- 1Department of Laboratory Medicine, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
- 2Department of Blood Transfusion, China-Japan Friendship Hospital, Beijing, China
- 3Department of Hematology, Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 4Department of Laboratory Medicine, The Second Affiliated Hospital, Guangzhou University of Chinese Medicine, Guangzhou, China
- 5Department of Laboratory Medicine, Guangdong Provincial Hospital of Chinese Medicine, Guangzhou, China
Background: This study aimed to examine the impact of an intervention carried out in 2011 to combat multi-drug resistance and outbreaks of imipenem-resistant Acinetobacter baumannii (IRAB), and to explore its resistance mechanism.
Methods: A total of 2572 isolates of A. baumannii, including 1673 IRAB isolates, were collected between 2007 and 2014. An intervention was implemented to control A. baumannii resistance and outbreaks. Antimicrobial susceptibility was tested by calculating minimal inhibitory concentrations (MICs), and outbreaks were typed using pulsed-field gel electrophoresis (PFGE). Resistance mechanisms were explored by polymerase chain reaction (PCR) and whole genome sequencing (WGS).
Results: Following the intervention in 2011, the resistance rates of A. baumannii to almost all tested antibiotics decreased, from 85.3 to 72.6% for imipenem, 100 to 80.8% for ceftriaxone, and 45.0 to 6.9% for tigecycline. The intervention resulted in a decrease in the number (seven to five), duration (8–3 months), and departments (five to three) affected by outbreaks; no outbreaks occurred in 2011. After the intervention, only blaAMPC (76.47 to 100%) and blaTEM–1 (75.74 to 96.92%) increased (P < 0.0001); whereas blaGES–1 (32.35 to 3.07%), blaPER–1 (21.32 to 1.54%), blaOXA–58 (60.29 to 1.54%), carO (37.50 to 7.69%), and adeB (9.56 to 3.08%) decreased (P < 0.0001). Interestingly, the frequency of class B β-lactamase genes decreased from 91.18% (blaSPM–1) and 61.03% (blaIMP–1) to 0%, while that of class D blaOXA–23 increased to 96.92% (P < 0.0001). WGS showed that the major PFGE types causing outbreaks each year (type 01, 11, 18, 23, 26, and 31) carried the same resistance genes (blaKPC–1, blaADC–25, blaOXA–66, and adeABC), AdeR-S mutations (G186V and A136V), and a partially blocked porin channel CarO. Meanwhile, plasmids harboring blaOXA–23 were found after the intervention.
Conclusion: The intervention was highly effective in reducing multi-drug resistance of A. baumannii and IRAB outbreaks in the long term. The resistance mechanisms of IRAB may involve genes encoding β-lactamases, efflux pump overexpression, outer membrane porin blockade, and plasmids; in particular, clonal spread of blaOXA–23 was the major cause of outbreaks. Similar interventions may also help reduce bacterial resistance rates and outbreaks in other hospitals.
Background
Acinetobacter baumannii is a life-threatening hospital pathogen, causing severe morbidity and mortality, particularly in intensive care units (ICUs) (Jain et al., 2019). Its rate of carbapenem resistance is surprisingly high (Singkham-In and Chatsuwan, 2018), and carbapenem-resistant strains of A. baumannii, especially imipenem-resistant A. baumannii (IRAB), have caused hospital outbreaks worldwide (Vauchel et al., 2019). To the best of our knowledge, many A. baumannii resistance studies have only reported the severity of IRAB resistance, such as mortality and outbreaks (Venditti et al., 2019; Wang et al., 2020). A few measures have been proposed in China (Yu et al., 2019); however, their impact was rarely monitored for a long period.
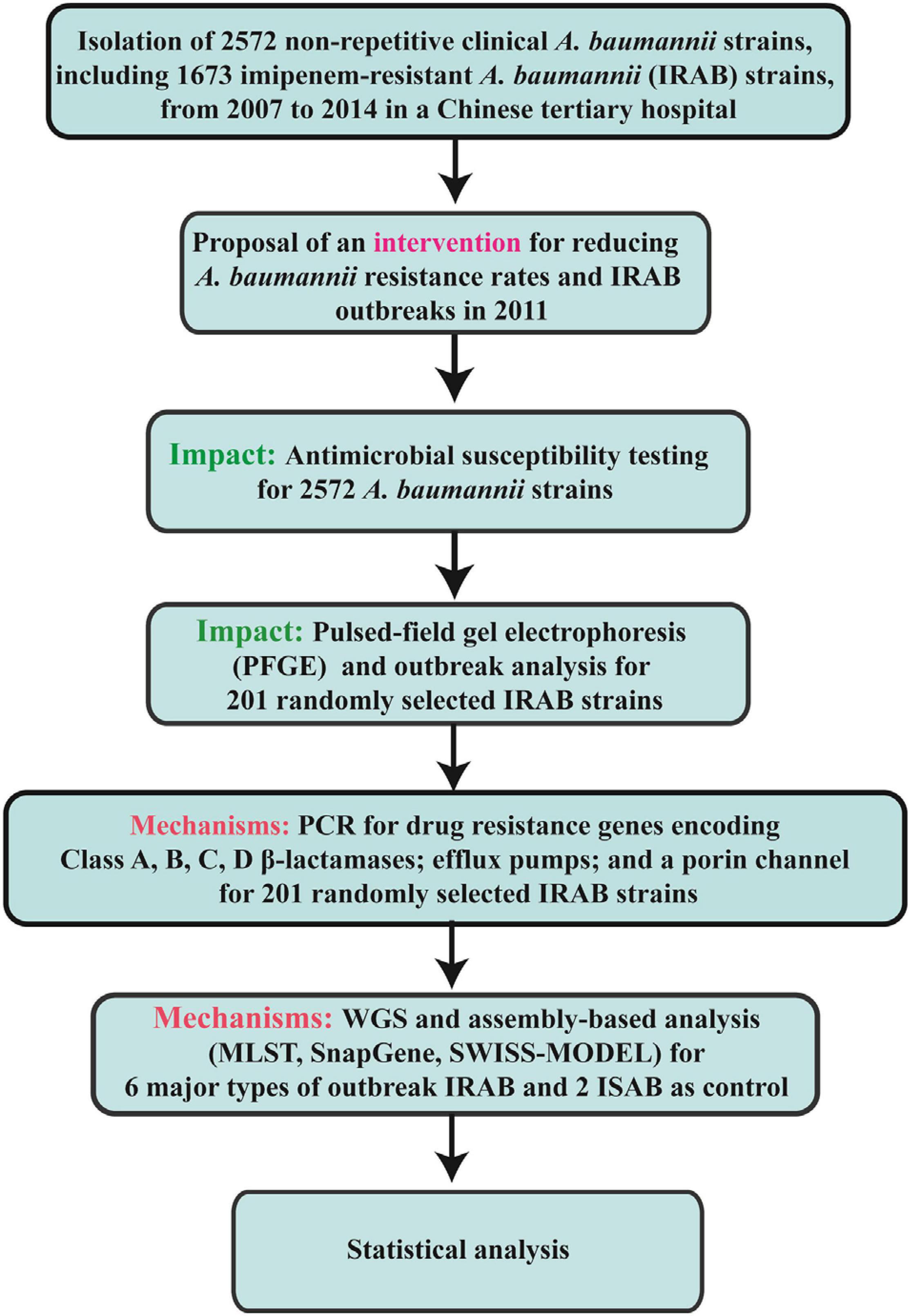
Hence, the aim of this study was to examine the efficacy of an intervention applied for the prevention and control of IRAB resistance and outbreaks. To discover changes before and after the intervention, we conducted an 8-year continuous survey of IRAB strains with respect to antibiotic resistance, outbreaks, detection of resistance genes, porin channels, and efflux pumps from 2007 to 2014 in a Chinese tertiary hospital.
We believe that similar interventions can help control infections and outbreaks in other hospitals as well, even for other pathogenic bacteria.
Materials and Methods
Isolation and Identification of Bacterial Strains
A total of 2572 non-repetitive A. baumannii strains, including 1673 IRAB strains, were recovered from samples collected from patients treated in different departments of The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China between January 2007 and December 2014, and identified using a Vitek2 compact automatic microbiological analysis system with GN colorimetric identification cards (bioMérieux, Marcy l’Etoile, France). These 2572 A. baumannii strains were tested for antibiotic resistance; among them, 201 IRAB strains were randomly selected by stratified random sampling method. Of these 201 IRAB strains, the first isolated from different patients each month were randomly selected, and among them a total of 25–26 strains were selected each year (2007–2014). This stratified random sampling method ensured that each IRAB strain had the same probability to be selected, and enabled to monitor statistically the major trends of occurrence of all isolates, reflected by those of the 201 IRAB strains.
These 201 IRAB strains were subjected to pulsed-field gel electrophoresis (PFGE), outbreak analyses, and PCR-based detection of resistance genes. Using whole genome sequencing (WGS), six major PFGE types of outbreak IRAB strains and two control imipenem-sensitive A. baumannii (ISAB) strains were sequenced. This study was approved by the Clinical Research and Ethics Committee of The First Affiliated Hospital of Sun Yat-sen University [2019(483)].
Antimicrobial Susceptibility Testing
Susceptibility testing was performed using AST-GN and AST-GN13 cards on the Vitek2 compact system (bioMérieux, Marcy l’Etoile, France). The microbroth dilution method was used in accordance with the standards published by the Clinical and Laboratory Standards Institute (CLSI). The test agents included imipenem, meropenem, gentamicin, tobramycin, ampicillin/sulbactam, piperacillin/tazobactam, levofloxacin, ciprofloxacin, cefepime, ceftazidime, ceftriaxone, sulfamethoxazole, furantoin, and tigecycline. Based on CLSI clinical breakpoints, isolates were designated as IRAB if imipenem MICs ≥ 8 μg/mL (2018; CLSI Document M100-S28). Quality control for susceptibility testing was performed with Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, and Escherichia coli ATCC 25922, which were purchased from the National Clinical Laboratory Center.
