- 1State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin, China
- 2College of Life Science, Northeast Forestry University, Harbin, China
- 3Administrative Office of the Summer Palace, Beijing Municipal Administration Center of Parks, Beijing, China
- 4The Center for Microbes, Development and Health (CMDH), Institut Pasteur of Shanghai, Chinese Academy of Sciences, Shanghai, China
- 5Forest Biotechnology Group, Department of Forestry and Environmental Resources, North Carolina State University, Raleigh, NC, United States
- 6Department of Plant and Microbial Biology, North Carolina State University, Raleigh, NC, United States
- 7Center for Biological Disaster Prevention and Control, National Forestry and Grassland Administration, Shenyang, China
- 8College of Biological Science and Technology, Hubei Minzu University, Enshi, China
- 9Nord Reet UG, Greifswald, Germany
- 10Institute of Marine Biotechnology e.V. (IMaB), Greifswald, Germany
Bursaphelenchus xylophilus, a plant parasitic nematode, is the causal agent of pine wilt, a devastating forest tree disease. Essentially, no efficient methods for controlling B. xylophilus and pine wilt disease have yet been developed. Enterobacter ludwigii AA4, isolated from the root of maize, has powerful nematocidal activity against B. xylophilus in a new in vitro dye exclusion test. The corrected mortality of the B. xylophilus treated by E. ludwigii AA4 or its cell extract reached 98.3 and 98.6%, respectively. Morphological changes in B. xylophilus treated with a cell extract from strain AA4 suggested that the death of B. xylophilus might be caused by an increased number of vacuoles in non-apoptotic cell death and the damage to tissues of the nematodes. In a greenhouse test, the disease index of the seedlings of Scots pine (Pinus sylvestris) treated with the cells of strain AA4 plus B. xylophilus or those treated by AA4 cell extract plus B. xylophilus was 38.2 and 30.3, respectively, was significantly lower than 92.5 in the control plants treated with distilled water and B. xylophilus. We created a sdaB gene knockout in strain AA4 by deleting the gene that was putatively encoding the beta-subunit of L-serine dehydratase through Red homologous recombination. The nematocidal and disease-suppressing activities of the knockout strain were remarkably impaired. Finally, we revealed a robust colonization of P. sylvestris seedling needles by E. ludwigii AA4, which is supposed to contribute to the disease-controlling efficacy of strain AA4. Therefore, E. ludwigii AA4 has significant potential to serve as an agent for the biological control of pine wilt disease caused by B. xylophilus.
Introduction
The pine wood nematode (PWN) Bursaphelenchus xylophilus causes serious damage to forest ecosystems and massive economic losses by inducing pine wilt disease (PWD) (Zhao et al., 2014; Lee et al., 2019; Guo et al., 2020). PWD may result in destruction of conifer forests and has long been a huge threat to Asian and European forestry for a long time (Kikuchi et al., 2011; Faria et al., 2015). As a pathogenic nematode native to North America (Li et al., 2015; Proenca et al., 2017), B. xylophilus, feeding on live trees and fungi colonizing dead or dying trees, is a migratory endoparasite transmitted by the insect vector Monochamus alternatus (Japanese pine sawyer beetle) (Kim et al., 2019, 2020). The annual economic cost of the PWN (Soliman et al., 2012) in the European Union (EU) alone is estimated at a billion euros for each of the past 22 years. The rapid death of pine trees infected by the PWN could be attributed to the dysfunction of the water-conducting system caused by the death of parenchyma cells. The secreted enzymes and surface coat proteins of B. xylophilus are involved in its pathogenicity (Futai, 2013; Nunes da Silva et al., 2015; Wen et al., 2021). The molecular mechanisms of PWN pathogenesis are still largely unknown, hindering the prospects for control of this pathogen and PWD. Chemical insecticides and nematicides used to control B. xylophilus by jet-sprays or trunk injections for decades (Qiu et al., 2019; Guo et al., 2020; Faria et al., 2021) have become a major social concern. More environmental-friendly strategies, such as beneficial microorganisms that suppress PWN, have recently gained more attention (Tian et al., 2007; Wu et al., 2013; Cai et al., 2022).
Biological control of plant parasitic nematodes (PPNs) using nematocidal bacteria or their metabolites, which are toxic to nematodes, is a potentially sustainable alternative to chemical nematicides (Haegeman et al., 2009; Kumar and Dara, 2021). Nematocidal prokaryotes mainly belonging to bacterial genera, such as Bacillus (Crickmore, 2005; Ponpandian et al., 2019; Park et al., 2020), Streptomyces (Kang et al., 2021), Serratia (Paiva et al., 2013; Nascimento et al., 2016; Abd El-Aal et al., 2021), Stenotrophomonas (Huang et al., 2009; Ponpandian et al., 2019), Pseudoduganella (Fang et al., 2019; Abd El-Aal et al., 2021), Novosphingobium (Topalović et al., 2020), Pasteuria (Tian et al., 2007), Pseudomonas (Chan et al., 2020), Enterobacter (Munif et al., 2000; Oh et al., 2018), and Curtobacterium (Kumar and Dara, 2021), are capable of suppressing PPNs by diverse modes of action, including parasitism (Wu et al., 2013; Proenca et al., 2017; Ponpandian et al., 2019), production of toxins (Abd El-Aal et al., 2021; Kahn et al., 2021), antibiotics plus enzymes (Yang et al., 2007; Huang et al., 2009), competition for nutrients (Proenca et al., 2019), and induction of systemic resistance of plants (Liang et al., 2019; Han et al., 2021). Within these nematocidal groups, some bacterial strains have exhibited efficient killing activity against the PWNs. Bacillus thuringiensis zjfc85 caused 90% mortality of B. xylophilus by producing a Cry protein named Cry5Ba3 (Kahn et al., 2021). Two Streptomyces strains did kill the hatched PWNs and affected egg hatching via biosynthesis of the toxic compounds, teleocidin B4, and spectinabilin, which effectively suppressed the development of PWD under field conditions (Kang et al., 2021). A 70 kD serine protease produced by Serratia sp. A88copa13 was majorly responsible for the toxicity of this PWN-killing strain (Paiva et al., 2013). Thus, the application of nematocidal bacteria is a promising strategy in suppressing PWD.
Enterobacter is an exceptionally diverse genus of bacteria found in various habitats in association with soil (El-Sayed et al., 2014; Habibi et al., 2019; Danish et al., 2020), plants (Park et al., 2015; Andres-Barrao et al., 2017; Sarkar et al., 2018), and animals, including humans (Mokracka et al., 2004; Peng et al., 2009). Many Enterobacter strains are characterized as plant beneficial bacteria. Such procaryotic microbes promote the growth of their plant hosts under favored or adverse abiotic conditions by producing indole-acetic acid (IAA) (Park et al., 2015; Srisuk et al., 2018; Habibi et al., 2019; Li et al., 2022), siderophores (Mokracka et al., 2004; Nurjadi et al., 2021; Li et al., 2022), hydrocyanic acid (Mpongwana et al., 2016; Mahdi et al., 2020; Javaheri Safa et al., 2021), salicylic acid (Kang et al., 2015) plus exopolysaccharides (Sayyed et al., 2015; Niu et al., 2018; Dhanya et al., 2021), solubilizing phosphate (Adhikari et al., 2020; Roslan et al., 2020; Aeron et al., 2021), by fixing nitrogen (Kämpfer et al., 2005; Peng et al., 2009; El-Sayed et al., 2014), reducing Na+ uptake (Um et al., 2017; Sarkar et al., 2018), and inducing the activity of enzymes with action as antioxidants (Andres-Barrao et al., 2017; Danish et al., 2020). At present, it has been demonstrated that some Enterobacter species exhibit inhibitory effects against the oomycete pathogen Pythium ultimum (Lohrke et al., 2002; Kageyama and Nelson, 2003; Windstam and Nelson, 2008), the fungal pathogens Fusarium moniliforme (Hinton and Bacon, 1995; Demirci et al., 2000; Rodrigues et al., 2018), F. oxysporum (El-Sayed et al., 2014; Del Barrio-Duque et al., 2019; Abdelshafy Mohamad et al., 2020), Aspergillus niger (Yadav et al., 2016; Kaushik et al., 2017; Batyrova et al., 2020), and Aspergillus flavus (Etcheverry et al., 2009; Mahmood et al., 2020), and the bacterial pathogen Ralstonia solanacearum (Sarkar and Chaudhuri, 2015; Yin et al., 2020; Zaki et al., 2021), indicating their protective efficacy on plants. A few Enterobacter strains exhibited significant nematocidal activity against the root-knot nematodes (Munif et al., 2000; El-Sayed et al., 2014; Oh et al., 2018). Enterobacter asburiae HK169 was able to reduce root gall formation rate by 66% and killed all juveniles of Meloidogyne incognita within 48 h (Oh et al., 2018). Similarly, an endophytic E. intermedius strain isolated from tomato roots remarkably decreased the number of root galls (Munif et al., 2000). Our knowledge of the molecular mechanisms underlying the inhibitory effects of Enterobacter strains against PPNs is still very limited.
In the present study, we identified a powerful PWN-killing bacterial strain, Enterobacter ludwigii AA4, which was previously isolated from maize roots (Niu et al., 2017) by screening the nematocidal activities of 374 bacterial strains against B. xylophilus using a new fluorescent staining-based PWN-killing activity test. E. ludwigii AA4 had a remarkable inhibitory effect against PWD under greenhouse conditions. After deleting the sdaB gene, presumed to encode the beta subunit of L-serine dehydratase, the nematocidal and disease-suppressing effects of the mutant strain were significantly reduced. The robust colonization of P. sylvestris seedling needles by E. ludwigii AA4, presumed to contribute to the disease-controlling efficacy of strain AA4, was quantified and compared to the strain carrying a sdaB gene deletion.
Materials and Methods
Microbial Strains and Growth Conditions
Bacterial strains kept frozen with 15% (v/v) glycerol at −80°C were streaked on Luria-Bertani (LB) agar plates and incubated at 30°C for 16 h. Then, a single colony of each strain was inoculated into 5 ml LB liquid medium and shaken at 30°C, 200 rpm for another 16 h. Antibiotics were supplemented where necessary at the following concentrations: kanamycin 50 μg/ml, tetracycline 50 μg/ml, gentamicin 25 μg/ml, streptomycin 50 μg/ml, and gentamicin 25 μg/ml (Table 1).
The spores of Botrytis cinerea were deposited at −80°C in 15% (v/v) glycerol. Ten microliters of fungal spores were dropped on a potato dextrose agar (PDA) plate and incubated at 25°C in the dark. When B. cinerea mycelia covered PDA plates, the fungal culture was used for inoculating B. xylophilus.
The Source and Culture of Pine Wood Nematodes
Cultures of the PWN, Bursaphelenchus xylophilus, were obtained from Dr. Hongtao Li at the Hebei Academy of Agriculture and Forestry Sciences and Dr. Kai Guo at Zhejiang A&F University. The fungus B. cinerea was purchased from the Shanghai Bioresource Collection Center (SHBCC).
Bursaphelenchus xylophilus was inoculated on a PDA culture of B. cinerea and incubated at 25°C in the dark until the fungal mycelia were completely consumed by B. xylophilus. Nematodes were collected using the modified Baermann funnel technique (Kitazume et al., 2018; Cesarz et al., 2019; Maehara et al., 2020) and washed with a mixture of 0.1% streptomycin sulfate and 0.002% actinone three times to remove surface microbial contaminants (Liu et al., 2016). Then, these nematodes were used for PWN-killing activity test and in planta biocontrol assays.
For microscopic image analysis of PWN morphology, about 10,000 nematodes were decanted into a burette containing 25 ml of 0.3% carboxymethyl cellulose (CMC) solution. After 12 h, the second-stage juveniles were collected from the top of the burette (Qiu et al., 2016, 2019). These worms were fed with B. cinerea. After 48 h, the L4 juveniles were collected in sterile water and washed with a mixture of 0.1% streptomycin sulfate and 0.002% actinone three times. These L4 juveniles were used for the image analysis.
Preparation of Cell Extracts of E. ludwigii AA4
Wild-type AA4, AA4ΔsdaB (derivative of wild-type AA4 that lacks sdaB), and C△sdaB (AA4ΔsdaB complemented with wild-type sdaB gene) kept frozen with 15% (v/v) glycerol at −80°C were streaked on LB agar plates with corresponding antibiotics (Table 1) and incubated at 30°C for 16 h. Then, a single colony of each strain was inoculated into 5 ml LB liquid medium and shaken at 30°C, 200 rpm for another 16 h. One milliliter of the culture of each strain was transferred into 50 ml LB liquid medium and shaken at 30°C, 200 rpm for 2–3 h until Optical Density at 600 nm (OD600) value reached 0.6–0.8 (early stationary phase). Cells were harvested by centrifuging at 9,100 rpm (8,000 × g) at 4°C for 20 min (Thermo, Multifuge X1R, Germany) and washed by phosphate-buffered saline (PBS). The cell precipitations were resuspended in 5 ml PBS before sonication. An ultrasound processor (Sonics, VCX130PB, United States) was used to sonicate the resuspended cell suspensions. The probe was immersed into the suspensions and sonicated the cells on ice for 60 min at a power of 130 W (pulse duration: 2 s on, 1 s off). The resulting cell lysates were centrifuged at 15,300 rpm (14,000 × g) for 30 min. The supernatant was collected and used as cell extracts. The concentration of total protein in cell extract was measured by using a biuret protein assay reagents kit (Solarbio, PC0010, China).
Fluorescent Staining-Based Pine Wood Nematode-Killing Activity Test
The nematocidal activity of the 43 efficient PWN-killing bacterial strains of AA4ΔsdaB and CΔsdaB was evaluated by the fluorescent staining-based PWN-killing activity test, while Bacillus pumilus YLT40 and Paenibacillus polymyxa M-1 were utilized for confirming the reliability of the assay. The cell density of bacterial culture suspension was adjusted to OD600 = 1.0 for each strain used in the test. Each well of the 96-well microplates was added with 40 μl of bacterial culture suspension and 20 μl of worm suspension containing 40–50 nematodes, while 40 μl of LB liquid medium or PBS and 20 μl of worm suspension was employed as negative control. Five wells were used for each treatment designated with the code of the strain listed in Supplementary Tables 1, 2. The inoculated microplates were incubated at 25°C in the dark for 24 h. Then, a plate washer (Biobase, BK-9622, China) was employed to change the liquid cultures with sterile water. 4’6-diamidino-2-phenylindole (DAPI, Solarbio, C0065, China) was used to stain the PWNs, and the final concentration of DAPI in each well was 5 μg/ml. After being stained for 1 h, the nematodes were observed by a confocal laser scanning microscope (Zeiss, LSM 800, Germany) using a excitation laser of 353 nm and collecting emission of 465 nm, where the dead worms displayed bright fluorescence while the live ones were dim.
To test the nematocidal activity of the cell extract of wild-type AA4, AA4ΔsdaB, and CΔsdaB, the extract containing total proteins of 5 mg/ml was mixed with 20 μl of worm suspension containing 40–50 nematodes in one well of a 96-well microplate. The remaining steps are the same as those for the assays with bacterial cell suspensions.
The mortality and corrected mortality of B. xylophilus were calculated as follows (Faria et al., 2013; Guo et al., 2016; Liu et al., 2019):
Two-tailed t-test (GrapPad Prism 8) was used for the statistical analysis. All these experiments above were repeated five times.
