- 1State Key Laboratory of Materials-Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing, China
- 2Jiangsu Key Laboratory of Marine Bioresources and Environment, Jiangsu Ocean University, Lianyungang, China
A novel multifunctional chitinase (CmChi3)-encoding gene was cloned from Chitinolyticbacter meiyuanensis and actively expressed in Escherichia coli. Sequence analysis showed that CmChi3 contains two glycoside hydrolase family 18 (GH18) catalytic domains and exhibited low identity with well-characterized chitinases. The optimum pH and temperature of purified recombinant CmChi3 were 6.0 and 50°C, respectively. CmChi3 exhibited strict substrate specificity of 4.1 U/mg toward colloidal chitin (CC) and hydrolyzed it to yield N-acetyl-D-glucosamine (GlcNAc) as the sole end product. An analysis of the hydrolysis products toward N-acetyl chitooligosaccharides (N-acetyl COSs) and CC substrates revealed that CmChi3 exhibits endochitinase, N-acetyl-β-d-glucosaminidase (NAGase), and transglycosylase (TGase) activities. Further studies revealed that the N-terminal catalytic domain of CmChi3 exhibited endo-acting and NAGase activities, while the C-terminal catalytic domain showed exo-acting and TGase activities. The hydrolytic properties and favorable environmental adaptations indicate that CmChi3 holds potential for commercial GlcNAc production from chitin.
Introduction
Chitin, a linear β-1,4-linked biopolymer of N-acetyl-D-glucosamine (GlcNAc), is the second most abundant renewable biomass in nature after cellulose (Kaur and Dhillon, 2015). Chitin is present as a structural constituent in the exoskeletons of arthropods, crustacean shells, and fungal cell walls (Wan and Tai, 2013). About 6–8 million tons of waste crab, shrimp, and lobster shells are produced and discarded annually worldwide, which results in wastage of resources and environmental problems (Yan and Chen, 2015). GlcNAc, which is the monomeric unit of chitin, exhibits many bioactivities and used widely in several fields, such as the food, pharmaceutical, agriculture, and fine chemical industries (Chen et al., 2010). Furthermore, GlcNAc contains nitrogen and is an ideal feedstock for manufacturing diverse intermediates of high value, such as 3-acetamido-5-acetylfuran (3A5AF) (Zang et al., 2021) and 5-hydroxymethylfurfural (5-HMF) (Zhou et al., 2020). Therefore, it would be economically and environmentally beneficial to produce GlcNAc from the abundant chitin resources, such as waste shrimp and crab shells.
Methods involving the conversion of chitin to GlcNAc have been widely explored to date (Husson et al., 2017; Zhang A. et al., 2021). In the commercial production of GlcNAc from chitin, acid hydrolysis at high temperatures is often used. However, with the increased awareness on environmental protection, chemical methods are not preferred as they are associated with pollution (Wei et al., 2017). Therefore, researchers have recently paid more attention to the enzymatic production of GlcNAc by the hydrolysis of chitin using chitin-degrading enzymes. This enzymatic process involves mild production conditions, provides high yields, and results in a product with high bioactivity (Chen et al., 2010).
Chitin-degrading enzymes, which are essential for chitin degradation, can be divided into endochitinase [randomly cleaves chitin at internal sites to release N-acetyl chitooligosaccharides (N-acetyl COSs)], exochitinase (hydrolyzes chitin oligosaccharides to liberate GlcNAc dimer), and N-acetyl-β-glucosaminidase (converts N-acetyl COSs to GlcNAc) (Dahiya et al., 2006). To efficiently convert chitin to GlcNAc, a multienzyme system containing at least one chitinase and one N-acetyl-β-glucosaminidase (NAGase) is often required. However, using multiple enzymes increases the costs, which limits their industrial application (Zhu et al., 2016). Therefore, identifying a multiple-function chitinase that can degrade chitin to GlcNAc and constructing a single-enzyme catalytic system are likely to be advantageous in reducing the cost and simplifying the conversion process. Several reports have shown that some chitinases have multiple catalytic functions and possess NAGase and chitinase activities (Fu et al., 2014; Zhang et al., 2018). However, these multifunctional enzymes always produce and accumulate GlcNAc dimer in the process of hydrolyzing chitin to prepare GlcNAc.
In our previous study, the chitinolytic bacterium Chitinolyticbacter meiyuanensis was isolated, and its extracellular chitin-degrading enzymes were found to efficiently hydrolyze chitin and produce GlcNAc as the sole product (Hao et al., 2011). Furthermore, several key chitinases and their coding genes were identified by combining whole-genome and peptide fingerprint analysis. Of these, a chitinase (ORF3769) containing two GH18 catalytic domains was observed to degrade chitin to GlcNAc without accumulating GlcNAc dimer. To the best of our knowledge, this is the first report on a chitinase with dual GH18 catalytic domains from Chitinolyticbacter species. However, its catalytic properties and specific hydrolysis pattern remain unclear.
In this study, the gene encoding ORF3769 (named CmChi3) was cloned from strain SYBC-H1 and heterologously expressed in Escherichia coli BL21(DE3). Sequence analysis, enzymatic properties, and hydrolysis pattern of CmChi3 were investigated. Furthermore, the individual GH18 catalytic domains of CmChi3 were studied to determine the hydrolysis mechanism. The findings were expected to provide a detailed understanding of CmChi3 and its usefulness in directly generating GlcNAc from chitin.
Materials and Methods
Chemicals
Chitin was purchased from Aladdin Reagent Co., Ltd. (Shanghai, China). The standards of N-acetyl COSs (purity: ≥95%) with a degree of polymerization between 2 and 6 were acquired from Qingdao Bozhi Biotechnology Co., Ltd. (Qingdao, China). Peptone and yeast extract were purchased from Oxoid Co., Ltd. (Beijing, China). All molecular reagents were purchased from TaKaRa Co., Ltd. (Dalian, China). Colloidal chitin (CC) was prepared as described by the method of Gao et al. (2015). Other chemicals and solvents used in this study were purchased from local suppliers and were of analytical grade.
Strains, Culture Conditions, and Plasmids
Chitinolyticbacter meiyuanensis SYBC-H1 (ATCC BAA-2140) used in this study was isolated and cultivated according to our previous study (Zhang et al., 2018). The E. coli strains, plasmids, and primers used in this study are listed in Supplementary Table 1. The E. coli strains were routinely cultivated aerobically at 37°C in Luria–Bertani (LB) medium (10 g/l tryptone, 5 g/l yeast extract, and 5 g/l NaCl). The transformants were selected on LB plates containing 50 mg/l of ampicillin.
Identification of Wild-Type CmChi3 From Chitinolyticbacter meiyuanensis SYBC-H1
Wild-type CmChi3 with chitinase activity was purified using chitinase–glycogen complex precipitation followed by autodigestion of the complex according to the method described in our previous study (Zhang et al., 2018). The corresponding protein (CmChi3) in the native polyacrylamide gel was sliced for peptide fingerprint analysis using the electrospray ionization quadrupole time-of-flight mass spectrometry (ESI-Q-TOF MS/MS) technique (PROTTECH, Inc., Suzhou, China). These masses were then compared with the theoretical values in Mascot website databases1 to discern the amino acid sequences of the peptide fragments. The sequences were then aligned with the genome of strain SYBC-H1 to find CmChi3 and its coding gene.
Gene Cloning
The genomic DNA of strain SYBC-H1 was used as the template for polymerase chain reaction (PCR) amplification. The plasmid templates with suitable combinations of primers listed in Supplementary Table 1 were used to generate CmChi3, N-terminal GH18 catalytic domain (CmChi3nGH18), and C-terminal catalytic domain (CmChi3cGH18). The PCR conditions were as follows: 2 min at 95°C, followed by 35 cycles of 95°C for 20 s, 55–62°C for 20 s, and 72°C for 30–90 s, with a final extension at 72°C for 5 min. After purifying by gel electrophoresis, the PCR products and the vectors were subjected to double digestion using restriction enzymes (NdeI and EcoRI; EcoRI and HindIII, respectively), followed by ligation using T4 DNA ligase (TaKaRa, Dalian, China). The recombinant plasmids were transformed into E. coli DH5α and sequenced by GenScript Biotech (Nanjing, China).
