- 1Department for Innovation in Biological, Agro-food and Forest systems (DIBAF), University of Tuscia, Viterbo, Italy
- 2Next-Generation Agronomics (NGA) Laboratory, Tarragona, Spain
- 3Department of Agricultural and Forestry Sciences (DAFNE), University of Tuscia, Viterbo, Italy
- 4Vivai Piante Battistini soc. agr. s.s., Cesena, Italy
- 5Council for Agricultural Research and Economics (CREA), Research Centre for Food and Nutrition, Rome, Italy
The species Pantoea agglomerans includes strains that are agronomically relevant for their growth-promoting or biocontrol traits. Molecular analysis demonstrated that the IPDC pathway involved in the conversion of tryptophan (Trp) to indole-3-acetic acid (IAA) is highly conserved among P. agglomerans strains at both gene and protein levels. Results also indicated that the promoter region controlling the inducible expression of ipdC gene differs from the model system Enterobacter cloacae, which is in accordance with the observation that P. agglomerans accumulates higher levels of IAA when cells are collected in the exponential phase of growth. To assess the potential applications of these microorganisms for IAA production, P. agglomerans C1, an efficient auxin-producer strain, was cultivated in 5 L fermenter so as to evaluate the effect of the medium formulation, the physiological state of the cells, and the induction timing on the volumetric productivity. Results demonstrated that higher IAA levels were obtained by using a saline medium amended with yeast extract and saccharose and by providing Trp, which acts both as a precursor and an inducer, to a culture in the exponential phase of growth. Untargeted metabolomic analysis revealed a significant effect of the carbon source on the exometabolome profile relative to IAA-related compounds and other plant bioactive signaling molecules. The IAA-enriched metabolites secreted in the culture medium by P. agglomerans C1 were used as plant biostimulants to run a series of trials at a large-scale nursery farm. Tests were carried out with in vitro and ex vitro systems following the regular protocols used for large-scale plant tree agamic propagation. Results obtained with 4,540 microcuttings of Prunus rootstock GF/677 and 1,080 plantlets of Corylus avellana L. showed that metabolites from strain C1 improved percentage of rooted-explant, number of adventitious root formation, plant survival, and quality of plant as vigor, with an increase in the leaf area between 17.5 and 42.7% compared to IBA-K (indole-3-butyric acid potassium salt)–treated plants.
Introduction
Indole-3-acetic acid (IAA) is the most abundant member of the auxin family of phytohormones, and its biosynthesis in plants and bacteria proceeds through distinct biosynthetic routes: both tryptophan (Trp)–dependent and Trp-independent pathways have been described (Eckardt, 2001; Ljung et al., 2002). Based on the distinct intermediates involved in Trp-dependent IAA biosynthesis, five different pathways have been characterized in bacteria, namely, indole-3-acetamide (IAM), indole-3-pyruvic acid (IPyA), indole-3-acetonitrile (IAN), tryptamine (TAM), and Trp side-chain oxidase pathway (Patten et al., 2013; Doktycz, 2019). Although the Trp-independent pathway is thought to occur in bacteria as well (Prinsen et al., 1993), no specific enzymes in this pathway have been characterized.
The IPyA pathway is operational in plant-beneficial bacteria, such as Azospirillum brasilense and representative members of Enterobacter cloacae complex and is subjected to extremely tight regulation. In this pathway, there is transamination of Trp to IPyA, followed by decarboxylation to indole-3-acetaldehyde (IAAld) by the enzyme indole-3-pyruvate decarboxylase (IPDC; EC 4.1.1.74), and then oxidation of IAAld to IAA. The key enzyme in this pathway, IPDC, is encoded by ipdC, and deletion or functional inactivation of ipdC gene affects IAA biosynthesis in some strains, such as Enterobacter ludwigii (formerly, E. cloacae) UW5, A. brasilense, Pantoea agglomerans 299R, and Pantoea species YR343 (Koga et al., 1991; Brandl and Lindow, 1997; Malhotra and Srivastava, 2008; Garcia et al., 2019). The IPDC genes code for polypeptides of approximately 550 amino acids in length, corresponding to a molecular mass of 60 kDa per subunit. The homotetrameric IPDC from E. ludwigii UW5, characterized both at biochemical and structural levels, has a molecular mass of 240 kDa and binds four molecules of the cofactors thiamine diphosphate (ThDP) and Mg2+ (Schutz et al., 2003). The ipdC gene from E. ludwigii UW5 is activated by the transcription factor TyrR and increases in response to the aromatic amino acid Trp, tyrosine, and phenylalanine (Ryu and Patten, 2008; Coulson and Patten, 2015). However, regulation varies across bacterial species: constitutive in Agrobacterium; regulated by specific transcriptional factors, such as RpoS or RpoN, in some Pseudomonas and Enterobacter strains; regulated by the global signal transduction system GacS/GacA that controls secondary metabolism in several plant-associated gram-negative bacteria (Spaepen et al., 2007).
Indole-3-acetic acid plays an important role in the regulation of growth and development of vascular plants, including cell division, cell extension, and cell differentiation (Kasahara, 2016). It specifically plays a crucial role in root initiation, apical dominance, tropisms, and senescence (Zhao, 2010). In addition, IAA is produced by plants, as well as by some beneficial bacteria in the rhizosphere, where it acts as a signaling molecule with significant effects on the communication between plants and microorganisms, and on plant growth (Spaepen and Vanderleyden, 2011; Duca et al., 2014).
In recent years, several studies have reported alternative approaches to the application of phytohormones and plant growth regulators, such as the use of symbiotic organisms and/or natural biostimulants from microbial and non-microbial organisms in agriculture systems and in tissue cultures, which are also environment friendly, as for microbial biostimulants (Ruzzi and Aroca, 2015; Orlikowska et al., 2017; Rouphael and Colla, 2018). Within this framework, the species P. agglomerans has drawn attention for its plant growth–promoting activity.
Classification of P. agglomerans into the biosecurity group 2 and the fact that sometimes this species causes human infections particularly in immunocompromised people prevent its utilization as bioinoculant in Europe (Dutkiewicz et al., 2016a; Büyükcam et al., 2018). However, increasing evidence has shown that selected members of the P. agglomerans species can have a great potential as plant growth–promoting bacteria (Paredes-Páliz et al., 2016a, b, 2017) and comprise strains that are agronomically relevant for their growth-promoting or biocontrol traits and have been increasingly regarded as ideal candidates among plant growth–promoting rhizobacteria to be used as a biocontrol agent (Dutkiewicz et al., 2016b).
In details, P. agglomerans strain C1, isolated from the phyllosphere of lettuce plants (Lactuca sativa L.) treated with plant-derived protein hydrolysates (Luziatelli et al., 2016), has been studied for its potential as novel biostimulant in sustainable agriculture. It has been specifically characterized for its heavy metal resistance and metabolic capacities (Luziatelli et al., 2019b, 2020), as well as for its ability to solubilize phosphate, to inhibit plant pathogens, to produce IAA and siderophores, and to improve the use of rock phosphates and the growth of corn (Zea mays L.) and tomato (Solanum lycopersicum L.) in pot experiments (Luziatelli et al., 2019a, 2020; Saia et al., 2020). In particular, when P. agglomerans strain C1 is grown onto medium rich in Trp, large amounts of IAA are produced, which makes it a natural biostimulant suitable to be used in the rooting phase of micropropagation instead of the synthetic auxins (Luziatelli et al., 2020).
Micropropagation allows a rapid multiplication of several plant species in large-scale clonal plants and is becoming a widespread technique for rootstocks and crops species propagation (Loberant et al., 2010). On the nursery farming scale, micropropagation has a strong economic impact, because in a relatively short time period, reduced space and growing controlled conditions are required independently of environmental conditions and season period (Akin-Idowu et al., 2009). Micropropagation starts, in fact, from a small part of selected elite plant and allows the production thousands of plants that can be produced in a continuous process (Pierik and Scholten, 1994). However, rhizogenesis and root growth are critical morphophysiological events for plant survival in ex vitro conditions and are achieved by auxin treatment of unrooted microshoots as the last phase of in vitro culture and/or the first phase of in vivo acclimatization phase.
In micropropagation of woody crops, it has been observed that, besides IAA, synthetic molecules with strong auxin activities can be used. Among them, indole-3-butyric acid (IBA) is the compound of choice, thanks to its higher stability in the culture medium (Nissen and Sutter, 1990; Bhatti and Jha, 2010). Moreover, it has been assumed that, among the metabolites making up the pool of excreted molecules (exometabolome), there are molecules that could have a synergistic activity with auxin and/or elicitor roles, acting on the regulation pathways of adventitious rooting events and quality of roots.
The study was designed to test the biostimulant properties of metabolites excreted by P. agglomerans strain C1 on root induction and plant development of some woody fruit crop species in large-scale nurseries, at the phase of in vitro and ex vitro rooting and acclimation.
Materials and Methods
Strain and Media
Pantoea agglomerans C1 is an environmental strain isolated from the phyllosphere of lettuce (Luziatelli et al., 2016, 2019a). Strain C1 was maintained as LB Lennox (Lennox, 1955)–glycerol 20% (vol/vol) stocks at −80°C and revived on LB broth at 30°C under agitation [180 revolutions/min (rpm)]. For production of IAA, the strain can be cultivated on both saline and rich media supplemented with Trp (4 mM). A total of six media were used: LB broth Lennox (per liter, tryptone 10 g, yeast extract 5 g, NaCl 5 g); saline M9-glucose medium (Maniatis et al., 1982); yeast extract sucrose (YES) and yeast extract glucose (YEG) medium [1 × M9 saline solution, yeast extract 5 g L–1, carbon source (sucrose or glucose), 5 g L–1]; and reinforced YES/YEG medium (rYES and rYEG medium containing double the amount of yeast extract, 10 g L–1).
Pathway and Gene Identification
As a preliminary step, a database of bacterial proteins that are associated to the five major IAA biosynthetic pathways was constructed following a comprehensive search of the literature. Then, the complete set of 4497 protein sequences encoded by P. agglomerans C1 (Luziatelli et al., 2020) was compared against this manually curated set of IAA biosynthetic genes using the Basic Local Alignment Search Tool (BLAST; Altschul et al., 1990). Multiple protein sequence alignments were generated using ClustalW algorithm (Thompson et al., 1994). Bacterial promoter prediction program, BPROM1, was used to identify the position of the promoter, that is, -10 box and -35 box in the input sequence. The PATRIC BLASTN and BLASTP services were used to perform homology searches against P. agglomerans genomes available at PATRIC Website2 using, as queries, nucleotide/protein sequences of strain C1 corresponding to (1) the 4 kb ipdC-containing region; (2) the 5′ upstream region of ipdC gene; (3) the genes encoding aminotransferase (peg.1678), IPDC (ipdC; peg.1955), and IAAld dehydrogenase (peg.576 and peg.879); (4) the deduced protein sequences of peg.1678, peg.1955, peg.576, and peg.879. The resulting hits were filtered at 100% query coverage and ≥ 80% of sequence identity.
Growth Conditions
Seed cultures, from a freeze glycerol stock of P. agglomerans strain C1, were inoculated into 500 Erlenmeyer flasks containing 50 mL of LB medium and incubated at 30°C with agitation (180 rpm).
Seed cultures in the late exponential phase of growth [optical density at 600 nm (OD600) of 4.5–4.8] were used to inoculate, with an initial OD600 of 0.1, 25 mL of production medium amended with Trp (4 mM), and growth was monitored by measuring the wet weight of cells. In addition, LB + Trp (4 mM) was used as reference media for IAA biosynthesis.
After 24 h of growth at 30°C and 180 rpm, cells were removed by centrifugation (10 min at 8,000 g), and the supernatant was filter-sterilized through a 0.22 μm filter and stored at -20°C until use.
Fermentation and Optimization of the Induction Conditions
To evaluate the relationship between the physiological state of the cells and the IAA production level, P. agglomerans strain C1 was grown in a BioFlo 120 bench-top stirred tank reactor (Eppendorf S.r.l., Milan, Italy) equipped with a 7.5-L vessel, two Rushton impellers, digital ISM probes for dissolved oxygen (DO), and pH (Mettler-Toledo S.p.A., Milan, Italy) and a platinum RTD (Pt100) sensor for the temperature. The growth was carried out in YES medium (5 L) at 30°C under aerobic conditions. The oxygen concentration was maintained greater than 20% of saturation (oscillation between 25 and 40%), blowing sterile air at an aeration rate of 0.2–1.5 (vol/vol/m), and by regulating the impellers speed from 150 rpm (initial condition) to 525 rpm (end of the exponential phase of growth). The initial pH of the medium was adjusted to 6.6 by addition of 1 M HCl or 2 M of NaOH, and growth was carried out without pH control.
