- 1Key Laboratory of Biotechnology and Bioengineering of State Ethnic Affairs Commission, Biomedical Research Center, Northwest Minzu University, Lanzhou, China
- 2College of Veterinary Medicine, Gansu Agricultural University, Lanzhou, China
- 3Menzies Health Institute Queensland, School of Pharmacy and Medical Science, Griffith University, Southport, QLD, Australia
Pseudorabies virus is a typical swine alphaherpesvirus, which can cause obvious neurological disorders and reproductive failure in pigs. It is capable of evading host antiviral immune response. However, the mechanism by which many PRV proteins assist the virus to evade innate immunity is not fully understood. This study identified PRV US3 protein as a crucial antagonistic viral factor that represses interferon beta (IFN-β) expression. A in-depth study showed that US3 protein restricted type I IFN production by targeting interferon regulatory factor 3 (IRF3), a key molecule required for type I IFN induction. Additionally, US3 protein interacted with IRF3, degraded its protein expression to block the phosphorylation of IRF3. These findings suggested a novel strategy utilized by PRV to inhibit IFN-β production and escape the host innate immunity.
Introduction
During a virus infection, host cellular recognition receptors (PRRs) recognize pathogen-associated molecular patterns (PAMPs) and trigger the induction of type I interferons (IFNs) and pro-inflammatory cytokines to restrict viral replication, clear up of infected cells, and further orchestrate the adaptive immune response to eradicate infected pathogens (Kawai and Akira, 2006; Carpenter et al., 2014; Beachboard and Horner, 2016; Chen et al., 2017). Among the PRRs, Cyclic GMP-AMP (cGAMP) synthase (cGAS) is a cytosolic DNA sensor and, when triggered, mounts a type I IFN response. Upon sensing pathogen DNA, cGAS catalyzes the synthesis of cGAMP, which activates the endoplasmic reticulum (ER)-anchored stimulator of interferon genes (STING). Stimulator of interferon genes then translocate from the ER to the Golgi apparatus to the recruit and phosphorylate TANK-binding kinase 1 (TBK1) and IκB kinase (IKK). These events then activate IRF3 and NF-κB to activate type I IFN production (Fitzgerald et al., 2003; Sharma et al., 2003; Sun et al., 2013; Xia et al., 2016).
To counteract the antiviral effects of cGAS-STING engagement, several DNA viruses, including Kaposi’s sarcoma-associated herpesvirus (Li et al., 2000; Ma et al., 2015; Wu et al., 2015), Herpes simplex virus 1 (Christensen et al., 2016; Su et al., 2016; Zhang et al., 2016; Zheng, 2018; Lin and Zheng, 2019; Zhu and Zheng, 2020), and Marek’s disease virus (Gao et al., 2019; Li et al., 2019), have evolved different evasion strategies. Pseudorabies virus (PRV), a member of the subfamily Alphaherpesvirinae of the family Herpesviridae (Mettenleiter, 2000), causes fatal fever and encephalomyelitis in pigs and susceptible animals (Sun et al., 2016). Although pigs are the natural host of PRV, other mammals, such as ruminants, carnivores, and rodents, are susceptible to PRV infection (Fonseca et al., 2010). Indeed, PRV infection is also known to cause human endophthalmitis in China (Ai et al., 2018; Fan et al., 2020; Liu et al., 2020; Wang et al., 2020). Importantly, PRV infection and the disease it causes have brought huge impact on economic for the swine industry.
Pseudorabies virus (PRV) is known to block type I IFN host antiviral responses. When PRV infects primary rat fibroblast cells, interferon-stimulated genes (ISGs) in these cells are suppressed (Brukman and Enquist, 2006b). The PRV glycoprotein gE/gI complex (Lamote et al., 2017), UL50 (Zhang et al., 2017), and EP0 (Brukman and Enquist, 2006a) can suppresses type I IFN host responses. Importantly, PRV UL13 inhibits cGAS-STING-mediated IFN-β production by phosphorylating IRF3 (Bo et al., 2020; Lv et al., 2020).
PRV protein kinase US3 has been shown to protect infected cells from apoptosis (Geenen et al., 2005; Qin et al., 2019). We reasoned that this could occur via exerting an uncharacterized antiviral evasion strategy and hypothesized that this could be occurring via the cGAS-STING pathway. US3 is a viral serine/threonine kinase, which is conserved in the alphaherpesvirus subfamily. Many studies indicated that Herpes simplex virus 1 (HSV-1) encoding US3 protein is involved in many processes during viral infection (Wagenaar et al., 1995; Leopardi et al., 1997; Reynolds et al., 2002; Cartier et al., 2003) and many other functions. Regarding how HSV-1 counteracts the host’s natural immune response, many studies showed that US3 could inhibit IFN-β (Wang et al., 2013; You et al., 2020) and Nuclear factor κB (NF-κB) (Wang et al., 2014) activation. Although HSV-1 US3 protein has multiple mechanisms for immune evasion, the immune evasion functions of PRV US3 are still poorly understood.
In this study, we found that PRV dampened IFN-β responses and that US3 protein impaired IFN-β production via degradation of IRF3. US3 also interacted with IRF3 and blocked its activation. Additionally, US3 knockdown partially recovered PRV infection-induced IRF3 degradation and IFN-β expression, suggesting PRV US3 could subvert antiviral innate immunity and evade host antiviral responses via a different mechanism compared to HSV-1 US3.
Materials and Methods
Cells and Viruses
The porcine kidney (PK15) cells were obtained from ATCC and cultured in DMEM supplemented with 10% new bovine serum (NBS) at 37°C in a 5% CO2 incubator. Pseudorabies Virus Bartha is an attenuated vaccine strain, obtained by extensive passaging of an Aujeszky strain isolated in Hungary (Christensen et al., 1992). Bartha-61 was propagated in BHK-21 cells, and the supernatants of infected cells were clarified and stored at −80°C.
Antibodies and Reagents
Anti-FLAG tag rabbit polyclonal antibody (D110005), Anti-cGAS rabbit polyclonal antibody (D163570), HRP (horseradish peroxidase)-conjugated Goat Anti-Rabbit IgG (D110058) and HRP-conjugated Goat Anti-Mouse IgG (D110087) were purchased from Sangon Biotech (Shanghai, China). HA tag Polyclonal antibody (51064-2-AP) and IRF3 Polyclonal antibody (11312-1-AP) were purchased from Proteintech (Wuhan, China); STING (D2P2F) Rabbit mAb (13647S), Phospho-IRF-3 (Ser386) (E7J8G) XP® Rabbit mAb antibody (37829S) and Myc-Tag (9B11) Mouse mAb (2276S) were bought from Cell Signaling Technology. Anti-HIST3H3 Polyclonal Antibody (K106623P) were purchased from Solarbio (Beijing, China). GAPDH Mouse Monoclonal Antibody (AF5009) and β-actin Mouse Monoclonal Antibody (AA128) were purchased from Beyotime Biotechnology (Shanghai, China). Anti-HSP90β antibody was purchased from Abbkine (ABP54794) (China).
TransStart® Top Green qPCR SuperMix (+Dye II) was purchased from Transgen (Beijing, China). Cell membrane/cytoplasm/nuclear membrane protein step extraction kit (BB-31042) was bought from BestBio (Shanghai, China). Lipofectamine 3000 was purchased from invitrogen. Chemical reagents polybrene (Millipore), puromycin and RNase inhibitor (Thermo), MG132 (Beyotime), chloroquine (CQ) (tlrl-chq, InvivoGen) and ISD (tlrl-isdc, InvivoGen) were bought from indicated manufactures.
Plasmids
Plasmids encoding HA-tagged cGAS and STING were constructed by molecular cloning methods. Myc tagged US3 plasmid was constructed in-house. All plasmids were verified by sequencing. The primer sequences used in this study are available upon request. pCMV-FLAG-TBK1, IRF3 constitutively active mutant IRF3/5D-FLAG and pCMV-FLAG-IRF3 expression plasmids were all constructed in-house.
Western Blotting
Cells were harvested and whole-cell extracts were prepared with lysis buffer buffer RIPA (Solarbio, Beijing, China). Cell extracts were subjected to 10% or 15% SDS-PAGE, and the separated proteins were transferred to PVDF membranes (Millipore). The PVDF membranes were incubated with specific primary and HRP-conjugated secondary antibodies. GAPDH or β-actin was served as a loading control. The proteins were detected using ECL Blotting Substrates (Bio-Rad, CA, United States).
