- 1National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, South China Agricultural University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, South China Agricultural University, Guangzhou, China
- 3Hackensack Meridian Health Center for Discovery and Innovation, Nutley, NJ, United States
Objectives: The emergence of mobile colistin resistance genes has compromised the efficacy of the last resort antibiotic, colistin, in clinical treatment. The mcr-2 gene was first identified in Belgium in association with the insertion sequence ISEc69. However, the molecular mechanisms of mcr-2 mobilization are not well understood.
Methods: To further explore the mobilization of mcr-2 gene via ISEc69, we constructed a conjugative plasmid that carries an intact composite transposon Tn7052. Transposition assays were performed by conjugation, the transposition sites were characterized by arbitrary primed PCR and DNA sequencing.
Results: In this study, we experimentally demonstrated that mcr-2 could be mobilized as a composite transposon Tn7052 and its transposition generated 8-bp AT-rich duplications in the host genome.
Conclusion: These results indicate that mcr-2 gene could be mobilized by ISEc69, the current investigations provide mechanistic insights in the transposition of mcr-2.
Introduction
The global dissemination of multidrug resistant bacteria, especially extended-spectrum cephalosporin and carbapenem resistant “ESKAPE” pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) become a significant threat to public health (Deslouches et al., 2015). Meanwhile, the identification of mobile colistin resistance genes (mcr) has compromised the efficacy of colistin as the last-resort antibiotics against these pathogens.
The mcr-2 is a novel plasmid-mediated colistin resistance gene that is 81% identical to mcr-1 and is associated with ISEc69 of the IS1595 superfamily. The mcr-2 was initially reported on a 35 kb IncX4 plasmid, pKP87-BE, flanked by directly oriented copies of ISEc69 (Partridge, 2017). Currently, the actual and reliable experimental data in regard to mechanism underlying the mcr-2 gene horizontal transfer are rarely reported. The transfer of antimicrobial resistance genes has been found to be catalyzed by specific mobile elements such as insertion element and integron. For instance, ISApl1, originally found in Actinobacillus pleuropneumoniae, is located upstream of the mcr-1 gene in the first described mcr-1-harboring IncI2 type plasmid pHNSHP45 (Liu et al., 2015; Ye et al., 2016) and is able to mediate mcr-1 mobilization (Poirel et al., 2017; He et al., 2019). Similarly, ISEc69, encoding a 217-amino-acid-long DDE-type transposase, belongs to the IS1595 family and is flanked by two inverted repeats, which share 16/19 nucleotide identity to inverted repeats from the IS1016C2 transposase. Directly downstream of mcr-2 is the pap2 (ORF) gene, encoding a PAP2 membrane-associated lipid phosphatase with 41% identity with Moraxella osloensis phosphatidic acid phosphatase. Tn7052(ISEc69-mcr-2-ORF-ISEc69) forms a composite transposon structure, and we suspected mcr-2 may be mobilized through transposition.
The aim of the current study was to experimentally determine whether ISEc69 could mobilize the mcr-2 gene and to identify the integration sites on bacterial genome. Preliminary experiments showed that the appearance of a circularized intermediate might accelerate the dissemination of mcr-2 (Sun et al., 2017). However, the mechanism of mcr-2 transfer has not yet to be studied, especially for its transposition. In this work, we focused on providing experimental evidence for the specific transposition of mcr-2 as well as the host transfer via conjugation and subsequent transposition into the host chromosome.