Sequence Analysis of CmChi3
Nucleotide and amino acid sequences were analyzed using the Snap Gene v.1.1.3 software and the ExPASy Protparam tool.2 The conserved domains and glycoside hydrolase (GH) family classification were identified using the SMART website.3 DNA and protein sequence alignments were performed using the National Center for Biotechnology Information (NCBI) server with programs BLASTN and BLASTP,4 respectively. Phylogenetic trees were constructed using the neighbor-joining algorithm in MEGA v.7.0 software and assessed using 1,000 bootstrap replications. The presence of a signal peptide and the enzyme location were analyzed using the SignalP v.5.0 server5 and the Gneg-mPLoc server v.2.0,6 respectively. Protein homologous sequence alignment was performed using ClustalX v.2.1 software and ESPript v.3.0.7 The three-dimensional (3D) structure of CmChi3 was predicted using I-TASSER.8
Gene Expression and Protein Purification
The positive clones were directly screened by colony PCR, transformed into E. coli BL21(DE3), inoculated in 10 ml LB medium containing 50 mg/l of ampicillin, and cultured at 37°C in a shaker (200 rpm). Once the optical density (OD600) of the culture broth reached 0.6–0.8, isopropyl-β-D-thio-galactopyranoside (IPTG) was added to a final concentration of 1 mM for induction, and the culture was further incubated at 18°C for 20 h. The cells were harvested by centrifugation (6,000 rpm for 10 min at 4°C); after which, they were resuspended in His-tag binding buffer (20 mM Tris–HCl, 500 mM NaCl, 20 mM imidazole, pH 8.0) and lysed by ultrasonication (JY92-IIN, Ningbo Xinzhi Biotechnology, Ltd., Ningbo, China). The cell debris was removed by centrifugation at 8,000 rpm for 30 min at 4°C, and the supernatant was retained as the crude enzyme. The recombinant proteins (chitinases) were purified using a fast protein liquid chromatography (FPLC) system (AKTA Pure 150; GE healthcare Co., Fairfield, CA, United States) with a Ni-nitrilotriacetic acid affinity chromatography (Ni-NTA) column (His Trap FF 5 ml). The target proteins were eluted with elution buffer (20 mM Tris–HCl, 500 mM NaCl, 250 mM imidazole, pH 8.0). The eluted fractions were passed through ultrafiltration tubes of 50 and 10 kDa (Millipore, Burlington, MA, United States) to remove imidazole with 50 mM phosphate buffer saline (PBS, pH 7.0) and concentrate the enzyme solution. All steps in the enzyme purification were performed at 4°C.
Determination of Enzymatic Activity
Activity assay toward the various substrates was performed using the 3,5-dinitrosalicylic acid (DNS) method (Breuil and Saddler, 1985). Unless otherwise indicated, the reaction mixture containing the suitably diluted enzyme and different polysaccharide substrates at a final concentration of 10 g/l in 100 mM sodium phosphate buffer (pH 6.0) was incubated for 30 min at 50°C. The absorbance was measured at 540 nm, and a standard curve was constructed to determine the amount of reducing sugars produced. One unit of chitinase activity was defined as the amount of enzyme required to produce 1 μmol of reducing sugar at 50°C in 1 min. All chitinase activities were assayed in triplicate, and the average enzyme activity with standard deviation was calculated.
Determination of Protein Concentration and Molecular Weight
Protein content was determined according to the method of Bradford (1976) using bovine serum albumin as the standard. The specific activity was expressed as units per milligram protein.
The molecular weights and purities of protein samples were analyzed by reductive sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 20 mM β-mercaptoethanol incubation. A premixed protein marker (TaKaRa Biotechnology Co., Ltd., Nanjing, China) containing 180−, 130−, 95−, 70−, 53−, 40−, 33−, 25−, 17−, and 10-kDa proteins was used as the molecular mass standards.
Enzymatic Characterization of Recombinant CmChi3
The optimal temperature for chitinase activity was determined over the range of 25–60°C in 100 mM sodium citrate buffer (pH 6.0) by using 10 g/l CC as the substrate. The thermostability of CmChi3 was determined by measuring the residual activity after pre-incubation of the purified enzyme in 100 mM sodium citrate buffer (pH 6.0) at 25–60°C in the absence of the substrate for 2 h. The optimal pH for the chitinase activity was assessed in several buffers at 50°C. The following buffers were used: 100 mM sodium citrate buffer, pH 3.0–6.0; 100 mM phosphate buffer, pH 6.0–8.0; Tris–HCl buffer, pH 8.0–9.0; and 0.4 mol/l glycine sodium hydroxide buffer, pH 9.0–10.0. For pH stability, the CmChi3 was pre-incubated in buffers with different pH at 4°C for 2 h, and the residual activities were determined using 10 g/l of CC as the substrate.
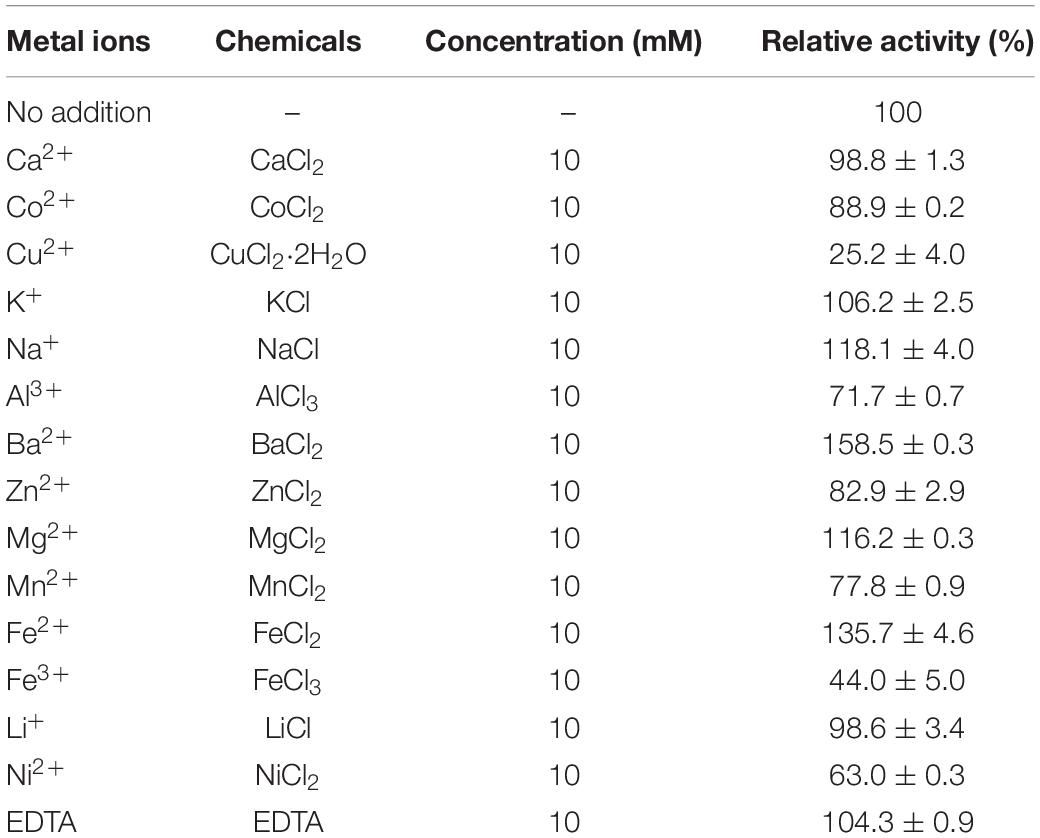
The effects of metal ions on the activity were also determined in this study. Purified CmChi3 was treated with 10 mM ethylenediaminetetraacetic acid (EDTA) for 5 h at 4°C and then dialyzed with 50 mM PBS (pH 7.0) to remove the EDTA. The activities were assayed as described previously and compared with that of an untreated enzyme solution incubated under similar conditions. For reactivation, the metal-free enzyme was incubated with metal ions (Ca2+, Cu2+, Co2+, K+, Na+, Al3+, Ba2+, Ni2+, Zn2+, Mg2+, Mn2+, Fe3+, and Fe2+) at a final concentration of 10 mM for 30 min, and the residual activity was determined. The activity prior to EDTA treatment was used as the control (100%).