Co-immunoprecipitation Assay
Cells were collected with a lysis buffer supplemented with a phosphatase inhibitor cocktail and incubated with anti-FLAG or anti-IRF3 antibody for 12 h at 4°. Then 10 μL of Protein G agarose slurry (Beyotime, China) was added to each lysate. After incubation for 4 h at 4°, the lysates were centrifuged at 2500 rpm for 5 min. The beads were collected and washed 5 times with ice-cold PBS. The precipitates were mixed with SDS buffer and boiled for 5 min at 95°. After centrifugation at 6000 rpm for 1 min, the supernatant was collected and used for western blot analysis.
RNA Extraction and Real-Time Quantitative PCR
mRNA levels was determined for IFN-β using relative qPCR. Cellular RNA was isolated and reverse-transcribed to cDNA. Methods were performed as previously described (Xie et al., 2020). Primers for RT-qPCR are available upon request.
CCK-8 Assay
Cell proliferation was determined using the CCK-8 assay. PK15 cells were seeded in 96-well plates overnight and then untreated or treated with MG132 or CQ. The proliferative ability of the cells was evaluated at 6 h and 12 h according to manufactures’ instruction.
Statistical Analysis
Measurements were compared using a one-way ANOVA. Statistical significance comparisons were calculated using a Student’s t-test in GraphPad Prism 7.0 software (La Jolla, CA, United States). Values are expressed in graph bars as the mean ± SD of at least three independent experiments, unless otherwise noted. Asterisks denote statistically significant differences (∗∗∗ p < 0.001, ∗∗ p < 0.01, and ∗ p < 0.05).
Results
Pseudorabies Virus US3 Protein Blocks IFN-β Activation
Given that HSV-1 US3 can prevent IFN-β activation during infection (Wang et al., 2013; You et al., 2020), we wondered if PRV US3 protein performed a similar function via interfering with an IFN-β pathway. ISD used in this study is a double-stranded DNA 60-mer oligonucleotide derived from the HSV-1 genome. PK15 cells were transfected with a US3 expression plasmid for 24 h before ISD transfection to determine the effect of PRV US3 on IFN-β production induced by ISD. RT-qPCR results showed ISD strongly activated IFN-β mRNA expression. However, the activation was remarkably decreased in the presence of Myc-US3 expression (Figure 1), suggesting PRV US3 expression inhibits the IFN-β activation in a dose-dependent manner.
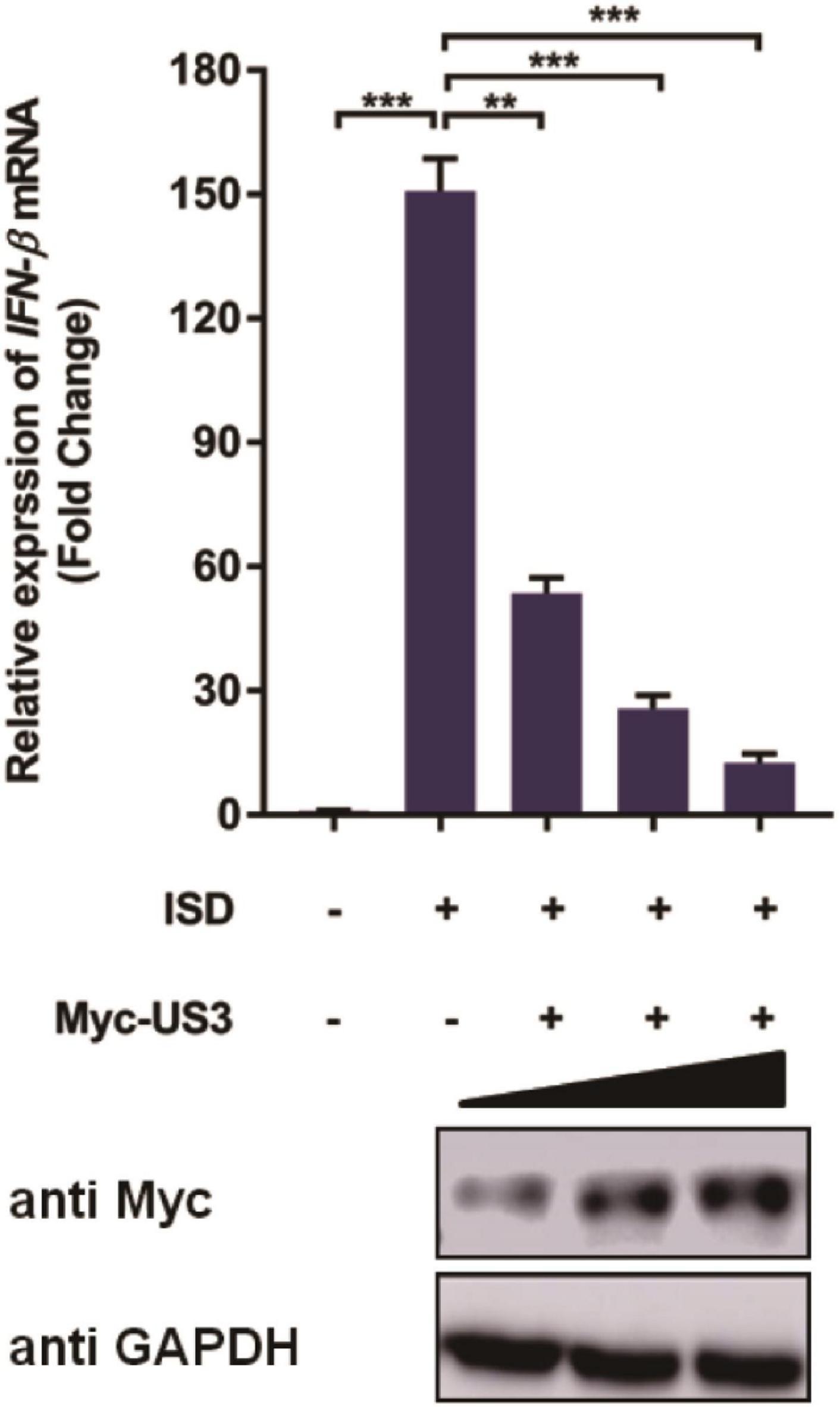
Figure 1. pCMV-Myc-US3 plasmid (0.2 μg, 0.5 μg, and 1.0 μg) was transfected into PK15 cells used Lipofectamine 3000. ISD (2 μg/mL) was transfected into above cells for 12 h before cells collection. US3 protein expression was detected by western blotting. An anti-Myc antibody was used and GAPDH served as the loading control. Then cellular RNA were extracted and cDNA was transcripted for IFN-β mRNA detection. Data were listed as mean ± SD from three independent experiments. Comparison between two groups was evaluated by unpaired Student’s t test. ** p < 0.01, *** p < 0.001.
IRF3 Might Be the Potential Target of Pseudorabies Virus US3 Protein
Type I IFN induction is mainly mediated by the cGAS-STING pathway during DNA virus infection. Cells were transfected with either plasmid encoding cGAS-STING signaling pathway adaptors TBK1, IRF3/5D, a constitutively active form of IRF3 containing five C-terminal substitutive Asp (D) residues (Ramos and Gale, 2011) and cGAS and STING proteins for 12 h before overexpressing US3 transiently for a further 24 h. Exogenous overexpression of any of these adaptor molecules significantly activated IFN-β expression. Interestingly, US3 protein expression repressed all of these adaptor molecules triggered IFN-β activation (Figures 2A–D). Thereby, we considered that IRF3 might be a targeting protein for US3 hindering the type I IFN pathway.
