- 1United States Department of Agriculture, Agricultural Research Service, National Center for Cool and Cold Water Aquaculture, Kearneysville, WV, United States
- 2Department of Chemistry and Biochemistry, University of Maryland, Baltimore County, Baltimore, MD, United States
Little is known about the underlying basis of serotype specificity among strains of Flavobacterium psychrophilum, the agent of rainbow trout fry syndrome and bacterial cold-water disease. The identification of different heat-stable O-serotypes among strains of this gram-negative pathogen does, however, suggest structural variations in the O-polysaccharide (O-PS) moiety of cell surface lipopolysaccharide (LPS). A trisaccharide composed of L-rhamnose (L-Rha), 2-acetamido-2-deoxy-L-fucose (L-FucNAc) and 2-acetamido-4-R-2,4-dideoxy-D-quinovose (D-Qui2NAc4NR), where R represents a dihydroxyhexanamido derivative, was previously identified as the repeating unit of Fp CSF259-93 O-PS. Interestingly, the O-PS gene cluster of this strain and that of Fp 950106-1/1, which belongs to a different O-serotype, are identical except for wzy, which encodes the putative polymerase that links trisaccharide repeats into O-PS chains. We have now found from results of glycosyl composition analysis and high-resolution nuclear magnetic resonance, that the linkage of D-Qui2NAc4NR to L-Rha, which is α1-2 for Fp CSF259-93 versus β1-3 for Fp 950106-1/1, is the only structural difference between O-PS from these strains. The corresponding difference in O-serotype specificity was established from the reactions of rabbit and trout anti-F. psychrophilum antibody with purified O-PS and LPS. Moreover, LPS-based differences in antigenicity were noted between strains with O-PS loci identical to those of Fp CSF259-93 or Fp 950106-1/1, except for the genes predicted to direct synthesis of different R-groups in Qui2NAc4NR. The findings provide a framework for defining the genetic basis of O-PS structure and antigenicity and suggest that the repertoire of F. psychrophilum O-serotypes extends beyond what is presently recognized from serological studies of this important fish pathogen.
Introduction
Flavobacterium psychrophilum, the agent of rainbow trout fry syndrome and bacterial cold-water disease, poses a serious threat to the salmonid aquaculture industry (Cipriano and Holt, 2005; Starliper, 2011). This threat is not from a single pathogenic entity but instead from several different host-specific serotypes (Mata et al., 2002; Izumi et al., 2003) and a wide range of multilocus sequence types (ST) (Nicolas et al., 2008). Accordingly, isolates of F. psychrophilum from diseased rainbow trout (Oncorhynchus mykiss) are most often identified as members of serotypes Fd or Th (Lorenzen and Olesen, 1997) and associated with STs that comprise clonal complex ST2/10 (Rochat et al., 2017) whereas other serotypes and STs are more frequently isolated from different species of salmon (Van Vliet et al., 2016) or non-salmonids such as ayu (Plecoglossus altivelis) (Fujiwara-Nagata et al., 2013). Different pathogen serotypes and STs are also commonly identified from the same outbreak of disease as well as from the same infected fish (Fujiwara-Nagata et al., 2013; Van Vliet et al., 2016; Ngo et al., 2017). Such outbreaks may play an important role in the ongoing evolution of F. psychrophilum by favoring the emergence of recombinants (Duchaud et al., 2018), such as those with altered serotypes. A genetic scheme for serotyping F. psychrophilum is needed to explore this possibility as well as other potential mechanisms of pathogenesis, host genetic resistance and immune protection.
The association of F. psychrophilum serotype specificity with heat stable O-antigens (Wakabayashi et al., 1994) suggests structural variations in the O-PS moiety of cell surface LPS. Previous structural characterization of F. psychrophilum (Fp) CSF259-93 O-PS, isolated following mild acid hydrolysis of LPS, revealed a trisaccharide repeating unit composed of L-Rha, L-FucNAc, and D-Qui2NAc4NR, where R is a 3,5-dihydroxyhexanoate derivative (MacLean et al., 2001). The corresponding locus for O-PS biosynthesis in this strain (Wiens et al., 2014) and related loci in 34 other strains of F. psychrophilum (Rochat et al., 2017) were tentatively identified by the presence of genes for synthesis of nucleotide-linked sugar precursors, glycosyltransferases and other proteins involved in polysaccharide biosynthesis. Importantly, those strains identified as serotype Fd or Th were also identified and distinguished by an allelic pair of genes for different putative O-antigen polymerases (Rochat et al., 2017). Variability in other O-PS genes, most notably those predicted to direct O-antigen R-group biosynthesis, was noted, however, among members of each serotype. Thus, F. psychrophilum serotypes Fd and Th, although clearly associated with different wzy alleles, were not associated with specific O-PS loci.
Synthesis of Fp CSF259-93 O-Ps by the well-studied Wzx/Wzy-dependent pathway (Islam and Lam, 2014) is expected to involve formation of the lipid-linked trisaccharide repeating unit on the inner surface of the cytoplasmic membrane, translocation of the saccharide moiety across the membrane by the action of a membrane-associated flippase (Wzx) and subsequent end-to-end polymerization of trisaccharide repeats by the action of a membrane-associated polymerase (Wzy). That the O-PS loci of Fp CSF259-93 and strains such as Fp 950106-1/1 (Rochat et al., 2017) are identical except for wzy suggests that the corresponding difference in O-PS structure is limited to the linkage between trisaccharide repeats. To test this hypothesis, we determined the structures of O-PS from these strains and antigenically compared each O-PS and LPS. In addition, we antigenically compared LPS from strains with O-PS loci that were either genetically identical to those of Fp 950106-1/1 or Fp CSF259-93 or non-identical with respect to the putative genes for synthesis of different R-groups in Qui2NAc4NR. The results support the notion that each genetically distinct O-PS locus is associated with a different LPS serotype.
Materials and Methods
Bacteria
The strains of F. psychrophilum used in the present study (Table 1) were stored as frozen stocks, cultured on plates of tryptone yeast extract salts (TYES) agar for 5 days at 15°C and handled using Biosafety Level 2 procedures as approved by the USDA/ARS North Atlantic Area Institutional Biosafety Committee. Genomic DNA was isolated using a cetyltrimethylammonium bromide (CTAB)/phenol-chloroform/ isoamyl alcohol procedure as described (Wilson, 2001) with minor modifications. Modifications included a final concentration of 170 μg/ml proteinase K during cell lysis and an additional RNase A treatment prior to final isopropanol precipitation of nucleic acids. Draft genome sequencing (∼90× coverage) was performed by The Sequencing Center (Fort Collins, CO, United States). DNA libraries were prepared using the Nextera XT Library Kit and sequenced using the MiniSeq System (Illumina) and the MiniSeq Reagent Kit, Mid Output (2 × 150 = 300 cycles, pair-end reads). Sequence data were assembled using default de novo assembly settings in Geneious software (v 11.1.2, Biomatters Ltd., New Zealand). GenBank accession numbers of annotated O-PS loci are listed in Table 1.
Lipopolysaccharide Preparation
Bacteria (8–16 gm wet weight) were harvested from TYES plates, washed twice by centrifugation in 20 mM Tris-Cl buffer (pH 7.5) containing 20 mm MgCl2, suspended to 20% (weight/vol.), and stored at −20°C prior to phenol-water extraction. Suspensions were thawed, supplemented with 100 μg/ml Proteinase K (Epicenter), 80 μg/ml RNase A (Qiagen), and 6 units/ml DNase I (Thermo Scientific), immediately disrupted using a Branson Sonifier Cell Disruptor 350 and incubated 1 h at 50°C to allow enzyme digestion. Digests were brought to 70°C and mixed with an equal volume of saturated phenol solution (pH 6.6) at the same temperature. The mixture was stirred vigorously for approximately 12 min, cooled on ice to 10°C and centrifuged to separate the phases, which were recovered and dialyzed against tap water followed by deionized water. The dialyzed phenol phase, which contained LPS as previously noted (LaFrentz et al., 2007), was concentrated by ultrafiltration above an Ultracel® 100 kDa membrane (EMD Millipore) and subjected to a second cycle of proteinase K and RNase A digestion, phenol-water extraction, dialysis and ultrafiltration as described above. Yields of phenol-phase LPS from Fp 950106-1/1 and Fp CSF259-93 were approximately 3 and 6 mg, respectively, per gram wet cells. Phenol phase LPS was hydrolyzed as described below to prepare O-PS or was further purified by HIC, which was performed following previously described protocols (Muck et al., 1999; Ren et al., 2012). For HIC, approximately 8 mg phenol-phase LPS was applied to columns (2.5 cm × 28 cm) of Butyl Sepharose 4 Fast Flow (GE Healthcare) in 0.7 M sodium acetate buffer (pH 4.5). Columns were rinsed with two column volumes of 0.7 M sodium acetate buffer to remove contaminating nucleic acid, two column volumes of 10% n-propanol to remove sodium acetate buffer, two column volumes of 30% n-propanol to elute bound LPS and two column volumes of water to remove n-propanol. The 30% n-propanol and water eluates were pooled, concentrated above a 100 kDa membrane, dialyzed against deionized water and stored at 4°C in the presence of a small drop of chloroform, which was added as preservative.