Substrate Specificity and Kinetic Parameters of Recombinant CmChi3
The substrate specificity was determined using 1% (w/v) solutions of various polysaccharides [CC, α-chitin powder, β-chitin powder, carboxymethyl cellulose (CMC), hemicellulose powder, and chitosan powder] under the optimum conditions. The amount of reducing sugars released from these polysaccharide substrates was estimated using the DNS method as described previously.
The kinetic parameters against CC under optimum conditions for recombinant CmChi3 were measured. The initial velocities were determined by incubating 20 μg of purified CmChi3 with CC of concentrations ranging from 1 to 20 g/l at 50°C in 1-ml reaction system (100 mM sodium citrate buffer, pH 6.0) for 30 min. The amount of reducing sugars released from these substrates was estimated using the DNS method as described previously. Km and Vmax values were obtained by Lineweaver–Burk plots (Price, 1985).
Hydrolysis Pattern of the Recombinant CmChi3, CmChi3nGH18, and CmChi3cGH18 Toward Colloidal Chitin
The reaction system (1 ml of 100 mM sodium citrate buffer, pH 6.0) contained CC (10 g/l) and 20 μg each of purified CmChi3, CmChi3nGH18, and CmChi3cGH18. The reaction was performed at 50°C for various time intervals. The samples were boiled for 5 min to stop the reaction.
Hydrolysis Pattern of the Recombinant CmChi3, CmChi3nGH18, and CmChi3cGH18 Toward N-Acetyl Chitooligosaccharides
Reaction mixtures containing purified enzymes (0.1 μg) and N-acetyl COSs (DP 2–6) at a final concentration of 10 g/l were incubated in a 20-μl volume of 100 mM sodium citrate buffer (pH 6.0) at 50°C for various time intervals. The reactions were stopped by boiling for 5 min.
High-Performance Liquid Chromatography Analysis of the Products
The products were detected using an Agilent 1260 series liquid chromatography system (Agilent Technologies, Santa Clara, CA, United States), according to our previous study (Zhang et al., 2018).
Nucleotide Sequence Accession Number
The sequence of the chitinase gene CmChi3 was deposited in the GenBank database under the accession number MZ559373.
Results and Discussion
Sequence Analysis of CmChi3
The CmChi3 gene (2,943 bp) encodes a protein of 980 amino acids with a calculated molecular mass and the predicted isoelectric (pI) point of 103 kDa and 6.64, respectively. The signal peptide prediction indicated the presence of a putative N-terminal signal peptide (residue 1–33) in the sequence of CmChi3, which signified that CmChi3 should be a secretory protein and explained the identification of CmChi3 in the fermentation broth of strain SYBC-H1.
According to the Carbohydrate-Active enZYmes (CAZy) database,9 chitinolytic enzymes are mainly divided into GH families 18, 19, and 20 based on amino acid sequence similarity. BLASTP analysis showed that CmChi3 belonged to GH family 18 (GH18) and shared the highest identity (74.52%) with the GH18 chitinase from Chitiniphilus eburneus (WP_136773598), followed by a hypothetical protein (73.57%) from Chitiniphilus shinanonensis (WP_018748580). However, these coding genes have not been expressed and studied. Among the characterized GH18 chitinases, CmChi3 showed the highest identity (54.42%) with the GH18 chitinase from Chromobacterium sp. C-61 (AAP88583) (Park et al., 2007), followed by the GH18 chitinase (39.48%) from Bacillus cereus (AAM48520) (Huang and Chen, 2005) and GH18 chitinase (27.46%) from Clostridium paraputrificum (BAA34922) (Morimoto et al., 1999). For chitinases containing two GH18 catalytic domains, ChiW (27.13%) from Paenibacillus sp. (BAM67143) (Itoh et al., 2013), and ChiB (25.86%) from Saccharophagus degradans 2–40 (DAA01334) (Howard et al., 2004), the putative GH18 chitinase with the highest similarity to CmChi3 and the verified GH18 chitinase were selected to construct the phylogenic tree, which also showed that CmChi3 exhibited a low sequence identity with most of the functionally characterized bacterial GH18 chitinases (Figure 1). Multiple alignments of the individual GH18 domains of CmChi3 with other GH18 chitinases from different sources indicated sequence DxDxE in CmChi3nGH18 and CmChi3cGH18 is the highly conserved amino acid sequence among the GH18 family members. DxDxE is a commonly conserved catalytic motif in GH18 family members, in which glutamic acid (E) residue is considered as proton donors in the catalytic process (Supplementary Figure 1; Dai et al., 2021).
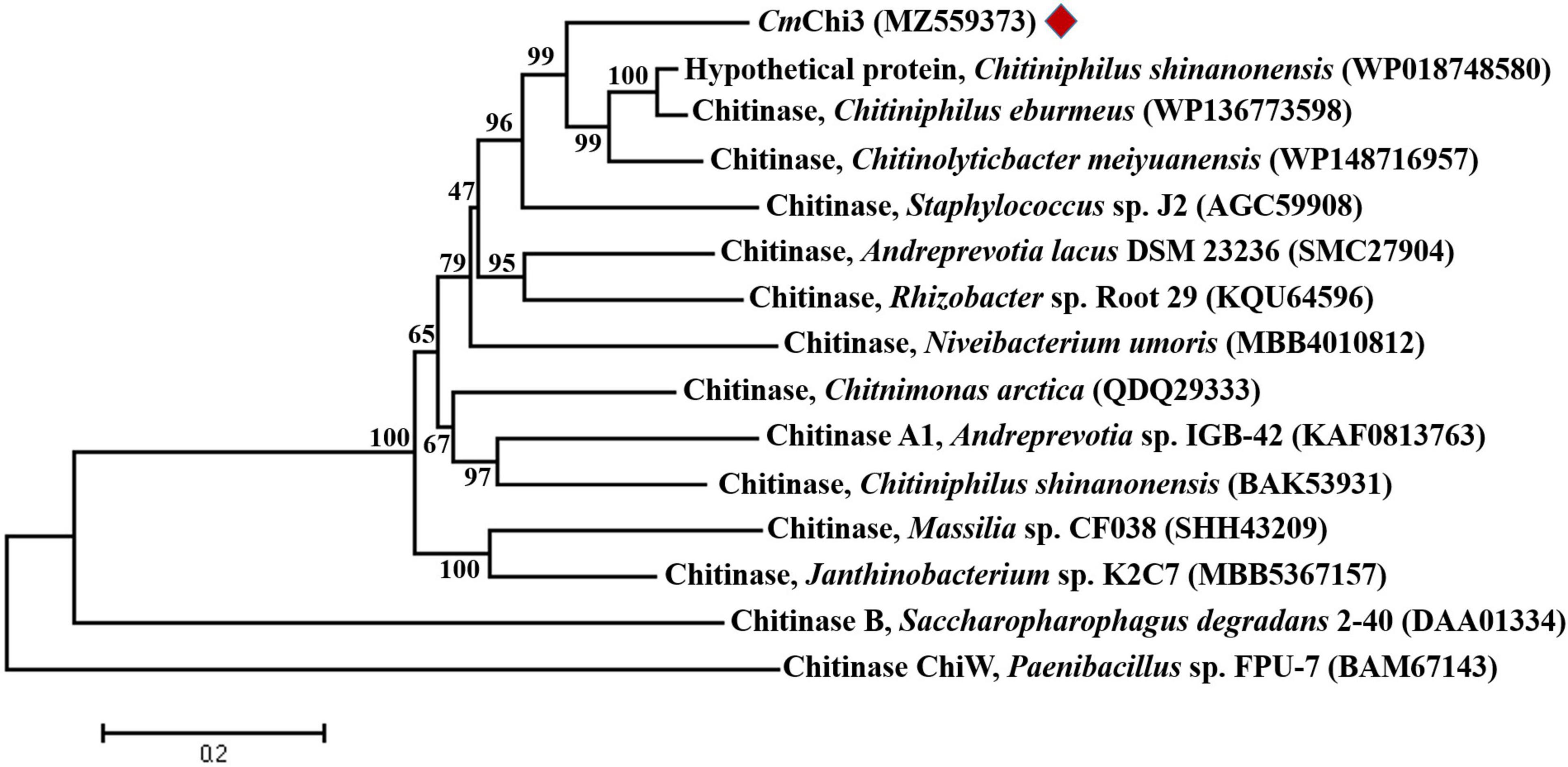
Figure 1. Phylogenetic relationships between chitinase CmChi3 and other bacterial chitinases from the GH family 18. The phylogenetic tree was constructed using the neighbor-joining algorithm based on the amino acid sequence alignment in MEGA v.7.0. The amino acid sequence of CmChi3 was aligned with those of the following proteins: GH18 chitinases from C. shinanonensis (WP018748580), C. eburmeus (WP136773598), C. meiyuanensis (WP148716957), Staphylococcus sp. J2 (AGC59908), A. lacus DSM 23236 (SMC27904), Rhizobacter sp. Root 29 (KQU64596), Niveibacterium umoris (MBB4010812), Chitnimonas arctica (QDQ29333), Andreprevotia sp. IGB-42 (KAF0813763), C. shinanonensis (BAK53931), Massilia sp. CF038 (SHH43209), Janthinobacterium sp. K2C7 (MBB5367157), Saccharopharophagus degradans 2–40 (DAA01334), and Paenibacillus sp. FPU-7 (BAM67143).