The reactor was inoculated at 5% (vol/vol) with an initial optical density (OD600) of 0.4, using an LB culture grown at 180 rpm and 30°C up to the late exponential phase. After 3, 3.5, 4, 4.5, 5, and 6 h of growth, 100 mL aliquots were collected from the fermenter, and the cells recovered by centrifugation (8,000 rpm for 10 min) were resuspended in fresh YES medium to obtain a final OD600 of 10 and used to inoculate shaken flasks containing YES medium (25 mL) amended with Trp (4 mM). All cultures were inoculated at the same initial OD600 of 0.5 and incubated at 180 rpm and 30°C for 18 h. For each time point, experiments were carried out in triplicate. To measure the accumulation of IAA in the culture medium, 1 mL samples were taken at intervals of 60 min for the first 2 h and at the end of the growth (18 h) and treated as reported before.
Spectrophotometric Determination of Indole Auxins
Auxin production was measured using Salkowski reagent as described previously (Luziatelli et al., 2020). In brief, 1 mL of filter-sterilized (0.22 μm) supernatant was added to 2 mL of Salkowski reagent (0.5 M FeCl3, 35% vol/vol HClO4), and the mixture was incubated at room temperature (in the dark) for 20 min. The presence of IAA and other indole auxins were detected measuring pink color development at 535 nm using a Cary 50 UV-Vis spectrophotometer (Agilent, Santa Clara, CA, United States). A series of IAA standard solutions of known concentrations were prepared to set up the calibration curve.
Metabolome Analysis by Quadrupole Time-of-Flight Liquid Chromatography–Mass Spectroscopy
For untargeted metabolomics, the sample (1 mL) was extracted in 5 mL of cold (−20°C) acidified (0.1% HCOOH) 80/20 methanol/water using an Ultra-Turrax Homogenizer (Ika T-25, Staufen, Germany), centrifuged at 1200 rpm and filtered through a 0.2 μm cellulose membrane. The analysis was carried out on an Agilent 6550 Q-TOF with ESI source, coupled with an Agilent 1290 UHPLC. A BEH C18 column from Waters (100 × 2.1 mm internal diameter, 1.7 μm) was used according to the procedure and gradient described in Tsugawa et al. (2019). Injection volume was 2 μL for all samples. A pooled quality control was obtained by mixing 10 μL of each sample and acquired in tandem mass spectroscopy mode using iterative function five consecutive times to increase the number of compounds with associate MS2 spectra. Blank filtering, alignment, and identification were accomplished using MS-DIAL (Tsugawa et al., 2015) and MS-FINDER (Tsugawa et al., 2016), with the procedure described by Blaženović et al. (2019).
The table with all compound peaks height was exported from MS-DIAL into MS-FLO (DeFelice et al., 2017) to reduce false positives and duplicates. Then, an internal developed workflow in R was employed for fold change and Benjamini-Hochberg corrected p-value, PLSDA analysis (Thévenot et al., 2015), and chemical enrichment analysis (Barupal et al., 2012).
Plant Inoculation
In vitro shoot tips of peach clonal rootstock “GF677” (Prunus persica × Prunus amygdalus) of 10 mm in length and in vitro shoot tips of hazelnut cv. “Fertile de Coutard” (Corylus avellana L.) of 15 mm in length were cultured on proliferation medium, containing a modified Quirin and Lepoivre (QL; Quoirin and Lepoivre, 1977) and Driver and Kuniyaki Walnut (DKW; Driver and Kuniyuki, 1984) basal salt solution, respectively, and enriched by 30 g L–1 sucrose and solidified by 6.8 g L–1 agar. The growth regulators added to the medium were as follows: 2.22 μM of 6-benzyladenine (BA), 0.05 μM of α-naphthalene acetic acid for the rootstock GF677; 10 μM of BA, 0.05 μM of IBA for the cv. Fertile de Coutard. The pH of the medium was adjusted to 6.3 ± 0.1 for the rootstock GF677, and 6.0 ± 0.1 for the cv. Fertile de Coutard, before addition of agar and sterilization at 120°C for 20 min. Shoots of rootstock GF677 were subcultured at a 4-week interval, while shoots of cv. Fertile de Coutard were subcultured at a 6-week interval, under 16 h light photoperiod, using white fluorescent lamps Philips TL-D 58/865-MASTER (Philips, Italia), at 40 μM m–2 s–1 photon flux at constant temperature of 23°C ± 1°C.
After the proliferation step, microcuttings were transferred for 15 days in Murashige and Skoog (MS) elongation medium (Murashige and Skoog, 1962), supplemented with 14 μM gibberellic acid (GA3) and 20 g L–1 sucrose. The medium was sterilized at 120°C (2 bars), for 20 min after addition of 6.8 g L–1 agar and pH titration to 6.5. Glass jars of 500 mL in volume, each containing 100 mL of culture medium, were used as culture vessels.
At the beginning of the acclimatization, before the transfer into the cell plug tray, plantlets were immersed for 10 s in an auxin solution containing either an appropriate dilution of IAA-enriched excretome from P. agglomerans strain C1 (indole auxins final concentration of 1 μM) or 10 μM IBA potassium salt (IBA-K). Ex vitro acclimatization started on April 2018 by transferring the treated plantlets into 360 cells plug tray (Jiffy, Netherlands), with a volume of each cell cavity of 7 cm3.
Trays were placed under controlled misting of the greenhouse (temperature 28°C ± 4°C, RH 90%), at natural photoperiod and photosynthetic active radiation varying daily between 300 and 500 μM m–2 s–1.
All on-farm trials, two with Prunus rootstock GF677 and one with hazelnut, were carried out in agreement with the company’s production cycle. In the first trial with rootstock GF677, 720 plants (two trays of 360 cells) were treated by dipping with secreted metabolites from strain C1, and an equal number of plants were treated with IBA-K solution. In the second trial, 3,780 plants (10.5 trays of 360 cells) were treated with C1 metabolites, and 1,800 plants (five trays of 360 cells) with IBA-K. In both experiments, after 10 days from the dip treatment, plants were treated again by spraying fine drops of the same solution until the complete wetting of the leaves was achieved.
For hazelnut adventitious rooting induction experiments, 1,440 plantlets were treated by dipping with secreted metabolites from strain C1, and 1,080 plantlets were treated with IBA-K.
On the 20th day of the ex vitro acclimatization, the percentage of rooted plantlets, the number of roots per plantlet, the root length, and the elongation of plantlet stem were recorded.
After 1 month, all plantlets were transferred into 60 preloaded cells plug tray (Jiffy), each cell plug with a volume of 117 cm3. Two weeks later, the survival ratio of plant was determined, and the total leaf area per plant was detected by Android “CANOPEO” application3 on a total of 600 randomly chosen plants (300 for control and 300 Pantoea-treated).
Since the beginning of the transfer in vivo, every 20 days, plants were fertilized with NUTRIGREEN AD (GREEN HAS ITALIA S.p.A.), through the fertigation system at the amount of 2 mL L–1.
Statistical Analysis
Statistically significant differences between the means were determined by the one-way analysis of variance using the SigmaStat 3.1 package (Systat Software Inc., San Jose, CA, United States).
Results
Identification of the IAA-Biosynthetic Genes
Genes encoding enzymes involved in IAA synthesis were identified in P. agglomerans C1 genome by BLASTx analysis using the sequences of 11 different enzymes associated with alternative IAA pathways, as queries (Table 1). This analysis revealed the presence, in strain C1, of the whole set of genes of only one of the five IAA pathways occurring in bacteria and plants: the indole-3-pyruvic acid (IPyA) pathway (Table 1 and Figure 1). Noteworthy, for each of the three enzymes involved in this pathway, the identity, at amino acid level, with sequences of other Pantoea strains was between 81 and 92% along the entire protein length (Table 1). For the same proteins, the identity between sequences from strain C1 and strains belonging to other genera, including Azospirillum, Pseudomonas, Enterobacter, Azospirillum, and Arthrobacter, varied between 25 and 56% (Table 1).
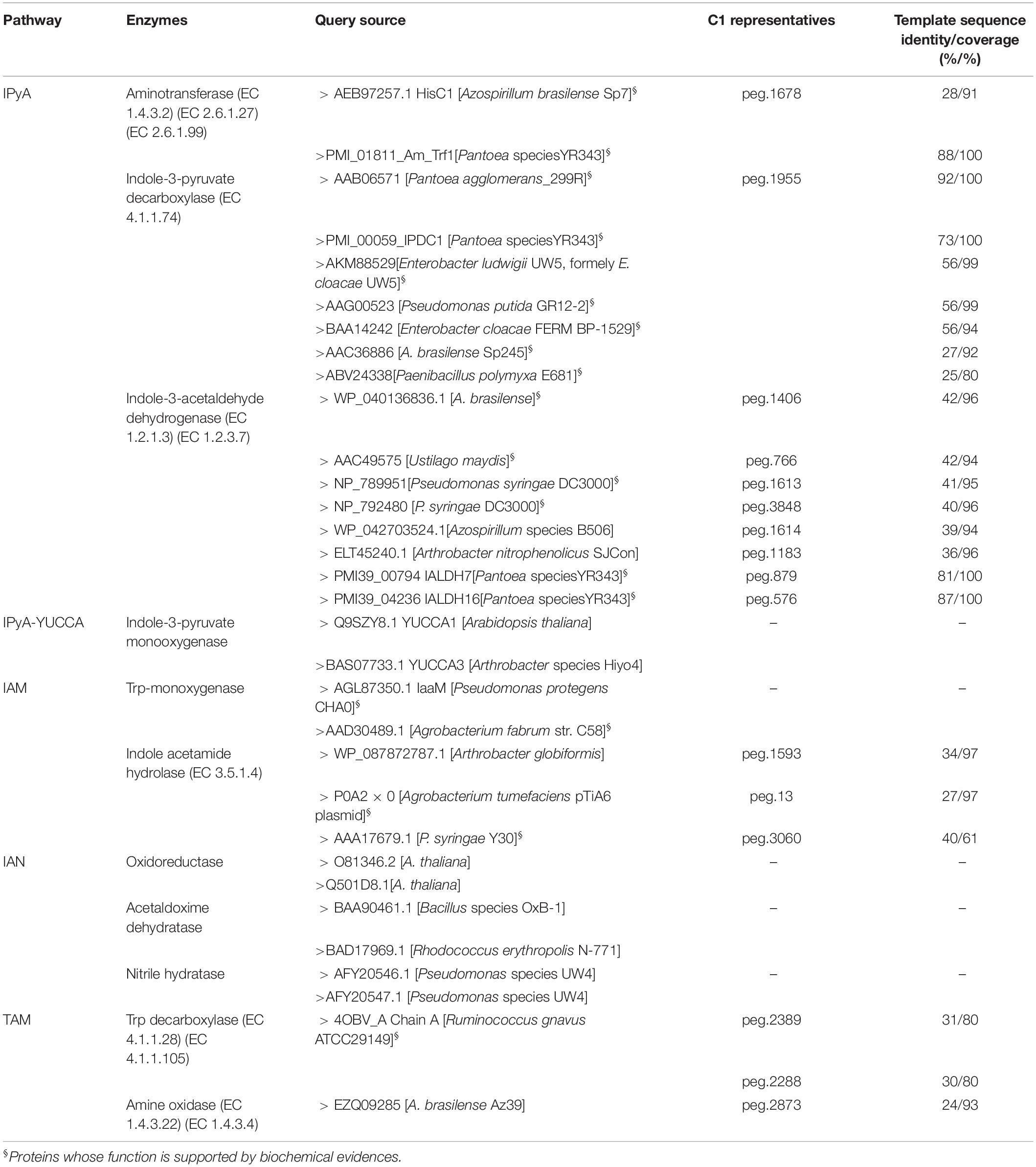
Table 1. Identification of protein encoding genes (peg) and enzymes involved in IAA biosynthesis in P. agglomerans strain C1 based on a systematic computational approach.
