- 1Department of Food Science, University of Copenhagen, Copenhagen, Denmark
- 2Department of Food Science and Technology, University of Energy and Natural Resources, Sunyani, Ghana
- 3Département Technologie Alimentaire, IRSAT/CNRST, Ouagadougou, Burkina Faso
- 4ESTCTPA, Université Nationale d’Agriculture, Ketou, Benin
Indigenous fermented food and beverages represent a valuable cultural heritage in sub-Saharan Africa, having one of the richest selections of fermented food products in the world. In many of these indigenous spontaneously fermented food and beverages, yeasts are of significant importance. Several factors including raw materials, processing methods, hygienic conditions as well as the interactions between yeasts and other commensal microorganisms have been shown to influence yeast species diversity and successions. Both at species and strain levels, successions take place due to the continuous change in intrinsic and extrinsic growth factors. The selection pressure from the microbial stress factors leads to niche adaptation and both yeast species and strains with traits deviating from those generally acknowledged in current taxonomic keys, have been isolated from indigenous sub-Saharan African fermented food products. Yeasts are important for flavor development, impact shelf life, and nutritional value and do, in some cases, even provide host-beneficial effects. In order to sustain and upgrade these traditional fermented products, it is quite important to obtain detailed knowledge on the microorganisms involved in the fermentations, their growth requirements and interactions. While other publications have reported on the occurrence of prokaryotes in spontaneously fermented sub-Saharan food and beverages, the present review focuses on yeasts considering their current taxonomic position, relative occurrence and successions, interactions with other commensal microorganisms as well as beneficial effects and importance in human diet. Additionally, the risk of opportunistic yeasts is discussed.
Yeasts as Contributors to Spontaneously Fermented Food and Beverages Produced in Sub-Saharan Africa
Fermentation is one of the oldest methodologies for food preservation and fermentation does therefore, globally play an important role in the processing of many indigenous food and beverages. Specifically, indigenous sub-Saharan Africa fermented food and beverages make up an import part of the daily diet in sub-Saharan African food culture, being of high nutritional value (Holzapfel, 2002). Fermented products are appreciated, not only due to the preservation and safety of these products but also because of their sensorial attributes (Holzapfel, 2002; Anukam and Reid, 2009). Moreover, several other beneficial effects of food fermentation have been reported including reduced loss of raw materials, reduced cooking time, prolonged shelf-life, enhanced bio-availability of micronutrients, and probiotic effects (Jespersen, 2003, 2005; Mufandaedza et al., 2006; Motlhanka et al., 2018). Most of these fermented products are produced in small-scale at small and medium-sized enterprises (SME) or at household level by spontaneous fermentation, sometimes including inoculation using back-slopping or repeated use of the same fermentation container (Holzapfel, 2002).
The most commonly used raw materials in indigenous sub-Saharan African fermented food and beverages are cereals, root tubers and legumes but other raw materials such as palm sap, milk, fruits, fish, and meat are also fermented (Jespersen, 2003; Ogunremi et al., 2017; Motlhanka et al., 2018). Indigenous fermented food and beverages are in general simple to process as they are typically based on a few ingredients and rely on simple processing methods (Marco et al., 2017). The nature of indigenous fermented food and beverages and the fact that they are easily accessible make these food systems ideal ecosystems for investigating the mechanisms of microbial community formation (Wolfe and Dutton, 2015). However, knowledge on the microbial ecology of indigenous sub-Saharan African fermented food and beverages lags behind.
The microorganisms involved in indigenous sub-Saharan African fermented food and beverages will predominantly originate from the raw materials and processing equipment as spontaneous fermentation or back-slopping is applied (Jespersen, 2003). The microbial consortium is complex, comprising a range of different microorganisms coexisting and interacting in many ways (Navarrete-Bolaños, 2012). Owing to their relatively high growth rate, lactic acid bacteria (LAB) will usually predominate the early stages of the fermentation, where after yeasts will increase and take part in the fermentation (Holzapfel, 2002).
The occurrence of yeasts in indigenous sub-Saharan African fermented food and beverage products has been studied for a range of end products. However, far fewer studies have examined the yeast dynamics during the fermentations. The yeast species dominating indigenous fermented food and beverages are those that are able to adapt to the changing intrinsic conditions caused by physicochemical changes, due to microbial activity (Navarrete-Bolaños, 2012). Species diversity is additionally influenced by different extrinsic factors related to the technological processing steps including length and temperature of fermentation, amount of water added, raw materials used, stirring, pasteurization as well as level of hygiene and sanitation (Jespersen, 2003; Achi and Ukwuru, 2015). Hence, a comprehensive understanding, linking intrinsic and extrinsic factors to microbial diversity and successions is of outmost importance for upgrading indigenous sub-Saharan African fermented food and beverages.
Several functional properties of yeasts have been reported for the processing of indigenous sub-Saharan African fermented food and beverages. These include fermentation of carbohydrates, flavor compound formation, stimulation of LAB, degradation of cyanogenic glycosides, production of tissue-degrading enzymes, binding and/or degradation of mycotoxins as well as probiotic properties (Jespersen, 2003, 2005; Omemu et al., 2007; Padonou et al., 2010; Achi and Ukwuru, 2015; Tamang et al., 2016). However, in most of the studied indigenous sub-Saharan African fermented food and beverages, the functional properties of the identified yeasts have not yet been extensively elucidated.
In general, information on yeasts in indigenous sub-Saharan African fermented food and beverages is very scattered, and though yeasts are considered very important for processing of local food and beverages, it is nearly impossible to get an overview on their identity, occurrence, impact and interactions. The aim of the present review is therefore to gather currently available scientific knowledge on yeasts involved in indigenous sub-Saharan African fermented food and beverages focusing on their current taxonomic position, species and strain diversity, microbial successions and interactions, as well as their impact on quality in terms of flavor, shelf life, safety and nutritional value. Reflections on health beneficial effects of yeasts, possible pathogenic traits as well as future technological improvements are likewise included. Although yeast species are also involved in the fermentation of cash crops such as coffee and cocoa (Masoud et al., 2004; Nielsen et al., 2005), these products are not included in the present survey.
Species Diversity and Strain Distribution
Taxonomic Methods for Identification of Yeasts
Methods used for identification of yeasts at species and strain levels can be classified roughly into two groups based on either phenotypic- or genotypic methodologies. Until recently, most publications dealing with yeasts in indigenous sub-Saharan African fermented food and beverages focused on identification based on phenotypic criteria. However, an enhanced availability of methods based on genotypic characterisation has changed the situation and current publications do to a greater extent base their identifications on genotypic methodologies. Below a brief overview on relevant methodologies for yeast identification is given.
Phenotypic methods include standard taxonomical tests, most recently described in the current taxonomic key edited by Kurtzman et al. (2011) as well as in a number of older publications such as the simplified identification method (SIM) described by Deak (2007). All of these methods are based on morphological and physiological characteristics (colony and cell morphology, growth conditions, assimilation and fermentation of carbohydrates as well as nitrogen compounds, osmotolerance, etc.). In the present survey, yeast species were reported for 43 indigenous sub-Saharan African fermented food and beverages, of which yeasts were identified by phenotypic approaches for 44% of the fermented products, as indicated in Tables 1–4. Identifications based only on phenotypic analyses can be doubtful as different species sometimes display very close morphological and physiological characteristics. Further, under certain conditions, strains might show different morphological and physiological traits, which deviate from the species description given in current taxonomic keys (Jespersen, 2003; Glover et al., 2005). While phenotypic characterisation provides useful information for a first grouping of isolates, it is nowadays recommended to include molecular identification analyses and vice versa (Kurtzman et al., 2015). Recently more advanced identification techniques based on phenotypic characteristics have been developed such as the matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (Pavlovic et al., 2014) and fourier transform infrared spectroscopy (FTIR) techniques (Wenning et al., 2002). However, these techniques have so far not been used for identification of yeasts from indigenous sub-Saharan African fermented food and beverages.
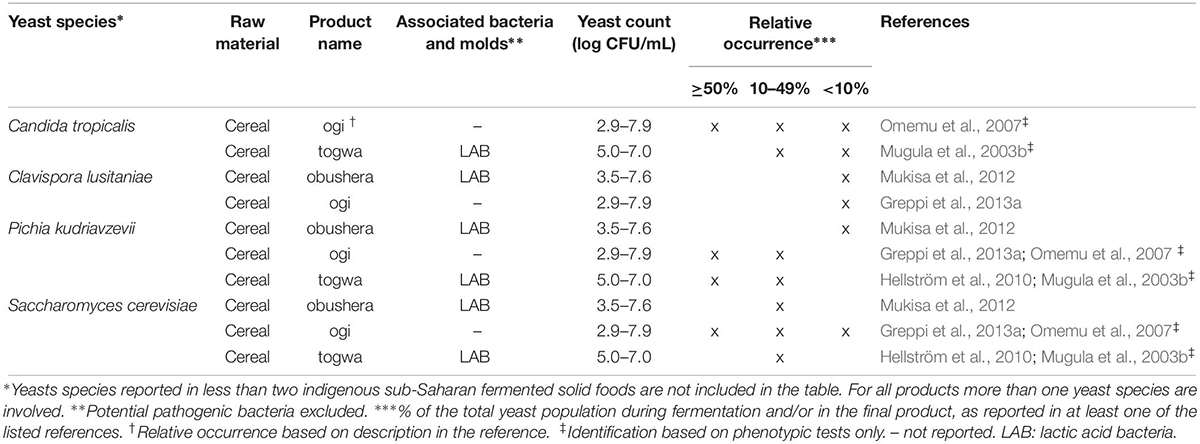
Table 2. Yeast species commonly identified in indigenous sub-Saharan African fermented non/low-alcoholic beverages.
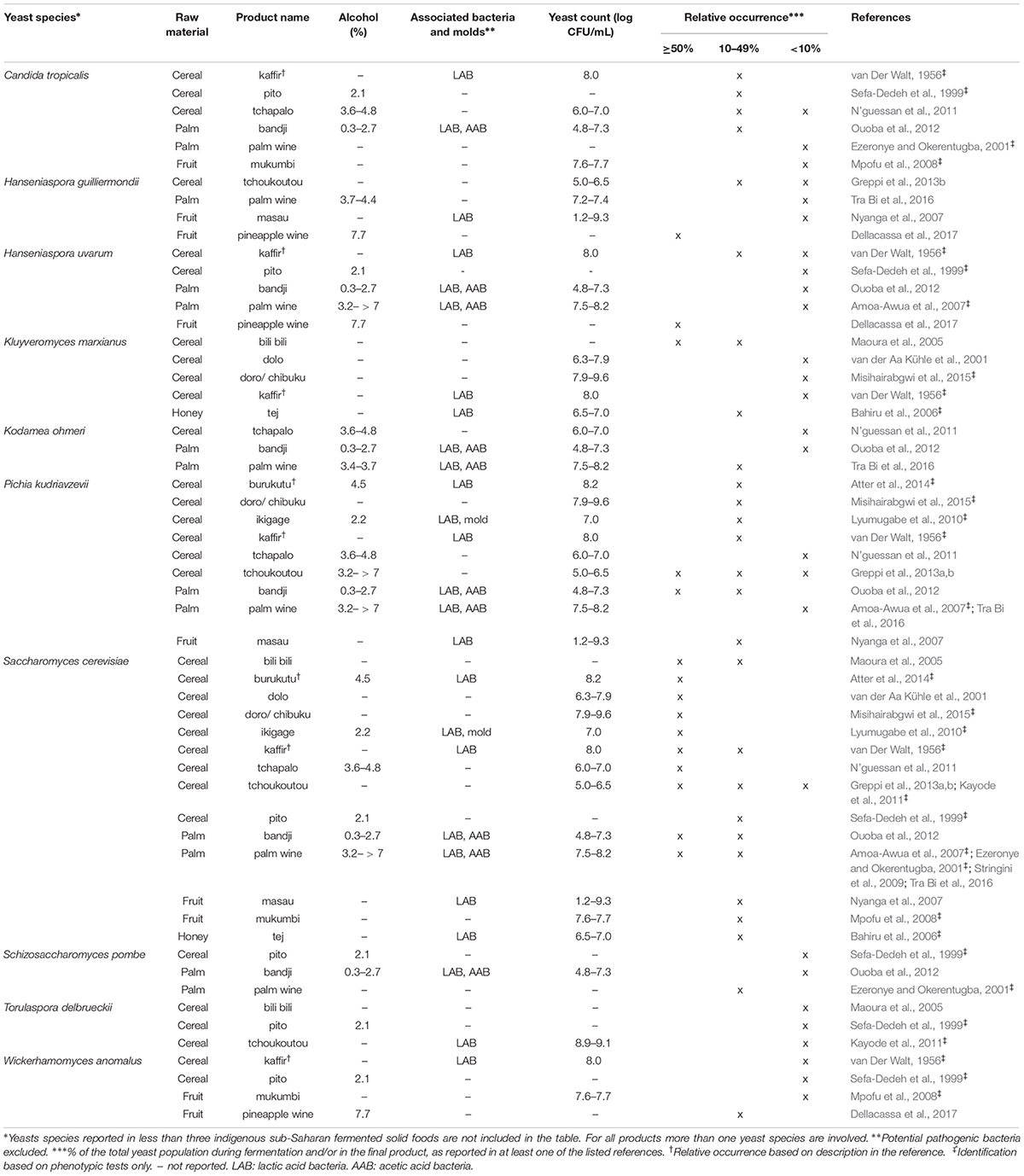
Table 3. Yeast species commonly identified in indigenoussub-Saharan African fermented alcoholic beverages.
