- 1School of Basic Medical Sciences, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 2Key Laboratory of Application of Ecology and Environmental Protection in Plateau Wetland of Sichuan, Xichang University, Xichang, China
- 3Research and Development Center, Guangdong Meilikang Bio-Sciences Ltd., Dongguan, China
- 4Dongguan Key Laboratory of Medical Bioactive Molecular Development and Translational Research, Guangdong Medical University, Dongguan, China
Berberine hydrochloride (BBR) is a natural product widely used in clinical medicine and animal production. It has a variety of antimicrobial effects, but its complex antimicrobial mechanism has not been clarified. This study aimed to discover the metabolic markers and gain a new perspective on the antibacterial mechanism of BBR. The effects of different inhibitory concentrations of BBR on the survival and growth of standard strain Staphylococcus aureus ATCC 25923 were analyzed by the bacteriostatic activity test. Differences in intracellular metabolites of S. aureus following 19 μg/ml BBR exposure for 1 h were investigated by combining non-targeted metabolomics techniques of gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS). The results showed that the minimum inhibitory concentration of BBR against S. aureus was 51 μg/ml. A total of 368 and 3,454 putative metabolites were identified by GC-MS and LC-MS analyses, respectively. Principal component analysis showed the separation of intracellular metabolite profiles between BBR-exposed samples and non-exposed controls. Pathway activity profiling analysis indicated a global inhibition of metabolisms by BBR exposure, while enhancement was also found in nucleic acid metabolism, amino sugar, and nucleotide sugar metabolism. Several metabolic markers were screened out mainly based on their variable importance of projection values. Two pyridine dicarboxylic acids were significantly downregulated, suggesting the reduction of stress resistance. The oxidized phospholipid (PHOOA-PE) was accumulated, while lipid antioxidant gamma-tocopherol was decreased, and farnesyl PP, the synthetic precursor of another antioxidant (staphyloxanthin), was decreased below the detection threshold. This evidence indicates that BBR reduced the antioxidant capacity of S. aureus. Accumulation of the precursors (UDP-GlcNAc, CDP-ribitol, and CDP-glycerol) and downregulation of the key metabolite D-Ala-D-Ala suggest the inhibition of cell wall synthesis, especially the peptidoglycan synthesis. Metabolites involved in the shikimate pathway (such as 3-dehydroshikimate) and downstream aromatic amino acid synthesis were disturbed. This study provides the first metabolomics information on the antibacterial mechanism of BBR against S. aureus. The key metabolic markers screened in this study suggest that the shikimate pathway, staphyloxanthin synthesis, and peptidoglycan biosynthesis are new directions for further study of BBR antibacterial mechanism in the future.
Introduction
Staphylococcus aureus is a common bacterial colonizer of human and a variety of animal species which can cause many infectious diseases, such as mastitis, endocarditis, septic arthritis, and sepsis. In the treatment of infectious diseases, antibiotics were initially highly effective, but they currently show several limitations, including widespread resistance, toxicity, and residues. S. aureus has developed resistance to various antibiotics including vancomycin (Ragunathan et al., 2018). We need to understand the resistance mechanism of microorganisms to existing antibiotics and develop antibiotics with new antimicrobial mechanisms (Anitha et al., 2016). As complementary or alternative drugs for the treatment or prevention of infection, natural products such as medicinal plants and their extracted components are a good choice (Herrmann et al., 2016; Newman and Cragg, 2016; Kokoska et al., 2019). Some natural compounds are considered to have multiple potential antimicrobial targets (Miryala et al., 2021, 2022). One of the natural anti-infective products is berberine, which is commonly used in clinical medicine, as well as in livestock production and aquaculture.
Berberine is an isoquinoline alkaloid found mainly in the form of hydrochloride (berberine hydrochloride) in many medicinal plants, such as Coptis chinensis, Phellodendron amurense, and Berberis vulgaris (Imenshahidi and Hosseinzadeh, 2016). It exhibits various pharmacological activities, including prominent anti-inflammatory and anti-infective effects, antagonizing a variety of pathogenic bacteria (including S. aureus), fungi, parasites, and even viruses (Shang et al., 2020). Berberine not only can exert an antimicrobial effect by regulating host immunity but also has a significant direct antimicrobial effect. Some experimental evidence shows that berberine alone or in combination with other agents showed obvious antimicrobial effects in vivo and in vitro (Aksoy et al., 2020; Huang et al., 2020; Xie et al., 2020; Bhatia et al., 2021). Berberine even has the potential to inhibit antibiotic-resistant bacteria (Morita et al., 2016; Zhang et al., 2020; Li et al., 2021), which gives it great potential in the development of new antibiotics. There are many studies on the antimicrobial activity of berberine, and it is found that berberine has a variety of antimicrobial effects. A multi-omics study suggests that berberine may interfere with the nucleic acid, cell wall, cell membrane transport, and motility functions of Escherichia coli and inhibit its metabolism (Karaosmanoglu et al., 2014). A recent study has shown that berberine can destroy the cell wall structure and cell membrane integrity of methicillin-resistant Staphylococcus aureus (MRSA) and play a synergistic antimicrobial role with clindamycin or rifamycin (Xia et al., 2022). In addition, a few studies have suggested other possible antimicrobial mechanisms of berberine, including the inhibition of biofilm formation, protein synthesis, and bacterial division (Boberek et al., 2010; Li et al., 2011). However, the pathway and potential targets of berberine causing these antibacterial effects are unclear. A more clear direction is needed to further study the complex antimicrobial molecular mechanism of berberine.
Analyzing the expression changes of specific genes in microorganisms is a common method to explain the antimicrobial effect of berberine. The intervention on gene expression level (including transcription and protein synthesis) may be indirect, so it is difficult to explain the antimicrobial mechanism of berberine. Metabolites are the final products of gene expression, and the metabolic alterations represent the exact physiological state of microorganisms (Dorries et al., 2014). Thus, the response of microorganisms to a given antibiotic stress at the metabolic level can help us better understand the antibacterial mechanisms. Several studies have used metabolomics technology to study the antimicrobial mechanism of antibiotics (Dorries et al., 2014; Schelli et al., 2017; Vemula et al., 2017) and natural products (Hussein et al., 2020; Shen et al., 2020; Tang et al., 2021; Chen et al., 2022). However, we have not read any report on the antimicrobial mechanism of berberine based on metabolomics. In this study, berberine hydrochloride (BBR) was used as the representative form of berberine, and we attempted to explore a new perspective of antimicrobial mechanism of berberine by using metabolomics technology. Two untargeted metabolomics techniques, namely, gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS), were combined to explore the antimicrobial mechanism of BBR against S. aureus through extensive screening of metabolic markers and analysis of related metabolic pathways.
Materials and Methods
Reagents, Bacterial Strain, and Culture Conditions
The BBR (HPLC ≥ 98%) and vancomycin (potency ≥ 900 μg/mg) were purchased from Desite Bio-Tech Co., Ltd. (Chengdu, CHN) and Solarbio Bio-Tech Co., Ltd. (Beijing, CHN), respectively, and were dissolved in sterilized ultra-pure water before use. The reagents used in the preparation of culture medium were purchased from Aoboxing Bio-Tech Co., Ltd. (Beijing, CHN). The standard strain S. aureus ATCC 25923 was purchased from the Biobw Culture Collection (Beijing, CHN). Before use, the strain was subcultured onto nutrient agar plates for 24 h at 37°C. Then, the single colony was selected and coated on a nutritious agar plate to grow for 7 h, and the culture would be used as a “seed” to cultivate working bacteria culture (WBC) for subsequent experiments. For the preparation of WBC, the “seed” was inoculated into the broth medium (composed of 1% tryptone, 0.3% beef extract powder, and 0.5% sodium chloride, pH ~ 7.4) with an initial inoculation dose of 107 CFU/ml and then cultured with shaking at 37°C and 120 rpm for ~2.5 h to enter the logarithmic growth phase (OD600 ~ 0.3).
MIC and MBC Test
The MIC value was determined by the improved broth dilution method to improve the test sensitivity (Surre et al., 2018). A small amount of phenol red (0.018 g/L) and glucose (0.5 %) were added to the broth for the test (containing 1% tryptone, 0.1% beef extract powder, 0.5% sodium chloride, pH ~ 7.4). The basic principle of the test is that bacteria grow to produce acid by fermenting glucose, which makes the broth turn yellow from the original red color. The operation method is similar to that of the traditional broth dilution method except that color change and turbidity change are combined as the basis for judging whether the bacteria grow or not. Generally speaking, the phenol red-containing broth added with a certain concentration of BBR was diluted in a series of gradients and used for the culture of S. aureus (~108 CFU/ml) at 37°C. The lowest BBR concentration without visible color change and turbidity change within 24 h was MIC. The bacterial cultures exposed to different concentrations of berberine for 24 h were coated on nutrient agar plates and cultured at 37°C for 24 h. The MBC was determined by the lowest berberine concentration without bacterial growth.