Domain structure prediction indicated that CmChi3 possesses two GH18 catalytic domains (residues 37–329 and 564–967) (Figure 2A). This result is contrary to most GH18 chitinases, which usually possess one GH18 catalytic domain. To date, few microbial chitinases with dual GH18 catalytic domains have been characterized. Meanwhile, accessory binding modules and the catalytic domain can improve the substrate accessibility of chitinases. CmChi3 also contains two family 5 carbohydrate-binding modules (residues 368–411 and 464–510), which showed that it might possess good binding abilities for the chitinous substrate. As shown in Figure 2, the 3D structure of CmChi3 was predicted based on the structure model from I-TASSER, which revealed that the amino acid sites F71, S112, D147, D149, E151, M217, D218, M265, W299, Y568, F600, D739, D741, E743, M813, Y815, D816, Y874, and W962 were the active sites of N- and C-terminal catalytic domains, respectively (Supplementary Figure 1). These amino acids possibly form active pockets.
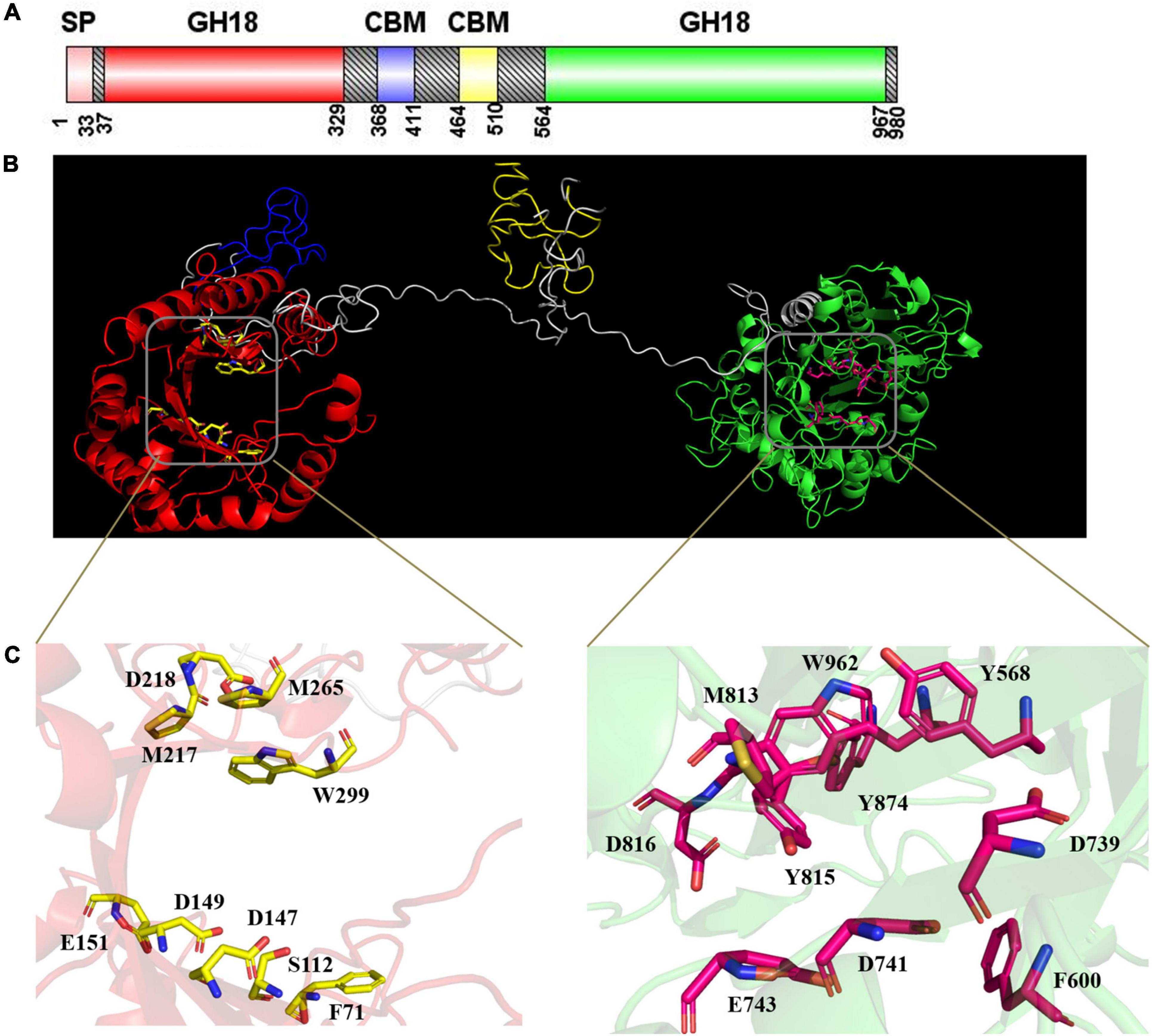
Figure 2. (A) Structural features of CmChi3. A putative signal sequence, two GH18 catalytic domains, and two carbohydrate-binding modules (CBMs) are indicated. This image was generated using IBS Illustrator (Liu et al., 2015). (B) The prediction of the 3D structure of CmChi3. Red indicates Glyco_18 domain; blue indicates CBM; yellow indicates CBM; green indicates Glyco_18 domain; gray indicates unknown region. (C) The active sites of CmChi3. D147, D149, and E151 were the active residues of the N-terminal catalytic domain; D739, D741, and E743 were the active residues of the C-terminal catalytic domain.
Cloning, Expression, and Purification of the Recombinant CmChi3
The gene encoding CmChi3 without the signal peptide sequence (2,844 bp) was successfully amplified and actively expressed in E. coli BL21(DE3). As shown in Supplementary Figure 2, the localization of CmChi3 was in the supernatant (lane 1) of the cell-free extract of the recombinant E. coli BL21(DE3)-(pCold I-CmChi3), indicating that CmChi3 was actively expressed. The recombinant CmChi3 with N-terminal His-tag was purified by NTA affinity chromatography (lane 2). The SDS-PAGE analysis showed that purified recombinant CmChi3 possesses a high purity with an approximate molecular weight of 110 kDa, which agrees with the 99.7 kDa calculated from the amino acid sequence (without signal peptide) containing the His-tag. The recombinant CmChi3 eluted with 250 mM imidazole from a Ni-NTA resin with a recovery yield of 49.1% (Supplementary Table 3).
Effects of Temperature and pH on the Enzymatic Activity and Stability of Recombinant CmChi3
The effect of temperature on CmChi3 activity was studied. The results showed that recombinant CmChi3 exhibited maximum activity at a temperature of around 50°C (Figure 3A). This result is comparable to that of some bacterial chitinases, such as CsChiE from C. shinanonensis (50°C) (Rani et al., 2020) and PbChi70 from Paenibacillus barengoltzii (55°C) (Yang et al., 2016). Few chitinases with dual GH18 domains are ChiW from Paenibacillus sp. (50°C) (Itoh et al., 2014), Tk-ChiA from Thermococcus kodakaraensis KOD1 (85°C) (Tanaka et al., 1999), and chitinase P1724 from a wetland soil metagenome (40°C) (Dai et al., 2021). Additionally, CmChi3 was stable and retained more than 90% activity for 2 h at temperatures below 40°C, but it was unstable at temperatures above 50°C.