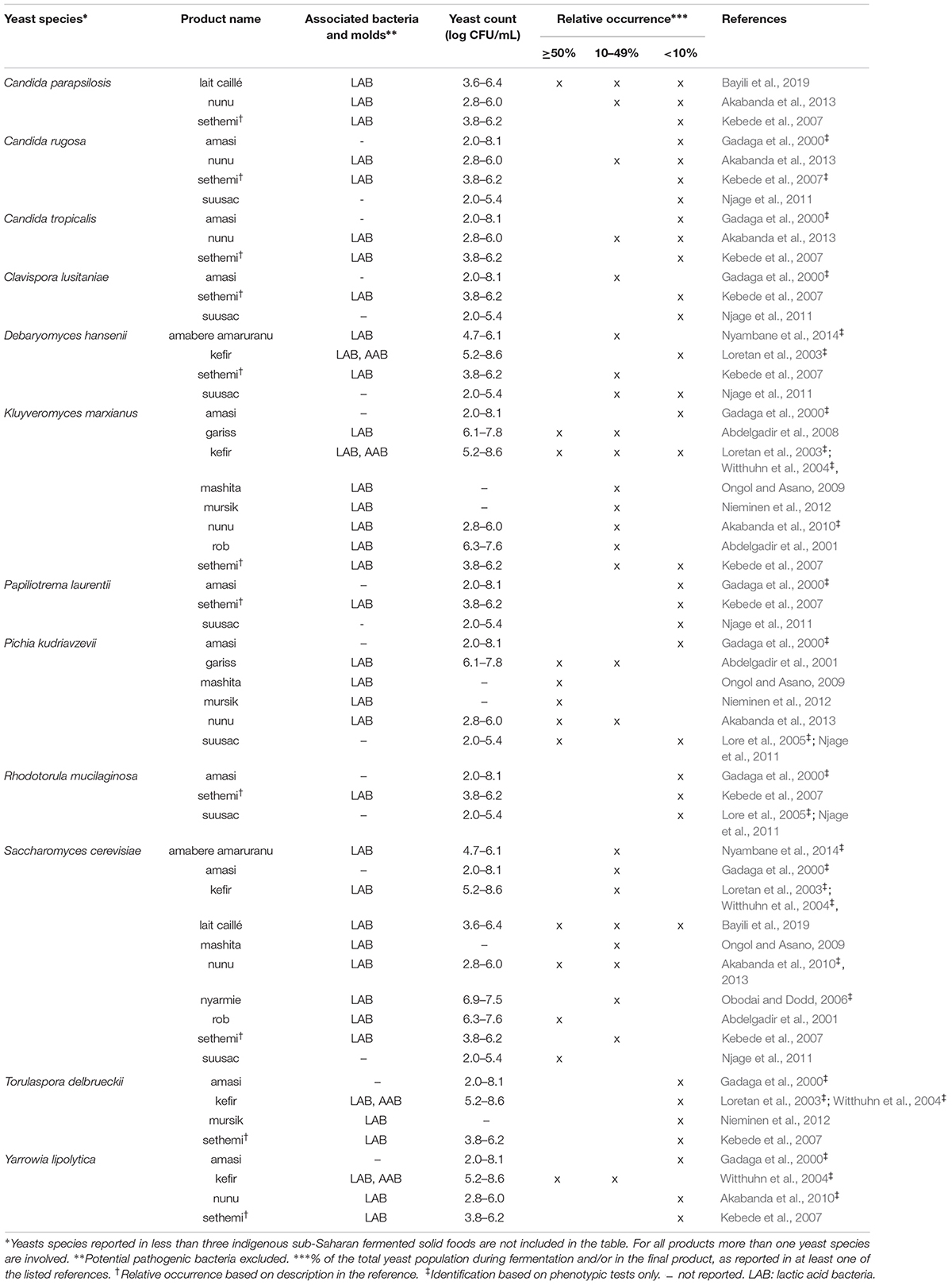
Table 4. Yeast species commonly identified in indigenous sub-Saharan African fermented dairy products.
Genotypic identification to species level is for yeasts to a great extent based on sequencing of the D1/D2 region of the 26S rRNA gene and to some extent based on sequencing of the internal transcribed spacer (ITS) regions (divided into ITS1 and ITS2 by the conserved 5.8S rRNA gene) (Jespersen, 2003; Daniel et al., 2014; Kurtzman et al., 2015). Genotypic identification to strain level is based on other specific molecular techniques including chromosome length polymorphism (CLP) analysis by pulse field gel electrophoresis (PFGE) (van der Aa Kühle et al., 2001; Glover et al., 2005; Maoura et al., 2005; Ezeronye and Legras, 2009), restriction fragment length polymorphism (RFLP) of the ITS regions using restriction enzymes such as HaeIII, EcoRI, HindIII, HinfI, DraI, HpaII, ScrFI, TaqI, etc. (Hayford and Jakobsen, 1999; van der Aa Kühle et al., 2001; Naumova et al., 2003; Glover et al., 2005; Maoura et al., 2005; Ezeronye and Legras, 2009; N’guessan et al., 2011), sequencing analyses of the ACT1 and/or COX2 genes (Glover et al., 2005; Ezeronye and Legras, 2009; N’guessan et al., 2011), microsatellite loci analyses (Naumova et al., 2003; Ezeronye and Legras, 2009; Stringini et al., 2009; Tapsoba et al., 2015; Tra Bi et al., 2016) as well as detection of specific loci such as the MAL loci distribution in Saccharomyces cerevisiae strains (van der Aa Kühle et al., 2001).
Yeast Species Diversity and Distribution
For each indigenous sub-Saharan African fermented food and beverage included in this review, the country of origin and raw material used can be found in Figure 1. Based on the current available literature, the fermented food and beverages have been classified into solid foods, non/low-alcoholic beverages, alcoholic beverages and fermented dairy products, in this review, even though it is acknowledged that other ways of classifying food fermentations exist (Steinkraus, 1997; Soro-Yao et al., 2014). The most frequently occurring yeast species are shown for the four groups in Tables 1–4, respectively. In order to use just one name for each yeast species, yeast species mentioned in Tables 1–4 and in the remaining parts of this review are only reported with their current taxonomic name, but mentioning the first time frequently reported former names, and for teleomorph (perfect) yeasts their anamorph (imperfect) name, if relevant. Differentiation between Hanseniaspora guilliermondii (anamorph Kloeckera apis) and Hanseniaspora opuntiae is not done, since these species cannot be distinguished by physiological criteria, and only the ITS region provide accurate identification as the D1/D2 region between the two species is highly conserved (Cadez et al., 2003).
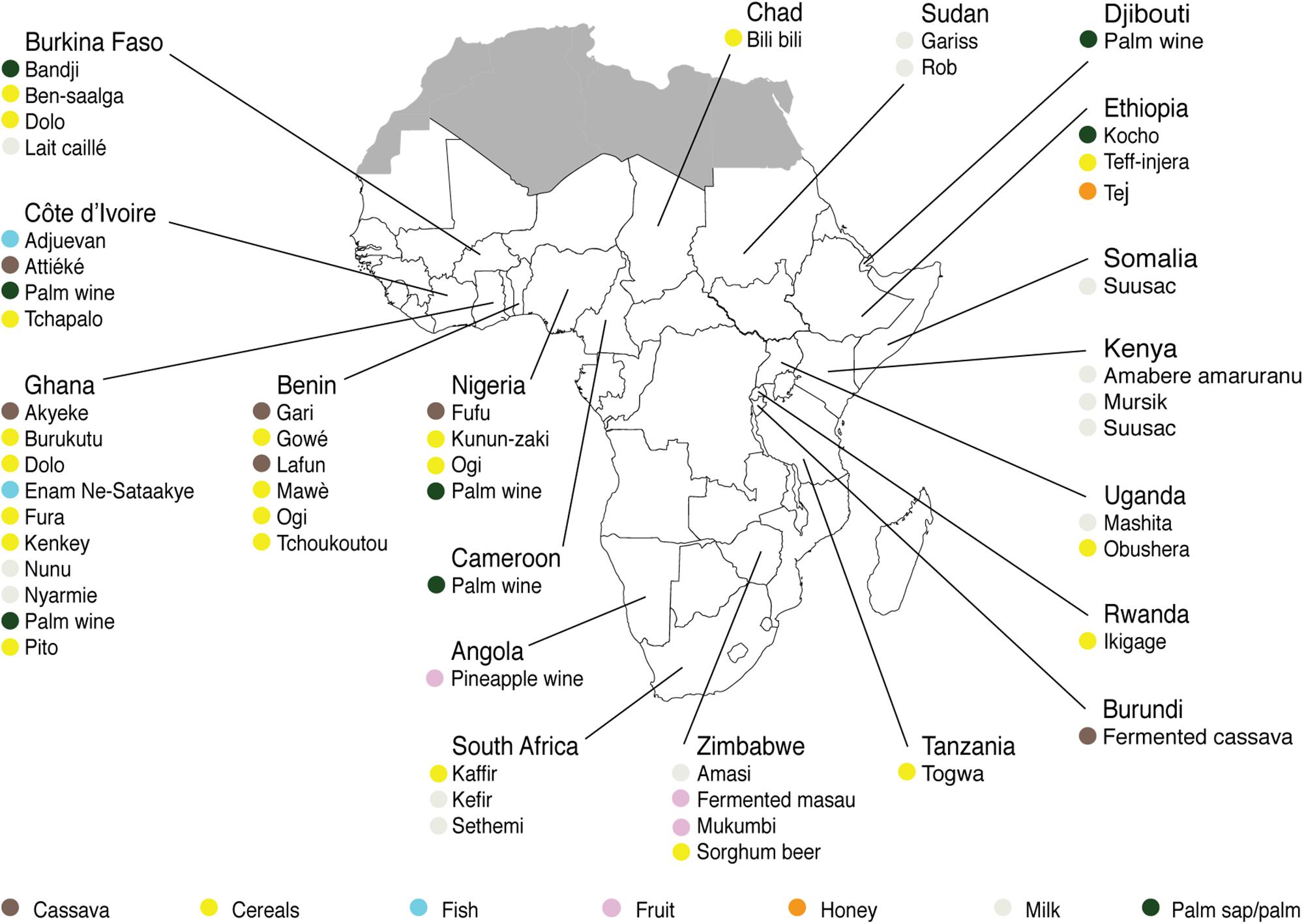
Figure 1. Origin and raw materials used for production of indigenous sub-Saharan African fermented food and beverages.
From the 43 indigenous sub-Saharan African fermented food and beverages investigated, a total of 98 different yeast species have been identified. S. cerevisiae was reported in 77% of the fermented products, being the predominant yeast species isolated, followed by Pichia kudriavzevii (f. Issatchenkia orientalis; anamorph Candida krusei) reported in 60%, Candida tropicalis in 47% and Kluyveromyces marxianus (anamorph Candida kefyr) in 44% of the fermented products.
Not surprisingly, S. cerevisiae is the most frequently occurring yeast across all indigenous sub-Saharan African fermented food and beverages, and moreover, reported as the dominant yeast species in many of the foods it is being isolated from. Hence, S. cerevisiae, plays a dominant role in fermentation of solid foods such as mawè (Greppi et al., 2013b) and kenkey (Halm et al., 1993; Table 1), as well as in the non/low-alcoholic beverage ogi (Omemu et al., 2007; Table 2). Expectedly, S. cerevisiae is the most frequently occurring yeast in alcoholic beverages and has been reported to dominate the fermentation in 93% of the indigenous sub-Saharan African alcoholic beverages. The majority of these alcoholic beverages are based on cereals (Table 3). However, S. cerevisiae also dominates in other alcoholic beverages such as palm wine based on sap (Amoa-Awua et al., 2007; Ezeronye and Legras, 2009; Stringini et al., 2009). S. cerevisiae has additionally been reported to dominate the fermentation of the fermented dairy products nunu (Akabanda et al., 2010, 2013), rob (Abdelgadir et al., 2001), and suusac (Njage et al., 2011; Table 4). In lower abundancies, S. cerevisiae has been reported in other indigenous sub-Saharan African fermented products such as the solid foods adjuevan (Clementine et al., 2012), fufu (Oyewole, 2001), and lafun (Padonou et al., 2009), and the non/low-alcoholic beverages obushera (Mukisa et al., 2012) and togwa (Mugula et al., 2003b; Hellström et al., 2010), as well as the fermented dairy products amabere amaruranu (Nyambane et al., 2014), amasi (Gadaga et al., 2000), kefir (Loretan et al., 2003; Witthuhn et al., 2004), mashita (Ongol and Asano, 2009), nyarmie (Obodai and Dodd, 2006) and sethemi (Kebede et al., 2007; Tables 1, 2, 4).
Interestingly, P. kudriavzevii quite often plays a significant role in the fermentation of indigenous sub-Saharan African food and beverages. Previous studies demonstrated that P. kudriavzevii dominates the fermentations of several solid fermented products such as fufu (Oyewole, 2001), fura (Pedersen et al., 2012), gowé (Greppi et al., 2013a), kenkey (Halm et al., 1993), and mawè (Greppi et al., 2013b; Houngbédji et al., 2018; Table 1). Likewise, P. kudriavzevii has been shown to dominate in the fermentation of non/low-alcoholic beverages such as ogi (Omemu et al., 2007) and togwa (Mugula et al., 2003b; Hellström et al., 2010; Table 2). In alcoholic cereal-based beverages P. kudriavzevii is present, though not being the dominant species, except in bandji (Ouoba et al., 2012) and tchoukoutou (Greppi et al., 2013a,b; Table 3). Further, P. kudriavzevii has been reported to dominate several fermented dairy products such as gariss (Abdelgadir et al., 2008), mashita (Ongol and Asano, 2009), mursik (Nieminen et al., 2012), nunu (Akabanda et al., 2013), and suusac (Lore et al., 2005; Table 4).
Likewise, C. tropicalis and K. marxianus are often found in indigenous sub-Saharan African fermented food and beverages. C. tropicalis is most frequently isolated from solid foods, being identified in 71% of the solid foods. C. tropicalis has been reported to dominate the fermentation of the solid foods akyeke (Obilie et al., 2003), attiéké (Coulin et al., 2006), fufu (Oyewole, 2001), and gowé (Greppi et al., 2013a). However, in most solid foods C. tropicalis does not dominate the fermentations (Table 1). C. tropicalis is not frequently associated with fermented dairy products and does only occur in low abundance in amasi (Gadaga et al., 2000), nunu (Gadaga et al., 2000; Akabanda et al., 2013), and sethemi (Kebede et al., 2007; Table 4). K. marxianus has been found to be dominant in the solid foods adjuevan (Clementine et al., 2012), gowé, (Greppi et al., 2013a), and mawè (Houngbédji et al., 2018; Table 1), as well as in the alcoholic beverage bili bili (Maoura et al., 2005; Table 3). Likewise, K. marxianus does mostly occur in fermented dairy products, being identified in 73% fermented dairy products, and is reported to dominate in the fermentation of the dairy products gariss (Abdelgadir et al., 2008), and kefir (Loretan et al., 2003; Witthuhn et al., 2004; Table 4). A novel yeast species, i.e., Hanseniaspora jakobsenii was isolated from bandji, a traditional fermented palm wine in Burkina Faso (Ouoba et al., 2015). The huge diversity of yeast species associated with indigenous sub-Saharan African fermented food and beverages underlines the high potential within the microbial heritage of these fermented food and beverages.
Successions of Yeasts During Fermentations
During spontaneous fermentation of food and beverages, a complex yeast population is often seen. The diversity and relative composition of the yeast population will usually vary along the fermentation and successions will take place (Fleet, 2007). The succession of yeast species depends on various intrinsic and extrinsic factors related to the food matrix including any microbial interactions. While certain species occur throughout the fermentation, other species might appear or disappear as the fermentation progresses (Navarrete-Bolaños, 2012). Generally, in the early stages of the spontaneous fermentations a large number of yeast species are present. These are often characterized by low tolerance to microbial stress factors as ethanol and organic acids. As the fermentation progresses and the acid and alcohol contents increase, the more stress tolerant yeast species take over resulting in far fewer species completing the fermentation (Ganucci et al., 2018). In order to understand fully the microbial successions occurring during spontaneous fermentations, it is important to get a comprehensive overview on the entire microbiota involved in the process.