O-Polysaccharide Preparation
Phenol-phase LPS (approximately 20 mg) was hydrolyzed as previously described (MacLean et al., 2001) in 6% glacial acetic acid for 2 h at 100°C to cleave O-PS from core-lipid A (Shaw, 1993). The hydrolysate was frozen at −78°C, thawed, and centrifuged (30 min × 20,000 g) at 4°C to pellet insoluble core-lipid A. The O-PS-containing supernatant was harvested, brought to pH 4.5 by addition of 3 M NaHAC buffer to a final concentration of 0.7 M and applied to a column (2.5 cm × 14 cm) of Butyl Sepharose 4 Fast Flow. O-PS passed through the column in 0.7 M NaHAC buffer and was recovered, concentrated to 6 ml above an Ultracel® 10 kDa Ultrafiltration membrane (EMD Millipore) and applied to a column (2.5 cm × 66 cm) of Sephacryl S-100 High Resolution (GE Healthcare) equilibrated with 20 mM Tris-buffered (pH 7.6) saline containing 0.02% sodium azide. O-PS emerged near the void volume of the column and was recovered, concentrated by ultrafiltration above a 10 kDa membrane, dialyzed against water to remove salt and lyophilized.
Biochemical Methods
The phenol-sulfuric acid assay (Masuko et al., 2005) for total carbohydrate was performed with L-rhamnose as standard. Values from this assay, which does not detect N-acetylated sugars, accounted for approximately 30% of the dry weight of purified LPS. Glycosyl composition analysis of O-PS samples (100–200 μg) was performed at the University of Georgia, Complex Carbohydrate Research Center by combined gas chromatography/mass spectrometry of the alditol acetates as described (Pena et al., 2012). Dried sample containing 20 μg of inositol as internal standard was hydrolyzed in 2 M trifluoroacetic acid (TFA) for 2 h in a sealed tube at 121°C, reduced with NaBD4, and acetylated using acetic anhydride/TFA. The resulting alditol acetates were analyzed on an Agilent 7890A GC interfaced to a 5975C MSD, electron impact ionization mode. Separation was performed on a 30 m Supelco SP-2331 bonded phase fused silica capillary column. A standard set of sugars was run to quantify detected monosaccharides. In addition, a sample of Pseudomonas sp. LPS containing bacillosamine was included to accurately identify the retention time of this residue.
NMR Spectroscopy
Nuclear magnetic resonance was performed following methods adapted from those used in studies of pneumococcal polysaccharides (Geno et al., 2017). O-PS samples (6 mg) were lyophilized twice from 99.8% D2O and dissolved in 99.996% D2O. NMR spectra were recorded at 55°C in Bruker Advance spectrometers running Topspin 3 software at 500 and 600 MHz. All proton and carbon chemical shifts were referenced relative to internal acetone using δ 1H = 2.225 ppm and δ 13C = 31.07 ppm. Multiplicity-edited HSQC was used to distinguish methylene from methine groups. The common homonuclear two-dimensional NMR methods of DQF-COSY, TOCSY and NOESY were augmented by the hybrid method HSQC-TOCSY, which was enhanced in the crowded carbohydrate spectra by high digital resolution in the indirect dimension (13C). HMBC and HSQMBC spectra were used to identify linkage positions and for residue assignments. All NMR data were processed by NMRpipe and NMRDraw (NMRScience) with analysis by NMRview (One Moon Scientific).
Rabbit and Trout Anti-F. psychrophilum Sera
Flavobacterium psychrophilum CSF259-93 or 950106-1/1 cells were harvested from TYES plates, washed three times with PBS, suspended in buffer, adjusted to an optical density of approximately 1.2 at 525 nm and stored at 4°C for a few days in the presence of 0.5% formalin, prepared by dilution of 10% Neutral Buffered Formalin (Thermo Fisher Scientific). Cell suspensions were shipped to Pacific Immunology for immunization of two rabbits per immunogen following the 13-week Antibody Production Protocol. Each animal received four subcutaneous injections of immunogen (0.25 ml for primary immunization followed by three 0.15 ml boosters) administered with an equal volume of Freund’s incomplete adjuvant. Rabbit antisera selected for study were those that gave the strongest cross reactions with heterologous LPS in immunodiffusion.
Year class 2017 ARS-Fp-R line rainbow trout (Leeds et al., 2010; Wiens et al., 2013) were maintained at the NCCCWA following Institutional Animal Care and Use Committee (Leetown, WV, United States) protocols (#98 and #139). Twenty fish per group, as part of a larger study, were immunized by intraperitoneal injection of 0.1 ml containing 80 μl immunogen (i.e., the washed Fp CSF259-93 or Fp 950106-1/1 cell suspensions described above) or PBS administered with 20% EMULSIGEN® (MVP adjuvants) as recommended by the manufacture. Each fish (mean body weight 162 g) received a single injection and was bled 33 days later (422 temperature degree days). Studies were conducted with pooled antisera against each immunogen prepared from the 20 fish in each group.
Immunological Methods
Dot blots were prepared by spotting serial dilutions of LPS or O-PS samples (1 μl/spot) on 0.45 μ HATF08250 membranes. Membranes were dried overnight, blocked 1 h in 20 mM Tris buffered saline (pH 7.5) containing 0.1% Tween 20 and 2% non-fat dry milk (blocking buffer), incubated 1 h with dilute (generally 1/15,000) rabbit anti-F. psychrophilum antiserum and 1 h with peroxidase-conjugated goat anti-rabbit Ig (Bio-Rad) prior to development using a Pierce DAB Substrate Kit (Thermo Fisher Scientific). The approximate sensitivity of this assay was 2 ng/ml for F. psychrophilum LPS and 25 ng/ml for O-PS. Immunodiffusion was performed in 1% agarose gel cast on GelBond®Film (Lonza) in sodium barbital buffered saline (pH 7.4) containing 0.1% Tween 20. Immunodiffusion wells were filled with rabbit anti-Fp 950106-1/1 antiserum, anti-Fp CSF259-93 antiserum that was concentrated fourfold using a Microcon-10 kDa Centrifugal Filter Unit (EMD Millipore) or solutions of O-PS or LPS at approximately 0.5 mg/ml. Gels were incubated overnight at 4°C to allow immunoprecipitation, soaked in buffer followed by water to remove soluble protein and salt, dried and stained (Crowle and Cline, 1977). Western blots were prepared from 12% acrylamide gels in pH 8.3 Tris glycine buffer containing 0.1% SDS that were transferred to nitrocellulose in Tris-glycine transfer buffer (pH 8.3) containing 20% methanol. Transfers were blocked overnight at 4°C in KPL Milk Diluent/Blocking solution (SeraCare), incubated 1 h with equivalent dilutions of rabbit anti-Fp CSF259-93 or anti-Fp 950106-1/1 antisera followed by 1 h with peroxidase-conjugated goat anti-rabbit Ig (Bio-Rad) prior to development with a Pierce DAB Substrate Kit.
ELISA was performed with Costar 3590 ELISA plates (Corning) coated overnight at 4°C with 50 ng/ml Fp CSF259-93 LPS or Fp 950106-1/1 LPS in Carbonate-Bicarbonate coating buffer (Sigma). LPS-coated plates were washed three times with TBS containing 0.1% Tween 20 (TBS-T20) immediately prior to use. ELISA inhibition reactions were set up in TBS containing 0.5% BSA (TBS-BSA) in U-bottom plates (120 μl/well) by adding rabbit anti-Fp CSF259-93 or anti-Fp 950106-1/1 antiserum, at a dilution sufficient for an OD450 of approximately 1.5 in ELISA, to serial twofold dilutions of LPS inhibitors. Dilutions of inhibitor were prepared from stock solutions of LPS at concentrations that ranged from 100 to 500 μg/ml, as determined from results of at least three phenol sulfuric acid assays (see above). Control reaction mixtures containing antibody alone or no antibody (for 0 and 100% inhibition of ELISA, respectively) were also included. Following overnight incubation at 4°C, reaction and control mixtures were transferred (100 μl/well) to a washed LPS-coated ELISA plate for 1 h incubation at room temperature. The ELISA plate was washed with TBS-T20, incubated 1 h with peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad) in TBS-BSA, washed and developed 30 min with KPL SureBlue TMB peroxidase substrate (SeraCare). Measurements of OD450 were made with an Epoch 2 Microplate reader (BioTec). Concentrations of LPS for 50% inhibition of ELISA ± standard errors were estimated from results of at least two independent experiments.
