- 1Department of Clinical Laboratory, Zhongnan Hospital, Wuhan University, Wuhan, China
- 2Department of Neurosurgery, Zhongnan Hospital, Wuhan University, Wuhan, China
The Coronavirus Disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has now become a global pandemic due to its high transmissibility. The unavoidable shortcomings of traditional diagnostic assay, including nucleic acid testing, diverse serological assays characterized by high-throughput and less workload, are playing a more and more crucial role to supplement the nucleic acid test. In this review, we summarize the dynamic change of the specific IgM, IgG, and IgA antibodies against SARS-CoV-2 as well as neutralizing antibodies and discuss the clinical utility and limitations of the different serological assays. SARS-CoV-2, a newly discovered virus, shows some unique pathogenetic and epidemiological characteristics that have not been completely understood so far. Currently, studies about the antibody responses against SARS-CoV-2 and the clinical utility of serological testing are increasing. It’s well suggested that the combination of serological tests and nucleic acid tests can cohesively improve the testing efficiency for identifying COVID-19 suspected patients.
Introduction
In December 2019, several cases of unknown pneumonia accompanied by respiratory syndromes were reported in Wuhan, Hubei, China (Chen N. et al., 2020). Subsequently, a novel coronavirus was identified in respiratory samples obtained from these patients with unknown pneumonia and the causative agent has currently been named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Guan et al., 2020). As of November 22, 2020, about 58 million confirmed cases have been reported worldwide with a total number of 1.3 million deaths (available from https://www.worldometers.info/coronavirus/?fbclid=IwAR2WhtIvODVjwxZEszArsVM0ypi0ZJvQ3SVjdnjuyl9ViV2IZPnIdKS5rto). Most COVID-19 patients show mild respiratory symptoms, while some severe cases might present acute respiratory distress syndrome (ARDS), septic shock, and multiple organ failure (Wang et al., 2020). It’s reported that the fatality rate of COVID-19 was approximately 4% (Zhang et al., 2020d), which is obviously lower than that of SARS (9.5%) and much lower than that of MERS (34.4%) (Petrosillo et al., 2020). Severe Acute Respiratory Syndrome (SARS) caused by SARS-CoV occurred during 2002–2003 and developed into a global pandemic with high infection rates among humans (Hu et al., 2017). MERS-CoV was first identified in Saudi Arabia and has now spread to the whole world with a high fatality rate, and it’s still not stopped yet (Bleibtreu et al., 2020).
SARS-CoV-2 is an enveloped, single-stranded RNA virus with oval or round particles that measure about 50–200 nm in diameter (Shereen et al., 2020). The major structural proteins of SARS-CoV-2 are the spike surface glycoprotein (S), a small envelope protein (E), matrix protein (M), and nucleocapsid protein (N), respectively. The M and E proteins play crucial roles in virus assembly (Malik, 2020). The spike protein (S) of coronavirus, a type I transmembrane glycoprotein, was found to have the ability of binding to the host cell surface receptor angiotensin-converting enzyme 2 (ACE2), followed by entering host cells (Wrapp et al., 2020). The N protein that binds to viral RNA is in the central role of the transcription and replication of RNA and can influence the cell cycle processes of host cells. It is widely accepted that IgM provides the first line of defense during microbial infections, before the generation of adaptive, high-affinity IgG responses that are important for long-lived immunity and immunological memory (Racine and Winslow, 2009). Both the S and N proteins are the important antigens of SARS-CoV-2 and many serological diagnostic tests have been developed based on the specific IgM and IgG antibodies against the S and the N proteins (Woo et al., 2005). Many publications have analyzed specific IgM, IgG, and IgA antibodies against the N and the S proteins in COVID-19 patients. Therefore, we summarize the kinetics of antibody response to SARS-CoV-2 in patients with COVID-19 including the specific IgM, IgG, and IgA against the S and the N proteins, to help us evaluate and employ the serological testing assays rapidly, and analyze the testing results reasonably, which help better contain the spread of the causative agent of SARS-CoV-2.
In this review, we first focus on the current knowledge regarding the humoral response to COVID-19 infection including the profiles of IgM, IgG, IgA, and neutralizing antibodies and then summarize the characteristics of antibodies dynamic change during the whole disease progression. We also discuss the diverse serological assays based on different antigens of SARS-CoV-2 and the limitations of the serodiagnosis. We strongly believe that those serological dynamic assays with high quality assurance system are crucial to facilitate better containment of the epidemic.
The Dynamic Change of Specific Antibody Against SARS-CoV-2 in Patients With COVID-19
The Seroconversion Time of Specific IgM and IgG Antibodies Against SARS-CoV-2
Previous studies revealed that the median seroconversion time of specific IgM and IgG against SARS-CoV-2 varies differently, ranging from 5 to 13 days, 11 to 14 days, respectively, after symptom onset (Gaddi et al., 2020; Guo L. et al., 2020; Long et al., 2020a; Sun et al., 2020; Xiang F. et al., 2020; Zhao et al., 2020). There exist three types of seroconversion including synchronous seroconversion of IgG and IgM (34.6%, 9/26), IgM seroconversion earlier than that of IgG (26.9%, 7/26), and IgM seroconversion later than that of IgG (38.5%, 10/26) according to a longitudinal observation (Long et al., 2020a). In our previous serological tests, the seroconversion of both IgM and IgG occurred at day 4 by colloidal gold-based immunochromatographic (ICG) strip among the confirmed cohort (Pan et al., 2020), which was 1 day earlier than that reported by Zhang et al. (2020c) using ELISA assay. The seroconversion of specific IgM and IgG varies against different types of antigens from SARS-CoV-2. More patients had earlier seroconversion for IgG than IgM against N proteins and RBD; the seroconversion of IgM in severe COVID-19 cases was delayed compared to IgG (Shen L. et al., 2020; To et al., 2020b). Thus, the delayed development of specific IgM antibodies could be a sign for severe patients with COVID-19.
By contrast, in SARS-CoV infections, specific IgM, IgG, or IgA antibodies against SARS-CoV were all tested negative until at least 3 days after symptoms onset and all positive after at least 19 days (Hsueh et al., 2004). In MERS infection, antibody response to MERS is commonly detected in weeks 2–3 after onset (Alshukairi et al., 2016; Corman et al., 2016).
The seroconversion of specific IgM and IgG antibodies against SARS-CoV-2 mostly turn positive in the second or third week after symptom onset and vary greatly when involved with different specific types of antigens. Consequently, we should pay more attention to the seroconversion time of specific IgM and IgG against SARS-CoV-2 in each different individual with COVID-19 to improve the accuracy and precision of serological assays.
The Dynamic Change of Specific IgM and IgG Antibodies in SARS-CoV-2 Infections Over the Whole Disease Progression
Our previous study conducted in Zhongnan Hospital of Wuhan University, China, showed that both IgM and IgG were first detected at day 4 and the positive rate of IgM and IgG were 11.1% and 3.6%, respectively, in patients of early stages; in intermediate and late stages, the positive rate remained about 75% and increased up to 96.8% for IgM and IgG, respectively (Pan et al., 2020). Patients confirmed with COVID-19 showed positive results of virus-specific IgM reaching a peak around 18–22 days post COVID-19 onset, specific IgG around 17–19 days (Long et al., 2020a; Xiang F. et al., 2020; Zhao et al., 2020). As shown in Figure 1, the seropositive percentages of specific IgM, IgG, and the combination of IgM and IgG in patients confirmed with COVID-19 varied over time (Pan et al., 2020). A study enrolled with 105 COVID-19 patients and 197 non-COVID-19 patients demonstrated that IgM reached peak within 15–21 days and slowly began to decline, while IgG peaked during 22–39 days and lasted for a longer time (Wu L. X. et al., 2020). Another study focused on the early humoral response to SARS-CoV-2 showed that IgM antibody level rose between days 8 and 14 but didn’t increase further between days 15 and 21 or after day 21; however, the IgG antibodies were detected on days 0–7, increased on days 8–14 and kept on rising until days 15–21, reaching a plateau by day 21 (Guo L. et al., 2020). Another study demonstrated that the level of IgM rose above the baseline level at day 6 (seroconversion time), peaked at around day 18, and fell to below the baseline level at about day 36, while IgG was seropositive by day 3, peaked at around 23 days, and then maintained at relatively high levels (Shu et al., 2020).