Pulsed-Field Gel Electrophoresis (PFGE)
The standard protocol of CDC PulseNet1 used to subtype Salmonella was used for bacterial DNA preparation for PFGE. Agarose plugs containing DNA were digested with 10 U of the restriction enzyme ApaI (TaKaRa, Ipswich, MA, United States) for 4 h. Electrophoresis was performed with a 1% SeaKem Gold® agarose gel (Lonza, Basel, Switzerland) in 0.5 × TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.3) using CHEF Mapper® XA (Bio-Rad Laboratories, Hercules, CA, United States) and alternating pulses at 120°, 6 V/cm, and 5–20 s for 18 h. Genomic DNA of standard Salmonella H9812 from the Centers for Disease Control and Prevention was digested with 50 U of XbaI and used as a molecular size marker. The interpretation of band patterning was performed according to the criteria of Tenover et al. (1995).
PCR for Identification of Drug Resistance Genes
Bacterial DNA was extracted from A. baumannii isolates by boiling at 100°C for 10 min. PCR was performed for antibiotic resistance-related genes using TaKaRa Ex Taq (Takara Bio Inc., Otsu, Japan) in a 50 μL reaction mixture containing 5 μL 10×buffer, 4 μL of a dNTPs mixture (2.5 mM), 0.25 μL Taq polymerase (5 U/μL), 1 μL forward primer (20 μM), 1 μL reverse primer (20 μM), 1 μL DNA template (20 ng/μL), and nuclease-free water. The PCR cycle consisted of denaturation at 93°C for 2 min, followed by 35cycles of 60 s at 93°C, annealing for 60 s at 55°C, and extension at 72°C for 60 s, with a final extension at 72°C for 10 min. An agarose gel (1%) was used to resolve and detect the PCR products. The primer sequences used for amplification of drug resistance genes are shown in Table 1.
Whole Genome Sequencing (WGS) and Assembly Based Analysis
Genomic DNA was extracted directly from six IRAB isolates of the major outbreak PFGE types and two ISAB isolates as control strains using the MiniBEST kit (Takara Bio Inc.). DNA libraries were prepared using QIAseq FX DNA Library Kits (QIAGEN, Hilden, Germany) and sequenced on an Illumina NextSeq 500 platform (Illumina, San Diego, CA, United States). Paired reads were assembled using SPAdes 3.13.0, and the resulting contigs were annotated in the Prokka website2. Assembled contigs were analyzed using the Pasteur Multilocus Sequence Typing (MLST) scheme3 and SnapGene 4.3.8.1. SWISS-MODEL4 was used for protein modeling.
Intervention Strategies
We applied a comprehensive intervention to reduce A. baumannii resistance rates and IRAB outbreaks in 2011. The intervention focused of five aspects: health-care workers, patients, antibiotic use, medical equipment, and environmental protection. The details of the measures are shown in Figure 1 and summarized as follows:
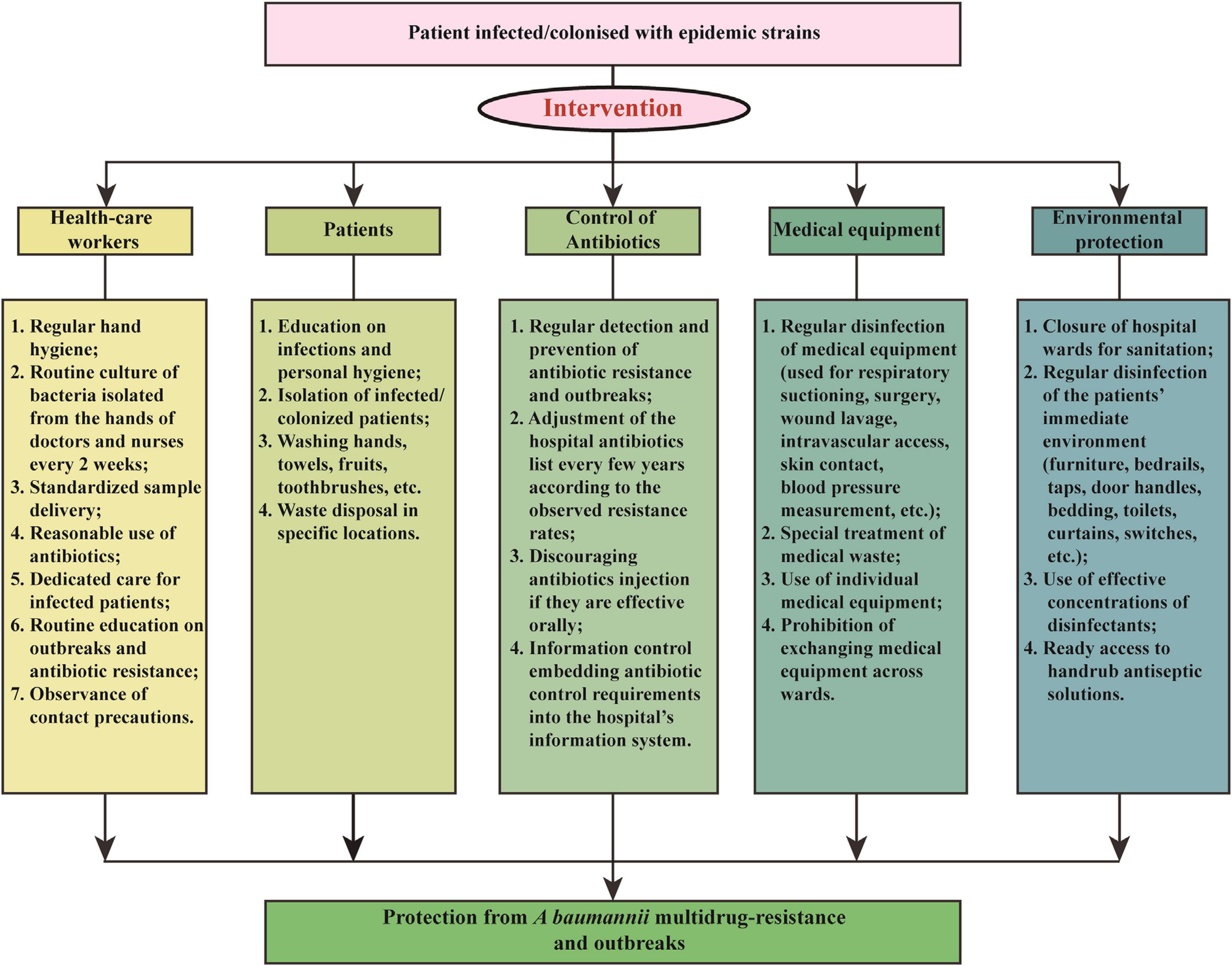
Figure 1. Multicomponent intervention to control A. baumannii resistance rates and imipenem-resistant A. baumannii outbreaks.
(1) Health-care workers: The hospital personnel received lectures on a regular basis about hand hygiene, standardized sample delivery, reasonable use of antibiotics, and dedicated care for infected patients, and routine culture of bacteria isolated from the hands of doctors and nurses every 2 weeks.
(2) Patients: Patients infected or colonized by outbreak bacteria were isolated. In addition, an enhanced personal hygiene, by washing hands, towels, fruits, toothbrushes, etc., and the disposal of waste at specific locations were deemed essential for the containment of A. baumannii transmission.
(3) Antibiotic use: Antimicrobials were used more effectively to prevent spread of harmful bacteria and outbreaks, through regular monitoring of antibiotic resistance and adjustment of the hospital antibiotics list every few years according to the observed resistance rates. For example, before the intervention the antibiotics in use were imipenem, meropenem, gentamicin, tobramycin, ampicillin/sulbactam, piperacillin/tazobactam, levofloxacin, ciprofloxacin, cefepime, ceftazidime, ceftriaxone, sulfamethoxazole, and furantoin; however, during the intervention of 2011, meropenem was removed from the hospital antibiotic list due to a rapidly increasing resistance rate, while tigecycline were added to the list. In addition, the injection of antibiotics was avoided if these were effective orally. Finally, an information control system embedding antibiotic control requirements into the hospital’s information system was also applied.
(4) Medical equipment: Special attention was paid to the regular disinfection of medical equipment, such as that used for respiratory suctioning, wound lavage, intravascular access, surgery, skin contact, blood pressure measurement, etc., with 75% ethyl alcohol on a daily basis. Additionally, the use of individual medical equipment and prohibition of exchanging medical equipment across wards also contributed to the control of A. baumannii outbreaks.
(5) Environmental protection: Hospital wards were carefully closed for sanitation and effective disinfection of the patients’ immediate environment, including furniture, bedrails, taps, door handles, bedding, toilets, curtains, switches, etc. Moreover, ready access to handrub antiseptic solutions increased the compliance to hand hygiene as well.
Statistical Analysis
Continuous variables are presented as means ± standard deviation (SD); categorical variables are shown as numbers and percentages. For continuous variables, comparisons between groups were analyzed by Mann–Whitney test or Student’s t-test. For categorical variables, comparisons between groups were analyzed by χ2 test. SPSS 19.0 (SPSS Inc., Chicago, IL, United States) was used for data analysis. Results displaying P < 0.05 were considered statistically significant. The flow chart of the current research methods are shown in Figure 2.