Antibacterial Effect Test
The WBC (OD600 ~ 0.3) was diluted with the same volume of broth medium containing BBR to give an approximate starting inoculum of 108 CFU/ml and to obtain final BBR concentrations of 1/4 × MIC, 3/8 × MIC, 1/2 × MIC, or MIC. The control culture was added with the same volume of broth medium without BBR. The cultures were incubated further for 8 h, and cell growth was monitored spectrophotometrically (OD600nm at 1 h intervals). Time kill tests of the above BBR concentrations were assessed by counting CFU (Lobritz et al., 2015). WBC was incubated in different concentrations of BBR at 37°C for 30 min. Bacterial cells were collected by centrifugation and washed two times with 0.85% sterile saline and then stained for 15 min using Live/Dead BacLight bacterial viability kit (Invitrogen, Eugene, OR, USA) according to the manufacturer's instructions and imaged by Image Xpress Micro Confocal (Molecular Devices LLC, Sunnyvale, CA).
Cultivation of Metabolic Samples
The BBR concentration (3/8 × MIC, 19 μg/ml) and exposure time (1 h) for the treatment of metabolic samples were determined by the result of time kill tests. The culture conditions were the same as the antibacterial effect test. Therefore, metabolic samples (group T1) were collected at the 1-h time point after being exposed to 19 μg/ml BBR. Two control groups (without BBR addition) collected at zero time point were treated as the initial control group (C0) and those collected 1 h later were treated as the growth control group (C1).
Intracellular Metabolites Extraction
Intracellular metabolites were extracted using cold methanol and chloroform (Stipetic et al., 2016). Briefly, after co-incubation with or without BBR, 80 ml of bacterial culture from each biological sample (six biological replicates for each group) was collected. The bacterial cells were harvested by 4°C cryogenic centrifugation at 6,000 rpm for 10 min, followed by cold phosphate buffer saline (PBS) washes. A total of 1 ml cold methanol:water (4:1, v/v) and 200 μl of chloroform were added to the cell pellet, and the mixtures were vortexed. After that, cells were broken up with an ultrasonic homogenizer (3 min, 500 W), and 20 μl of L-2-chlorophenylalanine (0.3 mg/ml) was added as the internal standard. Then, the mixture was extracted by ultrasonic for 20 min in ice-water bath and then centrifuged at 4°C (13,000 rpm) for 10 min. Finally, 200 μl of the supernatant for GC-MS (or 400 μl for LC-MS) was dried in a freeze concentration centrifugal dryer. Quality control sample (QC) was prepared by mixing an aliquot of all samples to be a pooled sample.
Untargeted Metabolomics Analyses
A Thermo Trace 1310/TSQ 9000 GC/MSD System was used for GC-MS analysis. DB-5MS fused-silica capillary column (30 m × 0.25 mm × 0.25 μm) was utilized to separate the derivative metabolites. For LC-MS analysis, the freeze-dried samples were re-extracted and analyzed by a Nexera UPLC system coupled with Q Exactive quadrupole-orbitrap mass spectrometer equipped with heated electrospray ionization (ESI) source. An ACQUITY UPLC HSS T3 column (1.8 μm, 2.1 × 100 mm) was employed in both positive and negative modes. Metabolome analysis was performed by Lu-Ming Biotech Co. Ltd. (Shanghai, China) [see details in the supporting information (Supplementary Text 1)].
Metabolome Data Processing
The MS-DIAL software was used to preprocess GC-MS data. Metabolite characterization is based on the LUG database (a self-built untarget database of GC-MS from Lu-Ming Bio). All peak signal intensities in each sample were segmented and normalized according to the internal standards with RSD > 0.3 after screening. LC-MS data were processed by software Progenesis QI V2.3. Main parameters of 5 ppm precursor tolerance, 10 ppm product tolerance, and 5% production threshold were applied. Compound identification was based on the precise mass-to-charge ratio (m/z), secondary fragments, and isotopic distribution using HMDB, Lipidmaps (V2.3), Metlin, EMDB, PMDB, and self-built databases to do qualitative analysis. Peaks with a missing value (ion intensity = 0) in more than 50% of groups were removed. Zero value was replaced by half of the minimum value. Compounds with resulting scores below 36 (out of 60) points were deemed to be inaccurate and removed.
Statistical Analyses
The data matrix of the metabolome was imported in R to carry out principle component analysis (PCA) and orthogonal partial least-squares-discriminant analysis (OPLS-DA). To prevent overfitting, 7-fold cross-validation and 200 response permutation testing (RPT) were carried out to evaluate the quality of the OPLS-DA models. Variable importance of projection (VIP) values obtained from the OPLS-DA model were used to rank the overall contribution of each variable to group discrimination. A two-tailed Student's T-test was further used to verify whether the metabolites of difference between the two groups were significant. P-value adjustment was performed using the Benjamini-Hochberg method (false discovery rate). Heatmaps (hierarchical clustering) were plotted by http://www.bioinformatics.com.cn, a free online platform for data analysis and visualization. Metabolomics datasets and metabolic pathway activities were correlated using the pathway activity profiling (PAPi) algorithm (Aggio, 2014). Only the significant metabolites (VIP > 1, FC > 2, P < 0.05 and adj. P < 0.05) were analyzed by the PAPi_1.8.0 package in R.
Results
Activity of BBR Against S. aureus
The MIC value of BBR against S. aureus strain ATCC 25923 was 51 μg/ml, and the MBC value was two times the value of MIC (102 μg/ml). When combined with vancomycin, BBR of 1/2 × MIC (25.5 μg/ml) halved the MIC value (2 μg/ml) of vancomycin against S. aureus strain ATCC 25923 (Supplementary Figure 1). Fluorescent staining of live and dead bacterial cells showed that BBR had a significant inhibitory effect on the survival of S. aureus (Supplementary Figure 2). BBR inhibited the growth of S. aureus in a concentration-dependent manner. No obvious growth trend was observed in S. aureus exposed to MIC and 1/2 × MIC of BBR, while moderate inhibition and mild inhibition were mediated by 3/8 × MIC and 1/4 × MIC, respectively (Supplementary Figure 3A). Based on the results of time-kill test (Supplementary Figure 3B), BBR concentration and exposure time for the treatment of metabolic samples were determined. The exposure time of 1 h was chosen because BBR had shown obvious bactericidal activity at this time (Supplementary Figure 3B); 3/8 × MIC (19 μg/ml) was a suitable exposure concentration to minimize cell death at the 1-h time point and to achieve an obvious bacteriostatic effect at later time points (Supplementary Figure 3B).
Metabolite Profiles
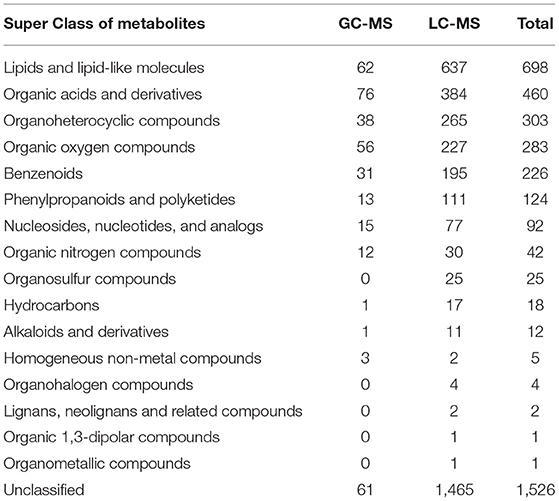
A total of 368 and 3,454 putative metabolites were identified, respectively, from GC-MS and LC-MS datasets. They were divided into 16 super classes including 2,296 classified metabolites and 1,526 unclassified metabolites (Table 1). PCA reviewed a significant separation between controls [initial control (C0) and growth control (C1)] and BBR-exposed group (T1) (Figures 1A–D). A pair of heatmaps demonstrated the characteristic profiles of T1 group (Figures 1E,F). OPLS-DA models showed the importance of each metabolite on group discrimination (Figures 1G–J). Model parameters indicated the excellent prediction ability of the models (Supplementary Table 1), and no overfitting was found (Supplementary Table 1, Supplementary Figure 4).