In general, two different approaches can be applied for analyzing the microbiota from complex foods. Presumptive yeasts can either be quantified by a culture-dependent approach, initially isolating the yeasts on appropriate media followed by species identification. Alternatively, the yeast community can be quantified by culture-independent methodologies such as DGGE (Ongol and Asano, 2009; Stringini et al., 2009; Clementine et al., 2012; Mukisa et al., 2012; Greppi et al., 2013b) or amplicon-based high-through put sequencing (Cocolin and Ercolini, 2015). Although accurate, culture-dependent methodologies are limited by the selective media used and will not be able to identify viable but non-culturable yeasts. On the other hand, culture-independent methods are unable to distinguish between living and dead microorganisms when quantifications are based on DNA (Cocolin and Ercolini, 2015). Combining both culture-dependent and -independent methodologies have been applied to study yeast diversity in mawè and tchoukoutou (Greppi et al., 2013b) as well as in palm wine (Stringini et al., 2009). Some few examples of yeasts successions in indigenous sub-Saharan African fermented food and beverages are given below.
Among the solid foods, microbial successions have been reported in the cereal-based mawè, in which P. kudriavzevii and K. marxianus have been shown to be present throughout the fermentation, i.e., 72 h (Greppi et al., 2013b; Houngbédji et al., 2018). In the study by Greppi et al. (2013b), S. cerevisiae was reported after 24 h and remained present until the end of the fermentation, while C. tropicalis, Wickerhamomyces anomalus (f. Hansenula anomala, Pichia anomala; anamorph Candida pelliculosa), Millerozyma farinosa (f. Pichia farinosa) and Rhodotorula mucilaginosa (f. Rhodotorula rubra) were reported only within 24 h of fermentation, though in low abundance. The opportunistic pathogenic yeast Candida glabrata was occasionally reported in high numbers during the initial stage of fermentation, i.e., 0–48 h, but disappeared at the end of the fermentation (Greppi et al., 2013b). In the solid food fufu based on cassava, P. kudriavzevii, C. tropicalis, Saturnispora saitoi (f. Pichia saitoi), S. cerevisiae, W. anomalus and Zygosaccharomyces bailii were isolated at the initial stage of the fermentation, i.e., 0–12 h (Oyewole, 2001). W. anomalus, S. saitoi, and S. cerevisiae disappeared within the first 48 h of fermentation, while P. kudriavzevii, C. tropicalis, and Z. bailii dominated the fermentation until the end, i.e., 96 h (Oyewole, 2001). In the solid food based on fermentation of enset palm into kocho, Galactomyces geotrichum (anamorph Geotrichum candidum), Pichia exigua and Pichia fermentans were reported as the most frequently isolated yeast species in fresh fermented kocho after 2–5 days of fermentation (Birmeta et al., 2019). As the fermentation progressed, G. geotrichum was still dominating after 3–4 months together with Candida cabralensis and Candida ethanolica. At the end of the fermentation (7–12 months) the dominant yeast species was Saturnispora silvae followed by Cyberlindnera jadinii (f. Pichia fabianii; anamorph Candida fabianii), while G. geotrichum only comprised a minor part of the yeasts (Birmeta et al., 2019).
Among the non/low-alcoholic beverages, microbial successions have been reported in the cereal-based ogi (Omemu et al., 2007). S. cerevisiae and Rhodotorula graminis dominated the steeping step. In the beginning of the fermentation, S. cerevisiae dominated, while the later part of the fermentation was dominated by P. kudriavzevii, C. tropicalis, Dipodascus fermentans (anamorph Geotrichum fermentans), and G. geotrichum (Omemu et al., 2007).
Among the alcoholic beverages, reports on microbial successions include the cereal-based tchoukoutou (Greppi et al., 2013b). Clavispora lusitaniae (anamorph Candida lusitaniae) followed by P. kudriavzevii was dominant for the first 4 h, while S. cerevisiae was found to dominate the fermentation from 4 h until the end of the fermentation, i.e., 12 h (Greppi et al., 2013b). According to Stringini et al. (2009), Saccharomycodes ludwigii was dominant at the beginning of palm wine fermentation. After 24 h S. cerevisiae increased in number while S. ludwigii decreased and could not be detected after 5 days of fermentation. Other yeast species as Z. bailii, Hanseniaspora uvarum, Candida parapsilosis, and Meyerozyma caribbica (anamorph Candida fermentati) were only reported during the first 3 days of fermentation, while S. cerevisiae was the only yeast species present from day 3–5 (Stringini et al., 2009).
Among the fermented dairy products, yeast successions was reported for nunu (Akabanda et al., 2013). Different successions occurred at the three production sites investigated. At one production site, only P. kudriavzevii and S. cerevisiae were reported in the fermentation. At the other two production sites, C. parapsilosis, Candida rugosa, and C. tropicalis were reported sporadically during the fermentation, with G. geotrichum making up a substantial part of the yeasts in the later stages of the fermentation at one of these sites (Akabanda et al., 2013). In the fermented dairy product lait caillé, C. parapsilosis was dominating the first 18 h of fermentation, while from 35 h until the end of the fermentation S. cerevisiae was dominating (Bayili et al., 2019).
Strain Diversity of Yeasts
Diversity is not only observed at species level, also at strain level significant diversity exists. The stressful microbial environments in fermented food and beverages result in a high selection pressure, which can lead to development of new strains better adapted to the fermentation process. As a consequence, species occurring in spontaneously fermented food and beverages might, over time, differentiate into populations of strains of the same species (Suzzi, 2011).
Several investigations have been carried out to understand the diversity of S. cerevisiae strains in indigenous sub-Saharan African fermented food and beverages. Especially in sub-Saharan African beers, strain diversities have been investigated. S. cerevisiae strains with atypical phenotypic and genotypic traits have been isolated from West African sorghum beer (van der Aa Kühle et al., 2001). Among the 40 isolates, only 78% were able to ferment sucrose and none were able to ferment raffinose or trehalose. Further, 55% of the isolates had assimilation patterns quite atypical of S. cerevisiae, where several of the strains were unable to assimilate raffinose and some trehalose (van der Aa Kühle et al., 2001). Significant variations in MAL loci composition between S. cerevisiae strains from West African sorghum beer and European brewing strains were observed (van der Aa Kühle et al., 2001; Jespersen, 2003). For all S. cerevisiae isolates from sorghum beer, MAL11 and MAL31 were identified as the only recognized MAL loci, while the isolates lacked several of the MAL loci usually present in brewer’s and baker’s yeasts. Additionally, 40% of the isolates had an undescribed MAL locus of approximately 950 kb, which seemed to be specifically linked to this group of isolates. Similarly, based on phenotypic characteristics, sequencing of the ITS1 region of the 26S rRNA gene and Southern hybridization analysis, Naumova et al. (2003) reported that S. cerevisiae strains isolated from sorghum beer in Ghana and Burkina Faso represent a divergent population as compared to the European type strain CBS 1171. Further, only SUC2 and RTM genes were detected in the sorghum beer strains, which is contrary to the European top and bottom fermenting brewing strains exhibiting multiple teleomeric SUC and RTM genes (Naumova et al., 2003). By whole genome sequencing of S. cerevisiae strains from around the world, distinct lineages correlating with geography, environmental niche and degree of human association have been reported in which, S. cerevisiae strains from African beers and palm wines were found to form a separate cluster (Gonçalves et al., 2016; Peter et al., 2018). Among the isolates, polyploidy (3–5n) was especially enriched in specific subpopulations, including the African beer and palm clades, which according to the authors strongly suggests that ploidy level is affected by human-related environments (Peter et al., 2018). Only observed among the West African wine strains, a specific type of inactivation of the PAD1 gene, involved in decarboxylation of ferulic acid to the phenolic off-flavor compound 4-vinylguaiacol, was reported (Gonçalves et al., 2016). Through inter-delta and microsatellite analyses, S. cerevisiae strains isolated from Nigerian palm wine were reported to form an autochthonous population genetically distant from strains isolated from Ghanaian sorghum beers (Ezeronye and Legras, 2009). In a study using microsatellite analysis to compare S. cerevisiae strains isolated from palm wines in Burkina Faso, Djibouti, Nigeria, and Côte d’Ivoire, S. cerevisiae strains were found to form different populations according to the source of isolation, suggesting that geography may explain the difference. Contrary, strains of S. cerevisiae from sorghum beers produced in Burkina Faso, Côte d’Ivoire and Ghana clustered closely together despite the country of isolation, which according to the authors might be linked to human migration and thereby distribution of yeasts for inoculation through back-slopping or as dried yeast harvested from previous brews (Tapsoba et al., 2015). Diversity between S. cerevisiae strains isolated from indigenous sub-Saharan African food and beverages has also been reported within properties such as tolerance to acidic stress (Halm et al., 2004; Kubo et al., 2014; Houngbédji et al., 2019), ability to bind aflatoxin B1 (Shetty et al., 2007), production of tissue degrading enzymes (Padonou et al., 2010), folate synthesis (Hjortmo et al., 2008a), and phytase activity (Greppi et al., 2015).
Very few studies have been carried out on strain diversity among non-conventional yeast species isolated from indigenous sub-Saharan African fermented food and beverages, though some exist on P. kudriavzevii. By pheno- and genotypic characterisation of strains isolated from fermented maize dough it was found that several strains of P. kudriavzevii were involved from the onset of the fermentation and remained dominant throughout the fermentation (Hayford and Jakobsen, 1999). Strain diversity among isolates have been studied in indigenous sub-Saharan African fermented foods as, e.g., gowé, mawè, ogi and tchoukoutou by use of (GTG)5 rep-PCR genomic fingerprint patterns analysis (Greppi et al., 2013a; Houngbédji et al., 2018). For all fermented food products, strain clusters of P. kudriavzevii were reported. In mawè, diversities among the strains were found to vary between different mawè doughs. Especially raw materials, processing method and production site had an influence on the strain composition. Moreover, some strains were reported only to be linked to a single production site (Houngbédji et al., 2018). Strain variations within K. marxianus from the sorghum beer, bili bili, were studied and 12 groups of K. marxianus strains were isolated from the souring step based on physiological characteristics and their PCR/RFLP-NTS2 profiles. Based on chromosome polymorphism analysis by PFGE the K. marxianus strains could be divided into three different karyotypes (Maoura et al., 2005). Strains of S. cerevisiae, P. kudriavzevii, K. marxianus, and C. glabrata isolated from the solid food mawè was influenced in a species and strain dependent manner upon exposure to the intrinsic stress factors of the fermented cereal doughs, mawè, i.e., low pH, ethanol, lactic acid and acetic acid (Houngbédji et al., 2019).
Yeasts and Their Interactions With the Commensal Microbiota
Microbial Interactions in Fermented Food and Beverages
Besides a high number of yeast species, several other groups of microorganisms take part in the spontaneous fermentation of indigenous sub-Saharan African fermented food and beverages. Not surprisingly, the predominant group of microorganisms is LAB, but also other groups of bacteria might occur (Tamang et al., 2016). The high heterogeneity in the composition of most raw materials for fermented food and beverages allows simultaneous occupation of multiple niches by specialized species or strains, e.g., through utilization of different carbon sources (Sieuwerts et al., 2008).
Microbial interactions are quite common in spontaneous fermentations and do often involve interactions between several groups of microorganisms, i.e., yeast-yeast, bacteria-bacteria, yeast-bacteria and yeast-mold. Several interactions will occur simultaneously resulting in an interrelationship between different species continuously shaping the microbial consortium, while the interactions then become significant for obtaining the desirable characteristics of these foods (Viljoen, 2006). Microbial interactions can be divided into five main classes, i.e., antagonism, competition, commensalism, mutualism and parasitism (Sieuwerts et al., 2008; Smid and Lacroix, 2013), and sometimes the interactions even depend on direct physical contact or signaling molecules (Nissen et al., 2003; Skandamis and Nychas, 2012; Johansen and Jespersen, 2017). In the following, some examples of interactions among yeasts and other groups of microorganisms in indigenous sub-Saharan African fermented food and beverages are given.
Yeast Interactions With Lactic Acid Bacteria (LAB)
Yeasts and LAB often co-exist during spontaneous fermentations. While yeasts can grow in a relatively simple medium, LAB are more fastidious and require more nutrients as, e.g., amino acids and vitamins (Viljoen, 2006; Ponomarova et al., 2017). The interactions between yeasts and LAB can be both synergistic and antagonistic, but are most often of mutual benefit (Romano et al., 2006). Mutualistic interactions between yeasts and LAB have been described for ogi, a non/low-alcoholic cereal-based beverage. While the yeasts will benefit from a decrease in pH by the acidification facilitated by the activity of the LAB, significant higher growth of Lactobacillus plantarum has been proved when co-cultured with either S. cerevisiae or especially P. kudriavzevii, indicating that these yeast species provide growth factors for the LAB (Omemu et al., 2007), most likely amino acids (Ponomarova et al., 2017). Mutualistic interactions have likewise been reported between species of LAB and yeasts originating from indigenous sub-Saharan African fermented milk products. When co-culturing Lactococcus lactis subsp. lactis biovar diacetylactis with K. marxianus in milk, the viability of L. lactis subsp. lactis biovar diacetylactis was enhanced, while K. marxianus could benefit from galactose, arising from lactose degradation by L. lactis subsp. lactis biovar diacetylactis (Gadaga et al., 2001a). Likewise, Lactobacillus paracasei subsp. paracasei reached significantly higher final counts when co-cultured in milk with especially K. marxianus, S. cerevisiae or Naumovozyma dairenensis (f. Saccharomyces dairenensis) (Gadaga et al., 2001b).
Eukaryotic Interactions
The ability of some yeast species to inhibit molds has been proven for many foods (van den Tempel and Jakobsen, 2000; Masoud et al., 2005; Andrade et al., 2014; Núñez et al., 2015). Despite the relevance for many indigenous sub-Saharan African food products, practically no publications exist on the antagonistic effect of yeasts against molds in these type of fermented products. However, it has been shown that initial high counts of Penicillium, Aspergillus, and Fusarium species were significantly reduced during the first 24 h of fermentation of maize dough for kenkey production (Jespersen et al., 1994). Recently, more complex interaction mechanisms involving cell-to-cell communication such as, e.g., quorum sensing (QS) have been described among yeast species (Johansen and Jespersen, 2017). QS could be an important factor in regulating yeast community structures in indigenous sub-Saharan African fermented food and beverages, though not reported yet. Another interesting antagonistic interaction occurring between yeast species is the production of killer toxins and antimicrobial peptides (AMPs) (Marquina et al., 2002). Even though no reports on killer toxin or AMP producing yeasts exist for indigenous sub-Saharan African alcoholic beverages, it is most likely that killer or AMP producing yeasts could be present and be a determinant for the composition of the yeast consortium, especially when other microbial stress factors such as ethanol and organic acids are limited.