Results
Reduction in Antimicrobial Resistance Rates After Intervention
The findings of the antibiotic susceptibility testing for all 2572 A. baumannii isolates are shown in Table 2. From 2007 to 2010, a total of 811 A. baumannii strains were recovered. The rates of resistance to various antibiotics increased every year (Figure 3A and Table 2). Specifically, the resistance rates increased rapidly from 13.8 to 57.2% for imipenem, from 11.0 to 53.1% for meropenem, and from 21.4 to 51.8% for piperacillin/tazobactam. In 2011, A. baumannii had the highest resistance rate to imipenem, reaching 85.3%; this trend was also observed for other antibiotics. Interestingly, after reaching its peak in 2011, with the implementation of the intervention in June 2011 (Figure 1), the rates of A. baumannii resistance to various antibiotics decreased every year (Figure 3A). Due to the rapidly increasing resistance rate, meropenem was removed from the hospital antibiotics list in 2011, while tigecycline were added to the list after the intervention. From 2011 to 2014, a total of 1761 A. baumannii strains were isolated for which the rate of imipenem resistance dropped from 85.3 to 72.6%, that of levofloxacin resistance from 74.9 to 42.5%, that of ceftazidime resistance from 85.4 to 51.0%, that of ceftriaxone resistance from 100.0 to 80.8%, and that of tigecycline resistance from 45.0 to 6.9%. Thus, the resistance rates of A. baumannii to almost all the antibiotics in use were reduced after the intervention. The results of antibiotic susceptibility testing for IRAB are shown in Table 2. Unlike A. baumannii, the resistance rates of IRAB hardly changed from 2007 to 2014 (Figure 3B and Table 2). A total of 1673 IRAB isolates were resistant to most antibiotics.
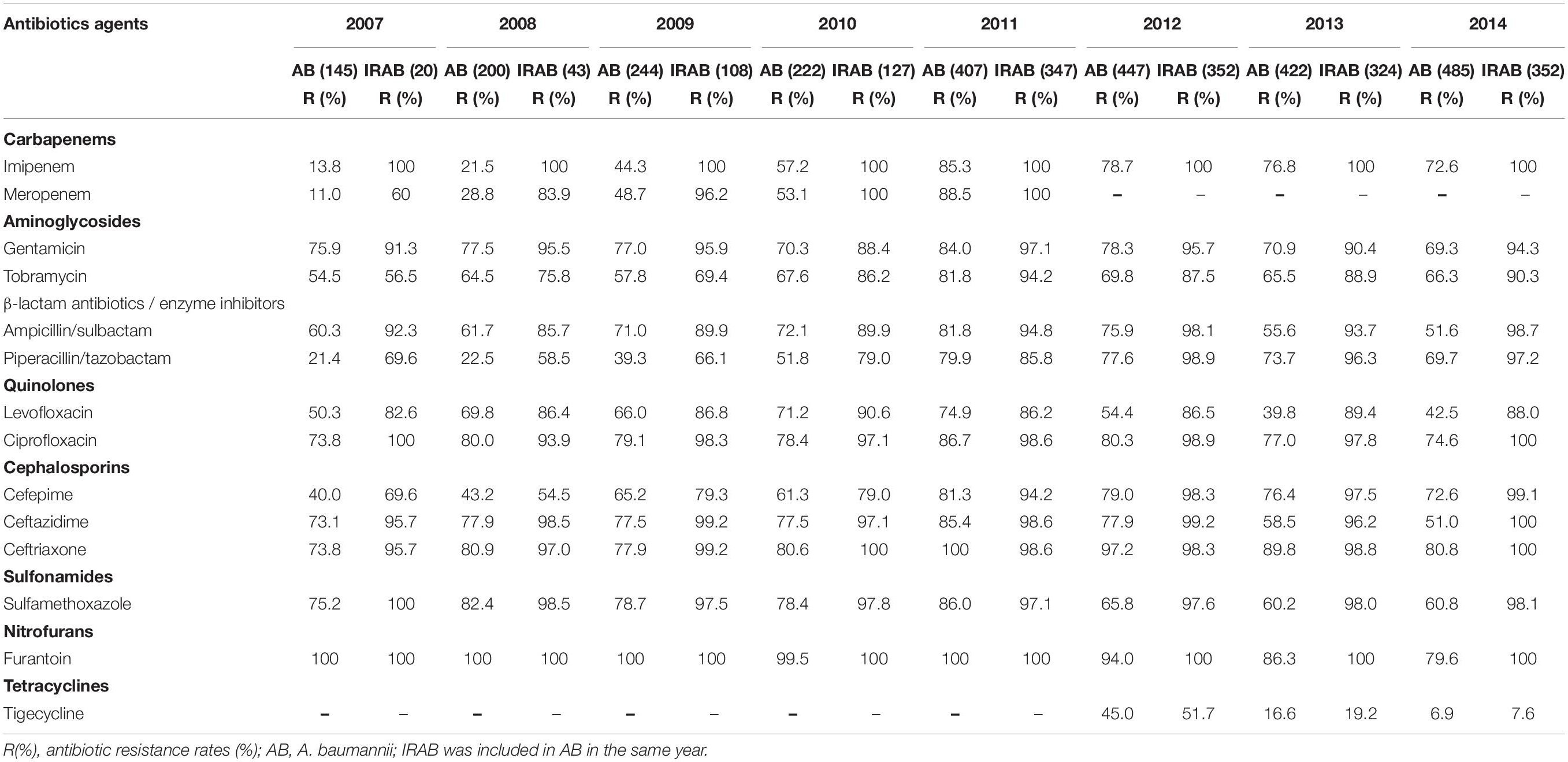
Table 2. Antibiotic resistance rates of 2572 A. baumannii stains including 1673 IRAB stains from 2007 to 2014.
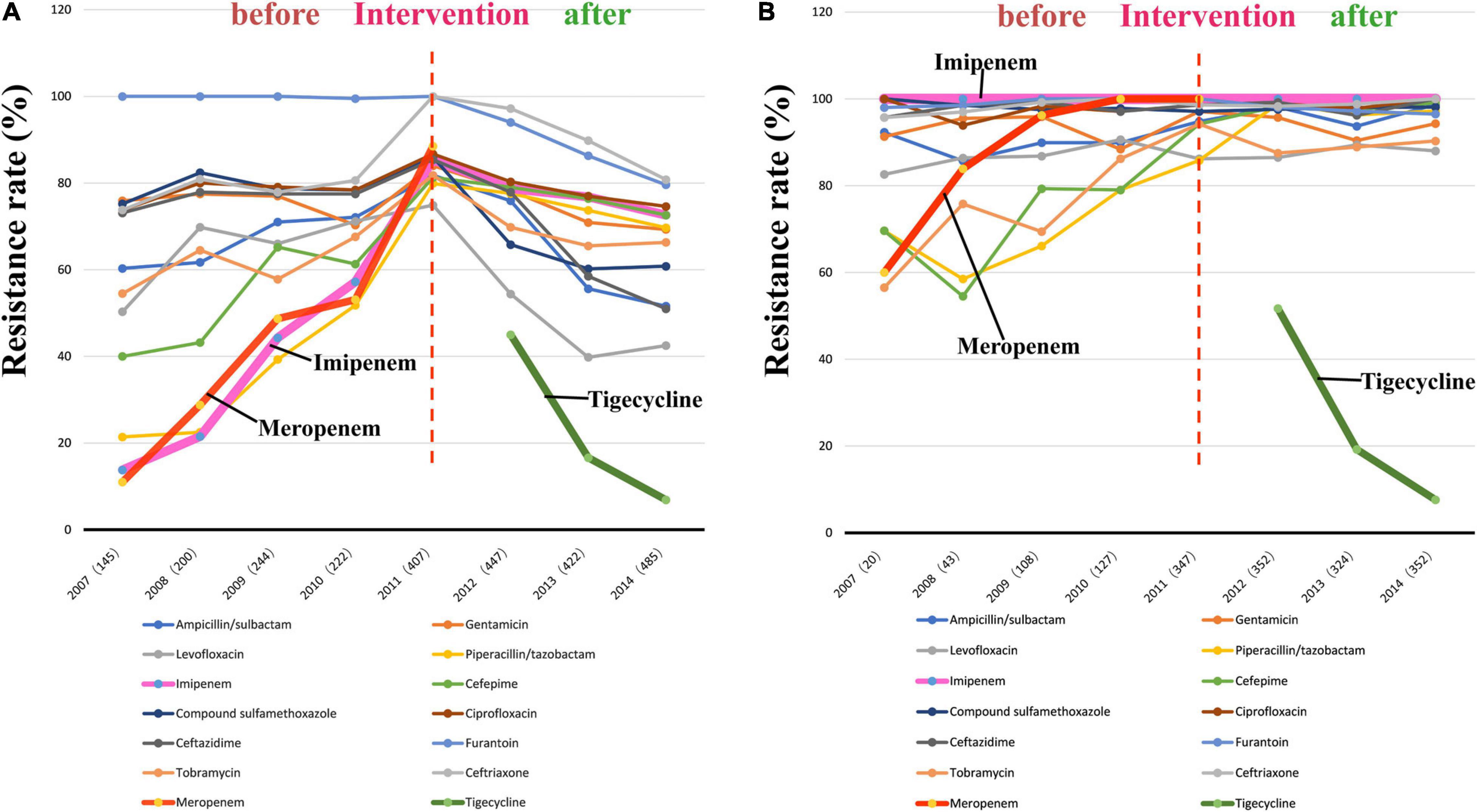
Figure 3. Changes in antibiotic resistance rates during 8 years spanningfrom before to after the intervention. (A) Antibiotic resistance rates of 2572 A. baumannii stains from 2007 to 2014; (B) Antibiotic resistance rates of 1673 IRAB stains from 2007 to 2014.