Z′factor, a combination of signal interval and variation, is a parameter used to evaluate the reliability of results obtained from an experiment for guiding a larger-scale experimentation. It is calculated as:
where σp and σn represent the standard deviations of positive control and negative control, respectively, and μp and μn represent the means of positive control and negative control, respectively. Z′factor was calculated by using the software Excel (Microsoft). The theoretical value of Z′factor is 1, and a Z′factor value between 0.5 and 1 indicates the outcomes of a given experiment is highly reliable, while a test of low reliability usually possesses a Z′factor value of less than 0.5 (Zhang X. D. et al., 2020).
Identification of the Efficient Bacterial Pine Wood Nematode-Killing Strains
Identification of the efficient bacterial PWN-killing strains were carried out by using 16S rRNA gene sequencing analysis. Genomic DNA was extracted from an overnight LB culture of each strain by utilizing a bacterial genomic DNA extraction kit (Tiangen, DP302-02, China) following the manufacturer’s protocol. The 16S rRNA gene was amplified in 25-μl PCR reactions using the universal primers 27F and 1492R (Table 2) (Monciardini et al., 2002; Niu et al., 2013; López and Alippi, 2019). The reaction mixture contained 1 μl of template DNA, 12.5 μl of master mix (Takara, RR350Q, Japan), 1 μl of each of the forward and reverse primers, and 9.5 μl of sterile deionized water. Amplifications were performed using a T100 Thermal Cycler (Bio-Rad, 621BR47532, United States) with the following cycle conditions: initial denaturation at 95°C for 5 min; 30 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 1.5 min; and a final extension at 72°C for 10 min. Amplicons were purified using a PCR purification kit (Omega, D6492-02, China) and sequenced using 27F primer at Tsingke Biotechnology Co., Ltd. DNA sequences were inspected for base-caller errors and were trimmed by removing any ambiguous trailing or leading bases using the software GAP4 in the STADEN Package1. The sequences were compared with those of the reference organisms by Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI) website2 and by Sequence Match at the Ribosomal Database Project (RDP) website3. The 16S rRNA gene sequences of efficient nematocidal strains were deposited in the GenBank database. The accession numbers were listed in Supplementary Table 1.
In planta Assays for Biocontrol Effect Against Pine Wilt Disease
The biological control effects of wild-type AA4, AA4ΔsdaB, and CΔsdaB were examined by using both 3-year-old and 1-month-old Scots pine (Pinus sylvestris) tree seedlings grown under greenhouse conditions. The 3-year-old seedlings were purchased from Longsheng nursery at Harbin. Ten milliliters of bacterial suspension (OD600 = 1.0) were inoculated on the 3-year-old seedlings by spraying. Then, a 2–4 cm silt was made on the surface of bark located at approximately 10–15 cm above the soil. One hundred microliters of PWN suspension containing about 5,000 worms were injected into the trunk. A small piece of sterilized cotton was fixed on the trunk to cover the silt (Xue et al., 2019). The plants were placed in a greenhouse under the following conditions: 16 h of light (day) and 8 h of dark (night), 25°C, and a relative humidity of 70%. Plants were watered periodically and maintained in the greenhouse. Thirty days after inoculation, the symptoms present on the seedlings were recorded with a camera (Canon, 80D, Japan). The severities of PWD were evaluated based on the disease ranks described in earlier studies (Qiu et al., 2016, 2019) as follows: In rank 0, all needles are green. In rank 1, less than a quarter of needles turn yellow. In rank 2, 25–75% of needles turn yellow. In rank 3, more than 75% of needles turn yellow and less than 50% of needles get wilted. In rank 4, more than 50% of the needles get wilted. Five plants were used for each of the two treatments designated as PWN (inoculation of seedlings with PWN alone) and PWN + AA4 (inoculation of seedlings with PWN jointly with wild-type AA4), respectively.
For the experimentations with 1-month-old Pinus sylvestris seedlings, the seeds were surface-sterilized by immersing in 5% KMnO4 for 60 min and placing in a growth chamber at 30°C in the light for 48 h. After germination, the seeds with roots were sown in the plant growth substrates prepared by mixing soil and vermiculite in a ratio of 3:1. Thirty days after emergence, 5 ml of bacterial suspensions (OD600 = 1.0) or cell extracts (containing total proteins of 5 mg/ml) of wild-type AA4, AA4ΔsdaB, and CΔsdaB were sprayed on the needles of Scots pine seedlings, respectively. In the meantime, 100 μl of PWN suspension containing approximately 3,000 worms were injected into the needles by using a syringe. A small piece of sterile cotton was fixed on the needles to cover the pinhole. The plants were incubated in greenhouse under the same condition on which the 3-year-old seedlings grew. The number of dead seedlings and symptoms were recorded on the ninth day after inoculation. Ten plants were used for each of the 11 treatments designated as PWN (inoculation of seedlings with PWN alone), PWN + ΔsdaB (inoculation of seedlings with PWN jointly with AA4ΔsdaB cell suspension), PWN + CΔsdaB (inoculation of seedlings with PWN jointly with AA4CΔsdaB cell suspension), PWN + WT (inoculation of seedlings with PWN jointly with wild-type AA4 cell suspension), PWN + ΔsdaBCE (inoculation of seedlings with PWN jointly with AA4ΔsdaB cell extract), PWN + CΔsdaBCE (inoculation of seedlings with PWN jointly with AA4CΔsdaB cell extract), PWN + WTCE (inoculation of seedlings with PWN jointly with wild-type AA4 cell extract), PWN + ΔsdaBS (inoculation of seedlings with PWN jointly with AA4ΔsdaB culture supernatant), PWN + CΔsdaBS (inoculation of seedlings with PWN jointly with AA4CΔsdaB culture supernatant), PWN + WTS (inoculation of seedlings with PWN jointly with wild-type AA4 culture supernatant), and DW (treatment with distilled water), respectively.
The severities of PWD were evaluated based on the disease ranks shown in Supplementary Figure 3. The disease severity indices were calculated as follows:
where Xi is the number of pine trees in each disease rank, Ai is the disease rank, X is the total number of pine trees, and Amax is the maximum rank (Qiu et al., 2019; Xue et al., 2019). Two-tailed t-test (GrapPad Prism 8) was used for the statistical analysis. These experiments were repeated three times.
Image Analysis of Pine Wood Nematode Morphology
The cell extract of E. ludwigii AA4 was prepared as above. Forty microliters of cell extract containing total proteins of 5 mg/ml and 20 μl of worm suspension containing 40–50 L4 juveniles of PWNs were added in a single well of the 96-well microplates. At the meantime, the combination of 40 μl of PBS and 20 μl of worm suspension was employed as negative control. The microplates were stored at 25°C in the dark for 24 h. Then, the worms were picked and put into 10 μl of PBS by injector pinhead. After being washed three times, the worms were transferred to a slide. Ten microliters of PBS were then dropped on the samples. The morphological changes of B. xylophilus were observed and recorded by a light microscope (Olympus, BX43, Japan).
Construction of Mutants of E. ludwigii AA4
The sdaB gene knock-out mutant of E. ludwigii AA4 was created by using the Red recombinase system (Datsenko and Wanner, 2000; Dahyot et al., 2020; Guérin et al., 2020). Primers were designed according to the information obtained from the whole genome sequence of strain AA4 (Niu et al., 2017). The upstream border sequence of the AA4 sdaB gene was amplified from AA4 chromosomal DNA using primers sdaB-H1up and sdaB-H1down (Table 2), while the downstream border sequence of sdaB was amplified with primers sdaB-H2up and sdaB-H2down (Table 2). A kanamycin resistance cassette, flanked by 23-bp-long sequences homologous with sdaB gene, was amplified from plasmid pKD4 DNA with primers Kup and Kdown (Table 2). Another fragment flanked by 22-bp-long sequences homologous with sdaB gene was amplified from plasmid pUC19 by using primers pUC19up and pUC19down (Table 2). The purified PCR products were assembled by Gibson DNA assembly technology, using a SoSoo Cloning Kit (Tsingke, T-TSV-S1, China) following the manufacturer’s instructions and then cloned into Escherichia coli DH5α. Clones containing the resulting recombinant pU19ΔsdaB vector were selected on LB agar supplemented with kanamycin. The recombinant plasmid pUC19ΔsdaB was isolated with a plasmid extraction kit (Omega, D6943-02, China) following the manufacturer’s instructions and used as template DNA for PCR amplification with the primers sdaB-H1up and sdaB-H2down (Table 2). After purification, the PCR products were introduced into the competent cells of AA4 carrying a Red helper plasmid pKD46 by electroporation (Bio-Rad, 411BR11661, United States) at 2.5 KV. The resulting transformants were selected on LB agar supplemented with kanamycin after incubation for 24 h at 30°C. Homologous recombination was confirmed by PCR and sequencing. Then, the elimination of pKD46 was performed by culturing the correct transformant by shaking at 37–42°C, 200 rpm overnight. The knock-out mutant can only grow at 30°C on the LB agar supplemented with kanamycin but not streptomycin.
To construct a complement strain of AA4ΔsdaB, the complete open reading frame of sdaB gene was amplified by PCR with primers sdaB-DF and sdaB-UR (Table 2) from the genomic DNA of strain AA4 and linked with the fragment flanked by 22-bp-long sequences homologous with sdaB gene and amplified from plasmid pBBR1 using primers sdaB-PBBR1-DF and sdaB-PBBR1-UR (Table 2) by the Gibson DNA assembly technology. The complement plasmid pBBR1-sdaB was introduced into the competent cells of AA4ΔsdaB by electroporation at 2.5 KV, and the resulting transformants were selected on LB agar supplemented with kanamycin and tetracycline after incubation at 30°C for 24 h. The complementation was then confirmed by PCR and sequencing.
To create a random transposon library, E. ludwigii AA4 was transformed with pDL1093 via conjugation at 30°C and outgrown at 42°C in the presence of kanamycin to induce transposition of miniTn10 (Duncan et al., 2018).
For the sake of constructing fluorescence protein-labeled AA4, plasmid pGFP78, obtained from Dr. Qi Wang at China Agricultural University, harboring the gene encoding for green fluorescent protein (GFP), was introduced into the competent cells of wild-type AA4, AA4ΔsdaB, or CΔsdaB by electroporation at 2.5 KV. The resulting transformants were selected on LB agar supplemented with tetracycline, or kanamycin and tetracycline, or a combination of kanamycin, tetracycline, and gentamicin where necessary after incubation at 30°C for 24 h. The correctness of the transformants were then confirmed by PCR, sequencing, and fluorescence microscopy.
Assay for L-Serine Dehydratase Activity
The cell extract of E. ludwigii AA4 was prepared as above, except washing and resuspending in the extraction buffer (50 mM K2HPO4, 2.5 mM serine protease inhibitors) (Velayudhan et al., 2004). Three aliquots of 1 ml of cell extract containing total proteins of 10 mg/ml was incubated with 500 μl of 300 mM L-serine at 37°C for 6 min. Then, the concentration of pyruvate was determined by using a pyruvate assay kit (Solarbio, BC2205, China). The L-serine hydrolase activity was calculated as:
where c is the concentration of pyruvate in μg/ml, v is the total reaction volume in ml, M is the relative molecular mass of pyruvate, T is the reaction time in minutes, and m is the weight of cell extract in mg.
Two-tailed t-test (GrapPad Prism 8) was used for the statistical analysis. This experiment was repeated five times.
Assays for Colonization of Scots Pine Needles
In order to investigate the ability of E. ludwigii AA4 to colonize Scots pine needles, 50 ml of LB cultures (OD600 = 1.0) of GFP-labeled wild-type AA4, AA4ΔsdaB, and CΔsdaB were sprayed on 21 1-month-old seedlings, respectively. The plants were placed in a greenhouse under the following conditions: 16 h of light (day) and 8 h of dark (night), 25°C, and a relative humidity of 70%. Three aliquots of 50 mg of needles were sampled at the 4th, 48th, 72th, 96th, 120th, and 144th, and 168th hour post inoculation, respectively, for bacterial quantification. The needles were crushed with 1 ml of PBS by using a sterile mortar. The resulting suspensions were diluted and spread on LB agar plates supplemented with tetracycline and incubated at 30°C for 16 h. The colony-forming unit (CFU) numbers were recorded. Two-tailed t-test (GraphPad Prism 8) was used for the statistical analysis. This experiment was repeated five times.
For image analysis, 48 h after inoculation, the needle slices were washed with PBS and transferred to a slide. Ten microliters of PBS were dropped on the samples. The colonization of needles by bacteria was visualized by a Confocal Laser Scanninc Microscopy (CLSM; Zeiss, LSM 800, Germany) using an excitation laser of 480 nm.
Results
Screening of Bacterial Strains for Nematocidal Activity Against Bursaphelenchus xylophilus
To find microorganisms possessing strong nematocidal activity against B. xylophilus for potential biological control of the PWD, we performed a PWN-killing test with 374 bacterial strains from our lab collection. First, we used an efficient PWN-killer Bacillus pumilus YLT40 and a moderate PWN-killing strain Paenibacillus polymyxa M-1 (Niu et al., 2013), identified previously (data not shown), as controls (Figure 1A) to verify if the PWN-Bacterium interaction system employed in this study was reliable for a large-scale screening. The corrected mortality rates of the nematode B. xylophilus treated by B. pumilus YLT40 and P. polymyxa M-1 were more than 85% and less than 25% (Figure 1A), respectively. We calculated a Z′factor score of 0.682 (Figure 1A) which indicated that the screening procedure utilized is sufficiently reliable for selecting powerful PWN-killing strains.
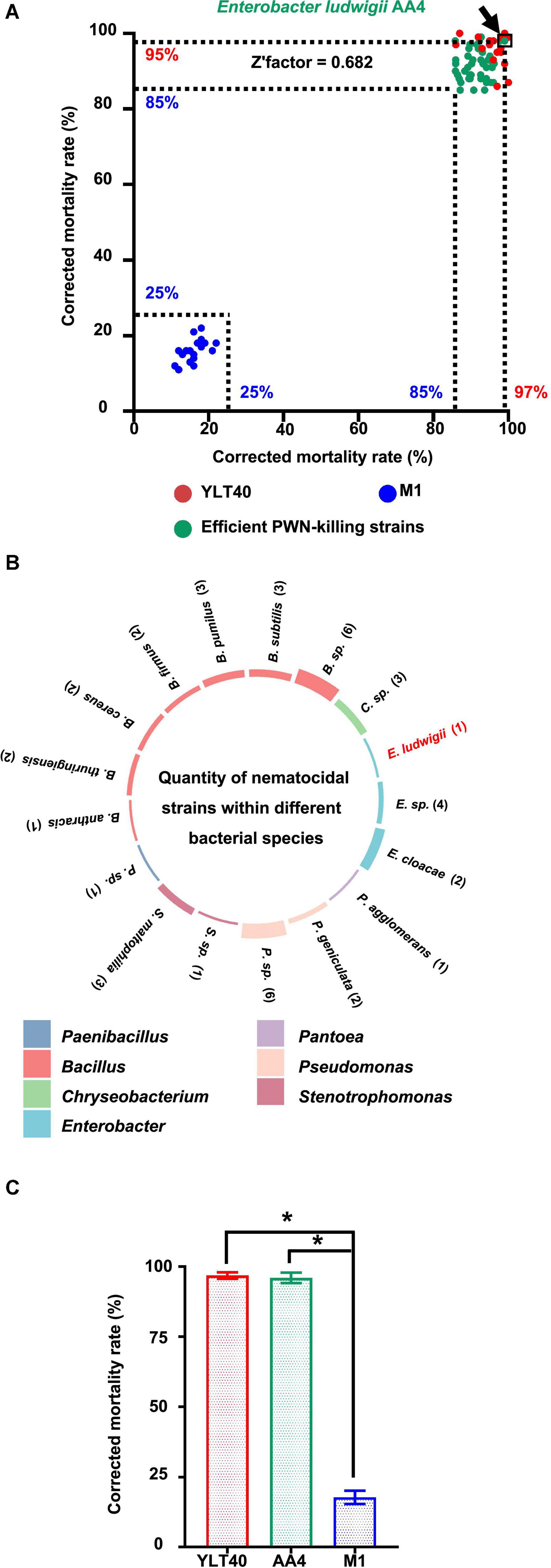
Figure 1. Screening and taxonomy of the nematocidal bacterial strains against pine wood nematodes (PWNs). (A) Screening of strong PWN-killing bacterial strains against Bursaphelenchus xylophilus. The green dots represent the 43 strong nematocidal strains causing the death of more than 85% of B. xylophilus treated with them, while the blue and red dots are bacterial cultures of Bacillus pumilus YLT40 and Paenibacillus polymyxa M-1 used as controls to confirm the reliability of the screening. The position of each dot was determined by the two values (showing on the x- and y-axis, respectively) of corrected mortality rate of PWNs calculated in two independent tests. The dot standing for Enterobacter ludwigii AA4 is framed and pointed out by an arrow. (B) The number of nematocidal strains within each bacterial species. The colors of the columns on the circle indicate the genus names of the 43 nematocidal strains, while histogram heights are consistent with the numbers (showing in the brackets) of the strains belonging to each bacterial species. (C) Nematocidal effect of E. ludwigii AA4 against PWNs. Asterisks indicate that differences among the means represented by the columns are statistically significant (*p < 0.0001). Two-tailed t-test (GrapPad Prism 8) was used for the analysis.
