 500 μl fresh broth culture (OD600 = 1.0) by using the Trizol reagent. Genomic DNA was eliminated by RNase-free DNase I treatment after the isolation procedure. Then 1 μg RNA sample was reverse transcribed to cDNA using the qScript cDNA SuperMix Kit (Quanta). Real-time qPCR was performed in the LightCycler (MBI Lab Equipment) using PerfeCTa SYBR Green FastMix Kit (Quanta). The fold change of relative expression was showed by the value of 2−ΔΔCt. The primers specific for exoS and the housekeeping gene proC (Savli et al., 2003) are shown in Supplementary Table S2.
T3SS Effector Secretion Assay
Pseudomonas aeruginosa strains were grown at 37°C in T3SS inducing conditions for 6 h. Following removal of the bacterial cells by centrifugation at 14,000 g, proteins in the supernatant were precipitated following addition of an equal volume of 100% trichloroacetic acid, washed with acetone, and pelleted. The proteins were re-suspended in sample buffer, separated by SDS-PAGE, and visualized with Coomassie stain (Goodman et al., 2004).
In vitro Expression and Purification of ArtR Protein
ArtR protein was expressed as a glutathione S-transferase (GST) tagged fusion protein using the pGEX protein expression system (GE Healthcare). The DNA fragment covering the entire artR gene was PCR amplified using primers pGEX-artR-S and pGEX-artR-A (Supplementary Table S2). The PCR product was then cloned into pGEX-4T-1 to generate pGEX-ArtR which was verified by sequencing. pGEX-ArtR was transferred into E. coli strain BL21(DE3)pLysS for ArtR protein production. BL21(DE3)pLysS carrying pGEX-ArtR was grown at 37°C in LB broth supplemented with 100 μg/ml Ap. When the culture reached an OD600 of 0.5–1.0, isopropyl β-D-thiogalactopyranoside (IPTG) was added at a final concentration of 0.5 mM to induce protein expression. The cells were harvested 3 h later by centrifugation. The pellets from 50 ml of culture were re-suspended in 6 ml PBS supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF) and 1 mg/ml lysozyme. Cells were ruptured by sonication using a Hielscher Sonifier. Cell debris was removed by centrifugation. The recombinant GST-ArtR protein was isolated from the supernatant using a high-affinity GST resin column (Genscript). Free glutathione in the eluted fractions was removed by dialysis and the purified protein was stored at −70°C. The purity of the protein samples was assessed by SDS-PAGE followed by Coomassie blue staining. Protein concentration was colorimetrically determined by using the Bradford protein assay.
ATPase Activity Assay
The ATPase activity of GST-ArtR protein was detected by measuring the amount of inorganic phosphate liberated from ATP using NaH2PO4 as a standard. Experiments were performed in a reaction buffer as described previously (Murat et al., 2006). Purified GST-ArtR protein (0–25 μg) was added into the reaction mixtures. The reaction was started by adding 2 mM ATP and terminated after 1 h by the addition of the Malachite Green reagent. After 5 min, absorbance was measured at 630 nm.
Subcellular Fractionation
Subcellular fractionation was performed as described previously with minor changes (Casabona et al., 2013). Fifty milliliters of overnight PAO1 culture was centrifuge for 10 min at 2,000 g. The cells were washed once with 1 ml of 50 mM Tris-HCl (pH 7.6) and re-suspended in 0.5 ml of 200 mM MgCl2, 50 mM Tris-HCl (pH 7.6). After incubation at 30°C for 30 min with gentle shaking, the cells were placed on ice for 5 min, and then incubated for 15 min at room temperature. Centrifuge the cells for 10 min at 8,000 g (4°C) and keep supernatant as periplasmic fraction. Wash the cell pellet in 1 ml of 50 mM Tris-HCl (pH 7.6) and centrifugation for 10 min at 8,000 g (4°C). Re-suspend the cell pellet in 0.5 ml of 50 mM Tris-HCl (pH 7.6) and sonicate twice for 30 s. Centrifuge at 2,000 g for 15 min to remove intact cells and cell debris and take supernatant as cytoplasm and membrane fraction. Transfer the supernatant into a tube compatible with ultracentrifugation and ultracentrifuge for 45 min at 120,000 g at 4°C. Harvest the supernatant as the cytoplasmic fraction. Resuspend the pellet corresponding to the total membrane fraction in the desired buffer. The ultrafiltration tube (Millipore) was used for enriching cell fragments, and the fractions then were used in subsequent experiments.