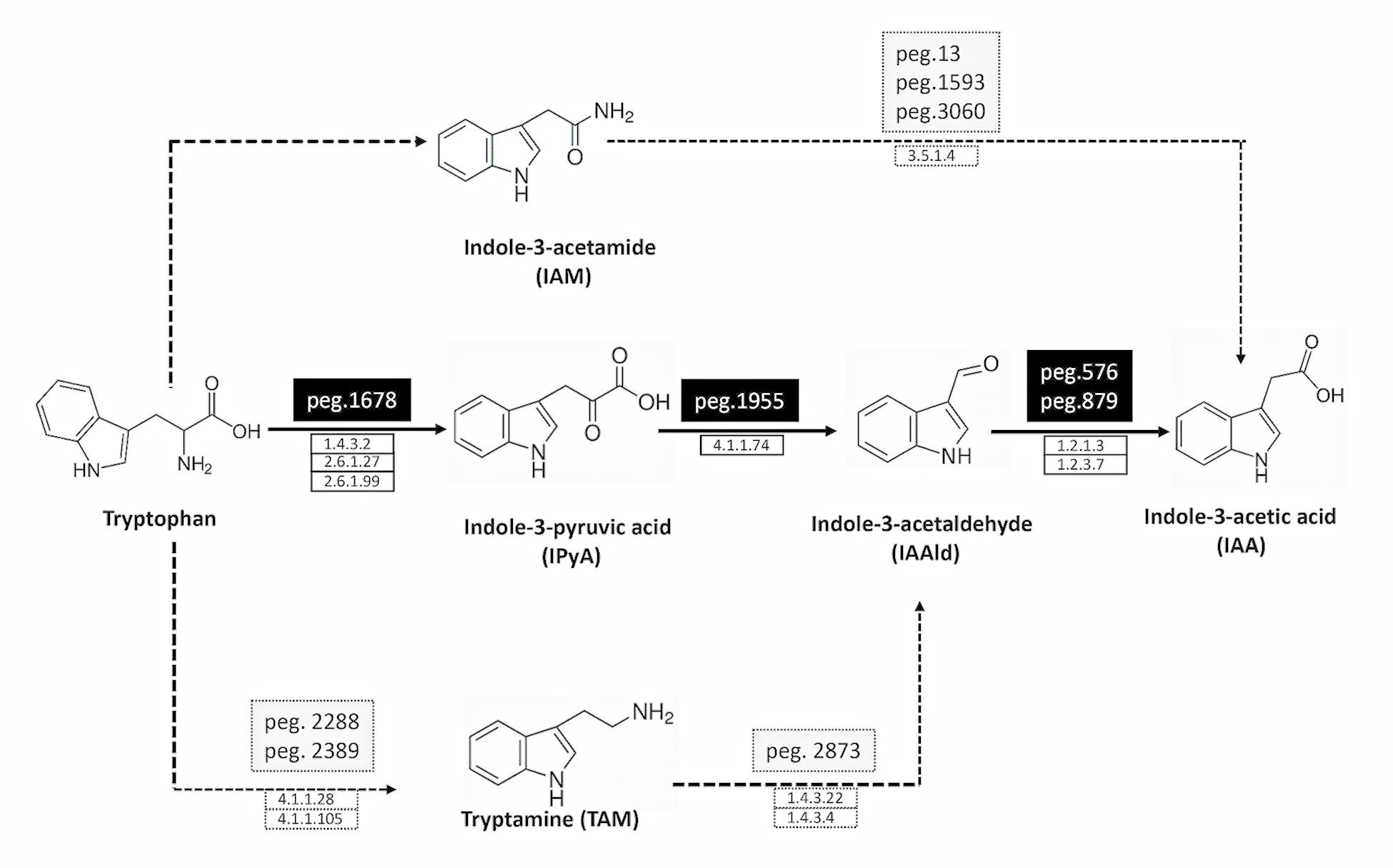
Figure 1. Deduced pathways for IAA biosynthesis in P. agglomerans strain C1. The solid line indicates the principal pathway for IAA biosynthesis, and the dotted ones the pathways that are not fully supported by molecular evidences (low identity with known proteins; see Table 1 for details). The names of the pathways indicate the name of their first products. The names of the protein encoding genes (peg) and the definition of the protein coding enzymes (EC number) are reported on the top and the bottom of the arrows, respectively.
Proteins with weak similarity to (i) indole acetamide hydrolase (pathway IAM) and (ii) Trp decarboxylase and amine oxidase (pathway TAM) were also identified (Table 1 and Figure 1). In contrast, no sequence related to Trp aminotransferase and YUCCA enzymes (pathway IPyA-YUCCA) or nitrilases (pathway IAN) was detected (Table 1).
Sequence Comparison of IPDC Proteins
BLAST analysis of the ThDP-binding indolepyruvate decarboxylase (IPDC EC 4.1.1.74) revealed (Table 1) that the deduced amino acid sequence encoded by C1_peg1955 from P. agglomerans C1 (IPDCPa_C1) shares a 92% of identity with IPDC from P. agglomerans (formerly Erwinia herbicola) strain 299R (IPDCPa_299R; Brandl and Lindow, 1996) and 73% of identity with IPDC1 from Pantoea species strain YR343 (IPDC1Psp_YR343; Garcia et al., 2019). To gain more information about IPDCPa_C1, a comparative analysis was carried out between this protein and IPCD from E. ludwigii (previously misidentified as Pseudomonas putida and subsequently as E. cloacae) strain UW5 (IPDCEc_UW5). The structural model of IPDCEc_UW5 has been published almost 10 years ago by Schutz et al. (2003), and the authors demonstrated that IPDCEc_UW5 is a homotetrameric enzyme in which each monomer has defined domains involved in the binding of both the substrate and the cofactors (Mg2+ and ThDP). Using the MULTALIN tool (Corpet, 1988), IPDC sequences from strain C1, E. ludwigii UW5 and other Pantoea strains were aligned as reported in Table 1. This analysis allowed demonstrating that several amino acids, which were found to be essential for the activity of IPDCEc_UW5, were conserved or conservatively replaced in all Pantoea strains (Figure 2). In detail, 90% of the active site residues of IPDCEc_UW5 are conserved in IPDCPa_C1, suggesting that the two enzymes might have a similar catalytic mechanism.

Figure 2. Sequence alignment of peg.1955 encoded protein from P. agglomerans strain C1 and functionally characterized IPDC from E. ludwigii and other Pantoea strains. Residues in IPDC sequence from E. ludwigii UW5 that have been identified by crystal structure analysis as being at the active site or involved in either ThDP or substrate binding are highlighted in black. In the alignment, the amino acid residues of these regions that are conserved among all or almost all different sequences are highlighted in black or gray.
Organization of the IPDC Coding Region
DNA sequence analysis of the genomic region surrounding the ipdC gene from P. agglomerans C1 revealed the presence of two ORFs encoding a 330-amino acid protein, annotated as L-glyceraldehyde 3-phosphate reductase (ORF1), and a 322-amino acid protein, with high homology to glucokinase, respectively (Figure 3). Interestingly, a similar genetic structure occurs in E. ludwigii UW5 (Coulson and Patten, 2015) and Pantoea species YR343 (Garcia et al., 2019; Figure 3), as well as in 45 of 50 P. agglomerans genomes sequenced so far. In the latter case, the identity over the entire 4 kb sequence of the ORF1-ipdC-ORF2 gene cluster was between 88 and 100%, with a mean value of 99% (Figure 4, lane A). Surprisingly, a high sequence identity (mean value = 98%) was also observed (Figure 4, lane Cg-Fg) comparing the DNA sequence of all P. agglomerans genes of the IPyA pathway, the genes encoding amino transferase, IPDC and IAAld decarboxylase (peg.1678, peg.1955, peg.576, and peg.879 in strain C1; Figure 1). The remarkable evolutionary conservation of the IPyA pathway among the members of the P. agglomerans species was also confirmed analyzing the variability at the level of protein sequence that varied between 1 and 2% and the kernel density plot (Figure 4, lane Cp-Fp). A similar observation was done analyzing the ipdC promoter region, which, as shown by the density of the data in the violin plot reported in Figure 4 (lane B), is more conserved than the IPCD coding sequence.
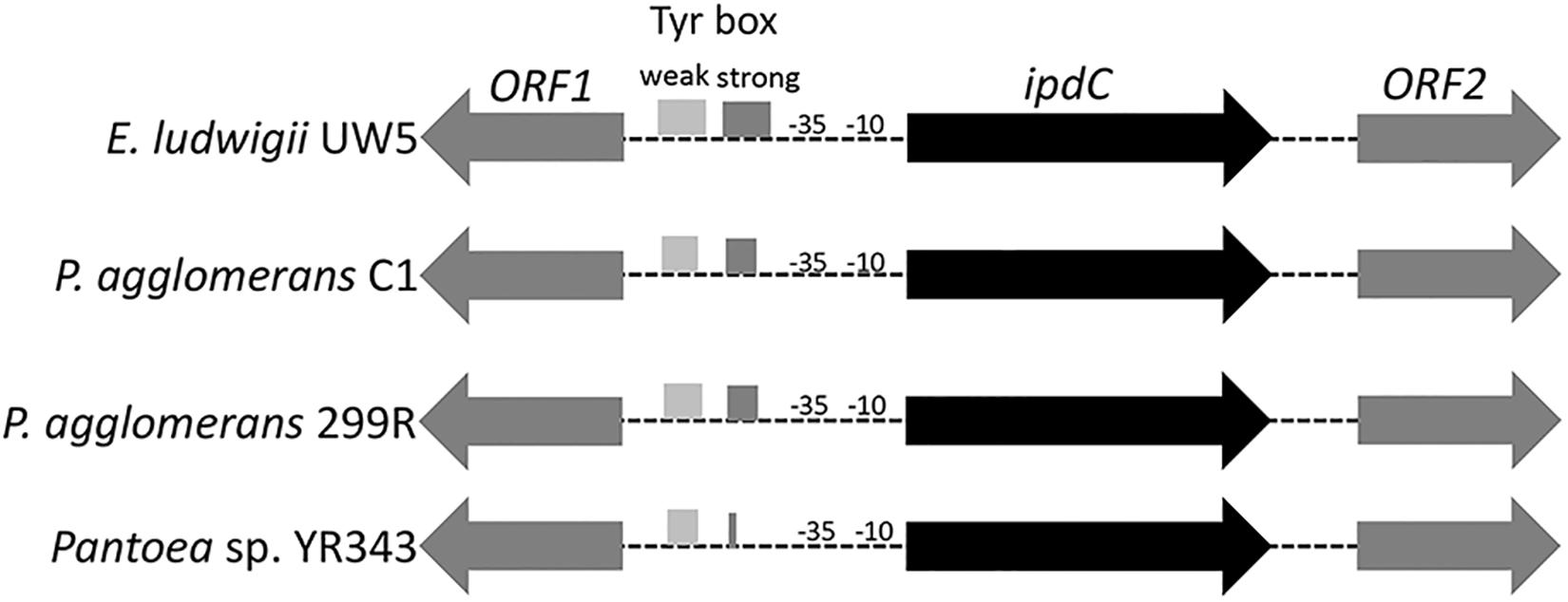
Figure 3. Organization of the chromosome region surrounding ipdC across different species. The boxes in the ipdC promoter region indicate the location of the sequences with partial similarity to the two TyrR binding boxes occurring in E. ludwigii EW5. The predicted -10 and -35 elements for ipdC gene are also shown.