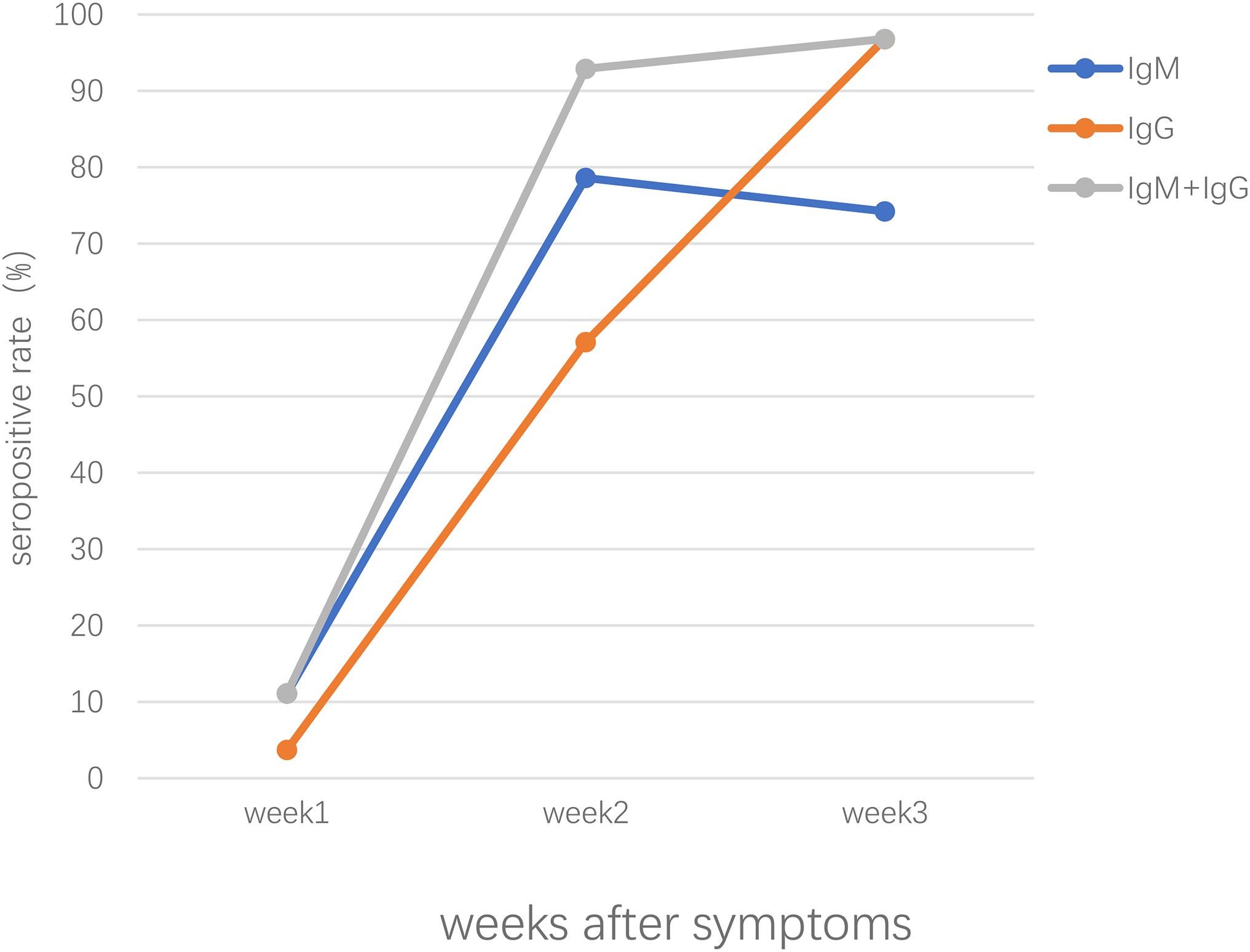
Figure 1. The seropositive rates of specific IgM, IgG, the combination of IgM and IgG antibody response in patients confirmed with SARS-CoV-2 infection after symptoms onset.
Among most non-ICU patients, total antibodies increased sharply since the first week and elevated during the next 2 weeks; specifically, N-IgM shared a similar dynamic pattern with N-IgG in the first 2 weeks in the same patient, IgM continued to increase until the third week and yet, the level of IgG exceeded IgM in the 2–3 week after onset, indicating that there was an IgM to IgG class switch (Sun et al., 2020; Zhao et al., 2020). Nevertheless, it’s the level of S-IgG that was much higher in COVID-19 patients with mild symptoms compared to another antigen and therefore it’s speculated that S-IgG plays a more important role in clearing the viruses and protecting patients (Sun et al., 2020). IgG antibodies generally keep positive for a long period time, whereas 3.2% (3/95) cases confirmed with COVID-19 showed IgG antibodies turning from positive to negative in just 5 days (Shu et al., 2020). It’s reported that 53.1% of mild cases were tested IgM negative, while only 14.3% negative in severe ones (Liu et al., 2020). Patients with severe COVID-19 tend to produce more robust humoral antibody response than those with mild ones. Other groups also reported that severe cases generally had an earlier SARS-CoV-2 specific IgM response and higher peak measurements compared with mild cases (Long et al., 2020a; Lynch et al., 2020; Perera et al., 2020; Zhao et al., 2020). Thus, the high titer of total Ab is probably the potential risk factor of worse clinical prognosis of COVID-19 patients. Notably, there have been reported that the seroconversion of mild COVID-19 cases took at least 4 weeks, which was longer than severe cases, and a small number of COVID-19 patients remained seronegative for Ab testing during the whole hospitalization period (Zhao et al., 2020). Guo X. et al. (2020) reported a case of mild COVID-19 with long virus-carrying time, which may be related to the weak production of virus-specific IgG and IgM, and recurrence of positive SARS-CoV-2 RNA occurred in stool specimens after discharge possibly due to intermittent virus shedding. Therefore, it’s necessary to apply multiple tests in order to make final correct diagnosis instead of just ignoring probable patients tested specific antibodies negative. What’s more, the serological assays tested positive for IgG and IgM were 81.1% (30/37) and 62.2% (23/37), respectively, in asymptomatic patients with COVID-19, which were obviously lower than 83.8% (31/37) for IgG and 78.4% (29/37) for IgM in the symptomatic COVID-19 patients (Long et al., 2020b).
The profiles of virus-specific antibodies vary in different individuals and further studies are needed to explore the uncertainties about the characteristics of immunological response to SARS-CoV-2.