Pulsed-Field Gel Electrophoresis and Outbreak Analysis From 2007 to 2014
Analysis of 201 IRAB strains by PFGE showed that there were seven outbreaks from January 2007 to December 2010 before intervention (Figure 4). Three outbreaks of type 01 occurred between January and December 2008, with a total of 13 cases in the medical ICU (MICU). In 2009 there were three outbreaks: an outbreak of type 06 in the respiratory and neurology departments involving a total of seven cases between April and December; an outbreak of type 11 in the neurology ICU (NICU) and neurology departments with 10 cases from April to December; and an outbreak of type 15 in the surgical ICU (SICU) with three cases from July to September. Furthermore, an outbreak of type 18 involving three cases occurred in MICU from July to September 2010. Almost all outbreak strains (98%) were isolated from sputum specimens. However, after the intervention was applied in 2011 (Figure 1), only five outbreaks were observed from January 2011 to December 2014 (Figure 4). Of these five outbreaks, four cases of type 23 outbreak occurred in the NICU and SICU from July to December 2012, seven cases of type 23 outbreak occurred in the MICU and NICU from April to June 2013, four cases of type 26 outbreak occurred in the MICU from July to December 2012 and three cases from January to March 2014; finally, three cases of type 31 outbreak occurred from October to December 2013 in the MICU and NICU. In summary, after the intervention, the frequency of IRAB outbreaks, the duration of each outbreak, and the number of outbreak strains decreased notably; in addition, there was no outbreak in 2011, and the major PFGE types of outbreak strains before and after the intervention were also different.
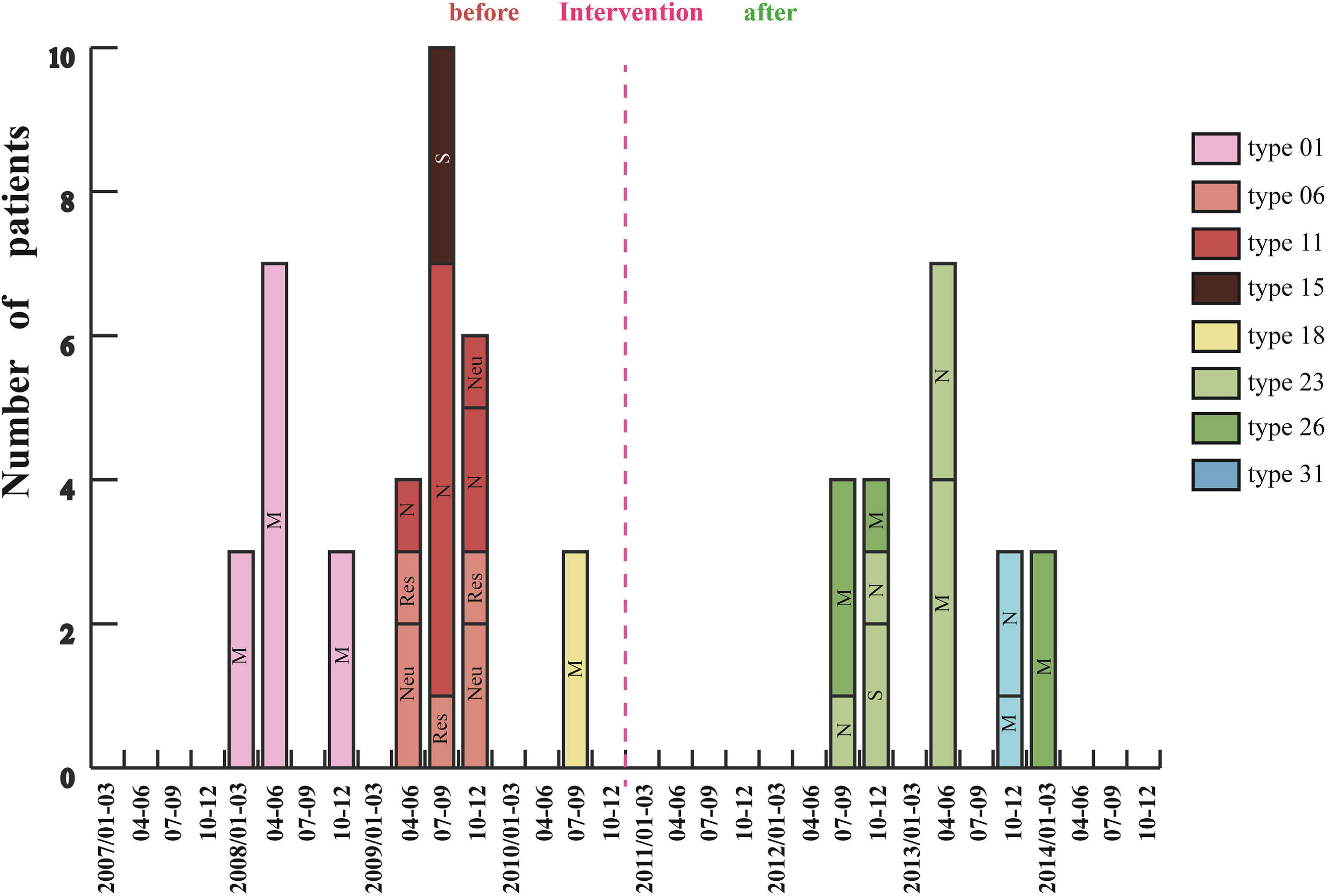
Figure 4. Changes in IRAB outbreaks across 8 years before and after the intervention. Each vertical bar represents an outbreak. M: MICU, Medical Intensive Care Unit; S: SICU, Surgical Intensive Care Unit; N: NICU, Neurology Intensive Care Unit; Neu, Neurology department; and Res, Respiratory department.
Distribution of Genes Encoding β-Lactamases, Porin Channels, and Efflux Pumps
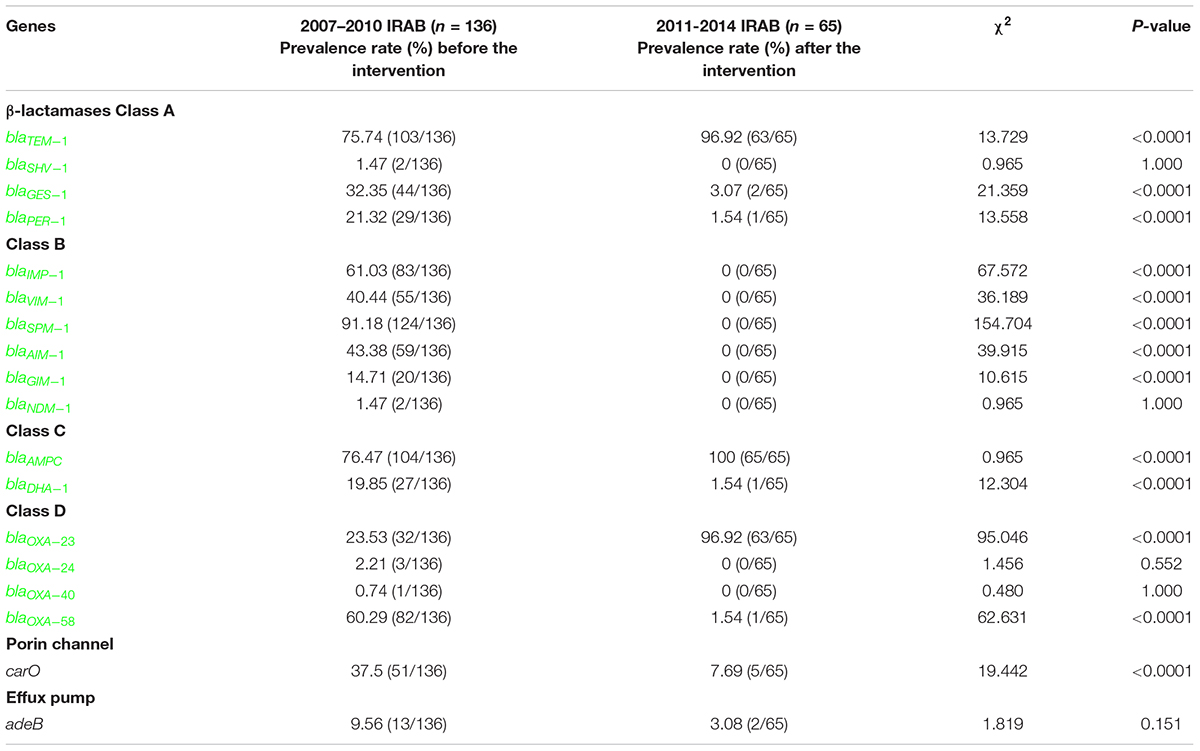
The distribution of genes encoding β-lactamases, porin channels, and efflux pumps in 201 randomly selected IRAB strains is shown in Table 3. Screening of resistance genes showed that before the intervention, from 2007 to 2010, the most prevalent β-lactamase-coding resistance gene was blaAMPC (76.47%) of class C, followed by blaTEM–1 (75.74%) of class A. Moreover, the IRAB isolates were found to harbor blaIMP–1 (61.03%), blaVIM–1 (40.44%), blaSPM–1 (91.18%), blaAIM–1 (43.38%), blaGIM–1 (14.71%), and blaNDM–1 (1.47%). However, the prevalence of the resistance genes from the 2011–2014 periods changed after the intervention. Among the IRAB strains, the prevalence of blaTEM–1 (96.92%), blaAMPC (100.00%), and blaOXA–23 (96.92%) had significantly increased (P < 0.001). Notably, the prevalence of the blaOXA–23 gene had significantly increased from 23.53% to 96.92% (P < 0.001), whereas that of blaGES–1 (3.07%), blaPER–1 (1.54%) of class A, blaDHA (1.54%) of class C, blaOXA–58 (1.54%) of class D, and carO (7.69%) had significantly decreased (P < 0.0001). In addition, the prevalence of class B genes, such as blaSPM–1, blaIMP–1, blaVIM–1, blaAIM–1, and blaGIM–1, decreased to negligible levels (P < 0.0001).