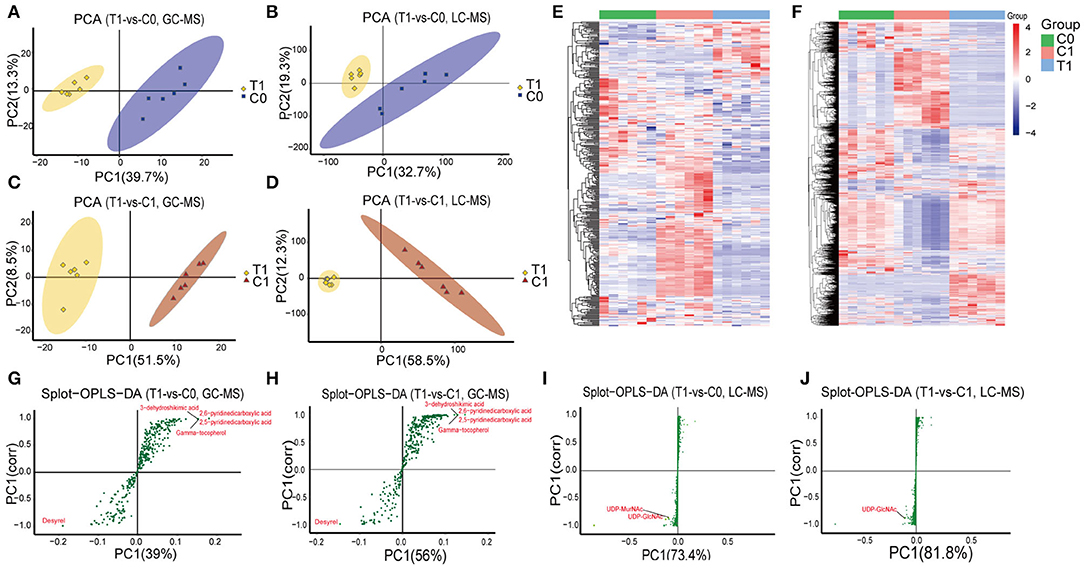
Figure 1. Overview of metabolomics analysis. (A–D) PCA analysis; (E, F) Heatmaps of identified metabolites; (G–J) Graphs of OPLS-DA model, points farther from the origin represent metabolites that contribute more to group discrimination. T1: BBR-exposed group; C0: initial control group; C1: growth control group.
Key Metabolic Pathways Affected by BBR
Metabolomics datasets and metabolic pathway activities were correlated using PAPi analysis. As compared with control groups, the activities of most metabolic pathways in the T1 group were downregulated (Figure 2, Supplementary Figure 5). The upregulated metabolic pathways were mainly nucleic acid metabolism, amino sugar, and nucleotide sugar metabolism, as well as glycan biosynthesis and metabolism. Similar trends can be seen in the volcanic maps (Supplementary Figure 6). The enhancement of nucleic acid metabolism activity involved the accumulation of nucleotide metabolites (Supplementary Figure 6). The upregulation of amino sugars, nucleotide sugars metabolism, and downstream glycan biosynthesis involved the accumulation of precursor metabolites of glycan biosynthesis, mainly UDP-N-acetylmuraminate (UDP-MurNAc), UDP-N-acetylglucosamine (UDP-GlcNAc), and UDP-N-acetyl-D-mannosamine (UDP-ManNAc) (Figure 3).
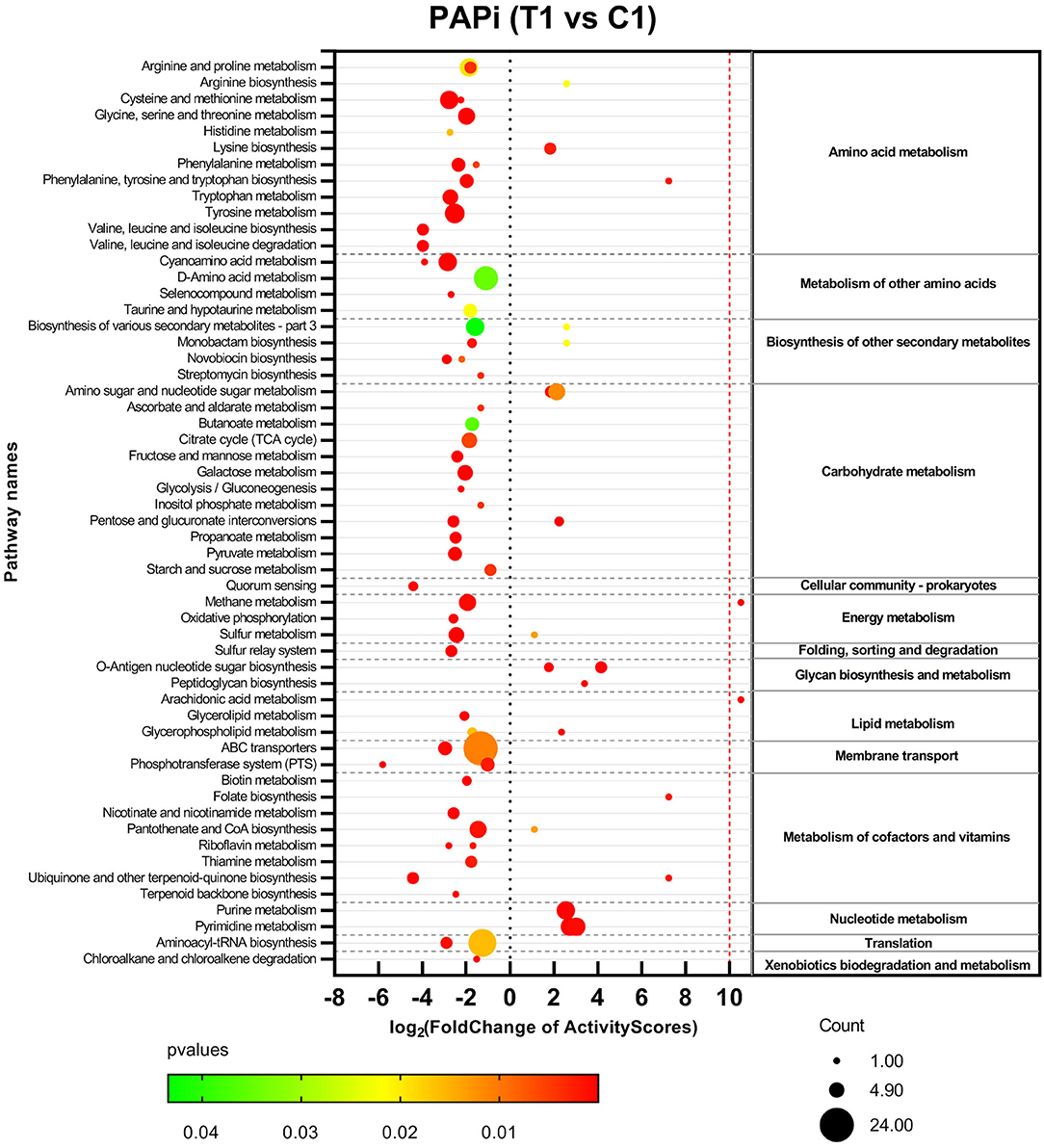
Figure 2. Activities profile of metabolic pathways in comparison with BBR-exposed group (T1) vs. growth control group (C1). Results from GC-MS and LC-MS datasets were merged. The color and size of bubbles represent the statistical significance and the number of metabolite species matched to the pathways, respectively.