By contrast, in SARS associated infections, nearly all SARS patients show virus-specific IgM and IgG antibody by week 3, which began to decrease in the 3–4 week after illness onset, and IgM remained at a low level in a few weeks, while the concentrations of IgG antibody that is 4∼8 times higher than that of IgM antibody, reaching its peak at 20 days after infection, and then maintained in a high level for a long time (Woo et al., 2004; Mo et al., 2005). There had been reported that an early MERS antibody response is correlated to the reduction of infection severity and the concentration of IgG antibody of most patients can reach a peak in the third week (Park et al., 2015).
The Development of the Secretory IgA Response May Play a Crucial Role in Preventing SARS-CoV-2 Infections
Current serological tests mainly revolve around the detections of specific IgM, IgG, or total immunoglobins. However, IgA plays a crucial part in mucosal immunity and is considered as the most powerful immunoglobin to fight infectious pathogens in respiratory and digestive systems, which can neutralize SARS-CoV-2 throughout virus entry (Chao et al., 2020). In a study, IgA seroconversion occurred in the first week in 3/4 (75%) patients after the disease onset, and the levels of IgA were persistently higher than IgM antibodies during the whole observation period, with a peak level at 20–22 days, which was later than that of IgM (Padoan et al., 2020b). Another study showed that the first seroconversion day of IgA was 2 days after onset of the initial symptoms and the levels of specific IgA noticeably increased about ∼2 weeks after the symptom onset and remained elevated for the following 14 days after seroconversion, which were significantly higher than those of IgM in both severe and non-severe patients (Yu et al., 2020). Guo L. et al. (2020) reported that both IgA and IgM increased between days 8 and 14 but did not rise further between days 15 and 21 or after day 21. It is similar to the observation in another study done about SARS-CoV associated infections that IgM and IgA shared similar dynamic patterns including seroconversion and antibody titers (Hsueh et al., 2004).
The Development of Neutralizing Antibodies (nAbs) During Infections by SARS-CoV-2
It’s reported that RBD-specific antibodies have greater potency to neutralize infection and thus the RBD of SARS-CoV-2 can serve as an important target for the development of potent and specific nAbs (Jiang et al., 2020). Wu F. et al. (2020) found that NAbs were detected in patients from days 4 to 6 and reached peak levels from days 10 to 15 after disease onset. What’s more, a SARS-CoV RBD-specific human neutralizing nAb, CR3022, could bind SARS-CoV-2 RBD with high affinity and recognize an epitope on the RBD that does not overlap with the ACE2-binding site while the neutralization capacity remains unclear (Tian et al., 2020). Whereas, findings in another study noted that high levels of nAbs may be the consequence of strong inflammation or innate immune response in older patients who developed higher nAbs titers and tend to have worse outcomes after SARS-CoV-2 infection (Wu F. et al., 2020). Therefore, the neutralizing antibodies maybe don’t necessarily play the protective roles in the illness as supposed. However, one pilot study on convalescent plasma with a high concentration of neutralizing antibodies can rapidly reduce the viral load and tends to improve clinical outcomes of patients infected with SARS-CoV-2 (Duan et al., 2020). To summarize, the therapeutic efficacy and effects of neutralizing antibodies need to be elucidated with further proofs and clinical evidence.
The Serological Tests for Specific IgM and IgG Antibodies
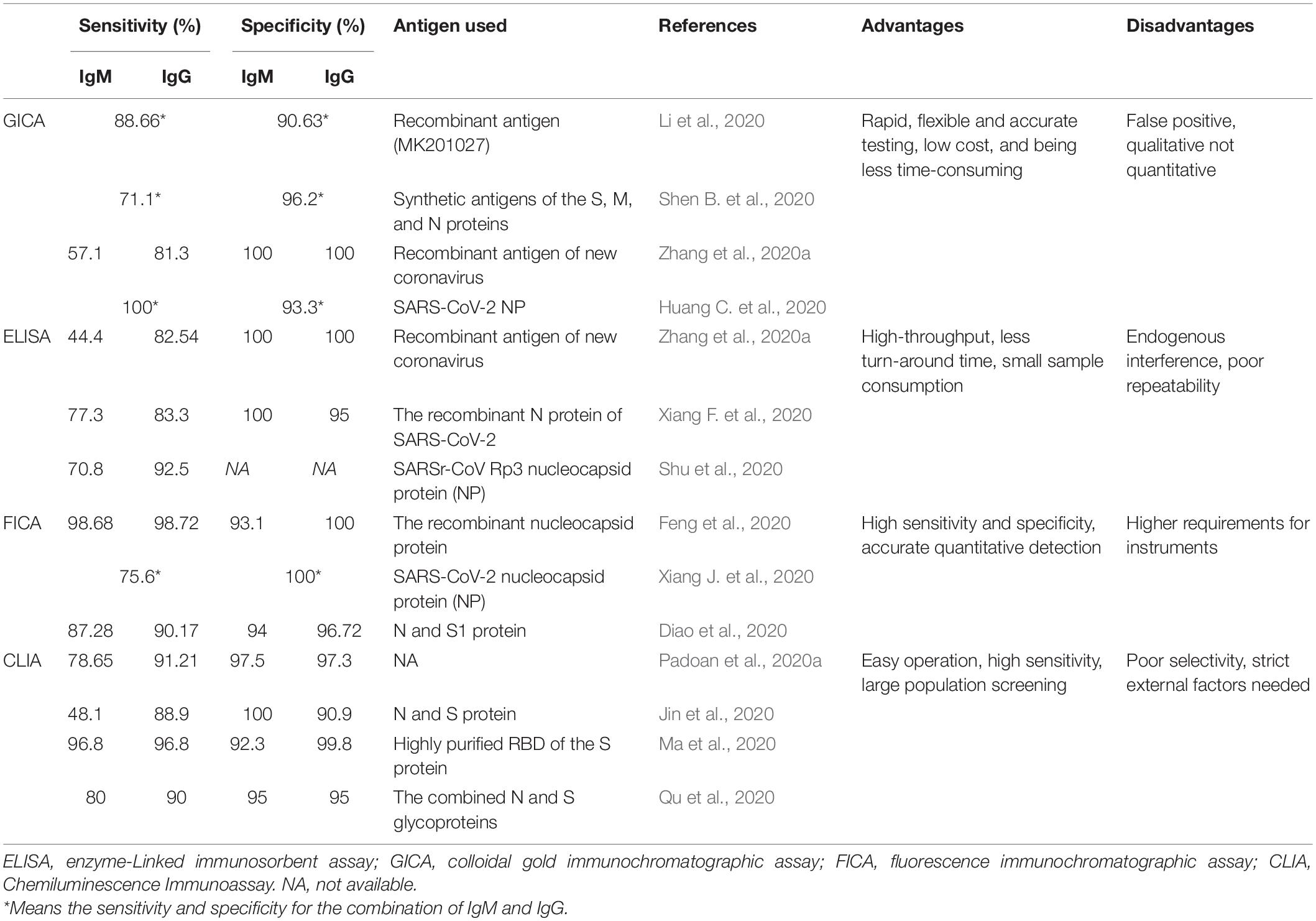
Many serological tests based on different antigens of SARS-CoV-2 have been developed in order to confirm suspected patients and to exclude patients infected with other respiratory viruses, thereby facilitating the control of this global pandemic. Some of the most commonly used immunoassays were listed in Table 1.