Whole Genome Sequencing (WGS) and Assembly Based Analysis
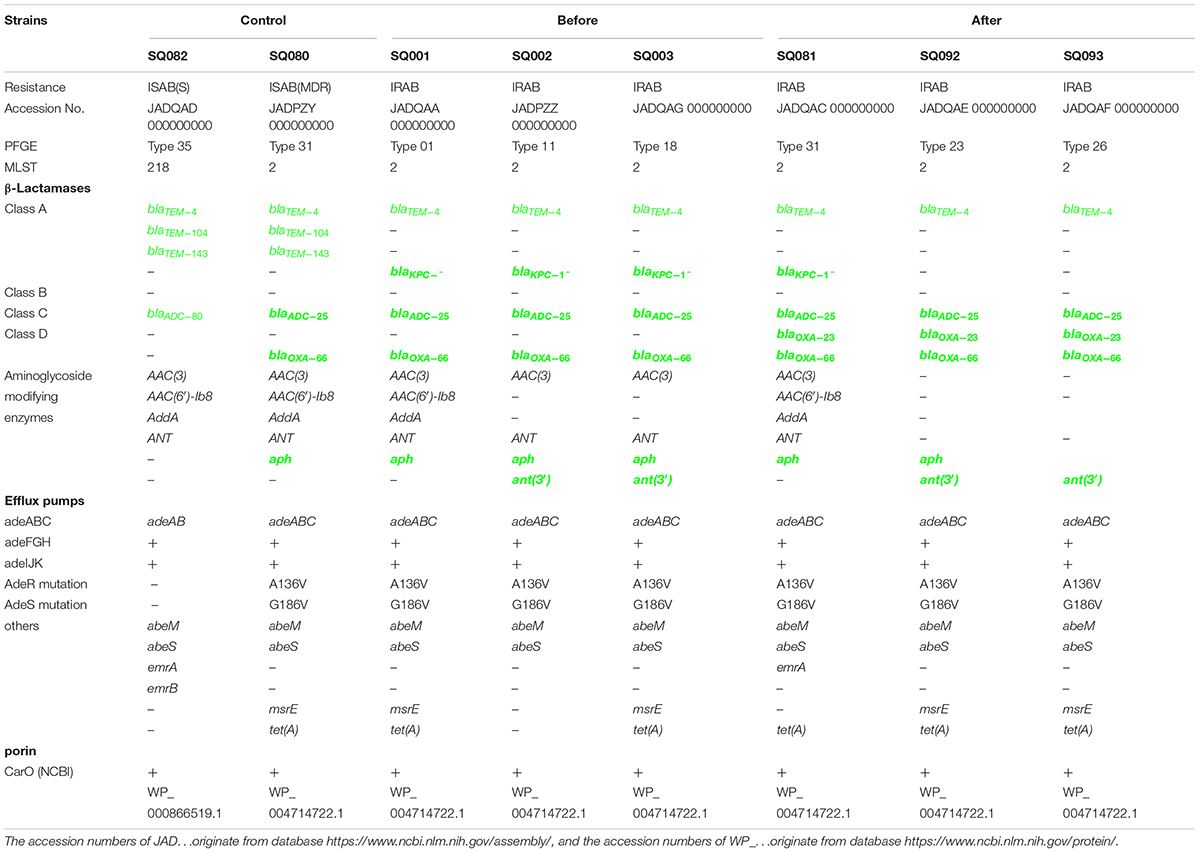
Whole genome sequencing was used to detect the major outbreak PFGE types (Figure 4) of IRAB strains before the intervention (SQ001-type 01, SQ002-type 11, and SQ003-type 18), after the intervention (SQ081-type 31, SQ092-type 23, and SQ093-type 26), and the control strains SQ082-ISAB (S) and SQ080-ISAB (MDR) to analyze the resistance mechanisms of IRAB. The control strain SQ082-ISAB (S) was sensitive to all antibiotics. SQ080-ISAB (MDR-AB) and SQ081-IRAB belonged to the same PFGE type 31; SQ080 was resistant to multi-antibiotics, but sensitive to imipenem, while SQ081 was resistant to both multi-antibiotics and imipenem.
The study of the virulence mechanism of outbreak strains by WGS revealed that these differed before and after the intervention (Table 4). For instance, the blaOXA–23 gene, coding for a class D β-lactamase, was present in IRAB (SQ081-type 31, SQ092-type 23, and SQ093-type 26) isolated after the intervention, but not in those IRAB isolates (SQ001-type 01, SQ002-type 11, and SQ003-type 18) that had been collected before the intervention. Second, the six IRAB isolates collected before (SQ001-type 01, SQ002-type 11, and SQ003-type 18) and after (SQ081-type 31, SQ092-type 23, and SQ093-type 26) the intervention showed some similarities in multiple drug resistance mechanisms, differing from ISAB (S) (SQ082): ① The six IRAB isolates were all ST2 by MLST analysis, while ISAB (S) (SQ082) was ST218; ② Compared to ISAB (S) (SQ082), these IRAB carried a wide variety of antibiotic enzymes: their β-lactamase genes included blaKPC–1 in class A, blaADC–25 in class C, and blaOXA–66 in class D; ③ Unlike ISAB(S) (SQ082), these IRAB displayed genes encoding aminoglycoside-modifying enzymes including ant(3′), ant(3′′), and aph; ④ Among efflux pumps, although the adeABC operon could be detected in IRAB, adeC could not be detected in ISAB(S) (SQ082). AdeR carried a A136V mutation, and AdeS a G186V mutation in IRAB, but AdeR-S showed no mutations in ISAB(S). IRAB also presented the genes msrE and tetA, encoding efflux pumps, while ISAB(S) (SQ082) did not; ⑤ With respect to the outer membrane porin permeability of antibiotics, the amino acid sequence of the CarO of IRAB (SQ001-type 01, SQ002-type 11, SQ003-type 18, SQ081-type 31, SQ092-type 23, and SQ093-type 26) matched that of the sequence under NCBI accession number WP_004714722.1, while that of ISAB(S) (SQ082) matched the sequence of the NCBI accession number WP_ 000866519.1. The SWISS-MODEL for CarO shows that the upper part of ATCC 19606 (Figure 5A) consists of a large channel formed by 14 β-sheets, while that of IRAB is formed by 8 β-sheets, leading to partial blockade of the channel (Figure 5B). The similarity between the CarO of IRAB and the CarO template 4fuv.1.A was 100% (Figure 5B), whereas the similarity between the CarO of ISAB (S) (SQ082) and the template 4fuv.1.A was only 75.93%; ⑥ Plasmid analysis revealed that the plasmid carried by IRAB (SQ081-type 31, SQ092-type 23, and SQ093-type 26) was similar to the plasmid pAC29b (NCBI registration number CP008851). The comparison of plasmids and contigs using the SnapGene software revealed that the IRAB plasmid covered the full length of blaOXA–23 on plasmid pAC29b, which contained the blaOXA–23 gene, indicating that blaOXA–23 was present on the plasmid of IRAB after the intervention (Figure 5C).