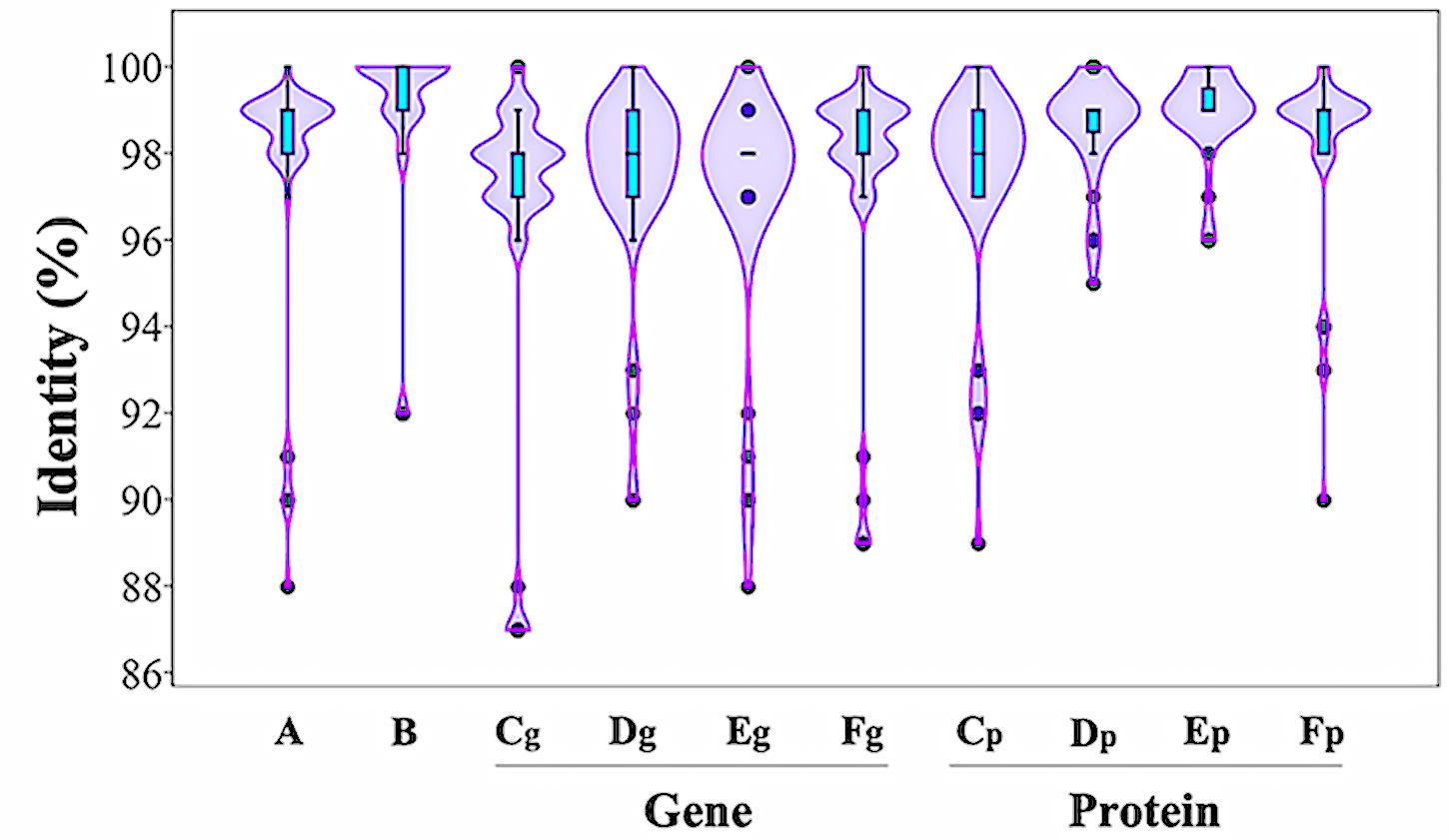
Figure 4. Distribution of sequence identity matches of P. agglomerans strain C1 to other P. agglomerans DNA and protein sequences related to IPyA pathway. The violin plots show the distribution of sequence identities of the ipdC gene cluster (A), the ipdC promoter region (B), the IPDC gene (Cg) and protein (Cp), the amino transferase gene (Dg) and protein (Dp), and the two IAAld dehydrogenase genes (Eg and Fg) and proteins (Ep and Ep).
Analysis of the ipdC Promoter
A search of functional motifs carried out with BPROM annotation package (Softberry Inc., Mount Kisco, NY, United States) revealed the presence, in the promoter region upstream of P. agglomerans C1 ipdC gene, of sequences that resemble to the Escherichia coli RpoD (s70) -10 and -35 elements, matching the E. coli consensus sequences at four of six nucleotides (-10), and five of six nucleotides (-35), respectively. Interestingly, the ipdC putative regulatory sites of C1 showed significant similarity with those predicted in the ipdC promoter of other Pantoea strains (YR343 and 299R) and E. ludwigii UW5 (Figure 5).
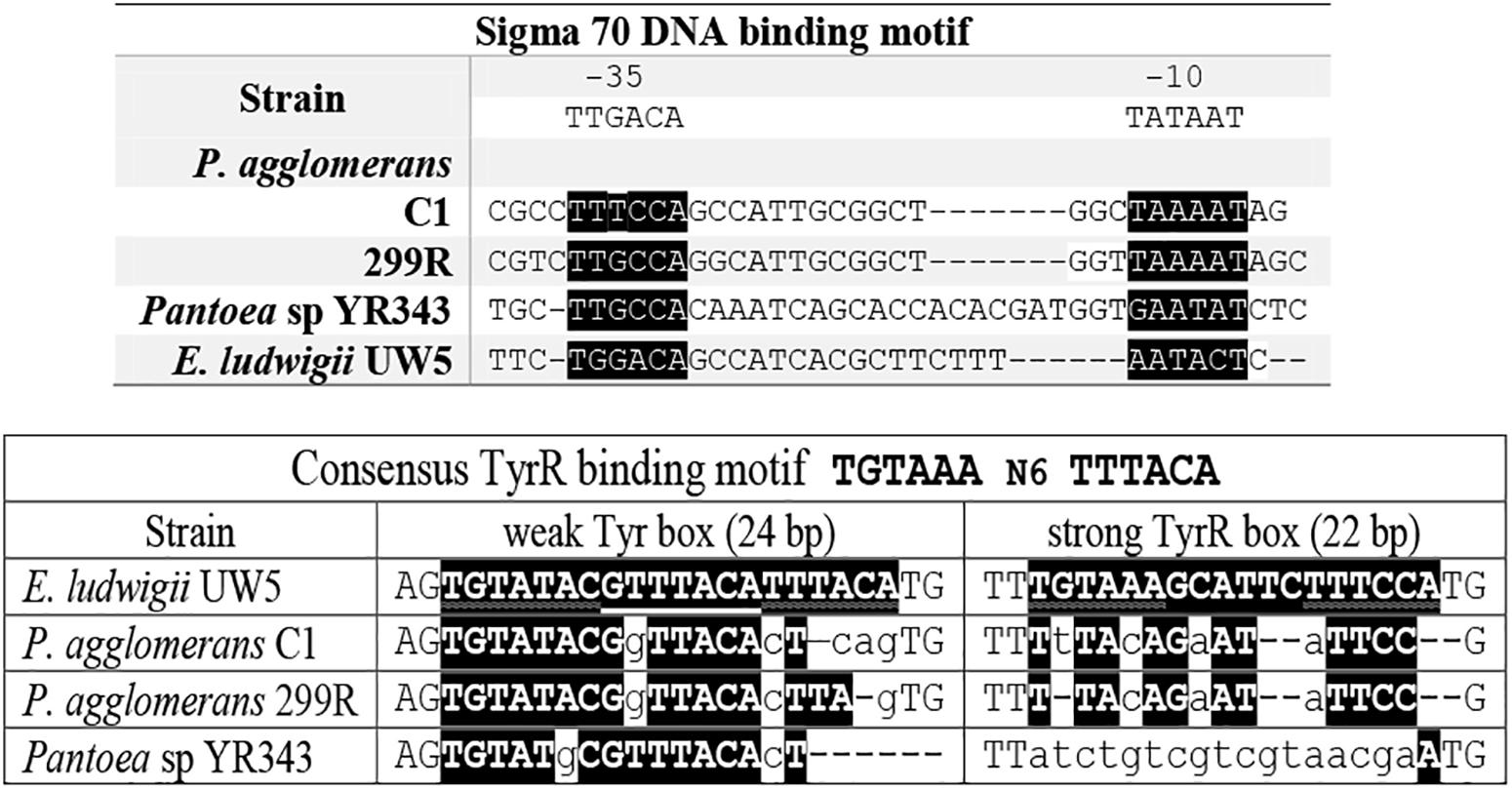
Figure 5. Alignment of relevant elements of the ipdC promoter sequence of E. ludwigii UW5, P. agglomerans C1, P. agglomerans 299R, and Pantoea species YR343. The conserved nucleotides in the -10 and -35 region (upper panel) and putative TyrR binding sites (lower panel) are highlighted in black.
Inspection of the 5′ untranslated region upstream from the C1_ipdC start codon showed no detectable cis-regulatory element such as the inverted repeats identified in the ipdC promoter of A. brasilense (Vande Broek et al., 2005) or the two 18-bp consensus sequences (weak and strong TyrR boxes) recognized by TyrR, which regulates the expression of the ipdC gene in E. ludwigii UW5 (Coulson and Patten, 2015). Sequences with weak similarity to the TyrR consensus motif (TGTAAA-N6-TTTACA) were found in the ipdC promoter region from Pantoea strains, but they do not meet the minimum molecular requirements for TyrR-mediated regulation: the presence of a strong TyrR box or a weak box with an adjacent strong box; the presence in the TyrR box of the G-C residues, essential for TyrR binding, spaced 14 bp apart (Figure 5). These evidences strongly support the hypothesis that the transcription of the ipdC gene in P. agglomerans is not controlled by the TyrR regulatory system.
Selection of the Growth Medium for IAA Production
In preliminary experiments, it was observed that, in contrast to LB, when P. agglomerans C1 cells were grown on saline M9-glucose medium without Trp, no basal level of IAA was produced. Providing Trp (as inducer and precursor) at 4 mM concentration, no significant difference was observed in the IAA final titer (54 ± 0.9 mg L–1) shifting from LB to M9-glucose medium. To further investigate the possibility to use a medium that has equivalent performances of LB but is free of animal-derived ingredients, the ability of C1 cells to grow and produce IAA on a simplified culture medium containing a saline phosphate buffer (M9 saline solution), a sugar, as a carbon source, and yeast extract, as a source of organic nitrogen, vitamins, and other growth factors, was investigated. For this purpose, glucose or sucrose, as a carbon source, was used alternatively, in combination with two concentrations of yeast extract (5 or 10 g L–1). All four media were amended with Trp (4 mM), and LB-Trp was used as a control.
Results reported in Figure 6 indicate that IAA production occurred in all tested conditions and, independently from the carbon source and the amount of yeast extract that was used, was higher in saline medium compared to the control medium (LB-Trp). In particular, the higher level of biomass (64.1 ± 3.1 g [wet weight] L–1) and IAA (120.5 ± 0.9 μg mL–1) were obtained cultivating the microorganism in the presence of both sucrose and yeast extract at a final concentration of 0.5% (wt/vol) (SYE medium; Figure 5). Surprisingly, an increase in the organic nitrogen content, with double the amount of yeast extract [from 0.5 to 1% (wt/vol)], did not affect IAA production (80.13 ± 0.11 μg mL–1 in rGYE and 88.1 ± 0.44 μg mL–1 in rSYE) and biomass yield (47.1 ± 2.5 or 48.7 ± 2.2 g [wet weight] L–1) (Figure 6).
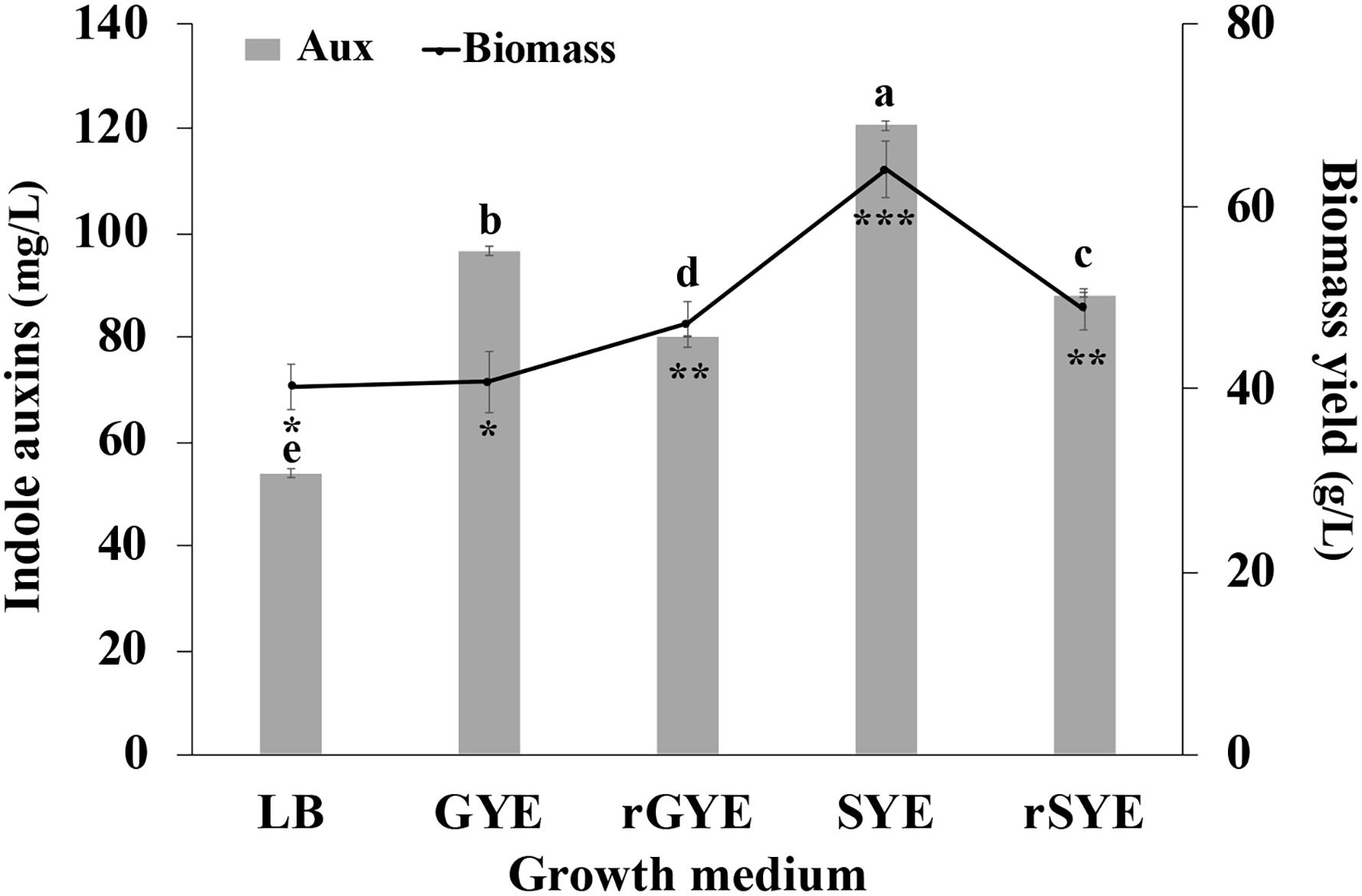
Figure 6. Effect of culture medium on IAA production (mg L–1) and biomass concentration (g [wet weight] L–1). LB, YEG, and YES contained 5 g L–1 of yeast extract; in reinforced media (rYEG and rYES) the concentration of this ingredient is doubled (10 g L–1). Differences in letters and symbols indicate that the values are significantly different (P > 0.05).