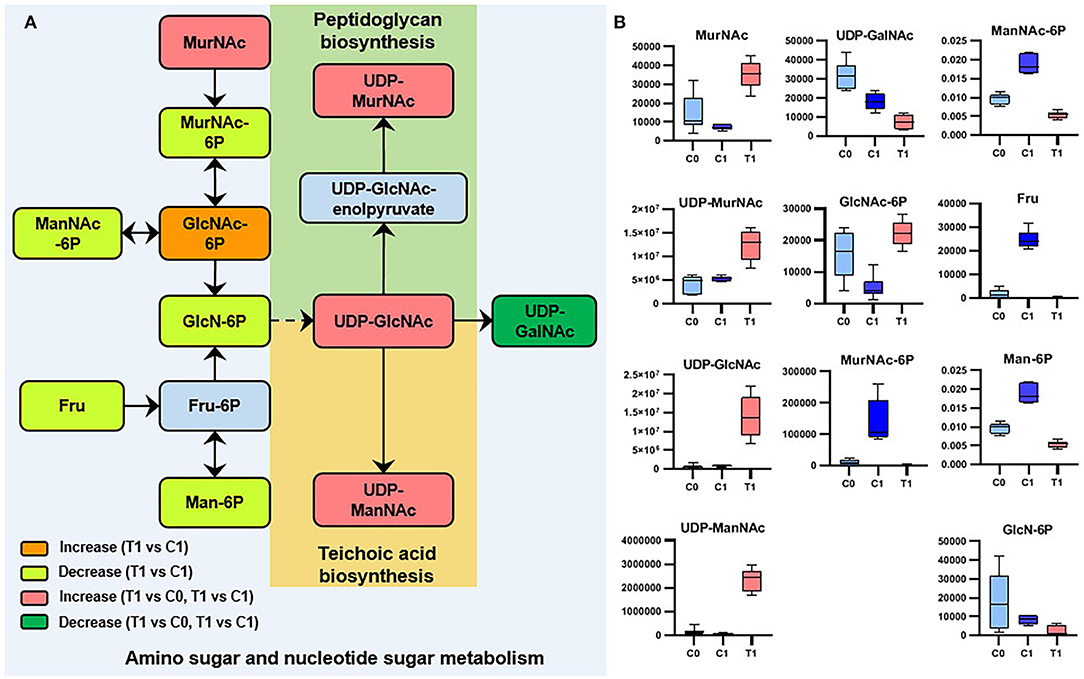
Figure 3. Significant metabolites involved in amino-sugar and sugar-nucleotide metabolism. (A) Relationship among metabolites. T1: BBR-exposed group; C0: initial control group; C1: growth control group. (B) Box plots for metabolites marked in A (|log2FC| > 1; P < 0.05; adj. P-value < 0.05).
The Metabolites of Cell Wall Biosynthesis Were Disturbed
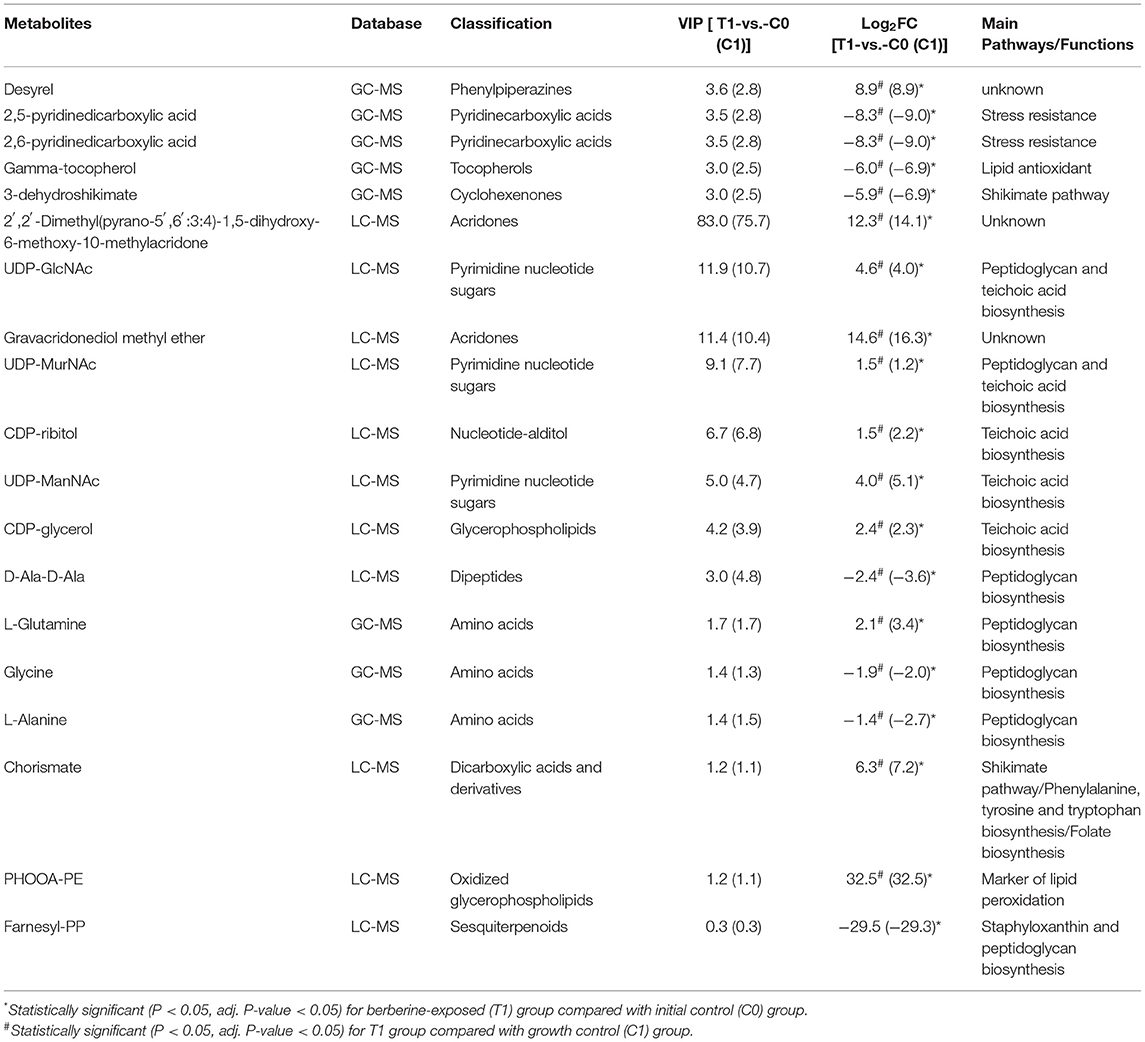
We analyzed the potential biological functions of the top five metabolites sorted by VIP values and the related metabolites (Table 2, Supplementary Figure 7). UDP-GlcNAc (a precursor of peptidoglycan and teichoic acid) was one of the VIP-top five metabolites in the LC-MS dataset and was significantly upregulated in the T1 group [log2FC = 4.6 (4.0)]. Six other significant metabolites involved in peptidoglycan biosynthesis were identified (Table 2), and their relationships are shown in Figure 4. Downregulated D-alanyl-D-alanine (D-Ala-D-Ala) [log2FC = −2.4 (−3.6)] restricts the upstream part of peptidoglycan biosynthesis pathway. Farnesyl diphosphate (farnesyl-PP), the precursor metabolite of undecaprenyl pyrophosphate (Und-PP) in the peptidoglycan biosynthesis pathway, was found to be decreased below the detection threshold in the T1 group, while it was detected in all the 12 samples of the control groups [log2FC = −29.5 (−29.3)]. L-alanine [log2FC = −1.4 (−2.7)] and glycine [log2FC = −1.9 (−2.0)] were downregulated, while L-glutamine was accumulated [log2FC = 2.1 (3.4)]. The precursors of teichoic acid biosynthesis (UDP-GlcNAc, UDP-ManNAc, CDP-ribitol, and CDP-glycerol) were also accumulated significantly (Table 2, Supplementary Figure 7).
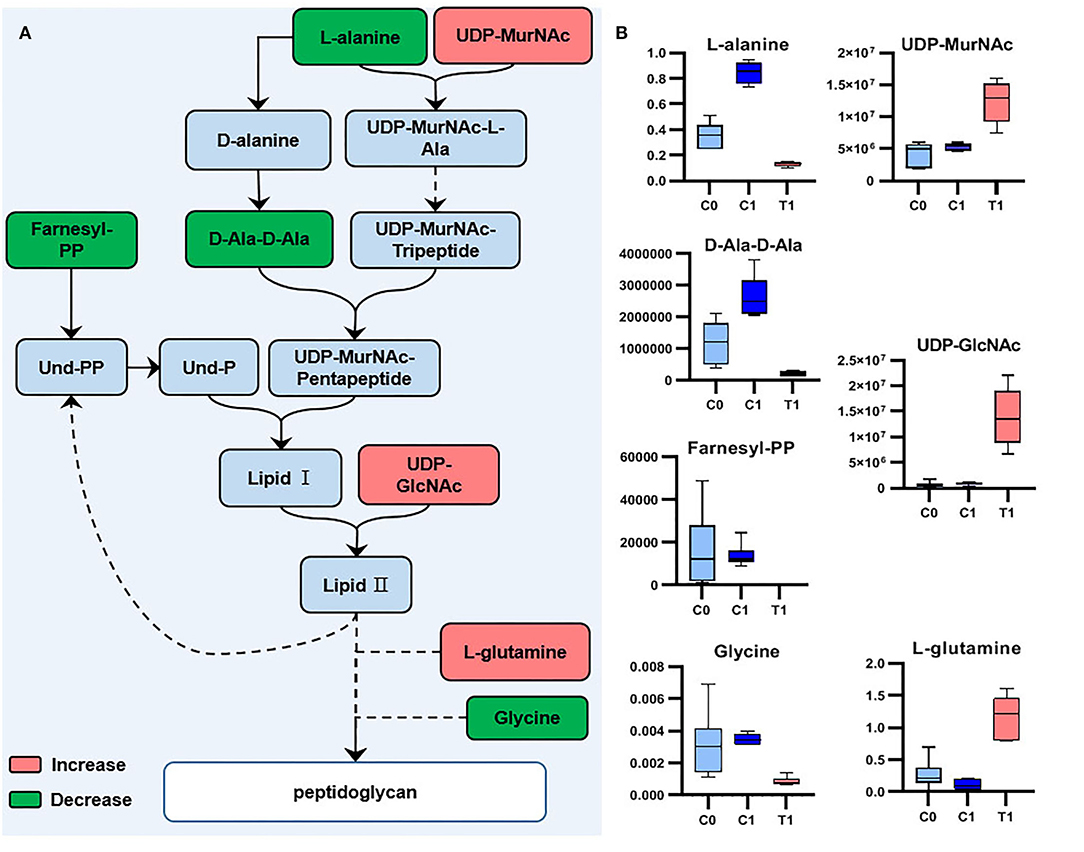
Figure 4. Significant metabolites involved in peptidoglycan biosynthesis. (A) Relationship among metabolites. (B) Box plots for significant metabolites (|log2FC| > 1; P < 0.05; adj. P-value < 0.05).