ELISA inhibition of rainbow trout antibody binding was performed as described above by incubating LPS-coated ELISA plates 1 h with reaction mixtures setup with equivalent dilutions (approximately 1/2500) of trout anti-Fp 950106-1/1 or anti-Fp CSF259-93 antiserum and decreasing amounts of LPS, 1 h with 0.1 μg/ml Warr’s 1–14 monoclonal antibody (DeLuca et al., 1983) against trout IgM and 1 h with peroxidase-labeled, affinity purified goat anti-mouse IgG (H+L) (KPL) prior to development with SureBlue TMB peroxidase substrate.
Results
Purification of F. psychrophilum LPS and O-PS
Following phenol-water extraction of F. psychrophilum cell lysates, virtually all antigenic activity (>95%) detected by dot immunoblotting was associated with LPS recovered from the dialyzed phenol phase. Contaminating nucleic acids, which accounted for 3–25% of the carbohydrate in LPS solutions, were readily removed by HIC performed with columns of Butyl-Sepharose. The capacity of such columns was, however, relatively low (i.e., approximately 50 μg F. psychrophilum LPS/ml gel), which prompted the use of phenol phase LPS, rather than HIC-purified LPS, as starting material for the preparation of O-PS. This shortcut proved useful as mild acid hydrolysis to cleave O-PS from core-Lipid A also cleaved acid-labile phosphodiester bonds, thereby allowing complete removal of contaminating nucleic acid during subsequent purification of O-PS. Starting with approximately 20 mg of phenol phase LPS, we obtained 8 mg Fp 950106-1/1 O-PS and 9 mg Fp CSF259-93 O-PS.
Structural Characterization of F. psychrophilum O-PS
The results from glycosyl composition analysis of Fp CSF259-93 O-PS and Fp 960106-1/1 O-PS were essentially identical. Each O-PS yielded prominent peaks of rhamnose and N-acetylfucosamine, in roughly equal proportions, along with a very small peak of bacillosamine (Supplementary Figure S1). Speculating that the low recovery of bacillosamine from Qui2NAc4NR, which was expected from previous studies of Fp CSF259-93 O-PS (MacLean et al., 2001), could be due to failure of amide hydrolysis, we attempted stronger acidic hydrolysis (6 N HCl for 4 h at 100°C) of O-PS samples as well as saponification with 4 M KOH for 4 h. Neither treatment improved the yield of bacillosamine. The presence of Qui2NAc4NR in both Fp CSF259-93 and Fp 960106 O-PS was, however, clearly established by NMR as described below.
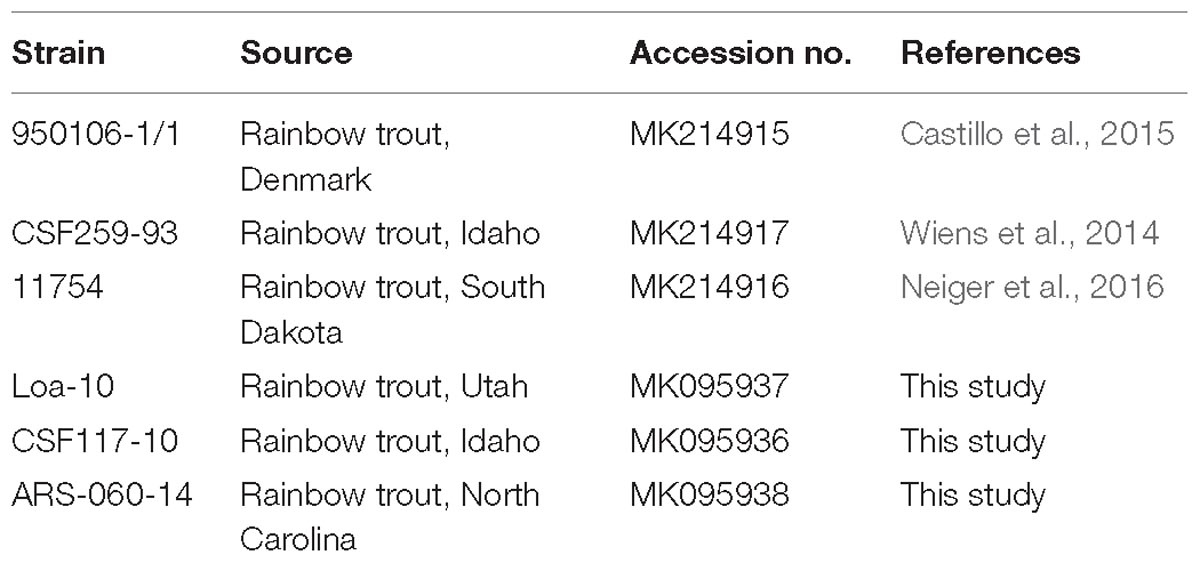
The 1H-13C HSQC spectrum of Fp CSF259-93 O-PS (Figure 1A) was independently assigned using the same letter designations (A, B, and C) for the three sugar residues as those used previously (MacLean et al., 2001). Starting from each anomeric C-H resonance, we used scalar coupling correlation by DQF-COSY to locate each H2 and HSQC for each C2. Using HSQC-TOCSY we identified the resonances of atoms 2, 3, 4, and 5 in combination with long-range 1H-13C scalar coupling by HMBC. Given the assignment of the C-H resonances of atoms 1 through 5 of each residue, the methyl groups of the 6-deoxy sugars were assigned by HSQC-TOCSY. Despite small differences 1H-13C chemical shifts, which presumably result from small differences in temperature and chemical shift referencing, our NMR assignment for Fp CSF259-93 O-PS (Table 1) is in complete agreement with the assignment of MacLean et al. (2001). Likewise, the previously determined glycosidic linkage positions of Fp CSF259-93 O-PS are the same as those determined by long-range 1H-13C coupling correlations in the present study.
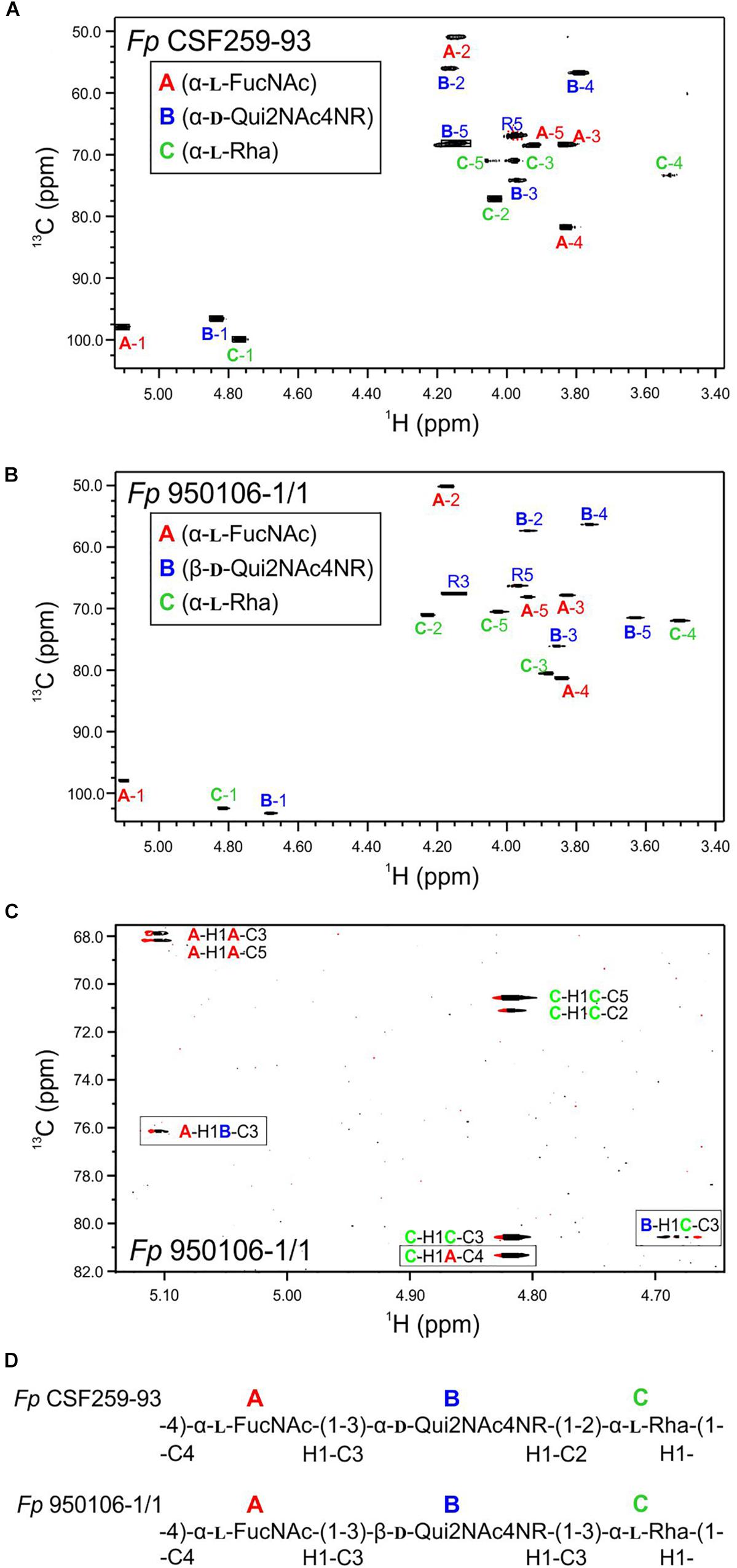
Figure 1. Multiplicity edited 1H-13C HSQC spectra recorded at 55°C: (A) Fp CSF259-93 O-PS; (B) Fp 950106-1/1 O-PS. (C) Phase-sensitive HSQMBC spectrum of Fp 950106-1/1 O-PS showing both intra- and inter-residue 1H-13C cross peaks; inter-residue cross peaks are identified by rectangles. (D) Structure of each O-PS deduced from 1H-13C HSQC and HMBC or phase-sensitive HSQMBC spectra.