We detected 43 efficient nematocidal bacterial strains exhibiting B. xylophilus mortality rates above 85% (Figure 1A, Supplementary Figure 1, and Supplementary Table 1). Based on 16S rRNA gene sequencing, we found that these bacterial strains belong to 21 species from seven genera, including Bacillus, Pseudomonas, Enterobacter, Chryseobacterium, Stenotrophomonas, Pantoea, and Paenibacillus (Figure 1B), all genera with known nematocidal activity. Among the seven genera, Bacillus harbored 19 nematocidal strains, while Pantoea and Paenibacillus possessed only one PWN-killing strain (Figure 1B). Interestingly, seven Enterobacter strains, including the plant beneficial bacterium E. ludwigii AA4 isolated from maize roots (Niu et al., 2017), exhibited high nematocidal activity against the PWN (Figure 1B). In addition, AA4 did kill almost all of the B. xylophilus clones (96%) in our assay (Figure 1C). In our collection, we found 43 strong PWN-killing strains involving E. ludwigii AA4. Next, we validated the PWN-killing function of strain AA4 in both in vitro and in planta experiments.
Biocontrol Effect of E. ludwigii AA4 Against B. xylophilus
To confirm the nematocidal effect of E. ludwigii AA4 against B. xylophilus, we developed a fluorescent dye-exclusion viability test to visualize PWN-killing activity. We used DAPI which differentially stains live and dead PWNs. Dead worms take up DAPI rapidly and show bright fluorescence, while the live ones are relatively non-fluorescent and look dim (Figure 2A).
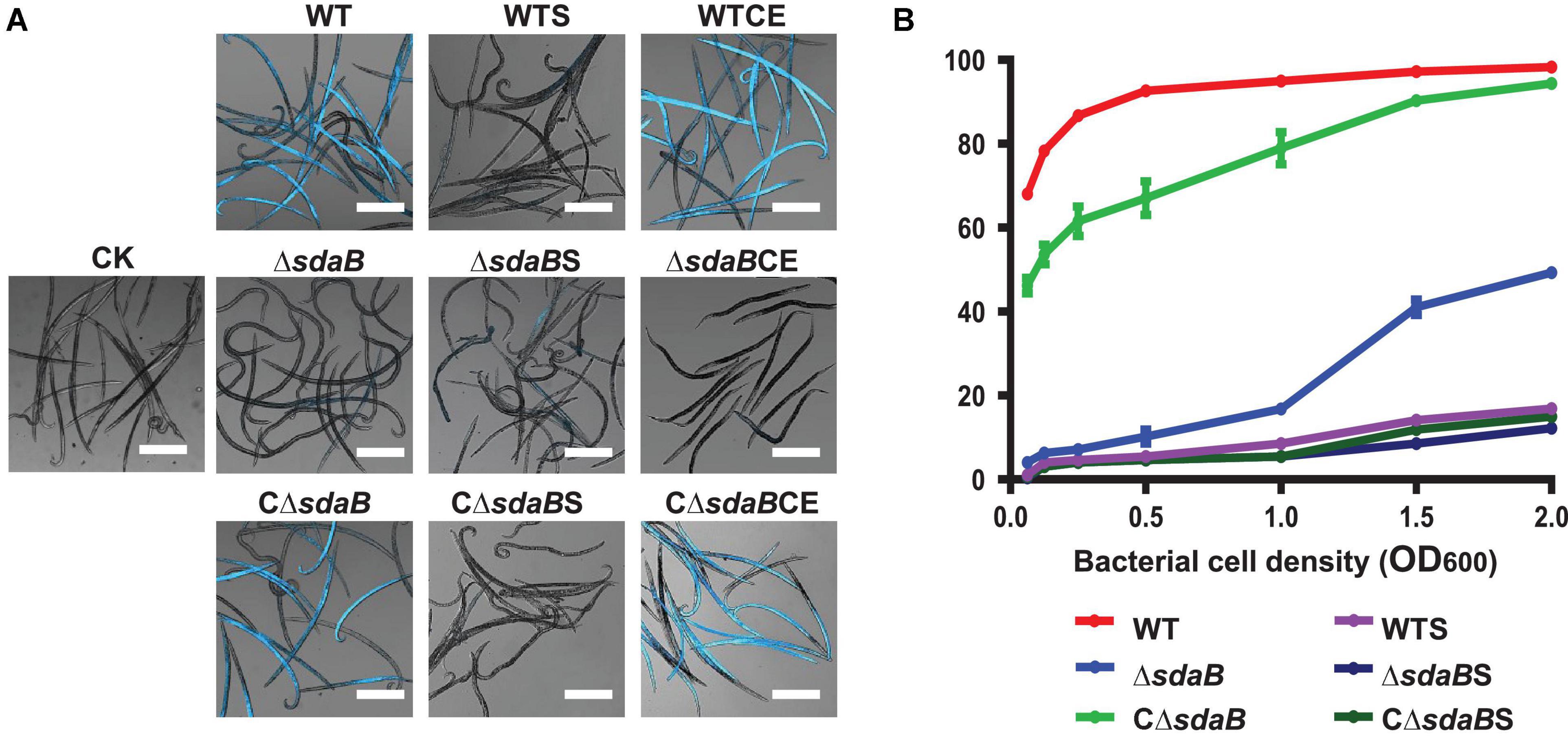
Figure 2. Nematocidal effect of E. ludwigii AA4 and its sdaB gene knock-out mutant against B. xylophilus. (A) Imaging analysis of the PWN-killing efficacy of wild-type E. ludwigii AA4 and AA4ΔsdaB against B. xylophilus. The dead nematodes showed cyan fluorescence when excited by 353 nm wavelength after being stained by 4′6-diamidino-2-phenylindole (DAPI), while the live worms stained by DAPI were dim under excitation with the same wavelength. These images were taken using a confocal laser scanning microscope (scale bars: 2 mm). (B) Mortality of the PWNs treated with bacterial cells or culture supernatants. B. xylophilus was co-cultured with cells of wild-type AA4 and AA4ΔsdaB or incubated with culture supernatants of wild-type AA4 and AA4ΔsdaB for 24 h, respectively. CK, sterile LB liquid medium; WT, cell culture of wild-type AA4; WTS, supernatant of wild-type AA4 culture; WTCE, cell extract of wild-type AA4; ΔsdaB, cell culture of AA4ΔsdaB; ΔsdaBS, supernatant of AA4ΔsdaB culture; ΔsdaBCE, cell extract of AA4ΔsdaB; CΔsdaB, cell culture of AA4ΔsdaB complemented with the wild-type sdaB gene; CΔsdaBS, supernatant of culture of AA4ΔsdaB complemented with the wild-type sdaB gene; CΔsdaBCE, cell extract of AA4ΔsdaB complemented with the wild-type sdaB gene.
In our assay, the majority (68%) of PWNs co-cultured with wild-type E. ludwigii AA4 cells in very low density (OD600 = 0.063) for 24 h were killed (Figure 2B). The corrected mortality rate of the nematodes treated with wild-type AA4 cells tends to increase with enhanced cell density. The percentage of dead worms dramatically increased from less than 70% to more than 92% when the OD600 value of the bacterial culture was raised from 0.063 to 0.5. The corrected mortality rate rose slowly to a maximum of 98.3% when the OD600 value of the cell suspension reached 2 (Figure 2B). Our visual test documented that nearly all the nematodes incubated with wild-type AA4 emitted intense fluorescent signals, while almost no fluorescence was detected without bacteria (Figure 2A). These results well corroborated the high mortality rate of B. xylophilus when exposed to wild-type AA4. E. ludwigii AA4 exhibited a remarkable in vitro PWN-killing effect.
We then investigated the disease-suppressing efficacy of E. ludwigii AA4 against PWD on 3-year-old Pinus sylvestris greenhouse-grown seedlings. Thirty days after inoculation, we found that the needles of the seedlings treated with PWNs together with wild-type AA4 were much greener and less wilted than those inoculated with B. xylophilus only (Figure 3A). The disease index of PWD on the P. sylvestris plants treated with PWNs together with wild-type AA4 (19.5) was significantly lower than that treated with B. xylophilus alone (86.9) (Figure 3B), indicating that the PWD severity was substantially reduced by treatment with wild-type AA4. E. ludwigii AA4 was capable of efficiently suppressing PWNs not only under in vitro conditions but also in in planta and was a strong bacterial biocontrol strain against PWD caused by B. xylophilus.

Figure 3. Biocontrol effect of E. ludwigii AA4 against PWD caused by B. xylophilus. (A) Symptoms on the 3-year-old Pinus sylvestris seedlings inoculated with PWN alone or PWN jointly with E. ludwigii AA4 (PWN + AA4). Photographs were taken 30 days after inoculation (Scale bars: 15 cm). (B) Severity of PWD on the 3-year-old P. sylvestris seedlings inoculated with PWN alone and PWN jointly with E. ludwigii AA4 (PWN + AA4). The asterisk indicates that differences among the means represented by the columns are statistically significant (*p < 0.0001). Two-tailed t-test (GrapPad Prism 8) was used for the analysis.
E. ludwigii AA4 Causes Morphological Defects in B. xylophilus
To further understand the PWN-killing effect of E. ludwigii AA4, culture supernatants and cell extracts were prepared for the DAPI-staining based nematocidal activity test. Treatment of B. xylophilus with the culture supernatant of wild-type AA4 did not affect the mortality rate of the nematode (Figure 2A). Wild-type AA4 cells did dramatically enhance fluorescence of the B. xylophilus nematodes (Figure 2A). This result was corroborated by the mortality rate of the PWNs, suggesting that the death rate of worms exposed to wild-type AA4 culture supernatant (less than 20%) was significantly less than that of B. xylophilus incubated with cells of wild-type AA4 (more than 60%) (Figure 2B). By contrast, we detected intense fluorescent signals from the worms treated with wild-type AA4 cell extract (Figure 4A) and found that more than 80% of PWNs were dead after being exposed to the extract (Figure 4A), indicating its strong nematocidal effect. Thus, our data indicates that the cell extract of wild-type AA4 rather than the culture supernatant contains the nematocidal activity.
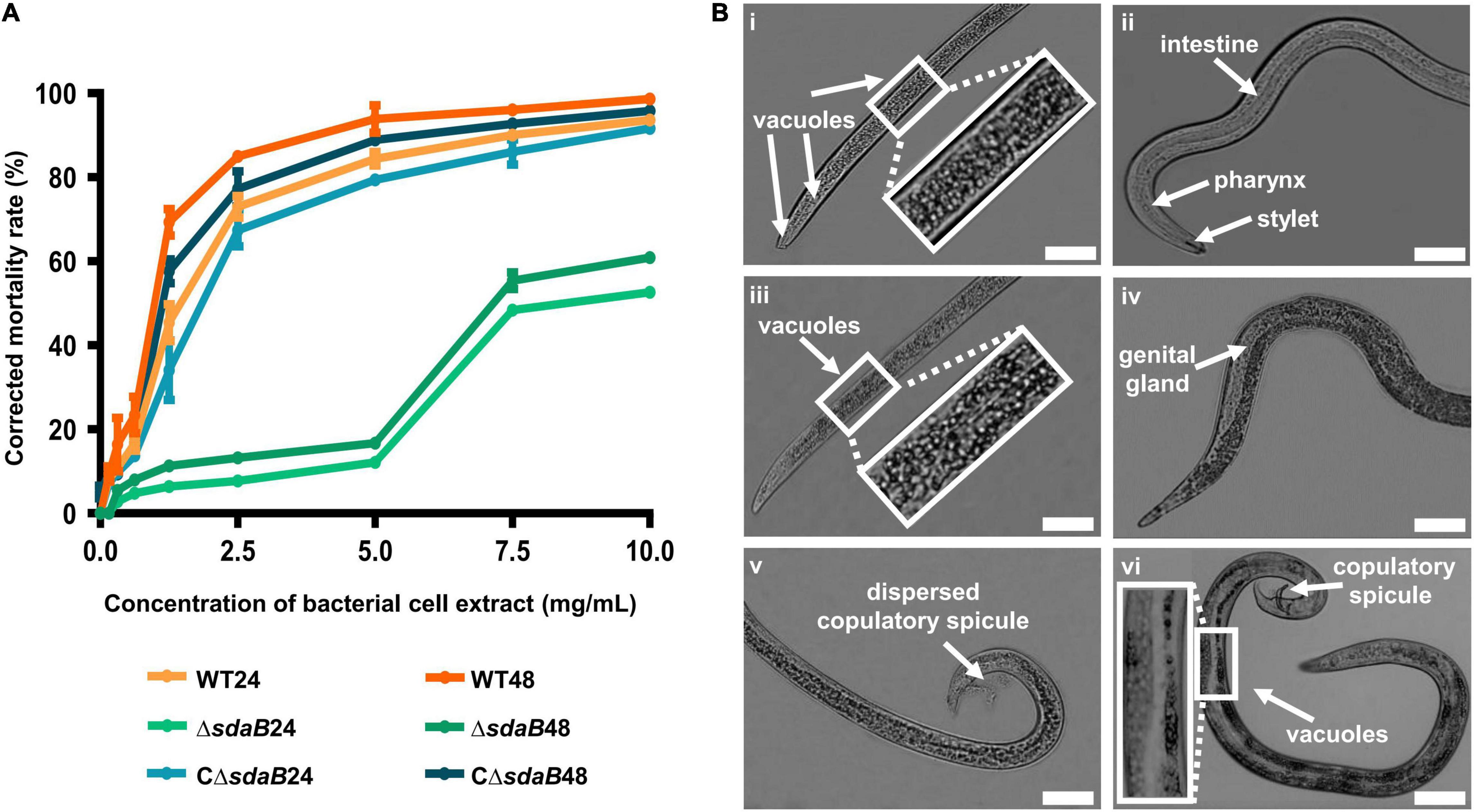
Figure 4. Nematocidal effect of cell extracts of E. ludwigii AA4 and its sdaB gene knock-out mutant against B. xylophilus. (A) Mortality of the B. xylophilus nematode treated with bacterial cell extracts. The worms were incubated with the cell extracts of wild-type AA4 and AA4ΔsdaB for 24 h or 48 h, respectively. WT24, PWNs treated with a cell extract of wild-type AA4 for 24 h; WT48, PWNs treated with a cell extract of wild-type AA4 for 48 h; ΔsdaB24, PWNs treated with a cell extract of AA4ΔsdaB for 24 h; ΔsdaB48, PWNs treated with a cell extract of AA4ΔsdaB for 48 h; CΔsdaB24, PWNs treated with a cell extract of AA4ΔsdaB complemented with a wild-type sdaB gene for 24 h; CΔsdaB48, PWNs treated with a cell extract of AA4ΔsdaB complemented with a wild-type sdaB gene for 48 h. (B) Morphological changes of B. xylophilus incubated with the cell extract of wild-type E. ludwigii AA4. The L4 stage worms exposed to a wild-type AA4 cell extract containing total proteins of 5 mg/mL for 24h, are shown in the left column, where the vacuoles in front (i) and back (iii) sections of worm bodies as well as dispersed copulatory spicule (v) are indicated by arrows. Non-treated L4 stage nematodes (ii,iv,vi) are presented in the right row, where the arrows indicate the intestine, pharynx, and stylet (ii), genital gland (iv), copulatory spicule and vacuoles (vi), respectively (scale bars: 1 mm).