Interestingly, there was no significant difference in the biomass yield when cells were grown on GYE or LB medium (40.8 ± 3.3 g [wet weight] L–1 and 40.3 ± 2.5 g [wet weight] L–1), but the IAA-specific productivity shifting from LB to GYE had approximately 1.8-fold increase (Figure 6). These results suggested that LB was not an optimal medium for IAA production by P. agglomerans strain C1.
Overall Impact of the Carbon Source on the Metabolites Secreted by P. agglomerans C1
Results obtained in shake-flask experiments indicated that, in contrast to previous finding obtained with P. agglomerans 299R (Brandl and Lindow, 1997), the production of IAA from strain C1 was affected by the carbon source. For this reason, the effect of the carbon source on the production of IAA and IAA-related compounds was studied in more detail by analyzing the exometabolome of P. agglomerans C1 using an untargeted metabolomics approach. Of the 528 features detected in the exhausted growth medium, when cells were cultured on YES or YEG amended with Trp, a total of 381 were more than twofold higher, and 94 were more than twofold lower when sucrose was used as a carbon source. ChemRICH analysis showed that a total of 58 metabolite clusters were significantly different (P > 0.05) between YES and YEG (Figure 7 and Supplementary Table S1). For 27 of these clusters, the differences arise from an increased level in all the compounds of the cluster, and for other 10 clusters, the enriched molecules represented 75–95% of the total compounds of the cluster. As shown in Figure 7, in sucrose-grown cultures, the classes of metabolites with the highest elevated level were as follows: dipeptides and cyclic peptides; triterpenes; compounds belonging to new clusters 1, 21, and 49. Clustering analysis also revealed significant differences in the concentration level of single metabolites belonging to the indole-oligopeptides clusters and to the flavonoids cluster. In the first two clusters, we observed an increase of some bisindole alkaloids, such as the guaiaflavine (140-fold); an increase of some monoindole alkaloids, such as the indole-3-carbinol (I3C; 4.3-fold) and the IAA (2.2-fold); an increase of some amide-linked-IAA-L-amino acid conjugates (IAA-aa), such as indole-3-acetyl-L-valine (39-fold) and indole-3-acetyl-L-leucine (2.4-fold); a threefold decrease of aldehyde derivatives, such as IAAld and 1H-indole-3-carboxaldehyde (I3A) (Supplementary Table S2). In the flavonoids cluster, a significant increase in N-containing flavonoids, such as phyllospadine (54-fold increase), was observed. These results clearly indicate that in P. agglomerans C1 the levels of secreted metabolites were significantly affected by the medium carbon source.
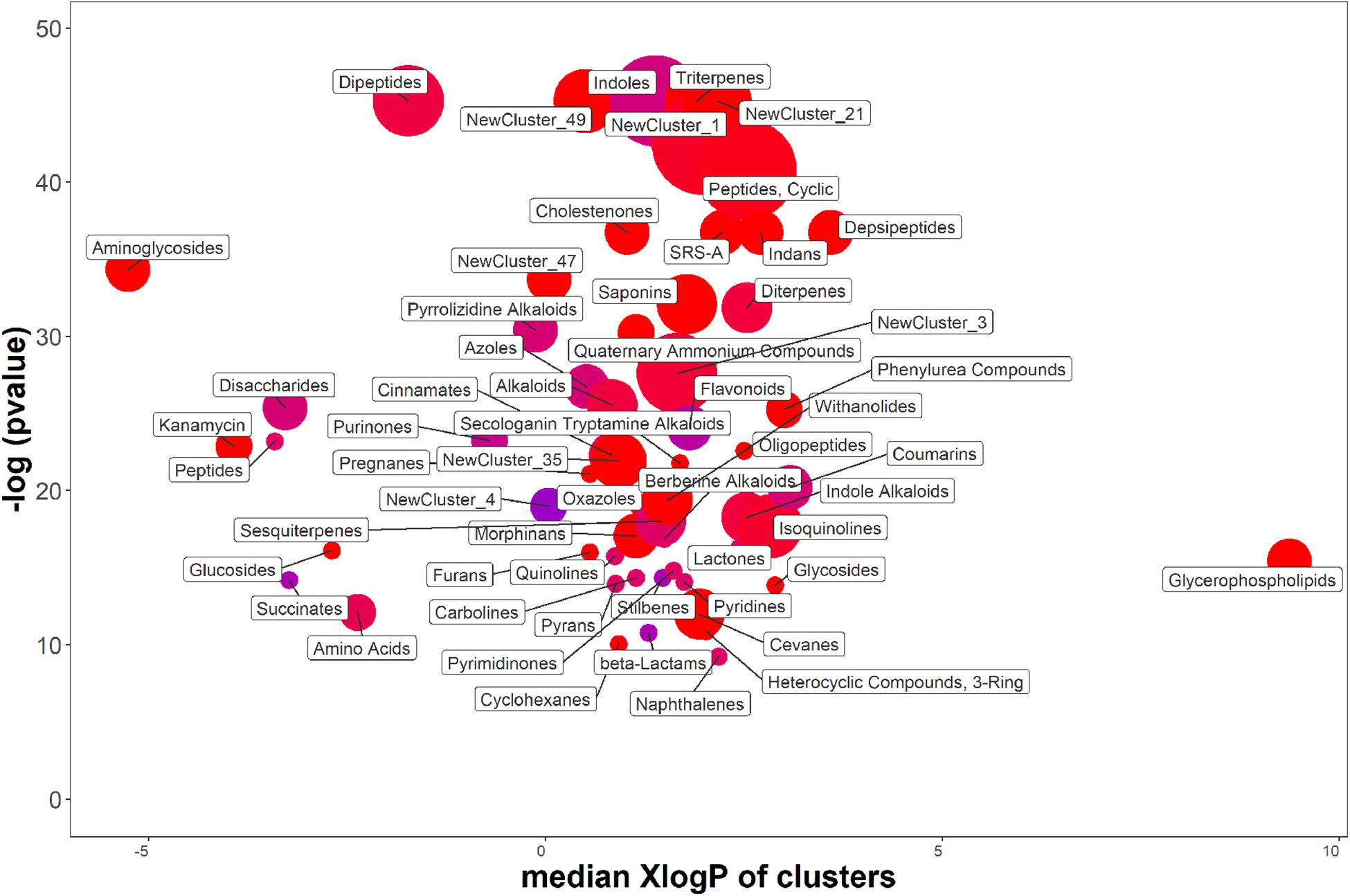
Figure 7. ChemRich analysis of exometabolome of P. agglomerans C1 grown on YES vs. YEG medium. Red clusters associated with higher outcomes, and the blue ones associated with lower outcomes.
Optimization of the Growth Conditions in Bench-Top Fermenter
In contrast with previous findings obtained with Enterobacter and Agrobacterium, in which the ipdC gene is under the control of TyrR and the accumulation of IAA occurred only after entrance in the stationary phase (Ryu and Patten, 2008), when P. agglomerans strain C1 was grown on LB medium, as well as on M9 glucose, no significant production of IAA was detected when Trp was provided to resting cells or to stationary-phase cultures. To better understand the link between the cell physiological state and the biosynthesis of IAA in P. agglomerans, strain C1 was grown under controlled bioreactor conditions ensuring optimum oxygen uptake and temperature control and high growth rates.
Analyzing the growth profile of C1 cultures grown at 30°C on YES medium under aerobic conditions (DO level > 20% of saturation), a lag phase of 30 min was observed followed by an acceleration phase (up to 1.5 h) during which the growth rate gradually increased (Figure 8). In the exponential phase, which reached its maximum at approximately 4 h, the specific growth rate was 1.63 h–1. Between the 4th and the 5.5th hour, the specific growth rate decreased to approximately 67% (from 1.63 to 0.53 h–1) and at approximately 5.5 h the culture entered in the stationary phase (Figure 8). Interestingly, during the first 3.5 h of growth, medium pH decreased slowly from 6.6 to 6.5, at a rate of approximately 0.028 unit per hour (Figure 8). In YES medium, when the culture reached the end of the exponentially phase (4 h), the pH started to decrease rapidly (at a rate of 0.87 unit per hour) and reached a minimum value of 6.22 at 5 h. Later, at the beginning of the stationary phase, the medium pH increased from 6.22 up to 6.34 and remained constant up to the end of the fermentation (Figure 8). Variations in the medium pH between the 3.5th and the 5.5th hour occurred concurrently with changes in the oxygen consumption rate that were controlled by DO-dependent modifications of the agitation speed and the airflow using a closed-loop system. As shown in Supplementary Figures S1B,C, the oxygen demand suddenly increased between 4 and 4.5 h (agitation speed increased > 10% in 30 min, from 400 to 450 rpm) and then remained stable up the beginning of the stationary phase (5.5 h).
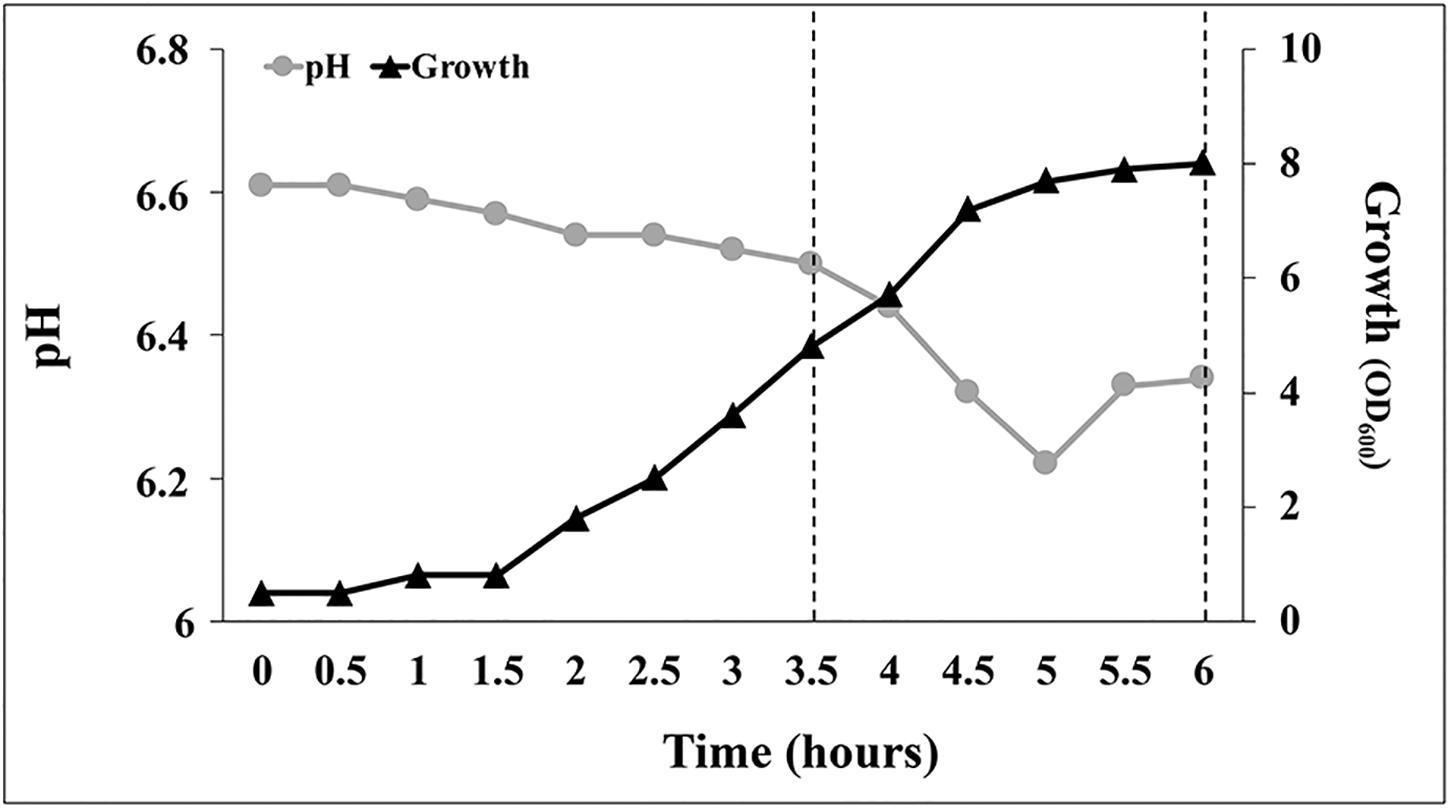
Figure 8. Growth pattern and pH evolution during batch scale tests in YES medium under aerobic conditions at 30°C. Data points are mean values of duplicate fermentations.