As noted for Fp CSF259-93 O-PS, the 1H-13C HSQC spectrum of Fp 950106-1/1 O-PS (Figure 1B) contained three peaks in the anomeric region, between 5.2 and 4.7 ppm in 1H and 103 to 97 ppm in 13C, thereby indicating the presence of three sugar residues in each polysaccharide. Although not shown in Figure 1, three peaks indicative of methyl groups of 6-deoxy sugars were also present in the region of 1.3–1.2 ppm in 1H and 16–18 ppm in 13C of both polymers (Table 1 and Supplementary Figure S2). Despite these similarities, differences between the spectra indicated two distinct structures.
To determine the distinct structure of Fp 950106-1/1 O-PS, we arbitrarily assigned the letters A, B, and C to the anomeric signals of this polysaccharide (Figure 1B). The anomeric signal of residue A at 5.106 and 97.79 ppm was correlated with A-H2 at 4.176 ppm with JH−1,H−2 = 3 Hz, which along with 1JC−H = 174 Hz indicates and α-configuration for residue A. A-H2 was also correlated with A-C2 at 50.20 ppm, a chemical shift typical of a 2-acetamido sugar. The assignments of A-3, A-4, and A-5 were identified by HSQC-TOCSY correlation with A-H1 and by long range C-H correlation with A-C3 and A-C5 (Figure 1C). The NMR data indicate that residue A of Fp 950106-1/1 O-PS, like that of Fp CSF259-93 O-PS, is α-FucNAc. Moreover, the signals from this residue in Figures 1A,B, which are labeled in red, were similar for the two polysaccharides.
A sharp anomeric resonance of residue C at 4.818 ppm in 1H and 102.48 ppm in 13C were correlated by DQF-COSY (Supplementary Figure S3) with C-H2 at 4.232 ppm, which in turn was correlated with C-C2 at 71.07 ppm. HSQC-TOCSY (Supplementary Figure S2) from C-H1 showed cross peaks with C-C2 and a very weak cross peak with C-C3 at 80.57 ppm. The DQF-COSY cross peak with C-H2 showed a small homonuclear coupling to C-H3 at 3.885 ppm, indicating that residue C is the rhamnose residue detected in the glycosyl composition analysis. HSQC-TOCSY from C-C3 and DQF-COSY identify C-H4 at 3.503 ppm and C-H5 at 4.027 ppm. The α-anomeric configuration of residue C (Rha) was indicated by 1JC−H = 169 Hz in coupled HSQC-TOCSY.
The anomeric resonance B-1 at 4.679 ppm in 1H and 103.28 in 13C shows a large homonuclear splitting (JH−1,H−2 = 7.5 Hz) and 1JCH = 162.5 Hz. These data both show that, in contrast to the case of Fp CSF259-93, Residue B in Fp 150106-1/1 O-PS has a β-anomeric configuration. B-H1 in the NMR spectrum of Fp 950106-1/1 O-PS was correlated with B-H2 at 3.939 ppm and HSQC showed that B-C2 is at 57.37 ppm for this acetamido sugar residue, as expected for Qui2NAc4NR, the third residue of Fp 950106-1/1 O-PS. The axial configuration of all the ring protons leads to HSQC-TOCSY correlation of B-H1 not only with B-C2 but also with B-C3 (76.14 ppm), B-C4 (56.34 ppm) and B-C5 (71.52 ppm). The corresponding 1H chemical shifts were assigned by DQF-COSY and HSQC. The resonances of all the 6-deoxy sugars were assigned by HSQC-TOCSY cross peaks between the well resolved 13C6 resonances with the 1H resonances of the sugar rings.
In addition to the assignment of the NMR signals of the sugar residues, we assigned the resonances of the 3,5-dihydroxyhexanoyl R-group in O-PS of both Fp CSF259-93 and Fp 950106-1/1. The assignment of R-group NMR spectra began at the terminal methyl group, R-6 at 1.212 ppm in 1H and 22.93 ppm in 13C. HSQC-TOCSY showed correlation between R-H6 and R-C5 at 66.34 ppm. We further observed HSQC-TOCSY correlation of R-C5 to the methylene group R-4 having R-H4 at 1.617 ppm and R-H4’ at 1.738 ppm. R-C4 was located by HSQC at 45.98 ppm. HSQC-TOCSY correlated R-H4 and 4′ to R-C3 at 67.60 ppm and DQF-COSY showed the correlation of R-H3, R-H4 and R-H4′. HSQC-TOCSY identified R-C2 at 44.83 ppm with R-H2 at 2.415 ppm.
To establish the amide linkage of the 3,5-dihydroxyhexanoyl group to the O-PS, we first assigned the amide linkages of the two N-Acetyl amino sugars to residues A and B. The acetyl methyl groups appear at 1.963 ppm in 1H and 23.01 ppm in 13C and at 2.053 ppm in 1H and 23.14 ppm in 13C. We used the carbonyl-selective HMBC experiment employing a soft 13C pulse in the 175 ppm region to detect 3-bond long-range 1H-13C coupling correlation between amide methyl protons and the carbonyl 13C atoms of the amide linkage. A cross peak observed between 2.059 ppm and 175.24 ppm was accompanied by a second cross peak at 4.176 ppm corresponding to H2 of residue A (α-FucNAc). A second cross peak seen between 1.963 ppm and 174.93 ppm had a further correlation to 3.934 ppm recognized as H2 of residue B (Qui2NAc4NR). There was a third group of cross peaks in carbonyl-selective HMBC at 175.01 ppm and 2.419 ppm, the chemical shift of the R-H2 protons of the hexanoyl amide linkage to the O-PS as well as to 3.764 ppm, which corresponds to B-H4, the 4N linkage position of the hexanoyl group in Qui2NAc4NR.
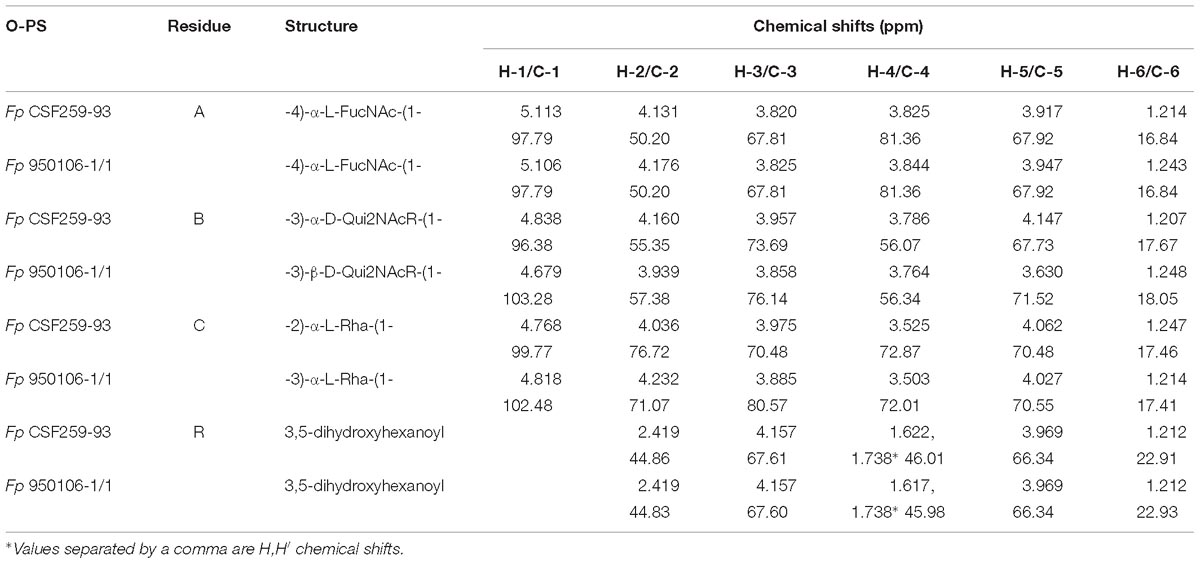
Table 2. Residue by residue comparison of HSQC 1H and 13C chemical shifts of Fp CSF259-95 and Fp 950106-1/1 O-PS.