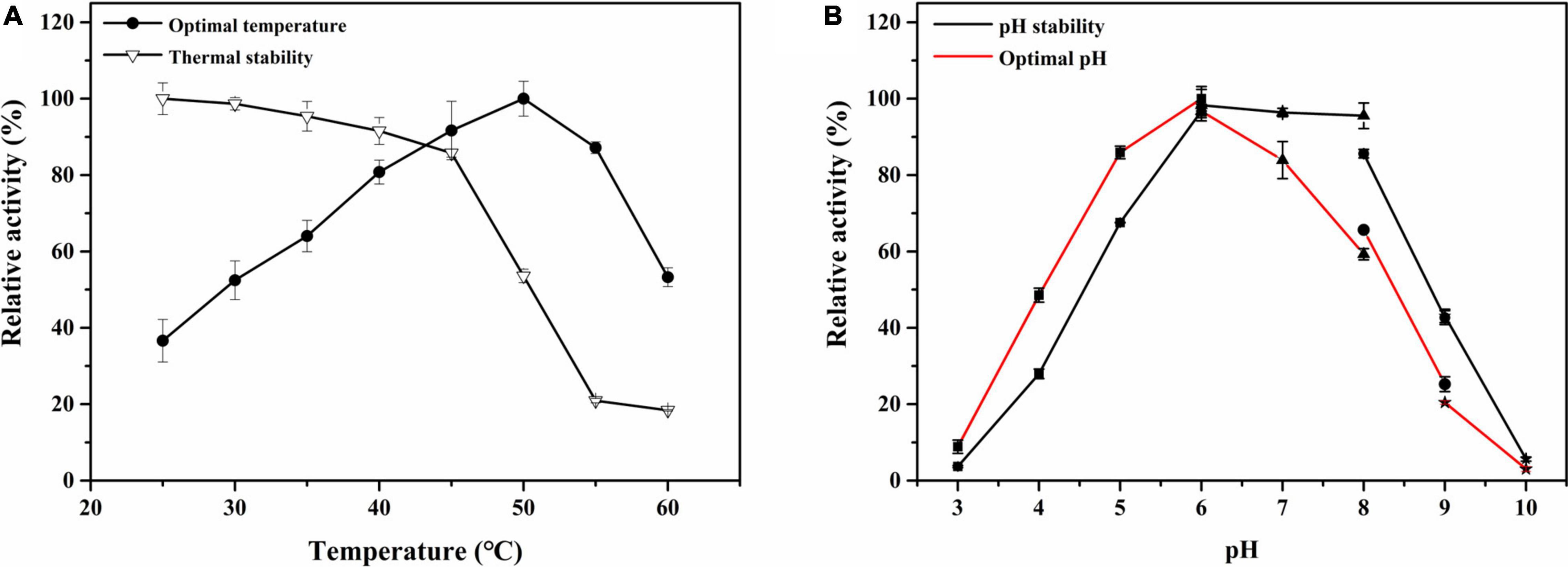
Figure 3. The effect of pH and temperature on the activity and stability of CmChi3. (A) Optimal temperature and thermal stability of the recombinant CmChi3. The optimum temperature was determined at different temperatures (25–60°C) in 100-mM sodium citrate (pH 6.0) (solid rounds). The residual activity was measured in 100-mM sodium citrate (pH 6.0) after the enzyme was treated for 2 h at different temperatures (open triangles) to determine the thermostability. (B) Optimal pH and pH stability of the recombinant CmChi3. The optimal pH was determined in various buffers within the pH range of 3.0–10.0 [filled square (■), sodium citrate buffer (pH 3.0–6.0); filled triangle (▲), phosphate buffer (pH 6.0–8.0); filled circle (●), Tris–HCl buffer (pH 8.0–9.0); and filled star (★), 0.4 mol/l glycine sodium hydroxide buffer (pH 9.0–10.0)] (red line). The enzyme was incubated at 4°C for 2 h with various pH buffers (black line) to determine pH stability, and the residual activities were measured.
The effects of pH on the recombinant CmChi3 were determined in the range of pH 3.0–10.0. As shown in Figure 3B, CmChi3 exhibited an optimum activity around pH 6.0. Meanwhile, the enzymatic activity of CmChi3 dropped more than 50% at pH ≤ 4.0 and pH ≥ 8.5 (Figure 3B). This result is similar to that of other bacterial chitinases, such as CsChiE from C. shinanonensis (pH 6.0) (Rani et al., 2020) and spChiD from Serratia proteamaculans (pH 6.0) (Purushotham and Podile, 2012). However, the optimum pH of chitinases from Microbispora sp. V2 (pH 3.0) (Nawani et al., 2002), Chitinibacter sp. GC72 (pH 6.8) (Gao et al., 2015), and Paenibacillus pasadenensis NCIM 5434 (pH 10.0) (Loni et al., 2014) was different from CmChi3. With regard to pH stability, CmChi3 retained more than 60% of its activity after storage at pH 5.0–8.5 for 2 h. This result is comparable to those of other chitinases from C. meiyuanensis, including CmChi1 (pH 5.2–8.2) (Zhang et al., 2018) and CmNAGase (pH 4.0–8.5) (Zhang et al., 2020a).
Effects of Metal Ions on the Activity of Recombinant CmChi3
The effects of metal ions on CmChi3 activity were investigated. All counter-ions of the used metal ions were Cl–. As shown in Table 1, EDTA did not affect the enzymatic activity, which indicates that CmChi3 is non-metal dependent. The activity was strongly inhibited by Cu2+, Ni2+, and Fe3+. The presence of Fe2+, Mg2+, Ba2+, and Na+ had a strengthening effect on CmChi3 activity. Previous works also have shown that Cu2+, Ni2+, and Fe3+ inhibit the activity of chitinases. For example, the chitinase ChiW from Paenibacillus sp. was inhibited by Cu2+ and Fe3+ (Itoh et al., 2014). The chitinase ChiA-Ba43 from Bacillus altitudinis KA15 was inhibited by Ni2+ (Asmani et al., 2020).
Substrate Specificity and Kinetic Parameters of Recombinant CmChi3
The hydrolysis ability of CmChi3 toward various insoluble polysaccharide substrates was investigated. Of the substrates tested (Table 2), CC was most effectively hydrolyzed by the CmChi3 with a specific activity of 4.1 U/mg, similar to the chitinase with dual catalytic domains, such as ChiW with a specific activity of 5.2 U/mg (Itoh et al., 2014). The activity was higher than that of most other reported chitinases, including chitinase from Streptomyces speibonae TKU048 (0.32 U/mg) (Thi Ngoc et al., 2019), chitinase CHI from Paenibacillus chitinolyticus (0.75 U/mg) (Liu C. et al., 2020), and Tk-ChiA from T. kodakaraensis KOD1 (2.4 U/mg) (Tanaka et al., 2001), but lower than that of PxChi52 from Paenibacillus xylanexedens (16 U/mg) (Zhang W. et al., 2021) and CmChi1 from C. meiyuanensis (15.3 U/mg) (Zhang et al., 2018). CmChi3 displayed low activities toward α-chitin (0.02 U/mg) and β-chitin powder (0.3 U/mg). No activity was found toward CMC, hemicellulose, and chitosan (Table 2).
Furthermore, the kinetic parameters of recombinant CmChi3 toward CC were investigated (Supplementary Table 2). The [s]-velocity plots of CC are shown in Supplementary Figure 3. The Km, kcat, and kcat/Km values were determined to be 7.53 ± 0.78 mg/ml, 9.08 ± 0.36 s–1, and 1.2 ± 0.11 ml/s/mg for CC (Supplementary Table 2).