Importance of Yeasts for Flavor
Yeasts as Important Contributors to Flavor Formation
Indigenous sub-Saharan African fermented food and beverages are well appreciated for their natural tastes and flavors (Holzapfel, 2002). The microorganisms involved during the fermentation are able to produce a wide variety of compounds, which give to fermented foods their typical sensorial attributes. Formation of flavor compounds by conversion of carbohydrates is one of the most studied functions of yeasts, especially in the brewing industry (Jespersen, 2003). The main flavor compounds produced by yeasts during fermentation include alcohols, esters, organic acids, aldehydes and sulfur compounds (Dzialo et al., 2017). Several factors including the yeast species, intra-species variations and culture conditions affect the concentrations of these compounds being produced during the fermentation, and hence the flavor of the final product (Nedović et al., 2015). The importance of yeasts for the flavor formation in indigenous sub-Saharan African fermented food and beverages has, however, not been investigated to the same extent as is seen for much more industrialized fermented products as, e.g., ale and lager beers. A few examples on how yeasts influence the flavor formation in different types of indigenous sub-Saharan African fermented food and beverages are given below and in Table 5.
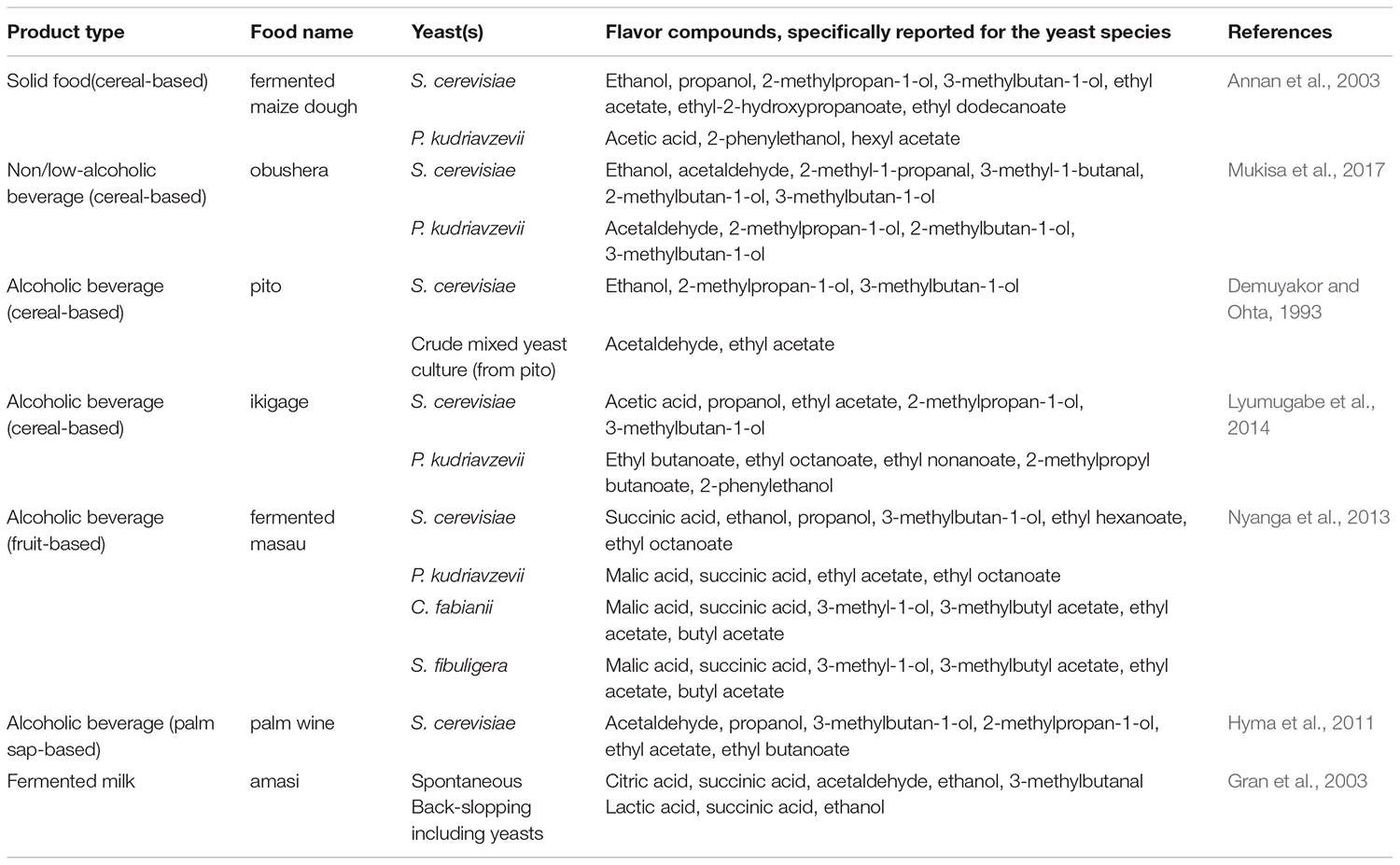
Table 5. Examples on the most common flavor compounds produced by yeasts in indigenous sub-Saharan African fermented food and beverages.
Yeast as Specific Producers of Flavor Compounds
For solid foods, the flavor profile of fermented maize doughs produced through spontaneous fermentation or by use of starter cultures of S. cerevisiae and P. kudriavzevii, were significantly affected by the microorganisms involved in the fermentation (Annan et al., 2003). At the end of the fermentation, i.e., 72 h, 64 flavor compounds were identified in the doughs including 20 alcohols, 22 carbonyls, 11 esters, seven acids, one furan and three phenolic compounds, of which 51 were volatile. In the spontaneous fermentation, higher levels of carbonyl compounds were reported. In fermentations with S. cerevisiae as starter culture higher amounts of esters and fusel alcohols were reported, whereas P. kudriavzevii as starter culture resulted in higher levels of acetic acid and lower levels of most volatiles (Annan et al., 2003).
For non/low-alcoholic beverages, flavor compounds have been studied in the cereal-based togwa and obushera (Mugula et al., 2003a). The volatile flavor compounds detected in togwa based on sorghum or maize included ethanol, acetaldehyde, diacetyl, acetoin, 2-methylpropanal, 2-methylbutanal, 3-methylbutanal, 2-methylpropan-1-ol, 2-methylbutan-1-ol and 3-methylbutan-1-ol, with ethanol being the predominant volatile compound. Starter cultures for togwa fermentation comprising either LAB or co-cultures of LAB and P. kudriavzevii were tested for their fermentation properties and flavor development. Especially the co-cultures of LAB and P. kudriavzevii were reported to enhance the production of malty volatile flavor compounds, which are descriptive for togwa flavor (Mugula et al., 2003a). Obushera beverages fermented with different starter cultures comprising LAB or co-cultures of LAB and S. cerevisiae were evaluated for their flavor profiles. The co-culture of LAB and S. cerevisiae resulted in reduced amounts of lactate and diacetyl, and higher amounts of acetaldehyde and malty compounds, resulting in a flavor profile close to that of spontaneously fermented obushera. Further, only single cultures of S. cerevisiae and P. kudriavzevii were able to produce 2-methyl-propan-1-ol, 2-methyl-butan-1-ol and 3-methyl-butan-1-ol in significant amounts (Mukisa et al., 2017).
Flavor compounds in indigenous sub-Saharan African fermented products have been studied in several alcoholic beverages. In the sorghum beer pito, flavor compounds reported included alcohols, esters, and ketones in varying amounts depending on whether a single S. cerevisiae strain or crude mixed yeast cultures were used (Demuyakor and Ohta, 1993). The mixed yeast culture was reported to give a flavor typical of pito, while the single culture of S. cerevisiae produced an atypical pito with a dry and slightly bitter taste (Demuyakor and Ohta, 1993). In the sorghum beer ikigage, 55 flavor compounds belonging to esters, alcohols, acids and carbonyl groups were detected, which could be linked to the microorganisms used (Lyumugabe et al., 2014). A mixed culture, including S. cerevisiae, P. kudriavzevii and Lactobacillus fermentum were reported to produce ikigage beer with flavor and mouth feel similar to spontaneously fermented ikigage characterized by high concentrations of certain alcohols (propanol, 2-methylpropan-1-ol and butane-2,3-diol), esters (ethyl acetate, 2-methylpropyl acetate, propyl acetate, ethyl 2-hydroxypropanoate and ethyl pentanoate), organic acids (acetic acid and heptanoic acid) and carbonyls (acetaldehyde). The majority of these compounds were produced by S. cerevisiae and/or P. kudriavzevii (Lyumugabe et al., 2014). In the alcoholic beverage made from masau (Ziziphus mauritania) fruits the best ethanol producing S. cerevisiae strains, and strains of the species P. kudriavzevii, Cyberlindnera fabianii (anamorph Candida fabianii; f. Pichia fabianii), and Saccharomycopsis fibuligera were tested for production of flavor compounds (Nyanga et al., 2013). The major volatiles detected were alcohols and esters along with trace amounts of organic acids and carbonyl compounds. The yeast species produced significantly different amounts of ethanol and other volatile compounds. The highest amounts of ethanol and ethyl esters were reported for S. cerevisiae, though strains of P. kudriavzevii, C. fabianii, and S. fibuligera produced the highest amounts of ethyl acetate. Likewise, Hyma et al. (2011) reported that S. cerevisiae strains originating from palm wine in West Africa resulted in a different flavor formation in fermented grape musts than S. cerevisiae strains from vineyards or laboratory strains. The palm wine strain did especially produce acetaldehyde, propanol, 3-methylbutan-1-ol, 2-methylpropan-1-ol, ethyl acetate, and ethyl butanoate (Hyma et al., 2011).
In the fermented milk product amasi, flavor compounds were detected for spontaneously fermented milk, milk fermented using back-slopping and milk fermented with starter cultures of LAB (Gran et al., 2003). Spontaneously fermented milk and milk fermented with back-slopping, containing LAB and yeasts, were reported to have higher amounts of succinate, ethanol, malty aldehydes and methyl alcohols than the fermentations using pure starter cultures of LAB. The high levels of malty compounds in the spontaneously fermented milk and in milk fermented using back-slopping were suggested to arise from yeasts, which thereby highly impact the flavor of amasi (Gran et al., 2003).
Nutritional Value and Health Benefits of Yeasts
The impact of fermentation on nutritional value and health benefits of fermented foods has been the focus of several studies, particularly focusing on bioavailability of micronutrients and probiotic properties. Some examples on the impact of yeasts in indigenous sub-Saharan African fermented food and beverages are given in Table 6.
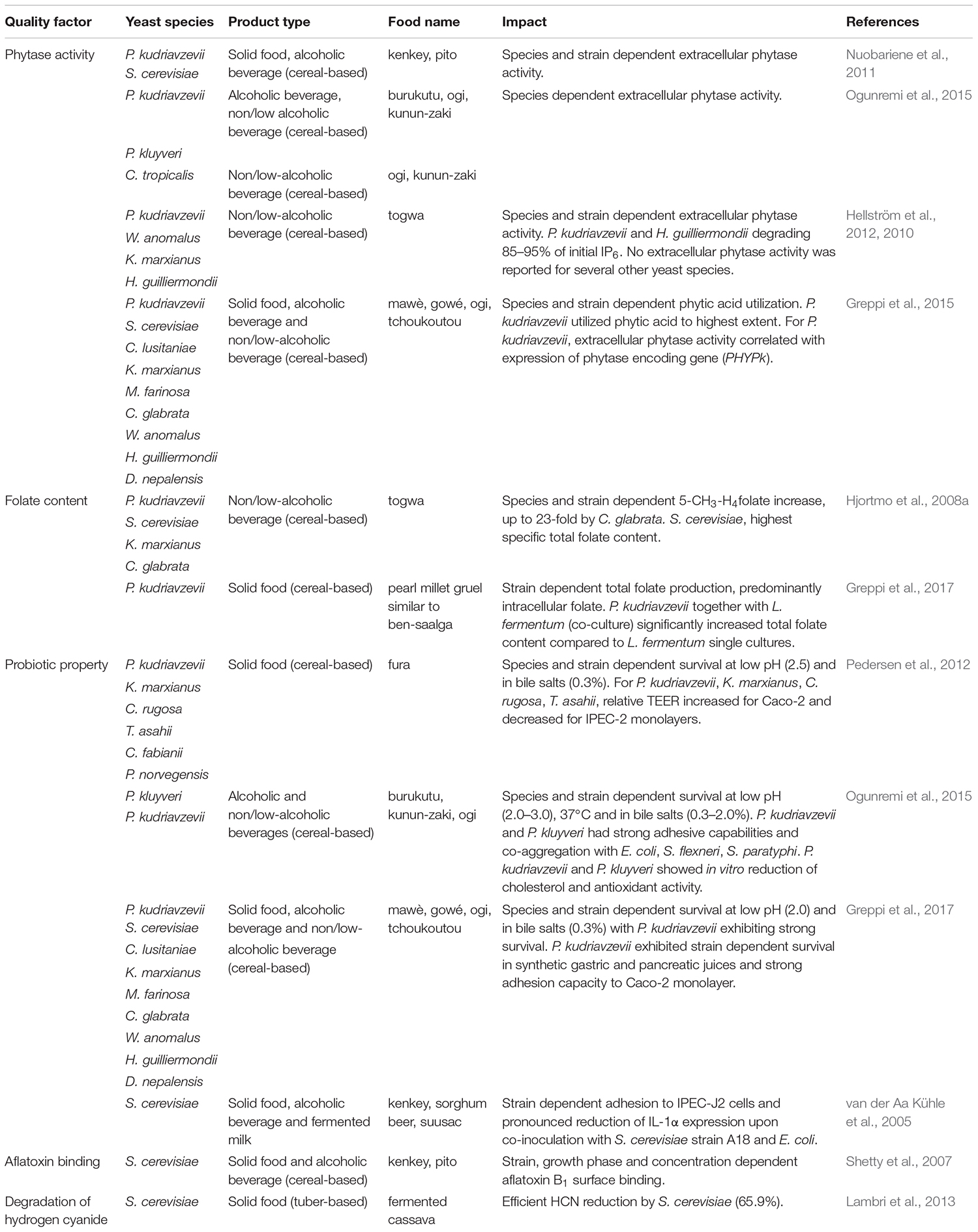
Table 6. Examples on health beneficial properties of yeasts isolated from indigenous sub-Saharan African fermented food and beverages.