Given the complete 1H and 13C assignments of each sugar residue (Table 2), saccharide linkages were readily determined from 1H-13C long range coupling correlations from phase-sensitive HSQMBC spectra. In the spectrum of Fp 950106 1/1 O-PS (Figure 1C), sugar linkage positions were indicated by a cross peak between A-H1 and B-C3, between B-H1 and C-C3 and between C-H1 and A-C4. In agreement with previous results of MacLean et al. (2001), the corresponding cross peaks in a long-range 1H-13C correlation spectrum of Fp CSF259-93 were between A-H1 and B-C3, between B-H1 and C-C2 and between C-H1 and A-C4 (Figure 1D). These findings indicate that the O-PS structures of Fp CSF259-93 and Fp 960106-1/1 are identical except for the linkage of D-Qui2NAc4NR to L-Rha, which is α1-2 for the former as opposed to β1-3 for the latter polysaccharide (Figure 1D).
Antigenic Characterization of F. psychrophilum O-PS and LPS
Immunodiffusion experiments performed with rabbit antisera against Fp 960106-1/1 and Fp CSF259-93 (Figure 2, wells a or b, respectively) indicated partial antigenic identity between O-PS from these strains (Figure 2A, red numbers) and complete identity between each O-PS and corresponding LPS (Figure 2A, black numbers). In similar experiments, reactions of complete identity were noted between LPS from strains with identical O-PS gene clusters (Figure 2B, wells 1 and 2; Figure 2C, wells 5 and 6) and partial identity between LPS from strains that contained the same wzy (i.e., wzy1 or wzy2) but different putative R-group genes (Figure 2B, compare well 3 to wells 1 and 2, Figure 2C, compare well 4 to wells 5 and 6). In one case, partial identity was also noted between homologous and heterologous LPS from strains that differed in both wzy and R-group genes (Figure 2C, well 3) while in the other case, the heterologous LPS failed to cross react (Figure 2B, well 4).
Figure 2. Immunodiffusion of Fp O-antigens: (A) comparison of O-PS and LPS of Fp 950106-1/1 and Fp CSF259-93; (B) comparison of LPS of Fp 950106-1/1 with LPS of other Fp strains; (C) comparison of LPS of Fp CSF259-93 with LPS of other Fp strains. Center wells contained (a) anti-Fp 950106-1/1 serum or (b) anti-Fp CSF259-93 serum. Outer wells contained (1) Fp 950106-1/1 LPS, (red 1) Fp 950106-1/1 O-PS, (2) Fp 11754 LPS, (3) Fp CSF117-10 LPS, (4) Fp ARS-060-14 LPS, (5) Fp Loa-10 LPS, (6) Fp CSF259-93 LPS or (red 6) Fp CSF259-93 O-PS. Genes that distinguish the O-PS loci of different Fp strains are labeled in panel (D).
Obvious differences in the immunoreactivity of LPS from different strains were also noted on Western transfers of LPS overlaid with equivalent dilutions of rabbit antiserum against Fp 960106-1/1 or Fp CSF259-93 (Figure 3). Thus, labeling of LPS on a transfer incubated with rabbit anti-Fp 950106-1/1 antiserum diluted 1/16,000 was much stronger for Fp 950106-1/1 and other strains that carried wzy1 (Figure 3A, lanes 1, 2 and 3) than for strains that carried wzy2 (lanes 4, 5 and 6). Reduced labeling of LPS due to differences in R-group genes (Figure 2D) was not seen among the former strains (compare lane 3 to lanes 1 and 2) but was seen among the later strains (compare lane 4 to lanes 5 and 6). Compared with Figure 3A, the opposite pattern of labeling appeared on an identical transfer overlaid with anti-Fp CSF259-93 serum diluted 1/400 (Figure 3B). This antiserum, in addition to labeling high molecular weight LPS, labeled a rapidly migrating band (Figure 3B) in the region expected for core-Lipid A (MacLachlan et al., 1993). Weak labeling of a band in the same region, although not present in Figure 3A, was noted on an identical transfer overlaid with a 1/400 dilution of rabbit anti-Fp 950106-1/1 antiserum (Supplementary Figure S4).
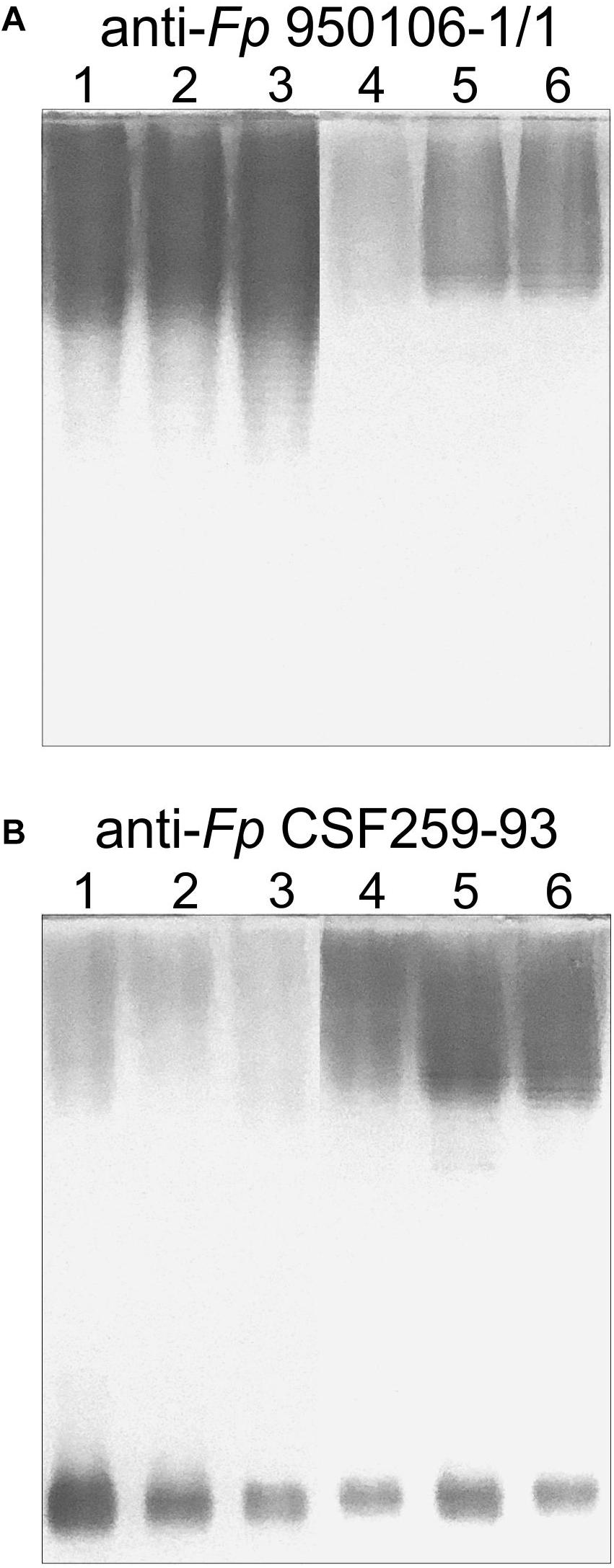
Figure 3. Western blots of LPS (0.5 μg/lane) developed with equivalent dilutions of (A) anti-Fp 950106-1/1 serum or (B) anti-Fp CSF259-93 serum. Number designations for LPS of different Fp strains are the same as in Figure 2: (1) Fp 950106-1/1; (2) Fp 11754; (3) Fp CSF117-10; (4) Fp ARS-060-14; (5) Fp Loa-10; and (6) Fp CSF259-93.
The importance of wzy as a genetic determinant of both rabbit and rainbow trout antibody specificity was evident from results of ELISA inhibition experiments performed with LPS from different strains (Table 3). Thus, concentrations of homologous LPS (i.e., LPS from either Fp 950106-1/1 and Fp 11754 or Fp CSF259-93 and Fp Loa-10) for 50% inhibition of either rabbit or rainbow trout antibody binding were in the 1–10 ng/ml range whereas concentrations of heterologous LPS (i.e., LPS from strains that differed only in wzy) for comparable inhibition were in the 2000–3000 ng/ml range for inhibition of rabbit antibody binding and greater than 50,000 ng/ml for inhibition of trout antibody binding (Table 3). In contrast with wzy, the contribution of R-group genes to immunological specificity varied between antisera. Thus, low concentrations of Fp CSF117-10 LPS (number 3 in Table 3 and Figure 2D) completely inhibited rainbow trout anti-Fp 950106-1/1 antibody binding whereas this LPS at concentrations that ranged from 10 to 5000 ng/ml only partially inhibited (approximately 60%) rabbit anti-Fp 950106-1/1 antibody binding. Similarly, intermediate concentrations of Fp ARS-060-10 LPS (number 4 in Table 3 and Figure 2D) completely inhibited rabbit anti-Fp CSF259-93 antibody binding while this LPS at concentrations that ranged from 10 to 50,000 ng/ml only partially inhibited (approximately 40%) rainbow trout anti-Fp CSF259-93 antibody binding (Table 3). Thus, the association of LPS antigenicity with R-group genes was evident from reactions of rainbow trout as well as rabbit antibodies.