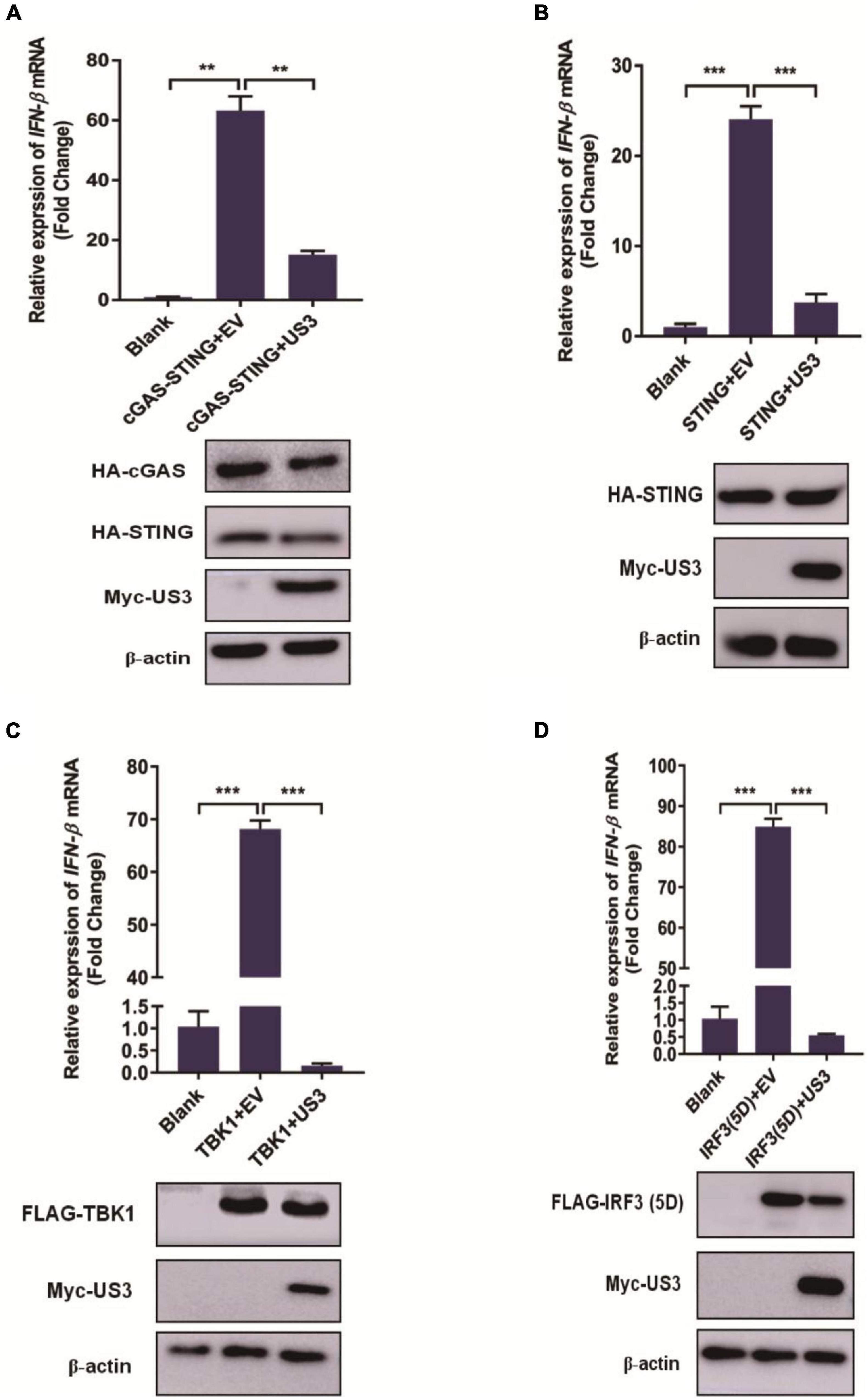
Figure 2. PK15 cells were cotransfected with empty vector (0.5 μg) or Myc-US3 (0.5 μg) plasmids and the indicated plasmids expressing cGAS (15 ng) + STING (2.5 ng) (A), STING (0.2 μg) (B), TBK1(0.2 μg) (C) or IRF3(5D) (0.2 μg) (D) for 24 h. Then cells were collected for total RNA extraction. IFN-β mRNA expression level was measured by RT-qPCR. Expression of various adaptor molecules and US3 protein was evaluated by western blotting. β-actin served as loading control. Data were listed as mean ± SD from three independent experiments. Comparison between two groups was evaluated by unpaired Student’s t test. ** p < 0.01, *** p < 0.001.
US3 Interacts With IRF3 and Degrades IRF3 Through the Proteasomal Pathway
IRF3 plays an extremely pivotal role in the induction of IFN in responding to viral infection. PRV US3 protein exhibited a remarkable inhibitory effect on IRF3 and its upstream adaptors (Figure 2), suggesting that it could target IRF3. PK15 cells were co-transfected with FLAG-IRF3 and Myc-US3 plasmids to test whether US3 interacted with IRF3. An anti-FLAG antibody was used to carry out co-immunoprecipitation (Co-IP) assay. As shown in Figure 3A, IRF3 coprecipitated with US3 protein, suggesting a direct interaction between US3 and IRF3 protein. To verify the interaction between US3 protein and endogenous IRF3, Co-IP was operated by transfecting PK15 cells with Myc-vector- or Myc-US3 expressing plasmids, anti-IRF3 antibody was used to detect and visualize IRF3 expression. As shown in Figure 3B, US3 protein was immunoprecipitated with endogenous IRF3.
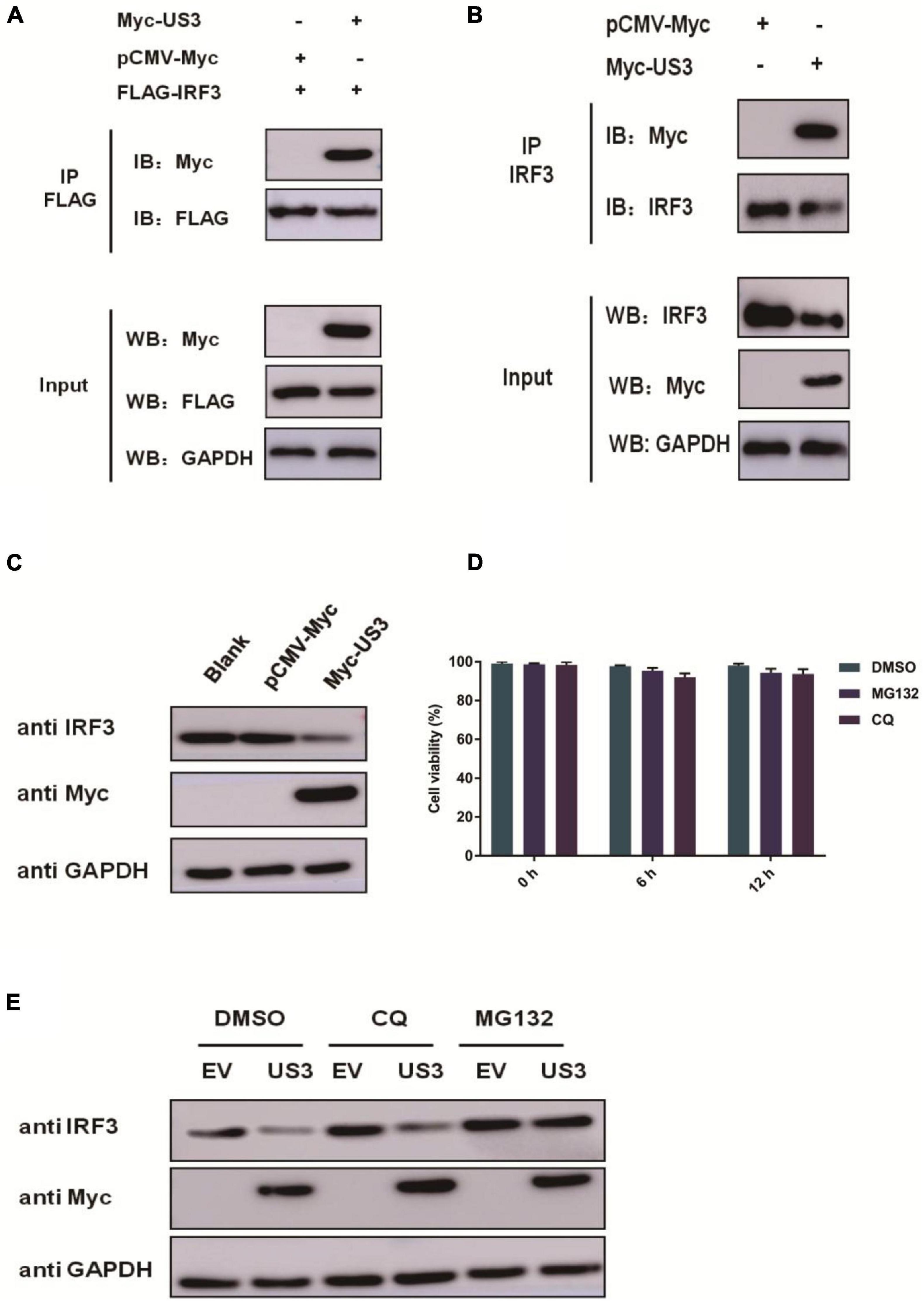
Figure 3. (A) PK15 cells were co-transfected with empty vector (0.5 μg) or Myc-US3 (0.5 μg) plasmids and FLAG-IRF3 (0.2 μg) plasmids for 30 h. The cells were then lysed and immunoprecipitated with an anti-Flag antibody. The whole-cell lysates (input) and immunoprecipitation (IP) complexes were analyzed using an anti-Myc, anti-FLAG or anti-GAPDH antibody by western blotting. (B) PK15 cells were transfected with empty vector (0.5 μg) or Myc-US3 (0.5 μg) plasmids for 30 h. The cells were lysed and immunoprecipitated with an anti-IRF3 antibody. The input and IP complexes were analyzed by western blotting using anti-IRF3, anti-Myc or anti-GAPDH antibodies. (C) PK15 cells were transfected with empty vector (0.5 μg) or Myc-US3 (0.5 μg) for 24 h, then cells were collected for western blotting using anti-IRF3, anti-Myc or anti-GAPDH antibodies. (D) Chemicals were previously tested for cytotoxicity at the concentrations used. PK15 cells treated with 7.5 μM MG132 or 50 μM CQ for 6 h and 12 h in 96 well plate. Cell viability was determined using the CCK8 reagent. Data were expressed as the mean ± SD from three independent experiments. (E) PK15 cells were transfected with Myc-US3 (0.5 μg) or empty vector (0.5 μg) for 24 h, then treated with lysosomal inhibitor MG132 (7.5 μM) or the lysosome inhibitor CQ(50 μM)for 6 h. DMSO treated cells served as vehicle control. Then cells were collected and immunoblotting for IRF3 and Myc. GAPDH served as a loading control.