Yeasts Affecting the Nutritional Value
In cereal-based foods, the presence of anti-nutritional factors such as the highly charged phytate limits the bioavailability of divalent ions through the process of chelation of cation minerals such as Fe2+, Zn2+, Ca2+, and Mg2+. The phytate complexes formed by chelation are insoluble at physiological pH and therefore, the divalent ions cannot be absorbed in the human intestine (Jespersen, 2003; Greppi et al., 2015). Microbial phytase activity has been reported for yeasts, particularly for P. kudriavzevii. Isolated from the solid fermented product kenkey (maize dough) and the alcoholic beverage pito (sorghum beer), strains of P. kudriavzevii and S. cerevisiae were reported to vary significantly in their extracellular phytase activity (Nuobariene et al., 2011). Similarly, strains of C. tropicalis, Pichia kluyveri and P. kudriavzevii isolated from burukutu (cereal beer), ogi and kunun-zaki (non/low-alcoholic beverages) were found to have high extracellular phytase activity (Ogunremi et al., 2015). Moreover, yeasts strains isolated from togwa, a non/low-alcoholic cereal-based fermented beverage were found to have significant phytase activities, with one strain of each of H. guilliermondii and P. kudriavzevii showing strong phytate degradation (≥95%) in a togwa model system (Hellström et al., 2010, 2012). Especially one strain of P. kudriavzevii (TY13) from togwa was shown to release substantial amounts of phytase enzyme to the environment. The extracellular phytase activity was found to be growth phase dependent, being highest in young growing cultures (Hellström et al., 2015). Phytase activity has also been reported for several different yeast species isolated from the solid foods mawè and gowé, the non/low-alcoholic beverage ogi and the alcoholic beverage tchoukoutou, with the highest phytase activity in P. kudriavzevii (Greppi et al., 2015). In the draft genome of P. kudriavzevii (M12) three phytase encoding genes were identified (Chan et al., 2012) and in Greppi et al. (2015), a correlation between the expression of these genes and extracellular phytase activity was established.
The production of folate (vitamin B9) by indigenous yeasts via fermentation is one way by which the nutritional value of cereal-based products can be improved. Unlike yeasts, the human body lacks the ability to synthesize folate and must therefore rely on sufficient intake from the diet (Hjortmo et al., 2008a). Yeasts isolated from indigenous sub-Saharan African fermented products have been evaluated for their capacity to produce folate in different studies. The production of folate by five yeast species originating from the non/low-alcoholic beverage togwa, i.e., C. glabrata, K. marxianus P. kudriavzevii, S. cerevisiae, and W. anomalus was determined in a model system of togwa by Hjortmo et al. (2008a). Folate production during fermentation were reported to vary depending on the yeast species and strain. Further, togwa fermented with yeasts contained 4 to 5-fold more folate than the un-inoculated control. Especially one S. cerevisiae strain (TY08) were shown to exhibit more than a 10-fold higher specific total folate content compared to other yeast strains (Hjortmo et al., 2008a). Besides species and strain dependent folate production, Hjortmo et al. (2008b) has shown that the specific folate content for S. cerevisiae vary extensively depending on cultivation medium, growth rate and growth state. Hence, controlling processing conditions could lead to significantly increased folate contents in the fermented food. In a food model mimicking the cereal-based gruel ben-saalga, total folate concentration was determined for co-cultures of L. fermentum and P. kudriavzevii as well as for LAB cultures alone. Here, fermentation of pearl millet with co-cultures of yeasts and LAB or LAB cultures alone were reported to increase the total folate concentration compared to unfermented pearl millet gruel. Furthermore, pearl millet gruels fermented with co-cultures of P. kudriavzevii and L. fermentum had significantly higher folate concentrations as compared to gruels fermented with LAB alone (Greppi et al., 2017).
Probiotic Properties of Yeasts Isolated From Indigenous Sub-Saharan African Fermented Food and Beverages
While a wide diversity of yeasts have been isolated from indigenous sub-Saharan African fermented foods, specific yeast strains with proven probiotic properties are sparsely reported. However, few studies have reported the potential probiotic properties of yeasts isolated from sub-Saharan African fermented foods (van der Aa Kühle et al., 2005; Pedersen et al., 2012; Ogunremi et al., 2015; Greppi et al., 2017). The properties of seven yeast species from the solid food fura, based on fermented millet, were investigated by Pedersen et al. (2012). All the examined yeast species could survive at conditions equivalent to the human gastro-intestinal tract (GIT), i.e., at low pH (2.5), and in 0.3% (w/v) bile salts at 37°C. Furthermore, strains of C. rugosa, K. marxianus, P. kudriavzevii and Trichosporon asahii were reported to be able to increase the relative trans-epithelial electrical resistance (TEER) of polarized monolayers of human intestinal epithelial cells (Caco-2) in a manner comparable to the approved human probiotic yeast S. cerevisiae var. boulardii (Pedersen et al., 2012). In a similar study of the probiotic properties of one P. kluyveri, three P. kudriavzevii and one C. tropicalis strains isolated from cereal-based alcoholic beverage burukutu, and the non/low-alcoholic beverages kunun-zaki and ogi, all yeast strains survived at low pH (2.0), and 0.3–2.0% (w/v) bile salts at 37°C, with strain-specific variabilities (Ogunremi et al., 2015). Yeasts isolated from burukutu, kunun-zaki and ogi were able to co-aggregate with pathogenic bacteria, with the highest co-aggregation reported for P. kudriavzevii and Escherichia coli (Ogunremi et al., 2015). Yeasts isolated from different fermented cereal products including the solid foods gowé and mawè, the non/low-alcoholic ogi, and the alcoholic beverage tchoukoutou were likewise shown to be able to survive low pH (2.0), and 0.3% (w/v) of bile salts at 37°C, with isolates of P. kudriavzevii having the best survival rate in synthetic gastric and pancreatic juices (Greppi et al., 2017). Similar results were reported for S. cerevisiae strains isolated from different indigenous sub-Saharan African fermented foods, including the solid food kenkey, the alcoholic beverages pito and dolo, and fermented dairy product suusac (van der Aa Kühle et al., 2005). Two of the S. cerevisiae strains were additionally shown to be able to adhere to the intestinal porcine epithelial cell line IPEC-J2. Co-inoculation of IPEC-J2 cells with the adhesive S. cerevisiae strain A18 from sorghum beer, were reported to lead to pronounced reduction of IL-1α expression upon infection with E. coli. This indicated that S. cerevisiae A18 were able to lower the pro-inflammatory cytokine IL-1α response in a manner similar to what was observed for the human probiotic yeast S. cerevisiae var. boulardii (van der Aa Kühle et al., 2005).
Reduction of Toxic Compounds by Yeasts Isolated From Indigenous Sub-Saharan African Fermented Foods
A strategy for reducing the deleterious effects of mycotoxins is biological decontamination by microorganisms, particularly by fermentation with LAB and yeasts. However, the ability of yeast isolates from indigenous sub-Saharan African fermented food to decontaminate or degrade mycotoxins is still sparsely investigated. Shetty et al. (2007) investigated the in vitro aflatoxin B1 binding abilities of 18 S. cerevisiae strains isolated from the solid food kenkey and the alcoholic beverage pito and reported that some strains could adsorb more than 40% of the added aflatoxin B1, and that the adsorption was strain specific but not dependent on the viability of the yeast strains.
Another group of toxic compounds, inherent to cassava tubers, are the cyanogenic glucosides linamarin and to a lesser extent lotaustralin, which have fatal consequences when consumed in unprocessed cassava. Fermentation has been widely documented as a way to enhance the safety, organoleptic and nutritional quality of many cassava-derived foods (Holzapfel, 2002), and S. cerevisiae has additionally been demonstrated to reduce the total cyanide content, e.g., in fermented cassava tubers from Burundi (Lambri et al., 2013).
Safety; Opportunistic Pathogenic Yeasts
As indigenous sub-Saharan African food and beverages predominantly are produced by spontaneous fermentation, the consumers may be exposed to large populations of different yeast species of often unknown origin (Ogunremi et al., 2017). Fortunately, unlike other microbial groups, food borne yeasts are generally not considered to be aggressive pathogens and are rarely associated with outbreaks of foodborne gastroenteritis, intoxications or other infections (Fleet, 2007; Ogunremi et al., 2017). Nonetheless, caution is needed particularly in selecting yeast strains for the development of starter cultures, as some species are capable of causing disease in especially immunocompromised individuals. Many species of the genus Candida are widely distributed in nature and have frequently been isolated from indigenous sub-Saharan African fermented products. Most of them are harmless, but some Candida spp. can take advantage of a locally or systemically impaired immune system to proliferate in the host and cause diseases generally termed “candidiasis” (Papon et al., 2013). C. albicans is the most frequently isolated agent of candidiasis, causing about 41–47% of total yeast infections worldwide (Pfaller and Castanheira, 2016). However, non-albicans candidiasis infections are also widespread with worldwide prevalence in invasive candidiasis as follows: C. glabrata (18–27%), C. parapsilosis (16–18%), C. tropicalis (9–11%), and C. krusei (1–3%) (Pfaller and Castanheira, 2016).
Generally, C. albicans is only found in few indigenous sub-Saharan African fermented foods, occurring in low numbers in the fermented milk products amabere amaruranu and sethemi (Kebede et al., 2007; Nyambane et al., 2014) as well as in the sorghum beer tchoukoutou (Kayode et al., 2011). C. glabrata has been reported in some indigenous sub-Saharan African fermented food and beverages, including gari, lafun, mawè, tchoukoutou, and togwa (Tables 1–3). Both yeasts are most probably occurring as a result of contamination during processing due to improper human handling. Contrary, C. tropicalis is frequently identified in indigenous sub-Saharan African fermented food and beverages, including akyeke, amasi, attiéké, bandji, fufu, fura, gari, gowé, kaffir, lafun, mawè, mukumbi, nunu, palm wine, pito, sethemi, tchapalo and teff-injera (Tables 1–4). Candida spp. have emerged as major agents of human mucosal, systemic and bloodstream yeast infections (Silva et al., 2012; Pfaller and Castanheira, 2016). Moreover, examples of resistance to well-known antifungal drugs have been reported, e.g., C. glabrata is particularly prone to develop resistance to fluconazole, a first-line antifungal treatment for yeast infections (Abbes et al., 2011), and resistance to fluconazole and other azoles appears to be increasing among clinical isolates of C. tropicalis (Pfaller and Castanheira, 2016). Consequently, further research is needed to optimize fermentation conditions to eliminate opportunistic pathogenic yeasts during processing and equally important, improved hygienic conditions need to be ensured in order to prevent cross-contamination from human handling during food processing.
As apparent from the above, P. kudriavzevii under different names, i.e., C. krusei and I. orientalis (Kudryavtsev, 1960; Kurtzman et al., 2008) is very often encountered in indigenous sub-Saharan African fermented food and beverages and plays key technological functions i.e., phytase production, flavor formation, etc. (Oyewole, 2001; Jespersen, 2003; Oguntoyinbo, 2008; Yadav et al., 2012; Greppi et al., 2015). It has recently been confirmed that C. krusei and P. kudriavzevii are the same species, and that there is no genetic distinction between clinical isolates of C. krusei and environmental isolates of P. kudriavzevii (Douglass et al., 2018). Traditionally, P. kudriavzevii has been considered as non-pathogenic due to its occurrence and technological role in many fermented food products. However, the species draws attention regarding the safety of its use in food processing due to its opportunistic pathogenic traits. Additionally, C. krusei (P. kudriavzevii) has been found to have innate resistance to widely used triazole antifungal drugs, particularly fluconazole, and has in some cases been reported to lead to mortality in immunocompromised individuals (Lamping et al., 2009; Garnacho-Montero et al., 2010). Consequently, C. krusei is not considered “generally regarded as safe” by the US Food and Drugs Administration nor included in the quality presumptive safety (QPS) list of the European Food Safety Authority (EFSA). Because of the taxonomic identity between P. kudriavzevii and C. krusei, the proven close relatedness with clinical isolates of C. krusei and the high resistance to fluconazole by the species, some strains of P. kudriavzevii could present potential health hazards to immunocompromised consumers (Douglass et al., 2018). As no studies have been carried out on the variability in potential pathogenic traits among strains of P. kudriavzevii isolated from indigenous sub-Saharan African food and beverages this topic still calls for investigation.
Future Perspectives for Up-Grading Indigenous Sub-Saharan African Fermented Food and Beverages
The market size of indigenous sub-Saharan African fermented food and beverages are growing, among others due to their ability to be used as convenient food by consumers. Additionally, fermentation is an affordable and sustainable way of processing that easily can be used to improve the quality and safety of food and beverages. Unfortunately, uncontrolled processing, handling and selling on streets may induce contamination and growth of harmful microorganisms. The challenges will be to develop better starter cultures targeted indigenous sub-Saharan African fermented food and beverages and to optimize the processing. To achieve this, several studies have been carried out to study the biochemical and microbiological changes which occur during processing including information on nutritional value and consumer preferences (Halm et al., 1996; Mugula et al., 2003b; Edema and Sanni, 2008; Padonou et al., 2010; Greffeuille et al., 2011; Akabanda et al., 2013; Houngbédji et al., 2018). Some of these studies have demonstrated that when the yeasts are combined in consortia it makes the fermented products more appealing. However, there is still a need to understand the organization of the consortium, the species-species interactions and species-environment interactions. In parallel, the need to select and develop starter cultures of non-pathogenic strains of yeasts with permissible levels of drug resistance for application in indigenous sub-Saharan African fermented food products is obvious.
To generate sufficient data for large-scale production in SMEs, optimal processing parameters such as raw material preparation as well as ways of microbial propagation and inoculation should be investigated further. Achievement of these challenges will help to develop these indigenous products as novel foods for an international market. The knowledge generated should be used to optimize processing in order to control the microbial community during fermentation for optimized quality, shelf life, safety, nutritional value and other health benefits. The success of indigenous sub-Saharan African fermented food and beverages is also based on the collection of appropriate information concerning consumers’ needs and expectations that are essential tools for building competitive advantage and long-term SMEs success in the market and for prevention of negative changes in product quality and acceptability, consumers’ complaints and product rejection.