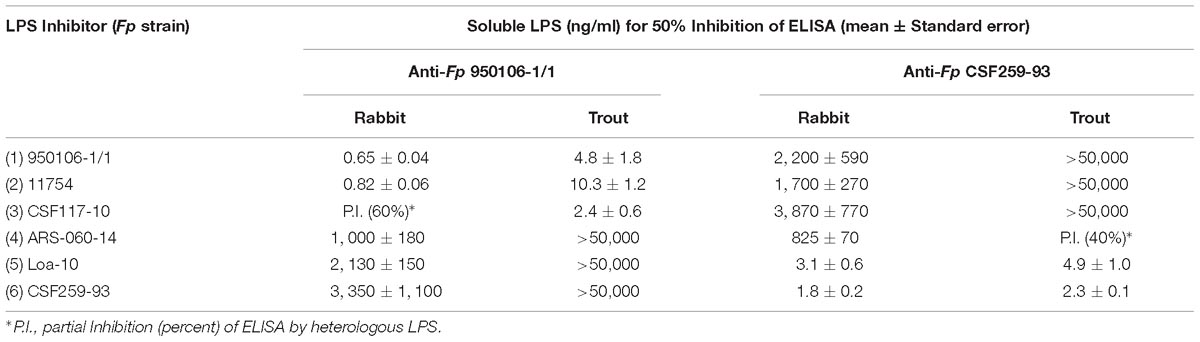
Table 3. ELISA Inhibition of rabbit and rainbow trout anti-Fp antibody binding to homologous LPS by LPS of different Fp strains.
Discussion
The present findings provide a framework for defining the genetic basis of O-PS structure and antigenicity. To facilitate discussion, gene names have been assigned to the 19 ORFs in the O-PS gene cluster of Fp 950106-1/1 (Figure 4) in accordance with recommendations of the Bacterial Polysaccharide Gene Database (Reeves et al., 1996), using established designations for certain pathway genes and wfp∗ designations for O-PS specific genes. That the O-PS structures of Fp 950106-1/1 and Fp CSF259-93 are identical except for the linkage of D-Qui2NAc4NR to L-Rha (Figure 1C) supports the presence of different O-antigen polymerases (i.e., Wzy1 and Wzy2) in serotype Fd and Th strains of F. psychrophilum (Rochat et al., 2017). Studies to genetically alter O-PS structure are underway to further establish the identity of wzy and obtain isogenic strains for studies of pathogenesis and immune protection. The presence of D-Qui2NAc4NR1 at the reducing end of putative O-PS biological repeating unit (Figure 4) indicates that the transfer of this residue to carrier lipid is the initial step in O-PS synthesis. This transfer can be attributed to wfpB, which encodes a homolog of PglC (Supplementary Table S1), the N,N′-diacetylbacillosamine-phosphotransferase of C. jejuni (Glover et al., 2006). Interestingly, the corresponding genes of Fp CSF117-10 and Fp ARS-060-14 (i.e., those designated wfpB) encode proteins that are virtually identical to WfpB of Fp 950106-1/1 except for the presence of distinct C-terminal sequences. Further studies are needed to determine whether WfpB of each former strain is tailored to a specific R-group (i.e., R2 or R3, respectively) in D-Qui2NAc4NR. The two remaining Fp 950106-1/1 genes for glycosyltransferases encode homologs of Escherichia coli O26 WbuA and WbuB (D’Souza et al., 2002). The WbuB homolog of F. psychrophilum has a different acceptor than WbuB of E. coli (i.e., D-Qui2NAc4NR vs. D-GlcNAc) and thus, has been designated WfpA in Figure 4. The WbuA designation has been retained, however, for the α-L-Rha transferase of F. psychrophilum as it has the same donor and acceptor as WbuA of E. coli. Following the WbuA-mediated transfer of α-L-Rha in F. psychrophilum, the lipid-linked trisaccharide is presumably flipped to the outer surface of the cytoplasmic membrane for β1-3 polymerization by Wzy1 of Fp 950106-1/1 (Figure 4) or α1-2 polymerization by Wzy2 of Fp CSF259-93.
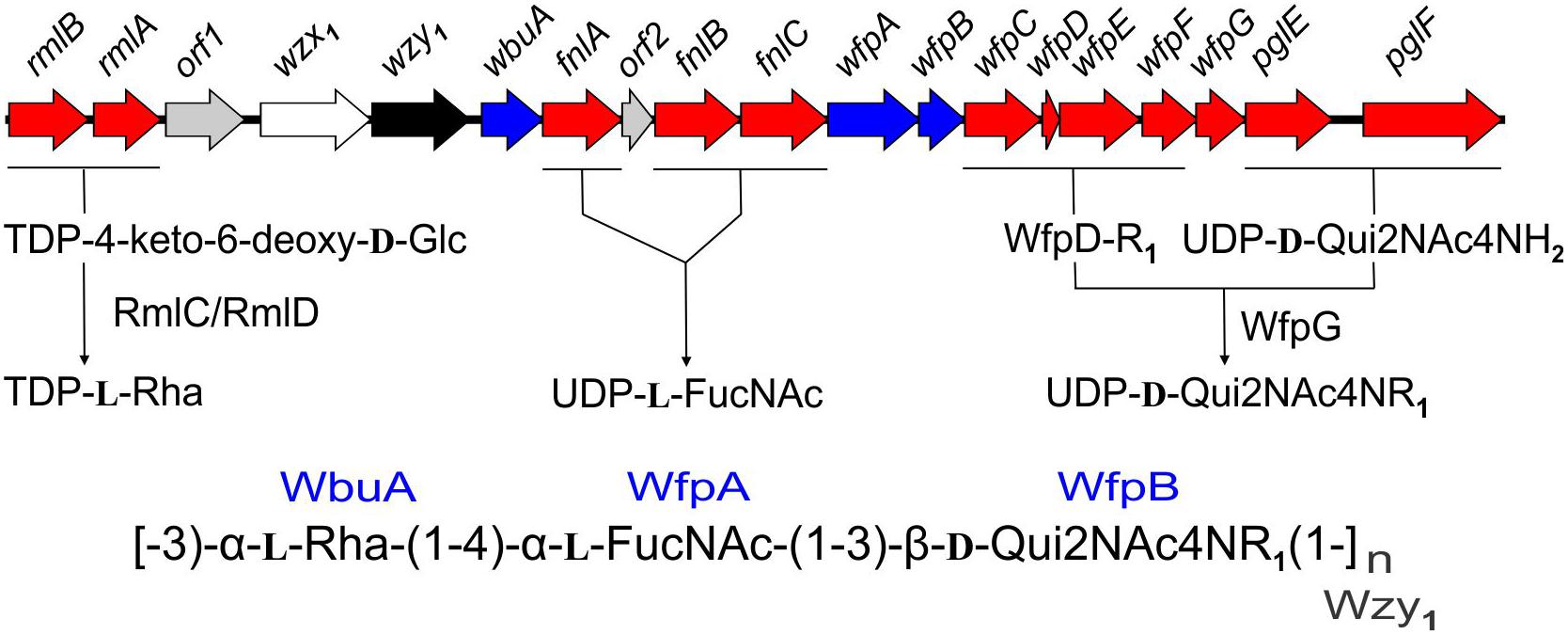
Figure 4. Proposed genetic basis of Fp 950106-1/1 O-PS structure showing genes for nucleotide sugar biosynthesis (red), glycosyl transferases (blue), polymerase (black), flippase (white), and genes of unknown function (gray), which are given orf designations.
The O-PS gene clusters of Fp 950106-1/1 (Figure 4) and Fp CSF259-93 share several genes for synthesis of nucleotide sugar precursors, including two of the four rml genes for conversion of α-D-Glc 1-phosphate to TDP-L-Rha (Samuel and Reeves, 2003); the other two rml genes (i.e., rmlC and rmlD) occur elsewhere in the chromosome. The O-PS gene cluster of Fp 950106-1/1 also contains three fln genes for conversion of UDP-D-GlcNAc to UDP-L-FucNAc and pglE and pglF for conversion of UDP-D-GlcNAc to UDP-D-Qui2NAc4NH2 (Kneidinger et al., 2003; Figure 4). In C. jejuni, which carries pglE and pglF, the additional presence of pglD for an acetyltransferase that N-acetylates UDP-D-Qui2NAc4NH2 results in formation of UDP-D-Qui2NAc4NAc (i.e., N,N′-diacetylbacillosamine) (Olivier et al., 2006). Similarly, the pglD-homolog designated wfpG in F. psychrophilum (Supplementary Table S1) encodes an acyltransferase that is predicted to transfer the R1 group from a putative O-PS-specific, acyl carrier protein encoded by wfpD to UDP-D-Qui2NAc4NH2 to form UDP-D-Qui2NAc4NR1 (Figure 4). The presence of an encoded acyl carrier protein clearly suggests synthesis of the R1-group (i.e., 3,5-dihydroxyhexanoyl) by a pathway that resembles type II fatty acid synthesis (Rock and Jackowski, 2002). Additional support for this possibility comes from the presence of adjacent wfpC and wfpE for different 3-ketoacyl-ACP synthases and wfpF for a 3-ketoacyl-ACP reductase (Supplementary Table S1). Fp CSF117-10 and Fp ARS-060-14 both lack genes for synthesis of the R1-group but each has a pglD homolog (wfpH or wfpI in Figure 2D and Supplementary Table S1) for an acyltransferase that is predicted to N-acylate UDP-D-Qui2NAc4NH2 to form UDP-D-Qui2NAc4NR2 or UDP-D-Qui2NAc4NR3, respectively. Structural characterization of O-PS from these strains is underway to identify the R-group in each polysaccharide.