Materials and Methods
Strains
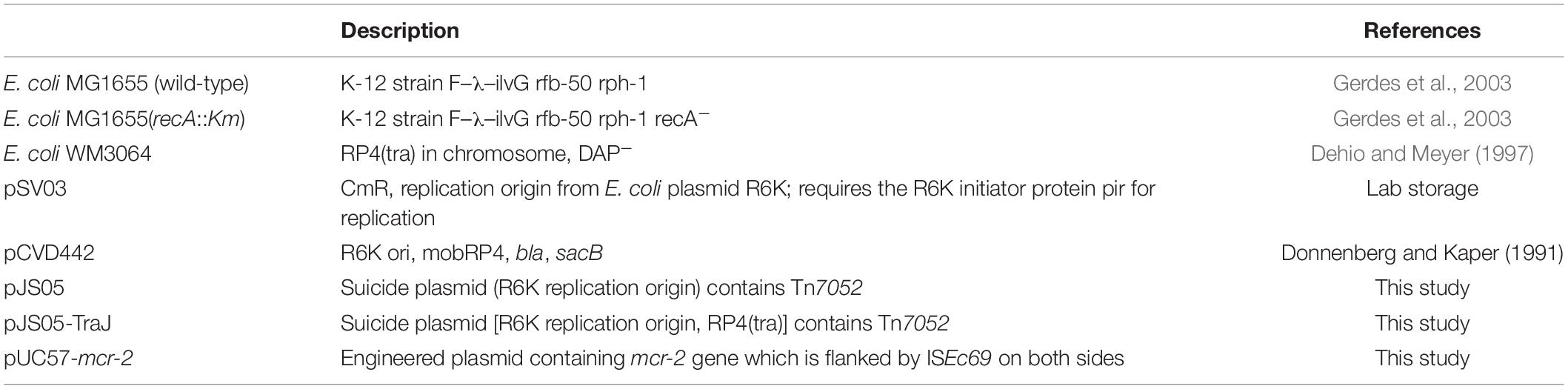
E. coli MG1655 (wild-type) and E. coli MG1655 (recA::Km) strains were used as recipients for transposition experiments as previously described (Gerdes et al., 2003). E. coli strain WM3064 (Dehio and Meyer, 1997) is a diaminopimelic acid (DAP) auxotroph strain, containing RP4 plasmid transfer machinery (McLaughlin and Ahmad, 1986) and the pir (Chen et al., 1998) plasmid maintenance sequence with an R6K (Rakowski and Filutowicz, 2013) ori. This strain was also used as a host for suicide plasmid bearing Tn7052. The strains and plasmids used in this study are listed in Table 1.
Plasmid Construction
We firstly constructed the suicide plasmid pJS05-Traj, containing the Tn7052 cassette, RP4oriT fragment from pCVD442 (Philippe et al., 2004) and R6K replication origin which relies on π protein, encoded by the pir gene. This suicide plasmid will only survive in a bacterial host with the pir gene (e.g., E. coli WM 3064), but won’t be able to replicate in other hosts. In brief, we synthesized a cassette in which the mcr-2 and orf genes are flanked by ISEc69 on both sides and cloned into the vector pUC57, generating the plasmid pUC57-mcr-2. R6K-BamHI and R6K-EcoRI primers (Table 2) were used to PCR amplify the backbone using pSV03 (He et al., 2019) as a template which contained the conditional replication origin R6K and CmR (chloramphenicol) resistance. The amplicon was sub-cloned into the BamHI and EcoRI restriction sites in pUC57 that contained Tn7052 to obtain recombinant plasmid pJS05 (Supplementary Figure 1A). The plasmid was electroporated into strain E. coli WM 3064 and clones were selected on Luria Bertani (LB) agar plates supplemented with 25 μg/ml chloramphenicol. The integrity of both ISEc69 and mcr-2 was confirmed by DNA sequencing. Traj-F and Traj-R primers (Table 2) were used to amplify RP4oriT fragment using pCVD442 as a template by PCR. The RP4oriT fragment was ligated into the SalI and EcoT22I sites of pJS05 giving rise to recombinant plasmid pJS05-Traj (Supplementary Figure 1B). The plasmid was electroporated into E. coli WM 3064 and selected on LB agar plates supplemented with 25 μg/ml chloramphenicol. The integrity of pJS05-Traj was confirmed by whole plasmid sequencing.
Transposition Assays
Transposition of the recombinant plasmids was examined using E. coli WM3064/pJS05-Traj as a donor and E. coli MG1655 (wild-type) and E. coli MG1655 (recA::Km) as recipients. Cells were cultured to mid-log phase (OD600 0.5) at 37°C in LB plus 300 μM DAP for donor, and 100 μL of each a donor and recipient were mixed and vortexed. The cells were then collected by centrifugation (16,100 × g for 30 s at 25°C) and suspended in 100 mL fresh LB containing 300 μM DAP and spread on LB agar plates containing 300 μM DAP and incubated at 37°C for 12 h. The cells were then recovered from the plates and suspended in 1 mL LB and aliquots were plated on LB agar containing 2 μg/ml colistin (without DAP) to select for transposons. Since the E. coli WM3064 is an auxotrophic strain whose growth relies on the supplementation of diaminopimelic acid (DAP) in the medium (Wang et al., 2015), and the plasmid pJS05-Traj can’t replicate in E. coli MG1655, the colonies that grow on LB agar plates containing 2 μg/ml colistin (without DAP) were likely the transposons. The presence of the full-length Tn7052 that integrated into the E. coli MG1655 genome was confirmed using PCR with primers ISEc69-F1 and IS69-R (Table 2). Transposition frequencies were calculated as the ratio of transposition events and the number of transformed cells in triplicate. The Pictogram program1 was used to illustrate the relative positions of transposition sites.