Conclusion and Perspectives
The present survey has proved that a significant number of yeast species are involved in the fermentation of indigenous sub-Saharan African food and beverages. Despite the fact that these fermented products play a significant role in the diet of sub-Saharan African inhabitants, they are not investigated to the same extent as their industrialized counterparts. Consequently, the potential of these products is not explored and even worse, their sustainability is under pressure. From a taxonomic point of view, it is clear that many of the yeast species found in these spontaneously fermented products could add significantly to our understanding of the evolution of yeast. Likewise, it becomes clear that these yeasts, to a greater extent, should be described in current taxonomic keys. Studying spontaneous fermentations offers a valuable scenario for understanding microbial interactions at various levels. Cross-kingdom interactions occur when yeasts and LAB evolve together. Yeasts acts as bio-controlling agents preventing the growth of pathogenic bacteria and mycotoxin producing molds, and at the intra-species level, an often overlooked battle is going on. As proved in the current review, strains within the same species often have different traits evolved as a consequence of their adaption to specific ecological niches. Strain variations are not only seen for industrialized processes such as in the production of barley beer and wine but similar strain variations exist within the yeast species involved in spontaneous fermentation of food and beverages in sub-Saharan Africa. This point toward an unexplored microbial heritage which could offer, not only valuable academic insight, but new enzymes, cell wall components, killer toxins, etc., with new interesting properties relevant for the biotechnological and biomedical sectors.
The predominant yeast species involved in the fermentation of indigenous sub-Saharan African fermented food and beverages are mostly harmless and do hardly ever cause any infections, contrary they are able to produce essential compounds as folate, enhance bioavailability by phytate degradation, degrade toxic compounds as linamarin, preventing uptake of toxins as, e.g., aflatoxin B1 in the human GIT, provide probiotic properties, etc. However, some species have been reported as having opportunistic pathogenic traits. This is especially of relevance for P. kudriavzevii, which according to the present survey, together with S. cerevisiae is the far most predominant yeast species in indigenous fermented sub-Saharan African food and beverages. As a paradox, P. kudriavzevii has also been proved, among several other yeast species, to be high in phytase and folate production, thereby offering a substantial improvement of spontaneously fermented foods in sub-Saharan Africa. The present survey calls for solid research to be conducted on strain variations in properties related to product quality and health concerns. C. glabrata, generally recognized as a human pathogen, does occasionally occur in indigenous fermented sub-Saharan African foods most likely due to unhygienic handling procedures. However, only limited knowledge exists on how to prevent the growth of this species during fermentation.
In conclusion, yeasts make a significant contribution to the fermentation of many food and beverages produced in sub-Saharan Africa. However, academic knowledge is still required, strain variations are still unexplored, methods for production of starter cultures need to be upgraded and methodologies for in-depth understanding of microbial interactions need to be developed. Only by going in-depth within these topics we can ensure the sustainability of these indigenous fermented products being very important not only for in-come generation but also for the nutrition of many people in sub-Saharan Africa.
Author Contributions
LJ designed the manuscript. All authors wrote the manuscript. PJ and LJ critically revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abbes, S., Sellami, H., Sellami, A., Makni, F., Mahfoudh, N., Makni, H., et al. (2011). Microsatellite analysis and susceptibility to FCZ of Candida glabrata invasive isolates in Sfax Hospital, Tunisia. Med. Mycol. 49, 10–15. doi: 10.3109/13693786.2010.493561
Abdelgadir, W., Nielsen, D. S., Hamad, S., and Jakobsen, M. (2008). A traditional Sudanese fermented camel’s milk product, Gariss, as a habitat of Streptococcus infantarius subsp. infantarius. Int. J. Food Microbiol. 127, 215–219. doi: 10.1016/j.ijfoodmicro.2008.07.008
Abdelgadir, W. S., Hamad, S. H., Møller, P. L., and Jakobsen, M. (2001). Characterisation of the dominant microbiota of Sudanese fermented milk Rob. Int. Dairy J. 11, 63–70. doi: 10.1016/S0958-6946(01)00042-5
Achi, O. K., and Ukwuru, M. (2015). Cereal-based fermented foods of africa as functional foods. Int. J. Microbiol. Appl. 2, 71–83. doi: 10.1007/978-3-319-54528-8_31-1
Akabanda, F., Owusu-Kwarteng, J., Glover, R. L. K., and Tano-Debrah, K. (2010). Microbiological characteristics of ghanaian traditional fermented milk product, Nunu. Nat. Sci. 8, 178–187.
Akabanda, F., Owusu-Kwarteng, J., Tano-Debrah, K., Glover, R. L. K., Nielsen, D. S., and Jespersen, L. (2013). Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiol. 34, 277–283. doi: 10.1016/j.fm.2012.09.025
Amoa-Awua, W. K., Sampson, E., and Tano-Debrah, K. (2007). Growth of yeasts, lactic and acetic acid bacteria in palm wine during tapping and fermentation from felled oil palm (Elaeis guineensis) in Ghana. J. Appl. Microbiol. 102, 599–606. doi: 10.1111/j.1365-2672.2006.03074.x
Andrade, M. J., Thorsen, L., Rodríguez, A., Córdoba, J. J., and Jespersen, L. (2014). Inhibition of ochratoxigenic moulds by Debaryomyces hansenii strains for biopreservation of dry-cured meat products. Int. J. Food Microbiol. 170, 70–77. doi: 10.1016/j.ijfoodmicro.2013.11.004
Annan, N. T., Poll, L., Sefa-Dedeh, S., Plahar, W. A., and Jakobsen, M. (2003). Volatile compounds produced by Lactobacillus fermentum, Saccharomyces cerevisiae and Candida krusei in single starter culture fermentations of Ghanaian maize dough. J. Appl. Microbiol. 94, 462–474. doi: 10.1046/j.1365-2672.2003.01852.x
Anukam, K. C., and Reid, G. (2009). African traditional fermented foods and probiotics. J. Med. Food 12, 1177–1184. doi: 10.1089/jmf.2008.0163
Asiedu, M., and Sanni, A. (2002). Chemical composition and microbiological changes during spontaneous and starter culture fermentation of Enam Ne-Setaakye, a West African fermented fish-carbohydrate product. Eur. Food Res. Technol. 215, 8–12. doi: 10.1007/s00217-002-0519-9
Assanvo, J. B., Agbo, G. N., Behi, Y. E. N., Coulin, P., and Farah, Z. (2006). Microflora of traditional starter made from cassava for “attiéké” production in Dabou (Côte d’Ivoire). Food Control 17, 37–41. doi: 10.1016/j.foodcont.2004.08.006
Atter, A., Obiri-Danso, K., and Amoa-Awua, W. K. A. (2014). Microbiological and chemical processes associated with the production of burukutu a traditional beer in Ghana. Int. Food Res. J. 21, 1769–1775.
Bahiru, B., Mehari, T., and Ashenafi, M. (2006). Yeast and lactic acid flora of tej, an indigenous Ethiopian honey wine: variations within and between production units. Food Microbiol. 23, 277–282. doi: 10.1016/j.fm.2005.05.007
Bayili, G. R., Johansen, P., Nielsen, D. S., Sawadogo-Lingani, H., Ouedraogo, G. A., Diawara, B., et al. (2019). Identification of the predominant microbiota during production of lait caillé, a spontaneously fermented milk product made in Burkina Faso. World J. Microb. Biot. 35:100. doi: 10.1007/s11274-019-2672-3
Birmeta, G., Bakeeva, A., and Passoth, V. (2019). Yeasts and bacteria associated with kocho, an Ethiopian fermented food produced from enset (Ensete ventricosum). Antonie Van Leeuwenhoek 112, 651–659. doi: 10.1007/s10482-018-1192-8
Cadez, N., Poot, G. A., Raspor, P., and Smith, M. T. (2003). Hanseniaspora meyeri sp. nov., Hanseniaspora clermontiae sp. nov., Hanseniaspora lachancei sp. nov. and Hanseniaspora opuntiae sp. nov., novel apiculate yeast species. Int. J. Syst. Evol. Micr. 53, 1671–1680. doi: 10.1099/ijs.0.02618-0
Chan, G. F., Gan, H. M., Ling, H. L., and Rashid, N. A. A. (2012). Genome sequence of Pichia kudriavzevii M12, a potential producer of bioethanol and phytase. Eukaryot. Cell 11, 1300–1301. doi: 10.1128/EC.00229-12
Clementine, K. A., Mohamed, C., Epiphane, K., David, B. K., Dje, M. K., and Montet, D. (2012). Identification of yeasts associated with the fermented fish, adjuevan, of Ivory Coast by using the molecular technique of PCR-denaturing gradient gel electrophoresis (DGGE). Afr. J. Microbiol. Res. 6, 4138–4145. doi: 10.5897/AJMR11.1391
Cocolin, L., and Ercolini, D. (2015). Zooming into food-associated microbial consortia: a ‘cultural’ evolution. Curr. Opin. Food Sci. 2, 43–50. doi: 10.1016/j.cofs.2015.01.003
Coulin, P., Farah, Z., Assanvo, J., Spillmann, H., and Puhan, Z. (2006). Characterisation of the microflora of attiéké, a fermented cassava product, during traditional small-scale preparation. Int. J. Food Microbiol. 106, 131–136. doi: 10.1016/j.ijfoodmicro.2005.06.012
Daniel, H. M., Lachance, M. A., and Kurtzman, C. P. (2014). On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie Van Leeuwenhoek 106, 67–84. doi: 10.1007/s10482-014-0170-z
Dellacassa, E., Trenchs, O., Fariña, L., Debernardis, F., Perez, G., Boido, E., et al. (2017). Pineapple (Ananas comosus L. Merr.) wine production in Angola: characterisation of volatile aroma compounds and yeast native flora. Int. J. Food Microbiol. 241, 161–167. doi: 10.1016/j.ijfoodmicro.2016.10.014
Demuyakor, B., and Ohta, Y. (1993). Characteristics of single and mixed culture fermentation of pito beer. J. Sci. Food Agr. 62, 401–408. doi: 10.1002/jsfa.2740620414
Desiye, A., and Abegaz, K. (2013). Isolation, characterization and identification of lactic acid bacteria and yeast involved in fermentation of Teff (EragrostisTef) batter. Int. J. Adv. Res. Biol. Sci. 1, 36–44.
Douglass, A. P., Offei, B., Braun-Galleani, S., Coughlan, A. Y., Martos, A. A. R., Ortiz-Merino, R. A., et al. (2018). Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: one species, four names. PLoS Pathog. 14:e1007138. doi: 10.1371/journal.ppat.1007138
Dzialo, M. C., Park, R., Steensels, J., Lievens, B., and Verstrepen, K. J. (2017). Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 41, S95–S128. doi: 10.1093/femsre/fux031
Edema, M. O., and Sanni, A. I. (2008). Functional properties of selected starter cultures for sour maize bread. Food Microbiol. 25, 616–625. doi: 10.1016/j.fm.2007.12.006
Edward, V. A., Egounlety, M., Huch, M., Van Zyl, P., Singh, S., Nesengani, N. D., et al. (2012). Isolation and screening of microorganisms from a gari fermentation process for starter culture development. Afr. J. Biotechnol. 11, 12865–12877. doi: 10.5897/AJB11.3672
Ezeronye, O. U., and Legras, J. L. (2009). Genetic analysis of Saccharomyces cerevisiae strains isolated from palm wine in eastern Nigeria. Comparison with other African strains. J. Appl. Microbiol. 106, 1569–1578. doi: 10.1111/j.1365-2672.2008.04118.x
Ezeronye, O. U., and Okerentugba, P. O. (2001). Genetic and physiological variants of yeast selected from palm wine. Mycopathologia 152, 85–89. doi: 10.1023/A:1012323721012
Fleet, G. H. (2007). Yeasts in foods and beverages: impact on product quality and safety. Curr. Opin. Biotech. 18, 170–175. doi: 10.1016/j.copbio.2007.01.010
Gadaga, T. H., Mutukumira, A. N., and Narvhus, J. A. (2000). Enumeration and identification of yeasts isolated from Zimbabwean traditional fermented milk. Int. Dairy J. 10, 459–466. doi: 10.1016/S0958-6946(00)00070-4
Gadaga, T. H., Mutukumira, A. N., and Narvhus, J. A. (2001a). Growth characteristics of Candida kefyr and two strains of Lactococcus lactis subsp. lactis isolated from Zimbabwean naturally fermented milk. Int. J. Food Microbiol. 70, 11–19. doi: 10.1016/S0168-1605(01)00501-3