Colloidal Gold Immunochromatographic Assay
Colloidal gold immunochromatographic assay (GICA) is point-of-care testing (POCT) applied for qualitative analysis of target antigen/antibody, which is a feasible method for the diagnosis of COVID-19 in primary hospitals and laboratories, especially in emergency situations with its advantages of straightforward operation, time-saving and clinically compatible steps, small sample consumption, and easy result interpretation (Kim et al., 2015; Li et al., 2019; Huang C. et al., 2020). The N-S recombinant protein-based GICA assay for testing SARS-CoV-2 specific IgM and IgG antibodies demonstrates the strip possesses high specificity and is highly reproducible in sample results (Shen L. et al., 2020). IgG/IgM Rapid Test, manufactured by Cellex Inc., on April 1, 2020, is an aid in the diagnosis of patients with suspected SARS-CoV-2 infections (FDA, 2020). Our previous research found that the positive rates of IgM and IgG were relatively low in the early stage (1–7 days from onset) of SARS-CoV-2 infection, and gradually increased in the intermediate stage (8–14 days from onset) and peaked at a late stage (more than 15 days), and the combination of IgM and IgG results dramatically increased the sensitivity of GICA tests (Pan et al., 2020). Other studies reported the sensitivity and specificity of the colloidal gold immunochromatography assay for SARS-CoV-2 specific IgM/IgG antibody using synthetic antigens of the S, M, and N proteins were 71.1 and 96.2%, respectively (Shen B. et al., 2020). Huang S. et al. (2020) found that the sensitivity of the combined GICA IgM and IgG detection was 75/91 (82.4%) and were negative for healthy controls with a specificity of 100%. Zhang et al. (2020b) evaluated and found that the eukaryotic expression spike proteins are more suitable than the prokaryotic expression nucleocapsid proteins for the proposed GICA.
In conclusion, GICA is a good diagnostic tool for large population screening and community surveillance for identification of SARS-CoV-2. It’s supplementary to the nucleic acid detection by RT-PCR and provides qualitative results compared to ELISA assay showing quantitative antibody titer (Gaddi et al., 2020).
Enzyme-Linked Immunosorbent Assay
Enzyme-linked immunosorbent assay (ELISA) is a common biomedical assay that utilizes enzyme-labeled antigens/antibodies to detect specific molecules in specimens and is featured by cost-saving, easy operation (Gan and Patel, 2013). IgM and IgG antibodies against SARS-CoV-2 could be detected in the middle and later stages of the disease by ELISA-based serology tests, and seroconversion of specific IgM and IgG antibodies was observed as early as the fourth day after the onset of symptoms by ELISA (Long et al., 2020a). In addition, among the patients confirmed with COVID-19, the sensitivity, specificity, PPV, NPV, and consistency rate of IgM were 77.3% (51/66), 100%, 100%, 80.0%, and 88.1%, respectively, and IgG was 83.3% (55/66), 95.0%, 94.8%, 83.8%, and 88.9%, respectively, using the recombinant N protein of SARS-CoV-2 (Xiang F. et al., 2020). Huang S. et al. (2020) reported the sensitivity of the combined ELISA IgM and IgG detection was 55/63 (87.3%) and had a specificity of 100%. Guo L. et al. (2020) revealed that the detection efficiency of IgM by ELISA was higher than that of qPCR after 5.5 days of symptom onset. Therefore, the combination of IgM ELISA assay with PCR can improve the detective efficacy to identify SARS-CoV-2 infection.
Recently, a microfluidic ELISA system to detect COVID-19 antibodies on a lab-on-chip platform was described and proposed by a research group. Furthermore, this device first separates plasma from whole blood using a microfluidic device and subsequently performs the detection of antibodies in the separated plasma using a semi-automated on-chip ELISA with high-quality plasma and minimal cell interference (Tripathi and Agrawal, 2020). Zhang et al. (2020c) detected specific IgG and IgM from human serum of COVID-19 patients by ELISA using the SARS-CoV-2 Rp3 nucleocapsid protein, which has 90% amino acid sequence homology to other SARS-related viruses. A diagnostic specificity of 100.0% and sensitivity of 98.31% was achieved in a newly established ELISA using 59 sera of infected or vaccinated animals including ferrets, raccoon dogs, hamsters, rabbits, chickens, cattle, and a cat and a total of 220 antibody-negative sera of the same animal species (Wernike et al., 2020). What’s more, Schöler et al. (2020) established a SARS-CoV-2 neutralization assay employing an in-cell ELISA (icELISA) approach. It allows rapid (<48 h in total, read-out in seconds) and automated quantification of COVID-19 infection in cell culture to evaluate the efficacy of NAbs and antiviral drugs using reagents and equipment present in most routine diagnostics departments (Schöler et al., 2020). A report on a microfluidic, multiplexed POC test based on multiple SARS-CoV-2 antigens—S, N, and RBD—shows good concordance with a live virus microneutralization assay and successfully tracked the longitudinal evolution of the antibody response in infected individuals (Heggestad et al., 2020).
Fluorescence Immunochromatographic Assay
Fluorescence immunochromatographic assay (FICA) designed for the detection of specific immunoglobulins in blood/serum is based on the combination of immunofluorescence technique and chromatography technology, showing high sensitivity, accurate quantitative detection, and good stability (Sotnikov et al., 2017).
It’s reported that the sensitivity and specificity of the FICA with Lanthanide, Eu (III) using the recombinant nucleocapsid protein as antigen were 98.72% and 100% (IgG), and 98.68% and 93.1% (IgM), respectively (Feng et al., 2020). In other studies, it’s concluded that fluorescence immunochromatography is much easier for quantitative detection and more sensitive and specific in serodiagnosis than the traditional colloidal gold immunochromatographic assay (Xie et al., 2014; Chen Z. et al., 2020).
Chemiluminescence Immunoassay
Chemiluminescence immunoassay (CLIA) is based on double-antibodies sandwich immunoassay to detect specific antibodies through amplified signals of chemical luminescence materials. With the features of easy operation and high sensitivity, it is in the central role of the early diagnosis of diseases and large population screening (Min et al., 2018). Padoan et al. (2020a) found that the sensitivities and specificities of CLIA were 78.65%, 97.5% (IgM), and 91.21%, 97.3% (IgG). Another study using N and S proteins calculated that the sensitivities and specificities of serum IgM and IgG antibodies were 48.1% and 100%, 88.9% and 90.9%, respectively (Jin et al., 2020). A systematic review and meta-analysis summarized that the pooled sensitivity and specificity of CLIA were 97.8% and 98%, respectively, and the sensitivity increased 3 weeks after symptom onset (Lisboa Bastos et al., 2020). Cai et al. (2020) developed a peptide-based chemiluminescent immunoassay using synthetic peptide antigens including the orf1a/b, S, and N proteins to detect the positive rate of immunoglobulin G and IgM, which were 71.4% and 57.2%, respectively, among patients infected with SARS-CoV-2. CLIA detecting antibodies against RBD is considered the best diagnostic test accuracy (Mekonnen et al., 2020). Thus, it is a reliable immunoassay for assessing the immunological response in patients infected with SARS-CoV-2 and the detective accuracy of SARS-CoV-2 can be enhanced by the combination of nucleotide assay RT-PCR.
Limitations About Serological Testing for COVID-19 Patients
The Duration of the Window Period of Specific IgM and IgG Antibodies Alter in Different Patients
Antibody responses to infection take days to weeks to be reliably detectable and the levels of those antibodies decrease over time. It’s reasonable that in early phases of SARS-CoV-2 infections, the levels of specific IgM and IgG antibodies are too low to be detected in the serum or plasma samples leading to false-negative results of serological tests. There is concern that virus-specific antibodies detection may miss cases due to a larger window of time for indirectly detecting SARS-CoV-2 (Udugama et al., 2020). More than 60% of infected individuals are seronegative (IgM or IgG) in the first 7 days (Zhao et al., 2020). IgM and IgG converted to negative around 36 days and over 50 days separately (Guo L. et al., 2020). Notably, there have been reports that the seroconversion of mild COVID-19 cases took at least 4 weeks, which was longer than severe cases, and a small number of COVID-19 patients remained seronegative for Ab testing during the whole hospitalization period (Zhao et al., 2020), and it’s assumed that the innate immune response of these mild patients cleared the virus before the humoral immune system produced any antibodies. Serology measures the host response to infection and is an indirect measure of infection that is best utilized retrospectively (Tang et al., 2020).