Arbitrary Primed PCR(AP-PCR) for Rapid Characterization of Transposon Insertion Sites
Transposon insertion sites in strain E. coli MG1655 (recA::Km) were identified by screening of 30 randomly selected colonies that were prepared from overnight cultures. DNA was extracted using the TIANamp Bacteria DNA Kit (Tiangen, Dalian, China). The right-side of the Tn7052 in the genome was confirmed by AP-PCR (Das et al., 2005). Briefly, DNAs from transposons were used as templates for the first-round PCR reactions. The arbitrary primers differed from their 3′ pentameric sequences (ABS1 to ABS3; Table 2) were paired in separate PCR reactions with the transposon-specific primer ISEc69-F1. The products of these reactions served as templates for second-round PCR reactions employing a nested transposon primer ISEc69-F2 and a primer composed of the common 5′ region from ABS1 to ABS3 (ABS; Table 2; Das et al., 2005). The identities of PCR products and associated genomic flanking regions were determined by DNA sequencing (Supplementary Figure 2). The left-side DNA sequences of Tn7052 in the genome were confirmed by standard PCR using primers TF1∼TF23 paired with CR-2U (Supplementary Table 1), respectively.
Results
The Transposition Frequencies of pJS05-Traj Into Two E. coli Strains
We identified the transposition abilities of the Tn7052 cassette by cloning it into a suicide plasmid that was then electroporated into the pir+ strain E. coli WM3064. This plasmid was transferred to recipient strains E. coli MG1655 (wild type) and E. coli MG1655 (recA::Km) by conjugation. The survival of the conjugants was contingent upon transposition of the selectable marker (mcr-2) into the host genome. The transposition frequencies of pJS05-Traj into E. coli MG1655 (wild type) and E coli MG1655 (recA::Km) strains were 4.09 × 10–6 and 3.53 × 10–6 per transformed cell, respectively (Table 3).
The Transposition Sites Preference of Tn7052 in the E. coli MG1655 (recA::Km) Genome
AP-PCR-based analysis identified 23 Tn7052 integration sites in the recipient chromosome among the 30 colonies. These events generated 8-bp sequence duplications at AT-rich regions with a high preference for insertion between T and A. The average AT contents at the 40 bp adjacent regions upstream and downstream of the 8-bp target sites were 63 and 67%, respectively (Figure 1A). In addition, the AT content at the target sites were 100% at positions in C1, C2, C7, and C8 and ranging from 83 to 91% at positions C3, C4, C5, and C6. Transposition outside the sequence duplicated target site (+2 to +10) ranged from 35 to 83% (Figure 1B).