Confocal Microscopy and Image Acquisition
Pseudomonas aeruginosa PAO1 carrying pME6032-artR-gfp was cultured with 0.5 mM IPTG at 37°C for 4 h. The cells were examined and imaged by confocal laser scanning microscopy, using a Nikon A1 operating system (Nikon, Japan).
Western Blot Analysis
Samples from subcellular fractionation were loaded and separated by 12% SDS-PAGE. The proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane and hybridized with a rabbit polyclonal ArtR antibody (Sangon Biotech) generated by using purified His6-ArtR protein. The signal was detected by the use of an ECL Plus kit (Amersham Biosciences).
For the test of ExoS and FLAG fusion proteins, overnight cultures of the tested strains were transformed into fresh LB medium (rsmA-FLAG) or with 5 mM EGTA and 20 mM MgCl2 (ExoS and exsA-FLAG) and cultivated to an OD600 of 1.0. Hundred microlitres of cultures were centrifuged and the pellets were re-suspended in 10 μl PBS. Bacterial cells were loaded and separated by 12% SDS-PAGE. The proteins were transferred onto a PVDF membrane and hybridized with a rabbit polyclonal ExoS antibody (generated in Dr. Shouguang Jin’s laboratory) or a mouse monoclonal FLAG antibody (Sigma). The signal was detected by an ECL Plus kit (Amersham Biosciences).
RNA-Seq and Data Analysis
Wild-type strain PAO1 and the ΔartR mutant were cultured in LB broth at 37°C and grown to mid-log-phage (OD600 = 1.0). Total RNA was isolated with an RNeasy Protect Bacteria minikit (Qiagen) and genomic DNA was eliminated by RNase-free DNase I treatment. After removing rRNA by using the MICROBExpress kit (Ambion), mRNA was used to generate the cDNA library according to the TruSeq RNA sample prep kit protocol (Illumina), which was then sequenced using the HiSeq 2000 system (Illumina). The sequence data were analyzed using a method described previously (Chua et al., 2014).
Statistical Analysis
Statistical analysis was performed using Student’s t-test to determine the statistical significance when applicable: NSp > 0.05; ∗p < 0.05; ∗∗p < 0.01, and ∗∗∗p < 0.001.
Data Availability
The NCBI database accession number is GSE127751.
Author Contributions
WK and KD designed the study. MD, RY, QL, HZ, WL, YZ, and HL contributed in the acquisition, analysis, and interpretation of the data. WK and KD wrote the manuscript.
Funding
This study was supported by grants from the National Science and Engineering Research Council of Canada (No. 402943-2011 RGPIN), NSFC (Nos. 31570131 and 31500111) and by IRT-15R55, NSFSP (2016JQ3013), FEDSP (15JK1706). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00560/full#supplementary-material
References
Becher, A., and Schweizer, H. P. (2000). Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29, 948–953. doi: 10.2144/00295bm04
Bhagirath, A. Y., Pydi, S. P., Li, Y., Lin, C., Kong, W. N., Chelikani, P., et al. (2017). Characterization of the direct interaction between hybrid sensor kinases PA1611 and RetS that controls biofilm formation and the type III secretion system in Pseudomonas aeruginosa. ACS Infect. Dis. 3, 162–175. doi: 10.1021/acsinfecdis.6b00153
Boel, G., Smith, P. C., Ning, W., Englander, M. T., Chen, B., Hashem, Y., et al. (2014). The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat. Struct. Mol. Biol. 21, 143–151. doi: 10.1038/nsmb.2740
Bouige, P., Laurent, D., Piloyan, L., and Dassa, E. (2002). Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr. Protein Pept. Sci. 3, 541–559. doi: 10.2174/1389203023380486
Brencic, A., and Lory, S. (2009). Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72, 612–632. doi: 10.1111/j.1365-2958.2009.06670.x
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x
Carlier, L., Haase, A. S., Burgos, Z. M. Y., Dassa, E., and Lequin, O. (2012a). Secondary structure and NMR resonance assignments of the C-terminal DNA-binding domain of Uup protein. Biomol. NMR Assign. 6, 197–200.