The combination of serological tests and nucleotide acid assays is essential to improve the sensitivity and specificity of clinical diagnosis for COVID-19.
The Cross-Reactivity Between SARS-CoV-2 and Other Endemic Human Coronaviruses
It’s widely accepted that cross-reactivity has a great impact on the sensitivity and specificity of the serological tests. A recent study has demonstrated negligible cross-reactivity from human 257 coronavirus, NL63, to SARS-CoV-2 (Amanat and Krammer, 2020). It’s reported that sera from the healthy donors were found to have SARS-CoV-2 neutralization activity comparable to that of samples from seropositive patients with COVID-19 (van der Heide, 2020). Lv et al. (2020) established models of mice infected with SARS-CoV-2 and their results showed that cross-reactivity in antibody binding to the spike protein is common, and cross-neutralization of the live viruses may be singular, revealing non-neutralizing antibody response to conserved epitopes in the spike protein. Long et al. (2020a) did observe in a study enrolled with a total of 285 patients with COVID-19 showed cross-reactivity to the nucleocapsid proteins of SARS-CoV-2. Therefore, other endemic human coronaviruses may still have impact on accurately diagnosing patients with real SARS-CoV-2 infections (Lee et al., 2020).
There Exist Substances Interfering With the Serological Tests Results
Endogenous components in the fingerstick blood or the plasma/serum of venous blood sample can have an effect on the immunoassays of specific antibodies. Notably, endogenous heterophilic antibodies are present in many serum samples to interfere with two-site ELISAs, thereby evoking false-positive signals as well as the human autoantibodies rheumatoid factors (Grebenchtchikov et al., 2002; Berth and Willaert, 2016). Hemolysis, the breakdown of erythrocytes with subsequent release of the intracellular contents, can frequently interfere with the serology tests, leading to either positive or a negative bias in test results (Snyder et al., 2004). For instance, three children diagnosed with Kawasaki disease in Hong Kong, China, with no communication contacts with COVID-19 patients, were tested positive against anti-RBD and anti-NP antibodies of SARS-CoV-2, yet negative for SARS-CoV-2 NPA PCR and neutralizing antibodies, indicating a false-positive antibodies result (To et al., 2020a). Another study found that the rate of positive antibodies against N-protein and S-RBD was 3.6 and 1.6%, respectively, among healthy and non-COVID-19 individuals, revealing that the positivity of N-protein and S-RBD don’t necessarily determine SARS-CoV-2 infection (McAndrews et al., 2020).
Discussion
The outbreak of Coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 made urgent and necessary the need for diagnostic tests that can identify COVID-19 patients. The dynamic profiles of specific IgM and IgG antibody against SARS-CoV-2 in patients with COVID-19 varies differently. The median time for seroconversion of specific IgM and IgG antibodies is about 9–14 days; however, early seroconversion has been reported at 3–5 days (Gaddi et al., 2020; Long et al., 2020a; Sun et al., 2020; Xiang F. et al., 2020; Zhao et al., 2020). It’s found that the IgM level lasted more than 1 month, indicating a prolonged stage of virus replication in SARS-CoV-2-infected patients, and the IgG levels generally increased only in the later stages of the disease (Dhama et al., 2020).
Apart from specific IgM and IgG antibodies, the secretory IgA is considered to be powerful in mucosal immunity to eliminate the invasive virus SARS-CoV-2 and the seroconversion of IgA occurred commonly earlier than other antibodies. High levels of neutralizing antibodies are assumed to be associated with the overreaction of innate immune response or strong inflammation and don’t necessarily play the protective role in SARS-CoV-2 infection. The therapeutic efficacy and effects of neutralizing antibodies need to be furthermore elucidated. The dynamic changes in the antibody level of COVID-19 patients can provide more clinical information about the illness progression so that medical staff can cope with COVID-19 patients in a more proper and efficient way. Testing with IgG positive indicates prior infections with the virus and does not necessarily mean protective immunity in infected individuals; what’s more, it’s very likely to be IgG positive while shedding virus as determined by molecular assays due to the timing of infection stages and serum/plasma sampling (Theel et al., 2020). In conclusion, at least three situations could happen in serological test results: (1) seropositive but negative for molecular genetic assay results reflecting clearance of an earlier, milder infection, (2) positive result from molecular genetic assays for SARS-CoV-2 infection are seronegative due to the lag in antibody production following infection, and (3) limitation in sensitivity and specificity of serological tests including false positive and negative results, which may have undesirable impact on the socioeconomic decisions and overall public confidence in the results (Carter et al., 2020). Strict quarantine and health surveillance should be taken for all COVID-19 patients, even discharged to prevent a potential virus spread. It is still blurred whether patients with COVID-19 infection would acquire permanent immunity to this disease after certain time, and further studies need to be done about this. In the aspect of detective assays, one of the drawbacks of serological tests is the limited sensitivity in the window period at early stages; choosing different target antigens also has impact on sensitivity and specificity of serological assays (Caruana et al., 2020). A vast number of serological tests have been developed on the market for the detection of SARS-CoV-2 infection. Lateral flow assay (LFAs) such as GICA and FICA often demonstrates lower sensitivity than ELISAs and CLIAs (Koczula and Gallotta, 2016). LFAs are powerful diagnostic tools for large scale population screening and community surveillance for identification of SARS-CoV-2. Numerous LFIA based rapid POC tests have been developed by several companies, which enable the detection of IgM and IgG antibodies produced in suspects in response to SARS-CoV-2 infection (Vashist, 2020). It does not require any instrument or trained staff and, thus, it can be employed at any place and time, especially in developing nations with limited healthcare resources and remote settings. The assay is ideal for primary healthcare workers for the rapid testing of COVID-19 suspects (Vashist, 2020). LFIA method could be helpful in assessing in short time the possible contagiousness of subjects who, due to work needs, cannot guarantee “social distancing” to avoid the spread of COVID-19 by symptomatic and above all by asymptomatic individuals (Noce et al., 2020). Both ELISAs and CLIAs are reliable to assess the immune response of patients infected with SARS-CoV-2 (Gaddi et al., 2020; Guo L. et al., 2020; Jin et al., 2020). In specific clinical situations, due to the limitations about serological testing, choosing proper serological tests can greatly improve the testing efficiency. In general, antibody detection for SARS-CoV-2 with the advantages of high-throughput, faster turn-around time, and less work labor can effectively compensate for the shortcomings of nucleic acid detection, and assist nucleic acid testing to confirm the diagnosis of suspected patients. Therefore, the combination of nucleic acid test and serological tests based on different antigen proteins play a crucial role to improve the diagnostic accuracy and better contain the spread of SARS-CoV-2.
In conclusion, antibody testing can be used as a supplement to the SARS-CoV-2 nucleic acid testing method to confirm and exclude COVID-19. The exact role of the antibody against SARS-CoV-2 in the clinical diagnosis, treatment, and prognosis judgment of COVID-19 needs further study on the pathogenic mechanism of SARS-CoV-2.