To elucidate the mode of action of the nematocidal effect of E. ludwigii AA4 against PWN, we choose its cell extract for investigating morphological variations of AA4-treated B. xylophilus. The L4 stage nematodes were incubated with wild-type AA4 cell extract. Afterward, the morphological changes of the worms were observed and recorded using an optical light microscope. Exposure of PWN to cell extract-induced formation of vast amounts of vacuoles filled the internal tissues of nematodes completely (Figures 4Bi,iii). Newly formed small vacuoles accumulate and progressively fuse into giant ones, leading to membrane rupture and death (Maltese and Overmeyer, 2014; Armenta and Dixon, 2020). This process is defined as methuosis (Cai et al., 2013; Chen et al., 2014; Nirmala and Lopus, 2020), a type of non-apoptotic cell death widely described in animals (Jang et al., 2016; Rajasekharan et al., 2017; Song et al., 2021). The death of B. xylophilus caused by AA4 might be attributed to methuosis induced by the damage to cell membranes.
In these vacuolated worms, some vital organs, e.g., intestine, pharynx, and genital glands, disappeared in the mass of bubbles. We also failed to capture a clear structure of stylet at the head of PWN treated with wild-type AA4 cell extract (Figures 4Bi,iii). The reproductive organ of male worms, the copulatory spicule, dispersed after being incubated with cell extract (Figure 4Bv). In contrast, we observed fewer vacuoles in the control nematodes exposed only to PBS. All the critical internal organs were intact and clearly visible in the control worm bodies (Figures 4Bii,iv,vi). Therefore, E. ludwigii AA4 appeared to destroy crucial internal organs of B. xylophilus through causing a series of morphological defects.
The sdaB Gene Is Involved in the Nematocidal Effect of E. ludwigii AA4 Against B. xylophilus
To resolve the molecular mechanisms underlying the strong nematocidal effect of E. ludwigii AA4, we characterized genes involved in its PWN-killing efficacy using transposon and PCR-targeted mutagenesis. First, by screening a miniTn10 transposon mutant library of strain AA4, we identified a mutant with greatly reduced nematocidal activity against B. xylophilus. We further analyzed the genomic site where the transposon was located and found a miniTn10 element that was inserted into an open reading frame (ORF) located between kilobase positions 3,956,912 and 3,955,545 (Supplementary Figure 4). This ORF was identified as the sdaB gene encoding the L-serine dehydratase beta subunit, indicating that the sdaB gene might be involved in the PWN-killing effect of AA4.
We next created a knock-out mutant of the sdaB gene by employing the Red (λ, β, exo)-mediated DNA homologous recombination system. The L-serine hydrolase activity of the sdaB gene knock-out deletion mutant AA4ΔsdaB was significantly lower than that of the wild-type AA4 or the AA4ΔsdaB mutant strain complemented with the sdaB wild-type gene (Figure 5B), which indicated that the interruption of the sdaB gene affected the biosynthesis of L-serine dehydratase. Then, we compared the PWN-killing activities of the wild-type AA4 and AA4ΔsdaB by using the fluorescent staining-based nematocidal activity assay (Figure 2A). Nearly no fluorescence could be detected in the worms treated with AA4ΔsdaB cells or their extracts, which confirmed the loss of nematocidal activity of the miniTn10 insertional mutant. Complementation of the mutant with a recombinant plasmid harboring sdaB partially restored the PWN-killing activity (Figure 2A). These results were corroborated by the nematode mortality curves, which documented that the corrected death rate of the PWNs treated by AA4ΔsdaB was significantly lower than those incubated with wild-type AA4, while the mortality of nematodes exposed to the mutant strain complemented with the sdaB wild-type gene increased remarkably (Figure 2B). Therefore, sdaB gene product has a major impact on the nematocidal activity of E. ludwigii AA4.
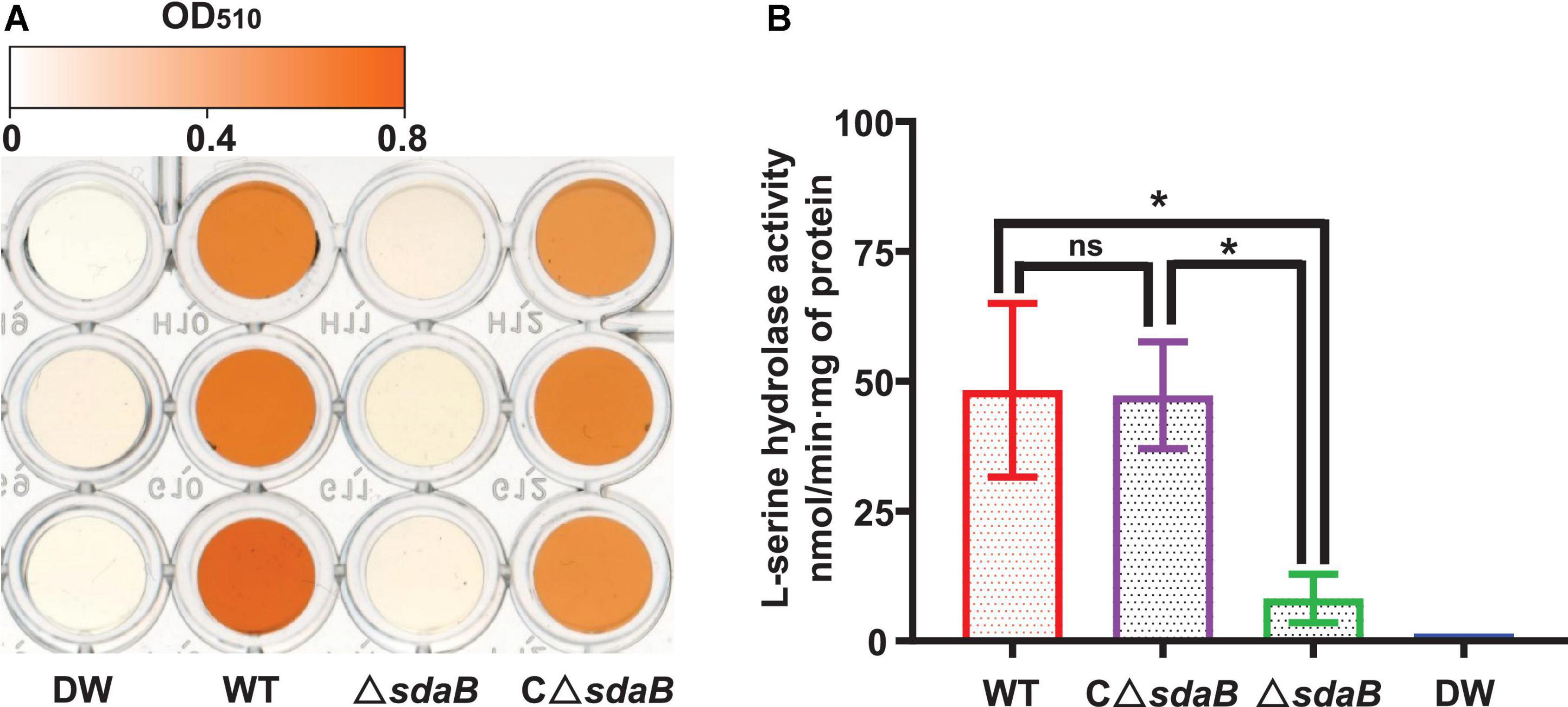
Figure 5. L-serine hydrolase activity of E. ludwigii AA4 and its sdaB gene knock-out mutant. (A) Colorimetrical quantification of pyruvate generated in a L-serine hydrolase activity assay. The color shown in each well of the microplate corresponds to the pyruvate concentration measured by a spectrophotometer at 520 nm. The values of optical density (OD) range from 0 (white) to 0.8 (orange). (B) Comparison of L-serine hydrolase activities of wild-type E. ludwigii AA4, its sdaB gene knock-out mutant, and AA4ΔsdaB complemented with a wild-type sdaB gene. DW, distilled water; WT, E. ludwigii AA4 wild-type strain; ΔsdaB, AA4 sdaB gene knock-out mutant; CΔsdaB, AA4ΔsdaB complemented with a wild-type sdaB gene. The asterisks indicate that differences among the means represented by the columns are statistically significant (*P < 0.0001). Two-tailed t-test (GrapPad Prism 8) was used for the analysis.
Then, we investigated the role of the sdaB gene product on the protective effect of E. ludwigii AA4 on P. sylvestris against PWD. Using a Pine-Nematode-Bacterium tripartite interaction system with 1-month-old P. sylvestris seedlings, a lower disease index was estimated when wild-type AA4 cells or their extracts were added to the system. Under these conditions, much less wilted and fewer chlorotic pine seedlings were observed compared to the control containing B. xylophilus alone (Figure 6B). We found more wilted and chlorotic seedlings when AA4ΔsdaB cells or their extracts were added instead of wild-type AA4 cells or their extracts. The number of diseased plants was reduced when the seedlings were treated with PWNs together with cells or cell extracts of the AA4ΔsdaB mutant strain complemented with the sdaB wild-type gene (Figure 6A). These findings were supported by disease severity analysis, where the disease index of plants treated with PWNs and AA4ΔsdaB mutant cells or cell extracts was significantly higher than that with PWNs treated with the cells or cell extracts of wild-type AA4 or the sdaB mutant complemented with the sdaB wild-type gene (Figure 6B). The indices of the PWN infected seedlings treated with cells or cell extracts of the AA4ΔsdaB mutant complemented with the wild-type sdaB gene and seedlings treated with PWNs plus the wild-type AA4 strain cells or cell extracts showed no significant differences. The culture supernatants of the wild-type AA4, the AA4ΔsdaB mutant strain, or AA4ΔsdaB mutant complemented with the sdaB wild-type gene not only exhibited nearly no nematocidal activity against B. xylophilus in vitro, but also failed to display biocontrol efficacy against PWD (Figure 6). The sdaB gene encoding the beta subunit of L-serine dehydratase, is highly related to the PWN-killing activity of E. ludwigii AA4 and may be a key molecular element controlling the interplay between strain AA4 and PWNs.
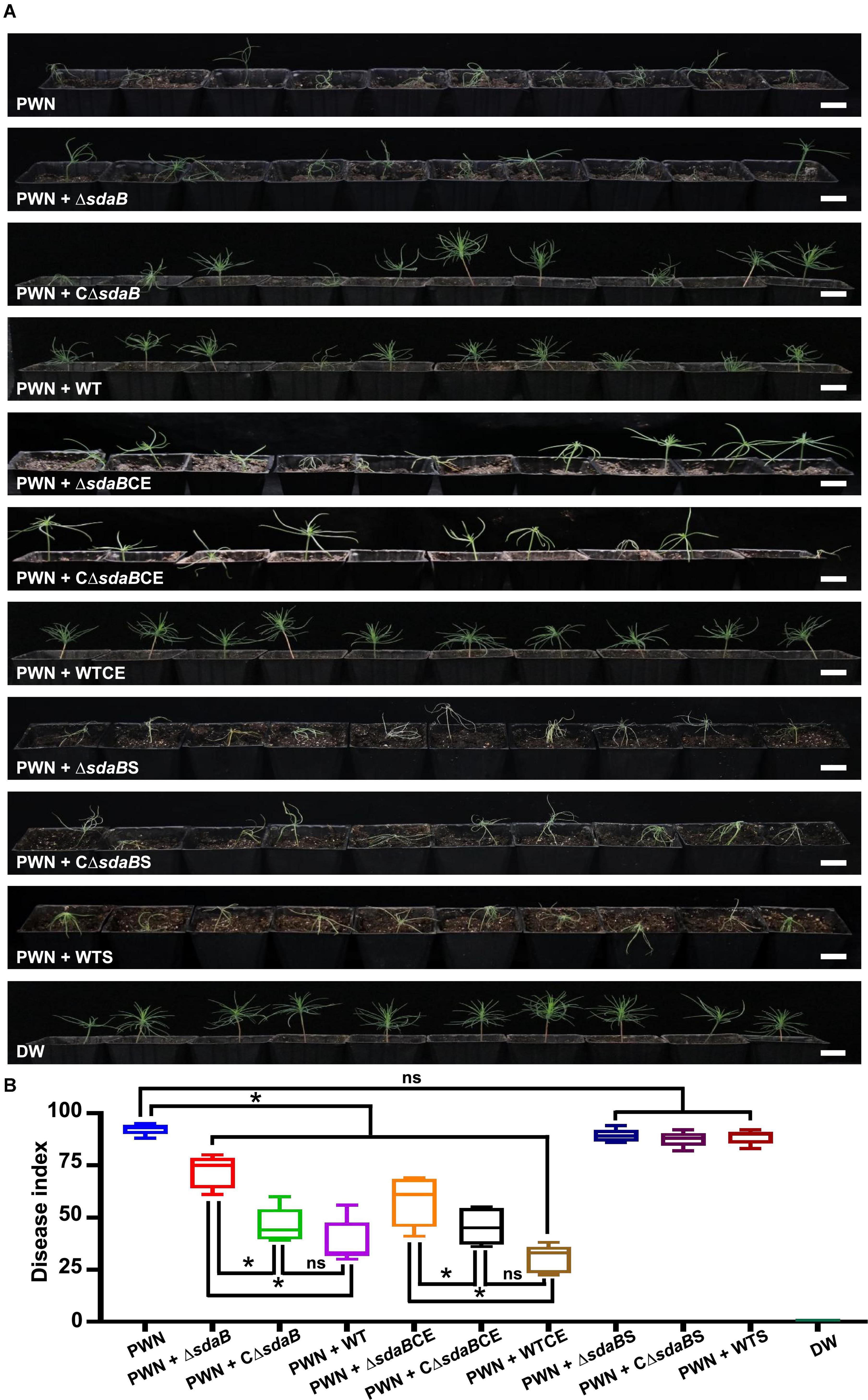
Figure 6. Disease-suppressing effect of E. ludwigii AA4 against PWD caused by B. xylophilus. (A) Biocontrol effect of E. ludwigii AA4 and its sdaB gene knock-out mutant against PWD on 1-month-old P. sylvestris seedlings. (B) Severity of PWD on 1-month-old P. sylvestris seedlings treated with PWN jointly with wild-type AA4 or AA4ΔsdaB. The horizontal bars within boxes represent the median. The tops and bottoms of boxes represent the 75th and 25th quartiles, respectively. The upper and lower whiskers extend from 75th quartiles to the maxima and from the 25th quartiles to the minima, respectively. The asterisks indicate that differences among the means represented by the boxes are statistically significant (*p ≤ 0.0345). Two-tailed t-test (GrapPad Prism 8) was used for the analysis. PWN, treatment with PWN alone; PWN + ΔsdaB, joint treatment with PWN and AA4ΔsdaB; PWN + CΔsdaB, joint treatment with PWN and AA4CΔsdaB; PWN + WT, joint treatment with PWN and wild-type AA4; PWN + ΔsdaBCE, joint treatment with PWN and AA4ΔsdaB cell extract; PWN + CΔsdaBCE, joint treatment with PWN and AA4CΔsdaB cell extract; PWN + WTCE, joint treatment with PWN and wild-type AA4 cell extract; PWN + ΔsdaBS, joint treatment with PWN and AA4ΔsdaB culture supernatant; PWN + CΔsdaBS, joint treatment with PWN and AA4CΔsdaB culture supernatant; PWN + WTS, joint treatment with PWN and wild-type AA4 culture supernatant; DW, treatment with distilled water (scale bars: 2 cm).