Hydrolysis Pattern of the Recombinant CmChi3
Colloidal chitin and N-acetyl chitooligosaccharides (DP 2–6) were used as hydrolytic substrates to evaluate the hydrolysis pattern of CmChi3. Our previous study confirmed that CmChi3 can completely convert CC solely to GlcNAc when using a high enzyme concentration (50 μg) (Zhang et al., 2020b). In this study, a low concentration of enzyme (20 μg) was used to study the specific intermediates of CC hydrolysis. As shown in Figure 4A, CmChi3 hydrolyzed CC to mainly produce GlcNAc, with a small amount of (GlcNAc)2 and (GlcNAc)3 in 30 min. Then, it yielded GlcNAc as the sole end product after 120 min, suggesting that CmChi3 possesses endochitinase and NAGase activities. Previously reported multifunctional chitinases CmChi1 from C. meiyuanensis (Zhang et al., 2018) and PbChi74 from P. barengoltzii (Fu et al., 2014) hydrolyzed CC to produce (GlcNAc)2 in the initial hydrolysis stage. Also, (GlcNAc)2 needed a longer reaction time to achieve full conversion to GlcNAc, which is different from our observation.
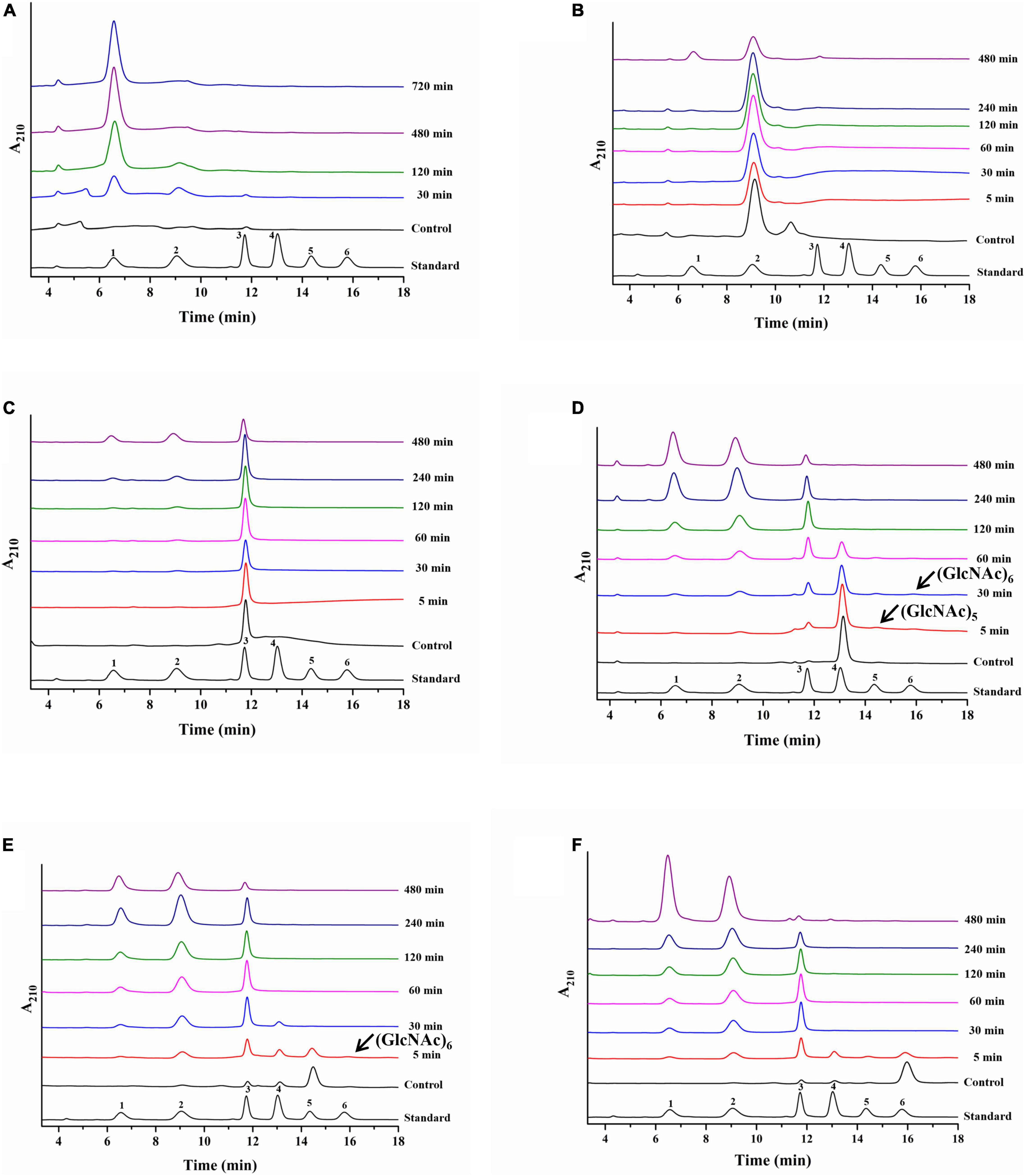
Figure 4. Hydrolysis pattern of CmChi3 toward CC and N-acetyl COSs. The reactions containing 20 μg CmChi3 and 10 g/l CC or 0.1 μg CmChi3 and 10 g/l (GlcNAc)2–6 were performed in sodium citrate buffer (pH 6.0) at 50°C. Aliquots were withdrawn at different time intervals and analyzed by HPLC. (A–F) The hydrolysis products from CC and (GlcNAc)2–6. Numbers 1–6 represent GlcNAc to (GlcNAc)6.
For (GlcNAc)2–6, CmChi3 slowly hydrolyzed (GlcNAc)2 into GlcNAc, showing that CmChi3 possesses NAGase activity. This result is similar to that of other studies (Liu Y. et al., 2020). However, (GlcNAc)2, (GlcNAc)2–3, (GlcNAc)2–4, and (GlcNAc)2–5 were each released from (GlcNAc)3, (GlcNAc)4, (GlcNAc)5, and (GlcNAc)6, respectively, which suggested that CmChi3 has endocleavage activities. Additionally, small amounts of (GlcNAc)5–6 and (GlcNAc)6 were observed when using (GlcNAc)4 and (GlcNAc)5 as substrates, respectively. This finding suggests that CmChi3 has a weak TGase activity toward (GlcNAc)4 and (GlcNAc)5 (Figure 4). Conclusively, CmChi3 is a multifunctional chitinase possessing endochitinase, NAGase, and TGase activities.
Hydrolysis Pattern of the Two GH18 Catalytic Domains, CmChi3nGH18 and CmChi3cGH18
Recently, several reports have shown that a single enzyme containing more than one catalytic domain may be particularly powerful in degrading polysaccharides, such as a cellulase from the thermophilic bacterium Caldicellulosiruptor bescii, which consists of GH9 and GH48 domains (Brunecky et al., 2013). Additionally, an enzyme from the actinobacterium Jonesia denitrificans contains AA10 lytic polysaccharide monooxygenase (LPMO) and GH18 catalytic domain (Mekasha et al., 2020). According to previous reports, chitinases with dual catalytic domains often possess different catalytic activities. For example, in the chitinase Tk-ChiA from T. kodakaraensis KOD1 (Tanaka et al., 2001) and chitinase ChiB from S. degradans 2–40 (Itoh et al., 2013), the N-terminal and C-terminal GH18 domains possessed exo- and endo-activities, respectively. In another study, Larsbrink et al. (2016) showed that chitinase ChiA from Flavobacterium johnsoniae comprised two GH18 domains, one of which was proposed to have a predominantly endo-activity. In contrast, another one was predicted to have an exo-acting activity (Larsbrink et al., 2016).
To determine the hydrolysis pattern of individual GH18 domains of CmChi3, N-terminal (CmChi3nGH18) and C-terminal (CmChi3cGH18) GH18 catalytic domains were successfully expressed and purified (Supplementary Figure 4). Then, the hydrolysis pattern of CmChi3nGH18 and CmChi3cGH18 toward CC and (GlcNAc)2–6 was analyzed. As shown in Supplementary Figure 5, CmChi3nGH18 hydrolyzed CC to produce (GlcNAc)1–3 (Supplementary Figure 5A) and hydrolyzed (GlcNAc)3–6 to (GlcNAc)1–2, (GlcNAc)1–3, (GlcNAc)1–4, and (GlcNAc)1–5, which showed that CmChi3nGH18 possesses endochitinase activity. Additionally, CmChi3nGH18 converted (GlcNAc)2 into GlcNAc (Supplementary Figure 5B), thus exhibiting NAGase activity.