The Metabolites Related to Stress Resistance and Antioxidant Activity Were Significantly Downregulated
The VIP-top five metabolites in GC-MS dataset were desyrel, 2,5-pyridinedicarboxylic acid, 2,6-pyridinedicarboxylic acid, gamma-tocopherol, and 3-dehydroshikimate (Table 2, Supplementary Figure 7). 2,5-pyridinedicarboxylic acid [log2FC = −8.3 (−9.0)] and 2,6-pyridinedicarboxylic acid [log2FC = −8.3 (−9.0)] were significantly decreased after BBR treatment. They belong to pyridine dicarboxylic acid, which is known to be abundant in bacterial spores and endows spores with stress resistance (Setlow, 2007; Ramirez-Guadiana et al., 2017). Gamma-tocopherol was downregulated in the T1 group [log2FC = −6.0 (−6.9)], which is known as a lipid vitamin and acts as an antioxidant (Kim et al., 2012). The decreased farnesyl-PP in the T1 group, as the precursor of Und-PP, is also the precursor of staphyloxanthin (another important antioxidant in S. aureus). The oxidized glycerol phospholipid (PHOOA-PE) was not detected in the control groups, but was accumulated in the T1 group (log2FC = 32.5).
The Metabolites Involved in the Shikimate Pathway and Biosynthesis of Aromatic Amino Acids Were Disturbed
The two metabolites involved in the shikimate pathway, 3-dehydroshikimate [log2FC = −5.9 (−6.9)] and chorismate [log2FC = 6.3 (7.2)], showed opposite regulatory trends. L-phenylalanine, L-tyrosine, and L-tryptophan, which were synthesized using chorismate as precursors, were downregulated (Figure 5).
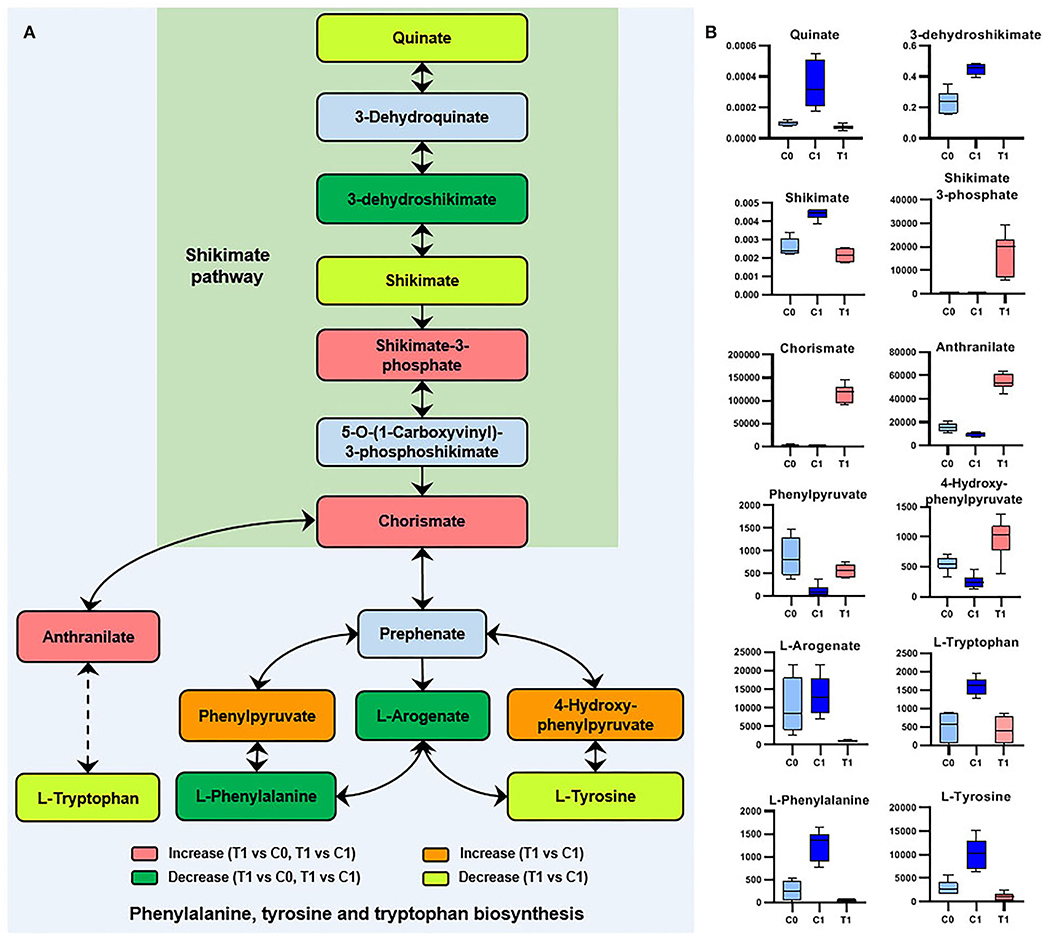
Figure 5. Significant metabolites involved in phenylalanine, tyrosine, and tryptophan biosynthesis. (A) Relationship among metabolites. T1: BBR-exposed group; C0: initial control group; C1: growth control group. (B) Box plots for metabolites marked in A (|log2FC| > 1; P < 0.05; adj. P-value < 0.05).
Discussion
According to the existing research, the antibacterial effect of berberine can be generally summarized as “multi-effect” (Karaosmanoglu et al., 2014). Berberine can inhibit the formation of biofilm (Zhang et al., 2020; Bhatia et al., 2021; Xu et al., 2021), destroy the cell wall and cell membrane (Karaosmanoglu et al., 2014; Zhang et al., 2020; Xia et al., 2022), inhibit bacterial cell division (Domadia et al., 2008), inhibit nucleic acid and protein synthesis (Du et al., 2020), and inhibit bacterial adhesion (Kang et al., 2015), etc.
This study revealed that BBR inhibited most of the metabolic activities of amino acid metabolism, carbon metabolism, and energy metabolism in S. aureus, while the accumulation of nucleotide metabolites suggested that the synthesis of downstream nucleic acid macromolecules may be inhibited (Figure 2). The metabolic changes in these pathways support the inhibitory effect of berberine on bacterial growth proposed by previous studies. However, these metabolic changes are not the unique antimicrobial properties of berberine, as exposure to other antibiotics also causes a general perturbation of bacterial metabolism (Dorries et al., 2014). Berberine and its derivatives inhibit bacterial division by inhibiting the cell division protein FtsZ (Boberek et al., 2010; Sun et al., 2014). Although the results of this study cannot directly verify the target of BBR inhibiting bacterial cell division, the extensive changes in bacterial metabolism caused by BBR may be the result of complex physiological regulation indirectly caused by the inhibition of bacterial cell division and other possible antibacterial activities of BBR.
The main metabolic markers screened in this study provide a new perspective for exploring the antibacterial mechanism of BBR, as follows.