Colonization of E. ludwigii AA4 on Needles of P. sylvestris
Efficient colonization of plant surfaces and tissues is fundamental for successful inhibition of pathogens by biological control agents. To investigate the ability of E. ludwigii AA4 to colonize P. sylvestris, we sprayed the needles with a GFP-labeled transgenic strain of AA4. We observed by CLSM that strain AA4 was capable of adhering to the surface of P. sylvestris needles by forming robust biofilms (Figure 7A). Although there was a dramatical reduction of the abundance of AA4 cells sticking to the needles, from around 1.9 × 107 CFU (colony-forming unit)/g (of pine needle fresh weight) 4 h after inoculation to 3.8 × 104 CFU/g 144 h after inoculation, the colonization rates of AA4 at days 6 and 7 (2.8 × 104 CFU/g) showed no significant difference (Figure 7B). This indicates that E. ludwigii AA4 can colonize the needles of P. sylvestris, presumably a precondition for its disease-controlling efficacy against PWD. AA4ΔsdaB cells were equally able to colonize pine needles compared to the wild-type AA4 (Figure 7A), demonstrating that the sdaB gene function is not needed for colonization. The biomass accumulation rate of both strains was at the same level along our 7-day-long experiment (Figure 7B). Therefore, E. ludwigii AA4 may be an efficient colonizer of pine needles, and that the eradication of sdaB gene did not affect its adherence to needles.
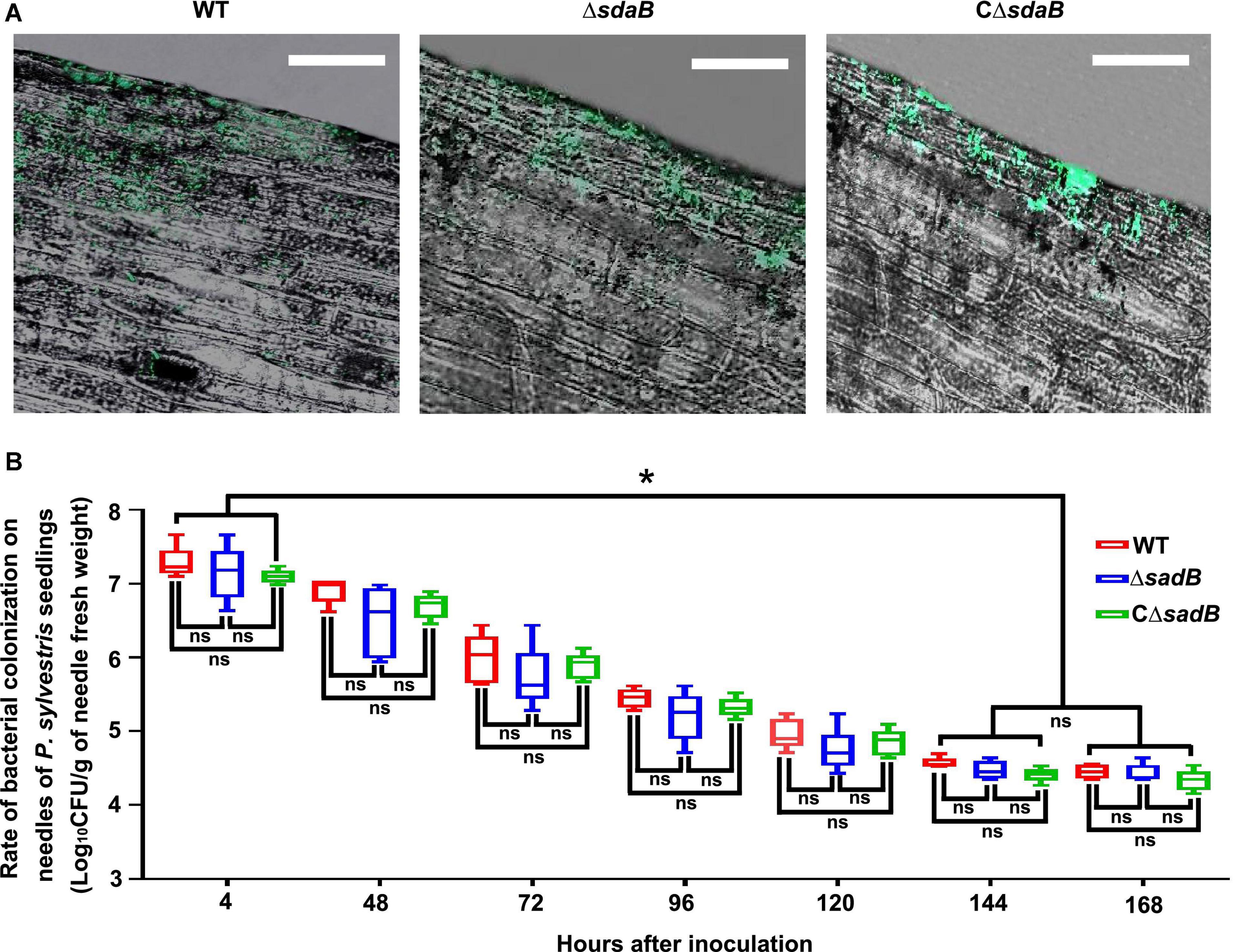
Figure 7. Colonization of E. ludwigii AA4 on needles of P. sylvestris seedlings. (A) Image analysis of needle colonization of green fluorescent protein (GFP)-labeled wild-type AA4, AA4 sdaB gene knock-out mutant, and AA4ΔsdaB complemented with a wild-type sdaB gene. These images were taken using a confocal laser scanning microscope at 48 h post inoculation (Scale bars: 20 μm). (B) The abundance of wild-type AA4, AA4 sdaB gene knock-out mutant, and AA4ΔsdaB complemented with a wild-type sdaB gene colonizing on the needles of P. sylvestris seedlings. WT, AA4 wild-type strain; ΔsdaB, AA4 sdaB gene knock-out mutant; CΔsdaB, AA4ΔsdaB complemented with a wild-type sdaB gene. The asterisks indicate that differences among the means represented by the columns are statistically significant (*p ≤ 0.0041). Two-tailed t-test (GrapPad Prism 8) was used for the analysis.
Discussion
Compared to the control of phytopathogens in agroecosystems, forest disease suppression puts more emphases on the sustainable management of pests due to the vital functionality of forests in maintaining the balance and health of their ecosystems (Nascimento et al., 2015; Proenca et al., 2017; Khan et al., 2020; Ozair et al., 2020). To this end, biological control of forest pathogens, including the PWNs, by beneficial microbes have been receiving increasing attention (Nunes da Silva et al., 2015; Ponpandian et al., 2019; Proenca et al., 2019). A considerable number of nematocidal microbial strains suppressing B. xylophilus have been identified and characterized. These strains are, mainly, species of Bacillus (Crickmore, 2005; Niu et al., 2011; Park et al., 2020), Streptomyces (Kang et al., 2021), Serratia (Paiva et al., 2013; Nascimento et al., 2016; Abd El-Aal et al., 2021), and Stenotrophomonas (Huang et al., 2009; Ponpandian et al., 2019).
We selected a PWN-killing Enterobacter ludwigii strain, designated as AA4 from a large-scale screening of nematocidal activity and inhibition of PWD (Figures 1C, 3). Representatives of the genus Enterobacter are able to inhibit a wide range of phytopathogens (Demirci et al., 2000; Kageyama and Nelson, 2003; Windstam and Nelson, 2008; Del Barrio-Duque et al., 2019; Batyrova et al., 2020), including plant pathogenic nematodes, such as root-knot nematodes (Munif et al., 2000; El-Sayed et al., 2014; Oh et al., 2018). To our knowledge, E. ludwigii AA4 is the first Enterobacter strain described as a nematocidal agent suppressing B. xylophilus. Our findings expand the knowledge of the biocontrol potential of the genus Enterobacter, one of the most expanding bacterial taxa in recent decades (Taghavi et al., 2010; Ibort et al., 2018; François et al., 2021).
Accurate and rapid determination of the mortality of nematodes is the key to efficient screening and selection of microbial strains with nematocidal activity. In earlier studies on PPN-bacterium interactions, the killing rate of nematodes was estimated mainly by the morphology of the worms, where rigid nematodes were considered dead, while the curved ones were considered alive (Liu et al., 2016, 2019; Ponpandian et al., 2019; Qiu et al., 2019). Such criteria do not always work well. We found that a significant number of apparently rigid nematodes could still move after being touched with a needle, suggesting that they were still alive. In addition, counting dead nematodes under a microscope is time consuming and tedious.
We developed a visual PWN-killing activity DAPI dye exclusion which is faster and more reliable (Figure 2). DAPI passes slowly through intact cell membranes of living nematodes, and thus preferentially stains the dead nematodes. Once entering the cells and binding to the minor groove of A-T-rich regions of DNA, DAPI fluorescence is greatly enhanced (Atale et al., 2014). As a result, dead worms display intense fluorescent signals. A similar method was employed in a previous study, where the dead Caenorhabditis elegans nematodes were stained with the fluorophore Sytox Orange (Natalia et al., 2013; Rajamuthiah et al., 2015; Xie et al., 2020). Applying fluorescence staining in the identification of PWN-killing bacterial strains may be amenable to high throughput or automated screening for biocontrol agents against other plant pathogenic nematodes.
In present study, we observed that the PWNs treated with cell extracts of wild-type E. ludwigii AA4 displayed dramatic morphological defects associated with the accumulation of numerous vacuoles, recognized as a sign of reversible cell injury (RCI) and methuosis, resulting in the destruction of internal organs of B. xylophilus needed for absorbing nutrients, pathogenicity, and reproduction (Figure 4). Similar changes in morphology were also reported in B. xylophilus incubated with indoles and abamectin (Rajasekharan et al., 2017). Another plant pathogenic nematode, Meloidogyne incognita, the causal agent of root knot in several crops, showed bubbling in its body when meeting toxic compounds as well (Jang et al., 2016). A large number of vacuoles were present in the transgenic C. elegans with an ectopically expressed and activated mek-1 gene (Koga et al., 2000). Intracytoplasmic vacuolation may commonly occur when nematodes live under stressful conditions, which can be triggered by fluid accumulation within the cytoplasm due to Ca2+ and water influx caused by RCI (Jurkowitz-Alexander et al., 1992; Basavappa et al., 1998; Rajasekharan et al., 2017). The death of B. xylophilus caused by AA4 might be attributed to methuosis induced by damage to the cell membrane caused by this nematocidal bacterial strain. The mechanisms underlying the PWN-killing function of E. ludwigii AA4 remains to be determined.
Some mechanisms of nematocidal activity against B. xylophilus have been recently documented, such as parasitization (Proenca et al., 2017; Ponpandian et al., 2019), production of toxins (Abd El-Aal et al., 2021; Kahn et al., 2021), antibiotics and destructive enzymes (Yang et al., 2007; Huang et al., 2009) or competing for nutrients (Proenca et al., 2019), and inducing systemic resistance of plants (Liang et al., 2019; Han et al., 2021). The molecular mechanisms underlying these PWN-killing effects are still largely unknown. In this work, we demonstrate that the sdaB gene encoding the beta subunit of L-serine dehydratase is involved in the nematocidal activity of E. ludwigii AA4 against PWNs. The deletion of this gene caused significant impairment in both L-serine hydrolase activity (Figure 5) and ability to inhibit PWN (Figures 2, 6), but not the growth of E. ludwigii AA4 in vitro or in planta (Supplementary Figure 2). L-serine dehydratase activity is widely found in diverse organisms. It specifically deaminates L-serine to produce pyruvate and ammonia (Xu et al., 2011; Grant, 2012; Thoden et al., 2014), and plays a critical role in maintaining amino acid homeostasis during shifts in nutrient availability (Velayudhan et al., 2004; Zhang et al., 2010; Chen et al., 2012). Until now, little or no data about the contribution of L-serine dehydratase to microbial nematocidal activities has been reported. L-serine is an important biomolecule, and excessive amounts of intracellular serine inhibit the biosynthesis of other amino acids (Zhang et al., 2018, 2019; Révora et al., 2020; Wang et al., 2020) essential to the production of certain bacterial toxins via the non-ribosomal peptide synthetase (NRPS) pathway (Cao et al., 2008; Soeriyadi et al., 2021).
Here, we assume that the sdaB gene product might control B. xylophilus by regulation of potential anti-nematode compounds in E. ludwigii AA4. Considering the tight relationship between L-serine and methylation reactions (Kalhan and Hanson, 2012; Datta et al., 2016; Bignell et al., 2018; Kriner and Subramaniam, 2020), such modulation may be linked to bacterial epigenetics. We noticed that the supernatant of strain AA4 cell culture has almost no effect (Figures 2, 6), which may be due to the low concentration of nematocidal compounds in the broth or suggests that the PWN-killing efficacy of AA4 might be contact-dependent. In future work, we will more deeply analyze the function of the sdaB gene in the nematocidal activity of E. ludwigii AA4.
Colonization of plant hosts is a necessary precondition for efficient pathogen inhibition by microbial biocontrol agents (Vurukonda et al., 2018; Niu et al., 2020; Zhang J. et al., 2020; Vandana et al., 2021). E. ludwigii AA4 was capable of colonizing the needles of P. sylvestris by forming sturdy biofilms (Figure 7), indicating its high affinity to pine needles, despite its presumed origin from herbaceous host plants. This finding underlines the great adaptability of the genus Enterobacter, which appears to be highly adaptable to diverse environments (Shoebitz et al., 2009; Singh et al., 2017; Shastry et al., 2020; Ranawat et al., 2021). These features may facilitate the development of an AA4-based biopesticide against B. xylophilus. Identification of genes and proteins involved in resistance to pests and pathogens is of long-term interest because such molecules represent the next generation of targets for creation of nematode resistant plants through genetic engineering or targeted mutagenesis. Our findings also pave the way for introducing agriculture biocontrol agents in forest disease management.