CmChi3cGH18 hydrolyzes CC solely into (GlcNAc)2 (Supplementary Figure 5G). For (GlcNAc)2–6, (GlcNAc)2 could not be hydrolyzed. When using (GlcNAc)3 as the substrate, the products were (GlcNAc)2 and GlcNAc. (GlcNAc)4 was mainly converted into (GlcNAc)2. When using (GlcNAc)5–6 as the substrate, (GlcNAc)2, (GlcNAc)3, and (GlcNAc)4 were generated (Supplementary Figure 5). These results showed that the enzyme has exochitinase activity. Also, CmChi3cGH18 synthesized higher N-acetyl-COSs (GlcNAc)5–6 from (GlcNAc)4 and (GlcNAc)6 from (GlcNAc)5, showing that it possesses TGase activity. This result also explains why little (GlcNAc)3 was observed from (GlcNAc)4 and (GlcNAc)6, (GlcNAc)4 was generated from (GlcNAc)5, and (GlcNAc)5 was obtained from (GlcNAc)6.
The above results showed that CmChi3nGH18 displayed endochitinase and NAGase activities and CmChi3cGH18 showed exochitinase and TGase activities. The results agree with those of CmChi3.
Conclusion
In this study, a novel chitinase (CmChi3) containing dual GH18 catalytic domains and two carbohydrate-binding modules from C. meiyuanensis was cloned, heterologously expressed in E. coli, and biochemically characterized. The analysis of the hydrolysis products revealed that CmChi3 is a multifunctional chitinase, which possesses exochitinase, NAGase, and TGase activities. Further studies on the individual catalytic domains of CmChi3 showed that N-terminal catalytic domain exhibited endo-acting and NAGase activities, while the C-terminal domain showed exo-acting and TGase activities. The multidomain structure of CmChi3 is expected to produce GlcNAc from chitin. This work provides a reference to chitinase with multifunctional domains and GlcNAc enzymatic production from chitin.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CW: writing-original draft preparation, formal analysis, and data curation. XC: investigation and methodology. NZ and YC: methodology. AZ: funding acquisition and writing-review and editing. KC and PO: conceptualization and supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Science Foundation for Young Scientists of China (Grant No. 21908101) and the Open-end Funds of Jiangsu Key Laboratory of Marine Bioresources and Environment (SH20201201).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.790301/full#supplementary-material
Footnotes
- ^ http://www.matrixscience.com
- ^ http://web.expasy.org/protparam/
- ^ http://smart.embl-heidelberg.de/
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://www.csbio.sjtu.edu.cn/bioinf/Gneg-multi/
- ^ http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi
- ^ https://zhanglab.ccmb.med.umich.edu/I-TASSER/
- ^ www.cazy.org/
References
Asmani, K.-L., Bouacem, K., Ouelhadj, A., Yahiaoui, M., Bechami, S., Mechri, S., et al. (2020). Biochemical and molecular characterization of an acido-thermostable endo-chitinase from Bacillus altitudinis KA15 for industrial degradation of chitinous waste. Carbohydr. Res. 495:108089. doi: 10.1016/j.carres.2020.108089
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Breuil, C., and Saddler, J. N. (1985). Comparison of the 3,5-dinitrosalicylic acid and Nelson-Somogyi methods of assaying for reducing sugars and determining cellulase activity. Enzyme Microb. Tech. 7, 327–332. doi: 10.1016/0141-0229(85)90111-5
Brunecky, R., Alahuhta, M., Xu, Q., Donohoe, B. S., Crowley, M. F., Kataeva, I. A., et al. (2013). Revealing nature’s cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342, 1513–1516. doi: 10.1126/science.1244273
Chen, J. K., Shen, C. R., and Liu, C. L. (2010). N-Acetylglucosamine: production and applications. Mar. Drugs 8, 2493–2516. doi: 10.3390/md8092493
Dahiya, N., Tewari, R., and Hoondal, G. S. (2006). Biotechnological aspects of chitinolytic enzymes: a review. Appl. Microbiol. Biotechnol. 71, 773–782. doi: 10.1007/s00253-005-0183-7
Dai, Y., Yang, F., Liu, X., and Wang, H. (2021). The discovery and characterization of a novel chitinase with dual catalytic domains from a Qinghai-Tibetan Plateau wetland soil metagenome. Int. J. Biol. Macromol. 188, 482–490. doi: 10.1016/j.ijbiomac.2021.07.153
Fu, X., Yan, Q., Yang, S., Yang, X., Guo, Y., and Jiang, Z. (2014). An acidic, thermostable exochitinase with beta-N-acetylglucosaminidase activity from Paenibacillus barengoltzii converting chitin to N-acetyl glucosamine. Biotechnol. Biofuels 7:174. doi: 10.1186/s13068-014-0174-y
Gao, C., Zhang, A., Chen, K., Hao, Z., Tong, J., and Ouyang, P. (2015). Characterization of extracellular chitinase from Chitinibacter sp. GC72 and its application in GlcNAc production from crayfish shell enzymatic degradation. Biochem. Eng. J. 97, 59–64. doi: 10.1016/j.bej.2015.02.010
Hao, Z. K., Cai, Y. J., Liao, X. R., Liang, X. H., Liu, J. Y., Fang, Z. Y., et al. (2011). Chitinolyticbacter meiyuanensis SYBC-H1(T), Gen. Nov., sp. Nov., a chitin-degrading bacterium isolated from soil. Curr. Microbiol. 62, 1732–1738. doi: 10.1007/s00284-011-9921-5
Howard, M. B., Ekborg, N. A., Taylor, L. E., Weiner, R. M., and Hutcheson, S. W. (2004). Chitinase B of “Microbulbifer degradans” 2-40 contains two catalytic domains with different chitinolytic activities. J. Bacteriol. 186, 1297–1303. doi: 10.1128/jb.186.5.1297-1303.2004
Huang, C. J., and Chen, C. Y. (2005). High-level expression and characterization of two chitinases, ChiCH and ChiCW, of Bacillus cereus 28-9 in Escherichia coli. Biochem. Biophys. Res. Commun. 327, 8–17. doi: 10.1016/j.bbrc.2004.11.140
Husson, E., Hadad, C., Huet, G., Laclef, S., Lesur, D., Lambertyn, V., et al. (2017). The effect of room temperature ionic liquids on the selective biocatalytic hydrolysis of chitin via sequential or simultaneous strategies. Green Chem. 19, 4122–4131. doi: 10.1039/c7gc01471f
Itoh, T., Hibi, T., Fujii, Y., Sugimoto, I., Fujiwara, A., Suzuki, F., et al. (2013). Cooperative degradation of chitin by extracellular and cell surface-expressed chitinases from Paenibacillus sp. Strain FPU-7. Appl. Microbiol. Biotechnol. 79, 7482–7490. doi: 10.1128/aem.02483-13
Itoh, T., Sugimoto, I., Hibi, T., Suzuki, F., Matsuo, K., Fujii, Y., et al. (2014). Overexpression, purification, and characterization of Paenibacillus cell surface-expressed chitinase ChiW with two catalytic domains. Biosci. Biotechnol. Biochem. 78, 624–634. doi: 10.1080/09168451.2014.891935
Kaur, S., and Dhillon, G. S. (2015). Recent trends in biological extraction of chitin from marine shell wastes: a review. Crit. Rev. Biotechnol. 35, 44–61. doi: 10.3109/07388551.2014.891935
Larsbrink, J., Zhu, Y., Kharade, S. S., Kwiatkowski, K. J., Eijsink, V. G. H., Koropatkin, N. M., et al. (2016). A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin. Biotechnol. Biofuels 9:260. doi: 10.1186/s13068-016-0674-z