Carlier, L., Haase, A. S., Burgos, Z. M. Y., Dassa, E., and Lequin, O. (2012b). The C-terminal domain of the Uup protein is a DNA-binding coiled coil motif. J. Struct. Biol. 180, 577–584. doi: 10.1016/j.jsb.2012.09.005
Casabona, M. G., Silverman, J. M., Sall, K. M., Boyer, F., Coute, Y., Poirel, J., et al. (2013). An ABC transporter and an outer membrane lipoprotein participate in posttranslational activation of type VI secretion in Pseudomonas aeruginosa. Environ. Microbiol. 15, 471–486. doi: 10.1111/j.1462-2920.2012.02816.x
Chen, B., Boel, G., Hashem, Y., Ning, W., Fei, J. Y., Wang, C., et al. (2014). EttA regulates translation by binding the ribosomal E site and restricting ribosome-tRNA dynamics. Nat. Struct. Mol. Biol. 21, 152–159. doi: 10.1038/nsmb.2741
Chua, S. L., Liu, Y., Yam, J. K. H., Chen, Y. C., Vejborg, R. M., Tan, B. G. C., et al. (2014). Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 5:4462. doi: 10.1038/ncomms5462
Coburn, B., Sekirov, I., and Finlay, B. B. (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20, 535–549. doi: 10.1128/CMR.00013-07
Davidson, A. L., Dassa, E., Orelle, C., and Chen, J. (2008). Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364. doi: 10.1128/MMBR.00031-07
de Aldana, C. R. V., Marton, M. J., and Hinnebusch, A. G. (1995). GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J. 14, 3184–3199. doi: 10.1002/j.1460-2075.1995.tb07321.x
Ditta, G., Stanfield, S., Corbin, D., and Helinski, D. R. (1980). Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 77, 7347–7351. doi: 10.1073/pnas.77.12.7347
Duan, K., Dammel, C., Stein, J., Rabin, H., and Surette, M. G. (2003). Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50, 1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x
Engel, J., and Balachandran, P. (2009). Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 12, 61–66. doi: 10.1016/j.mib.2008.12.007
Frank, D. W. (1997). The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26, 621–629. doi: 10.1046/j.1365-2958.1997.6251991.x
Friedman, L., and Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Ghosh, P. (2004). Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68, 771–795. doi: 10.1128/MMBR.68.4.771-795.2004
Goodman, A. L., Kulasekara, B., Rietsch, A., Boyd, D., Smith, R. S., and Lory, S. (2004). A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7, 745–754. doi: 10.1016/j.devcel.2004.08.020
Hoang, T. T., Karkhoff-Schweizer, R. A. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Hoang, T. T., Kutchma, A. J., Becher, A., and Schweizer, H. P. (2000). Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72. doi: 10.1006/plas.1999.1441
Hopkins, J. D., Clements, M., and Syvanen, M. (1983). New class of mutations in Escherichia coli (uup) that affect precise excision of insertion elements and bacteriophage Mu growth. J. Bacteriol. 153, 384–389.
Hovey, A. K., and Frank, D. W. (1995). Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 177, 4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995
Intile, P. J., Diaz, M. R., Urbanowski, M. L., Wolfgang, M. C., and Yahr, T. L. (2014). The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 196, 357–366. doi: 10.1128/JB.01199-13
Irie, Y., Starkey, M., Edwards, A. N., Wozniak, D. J., Romeo, T., and Parsek, M. R. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78, 158–172. doi: 10.1111/j.1365-2958.2010.07320.x
Kerr, I. D. (2004). Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem. Biophys. Res. Commun. 315, 166–173. doi: 10.1016/j.bbrc.2004.01.044
Kerr, K. G., and Snelling, A. M. (2009). Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 73, 338–344. doi: 10.1016/j.jhin.2009.04.020
Kong, W. N., Chen, L., Zhao, J. Q., Shen, T., Surette, M. G., Shen, L. X., et al. (2013). Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 88, 784–797. doi: 10.1111/mmi.12223
Kuhnau, S., Reyes, M., Sievertsen, A., Shuman, H. A., and Boos, W. (1991). The activities of the Escherichia coli MalK protein in maltose transport, regulation, and inducer exclusion can be separated by mutations. J. Bacteriol. 173, 2180–2186. doi: 10.1128/jb.173.7.2180-2186.1991
Kulasekara, H. D., Ventre, I., Kulasekara, B. R., Lazdunski, A., Filloux, A., and Lory, S. (2005). A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55, 368–380. doi: 10.1111/j.1365-2958.2004.04402.x
Kurahashi, K., Kajikawa, O., Sawa, T., Ohara, M., Gropper, M. A., Frank, D. W., et al. (1999). Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Invest. 104, 743–750. doi: 10.1172/JCI7124