Data Availability Statement
The data presented in the study are deposited in the Genbank repository, accession numbers CP018785, OM883853, OM899759-OM899796, OM899810, OM900025, and OM900026.
Author Contributions
BN and RB designed the project. YZ and BN wrote the manuscript. BN, RB, ZY, RS, and HS revised the manuscript. YZ and ZY performed the experiments. YZ, SW, DW, and BN analyzed the data. YC created the random transposon library. HW, HY, JP, and MP performed the in planta biocontrol assays. All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by National Science Foundation of China Grant No. 32071741 (to BN), Heilongjiang Provincial Science Foundation of China Grant No. YQ2019C002 (to BN), Beijing Municipal Administration Center of Parks Science and Technology Project Grant No. zx2021002 (to SW), Fundamental Research Funds for the Central Universities Grant No. 2572018AA36 (to ZY), Northeast Forestry University International Forest Biotech Center Project Grant (to BN), and Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Yaxin Shi for performing the screening of nematocidal bacterial strains, Hongtao Li, Kai Guo, and Ben Fan for offering the cultures of pinewood nematodes, Qi Wang for providing the pGFP78, Mingyue Wang for help with the construction of mutant strains, and members of the BN laboratory for the valuable advice.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.870519/full#supplementary-material
Supplementary Figure 1 | The corrected mortality of 43 efficient pine wood nematode (PWN)-killing bacterial strains. Bacillus pumilus YLT40 and Paenibacillus polymyxa M-1 were used as positive and negative controls, respectively. Asterisk indicates that differences among the means represented by the columns are statistically significant (*p < 0.0001). Two-tailed t-test (GrapPad Prism 8) was used for the analysis.
Supplementary Figure 2 | Growth curves of wild-type E. ludwigii AA4, its sdaB gene knock-out mutant, and AA4ΔsdaB complemented with a wild-type sdaB gene. WT, E. ludwigii AA4 wild-type strain; ΔsdaB, AA4 sdaB gene knock-out mutant; CΔsdaB, AA4ΔsdaB complemented with a wild-type sdaB gene; LB, sterile Luria-Bertani liquid medium.
Supplementary Figure 3 | Disease ranks of the pine wilt disease (PWD) caused by B. xylophilus under greenhouse conditions on day 9 after inoculation (scale bars: 20 μm). In rank 0, stems and needles are green and plump. In rank 1, yellowish speckles are presented on stems, and stems and needles appear slightly shriveled. In rank 2, stems turn yellow and shriveled, and less than 25% needles are partially wilted. In rank 3, stems turn yellow and shriveled, among which 25–50% of needles are partially wilted. In rank 4, stems turn brownish and completely shriveled, and more than 50% of needles are completely wilted and turn yellowish. W, the whole views of P. sylvestris seedlings; T, the top views of P. sylvestris seedlings; S, the stems of P. sylvestris seedlings; N, the needles of P. sylvestris seedlings.
Supplementary Figure 4 | The location of miniTn10 transposon on the genome sequence of E. ludwigii AA4. The site of miniTn10 transposon insertion is indicated by a black triangle.
Supplementary Table 1 | Corrected mortality rates for the 43 efficient pine wood nematode (PWN)-killing strains.
Footnotes
References
Abd El-Aal, E. M., Shahen, M., Sayed, S., Kesba, H., Ansari, M. J., El-Ashry, R. M., et al. (2021). In vivo and in vitro management of Meloidogyne incognita (Tylenchida: Heteroderidae) using rhizosphere bacteria, Pseudomonas spp. and Serratia spp. compared with oxamyl. Saudi J. Biol. Sci. 28, 4876–4883. doi: 10.1016/j.sjbs.2021.06.078
Abdelshafy Mohamad, O. A., Ma, J. B., Liu, Y. H., Zhang, D., Hua, S., Bhute, S., et al. (2020). Beneficial endophytic bacterial populations associated with medicinal plant Thymus vulgaris alleviate salt stress and confer resistance to Fusarium oxysporum. Front. Plant Sci. 11:47. doi: 10.3389/fpls.2020.00047
Adhikari, A., Lee, K. E., Khan, M. A., Kang, S. M., Adhikari, B., Imran, M., et al. (2020). Effect of silicate and phosphate solubilizing rhizobacterium Enterobacter ludwigii GAK2 on Oryza sativa L. under cadmium stress. J. Microbiol. Biotechnol. 30, 118–126. doi: 10.4014/jmb.1906.06010
Aeron, A., Dubey, R. C., and Maheshwari, D. K. (2021). Next-generation biofertilizers and novel biostimulants: documentation and validation of mechanism of endophytic plant growth-promoting rhizobacteria in tomato. Arch. Microbiol. 203, 3715–3726. doi: 10.1007/s00203-021-02344-0
Andres-Barrao, C., Lafi, F. F., Alam, I., Zelicourt, A., Eida, A. A., Bokhari, A., et al. (2017). Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Front. Microbiol. 8:2023. doi: 10.3389/fmicb.2017.02023
Armenta, D. A., and Dixon, S. J. (2020). Investigating nonapoptotic cell death using chemical biology approaches. Cell Chem. Biol. 27, 376–386. doi: 10.1016/j.chembiol.2020.03.005
Atale, N., Gupta, S., Yadav, U. C., and Rani, V. (2014). Cell-death assessment by fluorescent and nonfluorescent cytosolic and nuclear staining techniques. J. Microsc. 255, 7–19. doi: 10.1111/jmi.12133
Basavappa, S., Mobasheri, A., Errington, R., Huang, C. C., Al-Adawi, S., and Ellory, J. C. (1998). Inhibition of Na+, K+-ATPase activates swelling-induced taurine efflux in a human neuroblastoma cell line. J. Cell. Physiol. 174, 145–153. doi: 10.1002/(SICI)1097-4652(199802)174:2<145::AID-JCP1>3.0.CO;2-O
Batyrova, K. A., Khusnutdinova, A. N., Wang, P. H., Di Leo, R., Flick, R., Edwards, E. A., et al. (2020). Biocatalytic in Vitro and in Vivo FMN prenylation and (De)carboxylase activation. ACS Chem. Biol. 15, 1874–1882. doi: 10.1021/acschembio.0c00136
Bignell, D., Cheng, Z., and Bown, L. (2018). The coronafacoyl phytotoxins: structure, biosynthesis, regulation and biological activities. Antonie van Leeuwenhoek 111, 649–666. doi: 10.1007/s10482-017-1009-1
Cai, H., Liu, J., Fan, Q., and Li, X. (2013). Methuosis: a novel type of cell death. J. Southern Med. Univ. 33, 1844–1847.
Cai, S., Jia, J., He, C., Zeng, L., Fang, Y., Qiu, G., et al. (2022). Multi-omics of pine wood nematode pathogenicity associated with culturable associated microbiota through an artificial assembly approach. Front. Plant Sci. 12:798539. doi: 10.3389/fpls.2021.798539
Cao, L., Bandelac, G., Volgina, A., Korostoff, J., and DiRienzo, J. M. (2008). Role of aromatic amino acids in receptor binding activity and subunit assembly of the cytolethal distending toxin of Aggregatibacter actinomycetemcomitans. Infect. Immunity 76, 2812–2821. doi: 10.1128/IAI.00126-08
Cesarz, S., Eva Schulz, A., Beugnon, R., and Eisenhauer, N. (2019). Testing soil nematode extraction efficiency using different variations of the Baermann-funnel method. Soil Org. 91, 61–72. doi: 10.25674/so91201
Chan, S. Y., Liu, S. Y., Seng, Z., and Chua, S. L. (2020). Biofilm matrix disrupts nematode motility and predatory behavior. Int. Soc. Microb. Ecol. J. 15, 260–269. doi: 10.1038/s41396-020-00779-9
Chen, D., Song, M., Mohamad, O., and Yu, S. P. (2014). Inhibition of Na+/K+-ATPase induces hybrid cell death and enhanced sensitivity to chemotherapy in human glioblastoma cells. BMC Cancer 14:716. doi: 10.1186/1471-2407-14-716
Chen, S., Xu, X. L., and Grant, G. A. (2012). Allosteric activation and contrasting properties of L-serine dehydratase types 1 and 2. Biochemistry 51, 5320–5328. doi: 10.1021/bi300523p
Crickmore, N. (2005). Using worms to better understand how Bacillus thuringiensis kills insects. Trends Microbiol. 13, 347–350. doi: 10.1016/j.tim.2005.06.002
Dahyot, S., Oxaran, V., Niepceron, M., Dupart, E., Legris, S., Destruel, L., et al. (2020). Role of the LytSR two-component regulatory system in Staphylococcus lugdunensis biofilm formation and pathogenesis. Front. Microbiol. 11:39. doi: 10.3389/fmicb.2020.00039
Danish, S., Zafar-Ul-Hye, M., Mohsin, F., and Hussain, M. (2020). ACC-deaminase producing plant growth promoting rhizobacteria and biochar mitigate adverse effects of drought stress on maize growth. PLoS One 15:e0230615. doi: 10.1371/journal.pone.0230615
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Datta, A., Bhattacharyya, D., Singh, S., Ghosh, A., Schmidtchen, A., Malmsten, M., et al. (2016). Role of aromatic amino acids in lipopolysaccharide and membrane interactions of antimicrobial peptides for use in plant disease control. J. Biol. Chem. 291, 13301–13317. doi: 10.1074/jbc.M116.719575
Del Barrio-Duque, A., Ley, J., Samad, A., Antonielli, L., Sessitsch, A., and Compant, S. (2019). Beneficial endophytic bacteria-Serendipita indica interaction for crop enhancement and resistance to phytopathogens. Front. Microbiol. 10:2888. doi: 10.3389/fmicb.2019.02888
Demirci, F., Işcan, G., Güven, K., Kirimer, N., Demirci, B., and Başer, K. H. (2000). Antimicrobial activities of Ferulago essential oils. Z. Naturforsch. C J. Biosci. 55, 886–889. doi: 10.1515/znc-2000-11-1207
Dhanya, B. E., Athmika, and Rekha, P. D. (2021). Characterization of an exopolysaccharide produced by Enterobacter sp. YU16-RN5 and its potential to alleviate cadmium induced cytotoxicity in vitro. 3 Biotech 11:491. doi: 10.1007/s13205-021-03034-w
Duncan, M. C., Forbes, J. C., Nguyen, Y., Shull, L. M., Gillette, R. K., Lazinski, D. W., et al. (2018). Vibrio cholerae motility exerts drag force to impede attack by the bacterial predator Bdellovibrio bacteriovorus. Nat. Commun. 9:4757. doi: 10.1038/s41467-018-07245-3
El-Sayed, W. S., Akhkha, A., El-Naggar, M. Y., and Elbadry, M. (2014). In vitro antagonistic activity, plant growth promoting traits and phylogenetic affiliation of rhizobacteria associated with wild plants grown in arid soil. Front. Microbiol. 5:651. doi: 10.3389/fmicb.2014.00651
Etcheverry, M. G., Scandolara, A., Nesci, A., Vilas Boas Ribeiro, M. S., Pereira, P., and Battilani, P. (2009). Biological interactions to select biocontrol agents against toxigenic strains of Aspergillus flavus and Fusarium verticillioides from maize. Mycopathologia 167, 287–295. doi: 10.1007/s11046-008-9177-1
Fang, W., Qunqun, G., Linsong, W., Yi, M., Tingting, Z., and Ronggui, L. (2019). Nematicidal activities of bacterial volatiles from Pseudoduganella violaceinigra G5-3 and Novosphingobium pokkalii G8-2 against the pine wood nematode Bursaphelenchus xylophilus. Chiang Mai J. Sci. 46, 236–246.