Liu, C., Shen, N., Wu, J., Jiang, M., Shi, S., Wang, J., et al. (2020). Cloning, expression and characterization of a chitinase from Paenibacillus chitinolyticus strain UMBR 0002. PeerJ 8:e8964. doi: 10.7717/peerj.8964
Liu, W., Xie, Y., Ma, J., Luo, X., Nie, P., Zuo, Z., et al. (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. doi: 10.1093/bioinformatics/btv362
Liu, Y., Jiang, Z., Ma, J., Ma, S., Yan, Q., and Yang, S. (2020). Biochemical characterization and structural analysis of a beta-N-acetylglucosaminidase from Paenibacillus barengoltzii for efficient production of N-acetyl-D-glucosamine. J. Agric. Food Chem. 68, 5648–5657. doi: 10.1021/acs.jafc.9b08085
Loni, P. P., Patil, J. U., Phugare, S. S., and Bajekal, S. S. (2014). Purification and characterization of alkaline chitinase from Paenibacillus pasadenensis NCIM 5434. J. Basic Microb. 54, 1080–1089. doi: 10.1002/jobm.201300533
Mekasha, S., Tuveng, T. R., Askarian, F., Choudhary, S., Schmidt-Dannert, C., Niebisch, A., et al. (2020). A trimodular bacterial enzyme combining hydrolytic activity with oxidative glycosidic bond cleavage efficiently degrades chitin. J. Biol. Chem. 295, 9134–9146. doi: 10.1074/jbc.RA120.013040
Morimoto, K., Karita, S., Kimura, T., Sakka, K., and Ohmiya, K. (1999). Sequencing, expression, and transcription analysis of the Clostridium paraputrificum chiA gene encoding chitinase ChiA. Appl. Microbiol. Biotechnol. 51, 340–347. doi: 10.1007/s002530051400
Nawani, N. N., Kapadnis, B. P., Das, A. D., Rao, A. S., and Mahajan, S. K. (2002). Purification and characterization of a thermophilic and acidophilic chitinase from Microbispora sp. V2. J. Appl. Microbiol. 93, 965–975. doi: 10.1046/j.1365-2672.2002.01766.x
Park, S. K., Kim, C. W., Kim, H., Jung, J. S., and Harman, G. E. (2007). Cloning and high-level production of a chitinase from Chromobacterium sp and the role of conserved or nonconserved residues on its catalytic activity. Appl. Microbiol. Biotechnol. 74, 791–804. doi: 10.1007/s00253-006-0614-0
Price, N. C. (1985). The determination of Km values from Lineweaver-Burk plots. Biochem. Educ. 13:81. doi: 10.1016/0307-4412(85)90016-0
Purushotham, P., and Podile, A. R. (2012). Synthesis of long-chain chitooligosaccharides by a hypertransglycosylating processive endochitinase of Serratia proteamaculans 568. J. Bacteriol. 194, 4260–4271. doi: 10.1128/jb.06473-11
Rani, T. S., Madhuprakash, J., and Podile, A. R. (2020). Chitinase-E from Chitiniphilus shinanonensis generates chitobiose from chitin flakes. Int. J. Biol. Macromol. 163, 1037–1043. doi: 10.1016/j.ijbiomac.2020.07.052
Tanaka, T., Fujiwara, S., Nishikori, S., Fukui, T., Takagi, M., and Imanaka, T. (1999). A unique chitinase with dual active sites and triple substrate binding sites from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. Appl. Environ. Microb. 65, 5338–5344. doi: 10.1128/AEM.65.12.5338-5344.1999
Tanaka, T., Fukui, T., and Imanaka, T. (2001). Different cleavage specificities of the dual catalytic domains in chitinase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biol. Chem. 276, 35629–35635. doi: 10.1074/jbc.M105919200
Thi Ngoc, T., Chien Thang, D., Minh Trung, N., Van Bon, N., Thi Phuong Khanh, V., Anh Dzung, N., et al. (2019). An exochitinase with N-Acetyl-beta-glucosaminidase-like activity from shrimp head conversion by Streptomyces speibonae and its application in hydrolyzing beta-chitin powder to produce N-acetyl-d-glucosamine. Polymers (Basel) 11:1600. doi: 10.3390/polym11101600
Wan, A. C., and Tai, B. C. (2013). Chitin - a promising biomaterial for tissue engineering and stem cell technologies. Biotechnol. Adv. 31, 1776–1785. doi: 10.1016/j.biotechadv.2013.09.007
Wei, G., Zhang, A., Chen, K., and Ouyang, P. (2017). Enzymatic production of N-acetyl-D-glucosamine from crayfish shell wastes pretreated via high pressure homogenization. Carbohydr. Polym. 171, 236–241. doi: 10.1016/j.carbpol.2017.05.028
Yang, S., Fu, X., Yan, Q., Guo, Y., Liu, Z., and Jiang, Z. (2016). Cloning, expression, purification and application of a novel chitinase from a thermophilic marine bacterium Paenibacillus barengoltzii. Food Chem. 192, 1041–1048. doi: 10.1016/j.foodchem.2015.07.092
Zang, H., Lou, J., Jiao, S., Li, H., Du, Y., and Wang, J. (2021). Valorization of chitin derived N-acetyl-D-glucosamine into high valuable n-containing 3-acetamido-5-acetylfuran using pyridinium-based ionic liquids. J. Mol. Liquids 330:115667. doi: 10.1016/j.molliq.2021.115667
Zhang, A. L., He, Y. M., Wei, G. G., Zhou, J., Dong, W. L., Chen, K. Q., et al. (2018). Molecular characterization of a novel chitinase CmChi1 from Chitinolyticbacter meiyuanensis SYBC-H1 and its use in N-acetyl-D-glucosamine production. Biotechnol. Biofuels 11:179. doi: 10.1186/s13068-018-1169-x
Zhang, A., Mo, X., Zhou, N., Wang, Y., Wei, G., Chen, J., et al. (2020a). A novel bacterial beta-N-acetyl glucosaminidase from Chitinolyticbacter meiyuanensis possessing transglycosylation and reverse hydrolysis activities. Biotechnol. Biofuels 13:115. doi: 10.1186/s13068-020-01754-4
Zhang, A., Mo, X., Zhou, N., Wang, Y., Wei, G., Hao, Z., et al. (2020b). Identification of chitinolytic enzymes in Chitinolyticbacter meiyuanensis and mechanism of efficiently hydrolyzing chitin to N-Acetyl glucosamine. Front. Microbiol. 11:572053. doi: 10.3389/fmicb.2020.572053
Zhang, A., Wang, C., Chen, J., Wei, G., Zhou, N., Li, G., et al. (2021). Efficient enzymatic hydrolysis of chitin into N-acetyl glucosamine using alkali as a recyclable pretreatment reagent. Green Chem. 23, 3081–3089. doi: 10.1039/d1gc00818h
Zhang, W., Ma, J., Yan, Q., Jiang, Z., and Yang, S. (2021). Biochemical characterization of a novel acidic chitinase with antifungal activity from Paenibacillus xylanexedens Z2-4. Int. J. Biol. Macromol. 182, 1528–1536. doi: 10.1016/j.ijbiomac.2021.05.111
Zhou, D., Shen, D., Lu, W., Song, T., Wang, M., Feng, H., et al. (2020). Production of 5-hydroxymethylfurfural from chitin biomass: a review. Molecules 25:541. doi: 10.3390/molecules25030541
Keywords: chitin, CmChi3, dual functional domains, N-acetyl-D-glucosamine, hydrolysis pattern, chitinase
Citation: Wang C, Chen X, Zhou N, Chen Y, Zhang A, Chen K and Ouyang P (2022) Property and Function of a Novel Chitinase Containing Dual Catalytic Domains Capable of Converting Chitin Into N-Acetyl-D-Glucosamine. Front. Microbiol. 13:790301. doi: 10.3389/fmicb.2022.790301
Received: 06 October 2021; Accepted: 17 January 2022;
Published: 24 February 2022.
Edited by:
Saurabh Dhiman, South Dakota School of Mines and Technology, United StatesReviewed by:
Santhosh Pillai, Durban University of Technology, South AfricaPrabhu Ponnandy, University of Michigan, United States
Copyright © 2022 Wang, Chen, Zhou, Chen, Zhang, Chen and Ouyang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alei Zhang, zhangalei@njtech.edu.cn
 Chengyong Wang1
Chengyong Wang1 Alei Zhang
Alei Zhang Kequan Chen
Kequan Chen