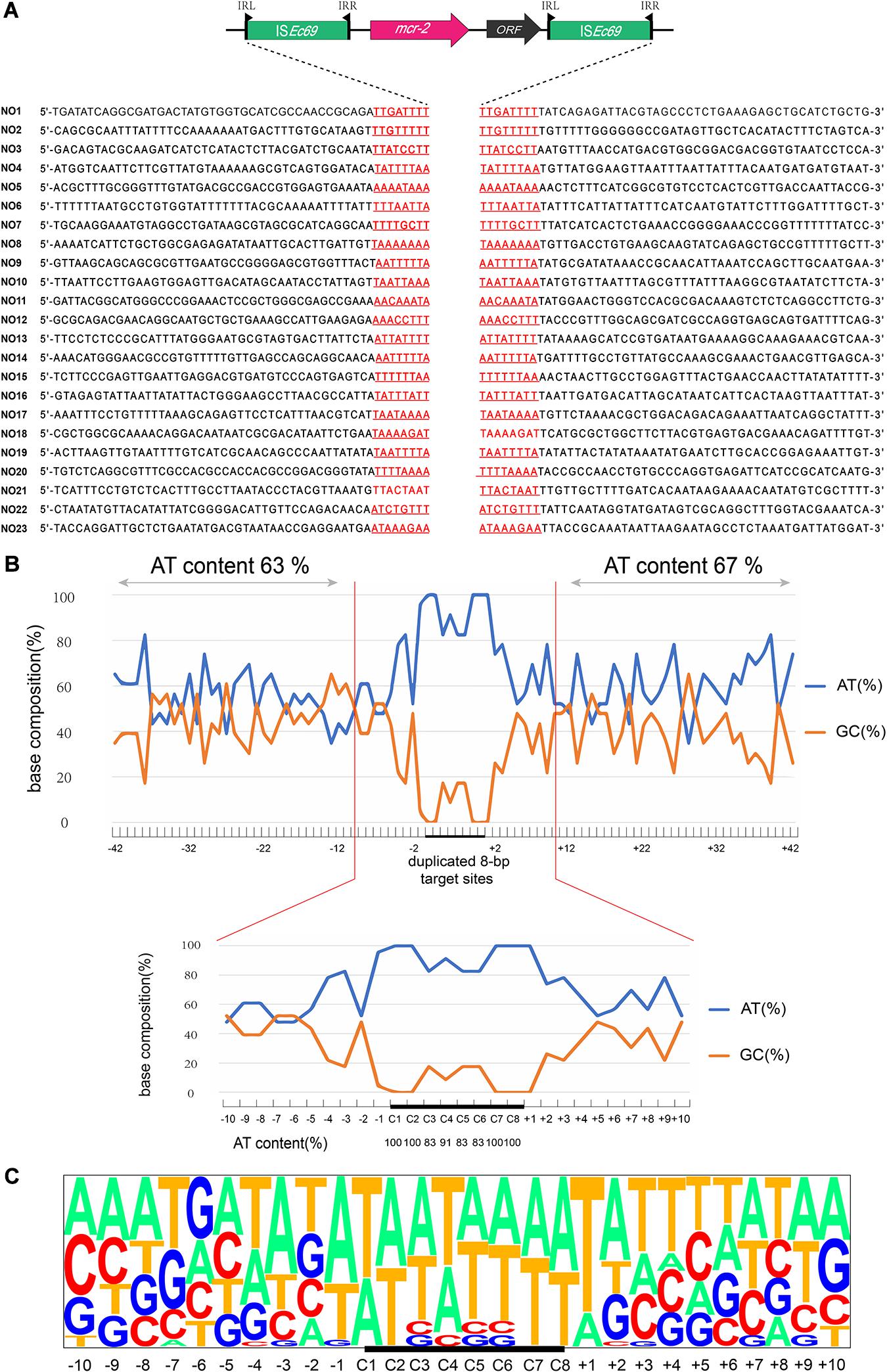
Figure 1. Target site analyses of Tn7052 transposons. (A) Molecular characterization of 23 transposition events of Tn7052 transposons in E. coli MG1655 (recA::Km). The duplicated 8-bp target sites are underlined in the context of the surrounding 42 nucleotides upstream and downstream from the target sites. (B) Statistical analyses of the 23 transposition sites. The percentage of AT and GC at each position from 42 nucleotides upstream to 42 nucleotides downstream of the target site are shown. The 8-bp duplicated target site (C1 to C8) are indicated by black bars. The AT and GC percentages of regions spanning positions –42 to –2 and positions +2 to +42 and that of the region spanning positions –2 to 2 are indicated in the upper and lower graphs, respectively. Relative nucleotide frequencies at each target site deduced from the 23 experimental transposition events shown in (A,C).
The Transposition Sites Position in the E. coli MG1655 (recA::Km) Genome
We further examined these 23 transposition events and calculated the relative frequencies of AT/GC and plotted for the region extending from 50 nucleotides upstream to 50 nucleotides downstream from the duplicated 8-bp target site (Figure 1C). In addition, the genomic locations of Tn7052 integration sites were further analyzed. The Tn7052 sites were all located in different single genes in the recipient chromosome (Supplementary Table 2).
Discussion
In this report, we illustrated a working model for the transposition of a mcr-2-containing cassette from a plasmid containing a unique transposon-like region. The mcr-2 gene conferring colistin resistance was identified on a plasmid vector (Xavier et al., 2016). However, its mobilization mechanism has not been confirmed.