Shikimate Pathway
The end product of the shikimate pathway (chorismate) was accumulated (Figure 5), indicating a disturbance in the biosynthesis of downstream aromatic amino acids and folate. In contrast, the intermediate metabolite (3-dehydroshikimate) of the shikimate pathway was significantly downregulated by BBR (Figure 5). Since being essential in bacteria and fungi, but absent from mammals, the shikimate pathway was suggested to be an antimicrobial target (Sadaka et al., 2018). Therefore, it is worthwhile to further study the interference mechanism of BBR on the bacterial shikimate pathway.
Antioxidant Capacity
Bactericidal antibiotics cause bacterial death due to oxidative damage by stimulating the generation of reactive oxygen species (ROS) (Kohanski et al., 2007), which is also applicable to the action mode of BBR (Du et al., 2020). In this study, some evidence suggests that BBR reduces the antioxidant capacity of S. aureus, such as the reduction of gamma-tocopherol and farnesyl-PP, and the accumulation of PHOOA-PE (Figure 6). Gamma-tocopherol reacts with ROS and protects unsaturated fatty acids from oxidation (Kim et al., 2012). Farnesyl-PP is a precursor metabolite of staphyloxanthin that acts as an antioxidant to protect S. aureus from being killed by ROS released by leukocytes (Clauditz et al., 2006). The downregulation of gamma-tocopherol and farnesyl-PP means the decrease in antioxidant capacity, and as a result, the oxidized glycerophospholipid (PHOOA-PE) was accumulated in the cells of S. aureus. Staphyloxanthin is the main pigment in S. aureus, and its accumulation is inhibited by BBR (Supplementary Figure 8). In addition, based on the transcriptome data of our related study (unpublished), the transcription levels of genes involved in staphyloxanthin biosynthesis were also downregulated by BBR (Supplementary Figure 9). The targeted perturbation of staphyloxanthin is expected to restore the sensitivity of drug-resistant strains to traditional antibiotics (Garcia-Fernandez et al., 2017; Hui et al., 2020). Therefore, the attractive potential of BBR to inhibit staphyloxanthin synthesis is another direction worthy of further study.
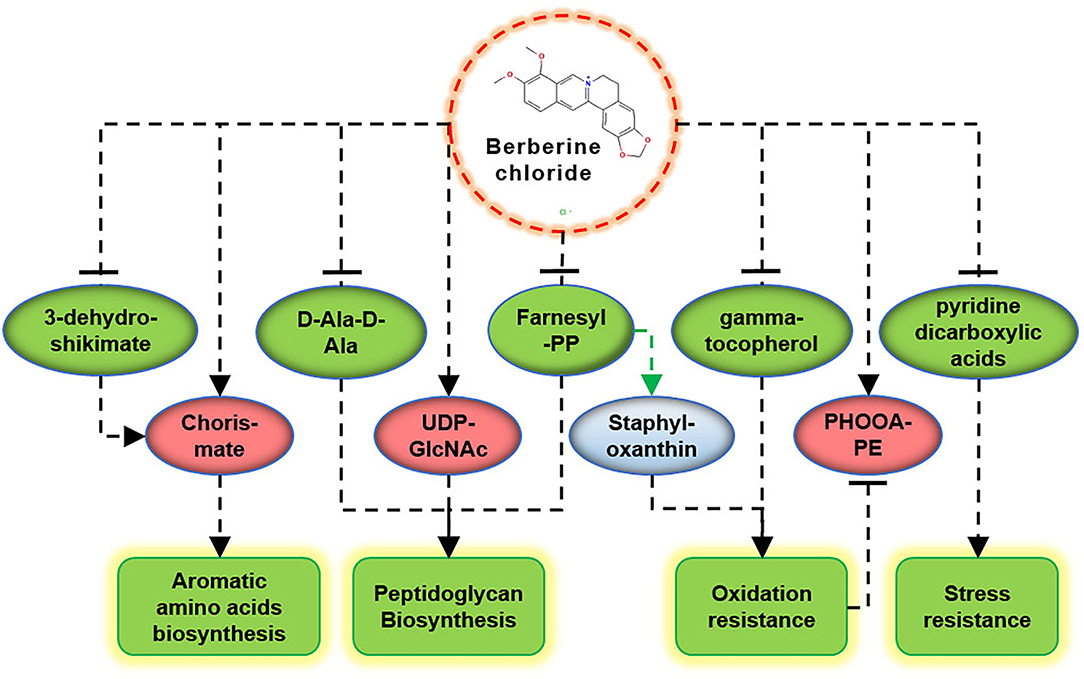
Figure 6. Schematic diagram of the key metabolites in S. aureus in response to BBR exposure. Upregulation (promoting) and downregulation (inhibiting) are shown in red and green colors, respectively.
The synthesis of farnesyl-PP is regulated by other upstream metabolic pathways (Lan et al., 2010). Since BBR interferes with most of the metabolic activities of S. aureus, its effect on the synthesis of farnesyl-PP is complex. As a key precursor metabolite that likes the biosynthesis of staphyloxanthin and peptidoglycan, farnesyl PP affects the abilities of antioxidation and cell wall synthesis, both of which are critical to the survival of S. aureus.
Cell Wall Synthesis
A recent study found that berberine can damage the cell walls of MRSA, but the mechanism is unknown (Xia et al., 2022). According to the metabolic responses of S. aureus to antibiotics with known targets, the precursor metabolites upstream would accumulate, while the level of metabolites downstream of the inhibited metabolic targets would decrease (Schelli et al., 2017; Vemula et al., 2017). In this study, besides farnesyl PP, the change of other metabolites also demonstrated that BBR act by impairing bacterial cell wall synthesis. The accumulation of precursor metabolites related to cell wall synthesis, such as UDP-GlcNAc, CDP-ribitol and CDP-glycerol, indicated that the downstream synthesis pathways of peptidoglycan and teichoic acid were inhibited. The level of D-Ala-D-Ala was significantly decreased, suggesting the inhibition of BBR to the early stage of peptidoglycan biosynthesis. The synthetic pathway of D-Ala-D-Ala is one of the targets of the cell wall-targeting antibiotics, such as D-cycloserine and D-boroalanine, which act through the inhibition of D-Ala-D-Ala-ligase (Vemula et al., 2017). Whether BBR has the same target as these two antibiotics needs further verification. Since BBR has synergistic antibacterial effects with other antibiotics such as clindamycin, rifamycin (Xia et al., 2022), and ampicillin (Dziedzic et al., 2015), and we confirmed the synergistic antibacterial effect of BBR combined with vancomycin (which also targets the synthesis of peptidoglycan) (Supplementary Figure 1), its inhibitory effect on the synthesis of peptidoglycan is an interesting discovery and has important value for further study.
Prospects
Some of the metabolites identified in this study, such as the pyridine dicarboxylic acids, have unclear metabolic pathways in S. aureus. Pyridine dicarboxylic acid is present in the spores of spore-producing bacteria in high concentrations and endorses the spores with stress-resistant properties (Setlow, 2007). The genes of dihydro-pyridine dicarboxylate synthases and reductase had been annotated in the genome of MRSA, and they were considered to have potential interactions with the penicillin-resistant binding protein (PBP2a) (Yuan et al., 2010). However, the properties of the substrates of these enzymes remain controversial (Devenish et al., 2010; Karsten et al., 2018). It is worth mentioning that gamma-tocopherol can be synthesized in plants and algae with farnesyl-PP as a precursor (56), although no study on its biosynthesis in S. aureus has been retrieved. It is predicted that the average number of metabolites of S. aureus ranges from more than 1,000 to more than 2,000, with a maximum of 4,416 (Renz and Drager, 2021). Among the more than 3,000 metabolites identified in this study, a considerable part of them should be products of unknown metabolic pathways, in addition to substances absorbed from the culture environment. As metabolic databases become richer, the number of metabolites accurately identified will increase. Therefore, the revelation of unknown metabolic pathways in the future will help us to deeply understand the complexity and intrinsic relationship of antibiotic effects on bacterial metabolism. In addition, when there is a clear research direction or determined metabolic pathway (as suggested in this study), the application of targeted metabolomics and other technologies that can accurately verify the specific antibacterial effect will further promote the research on the antibacterial mechanism of natural products such as BBR.
Conclusion
The key metabolic markers suggest that BBR plays antibacterial activities against S. aureus ATCC 25923 by inhibiting the cell wall biosynthesis, promoting oxidative damage, reducing stress resistance, and inhibiting the synthesis of aromatic amino acids (Figure 6). These results provide new directions of peptidoglycan biosynthesis, staphyloxanthin synthesis, and shikimate pathway to study the antibacterial molecular mechanism of BBR and help to promote its application in the field of anti-infective medicine.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
SW, KY, and NY conducted the experiments. SW, YH, and JN performed data analysis. SW and YG wrote the manuscript. SW and WD reviewed, edited the manuscript, and acquired funding. All authors made significant contributions to this article and have read and agreed to the final manuscript.