Indeed, overexpression of US3 decreased IRF3 protein expression (Figure 3C). US3 overexpressing cells were treated with proteasome inhibitor MG132 and the autophagy inhibitor chloroquine diphosphate (CQ) to explore the mechanism by which US3 decreases IRF3 expression. We confirmed that MG132 and CQ were not toxic on PK15 cells (Figure 3D). Immunoblotting analysis revealed that MG132 inhibited IRF3 degradation but not CQ, suggesting that degradation occurs through the ubiquitination-proteasomal, not autophagic pathway (Figure 3E).
US3 Protein Suppresses IRF3 Phosphorylation
The phosphorylation of IRF3 is required for the induction of IFNs. Pseudorabies Virus US3 protein blocks type I IFN production by targeting IRF3. The levels of ISD-induced IRF3 phosphorylation in the absence or presence of US3 protein were examined to investigate whether US3 protein affected the phosphorylation of IRF3. PK15 cells were transfected with Myc-US3 or empty vector plasmids along with ISD. ISD induced significant phosphorylation of IRF3 in both Myc-US3 and empty vector plasmid-transfected cells. Whether treated with MG132 or not, the phosphorylation level of IRF3 was markedly lower in the Myc-US3-transfected cells than in the empty vector-transfected cells (Figures 4A,B). These results indicated that PRV US3 protein abrogated IRF3 phosphorylation.
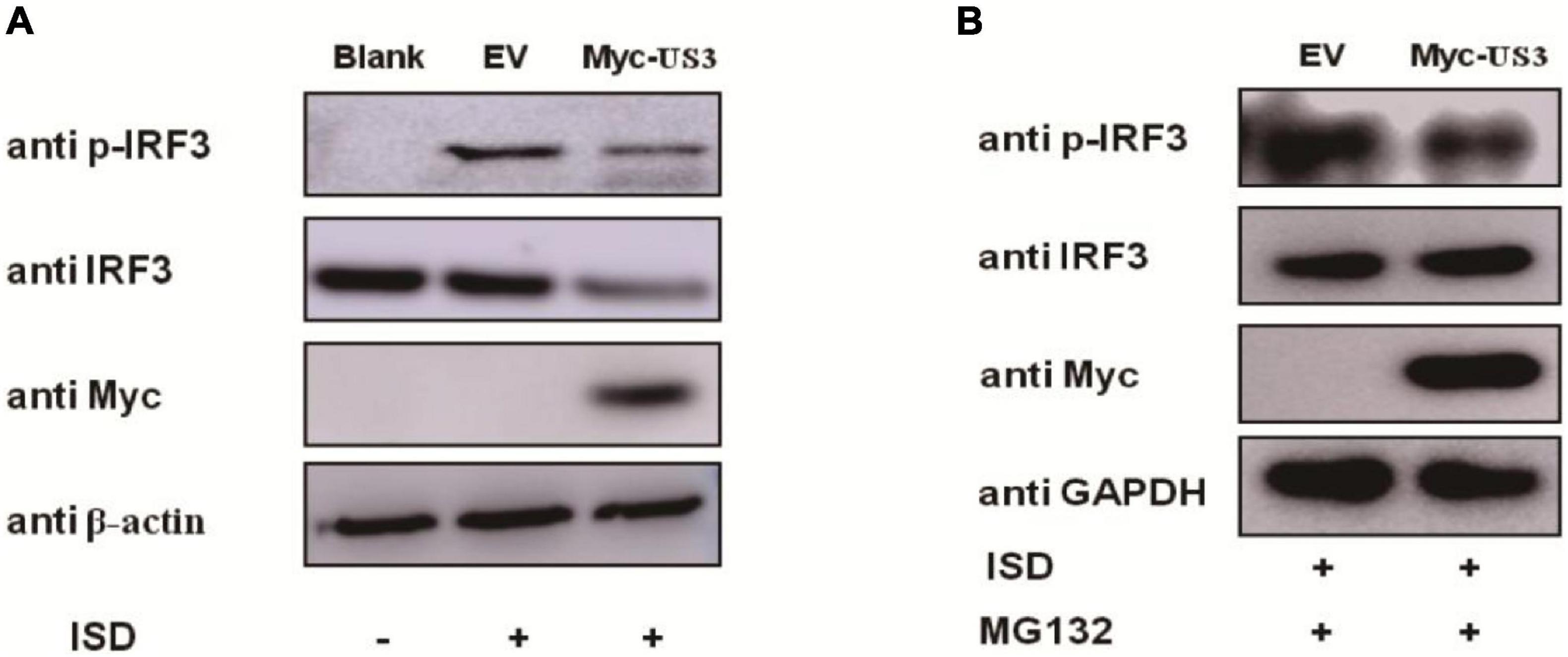
Figure 4. PK15 cells were transfected with empty vector (0.5 μg) or Myc-US3 (0.5 μg) plasmids for 24 h and then transfected with ISD (2 μg/mL) for 12 h. Cells were untreated (A) or treated (B) with 7.5 μM MG132 for another 6 h before collection. IRF3, phosphorylated IRF3 (p-IRF3), and US3 protein (Myc) expression was detected by immune blotting. GAPDH or β-actin served as loading control.
US3 Protein Blocks IRF3 Nuclear Translocation
IRF3 is a transcription factor that participated in type I IFN production, and its function is realized by transposition from the cytoplasm to the nucleus (Vandevenne et al., 2011). The phosphorylation of IRF3 causes its nuclear translocation. Our results showed that US3 interacts with IRF3 protein and inhibits its phosphorylation. PK15 cells were co-transfected with Myc-US3 expressing plasmids and ISD to investigate the effects of US3 protein on the nuclear translocation of IRF3. As US3 could degrade IRF3 expression through the proteasome pathway (Figure 3E), the above experimental cells were treated with MG132 for 6 h before collection. The distribution of IRF3 in cytoplasma or nucleus was detected by nuclear-cytoplasmic separation experiment. Compared with the empty vector-transfected group, in the US3 transfected group, most of the IRF3 protein remained in the cytoplasm, and only a small amount of IRF3 entered the nucleus (Figure 5). The result indicated that US3 protein inhibited ISD-induced nuclear translocation of IRF3 to prevent IFN-β production.