Although mcr-2 gene has not yet spread in the world, it only has only been identified once, on a IncX4 conjugative plasmid (Partridge, 2017). It is very important to study its transmission and transposable mechanism in advance. The transposition of the MCR-2 protein encoding gene is closely linked to ISEc69. Here, we used R6K ori to enable replication of a pJS05-Traj bearing Tn7052 element in E. coli WM3064. In addition, the origin of conjugative transfer (RP4oriT) was inserted to allow efficient transfer of the suicide plasmid to other E. coli hosts. This process was successful and implicates conjugational transfer as an effective mobilization method for mcr-2 into recipient strain chromosomal locations mediated by ISEc69 transposition. Interestingly, Tn7052 insertion sites duplicated an 8 bp AT-rich target site. In prokaryotic, the AT-rich region is generally the replication origin of bacterial chromosomes and plasmids (Rajewska et al., 2012). The transposition in the replication origin of bacterial chromosomes and plasmids may have a negative impact on bacterial and plasmid replication, especially on plasmid copy number and its stability. This may be one of the reasons for the low prevalence of mcr-2.
The 23 precise transposition sites were characterized by AP-PCR that is a lightweight and reliable tool for IS detection and transposon insertion site identification in bacterial genomes. Compared with previous digestion and inverse PCR strategies (Green and Sambrook, 2019), the method might be more convenient and efficient, and the cost is comparatively lower as well. The method is simple to perform and can be used in any laboratory that is equipped with PCR equipment for identifying transposase insertion sites in bacterial genomes. To further characterize the distribution of ISEc69 mediated mcr-2 transposition, we determined the insertion sites for 23 transposon events in E. coli MG1655(recA::Km) genome. We found that 11 Tn7052 transposition sites were randomly located into non-essential genes and 12 were inserted between two non-essential genes in the bacterial chromosome (Supplementary Table 2). This study only finds insertions in non-essential genes, maybe since insertions into essential genes are deleterious and may negatively impact growth. Therefore, they cannot be selected on plates and are unable to be identified.
Taken together, these results indicated that mcr-2 most likely was mobilized by an ISEc69 composite transposon, generating an 8-bp DR at TA-rich sites. Our work demonstrated that ISEc69 possessed the ability to transpose mcr-2 in E. coli that may act as a reservoir for the mcr-2 gene and contribute to its dissemination among different bacterial species. This is especially important for clinically relevant bacterial species such as Enterobacteriaceae family members. Compared with mcr-1, the prevalence of mcr-2 is relatively low. It may relate to the transposable capacity of ISEc69. The transposition experiments in this study were designed to investigate the mechanisms by which insertion sequences ISEc69 mediate the transfer of mcr-2 gene, including the insertion site’s preference and the transposition efficiency. Understanding these knowledges could help control the spread of mcr-2 gene.
Duo to very low prevalence of mcr-2 plasmid-mediated colistin, it is speculated that the distribution of mcr-2 between European countries might be related to geography or differences in veterinary practices between these regions, but further research is needed. In the future, more intensive and concrete work will focus on the other routes of mcr-2 gene transmission so as to elucidate pathways for blocking the spread of the mcr-2.
Accession number: The nucleotide sequence of Tn7052 has been deposited in GenBank under accession numbers: MW251710.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation. The authors declare that, all the experiments were performed strictly according to the guidelines of WHO (WHO-DURC). All samples including recombinant strains, plasmid, and their relevant storages are properly maintained during and after the research. These materials are currently kept with high biosafety standard and cannot be accessed without certain permission. The authors guaranteed all materials employed in this study will be only used for scientific purposes and no risk on spreading the antibiotic resistance to the naturally existing microorganisms.