Author Contributions
YF performed the data analyses and wrote the manuscript. YP contributed to the conception of the study. ZL and YL helped revise the draft and were responsible for the accuracy of this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by National Key Research and Development Program (Special Project for Public Security Risk Prevention and Emergency Technology Equipment), 2020YFC0847900.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alshukairi, A. N., Khalid, I., Ahmed, W. A., Dada, A. M., Bayumi, D. T., Malic, L. S., et al. (2016). Antibody Response and Disease Severity in Healthcare Worker MERS Survivors. Emerg. Infect. Dis. 22, 1113–1115. doi: 10.3201/eid2206.160010
Amanat, F., and Krammer, F. S. (2020). CoV-2 Vaccines: Status Report. Immunity 52, 583–589. doi: 10.1016/j.immuni.2020.03.007
Berth, M., and Willaert, S. (2016). Elimination of complement interference can improve the diagnostic performance of the VIDAS CMV IgG assay in acute cytomegalovirus infections. Diagn Microbiol. Infect. Dis. 85, 30–35. doi: 10.1016/j.diagmicrobio.2016.02.002
Bleibtreu, A., Bertine, M., Bertin, C., Houhou-Fidouh, N., and Visseaux, B. (2020). Focus on Middle East respiratory syndrome coronavirus (MERS-CoV). Med. Maladies infect. 50, 243–251. doi: 10.1016/j.medmal.2019.10.004
Cai, X. F., Chen, J., Li, Hu J, Long, Q. X., Deng, H. J., and Liu, P. (2020). A Peptide-Based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Coronavirus Disease 2019. J. Infect. Dis. 222, 189–193. doi: 10.1093/infdis/jiaa243
Carter, L. J., Garner, L. V., Smoot, J. W., Li, Y., Zhou, Q., Saveson, C. J., et al. (2020). Assay techniques and test development for COVID-19 diagnosis. ACS Publ. 6, 591–605. doi: 10.1021/acscentsci.0c00501
Caruana, G., Croxatto, A., Coste, A. T., Opota, O., Lamoth, F., Jaton, K., et al. (2020). Diagnostic strategies for SARS-CoV-2 infection and interpretation of microbiological results. Clin. Microbiol. Infect. 26, 1178–1182. doi: 10.1016/j.cmi.2020.06.019
Chao, Y. X., Rötzschke, O., and Tan, E. K. (2020). The role of IgA in COVID-19. Brain Behav. Immun. 87, 182–183. doi: 10.1016/j.bbi.2020.05.057
Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan. China 395, 507–513. doi: 10.1016/s0140-6736(20)30211-7
Chen, Z., Zhang, Z., Zhai, X., Li, Y., Lin, L., Zhao, H., et al. (2020). Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 92, 7226–7231. doi: 10.1021/acs.analchem.0c00784
Corman, V. M., Albarrak, A. M., Omrani, A. S., Albarrak, M. M., Farah, M. E., Almasri, M., et al. (2016). Viral Shedding and Antibody Response in 37 Patients With Middle East Respiratory Syndrome Coronavirus Infection. Clin. Infect. Dis. 62, 477–483. doi: 10.1093/cid/civ951
Dhama, K., Sharun, K., Tiwari, R., Sircar, S., Bhat, S., Malik, Y. S., et al. (2020). Coronavirus disease 2019–COVID-19. United States: ASM. doi: 10.20944/preprints202003.0001.v1
Diao, B., Wen, K., Zhang, J., Chen, J., Han, C., Chen, Y., et al. (2020). Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 5, S1198–S1743. doi: 10.1016/j.cmi.2020.09.057
Duan, K., Liu, B., Li, C., Zhang, H., Yu, T., Qu, J., et al. (2020). Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 117, 9490–9496. doi: 10.1073/pnas.2004168117
Feng, M., Chen, J., Xun, J., Dai, R., Zhao, W., Lu, H., et al. (2020). Development of a sensitive immunochromatographic method using lanthanide fluorescent microsphere for rapid serodiagnosis of COVID-19. ACS Sens 5, 2331–2337. doi: 10.1021/acssensors.0c00927
Gaddi, A. V., Capello, F., Aluigi, L., Antignani, P. L., Callegaro, A., Casu, G., et al. (2020). The Strategic Alliance between Clinical and Molecular Science in the War against SARS-CoV-2, with the Rapid-Diagnostics Test as an Indispensable Weapon for Front Line Doctors. Int. J. Mole. Sci. 21:12. doi: 10.3390/ijms21124446
Gan, S. D., and Patel, K. R. (2013). Enzyme immunoassay and enzyme-linked immunosorbent assay. J. Investig. Dermatol. 133:e12. doi: 10.1038/jid.2013.287
Grebenchtchikov, N., Sweep, C. G. J., Geurts-Moespot, A., Piffanelli, A., Foekens, J. A., and Benraad, T. J. (2002). An ELISA avoiding interference by heterophilic antibodies in the measurement of components of the plasminogen activation system in blood. J. Immunol. Methods 268, 219–231. doi: 10.1016/s0022-1759(02)00213-2
Guan, W. J., Ni, Z. Y., Hu, Y., Liang, W. H., Ou, C. Q., He, J. X., et al. (2020). Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 382, 1708–1720. doi: 10.1056/NEJMoa2002032
Guo, L., Ren, L., Yang, S., Xiao, M., Chang, D., Yang, F., et al. (2020). Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin. Infect. Dis. 71, 778–785. doi: 10.1093/cid/ciaa310
Guo, X., Zeng, L., Huang, Z., He, Y., Zhang, Z., and Zhong, Z. J. F. (2020). Longer Duration of SARS-CoV-2 Infection in a Case of Mild COVID-19 With Weak Production of the Specific IgM and IgG Antibodies. Front. Immunol. 11:1936. doi: 10.3389/fimmu.2020.01936
Heggestad, J. T., Kinnamon, D., Olson, L., Liu, J., Kelly, G., Wall, S., et al. (2020). Multiplexed, quantitative serological profiling of COVID-19 from a drop of blood by a point-of-care test. Version 1. medRxiv. Preprint. doi: 10.1101/2020.11.05.20226654
Hsueh, P. R., Huang, L. M., Chen, P. J., Kao, C. L., and Yang, P. C. (2004). Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 10, 1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x
Hu, B., Zeng, L. P., Yang, X. L., Ge, X. Y., Zhang, W., Li, B., et al. (2017). Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS pathogens. 13:e1006698. doi: 10.1371/journal.ppat.1006698
Huang, C., Wen, T., Shi, F.-J., Zeng, X.-Y., and Jiao, Y.-J. J. A. O. (2020). Rapid Detection of IgM Antibodies against the SARS-CoV-2 Virus via Colloidal Gold Nanoparticle-Based Lateral-Flow Assay. ACS Omega 5, 12550–12556. doi: 10.1021/acsomega.0c01554
Huang, S., Lin, C., Yan, M., Li, H., Liu, T., Michael, W., et al. (2020). Rapid Detection of COVID-19 by Serological Methods and the Evaluation of Diagnostic Efficacy of IgM and IgG. Clin. Lab. 66:11. doi: 10.7754/Clin.Lab.2020.200617
Jiang, S., Hillyer, C., and Du, L. (2020). Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 41, 355–359. doi: 10.1016/j.it.2020.03.007
Jin, Y., Wang, M., Zuo, Z., Fan, C., Ye, F., Cai, Z., et al. (2020). Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int. J. Infect. Dis. 94, 49–52. doi: 10.1016/j.ijid.2020.03.065
Kim, Y. R., Park, S. B., Fagutao, F. F., Nho, S. W., Jang, H. B., Cha, I. S., et al. (2015). Development of an immunochromatography assay kit for rapid detection of ranavirus. J. Virol. Methods 223, 33–39. doi: 10.1016/j.jviromet.2015.07.009
Koczula, K. M., and Gallotta, A. (2016). Lateral flow assays. Essays Biochem. 60, 111–120. doi: 10.1042/EBC20150012
Lee, C. Y.-P., Lin, R. T. P., Renia, L., and Ng, L. F. P. (2020). Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 11:879. doi: 10.3389/fimmu.2020.00879
Li, Z., Yi, Y., Luo, X., Xiong, N., Liu, Y., Li, S., et al. (2020). Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 13:10.1002/jmv.25727.