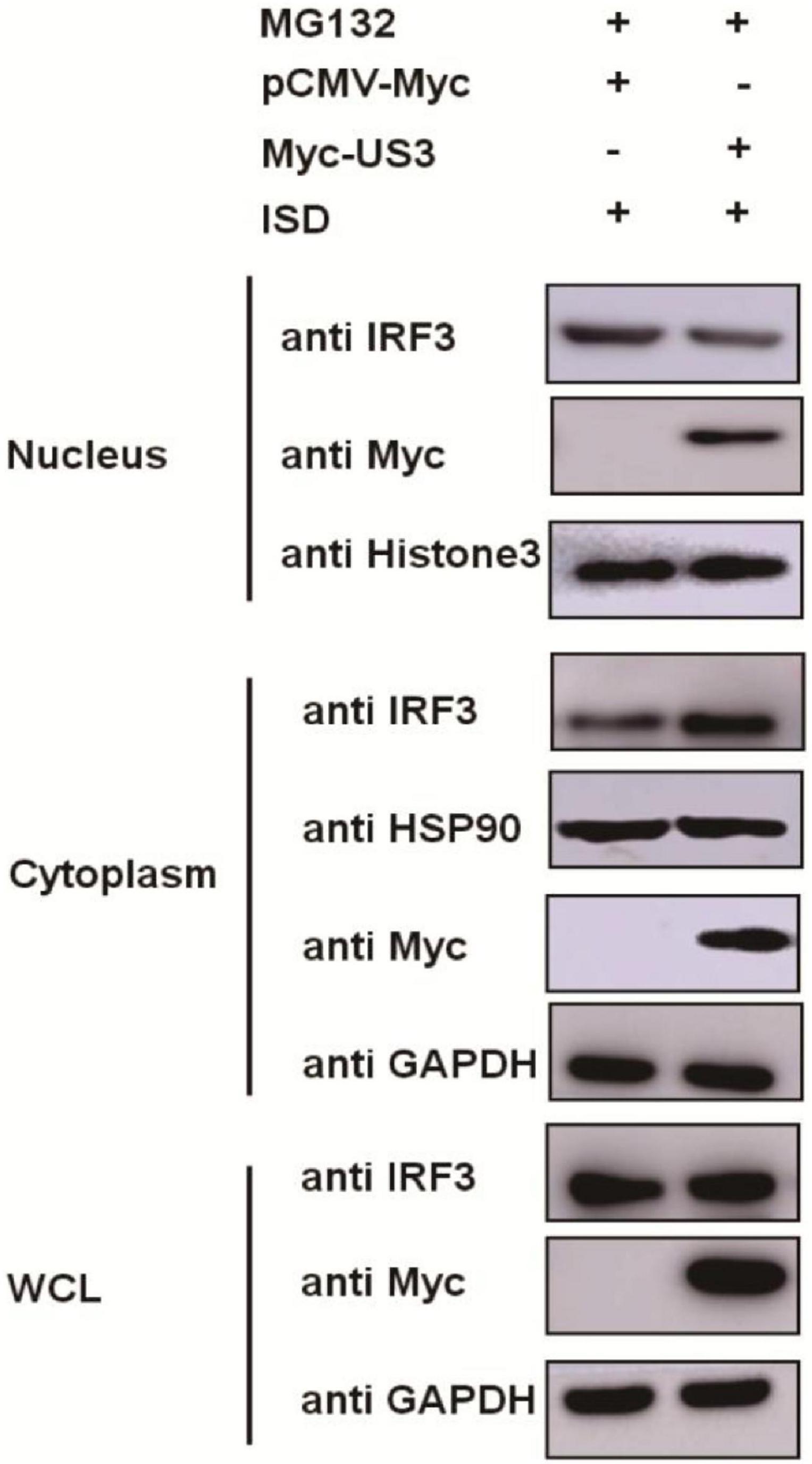
Figure 5. ISD (2 μg/mL) was transfected into PK15 cells in the presence of either pCMV-Myc (0.5 μg) or Myc-US3 (0.5 μg) for 24 h. Before collected, cells were treated with MG132 for 6 h. Then cytoplasmic and nuclear proteins were extracted and subjected to western blotting. Expression of IRF3 and Myc tagged US3 was detected with specific antibodies. HSP90 was used as a cytoplasmic protein marker, whereas Histone3 was used as a nuclear protein marker. GAPDH served as loading control.
Pseudorabies Virus Mutant Containing a US3 Knockdown Produces Higher Levels of Innate Immunity
To further determine US3 functions during a live PRV infection, two shRNAs targeted to US3 were designed and synthesized by Genechem (Shanghai, China). shRNAs targeted to US3 were transfected into PK15 cells following PRV infection. 12 h post-infection, cells were collected for RNA extraction. IFN-β mRNA expression was detected by RT-qPCR. Results showed in Figure 6A, IFN-β transcripts are present in significantly higher levels in cells transfected with shRNA targeting US3 than those transfected with shRNA-control. To verify whether this result is related to the reduced expression of US3, we examined the expression of US3 by western blotting and found that shRNA-US3 can effectively interfere with the expression of US3 (Figure 6B).
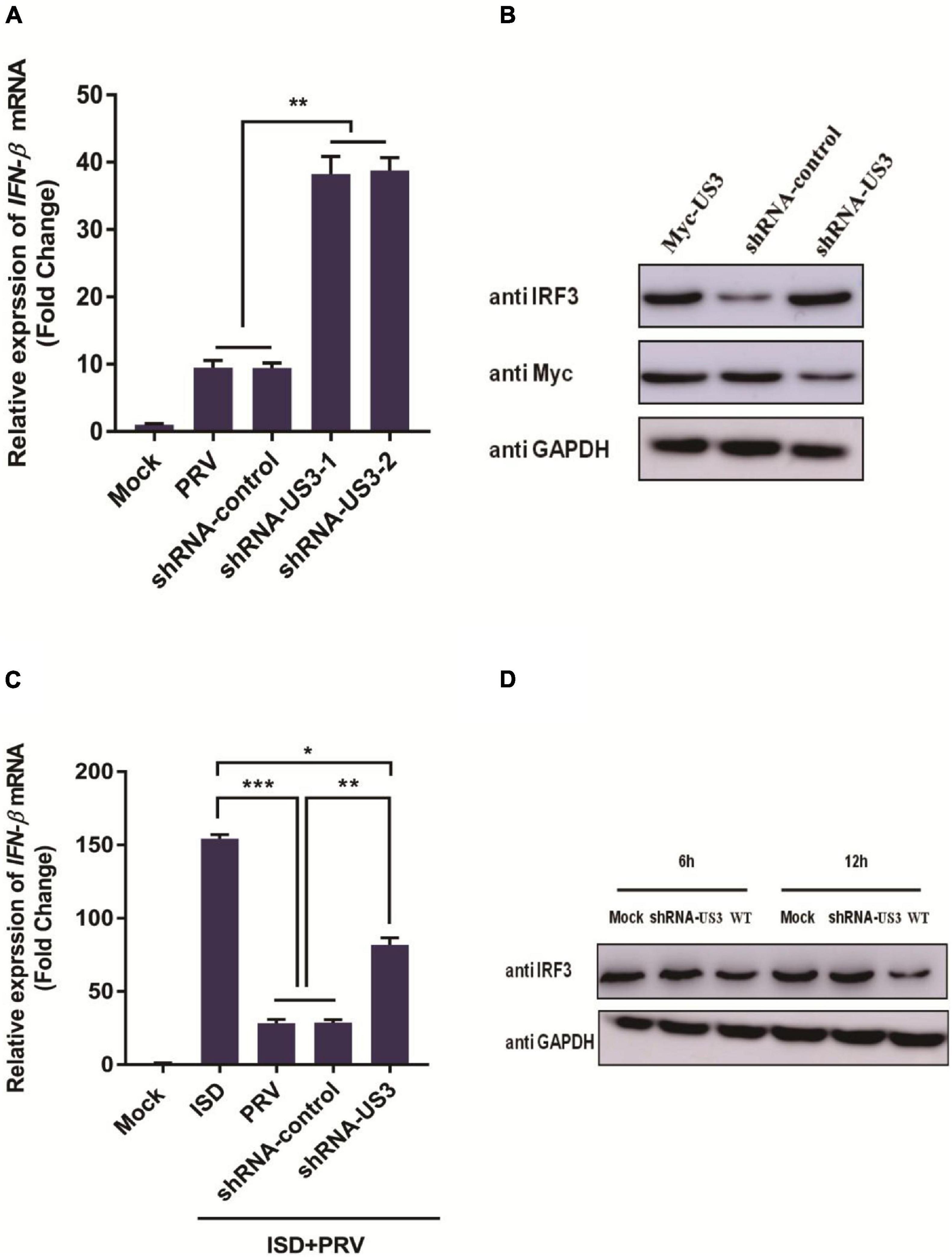
Figure 6. (A) PK15 cells were transfected with shRNA targeting US3 (0.5 μg) or control shRNA (0.5 μg) for 24 h. Then cells were infected with MOI of 1 PRV for 12 h. Cells were collected for cellular RNA extraction. cDNA was transcripted for IFN-β mRNA detection. Data were listed as mean ± SD from three independent experiments. Comparison between two groups were examined by unpaired Student’s t test. ** p < 0.01. (B) PK15 cells were transfected with shRNA-US3 (1 μg) or shRNA-control (1 μg) for 24h followed with Myc-US3 (0.5 μg) plasmid transfection. 24 h post transfection, cells were collected and lysed for IRF3 and Myc tagged US3 expression using specific antibodies. GAPDH served as loading control. (C) PK15 cells were transfected with shRNA-US3 (1 μg) or shRNA-control (1 μg) for 24 h followed with ISD (2 μg/mL) plasmid transfection. 12 h post transfection, above cells were uninfected or infected with MOI of 3 PRV for another 12 h. Cells were collected for total RNA extraction. 1 μg RNA was transcripted into cDNA for IFN-β mRNA detection. Data were expressed as mean ± SD from three independent experiments and were measured in technical duplicate. Comparisons between groups were performed by Student’s t test. * p < 0.05, ** p < 0.01, *** p < 0.001. (D) PK15 cells were infected with PRV Bartha strain (WT) and PRV-shRNA-US3 at 3 MOI. Endogenous IRF3 expression level was determined by western blotting at 6 h and 12 h post infection. GAPDH served as loading control.