Gadaga, T. H., Mutukumira, A. N., and Narvhus, J. A. (2001b). The growth and interaction of yeasts and lactic acid bacteria isolated from Zimbabwean naturally fermented milk in UHT milk. Int. J. Food Microbiol. 68, 21–32. doi: 10.1016/S0168-1605(01)00466-4
Ganucci, D., Guerrini, S., Mangani, S., Vincenzini, M., and Granchi, L. (2018). Quantifying the effects of ethanol and temperature on the fitness advantage of predominant Saccharomyces cerevisiae strains occurring in spontaneous wine fermentations. Front. Microbiol. 9:1563. doi: 10.3389/fmicb.2018.01563
Garnacho-Montero, J., Diaz-Martin, A., Garcia-Cabrera, E., Ruiz Perez de Pipaon, M., Hernandez-Caballero, C., Aznar-Martin, J., et al. (2010). Risk factors for fluconazole-resistant Candidemia. Antimicrob. Agents Chemother. 54, 3149–3154. doi: 10.1128/AAC.00479-10
Glover, R. L. K., Abaidoo, R. C., Jakobsen, M., and Jespersen, L. (2005). Biodiversity of Saccharomyces cerevisiae isolated from a survey of pito production sites in various parts of Ghana. Syst. Appl. Microbiol. 28, 755–761. doi: 10.1016/j.syapm.2005.05.003
Gonçalves, M., Pontes, A., Almeida, P., Barbosa, R., Serra, M., Libkind, D., et al. (2016). Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 26, 2750–2761. doi: 10.1016/j.cub.2016.08.040
Gran, H. M., Gadaga, H. T., and Narvhus, J. A. (2003). Utilization of various starter cultures in the production of Amasi, a Zimbabwean naturally fermented raw milk product. Int. J. Food Microbiol. 88, 19–28. doi: 10.1016/S0168-1605(03)00078-3
Greffeuille, V., Kayodé, A. P. P., Icard-Vernire, C., Gnimadi, M., Rochette, I., and Mouquet-Rivier, C. (2011). Changes in iron, zinc and chelating agents during traditional African processing of maize: effect of iron contamination on bioaccessibility. Food Chem. 126, 1800–1807. doi: 10.1016/j.foodchem.2010.12.087
Greppi, A., Krych, Ł, Costantini, A., Rantsiou, K., Hounhouigan, D. J., Arneborg, N., et al. (2015). Phytase-producing capacity of yeasts isolated from traditional African fermented food products and PHYPk gene expression of Pichia kudriavzevii strains. Int. J. Food Microbiol. 205, 81–89. doi: 10.1016/j.ijfoodmicro.2015.04.011
Greppi, A., Rantsiou, K., Padonou, W., Hounhouigan, J., Jespersen, L., Jakobsen, M., et al. (2013a). Determination of yeast diversity in ogi, mawè, gowé and tchoukoutou by using culture-dependent and -independent methods. Int. J. Food Microbiol. 165, 84–88. doi: 10.1016/j.ijfoodmicro.2013.05.005
Greppi, A., Rantisou, K., Padonou, W., Hounhouigan, J., Jespersen, L., Jakobsen, M., et al. (2013b). Yeast dynamics during spontaneous fermentation of mawè and tchoukoutou, two traditional products from Benin. Int. J. Food Microbiol. 165, 200–207. doi: 10.1016/j.ijfoodmicro.2013.05.004
Greppi, A., Saubade, F., Botta, C., Humblot, C., Guyot, J.-P., and Cocolin, L. (2017). Potential probiotic Pichia kudriavzevii strains and their ability to enhance folate content of traditional cereal-based African fermented food. Food Microbiol. 62, 169–177. doi: 10.1016/j.fm.2016.09.016
Halm, M., Hornbæk, T., Arneborg, N., Sefa-Dedeh, S., and Jespersen, L. (2004). Lactic acid tolerance determined by measurement of intracellular pH of single cells of Candida krusei and Saccharomyces cerevisiae isolated from fermented maize dough. Int. J. Food Microbiol. 94, 97–103. doi: 10.1016/j.ijfoodmicro.2003.12.019
Halm, M., Lillie, A., Sørensen, A. K., and Jakobsen, M. (1993). Microbiological and aromatic characteristics of fermented maize doughs for kenkey production in Ghana. Int. J. Food Microbiol. 19, 135–143. doi: 10.1016/0168-1605(93)90179-K
Halm, M., Osei-Yaw, A., Hayford, A., Kpodo, K. A., and Amoa-Awua, W. K. A. (1996). Experiences with the use of a starter culture in the fermentation of maize for “kenkey” production in Ghana. World J. Microb. Biot. 12, 531–536. doi: 10.1007/BF00419468
Hayford, A. E., and Jakobsen, M. (1999). Characterization of Candida krusei strains from spontaneously fermented maize dough by profiles of assimilation, chromosome profile, polymerase chain reaction and restriction endonuclease analysis. J. Appl. Microbiol. 87, 29–40. doi: 10.1046/j.1365-2672.1999.00786.x
Hellström, A., Qvirist, L., Svanberg, U., Veide Vilg, J., and Andlid, T. (2015). Secretion of non-cell-bound phytase by the yeast Pichia kudriavzevii TY13. J. Appl. Microbiol. 118, 1126–1136. doi: 10.1111/jam.12767
Hellström, A. M., Almgren, A., Carlsson, N.-G., Svanberg, U., and Andlid, T. A. (2012). Degradation of phytate by Pichia kudriavzevii TY13 and Hanseniaspora guilliermondii TY14 in Tanzanian togwa. Int. J. Food Microbiol. 153, 73–77. doi: 10.1016/j.ijfoodmicro.2011.10.018
Hellström, A. M., Vázques-Juárez, R., Svanberg, U., and Andlid, T. A. (2010). Biodiversity and phytase capacity of yeasts isolated from Tanzanian togwa. Int. J. Food Microbiol. 136, 352–358. doi: 10.1016/j.ijfoodmicro.2009.10.011
Hjortmo, S., Hellström, A. M., and Andlid, T. A. (2008a). Production of folates by yeasts in Tanzanian fermented togwa. FEMS Yeast Res. 8, 781–787. doi: 10.1111/j.1567-1364.2008.00398.x
Hjortmo, S., Patring, J., and Andlid, T. (2008b). Growth rate and medium composition strongly affect folate content in Saccharomyces cerevisiae. Int. J. Food Microbiol. 123, 93–100. doi: 10.1016/j.ijfoodmicro.2007.12.004
Holzapfel, W. H. (2002). Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 75, 197–212. doi: 10.1016/S0168-1605(01)00707-3
Houngbédji, M., Johansen, P., Padonou, S. W., Akissoé, N., Arneborg, N., Nielsen, D. S., et al. (2018). Occurrence of lactic acid bacteria and yeasts at species and strain level during spontaneous fermentation of mawè, a cereal dough produced in West Africa. Food Microbiol. 76, 267–278. doi: 10.1016/j.fm.2018.06.005
Houngbédji, M., Johansen, P., Padonou, S. W., Hounhouigan, D. J., Siegumfeldt, H., and Jespersen, L. (2019). Effects of intrinsic microbial stress factors on viability and physiological condition of yeasts isolated from spontaneously fermented cereal doughs. Int. J. Food Microbiol. 304, 75–88. doi: 10.1016/j.ijfoodmicro.2019.05.018
Hounhouigan, D. J., Nout, M. J. R., Nago, C. M., Houben, J. H., and Rombouts, F. M. (1994). Microbiological changes in mawè during natural fermentation. World J. Microb. Biot. 10, 410–413. doi: 10.1007/BF00144462
Hyma, K. E., Saerens, S. M., Verstrepen, K. J., and Fay, J. C. (2011). Divergence in wine characteristics produced by wild and domesticated strains of Saccharomyces cerevisiae. FEMS Yeast Res. 11, 540–551. doi: 10.1111/j.1567-1364.2011.00746.x
Jespersen, L. (2003). Occurrence and taxonomic characteristics of strains of Saccharomyces cerevisiae predominant in African indigenous fermented foods and beverages. FEMS Yeast Res. 3, 191–200. doi: 10.1016/S1567-1356(02)00185-X
Jespersen, L. (2005). “African fermented foods: role of yeasts,” in Food Fermentation, eds M. J. R. Nout, W. M. de Vos, and M. H. Zwietering (Wageningen: Wargening Acedemic Publishers), 45–52.
Jespersen, L., Halm, M., Kpodo, K., and Jakobsen, M. (1994). Significance of yeasts and moulds occurring in maize dough fermentation for “kenkey” production. Int. J. Food Microbiol. 24, 239–248. doi: 10.1016/0168-1605(94)90122-8
Johansen, P., and Jespersen, L. (2017). Impact of quorum sensing on the quality of fermented foods. Curr. Opin. Food Sci. 13, 16–25. doi: 10.1016/j.cofs.2017.01.001
Kayode, A. P. P., Vieira-Dalode, G., Linnemann, A. R., Kotchoni, S. O., Hounhouigan, D. J., van Boekel, M. A. J. S., et al. (2011). Diversity of yeasts involved in the fermentation of tchoukoutou, an opaque sorghum beer from Benin. Afr. J. Microbiol. Res. 5, 2737–2742. doi: 10.5897/AJMR11.546
Kebede, A., Viljoen, B. C., Gadaga, T. H., Narvhus, J. A., and Lourens-Hattingh, A. (2007). The effect of container type on the growth of yeast and lactic acid bacteria during production of Sethemi, South African spontaneously fermented milk. Food Res. Int. 40, 33–38. doi: 10.1016/j.foodres.2006.07.012
Kubo, R., Ohta, K., Funakawa, S., Kitabatake, N., Araki, S., and Izawa, S. (2014). Isolation of lactic acid-tolerant Saccharomyces cerevisiae from Cameroonian alcoholic beverage. J. Biosci. Bioeng. 118, 657–660. doi: 10.1016/j.jbiosc.2014.05.007
Kurtzman, C. P., Fell, J. W., and Boekhout, T. (2011). “Gene sequence analyses and other DNA-based methods for yeast species recognition,” in The Yeasts, eds C. P. Kurtzman, J. W. Fell, and T. Boekhout (Amsterdam: Elsevier).
Kurtzman, C. P., Mateo, R. Q., Kolecka, A., Theelen, B., Robert, V., and Boekhout, T. (2015). Advances in yeast systematics and phylogeny and their use as predictors of biotechnologically important metabolic pathways. FEMS Yeast Res. 15:fov050. doi: 10.1093/femsyr/fov050
Kurtzman, C. P., Robnett, C. J., and Basehoar-Powers, E. (2008). Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res. 8, 939–954. doi: 10.1111/j.1567-1364.2008.00419.x
Lambri, M., Fumi, M. D., Roda, A., and Faveri, D. M. D. (2013). Improved processing methods to reduce the total cyanide content of cassava roots from Burundi. Afr. J. Biotechnol. 12, 2685–2691.
Lamping, E., Ranchod, A., Nakamura, K., Tyndall, J. D. A., Niimi, K., Holmes, A. R., et al. (2009). Abc1p is a multidrug efflux transporter that tips the balance in favor of innate azole resistance in Candida krusei. Antimicrob. Agents Chemother. 53, 354–369. doi: 10.1128/AAC.01095-08
Lore, T. A., Mbugua, S. K., and Wangoh, J. (2005). Enumeration and identification of microflora in suusac, a Kenyan traditional fermented camel milk product. LWT Food Sci. Technol. 38, 125–130. doi: 10.1016/j.lwt.2004.05.008
Loretan, T., Mostert, J. F., and Viljoen, B. C. (2003). Microbial flora associated with South African household kefir. S. Afr. J. Sci. 99, 92–94.
Lyumugabe, F., Kamaliza, G., Bajyana, E., and Thonart, P. H. (2010). Microbiological and physico-chemical characteristic of Rwandese traditional beer “Ikigage.”. Afr. J. Biotechnol. 9, 4241–4246.
Lyumugabe, F., Uyisenga, J. P., Songa, E. B., and Thonart, P. (2014). Production of traditional sorghum beer “Ikigage” using Saccharomyces cerevisae, Lactobacillus fermentum and issatckenkia orientalis as starter cultures. Food Nutr. Sci. 5, 507–515. doi: 10.4236/fns.2014.56060
Maoura, N., Mbaiguinam, M., Ngyuen, H. D., Gaillardin, C., and Pourquie, J. (2005). Identification and typing of the yeast strains isolated from bili bili, a traditional sorghum beer of Chad. Afr. J. Biotechnol. 4, 646–656. doi: 10.5897/AJB2005.000-3117
Marco, M. L., Heeney, D., Binda, S., Cifelli, C. J., Cotter, P. D., Foligné, B., et al. (2017). Health benefits of fermented foods: microbiota and beyond. Curr. Opin. Biot. 44, 94–102. doi: 10.1016/j.copbio.2016.11.010
Marquina, D., Santos, A., and Peinado, J. (2002). Biology of killer yeasts. Int. Microbiol. 5, 65–71. doi: 10.1007/s10123-002-0066-z
Masoud, W., Cesar, L. B., Jespersen, L., and Jakobsen, M. (2004). Yeast involved in fermentation of Coffea arabica in East Africa determined by genotyping and by direct denaturating gradient gel electrophoresis. Yeast 21, 549–556. doi: 10.1002/yea.1124
Masoud, W., Poll, L., and Jakobsen, M. (2005). Influence of volatile compounds produced by yeasts predominant during processing of Coffea arabica in East Africa on growth and ochratoxin A (OTA) production by Aspergillus ochraceus. Yeast 22, 1133–1142. doi: 10.1002/yea.1304
Misihairabgwi, J. M., Kock, L., Pretorious, E., Pohl, C., and Zvauya, R. (2015). Characterisation of yeasts isolated from traditional opaque beer beverages brewed in Zimbabwean households. Afr. J. Microbiol. Res. 9, 549–556. doi: 10.5897/AJMR2014.7218
Motlhanka, K., Zhou, N., and Lebani, K. (2018). Microbial and chemical diversity of traditional non-cereal based alcoholic beverages of Sub-Saharan Africa. Beverages 4:36. doi: 10.3390/beverages4020036
Mpofu, A., Kock, J. L. F., Pretorious, E. E., Pohl, C. H., and Zvauya, R. (2008). Identification of yeasts isolated from mukumbi, a Zimbabwean traditional wine. J. Sustain. Dev. Afr. 10, 88–102.