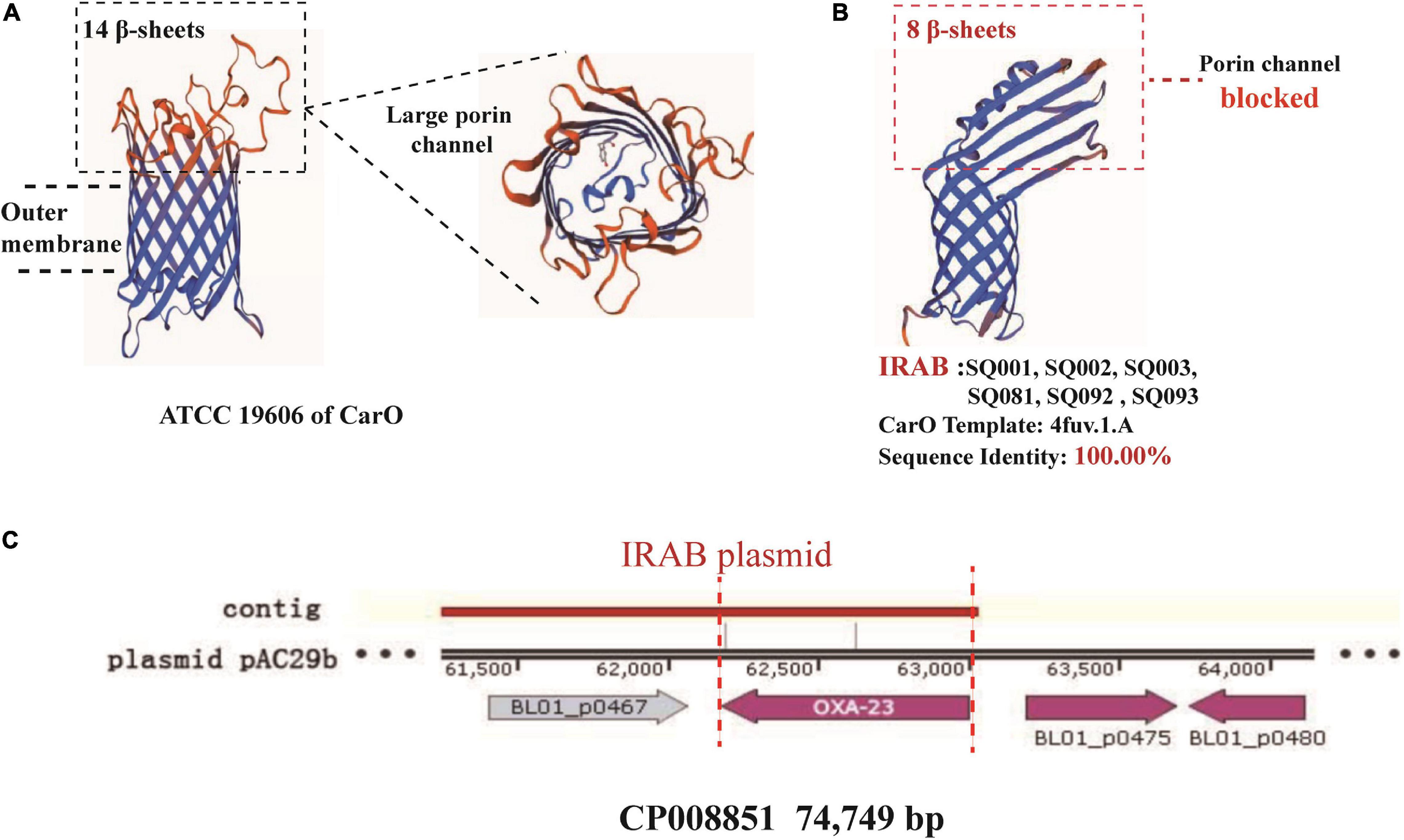
Figure 5. SWISS-model for porin permeability to imipenem and plasmid harboring blaOXA–23 in IRAB isolates. (A) 3D model for CarO of ATCC 19606 with 14 β-sheets, which form a large porin channel for imipenem to pass through; (B) 3D model for CarO of IRAB with 8 β-sheets, which block the porin channel to hinder the passage of imipenem; the sequence identity with the template 4fuv.1.A was 100%; (C) plasmid harboring blaOXA–23 in IRAB isolates.
Potential Resistance Mechanism of IRAB Before and After the Intervention
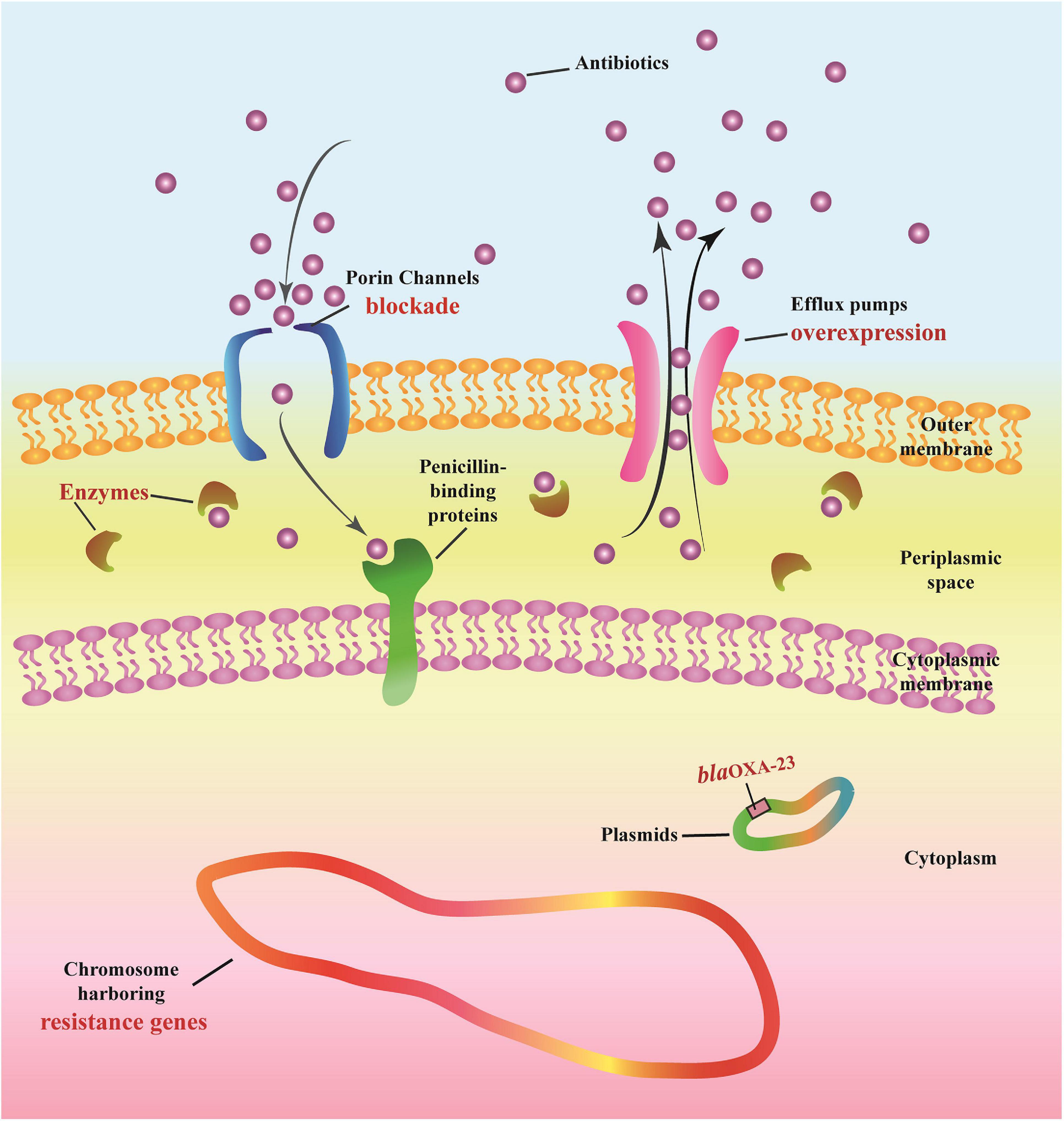
The following multi-drug resistance mechanisms were observed in IRAB isolates before and after the intervention. First, common mechanisms included (1) the partial blockade of the CarO porin channel, which reduced the antibiotic permeability of IRAB (Table 3 and Figure 5B); (2) the possible overexpression of the efflux pump AdeABC, owing to AdeR-S mutations (Tables 3, 4), which enhanced the antibiotic efflux capacity of IRAB; (3) the presence of enzymes residing in the periplasmic space, such as β-lactamases (encoded by blaTEM–1, blaTEM–4, blaGES–1, and blaPER–1 in class A; blaAMPC, blaDHA, and blaADC–25 in class C; and blaOXA–66 in class D) (Tables 3, 4) and aminoglycoside-modifying enzymes [encoded by ant(3′), ant(3′′), aph] (Table 4) which can degrade various antibiotics in IRAB. Second, WGS highlighted the occurrence of different mechanisms before and after intervention: in particular, among the enzymes residing in the periplasmic space, the types of β-lactamases changed, as blaOXA–23 in class D was only detected on the bacterial plasmid after the intervention (Table 4). Therefore, the plasmids harboring blaOXA–23 may be the major cause of IRAB outbreak after the intervention (Table 4 and Figures 4, 5C). The multi-drug resistance mechanisms of IRAB after the intervention are shown in Figure 6.
Imipenem-resistant Acinetobacter baumannii contains an outer membrane, periplasmic space, cytoplasmic membrane, cytoplasm, chromosome, and plasmids. Antibiotics must cross the outer membrane through porin channels (outer membrane proteins such as CarO) to reach the periplasmic space and then bind to the final targets of penicillin-binding proteins located at the level of the cytoplasmic membrane. Antibiotics can be degraded by enzymes (e.g., β-Lactamases of Class A, B, C, and D), and can also be actively expelled from the bacterial structure through efflux pumps (e.g., AdeABC and AdeR-S).
Impact of the Intervention
The intervention triggered many positive effects, that we describe as follows: ① the resistance rates of A. baumannii decreased after the intervention; ② the intervention resulted in a decrease in the number and duration of outbreaks, as well as in the number of departments affected by of outbreaks; ③ the frequency of most resistance genes decreased after the intervention; ④ the resistance mechanisms characterizing IRAB both before and after the intervention included porin channel blockade, efflux pump overexpression, and chromosomes harboring resistance genes and various enzymes; conversely, the frequent genes encoding β-lactamases passed from genes encoding class B β-lactamases to blaOXA–23 after the intervention.
Discussion
This Study Proposed a Systemic Intervention to Control A. baumannii Resistance and IRAB Outbreaks
Imipenem-resistant Acinetobacter baumannii is the most important group of bacteria associated with hospital-acquired infections and outbreaks worldwide (Xie et al., 2018). Several control measures have been applied against IRAB (Birgand et al., 2015, 2016), and some articles display various degrees of severity and prevalence of IRAB over many years (Du et al., 2019; Park et al., 2019). However, few studies similar to the present study have been published. In the present study, we proposed a systemic control intervention and also investigated the changes in severity and prevalence of resistance mechanisms of IRAB for a long time (8 years). The aim of this study was to characterize the impact of an intervention to control resistance and outbreaks of IRAB and to explore its resistance mechanisms through an 8-year survey. We hope that our results can help hospitals in other countries to prevent and control A. baumannii resistance and IRAB outbreaks.