Faria, J. M., Barbosa, P., Bennett, R. N., Mota, M., and Figueiredo, A. C. (2013). Bioactivity against Bursaphelenchus xylophilus: nematotoxics from essential oils, essential oils fractions and decoction waters. Phytochemistry 94, 220–228. doi: 10.1016/j.phytochem.2013.06.005
Faria, J. M., Sena, I., Vieira da Silva, I., Ribeiro, B., Barbosa, P., Ascensao, L., et al. (2015). In vitro co-cultures of Pinus pinaster with Bursaphelenchus xylophilus: a biotechnological approach to study pine wilt disease. Planta 241, 1325–1336. doi: 10.1007/s00425-015-2257-9
Faria, J., Barbosa, P., Vieira, P., Vicente, C., Figueiredo, A. C., and Mota, M. (2021). Phytochemicals as biopesticides against the pinewood nematode Bursaphelenchus xylophilus: a review on essential oils and their volatiles. Plants (Basel, Switzerland) 10:2614. doi: 10.3390/plants10122614
François, C. J., Batinovic, S., Petrovski, S., and Gendall, A. R. (2021). Draft genome sequence of Enterobacter asburiae NCR1, a plant growth-promoting rhizobacterium isolated from a cadmium-contaminated environment. Microbiol. Resour. Announc. 10:e0047821. doi: 10.1128/MRA.00478-21
Futai, K. (2013). Pine wood nematode, Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 51, 61–83. doi: 10.1146/annurev-phyto-081211-172910
Grant, G. A. (2012). Kinetic evidence of a noncatalytic substrate binding site that regulates activity in Legionella pneumophila L-serine dehydratase. Biochemistry 51, 6961–6967. doi: 10.1021/bi3008774
Guérin, F., Lallement, C., Goudergues, B., Isnard, C., Sanguinetti, M., Cacaci, M., et al. (2020). Landscape of in vivo fitness-associated genes of Enterobacter cloacae complex. Front. Microbiol. 11:1609. doi: 10.3389/fmicb.2020.01609
Guo, Q. Q., Du, G. C., Zhang, T. T., Wang, M. J., Wang, C., Qi, H. T., et al. (2020). Transcriptomic analysis of Bursaphelenchus xylophilus treated by a potential phytonematicide, punicalagin. J. Nematol. 52, 1–14. doi: 10.21307/jofnem-2020-001
Guo, Q., Du, G., He, H., Xu, H., Guo, D., and Li, R. (2016). Two nematicidal furocoumarins from Ficus carica L. leaves and their physiological effects on pine wood nematode (Bursaphelenchus xylophilus). Nat. Prod. Res. 30, 1969–1973. doi: 10.1080/14786419.2015.1094804
Habibi, S., Djedidi, S., Ohkama-Ohtsu, N., Sarhadi, W. A., Kojima, K., Rallos, R. V., et al. (2019). Isolation and screening of indigenous plant growth-promoting rhizobacteria from different rice cultivars in Afghanistan soils. Microbes Environ. 34, 347–355. doi: 10.1264/jsme2.ME18168
Haegeman, A., Vanholme, B., Jacob, J., Vandekerckhove, T., Claeys, M., Borgonie, G., et al. (2009). An endosymbiotic bacterium in a plant-parasitic nematode: member of a new Wolbachia supergroup. Int. J. Parasitol. 39, 1045–1054. doi: 10.1016/j.ijpara.2009.01.006
Han, G., Mannaa, M., Kim, N., Jeon, H. W., Jung, H., Lee, H. H., et al. (2021). Response of pine rhizosphere microbiota to foliar treatment with resistance-inducing bacteria against pine wilt disease. Microorganisms 9:688. doi: 10.3390/microorganisms9040688
Hinton, D. M., and Bacon, C. W. (1995). Enterobacter cloacae is an endophytic symbiont of corn. Mycopathologia 129, 117–125. doi: 10.1007/BF01103471
Huang, X., Liu, J., Ding, J., He, Q., Xiong, R., and Zhang, K. (2009). The investigation of nematocidal activity in Stenotrophomonas maltophilia G2 and characterization of a novel virulence serine protease. Can. J. Microbiol. 55, 934–942. doi: 10.1139/w09-045
Ibort, P., Molina, S., Ruiz-Lozano, J. M., and Aroca, R. (2018). Molecular insights into the involvement of a never ripe receptor in the interaction between two beneficial soil bacteria and tomato plants under well-watered and drought conditions. Mol. Plant Microbe Interact. 31, 633–650. doi: 10.1094/MPMI-12-17-0292-R
Jang, J. Y., Choi, Y. H., Shin, T. S., Kim, T. H., Shin, K. S., Park, H. W., et al. (2016). Biological control of Meloidogyne incognita by Aspergillus niger F22 producing oxalic acid. PLoS One 11:e0156230. doi: 10.1371/journal.pone.0156230
Javaheri Safa, Z., Olya, A., Zamani, M., Motalebi, M., Khalili, R., Haghbeen, K., et al. (2021). Biodegradation of cyanide to ammonia and carbon dioxide by an industrially valuable enzyme from the newly isolated Enterobacter zs. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 56, 1131–1137. doi: 10.1080/10934529.2021.1967653
Jurkowitz-Alexander, M. S., Altschuld, R. A., Hohl, C. M., Johnson, J. D., McDonald, J. S., Simmons, T. D., et al. (1992). Cell swelling, blebbing, and death are dependent on ATP depletion and independent of calcium during chemical hypoxia in a glial cell line (ROC-1). J. Neurochem. 59, 344–352. doi: 10.1111/j.1471-4159.1992.tb08910.x
Kageyama, K., and Nelson, E. B. (2003). Differential inactivation of seed exudate stimulation of Pythium ultimum sporangium germination by Enterobacter cloacae influences biological control efficacy on different plant species. Appl. Environ. Microbiol. 69, 1114–1120. doi: 10.1128/AEM.69.2.1114-1120.2003
Kahn, T. W., Duck, N. B., McCarville, M. T., Schouten, L. C., Schweri, K., Zaitseva, J., et al. (2021). A Bacillus thuringiensis Cry protein controls soybean cyst nematode in transgenic soybean plants. Nat. Commun. 12:3380. doi: 10.1038/s41467-021-23743-3
Kalhan, S. C., and Hanson, R. W. (2012). Resurgence of serine: an often neglected but indispensable amino Acid. J. Biol. Chem. 287, 19786–19791. doi: 10.1074/jbc.R112.357194
Kämpfer, P., Ruppel, S., and Remus, R. (2005). Enterobacter radicincitans sp. nov., a plant growth promoting species of the family Enterobacteriaceae. Syst. Appl. Microbiol. 28, 213–221. doi: 10.1016/j.syapm.2004.12.007
Kang, M. K., Kim, M. H., Liu, M. J., Jin, C. Z., Park, S. H., Lee, J., et al. (2021). Nematicidal activity of teleocidin B4 isolated from Streptomyces sp. against pine wood nematode, Bursaphelenchus xylophilus. Pest Manag. Sci. 77, 1607–1615. doi: 10.1002/ps.6095
Kang, S. M., Radhakrishnan, R., You, Y. H., Khan, A. L., Lee, K. E., Lee, J. D., et al. (2015). Enterobacter asburiae KE17 association regulates physiological changes and mitigates the toxic effects of heavy metals in soybean. Plant Biol. (Stuttgart, Germany). 17, 1013–1022. doi: 10.1111/plb.12341
Kaushik, C. P., Luxmi, R., Singh, D., and Kumar, A. (2017). Synthesis and antimicrobial evaluation of ester-linked 1,4-disubstituted 1,2,3-triazoles with a furyl/thienyl moiety. Mol. Divers. 21, 137–145. doi: 10.1007/s11030-016-9710-y
Khan, M. A., Ahmed, L., Mandal, P. K., Smith, R., and Haque, M. (2020). Modelling the dynamics of pine wilt disease with asymptomatic carriers and optimal control. Sci. Rep. 10:11412. doi: 10.1038/s41598-020-67090-7
Kikuchi, T., Cotton, J. A., Dalzell, J. J., Hasegawa, K., Kanzaki, N., McVeigh, P., et al. (2011). Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 7:e1002219. doi: 10.1371/journal.ppat.1002219
Kim, H. M., Jeong, S. G., Choi, I. S., Yang, J. E., Lee, K. H., Kim, J., et al. (2020). Mechanisms of insecticidal action of Mmetarhizium anisopliae on adult Japanese pine sawyer beetles (Monochamus alternatus).”. ACS Omega 5, 25312–25318. doi: 10.1021/acsomega.0c03585
Kim, J. C., Baek, S., Park, S. E., Kim, S., Lee, M. R., Jo, M., et al. (2019). Colonization of Metarhizium anisopliae on the surface of pine tree logs: a promising biocontrol strategy for the Japanese pine sawyer, Monochamus alternatus. Fungal Biol. 124, 125–134. doi: 10.1016/j.funbio.2019.12.006
Kitazume, H., Dayi, M., Tanaka, R., and Kikuchi, T. (2018). Assessment of the behaviour and survival of nematodes under low oxygen concentrations. PLoS One 13:e0197122. doi: 10.1371/journal.pone.0197122
Koga, M., Zwaal, R., Guan, K. L., Avery, L., and Ohshima, Y. (2000). A Caenorhabditis elegans MAP kinase kinase, MEK-1, is involved in stress responses. EMBO J. 19, 5148–5156. doi: 10.1093/emboj/19.19.5148
Kriner, M. A., and Subramaniam, A. R. (2020). The serine transporter SdaC prevents cell lysis upon glucose depletion in Escherichia coli. Microbiol. Open 9:e960. doi: 10.1002/mbo3.960
Kumar, K. K., and Dara, S. K. (2021). Fungal and bacterial endophytes as microbial control agents for plant-parasitic nematodes. Int. J. Environ. Res. Public Health 18:4269. doi: 10.3390/ijerph18084269
Lee, I. H., Han, H., Koh, Y. H., Kim, I. S., Lee, S. W., and Shim, D. (2019). Comparative transcriptome analysis of Pinus densiflora following inoculation with pathogenic (Bursaphelenchus xylophilus) or non-pathogenic nematodes (B. thailandae). Sci. Rep. 9:12180. doi: 10.1038/s41598-019-48660-w
Li, J., Zou, C., Xu, J., Ji, X., Niu, X., Yang, J., et al. (2015). Molecular mechanisms of nematode-nematophagous microbe interactions: basis for biological control of plant-parasitic nematodes. Annu. Rev. Phytopathol. 53, 67–95. doi: 10.1146/annurev-phyto-080614-120336
Li, Y., Mo, L., Zhou, X., Yao, Y., Ma, J., Liu, K., et al. (2022). Characterization of plant growth-promoting traits of Enterobacter sp. and its ability to promote cadmium/lead accumulation in Centella asiatica L. Environ. Sci. Pollut. Res. Int. 29, 4101–4115. doi: 10.1007/s11356-021-15948-2
Liang, L. M., Zou, C. G., Xu, J., and Zhang, K. Q. (2019). Signal pathways involved in microbe-nematode interactions provide new insights into the biocontrol of plant-parasitic nematodes. Philos. Trans. R. Soc. B Biol. Sci. 374:1767. doi: 10.1098/rstb.2018.0317
Liu, G., Lai, D., Liu, Q., Zhou, Z. L., and Liu, Z. L. (2016). Identification of nematicidal constituents of Notopterygium incisum rhizomes against Bursaphelenchus xylophilus and Meloidogyne incognita. Molecules 21:1276. doi: 10.3390/molecules21101276
Liu, M. J., Hwang, B. S., Jin, C. Z., Li, W. J., Park, D. J., Seo, S. T., et al. (2019). Screening, isolation and evaluation of a nematicidal compound from actinomycetes against the pine wood nematode, Bursaphelenchus xylophilus. Pest Manag. Sci. 75, 1585–1593. doi: 10.1002/ps.5272
Lohrke, S. M., Dery, P. D., Li, W., Reedy, R., Kobayashi, D. Y., and Roberts, D. R. (2002). Mutation of rpiA in Enterobacter cloacae decreases seed and root colonization and biocontrol of damping-off caused by Pythium ultimum on cucumber. Mol. Plant Microbe Interact. 15, 817–825. doi: 10.1094/MPMI.2002.15.8.817
López, A. C., and Alippi, A. M. (2019). Feasibility of using RFLP of PCR-amplified 16S rRNA gene(s) for rapid differentiation of isolates of aerobic spore-forming bacteria from honey. J. Microbiol. Methods 165:105690. doi: 10.1016/j.mimet.2019.105690
Maehara, N., Kanzaki, N., Aikawa, T., and Nakamura, K. (2020). Potential vector switching in the evolution of Bursaphelenchus xylophilus group nematodes (Nematoda: Aphelenchoididae). Ecol. Evol. 10, 14320–14329. doi: 10.1002/ece3.7033
Mahdi, I., Fahsi, N., Hafidi, M., Allaoui, A., and Biskri, L. (2020). Plant growth enhancement using rhizospheric halotolerant phosphate solubilizing bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 isolated from Chenopodium quinoa willd. Microorganisms 8:948. doi: 10.3390/microorganisms8060948
Mahmood, K., Akhter, Z., Asghar, M. A., Mirza, B., Ismail, H., Liaqat, F., et al. (2020). Synthesis, characterization and biological evaluation of novel benzimidazole derivatives. J. Biomol. Struct. Dyn. 38, 1670–1682. doi: 10.1080/07391102.2019.1617783
Maltese, W. A., and Overmeyer, J. H. (2014). Methuosis: nonapoptotic cell death associated with vacuolization of macropinosome and endosome compartments. Am. J. Pathol. 184, 1630–1642. doi: 10.1016/j.ajpath.2014.02.028
Mokracka, J., Koczura, R., and Kaznowski, A. (2004). Yersiniabactin and other siderophores produced by clinical isolates of Enterobacter spp. and Citrobacter spp. FEMS Immunol. Med. Microbiol. 40, 51–55. doi: 10.1016/S0928-8244(03)00276
Monciardini, P., Sosio, M., Cavaletti, L., Chiocchini, C., and Donadio, S. (2002). New PCR primers for the selective amplification of 16S rDNA from different groups of actinomycetes. FEMS Microbiol. Ecol. 42, 419–429. doi: 10.1111/j.1574-6941.2002.tb01031.x
Mpongwana, N., Ntwampe, S. K., Mekuto, L., Akinpelu, E. A., Dyantyi, S., and Mpentshu, Y. (2016). Isolation of high-salinity-tolerant bacterial strains, Enterobacter sp., Serratia sp., Yersinia sp., for nitrification and aerobic denitrification under cyanogenic conditions. Water Sci. Technol. 73, 2168–2175. doi: 10.2166/wst.2016.070
Munif, A., Hallmann, J., and Sikora, R. A. (2000). “Evaluation of the biocontrol activity of endophytic bacteria from tomato against Meloidogyne incignita,” in Proceedings of the Mededelingen Faculteit Landbouwkundige En Toegepaste Biologische Wetenschappen, Vol. 65, (Belgium: Universiteit Gent), 471–480.