Mufandaedza, J., Viljoen, B. C., Feresu, S. B., and Gadaga, T. H. (2006). Antimicrobial properties of lactic acid bacteria and yeast-LAB cultures isolated from traditional fermented milk against pathogenic Escherichia coli and Salmonella enteritidis strains. Int. J. Food Microbiol. 108, 147–152. doi: 10.1016/j.ijfoodmicro.2005.11.005
Mugula, J. K., Narvhus, J. A., and Sørhaug, T. (2003a). Use of starter cultures of lactic acid bacteria and yeasts in the preparation of togwa, a Tanzanian fermented food. Int. J. Food Microbiol. 83, 307–318. doi: 10.1016/S0168-1605(02)00386-0
Mugula, J. K., Nnko, S. A. M., Narvhus, J. A., and Sørhaug, T. (2003b). Microbiological and fermentation characteristics of togwa, a Tanzanian fermented food. Int. J. Food Microbiol. 80, 187–199. doi: 10.1016/S0168-1605(02)00141-1
Mukisa, I. M., Byaruhanga, Y. B., Muyanja, C. M. B. K., Langsrud, T., and Narvhus, J. A. (2017). Production of organic flavor compounds by dominant lactic acid bacteria and yeasts from Obushera, a traditional sorghum malt fermented beverage. Food Sci. Nutr. 5, 702–712. doi: 10.1002/fsn3.450
Mukisa, I. M., Porcellato, D., Byaruhanga, Y. B., Muyanja, C. M. B. K., Rudi, K., Langsrud, T., et al. (2012). The dominant microbial community associated with fermentation of Obushera (sorghum and millet beverages) determined by culture-dependent and culture-independent methods. Int. J. Food Microbiol. 160, 1–10. doi: 10.1016/j.ijfoodmicro.2012.09.023
Naumova, E., Korshunova, I., Jespersen, L., and Naumov, G. (2003). Molecular genetic identification of Saccharomyces cerevisiae sensu stricto strains from African sorghum beer. FEMS Yeast Res. 3, 177–184. doi: 10.1016/S1567-1356(02)00191-5
Navarrete-Bolaños, J. L. (2012). Improving traditional fermented beverages: how to evolve from spontaneous to directed fermentation: guidelines to design efficient directed fermentations based on spontaneous fermentations. Eng. Life Sci. 12, 410–418. doi: 10.1002/elsc.201100128
Nedović, V., Gibson, B., Mantzouridou, T. F., Bugarski, B., Djordjeviæ, V., Kaluševiæ, A., et al. (2015). Aroma formation by immobilized yeast cells in fermentation processes. Yeast 32, 173–216. doi: 10.1002/yea.3042
N’guessan, K. F., Brou, K., Jacques, N., Casaregola, S., and Dje, K. M. (2011). Identification of yeasts during alcoholic fermentation of tchapalo, a traditional sorghum beer from Côte d’Ivoire. Antonie Van Leeuwenhoek 99, 855–864. doi: 10.1007/s10482-011-9560-7
Nielsen, D. S., Hønholt, S., Tano-Debrah, K., and Jespersen, L. (2005). Yeast populations associated with Ghanaian cocoa fermentations analysed using denaturing gradient gel electrophoresis (DGGE). Yeast 22, 271–284. doi: 10.1002/yea.1207
Nieminen, T. T., Koskinen, K., Laine, P., Hultman, J., Säde, E., Paulin, L., et al. (2012). Comparison of microbial communities in marinated and unmarinated broiler meat by metagenomics. Int. J. Food Microbiol. 157, 142–149. doi: 10.1016/j.ijfoodmicro.2012.04.016
Nissen, P., Nielsen, D., and Arneborg, N. (2003). Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell-cell contact-mediated mechanism. Yeast 20, 331–341. doi: 10.1002/yea.965
Njage, P. M. K., Dolci, S., Jans, C., Wangoh, J., Lacroix, C., and Meile, L. (2011). Characterization of yeasts associated with camel milk using phenoypic and molecular identification techniques. Res. J. Microbiol. 6, 678–692. doi: 10.3923/jm.2011.678.692
Núñez, F., Lara, M. S., Peromingo, B., Delgado, J., Sánchez-Montero, L., and Andrade, M. J. (2015). Selection and evaluation of Debaryomyces hansenii isolates as potential bioprotective agents against toxigenic penicillia in dry-fermented sausages. Food Microbiol. 46, 114–120. doi: 10.1016/j.fm.2014.07.019
Nuobariene, L., Hansen, A. S., Jespersen, L., and Arneborg, N. (2011). Phytase-active yeasts from grain-based food and beer. J. Appl. Microbiol. 110, 1370–1380. doi: 10.1111/j.1365-2672.2011.04988.x
Nyambane, B., Thari, W. M., Wangoh, J., and Njage, P. M. K. (2014). Lactic acid bacteria and yeasts involved in the fermentation of amabere amaruranu, a Kenyan fermented milk. Food Sci. Nutr. 2, 692–699. doi: 10.1002/fsn3.162
Nyanga, L. K., Nout, M. J. R., Gadaga, T. H., Theelen, B., Boekhout, T., and Zwietering, M. H. (2007). Yeasts and lactic acid bacteria microbiota from masau (Ziziphus mauritiana) fruits and their fermented fruit pulp in Zimbabwe. Int. J. Food Microbiol. 120, 159–166. doi: 10.1016/j.ijfoodmicro.2007.06.021
Nyanga, L. K., Nout, M. J. R., Smid, E. J., Boekhout, T., and Zwietering, M. H. (2013). Fermentation characteristics of yeasts isolated from traditionally fermented masau (Ziziphus mauritiana) fruits. Int. J. Food Microbiol. 166, 426–432. doi: 10.1016/j.ijfoodmicro.2013.08.003
Obilie, E. M., Tano-Debrah, K., and Amoa-Awua, W. K. (2003). Microbial modification of the texture of grated cassava during fermentation into akyeke. Int. J. Food Microbiol. 89, 275–280. doi: 10.1016/S0168-1605(03)00294-0
Obodai, M., and Dodd, C. E. R. (2006). Characterization of dominant microbiota of a Ghanaian fermented milk product, nyarmie, by culture- and nonculture-based methods. J. Appl. Microbiol. 100, 1355–1363. doi: 10.1111/j.1365-2672.2006.02895.x
Ogunremi, O. R., Banwo, K., and Sanni, A. I. (2017). Starter-culture to improve the quality of cereal-based fermented foods: trends in selection and application. Curr. Opin. Food Sci. 13, 38–43. doi: 10.1016/j.cofs.2017.02.003
Ogunremi, O. R., Sanni, A. I., and Agrawal, R. (2015). Probiotic potentials of yeasts isolated from some cereal-based Nigerian traditional fermented food products. J. Appl. Microbiol. 119, 797–808. doi: 10.1111/jam.12875
Oguntoyinbo, F. A. (2008). Evaluation of diversity of Candida species isolated from fermented cassava during traditional small scale gari production in Nigeria. Food Control 19, 465–469. doi: 10.1016/j.foodcont.2007.05.010
Omemu, A. M., Oyewole, O. B., and Bankole, M. O. (2007). Significance of yeasts in the fermentation of maize for ogi production. Food Microbiol. 24, 571–576. doi: 10.1016/j.fm.2007.01.006
Ongol, M. P., and Asano, K. (2009). Main microorganisms involved in the fermentation of Ugandan ghee. Int. J. Food Microbiol. 133, 286–291. doi: 10.1016/j.ijfoodmicro.2009.06.003
Ouoba, L. I. I., Kando, C., Parkouda, C., Sawadogo-Lingani, H., Diawara, B., and Sutherland, J. P. (2012). The microbiology of Bandji, palm wine of Borassus akeassii from Burkina Faso: identification and genotypic diversity of yeasts, lactic acid and acetic acid bacteria. J. Appl. Microbiol. 113, 1428–1441. doi: 10.1111/jam.12014
Ouoba, L. I. I., Nielsen, D. S., Anyogu, A., Kando, C., Diawara, B., Jespersen, L., et al. (2015). Hanseniaspora jakobsenii sp. nov., a yeast isolated from Bandji, a traditional palm wine of Borassus akeassii. Int. J. Syst. Evol. Micr. 65, 3576–3579. doi: 10.1099/ijsem.0.000461
Oyewole, O. B. (2001). Characteristics and significance of yeasts’ involvement in cassava fermentation for ‘fufu’ production. Int. J. Food Microbiol. 65, 213–218. doi: 10.1016/S0168-1605(01)00431-7
Padonou, S. W., Hounhouigan, J. D., and Nago, M. C. (2009). Physical, chemical and microbiological characteristics of lafun produced in Benin. Afr. J. Biotechnol. 8, 3320–3325.
Padonou, S. W., Nielsen, D. S., Akissoe, N. H., Hounhouigan, J. D., Nago, M. C., and Jakobsen, M. (2010). Development of starter culture for improved processing of Lafun, an African fermented cassava food product: starter for Lafun processing. J. Appl. Microbiol. 109, 1402–1410. doi: 10.1111/j.1365-2672.2010.04769.x
Papon, N., Courdavault, V., Clastre, M., and Bennett, R. J. (2013). Emerging and emerged pathogenic Candida species: beyond the Candida albicans paradigm. PLoS Pathog. 9:e1003550. doi: 10.1371/journal.ppat.1003550
Pavlovic, M., Mewes, A., Maggipinto, M., Schmidt, W., Messelhäußer, U., Balsliemke, J., et al. (2014). MALDI-TOF MS based identification of food-borne yeast isolates. J. Microbiol. Methods 106, 123–128. doi: 10.1016/j.mimet.2014.08.021
Pedersen, L. L., Owusu-Kwarteng, J., Thorsen, L., and Jespersen, L. (2012). Biodiversity and probiotic potential of yeasts isolated from Fura, a West African spontaneously fermented cereal. Int. J. Food Microbiol. 159, 144–151. doi: 10.1016/j.ijfoodmicro.2012.08.016
Peter, J., De Chiara, M., Friedrich, A., Yue, J. X., Pflieger, D., Bergström, A., et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. doi: 10.1038/s41586-018-0030-5
Pfaller, M. A., and Castanheira, M. (2016). Nosocomial candidiasis: antifungal stewardship and the importance of rapid diagnosis. Med. Mycol. 54:myv076. doi: 10.1093/mmy/myv076
Ponomarova, O., Gabrielli, N., Sévin, D. C., Mülleder, M., Zirngibl, K., Bulyha, K., et al. (2017). Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 5, 345.e6–357.e6. doi: 10.1016/j.cels.2017.09.002
Romano, P., Capece, A., and Jespersen, L. (2006). “Taxonomic and Ecological Diversity of Food and Beverage Yeasts,” in Yeasts in Food and Beverages, eds A. Querol and G. Fleet (Berlin: Springer Berlin Heidelberg), 13–53. doi: 10.1007/978-3-540-28398-0_2
Sefa-Dedeh, S., Sanni, A. I., Tetteh, G., and Sakyi-Dawson, E. (1999). Yeasts in the traditional brewing of pito in Ghana. World J. Microb. Biot. 15, 593–597.
Shetty, P. H., Hald, B., and Jespersen, L. (2007). Surface binding of aflatoxin B1 by Saccharomyces cerevisiae strains with potential decontaminating abilities in indigenous fermented foods. Int. J. Food Microbiol. 113, 41–46. doi: 10.1016/j.ijfoodmicro.2006.07.013
Sieuwerts, S., De Bok, F. A. M., Hugenholtz, J., and Van Hylckama Vlieg, J. E. T. (2008). Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl. Environ. Microbiol. 74, 4997–5007. doi: 10.1128/AEM.00113-08
Silva, S., Negri, M., Henriques, M., Oliveira, R., Williams, D. W., and Azeredo, J. (2012). Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 36, 288–305. doi: 10.1111/j.1574-6976.2011.00278.x
Skandamis, P. N., and Nychas, G. J. E. (2012). Quorum sensing in the context of food microbiology. Appl. Environ. Microbiol. 78, 5473–5482. doi: 10.1128/AEM.00468-12
Smid, E. J., and Lacroix, C. (2013). Microbe-microbe interactions in mixed culture food fermentations. Curr. Opin. Biot. 24, 148–154. doi: 10.1016/j.copbio.2012.11.007
Soro-Yao, A. A., Brou, K., Amani, G., Thonart, P., and Dje, K. M. (2014). The use of lactic acid bacteria starter cultures during the processing of fermented cereal-based foods in West Africa: a review. Trop. Life Sci. Res. 25, 81–100.
Steinkraus, K. H. (1997). Classification of fermented foods: worldwide review of household fermentation techniques. Food Control 8, 311–317. doi: 10.1016/S0956-7135(97)00050-9
Stringini, M., Comitini, F., Taccari, M., and Ciani, M. (2009). Yeast diversity during tapping and fermentation of palm wine from Cameroon. Food Microbiol. 26, 415–420. doi: 10.1016/j.fm.2009.02.006
Suzzi, G. (2011). From wild strain to domesticated strain: the philosophy of microbial diversity in foods. Front. Microbiol. 2:169. doi: 10.3389/fmicb.2011.00169
Tamang, J. P., Shin, D. H., Jung, S. J., and Chae, S. W. (2016). Functional properties of microorganisms in fermented foods. Front. Microbiol. 7:578. doi: 10.3389/fmicb.2016.00578
Tapsoba, F., Legras, J. L., Savadogo, A., Dequin, S., and Traore, A. S. (2015). Diversity of Saccharomyces cerevisiae strains isolated from Borassus akeassii palm wines from Burkina Faso in comparison to other African beverages. Int. J. Food Microbiol. 211, 128–133. doi: 10.1016/j.ijfoodmicro.2015.07.010
Tra Bi, C. Y., N’guessan, F. K., Kouakou, C. A., Jacques, N., Casaregola, S., and Djè, M. K. (2016). Identification of yeasts isolated from raffia wine (Raphia hookeri) produced in Côte d’Ivoire and genotyping of Saccharomyces cerevisiae strains by PCR inter-delta. World J. Microb. Biot. 32, 1–9. doi: 10.1007/s11274-016-2095-3
van den Tempel, T., and Jakobsen, M. (2000). The technological characteristics of Debaryomyces hansenii and Yarrowia lipolytica and their potential as starter cultures for production of Danablu. Int. Dairy J. 10, 263–270. doi: 10.1016/S0958-6946(00)00053-4
van der Aa Kühle, A., Jesperen, L., Glover, R. L. K., Diawara, B., and Jakobsen, M. (2001). Identification and characterization of Saccharomyces cerevisiae strains isolated from West African sorghum beer. Yeast 18, 1069–1079. doi: 10.1002/yea.756
van der Aa Kühle, A., Skovgaard, K., and Jespersen, L. (2005). In vitro screening of probiotic properties of Saccharomyces cerevisiae var. boulardii and food-borne Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 101, 29–39. doi: 10.1016/j.ijfoodmicro.2004.10.039
van Der Walt, J. P. (1956). Kaffircorn malting and brewing studies. II.—Studies on the microbiology of Kaffir beer. J. Sci. Food Agr. 7, 105–113. doi: 10.1002/jsfa.2740070203
Vieira-Dalodé, G., Jespersen, L., Hounhouigan, J., Moller, P. L., Nago, C. M., and Jakobsen, M. (2007). Lactic acid bacteria and yeasts associated with gowé production from sorghum in Bénin. J. Appl. Microbiol. 103, 342–349. doi: 10.1111/j.1365-2672.2006.03252.x
Viljoen, C. B. (2006). “Yeast Ecological Interactions. Yeast – Yeast, Yeast – Bacteria, Yeast – Fungi Interactions and Yeasts as Biocontrol Agents,” in Yeast in Food and Beverages: The Yeast Handbook, eds A. Querol and G. H. Fleet (Berlin: Springer), 83–110. doi: 10.1007/978-3-540-28398-0_4
Wenning, M., Seiler, H., and Scherer, S. (2002). Fourier-transform infrared microspectroscopy, a novel and rapid tool for identification of yeasts. Appl. Environ. Microbiol. 68, 4717–4721. doi: 10.1128/AEM.68.10.4717-4721.2002
Witthuhn, R. C., Schoeman, T., and Britz, T. J. (2004). Isolation and characterization of the microbial population of different South African kefir grains. Int. J. Dairy Technol. 57, 33–37. doi: 10.1111/j.1471-0307.2004.00126.x
Wolfe, B. E., and Dutton, R. J. (2015). Fermented foods as experimentally tractable microbial ecosystems. Cell 161, 49–55. doi: 10.1016/j.cell.2015.02.034
Keywords: yeast, indigenous fermented foods, sub-Sahara Africa, species and strain diversity, yeast successions, microbial interaction, sensorial and nutritional quality, health benefits
Citation: Johansen PG, Owusu-Kwarteng J, Parkouda C, Padonou SW and Jespersen L (2019) Occurrence and Importance of Yeasts in Indigenous Fermented Food and Beverages Produced in Sub-Saharan Africa. Front. Microbiol. 10:1789. doi: 10.3389/fmicb.2019.01789
Received: 02 May 2019; Accepted: 19 July 2019;
Published: 06 August 2019.
Edited by:
Matthias Sipiczki, University of Debrecen, HungaryReviewed by:
Christian Ariel Lopes, Instituto de Investigacion y Desarrollo en Ingenieria de Procesos, Biotecnologia y Energias Alternativas (CONICET PROBIEN), ArgentinaRenata Vadkertiova, Slovak Academy of Sciences, Slovakia
Copyright © 2019 Johansen, Owusu-Kwarteng, Parkouda, Padonou and Jespersen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lene Jespersen, lj@food.ku.dk
 Pernille Greve Johansen
Pernille Greve Johansen James Owusu-Kwarteng
James Owusu-Kwarteng Charles Parkouda
Charles Parkouda S. Wilfrid Padonou
S. Wilfrid Padonou Lene Jespersen
Lene Jespersen