Next, we studied the effect of US3 knockdown expression on ISD-triggered IFN-β transcription. It was found that ISD can effectively stimulate the transcription of IFN-β, and PRV infection inhibited the expression of IFN-β at the mRNA level. However, in shRNA-US3 and ISD transfected cells, the inhibition of IFN-β expression by PRV was weakened (Figure 6C) but not completely disappeared, suggesting that other viral proteins may be involved in the process of resistance to host innate immunity. Altogether, these results further confirmed that US3 protein could antagonize the activation of IFN-β pathway.
To further study the expression of IRF3 protein in viral infection, PK15 cells were inoculated with 3 MOI PRV Bartha-61 strain (wild type, WT) or PRV-shRNA-US3 for 6 h and 12 h, respectively. Cells were collected for IRF3 detection. As in Figure 6D shown, levels of endogenous IRF3 in PRV-shRNA-US3 infected cells were markedly higher than in WT strain infected cells, suggesting that US3 helps to enable PRV degrade IRF3 and resistance to IFN-β signaling pathway.
Discussion
The innate immune system composes the first line of host, and protects hosts from viral infection. The capability of viruses to avoid and regulate host innate immunity response is of great importance for viral infection (Bowie and Unterholzner, 2008). As the cGAS-MITA-TBK1 axis plays an indispensable role in host defense against DNA viruses infection (Kato et al., 2017), the DNA viruses have developed numerous means to counteract this signaling pathway for replication and latent infection (Ma and Damania, 2016). This study showed that PK15 cells infection with PRV significantly suppressed type I IFN production. We also demonstrated the role of US3 in the IFN-β signaling pathway and revealed the mechanism used by PRV to antagonize host antiviral response.
The US3 protein is a multifunctional serine/threonine-protein kinase. US3 expression modulates a wide range of cellular processes, including virus nuclear egress, inhibition of apoptosis, reorganization of the cytoskeleton, and several immune modulators (Favoreel et al., 2005; Deruelle et al., 2007; Deruelle and Favoreel, 2011). In the current study, we showed that exogenous overexpression of PRV US3 inhibited cGAS-STING, TBK1, IRF3(5D), or ISD-triggered activation of IFN-β (Figure 2). Furthermore, US3 could interact with IRF3 and degrade the protein expression level of IRF3 (Figure 3).
In this study, IRF3 was recognized as a target of PRV US3 protein, through which it inhibited type I IFN production. IRF3 is a key regulator of IFN-β pathway. It can be phosphorylated by cellular and viral proteins, conducing to either the activation or suppression of IRF3 transcriptional activities. As a consequence, leading to increase or decrease of IFN-β production. Previous studies reported that several conserved herpes virus-encoded kinases might contribute be involved in anti-IFN function by suppressing the IRF3 pathway, such as HSV-1 UL13, HCMV UL97, MHV-68 ORF36, and the EBV BGLF4 kinase protein (Hwang et al., 2009). Here, we demonstrated that PRV US3 protein antagonized the IFN-β pathway by targeting IRF3, through degradation of its protein expression, inhibits IRF3 phosphorylated and nuclear translocation (Figures 3–5). There is a direct interaction between US3 and IRF3, and endogenous IRF3 levels are affected by US3, so phosphorylated IRF3 decreased maybe result from degradation of endogenous IRF3 by US3. To rule out this effect and explore more accurately the effect of US3 on IRF3 phosphorylation, we used MG132 to treat the cells transfected with Myc-US3 and ISD or empty vector and ISD. Results found that after treated with MG132, the total IRF3 in the cells did not decrease in the Myc-US3 transfection group, but the IRF3 phosphorylation level of cells transfected with Myc-US3 was significantly lower than that of cells transfected without empty vector (Figure 4B). These results further confirmed that US3 could inhibit the phosphorylation of IRF3.
To understand the role of US3 interaction with the IFN-β signaling pathway, we used the PRV-shRNA-US3 strain for further study. When knockdown US3 expression, its ability to inhibit IFN-β transcription was weakened (Figures 6A–C). These results indicate that US3 plays an important role in antagonizing innate immunity. US3 also influences IRF3 expression. IRF3 levels in PRV-shRNA-US3-infected cells are significantly higher than those in WT strain infected cells, indicating that a virus failure to express US3 has a weakened capability to prevent IRF3 and IFN-β activation.
In summary, our data demonstrated a possible mechanism that US3 antagonized IFN-β signaling pathway (Figure 7). US3 inhibited IFN-β production by targeting IRF3. There was a direct interaction between US3 and IRF3. Moreover, US3 degraded IRF3 protein level expression and blocked its activation. These findings suggested that PRV US3 could inhibit the IFN-β production and provide new insights into innate immune evasion by PRV.