The Intervention Reduced the Antibiotic Resistance Rate of A. baumannii and the Occurrence Rate of IRAB
Before the intervention, the drug resistance rate of A. baumannii increased every year, and the resistance rate for imipenem increased from 13.8% to 85.3%. During the intervention in 2011, meropenem was removed from the hospital antibiotics list due to a rapid growth of resistance rate, while tigecycline were added. After the intervention, the resistance rates of A. baumannii to various antibiotics decreased significantly every year. However, IRAB isolates were almost completely resistant to other antibiotics, and the resistance rates of IRAB to other antibiotics did not change significantly during the 8 years of the survey, which suggests that the intervention could effectively prevent and control the resistance rates of A. baumannii to various antibiotics and to imipenem, but had limited effect on A. baumannii that is already resistant to imipenem (IRAB). This could be due to the complex resistance mechanisms characterizing IRAB reported by Lee et al. (2017); indeed, once A. baumannii becomes IRAB, it might be difficult to reduce the drug resistance rates due to the emergence of complex resistance mechanisms to many antibiotics.
The Intervention Reduced the Frequency, Duration, and Isolate Number of IRAB Outbreaks
Pulsed-field gel electrophoresis analysis showed that the frequency of IRAB outbreaks before the intervention was 2–3 times per year, which decreased to almost once a year after the intervention. The duration of the IRAB outbreak was shortened from 8 to 3 months. The outbreaks occurred in the MICU, SICU, NICU, and respiratory and neurology departments before the intervention, but only in the MICU, SICU, and NICU after the intervention. Daniel et al. (2019) reported that MICU, SICU, and NICU were the key departments of IRAB outbreaks, consistent with our findings. The number of IRAB strains per outbreak was reduced from 11 to 3 isolates. Notably, there was no outbreak in 2011, the first year after implementing the intervention, which could be attributed to the replacements in the hospital antibiotic list and other control measures in the intervention. However, new IRAB strains that were resistant to new antibiotics and selection pressures caused new outbreaks after 2011. Therefore, regular replacement of antibiotics to prevent the emergence of new IRAB strains is also a key feature of this intervention.
PCR-Based Mechanistic Studies Showed That Changing Antibiotics in the Intervention May Lead to Changes in IRAB Resistance Mechanisms
Imipenem-resistant Acinetobacter baumannii resistance was the result of a combination of multiple resistance mechanisms. PCR analysis showed that genes encoding β-lactamases, such as blaAMPC and blaTEM–1were the major resistance genes of IRAB before and after the intervention. Before the intervention, blaSPM–1 and blaIMP–1 in class B were detected at a frequency of 91.18 and 61.03%, respectively, while blaOXA–23 in class D accounted for only 23.53%. However, after the intervention, the frequency of genes in class B decreased to 0.00%, but that of blaOXA–23 of class D increased to 96.92%, suggesting that there was clonal spread (or horizontal spread) of blaOXA–23 after 2011. Harboring the blaOXA–23 gene may have been one of the main resistance mechanisms of IRAB from 2011 to 2014. In the intervention, meropenem was replaced with tigecycline. Kulengowski et al. (2019) reported that blaVIM–1 of class B was related to meropenem resistance, while Savari et al. (2017) reported that blaOXA–23 and overexpression of the AdeABC efflux pump were associated with tigecycline resistance. Therefore, the significant differences in frequency between class B genes and blaOXA–23 before and after the intervention may be due to the replacement of antibiotics, which changed the selection pressure of A. baumannii, leading to changes in the drug resistance mechanism. The gene adeB encoding an efflux pump and the gene carO encoding a porin channel were also analyzed before and after the intervention. Therefore, multiple drug resistance mechanisms mediated widespread IRAB resistance.
WGS-Based Mechanistic Analysis Confirmed That IRAB Has a Combination of Resistance Mechanisms, and That Plasmids Harboring blaOXA–23 May Lead to New Outbreaks After the Intervention
Whole genome sequencing showed that IRAB had multiple drug resistance mechanisms compared to ISAB (S) (SQ080): (1) There are many types of enzymes for modification of antibiotics, such as β-lactamases and aminoglycoside-modifying enzymes. First, the production of β-lactamases is a major antibiotic resistance mechanism in A. baumannii; these enzymes inactivate β-lactam, and comprise four molecular classes: A, B, C, and D (Sawa et al., 2020). In this study, unlike ISAB (S)(SQ082), IRAB contained blaKPC –1 of class A, blaADC–25 of class C, and blaOXA–23, and blaOXA–66 of class D. Class A β-lactamases degrade early generation cephalosporins (Sawa et al., 2020); class C β-lactamases can confer resistance to cephamycins (cefoxitin and cefotetan), penicillins, and cephalosporins (Uddin et al., 2018); class D β-lactamases, also called OXAs (oxacillinases), hydrolyze oxacillin, and the presence of carbapenem-hydrolyzing class D β-lactamases is a major carbapenem resistance mechanism in A. baumannii (Stewart et al., 2019). Unlike the serine-dependent β-lactamases (classes A, C, and D), class B β-lactamases are metallo-β-lactamases that require zinc or another heavy metal for catalysis (Amin et al., 2019). However, genes of class B were not detected in IRAB or ISAB, indicating that class B β-lactamases did not cause IRAB resistance. Second, aminoglycoside-modifying enzymes are classified into acetyltransferases, adenyltransferases, and phosphotransferases (Sheikhalizadeh et al., 2017). In the present study, IRAB had ant(3′), ant(3′′), and aph genes, which conferred resistance to aminoglycosides. (2) Efflux pumps are associated with resistance against many different classes of antibiotics, such as imipenem and tigecycline, in A. baumannii (Knight et al., 2018). AdeABC is associated with aminoglycoside resistance and with decreasing susceptibility to tigecycline and fluoroquinolone antibiotics (Yoon et al., 2013). The adeABC gene was detected in IRAB, but adeC was not detected in ISAB, suggesting that efflux pumps of IRAB are more effective than those of ISAB. Notably, mutations in the AdeR-S two-component system lead to the overexpression of AdeABC (Yang et al., 2019). In IRAB, AdeR had a G186V mutation, and AdeS had an A136V mutation, but AdeR-S of ISAB had no mutation, suggesting overexpression of efflux pumps that are more effective than those of ISAB for antibiotic efflux. (3) To investigate the porin channel mechanism, the 3D structure of CarO was modeled using the SWISS-MODEL software. Compared to the structure of CarO from the standard strain ATCC 19606, it was found that the upper part of CarO from ATCC 19606 was a large channel formed by 14 β-sheets, while the upper part of CarO from IRAB in this study was only a small channel formed by 8 β-sheets; such change in the tertiary structure of the IRAB CarO suggested that the porin channel was partly blocked. In addition, the structure of CarO from IRAB elaborated in this study was 100% identical to that of the template 4fuv.1.A in SWISS-MODEL, representing a CarO channel with low permeability to imipenem. Zhu et al. (2019) reported that a partial blockade of the CarO porin channel can reduce the penetration of antibiotics into bacteria, leading to drug resistance, consistent with the results of this study. (4) Plasmid mechanistic analysis revealed that blaOXA–23 was present on the plasmid of IRAB after the intervention, and WGS showed that blaOXA–23 transmitted drug resistance through the plasmid and became the major reason for IRAB prevalence and outbreak after the intervention. Xiong et al. (2015) reported that changes in drug resistance mechanisms of bacteria may be due to different environmental selection pressure; therefore, with our intervention we might have induced a modification of the prevalent drug resistance mechanisms through a variety of control measures and adjustment of antibiotics use. For example, during the intervention meropenem, showing a fast-growing resistance rate, was discontinued while tigecycline was adopted. Such changes in environmental selection pressure may have reduced the prevalence of class B β-lactamases; consequently, blaOXA–23 began to spread and became the main epidemic factor after the intervention. This phenomenon could also explain why the outbreak did not occur throughout 2011 after applying the intervention, but started in July 2012: indeed, A. baumannii may have adapted, and major strains harboring the plasmid bearing blaOXA–23 may have been selected under the new environmental pressure throughout 2011. Therefore, the resistance to new antibiotics must be regularly monitored, and these should be replaced to prevent the emergence of new resistant strains in each new round of intervention. The present study demonstrated that strengthening the management of health-care workers, patients, medical equipment, and environmental protection is an effective measure against IRAB outbreaks. In addition, modifying the antibiotics list of the hospital according to the results of drug sensitivity is also an effective measure against IRAB resistance, for example by removing ampicillin, furantoin, and ceftriaxone to exhibit high resistance rates, and adding ceftazidime/avibactam (CZA). In fact, Norelle et al. (2018) reported that CZA exerted good inhibitory effects on bacteria carrying blaOXA–23.
Conclusion
In this study we proposed and applied a systemic intervention to control the antibiotic resistance of A. baumannii and the outbreak of IRAB, which included five aspects: health-care workers, patients, antibiotic use, medical equipment, and environmental protection. An 8-year survey has proven the effectiveness of the intervention, which led to a significant reduction in resistance rates; a decrease in the number, duration, and number of departments affected by outbreaks; and a reduced frequency of most resistance genes. The resistance mechanisms of IRAB include porin channel blockade, efflux pump overexpression, various enzymes, chromosomes harboring resistance genes, and blaOXA–23 on plasmids, which was the major reason for outbreaks after the intervention. As part of our intervention strategy, new rounds of interventions should be taken every few years to prevent the emergence of new resistant strains. We hope that this intervention strategy can help other countries and regions control the resistance of A. baumannii and outbreaks of IRAB.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
This study was approved by the Clinical Research and Ethics Committee of The First Affiliated Hospital of Sun Yat-sen University [2019(483)].