Following phenol-water extraction, we recovered F. psychrophilum LPS from the phenol phase whereas MacLean et al. (2001) recovered F. psychrophilum LPS from the clear phenol-water solution obtained by dilution of 50% hot phenol. Regardless of this difference, O-PS isolated in the present study has the same structure as that proposed previously. It is important to note, however, that while the presently determined structure is based solely on glycolyl composition (Supplementary Figure S1) and NMR data (Figure 1, Supplementary Figures S2, S3, and Table 2), the structure of MacLean et al. (2001) was derived from extensive characterization of the 3,5 dihydroxyhexanoyl R-group using HF solvolysis, methanolysis, chemical synthesis, mild acid hydrolysis, mass spectrometry and NMR spectroscopy and optical rotation to determine the absolute configuration of each sugar residue and the chiral centers in the 3,5 dihydroxy R-group. Comparable studies of Fp 950106-1/1 O-PS were not performed in the present study. We assume, however, that the structure of the R-group and the absolute configuration of sugar residues in this polysaccharide are the same as in Fp CSF259-93 O-PS since the O-PS loci of these strains are the same except for wzy.
The association of F. psychrophilum serotypes Fd and Th with the presence of wzy1 or wzy2, respectively, in the O-PS gene clusters of different strains is based on the use of serotyping reagents prepared against presently characterized Fp 950106-1, which is serotype Fd, and Fp DK002, which is serotype Th (Rochat et al., 2017). Importantly, the O-PS gene cluster of the later strain is identical to that of Fp CSF259-93. Based on the present findings, cross adsorption of antiserum against one strain with cells of the other is expected to remove antibodies against common epitopes, including those associated with R1-groups, and leave antibodies against specific epitopes associated with the wzy-dependent linkages that distinguish each O-PS. The present findings (Figures 2, 3 and Table 3) not only support the importance of wzy1 and wzy2 as genetic determinants of serological specificity but also show the influence of R-group genes. Thus, while antigenic identity was noted between LPS from strains with identical O-PS gene clusters, non-identity was seen between LPS from strains that differed in R-group genes (Figures 2B,C). The contribution of R-group genes to differences in LPS antigenicity was also evident in ELISA inhibition experiments performed with both rabbit antisera and with one of two pooled rainbow trout antisera (Table 3). Considered together, these findings indicate that the reactions of anti-Fd and anti-Th typing sera are serogroup (rather than serotype) specific and suggest that each genetically distinct O-PS locus is associated with a different LPS serotype. The latter suggestion is not without precedent. Indeed, the validity of genetically defined serotypes is well established from studies of several other pathogens, most notably the pneumococcus, where each of the over 90 capsular polysaccharide (CPS) serotypes identified by factor antisera (Henrichsen, 1995) has been associated with a distinct cps locus (Bentley et al., 2006). In the case of F. psychrophilum, comparison of O-PS loci from approximately 60 whole genome sequences suggests 17 different LPS serotypes, 15 of which have been described (Rochat et al., 2017).
The present and previous findings (Rochat et al., 2017) provide a firm basis for development of a comprehensive genetic scheme for serotyping F. psychrophilum based on serogroups that differ in wzy and serotypes on other genes that influence O-PS structure, such as those for R-group synthesis. Development of such a scheme would not reduce the need for further immunological studies, but instead would serve to focus such studies on antigenic characterization and comparison of genetically defined LPS serotypes as well as on the ability of different serotypes to afford cross protection in vaccine studies or cause disease in studies of host genetic resistance. Widely available PCR-based methods for accurate and comparable genetic serotyping of F. psychrophilum would also contribute to improved disease surveillance and thereby facilitate identification and tracking of virulent serotypes as well as novel serotypes that might arise by recombination during outbreaks of disease.
Ethics Statement
Animal studies were performed at Pacific Immunology following an approved animal protocol or at the NCCCWA following Institutional Animal Care and Use Committee (Leetown, WV, United States) protocols #98 and #139.
Author Contributions
JC and GW designed the study. JC isolated the LPS and O-PS, performed the immunological studies, and interpreted the genetic data. CB performed the NMR studies. GW was responsible for DNA sequencing and preparation of antisera. JC and CB drafted the manuscript. All authors edited and approved the final manuscript.
Funding
This work was supported by United States Department of Agriculture (USDA) Agricultural Research Service CRIS Project 1930-32000-006 “Integrated Research to Improve On-Farm Animal Health in Salmonid Aquaculture.” Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity employer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Keira Osbourn for isolating bacterial genomic DNA, Ryan Lipscomb for assistance in Western blotting experiments, Tim Leeds for breeding ARS-Fp-R line rainbow trout, Travis Moreland for fish husbandry, Ian Black for glycosyl composition analysis, which was supported by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, U.S. Department of Energy grant (DE-SC0015662) to Parastoo Azadi at the Complex Carbohydrate Research Center, Erin Bromage and USDA NIFA #2010-65121-20649 for supplying the Warr 1-14 antibody, Doug Call for providing Fp 950106-1/1, Scott LaPatra for providing Fp CSF117-10 and Fp CSF259-93, Regg Neiger for providing Fp 11754, and NIDCR, NIH for IT support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01041/full#supplementary-material
Abbreviations
DQF-COSY, double quantum filtered coherence spectroscopy; Fp, Flavobacterium psychrophilum; HIC, hydrophobic interaction chromatography; HMBC, heteronuclear multiple bond correlation; HSQC, heteronuclear single quantum coherence; HSQMBC, heteronuclear single quantum multiple bond correlation; LPS, lipopolysaccharide; NMR, nuclear magnetic resonance; NOESY, nuclear Overhauser effect spectroscopy; O-PS, O-polysaccharide; TOCSY, total coherence spectroscopy.
References
Bentley, S. D., Aanensen, D. M., Mavroidi, A., Saunders, D., Rabbinowitsch, E., Collins, M., et al. (2006). Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. doi: 10.1371/journal.pgen.0020031
Castillo, D., Christiansen, R. H., Dalsgaard, I., Madsen, L., and Middelboe, M. (2015). Bacteriophage resistance mechanisms in the fish pathogen Flavobacterium psychrophilum: linking genomic mutations to changes in bacterial virulence factors. Appl. Environ. Microbiol. 81, 1157–1167. doi: 10.1128/AEM.03699-14
Cipriano, R. C., and Holt, R. A. (2005). Flavobacterium Psychrophilum, the Cause of Bacterial Cold-Water Disease and Rainbow Trout Fry Syndrome. Kearneysville, WV: National Fish Health Research Laboratory.
Crowle, A. J., and Cline, L. J. (1977). An improved stain for immunodiffusion tests. J. Immunol. Methods 17, 379–381.
DeLuca, D., Wilson, M., and Warr, G. W. (1983). Lymphocyte heterogeneity in the trout, Salmo gairdneri, defined with monoclonal antibodies to IgM. Eur. J. Immunol. 13, 546–551. doi: 10.1002/eji.1830130706
D’Souza, J. M., Wang, L., and Reeves, P. (2002). Sequence of the Escherichia coli O26 O antigen gene cluster and identification of O26 specific genes. Gene 297, 123–127.
Duchaud, E., Rochat, T., Habib, C., Barbier, P., Loux, V., Guerin, C., et al. (2018). Genomic diversity and evolution of the fish pathogen Flavobacterium psychrophilum. Front. Microbiol. 9:138. doi: 10.3389/fmicb.2018.00138
Fujiwara-Nagata, E., Chantry-Darmon, C., Bernardet, J. F., Eguchi, M., Duchaud, E., and Nicolas, P. (2013). Population structure of the fish pathogen Flavobacterium psychrophilum at whole-country and model river levels in Japan. Vet. Res. 44:34. doi: 10.1186/1297-9716-44-34
Geno, K. A., Bush, C. A., Wang, M., Jin, C., Nahm, M. H., and Yang, J. (2017). WciG O-acetyltransferase functionality differentiates pneumococcal serotypes 35C and 42. J. Clin. Microbiol. 55, 2775–2784. doi: 10.1128/JCM.00822-17
Glover, K. J., Weerapana, E., Chen, M. M., and Imperiali, B. (2006). Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 45, 5343–5350. doi: 10.1021/bi0602056
Henrichsen, J. (1995). Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33, 2759–2762.