Li, Z. X., Zhao, F. P., Tang, T. T., Wang, M. M., Yu, X. L., Wang, R. C., et al. (2019). Development of a Colloidal Gold Immunochromatographic Strip Assay for Rapid Detection of Bovine Rotavirus. Viral. Immunol. 32, 393–401. doi: 10.1089/vim.2019.0071
Lisboa Bastos, M., Tavaziva, G., Abidi, S. K., Campbell, J. R., Haraoui, L. P., Johnston, J. C., et al. (2020). Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ 370:m2516. doi: 10.1136/bmj.m2516
Liu, Z.-L., Liu, Y., Wan, L.-G., Xiang, T.-X., Le, A.-P., Liu, P., et al. (2020). Antibody profiles in mild and severe cases of COVID-19. Clin. Chem. 66, 1102–1104. doi: 10.1093/clinchem/hvaa137
Long, Q.-X., Tang, X.-J., Shi, Q.-L., Li, Q., Deng, H.-J., Yuan, J., et al. (2020b). Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. 26, 1200–1204. doi: 10.1038/s41591-020-0965-6
Long, Q. X., Liu, B. Z., Deng, H. J., Wu, G. C., Deng, K., Chen, Y. K., et al. (2020a). Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 15, 845–848. doi: 10.1038/s41591-020-0897-1
Lv, H., Wu, N. C., Tsang, O. T., Yuan, M., Perera, R., Leung, W. S., et al. (2020). Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 31:107725. doi: 10.1016/j.celrep.2020.107725
Lynch, K. L., Whitman, J. D., Lacanienta, N. P., Beckerdite, E. W., Kastner, S. A., Shy, B. R., et al. (2020). Magnitude and kinetics of anti-SARS-CoV-2 antibody responses and their relationship to disease severity. Clin. Infect. Dis. 14:ciaa979. doi: 10.1101/2020.06.03.20121525
Ma, H., Zeng, W., He, H., Zhao, D., Jiang, D., Zhou, P., et al. (2020). Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol. Immunol. 17, 773–775. doi: 10.1038/s41423-020-0474-z
McAndrews, K. M., Dowlatshahi, D. P., Dai, J., Becker, L. M., Hensel, J., Snowden, L. M., et al. (2020). Heterogeneous antibodies against SARS-CoV-2 spike receptor binding domain and nucleocapsid with implications on COVID-19 immunity. JCI Insight. 11:879. doi: 10.1172/jci.insight.142386
Mekonnen, D., Mengist, H. M., Derbie, A., Nibret, E., Munshea, A., He, H., et al. (2020). Diagnostic accuracy of serological tests and kinetics of severe acute respiratory syndrome coronavirus 2 antibody: A systematic review and meta-analysis. Rev. Med. Virol. 5:e2181. doi: 10.1002/rmv.2181
Min, X. P., Fu, D., Zhang, J. Z., Zeng, J. T., Weng, Z. Y., Chen, W. D., et al. (2018). An automated microfluidic chemiluminescence immunoassay platform for quantitative detection of biomarkers. Biomed. Microdevices. 20:9. doi: 10.1007/s10544-018-0331-3
Mo, H. Y., Xu, J., Ren, X. L., Zeng, G. Q., Tan, Y. X., Chen, R. C., et al. (2005). Evaluation by indirect immunofluorescent assay and enzyme linked immunosorbent assay of the dynamic changes of serum antibody responses against severe acute respiratory syndrome coronavirus. Chin. Med. J. 118, 446–450.
Noce, A., Santoro, M. L., Marrone, G., D’Agostini, C., Amelio, I., Duggento, A., et al. (2020). Serological determinants of COVID-19. Biol. Direct. 15, 1–9. doi: 10.1186/s13062-020-00276-1
Padoan, A., Cosma, C., Sciacovelli, L., Faggian, D., Plebani, M. J. C. C., and Medicine, L. (2020b). Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin. Chem. Lab. Med. 1:32301749. doi: 10.1515/cclm-2020-0443
Padoan, A., Sciacovelli, L., Basso, D., Negrini, D., Zuin, S., Cosma, C., et al. (2020a). IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clin. Chim. Acta 507, 164–166. doi: 10.1016/j.cca.2020.04.026
Pan, Y., Li, X., Yang, G., Fan, J., Tang, Y., Zhao, J., et al. (2020). Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J. Infect. 81, e28–e32. doi: 10.1101/2020.03.13.20035428
Park, W. B., Perera, R., Choe, P. G., Lau, E. H. Y., Choi, S. J., Chun, J. Y., et al. (2015). Kinetics of Serologic Responses to MERS Coronavirus Infection in Humans, South Korea. Emerg. Infect. Dis. 21, 2186–2189. doi: 10.3201/eid2112.151421
Perera, R. A., Mok, C. K., Tsang, O. T., Lv, H., Ko, R. L., Wu, N. C., et al. (2020). Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). March 25:2000421. doi: 10.2807/1560-7917.ES.2020.25.16.2000421
Petrosillo, N., Viceconte, G., Ergonul, O., Ippolito, G., and Petersen, E. (2020). COVID-19, SARS and MERS: are they closely related? Clinical microbiology and infection : the official publication of the European Society of Clinical. Microbiol. Infect. Dis. 26, 729–734. doi: 10.1016/j.cmi.2020.03.026
Qu, J., Wu, C., Li, X., Zhang, G., Jiang, Z., Li, X., et al. (2020). Profile of immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 27:ciaa489. doi: 10.1093/cid/ciaa489
Racine, R., and Winslow, G. M. (2009). IgM in microbial infections: taken for granted? Immunol. Lett. 125, 79–85. doi: 10.1016/j.imlet.2009.06.003
Schöler, L., Le-Trilling, V. T. K., Eilbrecht, M., Mennerich, D., Anastasiou, O. E., Krawczyk, A., et al. (2020). A Novel In-Cell ELISA Assay Allows Rapid and Automated Quantification of SARS-CoV-2 to Analyze Neutralizing Antibodies and Antiviral Compounds. Front. Immunol. 11:573526. doi: 10.3389/fimmu.2020.573526
Shen, B., Zheng, Y., Zhang, X., Zhang, W., Wang, D., Jin, J., et al. (2020). Clinical evaluation of a rapid colloidal gold immunochromatography assay for SARS-Cov-2 IgM/IgG. Am. J. Transl. Res. 12, 1348–1354.