The growth of P. agglomerans strain C1 in bioreactor allowed increasing the biomass yield compared to shake-flask cultures of about sixfold, up to 292.5 ± 4.8 g [wet weight] L–1.
Influence of the Physiological State of the Inoculum on IAA Production
Cells collected at different time points during the growth in bioreactor were transferred in medium containing Trp, so as to evaluate the effect of the physiological state of the cells used for inoculation on the IAA production. The growth was carried out in shake flasks at 30°C, and the IAA production was monitored during the first 2 h and at the end of the growth. In all tested conditions, no significant difference was observed in the initial rate of IAA production that was approximately 42 ± 2 g L–1 h–1. Surprisingly, at the end of the growth, a significant influence of the inoculum was observed on the IAA titer that varied between 161.58 ± 4.91 (6 h old inoculum) and 263.33 ± 8.25 mg/L (4 h old inoculum; Figure 9).
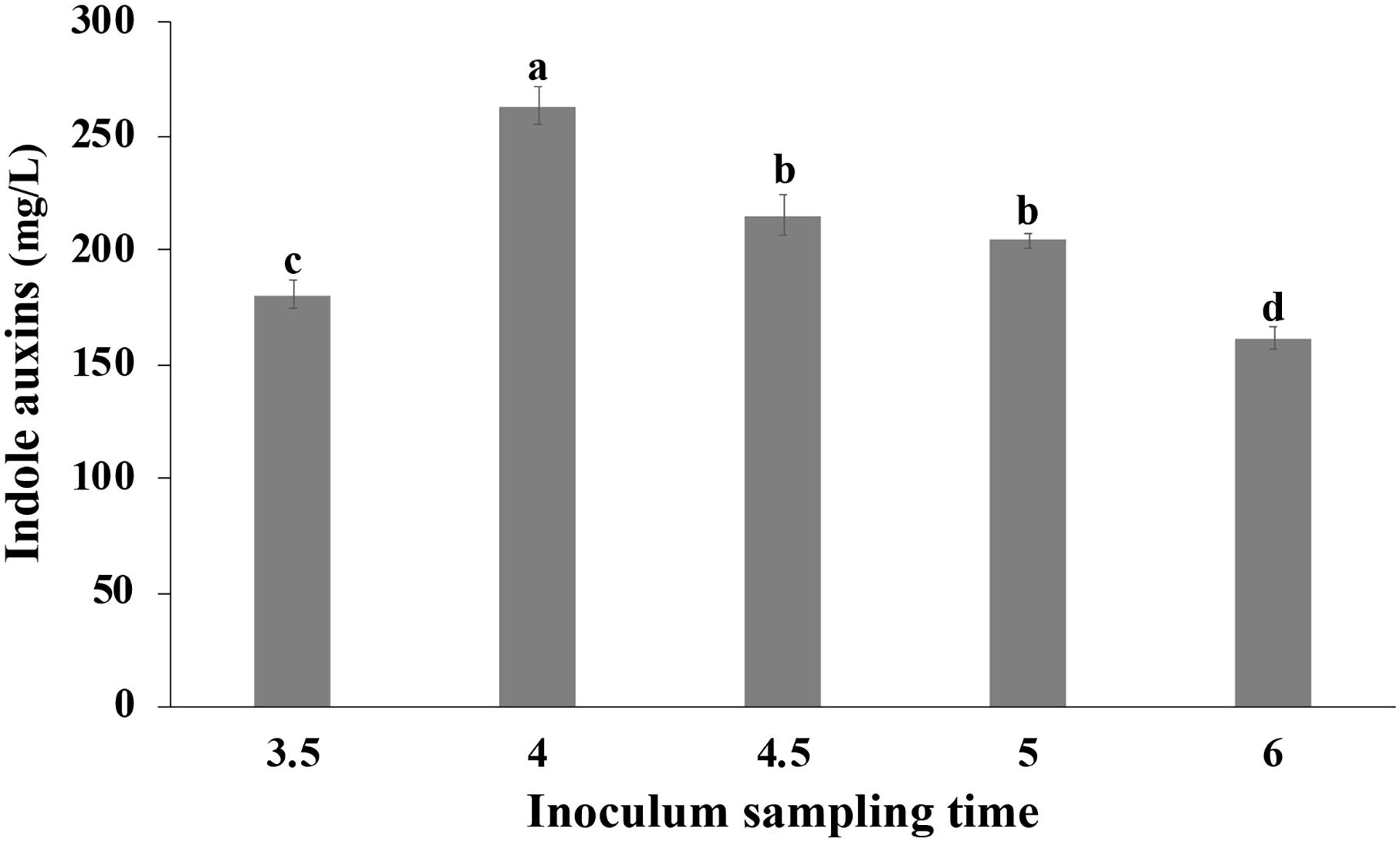
Figure 9. Effect of the physiological state of the cells used for inoculation on IAA production. Pantoea agglomerans strain C1 was cultivated on YES-tryptophan medium for 18 h, and cells used for the inoculum (OD = 0.5) were collected by the fermenter at the time points indicated on the x-axis. Differences in letters indicate that the values are significantly different (P > 0.05).
Use of IAA-Enriched Excretome From P. agglomerans C1 in Plant Nursery
The efficacy of the exhausted culture medium, containing IAA and other secreted metabolites from P. agglomerans strain C1, named IAA-EC1 (for IAA-enriched Excretome from strain C1) was tested on a total of 4,540 plants of Prunus rootstock GF/677 and 1,080 plants of hazelnut cv. Fertile de Coutard. The experiments were carried out in a large-scale nursery farm (Vivai Piante Battistini, Cesena, Italy) according to the protocols used for large agamic propagation of fruit crop trees.
Plant Survival Experiments
The inductive activity of adventitious rooting of exometabolites from strain C1 was tested in vitro on two sets of 760 plantlets of rootstock GF677 treated with either a 3 μM IBA-K solution or an appropriate volume of IAA-EC1 to achieve a final concentration of auxins of 1 μM. After 10 days of rooting stage, when plantlets were transferred to 360-cell plug tray, the percentage of survival was 85% for IAA-EC1–treated plants and 97% for IBA-treated plants; after 1 month of ex vitro acclimation, this value resulted 95% for IAA-EC1–treated plants and only 80% for IBA-treated plants. Taken together, these values indicate an overall increase of about 3% in plant survival when bacterial metabolites were used in alternative to IBA-K.
Ex vitro Experiments
After 20 days of ex vitro acclimatization, regardless of the inductive treatments, the percentage of rooting was 100%. However, the number of roots per rooted explant was significantly higher for IAA-EC1–treated plants (5.2 ± 0.5 roots per explant), compared to an average of 3.6 ± 0.5 of roots for IBA-K–treated plants. The quality of the roots also resulted improved after the treatment with IAA-EC (Figure 10). No significant difference was, instead, detected for the elongation of roots, which was approximately 14 mm for all plants, as well as for the plant survival percentage that, after 3 weeks from the beginning of the experiments, was 85% ± 1%. Finally, the average total leaf area per plant was higher in IAA-EC1–treated (115 ± 2 mm2) with respect to IBA-K–treated plants (102 ± 8 mm2).

Figure 10. Effect of different auxin treatments on morphology of rootstock GF677 roots. (A) IBA-K (3 μM); (B) diluted IAA-EC1 solution (IAA = 1 μM).
The inductive role of adventitious rooting by IAA-EC1 was also investigated on hazelnut microcuttings of cv. Fertile de Coutard using the same experimental protocol applied for Prunus rootstock. This experiment was carried out on two distinct pools of 1,080 and 1,440 binodal cuttings that were treated with IBA-K (3 μM solution) and IAA-EC1 (a 1:1000 diluted solution with an indole auxins final concentration of 1 μM), respectively. The percentage of rooted cuttings and number of roots per rooted explants resulted to be not significantly affected by the treatments, with the rooted ratio being between 64 and 75% and the value of root number being between 1.1 ± 0.1 and 1.3 ± 0.2 (Table 2). The root length was significantly affected by C1 metabolites and increased about 1.3-fold after treatment with IAA-EC1 compared to IBA-K (Table 2). A similar positive effect was also observed analyzing the stem elongation and the leaf area which both increase of about 1.4-fold in IAA-EC1–treated plants (Figure 11). It is worth mentioning that the treatment with C1 metabolites determined a development of adventitious roots that are not present in IBA-K–treated plants (Figure 12).
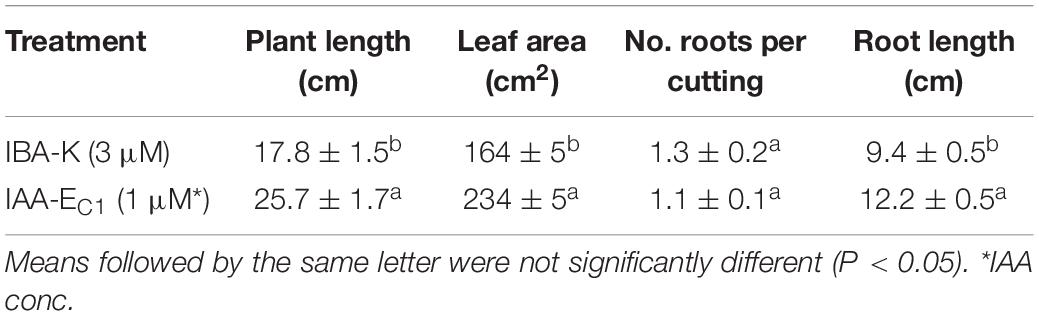
Table 2. Effect of different auxin treatments on plant length, leaf area, number of roots per cutting, and root length of hazelnut cv. “Fertile de Coutard.”