Nascimento, F. X., Espada, M., Barbosa, P., Rossi, M. J., Vicente, C. S., and Mota, M. (2016). Non-specific transient mutualism between the plant parasitic nematode, Bursaphelenchus xylophilus, and the opportunistic bacterium Serratia quinivorans BXF1, a plant-growth promoting pine endophyte with antagonistic effects. Environ. Microbiol. 18, 5265–5276. doi: 10.1111/1462-2920.13568
Nascimento, F. X., Hasegawa, K., Mota, M., and Vicente, C. S. (2015). Bacterial role in pine wilt disease development – review and future perspectives. Environ. Microbiol. Rep. 7, 51–63. doi: 10.1111/1758-2229.12202
Natalia, V. K., Daniel, R. K., Jonah, L., Carolina, W., Gary, R., and Frederick, M. A. (2013). Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 13, 406–416. doi: 10.1016/j.chom.2013.03.003
Nirmala, J. G., and Lopus, M. (2020). Cell death mechanisms in eukaryotes. Cell Biol. Toxicol. 36, 145–164. doi: 10.1007/s10565-019-09496-2
Niu, B., Paulson, J. N., Zheng, X., and Kolter, R. (2017). Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. U.S.A. 114, E2450–E2459. doi: 10.1073/pnas.1616148114
Niu, B., Vater, J., Rueckert, C., Blom, J., Lehmann, M., Ru, J. J., et al. (2013). Polymyxin P is the active principle in suppressing phytopathogenic Erwinia spp. by the biocontrol rhizobacterium Paenibacillus polymyxa M-1. BMC Microbiol. 13:137. doi: 10.1186/1471-2180-13-137
Niu, B., Wang, W., Yuan, Z., Sederoff, R. R., Sederoff, H., Chiang, V. L., et al. (2020). Microbial interactions within multiple-strain biological control agents impact soil-borne plant disease. Front. Microbiol. 11:585404. doi: 10.3389/fmicb.2020.585404
Niu, Q., Tian, Y., Zhang, L., Xu, X., Niu, X., Xia, Z., et al. (2011). Overexpression of the key virulence proteases Bace16 and Bae16 in Bacillus nematocidal B16 to improve its nematocidal activity. J. Mol. Microbiol. Biotechnol. 21, 130–137. doi: 10.1159/000332805
Niu, X., Song, L., Xiao, Y., and Ge, W. (2018). Drought-tolerant plant growth-promoting rhizobacteria associated with foxtail millet in a semi-arid agroecosystem and their potential in alleviating drought stress. Front. Microbiol. 8:2580. doi: 10.3389/fmicb.2017.02580
Nunes da Silva, M., Solla, A., Sampedro, L., Zas, R., and Vasconcelos, M. W. (2015). Susceptibility to the pinewood nematode (PWN) of four pine species involved in potential range expansion across Europe. Tree Physiol. 35, 987–999. doi: 10.1093/treephys/tpv046
Nurjadi, D., Kocer, K., Chanthalangsy, Q., Klein, S., Heeg, K., and Boutin, S. (2021). New delhi metallo-beta-lactamase facilitates the emergence of cefiderocol resistance in Enterobacter cloacae. Antimicrob. Agents Chemother. 66:e0201121. doi: 10.1128/AAC.02011-21
Oh, M., Han, J. W., Lee, C., Choi, G. J., and Kim, H. (2018). Nematicidal and plant growth-promoting activity of Enterobacter asburiae HK169: genome analysis provides insight into its biological activities. J. Microbiol. Biotechnol. 28, 968–975. doi: 10.4014/jmb.1801.01021
Ozair, M., Hussain, T., Awan, A. U., Aslam, A., Khan, R. A., Ali, F., et al. (2020). Bio-inspired analytical heuristics to study pine wilt disease model. Sci. Rep. 10:3534. doi: 10.1038/s41598-020-60088-1
Paiva, G. N., Proenca, D., Francisco, R., Verissimo, P., Santos, S. S., Fonseca, L., et al. (2013). Nematicidal bacteria associated to pinewood nematode produce extracellular proteases. PLoS One 8:e79705. doi: 10.1371/journal.pone.0079705
Park, A. R., Jeong, S. I., Jeon, H. W., Kim, J., Kim, N., Ha, M. T., et al. (2020). A diketopiperazine, Cyclo-(L-Pro-L-Ile), derived from Bacillus thuringiensis JCK-1233 controls pine wilt disease by elicitation of moderate hypersensitive reaction. Front. Plant Sci. 11:1023. doi: 10.3389/fpls.2020.01023
Park, J. M., Radhakrishnan, R., Kang, S. M., and Lee, I. J. (2015). IAA producing Enterobacter sp. I-3 as a potent bio-herbicide candidate for weed control: a special reference with lettuce growth inhibition. Indian J. Microbiol. 55, 207–212. doi: 10.1007/s12088-015-0515-y
Peng, G., Zhang, W., Luo, H., Xie, H., Lai, W., and Tan, Z. (2009). Enterobacter oryzae sp. nov., a nitrogen-fixing bacterium isolated from the wild rice species Oryza latifolia. Int. J. Syst. Evol. Microbiol. 59, 1650–1655. doi: 10.1099/ijs.0.005967-0
Ponpandian, L. N., Rim, S. O., Shanmugam, G., Jeon, J., Park, Y. H., Lee, S. K., et al. (2019). Phylogenetic characterization of bacterial endophytes from four Pinus species and their nematicidal activity against the pine wood nematode. Sci. Rep. 9:12457. doi: 10.1038/s41598-019-48745-6
Proenca, D. N., Grass, G., and Morais, P. V. (2017). Understanding pine wilt disease: roles of the pine endophytic bacteria and of the bacteria carried by the disease-causing pinewood nematode. Microbiol. Open 6:e00415. doi: 10.1002/mbo3.415
Proenca, D. N., Heine, T., Senges, C. H. R., Bandow, J. E., Morais, P. V., and Tischler, D. (2019). Bacterial metabolites produced under iron limitation kill pinewood nematode and attract Caenorhabditis elegans. Front. Microbiol.. 10:2166. doi: 10.3389/fmicb.2019.02166
Qiu, X. W., Wu, X. Q., Huang, L., and Ye, J. R. (2016). Influence of Bxpel1 gene silencing by dsRNA interference on the development and pathogenicity of the pine wood nematode, Bursaphelenchus xylophilus. Int. J. Mol. Sci. 17:125. doi: 10.3390/ijms17010125
Qiu, X., Yang, L., Ye, J., Wang, W., Zhao, T., Hu, H., et al. (2019). Silencing of cyp-33C9 gene affects the reproduction and pathogenicity of the pine wood nematode, Bursaphelenchus xylophilus. Int. J. Mol. Sci. 20:4520. doi: 10.3390/ijms20184520
Rajamuthiah, R., Jayamani, E., Majed, H., Conery, A. L., Kim, W., Kwon, B., et al. (2015). Antibacterial properties of 3-(phenylsulfonyl)-2-pyrazinecarbonitrile. Bioorg. Med. Chem. Lett. 25, 5203–5207. doi: 10.1016/j.bmcl.2015.09.066
Rajasekharan, S. K., Lee, J. H., Ravichandran, V., and Lee, J. (2017). Assessments of iodoindoles and abamectin as inducers of methuosis in pinewood nematode, Bursaphelenchus xylophilus. Sci. Rep. 7:6803. doi: 10.1038/s41598-017-07074-2
Ranawat, B., Mishra, S., and Singh, A. (2021). Enterobacter hormaechei (MF957335) enhanced yield, disease and salinity tolerance in tomato. Arch. Microbiol. 203, 2659–2667. doi: 10.1007/s00203-021-02226-5
Révora, V., Marchesini, M. I., and Comerci, D. J. (2020). Brucella abortus depends on l-Serine biosynthesis for intracellular proliferation. Infect. Immunity 88:e00840–19. doi: 10.1128/IAI.00840-19
Rodrigues, A. A., Araújo, M., Soares, R. S., Oliveira, B., Ribeiro, I., Sibov, S. T., et al. (2018). Isolation and prospection of diazotrophic rhizobacteria associated with sugarcane under organic management. Anais Acad. Bras. Cienc. 90, 3813–3829. doi: 10.1590/0001-3765201820180319
Roslan, M., Zulkifli, N. N., Sobri, Z. M., Zuan, A., Cheak, S. C., and Abdul Rahman, N. A. (2020). Seed biopriming with P- and K-solubilizing Enterobacter hormaechei sp. improves the early vegetative growth and the P and K uptake of okra (Abelmoschus esculentus) seedling. PLoS One 15:e0232860. doi: 10.1371/journal.pone.0232860
Sarkar, A., Ghosh, P. K., Pramanik, K., Mitra, S., Soren, T., Pandey, S., et al. (2018). A halotolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Res. Microbiol. 169, 20–32. doi: 10.1016/j.resmic.2017.08.005
Sarkar, S., and Chaudhuri, S. (2015). New report of additional enterobacterial species causing wilt in West Bengal, India. Can. J. Microbiol. 61, 477–486. doi: 10.1139/cjm-2015-0017
Sayyed, R. Z., Patel, P. R., and Shaikh, S. S. (2015). Plant growth promotion and root colonization by EPS producing Enterobacter sp. RZS5 under heavy metal contaminated soil. Indian J. Exp. Biol. 53, 116–123.
Shastry, R. P., Welch, M., Rai, V. R., Ghate, S. D., Sandeep, K., and Rekha, P. D. (2020). The whole-genome sequence analysis of Enterobacter cloacae strain Ghats1: insights into endophytic lifestyle-associated genomic adaptations. Arch. Microbiol. 202, 1571–1579. doi: 10.1007/s00203-020-01848-5
Shoebitz, M., Ribaudo, C. M., Pardo, M. A., Cantore, M. L., Ciampi, L., and Curá, J. A. (2009). Plant growth promoting properties of a strain of Enterobacter ludwigii isolated from Lolium perenne rhizosphere. Soil Biol. Biochem. 41, 1768–1774. doi: 10.1016/j.soilbio.2007.12.031
Singh, R. P., Runthala, A., Khan, S., and Jha, P. N. (2017). Quantitative proteomics analysis reveals the tolerance of wheat to salt stress in response to Enterobacter cloacae SBP-8. PLoS One 12:e0183513. doi: 10.1371/journal.pone.0183513
Soeriyadi, A. H., Ongley, S. E., Kehr, J. C., Pickford, R., Dittmann, E., and Neilan, B. A. (2021). Tailoring enzyme stringency masks the multispecificity of a Lyngbyatoxin (Indolactam Alkaloid) nonribosomal peptide synthetase. Chembiochem 23:e202100574. doi: 10.1002/cbic.202100574
Soliman, T., Mourits, M. C., Werf, W., Hengeveld, G. M., Robinet, C., and Lansink, A. G. (2012). Framework for modelling economic impacts of invasive species, applied to pine wood nematode in Europe. PLoS One 7:e45505. doi: 10.1371/journal.pone.0045505
Song, S., Zhang, Y., Ding, T., Ji, N., and Zhao, H. (2021). The dual role of macropinocytosis in cancers: promoting growth and inducing methuosis to participate in anticancer therapies as targets. Front. Oncol. 10:570108. doi: 10.3389/fonc.2020.570108
Srisuk, N., Sakpuntoon, V., and Nutaratat, P. (2018). Production of indole-3-acetic acid by Enterobacter sp. DMKU-RP206 using sweet whey as a low-cost feed stock. J. Microbiol. Biotechnol. 28, 1511–1516. doi: 10.4014/jmb.1804.04043
Taghavi, S., van der Lelie, D., Hoffman, A., Zhang, Y. B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 6:e1000943. doi: 10.1002/9781118297674.ch84
Thoden, J. B., Holden, H. M., and Grant, G. A. (2014). Structure of L-serine dehydratase from Legionella pneumophila: novel use of the C-terminal cysteine as an intrinsic competitive inhibitor. Biochemistry 53, 7615–7624. doi: 10.1021/bi501253w
Tian, B., Yang, J., and Qin, K. (2007). Bacteria used in the biological control of plant-parasitic nematodes: populations, mechanisms of action, and future prospects. FEMS Microbiol. Ecol. 61, 197–213. doi: 10.1111/j.1574-6941.2007.00349.x
Topalović, O., Hussain, M., and Heuer, H. (2020). Plants and associated soil microbiota cooperatively suppress plant-parasiticnematodes. Front. Microbiol. 11:313. doi: 10.3389/fmicb.2020.00313
Um, J., Kim, D. G., Jung, M. Y., Saratale, G. D., and Oh, M. K. (2017). Metabolic engineering of Enterobacter aerogenes for 2,3-butanediol production from sugarcane bagasse hydrolysate. Bioresour. Technol. 245, 1567–1574. doi: 10.1016/j.biortech.2017.05.166
Vandana, U. K., Rajkumari, J., Singha, L. P., Satish, L., Alavilli, H., Sudheer, P., et al. (2021). The endophytic microbiome as a hotspot of synergistic interactions, with prospects of plant growth promotion. Biology 10:101. doi: 10.3390/biology10020101
Velayudhan, J., Jones, M. A., Barrow, P. A., and Kelly, D. J. (2004). L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immunity 72, 260–268. doi: 10.1128/IAI.72.1.260-268.2004
Vurukonda, S., Giovanardi, D., and Stefani, E. (2018). Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 19:952. doi: 10.3390/ijms19040952
Wang, C., Wu, J., Shi, B., Shi, J., and Zhao, Z. (2020). Improving L-serine formation by Escherichia coli by reduced uptake of produced L-serine. Microb. Cell Fact. 19:66. doi: 10.1186/s12934-020-01323-2
Wen, T. Y., Wu, X. Q., Hu, L. J., Qiu, Y. J., Rui, L. Y., Zhang, X., et al. (2021). A novel pine wood nematode effector, BxSCD1, suppresses plant immunity and interacts with an ethylene-forming enzyme in pine. Mol. Plant Pathol. 22, 1399–1412. doi: 10.1111/mpp.13121
Windstam, S., and Nelson, E. B. (2008). Differential interference with Pythium ultimum sporangial activation and germination by Enterobacter cloacae in the corn and cucumber spermospheres. Appl. Environ. Microbiol. 74, 4285–4291. doi: 10.1128/AEM.00263-08
Wu, X. Q., Yuan, W. M., Tian, X. J., Fan, B., Fang, X., Ye, J. R., et al. (2013). Specific and functional diversity of endophytic bacteria from pine wood nematode Bursaphelenchus xylophilus with different virulence. Int. J. Biol. Sci. 9, 34–44. doi: 10.7150/ijbs.5071
Xie, F., Dai, S., Zhao, Y., Huang, P., Yu, S., Ren, B., et al. (2020). Generation of fluorinated amychelin siderophores against Pseudomonas aeruginosa infections by a combination of genome mining and mutasynthesis. Cell Chem. Biol. 27, 1532–1543.e6. doi: 10.1016/j.chembiol.2020.10.009
Xu, X. L., Chen, S., and Grant, G. A. (2011). Kinetic, mutagenic, and structural homology analysis of L-serine dehydratase from Legionella pneumophila. Arch. Biochem. Biophys. 515, 28–36. doi: 10.1016/j.abb.2011.08.005
Xue, Q., Xiang, Y., Wu, X. Q., and Li, M. J. (2019). Bacterial communities and virulence associated with pine wood nematode Bursaphelenchus xylophilus from different Pinus spp. Int. J. Mol. Sci. 20:3342. doi: 10.3390/ijms20133342
Yadav, M., Shukla, A. K., Srivastva, N., Upadhyay, S. N., and Dubey, S. K. (2016). Utilization of microbial community potential for removal of chlorpyrifos: a review. Crit. Rev. Biotechnol. 36, 727–742. doi: 10.3109/07388551.2015.1015958
Yang, J., Tian, B., Liang, L., and Zhang, K. (2007). Extracellular enzymes and the pathogenesis of nematophagous fungi. Appl. Microbiol. Biotechnol. 75, 21–31. doi: 10.1007/s00253-007-0881-4
Yin, L., Liu, B., Wang, H., Zhang, Y., Wang, S., Jiang, F., et al. (2020). The rhizosphere microbiome of Mikania micrantha provides insight into adaptation and invasion. Front. Microbiol. 11:1462. doi: 10.3389/fmicb.2020.01462
Zaki, S., Kamal, A., Ashmawy, N. A., and Shoeib, A. A. (2021). Nano-metals forming bacteria in Egypt. I. Synthesis, characterization and effect on some phytopathogenic bacteria in vitro. Sci. Rep. 11:12876. doi: 10.1038/s41598-021-92171-6
Zhang, J., Fu, B., Lin, Q., Riley, I. T., Ding, S., Chen, L., et al. (2020). Colonization of Beauveria bassiana 08F04 in root-zone soil and its biocontrol of cereal cyst nematode (Heterodera filipjevi). PLoS One 15:e0232770. doi: 10.1371/journal.pone.0232770
Zhang, X. D., Wang, D., Sun, S., and Zhang, H. (2020). Issues of Z-factor and an approach to avoid them for quality control in high-throughput screening studies. Bioinformatics 36, 22–23. doi: 10.1093/bioinformatics/btaa1049
Zhang, X., El-Hajj, Z. W., and Newman, E. (2010). Deficiency in L-serine deaminase interferes with one-carbon metabolism and cell wall synthesis in Escherichia coli K-12. J. Bacteriol. 192, 5515–5525. doi: 10.1128/JB.00748-10
Zhang, X., Xu, G., Shi, J., Koffas, M., and Xu, Z. (2018). Microbial production of l-Serine from renewable feedstocks. Trends Biotechnol. 36, 700–712. doi: 10.1016/j.tibtech.2018.02.001
Zhang, X., Zhang, D., Zhu, J., Liu, W., Xu, G., Zhang, X., et al. (2019). High-yield production of L-serine from glycerol by engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 46, 221–230. doi: 10.1007/s10295-018-2113-6
Keywords: sdaB, L-serine dehydratase, Enterobacter ludwigii, nematocidal activity, Bursaphelenchus xylophilus, pine wilt disease, methuosis
Citation: Zhao Y, Yuan Z, Wang S, Wang H, Chao Y, Sederoff RR, Sederoff H, Yan H, Pan J, Peng M, Wu D, Borriss R and Niu B (2022) Gene sdaB Is Involved in the Nematocidal Activity of Enterobacter ludwigii AA4 Against the Pine Wood Nematode Bursaphelenchus xylophilus. Front. Microbiol. 13:870519. doi: 10.3389/fmicb.2022.870519
Received: 06 February 2022; Accepted: 25 March 2022;
Published: 06 May 2022.
Edited by:
Jochen Fischer, Institut für Biotechnologie und Wirkstoff-Forschung (IBWF), GermanyReviewed by:
Kgabo Martha Pofu, University of Limpopo, South AfricaFlorence Fontaine, Université de Reims Champagne-Ardenne, France
Copyright © 2022 Zhao, Yuan, Wang, Wang, Chao, Sederoff, Sederoff, Yan, Pan, Peng, Wu, Borriss and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rainer Borriss, rainer.borriss@rz.hu-berlin.de; Ben Niu, ben_niu@nefu.edu.cn
†These authors have contributed equally to this work
 Yu Zhao1,2†
Yu Zhao1,2† Yanjie Chao
Yanjie Chao Ronald R. Sederoff
Ronald R. Sederoff Heike Sederoff
Heike Sederoff Di Wu
Di Wu Rainer Borriss
Rainer Borriss Ben Niu
Ben Niu