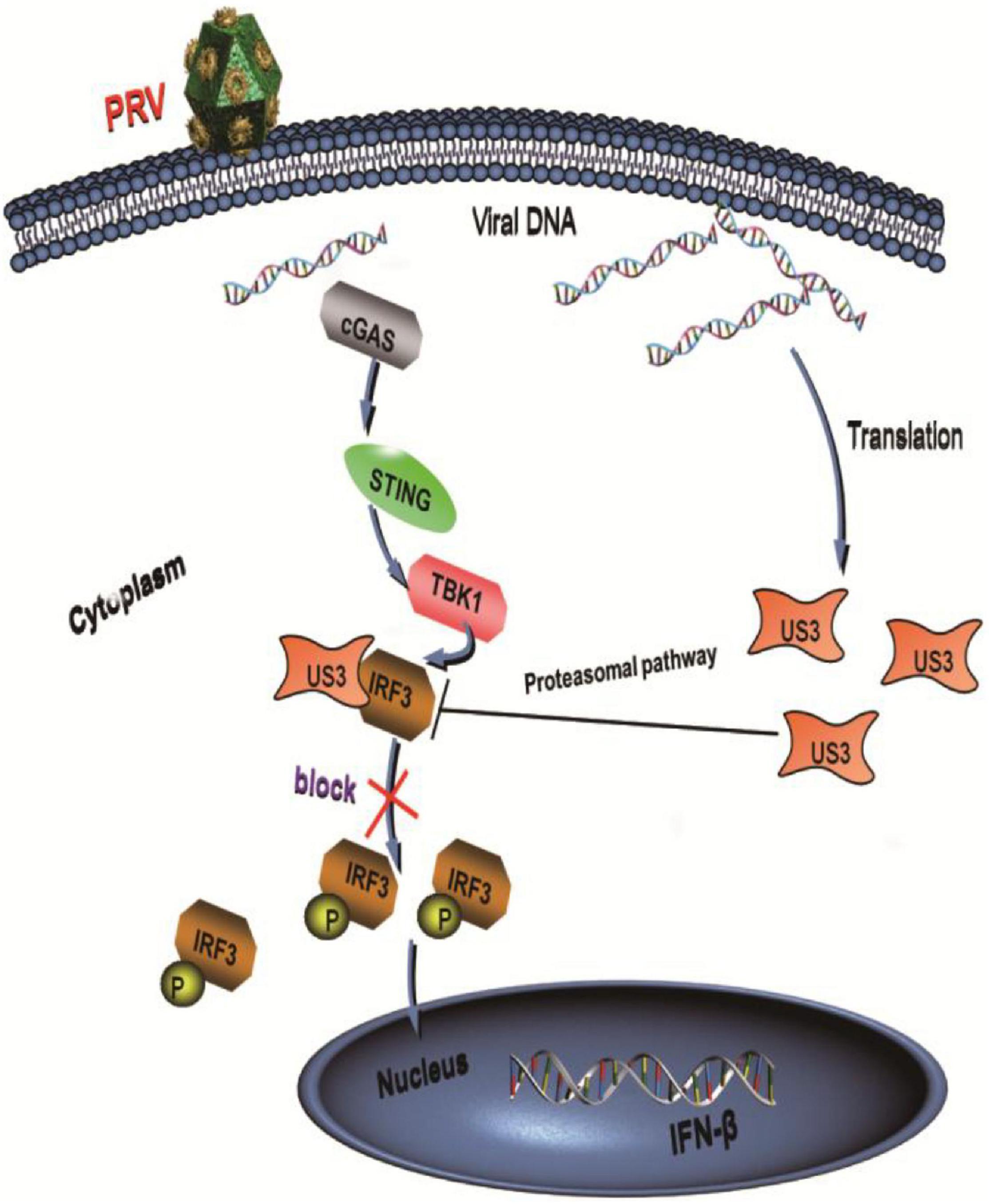
Figure 7. PRV protein kinase US3 interacts with IRF3 and degrades IRF3 protein expression through proteasome pathway. Additionally, US3 inhibits IRF3 phosphorylation and prevent its nuclear translocation, then negatively regulates IFN-β production.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
JX and XZ wrote the draft manuscript. JX and RF designed the experiment. JX, XZ, LC, YB, SX, and XL performed experiments and processed the data. AI, YZ, and RF revised and proofread the draft manuscript. RF supervised the entire process. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Program for Young Talent of SEAC [(2018)98], the Fundamental Research Funds for the Central Universities (31920190003, 31920200003, and 31920210051), Open Funds of the Biomedical Research Center from Northwest Minzu University (EB201801), and Gansu Youth Science and Technology Fund Project (20JR5RA501).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank YanDong Tang for the assistance in this research.
References
Ai, J., Weng, S., Cheng, Q., Cui, P., Li, Y., Wu, H., et al. (2018). Human endophthalmitis caused by pseudorabies virus infection, China, 2017. Emerging. Infect. Dis. 24, 1087–1090. doi: 10.3201/eid2406.171612
Beachboard, D. C., and Horner, S. M. (2016). Innate immune evasion strategies of DNA and RNA viruses. Curr. Opin. Microbiol. 32, 113–119. doi: 10.1016/j.mib.2016.05.015
Bo, Z., Miao, Y., Xi, R., Zhong, Q., Bao, C., Chen, H., et al. (2020). PRV UL13 inhibits cGAS-STING-mediated IFN-β production by phosphorylating IRF3. Vet. Res. 51:118. doi: 10.1186/s13567-020-00843-4
Bowie, A. G., and Unterholzner, L. (2008). Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8, 911–922. doi: 10.1038/nri2436
Brukman, A., and Enquist, L. W. (2006b). Suppression of the interferon-mediated innate immune response by pseudorabies virus. J. Virol. 80, 6345–6356. doi: 10.1128/JVI.00554-06
Brukman, A., and Enquist, L. W. (2006a). Pseudorabies virus EP0 protein counteracts an interferon-induced antiviral state in a species-specific manner. J. Virol. 80, 10871–10873. doi: 10.1128/JVI.01308-06
Carpenter, S., Ricci, E. P., Mercier, B. C., Moore, M. J., and Fitzgerald, K. A. (2014). Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 14, 361–376. doi: 10.1038/nri3682
Cartier, A., Komai, T., and Masucci, M. G. (2003). The Us3 protein kinase of herpes simplex virus 1 blocks apoptosis and induces phosporylation of the Bcl-2 family member Bad. Exp. Cell. Res. 291, 242–250. doi: 10.1016/s0014-4827(03)00375-6
Chen, N., Xia, P., Li, S., Zhang, T., Wang, T. T., and Zhu, J. (2017). RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life 69, 297–304. doi: 10.1002/iub.1625
Christensen, L. S., Medveczky, I., Strandbygaard, B. S., and Pejsak, Z. (1992). Characterization of field isolates of suid herpesvirus 1 (Aujeszky’s disease virus) as derivatives of attenuated vaccine strains. Arch. Virol. 124, 225–234. doi: 10.1007/BF01309804
Christensen, M. H., Jensen, S. B., Miettinen, J. J., Luecke, S., Prabakaran, T., Reinert, L. S., et al. (2016). HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO. J. 35, 1385–1399. doi: 10.15252/embj.201593458
Deruelle, M., Geenen, K., Nauwynck, H. J., and Favoreel, H. W. (2007). A point mutation in the putative ATP binding site of the pseudorabies virus US3 protein kinase prevents Bad phosphorylation and cell survival following apoptosis induction. Virus. Res. 128, 65–70. doi: 10.1016/j.virusres.2007.04.006
Deruelle, M. J., and Favoreel, H. W. (2011). Keep it in the subfamily: the conserved alphaherpesvirus US3 protein kinase. J. Gen. Virol. 92, 18–30. doi: 10.1099/vir.0.025593-0
Fan, S., Yuan, H., Liu, L., Li, H., Wang, S., Zhao, W., et al. (2020). Pseudorabies virus encephalitis in humans: a case series study. J. Neurovirol. 26, 556–564. doi: 10.1007/s13365-020-00855-y
Favoreel, H. W., van Minnebruggen, G., Adriaensen, D., and Nauwynck, H. J. (2005). Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an alphaherpesvirus are associated with enhanced spread. Proc. Natl. Acad. Sci. U. S. A. 102, 8990–8995. doi: 10.1073/pnas.0409099102
Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., et al. (2003). IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496. doi: 10.1038/ni921
Fonseca, A. A. Jr., Camargos, M. F., de Oliveira, A. M., Ciacci-Zanella, J. R., Patricio, M. A., et al. (2010). Molecular epidemiology of Brazilian pseudorabies viral isolates. Vet. Microbiol. 141, 238–245. doi: 10.1016/j.vetmic.2009.09.018
Gao, L., Li, K., Zhang, Y., Liu, Y., Liu, C., Zhang, Y., et al. (2019). Inhibition of DNA-sensing pathway by Marek’s disease virus VP23 protein through suppression of interferon regulatory factor 7 activation. J. Virol. 93, e01934–18. doi: 10.1128/JVI.01934-18
Geenen, K., Favoreel, H. W., Olsen, L., Enquist, L. W., and Nauwynck, H. J. (2005). The pseudorabies virus US3 protein kinase possesses anti-apoptotic activity that protects cells from apoptosis during infection and after treatment with sorbitol or staurosporine. Virology 331, 144–150. doi: 10.1016/j.virol.2004.10.027
Hwang, S., Kim, K. S., Flano, E., Wu, T. T., Tong, L. M., Park, A. N., et al. (2009). Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5, 166–178. doi: 10.1016/j.chom.2008.12.013
Kato, K., Omura, H., Ishitani, R., and Nureki, O. (2017). Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Annu. Rev. Biochem. 86, 541–566. doi: 10.1146/annurev-biochem-061516-044813
Kawai, T., and Akira, S. (2006). Innate immune recognition of viral infection. Nat. Immunol. 7, 131–137. doi: 10.1038/ni1303
Lamote, J., Kestens, M., Van Waesberghe, C., Delva, J., De Pelsmaeker, S., et al. (2017). The pseudorabies virus glycoprotein gE/gI complex suppresses type I interferon production by plasmacytoid dendritic cells. J. Virol. 91, e02276–16. doi: 10.1128/JVI.02276-16
Leopardi, R., Van Sant, C., and Roizman, B. (1997). The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. U. S. A. 94, 7891–7896. doi: 10.1073/pnas.94.15.7891
Li, K., Liu, Y., Xu, Z., Zhang, Y., Luo, D., and Gao, Y. (2019). Avian oncogenic herpesvirus antagonizes the cGAS–STING DNA-sensing pathway to mediate immune evasion. PLoS Pathog. 15:e1007999. doi: 10.1371/journal.ppat.1007999
Li, M., Damania, B., Alvarez, X., Ogryzko, V., Ozato, K., and Jung, J. U. (2000). Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol. Cell Biol. 20, 8254–8263. doi: 10.1128/mcb.20.21.8254-8263.200
Lin, Y., and Zheng, C. (2019). A tug of war: DNA-sensing antiviral innate immunity and herpes simplex virus type I infection. Front. Microbiol. 10:2627. doi: 10.3389/fmicb.2019.02627
Liu, Q., Wang, J., Xie, C., Ding, S., Yang, H., Guo, S., et al. (2020). A novel human acute encephalitis caused by pseudorabies virus variant strain. Clin. Infect. Dis. ciaa987. doi: 10.1093/cid/ciaa987 [Epub ahead of print].