Islam, S. T., and Lam, J. S. (2014). Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 60, 697–716. doi: 10.1139/cjm-2014-0595
Izumi, S., Liu, H., Aranishi, F., and Wakabayashi, H. (2003). A novel serotype of Flavobacterium psychrophilum detected using antiserum against an isolate from amago, Oncorhynchus masou rhodurus Jordan & Gilbert, in Japan. J. Fish Dis. 26, 677–680. doi: 10.1046/j.1365-2761.2003.00502.x
Kneidinger, B., O’Riordan, K., Li, J., Brisson, J. R., Lee, J. C., and Lam, J. S. (2003). Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-D-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5. Implications for the UDP-N-acetyl-L-fucosamine biosynthetic pathway. J. Biol. Chem. 278, 3615–3627. doi: 10.1074/jbc.M203867200
LaFrentz, B. R., Lindstrom, N. M., LaPatra, S. E., Call, D. R., and Cain, K. D. (2007). Electrophoretic and western blot analyses of the lipopolysaccharide and glycocalyx of Flavobacterium psychrophilum. Fish Shellfish Immunol. 23, 770–780. doi: 10.1016/j.fsi.2007.02.005
Leeds, T. D., Silverstein, J. T., Weber, G. M., Vallejo, R. L., Palti, Y., Rexroad, C. E., et al. (2010). Response to selection for bacterial cold water disease resistance in rainbow trout. J. Anim. Sci. 88, 1936–1946. doi: 10.2527/jas.2009-2538
Lorenzen, E., and Olesen, N. J. (1997). Characterization of isolates of Flavobacterium psychrophilum associated with coldwater disease or rainbow trout fry syndrome II: serological studies. Dis. Aquat. Organ. 31, 209–220. doi: 10.3354/dao031209
MacLachlan, P. R., Keenleyside, W. J., Dodgson, C., and Whitfield, C. (1993). Formation of the K30 (group I) capsule in Escherichia coli O9:K30 does not require attachment to lipopolysaccharide lipid A-core. J. Bacteriol. 175, 7515–7522.
MacLean, L. L., Vinogradov, E., Crump, E. M., Perry, M. B., and Kay, W. W. (2001). The structure of the lipopolysaccharide O-antigen produced by Flavobacterium psychrophilum (259-93). Eur. J. Biochem. 268, 2710–2716.
Masuko, T., Minami, A., Iwasaki, N., Majima, T., Nishimura, S., and Lee, Y. C. (2005). Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 339, 69–72. doi: 10.1016/j.ab.2004.12.001
Mata, M., Skarmeta, A., and Santos, Y. (2002). A proposed serotyping system for Flavobacterium psychrophilum. Lett. Appl. Microbiol. 35, 166–170. doi: 10.1046/j.1472-765X.2002.01157.x
Muck, A., Ramm, M., and Hamburger, M. (1999). Efficient method for preparation of highly purified lipopolysaccharides by hydrophobic interaction chromatography. J. Chromatogr. B Biomed. Sci. Appl. 732, 39–46.
Neiger, R., Thomas, M., Das, S., Barnes, M., Fletcher, B., Snekvik, K., et al. (2016). Draft Genome Sequences of three Flavobacterium psychrophilum strains isolated from coldwater disease outbreaks at three production hatcheries. Genome Announc. 4:e00035-16. doi: 10.1128/genomeA.00035-16
Ngo, T. P. H., Bartie, K. L., Thompson, K. D., Verner-Jeffreys, D. W., Hoare, R., and Adams, A. (2017). Genetic and serological diversity of Flavobacterium psychrophilum isolates from salmonids in United Kingdom. Vet. Microbiol. 201, 216–224. doi: 10.1016/j.vetmic.2017.01.032
Nicolas, P., Mondot, S., Achaz, G., Bouchenot, C., Bernardet, J. F., and Duchaud, E. (2008). Population structure of the fish-pathogenic bacterium Flavobacterium psychrophilum. Appl. Environ. Microbiol. 74, 3702–3709. doi: 10.1128/AEM.00244-08
Olivier, N. B., Chen, M. M., Behr, J. R., and Imperiali, B. (2006). In vitro biosynthesis of UDP-N,N’-diacetylbacillosamine by enzymes of the Campylobacter jejuni general protein glycosylation system. Biochemistry 45, 13659–13669. doi: 10.1021/bi061456h
Pena, M. J., Tuomivaara, S. T., Urbanowicz, B. R., O’Neill, M. A., and York, W. S. (2012). Methods for structural characterization of the products of cellulose- and xyloglucan-hydrolyzing enzymes. Methods Enzymol. 510, 121–139. doi: 10.1016/B978-0-12-415931-0.00007-0
Reeves, P. R., Hobbs, M., Valvano, M. A., Skurnik, M., Whitfield, C., Coplin, D., et al. (1996). Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4, 495–503.
Ren, K., Li, Y., Shi, F., and Wang, X. Y. (2012). Separation of lipopolysaccharides containing different fatty acid chains using hydrophobic interaction chromatography. Anal. Methods 4, 838–843. doi: 10.1039/c2ay05663a
Rochat, T., Fujiwara-Nagata, E., Calvez, S., Dalsgaard, I., Madsen, L., Calteau, A., et al. (2017). Genomic caracterization of Flavobacterium psychrophilum serotypes and development of a multiplex PCR-based serotyping scheme. Front. Microbiol. 8:1752. doi: 10.3389/fmicb.2017.01752
Rock, C. O., and Jackowski, S. (2002). Forty years of bacterial fatty acid synthesis. Biochem. Biophys. Res. Commun. 292, 1155–1166. doi: 10.1006/bbrc.2001.2022
Samuel, G., and Reeves, P. (2003). Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338, 2503–2519.
Shaw, D. H. (1993). “[3] Preparation of lipid A and polysaccharide from lipopolysaccharides,” in Methods in Carbohydrate Chemistry, eds J. N. BeMiller, R. L. Whistler, and D. H. Shaw (Hoboken, NJ: John Wiley & Sons, Inc),19–23.
Starliper, C. E. (2011). Bacterial coldwater disease of fishes caused by Flavobacterium psychrophilum. J. Adv. Res. 2, 97–108. doi: 10.1016/j.jare.2010.04.001
Van Vliet, D., Wiens, G. D., Loch, T. P., Nicolas, P., and Faisal, M. (2016). Genetic diversity of Flavobacterium psychrophilum isolates from three Oncorhynchus spp. in the United States, as revealed by multilocus sequence typing. Appl. Environ. Microbiol. 82, 3246–3255. doi: 10.1128/AEM.00411-16
Wakabayashi, H., Toyama, T., and Iida, T. (1994). A study on serotyping Cytophaga psychrophila isolated from fishes in Japan. Fish Pathol. 29, 101–104. doi: 10.3147/jsfp.29.101
Wiens, G. D., LaPatra, S. E., Welch, T. J., Evenhuis, J. P., Rexroad, C. E., and Leeds, T. D. (2013). On-farm performance of rainbow trout (Oncorhynchus mykiss) selectively bred for resistance to bacterial cold water disease: effect of rearing environment on survival phenotype. Aquaculture 388, 128–136. doi: 10.1016/j.aquaculture.2013.01.018
Wiens, G. D., LaPatra, S. E., Welch, T. J., Rexroad, C. III, Call, D. R., Cain, K. D., et al. (2014). Complete genome sequence of Flavobacterium psychrophilum strain CSF259-93, used to select rainbow trout for increased genetic resistance against bacterial cold water disease. Genome Announc. 2:e00889-14. doi: 10.1128/genomeA.00889-14
Keywords: Flavobacterium psychrophilum, fish pathogen, lipopolysaccharide, O-polysaccharide structure, O-polysaccharide genes, O-serotypes
Citation: Cisar JO, Bush CA and Wiens GD (2019) Comparative Structural and Antigenic Characterization of Genetically Distinct Flavobacterium psychrophilum O-Polysaccharides. Front. Microbiol. 10:1041. doi: 10.3389/fmicb.2019.01041
Received: 28 November 2018; Accepted: 25 April 2019;
Published: 08 May 2019.
Edited by:
Catherine Ayn Brissette, University of North Dakota, United StatesReviewed by:
Luiz R. Travassos, Federal University of São Paulo, BrazilBo Pang, Joint BioEnergy Institute and University of California, Berkeley, United States
Copyright © 2019 Cisar, Bush and Wiens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John O. Cisar, john.cisar@ars.usda.gov; john.cisar@nih.gov
 John O. Cisar
John O. Cisar C. Allen Bush2
C. Allen Bush2