Funding
This study was supported by the Sichuan Science and Technology Program (No. 2021YJ0112) and the Chengdu University of TCM Science Foundation (BSH2019010). The authors declare that this study received funding from Science and Technology Department of Sichuan Province and Chengdu University of TCM. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
JN was employed by Guangdong Meilikang Bio-Sciences Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Lu-Ming Biotech Co., Ltd. (Shanghai, China) for the sample detecting services. We also thank Yi-li Wang from the Innovative Institute of Chinese Medicine and Pharmacy, Chengdu University of Traditional Chinese Medicine, for her gracious assistance in the imaging technique of fluorescent staining.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.917414/full#supplementary-material
Supplementary Text 1. Details for untargeted metabolomics analyses.
Supplementary Figure 1. MIC test of BBR combined with vancomycin against S. aureus ATCC 25923.
Supplementary Figure 2. The effect of BBR on survival of S. aureus ATCC 25923.
Supplementary Figure 3. Growth curves (A) and the results of time kill tests (B) for S. aureus ATCC 25923 in the presence (1/4MIC, 3/8MIC, 1/2MIC or MIC) or absence (control) of berberine.
Supplementary Figure 4. Result of RPT.
Supplementary Figure 5. Activities change profile of metabolic pathways in comparison with berberine-exposed group (T1) vs. initial control group (C0).
Supplementary Figure 6. Volcano plots showing the trends of metabolites detected in the major metabolic pathways.
Supplementary Figure 7. Box plots for the major metabolic markers in S. aureus ATCC 25923 following berberine-exposure (T1) compared with non-exposed controls (C0, C1).
Supplementary Figure 8. Effect of BBR on pigment accumulation of S. aureus ATCC 25923.
Supplementary Figure 9. Expression profile of genes involved in staphyloxanthin biosynthesis in S. aureus ATCC 25923 responsive to berberine-exposure.
Supplementary Table 1. Parameters of OPLS-DA and RPT test.
References
Aggio, R. B.. (2014). Pathway activity profiling (PAPi): a tool for metabolic pathway analysis. Methods Mol. Biol. 1152, 233–250. doi: 10.1007/978-1-4939-0563-8_14
Aksoy, C. S., Avci, F. G., Ugurel, O. M., Atas, B., Sayar, N. A., and Sariyar Akbulut, B. (2020). Potentiating the activity of berberine for Staphylococcus aureus in a combinatorial treatment with thymol. Microb. Pathog. 149, 104542. doi: 10.1016/j.micpath.2020.104542
Anitha, P., Anbarasu, A., and Ramaiah, S. (2016). Gene network analysis reveals the association of important functional partners involved in antibiotic resistance: a report on an important pathogenic bacterium Staphylococcus aureus. Gene 575(2 Pt 1), 253–263. doi: 10.1016/j.gene.2015.08.068
Bhatia, E., Sharma, S., Jadhav, K., and Banerjee, R. (2021). Combinatorial liposomes of berberine and curcumin inhibit biofilm formation and intracellular methicillin resistant Staphylococcus aureus infections and associated inflammation. J. Mater. Chem. B 9, 864–875. doi: 10.1039/d0tb02036b
Boberek, J. M., Stach, J., and Good, L. (2010). Genetic evidence for inhibition of bacterial division protein FtsZ by berberine. PLoS ONE 5:e13745. doi: 10.1371/journal.pone.0013745
Chen, J., Zhang, J., Zhu, L., Qian, C., Tian, H., Zhao, Z., et al. (2022). Antibacterial activity of the essential oil from litsea cubeba against cutibacterium acnes and the investigations of its potential mechanism by gas chromatography-mass spectrometry metabolomics. Front. Microbiol. 13:823845. doi: 10.3389/fmicb.2022.823845
Clauditz, A., Resch, A., Wieland, K. P., Peschel, A., and Gotz, F. (2006). Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74, 4950–4953. doi: 10.1128/IAI.00204-06
Devenish, S. R., Blunt, J. W., and Gerrard, J. A. (2010). NMR studies uncover alternate substrates for dihydrodipicolinate synthase and suggest that dihydrodipicolinate reductase is also a dehydratase. J. Med. Chem. 53, 4808–4812. doi: 10.1021/jm100349s
Domadia, P. N., Bhunia, A., Sivaraman, J., Swarup, S., and Dasgupta, D. (2008). Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry 47, 3225–3234. doi: 10.1021/bi7018546
Dorries, K., Schlueter, R., and Lalk, M. (2014). Impact of antibiotics with various target sites on the metabolome of Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 7151–7163. doi: 10.1128/AAC.03104-14
Du, G. F., Le, Y. J., Sun, X., Yang, X. Y., and He, Q. Y. (2020). Proteomic investigation into the action mechanism of berberine against Streptococcus pyogenes. J. Proteomics 215, 103666. doi: 10.1016/j.jprot.2020.103666
Dziedzic, A., Wojtyczka, R. D., and Kubina, R. (2015). Inhibition of oral streptococci growth induced by the complementary action of berberine chloride and antibacterial compounds. Molecules 20, 13705–13724. doi: 10.3390/molecules200813705
Garcia-Fernandez, E., Koch, G., Wagner, R. M., Fekete, A., Stengel, S. T., Schneider, J., et al. (2017). Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 171, 1354–1367 e1320. doi: 10.1016/j.cell.2017.10.012
Herrmann, J., Lukezic, T., Kling, A., Baumann, S., Huttel, S., Petkovic, H., et al. (2016). Strategies for the discovery and development of new antibiotics from natural products: three case studies. Curr. Top. Microbiol. Immunol. 398, 339–363. doi: 10.1007/82_2016_498
Huang, X., Wang, P., Li, T., Tian, X., Guo, W., Xu, B., et al. (2020). Self-assemblies based on traditional medicine berberine and cinnamic acid for adhesion-induced inhibition multidrug-resistant Staphylococcus aureus. ACS Appl. Mater. Interfaces 12, 227–237. doi: 10.1021/acsami.9b17722
Hui, J., Dong, P. T., Liang, L., Mandal, T., Li, J., Ulloa, E. R., et al. (2020). Photo-disassembly of membrane microdomains revives conventional antibiotics against MRSA. Adv. Sci. 7, 1903117. doi: 10.1002/advs.201903117
Hussein, M., Karas, J. A., Schneider-Futschik, E. K., Chen, F., Swarbrick, J., Paulin, O. K. A., et al. (2020). The killing mechanism of teixobactin against methicillin-resistant Staphylococcus aureus: an untargeted metabolomics study. mSystems 5:e00077–20. doi: 10.1128/mSystems.00077-20
Imenshahidi, M., and Hosseinzadeh, H. (2016). Berberis vulgaris and berberine: an update review. Phytother. Res. 30, 1745–1764. doi: 10.1002/ptr.5693
Kang, S., Li, Z., Yin, Z., Jia, R., Song, X., Li, L., et al. (2015). The antibacterial mechanism of berberine against Actinobacillus pleuropneumoniae. Nat. Prod. Res. 29, 2203–2206. doi: 10.1080/14786419.2014.1001388
Karaosmanoglu, K., Sayar, N. A., Kurnaz, I. A., and Akbulut, B. S. (2014). Assessment of berberine as a multi-target antimicrobial: a multi-omics study for drug discovery and repositioning. OMICS 18, 42–53. doi: 10.1089/omi.2013.0100
Karsten, W. E., Nimmo, S. A., Liu, J., and Chooback, L. (2018). Identification of 2, 3-dihydrodipicolinate as the product of the dihydrodipicolinate synthase reaction from Escherichia coli. Arch. Biochem. Biophys. 653, 50–62. doi: 10.1016/j.abb.2018.06.011
Kim, S., Lee, E. H., Kim, S. H., Lee, S., and Lim, S. J. (2012). Comparison of three tocopherol analogs as an inhibitor of production of proinflammatory mediators in macrophages. J. Pharmacol. Sci. 118, 237–244. doi: 10.1254/jphs.11152fp
Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A., and Collins, J. J. (2007). A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810. doi: 10.1016/j.cell.2007.06.049