Author Contributions
JS designed this project. Y-ZH, T-FL, and BH performed the experiments. Y-ZH, T-FL, X-PLi, and JS analyzed the data. Y-ZH and GL made the figures. Y-ZH wrote this manuscript. JS, LC, and X-PLa edited and revised the manuscript. Y-HL coordinated the whole project. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0501300), the Program for Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (IRT13063IRT_17R39), National Natural Science Foundation of China (31972735), and the Graduate Student Overseas Study Program of South China Agricultural University (2018LHPY007).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.564973/full#supplementary-material
Footnotes
References
Chen, D., Feng, J., Kruger, R., Urh, M., Inman, R. B., and Filutowicz, M. (1998). Replication of R6K gamma origin in vitro: discrete start sites for DNA synthesis dependent on pi and its copy-up variants. J. Mol. Biol. 282, 775–787. doi: 10.1006/jmbi.1998.2055
Das, S., Noe, J. C., Paik, S., and Kitten, T. (2005). An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J. Microbiol. Methods 63, 89–94. doi: 10.1016/j.mimet.2005.02.011
Dehio, C., and Meyer, M. (1997). Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179, 538–540. doi: 10.1128/jb.179.2.538-540.1997
Deslouches, B., Steckbeck, J. D., Craigo, J. K., Doi, Y., Burns, J. L., and Montelaro, R. C. (2015). Engineered cationic antimicrobial peptides to overcome multidrug resistance by ESKAPE pathogens. Antimicrob. Agents Chemother. 59, 1329–1333. doi: 10.1128/aac.03937-14
Gerdes, S. Y., Scholle, M. D., Campbell, J. W., Balazsi, G., Ravasz, E., Daugherty, M. D., et al. (2003). Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185, 5673–5684. doi: 10.1128/jb.185.19.5673-5684.2003
Green, M. R., and Sambrook, J. (2019). Inverse polymerase chain reaction (PCR). Cold Spring Harb. Protoc. 2019:pdb.prot095166.
He, Y.-Z., Li, X.-P., Miao, Y.-Y., Lin, J., Sun, R.-Y., Wang, X.-P., et al. (2019). The ISApl12 dimer circular intermediate participates in mcr-1 transposition. Front. Microbiol. 10:15. doi: 10.3389/fmicb.2019.00015
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2015). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
McLaughlin, W., and Ahmad, M. H. (1986). Transfer of plasmids RP4 and R68.45 and chromosomal mobilization in cowpea rhizobia. Arch. Microbiol. 144, 408–441. doi: 10.1007/bf00409893
Partridge, S. R. (2017). mcr-2 in the IncX4 plasmid pKP37-BE is flanked by directly oriented copies of ISEc69. J. Antimicrob. Chemother. 72, 1533–1535.
Philippe, N., Alcaraz, J. P., Coursange, E., Geiselmann, J., and Schneider, D. (2004). Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51, 246–255. doi: 10.1016/j.plasmid.2004.02.003
Poirel, L., Kieffer, N., and Nordmann, P. (2017). In vitro study of ISApl1-mediated mobilization of the Colistin Resistance gene mcr-1. Antimicrob. Agents Chemother. 61:e00127-17.
Rajewska, M., Wegrzyn, K., and Konieczny, I. (2012). AT-rich region and repeated sequences – the essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 36, 408–434. doi: 10.1111/j.1574-6976.2011.00300.x
Rakowski, S. A., and Filutowicz, M. (2013). Plasmid R6K replication control. Plasmid 69, 231–242. doi: 10.1016/j.plasmid.2013.02.003
Sun, J., Xu, Y. C., Gao, R. S., Lin, J. X., Wei, W. H., Srinivas, S., et al. (2017). Deciphering MCR-2 Colistin Resistance. mBio 8:e00625-17.
Wang, P., Yu, Z., Li, B., Cai, X., Zeng, Z., Chen, X., et al. (2015). Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microb. Cell Fact. 14:11. doi: 10.1186/s12934-015-0194-8
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21:7.
Keywords: transposition mechanism, transposition sites, mcr-2 gene, ISEc69, colistin resistance
Citation: He Y-Z, Long T-F, He B, Li X-P, Li G, Chen L, Liao X-P, Liu Y-H and Sun J (2021) ISEc69-Mediated Mobilization of the Colistin Resistance Gene mcr-2 in Escherichia coli. Front. Microbiol. 11:564973. doi: 10.3389/fmicb.2020.564973
Received: 23 May 2020; Accepted: 18 December 2020;
Published: 12 January 2021.
Edited by:
Sebastian Guenther, University of Greifswald, GermanyCopyright © 2021 He, Long, He, Li, Li, Chen, Liao, Liu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Sun, jiansun@scau.edu.cn
 Yu-Zhang He
Yu-Zhang He Teng-Fei Long1,2
Teng-Fei Long1,2 Xing-Ping Li
Xing-Ping Li Xiao-Ping Liao
Xiao-Ping Liao Ya-Hong Liu
Ya-Hong Liu Jian Sun
Jian Sun