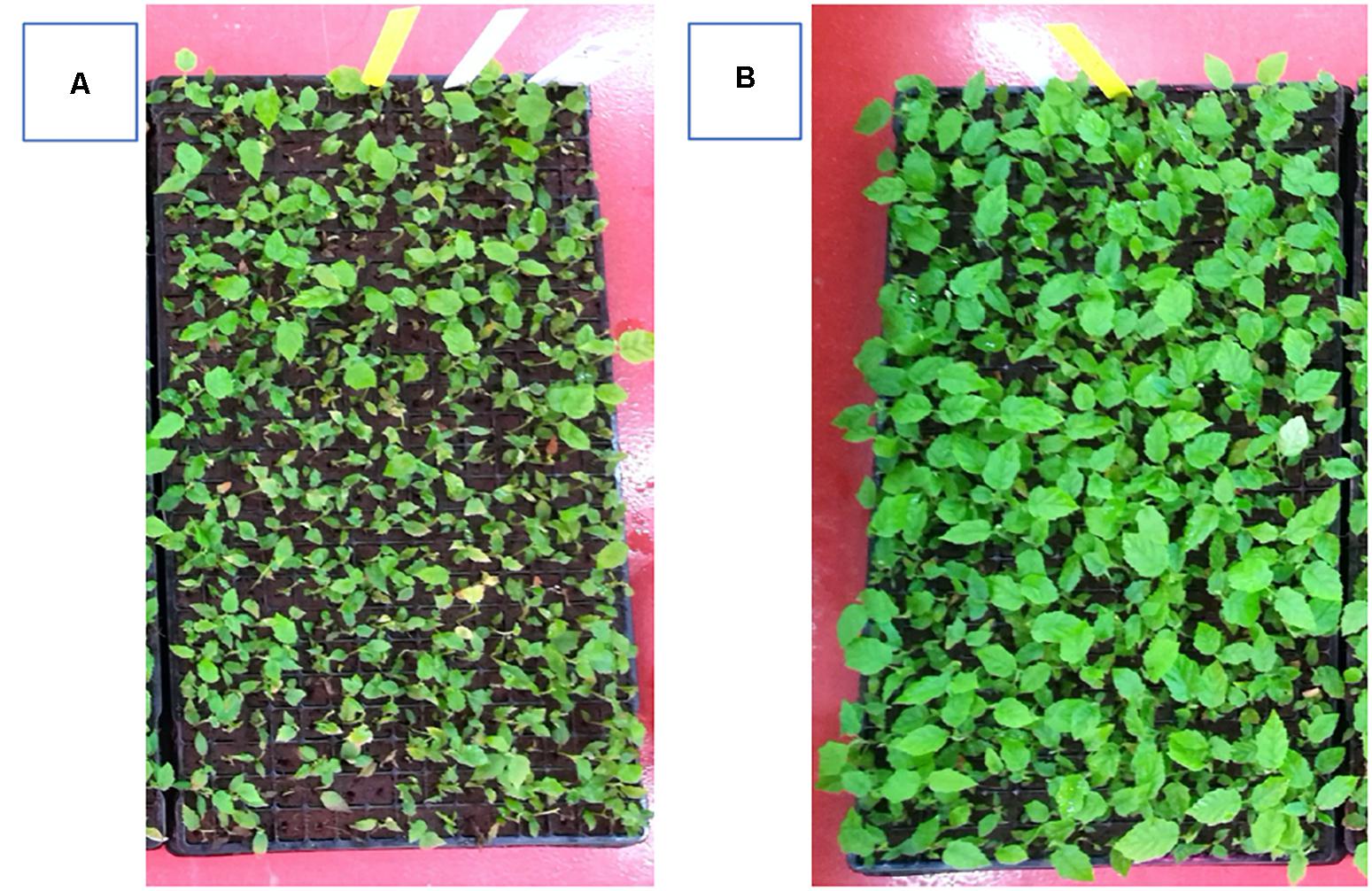
Figure 11. Effect of different auxin treatments on plant growth of Corylus avellana L. cv. “Fertile de Coutard.” (A) IBA-K (3 μM); (B) diluted IAA-EC1 solution (IAA = 1 μM).

Figure 12. Effect of different auxin treatments on morphology of Corylus avellana L. cv. “Fertile de Coutard” roots. (A) IBA-K (3 μM); (B) diluted IAA-EC1 solution (IAA = 1 μM).
Discussion
Indole-3-acetic acid production is widespread among plant growth–promoting bacteria and varies from species to species and also among strains belonging to the same species (Spaepen and Vanderleyden, 2011; Duca et al., 2014). Auxins and other plant growth–promoting metabolites obtained from non-pathogenic P. agglomerans strains can be successfully applied in agriculture systems as plant biostimulants. It has been demonstrated that P. agglomerans strain C1 produces IAA and siderophores, and the metabolites secreted by this strain can be utilized as biostimulants to improve the root surface area in tomato cuttings (Luziatelli et al., 2020). This strain is also able to solubilize phosphates and can improve the use of rock phosphates by corn (Zea mays L.) and tomato (S. lycopersicum L.) (Saia et al., 2020).
The application of comparative genetics, metabolomics, and fermentation technology in this study allowed analyzing in more detail the possibility of using this strain for production of a novel cell-free biostimulants. Whole-genome analysis demonstrated that P. agglomerans C1 has several genes connected with the production of IAA from Trp and all the genes of the IPyA pathway (Table 1 and Figure 1). In GenBank, there are several sequences encoding indolepyruvate decarboxylase, the key enzyme of the IPyA pathway, and homologous enzymes, but only in few cases that the corresponding proteins have been isolated and characterized for their biochemical properties. The 1,653 bp peg1955 gene from P. agglomerans C1 encodes a 551-amino acid protein, which shares high identity with well-characterized IPDC from other Pantoea strains: 92% identity with the IPDC from P. agglomerans 299R (Brandl and Lindow, 1996) and 73% identity with IPDC1 from Pantoea species YR343 (Garcia et al., 2019; Table 1 and Figure 2). Interestingly, the results obtained in this study demonstrated that, with few exceptions, in most of the sequenced genomes of isolates belonging to P. agglomerans species (45 of 50), the genes encoding enzymes of the IPyA pathway share an identity higher than 95% over the full-length of the sequence (Figure 4). This unusually high sequence conservation among bacteria occurring in diverse natural environments has never been reported before for IAA genes belonging to other species. This is a strong evidence of the evolutionary importance of this functional trait for the interaction between P. agglomerans and host plants.
Molecular data presented in this work also indicate that strain C1 produces IAA through the IPyA route, which is usually associated to beneficial bacteria, whereas the IAM pathway, linked to plant pathogens using auxin synthesis as a virulence factor (Patten et al., 2013), is not present. These observations encourage the exploitation of strain C1 and its metabolites as a plant growth promoter.
Interestingly, comparative genomics also allowed demonstrating that the non-coding region upstream of the ipdC gene is highly conserved among member of P. agglomerans species (Figure 4) and that this gene is not regulated by TyrR (Figures 3, 5). TyrR is a transcriptional regulator that controls the expression of a number of genes involved in the biosynthesis, catabolism, and transport of aromatic amino acids in E. coli (Pittard et al., 2005) and regulates expression of ipdC gene in E. ludwigii UW5 (Coulson and Patten, 2015). The ipdC promoter from strain UW5 contains two TyrR binding motifs, a strong (highly conserved) and a weak Tyr-box, which are not present in Pantoea strains (Figure 5). This result agrees with the observation that in P. agglomerans C1 the IAA production occurs in exponential phase of growth and not in stationary phase as in species in which ipdC gene is under the control of TyrR. The regulatory mechanism that control expression of the genes involved in IPyA pathway in P. agglomerans has not been elucidated yet, although there is evidence that multiple regulatory mechanisms may exist. For example, Brandl and Lindow (1997) demonstrated that in P. agglomerans strain 299R ipdC gene was expressed at low levels when cells were cultured in in vitro liquid cultures (independently from the presence of Trp or the growth phase of the culture) and was fully induced only when cells were grown on plants under water stress. In contrast, IAA production in P. agglomerans C1 is Trp-dependent and is significantly affected from the carbon source (Figure 6) and the physiological state of the cells (Figure 9). Moreover, cultivation in bench-top fermenter indicated that with strain C1 there is a correlation between the amount of IAA produced after induction with Trp and the preparation of the inoculum (Figure 9). This observation agrees with the findings of a previous work with recombinant E. coli cells, where the production of aromatic compounds in enteric bacteria is enhanced with cells in exponential phase of growth in which the availability of ATP is higher (Luziatelli et al., 2019c).
Untargeted metabolomics allowed demonstrating that P. agglomerans C1, on medium containing sucrose as a carbon source, produces, together with IAA, other IAA-related compounds, such as I3C and IAA-leucine, which can either modulate the effect of an excess of IAA (Katz et al., 2015) or increase the availability of this compound (Sanchez Carranza et al., 2016). In the exometabolome of P. agglomerans C1, other classes of compounds are also present, such as peptides and cyclopeptides that crosstalk with auxins or affect auxin transport or turnover.
Finally, the results obtained from studies run on a large scale on both one of the most diffuse Prunus rootstock and plant hazelnut confirmed that metabolites secreted by indole auxin–producing cells of P. agglomerans C1 have a high stimulating effect on adventitious rooting induction and adaption and plant growth in vitro. Interestingly, the excretome of strain C1 can also play relevant roles in root morphology (Figures 10, 12) and plant growth (Figure 11) in ex vitro conditions. It is worth noting that these effects, as shown by the control experiments with IBA-K, are only in part dependent on auxins and with IAA-EC1 can be achieved at a molar concentration of IAAequ threefold lower than the synthetic auxin (1 vs. 3 μM). Both in vitro and extra vitro procedures have shown that IAA-EC1 improved the performance and quality of micropropagated plant production. It should be emphasized that evaluation of the applicability of Pantoea metabolites in plant nursery industry was carried out following standard operation procedures used for large-scale production. Best practices for plant production rely on a careful control of microbial contamination of the production lines and equipment, the growing media, and the plant containers. In this specific context, the use of microbial inocula that can be beneficial for some plant cultures and not appropriate for others is always questionable and potentially dangerous. Under this respect, the use of liquid products, which are cell-free and contain only microbial metabolites, reduces contamination risks, is more compatible with the automation systems, and facilitates the adoption of this technology in plant nurseries.
Conclusion
In conclusion, the use of metabolites secreted by selected strains of P. agglomerans species can provide a valuable contribute for production of innovative biostimulants that can comply with current EU legislation. Further studies on gene expression will help to decipher the regulatory network that control IAA biosynthesis in P. agglomerans and provide more insight into the mechanisms by which Pantoea metabolites elicit plant growth promotion.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
FL, AF, PB, RM, and MR contributed to the conception and design of the study and wrote the first draft of the manuscript. FL, AF, PB, LG, RM, MM, MT, and MR contributed to define the methodology. FL, AF, PB, and MR contributed to software analysis and data validation. FL, AF, PB, LG, RM, MM, MT, FM, and MR contributed to investigation. FL, AF, RM, FM, and MR wrote and edited the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
LG was supported by a Ph.D. grant of Minister for Education, University and Research (MIUR), Department of Excellence project SAFE-Med (Law 232/2016). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
PB was employed by Next-Generation Agronomics Laboratory. MM was employed by the company Vivai Piante Battistini soc. agr. s.s.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the NGA Laboratory and Atens, Agrotecnologias Naturales S.L. (Spain) for helping us in the metabolomics analysis and metabolite annotation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01475/full#supplementary-material
Footnotes
- ^ http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb
- ^ https://patricbrc.org/
- ^ www.canopeoapp.com
References
Akin-Idowu, P. E., Ibitoye, D. O., and Ademoyegun, O. T. (2009). Tissue culture as a plant production technique for horticultural crops. African J. Biotechnol. 8, 3782–3788. doi: 10.5897/AJB2009.000-9374
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Barupal, D. K., Haldiya, P. K., Wohlgemuth, G., Kind, T., Kothari, S. L., Pinkerton, K. E., et al. (2012). MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinf. 13:99. doi: 10.1186/1471-2105-13-99
Bhatti, S., and Jha, G. (2010). Current trends and future prospects of biotechnological interventions through tissue culture in apple. Plant Cell. Rep. 29, 1215–1225. doi: 10.1007/s00299-010-0907-8
Blaženović, I., Kind, T., Sa, M. R., Ji, J., Vaniya, A., Wancewicz, B., et al. (2019). Structure annotation of all mass spectra in untargeted metabolomics. Anal. Chem. 91, 2155–2162. doi: 10.1021/acs.analchem.8b04698
Brandl, M. T., and Lindow, S. E. (1996). Cloning and characterization of a locus encoding an indolepyruvate decarboxylase involved in indole-3-acetic acid synthesis in Erwinia herbicola. Appl. Environ. Microbiol. 62, 4121–4128.
Brandl, M. T., and Lindow, S. E. (1997). Environmental signals modulate the expression of an indole-3-acetic acid biosynthetic gene in Erwinia herbicola. MPMI 10, 499–505. doi: 10.1094/MPMI.1997.10.4.499
Büyükcam, A., Tuncer, Ö, Gür, D., Sancak, B., Ceyhan, M., Cengiz, A. B., et al. (2018). Clinical and microbiological characteristics of Pantoea agglomerans infection in children. J. Infect. Public Health 11, 304–309. doi: 10.1016/j.jiph.2017.07.020
Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucl. Acids Res. 16, 10881–10890.