Lv, L., Cao, M. Z., Bai, J., Jin, L., Wang, X. W., Gao, Y. N., et al. (2020). PRV-encoded UL13 protein kinase acts as an antagonist of innate immunity by targeting IRF3-signaling pathways.Vet. Microbiol. 250:108860. doi: 10.1016/j.vetmic.2020.108860
Ma, Z., and Damania, B. (2016). The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 19, 150–158. doi: 10.1016/j.chom.2016.01.010
Ma, Z., Jacobs, S. R., West, J. A., Stopford, C., Zhang, Z., Davis, Z., et al. (2015). Modulation of the cGAS–STING DNA sensing pathway by gammaherpesviruses. Proc. Natl. Acad. Sci. U. S. A. 112, E4306–E4315. doi: 10.1073/pnas.1503831112
Mettenleiter, T. C. (2000). Aujeszky’s disease (pseudorabies) virus: the virus and molecular pathogenesis–state of the art, June 1999. Vet. Res. 31, 99–115. doi: 10.1051/vetres:2000110
Qin, C., Zhang, R., Lang, Y., Shao, A., Xu, A., and Feng, W. (2019). Bclaf1 critically regulates the type I interferon response and is degraded by alphaherpesvirus US3. PLoS Pathog. 15:e1007559. doi: 10.1371/journal.ppat.1007559
Ramos, H. J., and Gale, M. (2011). RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 1, 167–176. doi: 10.1016/j.coviro.2011.04.004
Reynolds, A. E., Wills, E. G., Roller, R. J., Ryckman, B. J., and Baines, J. D. (2002). Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76, 8939–8952. doi: 10.1128/jvi.76.17.8939-8952.2002
Sharma, S., tenOever, B. R., Grandvaux, N., Zhou, G. P., Lin, R., and Hiscott, J. (2003). Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151. doi: 10.1126/science.1081315
Su, C., Zhan, G., and Zheng, C. (2016). Evasion of host antiviral innate immunity by HSV-1, an update. Virol. J. 13:38. doi: 10.1186/s12985-016-0495-5
Sun, L., Wu, J., Du, F., Chen, X., and Chen, Z. J. (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. doi: 10.1126/science.1232458
Sun, Y., Luo, Y., Wang, C., Yuan, J., Li, N., Song, K., et al. (2016). Control of swine pseudorabies in China: opportunities and limitations. Vet. Microbiol. 183, 119–124. doi: 10.1016/j.vetmic.2015.12.008
Vandevenne, P., Lebrun, M., Ei Mjiyad, N., Ote, I., Di Valentin, E., Habraken, Y., et al. (2011). The varicella-zoster virus ORF47 kinase interferes with host innate immune response by inhibiting the activation of IRF3. PLoS One 6:e16870. doi: 10.1371/journal.pone.0016870
Wagenaar, F., Pol, J. M., Peeters, B., Gielkens, A. L., de Wind, N., and Kimman, T. G. (1995). The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76, 1851–1859. doi: 10.1099/0022-1317-76-7-1851
Wang, D., Tao, X., Fei, M., Chen, J., Guo, W., Li, P., et al. (2020). Human ncephalitis caused by pseudorabies virus infection: a case report. J. Neurovirol. 26, 442–448. doi: 10.1007/s13365-019-00822-2
Wang, K., Ni, L., Wang, S., and Zheng, C. (2014). Herpes simplex virus 1 protein kinase US3 hyperphosphorylates p65/RelA and dampens NF-κB activation. J. Virol. 88, 7941–7951. doi: 10.1128/JVI.03394-13
Wang, S., Wang, K., Lin, R., and Zheng, C. (2013). Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J. Virol. 87, 12814–12827. doi: 10.1128/JVI.02355-13
Wu, J., Li, W., Shao, Y., Avey, D., Fu, B., Gillen, J., et al. (2015). Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18, 333–344. doi: 10.1016/j.chom.2015.07.015
Xie, J., Bi, Y., Xu, S., Han, Y., Idris, A., Zhang, H., et al. (2020). Host antiviral protein IFITM2 restricts pseudorabies virus replication. Virus. Res. 287:198105. doi: 10.1016/j.virusres.2020.198105
Xia, P., Wang, S., Gao, P., Gao, G., and Fan, Z. (2016). DNA sensor cGAS-mediated immune recognition. Protein Cell. 7, 777–791. doi: 10.1007/s13238-016-0320-3
You, H., Lin, Y., Lin, F., Yang, M., Li, J., Zhang, R., et al. (2020). β-Catenin Is Required for the cGAS/STING Signaling Pathway but Antagonized by the Herpes Simplex Virus 1 US3 Protein. J. Virol. 94, e01847–19. doi: 10.1128/JVI.01847-19
Zhang, D., Su, C., and Zheng, C. (2016). Herpes simplex virus 1 serine protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J. Virol. 90, 5824–5829. doi: 10.1128/JVI.00186-16
Zhang, R., Xu, A., Qin, C., Zhang, Q., Chen, S., Lang, Y., et al. (2017). Pseudorabies virus dUTPase UL50 induces lysosomal degradation of type I interferon receptor 1 and antagonizes the alpha interferon response. J. Virol. 91, e01148–17. doi: 10.1128/JVI.01148-17
Zheng, C. (2018). Evasion of Cytosolic DNA-Stimulated Innate Immune Responses by Herpes Simplex Virus 1. J. Virol. 92, e00099–17. doi: 10.1128/JVI.00099-17
Keywords: pseudorabies virus, US3 protein, innate immunity, IRF3, IFN-β
Citation: Xie J, Zhang X, Chen L, Bi Y, Idris A, Xu S, Li X, Zhang Y and Feng R (2021) Pseudorabies Virus US3 Protein Inhibits IFN-β Production by Interacting With IRF3 to Block Its Activation. Front. Microbiol. 12:761282. doi: 10.3389/fmicb.2021.761282
Received: 19 August 2021; Accepted: 15 September 2021;
Published: 22 October 2021.
Edited by:
Chunfu Zheng, University of Calgary, CanadaReviewed by:
Chenhe Su, Wistar Institute, United StatesHongjuan You, Xuzhou Medical University, China
Copyright © 2021 Xie, Zhang, Chen, Bi, Idris, Xu, Li, Zhang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Zhang, zhychy@163.com; Ruofei Feng, fengruofei@xbmu.edu.cn
†These authors have contributed equally to this work
 Jingying Xie1,2†
Jingying Xie1,2† Adi Idris
Adi Idris Shujuan Xu
Shujuan Xu Yong Zhang
Yong Zhang Ruofei Feng
Ruofei Feng