Author Contributions
LC, PT, JZ, XY, YC, KL, PG, YC, ZW, PQ, and RC conducted the experiments. LC wrote the manuscript. CC and BH edited the manuscript. All the authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the National Natural Science Foundation of China (grants 81772249, 81672081, and 81871703).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Liang Chen from the Hackensack Meridian School of Medicine for his valuable guidance on sequencing data analysis.
Footnotes
- ^ http://www.cdc.gov/pulsenet/
- ^ http://www.vicbioinformatics.com/software.prokka.shtml
- ^ www.genomicepidemiology.org
- ^ https://swissmodel.expasy.org
References
Amin, M., Navidifar, T., Saleh, S. F., and Goodarzi, H. (2019). Association of the genes encoding metallo-beta-lactamase with the presence of integrons among multidrug-resistant clinical isolates of Acinetobacter baumannii. Infect. Drng Resist. 12, 1171–1180. doi: 10.2147/idr.s196575
Birgand, G., Johansson, A., Szilagyi, E., and Lucet, J. C. (2015). Overcoming the obstacles of implementing infection prevention and control guidelines. Clin. Microbiol. Infect. 21, 1067–1071. doi: 10.1016/j.cmi.2015.09.005
Birgand, G., Moore, L., Bourigault, C., Vella, V., Lepelletier, D., Holmes, A. H., et al. (2016). Measures to eradicate multidrug-resistant organism outbreaks: how much do they cost? Clin. Microbiol. Infect. 22, 161–162.
Daniel, M., Benoît, V., Marlène, S., Pascal, C., Michelle, T., Xavier, B., et al. (2019). Fourier-transform infrared spectroscopy can quickly type gram-negative bacilli responsible for hospital outbreaks. Front. Microbiol. 26:1440. doi: 10.3389/fmicb.2019.01440
Du, X., Xu, X., Yao, J., Deng, K., Chen, S., Shen, Z., et al. (2019). Predictors of mortality in patients infected with carbapenem-resistant Acinetobacter baumannii: a systematic review and meta-analysis. Am. J. Infect. Control 47, 1140–1145. doi: 10.1016/j.ajic.2019.03.003
Jain, M., Sharma, A., Sen, M. K., Rani, V., Gaind, R., and Suri, J. C. (2019). Phenotypic and molecular characterization of Acinetobacter baumannii isolates causing lower respiratory infections among ICU patients. Microb. Pathog. 128, 75–81. doi: 10.1016/j.micpath.2018.12.023
Knight, D. B., Rudin, S. D., Bonomo, R. A., and Rather, P. N. (2018). Acinetobacter nosocomialis: defining the role of efflux pumps in resistance to antimicrobial therapy, surface motility, and biofilm formation. Front. Microbiol. 9:1902. doi: 10.3389/fmicb.2018.01902
Kulengowski, B., Clark, J. A., and Burgess, D. S. (2019). Killing activity of meropenem in combination with amikacin against VIM- or KPC-producing Enterobacteriaceae that are susceptible, intermediate, or resistant to amikacin. Diagn. Microbiol. Infect. Dis. 93, 372–375. doi: 10.1016/j.diagmicrobio.2018.10.020
Lee, C. R., Lee, J. H., Park, M., Park, K. S., Bae, I. K., Kim, Y. B., et al. (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 7:55. doi: 10.3389/fcimb.2017.00055
Norelle, L. S., Sarah, L. B., and Benjamin, P. H. (2018). Ceftazidime/avibactam susceptibility by three different susceptibility testing methods in carbapenemase-producing Gram-negative bacteria from Australia. J. Antimicrob. Agents 52, 82–85. doi: 10.1016/j.ijantimicag.2018.02.017
Park, J. J., Seo, Y. B., Choi, Y. K., Kym, D., and Lee, J. (2019). Changes in the prevalence of causative pathogens isolated from severe burn patients from 2012 to 2017. Burns 46, 695–701. doi: 10.1016/j.burns.2019.09.008
Savari, M., Ekrami, A., Shoja, S., and Bahador, A. (2017). Plasmid borne Carbapenem-Hydrolyzing Class D beta-Lactamases (CHDLs) and AdeABC efflux pump conferring carbapenem-tigecycline resistance among Acinetobacter baumannii isolates harboring TnAbaRs. Microb. Pathog. 104, 310–317. doi: 10.1016/j.micpath.2017.01.045
Sawa, T., Kooguchi, K., and Moriyama, K. (2020). Molecular diversity of extended-spectrum beta-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 8:13.
Sheikhalizadeh, V., Hasani, A., Ahangarzadeh, R. M., Rahmati-Yamchi, M., Hasani, A., Ghotaslou, R., et al. (2017). Comprehensive study to investigate the role of various aminoglycoside resistance mechanisms in clinical isolates of Acinetobacter baumannii. J. Infect. Chemother. 23, 74–79. doi: 10.1016/j.jiac.2016.09.012
Singkham-In, U., and Chatsuwan, T. (2018). In vitro activities of carbapenems in combination with amikacin, colistin, or fosfomycin against carbapenem-resistant Acinetobacter baumannii clinical isolates. Diagn. Microbiol. Infect. Dis. 91, 169–174. doi: 10.1016/j.diagmicrobio.2018.01.008
Stewart, N. K., Smith, C. A., Antunes, N. T., Toth, M., and Vakulenko, S. B. (2019). Role of the hydrophobic bridge in the carbapenemase activity of class D beta-lactamases. Antimicrob. Agents Chemother. 63:e02191-18.
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995
Uddin, F., McHugh, T. D., Roulston, K., Platt, G., Khan, T. A., and Sohail, M. (2018). Detection of carbapenemases, AmpC and ESBL genes in Acinetobacter isolates from ICUs by DNA microarray. J. Microbiol. Methods 155, 19–23. doi: 10.1016/j.mimet.2018.11.004
Vauchel, T., Pirracchio, R., Chaussard, M., Lafaurie, M., Rouveau, M., Rousseau, C., et al. (2019). Impact of an Acinetobacter baumannii outbreak on kidney events in a burn unit: a targeted machine learning analysis. Am. J. Infect. Control 47, 435–438. doi: 10.1016/j.ajic.2018.09.010
Venditti, C., Vulcano, A., D’Arezzo, S., Gruber, C., Selleri, M., Antonini, M., et al. (2019). Epidemiological investigation of an Acinetobacter baumannii outbreak using core genome multilocus sequence typing. J. Glob. Antimicrob. Resist. 17, 245–249. doi: 10.1016/j.jgar.2018.11.027
Wang, Y. C., Ku, W. W., Yang, Y. S., Kao, C. C., Kang, F. Y., Kuo, S. C., et al. (2020). Is polymicrobial bacteremia an independent risk factor for mortality in Acinetobacter baumannii bacteremia? J. Clin. Med. 9:153. doi: 10.3390/jcm9010153
Xie, R. Q., Zhang, X. H. D., Zhao, Q., Peng, B., and Zheng, J. (2018). Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerg. Microbes Infect. 7:31.
Xiong, W. G., Sun, Y. X., Ding, X. Y., Wang, M. Z., and Zeng, Z. L. (2015). Selective pressure of antibiotics on ARGs and bacterial communities in manure-polluted freshwater-sediment microcosms. Front. Microbiol. 11:194. doi: 10.3389/fmicb.2015.00194
Yang, Y. S., Chen, H. Y., Hsu, W. J., Chou, Y. C., Perng, C. L., Shang, H. S., et al. (2019). Overexpression of AdeABC efflux pump associated with tigecycline resistance in clinical Acinetobacter nosocomialis isolates. Clin. Microbiol. Infect. 25, 511–512.
Yoon, E. J., Courvalin, P., and Grillot-Courvalin, C. (2013). RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeR-S mutations. Antimicrob. Agents Chemother. 57, 2989–2995. doi: 10.1128/aac.02556-12
Yu, X., Wang, R., Peng, W., Huang, H., Liu, G., Yang, Q., et al. (2019). Incidence, distribution and clinical relevance of microbial contamination of preservation solution in deceased kidney transplant recipients: a retrospective cohort study from China. Clin. Microbiol. Infect. 25, 595–600. doi: 10.1016/j.cmi.2018.12.040
Keywords: intervention, Acinetobacter baumannii, antibiotic resistance, outbreak, mechanism
Citation: Chen L, Tan P, Zeng J, Yu X, Cai Y, Liao K, Guo P, Chen Y, Wu Z, Qu P, Cai R, Chen C and Huang B (2021) Impact of an Intervention to Control Imipenem-Resistant Acinetobacter baumannii and Its Resistance Mechanisms: An 8-Year Survey. Front. Microbiol. 11:610109. doi: 10.3389/fmicb.2020.610109
Received: 25 September 2020; Accepted: 24 December 2020;
Published: 16 February 2021.
Edited by:
Santiago Castillo Ramírez, National Autonomous University of Mexico, MexicoReviewed by:
Jesus Silva-Sanchez, National Institute of Public Health, MexicoRayane Rafei, Lebanese University, Lebanon
Copyright © 2021 Chen, Tan, Zeng, Yu, Cai, Liao, Guo, Chen, Wu, Qu, Cai, Chen and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cha Chen, chencha906@163.com; Bin Huang, huangb3@mail.sysu.edu.cn
†These authors have contributed equally to this work
 Lida Chen1,2†
Lida Chen1,2† Jianming Zeng
Jianming Zeng Penghao Guo
Penghao Guo Cha Chen
Cha Chen Bin Huang
Bin Huang