Kokoska, L., Kloucek, P., Leuner, O., and Novy, P. (2019). Plant-derived products as antibacterial and antifungal agents in human health care. Curr. Med. Chem. 26, 5501–5541. doi: 10.2174/0929867325666180831144344
Lan, L., Cheng, A., Dunman, P. M., Missiakas, D., and He, C. (2010). Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J. Bacteriol. 192, 3068–3077. doi: 10.1128/JB.00928-09
Li, X., Song, Y., Wang, L., Kang, G., Wang, P., Yin, H., et al. (2021). A potential combination therapy of berberine hydrochloride with antibiotics against multidrug-resistant Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 11:660431. doi: 10.3389/fcimb.2021.660431
Li, Z., Garner, A. L., Gloeckner, C., Janda, K. D., and Carlow, C. K. (2011). Targeting the Wolbachia cell division protein FtsZ as a new approach for antifilarial therapy. PLoS Negl. Trop. Dis. 5:e1411. doi: 10.1371/journal.pntd.0001411
Lobritz, M. A., Belenky, P., Porter, C. B., Gutierrez, A., Yang, J. H., Schwarz, E. G., et al. (2015). Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U. S. A. 112, 8173–8180. doi: 10.1073/pnas.1509743112
Miryala, S. K., Basu, S., Naha, A., Debroy, R., Ramaiah, S., Anbarasu, A., et al. (2021). Identification of bioactive natural compounds as efficient inhibitors against Mycobacterium tuberculosis protein-targets: a molecular docking and molecular dynamics simulation study. J. Mol. Liq. 341, 117340. doi: 10.1016/j.molliq.2021.117340
Miryala, S. K., Basu, S., Naha, A., Debroy, R., Ramaiah, S., Anbarasu, A., et al. (2022). Datasets comprising the quality validations of simulated protein-ligand complexes and SYBYL docking scores of bioactive natural compounds as inhibitors of Mycobacterium tuberculosis protein-targets. Data in Brief 42, 108146. doi: 10.1016/j.dib.2022.108146
Morita, Y., Nakashima, K., Nishino, K., Kotani, K., Tomida, J., Inoue, M., et al. (2016). Berberine is a novel type efflux inhibitor which attenuates the MexXY-mediated aminoglycoside resistance in Pseudomonas aeruginosa. Front. Microbiol. 7:1223. doi: 10.3389/fmicb.2016.01223
Newman, D. J., and Cragg, G. M. (2016). Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661. doi: 10.1021/acs.jnatprod.5b01055
Ragunathan, A., Malathi, K., Ramaiah, S., and Anbarasu, A. (2018). FtsA as a cidal target for Staphylococcus aureus: molecular docking and dynamics studies. J. Cell. Biochem. doi: 10.1002/jcb.28049
Ramirez-Guadiana, F. H., Meeske, A. J., Rodrigues, C. D. A., Barajas-Ornelas, R. D. C., Kruse, A. C., and Rudner, D. Z. (2017). A two-step transport pathway allows the mother cell to nurture the developing spore in Bacillus subtilis. PLoS Genet. 13:e1007015. doi: 10.1371/journal.pgen.1007015
Renz, A., and Drager, A. (2021). Curating and comparing 114 strain-specific genome-scale metabolic models of Staphylococcus aureus. NPJ Syst. Biol. Appl. 7, 30. doi: 10.1038/s41540-021-00188-4
Sadaka, C., Ellsworth, E., Hansen, P. R., Ewin, R., Damborg, P., and Watts, J. L. (2018). Review on abyssomicins: inhibitors of the chorismate pathway and folate biosynthesis. Molecules 23, 1371. doi: 10.3390/molecules23061371
Schelli, K., Zhong, F., and Zhu, J. (2017). Comparative metabolomics revealing Staphylococcus aureus metabolic response to different antibiotics. Microb. Biotechnol. 10, 1764–1774. doi: 10.1111/1751-7915.12839
Setlow, P.. (2007). I will survive: DNA protection in bacterial spores. Trends Microbiol. 15, 172–180. doi: 10.1016/j.tim.2007.02.004
Shang, X. F., Yang, C. J., Morris-Natschke, S. L., Li, J. C., Yin, X. D., Liu, Y. Q., et al. (2020). Biologically active isoquinoline alkaloids covering 2014–2018. Med. Res. Rev. 40, 2212–2289. doi: 10.1002/med.21703
Shen, F., Ge, C., and Yuan, P. (2020). Metabolomics study reveals inhibition and metabolic dysregulation in Staphylococcus aureus Planktonic cells and biofilms induced by Carnosol. Front. Microbiol. 11:538572. doi: 10.3389/fmicb.2020.538572
Stipetic, L. H., Dalby, M. J., Davies, R. L., Morton, F. R., Ramage, G., and Burgess, K. E. (2016). A novel metabolomic approach used for the comparison of Staphylococcus aureus planktonic cells and biofilm samples. Metabolomics 12:75. doi: 10.1007/s11306-016-1002-0
Sun, N., Chan, F. Y., Lu, Y. J., Neves, M. A., Lui, H. K., Wang, Y., et al. (2014). Rational design of berberine-based FtsZ inhibitors with broad-spectrum antibacterial activity. PLoS ONE 9:e97514. doi: 10.1371/journal.pone.0097514
Surre, J., Canard, I., Bourne-Branchu, P., Courbiere, E., Franceschi, C., Chatellier, S., et al. (2018). Enhanced detection of carbapenemase-producing Enterobacteriaceae by an optimized phenol red assay. Diagn. Microbiol. Infect. Dis. 90, 11–17. doi: 10.1016/j.diagmicrobio.2017.09.005
Tang, C., Chen, J., Zhou, Y., Ding, P., He, G., Zhang, L., et al. (2021). Exploring antimicrobial mechanism of essential oil of Amomum villosum Lour through metabolomics based on gas chromatography-mass spectrometry in methicillin-resistant Staphylococcus aureus. Microbiol. Res. 242, 126608. doi: 10.1016/j.micres.2020.126608
Vemula, H., Ayon, N. J., Burton, A., and Gutheil, W. G. (2017). Antibiotic effects on methicillin-resistant Staphylococcus aureus cytoplasmic peptidoglycan intermediate levels and evidence for potential metabolite level regulatory loops. Antimicrob. Agents Chemother. 61, e02253–16. doi: 10.1128/AAC.02253-16
Xia, S., Ma, L., Wang, G., Yang, J., Zhang, M., Wang, X., et al. (2022). In vitro antimicrobial activity and the mechanism of berberine against methicillin-resistant Staphylococcus aureus isolated from bloodstream infection patients. Infect. Drug Resist. 15, 1933–1944. doi: 10.2147/idr.s357077
Xie, Y., Liu, X., and Zhou, P. (2020). In vitro antifungal effects of berberine against Candida spp. In Planktonic and biofilm conditions. Drug Des. Devel. Ther. 14, 87–101. doi: 10.2147/dddt.s230857
Xu, C., Wang, F., Huang, F., Yang, M., He, D., and Deng, L. (2021). Targeting effect of berberine on type I fimbriae of Salmonella Typhimurium and its effective inhibition of biofilm. Appl. Microbiol. Biotechnol. 105, 1563–1573. doi: 10.1007/s00253-021-11116-1
Yuan, W. W., Yang, J., Wang, D. M., Hu, Z., and Zhu, J. M. (2010). Screening of proteins interacting with PBP2a in methicillin-resistant Staphylococcus aureus. Acta Acad. Med. Militaris Tertiae 32, 749–753. doi: 10.16016/j.1000-5404.2010.08.009
Keywords: GC-MS untargeted metabolomics, LC-MS untargeted metabolomics, natural antimicrobial, metabolic markers, mechanism
Citation: Wu S, Yang K, Hong Y, Gong Y, Ni J, Yang N and Ding W (2022) A New Perspective on the Antimicrobial Mechanism of Berberine Hydrochloride Against Staphylococcus aureus Revealed by Untargeted Metabolomic Studies. Front. Microbiol. 13:917414. doi: 10.3389/fmicb.2022.917414
Received: 11 April 2022; Accepted: 09 June 2022;
Published: 13 July 2022.
Edited by:
Vinayak Singh, University of Cape Town, South AfricaReviewed by:
Anand Anbarasu, VIT University, IndiaKarishma R. Pardesi, Savitribai Phule Pune University, India
Copyright © 2022 Wu, Yang, Hong, Gong, Ni, Yang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijun Ding, Dingweijun@cdutcm.edu.cn
†These authors have contributed equally to this work and share first authorship
 Shu Wu
Shu Wu Kun Yang1†
Kun Yang1† Yanju Gong
Yanju Gong Jiajia Ni
Jiajia Ni Weijun Ding
Weijun Ding