Coulson, T. J., and Patten, C. L. (2015). Complete genome sequence of Enterobacter cloacae UW5, a rhizobacterium capable of high levels of indole-3-acetic acid production. Genome Announc. 3:e00843-15. doi: 10.1128/genomeA.00843-15
DeFelice, B. C., Mehta, S. S., Samra, S., Čajka, T., Wancewicz, B., Fahrmann, J. F., et al. (2017). Mass spectral feature list optimizer (MS-FLO): a tool to minimize false positive peak reports in untargeted liquid chromatography–mass spectroscopy (LC-MS) data processing. Anal. Chem. 89, 3250–3255. doi: 10.1021/acs.analchem.6b04372
Doktycz, M. J. (2019). Computationally guided discovery and experimental validation of indole-3-acetic acid synthesis pathways. ACS Chem. Biol. 14, 2867–2875. doi: 10.1021/acschembio.9b00725
Driver, J. A., and Kuniyuki, A. H. (1984). In vitro propagation of Paradox walnut rootstock. Hort. Sci. 19, 507–509.
Duca, D., Lorv, J., Patten, C. L., Rose, D., and Glick, B. R. (2014). Indole-3-acetic acid in plant-microbe interactions. Antonie Leeuwenhoek 106, 85–125. doi: 10.1007/s10482-013-0095-y
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., and Milanowski, J. (2016a). Pantoea agglomerans: a mysterious bacterium of evil and good. Part III. Deleterious effects: infections of humans, animals and plants. Ann. Agric. Environ. Med. 23, 197–205. doi: 10.5604/12321966.1203878
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., and Milanowski, J. (2016b). Pantoea agglomerans: a mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. 23, 206–222. doi: 10.5604/12321966.1203879
Eckardt, N. A. (2001). New insights into auxin biosynthesis. Plant Cell 13, 1–3. doi: 10.1105/tpc.13.1.1
Garcia, D. C., Cheng, X., Land, M. L., Standaert, R. F., Morrell-Falvey, J. L., Mitchel, J., et al. (2019). A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 16, 295–298. doi: 10.1038/s41592-019-0358-2
Kasahara, H. (2016). Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 80, 34–42. doi: 10.1080/09168451.2015.1086259
Katz, E., Nisani, S., Yadav, B. S., Woldemariam, M. G., Shai, B., Obolski, U., et al. (2015). The glucosinolate breakdown product indole-3-carbinol acts as an auxin antagonist in roots of Arabidopsis thaliana. Plant J. 82, 547–555. doi: 10.1111/tpj.12824
Koga, J., Adachi, T., and Hidaka, H. (1991). Molecular cloning of the gene for indolepyruvate decarboxylase from Enterobacter cloacae. Mol. Gen. Genet. 226, 10–16. doi: 10.1007/bf00273581
Lennox, S. (1955). Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1, 190–206. doi: 10.1016/0042-6822(55)90016-7
Ljung, K., Hull, A. K., Kowalczyk, M., Marchant, A., Celenza, J., Cohen, J. D., et al. (2002). Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 50, 309–332. doi: 10.1023/A:1015298812300
Loberant, B., Altman, A., and Smith, R. H. (2010). “Micropropagation of plants,” in Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology, ed. M. C. Flickinger, (New York, NY: Wiley), 1–17.
Luziatelli, F., Brunetti, L., Ficca, A. G., and Ruzzi, M. (2019c). Maximizing the efficiency of vanillin production by biocatalyst enhancement and process optimization. Front. Bioeng. Biotechnol. 7:279. doi: 10.3389/fbioe.2019.00279
Luziatelli, F., Ficca, A. G., Cardarelli, M. T., Melini, F., Cavalieri, A., and Ruzzi, M. (2020). Genome sequencing of Pantoea agglomerans C1 provides insights into molecular and genetic mechanisms of plant growth-promotion and tolerance to heavy metals. Microorganisms 8:153. doi: 10.3390/microorganisms8020153
Luziatelli, F., Ficca, A. G., Colla, G., Baldassarre Švecová, E., and Ruzzi, M. (2019a). Foliar application of vegetal derived bioactive compounds stimulates the growth of beneficialbacteria and enhances microbiome biodiversity in lettuce. Front. Plant Sci. 10:60. doi: 10.3389/fpls.2019.00060
Luziatelli, F., Ficca, A. G., Melini, F., and Ruzzi, M. (2019b). Genome sequence of the plant growth-promoting rhizobacterium (PGPR) Pantoea agglomerans C1. Microbiol. Resour. Announc. 8:e00828-19. doi: 10.1128/mra.00828-19
Luziatelli, F., Ficca, A. G., Colla, G., Švecová, E., and Ruzzi, M. (2016). Effects of a protein hydrolysate-based biostimulant and two micronutrient based fertilizers on plant growth and epiphytic bacterial population of lettuce. Acta Hortic. 1148, 43–48. doi: 10.17660/ActaHortic.2016
Malhotra, M., and Srivastava, S. (2008). Organization of the ipdC region regulates IAA levels in different Azospirillum brasilense strains: molecular and functional analysis of ipdC in strain SM. Environ Microbiol. 10, 1365–1373. doi: 10.1111/j.1462-2920.2007.01529.x
Maniatis, T., Fritsch, E. F., and Sambrook, J. (1982). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor.
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nissen, S. J., and Sutter, E. G. (1990). Stability of IAA and IBA in nutrient medium to several tissue culture procedures. Hort. Sci. 25, 800–802. doi: 10.21273/HORTSCI.25.7.800
Orlikowska, T., Nowak, K., and Reed, B. (2017). Bacteria in the plant tissue culture environment. Plant Cell Tiss. Organ. Cult. 128, 487–508. doi: 10.1007/s11240-016-1144-9
Paredes-Páliz, K. I., Caviedes, M. A., Doukkali, B., Mateos-Naranjo, E., Rodríguez-Llorente, I. D., and Pajuelo, E. (2016a). Screening beneficial rhizobacteria from Spartina maritima for phytoremediation of metal polluted salt marshes: comparison of gram-positive and gram-negative strains. Environ. Sci. Pollut. Res. 23, 19825–19837. doi: 10.1007/s11356-016-7184-1
Paredes-Páliz, K. I., Pajuelo, E., Doukkali, B., Caviedes, M. Á, Rodríguez-Llorente, I. D., and Mateos-Naranjo, E. (2016b). Bacterial inoculants for enhanced seed germination of Spartina densiflora: implications for restoration of metal polluted areas. Mar. Pollut. Bull. 110, 396–400. doi: 10.1016/j.marpolbul.2016.06.036
Paredes-Páliz, K. I., Mateos-Naranjo, E., Doukkali, B., Caviedes, M. A., Redondo-Gómez, S., Rodríguez-Llorente, I. D., et al. (2017). Modulation of Spartina densiflora plant growth and metal accumulation upon selective inoculation treatments: a comparison of gram negative and gram positive rhizobacteria. Mar. Pollut. Bull. 125, 77–85. doi: 10.1016/j.marpolbul.2017.07.072
Patten, C. L., Blakney, A. J., and Coulson, T. J. (2013). Activity, distribution and function of indole-3-acetic acid biosynthetic pathways in bacteria. Crit. Rev. Microbiol. 39, 395–415. doi: 10.3109/1040841X.2012.716819
Pierik, R. L. M., and Scholten, H. J. (1994). “Micropropagation, an essential tool in modern agriculture, horticulture, forestry and plant breeding,” in Plant Production on the Threshold of a New Century, Vol. 61, eds P. C. Struik, W. J. Vredenberg, J. A. Renkema, J. E. Parlevliet, and Develop Plant Soil Sci, (Dordrecht, DE: Springer).
Pittard, J., Camakaris, H., and Yang, J. (2005). The TyrR regulon. Mol. Microbiol. 55, 16–26. doi: 10.1111/j.1365-2958.2004.04385.x
Prinsen, E., Costacurta, A., Michiels, K., Vanderleyden, J., and Van Onckelen, H. (1993). Azospirillum brasilense indole-3-acetic acid biosynthesis: evidence for a non-tryptophan dependent pathway. Mol. Plant Microbe Interact. 6, 609–615. doi: 10.1046/j.1432-1327.1999.00033.x
Quoirin, M., and Lepoivre, P. (1977). Etudes de milieux adaptes aux cultures in vitro de Prunus. Acta Hortic. 78, 437–442.
Rouphael, Y., and Colla, G. (2018). Synergistic biostimulatory action: designing the next generation of plant biostimulants for sustainable agriculture. Front. Plant Sci. 9:1655. doi: 10.3389/fpls.2018.01655
Ruzzi, M., and Aroca, R. (2015). Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 196, 124–134. doi: 10.1016/j.scienta.2015.08.042
Ryu, R. J., and Patten, C. L. (2008). Aromatic amino acid-dependent expression of indole-3-pyruvate decarboxylase is regulated by TyrR in Enterobacter cloacae UW5. J. Bacteriol. 190, 7200–7208. doi: 10.1128/JB.00804-08
Saia, S., Aissa, E., Luziatelli, F., Ruzzi, M., Colla, G., Fiacca, A. G., et al. (2020). Growth-promoting bacteria and arbuscular mycorrhizal fungi differentially benefit tomato and corn depending upon the supplied form of phosphorus. Mycorrhiza 30, 133–147. doi: 10.1007/s00248-009-9531-y
Sanchez Carranza, A. P., Singh, A., Steinberger, K., Panigrahi, K., Palme, K., Dovzhenko, A., et al. (2016). Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci. Rep. 6:24212. doi: 10.1038/srep24212
Schutz, A., Golbik, R., Tittmann, K., Svergun, D. I, Koch, M. H., Hübner, G., et al. (2003). Studies on structure function relationships of indolepyruvate decarboxylase from Enterobacter cloacae, a key enzyme of the indole acetic acid pathway. Eur. J. Biochem. 270, 2322–2331. doi: 10.1046/j.1432-1033.2003.03602.x
Spaepen, S., and Vanderleyden, J. (2011). Auxin and plant-microbe interactions. CSH Perspect. Biol. 3:a001438. doi: 10.1101/cshperspect.a001438
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Thévenot, E. A., Roux, A., Xu, Y., Ezan, E., and Junot, C. (2015). Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 14, 3322–3335. doi: 10.1021/acs.jproteome.5b00354
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tsugawa, H., Cajka, T., Kind, T., Ma, Y., Higgins, B., Ikeda, K., et al. (2015). MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526. doi: 10.1038/nmeth.3393
Tsugawa, H., Kind, T., Nakabayashi, R., Yukihira, D., Tanaka, W., Cajka, T., et al. (2016). Hydrogen rearrangement rules: computational MS/MS Fragmentation and structure elucidation using MS-FINDER software. Anal. Chem. 88, 7946–7958. doi: 10.1021/acs.analchem.6b00770
Tsugawa, H., Nakabayashi, R., Mori, T., Yamada, Y., Takahashi, M., Rai, A., et al. (2019). A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 16, 295–298. doi: 10.1038/s41592-019-0358-2
Vande Broek, A., Gysegom, P., Ona, O., Hendrickx, N., Prinsen, E., Van Impe, J., et al. (2005). Transcriptional analysis of the Azospirillum brasilense indole-3-pyruvate decarboxylase gene and identification of a cis-acting sequence involved in auxin responsive expression. Mol. Plant–Microb. Interact. 18, 311–323. doi: 10.1094/MPMI-18-0311
Keywords: plant growth promotion, auxin, Pantoea, indole-3-pyruvate decarboxylase (ipdC) gene, culture condition, Q-TOF LC/MS, Prunus rootstock, Corylus avellana L.
Citation: Luziatelli F, Ficca AG, Bonini P, Muleo R, Gatti L, Meneghini M, Tronati M, Melini F and Ruzzi M (2020) A Genetic and Metabolomic Perspective on the Production of Indole-3-Acetic Acid by Pantoea agglomerans and Use of Their Metabolites as Biostimulants in Plant Nurseries. Front. Microbiol. 11:1475. doi: 10.3389/fmicb.2020.01475
Received: 31 March 2020; Accepted: 05 June 2020;
Published: 14 July 2020.
Edited by:
Francesco Vinale, University of Naples Federico II, ItalyReviewed by:
Sabrina Sarrocco, University of Pisa, ItalyEloisa Pajuelo, Universidad de Sevilla, Spain
Hassan Etesami, University of Tehran, Iran
Copyright © 2020 Luziatelli, Ficca, Bonini, Muleo, Gatti, Meneghini, Tronati, Melini and Ruzzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurizio Ruzzi, ruzzi@unitus.it
†These authors have contributed equally to this work
 Francesca Luziatelli
Francesca Luziatelli Anna Grazia Ficca
Anna Grazia Ficca Paolo Bonini
Paolo Bonini Rosario Muleo
Rosario Muleo Lorenzo Gatti
Lorenzo Gatti Massimiliano Meneghini4
Massimiliano Meneghini4 Michele Tronati
Michele Tronati Francesca Melini
Francesca Melini Maurizio Ruzzi
Maurizio Ruzzi