Li, K. W., Xu, C., Jin, Y. X., Sun, Z. Y., Liu, C., Shi, J., et al. (2013). SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. MBio 4:e419-13. doi: 10.1128/mBio.00419-13
Liang, H., Li, L., Dong, Z., Surette, M. G., and Duan, K. (2008). The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J. Bacteriol. 190, 6217–6227. doi: 10.1128/JB.00428-08
Liu, Z., Jacobs, M., Schaff, D. A., McCullen, C. A., and Binns, A. N. (2001). ChvD, a chromosomally encoded ATP-binding cassette transporter-homologous protein involved in regulation of virulence gene expression in Agrobacterium tumefaciens. J. Bacteriol. 183, 3310–3317. doi: 10.1128/JB.183.11.3310-3317.2001
Marsden, A. E., Intile, P. J., Schulmeyer, K. H., Simmons-Patterson, E. R., Urbanowski, M. L., Wolfgang, M. C., et al. (2016). Vfr directly activates exsA transcription to regulate expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 198, 1442–1450. doi: 10.1128/JB.00049-16
Murat, D., Bance, P., Callebaut, I., and Dassa, E. (2006). ATP hydrolysis is essential for the function of the Uup ATP-binding cassette ATPase in precise excision of transposons. J. Biol. Chem. 281, 6850–6859. doi: 10.1074/jbc.M509926200
Murat, D., Goncalves, L., and Dassa, E. (2008). Deletion of the Escherichia coliuup gene encoding a protein of the ATP binding cassette superfamily affects bacterial competitiveness. Res. Microbiol. 159, 671–677. doi: 10.1016/j.resmic.2008.09.004
Reddy, M., and Gowrishankar, J. (1997). Identification and characterization of ssb and uup mutants with increased frequency of precise excision of transposon Tn10 derivatives: nucleotide sequence of uup in Escherichia coli. J. Bacteriol. 179, 2892–2899. doi: 10.1128/jb.179.9.2892-2899.1997
Richards, M. J., Edwards, J. R., Culver, D. H., and Gaynes, R. P. (2000). Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21, 510–515. doi: 10.1086/501795
Roy-Burman, A., Savel, R. H., Racine, S., Swanson, B. L., Revadigar, N. S., Fujimoto, J., et al. (2001). Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183, 1767–1774. doi: 10.1086/320737
Savli, H., Karadenizli, A., Kolayli, F., Gundes, S., Ozbek, U., and Vahaboglu, H. (2003). Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52, 403–408. doi: 10.1099/jmm.0.05132-0
Schweizer, H. P. (1993). Two plasmids, X1918 and Z1918, for easy recovery of the xylE and lacZ reporter genes. Gene 134, 89–91. doi: 10.1016/0378-1119(93)90178-6
Shyamala, V., Baichwal, V., Beall, E., and Ames, G. F. (1991). Structure-function analysis of the histidine permease and comparison with cystic fibrosis mutations. J. Biol. Chem. 266, 18714–18719.
Simon, R., Priefer, U., and Pühler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1, 784–791. doi: 10.1038/nbt1183-784
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Tyzack, J. K., Wang, X., Belsham, G. J., and Proud, C. G. (2000). ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J. Biol. Chem. 275, 34131–34139. doi: 10.1074/jbc.M002868200
Vallis, A. J., Yahr, T. L., Barbieri, J. T., and Frank, D. W. (1999). Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect. Immun. 67, 914–920.
Vance, R. E., Rietsch, A., and Mekalanos, J. J. (2005). Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect. Immun. 73, 1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005
Wolfgang, M. C., Lee, V. T., Gilmore, M. E., and Lory, S. (2003). Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4, 253–263. doi: 10.1016/S1534-5807(03)00019-4
Yahr, T. L., Vallis, A. J., Hancock, M. K., Barbieri, J. T., and Frank, D. W. (1998). ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. U.S.A. 95, 13899–13904. doi: 10.1073/pnas.95.23.13899
Keywords: Pseudomonas aeruginosa, type III secretion system, PA4595, ArtR, REG sub-family, gene regulation
Citation: Kong W, Dong M, Yan R, Liang Q, Zhang H, Luo W, Zhang Y, Liang H and Duan K (2019) A Unique ATPase, ArtR (PA4595), Represses the Type III Secretion System in Pseudomonas aeruginosa. Front. Microbiol. 10:560. doi: 10.3389/fmicb.2019.00560
Received: 10 October 2018; Accepted: 05 March 2019;
Published: 21 March 2019.
Edited by:
Ilana Kolodkin-Gal, Weizmann Institute of Science, IsraelReviewed by:
Marianna Patrauchan, Oklahoma State University, United StatesSathish Kumar, Stanford University, United States
Copyright © 2019 Kong, Dong, Yan, Liang, Zhang, Luo, Zhang, Liang and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kangmin Duan, Kangmin.Duan@umanitoba.ca
†These authors have contributed equally to this work
 Weina Kong
Weina Kong Mengmeng Dong1†
Mengmeng Dong1† Rong Yan
Rong Yan Wei Luo
Wei Luo Yani Zhang
Yani Zhang Haihua Liang
Haihua Liang Kangmin Duan
Kangmin Duan