Shen, L., Wang, C., Zhao, J., Tang, X., Shen, Y., Lu, M., et al. (2020). Delayed specific IgM antibody responses observed among COVID-19 patients with severe progression. Emerg. Microb. Infect. 9, 1096–1101. doi: 10.1080/22221751.2020.1766382
Shereen, M. A., Khan, S., Kazmi, A., Bashir, N., and Siddique, R. (2020). C. O. V. I. D-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 24, 91–98. doi: 10.1016/j.jare.2020.03.005
Shu, H., Wang, S., Ruan, S., Wang, Y., Zhang, J., Yuan, Y., et al. (2020). Dynamic changes of antibodies to SARS-CoV-2 in COVID-19 patients at early stage of outbreak. Virol. Sin 27, 1–8. doi: 10.1007/s12250-020-00318-y
Snyder, J. A., Rogers, M. W., King, M. S., Phillips, J. C., Chapman, J. F., and Hammett-Stabler, C. A. (2004). The impact of hemolysis on Ortho-Clinical Diagnostic’s ECI and Roche’s elecsys immunoassay systems. Clin. Chim. Acta 348, 181–187. doi: 10.1016/j.cccn.2004.05.017
Sotnikov, D. V., Zherdev, A. V., and Dzantiev, B. B. (2017). Mathematical Model of Serodiagnostic Immunochromatographic Assay. Analyt. Chem. 89, 4419–4427. doi: 10.1021/acs.analchem.6b03635
Sun, B., Feng, Y., Mo, X., Zheng, P., Wang, Q., Li, P., et al. (2020). Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microb. Infect. 9, 940–948.
Tang, Y.-W., Schmitz, J. E., Persing, D. H., and Stratton, C. (2020). Laboratory diagnosis of COVID-19: current issues and challenges. J. Clin. Microbiol. 58, e512–e520.
Theel, E. S., Slev, P., Wheeler, S., Couturier, M. R., Wong, S. J., and Kadkhoda, K. (2020). The role of antibody testing for SARS-CoV-2: is there one? J. Clin. Microbiol. 58, e797–e720.
Tian, X., Li, C., Huang, A., Xia, S., Lu, S., Shi, Z., et al. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 9, 382–385. doi: 10.1080/22221751.2020.1729069
To, K. K., Chua, G. T., Kwok, K. L., Wong, J. S., Au, D. C. Y., Lam, Y. Y., et al. (2020a). False-positive SARS-CoV-2 serology in 3 children with Kawasaki disease. Diagn. Microbiol. Infect. Dis. 98, 115141. doi: 10.1016/j.diagmicrobio.2020.115141
To, K. K., Tsang, O. T., Leung, W. S., Tam, A. R., Wu, T. C., Lung, D. C., et al. (2020b). Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 20, 565–574. doi: 10.1016/s1473-3099(20)30196-1
Tripathi, S., and Agrawal, A. (2020). Blood Plasma Microfluidic Device: Aiming for the Detection of COVID-19 Antibodies Using an On-Chip ELISA Platform. Trans. Indian Natl. Acad. Eng. 10, 1–4.
Udugama, B., Kadhiresan, P., Kozlowski, H. N., Malekjahani, A., Osborne, M., Li, V. Y., et al. (2020). Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 14, 3822–3835. doi: 10.1021/acsnano.0c02624
Vashist, S. K. (2020). In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Scimago: Multidisciplinary Digital Publishing Institute.
Wang, B., Wang, L., Kong, X., Geng, J., Xiao, D., Ma, C., et al. (2020). Long-term coexistence of SARS-CoV-2 with antibody response in COVID-19 patients. J. Med. Virol. 92, 1684–1689. doi: 10.1002/jmv.25946
Wernike, K., Aebischer, A., Michelitsch, A., Hoffmann, D., Freuling, C., Balkema-Buschmann, A., et al. (2020). Multi-species ELISA for the detection of antibodies against SARS-CoV-2 in animals. Transbound Emerg. Dis. 2020:13926.
Woo, P. C., Lau, S. K., Wong, B. H., Tsoi, H. W., Fung, A. M., Kao, R. Y., et al. (2005). Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 43, 3054–3058. doi: 10.1128/jcm.43.7.3054-3058.2005
Woo, P. C. Y., Lau, S. K. P., Wong, B. H. L., Chan, K. H., Chu, C. M., Tsoi, H. W., et al. (2004). Longitudinal profile of immunoglobulin G (IgG), IgM, and TgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin. Diagn. Lab. Immunol. 11, 665–668. doi: 10.1128/cdli.11.4.665-668.2004
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263.
Wu, F., Liu, M., Wang, A., Lu, L., Wang, Q., Gu, C., et al. (2020). Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in Shanghai. China 180, 1356–1362. doi: 10.1001/jamainternmed.2020.4616
Wu, L. X., Wang, H., Gou, D., Fu, G., Wang, J., and Guo, B. (2020). Clinical significance of the serum IgM and IgG to SARS-CoV-2 in coronavirus disease-2019. New Jersey: Wiley, e23649.
Xiang, F., Wang, X., He, X., Peng, Z., Yang, B., Zhang, J., et al. (2020). Antibody detection and dynamic characteristics in patients with coronavirus disease 2019. Clin. Infect. Dis. 71, 1930–1934. doi: 10.1093/cid/ciaa461
Xiang, J., Yan, M., Li, H., Liu, T., Lin, C., Huang, S., et al. (2020). Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold-Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19). medRxiv.
Xie, Q. Y., Wu, Y. H., Xiong, Q. R., Xu, H. Y., Xiong, Y. H., Liu, K., et al. (2014). Advantages of fluorescent microspheres compared with colloidal gold as a label in immunochromatographic lateral flow assays. Biosen. Bioelectr. 54, 262–265. doi: 10.1016/j.bios.2013.11.002
Yu, H. Q., Sun, B. Q., Fang, Z. F., Zhao, J. C., Liu, X. Y., Li, Y. M., et al. (2020). Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur. Respir. J. 56:2001526.
Zhang, C., Zhou, L., Liu, H., Zhang, S., Tian, Y., Huo, J., et al. (2020a). Establishing a high sensitivity detection method for SARS-CoV-2 IgM/IgG and developing a clinical application of this method. 9, 2020–2029. doi: 10.1080/22221751.2020.1811161
Zhang, P., Gao, Q., Wang, T., Ke, Y., Mo, F., Jia, R., et al. (2020b). Development and evaluation of a serological test for diagnosis of COVID-19 with selected recombinant spike proteins. Eur. J. Clin. Microbiol. Infect. Dis. 12, 1–8.
Zhang, W., Du, R. H., Li, B., Zheng, X. S., Yang, X. L., Hu, B., et al. (2020c). Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 9, 386–389. doi: 10.1080/22221751.2020.1729071
Zhang, W., Zhao, Y., Zhang, F., Wang, Q., Li, T., Liu, Z., et al. (2020c). The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin. Immunol. 214:108393. doi: 10.1016/j.clim.2020.108393
Keywords: COVID-19, SARS-CoV-2, specific antibodies, dynamic change, serological assays
Citation: Fu Y, Pan Y, Li Z and Li Y (2021) The Utility of Specific Antibodies Against SARS-CoV-2 in Laboratory Diagnosis. Front. Microbiol. 11:603058. doi: 10.3389/fmicb.2020.603058
Received: 05 September 2020; Accepted: 01 December 2020;
Published: 13 January 2021.
Edited by:
Juarez Antonio Simões Quaresma, Evandro Chagas Institute, BrazilReviewed by:
Kuldeep Dhama, Indian Veterinary Research Institute (IVRI), IndiaLeticia Cedillo-Barron, Instituto Politécnico Nacional de México (CINVESTAV), Mexico
Copyright © 2021 Fu, Pan, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqiang Li, lizhiqiang@whu.edu.cn; Yirong Li, liyirong838@163.com
 Yu Fu1
Yu Fu1 Yunbao Pan
Yunbao Pan Zhiqiang Li
Zhiqiang Li Yirong Li
Yirong Li